Additional file. Exploring metabolic engineering design principles for the photosynthetic production of lactic acid by Synechocystis sp.
|
|
|
- Randall Chapman
- 6 years ago
- Views:
Transcription
1 Additional file Exploring metabolic engineering design principles for the photosynthetic production of lactic acid by Synechocystis sp. PCC6803 S. Andreas Angermayr 1, Aniek D. van der Woude 2, Danilo Correddu 1,2, Angie Vreugdenhil 2, Valeria Verrone 1, Klaas J. Hellingwerf 1,2 * Equal contribution 1 Molecular Microbial Physiology Group, Swammerdam Institute for Life Sciences, University of Amsterdam and Netherlands Institute of Systems Biology, The Netherlands 2 Photanol B.V., Roeterstraat 35, Amsterdam, The Netherlands * Corresponding author at: Swammerdam Institute for Life Sciences, Science Park 904, 1098 XH Amsterdam, The Netherlands, tel: , K.J.Hellingwerf@uva.nl
2 Figure S1. Growth and rate of lactic acid production in Synechocystis mutants. (A-I) OD 730 values were used as a proxy for growth (left ordinate). The rate of lactic acid production is given in mmol lactic acid/l/h (right ordinate). A 2 nd order polynomial regression was used to fit the growth data, to achieve smoothing. As introduced in [1] the grey bars indicate the linear growth phase (compare Figure S5) used to correlate enzymatic activity and production rates for the metabolic control analysis (MCA). Error bars represent the SD of biological replicates (n = 3), except for SAW001, SYW003, and SYW004 where n = 2; if error bars are not visible they are smaller than the data point symbol.
3 Figure S2. Non-linear regression employing a rectangular hyperbola (Michaelis-Menten fit) [2] to fit the relation between the relative activity of the heterologously expressed LDH and the resulting rate of lactic acid production. Prism 5 (GraphPad software) was used with least square optimization. Y=(B max *X)/(K d +X); B max =12.99 (± 3.291); K d =10.54 (±4.109); R 2 =
4 Figure S3. K M determination of the purified B. subtilis LDHs BsLDH WT and BsLDH V38R. The V38R mutation leads to an increase of LDH activity with NADPH as the co-factor. The K M for pyruvate does not change significantly due to this mutation. Errors and error bars represent the SEM (n = 3); if error bars are not visible they are smaller than the data point symbol.
5 Figure S4. Alignment of selected LDH enzymes. Protein sequences were aligned using ClustalW2. The highly conserved GXGXXG 19 D motif in the Rossmann fold is indicated with arrows. The valine or (iso)leucine that was changed into an arginine to change cofactor specificity is marked in yellow.
6 Figure S5. Schematic interpretation of the subsequent growth phases in a batch culture of Synechocystis. A typical Synechocystis batch culture in BG-11 medium, cultivated at moderate light intensity (for details: see the Material and Methods section) initially shows an exponential growth phase (exp.) which gradually transits into a linear growth phase, because of increased cellular self-shading. After this linear growth phase, cells enter the late growth phase, in which additional limitations (e.g. for carbon or phosphate [3]) take place. We have selected the linear growth phase as the reference phase for calculation of the production characteristics of batch cultures of Synechocystis.
7 Table S1. Primers used in this study. Name Sequence* Additional Information H1seq_F AATGGTCCCAAAATTGTC Colony PCR segregation at slr0168 insertion H2seq_R CTGTGGGTAGTAAACTGGC Colony PCR segregation at slr0168 insertion KanF_back_Seq TCCCGTTGAATATGGCTC Colony PCR slr0168 insertion collldh_seq_r AAGTCTTCGGCATCACCTTG Colony PCR Llldh insertion Synpyk seq R GGCCTGCATATCTTTAATGG Colony PCR Synpk insertion Efpyk seq R GGAATTTCAACACCCATGTC Colony PCR Efpk insertion ECpyk seq R GAGGCTTCCAAGATTTCGTC Colony PCR Ecpk insertion LLpyk seq R TCGGCGGCTTCAATAATTTC Colony PCR Llpk insertion SpecR_out_F CCGAGGCATAGACTGTACCC Colony PCR pdf insertion Ppc-H1-SacII-F CCCGAGCTCGTTGCCCTCATAGAGCACC ppc deletion Ppc-H1-NotI-R CCCGCGGCCGCGTACAAGATACTACGGGA ppc deletion TT Ppc-H2-XhoI-F CCCCTCGAGCGTGCGTTACCGTCGTTATTC ppc deletion Ppc-H2-KpnI-R CCCGGTACCCACGGTTATGCCCGGTTCTG ppc deletion dppc_check_f TTGGCCGGATAAACCAATAA Colony PCR ppc deletion PPC_check_R GATTCCGTTGTTGCTCGTTT Colony PCR ppc deletion ppc_back_r AGCTCCCCGCAGTAGTTCTT Colony PCR ppc deletion collldh_mt_l39r_f AAGAATTGGGTATTGTGGACCGGTTTAAA GAAAAAACCCAAGG Introduction L39R mutation in LlLDH collldh_mt_l39r_r CCTTGGGTTTTTTCTTTAAACCGGTCCACA ATACCCAATTCTT Introduction L39R mutation in LlLDH cobsldh_nde_r CATATGAACAAGCACGTCAATAAAGT Cloning of Bsldh in pet28b+ cobsldh_he_r GATATCAAGCTTCTGCAGCGG Cloning of Bsldh in pet28b+ cochgldh_nde_f AATCATATGAGCACCAAAGAGAAGTTAAT Cloning of Cgldh in pet28b+ cochgldh_he_r AATGATATCAAGCTTCTGCAGCGG Cloning of Cgldh in pet28b+ cobsldh_r38v_f ACGAGTTAGTTGTCATTGATGTAAACAAA Removal V38R mutation in GAAAAGGCTATGGG BsLDH cobsldh_r38v_r CCCATAGCCTTTTCTTTGTTTACATCAATG Removal V38R mutation in ACAACTAACTCGT BsLDH cochgldh_r53v_f ACGAATTAGCTATGGTGGATGTGATGGAA GACAAGTTGAAGGG Removal V53R mutation in ChLDH cochgldh_r53v_r CCCTTCAACTTGTCTTCCATCACATCCACC ATAGCTAATTCGT Removal V53R mutation in ChLDH T7_FW TAATACGACTCACTATAGGG Verification of pet28b+ T7_RV GCTAGTTATTGCTCAGCGG Verification of pet28b+ cobsldh_nde_f AATCATATGAACAAGCACGTCAATAAAGT Cloning of Bsldh G cobsldh_bam_r AATGGATCCTTAATTGACTTTCTGTTCAGC Cloning of Bsldh GAA cochgldh_nde_f AATCATATGAGCACCAAAGAGAAGTTAAT Cloning of Cgldh CTCC cochgldh_bam_r AATGGATCCTTACAAGGTCAATTCTTTTTG Cloning of Cgldh CACTC Str_Sal_F_R AATGTCGACGGTGATTGATTGAGCAAGCT TT Construction of phsh * anchoring restriction sites are underlined
8 Table S2. E. coli strains and plasmids used in this study. Strain/plasmid Precursor * Relevant Insert(s) or resulting genotype** Application Reference E. coli XL-1 blue - Tet R Standard cloning host Stratagene E. coli EPI400 - CopyCutter TM cells for cloning Epicentre E. coli BL-21 (DE3) - Protein production host Novagen phkh001 pbluescript II SK+ - Δslr0168::Kan R, Amp R Insertion at slr0168 [4] phkh020 phkh001 - Ptrc1:: Ll ldh co ::tt::kan R Strain SAA023 [1] phkh003 phkh001 - Ptrc1:: Ll ldh na ::tt::kan R Strain SAA025 [1] pyw001 phkh001 S Ptrc2:: Syn pk co ::tt::ptrc2:: Ll ldh co ::tt::kan R Strain SYW001 This study pyw002 phkh001 S Ptrc2:: Bs pk co ::tt::ptrc2:: Ll ldh co ::tt::kan R This study pyw003 phkh001 S Ptrc2:: Ef pk co ::tt::ptrc2:: Ll ldh co ::tt::kan R Strain SYW003 This study pyw004 phkh001 S Ptrc2:: Ec pk co ::tt::ptrc2:: Ll ldh co ::tt::kan R Strain SYW004 This study pyw005 phkh001 S Ptrc2:: Ll pk co ::tt::ptrc2:: Ll ldh co ::tt::kan R Strain SYW005 This study pyw010 phkh001 S Ptrc1:: Syn pk co ::tt::ptrc1:: Ll ldh co ::tt::kan R pdc001 This study paw001 pyw001 C tt::ptrc2:: Ll ldh co ::Kan R Strain SAW001 This study pav001 phkh020 C Ptrc1:: Ll ldh co L39R::tt::Kan R Strain SAV001 This study psb1ac3 - Amp R, Cm R Standard BioBrick plasmid backbone psb1ac3_tt psb1ac3 BBa_B0014 (tt), Amp R, Cm R Standard BioBrick plasmid pac_ldh psb1ac3 C Ptrc1:: Ll ldh na ::tt paa026 This study pdc001 pyw010 + pacldh C Ptrc1:: Llldh na ::tt::ptrc1:: Ll ldh co ::tt::kan R Strain SAA026 This study paa027 paa028 C Ptrc1:: Bs ldh co ::tt::kan R Strain SAA027 This study paa028 phkh001 S Ptrc1:: Bs ldh co V38R::tt::Kan R Strain SAA028 This study pet28b+ - Kan R Standard cloning and expression vector Novagen petbsldhw petbsldhm C Bsldh co His-Tag purification of BsLDH This study
9 petbsldhm paa028 C Bsldh co V38R His-Tag purification of BsLDH V38R This study paa029 paa030 C Ptrc1:: Cg ldh co ::tt::kan R Strain SAA029 This study paa030 phkh001 S Ptrc1:: Cg ldh co V53R::tt::Kan R Strain SAA030 This study paa035 phkh001 S Ptrc1:: Po ldh co I29R::tt::Kan R Strain SAA035 This study pdf_lac RSF1010 derivative - laci, Strep R Self-replicating plasmid [5] pdf_ldh pdf_lac C Ptrc2:: Ll ldh co ::tt, Strep R Strains SAW035, SAW039, SAW041 This study phsh phkh001 C Strep R Insertion at slr0168 This study paw010 pbluescript C Strep R homologous regions surrounding ppc ppc deletion This study * S: directly synthesized and sub-cloned at Genscript (NJ, USA), C: derived from cloning procedures, -: previously described, see reference. ** Tet R : tetracyclin resistance, Kan R : kanamycin resistance cassette, Strep R : streptomycin/spectinomycin resistance cassette, Amp R : ampicillin resistance cassette, Cm R : chloramphenicol resistance cassette, Ptrc1: trc1 promoter, Ptrc2: trc2 promoter, tt: transcriptional terminator (BBa_B0014), co: codon-optimized, na: native, Ll: L. lactis, Bs: B. subtilis, Syn: Synechocystis, Ec: E. coli, Ef: E. faecalis, Cg: C. gunnari, Po: P. ovale.
10 References 1. Angermayr SA, Hellingwerf KJ: On the use of metabolic control analysis in the optimization of cyanobacterial biosolar cell factories. J Phys Chem B 2013, 117: Fell DA: Metabolic control analysis: a survey of its theoretical and experimental development. Biochem J 1992, 286(Pt 2): Kim HW, Vannela R, Zhou C, Rittmann BE: Nutrient acquisition and limitation for the photoautotrophic growth of Synechocystis sp. PCC6803 as a renewable biomass source. Biotechnol Bioeng 2011, 108: Angermayr SA, Paszota M, Hellingwerf KJ: Engineering a cyanobacterial cell factory for production of lactic acid. Appl Environ Microbiol 2012, 78: Guerrero F, Carbonell V, Cossu M, Correddu D, Jones PR: Ethylene synthesis and regulated expression of recombinant protein in Synechocystis sp. PCC PLoS ONE 2012, 7:e50470.
SUPPLEMENTAL MATERIAL. Chirality matters: Synthesis and consumption of the D-enantiomer of lactic acid by Synechocystis sp.
 SUPPLEMENTAL MATERIAL Chirality matters: Synthesis and consumption of the D-enantiomer of lactic acid by Synechocystis sp. PCC 6803 S. Andreas Angermayr* a, Aniek D. van der Woude* b,#, Danilo Correddu
SUPPLEMENTAL MATERIAL Chirality matters: Synthesis and consumption of the D-enantiomer of lactic acid by Synechocystis sp. PCC 6803 S. Andreas Angermayr* a, Aniek D. van der Woude* b,#, Danilo Correddu
Exploring metabolic engineering design principles for the photosynthetic production of lactic acid by Synechocystis sp. PCC6803
 Angermayr et al. Biotechnology for Biofuels 2014, 7:99 RESEARCH Open Access Exploring metabolic engineering design principles for the photosynthetic production of lactic acid by Synechocystis sp. PCC6803
Angermayr et al. Biotechnology for Biofuels 2014, 7:99 RESEARCH Open Access Exploring metabolic engineering design principles for the photosynthetic production of lactic acid by Synechocystis sp. PCC6803
Swammerdam Institute Klaas J. Hellingwerf. On the use and optimization of Synechocystis PCC6803 as a bio-solar cell factory in a bio-based economy
 Swammerdam Institute Klaas J. Hellingwerf On the use and optimization of Synechocystis PCC6803 as a bio-solar cell factory in a bio-based economy Acknowledgements: S.A. Angermayr, P. Savakis, Dr. A. de
Swammerdam Institute Klaas J. Hellingwerf On the use and optimization of Synechocystis PCC6803 as a bio-solar cell factory in a bio-based economy Acknowledgements: S.A. Angermayr, P. Savakis, Dr. A. de
Supplementary Note 1. Enzymatic properties of the purified Syn BVR
 Supplementary Note 1. Enzymatic properties of the purified Syn BVR The expression vector pet15b-syn bvr allowed us to routinely prepare 15 mg of electrophoretically homogenous Syn BVR from 2.5 L of TB-medium
Supplementary Note 1. Enzymatic properties of the purified Syn BVR The expression vector pet15b-syn bvr allowed us to routinely prepare 15 mg of electrophoretically homogenous Syn BVR from 2.5 L of TB-medium
Supplementary Material. Increased heterocyst frequency by patn disruption in Anabaena leads to enhanced photobiological
 Supplementary Material Increased heterocyst frequency by patn disruption in Anabaena leads to enhanced photobiological hydrogen production at high light intensity and high cell density Applied Microbiology
Supplementary Material Increased heterocyst frequency by patn disruption in Anabaena leads to enhanced photobiological hydrogen production at high light intensity and high cell density Applied Microbiology
Reading Lecture 3: 24-25, 45, Lecture 4: 66-71, Lecture 3. Vectors. Definition Properties Types. Transformation
 Lecture 3 Reading Lecture 3: 24-25, 45, 55-66 Lecture 4: 66-71, 75-79 Vectors Definition Properties Types Transformation 56 VECTORS- Definition Vectors are carriers of a DNA fragment of interest Insert
Lecture 3 Reading Lecture 3: 24-25, 45, 55-66 Lecture 4: 66-71, 75-79 Vectors Definition Properties Types Transformation 56 VECTORS- Definition Vectors are carriers of a DNA fragment of interest Insert
Schematic representation of the endogenous PALB2 locus and gene-disruption constructs
 Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Molecular Biology Services. Make Research Easy
 Molecular Biology Services Make Research Easy Gene-on-Demand Technology Platform Any gene conceivable in any vector you desire GenScript s Gene-on-Demand technology platform combines patented OptimumGene
Molecular Biology Services Make Research Easy Gene-on-Demand Technology Platform Any gene conceivable in any vector you desire GenScript s Gene-on-Demand technology platform combines patented OptimumGene
Supplementary Figure 1 Performance analysis of RedLibs (version 0.2.3). Supplementary Figure 2
 Supplementary Figure 1 Performance analysis of RedLibs (version 0.2.3). The required calculation time for the evaluation of all sub-libraries with a given target size are plotted against the combinatorial
Supplementary Figure 1 Performance analysis of RedLibs (version 0.2.3). The required calculation time for the evaluation of all sub-libraries with a given target size are plotted against the combinatorial
To date, lactic acid has been produced with almost 100% conversion
 crossmark Chirality Matters: Synthesis and Consumption of the D-Enantiomer of Lactic Acid by Synechocystis sp. Strain PCC6803 S. Andreas Angermayr, a Aniek D. van der Woude, b Danilo Correddu, a,b Ramona
crossmark Chirality Matters: Synthesis and Consumption of the D-Enantiomer of Lactic Acid by Synechocystis sp. Strain PCC6803 S. Andreas Angermayr, a Aniek D. van der Woude, b Danilo Correddu, a,b Ramona
The GeneEditor TM in vitro Mutagenesis System: Site- Directed Mutagenesis Using Altered Beta-Lactamase Specificity
 Promega Notes Magazine Number 62, 1997, p. 02 The GeneEditor TM in vitro Mutagenesis System: Site- Directed Mutagenesis Using Altered Beta-Lactamase Specificity By Christine Andrews and Scott Lesley Promega
Promega Notes Magazine Number 62, 1997, p. 02 The GeneEditor TM in vitro Mutagenesis System: Site- Directed Mutagenesis Using Altered Beta-Lactamase Specificity By Christine Andrews and Scott Lesley Promega
Supporting Information
 Supporting Information Wiley-VCH 2007 69451 Weinheim, Germany A Biosynthetic Route to Dehydroalanine Containing Proteins Jiangyun Wang, Stefan M. Schiller and Peter G. Schultz* Department of Chemistry
Supporting Information Wiley-VCH 2007 69451 Weinheim, Germany A Biosynthetic Route to Dehydroalanine Containing Proteins Jiangyun Wang, Stefan M. Schiller and Peter G. Schultz* Department of Chemistry
7.1 Techniques for Producing and Analyzing DNA. SBI4U Ms. Ho-Lau
 7.1 Techniques for Producing and Analyzing DNA SBI4U Ms. Ho-Lau What is Biotechnology? From Merriam-Webster: the manipulation of living organisms or their components to produce useful usually commercial
7.1 Techniques for Producing and Analyzing DNA SBI4U Ms. Ho-Lau What is Biotechnology? From Merriam-Webster: the manipulation of living organisms or their components to produce useful usually commercial
Step 1: Digest vector with Reason for Step 1. Step 2: Digest T4 genomic DNA with Reason for Step 2: Step 3: Reason for Step 3:
 Biol/Chem 475 Spring 2007 Study Problems for Quiz 2 Quiz 2 (~50 pts) is scheduled for Monday May 14 It will cover all handouts and lab exercises to date except the handout/worksheet (yet to be distributed)
Biol/Chem 475 Spring 2007 Study Problems for Quiz 2 Quiz 2 (~50 pts) is scheduled for Monday May 14 It will cover all handouts and lab exercises to date except the handout/worksheet (yet to be distributed)
Supplementary Information
 Supplementary Information Supplementary Figures Figure S1. Study of mgtl translation in vitro. (A) Detection of 5 LR RNA using wild-type and anti-sd (91-95) substituted templates in a transcription-translation
Supplementary Information Supplementary Figures Figure S1. Study of mgtl translation in vitro. (A) Detection of 5 LR RNA using wild-type and anti-sd (91-95) substituted templates in a transcription-translation
Building DNA Libraries to Explore the Combinatorial Design Space. Dr. Yifan Li Senior Scientist GenScript USA Inc.
 Building DNA Libraries to Explore the Combinatorial Design Space Dr. Yifan Li Senior Scientist GenScript USA Inc. Outline Combinatorial Optimization for Metabolic Pathway Engineering Case Studies for Combinatorial
Building DNA Libraries to Explore the Combinatorial Design Space Dr. Yifan Li Senior Scientist GenScript USA Inc. Outline Combinatorial Optimization for Metabolic Pathway Engineering Case Studies for Combinatorial
Table S1. Bacterial strains (Related to Results and Experimental Procedures)
 Table S1. Bacterial strains (Related to Results and Experimental Procedures) Strain number Relevant genotype Source or reference 1045 AB1157 Graham Walker (Donnelly and Walker, 1989) 2458 3084 (MG1655)
Table S1. Bacterial strains (Related to Results and Experimental Procedures) Strain number Relevant genotype Source or reference 1045 AB1157 Graham Walker (Donnelly and Walker, 1989) 2458 3084 (MG1655)
Understanding the Cellular Mechanism of the Excess Microsporocytes I (EMSI) Gene. Andrew ElBardissi, The Pennsylvania State University
 Understanding the Cellular Mechanism of the Excess Microsporocytes I (EMSI) Gene Andrew ElBardissi, The Pennsylvania State University Abstract: Hong Ma, The Pennsylvania State University The Excess Microsporocytes
Understanding the Cellular Mechanism of the Excess Microsporocytes I (EMSI) Gene Andrew ElBardissi, The Pennsylvania State University Abstract: Hong Ma, The Pennsylvania State University The Excess Microsporocytes
Engineering E. coli for xylitol production during growth on xylose
 Engineering E. coli for xylitol production during growth on xylose Olubolaji Akinterinwa & Patrick C. Cirino IBE 2008 Annual Meeting, March 7th 008-17 Advances in Engineering Microbial Metabolism Xylitol
Engineering E. coli for xylitol production during growth on xylose Olubolaji Akinterinwa & Patrick C. Cirino IBE 2008 Annual Meeting, March 7th 008-17 Advances in Engineering Microbial Metabolism Xylitol
Bi 8 Lecture 4. Ellen Rothenberg 14 January Reading: from Alberts Ch. 8
 Bi 8 Lecture 4 DNA approaches: How we know what we know Ellen Rothenberg 14 January 2016 Reading: from Alberts Ch. 8 Central concept: DNA or RNA polymer length as an identifying feature RNA has intrinsically
Bi 8 Lecture 4 DNA approaches: How we know what we know Ellen Rothenberg 14 January 2016 Reading: from Alberts Ch. 8 Central concept: DNA or RNA polymer length as an identifying feature RNA has intrinsically
Confirming the Phenotypes of E. coli Strains
 Confirming the Phenotypes of E. coli Strains INTRODUCTION Before undertaking any experiments, we need to confirm that the phenotypes of the E. coli strains we intend to use in the planned experiments correspond
Confirming the Phenotypes of E. coli Strains INTRODUCTION Before undertaking any experiments, we need to confirm that the phenotypes of the E. coli strains we intend to use in the planned experiments correspond
CopyCutter EPI400 Electrocompetent E. coli CopyCutter EPI400 Chemically Competent E. coli CopyCutter Induction Solution
 CopyCutter EPI400 Electrocompetent E. coli CopyCutter EPI400 Chemically Competent E. coli CopyCutter Induction Solution Cat. Nos. C400EL10, C400CH10, and CIS40025 Available exclusively thru Lucigen. lucigen.com/epibio
CopyCutter EPI400 Electrocompetent E. coli CopyCutter EPI400 Chemically Competent E. coli CopyCutter Induction Solution Cat. Nos. C400EL10, C400CH10, and CIS40025 Available exclusively thru Lucigen. lucigen.com/epibio
GenBuilder TM Plus Cloning Kit User Manual
 GenBuilder TM Plus Cloning Kit User Manual Cat. No. L00744 Version 11242017 Ⅰ. Introduction... 2 I.1 Product Information... 2 I.2 Kit Contents and Storage... 2 I.3 GenBuilder Cloning Kit Workflow... 2
GenBuilder TM Plus Cloning Kit User Manual Cat. No. L00744 Version 11242017 Ⅰ. Introduction... 2 I.1 Product Information... 2 I.2 Kit Contents and Storage... 2 I.3 GenBuilder Cloning Kit Workflow... 2
Mycobacterium tuberculosis (Mtb) and Mycobacterium smegmatis (Msmeg) contain two ε (dnaq) exonuclease homologs.
 Supplementary Figure 1 Mycobacterium tuberculosis (Mtb) and Mycobacterium smegmatis (Msmeg) contain two ε (dnaq) exonuclease homologs. (a) Sequence alignment of the ε-exonuclease homologs from four different
Supplementary Figure 1 Mycobacterium tuberculosis (Mtb) and Mycobacterium smegmatis (Msmeg) contain two ε (dnaq) exonuclease homologs. (a) Sequence alignment of the ε-exonuclease homologs from four different
Supporting Information
 Supporting Information Gómez-Marín et al. 10.1073/pnas.1505463112 SI Materials and Methods Generation of BAC DK74B2-six2a::GFP-six3a::mCherry-iTol2. BAC clone (number 74B2) from DanioKey zebrafish BAC
Supporting Information Gómez-Marín et al. 10.1073/pnas.1505463112 SI Materials and Methods Generation of BAC DK74B2-six2a::GFP-six3a::mCherry-iTol2. BAC clone (number 74B2) from DanioKey zebrafish BAC
GenBuilder TM Plus Cloning Kit User Manual
 GenBuilder TM Plus Cloning Kit User Manual Cat.no L00744 Version 11242017 Ⅰ. Introduction... 2 I.1 Product Information... 2 I.2 Kit Contents and Storage... 2 I.3 GenBuilder Cloning Kit Workflow... 2 Ⅱ.
GenBuilder TM Plus Cloning Kit User Manual Cat.no L00744 Version 11242017 Ⅰ. Introduction... 2 I.1 Product Information... 2 I.2 Kit Contents and Storage... 2 I.3 GenBuilder Cloning Kit Workflow... 2 Ⅱ.
Stabilization of a virus-like particle and its application as a nanoreactor at physiological conditions
 Supporting Information Stabilization of a virus-like particle and its application as a nanoreactor at physiological conditions Lise Schoonen, b Sjors Maassen, b Roeland J. M. Nolte b and Jan C. M. van
Supporting Information Stabilization of a virus-like particle and its application as a nanoreactor at physiological conditions Lise Schoonen, b Sjors Maassen, b Roeland J. M. Nolte b and Jan C. M. van
Orange fluorescent proteins shift constructed from cyanobacteriochromes. chromophorylated with phycoerythrobilin. Kai-Hong Zhao 1 Ming Zhou 1, *
 Electronic Supplementary Material (ESI) for Photochemical. This journal is The Royal Society of Chemistry and Owner Societies 2014 Orange fluorescent proteins shift constructed from cyanobacteriochromes
Electronic Supplementary Material (ESI) for Photochemical. This journal is The Royal Society of Chemistry and Owner Societies 2014 Orange fluorescent proteins shift constructed from cyanobacteriochromes
Combinatory strategy for characterizing and understanding the ethanol synthesis pathway in cyanobacteria cell factories
 DOI 10.1186/s13068-015-0367-z RESEARCH Open Access Combinatory strategy for characterizing and understanding the ethanol synthesis pathway in cyanobacteria cell factories Guodong Luan 1,2, Yunjing Qi 4,
DOI 10.1186/s13068-015-0367-z RESEARCH Open Access Combinatory strategy for characterizing and understanding the ethanol synthesis pathway in cyanobacteria cell factories Guodong Luan 1,2, Yunjing Qi 4,
Supplemental Data. Li et al. (2015). Plant Cell /tpc
 Supplemental Data Supplemental Figure 1: Characterization of asr3 T-DNA knockout lines and complementation transgenic lines. (A) The scheme of At2G33550 (ASR3) with gray boxes indicating exons and dash
Supplemental Data Supplemental Figure 1: Characterization of asr3 T-DNA knockout lines and complementation transgenic lines. (A) The scheme of At2G33550 (ASR3) with gray boxes indicating exons and dash
Gateway Vectors for BiFC
 Gateway Vectors for BiFC 1. The enhanced YFP (EYFP) are used (Split EYFP). 2. The Fusion fusion gene is expressed by CaMV35S promoter. 3. The N- or C-terminal fragments of EYFP are fused subsequent to
Gateway Vectors for BiFC 1. The enhanced YFP (EYFP) are used (Split EYFP). 2. The Fusion fusion gene is expressed by CaMV35S promoter. 3. The N- or C-terminal fragments of EYFP are fused subsequent to
Supporting Information
 Supporting Information Development of a 2,4-Dinitrotoluene-Responsive Synthetic Riboswitch in E. coli cells Molly E. Davidson, Svetlana V. Harbaugh, Yaroslav G. Chushak, Morley O. Stone, Nancy Kelley-
Supporting Information Development of a 2,4-Dinitrotoluene-Responsive Synthetic Riboswitch in E. coli cells Molly E. Davidson, Svetlana V. Harbaugh, Yaroslav G. Chushak, Morley O. Stone, Nancy Kelley-
How Do You Clone a Gene?
 S-20 Edvo-Kit #S-20 How Do You Clone a Gene? Experiment Objective: The objective of this experiment is to gain an understanding of the structure of DNA, a genetically engineered clone, and how genes are
S-20 Edvo-Kit #S-20 How Do You Clone a Gene? Experiment Objective: The objective of this experiment is to gain an understanding of the structure of DNA, a genetically engineered clone, and how genes are
Mcbio 316: Exam 3. (10) 2. Compare and contrast operon vs gene fusions.
 Mcbio 316: Exam 3 Name (15) 1. Transposons provide useful tools for genetic analysis. List 5 different uses of transposon insertions. ANSWER: Many answers are possible, however, if multiple items on the
Mcbio 316: Exam 3 Name (15) 1. Transposons provide useful tools for genetic analysis. List 5 different uses of transposon insertions. ANSWER: Many answers are possible, however, if multiple items on the
Supplementary Figure 1 Autoaggregation of E. coli W3110 in presence of native
 Supplementary Figure 1 Autoaggregation of E. coli W3110 in presence of native Ag43 phase variation depends on Ag43, motility, chemotaxis and AI-2 sensing. Cells were grown to OD600 of 0.6 at 37 C and aggregation
Supplementary Figure 1 Autoaggregation of E. coli W3110 in presence of native Ag43 phase variation depends on Ag43, motility, chemotaxis and AI-2 sensing. Cells were grown to OD600 of 0.6 at 37 C and aggregation
Supporting Information. Sequence Independent Cloning and Posttranslational. Enzymatic Ligation
 Supporting Information Sequence Independent Cloning and Posttranslational Modification of Repetitive Protein Polymers through Sortase and Sfp-mediated Enzymatic Ligation Wolfgang Ott,,, Thomas Nicolaus,,
Supporting Information Sequence Independent Cloning and Posttranslational Modification of Repetitive Protein Polymers through Sortase and Sfp-mediated Enzymatic Ligation Wolfgang Ott,,, Thomas Nicolaus,,
A universal chassis plasmid pwh34 was first constructed to facilitate the
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 File name: Supplementary method Plasmids construction A universal chassis plasmid pwh34 was first constructed to facilitate the construction
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 File name: Supplementary method Plasmids construction A universal chassis plasmid pwh34 was first constructed to facilitate the construction
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than. SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with
 Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
Exam 2 Key - Spring 2008 A#: Please see us if you have any questions!
 Page 1 of 5 Exam 2 Key - Spring 2008 A#: Please see us if you have any questions! 1. A mutation in which parts of two nonhomologous chromosomes change places is called a(n) A. translocation. B. transition.
Page 1 of 5 Exam 2 Key - Spring 2008 A#: Please see us if you have any questions! 1. A mutation in which parts of two nonhomologous chromosomes change places is called a(n) A. translocation. B. transition.
Design. Construction. Characterization
 Design Construction Characterization DNA mrna (messenger) A C C transcription translation C A C protein His A T G C T A C G Plasmids replicon copy number incompatibility selection marker origin of replication
Design Construction Characterization DNA mrna (messenger) A C C transcription translation C A C protein His A T G C T A C G Plasmids replicon copy number incompatibility selection marker origin of replication
In vitro mutagenesis
 Core course BMS361N Genetic Engineering In vitro mutagenesis Prof. Narkunaraja Shanmugam Dept. Of Biomedical Science School of Basic Medical Sciences Bharathidasan University In vitro mutagenesis Many
Core course BMS361N Genetic Engineering In vitro mutagenesis Prof. Narkunaraja Shanmugam Dept. Of Biomedical Science School of Basic Medical Sciences Bharathidasan University In vitro mutagenesis Many
Bi 8 Lecture 5. Ellen Rothenberg 19 January 2016
 Bi 8 Lecture 5 MORE ON HOW WE KNOW WHAT WE KNOW and intro to the protein code Ellen Rothenberg 19 January 2016 SIZE AND PURIFICATION BY SYNTHESIS: BASIS OF EARLY SEQUENCING complex mixture of aborted DNA
Bi 8 Lecture 5 MORE ON HOW WE KNOW WHAT WE KNOW and intro to the protein code Ellen Rothenberg 19 January 2016 SIZE AND PURIFICATION BY SYNTHESIS: BASIS OF EARLY SEQUENCING complex mixture of aborted DNA
Genetic Engineering for Biofuels Production
 Genetic Engineering for Biofuels Production WSE 573 Spring 2013 Greeley Beck INTRODUCTION Alternative transportation fuels are needed in the United States because of oil supply insecurity, oil price increases,
Genetic Engineering for Biofuels Production WSE 573 Spring 2013 Greeley Beck INTRODUCTION Alternative transportation fuels are needed in the United States because of oil supply insecurity, oil price increases,
Chapter VI: Mutagenesis to Restore Chimera Function
 99 Chapter VI: Mutagenesis to Restore Chimera Function Introduction Identifying characteristics of functional and nonfunctional chimeras is one way to address the underlying reasons for why chimeric proteins
99 Chapter VI: Mutagenesis to Restore Chimera Function Introduction Identifying characteristics of functional and nonfunctional chimeras is one way to address the underlying reasons for why chimeric proteins
A Domain Swapping Study of Nano-Capsule Proteins. Rongli Fan, Aimee L. Boyle, Vee Vee Cheong, See Liang Ng, Brendan P. Orner*
 A Domain Swapping Study of Nano-Capsule Proteins Rongli Fan, Aimee L. Boyle, Vee Vee Cheong, See Liang Ng, Brendan P. Orner* Figure S1. Amino acid sequences of proteins used in this study. Grey shading
A Domain Swapping Study of Nano-Capsule Proteins Rongli Fan, Aimee L. Boyle, Vee Vee Cheong, See Liang Ng, Brendan P. Orner* Figure S1. Amino acid sequences of proteins used in this study. Grey shading
Programmable Sequence-Specific Transcriptional Regulation of Mammalian Genome Using Designer TAL Effectors
 Supplementary Information Programmable Sequence-Specific Transcriptional Regulation of Mammalian Genome Using Designer TAL Effectors Feng Zhang 1,2,3,5,7 *,±, Le Cong 2,3,4 *, Simona Lodato 5,6, Sriram
Supplementary Information Programmable Sequence-Specific Transcriptional Regulation of Mammalian Genome Using Designer TAL Effectors Feng Zhang 1,2,3,5,7 *,±, Le Cong 2,3,4 *, Simona Lodato 5,6, Sriram
Supplemental Fig. S1. Key to underlines: Key to amino acids:
 AspA-F1 AspA 1 MKQMETKGYGYFRKTKAYGLVCGIT--------------LAGALTLGTTSVSADDVTTLNPATNLTTLQTPPTADQTQLAHQAGQQSGELVSEVSNTEWD 86 SspB 1 MQKREV--FG-FRKSKVAKTLCGAV-LGAALIAIADQQVLADEVTETNSTANVAVTTTGNPATNLPEAQGEATEAASQSQAQAGSKDGALPVEVSADDLN
AspA-F1 AspA 1 MKQMETKGYGYFRKTKAYGLVCGIT--------------LAGALTLGTTSVSADDVTTLNPATNLTTLQTPPTADQTQLAHQAGQQSGELVSEVSNTEWD 86 SspB 1 MQKREV--FG-FRKSKVAKTLCGAV-LGAALIAIADQQVLADEVTETNSTANVAVTTTGNPATNLPEAQGEATEAASQSQAQAGSKDGALPVEVSADDLN
Applicazioni biotecnologiche
 Applicazioni biotecnologiche Analisi forense Sintesi di proteine ricombinanti Restriction Fragment Length Polymorphism (RFLP) Polymorphism (more fully genetic polymorphism) refers to the simultaneous occurrence
Applicazioni biotecnologiche Analisi forense Sintesi di proteine ricombinanti Restriction Fragment Length Polymorphism (RFLP) Polymorphism (more fully genetic polymorphism) refers to the simultaneous occurrence
Standards for Safety Assessments of Food Additives produced Using Genetically Modified Microorganisms
 Standards for Safety Assessments of Food Additives produced Using Genetically Modified Microorganisms (Food Safety Commission Decision of March 25, 2004) Chapter 1 General Provisions No. 1 Background on
Standards for Safety Assessments of Food Additives produced Using Genetically Modified Microorganisms (Food Safety Commission Decision of March 25, 2004) Chapter 1 General Provisions No. 1 Background on
Lab Book igem Stockholm Lysostaphin. Week 8
 Lysostaphin Week 8 Summarized below are the experiments conducted this week in chronological order. Click on the experiment name to view it. To go back to this summary, click Summary in the footer. Summary
Lysostaphin Week 8 Summarized below are the experiments conducted this week in chronological order. Click on the experiment name to view it. To go back to this summary, click Summary in the footer. Summary
! Intended Learning Outcomes:
 Introduction to Part Assembly: Building with BioBrick TM Objective: To simulate a standard method for assembling biological parts. Intended Learning Outcomes: 1. Students will be able to distinguish and
Introduction to Part Assembly: Building with BioBrick TM Objective: To simulate a standard method for assembling biological parts. Intended Learning Outcomes: 1. Students will be able to distinguish and
Computational Biology 2. Pawan Dhar BII
 Computational Biology 2 Pawan Dhar BII Lecture 1 Introduction to terms, techniques and concepts in molecular biology Molecular biology - a primer Human body has 100 trillion cells each containing 3 billion
Computational Biology 2 Pawan Dhar BII Lecture 1 Introduction to terms, techniques and concepts in molecular biology Molecular biology - a primer Human body has 100 trillion cells each containing 3 billion
Use of In-Fusion Cloning for Simple and Efficient Assembly of Gene Constructs No restriction enzymes or ligation reactions necessary
 No restriction enzymes or ligation reactions necessary Background The creation of genetic circuits and artificial biological systems typically involves the use of modular genetic components biological
No restriction enzymes or ligation reactions necessary Background The creation of genetic circuits and artificial biological systems typically involves the use of modular genetic components biological
SUPPLEMENTARY INFORMATION
 Ca 2+ /calmodulin Regulates Salicylic Acid-mediated Plant Immunity Liqun Du, Gul S. Ali, Kayla A. Simons, Jingguo Hou, Tianbao Yang, A.S.N. Reddy and B. W. Poovaiah * *To whom correspondence should be
Ca 2+ /calmodulin Regulates Salicylic Acid-mediated Plant Immunity Liqun Du, Gul S. Ali, Kayla A. Simons, Jingguo Hou, Tianbao Yang, A.S.N. Reddy and B. W. Poovaiah * *To whom correspondence should be
SUPPLEMENTARY MATERIAL
 SUPPLEMENTARY MATERIAL Engineering a pyridoxal '-phosphate supply for cadaverine production by using Escherichia coli whole-cell biocatalysis Weichao Ma 1,,, Weijia Cao 1,, Bowen Zhang 1,, Kequan Chen
SUPPLEMENTARY MATERIAL Engineering a pyridoxal '-phosphate supply for cadaverine production by using Escherichia coli whole-cell biocatalysis Weichao Ma 1,,, Weijia Cao 1,, Bowen Zhang 1,, Kequan Chen
1. Growth Models a) Using a Monod growth model, a line was fit to E. coli growth data as shown below in Figure 1. It was found that µ max
 ChE 170B. Midterm Examination 1 6 March 2009 1. Growth Models a) Using a Monod growth model, a line was fit to E. coli growth data as shown below in Figure 1. It was found that µ max was 1.2. The same
ChE 170B. Midterm Examination 1 6 March 2009 1. Growth Models a) Using a Monod growth model, a line was fit to E. coli growth data as shown below in Figure 1. It was found that µ max was 1.2. The same
Synthetic Biology. Sustainable Energy. Therapeutics Industrial Enzymes. Agriculture. Accelerating Discoveries, Expanding Possibilities. Design.
 Synthetic Biology Accelerating Discoveries, Expanding Possibilities Sustainable Energy Therapeutics Industrial Enzymes Agriculture Design Build Generate Solutions to Advance Synthetic Biology Research
Synthetic Biology Accelerating Discoveries, Expanding Possibilities Sustainable Energy Therapeutics Industrial Enzymes Agriculture Design Build Generate Solutions to Advance Synthetic Biology Research
Packaging of P22 DNA requires a pac site while packaging of lambda DNA requires a cos site. Briefly describe:
 1). (12 Points) Packaging of P22 DNA requires a pac site while packaging of lambda DNA requires a cos site. Briefly describe: 1. The mechanisms used by P22 and lambda to package DNA. P22 uses a headfull
1). (12 Points) Packaging of P22 DNA requires a pac site while packaging of lambda DNA requires a cos site. Briefly describe: 1. The mechanisms used by P22 and lambda to package DNA. P22 uses a headfull
Lecture 22: Molecular techniques DNA cloning and DNA libraries
 Lecture 22: Molecular techniques DNA cloning and DNA libraries DNA cloning: general strategy -> to prepare large quantities of identical DNA Vector + DNA fragment Recombinant DNA (any piece of DNA derived
Lecture 22: Molecular techniques DNA cloning and DNA libraries DNA cloning: general strategy -> to prepare large quantities of identical DNA Vector + DNA fragment Recombinant DNA (any piece of DNA derived
GenBuilder TM Cloning Kit User Manual
 GenBuilder TM Cloning Kit User Manual Cat.no L00701 Version 11242017 Ⅰ. Introduction... 2 I.1 Product Information... 2 I.2 Kit Contents and Storage... 2 I.3 GenBuilder Cloning Kit Workflow... 2 Ⅱ. DNA
GenBuilder TM Cloning Kit User Manual Cat.no L00701 Version 11242017 Ⅰ. Introduction... 2 I.1 Product Information... 2 I.2 Kit Contents and Storage... 2 I.3 GenBuilder Cloning Kit Workflow... 2 Ⅱ. DNA
7.02 Recombinant DNA Methods Spring 2005 Exam Study Questions Answer Key
 MIT Department of Biology 7.02 Experimental Biology & Communication, Spring 2005 7.02/10.702 Spring 2005 RDM Exam Study Questions 7.02 Recombinant DNA Methods Spring 2005 Exam Study Questions Answer Key
MIT Department of Biology 7.02 Experimental Biology & Communication, Spring 2005 7.02/10.702 Spring 2005 RDM Exam Study Questions 7.02 Recombinant DNA Methods Spring 2005 Exam Study Questions Answer Key
Arsenite oxidation regulator AioR regulates bacterial chemotaxis towards. arsenite in Agrobacterium tumefaciens GW4
 1 2 Arsenite oxidation regulator AioR regulates bacterial chemotaxis towards arsenite in Agrobacterium tumefaciens GW4 3 4 5 Kaixiang Shi 1, Xia Fan 1, Zixu Qiao 1, Yushan Han 1, Timothy R. McDermott 2,
1 2 Arsenite oxidation regulator AioR regulates bacterial chemotaxis towards arsenite in Agrobacterium tumefaciens GW4 3 4 5 Kaixiang Shi 1, Xia Fan 1, Zixu Qiao 1, Yushan Han 1, Timothy R. McDermott 2,
Your Protein Manufacturer
 Expression of Toxic Proteins in E. coli Your Protein Manufacturer Contents 1. Definition of Toxic Protein 2. Mechanisms of Protein Toxicity 3. Percentage of Protein Toxicity 4. Phenotypes of Protein Toxicity
Expression of Toxic Proteins in E. coli Your Protein Manufacturer Contents 1. Definition of Toxic Protein 2. Mechanisms of Protein Toxicity 3. Percentage of Protein Toxicity 4. Phenotypes of Protein Toxicity
Please sign below if you wish to have your grades posted by the last five digits of your SSN
 BIO 226R EXAM III (Sample) PRINT YOUR NAME Please sign below if you wish to have your grades posted by the last five digits of your Signature BIO 226R Exam III has 8 pages, and 26 questions. There are
BIO 226R EXAM III (Sample) PRINT YOUR NAME Please sign below if you wish to have your grades posted by the last five digits of your Signature BIO 226R Exam III has 8 pages, and 26 questions. There are
G0796 pacad5tretightmcssv40pa
 G0796 pacad5tretightmcssv40pa Plasmid Features: 6009bp Coordinates Feature ITR: 16-118 Ad5 wt 16-365 and 1240-3698 TRE-tight: 402-717 SV40: 802-1239 Ampicillin: 5806-4946 Antibiotic Resistance: Ampicillin
G0796 pacad5tretightmcssv40pa Plasmid Features: 6009bp Coordinates Feature ITR: 16-118 Ad5 wt 16-365 and 1240-3698 TRE-tight: 402-717 SV40: 802-1239 Ampicillin: 5806-4946 Antibiotic Resistance: Ampicillin
Molecular Biology Techniques Supporting IBBE
 Molecular Biology Techniques Supporting IBBE Jared Cartwright Protein Production Lab Head Contact Details: email jared.cartwright@york.ac.uk Phone 01904 328797 Presentation Aims Gene synthesis Cloning
Molecular Biology Techniques Supporting IBBE Jared Cartwright Protein Production Lab Head Contact Details: email jared.cartwright@york.ac.uk Phone 01904 328797 Presentation Aims Gene synthesis Cloning
Bio Course Syllabus Spring Biology Spring Biology Second Year Laboratory
 Biology 216 - Spring 2015 - Biology Second Year Laboratory Laboratory Sections. CRN 55840-2 W 2:00 pm 4:50 pm ISC 107, 302, 304 Instructors. Dr. Robert. D. Simon, Room ISC 358 Office Hours: W 10:00-12:00,
Biology 216 - Spring 2015 - Biology Second Year Laboratory Laboratory Sections. CRN 55840-2 W 2:00 pm 4:50 pm ISC 107, 302, 304 Instructors. Dr. Robert. D. Simon, Room ISC 358 Office Hours: W 10:00-12:00,
G0463 pscaavmcsbghpa MCS. Plasmid Features:
 3200 G0463 pscaavmcsbghpa Plasmid Features: Coordinates Feature 980-1085 AAV2 ITR 106bp (mutated ITR) 1110-1226 MCS 1227-1440 BgHpA 1453-1595 AAV2 ITR (143bp) 2496-3356 B-lactamase (Ampicillin) Antibiotic
3200 G0463 pscaavmcsbghpa Plasmid Features: Coordinates Feature 980-1085 AAV2 ITR 106bp (mutated ITR) 1110-1226 MCS 1227-1440 BgHpA 1453-1595 AAV2 ITR (143bp) 2496-3356 B-lactamase (Ampicillin) Antibiotic
Rotation Report Sample Version 3. Due Date: August 9, Analysis of the Guanine Nucleotide Exchange Activity of the S. cerevisiae Ats1 Protein
 Rotation Report Sample Version 3 Due Date: August 9, 1998 Analysis of the Guanine Nucleotide Exchange Activity of the S. cerevisiae Ats1 Protein Anita H. Corbett Advisor: Amy Jones Rotation 1 Abstract:
Rotation Report Sample Version 3 Due Date: August 9, 1998 Analysis of the Guanine Nucleotide Exchange Activity of the S. cerevisiae Ats1 Protein Anita H. Corbett Advisor: Amy Jones Rotation 1 Abstract:
Supplementary Information
 Supplementary Information Fluorescent protein choice The dual fluorescence channel ratiometric promoter characterization plasmid we designed for this study required a pair of fluorescent proteins with
Supplementary Information Fluorescent protein choice The dual fluorescence channel ratiometric promoter characterization plasmid we designed for this study required a pair of fluorescent proteins with
Ton van Maris Delft University of Technology Department of Biotechnology Section Industrial Microbiology Delft, the Netherlands
 INTACT EIB.10.008 INTegral engineering of ACetic acid Tolerance in yeast Ton van Maris Delft University of Technology Department of Biotechnology Section Industrial Microbiology Delft, the Netherlands
INTACT EIB.10.008 INTegral engineering of ACetic acid Tolerance in yeast Ton van Maris Delft University of Technology Department of Biotechnology Section Industrial Microbiology Delft, the Netherlands
Lecture 1. Basic Definitions and Nucleic Acids. Basic Definitions you should already know
 Lecture 1. Basic Definitions and Nucleic Acids Basic Definitions you should already know apple DNA: Deoxyribonucleic Acid apple RNA: Ribonucleic Acid apple mrna: messenger RNA: contains the genetic information(coding
Lecture 1. Basic Definitions and Nucleic Acids Basic Definitions you should already know apple DNA: Deoxyribonucleic Acid apple RNA: Ribonucleic Acid apple mrna: messenger RNA: contains the genetic information(coding
3 Designing Primers for Site-Directed Mutagenesis
 3 Designing Primers for Site-Directed Mutagenesis 3.1 Learning Objectives During the next two labs you will learn the basics of site-directed mutagenesis: you will design primers for the mutants you designed
3 Designing Primers for Site-Directed Mutagenesis 3.1 Learning Objectives During the next two labs you will learn the basics of site-directed mutagenesis: you will design primers for the mutants you designed
Supplementary Figure 1.
 Supplementary Figure 1. a c Percentage of targeted mutagenesis (TM) 8 6 4 2 0 1 2 3 4 Percentage of targeted gene insertion (TGI) 0.30 0,30 0.20 0,20 0.10 0,10 0.00 0,00 1 2 3 4 controls Mn17038 + sctrex2
Supplementary Figure 1. a c Percentage of targeted mutagenesis (TM) 8 6 4 2 0 1 2 3 4 Percentage of targeted gene insertion (TGI) 0.30 0,30 0.20 0,20 0.10 0,10 0.00 0,00 1 2 3 4 controls Mn17038 + sctrex2
Chapter 8. Microbial Genetics. Lectures prepared by Christine L. Case. Copyright 2010 Pearson Education, Inc.
 Chapter 8 Microbial Genetics Lectures prepared by Christine L. Case Structure and Function of Genetic Material Learning Objectives 8-1 Define genetics, genome, chromosome, gene, genetic code, genotype,
Chapter 8 Microbial Genetics Lectures prepared by Christine L. Case Structure and Function of Genetic Material Learning Objectives 8-1 Define genetics, genome, chromosome, gene, genetic code, genotype,
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/315/5819/1709/dc1 Supporting Online Material for RISPR Provides Acquired Resistance Against Viruses in Prokaryotes Rodolphe Barrangou, Christophe Fremaux, Hélène Deveau,
www.sciencemag.org/cgi/content/full/315/5819/1709/dc1 Supporting Online Material for RISPR Provides Acquired Resistance Against Viruses in Prokaryotes Rodolphe Barrangou, Christophe Fremaux, Hélène Deveau,
-7 ::(teto) ::(teto) 120. GFP membranes merge. overexposed. Supplemental Figure 1 (Marquis et al.)
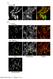 P spoiie -gfp -7 ::(teto) 120-91 ::(teto) 120 GFP membranes merge overexposed Supplemental Figure 1 (Marquis et al.) Figure S1 TetR-GFP is stripped off the teto array in sporulating cells that contain
P spoiie -gfp -7 ::(teto) 120-91 ::(teto) 120 GFP membranes merge overexposed Supplemental Figure 1 (Marquis et al.) Figure S1 TetR-GFP is stripped off the teto array in sporulating cells that contain
The plasmid shown to the right has an oriv and orit at the positions indicated, and is known to replicate bidirectionally.
 Name Microbial Genetics, BIO 410/510 2008 Exam II The plasmid shown to the right has an oriv and orit at the positions indicated, and is known to replicate bidirectionally. 1.) Indicate where replication
Name Microbial Genetics, BIO 410/510 2008 Exam II The plasmid shown to the right has an oriv and orit at the positions indicated, and is known to replicate bidirectionally. 1.) Indicate where replication
ET - Recombination. Introduction
 GENERAL & APPLIED GENETICS Geert Van Haute August 2003 ET - Recombination Introduction Homologous recombination is of importance to a variety of cellular processes, including the maintenance of genomic
GENERAL & APPLIED GENETICS Geert Van Haute August 2003 ET - Recombination Introduction Homologous recombination is of importance to a variety of cellular processes, including the maintenance of genomic
Single-cell gene expression analysis technologies and application
 2nd International Conference on Oceanography July 21-23, Las Vegas, Nevada, USA Single-cell gene expression analysis technologies and application Weiwen Zhang Laboratory of Synthetic Microbiology School
2nd International Conference on Oceanography July 21-23, Las Vegas, Nevada, USA Single-cell gene expression analysis technologies and application Weiwen Zhang Laboratory of Synthetic Microbiology School
Enhanced Arginase production: rocf
 Enhanced Arginase production: rocf Purpose and Justification: Bacillus subtilis produces urease, which catalyses the hydrolysis of urea into ammonium and carbonate. Since the cell wall of the bacteria
Enhanced Arginase production: rocf Purpose and Justification: Bacillus subtilis produces urease, which catalyses the hydrolysis of urea into ammonium and carbonate. Since the cell wall of the bacteria
PI NAME: Eleftherios Papoutsakis. Department of Chemical Engineering and the Delaware Biotechnology Institute, University of Delaware
 PI NAME: Eleftherios Papoutsakis Department of Chemical Engineering and the Delaware Biotechnology Institute, University of Delaware ONR award number: N000141010161 ONR Award Title: Engineering Complex
PI NAME: Eleftherios Papoutsakis Department of Chemical Engineering and the Delaware Biotechnology Institute, University of Delaware ONR award number: N000141010161 ONR Award Title: Engineering Complex
Chapter 4. Recombinant DNA Technology
 Chapter 4 Recombinant DNA Technology 5. Plasmid Cloning Vectors Plasmid Plasmids Self replicating Double-stranded Mostly circular DNA ( 500 kb) Linear : Streptomyces, Borrelia burgdorferi Replicon
Chapter 4 Recombinant DNA Technology 5. Plasmid Cloning Vectors Plasmid Plasmids Self replicating Double-stranded Mostly circular DNA ( 500 kb) Linear : Streptomyces, Borrelia burgdorferi Replicon
Generation of gene knockout vectors for Dictyostelium discoideum
 Generation of gene knockout vectors for Dictyostelium discoideum Instruction manual Last date of revision April 2012 2015 Version PR29-0001 PR29-0003 www.stargate.com This manual can be downloaded under
Generation of gene knockout vectors for Dictyostelium discoideum Instruction manual Last date of revision April 2012 2015 Version PR29-0001 PR29-0003 www.stargate.com This manual can be downloaded under
Cassette denotes the ORFs expressed by the expression cassette targeted by the PCR primers used to
 Stanyon, C.A., Limjindaporn, T., and Finley, Jr., R.L. Simultaneous transfer of open reading frames into several different expression vectors. Biotechniques, 35, 520-536, 2003. http://www.biotechniques.com
Stanyon, C.A., Limjindaporn, T., and Finley, Jr., R.L. Simultaneous transfer of open reading frames into several different expression vectors. Biotechniques, 35, 520-536, 2003. http://www.biotechniques.com
7. (10pts.) The diagram below shows the immunity region of phage l. The genes are:
 7. (10pts.) The diagram below shows the immunity region of phage l. The genes are: ci codes for ci repressor. The ci857 allele is temperature sensitive. cro codes for Cro protein. N codes for the transcription
7. (10pts.) The diagram below shows the immunity region of phage l. The genes are: ci codes for ci repressor. The ci857 allele is temperature sensitive. cro codes for Cro protein. N codes for the transcription
Utilising the native plasmid, pca2.4, from the cyanobacterium Synechocystis sp. strain PCC6803 as a cloning site for enhanced product production
 Armshaw et al. Biotechnol Biofuels (05) 8:0 DOI 0.86/s068-05-085-x Biotechnology for Biofuels METHODOLOGY Open Access Utilising the native plasmid, pca.4, from the cyanobacterium Synechocystis sp. strain
Armshaw et al. Biotechnol Biofuels (05) 8:0 DOI 0.86/s068-05-085-x Biotechnology for Biofuels METHODOLOGY Open Access Utilising the native plasmid, pca.4, from the cyanobacterium Synechocystis sp. strain
Supplemental Information. for. Structural and functional characterization of Pseudomonas aeruginosa AlgX: Role of AlgX in alginate acetylation
 Supplemental Information for Structural and functional characterization of Pseudomonas aeruginosa AlgX: Role of AlgX in alginate acetylation Laura M. Riley 1,#, Joel T. Weadge 1,#,$, Perrin Baker 1, Howard
Supplemental Information for Structural and functional characterization of Pseudomonas aeruginosa AlgX: Role of AlgX in alginate acetylation Laura M. Riley 1,#, Joel T. Weadge 1,#,$, Perrin Baker 1, Howard
Bio-Reagent Services. Custom Gene Services. Gateway to Smooth Molecular Biology! Your Innovation Partner in Drug Discovery!
 Bio-Reagent Services Custom Gene Services Gateway to Smooth Molecular Biology! Gene Synthesis Mutagenesis Mutant Libraries Plasmid Preparation sirna and mirna Services Large-scale DNA Sequencing GenPool
Bio-Reagent Services Custom Gene Services Gateway to Smooth Molecular Biology! Gene Synthesis Mutagenesis Mutant Libraries Plasmid Preparation sirna and mirna Services Large-scale DNA Sequencing GenPool
Chapter 10 (Part I) Gene Isolation and Manipulation
 Biology 234 J. G. Doheny Chapter 10 (Part I) Gene Isolation and Manipulation Practice Questions: Answer the following questions with one or two sentences. 1. From which types of organisms were most restriction
Biology 234 J. G. Doheny Chapter 10 (Part I) Gene Isolation and Manipulation Practice Questions: Answer the following questions with one or two sentences. 1. From which types of organisms were most restriction
Superposition and Synthetic Genetic Devices: Framework and Model System to Investigate Linearity in Escherichia coli
 Superposition and Synthetic Genetic Devices: Framework and Model System to Investigate Linearity in Escherichia coli Meghdad Hajimorad Electrical Engineering and Computer Sciences University of California
Superposition and Synthetic Genetic Devices: Framework and Model System to Investigate Linearity in Escherichia coli Meghdad Hajimorad Electrical Engineering and Computer Sciences University of California
Ligation Independent Cloning (LIC) Procedure
 Ligation Independent Cloning (LIC) Procedure Ligation Independent Cloning (LIC) LIC cloning allows insertion of DNA fragments without using restriction enzymes into specific vectors containing engineered
Ligation Independent Cloning (LIC) Procedure Ligation Independent Cloning (LIC) LIC cloning allows insertion of DNA fragments without using restriction enzymes into specific vectors containing engineered
Molecular Genetics Techniques. BIT 220 Chapter 20
 Molecular Genetics Techniques BIT 220 Chapter 20 What is Cloning? Recombinant DNA technologies 1. Producing Recombinant DNA molecule Incorporate gene of interest into plasmid (cloning vector) 2. Recombinant
Molecular Genetics Techniques BIT 220 Chapter 20 What is Cloning? Recombinant DNA technologies 1. Producing Recombinant DNA molecule Incorporate gene of interest into plasmid (cloning vector) 2. Recombinant
KOD -Plus- Mutagenesis Kit
 Instruction manual KOD -Plus- Mutagenesis Kit 0811 F0936K KOD -Plus- Mutagenesis Kit SMK-101 20 reactions Store at -20 C Contents [1] Introduction [2] Flow chart [3] Components [4] Notes [5] Protocol 1.
Instruction manual KOD -Plus- Mutagenesis Kit 0811 F0936K KOD -Plus- Mutagenesis Kit SMK-101 20 reactions Store at -20 C Contents [1] Introduction [2] Flow chart [3] Components [4] Notes [5] Protocol 1.
Technical University of Denmark. Written examination, 29 May 2012 Course name: Life Science. Course number: Aids allowed: Written material
 1 Technical University of Denmark Written examination, 29 May 2012 Course name: Life Science Course number: 27008 Aids allowed: Written material Exam duration: 4 hours Weighting: The exam set consists
1 Technical University of Denmark Written examination, 29 May 2012 Course name: Life Science Course number: 27008 Aids allowed: Written material Exam duration: 4 hours Weighting: The exam set consists
Δsig. ywa. yjbm. rela -re. rela
 A wt A. A, P rela -re la A/Δ yjbm A/Δ ywa C A, P hy-s igd, D (A, P IPTG hy-s igd, ) D D (+ IPTG ) D/Δ rela Figure S1 SigD B. Figure S1. SigD levels and swimming motility in (p)ppgpp synthetase mutants.
A wt A. A, P rela -re la A/Δ yjbm A/Δ ywa C A, P hy-s igd, D (A, P IPTG hy-s igd, ) D D (+ IPTG ) D/Δ rela Figure S1 SigD B. Figure S1. SigD levels and swimming motility in (p)ppgpp synthetase mutants.
Supplementary Information for: Engineered synthetic scaffolds for organising proteins within bacterial cytoplasms
 Lee et al. Supplementary Information 1 Supplementary Information for: Engineered synthetic scaffolds for organising proteins within bacterial cytoplasms Authors: Matthew J. Lee, 1 Judith Mantell, 2,3 Lorna
Lee et al. Supplementary Information 1 Supplementary Information for: Engineered synthetic scaffolds for organising proteins within bacterial cytoplasms Authors: Matthew J. Lee, 1 Judith Mantell, 2,3 Lorna
Supplementary
 Supplementary information Supplementary Material and Methods Plasmid construction The transposable element vectors for inducible expression of RFP-FUS wt and EGFP-FUS R521C and EGFP-FUS P525L were derived
Supplementary information Supplementary Material and Methods Plasmid construction The transposable element vectors for inducible expression of RFP-FUS wt and EGFP-FUS R521C and EGFP-FUS P525L were derived
