Supplementary Materials: A Toolbox of Genetically Encoded FRET-Based Biosensors for Rapid L-Lysine Analysis
|
|
|
- Jeffery Allison
- 5 years ago
- Views:
Transcription
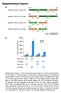
1 S1 of S9 Supplementary Materials: A Toolbox of Genetically Encoded FRET-Based Biosensors for Rapid L-Lysine Analysis Victoria Steffen, Julia Otten, Susann Engelmann, Andreas Radek, Michael Limberg, Bernd W. Koenig, Stephan Noack, Wolfgang Wiechert and Martina Pohl 1. Titration Curves of the Sensor Prototype with L-lysine, L-arginine, L-histidine, and L- glutamine Figure S1. Binding isotherms of the sensor prototype with L-lysine, L-arginine, L-histidine, and L- glutamine. Distortion of the titration curve >0.1 mm L-arginine and L-lysine, respectively, is due to the use of a crude sensor preparation. The results demonstrate that the sensor with the cplao-binding protein still has similar affinities for L-lysine (107 µm) and L-arginine (ca. 25 µm), whereas the affinity for L-histidine is much lower (>100 mm). 2. ph-dependent Binding Isotherms of the Sensor Prototype Figure S2. Binding isotherms of the sensor prototype in MOPS buffer with ph-values between ph 6.7 to 7.2. The solution of the sensor prototype was stored in 20 mm MOPS buffer, ph 7.3. This buffer was replaced by 20 mm MOPS buffer with the respective target ph via ultrafiltration. The recorded binding isotherms are shown in (a) and for better comparison the normalized binding isotherms are shown in (b).
2 S2 of S9 3. Isothermal Calorimetry Figure S3. Isothermal titration calorimetry data reflecting L-lysine binding to the binding protein cplao- BP (a) and the complete sensor prototype containing this binding protein (b). The upper two panels show baseline subtracted raw data for titration of 22 µm cplao-bp with 100 µm L-lysine (left) and 60 µm sensor prototype with 1 mm L-lysine (right). The lower panels display enthalpy changes per added mole of L- lysine as a function of the total molar ratio of lysine to protein in the calorimeter cell, reflecting binding isotherms. Only the sigmoidal isotherm in the left panel can be reliably fit to a binding model (single binding site, Kd = 1.5 µm, N = 0.36, ΔH = -58 kcal mol 1 ). Please note the different scaling of the y-axis in (a) and (b). 4. Protein Sequences from S. typhimurium and E. coli Figure S4. Comparison of the protein sequences derived from S. typhimurium and E. coli. The lysine binding proteins show high similarity. Homologous substitutions are marked in green, and non-homologous substitutions are marked in red. The amino acids involved in lysine binding are underlined. The sequence deleted during the circular permutation is marked in yellow. 5. Characteristic Mutations of the Fluorescent Proteins Relative to GFP ECFP: F64L/S65T/Y66W/N146I/M153T/V163A [1] Citrine: S65G/V68L/Q69M/S72A/T203Y/H231L [2]
3 S3 of S9 6. DNA and Protein Sequence of the Sensor Construct with cplao-bp The DNA sequence encoding the His-Tag is highlighted in bold, the sequence of CFP is marked blue, the recognition site of the restriction enzymes is underlined, the LAO-binding protein sequence is shown in green, and the Citrine sequence is shown in yellow. The same code was used for the protein sequence Nucleotide Sequence: ATGCGGGGTTCTCATCATCATCATCATCATGGTATGGCTGATACTCGCATTGGTGTAAC AATCTATAAGTCGGCTGGTATGGTGAGCAAGGGCGAGGAGCTGTTCACCGGGGTGGTGCCC ATCCTGGTCGAGCTGGACGGCGACGTAAACGGCCACAAGTTCAGCGTGTCCGGCGAGGGC GAGGGCGATGCCACCTACGGCAAGCTGACCCTGAAGTTCATCTGCACCACCGGCAAGCTG CCCGTGCCCTGGCCCACCCTCGTGACCACCCTGACCTGGGGCGTGCAGTGCTTCAGCCGCT ACCCCGACCACATGAAGCAGCACGACTTCTTCAAGTCCGCCATGCCCGAAGGCTACGTCCA GGAGCGCACCATCTTCTTCAAGGACGACGGCAACTACAAGACCCGCGCCGAGGTGAAGTT CGAGGGCGACACCCTGGTGAACCGCATCGAGCTGAAGGGCATCGACTTCAAGGAGGACGG CAACATCCTGGGGCACAAGCTGGAGTACAACTACATCAGCCACAACGTCTATATCACCGCC GACAAGCAGAAGAACGGCATCAAGGCCAACTTCAAGATCCGCCACAACATCGAGGACGG CAGCGTGCAGCTCGCCGACCACTACCAGCAGAACACCCCCATCGGCGACGGCCCCGTGCT GCTGCCCGACAACCACTACCTGAGCACCCAGTCCGCCCTGAGCAAAGACCCCAACGAGAA GCGCGATCACATGGTCCTGCTGGAGTTCGTGACCGCCGCCGGGATCGGATCCGGCACCGGT GTAGGGCTACGTAAAGATGATGCTGAACTGACGGCTGCCTTCAATAAGGCGCTTGGCGAGC TGCGTCAGGACGGCACCTACGACAAGATGGCGAAAAAGTATTTCGACTTTAATGTCTACGG TGACGGTGGCAGTGGAGGGAGCGGTGGAAGTGGCGGAAGCGCGCTACCGGAGACGGTAC GTATCGGAACCGATACCACCTACGCACCGTTCTCATCGAAAGATGCTAAAGGTGATTTTGTT GGCTTTGATATCGATCTCGGTAACGAGATGTGCAAACGGATGCAGGTGAAATGTACCTGGG TTGCCAGTGACTTTGACGCGCTGATCCCCTCACTGAAAGCGAAAAAAATCGACGCTATTAT TTCGTCGCTTTCCATTACCGATAAACGTCAGCAGGAGATTGCCTTCTCCGACAAGCTGTACG CCGCAGATTCTCGTTTGATTGCGGCCAAAGGTTCACCGATTCAGCCAACGCTGGATTCACTG AAAGGTAAACATGTTGGTGTGCTGCAGGGATCAACCCAGGAAGCTTACGCTAACGAGACC TGGCGTAGTAAAGGCGTGGATGTGGTGGCCTATGCCAACCAGGATTTGGTCTATTCCGATCT GGCTGCAGGACGTCTGGATGCTGCGTTACAAGATGAAGTTGCTGCCAGCGAAGGATTCCTC AAGCAACCTGCTGGTAAAGATTTCGCCTTTGCTGTCGACGAGCTGTTCACCGGGGTGGTGCC CATCCTGGTCGAGCTGGACGGCGACGTAAACGGCCACAAGTTCAGCGTGTCCGGCGAGGG CGAGGGCGATGCCACCTACGGCAAGCTGACCCTGAAGTTCATCTGCACCACCGGCAAGCT GCCCGTGCCCTGGCCCACCCTCGTGACCACCTTCGGCTACGGCCTGATGTGCTTCGCCCGCT ACCCCGACCACATGAAGCAGCACGACTTCTTCAAGTCCGCCATGCCCGAAGGCTACGTCCA GGAGCGCACCATCTTCTTCAAGGACGACGGCAACTACAAGACCCGCGCCGAGGTGAAGTT CGAGGGCGACACCCTGGTGAACCGCATCGAGCTGAAGGGCATCGACTTCAAGGAGGACGG CAACATCCTGGGGCACAAGCTGGAGTACAACTACAACAGCCACAACGTCTATATCATGGC CGACAAGCAGAAGAACGGCATCAAGGTGAACTTCAAGATCCGCCACAACATCGAGGACG GCAGCGTGCAGCTCGCCGACCACTACCAGCAGAACACCCCCATCGGCGACGGCCCCGTGC TGCTGCCCGACAACCACTACCTGAGCTACCAGTCCGCCCTGAGCAAAGACCCCAACGAGA AGCGCGATCACATGGTCCTGCTGGAGTTCGTGACCGCCGCCGGGATCACTCTCGGCATGGA CGAGCTGTACAAGTAA
4 S4 of S Protein Sequence MRGSHHHHHHGMADTRIGVTIYKSAGMVSKGEELFTGVVPILVELDGDVNGHKFSVSGEG EGDATYGKLTLKFICTTGKLPVPWPTLVTTLTWGVQCFSRYPDHMKQHDFFKSAMPEGYVQERT IFFKDDGNYKTRAEVKFEGDTLVNRIELKGIDFKEDGNILGHKLEYNYISHNVYITADKQKNGIKA NFKIRHNIEDGSVQLADHYQQNTPIGDGPVLLPDNHYLSTQSALSKDPNEKRDHMVLLEFVTAA GIGSGTGVGLRKDDAELTAAFNKALGELRQDGTYDKMAKKYFDFNVYGDGGSGGSGGSGGSAL PETVRIGTDTTYAPFSSKDAKGDFVGFDIDLGNEMCKRMQVKCTWVASDFDALIPSLKAKKIDAII SSLSITDKRQQEIAFSDKLYAADSRLIAAKGSPIQPTLDSLKGKHVGVLQGSTQEAYANETWRSKG VDVVAYANQDLVYSDLAAGRLDAALQDEVAASEGFLKQPAGKDFAFAVDELFTGVVPILVELD GDVNGHKFSVSGEGEGDATYGKLTLKFICTTGKLPVPWPTLVTTFGYGLMCFARYPDHMKQHD FFKSAMPEGYVQERTIFFKDDGNYKTRAEVKFEGDTLVNRIELKGIDFKEDGNILGHKLEYNYNS HNVYIMADKQKNGIKVNFKIRHNIEDGSVQLADHYQQNTPIGDGPVLLPDNHYLSYQSALSKDP NEKRDHMVLLEFVTAAGITLGMDELYK 7. Table S1 Table S1. Overview of the binding parameters of the toolbox sensors. Sensor 00 0F 0R F0 FF FR R0 RF RR R Rsat ΔR Kd (µm) 107 ± ± ± ± 3 3 ± ± ± 2 27 ± 4 Includes the FRET-ratios in the non-bound (R0) state and under saturating conditions (Rsat), the sensitivity (ΔR), and the affinity (Kd) of the sensor variants for lysine. 8. Table S2 Table S2. Overview of the binding parameters of the sensor prototype without additional linkers (00) in fresh medium and in culture supernatant (refers to Figure 5 in the main paper). Measurement System R0 Rsat ΔR Kd ph Fresh medium 0.63 ± ± ± 0.04 mm 7.0 Culture supernatant 0.93 ± ± ± 0.14 mm 7.5 MOPS buffer 1.70 ± ± ± 5 µm 7.3 The mean values of three independent measurements are shown. They were performed directly in the Biolector cultivation system in the Flowerplates at 1000 rpm and 30 C. For comparison, the values measured in MOPS buffer are shown. 9. Setup and Data Analysis of Sensor Application in Microscale Cultivation Experiments of a Lysine Producer In the following tables, the setup and analysis of the sensor application for L-lysine estimation in microtiter cultivation is explained in detail. See Table S3 for information about the plate layout. The fluorescence signals were measured constantly at λex = 430 ± 5 nm, λem = 468 ± 5 nm (ECFP) and λex = 430 ± 5 nm, λem = 532 ± 5 nm (Citrine), so the fluorescence intensity in each sample was recorded before each sampling and was subtracted from the measured fluorescence intensity in the presence of the sensor protein (Table S4). With the calculated FRET-ratios of the standards, the mean values and standard deviations of the in-plate calibration respective calibration lines were calculated (Table S5, Figure S5). Based thereon, the respective lysine concentrations in the samples were estimated (Table S6).
5 S1 of S9 10. Table S3 Well A = 0 h B = 4 h C = 8 h D = 12 h E = 16 h Table S3. Layout of the cultivation plate (Flowerplate ) Samples cultivation of L-lysine producing C. glutamicum DM1933 (4 wells per sample point) 0 mm L-lysine in-plate calibration Calibration Standards 1 mm L-lysine in-plate calibration 10 mm L-lysine in-plate calibration 100 mm L-lysine in-plate calibration F = 20 h Note: In each row, four wells were filled with culture broth (1 4) and four wells were filled with L-lysine standards for recalibration of the sensors depending on the ph and on the medium composition. Accordingly, the standards in row A were prepared with fresh media, whereas the standards in rows B F contained increasing amounts of the culture supernatant, which was prepared before with a wild-type strain of C. glutamicum. The calibration standards in row B were prepared with a mixture of 67% fresh medium and 33% culture supernatant and accordingly the standards in row C were prepared with a mixture of 33% fresh medium and 67% culture supernatant, whereas rows D, E, and F were prepared with 100% culture supernatant. 11. Table S4 Table S4. Raw data of the fluorescence measurement of the yellow and blue channel at sampling times. In-plate calibration data is shaded in gray, respectively. ECFP-Signal (λex = 430 ± 5 nm, λem = 468 ± 5 nm) Citrine-Signal (λex = 430 ± 5 nm, λem = 532 ± 5 nm) Well Number Raw Data Before Biosensor Addition Raw Data After Biosensor Addition Fluorescence Signal of the Sensor Minus Background Fluorescence of the Cultivation Broth Raw Data Before Biosensor Addition Raw Data After Biosensor Addition Fluorescence Signal of the Sensor Minus Background Fluorescence of the Cultivation Broth FRET-Ratio = Citrine/ECFP A
6 S2 of S9 A A A A A A B B B B B B B B C C C C C C C C D D D
7 S3 of S9 D D D D D E E E E E E E E F F F F F F F F
8 S1 of S9 12. Table S5 Table S5. FRET-ratios of the in-plate calibration with mean values and standard deviation. Sampling Time 0 mm L-lysine 1 mm L-lysine 10 mm L-lysine 100 mm L-lysine FRET-Ratios of the L-Lysine Standards 0 h 4 h 8 h 12 h 16 h 20 h Mean Value Standard Deviation Deviation in Percent Note: the saturation concentration of the sensor with L-lysine under the given conditions is > 10 mm but < 100 mm L-lysine and could not be resolved in the three-point calibration. Therefore, the calibration curves (Figure. S5) apparently increase continuously until 100 mm. 13. Figure S5 Figure S5. In-plate measured calibration curves. 14. Table S6 Table S6. Analysis of the culture broth using the sensor prototype. Sampling Time 0 h 4 h 8 h 12 h 16 h 20 h FRET-ratios of the culture broth Well Well Well Well Mean Value Standard Deviation (SD) Regression curve: y = a ln(x) + b
9 S2 of S9 a b L-lysine in culture broth 0 1 ± ± mm ± 15 mm ± 15 mm ± 12 ± 0.04 mm mm 0.01 mm 0.02 mm 0.3 mm mm References 1. Kremers, G.J.; Goedhart, J.; van Munster, E.B.; Gadella, T.W., Jr. Cyan and yellow super fluorescent proteins with improved brightness, protein folding, and FRET Forster radius. Biochemistry 2006, 45, Griesbeck, O.; Baird, G.S.; Campbell, R.E.; Zacharias, D.A.; Tsien, R.Y. Reducing the environmental sensitivity of yellow fluorescent protein. J. Biol. Chem. 2001, 276,
SUPPLEMENTARY INFORMATION. Supplementary Figures 1-8
 SUPPLEMENTARY INFORMATION Supplementary Figures 1-8 Supplementary Figure 1. TFAM residues contacting the DNA minor groove (A) TFAM contacts on nonspecific DNA. Leu58, Ile81, Asn163, Pro178, and Leu182
SUPPLEMENTARY INFORMATION Supplementary Figures 1-8 Supplementary Figure 1. TFAM residues contacting the DNA minor groove (A) TFAM contacts on nonspecific DNA. Leu58, Ile81, Asn163, Pro178, and Leu182
SUPPLEMENTARY INFORMATION. Tolerance of a knotted near infrared fluorescent protein to random circular permutation
 SUPPLEMENTARY INFORMATION Tolerance of a knotted near infrared fluorescent protein to random circular permutation Naresh Pandey 1,3, Brianna E. Kuypers 2,4, Barbara Nassif 1, Emily E. Thomas 1,3, Razan
SUPPLEMENTARY INFORMATION Tolerance of a knotted near infrared fluorescent protein to random circular permutation Naresh Pandey 1,3, Brianna E. Kuypers 2,4, Barbara Nassif 1, Emily E. Thomas 1,3, Razan
Challenges to measuring intracellular Ca 2+ Calmodulin: nature s Ca 2+ sensor
 Calcium Signals in Biological Systems Lecture 3 (2/9/0) Measuring intracellular Ca 2+ signals II: Genetically encoded Ca 2+ sensors Henry M. Colecraft, Ph.D. Challenges to measuring intracellular Ca 2+
Calcium Signals in Biological Systems Lecture 3 (2/9/0) Measuring intracellular Ca 2+ signals II: Genetically encoded Ca 2+ sensors Henry M. Colecraft, Ph.D. Challenges to measuring intracellular Ca 2+
Nature Structural & Molecular Biology: doi: /nsmb.2548
 Supplementary Figure 1. Structure of GltPhout. (a) Stereo view of a slice through a single GltPhout protomer shown in stick representation along with 2Fo-Fc and anomalous difference electron maps. The
Supplementary Figure 1. Structure of GltPhout. (a) Stereo view of a slice through a single GltPhout protomer shown in stick representation along with 2Fo-Fc and anomalous difference electron maps. The
Supplementary Figure 1. Design of linker truncation library between Nluc and either Venus or mneongreen
 1 2 3 Supplementary Figure 1. Design of linker truncation library between Nluc and either Venus or mneongreen 4 5 6 7 8 9 10 The cdna of C-terminally deleted FPs (mneongreen or Venus) mutants and N-terminally
1 2 3 Supplementary Figure 1. Design of linker truncation library between Nluc and either Venus or mneongreen 4 5 6 7 8 9 10 The cdna of C-terminally deleted FPs (mneongreen or Venus) mutants and N-terminally
Supplementary information. for the paper:
 Supplementary information for the paper: Screening for protein-protein interactions using Förster resonance energy transfer (FRET) and fluorescence lifetime imaging microscopy (FLIM) Anca Margineanu 1*,
