Life Sciences Reporting Summary
|
|
|
- Barbra Pope
- 6 years ago
- Views:
Transcription
1 Life Sciences Reporting Summary Corresponding author(s): Craig M. Powell Initial submission Revised version Final submission Nature Research wishes to improve the reproducibility of the work that we publish. This form is intended for publication with all accepted life science papers and provides structure for consistency and transparency in reporting. Every life science submission will use this form; some list items might not apply to an individual manuscript, but all fields must be completed for clarity. For further information on the points included in this form, see Reporting Life Sciences Research. For further information on Nature Research policies, including our data availability policy, see Authors & Referees and the Editorial Policy Checklist. Experimental design 1. Sample size Describe how sample size was determined. 2. Data exclusions Describe any data exclusions. 3. Replication Describe whether the experimental findings were reliably reproduced. 4. Randomization Describe how samples/organisms/participants were allocated into experimental groups. 5. Blinding Describe whether the investigators were blinded to group allocation during data collection and/or analysis. Sample size was chosen based on prior experience of the investigators with similar experiments previously published. The authors have published numerous peer reviewed papers demonstrating clear positive findings with similar sample sizes for the types of experiments included. Furthermore, for many experiments, the sample sizes are far above the standard in the field (example, behavioral experiments with N=20 or more are quite rare). No data points were excluded from analysis in any experiment depicted in this manuscript. While we did not attempt to independently replicate every experiment in this manuscript, several biological replicates are intrinsic to the manuscript. For example, the decrease in synaptic transmission in Kctd13 mutant mice was replicated under initial control conditions, then again once each in the vehicle controls for each of the two drugs tested, verifying that the initial finding was very reproducible (in both extracellular field recordings and in whole-cell patch clamp recordings). Loss of Kctd13 protein was reliably replicated. Beta-galactosidase staining was biologically replicated. Randomization was performed by blinding the investigator to genotype and allowing them to choose each subject blindly. No animals or slices were excluded from the analysis. Randomization was performed by blinding the investigator to genotype and allowing them to choose each subject blindly. For studies using pharmacological agents, the investigator was aware of the agent being used, but was not aware of the genotypes of the animals used. Investigators were not aware of the specific group to which an animal was assigned when doing the experiment or analysis until after completion. Note: all studies involving animals and/or human research participants must disclose whether blinding and randomization were used. nature research life sciences reporting summary June
2 6. Statistical parameters n/a For all figures and tables that use statistical methods, confirm that the following items are present in relevant figure legends (or in the Methods section if additional space is needed). Confirmed Software The exact sample size (n) for each experimental group/condition, given as a discrete number and unit of measurement (animals, litters, cultures, etc.) A description of how samples were collected, noting whether measurements were taken from distinct samples or whether the same sample was measured repeatedly A statement indicating how many times each experiment was replicated The statistical test(s) used and whether they are one- or two-sided (note: only common tests should be described solely by name; more complex techniques should be described in the Methods section) A description of any assumptions or corrections, such as an adjustment for multiple comparisons The test results (e.g. P values) given as exact values whenever possible and with confidence intervals noted A clear description of statistics including central tendency (e.g. median, mean) and variation (e.g. standard deviation, interquartile range) Clearly defined error bars Policy information about availability of computer code 7. Software Describe the software used to analyze the data in this study. See the web collection on statistics for biologists for further resources and guidance. Statistica, Excel, Graphpad Prism were used to analyze data in this study. For manuscripts utilizing custom algorithms or software that are central to the paper but not yet described in the published literature, software must be made available to editors and reviewers upon request. We strongly encourage code deposition in a community repository (e.g. GitHub). Nature Methods guidance for providing algorithms and software for publication provides further information on this topic. Materials and reagents Policy information about availability of materials 8. Materials availability Indicate whether there are restrictions on availability of unique materials or if these materials are only available for distribution by a for-profit company. All unique materials will be made available by the corresponding author on request. Should such requests become unduly burdensome, the corresponding author may deposit such materials in a widely accessible repository such as Jackson Laboratories or other Murine Mouse resources. nature research life sciences reporting summary June
3 9. Antibodies Describe the antibodies used and how they were validated for use in the system under study (i.e. assay and species). Western blot, antibodies, RhoA activity Fresh hippocampal tissue was isolated via rapid decapitation/dissection of males and females and lysed in ice-cold RIPA buffer (#89901, ThermoScientific, Waltham, MA) supplemented with Halt Protease & Phosphatase Inhibitor cocktail (#78446, ThermoScientific, Waltham, MA). Individual samples were sonicated, centrifuged at 4 C for 1 min at 1000 RPM, diluted in 4x Laemmli sample buffer containing β- mercaptoethanol (Bio-Rad, Hercules, CA), and incubated at 100 C for 5 min. Protein concentration was determined by BCA Protein Assay (#23225, ThermoScientific, Waltham, MA). Samples were diluted with sample buffer such that 5-25 μg of protein were loaded per lane onto Criterion TGX gels either 10% or 12% (Bio-Rad). Gels were run and then transferred to nitrocellulose membranes, blocked in Odyssey Blocking Buffer ( # , LI-COR, Lincoln, NE), and incubated with primary antibody overnight at 4 C. Membranes were washed in 5% TBS-T followed by a one-h secondary antibody incubation at room temperature using infrared fluorescence IRDye anti-rabbit (# , LI-COR, Lincoln, NE) or anti-mouse (# , LI-COR, Lincoln, NE) antibodies at 1:10,000. Proteins were visualized with Odyssey fc imager (LI-COR, Lincoln, NE). Signals were quantified using Image Studio Lite (LI-COR, Lincoln, NE), normalized to β-actin or gapdh and analyzed using Microsoft Excel and Graphpad Prism (La Jolla, CA). Antibodies used: KCTD13 (HPA043524, Atlas Antibodies, Stockholm, Sweden 1:400), RhoA (2117, Cell Signaling, Danvers, MA, 1:1000), actin (#MABT825, Sigma- Aldrich, St. Louis, MO, 1:50000). Both KCTD13 and RhoA antibody bands were verified using corresponding knockout brain tissue (RhoA KO brain tissue was a gift from Dr. Kim Tolias). Non-fluorescent immunohistochemistry: Mixed male and female mice were transcardially-perfused with 0.1 M PBS followed by 4% PFA (paraformaldehyde, NC , Fisher, Hampton, NH). Brains were removed, fixed with PFA for 24 h and dehydrated with 30% sucrose. Tissue was coronally sectioned in a series of ten (1:10) on a sliding microtome at 30 μm. Slide mounted sections were incubated with 3% of H2O2, 10% methanol in 0.1 M PBS and then blocked in 5% serum, 0.2% Triton X in 0.1 M PBS. Primary antibodies were diluted in the antibody solution (5% serum in 0.1 M PBS) and were applied overnight at room temperature. Slides were next incubated for 1 h with secondary antibodies (1:3000, Vector Labs, Burlingame, CA) followed by an avidin-biotin-hrp complex formed with VECTASTAIN ABC kit (PK-4000, Vector labs, Burlingame, CA). A 10-min exposure to Metal-Enhanced DAB (#34065, ThermoScientific, Waltham, MA) was performed to visualize the stain. Dehydration was performed with a series of 5-min ethanol dilutions (75%, 95%, 100%) and CitriSolv ( , Fisher, Hampton, NH) incubation prior to coverslipping with DPX mounting medium. For experiments involving BrdU staining, tissue was treated with 2 M HCL for 30 min and washed with 0.1 M Borax solution before blocking the endogenous peroxidase. Antibodies used: BrdU (1:400, #6326, Abcam, Cambridge, United Kingdom), Doublecortin (1:3000, #8066, Santa Cruz, Dallas, TX), Ki67 (1:400, MA , ThermoScientific, Waltham, MA). Fluorescent immunohistochemistry: 20-μm cryosectioned E15.5 brain sections or 30-μm slide-mounted adult brain sections were incubated in blocking solution (0.2 % Triton X, 10 % Serum, 1 % BSA in 0.1 M PBS) for 1 h. Primary antibodies were diluted in 5 % serum and 0.1 M PBS and were incubated overnight. Secondary antibodies (1:1000, Invitrogen, Carlsbad, CA) were applied for 1 h at room temperature. After extensive washing, sections were counterstained with DAPI (5 mg/ml, D3571, ThermoFisher, Waltham, MA) for 1 min. Prolong gold mounting media (ThermoFisher, #P36930) was used to secure coverslips. Antibodies used: GFP (1:1000, ab13970), Tbr1 (1:100, ab31940), Tbr2 (1:100, ab23345), Ctip2 (1:100, ab18465), Satb2 (1:10, ab92446, Abcam, Cambridge, United Kingdom), Cux1 (1:50, 13024, Santa Cruz, Dallas, TX), Tuj1 (1:1000, mms-435p, BioLegend San Diego, CA). nature research life sciences reporting summary June
