Supplemental Information. Structural Basis for the Versatile. and Methylation-Dependent Binding of CTCF to DNA
|
|
|
- Tobias Dickerson
- 6 years ago
- Views:
Transcription
1 Molecular ell, Volume 66 Supplemental Information Structural Basis for the Versatile and Methylation-Dependent Binding of TF to DNA Hideharu Hashimoto, Dongxue Wang, John R. Horton, Xing Zhang, Victor. orces, and Xiaodong heng
2 A DNA binding RNA binding htf ENP E SA B ZF1 ZF2 ZF Human 263 KKTFQELSYTPRRSNLDRHMKSHTDERPHKHLRAFRTVTLLRNHLNTHTTRPHKPDDMAFVTSELVRHRRYKHTHEKPFKSMDYASVEVSKLKRHIRSHTE 376 Mouse 263 KKTFQELSYTPRRSNLDRHMKSHTDERPHKHLRAFRTVTLLRNHLNTHTTRPHKPDDMAFVTSELVRHRRYKHTHEKPFKSMDYASVEVSKLKRHIRSHTE 376 Rat 263 KKTFQELSYTPRRSNLDRHMKSHTDERPHKHLRAFRTVTLLRNHLNTHTTRPHKPDDMAFVTSELVRHRRYKHTHEKPFKSMDYASVEVSKLKRHIRSHTE 376 ow 263 KKTFQELSYTPRRSNLDRHMKSHTDERPHKHLRAFRTVTLLRNHLNTHTTRPHKPDDMAFVTSELVRHRRYKHTHEKPFKSMDYASVEVSKLKRHIRSHTE 376 allus 263 KKTFQELSYTPRRSNLDRHMKSHTDERPHKHLRAFRTVTLLRNHLNTHTTRPHKPDDMAFVTSELVRHRRYKHTHEKPFKSMDYASVEVSKLKRHIRSHTE 376 Xenopus 261 KKTFQELSYTPRRSNLDRHMKSHTDERPHKHLRAFRTVTLLRNHLNTHTTRPHKPDDMAFVTSELVRHRRYKHTHEKPFKSMDYASVEVSKLKRHIRSHTE 374 Danio 303 KKTFQELSYTPRRSNLDRHMKSHTDERPHKHLRAFRTVTLLRNHLNTHTTRPHKTDDMAFVTSELVRHRRYKHTHEKPFKSMDYASVEVSKLKRHIRSHTE 416 htfl 254 KTFHDVMFTSSRMSSFNRHMKTHTSEKPHLHLLKTFRTVTLLRNHVNTHTTRPYKNDNMAFVTSELVRHRRYKHTHEKPFKSMKYASVEASKLKRHVRSHTE 367 Fly 291 HKYSPHPYTASKKFLITRHSRSHDVEPSFKSIERSFRSNVLQNHINTHMNKPHKKLESAFTTSELVRHTRYKHTKEKPHKTETYASVELTKLRRHMTHTE 404 ZF Human 377 RPFQSLSYASRDTYKLKRHMRTHSEKPYEYIHARFTQSTMKMHILQKHTENVAKFHPHDTVIARKSDLVHLRKQHSYIEQKKRYDAVFHERYALIQHQKSHKNE 492 Mouse 377 RPFQSLSYASRDTYKLKRHMRTHSEKPYEYIHARFTQSTMKMHILQKHTENVAKFHPHDTVIARKSDLVHLRKQHSYIEQKKRYDAVFHERYALIQHQKSHKNE 492 Rat 377 RPFQSLSYASRDTYKLKRHMRTHSEKPYEYIHARFTQSTMKMHILQKHTENVAKFHPHDTVIARKSDLVHLRKQHSYIEQKKRYDAVFHERYALIQHQKSHKNE 492 ow 377 RPFQSLSYASRDTYKLKRHMRTHSEKPYEYIHARFTQSTMKMHILQKHTENVAKFHPHDTVIARKSDLVHLRKQHSYIEQKKRYDAVFHERYALIQHQKSHKNE 492 allus 377 RPFQSLSYASRDTYKLKRHMRTHSEKPYEYIHARFTQSTMKMHILQKHTENVAKFHPHDTVIARKSDLVHLRKQHSYIEQKKRYDAVFHERYALIQHQKSHKNE 492 Xenopus 375 RPFQSLSYASRDTYKLKRHMRTHSEKPYEYIHARFTQSTMKMHILQKHTENVAKFHPHDTVIARKSDLVHLRKQHSYIEQKKRYDTVFHERYALIQHQKSHKNE 490 Danio 417 RPFQSLSYASRDTYKLKRHMRTHSEKPYEYIHARFTQSTMKMHILQKHTENVAKFHPHDTVIARKSDLVHLRKQHSYIEQRKRYDAVFHERYALIQHQKSHKNE 532 htfl 368 RPFQQSYASRDTYKLKRHMRTHSEKPYEHIHTRFTQSTMKIHILQKHENVPKYQPHATIIARKSDLRVHMRNLHAYSAAELKRYSAVFHERYALIQHQKTHKNE 483 Fly 405 RPYQPHTYASQDMFKLKRHMVIHTEKKYQDIKSRFTQSNSLKAHKLIHSVVDKPVFQNYPTTRKADLRVHIKHMHT-SDVPMTRRQQLPDRYQYKLHVKSHEE 519 ZF9 ZF10 ZF Human 493 KRFKDQDYARQERHMIMHKRTHTEKPYASHDKTFRQKQLLDMHFKRYHDPNFVPAAFVSKKTFTRRNTMARHADNAPDVEEN 588 Mouse 493 KRFKDQDYARQERHMIMHKRTHTEKPYASHDKTFRQKQLLDMHFKRYHDPNFVPAAFVSKKTFTRRNTMARHADNAPDVEEN 588 Rat 493 KRFKDQDYARQERHMIMHKRTHTEKPYASHDKTFRQKQLLDMHFKRYHDPNFVPAAFVSKKTFTRRNTMARHADNAPDVEEN 588 ow 493 KRFKDQDYARQERHMIMHKRTHTEKPYASHDKTFRQKQLLDMHFKRYHDPNFVPAAFVSKKTFTRRNTMARHADNAPDVEEN 588 allus 493 KRFKDQDYARQERHMVMHKRTHTEKPYASHDKTFRQKQLLDMHFKRYHDPNFVPAAFVSKKTFTRRNTMARHADNSLDEEN 588 Xenopus 491 KRFKDQEYARQERHMIMHKRTHTEKPYASHDKTFRQKQLLDMHFKRYHDPSFVPAAFVSKKTFTRRNTMSRHADNTPDTDEN 584 Danio 533 KRFKDQDYARQERHMVMHKRTHTEKPYASQEKTFRQKQLLDMHFRRYHDPNFVPTSFVTKKTFTRRNTMARHAENTMDSADEN 628 htfl 484 KRFKKHSYAKQERHMTAHIRTHTEKPFTLSNKFRQKQLLNAHFRKYHDANFIPTVYKSKKFSRWINLHRHSEKSEAKSAAS 579 Fly 520 KYSKLSYASVTQRHLASHMLIHLDEKPFHDQPQAFRQRQLLRRHMNLVHNEEYQPKLHKPSPREFTHKNLMRHMETHDDSANAREKRR 620 Human TF ZF Residues onstruct (px #) Protein yield (mg/l) ~ ~ (~25) (~25) ~ ~ ~ ~ ~ ~ ~ (~25) + K365T 1518 ~ (~25) (~25) (~25) (~25) rystals (kda) D PEPRE Human TF ZFs E F Signal tracks hr5 139,528 kb " 139,526 kb " 139,524 kb " H1-hES HeLa-S3 HEP2 A549 D20+ Dnd41 M12878 HME HSMM HSMMtube HUVE K562 Monocytes-D14+ NH-A NHDF-Ad NHEK NHLF Osteobl 5 -AAAA-3
3 Figure S1. The TF family, related to Figure 1 (A) Schematic representation of human TF. The N-terminal and -terminal domains of TF are known to interact with other proteins, for example, the centromeric protein ENP-E (Xiao et al., 2015) and the cohesin subunit SA2 (Xiao et al., 2011), respectively. In addition, TF binds to RNA (Kung et al., 2015; Saldana-Meyer et al., 2014). (B) Sequence alignment of the conserved 11-ZF region from human (NP_ ), mouse (NP_ ), rat (NP_ ), cow (NP_ ), allus (NP_ ), Xenopus (NP_ ), Danio (NP_ ), human TFL/BORIS (NP_ ) and fruit fly (NP_ ). Numbering of residues in human TF is shown above the sequence alignment. The sequence variations are highlighted in cyan. The four Zn-coordinating residues of each finger are highlighted with white letters against black. Side chains from three specific amino acids within each finger that make major groove contacts with the DNA bases of 3-bp triplet are highlighted in blue (for 5 base), red (for central base), and green (for 3 base). The first zinc-coordination His in each finger is assigned reference position 0. Residues prior to this, at positions 1 (blue), 4 (red), and 7 (green) form H-bonds with the exposed edges of the DNA bases in the major groove. (B) The expression constructs for human TF created in this study. () An 18% SDS-PAE showing examples of the purified ZF proteins used in this study. (E) The ORE sequence used for DNA binding assays and crystallization (indicated by a red arrow) is located at chromosome 5: , at a topologically associating domain (TAD) border. This site is constitutively bound in various cell types. Heat maps depicting observed intra-chromosomal contacts for chromosome 5: 139,000 kb -139,750 kb of in situ MboI Hi- data in M12878 cell (Rao et al., 2014). Reference genes, TF orientation, M12878 TF chip-seq data (Broad), and Rad21 chip-seq sequence (Stanford) are shown. Peaks are shown in blue squares, and contact domains are shown in yellow squares. (F) Signal tracks of TF hip-seq (Broad) in various cell types are shown for chromosome 5 (onsortium, 2012).
