Evaluating the Global CpG Methylation Status of Native DNA Utilizing a. Bipartite Split-Luciferase Sensor
|
|
|
- Adam Jones
- 5 years ago
- Views:
Transcription
1 Evaluating the Global CpG Methylation Status of Native DNA Utilizing a Bipartite Split-Luciferase Sensor Ahmed H. Badran, Jennifer L. Furman, Andrew S. Ma, Troy J. Comi, Jason R. Porter and Indraneel Ghosh* Department of Chemistry and Biochemistry, University of Arizona, Tucson, AZ * To whom correspondence should be addressed. ghosh@ .arizona.edu Supplementary Information Table of Contents: Table S1. Summary of Starting DNA-Binding Domain Constructs. Table S2. Summary of Synthesized Fusion Constructs. Table S3. Summary of Target Oligonucleotides. Figure S1. Interrogation of MBD-Fluc Translation Efficiency and MBD1/E2C Limit of Detection. Figure S2. Exogenous Methylation of DNA. Figure S3. Evaluation of Global HeLa Methylation. Supplementary Protocol. mcpg Distance Separation Calculation by CGContentAnalyzer. 1
2 Plasmid DNA Binding Domain Source I.M.A.G.E. No. potb7 MeCP2 flmecp2 (1-486) Open Biosystems puc57 MBD1 MBD1 (1-69) Genscript - pcmv-sport6 MBD2 flmbd2 (1-411) Open Biosystems puc57 MBD3 MBD3 (1-72) Genscript - potb7 MBD4 flmbd4 (1-580) Open Biosystems pcmv-sport6 ZBTB4 ZBTB4 ( ) Open Biosystems pbluescriptr ZBTB33 ZBTB33 ( ) Open Biosystems PAD1258 ZBTB38 ( ) Sasai et al. (2010) - pcr-bluntii-topo UHRF1 fluhrf1 (1-793) Open Biosystems pcmv-sport6 UHRF2 UHRF2 ( ) Open Biosystems pmal E2C E2C (1 172) Tan et al. (2004) - Table S1. Summary of Starting DNA-Binding Domain Constructs. The residues of each protein used in each construct are shown in parenthesis. Full-length proteins are designated fl. 2
3 Plasmid Fusion Protein DNA Binding Domain Fluc Fragment pcdna3.1(+) MeCP2-PWNFluc MeCP2-NFluc MeCP2 (77-167) NFluc (2-416) pcdna3.1(+) MBD1-PWNFluc MBD1-NFluc MBD1 (1-69) NFluc (2-416) pcdna3.1(+) MBD2-PWNFluc MBD2-NFluc MBD2 ( ) NFluc (2-416) pcdna3.1(+) MBD3-PWNFluc MBD3-NFluc MBD3 (1-72) NFluc (2-416) pcdna3.1(+) MBD4-PWNFluc MBD4-NFluc MBD4 (76-148) NFluc (2-416) pcdna3.1(+) ZBTB4-PWNFluc ZBTB4-NFluc ZBTB4 ( ) NFluc (2-416) pcdna3.1(+) ZBTB33-PWNFluc ZBTB33-NFluc ZBTB33 ( ) NFluc (2-416) pcdna3.1(+) ZBTB38-PWNFluc ZBTB38-NFluc ZBTB38 ( ) NFluc (2-416) pcdna3.1(+) UHRF1-PWNFluc UHRF1-NFluc UHRF1 ( ) NFluc (2-416) pcdna3.1(+) UHRF2-PWNFluc UHRF2-NFluc UHRF2 ( ) NFluc (2-416) pcdna3.1(+) PWCFluc-Zif268 PWCFluc-Zif268 Zif268 ( ) CFluc ( ) pdnc CFluc-Zif268 Zif268 ( ) CFluc ( ) pdnc MBD1-NFluc MBD1-NFluc MBD1 (1 69) NFluc (2-416) pdnc CFluc-E2C CFluc-E2C E2C (1 172) CFluc ( ) pdnc CFluc-MBD1 CFluc-MBD1 MBD1 (1 69) CFluc ( ) Table S2. Summary of Synthesized Fusion Constructs. The residues of each protein used in each construct are shown in parenthesis. 3
4 Target C2E(m) C2Z(u) C2Z(tm) C2Z(bm) C2Z(m) C6C(m) C21C(m) Sequence 5 -GCGTA m CGTACACTGCGGCTCCGGCCCCTACCG-3 3 -CGCATGC m ATGTGACGCCGAGGCCGGGGATGGC-5 5 -GCGTACGTACGCCCACGCCACCG-3 3 -CGCATGCATGCGGGTGCGGTGGC-5 5 -GCGTA m CGTACGCCCACGCCACCG-3 3 -CGCATGCATGCGGGTGCGGTGGC-5 5 -GCGTACGTACGCCCACGCCACCG-3 3 -CGCATGC m ATGCGGGTGCGGTGGC-5 5 -GCGTA m CGTACGCCCACGCCACCG-3 3 -CGCATGC m ATGCGGGTGCGGTGGC-5 5 -GATCA m CGATGGTA m CGACTAG-3 3 -CTAGTGC m TACCATGC m TGATC-5 5 -GCCTA m CGACTATCACCGCGGGTGATAGT m CGTAGGC-3 3 -CGGATGC m TGATAGTGGCGCCCACTATCAGC m ATCCG-5 Table S3. Summary of Target Oligonucleotides. The appropriate zinc finger sites are highlighted in blue, methylated CpG dinucleotides in red, and unmethylated CpG dinucleotides in green. All oligos were synthesized by Integrated DNA Technologies and HPLC purified. 4
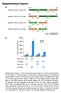
5 Figure S1. Interrogation of MBD-Fluc Translation Efficiency and MBD1/E2C Limit of Detection. a) 35 S- methionine labeled SDS-PAGE gel of in vitro translated MBD-NFluc fusion proteins. Two trials of the same experiment are shown. b) Relative luminescence in the presence of decreasing concentrations of the target, C2E(m), using the sensor comprising MBD1-NFluc and CFluc-E2C, shows a limit of detection (LOD) of 500 amols. 5
6 Figure S2. Exogenous Methylation of DNA. (a) Plasmid pet(u) was treated with a CpG methyltransferase to yield a fully methylated plasmid, pet(m). (b) Natively-methylated HeLa genomic DNA HeLa(n) was treated with a CpG methyltransferase to yield the fully methylated counterpart, HeLa(m). (c) HeLa(n) and HeLa(m) were digested with McrBC endonuclease, while pet(u) and pet(m) were digested with both BstUI and McrBC endonucleases. BstUI cleaves the symmetric 5'-CGCG-3 site only in the absence of methylation, while McrBC cleaves between two distant (G/A)mC sites separated by up to 3000 basepairs. BstUI fully degrades unmodified pet(u), while the fully methylated pet(m) is not digested. McrBC degrades HeLa(n), HeLa(m) and pet(m), indicative of CpG methylation. 6
7 Figure S3. Evaluation of Global HeLa Methylation. Genomic DNA extracted from 5-aza- 2 deoxycytidine- or vehicle-treated HeLa cells was digested with McrBC endonuclease. McrBC degrades vehicle-treated DNA the most, followed by 0.10 μm 5-aza-2 deoxycytidine and 1.0 μm 5- aza-2 deoxycytidine, indicating decreased genomic DNA methylation with the addition of 5-aza- 2 deoxycytidine. 7
8 Supplementary Protocol - CGContentAnalyzer Code (Java): import java.io.bufferedreader; import java.io.bufferedwriter; import java.io.filenotfoundexception; import java.io.filereader; import java.io.filewriter; import java.io.ioexception; import java.util.treemap; public class CGContentAnalyzer { private TreeMap<Integer, Integer> histogram; private String sequence; public CGContentAnalyzer(String filename) { StringBuilder temporarysequence = new StringBuilder(); try { histogram = new TreeMap<Integer, Integer>(); BufferedReader reader = new BufferedReader(new FileReader(fileName)); String nextline = reader.readline(); while (nextline!= null) { temporarysequence.append(nextline); nextline = reader.readline(); sequence = sequencebuilder(temporarysequence.tostring()); reader.close(); catch (FileNotFoundException fnfe) { fnfe.printstacktrace(); catch (IOException ioe) { ioe.printstacktrace(); private String sequencebuilder(string sequence) { StringBuilder result = new StringBuilder(); sequence = sequence.touppercase(); for (int index = 0; index < sequence.length(); index++) if (isvalidbase(sequence.charat(index)) == true) result.append(sequence.charat(index)); return result.tostring(); private boolean isvalidbase(char possiblebase) { return possiblebase == 'A' possiblebase == 'G' possiblebase == 'C' possiblebase == 'T'; public void run() { int CGcounter = 0; int temporarycounter = 0; int CGindex = 0; 8
9 2)) == false) { // find the first occurrence of a CG pair while ( CGindex < sequence.length() -1 && "CG".equals(sequence.substring(CGindex, CGindex + CGindex++; // rearrange circular plasmid to start indexing at a CG pair sequence = sequence.substring(cgindex) + sequence.substring(0, CGindex); System.out.println("Imported " + sequence.length() + " nucleotides"); for (int index = 0; index < sequence.length() - 1; index++) { // if a CG pair is found, the length between this and the last is // added to the histogram if ("CG".equals(sequence.substring(index, index + 2))) { index++; CGcounter++; if (histogram.containskey(temporarycounter)) { histogram.put(temporarycounter, histogram.get(temporarycounter) + 1); else { histogram.put(temporarycounter, 1); temporarycounter = 0; else temporarycounter++; // boundary condition to count the distance from the last CG pair temporarycounter++; if (histogram.containskey(temporarycounter)) { histogram.put(temporarycounter, histogram.get(temporarycounter) + 1); else { histogram.put(temporarycounter, 1); // remove the first CG pair encountered at onset histogram.put(0, histogram.get(0) - 1); try { BufferedWriter writer = new BufferedWriter(new FileWriter( "results.txt")); writer.write("number of CG's: " + CGcounter); writer.newline(); writer.write("length" + "\t" + "Occurences"); writer.newline(); for (int key : histogram.keyset()) { writer.write(key + "\t" + histogram.get(key)); writer.newline(); writer.close(); catch (FileNotFoundException fnfe) { fnfe.printstacktrace(); catch (IOException ioe) { ioe.printstacktrace(); 9
10 mcpg Distance Separation Calculation by CGContentAnalyzer. The function for determining the occurrences of CG in a plasmid was written in Java 6 and utilized via a simple consol interface. The function begins by constructing a CGContentAnalyzer object using the supplied plasmid file name. A histogram was stored as a TreeMap with the key representing the distance between CG pairs and the value of the number of occurrences at this length. Data was read into a StringBuilder, instead of a String, to decrease runtime due to multiple concatenation events. The temporary sequence is then converted to a String representing a sequence containing only A, C, G, T characters. This allows the user to analyze sequences containing line breaks and numbering of base pairs. The run method is then used to analyze the sequence generated in the constructor. The first step iterates over the sequence to find the first CG pair and wraps the sequence to ensure the start of the sequence is a CG pair. This is a valid operation as the sequences were circular plasmids. Next, the method iterates through the entire sequence detecting CG pairs. When a CG pair is found, the number of bases iterated since the last CG pair is recorded in the histogram, otherwise the counter for number of bases since the last CG event is incremented. After the sequence has been scanned completely, the final counter is added to the histogram to count the final CG separation. Next the histogram value at 0 is decremented to prevent counting the first CG twice. Finally the results are written to a text file for further analysis. 10
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.
 Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
SELECTED TECHNIQUES AND APPLICATIONS IN MOLECULAR GENETICS
 SELECTED TECHNIQUES APPLICATIONS IN MOLECULAR GENETICS Restriction Enzymes 15.1.1 The Discovery of Restriction Endonucleases p. 420 2 2, 3, 4, 6, 7, 8 Assigned Reading in Snustad 6th ed. 14.1.1 The Discovery
SELECTED TECHNIQUES APPLICATIONS IN MOLECULAR GENETICS Restriction Enzymes 15.1.1 The Discovery of Restriction Endonucleases p. 420 2 2, 3, 4, 6, 7, 8 Assigned Reading in Snustad 6th ed. 14.1.1 The Discovery
XXII DNA cloning and sequencing. Outline
 XXII DNA cloning and sequencing 1) Deriving DNA for cloning Outline 2) Vectors; forming recombinant DNA; cloning DNA; and screening for clones containing recombinant DNA [replica plating and autoradiography;
XXII DNA cloning and sequencing 1) Deriving DNA for cloning Outline 2) Vectors; forming recombinant DNA; cloning DNA; and screening for clones containing recombinant DNA [replica plating and autoradiography;
Nature Biotechnology: doi: /nbt.4198
 Supplementary Figure 1 Effect of DNA methylation on the base editing efficiency induced by BE3. (a) DNMT3-induced DNA methylation leads to the decreased base editing efficiency induced by BE3. Top panels,
Supplementary Figure 1 Effect of DNA methylation on the base editing efficiency induced by BE3. (a) DNMT3-induced DNA methylation leads to the decreased base editing efficiency induced by BE3. Top panels,
Shaping Epigenetics Discovery
 Shaping Epigenetics Discovery Kits, Assays, Controls, Inhibitors and Antibodies for DNA Methylation The life science business of Merck KGaA, Darmstadt, Germany operates as MilliporeSigma in the U.S. and
Shaping Epigenetics Discovery Kits, Assays, Controls, Inhibitors and Antibodies for DNA Methylation The life science business of Merck KGaA, Darmstadt, Germany operates as MilliporeSigma in the U.S. and
Suppl. Table S1. Characteristics of DHS regions analyzed by bisulfite sequencing. No. CpGs analyzed in the amplicon. Genomic location specificity
 Suppl. Table S1. Characteristics of DHS regions analyzed by bisulfite sequencing. DHS/GRE Genomic location Tissue specificity DHS type CpG density (per 100 bp) No. CpGs analyzed in the amplicon CpG within
Suppl. Table S1. Characteristics of DHS regions analyzed by bisulfite sequencing. DHS/GRE Genomic location Tissue specificity DHS type CpG density (per 100 bp) No. CpGs analyzed in the amplicon CpG within
Multiple choice questions (numbers in brackets indicate the number of correct answers)
 1 Multiple choice questions (numbers in brackets indicate the number of correct answers) February 1, 2013 1. Ribose is found in Nucleic acids Proteins Lipids RNA DNA (2) 2. Most RNA in cells is transfer
1 Multiple choice questions (numbers in brackets indicate the number of correct answers) February 1, 2013 1. Ribose is found in Nucleic acids Proteins Lipids RNA DNA (2) 2. Most RNA in cells is transfer
Practical solutions. for DNA methylation analysis
 DNA Methylation Analysis Tools Practical solutions for DNA methylation analysis Highly efficient and time-saving solutions Locus-specific and whole genome analysis Robust performance Innovative. Fast.
DNA Methylation Analysis Tools Practical solutions for DNA methylation analysis Highly efficient and time-saving solutions Locus-specific and whole genome analysis Robust performance Innovative. Fast.
Supplementary Figure S1 Supplementary Figure S2 Supplementary Figure S3. Supplementary Figure S4
 Supplementary Figure S1 Supplementary Figure S2 Supplementary Figure S3 Supplementary Figure S4 Supplementary Figure S5 Supplementary Figure S6 Supplementary Figure S7 Supplementary Figure S8 Supplementary
Supplementary Figure S1 Supplementary Figure S2 Supplementary Figure S3 Supplementary Figure S4 Supplementary Figure S5 Supplementary Figure S6 Supplementary Figure S7 Supplementary Figure S8 Supplementary
Identification of Putative Coding Regions in a 10kB Sequence of the E. coli 536 Genome. By Someone. Partner: Someone else
 1 Identification of Putative Coding Regions in a 10kB Sequence of the E. coli 536 Genome Introduction: By Someone Partner: Someone else The instructions required for the maintenance of life are encoded
1 Identification of Putative Coding Regions in a 10kB Sequence of the E. coli 536 Genome Introduction: By Someone Partner: Someone else The instructions required for the maintenance of life are encoded
Supporting Information
 Supporting Information Luminescence sensing for qualitative and quantitative detection of 5-methylcytosine Yushu Yuan,, Tingting Hong,, Yi Chen, Yafen Wang, Xueping Qiu, Fang Zheng, Xiaocheng Weng, *,
Supporting Information Luminescence sensing for qualitative and quantitative detection of 5-methylcytosine Yushu Yuan,, Tingting Hong,, Yi Chen, Yafen Wang, Xueping Qiu, Fang Zheng, Xiaocheng Weng, *,
Transcriptional Regulation of Pro-apoptotic Protein Kinase C-delta: Implications for Oxidative Stress-induced Neuronal Cell Death
 SUPPLEMENTAL DATA Transcriptional Regulation of Pro-apoptotic Protein Kinase C-delta: Implications for Oxidative Stress-induced Neuronal Cell Death Huajun Jin 1, Arthi Kanthasamy 1, Vellareddy Anantharam
SUPPLEMENTAL DATA Transcriptional Regulation of Pro-apoptotic Protein Kinase C-delta: Implications for Oxidative Stress-induced Neuronal Cell Death Huajun Jin 1, Arthi Kanthasamy 1, Vellareddy Anantharam
MCB 102 University of California, Berkeley August 11 13, Problem Set 8
 MCB 102 University of California, Berkeley August 11 13, 2009 Isabelle Philipp Handout Problem Set 8 The answer key will be posted by Tuesday August 11. Try to solve the problem sets always first without
MCB 102 University of California, Berkeley August 11 13, 2009 Isabelle Philipp Handout Problem Set 8 The answer key will be posted by Tuesday August 11. Try to solve the problem sets always first without
Contents... vii. List of Figures... xii. List of Tables... xiv. Abbreviatons... xv. Summary... xvii. 1. Introduction In vitro evolution...
 vii Contents Contents... vii List of Figures... xii List of Tables... xiv Abbreviatons... xv Summary... xvii 1. Introduction...1 1.1 In vitro evolution... 1 1.2 Phage Display Technology... 3 1.3 Cell surface
vii Contents Contents... vii List of Figures... xii List of Tables... xiv Abbreviatons... xv Summary... xvii 1. Introduction...1 1.1 In vitro evolution... 1 1.2 Phage Display Technology... 3 1.3 Cell surface
N173A. residue number
 a 0.3 Δω 1 ( 1 H Ν ) [ppm] 0-0.3 N173A -0.6 Y177A b Δω 3 ( 13 C α ) [ppm] 2 0-2 c Δω 3 ( 13 C β ) [ppm] 2 0-2 -4 Supplementary Figure 1 140 150 160 170 180 190 200 residue number Comparison of chemical
a 0.3 Δω 1 ( 1 H Ν ) [ppm] 0-0.3 N173A -0.6 Y177A b Δω 3 ( 13 C α ) [ppm] 2 0-2 c Δω 3 ( 13 C β ) [ppm] 2 0-2 -4 Supplementary Figure 1 140 150 160 170 180 190 200 residue number Comparison of chemical
SUPPLEMENTARY EXPEMENTAL PROCEDURES
 SUPPLEMENTARY EXPEMENTAL PROCEDURES Plasmids- Total RNAs were extracted from HeLaS3 cells and reverse-transcribed using Superscript III Reverse Transcriptase (Invitrogen) to obtain DNA template for the
SUPPLEMENTARY EXPEMENTAL PROCEDURES Plasmids- Total RNAs were extracted from HeLaS3 cells and reverse-transcribed using Superscript III Reverse Transcriptase (Invitrogen) to obtain DNA template for the
MethylMagnet mcpg DNA Isolation Kit
 MethylMagnet mcpg DNA Isolation Kit User Manual Version O 9-21-15 Catalog # MM101 Table of Contents Kit Components and Storage.. 2 MethylMagnet Work Flow 3 About MethylMagnet Proteins 4 MethylMagnet mcpg
MethylMagnet mcpg DNA Isolation Kit User Manual Version O 9-21-15 Catalog # MM101 Table of Contents Kit Components and Storage.. 2 MethylMagnet Work Flow 3 About MethylMagnet Proteins 4 MethylMagnet mcpg
Supplemental Information
 Supplemental Information ATP-dependent unwinding of U4/U6 snrnas by the Brr2 helicase requires the C-terminus of Prp8 Corina Maeder 1,3, Alan K. Kutach 1,2,3, and Christine Guthrie 1 1 Department of Biochemistry
Supplemental Information ATP-dependent unwinding of U4/U6 snrnas by the Brr2 helicase requires the C-terminus of Prp8 Corina Maeder 1,3, Alan K. Kutach 1,2,3, and Christine Guthrie 1 1 Department of Biochemistry
Deep sequencing reveals global patterns of mrna recruitment
 Supplementary information for: Deep sequencing reveals global patterns of mrna recruitment during translation initiation Rong Gao 1#*, Kai Yu 1#, Ju-Kui Nie 1,Teng-Fei Lian 1, Jian-Shi Jin 1, Anders Liljas
Supplementary information for: Deep sequencing reveals global patterns of mrna recruitment during translation initiation Rong Gao 1#*, Kai Yu 1#, Ju-Kui Nie 1,Teng-Fei Lian 1, Jian-Shi Jin 1, Anders Liljas
Supporting Information
 Supporting Information Wiley-VCH 2006 69451 Weinheim, Germany RNA ligands that distinguish metabolite-induced conformations in the TPP riboswitch Günter Mayer, Marie-Sophie L. Raddatz, Julia D. Grunwald,
Supporting Information Wiley-VCH 2006 69451 Weinheim, Germany RNA ligands that distinguish metabolite-induced conformations in the TPP riboswitch Günter Mayer, Marie-Sophie L. Raddatz, Julia D. Grunwald,
Basics of Recombinant DNA Technology Biochemistry 302. March 5, 2004 Bob Kelm
 Basics of Recombinant DNA Technology Biochemistry 302 March 5, 2004 Bob Kelm Applications of recombinant DNA technology Mapping and identifying genes (DNA cloning) Propagating genes (DNA subcloning) Modifying
Basics of Recombinant DNA Technology Biochemistry 302 March 5, 2004 Bob Kelm Applications of recombinant DNA technology Mapping and identifying genes (DNA cloning) Propagating genes (DNA subcloning) Modifying
Molecular Genetics Techniques. BIT 220 Chapter 20
 Molecular Genetics Techniques BIT 220 Chapter 20 What is Cloning? Recombinant DNA technologies 1. Producing Recombinant DNA molecule Incorporate gene of interest into plasmid (cloning vector) 2. Recombinant
Molecular Genetics Techniques BIT 220 Chapter 20 What is Cloning? Recombinant DNA technologies 1. Producing Recombinant DNA molecule Incorporate gene of interest into plasmid (cloning vector) 2. Recombinant
Experiment 5. Restriction Enzyme Digest and Plasmid Mapping. VY NGUYEN 26 February 2016
 Experiment 5 Restriction Enzyme Digest and Plasmid Mapping VY NGUYEN 26 February 2016 ABSTRACT 1. Understand the use of restriction enzymes as biotechnology tools 2. Become familiar with the principles
Experiment 5 Restriction Enzyme Digest and Plasmid Mapping VY NGUYEN 26 February 2016 ABSTRACT 1. Understand the use of restriction enzymes as biotechnology tools 2. Become familiar with the principles
MassArray Analytical Tools
 MassArray Analytical Tools Reid F. Thompson and John M. Greally April 30, 08 Contents Introduction Changes for MassArray in current BioC release 3 3 Optimal Amplicon Design 4 4 Conversion Controls 0 5
MassArray Analytical Tools Reid F. Thompson and John M. Greally April 30, 08 Contents Introduction Changes for MassArray in current BioC release 3 3 Optimal Amplicon Design 4 4 Conversion Controls 0 5
3 Designing Primers for Site-Directed Mutagenesis
 3 Designing Primers for Site-Directed Mutagenesis 3.1 Learning Objectives During the next two labs you will learn the basics of site-directed mutagenesis: you will design primers for the mutants you designed
3 Designing Primers for Site-Directed Mutagenesis 3.1 Learning Objectives During the next two labs you will learn the basics of site-directed mutagenesis: you will design primers for the mutants you designed
Computational Biology 2. Pawan Dhar BII
 Computational Biology 2 Pawan Dhar BII Lecture 1 Introduction to terms, techniques and concepts in molecular biology Molecular biology - a primer Human body has 100 trillion cells each containing 3 billion
Computational Biology 2 Pawan Dhar BII Lecture 1 Introduction to terms, techniques and concepts in molecular biology Molecular biology - a primer Human body has 100 trillion cells each containing 3 billion
Technical tips Session 4
 Technical tips Session 4 Biotinylation assay: Streptavidin is a small bacterial protein that binds with high affinity to the vitamin biotin. This streptavidin-biotin combination can be used to link molecules
Technical tips Session 4 Biotinylation assay: Streptavidin is a small bacterial protein that binds with high affinity to the vitamin biotin. This streptavidin-biotin combination can be used to link molecules
New Plant Breeding Technologies
 New Plant Breeding Technologies Ricarda A. Steinbrecher, PhD EcoNexus / ENSSER Berlin, 07 May 2015 r.steinbrecher@econexus.info distributed by EuropaBio What are the NPBTs? *RNAi *Epigenetic alterations
New Plant Breeding Technologies Ricarda A. Steinbrecher, PhD EcoNexus / ENSSER Berlin, 07 May 2015 r.steinbrecher@econexus.info distributed by EuropaBio What are the NPBTs? *RNAi *Epigenetic alterations
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/3/11/eaao1799/dc1 Supplementary Materials for Measuring quantitative effects of methylation on transcription factor DNA binding affinity Zheng Zuo, Basab Roy, Yiming
advances.sciencemag.org/cgi/content/full/3/11/eaao1799/dc1 Supplementary Materials for Measuring quantitative effects of methylation on transcription factor DNA binding affinity Zheng Zuo, Basab Roy, Yiming
Construct Design and Cloning Guide for Cas9-triggered homologous recombination
 Construct Design and Cloning Guide for Cas9-triggered homologous recombination Written by Dan Dickinson (ddickins@live.unc.edu) and last updated December 2013. Reference: Dickinson DJ, Ward JD, Reiner
Construct Design and Cloning Guide for Cas9-triggered homologous recombination Written by Dan Dickinson (ddickins@live.unc.edu) and last updated December 2013. Reference: Dickinson DJ, Ward JD, Reiner
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1137999/dc1 Supporting Online Material for Disrupting the Pairing Between let-7 and Enhances Oncogenic Transformation Christine Mayr, Michael T. Hemann, David P. Bartel*
www.sciencemag.org/cgi/content/full/1137999/dc1 Supporting Online Material for Disrupting the Pairing Between let-7 and Enhances Oncogenic Transformation Christine Mayr, Michael T. Hemann, David P. Bartel*
Methylamp Coupled DNA Isolation & Modification Kit
 Methylamp Coupled DNA Isolation & Modification Kit Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The Methylamp Coupled DNA Isolation & Modification Kit is very suitable for methylation research
Methylamp Coupled DNA Isolation & Modification Kit Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The Methylamp Coupled DNA Isolation & Modification Kit is very suitable for methylation research
GENETICS EXAM 3 FALL a) is a technique that allows you to separate nucleic acids (DNA or RNA) by size.
 Student Name: All questions are worth 5 pts. each. GENETICS EXAM 3 FALL 2004 1. a) is a technique that allows you to separate nucleic acids (DNA or RNA) by size. b) Name one of the materials (of the two
Student Name: All questions are worth 5 pts. each. GENETICS EXAM 3 FALL 2004 1. a) is a technique that allows you to separate nucleic acids (DNA or RNA) by size. b) Name one of the materials (of the two
SUPPLEMENTARY INFORMATION
 The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix Hideharu Hashimoto 1, John R. Horton 1, Xing Zhang 1, Magnolia Bostick 2, Steven Jacobsen 2,3, and Xiaodong Cheng 1 1 Department of Biochemistry,
The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix Hideharu Hashimoto 1, John R. Horton 1, Xing Zhang 1, Magnolia Bostick 2, Steven Jacobsen 2,3, and Xiaodong Cheng 1 1 Department of Biochemistry,
Chapter 13: Biotechnology
 Chapter Review 1. Explain why the brewing of beer is considered to be biotechnology. The United Nations defines biotechnology as any technological application that uses biological system, living organism,
Chapter Review 1. Explain why the brewing of beer is considered to be biotechnology. The United Nations defines biotechnology as any technological application that uses biological system, living organism,
Lecture 9. Eukaryotic gene regulation: DNA METHYLATION
 Lecture 9 Eukaryotic gene regulation: DNA METHYLATION Recap.. Eukaryotic RNA polymerases Core promoter elements General transcription factors Enhancers and upstream activation sequences Transcriptional
Lecture 9 Eukaryotic gene regulation: DNA METHYLATION Recap.. Eukaryotic RNA polymerases Core promoter elements General transcription factors Enhancers and upstream activation sequences Transcriptional
Supplementary Figure 1. NORAD expression in mouse (A) and dog (B). The black boxes indicate the position of the regions alignable to the 12 repeat
 Supplementary Figure 1. NORAD expression in mouse (A) and dog (B). The black boxes indicate the position of the regions alignable to the 12 repeat units in the human genome. Annotated transposable elements
Supplementary Figure 1. NORAD expression in mouse (A) and dog (B). The black boxes indicate the position of the regions alignable to the 12 repeat units in the human genome. Annotated transposable elements
Supplementary Figures
 Supplementary Figures a HindIII XbaI pcdna3.1/v5-his A_Nfluc-VVD 1-398 37-186 Nfluc Linker 1 VVD HindIII XhoI pcdna3.1/v5-his A_VVD-Nfluc 37-186 1-398 VVD Linker 2 Nfluc HindIII XbaI pcdna3.1/v5-his A_Cfluc-VVD
Supplementary Figures a HindIII XbaI pcdna3.1/v5-his A_Nfluc-VVD 1-398 37-186 Nfluc Linker 1 VVD HindIII XhoI pcdna3.1/v5-his A_VVD-Nfluc 37-186 1-398 VVD Linker 2 Nfluc HindIII XbaI pcdna3.1/v5-his A_Cfluc-VVD
Biotechnology Project VI lesson. 1 - Subcloning NRG1-III-β3 from pcr-blunt II-TOPO into the expression vector pegfp-c3
 26-11-2018 - Biotechnology Project VI lesson 1 - Subcloning NRG1-III-β3 from pcr-blunt II-TOPO into the expression vector pegfp-c3 2 - Subcloning NRG1-III-β3 from pcr-blunt II-TOPO or pegfp-c3 into the
26-11-2018 - Biotechnology Project VI lesson 1 - Subcloning NRG1-III-β3 from pcr-blunt II-TOPO into the expression vector pegfp-c3 2 - Subcloning NRG1-III-β3 from pcr-blunt II-TOPO or pegfp-c3 into the
Selected Techniques Part I
 1 Selected Techniques Part I Gel Electrophoresis Can be both qualitative and quantitative Qualitative About what size is the fragment? How many fragments are present? Is there in insert or not? Quantitative
1 Selected Techniques Part I Gel Electrophoresis Can be both qualitative and quantitative Qualitative About what size is the fragment? How many fragments are present? Is there in insert or not? Quantitative
Simple protocol for gene editing using GenCrisprTM Cas9 nuclease
 Simple protocol for gene editing using GenCrisprTM Cas9 nuclease Contents Protocol Step 1: Choose the target DNA sequence Step 2: Design sgrna Step 3: Preparation for sgrna 3.1 In vitro transcription of
Simple protocol for gene editing using GenCrisprTM Cas9 nuclease Contents Protocol Step 1: Choose the target DNA sequence Step 2: Design sgrna Step 3: Preparation for sgrna 3.1 In vitro transcription of
Supporting Information
 Supporting Information Mirror bisulfite sequencing a method for single-base resolution of hydroxymethylcytosine Darany Tan #, Tzu Hung Chung #, Xueguang Sun *, Xi-Yu Jia. School of Life Sciences, Guangzhou
Supporting Information Mirror bisulfite sequencing a method for single-base resolution of hydroxymethylcytosine Darany Tan #, Tzu Hung Chung #, Xueguang Sun *, Xi-Yu Jia. School of Life Sciences, Guangzhou
Review of previous work: Overview of previous work on DNA methylation and chromatin dynamics
 CONTENTS Preface Acknowledgements Lists of Commonly Used Abbreviations Review of previous work: Overview of previous work on DNA methylation and chromatin dynamics Introduction Enzymes involved in DNA
CONTENTS Preface Acknowledgements Lists of Commonly Used Abbreviations Review of previous work: Overview of previous work on DNA methylation and chromatin dynamics Introduction Enzymes involved in DNA
Understanding the Cellular Mechanism of the Excess Microsporocytes I (EMSI) Gene. Andrew ElBardissi, The Pennsylvania State University
 Understanding the Cellular Mechanism of the Excess Microsporocytes I (EMSI) Gene Andrew ElBardissi, The Pennsylvania State University Abstract: Hong Ma, The Pennsylvania State University The Excess Microsporocytes
Understanding the Cellular Mechanism of the Excess Microsporocytes I (EMSI) Gene Andrew ElBardissi, The Pennsylvania State University Abstract: Hong Ma, The Pennsylvania State University The Excess Microsporocytes
Chapter 6 - Molecular Genetic Techniques
 Chapter 6 - Molecular Genetic Techniques Two objects of molecular & genetic technologies For analysis For generation Molecular genetic technologies! For analysis DNA gel electrophoresis Southern blotting
Chapter 6 - Molecular Genetic Techniques Two objects of molecular & genetic technologies For analysis For generation Molecular genetic technologies! For analysis DNA gel electrophoresis Southern blotting
Recombinant DNA Technology
 Recombinant DNA Technology Common General Cloning Strategy Target DNA from donor organism extracted, cut with restriction endonuclease and ligated into a cloning vector cut with compatible restriction
Recombinant DNA Technology Common General Cloning Strategy Target DNA from donor organism extracted, cut with restriction endonuclease and ligated into a cloning vector cut with compatible restriction
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature09937 a Name Position Primersets 1a 1b 2 3 4 b2 Phenotype Genotype b Primerset 1a D T C R I E 10000 8000 6000 5000 4000 3000 2500 2000 1500 1000 800 Donor (D)
SUPPLEMENTARY INFORMATION doi:10.1038/nature09937 a Name Position Primersets 1a 1b 2 3 4 b2 Phenotype Genotype b Primerset 1a D T C R I E 10000 8000 6000 5000 4000 3000 2500 2000 1500 1000 800 Donor (D)
7.1 Techniques for Producing and Analyzing DNA. SBI4U Ms. Ho-Lau
 7.1 Techniques for Producing and Analyzing DNA SBI4U Ms. Ho-Lau What is Biotechnology? From Merriam-Webster: the manipulation of living organisms or their components to produce useful usually commercial
7.1 Techniques for Producing and Analyzing DNA SBI4U Ms. Ho-Lau What is Biotechnology? From Merriam-Webster: the manipulation of living organisms or their components to produce useful usually commercial
CRISPR RNA-guided activation of endogenous human genes
 CRISPR RNA-guided activation of endogenous human genes Morgan L Maeder, Samantha J Linder, Vincent M Cascio, Yanfang Fu, Quan H Ho, J Keith Joung Supplementary Figure 1 Comparison of VEGF activation induced
CRISPR RNA-guided activation of endogenous human genes Morgan L Maeder, Samantha J Linder, Vincent M Cascio, Yanfang Fu, Quan H Ho, J Keith Joung Supplementary Figure 1 Comparison of VEGF activation induced
Allele-specific locus binding and genome editing by CRISPR at the
 Supplementary Information Allele-specific locus binding and genome editing by CRISPR at the p6ink4a locus Toshitsugu Fujita, Miyuki Yuno, and Hodaka Fujii Supplementary Figure Legends Supplementary Figure
Supplementary Information Allele-specific locus binding and genome editing by CRISPR at the p6ink4a locus Toshitsugu Fujita, Miyuki Yuno, and Hodaka Fujii Supplementary Figure Legends Supplementary Figure
Site-directed Mutagenesis
 Site-directed Mutagenesis Applications Subtilisin (Met à Ala mutation resistant to oxidation) Fluorescent proteins Protein structure-function Substrate trapping mutants Identify regulatory regions/sequences
Site-directed Mutagenesis Applications Subtilisin (Met à Ala mutation resistant to oxidation) Fluorescent proteins Protein structure-function Substrate trapping mutants Identify regulatory regions/sequences
SUPPLEMENTARY INFORMATION
 Figure S1 Schematic of expressed protein ligation (EPL). Protein-αthioester is generated by thiolysis of the corresponding recombinant intein fusion protein. A native chemical ligation reaction is performed
Figure S1 Schematic of expressed protein ligation (EPL). Protein-αthioester is generated by thiolysis of the corresponding recombinant intein fusion protein. A native chemical ligation reaction is performed
Researchers use genetic engineering to manipulate DNA.
 Section 2: Researchers use genetic engineering to manipulate DNA. K What I Know W What I Want to Find Out L What I Learned Essential Questions What are the different tools and processes used in genetic
Section 2: Researchers use genetic engineering to manipulate DNA. K What I Know W What I Want to Find Out L What I Learned Essential Questions What are the different tools and processes used in genetic
Fisher (Fairlawn, NJ) and Sigma-Aldrich (St. Louis, MO) and were used without further. (Promega) and DpnI (New England Biolabs, Beverly, MA).
 175 Appendix III Chapter 4 Methods General. Unless otherwise noted, reagents were purchased from the commercial suppliers Fisher (Fairlawn, NJ) and Sigma-Aldrich (St. Louis, MO) and were used without further
175 Appendix III Chapter 4 Methods General. Unless otherwise noted, reagents were purchased from the commercial suppliers Fisher (Fairlawn, NJ) and Sigma-Aldrich (St. Louis, MO) and were used without further
Some types of Mutagenesis
 Mutagenesis What Is a Mutation? Genetic information is encoded by the sequence of the nucleotide bases in DNA of the gene. The four nucleotides are: adenine (A), thymine (T), guanine (G), and cytosine
Mutagenesis What Is a Mutation? Genetic information is encoded by the sequence of the nucleotide bases in DNA of the gene. The four nucleotides are: adenine (A), thymine (T), guanine (G), and cytosine
XactEdit Cas9 Nuclease with NLS User Manual
 XactEdit Cas9 Nuclease with NLS User Manual An RNA-guided recombinant endonuclease for efficient targeted DNA cleavage Catalog Numbers CE1000-50K, CE1000-50, CE1000-250, CE1001-250, CE1001-1000 Table of
XactEdit Cas9 Nuclease with NLS User Manual An RNA-guided recombinant endonuclease for efficient targeted DNA cleavage Catalog Numbers CE1000-50K, CE1000-50, CE1000-250, CE1001-250, CE1001-1000 Table of
Legend for Supplemental Figures and Tables
 Legend for Supplemental Figures and Tables Supplemental Fig. 1. Negative regulation of the CYP27B1 promoter in a ligand-dependent manner (A) OK-P cells were transfected with pcdna-trα, pcdna-trβ1 or pcdna3
Legend for Supplemental Figures and Tables Supplemental Fig. 1. Negative regulation of the CYP27B1 promoter in a ligand-dependent manner (A) OK-P cells were transfected with pcdna-trα, pcdna-trβ1 or pcdna3
Gene Expression Technology
 Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
Supplementary Figure 2
 Supplementary Figure 2 a SBS-C1 SBS-C2 SBS-C3 SBS-C4 SBS-C5 SBS-C6 SBS-C7 SBS-C8 SBS-C9 LCR CNS-1 CNS-2 Il5 Rad50 Il13 Il4 Kif3a Sept8 0 50 100 150 200kb Sau3AI 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17
Supplementary Figure 2 a SBS-C1 SBS-C2 SBS-C3 SBS-C4 SBS-C5 SBS-C6 SBS-C7 SBS-C8 SBS-C9 LCR CNS-1 CNS-2 Il5 Rad50 Il13 Il4 Kif3a Sept8 0 50 100 150 200kb Sau3AI 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17
Figure 1. Map of cloning vector pgem T-Easy (bacterial plasmid DNA)
 Texas A&M University-Corpus Christi CHEM4402 Biochemistry II Laboratory Laboratory 6: Ligation & Bacterial Transformation (Bring your text and laptop to class if you wish to work on your assignment during
Texas A&M University-Corpus Christi CHEM4402 Biochemistry II Laboratory Laboratory 6: Ligation & Bacterial Transformation (Bring your text and laptop to class if you wish to work on your assignment during
HPV E6 oncoprotein targets histone methyltransferases for modulating specific. Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu,
 1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
Diagnosis Sanger. Interpreting and Troubleshooting Chromatograms. Volume 1: Help! No Data! GENEWIZ Technical Support
 Diagnosis Sanger Interpreting and Troubleshooting Chromatograms GENEWIZ Technical Support DNAseq@genewiz.com Troubleshooting This troubleshooting guide is based on common issues seen from samples within
Diagnosis Sanger Interpreting and Troubleshooting Chromatograms GENEWIZ Technical Support DNAseq@genewiz.com Troubleshooting This troubleshooting guide is based on common issues seen from samples within
Sequence Analysis Lab Protocol
 Sequence Analysis Lab Protocol You will need this handout of instructions The sequence of your plasmid from the ABI The Accession number for Lambda DNA J02459 The Accession number for puc 18 is L09136
Sequence Analysis Lab Protocol You will need this handout of instructions The sequence of your plasmid from the ABI The Accession number for Lambda DNA J02459 The Accession number for puc 18 is L09136
GenBuilder TM Cloning Kit User Manual
 GenBuilder TM Cloning Kit User Manual Cat.no L00701 Version 11242017 Ⅰ. Introduction... 2 I.1 Product Information... 2 I.2 Kit Contents and Storage... 2 I.3 GenBuilder Cloning Kit Workflow... 2 Ⅱ. DNA
GenBuilder TM Cloning Kit User Manual Cat.no L00701 Version 11242017 Ⅰ. Introduction... 2 I.1 Product Information... 2 I.2 Kit Contents and Storage... 2 I.3 GenBuilder Cloning Kit Workflow... 2 Ⅱ. DNA
Chapter 9 Genetic Engineering
 Chapter 9 Genetic Engineering Biotechnology: use of microbes to make a protein product Recombinant DNA Technology: Insertion or modification of genes to produce desired proteins Genetic engineering: manipulation
Chapter 9 Genetic Engineering Biotechnology: use of microbes to make a protein product Recombinant DNA Technology: Insertion or modification of genes to produce desired proteins Genetic engineering: manipulation
Synthetic Biology. Sustainable Energy. Therapeutics Industrial Enzymes. Agriculture. Accelerating Discoveries, Expanding Possibilities. Design.
 Synthetic Biology Accelerating Discoveries, Expanding Possibilities Sustainable Energy Therapeutics Industrial Enzymes Agriculture Design Build Generate Solutions to Advance Synthetic Biology Research
Synthetic Biology Accelerating Discoveries, Expanding Possibilities Sustainable Energy Therapeutics Industrial Enzymes Agriculture Design Build Generate Solutions to Advance Synthetic Biology Research
Recitation CHAPTER 9 DNA Technologies
 Recitation CHAPTER 9 DNA Technologies DNA Cloning: General Scheme A cloning vector and eukaryotic chromosomes are separately cleaved with the same restriction endonuclease. (A single chromosome is shown
Recitation CHAPTER 9 DNA Technologies DNA Cloning: General Scheme A cloning vector and eukaryotic chromosomes are separately cleaved with the same restriction endonuclease. (A single chromosome is shown
Annotating the Genome (H)
 Annotating the Genome (H) Annotation principles (H1) What is annotation? In general: annotation = explanatory note* What could be useful as an annotation of a DNA sequence? an amino acid sequence? What
Annotating the Genome (H) Annotation principles (H1) What is annotation? In general: annotation = explanatory note* What could be useful as an annotation of a DNA sequence? an amino acid sequence? What
Identification and characterization of DNA aptamers specific for
 SUPPLEMENTARY INFORMATION Identification and characterization of DNA aptamers specific for phosphorylation epitopes of tau protein I-Ting Teng 1,, Xiaowei Li 1,, Hamad Ahmad Yadikar #, Zhihui Yang #, Long
SUPPLEMENTARY INFORMATION Identification and characterization of DNA aptamers specific for phosphorylation epitopes of tau protein I-Ting Teng 1,, Xiaowei Li 1,, Hamad Ahmad Yadikar #, Zhihui Yang #, Long
EpiMark Bisulfite Conversion Kit
 EPIGENETICS EpiMark Bisulfite Conversion Kit Instruction Manual NEB #E3318S 48 reactions Version 3.0 6/18 be INSPIRED drive DISCOVERY stay GENUINE Safety Information: The EpiMark Bisulfite Conversion Kit
EPIGENETICS EpiMark Bisulfite Conversion Kit Instruction Manual NEB #E3318S 48 reactions Version 3.0 6/18 be INSPIRED drive DISCOVERY stay GENUINE Safety Information: The EpiMark Bisulfite Conversion Kit
DNA METHYLATION NH 2 H 3. Solutions using bisulfite conversion and immunocapture Ideal for NGS, Sanger sequencing, Pyrosequencing, and qpcr
 NH 2 DNA METHYLATION H 3 C Solutions using bisulfite conversion and immunocapture Ideal for NGS, Sanger sequencing, Pyrosequencing, and qpcr NH 2 N N H PAGE 3 Understanding DNA Methylation DNA methylation
NH 2 DNA METHYLATION H 3 C Solutions using bisulfite conversion and immunocapture Ideal for NGS, Sanger sequencing, Pyrosequencing, and qpcr NH 2 N N H PAGE 3 Understanding DNA Methylation DNA methylation
Supplementary Online Material. An Expanded Eukaryotic Genetic Code 2QH, UK. * To whom correspondence should be addressed.
 Supplementary Online Material An Expanded Eukaryotic Genetic Code Jason W. Chin 1, T. Ashton Cropp, J. Christopher Anderson, Mridul Mukherji, Zhiwen Zhang and Peter G. Schultz* Department of Chemistry
Supplementary Online Material An Expanded Eukaryotic Genetic Code Jason W. Chin 1, T. Ashton Cropp, J. Christopher Anderson, Mridul Mukherji, Zhiwen Zhang and Peter G. Schultz* Department of Chemistry
GenBuilder TM Plus Cloning Kit User Manual
 GenBuilder TM Plus Cloning Kit User Manual Cat. No. L00744 Version 11242017 Ⅰ. Introduction... 2 I.1 Product Information... 2 I.2 Kit Contents and Storage... 2 I.3 GenBuilder Cloning Kit Workflow... 2
GenBuilder TM Plus Cloning Kit User Manual Cat. No. L00744 Version 11242017 Ⅰ. Introduction... 2 I.1 Product Information... 2 I.2 Kit Contents and Storage... 2 I.3 GenBuilder Cloning Kit Workflow... 2
CELL BIOLOGY - CLUTCH CH. 7 - GENE EXPRESSION.
 !! www.clutchprep.com CONCEPT: CONTROL OF GENE EXPRESSION BASICS Gene expression is the process through which cells selectively to express some genes and not others Every cell in an organism is a clone
!! www.clutchprep.com CONCEPT: CONTROL OF GENE EXPRESSION BASICS Gene expression is the process through which cells selectively to express some genes and not others Every cell in an organism is a clone
GenBuilder TM Plus Cloning Kit User Manual
 GenBuilder TM Plus Cloning Kit User Manual Cat.no L00744 Version 11242017 Ⅰ. Introduction... 2 I.1 Product Information... 2 I.2 Kit Contents and Storage... 2 I.3 GenBuilder Cloning Kit Workflow... 2 Ⅱ.
GenBuilder TM Plus Cloning Kit User Manual Cat.no L00744 Version 11242017 Ⅰ. Introduction... 2 I.1 Product Information... 2 I.2 Kit Contents and Storage... 2 I.3 GenBuilder Cloning Kit Workflow... 2 Ⅱ.
KAPA HiFi HotStart ReadyMix PCR Kit
 KAPA HiFi HotStart ReadyMix PCR Kit KR0370 v10.19 This provides product information and a detailed protocol for the KAPA HiFi HotStart ReadyMix PCR Kit. This document applies to the following kits: 07958919001,
KAPA HiFi HotStart ReadyMix PCR Kit KR0370 v10.19 This provides product information and a detailed protocol for the KAPA HiFi HotStart ReadyMix PCR Kit. This document applies to the following kits: 07958919001,
Fun with DNA polymerase
 Fun with DNA polymerase Why would we want to be able to make copies of DNA? Can you think of a situation where you have only a small amount and would like more? Enzymatic DNA synthesis To use DNA polymerase
Fun with DNA polymerase Why would we want to be able to make copies of DNA? Can you think of a situation where you have only a small amount and would like more? Enzymatic DNA synthesis To use DNA polymerase
A turn-on split-luciferase sensor for the direct detection of poly(adp-ribose) as a marker for DNA repair and cell death
 University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Cliff Stains Publications Published Research - Department of Chemistry 2011 A turn-on split-luciferase sensor for the direct
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Cliff Stains Publications Published Research - Department of Chemistry 2011 A turn-on split-luciferase sensor for the direct
Programmable and Multiparameter DNA-based Logic Platform For Cancer Recognition and Targeted Therapy
 Programmable and Multiparameter DNA-based Logic Platform For Cancer Recognition and Targeted Therapy Mingxu You,, Guizhi Zhu,, Tao Chen,, Michael J. Donovan and Weihong Tan, * Department of Chemistry and
Programmable and Multiparameter DNA-based Logic Platform For Cancer Recognition and Targeted Therapy Mingxu You,, Guizhi Zhu,, Tao Chen,, Michael J. Donovan and Weihong Tan, * Department of Chemistry and
7 Synthesizing the ykkcd Mutant Toxin Sensor RNA in vitro
 7 Synthesizing the ykkcd Mutant Toxin Sensor RNA in vitro 7.1 Learning Objective In the quest toward understanding how the ykkcd toxin sensor recognizes the antibiotic tetracycline you thus far designed
7 Synthesizing the ykkcd Mutant Toxin Sensor RNA in vitro 7.1 Learning Objective In the quest toward understanding how the ykkcd toxin sensor recognizes the antibiotic tetracycline you thus far designed
In silico analysis of complete bacterial genomes: PCR, AFLP-PCR, and endonuclease
 Bioinformatics Advance Access published January 29, 2004 In silico analysis of complete bacterial genomes: PCR, AFLP-PCR, and endonuclease restriction Joseba Bikandi*, Rosario San Millán, Aitor Rementeria,
Bioinformatics Advance Access published January 29, 2004 In silico analysis of complete bacterial genomes: PCR, AFLP-PCR, and endonuclease restriction Joseba Bikandi*, Rosario San Millán, Aitor Rementeria,
Chapter 19 Genetic Regulation of the Eukaryotic Genome. A. Bergeron AP Biology PCHS
 Chapter 19 Genetic Regulation of the Eukaryotic Genome A. Bergeron AP Biology PCHS 2 Do Now - Eukaryotic Transcription Regulation The diagram below shows five genes (with their enhancers) from the genome
Chapter 19 Genetic Regulation of the Eukaryotic Genome A. Bergeron AP Biology PCHS 2 Do Now - Eukaryotic Transcription Regulation The diagram below shows five genes (with their enhancers) from the genome
CRISPR-dCas9 mediated TET1 targeting for selective DNA demethylation at BRCA1 promoter
 CRISPR-dCas9 mediated TET1 targeting for selective DNA demethylation at BRCA1 promoter SUPPLEMENTARY DATA See Supplementary Sequence File: 1 Supplementary Figure S1: The total protein was extracted from
CRISPR-dCas9 mediated TET1 targeting for selective DNA demethylation at BRCA1 promoter SUPPLEMENTARY DATA See Supplementary Sequence File: 1 Supplementary Figure S1: The total protein was extracted from
SUPPLEMENTARY INFORMATION. Tolerance of a knotted near infrared fluorescent protein to random circular permutation
 SUPPLEMENTARY INFORMATION Tolerance of a knotted near infrared fluorescent protein to random circular permutation Naresh Pandey 1,3, Brianna E. Kuypers 2,4, Barbara Nassif 1, Emily E. Thomas 1,3, Razan
SUPPLEMENTARY INFORMATION Tolerance of a knotted near infrared fluorescent protein to random circular permutation Naresh Pandey 1,3, Brianna E. Kuypers 2,4, Barbara Nassif 1, Emily E. Thomas 1,3, Razan
Supplementary data. sienigma. F-Enigma F-EnigmaSM. a-p53
 Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
CRISPR/Cas9 Genome Editing: Transfection Methods
 CRISPR/ Genome Editing: Transfection Methods For over 20 years Mirus Bio has developed and manufactured high performance transfection products and technologies. That expertise is now being applied to the
CRISPR/ Genome Editing: Transfection Methods For over 20 years Mirus Bio has developed and manufactured high performance transfection products and technologies. That expertise is now being applied to the
Amplified segment of DNA can be purified from bacteria in sufficient quantity and quality for :
 Transformation Insertion of DNA of interest Amplification Amplified segment of DNA can be purified from bacteria in sufficient quantity and quality for : DNA Sequence. Understand relatedness of genes and
Transformation Insertion of DNA of interest Amplification Amplified segment of DNA can be purified from bacteria in sufficient quantity and quality for : DNA Sequence. Understand relatedness of genes and
Supplementary Information
 Supplementary Information Supplementary Figures Figure S1. Study of mgtl translation in vitro. (A) Detection of 5 LR RNA using wild-type and anti-sd (91-95) substituted templates in a transcription-translation
Supplementary Information Supplementary Figures Figure S1. Study of mgtl translation in vitro. (A) Detection of 5 LR RNA using wild-type and anti-sd (91-95) substituted templates in a transcription-translation
Molecular Biology Midterm Exam 2
 Molecular Biology Midterm Exam 2 1. The experiments by Frank Stahl and Matthew Messelson demonstrated that DNA strands separate during DNA replication. They showed that DNA replication is what kind of
Molecular Biology Midterm Exam 2 1. The experiments by Frank Stahl and Matthew Messelson demonstrated that DNA strands separate during DNA replication. They showed that DNA replication is what kind of
Impact of gdna Integrity on the Outcome of DNA Methylation Studies
 Impact of gdna Integrity on the Outcome of DNA Methylation Studies Application Note Nucleic Acid Analysis Authors Emily Putnam, Keith Booher, and Xueguang Sun Zymo Research Corporation, Irvine, CA, USA
Impact of gdna Integrity on the Outcome of DNA Methylation Studies Application Note Nucleic Acid Analysis Authors Emily Putnam, Keith Booher, and Xueguang Sun Zymo Research Corporation, Irvine, CA, USA
MCB 110 SP 2005 Second Midterm
 MCB110 SP 2005 Midterm II Page 1 ANSWER KEY MCB 110 SP 2005 Second Midterm Question Maximum Points Your Points I 40 II 40 III 36 IV 34 150 NOTE: This is my answer key (KC). The graders were given disgression
MCB110 SP 2005 Midterm II Page 1 ANSWER KEY MCB 110 SP 2005 Second Midterm Question Maximum Points Your Points I 40 II 40 III 36 IV 34 150 NOTE: This is my answer key (KC). The graders were given disgression
#FD µl (for 200 rxns) Expiry Date: Description. 1 ml of 10X FastDigest Green Buffer. Store at -20 C
 PRODUCT INFORMATION Thermo Scientific FastDigest SalI #FD0644 Lot: 5'...G T C G A C...3' 3'...C A G C T G...5' Supplied with: Store at -20 C 200 µl (for 200 rxns) Expiry Date: BSA included www.thermoscientific.com/onebio
PRODUCT INFORMATION Thermo Scientific FastDigest SalI #FD0644 Lot: 5'...G T C G A C...3' 3'...C A G C T G...5' Supplied with: Store at -20 C 200 µl (for 200 rxns) Expiry Date: BSA included www.thermoscientific.com/onebio
IDN1 and IDN2: two proteins required for de novo DNA methylation in Arabidopsis thaliana.
 IDN1 and IDN2: two proteins required for de novo DNA methylation in Arabidopsis thaliana. Israel Ausin, Todd C. Mockler, Joanne Chory, and Steven E. Jacobsen Col Ler nrpd1a rdr2 dcl3-1 ago4-1 idn1-1 idn2-1
IDN1 and IDN2: two proteins required for de novo DNA methylation in Arabidopsis thaliana. Israel Ausin, Todd C. Mockler, Joanne Chory, and Steven E. Jacobsen Col Ler nrpd1a rdr2 dcl3-1 ago4-1 idn1-1 idn2-1
Molecular Genetics II - Genetic Engineering Course (Supplementary notes)
 1 von 12 21.02.2015 15:13 Molecular Genetics II - Genetic Engineering Course (Supplementary notes) Figures showing examples of cdna synthesis (currently 11 figures) cdna is a DNA copy synthesized from
1 von 12 21.02.2015 15:13 Molecular Genetics II - Genetic Engineering Course (Supplementary notes) Figures showing examples of cdna synthesis (currently 11 figures) cdna is a DNA copy synthesized from
Intro to Microarray Analysis. Courtesy of Professor Dan Nettleton Iowa State University (with some edits)
 Intro to Microarray Analysis Courtesy of Professor Dan Nettleton Iowa State University (with some edits) Some Basic Biology Genes are DNA sequences that code for proteins. (e.g. gene lengths perhaps 1000
Intro to Microarray Analysis Courtesy of Professor Dan Nettleton Iowa State University (with some edits) Some Basic Biology Genes are DNA sequences that code for proteins. (e.g. gene lengths perhaps 1000
Computational Biology I LSM5191
 Computational Biology I LSM5191 Lecture 5 Notes: Genetic manipulation & Molecular Biology techniques Broad Overview of: Enzymatic tools in Molecular Biology Gel electrophoresis Restriction mapping DNA
Computational Biology I LSM5191 Lecture 5 Notes: Genetic manipulation & Molecular Biology techniques Broad Overview of: Enzymatic tools in Molecular Biology Gel electrophoresis Restriction mapping DNA
I. Gene Cloning & Recombinant DNA. Biotechnology: Figure 1: Restriction Enzyme Activity. Restriction Enzyme:
 I. Gene Cloning & Recombinant DNA Biotechnology: Figure 1: Restriction Enzyme Activity Restriction Enzyme: Most restriction enzymes recognize a single short base sequence, or Restriction Site. Restriction
I. Gene Cloning & Recombinant DNA Biotechnology: Figure 1: Restriction Enzyme Activity Restriction Enzyme: Most restriction enzymes recognize a single short base sequence, or Restriction Site. Restriction
Supplementary Methods Plasmid constructs
 Supplementary Methods Plasmid constructs. Mouse cdna encoding SHP-1, amplified from mrna of RAW264.7 macrophages with primer 5'cgtgcctgcccagacaaactgt3' and 5'cggaattcagacgaatgcccagatcacttcc3', was cloned
Supplementary Methods Plasmid constructs. Mouse cdna encoding SHP-1, amplified from mrna of RAW264.7 macrophages with primer 5'cgtgcctgcccagacaaactgt3' and 5'cggaattcagacgaatgcccagatcacttcc3', was cloned
Problem Set 8. Answer Key
 MCB 102 University of California, Berkeley August 11, 2009 Isabelle Philipp Online Document Problem Set 8 Answer Key 1. The Genetic Code (a) Are all amino acids encoded by the same number of codons? no
MCB 102 University of California, Berkeley August 11, 2009 Isabelle Philipp Online Document Problem Set 8 Answer Key 1. The Genetic Code (a) Are all amino acids encoded by the same number of codons? no
