A(+1) A( 7) RNA hairpin
|
|
|
- Gillian Allison
- 6 years ago
- Views:
Transcription
1 PP7 CP dimer A(+1) A( 7) RNA hairpin 5 3
2 Supplemental Figure 1. Sample electron density of RNA hairpin. Cartoon showing PP7 FG CP (wheat) bound to RNA hairpin (violet) showing 2Fo-Fc electron density (blue) calculated at 1σ.
3 0.008nM 250nM complex free Fraction Bound Kd,app = 1.6 ± 0.4nM [PP7 FG] M
4 Supplemental Figure 2. PP7 FG CP binds its translational operator with high affinity. Representative electrophoretic mobility shift assay shown with free 27nt RNA hairpin and RNA-protein complex labeled. Filled triangle in top panel represents PP7 FG CP concentration increasing by a factor of two per lane. A plot of the fraction of RNA bound as a function of PP7 FG concentration and its fit to the Hill equation is presented for the representative data. The K d,app based upon three independent measurements is 1.26±0.38nM.
5 a FG loop N c d e b b a f g α2 α1 C PP7 MS2
6 Supplemental Figure 3. Structure of PP7 FG CP dimer and its superposition with MS2 W82R CP. (a) Cartoon of PP7 FG CP dimer showing N-terminal β-hairpin (blue), five stranded β-sheet (wheat), truncated FG loop (green) and C-terminal α-helices (red) of one protomer with the other protomer shown in grey. (b) PP7 FG and MS2 W82R (1MSC) CPs have similar folds and superpose with an r.m.s.d. of 2.3Å for all C α atoms.
7 Supplemental Table 1. Data collection and refinement statistics PP7 FG PP7 FG-RNA Data collection Space group H3 C2 Cell dimensions a, b, c (Å) 155.0, 155.0, , 145.4, α, β, γ ( ) 90, 90, , 122.9, 90 Resolution (Å) ( ) ( ) R sym or R merge (0.475) (0.450) I / σi 36.8 (2.1) 18.7 (2.3) Completeness (%) (100.0) 98.6 (87.6) Redundancy 4.3 (3.9) 4.3 (4.0) Refinement Resolution (Å) No. reflections 35,670 82,206 R work / R free 15.7/ /23.6 No. atoms Protein 2,005 11,222 RNA NA 3,198 Water B-factors Protein RNA NA Water R.m.s. deviations Bond lengths (Å) Bond angles ( ) *Values in parentheses are for highest-resolution shell.
8 Structural basis for the co-evolution of a viral RNA-protein complex Jeffrey A. Chao, Yury Patskovsky, Steven C. Almo, Robert H. Singer Supplemental Data
9 Materials and Methods Protein and RNA preparation The pp7ct plasmid containing the full length PP7 coat protein gene was provided by D. Peabody (Albuquerque, New Mexico). Similar to the MS2 CP, the PP7 CP forms an obligate dimer that is the building block of the bacteriophage capsid and also responsible for translational repression of the replicase by binding an RNA stem-loop that contains the replicase start codon 1. These two functions can be separated for the MS2 CP because the FG loop, located between the sixth and seventh β-strands, that stabilizes capsid formation is not involved in RNA recognition 2-4. A PP7ΔFG truncation was constructed by PCR using the structure of the PP7 capsid as a guide that replaced loop residues with a BspEI restriction site (Ser-Gly) 5. The excised region includes the two cysteines that form inter-molecular disulphide bonds and covalently link the dimers at the capsid fivefold and quasi-sixfold axes 5. This construct was cloned into pet22ht as an N-terminal His 6 -tag followed by a TEV cleavage site 6. Recombinant protein was expressed in E. coli strain BL21(DE3) grown at 37 C by induction with 1mM IPTG at A600 = 0.7. Cell pellets were resuspended in lysis buffer (50mM sodium phosphate, 1M NaCl, 10mM imidazole, 10mM β-mercaptoethanol, ph 8.0) containing one Complete EDTA-free protease inhibitor tablet (Roche) and lysed by sonication. Cell debris was removed by centrifugation at 15,000g for 20 minutes at 4 C and the soluble His 6 -tagged protein was incubated with HIS-Select affinity gel (Sigma) equilibrated with lysis buffer for 1 hour at 4 C. The affinity gel was extensively washed with lysis buffer and protein was eluted with lysis buffer supplemented with 250mM imidazole. Elution fractions containing
10 His 6 - PP7ΔFG were pooled and dialyzed at 4 C against 25mM HEPES (ph 7.0), 25mM NaCl, 1mM DTT, 1mM EDTA. During dialysis, the His 6 -tag was removed by incubation with TEV protease leaving a residual Gly-Gly-Ser at the N-terminus of the PP7 FG construct. PP7ΔFG was further purified by cation exchange chromatography using a HiTrap SP HP column (GE). The resulting protein was dialyzed against 25mM HEPES (ph 7.0), 25mM NaCl, 1mM EDTA and concentrated to 1.3mM. Protein concentration was calculated by measuring the absorbance at 280nm using an extinction coefficient of 9970 M -1 cm -1 (0.733 mg ml -1 ) determined using the ProtParam server 7. RNAs used for crystallization and binding studies were prepared by chemical synthesis (Dharmacon) and purified by denaturing PAGE, deprotected, lyophilized, and stored according to the manufacturer s protocol. RNAs were resuspended in 25mM HEPES (ph 7.0), 25mM NaCl, 1mM EDTA. Annealing of the RNAs into hairpins was performed by incubation at 90ºC for 1 min followed by snap cooling of the RNA on ice for 30 minutes. Crystallization and structure determination of PP7ΔFG construct The free PP7ΔFG protein was crystallized using hanging-drop vapor diffusion at 22 C by mixing equal volumes of the protein and reservoir solution (1.1 M Na Malonate ph 7.0, 0.1 M HEPES ph 7.0, 0.5% v/v Jeffamine ED-2001 ph 7.0). Crystals were cryoprotected by soaking them in the reservoir solution supplemented with 25% glycerol before flash cooling in liquid nitrogen. Data were collected to 1.60Å resolution from a single crystal at the National Synchotron Light Source X29a beamline (Brookhaven, NY) at a wavelength of 1.008Å. The diffraction data were indexed, integrated and scaled with HKL
11 The structure of PP7ΔFG was determined by molecular replacement with Phaser using a monomer from the previously determined structure of PP7 bacteriophage capsid (1DWN) as a search model 5,9. The crystal contained two copies of the monomer corresponding to the expected dimer in the asymmetric unit. Rounds of refinement and model building were carried out with REFMAC5 and Coot 10,11. Non-crystallographic symmetry (NCS) restraints were applied during initial stages of refinement. Protein stereochemistry was checked using Molprobity 12. The final model contains residues (chain A) and 1-65: (chain B) using the residue numbering described for 1DWN with the GGS cloning artifact and initial methionine numbered as The non-native Ser and Gly generated during truncation of the FG loop are numbered as 67 and 68. Crystallization and structure determination of PP7ΔFG-RNA complex construct The PP7ΔFG-RNA complex was formed by equilibrating the protein and RNA hairpin (5 -GGCACAGAAGAUAUGGCUUCGUGCC-3 ) at a 2:1.1 molar ratio on ice for 30 minutes prior to crystallization trials. The final concentration of the complex used for crystallization was approximately 250µM. The number of base pairs in the lower helix of the RNA stem-loop was systematically varied and only the RNA construct exactly 25nts in length resulted in crystals of the complex. Crystals were obtained using sitting-drop vapor diffusion at 22 C by mixing equal volumes of the complex and reservoir solution (15% PEG 3350, 0.1M Na Citrate ph 5.5, 10% Hampton Crystal Screen 2 #25 (0.1M MES ph 6.5, 0.01M Cobaltous chloride hexahydrate, 1.8M Ammonium sulfate). Crystals were cryoprotected by soaking them in the reservoir solution supplemented with 30% glycerol before flash cooling in liquid nitrogen. Data
12 were collected to 2.44Å resolution from a single crystal as part of the Express Crystallography service at the Advanced Photon Source SGX-CAT beamline (Argonne, IL) at a wavelength of Å. The diffraction data were indexed, integrated and scaled with HKL The structure of the PP7ΔFG-RNA complex was determined by molecular replacement with Phaser using the PP7ΔFG dimer as a search model 9. The crystal contained six copies of the RNA-protein complex in the asymmetric unit. Initial electron density for the RNA after molecular replacement was of excellent quality and allowed unambiguous placement of all 25 nucleotides using X-fit 13. The RNA stems were found to stack on the two-fold axis explaining the RNA length dependence of crystallization. Early rounds of simulated-annealing refinement and model building were performed using CNS and X-fit 13,14. After all six RNA stem-loops were built, refinement and model building was continued using REFMAC5 and Coot 10,11. Tight NCS restraints were applied initially and were relaxed during iterative rounds of refinement. TLS groups were defined for all twelve PP7ΔFG monomers and the six RNA stem-loops. Protein stereochemistry and RNA geometry was checked using Molprobity 12. The final model contains residues (chain A), (chain B), 1-25 (chain C), (chain D), (chain E), 1-25 (chain F), (chain G), 1-21: (chain H), 1-25 (chain I), (chain J), -1-65: (chain K), 1-25 (chain L), (chain M), 1-21: (chain N), 1-25 (chain O), (chain P), 1-21:23-64: (chain Q) and 1-25 (chain R).
13 Electrophoretic mobility shift assay The complex between PP7 FG and a 27nt RNA hairpin (5 - GCGCACAGAAGAUAUGGCUUCGUGCGC-3 ) was monitored using an electrophoretic gel mobility shift assay. Fluorescein 5-thiosemicarbazide was used to 3 - end label the hairpin using a method adapted from Reines and Cantor 15,16. The RNA hairpin (100 pm) was equilibrated with a two-fold serial dilution of PP7 FG (250 nm to nm). The complex was equilibrated in buffer containing 10mM Tris ph 7.5, 25 mm NaCl, 0.1 mm EDTA, 0.01 mg ml -1 trna, 50 µg ml -1 heparin and 0.01% (v/v) IGEPAL CA630 for 2 hours. Prior to loading, a 6X loading dye (30% (v/v) glycerol and 0.01% (w/v) bromocresol green was added to each binding reaction. Protein-RNA complexes were resolved from free RNA by native polyacrylamide gel electrophoresis (6% (w/v) 29:1 acrylamide/bis-acrylamide, 0.5x TBE) at 22 C. Gels were run at 120V for 35 minutes and then scanned using a fluor-imager (GE, Typhoon 9400) with a blue laser at 488 nm for excitation. The fraction of bound RNA hairpin was determined and fit to a modified version of the Hill equation as previously described 17. References 1. Witherell, G.W., Gott, J.M. & Uhlenbeck, O.C. Specific interaction between RNA phage coat proteins and RNA. Prog Nucleic Acid Res Mol Biol 40, (1991). 2. Peabody, D.S. & Ely, K.R. Control of translational repression by protein-protein interactions. Nucleic Acids Res 20, (1992). 3. Ni, C.Z. et al. Crystal structure of the MS2 coat protein dimer: implications for RNA binding and virus assembly. Structure 3, (1995). 4. LeCuyer, K.A., Behlen, L.S. & Uhlenbeck, O.C. Mutants of the bacteriophage MS2 coat protein that alter its cooperative binding to RNA. Biochemistry 34, (1995). 5. Tars, K., Fridborg, K., Bundule, M. & Liljas, L. The three-dimensional structure of bacteriophage PP7 from Pseudomonas aeruginosa at 3.7-A resolution. Virology 272, (2000).
14 6. Chao, J.A. et al. Dual modes of RNA-silencing suppression by Flock House virus protein B2. Nat Struct Mol Biol 12, (2005). 7. Gasteiger, E. et al. (eds.). Protein Identification and Analysis Tools on the ExPASy Server, (Humana Press, 2005). 8. Otwinowski, Z. & Minor, W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology 276, (1997). 9. Read, R.J. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr D Biol Crystallogr 57, (2001). 10. Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, (2004). 11. Winn, M.D., Isupov, M.N. & Murshudov, G.N. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr 57, (2001). 12. Davis, I.W. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res (2007). 13. McRee, D.E. XtalView/Xfit--A versatile program for manipulating atomic coordinates and electron density. J Struct Biol 125, (1999). 14. Brunger, A.T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54, (1998). 15. Reines, S.A. & Cantor, C.R. New fluorescent hydrazide reagents for the oxidized 3'-terminus of RNA. Nucleic Acids Res 1, (1974). 16. Pagano, J.M., Farley, B.M., McCoig, L.M. & Ryder, S.P. Molecular basis of RNA recognition by the embryonic polarity determinant MEX-5. J Biol Chem 282, (2007). 17. Recht, M.I. & Williamson, J.R. Central domain assembly: thermodynamics and kinetics of S6 and S18 binding to an S15-RNA complex. J Mol Biol 313, (2001).
SUPPLEMENTAL DATA Experimental procedures
 SUPPLEMENTAL DATA Experimental procedures Cloning, protein production and purification. The DNA sequences corresponding to residues R10-S250 (DBD) and E258-T351 (IPT/TIG) in human EBF1 (gi:31415878), and
SUPPLEMENTAL DATA Experimental procedures Cloning, protein production and purification. The DNA sequences corresponding to residues R10-S250 (DBD) and E258-T351 (IPT/TIG) in human EBF1 (gi:31415878), and
Figure S2, related to Figure 1. Stereo images of the CarD/RNAP complex and. electrostatic potential surface representation of the CarD/RNAP interface
 Structure, Volume 21 Supplemental Information Structure of the Mtb CarD/RNAP -Lobes Complex Reveals the Molecular Basis of Interaction and Presents a Distinct DNA-Binding Domain for Mtb CarD Gulcin Gulten
Structure, Volume 21 Supplemental Information Structure of the Mtb CarD/RNAP -Lobes Complex Reveals the Molecular Basis of Interaction and Presents a Distinct DNA-Binding Domain for Mtb CarD Gulcin Gulten
PROCEDURE FOR USE NICKEL NTA Magnetic Agarose Beads (5%)
 1 AFFINITY HIS-TAG PURIFICATION PROCEDURE FOR USE NICKEL NTA Magnetic Agarose Beads (5%) DESCRIPTION Nickel NTA Magnetic Agarose Beads are products that allow rapid and easy small-scale purification of
1 AFFINITY HIS-TAG PURIFICATION PROCEDURE FOR USE NICKEL NTA Magnetic Agarose Beads (5%) DESCRIPTION Nickel NTA Magnetic Agarose Beads are products that allow rapid and easy small-scale purification of
AFFINITY HIS-TAG PURIFICATION
 DESCRIPTION Resins are products that allow batch or column purifications. This product is supplied as a suspension in 50% aqueous suspension containing 30 vol % ethanol. INSTRUCTIONS The resins are adapted
DESCRIPTION Resins are products that allow batch or column purifications. This product is supplied as a suspension in 50% aqueous suspension containing 30 vol % ethanol. INSTRUCTIONS The resins are adapted
Supporting Online Material. Av1 and Av2 were isolated and purified under anaerobic conditions according to
 Supporting Online Material Materials and Methods Av1 and Av2 were isolated and purified under anaerobic conditions according to published protocols (S1). Crystals of nf-, pcp- and adp-av2:av1 complexes
Supporting Online Material Materials and Methods Av1 and Av2 were isolated and purified under anaerobic conditions according to published protocols (S1). Crystals of nf-, pcp- and adp-av2:av1 complexes
Supporting Information
 Supporting Information Nishimura et al. 10.1073/pnas.1003553107 SI Text SI Materials and Methods. Plasmid construction. The cdnas of human Gα q were amplified and subcloned into pcmv5. Mutants of Gα q
Supporting Information Nishimura et al. 10.1073/pnas.1003553107 SI Text SI Materials and Methods. Plasmid construction. The cdnas of human Gα q were amplified and subcloned into pcmv5. Mutants of Gα q
Nickel-NTA Agarose Suspension
 Nickel-NTA Agarose Suspension Agarose beads for purification of His-tagged proteins Product No. A9735 Description Nickel-NTA Agarose Suspension is an agarose-based affinity chromatography resin allowing
Nickel-NTA Agarose Suspension Agarose beads for purification of His-tagged proteins Product No. A9735 Description Nickel-NTA Agarose Suspension is an agarose-based affinity chromatography resin allowing
INSTRUCTIONS The resins are adapted to work mainly in native conditions like denaturing.
 1 AFFINITY HIS-TAG PURIFICATION PROCEDURE FOR USE Nickel NTA Agarose Beads DESCRIPTION Resins are products that allow batch or column purifications. This product is supplied as a suspension in 50% aqueous
1 AFFINITY HIS-TAG PURIFICATION PROCEDURE FOR USE Nickel NTA Agarose Beads DESCRIPTION Resins are products that allow batch or column purifications. This product is supplied as a suspension in 50% aqueous
Supplemental Information. The structural basis of R Spondin recognition by LGR5 and RNF43
 Supplemental Information The structural basis of R Spondin recognition by LGR5 and RNF43 Po Han Chen 1, Xiaoyan Chen 1, Deyu Fang 2, Xiaolin He 1* 1 Department of Molecular Pharmacology and Biological
Supplemental Information The structural basis of R Spondin recognition by LGR5 and RNF43 Po Han Chen 1, Xiaoyan Chen 1, Deyu Fang 2, Xiaolin He 1* 1 Department of Molecular Pharmacology and Biological
AFFINITY HIS-TAG PURIFICATION
 DESCRIPTION Resins are products that allow batch or column purifications. This product is supplied as a suspension in 50% aqueous suspension containing 30 vol % ethanol. INSTRUCTIONS The resins are adapted
DESCRIPTION Resins are products that allow batch or column purifications. This product is supplied as a suspension in 50% aqueous suspension containing 30 vol % ethanol. INSTRUCTIONS The resins are adapted
AFFINITY HIS-TAG PURIFICATION
 DESCRIPTION Nickel NTA Agarose Cartridges 5ml are used for purification of histidine-tagged proteins in native or denaturing conditions. This cartridge can be used with an automated chromatography system,
DESCRIPTION Nickel NTA Agarose Cartridges 5ml are used for purification of histidine-tagged proteins in native or denaturing conditions. This cartridge can be used with an automated chromatography system,
Ni-NTA Agarose. User Manual. 320 Harbor Way South San Francisco, CA Phone: 1 (888) MCLAB-88 Fax: 1 (650)
 Ni-NTA Agarose User Manual 320 Harbor Way South San Francisco, CA 94080 Phone: 1 (888) MCLAB-88 Fax: 1 (650) 871-8796 www. Contents Introduction -----------------------------------------------------------------------
Ni-NTA Agarose User Manual 320 Harbor Way South San Francisco, CA 94080 Phone: 1 (888) MCLAB-88 Fax: 1 (650) 871-8796 www. Contents Introduction -----------------------------------------------------------------------
Nature Structural & Molecular Biology: doi: /nsmb.1969
 Supplementary Methods Structure determination All the diffraction data sets were collected on BL-41XU (using ADSC Quantum 315 HE CCD detector) at SPring8 (Harima, Japan) or on BL5A (using ADSC Quantum
Supplementary Methods Structure determination All the diffraction data sets were collected on BL-41XU (using ADSC Quantum 315 HE CCD detector) at SPring8 (Harima, Japan) or on BL5A (using ADSC Quantum
Steps in solving a structure. Diffraction experiment. Obtaining well-diffracting crystals. Three dimensional crystals
 Protein structure from X-ray diffraction Diffraction images: ciprocal space Protein, chemical structure: IALEFGPSLKMNE Conformation, 3D-structure: CRYST1 221.200 73.600 80.900 90.00 90.00 90.00 P 21 21
Protein structure from X-ray diffraction Diffraction images: ciprocal space Protein, chemical structure: IALEFGPSLKMNE Conformation, 3D-structure: CRYST1 221.200 73.600 80.900 90.00 90.00 90.00 P 21 21
Supporting Information
 Supporting Information Wiley-VCH 2006 69451 Weinheim, Germany Rolling-circle Amplification of a DNA Nanojunction Chenxiang Lin, Mingyi Xie, Julian J.L. Chen, Yan Liu and Hao Yan A. RCA replication of the
Supporting Information Wiley-VCH 2006 69451 Weinheim, Germany Rolling-circle Amplification of a DNA Nanojunction Chenxiang Lin, Mingyi Xie, Julian J.L. Chen, Yan Liu and Hao Yan A. RCA replication of the
GST Fusion Protein Purification Kit
 Glutathione Resin GST Fusion Protein Purification Kit Cat. No. L00206 Cat. No. L00207 Technical Manual No. TM0185 Version 01042012 Index 1. Product Description 2. Related Products 3. Purification Procedure
Glutathione Resin GST Fusion Protein Purification Kit Cat. No. L00206 Cat. No. L00207 Technical Manual No. TM0185 Version 01042012 Index 1. Product Description 2. Related Products 3. Purification Procedure
Stabilization of a virus-like particle and its application as a nanoreactor at physiological conditions
 Supporting Information Stabilization of a virus-like particle and its application as a nanoreactor at physiological conditions Lise Schoonen, b Sjors Maassen, b Roeland J. M. Nolte b and Jan C. M. van
Supporting Information Stabilization of a virus-like particle and its application as a nanoreactor at physiological conditions Lise Schoonen, b Sjors Maassen, b Roeland J. M. Nolte b and Jan C. M. van
Product. Ni-NTA His Bind Resin. Ni-NTA His Bind Superflow. His Bind Resin. His Bind Magnetic Agarose Beads. His Bind Column. His Bind Quick Resin
 Novagen offers a large variety of affinity supports and kits for the purification of recombinant proteins containing popular peptide fusion tags, including His Tag, GST Tag, S Tag and T7 Tag sequences.
Novagen offers a large variety of affinity supports and kits for the purification of recombinant proteins containing popular peptide fusion tags, including His Tag, GST Tag, S Tag and T7 Tag sequences.
Supporting Information
 Supporting Information Copper and zinc ions specifically promote non-amyloid aggregation of the highly stable human γ-d crystallin Liliana Quintanar, 1,* José A. Domínguez-Calva, 1 Eugene Serebryany, 2
Supporting Information Copper and zinc ions specifically promote non-amyloid aggregation of the highly stable human γ-d crystallin Liliana Quintanar, 1,* José A. Domínguez-Calva, 1 Eugene Serebryany, 2
XactEdit Cas9 Nuclease with NLS User Manual
 XactEdit Cas9 Nuclease with NLS User Manual An RNA-guided recombinant endonuclease for efficient targeted DNA cleavage Catalog Numbers CE1000-50K, CE1000-50, CE1000-250, CE1001-250, CE1001-1000 Table of
XactEdit Cas9 Nuclease with NLS User Manual An RNA-guided recombinant endonuclease for efficient targeted DNA cleavage Catalog Numbers CE1000-50K, CE1000-50, CE1000-250, CE1001-250, CE1001-1000 Table of
CAPZ PREPARATION BY BACTERIAL EXPRESSION (LYSIS BY DETERGENT, NOT SONICATION.) Soeno, Y. et al., J Mus. Res. Cell Motility 19:
 CAPZ PREPARATION BY BACTERIAL EXPRESSION (LYSIS BY DETERGENT, NOT SONICATION.) Soeno, Y. et al., J Mus. Res. Cell Motility 19:639-646 Notes before starting: *Bacterial strain BL21(DE) plys S transformed
CAPZ PREPARATION BY BACTERIAL EXPRESSION (LYSIS BY DETERGENT, NOT SONICATION.) Soeno, Y. et al., J Mus. Res. Cell Motility 19:639-646 Notes before starting: *Bacterial strain BL21(DE) plys S transformed
Viral RNAi suppressor reversibly binds sirna to. outcompete Dicer and RISC via multiple-turnover
 Supplementary Data Viral RNAi suppressor reversibly binds sirna to outcompete Dicer and RISC via multiple-turnover Renata A. Rawlings 1,2, Vishalakshi Krishnan 2 and Nils G. Walter 2 * 1 Biophysics and
Supplementary Data Viral RNAi suppressor reversibly binds sirna to outcompete Dicer and RISC via multiple-turnover Renata A. Rawlings 1,2, Vishalakshi Krishnan 2 and Nils G. Walter 2 * 1 Biophysics and
Jan 25, 05 His Bind Kit (Novagen)
 Jan 25, 05 His Bind Kit (Novagen) (1) Prepare 5ml of 1X Charge buffer (stock is 8X= 400mM NiSO4): 0.625ml of the stock + 4.375ml DH2O. (2) Prepare 13ml of 1X Binding buffer (stock is 8X = 40mM imidazole,
Jan 25, 05 His Bind Kit (Novagen) (1) Prepare 5ml of 1X Charge buffer (stock is 8X= 400mM NiSO4): 0.625ml of the stock + 4.375ml DH2O. (2) Prepare 13ml of 1X Binding buffer (stock is 8X = 40mM imidazole,
Supplemental Information. OprG Harnesses the Dynamics of its Extracellular. Loops to Transport Small Amino Acids across
 Structure, Volume 23 Supplemental Information OprG Harnesses the Dynamics of its Extracellular Loops to Transport Small Amino Acids across the Outer Membrane of Pseudomonas aeruginosa Iga Kucharska, Patrick
Structure, Volume 23 Supplemental Information OprG Harnesses the Dynamics of its Extracellular Loops to Transport Small Amino Acids across the Outer Membrane of Pseudomonas aeruginosa Iga Kucharska, Patrick
1. Bloomsbury BBSRC Centre for Structural Biology, Birkbeck College and University College London.
 Purification/Polishing of His-tagged proteins - Application of Centrifugal Vivapure Ion-exchange Membrane Devices to the Purification/Polishing of Histagged Background Multi-milligram quantities of highly
Purification/Polishing of His-tagged proteins - Application of Centrifugal Vivapure Ion-exchange Membrane Devices to the Purification/Polishing of Histagged Background Multi-milligram quantities of highly
Purification and crystallization of the bimagrumab Fab. The light- (residues 1 to 216, C216A
 SI Methods Purification and crystallization of the bimagrumab Fab. The light- (residues 1 to 216, C216A variant) and heavy-chain (residues 1 to 219, R212K variant) of the Bimagrumab Fab were cloned on
SI Methods Purification and crystallization of the bimagrumab Fab. The light- (residues 1 to 216, C216A variant) and heavy-chain (residues 1 to 219, R212K variant) of the Bimagrumab Fab were cloned on
Rapid GST Inclusion Body Solubilization and Renaturation Kit
 Product Manual Rapid GST Inclusion Body Solubilization and Renaturation Kit Catalog Number AKR-110 FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Bacteria are widely used for His
Product Manual Rapid GST Inclusion Body Solubilization and Renaturation Kit Catalog Number AKR-110 FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Bacteria are widely used for His
TECHNICAL BULLETIN. In Vitro Bacterial Split Fluorescent Protein Fold n Glow Solubility Assay Kits
 In Vitro Bacterial Split Fluorescent Protein Fold n Glow Solubility Assay Kits Catalog Numbers APPA001 In Vitro Bacterial Split GFP "Fold 'n' Glow" Solubility Assay Kit (Green) APPA008 In Vitro Bacterial
In Vitro Bacterial Split Fluorescent Protein Fold n Glow Solubility Assay Kits Catalog Numbers APPA001 In Vitro Bacterial Split GFP "Fold 'n' Glow" Solubility Assay Kit (Green) APPA008 In Vitro Bacterial
Supplemental Materials and Methods:
 Supplemental Materials and Methods: Cloning: Oligonucleotides used in the subcloning steps are listed in Supplemental Table 1. Human FANCI (isoform 1, KIAA1794) was subcloned from pcmv6-xl4 [FANCI] in
Supplemental Materials and Methods: Cloning: Oligonucleotides used in the subcloning steps are listed in Supplemental Table 1. Human FANCI (isoform 1, KIAA1794) was subcloned from pcmv6-xl4 [FANCI] in
Lecture 8: Affinity Chromatography-III
 Lecture 8: Affinity Chromatography-III Key words: Chromatography; Affinity chromatography; Protein Purification During this lecture, we shall be studying few more examples of affinity chromatography. The
Lecture 8: Affinity Chromatography-III Key words: Chromatography; Affinity chromatography; Protein Purification During this lecture, we shall be studying few more examples of affinity chromatography. The
AFFINITY GST PURIFICATION
 DESCRIPTION Glutathione Agarose Resin is used to purify recombinant derivatives of glutathione S-transferases or glutathione binding proteins. are products that allow batch or column purifications. Purification
DESCRIPTION Glutathione Agarose Resin is used to purify recombinant derivatives of glutathione S-transferases or glutathione binding proteins. are products that allow batch or column purifications. Purification
PRODUCT INFORMATION. Composition of SOC medium supplied :
 Product Name : Competent Cell BL21(DE3)pLysS Code No. : DS260 Size : 100 μl 10 Competency : > 5 10 7 cfu/μg (puc19) Supplied product : SOC medium, 1 ml 10 This product is for research use only Description
Product Name : Competent Cell BL21(DE3)pLysS Code No. : DS260 Size : 100 μl 10 Competency : > 5 10 7 cfu/μg (puc19) Supplied product : SOC medium, 1 ml 10 This product is for research use only Description
INSECT CELL/BACULOVIRUS PRODUCTION
 INSECT CELL/BACULOVIRUS PRODUCTION PEF # GENE NAME TRANSFER VECTOR BEVS MOLECULAR WEIGHT 2015-XXXX XXXX pbac1 flashbacultra TM 36.0 kda EXPRESSION METHOD OVERVIEW: Insect cells Spodoptera frugiperda (Sf9)
INSECT CELL/BACULOVIRUS PRODUCTION PEF # GENE NAME TRANSFER VECTOR BEVS MOLECULAR WEIGHT 2015-XXXX XXXX pbac1 flashbacultra TM 36.0 kda EXPRESSION METHOD OVERVIEW: Insect cells Spodoptera frugiperda (Sf9)
Masayoshi Honda, Jeehae Park, Robert A. Pugh, Taekjip Ha, and Maria Spies
 Molecular Cell, Volume 35 Supplemental Data Single-Molecule Analysis Reveals Differential Effect of ssdna-binding Proteins on DNA Translocation by XPD Helicase Masayoshi Honda, Jeehae Park, Robert A. Pugh,
Molecular Cell, Volume 35 Supplemental Data Single-Molecule Analysis Reveals Differential Effect of ssdna-binding Proteins on DNA Translocation by XPD Helicase Masayoshi Honda, Jeehae Park, Robert A. Pugh,
The preparation of native chromatin from cultured human cells.
 Native chromatin immunoprecipitation protocol The preparation of native chromatin from cultured human cells. All solutions need to be ice cold. Sucrose containing solutions must be made up fresh on the
Native chromatin immunoprecipitation protocol The preparation of native chromatin from cultured human cells. All solutions need to be ice cold. Sucrose containing solutions must be made up fresh on the
Protein Purification. igem TU/e 2016 Biomedical Engineering
 igem TU/e 2016 Biomedical Engineering Eindhoven University of Technology Room: Ceres 0.04 Den Dolech 2, 5612 AZ Eindhoven The Netherlands Tel. no. +1 50 247 55 59 2016.igem.org/Team:TU-Eindhoven Protein
igem TU/e 2016 Biomedical Engineering Eindhoven University of Technology Room: Ceres 0.04 Den Dolech 2, 5612 AZ Eindhoven The Netherlands Tel. no. +1 50 247 55 59 2016.igem.org/Team:TU-Eindhoven Protein
Nature Structural & Molecular Biology: doi: /nsmb.2548
 Supplementary Figure 1. Structure of GltPhout. (a) Stereo view of a slice through a single GltPhout protomer shown in stick representation along with 2Fo-Fc and anomalous difference electron maps. The
Supplementary Figure 1. Structure of GltPhout. (a) Stereo view of a slice through a single GltPhout protomer shown in stick representation along with 2Fo-Fc and anomalous difference electron maps. The
DNA Hybridization and Detection
 Chapter 6 DNA Hybridization and Detection Fluorescence Polarization Detection of DNA Hybridization........................................................ 6-2 Introduction.............................................................................................................
Chapter 6 DNA Hybridization and Detection Fluorescence Polarization Detection of DNA Hybridization........................................................ 6-2 Introduction.............................................................................................................
Crystallization and Preliminary X-Ray Analysis of the dsdna Bacteriophage HK97 Mature Empty Capsid
 VIROLOGY 243, 113 118 (1998) ARTICLE NO. VY989040 Crystallization and Preliminary X-Ray Analysis of the dsdna Bacteriophage HK97 Mature Empty Capsid William R. Wikoff,*, Robert L. Duda, Roger W. Hendrix,
VIROLOGY 243, 113 118 (1998) ARTICLE NO. VY989040 Crystallization and Preliminary X-Ray Analysis of the dsdna Bacteriophage HK97 Mature Empty Capsid William R. Wikoff,*, Robert L. Duda, Roger W. Hendrix,
AnaTag HiLyte Fluor 647 Protein Labeling Kit
 AnaTag HiLyte Fluor 647 Protein Labeling Kit Catalog # 72049 Kit Size 3 Conjugation Reactions This kit is optimized to conjugate HiLyte Fluor 647 SE to proteins (e.g., IgG). It provides ample materials
AnaTag HiLyte Fluor 647 Protein Labeling Kit Catalog # 72049 Kit Size 3 Conjugation Reactions This kit is optimized to conjugate HiLyte Fluor 647 SE to proteins (e.g., IgG). It provides ample materials
Diversity in DNA recognition by p53 revealed by crystal structures with Hoogsteen base pairs
 SUPPLEMENTARY INFORMATION Diversity in DNA recognition by p53 revealed by crystal structures with Hoogsteen base pairs Malka Kitayner 1, 3, Haim Rozenberg 1, 3, Remo Rohs 2, 3, Oded Suad 1, Dov Rabinovich
SUPPLEMENTARY INFORMATION Diversity in DNA recognition by p53 revealed by crystal structures with Hoogsteen base pairs Malka Kitayner 1, 3, Haim Rozenberg 1, 3, Remo Rohs 2, 3, Oded Suad 1, Dov Rabinovich
Fisher (Fairlawn, NJ) and Sigma-Aldrich (St. Louis, MO) and were used without further. (Promega) and DpnI (New England Biolabs, Beverly, MA).
 175 Appendix III Chapter 4 Methods General. Unless otherwise noted, reagents were purchased from the commercial suppliers Fisher (Fairlawn, NJ) and Sigma-Aldrich (St. Louis, MO) and were used without further
175 Appendix III Chapter 4 Methods General. Unless otherwise noted, reagents were purchased from the commercial suppliers Fisher (Fairlawn, NJ) and Sigma-Aldrich (St. Louis, MO) and were used without further
Structural Bioinformatics (C3210) DNA and RNA Structure
 Structural Bioinformatics (C3210) DNA and RNA Structure Importance of DNA/RNA 3D Structure Nucleic acids are essential materials found in all living organisms. Their main function is to maintain and transmit
Structural Bioinformatics (C3210) DNA and RNA Structure Importance of DNA/RNA 3D Structure Nucleic acids are essential materials found in all living organisms. Their main function is to maintain and transmit
Genetics and Genomics in Medicine Chapter 3. Questions & Answers
 Genetics and Genomics in Medicine Chapter 3 Multiple Choice Questions Questions & Answers Question 3.1 Which of the following statements, if any, is false? a) Amplifying DNA means making many identical
Genetics and Genomics in Medicine Chapter 3 Multiple Choice Questions Questions & Answers Question 3.1 Which of the following statements, if any, is false? a) Amplifying DNA means making many identical
Supporting Information
 Supporting Information Wiley-VCH 2006 69451 Weinheim, Germany RNA ligands that distinguish metabolite-induced conformations in the TPP riboswitch Günter Mayer, Marie-Sophie L. Raddatz, Julia D. Grunwald,
Supporting Information Wiley-VCH 2006 69451 Weinheim, Germany RNA ligands that distinguish metabolite-induced conformations in the TPP riboswitch Günter Mayer, Marie-Sophie L. Raddatz, Julia D. Grunwald,
2.5. Equipment and materials supplied by user PCR based template preparation Influence of temperature on in vitro EGFP synthesis 11
 Manual 15 Reactions LEXSY in vitro Translation Cell-free protein expression kit based on Leishmania tarentolae for PCR-based template generation Cat. No. EGE-2010-15 FOR RESEARCH USE ONLY. NOT INTENDED
Manual 15 Reactions LEXSY in vitro Translation Cell-free protein expression kit based on Leishmania tarentolae for PCR-based template generation Cat. No. EGE-2010-15 FOR RESEARCH USE ONLY. NOT INTENDED
AFFINITY HIS-TAG PURIFICATION
 DESCRIPTION Resins are products that allow batch or column purifications. This product is supplied as a suspension in 20% ethanol. INSTRUCTIONS The resins are adapted to work mainly in native conditions
DESCRIPTION Resins are products that allow batch or column purifications. This product is supplied as a suspension in 20% ethanol. INSTRUCTIONS The resins are adapted to work mainly in native conditions
Nanobody Library Selection by MACS
 Nanobody Library Selection by MACS Introduction We have written the protocol below with the goal of making steps of in vitro nanobody selection as clear and broadly applicable as possible, but it is important
Nanobody Library Selection by MACS Introduction We have written the protocol below with the goal of making steps of in vitro nanobody selection as clear and broadly applicable as possible, but it is important
Rapid GST Inclusion Body Solubilization and Renaturation Kit
 Product Manual Rapid GST Inclusion Body Solubilization and Renaturation Kit Catalog Number AKR-110 FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Bacteria are widely used for His
Product Manual Rapid GST Inclusion Body Solubilization and Renaturation Kit Catalog Number AKR-110 FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Bacteria are widely used for His
PURIFICATION AND PARTIAL CHARACTERIZATION OF L-asparaginase FROM MUTATED MNTG-7. Several techniques have been described for recovery and purification
 CHAPTER- V1 PURIFICATION AND PARTIAL CHARACTERIZATION OF L-asparaginase FROM MUTATED MNTG-7 Several techniques have been described for recovery and purification of L-asparaginase from different sources
CHAPTER- V1 PURIFICATION AND PARTIAL CHARACTERIZATION OF L-asparaginase FROM MUTATED MNTG-7 Several techniques have been described for recovery and purification of L-asparaginase from different sources
Electrophoresis. Ready-to-Run Buffers and Solutions
 Electrophoresis Ready-to-Run Buffers and Solutions Bio-Rad is a premier provider of buffers and premixed reagents for life science research. We offer a variety of different products for all your protein
Electrophoresis Ready-to-Run Buffers and Solutions Bio-Rad is a premier provider of buffers and premixed reagents for life science research. We offer a variety of different products for all your protein
Purification of (recombinant) proteins. Pekka Lappalainen, Institute of Biotechnology, University of Helsinki
 Purification of (recombinant) proteins Pekka Lappalainen, Institute of Biotechnology, University of Helsinki Physical properties of proteins that can be applied for purification -size -charge (isoelectric
Purification of (recombinant) proteins Pekka Lappalainen, Institute of Biotechnology, University of Helsinki Physical properties of proteins that can be applied for purification -size -charge (isoelectric
Supplemental Materials and Methods
 Supplemental Materials and Methods Proteins and reagents Proteins were purified as described previously: RecA, RecQ, and SSB proteins (Harmon and Kowalczykowski 1998); RecF protein (Morimatsu and Kowalczykowski
Supplemental Materials and Methods Proteins and reagents Proteins were purified as described previously: RecA, RecQ, and SSB proteins (Harmon and Kowalczykowski 1998); RecF protein (Morimatsu and Kowalczykowski
O-GlcNAcase Activity Assay
 O-GlcNAcase Activity Assay Prepared by Jen Groves and Junfeng Ma, The Johns Hopkins Unviersity School of Medicine Based on: Macauley MS et al., 2005. O-GlcNAcase uses substrate-assisted catalysis: kinetic
O-GlcNAcase Activity Assay Prepared by Jen Groves and Junfeng Ma, The Johns Hopkins Unviersity School of Medicine Based on: Macauley MS et al., 2005. O-GlcNAcase uses substrate-assisted catalysis: kinetic
Cloning small RNAs for Solexa Sequencing version 2.0 by Nelson Lau Page 1 of 5 (Modified from Solexa sequencing protocol from Bartel lab)
 Cloning small RNAs for Solexa Sequencing version 2.0 by Nelson Lau 09162008 Page 1 of 5 General Cloning Protocol: Gel-purification 1. Pour 1mm thick, urea denaturing 10% or 15% polyacrylamide gels, with
Cloning small RNAs for Solexa Sequencing version 2.0 by Nelson Lau 09162008 Page 1 of 5 General Cloning Protocol: Gel-purification 1. Pour 1mm thick, urea denaturing 10% or 15% polyacrylamide gels, with
Purification: Step 1. Lecture 11 Protein and Peptide Chemistry. Cells: Break them open! Crude Extract
 Purification: Step 1 Lecture 11 Protein and Peptide Chemistry Cells: Break them open! Crude Extract Total contents of cell Margaret A. Daugherty Fall 2003 Big Problem: Crude extract is not the natural
Purification: Step 1 Lecture 11 Protein and Peptide Chemistry Cells: Break them open! Crude Extract Total contents of cell Margaret A. Daugherty Fall 2003 Big Problem: Crude extract is not the natural
Purification: Step 1. Protein and Peptide Chemistry. Lecture 11. Big Problem: Crude extract is not the natural environment. Cells: Break them open!
 Lecture 11 Protein and Peptide Chemistry Margaret A. Daugherty Fall 2003 Purification: Step 1 Cells: Break them open! Crude Extract Total contents of cell Big Problem: Crude extract is not the natural
Lecture 11 Protein and Peptide Chemistry Margaret A. Daugherty Fall 2003 Purification: Step 1 Cells: Break them open! Crude Extract Total contents of cell Big Problem: Crude extract is not the natural
E.Z.N.A. Blood DNA Maxi Kit. D preps D preps
 E.Z.N.A. Blood DNA Maxi Kit D2492-00 2 preps D2492-03 50 preps April 2014 E.Z.N.A. Blood DNA Maxi Kit Table of Contents Introduction and Overview...2 Kit Contents/Storage and Stability...3 Preparing Reagents...4
E.Z.N.A. Blood DNA Maxi Kit D2492-00 2 preps D2492-03 50 preps April 2014 E.Z.N.A. Blood DNA Maxi Kit Table of Contents Introduction and Overview...2 Kit Contents/Storage and Stability...3 Preparing Reagents...4
HELIX TM HT-96 MD1-69
 HELIX TM HT-96 MD1-69 Crystallize a diverse range of nucleic acid topologies and molecular weights. This screen targets all types of DNA/RNA, triplexes, quadruplexes, pseudoknots, i-motifs, and large molecular
HELIX TM HT-96 MD1-69 Crystallize a diverse range of nucleic acid topologies and molecular weights. This screen targets all types of DNA/RNA, triplexes, quadruplexes, pseudoknots, i-motifs, and large molecular
E.Z.N.A. MicroElute Genomic DNA Kit. D preps D preps D preps
 E.Z.N.A. MicroElute Genomic DNA Kit D3096-00 5 preps D3096-01 50 preps D3096-02 200 preps December 2013 E.Z.N.A. MicroElute Genomic DNA Kit Table of Contents Introduction...2 Kit Contents/Storage and Stability...3
E.Z.N.A. MicroElute Genomic DNA Kit D3096-00 5 preps D3096-01 50 preps D3096-02 200 preps December 2013 E.Z.N.A. MicroElute Genomic DNA Kit Table of Contents Introduction...2 Kit Contents/Storage and Stability...3
Strep-Tactin XT Spin Column
 Strep-Tactin XT Spin Column Purification Protocol Last date of revision Last June date 2017 of revision June 2017 Version PR90-0001 Version PR90-0001 For research use only Important licensing information
Strep-Tactin XT Spin Column Purification Protocol Last date of revision Last June date 2017 of revision June 2017 Version PR90-0001 Version PR90-0001 For research use only Important licensing information
Table S1. Crystallographic Data and Refinement Statistics, Related to Experimental Procedures hsddb1-drddb2 CPD
 Data collection Table S1. Crystallographic Data and Refinement Statistics, Related to Experimental Procedures hsddb1-drddb2 CPD #1 hsddb1-drddb2 CPD #2 Values in parentheses are for the highest resolution
Data collection Table S1. Crystallographic Data and Refinement Statistics, Related to Experimental Procedures hsddb1-drddb2 CPD #1 hsddb1-drddb2 CPD #2 Values in parentheses are for the highest resolution
Protein Protein Interactions Methods and Applications
 METHODS IN MOLECULAR BIOLOGY TM TM Volume 261 Protein Protein Interactions Methods and Applications Edited by Haian Fu Fluorescence Gel Retardation Assay 155 10 Fluorescence Gel Retardation Assay to Detect
METHODS IN MOLECULAR BIOLOGY TM TM Volume 261 Protein Protein Interactions Methods and Applications Edited by Haian Fu Fluorescence Gel Retardation Assay 155 10 Fluorescence Gel Retardation Assay to Detect
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Conserved arginines on the rim of Hfq catalyze base pair formation and exchange Subrata Panja and Sarah A. Woodson T.C. Jenkins Department of Biophysics, Johns Hopkins University,
SUPPLEMENTARY INFORMATION Conserved arginines on the rim of Hfq catalyze base pair formation and exchange Subrata Panja and Sarah A. Woodson T.C. Jenkins Department of Biophysics, Johns Hopkins University,
Lecture Four. Molecular Approaches I: Nucleic Acids
 Lecture Four. Molecular Approaches I: Nucleic Acids I. Recombinant DNA and Gene Cloning Recombinant DNA is DNA that has been created artificially. DNA from two or more sources is incorporated into a single
Lecture Four. Molecular Approaches I: Nucleic Acids I. Recombinant DNA and Gene Cloning Recombinant DNA is DNA that has been created artificially. DNA from two or more sources is incorporated into a single
Protein Expression PURIFICATION & ANALYSIS. be INSPIRED drive DISCOVERY stay GENUINE
 Expression PURIFICATION & ANALYSIS be INSPIRED drive DISCOVERY stay GENUINE OVERVIEW Expression & Purification expression can be a very complex, multi-factorial process. Each protein requires a specific
Expression PURIFICATION & ANALYSIS be INSPIRED drive DISCOVERY stay GENUINE OVERVIEW Expression & Purification expression can be a very complex, multi-factorial process. Each protein requires a specific
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION A human XRCC4-XLF complex bridges DNA ends. Sara N. Andres 1, Alexandra Vergnes 2, Dejan Ristic 3, Claire Wyman 3, Mauro Modesti 2,4, and Murray Junop 2,4 1 Department of Biochemistry
SUPPLEMENTARY INFORMATION A human XRCC4-XLF complex bridges DNA ends. Sara N. Andres 1, Alexandra Vergnes 2, Dejan Ristic 3, Claire Wyman 3, Mauro Modesti 2,4, and Murray Junop 2,4 1 Department of Biochemistry
AVB Sepharose High Performance
 GE Healthcare Life Sciences Data file 28-927-54 AB Custom designed media AVB Sepharose High Performance AVB Sepharose High Performance is an affinity chromatography medium (resin) designed for the purification
GE Healthcare Life Sciences Data file 28-927-54 AB Custom designed media AVB Sepharose High Performance AVB Sepharose High Performance is an affinity chromatography medium (resin) designed for the purification
Introduction to Protein Purification
 Introduction to Protein Purification 1 Day 1) Introduction to Protein Purification. Input for Purification Protocol Development - Guidelines for Protein Purification Day 2) Sample Preparation before Chromatography
Introduction to Protein Purification 1 Day 1) Introduction to Protein Purification. Input for Purification Protocol Development - Guidelines for Protein Purification Day 2) Sample Preparation before Chromatography
Strep-Tactin Spin Column Purification Protocol
 Strep-Tactin Spin Column Purification Protocol Last date of revision February 2008 Version PR10-0005 IBA Headquarters IBA GmbH Rudolf-Wissell-Str. 28 D-37079 Göttingen Germany Tel: +49 (0) 551-50672-0
Strep-Tactin Spin Column Purification Protocol Last date of revision February 2008 Version PR10-0005 IBA Headquarters IBA GmbH Rudolf-Wissell-Str. 28 D-37079 Göttingen Germany Tel: +49 (0) 551-50672-0
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1133488/dc1 Supporting Online Material for An Inward-Facing Conformation of a Putative Metal-Chelate Type ABC Transporter H. W. Pinkett, A. T. Lee, P. Lum, K. P. Locher,
www.sciencemag.org/cgi/content/full/1133488/dc1 Supporting Online Material for An Inward-Facing Conformation of a Putative Metal-Chelate Type ABC Transporter H. W. Pinkett, A. T. Lee, P. Lum, K. P. Locher,
Ni-IDA/ Ni-NTA Agarose
 GENAON Ni-IDA/ Ni-NTA Agarose Purification of His-tagged Proteins Product Cat# Package size Ni-IDA Agarose 20mL 50% suspension S5353.0010 10mL Ni-IDA Agarose 100mL 50% suspension S5353.0050 50mL Ni-IDA
GENAON Ni-IDA/ Ni-NTA Agarose Purification of His-tagged Proteins Product Cat# Package size Ni-IDA Agarose 20mL 50% suspension S5353.0010 10mL Ni-IDA Agarose 100mL 50% suspension S5353.0050 50mL Ni-IDA
Amintra Affinity Resins
 Amintra Affinity Resins Ni-NTA Metal Chelate Affinity Resin Technical Data and Instruction Manual info@expedeon.com 2 TABLE OF CONTENTS TABLE OF CONTENTS 3 Introduction 4 Storage 4 Chemical compatibility
Amintra Affinity Resins Ni-NTA Metal Chelate Affinity Resin Technical Data and Instruction Manual info@expedeon.com 2 TABLE OF CONTENTS TABLE OF CONTENTS 3 Introduction 4 Storage 4 Chemical compatibility
Short Technical Reports
 Large-Scale Purification of a Stable Form of Recombinant Tobacco Etch Virus Protease BioTechniques 30:544-554 (March 2001) ABSTRACT Tobacco etch virus NIa proteinase (NIa- Pro) has become the enzyme of
Large-Scale Purification of a Stable Form of Recombinant Tobacco Etch Virus Protease BioTechniques 30:544-554 (March 2001) ABSTRACT Tobacco etch virus NIa proteinase (NIa- Pro) has become the enzyme of
Supplementary Figure 1. Pressure dependence of the self-cleavage reaction of the modified hairpin ribozyme. Time evolution of the modified hairpin
 Supplementary Figure 1. Pressure dependence of the self-cleavage reaction of the modified hairpin ribozyme. Time evolution of the modified hairpin ribozyme s fraction cleaved for the reaction in the presence
Supplementary Figure 1. Pressure dependence of the self-cleavage reaction of the modified hairpin ribozyme. Time evolution of the modified hairpin ribozyme s fraction cleaved for the reaction in the presence
Announcements. Next week s discussion will have a quiz on Chapter 3fg and Chapter 11ab Computer Lab (Chapter 11ab): 10/17 10/22
 Announcements Next week s discussion will have a quiz on Chapter 3fg and Chapter 11ab Computer Lab (Chapter 11ab): 10/17 10/22 SCI 162 will be open for 2 hours of each lab section to finish Chapter 3 Chapters
Announcements Next week s discussion will have a quiz on Chapter 3fg and Chapter 11ab Computer Lab (Chapter 11ab): 10/17 10/22 SCI 162 will be open for 2 hours of each lab section to finish Chapter 3 Chapters
Fisher BioReagents Buffers for Life Science Research
 Fisher BioReagents Buffers for Life Science Research The Water Makes a Difference. CellPURE Biological Buffers are of the highest quality (purity 99%). They are tested for low heavy metal content and the
Fisher BioReagents Buffers for Life Science Research The Water Makes a Difference. CellPURE Biological Buffers are of the highest quality (purity 99%). They are tested for low heavy metal content and the
In Vitro Monitoring of the Formation of Pentamers from the Monomer of GST Fused HPV 16 L1
 This journal is The Royal Society of Chemistry 213 In Vitro Monitoring of the Formation of Pentamers from the Monomer of GST Fused HPV 16 L1 Dong-Dong Zheng, a Dong Pan, a Xiao Zha, ac Yuqing Wu,* a Chunlai
This journal is The Royal Society of Chemistry 213 In Vitro Monitoring of the Formation of Pentamers from the Monomer of GST Fused HPV 16 L1 Dong-Dong Zheng, a Dong Pan, a Xiao Zha, ac Yuqing Wu,* a Chunlai
MOK. Media Optimization Kit
 MOK Media Optimization Kit The Media Optimization Kit determines the best medium formulation for maximizing accumulation of recombinant proteins expressed in E. coli, utilizing a series of Athena s superior
MOK Media Optimization Kit The Media Optimization Kit determines the best medium formulation for maximizing accumulation of recombinant proteins expressed in E. coli, utilizing a series of Athena s superior
Fisher BioReagents Buffers for Life Science Research
 Fisher BioReagents Buffers for Life Science Research The Water Makes a Difference. CellPURE Biological Buffers are of the highest quality (purity 99%). They are tested for low heavy metal content and the
Fisher BioReagents Buffers for Life Science Research The Water Makes a Difference. CellPURE Biological Buffers are of the highest quality (purity 99%). They are tested for low heavy metal content and the
Mechanism of Transcription Termination by RNA Polymerase III Utilizes a Non-template Strand Sequence-Specific Signal Element
 Molecular Cell Supplemental Information Mechanism of ranscription ermination by RNA Polymerase III Utilizes a Non-template Strand Sequence-Specific Signal Element Aneeshkumar G. Arimbasseri and Richard
Molecular Cell Supplemental Information Mechanism of ranscription ermination by RNA Polymerase III Utilizes a Non-template Strand Sequence-Specific Signal Element Aneeshkumar G. Arimbasseri and Richard
X-ray structures of fructosyl peptide oxidases revealing residues responsible for gating oxygen access in the oxidative half reaction
 X-ray structures of fructosyl peptide oxidases revealing residues responsible for gating oxygen access in the oxidative half reaction Tomohisa Shimasaki 1, Hiromi Yoshida 2, Shigehiro Kamitori 2 & Koji
X-ray structures of fructosyl peptide oxidases revealing residues responsible for gating oxygen access in the oxidative half reaction Tomohisa Shimasaki 1, Hiromi Yoshida 2, Shigehiro Kamitori 2 & Koji
1. Cross-linking and cell harvesting
 ChIP is a powerful tool that allows the specific matching of proteins or histone modifications to regions of the genome. Chromatin is isolated and antibodies to the antigen of interest are used to determine
ChIP is a powerful tool that allows the specific matching of proteins or histone modifications to regions of the genome. Chromatin is isolated and antibodies to the antigen of interest are used to determine
Multiple choice questions (numbers in brackets indicate the number of correct answers)
 1 Multiple choice questions (numbers in brackets indicate the number of correct answers) February 1, 2013 1. Ribose is found in Nucleic acids Proteins Lipids RNA DNA (2) 2. Most RNA in cells is transfer
1 Multiple choice questions (numbers in brackets indicate the number of correct answers) February 1, 2013 1. Ribose is found in Nucleic acids Proteins Lipids RNA DNA (2) 2. Most RNA in cells is transfer
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1153018/dc1 Supporting Online Material for A Cholesterol Biosynthesis Inhibitor Blocks Staphylococcus aureus Virulence Chia-I Liu, George Y. Liu, Yongcheng Song, Fenglin
www.sciencemag.org/cgi/content/full/1153018/dc1 Supporting Online Material for A Cholesterol Biosynthesis Inhibitor Blocks Staphylococcus aureus Virulence Chia-I Liu, George Y. Liu, Yongcheng Song, Fenglin
Optimizing Soluble Expression of Organophosphorus Acid Anhyrdolase (OPAA) Zingeber Udani BIOL 493
 Optimizing Soluble Expression of Organophosphorus Acid Anhyrdolase (OPAA) Zingeber Udani BIOL 493 Fall 2007 1 Abstract Organophosphorus acid anhydrolase (OPAA) is a proteolytic enzyme expressed by many
Optimizing Soluble Expression of Organophosphorus Acid Anhyrdolase (OPAA) Zingeber Udani BIOL 493 Fall 2007 1 Abstract Organophosphorus acid anhydrolase (OPAA) is a proteolytic enzyme expressed by many
Case 7 A Storage Protein From Seeds of Brassica nigra is a Serine Protease Inhibitor Last modified 29 September 2005
 Case 7 A Storage Protein From Seeds of Brassica nigra is a Serine Protease Inhibitor Last modified 9 September 005 Focus concept Purification of a novel seed storage protein allows sequence analysis and
Case 7 A Storage Protein From Seeds of Brassica nigra is a Serine Protease Inhibitor Last modified 9 September 005 Focus concept Purification of a novel seed storage protein allows sequence analysis and
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than. SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with
 Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
electrophoresis tech Methods Materials
 electrophoresis tech note 3176 Monitoring the Expression, Purification, and Processing of -Tagged Proteins Using the Experion Automated Electrophoresis System Xuemei He and William Strong, Bio-Rad Laboratories,
electrophoresis tech note 3176 Monitoring the Expression, Purification, and Processing of -Tagged Proteins Using the Experion Automated Electrophoresis System Xuemei He and William Strong, Bio-Rad Laboratories,
Nature Structural and Molecular Biology: doi: /nsmb.2937
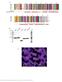 Supplementary Figure 1 Multiple sequence alignment of the CtIP N-terminal domain, purified CtIP protein constructs and details of the 2F o F c electron density map of CtIP-NTD. (a) Multiple sequence alignment,
Supplementary Figure 1 Multiple sequence alignment of the CtIP N-terminal domain, purified CtIP protein constructs and details of the 2F o F c electron density map of CtIP-NTD. (a) Multiple sequence alignment,
Protein Expression, Electrophoresis, His tag Purification
 Protein Expression, Electrophoresis, His tag Purification 2013 edition H.E. Schellhorn Day 2 Lecture 2- Protein Expression Protein over-expression and purification are key techniques in Molecular Biology
Protein Expression, Electrophoresis, His tag Purification 2013 edition H.E. Schellhorn Day 2 Lecture 2- Protein Expression Protein over-expression and purification are key techniques in Molecular Biology
Protocol for induction of expression and cell lysate production
 Protocol for induction of expression and cell lysate production AV-04 Doxycyclin induction and cell lysate 1.0 Introduction / Description This method is intended for the treatment of the previously transfected
Protocol for induction of expression and cell lysate production AV-04 Doxycyclin induction and cell lysate 1.0 Introduction / Description This method is intended for the treatment of the previously transfected
QS S Assist KINASE_MSA Kit
 QS S Assist KINASE_MSA Kit Description KINASE MSA Kit is designed for use in pharmacological assays for KINASE based on Off-chip mobility shift assay (MSA). This kit includes Assay Buffer, Termination
QS S Assist KINASE_MSA Kit Description KINASE MSA Kit is designed for use in pharmacological assays for KINASE based on Off-chip mobility shift assay (MSA). This kit includes Assay Buffer, Termination
Kinetics Review. Tonight at 7 PM Phys 204 We will do two problems on the board (additional ones than in the problem sets)
 Quiz 1 Kinetics Review Tonight at 7 PM Phys 204 We will do two problems on the board (additional ones than in the problem sets) I will post the problems with solutions on Toolkit for those that can t make
Quiz 1 Kinetics Review Tonight at 7 PM Phys 204 We will do two problems on the board (additional ones than in the problem sets) I will post the problems with solutions on Toolkit for those that can t make
E.Z.N.A. Blood DNA Midi Kit. D preps D preps
 E.Z.N.A. Blood DNA Midi Kit D3494-00 2 preps D3494-04 100 preps August 2013 E.Z.N.A. Blood DNA Midi Kit Table of Contents Introduction and Overview...2 Kit Contents/Storage and Stability...3 Preparing
E.Z.N.A. Blood DNA Midi Kit D3494-00 2 preps D3494-04 100 preps August 2013 E.Z.N.A. Blood DNA Midi Kit Table of Contents Introduction and Overview...2 Kit Contents/Storage and Stability...3 Preparing
1 ml gel corresponds to ml of 75% (v/v) Glutathione Agarose suspension.
 1 AFFINITY GST PURIFICATION Procedure for Use Glutathione Agarose 4 Resin DESCRIPTION Glutathione Agarose Resin is used to purify recombinant derivatives of glutathione S-transferases or glutathione binding
1 AFFINITY GST PURIFICATION Procedure for Use Glutathione Agarose 4 Resin DESCRIPTION Glutathione Agarose Resin is used to purify recombinant derivatives of glutathione S-transferases or glutathione binding
Bio-Scale Mini Profinity IMAC Cartridges, 1 and 5 ml. Instruction Manual. Catalog #
 Bio-Scale Mini Profinity IMAC Cartridges, 1 and 5 ml Instruction Manual Catalog # 732-4610 732-4612 732-4614 Table of Contents Section 1...Introduction...1 Section 2 Product Information...2 Section 3 Connection
Bio-Scale Mini Profinity IMAC Cartridges, 1 and 5 ml Instruction Manual Catalog # 732-4610 732-4612 732-4614 Table of Contents Section 1...Introduction...1 Section 2 Product Information...2 Section 3 Connection
Protein Expression Research Group (PERG) 2012 Study
 Protein Expression Research Group (PERG) 2012 Study Richard J Heath 1, Fei P Gao 2, Pamela Scott Adams 3, Cynthia Kinsland 4, James Bryson 5, Bo Xu 6, Thomas Neubert 7. 1 St Jude Children's Research Hospital,
Protein Expression Research Group (PERG) 2012 Study Richard J Heath 1, Fei P Gao 2, Pamela Scott Adams 3, Cynthia Kinsland 4, James Bryson 5, Bo Xu 6, Thomas Neubert 7. 1 St Jude Children's Research Hospital,
Wearing gloves is highly recommended when handling the kit contents.
 Warning: Wearing gloves is highly recommended when handling the kit contents. GeBAflex-tube is covered by the WO0190731 patent application assigned to Gene Bio-Application Ltd. GeBAflex-tubes are autoclaved
Warning: Wearing gloves is highly recommended when handling the kit contents. GeBAflex-tube is covered by the WO0190731 patent application assigned to Gene Bio-Application Ltd. GeBAflex-tubes are autoclaved
Structure formation and association of biomolecules. Prof. Dr. Martin Zacharias Lehrstuhl für Molekulardynamik (T38) Technische Universität München
 Structure formation and association of biomolecules Prof. Dr. Martin Zacharias Lehrstuhl für Molekulardynamik (T38) Technische Universität München Motivation Many biomolecules are chemically synthesized
Structure formation and association of biomolecules Prof. Dr. Martin Zacharias Lehrstuhl für Molekulardynamik (T38) Technische Universität München Motivation Many biomolecules are chemically synthesized