4 A -10 used in the crystallization ZF10 was not observed ZF9 ZF8 F B ZF2-7 ZF3 ZF2 ZF3-7 ZF3 H D -7 E -8
5 Figure S2. Summary of structural information of the human TF ZF DNA binding domain in complex with DNA derived from this study, related to Figure 2 and Table S1 (A) Structure of TF -10 in complex with DNA. The last finger (ZF10) was not visible. Each finger is colored as cyan (), blue (), green (), magenta (), orange (ZF8) and red (ZF9). (B) Structure of TF ZF 2-7 in complex with DNA. ZF2 (in grey) lies in the DNA major groove but does not make base-specific contacts with the DNA sequence used. (-E) Structures of TF ZF3-7 (panel ), -7 (panel D) and -8 (panel E) in complex with DNA, respectively. Vertical dashed line indicates all structures shown are aligned with common elements. (F) In the structure of ZF3-7 in complex with DNA, crystallographic phases were calculated by single-wavelength anomalous diffraction (SAD) from five zinc atoms, shown with the anomalous experimental electron density (grey mesh), contoured at 10σ above the mean, for the corresponding zinc atoms. (-H) The omit electron density (blue mesh), contoured at 2.0σ above the mean, is shown for the protein (panel ) or DNA (panel H).
6 Protein sequence of a particular TargetZnF DNA sequence Relative binding affinity protein sequence ofznfa particular target DNA sequence Zn Known 2H2-ZnF Structures A Name Zfp57 Zif268 (Egr1) WT1 2 (ZF2) (ZF1) (ZF3) (ZF2) () Klf4 (ZF2) Klf4 (E446D) TF B H2 PRSPEKFRDQSEVNRHLKVHQNKP APVESDRRFSRSDELTRHIRIHTQK PFADIRKFARSDERKRHTKIHLRQK QDFKDERRFSRSDQLKRHQRRHTVK SRWPSQKKFARSDELVRHHNMHQRNM HDWDWKFARSDELTRHYRKHTHR...D... () PFKSMDYASVEVSKLKRHIRSHTER () KFHPHDTVIARKSDLVHLRKQHSYI D E m > m > m > ca > 5m > m > m > N-5 F 5m > H Q369 Q369 E182 E535 E354 R178 5m 5m Zfp57 (PDB 4ZN) R351 5m Egr1 (PDB 4R2A) D446 E446 R511 5m Kaiso (PDB 4F6N) R443 Klf4 (PDB 4M9E) R443 Klf4 E446D (PDB 5KEA) 5m R366 WT1 (PDB 4R2E) 5ca R366 WT1 (PDB 4R2R) Figure S3. A recognition code for modified cytosine, related to Figure 3 (A) Sequence alignment of representative 2H2 ZF proteins known to interact with methylated (modified) cytosine. The amino acids at positions 1, 4, -7 or 8 and the primary DNA base recognition are highlighted with the same color code (blue, green, and red). In an early report of phage display study of Egr1/Zif268 (hoo and Klug, 1994), an aspartate (D) residue at the -4 position (rather than E in the wild type) shows a distinct preference for binding (unmodified) cytosine, whereas E appears to specify cytosine in wild-type Egr1/Zif268 (Pavletich and Pabo, 1991). This observation led hoo and Klug to comment that The physical basis for the interaction of aspartate/glutamate and cytosine is not yet clear, since hydrogen-bonding contacts between these groups have yet to be observed in zinc finger cocrystal structures (hoo and Klug, 1997). In recent studies, we and others showed that Egr1/Zif268 binds methylated DNA (Hashimoto et al., 2014; Zandarashvili et al., 2015). (B-E) An Arg-lu pair in Zfp57 (Liu et al., 2012), Egr1 (Hashimoto et al., 2014), Kaiso (BuckKoehntop et al., 2012), and Klf4 (Liu et al., 2014) recognizes a 5mp dinucleotide. In Kaiso, the Arg and lu residues are from two neighboring fingers (Liu et al., 2013). A negatively charged glutamate prevents the binding of 5ca; both carboxylate groups bear a full -1 charge and thus repel one another electrostatically. (F) In Klf4, substituting lu446 with aspartate (E446D) resulted in preference for unmodified cytosine (Hashimoto et al., 2016). (-H) A glutamine allows WT1 bind both 5m and 5ca (Hashimoto et al., 2014).
7 A B A model of htf2-9 DNA complex (this study) TFIIIA-DNA (PDB 1TF6) N-terminal finger ZF2 ZF1 ZF11-ZF10 ZF8 3 more -terminal ZFs Sequence non-specific interaction Sequence-specific contacts hip-exo Nakahashi et al. (2013) 5 Upstream Sequence non-specific interaction Sequence-specific contacts 15-bp core motif 8-bp gap Downstream 3? Figure S4. TF ZF8-9 spans across DNA phosphate backbone, related to Figure 4 (A-B) omparison of our model of TF ZF2-9 to an earlier example of transcription factor TFIIIA (Nolte et al., 1998) suggests that the -terminal fingers could span the greater length of the DNA duplexes without additional base specific interactions. () In a study by Nakahashi et al., in addition to the 15-bp ORE sequence, a second 5 upstream motif found at approximately 15% of TF-binding sites (Nakahashi et al., 2013). However, we were not able to confirm the binding of the -terminal ZF9-11 to DNA for the number of sequences tested (data not shown). In addition, our model does not explain how the 3 downstream DNA motif destabilizes TF occupancy (Nakahashi et al., 2013). One possible explanation is that other DNA binding protein(s) in the hip-exo experiments occupy the 5 upstream and/or 3 downstream sequences.
8 Table S1. Summary of X-ray data collection from SERAT 22-ID beamline and refinement statistics (*), related to Figures 2 and S2 TF ZF2-7 ZF3-7 ZF3-7 (M=5m) DNA (5-3 ) (3-5 ) AATATA TTATAT AATA TTAT AAMTA TTMAT AA TT TAA ATT TTAA AATT TTT AAA ATAAAAA TATTTTT PDB 5T0U 5KKQ 5T00 5K5H 5K5I 5K5J 5K5L 5UND - Wavelength (Å) Space roup P2 P1 P1 P P6 5 P P P2 1 P2 1 Unit cell (Å) 101.6, 41.0, , 44.9, , 44.9, , 52.8, , 45.1, , 58.9, , 52.4, , 73.8, , 74.3, 92.7 α, β, γ ( ) 90.0, 92.2, , 92.4, , 92.4, , 90, 90 90, 90, , 90, 90 90, 90, , 91.4, , 92.7, 90.0 Resolution (Å) ( ) ( ) ( ) ( ) ( ) ( ) ( ) ( ) ( ) a R merge (0.350) (0.549) (0.733) (0.593) (0.709) (0.726) (0.392) (0.458) (0.329) b <I/σI> 8.2 (2.3) 12.4 (1.6) 9.33 (2.43) (4.3) 27.7 (2.2) 27.3 (2.7) 14.9 (3.6) 14.9 (1.4) 12.9 (3.3) ompleteness (%) 99.3 (94.3) 92.6 (64.1) 98.7 (91.6) 84.1 (47.5) 99.3 (97.2) 98.7 (100.0) 98.7 (91.1) 99.8 (98.7) 100 (100) Redundancy 5.1 (4.7) 3.6 (1.8) 7.4 (7.0) 36.6 (22.0) 12.7 (4.1) 6.1 (6.2) 7.5 (4.8) 6.3 (5.4) 7.2 (7.2) 1/2, (/ 0.985) (0.637 / 0.882) (/ 0.960) (0.990 / 0.996) (0.647 / 0.886) (0.901 / 0.974) (0.980 / 0.995) (0.590 / 0.862) (0.898 / 0.973) Reflections (observed) 76, , , , ,071 87,503 63, ,009 79,994 Reflections (unique) 14,911 (1,018) 57,383 (4,006) 31,064 (2,903) 5,709 (219) 16,250 (1579) 14,353 (1419) 8,481 (755) 33,435 (3,301) 11,181 (1,102) Phasing MR-SAD Zn-SAD MR Zn-SAD Zn-SAD MR Zn-SAD MR MR Bijvoet pairs - 53,575-2,639 15,716-6,745 FOM Refinement Resolution (Å) Refinement No. Reflections 14,798 57,333 30,987 3,385 15,437 14,269 8,420 33,362 Not c R work/ d R free / / / / / / / / ontinued No. Atoms Protein DNA Zn Solvent B-factors (Å 2 ) Protein DNA Zn Solvent R.m.s. deviations Bond length (Å) Bond angles ( ) All atom clashscore Ramachandran (%) Favored Allowed β deviation * Values in parenthesis correspond to highest resolution shell; a R merge = Σ I - <I> /ΣI, where I is the observed intensity and <I> is the averaged intensity from multiple observations; b <I/σI> = averaged ratio of the intensity (I) to the error of the intensity (σi); c R work = Σ Fobs - Fcal /Σ Fobs, where Fobs and Fcal are the observed and calculated structure factors, respectively; d R free was calculated using a randomly chosen subset (5%) of the reflections not used in refinement.