4 10. Eukaryotic cell lines a. State the source of each eukaryotic cell line used. No eukaryotic cell lines were used. b. Describe the method of cell line authentication used. No eukaryotic cell lines were used. c. Report whether the cell lines were tested for mycoplasma contamination. d. If any of the cell lines used are listed in the database of commonly misidentified cell lines maintained by ICLAC, provide a scientific rationale for their use. Animals and human research participants No eukaryotic cell lines were used. No eukaryotic cell lines were used. Policy information about studies involving animals; when reporting animal research, follow the ARRIVE guidelines 11. Description of research animals Provide details on animals and/or animal-derived materials used in the study. Policy information about studies involving human research participants 12. Description of human research participants Describe the covariate-relevant population characteristics of the human research participants. Kctd13 mutants were created using targeted embryonic stem cells (ES line: VGB6, Strain: C57BL/6NTac) were obtained from the University of California, Davis Knockout Mouse Project (KOMP) Repository. These ES cells carry the targeting construct designed to replace the entire Mus musculus Kctd13 gene (from second reading frame to seventeenth base pair after stop codon of the gene, total 16,080 bp). This construct was created as a ZEN-UB1 cassette (assembled by combining a LacZ-p(A) gene and a hubcpro-neo-p(a) cassette flanked by 2 loxp sequences). The neo cassette provided a positive selection marker for the targeted ES clones. The LacZ-p(A) gene provided a reporter for Kctd13 promoter activity. ES clones were injected into blastocysts (background: Albino c57bl/6) to generate chimeras at the Transgenic Facility of University of Texas Southwestern Medical Center at Dallas. Resulting chimeric mice were bred with C57BL/6J mice to confirm germline transmission identified by polymerase chain reaction (PCR) with three primers as follows: forward (1): SD-sense (CTCGGATCTTTGAGGAGACAC), reverse: SD (TGTGGCTGATAGCACTGTCC), forward (2): NeoFwd (TCATTCTCAGTATTGTTTTGCC). WT DNA produced a 546 bp band. KI (knock-in targeting construct to replace Kctd13 gene) DNA produced a 408 bp band. To confirm correct targeting, genomic DNA from WT and KI mice were analyzed by Southern blotting with a probe that distinguished between the WT and KI Kctd13 alleles. The WT DNA resulted in a 5,790 bp band while KI DNA resulted in a 9,454 bp band (not shown). KI mice were further crossed with germline-cre expressing, B6.C-Tg(CMV-Cre), Cgn/J mice (The Jackson Laboratory, Bar Harbor, ME, background: C57BL/6J) to remove the hubcpro-neo-p(a) cassette. Successful removal of this selection cassette was identified by PCR using three primers as follows: forward: SU-sense-1 (GCGGTATTCTCCATCCACATGAACAAGG), reverse (1): SU-anti-1 (TGGGACTAGGGAGCCTGGAATGAACTG), reverse (2): LacZ-Rev (GCTGGCTTGGTCTGTCTGTCCTAGC). WT DNA produced a 659 bp band while KO (knockout Kctd13 and neo) DNA produced a 487 bp band (not shown). We also performed rt-pcr (reverse transcriptase-pcr) to confirm loss of Kctd13 mrna in the KO mice (forward: GTGAAGCTTCTACACAACCGCAG, reverse: AAGATGTTCAGTGTCTCCTCAAAG, band=267 bp). The resulting constitutive Kctd13 deletion mice were bred as het X het for all experiments. All animal care and use were approved by the UT Southwestern Medical Center Institutional Animal Care and Use Committee and are compliant with US government principles regarding the care and use of animals, Public Health Service Policy on Humane Care and Use of Laboratory Animals, Guide for the Care and Use of Laboratory Animals, and the Animal Welfare Act. Animal husbandry was performed in a facility accredited by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) International. The study did not involve human research participants. nature research life sciences reporting summary June
5 MRI Studies Reporting Summary Form fields will expand as needed. Please do not leave fields blank. Experimental design 1. Describe the experimental design. 2. Specify the number of blocks, trials or experimental units per session and/or subject, and specify the length of each trial or block (if trials are blocked) and interval between trials. 3. Describe how behavioral performance was measured. Corresponding author(s): Mice were scanned Ex-Vivo Craig M. Powell Initial submission Revised version Final submission A multi-channel 7.0 Tesla MRI scanner (Agilent Inc., Palo Alto, CA) was used to image the brains within the skulls. 16 custom-built solenoid coils were used to image the brains in parallel71,72. Two different age groups different sequences were required. In order to get the contrast required for image registration at p7, a 3-D diffusion weighted fast spin-echo sequence with an echo train length of 6 was used, with a TR of 270 ms, first TE of 30 ms, and a TE of 10 ms for the remaining 5 echoes, two averages, field-of-view 14 x 14 x 25 mm3 and a matrix size of 250 x 250 x 450 yielding an image with mm isotropic voxels. 6 high b-value images (b=2147 s/mm2) in 6 orthogonal directions were acquired. This was done to acquire an improved gray/white matter contrast at the young age. Total imaging time was 14 h. The high b-value images were averaged to create a single diffusion weighted image that had the contrast required for image registration. For the adult brains, a T2- weighted 3D fast spin-echo sequence was also used, with a cylindrical acquisition of k-space, and with a TR of 350 ms, and TEs of 12 ms per echo for 6 echoes, two averages, field-of-view of 20 x 20 x 25 mm3 and matrix size = 504 x 504 x 630 giving an image with mm isotropic voxels73. The current scan time required for this sequence is also ~14 hours. To visualize and compare any differences in the mouse brains the images from each age group are linearly (6 parameter followed by a 12 parameter) and non-linearly registered together separated by age group. We then used Deformation based morphometry to determine the volume difference between groups both regionally and voxelwise. Two scans were performed on Ex-Vivo Samples of two different age groups. A 3D - Diffusion Weighted FSE sequence was used to assess differences in the younger age group and a regular 3D FSE sequence was used for the adult group. As this was not an fmri study no block or trials per session and/or subject is required for this testing. for MRI experiments in this paper nature research flow cytometry reporting summary June
6 Acquisition 4. Imaging a. Specify the type(s) of imaging. Structural MRI b. Specify the field strength (in Tesla). Methods, page 30 A multi-channel 7.0 Tesla MRI scanner (Agilent Inc., Palo Alto, CA) c. Provide the essential sequence imaging parameters. Methods, page 30 A multi-channel 7.0 Tesla MRI scanner (Agilent Inc., Palo Alto, CA) was used to image the brains within the skulls. 16 custom-built solenoid coils were used to image the brains in parallel71,72. d. For diffusion MRI, provide full details of imaging parameters. For the two different age groups different sequences were required. In order to get the contrast required for image registration at p7, a 3-D diffusion weighted fast spin-echo sequence with an echo train length of 6 was used, with a TR of 270 ms, first TE of 30 ms, and a TE of 10 ms for the remaining 5 echoes, two averages, field-of-view 14 x 14 x 25 mm3 and a matrix size of 250 x 250 x 450 yielding an image with mm isotropic voxels. 6 high b-value images (b=2147 s/mm2) in 6 orthogonal directions were acquired. This was done to acquire an improved gray/white matter contrast at the young age. Total imaging time was 14 h. The high b-value images were averaged to create a single diffusion weighted image that had the contrast required for image registration. For the adult brains, a T2- weighted 3D fast spin-echo sequence was also used, with a cylindrical acquisition of k-space, and with a TR of 350 ms, and TEs of 12 ms per echo for 6 echoes, two averages, field-of-view of 20 x 20 x 25 mm3 and matrix size = 504 x 504 x 630 giving an image with mm isotropic voxels73. The current scan time required for this sequence is also ~14 hours. For the younger age group DTI was performed to increase the contrast between the gray and white matter at the young age when myelination is at its infancy. The parameters are as follows: a 3-D diffusion weighted fast spin-echo sequence with an echo train length of 6 was used, with a TR of 270 ms, first TE of 30 ms, and a TE of 10 ms for the remaining 5 echoes, two averages, field-of-view 14 x 14 x 25 mm3 and a matrix size of 250 x 250 x 450 yielding an image with mm isotropic voxels. 