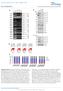
Supplementary Figure S1. MKRN1 depletion induces apoptosis via the caspase-8 pathway. kda h 72h 96h 6.8 % 22.5 % 21.
|
|
|
- Sherilyn Baker
- 6 years ago
- Views:
Transcription
1 Supplementry Figure S1. depletion indues poptosis vi the spse-8 pthwy. Atin h 72h 96h zvad 96h 5. % 6.8 % 8.7 % 4.3% simk1#5 9.5 % 22.5 % 39.6 % 11.8 % simk1#6 8. % 21.7 % 33.5 % 11. % 48h 72h 96h zvad 96h 3.1% simk1# AAD simk1# Supplementry Figure S1. depletion indues poptosis vi the spse-8 pthwy. () HeL ells were trnsfeted with 2 nm of ontrol sirna () or two kinds of sirna (simk1#5 or simk1#6) for 48 h. Expression levels of were mesured y western lot nlysis. () HeL ells were trnsfeted with 2 nm of the indited sirnas. z-vad-fmk (zvad, 2 µm) ws dded t 24 h post-trnsfetion. At 48, 72, nd 96 h posttrnsfetion, ells were stined with propidium iodide (PI) nd nlyzed y flow ytometry. The dt re summrized in Fig. 1. () HeL ells, trnsfeted nd treted with zvad s desried ove, were hrvested, immeditely stined with Annexin V nd 7-AAD, nd then nlyzed y flow ytometry. The perentges of the Annexin V/7-AAD- frtions re shown in Fig. 1. 1
2 Supplementry Figure S2. depletion indues ell deth in oth p53-positive nd p53-negtive ells. U2OS (p53 positive) simk1#5 simk1#6 H1299 (p53 negtive) simk1#5 simk1#6 48h 6.1% 15.3% 11.7% 48 h 3.6% 8.5% 6.5% 96h 6.4% 47.6%.2% 96 h 4.5% 31.8% 28.3% HCT116 p53/ HCT116 p53-/- simk1#5 simk1#6 simk1#5 simk1#6 48 h 8.8% 18.6% 14.8% 48 h 9.8% 15.9% 13.8% 96 h 11.6% 51.4% 32.3% 96 h 14.1% 33.9% 28.7% 6 Su G1 (%) simk1#5 simk1# h 96 h 48 h 96 h 48 h 96 h 48 h 96 h U2OS H1299 HCT116 p53/ HCT116 p53-/- Supplementry Figure S2. depletion indues ell deth in oth p53-positive nd p53-negtive ells. (, ) p53-positive (U2OS nd HCT116 p53/) nd p53-negtive (H1299 nd HCT116 p53-/-) ells were trnsfeted with 2 nm of ontrol sirna () or two kinds of sirna (simk1#5 or simk1#6). Then, 48 h or 96 h fter trnsfetion, ells were hrvested using trypsin, fixed in ethnol, stined with propidium iodide nd nlyzed y flow ytometry. The perentges of ells in the sug1 phse re shown in Supplementry Fig. S2. 2
3 Supplementry Figure S3. knokdown inreses poptosis upon nti-fs nd TRAIL tretment. 1 1 Cell Viility (%) shgfp shmk1#2 Cell Viility (%) shgfp shmk1#2 shmk1# αfs (ng/ml) shmk1# TRAIL (ng/ml) Supplementry Figure S3. knokdown inreses poptosis upon nti-fs nd TRAIL tretment. HeL ells stly expressing shgfp nd sh (shmk1#2 or shmk1#5) were treted with nti-fs ntiody (αfs) plus 5 µg/ml of CHX () or TRAIL () for 6 h t the indited onentrtions. Cell viility ws determined y mesuring intrellulr levels of ATP using the Cell Titer Glo Luminesent Cell Viility Assy kit. Dt re mens ± s.d.; n=3, P<.5, P<.1, P<.1 ompred to shgfp. 3
4 Supplementry Figure S4. depletion filittes morphologil hnges upon nti-fs nd TRAIL tretment. Csp8 inhiitor (C8i) αfs αfs/chx TRAIL TRAIL/CHX αfs/chx TRAIL/CHX αfs/chx TRAIL/CHX zvad Cell Viility (%) αfs/chx αfs/chx Ne-1 TRAIL TRAIL Ne-1 simk1#6 simk1#5 αfs αfs/chx TRAIL TRAIL/CHX αfs/chx TRAIL/CHX αfs/chx TRAIL/CHX simk1#6 simk1#5 simk1#5 simk1#6 DMSO Csp8 inhiitor zvad Supplementry Figure S4. depletion filittes morphologil hnges upon nti-fs nd TRAIL tretment. () The morphology of HeL ells with lted fter αfs (5 ng/ml) nd TRAIL (5 ng/ml) tretment for 3 h. CHX (5 µg/ml) ws used to enhne poptosis. Cells were pretreted with 2 µm sp8 inhiitor or zvad for 24 h. () HeL ells with sirna were treted s desried ove. After 6 h, ell viility ws mesured using the Cell Titer Glo Luminesent Cell Viility Assy kit. Dt re mens ± s.d.; n=3, P<.5, P<.1, P<.1 ompred to. () HeL ells with knoked-down were treted with αfs (5 ng/ml) plus CHX (5 µg/ml) or TRAIL (5 ng/ml) in the sene or presene of Ne-1 (3 µm) for 3 h. Morphologil hnges in ells undergoing ell deth were deteted using mirosope. 4
5 Supplementry Figure S5. Altion of inreses poptosis upon nti-fs nd TRAIL tretment. αfs/chx TRAIL Csp8 inhiitor (C8i) αfs/chx TRAIL simk1# AAD simk1# WB: Csp8 simk1 34 αfs/chx - - C8i C9i zvad TRAIL - - C8i C9i zvad p/p53 p/p41 WB: Csp9 WB: p18 Pro-sp9 Cleved -sp9 Pro-sp3 Csp3 17 Cleved -sp3 WB: Atin Cell Viility (%) NS simk1 - - C8i C9i zvad - C8i C9i zvad - αfs/chx TRAIL Supplementry Figure S5. Altion of inreses poptosis upon nti-fs nd TRAIL tretment. () -depleted HeL ells were treted with αfs (5 ng/ml) plus CHX (5 µg/ml) or TRAIL (1 ng/ml) lone for 3 h in the sene nd presene of 2 µm sp8 inhiitor. Cells were sujeted to Annexin V/7-AAD stining. () HeL ells, trnsfeted with 2 nm of or simk1#5, were treted with αfs (5 ng/ml) plus CHX (5 µg/ml) or TRAIL (5 ng/ml) lone for 3 h in the presene of 2 µm of C8i (spse-8 inhiitor), C9i (spse-9 inhiitor), or zvad (pn-spse inhiitor). Cells were hrvested nd spse levge ws deteted using the indited ntiodies. () HeL ells, trnsfeted nd treted s desried ove. After 6 h of deth stimuli, viility ws mesured using the Cell Titer Glo Luminesent Cell Viility Assy kit. Dt re mens ± s.d.; n=3, P<.5, P<.1, NS;non-sepifi. 5
6 Supplementry Figure S6. Arogtion of filittes spse-8 levge y induing rpid formtion of the DISC omplex. IP: Fs Input zvad shgfp shmk1 αfs/chx (h) WB: Fs WB: Fs zvad simk1 IP: Csp8 Input αfs/chx (h) IgG WB: Csp8 WB: Csp8 zvad simk1 IP: Csp8 Input TRAIL (h) IgG WB: Csp8 WB: Csp8 Supplementry Figure S6. Arogtion of filittes spse-8 levge y induing rpid formtion of the DISC omplex. () HeL ells in 1-mm-dimeter dishes were trnsfeted with sirnas. At 24 h post-trnsfetion, 1 µm zvad ws dded to ells. Cells were then inuted for n dditionl 24 h nd treted with αfs (5 ng/ml) nd CHX (5 µg/ml) s indited. Cell lystes were immunopreipitted with nti-fas ntiody, followed y western lotting with nti-fadd ntiody. (, ) HeL ells, trnsfeted with sirnas, were pretreted with zvad for 24 h. Cells were treted with αfs, CHX, nd TRAIL s indited. Cell lystes were immunopreipitted with nti-spse-8 ntiody, followed y western lot nlysis using nti-fadd ntiodies. Control IgG ws used s negtive ontrol. 6
7 Supplementry Figure S7. knokdown inreses TNF-α indued poptosis y filitting omplex II formtion. TNF TNF/CHX TNF Csp8 inhiitor TNF simk1#5 simk1# AAD simk1#6 simk1# Cell viility (%) shgfp shmk1#2 shmk1#5 TNF/CHX d TNF (h) WB: Csp8 WB: Csp3 WB: RIP1 simk e IP: Csp8 Input TNF (h) WB: RIP1 WB: Csp8 WB: RIP1 WB: Csp8 simk WB: Atin Supplementry Figure S7. knokdown inreses TNF-α indued poptosis y filitting omplex II formtion. () Morphologil hnges in HeL ells upon TNF-α tretment. HeL ells were trnsfeted with 2 nm sirnas for 48 h nd treted with 1 ng/ml of humn TNF-α (TNF) in the sene or presene of CHX (5 µg/ml). Cells were visulized y mirosopy. () Detetion of poptosis y Annexin V/7-AAD stining. HeL ells, trnsfeted nd treted with 1 ng/ml of TNF-α in the sene or presene of 2 µm spse-8 inhiitor, were hrvested using trypsin, wshed with PBS, stined with Annexin V nd 7-AAD, nd nlyzed y flow ytometry. () Viility of HeL ells with depletion upon TNF-α stimultion in the presene of CHX. Cells were trnsfeted nd treted s desried ove. Cell viility ws determined using the Cell Titer Glo Luminesent Cell Viility Assy kit. Dt re mens ± s.d.; n=3. (d) Western lot nlysis of HeL ells treted with TNF-α. HeL ells were trnsfeted with the indited sirnas nd stimulted with TNF-α. Cspse tivtion nd RIP1 levge were determined y western lotting using the indited ntiodies. (e) HeL ells were trnsfeted with sirnas. Twenty-four hours posttrnsfetion, 2 µm of z-vad-fmk ws dded to the ells for n dditionl 24 h. Cells were treted with 1 ng/ml of TNF-α s indited. Cell lystes were immunopreipitted with nti-spse-8 got ntiody, followed y western lot nlysis using nti-rip1 mouse ntiody. 7
8 Supplementry Figure S8. Degrdtion of FADD is not prevented y lysosoml inhiitor, while depletion inreses FADD stility. f. A1 CHX (h) FADD Atin simk1 CHX (h) FADD Atin Supplementry Figure S8. Degrdtion of FADD is not prevented y lysosoml inhiitor, while depletion inreses FADD stility. () HeL ells were treted with 4 µg/ml of CHX in the sene or presene of.2 µm filomyin A1 (f. A1). FADD protein stility ws determined y western lotting. () HeL ells, trnsiently trnsfeted with sirnas, were treted with 4 µg/ml of CHX. FADD protein stility ws determined y western lot nlysis. 8
9 Supplementry Figure S9. interts with FADD. IP : HA WCE IP : FLAG WCE HA-FADD FLAG- WB : FLAG FLAG- HA-FADD WB : HA WB : HA 37.8 WB : FLAG 61.5 IP: HA HA-FADDs FLAG- WB: HA DED Deth Domin 28 Binding - - WCE inding domin d IP:HA Input HA-FADD FLAG- H37E FLAG- WT WB: FLAG 61.5 WB: HA 37.8 Supplementry Figure S9. interts with FADD. (, ) Reiprol intertion etween FLAG- nd HA-FADD. HeL ells were trnsfeted with plsmids expressing FLAG- nd HA-FADD s indited. Cell lystes were immunopreipitted with nti-ha mouse ntiody () or nti-flag mouse ntiody (), followed y western lot nlysis using nti-ha rit nd nti-flag rit ntiodies. () Mpping of FADD domins responsile for the intertion. HeL ells were trnsfeted with the indited plsmids. Cell lystes were immunopreipitted with α-ha ntiody nd nlyzed using n α-flg ntiody. Shemti digrms of the FADD deletion mutnts re provided in the lower pnel. (d) Intertion etween the H37E mutnt nd FADD. 9
10 Supplementry Figure S1. Reintrodution of shr- into knokdown ells delys spse levge. αfs/chx TRAIL shr-h37e shr-wt shmk shgfp WB: Csp8 p/p53 p/p41 p18 WB: Csp3 Pro-sp3 Clevedsp3 Supplementry Figure S1. Reintrodution of shr- into knokdown ells delys spse levge. HeL/sh ells reonstituted with shr-wt or shr-h37e were treted with nti-fs (5 ng/ml) CHX (5 µg/ml) or TRAIL (1 ng/ml) for 3 h. Cspse-8 nd psse-3 levge ws nlyzed y western lotting. 1
11 Supplementry Figure S11. depletion filittes neroptosis under spse inhiition y enhning nerosome formtion in L929 ells. L929 ells mt mt/n mt/z mt/z/n 4.2% FADD RIP1 RIP3 Atin simmk1#1 simmk1# SSC Annexin V positive ells (%) simmk1#1 simmk1#2 Un mt mt/n mt/z mt/z/n d Cell Loss (%) simmk1#1 simmk1#2 Un mt mt/n mt/z mt/z/n Supplementry Figure S11. depletion filittes neroptosis under spse inhiition y enhning nerosome formtion in L929 ells. () L929 ells were trnsfeted with 5 nm of ontrol sirna () nd two kinds of mouse sirna (simmk1#1 or simmk1#2) for 72 h. Expression levels of, FADD, RIP1, nd RIP3 were mesured y western lotting. (, ) L929 ells were trnsfeted with 5 nm of the indited sirnas for 48 h. Cells were treted with the omintion of 5 ng/ml mtnf-α (mt), 1 µm z-vad-fmk (Z), nd 3 µm Ne-1 (N) s indited for 3 h. Cells undergoing neroptosis were deteted y Annexin V stining nd onfirmed y Ne-1 tretment. The men perentges of Annexin V-positive ells from three independent experiments performed in triplite re shown in (). Representtive dot plot dt re presented in (). Dt re mens ± s.d.; n=3, P<.5, P<.1, P<.1 ompred to. (d) L929 ells, trnsfeted with the indited sirnas nd treted s desried ove. Cell deth ws ssessed y mesuring intrellulr ATP levels using the Cell Titer Glo Luminesent Cell Viility Assy kit. Dt re mens ± s.d.; n=3, P<.1, P<.1 ompred to. 11
12 Supplementry Figure S12. depletion filittes neroptosis under spse inhiition y enhning nerosome formtion. shmk1 shgfp 72 simk1 #5 simk1 #6 FADD hrip1 hrip3 Atin shgfp shmk1 HT-29 ells Z lone T lone T/C T/C/Z T/C/Z/N T/C T/C/N T/C/Z T/C/Z/N d SSC Annexin V positive ells (%) 8 shgfp 6 shmk1 4 2 SSC e Cell loss (%) shgfp shmk1 f Input IP: Csp8 zvad T/C WB: hrip3 WB: hrip1 WB: Csp8 WB: hrip3 WB: hrip1 IgG shgfp shmk Supplementry Figure S12. depletion filittes neroptosis under spse inhiition y enhning nerosome formtion. () HT-29 ells were trnsfeted with 3 nm of ontrol sirna () nd two kinds of sirna (simk1#5 or simk1#6) for 48 h. Expression levels of, FADD, RIP1 nd RIP3 were mesured y western lot nlysis. () HT-29 ells stly expressing shgfp or sh were treted for 12 h s indited. Cells undergoing neroptosis were deteted y Annexin V stining nd onfirmed y Ne-1 tretment. Arevitions re s follows: T, TNFα (3 ng/ml); C, CHX (5 µg/ml); Z, z-vad-fmk (3 µm); N, Nerosttin-1 (3 µm). (, d) HT-29 ells stly expressing shgfp or sh were treted with omintion of TNFα (T), CHX (C), z-vad-fmk (Z), nd Ne-1 (N) s indited for 16 h. Neroptoti ell deth ws mesured y Annexin V stining. The men perentges of Annexin V-positive ells from three independent experiments performed in triplite re shown in () (Error rs indites s.d. P<.1, ompred to shgfp). Representtive dot plot dt re presented in (d). (e) Cell deth in HT-29 stle ell lines treted s desried ove ws ssessed using the Cell Titer Glo Luminesent Cell Viility Assy kit. Error rs indites s.d. n=3, P<.1, P<.1, ompred to shgfp. (f) HT-29 ells were treted with TNFα nd CHX (T/C) in the sene or presene of z-vad-fmk (Z) for 4 h. Cell lystes were immunopreipitted with ntispse-8 ntiodies, followed y western lot nlysis using nti-rip1 nd nti-hrip3 ntiodies. The sterisk () indites phosphorylted RIP3 s previously reported
13 Supplementry Figure S13. Knokdown of promotes neroptosis in FADD-independent mnner, ut FADD stiliztion upon knokdown inhiits neroptosis in HT-29 ells. T/C/Z T/C/Z/N Annexin V positive ells (%) shgfp shgfp shmk1 shfadd shfadd shmk1 shmk1 T/C/Z T/C/Z/N shfadd shmk1 6.1% shfadd SSC IP: hrip3 Input shgfp shfadd T/C/Z 4h WB: hrip1 WB: hrip3 WB: hrip1 WB: hrip3 IgG shmk1 shfadd shmk d simk1 12.2% sifadd sifadd simk1 SSC T/C/Z Supplementry Figure S13. Knokdown of promotes neroptosis in FADD-independent mnner, ut FADD stiliztion upon knokdown inhiits neroptosis in HT-29 ells. (, ) HT-29 ells stly expressing the indited shrnas were treted with 3 ng/ml humn TNF-α, 5 µg/ml CHX nd 3 µm z-vad-fmk (T/C/Z) in the sene nd presene of 3 µm Ne-1 (T/C/Z/N). Neroptoti ell deth ws mesured y Annexin V stining. The men perentges of Annexin V-positive ells from three independent experiments performed in triplite re shown in () (Error rs indites s.d. P<.1, ompred to shgfp). Representtive dot plot dt re presented in (). () HT-29 stle ell lines were treted with TNFα, CHX nd zvad (T/C/Z) for 4h s desried in Supplementry Fig. S12f. Cell lystes were immunopreipitted with nti-hrip3 ntiody, followed y western lot nlysis using nti- RIP1, nti-fadd, nd nti-hrip3 ntiodies. (d) HT-29 ells, trnsfeted with the indited sirnas (3 nm, eh) for 48 h, were treted with TNFα, CHX nd zvad (T/C/Z) for 12 h. Cells undergoing neroptosis were deteted y Annexin V stining. 13
14 Supplementry Figure S14. FADD stiliztion upon knokdown prevents neroptosis in L929 ells. simmk1 simfadd simfadd simmk1 5.9% mt/z/n mt/z SSC mt/z Mok HA-FADD 4.2% SSC mt/z/n Supplementry Figure S14. FADD stiliztion upon knokdown prevents neroptosis in L929 ells. () L929 ells were trnsfeted with the omintion of 3 nm mouse sirna #1 (simmk1) nd 3 nm mouse FADD sirna pool (simfadd) s indited. Control sirna ws used to keep the sirna onentrtion to 6 nm. Cells were treted with 5 ng/ml of mtnfα nd 1 µm of z-vad-fmk with or without 3 µm of Ne-1 (mt/z or mt/z/n) for 3 h. Cells undergoing neroptosis were deteted y Annexin V stining nd onfirmed y Ne-1 tretment. () L929 ells were trnsfeted with plsmid expressing HA-FADD. Cells were treted with mtnfα, z-vad-fmk, nd Ne-1 s indited for 4 h. Cells undergoing neroptosis were deteted y Annexin V stining nd onfirmed y Ne-1 tretment. 14
15 Supplementry Figure S15. is overexpressed in rest ner ell lines. d Reltive mount WB: Atin FADD MCF1A MCF7 T47D MDA-MB-468 BT2 ZR-75-1 SK-BR-3 JIMT Hs578T MBA-MB-231 e Reltive FADD protein ZR-75-1 T47D MCF7 MB-231 JIMT BT-2 Hs578T MB-468 SK-BR Reltive protein Cell Viility (%) MCF7 T47D MDA-MB-468 BT-2 Cell Viility (%) ZR-75-1 SK-BR-3 JIMT Hs578T MDA-MB-231 f g Cell Viility (%) TRAIL (ng/ml) TRAIL (ng/ml) IMR9 A549 H1299 H46 h i Cell Viility (%) TRAIL (ng/ml) TRAIL (ng/ml) RKO SW48 HCT116 WB: Atin WB: Atin 15
16 Supplementry Figure S15. is overexpressed in rest ner ell lines. () Expression of nd FADD in the norml rest ell line MCF1A nd the rest ner ell lines MCF7, T47D, SK-BR-3, MDA-MB-231, MDA-MB-468, Hs578T, BT-2, JIMT nd ZR-75-1 s determined y western lot nlysis. () Reltive expression levels of nd FADD in rest ner ell lines were quntified y densitometry, normlized to tin, nd indited in the grph. () An inverse orreltion etween nd FADD expression in rest ner ell lines, exluding MCF1A, is shown in the grph. (d, e) Brest ner ell lines were treted with inresing onentrtions of TRAIL for 24 h. Cell viility ws determined y mesuring intrellulr levels of ATP using the Cell Titer Glo Luminesent Cell Viility Assy kit. The results from three independent experiments re summrized in the grph. Dt re mens ± s.d. n=3. (f) Lung firolst (IMR9) nd ner ell lines (H46, H1299, nd A549) were treted with inresing onentrtion of TRAIL for 24 h. Cell viility ws determined s desried ove. The results from three independent experiments re summrized in the grph. Dt re mens ± s.d. n=3. (g) Expression of nd FADD in lung ell lines were determined y western lot nlysis. (h) The TRAIL-sensitivity of olon ner ell lines inluding RKO, SW48, nd HCT116 ws evluted s desried ove. The results from three independent experiments re summrized in the grph. Dt re mens ± s.d. n=3. (i) Expression of nd FADD in olon ner ell lines were evluted y western lot nlysis. 16
17 Supplementry Figure S16. Depletion of FADD rogtes TRAIL-medited poptosis in -knokdown ells. sicontrol simk1 sifadd sifadd simk1 TRAIL TRAIL sifadd pool simk WB: Csp8 34 p/p53 p/p41 17 p18 WB: Csp Pro-sp3 Clevedsp3 WB: Atin Supplementry Figure S16. Depletion of FADD rogtes TRAIL-medited poptosis in -knokdown ells. () MDA-MB-231 ells were trnsfeted with the omintion of 2 nm of sirna #5 (simk1) nd 2 nm FADD sirna pool (sifadd pool) s indited. Control sirna ws used to keep the sirna onentrtion to 4 nm. Forty-eight hours post-trnsfetion, ells were treted with 1 ng/ml TRAIL for 3 h. The morphologil hnges upon TRAIL tretment were visulized y mirosopy. () MDA-MB-231 ells were trnsfeted with sirnas nd treted with TRAIL s desried ove. Clevge of spse-8 nd -3 ws nlyzed y western lotting. 17
18 Supplementry Methods sirna-medited ltion of nd FADD Two kinds of flexitue sirnas (Hs 5; GGCGAAGCTGAGTCAAGAA nd Hs 6; (CG)GGATCCTCTCCAACTGCAA) otined from Qigen were resynthesized. FADD sirna (Hs_FADD_9, SI364916, (CA)GCGGGATCTCGTATCTTTA) ws purhsed nd resynthesized from Qigen nd used in the experiments presented in Fig. 3f. Mouse sirnas (sim#1; CAGGCGAAGCTGAGTCAAG nd sim#2; CAGAGGTCACAGCACATAA from Qigen (SI7337)) were synthesized y Qigen. Control sirna ws lso purhsed from Qigen-Xergon. ON-TARGET plus SMARTpool for humn FADD sirna (L- 38-) nd mouse FADD sirna (L-4488-) were purhsed from Dhrmon. HeL, MDA-MB-231, L929, nd HT-29 ells were trnsfeted with sirnas using Lipofetmine RNAiMx (Invitrogen) using the reversetrnsfetion method ording to the mnufturer s protool. Genertion of stle ell lines. Mission lentivirl shrna expression vetors for humn, humn FADD, nd ontrol GFP were purhsed from Sigm-Aldrih. sh#2 (TRCN4125, CCGGCCAGAGGTCACAGCACATAAAC TCGAGTTTATGTGCTGTGACCTCTGGTTTTTG), sh#5 (TRCN324883, CGGGTGTTGGATC ACTTGCTGAAACTCGAGTTTCAGCAAGTGATCCAACACTTTTTG), shfadd#1 (CCGGGTGCAGCATT TAACGTCATATCTCGAGATATGACGTTAAATGCTGCACTTTTTG), nd shfadd#2 (CCGGCATGGAAC TCAGACGCATCTACTCGAGTAGATGCGTCTGAGTTCCATGTTTTTG) were le to suppress endogenous expression nd were thus used in this study. 293FT ells were trnsfeted with plko.1 shrna-expressing vetor nd pkging vetors for 48 h, nd then superntnts were hrvested, filtrted, nd trnsferred to HeL nd MDA-MB-231 ells. shrna-expressing ells were seleted y puromyin tretment for 7 dys. For reonstitution, DNA ws suloned into the pbe-puro vetor, nd two ytosines of trgeted y shrna#2 were mutted into denine without hnging the mino id sequene. 293FT ells were trnsfeted with pbe-puro-shr- WT nd H37E or vetor lone in the presene of retrovirl pkging vetors for 48 h, nd then superntnts were hrvested, filtrted, nd trnsferred to HeL/sh#2 ells. Reverse trnsription (RT)-PCR nlysis Totl RNA ws extrted using Trizol regent (Invitrogen) ording to the mnufturer s instrution. DNA ws synthesized from 1 µg of totl RNA using Reverse Trnsriptse M-MLV (Tkr), mplified, nd then nlyzed using the QuntiTet SYBR Green PCR Kit nd rel-time PCR (Qigen) with the following primers: humn FADD, 5 -CAC 18
19 AGA CCA CCT GCT TCT GA-3 (forwrd) nd 5 -CTG GAC ACG GTT CCA ACT TT -3 (reverse); humn, 5 -GAG AAG GAC ATG GAG CTC TCA-5 (forwrd) nd 5 -CGC CTT GTT GCT CAT TGC CTC-3 (reverse). The primers used for glyerldehyde-3-phosphte dehydrogense were desried previously. 19
Supplementary Figure S1. Akaike et al.
 reltive expression of HIPK2 (HIPK2/GAPDH) Supplementry Figure S1. Akike et l. 1.25 ontrol HIPK2 #1 1 HIPK2 #2 HIPK2 3 U 0.75 0.5 0.25 0 ontrol : + + - HIPK2 #1: - - - + + - - - - HIPK2 #2: - - - - + +
reltive expression of HIPK2 (HIPK2/GAPDH) Supplementry Figure S1. Akike et l. 1.25 ontrol HIPK2 #1 1 HIPK2 #2 HIPK2 3 U 0.75 0.5 0.25 0 ontrol : + + - HIPK2 #1: - - - + + - - - - HIPK2 #2: - - - - + +
SUPPLEMENTARY INFORMATION
 S shrna S Viility, % of NT sirna trnsfete ells 1 1 Srmle NT sirna Csp-8 sirna RIP1 Atin L929 shrna sirna: Csp8 Atin L929: shrna Csp8 e Viility, % of NT sirna trnsfete ells NT sirna Csp-8 sirna M45 M45mutRHIM
S shrna S Viility, % of NT sirna trnsfete ells 1 1 Srmle NT sirna Csp-8 sirna RIP1 Atin L929 shrna sirna: Csp8 Atin L929: shrna Csp8 e Viility, % of NT sirna trnsfete ells NT sirna Csp-8 sirna M45 M45mutRHIM
SUPPLEMENTARY INFORMATION
 DOI: 1.138/n74 In the formt provided y the uthors nd unedited. d 331p 637p 9p 394p 48p 467p 22p 23p 489p 419p 3p 493p 332p 53p 39p 1 G1: 16.4% S: 73.1% G2/M: 1.5% 2 1 2 E-KO G1: 2.2% S: 7.7% G2/M: 9.1%
DOI: 1.138/n74 In the formt provided y the uthors nd unedited. d 331p 637p 9p 394p 48p 467p 22p 23p 489p 419p 3p 493p 332p 53p 39p 1 G1: 16.4% S: 73.1% G2/M: 1.5% 2 1 2 E-KO G1: 2.2% S: 7.7% G2/M: 9.1%
Gan et al., Supplemental Figure 1
 Gn et l., Supplementl Figure IB: DU45 Unp IB: py-68 IB: perk IB: ERK IB: Akt Unp DU45 DU45 ErB2 - EGF - EGF ErB2 75 75 Supplementl Figure. DU45 nd ells predominntly express nd re highly responsive to EGF.
Gn et l., Supplementl Figure IB: DU45 Unp IB: py-68 IB: perk IB: ERK IB: Akt Unp DU45 DU45 ErB2 - EGF - EGF ErB2 75 75 Supplementl Figure. DU45 nd ells predominntly express nd re highly responsive to EGF.
HeLa. CaSki. Supplementary Figure 1. Validation of the NKX6.1 expression in mrna level and
 Supplementry Figure. Li et l. Reltive expression ( / GAPDH ) 6 5 4 3 2 5 Reltive expression ( / GAPDH ) 4 3 2 Reltive expression ( / GAPDH ) 4 3 2 Reltive expression ( / GAPDH ) 4 3 2 Ve S Ve S2 S S2 Ve
Supplementry Figure. Li et l. Reltive expression ( / GAPDH ) 6 5 4 3 2 5 Reltive expression ( / GAPDH ) 4 3 2 Reltive expression ( / GAPDH ) 4 3 2 Reltive expression ( / GAPDH ) 4 3 2 Ve S Ve S2 S S2 Ve
sensitive VBSs Vh subdomains EF EF
 Tlin- Tlin-EGFP-His 2 3 2 3 ABD Mehno- ABD2 ABD3 sensitive VBSs DD 6xHis EGFP Vh sudomins EGFP-Vinulin EGFP 2 3 4 Vt EGFP-Vh EGFP 2 3 4 -tinin- CH CH2 SP SP SP SP EF EF mcherry--tinin- mcherry CH CH2 SP
Tlin- Tlin-EGFP-His 2 3 2 3 ABD Mehno- ABD2 ABD3 sensitive VBSs DD 6xHis EGFP Vh sudomins EGFP-Vinulin EGFP 2 3 4 Vt EGFP-Vh EGFP 2 3 4 -tinin- CH CH2 SP SP SP SP EF EF mcherry--tinin- mcherry CH CH2 SP
SUPPLEMENTARY INFORMATION
 Pulldown HeL lyste lyste lyste 25 5 75 5 5kD 37 25 5 MDDIFTQCREGNAVAVRLWLDNTENDLNQGDDHGFSPLHWACREGRSAVVEMLIMRGARINVMNRGDDTP LHLAASHGHRDIVQKLLQYKADINAVNEHGNVPLHYACFWGQDQVAEDLVANGALVSICNKYGEMPVDKA KAPLRELLRERAEKMGQNLNRIPYKDTFWKGTTRTRPRNGTLNKHSGIDFKQLNFLTKLNENHSGELWKG
Pulldown HeL lyste lyste lyste 25 5 75 5 5kD 37 25 5 MDDIFTQCREGNAVAVRLWLDNTENDLNQGDDHGFSPLHWACREGRSAVVEMLIMRGARINVMNRGDDTP LHLAASHGHRDIVQKLLQYKADINAVNEHGNVPLHYACFWGQDQVAEDLVANGALVSICNKYGEMPVDKA KAPLRELLRERAEKMGQNLNRIPYKDTFWKGTTRTRPRNGTLNKHSGIDFKQLNFLTKLNENHSGELWKG
SUPPLEMENTARY INFORMATION
 1 1 μm c d EGF + TPA + e f Intensity 1.8 1.6 1.4 1.2 1.8.6.4.2 2 4 8 2 4 8 (Hours) 2 4 6 8 1 Time (Hours) Reltive luciferse ctivity 4 3 2 1 + CAMEK1 FRE reporter Figure S1 inhiitor incresed protein expression
1 1 μm c d EGF + TPA + e f Intensity 1.8 1.6 1.4 1.2 1.8.6.4.2 2 4 8 2 4 8 (Hours) 2 4 6 8 1 Time (Hours) Reltive luciferse ctivity 4 3 2 1 + CAMEK1 FRE reporter Figure S1 inhiitor incresed protein expression
SUPPLEMENTARY INFORMATION
 doi:10.1038/nture10924 no tg Lsm1-my Lsm2-my Lsm5-my Lsm6-my Lsm7-my Lsm8-my no tg Lsm1-my Lsm2-my Lsm5-my Lsm6-my Lsm7-my Lsm8-my kd 220 120 100 80 60 50 40 30 20 SeeBlue Mrker Mgi Mrk no tg Sm1-my Smd3-my
doi:10.1038/nture10924 no tg Lsm1-my Lsm2-my Lsm5-my Lsm6-my Lsm7-my Lsm8-my no tg Lsm1-my Lsm2-my Lsm5-my Lsm6-my Lsm7-my Lsm8-my kd 220 120 100 80 60 50 40 30 20 SeeBlue Mrker Mgi Mrk no tg Sm1-my Smd3-my
a ATP release 4h after induction
 doi:1.138/nture9413 ATP relese 4h fter induction of poptosis (nm) 5 ATP 4 3 2 1 UV UV + zvad 1μM 3μM 5μM UV + 1μM 3μM 5μM UV + 18AGA 1μM 3μM 5μM UV + FFA HeL monolyer Scrpe Dye trnsfer HeL HeL-Cx43 HeL-Cx43
doi:1.138/nture9413 ATP relese 4h fter induction of poptosis (nm) 5 ATP 4 3 2 1 UV UV + zvad 1μM 3μM 5μM UV + 1μM 3μM 5μM UV + 18AGA 1μM 3μM 5μM UV + FFA HeL monolyer Scrpe Dye trnsfer HeL HeL-Cx43 HeL-Cx43
SUPPLEMENTARY INFORMATION
 doi: 1.138/nture77 c 2 2 1 15 1 1 5 1 1 5 5 129Sv 1.5 h IL-6 / HPRT 3 h 5 h 6 4 2 d 129Sv 4 4 3 3 2 1 1 8 6 4 2 e f g C57/BL6 Poly(dA-dT) Cell numer (normlized to medium control) 1 5 1 5 1 5 Fluorescence
doi: 1.138/nture77 c 2 2 1 15 1 1 5 1 1 5 5 129Sv 1.5 h IL-6 / HPRT 3 h 5 h 6 4 2 d 129Sv 4 4 3 3 2 1 1 8 6 4 2 e f g C57/BL6 Poly(dA-dT) Cell numer (normlized to medium control) 1 5 1 5 1 5 Fluorescence
SUPPLEMENTARY INFORMATION
 DOI: 1.138/nc2274 EpH4 Prentl -/MDCK EpH4 Prentl -/MDCK - FERM- Figure S1 (kd) delferm- (1-438)- c Prentl MDCK -/MDCK deljfr- Input Control IP IP Input Control IP IP d Control Control Figure S1 () Specificity
DOI: 1.138/nc2274 EpH4 Prentl -/MDCK EpH4 Prentl -/MDCK - FERM- Figure S1 (kd) delferm- (1-438)- c Prentl MDCK -/MDCK deljfr- Input Control IP IP Input Control IP IP d Control Control Figure S1 () Specificity
COS-1 cells transiently transfected with either HA hgr wt, HA hgr S211A or HA hgr S226A
 1 SUPPLEMENTRY FIGURES Fig. 1 & Specificity of the nti-p-s211 nd nti-p-s226 ntibodies COS-1 cells trnsiently trnsfected with either H hgr wt, H hgr S211 or H hgr S226 were treted with 1nM Dex for 1 hour.
1 SUPPLEMENTRY FIGURES Fig. 1 & Specificity of the nti-p-s211 nd nti-p-s226 ntibodies COS-1 cells trnsiently trnsfected with either H hgr wt, H hgr S211 or H hgr S226 were treted with 1nM Dex for 1 hour.
Interplay between NS3 protease and human La protein---- by Ray and Das Supplementary fig 1. NS3 pro
 Interply etween tese nd humn L protein---- y Ry nd Ds Supplementry fig 1 1 2 3 4 UV crosslinking ssy: α[ 32 P]UTP leled HCV IRES RNA ws UV-crosslinked to incresing concentrtions (0.1, 0.2 nd 0.4µM) in
Interply etween tese nd humn L protein---- y Ry nd Ds Supplementry fig 1 1 2 3 4 UV crosslinking ssy: α[ 32 P]UTP leled HCV IRES RNA ws UV-crosslinked to incresing concentrtions (0.1, 0.2 nd 0.4µM) in
Figure S1. Characterization of sirna uptake in HeLa cells.
 Supplementry Informtion The Rough Endoplsmti Retiulum is Centrl Nuletion Site of sirna-medited RNA Silening Luks Stlder, Wolf Heusermnn, Len Sokol, Domini Trojer, Joel Wirz, Justin Hen, Anj Fritzshe, Florin
Supplementry Informtion The Rough Endoplsmti Retiulum is Centrl Nuletion Site of sirna-medited RNA Silening Luks Stlder, Wolf Heusermnn, Len Sokol, Domini Trojer, Joel Wirz, Justin Hen, Anj Fritzshe, Florin
PML regulates p53 stability by sequestering Mdm2 to the nucleolus
 regultes stility y sequestering to the nuleolus Ros Bernrdi 1,Pier Polo Sglioni 1,3,Stephn Bergmnn 1,3,Henning F. Horn 2,Kren H. Vousden 2 nd Pier Polo Pndolfi 1,4 The promyeloyti leukemi () tumour-suppressor
regultes stility y sequestering to the nuleolus Ros Bernrdi 1,Pier Polo Sglioni 1,3,Stephn Bergmnn 1,3,Henning F. Horn 2,Kren H. Vousden 2 nd Pier Polo Pndolfi 1,4 The promyeloyti leukemi () tumour-suppressor
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nture11303 c Supplementry Figure 1: Genertion of INCB18424 persistent B/F3 Epor- V617F (EporVF) cells, which re cross resistnt to other inhiitors. : () Nïve EporVF
SUPPLEMENTARY INFORMATION doi:10.1038/nture11303 c Supplementry Figure 1: Genertion of INCB18424 persistent B/F3 Epor- V617F (EporVF) cells, which re cross resistnt to other inhiitors. : () Nïve EporVF
Won Jung, Seung Min An, Whaseon Lee-Kwon, Mario Chiong1, Sergio Lavandero1,2, and Hyug
 Supplementry Informtion suppresses IL-1-medited immunomodultion Soo Youn Choi, Hwn Hee Lee, Jun Ho Lee, Byeong Jin Ye, Eun Jin Yoo, Hyun Je Kng, Gyu Won Jung, Seung Min An, Whseon Lee-Kwon, Mrio Chiong1,
Supplementry Informtion suppresses IL-1-medited immunomodultion Soo Youn Choi, Hwn Hee Lee, Jun Ho Lee, Byeong Jin Ye, Eun Jin Yoo, Hyun Je Kng, Gyu Won Jung, Seung Min An, Whseon Lee-Kwon, Mrio Chiong1,
*** supplementary information. axon growth (µm/1h) 5 ng/ml NGF. 150 ng/ml NGF. 20 ng/ml NGF. n.s. n.s. n.s. DOI: /ncb1916
 DOI: 1.138/n1916 25 xon growth (µm/1h) 2 15 1 5 ng/ml NGF 5 ng/ml NGF 2 ng/ml NGF 1 ng/ml NGF 15 ng/ml NGF Figure S1 () Doseresponse urve for xon outgrowth in response to NGF. Speifi tivities of NGF from
DOI: 1.138/n1916 25 xon growth (µm/1h) 2 15 1 5 ng/ml NGF 5 ng/ml NGF 2 ng/ml NGF 1 ng/ml NGF 15 ng/ml NGF Figure S1 () Doseresponse urve for xon outgrowth in response to NGF. Speifi tivities of NGF from
melf4 Vec Fold induction ELF4 STK38 MAVS IFNa8- Luc
 IfnβmRNA() m h m h Type I IFN(unit/ml) Supplementry Informtion Supplementry Figures 3 7 3 2 2 35 1 1 mifnβ-lu hifnβ-lu Med HIV-1 EMCV SeV Poly(IC) Poly(dAdT) d e f g h 8 4 2 1 12 6 9 6 3 12 6 IFN2-Lu IFN4
IfnβmRNA() m h m h Type I IFN(unit/ml) Supplementry Informtion Supplementry Figures 3 7 3 2 2 35 1 1 mifnβ-lu hifnβ-lu Med HIV-1 EMCV SeV Poly(IC) Poly(dAdT) d e f g h 8 4 2 1 12 6 9 6 3 12 6 IFN2-Lu IFN4
SUPPLEMENTARY INFORMATION
 DOI: 0.038/n2228 HepG2 (ell pellet ml) Nuler Extrts (.8 mg) -epenent intertnts HepG2 (ell pellet 55.5 ml) Nuler Extrts (227.7 mg) glutthione sephrose F.T. glutthione sephrose F.T. oun F.T. oun F.T. -FXR
DOI: 0.038/n2228 HepG2 (ell pellet ml) Nuler Extrts (.8 mg) -epenent intertnts HepG2 (ell pellet 55.5 ml) Nuler Extrts (227.7 mg) glutthione sephrose F.T. glutthione sephrose F.T. oun F.T. oun F.T. -FXR
SUPPLEMENTARY INFORMATION
 doi:10.1038/nture12040 + + + Glc Gln Supplementry Figure 1., Reltive prolifertion of PDAC cell lines (8988T, Tu8902, Pnc1, Mipc2, PL45 nd MPnc96) nd low pssge primry humn PDAC cell lines (#1 nd #2) under
doi:10.1038/nture12040 + + + Glc Gln Supplementry Figure 1., Reltive prolifertion of PDAC cell lines (8988T, Tu8902, Pnc1, Mipc2, PL45 nd MPnc96) nd low pssge primry humn PDAC cell lines (#1 nd #2) under
Unprecedented inhibition of tubulin polymerization directed by gold nanoparticles inducing cell cycle arrest and apoptosis
 Eletroni supplementry informtion (ESI) for Nnosle Unpreedented inhiition of tuulin polymeriztion direted y gold nnoprtiles induing ell yle rrest nd poptosis Diptimn Choudhury,,e, Pulrjpilli Lourdu Xvier,,
Eletroni supplementry informtion (ESI) for Nnosle Unpreedented inhiition of tuulin polymeriztion direted y gold nnoprtiles induing ell yle rrest nd poptosis Diptimn Choudhury,,e, Pulrjpilli Lourdu Xvier,,
Hes6. PPARα. PPARγ HNF4 CD36
 SUPPLEMENTARY INFORMATION Supplementary Table Positions and Sequences of ChIP primers -63 AGGTCACTGCCA -79 AGGTCTGCTGTG Hes6-0067 GGGCAaAGTTCA ACOT -395 GGGGCAgAGTTCA PPARα -309 GGCTCAaAGTTCAaGTTCA CPTa
SUPPLEMENTARY INFORMATION Supplementary Table Positions and Sequences of ChIP primers -63 AGGTCACTGCCA -79 AGGTCTGCTGTG Hes6-0067 GGGCAaAGTTCA ACOT -395 GGGGCAgAGTTCA PPARα -309 GGCTCAaAGTTCAaGTTCA CPTa
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nture10177 MDYKDHDGDYKDHDIDYKDD DDKMAPKKKRKVGIHGVPAA MAERPFQCRICMRKFAQSGD LTRHTKIHTGEKPFQCRICM RNFSRSDVLSEHIRTHTGEK PFACDICGKKFADRSNRIKH TKIHTGSQKPFQCRICMRNF SRSDNLSEHIRTHTGEKPFA
SUPPLEMENTARY INFORMATION doi:10.1038/nture10177 MDYKDHDGDYKDHDIDYKDD DDKMAPKKKRKVGIHGVPAA MAERPFQCRICMRKFAQSGD LTRHTKIHTGEKPFQCRICM RNFSRSDVLSEHIRTHTGEK PFACDICGKKFADRSNRIKH TKIHTGSQKPFQCRICMRNF SRSDNLSEHIRTHTGEKPFA
Supplementary information
 PP GC B Epithelil cell Peyer s ptch B lymphocyte Dendritic cell Bsophil Neutrophil Mst cell Nturl killer cell T lymphocyte Supplementry informtion EAF2 medites germinl center B cell poptosis to suppress
PP GC B Epithelil cell Peyer s ptch B lymphocyte Dendritic cell Bsophil Neutrophil Mst cell Nturl killer cell T lymphocyte Supplementry informtion EAF2 medites germinl center B cell poptosis to suppress
Lineage-specific functions of Bcl6 in immunity and inflammation are mediated through distinct biochemical mechanisms
 Supplementry informtion for: Linege-specific functions of Bcl6 in immunity nd inflmmtion re medited through distinct iochemicl mechnisms Chunxin Hung, Kterin Htzi & Ari Melnick Division of Hemtology nd
Supplementry informtion for: Linege-specific functions of Bcl6 in immunity nd inflmmtion re medited through distinct iochemicl mechnisms Chunxin Hung, Kterin Htzi & Ari Melnick Division of Hemtology nd
Electronic supplementary information: High specific detection and near-infrared photothermal. therapy of lung cancer cells with high SERS active
 Electronic supplementry informtion: High specific detection nd ner-infrred phototherml therpy of lung cncer cells with high SERS ctive ptmer-silver-gold shell-core nnostructures Ping Wu, Yng Go, Yimei
Electronic supplementry informtion: High specific detection nd ner-infrred phototherml therpy of lung cncer cells with high SERS ctive ptmer-silver-gold shell-core nnostructures Ping Wu, Yng Go, Yimei
FL3-H::PI H2O2: %G1=66.3, %S=15.7, %G2=19.6. Cisplatin: %G1=66.9, %S=16.3, %G2=16.8 FL3-H::PI
 .8 mm HO, 8 h 66C14 4T1 MB468 MB31 ontrol FL4-H::ap annexin V HO ount ount ontrol: %G1=48., %S=3.7, %G=9.5 Dox: %G1=67.9, %S=15.3, %G=19.1 HO: %G1=66.3, %S=15.7, %G=19.6 Cisplatin: %G1=66.9, %S=16.3, %G=16.8
.8 mm HO, 8 h 66C14 4T1 MB468 MB31 ontrol FL4-H::ap annexin V HO ount ount ontrol: %G1=48., %S=3.7, %G=9.5 Dox: %G1=67.9, %S=15.3, %G=19.1 HO: %G1=66.3, %S=15.7, %G=19.6 Cisplatin: %G1=66.9, %S=16.3, %G=16.8
Jay S Desgrosellier, Leo A Barnes, David J Shields, Miller Huang, Steven K Lau, Nicolas Prévost, David Tarin, Sanford J Shattil and David A Cheresh
 Integrin αvβ3/c-src Oncogenic Unit Promotes Anchorge-independence nd Tumor Progression Jy S Desgrosellier, Leo A Brnes, Dvid J Shields, Miller Hung, Steven K Lu, Nicols Prévost, Dvid Trin, Snford J Shttil
Integrin αvβ3/c-src Oncogenic Unit Promotes Anchorge-independence nd Tumor Progression Jy S Desgrosellier, Leo A Brnes, Dvid J Shields, Miller Hung, Steven K Lu, Nicols Prévost, Dvid Trin, Snford J Shttil
Supplementary Figure 1A A404 Cells +/- Retinoic Acid
 Supplementary Figure 1A A44 Cells +/- Retinoic Acid 1 1 H3 Lys4 di-methylation SM-actin VEC cfos (-) RA (+) RA 14 1 1 8 6 4 H3 Lys79 di-methylation SM-actin VEC cfos (-) RA (+) RA Supplementary Figure
Supplementary Figure 1A A44 Cells +/- Retinoic Acid 1 1 H3 Lys4 di-methylation SM-actin VEC cfos (-) RA (+) RA 14 1 1 8 6 4 H3 Lys79 di-methylation SM-actin VEC cfos (-) RA (+) RA Supplementary Figure
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/nc2307 c No Jsplkinolide Jsplkinolide MDCK AP2-GFP Trnsferrin Merge BSC1 Trnsferrin fluorescence (.u.) MDCK - Jsplkinolide Jsplkinolide AP2-GFP fluorescence (.u.) Trnsferrin fluorescence (.u.)
DOI: 10.1038/nc2307 c No Jsplkinolide Jsplkinolide MDCK AP2-GFP Trnsferrin Merge BSC1 Trnsferrin fluorescence (.u.) MDCK - Jsplkinolide Jsplkinolide AP2-GFP fluorescence (.u.) Trnsferrin fluorescence (.u.)
Supplementary Figure 1. A TRPC5-like channel in the neurites and somata of aortic baroreceptor neurons.
 Supplementl Figure 1 c d e f Supplementry Figure 1. A TRPC5-like chnnel in the neurites nd somt of ortic roreceptor neurons. Cell-ttched ptch recordings from the neurite terminls (-c) nd the somt (d-f)
Supplementl Figure 1 c d e f Supplementry Figure 1. A TRPC5-like chnnel in the neurites nd somt of ortic roreceptor neurons. Cell-ttched ptch recordings from the neurite terminls (-c) nd the somt (d-f)
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
Heat-shock protein 70 inhibits apoptosis by preventing recruitment of procaspase-9 to the Apaf-1 apoptosome
 Het-shok protein 7 inhiits poptosis y preventing reruitment of prospse-9 to the poptosome Helen M. Beere*, Beni B. Wolf*, Kelvin Cin, Dik D. Mosser, Artin Mhoui*, Tomomi Kuwn*, Pnkj Tilor, Rihrd I. Morimoto,
Het-shok protein 7 inhiits poptosis y preventing reruitment of prospse-9 to the poptosome Helen M. Beere*, Beni B. Wolf*, Kelvin Cin, Dik D. Mosser, Artin Mhoui*, Tomomi Kuwn*, Pnkj Tilor, Rihrd I. Morimoto,
Fluorescence Intensities of. GFP-PAC-1 Strains
 DOI: 10.1038/ncb3168 Arbitrry Fluorescence Units 2500 2000 1500 1000 500 0 full length (1-4) Fluorescence Intensities of GFP-PAC-1 Strins ΔPH 392-838 575-4 GFP-PAC-1 Strins 2-610 1-574 b control c pc-1(3
DOI: 10.1038/ncb3168 Arbitrry Fluorescence Units 2500 2000 1500 1000 500 0 full length (1-4) Fluorescence Intensities of GFP-PAC-1 Strins ΔPH 392-838 575-4 GFP-PAC-1 Strins 2-610 1-574 b control c pc-1(3
Electronic Supplementary Information
 Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2017 Electronic Supplementary Information Dissecting binding of a β-barrel outer membrane
Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2017 Electronic Supplementary Information Dissecting binding of a β-barrel outer membrane
iaspp oncoprotein is a key inhibitor of p53 conserved from worm to human 12.5 no irradiation +100 J/m 2 UV germ-cell corpses / gonad arm
 onoprotein is key inhiitor of onserved from worm to humn 23 Nture Pulishing Group http://www.nture.om/nturegenetis Dniele Bergmshi 1 *, Yrden Smuels 1 *, Nigel J. O Neil 2 *, Giuseppe Triginte 1, Tim Crook
onoprotein is key inhiitor of onserved from worm to humn 23 Nture Pulishing Group http://www.nture.om/nturegenetis Dniele Bergmshi 1 *, Yrden Smuels 1 *, Nigel J. O Neil 2 *, Giuseppe Triginte 1, Tim Crook
Mammalian Sprouty4 suppresses Ras-independent ERK activation by binding to Raf1
 Mmmlin suppresses Rs-independent ERK tivtion y inding to Rf1 letters Atsuo Sski*, Tkhru Tketomi*, Reiko Kto*, Kzuko Seki*, Atsushi Nonmi*, Mik Sski*, Msmitsu Kuriym, Noki Sito, Msumi Shiuy nd Akihiko Yoshimur*
Mmmlin suppresses Rs-independent ERK tivtion y inding to Rf1 letters Atsuo Sski*, Tkhru Tketomi*, Reiko Kto*, Kzuko Seki*, Atsushi Nonmi*, Mik Sski*, Msmitsu Kuriym, Noki Sito, Msumi Shiuy nd Akihiko Yoshimur*
A functional genomic screen identifies a role for TAO1 kinase in spindle-checkpoint signalling
 A funtionl genomi sreen identifies role for kinse in spindle-hekpoint signlling Viji M. Drvim 1,5, Frnk Stegmeier 2,5, Grzegorz Nlep 3, Mthew E. Sow 3, Jing Chen 3, Anthony Ling 2, Gregory J. Hnnon 4,
A funtionl genomi sreen identifies role for kinse in spindle-hekpoint signlling Viji M. Drvim 1,5, Frnk Stegmeier 2,5, Grzegorz Nlep 3, Mthew E. Sow 3, Jing Chen 3, Anthony Ling 2, Gregory J. Hnnon 4,
Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microrna mir-21
 Negtive regultion of TLR vi trgeting of the proinflmmtory tumor suppressor y the mirorna mir-1 Frederik J Sheedy 1, Ev Plsson-MDermott 1, Elizeth J Hennessy 1, Cr Mrtin,, John J O Lery,, Qingguo Run, Derek
Negtive regultion of TLR vi trgeting of the proinflmmtory tumor suppressor y the mirorna mir-1 Frederik J Sheedy 1, Ev Plsson-MDermott 1, Elizeth J Hennessy 1, Cr Mrtin,, John J O Lery,, Qingguo Run, Derek
Application of Phi29 Motor prna for Targeted Therapeutic Delivery of sirna Silencing Metallothionein-IIA and Survivin in Ovarian Cancers
 originl rtile The Amerin Soiety of Gene & Cell Therpy Applition of Phi9 Motor prna for Trgeted Therpeuti Delivery of sirna Silening Metllothionein-IIA nd Survivin in Ovrin Cners Pheruz Trpore, Yi Shu,
originl rtile The Amerin Soiety of Gene & Cell Therpy Applition of Phi9 Motor prna for Trgeted Therpeuti Delivery of sirna Silening Metllothionein-IIA nd Survivin in Ovrin Cners Pheruz Trpore, Yi Shu,
Supplementary Figures
 Supplementary Figures Supplementary Fig. 1 Characterization of GSCs. a. Immunostaining of primary GSC spheres from GSC lines. Nestin (neural progenitor marker, red), TLX (green). Merged images of nestin,
Supplementary Figures Supplementary Fig. 1 Characterization of GSCs. a. Immunostaining of primary GSC spheres from GSC lines. Nestin (neural progenitor marker, red), TLX (green). Merged images of nestin,
The Wnt-5a-derived hexapeptide Foxy-5 inhibits breast cancer. metastasis in vivo by targeting cell motility
 SUPPLEMENTARY MATERIAL: The Wnt-5a-derived hexapeptide Foxy-5 inhibits breast cancer metastasis in vivo by targeting cell motility Annette Säfholm, Johanna Tuomela, Jeanette Rosenkvist, Janna Dejmek, Pirkko
SUPPLEMENTARY MATERIAL: The Wnt-5a-derived hexapeptide Foxy-5 inhibits breast cancer metastasis in vivo by targeting cell motility Annette Säfholm, Johanna Tuomela, Jeanette Rosenkvist, Janna Dejmek, Pirkko
Supporting Information
 Supporting Information Table S1. Oligonucleotide sequences used in this work Oligo DNA A B C D CpG-A CpG-B CpG-C CpG-D Sequence 5 ACA TTC CTA AGT CTG AAA CAT TAC AGC TTG CTA CAC GAG AAG AGC CGC CAT AGT
Supporting Information Table S1. Oligonucleotide sequences used in this work Oligo DNA A B C D CpG-A CpG-B CpG-C CpG-D Sequence 5 ACA TTC CTA AGT CTG AAA CAT TAC AGC TTG CTA CAC GAG AAG AGC CGC CAT AGT
Aneuploidy generates proteotoxic stress and DNA damage concurrently with p53-mediated post-mitotic apoptosis in SAC-impaired cells
 ARTICLE Reeived 3 Nov Aepted Jun 5 Pulished 6 Jul 5 DOI:.38/nomms8668 OPEN Aneuploidy genertes proteotoxi stress nd DNA dmge onurrently with p53-medited post-mitoti poptosis in SAC-impired ells Akihiro
ARTICLE Reeived 3 Nov Aepted Jun 5 Pulished 6 Jul 5 DOI:.38/nomms8668 OPEN Aneuploidy genertes proteotoxi stress nd DNA dmge onurrently with p53-medited post-mitoti poptosis in SAC-impired ells Akihiro
Supplemental Information. Human Senataxin Resolves RNA/DNA Hybrids. Formed at Transcriptional Pause Sites. to Promote Xrn2-Dependent Termination
 Supplemental Information Molecular Cell, Volume 42 Human Senataxin Resolves RNA/DNA Hybrids Formed at Transcriptional Pause Sites to Promote Xrn2-Dependent Termination Konstantina Skourti-Stathaki, Nicholas
Supplemental Information Molecular Cell, Volume 42 Human Senataxin Resolves RNA/DNA Hybrids Formed at Transcriptional Pause Sites to Promote Xrn2-Dependent Termination Konstantina Skourti-Stathaki, Nicholas
Breast cell lines with Combination Index (CI) and p53 mutation status
 Supplementary Table 1 Breast cell lines with Combination Index (CI) and p53 mutation status Cell Line CI p53 status a BT20 0.44 K132Q BT549 0.53 R249S CAL120 0.61 c.672+2t>g CAL51 0.73 wild-type CAMA1
Supplementary Table 1 Breast cell lines with Combination Index (CI) and p53 mutation status Cell Line CI p53 status a BT20 0.44 K132Q BT549 0.53 R249S CAL120 0.61 c.672+2t>g CAL51 0.73 wild-type CAMA1
PGRP negatively regulates NOD-mediated cytokine production in rainbow trout liver cells
 Supplementary Information for: PGRP negatively regulates NOD-mediated cytokine production in rainbow trout liver cells Ju Hye Jang 1, Hyun Kim 2, Mi Jung Jang 2, Ju Hyun Cho 1,2,* 1 Research Institute
Supplementary Information for: PGRP negatively regulates NOD-mediated cytokine production in rainbow trout liver cells Ju Hye Jang 1, Hyun Kim 2, Mi Jung Jang 2, Ju Hyun Cho 1,2,* 1 Research Institute
Supplementary. Table 1: Oligonucleotides and Plasmids. complementary to positions from 77 of the SRα '- GCT CTA GAG AAC TTG AAG TAC AGA CTG C
 Supplementary Table 1: Oligonucleotides and Plasmids 913954 5'- GCT CTA GAG AAC TTG AAG TAC AGA CTG C 913955 5'- CCC AAG CTT ACA GTG TGG CCA TTC TGC TG 223396 5'- CGA CGC GTA CAG TGT GGC CAT TCT GCT G
Supplementary Table 1: Oligonucleotides and Plasmids 913954 5'- GCT CTA GAG AAC TTG AAG TAC AGA CTG C 913955 5'- CCC AAG CTT ACA GTG TGG CCA TTC TGC TG 223396 5'- CGA CGC GTA CAG TGT GGC CAT TCT GCT G
SUPPLEMENTARY INFORMATION
 SUPPLEMENTRY INFORMTION DOI:./n7 d Tm d- d- Frequeny, e+ e+ kp e+ e+.e+ Ctegory # peks % peks % genome Fold Enrihment k upstrem TSS.7 9..77 Exon..7. ' UTR...7 ' UTR... Intron 79.
SUPPLEMENTRY INFORMTION DOI:./n7 d Tm d- d- Frequeny, e+ e+ kp e+ e+.e+ Ctegory # peks % peks % genome Fold Enrihment k upstrem TSS.7 9..77 Exon..7. ' UTR...7 ' UTR... Intron 79.
SUPPLEMENTARY INFORMATION
 doi:1.138/nture9816 control.1 µg/µl.1 µg/µl 1 µg/µl NH2 signl 2xFLAG PS Fc-tg CD4L [116-261] 55 274 146 c counts Stimultion of Rji cells Lysis nd 1 st IP with M2-eds (4 C) PreScission nd FLAG-peptide tretment
doi:1.138/nture9816 control.1 µg/µl.1 µg/µl 1 µg/µl NH2 signl 2xFLAG PS Fc-tg CD4L [116-261] 55 274 146 c counts Stimultion of Rji cells Lysis nd 1 st IP with M2-eds (4 C) PreScission nd FLAG-peptide tretment
Figure S1. Characterization of the irx9l-1 mutant. (A) Diagram of the Arabidopsis IRX9L gene drawn based on information from TAIR (the Arabidopsis
 1 2 3 4 5 6 7 8 9 10 11 12 Figure S1. Characterization of the irx9l-1 mutant. (A) Diagram of the Arabidopsis IRX9L gene drawn based on information from TAIR (the Arabidopsis Information Research). Exons
1 2 3 4 5 6 7 8 9 10 11 12 Figure S1. Characterization of the irx9l-1 mutant. (A) Diagram of the Arabidopsis IRX9L gene drawn based on information from TAIR (the Arabidopsis Information Research). Exons
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/10/494/eaan6284/dc1 Supplementary Materials for Activation of master virulence regulator PhoP in acidic ph requires the Salmonella-specific protein UgtL Jeongjoon
www.sciencesignaling.org/cgi/content/full/10/494/eaan6284/dc1 Supplementary Materials for Activation of master virulence regulator PhoP in acidic ph requires the Salmonella-specific protein UgtL Jeongjoon
Supplementary Figure 1. Localization of MST1 in RPE cells. Proliferating or ciliated HA- MST1 expressing RPE cells (see Fig. 5b for establishment of
 Supplementary Figure 1. Localization of MST1 in RPE cells. Proliferating or ciliated HA- MST1 expressing RPE cells (see Fig. 5b for establishment of the cell line) were immunostained for HA, acetylated
Supplementary Figure 1. Localization of MST1 in RPE cells. Proliferating or ciliated HA- MST1 expressing RPE cells (see Fig. 5b for establishment of the cell line) were immunostained for HA, acetylated
Rap1 Activation in Collagen Phagocytosis Is Dependent on Nonmuscle Myosin II-A
 Moleulr Biology of the ell Vol. 19, 532 546, Deemer 28 tivtion in ollgen Phgoytosis Is Dependent on Nonmusle Myosin II- Pmel D. ror,* Mry nne onti, Shoshn Rvid, Dvid B. Sks, ndrs Kpus, Roert S. delstein,
Moleulr Biology of the ell Vol. 19, 532 546, Deemer 28 tivtion in ollgen Phgoytosis Is Dependent on Nonmusle Myosin II- Pmel D. ror,* Mry nne onti, Shoshn Rvid, Dvid B. Sks, ndrs Kpus, Roert S. delstein,
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/nc2885 kd M ΔNZipA 66.4 55.6 ZipA 42.7 34.6 6x His NiNTA 27.0 c 1.,, 2. evnescent field supported memrne Supplementry Figure 1 Experimentl ssy. () Illustrtion of protein interctions (dpted
DOI: 10.1038/nc2885 kd M ΔNZipA 66.4 55.6 ZipA 42.7 34.6 6x His NiNTA 27.0 c 1.,, 2. evnescent field supported memrne Supplementry Figure 1 Experimentl ssy. () Illustrtion of protein interctions (dpted
Nod2-mediated recognition of the microbiota is critical for mucosal adjuvant activity of cholera toxin
 Supplementry informtion Nod2-medited recognition of the microiot is criticl for mucosl djuvnt ctivity of choler toxin Donghyun Kim 1,2, Yun-Gi Kim 1,2, Sng-Uk Seo 1,2, Dong-Je Kim 1,2, Nouhiko Kmd 3, Dve
Supplementry informtion Nod2-medited recognition of the microiot is criticl for mucosl djuvnt ctivity of choler toxin Donghyun Kim 1,2, Yun-Gi Kim 1,2, Sng-Uk Seo 1,2, Dong-Je Kim 1,2, Nouhiko Kmd 3, Dve
AKAP150 signaling complex promotes suppression of the M-current by muscarinic agonists
 AKAP150 signling omplex promotes suppression of the M-urrent y musrini gonists Noto Hoshi 1,2,Ji-Sheng Zhng 1,Miho Omki 3,Tkhiro Tkeuhi 1,Shigeru Yokoym 1,Niols Wnvereq 4, Lorene K Lngeerg 2,Yukio Yoned
AKAP150 signling omplex promotes suppression of the M-urrent y musrini gonists Noto Hoshi 1,2,Ji-Sheng Zhng 1,Miho Omki 3,Tkhiro Tkeuhi 1,Shigeru Yokoym 1,Niols Wnvereq 4, Lorene K Lngeerg 2,Yukio Yoned
Supplemental Figure S1
 Supplementl Figure S1 TG nrt1.5- Li et l., 1 nrt1.5- Lin et l., 8 F L CTGCCT R T 5'UTR 3'UTR 1 3 81p (k) nrt1.5- C nrt1.5- Supplementl Figure S1. Phenotypes of the T-DN insertion mutnts (this pper), nrt1.5-
Supplementl Figure S1 TG nrt1.5- Li et l., 1 nrt1.5- Lin et l., 8 F L CTGCCT R T 5'UTR 3'UTR 1 3 81p (k) nrt1.5- C nrt1.5- Supplementl Figure S1. Phenotypes of the T-DN insertion mutnts (this pper), nrt1.5-
Macmillan Publishers Limited. All rights reserved
 doi:1.138/nture12436 Pthogen loks host deth reeptor signlling y rginine GlNAyltion of deth domins Shn Li 1,2, Li Zhng 2,3, Qing Yo 1,2, Lin Li 2, N Dong 2, Jie Rong 4, Wenqing Go 2, Xiojun Ding 2, Liming
doi:1.138/nture12436 Pthogen loks host deth reeptor signlling y rginine GlNAyltion of deth domins Shn Li 1,2, Li Zhng 2,3, Qing Yo 1,2, Lin Li 2, N Dong 2, Jie Rong 4, Wenqing Go 2, Xiojun Ding 2, Liming
Supplemental Data Supplemental Figure 1.
 Supplemental Data Supplemental Figure 1. Silique arrangement in the wild-type, jhs, and complemented lines. Wild-type (WT) (A), the jhs1 mutant (B,C), and the jhs1 mutant complemented with JHS1 (Com) (D)
Supplemental Data Supplemental Figure 1. Silique arrangement in the wild-type, jhs, and complemented lines. Wild-type (WT) (A), the jhs1 mutant (B,C), and the jhs1 mutant complemented with JHS1 (Com) (D)
SUPPLEMENTARY INFORMATION
 doi:1.138/nture151 IL-1β (ng/ml) 15 1.5 1.5 LFn-FlA Lp _WT LFn-FlA Lp _3A b Cell deth (%) 1 8 6 4 2 LFn-FlA Lp _WT LFn-FlA Lp _3A Supplementry Figure 1. Effects of nthrx lethl fctor N- terminl domin-medited
doi:1.138/nture151 IL-1β (ng/ml) 15 1.5 1.5 LFn-FlA Lp _WT LFn-FlA Lp _3A b Cell deth (%) 1 8 6 4 2 LFn-FlA Lp _WT LFn-FlA Lp _3A Supplementry Figure 1. Effects of nthrx lethl fctor N- terminl domin-medited
Supplementary Methods Quantitative RT-PCR. For mrna, total RNA was prepared using TRIzol reagent (Invitrogen) and genomic DNA was eliminated with TURB
 Supplementary Methods Quantitative RT-PCR. For mrna, total RNA was prepared using TRIzol reagent (Invitrogen) and genomic DNA was eliminated with TURBO DNA-free Kit (Ambion). One µg of total RNA was reverse
Supplementary Methods Quantitative RT-PCR. For mrna, total RNA was prepared using TRIzol reagent (Invitrogen) and genomic DNA was eliminated with TURBO DNA-free Kit (Ambion). One µg of total RNA was reverse
Substrate elasticity provides mechanical signals for the expansion of hemopoietic stem and progenitor cells
 correction notice Nt. Biotechnol. 28, 1123 1128 (21); pulished online 3 Octoer 21; corrected fter print 27 April 211 Sustrte elsticity provides mechnicl signls for the expnsion of hemopoietic stem nd progenitor
correction notice Nt. Biotechnol. 28, 1123 1128 (21); pulished online 3 Octoer 21; corrected fter print 27 April 211 Sustrte elsticity provides mechnicl signls for the expnsion of hemopoietic stem nd progenitor
Add 5µl of 3N NaOH to DNA sample (final concentration 0.3N NaOH).
 Bisulfite Treatment of DNA Dilute DNA sample to 2µg DNA in 50µl ddh 2 O. Add 5µl of 3N NaOH to DNA sample (final concentration 0.3N NaOH). Incubate in a 37ºC water bath for 30 minutes. To 55µl samples
Bisulfite Treatment of DNA Dilute DNA sample to 2µg DNA in 50µl ddh 2 O. Add 5µl of 3N NaOH to DNA sample (final concentration 0.3N NaOH). Incubate in a 37ºC water bath for 30 minutes. To 55µl samples
Supplemental Data. mir156-regulated SPL Transcription. Factors Define an Endogenous Flowering. Pathway in Arabidopsis thaliana
 Cell, Volume 138 Supplemental Data mir156-regulated SPL Transcription Factors Define an Endogenous Flowering Pathway in Arabidopsis thaliana Jia-Wei Wang, Benjamin Czech, and Detlef Weigel Table S1. Interaction
Cell, Volume 138 Supplemental Data mir156-regulated SPL Transcription Factors Define an Endogenous Flowering Pathway in Arabidopsis thaliana Jia-Wei Wang, Benjamin Czech, and Detlef Weigel Table S1. Interaction
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:1.138/nture1371 As Brc11 (null) Brc5-13cK (conditioned llele) Reltive levels of mrna d C 1.2 1.8.6.4.2 Cereellum Ec Ev Ev As KO c Frequency Thymus KO KO 1 neo 11 Ec As 4 loxp
SUPPLEMENTARY INFORMATION doi:1.138/nture1371 As Brc11 (null) Brc5-13cK (conditioned llele) Reltive levels of mrna d C 1.2 1.8.6.4.2 Cereellum Ec Ev Ev As KO c Frequency Thymus KO KO 1 neo 11 Ec As 4 loxp
RESEARCH ARTICLE n.s.
 Cellular & Moleular Immunology (214), 12 ß 214 CSI and USTC. All rights reserved 1672-7681/14 $32. www.nature.om/mi a 5 RESEARCH ARTICLE TNF-a (pg/ml) 4 3 2 1 IL-6 (pg/ml) 15 1 5 IL-12p4 (pg/ml) 3 2 1
Cellular & Moleular Immunology (214), 12 ß 214 CSI and USTC. All rights reserved 1672-7681/14 $32. www.nature.om/mi a 5 RESEARCH ARTICLE TNF-a (pg/ml) 4 3 2 1 IL-6 (pg/ml) 15 1 5 IL-12p4 (pg/ml) 3 2 1
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature09732 Supplementary Figure 1: Depletion of Fbw7 results in elevated Mcl-1 abundance. a, Total thymocytes from 8-wk-old Lck-Cre/Fbw7 +/fl (Control) or Lck-Cre/Fbw7 fl/fl (Fbw7 KO) mice
doi:10.1038/nature09732 Supplementary Figure 1: Depletion of Fbw7 results in elevated Mcl-1 abundance. a, Total thymocytes from 8-wk-old Lck-Cre/Fbw7 +/fl (Control) or Lck-Cre/Fbw7 fl/fl (Fbw7 KO) mice
Supplementary Fig
 Supplementry Fig. 1 * 180-115- 82-64- 49-37- * 180-115- 85-64- 49-37- 26-26- Mem Cyt Mem Cyt Supplementry Fig.1 Specificity of nti-tie2 ntiodies. HUVECs were homogenized nd centrifuged t 400 000g to otin
Supplementry Fig. 1 * 180-115- 82-64- 49-37- * 180-115- 85-64- 49-37- 26-26- Mem Cyt Mem Cyt Supplementry Fig.1 Specificity of nti-tie2 ntiodies. HUVECs were homogenized nd centrifuged t 400 000g to otin
Figure S1 Yoo et al.
 doi:.38/nture6543 8 8 6 6 4 4 d Protoplsts Leves Reltive promoter ctivity (%) Reltive trnscript level 2 2 88 66 44 22 32 2 2 MKK-MYC MPK ctivity nti-mpk6 c ctr MKK - 4 5 4 5 MKK-MYC MPK3 ctivity MPK6 ctivity
doi:.38/nture6543 8 8 6 6 4 4 d Protoplsts Leves Reltive promoter ctivity (%) Reltive trnscript level 2 2 88 66 44 22 32 2 2 MKK-MYC MPK ctivity nti-mpk6 c ctr MKK - 4 5 4 5 MKK-MYC MPK3 ctivity MPK6 ctivity
WesternBright TM MCF and MCF-IR
 WesternBright TM MCF nd MCF-IR Quntittive, multi-color fluorescent Western lotting kits WesternBright MCF visile nd ner infrred (IR) fluorescent Western lotting kits llow the ssy of two proteins t once,
WesternBright TM MCF nd MCF-IR Quntittive, multi-color fluorescent Western lotting kits WesternBright MCF visile nd ner infrred (IR) fluorescent Western lotting kits llow the ssy of two proteins t once,
DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Ifna4 Control DAI Control DAI
 doi:1.138/nture613 (DLM-1/ZBP1) is ytosoli DNA sensor nd n tivtor of innte immune response Akinori Tkok 1,3, Zhiho Wng 1, Myoung Kwon hoi 1, Hideyuki Yni 1, Hideo Negishi 1, Ttsum Bn 1, Yn Lu 1, Mkoto
doi:1.138/nture613 (DLM-1/ZBP1) is ytosoli DNA sensor nd n tivtor of innte immune response Akinori Tkok 1,3, Zhiho Wng 1, Myoung Kwon hoi 1, Hideyuki Yni 1, Hideo Negishi 1, Ttsum Bn 1, Yn Lu 1, Mkoto
Supplementary Materials
 Supplementry Mterils Supplementry Figure 1 Supplementry Figure 2 in Fh deficient mice. Supplementry Figure 3 Supplementry Figure 4 Supplementry Figure 5 Supplementry Figure 6 Supplementry Figure 7 Supplementry
Supplementry Mterils Supplementry Figure 1 Supplementry Figure 2 in Fh deficient mice. Supplementry Figure 3 Supplementry Figure 4 Supplementry Figure 5 Supplementry Figure 6 Supplementry Figure 7 Supplementry
Supplementary Figure 1 PARP1 is involved in regulating the stability of mrnas from pro-inflammatory cytokine/chemokine mediators.
 Supplementary Figure 1 PARP1 is involved in regulating the stability of mrnas from pro-inflammatory cytokine/chemokine mediators. (a) A graphic depiction of the approach to determining the stability of
Supplementary Figure 1 PARP1 is involved in regulating the stability of mrnas from pro-inflammatory cytokine/chemokine mediators. (a) A graphic depiction of the approach to determining the stability of
Shigella IpgB1 promotes bacterial entry through the ELMO Dock180 machinery
 LETTERS promotes teril entry through the Dok18 mhinery Yutk Hn 1, Msto Suzuki 1, Kenji Ohy 1, Hiroki Iwi 1, Nozomi Ishijim 1, Anthony J. Koleske 3, Yoshinori Fukui 4 n Chihiro Sskw 1,2,5,6 use speil mehnism
LETTERS promotes teril entry through the Dok18 mhinery Yutk Hn 1, Msto Suzuki 1, Kenji Ohy 1, Hiroki Iwi 1, Nozomi Ishijim 1, Anthony J. Koleske 3, Yoshinori Fukui 4 n Chihiro Sskw 1,2,5,6 use speil mehnism
Mice Flow cytometry RT-MLPA analysis
 Mie Mie were used t 6 12 weeks of ge, unless stted otherwise, ge- nd sex-mthed within experiments nd were hndled in ordne with institutionl nd ntionl guidelines. All niml experiments were performed fter
Mie Mie were used t 6 12 weeks of ge, unless stted otherwise, ge- nd sex-mthed within experiments nd were hndled in ordne with institutionl nd ntionl guidelines. All niml experiments were performed fter
Endothelial Apelin-FGF Link Mediated by MicroRNAs 424 and 503 is Disrupted in Pulmonary Arterial Hypertension
 Endothelil Apelin-FGF Link Medited y MicroRNAs 424 nd 53 is Disrupted in Pulmonry Arteril Hypertension Jongmin Kim, Yujung Kng, Yoko Kojim, Jnet K. Lighthouse, Xioyue Hu, Michel A. Aldred, Dnielle L. McLen,
Endothelil Apelin-FGF Link Medited y MicroRNAs 424 nd 53 is Disrupted in Pulmonry Arteril Hypertension Jongmin Kim, Yujung Kng, Yoko Kojim, Jnet K. Lighthouse, Xioyue Hu, Michel A. Aldred, Dnielle L. McLen,
ΔPDD1 x ΔPDD1. ΔPDD1 x wild type. 70 kd Pdd1. Pdd3
 Supplemental Fig. S1 ΔPDD1 x wild type ΔPDD1 x ΔPDD1 70 kd Pdd1 50 kd 37 kd Pdd3 Supplemental Fig. S1. ΔPDD1 strains express no detectable Pdd1 protein. Western blot analysis of whole-protein extracts
Supplemental Fig. S1 ΔPDD1 x wild type ΔPDD1 x ΔPDD1 70 kd Pdd1 50 kd 37 kd Pdd3 Supplemental Fig. S1. ΔPDD1 strains express no detectable Pdd1 protein. Western blot analysis of whole-protein extracts
hcd1tg/hj1tg/ ApoE-/- hcd1tg/hj1tg/ ApoE+/+
 ApoE+/+ ApoE-/- ApoE-/- H&E (1x) Supplementary Figure 1. No obvious pathology is observed in the colon of diseased ApoE-/me. Colon samples were fixed in 1% formalin and laid out in Swiss rolls for paraffin
ApoE+/+ ApoE-/- ApoE-/- H&E (1x) Supplementary Figure 1. No obvious pathology is observed in the colon of diseased ApoE-/me. Colon samples were fixed in 1% formalin and laid out in Swiss rolls for paraffin
Legends for supplementary figures 1-3
 High throughput resistance profiling of Plasmodium falciparum infections based on custom dual indexing and Illumina next generation sequencing-technology Sidsel Nag 1,2 *, Marlene D. Dalgaard 3, Poul-Erik
High throughput resistance profiling of Plasmodium falciparum infections based on custom dual indexing and Illumina next generation sequencing-technology Sidsel Nag 1,2 *, Marlene D. Dalgaard 3, Poul-Erik
Supporting Information
 Supporting Information Transfection of DNA Cages into Mammalian Cells Email: a.turberfield@physics.ox.ac.uk Table of Contents Supporting Figure 1 DNA tetrahedra used in transfection experiments 2 Supporting
Supporting Information Transfection of DNA Cages into Mammalian Cells Email: a.turberfield@physics.ox.ac.uk Table of Contents Supporting Figure 1 DNA tetrahedra used in transfection experiments 2 Supporting
ARTICLES. Pyruvate kinase M2 is a phosphotyrosinebinding
 Vol 452 13 Mrh 2008 doi:10.1038/nture06667 ARTICLES Pyruvte kinse M2 is phosphotyrosineinding protein Hether R. Christofk 1, Mtthew G. Vnder Heiden 1,3, Ning Wu 1, John M. Asr 2,4 & Lewis C. Cntley 1,4
Vol 452 13 Mrh 2008 doi:10.1038/nture06667 ARTICLES Pyruvte kinse M2 is phosphotyrosineinding protein Hether R. Christofk 1, Mtthew G. Vnder Heiden 1,3, Ning Wu 1, John M. Asr 2,4 & Lewis C. Cntley 1,4
SUPPLEMENTARY INFORMATION
 NCS (ng/ml) Time (min) kd 500 500 0 30 0 30 IP: IP: 112 105 75 IB: ps407 IB: Mdm2 NCS + + IB: IB: tuulin IP input sup NCS (ng/ml) 50 100 500 Time (min) 0 15 30 60 120 15 30 60 120 15 30 60 120 IB: ps407
NCS (ng/ml) Time (min) kd 500 500 0 30 0 30 IP: IP: 112 105 75 IB: ps407 IB: Mdm2 NCS + + IB: IB: tuulin IP input sup NCS (ng/ml) 50 100 500 Time (min) 0 15 30 60 120 15 30 60 120 15 30 60 120 IB: ps407
Supplemental Materials and Methods
 Supplemental Materials and Methods Antibodies: Anti-SRF (cat# Sc-335) and anti-igf1r (sc-712) (Santa Cruz Biotech), and anti- ADAM-10 (14-6211) were from e-bioscience, anti-ku70 (cat# MS-329-P) (Labvision),
Supplemental Materials and Methods Antibodies: Anti-SRF (cat# Sc-335) and anti-igf1r (sc-712) (Santa Cruz Biotech), and anti- ADAM-10 (14-6211) were from e-bioscience, anti-ku70 (cat# MS-329-P) (Labvision),
Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and
 Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
Materials Protein synthesis kit. This kit consists of 24 amino acids, 24 transfer RNAs, four messenger RNAs and one ribosome (see below).
 Protein Synthesis Instructions The purpose of today s lab is to: Understand how a cell manufactures proteins from amino acids, using information stored in the genetic code. Assemble models of four very
Protein Synthesis Instructions The purpose of today s lab is to: Understand how a cell manufactures proteins from amino acids, using information stored in the genetic code. Assemble models of four very
SUPPLEMENTARY INFORMATION. Small molecule activation of the TRAIL receptor DR5 in human cancer cells
 SUPPLEMENTARY INFORMATION Small molecule activation of the TRAIL receptor DR5 in human cancer cells Gelin Wang 1*, Xiaoming Wang 2, Hong Yu 1, Shuguang Wei 1, Noelle Williams 1, Daniel L. Holmes 1, Randal
SUPPLEMENTARY INFORMATION Small molecule activation of the TRAIL receptor DR5 in human cancer cells Gelin Wang 1*, Xiaoming Wang 2, Hong Yu 1, Shuguang Wei 1, Noelle Williams 1, Daniel L. Holmes 1, Randal
SUPPLEMENTARY INFORMATION
 BRC repet RPA DSB RAD52 DSB Repir doi:1.138/nture9399 Gp Repir ssdna/dsdna junction ssdna/dsdna junction RPA Binding Resection RPA Binding Filment Formtion or or Filment Formtion DNA Piring DNA Piring
BRC repet RPA DSB RAD52 DSB Repir doi:1.138/nture9399 Gp Repir ssdna/dsdna junction ssdna/dsdna junction RPA Binding Resection RPA Binding Filment Formtion or or Filment Formtion DNA Piring DNA Piring
5 -triphosphate-sirna: turning gene silencing and Rig-I activation against melanoma
 28 Nture Pulishing Group http://www.nture.om/nturemediine -triphosphte-sirna: turning gene silening nd Rig-I tivtion ginst melnom Hendrik Poek 1 3,12, Roert Besh 4,12, Cornelius Mihoefer 2,3,12, Mrel Renn,12,
28 Nture Pulishing Group http://www.nture.om/nturemediine -triphosphte-sirna: turning gene silening nd Rig-I tivtion ginst melnom Hendrik Poek 1 3,12, Roert Besh 4,12, Cornelius Mihoefer 2,3,12, Mrel Renn,12,
Meganuclease-mediated Virus Self-cleavage Facilitates Tumor-specific Virus Replication
 originl rtile The Amerin Soiety of Gene & Cell Therpy Megnulese-medited Virus Self-levge Filittes Tumor-speifi Virus Replition Engin Gürlevik 1, Peter Shhe 1, Anneliese Goez 1, Arnold Kloos 1, Normn Woller
originl rtile The Amerin Soiety of Gene & Cell Therpy Megnulese-medited Virus Self-levge Filittes Tumor-speifi Virus Replition Engin Gürlevik 1, Peter Shhe 1, Anneliese Goez 1, Arnold Kloos 1, Normn Woller
SUPPLEMENTARY INFORMATION
 doi:10.1038/nture09470 prmt5-1 prmt5-2 Premture Stop Codon () GGA TGA PRMT5 Hypocotyl Length (Reltive to Drk) 0.5 0.3 0.1 ** 30 *** c prmt5-1 d ** *** 150 prmt5-2 ** *** 28 100 26 24 50 prmt5-1 prmt5-2
doi:10.1038/nture09470 prmt5-1 prmt5-2 Premture Stop Codon () GGA TGA PRMT5 Hypocotyl Length (Reltive to Drk) 0.5 0.3 0.1 ** 30 *** c prmt5-1 d ** *** 150 prmt5-2 ** *** 28 100 26 24 50 prmt5-1 prmt5-2
LETTER. SEC14L2 enables pan-genotype HCV replication in cell culture
 LETTER doi:.8/nture899 enles pn-genotype HCV replition in ell ulture Mohsn Seed, Ursul Andreo, Hyo-Young Chung, Christine Espiritu, Andre D. Brnh, Jose M. Silv & Chrles M. Rie Sine its disovery in 989,
LETTER doi:.8/nture899 enles pn-genotype HCV replition in ell ulture Mohsn Seed, Ursul Andreo, Hyo-Young Chung, Christine Espiritu, Andre D. Brnh, Jose M. Silv & Chrles M. Rie Sine its disovery in 989,
Cysteine methylation disrupts ubiquitin-chain sensing in NF-kB activation. NF-κB activation (fold change) NleE TRAF p-ikk /β.
 doi:10.1038/nture10690 Cysteine methyltion disrupts uiquitin-hin sensing in NF-kB tivtion Li Zhng 1,2, Xiojun Ding 2, Jixin Cui 2,HoXu 2, Jing Chen 2, Yi-Nn Gong 2, Liyn Hu 2, Yn Zhou 2, Jinning Ge 2,
doi:10.1038/nture10690 Cysteine methyltion disrupts uiquitin-hin sensing in NF-kB tivtion Li Zhng 1,2, Xiojun Ding 2, Jixin Cui 2,HoXu 2, Jing Chen 2, Yi-Nn Gong 2, Liyn Hu 2, Yn Zhou 2, Jinning Ge 2,
An engineered tryptophan zipper-type peptide as a molecular recognition scaffold
 SUPPLEMENTARY MATERIAL An engineered tryptophan zipper-type peptide as a molecular recognition scaffold Zihao Cheng and Robert E. Campbell* Supplementary Methods Library construction for FRET-based screening
SUPPLEMENTARY MATERIAL An engineered tryptophan zipper-type peptide as a molecular recognition scaffold Zihao Cheng and Robert E. Campbell* Supplementary Methods Library construction for FRET-based screening
SUPPLEMENTARY INFORMATION
 SI Fig. PrpS is single copy gene k 3. 9... EcoRV EcoRV k 5 BmH Pst c well k HindIII HindIII HindIII.3.5 3.. Southern lots of Ppver genomic DNA from plnts with SS8 hplotypes, hyridized with PrpS proe..
SI Fig. PrpS is single copy gene k 3. 9... EcoRV EcoRV k 5 BmH Pst c well k HindIII HindIII HindIII.3.5 3.. Southern lots of Ppver genomic DNA from plnts with SS8 hplotypes, hyridized with PrpS proe..
Disease and selection in the human genome 3
 Disease and selection in the human genome 3 Ka/Ks revisited Please sit in row K or forward RBFD: human populations, adaptation and immunity Neandertal Museum, Mettman Germany Sequence genome Measure expression
Disease and selection in the human genome 3 Ka/Ks revisited Please sit in row K or forward RBFD: human populations, adaptation and immunity Neandertal Museum, Mettman Germany Sequence genome Measure expression
Supplemental Figure Legends:
 Supplemental Figure Legends: Fig S1. GFP-ABRO1 localization. U2OS cells were infected with retrovirus expressing GFP- ABRO1. The cells were fixed with 3.6% formaldehyde and stained with antibodies against
Supplemental Figure Legends: Fig S1. GFP-ABRO1 localization. U2OS cells were infected with retrovirus expressing GFP- ABRO1. The cells were fixed with 3.6% formaldehyde and stained with antibodies against
Arabidopsis actin depolymerizing factor AtADF4 mediates defense signal transduction triggered by the Pseudomonas syringae effector AvrPphB
 Arabidopsis actin depolymerizing factor mediates defense signal transduction triggered by the Pseudomonas syringae effector AvrPphB Files in this Data Supplement: Supplemental Table S1 Supplemental Table
Arabidopsis actin depolymerizing factor mediates defense signal transduction triggered by the Pseudomonas syringae effector AvrPphB Files in this Data Supplement: Supplemental Table S1 Supplemental Table
