Supplementary Figure 1. Study design of a multi-stage GWAS of gout.
|
|
|
- Alexis Lawson
- 5 years ago
- Views:
Transcription
1 Supplementary Figure 1. Study design of a multi-stage GWAS of gout.
2 Supplementary Figure 2. Plot of the first two principal components from the analysis of the genome-wide study (after QC) combined with HapMap data. The samples disease status and the ethnicity of the HapMap samples are indicated by color. Briefly, Controls are red circles (n = 1,848); Cases are blue circles (n = 1,255); HapMap Chinese (CHB) samples are pink circles (n = 45), Japanese (JPT) samples are yellow circles (n = 45), European (CEU) samples are purple circles (n = 90) and African (YRI) samples are orange circles (n = 90).
3 C controls cases C1 Supplementary Figure 3. Plot of the first two principal components from the analysis of the genome-wide study (after QC). Controls are red circles (n = 1,848) and Cases are blue circles (n = 1,255).
4 Supplementary Figure 4. Quantile-Quantile (Q-Q) plot of the discovery data.
5 a) Genotyped b) Genotyped and Imputed (autosomes) Supplementary Figure 5. Plot of genome-wide association result for the discovery data. Chromosomes are delineated by alternating colors, as labeled on the x-axis. The y-axis shows the log10 P-values (using logistic regression).
6 Supplementary Table 1. Description of the discovery and replication phases samples. N Location %Female Age (years±s.d.) Uric Acid (umol l -1 ±s.d.) Discovery Phase Cases 1255 Shandong Province ± ±107.6 Controls 1848 Northern China a ± Replication Phase 1 Cases 814 Shandong Province ± ±110.2 Controls 1414 Shandong Province ± ±48.5 Replication Phase 2 Cases 882 Shandong Province ± ±110.6 Controls 1895 Shandong Province ± ±46.6 Replication Phase 3 Cases 996 Northern China a ± ±126.8 Controls 786 Northern China a ± ±50.9 Cases 328 Sichuan Province ± ±135.4 Controls 329 Sichuan Province ± ±41.6 Female cohort
7 N Location %Female Age (years±s.d.) Uric Acid (umol l -1 ±s.d.) Cases 215 Shandong Province (Mainly) ± ±120.2 Controls 541 Shandong Province (Mainly) ± ±39.3 Hyperuricemia cohort 1644 Shandong Province (Mainly) ± ±56.2 a Northern China: Shandong, Heilongjiang, Shanxi, Hebei, and Beijing.
8 Supplementary Table 2. Results for the SNPs shown P<5e-5 in the discovery phase (GWAS) within the loci identified in the previous GWASs. Chr. SNP Position A1 Freq. OR a P a 4 rs T E-06 4 rs T E-05 4 rs T E-05 4 rs T E-05 4 rs A E-05 4 rs A E-05 4 rs A E-06 4 rs G E-06 4 rs C E-06 4 rs T E-10 4 chr4_ G E-06 4 rs A E rs T E rs T E-05 Position, based on hg18; A1, minor allele; Freq., frequency of A1 in controls; OR, odds ratio; The minor allele was the coded allele; P, P value. a The OR and P values are PCA-adjusted (using logistic regression).
9 Supplementary Table 3. Results of the follow-up phase II (REP 2) and GWAS-REP1-REP2 meta-analysis for the 13 replication SNPs. Chr. SNP Position A1 A2 REP2 (882 cases and 1895 controls) GWAS-REP1-REP2-META F_A F_U P a OR a P b OR b Q I 1 rs G T E E rs C T E E rs A G E E rs G C E E rs A G E E rs T C E E rs G A E E rs G A E E rs G A E E rs C T E E rs A G E E rs T C E E rs T C E E SD, Shan Dong; SC, Si Chuan; Position, based on hg18; A1, minor allele; A2, major allele; F_A, frequency of the minor allele in cases; F_U, frequency of the minor allele in controls; OR, odds ratio; The minor allele was the coded allele; P, P value; Q, p-value for Cochrane's Q statistic; I 2, I 2 heterogeneity index (0-100). a The OR and P values are based on the logistic regression. b The OR and P values are based on the meta-analysis under fixed-effects model.
10 Supplementary Table 4. Results of the follow-up phase III (REP 3) for the 4 replication SNPs. Chr. SNP Position A1 A2 REP3-META REP3-NC (996 cases and 786 controls) REP3-SC (328 cases and 329 controls) P a OR a Q I F_A F_U P b OR b F_A F_U P b OR b 9 rs G A 1.46E rs G A 2.79E rs T C 2.38E rs T C 7.38E E NC, Northern China; SC, Si Chuan; Position, based on hg18; A1, minor allele; A2, major allele; F_A, frequency of the minor allele in cases; F_U, frequency of the minor allele in controls; OR, odds ratio; The minor allele was the coded allele; P, P value; Q, p-value for Cochrane's Q statistic; I 2, I 2 heterogeneity index (0-100). a The OR and P values are based on the meta-analysis under fixed-effects model. b The OR and P values are based on the logistic regression.
11 Supplementary Table 5. Association results without and with adjustment for egfr in a subset sample for the four genome-wide significant SNPs. Chr. SNP Position A1 Without adjustment for egfr With adjustment for egfr OR P OR P 9 rs G E E rs G E E rs T E E rs T E E-06 Position, based on hg18; A1, minor allele; OR, odds ratio; The minor allele was the coded allele; P, P value; egfr, estimated glomerular filtration rate. The OR and P values are based on the logistic regression.
12 Supplementary Table 6. Results of the female cohort for the four genome-wide significant SNPs. Chr. SNP Position A1 F_A F_U A2 P OR 9 rs G A rs G A rs T C rs T C Position, based on hg18; A1, minor allele; A2, major allele; F_A, frequency of the minor allele in cases; F_U, frequency of the minor allele in controls; OR, odds ratio; The minor allele was the coded allele; P, P value. The OR and P values are based on the logistic regression.
13 Supplementary Table 7. Summary of genomic annotation by HaploReg v2 for the genome wide significant loci. CHR SNP LD r² D' Promoter Enhancer DNAse Proteins bound Motifs changed 9 rs CD34.MBP cell types H1-hESC,CD34+_Mobilized,WERI-Rb-1 CTCF 4 altered motifs 9 rs CD34.MBP cell types 6 cell types CTCF GR 9 rs cell types Egr-1 9 rs CD34.C 7 altered motifs 9 rs CD34.C AP-1,CEBPB,PEBP 9 rs DMRT2,HDAC2,RXR::LXR 9 rs rs LUN-1,PLZF 11 rs cell types 10 altered motifs 11 rs cell types ips 4 altered motifs 17 rs cell types HMEC,Caco-2 4 altered motifs 17 rs Egr-1,Mtf1,Pax-4 CHR, chromosome; the LD information were derived from the 1000 Genomes Project ASI data for the associated SNP (marked in bold) and its surrogates. Promoter, Enhancer, DNAse, Proteins bound and Motifs changed demonstrated evidence of histone modifications, DNase hypersensitivity sites or transcription factor occupancy as shown by the HaploReg v2 analysis.
14 Supplementary Table 8. LD of the gout associated SNP and the reported T2D associated SNPs in KCNQ1 region. Gout associated SNP Urate associated SNP Distance r 2 D rs rs rs rs rs rs rs rs rs rs rs rs rs rs The r2 and D values were estimated from the 1000Genome Asian dataset.
15 Supplementary Table 9. Power Analyses at P < 5e-5 for the GWAS discovery stage. OR % 0% 1% 1% 1% 1% 1% 0% % 3% 11% 18% 22% 20% 8% 3% % 9% 28% 41% 46% 41% 18% 6% % 19% 50% 66% 70% 64% 32% 11% % 34% 72% 84% 87% 81% 47% 19% % 52% 87% 94% 95% 92% 61% 27% % 82% 98% 100% 100% 99% 82% 45% Power figures at representative and relevant ORs (ORs of 1.10 to 1.50) and allele frequencies (0.05 to 0.85) are displayed for the Stage 1 GWAS (discovery) analysis. The powers over 80% were indicated in bold.
Supplementary Figure 1 a
 Supplementary Figure 1 a b GWAS second stage log 10 observed P 0 2 4 6 8 10 12 0 1 2 3 4 log 10 expected P rs3077 (P hetero =0.84) GWAS second stage (BBJ, Japan) First replication (BBJ, Japan) Second replication
Supplementary Figure 1 a b GWAS second stage log 10 observed P 0 2 4 6 8 10 12 0 1 2 3 4 log 10 expected P rs3077 (P hetero =0.84) GWAS second stage (BBJ, Japan) First replication (BBJ, Japan) Second replication
Genome-wide association study identifies five loci associated with susceptibility to pancreatic cancer in Chinese populations. Supplementary Materials
 Genome-wide association study identifies five loci associated with susceptibility to pancreatic cancer in Chinese populations Supplementary Materials Chen Wu 1, 22, Xiaoping Miao 2, 22, Liming Huang 1,
Genome-wide association study identifies five loci associated with susceptibility to pancreatic cancer in Chinese populations Supplementary Materials Chen Wu 1, 22, Xiaoping Miao 2, 22, Liming Huang 1,
Nature Genetics: doi: /ng.3143
 Supplementary Figure 1 Quantile-quantile plot of the association P values obtained in the discovery sample collection. The two clear outlying SNPs indicated for follow-up assessment are rs6841458 and rs7765379.
Supplementary Figure 1 Quantile-quantile plot of the association P values obtained in the discovery sample collection. The two clear outlying SNPs indicated for follow-up assessment are rs6841458 and rs7765379.
Nature Genetics: doi: /ng Supplementary Figure 1. Eigenvector plots for the three GWAS including subpopulations from the NCI scan.
 Supplementary Figure 1 Eigenvector plots for the three GWAS including subpopulations from the NCI scan. The NCI subpopulations are as follows: NITC, Nutrition Intervention Trial Cohort; SHNX, Shanxi Cancer
Supplementary Figure 1 Eigenvector plots for the three GWAS including subpopulations from the NCI scan. The NCI subpopulations are as follows: NITC, Nutrition Intervention Trial Cohort; SHNX, Shanxi Cancer
Supplementary Fig. 1. Location of top two candidemia associated SNPs in CD58 gene
 Supplementary Figures Supplementary Fig. 1. Location of top two candidemia associated SNPs in CD58 gene locus. The region encompass CD58 and three long non-coding RNAs (RP4-655J12.4, RP5-1086K13.1 and
Supplementary Figures Supplementary Fig. 1. Location of top two candidemia associated SNPs in CD58 gene locus. The region encompass CD58 and three long non-coding RNAs (RP4-655J12.4, RP5-1086K13.1 and
Genome-wide association study identifies a susceptibility locus for HCVinduced hepatocellular carcinoma. Supplementary Information
 Genome-wide association study identifies a susceptibility locus for HCVinduced hepatocellular carcinoma Vinod Kumar 1,2, Naoya Kato 3, Yuji Urabe 1, Atsushi Takahashi 2, Ryosuke Muroyama 3, Naoya Hosono
Genome-wide association study identifies a susceptibility locus for HCVinduced hepatocellular carcinoma Vinod Kumar 1,2, Naoya Kato 3, Yuji Urabe 1, Atsushi Takahashi 2, Ryosuke Muroyama 3, Naoya Hosono
Supplementary Figure 2.Quantile quantile plots (QQ) of the exome sequencing results Chi square was used to test the association between genetic
 SUPPLEMENTARY INFORMATION Supplementary Figure 1.Description of the study design The samples in the initial stage (China cohort, exome sequencing) including 216 AMD cases and 1,553 controls were from the
SUPPLEMENTARY INFORMATION Supplementary Figure 1.Description of the study design The samples in the initial stage (China cohort, exome sequencing) including 216 AMD cases and 1,553 controls were from the
Supplementary Figure 1. Linkage disequilibrium (LD) at the CDKN2A locus
 rs3731249 rs3731217 Supplementary Figure 1. Linkage disequilibrium (LD) at the CDKN2A locus. Minimal correlation was observed (r 2 =0.0007) in Hapmap CEU individuals between B ALL risk variants rs3731249
rs3731249 rs3731217 Supplementary Figure 1. Linkage disequilibrium (LD) at the CDKN2A locus. Minimal correlation was observed (r 2 =0.0007) in Hapmap CEU individuals between B ALL risk variants rs3731249
Supplementary Online Content
 Supplementary Online Content Lee JH, Cheng R, Barral S, Reitz C, Medrano M, Lantigua R, Jiménez-Velazquez IZ, Rogaeva E, St. George-Hyslop P, Mayeux R. Identification of novel loci for Alzheimer disease
Supplementary Online Content Lee JH, Cheng R, Barral S, Reitz C, Medrano M, Lantigua R, Jiménez-Velazquez IZ, Rogaeva E, St. George-Hyslop P, Mayeux R. Identification of novel loci for Alzheimer disease
Supplementary Information
 Supplementary Information Two new susceptibility loci for Kawasaki disease identified through genome-wide association analysis Yi-Ching Lee 1,2, Ho-Chang Kuo 3,4, Jeng-Sheng Chang,Luan-Yin Chang 6,1, Li-Min
Supplementary Information Two new susceptibility loci for Kawasaki disease identified through genome-wide association analysis Yi-Ching Lee 1,2, Ho-Chang Kuo 3,4, Jeng-Sheng Chang,Luan-Yin Chang 6,1, Li-Min
A genome-wide association study in Han Chinese identifies new susceptibility loci for. ankylosing spondylitis. Supplementary Materials
 A genome-wide association study in Han Chinese identifies new susceptibility loci for ankylosing spondylitis Supplementary Materials Zhiming Lin 1,24, Jin-Xin Bei 2,24, Meixin Shen 3, Qiuxia Li 1, Zetao
A genome-wide association study in Han Chinese identifies new susceptibility loci for ankylosing spondylitis Supplementary Materials Zhiming Lin 1,24, Jin-Xin Bei 2,24, Meixin Shen 3, Qiuxia Li 1, Zetao
Single Nucleotide Polymorphisms (SNPs)
 Single Nucleotide Polymorphisms (SNPs) Sequence variations Single nucleotide polymorphisms Insertions/deletions Copy number variations (large: >1kb) Variable (short) number tandem repeats Single Nucleotide
Single Nucleotide Polymorphisms (SNPs) Sequence variations Single nucleotide polymorphisms Insertions/deletions Copy number variations (large: >1kb) Variable (short) number tandem repeats Single Nucleotide
Supplementary Material
 Supplementary Material Supplementary Table 1: Luciferase assay primer sequences rs7090445 Forward 5-3 AATCCCAGGTGCTTATGGACA rs7090445 Reverse 5-3 TGAGCCGAGATTGCACCATT rs7896246 Forward 5-3 GTGTCCAGGAACTCCCAAGG
Supplementary Material Supplementary Table 1: Luciferase assay primer sequences rs7090445 Forward 5-3 AATCCCAGGTGCTTATGGACA rs7090445 Reverse 5-3 TGAGCCGAGATTGCACCATT rs7896246 Forward 5-3 GTGTCCAGGAACTCCCAAGG
Genetic Variation and Genome- Wide Association Studies. Keyan Salari, MD/PhD Candidate Department of Genetics
 Genetic Variation and Genome- Wide Association Studies Keyan Salari, MD/PhD Candidate Department of Genetics How many of you did the readings before class? A. Yes, of course! B. Started, but didn t get
Genetic Variation and Genome- Wide Association Studies Keyan Salari, MD/PhD Candidate Department of Genetics How many of you did the readings before class? A. Yes, of course! B. Started, but didn t get
A genome wide association study of metabolic traits in human urine
 Supplementary material for A genome wide association study of metabolic traits in human urine Suhre et al. CONTENTS SUPPLEMENTARY FIGURES Supplementary Figure 1: Regional association plots surrounding
Supplementary material for A genome wide association study of metabolic traits in human urine Suhre et al. CONTENTS SUPPLEMENTARY FIGURES Supplementary Figure 1: Regional association plots surrounding
Nature Genetics: doi: /ng Supplementary Figure 1
 Supplementary Figure 1 Flowchart illustrating the three complementary strategies for gene prioritization used in this study.. Supplementary Figure 2 Flow diagram illustrating calcaneal quantitative ultrasound
Supplementary Figure 1 Flowchart illustrating the three complementary strategies for gene prioritization used in this study.. Supplementary Figure 2 Flow diagram illustrating calcaneal quantitative ultrasound
Supplementary Figures
 1 Supplementary Figures exm26442 2.40 2.20 2.00 1.80 Norm Intensity (B) 1.60 1.40 1.20 1 0.80 0.60 0.40 0.20 2 0-0.20 0 0.20 0.40 0.60 0.80 1 1.20 1.40 1.60 1.80 2.00 2.20 2.40 2.60 2.80 Norm Intensity
1 Supplementary Figures exm26442 2.40 2.20 2.00 1.80 Norm Intensity (B) 1.60 1.40 1.20 1 0.80 0.60 0.40 0.20 2 0-0.20 0 0.20 0.40 0.60 0.80 1 1.20 1.40 1.60 1.80 2.00 2.20 2.40 2.60 2.80 Norm Intensity
A genome-wide association study identifies multiple susceptibility loci for chronic lymphocytic leukemia
 A genome-wide association study identifies multiple susceptibility loci for chronic lymphocytic leukemia Helen E Speedy 1*, Maria Chiara Di Bernardo 1*, Georgina P Sava 1, Martin J S Dyer 2, Amy Holroyd
A genome-wide association study identifies multiple susceptibility loci for chronic lymphocytic leukemia Helen E Speedy 1*, Maria Chiara Di Bernardo 1*, Georgina P Sava 1, Martin J S Dyer 2, Amy Holroyd
Supplementary Table 1. Idd13 candidate interval supporting human LTC-ICs.
 Supplementary Table 1. Idd13 candidate interval supporting human LTC-ICs. Chr Start position Genomic marker EnsEMBL gene ID Gene symbol Primer 1 (5 3 ) Primer 2 (5 3 ) 2 128675293 ENSMUSG00000027387 Zc3h8
Supplementary Table 1. Idd13 candidate interval supporting human LTC-ICs. Chr Start position Genomic marker EnsEMBL gene ID Gene symbol Primer 1 (5 3 ) Primer 2 (5 3 ) 2 128675293 ENSMUSG00000027387 Zc3h8
Genome-wide association study identifies multiple susceptibility loci for pulmonary fibrosis
 correction notice Nat. Genet. 45, 613 620 (2013); published online 14 April 2013; corrected online 1 October 2013 Genome-wide association study identifies multiple susceptibility loci for pulmonary fibrosis
correction notice Nat. Genet. 45, 613 620 (2013); published online 14 April 2013; corrected online 1 October 2013 Genome-wide association study identifies multiple susceptibility loci for pulmonary fibrosis
Redefine what s possible with the Axiom Genotyping Solution
 Redefine what s possible with the Axiom Genotyping Solution From discovery to translation on a single platform The Axiom Genotyping Solution enables enhanced genotyping studies to accelerate your research
Redefine what s possible with the Axiom Genotyping Solution From discovery to translation on a single platform The Axiom Genotyping Solution enables enhanced genotyping studies to accelerate your research
S G. Design and Analysis of Genetic Association Studies. ection. tatistical. enetics
 S G ection ON tatistical enetics Design and Analysis of Genetic Association Studies Hemant K Tiwari, Ph.D. Professor & Head Section on Statistical Genetics Department of Biostatistics School of Public
S G ection ON tatistical enetics Design and Analysis of Genetic Association Studies Hemant K Tiwari, Ph.D. Professor & Head Section on Statistical Genetics Department of Biostatistics School of Public
A candidate gene study of the type I interferon pathway implicates IKBKE and IL8 as risk loci for SLE
 A candidate gene study of the type I interferon pathway implicates IKBKE and IL8 as risk loci for SLE Johanna K. Sandling, Sophie Garnier, Snaevar Sigurdsson, Chuan Wang, Gunnel Nordmark, Iva Gunnarsson,
A candidate gene study of the type I interferon pathway implicates IKBKE and IL8 as risk loci for SLE Johanna K. Sandling, Sophie Garnier, Snaevar Sigurdsson, Chuan Wang, Gunnel Nordmark, Iva Gunnarsson,
Bioinformatic Analysis of SNP Data for Genetic Association Studies EPI573
 Bioinformatic Analysis of SNP Data for Genetic Association Studies EPI573 Mark J. Rieder Department of Genome Sciences mrieder@u.washington washington.edu Epidemiology Studies Cohort Outcome Model to fit/explain
Bioinformatic Analysis of SNP Data for Genetic Association Studies EPI573 Mark J. Rieder Department of Genome Sciences mrieder@u.washington washington.edu Epidemiology Studies Cohort Outcome Model to fit/explain
Data Sources and Biobanks in the Asia-Pacific Region. Wei Zhou, MD, Ph.D. Department of Epidemiology, Merck Research Laboratories October 23, 2014
 Data Sources and Biobanks in the Asia-Pacific Region Wei Zhou, MD, Ph.D. Department of Epidemiology, Merck Research Laboratories October 23, 2014 1 Disclosures Wei Zhou is currently an employee of Merck
Data Sources and Biobanks in the Asia-Pacific Region Wei Zhou, MD, Ph.D. Department of Epidemiology, Merck Research Laboratories October 23, 2014 1 Disclosures Wei Zhou is currently an employee of Merck
Population stratification. Background & PLINK practical
 Population stratification Background & PLINK practical Variation between, within populations Any two humans differ ~0.1% of their genome (1 in ~1000bp) ~8% of this variation is accounted for by the major
Population stratification Background & PLINK practical Variation between, within populations Any two humans differ ~0.1% of their genome (1 in ~1000bp) ~8% of this variation is accounted for by the major
Genotype quality control with plinkqc Hannah Meyer
 Genotype quality control with plinkqc Hannah Meyer 219-3-1 Contents Introduction 1 Per-individual quality control....................................... 2 Per-marker quality control.........................................
Genotype quality control with plinkqc Hannah Meyer 219-3-1 Contents Introduction 1 Per-individual quality control....................................... 2 Per-marker quality control.........................................
Population description. 103 CHB Han Chinese in Beijing, China East Asian EAS. 104 JPT Japanese in Tokyo, Japan East Asian EAS
 1 Supplementary Table 1 Description of the 1000 Genomes Project Phase 3 representing 2504 individuals from 26 different global populations that are assigned to five super-populations Number of individuals
1 Supplementary Table 1 Description of the 1000 Genomes Project Phase 3 representing 2504 individuals from 26 different global populations that are assigned to five super-populations Number of individuals
I/O Suite, VCF (1000 Genome) and HapMap
 I/O Suite, VCF (1000 Genome) and HapMap Hin-Tak Leung April 13, 2013 Contents 1 Introduction 1 1.1 Ethnic Composition of 1000G vs HapMap........................ 2 2 1000 Genome vs HapMap YRI (Africans)
I/O Suite, VCF (1000 Genome) and HapMap Hin-Tak Leung April 13, 2013 Contents 1 Introduction 1 1.1 Ethnic Composition of 1000G vs HapMap........................ 2 2 1000 Genome vs HapMap YRI (Africans)
DNA Collection. Data Quality Control. Whole Genome Amplification. Whole Genome Amplification. Measure DNA concentrations. Pros
 DNA Collection Data Quality Control Suzanne M. Leal Baylor College of Medicine sleal@bcm.edu Copyrighted S.M. Leal 2016 Blood samples For unlimited supply of DNA Transformed cell lines Buccal Swabs Small
DNA Collection Data Quality Control Suzanne M. Leal Baylor College of Medicine sleal@bcm.edu Copyrighted S.M. Leal 2016 Blood samples For unlimited supply of DNA Transformed cell lines Buccal Swabs Small
Haplotypes, linkage disequilibrium, and the HapMap
 Haplotypes, linkage disequilibrium, and the HapMap Jeffrey Barrett Boulder, 2009 LD & HapMap Boulder, 2009 1 / 29 Outline 1 Haplotypes 2 Linkage disequilibrium 3 HapMap 4 Tag SNPs LD & HapMap Boulder,
Haplotypes, linkage disequilibrium, and the HapMap Jeffrey Barrett Boulder, 2009 LD & HapMap Boulder, 2009 1 / 29 Outline 1 Haplotypes 2 Linkage disequilibrium 3 HapMap 4 Tag SNPs LD & HapMap Boulder,
Analysis of genome-wide genotype data
 Analysis of genome-wide genotype data Acknowledgement: Several slides based on a lecture course given by Jonathan Marchini & Chris Spencer, Cape Town 2007 Introduction & definitions - Allele: A version
Analysis of genome-wide genotype data Acknowledgement: Several slides based on a lecture course given by Jonathan Marchini & Chris Spencer, Cape Town 2007 Introduction & definitions - Allele: A version
Supplementary Figure 1. Quantile quantile plot for the combined analysis of cohorts 1 and 2.
 Supplementary Figure 1 Quantile quantile plot for the combined analysis of cohorts 1 and 2. Quantile quantile plot of the observed log 10 (P values) versus the expectation under the null hypothesis. Data
Supplementary Figure 1 Quantile quantile plot for the combined analysis of cohorts 1 and 2. Quantile quantile plot of the observed log 10 (P values) versus the expectation under the null hypothesis. Data
Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C
 CORRECTION NOTICE Nat. Genet. 47, 598 606 (2015) Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C Borbala Mifsud, Filipe Tavares-Cadete, Alice N Young, Robert Sugar,
CORRECTION NOTICE Nat. Genet. 47, 598 606 (2015) Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C Borbala Mifsud, Filipe Tavares-Cadete, Alice N Young, Robert Sugar,
Petar Pajic 1 *, Yen Lung Lin 1 *, Duo Xu 1, Omer Gokcumen 1 Department of Biological Sciences, University at Buffalo, Buffalo, NY.
 The psoriasis associated deletion of late cornified envelope genes LCE3B and LCE3C has been maintained under balancing selection since Human Denisovan divergence Petar Pajic 1 *, Yen Lung Lin 1 *, Duo
The psoriasis associated deletion of late cornified envelope genes LCE3B and LCE3C has been maintained under balancing selection since Human Denisovan divergence Petar Pajic 1 *, Yen Lung Lin 1 *, Duo
Population differentiation analysis of 54,734 European Americans reveals independent evolution of ADH1B gene in Europe and East Asia
 Population differentiation analysis of 54,734 European Americans reveals independent evolution of ADH1B gene in Europe and East Asia Kevin Galinsky Harvard T. H. Chan School of Public Health American Society
Population differentiation analysis of 54,734 European Americans reveals independent evolution of ADH1B gene in Europe and East Asia Kevin Galinsky Harvard T. H. Chan School of Public Health American Society
Human Genetics and Gene Mapping of Complex Traits
 Human Genetics and Gene Mapping of Complex Traits Advanced Genetics, Spring 2015 Human Genetics Series Thursday 4/02/15 Nancy L. Saccone, nlims@genetics.wustl.edu ancestral chromosome present day chromosomes:
Human Genetics and Gene Mapping of Complex Traits Advanced Genetics, Spring 2015 Human Genetics Series Thursday 4/02/15 Nancy L. Saccone, nlims@genetics.wustl.edu ancestral chromosome present day chromosomes:
Genotype Prediction with SVMs
 Genotype Prediction with SVMs Nicholas Johnson December 12, 2008 1 Summary A tuned SVM appears competitive with the FastPhase HMM (Stephens and Scheet, 2006), which is the current state of the art in genotype
Genotype Prediction with SVMs Nicholas Johnson December 12, 2008 1 Summary A tuned SVM appears competitive with the FastPhase HMM (Stephens and Scheet, 2006), which is the current state of the art in genotype
SUPPLEMENTAL MATERIAL
 SUPPLEMENTAL MATERIAL Supplementary Table 1: RT-qPCR primer sequences. Sequences are shown from 5 to 3 direction; all primers are designed using mouse genome as reference. 36B4-F; TGAAGCAAAGGAAGAGTCGGAGGA
SUPPLEMENTAL MATERIAL Supplementary Table 1: RT-qPCR primer sequences. Sequences are shown from 5 to 3 direction; all primers are designed using mouse genome as reference. 36B4-F; TGAAGCAAAGGAAGAGTCGGAGGA
4.1.1 Association of SNP variants within PARK2-PACRG gene regulatory region with leprosy susceptibility sharing chromosomal region 6q26
 4.1 GENETIC VARIATIONS IN PARK2 AND PACRG GENE REGULATORY REGIONS AND THEIR INTERACTION WITH IMPORTANT IMMUNO-REGULATORY GENES IN THE OUTCOME OF LEPROSY PARK2 and PACRG gene regulatory region was saturated
4.1 GENETIC VARIATIONS IN PARK2 AND PACRG GENE REGULATORY REGIONS AND THEIR INTERACTION WITH IMPORTANT IMMUNO-REGULATORY GENES IN THE OUTCOME OF LEPROSY PARK2 and PACRG gene regulatory region was saturated
Wu et al., Determination of genetic identity in therapeutic chimeric states. We used two approaches for identifying potentially suitable deletion loci
 SUPPLEMENTARY METHODS AND DATA General strategy for identifying deletion loci We used two approaches for identifying potentially suitable deletion loci for PDP-FISH analysis. In the first approach, we
SUPPLEMENTARY METHODS AND DATA General strategy for identifying deletion loci We used two approaches for identifying potentially suitable deletion loci for PDP-FISH analysis. In the first approach, we
Supplementary Figures
 Supplementary Figures 1 Supplementary Figure 1. Analyses of present-day population differentiation. (A, B) Enrichment of strongly differentiated genic alleles for all present-day population comparisons
Supplementary Figures 1 Supplementary Figure 1. Analyses of present-day population differentiation. (A, B) Enrichment of strongly differentiated genic alleles for all present-day population comparisons
Statistical Tools for Predicting Ancestry from Genetic Data
 Statistical Tools for Predicting Ancestry from Genetic Data Timothy Thornton Department of Biostatistics University of Washington March 1, 2015 1 / 33 Basic Genetic Terminology A gene is the most fundamental
Statistical Tools for Predicting Ancestry from Genetic Data Timothy Thornton Department of Biostatistics University of Washington March 1, 2015 1 / 33 Basic Genetic Terminology A gene is the most fundamental
Derrek Paul Hibar
 Derrek Paul Hibar derrek.hibar@ini.usc.edu Obtain the ADNI Genetic Data Quality Control Procedures Missingness Testing for relatedness Minor allele frequency (MAF) Hardy-Weinberg Equilibrium (HWE) Testing
Derrek Paul Hibar derrek.hibar@ini.usc.edu Obtain the ADNI Genetic Data Quality Control Procedures Missingness Testing for relatedness Minor allele frequency (MAF) Hardy-Weinberg Equilibrium (HWE) Testing
Genotyping Technology How to Analyze Your Own Genome Fall 2013
 Genotyping Technology 02-223 How to nalyze Your Own Genome Fall 2013 HapMap Project Phase 1 Phase 2 Phase 3 Samples & POP panels Genotyping centers Unique QC+ SNPs 269 samples (4 populations) HapMap International
Genotyping Technology 02-223 How to nalyze Your Own Genome Fall 2013 HapMap Project Phase 1 Phase 2 Phase 3 Samples & POP panels Genotyping centers Unique QC+ SNPs 269 samples (4 populations) HapMap International
Understanding genetic association studies. Peter Kamerman
 Understanding genetic association studies Peter Kamerman Outline CONCEPTS UNDERLYING GENETIC ASSOCIATION STUDIES Genetic concepts: - Underlying principals - Genetic variants - Linkage disequilibrium -
Understanding genetic association studies Peter Kamerman Outline CONCEPTS UNDERLYING GENETIC ASSOCIATION STUDIES Genetic concepts: - Underlying principals - Genetic variants - Linkage disequilibrium -
Nature Genetics: doi: /ng Supplementary Figure 1. H3K27ac HiChIP enriches enhancer promoter-associated chromatin contacts.
 Supplementary Figure 1 H3K27ac HiChIP enriches enhancer promoter-associated chromatin contacts. (a) Schematic of chromatin contacts captured in H3K27ac HiChIP. (b) Loop call overlap for cohesin HiChIP
Supplementary Figure 1 H3K27ac HiChIP enriches enhancer promoter-associated chromatin contacts. (a) Schematic of chromatin contacts captured in H3K27ac HiChIP. (b) Loop call overlap for cohesin HiChIP
Supplementary table 1. Study design
 Supplementary table 1. Study design Population GWAS genotyping platform N Case/Controls After genotyping quality controls Genotyped SNPs Analyzed SNPs (overlapping between populations) Statistical Power*
Supplementary table 1. Study design Population GWAS genotyping platform N Case/Controls After genotyping quality controls Genotyped SNPs Analyzed SNPs (overlapping between populations) Statistical Power*
The HapMap Project and Haploview
 The HapMap Project and Haploview David Evans Ben Neale University of Oxford Wellcome Trust Centre for Human Genetics Human Haplotype Map General Idea: Characterize the distribution of Linkage Disequilibrium
The HapMap Project and Haploview David Evans Ben Neale University of Oxford Wellcome Trust Centre for Human Genetics Human Haplotype Map General Idea: Characterize the distribution of Linkage Disequilibrium
The Whole Genome TagSNP Selection and Transferability Among HapMap Populations. Reedik Magi, Lauris Kaplinski, and Maido Remm
 The Whole Genome TagSNP Selection and Transferability Among HapMap Populations Reedik Magi, Lauris Kaplinski, and Maido Remm Pacific Symposium on Biocomputing 11:535-543(2006) THE WHOLE GENOME TAGSNP SELECTION
The Whole Genome TagSNP Selection and Transferability Among HapMap Populations Reedik Magi, Lauris Kaplinski, and Maido Remm Pacific Symposium on Biocomputing 11:535-543(2006) THE WHOLE GENOME TAGSNP SELECTION
Genome-wide analyses in admixed populations: Challenges and opportunities
 Genome-wide analyses in admixed populations: Challenges and opportunities E-mail: esteban.parra@utoronto.ca Esteban J. Parra, Ph.D. Admixed populations: an invaluable resource to study the genetics of
Genome-wide analyses in admixed populations: Challenges and opportunities E-mail: esteban.parra@utoronto.ca Esteban J. Parra, Ph.D. Admixed populations: an invaluable resource to study the genetics of
Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Supplementary information
 Fast and accurate genotype imputation in genome-wide association studies through pre-phasing Supplementary information Bryan Howie 1,6, Christian Fuchsberger 2,6, Matthew Stephens 1,3, Jonathan Marchini
Fast and accurate genotype imputation in genome-wide association studies through pre-phasing Supplementary information Bryan Howie 1,6, Christian Fuchsberger 2,6, Matthew Stephens 1,3, Jonathan Marchini
H3A - Genome-Wide Association testing SOP
 H3A - Genome-Wide Association testing SOP Introduction File format Strand errors Sample quality control Marker quality control Batch effects Population stratification Association testing Replication Meta
H3A - Genome-Wide Association testing SOP Introduction File format Strand errors Sample quality control Marker quality control Batch effects Population stratification Association testing Replication Meta
Comparative eqtl analyses within and between seven tissue types suggest mechanisms underlying cell type specificity of eqtls
 Comparative eqtl analyses within and between seven tissue types suggest mechanisms underlying cell type specificity of eqtls, Duke University Christopher D Brown, University of Pennsylvania November 9th,
Comparative eqtl analyses within and between seven tissue types suggest mechanisms underlying cell type specificity of eqtls, Duke University Christopher D Brown, University of Pennsylvania November 9th,
Blood Pressure and Hypertension Genetics
 Blood Pressure and Hypertension Genetics Yong Huo, M.D. Wei Gao, M.D. Yan Zhang, M.D. Santhi K. Ganesh, M.D. Outline Blood pressure and hypertension in China Update on genetics of blood pressure BP/HTN
Blood Pressure and Hypertension Genetics Yong Huo, M.D. Wei Gao, M.D. Yan Zhang, M.D. Santhi K. Ganesh, M.D. Outline Blood pressure and hypertension in China Update on genetics of blood pressure BP/HTN
SeattleSNPs Interactive Tutorial: Database Inteface Entrez, dbsnp, HapMap, Perlegen
 SeattleSNPs Interactive Tutorial: Database Inteface Entrez, dbsnp, HapMap, Perlegen The tutorial is designed to take you through the steps necessary to access SNP data from the primary database resources:
SeattleSNPs Interactive Tutorial: Database Inteface Entrez, dbsnp, HapMap, Perlegen The tutorial is designed to take you through the steps necessary to access SNP data from the primary database resources:
Robust Prediction of Expression Differences among Human Individuals Using Only Genotype Information
 Robust Prediction of Expression Differences among Human Individuals Using Only Genotype Information Ohad Manor 1,2, Eran Segal 1,2 * 1 Department of Computer Science and Applied Mathematics, Weizmann Institute
Robust Prediction of Expression Differences among Human Individuals Using Only Genotype Information Ohad Manor 1,2, Eran Segal 1,2 * 1 Department of Computer Science and Applied Mathematics, Weizmann Institute
SUPPLEMENTARY INFORMATION. Common variants in TMPRSS6 are associated with iron status and erythrocyte volume
 SUPPLEMENTARY INFORMATION Common variants in TMPRSS6 are associated with iron status and erythrocyte volume Beben Benyamin, Manuel A. R. Ferreira, Gonneke Willemsen, Scott Gordon, Rita P. S. Middelberg,
SUPPLEMENTARY INFORMATION Common variants in TMPRSS6 are associated with iron status and erythrocyte volume Beben Benyamin, Manuel A. R. Ferreira, Gonneke Willemsen, Scott Gordon, Rita P. S. Middelberg,
1: Categorical phenotyping protocol for the pinna traits examined
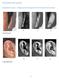 SUPPLEMENTARY FIGURES Supplementary Figure 1: Categorical phenotyping protocol for the pinna traits examined 1. Ear Protrusion 2. Lobe Attachment 1 3. Lobe Size 4. Antitragus Size 5. Tragus Size 2 6. Helix
SUPPLEMENTARY FIGURES Supplementary Figure 1: Categorical phenotyping protocol for the pinna traits examined 1. Ear Protrusion 2. Lobe Attachment 1 3. Lobe Size 4. Antitragus Size 5. Tragus Size 2 6. Helix
Genome-Wide Association Studies. Ryan Collins, Gerissa Fowler, Sean Gamberg, Josselyn Hudasek & Victoria Mackey
 Genome-Wide Association Studies Ryan Collins, Gerissa Fowler, Sean Gamberg, Josselyn Hudasek & Victoria Mackey Introduction The next big advancement in the field of genetics after the Human Genome Project
Genome-Wide Association Studies Ryan Collins, Gerissa Fowler, Sean Gamberg, Josselyn Hudasek & Victoria Mackey Introduction The next big advancement in the field of genetics after the Human Genome Project
Runs of Homozygosity Analysis Tutorial
 Runs of Homozygosity Analysis Tutorial Release 8.7.0 Golden Helix, Inc. March 22, 2017 Contents 1. Overview of the Project 2 2. Identify Runs of Homozygosity 6 Illustrative Example...............................................
Runs of Homozygosity Analysis Tutorial Release 8.7.0 Golden Helix, Inc. March 22, 2017 Contents 1. Overview of the Project 2 2. Identify Runs of Homozygosity 6 Illustrative Example...............................................
What Can the Epigenome Teach Us About Cellular States and Diseases?
 What Can the Epigenome Teach Us About Cellular States and Diseases? (a computer scientist s view) Luca Pinello Outline Epigenetic: the code over the code What can we learn from epigenomic data? Resources
What Can the Epigenome Teach Us About Cellular States and Diseases? (a computer scientist s view) Luca Pinello Outline Epigenetic: the code over the code What can we learn from epigenomic data? Resources
Release Notes. JMP Genomics. Version 3.1
 JMP Genomics Version 3.1 Release Notes Creativity involves breaking out of established patterns in order to look at things in a different way. Edward de Bono JMP. A Business Unit of SAS SAS Campus Drive
JMP Genomics Version 3.1 Release Notes Creativity involves breaking out of established patterns in order to look at things in a different way. Edward de Bono JMP. A Business Unit of SAS SAS Campus Drive
Human Populations: History and Structure
 Human Populations: History and Structure In the paper Novembre J, Johnson, Bryc K, Kutalik Z, Boyko AR, Auton A, Indap A, King KS, Bergmann A, Nelson MB, Stephens M, Bustamante CD. 2008. Genes mirror geography
Human Populations: History and Structure In the paper Novembre J, Johnson, Bryc K, Kutalik Z, Boyko AR, Auton A, Indap A, King KS, Bergmann A, Nelson MB, Stephens M, Bustamante CD. 2008. Genes mirror geography
Human Genetics and Gene Mapping of Complex Traits
 Human Genetics and Gene Mapping of Complex Traits Advanced Genetics, Spring 2018 Human Genetics Series Thursday 4/5/18 Nancy L. Saccone, Ph.D. Dept of Genetics nlims@genetics.wustl.edu / 314-747-3263 What
Human Genetics and Gene Mapping of Complex Traits Advanced Genetics, Spring 2018 Human Genetics Series Thursday 4/5/18 Nancy L. Saccone, Ph.D. Dept of Genetics nlims@genetics.wustl.edu / 314-747-3263 What
Genome Wide Association Studies
 Genome Wide Association Studies Liz Speliotes M.D., Ph.D., M.P.H. Instructor of Medicine and Gastroenterology Massachusetts General Hospital Harvard Medical School Fellow Broad Institute Outline Introduction
Genome Wide Association Studies Liz Speliotes M.D., Ph.D., M.P.H. Instructor of Medicine and Gastroenterology Massachusetts General Hospital Harvard Medical School Fellow Broad Institute Outline Introduction
PLINK gplink Haploview
 PLINK gplink Haploview Whole genome association software tutorial Shaun Purcell Center for Human Genetic Research, Massachusetts General Hospital, Boston, MA Broad Institute of Harvard & MIT, Cambridge,
PLINK gplink Haploview Whole genome association software tutorial Shaun Purcell Center for Human Genetic Research, Massachusetts General Hospital, Boston, MA Broad Institute of Harvard & MIT, Cambridge,
emerge-ii site report Vanderbilt
 emerge-ii site report Vanderbilt 29 June 2015 Vanderbilt activities emerge II PGx implementation locally and emerge-pgx SCN5A/KCNH2 project provider attitudes Phenotype contributions Methods development
emerge-ii site report Vanderbilt 29 June 2015 Vanderbilt activities emerge II PGx implementation locally and emerge-pgx SCN5A/KCNH2 project provider attitudes Phenotype contributions Methods development
Human Population Differentiation Is Strongly Correlated with Local Recombination Rate
 Human Population Differentiation Is Strongly Correlated with Local Recombination Rate Alon Keinan 1,2,3 *, David Reich 1,2 1 Department of Genetics, Harvard Medical School, Boston, Massachusetts, United
Human Population Differentiation Is Strongly Correlated with Local Recombination Rate Alon Keinan 1,2,3 *, David Reich 1,2 1 Department of Genetics, Harvard Medical School, Boston, Massachusetts, United
Figure S1. nuclear extracts. HeLa cell nuclear extract. Input IgG IP:ORC2 ORC2 ORC2. MCM4 origin. ORC2 occupancy
 A nuclear extracts B HeLa cell nuclear extract Figure S1 ORC2 (in kda) 21 132 7 ORC2 Input IgG IP:ORC2 32 ORC C D PRKDC ORC2 occupancy Directed against ORC2 C-terminus (sc-272) MCM origin 2 2 1-1 -1kb
A nuclear extracts B HeLa cell nuclear extract Figure S1 ORC2 (in kda) 21 132 7 ORC2 Input IgG IP:ORC2 32 ORC C D PRKDC ORC2 occupancy Directed against ORC2 C-terminus (sc-272) MCM origin 2 2 1-1 -1kb
Designing Genome-Wide Association Studies: Sample Size, Power, Imputation, and the Choice of Genotyping Chip
 : Sample Size, Power, Imputation, and the Choice of Genotyping Chip Chris C. A. Spencer., Zhan Su., Peter Donnelly ", Jonathan Marchini " * Department of Statistics, University of Oxford, Oxford, United
: Sample Size, Power, Imputation, and the Choice of Genotyping Chip Chris C. A. Spencer., Zhan Su., Peter Donnelly ", Jonathan Marchini " * Department of Statistics, University of Oxford, Oxford, United
CONTRACTING ORGANIZATION: Icahn School of Medicine at Mount Sinai New York, NY 10029
 AWARD NUMBER: W81XWH-14-1-0399 TITLE: Molecular & Genetic Investigation of Tau in Chronic Traumatic Encephalopathy (Log No. 13267017) PRINCIPAL INVESTIGATOR: John F. Crary, MD-PhD CONTRACTING ORGANIZATION:
AWARD NUMBER: W81XWH-14-1-0399 TITLE: Molecular & Genetic Investigation of Tau in Chronic Traumatic Encephalopathy (Log No. 13267017) PRINCIPAL INVESTIGATOR: John F. Crary, MD-PhD CONTRACTING ORGANIZATION:
Cufflinks Scripture Both. GENCODE/ UCSC/ RefSeq 0.18% 0.92% Scripture
 Cabili_SuppFig_1 a 1.9.8.7.6.5.4.3.2.1 Partially compatible % Partially recovered RefSeq coding Fully compatible % Fully recovered RefSeq coding Cufflinks Scripture Both b 18.5% GENCODE/ UCSC/ RefSeq Cufflinks
Cabili_SuppFig_1 a 1.9.8.7.6.5.4.3.2.1 Partially compatible % Partially recovered RefSeq coding Fully compatible % Fully recovered RefSeq coding Cufflinks Scripture Both b 18.5% GENCODE/ UCSC/ RefSeq Cufflinks
Author's response to reviews
 Author's response to reviews Title: A pooling-based genome-wide analysis identifies new potential candidate genes for atopy in the European Community Respiratory Health Survey (ECRHS) Authors: Francesc
Author's response to reviews Title: A pooling-based genome-wide analysis identifies new potential candidate genes for atopy in the European Community Respiratory Health Survey (ECRHS) Authors: Francesc
Supplementary Information
 A rare variant in MYH6 is associated with high risk of sick sinus syndrome Hilma Holm 1,8, Daniel F. Gudbjartsson 1,8, Patrick Sulem 1, Gisli Masson 1, Hafdis Th. Helgadottir 1, Carlo Zanon 1, Olafur Th.
A rare variant in MYH6 is associated with high risk of sick sinus syndrome Hilma Holm 1,8, Daniel F. Gudbjartsson 1,8, Patrick Sulem 1, Gisli Masson 1, Hafdis Th. Helgadottir 1, Carlo Zanon 1, Olafur Th.
PERSPECTIVES. A gene-centric approach to genome-wide association studies
 PERSPECTIVES O P I N I O N A gene-centric approach to genome-wide association studies Eric Jorgenson and John S. Witte Abstract Genic variants are more likely to alter gene function and affect disease
PERSPECTIVES O P I N I O N A gene-centric approach to genome-wide association studies Eric Jorgenson and John S. Witte Abstract Genic variants are more likely to alter gene function and affect disease
Human Genetics and Gene Mapping of Complex Traits
 Human Genetics and Gene Mapping of Complex Traits Advanced Genetics, Spring 2017 Human Genetics Series Tuesday 4/10/17 Nancy L. Saccone, nlims@genetics.wustl.edu ancestral chromosome present day chromosomes:
Human Genetics and Gene Mapping of Complex Traits Advanced Genetics, Spring 2017 Human Genetics Series Tuesday 4/10/17 Nancy L. Saccone, nlims@genetics.wustl.edu ancestral chromosome present day chromosomes:
Department of Psychology, Ben Gurion University of the Negev, Beer Sheva, Israel;
 Polygenic Selection, Polygenic Scores, Spatial Autocorrelation and Correlated Allele Frequencies. Can We Model Polygenic Selection on Intellectual Abilities? Davide Piffer Department of Psychology, Ben
Polygenic Selection, Polygenic Scores, Spatial Autocorrelation and Correlated Allele Frequencies. Can We Model Polygenic Selection on Intellectual Abilities? Davide Piffer Department of Psychology, Ben
Multi-SNP Models for Fine-Mapping Studies: Application to an. Kallikrein Region and Prostate Cancer
 Multi-SNP Models for Fine-Mapping Studies: Application to an association study of the Kallikrein Region and Prostate Cancer November 11, 2014 Contents Background 1 Background 2 3 4 5 6 Study Motivation
Multi-SNP Models for Fine-Mapping Studies: Application to an association study of the Kallikrein Region and Prostate Cancer November 11, 2014 Contents Background 1 Background 2 3 4 5 6 Study Motivation
Genome-wide association studies (GWAS) Part 1
 Genome-wide association studies (GWAS) Part 1 Matti Pirinen FIMM, University of Helsinki 03.12.2013, Kumpula Campus FIMM - Institiute for Molecular Medicine Finland www.fimm.fi Published Genome-Wide Associations
Genome-wide association studies (GWAS) Part 1 Matti Pirinen FIMM, University of Helsinki 03.12.2013, Kumpula Campus FIMM - Institiute for Molecular Medicine Finland www.fimm.fi Published Genome-Wide Associations
Appendix 5: Details of statistical methods in the CRP CHD Genetics Collaboration (CCGC) [posted as supplied by
 Appendix 5: Details of statistical methods in the CRP CHD Genetics Collaboration (CCGC) [posted as supplied by author] Statistical methods: All hypothesis tests were conducted using two-sided P-values
Appendix 5: Details of statistical methods in the CRP CHD Genetics Collaboration (CCGC) [posted as supplied by author] Statistical methods: All hypothesis tests were conducted using two-sided P-values
Human Population Differentiation is Strongly Correlated With Local Recombination Rate
 Human Population Differentiation is Strongly Correlated With Local Recombination Rate The Harvard community has made this article openly available. Please share how this access benefits you. Your story
Human Population Differentiation is Strongly Correlated With Local Recombination Rate The Harvard community has made this article openly available. Please share how this access benefits you. Your story
EPIB 668 Genetic association studies. Aurélie LABBE - Winter 2011
 EPIB 668 Genetic association studies Aurélie LABBE - Winter 2011 1 / 71 OUTLINE Linkage vs association Linkage disequilibrium Case control studies Family-based association 2 / 71 RECAP ON GENETIC VARIANTS
EPIB 668 Genetic association studies Aurélie LABBE - Winter 2011 1 / 71 OUTLINE Linkage vs association Linkage disequilibrium Case control studies Family-based association 2 / 71 RECAP ON GENETIC VARIANTS
Finding Enrichments of Functional Annotations for Disease-Associated Single- Nucleotide Polymorphisms
 Finding Enrichments of Functional Annotations for Disease-Associated Single- Nucleotide Polymorphisms Steven Homberg Mentor: Dr. Luke Ward Third Annual MIT PRIMES Conference, May 18-19 2013 Motivation
Finding Enrichments of Functional Annotations for Disease-Associated Single- Nucleotide Polymorphisms Steven Homberg Mentor: Dr. Luke Ward Third Annual MIT PRIMES Conference, May 18-19 2013 Motivation
Imputation. Genetics of Human Complex Traits
 Genetics of Human Complex Traits GWAS results Manhattan plot x-axis: chromosomal position y-axis: -log 10 (p-value), so p = 1 x 10-8 is plotted at y = 8 p = 5 x 10-8 is plotted at y = 7.3 Advanced Genetics,
Genetics of Human Complex Traits GWAS results Manhattan plot x-axis: chromosomal position y-axis: -log 10 (p-value), so p = 1 x 10-8 is plotted at y = 8 p = 5 x 10-8 is plotted at y = 7.3 Advanced Genetics,
THE HEALTH AND RETIREMENT STUDY: GENETIC DATA UPDATE
 : GENETIC DATA UPDATE April 30, 2014 Biomarker Network Meeting PAA Jessica Faul, Ph.D., M.P.H. Health and Retirement Study Survey Research Center Institute for Social Research University of Michigan HRS
: GENETIC DATA UPDATE April 30, 2014 Biomarker Network Meeting PAA Jessica Faul, Ph.D., M.P.H. Health and Retirement Study Survey Research Center Institute for Social Research University of Michigan HRS
Lecture 3: Introduction to the PLINK Software. Summer Institute in Statistical Genetics 2015
 Lecture 3: Introduction to the PLINK Software Timothy Thornton and Michael Wu Summer Institute in Statistical Genetics 2015 1 / 1 PLINK Overview PLINK is a free, open-source whole genome association analysis
Lecture 3: Introduction to the PLINK Software Timothy Thornton and Michael Wu Summer Institute in Statistical Genetics 2015 1 / 1 PLINK Overview PLINK is a free, open-source whole genome association analysis
Lecture 3: Introduction to the PLINK Software. Summer Institute in Statistical Genetics 2017
 Lecture 3: Introduction to the PLINK Software Instructors: Timothy Thornton and Michael Wu Summer Institute in Statistical Genetics 2017 1 / 20 PLINK Overview PLINK is a free, open-source whole genome
Lecture 3: Introduction to the PLINK Software Instructors: Timothy Thornton and Michael Wu Summer Institute in Statistical Genetics 2017 1 / 20 PLINK Overview PLINK is a free, open-source whole genome
Axiom mydesign Custom Array design guide for human genotyping applications
 TECHNICAL NOTE Axiom mydesign Custom Genotyping Arrays Axiom mydesign Custom Array design guide for human genotyping applications Overview In the past, custom genotyping arrays were expensive, required
TECHNICAL NOTE Axiom mydesign Custom Genotyping Arrays Axiom mydesign Custom Array design guide for human genotyping applications Overview In the past, custom genotyping arrays were expensive, required
Chromatin signature identifies monoallelic gene expression across mammalian cell types
 Chromatin signature identifies monoallelic gene expression across mammalian cell types Anwesha Nag* 1, Sébastien Vigneau* 1, Virginia Savova*, Lillian M. Zwemer*, Alexander A. Gimelbrant* 2 * Department
Chromatin signature identifies monoallelic gene expression across mammalian cell types Anwesha Nag* 1, Sébastien Vigneau* 1, Virginia Savova*, Lillian M. Zwemer*, Alexander A. Gimelbrant* 2 * Department
Nature Genetics: doi: /ng Supplementary Figure 1
 Supplementary Figure 1 Processing of mutations and generation of simulated controls. On the left, a diagram illustrates the manner in which covariate-matched simulated mutations were obtained, filtered
Supplementary Figure 1 Processing of mutations and generation of simulated controls. On the left, a diagram illustrates the manner in which covariate-matched simulated mutations were obtained, filtered
Analyzing Affymetrix GeneChip SNP 6 Copy Number Data in Partek
 Analyzing Affymetrix GeneChip SNP 6 Copy Number Data in Partek This example data set consists of 20 selected HapMap samples, representing 10 females and 10 males, drawn from a mixed ethnic population of
Analyzing Affymetrix GeneChip SNP 6 Copy Number Data in Partek This example data set consists of 20 selected HapMap samples, representing 10 females and 10 males, drawn from a mixed ethnic population of
Supplementary Methods 2. Supplementary Table 1: Bottleneck modeling estimates 5
 Supplementary Information Accelerated genetic drift on chromosome X during the human dispersal out of Africa Keinan A, Mullikin JC, Patterson N, and Reich D Supplementary Methods 2 Supplementary Table
Supplementary Information Accelerated genetic drift on chromosome X during the human dispersal out of Africa Keinan A, Mullikin JC, Patterson N, and Reich D Supplementary Methods 2 Supplementary Table
Nature Methods: doi: /nmeth.4396
 Supplementary Figure 1 Comparison of technical replicate consistency between and across the standard ATAC-seq method, DNase-seq, and Omni-ATAC. (a) Heatmap-based representation of ATAC-seq quality control
Supplementary Figure 1 Comparison of technical replicate consistency between and across the standard ATAC-seq method, DNase-seq, and Omni-ATAC. (a) Heatmap-based representation of ATAC-seq quality control
Supplementary Materials
 Supplementary Materials Genome-wide association study identifies 1p36.22 as a new susceptibility locus for hepatocellular carcinoma in chronic hepatitis B virus carriers Hongxing Zhang 1, Yun Zhai 1, Zhibin
Supplementary Materials Genome-wide association study identifies 1p36.22 as a new susceptibility locus for hepatocellular carcinoma in chronic hepatitis B virus carriers Hongxing Zhang 1, Yun Zhai 1, Zhibin
Efficient Genomewide Selection of PCA-Correlated tsnps for Genotype Imputation
 Efficient Genomewide Selection of PCA-Correlated tsnps for Genotype Imputation Asif Javed 1,2, Petros Drineas 2, Michael W. Mahoney 3 and Peristera Paschou 4 1 Computational Biology Center, IBM T. J. Watson
Efficient Genomewide Selection of PCA-Correlated tsnps for Genotype Imputation Asif Javed 1,2, Petros Drineas 2, Michael W. Mahoney 3 and Peristera Paschou 4 1 Computational Biology Center, IBM T. J. Watson
Supplementary Methods
 Supplementary Methods 1. Data We used recombination data from six recent studies of human pedigrees, including one that we previously analyzed 1 and five datasets released publicly. Table 1 presents these
Supplementary Methods 1. Data We used recombination data from six recent studies of human pedigrees, including one that we previously analyzed 1 and five datasets released publicly. Table 1 presents these
Supplementary Note: Detecting population structure in rare variant data
 Supplementary Note: Detecting population structure in rare variant data Inferring ancestry from genetic data is a common problem in both population and medical genetic studies, and many methods exist to
Supplementary Note: Detecting population structure in rare variant data Inferring ancestry from genetic data is a common problem in both population and medical genetic studies, and many methods exist to
mrna Sequencing Quality Control (V6)
 mrna Sequencing Quality Control (V6) Notes: the following analyses are based on 8 adult brains sequenced in USC and Yale 1. Error Rates The error rates of each sequencing cycle are reported for 120 tiles
mrna Sequencing Quality Control (V6) Notes: the following analyses are based on 8 adult brains sequenced in USC and Yale 1. Error Rates The error rates of each sequencing cycle are reported for 120 tiles