6 high b-value images (b=2147 s/mm2) in 6 orthogonal directions were acquired. The images used for registration and analysis were an average of the high b-value images (in the 6 orthogonal directions). 5. State area of acquisition. The whole brain was imaged at both the younger and older age groups. We used pre-existing atlases that included full brain coverage. For the older age group this included 159 different regions encompassing the entire brain volume. For the younger age group the atlas included 59 different regions again covering the entire brain volume. There are further delineations in the older age atlas for the cortex and cerebellum which accounts for the different number of regions between two groups. Preprocessing 6. Describe the software used for preprocessing. All of our software is home-built using a combination of R, Perl, Python and C. The only preprocessing that is used prior to the registration is a distortion correction, which is used to eliminate the distortions created by an inhomgeneous magnetic field. nature research flow cytometry reporting summary June
7 7. Normalization a. If data were normalized/standardized, describe the approach(es). b. Describe the template used for normalization/ transformation. 8. Describe your procedure for artifact and structured noise removal. 9. Define your software and/or method and criteria for volume censoring, and state the extent of such censoring. Statistical modeling & inference 10. Define your model type and settings. For regional measurements p-values are calculated using a two tailed t- test and voxelwise measurements we are using a linear model to determin differences in volume. These are both controlled for multiple comparisons using the false discovery rate. See Methods Page Specify the precise effect tested. The Jacobian determinants are used after deformation based morphometry to determined significant differences in the voxelwise measurements. Standard two-tailed t-tests are used on regional volumes either measured as absolute volume in mm^3 or relative volume as a % total brain volume. Methods, page 30 & Extended Data page Analysis a. Specify whether analysis is whole brain or ROI-based. The whole brain is examined voxelwise as well as a regional ROI assessment encompassing the whole brain volume. b. If ROI-based, describe how anatomical locations were determined. 13. State the statistic type for inference. (See Eklund et al ) 14. Describe the type of correction and how it is obtained for multiple comparisons. 15. Connectivity a. For functional and/or effective connectivity, report the measures of dependence used and the model details. b. For graph analysis, report the dependent variable and functional connectivity measure. Pre existing atlases were used to calculate the volume of 159 different regions throughout the whole brain volume. Please see references: Dorr et al. NeuroImage 2008 Ullman et al. NeuroImage 2013 Steadman et al. Autism Research 2012 T-Values See Methods, page 30 & Extended Data page 49 We used the False Discovery Rate to control for Multiple comparisons. Please see reference Genovese et al See Methods, page 30 & Extended Data page 49 nature research flow cytometry reporting summary June
8 16. For multivariate modeling and predictive analysis, specify independent variables, features extraction and dimension reduction, model, training and evaluation metrics. The neuroanatomy was assessed and volume was measured as either absolute volume in mm3 or relative volume (% total brain volume). Methods, page 30 & Extended Data page 49. Supplemental Data Table S2 & Supplemental Data Table S3. nature research flow cytometry reporting summary June
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION DOI: 10.1038/ncb3575 In the format provided by the authors and unedited. Supplementary Figure 1 Validation of key reagents and assays a, top, IHC with antibody recognizing specifically
SUPPLEMENTARY INFORMATION DOI: 10.1038/ncb3575 In the format provided by the authors and unedited. Supplementary Figure 1 Validation of key reagents and assays a, top, IHC with antibody recognizing specifically
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling
 Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
SANTA CRUZ BIOTECHNOLOGY, INC.
 TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
Experimental Protocol for Multiplex Fluorescent Blotting Using the ChemiDoc MP Imaging System
 Experimental Protocol for Multiplex Fluorescent Blotting Using the ChemiDoc MP Imaging System Protocol Bulletin 6570 Stefanie L. Ritter and Donald G. Rainie, Deparment of Behavioral Neuroscience and Psychiatric
Experimental Protocol for Multiplex Fluorescent Blotting Using the ChemiDoc MP Imaging System Protocol Bulletin 6570 Stefanie L. Ritter and Donald G. Rainie, Deparment of Behavioral Neuroscience and Psychiatric
Segments of the obstructed intestinal loops were fixed in 4% paraformaldehyde
 Supplementary text Supplementary materials and methods Histopathological examination Segments of the obstructed intestinal loops were fixed in 4% paraformaldehyde (PFA) and embedded in paraffin wax with
Supplementary text Supplementary materials and methods Histopathological examination Segments of the obstructed intestinal loops were fixed in 4% paraformaldehyde (PFA) and embedded in paraffin wax with
Figure S2. Response of mouse ES cells to GSK3 inhibition. Mentioned in discussion
 Stem Cell Reports, Volume 1 Supplemental Information Robust Self-Renewal of Rat Embryonic Stem Cells Requires Fine-Tuning of Glycogen Synthase Kinase-3 Inhibition Yaoyao Chen, Kathryn Blair, and Austin
Stem Cell Reports, Volume 1 Supplemental Information Robust Self-Renewal of Rat Embryonic Stem Cells Requires Fine-Tuning of Glycogen Synthase Kinase-3 Inhibition Yaoyao Chen, Kathryn Blair, and Austin
RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by. 5 AGACACAAACACCAUUGUCACACUCCACAGC; Rand-2 OMe,
 Materials and methods Oligonucleotides and DNA constructs RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by Dharmacon Inc. (Lafayette, CO). The sequences were: 122-2 OMe, 5
Materials and methods Oligonucleotides and DNA constructs RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by Dharmacon Inc. (Lafayette, CO). The sequences were: 122-2 OMe, 5
Technical Note Detection of post-immunoprecipitation proteins by Western blot using the Quick Western Kit IRDye 680RD
 Technical Note Detection of post-immunoprecipitation proteins by Western blot using the Quick Western Kit IRDye 680RD Developed for: Aerius, Odyssey Classic, Odyssey CLx and Odyssey Sa Imaging Systems
Technical Note Detection of post-immunoprecipitation proteins by Western blot using the Quick Western Kit IRDye 680RD Developed for: Aerius, Odyssey Classic, Odyssey CLx and Odyssey Sa Imaging Systems
Technical Note. Housekeeping Protein Validation Protocol
 Technical Note Housekeeping Protein Validation Protocol Published March 2017. The most recent version of this Technical Note is posted at licor.com/bio/support. Visit us on protocols.io! Explore an interactive
Technical Note Housekeeping Protein Validation Protocol Published March 2017. The most recent version of this Technical Note is posted at licor.com/bio/support. Visit us on protocols.io! Explore an interactive
TrkB knockdown cell lines (i.e., BBM1-KD and 361-KD cells) were prepared by
 Supplemental Information Cell Culture TrkB knockdown cell lines (i.e., BBM1-KD and 361-KD cells) were prepared by transducing BBM1 or 361 cells with a lentivirus encoding shrna for TrkB. Transduction was
Supplemental Information Cell Culture TrkB knockdown cell lines (i.e., BBM1-KD and 361-KD cells) were prepared by transducing BBM1 or 361 cells with a lentivirus encoding shrna for TrkB. Transduction was
Confocal immunofluorescence microscopy
 Confocal immunofluorescence microscopy HL-6 and cells were cultured and cytospun onto glass slides. The cells were double immunofluorescence stained for Mt NPM1 and fibrillarin (nucleolar marker). Briefly,
Confocal immunofluorescence microscopy HL-6 and cells were cultured and cytospun onto glass slides. The cells were double immunofluorescence stained for Mt NPM1 and fibrillarin (nucleolar marker). Briefly,
GM130 Is Required for Compartmental Organization of Dendritic Golgi Outposts
 Current Biology, Volume 24 Supplemental Information GM130 Is Required for Compartmental Organization of Dendritic Golgi Outposts Wei Zhou, Jin Chang, Xin Wang, Masha G. Savelieff, Yinyin Zhao, Shanshan
Current Biology, Volume 24 Supplemental Information GM130 Is Required for Compartmental Organization of Dendritic Golgi Outposts Wei Zhou, Jin Chang, Xin Wang, Masha G. Savelieff, Yinyin Zhao, Shanshan
Introducing new DNA into the genome requires cloning the donor sequence, delivery of the cloned DNA into the cell, and integration into the genome.
