Algorithms for Bioinformatics
|
|
|
- Melinda Walsh
- 6 years ago
- Views:
Transcription
1 Algorithms for Bioinformatics Compressive Genomics Ulf Leser
2 Content of this Lecture Next Generation Sequencing Sequence compression Approximate search in compressed genomes Using multiple references This lecture is not part of the examination Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/2016 2
3 Large Scale Sequencing Projects samples: To obtain a comprehensive description of genomic, transcriptomic and epigenomic changes in 50 different tumor types and/or subtypes which are of clinical and societal importance across the globe. Genomics England is creating a lasting legacy for patients, the NHS and the UK economy through the sequencing of 100,000 genomes: the 100,000 Genomes Project. The Veterans Affairs (VA) Office of Research and Development is launching the Million Veteran Program (MVP). The goal of MVP is to better understand how genes affect health and illness in order to improve health care. Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/2016 3
4 Next Generation Sequencing New generation of sequencers since ~2005 Illumina, Solexa, 454, Solid, Much higher throughput ~15 TB raw data in 3-5 days ~600 GB processed data/week Cost for sequencing a genome down to ~2.000 USD 3 rd generation sequencers Single molecule sequencing A (human) genome in a day Sequence every human Sequence different cells in every human Illumina HiSeq DNAVision Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/2016 4
5 Latest 600GB / day, genomes per year $1,000 genome at 30x coverage Amortized over 18,000 genomes per year over four-year period (Not cheap) Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/2016 5
6 Data Tsunami Stein, L. D. (2010). Genome Biol Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/2016 6
7 The real Cost of Genomic Sequencing Sboner, A. (2011). The real cost of sequencing: higher than you think! Genome Biology 2011 Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/2016 7
8 New Problems Need to process huge amounts of data with complex pipelines Terabytes per week Pipelines with dozens of steps Need to store huge amounts of sequence data (Hundreds of) thousands of genomes Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/2016 8
9 Content of this Lecture Next Generation Sequencing Sequence compression Referential compression Four issues Approximate search in compressed genomes Using multiple references Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/2016 9
10 Compressing Genomes Four basic techniques (lossless) Bit packing Statistical compression Dictionary-based Referential compression Criteria for compression methods Compression ratio Compression speed / decompression speed Analyzing (searching) compressed data Compressing reads is another topic Quality information, non-standard bases, short strings, Another big topic: Lossy compression Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
11 1. Bit Packing A genome consists of 4 different bases Representing one bases thus requires 2 bits only One byte four bases Compression ratio (compared to ASCII / FASTA): 1:4 Advantages: Fast, universal, simple Disadvantage: Low compression ratio Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
12 2. Statistical Compression Idea: Don t use the same number of bits for every char Frequent characters are represented with less bits Useful for larger alphabets with large differences in character frequencies Can be extended to q-grams But: DNA q-gram are roughly equally frequent Disadvantage: Low compression ratio (~1:5) Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
13 3. Dictionary-based Compression Idea: Represent frequent substrings with short codes Ziv-Lempel-Welch: Find most frequent substrings online Stored in a dictionary Index in dictionary is used as code Can be compressed Trade-Off: Dictionary-size, compression speed, compression ratio Useful when large diffs in frequency of substrings exist Recurring patterns: Images, language, tables, Disadvantage: Low compression ratio (for DNA, ~1:4-6) Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
14 4. Referential Compression For NGS data, we usually know the reference genome Idea: Use reference as (external, predefined) dictionary Sequences are represented as lists of referential match entries: (start, length, mismatch) Issues Find long matches fast Trade-off: Long matches: ratio++; faster compression: ratio Efficient coding of RMEs Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
15 Greedy Algorithm Compression rate for human chromosomes: ~1:60 Compression speed for human chromosomes : 80 MB/s Main memory usage: ~4*size(ref)+size(s) Using DNA-optimized compressed suffix trees for reference Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
16 Content of this Lecture Next Generation Sequencing Sequence compression Referential compression Four issues in referential compression Approximate search in compressed genomes Using multiple references Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
17 Issues Compact encoding of RMEs Main memory usage Faster compression / decompression Which reference? General: Balancing the trade-off between compression ratio and compression speed Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
18 1. Encoding RME s Very frequent: Series of consecutive matches with SNVs in between Rare! (1000,5,A), (1006,12,C), (1019,4,A), (1024,20,C), (1045,8,B), (9453,25,C), Improvement: Delta encoding (with/out default stepsize) (1000,5,A), (1006,12,C), (1019,4,A), (1024,20,C), (1045,8,B), (9453,25,C), (1000,5,A), (+6,12,C), (+13,4,A), (+5,20,C), (+21,8,B), (9453,25,C), (1000,5,A), (+0,12,C), (+0,4,A), (+0,20,C), (+0,8,B), (9453,25,C), Large impact on compression ratio Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
19 2. Improving Main Memory Usage Best (compressed) suffix tree libraries need ~3-4*n space Observation: We often find matches in sequential regions Can be exploited to save main memory Partition reference and input into blocks (e.g. 5MB) Keep one (indexed) block each at a time in main memory Search other reference blocks only when no good match is found Switching blocks is costly: Avoid Even if this means less optimal compression Typical: Threshold on minimal length of RMEs; otherwise switch Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
20 Memory / compression speed / compression ratio Evaluation for human chromosome 1 Small blocks: Frequent block changes, bad ratio Blocks larger than ~100MB: No further improvements Compression/decompression requires only ~500MB for dictionary Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
21 3. Improving compression speed Runtime dominated by looking up prefixes in the compressed suffix tree (CST++) Decoding the compressed suffix tree structure costs time Maximal throughput: ~ lookups / sec Improvement: Local matching Search next RME near previous RME directly in the reference Ignoring the index Accept best next match iff RME sufficiently long Speed-up by a factor of ~5-10 Also improves compression ratio Next matches close to previous ones effective delta encoding But may not find longest RME Evaluation: Overall space reduction Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
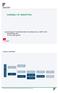
22 Results: Ratio (Data: 1000 Genomes project) Overall compression ratio: ~1:400 Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
23 Results: Speed Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
24 4. Which reference to use? Given a set of genomes: Which should be the reference? Similarity to reference is key to high compression rates Compressing Human against Mouse: Disaster Similarity in non-coding region is low Exhaustive reference selection is very time consuming (took 6 days for 1092 * 1091 H-22) Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
25 Two Alternatives Heuristic-based reference selection Define a heuristic for the similarity of two sequences For instance: Compute best reference based on small sample Use any other fast similarity estimation method Pick the sequence most similar to all other sequences according to this heuristic Better: Build your own reference Reference rewriting Given a reference, rewrite it in order to obtain higher compression ratios Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
26 Selection versus Rewriting Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
27 Selection versus Rewriting Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
28 Fresco: Comparative Evaluation Second Order Compression: Compress RME sets All sequences are similar to each other Thus, different sequences produce very similar RME lists Idea: Compress (using meta referential compression) Best algorithms today [Deorowisc 2015, GDC-2] Compression ratio 1:9500 7TB FASTA compressed to 700MB Speed: 200MB/sec (beware: measured on different hardware) Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
29 Content of this Lecture Next Generation Sequencing Sequence compression Referential compression Four issues Approximate search in compressed genomes Using multiple references Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
30 K-Approximate Matching (k-difference Mathcing) Given a collection of (referentially compressed) sequences S, find all k-approximate matches of a query q Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
31 Example Application: Translational Medicine Modern cancer drugs depend on genotype of patients Genotype: Mutations in certain cancer genes Clinics sequence thousands of human genomes Given a set of patient genomes C with known outcome and the sequence of a cancer gene g in a new patient q what is the most similar occurrence of g in C? Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
32 Example Application: Translational Medicine Modern cancer drugs depend on genotype of patients Genotype: Mutations in certain cancer genes Clinics sequence thousands of human genomes Given a set of patient genomes C with known outcome and the sequence of a cancer gene g in a new patient q what is the most similar occurrence of g in C? 1000 genomes -> ~3TB data Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
33 Storing Similar Strings Popular idea: Referential Compression Choose a reference string p from C When adding a new string s, only store differences between s and p p: Kohala Coast-Hawaii s 2 : Kohala Cost s 3 : Koala Coast/Hawaii s 2 : (p,0,9,s),(p,11,1,_) s 3 : (p,0,2,a),(p,4,8,/),(p,13,6,_) RME: Referential Match Entries Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
34 RCSI: Approx. matching in ref. compressed genomes Key idea: Find matches in all compressed sequences simultaneously by searching the reference Store reference as suffix tree Search using standard BYP-algorithm For every match, find all RME completely containing the match Build an interval tree over all RMEs If RME X contains match, only children of X may contain other matches Problem: Matches not contained in the reference AAA ACAA AAACT ACAAACTG CGGACAAACTGACGTTCGACG Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
35 RCSI Approach Fix maximal query length q max and maximal k max Compute overlap sequences One for every mismatch leading to two consecutive RMEs How long must these overlaps be? Answer: 2* q max + k max Very conservative estimation, guaranteed to not loose any match Set of overlap sequences is indexed as well This index is searched using BYP Additional to reference Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
36 RCSI: Architecture Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
37 Evaluation: Indexing time Indexing one genome: ~30 sec Indexing 1000 genomes: ~8 hours Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
38 Evaluation: Approx. search in 1000 genomes ( q [120,170]) Until k=5, almost all queries finish in <10ms For k=1, almost all queries finish in <1ms Outliers: Queries from repetitive regions Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
39 Competitors GC open source code lacks important preprocessing step We could only compare using the data from GC paper RCSI between 10 and 100 times faster And computes all results Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
40 Content of this Lecture Next Generation Sequencing Sequence compression Referential compression Four issues Approximate search in compressed genomes Using multiple references Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
41 Collections of Similar Strings Often (not always): Strings are similar to each other All human genomes are 99% identical All mammal genomes are >90% identical All elements of a Wikipedia revision histories are highly similar Elements of version histories are very similar (SVN, subversion, ) Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
42 Heterogeneous String Collections s 3 : s 4 : p: Kohala Coast-Hawaii s 2 : Kohala Cost Koala Coast/Hawaii Islands Kohala Coast-Hawaii Islands s 5 : Orchid Island s 6 : Orchied Island Kohala Coast-Hawaii compressed against Kohala Cost Kohala Coast/Hawaii Islands Koala Coast/Hawaii Islands Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
43 Heterogeneous String Collections s 3 : s 4 : p: Kohala Coast-Hawaii s 2 : Kohala Cost Koala Coast/Hawaii Islands Kohala Coast-Hawaii Islands s 5 : Orchid Island s 6 : Orchied Island Kohala Coast-Hawaii Orchid Island Kohala Cost Kohala Coast/Hawaii Islands Orchied Island Koala Coast/Hawaii Islands Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
44 Novel Idea: Use Multiple References Strings are compressed against different references Challenge: Which are the best references? Kohala Coast-Hawaii Orchid Island Kohala Cost Koala Coast/Hawaii Islands Orchied Island Kohala Coast/Hawaii Islands Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
45 Novel Idea: Allow Hierarchical Compressions Compression dependencies can form hierarchies Challenge: Which is the best parent? Kohala Coast-Hawaii Orchid Island Kohala Cost Koala Coast/Hawaii Islands Orchied Island Kohala Coast/Hawaii Islands Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
46 Novel Idea: Compress against Multiple References Strings are compressed against multiple other strings Challenge: Which is the best set of parents? Kohala Coast-Hawaii Orchid Island Kohala Cost Koala Coast/Hawaii Islands Orchied Island Kohala Coast/Hawaii Islands Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
47 MRSCI: Multiple References Compression Challenges during compression Which strings should be references and how many? How can we efficiently find good parents? What is the optimal compression hierarchy? How to perform k-approximate search in a multi-reference compression hierarchy? Findings Proof that finding an optimal compression hierarchy is NP-hard Three heuristics to build increasingly complex CHs Increasingly better compression rates Moderate increase in indexing time, roughly same search speed Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
48 Overview RCSI: VLDB 2014 CPart CForest CDAG disallowed; heuristic solutions Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
49 CPart: Using Multiple References Iteratively and greedily compress strings from C Choose first string as first reference p, set P={p} Compress all other strings s one-by-one Find reference p from P most similar to s If p and s are sufficiently similar compress s against p If not, add s to P (new reference, new root) Needs fast method for assessing string similarity Essentially performs a greedy clustering of C Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
50 Competitors Sweet spot: Strong and fast compression, fast search Two classes of competitors Pure indexer: ESA, CST: Large memory footprint, fast search Pure compressors: Strong compression, slow search Variations we built: Compressors with additional search indexes RLZ / Tong after modification: irlz, itong Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
51 Evaluation: Indexing WikiPedia Revisions Wikipedia Helsinki, ~3K versions 577 MB Wikipedia GW Bush, ~45K versions 1400 MB CDAG strongest of index-based, almost as small as best CDAG (or CPart) are fastest (2-4 times faster than itong) Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
52 Evaluation: Searching (HEL) 5000 random queries, length char ESA fastest in search All compressing method perform roughly the same Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
53 Evaluation: Large Datasets Human chromosome 21, up to 640 versions, up to 51GB Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
54 Conclusions Referential compression beats standard compression tools by orders of magnitude for highly-similar sequences (w.r.t. storage and speed) Inherent trade-off between compression ratio and de- /compression speed Given a referential index, some (many?) string matching problems can be solved much more efficiently ample room for further research Compressive genomics Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
55 References Deorowicz, Sebastian, Agnieszka Danek, and Marcin Niemiec. "GDC 2: Compression of large collections of genomes." arxiv preprint arxiv: (2015). Wandelt, S. and U. Leser (2014). MRCSI: Compressing and Searching String Collections with Multiple References. PVLDB. Kona, Hawaii. Wandelt, S. and U. Leser (2013). "FRESCO: Referential Compression of Highly-Similar sequences." Transactions on Computational Biology and Bioinformatics 10(5): Wandelt, S. and U. Leser (2012). "Adaptive efficient compression of genomes." Algorithms for Molecular Biology 7(30). Wandelt, S., J. Starlinger, M. Bux and U. Leser (2013). RCSI: Scalable similarity search in thousand(s) of genome s. PVLDB, Hangzhou, China. Wandelt, S., Rheinländer, A., Bux, M., Thalheim, L., Haldemann, B. and Leser, U. (2012). "Data Management Challenges in Next Generation Sequencing." Datenbank Spektrum Ulf Leser: Algorithms for Bioinformatics, Winter Semester 2015/
Computational Challenges of Medical Genomics
 Talk at the VSC User Workshop Neusiedl am See, 27 February 2012 [cbock@cemm.oeaw.ac.at] http://medical-epigenomics.org (lab) http://www.cemm.oeaw.ac.at (institute) Introducing myself to Vienna s scientific
Talk at the VSC User Workshop Neusiedl am See, 27 February 2012 [cbock@cemm.oeaw.ac.at] http://medical-epigenomics.org (lab) http://www.cemm.oeaw.ac.at (institute) Introducing myself to Vienna s scientific
Introduction to Bioinformatics
 Introduction to Bioinformatics Richard Corbett Canada s Michael Smith Genome Sciences Centre Vancouver, British Columbia June 28, 2017 Our mandate is to advance knowledge about cancer and other diseases
Introduction to Bioinformatics Richard Corbett Canada s Michael Smith Genome Sciences Centre Vancouver, British Columbia June 28, 2017 Our mandate is to advance knowledge about cancer and other diseases
Machine Learning. HMM applications in computational biology
 10-601 Machine Learning HMM applications in computational biology Central dogma DNA CCTGAGCCAACTATTGATGAA transcription mrna CCUGAGCCAACUAUUGAUGAA translation Protein PEPTIDE 2 Biological data is rapidly
10-601 Machine Learning HMM applications in computational biology Central dogma DNA CCTGAGCCAACTATTGATGAA transcription mrna CCUGAGCCAACUAUUGAUGAA translation Protein PEPTIDE 2 Biological data is rapidly
NEXT GENERATION SEQUENCING. Farhat Habib
 NEXT GENERATION SEQUENCING HISTORY HISTORY Sanger Dominant for last ~30 years 1000bp longest read Based on primers so not good for repetitive or SNPs sites HISTORY Sanger Dominant for last ~30 years 1000bp
NEXT GENERATION SEQUENCING HISTORY HISTORY Sanger Dominant for last ~30 years 1000bp longest read Based on primers so not good for repetitive or SNPs sites HISTORY Sanger Dominant for last ~30 years 1000bp
Accelerate High Throughput Analysis for Genome Sequencing with GPU
 Accelerate High Throughput Analysis for Genome Sequencing with GPU ATIP - A*CRC Workshop on Accelerator Technologies in High Performance Computing May 7-10, 2012 Singapore BingQiang WANG, Head of Scalable
Accelerate High Throughput Analysis for Genome Sequencing with GPU ATIP - A*CRC Workshop on Accelerator Technologies in High Performance Computing May 7-10, 2012 Singapore BingQiang WANG, Head of Scalable
Compression and Integration of Genomic Variants Into Smart EHR Systems
 Compression and Integration of Genomic Variants Into Smart EHR Systems Andrew Gritsevskiy and Adithya Vellal Mentor: Dr. Gil Alterovitz 6 th Annual PRIMES Conference May 22 2016 An Introduction to Genomic
Compression and Integration of Genomic Variants Into Smart EHR Systems Andrew Gritsevskiy and Adithya Vellal Mentor: Dr. Gil Alterovitz 6 th Annual PRIMES Conference May 22 2016 An Introduction to Genomic
Illumina (Solexa) Throughput: 4 Tbp in one run (5 days) Cheapest sequencing technology. Mismatch errors dominate. Cost: ~$1000 per human genme
 Illumina (Solexa) Current market leader Based on sequencing by synthesis Current read length 100-150bp Paired-end easy, longer matepairs harder Error ~0.1% Mismatch errors dominate Throughput: 4 Tbp in
Illumina (Solexa) Current market leader Based on sequencing by synthesis Current read length 100-150bp Paired-end easy, longer matepairs harder Error ~0.1% Mismatch errors dominate Throughput: 4 Tbp in
Genome Reassembly From Fragments. 28 March 2013 OSU CSE 1
 Genome Reassembly From Fragments 28 March 2013 OSU CSE 1 Genome A genome is the encoding of hereditary information for an organism in its DNA The mathematical model of a genome is a string of character,
Genome Reassembly From Fragments 28 March 2013 OSU CSE 1 Genome A genome is the encoding of hereditary information for an organism in its DNA The mathematical model of a genome is a string of character,
Read Mapping and Variant Calling. Johannes Starlinger
 Read Mapping and Variant Calling Johannes Starlinger Application Scenario: Personalized Cancer Therapy Different mutations require different therapy Collins, Meredith A., and Marina Pasca di Magliano.
Read Mapping and Variant Calling Johannes Starlinger Application Scenario: Personalized Cancer Therapy Different mutations require different therapy Collins, Meredith A., and Marina Pasca di Magliano.
Dipping into Guacamole. Tim O Donnell & Ryan Williams NYC Big Data Genetics Meetup Aug 11, 2016
 Dipping into uacamole Tim O Donnell & Ryan Williams NYC Big Data enetics Meetup ug 11, 2016 Who we are: Hammer Lab Computational lab in the department of enetics and enomic Sciences at Mount Sinai Principal
Dipping into uacamole Tim O Donnell & Ryan Williams NYC Big Data enetics Meetup ug 11, 2016 Who we are: Hammer Lab Computational lab in the department of enetics and enomic Sciences at Mount Sinai Principal
De novo meta-assembly of ultra-deep sequencing data
 De novo meta-assembly of ultra-deep sequencing data Hamid Mirebrahim 1, Timothy J. Close 2 and Stefano Lonardi 1 1 Department of Computer Science and Engineering 2 Department of Botany and Plant Sciences
De novo meta-assembly of ultra-deep sequencing data Hamid Mirebrahim 1, Timothy J. Close 2 and Stefano Lonardi 1 1 Department of Computer Science and Engineering 2 Department of Botany and Plant Sciences
Basics of RNA-Seq. (With a Focus on Application to Single Cell RNA-Seq) Michael Kelly, PhD Team Lead, NCI Single Cell Analysis Facility
 2018 ABRF Meeting Satellite Workshop 4 Bridging the Gap: Isolation to Translation (Single Cell RNA-Seq) Sunday, April 22 Basics of RNA-Seq (With a Focus on Application to Single Cell RNA-Seq) Michael Kelly,
2018 ABRF Meeting Satellite Workshop 4 Bridging the Gap: Isolation to Translation (Single Cell RNA-Seq) Sunday, April 22 Basics of RNA-Seq (With a Focus on Application to Single Cell RNA-Seq) Michael Kelly,
What is Bioinformatics? Bioinformatics is the application of computational techniques to the discovery of knowledge from biological databases.
 What is Bioinformatics? Bioinformatics is the application of computational techniques to the discovery of knowledge from biological databases. Bioinformatics is the marriage of molecular biology with computer
What is Bioinformatics? Bioinformatics is the application of computational techniques to the discovery of knowledge from biological databases. Bioinformatics is the marriage of molecular biology with computer
Efficient compression of genomic sequences
 Efficient compression of genomic sequences Diogo Pratas, Armando J. Pinho, and Paulo J. S. G. Ferreira IEETA - Institute of Electronics and Informatics Engineering of Aveiro DETI - Department of Electronics,
Efficient compression of genomic sequences Diogo Pratas, Armando J. Pinho, and Paulo J. S. G. Ferreira IEETA - Institute of Electronics and Informatics Engineering of Aveiro DETI - Department of Electronics,
Next Generation Sequencing. Tobias Österlund
 Next Generation Sequencing Tobias Österlund tobiaso@chalmers.se NGS part of the course Week 4 Friday 13/2 15.15-17.00 NGS lecture 1: Introduction to NGS, alignment, assembly Week 6 Thursday 26/2 08.00-09.45
Next Generation Sequencing Tobias Österlund tobiaso@chalmers.se NGS part of the course Week 4 Friday 13/2 15.15-17.00 NGS lecture 1: Introduction to NGS, alignment, assembly Week 6 Thursday 26/2 08.00-09.45
Accelerating Genomic Computations 1000X with Hardware
 Accelerating Genomic Computations 1000X with Hardware Yatish Turakhia EE PhD candidate Stanford University Prof. Bill Dally (Electrical Engineering and Computer Science) Prof. Gill Bejerano (Computer Science,
Accelerating Genomic Computations 1000X with Hardware Yatish Turakhia EE PhD candidate Stanford University Prof. Bill Dally (Electrical Engineering and Computer Science) Prof. Gill Bejerano (Computer Science,
Compressing Population DNA Sequences Using Multiple Reference Sequences
 Compressing Population DNA Sequences Using Multiple Reference Sequences Kin-On Cheng, Ngai-Fong Law and Wan-Chi Siu Dept of Electronic and Information Engineering, The Hong Kong Polytechnic University,
Compressing Population DNA Sequences Using Multiple Reference Sequences Kin-On Cheng, Ngai-Fong Law and Wan-Chi Siu Dept of Electronic and Information Engineering, The Hong Kong Polytechnic University,
Chapter 2: Access to Information
 Chapter 2: Access to Information Outline Introduction to biological databases Centralized databases store DNA sequences Contents of DNA, RNA, and protein databases Central bioinformatics resources: NCBI
Chapter 2: Access to Information Outline Introduction to biological databases Centralized databases store DNA sequences Contents of DNA, RNA, and protein databases Central bioinformatics resources: NCBI
A framework for a general- purpose sequence compression pipeline: a centroid based compression
 A framework for a general- purpose sequence compression pipeline: a centroid based compression Liqing Zhang 1, Daniel Nasko 2, Martin Tříska 3, Harold Garner 4 1. Department of Computer Science, Programs
A framework for a general- purpose sequence compression pipeline: a centroid based compression Liqing Zhang 1, Daniel Nasko 2, Martin Tříska 3, Harold Garner 4 1. Department of Computer Science, Programs
Review of whole genome methods
 Review of whole genome methods Suffix-tree based MUMmer, Mauve, multi-mauve Gene based Mercator, multiple orthology approaches Dot plot/clustering based MUMmer 2.0, Pipmaker, LASTZ 10/3/17 0 Rationale:
Review of whole genome methods Suffix-tree based MUMmer, Mauve, multi-mauve Gene based Mercator, multiple orthology approaches Dot plot/clustering based MUMmer 2.0, Pipmaker, LASTZ 10/3/17 0 Rationale:
Introduction. CS482/682 Computational Techniques in Biological Sequence Analysis
 Introduction CS482/682 Computational Techniques in Biological Sequence Analysis Outline Course logistics A few example problems Course staff Instructor: Bin Ma (DC 3345, http://www.cs.uwaterloo.ca/~binma)
Introduction CS482/682 Computational Techniques in Biological Sequence Analysis Outline Course logistics A few example problems Course staff Instructor: Bin Ma (DC 3345, http://www.cs.uwaterloo.ca/~binma)
Computational Challenges in Life Sciences Research Infrastructures
 Computational Challenges in Life Sciences Research Infrastructures Alvis Brazma European Bioinformatics Institute European Molecular Biology Laboratory European Bioinformatics Institute (EBI) EBI is in
Computational Challenges in Life Sciences Research Infrastructures Alvis Brazma European Bioinformatics Institute European Molecular Biology Laboratory European Bioinformatics Institute (EBI) EBI is in
Whole Genome Sequencing in Cancer Diagnostics (research) Nederlandse Pathologiedagen 19 & 20 November 2015
 Whole Genome Sequencing in Cancer Diagnostics (research) Nederlandse Pathologiedagen 19 & 20 November 2015 Dr. I.J. Nijman Disclosure slide (Potential) conflict of interest None For this meeting relevant
Whole Genome Sequencing in Cancer Diagnostics (research) Nederlandse Pathologiedagen 19 & 20 November 2015 Dr. I.J. Nijman Disclosure slide (Potential) conflict of interest None For this meeting relevant
Genomic Technologies. Michael Schatz. Feb 1, 2018 Lecture 2: Applied Comparative Genomics
 Genomic Technologies Michael Schatz Feb 1, 2018 Lecture 2: Applied Comparative Genomics Welcome! The primary goal of the course is for students to be grounded in theory and leave the course empowered to
Genomic Technologies Michael Schatz Feb 1, 2018 Lecture 2: Applied Comparative Genomics Welcome! The primary goal of the course is for students to be grounded in theory and leave the course empowered to
Bioinformatics : Gene Expression Data Analysis
 05.12.03 Bioinformatics : Gene Expression Data Analysis Aidong Zhang Professor Computer Science and Engineering What is Bioinformatics Broad Definition The study of how information technologies are used
05.12.03 Bioinformatics : Gene Expression Data Analysis Aidong Zhang Professor Computer Science and Engineering What is Bioinformatics Broad Definition The study of how information technologies are used
Sequencing technologies. Jose Blanca COMAV institute bioinf.comav.upv.es
 Sequencing technologies Jose Blanca COMAV institute bioinf.comav.upv.es Outline Sequencing technologies: Sanger 2nd generation sequencing: 3er generation sequencing: 454 Illumina SOLiD Ion Torrent PacBio
Sequencing technologies Jose Blanca COMAV institute bioinf.comav.upv.es Outline Sequencing technologies: Sanger 2nd generation sequencing: 3er generation sequencing: 454 Illumina SOLiD Ion Torrent PacBio
Data Retrieval from GenBank
 Data Retrieval from GenBank Peter J. Myler Bioinformatics of Intracellular Pathogens JNU, Feb 7-0, 2009 http://www.ncbi.nlm.nih.gov (January, 2007) http://ncbi.nlm.nih.gov/sitemap/resourceguide.html Accessing
Data Retrieval from GenBank Peter J. Myler Bioinformatics of Intracellular Pathogens JNU, Feb 7-0, 2009 http://www.ncbi.nlm.nih.gov (January, 2007) http://ncbi.nlm.nih.gov/sitemap/resourceguide.html Accessing
SYMPOSIUM March 22-23, 2018
 Bigger and Better Data Lessons from Frontlines of Precision Medicine Getting Your Transformation Right Frank Lee PhD IBM Global Industry Leader for Systems Group SYMPOSIUM March 22-23, 2018 5th Annual
Bigger and Better Data Lessons from Frontlines of Precision Medicine Getting Your Transformation Right Frank Lee PhD IBM Global Industry Leader for Systems Group SYMPOSIUM March 22-23, 2018 5th Annual
A New Approach of Protein Sequence Compression using Repeat Reduction and ASCII Replacement
 IOSR Journal of Computer Engineering (IOSR-JCE) e-issn: 2278-0661, p- ISSN: 2278-8727Volume 10, Issue 5 (Mar. - Apr. 2013), PP 46-51 A New Approach of Protein Sequence Compression using Repeat Reduction
IOSR Journal of Computer Engineering (IOSR-JCE) e-issn: 2278-0661, p- ISSN: 2278-8727Volume 10, Issue 5 (Mar. - Apr. 2013), PP 46-51 A New Approach of Protein Sequence Compression using Repeat Reduction
High-Throughput Bioinformatics: Re-sequencing and de novo assembly. Elena Czeizler
 High-Throughput Bioinformatics: Re-sequencing and de novo assembly Elena Czeizler 13.11.2015 Sequencing data Current sequencing technologies produce large amounts of data: short reads The outputted sequences
High-Throughput Bioinformatics: Re-sequencing and de novo assembly Elena Czeizler 13.11.2015 Sequencing data Current sequencing technologies produce large amounts of data: short reads The outputted sequences
Vertical DNA Sequences Compression Algorithm Based on Hexadecimal Representation
 , October 21-23, 2015, San Francisco, USA Vertical DNA Sequences Compression Algorithm Based on Hexadecimal Representation Bacem Saada, Jing Zhang Abstract The number of DNA sequences stored in databases
, October 21-23, 2015, San Francisco, USA Vertical DNA Sequences Compression Algorithm Based on Hexadecimal Representation Bacem Saada, Jing Zhang Abstract The number of DNA sequences stored in databases
ALGORITHMS IN BIO INFORMATICS. Chapman & Hall/CRC Mathematical and Computational Biology Series A PRACTICAL INTRODUCTION. CRC Press WING-KIN SUNG
 Chapman & Hall/CRC Mathematical and Computational Biology Series ALGORITHMS IN BIO INFORMATICS A PRACTICAL INTRODUCTION WING-KIN SUNG CRC Press Taylor & Francis Group Boca Raton London New York CRC Press
Chapman & Hall/CRC Mathematical and Computational Biology Series ALGORITHMS IN BIO INFORMATICS A PRACTICAL INTRODUCTION WING-KIN SUNG CRC Press Taylor & Francis Group Boca Raton London New York CRC Press
A Survey on Data Compression Methods for Biological Sequences
 Article A Survey on Data Compression Methods for Biological Sequences Morteza Hosseini *, Diogo Pratas and Armando J. Pinho Institute of Electronics and Informatics Engineering of Aveiro/Department of
Article A Survey on Data Compression Methods for Biological Sequences Morteza Hosseini *, Diogo Pratas and Armando J. Pinho Institute of Electronics and Informatics Engineering of Aveiro/Department of
axe Documentation Release g6d4d1b6-dirty Kevin Murray
 axe Documentation Release 0.3.2-5-g6d4d1b6-dirty Kevin Murray Jul 17, 2017 Contents 1 Axe Usage 3 1.1 Inputs and Outputs..................................... 4 1.2 The barcode file......................................
axe Documentation Release 0.3.2-5-g6d4d1b6-dirty Kevin Murray Jul 17, 2017 Contents 1 Axe Usage 3 1.1 Inputs and Outputs..................................... 4 1.2 The barcode file......................................
Compressing the Human Genome against a reference
 Compressing the Human Genome against a reference Pragya Pande prpande@cs.stonybrook.edu Stony Brook University Dhruv Matani dmatani@cs.stonybrook.edu Stony Brook University December 8, 2011 Abstract We
Compressing the Human Genome against a reference Pragya Pande prpande@cs.stonybrook.edu Stony Brook University Dhruv Matani dmatani@cs.stonybrook.edu Stony Brook University December 8, 2011 Abstract We
Introduction to Bioinformatics. Ulf Leser
 Introduction to Bioinformatics Ulf Leser Bioinformatics 25.4.2003 50. Jubiläum der Entdeckung der Doppelhelix durch Watson/Crick 14.4.2003 Humanes Genom zu 99% sequenziert mit 99.99% Genauigkeit 2008 Genom
Introduction to Bioinformatics Ulf Leser Bioinformatics 25.4.2003 50. Jubiläum der Entdeckung der Doppelhelix durch Watson/Crick 14.4.2003 Humanes Genom zu 99% sequenziert mit 99.99% Genauigkeit 2008 Genom
LARGE DATA AND BIOMEDICAL COMPUTATIONAL PIPELINES FOR COMPLEX DISEASES
 1 LARGE DATA AND BIOMEDICAL COMPUTATIONAL PIPELINES FOR COMPLEX DISEASES Ezekiel Adebiyi, PhD Professor and Head, Covenant University Bioinformatics Research and CU NIH H3AbioNet node Covenant University,
1 LARGE DATA AND BIOMEDICAL COMPUTATIONAL PIPELINES FOR COMPLEX DISEASES Ezekiel Adebiyi, PhD Professor and Head, Covenant University Bioinformatics Research and CU NIH H3AbioNet node Covenant University,
GENETIC ALGORITHMS. Narra Priyanka. K.Naga Sowjanya. Vasavi College of Engineering. Ibrahimbahg,Hyderabad.
