NIH Public Access Author Manuscript Proc IEEE Int Symp Biomed Imaging. Author manuscript; available in PMC 2011 July 29.
|
|
|
- Olivia Rose
- 5 years ago
- Views:
Transcription
1 NIH Public Access Author Manuscript Published in final edited form as: Proc IEEE Int Symp Biomed Imaging June 9; 2011(March April ): doi: /ISBI AUTOMATED ESTIMATION OF MICROTUBULE MODEL PARAMETERS FROM 3-D LIVE CELL MICROSCOPY IMAGES Aabid Shariff 1, Robert F. Murphy 1,2,3,5, and Gustavo K. Rohde 1,2,4 1 Lane Center for Computational Biology and Center for Bioimage Informatics, Carnegie Mellon University, Pittsburgh, PA 2 Department of Biomedical Engineering, Carnegie Mellon University, Pittsburgh, PA 3 Department of Biological Sciences and Machine Learning, Carnegie Mellon University, Pittsburgh, PA 4 Electrical & Computer Engineering Department, Carnegie Mellon University, Pittsburgh, PA 5 External Senior Fellow, Freiburg Institute for Advanced Studies, Freiburg, Germany Abstract While basic principles of microtubule organization are well understood, much remains to be learned about the extent and significance of variation in that organization among cell types and conditions. Large numbers of images of microtubule distributions for many cell types can be readily obtained by high throughput fluorescence microscopy but direct estimation of the parameters underlying the organization is problematic because it is difficult to resolve individual microtubules present at the microtubule-organizing center or at regions of high crossover. Previously, we developed an indirect, generative model-based approach that can estimate such spatial distribution parameters as the number and mean length of microtubules. In order to validate this approach, we have applied it to 3D images of NIH 3T3 cells expressing fluorescently-tagged tubulin in the presence and absence of the microtubule depolymerizing drug nocodazole. We describe here the first application of our inverse modeling approach to live cell images and demonstrate that it yields estimates consistent with expectations. Index Terms Microtubules; parameter estimation; nocodazole; NIH 3T3 cells; generative models 1. INTRODUCTION Microtubules play a critical role in many cellular processes and are a target of drugs used to treat cancer. The spatial distributions of microtubules are such that the density is very high close the centrosomal region and often very low at the lamellipodial region of the cell. High throughput image acquisition methods such as fluorescence microscopy can acquire images of an intact microtubule network, but their current resolution is not high enough to trace all individual microtubules in intact cells. Currently, methods and validation sets have been generated on portions of microtubules that are clearly distinguishable, accounting for only a small fraction of the total microtubules in an intact cell [1,2]. While these likely suffice for studying dynamics of microtubules that reach the lamellipodium, they do not allow construction of whole cell models.
2 Shariff et al. Page 2 We have previously described a generative model of microtubules and developed an indirect method of estimating its parameters [3]. Since whole cell images with known parameters were not available, we tested the ability of the method to accurately estimate model parameters using synthetic images generated using the model. These tests revealed a low error in estimation but estimates for real images could only be described as generally consistent with current knowledge. Here we describe estimation of microtubule model parameters from 3D fluorescence microscopy images of live cells under conditions in which changes in those parameters are expected. This was done by acquiring images of living NIH 3T3 cells expressing fluorescently-tagged tubulin in the presence and absence of nocodazole, a drug that is known to depolymerize microtubules [4]. 2. DATA ACQUISITION d NIH 3T3 dataset NIH 3T3 cells expressing EGFP-tagged alpha tubulin generated using CD-tagging [5] were cultured in DMEM supplemented with 10% Fetal Calf Serum and 100 U/ml penicillin and 100 ug/ml streptomycin. The cells were grown to 80% confluency. On the day of imaging, the media was changed to Opti-MEM and a final concentration of 0.5 ug/ml of Hoechst was added to label nuclei. The dish was incubated for at least 3 h in a CO2 incubator and then placed in a heated chamber that was maintained at 37 C throughout image acquisition. 3D images were acquired using a Zeiss LSM 510 confocal fluorescence microscope. The spacing between voxels was 0.09 microns in the focal plane and 0.48 microns along the axial dimension. 3D images of five different cells were acquired at 0, 10, 20, 30, 40 min after addition of nocodozale or buffer. Due to photobleaching, full 3D images could not be acquired for the same cell at each time point, and therefore different cells were imaged at each time point (only interphase cells were selected) Fluorescent bead acquisition 3. METHODS As our modeling approach requires a model of the point spread function of the microscope used for acquisition, we generated an empirical estimate of the function using 20 nm fluorescent beads (488 nm absorption). 0.1 ml of a suspension of beads in optimem was placed on a clean glass slide and quickly covered by a coverslip. 3D images were acquired as above Generative model of microtubules The generative model of polymerized tubulin distribution previously described for HeLa cells [3] was applied to NIH 3T3 with only minor modifications. While the plasma membrane position for HeLa images was estimated using a fluorescence channel showing total cell protein, this channel was not available in the 3T3 images. The tubulin image itself was therefore used for this purpose since the presence of free tubulin allowed for a reliable estimate of cell boundaries Point spread function 3D images of beads were segmented into individual bead regions using Ridler-Calvard thresholding and registered using the 3D centroid of the bead. The beads were then averaged to estimate the point spread function.
3 Shariff et al. Page Free tubulin distribution estimation and generation Our previous generative model only took into account polymerized tubulin because the images were acquired by immunofluorescence staining of fixed cells lacking appreciable free tubulin. This is because permeabilization of cells with detergents like Triton-X to allow antibody penetration causes most of the free tubulin to diffuse away. However, live cell imaging of fluorescently-tagged tubulin detects both free tubulin monomers and polymerized microtubules. We therefore extend the previous model to account for free tubulin by estimating histograms of free tubulin intensities h(reg, nz) for each nuclear or cytoplasmic region and for each 2D slice number nz. Free tubulin regions in each of the 2D slices was estimated by first detecting and removing the polymerized tubulin regions, as follows. The input image was blurred using a Gaussian filter with standard deviation of 3, and the resulting image was subtracted from the input image. The subtracted image was binarized to separate zero and nonzero pixels. Since the binary image has small clusters of disconnected objects seemingly forming microtubule fibers, the binary image is blurred again to connect objects that are close to each other. This operation was performed using a Gaussian filter with standard deviation of 2. The resulting image was again binarized. This ad hoc approach resulted in a reasonable definition of microtubules (as shown in Fig. 1). In order to generate free tubulin images for simulations, the histograms h(reg, nz) were sampled to generate the corresponding distribution of free tubulin in all regions of the cell, f(x) Tubulin Image Formation Here we describe the tubulin fluorescence image formation used for generating simulated images. Let I(x) be the tubulin fluorescence image. Let p(x) and f(x) be the polymerized tubulin and free tubulin images respectively. Let * denote a 3D convolution. Then, I(x) = psf *[p(x) + f(x)], where psf is the point spread function of the imaging system (estimated as above). This can be written as: where p (x) is the model generated in pixel coordinates by the generative model for a given set of parameters and λ is the scaling factor that matches the single polymerized tubulin intensities in the simulated images to the real images (see below). Let f 2 ( x) = psf * f(x). Equation (1) then becomes: Hence, for a given set of parameters θ, I(x θ) can be generated. For a given set of parameters, the amount of free tubulin was adjusted by scaling f 2 (x) according to the total amounts (total intensity) available (see Figure 2 for an example). 3.5 Single microtubule intensity estimation The intensity of a single microtubule was estimated from the 2D slice and region just below the nucleus of the cell. The reason for this is that the microtubules (if present) in this region have a very minimal overlap and are generally traceable. λ was defined as: (1)
4 Shariff et al. Page Library generation where ϕ[.] is the single microtubule intensity in the real (R) and simulated (S) images. ϕ[p R (x)] was estimated by averaging tubular pixel values and subtracting out the average free tubulin pixel values. The tubular pixel regions were detected using the method described by Frangi et al. [6] (see Figure 3 for an example). The remaining regions were assumed to be free tubulin. ϕ[p S (x)] was estimated directly from generated polymerized tubulin images p(x). λ was estimated from many images across the dataset and a single average value λ was used. As described in [3], a library of simulated images was generated for all combinations of discrete values of the four parameters: Number of microtubules = 0, 5, 20, 40, 60, 80, 100, 120, 140, 160, 180, 200, 220 Mean of length distribution (μ) = 5, 20, 40, 60, 80, 100, 120, 140, 160, 180, 200, 220 microns Coefficient of variation of length = 0, 0.1, 0.2, 0.3 Collinearity (cos α) = 0.97, 0.984, 0.992, 0.996, Feature selection and matching 4. RESULTS As described previously [3], parameters are indirectly estimated by choosing the synthetic image from the library that is most similar to a given real image. This choice is made using numerical features calculated to describe the fluorescence distributions, and a critical component of this approach is the choice of features and distance function. We describe here a feature selection method to include in the distance function using training data. Cells corresponding to the 40-min time point do not appear to have polymerized tubulin. Therefore features were selected so as to minimize the normalized Euclidean distance in feature space between 4 images of the 40-min time point of nocodazole treated cells and simulated images for 0 microtubules (only free tubulin). 3D confocal microscopy images were acquired at five different time points in the presence and absence of nocodazole, keeping all imaging parameters fixed. Figure 4 shows an example set of such images for various times of treatment with nocodazole. Cells treated with nocodazole for 40 min appear to have all of their microtubules depolymerized. All but one of the five images at this time point were therefore used to train the feature selection approach, and the features selected were used to estimate model parameters from all the images except the ones that were used for training. This procedure was repeated by holding out each image in turn (five-fold cross-validation). Figure 5 shows the parameter estimates averaged over the five folds and the five replicates per time point. Hence all points are averaged over 25 (5 folds 5 replicates) except that the last time point is averaged over five folds only. The number and mean of length distribution for nocodazole-treated cells decrease as a function of time, but in the control case, these parameters do not show a decreasing trend. The standard deviation error bars are very large in some of the points. This is because the parameters are averaged over different cells that are likely to have varying numbers and lengths of microtubules because of their varying sizes. However, there is a clear decrease in the number and mean of the length from the first and last time points in the nocodazole treated case as opposed to the untreated case.
5 Shariff et al. Page 5 5. CONCLUSIONS AND DISCUSSION Acknowledgments References We have validated a microtubule distribution estimation system by estimating parameters from an image set of live cells. The estimated parameters follow the expected trend: cells treated with nocodazole tend to have less polymerized tubulin. Future work will include improving many of the image processing routines to achieve higher efficiency and robustness, as well as exploring the dependence of the estimates on the accuracy of the point spread function. In future, we plan to estimate parameters from different cell types. We also plan to build generative models of organelles (such as lysosomes or mitochondria) whose distribution may be conditioned on the microtubule model. Ultimately, we seek to build models in a hierarchical, conditional manner so that models of all cell components can be constructed by automated learning from cell images. This work was supported in part by NIH grants GM and GM Gelasca, ED.; Byun, J.; Obara, B., et al. Benchmark for evaluating biological image analysis tools. Workshop on Bio-Image Informatics: Biological Imaging, Computer Vision and Data Mining; Santa Barbara, CA: Center for Bio-Image Informatics, UCSB; Sargin, ME.; Altinok, A.; Kiris, E., et al. Tracing Microtubules in Live Cell Images. Proc. IEEE Int. Symp. Biomed. Imaging; p Shariff A, Murphy RF, Rohde GK. A generative model of microtubule distributions, and indirect estimation of its parameters from fluorescence microscopy images. Cytometry A. May; (5): [PubMed: ] 4. Solomon F. Neuroblastoma cells recapitulate their detailed neurite morphologies after reversible microtubule disassembly. Cell. Sep; (2): [PubMed: ] 5. Jarvik JW, Fisher GW, Shi C, et al. In vivo functional proteomics: mammalian genome annotation using CD-tagging. Biotechniques. Oct; (4): , passim. [PubMed: ] 6. Frangi, AF.; Niessen, WJ.; Vincken, KL., et al. Multiscale Vessel Enhancement Filtering. In: Wells, W.; Colchester, A.; Delp, S., editors. Lecture Notes in Computer Science; Medical Image Computing and Computer-Assisted Interventation MICCAI 98; Berlin/Heidelberg: Springer; 1998.
6 Shariff et al. Page 6 Fig. 1. (A) 2D slice from a 3D image stack of a cell untreated with nocodazole. (B) Removal of polymerized tubulin (C) Regeneration of free tubulin distribution by sampling from free tubulin intensity histograms estimated from (B).
7 Shariff et al. Page 7 Fig. 2. A 2D slice in the 3D stack of a simulated image. The image was generated with the number of microtubules set to 100, the mean of the length distribution to 60 microns, the standard deviation of length to 6 microns and the collinearity to
8 Shariff et al. Page 8 Fig. 3. Single microtubule intensity detection on microtubules in a slice just below the nucleus. The tubulin image is shown in blue and the points identified as showing a single microtubule are marked in red.
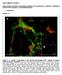
9 Shariff et al. Page 9 Fig. 4. Example images of NIH 3T3 cells expressing EGFP-tagged alpha-tubulin at various time points after addition of 20 um nocodazole (from left to right, 0, 10, 20, 30, and 40 min).
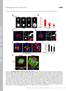
10 Shariff et al. Page 10 Fig. 5. Parameter estimates of the number (A) and mean length (B) averaged over different folds and repetitions.
Estimating Microtubule Distributions from 2D Immunofluorescence Microscopy Images Reveals Differences among Human Cultured Cell Lines
 Estimating Microtubule Distributions from 2D Immunofluorescence Microscopy Images Reveals Differences among Human Cultured Cell Lines Jieyue Li 1,3., Aabid Shariff 1,2., Mikaela Wiking 7, Emma Lundberg
Estimating Microtubule Distributions from 2D Immunofluorescence Microscopy Images Reveals Differences among Human Cultured Cell Lines Jieyue Li 1,3., Aabid Shariff 1,2., Mikaela Wiking 7, Emma Lundberg
Automating Biomedical Research Through Machine Learning
 Automating Biomedical Research Through Machine Learning Robert F Murphy Ray & Stephanie Lane Professor of Computational Biology and Professor of Biological Sciences, Biomedical Engineering and Machine
Automating Biomedical Research Through Machine Learning Robert F Murphy Ray & Stephanie Lane Professor of Computational Biology and Professor of Biological Sciences, Biomedical Engineering and Machine
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION WWW.NATURE.COM/NATURECELLBIOLOGY 1 SUPPLEMENTARY INFORMATION 2 WWW.NATURE.COM/NATURECELLBIOLOGY SUPPLEMENTARY INFORMATION WWW.NATURE.COM/NATURECELLBIOLOGY 3 SUPPLEMENTARY INFORMATION
SUPPLEMENTARY INFORMATION WWW.NATURE.COM/NATURECELLBIOLOGY 1 SUPPLEMENTARY INFORMATION 2 WWW.NATURE.COM/NATURECELLBIOLOGY SUPPLEMENTARY INFORMATION WWW.NATURE.COM/NATURECELLBIOLOGY 3 SUPPLEMENTARY INFORMATION
ab CytoPainter ER Staining Kit Red Fluorescence
 ab139482 CytoPainter ER Staining Kit Red Fluorescence Instructions for Use Designed to detect Human endoplasmic reticulum by microscopy. This product is for research use only and is not intended for diagnostic
ab139482 CytoPainter ER Staining Kit Red Fluorescence Instructions for Use Designed to detect Human endoplasmic reticulum by microscopy. This product is for research use only and is not intended for diagnostic
ab CytoPainter ER Staining Kit Red Fluorescence
 ab139482 CytoPainter ER Staining Kit Red Fluorescence Instructions for Use Designed to detect Human endoplasmic reticulum by microscopy. This product is for research use only and is not intended for diagnostic
ab139482 CytoPainter ER Staining Kit Red Fluorescence Instructions for Use Designed to detect Human endoplasmic reticulum by microscopy. This product is for research use only and is not intended for diagnostic
Supplementary Methods
 Supplementary Methods Antibodies For immunocytochemistry, the following antibodies were used: mouse anti-γ-h2ax (Upstate), rabbit anti-γ-h2ax (Abcam), rabbit anti-53bp1 (Novus), mouse anti-atm-phosphoserine1981
Supplementary Methods Antibodies For immunocytochemistry, the following antibodies were used: mouse anti-γ-h2ax (Upstate), rabbit anti-γ-h2ax (Abcam), rabbit anti-53bp1 (Novus), mouse anti-atm-phosphoserine1981
Supporting Information
 Supporting Information Stavru et al. 0.073/pnas.357840 SI Materials and Methods Immunofluorescence. For immunofluorescence, cells were fixed for 0 min in 4% (wt/vol) paraformaldehyde (Electron Microscopy
Supporting Information Stavru et al. 0.073/pnas.357840 SI Materials and Methods Immunofluorescence. For immunofluorescence, cells were fixed for 0 min in 4% (wt/vol) paraformaldehyde (Electron Microscopy
Fast, three-dimensional super-resolution imaging of live cells
 Nature Methods Fast, three-dimensional super-resolution imaging of live cells Sara A Jones, Sang-Hee Shim, Jiang He & Xiaowei Zhuang Supplementary Figure 1 Supplementary Figure 2 Supplementary Figure 3
Nature Methods Fast, three-dimensional super-resolution imaging of live cells Sara A Jones, Sang-Hee Shim, Jiang He & Xiaowei Zhuang Supplementary Figure 1 Supplementary Figure 2 Supplementary Figure 3
3D Transfection System
 3D Transfection System Why 3D Technology? Introduction to Three Dimensional (3D) Cell Culture The goal of three-dimensional (3D) cell culture is to eliminate the stress and artificial responses cells experience
3D Transfection System Why 3D Technology? Introduction to Three Dimensional (3D) Cell Culture The goal of three-dimensional (3D) cell culture is to eliminate the stress and artificial responses cells experience
EXTRACTING AND STRUCTURING SUBCELLULAR LOCATION INFORMATION FROM ON-LINE JOURNAL ARTICLES: THE SUBCELLULAR LOCATION IMAGE FINDER
 EXTRACTING AND STRUCTURING SUBCELLULAR LOCATION INFORMATION FROM ON-LINE JOURNAL ARTICLES: THE SUBCELLULAR LOCATION IMAGE FINDER Robert F. Murphy murphy@cmu.edu Zhenzhen Kou zkou@andrew.cmu.edu Juchang
EXTRACTING AND STRUCTURING SUBCELLULAR LOCATION INFORMATION FROM ON-LINE JOURNAL ARTICLES: THE SUBCELLULAR LOCATION IMAGE FINDER Robert F. Murphy murphy@cmu.edu Zhenzhen Kou zkou@andrew.cmu.edu Juchang
CytoPainter Golgi Staining Kit Green Fluorescence
 ab139483 CytoPainter Golgi Staining Kit Green Fluorescence Instructions for Use Designed for the detection of Golgi bodies by microscopy This product is for research use only and is not intended for diagnostic
ab139483 CytoPainter Golgi Staining Kit Green Fluorescence Instructions for Use Designed for the detection of Golgi bodies by microscopy This product is for research use only and is not intended for diagnostic
JCB. Supplemental material THE JOURNAL OF CELL BIOLOGY. Prospéri et al.,
 Supplemental material JCB Prospéri et al., http://www.jcb.org/cgi/content/full/jcb.201501018/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Myo1b Tail interacts with YFP-EphB2 coated beads and genistein inhibits
Supplemental material JCB Prospéri et al., http://www.jcb.org/cgi/content/full/jcb.201501018/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Myo1b Tail interacts with YFP-EphB2 coated beads and genistein inhibits
Fluorescence Light Microscopy for Cell Biology
 Fluorescence Light Microscopy for Cell Biology Why use light microscopy? Traditional questions that light microscopy has addressed: Structure within a cell Locations of specific molecules within a cell
Fluorescence Light Microscopy for Cell Biology Why use light microscopy? Traditional questions that light microscopy has addressed: Structure within a cell Locations of specific molecules within a cell
Supporting Online Material Material and Methods
 Supporting Online Material Material and Methods Live-cell DIC recordings PtK 1 cells (ATCC, Manassas, VA) were filmed on a Nikon (Nikon Instruments, Melville, NY) inverted microscope equipped with 60x
Supporting Online Material Material and Methods Live-cell DIC recordings PtK 1 cells (ATCC, Manassas, VA) were filmed on a Nikon (Nikon Instruments, Melville, NY) inverted microscope equipped with 60x
Imaging of BacMam Transfected U-2 OS Cells
 A p p l i c a t i o n N o t e Imaging of BacMam Transfected U-2 OS Cells Optimization of Transfection Conditions Using the Cytation 3 Multi- Mode Reader and Gen5 Data Analysis Software Paul Held Ph. D.
A p p l i c a t i o n N o t e Imaging of BacMam Transfected U-2 OS Cells Optimization of Transfection Conditions Using the Cytation 3 Multi- Mode Reader and Gen5 Data Analysis Software Paul Held Ph. D.
CytoPainter Nucleolar Staining Kit - Green Fluorescence
 ab139475 CytoPainter Nucleolar Staining Kit - Green Fluorescence Instructions for Use Specifically designed for visualizing microscopically nucleoli in living cells. This product is for research use only
ab139475 CytoPainter Nucleolar Staining Kit - Green Fluorescence Instructions for Use Specifically designed for visualizing microscopically nucleoli in living cells. This product is for research use only
Immunofluorescence Confocal Microscopy of 3D Cultures Grown on Alvetex
 Immunofluorescence Confocal Microscopy of 3D Cultures Grown on Alvetex 1.0. Introduction Immunofluorescence uses the recognition of cellular targets by fluorescent dyes or antigen-specific antibodies coupled
Immunofluorescence Confocal Microscopy of 3D Cultures Grown on Alvetex 1.0. Introduction Immunofluorescence uses the recognition of cellular targets by fluorescent dyes or antigen-specific antibodies coupled
A Supersandwich Fluorescence in Situ Hybridization (SFISH) Strategy. for Highly Sensitive and Selective mrna Imaging in Tumor Cells
 Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2015 Electronic Supplementary Information (ESI) A Supersandwich Fluorescence in Situ Hybridization (SFISH)
Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2015 Electronic Supplementary Information (ESI) A Supersandwich Fluorescence in Situ Hybridization (SFISH)
SURFACE ENHANCED RAMAN SCATTERING NANOPARTICLES AS AN ALTERNATIVE TO FLUORESCENT PROBES AN EVALUATION
 APPLICATION NOTE SURFACE ENHANCED RAMAN SCATTERING NANOPARTICLES AS AN ALTERNATIVE TO FLUORESCENT PROBES AN EVALUATION Summary: Interest in using nanoparticles specifically, Surface Enhanced Raman Scattering
APPLICATION NOTE SURFACE ENHANCED RAMAN SCATTERING NANOPARTICLES AS AN ALTERNATIVE TO FLUORESCENT PROBES AN EVALUATION Summary: Interest in using nanoparticles specifically, Surface Enhanced Raman Scattering
SONOMA STATE UNIVERSITY DEPARTMENT OF BIOLOGY BIOLOGY 344: CELL BIOLOGY Fall 2013
 SONOMA STATE UNIVERSITY DEPARTMENT OF BIOLOGY BIOLOGY 344: CELL BIOLOGY Fall 2013 Instructor Murali C. Pillai, PhD Office 214 Darwin Hall Telephone (707) 664-2981 E-mail pillai@sonoma.edu Website www.sonoma.edu/users/p/pillai
SONOMA STATE UNIVERSITY DEPARTMENT OF BIOLOGY BIOLOGY 344: CELL BIOLOGY Fall 2013 Instructor Murali C. Pillai, PhD Office 214 Darwin Hall Telephone (707) 664-2981 E-mail pillai@sonoma.edu Website www.sonoma.edu/users/p/pillai
Organelle Atlas APPENDIX TO CHAPTER 4 4A.1
 APPENDIX TO CHAPTER 4 This atlas contains images of cellular organelles visualized using a wide variety of reagents and techniques and demonstrates the diversity of methods for exploring the cell. The
APPENDIX TO CHAPTER 4 This atlas contains images of cellular organelles visualized using a wide variety of reagents and techniques and demonstrates the diversity of methods for exploring the cell. The
SUPPLEMENTARY INFORMATION
 Nanomechanical mapping of first binding steps of a virus to animal cells David Alsteens, Richard Newton, Rajib Schubert, David Martinez-Martin, Martin Delguste, Botond Roska and Daniel J. Müller This PDF
Nanomechanical mapping of first binding steps of a virus to animal cells David Alsteens, Richard Newton, Rajib Schubert, David Martinez-Martin, Martin Delguste, Botond Roska and Daniel J. Müller This PDF
SUPPLEMENTAL INFORMATION: 1. Supplemental methods 2. Supplemental figure legends 3. Supplemental figures
 Supplementary Material (ESI) for Lab on a Chip This journal is The Royal Society of Chemistry 2008 A microfluidics-based turning assay reveals complex growth cone responses to integrated gradients of substrate-bound
Supplementary Material (ESI) for Lab on a Chip This journal is The Royal Society of Chemistry 2008 A microfluidics-based turning assay reveals complex growth cone responses to integrated gradients of substrate-bound
ApoTrack Cytochrome c Apoptosis ICC Antibody Kit: 2 color immunocytochemistry of cytochrome c and mitochondria.
 PROTOCOL ApoTrack Cytochrome c Apoptosis ICC Antibody Kit 1850 Millrace Drive, Suite 3A Eugene, Oregon 97403 MSA07 Rev.1 DESCRIPTION ApoTrack Cytochrome c Apoptosis ICC Antibody Kit: 2 color immunocytochemistry
PROTOCOL ApoTrack Cytochrome c Apoptosis ICC Antibody Kit 1850 Millrace Drive, Suite 3A Eugene, Oregon 97403 MSA07 Rev.1 DESCRIPTION ApoTrack Cytochrome c Apoptosis ICC Antibody Kit: 2 color immunocytochemistry
Lab Module 7: Cell Adhesion
 Lab Module 7: Cell Adhesion Tissues are made of cells and materials secreted by cells that occupy the spaces between the individual cells. This material outside of cells is called the Extracellular Matrix
Lab Module 7: Cell Adhesion Tissues are made of cells and materials secreted by cells that occupy the spaces between the individual cells. This material outside of cells is called the Extracellular Matrix
Performance of cell viability and cytotoxicity assays on the IN Cell Analyzer 3000
 GE Healthcare Application Note 28-4070-51 AA IN Cell Analyzer 3000 Performance of cell viability and cytotoxicity assays on the IN Cell Analyzer 3000 Key words: cell-based assay viability cytotoxicity
GE Healthcare Application Note 28-4070-51 AA IN Cell Analyzer 3000 Performance of cell viability and cytotoxicity assays on the IN Cell Analyzer 3000 Key words: cell-based assay viability cytotoxicity
ApoTrack Cytochrome c Apoptosis ICC Antibody
 ab110417 ApoTrack Cytochrome c Apoptosis ICC Antibody Instructions for Use For the Immunocytochemistry analysis of cytochrome c and a mitochondrial marker (Complex Vα) in apoptotic cells and nonapoptotic
ab110417 ApoTrack Cytochrome c Apoptosis ICC Antibody Instructions for Use For the Immunocytochemistry analysis of cytochrome c and a mitochondrial marker (Complex Vα) in apoptotic cells and nonapoptotic
204 Part 3.3 SUMMARY INTRODUCTION
 204 Part 3.3 Chapter # METHODOLOGY FOR BUILDING OF COMPLEX WORKFLOWS WITH PROSTAK PACKAGE AND ISIMBIOS Matveeva A. *, Kozlov K., Samsonova M. Department of Computational Biology, Center for Advanced Studies,
204 Part 3.3 Chapter # METHODOLOGY FOR BUILDING OF COMPLEX WORKFLOWS WITH PROSTAK PACKAGE AND ISIMBIOS Matveeva A. *, Kozlov K., Samsonova M. Department of Computational Biology, Center for Advanced Studies,
Supplemental material JCB. Pitaval et al., Cell shape and ciliogenesis Pitaval et al.
 Supplemental material THE JOURNAL OF CELL BIOLOGY Pitaval et al., http://www.jcb.org/cgi/content/full/jcb.201004003/dc1 S1 Figure S1. Cell cycle exit analysis. (A) Experimental procedure. RPE1 cells were
Supplemental material THE JOURNAL OF CELL BIOLOGY Pitaval et al., http://www.jcb.org/cgi/content/full/jcb.201004003/dc1 S1 Figure S1. Cell cycle exit analysis. (A) Experimental procedure. RPE1 cells were
A synthetic recombinase-based feedback loop results in robust expression Supporting Information
 A synthetic recombinase-based feedback loop results in robust expression Supporting Information 1 Fig. S1: Control experiments performed to demonstrate the expected function of our closed-loop feedback
A synthetic recombinase-based feedback loop results in robust expression Supporting Information 1 Fig. S1: Control experiments performed to demonstrate the expected function of our closed-loop feedback
Title: Large-Scale Automated Analysis of Location Patterns in Randomly-Tagged 3T3 Cells
 Editorial Manager(tm) for Annals of Biomedical Engineering Manuscript Draft Manuscript Number: ABMER1 Title: Large-Scale Automated Analysis of Location Patterns in Randomly-Tagged T Cells Article Type:
Editorial Manager(tm) for Annals of Biomedical Engineering Manuscript Draft Manuscript Number: ABMER1 Title: Large-Scale Automated Analysis of Location Patterns in Randomly-Tagged T Cells Article Type:
ApoTrack Cytochrome c Apoptosis ICC Antibody Kit
 ab110417 ApoTrack Cytochrome c Apoptosis ICC Antibody Kit Instructions for Use For the Immunocytochemistry analysis of cytochrome c and a mitochondrial marker (Complex Vα) in apoptotic cells and non-apoptotic
ab110417 ApoTrack Cytochrome c Apoptosis ICC Antibody Kit Instructions for Use For the Immunocytochemistry analysis of cytochrome c and a mitochondrial marker (Complex Vα) in apoptotic cells and non-apoptotic
Super Resolution Imaging Solution Provider. Imaging Future
 Super Resolution Imaging Solution Provider Imaging Future Imaging Solution More Than Equipment NanoBioImaging(NBI) is the Industrial Partner of HKUST Super Resolution Imaging Center (SRIC). NBI aims to
Super Resolution Imaging Solution Provider Imaging Future Imaging Solution More Than Equipment NanoBioImaging(NBI) is the Industrial Partner of HKUST Super Resolution Imaging Center (SRIC). NBI aims to
PROTOCOL. Live Cell Imaging of 3D Cultures Grown on Alvetex Scaffold Using Confocal Microscopy. Introduction
 Page 1 Introduction Live cell imaging allows real time monitoring of cell shape and form during cell differentiation, proliferation and migration. Alvetex Scaffold offers a unique ability to study cell
Page 1 Introduction Live cell imaging allows real time monitoring of cell shape and form during cell differentiation, proliferation and migration. Alvetex Scaffold offers a unique ability to study cell
High content imaging and applications
 High content imaging and applications Nick Dolman Ph.D. Senior Staff Scientist Biosciences Division- Thermo Fisher Scientific For Research Use Only. Not for use in diagnostic procedures. The world leader
High content imaging and applications Nick Dolman Ph.D. Senior Staff Scientist Biosciences Division- Thermo Fisher Scientific For Research Use Only. Not for use in diagnostic procedures. The world leader
ab CytoPainter Mitochondrial Staining Kit NIR Fluorescence
 ab176747 CytoPainter Mitochondrial Staining Kit NIR Fluorescence Instructions for Use For staining Mitochondria in live cells with our proprietary NIR probe. This product is for research use only and is
ab176747 CytoPainter Mitochondrial Staining Kit NIR Fluorescence Instructions for Use For staining Mitochondria in live cells with our proprietary NIR probe. This product is for research use only and is
THE BASICS OF IMMUNOHISTOCHEMISTRY
 THE BASICS OF IMMUNOHISTOCHEMISTRY Introduction Immunohistochemistry (IHC) identifies specific tissue components by means of a specific antigen/antibody reaction tagged with a visible label. IHC makes
THE BASICS OF IMMUNOHISTOCHEMISTRY Introduction Immunohistochemistry (IHC) identifies specific tissue components by means of a specific antigen/antibody reaction tagged with a visible label. IHC makes
Spectral Separation of Multifluorescence Labels with the LSM 510 META
 Microscopy from Carl Zeiss Spectral Separation of Multifluorescence Labels with the LSM 510 META Indians living in the South American rain forest can distinguish between almost 200 hues of green in their
Microscopy from Carl Zeiss Spectral Separation of Multifluorescence Labels with the LSM 510 META Indians living in the South American rain forest can distinguish between almost 200 hues of green in their
The CQ1 Confocal Quantitative Image Cytometer and its Application to Biological Measurement
 The CQ1 Confocal Quantitative Image Cytometer and its Application to Biological Measurement Hirofumi Sakashita *1 Koji Ohashi *1 Kazuo Ozawa *2 ohei Tsubouchi *1 The CQ1 confocal quantitative image cytometer,
The CQ1 Confocal Quantitative Image Cytometer and its Application to Biological Measurement Hirofumi Sakashita *1 Koji Ohashi *1 Kazuo Ozawa *2 ohei Tsubouchi *1 The CQ1 confocal quantitative image cytometer,
Brightfield and Fluorescence Imaging using 3D PrimeSurface Ultra-Low Attachment Microplates
 A p p l i c a t i o n N o t e Brightfield and Fluorescence Imaging using 3D PrimeSurface Ultra-Low Attachment Microplates Brad Larson, BioTek Instruments, Inc., Winooski, VT USA Anju Dang, S-BIO, Hudson,
A p p l i c a t i o n N o t e Brightfield and Fluorescence Imaging using 3D PrimeSurface Ultra-Low Attachment Microplates Brad Larson, BioTek Instruments, Inc., Winooski, VT USA Anju Dang, S-BIO, Hudson,
EXPERIMENT 2 Cell Fractionation and DNA Isolation
 EXPERIMENT 2 Cell Fractionation and DNA Isolation Background Information A. The Eukaryotic Cell Cells are frequently classified into two basic types: prokaryotic and eukar yotic. The prokaryotic cell,
EXPERIMENT 2 Cell Fractionation and DNA Isolation Background Information A. The Eukaryotic Cell Cells are frequently classified into two basic types: prokaryotic and eukar yotic. The prokaryotic cell,
BIO 315 Lab Exam I. Section #: Name:
 Section #: Name: Also provide this information on the computer grid sheet given to you. (Section # in special code box) BIO 315 Lab Exam I 1. In labeling the parts of a standard compound light microscope
Section #: Name: Also provide this information on the computer grid sheet given to you. (Section # in special code box) BIO 315 Lab Exam I 1. In labeling the parts of a standard compound light microscope
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Ono et al., http://www.jcb.org/cgi/content/full/jcb.201208008/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Chromatin-binding properties of MCM2 and condensin II during
Supplemental material Ono et al., http://www.jcb.org/cgi/content/full/jcb.201208008/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Chromatin-binding properties of MCM2 and condensin II during
Imaging of Cells using fluorescents dyes. By: Josué A. Benjamín Rivera September 27, 2018
 Imaging of Cells using fluorescents dyes By: Josué A. Benjamín Rivera September 27, 2018 1 History Sir William Henry Perkin BRITISH CHEMIST In 1856, at the age of 18, William Henry Perkin set out with
Imaging of Cells using fluorescents dyes By: Josué A. Benjamín Rivera September 27, 2018 1 History Sir William Henry Perkin BRITISH CHEMIST In 1856, at the age of 18, William Henry Perkin set out with
Contents. SCHOOL of FLUORESCENCE. For more information, go to lifetechnologies.com/imagingbasics
 MPSF educator packet This packet contains illustrations and figures from the Molecular Probes School of Fluorescence website. They illustrate concepts from the basic physical properties that underlie fluorescence
MPSF educator packet This packet contains illustrations and figures from the Molecular Probes School of Fluorescence website. They illustrate concepts from the basic physical properties that underlie fluorescence
Supplementary Protocol. sirna transfection methodology and performance
 Supplementary Protocol sirna transfection methodology and performance sirna oligonucleotides, DNA construct and cell line. Chemically synthesized 21 nt RNA duplexes were obtained from Ambion Europe, Ltd.
Supplementary Protocol sirna transfection methodology and performance sirna oligonucleotides, DNA construct and cell line. Chemically synthesized 21 nt RNA duplexes were obtained from Ambion Europe, Ltd.
Electronic Supplementary Information
 Electronic Supplementary Material (ESI) for Integrative Biology. This journal is The Royal Society of Chemistry 2015 Electronic Supplementary Information Table S1. Definition of quantitative cellular features
Electronic Supplementary Material (ESI) for Integrative Biology. This journal is The Royal Society of Chemistry 2015 Electronic Supplementary Information Table S1. Definition of quantitative cellular features
ER-ID Red Assay Kit (GFP-Certified )
 Enabling Discovery in Life Science ER-ID Red Assay Kit (GFP-Certified ) for detection of endoplasmic reticulum by microscopy Instruction Manual Cat. No. ENZ-51026-K500 500 assays For research use only.
Enabling Discovery in Life Science ER-ID Red Assay Kit (GFP-Certified ) for detection of endoplasmic reticulum by microscopy Instruction Manual Cat. No. ENZ-51026-K500 500 assays For research use only.
ab CytoPainter Lysosome/ER/Nuclear Staining Reagent (Fluorometric)
 ab139488 CytoPainter Lysosome/ER/Nuclear Staining Reagent (Fluorometric) Instructions for Use Designed to detect lysosomes, endoplasmic reticulum and nucleic acids by microscopy. This product is for research
ab139488 CytoPainter Lysosome/ER/Nuclear Staining Reagent (Fluorometric) Instructions for Use Designed to detect lysosomes, endoplasmic reticulum and nucleic acids by microscopy. This product is for research
Comparison of Oridonin Cytotoxicity in U-2 OS and HepG2 Cells
 A p p l i c a t i o n N o t e Comparison of Oridonin Cytotoxicity in U-2 OS and HepG2 Cells Using the BioSpa 8 to Manage Repeated Reagent Additions to Live Cells Paul Held, PhD, Laboratory Manager, Applications
A p p l i c a t i o n N o t e Comparison of Oridonin Cytotoxicity in U-2 OS and HepG2 Cells Using the BioSpa 8 to Manage Repeated Reagent Additions to Live Cells Paul Held, PhD, Laboratory Manager, Applications
Supporting Information. ribosome-mrna interactions in single cells. Kelly S. Burke, Katie A. Antilla, and David A. Tirrell*
 Supporting Information A fluorescence in situ hybridization method to quantify mrna translation by visualizing ribosome-mrna interactions in single cells. Kelly S. Burke, Katie A. Antilla, and David A.
Supporting Information A fluorescence in situ hybridization method to quantify mrna translation by visualizing ribosome-mrna interactions in single cells. Kelly S. Burke, Katie A. Antilla, and David A.
Plasmid DNA transfection of SW480 human colorectal cancer cells with the Biontex K2 Transfection System
 Plasmid DNA transfection of human colorectal cancer cells with the Biontex K2 Transfection System Stephanie Hehlgans and Franz Rödel, Department of Radiotherapy and Oncology, Goethe- University Frankfurt,
Plasmid DNA transfection of human colorectal cancer cells with the Biontex K2 Transfection System Stephanie Hehlgans and Franz Rödel, Department of Radiotherapy and Oncology, Goethe- University Frankfurt,
Analysis and multiplexing of an automated, high-content, AKT activation assay using IN Cell Analyzer 2000
 GE Healthcare Application note 28-9545-14 AA IN Cell Analyzer 2000 Analysis and multiplexing of an automated, high-content, AKT activation assay using IN Cell Analyzer 2000 Key words: IN Cell Analyzer
GE Healthcare Application note 28-9545-14 AA IN Cell Analyzer 2000 Analysis and multiplexing of an automated, high-content, AKT activation assay using IN Cell Analyzer 2000 Key words: IN Cell Analyzer
Supplementary material to Alterations in the properties of the cell membrane due to glycosphingolipid accumulation in a model of Gaucher disease
 Supplementary material to Alterations in the properties of the cell membrane due to glycosphingolipid accumulation in a model of Gaucher disease Gyula Batta, Lilla Soltész, Tamás Kovács, Tamás Bozó, Zoltán
Supplementary material to Alterations in the properties of the cell membrane due to glycosphingolipid accumulation in a model of Gaucher disease Gyula Batta, Lilla Soltész, Tamás Kovács, Tamás Bozó, Zoltán
SUPPLEMENTARY INFORMATION
 a 14 12 Densitometry (AU) 1 8 6 4 2 t b 16 NMHC-IIA GAPDH NMHC-IIB Densitometry (AU) 14 12 1 8 6 4 2 1 nm 1 nm 1 nm 1 nm sirna 1 nm 1 nm Figure S1 S4 Quantification of protein levels. (a) The microtubule
a 14 12 Densitometry (AU) 1 8 6 4 2 t b 16 NMHC-IIA GAPDH NMHC-IIB Densitometry (AU) 14 12 1 8 6 4 2 1 nm 1 nm 1 nm 1 nm sirna 1 nm 1 nm Figure S1 S4 Quantification of protein levels. (a) The microtubule
< Supporting Information >
 SUPPORTING INFORMATION 1 < Supporting Information > Discovery of autophagy modulators through the construction of high-content screening platform via monitoring of lipid droplets Sanghee Lee, Eunha Kim,
SUPPORTING INFORMATION 1 < Supporting Information > Discovery of autophagy modulators through the construction of high-content screening platform via monitoring of lipid droplets Sanghee Lee, Eunha Kim,
0.5% Triton X-100 for 5 min at room temperature. Fixed and permeabilized cells were
 1 Supplementary Methods Immunohistochemistry EBC-1 cells were fixed in 4% paraformaldehyde for 15 min at room temperature, followed by 0.5% Triton X-100 for 5 min at room temperature. Fixed and permeabilized
1 Supplementary Methods Immunohistochemistry EBC-1 cells were fixed in 4% paraformaldehyde for 15 min at room temperature, followed by 0.5% Triton X-100 for 5 min at room temperature. Fixed and permeabilized
Confocal immunofluorescence microscopy
 Confocal immunofluorescence microscopy HL-6 and cells were cultured and cytospun onto glass slides. The cells were double immunofluorescence stained for Mt NPM1 and fibrillarin (nucleolar marker). Briefly,
Confocal immunofluorescence microscopy HL-6 and cells were cultured and cytospun onto glass slides. The cells were double immunofluorescence stained for Mt NPM1 and fibrillarin (nucleolar marker). Briefly,
SUPPLEMENTARY INFORMATION FIGURE 1 - 1
 SUPPLEMENTARY INFORMATION FIGURE 1-1 SUPPLEMENTARY INFORMATION FIGURE 2-2 SUPPLEMENTARY INFORMATION METHODS GST-Pull-Down. Cultures of E. Coli (BL21) were transformed with pgex (Clontech) and pgex recombinant
SUPPLEMENTARY INFORMATION FIGURE 1-1 SUPPLEMENTARY INFORMATION FIGURE 2-2 SUPPLEMENTARY INFORMATION METHODS GST-Pull-Down. Cultures of E. Coli (BL21) were transformed with pgex (Clontech) and pgex recombinant
ER stress and autophagy: new players in the mechanism of action and drug resistance of SUPPLEMENTAL DATA
 ER stress and autophagy: new players in the mechanism of action and drug resistance of the cyclin-dependent kinase inhibitor SUPPLEMENTAL DATA METHODS Confocal immunofluorescence microscopy of fixed cells:
ER stress and autophagy: new players in the mechanism of action and drug resistance of the cyclin-dependent kinase inhibitor SUPPLEMENTAL DATA METHODS Confocal immunofluorescence microscopy of fixed cells:
M X 500 µl. M X 1000 µl
 GeneGlide TM sirna Transfection Reagent (Catalog # M1081-300, -500, -1000; Store at 4 C) I. Introduction: BioVision s GeneGlide TM sirna Transfection reagent is a cationic proprietary polymer/lipid formulation,
GeneGlide TM sirna Transfection Reagent (Catalog # M1081-300, -500, -1000; Store at 4 C) I. Introduction: BioVision s GeneGlide TM sirna Transfection reagent is a cationic proprietary polymer/lipid formulation,
Plasmid DNA transfection of LN-229 human glioblastoma cells with the Biontex K2 Transfection System
 Plasmid DNA transfection of human glioblastoma cells with the Biontex K2 Transfection System Stephanie Hehlgans and Franz Rödel, Department of Radiotherapy and Oncology, Goethe- University Frankfurt, Theodor-Stern-Kai
Plasmid DNA transfection of human glioblastoma cells with the Biontex K2 Transfection System Stephanie Hehlgans and Franz Rödel, Department of Radiotherapy and Oncology, Goethe- University Frankfurt, Theodor-Stern-Kai
A legacy of innovation and discovery
 A legacy of innovation and discovery CellInsight CX7 LZR High Content Analysis Platform Quantifiably brilliant data Since the introduction of Thermo Scientific ArrayScan High Content Analysis (HCA) Readers
A legacy of innovation and discovery CellInsight CX7 LZR High Content Analysis Platform Quantifiably brilliant data Since the introduction of Thermo Scientific ArrayScan High Content Analysis (HCA) Readers
An automated image processing routine for segmentation of cell cytoplasms in high-resolution autofluorescence images
 An automated image processing routine for segmentation of cell cytoplasms in high-resolution autofluorescence images Alex J. Walsh a, Melissa C. Skala *a a Department of Biomedical Engineering, Vanderbilt
An automated image processing routine for segmentation of cell cytoplasms in high-resolution autofluorescence images Alex J. Walsh a, Melissa C. Skala *a a Department of Biomedical Engineering, Vanderbilt
Supplemental Information. Histo-Cytometry: A Method for Highly Multiplex. Quantitative Tissue Imaging Analysis Applied to
 Immunity, Volume 37 Supplemental Information Histo-Cytometry: A Method for Highly Multiplex Quantitative Tissue Imaging Analysis Applied to Dendritic Cell Subset Microanatomy in Lymph Nodes Michael Y.
Immunity, Volume 37 Supplemental Information Histo-Cytometry: A Method for Highly Multiplex Quantitative Tissue Imaging Analysis Applied to Dendritic Cell Subset Microanatomy in Lymph Nodes Michael Y.
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/NCHEM.1805 Visualization and Selective Chemical Targeting of RNA G-quadruplex Structures in the Cytoplasm of Human Cells Giulia Biffi 1, Marco Di Antonio 2, David Tannahill 1 and Shankar Balasubramanian
DOI: 10.1038/NCHEM.1805 Visualization and Selective Chemical Targeting of RNA G-quadruplex Structures in the Cytoplasm of Human Cells Giulia Biffi 1, Marco Di Antonio 2, David Tannahill 1 and Shankar Balasubramanian
BIO 315 Lab Exam I. Section #: Name:
 Section #: Name: Also provide this information on the computer grid sheet given to you. (Section # in special code box) BIO 315 Lab Exam I 1. In labeling the parts of a standard compound light microscope
Section #: Name: Also provide this information on the computer grid sheet given to you. (Section # in special code box) BIO 315 Lab Exam I 1. In labeling the parts of a standard compound light microscope
Monitoring Cell Cycle Progression in Cancer Cells
 A p p l i c a t i o n N o t e Monitoring Cell Cycle Progression in Cancer Cells Using Nuclear Staining to Assess Cellular DNA Content Paul Held, Ph.D., Laboratory Manager, Applications Department, BioTek
A p p l i c a t i o n N o t e Monitoring Cell Cycle Progression in Cancer Cells Using Nuclear Staining to Assess Cellular DNA Content Paul Held, Ph.D., Laboratory Manager, Applications Department, BioTek
Actin cap associated focal adhesions and their distinct role in cellular mechanosensing
 Actin cap associated focal adhesions and their distinct role in cellular mechanosensing Dong-Hwee Kim 1,2, Shyam B. Khatau 1,2, Yunfeng Feng 1,3, Sam Walcott 1,4, Sean X. Sun 1,2,4, Gregory D. Longmore
Actin cap associated focal adhesions and their distinct role in cellular mechanosensing Dong-Hwee Kim 1,2, Shyam B. Khatau 1,2, Yunfeng Feng 1,3, Sam Walcott 1,4, Sean X. Sun 1,2,4, Gregory D. Longmore
Beta3 integrin promotes long-lasting activation and polarization of Vascular Endothelial Growth Factor Receptor 2 by immobilized ligand
 SUPPLEMENTAL FIGURES Beta3 integrin promotes long-lasting activation and polarization of Vascular Endothelial Growth Factor Receptor 2 by immobilized ligand C. Ravelli et al. FIGURE S. I Figure S. I: Gremlin
SUPPLEMENTAL FIGURES Beta3 integrin promotes long-lasting activation and polarization of Vascular Endothelial Growth Factor Receptor 2 by immobilized ligand C. Ravelli et al. FIGURE S. I Figure S. I: Gremlin
Supplementary Materials and Methods
 Supplementary Materials and Methods Reagents Supplementary Material (ESI) for Lab on a Chip RPMI medium, FBS, HEPES buffer solution, sodium pyruvate, penicillin, and streptomycin were obtained from Biological
Supplementary Materials and Methods Reagents Supplementary Material (ESI) for Lab on a Chip RPMI medium, FBS, HEPES buffer solution, sodium pyruvate, penicillin, and streptomycin were obtained from Biological
A fluorescent probe for cysteine depalmitoylation reveals dynamic APT signaling
 SUPPLEMENTARY INFRMATIN A fluorescent probe for cysteine depalmitoylation reveals dynamic APT signaling Rahul S. Kathayat 1, Pablo D. Elvira 1, Bryan C. Dickinson 1 * 1 Department of Chemistry, The University
SUPPLEMENTARY INFRMATIN A fluorescent probe for cysteine depalmitoylation reveals dynamic APT signaling Rahul S. Kathayat 1, Pablo D. Elvira 1, Bryan C. Dickinson 1 * 1 Department of Chemistry, The University
PCCS Growth Media, Cell Tagging, Cell Separation Final Assignment. Igneris Rosado-Erazo. Panama College of Cell Science
 Running Head: Growth Media, Cell Tagging, Cell Separation PCCS Growth Media, Cell Tagging, Cell Separation Final Assignment Igneris Rosado-Erazo Panama College of Cell Science In partial fulfillment of
Running Head: Growth Media, Cell Tagging, Cell Separation PCCS Growth Media, Cell Tagging, Cell Separation Final Assignment Igneris Rosado-Erazo Panama College of Cell Science In partial fulfillment of
Microtubules. Forms a sheet of protofilaments that folds to a tube Both tubulins bind GTP
 Microtubules Polymerize from a-tubulin/ß-tubulin dimers Hollow tube of 13 protofilaments In vitro- filament number varies In vivo- always 13 protofilaments Forms a sheet of protofilaments that folds to
Microtubules Polymerize from a-tubulin/ß-tubulin dimers Hollow tube of 13 protofilaments In vitro- filament number varies In vivo- always 13 protofilaments Forms a sheet of protofilaments that folds to
Pre-made Lentiviral Particles for nuclear permeant CRE recombinase expression
 Pre-made Lentiviral Particles for nuclear permeant CRE recombinase expression Cat# Product Name Amounts* LVP336 NLS-CRE (Bsd) LVP336-PBS NLS-CRE (Bsd), in vivo ready LVP339 NLS-CRE (Puro) LVP339-PBS NLS-CRE
Pre-made Lentiviral Particles for nuclear permeant CRE recombinase expression Cat# Product Name Amounts* LVP336 NLS-CRE (Bsd) LVP336-PBS NLS-CRE (Bsd), in vivo ready LVP339 NLS-CRE (Puro) LVP339-PBS NLS-CRE
Supplemental Information. A Versatile Tool for Live-Cell Imaging. and Super-Resolution Nanoscopy Studies. of HIV-1 Env Distribution and Mobility
 Cell Chemical Biology, Volume 24 Supplemental Information A Versatile Tool for Live-Cell Imaging and Super-Resolution Nanoscopy Studies of HIV-1 Env Distribution and Mobility Volkan Sakin, Janina Hanne,
Cell Chemical Biology, Volume 24 Supplemental Information A Versatile Tool for Live-Cell Imaging and Super-Resolution Nanoscopy Studies of HIV-1 Env Distribution and Mobility Volkan Sakin, Janina Hanne,
Localization Microscopy
 Localization Microscopy Theory, Sample Prep & Practical Considerations Patrina Pellett & Ann McEvoy Applications Scientist GE Healthcare, Cell Technologies May 27 th, 2015 Localization Microscopy Talk
Localization Microscopy Theory, Sample Prep & Practical Considerations Patrina Pellett & Ann McEvoy Applications Scientist GE Healthcare, Cell Technologies May 27 th, 2015 Localization Microscopy Talk
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rodríguez-Fraticelli et al., http://www.jcb.org/cgi/content/full/jcb.201203075/dc1 Figure S1. Cell spreading and lumen formation in confined
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rodríguez-Fraticelli et al., http://www.jcb.org/cgi/content/full/jcb.201203075/dc1 Figure S1. Cell spreading and lumen formation in confined
MEFs were treated with the indicated concentrations of LLOMe for three hours, washed
 Supplementary Materials and Methods Cell Fractionation MEFs were treated with the indicated concentrations of LLOMe for three hours, washed with ice-cold PBS, collected by centrifugation, and then homogenized
Supplementary Materials and Methods Cell Fractionation MEFs were treated with the indicated concentrations of LLOMe for three hours, washed with ice-cold PBS, collected by centrifugation, and then homogenized
Supplementary Figure 1 Pfn1, but not other Pfn isoforms are expressed in
 Supplementary Figure 1 Pfn1, but not other Pfn isoforms are expressed in platelets. (a) RT-PCR of Pfn isoforms in control mouse platelets, Pfn1 -/- platelets and control heart. Expected band size for Pfn1
Supplementary Figure 1 Pfn1, but not other Pfn isoforms are expressed in platelets. (a) RT-PCR of Pfn isoforms in control mouse platelets, Pfn1 -/- platelets and control heart. Expected band size for Pfn1
B. ADM: C. D. Apoptosis: 1.68% 2.99% 1.31% Figure.S1,Li et al. number. invaded cells. HuH7 BxPC-3 DLD-1.
 A. - Figure.S1,Li et al. B. : - + - + - + E-cadherin CK19 α-sma vimentin β -actin C. D. Apoptosis: 1.68% 2.99% 1.31% - : - + - + - + Apoptosis: 48.33% 45.32% 44.59% E. invaded cells number 400 300 200
A. - Figure.S1,Li et al. B. : - + - + - + E-cadherin CK19 α-sma vimentin β -actin C. D. Apoptosis: 1.68% 2.99% 1.31% - : - + - + - + Apoptosis: 48.33% 45.32% 44.59% E. invaded cells number 400 300 200
SUPPLEMENTARY INFORMATION FILE
 SUPPLEMENTARY INFORMATION FILE Existence of a microrna pathway in anucleate platelets Patricia Landry, Isabelle Plante, Dominique L. Ouellet, Marjorie P. Perron, Guy Rousseau & Patrick Provost 1. SUPPLEMENTARY
SUPPLEMENTARY INFORMATION FILE Existence of a microrna pathway in anucleate platelets Patricia Landry, Isabelle Plante, Dominique L. Ouellet, Marjorie P. Perron, Guy Rousseau & Patrick Provost 1. SUPPLEMENTARY
Automated Digital Microscopy
 A p p l i c a t i o n G u i d e Peter Banks, Ph.D. and Peter J. Brescia, Applications Department, BioTek Instruments, Inc., Winooski, VT Table of Contents Introduction ----------------------------------------------------------------------------------------------------------------------
A p p l i c a t i o n G u i d e Peter Banks, Ph.D. and Peter J. Brescia, Applications Department, BioTek Instruments, Inc., Winooski, VT Table of Contents Introduction ----------------------------------------------------------------------------------------------------------------------
DNA damage - AP site- Assay Kit (Cell-Based)
 ab133076 DNA damage - AP site- Assay Kit (Cell-Based) Instructions for Use For the detection of aldehyde sites in cells. This product is for research use only and is not intended for diagnostic use. 1
ab133076 DNA damage - AP site- Assay Kit (Cell-Based) Instructions for Use For the detection of aldehyde sites in cells. This product is for research use only and is not intended for diagnostic use. 1
Accessorize Your Imaging. Ian Clements Invitrogen Corp
 Accessorize Your Imaging Ian Clements Invitrogen Corp Imaging Tools and Accessories Antifade Reagents Prolong Gold SlowFade Gold Image-iT FX Signal Enhancer FocalCheck Microscope Test Slides Fluorescent
Accessorize Your Imaging Ian Clements Invitrogen Corp Imaging Tools and Accessories Antifade Reagents Prolong Gold SlowFade Gold Image-iT FX Signal Enhancer FocalCheck Microscope Test Slides Fluorescent
SUPPLEMENTARY NOTE 3. Supplememtary Note 3, Wehr et al., Monitoring Regulated Protein-Protein Interactions Using Split-TEV 1
 SUPPLEMENTARY NOTE 3 Fluorescent Proteolysis-only TEV-Reporters We generated TEV reporter that allow visualizing TEV activity at the membrane and in the cytosol of living cells not relying on the addition
SUPPLEMENTARY NOTE 3 Fluorescent Proteolysis-only TEV-Reporters We generated TEV reporter that allow visualizing TEV activity at the membrane and in the cytosol of living cells not relying on the addition
Impedance Array Studies of Mammalian Cell Growth
 Impedance Array Studies of Mammalian Cell Growth Duc D. Nguyen and Michael M. Domach Department of Chemical Engineering, Carnegie Mellon University 5000 Forbes Avenue, Pittsburgh, PA 15217 Xiaoqiu Huang
Impedance Array Studies of Mammalian Cell Growth Duc D. Nguyen and Michael M. Domach Department of Chemical Engineering, Carnegie Mellon University 5000 Forbes Avenue, Pittsburgh, PA 15217 Xiaoqiu Huang
NucView TM 488 Caspase-3 Assay Kit for Live Cells
 NucView TM 488 Caspase-3 Assay Kit for Live Cells Catalog Number: 30029 (100-500 assays) Contact Information Address: Biotium, Inc. 3423 Investment Blvd. Suite 8 Hayward, CA 94545 USA Telephone: (510)
NucView TM 488 Caspase-3 Assay Kit for Live Cells Catalog Number: 30029 (100-500 assays) Contact Information Address: Biotium, Inc. 3423 Investment Blvd. Suite 8 Hayward, CA 94545 USA Telephone: (510)
Image Analysis for Calculation of the Toxicity Degree of Cells in Phase Contrast Microscopy Images
 Image Analysis for Calculation of the Toxicity Degree of Cells in Phase Contrast Microscopy Images M. Athelogou 1, M. Eblenkamp 2, G. Schmidt 1, F. Novotny 2, E. Wintermantel 2, G. Binnig 1 1 Definiens
Image Analysis for Calculation of the Toxicity Degree of Cells in Phase Contrast Microscopy Images M. Athelogou 1, M. Eblenkamp 2, G. Schmidt 1, F. Novotny 2, E. Wintermantel 2, G. Binnig 1 1 Definiens
CHAPTER-6 HISTOGRAM AND MORPHOLOGY BASED PAP SMEAR IMAGE SEGMENTATION
 CHAPTER-6 HISTOGRAM AND MORPHOLOGY BASED PAP SMEAR IMAGE SEGMENTATION 6.1 Introduction to automated cell image segmentation The automated detection and segmentation of cell nuclei in Pap smear images is
CHAPTER-6 HISTOGRAM AND MORPHOLOGY BASED PAP SMEAR IMAGE SEGMENTATION 6.1 Introduction to automated cell image segmentation The automated detection and segmentation of cell nuclei in Pap smear images is
Long-term dynamics of CA1 hippocampal place codes
 Long-term dynamics of CA1 hippocampal place codes Yaniv Ziv, Laurie D. Burns, Eric D. Cocker, Elizabeth O. Hamel, Kunal K. Ghosh, Lacey J. Kitch, Abbas El Gamal, and Mark J. Schnitzer Supplementary Fig.
Long-term dynamics of CA1 hippocampal place codes Yaniv Ziv, Laurie D. Burns, Eric D. Cocker, Elizabeth O. Hamel, Kunal K. Ghosh, Lacey J. Kitch, Abbas El Gamal, and Mark J. Schnitzer Supplementary Fig.
Input. Pulldown GST. GST-Ahi1
 A kd 250 150 100 75 50 37 25 -GST-Ahi1 +GST-Ahi1 kd 148 98 64 50 37 22 GST GST-Ahi1 C kd 250 148 98 64 50 Control Input Hap1A Hap1 Pulldown GST GST-Ahi1 Hap1A Hap1 Hap1A Hap1 Figure S1. (A) Ahi1 western
A kd 250 150 100 75 50 37 25 -GST-Ahi1 +GST-Ahi1 kd 148 98 64 50 37 22 GST GST-Ahi1 C kd 250 148 98 64 50 Control Input Hap1A Hap1 Pulldown GST GST-Ahi1 Hap1A Hap1 Hap1A Hap1 Figure S1. (A) Ahi1 western
ab CytoPainter Mitochondrial Staining Kit - Green Fluorescence
 ab112143 CytoPainter Mitochondrial Staining Kit - Green Fluorescence Instructions for Use For staining Mitochondria in live cells using our proprietary green fluorescence probe This product is for research
ab112143 CytoPainter Mitochondrial Staining Kit - Green Fluorescence Instructions for Use For staining Mitochondria in live cells using our proprietary green fluorescence probe This product is for research
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
Fluorescence Nanoscopy
 Fluorescence Nanoscopy Keith A. Lidke University of New Mexico panda3.phys.unm.edu/~klidke/index.html Optical Microscopy http://en.wikipedia.org/wiki/k%c3%b6hler_illumination 30 µm Fluorescent Probes Michalet
Fluorescence Nanoscopy Keith A. Lidke University of New Mexico panda3.phys.unm.edu/~klidke/index.html Optical Microscopy http://en.wikipedia.org/wiki/k%c3%b6hler_illumination 30 µm Fluorescent Probes Michalet
Supplementary Information
 Supplementary Information Figure S1. Transiently overexpressed ECFP-TIAF1/EYFP-TIAF1 causes apoptosis of Mv1Lu cells. Mv1Lu cells were transfected by electroporation with ECFP, EYFP, ECFP-TIAF1, EYFP-
Supplementary Information Figure S1. Transiently overexpressed ECFP-TIAF1/EYFP-TIAF1 causes apoptosis of Mv1Lu cells. Mv1Lu cells were transfected by electroporation with ECFP, EYFP, ECFP-TIAF1, EYFP-
Marilyn G. Rimando, Hao-Hsiang Wu, Yu-An Liu, Chien-Wei Lee, Shu-Wen Kuo, Yin-
 Supplementary Information Glucocorticoid receptor and Histone Deacetylase 6 mediate the differential effect of dexamethasone during osteogenesis of Mesenchymal stromal cells (MSCs) Marilyn G. Rimando,
Supplementary Information Glucocorticoid receptor and Histone Deacetylase 6 mediate the differential effect of dexamethasone during osteogenesis of Mesenchymal stromal cells (MSCs) Marilyn G. Rimando,
Confocal Microscopy Analyzes Cells
 Choosing Filters for Fluorescence A Laurin Publication Photonic Solutions for Biotechnology and Medicine November 2002 Confocal Microscopy Analyzes Cells Reprinted from the November 2002 issue of Biophotonics
Choosing Filters for Fluorescence A Laurin Publication Photonic Solutions for Biotechnology and Medicine November 2002 Confocal Microscopy Analyzes Cells Reprinted from the November 2002 issue of Biophotonics
λ N -GFP: an RNA reporter system for live-cell imaging
 λ N -GFP: an RNA reporter system for live-cell imaging Nathalie Daigle & Jan Ellenberg Supplementary Figures and Text: Supplementary Figure 1 localization in the cytoplasm. 4 λ N22-3 megfp-m9 serves as
λ N -GFP: an RNA reporter system for live-cell imaging Nathalie Daigle & Jan Ellenberg Supplementary Figures and Text: Supplementary Figure 1 localization in the cytoplasm. 4 λ N22-3 megfp-m9 serves as
Analysis of cytoskeletal features in dried and living cells and slow dynamic experiments using Atomic Force Microscopy
 Analysis of cytoskeletal features in dried and living cells and slow dynamic experiments using Atomic Force Microscopy Author Watson, Gregory, Watson, Jolanta Published 008 Journal Title Journal of Physics:
Analysis of cytoskeletal features in dried and living cells and slow dynamic experiments using Atomic Force Microscopy Author Watson, Gregory, Watson, Jolanta Published 008 Journal Title Journal of Physics:
