Maria Spies, Mark S. Dillingham, and Stephen C. Kowalczykowski. Sections of Microbiology, and of Molecular and Cellular Biology, Center for
|
|
|
- Donald Atkins
- 6 years ago
- Views:
Transcription
1 DNA HELICASES Maria Spies, Mark S. Dillingham, and Stephen C. Kowalczykowski Sections of Microbiology, and of Molecular and Cellular Biology, Center for Genetics and Development, University of California, Davis. In all organisms, genetic information is locked within a double helix formed by the two antiparallel deoxyribonucleic acid (DNA) strands. Although double-stranded DNA (dsdna) is suitable for secure information storage, hydrogen bonds formed between complementary bases impair readout of this information by the cellular machineries that frequently require single-stranded DNA (ssdna) as the template. The unwinding of dsdna into ssdna, a function critical for virtually every aspect of cellular DNA metabolism including replication, recombination and repair, is catalyzed by a ubiquitous class of enzymes called DNA helicases. First identified in the 1970s, DNA helicases are motor proteins that convert chemical energy into mechanical work. Chemical energy is derived from the hydrolysis of adenosine triphosphate (ATP) or other nucleoside triphosphates, and is coupled to mechanical work during at least two important steps within the helicase reaction cycle (Fig. 1): (1) the unidirectional translocation of the enzyme along the DNA molecule, and (2) the separation of the DNA duplex. Classifications On the basis of amino acid sequence comparisons, helicases are divided into the three large superfamilies and two smaller families that are illustrated schematically in 1
2 Fig. 2. Moreover, these analyses identified several protein signature motifs that are indicative of nucleic acid strand-separation activity and/or the capacity to translocate along DNA. Biochemical and structural data show that helicases can function as monomers, dimers, and multimers (predominantly hexamers) and that they can be further classified on the basis of their requirement: dsdna, dsrna, or DNA-RNA hybrid. Superfamilies 1 (SF1) and 2 (SF2) include thousands of DNA and RNA helicases. SF1 and SF2 enzymes are primarily monomeric and dimeric helicases that unwind DNA with either 3 5 or 5 3 polarity (see below). Domains 1A and 2B in SF1 helicases and domains 1 and 2 in SF2 helicases contain similar sets of seven helicase signature motifs that comprise the basic structural core of these enzymes (Fig. 2). This core is responsible for both nucleotide binding and hydrolysis, and it includes a repeat of either of two structural domains: the RecA-fold in SF1 helicases, or the related adenylate kinase-fold in SF2 helicases. Variation at the N- and C-termini, as well as the insertions between the core domains, give rise to differences in the substrate specificity or to additional enzyme activities. Motifs I and II (Walker A {GxGKT} and B {DExo/H} motifs, respectively) are the most conserved among all helicases. Superfamily 3 (SF3) helicases are found in small DNA and RNA viruses. In contrast to SF1 and SF2 helicases, each subunit of these enzymes comprises a single vaaa + - fold that is responsible for nucleotide binding and hydrolysis. The smaller superfamilies 4 and 5 consist primarily of 5 3 hexameric helicases related either to the E. coli DnaB helicase (SF4), or to the transcription termination factor Rho (SF5). The nucleotide binding and hydrolysis core of these enzymes possesses the highest degree of similarity to that of F 1 ATPase. 2
3 Directional translocation Helicases are further classified by their polarity of unwinding as either 3' 5' or 5' 3' enzymes. This polarity refers to an experimentally defined requirement for a region of ssdna of the given polarity adjacent to the dsdna to be unwound to act as a loading site for the helicase. It is now widely accepted that the unwinding polarity relates to a directional bias in translocation on ssdna. For example, the enzyme depicted in Fig. 1 is a 3' 5' helicase. Upon binding to the ssdna, it starts moving toward the 5' end of the loading strand, which brings the enzyme to the ssdna-dsdna junction; subsequent translocation along the ssdna, coupled to either physical or electrostatic destabilization of the dsdna, results in strand separation in the duplex portion of the substrate. Conversely, a 5 3 helicase translocates toward the 3 end of the loading strand and, therefore, requires a 5 -terminated region of ssdna to unwind a DNA duplex. Interestingly, high-resolution structural data suggest that the helicase signature motifs are not essential for the duplex DNA separation per se, but for the ATP-dependent unidirectional motion of the helicases on the DNA lattice. Consequently, it is proposed that the helicase signature motifs define a modular structure for a DNA motor, while the additional domains, which may vary from one protein to another, are responsible for either modulating DNA unwinding, mediating protein-protein interactions, or determining structure specificity. Although the classical substrate for a DNA helicase depicted in Fig. 1 is duplex DNA adjacent to a region of ssdna, some enzymes preferentially unwind more complicated structures, such as flaps, forks, bubbles, or threeand four-way junctions. 3
4 DNA translocation can also be characterized in terms of processivity, which is defined as the probability of forward translocation at each base pair opening cycle. For a highly processive enzyme, stepping forward is much more likely than dissociation. Consequently, an enzyme that never dissociates after each step has a processivity equal to one (which is a physically unrealistic situation where the enzyme is infinitely processive), whereas an enzyme that dissociates after each step has a processivity equal to zero (and is referred to as being dispersive or distributive). The average number of base pairs unwound (or translocated) by an enzyme in a single binding event (N) relates to the processivity (P) as N = 1/(1-P). For example, UvrD helicase, whose processivity is 0.98, unwinds on average 50 base pairs. On the other hand, the highly processive RecBCD enzyme (P = ) unwinds on average 30,000 base pairs before dissociation. Step size Though seemingly simple, one of the more poorly defined parameters for DNA helicase behavior is the step size. The step size has been defined in at least three different ways: (1) the number of base pairs that the enzyme unwinds upon hydrolyzing one ATP molecule under its most efficient reaction conditions; (2) as a physical step size, which is a measure of the helicase s ability to step over a discontinuity in the DNA; and (3) as a distance associated with a kinetically rate-limiting step. The parameters obtained using these three methods are not necessarily identical. For example, RecBCD enzyme hydrolyses about 2 molecules of ATP per base pair unwound, but has a kinetic step size of six base pairs. The physical step size of the related RecBC enzyme was determined to be 23 base pairs. 4