 GENETIC ALGORITHMS Narra Priyanka K.Naga Sowjanya Vasavi College of Engineering. Ibrahimbahg,Hyderabad mynameissowji@yahoo.com priyankanarra@yahoo.com Abstract Genetic algorithms are a part of evolutionary
GENETIC ALGORITHMS Narra Priyanka K.Naga Sowjanya Vasavi College of Engineering. Ibrahimbahg,Hyderabad mynameissowji@yahoo.com priyankanarra@yahoo.com Abstract Genetic algorithms are a part of evolutionary
An Analytical Upper Bound on the Minimum Number of. Recombinations in the History of SNP Sequences in Populations
 An Analytical Upper Bound on the Minimum Number of Recombinations in the History of SNP Sequences in Populations Yufeng Wu Department of Computer Science and Engineering University of Connecticut Storrs,
An Analytical Upper Bound on the Minimum Number of Recombinations in the History of SNP Sequences in Populations Yufeng Wu Department of Computer Science and Engineering University of Connecticut Storrs,
Introduction to Microarray Data Analysis and Gene Networks. Alvis Brazma European Bioinformatics Institute
 Introduction to Microarray Data Analysis and Gene Networks Alvis Brazma European Bioinformatics Institute A brief outline of this course What is gene expression, why it s important Microarrays and how
Introduction to Microarray Data Analysis and Gene Networks Alvis Brazma European Bioinformatics Institute A brief outline of this course What is gene expression, why it s important Microarrays and how
Time Series Motif Discovery
 Time Series Motif Discovery Bachelor s Thesis Exposé eingereicht von: Jonas Spenger Gutachter: Dr. rer. nat. Patrick Schäfer Gutachter: Prof. Dr. Ulf Leser eingereicht am: 10.09.2017 Contents 1 Introduction
Time Series Motif Discovery Bachelor s Thesis Exposé eingereicht von: Jonas Spenger Gutachter: Dr. rer. nat. Patrick Schäfer Gutachter: Prof. Dr. Ulf Leser eingereicht am: 10.09.2017 Contents 1 Introduction
Introduction to Bioinformatics. Ulf Leser
 Introduction to Bioinformatics Ulf Leser Bioinformatics 25.4.2003 50. Jubiläum der Entdeckung der Doppelhelix durch Watson/Crick 14.4.2003 Humanes Genom zu 99% sequenziert mit 99.99% Genauigkeit 2008 Genom
Introduction to Bioinformatics Ulf Leser Bioinformatics 25.4.2003 50. Jubiläum der Entdeckung der Doppelhelix durch Watson/Crick 14.4.2003 Humanes Genom zu 99% sequenziert mit 99.99% Genauigkeit 2008 Genom
Illumina Sequencing Error Profiles and Quality Control
 Illumina Sequencing Error Profiles and Quality Control RNA-seq Workflow Biological samples/library preparation Sequence reads FASTQC Adapter Trimming (Optional) Splice-aware mapping to genome Counting
Illumina Sequencing Error Profiles and Quality Control RNA-seq Workflow Biological samples/library preparation Sequence reads FASTQC Adapter Trimming (Optional) Splice-aware mapping to genome Counting
Users? Data. Implications for HPC in Biology Dr Thomas R Connor School of Biosciences, Cardiff University
 Users? Data Compute Network Implications for HPC in Biology Dr Thomas R Connor School of Biosciences, Cardiff University connortr@cardiff.ac.uk ; @tomrconnor The Bigger Picture 200 Million people have
Users? Data Compute Network Implications for HPC in Biology Dr Thomas R Connor School of Biosciences, Cardiff University connortr@cardiff.ac.uk ; @tomrconnor The Bigger Picture 200 Million people have
The Genomic Transformation of Health
 The Genomic Transformation of Health or: an introduction to the potential of genomics in healthcare Dr Tom Connor School of Biosciences and Systems Immunity University Research Institute Cardiff University
The Genomic Transformation of Health or: an introduction to the potential of genomics in healthcare Dr Tom Connor School of Biosciences and Systems Immunity University Research Institute Cardiff University
Genomic DNA ASSEMBLY BY REMAPPING. Course overview
 ASSEMBLY BY REMAPPING Laurent Falquet, The Bioinformatics Unravelling Group, UNIFR & SIB MA/MER @ UniFr Group Leader @ SIB Course overview Genomic DNA PacBio Illumina methylation de novo remapping Annotation
ASSEMBLY BY REMAPPING Laurent Falquet, The Bioinformatics Unravelling Group, UNIFR & SIB MA/MER @ UniFr Group Leader @ SIB Course overview Genomic DNA PacBio Illumina methylation de novo remapping Annotation
Development of Cache Oblivious Based Fast Multiple Longest Common Subsequence Technique(CMLCS) for Biological Sequences Prediction
 Research Journal of Applied Sciences, Engineering and Technology 4(9): 1198-1204, 2012 ISSN: 2040-7467 Maxwell Scientific Organization, 2012 Submitted: January 11, 2012 Accepted: February 06, 2012 Published:
Research Journal of Applied Sciences, Engineering and Technology 4(9): 1198-1204, 2012 ISSN: 2040-7467 Maxwell Scientific Organization, 2012 Submitted: January 11, 2012 Accepted: February 06, 2012 Published:
Basic Local Alignment Search Tool
 14.06.2010 Table of contents 1 History History 2 global local 3 Score functions Score matrices 4 5 Comparison to FASTA References of BLAST History the program was designed by Stephen W. Altschul, Warren
14.06.2010 Table of contents 1 History History 2 global local 3 Score functions Score matrices 4 5 Comparison to FASTA References of BLAST History the program was designed by Stephen W. Altschul, Warren
Protein-Protein-Interaction Networks. Ulf Leser, Samira Jaeger
 Protein-Protein-Interaction Networks Ulf Leser, Samira Jaeger This Lecture Protein-protein interactions Characteristics Experimental detection methods Databases Protein-protein interaction networks Ulf
Protein-Protein-Interaction Networks Ulf Leser, Samira Jaeger This Lecture Protein-protein interactions Characteristics Experimental detection methods Databases Protein-protein interaction networks Ulf
ACCELERATING GENOMIC ANALYSIS ON THE CLOUD. Enabling the PanCancer Analysis of Whole Genomes (PCAWG) consortia to analyze thousands of genomes
 ACCELERATING GENOMIC ANALYSIS ON THE CLOUD Enabling the PanCancer Analysis of Whole Genomes (PCAWG) consortia to analyze thousands of genomes Enabling the PanCancer Analysis of Whole Genomes (PCAWG) consortia
ACCELERATING GENOMIC ANALYSIS ON THE CLOUD Enabling the PanCancer Analysis of Whole Genomes (PCAWG) consortia to analyze thousands of genomes Enabling the PanCancer Analysis of Whole Genomes (PCAWG) consortia
Advisors: Prof. Louis T. Oliphant Computer Science Department, Hiram College.
