Characterization of Star and its interactions with sevenless and EGF receptor during photoreceptor cell development in Drosophila
|
|
|
- Priscilla Collins
- 6 years ago
- Views:
Transcription
1 Development 120, (1994) Printed in Great Britain The Company of Biologists Limited Characterization of Star and its interactions with sevenless and EGF receptor during photoreceptor cell development in Drosophila Alex L. Kolodkin, 1, * Amanda T. Pickup, 2, * David M. Lin, 1 Corey S. Goodman 1 and Utpal Banerjee 2, 1 Howard Hughes Medical Institute, Department of Molecular and Cell Biology, LSA 519, University of California, Berkeley, CA 94720, USA 2 Department of Biology, Molecular Biology Institute and Brain Research Institute, University of California, Los Angeles, CA 90024, USA *These two authors have contributed equally to this project Author for correspondence SUMMARY Loss-of-function mutations in Star impart a dominant rough eye phenotype and, when homozygous, are embryonic lethal with ventrolateral cuticular defects. We have cloned the Star gene and show that it encodes a novel protein with a putative transmembrane domain. Star transcript is expressed in a dynamic pattern in the embryo including in cells of the ventral midline. In the larval eye disc, Star is expressed first at the morphogenetic furrow, then in the developing R2, R5 and R8 cells as well as in the posterior clusters of the disc in additional R cells. Star interacts with Drosophila EGF receptor in the eye and mosaic analysis of Star in the larval eye disc reveals that homozygous Star patches contain no developing R cells. Taken together with the expression pattern at the morphogenetic furrow, these results demonstrate an early role for Star in photoreceptor development. Additionally, lossof-function mutations in Star act as suppressors of R7 development in a sensitized genetic background involving the Son of sevenless (Sos) locus, and overexpression of Star enhances R7 development in this genetic background. Based on the genetic interactions with Sos, we suggest that Star also has a later role in photoreceptor development including the recruitment of the R7 cell through the sevenless pathway. Key words: Drosophila melanogaster, eye development, spitz group genes, neuronal determination, embryonic midline INTRODUCTION Signal transduction pathways have been shown to mediate many determinative events that underlie development. Cell fate changes and directed cell movements often require intercellular and intracellular signaling events in order to enact developmental programs. Analysis of these pathways in Drosophila and Caenorhabditis elegans has resulted in the identification and detailed characterization of several molecules that function in signal transduction events during development (reviewed in Greenwald and Rubin, 1992). Genetic analysis affords the opportunity to identify additional members of these pathways, and we describe here the involvement of the Star gene in two different signaling pathways in Drosophila eye development. The Star mutation was isolated by Bridges and Morgan (1919) on the basis of its dominant rough eye phenotype. The regular array of facets in the eye was found to be disrupted. Subsequently, Lewis (1945) described the chromosomal region that includes Star and the neighboring asteroid rough eye mutation. Star and asteroid mutations show strong interactions, fail to complement one another for the eye phenotype, map extremely close to each other, and yet are separable by rare recombination events. It therefore remained an open question as to whether they are mutations in the same gene. A role for Star during embryonic development was first suggested by Mayer and Nusslein-Volhard (1988), who showed that this gene is involved in dorsoventral pattern formation in the embryo. Based on their phenotypic similarities, a group of genes including Star (S), spitz (spi), rhomboid (rho) and pointed (pnt) were called the 'spitz group' genes. Mutations in any of these loci cause embryonic lethality and share similar pattern defects in the derivatives of the ventral ectoderm. In addition to cuticular defects (Kim and Crews, 1993), mutant embryos also display severe central nervous system (CNS) abnormalities. These defects are caused by the improper positioning or development of the cells along the ventral midline of the embryo. In the case of rho, spi and S, the anterior and posterior commissures are fused, owing to the failure of the midline glial cells to differentiate and migrate properly (Klämbt et al., 1991). Star and other members of the spitz group genes are also involved in the development of the female germ line and in the formation of a subset of the sensory organs of the peripheral nervous system (PNS) (Mayer and Nusslein-Volhard, 1988; Rutledge et al., 1992; Bier et al.,1990). The discovery that spitz encodes an EGF-like molecule has
2 1732 A. Kolodkin and others led to the suggestion that the spitz group genes may participate in a signaling pathway that includes the Drosophila EGF receptor (Egfr) (Rutledge et al., 1992). In fact, lethal loss-offunction alleles, Egfr flb, have ventrolateral defects that are hallmarks of the spitz group genes (Schejter and Shilo, 1989). In addition, dosage-sensitive interactions between other Egfr alleles and the spitz group genes have been found. For example, the wing phenotype of the viable rho VE mutation is suppressed by Egfr Elp alleles (Sturtevant et al., 1993), and Star enhances the wing phenotype of Egfr top mutations (J. Price and T. Schüpbach, personal communication). Here we present results that are consistent with the idea that Star and Egfr participate in a common signal transduction pathway. While the Drosophila embryo provides an ideal system for the analysis of mutants affecting pattern formation, the compound eye offers some advantages for studying genes that have a wide spectrum of developmental functions. This is due to the ease with which developmental events can be studied at the level of identified cells and the availability of sensitized genetic systems, allowing for the functions of these genes to be separated into distinct pathways (Rogge et al., 1991; Simon et al., 1991; Dickson et al., 1993). Two signal transduction pathways affecting photoreceptor cell development in the eye have been studied in great detail (reviewed in Banerjee and Zipursky, 1990; Rubin, 1991). One involves the Sevenless tyrosine kinase receptor (Hafen et al., 1987; Banerjee et al., 1987a) and the other the EGF tyrosine kinase receptor. The Egfr gene product is expressed in developing eye discs (Zak and Shilo, 1992; Baker and Rubin, 1992) and the proper expression of this gene is important for determining the spacing of the neuronal clusters (Baker and Rubin, 1989). Each of these clusters contains eight photoreceptor cells (R cells) called R1-R8. The clusters develop at a morphogenetic front, called the furrow, and are spaced at fixed distances, ensuring the final development of a smooth eye with the facets (also called ommatidia) in regular hexagonal arrays. In dominant Ellipse (Egfr Elp ) mutants, this regular spacing is disrupted (Baker and Rubin, 1992). The photoreceptors R8, R2 and R5 are the first cells to differentiate from amongst a large rosette of cells (Wolff and Ready, 1991). This is followed by the pairwise differentiation of R3 and R4, then R1 and R6. The R7 cell is the last photoreceptor to differentiate in each cluster (Tomlinson and Ready, 1987). Detailed molecular and genetic analysis has established that the neuronal fate of R7 is induced by the previously determined R8 cell. This involves the signaling molecule, Boss, which is expressed on the surface of R8, and the tyrosine kinase receptor Sevenless, expressed on the R7 precursor membrane (Reinke and Zipursky, 1988; Hart et al., 1990; Kramer et al., 1991; Cagan et al., 1992). Sevenless protein is expressed, but not activated, in cells other than R7 (Banerjee et al., 1987b; Tomlinson et al., 1987; Van Vactor et al., 1991). The Sevenless and EGF signaling pathways share many downstream components including Ras1 and Sos (Simon et al., 1991; Bonfini et al., 1992). Mosaic analysis shows that Star functions in R cell development (Heberlein and Rubin, 1991), since homozygous patches of Star alleles produce scars in the adult eye. Interestingly, the ommatidia along the mosaic patch boundary that have followed a normal course of development require wild-type Star function in the R2, R5 and R8 cells. Thus, the function of Star is only required in these three early differentiating cells for the ommatidium to develop normally. In this paper, we report the molecular characterization of the Star gene, its expression in the embryo and a detailed analysis of its role in photoreceptor cell development. Based on various genetic interactions, the spatial expression pattern of the Star transcript, and a clonal analysis of Star in the eye disc, we conclude that normal Star function is important for the early signaling processes that cause R2, R5 and R8 to develop and also for the later Sevenless-mediated pathway that promotes the development of the R7 cell. MATERIALS AND METHODS Screen for second site suppressors of Sos JC2 sev E4 /Y; Sos JC2 /Sos JC2 males were mutagenized with X-irradiation (Grigliatti, 1986) and mated to sev E4 /sev E4 ;CyO/Tft females. A primary screening was done in which flies were allowed to choose between green and UV light (Banerjee et al., 1987a). In this test, suppressors of Sos JC2 with no R7 cells are expected to choose visible light. However, owing to the incomplete penetrance of suppression in sev E4 ;Sos JC2 flies, an 8% level of false positives were found amongst all the mutagenised flies. This method was used as a prescreen to enrich for mutants. All flies choosing visible light were individually mated to sev E4 ; Cyo/Tft females. Their progeny were then screened by the deep pseudopupil method (see Banerjee et al., 1987a) to look for true second site suppressors of R7 development. Mapping of S 104E S 104E was mapped meiotically using the multiply marked chromosome al,dp,b,pr,c,px,sp (obtained from the Indiana Stock Center). The lethality was further mapped using a series of deficiencies (also from the Indiana Stock Center) in the region between al and dp. Star P allele The enhancer trap line S F126 (which was obtained from Y. Hiromi) contains a P element, inserted at the cytological location 21E1-2. This P element is designated FZ and includes the E. coli lacz reporter gene (Mlodzik and Hiromi, 1992). The transposon was mobilized by crossing to flies carrying a stable source of transposase activity (Robertson et al., 1988) and identifying excision events by loss of the rosy + marker. These lines were tested for reversion of the Star phenotype and for presence of the ast phenotype. cdna isolation, sequence analysis and northern analysis A phage genomic library was made in λdash (Stratagene) using DNA from the S F126 line and was subsequently screened using a P-elementspecific probe to isolate flanking genomic DNA. This was used to construct a genomic walk of wild-type DNA of approximately 35 kb. A genomic fragment ~3 kb from the site of the P-element insertion was used to screen clones from a Drosophila embryonic cdna library (Zinn et al., 1988). 20 clones were recovered and one 4 kb clone (called K2C) was sequenced using the dideoxy chain termination method (Sanger et al., 1977) and Sequenase (US Biochemical Corp.). Templates were made from M13 mp10 vectors containing inserts generated by sonication of plasmid clones. K2C was completely sequenced on both strands; oligonucleotides and double strand sequencing of plasmid DNA (Sambrook et al., 1989) were used to fill gaps. The predicted protein sequence was analyzed using the FASTDB and BLASTP programs (Intelligenetics). For northern analysis, total RNA was prepared from staged Drosophila embryos, loaded (40 µg/lane) onto formaldehyde-containing 1% agarose gels
3 Star and its interactions with sevenless and EGF receptor in Drosophila 1733 and blotted onto GeneScreen Plus membranes. The filters were subsequently hybridized with 32 P-labeled probes made from the Star cdna, K2C. Construction of Star hsp70-cdna vector and rescue experiment A 2865 bp SnaB1-DraI restriction fragment from K2C, containing the entire 1796 bp Star ORF, 347 bp of 3 untranslated sequences, and 722 bp of 5 untranslated sequences was cloned into a modified Bluescript (Stratagene) vector that allows for the Star sequences to be isolated as a single KpnI restriction fragment. This KpnI fragment was cloned into the Carnegie 20-derived hsp70 vector HT-4 (Schneuwly et al., 1987) and transformed into flies (Spradling, 1986) to give the S hs.8 and S hs.7 lines. To assay rescue of Star by the hsp70 cdna construct, the S hs.8 insertion was first localized to 47A by standard chromosome in situ hybridization (Ashburner, 1989). S 104E,dp,S hs.8 recombinant chromosomes were produced by mating S 104E,dp,b, pr/sm6a flies to al, S hs.8 /Cyo flies. Recombinant males were identified by backcrossing to a recessively marked chromosome; al,dp,b,pr,c,px,sp and 15 independent recombinants were balanced over SM6a. Brother, sister matings of S 104E,dp,S hs.8 /SM6a flies from each line were made to generate flies of a S 104E,dp,S hs.8 /S 104E,dp,S hs.8 genotype. These flies were viable and had normal eyes, indicating a rescue of S 104E by S hs.8. All recombinant chromosomes were crossed to S 104E and ast 1 to confirm the presence of Star. The recombinant chromosomes were all lethal over S 104E and had mild rough eyes over ast 1. The presence of S hs.8 on the recombinant chromosome was determined by scoring for the linked ry + marker on the hsp70-cdna insertion, and by its ability to enhance R7 development in a sev E4 /Y; Sos JC2 /+ background. An analagous genetic scheme was used to recover ast 1,dp,S hs.8 flies. The presence of ast 1 on the recombinant chromosomes was confirmed by backcrossing to ast 1 ; in all 17 cases this gave a rough eye. Two copies of the hsp70 K2C construct rescued the ast 1 /ast 1 eye phenotype. sevenless enhancer-driven Star cdna A 4 kb EcoRI fragment containing the entire Star cdna was cloned into the Bluescript vector. The insert was excised as a KpnI-NotI fragment and then cloned into the pbd365 vector (a gift from Barry Dickson and Ernst Hafen) which has two sevenless enhancers linked to the hsp70 promoter and this plasmid was transformed into flies. Eleven independently isolated transformant lines were named se- Star1 through se-star11. Whole-mount in situ analysis of embryos and eye discs Whole-mount in situ analysis of Star transcripts in both embryos and eye discs was performed essentially as described (Kopczynski and Muskavitch, 1992), with minor modifications to increase sensitivity (Kopczynski and Goodman, unpublished data). Generating flp patches in the larval eye disc To produce patches of homozygous S 1 in the eye disc (Golic, 1991), virgins from a stock with an FRT element inserted at 33, proximal to Star, were crossed to males from a third chromosome FLP stock (with the FLP recombinase on the Mkrs balancer chromosome, marked with Sb). Female progeny, of the genotype FRT/+; FLP, Sb/+ were mated with males from a recombinant S 1, FRT/SM6a line hours after egg laying, larvae from this cross were heat shocked in a waterbath for 90 minutes at 38.5 C. Eye discs from late third instar larvae were dissected out and stained with mab22c10 (provided by Seymour Benzer) as described in Kramer et al. (1991). From this cross, 1/8th of the progeny are expected to have the genotype FRT/ S 1, FRT; Flp, Sb/+, which allows a FLP-induced recombination event to occur. 600 discs were stained and 60 of them contained patches. We estimate that under these conditions recombination events are occuring at a rate of about 80% of the above genotype. RESULTS Star alleles can be isolated in a sensitized genetic screen Mutations affecting the development of the R7 cell in the eye can be detected in any one of several sensitized genetic backgrounds (Simon et al., 1991; Bonfini et al., 1992; Dickson et al., 1993). Our screen utilizes a dominant mutation in the Son of sevenless (Sos) gene, called Sos JC2, which partially suppresses the mutant phenotype of the sev E4 allele of sevenless (Rogge et al., 1991; Bonfini et al., 1992). In sev E4 /Y; Sos JC2 /+ flies, about 16% of the ommatidia have R7 cells, in contrast to 0% in sev E4 alone. The number of R7 cells developing in this genetic background is sensitive to the dosage of other genes in the pathway. As shown in Table 1, loss of one copy of boss or Ras1, genes participating upstream and downstream of Sos respectively, completely eliminates this suppression (Bonfini et al., 1992). Similarly, the loss of one copy of a gene encoding a molecule with an inhibitory function, the GAP1 gene (Gaul et al., 1992), causes a larger percentage of the ommatidia to develop R7 cells in this genetic background (Rogge et al., 1992). The effect of the loss of one copy of other genes on the suppression level in sev E4 /Y; Sos JC2 /+ flies allowed us to devise a screening technique to identify suppressors and enhancers of R7 formation in this double mutant background. One of the mutants isolated in such a screen was called 104E. When one copy of 104E is placed in a sev E4 /Y; Sos JC2 /+ background, the percentage of ommatidia with R7 cells (the Table 1. Mutations in genes belonging to the Sevenless pathway can be detected as suppressors or enhancers of Sos JC2 Ommatidia with Total ommatidia Genotype central cells (%) counted sev E4 /Y; Sos JC2 / sev E4 /Y; Sos JC2 /+; boss 1 / sev E4 /Y; Sos JC2, +/+, Df(Ras1) sev E4 /Y; Sos JC2 /+; Gap1 sxt / Individual ommatidia were scored for the presence of R7 by using the optical technique of pseudopupil (Franceschini and Kirshfeld, 1971). boss 1 is a null allele of boss (Hart et al., 1990). The deficiency, Df (3R)by10, which includes Ras1 was used. Gap1 sxt is a P allele of the Gap1 gene called sextra (Rogge et al., 1992). Table 2. Star and asteroid mutations are suppressors of Sos JC2 Ommatidia with Total ommatidia Genotype central cells (%) counted sev E4 /Y; Sos JC2 / sev E4 /Y; S 104E, Sos JC2 /+, sev E4 /Y; +, Sos JC2 /S 54, sev E4 /Y; +, Sos JC2 /S 11N23, sev E4 /Y; +, Sos JC2 /S 1, sev E4 /Y; +, Sos JC2 /ast 1, sev E4 /Y; +, Sos JC2 /ast 4, S 1, S 54 and S 11N23 are presumed loss-of-function alleles of Star; ast 1 and ast 4 are loss-of-function asteroid alleles.
4 1734 A. Kolodkin and others level of suppression) drops from the 16% found in sev E4 /Y; Sos JC2 /+ flies to 0% (Table 2). The 104E mutation has a dominant rough eye. This dominant phenotype was mapped by standard recombination between al and dp on the 2nd chromosome. 104E is homozygous lethal and the lethality maps to the same region as the dominant rough eye phenotype. The map position and phenotypes are consistent with 104E being a mutant allele of the Star gene. This was confirmed by standard complementation tests; 104E is completely lethal when placed over the S 54, S 11N23 and S 1 alleles of Star. When these alleles of Star were placed into the sensitized background, they also lower the suppression level (Table 2). The asteroid mutation Fig. 1. Genetic interactions of Star with asteroid, Sos and Egfr flb. (A-D and I-L) Scanning electron micrographs (SEMs); bar=66.7 µm. (E-H and M-P) Light microscope sections (bar=1 µm), of adult eyes. (A,E) Wild type. The regular hexagonal array of facets is seen. The dark structures are rhabdomeres of the photoreceptor (R) cells. At this level of section, rhabdomeres of outer R1-6 cells surround the central R7 rhabdomere. The rhabdomere of R8 is proximal to R7 and is therefore not seen in this section. (B,F) S 1 /+. The regular arrangement of facets is disrupted, resulting in a slightly rough eye. In sections, most of the ommatidia appear wild type, with only occasional examples of ommatidia that are missing R cells. (C,G) ast 1 /ast 1. The eye is quite rough and smaller than in Star/+. In sections, most ommatidia are found to be wild type, with a few cases in which the R cells are missing. (D,H) S 1 /ast 1. The eye is very reduced in size, containing few facets. The defects in external morphology, and also in ommatidial organization, are much more severe than in S 1 /+ or ast 1 /ast 1 alone. The majority of ommatidia are missing one or more R cells. Ommatidia are often separated by islands that contain only pigment cells. (I,M) S 54 /+. The phenotype is similar but slightly weaker than S 1 /+. (J,N) S 54, +/+, Sos x122. The eye is slightly rougher than S 54 /+. The section shows that many more ommatidia are missing central R7 cells and/or outer R1-6 cells than in S 54 /+. Also see Table 3. (K,O) flb 2L65 /+. The eye looks wild type, both externally and in sections. (L,P) +, Egfr flb /S 54, +. The eye phenotypes of S 54 /+ and Egfr flb /+ are synergistically enhanced in this double mutant combination. Externally, the eye is rougher than either S 54 /+ or Egfr flb /+. In sections, many ommatidia have abnormal numbers of R cells (see text for
5 Star and its interactions with sevenless and EGF receptor in Drosophila 1735 was placed into this sensitized genetic background. The ast 1 and the ast 4 mutations lower the suppression level to 1 and 3% respectively. Thus, in the Sos JC2 suppression paradigm, ast alleles have the same effect as Star (Table 2). Adult eye phenotype of Star and asteroid A dominant rough eye phenotype is seen in Star (compare Fig. 1A and B). This external roughness is likely to be largely due to defects in non-photoreceptor cells, like cone and pigment cells. Additionally, the weak external roughness is reflected in rather mild defects in photoreceptor organization. We analyzed these defects further by cutting sections tangential to the ommatidia. As shown in Fig. 1F, most of the ommatidia are wild type. In a small, but reproducible fraction of ommatidia, either R7 or the outer R1-R6 type cells fail to develop (Table 3). All asteroid mutations are viable and recessive, and have rough eyes with minor defects in ommatidial assembly (Fig. 1C,G). In S /ast transheterozygotes, the S /+ eye phenotype is greatly enhanced; the eyes are reduced in size and sections show that many more ommatidia are missing R cells (Fig. 1D,H). Genetic interactions suggest a role for Star in R cell development As discussed above, S /+ flies have mild disruptions in their ommatidial organization. Table 3 shows that, while one copy of a Sos loss-of-function mutation, Sos x122 /+, shows no phenotype, in a S, Sos + /S +, Sos x122 fly, significantly increased numbers of ommatidia are missing R cells as compared to S alone (Fig. 1M,N; Table 3). As shown in Table 3, 45% of the ommatidia in these flies are defective. Of these, 21% of the ommatidia fail to develop R7 cells, even though these flies are wild type for the sevenless and boss loci and 40% of the ommatidia lack one or more of the outer R1-6 cells. At basal planes of section, an R8 cell is found to develop normally in each ommatidium (not shown). In summary, Star not only interacts with the gain-of-function allele (Sos JC2 ) in a sevenless background, but also with a loss-of-function allele (Sos x122 ) in an otherwise wild-type background and, in each case, the development of R7 cells is affected. In addition, an effect on the development of R1-6 can be seen in the interaction with the loss-of-function allele of Sos. To study further the role of Star in eye development, we combined Star with mutations in the Drosophila EGF receptor. We detected a synergistic interaction in the eye between strong alleles of Star and the gain-of-function allele Egfr Elp (Fig. 2). When compared with an Elp 1 /+ eye (Fig. 2B) or a S 1 /+ eye (Fig. 2A), flies of a S 1,+/Elp,+ genotype have a dramatically reduced eye containing only a small number of facets (Fig. 2C). Furthermore, when the Elp allele is combined with one copy of a chromosome that is deleted for both Star and asteroid, Df(2L)ast 2, the reduced eye phenotype is much more pronounced (Fig. 2E). These results show that the Egfr Elp mutation interacts dominantly with loss-of-function alleles of both Star and asteroid. As shown in Fig. 2F, no interaction is seen in the eye when Elp is combined with one copy of S hs.8 in which the Star cdna is overexpressed using the heat shock promoter (see below). In addition to the interactions between Star and Ellipse, we also observed dominant interactions in the eye between Star and loss-of-function mutations in Egfr. In combination with Egfr flb alleles, we observed a significant enhancement of the S /+ phenotype. Over 40% of the ommatidia in S, +/+, Egfr flb flies contain less than the wild-type complement of R cells (Figs 1O, P). This is a synergistic effect, since only 9% of the ommatidia are affected in Star/+ and all Egfr flb /+ ommatidia are wild type. This suggests that Star functions with the EGF receptor in determining the early events of R cell development. The above results are consistent with those from previous studies suggesting that Star and the EGF receptor function together in the development of a variety of tissue types (Sturtevant et al., 1993; J. Price and T. Schüpbach, personal communication). The involvement of Star in the Sevenless and EGFR pathways is consistent with the observations of Heberlein et al. (1993), who have shown that Star interacts with Ras 1. Cloning the Star gene During the course of a mutagenesis designed to identify mutations that affect the embryonic CNS, a lethal P-element insertion (designated S F126 ) exhibiting defects characteristic of the spitz group genes was isolated. Embryos homozygous for S F126 exhibit CNS and cuticle defects characteristic of Star mutations, including deletion of medial portions of the denticle bands and a narrowing of the ventral nerve chord. Standard complementation tests were performed using several Star alleles, establishing S F126 as an allele of Star (data not shown). The cytological location of the insertion in S F126 is 21E1-2, the location of the Star locus. We confirmed that the S F126 phenotype is due to the P-element insertion by mobilizing it (see Materials and Methods) and isolating ry revertants. Out of 22 revertants, 7 were S +, 14 were S (exhibiting both eye and embryonic S phenotypes), and one was phenotypically ast. Taken together, these data show that S F126 is an allele of Star. To identify the Star transcript, genomic DNA flanking the S F126 insertion was isolated and used to conduct a genomic walk encompassing about 35 kb (see Materials and Methods). Table 3. Star interacts with a loss-of-function mutation in Sos Ommatidia with one Ommatidia Ommatidia or more outer with R7 with both R7 cells (R1-R6) missing, but and one or more Total % of Total number missing, but with normal outer (R1-R6) cells defective of ommatidia Genotype with normal R7 (%) R1-R6 (%) missing (%) ommatidia counted sev + /Y; S 54 / sev + /Y; Sos x122 / sev + /Y; +, Sos x122 /S 54, Sos x122 is a lethal loss of function allele of Sos (Rogge et al., 1991).
6 1736 A. Kolodkin and others Genomic sequences were in turn used to probe northern blots, identifying a 4 kb transcript contained within this walk (Fig. 3C). The S F126 insertion is located within the downstream intron of this transcription unit (Fig. 3C), defining this transcript as a good candidate to encode the Star protein. The expression of this transcript is temporally regulated during embryonic development (Fig. 3B), showing peak expression between 10 and 14 hours after egg laying. The onset of Star expression is consistent with the expected temporal requirements for the Star protein as deduced from the observed cuticle and CNS defects in mutant embryos (Mayer and Nusslein- Volhard, 1988; Klämbt et al., 1991). We used the genomic sequences that cross-hybridized to the putative Star transcript to isolate several cdnas from an embryonic Drosophila cdna library (Zinn et al., 1988), all of which showed cross hybridization at high stringency. The longest cdna, called K2C, is 4 kb in length, the same size as the transcript disrupted by the S F126 insertion. This cdna contains a poly(a) tract 18 nucleotides 3 from a polyadenylation signal, and primer extension analysis shows the 5 end of this sequence to be the site of transcription initiation (data not shown). The genomic region encoding the K2C cdna is large, encompassing about 27 kb, and the extent of the controlling region is unknown. To determine the ability of this cdna to rescue the Star phenotype, we made an hsp70 promoterconstruct containing the K2C cdna. This construct was transformed into flies using P-element-mediated transformation (Spradling, 1986). Two independent transformant lines, Star hs.8 and Star hs.7 were isolated. Both of these insertions map to the second chromosome and are homozygous lethal. The Star hs.7 /Star hs.8 genotype is also lethal, suggesting that the recessive lethality is due to the overexpression of the K2C cdna and not due to the sites of the insertions. A small number of escapers can be recovered from both lines, and they have rough eyes when raised without heat shock at room temperature (Fig. 4A), with 24% of the ommatidia showing extra R7-like cells. To rescue the Star phenotype with the K2C cdna, we used the Star hs.8 insertion. In these experiments, the flies were not heat shocked and the effects observed are probably due to the influence of an unknown enhancer element on the hsp70 promoter. We first established that the transformant has a single insertion of the hsp70-k2c construct mapping to the 47A band on the right arm of the second chromosome. Fifteen independent recombinants were isolated, each with a copy of S 104E and the S hs.8 insertion on the same chromosome. All the recombinants are homozygous viable and have wild-type eyes (Fig. 4B,F). Since both Star homozygotes and Star hs.8 homozygotes are lethal, this represents a reciprocal rescue, presumably because the loss of Star function in S /S balances out the overexpression of Star protein in the S hs.8 /S hs.8 genotype. The rescue of Star embryonic lethality by the K2C cdna shows that K2C encodes a Star transcript. The S hs.8 insert was also recombined onto a chromosome containing the ast 1 mutation. When homozygous, the ast 1, S hs.8 /ast 1, S hs.8 flies are viable and have normal eyes, as seen both externally (Fig. 4D) and in sections (Fig. 4H). Thus, the S hs.8 construct rescues the asteroid rough eye phenotype, as well as the star lethality. These data are consistent with asteroid representing a viable class of mutant alleles in the Star gene and show that the K2C cdna corresponds to this gene. Sequence analysis of the Star cdna The K2C cdna was sequenced in its entirety (see Materials and Methods) and was found to contain a single large open Fig. 2. Genetic interactions of Star and asteroid with Egfr Elp. (A-F) SEMs of adult eyes; bar=66.7µm. (A) S 1 /+. The eye is moderately rough. (B) Elp 1 /+. The eye is quite rough and slightly smaller than a wild-type eye (see Fig. 1A). (C) S 1, +/+,Elp 1. The eye is small and contains fewer facets than in S 1 /+ or Elp 1 /+ eyes. (D) Df(2L)ast 2/+. The breakpoints of this deficiency, which eliminates both Star and asteroid function, are 21D1-2; 22B2-3. The eye is mildly rough. (E) Df(2L)ast2, +/+, Elp 1. The eye is much smaller and rougher than Df(2L)ast2/+ or Elp 1 /+ alone and has a more extreme phenotype than S 1,+/+,Elp 1 (compare with 2C). (F) S hs.8,+/elp 1. The eye is indistinguishable from Elp 1 /+ alone (compare with 2B).
7 Star and its interactions with sevenless and EGF receptor in Drosophila 1737 reading frame (ORF), 1.8 kb in length, with approximately 1 kb each of both 5 and 3 untranslated sequences. Conceptual translation of this ORF is shown in Fig. 3A. It encodes a polypeptide of 598 amino acid residues, with a calculated molecular mass of 66 kd. The Star ORF shows no sequence similarity, with the exception of an opa repeat, a histidine-rich region, and a glycine rich region, when compared with other proteins in the PD2 data base using BLASTP (Altschul et al., 1990). The ORF does not contain a signal sequence, however it does contain a sequence starting at amino acid 280, that by hydropathy analysis (Fig. 3A,D) is an excellent candidate for a transmembrane spanning sequence. This sequence contains 22 hydrophobic residues, no internal proline residues, and is flanked by charged amino acids. This putative transmembrane sequence is located in the middle of the ORF and appears to separate the protein into a highly hydrophilic amino-terminal Fig. 3. Molecular analysis of Star. (A) Predicted amino acid sequence of the Star protein based on cdna sequence analysis (nucleotide sequence not shown but entered in data base). The putative transmembrane domain is underlined. Potential N-linked glycosylation sites are noted with closed circles. The boundaries of the putative PEST sequences are marked by carets (residues and residues ). The beginning of the 7 residue domain containing alternating positive and negative charges is marked with an open circle. (B) Northern analysis of the Star transcript during embryonic development. Lanes 1: 0-6 hours; 2: 6-10 hours; 3: hours; 4: hours. Equal amounts of total RNA were used in each lane to generate a developmental northern blot that was hybridized with a probe made from the Star cdna, K2C. (C) Genomic organization of the Star locus. The restriction map of genomic DNA that encodes the Star transcript and the site of the P-element insertion causing the S F126 mutation are shown. The orientation of the P element is indicated by the direction of transcription of the β- galactosidase gene (shown by an arrow). R, EcoR1; S, Sal1; ry, rosy. (D) Hydropathy analysis of the predicted sequence of the Star protein. Numbering of residues is the same as in A. Regions below the line indicate relative hydrophobicity and regions above the line relative hydrophilicity. The putative transmembrane domain is marked with an arrow.
8 1738 A. Kolodkin and others Fig. 4. Rescue of Star and asteroid eye phenotypes by the K2C cdna. (A-D) SEMs (bar=66.7 µm) and (E-H) light microscope distal sections (bar=1 µm), of adult eyes. (A,E) S hs.8 /S hs.8. The S hs.8 /S hs.8 genotype causes recessive lethality. A rare escaper is shown here that has a rough eye. The section shows that most of the ommatidia are normal; in a small number of cases, there are extra R7-like cells. (B,F) S 104E, dp, S hs.8 /S 104E, dp, S hs.8. These flies are fully viable. The eye looks wild type both externally and in sections, showing a rescue of the roughness associated with S 104E and S hs.8. (C,G) ast 1 /ast 1. The eye is reduced in size and very rough, but the ommatidia that do develop usually have the normal complement of R cells. (D,H) ast 1, dp, S hs.8 /ast 1, dp, S hs.8. The external morphology and ommatidial structure are completely wild type, showing full rescue of the ast 1 rough eye phenotype by the K2C cdna. domain of 279 amino acid residues, and a carboxy-terminal domain of 296 residues that is relatively neutral with respect to hydrophilicity (Fig. 3D). These characteristics are consistent with the ORF encoding a type II membrane protein in which the amino-terminal end is cytoplasmic and the residues carboxy-terminal to the transmembrane domain are extracellularly located. Northern analysis utilizing equal amounts of total RNA, free polysome-associated RNA and membrane-bound polysome-associated RNA shows a large enrichment for the Star transcript in RNA associated with membrane-bound polysomes, suggesting that the Star transcript is localized in a manner consistent with it encoding a membrane protein (data not shown). There are five potential sites for N-linked glycosylation (Fig. 3A), two amino-terminal and three carboxyterminal to the transmembrane domain. In the putative aminoterminal cytoplasmic domain, there are two PEST sequences thought to be associated with proteins that have short half lives (Rogers et al., 1986). This cytoplasmic domain also contains a stretch of alternating positive and negative charges (Fig. 3A). This is similar to sequences in c-fos and N-myc leucine zippers proposed to form an alpha-helix (Kohl et al., 1986; Landschultz et al., 1988). Alternating positively and negatively charged amino acids are also found in the cytoplasmic domain of the Toll product (Hashimoto et al., 1988) and the proposed cytoplasmic domain of the Rhomboid protein (Bier et al., 1990). Expression of Star in the embryo and the eye disc The embryonic expression of the Star transcript, as determined by whole-mount in situ hybridization of digoxigenin (-DIG)- labeled RNA probes, is temporally and spatially dynamic (Fig. 5). We first detect expression in the early blastoderm (~stage 4; staging as in Campos-Ortega and Hartenstein, 1985) in a longitudinal ventrolateral domain that is approximately 7-9 cells wide (not shown). As gastrulation proceeds (stage 7) the transcript localization is seen to move ventrally and becomes prominent in a row of cells approximately 5-7 cells wide on either side of the ventral midline (Fig. 5A,B). During the initial stages of germ-band elongation, the two lateral bands of the Star transcript form a single band along the ventral midline that initially is 6-10 cells in width but becomes narrower, so that by stage 8 a single darkly stained band 2-4 cells wide can be seen, with some reduced expression evident in the cells just lateral to the midline. As germband elongation nears completion (stage 9-10; Fig. 5C,E), the midline expression becomes further refined and is elevated in a single band of cells, 1-2 cells wide, that corresponds to the mesectoderm. At this time, dorsoventral epidermal stripes, faintly visible in early stage 7, are more prominent and appear darkest in the central portion of each segment. These stripes become more intense by stage (Fig. 5D), and then decay so that by stage they are not visible. As germband retraction proceeds, the midline Star expression is further refined to small clusters of cells (not shown) that appear to be in the position of the midline glial cells (Klämbt et al., 1991); however, certain identification of these cells (and also lateral body wall cells in the position of PNS organs) will require antibodies to the Star protein. Star expression is also seen in cells of the embryonic brain in the location of the optic lobe anlagen (Campos-Ortega and Hartenstein, 1985), starting at stage 12 and seen more clearly in stage 15 embryos (Fig. 5F). In addition to the tissues described above, Star expression is seen at much lower levels in many other
9 Star and its interactions with sevenless and EGF receptor in Drosophila 1739 tissues between stages 4 and 15. This more general expression is absent when a sense-strand riboprobe is used (not shown). The results of hybridizing DIG-labeled RNA probes to late third instar eye discs are shown in Fig. 6. High levels of the Star transcript are expressed at the morphogenetic furrow in a very narrow band that is only one cluster wide (Fig. 6A,B). The expression is seen in clusters of cells that are regularly spaced, with unstained cells in between. While these groups of cells resemble the early developing photoreceptor clusters, the resolution of this technique does not allow us to make assignments of this expression pattern to individual cell types. Immediately posterior to the morphogenetic furrow, the level of Star transcript drops quite dramatically for three to four columns before being expressed at high levels again in the developing photoreceptor cell clusters (Fig. 6A,B). Initially the transcript is strongly expressed in three cells, which are likely to be R2, R5 and R8 (Fig. 6D). Later in development, other cells in addition to R2, R5 and R8 express Star (Fig. 6E); however, the resolution of this technique does not allow us to determine unambiguously which cells in more mature posterior clusters are expressing the transcript. unlikely to be a consequence of extra R8 cells or of boss misexpression. Since a small change in the level of Boss expression will not be detectable in our assay, we cannot rule out the possibility that the effect on R7 development is indirectly through controlling levels of Boss. The enhancement of the sev E4 /Y;Sos JC2 /+ eye phenotype and the suppression of S 1 /S 1 lethality by S hs.8 are effects seen without the application of heat shock. We assume that this construct is under the influence of an unknown enhancer element causing high levels of expression at 25 C. To express Star cdna using a defined enhancer element, we also isolated eleven independent transformants, se-s1 to se-s11, in which Star is overexpressed in cells where Sevenless is normally expressed using two sevenless enhancers fused to the hsp70 promoter. In contrast to the hs.8 transformant Overexpressed Star facilitates R7 development To study further the role of Star in R7 development, we crossed a single copy of the Star hs.8 insert into the sensitized sev E4 /Y; Sos JC2 /+ background. As shown in Fig. 7D and in Table 4, this construct dominantly enhances R7 development, raising the level of suppression from 17% to 73%. In 13% of these ommatidia an extra R7-like cell is seen. We have not yet established the developmental origin of these extra R7 cells. R8 cells develop normally in this genetic background. Moreover, S hs.8 /CyO larval eye discs stained with an antibody against Boss, exhibit no change in the expression pattern of Boss protein (not shown). Thus, the enhancement of the sev E4 /Y;Sos JC2 /+ phenotype caused by overexpression of the Star gene is Fig. 5. Embryonic expression of the Star transcript. Whole-mount in situ analysis using DIG-labeled Star cdna probes. Anterior is to the left. (A,B) Stage 7 (ventral view; staging as in Campos-Ortega and Hartenstein, 1985) embryo showing Star transcript localization restricted to a row of cells approximately 5-7 cells wide on either side of the ventral midline (arrowheads). (C,E) Stage 9-10 embryo (ventral view) showing midline Star expression restricted to a single row of cells, 1-2 cells wide (arrows), that corresponds to the mesectoderm. Dorsoventral epidermal stripes, visible on either side of the midline (arrowheads), appear restricted to the central portion of each segment. (D) Stage 11 embryo, lateral view, showing segmentally repeated epidermal stripes (arrowhead). (F) Star expression in the optic lobe anlagen of the embryonic brain (arrowhead) as seen in a lateral view of a stage 15 embryo. Bar: (A-D),F: 50µm; E: 25µm.
10 1740 A. Kolodkin and others described above, all of the se-s lines are homozygous viable, consistent with the expression being controlled by the eyespecific sevenless enhancer (se). Ten of the lines, se-s1 to se- S10, show a rough eye when two copies of the insertion are present. For se-s1 to se-s9 a single copy of the insertion gives rise to an eye with wild-type appearance, both externally and in sections. For se-s10, the eye is mildly rough when one copy of the insertion is present. Since the phenotypes for these lines are qualitatively similar to one another, they were analyzed further. The eleventh line, se-s11, shows no roughness, either in one or two copies. Since the phenotype of this atypical line may be influenced by position effects or is due to its site of insertion, it is not described further in this paper. The external eye morphology due to one of the ten typical insertions is shown in Fig. 8A and B. The roughness of these eyes is likely to be due to defects in cone and pigment cells. The defects in photoreceptor cell development can be seen in sections. Qualitatively the phenotypes are similar in all lines, but quantitatively they are quite variable (compare Fig. 8C with D). In the weakest line, se-s3, 99% (n=460) of the ommatidia are wild type and 1% of the ommatidia show an extra cell with either outer cell or R7-like morphology (Fig. 8C). In the strongest insertion, se-s10, 26% of the ommatidia contain extra cells with R7-like morphology, while 27% of the ommatidia have extra cells with R1-6 morphology (n=273). In order to determine the effects of the se-s insertions on R7 development, these lines were crossed into the sensitized sev E4 /Y; Sos JC2 /+ background. All six lines tested act as enhancers of R7 cell development (see Table 5). While we cannot rule out that some of this enhancement is due to leaky expression from the hsp70 promoter in R8 cells, the fact that all these se-s lines act as enhancers suggests to us that expression due to the sevenless enhancer as lead to the development of R7 cells. These results will be further confirmed using a sevenless promoter construct now available (Fortini et al., 1993). number of primary causes. To identify the primary cause for this defect, we made homozygous mutant patches of the strong S 1 allele in the larval eye disc. We used the Flp recombinase method (Golic, 1991) to generate homozygous mutant clones at high frequencies. The eye discs were stained using the neuronspecific antibody mab22c10 and 60 independent homozygous clones, with no staining were identified. Patches were generated at an estimated frequency of 80% (see Materials and Methods for details). As shown in Fig. 9, the phenotype of these clones is a complete absence of staining with mab22c10. The mosaic clones were also double stained with mab22c10 and an antibody against Boss which is specific to the R8 cell. As expected, the mutant clones do not stain with the Boss antibody either (not shown). Defects of this nature are never seen in wildtype controls stained with mab22c10 or Boss antibody. This result suggests that neuronal development does not take place in the eye disc if Star gene function is missing. Similar observations have been made by Heberlein et al. (1993) who have further shown evidence for cell death when Star function is lacking. DISCUSSION In this paper, we report the cloning of the Star gene by P- element tagging. Excision of the P element gives rise to both Star function is necessary for early ommatidial assembly For most essential genes, a later function in photoreceptor cell development can be assessed by analyzing mosaic patches of mutant tissue in the adult eye. This is not possible for Star, since homozygous patches scored in the adult eye are seen as scars containing no photoreceptor cells (Heberlein and Rubin, 1988). This is a terminal phenotype which could have any Fig. 6. Expression of the Star transcript in third instar larval eye discs. Whole-mount in situ analysis on eye discs; posterior to the left. (A,B) Two different examples of eye discs stained in situ with the Star K2C riboprobe are shown. In both cases, a narrow band of signal is seen at the morphogenetic furrow (arrows). The staining is in discrete clusters spaced at regular intervals. The expression level drops sharply posterior to the furrow and rises again 3-4 columns later in developing clusters. (C) No signal is seen in control discs hybridized to a sense RNA probe. (D) The second wave of Star expression is restricted to three cells in each developing cluster, corresponding to the positions of R2, R5 and R8 in the developing clusters. The disc shown in this panel is young, and only one column of clusters shows Star expression. (E) In an older disc, several columns of clusters show Star expression in three cells only (empty arrow). In more mature clusters, more than three cells (solid arrow) express Star. The resolution of this technique does not allow us to unambiguously identify the cells in the more mature clusters.
11 Star and its interactions with sevenless and EGF receptor in Drosophila 1741 Star and asteroid mutants. A 4 kb cdna from the region of the insertion rescues the embryonic lethality of Star mutants and the eye phenotype of asteroid mutants. Taken together, the rescue and excision results imply that asteroid and Star are different classes of mutations in the same gene. The Star gene encodes a 4 kb transcript, which is expressed at the morphogenetic furrow and in the posterior developing clusters in third instar eye discs. In embryos, this transcript is expressed in several tissues, most notably in the mesectoderm and some of its derivatives. The conceptual translation product of the gene suggests that it could encode a novel type II membrane protein, with a hydrophilic intracellular domain and an extracellular domain that is neutral with respect to hydrophilicity. Internal hydrophobic domains are known to mediate membrane insertion of type II membrane proteins, resulting in the hydrophilic amino terminus facing the cytoplasm and an exoplasmic carboxy terminus (Spiess and Lodish, 1986). The charge distribution in the amino terminus of the Star protein is consistent with this interpretation (reviewed in Dalbey, 1990). The amino terminus, particularly the residues adjacent to the putative transmembrane domain, are highly positively charged. The function of Star is fairly widespread during Drosophila Table 4. S hs.7 and S hs.8 enhance Sos JC2 Ommatidia with Total ommatidia Genotype central cells (%) counted sev E4 /Y; Sos JC2 / sev E4 /Y; Sos JC2 /+, S hs sev E4 /Y; Sos JC2 /+, S hs development. Detailed analysis of Star function in embryonic CNS development has established its importance in the correct differentiation of midline glia (Klämbt et al.,1991). Star also functions in germ line, embryonic PNS and establishment of embryonic dorsoventral patterning (Mayer and Nusslein- Volhard, 1988; Rutledge et al., 1992; Bier et al., 1990). A genetically sensitized screening technique, involving a dominant allele of the Son of sevenless gene, allowed us to characterize a role for Star in photoreceptor cell development. This technique utilizes the appearance of R7 cells in a genetic background where the components of the sevenless signaling system are present in limiting amounts. A twofold change in the dose of many gene products involved in R7 development can be detected. A particularly useful feature of this assay is that it can identify genes specific to the development of R7 (e.g. boss), as well as genes whose function is not limited to the development of R7 (e.g. Ras1). Thus, molecules with widespread roles in development can be isolated, and their particular role in R7 development can be determined. A unique feature of this screen is the fact that components upstream and downstream of Sos can be detected. Furthermore, this assay can detect mutations in a single copy of these genes and therefore is able to identify recessive lethal mutations. Since the Sevenless and EGF receptors have been placed in genetically defined pathways that share several downstream molecules (Simon et al., 1991; Bonfini et al., 1992), it is not surprising that many of the genes identified in this screen participate in both pathways. All of the above features were important for us to be able to isolate alleles of Star in this screen. Mutations in Star are embryonic lethal, implying Star functions early in development. In the eye, mosaic analysis showed that Star function is genetically required in R2, R5 and R8 (Heberlein and Rubin, 1991), suggesting that its role in R7 development is likely to be non-autonomous and upstream of the Sevenless receptor. Fig. 7. S hs.8 promotes R7 development. (Bar=1 µm). (A) S hs.8 /+. In heterozygotes, containing only one copy of the K2C cdna insert, all ommatidia are wild type. (B) S hs.8 /S hs.8. In escapers of this genotype, 24% of the ommatidia contain extra R7-like cells. (C) sev E4 ; Sos JC2 /+. In this genetic background, 16% of the ommatidia contain a R7 cell. (D) sev E4 ; Sos JC2 /+, S hs.8. In the sensitized genetic background, 73% of the ommatidia develop R7 cells. This is a significant increase over the percentage of R7 cells seen in C (see Table 4). Table 5. sevenless enhancer-star cdna lines enhance Sos JC2 Ommatidia with Total ommatidia Genotype central cells (%) counted sev E4 /Y; Sos JC2 / sev E4 /Y; Sos JC2 /+, se-s sev E4 /Y; Sos JC2 /+, se-s sev E4 /Y; Sos JC2 /+, se-s sev E4 /Y; Sos JC2 /+; se-s6/ sev E4 /Y; Sos JC2 /+; se-s7/ sev E4 /Y; Sos JC2 /+; se-s10/ The levels of suppression are determined by the pseudopupil method, counting at least 2000 ommatidia for each line as in Table 1, except for the se-s10 transformant. This line gives dominant rough eyes which makes counting by pseudopupil inaccurate. The number of R7 cells in the eyes of sev E4 /Y; Sos JC2 /+; se-s10/+ flies was determined from sections of adult eyes.
12 1742 A. Kolodkin and others However, other possibilities are discussed later. Star also interacts with the EGF receptor, both in the eye and in other tissues, suggesting that it plays a more general role in allowing cells to interact with each other in different contexts. We have found, using mosaic analysis in developing eye discs, that loss of Star function at the furrow prevents R cells from developing. This implies that Star is required at an early step in the establishment of the identity of R8, R2 and R5. A construct containing the Star cdna under the control of the heat-shock promoter rescues the embryonic lethality caused by Star mutations. Embryonic defects seen in Star mutants include deletion of structures along the ventral midline resulting in loss of medial portions of the denticle belts, deletions of certain components of the PNS, and CNS midline defects. Though our analysis of Star expression is limited by the resolution of the whole-mount in situ technique, the pattern of expression that we observe is consistent with the phenotypes seen in Star mutations. The Star transcript is first seen in longitudinal lateral stripes that move ventrally during early embryonic development and resolve, by stage 9-10, into a single stripe 1-2 cells wide along the ventral midline. The Star CNS defect results in fusion of the two commissures, and analysis of the midline glia that appear to be essential for proper separation of the commissural tracts reveals that, in Star mutant embryos, these glia do develop, but subsequently fail to migrate and die (Klämbt et al., 1991). Star is expressed in the mesectoderm, which gives rise to the midline glia, and later during development in cells along the midline that are likely to be the midline glia. The additional early expression of Star in cells lateral to the mesectoderm and later expression in epidermal stripes, in addition to weak expression in the region of the lateral PNS organs, is also expected given the embryonic phenotypes associated with Star mutations (Kim and Crews, 1993); however, antibodies to the Star protein will be needed to characterize more fully these aspects of Star expression. the sequences of Star and Rhomboid share certain characteristics. Both are putative integral membrane proteins with no signal sequence, contain highly charged amino-terminal segments and contain PEST sequences. Additionally, both Star and rho are expressed in overlapping embryonic tissues, most significantly in the mesectoderm and in similar patterns in the third instar larval eye disc. In the eye, the Rhomboid protein is strongly expressed in R2, R5 and R8 (Freeman et al., 1992), the same cells in which Star function is required for correct ommatidial assembly and in which high levels of Star expression are seen. When we tested strong rhomboid mutations in the sensitized sev E4 ; Sos JC2 background, there was no significant effect on the level of suppression (data not shown). However, since rhomboid may have a redundant role in photoreceptor development (Freeman et al., 1992), this does not rule out a function for rho in R7 determination. Overexpression of either Star or rhomboid can give rise to supernumerary photoreceptor cells with outer cell morphology (Fig. 8C,D; Freeman et al., 1992). While the expression pattern in the eye disc and the phenotypes of the overexpressed gene products suggests that Star and Rhomboid may function in common processes in eye development, it remains to be seen whether they are members of the same pathway. An early and a late role for Star in eye development Based on our genetic analysis, we propose that Star function is necessary for the determination of R cell fate, both early at Star and rhomboid share common features Within the spitz group genes, only rhomboid and Star have so far been implicated in eye development. These two genes also seem to have similar functions in other tissues. In the embryonic CNS, both rhomboid and Star affect the development of the midline glial cells (Klämbt et al., 1991). In the embryonic PNS, both mutants have similar reductions in the number of specific sensory organs (Mayer and Nusslein-Volhard, 1988; Bier et al., 1990). Both Star and rhomboid function in wing development, since Star strongly enhances the wing phenotype of the rho VE allele. Based on this result and other genetic interactions, Sturtevant et al. (1993) have proposed that Star and Rhomboid may interact directly in the wing imaginal disc to enhance a ventrolateral signal which controls longitudinal wing vein development. Also, Fig. 8. Star cdna expressed under the control of the sevenless enhancer promotes supernumerary R cell development. (A,C) SEMs of adult eyes; bar=66.7 µm. (B,D) Light microscope sections of adult eyes; bar=1 µm. (A) A single copy of se-s3 leads to a wild-type eye. (C) In two copies, se-s3/se-s3 gives a mild rough eye. (B) se- S4/sE-S4. This is an example of a transformant with a mild phenotype. This section, at the R7 plane, shows one example of an ommatidium with an extra outer cell (arrow). The position of this extra cell resembles the position of the extra outer cells seen when rhomboid is expressed using the sevenless enhancer (Freeman et al. 1992). It is likely that this cell results from the transformation of a mystery cell into a R cell. (D) se- S3/sE-S3. An example of an insertion with a strong eye phenotype. Sections taken at the R7 plane show ommatidia with extra outer cells (arrow) as well as those with extra R7- like cells (arrowheads).
13 Star and its interactions with sevenless and EGF receptor in Drosophila 1743 Fig. 9. Mosaic analysis of Flp-induced S 1 /S 1 patches in the larval eye disc. All eye discs were stained with the neuron-specific marker mab 22C10. (A,B) Two different examples of clones homozygous for the strong S 1 allele. The arrowheads delimit the boundaries of the clones. The clones do not stain with mab22c10, indicating that cells of a S 1 /S 1 genotype fail to develop as photoreceptors. (C) The same clone as in B, at a higher magnification, showing that there are single cells that stain with mab22c10 along the mosaic boundary, but cells within the patch are unstained. the morphogenetic furrow and also later, when the R7 cell identity is established. The early expression of Star in R8, R2 and R5 at the furrow and the later expression in more cells in the developing clusters is consistent with this notion. Mosaic analysis in the adult eye demonstrates that Star is required in the first three photoreceptor cells to develop (Heberlein and Rubin, 1991). The early role of Star in R cell development was further substantiated by generating mosaic patches in the larval eye disc. We found that in the absence of Star function, R2, R5 and R8 fail to develop and, perhaps as a consequence, the rest of the R cells also do not develop. It could be argued that Star has no role to play in the development of R7, instead its only role is in the earlier step when R8 identity is established. In this model, Star ceases to function once R8 development is complete and any effects on R7 development are simply a consequence of the R8 cell not developing at all. Our results do not support such a view. While it is certainly true that mosaic analysis suggests that complete loss of Star function will prevent R8 cells from developing, other genetic tests clearly show that in a wild-type fly, the function of Star is needed at a later stage when R7 cells develop. One line of evidence for this is the effect of increased levels of Star on the development of R7 cells in the sensitized genetic background provided by the sev E4 ; Sos JC2 double mutant. In this background, the R8 cell is normal, but the sevenless-mediated developmental signal is attenuated. When the level of Star is increased, the number of ommatidia with an R7 cell goes up dramatically. This process is dependent on Sos and sevenless, and is therefore utilizing the normal induction process that leads to the development of the R7 cell. Furthermore, while Star /+ and Sos /+ flies have negligible defects in R7 development, a synergistic effect is seen in S, +/+, Sos ommatidia, which often fail to develop R7, even though an R8 cell develops in every ommatidium. Taken together, the results from genetic backgrounds involving gain or loss of function of Star lead us to conclude that an alteration in the level of Star protein not only affects the early determination of R8, but also subsequently affects the proper development of the R7 cell. Mosaic analysis in adult eyes has suggested a role for Star in R2, R5 and R8 for proper ommatidial assembly (Heberlein and Rubin, 1991). The simplest conclusion from the mosaic analysis is that Star function is required non-autonomously in the R8 cell for the R7 cell to develop. The results from this study suggest that the role of Star may be more complex. While early developing clusters express Star in R2, R5 and R8, in mature photoreceptor cell clusters, Star is expressed in other R cells. Also, overexpression of Star in the se-star constructs promotes R7 development, although the sevenless enhancer does not give expression in R8, R2 and R5. Two hypotheses can be proposed. Star may have a redundant, but autonomous function in R7, not evident in adult mosaic patches, and only becoming apparent in a sensitized genetic background where the R7 developmental signal is attenuated. Alternatively, Star function could be required in both R7 and R8, but loss of Star function from either one of these cells is not detrimental to the development of R7. Thus, in the mosaic patches, Star R7 cells can develop if R8 is Star +. Since the early role of Star in R8 cells is essential for neuronal development, an ommatidium with a mutant R8 would never initiate a normal cluster. Star functions in two different signal transduction pathways We have found that Star interacts genetically with mutations in EGFR and Sevenless, both of which function in the eye. We hypothesize that the early role of Star is important for R8 development and is mediated by the participation of Star in the EGF receptor pathway and that the late role of Star, important for R7 induction, is mediated by an involvement in the Sevenless pathway. Star is also involved in many aspects of embryonic development that require the EGFR, including the development of structures that derive from the ventral midline. A likely function for Star could be enhancing or amplifying signals exchanged by different signal transduction systems, thereby enabling a cell to acquire its differentiated state. Future biochemical analysis will reveal if this model is indeed correct. We are grateful to Ulrike Heberlein and Gerald Rubin, and to Ethan Bier for communicating unpublished results. We thank Yasushi Hiromi for the P insertion line S F126, Eric Schwartz for Star and asteroid stocks, Barry Dickson and Ernst Hafen for the sevenless enhancer construct, Lily Jan and Yuh N. Jan for the FRT and Flp
Enhancer genetics Problem set E
 Enhancer genetics Problem set E How are specific cell types generated during development? There are thought to be 10,000 different types of neurons in the human nervous system. How are all of these neural
Enhancer genetics Problem set E How are specific cell types generated during development? There are thought to be 10,000 different types of neurons in the human nervous system. How are all of these neural
Trasposable elements: Uses of P elements Problem set B at the end
 Trasposable elements: Uses of P elements Problem set B at the end P-elements have revolutionized the way Drosophila geneticists conduct their research. Here, we will discuss just a few of the approaches
Trasposable elements: Uses of P elements Problem set B at the end P-elements have revolutionized the way Drosophila geneticists conduct their research. Here, we will discuss just a few of the approaches
a) Stock 4: 126 flies total: 120 are P[w+]: 64 are CyO; 56 are TM3; all are females. 132 are w-: 68 are CyO; 64 are TM3; all are males.
![a) Stock 4: 126 flies total: 120 are P[w+]: 64 are CyO; 56 are TM3; all are females. 132 are w-: 68 are CyO; 64 are TM3; all are males. a) Stock 4: 126 flies total: 120 are P[w+]: 64 are CyO; 56 are TM3; all are females. 132 are w-: 68 are CyO; 64 are TM3; all are males.](/thumbs/88/116538069.jpg) Drosophila Problem Set 2012 1) You have created P element transformants of a construct that contains the mini-white gene, which confers an orange eye color in a homozygous white mutant background. For
Drosophila Problem Set 2012 1) You have created P element transformants of a construct that contains the mini-white gene, which confers an orange eye color in a homozygous white mutant background. For
a) Stock 4: 252 flies total: 120 are P[w+]: 64 are CyO; 56 are TM3; all are females. 132 are w-: 68 are CyO; 64 are TM3; all are males.
![a) Stock 4: 252 flies total: 120 are P[w+]: 64 are CyO; 56 are TM3; all are females. 132 are w-: 68 are CyO; 64 are TM3; all are males. a) Stock 4: 252 flies total: 120 are P[w+]: 64 are CyO; 56 are TM3; all are females. 132 are w-: 68 are CyO; 64 are TM3; all are males.](/thumbs/91/106088469.jpg) Drosophila Problem Set 2018 1) You have created P element transformants of a construct that contains the mini-white gene, which confers an orange eye color in a homozygous white mutant background. For
Drosophila Problem Set 2018 1) You have created P element transformants of a construct that contains the mini-white gene, which confers an orange eye color in a homozygous white mutant background. For
Enhancers mutations that make the original mutant phenotype more extreme. Suppressors mutations that make the original mutant phenotype less extreme
 Interactomics and Proteomics 1. Interactomics The field of interactomics is concerned with interactions between genes or proteins. They can be genetic interactions, in which two genes are involved in the
Interactomics and Proteomics 1. Interactomics The field of interactomics is concerned with interactions between genes or proteins. They can be genetic interactions, in which two genes are involved in the
Map-Based Cloning of Qualitative Plant Genes
 Map-Based Cloning of Qualitative Plant Genes Map-based cloning using the genetic relationship between a gene and a marker as the basis for beginning a search for a gene Chromosome walking moving toward
Map-Based Cloning of Qualitative Plant Genes Map-based cloning using the genetic relationship between a gene and a marker as the basis for beginning a search for a gene Chromosome walking moving toward
Supplementary Methods
 Supplementary Methods MARCM-based forward genetic screen We used ethylmethane sulfonate (EMS) to mutagenize flies carrying FRT 2A and FRT 82B transgenes, which are sites for the FLP-mediated recombination
Supplementary Methods MARCM-based forward genetic screen We used ethylmethane sulfonate (EMS) to mutagenize flies carrying FRT 2A and FRT 82B transgenes, which are sites for the FLP-mediated recombination
A) (5 points) As the starting step isolate genomic DNA from
 GS Final Exam Spring 00 NAME. bub ts is a recessive temperature sensitive mutation in yeast. At º C bub ts cells grow normally, but at º C they die. Use the information below to clone the wild-type BUB
GS Final Exam Spring 00 NAME. bub ts is a recessive temperature sensitive mutation in yeast. At º C bub ts cells grow normally, but at º C they die. Use the information below to clone the wild-type BUB
Lecture 20: Drosophila melanogaster
 Lecture 20: Drosophila melanogaster Model organisms Polytene chromosome Life cycle P elements and transformation Embryogenesis Read textbook: 732-744; Fig. 20.4; 20.10; 20.15-26 www.mhhe.com/hartwell3
Lecture 20: Drosophila melanogaster Model organisms Polytene chromosome Life cycle P elements and transformation Embryogenesis Read textbook: 732-744; Fig. 20.4; 20.10; 20.15-26 www.mhhe.com/hartwell3
Additional Practice Problems for Reading Period
 BS 50 Genetics and Genomics Reading Period Additional Practice Problems for Reading Period Question 1. In patients with a particular type of leukemia, their leukemic B lymphocytes display a translocation
BS 50 Genetics and Genomics Reading Period Additional Practice Problems for Reading Period Question 1. In patients with a particular type of leukemia, their leukemic B lymphocytes display a translocation
Integrins are the receptors than mediate adhesion between the dorsal and ventral surfaces of the wing.
 Problem set 8 answers 1. Integrins are alpha/beta heterodimeric receptors that bind to molecules in the extracellular matrix. A primary role of integrins is in adhesion, ensuring that cells attach to the
Problem set 8 answers 1. Integrins are alpha/beta heterodimeric receptors that bind to molecules in the extracellular matrix. A primary role of integrins is in adhesion, ensuring that cells attach to the
Before starting, write your name on the top of each page Make sure you have all pages
 Biology 105: Introduction to Genetics Name Student ID Before starting, write your name on the top of each page Make sure you have all pages You can use the back-side of the pages for scratch, but we will
Biology 105: Introduction to Genetics Name Student ID Before starting, write your name on the top of each page Make sure you have all pages You can use the back-side of the pages for scratch, but we will
Enzyme that uses RNA as a template to synthesize a complementary DNA
 Biology 105: Introduction to Genetics PRACTICE FINAL EXAM 2006 Part I: Definitions Homology: Comparison of two or more protein or DNA sequence to ascertain similarities in sequences. If two genes have
Biology 105: Introduction to Genetics PRACTICE FINAL EXAM 2006 Part I: Definitions Homology: Comparison of two or more protein or DNA sequence to ascertain similarities in sequences. If two genes have
1a. What is the ratio of feathered to unfeathered shanks in the offspring of the above cross?
 1. Whether or not the shanks of chickens contains feathers is due to two independently assorting genes. Individuals have unfeathered shanks when they are homozygous for recessive genes at two loci; the
1. Whether or not the shanks of chickens contains feathers is due to two independently assorting genes. Individuals have unfeathered shanks when they are homozygous for recessive genes at two loci; the
Lecture 20: Drosophila embryogenesis
 Lecture 20: Drosophila embryogenesis Mitotic recombination/clonal analyses Embrygenesis Four classes of genes: Maternal genes Gap genes Pair-rule genes Segment polarity genes Homeotic genes Read 140-141;
Lecture 20: Drosophila embryogenesis Mitotic recombination/clonal analyses Embrygenesis Four classes of genes: Maternal genes Gap genes Pair-rule genes Segment polarity genes Homeotic genes Read 140-141;
Understanding the Cellular Mechanism of the Excess Microsporocytes I (EMSI) Gene. Andrew ElBardissi, The Pennsylvania State University
 Understanding the Cellular Mechanism of the Excess Microsporocytes I (EMSI) Gene Andrew ElBardissi, The Pennsylvania State University Abstract: Hong Ma, The Pennsylvania State University The Excess Microsporocytes
Understanding the Cellular Mechanism of the Excess Microsporocytes I (EMSI) Gene Andrew ElBardissi, The Pennsylvania State University Abstract: Hong Ma, The Pennsylvania State University The Excess Microsporocytes
Biology 105: Introduction to Genetics PRACTICE FINAL EXAM Part I: Definitions. Homology: Reverse transcriptase. Allostery: cdna library
 Biology 105: Introduction to Genetics PRACTICE FINAL EXAM 2006 Part I: Definitions Homology: Reverse transcriptase Allostery: cdna library Transformation Part II Short Answer 1. Describe the reasons for
Biology 105: Introduction to Genetics PRACTICE FINAL EXAM 2006 Part I: Definitions Homology: Reverse transcriptase Allostery: cdna library Transformation Part II Short Answer 1. Describe the reasons for
TRANSGENIC ANIMALS. -transient transfection of cells -stable transfection of cells. - Two methods to produce transgenic animals:
 TRANSGENIC ANIMALS -transient transfection of cells -stable transfection of cells - Two methods to produce transgenic animals: 1- DNA microinjection - random insertion 2- embryonic stem cell-mediated gene
TRANSGENIC ANIMALS -transient transfection of cells -stable transfection of cells - Two methods to produce transgenic animals: 1- DNA microinjection - random insertion 2- embryonic stem cell-mediated gene
1a. What is the ratio of feathered to unfeathered shanks in the offspring of the above cross?
 Problem Set 5 answers 1. Whether or not the shanks of chickens contains feathers is due to two independently assorting genes. Individuals have unfeathered shanks when they are homozygous for recessive
Problem Set 5 answers 1. Whether or not the shanks of chickens contains feathers is due to two independently assorting genes. Individuals have unfeathered shanks when they are homozygous for recessive
Genome manipulation by homologous recombination in Drosophila Xiaolin Bi and Yikang S. Rong Date received (in revised form): 9th May 2003
 Xiaolin Bi is a post doctoral research fellow at the Laboratory of Molecular Cell Biology, National Cancer Institute, National Institutes of Health, Bethesda, Maryland, USA. Yikang S. Rong is the principal
Xiaolin Bi is a post doctoral research fellow at the Laboratory of Molecular Cell Biology, National Cancer Institute, National Institutes of Health, Bethesda, Maryland, USA. Yikang S. Rong is the principal
(i) A trp1 mutant cell took up a plasmid containing the wild type TRP1 gene, which allowed that cell to multiply and form a colony
 1. S. pombe is a distant relative of baker s yeast (which you used in quiz section). Wild type S. pombe can grow on plates lacking tryptophan (-trp plates). A mutant has been isolated that cannot grow
1. S. pombe is a distant relative of baker s yeast (which you used in quiz section). Wild type S. pombe can grow on plates lacking tryptophan (-trp plates). A mutant has been isolated that cannot grow
The Role of Star in the Production of an Activated Ligand for the EGF Receptor Signaling Pathway
 Developmental Biology 205, 254 259 (1999) Article ID dbio.1998.9119, available online at http://www.idealibrary.com on The Role of Star in the Production of an Activated Ligand for the EGF Receptor Signaling
Developmental Biology 205, 254 259 (1999) Article ID dbio.1998.9119, available online at http://www.idealibrary.com on The Role of Star in the Production of an Activated Ligand for the EGF Receptor Signaling
BSCI410-Liu/Spring 06 Exam #1 Feb. 23, 06
 Your Name: Your UID# 1. (20 points) Match following mutations with corresponding mutagens (X-RAY, Ds transposon excision, UV, EMS, Proflavin) a) Thymidine dimmers b) Breakage of DNA backbone c) Frameshift
Your Name: Your UID# 1. (20 points) Match following mutations with corresponding mutagens (X-RAY, Ds transposon excision, UV, EMS, Proflavin) a) Thymidine dimmers b) Breakage of DNA backbone c) Frameshift
No, because expression of the P elements and hence transposase in suppressed in the F1.
 Problem set B 1. A wild-type ry+ (rosy) gene was introduced into a ry mutant using P element-mediated gene transformation, and a strain containing a stable ry+ gene was established. If a transformed male
Problem set B 1. A wild-type ry+ (rosy) gene was introduced into a ry mutant using P element-mediated gene transformation, and a strain containing a stable ry+ gene was established. If a transformed male
Chapter 14 Regulation of Transcription
 Chapter 14 Regulation of Transcription Cis-acting sequences Distance-independent cis-acting elements Dissecting regulatory elements Transcription factors Overview transcriptional regulation Transcription
Chapter 14 Regulation of Transcription Cis-acting sequences Distance-independent cis-acting elements Dissecting regulatory elements Transcription factors Overview transcriptional regulation Transcription
Intragenic suppressors: true revertants
 Suppressor Genetics: intragenic and informational suppressors We have covered one approach used in genetics to study gene function: classical forward genetics where an investigator selects or screens for
Suppressor Genetics: intragenic and informational suppressors We have covered one approach used in genetics to study gene function: classical forward genetics where an investigator selects or screens for
Lecture 8: Transgenic Model Systems and RNAi
 Lecture 8: Transgenic Model Systems and RNAi I. Model systems 1. Caenorhabditis elegans Caenorhabditis elegans is a microscopic (~1 mm) nematode (roundworm) that normally lives in soil. It has become one
Lecture 8: Transgenic Model Systems and RNAi I. Model systems 1. Caenorhabditis elegans Caenorhabditis elegans is a microscopic (~1 mm) nematode (roundworm) that normally lives in soil. It has become one
Genetics - Problem Drill 19: Dissection of Gene Function: Mutational Analysis of Model Organisms
 Genetics - Problem Drill 19: Dissection of Gene Function: Mutational Analysis of Model Organisms No. 1 of 10 1. The mouse gene knockout is based on. (A) Homologous recombination (B) Site-specific recombination
Genetics - Problem Drill 19: Dissection of Gene Function: Mutational Analysis of Model Organisms No. 1 of 10 1. The mouse gene knockout is based on. (A) Homologous recombination (B) Site-specific recombination
Sydney Brenner: Present
 Sydney Brenner: 1927 - Present Born in the small town of Germiston, South Africa Won a scholarship to the University of Witwatersrand at the age of 15 Received his PhD from Exeter College, Oxford Made
Sydney Brenner: 1927 - Present Born in the small town of Germiston, South Africa Won a scholarship to the University of Witwatersrand at the age of 15 Received his PhD from Exeter College, Oxford Made
Motivation From Protein to Gene
 MOLECULAR BIOLOGY 2003-4 Topic B Recombinant DNA -principles and tools Construct a library - what for, how Major techniques +principles Bioinformatics - in brief Chapter 7 (MCB) 1 Motivation From Protein
MOLECULAR BIOLOGY 2003-4 Topic B Recombinant DNA -principles and tools Construct a library - what for, how Major techniques +principles Bioinformatics - in brief Chapter 7 (MCB) 1 Motivation From Protein
GENOME 371, Problem Set 6
 GENOME 371, Problem Set 6 1. S. pombe is a distant relative of baker s yeast (which you used in quiz section). Wild type S. pombe can grow on plates lacking tryptophan (-trp plates). A mutant has been
GENOME 371, Problem Set 6 1. S. pombe is a distant relative of baker s yeast (which you used in quiz section). Wild type S. pombe can grow on plates lacking tryptophan (-trp plates). A mutant has been
[Presented by: Andrew Howlett, Cruise Slater, Mahmud Hasan, Greg Dale]
![[Presented by: Andrew Howlett, Cruise Slater, Mahmud Hasan, Greg Dale] [Presented by: Andrew Howlett, Cruise Slater, Mahmud Hasan, Greg Dale]](/thumbs/96/128035026.jpg) Mutational Dissection [Presented by: Andrew Howlett, Cruise Slater, Mahmud Hasan, Greg Dale] Introduction What is the point of Mutational Dissection? It allows understanding of normal biological functions
Mutational Dissection [Presented by: Andrew Howlett, Cruise Slater, Mahmud Hasan, Greg Dale] Introduction What is the point of Mutational Dissection? It allows understanding of normal biological functions
Sol narae (Sona) is a Drosophila ADAMTS involved in Wg signaling
 Sol narae (Sona) is a Drosophila ADAMTS involved in Wg signaling Go-Woon Kim, Jong-Hoon Won, Ok-Kyung Lee, Sang-Soo Lee, Jeong-Hoon Han, Orkhon Tsogtbaatar, Sujin Nam, Yeon Kim, and Kyung-Ok Cho* Supplementary
Sol narae (Sona) is a Drosophila ADAMTS involved in Wg signaling Go-Woon Kim, Jong-Hoon Won, Ok-Kyung Lee, Sang-Soo Lee, Jeong-Hoon Han, Orkhon Tsogtbaatar, Sujin Nam, Yeon Kim, and Kyung-Ok Cho* Supplementary
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10810 Supplementary Fig. 1: Mutation of the loqs gene leads to shortened lifespan and adult-onset brain degeneration. a. Northern blot of control and loqs f00791 mutant flies. loqs f00791
doi:10.1038/nature10810 Supplementary Fig. 1: Mutation of the loqs gene leads to shortened lifespan and adult-onset brain degeneration. a. Northern blot of control and loqs f00791 mutant flies. loqs f00791
beta-galactosidase (A420) ctrl
 0.6 beta-galactosidase (A420) 0.4 0.2 ctrl 2A 22A 35B 36B 58A 64A 68E 75C 86Fa 86Fb 102F Fig. S1. Levels of induced expression at various attp sites. The induced β-galactosidase activity was measured from
0.6 beta-galactosidase (A420) 0.4 0.2 ctrl 2A 22A 35B 36B 58A 64A 68E 75C 86Fa 86Fb 102F Fig. S1. Levels of induced expression at various attp sites. The induced β-galactosidase activity was measured from
4 Mutant Hunts - To Select or to Screen (Perhaps Even by Brute Force)
 Genetic Techniques for Biological Research Corinne A. Michels Copyright q 2002 John Wiley & Sons, Ltd ISBNs: 0-471-89921-6 (Hardback); 0-470-84662-3 (Electronic) 4 Mutant Hunts - To Select or to Screen
Genetic Techniques for Biological Research Corinne A. Michels Copyright q 2002 John Wiley & Sons, Ltd ISBNs: 0-471-89921-6 (Hardback); 0-470-84662-3 (Electronic) 4 Mutant Hunts - To Select or to Screen
Genetic Transformation of Drosophila with Transposable Element Vectors SCIENCE, VOL. 218, 22 OCTOBER 1982
 Genetic Transformation of Drosophila with Transposable Element Vectors SCIENCE, VOL. 218, 22 OCTOBER 1982 Transposition of Cloned P Elements into Drosophila Germ Line Chromosomes SCIENCE, VOL. 218, 22
Genetic Transformation of Drosophila with Transposable Element Vectors SCIENCE, VOL. 218, 22 OCTOBER 1982 Transposition of Cloned P Elements into Drosophila Germ Line Chromosomes SCIENCE, VOL. 218, 22
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/310/5753/1487/dc1 Supporting Online Material for Stem Cell Self-Renewal Controlled by Chromatin Remodeling Factors Rongwen Xi and Ting Xie* *To whom correspondence should
www.sciencemag.org/cgi/content/full/310/5753/1487/dc1 Supporting Online Material for Stem Cell Self-Renewal Controlled by Chromatin Remodeling Factors Rongwen Xi and Ting Xie* *To whom correspondence should
7.03 Final Exam Review 12/19/2006
 7.03 Final Exam Review 12/19/2006 1. You have been studying eye color mutations in Drosophila, which normally have red eyes. White eyes is a recessive mutant trait that is caused by w, a mutant allele
7.03 Final Exam Review 12/19/2006 1. You have been studying eye color mutations in Drosophila, which normally have red eyes. White eyes is a recessive mutant trait that is caused by w, a mutant allele
Melton, D.W. (1994) Gene targeting in the mouse. Bioessays 16:633-8
 Reverse genetics - Knockouts Paper to read for this section : Melton, D.W. (1994) Gene targeting in the mouse. Bioessays 16:633-8 Up until now, we ve concentrated on ways to get a cloned gene. We ve seen
Reverse genetics - Knockouts Paper to read for this section : Melton, D.W. (1994) Gene targeting in the mouse. Bioessays 16:633-8 Up until now, we ve concentrated on ways to get a cloned gene. We ve seen
Concepts: What are RFLPs and how do they act like genetic marker loci?
 Restriction Fragment Length Polymorphisms (RFLPs) -1 Readings: Griffiths et al: 7th Edition: Ch. 12 pp. 384-386; Ch.13 pp404-407 8th Edition: pp. 364-366 Assigned Problems: 8th Ch. 11: 32, 34, 38-39 7th
Restriction Fragment Length Polymorphisms (RFLPs) -1 Readings: Griffiths et al: 7th Edition: Ch. 12 pp. 384-386; Ch.13 pp404-407 8th Edition: pp. 364-366 Assigned Problems: 8th Ch. 11: 32, 34, 38-39 7th
7.012 Problem Set 5. Question 1
 Name Section 7.012 Problem Set 5 Question 1 While studying the problem of infertility, you attempt to isolate a hypothetical rabbit gene that accounts for the prolific reproduction of rabbits. After much
Name Section 7.012 Problem Set 5 Question 1 While studying the problem of infertility, you attempt to isolate a hypothetical rabbit gene that accounts for the prolific reproduction of rabbits. After much
Problem set D answers
 Problem set D answers 1. You find a mouse with no tail. In order to determine whether this mouse carries a new mutation, you cross it to a normal mouse. All the F1 progeny of this cross are wild type.
Problem set D answers 1. You find a mouse with no tail. In order to determine whether this mouse carries a new mutation, you cross it to a normal mouse. All the F1 progeny of this cross are wild type.
Biology 201 (Genetics) Exam #3 120 points 20 November Read the question carefully before answering. Think before you write.
 Name KEY Section Biology 201 (Genetics) Exam #3 120 points 20 November 2006 Read the question carefully before answering. Think before you write. You will have up to 50 minutes to take this exam. After
Name KEY Section Biology 201 (Genetics) Exam #3 120 points 20 November 2006 Read the question carefully before answering. Think before you write. You will have up to 50 minutes to take this exam. After
Test Bank for Molecular Cell Biology 7th Edition by Lodish
 Test Bank for Molecular Cell Biology 7th Edition by Lodish Link download full: http://testbankair.com/download/test-bank-formolecular-cell-biology-7th-edition-by-lodish/ Chapter 5 Molecular Genetic Techniques
Test Bank for Molecular Cell Biology 7th Edition by Lodish Link download full: http://testbankair.com/download/test-bank-formolecular-cell-biology-7th-edition-by-lodish/ Chapter 5 Molecular Genetic Techniques
BIO 304 Fall 2000 Exam II Name: ID #: 1. Fill in the blank with the best answer from the provided word bank. (2 pts each)
 1. Fill in the blank with the best answer from the provided word bank. (2 pts each) incomplete dominance conditional mutation penetrance expressivity pleiotropy Southern blotting hybridization epistasis
1. Fill in the blank with the best answer from the provided word bank. (2 pts each) incomplete dominance conditional mutation penetrance expressivity pleiotropy Southern blotting hybridization epistasis
BIO 304 Genetics (Fall 2003) Exam #2 Name KEY SSN
 BIO 304 Genetics (Fall 2003) Exam #2 Name KEY SSN transformation conditional mutation penetrance expressivity Southern blotting hybridization epistasis co-dominance nonsense mutation translocation amplification
BIO 304 Genetics (Fall 2003) Exam #2 Name KEY SSN transformation conditional mutation penetrance expressivity Southern blotting hybridization epistasis co-dominance nonsense mutation translocation amplification
Supplementary Information
 Supplementary Information MicroRNA-212/132 family is required for epithelial stromal interactions necessary for mouse mammary gland development Ahmet Ucar, Vida Vafaizadeh, Hubertus Jarry, Jan Fiedler,
Supplementary Information MicroRNA-212/132 family is required for epithelial stromal interactions necessary for mouse mammary gland development Ahmet Ucar, Vida Vafaizadeh, Hubertus Jarry, Jan Fiedler,
3. human genomics clone genes associated with genetic disorders. 4. many projects generate ordered clones that cover genome
 Lectures 30 and 31 Genome analysis I. Genome analysis A. two general areas 1. structural 2. functional B. genome projects a status report 1. 1 st sequenced: several viral genomes 2. mitochondria and chloroplasts
Lectures 30 and 31 Genome analysis I. Genome analysis A. two general areas 1. structural 2. functional B. genome projects a status report 1. 1 st sequenced: several viral genomes 2. mitochondria and chloroplasts
SUPPORTING ONLINE MATERIAL
 SUPPORTING ONLINE MATERIAL MATERIAL AND METHODS Fly stocks All Drosophila stocks were grown at 25 C on yeast-containing corn meal/molasses medium. The nanos (nos) alleles nos 18 and nos 53 have been previously
SUPPORTING ONLINE MATERIAL MATERIAL AND METHODS Fly stocks All Drosophila stocks were grown at 25 C on yeast-containing corn meal/molasses medium. The nanos (nos) alleles nos 18 and nos 53 have been previously
BSCI410-Liu/Spring 09/Feb 26 Exam #1 Your name:
 1. (20 points) Give the name of a mutagen that could cause the following damages to DNA: a) Thymidine dimers UV b) Breakage of DNA backbone X-Ray c) 2 bp insertion (frameshift mutation) proflavin, acridine
1. (20 points) Give the name of a mutagen that could cause the following damages to DNA: a) Thymidine dimers UV b) Breakage of DNA backbone X-Ray c) 2 bp insertion (frameshift mutation) proflavin, acridine
All organisms respond to cellular stress by shutting off the regular protein synthesis and
 1. INTRODUCTION All organisms respond to cellular stress by shutting off the regular protein synthesis and starting the synthesis of a new set of proteins called heat shock proteins or more generally,
1. INTRODUCTION All organisms respond to cellular stress by shutting off the regular protein synthesis and starting the synthesis of a new set of proteins called heat shock proteins or more generally,
Analysis of genetic mosaics in developing and adult Drosophila tissues
 Development 117, 1223-1237 (1993) Printed in Great Britain The Company of Biologists Limited 1993 1223 Analysis of genetic mosaics in developing and adult Drosophila tissues Tian Xu and Gerald M. Rubin
Development 117, 1223-1237 (1993) Printed in Great Britain The Company of Biologists Limited 1993 1223 Analysis of genetic mosaics in developing and adult Drosophila tissues Tian Xu and Gerald M. Rubin
Name AP Biology Mrs. Laux Take home test #11 on Chapters 14, 15, and 17 DUE: MONDAY, DECEMBER 21, 2009
 MULTIPLE CHOICE QUESTIONS 1. Inducible genes are usually actively transcribed when: A. the molecule degraded by the enzyme(s) is present in the cell. B. repressor molecules bind to the promoter. C. lactose
MULTIPLE CHOICE QUESTIONS 1. Inducible genes are usually actively transcribed when: A. the molecule degraded by the enzyme(s) is present in the cell. B. repressor molecules bind to the promoter. C. lactose
Genome 371, 12 February 2010, Lecture 10. Analyzing mutants. Drosophila transposon mutagenesis. Genetics of cancer. Cloning genes
 Genome 371, 12 February 2010, Lecture 10 Analyzing mutants Drosophila transposon mutagenesis Genetics of cancer Cloning genes Suppose you are setting up a transposon mutagenesis screen in fruit flies starting
Genome 371, 12 February 2010, Lecture 10 Analyzing mutants Drosophila transposon mutagenesis Genetics of cancer Cloning genes Suppose you are setting up a transposon mutagenesis screen in fruit flies starting
Gene Expression: Transcription
 Gene Expression: Transcription The majority of genes are expressed as the proteins they encode. The process occurs in two steps: Transcription = DNA RNA Translation = RNA protein Taken together, they make
Gene Expression: Transcription The majority of genes are expressed as the proteins they encode. The process occurs in two steps: Transcription = DNA RNA Translation = RNA protein Taken together, they make
MUTANT: A mutant is a strain that has suffered a mutation and exhibits a different phenotype from the parental strain.
 OUTLINE OF GENETICS LECTURE #1 A. TERMS PHENOTYPE: Phenotype refers to the observable properties of an organism, such as morphology, growth rate, ability to grow under different conditions or media. For
OUTLINE OF GENETICS LECTURE #1 A. TERMS PHENOTYPE: Phenotype refers to the observable properties of an organism, such as morphology, growth rate, ability to grow under different conditions or media. For
Genetics C Examination #3 - December 7, 1999
 Genetics C3032 - Examination #3 - December 7, 1999 General instructions: Don't Panic. Be sure your name is on every page and that you write your exam in ink. Answer the questions in the space provided.
Genetics C3032 - Examination #3 - December 7, 1999 General instructions: Don't Panic. Be sure your name is on every page and that you write your exam in ink. Answer the questions in the space provided.
Schematic representation of the endogenous PALB2 locus and gene-disruption constructs
 Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Genetics Lecture Notes Lectures 6 9
 Genetics Lecture Notes 7.03 2005 Lectures 6 9 Lecture 6 Until now our analysis of genes has focused on gene function as determined by phenotype differences brought about by different alleles or by a direct
Genetics Lecture Notes 7.03 2005 Lectures 6 9 Lecture 6 Until now our analysis of genes has focused on gene function as determined by phenotype differences brought about by different alleles or by a direct
Biology 163 Laboratory in Genetics, Final Exam, Dec. 10, 2005
 1 Biology 163 Laboratory in Genetics, Final Exam, Dec. 10, 2005 Honor Pledge: I have neither given nor received any unauthorized help on this exam: Name Printed: Signature: 1. (2 pts) If you see the following
1 Biology 163 Laboratory in Genetics, Final Exam, Dec. 10, 2005 Honor Pledge: I have neither given nor received any unauthorized help on this exam: Name Printed: Signature: 1. (2 pts) If you see the following
Genome annotation & EST
 Genome annotation & EST What is genome annotation? The process of taking the raw DNA sequence produced by the genome sequence projects and adding the layers of analysis and interpretation necessary
Genome annotation & EST What is genome annotation? The process of taking the raw DNA sequence produced by the genome sequence projects and adding the layers of analysis and interpretation necessary
Problem Set 4
 7.016- Problem Set 4 Question 1 Arginine biosynthesis is an example of multi-step biochemical pathway where each step is catalyzed by a specific enzyme (E1, E2 and E3) as is outlined below. E1 E2 E3 A
7.016- Problem Set 4 Question 1 Arginine biosynthesis is an example of multi-step biochemical pathway where each step is catalyzed by a specific enzyme (E1, E2 and E3) as is outlined below. E1 E2 E3 A
Segment polarity gene interactions modulate epidermal patterning in Drosophila embryos
 Development 119, 501-517 (1993) Printed in Great Britain The Company of Biologists Limited 1993 501 Segment polarity gene interactions modulate epidermal patterning in Drosophila embryos Amy Bejsovec*
Development 119, 501-517 (1993) Printed in Great Britain The Company of Biologists Limited 1993 501 Segment polarity gene interactions modulate epidermal patterning in Drosophila embryos Amy Bejsovec*
7.06 Problem Set #3, Spring 2005
 7.06 Problem Set #3, Spring 2005 1. The Drosophila compound eye is composed of about 800 units called ommatidia. Each ommatidium contains eight photoreceptor neurons (R1 through R8), which develop in a
7.06 Problem Set #3, Spring 2005 1. The Drosophila compound eye is composed of about 800 units called ommatidia. Each ommatidium contains eight photoreceptor neurons (R1 through R8), which develop in a
Please sign below if you wish to have your grades posted by the last five digits of your SSN
 BIO 226R EXAM III (Sample) PRINT YOUR NAME Please sign below if you wish to have your grades posted by the last five digits of your Signature BIO 226R Exam III has 8 pages, and 26 questions. There are
BIO 226R EXAM III (Sample) PRINT YOUR NAME Please sign below if you wish to have your grades posted by the last five digits of your Signature BIO 226R Exam III has 8 pages, and 26 questions. There are
ksierzputowska.com Research Title: Using novel TALEN technology to engineer precise mutations in the genome of D. melanogaster
 Research Title: Using novel TALEN technology to engineer precise mutations in the genome of D. melanogaster Research plan: Specific aims: 1. To successfully engineer transgenic Drosophila expressing TALENs
Research Title: Using novel TALEN technology to engineer precise mutations in the genome of D. melanogaster Research plan: Specific aims: 1. To successfully engineer transgenic Drosophila expressing TALENs
BIO 202 Midterm Exam Winter 2007
 BIO 202 Midterm Exam Winter 2007 Mario Chevrette Lectures 10-14 : Question 1 (1 point) Which of the following statements is incorrect. a) In contrast to prokaryotic DNA, eukaryotic DNA contains many repetitive
BIO 202 Midterm Exam Winter 2007 Mario Chevrette Lectures 10-14 : Question 1 (1 point) Which of the following statements is incorrect. a) In contrast to prokaryotic DNA, eukaryotic DNA contains many repetitive
Figure S1. Verification of ihog Mutation by Protein Immunoblotting Figure S2. Verification of ihog and boi
 Figure S1. Verification of ihog Mutation by Protein Immunoblotting Extracts from S2R+ cells, embryos, and adults were analyzed by immunoprecipitation and immunoblotting with anti-ihog antibody. The Ihog
Figure S1. Verification of ihog Mutation by Protein Immunoblotting Extracts from S2R+ cells, embryos, and adults were analyzed by immunoprecipitation and immunoblotting with anti-ihog antibody. The Ihog
Selected Techniques Part I
 1 Selected Techniques Part I Gel Electrophoresis Can be both qualitative and quantitative Qualitative About what size is the fragment? How many fragments are present? Is there in insert or not? Quantitative
1 Selected Techniques Part I Gel Electrophoresis Can be both qualitative and quantitative Qualitative About what size is the fragment? How many fragments are present? Is there in insert or not? Quantitative
R1 12 kb R1 4 kb R1. R1 10 kb R1 2 kb R1 4 kb R1
 Bcor101 Sample questions Midterm 3 1. The maps of the sites for restriction enzyme EcoR1 (R1) in the wild type and mutated cystic fibrosis genes are shown below: Wild Type R1 12 kb R1 4 kb R1 _ _ CF probe
Bcor101 Sample questions Midterm 3 1. The maps of the sites for restriction enzyme EcoR1 (R1) in the wild type and mutated cystic fibrosis genes are shown below: Wild Type R1 12 kb R1 4 kb R1 _ _ CF probe
GENETICS EXAM 3 FALL a) is a technique that allows you to separate nucleic acids (DNA or RNA) by size.
 Student Name: All questions are worth 5 pts. each. GENETICS EXAM 3 FALL 2004 1. a) is a technique that allows you to separate nucleic acids (DNA or RNA) by size. b) Name one of the materials (of the two
Student Name: All questions are worth 5 pts. each. GENETICS EXAM 3 FALL 2004 1. a) is a technique that allows you to separate nucleic acids (DNA or RNA) by size. b) Name one of the materials (of the two
Lac Operon contains three structural genes and is controlled by the lac repressor: (1) LacY protein transports lactose into the cell.
 Regulation of gene expression a. Expression of most genes can be turned off and on, usually by controlling the initiation of transcription. b. Lactose degradation in E. coli (Negative Control) Lac Operon
Regulation of gene expression a. Expression of most genes can be turned off and on, usually by controlling the initiation of transcription. b. Lactose degradation in E. coli (Negative Control) Lac Operon
GENETICS - CLUTCH CH.15 GENOMES AND GENOMICS.
 !! www.clutchprep.com CONCEPT: OVERVIEW OF GENOMICS Genomics is the study of genomes in their entirety Bioinformatics is the analysis of the information content of genomes - Genes, regulatory sequences,
!! www.clutchprep.com CONCEPT: OVERVIEW OF GENOMICS Genomics is the study of genomes in their entirety Bioinformatics is the analysis of the information content of genomes - Genes, regulatory sequences,
Using mutants to clone genes
 Using mutants to clone genes Objectives: 1. What is positional cloning? 2. What is insertional tagging? 3. How can one confirm that the gene cloned is the same one that is mutated to give the phenotype
Using mutants to clone genes Objectives: 1. What is positional cloning? 2. What is insertional tagging? 3. How can one confirm that the gene cloned is the same one that is mutated to give the phenotype
Applicazioni biotecnologiche
 Applicazioni biotecnologiche Analisi forense Sintesi di proteine ricombinanti Restriction Fragment Length Polymorphism (RFLP) Polymorphism (more fully genetic polymorphism) refers to the simultaneous occurrence
Applicazioni biotecnologiche Analisi forense Sintesi di proteine ricombinanti Restriction Fragment Length Polymorphism (RFLP) Polymorphism (more fully genetic polymorphism) refers to the simultaneous occurrence
Molecular Cell Biology - Problem Drill 11: Recombinant DNA
 Molecular Cell Biology - Problem Drill 11: Recombinant DNA Question No. 1 of 10 1. Which of the following statements about the sources of DNA used for molecular cloning is correct? Question #1 (A) cdna
Molecular Cell Biology - Problem Drill 11: Recombinant DNA Question No. 1 of 10 1. Which of the following statements about the sources of DNA used for molecular cloning is correct? Question #1 (A) cdna
Lecture Four. Molecular Approaches I: Nucleic Acids
 Lecture Four. Molecular Approaches I: Nucleic Acids I. Recombinant DNA and Gene Cloning Recombinant DNA is DNA that has been created artificially. DNA from two or more sources is incorporated into a single
Lecture Four. Molecular Approaches I: Nucleic Acids I. Recombinant DNA and Gene Cloning Recombinant DNA is DNA that has been created artificially. DNA from two or more sources is incorporated into a single
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Wang et al., http://www.jcb.org/cgi/content/full/jcb.201405026/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Generation and characterization of unc-40 alleles. (A and
Supplemental material Wang et al., http://www.jcb.org/cgi/content/full/jcb.201405026/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Generation and characterization of unc-40 alleles. (A and
fga phenotypes were also analyzed in the tracheal system, the organ responsible for
 SUPPLEMENTARY TEXT Tracheal defects fga phenotypes were also analyzed in the tracheal system, the organ responsible for oxygen delivery in the fly. In fga homozygous mutant alleles, we observed defects
SUPPLEMENTARY TEXT Tracheal defects fga phenotypes were also analyzed in the tracheal system, the organ responsible for oxygen delivery in the fly. In fga homozygous mutant alleles, we observed defects
Lecture 2: Biology Basics Continued. Fall 2018 August 23, 2018
 Lecture 2: Biology Basics Continued Fall 2018 August 23, 2018 Genetic Material for Life Central Dogma DNA: The Code of Life The structure and the four genomic letters code for all living organisms Adenine,
Lecture 2: Biology Basics Continued Fall 2018 August 23, 2018 Genetic Material for Life Central Dogma DNA: The Code of Life The structure and the four genomic letters code for all living organisms Adenine,
Mos1 insertion. MosTIC protocol-11/2006. repair template - Mos1 transposase expression - Mos1 excision - DSB formation. homolog arm.
 MosTIC (Mos1 excision induced Transgene Instructed gene Conversion) Valérie Robert (vrobert@biologie.ens.fr) and Jean-Louis Bessereau (jlbesse@biologie.ens.fr) (November 2006) Introduction: MosTIC (Robert
MosTIC (Mos1 excision induced Transgene Instructed gene Conversion) Valérie Robert (vrobert@biologie.ens.fr) and Jean-Louis Bessereau (jlbesse@biologie.ens.fr) (November 2006) Introduction: MosTIC (Robert
Authors: Vivek Sharma and Ram Kunwar
 Molecular markers types and applications A genetic marker is a gene or known DNA sequence on a chromosome that can be used to identify individuals or species. Why we need Molecular Markers There will be
Molecular markers types and applications A genetic marker is a gene or known DNA sequence on a chromosome that can be used to identify individuals or species. Why we need Molecular Markers There will be
3. Translation. 2. Transcription. 1. Replication. and functioning through their expression in. Genes are units perpetuating themselves
 Central Dogma Genes are units perpetuating themselves and functioning through their expression in the form of proteins 1 DNA RNA Protein 2 3 1. Replication 2. Transcription 3. Translation Spring 2002 21
Central Dogma Genes are units perpetuating themselves and functioning through their expression in the form of proteins 1 DNA RNA Protein 2 3 1. Replication 2. Transcription 3. Translation Spring 2002 21
Chapter 20 DNA Technology & Genomics. If we can, should we?
 Chapter 20 DNA Technology & Genomics If we can, should we? Biotechnology Genetic manipulation of organisms or their components to make useful products Humans have been doing this for 1,000s of years plant
Chapter 20 DNA Technology & Genomics If we can, should we? Biotechnology Genetic manipulation of organisms or their components to make useful products Humans have been doing this for 1,000s of years plant
1. You are studying three autosomal recessive mutations in the fruit fly Drosophila
 Exam Questions from Exam 1 Basic Genetic Tests, Setting up and Analyzing Crosses, and Genetic Mapping 1. You are studying three autosomal recessive mutations in the fruit fly Drosophila melanogaster. Flies
Exam Questions from Exam 1 Basic Genetic Tests, Setting up and Analyzing Crosses, and Genetic Mapping 1. You are studying three autosomal recessive mutations in the fruit fly Drosophila melanogaster. Flies
Supplemental Information. Autoregulatory Feedback Controls. Sequential Action of cis-regulatory Modules. at the brinker Locus
 Developmental Cell, Volume 26 Supplemental Information Autoregulatory Feedback Controls Sequential Action of cis-regulatory Modules at the brinker Locus Leslie Dunipace, Abbie Saunders, Hilary L. Ashe,
Developmental Cell, Volume 26 Supplemental Information Autoregulatory Feedback Controls Sequential Action of cis-regulatory Modules at the brinker Locus Leslie Dunipace, Abbie Saunders, Hilary L. Ashe,
12 Interaction of Genes
 12 Interaction of Genes Yeast genetics has been particularly amenable for identifying and characterizing gene products that directly or indirectly interact with each other, especially when two mutations
12 Interaction of Genes Yeast genetics has been particularly amenable for identifying and characterizing gene products that directly or indirectly interact with each other, especially when two mutations
SUPPLEMENTARY INFORMATION
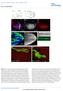 DOI: 10.1038/ncb2806 Supplemental Material: Figure S1 Wnt4 PCP phenotypes are not mediated through canonical Wnt signaling. (a-a ) Illustration of wg, dwnt4 and dwnt6 expression patterns in wing (a) and
DOI: 10.1038/ncb2806 Supplemental Material: Figure S1 Wnt4 PCP phenotypes are not mediated through canonical Wnt signaling. (a-a ) Illustration of wg, dwnt4 and dwnt6 expression patterns in wing (a) and
Biotechnology. Chapter 20. Biology Eighth Edition Neil Campbell and Jane Reece. PowerPoint Lecture Presentations for
 Chapter 20 Biotechnology PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece Lectures by Chris Romero, updated by Erin Barley with contributions from Joan Sharp Copyright
Chapter 20 Biotechnology PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece Lectures by Chris Romero, updated by Erin Barley with contributions from Joan Sharp Copyright
Molecular Genetics Techniques. BIT 220 Chapter 20
 Molecular Genetics Techniques BIT 220 Chapter 20 What is Cloning? Recombinant DNA technologies 1. Producing Recombinant DNA molecule Incorporate gene of interest into plasmid (cloning vector) 2. Recombinant
Molecular Genetics Techniques BIT 220 Chapter 20 What is Cloning? Recombinant DNA technologies 1. Producing Recombinant DNA molecule Incorporate gene of interest into plasmid (cloning vector) 2. Recombinant
Supplementary Methods
 Caussinus and Gonzalez Supplementary Methods Mitotic clones Clones of NBs homozygous for numb 03235, mira ZZ176, pros 17 or apkc k06403 were generated by FLP/FRT mediated mitotic recombination (1). As
Caussinus and Gonzalez Supplementary Methods Mitotic clones Clones of NBs homozygous for numb 03235, mira ZZ176, pros 17 or apkc k06403 were generated by FLP/FRT mediated mitotic recombination (1). As
E. Incorrect! The four different DNA nucleotides follow a strict base pairing arrangement:
 AP Biology - Problem Drill 10: Molecular and Human Genetics Question No. 1 of 10 Instructions: (1) Read the problem and answer choices carefully, (2) Work the problems on paper as 1. Which of the following
AP Biology - Problem Drill 10: Molecular and Human Genetics Question No. 1 of 10 Instructions: (1) Read the problem and answer choices carefully, (2) Work the problems on paper as 1. Which of the following
Review Quizzes Chapters 11-16
 Review Quizzes Chapters 11-16 1. In pea plants, the allele for smooth seeds (S) is dominant over the allele for wrinkled seeds (s). In an experiment, when two hybrids are crossed, what percent of the offspring
Review Quizzes Chapters 11-16 1. In pea plants, the allele for smooth seeds (S) is dominant over the allele for wrinkled seeds (s). In an experiment, when two hybrids are crossed, what percent of the offspring
SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURES
 SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURES Supplementary Figure 1. Generation of inducible BICD2 knock-out mice. A) The mouse BICD2 locus and gene targeting constructs. To generate an inducible Bicd2
SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURES Supplementary Figure 1. Generation of inducible BICD2 knock-out mice. A) The mouse BICD2 locus and gene targeting constructs. To generate an inducible Bicd2
Problem Set 8. Answer Key
 MCB 102 University of California, Berkeley August 11, 2009 Isabelle Philipp Online Document Problem Set 8 Answer Key 1. The Genetic Code (a) Are all amino acids encoded by the same number of codons? no
MCB 102 University of California, Berkeley August 11, 2009 Isabelle Philipp Online Document Problem Set 8 Answer Key 1. The Genetic Code (a) Are all amino acids encoded by the same number of codons? no
Q1 (1 point): Explain why a lettuce leaf wilts when it is placed in a concentrated salt solution.
 Short questions 1 point per question. Q1 (1 point): Explain why a lettuce leaf wilts when it is placed in a concentrated salt solution. Q2 (1 point): Put a cross by the correct answer(s) below. The Na
Short questions 1 point per question. Q1 (1 point): Explain why a lettuce leaf wilts when it is placed in a concentrated salt solution. Q2 (1 point): Put a cross by the correct answer(s) below. The Na
RFLP Method - Restriction Fragment Length Polymorphism
 RFLP Method - Restriction Fragment Length Polymorphism RFLP (often pronounced "rif lip", as if it were a word) is a method used by molecular biologists to follow a particular sequence of DNA as it is passed
RFLP Method - Restriction Fragment Length Polymorphism RFLP (often pronounced "rif lip", as if it were a word) is a method used by molecular biologists to follow a particular sequence of DNA as it is passed
b number of motif occurrences
 supplementary information DOI: 1.138/ncb194 a b number of motif occurrences number of motif occurrences number of motif occurrences I II III IV V X TTGGTCAGTGCA 3 8 23 4 13 239 TTGGTCAGTGCT 7 5 8 1 3 83
supplementary information DOI: 1.138/ncb194 a b number of motif occurrences number of motif occurrences number of motif occurrences I II III IV V X TTGGTCAGTGCA 3 8 23 4 13 239 TTGGTCAGTGCT 7 5 8 1 3 83
7 Gene Isolation and Analysis of Multiple
 Genetic Techniques for Biological Research Corinne A. Michels Copyright q 2002 John Wiley & Sons, Ltd ISBNs: 0-471-89921-6 (Hardback); 0-470-84662-3 (Electronic) 7 Gene Isolation and Analysis of Multiple
Genetic Techniques for Biological Research Corinne A. Michels Copyright q 2002 John Wiley & Sons, Ltd ISBNs: 0-471-89921-6 (Hardback); 0-470-84662-3 (Electronic) 7 Gene Isolation and Analysis of Multiple
