Identification of Microprotein-Protein Interactions via APEX Tagging
|
|
|
- Samantha Russell
- 6 years ago
- Views:
Transcription
1 Supporting Information Identification of Microprotein-Protein Interactions via APEX Tagging Qian Chu, Annie Rathore,, Jolene K. Diedrich,, Cynthia J. Donaldson, John R. Yates III, and Alan Saghatelian *, The Salk Institute for Biological Studies, Clayton Foundation Laboratories for Peptide Biology, N. Torrey Pines Rd, La Jolla, CA 92037, USA Division of Biological Sciences, University of California San Diego, 9500 Gilman Dr., La Jolla, CA 92093, USA Department of Chemical Physiology, The Scripps Research Institute, N. Torrey Pines Rd, La Jolla, CA 92037, USA * To whom correspondence should be addressed. asaghatelian@salk.edu (A.S.) S1
2 Experimental Section Mass Spectrometry and Data Analysis The digested samples were analyzed on a Q Exactive mass spectrometer (Thermo). The digest was injected directly onto a 30cm, 75µm ID column packed with BEH 1.7µm C18 resin (Waters). Samples were separated at a flow rate of 200nl/min on a nlc 1000 (Thermo). Buffer A and B were 0.1% formic acid in water and acetonitrile, respectively. A gradient of 5-40%B over 110min, an increase to 50%B over 10min, an increase to 90%B over another 10min and held at 90%B for a final 10min of washing was used for 140min total run time. Column was re-equilibrated with 20µl of buffer A prior to the injection of sample. Peptides were eluted directly from the tip of the column and nanosprayed directly into the mass spectrometer by application of 2.5kV voltage at the back of the column. The Q Exactive was operated in a data dependent mode. Full MS 1 scans were collected in the Orbitrap at 70K resolution with a mass range of 400 to 1800 m/z and an AGC target of 5e 6. The ten most abundant ions per scan were selected for MS/MS analysis with HCD fragmentation of 25NCE, an AGC target of 5e 6 and minimum intensity of 4e 3. Maximum fill times were set to 60ms and 120ms for MS and MS/MS scans respectively. Quadrupole isolation of 2.0m/z was used, dynamic exclusion was set to 15 sec and unassigned charge states were excluded. Protein and peptide identification were done with Integrated Proteomics Pipeline IP2 (Integrated Proteomics Applications). Tandem mass spectra were extracted from raw files using RawConverter 1 and searched with ProLuCID 2 against human UniProt database appended with microprotein sequences. The search space included all fullytryptic and half-tryptic peptide candidates with maximum of two missed cleavages. Carbamidomethylation on cysteine was counted as a static modification. Data was searched with 50 ppm precursor ion tolerance and 50 ppm fragment ion tolerance. Data was filtered to 10 ppm precursor ion tolerance post search. Identified proteins were filtered using DTASelect 3 and utilizing a target-decoy database search strategy to control the false discovery rate to 1% at the protein level. S2
3 Materials Primary antibodies: rabbit anti-myc tag (#2272, Cell Signaling Technology), mouse anti- FLAG (F1804, Sigma), rabbit anti-flag (#2368, Cell signaling Technology), anti-biotin, HRP-linked antibody (#7075, Cell Signaling Technology), rabbit anti-ku70 (#4104, Cell Signaling Technology) and rabbit anti-ku80 (#2180, Cell Signaling Technology), mouse anti-ha (H9658, Sigma). Secondary antibodies: goat anti-rabbit IRDye 800CW ( , LI-COR), goat antimouse IRDye 800CW ( , LI-COR), Goat anti-mouse Alexa Fluor 647 (A21235, Life Technologies), Goat anti-rabbit Alexa Fluor 488 (A11008, Life Technologies). Plasmids pcdna3-mri-1 was generated as previously described 4. pcdna3-apex-nes was a gift from Alice Ting (Addgene plasmid #49386). pcdna3-flag-apex was generated by inserting a stop codon before NES sequence using QuikChange II kit (Agilent Technologies). APEX coding sequence was subcloned into pcdna3.1(+) vector with an N-terminal myc-tag to generate pcdna3.1-myc-apex construct. C11orf98 microprotein coding sequence was obtained by PCR amplification of HEK293T cdna pool. FLAG- MRI-APEX, C11orf98-APEX-myc and myc-apex-c11orf98 were amplified by overlap extension PCR and subcloned into pcdna3.1(+) vector. NPM1 cdna (BC050628) was obtained from MGC cdna library at the Salk Institute, and subcloned into pcdna3.1(+) vector with a C-terminal HA tag. S3
4 Figure S1. Loading control of APEX and MRI-APEX FLAG immunoprecipitation (Figure 2B). Anti-FLAG immunoprecipitation of HEK293T cells expressing APEX or MRI-APEX. Eluted proteins were separated by SDS-PAGE and visualized by Coomassie staining. (WCL = whole cell lysate) S4
5 Figure S2. Biotinylated proteomes of untransfected, APEX and MRI-APEX transfected cells. HEK293T cells were transfected with FLAG-MRI-APEX or APEX control construct. 24 hours post-transfection, cell culture medium was changed to fresh growth media containing 500 µm biotin-tyramide. After incubation for 30 min, H 2 O 2 was added at a final concentration of 1 mm and treated for 1 min. Cells were then lysed and biotinylated proteins were enriched by streptavidin beads and analyzed by Western blotting using anti-biotin antibody, A) short exposure and B) long exposure. Biotin-ladder was purchase from Cell Signaling #7727. S5
6 Figure S3. Loading control of MRI-APEX biotinylation (Figure 2C and S2). HEK293T cells were transfected with FLAG-MRI-APEX or APEX control construct. 24 hours post-transfection, cell culture medium was changed to fresh growth media containing 500 µm biotin-tyramide. After incubation for 30 min, H 2 O 2 was added at a final concentration of 1 mm and treated for 1 min. Cells were then lysed and biotinylated proteins were enriched by streptavidin beads and analyzed by Coomassie staining. S6
7 Figure S4. Schematic illustration of APEX control and C11orf98-APEX constructs. S7
8 Figure S5. The majority of proteins enriched by APEX-C11orf98 and C11orf98- APEX are reported to have a nuclear localization. S8
9 Figure S6. Loading control of C11orf98-APEX and APEX-C11orf98 biotinylation (Figure 4A). HEK293T cells were transfected with C11orf98-APEX, APEX-C11orf98 or APEX control construct. 24 hours post-transfection, cell culture medium was changed to fresh growth media containing 500 µm biotin-tyramide. After incubation for 30 min, H 2 O 2 was added at a final concentration of 1 mm and treated for 1 min. Cells were then lysed and biotinylated proteins were enriched by streptavidin beads and analyzed by Coomassie staining. S9
10 Figure S7. Validation of C11orf98 interaction with NPM1 and NCL by FLAG immunoprecipitation. HEK293T cells were transfected with C11orf98-FLAG or pcdna3.1(+) empty vector. 48 hours post-transfection, cells were harvested and cell lysates were subjected to FLAG immunoprecipitation by anti-flag M2 affinity gel. Bound proteins were eluted by incubating the beads with 3 FLAG peptide for 1 hour and analyzed by Western blotting with indicated antibodies. S10
11 Figure S8. Loading control of C11orf98-FLAG FLAG immunoprecipitation (Figure S7). Anti-FLAG immunoprecipitation of HEK293T cells expressing C11orf98-FLAG or pcdna3.1 control vector. Eluted proteins were separated by SDS-PAGE and visualized by Coomassie staining. (WCL = whole cell lysate) S11
12 Figure S9. Loading control of reciprocal anti-ha immunoprecipitation (Figure 4B). Reciprocal anti-ha immunoprecipitation of NPM1-HA from HEK293T cells co-expressing C11orf98-FLAG, with cells expressing C11orf98-FLAG alone as a control. Eluted proteins were analyzed by Coomassie staining. S12
13 Figure S10. Validation of C11orf98 interaction with NPM1 and NCL by reciprocal immunoprecipitation. HEK293T cells were co-transfected with C11orf98-FLAG and NPM1-HA. 48 hours post-transfection, cell lysates were subjected to immunoprecipitation using anti-ha agarose beads (mouse IgG beads as control). Bound proteins were eluted and separated by SDS-PAGE. NPM1 and C11orf98 proteins were visualized by Western blotting using anti-ha and anti-flag antibodies. S13
14 Figure S11. Loading control of reciprocal anti-ha immunoprecipitation (Figure S10). HEK293T cells were co-transfected with C11orf98-FLAG and NPM1-HA. 48 hours post-transfection, cell lysates were subjected to immunoprecipitation using anti-ha agarose beads (mouse IgG beads as control). Bound proteins were eluted and separated by SDS-PAGE. NPM1 and C11orf98 proteins were visualized by Coomassie staining. S14
15 Figure S12. C11orf98 microprotein was identified in public protein interaction database. Searching IP-MS raw data of NPM1 (from Mann s dataset and Gygi s Bioplex) and NCL (from Gygi s Bioplex) revealed multiple A) spectral counts and B) peptide fragments of C11orf98 microprotein. S15
16 References: (1) He, L.; Diedrich, J.; Chu, Y. Y. and Yates, J. R., 3rd (2015) Extracting Accurate Precursor Information for Tandem Mass Spectra by RawConverter. Anal. Chem. 87, (2) Xu, T.; Park, S. K.; Venable, J. D.; Wohlschlegel, J. A.; Diedrich, J. K.; Cociorva, D.; Lu, B.; Liao, L.; Hewel, J.; Han, X.; Wong, C. C.; Fonslow, B.; Delahunty, C.; Gao, Y.; Shah, H. and Yates, J. R., 3rd (2015) ProLuCID: An improved SEQUEST-like algorithm with enhanced sensitivity and specificity. J. Proteomics 129, (3) Tabb, D. L.; McDonald, W. H. and Yates, J. R., 3rd (2002) DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1, (4) Slavoff, S. A.; Heo, J.; Budnik, B. A.; Hanakahi, L. A. and Saghatelian, A. (2014) A human short open reading frame (sorf)-encoded polypeptide that stimulates DNA end joining. J. Biol. Chem. 289, S16
SUPPLEMENTARY INFORMATION
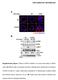 SUPPLEMENTARY INFORMATION Supplementary Figure 1 Effect of ROCK inhibition on lumen abnormality in MDCK cysts. (A) MDCK cells as indicated cultured in Matrigel were treated with and without Y27632 (10
SUPPLEMENTARY INFORMATION Supplementary Figure 1 Effect of ROCK inhibition on lumen abnormality in MDCK cysts. (A) MDCK cells as indicated cultured in Matrigel were treated with and without Y27632 (10
Filter-based Protein Digestion (FPD): A Detergent-free and Scaffold-based Strategy for TMT workflows
 Supporting Information Filter-based Protein Digestion (FPD): A Detergent-free and Scaffold-based Strategy for TMT workflows Ekaterina Stepanova 1, Steven P. Gygi 1, *, Joao A. Paulo 1, * 1 Department of
Supporting Information Filter-based Protein Digestion (FPD): A Detergent-free and Scaffold-based Strategy for TMT workflows Ekaterina Stepanova 1, Steven P. Gygi 1, *, Joao A. Paulo 1, * 1 Department of
Objective. Introduction. IP assisted LC/MS/MS making study protein complexes easy. Jon Hao 1, Yi Liu 1, Xiaozhi Ren 2, and King-Wai Yau 2
 IP assisted LC/MS/MS making study protein complexes easy Jon Hao 1, Yi Liu 1, Xiaozhi Ren 2, and King-Wai Yau 2 1 Poochon Scientific, Frederick, Maryland 21701 2 Department of Neuroscience, Johns Hopkins
IP assisted LC/MS/MS making study protein complexes easy Jon Hao 1, Yi Liu 1, Xiaozhi Ren 2, and King-Wai Yau 2 1 Poochon Scientific, Frederick, Maryland 21701 2 Department of Neuroscience, Johns Hopkins
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. FANCD2 Western blot. Lane 1 marker, lane 2 LN9SV, lane 3 LN9SV treated with hydroxyurea (HU), lane 4 GM6914, lane 5 GM6914 treated with HU, lane 6 VU697L,
Supplementary Figures Supplementary Figure 1. FANCD2 Western blot. Lane 1 marker, lane 2 LN9SV, lane 3 LN9SV treated with hydroxyurea (HU), lane 4 GM6914, lane 5 GM6914 treated with HU, lane 6 VU697L,
Supplementary information, Figure S1A ShHTL7 interacted with MAX2 but not another F-box protein COI1.
 GR24 (μm) 0 20 0 20 GST-ShHTL7 anti-gst His-MAX2 His-COI1 PVDF staining Supplementary information, Figure S1A ShHTL7 interacted with MAX2 but not another F-box protein COI1. Pull-down assays using GST-ShHTL7
GR24 (μm) 0 20 0 20 GST-ShHTL7 anti-gst His-MAX2 His-COI1 PVDF staining Supplementary information, Figure S1A ShHTL7 interacted with MAX2 but not another F-box protein COI1. Pull-down assays using GST-ShHTL7
A highly sensitive and robust 150 µm column to enable high-throughput proteomics
 APPLICATION NOTE 21744 Robust LC Separation Optimized MS Acquisition Comprehensive Data Informatics A highly sensitive and robust 15 µm column to enable high-throughput proteomics Authors Xin Zhang, 1
APPLICATION NOTE 21744 Robust LC Separation Optimized MS Acquisition Comprehensive Data Informatics A highly sensitive and robust 15 µm column to enable high-throughput proteomics Authors Xin Zhang, 1
Supplementary Figure 1. Immunoprecipitation of synthetic SUMOm-remnant peptides using UMO monoclonal antibody. (a) LC-MS analyses of tryptic
 Supplementary Figure 1. Immunoprecipitation of synthetic SUMOm-remnant peptides using UMO 1-7-7 monoclonal antibody. (a) LC-MS analyses of tryptic digest from HEK293 cells spiked with 6 SUMOmremnant peptides
Supplementary Figure 1. Immunoprecipitation of synthetic SUMOm-remnant peptides using UMO 1-7-7 monoclonal antibody. (a) LC-MS analyses of tryptic digest from HEK293 cells spiked with 6 SUMOmremnant peptides
Automated platform of µlc-ms/ms using SAX trap column for
 Analytical and Bioanalytical Chemistry Electronic Supplementary Material Automated platform of µlc-ms/ms using SAX trap column for highly efficient phosphopeptide analysis Xionghua Sun, Xiaogang Jiang
Analytical and Bioanalytical Chemistry Electronic Supplementary Material Automated platform of µlc-ms/ms using SAX trap column for highly efficient phosphopeptide analysis Xionghua Sun, Xiaogang Jiang
Nature Methods: doi: /nmeth Supplementary Figure 1
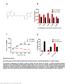 Supplementary Figure 1 Enrichment with anti-biotin antibody significantly increases detection of biotinylated peptides in complex samples. (A) Structure of NHS-biotin (B) Results of spike-in studies. Bar
Supplementary Figure 1 Enrichment with anti-biotin antibody significantly increases detection of biotinylated peptides in complex samples. (A) Structure of NHS-biotin (B) Results of spike-in studies. Bar
The effect of simulated microgravity on the Brassica napus seedling proteome
 10.1071/FP16378_AC CSIRO 2018 Supplementary Material: Functional Plant Biology, 2018, 45(4), 440 452. Supplementary Material The effect of simulated microgravity on the Brassica napus seedling proteome
10.1071/FP16378_AC CSIRO 2018 Supplementary Material: Functional Plant Biology, 2018, 45(4), 440 452. Supplementary Material The effect of simulated microgravity on the Brassica napus seedling proteome
Supplementary Fig. 1 Identification of Nedd4 as an IRS-2-associated protein in camp-treated FRTL-5 cells.
 Supplementary Fig. 1 Supplementary Fig. 1 Identification of Nedd4 as an IRS-2-associated protein in camp-treated FRTL-5 cells. (a) FRTL-5 cells were treated with 1 mm dibutyryl camp for 24 h, and the lysates
Supplementary Fig. 1 Supplementary Fig. 1 Identification of Nedd4 as an IRS-2-associated protein in camp-treated FRTL-5 cells. (a) FRTL-5 cells were treated with 1 mm dibutyryl camp for 24 h, and the lysates
Appendix. Table of contents
 Appendix Table of contents 1. Appendix figures 2. Legends of Appendix figures 3. References Appendix Figure S1 -Tub STIL (41) DVFFYQADDEHYIPR (55) (64) AVLLDLEPR (72) 1 451aa (1071) YLNENQLSQLSVTR (1084)
Appendix Table of contents 1. Appendix figures 2. Legends of Appendix figures 3. References Appendix Figure S1 -Tub STIL (41) DVFFYQADDEHYIPR (55) (64) AVLLDLEPR (72) 1 451aa (1071) YLNENQLSQLSVTR (1084)
ENCODE DCC Antibody Validation Document
 ENCODE DCC Antibody Validation Document Date of Submission 09/12/12 Name: Trupti Kawli Email: trupti@stanford.edu Lab Snyder Antibody Name: SREBP1 (sc-8984) Target: SREBP1 Company/ Source: Santa Cruz Biotechnology
ENCODE DCC Antibody Validation Document Date of Submission 09/12/12 Name: Trupti Kawli Email: trupti@stanford.edu Lab Snyder Antibody Name: SREBP1 (sc-8984) Target: SREBP1 Company/ Source: Santa Cruz Biotechnology
Supplementary Figure S1 Purification of deubiquitinases HEK293 cells were transfected with the indicated DUB-expressing plasmids.
 Supplementary Figure S1 Purification of deubiquitinases HEK293 cells were transfected with the indicated DUB-expressing plasmids. The cells were harvested 72 h after transfection. FLAG-tagged deubiquitinases
Supplementary Figure S1 Purification of deubiquitinases HEK293 cells were transfected with the indicated DUB-expressing plasmids. The cells were harvested 72 h after transfection. FLAG-tagged deubiquitinases
Supplementary Tables. Note: Open-pFind is embedded as the default open search workflow of the pfind tool. Nature Biotechnology: doi: /nbt.
 Supplementary Tables Supplementary Table 1. Detailed information for the six datasets used in this study Dataset Mass spectrometer # Raw files # MS2 scans Reference Dong-Ecoli-QE Q Exactive 5 202,452 /
Supplementary Tables Supplementary Table 1. Detailed information for the six datasets used in this study Dataset Mass spectrometer # Raw files # MS2 scans Reference Dong-Ecoli-QE Q Exactive 5 202,452 /
High Resolution Accurate Mass Peptide Quantitation on Thermo Scientific Q Exactive Mass Spectrometers. The world leader in serving science
 High Resolution Accurate Mass Peptide Quantitation on Thermo Scientific Q Exactive Mass Spectrometers The world leader in serving science Goals Explore the capabilities of High Resolution Accurate Mass
High Resolution Accurate Mass Peptide Quantitation on Thermo Scientific Q Exactive Mass Spectrometers The world leader in serving science Goals Explore the capabilities of High Resolution Accurate Mass
ENCODE DCC Antibody Validation Document
 ENCODE DCC Antibody Validation Document Date of Submission 06/15/2012 Name: Trupti Kawli Email: trupti@stanford.edu Lab Snyder Antibody Name: CDP (sc6327) Target: CDP Company/ Source: Santa Cruz Catalog
ENCODE DCC Antibody Validation Document Date of Submission 06/15/2012 Name: Trupti Kawli Email: trupti@stanford.edu Lab Snyder Antibody Name: CDP (sc6327) Target: CDP Company/ Source: Santa Cruz Catalog
Supplementary Online Material. An Expanded Eukaryotic Genetic Code 2QH, UK. * To whom correspondence should be addressed.
 Supplementary Online Material An Expanded Eukaryotic Genetic Code Jason W. Chin 1, T. Ashton Cropp, J. Christopher Anderson, Mridul Mukherji, Zhiwen Zhang and Peter G. Schultz* Department of Chemistry
Supplementary Online Material An Expanded Eukaryotic Genetic Code Jason W. Chin 1, T. Ashton Cropp, J. Christopher Anderson, Mridul Mukherji, Zhiwen Zhang and Peter G. Schultz* Department of Chemistry
Cell Signaling Technology
 Cell Signaling Technology PTMScan Direct: Multipathway v2.0 Proteomics Service Group January 14, 2013 PhosphoScan Deliverables Project Overview Methods PTMScan Direct: Multipathway V2.0 (Tables 1,2) Qualitative
Cell Signaling Technology PTMScan Direct: Multipathway v2.0 Proteomics Service Group January 14, 2013 PhosphoScan Deliverables Project Overview Methods PTMScan Direct: Multipathway V2.0 (Tables 1,2) Qualitative
Executing T-REX in mammalian cells
 Supplementary Figure 1 Executing T-REX in mammalian cells HEK-293 cells cultured (a) in 2x 55 cm 2 adherent cell culture plates, and (b and c) in a 48-well multi-well adherent cell culture plate. No cover
Supplementary Figure 1 Executing T-REX in mammalian cells HEK-293 cells cultured (a) in 2x 55 cm 2 adherent cell culture plates, and (b and c) in a 48-well multi-well adherent cell culture plate. No cover
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Han et al., http://www.jcb.org/cgi/content/full/jcb.201311007/dc1 Figure S1. SIVA1 interacts with PCNA. (A) HEK293T cells were transiently
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Han et al., http://www.jcb.org/cgi/content/full/jcb.201311007/dc1 Figure S1. SIVA1 interacts with PCNA. (A) HEK293T cells were transiently
This is the author's accepted version of the manuscript.
 This is the author's accepted version of the manuscript. The definitive version is published in Nature Communications Online Edition: 2015/4/16 (Japan time), doi:10.1038/ncomms7780. The final version published
This is the author's accepted version of the manuscript. The definitive version is published in Nature Communications Online Edition: 2015/4/16 (Japan time), doi:10.1038/ncomms7780. The final version published
Antibodies against PCNA were previously described [1]. To deplete PCNA from Xenopus egg
![Antibodies against PCNA were previously described [1]. To deplete PCNA from Xenopus egg Antibodies against PCNA were previously described [1]. To deplete PCNA from Xenopus egg](/thumbs/84/90687567.jpg) Supplementary information Supplementary methods PCNA antibody and immunodepletion Antibodies against PCNA were previously described [1]. To deplete PCNA from Xenopus egg extracts, one volume of protein
Supplementary information Supplementary methods PCNA antibody and immunodepletion Antibodies against PCNA were previously described [1]. To deplete PCNA from Xenopus egg extracts, one volume of protein
Spectral Counting Approaches and PEAKS
 Spectral Counting Approaches and PEAKS INBRE Proteomics Workshop, April 5, 2017 Boris Zybailov Department of Biochemistry and Molecular Biology University of Arkansas for Medical Sciences 1. Introduction
Spectral Counting Approaches and PEAKS INBRE Proteomics Workshop, April 5, 2017 Boris Zybailov Department of Biochemistry and Molecular Biology University of Arkansas for Medical Sciences 1. Introduction
Computing with large data sets
 Computing with large data sets Richard Bonneau, spring 2009 Lecture 14 (week 8): genomics 1 Central dogma Gene expression DNA RNA Protein v22.0480: computing with data, Richard Bonneau Lecture 14 places
Computing with large data sets Richard Bonneau, spring 2009 Lecture 14 (week 8): genomics 1 Central dogma Gene expression DNA RNA Protein v22.0480: computing with data, Richard Bonneau Lecture 14 places
Title: A Human Microprotein that Interacts with the mrna Decapping Complex
 Supplementary Information Title: A Human Microprotein that Interacts with the mrna Decapping Complex Authors: Nadia G. D Lima 1, Jiao Ma 2, *, Lauren Winkler 1, *, Qian Chu 2, Ken H. Loh 3, Elizabeth O.
Supplementary Information Title: A Human Microprotein that Interacts with the mrna Decapping Complex Authors: Nadia G. D Lima 1, Jiao Ma 2, *, Lauren Winkler 1, *, Qian Chu 2, Ken H. Loh 3, Elizabeth O.
Application Note. Kwasi Antwi, Amanda Ribar, Urban A. Kiernan, and Eric E. Niederkofler Thermo Fisher Scientific, Tempe, Arizona
 Analysis of an Antibody Drug Conjugate using MSIA D.A.R.T. S. Technology, an Integral Part of Ligand Binding Mass Spectrometric Immunoassay (LB-MSIA) Workflow Application Note Kwasi Antwi, Amanda Ribar,
Analysis of an Antibody Drug Conjugate using MSIA D.A.R.T. S. Technology, an Integral Part of Ligand Binding Mass Spectrometric Immunoassay (LB-MSIA) Workflow Application Note Kwasi Antwi, Amanda Ribar,
Click-&-Go TM Protein Enrichment Kit *for capture alkyne-modified proteins*
 Click-&-Go TM Protein Enrichment Kit *for capture alkyne-modified proteins* Product No. 1039 Introduction The Click-&-Go TM Protein Enrichment Kit is an efficient, biotin-streptavidin free tool for capture
Click-&-Go TM Protein Enrichment Kit *for capture alkyne-modified proteins* Product No. 1039 Introduction The Click-&-Go TM Protein Enrichment Kit is an efficient, biotin-streptavidin free tool for capture
A Highly Accurate Mass Profiling Approach to Protein Biomarker Discovery Using HPLC-Chip/ MS-Enabled ESI-TOF MS
 Application Note PROTEOMICS METABOLOMICS GENOMICS INFORMATICS GLYILEVALCYSGLUGLNALASERLEUASPARG CYSVALLYSPROLYSPHETYRTHRLEUHISLYS A Highly Accurate Mass Profiling Approach to Protein Biomarker Discovery
Application Note PROTEOMICS METABOLOMICS GENOMICS INFORMATICS GLYILEVALCYSGLUGLNALASERLEUASPARG CYSVALLYSPROLYSPHETYRTHRLEUHISLYS A Highly Accurate Mass Profiling Approach to Protein Biomarker Discovery
Supplementary data. sienigma. F-Enigma F-EnigmaSM. a-p53
 Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Supplementary information
 Supplementary information Table of Content: Supplementary Results... 2 Supplementary Figure S1: Experimental validation of AP-MS results by coimmunprecipitation Western blot analysis.... 3 Supplementary
Supplementary information Table of Content: Supplementary Results... 2 Supplementary Figure S1: Experimental validation of AP-MS results by coimmunprecipitation Western blot analysis.... 3 Supplementary
Supporting Information. for. Development and Application of a Quantitative Multiplexed Small GTPase Activity. Assay Using Targeted Proteomics
 Supporting Information for Development and Application of a Quantitative Multiplexed Small GTPase Activity Assay Using Targeted Proteomics Cheng-Cheng Zhang, 1 Ru Li, 1,2 Honghui Jiang, 1,2 Shujun Lin
Supporting Information for Development and Application of a Quantitative Multiplexed Small GTPase Activity Assay Using Targeted Proteomics Cheng-Cheng Zhang, 1 Ru Li, 1,2 Honghui Jiang, 1,2 Shujun Lin
SANTA CRUZ BIOTECHNOLOGY, INC.
 TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
Supplementary Information: Materials and Methods. GST and GST-p53 were purified according to standard protocol after
 Supplementary Information: Materials and Methods Recombinant protein expression and in vitro kinase assay. GST and GST-p53 were purified according to standard protocol after induction with.5mm IPTG for
Supplementary Information: Materials and Methods Recombinant protein expression and in vitro kinase assay. GST and GST-p53 were purified according to standard protocol after induction with.5mm IPTG for
Perform reproducible immunoprecipitation in less than 40 minutes
 Perform reproducible immunoprecipitation in less than 40 minutes Dynabeads products Immunoprecipitation made easy with low nonspecific binding, high yield, and reproducibility Magnetic beads have become
Perform reproducible immunoprecipitation in less than 40 minutes Dynabeads products Immunoprecipitation made easy with low nonspecific binding, high yield, and reproducibility Magnetic beads have become
Strategies for Quantitative Proteomics. Atelier "Protéomique Quantitative" La Grande Motte, France - June 26, 2007
 Strategies for Quantitative Proteomics Atelier "Protéomique Quantitative", France - June 26, 2007 Bruno Domon, Ph.D. Institut of Molecular Systems Biology ETH Zurich Zürich, Switzerland OUTLINE Introduction
Strategies for Quantitative Proteomics Atelier "Protéomique Quantitative", France - June 26, 2007 Bruno Domon, Ph.D. Institut of Molecular Systems Biology ETH Zurich Zürich, Switzerland OUTLINE Introduction
ProteinPilot Report for ProteinPilot Software
 ProteinPilot Report for ProteinPilot Software Detailed Analysis of Protein Identification / Quantitation Results Automatically Sean L Seymour, Christie Hunter SCIEX, USA Powerful mass spectrometers like
ProteinPilot Report for ProteinPilot Software Detailed Analysis of Protein Identification / Quantitation Results Automatically Sean L Seymour, Christie Hunter SCIEX, USA Powerful mass spectrometers like
SUPPLEMENTARY INFORMATION
 (Supplementary Methods and Materials) GST pull-down assay GST-fusion proteins Fe65 365-533, and Fe65 538-700 were expressed in BL21 bacterial cells and purified with glutathione-agarose beads (Sigma).
(Supplementary Methods and Materials) GST pull-down assay GST-fusion proteins Fe65 365-533, and Fe65 538-700 were expressed in BL21 bacterial cells and purified with glutathione-agarose beads (Sigma).
iprg-2016 Proteome Informatics Research Group Study: Inferring Proteoforms from Bottom-up Proteomics Data
 iprg-2016 Proteome Informatics Research Group Study: Inferring Proteoforms from Bottom-up Proteomics Data The ABRF iprg Research Group October 10, 2016 Dear iprg 2016 Study Participant, Thank you for participating
iprg-2016 Proteome Informatics Research Group Study: Inferring Proteoforms from Bottom-up Proteomics Data The ABRF iprg Research Group October 10, 2016 Dear iprg 2016 Study Participant, Thank you for participating
Separose 4 Fast Flow beads stored in 100% isopropanol (Amersham. Bioscience) were washed four times with 1 mm HCl and twice with coupling buffer
 Supporting Information Materials and Methods Affinity chromatography. Three-hundred microliters of N-hydroxysuccinimideactivated Separose 4 Fast Flow beads stored in 100% isopropanol (Amersham Bioscience)
Supporting Information Materials and Methods Affinity chromatography. Three-hundred microliters of N-hydroxysuccinimideactivated Separose 4 Fast Flow beads stored in 100% isopropanol (Amersham Bioscience)
Figure S1. Sequence alignments of ATRIP and ATR TopBP1 interacting regions.
 A H. sapiens 204 TKLQTS--ERANKLAAPSVSH VSPRKNPSVVIKPEACS-PQFGKTSFPTKESFSANMS LP 259 B. taurus 201 TKLQSS--ERANKLAVPTVSH VSPRKSPSVVIKPEACS-PQFGKPSFPTKESFSANKS LP 257 M. musculus 204 TKSQSN--GRTNKPAAPSVSH
A H. sapiens 204 TKLQTS--ERANKLAAPSVSH VSPRKNPSVVIKPEACS-PQFGKTSFPTKESFSANMS LP 259 B. taurus 201 TKLQSS--ERANKLAVPTVSH VSPRKSPSVVIKPEACS-PQFGKPSFPTKESFSANKS LP 257 M. musculus 204 TKSQSN--GRTNKPAAPSVSH
Supplementary Figure S1 Supplementary Figure S2 Supplementary Figure S3. Supplementary Figure S4
 Supplementary Figure S1 Supplementary Figure S2 Supplementary Figure S3 Supplementary Figure S4 Supplementary Figure S5 Supplementary Figure S6 Supplementary Figure S7 Supplementary Figure S8 Supplementary
Supplementary Figure S1 Supplementary Figure S2 Supplementary Figure S3 Supplementary Figure S4 Supplementary Figure S5 Supplementary Figure S6 Supplementary Figure S7 Supplementary Figure S8 Supplementary
SUPPLEMENTAL MATERIAL. Supplemental Methods:
 SUPPLEMENTAL MATERIAL Supplemental Methods: Immunoprecipitation- As we described but with some modifications [22]. As part of another ongoing project, lysate from human umbilical vein endothelial cells
SUPPLEMENTAL MATERIAL Supplemental Methods: Immunoprecipitation- As we described but with some modifications [22]. As part of another ongoing project, lysate from human umbilical vein endothelial cells
Algorithm for Matching Additional Spectra
 Improved Methods for Comprehensive Sample Analysis Using Protein Prospector Peter R. Baker 1, Katalin F. Medzihradszky 1 and Alma L. Burlingame 1 1 Mass Spectrometry Facility, Dept. of Pharmaceutical Chemistry,
Improved Methods for Comprehensive Sample Analysis Using Protein Prospector Peter R. Baker 1, Katalin F. Medzihradszky 1 and Alma L. Burlingame 1 1 Mass Spectrometry Facility, Dept. of Pharmaceutical Chemistry,
Click Chemistry Capture Kit
 http://www.clickchemistrytools.com tel: 480 584 3340 fax: 866 717 2037 Click Chemistry Capture Kit Product No. 1065 Introduction The Click Chemistry Capture Kit provides all the necessary reagents* to
http://www.clickchemistrytools.com tel: 480 584 3340 fax: 866 717 2037 Click Chemistry Capture Kit Product No. 1065 Introduction The Click Chemistry Capture Kit provides all the necessary reagents* to
Application Note 639. Daniel Lopez-Ferrer 1, Michael Blank 1, Stephan Meding 2, Aran Paulus 1, Romain Huguet 1, Remco Swart 2, Andreas FR Huhmer 1
 Pushing the Limits of Bottom-Up Proteomics with State-Of-The-Art Capillary UHPLC and Orbitrap Mass Spectrometry for Reproducible Quantitation of Proteomes Daniel Lopez-Ferrer 1, Michael Blank 1, Stephan
Pushing the Limits of Bottom-Up Proteomics with State-Of-The-Art Capillary UHPLC and Orbitrap Mass Spectrometry for Reproducible Quantitation of Proteomes Daniel Lopez-Ferrer 1, Michael Blank 1, Stephan
Supplementary Fig. 1. S-1. Supplementary Fig. 2. S-2. Supplementary Fig. 3. S-3. Supplementary Fig. 4. S-4. Supplementary Fig. 5.
 Supplementary Information - An IonStar experimental strategy for MS1 ion current-based quantification using ultra-high-field Orbitrap: reproducible, in-depth and accurate protein measurement in larger
Supplementary Information - An IonStar experimental strategy for MS1 ion current-based quantification using ultra-high-field Orbitrap: reproducible, in-depth and accurate protein measurement in larger
Isotopic Resolution of Chromatographically Separated IdeS Subunits Using the X500B QTOF System
 Isotopic Resolution of Chromatographically Separated IdeS Subunits Using the X500B QTOF System Fan Zhang 2, Sean McCarthy 1 1 SCIEX, MA, USA, 2 SCIEX, CA USA Analysis of protein subunits using high resolution
Isotopic Resolution of Chromatographically Separated IdeS Subunits Using the X500B QTOF System Fan Zhang 2, Sean McCarthy 1 1 SCIEX, MA, USA, 2 SCIEX, CA USA Analysis of protein subunits using high resolution
Introduction. Benefits of the SWATH Acquisition Workflow for Metabolomics Applications
 SWATH Acquisition Improves Metabolite Coverage over Traditional Data Dependent Techniques for Untargeted Metabolomics A Data Independent Acquisition Technique Employed on the TripleTOF 6600 System Zuzana
SWATH Acquisition Improves Metabolite Coverage over Traditional Data Dependent Techniques for Untargeted Metabolomics A Data Independent Acquisition Technique Employed on the TripleTOF 6600 System Zuzana
Rapid Peptide Mapping via Automated Integration of On-line Digestion, Separation and Mass Spectrometry for the Analysis of Therapeutic Proteins
 Rapid Peptide Mapping via Automated Integration of On-line Digestion, Separation and Mass Spectrometry for the Analysis of Therapeutic Proteins Esther Lewis, 1 Kevin Meyer, 2 Zhiqi Hao, 1 Nick Herold,
Rapid Peptide Mapping via Automated Integration of On-line Digestion, Separation and Mass Spectrometry for the Analysis of Therapeutic Proteins Esther Lewis, 1 Kevin Meyer, 2 Zhiqi Hao, 1 Nick Herold,
SUPPLEMENTARY INFORMATION
 The Supplementary Information (SI) Methods Cell culture and transfections H1299, U2OS, 293, HeLa cells were maintained in DMEM medium supplemented with 10% fetal bovine serum. H1299 and 293 cells were
The Supplementary Information (SI) Methods Cell culture and transfections H1299, U2OS, 293, HeLa cells were maintained in DMEM medium supplemented with 10% fetal bovine serum. H1299 and 293 cells were
Supplemental Materials and Methods
 Supplemental Materials and Methods In situ hybridization In situ hybridization analysis of HFE2 and genin mrna in rat liver tissues was performed as previously described (1). Briefly, the digoxigenin-labeled
Supplemental Materials and Methods In situ hybridization In situ hybridization analysis of HFE2 and genin mrna in rat liver tissues was performed as previously described (1). Briefly, the digoxigenin-labeled
Combination of Isobaric Tagging Reagents and Cysteinyl Peptide Enrichment for In-Depth Quantification
 Combination of Isobaric Tagging Reagents and Cysteinyl Peptide Enrichment for In-Depth Quantification Protein Expression Analysis using the TripleTOF 5600 System and itraq Reagents Vojtech Tambor 1, Christie
Combination of Isobaric Tagging Reagents and Cysteinyl Peptide Enrichment for In-Depth Quantification Protein Expression Analysis using the TripleTOF 5600 System and itraq Reagents Vojtech Tambor 1, Christie
Lecture 8: Affinity Chromatography-III
 Lecture 8: Affinity Chromatography-III Key words: Chromatography; Affinity chromatography; Protein Purification During this lecture, we shall be studying few more examples of affinity chromatography. The
Lecture 8: Affinity Chromatography-III Key words: Chromatography; Affinity chromatography; Protein Purification During this lecture, we shall be studying few more examples of affinity chromatography. The
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3363 Supplementary Figure 1 Several WNTs bind to the extracellular domains of PKD1. (a) HEK293T cells were co-transfected with indicated plasmids. Flag-tagged proteins were immunoprecipiated
DOI: 10.1038/ncb3363 Supplementary Figure 1 Several WNTs bind to the extracellular domains of PKD1. (a) HEK293T cells were co-transfected with indicated plasmids. Flag-tagged proteins were immunoprecipiated
A Complete Workflow Solution for Intact Monoclonal Antibody Characterization Using a New High-Performance Benchtop Quadrupole- Orbitrap LC-MS/MS
 A Complete Workflow Solution for Intact Monoclonal Antibody Characterization Using a New High-Performance Benchtop Quadrupole- Orbitrap LC-MS/MS Zhiqi Hao, 1 Yi Zhang, 1 David Horn, 1 Seema Sharma, 1 Shiaw-Lin
A Complete Workflow Solution for Intact Monoclonal Antibody Characterization Using a New High-Performance Benchtop Quadrupole- Orbitrap LC-MS/MS Zhiqi Hao, 1 Yi Zhang, 1 David Horn, 1 Seema Sharma, 1 Shiaw-Lin
Vivapure Anti-HSA/IgG Kits for Human Albumin and Human Albumin/IgG Depletion
 Vivapure Anti-HSA/IgG Kits for Human and Human /IgG Depletion Fisher Scientific Vivapure Anti-HSA/IgG Kits for Human and Human /IgG Depletion Introduction The Vivapure Anti-HSA and Anti-HSA/IgG kits are
Vivapure Anti-HSA/IgG Kits for Human and Human /IgG Depletion Fisher Scientific Vivapure Anti-HSA/IgG Kits for Human and Human /IgG Depletion Introduction The Vivapure Anti-HSA and Anti-HSA/IgG kits are
Highly Confident Peptide Mapping of Protein Digests Using Agilent LC/Q TOFs
 Technical Overview Highly Confident Peptide Mapping of Protein Digests Using Agilent LC/Q TOFs Authors Stephen Madden, Crystal Cody, and Jungkap Park Agilent Technologies, Inc. Santa Clara, California,
Technical Overview Highly Confident Peptide Mapping of Protein Digests Using Agilent LC/Q TOFs Authors Stephen Madden, Crystal Cody, and Jungkap Park Agilent Technologies, Inc. Santa Clara, California,
Strategies in proteomics
 Strategies in proteomics Systems biology - understand cellpathways, network, and complex interacting (includes Genomics, Proteomics, Metabolomics..) Biological processes - characterize protein complexes,
Strategies in proteomics Systems biology - understand cellpathways, network, and complex interacting (includes Genomics, Proteomics, Metabolomics..) Biological processes - characterize protein complexes,
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling
 Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Product. Ni-NTA His Bind Resin. Ni-NTA His Bind Superflow. His Bind Resin. His Bind Magnetic Agarose Beads. His Bind Column. His Bind Quick Resin
 Novagen offers a large variety of affinity supports and kits for the purification of recombinant proteins containing popular peptide fusion tags, including His Tag, GST Tag, S Tag and T7 Tag sequences.
Novagen offers a large variety of affinity supports and kits for the purification of recombinant proteins containing popular peptide fusion tags, including His Tag, GST Tag, S Tag and T7 Tag sequences.
Use of a Label-Free Quantitative Platform Based on MS/MS Average TIC to Calculate Dynamics of Protein Complexes in Insulin Signaling
 ABRF 2009 SELECTED PRESENTATIONS Use of a Label-Free Quantitative Platform Based on MS/MS Average TIC to Calculate Dynamics of Protein Complexes in Insulin Signaling Xuemei Yang, 1 Adam Friedman, 2 Shailender
ABRF 2009 SELECTED PRESENTATIONS Use of a Label-Free Quantitative Platform Based on MS/MS Average TIC to Calculate Dynamics of Protein Complexes in Insulin Signaling Xuemei Yang, 1 Adam Friedman, 2 Shailender
Supplemental Online Material. The mouse embryonic fibroblast cell line #10 derived from β-arrestin1 -/- -β-arrestin2 -/-
 #1074683s 1 Supplemental Online Material Materials and Methods Cell lines and tissue culture The mouse embryonic fibroblast cell line #10 derived from β-arrestin1 -/- -β-arrestin2 -/- knock-out animals
#1074683s 1 Supplemental Online Material Materials and Methods Cell lines and tissue culture The mouse embryonic fibroblast cell line #10 derived from β-arrestin1 -/- -β-arrestin2 -/- knock-out animals
APPLICATION SPOTLIGHT INTRODUCTION NICOLAS L. TAYLOR
 APPLICATION SPOTLIGHT Measuring protein abundance by peptide selected reaction monitoring () mass spectrometry in knock-out organisms when antibodies are unavailable/ ineffective - An example from plant
APPLICATION SPOTLIGHT Measuring protein abundance by peptide selected reaction monitoring () mass spectrometry in knock-out organisms when antibodies are unavailable/ ineffective - An example from plant
Bcl-2 family member Bcl-G is not a pro-apoptotic BH3-only protein
 Bcl-2 family member Bcl-G is not a pro-apoptotic BH3-only protein Maybelline Giam 1,2, Toru Okamoto 1,2,3, Justine D. Mintern 1,2,4, Andreas Strasser 1,2 and Philippe Bouillet 1, 2 1 The Walter and Eliza
Bcl-2 family member Bcl-G is not a pro-apoptotic BH3-only protein Maybelline Giam 1,2, Toru Okamoto 1,2,3, Justine D. Mintern 1,2,4, Andreas Strasser 1,2 and Philippe Bouillet 1, 2 1 The Walter and Eliza
Nature Biotechnology: doi: /nbt Supplementary Figure 1. The workflow of Open-pFind.
 Supplementary Figure 1 The workflow of Open-pFind. The MS data are first preprocessed by pparse, and then the MS/MS data are searched by the open search module. Next, the MS/MS data are re-searched by
Supplementary Figure 1 The workflow of Open-pFind. The MS data are first preprocessed by pparse, and then the MS/MS data are searched by the open search module. Next, the MS/MS data are re-searched by
Supporting Information
 Supporting Information Deng et al. 10.1073/pnas.1515692112 SI Materials and Methods FPLC. All fusion proteins were expressed and purified through a three-step FPLC purification protocol, as described (20),
Supporting Information Deng et al. 10.1073/pnas.1515692112 SI Materials and Methods FPLC. All fusion proteins were expressed and purified through a three-step FPLC purification protocol, as described (20),
1. Cross-linking and cell harvesting
 ChIP is a powerful tool that allows the specific matching of proteins or histone modifications to regions of the genome. Chromatin is isolated and antibodies to the antigen of interest are used to determine
ChIP is a powerful tool that allows the specific matching of proteins or histone modifications to regions of the genome. Chromatin is isolated and antibodies to the antigen of interest are used to determine
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11070 Supplementary Figure 1 Purification of FLAG-tagged proteins. a, Purification of FLAG-RNF12 by FLAG-affinity from nuclear extracts of wild-type (WT) and two FLAG- RNF12 transgenic
doi:10.1038/nature11070 Supplementary Figure 1 Purification of FLAG-tagged proteins. a, Purification of FLAG-RNF12 by FLAG-affinity from nuclear extracts of wild-type (WT) and two FLAG- RNF12 transgenic
supplementary information
 DOI: 10.1038/ncb2116 Figure S1 CDK phosphorylation of EZH2 in cells. (a) Comparison of candidate CDK phosphorylation sites on EZH2 with known CDK substrates by multiple sequence alignments. (b) CDK1 and
DOI: 10.1038/ncb2116 Figure S1 CDK phosphorylation of EZH2 in cells. (a) Comparison of candidate CDK phosphorylation sites on EZH2 with known CDK substrates by multiple sequence alignments. (b) CDK1 and
Analysis of Monoclonal Antibodies and Their Fragments by Size Exclusion Chromatography Coupled with an Orbitrap Mass Spectrometer
 Analysis of Monoclonal Antibodies and Their Fragments by Size Exclusion Chromatography Coupled with an Orbitrap Mass Spectrometer Shanhua Lin, 1 Hongxia Wang, 2 Zhiqi Hao, 2 Patrick Bennett, 2 and Xiaodong
Analysis of Monoclonal Antibodies and Their Fragments by Size Exclusion Chromatography Coupled with an Orbitrap Mass Spectrometer Shanhua Lin, 1 Hongxia Wang, 2 Zhiqi Hao, 2 Patrick Bennett, 2 and Xiaodong
Simultaneous Quantitation of a Monoclonal Antibody and Two Proteins in Human Plasma by High Resolution and Accurate Mass Measurements
 Simultaneous Quantitation of a Monoclonal Antibody and Two Proteins in Human Plasma by High Resolution and Accurate Mass Measurements Paul-Gerhard Lassahn 1, Kai Scheffler 2, Myriam Demant 3, Nathanael
Simultaneous Quantitation of a Monoclonal Antibody and Two Proteins in Human Plasma by High Resolution and Accurate Mass Measurements Paul-Gerhard Lassahn 1, Kai Scheffler 2, Myriam Demant 3, Nathanael
Technical Note Detection of post-immunoprecipitation proteins by Western blot using the Quick Western Kit IRDye 680RD
 Technical Note Detection of post-immunoprecipitation proteins by Western blot using the Quick Western Kit IRDye 680RD Developed for: Aerius, Odyssey Classic, Odyssey CLx and Odyssey Sa Imaging Systems
Technical Note Detection of post-immunoprecipitation proteins by Western blot using the Quick Western Kit IRDye 680RD Developed for: Aerius, Odyssey Classic, Odyssey CLx and Odyssey Sa Imaging Systems
Myers Lab ChIP-seq Protocol v Modified January 10, 2014
 Myers Lab ChIP-seq Protocol V011014 1 Contact information: Dr. Florencia Pauli Behn HudsonAlpha Institute for Biotechnology 601 Genome Way Huntsville, AL 35806 Telephone: 256-327-5229 Email: fpauli@hudsonalpha.org
Myers Lab ChIP-seq Protocol V011014 1 Contact information: Dr. Florencia Pauli Behn HudsonAlpha Institute for Biotechnology 601 Genome Way Huntsville, AL 35806 Telephone: 256-327-5229 Email: fpauli@hudsonalpha.org
Investigating Biological Variation of Liver Enzymes in Human Hepatocytes
 Investigating Biological Variation of Liver Enzymes in Human Hepatocytes SWATH Acquisition on the TripleTOF Systems Xu Wang 1, Hui Zhang 2, Christie Hunter 1 1 SCIEX, USA, 2 Pfizer, USA MS/MS ALL with
Investigating Biological Variation of Liver Enzymes in Human Hepatocytes SWATH Acquisition on the TripleTOF Systems Xu Wang 1, Hui Zhang 2, Christie Hunter 1 1 SCIEX, USA, 2 Pfizer, USA MS/MS ALL with
hnrnp C promotes APP translation by competing with FMRP for APP mrna recruitment to P bodies
 hnrnp C promotes APP translation by competing with for APP mrna recruitment to P bodies Eun Kyung Lee 1, Hyeon Ho Kim 1, Yuki Kuwano 1, Kotb Abdelmohsen 1, Subramanya Srikantan 1, Sarah S. Subaran 2, Marc
hnrnp C promotes APP translation by competing with for APP mrna recruitment to P bodies Eun Kyung Lee 1, Hyeon Ho Kim 1, Yuki Kuwano 1, Kotb Abdelmohsen 1, Subramanya Srikantan 1, Sarah S. Subaran 2, Marc
Supplementary Figure 1. GST pull-down analysis of the interaction of GST-cIAP1 (A, B), GSTcIAP1
 Legends Supplementary Figure 1. GST pull-down analysis of the interaction of GST- (A, B), GST mutants (B) or GST- (C) with indicated proteins. A, B, Cell lysate from untransfected HeLa cells were loaded
Legends Supplementary Figure 1. GST pull-down analysis of the interaction of GST- (A, B), GST mutants (B) or GST- (C) with indicated proteins. A, B, Cell lysate from untransfected HeLa cells were loaded
Sarker et al. Supplementary Material. Subcellular Fractionation
 Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
Peptide enrichment and fractionation
 Protein sample preparation Successful analysis of low-abundance proteins and/or the identification of posttranslationally modified peptides often require several steps: enrichment, fractionation, and/or
Protein sample preparation Successful analysis of low-abundance proteins and/or the identification of posttranslationally modified peptides often require several steps: enrichment, fractionation, and/or
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.
 Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
PROTÉOMIQUE. Application de la protéomique à la cartographie de l interactome et la découverte de biomarqueurs. BENOIT COULOMBE, PhD
 PROTÉOMIQUE Application de la protéomique à la cartographie de l interactome et la découverte de biomarqueurs BENOIT COULOMBE, PhD Laboratoire de protéomique & transcription génique BCM2003, mars 2012
PROTÉOMIQUE Application de la protéomique à la cartographie de l interactome et la découverte de biomarqueurs BENOIT COULOMBE, PhD Laboratoire de protéomique & transcription génique BCM2003, mars 2012
Application Note. Authors. Abstract. Ravindra Gudihal Agilent Technologies India Pvt. Ltd. Bangalore, India
 Quantitation of Oxidation Sites on a Monoclonal Antibody Using an Agilent 1260 Infinity HPLC-Chip/MS System Coupled to an Accurate-Mass 6520 Q-TOF LC/MS Application Note Authors Ravindra Gudihal Agilent
Quantitation of Oxidation Sites on a Monoclonal Antibody Using an Agilent 1260 Infinity HPLC-Chip/MS System Coupled to an Accurate-Mass 6520 Q-TOF LC/MS Application Note Authors Ravindra Gudihal Agilent
Supplementary Method 1 - Protein biotinylation
 Supplementary Method 1 - Protein biotinylation For the immobilization on SA beads, target proteins need to be biotinylated. In general, protein biotinylation can be done either enzymatically or chemically.
Supplementary Method 1 - Protein biotinylation For the immobilization on SA beads, target proteins need to be biotinylated. In general, protein biotinylation can be done either enzymatically or chemically.
Supplementary Information: Chemical proteomics reveals ADP-ribosylation of small GTPases during oxidative stress
 Supplementary Information: Chemical proteomics reveals ADP-ribosylation of small GTPases during oxidative stress Nathan P. Westcott 1, Joseph P. Fernandez 2, Henrik Molina 2 and Howard C. Hang 1,* 1 Laboratory
Supplementary Information: Chemical proteomics reveals ADP-ribosylation of small GTPases during oxidative stress Nathan P. Westcott 1, Joseph P. Fernandez 2, Henrik Molina 2 and Howard C. Hang 1,* 1 Laboratory
Newsletter Issue 7 One-STrEP Analysis of Protein:Protein-Interactions
 www.iba-biotagnology.com Newsletter Issue 7 One-STrEP Analysis of Protein:Protein-Interactions Strep-tag and One-STrEP-tag PPI Analysis with the co-precipitation/ mass spectrometry approach 3 Background
www.iba-biotagnology.com Newsletter Issue 7 One-STrEP Analysis of Protein:Protein-Interactions Strep-tag and One-STrEP-tag PPI Analysis with the co-precipitation/ mass spectrometry approach 3 Background
Stabilization of a virus-like particle and its application as a nanoreactor at physiological conditions
 Supporting Information Stabilization of a virus-like particle and its application as a nanoreactor at physiological conditions Lise Schoonen, b Sjors Maassen, b Roeland J. M. Nolte b and Jan C. M. van
Supporting Information Stabilization of a virus-like particle and its application as a nanoreactor at physiological conditions Lise Schoonen, b Sjors Maassen, b Roeland J. M. Nolte b and Jan C. M. van
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3562 In the format provided by the authors and unedited. Supplementary Figure 1 Glucose deficiency induced FH-ATF2 interaction. In b-m, immunoblotting or immunoprecipitation analyses were
DOI: 10.1038/ncb3562 In the format provided by the authors and unedited. Supplementary Figure 1 Glucose deficiency induced FH-ATF2 interaction. In b-m, immunoblotting or immunoprecipitation analyses were
Center for Mass Spectrometry and Proteomics Phone (612) (612)
 Outline Database search types Peptide Mass Fingerprint (PMF) Precursor mass-based Sequence tag Results comparison across programs Manual inspection of results Terminology Mass tolerance MS/MS search FASTA
Outline Database search types Peptide Mass Fingerprint (PMF) Precursor mass-based Sequence tag Results comparison across programs Manual inspection of results Terminology Mass tolerance MS/MS search FASTA
Stable Isotope Labeling with Amino Acids in Cell Culture. Thermo Scientific Pierce SILAC Protein Quantitation Kits
 Stable Isotope Labeling with Amino Acids in Cell Culture Thermo Scientific Pierce SILAC Protein Quantitation Kits Quantitative Analysis of Differential Protein Expression Stable isotope labeling using
Stable Isotope Labeling with Amino Acids in Cell Culture Thermo Scientific Pierce SILAC Protein Quantitation Kits Quantitative Analysis of Differential Protein Expression Stable isotope labeling using
Really high sensitivity mass spectrometry and Discovery and analysis of protein complexes
 Really high sensitivity mass spectrometry and Discovery and analysis of protein complexes The PRIME lab and AMS Importance of protein complexes in biology Methods for isolation of protein complexes In
Really high sensitivity mass spectrometry and Discovery and analysis of protein complexes The PRIME lab and AMS Importance of protein complexes in biology Methods for isolation of protein complexes In
supplementary information
 DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
Supplementary methods
 Supplementary methods Cell culture, infection, transfection, and RNA interference HEK293 cells and its derivatives were grown in DMEM supplemented with 10% FBS. Various constructs were introduced into
Supplementary methods Cell culture, infection, transfection, and RNA interference HEK293 cells and its derivatives were grown in DMEM supplemented with 10% FBS. Various constructs were introduced into
Protein Reports CPTAC Common Data Analysis Pipeline (CDAP)
 Protein Reports CPTAC Common Data Analysis Pipeline (CDAP) v. 4/13/2015 Summary The purpose of this document is to describe the protein reports generated as part of the CPTAC Common Data Analysis Pipeline
Protein Reports CPTAC Common Data Analysis Pipeline (CDAP) v. 4/13/2015 Summary The purpose of this document is to describe the protein reports generated as part of the CPTAC Common Data Analysis Pipeline
ChIP protocol Chromatin fragmentation using the Covaris S2 sonicator by Ethan Ford (version 12/1/11) X- link Cells
 ChIP protocol Chromatin fragmentation using the Covaris S2 sonicator by Ethan Ford (version 12/1/11) X- link Cells 1. Grow six 15 cm plates of HeLa cells to 90% confluency. 2. Remove media and add wash
ChIP protocol Chromatin fragmentation using the Covaris S2 sonicator by Ethan Ford (version 12/1/11) X- link Cells 1. Grow six 15 cm plates of HeLa cells to 90% confluency. 2. Remove media and add wash
Gα i Activation Assay Kit
 A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Gα i Activation Assay Kit Catalog Number 80301 20 assays NewEast Biosciences, Inc 1 Table of Content Product
A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Gα i Activation Assay Kit Catalog Number 80301 20 assays NewEast Biosciences, Inc 1 Table of Content Product
Liver Mitochondria Proteomics Employing High-Resolution MS Technology
 Liver Mitochondria Proteomics Employing High-Resolution MS Technology Jenny Ho, 1 Loïc Dayon, 2 John Corthésy, 2 Umberto De Marchi, 2 Antonio Núñez, 2 Andreas Wiederkehr, 2 Rosa Viner, 3 Michael Blank,
Liver Mitochondria Proteomics Employing High-Resolution MS Technology Jenny Ho, 1 Loïc Dayon, 2 John Corthésy, 2 Umberto De Marchi, 2 Antonio Núñez, 2 Andreas Wiederkehr, 2 Rosa Viner, 3 Michael Blank,
Smartube Benchmark of Nano-LC
 Smartube Benchmark of Nano-LC The state of the art Smartube is a high performance chromatographic column product line. It is made for high efficiency, high selectivity microscale separations. Compared
Smartube Benchmark of Nano-LC The state of the art Smartube is a high performance chromatographic column product line. It is made for high efficiency, high selectivity microscale separations. Compared
Nguyen et al., Supplemental Figures, Methods and References Supplemental Figures
 Nguyen et al., Supplemental Figures, Methods and References Supplemental Figures Supplemental Figure 1. Additional analyses of genome-wide shrna screen for liver colonization and survival in culture (A)
Nguyen et al., Supplemental Figures, Methods and References Supplemental Figures Supplemental Figure 1. Additional analyses of genome-wide shrna screen for liver colonization and survival in culture (A)
Separation of Native Monoclonal Antibodies and Identification of Charge Variants:
 Separation of Native Monoclonal Antibodies and Identification of Charge Variants: Teamwork of the Agilent 31 OFFGEL Fractionator, Agilent 21 Bioanalyzer and Agilent LC/MS Systems Application Note Biosimilar
Separation of Native Monoclonal Antibodies and Identification of Charge Variants: Teamwork of the Agilent 31 OFFGEL Fractionator, Agilent 21 Bioanalyzer and Agilent LC/MS Systems Application Note Biosimilar
Thermo Scientific Ligand Binding Mass Spectrometric Immunoassay (LB-MSIA)
 INSTRUCTIONS FOR USE OF PRODUCTS Thermo Scientific Ligand Binding Mass Spectrometric Immunoassay (LB-MSIA) Demonstration of the LB-MSIA Protocol for the Analysis of Intact Adalimumab from Mouse Plasma
INSTRUCTIONS FOR USE OF PRODUCTS Thermo Scientific Ligand Binding Mass Spectrometric Immunoassay (LB-MSIA) Demonstration of the LB-MSIA Protocol for the Analysis of Intact Adalimumab from Mouse Plasma
