Really high sensitivity mass spectrometry and Discovery and analysis of protein complexes
|
|
|
- Clementine Grant
- 5 years ago
- Views:
Transcription
1 Really high sensitivity mass spectrometry and Discovery and analysis of protein complexes The PRIME lab and AMS Importance of protein complexes in biology Methods for isolation of protein complexes In solution On a chip In a gel (Paul Brookes) Analysis of protein complexes
2 Purdue Rare Isotope Measurement Lab Accelerator mass spectrometry for rare isotopes, 10 Be, 14 C, 26 Al, 36 Cl, 41 Ca, 129 I
3 Accelerator in PRIME Lab Dr. David Elmore next to 10 MV accelerator
4 Inside Accelerator in PRIME Lab If an animal is given 50 nci of a 14 C-labeled compound and 0.01% is absorbed and reaches the brain, then 20 mg of tissue is sufficient to provide enough signal to give a 1000:1 signalto-noise ratio
5 Collapse of the single target paradigm Old paradigm Diseases are due to single genes - by knocking out the gene, or designing specific inhibitors to its protein, disease can be cured But the gene KO mouse didn t notice the loss of the gene New paradigm We have to understand gene and protein networks - proteins don t act alone - effective systems have built in redundancy
6 Proteins aren t random in cells So, who s binding to whom?
7 Proteins don t act alone Peptidomimetic targets Signal transduction complex lying in anticipation
8 How to discover protein brotherhoods Old method: Yeast 2-hybrid screen New method: Recover protein complexes SDS-PAGE IEF/SDS-PAGE
9 Affinity isolation of EGF-responsive proteins Pandey et al., PNAS 97: (2000) EGF-induced tyrosine phosphorylation in HeLa cells. Serum-deprived HeLa S3 cells (5 x 10 9 ) were either left untreated or treated with 1 mg/ml EGF for 5 min. Cleared cell lysates were immunoprecipitated with a mixture of monoclonal antiphosphotyrosine antibodies, washed, and resolved by SDS/PAGE. The gel was then silver-stained. Numbers indicate the positions of the bands that were excised for enzymatic digestion by trypsin and subsequent mass spectrometric analysis.
10 EGF-stimulated, tyrosine-phosphorylated proteins identified by mass spec EGF Hrs Eps15 HD Vav-2 EGF-R Shc p62 p85 Grb-2 Cbl See protein interactions at
11 Affinity methods for recovering complexes streptavidin antibody biotin glutathione GST Multiprotein complex
12 Recovering a ribosomal protein complex In A, the proteins pulled down by untagged (-) and tagged (+) Nop7p were analyzed by SDS-PAGE In B, these proteins were separated by reverse-phase HPLC and were subjected to trypsin fingerprint analysis by MALDI-TOF Harnpicharnchai et al., Mol. Cell 8: 505 (2001)
13 Affinity purification of nucleoporin interaction proteins Allen et al.,j Biol Chem 276: (2001)
14 Tap-Tag isolation of protein complexes Gavin et al.,nature 415: 141 (2002)
15 Validation of partners in protein complexes - cross correlation analysis Gavin et al.,nature 415, 141 (2002)
16 Summary of protein complexes discovered in yeast by the Tap-Tag method Gavin et al., Nature 415, 141 (2002)
17 Comparison of the effectiveness of protein-protein interaction methods Von Mering et al., Nature 417: 399 (2002)
18 Analysis of bridged protein complexes Back et al., Anal Chem 74:4417 (2002) Digestion of chemically linked proteins results in a bridged peptide. If the hydrolysis is carried out in H 2 O 18, then there will be four O 18 and hence the MW will increase by 8
19 Tandem MS of bridged peptide Note the increase in the Arg fragment (m/z 175) to m/z 179 Back et al., Anal Chem 74:4417 (2002)
20 Surface enhanced laser desorption ionization (SELDI) Selective binding of proteins to the surface of the chip - add matrix and analyze by MALDI- TOF-MS hydrophobic Future: Ab or protein coated onto chip positive negative
21 Spotted array of 80% of the yeast proteome 6566 protein samples representing 5800 unique proteins were spotted in duplicate on a single nickel-coated microscope slide. The slide was probed with anti-gst. Zhu & Snyder, Science 293, 2101 (2001)
22 Application of protein chip to calmodulin binding and lipid binding proteins A. Positive signals in duplicate (green) are in the bottom row of each panel; the top row shows the amounts of the yeast protein preparations probed with anti- GST (red). Zhu & Snyder, Science 293, 2101 (2001) B. A putative calmodulin-binding motif. Fourteen of 39 positive proteins share a motif whose consensus is (I/L)QXK(K/X)GB, where X is any residue and B is a basic residue. The size of the letter indicates the relative frequency of the amino acid indicated
Proteomics and some of its Mass Spectrometric Applications
 Proteomics and some of its Mass Spectrometric Applications What? Large scale screening of proteins, their expression, modifications and interactions by using high-throughput approaches 2 1 Why? The number
Proteomics and some of its Mass Spectrometric Applications What? Large scale screening of proteins, their expression, modifications and interactions by using high-throughput approaches 2 1 Why? The number
ENCODE DCC Antibody Validation Document
 ENCODE DCC Antibody Validation Document Date of Submission 09/12/12 Name: Trupti Kawli Email: trupti@stanford.edu Lab Snyder Antibody Name: SREBP1 (sc-8984) Target: SREBP1 Company/ Source: Santa Cruz Biotechnology
ENCODE DCC Antibody Validation Document Date of Submission 09/12/12 Name: Trupti Kawli Email: trupti@stanford.edu Lab Snyder Antibody Name: SREBP1 (sc-8984) Target: SREBP1 Company/ Source: Santa Cruz Biotechnology
Computing with large data sets
 Computing with large data sets Richard Bonneau, spring 2009 Lecture 14 (week 8): genomics 1 Central dogma Gene expression DNA RNA Protein v22.0480: computing with data, Richard Bonneau Lecture 14 places
Computing with large data sets Richard Bonneau, spring 2009 Lecture 14 (week 8): genomics 1 Central dogma Gene expression DNA RNA Protein v22.0480: computing with data, Richard Bonneau Lecture 14 places
Supplementary Fig. 1 Identification of Nedd4 as an IRS-2-associated protein in camp-treated FRTL-5 cells.
 Supplementary Fig. 1 Supplementary Fig. 1 Identification of Nedd4 as an IRS-2-associated protein in camp-treated FRTL-5 cells. (a) FRTL-5 cells were treated with 1 mm dibutyryl camp for 24 h, and the lysates
Supplementary Fig. 1 Supplementary Fig. 1 Identification of Nedd4 as an IRS-2-associated protein in camp-treated FRTL-5 cells. (a) FRTL-5 cells were treated with 1 mm dibutyryl camp for 24 h, and the lysates
11/22/13. Proteomics, functional genomics, and systems biology. Biosciences 741: Genomics Fall, 2013 Week 11
 Proteomics, functional genomics, and systems biology Biosciences 741: Genomics Fall, 2013 Week 11 1 Figure 6.1 The future of genomics Functional Genomics The field of functional genomics represents the
Proteomics, functional genomics, and systems biology Biosciences 741: Genomics Fall, 2013 Week 11 1 Figure 6.1 The future of genomics Functional Genomics The field of functional genomics represents the
MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1. Lecture 25: Mass Spectrometry Applications
![MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1. Lecture 25: Mass Spectrometry Applications MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1. Lecture 25: Mass Spectrometry Applications](/thumbs/74/71368870.jpg) MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1 Lecture 25: Mass Spectrometry Applications Measuring Protein Abundance o ICAT o DIGE Identifying Post-Translational Modifications Protein-protein
MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1 Lecture 25: Mass Spectrometry Applications Measuring Protein Abundance o ICAT o DIGE Identifying Post-Translational Modifications Protein-protein
This is the author's accepted version of the manuscript.
 This is the author's accepted version of the manuscript. The definitive version is published in Nature Communications Online Edition: 2015/4/16 (Japan time), doi:10.1038/ncomms7780. The final version published
This is the author's accepted version of the manuscript. The definitive version is published in Nature Communications Online Edition: 2015/4/16 (Japan time), doi:10.1038/ncomms7780. The final version published
Experimental Techniques 2
 Experimental Techniques 2 High-throughput interaction detection Yeast two-hybrid - pairwise organisms as machines to learn about organisms yeast, worm, fly, human,... low intersection between repeated
Experimental Techniques 2 High-throughput interaction detection Yeast two-hybrid - pairwise organisms as machines to learn about organisms yeast, worm, fly, human,... low intersection between repeated
Proteomics. Proteomics is the study of all proteins within organism. Challenges
 Proteomics Proteomics is the study of all proteins within organism. Challenges 1. The proteome is larger than the genome due to alternative splicing and protein modification. As we have said before we
Proteomics Proteomics is the study of all proteins within organism. Challenges 1. The proteome is larger than the genome due to alternative splicing and protein modification. As we have said before we
Chapter 9 Proteomics. From genomics to proteomics
 Chapter 9 Proteomics From genomics to proteomics From structural genomics, functional genomics to proteomics http://www.nature.com/scitable/topicpage/the-proteome-discovering-the-structure-and-function-613
Chapter 9 Proteomics From genomics to proteomics From structural genomics, functional genomics to proteomics http://www.nature.com/scitable/topicpage/the-proteome-discovering-the-structure-and-function-613
2D separations and analysis of proteins in biological samples
 January 18, 2005 2D separations and analysis of proteins in biological samples Helen Kim 934-3880 helenkim@uab.edu McCallum Building, room 460 Jan 18, 2005 HelenKim/UAB/PharmTox 1 Learning objectives 2-D
January 18, 2005 2D separations and analysis of proteins in biological samples Helen Kim 934-3880 helenkim@uab.edu McCallum Building, room 460 Jan 18, 2005 HelenKim/UAB/PharmTox 1 Learning objectives 2-D
Enhancers mutations that make the original mutant phenotype more extreme. Suppressors mutations that make the original mutant phenotype less extreme
 Interactomics and Proteomics 1. Interactomics The field of interactomics is concerned with interactions between genes or proteins. They can be genetic interactions, in which two genes are involved in the
Interactomics and Proteomics 1. Interactomics The field of interactomics is concerned with interactions between genes or proteins. They can be genetic interactions, in which two genes are involved in the
27041, Week 02. Review of Week 01
 27041, Week 02 Review of Week 01 The human genome sequencing project (HGP) 2 CBS, Department of Systems Biology Systems Biology and emergent properties 3 CBS, Department of Systems Biology Different model
27041, Week 02 Review of Week 01 The human genome sequencing project (HGP) 2 CBS, Department of Systems Biology Systems Biology and emergent properties 3 CBS, Department of Systems Biology Different model
Lecture 5: 8/31. CHAPTER 5 Techniques in Protein Biochemistry
 Lecture 5: 8/31 CHAPTER 5 Techniques in Protein Biochemistry Chapter 5 Outline The proteome is the entire set of proteins expressed and modified by a cell under a particular set of biochemical conditions.
Lecture 5: 8/31 CHAPTER 5 Techniques in Protein Biochemistry Chapter 5 Outline The proteome is the entire set of proteins expressed and modified by a cell under a particular set of biochemical conditions.
