Mechanism-Based Inhibitors of Deoxythymidine Monophosphate Synthesis as Antineoplastic Agents
|
|
|
- Ami Gilmore
- 6 years ago
- Views:
Transcription
1 Mechanism-Based Inhibitors of Deoxythymidine Monophosphate Synthesis as Antineoplastic Agents Victoria F. Roche 1 School of Pharmacy and Allied Health Professions, Creighton University, Omaha NE INTRODUCTION 5-Fluorouracil and methotrexate are antimetabolites widely used in the treatment of neoplastic disease. They act by inactivating the two enzymes involved in the biosynthesis of the pyrimidine nucleotide 2 -deoxythymidine 5 -monophosphate, namely thymidylate synthase and dihydrofolate reductase. The three dimensional structure of each enzyme is known and the mechanisms of the reactions they catalyze are well understood. This allows the medicinal chemistry professor the opportunity to engage students in a mechanism-based discussion of how the structure of each antimetabolite permits selective enzyme targeting and inhibition. This article will provide an overview of the structure and function of the thymidylate synthase and dihydrofolate reductase enzymes, take a mechanistic look at the structureactivity relationships of 5-fluorouracil, methotrexate and their clinically important analogs, and highlight some of the newer pyrimidine antagonist molecules recently reported in 1 Associate Professor of Pharmaceutical Sciences. the literature. A brief discussion of the role of antifolates in inhibiting purine nucleotide biosynthesis is provided. Among the more widely used antineoplastic agents in the anticancer armamentarium are the pyrimidine antimetabolites 5-fluorouracil (5-FU) and methotrexate. Distinctly different in structure, these two pyrimidine antagonists have different enzymatic sites of action, but both ultimately inhibit the rate-limiting enzyme in the dtmp synthesis pathway, thymidylate synthase. 5-FU is a close structural analog of the endogenous substrate deoxyuridylic acid, and inhibits this sulfhydryl enzyme directly. The antifolate methotrexate, through a direct inhibition of dihydrofolate reductase, induces a feedback inhibition of the synthase. The structures of both thymidylate synthase and dihydrofolate reductase enzymes have been well characterized, and the mechanisms of the reactions they catalyze are well understood. Thus a discussion of the action of these two drug structures provides a great opportunity to demonstrate to students the importance of chemistry to the rational (and therefore efficient) design of therapeutic agents, and to the true understanding of the mechanism of drug action. 196 American Journal of Pharmaceutical Education Vol. 58, Summer 1994
2 Fig. 1. Endogenous substrates and cofactors in pyrimidine and purine nucleotide biosynthesis. SYNTHESIS OF DEOXYTHYMIDINEMONOPHOSPHATE (dtmp) The targeting of the dtmp synthesis pathway in the treatment of selected neoplasms is a logical one, since cells deficient in this nucleotide cannot provide the 2'- deoxythymidine-5'-triphosphate (dttp) which, along with dctp, serves as a primary pyrimidine substrate for DNA polymerase(l). The substrate for dtmp synthesis is the 5- desmethyl analog 2'-deoxyuridine 5'-monophosphate (also known as deoxyuridylic acid or dump). This pyrimidine nucleotide is generated from orotidylic acid (also a pyrimidine nucleotide) through the action of several enzymes, including orotidylate decarboxylase (which provides UMP), UMP kinase (which provides UDP), ribonucleotide reductase (which provides dudp), and a phosphatase (which provides dump)(l,2). A second route to this key intermediate involves the hydrolysis and ribonucleotide reductase-' mediated reduction of CTP to form dcdp, which is then converted by the action of phosphatase and deaminase enzymes to dump. In yet another biosynthetic pathway, dutp, which forms through deamination of dctp, can hydrolyze via a pyrophosphatase to form dump(l). dump undergoes a methylation reaction catalyzed by thymidylate synthase to form dtmp, a process which is significantly accelerated in cancer and other rapidly proliferating cells. 5,10-Methylenetetrahydrofolate serves as the cofactor which will donate the one carbon unit to the substrate. The structures of the nucleotide substrate and folate cofactor are found in Figure 1. The complete reaction sequence ateo requires the enzyme dihydrofolate reductase. The reaction mechanism is depicted in Scheme 1. In brief, the synthase enzyme binds both dump substrate and folate cofactor, resulting in a reversible ternary complex. Properly positioned, the nucleophilic sulfhydryl group of thymidylate synthase is attracted to the electrophilic C 6 of the dump substrate. The sulfhydryl group Scheme 1. Synthesis of 2'deoxythymidine-5'-phosphate (dtmp). attacks in a Michael fashion to displace the electrons of the double bond, which subsequently attack the electrophilic methylene group of the cofactor. Students are expected to recognize the inductive and resonance effects which promote the electron deficient character of both of these electrophilic targets. This second attack, which breaks the bond between the methylene group and N 10 of the cofactor(3), results in the joining of enzyme, substrate and cofactor into a covalent ternary complex. This covalent complex undergoes oxidative decomposition to product (dtmp) only through the transfer of the dump C 5 hydrogen atom to a basic center of the cofactor. Once this abstraction occurs, the methylene unit is released to the substrate, the cofactor oxidizes elimination, and a resultant hydride shift from cofactor to the substrate's exocyclic methylene unit provides the 5-CH 3 group of the product dtmp. The original electron flow pattern reverses to regenerate the pyrimidine olefinic linkage and release free thymidylate synthase(l). Left behind to fend for itself is the oxidized cofactor, dihydrofolate. In order to participate in another dtmp synthesis cycle, this molecule must convert to tetrahydrofolate via the enzyme dihydrofolate reductase, and submit to methylene insertion by serine with the assistance of pyridoxal phosphate and serine hydroxymethylase(4). THE THYMIDYLATE SYNTHASE TARGET The rate limiting reaction in the dtmp synthesis sequence is the formation of the covalent enzyme-substrate-cofactor ternary complex catalyzed by thymidylate synthase. The American Journal of Pharmaceutical Education Vol. 58, Summer
3 structure of the enzyme isolated from Lactobacillus casei was elucidated in 1987 by Hardy, et al.(5) and has been subsequently well characterized by others. Thymidylate synthase is the most highly conserved enzyme known(6), and this fact supports the involvement of multiple residues in the catalysis reaction(7). Through crystallographic analysis investigators have shown that the active site is a large, open cavity which accommodates both substrate and cofac-tor. When the cof actor binds, conf ormational changes in the C- terminus cause this cavity to close down. This facilitates the orientation of reaction participants and the subsequent production of dtmp(8,9). Complementarity of the faces of the pteridine portion of the cof actor and the pyrimidine ring of the substrate ensures proper placement for ternary complex stability(10). The synthase enzyme is dimeric, and binds substrate and cofactor equally, but sequentially, to both active sites. Key active site residues which interact with substrate through hydrogen bonding include Arg23 (which binds with the 5 - phosphate), Tyr261 and His259 (which bind with the 3 - OH), Asn229 (which binds with the pyrimidine N 3 -H) and, of course, Cysl98 (the nucleophile which attacks C 6 of dump). A critical active site conformation-maintaining role has been proposed for the highly conserved Tyr33, as mutation to His33 in the mammalian enzyme results in a 3-4 fold increase in resistance to 5-FU(11). Ordered molecules of water in the active site cavity are important in promoting enzyme specificity. One water molecule, activated by His l99, forms a hydrogen bond with the C 4 oxygen atom of dump(12). As indicated above, the tetrahydrofolate cofactor also binds to thymidylate synthase, and does so most effectively in the natural 6R, polyglutamated form. Monoglutamated folate is selectively transported into cells where a glutamyl transferase enzyme (also known as folyl polyglutamyl synthase) adds several (most commonly 3-6) additional γ- glutamate units to the original residue. This not only dramatically enhances affinity for the synthase enzyme, but also augments the catalytic capability of the synthase(13). The folate binding loop of thymidylate synthase includes residues and contains several basic amino acids, among them Lys50 and His53. With the assistance of highly ordered water molecules, these residues anchor the first Glu residue of the polyglutamate side chain of the cofactor(7,13). The oxygen atoms of Leu224 and Ile310, as well as Asp221, are also involved in cofactor anchoring. Diffuse electrostatic interactions hold subsequent Glu residues to the synthase enzyme. In studies withsynthase isolated from E. coli, Kamb et al. found that the first glutamate residue of the 5,10- methylenetetrahydrofolate cofactor is anchored rigidly to the synthase, with each successive glutamate being more and more disordered. The reduction in K i contributed by each glutamate residue in the chain is highly species specific. Citing the fact that the region of the synthase enzyme which binds the polyglutamate chain of the cofactor is less highly conserved than the active dump-binding site, Kamb and coworkers hypothesize that more species-selective dtmp synthesis inhibitors might be generated by targeting the polyglutamate binding area as a drug receptor(13). While not a part of the active site or folate binding loop, the C-terminal Val316 is critical in the catalytic action of thymidylate synthase. Studies with L. casei synthase have shown that, when substrate and cofactor are bound, a significant conformational change moves the flexible C-terminus approximately 4 angstroms. This forms a tight cover or lid over the active site. In this conformation, the C-terminal Val316 interacts hydrophobically with Thr24, and through hydrogen bonds with Arg23 and Trp85 residues on the opposite side of the binding pocket(14). As previously mentioned, this closing down of the active site aligns all dtmp synthesis players for effective covalent ternary complex formation. Synthase mutants lacking Val316 retain the ability to bind substrate, albeit in a different conformation from the wild type enzyme(14), but lack the ability to catalyze dtmp synthesis. This lack of catalytic capability is presumed due to an approximate 40 fold decrease in folate affinity, and difficulty in forming the covalent ternary complex(9,15). 5-FLUOROURACIL AND RELATED ANALOGS: A CASE OF CHEMICAL ENTRAPMENT The prototypical substrate-based inhibitor of thymidylate synthase is 5-fluorouracil (Figure 1) Once students have a clear understanding of the mechanisms involved in dump binding and subsequent conversion to dtmp via the synthase enzyme, the elegant rationale of the simple replacement of fluorine for hydrogen in dump to produce this antimetabolite becomes clear. Originally conceived in 1957 by Heidelberger(16), the mechanism of dtmp synthesis inhibition by 5-FU represents chemically-based enzyme entrapment at its finest. No self-respecting enzyme that naturally binds substrate as nucleotide would be fooled by a drug structure missing the large phosphorylated deoxyribose unit. Thus, 5- FU must be activated by conversion to 5-fluoro-2 - deoxyuridylic acid (5-FdUMP). This occurs in vivo through a number of pathways(17,18). In one, a phosphorylated ribose unit is transferred to N 1 of 5-FU through the action of orotate phosphoribosyl transferase and 5-phosphoribosyl- 1-pyrophosphate to form 5-FUMP. UMP kinase converts this nucleotide to the diphosphorylated derivative, 5-FUDP, which is then vulnerable to reduction by ribonucleotide reductase, forming 5-FdUDP. Hydrolysis of 5-FdUDP by a phosphatase yields the desired 5-FdUMP. Alternatively, ribose-1-phosphate can react with 5-FU with enzymatic assistance from uridine phosphorylase to form the fluorinated nucleoside. Uridine kinase phosphorylation provides 5-FUMP, which can follow the pathway described above to yield 5-FdUMP. In yet another pathway, 5-FU is converted to 5-FdUMP through the action of thymidine phosphorylase and thymidine kinase. Once activated, the drug molecule is ready for the chemical sting, and the properties of the 5-fluoro group (the only functional group distinguishing natural from false substrate) will be solely responsible for setting the trap. As a small atom (about the size of the endogenous C 5 -hydrogen), fluorine provides no steric hindrance to binding in the active site cavity of the synthase. As the most electronegative atom known, its negative inductive effects dramatically increase the electrophilic character of the adjacent C 6, which is the target for attack by the nucleophilic synthase Cysl98. The result is a very rapid nucleophilic attack by the enzyme, with the resultant formation of a highly stable covalent ternary enzyme-substrate-cofactor complex. At this point the drug has directly and permanently inhibited the synthase enzyme, since lack of an abstractable C 5 -hydrogen means 198 American Journal of Pharmaceutical Education Vol. 58, Summer 1994
4 Fig. 2. Fluoropyrimidine inhibitors of thymidylate synthase. the covalent ternary complex will be unable to undergo oxidative breakdown. No breakdown, no product and, more importantly, no regeneration of thymidylate synthase. Consequently, there will be no conversion of endogenous molecules of dump to dtmp by the inhibited enzyme, and the cell must generate new thymidylate synthase de novo for DNA production. Until new enzyme is synthesized, DNA production ceases which, of course, is the therapeutic goal. Supporting the unique chemical role of the fluorine atom in halting dtmp synthesis is the almost total lack of synthase inhibiting action of other 5-halogenated dump analogs. Chlorine, bromine and iodine are all bulkier and less electronegative than fluorine. Therefore, they interfere significantly with substrate anchoring, and promote a less electrophilic C 6 than the fluorinated analog(1). Other fluoropyrimidine structures have demonstrated effective inhibition of the synthase enzyme (Figure 2). The deoxyribonucleoside version of 5-FU, (5-fluorodeoxyuridine or floxuridine), is commercially available. Since this molecule must be phosphorylated by uridine kinase to the same active false nucleotide substrate as is formed with 5-FU, it has a similar activity profile. The 5-trifluoromethyl version of floxuridine (trifluridine) was another brainchild of Heidelberger(16). After thymidine kinase-catalyzed phosphorylation to give the nucleotide, the electron withdrawing trifluoromethyl group promotes rapid nucleophilic attack at adjacent C 6 by the synthase Cys198. This results in a displacement of HF and generates a highly reactive difluoromethylene unit. This reactive intermediate then presumably interacts covalently with the cofactor to irreversibly inhibit thymidylate synthase(4). Tegafer, the 1- tetrahydrofuranyl analog of 5-FU is slowly cleaved, possibly through thymidine phosphorylase, to 5-FU(1). Another unique 5-FUprodrug is 5-fluoropyrimidin-4(1H)-one, which is oxidized in vivo to 5-FU by xanthine oxidase(1). Other nonpyrimidine based direct thymidylate synthase inhibitors are being reported in the literature (Figure 3). 4',5'-Dichloro, dibromo, and diiodo derivatives of pyridoxine (vitamin B 6 ) have demonstrated an irreversible inhibition of synthase. Pretreatment with 10µmol dump completely protects against enzyme inactivation, which indicates inhibitor interaction at the active catalytic site(19). Stoichet and coworkers, through the use of a molecular Fig. 3. Nonpyrimidine-based inhibitors of thymidylate synthase. docking computer program, identified unique phenolphthalein inhibitors which bind to an area of the synthase active site that is distinct from the area which binds substratebased inhibitor molecules. The tetraiodo analog provided a synthase IC 50 of 3µM(20).6,7-Imidazotetrahydroquinolines conceptualized by iterative ligand design to occupy the folate ligand binding pocket of thymidylate synthase have recently been synthesized(21). Webber et al. reported the synthesis of 5-(arylthio)quinazolin-4-ones which were designed by repetitive crystallographic analysis to bind to the folate binding area of the synthase(22). Other quinoline and quinazoline antifolates, ICID1694 for example, have demonstrated effectiveness as inhibitors of thymidylate synthase(23-25). The 2-CH 3 group of the quinazoline synthase inhibitors provides increased enzymatic specificity compared to 2-NH 2 precursor antagonist molecules. In addition, better solubility properties result in lower nephrotoxicity. The better inhibition of cell growth observed from the 2-CH 3 quinazolines is presumably due to more effective polyglutamation(26). THE DIHYDROFOLATE REDUCTASE TARGET The family of isoenzymes known as the dihydrofolate reductases (DHFRs) have been extensively characterized. DHFRs are relatively small proteins (approximately 20,000 Da), and key active site residues of enzymes isolated from mammalian and bacterial sources are highly conserved. The tetrahydrofolates they produce accept single carbon units in various forms, and are of considerable biological importance as one carbon donors to numerous endogenous substrates(27). DHFR requires NADPH as cofactor to reduce natural folates and antifolates. Catalytic reduction involves the formation of a ternary complex of enzyme, folate and American Journal of Pharmaceutical Education Vol.58, Summer
5 levels of folyl polyglutamyl synthase should respond better to methotrexate or other antifolates which do not require polyglutamation for high affinity binding to DHFR. The key residues of the active site of Echerichia coli derived DHFR which bind the natural substrate dihydrofolate (DHF) have been identified. Primary among them is Asp27 which donates a proton to N 5 of DHF and initiates the reduction reaction. Significantly, N 5 is the most basic nitrogen atom in the substrate. The ion-ion bond formed between Asp27 and N 5 is complemented by hydrogen bonds between Arg57 and the α-carboxyl group, and between Thr43 and the 2-NH 2 of DHF. As might be anticipated, stereochemistry of the glutamate side chain is important to activity, with the natural L isomer showing a 10 fold increase in potency over the unnatural D form(33). The carbons of the pteridine and phenyl rings bind hydrophobically to Ile5, Ala7, Phe31, and to Leu28 and Ile50 respectively. Both rings bind through hydrophobic bonds to Ile94(27,34). Fig. 4. Classical and nonclassical inhibitors of dihydrofolate reductase. cofactor. The order of binding of substrate and cofactor to DHFR is random, but the anchoring of one entity greatly facilitates the rapid binding of the other. Once bound, the C 4 of NADPH stereospecifically transfers hydride to the substrate, a process which is promoted by the protonation of the N 5 of the folate by Asp27(27,28). Anionic polyglutamate chains are attached enzymatically to natural folates and synthetic antifolates, which ensures their intracellular localization(13). While some folate polyglutamates demonstrate enhanced affinity for DHFR compared to monoglutamated analogs, the effect is generally much lower (less than 10 fold) than observed with other folate-binding enzymes. In classical antifolates like methotrexate, this diminished effect is believed due to the very strong, essentially stoichiometric, binding to the reductase enzyme provided by the amino groups at positions 2 and 4. Interestingly, in studies with recombinant human DHFR, Rosowsky et al. found that affinity for DHFR of 2-desamino- 2-methylaminopterin modified with three or four γ-glutamyl residues was two orders of magnitude higher than the parent antifolate. This observation caused these investigators to propose a domain in the distal region of the active site which might contain cationic Lys or Arg residues that could interact electrostatically with the polyglutamate anions of antifolates(29). Many investigators have noted that folyl polyglutamyl synthase activity is higher in certain tumor cells than in normal, but rapidly proliferating, cells(30-32). Thus the development of antifolates which require polyglutamation prior to anchoring to their target enzyme could permit more selective, and therefore safer, antifolate chemotherapy than is currently available(29). On the flip side, tumors with low METHOTREXATE AND OTHER CLASSICAL DHFR INHIBITORS Methotrexate (Figure 4) is termed a classical antifolate because it retains the p-aminobenzoylglutamic acid side chain of natural DHF. The structural distinctions between this antifolate and the natural folate include the replacement of the 4-oxo group of DHF with an amino group, the presence of a 7,8-double bond, and the addition of a methyl group at N 10. However these slight structural changes result in highly significant changes in reductase binding and activity. While the electron withdrawing 4-oxo group of DHF decreases lone pair availability at N 1 (recall N 5 is the most basic nitrogen of DHF), the 4-amino group of methotrexate can donate electrons to N 1 through the pi electron system, which increases its basic strength over 1000 fold. N 1 is the most basic nitrogen of methotrexate, and is sought out by Asp27 at the expense of N 5. This ion-ion anchoring between Asp27 and N 1 alters the pteridine ring orientation 180 compared to the dihydropteridine ring of DHF. The binding that occurs between the rest of the methotrexate structure and the DHFR active site is believed to parallel that observed with DHF(27). The importance of unhindered hydrogen bonds between the NH 2 groups at C 2 and C 4 is emphasized by the significant loss of DHFR inhibiting potency when these groups are alkylated(35). Methotrexate can undergo polyglutamation in vivo, but this structural modification produces only a modest enhancement of DHFR affinity at best. It is interesting to note that, while the polyglutamate analogs of methotrexate are capable of directly inhibiting thymidylate synthase at commonly achieved intracellular concentrations, the affinity for the synthase is two to four orders of magnitude below that observed for the primary target, DHFR(36). Cody et al. have recently determined the crystal structure of a recombinant human DHFR-NADPHmethotrexate-γ-tetrazole complex to 2.3 angstrom resolution(37). In their complex, N 1 is protonated by Glu30, and the methotrexate 2-amino group is hydrogen bonded to a water molecule which also interacts with Glu30 and Thr136. The 4-amino group binds with Ile7 and Tyr121, and the α- carboxylate of the glutamate side chain forms a bridge with Arg American Journal of Pharmaceutical Education Vol. 58, Summer 1994
6 While technically a competitive antagonist of DHF, methotrexate is so tightly bound as a result of the N 1 -enzyme interaction that it appears noncompetitive or pseudoirreversible (4). While K i s will differ with the DHFR source, values in the 4-7 picomolar range are common(27,38). These K i values reflect the affinity of methotrexate for DHFR in the presence of bound NADPH. The route from initially reversible to pseudoirreversible binding has been proposed to involve a slow isomerization of the antifolate, which could induce subtle changes in the conformation of active site amino acids and associated water molecules(39). Interestingly, whether single or multiple conformations exist when the DHFR enzyme is initially bound to substrate or cofactor (binary complexes) appears to be species dependent(40). The ternary complexes studied in E. coli maintain a single conformation. Armed with an understanding of the differences in substrate chemistry and DHFR binding induced by the structural differences in DHF and methotrexate, the mechanism of DHFR inhibition by this antifolate becomes clear. Since N 5 of methotrexate will not be protonated by DHFR residues, reduction of the 5,6-double bond to form tetrahydrofolate does not occur. The presence of the methotrexate 7,8-double bond, which provides for a totally aromatic pteridine ring, should also retard reduction attempts. The net result is that the antifolate structure anchors very tightly to DHFR through protonated N 1, and the C 2 and C 4 amino groups, but will not react with it, thus there is no release of reduced product nor regeneration of free enzyme. As a permanent occupant of the DHFR active site, methotrexate keeps the natural byproduct of dtmp synthesis, dihydrofolate, from binding to the reductase. Therefore, no reduced tetrahydrofolate can form which, in turn, means that no 5,10-methylenetetrahydrofolate cofactor can be synthesized. More importantly, the buildup of DHF in the cell signals the rate-limiting thymidylate synthase enzyme via a feedback mechanism to shut down, and dtmp synthesis (and therefore DNA synthesis) grinds to a halt. In terms of cancer chemotherapy, mission accomplished. The structures of some classical and nonclassical DHFR inhibitors are provided in Figure 4. The classical antifolate most closely related to methotrexate is the N 10 desmethyl analog aminopterin. This molecule, while effective as an antileukemic agent, is decidedly more toxic than methotrexate(27), and has not been developed for clinical use. However 10-ethyl-lO-deazaaminopterin (10-EDAM) has shown promise as a therapeutic agent in a non-small cell lung tumor trial(41,42). This molecule appears to have affinity for DHFR comparable to methotrexate (K i = 2-3 pm), but is less toxic due to favorable uptake and polyglutamation in tumor cells(26) coupled with rapid clearance from susceptible healthy tissues(27). Rosowsky(26) provides a review of the favorable results obtained with 10- EDAM in patients with advanced cancer in Phase I and II clinical trials. Some deaza-and dideazatetrahydrofolate analogs, among them the nonclassical antifolate trimetrexate (Figure 4), have been investigated for reductase inhibiting activity. Trimetrexate is a 5-deazafolate analog with a trimethoxy substituted phenyl ring. Because it lacks the p- aminobenzoylglutamic acid side chain it is not polyglutamated in vivo. Thus it may show enhanced efficiency in tumors resistant to methotrexate due to folyl polyglugamyl synthase deficiency, as long as drug transport mechanisms, DHFR binding affinity and thymidylate synthase levels are not impaired(43). While not currently marketed as an antineoplastic agent, this nonclassical antifolate is available for the treatment of Pneumocystis carinii infections. Mono-and diester derivatives of the methotrexate glutamate carboxylate groups have been evaluated for DHFR inhibiting activity. The rationale behind glutamate esterification is enhancement of cell membrane penetration. While generally not as potent as methotrexate, the esters do retain DHFR inhibiting action, with potency increasing directly with ester lipophilicity up to a maximum chain length of 4-5 carbons(44). The α, γ- dibutyl ester was less active than either monobutyl ester or methotrexate when assayed with rabbit liver DHFR. In the rabbit liver DHFR system, the two monobutyl esters were of equal potency, but when the DHFR was obtained from murine leukemia (L1210) cells, a 10 fold potency increase of the γ-butyl ester over the a-butyl derivative was noted. The potency of the γ-butyl ester was comparable with methotrexate in this system(45,46). Capitalizing on the pseudoirreversible methotrexate- DHFR interaction in a unique way, Hawkins et al. have designed a novel system for delivering radionuclides to tumor cells(47). In this in vitro study, a nontoxic, bivalent alpha-transferrin monoclonal antibody targeted to human K562 erythroleukemia cells was conjugated to recombinant human DHFR and administered. Then a methotrexate analog labelled with m In was administered, and highly specific delivery of nuclide to the tumor target was demonstrated. PURINE SYNTHESIS INHIBITION BY ANTIFOLATES: THE GAR FORMYLTRANSFERASE TARGET While peripheral to a discussion of mechanism-based pyrimidine antagonists, it is important to note that antifolates such as methotrexate have the ability to inhibit purine, as well as pyrimidine, nucleotide biosynthesis. The two folaterequiring enzymes in the purine synthesis pathway are glycinamide ribonucleotide formyltransferase (also known as GAR transformylase, GART and GARFT) and aminoimidazolecarboxamide ribonucleotide formyltransferase (AICARFT). The single carbon-donating folate cofactor required for both formylation reactions is 10- formyltetrahydrofolate. This folate is produced from the thymidylate synthase cofactor 5,10-methylenetetrahydrofolate through oxidation. GARFT catalyzes the transfer of the 10-formyl group of 10-formyltetrahydrofolate to the substrate glycinamide ribonucleotide to generate the purine nucleotide intermediate American Journal of Pharmaceutical Education Vol. 58, Summer
7 formylglycinamide ribonucleotide and tetrahydrofolate. AICARFT catalyzes a later formylation of an aminoimidazolecarboxamide ribonucleotide to produce 5- formylaminoimidazole-4-carboxamide ribonucleotide, the immediate precursor to inosine. The structures of these important substrates and the formylated folate cofactor are provided in Figure 1. In studies with mammalian GARFT, Caperelli demonstrated a substrate: cofactor stoichiometry of 1:1(48). While methotrexate itself has a relatively low affinity for these purine synthesis enzymes, polyglutamate analogs of methotrexate anchor much more strongly (29,49,50). Antineoplastic agents which more selectively antagonize purine biosynthesis through GARFT inhibition can be found in 5,10-dideaza analogs of tetrahydrofolate(51-53). Racemic 5,10-dideazatetrahydrofolic acid, also known as DDATHF or lometrexol, inhibits GARFT with a K i of 39nM. A 100 fold decrease in K i is noted in the derivative containing 5 γ-glutamate residues(26). While the two enantiomers have similar enzymatic activity profiles and IC 50 values, the 6R isomer was selected for clinical evaluation(26). Alkylation at C 5 or C 10 in the dideazatetrahydrofolate analogs often decreases affinity for GARFT, but may enhance transport into tumor cells(53,54). Preliminary data from lometrexol Phase I clinical trials have been promising. However the side effects of myelosuppression and anemia proved problematic(26,55-57). Investigators found that the cumulative toxicity induced by lometrexol could be significantly attenuated or reversed with the concomitant administration of folic acid or leucovorin (5-formyltetrahydrofolic acid). A discussion of other new antifolate structures currently in preclinical development is provided by Rosowsky(26). In summary, the topic of nucleotide antagonism in the treatment of neoplastic disease provides the medicinal chemistry instructor with an excellent opportunity to discuss rational, mechanistic approaches to drug design and action. The number of antifolate structures which have been designed, synthesized and evaluated for selective inhibition of key enzymes in the pyrimidine and purine biosynthetic pathways continues to burgeon(58), and many new therapeutically useful compounds are on the clinical horizon. And, while fewer in number, the fluoropyrimidine-based inhibitors of dtmp synthesis dramatically reinforce that an understanding of the chemical properties and behavior of endogenous macromolecules and drug structures is the key to interpreting and predicting drug action. Am. J. Pharm. Educ., 58, (1994); received 2/1/94, accepted 4/13/94. References (1) Hobbs, J.B., Purine and pyrimidine targets in Comprehensive Medicinal Chemistry, The Rational Design, Mechanistic Study and Therapeutic Application of Chemical Compounds, Volume 2. (edit. Sammes, P.G.) Pergamon Pres s, New York NY (1990) pp (2) Stryer, L., Biosynthesis of nucleotides in Biochemistry, Third Edition, Freeman and Company, New York NY (1988) pp (3) Pellino, A.M. and Danenberg, P.V., Evidence from chemical degradation studies for a covalent bond from 5-fluoro-2 -deoxyuridylate to N-5 of tetrahydrofolate in the ternary complex of thymidylate synthase-5- fluoro-2 -deoxyuridylate-5,10-methylenetetrahydrofolate, J. Biol. Chem., 260, (1985). (4) Remers, W.A., Antineoplastic agents in Wilson and Gisvolds Textbook of Organic Medicinal and Pharmaceutical Chemistry, Ninth Edition, (edits. Delgado, J.N and Remers, W.A.) J.B. Lippincott Company, Philadelphia (1991) pp (5)Hardy, L.W., Finer-Moore, J.S., Montfort, W.R., Jones, M.O., Santi, D.V. and Stroud, R.M., Atomic structure of thymidylate synthase: Target for rational drug design, Science, 235, (1987). (6) Perry, K.M., Fauman, E.B., Finer-Moore, J.S., Montfort, W.R., Maley, G.F., Maley, F. and Stroud, R.M. Plastic adaptation toward mutations in proteins: Structural comparison of thymidylate synthase. Proteins, 8, (1990). (7) Finer-Moore, J.S., Fauman, E.B., Foster, P.G., Perry, K.M., Santi, D.V. and Stroud, R.M., Refined structures of substrate-bound and phosphate-bound thymidylate synthase from Lactobadllus casei, J. Mol. Biol., 232, (1993). (8) Stroud, R.M. and Finer-Moore, J.S., Stereochemistry of a multistep/ bipartite methyltransfer reaction: Thymidylate synthase, FASEB. 7, (1993). (9) Carreras, C.W., Climie, S.C. and Santi, D.V., Thymidylate synthase with a C-terminal deletion catalyzes partial reactions, but is unable to catalyze thymidylate formation, Biochemistry, 31, (1992). (10) Finer-Moore, J.S., Montfort, W.R. and Stroud, R., Pairwise specificity and sequential binding in enzyme catalysis: Thymidylate synthase, ibid., 29, (1990). (11) Hughey, C.T., Barbour, K.W., Berger, F.G. and Berger, S.H., Functional effects of a naturally-occuring amino acid substitution in human thymidylate synthase, Mol. Pharmacol., 44, (1993). (12) Liu, L. and Santi, D.V., Exclusion of 2 -deoxycytidine 5 -monophosphate by asparagine 229 of thymidylate synthase, Biochemistry, 32, (1993). (13) Kamb, A., Finer-Moore, J.S., Calvert, A.H. and Stroud, R.M., Structural basis for recognition of polyglutamyl folates by thymidylate synthase, ibid., 31, (1992). (14) Perry, K.M., Carreras, C.W., Chang, L.C., Santi, D.V. and Stroud, R.M., Structures of thymidylate synthase with a C-terminal deletion: Role of the C-terminus in alignment of 2 -deoxyuridine 5 -monophosphateand5,10-methylenetetrahydrofolate, ibid., 32, (1993). (15) Cisneros, R.J., Zapf, J.W. and Dunlap, R.B., Studies of 5- fluorodeoxyuridine 5 -monophosphate binding to carboxypeptidase A-inactivated thymidylate synthase from Lactobadllus casei, J. Biochem., 268, (1993). (16) Heidelberger, C, Fluorinated pyrimidines and their nucleosides, Handbook Exper. Pharmacol., 38,193(1975). (17) Chabner, B.A., Pyrimidine antagonists, in Pharmacologic Principles of Cancer Treatment. W.B. Saunders Company, Philadelphia (1982) p (18) Calabresi, P. and Chabner, B.A., Chemotherapy of neoplastic diseases in Goodman and Gilman s The Pharmacological Basis of Therapeutics, Eighth Edition (edits., Gilman, A.G., Rail, T.W., Nies, A.S, Taylor, P.) McGraw-Hill, Inc., New York NY (1993) pp (19) Huang, S., Parish, E.J. and Aull, J.L., Irreversible inhibition of thymidylate synthase by pyridoxine (B 6 ) analogs. J. Enzym. Inhib., 6, (1992). (20) Schoichet, B.K., Stroud, R.M., Santi, D.V., Kuntz, I.D. and Perry, K.M., Structure-based discovery of inhibitors of thymidylate synthase, Science, 259, (1993). (21) Reich, S.H. et al., Design and synthesis of novel 6,7- imidazoltetrahydroquinoline inhibitors tf thymidylate synthase using iterative protein crystal structure analysis, J. Med. Chem., 35, (1992). (22) Webber, S.E. et al., Design of thymidylate synthase inhibitors using protein crystal structures: The synthesis and biological evaluation of a novel class of 5-substituted quinazolinones, ibid., 36, (1993). (23) Warner, P., et al., Quinoline antifolate thymidylate synthase inhibitors: Variation of the C2-and C4-substitutents, ibid., 35, (1992). (24) Thornton, TJ. et al., Quinazoline antifolate thymidylate synthase inhibitors, ibid., 35, (1992). (25) Bisset, G.M.F., Pawelczak, K., Jackman, A.L. and Calvert, A.H., Synthesis and thymidylate synthase inhibitory activity of the poly-γglutamyl conjugates of N-[5-[N-(3,4,-dihydro-2-methyl-4- oxoquinazolin-6-ylmethyl)-n-methylamino]2-thenoyi]-l-glutamic acid (ICI D1694) and Other Quinazoline antifolates, ibid., 35, (1992). (26) Rosowsky, A., Development of antifolate analogs as anticancer agents, Am. J. Pharm. Educ., 56, (1992). (27) McCormack,J.J., Reductases, in Comprehensive Medicinal Chemistry, The Rational Design, Mechanistic Study and Therapeutic Application of Chemical Compounds, Volume 2, (edit. Sammes, P.G.) Pergamon Press, New York NY (1990) pp (28) Coward, J.K., Parameswaran, K.N., Cashmore, A.R. and Bertino, J.R., 7,8-Dihydropteroyl oligo-gamma-l-glutamates: Synthesis and 202 American Journal of Pharmaceutical Education Vol. 58, Summer 1994
8 kinetic studies with purified dihydrofolate reductase from mammalian sources, Biochemistry, 13, (1974). (29) Rosowsky, A., et al., Biochemical and biological studies on 2- desamino-2-methylaminopterin, an antifolate the polyglutamates of which are more potent than the monoglutatmate against three key enzymes of folate metabolism, Cancer Res., 52, (1992). (30) Poser, R.G., Sirotnak, F.M. and Chello, P.L., Differential synthesis of methotrexate polyglutamates in normal proliferative and neoplastic tissues in vivo, ibid., 41, (1981). (31) Fabre, I., Fabre, G. and Goldman, I.D., Polyglutamation, an important element in methotrexate cytotoxicity and selectivity in tumor versus murine granulocyte progenitor cells in vitro, ibid., 44, (1984). (32) Runberger, B.G.,Barrueco,J.R. and Sirotnak, F.M., Differing specificities for 4-aminofolate analogues of folyl polyglutamyl synthetase from tumors and proliferative intestinal epithelium of the mouse with significance for selective antitumor action, ibid., 50, (1990). (33) Cramer, S.M., Schornagel, J.H., Kalghatgi, K.K., Bertino, J.R. and Horvath, C., Occurnace and significance of D-methotrexate as a contaminant ofcommerical methotrexate, ibid., 44, (1984). (34) Roth, B., Selective inhibitors of bacterial dihydrofolate reductase: Structure-activitity relationships, Handbook of Exp. Pharmacol.,64, (1983). (35) Baker, B.R., Design of Active Site Directed Irreversivle Enzyme Inhibitors, John Wiley & Sons, New York NY (1967) p (36) Rhee, M.S., Coward, J.K. and Galivan, J., Depletion of 5,10- methylenetetrahydrofolate and 10-formyltetrahydrofolate by methotrexate in cultured hepatoma cells, Mol. Pharmacol., 42, (1992). (37) Cody, V., Luft, J.R., Ciszak, E., Kalman, T.I. and Freisheim, J.H., Crystal structure determination at 2.3 angstrom of recombinant human dihydrofolate reductase ternary complex with NADPH and methotrexate-γ-tetrazole, Anticancer Drug Des., 7, (1992). (38) Jackson, R.C., Hart, L.I. and Harrap. K.R., Intrinsic resistance to methotrexate of cultured mammalian cells in relation to the inhibition kinetics of their dihydrofolate reductases, Cancer Res., 36, (1976). (39) Blakley, R.L. and Cocco, L., Role of isomerization of initial complexes in the binding of inhibitors to dihydrofolate reductase, Biochemistry, 24, (1985). (40) Cheung, H.T.A., Birdsall, B. and Feeney, J., 13 C-NMR studies of complexes of Escherichia coli dihydrofolate reductase formed with methotrexate and with folic acid, FEBS, 312, (1992). (41) DeGraw, J.I., Brown, V.H., Tagawa, FL, Kisliuk, R.L., Gaumont, Y. and Sirotnak, F.M., Synthesis and antitumor activity of 10-alkyl-10- deazaminopterins. A convenient synthesis of 10-deazaminopterin, J. Med. Chem., 25, (1982). (42) Shum, K.Y., Kris, M.G., Gralla, R.J., Burke, M.T., Marks, L.D. and Heelan, R.T., Phase II study of 10-ethyl-10-deazaaminopterin in patients with Stage III and IV non-small-cell lung cancer, J. Clin. Oncol., 6, (1988). (43) Koizumi, S. and Allegra, C.J., Enzyme studies of methotrexateresistant human leukemic cell (K562) subclones. Leuk. Res., 16, (1992). (44) Johns, D.G., Farquhar, D.. Chabner, B.A., Wolpert, M.K. and Adamson, R.H., Antineoplastic activity of lipid-soluble dialkyl esters of methotrexate, Experientia, 29, (1973). (45) Rosowsky, A., Beardsley, G.P., Ensminger, W.D., Lazarus, H. and Yu, C-S., Methotrexate analogues. 11. Unambiguous chemical synthesis and in vitro biological evaluation of α-and γ-monoesters as potential prodrugs, J. Med. Chem., 21, (1978). (46) Rosowsky, A. et al., Methotrexate analogues. 25. Chemical and biological studies on the γ-t-butyl esters of methotrexate and aminopterin, ibid., 28, (1985). (47) Hawkins, G.A. et al., Delivery of radionuclides to pretargeted monoclonal antibodies using dihydrofolate reductase and methotrexate, Cancer Res., 53(10 Suppl), (1993). (48) Caperelli, C.A., Mammalian glycinamide ribonucleotide transformylase. Kinetic mechanism, and associated de novo purine biosynthetic activities, J. Biol. Chem., 264, (1989). (49) Baram, J., Chabner, B.A., Drake, J.C., Fitzhugh, A.L., Sholar and P.W., Allegra, C.J., Identification and biochemical properties of 10- formyldihydrofolate, a novel folate found in methotrexate-treated cells, ibid., 263, (1988). (50) Allegra, C.J., Drake, J.C., Jolivet, J. and Chabner, B.A., Inhibition of phosphoribosyl-aminoimidazolecarboxamide transformylase by methotrexate and dihydrofolic acid polyglutamates, Proc. Natl. Acad., Sci, USA, 82, (1985). (51) Chen, P., et al., Crystal structure of glycinamide ribonucleotide transformylase from Escherichia coli at 3.0 angstrom resolution, J. Mol. Biol. 227, (1992). (52) Beardsley, G.P., Moroson, B.A., Taylor, E.C. and Moran, R.G., A new folate antimetabolite, 5,10-dideaza-5,6,7,8-tetrahydrofolate is a potent inhibitor of de novo purine synthesis, J. Biol. Chem., 264, (1989). (53) Piper, J.R. et al., Synthesis and antifolate activity of 5-methyl-5,10- dideaza analogues of aminopterin and folic acid and an alternative synthesis of 5,10-dideazatetrahydrofolic acid, a potent inhibitor of glycinamide ribonucleotide formyltransferase, J. Med. Chem., 31, (1988). (54) DeGraw, J.I., Christie, P.H., Kisliuk, R.L., Gaumont, Y. and Sirotnak, F.M., Synthesis and antifolate properties of 10-alkyl-5,10-dideaza analogues of methotrexate and tetrahydrofolic acid, ibid., 33, (1990). (55) Muggia, F. et al., Phase I clinical trial of weekly 5,10- dideazatetrahydrofolate (LY , DDATHF-B), Proc. Am. Assoc. Clin. Oncol., 9, 74(1990). (56) Young, C.W. et al., Improved clinical tolerance of lometrexol with oral folic acid, Proc. Am. Assoc. Cancer Res., 33, 406(1992). (57) Cole, J.T., Gralla, R.J., Kardinal, C.G. and Rivera, N.P., Lometrexol (DD AHF): Phase I trial of a weekly schedule of this new antifolate, ibid., 33, 413(1992). (58) Rosowsky, A., Chemical and biological activity of antifolates, Prog. Med. Chem., 26, 1-252(1989). American Journal of Pharmaceutical Education Vol. 58, Summer
Grover L. Waldrop From the Division of Biochemistry and Molecular Biology, Louisiana State University, Baton Rouge, Louisiana 70803
 Q 2009 by The International Union of Biochemistry and Molecular Biology BIOCHEMISTRY AND MOLECULAR BIOLOGY EDUCATION Vol. 37, No. 1, pp. 11 15, 2009 Articles A Qualitative Approach to Enzyme Inhibition
Q 2009 by The International Union of Biochemistry and Molecular Biology BIOCHEMISTRY AND MOLECULAR BIOLOGY EDUCATION Vol. 37, No. 1, pp. 11 15, 2009 Articles A Qualitative Approach to Enzyme Inhibition
Lipid Metabolism. Lipid Metabolism. Lipid Metabolism
 Chem 352 - Lecture 10 Lipid, Amino Acid, and Nucleotide Metabolism Part III: Nucleotide Metabolism 1 Lipid Metabolism 2-1 Chem 352, Lecture 10, Part I: Lipid Metabolism 2 Lipid Metabolism 2-2 Question:
Chem 352 - Lecture 10 Lipid, Amino Acid, and Nucleotide Metabolism Part III: Nucleotide Metabolism 1 Lipid Metabolism 2-1 Chem 352, Lecture 10, Part I: Lipid Metabolism 2 Lipid Metabolism 2-2 Question:
Student Name Biochem. 461 Exam 1 Key, September 23, 2010
 Student Name Biochem. 461 Exam 1 Key, September 23, 2010 (100 POINTS TOTAL) ANSWER ALL QUESTIONS IN THE SPACES PROVIDED [10 pts] We will learn in Chapter 28 that ribonuclosides and deoxyribonucleosides
Student Name Biochem. 461 Exam 1 Key, September 23, 2010 (100 POINTS TOTAL) ANSWER ALL QUESTIONS IN THE SPACES PROVIDED [10 pts] We will learn in Chapter 28 that ribonuclosides and deoxyribonucleosides
Enzymes Part III: regulation I. Dr. Mamoun Ahram Summer, 2017
 Enzymes Part III: regulation I Dr. Mamoun Ahram Summer, 2017 Mechanisms of regulation Expression of isoenzymes Regulation of enzymatic activity Inhibitors Conformational changes Allostery Modulators Reversible
Enzymes Part III: regulation I Dr. Mamoun Ahram Summer, 2017 Mechanisms of regulation Expression of isoenzymes Regulation of enzymatic activity Inhibitors Conformational changes Allostery Modulators Reversible
Nucleotide Metabolism
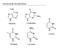 Nucleotide Metabolism Adenine Guanidine Uracil Thymine Cytosine Last Week Nucleotide Metabolism - Mostly, we think of nucleotides as being part of RNA/DNA, but now that we ve studied metabolism, we know
Nucleotide Metabolism Adenine Guanidine Uracil Thymine Cytosine Last Week Nucleotide Metabolism - Mostly, we think of nucleotides as being part of RNA/DNA, but now that we ve studied metabolism, we know
Nucleotide Metabolism
 Nucleotide Metabolism Nucleotide Synthesis De Novo Pathway Synthesize purine and pyrimidine nucleotides from low M.W. precursors Salvage Pathway synthesize nucleotides from nucleosides of nucleobases NB:
Nucleotide Metabolism Nucleotide Synthesis De Novo Pathway Synthesize purine and pyrimidine nucleotides from low M.W. precursors Salvage Pathway synthesize nucleotides from nucleosides of nucleobases NB:
I. NUCLEOTIDES. Nitrogenous bases are frequently represented by single letter abbreviations. The abbreviations for several bases are shown below:
 Biochemistry II Nucleotide Metabolism I. NUCLEOTIDES Nitrogenous bases are frequently represented by single letter abbreviations. The abbreviations for several bases are shown below: Common Nitrogenous
Biochemistry II Nucleotide Metabolism I. NUCLEOTIDES Nitrogenous bases are frequently represented by single letter abbreviations. The abbreviations for several bases are shown below: Common Nitrogenous
number Done by Corrected by Doctor
 number 34 Done by Abdulrahman Alhanbali Corrected by Mohammad Mahmoud Tarabeih Doctor Diala Nucleotide metabolism 1 P age In this lecture we will talk about nucleotides; their structures, the synthesis
number 34 Done by Abdulrahman Alhanbali Corrected by Mohammad Mahmoud Tarabeih Doctor Diala Nucleotide metabolism 1 P age In this lecture we will talk about nucleotides; their structures, the synthesis
number Done by Corrected by Doctor Diala
 number 34 Done by Abdulrahman Alhanbali Corrected by Mohammad Mahmoud Tarabeih Doctor Diala 1 P a g e Nucleotide metabolism In this lecture we will talk about nucleotides; their structures, the synthesis
number 34 Done by Abdulrahman Alhanbali Corrected by Mohammad Mahmoud Tarabeih Doctor Diala 1 P a g e Nucleotide metabolism In this lecture we will talk about nucleotides; their structures, the synthesis
Biosynthesis of nucleotides
 Biosynthesis of nucleotides De novo pathways De novo purine nucleotide synthesis Regulation of purine biosynthesis De novo pyrimidine nucleotides biosynthesis Salvage pathways Lesch-Nyhan syndrome Why
Biosynthesis of nucleotides De novo pathways De novo purine nucleotide synthesis Regulation of purine biosynthesis De novo pyrimidine nucleotides biosynthesis Salvage pathways Lesch-Nyhan syndrome Why
Chapter 3 Nucleic Acids, Proteins, and Enzymes
 3 Nucleic Acids, Proteins, and Enzymes Chapter 3 Nucleic Acids, Proteins, and Enzymes Key Concepts 3.1 Nucleic Acids Are Informational Macromolecules 3.2 Proteins Are Polymers with Important Structural
3 Nucleic Acids, Proteins, and Enzymes Chapter 3 Nucleic Acids, Proteins, and Enzymes Key Concepts 3.1 Nucleic Acids Are Informational Macromolecules 3.2 Proteins Are Polymers with Important Structural
Nucleic Acids, Proteins, and Enzymes
 3 Nucleic Acids, Proteins, and Enzymes Chapter 3 Nucleic Acids, Proteins, and Enzymes Key Concepts 3.1 Nucleic Acids Are Informational Macromolecules 3.2 Proteins Are Polymers with Important Structural
3 Nucleic Acids, Proteins, and Enzymes Chapter 3 Nucleic Acids, Proteins, and Enzymes Key Concepts 3.1 Nucleic Acids Are Informational Macromolecules 3.2 Proteins Are Polymers with Important Structural
Proteins Amides from Amino Acids
 Chapter 26 and Chapter 28 Proteins Amides from Amino Acids Amino acids contain a basic amino group and an acidic carboxyl group Joined as amides between the ¾NH 2 of one amino acid and the ¾CO 2 H to the
Chapter 26 and Chapter 28 Proteins Amides from Amino Acids Amino acids contain a basic amino group and an acidic carboxyl group Joined as amides between the ¾NH 2 of one amino acid and the ¾CO 2 H to the
NANYANG TECHNOLOGICAL UNIVERSITY SEMESTER I EXAMINATION CBC922 Medicinal Chemistry. NOVEMBER TIME ALLOWED: 120 min
 AYAG TECLGICAL UIVERSITY SEMESTER I EXAMIATI 2006-2007 CBC922 Medicinal Chemistry VEMBER 2006 - TIME ALLWED 120 min ISTRUCTIS T CADIDATES 1. This examination paper contains TW (2) parts and comprises SIX
AYAG TECLGICAL UIVERSITY SEMESTER I EXAMIATI 2006-2007 CBC922 Medicinal Chemistry VEMBER 2006 - TIME ALLWED 120 min ISTRUCTIS T CADIDATES 1. This examination paper contains TW (2) parts and comprises SIX
Gary Ketner, PhD Johns Hopkins University. Treatment of Infectious Disease: Drugs and Drug Resistance
 This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike License. Your use of this material constitutes acceptance of that license and the conditions of use of materials on this
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike License. Your use of this material constitutes acceptance of that license and the conditions of use of materials on this
BC 367, Exam 2 November 13, Part I. Multiple Choice (3 pts each)- Please circle the single best answer.
 Name BC 367, Exam 2 November 13, 2008 Part I. Multiple Choice (3 pts each)- Please circle the single best answer. 1. The enzyme pyruvate dehydrogenase catalyzes the following reaction. What kind of enzyme
Name BC 367, Exam 2 November 13, 2008 Part I. Multiple Choice (3 pts each)- Please circle the single best answer. 1. The enzyme pyruvate dehydrogenase catalyzes the following reaction. What kind of enzyme
6-Foot Mini Toober Activity
 Big Idea The interaction between the substrate and enzyme is highly specific. Even a slight change in shape of either the substrate or the enzyme may alter the efficient and selective ability of the enzyme
Big Idea The interaction between the substrate and enzyme is highly specific. Even a slight change in shape of either the substrate or the enzyme may alter the efficient and selective ability of the enzyme
Nucleotide Metabolism Biochemistry by Lippincott pp
 Nucleotide Metabolism Biochemistry by Lippincott pp 291-306 Metabolism: CONCEPT Ø Metabolism is the totality of an organism s chemical reactions. Ø A metabolic pathway begins with a specific molecule and
Nucleotide Metabolism Biochemistry by Lippincott pp 291-306 Metabolism: CONCEPT Ø Metabolism is the totality of an organism s chemical reactions. Ø A metabolic pathway begins with a specific molecule and
GENERAL NUCLEIC ACID BIOCHEMISTRY 461, FALL A two (2) unit course for non-biochemistry majors only.
 GENERAL NUCLEIC ACID BIOCHEMISTRY 461, FALL 2010 A two (2) unit course for non-biochemistry majors only. PREREQUISITES: Biology 181 Organic Chemistry (Chem 241a,b) Concurrent/previous registration in Biochemistry
GENERAL NUCLEIC ACID BIOCHEMISTRY 461, FALL 2010 A two (2) unit course for non-biochemistry majors only. PREREQUISITES: Biology 181 Organic Chemistry (Chem 241a,b) Concurrent/previous registration in Biochemistry
Practice Problems on Pharmaceutical Drugs
 Drugs.1 Practice Problems on Pharmaceutical Drugs 1. In the United States, the pharmaceutical-drug approval process is regulated by the FDA. Which organization in Canada performs the same function as the
Drugs.1 Practice Problems on Pharmaceutical Drugs 1. In the United States, the pharmaceutical-drug approval process is regulated by the FDA. Which organization in Canada performs the same function as the
RNA Expression of the information in a gene generally involves production of an RNA molecule transcribed from a DNA template. RNA differs from DNA
 RNA Expression of the information in a gene generally involves production of an RNA molecule transcribed from a DNA template. RNA differs from DNA that it has a hydroxyl group at the 2 position of the
RNA Expression of the information in a gene generally involves production of an RNA molecule transcribed from a DNA template. RNA differs from DNA that it has a hydroxyl group at the 2 position of the
36. The double bonds in naturally-occuring fatty acids are usually isomers. A. cis B. trans C. both cis and trans D. D- E. L-
 36. The double bonds in naturally-occuring fatty acids are usually isomers. A. cis B. trans C. both cis and trans D. D- E. L- 37. The essential fatty acids are A. palmitic acid B. linoleic acid C. linolenic
36. The double bonds in naturally-occuring fatty acids are usually isomers. A. cis B. trans C. both cis and trans D. D- E. L- 37. The essential fatty acids are A. palmitic acid B. linoleic acid C. linolenic
6 Enzymes II W. H. Freeman and Company
 6 Enzymes II 2017 W. H. Freeman and Company The role of an enzyme in an enzyme-catalyzed reaction is to: A. bind a transition state intermediate, such that it cannot be converted back to substrate. B.
6 Enzymes II 2017 W. H. Freeman and Company The role of an enzyme in an enzyme-catalyzed reaction is to: A. bind a transition state intermediate, such that it cannot be converted back to substrate. B.
Nanobiotechnology. Place: IOP 1 st Meeting Room Time: 9:30-12:00. Reference: Review Papers. Grade: 50% midterm, 50% final.
 Nanobiotechnology Place: IOP 1 st Meeting Room Time: 9:30-12:00 Reference: Review Papers Grade: 50% midterm, 50% final Midterm: 5/15 History Atom Earth, Air, Water Fire SEM: 20-40 nm Silver 66.2% Gold
Nanobiotechnology Place: IOP 1 st Meeting Room Time: 9:30-12:00 Reference: Review Papers Grade: 50% midterm, 50% final Midterm: 5/15 History Atom Earth, Air, Water Fire SEM: 20-40 nm Silver 66.2% Gold
For more information about how to cite these materials visit
 Author(s): Dr. Robert Lyons, 2009 License: Unless otherwise noted, this material is made available under the terms of the Creative Commons Attribution Share Alike 3.0 License: http:// creativecommons.org/licenses/by-sa/3.0/
Author(s): Dr. Robert Lyons, 2009 License: Unless otherwise noted, this material is made available under the terms of the Creative Commons Attribution Share Alike 3.0 License: http:// creativecommons.org/licenses/by-sa/3.0/
Biochem for the Boards 2nd Edition
 Biochem for the Boards! Biochem for the Boards 2nd Edition Active Learning Workbook for the BDE 1, DDS, MA Bonus Chapter ucleotide Metabolism Socrates Publishing Co. ew York, Y Page 1 ucleotide Metabolism
Biochem for the Boards! Biochem for the Boards 2nd Edition Active Learning Workbook for the BDE 1, DDS, MA Bonus Chapter ucleotide Metabolism Socrates Publishing Co. ew York, Y Page 1 ucleotide Metabolism
Zool 3200: Cell Biology Exam 3 3/6/15
 Name: Trask Zool 3200: Cell Biology Exam 3 3/6/15 Answer each of the following questions in the space provided; circle the correct answer or answers for each multiple choice question and circle either
Name: Trask Zool 3200: Cell Biology Exam 3 3/6/15 Answer each of the following questions in the space provided; circle the correct answer or answers for each multiple choice question and circle either
 The replication of DNA Kornberg 1957 Meselson and Stahl 1958 Cairns 1963 Okazaki 1968 DNA Replication The driving force for DNA synthesis. The addition of a nucleotide to a growing polynucleotide
The replication of DNA Kornberg 1957 Meselson and Stahl 1958 Cairns 1963 Okazaki 1968 DNA Replication The driving force for DNA synthesis. The addition of a nucleotide to a growing polynucleotide
The Stringent Response
 The Stringent Response When amino acids are limiting a response is triggered to shut down a wide range of biosynthetic processes This process is called the Stringent Response It results in the synthesis
The Stringent Response When amino acids are limiting a response is triggered to shut down a wide range of biosynthetic processes This process is called the Stringent Response It results in the synthesis
1) The penicillin family of antibiotics, discovered by Alexander Fleming in 1928, has the following general structure: O O
 ame: TF ame: LS1a Fall 06 Problem Set #3 Due Friday 10/13 at noon in your TF s drop box on the 2 nd floor of the Science Center All questions including the (*extra*) ones should be turned in 1) The penicillin
ame: TF ame: LS1a Fall 06 Problem Set #3 Due Friday 10/13 at noon in your TF s drop box on the 2 nd floor of the Science Center All questions including the (*extra*) ones should be turned in 1) The penicillin
AP Biology Book Notes Chapter 3 v Nucleic acids Ø Polymers specialized for the storage transmission and use of genetic information Ø Two types DNA
 AP Biology Book Notes Chapter 3 v Nucleic acids Ø Polymers specialized for the storage transmission and use of genetic information Ø Two types DNA Encodes hereditary information Used to specify the amino
AP Biology Book Notes Chapter 3 v Nucleic acids Ø Polymers specialized for the storage transmission and use of genetic information Ø Two types DNA Encodes hereditary information Used to specify the amino
Nucleotide Metabolism December 12-17, 2008 Dr. Robert Lyons. See: for supplementary course materials.
 1 2 ucleotide Metabolism December 12-17, 2008 Dr. Robert Lyons See: http://seqcore.brcf.med.umich.edu/mcb500 for supplementary course materials. Medical relevance of nucleotide pathways Critical in cancer
1 2 ucleotide Metabolism December 12-17, 2008 Dr. Robert Lyons See: http://seqcore.brcf.med.umich.edu/mcb500 for supplementary course materials. Medical relevance of nucleotide pathways Critical in cancer
Drug DNA interaction. Modeling DNA ligand interaction of intercalating ligands
 Drug DNA interaction DNA as carrier of genetic information is a major target for drug interaction because of the ability to interfere with transcription (gene expression and protein synthesis) and DNA
Drug DNA interaction DNA as carrier of genetic information is a major target for drug interaction because of the ability to interfere with transcription (gene expression and protein synthesis) and DNA
BIO 311C Spring Lecture 16 Monday 1 March
 BIO 311C Spring 2010 Lecture 16 Monday 1 March Review Primary Structure of a portion of a polypeptide chain backbone of Polypeptide chain R-groups of amino acids Native conformation of a dimeric protein,
BIO 311C Spring 2010 Lecture 16 Monday 1 March Review Primary Structure of a portion of a polypeptide chain backbone of Polypeptide chain R-groups of amino acids Native conformation of a dimeric protein,
From mechanism to medicne
 From mechanism to medicne a look at proteins and drug design Chem 342 δ δ δ+ M 2009 δ+ δ+ δ M Drug Design - an Iterative Approach @ DSU Structural Analysis of Receptor Structural Analysis of Ligand-Receptor
From mechanism to medicne a look at proteins and drug design Chem 342 δ δ δ+ M 2009 δ+ δ+ δ M Drug Design - an Iterative Approach @ DSU Structural Analysis of Receptor Structural Analysis of Ligand-Receptor
Feedback D. Incorrect! No, although this is a correct characteristic of RNA, this is not the best response to the questions.
 Biochemistry - Problem Drill 23: RNA No. 1 of 10 1. Which of the following statements best describes the structural highlights of RNA? (A) RNA can be single or double stranded. (B) G-C pairs have 3 hydrogen
Biochemistry - Problem Drill 23: RNA No. 1 of 10 1. Which of the following statements best describes the structural highlights of RNA? (A) RNA can be single or double stranded. (B) G-C pairs have 3 hydrogen
Questions from chapters in the textbook that are relevant for the final exam
 Questions from chapters in the textbook that are relevant for the final exam Chapter 9 Replication of DNA Question 1. Name the two substrates for DNA synthesis. Explain why each is necessary for DNA synthesis.
Questions from chapters in the textbook that are relevant for the final exam Chapter 9 Replication of DNA Question 1. Name the two substrates for DNA synthesis. Explain why each is necessary for DNA synthesis.
Protein Structure/Function Relationships
 Protein Structure/Function Relationships W. M. Grogan, Ph.D. OBJECTIVES 1. Describe and cite examples of fibrous and globular proteins. 2. Describe typical tertiary structural motifs found in proteins.
Protein Structure/Function Relationships W. M. Grogan, Ph.D. OBJECTIVES 1. Describe and cite examples of fibrous and globular proteins. 2. Describe typical tertiary structural motifs found in proteins.
Biochemistry study of the molecular basis of life
 Biochemistry : An Introduction Biochemistry study of the molecular basis of life n Study of the chemistry of living organisms Studies organic molecules & organic reactions in living organisms n Living
Biochemistry : An Introduction Biochemistry study of the molecular basis of life n Study of the chemistry of living organisms Studies organic molecules & organic reactions in living organisms n Living
Protein analysis. Dr. Mamoun Ahram Summer semester, Resources This lecture Campbell and Farrell s Biochemistry, Chapters 5
 Protein analysis Dr. Mamoun Ahram Summer semester, 2015-2016 Resources This lecture Campbell and Farrell s Biochemistry, Chapters 5 Bases of protein separation Proteins can be purified on the basis Solubility
Protein analysis Dr. Mamoun Ahram Summer semester, 2015-2016 Resources This lecture Campbell and Farrell s Biochemistry, Chapters 5 Bases of protein separation Proteins can be purified on the basis Solubility
Current Status and Future Prospects p. 46 Acknowledgements p. 46 References p. 46 Hammerhead Ribozyme Crystal Structures and Catalysis
 Ribozymes and RNA Catalysis: Introduction and Primer What are Ribozymes? p. 1 What is the Role of Ribozymes in Cells? p. 1 Ribozymes Bring about Significant Rate Enhancements p. 4 Why Study Ribozymes?
Ribozymes and RNA Catalysis: Introduction and Primer What are Ribozymes? p. 1 What is the Role of Ribozymes in Cells? p. 1 Ribozymes Bring about Significant Rate Enhancements p. 4 Why Study Ribozymes?
 Problem: The GC base pairs are more stable than AT base pairs. Why? 5. Triple-stranded DNA was first observed in 1957. Scientists later discovered that the formation of triplestranded DNA involves a type
Problem: The GC base pairs are more stable than AT base pairs. Why? 5. Triple-stranded DNA was first observed in 1957. Scientists later discovered that the formation of triplestranded DNA involves a type
Investigating the interactions of FCS-304 and other Monoamine Oxidase (MAO) A Inhibitors Using Molecular Docking
 Available online at www.scholarsresearchlibrary.com J. Comput. Method. Mol. Design, 2011, 1 (3): 24-28 (http://scholarsresearchlibrary.com/archive.html) ISSN : 2231-3176 CODEN (USA): JCMMDA Investigating
Available online at www.scholarsresearchlibrary.com J. Comput. Method. Mol. Design, 2011, 1 (3): 24-28 (http://scholarsresearchlibrary.com/archive.html) ISSN : 2231-3176 CODEN (USA): JCMMDA Investigating
Bioreaction Kinetics Seungwook Kim Chem. & Bio. Eng.
 Bioreaction Kinetics 2 2004 Seungwook Kim Chem. & Bio. Eng. Reference Chemistry and the Living Organism, 6 th edition, Molly M. Bloomfield, Lawrence J. Stephens, John Wiley & Sons, Inc. Enzymes 1. Most
Bioreaction Kinetics 2 2004 Seungwook Kim Chem. & Bio. Eng. Reference Chemistry and the Living Organism, 6 th edition, Molly M. Bloomfield, Lawrence J. Stephens, John Wiley & Sons, Inc. Enzymes 1. Most
Solutions to 7.02 Quiz II 10/27/05
 Solutions to 7.02 Quiz II 10/27/05 Class Average = 83 Standard Deviation = 9 Range Grade % 87-100 A 43 74-86 B 39 55-73 C 17 > 54 D 1 Question 1 (56 points) While studying deep sea bacteria, you discover
Solutions to 7.02 Quiz II 10/27/05 Class Average = 83 Standard Deviation = 9 Range Grade % 87-100 A 43 74-86 B 39 55-73 C 17 > 54 D 1 Question 1 (56 points) While studying deep sea bacteria, you discover
Chemistry 1050 Exam 4 Study Guide
 Chapter 19 Chemistry 1050 Exam 4 Study Guide 19.1 and 19.2 Know there are 20 common amino acids that can polymerize into proteins. Know why amino acids are called alpha amino acids. Identify the charges
Chapter 19 Chemistry 1050 Exam 4 Study Guide 19.1 and 19.2 Know there are 20 common amino acids that can polymerize into proteins. Know why amino acids are called alpha amino acids. Identify the charges
Chemistry 1120 Exam 3 Study Guide
 Chemistry 1120 Exam 3 Study Guide Chapter 9 9.1 and 9.2 Know there are 20 common amino acids that can polymerize into proteins. Know why amino acids are called alpha amino acids. Identify the charges of
Chemistry 1120 Exam 3 Study Guide Chapter 9 9.1 and 9.2 Know there are 20 common amino acids that can polymerize into proteins. Know why amino acids are called alpha amino acids. Identify the charges of
Drug Design 2. Overview. Drug Discovery Pipeline. Drug Design. Oliver Kohlbacher Winter 2009/ Success Stories
 Drug Design 2 Oliver Kohlbacher Winter 2009/2010 13. Success Stories Abt. Simulation biologischer Systeme WSI/ZBIT, Eberhard Karls Universität Tübingen Overview Drug Discovery Pipeline reloaded Successful
Drug Design 2 Oliver Kohlbacher Winter 2009/2010 13. Success Stories Abt. Simulation biologischer Systeme WSI/ZBIT, Eberhard Karls Universität Tübingen Overview Drug Discovery Pipeline reloaded Successful
Organic Pharmaceutical Chemistry: Prodrugs
 Organic Pharmaceutical Chemistry IV 1st Semester, Year 5 (2016-2017) Lecture 1 Organic Pharmaceutical Chemistry: Prodrugs Dr Asim A. Balakit Pharmaceutical Chemistry Dept., College of Pharmacy, Babylon
Organic Pharmaceutical Chemistry IV 1st Semester, Year 5 (2016-2017) Lecture 1 Organic Pharmaceutical Chemistry: Prodrugs Dr Asim A. Balakit Pharmaceutical Chemistry Dept., College of Pharmacy, Babylon
Amino Acids and Proteins
 Various Functions of Proteins SB203 Amino Acids and Proteins Jirundon Yuvaniyama, Ph.D. Department of Biochemistry Faculty of Science Mahidol University Enzymes Transport proteins utrient and storage proteins
Various Functions of Proteins SB203 Amino Acids and Proteins Jirundon Yuvaniyama, Ph.D. Department of Biochemistry Faculty of Science Mahidol University Enzymes Transport proteins utrient and storage proteins
Supplementary Note 1. Enzymatic properties of the purified Syn BVR
 Supplementary Note 1. Enzymatic properties of the purified Syn BVR The expression vector pet15b-syn bvr allowed us to routinely prepare 15 mg of electrophoretically homogenous Syn BVR from 2.5 L of TB-medium
Supplementary Note 1. Enzymatic properties of the purified Syn BVR The expression vector pet15b-syn bvr allowed us to routinely prepare 15 mg of electrophoretically homogenous Syn BVR from 2.5 L of TB-medium
Chapter 5: Microbial Metabolism (Part I)
 Chapter 5: Microbial Metabolism (Part I) Microbial Metabolism Metabolism refers to all chemical reactions that occur within a living organism. These chemical reactions are generally of two types: Catabolic:
Chapter 5: Microbial Metabolism (Part I) Microbial Metabolism Metabolism refers to all chemical reactions that occur within a living organism. These chemical reactions are generally of two types: Catabolic:
Unless otherwise noted, the content of this course material is licensed under a Creative Commons Attribution Share Alike 3.0 License.
 Unless otherwise noted, the content of this course material is licensed under a Creative Commons Attribution Share Alike 3.0 License. Copyright 2007, Robert Lyons. The following information is intended
Unless otherwise noted, the content of this course material is licensed under a Creative Commons Attribution Share Alike 3.0 License. Copyright 2007, Robert Lyons. The following information is intended
Examination I PHRM 836 Biochemistry for Pharmaceutical Sciences II September 29, 2015
 Examination I PHRM 836 Biochemistry for Pharmaceutical Sciences II September 29, 2015 PHRM 836 Exam I - 1 Name: Instructions 1. Check your exam to make certain that it has 9 pages including this cover
Examination I PHRM 836 Biochemistry for Pharmaceutical Sciences II September 29, 2015 PHRM 836 Exam I - 1 Name: Instructions 1. Check your exam to make certain that it has 9 pages including this cover
A. Incorrect! Enzymes are not altered or consumed by the reactions they catalyze.
 CLEP Biology - Problem Drill 04: Enzymes and Cellular Metabolism No. 1 of 10 1. Which of the following statements about enzymes is correct? (A) Enzymes are consumed in a reaction. (B) Enzymes act by lowering
CLEP Biology - Problem Drill 04: Enzymes and Cellular Metabolism No. 1 of 10 1. Which of the following statements about enzymes is correct? (A) Enzymes are consumed in a reaction. (B) Enzymes act by lowering
Protocol S1: Supporting Information
 Protocol S1: Supporting Information Basis for the specificity of the kinase domain of Abl for peptide substrates The crystal structures reported in this work were obtained using two different ATP analog-peptide
Protocol S1: Supporting Information Basis for the specificity of the kinase domain of Abl for peptide substrates The crystal structures reported in this work were obtained using two different ATP analog-peptide
Biochemistry 302, February 11, 2004 Exam 1 (100 points) 1. What form of DNA is shown on this Nature Genetics cover? Z-DNA or left-handed DNA
 1 Biochemistry 302, February 11, 2004 Exam 1 (100 points) Name I. Structural recognition (very short answer, 2 points each) 1. What form of DNA is shown on this Nature Genetics cover? Z-DNA or left-handed
1 Biochemistry 302, February 11, 2004 Exam 1 (100 points) Name I. Structural recognition (very short answer, 2 points each) 1. What form of DNA is shown on this Nature Genetics cover? Z-DNA or left-handed
ENZYME SCIENCE AND ENGINEERING PROF. SUBHASH CHAND DEPARTMENT OF BIOCHEMICAL ENGINEERING AND BIOTECHNOLOGY IIT DELHI LECTURE 5
 ENZYME SCIENCE AND ENGINEERING PROF. SUBHASH CHAND DEPARTMENT OF BIOCHEMICAL ENGINEERING AND BIOTECHNOLOGY IIT DELHI LECTURE 5 SPECIFICITY OF ENZYME ACTION You may recall that earlier we have outlined
ENZYME SCIENCE AND ENGINEERING PROF. SUBHASH CHAND DEPARTMENT OF BIOCHEMICAL ENGINEERING AND BIOTECHNOLOGY IIT DELHI LECTURE 5 SPECIFICITY OF ENZYME ACTION You may recall that earlier we have outlined
Asparagine 229 Mutants of Thymidylate Synthase Catalyze the Methylation of 3-Methyl-2 -deoxyuridine 5 -Monophosphate
 3944 Biochemistry 1996, 35, 3944-3949 Asparagine 229 Mutants of Thymidylate Synthase Catalyze the Methylation of 3-Methyl-2 -deoxyuridine 5 -Monophosphate Paola M. Costi, Lu Liu, Janet S. Finer-Moore,
3944 Biochemistry 1996, 35, 3944-3949 Asparagine 229 Mutants of Thymidylate Synthase Catalyze the Methylation of 3-Methyl-2 -deoxyuridine 5 -Monophosphate Paola M. Costi, Lu Liu, Janet S. Finer-Moore,
SCBC203 Cell Division & DNA Synthesis. Assoc. Prof. Rutaiwan Tohtong Department of Biochemistry Faculty of Science PR318
 SCBC203 Cell Division & DNA Synthesis Assoc. Prof. Rutaiwan Tohtong Department of Biochemistry Faculty of Science PR318 Rutaiwan.toh@mahidol.ac.th 1 Objectives 1. Explain the biological roles & significance
SCBC203 Cell Division & DNA Synthesis Assoc. Prof. Rutaiwan Tohtong Department of Biochemistry Faculty of Science PR318 Rutaiwan.toh@mahidol.ac.th 1 Objectives 1. Explain the biological roles & significance
Biomolecules: nucleotides
 Biomolecules: nucleotides Nucleotides and nucleic acids Nucleic acids are linear hetero-polymers adapted to maintain and transmit the genetic information Depending on their sugar moiety they can be classified
Biomolecules: nucleotides Nucleotides and nucleic acids Nucleic acids are linear hetero-polymers adapted to maintain and transmit the genetic information Depending on their sugar moiety they can be classified
Paper : 03 Structure and Function of Biomolecules II Module: 02 Nucleosides, Nucleotides and type of Nucleic Acids
 Paper : 03 Structure and Function of Biomolecules II Module: 02 Nucleosides, Nucleotides and type of Nucleic Acids Principal Investigator Paper Coordinators Prof. Sunil Kumar Khare, Professor, Department
Paper : 03 Structure and Function of Biomolecules II Module: 02 Nucleosides, Nucleotides and type of Nucleic Acids Principal Investigator Paper Coordinators Prof. Sunil Kumar Khare, Professor, Department
RNA is a single strand molecule composed of subunits called nucleotides joined by phosphodiester bonds.
 The Versatility of RNA Primary structure of RNA RNA is a single strand molecule composed of subunits called nucleotides joined by phosphodiester bonds. Each nucleotide subunit is composed of a ribose sugar,
The Versatility of RNA Primary structure of RNA RNA is a single strand molecule composed of subunits called nucleotides joined by phosphodiester bonds. Each nucleotide subunit is composed of a ribose sugar,
Chapter 17 Nucleic Acids and Protein Synthesis
 Chapter 17 Nucleic Acids and Protein Synthesis Nucleic Acids Nucleic acids are the components that make up the genetic material DNA (deoxyribonucleic acid). DNA is a macromolecule which contains all the
Chapter 17 Nucleic Acids and Protein Synthesis Nucleic Acids Nucleic acids are the components that make up the genetic material DNA (deoxyribonucleic acid). DNA is a macromolecule which contains all the
Fig. 1. A schematic illustration of pipeline from gene to drug : integration of virtual and real experiments.
 1 Integration of Structure-Based Drug Design and SPR Biosensor Technology in Discovery of New Lead Compounds Alexis S. Ivanov Institute of Biomedical Chemistry RAMS, 10, Pogodinskaya str., Moscow, 119121,
1 Integration of Structure-Based Drug Design and SPR Biosensor Technology in Discovery of New Lead Compounds Alexis S. Ivanov Institute of Biomedical Chemistry RAMS, 10, Pogodinskaya str., Moscow, 119121,
Precluding uracil from DNA Dmitry G Vassylyev 1 * and Kosuke Morikawa 2 *
 Minireview 1381 Precluding uracil from DNA Dmitry G Vassylyev 1 * and Kosuke Morikawa 2 * Two enzymes, dutp pyrophosphatase and uracil-dna glycosylase, prevent the misincorporation of uracil into the genome
Minireview 1381 Precluding uracil from DNA Dmitry G Vassylyev 1 * and Kosuke Morikawa 2 * Two enzymes, dutp pyrophosphatase and uracil-dna glycosylase, prevent the misincorporation of uracil into the genome
Chem 250 Answer Key In-class Quiz #3v1
 age 1 of 6 Quiz #3 ame. Chem 250 Answer Key In-class Quiz #3v1 This exam is composed of 20 questions. lease scan them all before starting. As discussed in the course syllabus, honesty and integrity are
age 1 of 6 Quiz #3 ame. Chem 250 Answer Key In-class Quiz #3v1 This exam is composed of 20 questions. lease scan them all before starting. As discussed in the course syllabus, honesty and integrity are
Each enzyme has a unique 3-D shape and recognizes and binds only the specific substrate of a reaction.
 1 Enzyme = protein molecule that serves as a biological catalyst. allow life to go on. speed up and regulate metabolic reactions. Catalyst= a chemical that speeds up the rate of a reaction without itself
1 Enzyme = protein molecule that serves as a biological catalyst. allow life to go on. speed up and regulate metabolic reactions. Catalyst= a chemical that speeds up the rate of a reaction without itself
Enzymes. Most known enzymes are large globular proteins.
 Chapter 10 Enzymes Enzymes Most known enzymes are large globular proteins. There are 27,383 proteins in the human genome. 12,000 have unknown functions and 6,000 behave as enzymes. Enzymes are necessary
Chapter 10 Enzymes Enzymes Most known enzymes are large globular proteins. There are 27,383 proteins in the human genome. 12,000 have unknown functions and 6,000 behave as enzymes. Enzymes are necessary
Rajan Sankaranarayanan
 Proofreading during translation of the genetic code Rajan Sankaranarayanan Centre for Cellular and Molecular Biology (CCMB) Council for Scientific and Industrial Research (CSIR) Hyderabad, INDIA Accuracy
Proofreading during translation of the genetic code Rajan Sankaranarayanan Centre for Cellular and Molecular Biology (CCMB) Council for Scientific and Industrial Research (CSIR) Hyderabad, INDIA Accuracy
Antibody-Antigen recognition. Structural Biology Weekend Seminar Annegret Kramer
 Antibody-Antigen recognition Structural Biology Weekend Seminar 10.07.2005 Annegret Kramer Contents Function and structure of antibodies Features of antibody-antigen interfaces Examples of antibody-antigen
Antibody-Antigen recognition Structural Biology Weekend Seminar 10.07.2005 Annegret Kramer Contents Function and structure of antibodies Features of antibody-antigen interfaces Examples of antibody-antigen
NMR in Drug Metabolism Extraction NMR - Keeping up with the Pace of Drug Discovery. Dr Ute Gerhard
 F NMR in Drug Metabolism Extraction NMR - Keeping up with the Pace of Drug Discovery Dr Ute Gerhard The Early Days Eat plant Feel Worse Feel Better Try again Drug! The Golden Age Make molecule Feed to
F NMR in Drug Metabolism Extraction NMR - Keeping up with the Pace of Drug Discovery Dr Ute Gerhard The Early Days Eat plant Feel Worse Feel Better Try again Drug! The Golden Age Make molecule Feed to
Algorithms in Bioinformatics ONE Transcription Translation
 Algorithms in Bioinformatics ONE Transcription Translation Sami Khuri Department of Computer Science San José State University sami.khuri@sjsu.edu Biology Review DNA RNA Proteins Central Dogma Transcription
Algorithms in Bioinformatics ONE Transcription Translation Sami Khuri Department of Computer Science San José State University sami.khuri@sjsu.edu Biology Review DNA RNA Proteins Central Dogma Transcription
7.014 Quiz II Handout
 7.014 Quiz II Handout Quiz II: Wednesday, March 17 12:05-12:55 54-100 **This will be a closed book exam** Quiz Review Session: Friday, March 12 7:00-9:00 pm room 54-100 Open Tutoring Session: Tuesday,
7.014 Quiz II Handout Quiz II: Wednesday, March 17 12:05-12:55 54-100 **This will be a closed book exam** Quiz Review Session: Friday, March 12 7:00-9:00 pm room 54-100 Open Tutoring Session: Tuesday,
Examination I PHRM 836 Biochemistry for Pharmaceutical Sciences II September 29, 2015
 Examination I PHRM 836 Biochemistry for Pharmaceutical Sciences II September 29, 2015 PHRM 836 Exam I - 1 Name: Instructions 1. Check your exam to make certain that it has 9 pages including this cover
Examination I PHRM 836 Biochemistry for Pharmaceutical Sciences II September 29, 2015 PHRM 836 Exam I - 1 Name: Instructions 1. Check your exam to make certain that it has 9 pages including this cover
Chem Lecture 5 Catalytic Strategies
 Chem 452 - Lecture 5 Catalytic Strategies 111026 Enzymes have evolved an array of different strategies or enhancing the power and specificity of the reactions they catalyze. For numerous enzymes the details
Chem 452 - Lecture 5 Catalytic Strategies 111026 Enzymes have evolved an array of different strategies or enhancing the power and specificity of the reactions they catalyze. For numerous enzymes the details
Figure A summary of spontaneous alterations likely to require DNA repair.
 DNA Damage Figure 5-46. A summary of spontaneous alterations likely to require DNA repair. The sites on each nucleotide that are known to be modified by spontaneous oxidative damage (red arrows), hydrolytic
DNA Damage Figure 5-46. A summary of spontaneous alterations likely to require DNA repair. The sites on each nucleotide that are known to be modified by spontaneous oxidative damage (red arrows), hydrolytic
Basic concepts of molecular biology
 Basic concepts of molecular biology Gabriella Trucco Email: gabriella.trucco@unimi.it Life The main actors in the chemistry of life are molecules called proteins nucleic acids Proteins: many different
Basic concepts of molecular biology Gabriella Trucco Email: gabriella.trucco@unimi.it Life The main actors in the chemistry of life are molecules called proteins nucleic acids Proteins: many different
STRUCTURAL BIOLOGY. α/β structures Closed barrels Open twisted sheets Horseshoe folds
 STRUCTURAL BIOLOGY α/β structures Closed barrels Open twisted sheets Horseshoe folds The α/β domains Most frequent domain structures are α/β domains: A central parallel or mixed β sheet Surrounded by α
STRUCTURAL BIOLOGY α/β structures Closed barrels Open twisted sheets Horseshoe folds The α/β domains Most frequent domain structures are α/β domains: A central parallel or mixed β sheet Surrounded by α
Molecular Docking Study of Some Novel Nitroimidazo[1,2-b]pyridazine
![Molecular Docking Study of Some Novel Nitroimidazo[1,2-b]pyridazine Molecular Docking Study of Some Novel Nitroimidazo[1,2-b]pyridazine](/thumbs/95/125396186.jpg) DOI:10.7598/cst2016.1227 Chemical Science Transactions ISSN:2278-3458 2016, 5(3), 700-710 RESEARCH ARTICLE Molecular Docking Study of Some Novel Nitroimidazo[1,2-b]pyridazine based Heterocyclics on Methicillin
DOI:10.7598/cst2016.1227 Chemical Science Transactions ISSN:2278-3458 2016, 5(3), 700-710 RESEARCH ARTICLE Molecular Docking Study of Some Novel Nitroimidazo[1,2-b]pyridazine based Heterocyclics on Methicillin
Metabolism BIOL 3702: Chapter 10
 Metabolism BIOL 3702: Chapter 10 Introduction to Metabolism u Metabolism is the sum total of all the chemical reactions occurring in a cell u Two major parts of metabolism: v Catabolism Ø Large, more complex
Metabolism BIOL 3702: Chapter 10 Introduction to Metabolism u Metabolism is the sum total of all the chemical reactions occurring in a cell u Two major parts of metabolism: v Catabolism Ø Large, more complex
Metabolic collateral vulnerabilities of MTAP-deleted cancers as therapeutic opportunities Keystone on Tumor Metabolism 2017
 Metabolic collateral vulnerabilities of MTAP-deleted cancers as therapeutic opportunities Keystone on Tumor Metabolism 2017 5 9 March 2017 Whistler, Canada The Challenge: Identifying Precision Medicine
Metabolic collateral vulnerabilities of MTAP-deleted cancers as therapeutic opportunities Keystone on Tumor Metabolism 2017 5 9 March 2017 Whistler, Canada The Challenge: Identifying Precision Medicine
Peptide libraries: applications, design options and considerations. Laura Geuss, PhD May 5, 2015, 2:00-3:00 pm EST
 Peptide libraries: applications, design options and considerations Laura Geuss, PhD May 5, 2015, 2:00-3:00 pm EST Overview 1 2 3 4 5 Introduction Peptide library basics Peptide library design considerations
Peptide libraries: applications, design options and considerations Laura Geuss, PhD May 5, 2015, 2:00-3:00 pm EST Overview 1 2 3 4 5 Introduction Peptide library basics Peptide library design considerations
BIOCHEMISTRY Nucleic Acids
 BIOCHEMISTRY Nucleic Acids BIOB111 CHEMISTRY & BIOCHEMISTRY Session 17 Session Plan Types of Nucleic Acids Nucleosides Nucleotides Primary Structure of Nucleic Acids DNA Double Helix DNA Replication Types
BIOCHEMISTRY Nucleic Acids BIOB111 CHEMISTRY & BIOCHEMISTRY Session 17 Session Plan Types of Nucleic Acids Nucleosides Nucleotides Primary Structure of Nucleic Acids DNA Double Helix DNA Replication Types
Chapter Twelve Protein Synthesis: Translation of the Genetic Message
 Mary K. Campbell Shawn O. Farrell international.cengage.com/ Chapter Twelve Protein Synthesis: Translation of the Genetic Message Paul D. Adams University of Arkansas 1 Translating the Genetic Message
Mary K. Campbell Shawn O. Farrell international.cengage.com/ Chapter Twelve Protein Synthesis: Translation of the Genetic Message Paul D. Adams University of Arkansas 1 Translating the Genetic Message
Ch 10 Molecular Biology of the Gene
 Ch 10 Molecular Biology of the Gene For Next Week Lab -Hand in questions from 4 and 5 by TUES in my mailbox (Biology Office) -Do questions for Lab 6 for next week -Lab practical next week Lecture Read
Ch 10 Molecular Biology of the Gene For Next Week Lab -Hand in questions from 4 and 5 by TUES in my mailbox (Biology Office) -Do questions for Lab 6 for next week -Lab practical next week Lecture Read
Natural Products and Drug Discovery
 Natural Products and Drug Discovery Secondary metabolism Building blocks & Phytochemicals Drug discovery 26-Oct-18 2 Secondary metabolism: Biochemical reactions derived from primary metabolic pathways
Natural Products and Drug Discovery Secondary metabolism Building blocks & Phytochemicals Drug discovery 26-Oct-18 2 Secondary metabolism: Biochemical reactions derived from primary metabolic pathways
Fluorescent Nucleotides: A powerful toolbox for labeling of biological macromolecules
 Fluorescent Nucleotides: A powerful toolbox for labeling of biological macromolecules July 5 th 8 th 2012 Jena Bioscience GmbH Loebstedter Str. 80 07749 Jena, Germany Tel.: +49-3641-628-5000 Fax: +49-3641-628-5100
Fluorescent Nucleotides: A powerful toolbox for labeling of biological macromolecules July 5 th 8 th 2012 Jena Bioscience GmbH Loebstedter Str. 80 07749 Jena, Germany Tel.: +49-3641-628-5000 Fax: +49-3641-628-5100
Dr. R. Sankar, BSE 631 (2018)
 Pauling, Corey and Branson Diffraction of DNA http://www.nature.com/scitable/topicpage/dna-is-a-structure-that-encodes-biological-6493050 In short, stereochemistry is important in determining which helices
Pauling, Corey and Branson Diffraction of DNA http://www.nature.com/scitable/topicpage/dna-is-a-structure-that-encodes-biological-6493050 In short, stereochemistry is important in determining which helices
RNA synthesis/transcription I Biochemistry 302. February 6, 2004 Bob Kelm
 RNA synthesis/transcription I Biochemistry 302 February 6, 2004 Bob Kelm Overview of RNA classes Messenger RNA (mrna) Encodes protein Relatively short half-life ( 3 min in E. coli, 30 min in eukaryotic
RNA synthesis/transcription I Biochemistry 302 February 6, 2004 Bob Kelm Overview of RNA classes Messenger RNA (mrna) Encodes protein Relatively short half-life ( 3 min in E. coli, 30 min in eukaryotic
Experimental Design. Margaret A. Daugherty. Fall Michaelis Menton Kinetics and Inhibition. Lecture 14: Enzymes & Kinetics III E + S ES E + P
 Lecture 14: Enzymes & Kinetics III Michaelis Menton Kinetics and Inhibition Margaret A. Daugherty Fall 2003 Experimental Design k 1 k cat E + S ES E + P k -1 I want to measure the reactivity of my enzyme
Lecture 14: Enzymes & Kinetics III Michaelis Menton Kinetics and Inhibition Margaret A. Daugherty Fall 2003 Experimental Design k 1 k cat E + S ES E + P k -1 I want to measure the reactivity of my enzyme
SYNTHESIS OF UNNATURAL TRYPTOPHAN ANALOGS TO INVESTIGATE THE MOLECULAR RECOGNITION OF METHYLATED LYSINE BY THE HP1 CHROMODOMAIN
 SYNTHESIS OF UNNATURAL TRYPTOPHAN ANALOGS TO INVESTIGATE THE MOLECULAR RECOGNITION OF METHYLATED LYSINE BY THE HP1 CHROMODOMAIN M. Kaitlyn Tsai Honors Thesis May 2016 ABSTRACT DNA is wrapped around a histone
SYNTHESIS OF UNNATURAL TRYPTOPHAN ANALOGS TO INVESTIGATE THE MOLECULAR RECOGNITION OF METHYLATED LYSINE BY THE HP1 CHROMODOMAIN M. Kaitlyn Tsai Honors Thesis May 2016 ABSTRACT DNA is wrapped around a histone
Model Peptides Reveal Specificity of IV -Acetyltransferase from Saccharomyces cerevisiae*
 THE JOURNAL OF B~OLOCICAL CHEMISTRY 0 1990 by The American Society for Biochemistry and Molecular Biology, Inc. Vol. 265, No. 20, Issue of July 15, pp. 11576-11580,19!30 Printed in U.S. A. Model Peptides
THE JOURNAL OF B~OLOCICAL CHEMISTRY 0 1990 by The American Society for Biochemistry and Molecular Biology, Inc. Vol. 265, No. 20, Issue of July 15, pp. 11576-11580,19!30 Printed in U.S. A. Model Peptides
Highlights Translation I
 Highlights Translation I 1. Prokaryotic ribosomes are a 70S multi-protein, multi-rna complex, consisting of a large 50S subunit (31 proteins + 23S rrna + 5S rrna) and a smaller 30S subunit (21 proteins
Highlights Translation I 1. Prokaryotic ribosomes are a 70S multi-protein, multi-rna complex, consisting of a large 50S subunit (31 proteins + 23S rrna + 5S rrna) and a smaller 30S subunit (21 proteins
CHAPTER 8 Nucleotides and Nucleic Acids
 CHAPTER 8 Nucleotides and Nucleic Acids Key topics Biological function of nucleotides and nucleic acids Structures of common nucleotides Structure of double-stranded DNA Structures of ribonucleic acids
CHAPTER 8 Nucleotides and Nucleic Acids Key topics Biological function of nucleotides and nucleic acids Structures of common nucleotides Structure of double-stranded DNA Structures of ribonucleic acids
Four levels of protein Structure
 Proteins (polypeptides) Four levels of protein Structure Primary Structure (1 structure): Secondary Structure (2 structure): Tertiary Structure (3 structure): Quaternary Structure (4 structure): Proteins
Proteins (polypeptides) Four levels of protein Structure Primary Structure (1 structure): Secondary Structure (2 structure): Tertiary Structure (3 structure): Quaternary Structure (4 structure): Proteins
CHEM 4420 Exam I Spring 2013 Page 1 of 6
 CHEM 4420 Exam I Spring 2013 Page 1 of 6 Name Use complete sentences when requested. There are 100 possible points on this exam. The multiple choice questions are worth 2 points each. All other questions
CHEM 4420 Exam I Spring 2013 Page 1 of 6 Name Use complete sentences when requested. There are 100 possible points on this exam. The multiple choice questions are worth 2 points each. All other questions
Chemistry 5.07 Problem Set #3 - Enzyme catalysis and kinetics
 Chemistry 5.07 Problem Set #3 - Enzyme catalysis and kinetics Problem 1. Interleukin 1 β (IL-1 β) has been implicated in the pathogenesis of acute chronic inflammatory diseases including septic shock,
Chemistry 5.07 Problem Set #3 - Enzyme catalysis and kinetics Problem 1. Interleukin 1 β (IL-1 β) has been implicated in the pathogenesis of acute chronic inflammatory diseases including septic shock,
A Novel dcmp Methylase by Engineering Thymidylate Synthase
 Biochemistry 1997, 36, 15909-15917 15909 A Novel dcmp Methylase by Engineering Thymidylate Synthase Sanjay Agarwalla, Sherry LaPorte, Lu Liu, Janet Finer-Moore, Robert M. Stroud, and Daniel V. Santi* Department
Biochemistry 1997, 36, 15909-15917 15909 A Novel dcmp Methylase by Engineering Thymidylate Synthase Sanjay Agarwalla, Sherry LaPorte, Lu Liu, Janet Finer-Moore, Robert M. Stroud, and Daniel V. Santi* Department
Lecture 7: 9/7. CHAPTER 7 Kinetics and Regulation
 Lecture 7: 9/7 CHAPTER 7 Kinetics and Regulation Chapter 7 Outline The rate or velocity of an enzymatic reaction Consider a simple reaction: The velocity or rate of the reaction is determined by measuring
Lecture 7: 9/7 CHAPTER 7 Kinetics and Regulation Chapter 7 Outline The rate or velocity of an enzymatic reaction Consider a simple reaction: The velocity or rate of the reaction is determined by measuring