 Author: Sulochana Bramhacharya Affiliation: Hiram College, Hiram OH. Address: P.O.B 1257 Hiram, OH 44234 Email: bramhacharyas1@my.hiram.edu ACM number: 8983027 Category: Undergraduate research Advisors:
Author: Sulochana Bramhacharya Affiliation: Hiram College, Hiram OH. Address: P.O.B 1257 Hiram, OH 44234 Email: bramhacharyas1@my.hiram.edu ACM number: 8983027 Category: Undergraduate research Advisors:
Challenges in precision medicine: From sequencing to big data processing Mikel Hernaez
 Challenges in precision medicine: From sequencing to big data processing Mikel Hernaez Torino, July 19th 2017 Tonight I m launching a new Precision Medicine Initiative to bring us closer to curing diseases
Challenges in precision medicine: From sequencing to big data processing Mikel Hernaez Torino, July 19th 2017 Tonight I m launching a new Precision Medicine Initiative to bring us closer to curing diseases
Computational Intelligence Lecture 20:Intorcution to Genetic Algorithm
 Computational Intelligence Lecture 20:Intorcution to Genetic Algorithm Farzaneh Abdollahi Department of Electrical Engineering Amirkabir University of Technology Fall 2012 Farzaneh Abdollahi Computational
Computational Intelligence Lecture 20:Intorcution to Genetic Algorithm Farzaneh Abdollahi Department of Electrical Engineering Amirkabir University of Technology Fall 2012 Farzaneh Abdollahi Computational
Genetics and Bioinformatics
 Genetics and Bioinformatics Kristel Van Steen, PhD 2 Montefiore Institute - Systems and Modeling GIGA - Bioinformatics ULg kristel.vansteen@ulg.ac.be Lecture 1: Setting the pace 1 Bioinformatics what s
Genetics and Bioinformatics Kristel Van Steen, PhD 2 Montefiore Institute - Systems and Modeling GIGA - Bioinformatics ULg kristel.vansteen@ulg.ac.be Lecture 1: Setting the pace 1 Bioinformatics what s
Accelerating Motif Finding in DNA Sequences with Multicore CPUs
 Accelerating Motif Finding in DNA Sequences with Multicore CPUs Pramitha Perera and Roshan Ragel, Member, IEEE Abstract Motif discovery in DNA sequences is a challenging task in molecular biology. In computational
Accelerating Motif Finding in DNA Sequences with Multicore CPUs Pramitha Perera and Roshan Ragel, Member, IEEE Abstract Motif discovery in DNA sequences is a challenging task in molecular biology. In computational
Protein design. CS/CME/Biophys/BMI 279 Oct. 20 and 22, 2015 Ron Dror
 Protein design CS/CME/Biophys/BMI 279 Oct. 20 and 22, 2015 Ron Dror 1 Optional reading on course website From cs279.stanford.edu These reading materials are optional. They are intended to (1) help answer
Protein design CS/CME/Biophys/BMI 279 Oct. 20 and 22, 2015 Ron Dror 1 Optional reading on course website From cs279.stanford.edu These reading materials are optional. They are intended to (1) help answer
Course Presentation. Ignacio Medina Presentation
 Course Index Introduction Agenda Analysis pipeline Some considerations Introduction Who we are Teachers: Marta Bleda: Computational Biologist and Data Analyst at Department of Medicine, Addenbrooke's Hospital
Course Index Introduction Agenda Analysis pipeline Some considerations Introduction Who we are Teachers: Marta Bleda: Computational Biologist and Data Analyst at Department of Medicine, Addenbrooke's Hospital
Introduction to Bioinformatics Finish. Johannes Starlinger
 Introduction to Bioinformatics Finish Johannes Starlinger This Lecture Genomics Sequencing Gene prediction Evolutionary relationships Motifs - TFBS Transcriptomics Alignment Proteomics Structure prediction
Introduction to Bioinformatics Finish Johannes Starlinger This Lecture Genomics Sequencing Gene prediction Evolutionary relationships Motifs - TFBS Transcriptomics Alignment Proteomics Structure prediction
ISO/IEC JTC 1/SC 29/WG 11 N15527 Warsaw, CH June Introduction
 INTERNATIONAL ORGANISATION FOR STANDARDISATION ORGANISATION INTERNATIONALE DE NORMALISATION ISO/IEC JTC 1/SC 29/WG 11 CODING OF MOVING PICTURES AND AUDIO ISO/IEC JTC 1/SC 29/WG 11 N15527 Warsaw, CH June
INTERNATIONAL ORGANISATION FOR STANDARDISATION ORGANISATION INTERNATIONALE DE NORMALISATION ISO/IEC JTC 1/SC 29/WG 11 CODING OF MOVING PICTURES AND AUDIO ISO/IEC JTC 1/SC 29/WG 11 N15527 Warsaw, CH June
Using Test Oracles in Automation. Topics
 Using Test Oracles in Automation Software Test Automation Conference Spring 2003 Douglas Hoffman Software Quality Methods, LLC. 24646 Heather Heights Place Saratoga, California 95070-9710 Phone 408-741-4830
Using Test Oracles in Automation Software Test Automation Conference Spring 2003 Douglas Hoffman Software Quality Methods, LLC. 24646 Heather Heights Place Saratoga, California 95070-9710 Phone 408-741-4830
Experimental Design Microbial Sequencing
 Experimental Design Microbial Sequencing Matthew L. Settles Genome Center Bioinformatics Core University of California, Davis settles@ucdavis.edu; bioinformatics.core@ucdavis.edu General rules for preparing
Experimental Design Microbial Sequencing Matthew L. Settles Genome Center Bioinformatics Core University of California, Davis settles@ucdavis.edu; bioinformatics.core@ucdavis.edu General rules for preparing
Accelerate precision medicine with Microsoft Genomics
 Accelerate precision medicine with Microsoft Genomics Copyright 2018 Microsoft, Inc. All rights reserved. This content is for informational purposes only. Microsoft makes no warranties, express or implied,
Accelerate precision medicine with Microsoft Genomics Copyright 2018 Microsoft, Inc. All rights reserved. This content is for informational purposes only. Microsoft makes no warranties, express or implied,
Exploring Long DNA Sequences by Information Content
 Exploring Long DNA Sequences by Information Content Trevor I. Dix 1,2, David R. Powell 1,2, Lloyd Allison 1, Samira Jaeger 1, Julie Bernal 1, and Linda Stern 3 1 Faculty of I.T., Monash University, 2 Victorian
Exploring Long DNA Sequences by Information Content Trevor I. Dix 1,2, David R. Powell 1,2, Lloyd Allison 1, Samira Jaeger 1, Julie Bernal 1, and Linda Stern 3 1 Faculty of I.T., Monash University, 2 Victorian
Lecture 2: Central Dogma of Molecular Biology & Intro to Programming
 Lecture 2: Central Dogma of Molecular Biology & Intro to Programming Central Dogma of Molecular Biology Proteins: workhorse molecules of biological systems Proteins are synthesized from the genetic blueprints
Lecture 2: Central Dogma of Molecular Biology & Intro to Programming Central Dogma of Molecular Biology Proteins: workhorse molecules of biological systems Proteins are synthesized from the genetic blueprints
QIAGEN s NGS Solutions for Biomarkers NGS & Bioinformatics team QIAGEN (Suzhou) Translational Medicine Co.,Ltd
 QIAGEN s NGS Solutions for Biomarkers NGS & Bioinformatics team QIAGEN (Suzhou) Translational Medicine Co.,Ltd 1 Our current NGS & Bioinformatics Platform 2 Our NGS workflow and applications 3 QIAGEN s
QIAGEN s NGS Solutions for Biomarkers NGS & Bioinformatics team QIAGEN (Suzhou) Translational Medicine Co.,Ltd 1 Our current NGS & Bioinformatics Platform 2 Our NGS workflow and applications 3 QIAGEN s
Next-Generation Sequencing Services à la carte
 Next-Generation Sequencing Services à la carte www.seqme.eu ngs@seqme.eu SEQme 2017 All rights reserved The trademarks and names of other companies and products mentioned in this brochure are the property
Next-Generation Sequencing Services à la carte www.seqme.eu ngs@seqme.eu SEQme 2017 All rights reserved The trademarks and names of other companies and products mentioned in this brochure are the property
High-throughput DNA sequence data compression Zexuan Zhu, Yongpeng Zhang, Zhen Ji, Shan He and Xiao Yang
 BRIEFINGS IN BIOINFORMATICS. VOL 16. NO 1. 1^15 Advance Access published on 3 December 2013 doi:10.1093/bib/bbt087 High-throughput DNA sequence data compression Zexuan Zhu, Yongpeng Zhang, Zhen Ji, Shan
BRIEFINGS IN BIOINFORMATICS. VOL 16. NO 1. 1^15 Advance Access published on 3 December 2013 doi:10.1093/bib/bbt087 High-throughput DNA sequence data compression Zexuan Zhu, Yongpeng Zhang, Zhen Ji, Shan
Introduction to Bioinformatics and Gene Expression Technologies
 Introduction to Bioinformatics and Gene Expression Technologies Utah State University Fall 2017 Statistical Bioinformatics (Biomedical Big Data) Notes 1 1 Vocabulary Gene: hereditary DNA sequence at a
Introduction to Bioinformatics and Gene Expression Technologies Utah State University Fall 2017 Statistical Bioinformatics (Biomedical Big Data) Notes 1 1 Vocabulary Gene: hereditary DNA sequence at a
Introduction to Bioinformatics and Gene Expression Technologies
 Vocabulary Introduction to Bioinformatics and Gene Expression Technologies Utah State University Fall 2017 Statistical Bioinformatics (Biomedical Big Data) Notes 1 Gene: Genetics: Genome: Genomics: hereditary
Vocabulary Introduction to Bioinformatics and Gene Expression Technologies Utah State University Fall 2017 Statistical Bioinformatics (Biomedical Big Data) Notes 1 Gene: Genetics: Genome: Genomics: hereditary
CS681: Advanced Topics in Computational Biology
 CS681: Advanced Topics in Computational Biology Can Alkan EA224 calkan@cs.bilkent.edu.tr Week 3, Lectures 2-3 http://www.cs.bilkent.edu.tr/~calkan/teaching/cs681/ Features of NGS data Short sequence reads
CS681: Advanced Topics in Computational Biology Can Alkan EA224 calkan@cs.bilkent.edu.tr Week 3, Lectures 2-3 http://www.cs.bilkent.edu.tr/~calkan/teaching/cs681/ Features of NGS data Short sequence reads
Protein-Protein-Interaction Networks. Ulf Leser, Samira Jaeger
 Protein-Protein-Interaction Networks Ulf Leser, Samira Jaeger This Lecture Protein-protein interactions Characteristics Experimental detection methods Databases Biological networks Ulf Leser: Introduction
Protein-Protein-Interaction Networks Ulf Leser, Samira Jaeger This Lecture Protein-protein interactions Characteristics Experimental detection methods Databases Biological networks Ulf Leser: Introduction
Scottish Genomes Project: Taking Genomics into Healthcare. Zosia Miedzybrodzka
 Scottish Genomes Project: Taking Genomics into Healthcare Zosia Miedzybrodzka DNA GENOME IS ALL OF OUR DNA: CODING & NON-CODING Sequencing of Whole Genomes at Scale in Scotland Edinburgh Genomics Sequencing
Scottish Genomes Project: Taking Genomics into Healthcare Zosia Miedzybrodzka DNA GENOME IS ALL OF OUR DNA: CODING & NON-CODING Sequencing of Whole Genomes at Scale in Scotland Edinburgh Genomics Sequencing
Introduction to Bioinformatics
 Introduction to Bioinformatics Dr. Taysir Hassan Abdel Hamid Lecturer, Information Systems Department Faculty of Computer and Information Assiut University taysirhs@aun.edu.eg taysir_soliman@hotmail.com
Introduction to Bioinformatics Dr. Taysir Hassan Abdel Hamid Lecturer, Information Systems Department Faculty of Computer and Information Assiut University taysirhs@aun.edu.eg taysir_soliman@hotmail.com
Chapter 7. DNA Microarrays
 Bioinformatics III Structural Bioinformatics and Genome Analysis Chapter 7. DNA Microarrays 7.9 Next Generation Sequencing 454 Sequencing Solexa Illumina Solid TM System Sequencing Process of determining
Bioinformatics III Structural Bioinformatics and Genome Analysis Chapter 7. DNA Microarrays 7.9 Next Generation Sequencing 454 Sequencing Solexa Illumina Solid TM System Sequencing Process of determining
From Variants to Pathways: Agilent GeneSpring GX s Variant Analysis Workflow
 From Variants to Pathways: Agilent GeneSpring GX s Variant Analysis Workflow Technical Overview Import VCF Introduction Next-generation sequencing (NGS) studies have created unanticipated challenges with
From Variants to Pathways: Agilent GeneSpring GX s Variant Analysis Workflow Technical Overview Import VCF Introduction Next-generation sequencing (NGS) studies have created unanticipated challenges with
Francisco García Quality Control for NGS Raw Data
 Contents Data formats Sequence capture Fasta and fastq formats Sequence quality encoding Quality Control Evaluation of sequence quality Quality control tools Identification of artifacts & filtering Practical
Contents Data formats Sequence capture Fasta and fastq formats Sequence quality encoding Quality Control Evaluation of sequence quality Quality control tools Identification of artifacts & filtering Practical
Next Generation Sequencing (NGS) Market Size, Growth and Trends ( )
 Next Generation Sequencing (NGS) Market Size, Growth and Trends (2014-2020) July, 2017 4 th edition Information contained in this market report is believed to be reliable at the time of publication. DeciBio
Next Generation Sequencing (NGS) Market Size, Growth and Trends (2014-2020) July, 2017 4 th edition Information contained in this market report is believed to be reliable at the time of publication. DeciBio
CapSel GA Genetic Algorithms.
 CapSel GA - 01 Genetic Algorithms keppens@rijnh.nl Typical usage: optimization problems both minimization and maximization of complicated functions completely standard problem with non-standard solution
CapSel GA - 01 Genetic Algorithms keppens@rijnh.nl Typical usage: optimization problems both minimization and maximization of complicated functions completely standard problem with non-standard solution
Bio nformatics. Lecture 2. Saad Mneimneh
 Bio nformatics Lecture 2 RFLP: Restriction Fragment Length Polymorphism A restriction enzyme cuts the DNA molecules at every occurrence of a particular sequence, called restriction site. For example, HindII
Bio nformatics Lecture 2 RFLP: Restriction Fragment Length Polymorphism A restriction enzyme cuts the DNA molecules at every occurrence of a particular sequence, called restriction site. For example, HindII
Reveal Motif Patterns from Financial Stock Market
 Reveal Motif Patterns from Financial Stock Market Prakash Kumar Sarangi Department of Information Technology NM Institute of Engineering and Technology, Bhubaneswar, India. Prakashsarangi89@gmail.com.
Reveal Motif Patterns from Financial Stock Market Prakash Kumar Sarangi Department of Information Technology NM Institute of Engineering and Technology, Bhubaneswar, India. Prakashsarangi89@gmail.com.
Feature Selection of Gene Expression Data for Cancer Classification: A Review
 Available online at www.sciencedirect.com ScienceDirect Procedia Computer Science 50 (2015 ) 52 57 2nd International Symposium on Big Data and Cloud Computing (ISBCC 15) Feature Selection of Gene Expression
Available online at www.sciencedirect.com ScienceDirect Procedia Computer Science 50 (2015 ) 52 57 2nd International Symposium on Big Data and Cloud Computing (ISBCC 15) Feature Selection of Gene Expression
NUCLEIC ACIDS. DNA (Deoxyribonucleic Acid) and RNA (Ribonucleic Acid): information storage molecules made up of nucleotides.
 NUCLEIC ACIDS DNA (Deoxyribonucleic Acid) and RNA (Ribonucleic Acid): information storage molecules made up of nucleotides. Base Adenine Guanine Cytosine Uracil Thymine Abbreviation A G C U T DNA RNA 2
NUCLEIC ACIDS DNA (Deoxyribonucleic Acid) and RNA (Ribonucleic Acid): information storage molecules made up of nucleotides. Base Adenine Guanine Cytosine Uracil Thymine Abbreviation A G C U T DNA RNA 2
Imaging informatics computer assisted mammogram reading Clinical aka medical informatics CDSS combining bioinformatics for diagnosis, personalized
 1 2 3 Imaging informatics computer assisted mammogram reading Clinical aka medical informatics CDSS combining bioinformatics for diagnosis, personalized medicine, risk assessment etc Public Health Bio
1 2 3 Imaging informatics computer assisted mammogram reading Clinical aka medical informatics CDSS combining bioinformatics for diagnosis, personalized medicine, risk assessment etc Public Health Bio
Bioinformatics Support of Genome Sequencing Projects. Seminar in biology
 Bioinformatics Support of Genome Sequencing Projects Seminar in biology Introduction The Big Picture Biology reminder Enzyme for DNA manipulation DNA cloning DNA mapping Sequencing genomes Alignment of
Bioinformatics Support of Genome Sequencing Projects Seminar in biology Introduction The Big Picture Biology reminder Enzyme for DNA manipulation DNA cloning DNA mapping Sequencing genomes Alignment of
Synthetic Viruses Targeting Cancer
 Synthetic Viruses Targeting Cancer Andrew Hessel September 7, 2007 SENS 3, Cambridge, UK Why is a new strategy necessary? Breast cancer remains a significant cause of illness and death The treatment options
Synthetic Viruses Targeting Cancer Andrew Hessel September 7, 2007 SENS 3, Cambridge, UK Why is a new strategy necessary? Breast cancer remains a significant cause of illness and death The treatment options
Sequencing technologies. Jose Blanca COMAV institute bioinf.comav.upv.es
 Sequencing technologies Jose Blanca COMAV institute bioinf.comav.upv.es Outline Sequencing technologies: Sanger 2nd generation sequencing: 3er generation sequencing: 454 Illumina SOLiD Ion Torrent PacBio
Sequencing technologies Jose Blanca COMAV institute bioinf.comav.upv.es Outline Sequencing technologies: Sanger 2nd generation sequencing: 3er generation sequencing: 454 Illumina SOLiD Ion Torrent PacBio
DNA. Clinical Trials. Research RNA. Custom. Reports CLIA CAP GCP. Tumor Genomic Profiling Services for Clinical Trials
 Tumor Genomic Profiling Services for Clinical Trials Custom Reports DNA RNA Focused Gene Sets Clinical Trials Accuracy and Content Enhanced NGS Sequencing Extended Panel, Exomes, Transcriptomes Research
Tumor Genomic Profiling Services for Clinical Trials Custom Reports DNA RNA Focused Gene Sets Clinical Trials Accuracy and Content Enhanced NGS Sequencing Extended Panel, Exomes, Transcriptomes Research
We begin with a high-level overview of sequencing. There are three stages in this process.
 Lecture 11 Sequence Assembly February 10, 1998 Lecturer: Phil Green Notes: Kavita Garg 11.1. Introduction This is the first of two lectures by Phil Green on Sequence Assembly. Yeast and some of the bacterial
Lecture 11 Sequence Assembly February 10, 1998 Lecturer: Phil Green Notes: Kavita Garg 11.1. Introduction This is the first of two lectures by Phil Green on Sequence Assembly. Yeast and some of the bacterial
Sequencing technologies. Jose Blanca COMAV institute bioinf.comav.upv.es
 Sequencing technologies Jose Blanca COMAV institute bioinf.comav.upv.es Outline Sequencing technologies: Sanger 2nd generation sequencing: 3er generation sequencing: 454 Illumina SOLiD Ion Torrent PacBio
Sequencing technologies Jose Blanca COMAV institute bioinf.comav.upv.es Outline Sequencing technologies: Sanger 2nd generation sequencing: 3er generation sequencing: 454 Illumina SOLiD Ion Torrent PacBio
Next Generation Sequences & Chloroplast Assembly. 8 June, 2012 Jongsun Park
 Next Generation Sequences & Chloroplast Assembly 8 June, 2012 Jongsun Park Table of Contents 1 History of Sequencing Technologies 2 Genome Assembly Processes With NGS Sequences 3 How to Assembly Chloroplast
Next Generation Sequences & Chloroplast Assembly 8 June, 2012 Jongsun Park Table of Contents 1 History of Sequencing Technologies 2 Genome Assembly Processes With NGS Sequences 3 How to Assembly Chloroplast
Optimisation and Operations Research
 Optimisation and Operations Research Lecture 17: Genetic Algorithms and Evolutionary Computing Matthew Roughan http://www.maths.adelaide.edu.au/matthew.roughan/ Lecture_notes/OORII/
Optimisation and Operations Research Lecture 17: Genetic Algorithms and Evolutionary Computing Matthew Roughan http://www.maths.adelaide.edu.au/matthew.roughan/ Lecture_notes/OORII/
ROAD TO STATISTICAL BIOINFORMATICS CHALLENGE 1: MULTIPLE-COMPARISONS ISSUE
 CHAPTER1 ROAD TO STATISTICAL BIOINFORMATICS Jae K. Lee Department of Public Health Science, University of Virginia, Charlottesville, Virginia, USA There has been a great explosion of biological data and
CHAPTER1 ROAD TO STATISTICAL BIOINFORMATICS Jae K. Lee Department of Public Health Science, University of Virginia, Charlottesville, Virginia, USA There has been a great explosion of biological data and
The Expanded Illumina Sequencing Portfolio New Sample Prep Solutions and Workflow
 The Expanded Illumina Sequencing Portfolio New Sample Prep Solutions and Workflow Marcus Hausch, Ph.D. 2010 Illumina, Inc. All rights reserved. Illumina, illuminadx, Solexa, Making Sense Out of Life, Oligator,
The Expanded Illumina Sequencing Portfolio New Sample Prep Solutions and Workflow Marcus Hausch, Ph.D. 2010 Illumina, Inc. All rights reserved. Illumina, illuminadx, Solexa, Making Sense Out of Life, Oligator,
M a x i m i z in g Value from NGS Analytics in t h e E n terprise
 Global Headquarters: 5 Speen Street Framingham, MA 01701 USA P.508.935.4445 F.508.988.7881 www.idc-hi.com M a x i m i z in g Value from NGS Analytics in t h e E n terprise C U S T O M I N D U S T R Y B
Global Headquarters: 5 Speen Street Framingham, MA 01701 USA P.508.935.4445 F.508.988.7881 www.idc-hi.com M a x i m i z in g Value from NGS Analytics in t h e E n terprise C U S T O M I N D U S T R Y B
Keywords: Clone, Probe, Entropy, Oligonucleotide, Algorithm
 Probe Selection Problem: Structure and Algorithms Daniel CAZALIS and Tom MILLEDGE and Giri NARASIMHAN School of Computer Science, Florida International University, Miami FL 33199, USA. ABSTRACT Given a
Probe Selection Problem: Structure and Algorithms Daniel CAZALIS and Tom MILLEDGE and Giri NARASIMHAN School of Computer Science, Florida International University, Miami FL 33199, USA. ABSTRACT Given a
Bioinformatics Tools. Stuart M. Brown, Ph.D Dept of Cell Biology NYU School of Medicine
 Bioinformatics Tools Stuart M. Brown, Ph.D Dept of Cell Biology NYU School of Medicine Bioinformatics Tools Stuart M. Brown, Ph.D Dept of Cell Biology NYU School of Medicine Overview This lecture will
Bioinformatics Tools Stuart M. Brown, Ph.D Dept of Cell Biology NYU School of Medicine Bioinformatics Tools Stuart M. Brown, Ph.D Dept of Cell Biology NYU School of Medicine Overview This lecture will
Introduction and Public Sequence Databases. BME 110/BIOL 181 CompBio Tools
 Introduction and Public Sequence Databases BME 110/BIOL 181 CompBio Tools Todd Lowe March 29, 2011 Course Syllabus: Admin http://www.soe.ucsc.edu/classes/bme110/spring11 Reading: Chapters 1, 2 (pp.29-56),
Introduction and Public Sequence Databases BME 110/BIOL 181 CompBio Tools Todd Lowe March 29, 2011 Course Syllabus: Admin http://www.soe.ucsc.edu/classes/bme110/spring11 Reading: Chapters 1, 2 (pp.29-56),
Gene Identification in silico
 Gene Identification in silico Nita Parekh, IIIT Hyderabad Presented at National Seminar on Bioinformatics and Functional Genomics, at Bioinformatics centre, Pondicherry University, Feb 15 17, 2006. Introduction
Gene Identification in silico Nita Parekh, IIIT Hyderabad Presented at National Seminar on Bioinformatics and Functional Genomics, at Bioinformatics centre, Pondicherry University, Feb 15 17, 2006. Introduction
College of information technology Department of software
 University of Babylon Undergraduate: third class College of information technology Department of software Subj.: Application of AI lecture notes/2011-2012 ***************************************************************************
University of Babylon Undergraduate: third class College of information technology Department of software Subj.: Application of AI lecture notes/2011-2012 ***************************************************************************
Bioinformatics and Life Sciences Standards and Programming for Heterogeneous Architectures
 Bioinformatics and Life Sciences Standards and Programming for Heterogeneous Architectures Eric Stahlberg Ph.D. (SAIC-Frederick contractor) stahlbergea@mail.nih.gov SIAM Conference on Parallel Processing
Bioinformatics and Life Sciences Standards and Programming for Heterogeneous Architectures Eric Stahlberg Ph.D. (SAIC-Frederick contractor) stahlbergea@mail.nih.gov SIAM Conference on Parallel Processing
