Supplementary Figures
|
|
|
- Beryl Preston
- 6 years ago
- Views:
Transcription
1 Supplementary Figures a HindIII XbaI pcdna3.1/v5-his A_Nfluc-VVD Nfluc Linker 1 VVD HindIII XhoI pcdna3.1/v5-his A_VVD-Nfluc VVD Linker 2 Nfluc HindIII XbaI pcdna3.1/v5-his A_Cfluc-VVD Cfluc Linker VVD HindIII XhoI pcdna3.1/v5-his A_VVD-Cfluc VVD Linker 2 Cfluc Linker 1: LEGGSGGSGGSG! Linker 2: GGSGGSGGGS! b 10000! Bioluminescence (RLU) 8000! 6000! 4000! 2000! 0! Dark! BL! Nfluc-VVD:! +! +!!! VVD-Nfluc:!!! +! +! Cfluc-VVD:! +!! +!! VVD-Cfluc:!! +!! +! Supplementary Figure 1. A bioluminescence assay system for monitoring dimerization of VVD based on complementation of split-firefly luciferase fragments. (a) Schematic representations of domain structures of the fusion proteins tested in Supplementary Figure 1b. An N-half fragment (Nfluc: residues 1 398) and a C-half fragment (Cfluc: residues ) of a split-firefly luciferase were separately tagged with VVD with different orientations. (b) Detecting blue light-dependent dimerization of VVD with split luciferase fragments. COS-7 cells co-expressing Nfluc-VVD or VVD-Nfluc along with Cfluc-VVD or VVD-Cfluc were irradiated with blue light (470 ± 20 nm, 3 mw cm -2 ) for 30 s. The pair of Nfluc-VVD and VVD-Cfluc produced a larger bioluminescence signal than others in response to blue light. RLU, relative light unit. The result is means ± S.D. of three independent measurements.
2 a 2500! Bioluminescence (RLU) 2000! 1500! 1000! 500! 0! Dark! 1! 3! 10! 30! 60! 180! 600! Irradiation time (s) b 3000! Bioluminescence (RLU) 2500! 2000! 1500! 1000! 500! 0! Dark! 0.1! 0.5! 1! 2! 3! 4! 5! 6! Irradiation intensity (mw/cm 2 ) Supplementary Figure 2. Optimizing conditions for the irradiation to induce the VVD dimerization. (a) COS-7 cells expressing Nfluc-VVD and VVD-Cfluc were irradiated with blue light (470 ± 20 nm, 3 mw cm -2 ) for 1, 3, 10, 30, 60, 180 and 600 s, respectively. The bioluminescence intensity reached the maximum level upon blue light irradiation with a power density at 3 mw cm -2 for 30 s. (b) COS-7 cells expressing Nfluc-VVD and VVD-Cfluc were irradiated for 30 s with 0.1, 0.5, 1, 2, 3, 4, 5 and 6 mw cm -2 blue light, respectively. The bioluminescence intensity reached the maximum level upon blue light irradiation with a power density at 3 mw cm -2 for 30 s. RLU, relative light unit. The results are means ± S.D. of three independent measurements.!
3 a Bioluminescence (RLU) 5000! 4000! 3000! 2000! 1000! 1.0 0! Residue:! 47! Nfluc-X:! X-Cfluc: Dark! BL! ! 49! 50! 51! ! 53! 54! 55! ! VVD! C71S! VVD! C71S! b Bioluminescence (RLU) 1200! 1000! 800! 600! 400! 200! 1.1 Dark! BL! c Fold induction (right/dark) 10! 8! 6! 4! 2! E/ R/ 0! Residue:! 47! Nfluc-X:! R! Y-Cfluc: 48! R! 50! R! 51! R! 52! R! 54! R! 55! R! 0! Residue:! 47! 48! 50! 51! 52! 54! 55! d Bioluminescence (RLU) 10000! 8000! 6000! 4000! 2000! ! Nfluc-X:! K! Y(X)-Cfluc: K! 16.4 R! R! D! D! Dark! BL! K! D! K! R! D! R! D! K! K! D! R! 24.7 VVD! R! VVD! e Nfluc-X D! R! K! X(Y)-Cfluc K! R! D! Fold induction (right/dark)
4 f Bioluminescence (RLU) 6000! 5000! 4000! 3000! 2000! 1000! 0! Nfluc-X:! K! Y(X)-Cfluc: K! Dark! BL! *! R! R! *! D! D! K! D! K! *! 6.0 R! D! R! D! K! K! D! R! 32.3 VVD! R! VVD! g Nfluc-X D! R! K! X(Y)-Cfluc K! R! D! Fold induction (right/dark) h Bioluminescence (RLU) 8000! 7000! 6000! 5000! 4000! 3000! 2000! 1000! 0! Nfluc-X:! X-Cfluc: RK! RK! Dark! BL! **! RR! RD! RR! RD! R R 1.4 DK! DK! 1.4 DR! DD! DR! DD! D D R_! R_! 31.6 D_! VVD! D_! VVD! i Bioluminescence (RLU) 8000! 7000! 6000! 5000! 4000! 3000! 2000! 1000! 0! Nfluc-X:! RK! Y(X)-Cfluc: DK! Dark! BL! ! 8.5 RK! RK! RK! RR! RR! RR! RR! RD! RD! RD! RD! DR! DD! D DK! DR! DD! D DK! DR! DD! D R R R DK! DR! DD! R VVD! D VVD!
5 j Bioluminescence (RLU) 8000! 7000! 6000! 5000! 4000! 3000! 2000! 1000! 1.9 0! Nfluc-X:! R_! Y(X)-Cfluc: DK! Dark! BL! 1.9 R_! DR! R_! DD! R_! RK! RR! RD! R D D_! D_! D_! D_! R_! VVD! D_! VVD! k Nfluc-X R_! R RD! RR! RK! Y-Cfluc DK! DR! DD! D D_! Fold induction (right/dark) l Bioluminescence (RLU) 12000! 10000! 8000! 6000! 4000! 2000! 13.3 Dark! BL! **! ! Nfluc-X:! RR! Y-Cfluc: DH! RR! RR! RR! RR! RR! RR! RR! RR! RR! RR! RR! DN! DQ! DS! DT! DY! DF! DW! DA! DV! DL! DI! RR! RR! RR! DP! D_! DC! RR! VVD! DG! VVD! m Bioluminescence (RLU) 12000! 10000! 8000! 6000! 4000! 2000! 0! 1.7 Nfluc-X:! DH! X-Cfluc: DH! Dark! BL! DN! DQ! DS! DT! DY! DF! DW! DA! DV! DL! DN! DQ! DS! DT! DY! DF! DW! DA! DV! DL! DI! DI! DP! D_! DC! DP! D_! DC! **! 28.2 DG! VVD! DG! VVD!
