Supplementary Information for: Engineered synthetic scaffolds for organising proteins within bacterial cytoplasms
|
|
|
- Cory Stanley
- 5 years ago
- Views:
Transcription
1 Lee et al. Supplementary Information 1 Supplementary Information for: Engineered synthetic scaffolds for organising proteins within bacterial cytoplasms Authors: Matthew J. Lee, 1 Judith Mantell, 2,3 Lorna Hodgson, 2 Dominic Alibhai, 2 Jordan M Fletcher, 4 Ian R. Brown, 1 Stefanie Frank, 5 Wei-Feng Xue, 1 Paul Verkade, 2,3,6 Derek N Woolfson, 2,4,6 * Martin J Warren 1 *. 1 Industrial Biotechnology Centre, School of Biosciences, University of Kent, Canterbury CT2 7NJ, UK 2 School of Biochemistry, University of Bristol, Medical Sciences Building, University Walk, Bristol BS8 1TD, UK. 3 Wolfson Bioimaging Facility, Medical Sciences Building, University Walk, Bristol BS8 1TD, UK. 4 School of Chemistry, University of Bristol, Cantock's Close, Bristol BS8 1TS, UK. 5 Department of Biochemical Engineering, University College London, Bernard Katz Building, Gordon Street, London WC1E 6BT, UK. 6 BrisSynBio, Life Sciences Building, Tyndall Avenue, Bristol BS8 1TQ, UK. * Correspondence to: Martin J Warren (M.J. Warren@kent.ac.uk) and Derek N Woolfson (D.N.Woolfson@bristol.ac.uk)
2 Lee et al. Supplementary Information 2 Supplementary Fig. 1. Construction of a bacterial cytoscaffold. Hexagonally arrayed, tubular filaments of the PduA* protein (red) carrying half of a de novo designed heterodimeric coiled coil (blue).
3 Lee et al. Supplementary Information 3 Supplementary Fig. 2. TEM analysis of thin sectioned E. coli cells transformed with plyss- PduA*. Scale bar shows 200 nm
4 Lee et al. Supplementary Information 4 Supplementary Fig. 3. TEM analysis of E. coli cells transformed with C-PduA*. Scale bar shows 200 nm.
5 Lee et al. Supplementary Information 5 Supplementary Fig. 4. Western blot analysis of E. coli cells transformed with varying PduA* constructs. Total lysates were analyzed by SDS-PAGE and subsequently western blotted with an anti-pdua* primary antibody in comparison to a molecular weight standard. Cell densities were normalized to an OD 600 of 5 for loading.
6 Lee et al. Supplementary Information 6 Supplementary Fig. 5. Time course of plyss-cc-di-b-pdua* induction with 400 µm IPTG in BL21 * (DE3) cells. (a) pre-induction (b) 1h post induction (c) 2h post induction (d) 3h post induction (e) 4h post induction (f) 6h post induction (g) 20h post induction (h) Control strain transformed with an empty plyss vector. Scale bars show 200 nm
7 Lee et al. Supplementary Information 7 Supplementary Fig. 6. SDS-PAGE of CC-Di-B-PduA* purification. Lane 1 lysate (2 μl), lane 2 supernatant after first centrifugation (2 μl), lane 3 pellet after first centrifugation (5 μl), lane 4 pellet after second centrifugation (5 μl), lane 5 supernatant after second centrifugation (5 μl), lane 6 supernatant after third centrifugation (5 μl), lane 7 pellet after third centrifugation (5 μl), lane 8 supernatant after final centrifugation (5 μl), lane 9 Pellet after final centrifugation (5 μl).
8 Lee et al. Supplementary Information 8 Supplementary Fig. 7. TEM (a), AFM height (b), PeakForce (c) and height profile corresponding to line 1 (panel B) (d) analysis of purified CC-Di-B-PduA* nanotubes.
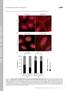
9 Lee et al. Supplementary Information 9 Supplementary Fig. 8. Confocal analysis of BL21 * (DE3) cells expressing CC-Di-B-PduA* or a control empty vector (plyss) with citrine tagged with CC-Di-A or CC-Di-B or a control C- citrine.
10 Lee et al. Supplementary Information 10 Supplementary Fig. 9. Correlative Light Electron Microscopy of E. coli cells expressing CC- Di-B-PduA* and C-GFP.
11 Lee et al. Supplementary Information 11 Supplementary Fig. 10. (a) Growth curves of strains producing ethanol and control strains shown as OD 600 against time (hours). (b) In vivo ethanol production. Graph shows ethanol content of the growth medium over time. E. coli strain transformed with empty plasmids (pet14b and plyss) ( ), strain producing CC-Di-B-PduA* only ( ), strain producing CC-Di- A-Pdc and CC-Di-A-Adh ( ), strain producing CC-Di-A-Pdc, CC-Di-A-Adh and CC-Di-B- PduA* ( ). Data points represent an average of three independent experiments; standard deviations are represented by error bars.
12 Lee et al. Supplementary Information 12 Supplementary Fig. 11. Western blot analysis of strains producing ethanol and control strain in comparison to a molecular weight marker (M). Total lysates at 24h were adjusted to the same OD 600 (1.6) and analyzed by SDS-PAGE and subsequently western blotted with an anti-polyhistidine primary antibody. (1) E. coli strain transformed with empty plasmids (pet14b and plyss), (2) strain producing CC-Di-B-PduA* only, (3) strain producing CC-Di- A-Pdc and CC-Di-A-Adh, (4) strain producing CC-Di-A-Pdc, CC-Di-A-Adh and CC-Di-B- PduA*.
13 Lee et al. Supplementary Information 13 Supplementary Fig. 12. TEM analysis of ethanol producing strains after 24 hours. (a) E. coli strain transformed with empty plasmids (pet14b and plyss), (b) strain producing CC- Di-B-PduA* only, (c) strain producing CC-Di-A-Pdc and CC-Di-A-Adh, (d) strain producing CC-Di-A-Pdc, CC-Di-A-Adh and CC-Di-B-PduA*.
14 Lee et al. Supplementary Information 14 Supplementary Fig. 13. Confocal image of strain expressing C citrine MinD; scale bar is 5 µm.
15 Lee et al. Supplementary Information 15 Supplementary Fig. 14. TEM micrographs of strains producing CC-Di-B PduA* only (a); CC-Di-B PduA* plus the C citrine MinD control (b), arrows indicate transverse filaments. Zoom in of areas 1 and 2 in panels a and b respectively (c and d).