Supplementary information for the paper: Screening for protein-protein interactions using Förster resonance energy transfer (FRET) and fluorescence lifetime imaging microscopy (FLIM) Anca Margineanu 1*,
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Conserved arginines on the rim of Hfq catalyze base pair formation and exchange Subrata Panja and Sarah A. Woodson T.C. Jenkins Department of Biophysics, Johns Hopkins University,
SUPPLEMENTARY INFORMATION Conserved arginines on the rim of Hfq catalyze base pair formation and exchange Subrata Panja and Sarah A. Woodson T.C. Jenkins Department of Biophysics, Johns Hopkins University,
Evaluation of Cu(I) Binding to the E2 Domain of the Amyloid. Precursor Protein A Lesson in Quantification of Metal Binding to
 Electronic Supplementary Material (ESI) for Metallomics. This journal is The Royal Society of Chemistry 2017 Evaluation of Cu(I) Binding to the E2 Domain of the Amyloid Precursor Protein A Lesson in Quantification
Electronic Supplementary Material (ESI) for Metallomics. This journal is The Royal Society of Chemistry 2017 Evaluation of Cu(I) Binding to the E2 Domain of the Amyloid Precursor Protein A Lesson in Quantification
Aminoacid change in chromophore. PIN3::PIN3-GFP GFP S65 (no change) (5.9) (Kneen et al., 1998)
 Supplemental Table. Table S1. Fluorophore characteristics of used fluorescent proteins, Related to Figure 2 and 3. Transgenic line Fluprescent marker Aminoacid change in chromophore pk(a) Ref. PIN3::PIN3-GFP
Supplemental Table. Table S1. Fluorophore characteristics of used fluorescent proteins, Related to Figure 2 and 3. Transgenic line Fluprescent marker Aminoacid change in chromophore pk(a) Ref. PIN3::PIN3-GFP
Masayoshi Honda, Jeehae Park, Robert A. Pugh, Taekjip Ha, and Maria Spies
 Molecular Cell, Volume 35 Supplemental Data Single-Molecule Analysis Reveals Differential Effect of ssdna-binding Proteins on DNA Translocation by XPD Helicase Masayoshi Honda, Jeehae Park, Robert A. Pugh,
Molecular Cell, Volume 35 Supplemental Data Single-Molecule Analysis Reveals Differential Effect of ssdna-binding Proteins on DNA Translocation by XPD Helicase Masayoshi Honda, Jeehae Park, Robert A. Pugh,
A quantitative investigation of linker histone interactions with nucleosomes and chromatin
 White et al Supplementary figures and figure legends A quantitative investigation of linker histone interactions with nucleosomes and chromatin 1 Alison E. White, Aaron R. Hieb, and 1 Karolin Luger 1 White
White et al Supplementary figures and figure legends A quantitative investigation of linker histone interactions with nucleosomes and chromatin 1 Alison E. White, Aaron R. Hieb, and 1 Karolin Luger 1 White
Creation of circularly permutated yellow fluorescent proteins using fluorescence screening and a tandem fusion template
 Biotechnology Letters (2006) 28: 471 475 Ó Springer 2006 DOI 10.1007/s10529-006-0007-6 Creation of circularly permutated yellow fluorescent proteins using fluorescence screening and a tandem fusion template
Biotechnology Letters (2006) 28: 471 475 Ó Springer 2006 DOI 10.1007/s10529-006-0007-6 Creation of circularly permutated yellow fluorescent proteins using fluorescence screening and a tandem fusion template
Green Fluorescent Protein. Avinash Bayya Varun Maturi Nikhileswar Reddy Mukkamala Aravindh Subhramani
 Green Fluorescent Protein Avinash Bayya Varun Maturi Nikhileswar Reddy Mukkamala Aravindh Subhramani Introduction The Green fluorescent protein (GFP) was first isolated from the Jellyfish Aequorea victoria,
Green Fluorescent Protein Avinash Bayya Varun Maturi Nikhileswar Reddy Mukkamala Aravindh Subhramani Introduction The Green fluorescent protein (GFP) was first isolated from the Jellyfish Aequorea victoria,
Supplementary Information
 Supplementary Information Fluorescent protein choice The dual fluorescence channel ratiometric promoter characterization plasmid we designed for this study required a pair of fluorescent proteins with
Supplementary Information Fluorescent protein choice The dual fluorescence channel ratiometric promoter characterization plasmid we designed for this study required a pair of fluorescent proteins with
Supplementary Figure 1. WT ph 4.5. γ (ion s
 Supplementary Figure 1 a WT ph 4.5 b 10 5 γ (ion s 1 ) 10 4 10 3 10 2 WT Y445A EA/YA E148A µcal s 1 Supplementary Figure 1 ph dependence of Cl transport and binding. a) Cl transport rates at ph 7.5 (white)
Supplementary Figure 1 a WT ph 4.5 b 10 5 γ (ion s 1 ) 10 4 10 3 10 2 WT Y445A EA/YA E148A µcal s 1 Supplementary Figure 1 ph dependence of Cl transport and binding. a) Cl transport rates at ph 7.5 (white)
Supplementary Figures
 Supplementary Figures a HindIII XbaI pcdna3.1/v5-his A_Nfluc-VVD 1-398 37-186 Nfluc Linker 1 VVD HindIII XhoI pcdna3.1/v5-his A_VVD-Nfluc 37-186 1-398 VVD Linker 2 Nfluc HindIII XbaI pcdna3.1/v5-his A_Cfluc-VVD
Supplementary Figures a HindIII XbaI pcdna3.1/v5-his A_Nfluc-VVD 1-398 37-186 Nfluc Linker 1 VVD HindIII XhoI pcdna3.1/v5-his A_VVD-Nfluc 37-186 1-398 VVD Linker 2 Nfluc HindIII XbaI pcdna3.1/v5-his A_Cfluc-VVD
Donor DNA Utilization during Gene Targeting with Zinc- finger Nucleases
 Donor DNA Utilization during Gene Targeting with Zinc- finger Nucleases Kelly J. Beumer, Jonathan K. Trautman, Kusumika Mukherjee and Dana Carroll Department of Biochemistry University of Utah School of
Donor DNA Utilization during Gene Targeting with Zinc- finger Nucleases Kelly J. Beumer, Jonathan K. Trautman, Kusumika Mukherjee and Dana Carroll Department of Biochemistry University of Utah School of
An engineered tryptophan zipper-type peptide as a molecular recognition scaffold
 SUPPLEMENTARY MATERIAL An engineered tryptophan zipper-type peptide as a molecular recognition scaffold Zihao Cheng and Robert E. Campbell* Supplementary Methods Library construction for FRET-based screening
SUPPLEMENTARY MATERIAL An engineered tryptophan zipper-type peptide as a molecular recognition scaffold Zihao Cheng and Robert E. Campbell* Supplementary Methods Library construction for FRET-based screening
Microscale Thermophoresis
 AN INTRODUCTION Microscale Thermophoresis A sensitive method to detect and quantify molecular interactions OCTOBER 2012 CHAPTER 1 Overview Microscale thermophoresis (MST) is a new method that enables the
AN INTRODUCTION Microscale Thermophoresis A sensitive method to detect and quantify molecular interactions OCTOBER 2012 CHAPTER 1 Overview Microscale thermophoresis (MST) is a new method that enables the
HEK293T. Fig. 1 in the
 Supplementary Information Supplementary Figure 1 Zinc uptake assay of hzip4 and hzip4-δecd transiently expressed in HEK293T cells. The results of one representative e experiment are shown in Fig. 1 in
Supplementary Information Supplementary Figure 1 Zinc uptake assay of hzip4 and hzip4-δecd transiently expressed in HEK293T cells. The results of one representative e experiment are shown in Fig. 1 in
A quantitative protocol for intensity-based live cell FRET imaging.
 A quantitative protocol for intensity-based live cell FRET imaging. Kaminski CF, Rees EJ, Schierle GS. Methods Mol Biol. 2014; 1076:445-454. Department of Chemical Engineering and Biotechnology, Pembroke
A quantitative protocol for intensity-based live cell FRET imaging. Kaminski CF, Rees EJ, Schierle GS. Methods Mol Biol. 2014; 1076:445-454. Department of Chemical Engineering and Biotechnology, Pembroke
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Supplementary figures Supplementary Figure 1: Suv39h1, but not Suv39h2, promotes HP1α sumoylation in vivo. In vivo HP1α sumoylation assay. Top: experimental scheme. Middle: we
SUPPLEMENTARY INFORMATION Supplementary figures Supplementary Figure 1: Suv39h1, but not Suv39h2, promotes HP1α sumoylation in vivo. In vivo HP1α sumoylation assay. Top: experimental scheme. Middle: we
Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product.
 Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplementary Note 1. Enzymatic properties of the purified Syn BVR
 Supplementary Note 1. Enzymatic properties of the purified Syn BVR The expression vector pet15b-syn bvr allowed us to routinely prepare 15 mg of electrophoretically homogenous Syn BVR from 2.5 L of TB-medium
Supplementary Note 1. Enzymatic properties of the purified Syn BVR The expression vector pet15b-syn bvr allowed us to routinely prepare 15 mg of electrophoretically homogenous Syn BVR from 2.5 L of TB-medium
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature11500 Figure S1: The sector in the PDZ domain family. Sectors are defined as groups of statistically coevolving amino acid positions in a protein family and
SUPPLEMENTARY INFORMATION doi:10.1038/nature11500 Figure S1: The sector in the PDZ domain family. Sectors are defined as groups of statistically coevolving amino acid positions in a protein family and
BoTest A/E Botulinum Neurotoxin Detection Kit- Detailed Description
 Figure 1. BoTest A/E Reporter. (A) BioSentinel s BoTest A/E Reporter. CFP and YFP are connected by SNAP-25 (amino acids 141-206). The cleavage sites for each BoNT sero-type are indicated. (B) A FRET assay
Figure 1. BoTest A/E Reporter. (A) BioSentinel s BoTest A/E Reporter. CFP and YFP are connected by SNAP-25 (amino acids 141-206). The cleavage sites for each BoNT sero-type are indicated. (B) A FRET assay
Quantitative analysis of recombination in YFP and CFP gene of FRET biosensor induced by lentiviral or retroviral gene transfer.
 Supplementary Information: Quantitative analysis of recombination in and gene of FRET biosensor induced by lentiviral or retroviral gene transfer. Akira T. Komatsubara, Michiyuki Matsuda,, and Kazuhiro
Supplementary Information: Quantitative analysis of recombination in and gene of FRET biosensor induced by lentiviral or retroviral gene transfer. Akira T. Komatsubara, Michiyuki Matsuda,, and Kazuhiro
Supplementary Materials. Signal recognition particle binds to translating ribosomes before emergence of a signal anchor sequence
 Supplementary Materials Signal recognition particle binds to translating ribosomes before emergence of a signal anchor sequence Evan Mercier, Wolf Holtkamp, Marina V. Rodnina, Wolfgang Wintermeyer* Department
Supplementary Materials Signal recognition particle binds to translating ribosomes before emergence of a signal anchor sequence Evan Mercier, Wolf Holtkamp, Marina V. Rodnina, Wolfgang Wintermeyer* Department
MST Starting Guide Monolith NT.115
 MST Starting Guide Monolith NT.115 Contents 1. How to design an experiment 2. Before you start 3. Assay Setup Pretests 4. Assay Setup 5. MST-experiment using temperature control 6. Data Interpretation
MST Starting Guide Monolith NT.115 Contents 1. How to design an experiment 2. Before you start 3. Assay Setup Pretests 4. Assay Setup 5. MST-experiment using temperature control 6. Data Interpretation
Figure S1, related to Figure 1. Characterization of biosensor behavior in vivo.
 Developmental Cell, Volume 23 Supplemental Information Separase Biosensor Reveals that Cohesin Cleavage Timing Depends on Phosphatase PP2A Cdc55 Regulation Gilad Yaakov, Kurt Thorn, and David O. Morgan
Developmental Cell, Volume 23 Supplemental Information Separase Biosensor Reveals that Cohesin Cleavage Timing Depends on Phosphatase PP2A Cdc55 Regulation Gilad Yaakov, Kurt Thorn, and David O. Morgan
Supplemental Experimental Procedures
 Supplemental Experimental Procedures Cloning, expression and purification of untagged P C, Q D -N 0123, Q D- N 012 and Q D -N 01 - The DNA sequence encoding non tagged P C (starting at the Arginine 57
Supplemental Experimental Procedures Cloning, expression and purification of untagged P C, Q D -N 0123, Q D- N 012 and Q D -N 01 - The DNA sequence encoding non tagged P C (starting at the Arginine 57
Tag-lite Tachykinin NK1 labeled Cells, ready-to-use (transformed & labeled), 200 tests* (Part# C1TT1NK1)
 TAG - L I T E Tag-lite Tachykinin NK1 Receptor Ligand Binding Assay protocol Tachykinin NK1 labeled cells for: 200 tests Part#: C1TT1NK1 Lot#: 03-1 March 2016 (See expiration date on package label) Store
TAG - L I T E Tag-lite Tachykinin NK1 Receptor Ligand Binding Assay protocol Tachykinin NK1 labeled cells for: 200 tests Part#: C1TT1NK1 Lot#: 03-1 March 2016 (See expiration date on package label) Store
TECHNICAL BULLETIN. In Vitro Bacterial Split Fluorescent Protein Fold n Glow Solubility Assay Kits
 In Vitro Bacterial Split Fluorescent Protein Fold n Glow Solubility Assay Kits Catalog Numbers APPA001 In Vitro Bacterial Split GFP "Fold 'n' Glow" Solubility Assay Kit (Green) APPA008 In Vitro Bacterial
In Vitro Bacterial Split Fluorescent Protein Fold n Glow Solubility Assay Kits Catalog Numbers APPA001 In Vitro Bacterial Split GFP "Fold 'n' Glow" Solubility Assay Kit (Green) APPA008 In Vitro Bacterial
Supplementary Materials. for. array reveals biophysical and evolutionary landscapes
 Supplementary Materials for Quantitative analysis of RNA- protein interactions on a massively parallel array reveals biophysical and evolutionary landscapes Jason D. Buenrostro 1,2,4, Carlos L. Araya 1,4,
Supplementary Materials for Quantitative analysis of RNA- protein interactions on a massively parallel array reveals biophysical and evolutionary landscapes Jason D. Buenrostro 1,2,4, Carlos L. Araya 1,4,
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Genome-scale engineering of Saccharomyces cerevisiae with single nucleotide precision Zehua Bao, 1 Mohammad HamediRad, 2 Pu Xue, 2 Han Xiao, 1,7 Ipek Tasan, 3 Ran Chao, 2 Jing
SUPPLEMENTARY INFORMATION Genome-scale engineering of Saccharomyces cerevisiae with single nucleotide precision Zehua Bao, 1 Mohammad HamediRad, 2 Pu Xue, 2 Han Xiao, 1,7 Ipek Tasan, 3 Ran Chao, 2 Jing
Investigation into the Amino Acid Characteristic of Intrinsically Disordered Proteins
 University of Wyoming Wyoming Scholars Repository Honors Theses AY 17/18 Undergraduate Honors Theses Spring 5-11-2018 Investigation into the Amino Acid Characteristic of Intrinsically Disordered Proteins
University of Wyoming Wyoming Scholars Repository Honors Theses AY 17/18 Undergraduate Honors Theses Spring 5-11-2018 Investigation into the Amino Acid Characteristic of Intrinsically Disordered Proteins
Visualizing mechanical tension across membrane receptors with a fluorescent sensor
 Nature Methods Visualizing mechanical tension across membrane receptors with a fluorescent sensor Daniel R. Stabley, Carol Jurchenko, Stephen S. Marshall, Khalid S. Salaita Supplementary Figure 1 Fabrication
Nature Methods Visualizing mechanical tension across membrane receptors with a fluorescent sensor Daniel R. Stabley, Carol Jurchenko, Stephen S. Marshall, Khalid S. Salaita Supplementary Figure 1 Fabrication
Supporting Information
 Supporting Information Direct Selection of Fluorescence-Enhancing RNA Aptamers Michael Gotrik, Gurpreet Sekhon, Saumya Saurabh, Margaret Nakamoto, Michael Eisenstein, H. Tom Soh Supplementary Figures Figure
Supporting Information Direct Selection of Fluorescence-Enhancing RNA Aptamers Michael Gotrik, Gurpreet Sekhon, Saumya Saurabh, Margaret Nakamoto, Michael Eisenstein, H. Tom Soh Supplementary Figures Figure
University of Groningen
 University of Groningen Coupling dttp Hydrolysis with DNA Unwinding by the DNA Helicase of Bacteriophage T7 Satapathy, Ajit K.; Kulczyk, Arkadiusz W.; Ghosh, Sharmistha; van Oijen, Antonius; Richardson,
University of Groningen Coupling dttp Hydrolysis with DNA Unwinding by the DNA Helicase of Bacteriophage T7 Satapathy, Ajit K.; Kulczyk, Arkadiusz W.; Ghosh, Sharmistha; van Oijen, Antonius; Richardson,
Supplemental Information
 Supplemental Information ATP-dependent unwinding of U4/U6 snrnas by the Brr2 helicase requires the C-terminus of Prp8 Corina Maeder 1,3, Alan K. Kutach 1,2,3, and Christine Guthrie 1 1 Department of Biochemistry
Supplemental Information ATP-dependent unwinding of U4/U6 snrnas by the Brr2 helicase requires the C-terminus of Prp8 Corina Maeder 1,3, Alan K. Kutach 1,2,3, and Christine Guthrie 1 1 Department of Biochemistry
Phage Antibody Selection With Reichert SPR System
 Phage Antibody Selection With Reichert SPR System Reichert Technologies Webinar April 4, 2016 Mark A. Sullivan, Ph.D. Department of Microbiology and Immunology University of Rochester Presentation Outline
Phage Antibody Selection With Reichert SPR System Reichert Technologies Webinar April 4, 2016 Mark A. Sullivan, Ph.D. Department of Microbiology and Immunology University of Rochester Presentation Outline
TITLE: Restoration of Wild-Type Activity to Mutant p53 in Prostate Cancer: A Novel Therapeutic Approach
 AD Award Number: W81XWH-05-1-0109 TITLE: Restoration of Wild-Type Activity to Mutant p53 in Prostate Cancer: A Novel Therapeutic Approach PRINCIPAL INVESTIGATOR: James Manfredi, Ph.D. CONTRACTING ORGANIZATION:
AD Award Number: W81XWH-05-1-0109 TITLE: Restoration of Wild-Type Activity to Mutant p53 in Prostate Cancer: A Novel Therapeutic Approach PRINCIPAL INVESTIGATOR: James Manfredi, Ph.D. CONTRACTING ORGANIZATION:
Supplementary Information Titles
 Supplementary Information Titles Please list each supplementary item and its title or caption, in the order shown below. Note that we do NOT copy edit or otherwise change supplementary information, and
Supplementary Information Titles Please list each supplementary item and its title or caption, in the order shown below. Note that we do NOT copy edit or otherwise change supplementary information, and
Chapter VI: Mutagenesis to Restore Chimera Function
 99 Chapter VI: Mutagenesis to Restore Chimera Function Introduction Identifying characteristics of functional and nonfunctional chimeras is one way to address the underlying reasons for why chimeric proteins
99 Chapter VI: Mutagenesis to Restore Chimera Function Introduction Identifying characteristics of functional and nonfunctional chimeras is one way to address the underlying reasons for why chimeric proteins
Click chemistry for advanced drug discovery applications of human protein kinase CK2
 Click chemistry for advanced drug discovery applications of human protein kinase CK2 Christian Nienberg 1, *, Anika Retterath 1, Kira Sophie Becher 2, Henning D. Mootz 2 and Joachim Jose 1 1 Institut für
Click chemistry for advanced drug discovery applications of human protein kinase CK2 Christian Nienberg 1, *, Anika Retterath 1, Kira Sophie Becher 2, Henning D. Mootz 2 and Joachim Jose 1 1 Institut für
SUPPLEMENTARY MATERIAL
 SUPPLEMENTARY MATERIAL Materials and Methods Circular dichroism (CD) spectroscopy. Far ultraviolet (UV) CD spectra of apo- and holo- CaM and the CaM mutants were recorded on a Jasco J-715 spectropolarimeter
SUPPLEMENTARY MATERIAL Materials and Methods Circular dichroism (CD) spectroscopy. Far ultraviolet (UV) CD spectra of apo- and holo- CaM and the CaM mutants were recorded on a Jasco J-715 spectropolarimeter
Supplementary Figure S1. Np95 interacts with de novo methyltransferases Dnmt3a and 3b. (A) Co-immunoprecipitation of endogenous Dnmt3a2 (left and
 Supplementary Figure S1. Np95 interacts with de novo methyltransferases Dnmt3a and 3b. (A) Co-immunoprecipitation of endogenous Dnmt3a2 (left and right), Dnmt3b isoforms (left) and Dnmt1 (right) with GFP-Np95
Supplementary Figure S1. Np95 interacts with de novo methyltransferases Dnmt3a and 3b. (A) Co-immunoprecipitation of endogenous Dnmt3a2 (left and right), Dnmt3b isoforms (left) and Dnmt1 (right) with GFP-Np95
Orange fluorescent proteins shift constructed from cyanobacteriochromes. chromophorylated with phycoerythrobilin. Kai-Hong Zhao 1 Ming Zhou 1, *
 Electronic Supplementary Material (ESI) for Photochemical. This journal is The Royal Society of Chemistry and Owner Societies 2014 Orange fluorescent proteins shift constructed from cyanobacteriochromes
Electronic Supplementary Material (ESI) for Photochemical. This journal is The Royal Society of Chemistry and Owner Societies 2014 Orange fluorescent proteins shift constructed from cyanobacteriochromes
Supplementary Information
 Supplementary Information Experimental procedures Identifying the viable circular permutation sites of E. coli EPSPS Bioinformatics prediction and E. coli complementation assay were performed together
Supplementary Information Experimental procedures Identifying the viable circular permutation sites of E. coli EPSPS Bioinformatics prediction and E. coli complementation assay were performed together
Protein-Protein Interactions II
 Biochemistry 412 Protein-Protein Interactions II March 28, 2008 Delano (2002) Curr. Opin. Struct. Biol. 12, 14. Some ways that mutations can destabilize protein-protein interactions Delano (2002) Curr.
Biochemistry 412 Protein-Protein Interactions II March 28, 2008 Delano (2002) Curr. Opin. Struct. Biol. 12, 14. Some ways that mutations can destabilize protein-protein interactions Delano (2002) Curr.
PHF20 is an effector protein of p53 double lysine methylation
 SUPPLEMENTARY INFORMATION PHF20 is an effector protein of p53 double lysine methylation that stabilizes and activates p53 Gaofeng Cui 1, Sungman Park 2, Aimee I Badeaux 3, Donghwa Kim 2, Joseph Lee 1,
SUPPLEMENTARY INFORMATION PHF20 is an effector protein of p53 double lysine methylation that stabilizes and activates p53 Gaofeng Cui 1, Sungman Park 2, Aimee I Badeaux 3, Donghwa Kim 2, Joseph Lee 1,
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Moutin et al., http://www.jcb.org/cgi/content/full/jcb.201110101/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Tagged Homer1a and Homer are functional and display different
Supplemental material Moutin et al., http://www.jcb.org/cgi/content/full/jcb.201110101/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Tagged Homer1a and Homer are functional and display different
Microbial Cell Factories. Open Access RESEARCH. Tobias Ladner, Markus Held, David Flitsch, Mario Beckers and Jochen Büchs *
 DOI 10.1186/s12934-016-0608-2 Microbial Cell Factories RESEARCH Open Access Quasi continuous parallel online scattered light, fluorescence and dissolved oxygen tension measurement combined with monitoring
DOI 10.1186/s12934-016-0608-2 Microbial Cell Factories RESEARCH Open Access Quasi continuous parallel online scattered light, fluorescence and dissolved oxygen tension measurement combined with monitoring
TAG-LITE RECEPTOR LIGAND BINDING ASSAY
 TAG - L I T E TAG-LITE RECEPTOR LIGAND BINDING ASSAY PROTOCOL ASSAY PRINCIPLE The Tag-lite Ligand Binding Assay is a homogeneous alternative to radio ligand binding assays for HTS and compound profiling.
TAG - L I T E TAG-LITE RECEPTOR LIGAND BINDING ASSAY PROTOCOL ASSAY PRINCIPLE The Tag-lite Ligand Binding Assay is a homogeneous alternative to radio ligand binding assays for HTS and compound profiling.
Supporting Information
 Supporting Information Wiley-VCH 2006 69451 Weinheim, Germany RNA ligands that distinguish metabolite-induced conformations in the TPP riboswitch Günter Mayer, Marie-Sophie L. Raddatz, Julia D. Grunwald,
Supporting Information Wiley-VCH 2006 69451 Weinheim, Germany RNA ligands that distinguish metabolite-induced conformations in the TPP riboswitch Günter Mayer, Marie-Sophie L. Raddatz, Julia D. Grunwald,
SUPPLEMENTARY FIGURES and LEGENDS
 DARPins recognizing the tumor-associated antigen EpCAM selected by phage and ribosome display and engineered for multivalency Nikolas Stefan 1, 3, Patricia Martin-Killias 1, 3, Sascha Wyss-Stoeckle 1,
DARPins recognizing the tumor-associated antigen EpCAM selected by phage and ribosome display and engineered for multivalency Nikolas Stefan 1, 3, Patricia Martin-Killias 1, 3, Sascha Wyss-Stoeckle 1,
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature08627 Supplementary Figure 1. DNA sequences used to construct nucleosomes in this work. a, DNA sequences containing the 601 positioning sequence (blue)24 with a PstI restriction site
doi: 10.1038/nature08627 Supplementary Figure 1. DNA sequences used to construct nucleosomes in this work. a, DNA sequences containing the 601 positioning sequence (blue)24 with a PstI restriction site
Sequence Determinants of a Conformational Switch in a Protein
 Sequence Determinants of a Conformational Switch in a Protein Thomas A. Anderson, Matthew H. J. Cordes, and Robert T. Sauer Kyle Skalenko 4/17/09 Introduction Protein folding is guided by the following
Sequence Determinants of a Conformational Switch in a Protein Thomas A. Anderson, Matthew H. J. Cordes, and Robert T. Sauer Kyle Skalenko 4/17/09 Introduction Protein folding is guided by the following
BS 50 Genetics and Genomics Week of Oct 24
 BS 50 Genetics and Genomics Week of Oct 24 Additional Practice Problems for Section Question 1: The following table contains a list of statements that apply to replication, transcription, both, or neither.
BS 50 Genetics and Genomics Week of Oct 24 Additional Practice Problems for Section Question 1: The following table contains a list of statements that apply to replication, transcription, both, or neither.
fibrils, however, oligomeric structures and amorphous protein aggregates were
 Supplementary Figure 1: Effect of equimolar EGCG on αs and Aβ aggregate formation. A D B E αs C F Figure S1: (A-C) Analysis of EGCG treated αs (1 µm) aggregation reactions by EM. A 1:1 molar ratio of EGCG
Supplementary Figure 1: Effect of equimolar EGCG on αs and Aβ aggregate formation. A D B E αs C F Figure S1: (A-C) Analysis of EGCG treated αs (1 µm) aggregation reactions by EM. A 1:1 molar ratio of EGCG
The Green Fluorescent Protein. w.chem.uwec.edu/chem412_s99/ppt/green.ppt
 The Green Fluorescent Protein w.chem.uwec.edu/chem412_s99/ppt/green.ppt www.chem.uwec.edu/chem412_s99/ppt/green.ppt Protein (gene) is from a jellyfish: Aequorea victoria www.chem.uwec.edu/chem412_s99/ppt/green.ppt
The Green Fluorescent Protein w.chem.uwec.edu/chem412_s99/ppt/green.ppt www.chem.uwec.edu/chem412_s99/ppt/green.ppt Protein (gene) is from a jellyfish: Aequorea victoria www.chem.uwec.edu/chem412_s99/ppt/green.ppt
Biochemistry 111. Carl Parker x A Braun
 Biochemistry 111 Carl Parker x6368 101A Braun csp@caltech.edu Central Dogma of Molecular Biology DNA-Dependent RNA Polymerase Requires a DNA Template Synthesizes RNA in a 5 to 3 direction Requires ribonucleoside
Biochemistry 111 Carl Parker x6368 101A Braun csp@caltech.edu Central Dogma of Molecular Biology DNA-Dependent RNA Polymerase Requires a DNA Template Synthesizes RNA in a 5 to 3 direction Requires ribonucleoside
Supplementary Information. Arrays of Individual DNA Molecules on Nanopatterned Substrates
 Supplementary Information Arrays of Individual DNA Molecules on Nanopatterned Substrates Roland Hager, Alma Halilovic, Jonathan R. Burns, Friedrich Schäffler, Stefan Howorka S1 Figure S-1. Characterization
Supplementary Information Arrays of Individual DNA Molecules on Nanopatterned Substrates Roland Hager, Alma Halilovic, Jonathan R. Burns, Friedrich Schäffler, Stefan Howorka S1 Figure S-1. Characterization
Supplementary Information. A superfolding Spinach2 reveals the dynamic nature of. trinucleotide repeat RNA
 Supplementary Information A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat RNA Rita L. Strack 1, Matthew D. Disney 2 & Samie R. Jaffrey 1 1 Department of Pharmacology, Weill Medical
Supplementary Information A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat RNA Rita L. Strack 1, Matthew D. Disney 2 & Samie R. Jaffrey 1 1 Department of Pharmacology, Weill Medical
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Contents: Supplementary Figure 1. Additional structural and binding data for designed tuim peptides. Supplementary Figure 2. Subcellular localization patterns of designed tuim
SUPPLEMENTARY INFORMATION Contents: Supplementary Figure 1. Additional structural and binding data for designed tuim peptides. Supplementary Figure 2. Subcellular localization patterns of designed tuim
Event COT102 Cotton. Real-time, Event-specific Polymerase Chain Reaction Method
 Event COT102 Cotton Real-time, Event-specific Polymerase Chain Reaction Method Syngenta Crop Protection, LLC 3054 East Cornwallis Road PO Box 12257 Research Triangle Park, NC 27709-2257, USA For technical
Event COT102 Cotton Real-time, Event-specific Polymerase Chain Reaction Method Syngenta Crop Protection, LLC 3054 East Cornwallis Road PO Box 12257 Research Triangle Park, NC 27709-2257, USA For technical
DNA supercoiling, a critical signal regulating the basal expression of the lac operon in Escherichia coli
 Supplementary Information DNA supercoiling, a critical signal regulating the basal expression of the lac operon in Escherichia coli Geraldine Fulcrand 1,2, Samantha Dages 1,2, Xiaoduo Zhi 1,2, Prem Chapagain
Supplementary Information DNA supercoiling, a critical signal regulating the basal expression of the lac operon in Escherichia coli Geraldine Fulcrand 1,2, Samantha Dages 1,2, Xiaoduo Zhi 1,2, Prem Chapagain
Designed BH3 peptides with high affinity and specificity for
 Designed BH3 peptides with high affinity and specificity for targeting Mcl-1 in cells Glenna Wink Foight a, Jeremy A. Ryan b, Stefano Gullá a,c, Anthony Letai b,d and Amy E. Keating a * Author affiliations:
Designed BH3 peptides with high affinity and specificity for targeting Mcl-1 in cells Glenna Wink Foight a, Jeremy A. Ryan b, Stefano Gullá a,c, Anthony Letai b,d and Amy E. Keating a * Author affiliations:
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Structure and mechanism of a canonical poly(adp-ribose) glycohydrolase Mark S. Dunstan 1$, Eva Barkauskaite 2$, Pierre Lafite 3, Claire E. Knezevic 4, Amy Brassington 1, Marijan
SUPPLEMENTARY INFORMATION Structure and mechanism of a canonical poly(adp-ribose) glycohydrolase Mark S. Dunstan 1$, Eva Barkauskaite 2$, Pierre Lafite 3, Claire E. Knezevic 4, Amy Brassington 1, Marijan
Comparison of different methods for purification analysis of a green fluorescent Strep-tag fusion protein. Application
 Comparison of different methods for purification analysis of a green fluorescent Strep-tag fusion protein Application Petra Sebastian Meike Kuschel Stefan Schmidt Abstract This Application Note describes
Comparison of different methods for purification analysis of a green fluorescent Strep-tag fusion protein Application Petra Sebastian Meike Kuschel Stefan Schmidt Abstract This Application Note describes
Conformation of the Mineralocorticoid Receptor N- terminal Domain: Evidence for Induced and Stable Structure
 ME-10-0005 Conformation of the Mineralocorticoid Receptor N- terminal Domain: Evidence for Induced and Stable Structure Katharina Fischer 1, Sharon M. Kelly 2, Kate Watt 1, Nicholas C. Price 2 and Iain
ME-10-0005 Conformation of the Mineralocorticoid Receptor N- terminal Domain: Evidence for Induced and Stable Structure Katharina Fischer 1, Sharon M. Kelly 2, Kate Watt 1, Nicholas C. Price 2 and Iain
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Supplementary Figure 1 Speed and run length properties of dynein mutants. Velocity (a,b) and run length (c,d) histograms for SRS/BLH motors of different stalk length and same
SUPPLEMENTARY INFORMATION Supplementary Figure 1 Speed and run length properties of dynein mutants. Velocity (a,b) and run length (c,d) histograms for SRS/BLH motors of different stalk length and same
For quantitative detection of mouse IGF-1 in serum, body fluids, tissue lysates or cell culture supernatants.
 Mouse IGF-1 ELISA Kit (migf-1-elisa) Cat. No. EK0378 96 Tests in 8 x 12 divisible strips Background Insulin-like growth factor 1 (IGF-1), also known as somatomedin C, is a polypeptide protein hormone similar
Mouse IGF-1 ELISA Kit (migf-1-elisa) Cat. No. EK0378 96 Tests in 8 x 12 divisible strips Background Insulin-like growth factor 1 (IGF-1), also known as somatomedin C, is a polypeptide protein hormone similar
Adapta Assay Setup Guide on the BioTek Instruments Synergy 2 Multi-Mode Microplate Reader
 Page 1 of 19 Adapta Assay Setup Guide on the BioTek Instruments Synergy 2 Multi-Mode Microplate Reader NOTE: The BioTek Instruments Synergy 2 Multi-Mode Microplate Reader was tested for compatibility with
Page 1 of 19 Adapta Assay Setup Guide on the BioTek Instruments Synergy 2 Multi-Mode Microplate Reader NOTE: The BioTek Instruments Synergy 2 Multi-Mode Microplate Reader was tested for compatibility with
Identification and characterization of DNA aptamers specific for
 SUPPLEMENTARY INFORMATION Identification and characterization of DNA aptamers specific for phosphorylation epitopes of tau protein I-Ting Teng 1,, Xiaowei Li 1,, Hamad Ahmad Yadikar #, Zhihui Yang #, Long
SUPPLEMENTARY INFORMATION Identification and characterization of DNA aptamers specific for phosphorylation epitopes of tau protein I-Ting Teng 1,, Xiaowei Li 1,, Hamad Ahmad Yadikar #, Zhihui Yang #, Long
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Domain architecture and conformational states of the decapping complex, as revealed by structural studies. (a) Domain organization of Schizosaccharomyces pombe (Sp) and Saccharomyces
Supplementary Figure 1 Domain architecture and conformational states of the decapping complex, as revealed by structural studies. (a) Domain organization of Schizosaccharomyces pombe (Sp) and Saccharomyces
Packaging of P22 DNA requires a pac site while packaging of lambda DNA requires a cos site. Briefly describe:
 1). (12 Points) Packaging of P22 DNA requires a pac site while packaging of lambda DNA requires a cos site. Briefly describe: 1. The mechanisms used by P22 and lambda to package DNA. P22 uses a headfull
1). (12 Points) Packaging of P22 DNA requires a pac site while packaging of lambda DNA requires a cos site. Briefly describe: 1. The mechanisms used by P22 and lambda to package DNA. P22 uses a headfull
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10016 Supplementary discussion on binding site density for protein complexes on the surface: The density of biotin sites on the chip is ~10 3 biotin-peg per µm 2. The biotin sites are
doi:10.1038/nature10016 Supplementary discussion on binding site density for protein complexes on the surface: The density of biotin sites on the chip is ~10 3 biotin-peg per µm 2. The biotin sites are
Rat IGF-1 ELISA Kit (rigf-1-elisa)
 Rat IGF-1 ELISA Kit (rigf-1-elisa) Cat. No. EK0377 96 Tests in 8 x 12 divisible strips Background Insulin-like growth factor 1 (IGF-1), also known as somatomedin C, is a polypeptide protein hormone similar
Rat IGF-1 ELISA Kit (rigf-1-elisa) Cat. No. EK0377 96 Tests in 8 x 12 divisible strips Background Insulin-like growth factor 1 (IGF-1), also known as somatomedin C, is a polypeptide protein hormone similar
Before You Begin. Calibration Protocols
 Before You Begin Read through this entire protocol sheet carefully before you start your experiment and prepare any materials you may need. Calibration Protocols 1. OD 600 Reference point You will use
Before You Begin Read through this entire protocol sheet carefully before you start your experiment and prepare any materials you may need. Calibration Protocols 1. OD 600 Reference point You will use
Cell-Free Protein Expression Kit
 Cell-Free Protein Expression Kit Handbook Version v.01, January 2018 Cat# 507024 (Sigma 70 Master Mix Kit, 24 Rxns) Cat# 507096 (Sigma 70 Master Mix Kit, 96 Rxns) Please refer to this product in your publication
Cell-Free Protein Expression Kit Handbook Version v.01, January 2018 Cat# 507024 (Sigma 70 Master Mix Kit, 24 Rxns) Cat# 507096 (Sigma 70 Master Mix Kit, 96 Rxns) Please refer to this product in your publication
Supporting Information
 Supporting Information Supplementary Figure-1 Results of Phage Display Screening Against TNT The sequences seen in Supplementary Figure-1 represent the phages sequenced after the third and fourth rounds
Supporting Information Supplementary Figure-1 Results of Phage Display Screening Against TNT The sequences seen in Supplementary Figure-1 represent the phages sequenced after the third and fourth rounds
The use of surface plasmon resonance to guide the choice of nucleic acid oligomers for co-crystallisation with proteins
 The use of surface plasmon resonance to guide the choice of nucleic acid oligomers for co-crystallisation with proteins Clare Stevenson John Innes Centre, Norwich, U.K. clare.stevenson@jic.ac.uk RAMC,
The use of surface plasmon resonance to guide the choice of nucleic acid oligomers for co-crystallisation with proteins Clare Stevenson John Innes Centre, Norwich, U.K. clare.stevenson@jic.ac.uk RAMC,