5 Accessory factors Once a helicase has separated the strands of dsdna, spontaneous reannealing of the duplex can be avoided if the nascent ssdna strands are trapped by a single-stranded DNA binding protein or by other factors that pass the unwound nucleic acid intermediate to the next step of a reaction pathway (Fig. 1). While ssdna binding proteins have frequently been shown to stimulate helicase activity in vitro, helicase activity is also stimulated by other accessory factors that increase the rate or processivity of unwinding (such as processivity factors, clamp loaders, polymerases, etc.). The primary replicative helicase of Escherichia coli, DnaB, is a good example of a helicase that acts poorly in isolation but, as part of the replisome (the DNA synthesis machinery of the cell), DnaB efficiently separates the DNA strands at the replication fork. The rate of movement of the replication machinery at the fork is coordinated by an interaction between DnaB and DNA polymerase (the enzyme that synthesizes the complementary strand of DNA) that enhances its translocation rate by almost 30-fold to 1000 base pairs per second. Single-molecule analysis of DNA helicases Until recently, virtually all biochemical data on helicases were derived from conventional bulk-phase techniques, which observe the population-averaged properties of molecular ensembles. Direct observation of dsdna unwinding by a single helicase enzyme has the potential to uncover valuable information on protein and DNA dynamics that may be obscured in bulk-phase assays. Two of these techniques were used to visualize translocation of a single molecule of RecBCD, a multifunctional enzyme involved in the repair of dsdna breaks by homologous recombination. RecBCD is an 5
6 exceptionally fast and processive helicase that can unwind, on average, 30,000 bp of dsdna per binding event, at a rate of 1000 bp/s, while simultaneously degrading the ssdna products of its helicase activity. In one approach, a device called an optical trap is used to manipulate individual, fluorescently labeled DNA molecules and to visualize their unwinding and degradation by RecBCD enzyme. In the presence of ATP, RecBCD enzyme unwinds the dsdna, which is observed as a progressive shortening of the fluorescently labeled DNA molecule. Recently (2003), this technique was used to demonstrate that RecBCD enzyme pauses for several seconds at a specific DNA sequence called Chi, which is a hotspot for DNA recombination and then continues but at a reduced translocation speed. This finding nicely illustrates the ability of single molecule studies to discover new behavior, as this phenomenon had not been detected in conventional bulk-phase kinetic analyses. An alternative single-molecule approach uses light microscopy to follow translocation of a biotin-tagged RecBCD enzyme bound to a streptavidin-coated polystyrene bead in the tethered particle motion experiment. The two single-molecule experiments are different yet complementary: the tethered particle motion experiment directly measures translocation, whereas the optical trap method measures dsdna unwinding. Together, these studies provide additional powerful evidence for the coupling of DNA strand separation with movement of the helicase protein on its substrate lattice. Both single-molecule visualization methods show that RecBCD translocates unidirectionally and processively on dsdna, with each molecule moving at a constant rate (within the limit of experimental detection). Although the average translocation rate is similar to that derived from bulk measurements, considerable variation is observed in 6
7 the translocation rate of individual RecBCD enzymes. This surprising observation is another example of the kind of information that is accessible only by single-molecule studies. To study helicases that have a low processivity, still other single-molecule strategies were developed. In one such strategy, fluorescence resonance energy transfer (FRET) is used to detect the separation of strands of individual base-pairs in a DNA molecule. By following the temporal fluctuations of the FRET signal from a single molecule, the kinetics of complex biochemical reaction that are unsynchronized can be interpreted more clearly that when performing bulk-phase FRET experiments. Yet another technique for detecting helicase action at the single-molecule level measures changes in the extension of a DNA molecule as a consequence of unwinding by the UvrD helicase. At forces above 5 pn, ssdna is longer than dsdna, and so unwinding results in an increase in the DNA length. This technique allows determination of the individual helicase rate ( 40 bp s -1, which is 3-fold higher than bulk-phase estimates), the average number of base pairs unwound per binding event (240 bps), and the step size (6 bp per enzymatic cycle). Surprisingly, in some cases UvrD helicase is observed to switch strands and translocate on the opposite strand resulting in slow rezipping of the unwound DNA. Conclusion Although considerable progress has been made in our understanding of the molecular mechanisms of DNA helicases, many questions remain to be answered. The new techniques of single molecule detection will play an important role in this endeavor. 7
8 Perhaps the next big challenges in this field are to understand how these DNA motors are incorporated into, and function within, the context of macromolecular machines to orchestrate complex DNA processing events. See also: Adenosine triphosphate (ATP); Deoxyribonucleic acid (DNA); Enzyme; Molecular biology; Nucleoprotein Bibliography B. Alberts et al., Molecular Biology of the Cell, 3d ed. (II, Chap. 6), Garland Publishing, New York, 1994 H. Lodish et al., Molecular Cell Biology, 4th ed. (Chap. 12), W H Freeman, New York, 2000 P. Soultanas and D. B. Wigley, Unwinding the "Gordian Knot" of helicase action, Trends Biochem. Sci., 26(1):47-54, 2001 Additional Reading P. R. Bianco et al., Processive translocation and DNA unwinding by individual RecBCD enzyme molecules, Nature, 409(18): , 2001 Spies, M., Bianco, P.R., Dillingham, M.S., Handa, N., Baskin, R.J. and Kowalczykowski, S.C. (2003) A molecular throttle: The recombination hotspot, χ, controls DNA translocation by the RecBCD helicase. Cell, 114, K. M. Dohoney and J. Gelles, χ-sequence recognition and DNA translocation by single RecBCD helicase/nuclease molecules, Nature, 409(18): ,
9 Ha, T., Rasnik, I., Cheng, W., Babcock, H.P., Gauss, G.H., Lohman, T.M. and Chu, S. (2002) Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature, 419, Dessinges, M.-N., Lionnet, T., Xi, X.G., Bensimon, D., Croquette, V. (2004) Single molecule assay reveals strand switching and enhanced processivity of UvrD. Proc. Nat. Ac. Sci., 101, A movie of translocation/unwinding by RecBCD helicase along fluorescently labeled DNA molecule can be viewed at Brownian motion of DNA-tethered beads can be viewed at Figure legends Fig. 1 Schematic representation of DNA helicase action. The helicase enzyme translocates along the DNA molecule and separates the strands. Energy for this unfavorable reaction is coupled to the hydrolysis of adenosine triphosphate (ATP) to adenosine diphosphate (ADP) and inorganic phosphate (P i ). In the presence of a singlestranded DNA binding protein, reannealing of the DNA duplex is prevented. The helicase depicted here displays a 3' 5' polarity, tracking unidirectionally along the lower of the two DNA strands in the duplex (the loading strand). 9
10 Fig. 2. Five major superfamilies of DNA helicases. The conserved signature motifs are shown relative to the modular domains (shown as solid blocks) of helicases that belong to 5 major families. The motifs that are characteristic of all helicases within a particular superfamily are shown in black, while the motifs characteristic only to subsets of the SF2 helicases are represented by a hashed block. The areas containing large insertions that define additional enzymatic activities are indicated by dashed lines. The oligomeric state and translocation polarity for each superfamily is indicated on the right of the block diagram. 10
11 n(atp) n(adp + P i ) trapping re-annealing Fig. 1.
12 Oligomeric state Translocation SF1 RecA-fold RecA-fold Domains: N Motifs: 1A 1B 1A 2A 2B 2A I Ia II III IV VVI C Monomers*, dimers SF2 Adenylate kinase-fold Adenylate kinase-fold Domains: N Motifs: Q I IaIb 1 2 II III IV VVI C Monomers, dimers, oligomers SF3 (Origin DNA interaction) N Motifs: vaaa + -fold A B B C C (Zn-finger domain) Hexamers, double hexamers 3 5 SF4 (DnaB-like) RecA-fold Domains: N Motifs: Primase 1 (A) Helicase 1a 2 3 (B) 4 C Hexamers 5 3 SF5 (Rho( Rho-like) F 1 ATPase-fold N Motifs: I Ia II III IV (R loop) (Q loop) C Hexamers 5 3 Fig. 2.