Suppl. Figure 1: RCC1 sequence and sequence alignments. (a) Amino acid
 Supplementary Figures Suppl. Figure 1: RCC1 sequence and sequence alignments. (a) Amino acid sequence of Drosophila RCC1. Same colors are for Figure 1 with sequence of β-wedge that interacts with Ran in
Supplementary Figures Suppl. Figure 1: RCC1 sequence and sequence alignments. (a) Amino acid sequence of Drosophila RCC1. Same colors are for Figure 1 with sequence of β-wedge that interacts with Ran in
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature06147 SUPPLEMENTARY INFORMATION Figure S1 The genomic and domain structure of Dscam. The Dscam gene comprises 24 exons, encoding a signal peptide (SP), 10 IgSF domains, 6 fibronectin
doi: 10.1038/nature06147 SUPPLEMENTARY INFORMATION Figure S1 The genomic and domain structure of Dscam. The Dscam gene comprises 24 exons, encoding a signal peptide (SP), 10 IgSF domains, 6 fibronectin
SUPPLEMENTARY INFORMATION
 Supplementary Table 1. Crystallographic statistics CRM1-SNUPN complex Space group P6 4 22 a=b=250.4, c=190.4 Data collection statistics: CRM1-selenomethionine SNUPN MAD data Peak Inflection Remote Native
Supplementary Table 1. Crystallographic statistics CRM1-SNUPN complex Space group P6 4 22 a=b=250.4, c=190.4 Data collection statistics: CRM1-selenomethionine SNUPN MAD data Peak Inflection Remote Native
Supplementary materials for Structure of an open clamp type II topoisomerase-dna complex provides a mechanism for DNA capture and transport
 Supplementary materials for Structure of an open clamp type II topoisomerase-dna complex provides a mechanism for DNA capture and transport Ivan Laponogov 1,2, Dennis A. Veselkov 1, Isabelle M-T. Crevel
Supplementary materials for Structure of an open clamp type II topoisomerase-dna complex provides a mechanism for DNA capture and transport Ivan Laponogov 1,2, Dennis A. Veselkov 1, Isabelle M-T. Crevel
Nature Structural & Molecular Biology: doi: /nsmb.3428
 Supplementary Figure 1 Biochemical characterization of the monou and oligou activity switch of TUT4(7). (a) Mouse TUT4 and human TUT7 were assayed for monou and Lin28-dependent oligou addition activities
Supplementary Figure 1 Biochemical characterization of the monou and oligou activity switch of TUT4(7). (a) Mouse TUT4 and human TUT7 were assayed for monou and Lin28-dependent oligou addition activities
SUPPLEMENTARY INFORMATION
 Molecular basis of RNA-dependent RNA polymerase II activity Elisabeth Lehmann, Florian Brueckner, and Patrick Cramer Gene Center Munich and Center for integrated Protein Science CiPS M, Department of Chemistry
Molecular basis of RNA-dependent RNA polymerase II activity Elisabeth Lehmann, Florian Brueckner, and Patrick Cramer Gene Center Munich and Center for integrated Protein Science CiPS M, Department of Chemistry
X-ray structures of fructosyl peptide oxidases revealing residues responsible for gating oxygen access in the oxidative half reaction
 X-ray structures of fructosyl peptide oxidases revealing residues responsible for gating oxygen access in the oxidative half reaction Tomohisa Shimasaki 1, Hiromi Yoshida 2, Shigehiro Kamitori 2 & Koji
X-ray structures of fructosyl peptide oxidases revealing residues responsible for gating oxygen access in the oxidative half reaction Tomohisa Shimasaki 1, Hiromi Yoshida 2, Shigehiro Kamitori 2 & Koji
SUPPLEMENTARY INFORMATION
 This supplementary information is an extension of the letter with the same title and includes further discussion on the comparison of our designed Fe B Mb (computer model and crystal structure) with the
This supplementary information is an extension of the letter with the same title and includes further discussion on the comparison of our designed Fe B Mb (computer model and crystal structure) with the
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than. SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with
 Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
SUPPLEMENTARY INFORMATION. Reengineering Protein Interfaces Yields Copper-Inducible Ferritin Cage Assembly
 SUPPLEMENTARY INFORMATION Reengineering Protein Interfaces Yields Copper-Inducible Ferritin Cage Assembly Dustin J. E. Huard, Kathleen M. Kane and F. Akif Tezcan* Department of Chemistry and Biochemistry,
SUPPLEMENTARY INFORMATION Reengineering Protein Interfaces Yields Copper-Inducible Ferritin Cage Assembly Dustin J. E. Huard, Kathleen M. Kane and F. Akif Tezcan* Department of Chemistry and Biochemistry,
Hmwk # 8 : DNA-Binding Proteins : Part II
 The purpose of this exercise is : Hmwk # 8 : DNA-Binding Proteins : Part II 1). to examine the case of a tandem head-to-tail homodimer binding to DNA 2). to view a Zn finger motif 3). to consider the case
The purpose of this exercise is : Hmwk # 8 : DNA-Binding Proteins : Part II 1). to examine the case of a tandem head-to-tail homodimer binding to DNA 2). to view a Zn finger motif 3). to consider the case
Supplementary Table 1. DNA sequence synthesized to express the Zika virus NS5 protein.
 Supplementary Table 1. DNA sequence synthesized to express the Zika virus NS5 protein. MR766 NS5 sequence 1 ACAGAGAACAGATTGGTGGTGGAGGTGGGACGGGAGAGACTCTGGGAGAGAAGTGGAAAG 61 CTCGTCTGAATCAGATGTCGGCCCTGGAGTTCTACTCTTATAAAAAGTCAGGTATCACTG
Supplementary Table 1. DNA sequence synthesized to express the Zika virus NS5 protein. MR766 NS5 sequence 1 ACAGAGAACAGATTGGTGGTGGAGGTGGGACGGGAGAGACTCTGGGAGAGAAGTGGAAAG 61 CTCGTCTGAATCAGATGTCGGCCCTGGAGTTCTACTCTTATAAAAAGTCAGGTATCACTG
Chapter 14 Regulation of Transcription
 Chapter 14 Regulation of Transcription Cis-acting sequences Distance-independent cis-acting elements Dissecting regulatory elements Transcription factors Overview transcriptional regulation Transcription
Chapter 14 Regulation of Transcription Cis-acting sequences Distance-independent cis-acting elements Dissecting regulatory elements Transcription factors Overview transcriptional regulation Transcription
SUPPLEMENTARY INFORMATION
 Structure of a tyrosyl-trna synthetase splicing factor bound to a group I intron RNA Paul J. Paukstelis 1, Jui-Hui Chen 2, Elaine Chase 2, Alan M. Lambowitz 1,*, and Barbara L. Golden 2,*,. 1 Institute
Structure of a tyrosyl-trna synthetase splicing factor bound to a group I intron RNA Paul J. Paukstelis 1, Jui-Hui Chen 2, Elaine Chase 2, Alan M. Lambowitz 1,*, and Barbara L. Golden 2,*,. 1 Institute
Supplementary Information. Structural basis for duplex RNA recognition and cleavage by A.
 Supplementary Information Structural asis for duplex RNA recognition and cleavage y A. fulgidus C3PO Eneida arizotto 1, Edward D Lowe 1 & James S Parker 1 1 Department of Biochemistry University of Oxford
Supplementary Information Structural asis for duplex RNA recognition and cleavage y A. fulgidus C3PO Eneida arizotto 1, Edward D Lowe 1 & James S Parker 1 1 Department of Biochemistry University of Oxford
Molecular design principles underlying β-strand swapping. in the adhesive dimerization of cadherins
 Supplementary information for: Molecular design principles underlying β-strand swapping in the adhesive dimerization of cadherins Jeremie Vendome 1,2,3,5, Shoshana Posy 1,2,3,5,6, Xiangshu Jin, 1,3 Fabiana
Supplementary information for: Molecular design principles underlying β-strand swapping in the adhesive dimerization of cadherins Jeremie Vendome 1,2,3,5, Shoshana Posy 1,2,3,5,6, Xiangshu Jin, 1,3 Fabiana
Supplementary Information for. Structure of human tyrosylprotein sulfotransferase-2 reveals the mechanism of protein tyrosine sulfation reaction
 Supplementary Information for Structure of human tyrosylprotein sulfotransferase-2 reveals the mechanism of protein tyrosine sulfation reaction Takamasa Teramoto, Yukari Fujikawa, Yoshirou Kawaguchi, Katsuhisa
Supplementary Information for Structure of human tyrosylprotein sulfotransferase-2 reveals the mechanism of protein tyrosine sulfation reaction Takamasa Teramoto, Yukari Fujikawa, Yoshirou Kawaguchi, Katsuhisa
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature10258 Supplementary Figure 1 Reconstitution of the CENP-A nucleosome with recombinant human histones H2A, H2B, H4, and CENP-A. a, Purified recombinant human
SUPPLEMENTARY INFORMATION doi:10.1038/nature10258 Supplementary Figure 1 Reconstitution of the CENP-A nucleosome with recombinant human histones H2A, H2B, H4, and CENP-A. a, Purified recombinant human
C2H2 zinc finger proteins greatly expand the human regulatory lexicon
 Supplementary Information for: C2H2 zinc finger proteins greatly expand the human regulatory lexicon Hamed S. Najafabadi*,1, Sanie Mnaimneh*,1, Frank W. Schmitges*,1, Michael Garton 1, Kathy N. Lam 2,
Supplementary Information for: C2H2 zinc finger proteins greatly expand the human regulatory lexicon Hamed S. Najafabadi*,1, Sanie Mnaimneh*,1, Frank W. Schmitges*,1, Michael Garton 1, Kathy N. Lam 2,
Chapter 8 DNA Recognition in Prokaryotes by Helix-Turn-Helix Motifs
 Chapter 8 DNA Recognition in Prokaryotes by Helix-Turn-Helix Motifs 1. Helix-turn-helix proteins 2. Zinc finger proteins 3. Leucine zipper proteins 4. Beta-scaffold factors 5. Others λ-repressor AND CRO
Chapter 8 DNA Recognition in Prokaryotes by Helix-Turn-Helix Motifs 1. Helix-turn-helix proteins 2. Zinc finger proteins 3. Leucine zipper proteins 4. Beta-scaffold factors 5. Others λ-repressor AND CRO
The Cys 2. His 2. Research Article 451
 Research Article 451 High-resolution structures of variant Zif268 DNA complexes: implications for understanding zinc finger DNA recognition Monicia Elrod-Erickson 1, Timothy E Benson 1 and Carl O Pabo
Research Article 451 High-resolution structures of variant Zif268 DNA complexes: implications for understanding zinc finger DNA recognition Monicia Elrod-Erickson 1, Timothy E Benson 1 and Carl O Pabo
SUPPLEMENTARY INFORMATION
 The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix Hideharu Hashimoto 1, John R. Horton 1, Xing Zhang 1, Magnolia Bostick 2, Steven Jacobsen 2,3, and Xiaodong Cheng 1 1 Department of Biochemistry,
The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix Hideharu Hashimoto 1, John R. Horton 1, Xing Zhang 1, Magnolia Bostick 2, Steven Jacobsen 2,3, and Xiaodong Cheng 1 1 Department of Biochemistry,
Supplementary Fig. 1. Initial electron density maps for the NOX-D20:mC5a complex obtained after SAD-phasing. (a) Initial experimental electron
 Supplementary Fig. 1. Initial electron density maps for the NOX-D20:mC5a complex obtained after SAD-phasing. (a) Initial experimental electron density map obtained after SAD-phasing and density modification
Supplementary Fig. 1. Initial electron density maps for the NOX-D20:mC5a complex obtained after SAD-phasing. (a) Initial experimental electron density map obtained after SAD-phasing and density modification
has only one nucleotide, U20, between G19 and A21, while trna Glu CUC has two
 SPPLEMENTRY INFORMTION doi:1.138/nature9411 Supplementary Discussion The structural characteristics of trn Gln G in comparison to trn Glu 16 in trn Gln G is directed towards the G19 56 pair, or the outer
SPPLEMENTRY INFORMTION doi:1.138/nature9411 Supplementary Discussion The structural characteristics of trn Gln G in comparison to trn Glu 16 in trn Gln G is directed towards the G19 56 pair, or the outer
Supplementary Online Material. Structural mimicry in transcription regulation of human RNA polymerase II by the. DNA helicase RECQL5
 Supplementary Online Material Structural mimicry in transcription regulation of human RNA polymerase II by the DNA helicase RECQL5 Susanne A. Kassube, Martin Jinek, Jie Fang, Susan Tsutakawa and Eva Nogales
Supplementary Online Material Structural mimicry in transcription regulation of human RNA polymerase II by the DNA helicase RECQL5 Susanne A. Kassube, Martin Jinek, Jie Fang, Susan Tsutakawa and Eva Nogales
Table S1. Crystallographic Data and Refinement Statistics, Related to Experimental Procedures hsddb1-drddb2 CPD
 Data collection Table S1. Crystallographic Data and Refinement Statistics, Related to Experimental Procedures hsddb1-drddb2 CPD #1 hsddb1-drddb2 CPD #2 Values in parentheses are for the highest resolution
Data collection Table S1. Crystallographic Data and Refinement Statistics, Related to Experimental Procedures hsddb1-drddb2 CPD #1 hsddb1-drddb2 CPD #2 Values in parentheses are for the highest resolution
Gene regulation II Biochemistry 302. Bob Kelm March 1, 2004
 Gene regulation II Biochemistry 302 Bob Kelm March 1, 2004 Lessons to learn from bacteriophage λ in terms of transcriptional regulation Similarities to E. coli Cis-elements (operator elements) are adjacent
Gene regulation II Biochemistry 302 Bob Kelm March 1, 2004 Lessons to learn from bacteriophage λ in terms of transcriptional regulation Similarities to E. coli Cis-elements (operator elements) are adjacent
SUPPLEMENTARY INFORMATION. Supplementary Figures 1-8
 SUPPLEMENTARY INFORMATION Supplementary Figures 1-8 Supplementary Figure 1. TFAM residues contacting the DNA minor groove (A) TFAM contacts on nonspecific DNA. Leu58, Ile81, Asn163, Pro178, and Leu182
SUPPLEMENTARY INFORMATION Supplementary Figures 1-8 Supplementary Figure 1. TFAM residues contacting the DNA minor groove (A) TFAM contacts on nonspecific DNA. Leu58, Ile81, Asn163, Pro178, and Leu182
1 Najafabadi, H. S. et al. C2H2 zinc finger proteins greatly expand the human regulatory lexicon. Nat Biotechnol doi: /nbt.3128 (2015).
 F op-scoring motif Optimized motifs E Input sequences entral 1 bp region Dinucleotideshuffled seqs B D ll B1H-R predicted motifs Enriched B1H- R predicted motifs L!=!7! L!=!6! L!=5! L!=!4! L!=!3! L!=!2!
F op-scoring motif Optimized motifs E Input sequences entral 1 bp region Dinucleotideshuffled seqs B D ll B1H-R predicted motifs Enriched B1H- R predicted motifs L!=!7! L!=!6! L!=5! L!=!4! L!=!3! L!=!2!