 Key Terms Chapter 32: Genetic Engineering Cloning describes propagation of a DNA sequence by incorporating it into a hybrid construct that can be replicated in a host cell. A cloning vector is a plasmid
Key Terms Chapter 32: Genetic Engineering Cloning describes propagation of a DNA sequence by incorporating it into a hybrid construct that can be replicated in a host cell. A cloning vector is a plasmid
Ramp1 EPD0843_4_B11. EUCOMM/KOMP-CSD Knockout-First Genotyping
 EUCOMM/KOMP-CSD Knockout-First Genotyping Introduction The majority of animals produced from the EUCOMM/KOMP-CSD ES cell resource contain the Knockout-First-Reporter Tagged Insertion allele. As well as
EUCOMM/KOMP-CSD Knockout-First Genotyping Introduction The majority of animals produced from the EUCOMM/KOMP-CSD ES cell resource contain the Knockout-First-Reporter Tagged Insertion allele. As well as
Usp14 EPD0582_2_G09. EUCOMM/KOMP-CSD Knockout-First Genotyping
 EUCOMM/KOMP-CSD Knockout-First Genotyping Introduction The majority of animals produced from the EUCOMM/KOMP-CSD ES cell resource contain the Knockout-First-Reporter Tagged Insertion allele. As well as
EUCOMM/KOMP-CSD Knockout-First Genotyping Introduction The majority of animals produced from the EUCOMM/KOMP-CSD ES cell resource contain the Knockout-First-Reporter Tagged Insertion allele. As well as
Isolation, culture, and transfection of primary mammary epithelial organoids
 Supplementary Experimental Procedures Isolation, culture, and transfection of primary mammary epithelial organoids Primary mammary epithelial organoids were prepared from 8-week-old CD1 mice (Charles River)
Supplementary Experimental Procedures Isolation, culture, and transfection of primary mammary epithelial organoids Primary mammary epithelial organoids were prepared from 8-week-old CD1 mice (Charles River)
To isolate single GNS 144 cell clones, cells were plated at a density of 1cell/well
 Supplemental Information: Supplemental Methods: Cell culture To isolate single GNS 144 cell clones, cells were plated at a density of 1cell/well in 96 well Primaria plates in GNS media and incubated at
Supplemental Information: Supplemental Methods: Cell culture To isolate single GNS 144 cell clones, cells were plated at a density of 1cell/well in 96 well Primaria plates in GNS media and incubated at
IKK is a therapeutic target in KRAS-induced lung cancer with disrupted p53 activity
 IKK is a therapeutic target in KRAS-induced lung cancer with disrupted p5 activity H6 5 5 H58 A59 H6 H58 A59 anti-ikkα anti-ikkβ anti-panras anti-gapdh anti-ikkα anti-ikkβ anti-panras anti-gapdh anti-ikkα
IKK is a therapeutic target in KRAS-induced lung cancer with disrupted p5 activity H6 5 5 H58 A59 H6 H58 A59 anti-ikkα anti-ikkβ anti-panras anti-gapdh anti-ikkα anti-ikkβ anti-panras anti-gapdh anti-ikkα
Cell culture and drug treatment. Lineage - Sca-1+ CD31+ EPCs were cultured on
 Supplemental Material Detailed Methods Cell culture and drug treatment. Lineage - Sca-1+ CD31+ EPCs were cultured on 5µg/mL human fibronectin coated plates in DMEM supplemented with 10% FBS and penicillin/streptomycin
Supplemental Material Detailed Methods Cell culture and drug treatment. Lineage - Sca-1+ CD31+ EPCs were cultured on 5µg/mL human fibronectin coated plates in DMEM supplemented with 10% FBS and penicillin/streptomycin
Supplemental Materials. Matrix Proteases Contribute to Progression of Pelvic Organ Prolapse in Mice and Humans
 Supplemental Materials Matrix Proteases Contribute to Progression of Pelvic Organ Prolapse in Mice and Humans Madhusudhan Budatha, Shayzreen Roshanravan, Qian Zheng, Cecilia Weislander, Shelby L. Chapman,
Supplemental Materials Matrix Proteases Contribute to Progression of Pelvic Organ Prolapse in Mice and Humans Madhusudhan Budatha, Shayzreen Roshanravan, Qian Zheng, Cecilia Weislander, Shelby L. Chapman,
PARP-1 (cleaved) Human In-Cell ELISA Kit (IR)
 ab110215 PARP-1 (cleaved) Human In-Cell ELISA Kit (IR) Instructions for Use For the quantitative measurement of Human PARP-1 (cleaved) concentrations in cultured adherent and suspension cells. This product
ab110215 PARP-1 (cleaved) Human In-Cell ELISA Kit (IR) Instructions for Use For the quantitative measurement of Human PARP-1 (cleaved) concentrations in cultured adherent and suspension cells. This product
For identifying inhibitors and activators of mitochondrial biogenesis in adherent cultured cells.
 ab110216 MitoBiogenesis TM In-Cell ELISA Kit (IR) Instructions for Use For identifying inhibitors and activators of mitochondrial biogenesis in adherent cultured cells. This product is for research use
ab110216 MitoBiogenesis TM In-Cell ELISA Kit (IR) Instructions for Use For identifying inhibitors and activators of mitochondrial biogenesis in adherent cultured cells. This product is for research use
In-Cell Western Kits I and II
 Odyssey and Aerius Infrared Imaging Systems In-Cell Western Assay Kits I and II Published November, 2006. The most recent version of this protocol is posted at http://biosupport.licor.com/protocols.jsp
Odyssey and Aerius Infrared Imaging Systems In-Cell Western Assay Kits I and II Published November, 2006. The most recent version of this protocol is posted at http://biosupport.licor.com/protocols.jsp
ab Ran Activation Assay Kit
 ab173247 Ran Activation Assay Kit Instructions for Use For the simple and fast measurement of Ran activation. This product is for research use only and is not intended for diagnostic use. Version 1 Last
ab173247 Ran Activation Assay Kit Instructions for Use For the simple and fast measurement of Ran activation. This product is for research use only and is not intended for diagnostic use. Version 1 Last
ab BrdU Immunohistochemistry Kit
 ab125306 - BrdU Immunohistochemistry Kit Instructions for Use For the detection and localization of bromodeoxyuridine incorporated into newly synthesized DNA of actively proliferating cells. This product
ab125306 - BrdU Immunohistochemistry Kit Instructions for Use For the detection and localization of bromodeoxyuridine incorporated into newly synthesized DNA of actively proliferating cells. This product
Multiplex Fluorescent Western Blot Starter Kit for the Bio- Rad ChemiDoc MP
 Page 1 of 7 INSTRUCTIONS: Z-310 Multiplex Fluorescent Western Blot Starter Kit for the Bio- Rad ChemiDoc MP Rockland Immunochemicals and Bio-Rad Laboratories have jointly developed an easy to use multiplex
Page 1 of 7 INSTRUCTIONS: Z-310 Multiplex Fluorescent Western Blot Starter Kit for the Bio- Rad ChemiDoc MP Rockland Immunochemicals and Bio-Rad Laboratories have jointly developed an easy to use multiplex
Strep-tag detection in Western blots
 Strep-tag detection in Western blots General protocol for the detection of Strep-tag fusion proteins Last date of revision April 2012 Version PR07-0010 www.strep-tag.com For research use only Important
Strep-tag detection in Western blots General protocol for the detection of Strep-tag fusion proteins Last date of revision April 2012 Version PR07-0010 www.strep-tag.com For research use only Important
In-Cell Western Assay
 In-Cell Western Assay Complete Sample Protocol for Measuring IC 50 of Inhibitor U0126 in NIH3T3 Responding to Acidic Fibroblast Growth Factor (afgf-1) Developed for: Aerius, Odyssey Classic, Odyssey CLx,
In-Cell Western Assay Complete Sample Protocol for Measuring IC 50 of Inhibitor U0126 in NIH3T3 Responding to Acidic Fibroblast Growth Factor (afgf-1) Developed for: Aerius, Odyssey Classic, Odyssey CLx,
TRANSGENIC TECHNOLOGIES: Gene-targeting
 TRANSGENIC TECHNOLOGIES: Gene-targeting Reverse Genetics Wild-type Bmp7 -/- Forward Genetics Phenotype Gene or Mutations First Molecular Analysis Second Reverse Genetics Gene Phenotype or Molecular Analysis
TRANSGENIC TECHNOLOGIES: Gene-targeting Reverse Genetics Wild-type Bmp7 -/- Forward Genetics Phenotype Gene or Mutations First Molecular Analysis Second Reverse Genetics Gene Phenotype or Molecular Analysis
Mouse Engineering Technology. Musculoskeletal Research Center 2016 Summer Educational Series David M. Ornitz Department of Developmental Biology
 Mouse Engineering Technology Musculoskeletal Research Center 2016 Summer Educational Series David M. Ornitz Department of Developmental Biology Core service and new technologies Mouse ES core Discussions
Mouse Engineering Technology Musculoskeletal Research Center 2016 Summer Educational Series David M. Ornitz Department of Developmental Biology Core service and new technologies Mouse ES core Discussions
Beta3 integrin promotes long-lasting activation and polarization of Vascular Endothelial Growth Factor Receptor 2 by immobilized ligand
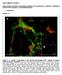 SUPPLEMENTAL FIGURES Beta3 integrin promotes long-lasting activation and polarization of Vascular Endothelial Growth Factor Receptor 2 by immobilized ligand C. Ravelli et al. FIGURE S. I Figure S. I: Gremlin
SUPPLEMENTAL FIGURES Beta3 integrin promotes long-lasting activation and polarization of Vascular Endothelial Growth Factor Receptor 2 by immobilized ligand C. Ravelli et al. FIGURE S. I Figure S. I: Gremlin
Supplemental information
 Supplemental information - Control samples (200 subjects) - Immunohistochemistry of rat brain - Immunocytochemistry on neuronal cultures - Immunocompetition assay - Immunoprecipitation - Immunocytochemistry
Supplemental information - Control samples (200 subjects) - Immunohistochemistry of rat brain - Immunocytochemistry on neuronal cultures - Immunocompetition assay - Immunoprecipitation - Immunocytochemistry
Figure S1. Figure S2. Figure S3 HB Anti-FSP27 (COOH-terminal peptide) Ab. Anti-GST-FSP27(45-127) Ab.
 / 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
/ 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1137999/dc1 Supporting Online Material for Disrupting the Pairing Between let-7 and Enhances Oncogenic Transformation Christine Mayr, Michael T. Hemann, David P. Bartel*
www.sciencemag.org/cgi/content/full/1137999/dc1 Supporting Online Material for Disrupting the Pairing Between let-7 and Enhances Oncogenic Transformation Christine Mayr, Michael T. Hemann, David P. Bartel*
LI-COR, Odyssey, Aerius, IRDye and In-Cell Western are registered trademarks or trademarks of LI-COR Biosciences Inc.
 PROTOCOL PARP-1 (cleaved) In-Cell ELISA Kit (IR) 185 Millrace Drive, Suite 3A Eugene, Oregon 9743 MSA43 Rev. DESCRIPTION An In-Cell ELISA kit for measuring cleaved PARP-1 (89kDa fragment) in human adherent
PROTOCOL PARP-1 (cleaved) In-Cell ELISA Kit (IR) 185 Millrace Drive, Suite 3A Eugene, Oregon 9743 MSA43 Rev. DESCRIPTION An In-Cell ELISA kit for measuring cleaved PARP-1 (89kDa fragment) in human adherent
1. Cross-linking and cell harvesting
 ChIP is a powerful tool that allows the specific matching of proteins or histone modifications to regions of the genome. Chromatin is isolated and antibodies to the antigen of interest are used to determine
ChIP is a powerful tool that allows the specific matching of proteins or histone modifications to regions of the genome. Chromatin is isolated and antibodies to the antigen of interest are used to determine
This Document Contains:
 This Document Contains: 1. In-Cell Western Protocol II. Cell Seeding and Stimulation Supplemental Protocol III. Complete Assay Example: Detailing the Seeding, Stimulation and Detection of the A431 Cellular
This Document Contains: 1. In-Cell Western Protocol II. Cell Seeding and Stimulation Supplemental Protocol III. Complete Assay Example: Detailing the Seeding, Stimulation and Detection of the A431 Cellular
Supplemental Data. LMO4 Controls the Balance between Excitatory. and Inhibitory Spinal V2 Interneurons
 Neuron, Volume 61 Supplemental Data LMO4 Controls the Balance between Excitatory and Inhibitory Spinal V2 Interneurons Kaumudi Joshi, Seunghee Lee, Bora Lee, Jae W. Lee, and Soo-Kyung Lee Supplemental
Neuron, Volume 61 Supplemental Data LMO4 Controls the Balance between Excitatory and Inhibitory Spinal V2 Interneurons Kaumudi Joshi, Seunghee Lee, Bora Lee, Jae W. Lee, and Soo-Kyung Lee Supplemental
sirna Transfection Into Primary Neurons Using Fuse-It-siRNA
 sirna Transfection Into Primary Neurons Using Fuse-It-siRNA This Application Note describes a protocol for sirna transfection into sensitive, primary cortical neurons using Fuse-It-siRNA. This innovative
sirna Transfection Into Primary Neurons Using Fuse-It-siRNA This Application Note describes a protocol for sirna transfection into sensitive, primary cortical neurons using Fuse-It-siRNA. This innovative
ab Mouse and Rabbit Specific HRP/DAB (ABC) Detection IHC Kit
 ab64264 - Mouse and Rabbit Specific HRP/DAB (ABC) Detection IHC Kit Instructions for Use For the detection of a specific antibody bound to an antigen in tissue sections. This product is for research use
ab64264 - Mouse and Rabbit Specific HRP/DAB (ABC) Detection IHC Kit Instructions for Use For the detection of a specific antibody bound to an antigen in tissue sections. This product is for research use
Life Sciences Reporting Summary
 Life Sciences Reporting Summary Corresponding author(s): George Q. Daley Initial submission Revised version Final submission Nature Research wishes to improve the reproducibility of the work that we publish.
Life Sciences Reporting Summary Corresponding author(s): George Q. Daley Initial submission Revised version Final submission Nature Research wishes to improve the reproducibility of the work that we publish.
Initial genotyping of all new litters was performed by Transnetyx (Memphis,
 SUPPLEMENTAL INFORMATION MURILLO ET AL. SUPPLEMENTAL EXPERIMENTAL PROCEDURES Mouse Genotyping Initial genotyping of all new litters was performed by Transnetyx (Memphis, USA). Assessment of LoxPpα recombination
SUPPLEMENTAL INFORMATION MURILLO ET AL. SUPPLEMENTAL EXPERIMENTAL PROCEDURES Mouse Genotyping Initial genotyping of all new litters was performed by Transnetyx (Memphis, USA). Assessment of LoxPpα recombination
Orexin A (HUMAN, MOUSE, RAT, PORCINE, OVINE,
 Orexin A (HUMAN, MOUSE, RAT, PORCINE, OVINE, BOVINE) Western Blot Kit Protocol (Catalog #WBK-003-30) PHOENIX PHARMACEUTICALS, INC. TABLE OF CONTENTS 1. Kit Contents...2 2. Storage...2 3. Introduction...3
Orexin A (HUMAN, MOUSE, RAT, PORCINE, OVINE, BOVINE) Western Blot Kit Protocol (Catalog #WBK-003-30) PHOENIX PHARMACEUTICALS, INC. TABLE OF CONTENTS 1. Kit Contents...2 2. Storage...2 3. Introduction...3
supplementary information
 DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
Respiratory distress and early neonatal lethality in Hspa4l/Hspa4 double mutant mice
 Respiratory distress and early neonatal lethality in Hspa4l/Hspa4 double mutant mice Belal A. Mohamed, Amal Z. Barakat, Torsten Held, Manar Elkenani, Christian Mühlfeld, Jörg Männer, and Ibrahim M. Adham
Respiratory distress and early neonatal lethality in Hspa4l/Hspa4 double mutant mice Belal A. Mohamed, Amal Z. Barakat, Torsten Held, Manar Elkenani, Christian Mühlfeld, Jörg Männer, and Ibrahim M. Adham
SOX2 antibody - Embryonic Stem Cell Marker (ab15830) datasheet
 -Embryonic Stem Cell Marker (ab15830) 1/8 ページ d Products: Stem Cells >> Germline Stem Cells >> Embryonic Germ Cells SOX2 antibody - Embryonic Stem Cell Marker (ab15830) datasheet Product Name Product type
-Embryonic Stem Cell Marker (ab15830) 1/8 ページ d Products: Stem Cells >> Germline Stem Cells >> Embryonic Germ Cells SOX2 antibody - Embryonic Stem Cell Marker (ab15830) datasheet Product Name Product type
ab Mouse and Rabbit AP/Fast-Red (ABC) Detection IHC Kit
 ab128967 - Mouse and Rabbit AP/Fast-Red (ABC) Detection IHC Kit Instructions for Use For the detection of a specific antibody bound to an antigen in tissue sections. This product is for research use only
ab128967 - Mouse and Rabbit AP/Fast-Red (ABC) Detection IHC Kit Instructions for Use For the detection of a specific antibody bound to an antigen in tissue sections. This product is for research use only
Parthanatos mediates AIMP2-activated age-dependent dopaminergic neuronal loss
 SUPPLEMENTARY INFORMATION Parthanatos mediates AIMP2-activated age-dependent dopaminergic neuronal loss Yunjong Lee, Senthilkumar S. Karuppagounder, Joo-Ho Shin, Yun-Il Lee, Han Seok Ko, Debbie Swing,
SUPPLEMENTARY INFORMATION Parthanatos mediates AIMP2-activated age-dependent dopaminergic neuronal loss Yunjong Lee, Senthilkumar S. Karuppagounder, Joo-Ho Shin, Yun-Il Lee, Han Seok Ko, Debbie Swing,
Supplementary Figure 1. Isolation of GFPHigh cells.
 Supplementary Figure 1. Isolation of GFP High cells. (A) Schematic diagram of cell isolation based on Wnt signaling activity. Colorectal cancer (CRC) cell lines were stably transduced with lentivirus encoding
Supplementary Figure 1. Isolation of GFP High cells. (A) Schematic diagram of cell isolation based on Wnt signaling activity. Colorectal cancer (CRC) cell lines were stably transduced with lentivirus encoding
BrdU IHC Kit. For the detection and localization of bromodeoxyuridine incorporated into newly synthesized DNA of actively proliferating cells
 K-ASSAY BrdU IHC Kit For the detection and localization of bromodeoxyuridine incorporated into newly synthesized DNA of actively proliferating cells Cat. No. KT-077 For Research Use Only. Not for Use in
K-ASSAY BrdU IHC Kit For the detection and localization of bromodeoxyuridine incorporated into newly synthesized DNA of actively proliferating cells Cat. No. KT-077 For Research Use Only. Not for Use in
Plasmid DNA transfection of SW480 human colorectal cancer cells with the Biontex K2 Transfection System
 Plasmid DNA transfection of human colorectal cancer cells with the Biontex K2 Transfection System Stephanie Hehlgans and Franz Rödel, Department of Radiotherapy and Oncology, Goethe- University Frankfurt,
Plasmid DNA transfection of human colorectal cancer cells with the Biontex K2 Transfection System Stephanie Hehlgans and Franz Rödel, Department of Radiotherapy and Oncology, Goethe- University Frankfurt,
Autophagy Assays (LC3B immunofluorescence, LC3B western blot, acridine orange assay) Xin Zhang and Qingsong Liu *
 Autophagy Assays (LC3B immunofluorescence, LC3B western blot, acridine orange assay) Xin Zhang and Qingsong Liu * High Magnetic Field Laboratory, Chinese Academy of Sciences, Hefei, Anhui, China *For correspondence:
Autophagy Assays (LC3B immunofluorescence, LC3B western blot, acridine orange assay) Xin Zhang and Qingsong Liu * High Magnetic Field Laboratory, Chinese Academy of Sciences, Hefei, Anhui, China *For correspondence:
MitoBiogenesis In-Cell ELISA Kit (Colorimetric)
 PROTOCOL MitoBiogenesis In-Cell ELISA Kit (Colorimetric) DESCRIPTION 1850 Millrace Drive, Suite 3A Eugene, Oregon 97403 MS643 Rev.2 For identifying inhibitors and activators of mitochondrial biogenesis
PROTOCOL MitoBiogenesis In-Cell ELISA Kit (Colorimetric) DESCRIPTION 1850 Millrace Drive, Suite 3A Eugene, Oregon 97403 MS643 Rev.2 For identifying inhibitors and activators of mitochondrial biogenesis
ASPP1 Fw GGTTGGGAATCCACGTGTTG ASPP1 Rv GCCATATCTTGGAGCTCTGAGAG
 Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
Week 1: Protein isolation and quantification
 Week 1: Protein isolation and quantification Objective The objective of this lab exercise is to obtain protein samples from fruit fly larvae, BCS and FBS, all of which are then quantitated in the preparation
Week 1: Protein isolation and quantification Objective The objective of this lab exercise is to obtain protein samples from fruit fly larvae, BCS and FBS, all of which are then quantitated in the preparation
Supplementary Materials for
 www.sciencetranslationalmedicine.org/cgi/content/full/8/332/332ra44/dc1 Supplementary Materials for Neuronal heparan sulfates promote amyloid pathology by modulating brain amyloid- clearance and aggregation
www.sciencetranslationalmedicine.org/cgi/content/full/8/332/332ra44/dc1 Supplementary Materials for Neuronal heparan sulfates promote amyloid pathology by modulating brain amyloid- clearance and aggregation
Overview of Solulink Products. June 2011
 Overview of Solulink Products June 2011 1 Who we are Established in 2003, Solulink develops, patents, manufactures, and sells consumables to over 1,000 customers in life science, diagnostic, and pharmaceutical
Overview of Solulink Products June 2011 1 Who we are Established in 2003, Solulink develops, patents, manufactures, and sells consumables to over 1,000 customers in life science, diagnostic, and pharmaceutical
Cold Fusion Cloning Kit. Cat. #s MC100A-1, MC101A-1. User Manual
 Fusion Cloning technology Cold Fusion Cloning Kit Store the master mixture and positive controls at -20 C Store the competent cells at -80 C. (ver. 120909) A limited-use label license covers this product.
Fusion Cloning technology Cold Fusion Cloning Kit Store the master mixture and positive controls at -20 C Store the competent cells at -80 C. (ver. 120909) A limited-use label license covers this product.
western blotting tech
 western blotting tech note 6148 Transfer of High Molecular Weight Proteins to Membranes: A Comparison of Transfer Efficiency Between Blotting Systems Nik Chmiel, Bio-Rad Laboratories, Inc., 6000 James
western blotting tech note 6148 Transfer of High Molecular Weight Proteins to Membranes: A Comparison of Transfer Efficiency Between Blotting Systems Nik Chmiel, Bio-Rad Laboratories, Inc., 6000 James
Transfection of CRISPR/Cas9 Nuclease NLS ribonucleoprotein (RNP) into adherent mammalian cells using Lipofectamine RNAiMAX
 Transfection of CRISPR/Cas9 Nuclease NLS ribonucleoprotein (RNP) into adherent mammalian cells using Lipofectamine RNAiMAX INTRODUCTION The CRISPR/Cas genome editing system consists of a single guide RNA
Transfection of CRISPR/Cas9 Nuclease NLS ribonucleoprotein (RNP) into adherent mammalian cells using Lipofectamine RNAiMAX INTRODUCTION The CRISPR/Cas genome editing system consists of a single guide RNA
MMLV Reverse Transcriptase 1st-Strand cdna Synthesis Kit
 MMLV Reverse Transcriptase 1st-Strand cdna Synthesis Kit Cat. No. MM070150 Available exclusively thru Lucigen. lucigen.com/epibio www.lucigen.com MA265E MMLV Reverse Transcriptase 1st-Strand cdna Synthesis
MMLV Reverse Transcriptase 1st-Strand cdna Synthesis Kit Cat. No. MM070150 Available exclusively thru Lucigen. lucigen.com/epibio www.lucigen.com MA265E MMLV Reverse Transcriptase 1st-Strand cdna Synthesis
Product Information. Before you begin. Component A 1 vial of 30 ul vial of 300 ul each Glycerol. Tris
 Glowing Products for Science Mix-n-Stain Antibody Labeling Kits Size: 1 labeling per kit Storage: -20 o C Stability: Stable for at least 1 year from date of receipt when stored as recommended. Components:
Glowing Products for Science Mix-n-Stain Antibody Labeling Kits Size: 1 labeling per kit Storage: -20 o C Stability: Stable for at least 1 year from date of receipt when stored as recommended. Components:
ab Hypoxic Response Human Flow Cytometry Kit
 ab126585 Hypoxic Response Human Flow Cytometry Kit Instructions for Use For measuring protein levels by flow cytometry: hypoxia-inducible factor 1-alpha (HIF1A) and BCL2/adenovirus E1B 19 kda proteininteracting
ab126585 Hypoxic Response Human Flow Cytometry Kit Instructions for Use For measuring protein levels by flow cytometry: hypoxia-inducible factor 1-alpha (HIF1A) and BCL2/adenovirus E1B 19 kda proteininteracting
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/317/5837/477/dc1 Supporting Online Material for AAV Vector Integration Sites in Mouse Hepatocellular Carcinoma Anthony Donsante, Daniel G. Miller, Yi Li, Carole Vogler,
www.sciencemag.org/cgi/content/full/317/5837/477/dc1 Supporting Online Material for AAV Vector Integration Sites in Mouse Hepatocellular Carcinoma Anthony Donsante, Daniel G. Miller, Yi Li, Carole Vogler,
Intestinal Epithelial Cell-Specific Deletion of PLD2 Alleviates DSS-Induced Colitis by. Regulating Occludin
 Intestinal Epithelial Cell-Specific Deletion of PLD2 Alleviates DSS-Induced Colitis by Regulating Occludin Chaithanya Chelakkot 1,ǂ, Jaewang Ghim 2,3,ǂ, Nirmal Rajasekaran 4, Jong-Sun Choi 5, Jung-Hwan
Intestinal Epithelial Cell-Specific Deletion of PLD2 Alleviates DSS-Induced Colitis by Regulating Occludin Chaithanya Chelakkot 1,ǂ, Jaewang Ghim 2,3,ǂ, Nirmal Rajasekaran 4, Jong-Sun Choi 5, Jung-Hwan
ab TripleStain IHC Kit: M&M&R on human tissue (DAB, Red/AP & DAB/Ni)
 ab183287 TripleStain IHC Kit: M&M&R on human tissue (DAB, Red/AP & DAB/Ni) Instructions for Use For the detection of Rabbit and Mouse Primary antibodies on Human tissue or cell samples. This product is
ab183287 TripleStain IHC Kit: M&M&R on human tissue (DAB, Red/AP & DAB/Ni) Instructions for Use For the detection of Rabbit and Mouse Primary antibodies on Human tissue or cell samples. This product is
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10163 Supplementary Table 1 Efficiency of vector construction. Process wells recovered efficiency (%) Recombineering* 480 461 96 Intermediate plasmids 461 381 83 Recombineering efficiency
doi:10.1038/nature10163 Supplementary Table 1 Efficiency of vector construction. Process wells recovered efficiency (%) Recombineering* 480 461 96 Intermediate plasmids 461 381 83 Recombineering efficiency
Supplementary Figure 1: Expression of RNF8, HERC2 and NEURL4 in the cerebellum and knockdown of RNF8 by RNAi (a) Lysates of the cerebellum from rat
 Supplementary Figure 1: Expression of RNF8, HERC2 and NEURL4 in the cerebellum and knockdown of RNF8 by RNAi (a) Lysates of the cerebellum from rat pups at P6, P14, P22, P30 and adult (A) rats were subjected
Supplementary Figure 1: Expression of RNF8, HERC2 and NEURL4 in the cerebellum and knockdown of RNF8 by RNAi (a) Lysates of the cerebellum from rat pups at P6, P14, P22, P30 and adult (A) rats were subjected
Supporting Information
 Supporting Information Su et al. 10.1073/pnas.1211604110 SI Materials and Methods Cell Culture and Plasmids. Tera-1 and Tera-2 cells (ATCC: HTB- 105/106) were maintained in McCoy s 5A medium with 15% FBS
Supporting Information Su et al. 10.1073/pnas.1211604110 SI Materials and Methods Cell Culture and Plasmids. Tera-1 and Tera-2 cells (ATCC: HTB- 105/106) were maintained in McCoy s 5A medium with 15% FBS
GeneChip Eukaryotic Small Sample Target Labeling Assay Version II *
 GENE EXPRESSION MONITORING TECHNICAL NOTE GeneChip Eukaryotic Small Sample Target Labeling Assay Version II * Introduction There is an overwhelming and continuing demand for a well characterized protocol
GENE EXPRESSION MONITORING TECHNICAL NOTE GeneChip Eukaryotic Small Sample Target Labeling Assay Version II * Introduction There is an overwhelming and continuing demand for a well characterized protocol
1. Paraffin section slides can be stored at room temperature for a long time.
 Immunohistochemistry (IHC) Protocols Immunohistochemistry (IHC) Protocol of Paraffin Section 1. Fix dissected tissues with 10% formalin for no less than 48 hours at room temperature. Inadequately fixation
Immunohistochemistry (IHC) Protocols Immunohistochemistry (IHC) Protocol of Paraffin Section 1. Fix dissected tissues with 10% formalin for no less than 48 hours at room temperature. Inadequately fixation
Dolphin-Chemi Plus. Aim: To visualise and evaluate the performance of chemiluminescent immunoblots using Wealtec s Dolphin-Chemi plus image system
 Application Note 03 Dolphin-Chemi plus 8/22/2007 Dolphin-Chemi Plus Aim: To visualise and evaluate the performance of chemiluminescent immunoblots using Wealtec s Dolphin-Chemi plus image system INTRODUCTION
Application Note 03 Dolphin-Chemi plus 8/22/2007 Dolphin-Chemi Plus Aim: To visualise and evaluate the performance of chemiluminescent immunoblots using Wealtec s Dolphin-Chemi plus image system INTRODUCTION
Molecular Cell Biology - Problem Drill 11: Recombinant DNA
 Molecular Cell Biology - Problem Drill 11: Recombinant DNA Question No. 1 of 10 1. Which of the following statements about the sources of DNA used for molecular cloning is correct? Question #1 (A) cdna
Molecular Cell Biology - Problem Drill 11: Recombinant DNA Question No. 1 of 10 1. Which of the following statements about the sources of DNA used for molecular cloning is correct? Question #1 (A) cdna
SensoLyte Anti-alpha-Synuclein Quantitative ELISA Kit (Human/Mouse/Rat) *Colorimetric*
 Catalog # Kit Size SensoLyte Anti-alpha-Synuclein Quantitative ELISA Kit (Human/Mouse/Rat) *Colorimetric* AS-55550 One 96-well strip plate This kit is optimized to detect human/mouse/rat alpha-synuclein
Catalog # Kit Size SensoLyte Anti-alpha-Synuclein Quantitative ELISA Kit (Human/Mouse/Rat) *Colorimetric* AS-55550 One 96-well strip plate This kit is optimized to detect human/mouse/rat alpha-synuclein
Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna
 Supplemental Materials and Methods Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna (Mission shrna, Sigma) against mouse MMP14 were transfected into HEK293 cells using FuGene6
Supplemental Materials and Methods Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna (Mission shrna, Sigma) against mouse MMP14 were transfected into HEK293 cells using FuGene6
Transfection of Mouse ES Cells and Mouse ips cells using the Stemfect 2.0 -mesc Transfection Reagent
 APPLICATION NOTE Page 1 Transfection of Mouse ES Cells and Mouse ips cells using the Stemfect 2.0 -mesc Transfection Reagent Authors: Amelia L. Cianci 1, Xun Cheng 1 and Kerry P. Mahon 1,2 1 Stemgent Inc.,
APPLICATION NOTE Page 1 Transfection of Mouse ES Cells and Mouse ips cells using the Stemfect 2.0 -mesc Transfection Reagent Authors: Amelia L. Cianci 1, Xun Cheng 1 and Kerry P. Mahon 1,2 1 Stemgent Inc.,
FFPE DNA Extraction kit
 FFPE DNA Extraction kit Instruction Manual FFPE DNA Extraction kit Cat. No. C20000030, Format 50 rxns Version 1 / 14.08.13 Content Introduction 4 Overview of FFPE DNA extraction workflow 4 Kit contents
FFPE DNA Extraction kit Instruction Manual FFPE DNA Extraction kit Cat. No. C20000030, Format 50 rxns Version 1 / 14.08.13 Content Introduction 4 Overview of FFPE DNA extraction workflow 4 Kit contents
Genetics - Problem Drill 19: Dissection of Gene Function: Mutational Analysis of Model Organisms
 Genetics - Problem Drill 19: Dissection of Gene Function: Mutational Analysis of Model Organisms No. 1 of 10 1. The mouse gene knockout is based on. (A) Homologous recombination (B) Site-specific recombination
Genetics - Problem Drill 19: Dissection of Gene Function: Mutational Analysis of Model Organisms No. 1 of 10 1. The mouse gene knockout is based on. (A) Homologous recombination (B) Site-specific recombination
Fisher (Fairlawn, NJ) and Sigma-Aldrich (St. Louis, MO) and were used without further. (Promega) and DpnI (New England Biolabs, Beverly, MA).
 175 Appendix III Chapter 4 Methods General. Unless otherwise noted, reagents were purchased from the commercial suppliers Fisher (Fairlawn, NJ) and Sigma-Aldrich (St. Louis, MO) and were used without further
175 Appendix III Chapter 4 Methods General. Unless otherwise noted, reagents were purchased from the commercial suppliers Fisher (Fairlawn, NJ) and Sigma-Aldrich (St. Louis, MO) and were used without further
FACTORS CONTRIBUTING TO VARIABILITY IN DNA MICROARRAY RESULTS: THE ABRF MICROARRAY RESEARCH GROUP 2002 STUDY
 FACTORS CONTRIBUTING TO VARIABILITY IN DNA MICROARRAY RESULTS: THE ABRF MICROARRAY RESEARCH GROUP 2002 STUDY K. L. Knudtson 1, C. Griffin 2, A. I. Brooks 3, D. A. Iacobas 4, K. Johnson 5, G. Khitrov 6,
FACTORS CONTRIBUTING TO VARIABILITY IN DNA MICROARRAY RESULTS: THE ABRF MICROARRAY RESEARCH GROUP 2002 STUDY K. L. Knudtson 1, C. Griffin 2, A. I. Brooks 3, D. A. Iacobas 4, K. Johnson 5, G. Khitrov 6,
Quantitative analysis of PCR fragments with the Agilent 2100 Bioanalyzer. Application Note. Odilo Mueller. Abstract
 Quantitative analysis of PCR fragments with the Agilent 2100 Bioanalyzer Application Note Odilo Mueller Abstract This application note describes how the Agilent Technologies 2100 bioanalyzer can be used
Quantitative analysis of PCR fragments with the Agilent 2100 Bioanalyzer Application Note Odilo Mueller Abstract This application note describes how the Agilent Technologies 2100 bioanalyzer can be used
Theoretical cloning project
 Theoretical cloning project Needed to get credits Make it up yourself, don't copy Possible to do in groups of 2-4 students If you need help or an idea, ask! If you have no idea what to clone, I can give
Theoretical cloning project Needed to get credits Make it up yourself, don't copy Possible to do in groups of 2-4 students If you need help or an idea, ask! If you have no idea what to clone, I can give
Note that steps 1-16 are identical to steps 1-16 in the immunostaining protocol, except 0.5X PBtween is used in this protocol rather than 0.5X PBT.
 Embryo In-situ Hybridization: This protocol is based closely on an immunostaining protocol for Hypsibius dujardini (Gabriel and Goldstein 2007) and available in-situ protocols for both insects (Tomoysu
Embryo In-situ Hybridization: This protocol is based closely on an immunostaining protocol for Hypsibius dujardini (Gabriel and Goldstein 2007) and available in-situ protocols for both insects (Tomoysu
ab Optiblot Fluorescent Western Blot Kit
 ab133410 Optiblot Fluorescent Western Blot Kit Instructions for Use For quantitative, multi-color fluorescent Western blotting. This product is for research use only and is not intended for diagnostic
ab133410 Optiblot Fluorescent Western Blot Kit Instructions for Use For quantitative, multi-color fluorescent Western blotting. This product is for research use only and is not intended for diagnostic
Cell Stem Cell, volume 13 Supplemental Information
 Cell Stem Cell, volume 13 Supplemental Information Whole-Genome Bisulfite Sequencing of Two Distinct Interconvertible DNA Methylomes of Mouse Embryonic Stem Cells Ehsan Habibi, Arie B. Brinkman, Julia
Cell Stem Cell, volume 13 Supplemental Information Whole-Genome Bisulfite Sequencing of Two Distinct Interconvertible DNA Methylomes of Mouse Embryonic Stem Cells Ehsan Habibi, Arie B. Brinkman, Julia
Non-Organic-Based Isolation of Mammalian microrna using Norgen s microrna Purification Kit
 Application Note 13 RNA Sample Preparation Non-Organic-Based Isolation of Mammalian microrna using Norgen s microrna Purification Kit B. Lam, PhD 1, P. Roberts, MSc 1 Y. Haj-Ahmad, M.Sc., Ph.D 1,2 1 Norgen
Application Note 13 RNA Sample Preparation Non-Organic-Based Isolation of Mammalian microrna using Norgen s microrna Purification Kit B. Lam, PhD 1, P. Roberts, MSc 1 Y. Haj-Ahmad, M.Sc., Ph.D 1,2 1 Norgen
Tissue & Cell Genomic DNA Purification Kit. Cat. #:DP021/ DP Size:50/150 reactions Store at RT For research use only
 Tissue & Cell Genomic DNA Purification Kit Cat. #:DP021/ DP021-150 Size:50/150 reactions Store at RT For research use only 1 Description: The Tissue & Cell Genomic DNA Purification Kit provides a rapid,
Tissue & Cell Genomic DNA Purification Kit Cat. #:DP021/ DP021-150 Size:50/150 reactions Store at RT For research use only 1 Description: The Tissue & Cell Genomic DNA Purification Kit provides a rapid,
Protein Translation Study Label Protein with S35 Methionine in Cells Salma Hasan and Isabelle Plo *
 Protein Translation Study Label Protein with S35 Methionine in Cells Salma Hasan and Isabelle Plo * INSERM U1009, Gustave Roussy, Villejuif, France *For correspondence: isabelle.plo@gustaveroussy.fr [Abstract]
Protein Translation Study Label Protein with S35 Methionine in Cells Salma Hasan and Isabelle Plo * INSERM U1009, Gustave Roussy, Villejuif, France *For correspondence: isabelle.plo@gustaveroussy.fr [Abstract]
*Corresponding author. Tel: ;
 1 SUPPLEMENTARY DATA 2 3 4 5 6 7 8 9 10 11 Integrin 2 1 in nonactivated conformation can induce focal adhesion kinase signaling Maria Salmela 1, Johanna Jokinen 1,2, Silja Tiitta 1, Pekka Rappu 1, Holland
1 SUPPLEMENTARY DATA 2 3 4 5 6 7 8 9 10 11 Integrin 2 1 in nonactivated conformation can induce focal adhesion kinase signaling Maria Salmela 1, Johanna Jokinen 1,2, Silja Tiitta 1, Pekka Rappu 1, Holland
For the rapid isolation of Mitochondrial DNA in various cell and tissue samples.
 ab65321 Mitochondrial DNA Isolation Kit Instructions for Use For the rapid isolation of Mitochondrial DNA in various cell and tissue samples. This product is for research use only and is not intended for
ab65321 Mitochondrial DNA Isolation Kit Instructions for Use For the rapid isolation of Mitochondrial DNA in various cell and tissue samples. This product is for research use only and is not intended for
Mitochondrial DNA Isolation Kit
 Mitochondrial DNA Isolation Kit Catalog Number KA0895 50 assays Version: 03 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 General Information... 4 Materials
Mitochondrial DNA Isolation Kit Catalog Number KA0895 50 assays Version: 03 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 General Information... 4 Materials
Nature Methods: doi: /nmeth Supplementary Figure 1. Retention of RNA with LabelX.
 Supplementary Figure 1 Retention of RNA with LabelX. (a) Epi-fluorescence image of single molecule FISH (smfish) against GAPDH on HeLa cells expanded without LabelX treatment. (b) Epi-fluorescence image
Supplementary Figure 1 Retention of RNA with LabelX. (a) Epi-fluorescence image of single molecule FISH (smfish) against GAPDH on HeLa cells expanded without LabelX treatment. (b) Epi-fluorescence image
In-Gel Western Detection Using Near-Infrared Fluorescence
 In-Gel Western Detection Using Near-Infrared Fluorescence Developed for: Aerius, and Odyssey Family of Imagers Please refer to your manual to confirm that this protocol is appropriate for the applications
In-Gel Western Detection Using Near-Infrared Fluorescence Developed for: Aerius, and Odyssey Family of Imagers Please refer to your manual to confirm that this protocol is appropriate for the applications
Blot: a spot or stain, especially of ink on paper.
 Blotting technique Blot: a spot or stain, especially of ink on paper. 2/27 In molecular biology and genetics, a blot is a method of transferring proteins, DNA or RNA, onto a carrier (for example, a nitrocellulose,pvdf
Blotting technique Blot: a spot or stain, especially of ink on paper. 2/27 In molecular biology and genetics, a blot is a method of transferring proteins, DNA or RNA, onto a carrier (for example, a nitrocellulose,pvdf
FOR RESEARCH USE ONLY. NOT FOR HUMAN OR DIAGNOSTIC USE.
 Instruction manual RNA-direct SYBR Green Realtime PCR Master Mix 0810 F0930K RNA-direct SYBR Green Realtime PCR Master Mix Contents QRT-201T QRT-201 0.5mLx2 0.5mLx5 Store at -20 C, protected from light
Instruction manual RNA-direct SYBR Green Realtime PCR Master Mix 0810 F0930K RNA-direct SYBR Green Realtime PCR Master Mix Contents QRT-201T QRT-201 0.5mLx2 0.5mLx5 Store at -20 C, protected from light
Myers Lab ChIP-seq Protocol v Modified January 10, 2014
 Myers Lab ChIP-seq Protocol V011014 1 Contact information: Dr. Florencia Pauli Behn HudsonAlpha Institute for Biotechnology 601 Genome Way Huntsville, AL 35806 Telephone: 256-327-5229 Email: fpauli@hudsonalpha.org
Myers Lab ChIP-seq Protocol V011014 1 Contact information: Dr. Florencia Pauli Behn HudsonAlpha Institute for Biotechnology 601 Genome Way Huntsville, AL 35806 Telephone: 256-327-5229 Email: fpauli@hudsonalpha.org
ab Ubiquitylation Assay Kit (HeLa lysate-based)
 ab139471 Ubiquitylation Assay Kit (HeLa lysate-based) Instructions for Use For the generation of ubiquitin-conjugated lysate proteins This product is for research use only and is not intended for diagnostic
ab139471 Ubiquitylation Assay Kit (HeLa lysate-based) Instructions for Use For the generation of ubiquitin-conjugated lysate proteins This product is for research use only and is not intended for diagnostic
Whole Mount IHC Protocol
 Whole Mount IHC Protocol Authors: Ruth Sullivan, Ryan Trevena and Kyle Wegner Creation Date: 03/17/2016 All steps should be conducted with gentle agitation on an orbital shaker, unless otherwise instructed.
Whole Mount IHC Protocol Authors: Ruth Sullivan, Ryan Trevena and Kyle Wegner Creation Date: 03/17/2016 All steps should be conducted with gentle agitation on an orbital shaker, unless otherwise instructed.
Gene Expression Technology
 Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
Electrophoresis and transfer
 Electrophoresis and transfer Electrophoresis Cation = positively charged ion, it moves toward the cathode (-) Anion = negatively charged ion, it moves toward the anode (+) Amphoteric substance = can have
Electrophoresis and transfer Electrophoresis Cation = positively charged ion, it moves toward the cathode (-) Anion = negatively charged ion, it moves toward the anode (+) Amphoteric substance = can have