Universal Methyltransferase Activity Assay Kit
 ab139433 Universal Methyltransferase Activity Assay Kit Instructions for Use A complete kit for the screening of candidate compounds that may alter normal Methyltransferase activity. This product is for
ab139433 Universal Methyltransferase Activity Assay Kit Instructions for Use A complete kit for the screening of candidate compounds that may alter normal Methyltransferase activity. This product is for
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Multiple sequence alignments of four Swi2/Snf2 subfamily proteins, ScChd1, SsoRad54 and the RNA helicase Vasa. The sequence alignments of the Swi2/Snf2 subfamily proteins, ScChd1
Supplementary Figure 1 Multiple sequence alignments of four Swi2/Snf2 subfamily proteins, ScChd1, SsoRad54 and the RNA helicase Vasa. The sequence alignments of the Swi2/Snf2 subfamily proteins, ScChd1
Chapter 4 Fluorescence Resonance Energy Transfer (FRET) by Minor Groove-Associated Cyanine-Polyamide Conjugates
 Chapter 4 Fluorescence Resonance Energy Transfer (FRET) by Minor Groove-Associated Cyanine-Polyamide Conjugates The work described in this chapter was accomplished in collaboration with V. Rucker (Dervan
Chapter 4 Fluorescence Resonance Energy Transfer (FRET) by Minor Groove-Associated Cyanine-Polyamide Conjugates The work described in this chapter was accomplished in collaboration with V. Rucker (Dervan
with pentaglycine (GGGGG) peptide motifs. Reaction conditions: (1) hexamethylene
 Supplementary Figure 1. Sequential surface reaction scheme to modify polyurethane catheters with pentaglycine (GGGGG) peptide motifs. Reaction conditions: (1) hexamethylene diisocyanate/triethylamine;
Supplementary Figure 1. Sequential surface reaction scheme to modify polyurethane catheters with pentaglycine (GGGGG) peptide motifs. Reaction conditions: (1) hexamethylene diisocyanate/triethylamine;
Protein Sequence-Structure-Function Relationship: Testing KE-50 Modification on Recombinant Green Fluorescent Protein (AcGFP)
 The University of Akron IdeaExchange@UAkron Honors Research Projects The Dr. Gary B. and Pamela S. Williams Honors College Spring 2016 Protein Sequence-Structure-Function Relationship: Testing KE-50 Modification
The University of Akron IdeaExchange@UAkron Honors Research Projects The Dr. Gary B. and Pamela S. Williams Honors College Spring 2016 Protein Sequence-Structure-Function Relationship: Testing KE-50 Modification
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature09937 a Name Position Primersets 1a 1b 2 3 4 b2 Phenotype Genotype b Primerset 1a D T C R I E 10000 8000 6000 5000 4000 3000 2500 2000 1500 1000 800 Donor (D)
SUPPLEMENTARY INFORMATION doi:10.1038/nature09937 a Name Position Primersets 1a 1b 2 3 4 b2 Phenotype Genotype b Primerset 1a D T C R I E 10000 8000 6000 5000 4000 3000 2500 2000 1500 1000 800 Donor (D)
Solutions to 7.02 Quiz II 10/27/05
 Solutions to 7.02 Quiz II 10/27/05 Class Average = 83 Standard Deviation = 9 Range Grade % 87-100 A 43 74-86 B 39 55-73 C 17 > 54 D 1 Question 1 (56 points) While studying deep sea bacteria, you discover
Solutions to 7.02 Quiz II 10/27/05 Class Average = 83 Standard Deviation = 9 Range Grade % 87-100 A 43 74-86 B 39 55-73 C 17 > 54 D 1 Question 1 (56 points) While studying deep sea bacteria, you discover
Coleman et al., Supplementary Figure 1
 Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Directed Evolution Methods for Protein Engineering
 Directed Evolution Methods for Protein Engineering XXX Biosciences, Life Sciences Solutions, Geneart AG, Regensburg, Germany The world leader in serving science 1 Directed Evolution 2 Random mutagenesis
Directed Evolution Methods for Protein Engineering XXX Biosciences, Life Sciences Solutions, Geneart AG, Regensburg, Germany The world leader in serving science 1 Directed Evolution 2 Random mutagenesis
Lecture 8: Affinity Chromatography-III
 Lecture 8: Affinity Chromatography-III Key words: Chromatography; Affinity chromatography; Protein Purification During this lecture, we shall be studying few more examples of affinity chromatography. The
Lecture 8: Affinity Chromatography-III Key words: Chromatography; Affinity chromatography; Protein Purification During this lecture, we shall be studying few more examples of affinity chromatography. The
Basic concepts of molecular biology
 Basic concepts of molecular biology Gabriella Trucco Email: gabriella.trucco@unimi.it Life The main actors in the chemistry of life are molecules called proteins nucleic acids Proteins: many different
Basic concepts of molecular biology Gabriella Trucco Email: gabriella.trucco@unimi.it Life The main actors in the chemistry of life are molecules called proteins nucleic acids Proteins: many different
University of Bristol - Explore Bristol Research
 Butterer, A., Pernstich, C., Smith, R. M., Sobott, F., Szczelkun, M. D., & Tóth, J. (2014). Type III restriction endonucleases are heterotrimeric: comprising one helicase-nuclease subunit and a dimeric
Butterer, A., Pernstich, C., Smith, R. M., Sobott, F., Szczelkun, M. D., & Tóth, J. (2014). Type III restriction endonucleases are heterotrimeric: comprising one helicase-nuclease subunit and a dimeric
P4EU Heidelberg 2016, June On-column protein cleavage: The Profinity and bdsumo systems. Tamar Unger.
 P4EU Heidelberg 2016, June 15-16 On-column protein cleavage: The Profinity and bdsumo systems Tamar Unger www.weizmann.ac.il/ispc Outline of talk: Description of the Profinity System; Principles, vectors,
P4EU Heidelberg 2016, June 15-16 On-column protein cleavage: The Profinity and bdsumo systems Tamar Unger www.weizmann.ac.il/ispc Outline of talk: Description of the Profinity System; Principles, vectors,
Supplementary Information. Human Antibody-Based Chemically Induced Dimerizers for Cell Therapeutic Applications
 Supplementary Information Human Antibody-Based Chemically Induced Dimerizers for Cell Therapeutic Applications Zachary B Hill 1,4, Alexander J Martinko 1,2,4, Duy P Nguyen 1 & James A Wells *1,3 1 Department
Supplementary Information Human Antibody-Based Chemically Induced Dimerizers for Cell Therapeutic Applications Zachary B Hill 1,4, Alexander J Martinko 1,2,4, Duy P Nguyen 1 & James A Wells *1,3 1 Department
LanthaScreen Europium Assay Setup Guide on the BMG LABTECH CLARIOstar Microplate Readers
 Page 1 of 20 LanthaScreen Europium Assay Setup Guide on the BMG LABTECH CLARIOstar Microplate Readers The BMG LABTECH CLARIOstar Microplate Readers were tested for compatibility with LanthaScreen Eu Kinase
Page 1 of 20 LanthaScreen Europium Assay Setup Guide on the BMG LABTECH CLARIOstar Microplate Readers The BMG LABTECH CLARIOstar Microplate Readers were tested for compatibility with LanthaScreen Eu Kinase
igem 2018 InterLab Study Protocol Before You Begin
 igem 2018 InterLab Study Protocol Before You Begin Read through this entire protocol sheet carefully before you start your experiment and prepare any materials you may need. In order to improve reproducibility,
igem 2018 InterLab Study Protocol Before You Begin Read through this entire protocol sheet carefully before you start your experiment and prepare any materials you may need. In order to improve reproducibility,
Supplementary Information for
 Supplementary Information for Conformational landscapes of DNA polymerase I and mutator derivatives establish fidelity checkpoints for nucleotide insertion Hohlbein et al. 1 Supplementary Figure S1. Wt
Supplementary Information for Conformational landscapes of DNA polymerase I and mutator derivatives establish fidelity checkpoints for nucleotide insertion Hohlbein et al. 1 Supplementary Figure S1. Wt