 The replication of DNA Kornberg 1957 Meselson and Stahl 1958 Cairns 1963 Okazaki 1968 DNA Replication The driving force for DNA synthesis. The addition of a nucleotide to a growing polynucleotide
The replication of DNA Kornberg 1957 Meselson and Stahl 1958 Cairns 1963 Okazaki 1968 DNA Replication The driving force for DNA synthesis. The addition of a nucleotide to a growing polynucleotide
RNA Expression of the information in a gene generally involves production of an RNA molecule transcribed from a DNA template. RNA differs from DNA
 RNA Expression of the information in a gene generally involves production of an RNA molecule transcribed from a DNA template. RNA differs from DNA that it has a hydroxyl group at the 2 position of the
RNA Expression of the information in a gene generally involves production of an RNA molecule transcribed from a DNA template. RNA differs from DNA that it has a hydroxyl group at the 2 position of the
DNA Replication II Biochemistry 302. January 25, 2006
 DNA Replication II Biochemistry 302 January 25, 2006 Following in Dad s footsteps Original A. Kornberg E. coli DNA Pol I is a lousy replicative enzyme. 400 molecules/cell but ~2 replication forks/cell
DNA Replication II Biochemistry 302 January 25, 2006 Following in Dad s footsteps Original A. Kornberg E. coli DNA Pol I is a lousy replicative enzyme. 400 molecules/cell but ~2 replication forks/cell
DNA Replication II Biochemistry 302. Bob Kelm January 28, 2004
 DNA Replication II Biochemistry 302 Bob Kelm January 28, 2004 Conceptual model for proofreading based on kinetic considerations Fig. 24.44 stalling transient melting exonuclease site occupancy Following
DNA Replication II Biochemistry 302 Bob Kelm January 28, 2004 Conceptual model for proofreading based on kinetic considerations Fig. 24.44 stalling transient melting exonuclease site occupancy Following
Chapter 11 DNA Replication and Recombination
 Chapter 11 DNA Replication and Recombination Copyright Copyright 2009 Pearson 2009 Pearson Education, Education, Inc. Inc. 11.1 DNA is reproduced by Semiconservative Replication The complementarity of
Chapter 11 DNA Replication and Recombination Copyright Copyright 2009 Pearson 2009 Pearson Education, Education, Inc. Inc. 11.1 DNA is reproduced by Semiconservative Replication The complementarity of
III. Detailed Examination of the Mechanism of Replication A. Initiation B. Priming C. Elongation D. Proofreading and Termination
 Outline for Replication I. General Features of Replication A. Semi-Conservative B. Starts at Origin C. Bidirectional D. Semi-Discontinuous II. Proteins and Enzymes of Replication III. Detailed Examination
Outline for Replication I. General Features of Replication A. Semi-Conservative B. Starts at Origin C. Bidirectional D. Semi-Discontinuous II. Proteins and Enzymes of Replication III. Detailed Examination
RNA synthesis/transcription I Biochemistry 302. February 6, 2004 Bob Kelm
 RNA synthesis/transcription I Biochemistry 302 February 6, 2004 Bob Kelm Overview of RNA classes Messenger RNA (mrna) Encodes protein Relatively short half-life ( 3 min in E. coli, 30 min in eukaryotic
RNA synthesis/transcription I Biochemistry 302 February 6, 2004 Bob Kelm Overview of RNA classes Messenger RNA (mrna) Encodes protein Relatively short half-life ( 3 min in E. coli, 30 min in eukaryotic
Proposed Models of DNA Replication. Conservative Model. Semi-Conservative Model. Dispersive model
 5.2 DNA Replication Cell Cycle Life cycle of a cell Cells can reproduce Daughter cells receive an exact copy of DNA from parent cell DNA replication happens during the S phase Proposed Models of DNA Replication
5.2 DNA Replication Cell Cycle Life cycle of a cell Cells can reproduce Daughter cells receive an exact copy of DNA from parent cell DNA replication happens during the S phase Proposed Models of DNA Replication
University of Groningen. Single-molecule studies of DNA replication Geertsema, Hylkje
 University of Groningen Single-molecule studies of DNA replication Geertsema, Hylkje IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's PDF) if you wish to cite from it. Please
University of Groningen Single-molecule studies of DNA replication Geertsema, Hylkje IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's PDF) if you wish to cite from it. Please
University of Groningen. Single-molecule studies of DNA replication Geertsema, Hylkje
 University of Groningen Single-molecule studies of DNA replication Geertsema, Hylkje IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's PDF) if you wish to cite from it. Please
University of Groningen Single-molecule studies of DNA replication Geertsema, Hylkje IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's PDF) if you wish to cite from it. Please
DNA Replication II Biochemistry 302. Bob Kelm January 26, 2005
 DNA Replication II Biochemistry 302 Bob Kelm January 26, 2005 Following in Dad s footsteps Original A. Kornberg E. coli DNA Pol I is a lousy replicative enzyme. 400 molecules/cell but ~2 replication forks/cell
DNA Replication II Biochemistry 302 Bob Kelm January 26, 2005 Following in Dad s footsteps Original A. Kornberg E. coli DNA Pol I is a lousy replicative enzyme. 400 molecules/cell but ~2 replication forks/cell
Questions from chapters in the textbook that are relevant for the final exam
 Questions from chapters in the textbook that are relevant for the final exam Chapter 9 Replication of DNA Question 1. Name the two substrates for DNA synthesis. Explain why each is necessary for DNA synthesis.
Questions from chapters in the textbook that are relevant for the final exam Chapter 9 Replication of DNA Question 1. Name the two substrates for DNA synthesis. Explain why each is necessary for DNA synthesis.
Genetic Information: DNA replication
 Genetic Information: DNA replication Umut Fahrioglu, PhD MSc DNA Replication Replication of DNA is vital to the transmission of genomes and the genes they contain from one cell generation to the other.
Genetic Information: DNA replication Umut Fahrioglu, PhD MSc DNA Replication Replication of DNA is vital to the transmission of genomes and the genes they contain from one cell generation to the other.
The Size and Packaging of Genomes
 DNA Replication The Size and Packaging of Genomes Vary greatly in size Ø Smallest viruses- 4 or 5 genes Ø Escherichia coli- 4,288 genes Ø Human cell- 20,000 to 25,000 genes E. coli 4 million base pairs
DNA Replication The Size and Packaging of Genomes Vary greatly in size Ø Smallest viruses- 4 or 5 genes Ø Escherichia coli- 4,288 genes Ø Human cell- 20,000 to 25,000 genes E. coli 4 million base pairs
Replication. Obaidur Rahman
 Replication Obaidur Rahman DIRCTION OF DNA SYNTHESIS How many reactions can a DNA polymerase catalyze? So how many reactions can it catalyze? So 4 is one answer, right, 1 for each nucleotide. But what
Replication Obaidur Rahman DIRCTION OF DNA SYNTHESIS How many reactions can a DNA polymerase catalyze? So how many reactions can it catalyze? So 4 is one answer, right, 1 for each nucleotide. But what
Nucleic acids deoxyribonucleic acid (DNA) ribonucleic acid (RNA) nucleotide
 Nucleic Acids Nucleic acids are molecules that store information for cellular growth and reproduction There are two types of nucleic acids: - deoxyribonucleic acid (DNA) and ribonucleic acid (RNA) These
Nucleic Acids Nucleic acids are molecules that store information for cellular growth and reproduction There are two types of nucleic acids: - deoxyribonucleic acid (DNA) and ribonucleic acid (RNA) These
3.A.1 DNA and RNA: Structure and Replication
 3.A.1 DNA and RNA: Structure and Replication Each DNA polymer is made of Nucleotides (monomer) which are made of: a) Phosphate group: Negatively charged and polar b) Sugar: deoxyribose- a 5 carbon sugar
3.A.1 DNA and RNA: Structure and Replication Each DNA polymer is made of Nucleotides (monomer) which are made of: a) Phosphate group: Negatively charged and polar b) Sugar: deoxyribose- a 5 carbon sugar
DNA REPLICATION. Third Stage. Lec. 12 DNA Replication. Lecture No.: 12. A. Watson & Crick (1952) C. Cairns (1963) autoradiographic experiment
 Lec. 12 DNA Replication A. Watson & Crick (1952) Proposed a model where hydrogen bonds break, the two strands separate, and DNA synthesis occurs semi-conservatively in the same net direction. While a straightforward
Lec. 12 DNA Replication A. Watson & Crick (1952) Proposed a model where hydrogen bonds break, the two strands separate, and DNA synthesis occurs semi-conservatively in the same net direction. While a straightforward
NUCLEIC ACID METABOLISM. Omidiwura, B.R.O
 NUCLEIC ACID METABOLISM Omidiwura, B.R.O Nucleic Acids Nucleic acids are molecules that store information for cellular growth and reproduction There are two types of nucleic acids: - deoxyribonucleic acid
NUCLEIC ACID METABOLISM Omidiwura, B.R.O Nucleic Acids Nucleic acids are molecules that store information for cellular growth and reproduction There are two types of nucleic acids: - deoxyribonucleic acid
DNA Recombination Biochemistry 302. Bob Kelm February 2, 2005
 DNA Recombination Biochemistry 302 Bob Kelm February 2, 2005 Homologous recombination: Why is it advantageous to a cell? Bacterial cell Information exchange during bacterial conjugation (mating) between
DNA Recombination Biochemistry 302 Bob Kelm February 2, 2005 Homologous recombination: Why is it advantageous to a cell? Bacterial cell Information exchange during bacterial conjugation (mating) between
Molecular Biology: General Theory
 Molecular Biology: General Theory Author: Dr Darshana Morar Licensed under a Creative Commons Attribution license. DNA REPLICATION DNA replication is the process of duplicating the DNA sequence in the
Molecular Biology: General Theory Author: Dr Darshana Morar Licensed under a Creative Commons Attribution license. DNA REPLICATION DNA replication is the process of duplicating the DNA sequence in the
Molecular Biology: General Theory
 Molecular Biology: General Theory Author: Dr Darshana Morar Licensed under a Creative Commons Attribution license. DNA REPLICATION DNA replication is the process of duplicating the DNA sequence in the
Molecular Biology: General Theory Author: Dr Darshana Morar Licensed under a Creative Commons Attribution license. DNA REPLICATION DNA replication is the process of duplicating the DNA sequence in the
The replication forks Summarising what we know:
 When does replication occur? MBLG1001 lecture 10 Replication the once in a lifetime event! Full blown replication only occurs once, just before cell division BUT the DNA template is constantly being repaired.
When does replication occur? MBLG1001 lecture 10 Replication the once in a lifetime event! Full blown replication only occurs once, just before cell division BUT the DNA template is constantly being repaired.
Tutorial Week #9 Page 1 of 11
 Tutorial Week #9 Page 1 of 11 Tutorial Week #9 DNA Replication Before the tutorial: Read ECB Chapter 6 p195-207, and review your lecture notes Read this tutorial and create a table of definitions and functions
Tutorial Week #9 Page 1 of 11 Tutorial Week #9 DNA Replication Before the tutorial: Read ECB Chapter 6 p195-207, and review your lecture notes Read this tutorial and create a table of definitions and functions
The Structure of DNA
 The Structure of DNA Questions to Ponder 1) How is the genetic info copied? 2) How does DNA store the genetic information? 3) How is the genetic info passed from generation to generation? The Structure
The Structure of DNA Questions to Ponder 1) How is the genetic info copied? 2) How does DNA store the genetic information? 3) How is the genetic info passed from generation to generation? The Structure
Fork sensing and strand switching control antagonistic activities of. RecQ helicases. Supplementary Information
 Fork sensing and strand switching control antagonistic activities of RecQ helicases Daniel Klaue, Daniela Kobbe, Felix Kemmerich, Alicja Kozikowska, Holger Puchta, Ralf Seidel Supplementary Information
Fork sensing and strand switching control antagonistic activities of RecQ helicases Daniel Klaue, Daniela Kobbe, Felix Kemmerich, Alicja Kozikowska, Holger Puchta, Ralf Seidel Supplementary Information
Chapter 12. DNA Replication and Recombination
 Chapter 12 DNA Replication and Recombination I. DNA replication Three possible modes of replication A. Conservative entire original molecule maintained B. Semiconservative one strand is template for new
Chapter 12 DNA Replication and Recombination I. DNA replication Three possible modes of replication A. Conservative entire original molecule maintained B. Semiconservative one strand is template for new
Answers to Module 1. An obligate aerobe is an organism that has an absolute requirement of oxygen for growth.
 Answers to Module 1 Short Answers 1) What is an obligate aerobe? An obligate aerobe is an organism that has an absolute requirement of oxygen for growth. What about facultative anaerobe? 2) Distinguish
Answers to Module 1 Short Answers 1) What is an obligate aerobe? An obligate aerobe is an organism that has an absolute requirement of oxygen for growth. What about facultative anaerobe? 2) Distinguish
Chapter Twelve: DNA Replication and Recombination
 This is a document I found online that is based off of the fourth version of your book. Not everything will apply to the upcoming exam so you ll have to pick out what you thing is important and applicable.
This is a document I found online that is based off of the fourth version of your book. Not everything will apply to the upcoming exam so you ll have to pick out what you thing is important and applicable.
Welcome to Class 18! Lecture 18: Outline and Objectives. Replication is semiconservative! Replication: DNA DNA! Introductory Biochemistry!
 Lecture 18: Outline and Objectives Welcome to Class 18! Introductory Biochemistry! l DNA Replication! l DNA polymerase! l the enzymatic reaction! l proofreading and accuracy! l DNA synthesis! l origins
Lecture 18: Outline and Objectives Welcome to Class 18! Introductory Biochemistry! l DNA Replication! l DNA polymerase! l the enzymatic reaction! l proofreading and accuracy! l DNA synthesis! l origins
Lecture 13. Motor Proteins I
 Lecture 13 Motor Proteins I Introduction: The study of motor proteins has become a major focus in cell and molecular biology. Motor proteins are very interesting because they do what no man-made engines
Lecture 13 Motor Proteins I Introduction: The study of motor proteins has become a major focus in cell and molecular biology. Motor proteins are very interesting because they do what no man-made engines
Transcription. By : Lucia Dhiantika Witasari M.Biotech., Apt
 Transcription By : Lucia Dhiantika Witasari M.Biotech., Apt REGULATION OF GENE EXPRESSION 11/26/2010 2 RNA Messenger RNAs (mrnas) encode the amino acid sequence of one or more polypeptides specified by
Transcription By : Lucia Dhiantika Witasari M.Biotech., Apt REGULATION OF GENE EXPRESSION 11/26/2010 2 RNA Messenger RNAs (mrnas) encode the amino acid sequence of one or more polypeptides specified by
Enzymes used in DNA Replication
 Enzymes used in DNA Replication This document holds the enzymes used in DNA replication, their pictorial representation and functioning. DNA polymerase: DNA polymerase is the chief enzyme of DNA replication.
Enzymes used in DNA Replication This document holds the enzymes used in DNA replication, their pictorial representation and functioning. DNA polymerase: DNA polymerase is the chief enzyme of DNA replication.
DNA Metabolism. I. DNA Replication. A. Template concept: 1. How can you make a copy of a molecule? 2. Complementary Hydrogen bonding
 DNA Metabolism I. DNA Replication A. Template concept: 1. How can you make a copy of a molecule? 2. Complementary Hydrogen bonding B. DNA replication follows a set of fundamental rules 1. Semiconservative
DNA Metabolism I. DNA Replication A. Template concept: 1. How can you make a copy of a molecule? 2. Complementary Hydrogen bonding B. DNA replication follows a set of fundamental rules 1. Semiconservative
Transcription in Prokaryotes. Jörg Bungert, PhD Phone:
 Transcription in Prokaryotes Jörg Bungert, PhD Phone: 352-273-8098 Email: jbungert@ufl.edu Objectives Understand the basic mechanism of transcription. Know the function of promoter elements and associating
Transcription in Prokaryotes Jörg Bungert, PhD Phone: 352-273-8098 Email: jbungert@ufl.edu Objectives Understand the basic mechanism of transcription. Know the function of promoter elements and associating
IDTutorial: DNA Replication
 IDTutorial: DNA Replication Introduction In their report announcing the structure of the DNA molecule, Watson and Crick (Nature, 171: 737-738, 1953) observe, It has not escaped our notice that the specific
IDTutorial: DNA Replication Introduction In their report announcing the structure of the DNA molecule, Watson and Crick (Nature, 171: 737-738, 1953) observe, It has not escaped our notice that the specific
Chapter 11 Homologous Recombination at the Molecular Level. 吳彰哲 (Chang-Jer Wu) Department of Food Science National Taiwan Ocean University
 Chapter 11 Homologous Recombination at the Molecular Level 吳彰哲 (Chang-Jer Wu) Department of Food Science National Taiwan Ocean University 1 Introduction All DNA is recombinant DNA During meiosis homologous
Chapter 11 Homologous Recombination at the Molecular Level 吳彰哲 (Chang-Jer Wu) Department of Food Science National Taiwan Ocean University 1 Introduction All DNA is recombinant DNA During meiosis homologous
Supplementary material of: Human Upf1 is a highly processive RNA helicase and translocase with RNP remodelling activities
 Supplementary material of: Human Upf1 is a highly processive RNA helicase and translocase with RNP remodelling activities By Francesca Fiorini, Debjani Bagchi, Hervé Le Hir and Vincent Croquette Supplementary
Supplementary material of: Human Upf1 is a highly processive RNA helicase and translocase with RNP remodelling activities By Francesca Fiorini, Debjani Bagchi, Hervé Le Hir and Vincent Croquette Supplementary
Chapter 12: Molecular Biology of the Gene
 Biology Textbook Notes Chapter 12: Molecular Biology of the Gene p. 214-219 The Genetic Material (12.1) - Genetic Material must: 1. Be able to store information that pertains to the development, structure,
Biology Textbook Notes Chapter 12: Molecular Biology of the Gene p. 214-219 The Genetic Material (12.1) - Genetic Material must: 1. Be able to store information that pertains to the development, structure,
MIDTERM I NAME: Student ID Number:
 MIDTERM I NAME: Student ID Number: Question Maximum Points Your Points I 35 II 29 III 30 IV 32 V 24 150 Please write your name/student ID number on each of the following five pages. This exam must be written
MIDTERM I NAME: Student ID Number: Question Maximum Points Your Points I 35 II 29 III 30 IV 32 V 24 150 Please write your name/student ID number on each of the following five pages. This exam must be written
Initiation of Replication. This section will begin with a brief discussion of chromosomal DNA sequences that sponsor the initiation of replication.
 Initiation of Replication This section will begin with a brief discussion of chromosomal DNA sequences that sponsor the initiation of replication. Replicons and origins of replication The unit of DNA replication
Initiation of Replication This section will begin with a brief discussion of chromosomal DNA sequences that sponsor the initiation of replication. Replicons and origins of replication The unit of DNA replication
Plant Molecular and Cellular Biology Lecture 4: E. coli DNA Replicase Structure & Function. Gary Peter
 Plant Molecular and Cellular Biology Lecture 4: E. coli DNA Replicase Structure & Function Gary Peter Learning Objectives 1. List and explain the mechanisms by which E. coli DNA is replicated 2. Describe
Plant Molecular and Cellular Biology Lecture 4: E. coli DNA Replicase Structure & Function Gary Peter Learning Objectives 1. List and explain the mechanisms by which E. coli DNA is replicated 2. Describe
Syllabus for GUTS Lecture on DNA and Nucleotides
 Syllabus for GUTS Lecture on DNA and Nucleotides I. Introduction. DNA is the instruction manual for how to build a living organism here on earth. The instructions in DNA are propagated to future generations
Syllabus for GUTS Lecture on DNA and Nucleotides I. Introduction. DNA is the instruction manual for how to build a living organism here on earth. The instructions in DNA are propagated to future generations
DNA Replication I Biochemistry 302. Bob Kelm January 24, 2005
 DNA Replication I Biochemistry 302 Bob Kelm January 24, 2005 Watson Crick prediction: Each stand of parent DNA serves as a template for synthesis of a new complementary daughter strand Fig. 4.12 Proof
DNA Replication I Biochemistry 302 Bob Kelm January 24, 2005 Watson Crick prediction: Each stand of parent DNA serves as a template for synthesis of a new complementary daughter strand Fig. 4.12 Proof
DNA: Structure & Replication
 DNA Form & Function DNA: Structure & Replication Understanding DNA replication and the resulting transmission of genetic information from cell to cell, and generation to generation lays the groundwork
DNA Form & Function DNA: Structure & Replication Understanding DNA replication and the resulting transmission of genetic information from cell to cell, and generation to generation lays the groundwork
Nucleic Acid Structure:
 Nucleic Acid Structure: Purine and Pyrimidine nucleotides can be combined to form nucleic acids: 1. Deoxyribonucliec acid (DNA) is composed of deoxyribonucleosides of! Adenine! Guanine! Cytosine! Thymine
Nucleic Acid Structure: Purine and Pyrimidine nucleotides can be combined to form nucleic acids: 1. Deoxyribonucliec acid (DNA) is composed of deoxyribonucleosides of! Adenine! Guanine! Cytosine! Thymine
BIOCHEMISTRY REVIEW. Overview of Biomolecules. Chapter 11 DNA Replication
 BIOCHEMISTRY REVIEW Overview of Biomolecules Chapter 11 DNA Replication 2 3 4 5 6 7 8 9 Are You Getting It?? Which characteristics will be part of semi-conservative replication? (multiple answers) a) The
BIOCHEMISTRY REVIEW Overview of Biomolecules Chapter 11 DNA Replication 2 3 4 5 6 7 8 9 Are You Getting It?? Which characteristics will be part of semi-conservative replication? (multiple answers) a) The
Covalently bonded sugar-phosphate backbone with relatively strong bonds keeps the nucleotides in the backbone connected in the correct sequence.
 Unit 14: DNA Replication Study Guide U7.1.1: DNA structure suggested a mechanism for DNA replication (Oxford Biology Course Companion page 347). 1. Outline the features of DNA structure that suggested
Unit 14: DNA Replication Study Guide U7.1.1: DNA structure suggested a mechanism for DNA replication (Oxford Biology Course Companion page 347). 1. Outline the features of DNA structure that suggested
Technical tips Session 4
 Technical tips Session 4 Biotinylation assay: Streptavidin is a small bacterial protein that binds with high affinity to the vitamin biotin. This streptavidin-biotin combination can be used to link molecules
Technical tips Session 4 Biotinylation assay: Streptavidin is a small bacterial protein that binds with high affinity to the vitamin biotin. This streptavidin-biotin combination can be used to link molecules
Chapters 31-32: Ribonucleic Acid (RNA)
 Chapters 31-32: Ribonucleic Acid (RNA) Short segments from the transcription, processing and translation sections of each chapter Slide 1 RNA In comparison with DNA RNA utilizes uracil in place of thymine
Chapters 31-32: Ribonucleic Acid (RNA) Short segments from the transcription, processing and translation sections of each chapter Slide 1 RNA In comparison with DNA RNA utilizes uracil in place of thymine
RNA does not adopt the classic B-DNA helix conformation when it forms a self-complementary double helix
 Reason: RNA has ribose sugar ring, with a hydroxyl group (OH) If RNA in B-from conformation there would be unfavorable steric contact between the hydroxyl group, base, and phosphate backbone. RNA structure
Reason: RNA has ribose sugar ring, with a hydroxyl group (OH) If RNA in B-from conformation there would be unfavorable steric contact between the hydroxyl group, base, and phosphate backbone. RNA structure
ARUNAI ACADEMY FOR PG TRB-BOTANY DHARMAPURI REPLICATION - ENZYMES.
 ARUNAI ACADEMY FOR PG TRB-BOTANY DHARMAPURI.9500244679 REPLICATION - ENZYMES DNA HELICASE Sparation of two strands- DNA helicase enzyme functions Unwinds DNA. DNA double helix by breaking the hydrogen
ARUNAI ACADEMY FOR PG TRB-BOTANY DHARMAPURI.9500244679 REPLICATION - ENZYMES DNA HELICASE Sparation of two strands- DNA helicase enzyme functions Unwinds DNA. DNA double helix by breaking the hydrogen
DNA Replication semiconservative replication conservative replication dispersive replication DNA polymerase
 DNA Replication DNA Strands are templates for DNA synthesis: Watson and Crick suggested that the existing strands of DNA served as a template for the producing of new strands, with bases being added to
DNA Replication DNA Strands are templates for DNA synthesis: Watson and Crick suggested that the existing strands of DNA served as a template for the producing of new strands, with bases being added to
All This For Four Letters!?! DNA and Its Role in Heredity
 All This For Four Letters!?! DNA and Its Role in Heredity What Is the Evidence that the Gene Is DNA? By the 1920s, it was known that chromosomes consisted of DNA and proteins. A new dye stained DNA and
All This For Four Letters!?! DNA and Its Role in Heredity What Is the Evidence that the Gene Is DNA? By the 1920s, it was known that chromosomes consisted of DNA and proteins. A new dye stained DNA and
DNA REPLICATION. DNA structure. Semiconservative replication. DNA structure. Origin of replication. Replication bubbles and forks.
 DNA REPLICATION 5 4 Phosphate 3 DNA structure Nitrogenous base 1 Deoxyribose 2 Nucleotide DNA strand = DNA polynucleotide 2004 Biology Olympiad Preparation Program 2 2004 Biology Olympiad Preparation Program
DNA REPLICATION 5 4 Phosphate 3 DNA structure Nitrogenous base 1 Deoxyribose 2 Nucleotide DNA strand = DNA polynucleotide 2004 Biology Olympiad Preparation Program 2 2004 Biology Olympiad Preparation Program
Repetitive shuttling of a motor protein on DNA
 Vol 437 27 October 2005 doi:10.1038/nature04049 Repetitive shuttling of a motor protein on DNA Sua Myong 1, Ivan Rasnik 1, Chirlmin Joo 1, Timothy M. Lohman 3 & Taekjip Ha 1,2 Many helicases modulate recombination,
Vol 437 27 October 2005 doi:10.1038/nature04049 Repetitive shuttling of a motor protein on DNA Sua Myong 1, Ivan Rasnik 1, Chirlmin Joo 1, Timothy M. Lohman 3 & Taekjip Ha 1,2 Many helicases modulate recombination,
Replication, Transcription, and Translation
 Replication, Transcription, and Translation Information Flow from DNA to Protein The Central Dogma of Molecular Biology Replication is the copying of DNA in the course of cell division. Transcription is
Replication, Transcription, and Translation Information Flow from DNA to Protein The Central Dogma of Molecular Biology Replication is the copying of DNA in the course of cell division. Transcription is
DNA Replication Query Query
 DNA Replication 1. Query: How is the origin opened during the initiation step of replication of E. coli DNA? What is the role of negative supercoiling in this process? How does the DnaA protein help the
DNA Replication 1. Query: How is the origin opened during the initiation step of replication of E. coli DNA? What is the role of negative supercoiling in this process? How does the DnaA protein help the
DNA Recombination II Biochemistry 302. Bob Kelm February 4, 2004
 DNA Recombination II Biochemistry 302 Bob Kelm February 4, 2004 Core interactions at junction crossover (A 6 C 7 C 8 or R 6 -C 7 -Y 8 motif) parallel stacked-x WC H-bonds red dots ACC H-bonds blue dots
DNA Recombination II Biochemistry 302 Bob Kelm February 4, 2004 Core interactions at junction crossover (A 6 C 7 C 8 or R 6 -C 7 -Y 8 motif) parallel stacked-x WC H-bonds red dots ACC H-bonds blue dots
Expression of the genome. Books: 1. Molecular biology of the gene: Watson et al 2. Genetics: Peter J. Russell
 Expression of the genome Books: 1. Molecular biology of the gene: Watson et al 2. Genetics: Peter J. Russell 1 Transcription 1. Francis Crick (1956) named the flow of information from DNA RNA protein the
Expression of the genome Books: 1. Molecular biology of the gene: Watson et al 2. Genetics: Peter J. Russell 1 Transcription 1. Francis Crick (1956) named the flow of information from DNA RNA protein the
CELL BIOLOGY: DNA. Generalized nucleotide structure: NUCLEOTIDES: Each nucleotide monomer is made up of three linked molecules:
 BIOLOGY 12 CELL BIOLOGY: DNA NAME: IMPORTANT FACTS: Nucleic acids are organic compounds found in all living cells and viruses. Two classes of nucleic acids: 1. DNA = ; found in the nucleus only. 2. RNA
BIOLOGY 12 CELL BIOLOGY: DNA NAME: IMPORTANT FACTS: Nucleic acids are organic compounds found in all living cells and viruses. Two classes of nucleic acids: 1. DNA = ; found in the nucleus only. 2. RNA
Storage and Expression of Genetic Information
 Storage and Expression of Genetic Information 29. DNA structure, Replication and Repair ->Ch 25. DNA metabolism 30. RNA Structure, Synthesis and Processing ->Ch 26. RNA metabolism 31. Protein Synthesis
Storage and Expression of Genetic Information 29. DNA structure, Replication and Repair ->Ch 25. DNA metabolism 30. RNA Structure, Synthesis and Processing ->Ch 26. RNA metabolism 31. Protein Synthesis
Requirements for the Genetic Material
 Requirements for the Genetic Material 1. Replication Reproduced and transmitted faithfully from cell to cell-generation to generation. 2. Information Storage Biologically useful information in a stable
Requirements for the Genetic Material 1. Replication Reproduced and transmitted faithfully from cell to cell-generation to generation. 2. Information Storage Biologically useful information in a stable
CH 4 - DNA. DNA = deoxyribonucleic acid. DNA is the hereditary substance that is found in the nucleus of cells
 CH 4 - DNA DNA is the hereditary substance that is found in the nucleus of cells DNA = deoxyribonucleic acid» its structure was determined in the 1950 s (not too long ago).» scientists were already investigating
CH 4 - DNA DNA is the hereditary substance that is found in the nucleus of cells DNA = deoxyribonucleic acid» its structure was determined in the 1950 s (not too long ago).» scientists were already investigating
DNA Structure. DNA: The Genetic Material. Chapter 14
 DNA: The Genetic Material Chapter 14 DNA Structure DNA is a nucleic acid. The building blocks of DNA are nucleotides, each composed of: a 5-carbon sugar called deoxyribose a phosphate group (PO 4 ) a nitrogenous
DNA: The Genetic Material Chapter 14 DNA Structure DNA is a nucleic acid. The building blocks of DNA are nucleotides, each composed of: a 5-carbon sugar called deoxyribose a phosphate group (PO 4 ) a nitrogenous
GENETICS - CLUTCH CH.8 DNA REPLICATION.
 !! www.clutchprep.com CONCEPT: SEMICONSERVATIVE REPLICATION Before replication was understood, there were three of how DNA is replicated Conservative replication states that after replication, there is
!! www.clutchprep.com CONCEPT: SEMICONSERVATIVE REPLICATION Before replication was understood, there were three of how DNA is replicated Conservative replication states that after replication, there is
DNA vs. RNA DNA: deoxyribonucleic acid (double stranded) RNA: ribonucleic acid (single stranded) Both found in most bacterial and eukaryotic cells RNA
 DNA Replication DNA vs. RNA DNA: deoxyribonucleic acid (double stranded) RNA: ribonucleic acid (single stranded) Both found in most bacterial and eukaryotic cells RNA molecule can assume different structures
DNA Replication DNA vs. RNA DNA: deoxyribonucleic acid (double stranded) RNA: ribonucleic acid (single stranded) Both found in most bacterial and eukaryotic cells RNA molecule can assume different structures
Please write your name or student ID number on every page.
 MCB 110 First Exam A TOTAL OF SIX PAGES NAME: Student ID Number: Question Maximum Points Your Points I 30 II 30 III 30 IV 38 V 22 150 Please write your name or student ID number on every page. This exam
MCB 110 First Exam A TOTAL OF SIX PAGES NAME: Student ID Number: Question Maximum Points Your Points I 30 II 30 III 30 IV 38 V 22 150 Please write your name or student ID number on every page. This exam
Biochemistry 111. Carl Parker x A Braun
 Biochemistry 111 Carl Parker x6368 101A Braun csp@caltech.edu Central Dogma of Molecular Biology DNA-Dependent RNA Polymerase Requires a DNA Template Synthesizes RNA in a 5 to 3 direction Requires ribonucleoside
Biochemistry 111 Carl Parker x6368 101A Braun csp@caltech.edu Central Dogma of Molecular Biology DNA-Dependent RNA Polymerase Requires a DNA Template Synthesizes RNA in a 5 to 3 direction Requires ribonucleoside
Towards DNA-AddAB Interactions In Magnetic Tweezers
 Towards DNA-AddAB Interactions In Magnetic Tweezers M. A. Otte September 2007 - February 2008 Supervisors: Dr. F. Moreno - Herrero Dr. N. H. Dekker Centro de Investigacion de Nanociencia y Nanotecnologia
Towards DNA-AddAB Interactions In Magnetic Tweezers M. A. Otte September 2007 - February 2008 Supervisors: Dr. F. Moreno - Herrero Dr. N. H. Dekker Centro de Investigacion de Nanociencia y Nanotecnologia
MCB 110 Spring 2017 Exam 1 SIX PAGES
 MCB 110 Spring 2017 Exam 1 SIX PAGES NAME: SID Number: Question Maximum Points Your Points I 28 II 32 III 32 IV 30 V 28 150 PLEASE WRITE your NAME or SID number on each page. This exam must be written
MCB 110 Spring 2017 Exam 1 SIX PAGES NAME: SID Number: Question Maximum Points Your Points I 28 II 32 III 32 IV 30 V 28 150 PLEASE WRITE your NAME or SID number on each page. This exam must be written
Student name ID # Second Mid Term Exam, Biology 2020, Spring 2002 Scores Total
 Second Mid Term Exam, Biology 2020, Spring 2002 Scores 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. 11. 12. 13. 14. 15. 16. 17. 18. 19. 20. 21. Total 1 1. Matching (7 pts). Each answer is used exactly once Helicase
Second Mid Term Exam, Biology 2020, Spring 2002 Scores 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. 11. 12. 13. 14. 15. 16. 17. 18. 19. 20. 21. Total 1 1. Matching (7 pts). Each answer is used exactly once Helicase
BIOLOGY 101. CHAPTER 16: The Molecular Basis of Inheritance: Life s Operating Instructions
 BIOLOGY 101 CHAPTER 16: The Molecular Basis of Inheritance: Life s Operating Instructions Life s Operating Instructions CONCEPTS: 16.1 DNA is the genetic material 16.2 Many proteins work together in DNA
BIOLOGY 101 CHAPTER 16: The Molecular Basis of Inheritance: Life s Operating Instructions Life s Operating Instructions CONCEPTS: 16.1 DNA is the genetic material 16.2 Many proteins work together in DNA
Exam 2 Key - Spring 2008 A#: Please see us if you have any questions!
 Page 1 of 5 Exam 2 Key - Spring 2008 A#: Please see us if you have any questions! 1. A mutation in which parts of two nonhomologous chromosomes change places is called a(n) A. translocation. B. transition.
Page 1 of 5 Exam 2 Key - Spring 2008 A#: Please see us if you have any questions! 1. A mutation in which parts of two nonhomologous chromosomes change places is called a(n) A. translocation. B. transition.
Fig. 16-7a. 5 end Hydrogen bond 3 end. 1 nm. 3.4 nm nm
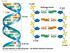 Fig. 16-7a end Hydrogen bond end 1 nm 3.4 nm 0.34 nm (a) Key features of DNA structure end (b) Partial chemical structure end Fig. 16-8 Adenine (A) Thymine (T) Guanine (G) Cytosine (C) Concept 16.2: Many
Fig. 16-7a end Hydrogen bond end 1 nm 3.4 nm 0.34 nm (a) Key features of DNA structure end (b) Partial chemical structure end Fig. 16-8 Adenine (A) Thymine (T) Guanine (G) Cytosine (C) Concept 16.2: Many
DNA Replication and Repair
 DN Replication and Repair http://hyperphysics.phy-astr.gsu.edu/hbase/organic/imgorg/cendog.gif DN Replication genetic information is passed on to the next generation semi-conservative Parent molecule with
DN Replication and Repair http://hyperphysics.phy-astr.gsu.edu/hbase/organic/imgorg/cendog.gif DN Replication genetic information is passed on to the next generation semi-conservative Parent molecule with
Transcription & post transcriptional modification
 Transcription & post transcriptional modification Transcription The synthesis of RNA molecules using DNA strands as the templates so that the genetic information can be transferred from DNA to RNA Similarity
Transcription & post transcriptional modification Transcription The synthesis of RNA molecules using DNA strands as the templates so that the genetic information can be transferred from DNA to RNA Similarity
Unit IX Problem 3 Genetics: Basic Concepts in Molecular Biology
 Unit IX Problem 3 Genetics: Basic Concepts in Molecular Biology - The central dogma (principle) of molecular biology: Information from DNA are transcribed to mrna which will be further translated to synthesize
Unit IX Problem 3 Genetics: Basic Concepts in Molecular Biology - The central dogma (principle) of molecular biology: Information from DNA are transcribed to mrna which will be further translated to synthesize
Biological Nanomachines
 Biological Nanomachines Yann R Chemla Dept. of Physics, University of Illinois at Urbana Champaign Saturday Physics for Everyone, Sept. 14, 2013 Biophysics at Illinois Part I: WHAT IS BIOPHYSICS? Physicists
Biological Nanomachines Yann R Chemla Dept. of Physics, University of Illinois at Urbana Champaign Saturday Physics for Everyone, Sept. 14, 2013 Biophysics at Illinois Part I: WHAT IS BIOPHYSICS? Physicists
Gene Expression- Protein Synthesis
 Gene Expression- Protein Synthesis Protein synthesis is the key to expression of biological information - Structural Protein: bones, cartilage - Contractile Proteins: myosin, actin - Enzymes - Transport
Gene Expression- Protein Synthesis Protein synthesis is the key to expression of biological information - Structural Protein: bones, cartilage - Contractile Proteins: myosin, actin - Enzymes - Transport
The flow of Genetic information
 The flow of Genetic information http://highered.mcgrawhill.com/sites/0072507470/student_view0/chapter3/animation dna_replication quiz_1_.html 1 DNA Replication DNA is a double-helical molecule Watson and
The flow of Genetic information http://highered.mcgrawhill.com/sites/0072507470/student_view0/chapter3/animation dna_replication quiz_1_.html 1 DNA Replication DNA is a double-helical molecule Watson and
Biochemistry 302, February 11, 2004 Exam 1 (100 points) 1. What form of DNA is shown on this Nature Genetics cover? Z-DNA or left-handed DNA
 1 Biochemistry 302, February 11, 2004 Exam 1 (100 points) Name I. Structural recognition (very short answer, 2 points each) 1. What form of DNA is shown on this Nature Genetics cover? Z-DNA or left-handed
1 Biochemistry 302, February 11, 2004 Exam 1 (100 points) Name I. Structural recognition (very short answer, 2 points each) 1. What form of DNA is shown on this Nature Genetics cover? Z-DNA or left-handed
Quiz 9. Work Song. Speed Bump. Theme Music: Cannonball Adderly. Cartoon: DaveCoverley. Physics /23/11. Prof. E. F. Redish 1
 Physics 131 11/23/11 November 23, 2011 Physics 131 Prof. E. F. Redish heme Music: Cannonball Adderly Work Song Cartoon: DaveCoverley Speed Bump 11/23/11 Physics 131 1 Quiz 9 9.1 9.2 9.3 A 32% 26% 68% B
Physics 131 11/23/11 November 23, 2011 Physics 131 Prof. E. F. Redish heme Music: Cannonball Adderly Work Song Cartoon: DaveCoverley Speed Bump 11/23/11 Physics 131 1 Quiz 9 9.1 9.2 9.3 A 32% 26% 68% B
STRUCTURE OF RNA. Long unbranched,single stranded polymer of ribonucleotide units. A ribonucleotide unit has: 5-Carbon ribose sugar.
 STRUCTURE OF RNA & REPLICATION BY:HIMANSHU LATAWA BIOLOGY LECTURER G.G.S.S.SIRHIND MANDI anshu223@gmail.com 9815543311 STRUCTURE OF RNA Long unbranched,single stranded polymer of ribonucleotide units.
STRUCTURE OF RNA & REPLICATION BY:HIMANSHU LATAWA BIOLOGY LECTURER G.G.S.S.SIRHIND MANDI anshu223@gmail.com 9815543311 STRUCTURE OF RNA Long unbranched,single stranded polymer of ribonucleotide units.
DNA Model Stations. For the following activity, you will use the following DNA sequence.
 Name: DNA Model Stations DNA Replication In this lesson, you will learn how a copy of DNA is replicated for each cell. You will model a 2D representation of DNA replication using the foam nucleotide pieces.
Name: DNA Model Stations DNA Replication In this lesson, you will learn how a copy of DNA is replicated for each cell. You will model a 2D representation of DNA replication using the foam nucleotide pieces.
Chapter 8 Lecture Outline. Transcription, Translation, and Bioinformatics
 Chapter 8 Lecture Outline Transcription, Translation, and Bioinformatics Replication, Transcription, Translation n Repetitive processes Build polymers of nucleotides or amino acids n All have 3 major steps
Chapter 8 Lecture Outline Transcription, Translation, and Bioinformatics Replication, Transcription, Translation n Repetitive processes Build polymers of nucleotides or amino acids n All have 3 major steps
MCB 110 SP 2005 Second Midterm
 MCB110 SP 2005 Midterm II Page 1 ANSWER KEY MCB 110 SP 2005 Second Midterm Question Maximum Points Your Points I 40 II 40 III 36 IV 34 150 NOTE: This is my answer key (KC). The graders were given disgression
MCB110 SP 2005 Midterm II Page 1 ANSWER KEY MCB 110 SP 2005 Second Midterm Question Maximum Points Your Points I 40 II 40 III 36 IV 34 150 NOTE: This is my answer key (KC). The graders were given disgression
MIDTERM I NAME: Student ID Number: I 32 II 33 III 24 IV 30 V
 MIDTERM I NAME: Student ID Number: Question Maximum Points Your Points I 32 II 33 III 24 IV 30 V 31 150 Please write your name/student ID number on each of the following five pages. This exam must be written
MIDTERM I NAME: Student ID Number: Question Maximum Points Your Points I 32 II 33 III 24 IV 30 V 31 150 Please write your name/student ID number on each of the following five pages. This exam must be written
1. True or False? At the DNA level, recombination is initiated by a single stranded break in a DNA molecule.
 1. True or False? At the DNA level, recombination is initiated by a single stranded break in a DNA molecule. 2. True or False? Dideoxy sequencing is a chain initiation method of DNA sequencing. 3. True
1. True or False? At the DNA level, recombination is initiated by a single stranded break in a DNA molecule. 2. True or False? Dideoxy sequencing is a chain initiation method of DNA sequencing. 3. True
Zoo-342 Molecular biology Lecture 2. DNA replication
 Zoo-342 Molecular biology Lecture 2 DNA replication DNA replication DNA replication is the process in which one doubled-stranded DNA molecule is used to create two double-stranded molecules with identical
Zoo-342 Molecular biology Lecture 2 DNA replication DNA replication DNA replication is the process in which one doubled-stranded DNA molecule is used to create two double-stranded molecules with identical
BIOCHEMISTRY REVIEW. Overview of Biomolecules. Chapter 12 Transcription
 BIOCHEMISTRY REVIEW Overview of Biomolecules Chapter 12 Transcription 2 3 4 5 Are You Getting It?? Which are general characteristics of transcription? (multiple answers) a) An entire DNA molecule is transcribed
BIOCHEMISTRY REVIEW Overview of Biomolecules Chapter 12 Transcription 2 3 4 5 Are You Getting It?? Which are general characteristics of transcription? (multiple answers) a) An entire DNA molecule is transcribed
DNA is the genetic material. DNA structure. Chapter 7: DNA Replication, Transcription & Translation; Mutations & Ames test
 DNA is the genetic material Chapter 7: DNA Replication, Transcription & Translation; Mutations & Ames test Dr. Amy Rogers Bio 139 General Microbiology Hereditary information is carried by DNA Griffith/Avery
DNA is the genetic material Chapter 7: DNA Replication, Transcription & Translation; Mutations & Ames test Dr. Amy Rogers Bio 139 General Microbiology Hereditary information is carried by DNA Griffith/Avery
DNA, RNA, Replication and Transcription
 Harriet Wilson, Lecture Notes Bio. Sci. 4 - Microbiology Sierra College DNA, RNA, Replication and Transcription The metabolic processes described earlier (glycolysis, cellular respiration, photophosphorylation,
Harriet Wilson, Lecture Notes Bio. Sci. 4 - Microbiology Sierra College DNA, RNA, Replication and Transcription The metabolic processes described earlier (glycolysis, cellular respiration, photophosphorylation,
DNA Transcription. Visualizing Transcription. The Transcription Process
 DNA Transcription By: Suzanne Clancy, Ph.D. 2008 Nature Education Citation: Clancy, S. (2008) DNA transcription. Nature Education 1(1) If DNA is a book, then how is it read? Learn more about the DNA transcription
DNA Transcription By: Suzanne Clancy, Ph.D. 2008 Nature Education Citation: Clancy, S. (2008) DNA transcription. Nature Education 1(1) If DNA is a book, then how is it read? Learn more about the DNA transcription
CHAPTER 22: Nucleic Acids & Protein Synthesis. General, Organic, & Biological Chemistry Janice Gorzynski Smith
 CHAPTER 22: Nucleic Acids & Protein Synthesis General, rganic, & Biological Chemistry Janice Gorzynski Smith CHAPTER 22: Nucleic Acids & Protein Synthesis Learning bjectives: q Nucleosides & Nucleo@des:
CHAPTER 22: Nucleic Acids & Protein Synthesis General, rganic, & Biological Chemistry Janice Gorzynski Smith CHAPTER 22: Nucleic Acids & Protein Synthesis Learning bjectives: q Nucleosides & Nucleo@des:
Fidelity of DNA polymerase
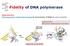 Fidelity of DNA polymerase Shape selectivity: DNA polymerase's conformational change for determination of fidelity for each nucleotide Induced fit: Structure determines function Matched nucleotide Fidelity
Fidelity of DNA polymerase Shape selectivity: DNA polymerase's conformational change for determination of fidelity for each nucleotide Induced fit: Structure determines function Matched nucleotide Fidelity
The Genetic Material. The Genetic Material. The Genetic Material. DNA: The Genetic Material. Chapter 14
 DNA: Chapter 14 Frederick Griffith, 1928 studied Streptococcus pneumoniae, a pathogenic bacterium causing pneumonia there are 2 strains of Streptococcus: - S strain is virulent - R strain is nonvirulent
DNA: Chapter 14 Frederick Griffith, 1928 studied Streptococcus pneumoniae, a pathogenic bacterium causing pneumonia there are 2 strains of Streptococcus: - S strain is virulent - R strain is nonvirulent
Name: - Bio A.P. DNA Replication & Protein Synthesis
 Name: - Bio A.P. DNA Replication & Protein Synthesis 1 ESSENTIAL KNOWLEDGE Big Idea 3: Living Systems store, retrieve, transmit and respond to information critical to living systems Enduring Understanding:
Name: - Bio A.P. DNA Replication & Protein Synthesis 1 ESSENTIAL KNOWLEDGE Big Idea 3: Living Systems store, retrieve, transmit and respond to information critical to living systems Enduring Understanding:
