Methoden zur Analyse von Transkriptionsfaktoren. Seminar: BCII, Lausen
|
|
|
- Roland Lindsey
- 5 years ago
- Views:
Transcription
1 Methoden zur Analyse von Transkriptionsfaktoren Seminar: BCII, Lausen
2 Gene expression: from transcription to translation Orphanides G, Reinberg D.Cell Feb 22;108(4):
3 Schematic of a gene regulatory region Promoter - Core promoter - Proximal promoter elements Distal Regulatory Elements - Silencer - Enhancer - Insulator - Locus control region
4 Gene regulation is more complex in higher organisms Yeast 300 TF 6000 Genes Drosophila 1000 TF Genes C.elegans 1000 TF Genes Human 3000 TF Genes Yeast Enhancer are rare Drosophila Several Enhancers/Gene Levine and Tijan, Nature 424, 2003
5 The eukaryotic transcriptional machinery Core Promoter - TATA-box - TFIID complex binds to the TATA box - TSS, Transcription start site PIC, Transcription preinitiation complex - General Transcription Factors (GTF) - The PIC allows only basic transcription Activator (or Repressor) - DNA-binding protein with an AD - Greatly enhances transcription Coactivator (Corepressor) - Has no direct DNA-binding activity
6 Core promoter elements BRE: TFIIB-recognition element (25% of all promoters) TATA: TBP (TFIID) complex binding (15%) Inr: Initiator element (50%) MTE: Motif Ten Element (?) DPE: Downstream Promoter Element (25 %) DCE: Downstream Core Element (?) 25% of all promoters do not have a TATA, Inr, DPE or BRE site
7 Distal transcriptional regulatory elements Enhancers - Increase transcription - Cluster of TFBS - Function independent of their orientation and position - Act cell type specific - DNA-looping model Silencer - Binding sites for repressors Insulator - Can block enhancer promoter communication - Can prevent the spread of repressive chromatin Locus Control Region - Group of regulatory elements - Regulate a single gene or a gene cluster
8
9
10 The ENCODE (ENCyclopedia Of DNA Elements) Project analyses genes and regulatory elements 30 Mb of the human genome (1%) have been selected - regions of up to 2 Mb - presents of characterized genes and elements - availability of comparative sequence data - random choice (50%) The ENCODE members focus on the choosen genomic regions - compare large scale data - focus on a number of cell lines - sequence the ENCODE region of other species - publish results in a comprehensive way 2003 focus on the ENCODE region Develop new methods Now, whole genome analysis!!
11 Functional genomic elements being identified by the ENCODE project Methods used
12
13 Gene specific transcription factors Expressed in the cell where the target gene is expressed They recognize a specific DNA sequence Are often activated by signalling: e.g. phosphorylation, hormones Have or recruit chromatin modifying function: e.g. acetylation Are regulated by: Ligands, Modification, Concentration, DNA-Affinity, Cofactors, Dimerisation
14 The RUNX1 is a cell type specific transcription factor important for hematopoietic stem cell development A diagram of RUNX1 protein with functional domains. RD: Runt domain, AD: transcription activation domain, TE1, TE2 and TE3: subelements within AD, ID: transcription inhibition domain; NLS: nuclear localization signal, NRDBn and NRDBc: negative regulatory region for DNA binding in N-terminal (n) and C-terminal (c). NRHn and NRHc: negative regulatory region for heterodimerization in N-terminal (n) and C-terminal (c). NMTS: nuclear matrix targeting signal. Ito Y. Oncogene May 24;23(24):
15 Identification and Characterisation of Regulatory Elements
16 Identification of regulatory sequences by homology mapping High homology between organisms in coding regions Regulatory Sequences are conserved => Enhancer
17 ACCCTXGXXCCXTGAATTGCAGAXCTGTTGTGTTGGTTTXTGACCATCTGCCCATTCTTCCTGTTATGACAXAGCTTGT CANNTG Conserved E-Box
18 Distribution of binding sites across the genome Jothi et al. Nucleic Acids Research 36(16) , 2008
19 Reporter Gene Assay for the Analysis of Regulatory Elements and Transcription Factor Activity
20 Enhancer/Silencer TF: Transcription factor TF TATA TF binding site TATA TF binding site Enhancer/Silencer Exon1 Intron Luciferase GFP LacZ CAT Exon2
21 Transfection of Reporter Plasmid and TF-Expression Plasmid Transient luciferase reportergene assay TF is expresssed TF activates reporter Reporter gene is expressed Cell extract Measurement of luciferase activity
22 Luciferase reporter assay identifies a transcription factor binding site TATA TF Bindestelle Luciferase Relative Luciferase Activity + TF TATA Luciferase +++ TF TATA Luciferase +
23 Analysis of transcription factor functions by luciferase reportergene assay (examples) TATA Luciferase TATA Luciferase Deletion R=>A TATA Luciferase Mutation CoF TATA Luciferase Co-Expression
24 LacZ reporter is driven by an identified regulatory element that targets LacZ expression to specific tissues Kappel A, Rönicke V, Damert A, Flamme I, Risau W, Breier G. Blood Jun 15;93(12):
25 Analysis of Transcription Factor Binding to DNA
26 Oligopulldown biotinylated oligo lysate TF TF Strp pcdna3 Tal1 Ε47 Ε47 + Tal1 α-flag streptavidin beads SDS-PAGE Ab against TF
27 Bandshift Assay, Gelshift EMSA: Electrophoretic mobility shift assay Aims Sequence spezific binding of TF Identification of binding sequence for a TF Identification of a TF that binds to a sequence Characterisation of TF Material Native Polyacrylamid Gel Radioactiv labeled oligonucleotid Nuclear extracts oder purified TF (Antibody)
28 Examples for the use of the EMSA method super shift shift Competition with cold Oligo Lausen et al. Nucleic Acids Research 28(2) , 2000
29 Chromatin Immunoprecipitation Analysis of in vivo TF binding on DNA Binding to a known region of genomic DNA PCR Binding to an unknown DNA sequence Cloning DNA-Array ChIP-sequencing
30 Direct cloning Sequencing Massie CE, Mills IG. ChIPping away at gene regulation. EMBO Rep Apr;9(4):
31 Detection of transcription factor binding to a known genomic region by ChIP PCR Primer P Primer US 1. Tube with Input DNA 2. Tube with DNA ChIPd with control-antibody 3. Tube with DNA ChIPd with TF-Antibody Enhancer/Silencer TF Enhancer/Silencer TATA Exon1 Intron Exon2 Primer Pair US Primer Pair P
32 ChIP on Chip Bioinformatic Bulyka, Current Opinion in Biotechnology Volume 17, Issue 4,
Chapter 17 Lecture. Concepts of Genetics. Tenth Edition. Regulation of Gene Expression in Eukaryotes
 Chapter 17 Lecture Concepts of Genetics Tenth Edition Regulation of Gene Expression in Eukaryotes Chapter Contents 17.1 Eukaryotic Gene Regulation Can Occur at Any of the Steps Leading from DNA to Protein
Chapter 17 Lecture Concepts of Genetics Tenth Edition Regulation of Gene Expression in Eukaryotes Chapter Contents 17.1 Eukaryotic Gene Regulation Can Occur at Any of the Steps Leading from DNA to Protein
Enhancers. Activators and repressors of transcription
 Enhancers Can be >50 kb away from the gene they regulate. Can be upstream from a promoter, downstream from a promoter, within an intron, or even downstream of the final exon of a gene. Are often cell type
Enhancers Can be >50 kb away from the gene they regulate. Can be upstream from a promoter, downstream from a promoter, within an intron, or even downstream of the final exon of a gene. Are often cell type
Transcriptional Regulation in Eukaryotes
 Transcriptional Regulation in Eukaryotes Concepts, Strategies, and Techniques Michael Carey Stephen T. Smale COLD SPRING HARBOR LABORATORY PRESS NEW YORK 2000 Cold Spring Harbor Laboratory Press, 0-87969-537-4
Transcriptional Regulation in Eukaryotes Concepts, Strategies, and Techniques Michael Carey Stephen T. Smale COLD SPRING HARBOR LABORATORY PRESS NEW YORK 2000 Cold Spring Harbor Laboratory Press, 0-87969-537-4
Transcriptional Regulation (Gene Regulation)
 Experimental Techniques in Biomedical Sciences 의생명과학실험기법 Transcriptional Regulation (Gene Regulation) 4/17/13 Jeong Hoon Kim (jeongkim@skku.edu) Department of Health Sciences and Technology, SKKU Graduate
Experimental Techniques in Biomedical Sciences 의생명과학실험기법 Transcriptional Regulation (Gene Regulation) 4/17/13 Jeong Hoon Kim (jeongkim@skku.edu) Department of Health Sciences and Technology, SKKU Graduate
Complex Transcription Machinery
 Complex Transcription Machinery Subunits of the Basal Txn Apparatus Stepwise Assembly of the Pre-initiation Complex Interplay of Activators, Co-regulators and RNA Polymerase at the Promoter Divide and
Complex Transcription Machinery Subunits of the Basal Txn Apparatus Stepwise Assembly of the Pre-initiation Complex Interplay of Activators, Co-regulators and RNA Polymerase at the Promoter Divide and
Chromatographic Separation of the three forms of RNA Polymerase II.
 Chromatographic Separation of the three forms of RNA Polymerase II. α-amanitin α-amanitin bound to Pol II Function of the three enzymes. Yeast Pol II. RNA Polymerase Subunit Structures 10-7 Subunit structure.
Chromatographic Separation of the three forms of RNA Polymerase II. α-amanitin α-amanitin bound to Pol II Function of the three enzymes. Yeast Pol II. RNA Polymerase Subunit Structures 10-7 Subunit structure.
Eukaryotic Transcription
 Eukaryotic Transcription I. Differences between eukaryotic versus prokaryotic transcription. II. (core vs holoenzyme): RNA polymerase II - Promotor elements. - General Pol II transcription factors (GTF).
Eukaryotic Transcription I. Differences between eukaryotic versus prokaryotic transcription. II. (core vs holoenzyme): RNA polymerase II - Promotor elements. - General Pol II transcription factors (GTF).
Transcription Regulation And Gene Expression in Eukaryotes FS 2016 Graduate Course G2 P Matthias and RG Clerc Pharmazentrum Hörsaal 2 16h15-18h00
 Transcription Regulation And Gene Expression in Eukaryotes FS 2016 Graduate Course G2 P Matthias and RG Clerc Pharmazentrum Hörsaal 2 16h15-18h00 The general problem RG Clerc March 2, 2016 RNA Transcription
Transcription Regulation And Gene Expression in Eukaryotes FS 2016 Graduate Course G2 P Matthias and RG Clerc Pharmazentrum Hörsaal 2 16h15-18h00 The general problem RG Clerc March 2, 2016 RNA Transcription
Differential Gene Expression
 Biology 4361 Developmental Biology Differential Gene Expression September 28, 2006 Chromatin Structure ~140 bp ~60 bp Transcriptional Regulation: 1. Packing prevents access CH 3 2. Acetylation ( C O )
Biology 4361 Developmental Biology Differential Gene Expression September 28, 2006 Chromatin Structure ~140 bp ~60 bp Transcriptional Regulation: 1. Packing prevents access CH 3 2. Acetylation ( C O )
CONTENTS. LIST OF FIGURES... i. LIST OF TABLES... iii. SUMMARY OF THE WORK DONE... vi
 CONTENTS SECTION ~ LIST OF FIGURES... i LIST OF TABLES... iii LIST OF ABBREVIATIONS USED... ~... iv SUMMARY OF THE WORK DONE... vi 1. INTROD UCTION... :.................................... 1 1.1. AN OVERVIEW
CONTENTS SECTION ~ LIST OF FIGURES... i LIST OF TABLES... iii LIST OF ABBREVIATIONS USED... ~... iv SUMMARY OF THE WORK DONE... vi 1. INTROD UCTION... :.................................... 1 1.1. AN OVERVIEW
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.
 Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
Chapter 14 Regulation of Transcription
 Chapter 14 Regulation of Transcription Cis-acting sequences Distance-independent cis-acting elements Dissecting regulatory elements Transcription factors Overview transcriptional regulation Transcription
Chapter 14 Regulation of Transcription Cis-acting sequences Distance-independent cis-acting elements Dissecting regulatory elements Transcription factors Overview transcriptional regulation Transcription
Transcription Eukaryotic Cells
 Transcription Eukaryotic Cells Packet #20 1 Introduction Transcription is the process in which genetic information, stored in a strand of DNA (gene), is copied into a strand of RNA. Protein-encoding genes
Transcription Eukaryotic Cells Packet #20 1 Introduction Transcription is the process in which genetic information, stored in a strand of DNA (gene), is copied into a strand of RNA. Protein-encoding genes
Eukaryotic Gene Expression Prof. P. N. Rangarajan Department of Biochemistry Indian Institute of Science, Bangalore
 Eukaryotic Gene Expression Prof. P. N. Rangarajan Department of Biochemistry Indian Institute of Science, Bangalore Module No. # 01 Lecture No. # 04 Gene Regulation in Eukaryotes: Proximal and Distal Promoter
Eukaryotic Gene Expression Prof. P. N. Rangarajan Department of Biochemistry Indian Institute of Science, Bangalore Module No. # 01 Lecture No. # 04 Gene Regulation in Eukaryotes: Proximal and Distal Promoter
Supplementary data. sienigma. F-Enigma F-EnigmaSM. a-p53
 Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Molecular Biology (BIOL 4320) Exam #1 March 12, 2002
 Molecular Biology (BIOL 4320) Exam #1 March 12, 2002 Name KEY SS# This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses after the question number.
Molecular Biology (BIOL 4320) Exam #1 March 12, 2002 Name KEY SS# This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses after the question number.
Finding Eukaryotic Genes Computationally
 Gene Identification Finding Eukaryotic Genes Computationally ü Content-based Methods GC content, hexamer repeats, composition statistics, codon frequencies ü Site-based Methods donor sites, acceptor sites,
Gene Identification Finding Eukaryotic Genes Computationally ü Content-based Methods GC content, hexamer repeats, composition statistics, codon frequencies ü Site-based Methods donor sites, acceptor sites,
CELL BIOLOGY - CLUTCH CH. 7 - GENE EXPRESSION.
 !! www.clutchprep.com CONCEPT: CONTROL OF GENE EXPRESSION BASICS Gene expression is the process through which cells selectively to express some genes and not others Every cell in an organism is a clone
!! www.clutchprep.com CONCEPT: CONTROL OF GENE EXPRESSION BASICS Gene expression is the process through which cells selectively to express some genes and not others Every cell in an organism is a clone
Differential Gene Expression
 Biology 4361 Developmental Biology Differential Gene Expression June 19, 2008 Differential Gene Expression Overview Chromatin structure Gene anatomy RNA processing and protein production Initiating transcription:
Biology 4361 Developmental Biology Differential Gene Expression June 19, 2008 Differential Gene Expression Overview Chromatin structure Gene anatomy RNA processing and protein production Initiating transcription:
transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected with the wwp-luc reporter, and FLAG-tagged FHL1,
 Supplementary Data Supplementary Figure Legends Supplementary Figure 1 FHL-mediated TGFβ-responsive reporter transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected
Supplementary Data Supplementary Figure Legends Supplementary Figure 1 FHL-mediated TGFβ-responsive reporter transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected
Gene expression analysis. Biosciences 741: Genomics Fall, 2013 Week 5. Gene expression analysis
 Gene expression analysis Biosciences 741: Genomics Fall, 2013 Week 5 Gene expression analysis From EST clusters to spotted cdna microarrays Long vs. short oligonucleotide microarrays vs. RT-PCR Methods
Gene expression analysis Biosciences 741: Genomics Fall, 2013 Week 5 Gene expression analysis From EST clusters to spotted cdna microarrays Long vs. short oligonucleotide microarrays vs. RT-PCR Methods
Activation of a Floral Homeotic Gene in Arabidopsis
 Activation of a Floral Homeotic Gene in Arabidopsis By Maximiliam A. Busch, Kirsten Bomblies, and Detlef Weigel Presentation by Lis Garrett and Andrea Stevenson http://ucsdnews.ucsd.edu/archive/graphics/images/image5.jpg
Activation of a Floral Homeotic Gene in Arabidopsis By Maximiliam A. Busch, Kirsten Bomblies, and Detlef Weigel Presentation by Lis Garrett and Andrea Stevenson http://ucsdnews.ucsd.edu/archive/graphics/images/image5.jpg
Eukaryotic & Prokaryotic Transcription. RNA polymerases
 Eukaryotic & Prokaryotic Transcription RNA polymerases RNA Polymerases A. E. coli RNA polymerase 1. core enzyme = ββ'(α)2 has catalytic activity but cannot recognize start site of transcription ~500,000
Eukaryotic & Prokaryotic Transcription RNA polymerases RNA Polymerases A. E. coli RNA polymerase 1. core enzyme = ββ'(α)2 has catalytic activity but cannot recognize start site of transcription ~500,000
Lecture 5: Regulation
 Machine Learning in Computational Biology CSC 2431 Lecture 5: Regulation Instructor: Anna Goldenberg Central Dogma of Biology Transcription DNA RNA protein Process of producing RNA from DNA Constitutive
Machine Learning in Computational Biology CSC 2431 Lecture 5: Regulation Instructor: Anna Goldenberg Central Dogma of Biology Transcription DNA RNA protein Process of producing RNA from DNA Constitutive
Differential Gene Expression
 Developmental Biology Biology 4361 Differential Gene Expression October 13, 2005 core transcription initiation site 5 promoter 3 TATAT +1 upstream downstream Basal transcription factors (eukaryotes) TFIID
Developmental Biology Biology 4361 Differential Gene Expression October 13, 2005 core transcription initiation site 5 promoter 3 TATAT +1 upstream downstream Basal transcription factors (eukaryotes) TFIID
Another groups asks with what does Gal4- VP16 interact?
 Another groups asks with what does Gal4- VP16 interact? DNA attached to agarose beads Now add components in steps Ask if the recruitment of a component requires that Gal4-VP16 be present. Primer extension
Another groups asks with what does Gal4- VP16 interact? DNA attached to agarose beads Now add components in steps Ask if the recruitment of a component requires that Gal4-VP16 be present. Primer extension
GENETICS - CLUTCH CH.10 TRANSCRIPTION.
 !! www.clutchprep.com CONCEPT: OVERVIEW OF TRANSCRIPTION Transcription is the process of using DNA as a template to RNA RNA polymerase is the enzyme that transcribes DNA - There are many different types
!! www.clutchprep.com CONCEPT: OVERVIEW OF TRANSCRIPTION Transcription is the process of using DNA as a template to RNA RNA polymerase is the enzyme that transcribes DNA - There are many different types
Nature Genetics: doi: /ng Supplementary Figure 1. ChIP-seq genome browser views of BRM occupancy at previously identified BRM targets.
 Supplementary Figure 1 ChIP-seq genome browser views of BRM occupancy at previously identified BRM targets. Gene structures are shown underneath each panel. Supplementary Figure 2 pref6::ref6-gfp complements
Supplementary Figure 1 ChIP-seq genome browser views of BRM occupancy at previously identified BRM targets. Gene structures are shown underneath each panel. Supplementary Figure 2 pref6::ref6-gfp complements
nature methods A paired-end sequencing strategy to map the complex landscape of transcription initiation
 nature methods A paired-end sequencing strategy to map the complex landscape of transcription initiation Ting Ni, David L Corcoran, Elizabeth A Rach, Shen Song, Eric P Spana, Yuan Gao, Uwe Ohler & Jun
nature methods A paired-end sequencing strategy to map the complex landscape of transcription initiation Ting Ni, David L Corcoran, Elizabeth A Rach, Shen Song, Eric P Spana, Yuan Gao, Uwe Ohler & Jun
Motivation From Protein to Gene
 MOLECULAR BIOLOGY 2003-4 Topic B Recombinant DNA -principles and tools Construct a library - what for, how Major techniques +principles Bioinformatics - in brief Chapter 7 (MCB) 1 Motivation From Protein
MOLECULAR BIOLOGY 2003-4 Topic B Recombinant DNA -principles and tools Construct a library - what for, how Major techniques +principles Bioinformatics - in brief Chapter 7 (MCB) 1 Motivation From Protein
Control of Eukaryotic Gene Expression (Learning Objectives)
 Control of Eukaryotic Gene Expression (Learning Objectives) 1. Compare and contrast chromatin and chromosome: composition, proteins involved and level of packing. Explain the structure and function of
Control of Eukaryotic Gene Expression (Learning Objectives) 1. Compare and contrast chromatin and chromosome: composition, proteins involved and level of packing. Explain the structure and function of
The RNA Polymerase II General Transcription Machinery Prof. Michael Hampsey
 The RNA Polymerase II General Transcription Machinery Michael Hampsey, PhD Robert Wood Johnson Medical School Piscataway, New Jersey 1 Central Dogma of Molecular Biology DNA Stores genetic information
The RNA Polymerase II General Transcription Machinery Michael Hampsey, PhD Robert Wood Johnson Medical School Piscataway, New Jersey 1 Central Dogma of Molecular Biology DNA Stores genetic information
However, only a fraction of these genes are transcribed in an individual cell at any given time.
 All cells in an organism contain the same set of genes. However, only a fraction of these genes are transcribed in an individual cell at any given time. It is the pattern of gene expression that determines
All cells in an organism contain the same set of genes. However, only a fraction of these genes are transcribed in an individual cell at any given time. It is the pattern of gene expression that determines
Transcriptomics. Marta Puig Institut de Biotecnologia i Biomedicina Universitat Autònoma de Barcelona
 Transcriptomics Marta Puig Institut de Biotecnologia i Biomedicina Universitat Autònoma de Barcelona Central dogma of molecular biology Central dogma of molecular biology Genome Complete DNA content of
Transcriptomics Marta Puig Institut de Biotecnologia i Biomedicina Universitat Autònoma de Barcelona Central dogma of molecular biology Central dogma of molecular biology Genome Complete DNA content of
Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and
 Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
17.5 Eukaryotic Transcription Initiation Is Regulated by Transcription Factors That Bind to Cis-Acting Sites
 17.5 Eukaryotic Transcription Initiation Is Regulated by Transcription Factors That Bind to Cis-Acting Sites 1 Section 17.5 Transcription regulatory proteins, transcription factors, target cis-acting sites
17.5 Eukaryotic Transcription Initiation Is Regulated by Transcription Factors That Bind to Cis-Acting Sites 1 Section 17.5 Transcription regulatory proteins, transcription factors, target cis-acting sites
Chapter 25: Regulating Eukaryotic Transcription The Ligand Responsive Activators
 Chapter 25: Regulating Eukaryotic Transcription The Ligand Responsive Activators At least 5 potential gene expression control points Superfamily of Gene Regulators Activation of gene structure Initiation
Chapter 25: Regulating Eukaryotic Transcription The Ligand Responsive Activators At least 5 potential gene expression control points Superfamily of Gene Regulators Activation of gene structure Initiation
Structure/function relationship in DNA-binding proteins
 PHRM 836 September 22, 2015 Structure/function relationship in DNA-binding proteins Devlin Chapter 8.8-9 u General description of transcription factors (TFs) u Sequence-specific interactions between DNA
PHRM 836 September 22, 2015 Structure/function relationship in DNA-binding proteins Devlin Chapter 8.8-9 u General description of transcription factors (TFs) u Sequence-specific interactions between DNA
Transcriptional Regulation
 Experimental Techniques in Biomedical Sciences 의생명과학실험기법 Transcriptional Regulation (Gene Regulation) 3/14/12 Jeong Hoon Kim (jeongkim@skku.edu) Department of Health Sciences and Technology, SKKU Graduate
Experimental Techniques in Biomedical Sciences 의생명과학실험기법 Transcriptional Regulation (Gene Regulation) 3/14/12 Jeong Hoon Kim (jeongkim@skku.edu) Department of Health Sciences and Technology, SKKU Graduate
Binding of the Thyroid Hormone Receptor to a Negative Element in the Basal Growth Hormone Promoter Is Associated with Histone Acetylation*
 THE JOURNAL OF BIOLOGICAL CHEMISTRY Vol. 278, No. 41, Issue of October 10, pp. 39383 39391, 2003 2003 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in U.S.A. Binding of the
THE JOURNAL OF BIOLOGICAL CHEMISTRY Vol. 278, No. 41, Issue of October 10, pp. 39383 39391, 2003 2003 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in U.S.A. Binding of the
30 Gene expression: Transcription
 30 Gene expression: Transcription Gene structure. o Exons coding region of DNA. o Introns non-coding region of DNA. o Introns are interspersed between exons of a single gene. o Promoter region helps enzymes
30 Gene expression: Transcription Gene structure. o Exons coding region of DNA. o Introns non-coding region of DNA. o Introns are interspersed between exons of a single gene. o Promoter region helps enzymes
M1 - Biochemistry. Nucleic Acid Structure II/Transcription I
 M1 - Biochemistry Nucleic Acid Structure II/Transcription I PH Ratz, PhD (Resources: Lehninger et al., 5th ed., Chapters 8, 24 & 26) 1 Nucleic Acid Structure II/Transcription I Learning Objectives: 1.
M1 - Biochemistry Nucleic Acid Structure II/Transcription I PH Ratz, PhD (Resources: Lehninger et al., 5th ed., Chapters 8, 24 & 26) 1 Nucleic Acid Structure II/Transcription I Learning Objectives: 1.
Liu, Yang (2012) The characterization of a novel abscission-related gene in Arabidopsis thaliana. PhD thesis, University of Nottingham.
 Liu, Yang (2012) The characterization of a novel abscission-related gene in Arabidopsis thaliana. PhD thesis, University of Nottingham. Access from the University of Nottingham repository: http://eprints.nottingham.ac.uk/12529/4/thesis_part_3_final.pdf
Liu, Yang (2012) The characterization of a novel abscission-related gene in Arabidopsis thaliana. PhD thesis, University of Nottingham. Access from the University of Nottingham repository: http://eprints.nottingham.ac.uk/12529/4/thesis_part_3_final.pdf
Non-coding Function & Variation, MPRAs II. Mike White Bio /5/18
 Non-coding Function & Variation, MPRAs II Mike White Bio 5488 3/5/18 MPRA Review Problem 1: Where does your CRE DNA come from? DNA synthesis Genomic fragments Targeted regulome capture Problem 2: How do
Non-coding Function & Variation, MPRAs II Mike White Bio 5488 3/5/18 MPRA Review Problem 1: Where does your CRE DNA come from? DNA synthesis Genomic fragments Targeted regulome capture Problem 2: How do
Estradiol-Estrogen Receptor α Mediates the Expression of the CXXC5 Gene through the Estrogen Response Element-Dependent Signaling Pathway
 Estradiol-Estrogen Receptor α Mediates the Expression of the CXXC5 Gene through the Estrogen Response Element-Dependent Signaling Pathway Pelin Yaşar, Gamze Ayaz and Mesut Muyan SUPPLEMENTARY INFORMATION
Estradiol-Estrogen Receptor α Mediates the Expression of the CXXC5 Gene through the Estrogen Response Element-Dependent Signaling Pathway Pelin Yaşar, Gamze Ayaz and Mesut Muyan SUPPLEMENTARY INFORMATION
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11070 Supplementary Figure 1 Purification of FLAG-tagged proteins. a, Purification of FLAG-RNF12 by FLAG-affinity from nuclear extracts of wild-type (WT) and two FLAG- RNF12 transgenic
doi:10.1038/nature11070 Supplementary Figure 1 Purification of FLAG-tagged proteins. a, Purification of FLAG-RNF12 by FLAG-affinity from nuclear extracts of wild-type (WT) and two FLAG- RNF12 transgenic
Bi8 Lecture 19. Review and Practice Questions March 8th 2016
 Bi8 Lecture 19 Review and Practice Questions March 8th 2016 Common Misconceptions Where Words Matter Common Misconceptions DNA vs RNA vs Protein Su(H) vs Su(H) Replication Transcription Transcribed vs
Bi8 Lecture 19 Review and Practice Questions March 8th 2016 Common Misconceptions Where Words Matter Common Misconceptions DNA vs RNA vs Protein Su(H) vs Su(H) Replication Transcription Transcribed vs
Chapter 1. from genomics to proteomics Ⅱ
 Proteomics Chapter 1. from genomics to proteomics Ⅱ 1 Functional genomics Functional genomics: study of relations of genomics to biological functions at systems level However, it cannot explain any more
Proteomics Chapter 1. from genomics to proteomics Ⅱ 1 Functional genomics Functional genomics: study of relations of genomics to biological functions at systems level However, it cannot explain any more
Enhancers mutations that make the original mutant phenotype more extreme. Suppressors mutations that make the original mutant phenotype less extreme
 Interactomics and Proteomics 1. Interactomics The field of interactomics is concerned with interactions between genes or proteins. They can be genetic interactions, in which two genes are involved in the
Interactomics and Proteomics 1. Interactomics The field of interactomics is concerned with interactions between genes or proteins. They can be genetic interactions, in which two genes are involved in the
Transcriptional Regulation
 Experimental Techniques in Biomedical Sciences 의생명과학연구기법 Transcriptional Regulation (Gene Regulation) 4/2/16 Jeong Hoon Kim (jeongkim@skku.edu) edu) Department of Health Sciences and Technology, SKKU Graduate
Experimental Techniques in Biomedical Sciences 의생명과학연구기법 Transcriptional Regulation (Gene Regulation) 4/2/16 Jeong Hoon Kim (jeongkim@skku.edu) edu) Department of Health Sciences and Technology, SKKU Graduate
CHAPTER 13 LECTURE SLIDES
 CHAPTER 13 LECTURE SLIDES Prepared by Brenda Leady University of Toledo To run the animations you must be in Slideshow View. Use the buttons on the animation to play, pause, and turn audio/text on or off.
CHAPTER 13 LECTURE SLIDES Prepared by Brenda Leady University of Toledo To run the animations you must be in Slideshow View. Use the buttons on the animation to play, pause, and turn audio/text on or off.
Nature Genetics: doi: /ng.3556 INTEGRATED SUPPLEMENTARY FIGURE TEMPLATE. Supplementary Figure 1
 INTEGRATED SUPPLEMENTARY FIGURE TEMPLATE Supplementary Figure 1 REF6 expression in transgenic lines. (a,b) Expression of REF6 in REF6-HA ref6 and REF6ΔZnF-HA ref6 plants detected by RT qpcr (a) and immunoblot
INTEGRATED SUPPLEMENTARY FIGURE TEMPLATE Supplementary Figure 1 REF6 expression in transgenic lines. (a,b) Expression of REF6 in REF6-HA ref6 and REF6ΔZnF-HA ref6 plants detected by RT qpcr (a) and immunoblot
Bioinformatics of Transcriptional Regulation
 Bioinformatics of Transcriptional Regulation Carl Herrmann IPMB & DKFZ c.herrmann@dkfz.de Wechselwirkung von Maßnahmen und Auswirkungen Einflussmöglichkeiten in einem Dialog From genes to active compounds
Bioinformatics of Transcriptional Regulation Carl Herrmann IPMB & DKFZ c.herrmann@dkfz.de Wechselwirkung von Maßnahmen und Auswirkungen Einflussmöglichkeiten in einem Dialog From genes to active compounds
CLASS 3.5: 03/29/07 EUKARYOTIC TRANSCRIPTION I: PROMOTERS AND ENHANCERS
 CLASS 3.5: 03/29/07 EUKARYOTIC TRANSCRIPTION I: PROMOTERS AND ENHANCERS A. Promoters and Polymerases (RNA pols): 1. General characteristics - Initiation of transcription requires a. Transcription factors
CLASS 3.5: 03/29/07 EUKARYOTIC TRANSCRIPTION I: PROMOTERS AND ENHANCERS A. Promoters and Polymerases (RNA pols): 1. General characteristics - Initiation of transcription requires a. Transcription factors
Chapter 18: Regulation of Gene Expression. 1. Gene Regulation in Bacteria 2. Gene Regulation in Eukaryotes 3. Gene Regulation & Cancer
 Chapter 18: Regulation of Gene Expression 1. Gene Regulation in Bacteria 2. Gene Regulation in Eukaryotes 3. Gene Regulation & Cancer Gene Regulation Gene regulation refers to all aspects of controlling
Chapter 18: Regulation of Gene Expression 1. Gene Regulation in Bacteria 2. Gene Regulation in Eukaryotes 3. Gene Regulation & Cancer Gene Regulation Gene regulation refers to all aspects of controlling
Practice Exam A. Briefly describe how IL-25 treatment might be able to help this responder subgroup of liver cancer patients.
 Practice Exam 2007 1. A special JAK-STAT signaling system (JAK5-STAT5) was recently identified in which a gene called TS5 becomes selectively transcribed and expressed in the liver upon induction by a
Practice Exam 2007 1. A special JAK-STAT signaling system (JAK5-STAT5) was recently identified in which a gene called TS5 becomes selectively transcribed and expressed in the liver upon induction by a
(c) 2014 Dr. Alice Heicklen & Dr. Deborah Mowshowitz, Columbia University, New York, NY. Last update 02/26/ :57 PM
 C2006/F2402 '14 OUTLINE OF LECTURE #11 (c) 2014 Dr. Alice Heicklen & Dr. Deborah Mowshowitz, Columbia University, New York, NY. Last update 02/26/2014 12:57 PM Handouts: 10C -- Typical Eukaryotic Gene,
C2006/F2402 '14 OUTLINE OF LECTURE #11 (c) 2014 Dr. Alice Heicklen & Dr. Deborah Mowshowitz, Columbia University, New York, NY. Last update 02/26/2014 12:57 PM Handouts: 10C -- Typical Eukaryotic Gene,
Gene Regulation Biology
 Gene Regulation Biology Potential and Limitations of Cell Re-programming in Cancer Research Eric Blanc KCL April 13, 2010 Eric Blanc (KCL) Gene Regulation Biology April 13, 2010 1 / 21 Outline 1 The Central
Gene Regulation Biology Potential and Limitations of Cell Re-programming in Cancer Research Eric Blanc KCL April 13, 2010 Eric Blanc (KCL) Gene Regulation Biology April 13, 2010 1 / 21 Outline 1 The Central
- all cells express large number of the same genes - housekeeping or common genes - many cells also express cell-type-specific genes
 Developmental Biology - Biology 4361 Lecture 8 - Differential Gene Expression October 13, 2005 The principle of genomic equivalence states that all cells in a developing organism have the same genetic
Developmental Biology - Biology 4361 Lecture 8 - Differential Gene Expression October 13, 2005 The principle of genomic equivalence states that all cells in a developing organism have the same genetic
Long-range gene regulation
 Long-range gene regulation Short Course in Medical Genetics Melbourne, June 2011 Question Why should long-range regulation of gene expression be of interest to Clinical Scientists and Pathologists interested
Long-range gene regulation Short Course in Medical Genetics Melbourne, June 2011 Question Why should long-range regulation of gene expression be of interest to Clinical Scientists and Pathologists interested
BS 50 Genetics and Genomics Week of Oct 24
 BS 50 Genetics and Genomics Week of Oct 24 Additional Practice Problems for Section Question 1: The following table contains a list of statements that apply to replication, transcription, both, or neither.
BS 50 Genetics and Genomics Week of Oct 24 Additional Practice Problems for Section Question 1: The following table contains a list of statements that apply to replication, transcription, both, or neither.
Gene regulation V Biochemistry 302. March 6, 2006
 Gene regulation V Biochemistry 302 March 6, 2006 Common structural motifs associated with transcriptional regulatory proteins Helix-turn-helix Prokaryotic repressors and activators Eukaryotic homeodomain
Gene regulation V Biochemistry 302 March 6, 2006 Common structural motifs associated with transcriptional regulatory proteins Helix-turn-helix Prokaryotic repressors and activators Eukaryotic homeodomain
Promoter Architectures and Developmental Gene Regulation
 Promoter Architectures and Developmental Gene Regulation Vanja Haberle a,b,1 and Boris Lenhard a,* a Institute of Clinical Sciences and MRC Clinical Sciences Center, Faculty of Medicine, Imperial College
Promoter Architectures and Developmental Gene Regulation Vanja Haberle a,b,1 and Boris Lenhard a,* a Institute of Clinical Sciences and MRC Clinical Sciences Center, Faculty of Medicine, Imperial College
DNA Transcription. Dr Aliwaini
 DNA Transcription 1 DNA Transcription-Introduction The synthesis of an RNA molecule from DNA is called Transcription. All eukaryotic cells have five major classes of RNA: ribosomal RNA (rrna), messenger
DNA Transcription 1 DNA Transcription-Introduction The synthesis of an RNA molecule from DNA is called Transcription. All eukaryotic cells have five major classes of RNA: ribosomal RNA (rrna), messenger
EpiQuik Methyl-CpG Binding Domain Protein 2 ChIP Kit
 EpiQuik Methyl-CpG Binding Domain Protein 2 ChIP Kit Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Methyl-CpG Binding Domain Protein 2 ChIP Kit is suitable for combining the
EpiQuik Methyl-CpG Binding Domain Protein 2 ChIP Kit Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Methyl-CpG Binding Domain Protein 2 ChIP Kit is suitable for combining the
Online Supplementary Information
 Online Supplementary Information NLRP4 negatively regulates type I interferon signaling by targeting TBK1 for degradation via E3 ubiquitin ligase DTX4 Jun Cui 1,4,6,7, Yinyin Li 1,5,6,7, Liang Zhu 1, Dan
Online Supplementary Information NLRP4 negatively regulates type I interferon signaling by targeting TBK1 for degradation via E3 ubiquitin ligase DTX4 Jun Cui 1,4,6,7, Yinyin Li 1,5,6,7, Liang Zhu 1, Dan
Applied Bioinformatics - Lecture 16: Transcriptomics
 Applied Bioinformatics - Lecture 16: Transcriptomics David Hendrix Oregon State University Feb 15th 2016 Transcriptomics High-throughput Sequencing (deep sequencing) High-throughput sequencing (also
Applied Bioinformatics - Lecture 16: Transcriptomics David Hendrix Oregon State University Feb 15th 2016 Transcriptomics High-throughput Sequencing (deep sequencing) High-throughput sequencing (also
ChampionChIP Quick, High Throughput Chromatin Immunoprecipitation Assay System
 ChampionChIP Quick, High Throughput Chromatin Immunoprecipitation Assay System Liyan Pang, Ph.D. Application Scientist 1 Topics to be Covered Introduction What is ChIP-qPCR? Challenges Facing Biological
ChampionChIP Quick, High Throughput Chromatin Immunoprecipitation Assay System Liyan Pang, Ph.D. Application Scientist 1 Topics to be Covered Introduction What is ChIP-qPCR? Challenges Facing Biological
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Endogenous gene tagging to study subcellular localization and chromatin binding. a, b, Schematic of experimental set-up to endogenously tag RNAi factors using the CRISPR Cas9 technology,
Supplementary Figure 1 Endogenous gene tagging to study subcellular localization and chromatin binding. a, b, Schematic of experimental set-up to endogenously tag RNAi factors using the CRISPR Cas9 technology,
BS1940 Course Topics Fall 2001 Drs. Hatfull and Arndt
 BS1940 Course Topics Fall 2001 Drs. Hatfull and Arndt Introduction to molecular biology Combining genetics, biochemistry, structural chemistry Information flow in biological systems: The Central Dogma
BS1940 Course Topics Fall 2001 Drs. Hatfull and Arndt Introduction to molecular biology Combining genetics, biochemistry, structural chemistry Information flow in biological systems: The Central Dogma
3.1 The Role of the Enzymatic Activity of Sir2 for Efficient. The Sir proteins are recruited to DNA by site-specific DNA-binding proteins and
 35 3 RESULTS 3.1 The Role of the Enzymatic Activity of Sir2 for Efficient Association of the SIR Complex with DNA The Sir proteins are recruited to DNA by site-specific DNA-binding proteins and subsequently
35 3 RESULTS 3.1 The Role of the Enzymatic Activity of Sir2 for Efficient Association of the SIR Complex with DNA The Sir proteins are recruited to DNA by site-specific DNA-binding proteins and subsequently
Contents... vii. List of Figures... xii. List of Tables... xiv. Abbreviatons... xv. Summary... xvii. 1. Introduction In vitro evolution...
 vii Contents Contents... vii List of Figures... xii List of Tables... xiv Abbreviatons... xv Summary... xvii 1. Introduction...1 1.1 In vitro evolution... 1 1.2 Phage Display Technology... 3 1.3 Cell surface
vii Contents Contents... vii List of Figures... xii List of Tables... xiv Abbreviatons... xv Summary... xvii 1. Introduction...1 1.1 In vitro evolution... 1 1.2 Phage Display Technology... 3 1.3 Cell surface
Chapter 3. DNA, RNA, and Protein Synthesis
 Chapter 3. DNA, RNA, and Protein Synthesis 4. Transcription Gene Expression Regulatory region (promoter) 5 flanking region Upstream region Coding region 3 flanking region Downstream region Transcription
Chapter 3. DNA, RNA, and Protein Synthesis 4. Transcription Gene Expression Regulatory region (promoter) 5 flanking region Upstream region Coding region 3 flanking region Downstream region Transcription
Overlap with exam 1 begins
 Exam 2 begins here Overlap with exam 1 begins Another groups asks with what does Gal4- VP16 interact? DNA attached to agarose beads Now add components in steps Ask if the recruitment of a component requires
Exam 2 begins here Overlap with exam 1 begins Another groups asks with what does Gal4- VP16 interact? DNA attached to agarose beads Now add components in steps Ask if the recruitment of a component requires
Document S1. Supplemental Experimental Procedures and Three Figures (see next page)
 Supplemental Data Document S1. Supplemental Experimental Procedures and Three Figures (see next page) Table S1. List of Candidate Genes Identified from the Screen. Candidate genes, corresponding dsrnas
Supplemental Data Document S1. Supplemental Experimental Procedures and Three Figures (see next page) Table S1. List of Candidate Genes Identified from the Screen. Candidate genes, corresponding dsrnas
Section C: The Control of Gene Expression
 Section C: The Control of Gene Expression 1. Each cell of a multicellular eukaryote expresses only a small fraction of its genes 2. The control of gene expression can occur at any step in the pathway from
Section C: The Control of Gene Expression 1. Each cell of a multicellular eukaryote expresses only a small fraction of its genes 2. The control of gene expression can occur at any step in the pathway from
TRANSGENIC ANIMALS. -transient transfection of cells -stable transfection of cells. - Two methods to produce transgenic animals:
 TRANSGENIC ANIMALS -transient transfection of cells -stable transfection of cells - Two methods to produce transgenic animals: 1- DNA microinjection - random insertion 2- embryonic stem cell-mediated gene
TRANSGENIC ANIMALS -transient transfection of cells -stable transfection of cells - Two methods to produce transgenic animals: 1- DNA microinjection - random insertion 2- embryonic stem cell-mediated gene
Engineering splicing factors with designed specificities
 nature methods Engineering splicing factors with designed specificities Yang Wang, Cheom-Gil Cheong, Traci M Tanaka Hall & Zefeng Wang Supplementary figures and text: Supplementary Figure 1 Supplementary
nature methods Engineering splicing factors with designed specificities Yang Wang, Cheom-Gil Cheong, Traci M Tanaka Hall & Zefeng Wang Supplementary figures and text: Supplementary Figure 1 Supplementary
Partial list of differentially expressed genes from cdna microarray, comparing MUC18-
 Supplemental Figure legends Table-1 Partial list of differentially expressed genes from cdna microarray, comparing MUC18- silenced and NT-transduced A375SM cells. Supplemental Figure 1 Effect of MUC-18
Supplemental Figure legends Table-1 Partial list of differentially expressed genes from cdna microarray, comparing MUC18- silenced and NT-transduced A375SM cells. Supplemental Figure 1 Effect of MUC-18
DIAMANTINA INSTITUTE for Cancer, Immunology and Metabolic Medicine
 DIAMANTINA INSTITUTE for Cancer, Immunology and Metabolic Medicine Defining MYB Transcriptional Network by Genome-wide Chromatin Occupancy Profiling (ChIP-Seq) 2010 E.Glazov, L. Zhao Transcription Factors:
DIAMANTINA INSTITUTE for Cancer, Immunology and Metabolic Medicine Defining MYB Transcriptional Network by Genome-wide Chromatin Occupancy Profiling (ChIP-Seq) 2010 E.Glazov, L. Zhao Transcription Factors:
Table S1. Primers used in RT-PCR studies (all in 5 to 3 direction)
 Table S1. Primers used in RT-PCR studies (all in 5 to 3 direction) Epo Fw CTGTATCATGGACCACCTCGG Epo Rw TGAAGCACAGAAGCTCTTCGG Jak2 Fw ATCTGACCTTTCCATCTGGGG Jak2 Rw TGGTTGGGTGGATACCAGATC Stat5A Fw TTACTGAAGATCAAGCTGGGG
Table S1. Primers used in RT-PCR studies (all in 5 to 3 direction) Epo Fw CTGTATCATGGACCACCTCGG Epo Rw TGAAGCACAGAAGCTCTTCGG Jak2 Fw ATCTGACCTTTCCATCTGGGG Jak2 Rw TGGTTGGGTGGATACCAGATC Stat5A Fw TTACTGAAGATCAAGCTGGGG
Computational Biology I LSM5191 (2003/4)
 Computational Biology I LSM5191 (2003/4) Aylwin Ng, D.Phil Lecture Notes: Transcriptome: Molecular Biology of Gene Expression I Flow of information: DNA to polypeptide DNA Start Exon1 Intron Exon2 Termination
Computational Biology I LSM5191 (2003/4) Aylwin Ng, D.Phil Lecture Notes: Transcriptome: Molecular Biology of Gene Expression I Flow of information: DNA to polypeptide DNA Start Exon1 Intron Exon2 Termination
Supplemental Figure legends Figure S1. (A) (B) (C) (D) Figure S2. Figure S3. (A-E) Figure S4. Figure S5. (A, C, E, G, I) (B, D, F, H, Figure S6.
 Supplemental Figure legends Figure S1. Map-based cloning and complementation testing for ZOP1. (A) ZOP1 was mapped to a ~273-kb interval on Chromosome 1. In the interval, a single-nucleotide G to A substitution
Supplemental Figure legends Figure S1. Map-based cloning and complementation testing for ZOP1. (A) ZOP1 was mapped to a ~273-kb interval on Chromosome 1. In the interval, a single-nucleotide G to A substitution
Supplemental Information. Autoregulatory Feedback Controls. Sequential Action of cis-regulatory Modules. at the brinker Locus
 Developmental Cell, Volume 26 Supplemental Information Autoregulatory Feedback Controls Sequential Action of cis-regulatory Modules at the brinker Locus Leslie Dunipace, Abbie Saunders, Hilary L. Ashe,
Developmental Cell, Volume 26 Supplemental Information Autoregulatory Feedback Controls Sequential Action of cis-regulatory Modules at the brinker Locus Leslie Dunipace, Abbie Saunders, Hilary L. Ashe,
32 Gene regulation in Eukaryotes Lecture Outline 11/28/05. Gene Regulation in Prokaryotes and Eukarykotes
 3 Gene regulation in Eukaryotes Lecture Outline /8/05 Gene regulation in eukaryotes Chromatin remodeling More kinds of control elements Promoters, Enhancers, and Silencers Combinatorial control Cell-specific
3 Gene regulation in Eukaryotes Lecture Outline /8/05 Gene regulation in eukaryotes Chromatin remodeling More kinds of control elements Promoters, Enhancers, and Silencers Combinatorial control Cell-specific
Synthetic cells: do bacteria need all its genes? No.
 NO NEED TO REFER TO THE SLIDES. بسم هللا الرحمن الرحيم Do we need all the non coding regions of the DNA? Two weeks ago, they discovered that the genome of a plant is very small (recall that plant genome
NO NEED TO REFER TO THE SLIDES. بسم هللا الرحمن الرحيم Do we need all the non coding regions of the DNA? Two weeks ago, they discovered that the genome of a plant is very small (recall that plant genome
EpiQuik Tissue Methyl-CpG Binding Domain Protein 2 ChIP Kit
 EpiQuik Tissue Methyl-CpG Binding Domain Protein 2 ChIP Kit Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Tissue Methyl-CpG Binding Domain Protein 2 ChIP Kit is suitable for
EpiQuik Tissue Methyl-CpG Binding Domain Protein 2 ChIP Kit Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Tissue Methyl-CpG Binding Domain Protein 2 ChIP Kit is suitable for
We can now identify three major pathways of information flow in the cell (in replication, information passes from one DNA molecule to other DNA
 1 We can now identify three major pathways of information flow in the cell (in replication, information passes from one DNA molecule to other DNA molecules; in transcription, information passes from DNA
1 We can now identify three major pathways of information flow in the cell (in replication, information passes from one DNA molecule to other DNA molecules; in transcription, information passes from DNA
ChIP grade antibodies: selection and validation. Rachel Imoberdorf, PhD Senior Development Scientist
 ChIP grade antibodies: selection and validation Rachel Imoberdorf, PhD Senior Development Scientist Introduction Antibody selection Antibody validation ChIP at www.abcam.com Introduction Chromatin Immunoprecipitation
ChIP grade antibodies: selection and validation Rachel Imoberdorf, PhD Senior Development Scientist Introduction Antibody selection Antibody validation ChIP at www.abcam.com Introduction Chromatin Immunoprecipitation
Introduction. Matıas Montes 1, *, Sandra Moreira-Ramos 1, *, Diego A. Rojas 2, *, Fabiola Urbina 1, Norbert F. K aufer 3 and Edio Maldonado 1
 RNA polymerase II components and Rrn7 form a preinitiation complex on the HomolD box to promote ribosomal protein gene expression in Schizosaccharomyces pombe Matıas Montes 1, *, Sandra Moreira-Ramos 1,
RNA polymerase II components and Rrn7 form a preinitiation complex on the HomolD box to promote ribosomal protein gene expression in Schizosaccharomyces pombe Matıas Montes 1, *, Sandra Moreira-Ramos 1,
Transcription & post transcriptional modification
 Transcription & post transcriptional modification Transcription The synthesis of RNA molecules using DNA strands as the templates so that the genetic information can be transferred from DNA to RNA Similarity
Transcription & post transcriptional modification Transcription The synthesis of RNA molecules using DNA strands as the templates so that the genetic information can be transferred from DNA to RNA Similarity
Supplementary Figure 2
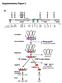 Supplementary Figure 2 a SBS-C1 SBS-C2 SBS-C3 SBS-C4 SBS-C5 SBS-C6 SBS-C7 SBS-C8 SBS-C9 LCR CNS-1 CNS-2 Il5 Rad50 Il13 Il4 Kif3a Sept8 0 50 100 150 200kb Sau3AI 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17
Supplementary Figure 2 a SBS-C1 SBS-C2 SBS-C3 SBS-C4 SBS-C5 SBS-C6 SBS-C7 SBS-C8 SBS-C9 LCR CNS-1 CNS-2 Il5 Rad50 Il13 Il4 Kif3a Sept8 0 50 100 150 200kb Sau3AI 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17
8Br-cAMP was purchased from Sigma (St. Louis, MO). Silencer Negative Control sirna #1 and
 1 Supplemental information 2 3 Materials and Methods 4 Reagents and animals 5 8Br-cAMP was purchased from Sigma (St. Louis, MO). Silencer Negative Control sirna #1 and 6 Silencer Select Pre-designed sirna
1 Supplemental information 2 3 Materials and Methods 4 Reagents and animals 5 8Br-cAMP was purchased from Sigma (St. Louis, MO). Silencer Negative Control sirna #1 and 6 Silencer Select Pre-designed sirna
Comparative Analysis of Argonaute-Dependent Small RNA Pathways in Drosophila
 Molecular Cell, Volume 32 Supplemental Data Comparative Analysis of Argonaute-Dependent Small RNA Pathways in Drosophila Rui Zhou, Ikuko Hotta, Ahmet M. Denli, Pengyu Hong, Norbert Perrimon, and Gregory
Molecular Cell, Volume 32 Supplemental Data Comparative Analysis of Argonaute-Dependent Small RNA Pathways in Drosophila Rui Zhou, Ikuko Hotta, Ahmet M. Denli, Pengyu Hong, Norbert Perrimon, and Gregory
Technical tips Session 4
 Technical tips Session 4 Biotinylation assay: Streptavidin is a small bacterial protein that binds with high affinity to the vitamin biotin. This streptavidin-biotin combination can be used to link molecules
Technical tips Session 4 Biotinylation assay: Streptavidin is a small bacterial protein that binds with high affinity to the vitamin biotin. This streptavidin-biotin combination can be used to link molecules
Chapter 9-II - Transcriptional Control of Gene Expression
 Chapter 9-II - Transcriptional Control of Gene Expression Transcriptional Control of Gene Expression 9.3 RNA Polymerase II Promoters and General Transcription Factors Three types of promoter sequences
Chapter 9-II - Transcriptional Control of Gene Expression Transcriptional Control of Gene Expression 9.3 RNA Polymerase II Promoters and General Transcription Factors Three types of promoter sequences
HPV E6 oncoprotein targets histone methyltransferases for modulating specific. Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu,
 1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
Announcement Structure Analysis
 Announcement Structure Analysis BSC 4439/BSC 5436: Biomedical Informatics: Structure Analysis Spring 2019, CB117 Monday and Wednesday 12:00 1:15pm Office hour: Monday and Wednesday 1:15 2pm Topics include
Announcement Structure Analysis BSC 4439/BSC 5436: Biomedical Informatics: Structure Analysis Spring 2019, CB117 Monday and Wednesday 12:00 1:15pm Office hour: Monday and Wednesday 1:15 2pm Topics include
Selected Techniques Part I
 1 Selected Techniques Part I Gel Electrophoresis Can be both qualitative and quantitative Qualitative About what size is the fragment? How many fragments are present? Is there in insert or not? Quantitative
1 Selected Techniques Part I Gel Electrophoresis Can be both qualitative and quantitative Qualitative About what size is the fragment? How many fragments are present? Is there in insert or not? Quantitative
Introduction to genome biology
 Introduction to genome biology Lisa Stubbs Deep transcritpomes for traditional model species from ENCODE (and modencode) Deep RNA-seq and chromatin analysis on 147 human cell types, as well as tissues,
Introduction to genome biology Lisa Stubbs Deep transcritpomes for traditional model species from ENCODE (and modencode) Deep RNA-seq and chromatin analysis on 147 human cell types, as well as tissues,
