Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
|
|
|
- Osborne Rogers
- 6 years ago
- Views:
Transcription
1 Supplementary Figure 1 Origin use and efficiency are similar among WT, rrm3, pif1-m2, and pif1-m2; rrm3 strains. A. Analysis of fork progression around confirmed and likely origins (from cerevisiae.oridb.org). The percent of forks moving from left to right was calculated by the proportion of reads mapping to the Crick strand for each 100bp bin around the annotated origin of replication averaged across all sites (n=265). The origin signal is an ascending slope in this graph (compare to Figure 2A(i) and C). B. Scatter plot of origin usage at all confirmed and likely origins. The origin signal was calculated at each site by the difference between the percent of forks moving right to left upstream (+1-3kb) and downstream (-1-3kb) of the site, similar to the termination signal described in Figure 2, but with the opposite polarity.
2 Supplementary Figure 2 Further analysis of Okazaki-fragment termini in pif1 and rrm3 mutant strains. A. Distribution of Okazaki fragment 3 termini around consensus nucleosome dyads (Jiang & Pugh, Genome Biology 10, R109) as shown for 5 -ends in Fig 1B. B. rrm3, pif1-m2, and pif1-m2;-rrm3 S. cerevisiae strains generate fully ligatable, nucleosome-sized Okazaki fragments similar to those observed in wild-type cells. Genomic DNA was prepared and labeled as described in the Materials and Methods. C. Model for Pif1 and Rrm3 activity on the lagging strand. Histones are redeposited on the nascent lagging strand through the action of Caf1 (blue) as the lagging strand is being synthesized. Either Pif1 or Rrm3 (red) is required for normal DNA Pol (green) processivity, with Okazaki fragment ends enriched around nucleosome dyads as shown in Figure 1B. In the absence of both Pif1 and Rrm3, mature Okazaki fragment ends are enriched upstream of the nucleosome dyads.
3 Supplementary Figure 3
4 tdnas are the predominant sites of fork stalling genome wide. A. Fork progression of rightward (upper) and leftward (lower) moving forks at tdnas selected for further analysis (n=93; rightward n=51, leftward n=42; see Materials and Methods). The percent of forks moving left-to-right was calculated by the percent of reads mapping to the Crick strand. B. Fork progression at all tdnas with an origin in the analysis window in the WT (black) and pif1-m2;rrm3 (purple) mutant cells. The percent of forks moving left-to-right was calculated as described in the Materials and Methods with a correction for the slight difference in origin efficiency in the pif1-m2;rrm3 mutant (see Figure S1). The ascending slope in the upstream region is the origin signal (see Figure S1A). C. Fork progression at all tdnas without an origin within the analysis window as shown in part B. D. Fork progression at tdnas without an origin in the analysis window and without an origin-proximal tdna plotted as in part B, and corrected for the slight difference in origin efficiency. E-G. Random sites with an origin within the analysis window (E), without an origin in the analysis window (F), and without an tdna in the analysis window or proximal to the nearest upstream origin of replication (G) plotted as in part B with a correction for the slight difference in origin efficiency. H. Grand mean ± SD from three independent experiments of the change in replication direction, interpreted as indicative of replisome stalling, at random sites (n=381), random sites that exclude tdnas in the window of analysis (n=292), and tdnas (n=93) for the indicated strains. Significance was determined by Monte Carlo resampling; *** indicates a p<0.0001; n/s indicates p>0.05.
5 Supplementary Figure 4 Fork arrest is a feature common to tdnas. A. Heatmap showing the difference in percent forks moving left to right (calculated by the percent reads mapping to the Crick strand) between the WT and the pif1-m2 rrm3 double mutant. tdnas were sorted by their average fork direction (see Materials and Methods), and data were binned to 100bp. Negative values (i.e. pif1-m2 rrm3 forks more leftward moving than wild type) were visualized in yellow while positive values (i.e. pif1-m2;rrm3 forks more rightward moving than wild type) were visualized in blue. Heatmap was constructed with Gitools. B. Scatter plot of all 93 tdnas without a nearby origin or sequence gap. The change in replication direction was calculated for the same site in the WT and pif1-m2;rrm3 strains. Black dots are the WT plotted against the WT data and therefore give a line with the slope of 1. The pif1-m2;rrm3 change in replication direction divided by the change in the WT strain is plotted in purple. C. Reproducibility of tdna arrest signal from biological replicate 1 to biological replicate 2. The change in replication direction from dataset 1 and dataset 2 at the 93 tdnas included in our analysis. Each site was plotted four times, once for each strain, and the data were colored by strain. R 2 -values for the correlation between datasets for each strain were calculated using Graphpad Prism.
6 Supplementary Figure 5
7 Lack of robust stalling or arrest signal at RNA Pol II genes and G-quadruplex-forming sequences A. Grand mean ± SD for three independent experiments of the change in replication direction, interpreted as indicative of replisome arrest, at random sites (n=381), G-quadruplex forming sequences (Capra, J. A. et al. PLoS Comput Biol 6, e (2010)) (n=180), Highly transcribed RNA Pol II genes (Pelechano, V., Wei, W. & Steinmetz, L. M. Nature 497, (2013)) (n=207), ribosomal protein genes (n=56), and tdnas (n=93) as calculated in Figure 2E. In this analysis, random sites, G-quadruplex sequences, and RNA pol II genes with a tdna in the analysis window were not removed (compare to Figure 4A). Significance was determined by Monte Carlo resampling; *** indicates a p<0.0001; * indicates < p < n/s indicates p>0.05. B. Scatter plot of the change in replication direction at individual sites for all G-quadruplex forming sequences (n=180), highly transcribed RNA pol II genes (n=207), and tdnas (n=93) for the WT (gray) and pif1-m2;rrm3δ strain (purple). Sites are as defined in part A. Individual termination signals were calculated at each site as described in Figure 2 and the sites used were described in part A. The mean of each dataset is depicted as a black bar. C. Grand mean ± SD of the change in replication direction from three independent experiments at different subsets of G-quadruplex sites previously shown to stall the fork or bind to Pif1(Paeschke, K., Capra, J. A. & Zakian, V. A. Cell 145, (2011); Paeschke, K. et al. Nature 497, (2013)) (Pol2-pause n=53; Pol2-pause without tdna in window n=43; Pif1-binding peak n=19; Pif1- binding peak no tdna in window n=14). Significance was determined by Monte Carlo resampling; *** indicates a p<0.0001; * indicates < p < n/s indicates p>0.05. D. Addition of Hydroxyurea (HU, 25mM) to the growth media, which slows replication forks by depleting the dntp pool, did not increase specific stalling at G-quadruplex forming structures or tdna sites. Grand mean ± SD of three independent experiments for replisome arrest was plotted for G-quadruplex forming sites (n=180) and tdnas (n=93) as in part A. Significance was determined by Monte Carlo resampling; *** indicates a p< n/s indicates p>0.05. E. Selection of a subset of G-quadruplexes and random sites with a significant stalling in the pif1-m2;rrm3δ double mutant. For both the G-quadruplex forming sequences (n=46, p<0.0001) and the random sites (n=43, p<0.0001), we were able to select a subset of the sites that shows significant stalling. Significance was determined by Monte Carlo resampling; *** indicates a p<0.0001; * indicates < p < n/s indicates p>0.05.
8 Supplementary Figure 6 Direction of transcription at RNA Pol II genes and the strand of the G-quadruplex structure do not affect fork stalling or arrest at these sites. A. Grand mean ± SD from three independent experiments of the change in replication direction, interpreted as indicative of replisome arrest, at RNA Pol II genes and G-quadruplex sequences binned by direction of replication (see Materials and Methods). Significance was determined by Monte Carlo resampling; n/s indicates p>0.05.
SUPPLEMENTARY INFORMATION
 doi:1.138/nature1895 i Core histones CAF-1 v ii Reb1 CAF-1 Fen1 vi Mature Okazaki fragment iii PCNA Reb1 iv vii Mature Okazaki fragment Reb1 Mature Okazaki fragments Figure S1. Model for nucleosome-mediated
doi:1.138/nature1895 i Core histones CAF-1 v ii Reb1 CAF-1 Fen1 vi Mature Okazaki fragment iii PCNA Reb1 iv vii Mature Okazaki fragment Reb1 Mature Okazaki fragments Figure S1. Model for nucleosome-mediated
Nature Structural and Molecular Biology: doi: /nsmb Supplementary Figure 1. Validation of CDK9-inhibitor treatment.
 Supplementary Figure 1 Validation of CDK9-inhibitor treatment. (a) Schematic of GAPDH with the middle of the amplicons indicated in base pairs. The transcription start site (TSS) and the terminal polyadenylation
Supplementary Figure 1 Validation of CDK9-inhibitor treatment. (a) Schematic of GAPDH with the middle of the amplicons indicated in base pairs. The transcription start site (TSS) and the terminal polyadenylation
Prokaryotic Physiology. March 3, 2017
 1. (10 pts) Explain the replication of both strands of DNA in prokaryotes. At a minimum explain the direction of synthesis, synthesis of the leading and lagging strand, separation of the strands and the
1. (10 pts) Explain the replication of both strands of DNA in prokaryotes. At a minimum explain the direction of synthesis, synthesis of the leading and lagging strand, separation of the strands and the
Simultaneous profiling of transcriptome and DNA methylome from a single cell
 Additional file 1: Supplementary materials Simultaneous profiling of transcriptome and DNA methylome from a single cell Youjin Hu 1, 2, Kevin Huang 1, 3, Qin An 1, Guizhen Du 1, Ganlu Hu 2, Jinfeng Xue
Additional file 1: Supplementary materials Simultaneous profiling of transcriptome and DNA methylome from a single cell Youjin Hu 1, 2, Kevin Huang 1, 3, Qin An 1, Guizhen Du 1, Ganlu Hu 2, Jinfeng Xue
SUPPLEMENTARY INFORMATION
 AS-NMD modulates FLM-dependent thermosensory flowering response in Arabidopsis NATURE PLANTS www.nature.com/natureplants 1 Supplementary Figure 1. Genomic sequence of FLM along with the splice sites. Sequencing
AS-NMD modulates FLM-dependent thermosensory flowering response in Arabidopsis NATURE PLANTS www.nature.com/natureplants 1 Supplementary Figure 1. Genomic sequence of FLM along with the splice sites. Sequencing
Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C
 CORRECTION NOTICE Nat. Genet. 47, 598 606 (2015) Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C Borbala Mifsud, Filipe Tavares-Cadete, Alice N Young, Robert Sugar,
CORRECTION NOTICE Nat. Genet. 47, 598 606 (2015) Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C Borbala Mifsud, Filipe Tavares-Cadete, Alice N Young, Robert Sugar,
Figure S1: NUN preparation yields nascent, unadenylated RNA with a different profile from Total RNA.
 Summary of Supplemental Information Figure S1: NUN preparation yields nascent, unadenylated RNA with a different profile from Total RNA. Figure S2: rrna removal procedure is effective for clearing out
Summary of Supplemental Information Figure S1: NUN preparation yields nascent, unadenylated RNA with a different profile from Total RNA. Figure S2: rrna removal procedure is effective for clearing out
ChIP-Seq Data Analysis. J Fass UCD Genome Center Bioinformatics Core Wednesday December 17, 2014
 ChIP-Seq Data Analysis J Fass UCD Genome Center Bioinformatics Core Wednesday December 17, 2014 What s the Question? Where do Transcription Factors (TFs) bind genomic DNA 1? (Where do other things bind
ChIP-Seq Data Analysis J Fass UCD Genome Center Bioinformatics Core Wednesday December 17, 2014 What s the Question? Where do Transcription Factors (TFs) bind genomic DNA 1? (Where do other things bind
Storage and Expression of Genetic Information
 Storage and Expression of Genetic Information 29. DNA structure, Replication and Repair ->Ch 25. DNA metabolism 30. RNA Structure, Synthesis and Processing ->Ch 26. RNA metabolism 31. Protein Synthesis
Storage and Expression of Genetic Information 29. DNA structure, Replication and Repair ->Ch 25. DNA metabolism 30. RNA Structure, Synthesis and Processing ->Ch 26. RNA metabolism 31. Protein Synthesis
IN E. COLI WHAT IS THE FUNCTION OF DNA POLYMERASE III
 10 January, 2018 IN E. COLI WHAT IS THE FUNCTION OF DNA POLYMERASE III Document Filetype: PDF 312 KB 0 IN E. COLI WHAT IS THE FUNCTION OF DNA POLYMERASE III The actual replication enzyme in E. Both will
10 January, 2018 IN E. COLI WHAT IS THE FUNCTION OF DNA POLYMERASE III Document Filetype: PDF 312 KB 0 IN E. COLI WHAT IS THE FUNCTION OF DNA POLYMERASE III The actual replication enzyme in E. Both will
ChIP-Seq Data Analysis. J Fass UCD Genome Center Bioinformatics Core Wednesday 15 June 2015
 ChIP-Seq Data Analysis J Fass UCD Genome Center Bioinformatics Core Wednesday 15 June 2015 What s the Question? Where do Transcription Factors (TFs) bind genomic DNA 1? (Where do other things bind DNA
ChIP-Seq Data Analysis J Fass UCD Genome Center Bioinformatics Core Wednesday 15 June 2015 What s the Question? Where do Transcription Factors (TFs) bind genomic DNA 1? (Where do other things bind DNA
ChIP-Seq Tools. J Fass UCD Genome Center Bioinformatics Core Wednesday September 16, 2015
 ChIP-Seq Tools J Fass UCD Genome Center Bioinformatics Core Wednesday September 16, 2015 What s the Question? Where do Transcription Factors (TFs) bind genomic DNA 1? (Where do other things bind DNA or
ChIP-Seq Tools J Fass UCD Genome Center Bioinformatics Core Wednesday September 16, 2015 What s the Question? Where do Transcription Factors (TFs) bind genomic DNA 1? (Where do other things bind DNA or
Why does this matter?
 Background Why does this matter? Better understanding of how the nucleosome affects transcription Important for understanding the nucleosome s role in gene expression Treats each component and region of
Background Why does this matter? Better understanding of how the nucleosome affects transcription Important for understanding the nucleosome s role in gene expression Treats each component and region of
% Viability. isw2 ino isw2 ino isw2 ino isw2 ino mM HU 4-NQO CPT
 a Drug concentration b 1.3% MMS nhp1 nhp1 8 nhp1 mag1.5% MMS.3% MMS nhp1 nhp1 ino8 9 ino8 9 % Viability 4.5% MMS ino8 9 ino8 9 2.5.1.15 % MMS c d nhp1 nhp1 nhp1 nhp1 nhp1 nhp1 Control (YPD) γ IR (1 gy)
a Drug concentration b 1.3% MMS nhp1 nhp1 8 nhp1 mag1.5% MMS.3% MMS nhp1 nhp1 ino8 9 ino8 9 % Viability 4.5% MMS ino8 9 ino8 9 2.5.1.15 % MMS c d nhp1 nhp1 nhp1 nhp1 nhp1 nhp1 Control (YPD) γ IR (1 gy)
Chapter 11 DNA Replication and Recombination
 Chapter 11 DNA Replication and Recombination Copyright Copyright 2009 Pearson 2009 Pearson Education, Education, Inc. Inc. 11.1 DNA is reproduced by Semiconservative Replication The complementarity of
Chapter 11 DNA Replication and Recombination Copyright Copyright 2009 Pearson 2009 Pearson Education, Education, Inc. Inc. 11.1 DNA is reproduced by Semiconservative Replication The complementarity of
DNA Replication in Prokaryotes and Eukaryotes
 DNA Replication in Prokaryotes and Eukaryotes 1. Overall mechanism 2. Roles of Polymerases & other proteins 3. More mechanism: Initiation and Termination 4. Mitochondrial DNA replication DNA replication
DNA Replication in Prokaryotes and Eukaryotes 1. Overall mechanism 2. Roles of Polymerases & other proteins 3. More mechanism: Initiation and Termination 4. Mitochondrial DNA replication DNA replication
AD BD TOC1. Supplementary Figure 1: Yeast two-hybrid assays showing the interaction between
 AD X BD TOC1 AD BD X PIFΔAD PIF TOC1 TOC1 PIFΔAD PIF N TOC1 TOC1 C1 PIFΔAD PIF C1 TOC1 TOC1 C PIFΔAD PIF C TOC1 Supplementary Figure 1: Yeast two-hybrid assays showing the interaction between PIF and TOC1
AD X BD TOC1 AD BD X PIFΔAD PIF TOC1 TOC1 PIFΔAD PIF N TOC1 TOC1 C1 PIFΔAD PIF C1 TOC1 TOC1 C PIFΔAD PIF C TOC1 Supplementary Figure 1: Yeast two-hybrid assays showing the interaction between PIF and TOC1
SUPPLEMENTARY INFORMATION
 SUPPLEMENRY INFORMION doi:.38/nature In vivo nucleosome mapping D4+ Lymphocytes radient-based and I-bead cell sorting D8+ Lymphocytes ranulocytes Lyse the cells Isolate and sequence mononucleosome cores
SUPPLEMENRY INFORMION doi:.38/nature In vivo nucleosome mapping D4+ Lymphocytes radient-based and I-bead cell sorting D8+ Lymphocytes ranulocytes Lyse the cells Isolate and sequence mononucleosome cores
DNA Metabolism. I. DNA Replication. A. Template concept: 1. How can you make a copy of a molecule? 2. Complementary Hydrogen bonding
 DNA Metabolism I. DNA Replication A. Template concept: 1. How can you make a copy of a molecule? 2. Complementary Hydrogen bonding B. DNA replication follows a set of fundamental rules 1. Semiconservative
DNA Metabolism I. DNA Replication A. Template concept: 1. How can you make a copy of a molecule? 2. Complementary Hydrogen bonding B. DNA replication follows a set of fundamental rules 1. Semiconservative
Epigenetic control of tomato fruit development. Key Laboratories of Molecular Epigenetics of MOE, Northeast Normal University, Changchun, China
 Epigenetic control of tomato fruit development Silin Zhong 1,2*,, Zhangjun Fei 1,3,*,, Yun-Ru Chen 1, Yi Zheng 1, Mingyun Huang 1, Julia Vrebalov 1, Ryan McQuinn 1, Nigel Gapper 1, Bao Liu 2, Jenny Xiang
Epigenetic control of tomato fruit development Silin Zhong 1,2*,, Zhangjun Fei 1,3,*,, Yun-Ru Chen 1, Yi Zheng 1, Mingyun Huang 1, Julia Vrebalov 1, Ryan McQuinn 1, Nigel Gapper 1, Bao Liu 2, Jenny Xiang
BS 50 Genetics and Genomics Week of Oct 24
 BS 50 Genetics and Genomics Week of Oct 24 Additional Practice Problems for Section Question 1: The following table contains a list of statements that apply to replication, transcription, both, or neither.
BS 50 Genetics and Genomics Week of Oct 24 Additional Practice Problems for Section Question 1: The following table contains a list of statements that apply to replication, transcription, both, or neither.
P&TIT: A computer tool for predicting prototypical transcription promoter and terminator elements by conserved motifs
 P&TIT: A computer tool for predicting prototypical transcription promoter and terminator elements by conserved motifs Marco Di Salvo a, Eva Pinatel b, Gianluca De Bellis b, Adelfia Talà a, Clelia Peano
P&TIT: A computer tool for predicting prototypical transcription promoter and terminator elements by conserved motifs Marco Di Salvo a, Eva Pinatel b, Gianluca De Bellis b, Adelfia Talà a, Clelia Peano
d. reading a DNA strand and making a complementary messenger RNA
 Biol/ MBios 301 (General Genetics) Spring 2003 Second Midterm Examination A (100 points possible) Key April 1, 2003 10 Multiple Choice Questions-4 pts. each (Choose the best answer) 1. Transcription involves:
Biol/ MBios 301 (General Genetics) Spring 2003 Second Midterm Examination A (100 points possible) Key April 1, 2003 10 Multiple Choice Questions-4 pts. each (Choose the best answer) 1. Transcription involves:
Galaxy Platform For NGS Data Analyses
 Galaxy Platform For NGS Data Analyses Weihong Yan wyan@chem.ucla.edu Collaboratory Web Site http://qcb.ucla.edu/collaboratory http://collaboratory.lifesci.ucla.edu Workshop Outline ü Day 1 UCLA galaxy
Galaxy Platform For NGS Data Analyses Weihong Yan wyan@chem.ucla.edu Collaboratory Web Site http://qcb.ucla.edu/collaboratory http://collaboratory.lifesci.ucla.edu Workshop Outline ü Day 1 UCLA galaxy
SUPPLEMENTARY INFORMATION
 DOI: 1.1/ncb2918 Supplementary Figure 1 (a) Biotin-dUTP labelling does not affect S phase progression. Cells synchronized in mid-s phase were labelled with biotindutp during a 5 min hypotonic shift (red)
DOI: 1.1/ncb2918 Supplementary Figure 1 (a) Biotin-dUTP labelling does not affect S phase progression. Cells synchronized in mid-s phase were labelled with biotindutp during a 5 min hypotonic shift (red)
(A) Extrachromosomal DNA (B) RNA found in bacterial cells (C) Is part of the bacterial chromosome (D) Is part of the eukaryote chromosome
 Microbiology - Problem Drill 07: Microbial Genetics and Biotechnology No. 1 of 10 1. A plasmid is? (A) Extrachromosomal DNA (B) RNA found in bacterial cells (C) Is part of the bacterial chromosome (D)
Microbiology - Problem Drill 07: Microbial Genetics and Biotechnology No. 1 of 10 1. A plasmid is? (A) Extrachromosomal DNA (B) RNA found in bacterial cells (C) Is part of the bacterial chromosome (D)
Differential Gene Expression
 Biology 4361 Developmental Biology Differential Gene Expression September 28, 2006 Chromatin Structure ~140 bp ~60 bp Transcriptional Regulation: 1. Packing prevents access CH 3 2. Acetylation ( C O )
Biology 4361 Developmental Biology Differential Gene Expression September 28, 2006 Chromatin Structure ~140 bp ~60 bp Transcriptional Regulation: 1. Packing prevents access CH 3 2. Acetylation ( C O )
Genetic Information: DNA replication
 Genetic Information: DNA replication Umut Fahrioglu, PhD MSc DNA Replication Replication of DNA is vital to the transmission of genomes and the genes they contain from one cell generation to the other.
Genetic Information: DNA replication Umut Fahrioglu, PhD MSc DNA Replication Replication of DNA is vital to the transmission of genomes and the genes they contain from one cell generation to the other.
DNA vs. RNA DNA: deoxyribonucleic acid (double stranded) RNA: ribonucleic acid (single stranded) Both found in most bacterial and eukaryotic cells RNA
 DNA Replication DNA vs. RNA DNA: deoxyribonucleic acid (double stranded) RNA: ribonucleic acid (single stranded) Both found in most bacterial and eukaryotic cells RNA molecule can assume different structures
DNA Replication DNA vs. RNA DNA: deoxyribonucleic acid (double stranded) RNA: ribonucleic acid (single stranded) Both found in most bacterial and eukaryotic cells RNA molecule can assume different structures
DNA Transcription. Dr Aliwaini
 DNA Transcription 1 DNA Transcription-Introduction The synthesis of an RNA molecule from DNA is called Transcription. All eukaryotic cells have five major classes of RNA: ribosomal RNA (rrna), messenger
DNA Transcription 1 DNA Transcription-Introduction The synthesis of an RNA molecule from DNA is called Transcription. All eukaryotic cells have five major classes of RNA: ribosomal RNA (rrna), messenger
Supplementary Figure 1: sgrna library generation and the length of sgrnas for the functional screen. (a) A diagram of the retroviral vector for sgrna
 Supplementary Figure 1: sgrna library generation and the length of sgrnas for the functional screen. (a) A diagram of the retroviral vector for sgrna expression. It contains a U6-promoter-driven sgrna
Supplementary Figure 1: sgrna library generation and the length of sgrnas for the functional screen. (a) A diagram of the retroviral vector for sgrna expression. It contains a U6-promoter-driven sgrna
Molecular Biology (2)
 Molecular Biology (2) DNA replication Mamoun Ahram, PhD Second semester, 2018-2019 Resources This lecture Cooper, pp. 191-207 2 Some basic information The entire DNA content of the cell is known as genome.
Molecular Biology (2) DNA replication Mamoun Ahram, PhD Second semester, 2018-2019 Resources This lecture Cooper, pp. 191-207 2 Some basic information The entire DNA content of the cell is known as genome.
Answers to Module 1. An obligate aerobe is an organism that has an absolute requirement of oxygen for growth.
 Answers to Module 1 Short Answers 1) What is an obligate aerobe? An obligate aerobe is an organism that has an absolute requirement of oxygen for growth. What about facultative anaerobe? 2) Distinguish
Answers to Module 1 Short Answers 1) What is an obligate aerobe? An obligate aerobe is an organism that has an absolute requirement of oxygen for growth. What about facultative anaerobe? 2) Distinguish
DNA Replication II Biochemistry 302. January 25, 2006
 DNA Replication II Biochemistry 302 January 25, 2006 Following in Dad s footsteps Original A. Kornberg E. coli DNA Pol I is a lousy replicative enzyme. 400 molecules/cell but ~2 replication forks/cell
DNA Replication II Biochemistry 302 January 25, 2006 Following in Dad s footsteps Original A. Kornberg E. coli DNA Pol I is a lousy replicative enzyme. 400 molecules/cell but ~2 replication forks/cell
Supplemental Figure 1.
 Supplemental Data. Charron et al. Dynamic landscapes of four histone modifications during de-etiolation in Arabidopsis. Plant Cell (2009). 10.1105/tpc.109.066845 Supplemental Figure 1. Immunodetection
Supplemental Data. Charron et al. Dynamic landscapes of four histone modifications during de-etiolation in Arabidopsis. Plant Cell (2009). 10.1105/tpc.109.066845 Supplemental Figure 1. Immunodetection
DNA Replication II Biochemistry 302. Bob Kelm January 28, 2004
 DNA Replication II Biochemistry 302 Bob Kelm January 28, 2004 Conceptual model for proofreading based on kinetic considerations Fig. 24.44 stalling transient melting exonuclease site occupancy Following
DNA Replication II Biochemistry 302 Bob Kelm January 28, 2004 Conceptual model for proofreading based on kinetic considerations Fig. 24.44 stalling transient melting exonuclease site occupancy Following
S156AT168AY175A (AAA) were purified as GST-fusion proteins and incubated with GSTfused
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 Supplemental Materials Supplemental Figure S1 (a) Phenotype of the wild type and grik1-2 grik2-1 plants after 8 days in darkness.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 Supplemental Materials Supplemental Figure S1 (a) Phenotype of the wild type and grik1-2 grik2-1 plants after 8 days in darkness.
BIOL3090 Final Exam Summary, Pete s Lectures. Lecture 1: DNA Replication
 BIOL3090 Final Exam Summary, Pete s Lectures Lecture 1: DNA Replication - The five rules of DNA replication: 1. DNA is made of two complementary strands (always) 2. DNA replication is semiconservative
BIOL3090 Final Exam Summary, Pete s Lectures Lecture 1: DNA Replication - The five rules of DNA replication: 1. DNA is made of two complementary strands (always) 2. DNA replication is semiconservative
DNA replication: Enzymes link the aligned nucleotides by phosphodiester bonds to form a continuous strand.
 DNA replication: Copying genetic information for transmission to the next generation Occurs in S phase of cell cycle Process of DNA duplicating itself Begins with the unwinding of the double helix to expose
DNA replication: Copying genetic information for transmission to the next generation Occurs in S phase of cell cycle Process of DNA duplicating itself Begins with the unwinding of the double helix to expose
Expression of the genome. Books: 1. Molecular biology of the gene: Watson et al 2. Genetics: Peter J. Russell
 Expression of the genome Books: 1. Molecular biology of the gene: Watson et al 2. Genetics: Peter J. Russell 1 Transcription 1. Francis Crick (1956) named the flow of information from DNA RNA protein the
Expression of the genome Books: 1. Molecular biology of the gene: Watson et al 2. Genetics: Peter J. Russell 1 Transcription 1. Francis Crick (1956) named the flow of information from DNA RNA protein the
RNA POLYMERASE FUNCTIONS E-BOOK
 08 March, 2018 RNA POLYMERASE 1 2 3 FUNCTIONS E-BOOK Document Filetype: PDF 431.06 KB 0 RNA POLYMERASE 1 2 3 FUNCTIONS E-BOOK It catalyzes the transcription of DNA to synthesize precursors of mrna and
08 March, 2018 RNA POLYMERASE 1 2 3 FUNCTIONS E-BOOK Document Filetype: PDF 431.06 KB 0 RNA POLYMERASE 1 2 3 FUNCTIONS E-BOOK It catalyzes the transcription of DNA to synthesize precursors of mrna and
Nature Structural and Molecular Biology: doi: /nsmb.2847
 Supplementary Figure 1 Specificity coefficients and generality of the TRM code. (A) The ranking of TRM specificity is shown based on the percent enrichment of the dominant base at position +2 multiplied
Supplementary Figure 1 Specificity coefficients and generality of the TRM code. (A) The ranking of TRM specificity is shown based on the percent enrichment of the dominant base at position +2 multiplied
Zool 3200: Cell Biology Exam 2 2/20/15
 Name: TRASK Zool 3200: Cell Biology Exam 2 2/20/15 Answer each of the following short and longer answer questions in the space provided; circle the BEST answer or answers for each multiple choice question
Name: TRASK Zool 3200: Cell Biology Exam 2 2/20/15 Answer each of the following short and longer answer questions in the space provided; circle the BEST answer or answers for each multiple choice question
SUPPLEMENTARY INFORMATION
 Supplementary Discussion Interestingly, a recent study demonstrated that knockdown of Tet1 alone would lead to dysregulation of differentiation genes, but only minor defects in ES maintenance and no change
Supplementary Discussion Interestingly, a recent study demonstrated that knockdown of Tet1 alone would lead to dysregulation of differentiation genes, but only minor defects in ES maintenance and no change
TECH NOTE Pushing the Limit: A Complete Solution for Generating Stranded RNA Seq Libraries from Picogram Inputs of Total Mammalian RNA
 TECH NOTE Pushing the Limit: A Complete Solution for Generating Stranded RNA Seq Libraries from Picogram Inputs of Total Mammalian RNA Stranded, Illumina ready library construction in
TECH NOTE Pushing the Limit: A Complete Solution for Generating Stranded RNA Seq Libraries from Picogram Inputs of Total Mammalian RNA Stranded, Illumina ready library construction in
Name Genotype Reference
 Supplemental Data Supplemental Table 1 S. cerevisiae strains. Name Genotype Reference RMY200 UKY403 W303-1a MATa ade2-101 (och) his3 200 lys2-801 (amp) trp1 901 ura3-52 hht1,hhf1::leu2 hht2,hhf2::his3
Supplemental Data Supplemental Table 1 S. cerevisiae strains. Name Genotype Reference RMY200 UKY403 W303-1a MATa ade2-101 (och) his3 200 lys2-801 (amp) trp1 901 ura3-52 hht1,hhf1::leu2 hht2,hhf2::his3
Nature Methods: doi: /nmeth Supplementary Figure 1. Pilot CrY2H-seq experiments to confirm strain and plasmid functionality.
 Supplementary Figure 1 Pilot CrY2H-seq experiments to confirm strain and plasmid functionality. (a) RT-PCR on HIS3 positive diploid cell lysate containing known interaction partners AT3G62420 (bzip53)
Supplementary Figure 1 Pilot CrY2H-seq experiments to confirm strain and plasmid functionality. (a) RT-PCR on HIS3 positive diploid cell lysate containing known interaction partners AT3G62420 (bzip53)
of NOBOX. Supplemental Figure S1F shows the staining profile of LHX8 antibody (Abcam, ab41519), which is a
 SUPPLEMENTAL MATERIALS AND METHODS LHX8 Staining During the course of this work, it came to our attention that the NOBOX antibody used in this study had been discontinued. There are a number of other nuclear
SUPPLEMENTAL MATERIALS AND METHODS LHX8 Staining During the course of this work, it came to our attention that the NOBOX antibody used in this study had been discontinued. There are a number of other nuclear
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Endogenous gene tagging to study subcellular localization and chromatin binding. a, b, Schematic of experimental set-up to endogenously tag RNAi factors using the CRISPR Cas9 technology,
Supplementary Figure 1 Endogenous gene tagging to study subcellular localization and chromatin binding. a, b, Schematic of experimental set-up to endogenously tag RNAi factors using the CRISPR Cas9 technology,
Genes found in the genome include protein-coding genes and non-coding RNA genes. Which nucleotide is not normally found in non-coding RNA genes?
 Midterm Q Genes found in the genome include protein-coding genes and non-coding RNA genes Which nucleotide is not normally found in non-coding RNA genes? G T 3 A 4 C 5 U 00% Midterm Q Which of the following
Midterm Q Genes found in the genome include protein-coding genes and non-coding RNA genes Which nucleotide is not normally found in non-coding RNA genes? G T 3 A 4 C 5 U 00% Midterm Q Which of the following
Deep sequencing reveals global patterns of mrna recruitment
 Supplementary information for: Deep sequencing reveals global patterns of mrna recruitment during translation initiation Rong Gao 1#*, Kai Yu 1#, Ju-Kui Nie 1,Teng-Fei Lian 1, Jian-Shi Jin 1, Anders Liljas
Supplementary information for: Deep sequencing reveals global patterns of mrna recruitment during translation initiation Rong Gao 1#*, Kai Yu 1#, Ju-Kui Nie 1,Teng-Fei Lian 1, Jian-Shi Jin 1, Anders Liljas
Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) (b)
 Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) A schematic overview of the production and amplification of a single pirna from a transposon transcript. The
Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) A schematic overview of the production and amplification of a single pirna from a transposon transcript. The
Requirements for the Genetic Material
 Requirements for the Genetic Material 1. Replication Reproduced and transmitted faithfully from cell to cell-generation to generation. 2. Information Storage Biologically useful information in a stable
Requirements for the Genetic Material 1. Replication Reproduced and transmitted faithfully from cell to cell-generation to generation. 2. Information Storage Biologically useful information in a stable
pej605 pej414 containing 81 bp downstream and 579 bp This study
 SUPPLEMENTARY DATA Table S. Details of plasmids used in this study. Plasmid Description Reference or source pfm8 Protein expression vector based on pet5b containing (0) His-tagged lexa. pcr 4-TOPO Cloning
SUPPLEMENTARY DATA Table S. Details of plasmids used in this study. Plasmid Description Reference or source pfm8 Protein expression vector based on pet5b containing (0) His-tagged lexa. pcr 4-TOPO Cloning
Gene Expression Transcription/Translation Protein Synthesis
 Gene Expression Transcription/Translation Protein Synthesis 1. Describe how genetic information is transcribed into sequences of bases in RNA molecules and is finally translated into sequences of amino
Gene Expression Transcription/Translation Protein Synthesis 1. Describe how genetic information is transcribed into sequences of bases in RNA molecules and is finally translated into sequences of amino
Applied Bioinformatics - Lecture 16: Transcriptomics
 Applied Bioinformatics - Lecture 16: Transcriptomics David Hendrix Oregon State University Feb 15th 2016 Transcriptomics High-throughput Sequencing (deep sequencing) High-throughput sequencing (also
Applied Bioinformatics - Lecture 16: Transcriptomics David Hendrix Oregon State University Feb 15th 2016 Transcriptomics High-throughput Sequencing (deep sequencing) High-throughput sequencing (also
You Should Be Able To
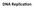 DNA Replica,on You Should Be Able To 1. Describe the func9on of: DNA POL1, DNA POL3, Sliding Clamp, SSBPs, Ligase, Topoisomerase, Helicase, Primase 2. Describe DNA synthesis on the leading and lagging
DNA Replica,on You Should Be Able To 1. Describe the func9on of: DNA POL1, DNA POL3, Sliding Clamp, SSBPs, Ligase, Topoisomerase, Helicase, Primase 2. Describe DNA synthesis on the leading and lagging
Written by: Prof. Brian White
 Molecular Biology II: DNA Transcription Written by: Prof. Brian White Learning Goals: To work with a physical model of DNA and RNA in order to help you to understand: o rules for both DNA & RNA structure
Molecular Biology II: DNA Transcription Written by: Prof. Brian White Learning Goals: To work with a physical model of DNA and RNA in order to help you to understand: o rules for both DNA & RNA structure
Chapter 24: Promoters and Enhancers
 Chapter 24: Promoters and Enhancers A typical gene transcribed by RNA polymerase II has a promoter that usually extends upstream from the site where transcription is initiated the (#1) of transcription
Chapter 24: Promoters and Enhancers A typical gene transcribed by RNA polymerase II has a promoter that usually extends upstream from the site where transcription is initiated the (#1) of transcription
Wednesday, November 22, 17. Exons and Introns
 Exons and Introns Introns and Exons Exons: coded regions of DNA that get transcribed and translated into proteins make up 5% of the genome Introns and Exons Introns: non-coded regions of DNA Must be removed
Exons and Introns Introns and Exons Exons: coded regions of DNA that get transcribed and translated into proteins make up 5% of the genome Introns and Exons Introns: non-coded regions of DNA Must be removed
Delve AP Biology Lecture 7: 10/30/11 Melissa Ko and Anne Huang
 Today s Agenda: I. DNA Structure II. DNA Replication III. DNA Proofreading and Repair IV. The Central Dogma V. Transcription VI. Post-transcriptional Modifications Delve AP Biology Lecture 7: 10/30/11
Today s Agenda: I. DNA Structure II. DNA Replication III. DNA Proofreading and Repair IV. The Central Dogma V. Transcription VI. Post-transcriptional Modifications Delve AP Biology Lecture 7: 10/30/11
Supplementary Tables. Primers and probes. Target Primer / probe sequences Chemistry Human HBB F: AACTGTGTTCACTAGCAACCTCAAA
 Supplementary Tables Primers and probes Target Primer / probe sequences Chemistry H F: AACTGTGTTCACTAGCAACCTCAAA promoter R: ACAGGGCAGTAACGGCAGACT H Downstream Actb alpha (162) alpha (162.) (Pro) (16)
Supplementary Tables Primers and probes Target Primer / probe sequences Chemistry H F: AACTGTGTTCACTAGCAACCTCAAA promoter R: ACAGGGCAGTAACGGCAGACT H Downstream Actb alpha (162) alpha (162.) (Pro) (16)
SSA Signal Search Analysis II
 SSA Signal Search Analysis II SSA other applications - translation In contrast to translation initiation in bacteria, translation initiation in eukaryotes is not guided by a Shine-Dalgarno like motif.
SSA Signal Search Analysis II SSA other applications - translation In contrast to translation initiation in bacteria, translation initiation in eukaryotes is not guided by a Shine-Dalgarno like motif.
Nature Biotechnology: doi: /nbt Supplementary Figure 1. sndrop-seq overview.
 Supplementary Figure 1 sndrop-seq overview. A. sndrop-seq method showing modifications needed to process nuclei, including bovine serum albumin (BSA) coating and droplet heating to ensure complete nuclear
Supplementary Figure 1 sndrop-seq overview. A. sndrop-seq method showing modifications needed to process nuclei, including bovine serum albumin (BSA) coating and droplet heating to ensure complete nuclear
The Size and Packaging of Genomes
 DNA Replication The Size and Packaging of Genomes Vary greatly in size Ø Smallest viruses- 4 or 5 genes Ø Escherichia coli- 4,288 genes Ø Human cell- 20,000 to 25,000 genes E. coli 4 million base pairs
DNA Replication The Size and Packaging of Genomes Vary greatly in size Ø Smallest viruses- 4 or 5 genes Ø Escherichia coli- 4,288 genes Ø Human cell- 20,000 to 25,000 genes E. coli 4 million base pairs
Molecular Biology: General Theory
 Molecular Biology: General Theory Author: Dr Darshana Morar Licensed under a Creative Commons Attribution license. DNA REPLICATION DNA replication is the process of duplicating the DNA sequence in the
Molecular Biology: General Theory Author: Dr Darshana Morar Licensed under a Creative Commons Attribution license. DNA REPLICATION DNA replication is the process of duplicating the DNA sequence in the
Molecular Biology: General Theory
 Molecular Biology: General Theory Author: Dr Darshana Morar Licensed under a Creative Commons Attribution license. DNA REPLICATION DNA replication is the process of duplicating the DNA sequence in the
Molecular Biology: General Theory Author: Dr Darshana Morar Licensed under a Creative Commons Attribution license. DNA REPLICATION DNA replication is the process of duplicating the DNA sequence in the
Zhang et al., RepID facilitates replication Initiation. Supplemental Information:
 Supplemental Information: a b 1 Supplementary Figure 1 (a) DNA sequence of all the oligonucleotides used in this study. Only one strand is shown. The unshaded nucleotide sequences show changes from the
Supplemental Information: a b 1 Supplementary Figure 1 (a) DNA sequence of all the oligonucleotides used in this study. Only one strand is shown. The unshaded nucleotide sequences show changes from the
Diversity of Eukaryotic DNA Replication Origins Revealed by Genome-Wide Analysis of Chromatin Structure
 Diversity of Eukaryotic DNA Replication Origins Revealed by Genome-Wide Analysis of Chromatin Structure Nicolas M. Berbenetz 1,2, Corey Nislow 1,2 *, Grant W. Brown 2,3 * 1 Department of Molecular Genetics,
Diversity of Eukaryotic DNA Replication Origins Revealed by Genome-Wide Analysis of Chromatin Structure Nicolas M. Berbenetz 1,2, Corey Nislow 1,2 *, Grant W. Brown 2,3 * 1 Department of Molecular Genetics,
7.014 Quiz II Handout
 7.014 Quiz II Handout Quiz II: Wednesday, March 17 12:05-12:55 54-100 **This will be a closed book exam** Quiz Review Session: Friday, March 12 7:00-9:00 pm room 54-100 Open Tutoring Session: Tuesday,
7.014 Quiz II Handout Quiz II: Wednesday, March 17 12:05-12:55 54-100 **This will be a closed book exam** Quiz Review Session: Friday, March 12 7:00-9:00 pm room 54-100 Open Tutoring Session: Tuesday,
Fidelity of DNA polymerase
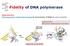 Fidelity of DNA polymerase Shape selectivity: DNA polymerase's conformational change for determination of fidelity for each nucleotide Induced fit: Structure determines function Matched nucleotide Fidelity
Fidelity of DNA polymerase Shape selectivity: DNA polymerase's conformational change for determination of fidelity for each nucleotide Induced fit: Structure determines function Matched nucleotide Fidelity
Unit IX Problem 3 Genetics: Basic Concepts in Molecular Biology
 Unit IX Problem 3 Genetics: Basic Concepts in Molecular Biology - The central dogma (principle) of molecular biology: Information from DNA are transcribed to mrna which will be further translated to synthesize
Unit IX Problem 3 Genetics: Basic Concepts in Molecular Biology - The central dogma (principle) of molecular biology: Information from DNA are transcribed to mrna which will be further translated to synthesize
DNA Structure. DNA: The Genetic Material. Chapter 14
 DNA: The Genetic Material Chapter 14 DNA Structure DNA is a nucleic acid. The building blocks of DNA are nucleotides, each composed of: a 5-carbon sugar called deoxyribose a phosphate group (PO 4 ) a nitrogenous
DNA: The Genetic Material Chapter 14 DNA Structure DNA is a nucleic acid. The building blocks of DNA are nucleotides, each composed of: a 5-carbon sugar called deoxyribose a phosphate group (PO 4 ) a nitrogenous
Supplementary Information. Synergistic action of RNA polymerases in overcoming the nucleosomal barrier
 Supplementary Information Synergistic action of RNA polymerases in overcoming the nucleosomal barrier Jing Jin, Lu Bai, Daniel S. Johnson, Robert M. Fulbright, Maria L. Kireeva, Mikhail Kashlev, Michelle
Supplementary Information Synergistic action of RNA polymerases in overcoming the nucleosomal barrier Jing Jin, Lu Bai, Daniel S. Johnson, Robert M. Fulbright, Maria L. Kireeva, Mikhail Kashlev, Michelle
The replication forks Summarising what we know:
 When does replication occur? MBLG1001 lecture 10 Replication the once in a lifetime event! Full blown replication only occurs once, just before cell division BUT the DNA template is constantly being repaired.
When does replication occur? MBLG1001 lecture 10 Replication the once in a lifetime event! Full blown replication only occurs once, just before cell division BUT the DNA template is constantly being repaired.
File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description:
 File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description: Supplementary Figure 1. dcas9-mq1 fusion protein induces de novo
File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description: Supplementary Figure 1. dcas9-mq1 fusion protein induces de novo
Supplementary Information: Conserved nucleosome positioning defines replication origins
 Supplementary Information: Conserved nucleosome positioning defines replication origins Matthew L. Eaton 1, Kyriaki Galani 2, Sukhyun Kang 2, Stephen P. Bell 2, and David M. MacAlpine 1 1 Department of
Supplementary Information: Conserved nucleosome positioning defines replication origins Matthew L. Eaton 1, Kyriaki Galani 2, Sukhyun Kang 2, Stephen P. Bell 2, and David M. MacAlpine 1 1 Department of
Supplementary Figure 1 Strategy for parallel detection of DHSs and adjacent nucleosomes
 Supplementary Figure 1 Strategy for parallel detection of DHSs and adjacent nucleosomes DNase I cleavage DNase I DNase I digestion Sucrose gradient enrichment Small Large F1 F2...... F9 F1 F1 F2 F3 F4
Supplementary Figure 1 Strategy for parallel detection of DHSs and adjacent nucleosomes DNase I cleavage DNase I DNase I digestion Sucrose gradient enrichment Small Large F1 F2...... F9 F1 F1 F2 F3 F4
TECH NOTE Stranded NGS libraries from FFPE samples
 TECH NOTE Stranded NGS libraries from FFPE samples Robust performance with extremely degraded FFPE RNA (DV 200 >25%) Consistent library quality across a range of input amounts (5 ng 50 ng) Compatibility
TECH NOTE Stranded NGS libraries from FFPE samples Robust performance with extremely degraded FFPE RNA (DV 200 >25%) Consistent library quality across a range of input amounts (5 ng 50 ng) Compatibility
Welcome to Class 18! Lecture 18: Outline and Objectives. Replication is semiconservative! Replication: DNA DNA! Introductory Biochemistry!
 Lecture 18: Outline and Objectives Welcome to Class 18! Introductory Biochemistry! l DNA Replication! l DNA polymerase! l the enzymatic reaction! l proofreading and accuracy! l DNA synthesis! l origins
Lecture 18: Outline and Objectives Welcome to Class 18! Introductory Biochemistry! l DNA Replication! l DNA polymerase! l the enzymatic reaction! l proofreading and accuracy! l DNA synthesis! l origins
Chapter 19 Genetic Regulation of the Eukaryotic Genome. A. Bergeron AP Biology PCHS
 Chapter 19 Genetic Regulation of the Eukaryotic Genome A. Bergeron AP Biology PCHS 2 Do Now - Eukaryotic Transcription Regulation The diagram below shows five genes (with their enhancers) from the genome
Chapter 19 Genetic Regulation of the Eukaryotic Genome A. Bergeron AP Biology PCHS 2 Do Now - Eukaryotic Transcription Regulation The diagram below shows five genes (with their enhancers) from the genome
Box 7905, Engineering Building I, North Carolina State University, Raleigh, NC 27695, Phone: Fax:
 Inefficient ribosomal skipping enables simultaneous secretion and display of proteins in Saccharomyces cerevisiae Carlos A. Cruz-Teran 1, Karthik Tiruthani 1, Adam Mischler, and Balaji M. Rao * Department
Inefficient ribosomal skipping enables simultaneous secretion and display of proteins in Saccharomyces cerevisiae Carlos A. Cruz-Teran 1, Karthik Tiruthani 1, Adam Mischler, and Balaji M. Rao * Department
Molecular Cell Biology - Problem Drill 11: Recombinant DNA
 Molecular Cell Biology - Problem Drill 11: Recombinant DNA Question No. 1 of 10 1. Which of the following statements about the sources of DNA used for molecular cloning is correct? Question #1 (A) cdna
Molecular Cell Biology - Problem Drill 11: Recombinant DNA Question No. 1 of 10 1. Which of the following statements about the sources of DNA used for molecular cloning is correct? Question #1 (A) cdna
Nature Methods: doi: /nmeth.4396
 Supplementary Figure 1 Comparison of technical replicate consistency between and across the standard ATAC-seq method, DNase-seq, and Omni-ATAC. (a) Heatmap-based representation of ATAC-seq quality control
Supplementary Figure 1 Comparison of technical replicate consistency between and across the standard ATAC-seq method, DNase-seq, and Omni-ATAC. (a) Heatmap-based representation of ATAC-seq quality control
Supplementary Table 1: Oligo designs. A list of ATAC-seq oligos used for PCR.
 Ad1_noMX: Ad2.1_TAAGGCGA Ad2.2_CGTACTAG Ad2.3_AGGCAGAA Ad2.4_TCCTGAGC Ad2.5_GGACTCCT Ad2.6_TAGGCATG Ad2.7_CTCTCTAC Ad2.8_CAGAGAGG Ad2.9_GCTACGCT Ad2.10_CGAGGCTG Ad2.11_AAGAGGCA Ad2.12_GTAGAGGA Ad2.13_GTCGTGAT
Ad1_noMX: Ad2.1_TAAGGCGA Ad2.2_CGTACTAG Ad2.3_AGGCAGAA Ad2.4_TCCTGAGC Ad2.5_GGACTCCT Ad2.6_TAGGCATG Ad2.7_CTCTCTAC Ad2.8_CAGAGAGG Ad2.9_GCTACGCT Ad2.10_CGAGGCTG Ad2.11_AAGAGGCA Ad2.12_GTAGAGGA Ad2.13_GTCGTGAT
MODULE 5: TRANSLATION
 MODULE 5: TRANSLATION Lesson Plan: CARINA ENDRES HOWELL, LEOCADIA PALIULIS Title Translation Objectives Determine the codons for specific amino acids and identify reading frames by looking at the Base
MODULE 5: TRANSLATION Lesson Plan: CARINA ENDRES HOWELL, LEOCADIA PALIULIS Title Translation Objectives Determine the codons for specific amino acids and identify reading frames by looking at the Base
الحمد هلل رب العالميه الذي هداوا لهذا وما كىا لىهتدي لىال أن هداوا اهلل والصالة والسالم على أشزف األوبياء. 222Cell Biolgy 1
 الحمد هلل رب العالميه الذي هداوا لهذا وما كىا لىهتدي لىال أن هداوا اهلل والصالة والسالم على أشزف األوبياء 222Cell Biolgy 1 Lecture 14 222Cell Biolgy 2 DNA replication DNA replication is a semi-conservative
الحمد هلل رب العالميه الذي هداوا لهذا وما كىا لىهتدي لىال أن هداوا اهلل والصالة والسالم على أشزف األوبياء 222Cell Biolgy 1 Lecture 14 222Cell Biolgy 2 DNA replication DNA replication is a semi-conservative
M1 - Biochemistry. Nucleic Acid Structure II/Transcription I
 M1 - Biochemistry Nucleic Acid Structure II/Transcription I PH Ratz, PhD (Resources: Lehninger et al., 5th ed., Chapters 8, 24 & 26) 1 Nucleic Acid Structure II/Transcription I Learning Objectives: 1.
M1 - Biochemistry Nucleic Acid Structure II/Transcription I PH Ratz, PhD (Resources: Lehninger et al., 5th ed., Chapters 8, 24 & 26) 1 Nucleic Acid Structure II/Transcription I Learning Objectives: 1.
Activation of a Floral Homeotic Gene in Arabidopsis
 Activation of a Floral Homeotic Gene in Arabidopsis By Maximiliam A. Busch, Kirsten Bomblies, and Detlef Weigel Presentation by Lis Garrett and Andrea Stevenson http://ucsdnews.ucsd.edu/archive/graphics/images/image5.jpg
Activation of a Floral Homeotic Gene in Arabidopsis By Maximiliam A. Busch, Kirsten Bomblies, and Detlef Weigel Presentation by Lis Garrett and Andrea Stevenson http://ucsdnews.ucsd.edu/archive/graphics/images/image5.jpg
Figure S1. nuclear extracts. HeLa cell nuclear extract. Input IgG IP:ORC2 ORC2 ORC2. MCM4 origin. ORC2 occupancy
 A nuclear extracts B HeLa cell nuclear extract Figure S1 ORC2 (in kda) 21 132 7 ORC2 Input IgG IP:ORC2 32 ORC C D PRKDC ORC2 occupancy Directed against ORC2 C-terminus (sc-272) MCM origin 2 2 1-1 -1kb
A nuclear extracts B HeLa cell nuclear extract Figure S1 ORC2 (in kda) 21 132 7 ORC2 Input IgG IP:ORC2 32 ORC C D PRKDC ORC2 occupancy Directed against ORC2 C-terminus (sc-272) MCM origin 2 2 1-1 -1kb
Next- genera*on Sequencing. Lecture 13
 Next- genera*on Sequencing Lecture 13 ChIP- seq Applica*ons iden%fy sequence varia%ons DNA- seq Iden%fy Pathogens RNA- seq Kahvejian et al, 2008 Protein-DNA interaction DNA is the informa*on carrier of
Next- genera*on Sequencing Lecture 13 ChIP- seq Applica*ons iden%fy sequence varia%ons DNA- seq Iden%fy Pathogens RNA- seq Kahvejian et al, 2008 Protein-DNA interaction DNA is the informa*on carrier of
DNA metabolism. DNA Replication DNA Repair DNA Recombination
 DNA metabolism DNA Replication DNA Repair DNA Recombination Chutima Talabnin Ph.D. School of Biochemistry,Institute of Science, Suranaree University of Technology Central Dogma or Flow of genetic information
DNA metabolism DNA Replication DNA Repair DNA Recombination Chutima Talabnin Ph.D. School of Biochemistry,Institute of Science, Suranaree University of Technology Central Dogma or Flow of genetic information
mrna Sequencing Quality Control (V6)
 mrna Sequencing Quality Control (V6) Notes: the following analyses are based on 8 adult brains sequenced in USC and Yale 1. Error Rates The error rates of each sequencing cycle are reported for 120 tiles
mrna Sequencing Quality Control (V6) Notes: the following analyses are based on 8 adult brains sequenced in USC and Yale 1. Error Rates The error rates of each sequencing cycle are reported for 120 tiles
Chapter 8. Microbial Genetics. Lectures prepared by Christine L. Case. Copyright 2010 Pearson Education, Inc.
 Chapter 8 Microbial Genetics Lectures prepared by Christine L. Case Structure and Function of Genetic Material Learning Objectives 8-1 Define genetics, genome, chromosome, gene, genetic code, genotype,
Chapter 8 Microbial Genetics Lectures prepared by Christine L. Case Structure and Function of Genetic Material Learning Objectives 8-1 Define genetics, genome, chromosome, gene, genetic code, genotype,
Browser Exercises - I. Alignments and Comparative genomics
 Browser Exercises - I Alignments and Comparative genomics 1. Navigating to the Genome Browser (GBrowse) Note: For this exercise use http://www.tritrypdb.org a. Navigate to the Genome Browser (GBrowse)
Browser Exercises - I Alignments and Comparative genomics 1. Navigating to the Genome Browser (GBrowse) Note: For this exercise use http://www.tritrypdb.org a. Navigate to the Genome Browser (GBrowse)
Nature Immunology: doi: /ni.3694
 Supplementary Figure 1 Expression of Bhlhe41 and Bhlhe40 in B cell development and mature B cell subsets. (a) Scatter plot showing differential expression of genes between splenic B-1a cells and follicular
Supplementary Figure 1 Expression of Bhlhe41 and Bhlhe40 in B cell development and mature B cell subsets. (a) Scatter plot showing differential expression of genes between splenic B-1a cells and follicular
Zoo-342 Molecular biology Lecture 2. DNA replication
 Zoo-342 Molecular biology Lecture 2 DNA replication DNA replication DNA replication is the process in which one doubled-stranded DNA molecule is used to create two double-stranded molecules with identical
Zoo-342 Molecular biology Lecture 2 DNA replication DNA replication DNA replication is the process in which one doubled-stranded DNA molecule is used to create two double-stranded molecules with identical
Charles Girardot, Furlong Lab. MACS, CisGenome, SISSRs and other peak calling algorithms: differences and practical use
 Charles Girardot, Furlong Lab MACS, CisGenome, SISSRs and other peak calling algorithms: differences and practical use ChIP-Seq signal properties Only 5 ends of ChIPed fragments are sequenced Shifted read
Charles Girardot, Furlong Lab MACS, CisGenome, SISSRs and other peak calling algorithms: differences and practical use ChIP-Seq signal properties Only 5 ends of ChIPed fragments are sequenced Shifted read
BIOLOGY 205 Midterm II - 19 February Each of the following statements are correct regarding Eukaryotic genes and genomes EXCEPT?
 BIOLOGY 205 Midterm II - 19 February 1999 Name Multiple choice questions 4 points each (Best 12 out of 13). 1. Each of the following statements are correct regarding Eukaryotic genes and genomes EXCEPT?
BIOLOGY 205 Midterm II - 19 February 1999 Name Multiple choice questions 4 points each (Best 12 out of 13). 1. Each of the following statements are correct regarding Eukaryotic genes and genomes EXCEPT?
Supplementary Materials. for. array reveals biophysical and evolutionary landscapes
 Supplementary Materials for Quantitative analysis of RNA- protein interactions on a massively parallel array reveals biophysical and evolutionary landscapes Jason D. Buenrostro 1,2,4, Carlos L. Araya 1,4,
Supplementary Materials for Quantitative analysis of RNA- protein interactions on a massively parallel array reveals biophysical and evolutionary landscapes Jason D. Buenrostro 1,2,4, Carlos L. Araya 1,4,