Proteomics. Areas of Application for Proteomics Most Commonly Used Proteomics Techniques: Limitations: Examples
 Proteomics Areas of Application for Proteomics Most Commonly Used Proteomics Techniques: Antibody arrays Protein activity arrays 2-D gels ICAT technology SELDI Limitations: protein sources surfaces and
Proteomics Areas of Application for Proteomics Most Commonly Used Proteomics Techniques: Antibody arrays Protein activity arrays 2-D gels ICAT technology SELDI Limitations: protein sources surfaces and
Areas of Application for Proteomics Most Commonly Used Proteomics Techniques:
 Proteomics Areas of Application for Proteomics Most Commonly Used Proteomics Techniques: Antibody arrays Protein activity arrays 2-D gels ICAT technology SELDI Limitations: Examples protein sources surfaces
Proteomics Areas of Application for Proteomics Most Commonly Used Proteomics Techniques: Antibody arrays Protein activity arrays 2-D gels ICAT technology SELDI Limitations: Examples protein sources surfaces
Post-translational modification
 Protein expression Western blotting, is a widely used and accepted technique to detect levels of protein expression in a cell or tissue extract. This technique measures protein levels in a biological sample
Protein expression Western blotting, is a widely used and accepted technique to detect levels of protein expression in a cell or tissue extract. This technique measures protein levels in a biological sample
Proteomics. Areas of Application for Proteomics. Most Commonly Used Proteomics Techniques: Limitations: Examples
 Proteomics Areas of Application for Proteomics Most Commonly Used Proteomics Techniques: Antibody arrays Protein activity arrays 2-D gels ICAT technology SELDI Limitations: protein sources surfaces and
Proteomics Areas of Application for Proteomics Most Commonly Used Proteomics Techniques: Antibody arrays Protein activity arrays 2-D gels ICAT technology SELDI Limitations: protein sources surfaces and
Identification of human serum proteins detectable after Albumin removal with Vivapure Anti-HSA Kit
 Andreas Kocourek, Pieter Eyckerman, Robert Zeidler, Pascal Bolon and Birgit Thome-Kromer Introduction The analysis of the complete proteome is a major interest of many researchers. Of particular importance
Andreas Kocourek, Pieter Eyckerman, Robert Zeidler, Pascal Bolon and Birgit Thome-Kromer Introduction The analysis of the complete proteome is a major interest of many researchers. Of particular importance
Strategies in proteomics
 Strategies in proteomics Systems biology - understand cellpathways, network, and complex interacting (includes Genomics, Proteomics, Metabolomics..) Biological processes - characterize protein complexes,
Strategies in proteomics Systems biology - understand cellpathways, network, and complex interacting (includes Genomics, Proteomics, Metabolomics..) Biological processes - characterize protein complexes,
SGN-6106 Computational Systems Biology I
 SGN-6106 Computational Systems Biology I A View of Modern Measurement Systems in Cell Biology Kaisa-Leena Taattola 21.2.2007 1 The cell a complex system (Source: Lehninger Principles of Biochemistry 4th
SGN-6106 Computational Systems Biology I A View of Modern Measurement Systems in Cell Biology Kaisa-Leena Taattola 21.2.2007 1 The cell a complex system (Source: Lehninger Principles of Biochemistry 4th
Comparability Analysis of Protein Therapeutics by Bottom-Up LC-MS with Stable Isotope-Tagged Reference Standards
 Comparability Analysis of Protein Therapeutics by Bottom-Up LC-MS with Stable Isotope-Tagged Reference Standards 16 September 2011 Abbott Bioresearch Center, Worcester, MA USA Manuilov, A. V., C. H. Radziejewski
Comparability Analysis of Protein Therapeutics by Bottom-Up LC-MS with Stable Isotope-Tagged Reference Standards 16 September 2011 Abbott Bioresearch Center, Worcester, MA USA Manuilov, A. V., C. H. Radziejewski
Proteomics And Cancer Biomarker Discovery. Dr. Zahid Khan Institute of chemical Sciences (ICS) University of Peshawar. Overview. Cancer.
 Proteomics And Cancer Biomarker Discovery Dr. Zahid Khan Institute of chemical Sciences (ICS) University of Peshawar Overview Proteomics Cancer Aims Tools Data Base search Challenges Summary 1 Overview
Proteomics And Cancer Biomarker Discovery Dr. Zahid Khan Institute of chemical Sciences (ICS) University of Peshawar Overview Proteomics Cancer Aims Tools Data Base search Challenges Summary 1 Overview
ENCODE DCC Antibody Validation Document
 ENCODE DCC Antibody Validation Document Date of Submission 06/15/2012 Name: Trupti Kawli Email: trupti@stanford.edu Lab Snyder Antibody Name: CDP (sc6327) Target: CDP Company/ Source: Santa Cruz Catalog
ENCODE DCC Antibody Validation Document Date of Submission 06/15/2012 Name: Trupti Kawli Email: trupti@stanford.edu Lab Snyder Antibody Name: CDP (sc6327) Target: CDP Company/ Source: Santa Cruz Catalog
Advances in analytical biochemistry and systems biology: Proteomics
 Advances in analytical biochemistry and systems biology: Proteomics Brett Boghigian Department of Chemical & Biological Engineering Tufts University July 29, 2005 Proteomics The basics History Current
Advances in analytical biochemistry and systems biology: Proteomics Brett Boghigian Department of Chemical & Biological Engineering Tufts University July 29, 2005 Proteomics The basics History Current
Zakopane, April, 2011
 Clinical Proteomics: A Technology to shape the future? Zakopane, April, 2011 Protein Chemistry/Proteomics/Peptide Synthesis and Array Unit Institute of Biomedicine/Anatomy Biomedicum Helsinki, 00014 University
Clinical Proteomics: A Technology to shape the future? Zakopane, April, 2011 Protein Chemistry/Proteomics/Peptide Synthesis and Array Unit Institute of Biomedicine/Anatomy Biomedicum Helsinki, 00014 University
Biology: the basis for smart proteomic approaches to protein analysis
 QuickTime and a TIFF (Uncompressed) decompressor are needed to see this picture. Biology: the basis for smart proteomic approaches to protein analysis Helen Kim, PhD Department of Pharmacology & Toxicology
QuickTime and a TIFF (Uncompressed) decompressor are needed to see this picture. Biology: the basis for smart proteomic approaches to protein analysis Helen Kim, PhD Department of Pharmacology & Toxicology
MagSi Beads. Magnetic Silica Beads for Life Science and Biotechnology study
 MagSi Beads Magnetic Silica Beads for Life Science and Biotechnology study MagnaMedics Diagnostics B.V. / Rev. 9.2 / 2012 Wide range of products for numerous applications MagnaMedics separation solutions
MagSi Beads Magnetic Silica Beads for Life Science and Biotechnology study MagnaMedics Diagnostics B.V. / Rev. 9.2 / 2012 Wide range of products for numerous applications MagnaMedics separation solutions
The BioPharmaSpec Approach : Mass Spectrometry Based Host Cell Protein Identification and Quantitation
 The BioPharmaSpec Approach : Mass Spectrometry Based Host Cell Protein Identification and Quantitation 1. Introduction As part of the development of any biopharmaceutical product, the impurities present
The BioPharmaSpec Approach : Mass Spectrometry Based Host Cell Protein Identification and Quantitation 1. Introduction As part of the development of any biopharmaceutical product, the impurities present
Kinetics Review. Tonight at 7 PM Phys 204 We will do two problems on the board (additional ones than in the problem sets)
 Quiz 1 Kinetics Review Tonight at 7 PM Phys 204 We will do two problems on the board (additional ones than in the problem sets) I will post the problems with solutions on Toolkit for those that can t make
Quiz 1 Kinetics Review Tonight at 7 PM Phys 204 We will do two problems on the board (additional ones than in the problem sets) I will post the problems with solutions on Toolkit for those that can t make
Biomarker Discovery using Surface Plasmon Resonance Imaging
 F e a t u r e A r t i c l e Feature Article Biomarker Discovery using Surface Plasmon Resonance Imaging Elodie LY-MORIN, Sophie BELLON, Géraldine MÉLIZZI, Chiraz FRYDMAN Surface Plasmon Resonance (SPR)
F e a t u r e A r t i c l e Feature Article Biomarker Discovery using Surface Plasmon Resonance Imaging Elodie LY-MORIN, Sophie BELLON, Géraldine MÉLIZZI, Chiraz FRYDMAN Surface Plasmon Resonance (SPR)
Description of supplementary material file
 Description of supplementary material file In the supplementary results we show that the VHL-fibronectin interaction is indirect, mediated by fibronectin binding to COL4A2. This provides additional information
Description of supplementary material file In the supplementary results we show that the VHL-fibronectin interaction is indirect, mediated by fibronectin binding to COL4A2. This provides additional information
Reducing dynamic range with Proteominer
 Reducing dynamic range with Proteominer Dynamic Range of the Plasma Proteome Albumin Apolipo-A1 Apolipo-B AGP Lipoprotein A IgG Transferrin Fibrinogen IgA α2 macroglobulin IgM α1-at C3 Comp factor Haptoglobulin
Reducing dynamic range with Proteominer Dynamic Range of the Plasma Proteome Albumin Apolipo-A1 Apolipo-B AGP Lipoprotein A IgG Transferrin Fibrinogen IgA α2 macroglobulin IgM α1-at C3 Comp factor Haptoglobulin
Newsletter Issue 7 One-STrEP Analysis of Protein:Protein-Interactions
 www.iba-biotagnology.com Newsletter Issue 7 One-STrEP Analysis of Protein:Protein-Interactions Strep-tag and One-STrEP-tag PPI Analysis with the co-precipitation/ mass spectrometry approach 3 Background
www.iba-biotagnology.com Newsletter Issue 7 One-STrEP Analysis of Protein:Protein-Interactions Strep-tag and One-STrEP-tag PPI Analysis with the co-precipitation/ mass spectrometry approach 3 Background
BIOLOGY 429 PROTEOMICS LABORATORY Fall 2004
 Dr. Karin Sauer Dr. Anna Tan-Wilson BIOLOGY 429 PROTEOMICS LABORATORY Fall 2004 COURSE BULLETIN DESCRIPTION: Proteomics techniques for the analysis of global changes in protein patterns. Methods include
Dr. Karin Sauer Dr. Anna Tan-Wilson BIOLOGY 429 PROTEOMICS LABORATORY Fall 2004 COURSE BULLETIN DESCRIPTION: Proteomics techniques for the analysis of global changes in protein patterns. Methods include
2D Gel PhosphoTyrosine Western blotting. Nancy Kendrick, Jon Johansen & Matt Hoelter, Kendrick Labs Inc
 2D Gel PhosphoTyrosine Western blotting Nancy Kendrick, Jon Johansen & Matt Hoelter, Kendrick Labs Inc www.kendricklabs.com Talk Outline Kendrick Labs, 2DE Importance of focusing on protein subsets P-Tyrosine
2D Gel PhosphoTyrosine Western blotting Nancy Kendrick, Jon Johansen & Matt Hoelter, Kendrick Labs Inc www.kendricklabs.com Talk Outline Kendrick Labs, 2DE Importance of focusing on protein subsets P-Tyrosine
Module 1 overview PRESIDENTʼS DAY
 1 Module 1 overview lecture lab 1. Introduction to the module 1. Start-up protein eng. 2. Rational protein design 2. Site-directed mutagenesis 3. Fluorescence and sensors 3. DNA amplification PRESIDENTʼS
1 Module 1 overview lecture lab 1. Introduction to the module 1. Start-up protein eng. 2. Rational protein design 2. Site-directed mutagenesis 3. Fluorescence and sensors 3. DNA amplification PRESIDENTʼS
Proteomics. Manickam Sugumaran. Department of Biology University of Massachusetts Boston, MA 02125
 Proteomics Manickam Sugumaran Department of Biology University of Massachusetts Boston, MA 02125 Genomic studies produced more than 75,000 potential gene sequence targets. (The number may be even higher
Proteomics Manickam Sugumaran Department of Biology University of Massachusetts Boston, MA 02125 Genomic studies produced more than 75,000 potential gene sequence targets. (The number may be even higher
Meso Scale Discovery Applications
 A Suite of Assays to Detect hosphorylated Receptor Tyrosine Kinases Associated with Neoplasia Timothy S. Schaefer, Jane Zhang, Sean Higgins, Laura K. Schaefer, Robert M. Umek and Jacob N. Wohlstadter Reversible
A Suite of Assays to Detect hosphorylated Receptor Tyrosine Kinases Associated with Neoplasia Timothy S. Schaefer, Jane Zhang, Sean Higgins, Laura K. Schaefer, Robert M. Umek and Jacob N. Wohlstadter Reversible
Laboratory Water Quality Affects Protein Separation by 2D Gel Electrophoresis
 Laboratory Water Quality Affects Protein Separation by 2D Gel Electrophoresis 2D gel electrophoresis remains a dominant technique in proteomics. Obtaining high quality gels requires careful and tedious
Laboratory Water Quality Affects Protein Separation by 2D Gel Electrophoresis 2D gel electrophoresis remains a dominant technique in proteomics. Obtaining high quality gels requires careful and tedious
SILAC Protein Identification (ID) and Quantitation Kits
 SILAC Protein Identification (ID) and Quantitation Kits For identifying and quantifying phosphoproteins and membrane proteins Catalog no. SP10001, SM10002, SP10005, SM10006 MS10030, MS10031, MS10032, MS10033
SILAC Protein Identification (ID) and Quantitation Kits For identifying and quantifying phosphoproteins and membrane proteins Catalog no. SP10001, SM10002, SP10005, SM10006 MS10030, MS10031, MS10032, MS10033
Lecture 8: Affinity Chromatography-III
 Lecture 8: Affinity Chromatography-III Key words: Chromatography; Affinity chromatography; Protein Purification During this lecture, we shall be studying few more examples of affinity chromatography. The
Lecture 8: Affinity Chromatography-III Key words: Chromatography; Affinity chromatography; Protein Purification During this lecture, we shall be studying few more examples of affinity chromatography. The
Solutions to 7.02 Quiz II 10/27/05
 Solutions to 7.02 Quiz II 10/27/05 Class Average = 83 Standard Deviation = 9 Range Grade % 87-100 A 43 74-86 B 39 55-73 C 17 > 54 D 1 Question 1 (56 points) While studying deep sea bacteria, you discover
Solutions to 7.02 Quiz II 10/27/05 Class Average = 83 Standard Deviation = 9 Range Grade % 87-100 A 43 74-86 B 39 55-73 C 17 > 54 D 1 Question 1 (56 points) While studying deep sea bacteria, you discover
Protein microarray and proteomics
 17 2 2005 4 Chinese Bulletin of Life Sciences Vol. 17, No. 2 Apr., 2005 1004-0374(2005)02-0132-05 510515 ; Q503;Q51 A Protein microarray and proteomics FEI Jia, MA Wen-Li*, ZHENG Wen-Ling (Institute of
17 2 2005 4 Chinese Bulletin of Life Sciences Vol. 17, No. 2 Apr., 2005 1004-0374(2005)02-0132-05 510515 ; Q503;Q51 A Protein microarray and proteomics FEI Jia, MA Wen-Li*, ZHENG Wen-Ling (Institute of
Exam MOL3007 Functional Genomics
 Faculty of Medicine Department of Cancer Research and Molecular Medicine Exam MOL3007 Functional Genomics Tuesday May 29 th 9.00-13.00 ECTS credits: 7.5 Number of pages (included front-page): 5 Supporting
Faculty of Medicine Department of Cancer Research and Molecular Medicine Exam MOL3007 Functional Genomics Tuesday May 29 th 9.00-13.00 ECTS credits: 7.5 Number of pages (included front-page): 5 Supporting
Develop Better Assays for Every Human Protein. OriGene Technologies, Inc.
 Develop Better Assays for Every Human Protein OriGene Technologies, Inc. About OriGene Started in1996 with the goal to create largest commercial collection of full-length human cdnas Develop technologies
Develop Better Assays for Every Human Protein OriGene Technologies, Inc. About OriGene Started in1996 with the goal to create largest commercial collection of full-length human cdnas Develop technologies
Application Note TOF/MS
 Application Note TOF/MS New Level of Confidence for Protein Identification: Results Dependent Analysis and Peptide Mass Fingerprinting Using the 4700 Proteomics Discovery System Purpose The Applied Biosystems
Application Note TOF/MS New Level of Confidence for Protein Identification: Results Dependent Analysis and Peptide Mass Fingerprinting Using the 4700 Proteomics Discovery System Purpose The Applied Biosystems
Accelerate mab Characterization Using Automated Sample Prep
 Accelerate mab Characterization Using Automated Sample Prep David Knorr, Ph.D. Automation Solutions Ning Tang, Ph.D. LC/MS 15 February 2012 Page 1 Protein Sample Processing Workflows Glycan Profiling Biological
Accelerate mab Characterization Using Automated Sample Prep David Knorr, Ph.D. Automation Solutions Ning Tang, Ph.D. LC/MS 15 February 2012 Page 1 Protein Sample Processing Workflows Glycan Profiling Biological
supplementary information
 DOI: 10.1038/ncb2116 Figure S1 CDK phosphorylation of EZH2 in cells. (a) Comparison of candidate CDK phosphorylation sites on EZH2 with known CDK substrates by multiple sequence alignments. (b) CDK1 and
DOI: 10.1038/ncb2116 Figure S1 CDK phosphorylation of EZH2 in cells. (a) Comparison of candidate CDK phosphorylation sites on EZH2 with known CDK substrates by multiple sequence alignments. (b) CDK1 and
Objective. Introduction. IP assisted LC/MS/MS making study protein complexes easy. Jon Hao 1, Yi Liu 1, Xiaozhi Ren 2, and King-Wai Yau 2
 IP assisted LC/MS/MS making study protein complexes easy Jon Hao 1, Yi Liu 1, Xiaozhi Ren 2, and King-Wai Yau 2 1 Poochon Scientific, Frederick, Maryland 21701 2 Department of Neuroscience, Johns Hopkins
IP assisted LC/MS/MS making study protein complexes easy Jon Hao 1, Yi Liu 1, Xiaozhi Ren 2, and King-Wai Yau 2 1 Poochon Scientific, Frederick, Maryland 21701 2 Department of Neuroscience, Johns Hopkins
SUPPLEMENTARY INFORMATION
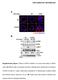 SUPPLEMENTARY INFORMATION Supplementary Figure 1 Effect of ROCK inhibition on lumen abnormality in MDCK cysts. (A) MDCK cells as indicated cultured in Matrigel were treated with and without Y27632 (10
SUPPLEMENTARY INFORMATION Supplementary Figure 1 Effect of ROCK inhibition on lumen abnormality in MDCK cysts. (A) MDCK cells as indicated cultured in Matrigel were treated with and without Y27632 (10
Develop Better Assays for Every Human Protein
 Develop Better Assays for Every Human Protein OriGene overview Application of transfected over-expression lysates Application of purified recombinant human proteins About OriGene Started in1996 with the
Develop Better Assays for Every Human Protein OriGene overview Application of transfected over-expression lysates Application of purified recombinant human proteins About OriGene Started in1996 with the
PROTEOMICS. Timothy Palzkill Baylor College of Medicine. KLUWER ACADEMIC PUBLISHERS New York / Boston / Dordrecht / London / Moscow
 PROTEOMICS PROTEOMICS by Timothy Palzkill Baylor College of Medicine KLUWER ACADEMIC PUBLISHERS New York / Boston / Dordrecht / London / Moscow ebook ISBN: 0-306-46895-6 Print ISBN: 0-792-37565-3 2002
PROTEOMICS PROTEOMICS by Timothy Palzkill Baylor College of Medicine KLUWER ACADEMIC PUBLISHERS New York / Boston / Dordrecht / London / Moscow ebook ISBN: 0-306-46895-6 Print ISBN: 0-792-37565-3 2002
12/11/12. Dr. Sanjeeva Srivastava. IIT Bombay 2. Interactomics Yeast Two-Hybrid Immunoprecipitation Protein microarrays. Proteomics Course NPTEL
 Dr. Sanjeeva Srivastava IIT Bombay Interactomics Yeast Two-Hybrid Immunoprecipitation Protein microarrays IIT Bombay 2 1 IIT Bombay Tremendous effect of interactions IIT Bombay 4 2 Complex formation Protein
Dr. Sanjeeva Srivastava IIT Bombay Interactomics Yeast Two-Hybrid Immunoprecipitation Protein microarrays IIT Bombay 2 1 IIT Bombay Tremendous effect of interactions IIT Bombay 4 2 Complex formation Protein
Supplementary methods Shoc2 In Vitro Ubiquitination Assay
 Supplementary methods Shoc2 In Vitro Ubiquitination Assay 35 S-labelled Shoc2 was prepared using a TNT quick Coupled transcription/ translation System (Promega) as recommended by manufacturer. For the
Supplementary methods Shoc2 In Vitro Ubiquitination Assay 35 S-labelled Shoc2 was prepared using a TNT quick Coupled transcription/ translation System (Promega) as recommended by manufacturer. For the
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or
 Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
Thermo Scientific GTPase Research Tools
 Thermo Scientific Research Tools Active Pull-Down Assays We offer two different tools to study biology, one for active monitoring and one for global profiling. The Thermo Scientific Pierce Active Pull-Down
Thermo Scientific Research Tools Active Pull-Down Assays We offer two different tools to study biology, one for active monitoring and one for global profiling. The Thermo Scientific Pierce Active Pull-Down
SUPPLEMENTAL MATERIAL. Supplemental Methods:
 SUPPLEMENTAL MATERIAL Supplemental Methods: Immunoprecipitation- As we described but with some modifications [22]. As part of another ongoing project, lysate from human umbilical vein endothelial cells
SUPPLEMENTAL MATERIAL Supplemental Methods: Immunoprecipitation- As we described but with some modifications [22]. As part of another ongoing project, lysate from human umbilical vein endothelial cells
Genome Biology and Biotechnology
 Genome Biology and Biotechnology 10. The proteome Prof. M. Zabeau Department of Plant Systems Biology Flanders Interuniversity Institute for Biotechnology (VIB) University of Gent International course
Genome Biology and Biotechnology 10. The proteome Prof. M. Zabeau Department of Plant Systems Biology Flanders Interuniversity Institute for Biotechnology (VIB) University of Gent International course
Peptide libraries: applications, design options and considerations. Laura Geuss, PhD May 5, 2015, 2:00-3:00 pm EST
 Peptide libraries: applications, design options and considerations Laura Geuss, PhD May 5, 2015, 2:00-3:00 pm EST Overview 1 2 3 4 5 Introduction Peptide library basics Peptide library design considerations
Peptide libraries: applications, design options and considerations Laura Geuss, PhD May 5, 2015, 2:00-3:00 pm EST Overview 1 2 3 4 5 Introduction Peptide library basics Peptide library design considerations
Supplementary Information
 Supplementary Information Peroxiredoxin-2 and STAT3 form a redox relay for H 2 O 2 signaling Mirko C. Sobotta 1, Willy Liou 1, Sarah Stöcker 1, Deepti Talwar 1, Michael Oehler 1, Thomas Ruppert 2, Annette
Supplementary Information Peroxiredoxin-2 and STAT3 form a redox relay for H 2 O 2 signaling Mirko C. Sobotta 1, Willy Liou 1, Sarah Stöcker 1, Deepti Talwar 1, Michael Oehler 1, Thomas Ruppert 2, Annette
Expression Proteomics: principles and examples of application
 Expression Proteomics: principles and examples of application Miguel Teixeira IBB Institute for Biotechnology and Bioengineering, Centro de Eng. Biológica e Química, IST Functional Genomics and Bioinformatics
Expression Proteomics: principles and examples of application Miguel Teixeira IBB Institute for Biotechnology and Bioengineering, Centro de Eng. Biológica e Química, IST Functional Genomics and Bioinformatics
Antibody Profiling on Invitrogen ProtoArray High-Density Protein Microarrays
 1 Antibody Profiling on Invitrogen ProtoArray High-Density Protein Microarrays The application of antibodies in research and development, in vitro/in vivo diagnostics, or therapeutics requires that antibodies
1 Antibody Profiling on Invitrogen ProtoArray High-Density Protein Microarrays The application of antibodies in research and development, in vitro/in vivo diagnostics, or therapeutics requires that antibodies
Supplementary Information: Materials and Methods. GST and GST-p53 were purified according to standard protocol after
 Supplementary Information: Materials and Methods Recombinant protein expression and in vitro kinase assay. GST and GST-p53 were purified according to standard protocol after induction with.5mm IPTG for
Supplementary Information: Materials and Methods Recombinant protein expression and in vitro kinase assay. GST and GST-p53 were purified according to standard protocol after induction with.5mm IPTG for
Separose 4 Fast Flow beads stored in 100% isopropanol (Amersham. Bioscience) were washed four times with 1 mm HCl and twice with coupling buffer
 Supporting Information Materials and Methods Affinity chromatography. Three-hundred microliters of N-hydroxysuccinimideactivated Separose 4 Fast Flow beads stored in 100% isopropanol (Amersham Bioscience)
Supporting Information Materials and Methods Affinity chromatography. Three-hundred microliters of N-hydroxysuccinimideactivated Separose 4 Fast Flow beads stored in 100% isopropanol (Amersham Bioscience)
TITLE: Characterization of Odin, a Novel Inhibitory Molecule, in EGF Receptor Signaling
 AD Award Number: W81XWH-05-1-0304 TITLE: Characterization of Odin, a Novel Inhibitory Molecule, in EGF Receptor Signaling PRINCIPAL INVESTIGATOR: Jun Zhong Akhilesh Pandey CONTRACTING ORGANIZATION: Johns
AD Award Number: W81XWH-05-1-0304 TITLE: Characterization of Odin, a Novel Inhibitory Molecule, in EGF Receptor Signaling PRINCIPAL INVESTIGATOR: Jun Zhong Akhilesh Pandey CONTRACTING ORGANIZATION: Johns
Quantitative mass spec based proteomics
 Quantitative mass spec based proteomics Tuula Nyman Institute of Biotechnology tuula.nyman@helsinki.fi THE PROTEOME The complete protein complement expressed by a genome or by a cell or a tissue type (M.
Quantitative mass spec based proteomics Tuula Nyman Institute of Biotechnology tuula.nyman@helsinki.fi THE PROTEOME The complete protein complement expressed by a genome or by a cell or a tissue type (M.
NPTEL VIDEO COURSE PROTEOMICS PROF. SANJEEVA SRIVASTAVA
 LECTURE-26 Interactomics: Yeast two-hybrid immunoprecipitation protein microarrays TRANSCRIPT Welcome to the proteomics course. Today we will talk about interactomics, which is studying interactions of
LECTURE-26 Interactomics: Yeast two-hybrid immunoprecipitation protein microarrays TRANSCRIPT Welcome to the proteomics course. Today we will talk about interactomics, which is studying interactions of
Strategies for Quantitative Proteomics. Atelier "Protéomique Quantitative" La Grande Motte, France - June 26, 2007
 Strategies for Quantitative Proteomics Atelier "Protéomique Quantitative", France - June 26, 2007 Bruno Domon, Ph.D. Institut of Molecular Systems Biology ETH Zurich Zürich, Switzerland OUTLINE Introduction
Strategies for Quantitative Proteomics Atelier "Protéomique Quantitative", France - June 26, 2007 Bruno Domon, Ph.D. Institut of Molecular Systems Biology ETH Zurich Zürich, Switzerland OUTLINE Introduction
NPTEL
 NPTEL Syllabus Proteomics: Principles and Techniques - Video course COURSE OUTLINE An introduction to proteomics: Basics of protein structure and function, An overview of systems biology, Evolution from
NPTEL Syllabus Proteomics: Principles and Techniques - Video course COURSE OUTLINE An introduction to proteomics: Basics of protein structure and function, An overview of systems biology, Evolution from
HPV E6 oncoprotein targets histone methyltransferases for modulating specific. Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu,
 1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
Top down proteomics approaches: (a) to monitor protein purification; (b) to resolve PTM isoforms and protein complexes
 Top down proteomics approaches: (a) to monitor protein purification; (b) to resolve PTM isoforms and protein complexes Jan 11, 2013 BMG744 Helen Kim Department of Pharmacology and Toxicology Targeted Metabolomics
Top down proteomics approaches: (a) to monitor protein purification; (b) to resolve PTM isoforms and protein complexes Jan 11, 2013 BMG744 Helen Kim Department of Pharmacology and Toxicology Targeted Metabolomics
Protein Microarrays?
 Protein Microarrays? Protein Chemistry/Proteomics Unit and the Neuroscience Program,Biomedicum Helsinki E-Mail: marc.baumann@helsinki.fi (http://research.med.helsinki.fi/corefacilities/proteinchem) What
Protein Microarrays? Protein Chemistry/Proteomics Unit and the Neuroscience Program,Biomedicum Helsinki E-Mail: marc.baumann@helsinki.fi (http://research.med.helsinki.fi/corefacilities/proteinchem) What
Lecture 22 Eukaryotic Genes and Genomes III
 Lecture 22 Eukaryotic Genes and Genomes III In the last three lectures we have thought a lot about analyzing a regulatory system in S. cerevisiae, namely Gal regulation that involved a hand full of genes.
Lecture 22 Eukaryotic Genes and Genomes III In the last three lectures we have thought a lot about analyzing a regulatory system in S. cerevisiae, namely Gal regulation that involved a hand full of genes.
Androgen Receptor (Phospho-Tyr363) Colorimetric Cell-Based ELISA Kit. Catalog #: OKAG02138
 Androgen Receptor (Phospho-Tyr363) Colorimetric Cell-Based ELISA Kit Catalog #: OKAG02138 Please read the provided manual entirely prior to use as suggested experimental protocols may have changed. Research
Androgen Receptor (Phospho-Tyr363) Colorimetric Cell-Based ELISA Kit Catalog #: OKAG02138 Please read the provided manual entirely prior to use as suggested experimental protocols may have changed. Research
INOS. Colorimetric Cell-Based ELISA Kit. Catalog #: OKAG00807
 INOS Colorimetric Cell-Based ELISA Kit Catalog #: OKAG00807 Please read the provided manual entirely prior to use as suggested experimental protocols may have changed. Research Purposes Only. Not Intended
INOS Colorimetric Cell-Based ELISA Kit Catalog #: OKAG00807 Please read the provided manual entirely prior to use as suggested experimental protocols may have changed. Research Purposes Only. Not Intended
Proteomics and Mass Spectrometry
 Molecular Cell Biology Lecture. Oct. 18, 2018 Proteomics and Mass Spectrometry Ron Bose, MD PhD Biochemistry and Molecular Cell Biology Programs Lab: Couch Research Building (4515 McKinley), 3 rd floor
Molecular Cell Biology Lecture. Oct. 18, 2018 Proteomics and Mass Spectrometry Ron Bose, MD PhD Biochemistry and Molecular Cell Biology Programs Lab: Couch Research Building (4515 McKinley), 3 rd floor
Protein Purification Products. Complete Solutions for All of Your Protein Purification Applications
 Protein Purification Products Complete Solutions for All of Your Protein Purification Applications FLAG-Tagged Protein Products EXPRESS with the pcmv-dykddddk Vector Set Fuse your protein of interest to
Protein Purification Products Complete Solutions for All of Your Protein Purification Applications FLAG-Tagged Protein Products EXPRESS with the pcmv-dykddddk Vector Set Fuse your protein of interest to
MSD Immuno-Dot-Blot Assays. A division of Meso Scale Diagnostics, LLC.
 MSD Immuno-Dot-Blot Assays Example: High Throughput Western Blots Replacements Traditional Western Blots High content Molecular weight and immunoreactivity Labor and protein intensive Inherently low throughput
MSD Immuno-Dot-Blot Assays Example: High Throughput Western Blots Replacements Traditional Western Blots High content Molecular weight and immunoreactivity Labor and protein intensive Inherently low throughput
User Rules & Regulations Core Facility for Mass Spectrometry and Proteomics (CFMP) at the Centre for Molecular Biology of Heidelberg University (ZMBH)
 User Rules & Regulations Core Facility for Mass Spectrometry and Proteomics (CFMP) at the Centre for Molecular Biology of Heidelberg University (ZMBH) Accepted on 30. January 2017 1 Definition and aims
User Rules & Regulations Core Facility for Mass Spectrometry and Proteomics (CFMP) at the Centre for Molecular Biology of Heidelberg University (ZMBH) Accepted on 30. January 2017 1 Definition and aims
Chapter 3. Exploring Proteins and Proteomes. Dr. Jaroslava Miksovska
 Chapter 3 Exploring Proteins and Proteomes Dr. Jaroslava Miksovska Chapter objectives: Protein purification and isolation techniques Protein sequencing Immunology for protein identification Protein identification
Chapter 3 Exploring Proteins and Proteomes Dr. Jaroslava Miksovska Chapter objectives: Protein purification and isolation techniques Protein sequencing Immunology for protein identification Protein identification
Molecular Techniques. 3 Goals in Molecular Biology. Nucleic Acids: DNA and RNA. Disclaimer Nucleic Acids Proteins
 Molecular Techniques Disclaimer Nucleic Acids Proteins Houpt, CMN, 9-30-11 3 Goals in Molecular Biology Identify All nucleic acids (and proteins) are chemically identical in aggregate - need to identify
Molecular Techniques Disclaimer Nucleic Acids Proteins Houpt, CMN, 9-30-11 3 Goals in Molecular Biology Identify All nucleic acids (and proteins) are chemically identical in aggregate - need to identify
Qualification of Antibody- Cross-Reactivity Using HuProt TM Protein Microarray Version 3.0
 Qualification of Antibody- Cross-Reactivity Using HuProt TM Protein Microarray Version 3.0 Contents Cynthia French, Krista Trappl-Kimmons, Arlo Randall, Gary Hermanson and Tim Le October 2017 Introduction...
Qualification of Antibody- Cross-Reactivity Using HuProt TM Protein Microarray Version 3.0 Contents Cynthia French, Krista Trappl-Kimmons, Arlo Randall, Gary Hermanson and Tim Le October 2017 Introduction...
Bioinformatics (Lec 19) Picture Copyright: the National Museum of Health
 3/29/05 1 Picture Copyright: AccessExcellence @ the National Museum of Health PCR 3/29/05 2 Schematic outline of a typical PCR cycle Target DNA Primers dntps DNA polymerase 3/29/05 3 Gel Electrophoresis
3/29/05 1 Picture Copyright: AccessExcellence @ the National Museum of Health PCR 3/29/05 2 Schematic outline of a typical PCR cycle Target DNA Primers dntps DNA polymerase 3/29/05 3 Gel Electrophoresis
Center for Mass Spectrometry and Proteomics Phone (612) (612)
 Outline Database search types Peptide Mass Fingerprint (PMF) Precursor mass-based Sequence tag Results comparison across programs Manual inspection of results Terminology Mass tolerance MS/MS search FASTA
Outline Database search types Peptide Mass Fingerprint (PMF) Precursor mass-based Sequence tag Results comparison across programs Manual inspection of results Terminology Mass tolerance MS/MS search FASTA
Microarray Industry Products
 Via Nicaragua, 12-14 00040 Pomezia (Roma) Phone: +39 06 91601628 Fax: +39 06 91612477 info@lifelinelab.com www.lifelinelab.com Microarray Industry Products Page 10 NBT / BCPIP Chromogenic phosphatase
Via Nicaragua, 12-14 00040 Pomezia (Roma) Phone: +39 06 91601628 Fax: +39 06 91612477 info@lifelinelab.com www.lifelinelab.com Microarray Industry Products Page 10 NBT / BCPIP Chromogenic phosphatase
Stable Isotope Labeling with Amino Acids in Cell Culture. Thermo Scientific Pierce SILAC Protein Quantitation Kits
 Stable Isotope Labeling with Amino Acids in Cell Culture Thermo Scientific Pierce SILAC Protein Quantitation Kits Quantitative Analysis of Differential Protein Expression Stable isotope labeling using
Stable Isotope Labeling with Amino Acids in Cell Culture Thermo Scientific Pierce SILAC Protein Quantitation Kits Quantitative Analysis of Differential Protein Expression Stable isotope labeling using
sample preparation Accessing Low-Abundance Proteins in Serum and Plasma With a Novel, Simple Enrichment and Depletion Method tech note 5632
 sample preparation tech note 5632 Accessing Low-Abundance Proteins in Serum and Plasma With a Novel, Simple Enrichment and Depletion Method Aran Paulus, Steve Freeby, Katrina Academia, Vanitha Thulasiraman,
sample preparation tech note 5632 Accessing Low-Abundance Proteins in Serum and Plasma With a Novel, Simple Enrichment and Depletion Method Aran Paulus, Steve Freeby, Katrina Academia, Vanitha Thulasiraman,