16 Lee et al. Supplementary Information 16 Supplementary Tables Supplementary Table 1. Strains used in this study Strain Genotype Source JM109 enda1, reca1, gyra96, thi, hsdr17 (rk, mk+), rela1, supe44, Δ(lac-proAB), [F, trad36, proab, laqiqzδm15] Promega BL21 * (DE3) F ompt hsdsb (rb mb ) gal dcm (DE3) Novagen Supplementary Table 2. DNA synthesized for this study Name CC-Di-A CC-Di-B Control DNA / translated sequence (coiled-coil peptides underlined) TCTAGAAATAATTTTGTTTAACTTTAAGAAGGAGATATATGGGTAGCG AAATTGCCGCGCTGGAAAAAGAAAACGCTGCCCTGGAGCAGGAAAT CGCGGCACTGGAGCAGGGTAGCGGCAGCGGTAGCACCATGGGCAG CAGCCATCATCATCATCATCACAGCAGCGGCCTGGTGCCGCGCGGC AGCCATATG MGSEIAALEKENAALEQEIAALEQGSGSGSTMGSSHHHHHHSSGLVPR GSHM TCTAGAAATAATTTTGTTTAACTTTAAGAAGGAGATATATGGGTAGCA AAATCGCCGCACTGAAAAAGAAAAACGCTGCGCTGAAACAGAAAATT GCCGCGCTGAAACAGGGTAGCGGCAGCGGTAGCACCATGGGCAGC AGCCATCATCATCATCATCACAGCAGCGGCCTGGTGCCGCGCGGCA GCCATATG MGSKIAALKKKNAALKQKIAALKQGSGSGSTMGSSHHHHHHSSGLVPR GSHM TCTAGAAATAATTTTGTTTAACTTTAAGAAGGAGATATATGGGTAGCG GTAGCGGCAGCGGTAGCACCATGGGCAGCAGCCATCATCATCATCA TCACAGCAGCGGCCTGGTGCCGCGCGGCAGCCATATG MGSGSGSGSTMGSSHHHHHHSSGLVPRGSHM MinD ACTAGTGGTTCGGGATCGCCGTTGCAAGTCTTGGAGGAACAAAACA AAGGGATGATGGCTAAAATCAAATCATTTTTCGGTGTACGTTCCTAA CCTAGGTTTGGATCCGGCTGCTAACAAAGCCCGAAAGGAAGCTGAG TTGGCTGCTGCCACCGCTGAGC TSGSGSPLQVLEEQNKGMMAKIKSFFGVRS
17 Lee et al. Supplementary Information 17 Supplementary Table 3. Plasmids Used in this study Plasmid name Description Source pet14b Overexpression vector containing N-terminal hexahistidine-tag, modified to include an SpeI site 5 of BamHI Novagen plyss Basal expression suppressor Novagen plyss_2 pet3a_pdua* plyss_pdua* pet_cc_di_a pet_cc_di_b pet_c pet_cc_di_a_pdua* pet_cc_di_b_pdua* pet_c_pdua* pet-tbad-coba pet3a-mcherry- PduABB JKNU pet-tbad-mcherry- PduABJKNU. plyss-mcherry- PduABB plyss-prha-mcherry- PduAB plyss_tbad_mcherry _PduABJKNU plyss_cc_di_a_pdu A* Basal expression suppressor containing the T7 promoter-mcs-t7 terminator cassette from pet14b PCR product of pdua* ligated into the NdeI and SpeI sites of pet3a (4 bp removed from 3' end of pdua, this allowed non-coding DNA from the vector to be translated) NdeI/SpeI fragment of pet3a_pdua* ligated into NdeI/SpeI sites of plyss_2 Synthesised CC_Di_A gene fragment cloned into XbaI/NdeI sites of pet14b Synthesised CC_Di_B gene fragment cloned into XbaI/NdeI sites of pet14b Synthesised control (C) gene fragment cloned into XbaI/NdeI sites of pet14b NdeI/SpeI fragment of plyss_pdua* cloned into NdeI/SpeI sites of pet_cc_di_a NdeI/SpeI fragment of plyss_pdua* cloned into NdeI/SpeI sites of pet_cc_di_b NdeI/SpeI fragment of plyss_pdua* cloned into NdeI/SpeI sites of pet_c XbaI/HindIII fragment of pet3a-mcherry- PduABJKNU cloned into XbaI/HindIII sites of pet-tbad-coba Exponential Megapriming PCR (EMP) and splicing by Overlap Extension (SOE) to insert BglII site and prha promoter at former T7 promoter site of plyss-mcherry-pduabb BglII/SpeI fragment of pet-tbad-mcherry- PduABJKNU cloned into BglII/SpeI sites of plyss-prha-mcherry-pduab BglII/SpeI fragment of pet_cc_di_a_pdua* cloned into BglII/SpeI sites of Parsons et al., 2008 Parsons et al., 2010 Gift from E. Deery Parsons et al., 2010 Parsons et al., 2010
18 Lee et al. Supplementary Information 18 plyss_cc_di_b_pdu A* plyss_c_pdua* pet_cc_di_a_citrine pet_cc_di_b_citrine pet_c_citrine pet_cc_di_a_mcherr y pet_cc_di_b_mcherr y pet_c_mcherry pet_cc_di_a_citrine _CC_Di_A_mCherry pet_c_citrine_c_mc herry pet23b-pdc pet23b-adh pet_cc_di_a_pdc pet_cc_di_a_adh pet_cc_di_a_adh_c C_Di_A_Pdc plyss_tbad_mcherry_pduabjknu BglII/SpeI fragment of pet_cc_di_b_pdua* cloned into BglII/SpeI sites of plyss_tbad_mcherry_pduabjknu BglII/SpeI fragment of pet_c_pdua* cloned into BglII/SpeI sites of plyss_tbad_mcherry_pduabjknu Citrine PCR product ligated into NdeI/SpeI sites of pet_cc_di_a Citrine PCR product ligated into NdeI/SpeI sites of pet_cc_di_b Citrine PCR product ligated into NdeI/SpeI sites of pet_c mcherry PCR product ligated into NdeI/SpeI sites of pet_cc_di_a mcherry PCR product ligated into NdeI/SpeI sites of pet_cc_di_b mcherry PCR product ligated into NdeI/SpeI sites of pet_c XbaI/EcoRI fragment from pet_cc_di_a_mcherry ligated into SpeI/EcoRI sites of pet_cc_di_a_citrine XbaI/EcoRI fragment from pet_c_mcherry ligated into SpeI/EcoRI sites of pet_c_citrine NdeI/EcoRI fragment of pet23b-pdc cloned into NdeI/EcoRI sites of pet_cc_di_a NdeI/EcoRI fragment of pet23b-adh cloned into NdeI/EcoRI sites of pet_cc_di_a XbaI/EcoRI fragment from pet_cc_di_a_pdc ligated into SpeI/EcoRI sites of pet_cc_di_a_adh Lawrence et al., 2014 Lawrence et al., 2014
19 Lee et al. Supplementary Information 19 Supplementary Table 4. Oligonucleotides used in this study, restriction sites are underlined Name Sequence 5 3 Citrine_NdeI_FW CACCATATGGTGAGCAAGGGCGAGGAGC Citrine_SpeI_RV CACACTAGTTTACTTGTACAGCTCGTCC mcherry_ndei_fw CAGCATATGGTGAGCAAGGGCGAGG mcherry_spei_rv GACACTAGTTTACTTGTACAGCTCGTCCATGC Citrine_SpeI_NoStop_RV CACACTAGTCTTGTACAGCTCGTCC
20 Lee et al. Supplementary Information 20 Supplementary Movies Supplementary Video 1. Tomography of CC-Di-B-PduA filaments and automated microtubule tracing. Supplementary Video 2. Refined tracing model on a small area of the tomogram shown in movie S1.
Construction of a tunable multi-enzyme-coordinate expression. system for biosynthesis of chiral drug intermediates
 Construction of a tunable multi-enzyme-coordinate expression system for biosynthesis of chiral drug intermediates Wei Jiang 1, 2 1, 2, 3, Baishan Fang 1 Department of Chemical and Biochemical Engineering,
Construction of a tunable multi-enzyme-coordinate expression system for biosynthesis of chiral drug intermediates Wei Jiang 1, 2 1, 2, 3, Baishan Fang 1 Department of Chemical and Biochemical Engineering,
Arsenite oxidation regulator AioR regulates bacterial chemotaxis towards. arsenite in Agrobacterium tumefaciens GW4
 1 2 Arsenite oxidation regulator AioR regulates bacterial chemotaxis towards arsenite in Agrobacterium tumefaciens GW4 3 4 5 Kaixiang Shi 1, Xia Fan 1, Zixu Qiao 1, Yushan Han 1, Timothy R. McDermott 2,
1 2 Arsenite oxidation regulator AioR regulates bacterial chemotaxis towards arsenite in Agrobacterium tumefaciens GW4 3 4 5 Kaixiang Shi 1, Xia Fan 1, Zixu Qiao 1, Yushan Han 1, Timothy R. McDermott 2,
Supporting Information: DNA-mediated signaling by proteins with 4Fe-4S clusters is necessary for genomic integrity
 Supporting Information: DNA-mediated signaling by proteins with 4Fe-4S clusters is necessary for genomic integrity Michael A. Grodick, 1 Helen M. Segal, 1 Theodore J. Zwang, 1 and Jacqueline K. Barton
Supporting Information: DNA-mediated signaling by proteins with 4Fe-4S clusters is necessary for genomic integrity Michael A. Grodick, 1 Helen M. Segal, 1 Theodore J. Zwang, 1 and Jacqueline K. Barton
Fig. S1. CrgA intracellular levels in M. tuberculosis. Ten and twenty micrograms of
 Supplementary data Fig. S1. CrgA intracellular levels in M. tuberculosis. Ten and twenty micrograms of cell free protein lysates from WT M. tuberculosis (Rv) together with various known concentrations
Supplementary data Fig. S1. CrgA intracellular levels in M. tuberculosis. Ten and twenty micrograms of cell free protein lysates from WT M. tuberculosis (Rv) together with various known concentrations
Figure legends. Fig. S1. SDS-PAGE analyses of recombinant OcSus1 (panel A), OcSus2 (panel B)
 Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2016 Figure legends Fig. S1. SDS-PAGE analyses of recombinant OcSus1 (panel A), OcSus2 (panel B)
Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2016 Figure legends Fig. S1. SDS-PAGE analyses of recombinant OcSus1 (panel A), OcSus2 (panel B)
Hetero-Stagger PCR Cloning Kit
 Product Name: Code No: Size: DynaExpress Hetero-Stagger PCR Cloning Kit DS150 20 reactions Kit Components: Box 1 (-20 ) phst-1 Vector, linearized Annealing Buffer Ligase Mixture phst Forward Sequence Primer
Product Name: Code No: Size: DynaExpress Hetero-Stagger PCR Cloning Kit DS150 20 reactions Kit Components: Box 1 (-20 ) phst-1 Vector, linearized Annealing Buffer Ligase Mixture phst Forward Sequence Primer
Stabilization of a virus-like particle and its application as a nanoreactor at physiological conditions
 Supporting Information Stabilization of a virus-like particle and its application as a nanoreactor at physiological conditions Lise Schoonen, b Sjors Maassen, b Roeland J. M. Nolte b and Jan C. M. van
Supporting Information Stabilization of a virus-like particle and its application as a nanoreactor at physiological conditions Lise Schoonen, b Sjors Maassen, b Roeland J. M. Nolte b and Jan C. M. van
pgm-t Cloning Kit Cat. # : GVT202 Size : 20 Reactions Store at -20
 pgm-t Cloning Kit Cat. # : GVT202 Size : 20 Reactions Store at -20 1 Kit Contents Contents pgm-t Cloning Kit pgm-t Vector (50 ng/μl) 20 μl T4 DNA Ligase (3 U/μl) 20 μl 10X T4 DNA Ligation Buffer 30 μl
pgm-t Cloning Kit Cat. # : GVT202 Size : 20 Reactions Store at -20 1 Kit Contents Contents pgm-t Cloning Kit pgm-t Vector (50 ng/μl) 20 μl T4 DNA Ligase (3 U/μl) 20 μl 10X T4 DNA Ligation Buffer 30 μl
SUPPLEMENTARY MATERIAL
 SUPPLEMENTARY MATERIAL Engineering a pyridoxal '-phosphate supply for cadaverine production by using Escherichia coli whole-cell biocatalysis Weichao Ma 1,,, Weijia Cao 1,, Bowen Zhang 1,, Kequan Chen
SUPPLEMENTARY MATERIAL Engineering a pyridoxal '-phosphate supply for cadaverine production by using Escherichia coli whole-cell biocatalysis Weichao Ma 1,,, Weijia Cao 1,, Bowen Zhang 1,, Kequan Chen
Figure S1. Biosynthetic pathway of GDP-PerNAc. Mass spectrum of purified GDP-PerNAc. Nature Protocols: doi: /nprot
 Synthesis of GDP-PerNAc In E. coli O157, the biosynthesis of GDP- -N-acetyl-D-perosamine (GDP-PerNAc) involves three enzymes (Fig. S1). GDP-D-Mannose is converted by GDP-mannose-4,6-dehydratase (GMD) into
Synthesis of GDP-PerNAc In E. coli O157, the biosynthesis of GDP- -N-acetyl-D-perosamine (GDP-PerNAc) involves three enzymes (Fig. S1). GDP-D-Mannose is converted by GDP-mannose-4,6-dehydratase (GMD) into
Supplementary Information
 Supplementary Information Creation of artificial cellulosomes on DNA scaffolds by zinc finger protein-guided assembly Qing Sun, a Bhawna Madan, b Shen-Long Tsai, c Matthew P. DeLisa d and Wilfred Chen*
Supplementary Information Creation of artificial cellulosomes on DNA scaffolds by zinc finger protein-guided assembly Qing Sun, a Bhawna Madan, b Shen-Long Tsai, c Matthew P. DeLisa d and Wilfred Chen*
A plot of viable bacterial cell count (CFU/ml) against the OD 600 reading based
 RESULTS 4.1 H. pylori and MKN28 cells 4.1.1 H. pylori 4.1.1.1 Enumeration of H. pylori A plot of viable bacterial cell count (CFU/ml) against the OD 600 reading based on different dilutions of a 3-day-old
RESULTS 4.1 H. pylori and MKN28 cells 4.1.1 H. pylori 4.1.1.1 Enumeration of H. pylori A plot of viable bacterial cell count (CFU/ml) against the OD 600 reading based on different dilutions of a 3-day-old
SUPPLEMENTARY DATA: Supplementary Figures:
 SUPPLEMENTARY DATA: Supplementary Figures: Supplementary Fig. S1: In vivo recombinant Mtb reporter assay: Dependence of mcherry expression as a function of E. coli growth: A. Fluorescent intensity of mcherry
SUPPLEMENTARY DATA: Supplementary Figures: Supplementary Fig. S1: In vivo recombinant Mtb reporter assay: Dependence of mcherry expression as a function of E. coli growth: A. Fluorescent intensity of mcherry
forms oligomeric ring-shaped structures and interacts with the motor ATPase FlaI " by Banerjee et al.
 Supplementary material to " FlaX, a unique component of the crenarchaeal archaellum, forms oligomeric ring-shaped structures and interacts with the motor ATPase FlaI " by Banerjee et al. Experimental procedures
Supplementary material to " FlaX, a unique component of the crenarchaeal archaellum, forms oligomeric ring-shaped structures and interacts with the motor ATPase FlaI " by Banerjee et al. Experimental procedures
Strain, plasmid, primer or dsdna fragment Relevant genotype, description, or sequence
 Supplemental Tables Table S1. Strains, Plasmids, Primers and dsdna fragments used in this study Strain, plasmid, primer or dsdna fragment Relevant genotype, description, or sequence Reference or Source
Supplemental Tables Table S1. Strains, Plasmids, Primers and dsdna fragments used in this study Strain, plasmid, primer or dsdna fragment Relevant genotype, description, or sequence Reference or Source
Step 1: Digest vector with Reason for Step 1. Step 2: Digest T4 genomic DNA with Reason for Step 2: Step 3: Reason for Step 3:
 Biol/Chem 475 Spring 2007 Study Problems for Quiz 2 Quiz 2 (~50 pts) is scheduled for Monday May 14 It will cover all handouts and lab exercises to date except the handout/worksheet (yet to be distributed)
Biol/Chem 475 Spring 2007 Study Problems for Quiz 2 Quiz 2 (~50 pts) is scheduled for Monday May 14 It will cover all handouts and lab exercises to date except the handout/worksheet (yet to be distributed)
PRODUCT INFORMATION. Composition of SOC medium supplied :
 Product Name : Competent Cell BL21(DE3)pLysS Code No. : DS260 Size : 100 μl 10 Competency : > 5 10 7 cfu/μg (puc19) Supplied product : SOC medium, 1 ml 10 This product is for research use only Description
Product Name : Competent Cell BL21(DE3)pLysS Code No. : DS260 Size : 100 μl 10 Competency : > 5 10 7 cfu/μg (puc19) Supplied product : SOC medium, 1 ml 10 This product is for research use only Description
Supporting Information
 Supporting Information Risser and Callahan 10.1073/pnas.0909152106 SI Text Plasmid Construction. Plasmid pdr211 is a mobilizable shuttle vector containing P pete -pats. A fragment containing P pete was
Supporting Information Risser and Callahan 10.1073/pnas.0909152106 SI Text Plasmid Construction. Plasmid pdr211 is a mobilizable shuttle vector containing P pete -pats. A fragment containing P pete was
(phosphatase tensin) domain is shown in dark gray, the FH1 domain in black, and the
 Supplemental Figure 1. Predicted Domain Organization of the AFH14 Protein. (A) Schematic representation of the predicted domain organization of AFH14. The PTEN (phosphatase tensin) domain is shown in dark
Supplemental Figure 1. Predicted Domain Organization of the AFH14 Protein. (A) Schematic representation of the predicted domain organization of AFH14. The PTEN (phosphatase tensin) domain is shown in dark
NZYGene Synthesis kit
 Kit components Component Concentration Amount NZYGene Synthesis kit Catalogue number: MB33901, 10 reactions GS DNA Polymerase 1U/ μl 30 μl Reaction Buffer for GS DNA Polymerase 10 150 μl dntp mix 2 mm
Kit components Component Concentration Amount NZYGene Synthesis kit Catalogue number: MB33901, 10 reactions GS DNA Polymerase 1U/ μl 30 μl Reaction Buffer for GS DNA Polymerase 10 150 μl dntp mix 2 mm
Grb2-Mediated Alteration in the Trafficking of AβPP: Insights from Grb2-AICD Interaction
 Journal of Alzheimer s Disease 20 (2010) 1 9 1 IOS Press Supplementary Material Grb2-Mediated Alteration in the Trafficking of AβPP: Insights from Grb2-AICD Interaction Mithu Raychaudhuri and Debashis
Journal of Alzheimer s Disease 20 (2010) 1 9 1 IOS Press Supplementary Material Grb2-Mediated Alteration in the Trafficking of AβPP: Insights from Grb2-AICD Interaction Mithu Raychaudhuri and Debashis
Applied Microbiology and Biotechnology. De-bugging and maximizing plant cytochrome P450 production in Escherichia coli with C-terminal GFP-fusions
 Applied Microbiology and Biotechnology De-bugging and maximizing plant cytochrome P450 production in Escherichia coli with C-terminal GFP-fusions Ulla Christensen 1, Dario V. Albacete 1, Karina M. Søgaard
Applied Microbiology and Biotechnology De-bugging and maximizing plant cytochrome P450 production in Escherichia coli with C-terminal GFP-fusions Ulla Christensen 1, Dario V. Albacete 1, Karina M. Søgaard
Supporting information
 Supporting information Construction of strains and plasmids To create ptc67, a PCR product obtained with primers cc2570-162f (gcatgggcaagcttgaggacggcgtcatgt) and cc2570+512f (gaggccgtggtaccatagaggcgggcg),
Supporting information Construction of strains and plasmids To create ptc67, a PCR product obtained with primers cc2570-162f (gcatgggcaagcttgaggacggcgtcatgt) and cc2570+512f (gaggccgtggtaccatagaggcgggcg),
Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product.
 Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
BL21(DE3) expression competent cell pack DS265. Component Code No. Contents Competent cell BL21(DE3) DS250 5 tubes (100 μl/tube)
 Product Name : Code No. : Kit Component : BL21(DE3) expression competent cell pack DS265 Component Code No. Contents Competent cell BL21(DE3) DS250 5 tubes (100 μl/tube) transfromation efficiency: 5 10
Product Name : Code No. : Kit Component : BL21(DE3) expression competent cell pack DS265 Component Code No. Contents Competent cell BL21(DE3) DS250 5 tubes (100 μl/tube) transfromation efficiency: 5 10
Supplemental Fig. S1. Key to underlines: Key to amino acids:
 AspA-F1 AspA 1 MKQMETKGYGYFRKTKAYGLVCGIT--------------LAGALTLGTTSVSADDVTTLNPATNLTTLQTPPTADQTQLAHQAGQQSGELVSEVSNTEWD 86 SspB 1 MQKREV--FG-FRKSKVAKTLCGAV-LGAALIAIADQQVLADEVTETNSTANVAVTTTGNPATNLPEAQGEATEAASQSQAQAGSKDGALPVEVSADDLN
AspA-F1 AspA 1 MKQMETKGYGYFRKTKAYGLVCGIT--------------LAGALTLGTTSVSADDVTTLNPATNLTTLQTPPTADQTQLAHQAGQQSGELVSEVSNTEWD 86 SspB 1 MQKREV--FG-FRKSKVAKTLCGAV-LGAALIAIADQQVLADEVTETNSTANVAVTTTGNPATNLPEAQGEATEAASQSQAQAGSKDGALPVEVSADDLN
Table S1. Bacterial strains (Related to Results and Experimental Procedures)
 Table S1. Bacterial strains (Related to Results and Experimental Procedures) Strain number Relevant genotype Source or reference 1045 AB1157 Graham Walker (Donnelly and Walker, 1989) 2458 3084 (MG1655)
Table S1. Bacterial strains (Related to Results and Experimental Procedures) Strain number Relevant genotype Source or reference 1045 AB1157 Graham Walker (Donnelly and Walker, 1989) 2458 3084 (MG1655)
Supplemental Data. Liu et al. (2013). Plant Cell /tpc
 Supplemental Figure 1. The GFP Tag Does Not Disturb the Physiological Functions of WDL3. (A) RT-PCR analysis of WDL3 expression in wild-type, WDL3-GFP, and WDL3 (without the GFP tag) transgenic seedlings.
Supplemental Figure 1. The GFP Tag Does Not Disturb the Physiological Functions of WDL3. (A) RT-PCR analysis of WDL3 expression in wild-type, WDL3-GFP, and WDL3 (without the GFP tag) transgenic seedlings.
Genlantis A division of Gene Therapy Systems, Inc Telesis Court San Diego, CA USA Telephone: or (US toll free)
 TurboCells BL21(DE3) TurboCells BL21(DE3)pLysS Chemically Competent E. coli Instruction Manual Catalog Numbers C302020 C303020 A division of Gene Therapy Systems, Inc. 10190 Telesis Court San Diego, CA
TurboCells BL21(DE3) TurboCells BL21(DE3)pLysS Chemically Competent E. coli Instruction Manual Catalog Numbers C302020 C303020 A division of Gene Therapy Systems, Inc. 10190 Telesis Court San Diego, CA
LysoTracker Red DND-99 (Invitrogen) was used as a marker of lysosome or acidic
 information MATERIAL AND METHODS Lysosome staining LysoTracker Red DND-99 (Invitrogen) was used as a marker of lysosome or acidic compartments, according to the manufacturer s protocol. Plasmid independent
information MATERIAL AND METHODS Lysosome staining LysoTracker Red DND-99 (Invitrogen) was used as a marker of lysosome or acidic compartments, according to the manufacturer s protocol. Plasmid independent
Certificate of Analysis
 Certificate of Analysis pet6xhn Expression Vector Set Contents Product Information... 1 pet6xhn-n, pet6xhn-c, and pet6xhn-gfpuv Vector Information... 2 Location of Features... 4 Additional Information...
Certificate of Analysis pet6xhn Expression Vector Set Contents Product Information... 1 pet6xhn-n, pet6xhn-c, and pet6xhn-gfpuv Vector Information... 2 Location of Features... 4 Additional Information...
pgm-t Cloning Kit For direct cloning of PCR products generated by Taq DNA polymerases For research use only Cat. # : GVT202 Size : 20 Reactions
 pgm-t Cloning Kit For direct cloning of PCR products generated by Taq DNA polymerases Cat. # : GVT202 Size : 20 Reactions Store at -20 For research use only 1 pgm-t Cloning Kit Cat. No.: GVT202 Kit Contents
pgm-t Cloning Kit For direct cloning of PCR products generated by Taq DNA polymerases Cat. # : GVT202 Size : 20 Reactions Store at -20 For research use only 1 pgm-t Cloning Kit Cat. No.: GVT202 Kit Contents
Supporting Information
 Supporting Information Development of a 2,4-Dinitrotoluene-Responsive Synthetic Riboswitch in E. coli cells Molly E. Davidson, Svetlana V. Harbaugh, Yaroslav G. Chushak, Morley O. Stone, Nancy Kelley-
Supporting Information Development of a 2,4-Dinitrotoluene-Responsive Synthetic Riboswitch in E. coli cells Molly E. Davidson, Svetlana V. Harbaugh, Yaroslav G. Chushak, Morley O. Stone, Nancy Kelley-
Contents... vii. List of Figures... xii. List of Tables... xiv. Abbreviatons... xv. Summary... xvii. 1. Introduction In vitro evolution...
 vii Contents Contents... vii List of Figures... xii List of Tables... xiv Abbreviatons... xv Summary... xvii 1. Introduction...1 1.1 In vitro evolution... 1 1.2 Phage Display Technology... 3 1.3 Cell surface
vii Contents Contents... vii List of Figures... xii List of Tables... xiv Abbreviatons... xv Summary... xvii 1. Introduction...1 1.1 In vitro evolution... 1 1.2 Phage Display Technology... 3 1.3 Cell surface
Supplementary Material. Increased heterocyst frequency by patn disruption in Anabaena leads to enhanced photobiological
 Supplementary Material Increased heterocyst frequency by patn disruption in Anabaena leads to enhanced photobiological hydrogen production at high light intensity and high cell density Applied Microbiology
Supplementary Material Increased heterocyst frequency by patn disruption in Anabaena leads to enhanced photobiological hydrogen production at high light intensity and high cell density Applied Microbiology
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Impaired ITS2 processing results in mislocalization of foot-factors to the cytoplasm. (a) Schematic representation of ITS2 processing. The endonuclease Las1 initiates ITS2 processing
Supplementary Figure 1 Impaired ITS2 processing results in mislocalization of foot-factors to the cytoplasm. (a) Schematic representation of ITS2 processing. The endonuclease Las1 initiates ITS2 processing
Supplementary information. Interplay between Penicillin-binding proteins and SEDS proteins promotes. bacterial cell wall synthesis
 Supplementary information Interplay between Penicillin-binding proteins and SEDS proteins promotes bacterial cell wall synthesis Sophie Leclercq 1, Adeline Derouaux 1, Samir Olatunji 1, Claudine Fraipont
Supplementary information Interplay between Penicillin-binding proteins and SEDS proteins promotes bacterial cell wall synthesis Sophie Leclercq 1, Adeline Derouaux 1, Samir Olatunji 1, Claudine Fraipont
Supplementary Information
 Supplementary Information Supplementary Figures Figure S1. Study of mgtl translation in vitro. (A) Detection of 5 LR RNA using wild-type and anti-sd (91-95) substituted templates in a transcription-translation
Supplementary Information Supplementary Figures Figure S1. Study of mgtl translation in vitro. (A) Detection of 5 LR RNA using wild-type and anti-sd (91-95) substituted templates in a transcription-translation
SUPPLEMENTARY INFORMATION. Tolerance of a knotted near infrared fluorescent protein to random circular permutation
 SUPPLEMENTARY INFORMATION Tolerance of a knotted near infrared fluorescent protein to random circular permutation Naresh Pandey 1,3, Brianna E. Kuypers 2,4, Barbara Nassif 1, Emily E. Thomas 1,3, Razan
SUPPLEMENTARY INFORMATION Tolerance of a knotted near infrared fluorescent protein to random circular permutation Naresh Pandey 1,3, Brianna E. Kuypers 2,4, Barbara Nassif 1, Emily E. Thomas 1,3, Razan
Mycobacterium tuberculosis (Mtb) and Mycobacterium smegmatis (Msmeg) contain two ε (dnaq) exonuclease homologs.
 Supplementary Figure 1 Mycobacterium tuberculosis (Mtb) and Mycobacterium smegmatis (Msmeg) contain two ε (dnaq) exonuclease homologs. (a) Sequence alignment of the ε-exonuclease homologs from four different
Supplementary Figure 1 Mycobacterium tuberculosis (Mtb) and Mycobacterium smegmatis (Msmeg) contain two ε (dnaq) exonuclease homologs. (a) Sequence alignment of the ε-exonuclease homologs from four different
Design. Construction. Characterization
 Design Construction Characterization DNA mrna (messenger) A C C transcription translation C A C protein His A T G C T A C G Plasmids replicon copy number incompatibility selection marker origin of replication
Design Construction Characterization DNA mrna (messenger) A C C transcription translation C A C protein His A T G C T A C G Plasmids replicon copy number incompatibility selection marker origin of replication
Lab Book igem Stockholm Lysostaphin. Week 6
 Lysostaphin Week 6 Summarized below are the experiments conducted this week in chronological order. Click on the experiment name to view it. To go back to this summary, click Summary in the footer. Summary
Lysostaphin Week 6 Summarized below are the experiments conducted this week in chronological order. Click on the experiment name to view it. To go back to this summary, click Summary in the footer. Summary
SUPPLEMENTARY EXPEMENTAL PROCEDURES
 SUPPLEMENTARY EXPEMENTAL PROCEDURES Plasmids- Total RNAs were extracted from HeLaS3 cells and reverse-transcribed using Superscript III Reverse Transcriptase (Invitrogen) to obtain DNA template for the
SUPPLEMENTARY EXPEMENTAL PROCEDURES Plasmids- Total RNAs were extracted from HeLaS3 cells and reverse-transcribed using Superscript III Reverse Transcriptase (Invitrogen) to obtain DNA template for the
The Expression of Recombinant Sheep Prion Protein (RecShPrPC) and its Detection Using Western Blot and Immuno-PCR
 The Expression of Recombinant Sheep Prion Protein (RecShPrPC) and its Detection Using Western Blot and Immuno-PCR S. Thomas, C. S. Fernando, J. Roach, U. DeSilva and C. A. Mireles DeWitt The objective
The Expression of Recombinant Sheep Prion Protein (RecShPrPC) and its Detection Using Western Blot and Immuno-PCR S. Thomas, C. S. Fernando, J. Roach, U. DeSilva and C. A. Mireles DeWitt The objective
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Supplementary figure 1: List of primers/oligonucleotides used in this study. 1 Supplementary figure 2: Sequences and mirna-targets of i) mcherry expresses in transgenic fish used
SUPPLEMENTARY INFORMATION Supplementary figure 1: List of primers/oligonucleotides used in this study. 1 Supplementary figure 2: Sequences and mirna-targets of i) mcherry expresses in transgenic fish used
Supplemental Information
 FOOT-AND-MOUTH DISEASE VIRUS PROTEIN 2C IS A HEXAMERIC AAA+ PROTEIN WITH A COORDINATED ATP HYDROLYSIS MECHANISM. Trevor R. Sweeney 1, Valentina Cisnetto 1, Daniel Bose 2, Matthew Bailey 3, Jon R. Wilson
FOOT-AND-MOUTH DISEASE VIRUS PROTEIN 2C IS A HEXAMERIC AAA+ PROTEIN WITH A COORDINATED ATP HYDROLYSIS MECHANISM. Trevor R. Sweeney 1, Valentina Cisnetto 1, Daniel Bose 2, Matthew Bailey 3, Jon R. Wilson
SUPPLEMENTARY INFORMATION
 Supplementary Table 1 SaPIbov1 complementation analysis. No CdCl 2 5 µm CdCl 2 Strain Phage Plasmid expressing SaPIbov1 titre a Phage titre b SaPIbov1 titre a Phage titre b JP1794 φ11-2.0 x 10 7 6.4 x
Supplementary Table 1 SaPIbov1 complementation analysis. No CdCl 2 5 µm CdCl 2 Strain Phage Plasmid expressing SaPIbov1 titre a Phage titre b SaPIbov1 titre a Phage titre b JP1794 φ11-2.0 x 10 7 6.4 x
Development of Immunogens to Protect Against Turkey Cellulitis. Douglas. N. Foster and Robyn Gangl. Department of Animal Science
 Development of Immunogens to Protect Against Turkey Cellulitis Douglas. N. Foster and Robyn Gangl Department of Animal Science University of Minnesota St. Paul, MN 55108 Introduction Clostridial dermatitis
Development of Immunogens to Protect Against Turkey Cellulitis Douglas. N. Foster and Robyn Gangl Department of Animal Science University of Minnesota St. Paul, MN 55108 Introduction Clostridial dermatitis
Supplementary data. sienigma. F-Enigma F-EnigmaSM. a-p53
 Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Human Cell-Free Protein Expression System
 Cat. # 3281 For Research Use Human Cell-Free Protein Expression System Product Manual Table of Contents I. Description... 3 II. Components... 3 III. Materials Required but not Provided... 4 IV. Storage...
Cat. # 3281 For Research Use Human Cell-Free Protein Expression System Product Manual Table of Contents I. Description... 3 II. Components... 3 III. Materials Required but not Provided... 4 IV. Storage...
SUPPLEMENTARY INFORMATION
 Clustering of a kinesin-14 motor enables processive retrograde microtubule-based transport in plants NATURE PLANTS www.nature.com/natureplants 1 2 NATURE PLANTS www.nature.com/natureplants NATURE PLANTS
Clustering of a kinesin-14 motor enables processive retrograde microtubule-based transport in plants NATURE PLANTS www.nature.com/natureplants 1 2 NATURE PLANTS www.nature.com/natureplants NATURE PLANTS
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling
 Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Different Upstream Activators Stimulate Transcription Through Distinct Coactivator Complexes
 Supplemental Material for Different Upstream Activators Stimulate Transcription Through Distinct Coactivator Complexes Dong-ki Lee, Soyoun Kim, and John T. Lis Due to be published as a Research Communication
Supplemental Material for Different Upstream Activators Stimulate Transcription Through Distinct Coactivator Complexes Dong-ki Lee, Soyoun Kim, and John T. Lis Due to be published as a Research Communication
Supporting Information
 Supporting Information Ni/NiO Core/shell Nanoparticles for Selective Binding and Magnetic Separation of Histidine-Tagged Proteins In Su Lee,, No Hyun Lee,, Jongnam Park,, Byung Hyo Kim,, Yong-Weon Yi,
Supporting Information Ni/NiO Core/shell Nanoparticles for Selective Binding and Magnetic Separation of Histidine-Tagged Proteins In Su Lee,, No Hyun Lee,, Jongnam Park,, Byung Hyo Kim,, Yong-Weon Yi,
TECHNICAL BULLETIN. In Vitro Bacterial Split Fluorescent Protein Fold n Glow Solubility Assay Kits
 In Vitro Bacterial Split Fluorescent Protein Fold n Glow Solubility Assay Kits Catalog Numbers APPA001 In Vitro Bacterial Split GFP "Fold 'n' Glow" Solubility Assay Kit (Green) APPA008 In Vitro Bacterial
In Vitro Bacterial Split Fluorescent Protein Fold n Glow Solubility Assay Kits Catalog Numbers APPA001 In Vitro Bacterial Split GFP "Fold 'n' Glow" Solubility Assay Kit (Green) APPA008 In Vitro Bacterial
To clone eif4a fragments into SalI-NotI sites of pacycduet-1 (pmb09); and petduet-1
 Supplementary Methods DNA constructs and strains To clone eif4a fragments into SalI-NotI sites of pacycduet-1 (pmb09); and petduet-1 (Novagen) (pmb11), we PCR-amplified each domain from the mouse eif4a
Supplementary Methods DNA constructs and strains To clone eif4a fragments into SalI-NotI sites of pacycduet-1 (pmb09); and petduet-1 (Novagen) (pmb11), we PCR-amplified each domain from the mouse eif4a
Nature Biotechnology: doi: /nbt.4166
 Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
U937 macrophages Human monocytic cell line ATCC CRL Plasmids Legionella vector for expression of mcherry from a ptac promoter, X.
 "#$%&'("#$%&'()&(*+,&)-'*).)/*'($0'))$.*1 2(,1%/,/3&($4/(5$1+'6&,'(75%-&$'5('))058(9 Strain or plasmid Relevant properties Reference E. coli XL1-Blue reca1 enda1 gyra96 thi-1 hsdr17 supe44 rela1 lac [F
"#$%&'("#$%&'()&(*+,&)-'*).)/*'($0'))$.*1 2(,1%/,/3&($4/(5$1+'6&,'(75%-&$'5('))058(9 Strain or plasmid Relevant properties Reference E. coli XL1-Blue reca1 enda1 gyra96 thi-1 hsdr17 supe44 rela1 lac [F
Supplemental Information
 Supplemental Information ATP-dependent unwinding of U4/U6 snrnas by the Brr2 helicase requires the C-terminus of Prp8 Corina Maeder 1,3, Alan K. Kutach 1,2,3, and Christine Guthrie 1 1 Department of Biochemistry
Supplemental Information ATP-dependent unwinding of U4/U6 snrnas by the Brr2 helicase requires the C-terminus of Prp8 Corina Maeder 1,3, Alan K. Kutach 1,2,3, and Christine Guthrie 1 1 Department of Biochemistry
A novel therapeutic strategy to rescue the immune effector function of proteolyticallyinactivated
 A novel therapeutic strategy to rescue the immune effector function of proteolyticallyinactivated cancer therapeutic antibodies Supplemental Data File in this Data Supplement: Supplementary Figure 1-4
A novel therapeutic strategy to rescue the immune effector function of proteolyticallyinactivated cancer therapeutic antibodies Supplemental Data File in this Data Supplement: Supplementary Figure 1-4
Supporting Information
 Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2016 Supporting Information An Ion Signal Responsive Dynamic Protein Nano-spring Constructed by High
Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2016 Supporting Information An Ion Signal Responsive Dynamic Protein Nano-spring Constructed by High
mod-1::mcherry unc-47::gfp RME ser-4::gfp vm2 vm2 vm2 vm2 VNC
 A mod-1::mcherry unc-47::gfp RME B ser-4::gfp vm2 vm2 VNC vm2 vm2 VN Figure S1 Further details of mod- 1 and ser- 4 reporter expression patterns. (A) Adult head region showing mod- 1::mCherry and unc-
A mod-1::mcherry unc-47::gfp RME B ser-4::gfp vm2 vm2 VNC vm2 vm2 VN Figure S1 Further details of mod- 1 and ser- 4 reporter expression patterns. (A) Adult head region showing mod- 1::mCherry and unc-
Rotation Report Sample Version 3. Due Date: August 9, Analysis of the Guanine Nucleotide Exchange Activity of the S. cerevisiae Ats1 Protein
 Rotation Report Sample Version 3 Due Date: August 9, 1998 Analysis of the Guanine Nucleotide Exchange Activity of the S. cerevisiae Ats1 Protein Anita H. Corbett Advisor: Amy Jones Rotation 1 Abstract:
Rotation Report Sample Version 3 Due Date: August 9, 1998 Analysis of the Guanine Nucleotide Exchange Activity of the S. cerevisiae Ats1 Protein Anita H. Corbett Advisor: Amy Jones Rotation 1 Abstract:
BIOCHEMISTRY 551: BIOCHEMICAL METHODS SYLLABUS
 BIOCHEMISTRY 551: BIOCHEMICAL METHODS SYLLABUS Course Description: Biochemistry 551 is an integrated lecture, lab and seminar course that covers biochemistry-centered theory and techniques. The course
BIOCHEMISTRY 551: BIOCHEMICAL METHODS SYLLABUS Course Description: Biochemistry 551 is an integrated lecture, lab and seminar course that covers biochemistry-centered theory and techniques. The course
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Feng et al., http://www.jcb.org/cgi/content/full/jcb.201408079/dc1 Figure S1. A modest elevation of disulfide-bonded K14 in primary mouse
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Feng et al., http://www.jcb.org/cgi/content/full/jcb.201408079/dc1 Figure S1. A modest elevation of disulfide-bonded K14 in primary mouse
A fail-safe mechanism in the septal ring assembly pathway generated by the sequential recruitment of cell separation amidases and their activators
 Supplemental Information for: A fail-safe mechanism in the septal ring assembly pathway generated by the sequential recruitment of cell separation amidases and their activators Nick T. Peters, Thuy Dinh,
Supplemental Information for: A fail-safe mechanism in the septal ring assembly pathway generated by the sequential recruitment of cell separation amidases and their activators Nick T. Peters, Thuy Dinh,
Supplementary Figure S1 Complementarity between VR-RNA and cola mrna 5 UTR is important for cola regulation. (A) Mutation sites within VR-RNA-colA
 Supplementary Figure S1 Complementarity between VR-RNA and cola mrna 5 UTR is important for cola regulation. (A) Mutation sites within VR-RNA-colA RNA duplex. Mutation sites in cola mrna 5 UTR or VR-RNA
Supplementary Figure S1 Complementarity between VR-RNA and cola mrna 5 UTR is important for cola regulation. (A) Mutation sites within VR-RNA-colA RNA duplex. Mutation sites in cola mrna 5 UTR or VR-RNA
Engineering splicing factors with designed specificities
 nature methods Engineering splicing factors with designed specificities Yang Wang, Cheom-Gil Cheong, Traci M Tanaka Hall & Zefeng Wang Supplementary figures and text: Supplementary Figure 1 Supplementary
nature methods Engineering splicing factors with designed specificities Yang Wang, Cheom-Gil Cheong, Traci M Tanaka Hall & Zefeng Wang Supplementary figures and text: Supplementary Figure 1 Supplementary
Optimization of gene expression cassette for M. gryphiswaldense
 Optimization of gene expression cassette for M. gryphiswaldense A) 325 bp P mamdc 270 bp 170 bp 102 bp 45 bp egfp Gradual truncation P mamdc45 egfp minimal transcriptionally active promoter B) P mamdc45
Optimization of gene expression cassette for M. gryphiswaldense A) 325 bp P mamdc 270 bp 170 bp 102 bp 45 bp egfp Gradual truncation P mamdc45 egfp minimal transcriptionally active promoter B) P mamdc45
At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in
 Supplementary Materials and Methods Barrier function assays At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in acidic X-gal mix (100 mm phosphate buffer at ph4.3, 3 mm
Supplementary Materials and Methods Barrier function assays At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in acidic X-gal mix (100 mm phosphate buffer at ph4.3, 3 mm
Supporting Information
 Supporting Information Massie et al. 10.1073/pnas.1115663109 SI Materials and Methods Construction of DGC Overexpression Plasmids. The overexpression plasmids for each V. cholerae DGC were constructed
Supporting Information Massie et al. 10.1073/pnas.1115663109 SI Materials and Methods Construction of DGC Overexpression Plasmids. The overexpression plasmids for each V. cholerae DGC were constructed
Supplementary Figure 1 Performance analysis of RedLibs (version 0.2.3). Supplementary Figure 2
 Supplementary Figure 1 Performance analysis of RedLibs (version 0.2.3). The required calculation time for the evaluation of all sub-libraries with a given target size are plotted against the combinatorial
Supplementary Figure 1 Performance analysis of RedLibs (version 0.2.3). The required calculation time for the evaluation of all sub-libraries with a given target size are plotted against the combinatorial
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.
 Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
TIANpure Mini Plasmid Kit
 TIANpure Mini Plasmid Kit For purification of molecular biology grade DNA www.tiangen.com PP080822 TIANpure Mini Plasmid Kit Kit Contents Storage Contents RNaseA (10 mg/ml) Buffer BL Buffer P1 Buffer P2
TIANpure Mini Plasmid Kit For purification of molecular biology grade DNA www.tiangen.com PP080822 TIANpure Mini Plasmid Kit Kit Contents Storage Contents RNaseA (10 mg/ml) Buffer BL Buffer P1 Buffer P2
Optically Controlled Reversible Protein Hydrogels Based on Photoswitchable Fluorescent Protein Dronpa. Electronic Supplementary Information
 Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2017 Optically Controlled Reversible Protein Hydrogels Based on Photoswitchable Fluorescent Protein
Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2017 Optically Controlled Reversible Protein Hydrogels Based on Photoswitchable Fluorescent Protein
Biotechnology and Energy Conservation. Prof. Dr.oec.troph. Ir. Krishna Purnawan Candra, M.S. Program Magister Ilmu Lingkungan Universitas Mulawarman
 Biotechnology and Energy Conservation Prof. Dr.oec.troph. Ir. Krishna Purnawan Candra, M.S. Program Magister Ilmu Lingkungan Universitas Mulawarman 12 th Lecture Genetic Engineering The Aim: Students can
Biotechnology and Energy Conservation Prof. Dr.oec.troph. Ir. Krishna Purnawan Candra, M.S. Program Magister Ilmu Lingkungan Universitas Mulawarman 12 th Lecture Genetic Engineering The Aim: Students can
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than. SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with
 Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
CHAPTER 9 DNA Technologies
 CHAPTER 9 DNA Technologies Recombinant DNA Artificially created DNA that combines sequences that do not occur together in the nature Basis of much of the modern molecular biology Molecular cloning of genes
CHAPTER 9 DNA Technologies Recombinant DNA Artificially created DNA that combines sequences that do not occur together in the nature Basis of much of the modern molecular biology Molecular cloning of genes
yellow protein turn off/on label (POOL) , Republic of Korea The PYP gene was subcloned in the pqe80l vector using EZ-Cloning (enzynomics ).
 Supporting Information High-throughput instant quantification of protein expression and purity based on photoactive yellow protein turn off/on label (POOL) Youngmin Kim 1,2, Prabhakar Ganesan 1,2 and Hyotcherl
Supporting Information High-throughput instant quantification of protein expression and purity based on photoactive yellow protein turn off/on label (POOL) Youngmin Kim 1,2, Prabhakar Ganesan 1,2 and Hyotcherl
RTS 100 E. coli LinTempGenSet, Histag
 RTS 100 E. coli LinTempGenSet, Histag Manual For rapid production of linear expression templates using PCR RTS 100 E. coli LinTempGenSet, His-tag Manual, October, 2009 2009 5 PRIME, all rights reserved.
RTS 100 E. coli LinTempGenSet, Histag Manual For rapid production of linear expression templates using PCR RTS 100 E. coli LinTempGenSet, His-tag Manual, October, 2009 2009 5 PRIME, all rights reserved.
FtsEX is required for CwlO peptidoglycan hydrolase activity during cell wall elongation in Bacillus subtilis
 FtsEX is required for CwlO peptidoglycan hydrolase activity during cell wall elongation in Bacillus subtilis Jeffrey Meisner, Paula Montero Llopis, Lok-To Sham, Ethan Garner, Thomas G. Bernhardt, David
FtsEX is required for CwlO peptidoglycan hydrolase activity during cell wall elongation in Bacillus subtilis Jeffrey Meisner, Paula Montero Llopis, Lok-To Sham, Ethan Garner, Thomas G. Bernhardt, David
SUPPORTING INFORMATION
 SUPPORTING INFORMATION Polyketide Ring Expansion Mediated by a Thioesterase, Chain Elongation and Cyclization, in Azinomycin Biosynthesis: Characterization of AziB and AziG Shogo Mori, [1] Dinesh Simkhada,
SUPPORTING INFORMATION Polyketide Ring Expansion Mediated by a Thioesterase, Chain Elongation and Cyclization, in Azinomycin Biosynthesis: Characterization of AziB and AziG Shogo Mori, [1] Dinesh Simkhada,
Supporting Information. Sequence Independent Cloning and Posttranslational. Enzymatic Ligation
 Supporting Information Sequence Independent Cloning and Posttranslational Modification of Repetitive Protein Polymers through Sortase and Sfp-mediated Enzymatic Ligation Wolfgang Ott,,, Thomas Nicolaus,,
Supporting Information Sequence Independent Cloning and Posttranslational Modification of Repetitive Protein Polymers through Sortase and Sfp-mediated Enzymatic Ligation Wolfgang Ott,,, Thomas Nicolaus,,
Molecular Biology Techniques Supporting IBBE
 Molecular Biology Techniques Supporting IBBE Jared Cartwright Protein Production Lab Head Contact Details: email jared.cartwright@york.ac.uk Phone 01904 328797 Presentation Aims Gene synthesis Cloning
Molecular Biology Techniques Supporting IBBE Jared Cartwright Protein Production Lab Head Contact Details: email jared.cartwright@york.ac.uk Phone 01904 328797 Presentation Aims Gene synthesis Cloning
Supplementary table captions
 Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 06 Table S Strains and plasmids used in the present study Supplementary table captions Table S Primers
Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 06 Table S Strains and plasmids used in the present study Supplementary table captions Table S Primers
Figure 1. Map of cloning vector pgem T-Easy (bacterial plasmid DNA)
 Texas A&M University-Corpus Christi CHEM4402 Biochemistry II Laboratory Laboratory 6: Ligation & Bacterial Transformation (Bring your text and laptop to class if you wish to work on your assignment during
Texas A&M University-Corpus Christi CHEM4402 Biochemistry II Laboratory Laboratory 6: Ligation & Bacterial Transformation (Bring your text and laptop to class if you wish to work on your assignment during
Genome Sequence Assembly
 Genome Sequence Assembly Learning Goals: Introduce the field of bioinformatics Familiarize the student with performing sequence alignments Understand the assembly process in genome sequencing Introduction:
Genome Sequence Assembly Learning Goals: Introduce the field of bioinformatics Familiarize the student with performing sequence alignments Understand the assembly process in genome sequencing Introduction:
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Moutin et al., http://www.jcb.org/cgi/content/full/jcb.201110101/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Tagged Homer1a and Homer are functional and display different
Supplemental material Moutin et al., http://www.jcb.org/cgi/content/full/jcb.201110101/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Tagged Homer1a and Homer are functional and display different
SAS6-like protein in Plasmodium indicates that conoid-associated apical complex proteins persist in invasive stages within the mosquito vector
 SAS6-like protein in Plasmodium indicates that conoid-associated apical complex proteins persist in invasive stages within the mosquito vector Richard J. Wall 1,#, Magali Roques 1,+, Nicholas J. Katris
SAS6-like protein in Plasmodium indicates that conoid-associated apical complex proteins persist in invasive stages within the mosquito vector Richard J. Wall 1,#, Magali Roques 1,+, Nicholas J. Katris
ChIP DNA Clean & Concentrator Catalog Nos. D5201 & D5205
 Page 0 INSTRUCTION MANUAL ChIP DNA Clean & Concentrator Catalog Nos. D5201 & D5205 Highlights Quick (2 minute) recovery of ultra-pure DNA from chromatin immunoprecipitation (ChIP) assays, cell lysates,
Page 0 INSTRUCTION MANUAL ChIP DNA Clean & Concentrator Catalog Nos. D5201 & D5205 Highlights Quick (2 minute) recovery of ultra-pure DNA from chromatin immunoprecipitation (ChIP) assays, cell lysates,
Supplemental Information. Pacer Mediates the Function of Class III PI3K. and HOPS Complexes in Autophagosome. Maturation by Engaging Stx17
 Molecular Cell, Volume 65 Supplemental Information Pacer Mediates the Function of Class III PI3K and HOPS Complexes in Autophagosome Maturation by Engaging Stx17 Xiawei Cheng, Xiuling Ma, Xianming Ding,
Molecular Cell, Volume 65 Supplemental Information Pacer Mediates the Function of Class III PI3K and HOPS Complexes in Autophagosome Maturation by Engaging Stx17 Xiawei Cheng, Xiuling Ma, Xianming Ding,
Supplementary Figure 1 Structural modeling and purification of V. cholerae ABH. (a) The migration of the purified rabh and catalytically inactive
 Supplementary Figure 1 Structural modeling and purification of V. cholerae ABH. (a) The migration of the purified rabh and catalytically inactive variants rabhs, rabhd, and rabhh are shown on 12% SDS-PAGE
Supplementary Figure 1 Structural modeling and purification of V. cholerae ABH. (a) The migration of the purified rabh and catalytically inactive variants rabhs, rabhd, and rabhh are shown on 12% SDS-PAGE
3 -end. Sau3A. 3 -end TAA TAA. ~9.1kb SalI TAA. ~12.6kb. HindШ ATG 5,860bp TAA. ~12.7kb
 Supplemental Data. Chen et al. (2008). Badh2, encoding betaine aldehyde dehydrogenase, inhibits the biosynthesis of 2-acetyl-1-pyrroline, a major component in rice fragrance. A pcam-cah/cah Sau3A Promoter
Supplemental Data. Chen et al. (2008). Badh2, encoding betaine aldehyde dehydrogenase, inhibits the biosynthesis of 2-acetyl-1-pyrroline, a major component in rice fragrance. A pcam-cah/cah Sau3A Promoter
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
CT was cloned from Vibrio cholera DNA provided by Ontario Public Health. laboratory using CTFP/CTRP primer set and High-fidelity DNA polymerase
 Supplementary Material Construction of Mutated CT and VT 1. Cloning and inactivation of CT and VT CT was cloned from Vibrio cholera DNA provided by Ontario Public Health laboratory using CTFP/CTRP primer
Supplementary Material Construction of Mutated CT and VT 1. Cloning and inactivation of CT and VT CT was cloned from Vibrio cholera DNA provided by Ontario Public Health laboratory using CTFP/CTRP primer
FGF9 monomer/dimer equilibrium regulates. extracellular matrix affinity and tissue diffusion
 SUPPLEMENTARY INFORMATGION FGF9 monomer/dimer equilibrium regulates extracellular matrix affinity and tissue diffusion Masayo Harada, Hirotaka Murakami, Akihiko Okawa, Noriaki Okimoto, Shuichi Hiraoka,
SUPPLEMENTARY INFORMATGION FGF9 monomer/dimer equilibrium regulates extracellular matrix affinity and tissue diffusion Masayo Harada, Hirotaka Murakami, Akihiko Okawa, Noriaki Okimoto, Shuichi Hiraoka,
Transport of Potato Lipoxygenase into the Vacuole Larsen, Mia Kruse Guldstrand; Welinder, Karen Gjesing; Jørgensen, Malene
 Aalborg Universitet Transport of Potato Lipoxygenase into the Vacuole Larsen, Mia Kruse Guldstrand; Welinder, Karen Gjesing; Jørgensen, Malene Publication date: 2009 Document Version Publisher's PDF, also
Aalborg Universitet Transport of Potato Lipoxygenase into the Vacuole Larsen, Mia Kruse Guldstrand; Welinder, Karen Gjesing; Jørgensen, Malene Publication date: 2009 Document Version Publisher's PDF, also
Lab Book igem Stockholm Lysostaphin. Week 9
 Lysostaphin Week 9 Summarized below are the experiments conducted this week in chronological order. Click on the experiment name to view it. To go back to this summary, click Summary in the footer. Summary
Lysostaphin Week 9 Summarized below are the experiments conducted this week in chronological order. Click on the experiment name to view it. To go back to this summary, click Summary in the footer. Summary
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION 1 Supplementary Figure S1. Proportion of HEK293T transfectants showing fluorescent foci after heat sho Blinded slides were counted for the appearance of fluorescent foci of tagrfp-t
SUPPLEMENTARY INFORMATION 1 Supplementary Figure S1. Proportion of HEK293T transfectants showing fluorescent foci after heat sho Blinded slides were counted for the appearance of fluorescent foci of tagrfp-t
Molecular Biology. Manufactured by. Online Ordering Available
 Molecular Biology Manufactured by Online Ordering Available Molecular Biology Table of Contents 1 Bacterial Culture Media Powder 2 Bacterial Culture Agar Plates 2 Bacterial Culture Agar Heat and Pour Format
Molecular Biology Manufactured by Online Ordering Available Molecular Biology Table of Contents 1 Bacterial Culture Media Powder 2 Bacterial Culture Agar Plates 2 Bacterial Culture Agar Heat and Pour Format