6 Supplementary Figure 3. Electrostatic interactions-based discrimination of the hetero-dimer from the homo-dimer. (a) Homo-dimerization of VVD variants, having single substitutions with glutamic acid at amino acid residues from 47 to 56, respectively. Nfluc-X and X(Y)-Cfluc (X and Y stand for VVD variants) were co-expressed in COS-7 cells. The cells were irradiated with blue light (470 ± 20 nm, 3 mw cm -2 ) for 30 s, and bioluminescence measurements of the live cells were then performed. The fold induction (light/dark) of bioluminescence under the light condition to that under the dark condition is shown above each white bar. (b) Hetero-dimerization of each of the arginine variants and each of the glutamic acid variants, both having single substitutions at amino acid residue 47, 48, 50, 51, 52, 54 or 55. (c) A comparison of the fold induction (light/dark) between the homo-dimerization (closed circle) and the hetero-dimerization (open circle) of the VVD variants having single substitutions of amino acid residue 47, 48, 50, 51, 52, 54 or 55. The single substitution of Ile52 or Met55 was more effective to differentiate the fold inductions (right/dark) between the hetero-dimerization and the homo-dimerization than others. (d, f) Homo-dimerization and hetero-dimerization of VVD variants having single substitution with a charged amino acid at amino acid residue 55 (d) or 52 (f). In the case of Met55, its single substitution with a negatively charged amino acid suppressed homo-dimerization of the VVD variants, and hetero-dimerization was promoted between the variants with a negatively charged amino acid and those with a positively charged amino acid (d). However, single substitution of Met55 with a positively charged amino acid did not inhibit homo-dimerization of the VVD variants. This is in contrast to the case of substitution of Ile52 (f). The single substitution of Ile52 suppressed not only homo-dimerization of the VVD variants with a negatively charged amino acid but also that with a positively charged amino acid. In addition, the single substitution of Ile52 was effective to promote hetero-dimerization of the VVD variants with a negatively charged amino acid and those with a positively charged amino acid. Among the VVD variants with single substitutions of Ile52, the pair of the arginine variant (I52R) and the aspartic acid variant (I52D) was more effective to promote the heterodimerization than others. (e, g) Heat maps of the fold induction (light/dark) shown in Supplementary Figure 3d,f, respectively. (h) Homo-dimerization of VVD variants having double substitutions with a charged amino acid at amino acid residues 52 and 55. Two-letter code with R, K, D and E represents each of VVD variants with arginine, lysine, aspartic acid and glutamic acid at amino acid residues 52 and 55 (e.g., RK stands for I52R/M55K). I52R and I52D, having no substitution at amino acid residue 55, are depicted as R_ and D_, respectively. (i) Heterodimerization of VVD variants having double substitutions with a charged amino acid at amino acid residues 52 and 55. The double substitutions of both the residues 52 and 55 with a charged amino acid were not effective for the hetero-dimerization. (j) Hetero-dimerization between I52R or I52D and each of VVD variants having substitution with a charged amino acid at amino acid residue 55 of I52D or I52R. We found that I52R/M55R with arginine substitutions of both the residues 52 and 55 efficiently formed a hetero-dimer with I52D having no substitution of Met55. This implies that as the residue 55 of VVD variants having the I52D substitution, uncharged amino acids may be more promising compared with charged amino acids to achieve efficient hetero-dimerization with I52R/ M55R. Thus we substituted Met55 of I52D with other fifteen uncharged amino acids, respectively, and examined the hetero-dimerization with I52R/M55R and the homo-dimerization in Supplementary Figure 3l,m. (k) A heat map of the fold induction shown in Supplementary Figure 3i,j. (l) Hetero-dimerization between I52R/M55R and each of VVD variants having substitution with an uncharged amino acid at amino acid residue 55 of I52D. (m) Homo-dimerization of VVD variants with double substitutions with an uncharged amino acid at amino acid residues 52 and 55. The results are means ± S.D. of three independent measurements. Single asterisks highlight I52R and I52D. Double asterisks highlight I52R/M55R and I52D/M52G, renamed pmag and nmag, respectively.!
7 1! BL!! Normalized intensity 0.8! 0.6! 0.4! 0.2! 0! -1! 0! 1! 2! 3! 4! 5! 6! 7! 8! 9! 10! 11! Time (h) Supplementary Figure 4. Dissociation kinetics of the pmag nmag hetero-dimer (black) and the VVD VVD homo-dimer (orange). COS-7 cells expressing the pair of Nfluc-pMag and nmag-cfluc or that of Nfluc-VVD and VVD-Cfluc were irradiated with blue light (470 ± 20 nm, 3 mw cm -2 ) for 30 s. The homo-dimer of VVD dissociated with half-life t 1/2 of 2.0 h (rate constant, ), similar to the pmag nmag hetero-dimer (t 1/2, 1.8 h, rate constant, ). The result represents typical experiments from three independent measurements.!
8 14000! Bioluminescence (RLU) 12000! 10000! 8000! 6000! 4000! 2000! 0! Dark! BL! #1! #2! #3! #4! #5! #6! #7! #8! #9! #1! #2! #3! #4! #5! #6! #7! #8! #9! Nfluc-X! pmag! pmag! pmagfast1! pmagfast2! pmaghigh1! nmag! nmagfast1! nmagfast2! nmaghigh1! X(Y)-Cfluc! nmag! pmag! pmagfast1! pmagfast2! pmaghigh1! nmag! nmagfast1! nmagfast2! nmaghigh1! Supplementary Figure 5. Homo-dimerization of pmag variants and that of nmag variants. COS-7 cells expressing the Nfluc-X and X(Y)-Cfluc (X and Y stand for any one of Magnet variants shown in the figure) were irradiated with blue light (470 ± 20 nm, 3 mw cm -2 ) for 30 s. The pmag variants and the nmag variants do not efficiently form their homodimers upon blue light irradiation. The results are means ± S.D. of three independent measurements.!
9 Activation! CAAX! PM! EGFP! BL PA-Rac1(Small GTPase)! BL Magnets Dark GDP GTP Tiam1! irfp! Supplementary Figure 6. A schematic diagram of a Magnet-based photoactivatable actuator for a small GTPase (Rac1). Upon blue light irradiation, a functional domain is recruited to the plasma membrane with the help of the present paired photoswitches, Magnets (red and blue). Here we demonstrated photoactivatable Rac1 using pmagfast2(3x) (red) and nmaghigh1 (blue). To develop photoactivatable Rac1, we used a guanine nucleotide exchanging factor (GEF) domain (residues ) from mouse Tiam1.!
10 a Tiam1-pMagFast2(3x)-iRFP! + nmaghigh1-egfp-caax! + Lifeact-mCherry! Dark! Lifeact-mCherry! BL! Expanded area! Contracted area! Overlapped area! Cell 1 Cell 2 Cell 3
11 b Tiam1-pMagFast2(3x)-iRFP! + nmaghigh1-egfp-caax! + Lifeact-mCherry! + Rac1 inhibitor (100 μm)!! Lifeact-mCherry! Dark! Cell 1 Cell 2 Cell 3 BL! Expanded area! Contracted area! Overlapped area!
12 c Tiam1-pMagFast2(3x)-iRFP! + Lifeact-mCherry! Dark! Lifeact-mCherry! BL! Expanded area! Contracted area! Overlapped area! Cell 1 Cell 2 Cell 3
13 d Tiam1-pMagFast2(3x)-iRFP! + nmaghigh1-egfp-caax! + Lifeact-mCherry! Dark (0 min)! Lifeact-mCherry! Dark (20 min)! Expanded area! Contracted area! Overlapped area! Cell 1 Cell 2 Cell 3
14 e 1000! *** *** Expanded area (μm 2 ) 800! 600! 400! 200! 0! -200! Tiam1-pMagFast2(3x)-iRFP!! +! +! +! +! nmaghigh1-egfp-caax!!! +! +! +! BL irradiation! +! +!! +! +! Rac1 Inhibitor (100 μm)!!!!! +! n =! 10! 10! 10! 10! 10! Supplementary Figure 7. Magnet-based photoactivatable Rac1. (a) Fluorescence images of COS-7 cells expressing nmaghigh1-egfp-caax, Tiam1-pMagFast2(3x)-iRFP and Lifeact-mCherry before and 20 min after irradiation with blue light (Left and middle panels). Right panels show merged images of expanded area (cyan), contracted area (red) and overlapped area (white). (b) A potent inhibitor for Rac1-specific GEF Tiam1 (100 μm) blocked the blue light-dependent cell expansion. (c) Fluorescence images of COS-7 cells expressing Tiam1-pMagFast2(3x)-iRFP and Lifeact-mCherry before and 20 min after irradiation with blue light. (d) Fluorescence images of COS-7 cells expressing nmaghigh1- EGFP-CAAX, Tiam1-pMagFast2(3x)-iRFP and Lifeact-mCherry at initiation time (0 min) and after elapsed time of 20 min in the dark condition. (e) Quantification of expanded area of the COS-7 cells expressing Magnet-based photoactivatable Rac1. The expanded area was calculated by subtracting cell area before blue light irradiation and from that after irradiation. Blue light irradiation was completed with a Multi-line Argon laser (488 nm) at 0.2 mw. Scale bars of all images, 10 μm.!
15 Protein! A! Protein! B! Electrostatic interactions! (Dimer interfaces) OFF Kinetic mutations! (Cofactor-binding domains)! Light FASTER dissociation Dark Protein! A! Protein! B! Selective HETERO dimerization ENHANCED efficiency ON Supplementary Figure 8. Re-engineering of the nature of a fungal photoreceptor has created pairs of distinct photoswitches with selective hetero-dimerization properties, substantially enhanced dimerization efficiencies and accelerated dissociation kinetics, allowing spatially and temporally precise control of cellular proteins with blue light.!
16 HindIII XbaI pcdna3.1/v5-his A_pMag-mCherry pmag L1 mcherry HindIII XbaI pcdna3.1/v5-his A_pMag(2x)-mCherry pmag L2 pmag L1 mcherry HindIII XbaI pcdna3.1/v5-his A_pMag(3x)-mCherry pmag pmag pmag L2 L2 L1 mcherry HindIII XhoI pcdna3.1/v5-his A_nMagHigh1-mKikGR-CAAX High1 L3 mkikgr L4 CAAX1 HindIII XbaI pcdna3.1_pmagfast2(3x)-irfp Fast2 Fast2 Fast2 L2 L2 L5 irfp EcoRI XbaI pcdna3.1_nmaghigh1-egfp-caax High1 L6 EGFP CAAX2 HindIII XbaI pcdna3.1_ish2-pmag(3x)-irfp ish2 L7 pmag L2 pmag L2 pmag L5 irfp HindIII XbaI pcdna3.1_ish2-pmagfast2(3x)-irfp ish2 L7 Fast2 Fast2 Fast2 L2 L2 L5 irfp HindIII XbaI pcdna3.1_tiam1-pmagfast2(3x)-irfp Tiam1 L6 Fast2 Fast2 Fast2 L2 L2 L5 irfp pcdna3.1_aktph-mcherry pcdna3.1_lifeact-mcherry HindIII XbaI AktPH mcherry! L8 HindIII XbaI mcherry! Lifeact L9 L1: GS! L2: GSGGSGGSGGSG! L3: GGSGGSGGGS! L4: GGSGG! L5: GSGGSGGSGGSGGTGGSGGSGGSG! L6: GGSGGSGGSGGSGGGS! L7: GSGGSGGSGGSGGSGGGS! L8: EFGGSGGGGSGGGGSGGRS! L9: GDPPVAT!! CAAX 1: QGCMGLPCVVM! CAAX 2: KKKKKKSKTKCVIM! Lifeact: MGVADLIKKFESISKEE Supplementary Figure 9. Schematic representations of domain structures of constructs used in this study. L1-9, linker domains; CAAX1, a CAAX box motif (QGCMGLPCVVM) derived from C-terminus of N-Ras; CAAX2; a CAAX box motif (KKKKKKSKTKCVIM) derived from C-terminus of K-Ras; ish2, an inter SH2 domain (residues ) from bovine p85alpha-pi3kinase; Tiam1, a guanine nucleotide exchanging factor (GEF) domain (residues ) from mouse Tiam1; AktPH, a PH domain (residues 1-164) derived from human Akt1.!
17 a
18
19 b
20
21 c
22 d
23 e
24 f
25
26 i
27
28 j
29 k
30
31 l
32 m
33
34 n
35
36 o
37
38 p
39 q
40 Supplementary Figure 10. List of DNA sequences and amino acid sequences of constructs used in this study. Dark green; Nfluc and Cfluc, Orange; VVD, Red; pmag and pmagfast2, Magenta; mcherry, Blue; nmaghigh1, Light green, mkikgr and EGFP, Gray; CAAX, Dark red; irfp, Light blue; ish2 domain and Tiam1 domain, Purple; AktPH, Brown; Lifeact, Black; Start codon, Stop codon, linkers and histidine tag.!
41 Supplementary Table
42
43 Supplementary Movies Supplementary Movie 1. Blue light-dependent translocation of pmagfast2(3x)-irfp to the plasma membrane (PM) and its fast dissociation when the blue light is turned off. nmaghigh1 is tethered to the PM of the NIH3T3 cell. Imaging was conducted at 37 C with a 63 oil objective on the stage of a LSM 710 confocal laser scanning microscope. Fluorescence images of irfp were taken using a HeNe laser (633 nm). Scale bar, 10 μm.!! Supplementary Movie 2. Blue light-dependent morphological changes in the PM of a COS-7 cell expressing ish2-pmag(3x)-irfp and nmaghigh1-egfp-caax. Lifeact-mCherry is expressed to observe actin polymerization in the cell. Imaging was conducted at room temperature with a 63 oil objective on the stage of a LSM 710 confocal laser scanning microscope. Fluorescence images of mcherry were taken using a HeNe laser (543 nm). Scale bar, 10 μm.!
Near-infrared optogenetic pair for protein regulation and spectral multiplexing
 Supplementary Information Near-infrared optogenetic pair for protein regulation and spectral multiplexing Taras A. Redchuk 1, Evgeniya S. Omelina 1, Konstantin G. Chernov 1 and Vladislav V. Verkhusha 1,2
Supplementary Information Near-infrared optogenetic pair for protein regulation and spectral multiplexing Taras A. Redchuk 1, Evgeniya S. Omelina 1, Konstantin G. Chernov 1 and Vladislav V. Verkhusha 1,2
Nature Methods: doi: /nmeth Supplementary Figure 1. Schematics of SAM, dcas9-suntag and dcas9-vpr.
 Supplementary Figure 1 Schematics of SAM, dcas9-suntag and dcas9-vpr. (a) Schematics of SAM. SAM consists of two chimeric proteins, dcas9 fused with VP64 and MS2-coat protein fused with p65 and HSF1 activator
Supplementary Figure 1 Schematics of SAM, dcas9-suntag and dcas9-vpr. (a) Schematics of SAM. SAM consists of two chimeric proteins, dcas9 fused with VP64 and MS2-coat protein fused with p65 and HSF1 activator
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than. SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with
 Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
Supporting Information Defined Bilayer Interactions of DNA Nanopores Revealed with a Nuclease-Based Nanoprobe Strategy
 Supporting Information Defined Bilayer Interactions of DNA Nanopores Revealed with a Nuclease-Based Nanoprobe Strategy Jonathan R. Burns* & Stefan Howorka* 1 Contents 1. Design of DNA nanopores... 3 1.1.
Supporting Information Defined Bilayer Interactions of DNA Nanopores Revealed with a Nuclease-Based Nanoprobe Strategy Jonathan R. Burns* & Stefan Howorka* 1 Contents 1. Design of DNA nanopores... 3 1.1.
Chapter One. Construction of a Fluorescent α5 Subunit. Elucidation of the unique contribution of the α5 subunit is complicated by several factors
 4 Chapter One Construction of a Fluorescent α5 Subunit The significance of the α5 containing nachr receptor (α5* receptor) has been a challenging question for researchers since its characterization by
4 Chapter One Construction of a Fluorescent α5 Subunit The significance of the α5 containing nachr receptor (α5* receptor) has been a challenging question for researchers since its characterization by
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2271 Supplementary Figure a! WM266.4 mock WM266.4 #7 sirna WM266.4 #10 sirna SKMEL28 mock SKMEL28 #7 sirna SKMEL28 #10 sirna WM1361 mock WM1361 #7 sirna WM1361 #10 sirna 9 WM266. WM136
DOI: 10.1038/ncb2271 Supplementary Figure a! WM266.4 mock WM266.4 #7 sirna WM266.4 #10 sirna SKMEL28 mock SKMEL28 #7 sirna SKMEL28 #10 sirna WM1361 mock WM1361 #7 sirna WM1361 #10 sirna 9 WM266. WM136
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1. Activation capacity of tettale-ad compared to tet trans-activator (ttas) using different teto variants. (A) HeLa cells were co-transfected with activation
Supplementary Figure 1 Supplementary Figure 1. Activation capacity of tettale-ad compared to tet trans-activator (ttas) using different teto variants. (A) HeLa cells were co-transfected with activation
J. Cell Sci. 128: doi: /jcs : Supplementary Material. Supplemental Figures. Journal of Cell Science Supplementary Material
 Supplemental Figures Figure S1. Trio controls endothelial barrier function. (A) TagRFP-shTrio constructs were expressed in ECs. Western blot shows efficient Trio knockdown in TagRFP-expressing ECs. (B)
Supplemental Figures Figure S1. Trio controls endothelial barrier function. (A) TagRFP-shTrio constructs were expressed in ECs. Western blot shows efficient Trio knockdown in TagRFP-expressing ECs. (B)
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Wang et al., http://www.jcb.org/cgi/content/full/jcb.201405026/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Generation and characterization of unc-40 alleles. (A and
Supplemental material Wang et al., http://www.jcb.org/cgi/content/full/jcb.201405026/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Generation and characterization of unc-40 alleles. (A and
Supplementary Figure 1. Serial deletion mutants of BLITz.
 Supplementary Figure 1 Serial deletion mutants of BLITz. (a) Design of TEVseq insertion into J -helix. C-terminal end of J -helix was serially deleted and replaced by TEVseq. TEV cleavage site is labeled
Supplementary Figure 1 Serial deletion mutants of BLITz. (a) Design of TEVseq insertion into J -helix. C-terminal end of J -helix was serially deleted and replaced by TEVseq. TEV cleavage site is labeled
Nature Biotechnology: doi: /nbt Supplementary Figure 1
 Supplementary Figure 1 The mass accuracy of fragment ions is important for peptide recovery in wide-tolerance searches. The same data as in Figure 1B was searched with varying fragment ion tolerances (FIT).
Supplementary Figure 1 The mass accuracy of fragment ions is important for peptide recovery in wide-tolerance searches. The same data as in Figure 1B was searched with varying fragment ion tolerances (FIT).
embryos. Asterisk represents loss of or reduced expression. Brackets represent
 Supplemental Figures Supplemental Figure 1. tfec expression is highly enriched in tail endothelial cells (A- B) ISH of tfec at 15 and 16hpf in WT embryos. (C- D) ISH of tfec at 36 and 38hpf in WT embryos.
Supplemental Figures Supplemental Figure 1. tfec expression is highly enriched in tail endothelial cells (A- B) ISH of tfec at 15 and 16hpf in WT embryos. (C- D) ISH of tfec at 36 and 38hpf in WT embryos.
JCB. Supplemental material THE JOURNAL OF CELL BIOLOGY. Hong et al.,
 Supplemental material JCB Hong et al., http://www.jcb.org/cgi/content/full/jcb.201412127/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Analysis of purified proteins by SDS-PAGE and pull-down assays. (A) Coomassie-stained
Supplemental material JCB Hong et al., http://www.jcb.org/cgi/content/full/jcb.201412127/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Analysis of purified proteins by SDS-PAGE and pull-down assays. (A) Coomassie-stained
Supplementatry Fig 1. Domain structure, biophysical characterisation and electron microscopy of a TD. (a) XTACC3/Maskin and XMAP215/chTOG domain
 Supplementatry Fig 1. Domain structure, biophysical characterisation and electron microscopy of a TD. (a) XTACC3/Maskin and XMAP215/chTOG domain architecture. Various C-terminal fragments were cloned and
Supplementatry Fig 1. Domain structure, biophysical characterisation and electron microscopy of a TD. (a) XTACC3/Maskin and XMAP215/chTOG domain architecture. Various C-terminal fragments were cloned and
Supplementary Information
 Supplementary Information Supplementary Figure 1: Over-expression of CD300f in NIH3T3 cells enhances their capacity to phagocytize AC. (a) NIH3T3 cells were stably transduced by EV, CD300f WT or CD300f
Supplementary Information Supplementary Figure 1: Over-expression of CD300f in NIH3T3 cells enhances their capacity to phagocytize AC. (a) NIH3T3 cells were stably transduced by EV, CD300f WT or CD300f
Supplementary Materials: A Toolbox of Genetically Encoded FRET-Based Biosensors for Rapid L-Lysine Analysis
 S1 of S9 Supplementary Materials: A Toolbox of Genetically Encoded FRET-Based Biosensors for Rapid L-Lysine Analysis Victoria Steffen, Julia Otten, Susann Engelmann, Andreas Radek, Michael Limberg, Bernd
S1 of S9 Supplementary Materials: A Toolbox of Genetically Encoded FRET-Based Biosensors for Rapid L-Lysine Analysis Victoria Steffen, Julia Otten, Susann Engelmann, Andreas Radek, Michael Limberg, Bernd
Supplementary Table 1: List of CH3 domain interface residues in the first chain (A) and
 Supplementary Tables Supplementary Table 1: List of CH3 domain interface residues in the first chain (A) and their side chain contacting residues in the second chain (B) a Interface Res. in Contacting
Supplementary Tables Supplementary Table 1: List of CH3 domain interface residues in the first chain (A) and their side chain contacting residues in the second chain (B) a Interface Res. in Contacting
Supplementary Figure 1. Espn-1 knockout characterization. (a) The predicted recombinant Espn-1 -/- allele was detected by PCR of the left (5 )
 Supplementary Figure 1. Espn-1 knockout characterization. (a) The predicted recombinant Espn-1 -/- allele was detected by PCR of the left (5 ) homologous recombination arm (left) and of the right (3 )
Supplementary Figure 1. Espn-1 knockout characterization. (a) The predicted recombinant Espn-1 -/- allele was detected by PCR of the left (5 ) homologous recombination arm (left) and of the right (3 )
HEK293T. Fig. 1 in the
 Supplementary Information Supplementary Figure 1 Zinc uptake assay of hzip4 and hzip4-δecd transiently expressed in HEK293T cells. The results of one representative e experiment are shown in Fig. 1 in
Supplementary Information Supplementary Figure 1 Zinc uptake assay of hzip4 and hzip4-δecd transiently expressed in HEK293T cells. The results of one representative e experiment are shown in Fig. 1 in
Supplemental Data. Farmer et al. (2010) Plant Cell /tpc
 Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
Oligonucleotides were purchased from Eurogentec, purified by denaturing gel electrophoresis
 SUPPLEMENRY INFORMION Purification of probes and Oligonucleotides sequence Oligonucleotides were purchased from Eurogentec, purified by denaturing gel electrophoresis and recovered by electroelution. Labelling
SUPPLEMENRY INFORMION Purification of probes and Oligonucleotides sequence Oligonucleotides were purchased from Eurogentec, purified by denaturing gel electrophoresis and recovered by electroelution. Labelling
Supplementary Figure 1. Design of linker truncation library between Nluc and either Venus or mneongreen
 1 2 3 Supplementary Figure 1. Design of linker truncation library between Nluc and either Venus or mneongreen 4 5 6 7 8 9 10 The cdna of C-terminally deleted FPs (mneongreen or Venus) mutants and N-terminally
1 2 3 Supplementary Figure 1. Design of linker truncation library between Nluc and either Venus or mneongreen 4 5 6 7 8 9 10 The cdna of C-terminally deleted FPs (mneongreen or Venus) mutants and N-terminally
Coleman et al., Supplementary Figure 1
 Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
SUPPLEMENTARY INFORMATION. Supplementary Figures 1-8
 SUPPLEMENTARY INFORMATION Supplementary Figures 1-8 Supplementary Figure 1. TFAM residues contacting the DNA minor groove (A) TFAM contacts on nonspecific DNA. Leu58, Ile81, Asn163, Pro178, and Leu182
SUPPLEMENTARY INFORMATION Supplementary Figures 1-8 Supplementary Figure 1. TFAM residues contacting the DNA minor groove (A) TFAM contacts on nonspecific DNA. Leu58, Ile81, Asn163, Pro178, and Leu182
Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product.
 Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Electronic Supplementary Information
 Electronic Supplementary Material (ESI) for Integrative Biology. This journal is The Royal Society of Chemistry 2015 Electronic Supplementary Information Table S1. Definition of quantitative cellular features
Electronic Supplementary Material (ESI) for Integrative Biology. This journal is The Royal Society of Chemistry 2015 Electronic Supplementary Information Table S1. Definition of quantitative cellular features
Supplementary Figure 1. Nature Structural & Molecular Biology: doi: /nsmb.3494
 Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10016 Supplementary discussion on binding site density for protein complexes on the surface: The density of biotin sites on the chip is ~10 3 biotin-peg per µm 2. The biotin sites are
doi:10.1038/nature10016 Supplementary discussion on binding site density for protein complexes on the surface: The density of biotin sites on the chip is ~10 3 biotin-peg per µm 2. The biotin sites are
Nature Structural & Molecular Biology: doi: /nsmb.3018
 Supplementary Figure 1 Validation of genetic complementation assay in Bmal1 / Per2 Luc fibroblasts. (a) Only Bmal1, not Bmal2, rescues circadian rhythms from cells. Cells expressing various Bmal constructs
Supplementary Figure 1 Validation of genetic complementation assay in Bmal1 / Per2 Luc fibroblasts. (a) Only Bmal1, not Bmal2, rescues circadian rhythms from cells. Cells expressing various Bmal constructs
Phi29 Scaffold Has a Helix-Loop-Helix Motif and a Disordered Tail. Marc C Morais et al., Nature Structural Biology 2003
 Phi29 Scaffold Has a Helix-Loop-Helix Motif and a Disordered Tail Marc C Morais et al., Nature Structural Biology 2003 Free scaffold 13+ 10 min 3 min 1 min 40 sec 100 % 0 100 % 0 100 % 0 100 % 0 Partially
Phi29 Scaffold Has a Helix-Loop-Helix Motif and a Disordered Tail Marc C Morais et al., Nature Structural Biology 2003 Free scaffold 13+ 10 min 3 min 1 min 40 sec 100 % 0 100 % 0 100 % 0 100 % 0 Partially
Hmwk # 8 : DNA-Binding Proteins : Part II
 The purpose of this exercise is : Hmwk # 8 : DNA-Binding Proteins : Part II 1). to examine the case of a tandem head-to-tail homodimer binding to DNA 2). to view a Zn finger motif 3). to consider the case
The purpose of this exercise is : Hmwk # 8 : DNA-Binding Proteins : Part II 1). to examine the case of a tandem head-to-tail homodimer binding to DNA 2). to view a Zn finger motif 3). to consider the case
Supplementary Figures
 Supplementary Figures Supplementary Figure S1. Inhibition kinetics of p53/hdm2 binding induced by Nutlin 3 on live cells. Reproducibility of the p53/hdm2 binding disruption kinetics induced by Nutlin 3
Supplementary Figures Supplementary Figure S1. Inhibition kinetics of p53/hdm2 binding induced by Nutlin 3 on live cells. Reproducibility of the p53/hdm2 binding disruption kinetics induced by Nutlin 3
Supplementary Figure S1. Np95 interacts with de novo methyltransferases Dnmt3a and 3b. (A) Co-immunoprecipitation of endogenous Dnmt3a2 (left and
 Supplementary Figure S1. Np95 interacts with de novo methyltransferases Dnmt3a and 3b. (A) Co-immunoprecipitation of endogenous Dnmt3a2 (left and right), Dnmt3b isoforms (left) and Dnmt1 (right) with GFP-Np95
Supplementary Figure S1. Np95 interacts with de novo methyltransferases Dnmt3a and 3b. (A) Co-immunoprecipitation of endogenous Dnmt3a2 (left and right), Dnmt3b isoforms (left) and Dnmt1 (right) with GFP-Np95
Dimeric WH2 domains in Vibrio VopF promote actin filament barbed end uncapping and assisted elongation
 Dimeric WH2 domains in Vibrio VopF promote actin filament barbed end uncapping and assisted elongation Julien Pernier, Jozsef Orban 1, Balendu Sankara Avvaru, Antoine Jégou, Guillaume Romet- Lemonne, Bérengère
Dimeric WH2 domains in Vibrio VopF promote actin filament barbed end uncapping and assisted elongation Julien Pernier, Jozsef Orban 1, Balendu Sankara Avvaru, Antoine Jégou, Guillaume Romet- Lemonne, Bérengère
Supplemental Figure 1 Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical
 Supplemental Figure Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical in all six REEPs are highlighted in green. Additional
Supplemental Figure Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical in all six REEPs are highlighted in green. Additional
AD BD TOC1. Supplementary Figure 1: Yeast two-hybrid assays showing the interaction between
 AD X BD TOC1 AD BD X PIFΔAD PIF TOC1 TOC1 PIFΔAD PIF N TOC1 TOC1 C1 PIFΔAD PIF C1 TOC1 TOC1 C PIFΔAD PIF C TOC1 Supplementary Figure 1: Yeast two-hybrid assays showing the interaction between PIF and TOC1
AD X BD TOC1 AD BD X PIFΔAD PIF TOC1 TOC1 PIFΔAD PIF N TOC1 TOC1 C1 PIFΔAD PIF C1 TOC1 TOC1 C PIFΔAD PIF C TOC1 Supplementary Figure 1: Yeast two-hybrid assays showing the interaction between PIF and TOC1
Supplementary Table, Figures and Videos
 Supplementary Table, Figures and Videos Table S1. Oligonucleotides used for different approaches. (A) RT-qPCR study. (B) qpcr study after ChIP assay. (C) Probes used for EMSA. Figure S1. Notch activation
Supplementary Table, Figures and Videos Table S1. Oligonucleotides used for different approaches. (A) RT-qPCR study. (B) qpcr study after ChIP assay. (C) Probes used for EMSA. Figure S1. Notch activation
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Conserved arginines on the rim of Hfq catalyze base pair formation and exchange Subrata Panja and Sarah A. Woodson T.C. Jenkins Department of Biophysics, Johns Hopkins University,
SUPPLEMENTARY INFORMATION Conserved arginines on the rim of Hfq catalyze base pair formation and exchange Subrata Panja and Sarah A. Woodson T.C. Jenkins Department of Biophysics, Johns Hopkins University,
Supplementary information. for the paper:
 Supplementary information for the paper: Screening for protein-protein interactions using Förster resonance energy transfer (FRET) and fluorescence lifetime imaging microscopy (FLIM) Anca Margineanu 1*,
Supplementary information for the paper: Screening for protein-protein interactions using Förster resonance energy transfer (FRET) and fluorescence lifetime imaging microscopy (FLIM) Anca Margineanu 1*,
Supplementary Figure 1 Collision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK2, PPK3 and PPK4 respectively.
 Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
Schematic representation of the endogenous PALB2 locus and gene-disruption constructs
 Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
SUPPLEMENTARY INFORMATION
 a 14 12 Densitometry (AU) 1 8 6 4 2 t b 16 NMHC-IIA GAPDH NMHC-IIB Densitometry (AU) 14 12 1 8 6 4 2 1 nm 1 nm 1 nm 1 nm sirna 1 nm 1 nm Figure S1 S4 Quantification of protein levels. (a) The microtubule
a 14 12 Densitometry (AU) 1 8 6 4 2 t b 16 NMHC-IIA GAPDH NMHC-IIB Densitometry (AU) 14 12 1 8 6 4 2 1 nm 1 nm 1 nm 1 nm sirna 1 nm 1 nm Figure S1 S4 Quantification of protein levels. (a) The microtubule
Supplementary Figure S1. Binding of HSA mutants to hfcrn. (a) The levels of titrated amounts of HSA
 Supplementary Figure S1. Binding of HSA mutants to hfcrn. (a) The levels of titrated amounts of HSA variants (5.0-0.002 μg/ml) directly coated in the wells at ph 6.0 were controlled using a horseradish
Supplementary Figure S1. Binding of HSA mutants to hfcrn. (a) The levels of titrated amounts of HSA variants (5.0-0.002 μg/ml) directly coated in the wells at ph 6.0 were controlled using a horseradish
TITLE: Elucidating Mechanisms of Farnesyltransferase Inhibitor Action and Resistance in Breast Cancer by Bioluminescence Imaging
 AD Award Number: W81XWH-07-1-0426 TITLE: Elucidating Mechanisms of Farnesyltransferase Inhibitor Action and Resistance in Breast Cancer by Bioluminescence Imaging PRINCIPAL INVESTIGATOR: David Piwnica-Worms,
AD Award Number: W81XWH-07-1-0426 TITLE: Elucidating Mechanisms of Farnesyltransferase Inhibitor Action and Resistance in Breast Cancer by Bioluminescence Imaging PRINCIPAL INVESTIGATOR: David Piwnica-Worms,
Supplementary Information for. Structure of human tyrosylprotein sulfotransferase-2 reveals the mechanism of protein tyrosine sulfation reaction
 Supplementary Information for Structure of human tyrosylprotein sulfotransferase-2 reveals the mechanism of protein tyrosine sulfation reaction Takamasa Teramoto, Yukari Fujikawa, Yoshirou Kawaguchi, Katsuhisa
Supplementary Information for Structure of human tyrosylprotein sulfotransferase-2 reveals the mechanism of protein tyrosine sulfation reaction Takamasa Teramoto, Yukari Fujikawa, Yoshirou Kawaguchi, Katsuhisa
Expanded View Figures
 EMO Molecular Medicine nolactone inhibits NRG1-ER4 signaling Michael Wehr et al Expanded View Figures Figure EV1. co-culture assay based on the split TEV technique to monitor NRG1-ER4 signaling activity.
EMO Molecular Medicine nolactone inhibits NRG1-ER4 signaling Michael Wehr et al Expanded View Figures Figure EV1. co-culture assay based on the split TEV technique to monitor NRG1-ER4 signaling activity.
In vitro EGFP-CALI comprehensive analysis. Liang CAI. David Humphrey. Jessica Wilson. Ken Jacobson. Dept. of Cell and Developmental Biology
 In vitro EGFP-CALI comprehensive analysis Liang CAI David Humphrey Jessica Wilson Ken Jacobson Dept. of Cell and Developmental Biology 1/15 Liang s Rotation in Ken s Lab, Sep-Nov, 2003 1. CALI background
In vitro EGFP-CALI comprehensive analysis Liang CAI David Humphrey Jessica Wilson Ken Jacobson Dept. of Cell and Developmental Biology 1/15 Liang s Rotation in Ken s Lab, Sep-Nov, 2003 1. CALI background
Monitoring Protein:Protein Interactions in Living Cells Using NanoBRET Technology. Danette L. Daniels, Ph.D. Group Leader, Functional Proteomics
 Monitoring Protein:Protein Interactions in Living Cells Using NanoBRET Technology Danette L. Daniels, Ph.D. Group Leader, Functional Proteomics Proteomics and studying dynamic interactions AP-MS Proteomics
Monitoring Protein:Protein Interactions in Living Cells Using NanoBRET Technology Danette L. Daniels, Ph.D. Group Leader, Functional Proteomics Proteomics and studying dynamic interactions AP-MS Proteomics
Supplementary Figure 1. Supporting data for Figure 1
 Supplementary Figure 1 Supporting data for Figure 1 (A) Schematic of BLI assay used to measure dissociation. 5 monobiotinylated substrate DNA (identical to λ1, Figure 2) is associated with streptavidin-coated
Supplementary Figure 1 Supporting data for Figure 1 (A) Schematic of BLI assay used to measure dissociation. 5 monobiotinylated substrate DNA (identical to λ1, Figure 2) is associated with streptavidin-coated
λ N -GFP: an RNA reporter system for live-cell imaging
 λ N -GFP: an RNA reporter system for live-cell imaging Nathalie Daigle & Jan Ellenberg Supplementary Figures and Text: Supplementary Figure 1 localization in the cytoplasm. 4 λ N22-3 megfp-m9 serves as
λ N -GFP: an RNA reporter system for live-cell imaging Nathalie Daigle & Jan Ellenberg Supplementary Figures and Text: Supplementary Figure 1 localization in the cytoplasm. 4 λ N22-3 megfp-m9 serves as
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Domain architecture and conformational states of the decapping complex, as revealed by structural studies. (a) Domain organization of Schizosaccharomyces pombe (Sp) and Saccharomyces
Supplementary Figure 1 Domain architecture and conformational states of the decapping complex, as revealed by structural studies. (a) Domain organization of Schizosaccharomyces pombe (Sp) and Saccharomyces
Cancer cells that survive radiation therapy acquire HIF-1 activity and translocate toward tumor blood vessels Supplementary Information
 Cancer cells that survive radiation therapy acquire HIF-1 activity and translocate toward tumor blood vessels Supplementary Information 1. Supplementary Figure S1-S10: Pages 2-11 2. Supplementary References:
Cancer cells that survive radiation therapy acquire HIF-1 activity and translocate toward tumor blood vessels Supplementary Information 1. Supplementary Figure S1-S10: Pages 2-11 2. Supplementary References:
Table S1. List of DNA constructs and primers, part 1 Construct
 SUPPLEMENTARY TABLES: Table S1. List of DNA constructs and primers, part 1 Construct Comment (Expressed protein) Vector, PCR primers and template or source of Restriction Sites name Resistance construct
SUPPLEMENTARY TABLES: Table S1. List of DNA constructs and primers, part 1 Construct Comment (Expressed protein) Vector, PCR primers and template or source of Restriction Sites name Resistance construct
Supplementary Material
 Supplementary Material Supplementary Table 1. Definitions of variants described in this work. Protein variant Substitutions and modifications relative to parent mtfp1 a = cfp484 b + delete all residues
Supplementary Material Supplementary Table 1. Definitions of variants described in this work. Protein variant Substitutions and modifications relative to parent mtfp1 a = cfp484 b + delete all residues
Supplemental Data. Guo et al. (2015). Plant Cell /tpc
 Supplemental Figure 1. The Mutant exb1-d Displayed Pleiotropic Phenotypes and Produced Branches in the Axils of Cotyledons. (A) Branches were developed in exb1-d but not in wild-type plants. (B) and (C)
Supplemental Figure 1. The Mutant exb1-d Displayed Pleiotropic Phenotypes and Produced Branches in the Axils of Cotyledons. (A) Branches were developed in exb1-d but not in wild-type plants. (B) and (C)
Supplemental Data. Na Xu et al. (2016). Plant Cell /tpc
 Supplemental Figure 1. The weak fluorescence phenotype is not caused by the mutation in At3g60240. (A) A mutation mapped to the gene At3g60240. Map-based cloning strategy was used to map the mutated site
Supplemental Figure 1. The weak fluorescence phenotype is not caused by the mutation in At3g60240. (A) A mutation mapped to the gene At3g60240. Map-based cloning strategy was used to map the mutated site
Caspase-2 BiFC plasmids
 Caspase-2 BiFC plasmids Caspase-2 BiFC - background Bimolecular Fluorescence Complementation (BiFC) describes the use of split fluorescent proteins to measure protein-protein interactions in cells. The
Caspase-2 BiFC plasmids Caspase-2 BiFC - background Bimolecular Fluorescence Complementation (BiFC) describes the use of split fluorescent proteins to measure protein-protein interactions in cells. The
Figure legends for supplement
 Figure legends for supplement Supplemental Figure 1 Characterization of purified and recombinant proteins Relevant fractions related the final stage of the purification protocol(bingham et al., 1998; Toba
Figure legends for supplement Supplemental Figure 1 Characterization of purified and recombinant proteins Relevant fractions related the final stage of the purification protocol(bingham et al., 1998; Toba
Nature Biotechnology: doi: /nbt Supplementary Figure 1
 Supplementary Figure 1 Generation of the AARE-Gene system construct. (a) Position and sequence alignment of AAREs extracted from human Trb3, Chop or Atf3 promoters. AARE core sequences are boxed in grey.
Supplementary Figure 1 Generation of the AARE-Gene system construct. (a) Position and sequence alignment of AAREs extracted from human Trb3, Chop or Atf3 promoters. AARE core sequences are boxed in grey.
Single-Molecule Imaging Reveals Dynamics of CREB Transcription Factor Bound to Its Target Sequence
 Supplementary Information Single-Molecule Imaging Reveals Dynamics of CREB Transcription Factor Bound to Its Target Sequence Noriyuki Sugo, Masatoshi Morimatsu, Yoshiyuki Arai, Yoshinori Kousoku, Aya Ohkuni,
Supplementary Information Single-Molecule Imaging Reveals Dynamics of CREB Transcription Factor Bound to Its Target Sequence Noriyuki Sugo, Masatoshi Morimatsu, Yoshiyuki Arai, Yoshinori Kousoku, Aya Ohkuni,
Nature Neuroscience: doi: /nn Supplementary Figure 1
 Supplementary Figure 1 PCR-genotyping of the three mouse models used in this study and controls for behavioral experiments after semi-chronic Pten inhibition. a-c. DNA from App/Psen1 (a), Pten tg (b) and
Supplementary Figure 1 PCR-genotyping of the three mouse models used in this study and controls for behavioral experiments after semi-chronic Pten inhibition. a-c. DNA from App/Psen1 (a), Pten tg (b) and
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
Supplemental Data. Cui et al. (2012). Plant Cell /tpc a b c d. Stem UBC32 ACTIN
 A Root Stem Leaf Flower Silique Senescence leaf B a b c d UBC32 ACTIN C * Supplemental Figure 1. Expression Pattern and Protein Sequence of UBC32 Homologues in Yeast, Human, and Arabidopsis. (A) Expression
A Root Stem Leaf Flower Silique Senescence leaf B a b c d UBC32 ACTIN C * Supplemental Figure 1. Expression Pattern and Protein Sequence of UBC32 Homologues in Yeast, Human, and Arabidopsis. (A) Expression
Mitotic cell rounding and epithelial thinning regulate lumen
 Mitotic cell rounding and epithelial thinning regulate lumen growth and shape Esteban Hoijman, Davide Rubbini, Julien Colombelli and Berta Alsina SUPPLEMENTARY FIGURES Supplementary Figure 1 (a-c) Localization
Mitotic cell rounding and epithelial thinning regulate lumen growth and shape Esteban Hoijman, Davide Rubbini, Julien Colombelli and Berta Alsina SUPPLEMENTARY FIGURES Supplementary Figure 1 (a-c) Localization
Methanol fixation allows better visualization of Kal7. To compare methods for
 Supplementary Data Methanol fixation allows better visualization of Kal7. To compare methods for visualizing Kal7 in dendrites, mature cultures of dissociated hippocampal neurons (DIV21) were fixed with
Supplementary Data Methanol fixation allows better visualization of Kal7. To compare methods for visualizing Kal7 in dendrites, mature cultures of dissociated hippocampal neurons (DIV21) were fixed with
Golgi are located near the MTOC while the ER is spread throughout the cytoplasm, so which of the following is probably true?
 Golgi are located near the MTOC while the ER is spread throughout the cytoplasm, so which of the following is probably true? A. COPII and COPI vesicles are transported with dynein. B. COPII and COPI vesicles
Golgi are located near the MTOC while the ER is spread throughout the cytoplasm, so which of the following is probably true? A. COPII and COPI vesicles are transported with dynein. B. COPII and COPI vesicles
Expanded View Figures
 EMO reports rystal structure of Mis18 Yippee-like domain Lakxmi Subramanian et al Expanded View Figures Figure EV1. Structural characterization of the N-terminal Yippee-like globular domain of spmis18.
EMO reports rystal structure of Mis18 Yippee-like domain Lakxmi Subramanian et al Expanded View Figures Figure EV1. Structural characterization of the N-terminal Yippee-like globular domain of spmis18.
Supplement Figure S1:
 Supplement Figure S1: A, Sequence of Xcadherin-11 Morpholino 1 (Xcad-11MO) and 2 (Xcad-11 MO2) and control morpholino in comparison to the Xcadherin-11 sequence. The Xcad-11MO binding sequence spans the
Supplement Figure S1: A, Sequence of Xcadherin-11 Morpholino 1 (Xcad-11MO) and 2 (Xcad-11 MO2) and control morpholino in comparison to the Xcadherin-11 sequence. The Xcad-11MO binding sequence spans the
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Moutin et al., http://www.jcb.org/cgi/content/full/jcb.201110101/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Tagged Homer1a and Homer are functional and display different
Supplemental material Moutin et al., http://www.jcb.org/cgi/content/full/jcb.201110101/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Tagged Homer1a and Homer are functional and display different
Aoki et al.,
 JCB: SUPPLEMENTAL MATERIAL Aoki et al., http://www.jcb.org/cgi/content/full/jcb.200609017/dc1 Supplemental materials and methods Calculation of the raw number of translocated proteins First, the average
JCB: SUPPLEMENTAL MATERIAL Aoki et al., http://www.jcb.org/cgi/content/full/jcb.200609017/dc1 Supplemental materials and methods Calculation of the raw number of translocated proteins First, the average
SUPPLEMENTARY INFORMATION
 Results Construct purification and coupling. Two A1-GP1bα ReaLiSM constructs, with and without cysteine residues near the N and C-termini (Fig. S2a), were expressed and purified by Ni affinity chromatography
Results Construct purification and coupling. Two A1-GP1bα ReaLiSM constructs, with and without cysteine residues near the N and C-termini (Fig. S2a), were expressed and purified by Ni affinity chromatography
Expanded View Figures
 Expanded View Figures Figure EV1. AM3-CLE45 control experiments and bam3 alleles. A Relative primary root length of indicated genotypes at 9 dag, in response to increasing amounts of CLE45 in the media.
Expanded View Figures Figure EV1. AM3-CLE45 control experiments and bam3 alleles. A Relative primary root length of indicated genotypes at 9 dag, in response to increasing amounts of CLE45 in the media.
Supplemental figures (1-9) Gu et al. ADF/Cofilin-Mediated Actin Dynamics Regulate AMPA Receptor Trafficking during Synaptic Plasticity
 Supplemental figures (1-9) Gu et al. ADF/Cofilin-Mediated Actin Dynamics Regulate AMPA Receptor Trafficking during Synaptic Plasticity Supplemental figure 1. The quantification method to determine the
Supplemental figures (1-9) Gu et al. ADF/Cofilin-Mediated Actin Dynamics Regulate AMPA Receptor Trafficking during Synaptic Plasticity Supplemental figure 1. The quantification method to determine the
Confocal bioluminescence imaging for living tissues with a caged. substrate of luciferin
 Supporting Information Confocal bioluminescence imaging for living tissues with a caged substrate of luciferin Mitsuru Hattori 1, Genki Kawamura 1, Ryosuke Kojima 2, Mako kamiya 3, Yasuteru Urano 2,3 &
Supporting Information Confocal bioluminescence imaging for living tissues with a caged substrate of luciferin Mitsuru Hattori 1, Genki Kawamura 1, Ryosuke Kojima 2, Mako kamiya 3, Yasuteru Urano 2,3 &
Supplementary Information. Small Molecule-Induced Domain Swapping as a Mechanism for Controlling Protein Function and Assembly
 Supplementary Information Small Molecule-Induced Domain Swapping as a Mechanism for Controlling Protein Function and Assembly Joshua M. Karchin, Jeung-Hoi Ha, Kevin E. Namitz, Michael S. Cosgrove, and
Supplementary Information Small Molecule-Induced Domain Swapping as a Mechanism for Controlling Protein Function and Assembly Joshua M. Karchin, Jeung-Hoi Ha, Kevin E. Namitz, Michael S. Cosgrove, and
Biochemical characteristics and gene expression profiles of two. paralogous luciferases from the Japanese firefly Pyrocoelia
 Electronic Supplementary Material (ESI) for Photochemical & Photobiological Sciences. This journal is The Royal Society of Chemistry and Owner Societies 0 Electronic Supplementary Material (ESI) for Photochemical
Electronic Supplementary Material (ESI) for Photochemical & Photobiological Sciences. This journal is The Royal Society of Chemistry and Owner Societies 0 Electronic Supplementary Material (ESI) for Photochemical
High-resolution Identification and Separation of Living Cell Types by Multiple microrna-responsive Synthetic mrnas
 Supplementary information High-resolution Identification and Separation of Living Cell Types by Multiple microrna-responsive Synthetic mrnas Kei Endo *, Karin Hayashi, Hirohide Saito * Contents: 1. Supplementary
Supplementary information High-resolution Identification and Separation of Living Cell Types by Multiple microrna-responsive Synthetic mrnas Kei Endo *, Karin Hayashi, Hirohide Saito * Contents: 1. Supplementary
Supplemental Data. Wang et al. (2017). Plant Cell /tpc NIP1;2 NIP7;1
 Supplemental Data. Wang et al. (217). Plant Cell 1.115/tpc.16.825 NIP1;1 NIP1;2 NIP2;1 NIP3;1 NIP4;1 NIP5;1 NIP6;1 NIP7;1 Supplemental Figure 1. Distinct Localization of NIPs in Root Epidermal Cells. (Supports
Supplemental Data. Wang et al. (217). Plant Cell 1.115/tpc.16.825 NIP1;1 NIP1;2 NIP2;1 NIP3;1 NIP4;1 NIP5;1 NIP6;1 NIP7;1 Supplemental Figure 1. Distinct Localization of NIPs in Root Epidermal Cells. (Supports
SUPPLEMENTARY INFORMATION. Evolution of a split RNA polymerase as a versatile biosensor platform
 SUPPLEMENTARY INFORMATION Evolution of a split RNA polymerase as a versatile iosensor platform Jinyue Pu, Julia Zinkus-Boltz, Bryan C. Dickinson Department of Chemistry, The University of Chicago, Chicago,
SUPPLEMENTARY INFORMATION Evolution of a split RNA polymerase as a versatile iosensor platform Jinyue Pu, Julia Zinkus-Boltz, Bryan C. Dickinson Department of Chemistry, The University of Chicago, Chicago,
Supplementary Table 1. DNA sequence synthesized to express the Zika virus NS5 protein.
 Supplementary Table 1. DNA sequence synthesized to express the Zika virus NS5 protein. MR766 NS5 sequence 1 ACAGAGAACAGATTGGTGGTGGAGGTGGGACGGGAGAGACTCTGGGAGAGAAGTGGAAAG 61 CTCGTCTGAATCAGATGTCGGCCCTGGAGTTCTACTCTTATAAAAAGTCAGGTATCACTG
Supplementary Table 1. DNA sequence synthesized to express the Zika virus NS5 protein. MR766 NS5 sequence 1 ACAGAGAACAGATTGGTGGTGGAGGTGGGACGGGAGAGACTCTGGGAGAGAAGTGGAAAG 61 CTCGTCTGAATCAGATGTCGGCCCTGGAGTTCTACTCTTATAAAAAGTCAGGTATCACTG
Quantitative analysis of recombination in YFP and CFP gene of FRET biosensor induced by lentiviral or retroviral gene transfer.
 Supplementary Information: Quantitative analysis of recombination in and gene of FRET biosensor induced by lentiviral or retroviral gene transfer. Akira T. Komatsubara, Michiyuki Matsuda,, and Kazuhiro
Supplementary Information: Quantitative analysis of recombination in and gene of FRET biosensor induced by lentiviral or retroviral gene transfer. Akira T. Komatsubara, Michiyuki Matsuda,, and Kazuhiro
i-stop codon positions in the mcherry gene
 Supplementary Figure 1 i-stop codon positions in the mcherry gene The grnas (green) that can potentially generate stop codons from Trp (63 th and 98 th aa, upper panel) and Gln (47 th and 114 th aa, bottom
Supplementary Figure 1 i-stop codon positions in the mcherry gene The grnas (green) that can potentially generate stop codons from Trp (63 th and 98 th aa, upper panel) and Gln (47 th and 114 th aa, bottom
Post-expansion antibody delivery, after epitope-preserving homogenization.
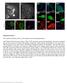 Supplementary Figure 1 Post-expansion antibody delivery, after epitope-preserving homogenization. (a, b) Wide-field fluorescence images of Thy1-YFP-expressing mouse brain hemisphere slice before expansion
Supplementary Figure 1 Post-expansion antibody delivery, after epitope-preserving homogenization. (a, b) Wide-field fluorescence images of Thy1-YFP-expressing mouse brain hemisphere slice before expansion
The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit
 Cell Reports, Volume 5 Supplemental Information The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit Andrey Poleshko, Katelyn M. Mansfield, Caroline
Cell Reports, Volume 5 Supplemental Information The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit Andrey Poleshko, Katelyn M. Mansfield, Caroline
transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected with the wwp-luc reporter, and FLAG-tagged FHL1,
 Supplementary Data Supplementary Figure Legends Supplementary Figure 1 FHL-mediated TGFβ-responsive reporter transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected
Supplementary Data Supplementary Figure Legends Supplementary Figure 1 FHL-mediated TGFβ-responsive reporter transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected
Supporting Information
 Supporting Information Powell and Xu 10.1073/pnas.0807274105 SI Methods Design of Estrogen Receptor (ER) Fusion Constructs for Use in Bioluminescence Resonance Energy Transfer (BRET) Assay. Because the
Supporting Information Powell and Xu 10.1073/pnas.0807274105 SI Methods Design of Estrogen Receptor (ER) Fusion Constructs for Use in Bioluminescence Resonance Energy Transfer (BRET) Assay. Because the
No wash 2 Washes 2 Days ** ** IgG-bead phagocytosis (%)
 Supplementary Figures Supplementary Figure 1. No wash 2 Washes 2 Days Tat Control ** ** 2 4 6 8 IgG-bead phagocytosis (%) Supplementary Figure 1. Reversibility of phagocytosis inhibition by Tat. Human
Supplementary Figures Supplementary Figure 1. No wash 2 Washes 2 Days Tat Control ** ** 2 4 6 8 IgG-bead phagocytosis (%) Supplementary Figure 1. Reversibility of phagocytosis inhibition by Tat. Human
Supplementary Figure 1. Processing and quality control for recombinant proteins.
 Supplementary Figure 1 Processing and quality control for recombinant proteins. (a) Schematic representation of processing of a recombinant protein library. Recombinant proteins were generated individually.
Supplementary Figure 1 Processing and quality control for recombinant proteins. (a) Schematic representation of processing of a recombinant protein library. Recombinant proteins were generated individually.
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified
 Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Supplemental Data. Furlan et al. Plant Cell (2017) /tpc
 Supplemental Data. Furlan et al. Plant Cell (0) 0.0/tpc..00. Supplemental Data. Furlan et al. Plant Cell (0) 0.0/tpc..00. Supplemental Data. Furlan et al. Plant Cell (0) 0.0/tpc..00. Supplemental Figure.
Supplemental Data. Furlan et al. Plant Cell (0) 0.0/tpc..00. Supplemental Data. Furlan et al. Plant Cell (0) 0.0/tpc..00. Supplemental Data. Furlan et al. Plant Cell (0) 0.0/tpc..00. Supplemental Figure.
Problem Set #2
 20.320 Problem Set #2 Due on September 30rd, 2011 at 11:59am. No extensions will be granted. General Instructions: 1. You are expected to state all of your assumptions, and provide step-by-step solutions
20.320 Problem Set #2 Due on September 30rd, 2011 at 11:59am. No extensions will be granted. General Instructions: 1. You are expected to state all of your assumptions, and provide step-by-step solutions
Basic concepts of molecular biology
 Basic concepts of molecular biology Gabriella Trucco Email: gabriella.trucco@unimi.it Life The main actors in the chemistry of life are molecules called proteins nucleic acids Proteins: many different
Basic concepts of molecular biology Gabriella Trucco Email: gabriella.trucco@unimi.it Life The main actors in the chemistry of life are molecules called proteins nucleic acids Proteins: many different
Fast, three-dimensional super-resolution imaging of live cells
 Nature Methods Fast, three-dimensional super-resolution imaging of live cells Sara A Jones, Sang-Hee Shim, Jiang He & Xiaowei Zhuang Supplementary Figure 1 Supplementary Figure 2 Supplementary Figure 3
Nature Methods Fast, three-dimensional super-resolution imaging of live cells Sara A Jones, Sang-Hee Shim, Jiang He & Xiaowei Zhuang Supplementary Figure 1 Supplementary Figure 2 Supplementary Figure 3
Conformational changes in IgE contribute to its. uniquely slow dissociation rate from receptor FcεRI
 Conformational changes in IgE contribute to its uniquely slow dissociation rate from receptor FcεRI M.D. Holdom, A.M. Davies, J.E. Nettleship, S.C. Bagby, B. Dhaliwal, E. Girardi, J. Hunt, H.J. Gould,
Conformational changes in IgE contribute to its uniquely slow dissociation rate from receptor FcεRI M.D. Holdom, A.M. Davies, J.E. Nettleship, S.C. Bagby, B. Dhaliwal, E. Girardi, J. Hunt, H.J. Gould,
Masayoshi Honda, Jeehae Park, Robert A. Pugh, Taekjip Ha, and Maria Spies
 Molecular Cell, Volume 35 Supplemental Data Single-Molecule Analysis Reveals Differential Effect of ssdna-binding Proteins on DNA Translocation by XPD Helicase Masayoshi Honda, Jeehae Park, Robert A. Pugh,
Molecular Cell, Volume 35 Supplemental Data Single-Molecule Analysis Reveals Differential Effect of ssdna-binding Proteins on DNA Translocation by XPD Helicase Masayoshi Honda, Jeehae Park, Robert A. Pugh,
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
HBV RNA pre-genome encodes specific motifs that mediate interactions with the viral core protein that promote nucleocapsid assembly
 In the format provided by the authors and unedited. SUPPLEMENTARY INFORMATION Nikesh Patel *, Simon J. White *, Rebecca F Thompson, Richard Bingham 1, Eva U. Weiß 1, Daniel P. Maskell, Adam Zlotnick 2,
In the format provided by the authors and unedited. SUPPLEMENTARY INFORMATION Nikesh Patel *, Simon J. White *, Rebecca F Thompson, Richard Bingham 1, Eva U. Weiß 1, Daniel P. Maskell, Adam Zlotnick 2,
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Contents: Supplementary Figure 1. Additional structural and binding data for designed tuim peptides. Supplementary Figure 2. Subcellular localization patterns of designed tuim
SUPPLEMENTARY INFORMATION Contents: Supplementary Figure 1. Additional structural and binding data for designed tuim peptides. Supplementary Figure 2. Subcellular localization patterns of designed tuim
Orange fluorescent proteins shift constructed from cyanobacteriochromes. chromophorylated with phycoerythrobilin. Kai-Hong Zhao 1 Ming Zhou 1, *
 Electronic Supplementary Material (ESI) for Photochemical. This journal is The Royal Society of Chemistry and Owner Societies 2014 Orange fluorescent proteins shift constructed from cyanobacteriochromes
Electronic Supplementary Material (ESI) for Photochemical. This journal is The Royal Society of Chemistry and Owner Societies 2014 Orange fluorescent proteins shift constructed from cyanobacteriochromes
Background for Schizophrenia Research Article. Sekar A, et al., Schizophrenia risk from complex variation of complement component 4, Nature, 2016
 Background for Schizophrenia Research Article Sekar A, et al., Schizophrenia risk from complex variation of complement component 4, Nature, 2016 Topics The Major Histocompatibility Complex (MHC) region
Background for Schizophrenia Research Article Sekar A, et al., Schizophrenia risk from complex variation of complement component 4, Nature, 2016 Topics The Major Histocompatibility Complex (MHC) region