Structure/function relationship in DNA-binding proteins
 PHRM 836 September 22, 2015 Structure/function relationship in DNA-binding proteins Devlin Chapter 8.8-9 u General description of transcription factors (TFs) u Sequence-specific interactions between DNA
PHRM 836 September 22, 2015 Structure/function relationship in DNA-binding proteins Devlin Chapter 8.8-9 u General description of transcription factors (TFs) u Sequence-specific interactions between DNA
HEK293T. Fig. 1 in the
 Supplementary Information Supplementary Figure 1 Zinc uptake assay of hzip4 and hzip4-δecd transiently expressed in HEK293T cells. The results of one representative e experiment are shown in Fig. 1 in
Supplementary Information Supplementary Figure 1 Zinc uptake assay of hzip4 and hzip4-δecd transiently expressed in HEK293T cells. The results of one representative e experiment are shown in Fig. 1 in
Basic concepts of molecular biology
 Basic concepts of molecular biology Gabriella Trucco Email: gabriella.trucco@unimi.it Life The main actors in the chemistry of life are molecules called proteins nucleic acids Proteins: many different
Basic concepts of molecular biology Gabriella Trucco Email: gabriella.trucco@unimi.it Life The main actors in the chemistry of life are molecules called proteins nucleic acids Proteins: many different
Structural Analysis of the EGR Family of Transcription Factors: Templates for Predicting Protein DNA Interactions
 Introduction Structural Analysis of the EGR Family of Transcription Factors: Templates for Predicting Protein DNA Interactions Jamie Duke, Rochester Institute of Technology Mentor: Carlos Camacho, University
Introduction Structural Analysis of the EGR Family of Transcription Factors: Templates for Predicting Protein DNA Interactions Jamie Duke, Rochester Institute of Technology Mentor: Carlos Camacho, University
Supplemental Information Molecular Cell, Volume 41
 Supplemental Information Molecular Cell, Volume 41 Molecular Mechanisms for the RNA-Dependent ATPase Activity of Upf1 and Its Regulation by Upf2 Sutapa Chakrabarti, Uma Jayachandran, Fabien Bonneau, Francesca
Supplemental Information Molecular Cell, Volume 41 Molecular Mechanisms for the RNA-Dependent ATPase Activity of Upf1 and Its Regulation by Upf2 Sutapa Chakrabarti, Uma Jayachandran, Fabien Bonneau, Francesca
The Adhesion of Lactobacillus salivarius REN to a Human Intestinal Epithelial
 The Adhesion of Lactobacillus salivarius REN to a Human Intestinal Epithelial Cell Line Requires S-layer Proteins Ran Wang 1#, Lun Jiang 1#, Ming Zhang 2, Liang Zhao 1, Yanling Hao 1, Huiyuan Guo 1, Yue
The Adhesion of Lactobacillus salivarius REN to a Human Intestinal Epithelial Cell Line Requires S-layer Proteins Ran Wang 1#, Lun Jiang 1#, Ming Zhang 2, Liang Zhao 1, Yanling Hao 1, Huiyuan Guo 1, Yue
Supplemental Material Liu et al.
 Supplemental Material Liu et al. Supplemental Methods Protein expression, purification Recombinant etud11 (a.a 2344-2515) and Tud7-11 (a.a. 1617-2515) proteins (Tud7-11) were expressed in the Rosetta strain
Supplemental Material Liu et al. Supplemental Methods Protein expression, purification Recombinant etud11 (a.a 2344-2515) and Tud7-11 (a.a. 1617-2515) proteins (Tud7-11) were expressed in the Rosetta strain
Structural Bioinformatics (C3210) DNA and RNA Structure
 Structural Bioinformatics (C3210) DNA and RNA Structure Importance of DNA/RNA 3D Structure Nucleic acids are essential materials found in all living organisms. Their main function is to maintain and transmit
Structural Bioinformatics (C3210) DNA and RNA Structure Importance of DNA/RNA 3D Structure Nucleic acids are essential materials found in all living organisms. Their main function is to maintain and transmit
DNA & DNA : Protein Interactions BIBC 100
 DNA & DNA : Protein Interactions BIBC 100 Sequence = Information Alphabet = language L,I,F,E LIFE DNA = DNA code A, T, C, G CAC=Histidine CAG=Glutamine GGG=Glycine Protein = Protein code 20 a.a. LIVE EVIL
DNA & DNA : Protein Interactions BIBC 100 Sequence = Information Alphabet = language L,I,F,E LIFE DNA = DNA code A, T, C, G CAC=Histidine CAG=Glutamine GGG=Glycine Protein = Protein code 20 a.a. LIVE EVIL
Biochemistry 111. Carl Parker x A Braun
 Biochemistry 111 Carl Parker x6368 101A Braun csp@caltech.edu Central Dogma of Molecular Biology DNA-Dependent RNA Polymerase Requires a DNA Template Synthesizes RNA in a 5 to 3 direction Requires ribonucleoside
Biochemistry 111 Carl Parker x6368 101A Braun csp@caltech.edu Central Dogma of Molecular Biology DNA-Dependent RNA Polymerase Requires a DNA Template Synthesizes RNA in a 5 to 3 direction Requires ribonucleoside
RNP purification, components and activity.
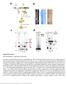 Supplementary Figure 1 RNP purification, components and activity. (a) Intein-mediated RNP production and purification. The Ll.LtrB intron RNA (red) (Exons E1 and E2 in green) and associated intron encoded
Supplementary Figure 1 RNP purification, components and activity. (a) Intein-mediated RNP production and purification. The Ll.LtrB intron RNA (red) (Exons E1 and E2 in green) and associated intron encoded
Nature Structural & Molecular Biology: doi: /nsmb.2307
 A novel locally-closed conformation of a bacterial pentameric proton-gated ion channel Marie S. Prevost*, Ludovic Sauguet*, Hugues Nury, Catherine Van Renterghem, Christèle Huon, Frederic Poitevin, Marc
A novel locally-closed conformation of a bacterial pentameric proton-gated ion channel Marie S. Prevost*, Ludovic Sauguet*, Hugues Nury, Catherine Van Renterghem, Christèle Huon, Frederic Poitevin, Marc
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10963 Supplementary Table 1 Data collection, phasing and refinement statistics. Crystal Native Derivative-1 (OsO 4 ) Derivative-2 (Orange-Pt) Data collection Space group C2 C2 C2 Cell
doi:10.1038/nature10963 Supplementary Table 1 Data collection, phasing and refinement statistics. Crystal Native Derivative-1 (OsO 4 ) Derivative-2 (Orange-Pt) Data collection Space group C2 C2 C2 Cell
Supplemental Material
 Supplemental Material Molecular basis for oncohistone H3 recognition by SETD2 methyltransferase Shuang Yang, 1, 2 Xiangdong Zheng, 1, 2, 3 Chao Lu, 4 Guo-Min Li, 2 C. David Allis, 4 1, 2, 3, 5* and Haitao
Supplemental Material Molecular basis for oncohistone H3 recognition by SETD2 methyltransferase Shuang Yang, 1, 2 Xiangdong Zheng, 1, 2, 3 Chao Lu, 4 Guo-Min Li, 2 C. David Allis, 4 1, 2, 3, 5* and Haitao
Protocol S1: Supporting Information
 Protocol S1: Supporting Information Basis for the specificity of the kinase domain of Abl for peptide substrates The crystal structures reported in this work were obtained using two different ATP analog-peptide
Protocol S1: Supporting Information Basis for the specificity of the kinase domain of Abl for peptide substrates The crystal structures reported in this work were obtained using two different ATP analog-peptide
The Skap-hom Dimerization and PH Domains Comprise
 Molecular Cell, Volume 32 Supplemental Data The Skap-hom Dimerization and PH Domains Comprise a 3 -Phosphoinositide-Gated Molecular Switch Kenneth D. Swanson, Yong Tang, Derek F. Ceccarelli, Florence Poy,
Molecular Cell, Volume 32 Supplemental Data The Skap-hom Dimerization and PH Domains Comprise a 3 -Phosphoinositide-Gated Molecular Switch Kenneth D. Swanson, Yong Tang, Derek F. Ceccarelli, Florence Poy,
Figure 1. FasterDB SEARCH PAGE corresponding to human WNK1 gene. In the search page, gene searching, in the mouse or human genome, can be done: 1- By
 1 2 3 Figure 1. FasterD SERCH PGE corresponding to human WNK1 gene. In the search page, gene searching, in the mouse or human genome, can be done: 1- y keywords (ENSEML ID, HUGO gene name, synonyms or
1 2 3 Figure 1. FasterD SERCH PGE corresponding to human WNK1 gene. In the search page, gene searching, in the mouse or human genome, can be done: 1- y keywords (ENSEML ID, HUGO gene name, synonyms or
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Multiple sequence alignments of four Swi2/Snf2 subfamily proteins, ScChd1, SsoRad54 and the RNA helicase Vasa. The sequence alignments of the Swi2/Snf2 subfamily proteins, ScChd1
Supplementary Figure 1 Multiple sequence alignments of four Swi2/Snf2 subfamily proteins, ScChd1, SsoRad54 and the RNA helicase Vasa. The sequence alignments of the Swi2/Snf2 subfamily proteins, ScChd1
Nature Genetics: doi: /ng.3556 INTEGRATED SUPPLEMENTARY FIGURE TEMPLATE. Supplementary Figure 1
 INTEGRATED SUPPLEMENTARY FIGURE TEMPLATE Supplementary Figure 1 REF6 expression in transgenic lines. (a,b) Expression of REF6 in REF6-HA ref6 and REF6ΔZnF-HA ref6 plants detected by RT qpcr (a) and immunoblot
INTEGRATED SUPPLEMENTARY FIGURE TEMPLATE Supplementary Figure 1 REF6 expression in transgenic lines. (a,b) Expression of REF6 in REF6-HA ref6 and REF6ΔZnF-HA ref6 plants detected by RT qpcr (a) and immunoblot
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Domain architecture and conformational states of the decapping complex, as revealed by structural studies. (a) Domain organization of Schizosaccharomyces pombe (Sp) and Saccharomyces
Supplementary Figure 1 Domain architecture and conformational states of the decapping complex, as revealed by structural studies. (a) Domain organization of Schizosaccharomyces pombe (Sp) and Saccharomyces
Supplementary Information For. A genetically encoded tool for manipulation of NADP + /NADPH in living cells
 Supplementary Information For A genetically encoded tool for manipulation of NADP + /NADPH in living cells Valentin Cracan 1,2,3, Denis V. Titov 1,2,3, Hongying Shen 1,2,3, Zenon Grabarek 1* and Vamsi
Supplementary Information For A genetically encoded tool for manipulation of NADP + /NADPH in living cells Valentin Cracan 1,2,3, Denis V. Titov 1,2,3, Hongying Shen 1,2,3, Zenon Grabarek 1* and Vamsi
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Supplementary Figure 1: Function of MICAL1 and dmical in cytokinesis (a) HeLa transfected with GFP- MICAL1 (green) were stained with Aurora B (red). Scale bars, 10 µm. (b) Western
SUPPLEMENTARY INFORMATION Supplementary Figure 1: Function of MICAL1 and dmical in cytokinesis (a) HeLa transfected with GFP- MICAL1 (green) were stained with Aurora B (red). Scale bars, 10 µm. (b) Western
TALENs (Transcription Activator-Like Effector Nucleases)
 TALENs (Transcription Activator-Like Effector Nucleases) The fundamental rationale between TALENs and ZFNs is similar, namely, combine a sequencespecific DNA-binding peptide domain with a nuclease domain
TALENs (Transcription Activator-Like Effector Nucleases) The fundamental rationale between TALENs and ZFNs is similar, namely, combine a sequencespecific DNA-binding peptide domain with a nuclease domain
Antibody-Antigen recognition. Structural Biology Weekend Seminar Annegret Kramer
 Antibody-Antigen recognition Structural Biology Weekend Seminar 10.07.2005 Annegret Kramer Contents Function and structure of antibodies Features of antibody-antigen interfaces Examples of antibody-antigen
Antibody-Antigen recognition Structural Biology Weekend Seminar 10.07.2005 Annegret Kramer Contents Function and structure of antibodies Features of antibody-antigen interfaces Examples of antibody-antigen
Structure of DNMT1-DNA Complex Reveals a Role for Autoinhibition in Maintenance DNA Methylation
 www.sciencemag.org/cgi/content/full/science.1195380/dc1 Supporting Online Material for Structure of DNMT1-DNA Complex Reveals a Role for Autoinhibition in Maintenance DNA Methylation Jikui Song, Olga Rechkoblit,
www.sciencemag.org/cgi/content/full/science.1195380/dc1 Supporting Online Material for Structure of DNMT1-DNA Complex Reveals a Role for Autoinhibition in Maintenance DNA Methylation Jikui Song, Olga Rechkoblit,
Supplemental Figure 1 Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical
 Supplemental Figure Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical in all six REEPs are highlighted in green. Additional
Supplemental Figure Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical in all six REEPs are highlighted in green. Additional
Solutions to 7.02 Quiz II 10/27/05
 Solutions to 7.02 Quiz II 10/27/05 Class Average = 83 Standard Deviation = 9 Range Grade % 87-100 A 43 74-86 B 39 55-73 C 17 > 54 D 1 Question 1 (56 points) While studying deep sea bacteria, you discover
Solutions to 7.02 Quiz II 10/27/05 Class Average = 83 Standard Deviation = 9 Range Grade % 87-100 A 43 74-86 B 39 55-73 C 17 > 54 D 1 Question 1 (56 points) While studying deep sea bacteria, you discover
CSE : Computational Issues in Molecular Biology. Lecture 19. Spring 2004
 CSE 397-497: Computational Issues in Molecular Biology Lecture 19 Spring 2004-1- Protein structure Primary structure of protein is determined by number and order of amino acids within polypeptide chain.
CSE 397-497: Computational Issues in Molecular Biology Lecture 19 Spring 2004-1- Protein structure Primary structure of protein is determined by number and order of amino acids within polypeptide chain.
Binding Studies with Mutants of Zif268
 THE JOURNAL OF BIOLOGICAL CHEISTRY Vol. 274, No. 27, Issue of July 2, pp. 19281 19285, 1999 1999 by The American Society for Biochemistry and olecular Biology, Inc. Printed in U.S.A. Binding Studies with
THE JOURNAL OF BIOLOGICAL CHEISTRY Vol. 274, No. 27, Issue of July 2, pp. 19281 19285, 1999 1999 by The American Society for Biochemistry and olecular Biology, Inc. Printed in U.S.A. Binding Studies with
SUPPLEMENTARY INFORMATION. Determinants of laminin polymerization revealed. by the structure of the α5 chain amino-terminal region
 SUPPLEMENTARY INFORMATION Determinants of laminin polymerization revealed by the structure of the α5 chain amino-terminal region Sadaf-Ahmahni Hussain 1, Federico Carafoli 1 & Erhard Hohenester 1 1 Department
SUPPLEMENTARY INFORMATION Determinants of laminin polymerization revealed by the structure of the α5 chain amino-terminal region Sadaf-Ahmahni Hussain 1, Federico Carafoli 1 & Erhard Hohenester 1 1 Department
SUPPLEMENTARY INFORMATION. Tolerance of a knotted near infrared fluorescent protein to random circular permutation
 SUPPLEMENTARY INFORMATION Tolerance of a knotted near infrared fluorescent protein to random circular permutation Naresh Pandey 1,3, Brianna E. Kuypers 2,4, Barbara Nassif 1, Emily E. Thomas 1,3, Razan
SUPPLEMENTARY INFORMATION Tolerance of a knotted near infrared fluorescent protein to random circular permutation Naresh Pandey 1,3, Brianna E. Kuypers 2,4, Barbara Nassif 1, Emily E. Thomas 1,3, Razan
Nature Genetics: doi: /ng Supplementary Figure 1. ChIP-seq genome browser views of BRM occupancy at previously identified BRM targets.
 Supplementary Figure 1 ChIP-seq genome browser views of BRM occupancy at previously identified BRM targets. Gene structures are shown underneath each panel. Supplementary Figure 2 pref6::ref6-gfp complements
Supplementary Figure 1 ChIP-seq genome browser views of BRM occupancy at previously identified BRM targets. Gene structures are shown underneath each panel. Supplementary Figure 2 pref6::ref6-gfp complements
GENES AND CHROMOSOMES V. Lecture 7. Biology Department Concordia University. Dr. S. Azam BIOL 266/
 1 GENES AND CHROMOSOMES V Lecture 7 BIOL 266/4 2014-15 Dr. S. Azam Biology Department Concordia University 2 CELL NUCLEUS AND THE CONTROL OF GENE EXPRESSION An Overview of Gene Regulation in Eukaryotes
1 GENES AND CHROMOSOMES V Lecture 7 BIOL 266/4 2014-15 Dr. S. Azam Biology Department Concordia University 2 CELL NUCLEUS AND THE CONTROL OF GENE EXPRESSION An Overview of Gene Regulation in Eukaryotes
Supplementary Figure 1.
 Supplementary Figure 1. Assessment of quaternary structure of soluble RSV F proteins. Soluble variants of F proteins from A2 and B1 RSV strains were expressed in HEK293 cells. The cell culture supernatants
Supplementary Figure 1. Assessment of quaternary structure of soluble RSV F proteins. Soluble variants of F proteins from A2 and B1 RSV strains were expressed in HEK293 cells. The cell culture supernatants
CS 4491/CS 7990 SPECIAL TOPICS IN BIOINFORMATICS
 1 CS 4491/CS 7990 SPECIAL TOPICS IN BIOINFORMATICS * Some contents are adapted from Dr. Jean Gao at UT Arlington Mingon Kang, PhD Computer Science, Kennesaw State University 2 Genetics The discovery of
1 CS 4491/CS 7990 SPECIAL TOPICS IN BIOINFORMATICS * Some contents are adapted from Dr. Jean Gao at UT Arlington Mingon Kang, PhD Computer Science, Kennesaw State University 2 Genetics The discovery of
Basic concepts of molecular biology
 Basic concepts of molecular biology Gabriella Trucco Email: gabriella.trucco@unimi.it What is life made of? 1665: Robert Hooke discovered that organisms are composed of individual compartments called cells
Basic concepts of molecular biology Gabriella Trucco Email: gabriella.trucco@unimi.it What is life made of? 1665: Robert Hooke discovered that organisms are composed of individual compartments called cells
BMB/Bi/Ch 170 Fall 2017 Problem Set 1: Proteins I
 BMB/Bi/Ch 170 Fall 2017 Problem Set 1: Proteins I Please use ray-tracing feature for all the images you are submitting. Use either the Ray button on the right side of the command window in PyMOL or variations
BMB/Bi/Ch 170 Fall 2017 Problem Set 1: Proteins I Please use ray-tracing feature for all the images you are submitting. Use either the Ray button on the right side of the command window in PyMOL or variations
Chapter 25: Regulating Eukaryotic Transcription The Ligand Responsive Activators
 Chapter 25: Regulating Eukaryotic Transcription The Ligand Responsive Activators At least 5 potential gene expression control points Superfamily of Gene Regulators Activation of gene structure Initiation
Chapter 25: Regulating Eukaryotic Transcription The Ligand Responsive Activators At least 5 potential gene expression control points Superfamily of Gene Regulators Activation of gene structure Initiation
3.1 Storing Information. Polypeptides. Protein Structure. Helical Secondary Structure. Types of Protein Structure. Molecular Biology Fourth Edition
 Lecture PowerPoint to accompany Molecular Biology Fourth Edition Robert F. Weaver Chapter 3 An Introduction to Gene Function 3.1 Storing Information Producing a protein from DNA information involves both
Lecture PowerPoint to accompany Molecular Biology Fourth Edition Robert F. Weaver Chapter 3 An Introduction to Gene Function 3.1 Storing Information Producing a protein from DNA information involves both
Recognition of transcription factors in complex with operator DNA at atomic resolution
 Recognition of transcription factors in complex with operator DNA at atomic resolution Authors: Y.N. Chirgadze 1 *, R.V. Polozov 2 1 Institute of Protein Research, Russian Academy of Sciences, Pushchino
Recognition of transcription factors in complex with operator DNA at atomic resolution Authors: Y.N. Chirgadze 1 *, R.V. Polozov 2 1 Institute of Protein Research, Russian Academy of Sciences, Pushchino
Structure determination and activity manipulation of the turfgrass ABA receptor FePYR1
 Supplemental information for: Structure determination and activity manipulation of the turfgrass AA receptor FePYR1 Zhizhong Ren 1, 2,#, Zhen Wang 1, 2,#, X Edward Zhou 3, Huazhong Shi 4, Yechun Hong 1,
Supplemental information for: Structure determination and activity manipulation of the turfgrass AA receptor FePYR1 Zhizhong Ren 1, 2,#, Zhen Wang 1, 2,#, X Edward Zhou 3, Huazhong Shi 4, Yechun Hong 1,
A 1. How many nitrogen atoms are found in the backbone of each amino acid? A. 1 B. 2 C. 3 D. 4
 Your Pre Build Model should have been impounded the morning of the competition. You may pick up your Pre Build model at the end of the competition after all models have been scored. Unclaimed models will
Your Pre Build Model should have been impounded the morning of the competition. You may pick up your Pre Build model at the end of the competition after all models have been scored. Unclaimed models will
Bioinformatics. ONE Introduction to Biology. Sami Khuri Department of Computer Science San José State University Biology/CS 123A Fall 2012
 Bioinformatics ONE Introduction to Biology Sami Khuri Department of Computer Science San José State University Biology/CS 123A Fall 2012 Biology Review DNA RNA Proteins Central Dogma Transcription Translation
Bioinformatics ONE Introduction to Biology Sami Khuri Department of Computer Science San José State University Biology/CS 123A Fall 2012 Biology Review DNA RNA Proteins Central Dogma Transcription Translation
Not The Real Exam Just for Fun! Chemistry 391 Fall 2018
 Not The Real Exam Just for Fun! Chemistry 391 Fall 2018 Do not open the exam until ready to begin! Rules of the Game: This is a take-home Exam. The exam is due on Thursday, October 11 th at 9 AM. Otherwise,
Not The Real Exam Just for Fun! Chemistry 391 Fall 2018 Do not open the exam until ready to begin! Rules of the Game: This is a take-home Exam. The exam is due on Thursday, October 11 th at 9 AM. Otherwise,
Supplemental Figure legends Figure S1. (A) (B) (C) (D) Figure S2. Figure S3. (A-E) Figure S4. Figure S5. (A, C, E, G, I) (B, D, F, H, Figure S6.
 Supplemental Figure legends Figure S1. Map-based cloning and complementation testing for ZOP1. (A) ZOP1 was mapped to a ~273-kb interval on Chromosome 1. In the interval, a single-nucleotide G to A substitution
Supplemental Figure legends Figure S1. Map-based cloning and complementation testing for ZOP1. (A) ZOP1 was mapped to a ~273-kb interval on Chromosome 1. In the interval, a single-nucleotide G to A substitution
SUPPLEMENTARY INFORMATION
 Results Construct purification and coupling. Two A1-GP1bα ReaLiSM constructs, with and without cysteine residues near the N and C-termini (Fig. S2a), were expressed and purified by Ni affinity chromatography
Results Construct purification and coupling. Two A1-GP1bα ReaLiSM constructs, with and without cysteine residues near the N and C-termini (Fig. S2a), were expressed and purified by Ni affinity chromatography
Roadmap. The Cell. Introduction to Molecular Biology. DNA RNA Protein Central dogma Genetic code Gene structure Human Genome
 Introduction to Molecular Biology Lodish et al Ch1-4 http://www.ncbi.nlm.nih.gov/books EECS 458 CWRU Fall 2004 DNA RNA Protein Central dogma Genetic code Gene structure Human Genome Roadmap The Cell Lodish
Introduction to Molecular Biology Lodish et al Ch1-4 http://www.ncbi.nlm.nih.gov/books EECS 458 CWRU Fall 2004 DNA RNA Protein Central dogma Genetic code Gene structure Human Genome Roadmap The Cell Lodish
Nucleic acids. How DNA works. DNA RNA Protein. DNA (deoxyribonucleic acid) RNA (ribonucleic acid) Central Dogma of Molecular Biology
 Nucleic acid chemistry and basic molecular theory Nucleic acids DNA (deoxyribonucleic acid) RNA (ribonucleic acid) Central Dogma of Molecular Biology Cell cycle DNA RNA Protein Transcription Translation
Nucleic acid chemistry and basic molecular theory Nucleic acids DNA (deoxyribonucleic acid) RNA (ribonucleic acid) Central Dogma of Molecular Biology Cell cycle DNA RNA Protein Transcription Translation
Genomics and Gene Recognition Genes and Blue Genes
 Genomics and Gene Recognition Genes and Blue Genes November 1, 2004 Prokaryotic Gene Structure prokaryotes are simplest free-living organisms studying prokaryotes can give us a sense what is the minimum
Genomics and Gene Recognition Genes and Blue Genes November 1, 2004 Prokaryotic Gene Structure prokaryotes are simplest free-living organisms studying prokaryotes can give us a sense what is the minimum
Structure-Guided Deimmunization CMPS 3210
 Structure-Guided Deimmunization CMPS 3210 Why Deimmunization? Protein, or biologic therapies are proving to be useful, but can be much more immunogenic than small molecules. Like a drug compound, a biologic
Structure-Guided Deimmunization CMPS 3210 Why Deimmunization? Protein, or biologic therapies are proving to be useful, but can be much more immunogenic than small molecules. Like a drug compound, a biologic
A Molecular Threading Mechanism Underlies Jumonji Lysine Demethylase KDM2A Regulation of Methylated H3K36. Supplementary Materials
 A Molecular Threading Mechanism Underlies Jumonji Lysine Demethylase KDM2A Regulation of Methylated H3K36 Zhongjun Cheng 1, Peggie Cheung 2, Alex J. Kuo 2, Erik T. Yukl 3, Carrie M. Wilmot 3, Or Gozani
A Molecular Threading Mechanism Underlies Jumonji Lysine Demethylase KDM2A Regulation of Methylated H3K36 Zhongjun Cheng 1, Peggie Cheung 2, Alex J. Kuo 2, Erik T. Yukl 3, Carrie M. Wilmot 3, Or Gozani
Hoffmann et al., http ://www.jcb.org /cgi /content /full /jcb /DC1
 Supplemental material JCB Hoffmann et al., http ://www.jcb.org /cgi /content /full /jcb.201506071 /DC1 THE JOU RNAL OF CELL BIO LOGY Figure S1. TRA IP harbors a PCNA-binding PIP box required for its recruitment
Supplemental material JCB Hoffmann et al., http ://www.jcb.org /cgi /content /full /jcb.201506071 /DC1 THE JOU RNAL OF CELL BIO LOGY Figure S1. TRA IP harbors a PCNA-binding PIP box required for its recruitment
Supplementatry Fig 1. Domain structure, biophysical characterisation and electron microscopy of a TD. (a) XTACC3/Maskin and XMAP215/chTOG domain
 Supplementatry Fig 1. Domain structure, biophysical characterisation and electron microscopy of a TD. (a) XTACC3/Maskin and XMAP215/chTOG domain architecture. Various C-terminal fragments were cloned and
Supplementatry Fig 1. Domain structure, biophysical characterisation and electron microscopy of a TD. (a) XTACC3/Maskin and XMAP215/chTOG domain architecture. Various C-terminal fragments were cloned and
Supplementary Figure S1. Binding of HSA mutants to hfcrn. (a) The levels of titrated amounts of HSA
 Supplementary Figure S1. Binding of HSA mutants to hfcrn. (a) The levels of titrated amounts of HSA variants (5.0-0.002 μg/ml) directly coated in the wells at ph 6.0 were controlled using a horseradish
Supplementary Figure S1. Binding of HSA mutants to hfcrn. (a) The levels of titrated amounts of HSA variants (5.0-0.002 μg/ml) directly coated in the wells at ph 6.0 were controlled using a horseradish
Supplemental Information. Autoregulatory Feedback Controls. Sequential Action of cis-regulatory Modules. at the brinker Locus
 Developmental Cell, Volume 26 Supplemental Information Autoregulatory Feedback Controls Sequential Action of cis-regulatory Modules at the brinker Locus Leslie Dunipace, Abbie Saunders, Hilary L. Ashe,
Developmental Cell, Volume 26 Supplemental Information Autoregulatory Feedback Controls Sequential Action of cis-regulatory Modules at the brinker Locus Leslie Dunipace, Abbie Saunders, Hilary L. Ashe,
Specificity: Induced Fit
 Specificity: Induced Fit Conformational changes may occur upon ligand binding (Daniel Koshland in 1958) This adaptation is called the induced fit Induced fit allows for tighter binding of the ligand Induced
Specificity: Induced Fit Conformational changes may occur upon ligand binding (Daniel Koshland in 1958) This adaptation is called the induced fit Induced fit allows for tighter binding of the ligand Induced
Transcription steps. Transcription steps. Eukaryote RNA processing
 Transcription steps Initiation at 5 end of gene binding of RNA polymerase to promoter unwinding of DNA Elongation addition of nucleotides to 3 end rules of base pairing requires Mg 2+ energy from NTP substrates
Transcription steps Initiation at 5 end of gene binding of RNA polymerase to promoter unwinding of DNA Elongation addition of nucleotides to 3 end rules of base pairing requires Mg 2+ energy from NTP substrates
File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description:
 File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description: Supplementary Figure 1. dcas9-mq1 fusion protein induces de novo
File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description: Supplementary Figure 1. dcas9-mq1 fusion protein induces de novo
Figure S1
 Supplementary Figure 1 The distribution of chlorophyll containing complexes eluted from DEAE-cellulose column in a sucrose gradient tube Six pigment-containing bands were resolved and identified as: B1,
Supplementary Figure 1 The distribution of chlorophyll containing complexes eluted from DEAE-cellulose column in a sucrose gradient tube Six pigment-containing bands were resolved and identified as: B1,
Laboratory 3. Molecular Genetics
 Laboratory 3. Molecular Genetics I. The Basics of DNA Structure DNA molecules are composed of small building blocks called nucleotides: A. Each DNA nucleotide is composed of three smaller molecules bonded
Laboratory 3. Molecular Genetics I. The Basics of DNA Structure DNA molecules are composed of small building blocks called nucleotides: A. Each DNA nucleotide is composed of three smaller molecules bonded
Supplemental Data. Guo et al. (2015). Plant Cell /tpc
 Supplemental Figure 1. The Mutant exb1-d Displayed Pleiotropic Phenotypes and Produced Branches in the Axils of Cotyledons. (A) Branches were developed in exb1-d but not in wild-type plants. (B) and (C)
Supplemental Figure 1. The Mutant exb1-d Displayed Pleiotropic Phenotypes and Produced Branches in the Axils of Cotyledons. (A) Branches were developed in exb1-d but not in wild-type plants. (B) and (C)
M1 - Biochemistry. Nucleic Acid Structure II/Transcription I
 M1 - Biochemistry Nucleic Acid Structure II/Transcription I PH Ratz, PhD (Resources: Lehninger et al., 5th ed., Chapters 8, 24 & 26) 1 Nucleic Acid Structure II/Transcription I Learning Objectives: 1.
M1 - Biochemistry Nucleic Acid Structure II/Transcription I PH Ratz, PhD (Resources: Lehninger et al., 5th ed., Chapters 8, 24 & 26) 1 Nucleic Acid Structure II/Transcription I Learning Objectives: 1.
Activation of a Floral Homeotic Gene in Arabidopsis
 Activation of a Floral Homeotic Gene in Arabidopsis By Maximiliam A. Busch, Kirsten Bomblies, and Detlef Weigel Presentation by Lis Garrett and Andrea Stevenson http://ucsdnews.ucsd.edu/archive/graphics/images/image5.jpg
Activation of a Floral Homeotic Gene in Arabidopsis By Maximiliam A. Busch, Kirsten Bomblies, and Detlef Weigel Presentation by Lis Garrett and Andrea Stevenson http://ucsdnews.ucsd.edu/archive/graphics/images/image5.jpg
DNA. Branden & Tooze, Ch. 7 Deoxyribose nucleic acids are made of three parts
 DNA Branden & Tooze, Ch. 7 Deoxyribose nucleic acids are made of three parts base: adenine, cytosine, guanine, thymine sugar: deoxyribose phosphate: will form the phosphate backbone wide narrow DNA binding
DNA Branden & Tooze, Ch. 7 Deoxyribose nucleic acids are made of three parts base: adenine, cytosine, guanine, thymine sugar: deoxyribose phosphate: will form the phosphate backbone wide narrow DNA binding
human Cdc45 Figure 1c. (c)
 1 Details of the refined crystallographic model of human Cdc45 and comparison of its active-site region with that of bacterial RecJ. (a) Stereo view of a representative example of the final 2F o -F c electron
1 Details of the refined crystallographic model of human Cdc45 and comparison of its active-site region with that of bacterial RecJ. (a) Stereo view of a representative example of the final 2F o -F c electron
Discovering gene regulatory control using ChIP-chip and ChIP-seq. Part 1. An introduction to gene regulatory control, concepts and methodologies
 Discovering gene regulatory control using ChIP-chip and ChIP-seq Part 1 An introduction to gene regulatory control, concepts and methodologies Ian Simpson ian.simpson@.ed.ac.uk http://bit.ly/bio2links
Discovering gene regulatory control using ChIP-chip and ChIP-seq Part 1 An introduction to gene regulatory control, concepts and methodologies Ian Simpson ian.simpson@.ed.ac.uk http://bit.ly/bio2links
Supplemental Figure 1 A
 Supplemental Figure 1 A Supplemental Data. Han et al. (2016). Plant Cell 10.1105/tpc.15.00997 BamHI -1616 bp SINE repeats SalI ATG SacI +1653 bp HindIII LB HYG p35s pfwa LUC Tnos RB pfwa-bamhi-f: CGGGATCCCGCCTTTCTCTTCCTCATCTGC
Supplemental Figure 1 A Supplemental Data. Han et al. (2016). Plant Cell 10.1105/tpc.15.00997 BamHI -1616 bp SINE repeats SalI ATG SacI +1653 bp HindIII LB HYG p35s pfwa LUC Tnos RB pfwa-bamhi-f: CGGGATCCCGCCTTTCTCTTCCTCATCTGC
All Rights Reserved. U.S. Patents 6,471,520B1; 5,498,190; 5,916, North Market Street, Suite CC130A, Milwaukee, WI 53202
 Secondary Structure In the previous protein folding activity, you created a hypothetical 15-amino acid protein and learned that basic principles of chemistry determine how each protein spontaneously folds
Secondary Structure In the previous protein folding activity, you created a hypothetical 15-amino acid protein and learned that basic principles of chemistry determine how each protein spontaneously folds
Algorithms in Bioinformatics ONE Transcription Translation
 Algorithms in Bioinformatics ONE Transcription Translation Sami Khuri Department of Computer Science San José State University sami.khuri@sjsu.edu Biology Review DNA RNA Proteins Central Dogma Transcription
Algorithms in Bioinformatics ONE Transcription Translation Sami Khuri Department of Computer Science San José State University sami.khuri@sjsu.edu Biology Review DNA RNA Proteins Central Dogma Transcription
Key Area 1.3: Gene Expression
 Key Area 1.3: Gene Expression RNA There is a second type of nucleic acid in the cell, called RNA. RNA plays a vital role in the production of protein from the code in the DNA. What is gene expression?
Key Area 1.3: Gene Expression RNA There is a second type of nucleic acid in the cell, called RNA. RNA plays a vital role in the production of protein from the code in the DNA. What is gene expression?
Genetic Parts in Bacterial Gene Expression
 Genetic Parts in Bacterial Gene Expression The Focus Promoters Operators Transcription Factors Transcriptional Terminators Ribosome Binding Sites An Abstract Annotation We ll Pay Particular Attention To:
Genetic Parts in Bacterial Gene Expression The Focus Promoters Operators Transcription Factors Transcriptional Terminators Ribosome Binding Sites An Abstract Annotation We ll Pay Particular Attention To:
