Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Tables
|
|
|
- Neil Ellis
- 5 years ago
- Views:
Transcription
1 Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Tables Title of file for HTML: Supplementary Data 1 Description: Complete mass isotopomer distribution of data displayed in Fig 6d and Fig S7d from U- 13C-proline after 24hrs. All data are presented as mean ± SEM. n=3 replicates per condition. M represents unlabelled molecules with each increasing number representing additional 13C-labelled carbons.
2 PDAC Normal pancreas #1 #6 #1 #14 #2 #7 #2 #15 #3 #8 #3 #16 #4 #9 #12 #17 #5 #1 #13 #18 #11 Supplementary Figure 1 PRODH1 is expressed in human PDAC samples. Staining of PRODH1 by IHC on human tissue macroarray (TMA) containing PDAC (n=1), adjacent normal pancreas (n=3, #1 to #3) and normal pancreas from PDAC-free patients (n=8, #4 to #11). Quantification of PRODH1 expression in TMA samples was determined by densitometry using Image J software (NIH) with the same threshold for all images. Scale bar: µm.
3 49.2 +/- 13.4% /- 11.3% /- 9.4% Complete mm Glc mm Gln 8 +/- 14.5% 8 +/- 15.3% /- 9.2% /- % 1% O2 (48hrs) 1 b * Complete mm Glc f mm Gln Col IV/DAPI Merge mm Gln /- 2.8% /- 1.4% mm Glc, 1% O 2 Lamp1/DAPI mm Glc Col IV/DAPI / Complete Col IV DQ positive cells (%) Col I DQ positive cells (%) e 1% O2 (48hrs) Collagen IV DQ fluorescein intensity c Col I mean fluorescence intensity /- 4.5% b 4 * 3 1% O2 (48hrs) c c 2 * 1 Complete mm Glc d Col IV mean fluorescence intensity /- 8.2% mm Gln /- 8.9% mm Glc Positive cells (%) Complete Collagen I DQ fluorescein intensity Positive cells (%) a mm Gln 1% O2 (48hrs) Complete mm Glc mm Gln, 1% O 2 Lamp1/DAPI mm Gln Merge Supplementary Figure 2 PDAC cells take up and degrade collagen after glucose or glutamine deprivation under hypoxia. (a) and (b) Representative flow cytometry profiles (upper panels) and charts (lower panels) representing the percentage of PK4A cells taking up collagen I (a) or IV (b) DQ. Mean (c) Col I or (d) Col IV DQ fluorescence intensity (MFI) in PK4A cells cultured during 48hrs under hypoxia (1% O2) as in Fig. 3a-b. Data represent mean ± SEM (n=3 independent experiments). P values, indicated by asterisks, show statistical significance relative to respective culture condition without (* p<5), while letters indicate statistical significance relative to complete media without (c p<5, b p<1). Two-tailed unpaired Student s t-test. (e) and (f) Representative co-staining of Col IV DQ and Lamp1 lysosomal marker in PK4A cells cultured in glucose-free (e) or glutamine-free (f) media under hypoxic conditions with or without. Insets show magnification of Col IV, Lamp1 or Col IV/Lamp1 (merge) PKA4 cells in each condition. Images are representative of n=3 independent experiments. Scale bar: µm.
4 a Relative Mrc2 expression (fold) Control *** PDAC b uparap/ Endo18 Amido Black Control PDAC -167 c Murine Human uparap/endo18 Supplementary Figure 3 MRC2/Endo18 is expressed by PDAC tumour cells. (a) mrna levels of the receptor uparap/endo18 (Mrc2 gene) measured by quantitative RT-PCR in PDAC from PKI mice (n=3) versus control pancreases from KI mice (n=3). Data are presented as mean ± SEM. *** p<1; two-tailed unpaired Student s t-test. (b) Protein levels of uparap/endo18 in PDAC and control pancreas from PKI and KI mice, respectively (upper panel). Marker size in is indicated. Total protein stained with Amido black were used as loading control (lower panel). Uncropped images of blots are shown in Supplementary Fig. 1. (c) Representative uparap/endo18 staining in human and murine PDAC sections (n=4 patients and n=3 PKI mice). Scale bar: 1 µm.
5 a mm b mm Uncoated Col I-coated Uncoated Col I-coated Supplementary Figure 4 Collagen promotes PDAC cell survival. (a) and (b) Survival of PK4A cells cultured in glucose-free media on uncoated or coated-collagen I plates. Representative images of unstained cells (a) and cells stained with crystal violet (b) after 96hrs of treatment (n=2 independent experiments). Scale bar: 1, µm (a: upper images), 2 µm (a: lower images) and 5 mm (b).
6 Percent of Total Carbon mM Total Acc ounting 5mM mm mm Glutamine Non-Polar Protein Polar RNA DNA Percent of Total Carbon mM 5mM Protein mm mm Glutamine Percent of Total Carbon mM Nucleotides 5mM mm mm Glutamine RNA DNA Percent of Total Carbon mM Non-polar 5mM mm mm Glutamine Supplementary Figure 5 Accounting of proline-derived carbon contributions to different cellular macromolecules. Relative contributions of proline to proteins, nucleotides (RNA, DNA) and non-polar (lipids) fractions determined under different nutrient conditions. Each bar represents the average of n=3 biological replicates ± SEM.
7 a concentration (mm) Time (hrs) Lactate concentration (mm) *** **** **** **** Time (hrs) 25mM 5mM 1mM mm b Time 1hr 3hrs 6hrs 24hrs Glutamine 4mM 2mM.5mM 4mM 2mM.5mM 4mM 2mM.5mM 4mM 2mM.5mM M SEM M SEM M SEM M SEM Supplementary Figure 6 Glutamine depletion does not change relative glucose contribution to lactate production. (a) uptake (left panel) and lactate production (right panel) by subconfluent PK4A cells cultured under gradual glucose deprivation (25mM, 5mM, 1mM and mm) at 8, 14 and 24hrs. Data are mean ± SEM, n=4 independent experiments. P value indicated by asterisks is relative to lactate value in 25mM glucose media at each time point. *** p< 1, **** p< 1; two-way ANOVA followed by Bonferroni post-hoc test. (b) Complete mass isotopomer distribution (MID) of lactate from U-13C-glucose at 1, 3, 6 and 24hrs under a gradual decline in glutamine concentration. All data are presented as mean ± SEM. n=3 replicates per condition.
8 a MEF Collagen I Fibronectin Hsp9 ECM MEF ECM MEF ECM kd 2 1 MEF ECM kd b ECM - Acid Hydrolysis Glu+Gln Hydroxyproline Proline c Short Exposure 4 Long Exposure 1 M M1 M2 M3 M4 M5 Mass Isotopomer Distribution % of Total Amino Acids Pool ECM DMEM DMEM +1% FBS d Ala Arg Asp Glu +Asn +Gln Gly Ile Leu Lys Met Phe Pro Ser Thr Tyr Val Amino Acid Aspartate M M1 M2 M3 M4 Pyruvate M M1 M2 M3 CO 2 Malate M M1 M2 M3 M4 Acetyl-CoA Citrate M M1 M2 M3 M4 M5 M6 TCA Cycle mM 5mM mm mm Glutamine akg CO M M1 M2 M3 M4 M5 25mM 5mM mm mm Glutamine M Proline M5 Glutamate M M1 M2 M3 M4 M5 Supplementary Figure 7 Fumarate M M1 M2 M3 M4 Succinate M M1 M2 M3 M4 CO 2 Glutamine M M1 M2 M3 M4 M5 ECM is proline-rich and essential amino acid poor and proline supplies the TCA cycle. (a) Western blot of primary MEFs and de-cellularised ECM indicating successful de-cellularisation of ECM prior to PK4A cells plating. Uncropped blots are representative of n=2 independent experiments. (b) Full mass isotopomer distribution of labelled ECM after acid hydrolysis. Data is from n=1 experiment and is representative of n=2 experiments. M represents unlabelled molecules with each increasing number representing additional 13 C-labelled carbons. (c) Percent of total protein pool of each amino acid following acid hydrolysis of fibroblast-synthesized ECM, DMEM and DMEM + 1% FBS. (d) Full GCMS tracing analysis of U- 13 C-proline into central carbon metabolism in PDAC cells after 24hrs under indicated nutrient conditions. M represents unlabelled molecules with each increasing number representing another labelled carbon.
9 a Proline Time 25mM + 4mM Gln 5mM + 4mM Gln mm + 4mM Gln 25mM + mm Gln O2 21% 1% 21% 1% 21% 1% 21% 1% M SEM M SEM M SEM M SEM M SEM M SEM Glutamate Time 25mM + 4mM Gln 5mM + 4mM Gln mm + 4mM Gln 25mM + mm Gln O2 21% 1% 21% 1% 21% 1% 21% 1% M SEM M SEM M SEM M SEM M SEM M SEM b akg Time 25mM + 4mM Gln 5mM + 4mM Gln mm + 4mM Gln 25mM + mm Gln O2 21% 1% 21% 1% 21% 1% 21% 1% M SEM M SEM M SEM M SEM M SEM M SEM Citrate akg Succinate mM Aspartate Fumarate Malate Proline Glutamine Glutamate DHP + DHP Citrate akg Succinate mM Glutamine Aspartate Fumarate Malate Proline Glutamine Glutamate Supplementary Figure 8 Hypoxia does not influence proline conversion to glutamate or alpha-ketoglutarate but DHP inhibits metabolism of proline to TCA. (a) Complete mass isotopomer distribution of proline, glutamate and alpha-ketoglutarate (α-kg) from U- 13 C-proline after 24hrs at the indicated conditions in normoxia (21% O 2 ) and hypoxia (1% O 2 ). All data are presented as mean ± SEM. n=3 replicates per condition. M represents unlabelled molecules with each increasing number representing additional 13 C-labelled carbons. (b) Fractional labelling of proline, glutamine and TCA intermediate from U- 13 C-proline after 24hrs at 5mM glucose (left charts) or.5mm glutamine (right charts) with or without DHP 1mM. All data presented as mean ± SEM. n=3 replicates per condition.
10 PRODH1 _clone 9 _clone 3 a b sg-control _clone 9 sg-control - Complete _clone 3 β-actin - PRODH1 (Relative expression) sg-control * * _clone 9 _clone 3 Colony-forming area 1 1 sg-control * * _clone 9 _clone 3 Supplementary Figure 9 Prodh1 gene-editing in PK4A cells using the CRISPR-Cas9 system. (a) Illustration and quantification (upper and lower panels, respectively) of PRODH1 levels in sg-control and _clone9 and _clone3 PK4A cells revealed by western blot analysis (marker size indicated in ). Protein levels in each cell line are normalised to β-actin. Data are expressed as mean ± SEM (n=2 independent experiments). Uncropped images of blots are shown in supplementary Fig. 1. * p<5, two-tailed unpaired Student s t-test. (b) Representative clonogenic assay (upper panel) and quantification of sg-control and _clone 9 and _clone 3 PK4A cells colony-forming area (lower panel) in complete media. Data are mean ± SEM (n=3 independent experiments). * p<5, two-tailed unpaired Student s t-test. Scale bar: 5 mm
11 Fig 1e Complete hrs 48hrs Prolidase Fig 2d Control 2 3 PDAC Control PDAC Control PDAC P4HA3 PRODH1 Amido black Fig 4b mm.5 mm Glutamine -ColIV +ColIV -ColIV +ColIV perk1/2 perk1/2 ERK1/2 ERK1/2 mm -ColIV +ColIV.5 mm Glutamine -ColIV +ColIV pt389-p7s6k pt389-p7s6k Supplementary Figure 1a
12 mm -ColIV +ColIV.5 mm Glutamine -ColIV + ColIV p7s6k p7s6k Fig 4c Supp. Fig 3b 148 mm 24hrs 48hrs mm Glutamine 24hrs 48hrs 1 Control 2 3 PDAC uparap/endo18 Prolidase Amido black Supp. Fig 9a KDa sg-control clone 9 clone 3 KDa sg-control clone 9 clone 3 PRODH1 β-actin Supplementary Figure 1b
13 Supplementary Table 1. Primer sequences (mus musculus) used to determine transcript expression profiles by real-time PCR Gene name Forward primer (5'-->3') Reverse primer (5'-->3') RefSeq mrna pepd GGGAAAGATCCATTCCAAGG CACTGCCACTGTCTGTGTTG NM_882 mmp2 CCAGATCACATACAGGATCATT CCATCATGGATTCGAGAAAA NM_861 mmp9 CAGCTGGCAGAGGCATACTT TTCTGAAGCATCAGCAAAGC NM_ mmp13 GGGACTAAAGAACATGGTGACT AGCCTTTGGAACTGCTTGTC NM_867.2 b4 GCTGATGGGCAAGAACACCA CCCAAAGCCTGGAAGAAGGA NM_ endo18 GCCCCATCAAGAGTAACGAC CGTGATACTCAGCAAGTCTGC NM_ Supplementary Table 2. Amino Acid Fragments Used for Isotope Quantification Metabolite Carbons a Formula b Mass (m/z) KG C 14 H 28 O 5 NSi Ala 23 C 1 H 26 ONSi Ala 123 C 11 H 26 O 2 NSi 2 26 Arg C 17 H 38 N 3 Si 2 34 Arg C 2 H 44 O 2 N 3 Si Asp 12 C 14 H 32 O 2 NSi 2 32 Asp 234 C 17 H 4 O 3 NSi 3 39 Asp 1234 C 18 H 4 O 4 NSi Cit C 2 H 39 O 6 Si Cit C 26 H 55 O 7 Si Fum 1234 C 12 H 23 O 4 NSi Gln C 19 H 43 O 3 N 2 Si Glu 2345 C 16 H O 2 NSi 2 33 Glu C 19 H 42 O 4 NSi Gly 2 C 9 H 24 ONSi Gly 12 C 1 H 24 O 2 NSi Ile C 11 H 26 NSi 2 Ile C 13 H 32 ONSi Ile C 14 H 32 O 2 NSi 2 32 Lac 123 C 11 H 25 O 3 Si Leu C 11 H 26 NSi 2 Leu C 13 H 32 ONSi Leu C 14 H 32 O 2 NSi 2 32 Lys C 17 H 41 N 2 Si Lys C 2 H 47 O 2 N 2 Si Mal 1234 C 18 H 39 O 5 Si Met 2345 C 1 H 24 NSiS 218 Met 2345 C 12 H 3 ONSi 2 S 292 Met C 13 H 3 O 2 NSi 2 S 32 Phe C 14 H 24 NSi 234 Phe C 14 H 32 O 2 NSi 2 38 Phe C 17 H 3 O 2 NSi 2 3 Pro C 16 H O 2 NSi 2 33 (Hydroxy)Pro C 19 H 45 O 3 NSi Pyr 123 C 6 H 12 O 3 NSi 174 Suc 1234 C 12 H 25 O 4 Si Ser 23 C 14 H 34 ONSi Ser 12 C 14 H 32 O 2 NSi 2 32 Ser 23 C 16 H 4 O 2 NSi 3 2 1
14 Ser 123 C 17 H 4 O 3 NSi 3 39 Thr 234 C 17 H 42 O 2 NSi Thr 1234 C 18 H 42 O 3 NSi 3 44 Tyr C 2 H 38 ONSi 2 3 Val 2345 C 12 H 3 ONSi 2 26 Val C 13 H 3 O 2 NSi a = Carbons indicates the carbons present in the derivative that is measured via GC-MS. b = chemical formula of the derivative measured via GC-MS (derivatization process described in the method section) 2
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature12138 Supplementary Figure 1. Knockdown of KRAS leads to a reduction in macropinocytosis. (a) KRAS knockdown in MIA PaCa-2 cells expressing KRASspecific shrnas
SUPPLEMENTARY INFORMATION doi:10.1038/nature12138 Supplementary Figure 1. Knockdown of KRAS leads to a reduction in macropinocytosis. (a) KRAS knockdown in MIA PaCa-2 cells expressing KRASspecific shrnas
466 Asn (N) to Ala (A) Generate beta dimer Interface
 Table S1: Amino acid changes to the HexA α-subunit to convert the dimer interface from α to β and to introduce the putative GM2A binding surface from β- onto the α- subunit Residue position (α-numbering)
Table S1: Amino acid changes to the HexA α-subunit to convert the dimer interface from α to β and to introduce the putative GM2A binding surface from β- onto the α- subunit Residue position (α-numbering)
36. The double bonds in naturally-occuring fatty acids are usually isomers. A. cis B. trans C. both cis and trans D. D- E. L-
 36. The double bonds in naturally-occuring fatty acids are usually isomers. A. cis B. trans C. both cis and trans D. D- E. L- 37. The essential fatty acids are A. palmitic acid B. linoleic acid C. linolenic
36. The double bonds in naturally-occuring fatty acids are usually isomers. A. cis B. trans C. both cis and trans D. D- E. L- 37. The essential fatty acids are A. palmitic acid B. linoleic acid C. linolenic
Basic concepts of molecular biology
 Basic concepts of molecular biology Gabriella Trucco Email: gabriella.trucco@unimi.it Life The main actors in the chemistry of life are molecules called proteins nucleic acids Proteins: many different
Basic concepts of molecular biology Gabriella Trucco Email: gabriella.trucco@unimi.it Life The main actors in the chemistry of life are molecules called proteins nucleic acids Proteins: many different
 Problem: The GC base pairs are more stable than AT base pairs. Why? 5. Triple-stranded DNA was first observed in 1957. Scientists later discovered that the formation of triplestranded DNA involves a type
Problem: The GC base pairs are more stable than AT base pairs. Why? 5. Triple-stranded DNA was first observed in 1957. Scientists later discovered that the formation of triplestranded DNA involves a type
Algorithms in Bioinformatics ONE Transcription Translation
 Algorithms in Bioinformatics ONE Transcription Translation Sami Khuri Department of Computer Science San José State University sami.khuri@sjsu.edu Biology Review DNA RNA Proteins Central Dogma Transcription
Algorithms in Bioinformatics ONE Transcription Translation Sami Khuri Department of Computer Science San José State University sami.khuri@sjsu.edu Biology Review DNA RNA Proteins Central Dogma Transcription
11 questions for a total of 120 points
 Your Name: BYS 201, Final Exam, May 3, 2010 11 questions for a total of 120 points 1. 25 points Take a close look at these tables of amino acids. Some of them are hydrophilic, some hydrophobic, some positive
Your Name: BYS 201, Final Exam, May 3, 2010 11 questions for a total of 120 points 1. 25 points Take a close look at these tables of amino acids. Some of them are hydrophilic, some hydrophobic, some positive
Biochemistry and Cell Biology
 Biochemistry and Cell Biology Monomersare simple molecules that can be linked into chains. (Nucleotides, Amino acids, monosaccharides) Polymersare the long chains of monomers. (DNA, RNA, Cellulose, Protein,
Biochemistry and Cell Biology Monomersare simple molecules that can be linked into chains. (Nucleotides, Amino acids, monosaccharides) Polymersare the long chains of monomers. (DNA, RNA, Cellulose, Protein,
De novo sequencing in the identification of mass data. Wang Quanhui Liu Siqi Beijing Institute of Genomics, CAS
 De novo sequencing in the identification of mass data Wang Quanhui Liu Siqi Beijing Institute of Genomics, CAS The difficulties in mass data analysis Although the techniques of genomic sequencing are being
De novo sequencing in the identification of mass data Wang Quanhui Liu Siqi Beijing Institute of Genomics, CAS The difficulties in mass data analysis Although the techniques of genomic sequencing are being
Zool 3200: Cell Biology Exam 3 3/6/15
 Name: Trask Zool 3200: Cell Biology Exam 3 3/6/15 Answer each of the following questions in the space provided; circle the correct answer or answers for each multiple choice question and circle either
Name: Trask Zool 3200: Cell Biology Exam 3 3/6/15 Answer each of the following questions in the space provided; circle the correct answer or answers for each multiple choice question and circle either
Nature Biotechnology: doi: /nbt Supplementary Figure 1
 Supplementary Figure 1 Generation of the AARE-Gene system construct. (a) Position and sequence alignment of AAREs extracted from human Trb3, Chop or Atf3 promoters. AARE core sequences are boxed in grey.
Supplementary Figure 1 Generation of the AARE-Gene system construct. (a) Position and sequence alignment of AAREs extracted from human Trb3, Chop or Atf3 promoters. AARE core sequences are boxed in grey.
WT Day 90 after injections
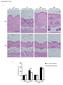 Supplementary Figure 1 a Day 1 after injections Day 9 after injections Klf5 +/- Day 1 after injections Klf5 +/- Day 9 after injections BLM PBS b Day 1 after injections Dermal thickness (μm) 3 1 Day 9 after
Supplementary Figure 1 a Day 1 after injections Day 9 after injections Klf5 +/- Day 1 after injections Klf5 +/- Day 9 after injections BLM PBS b Day 1 after injections Dermal thickness (μm) 3 1 Day 9 after
Station 1: DNA Structure Use the figure above to answer each of the following questions. 1.This is the subunit that DNA is composed of. 2.
 1. Station 1: DNA Structure Use the figure above to answer each of the following questions. 1.This is the subunit that DNA is composed of. 2.This subunit is composed of what 3 parts? 3.What molecules make
1. Station 1: DNA Structure Use the figure above to answer each of the following questions. 1.This is the subunit that DNA is composed of. 2.This subunit is composed of what 3 parts? 3.What molecules make
Materials Protein synthesis kit. This kit consists of 24 amino acids, 24 transfer RNAs, four messenger RNAs and one ribosome (see below).
 Protein Synthesis Instructions The purpose of today s lab is to: Understand how a cell manufactures proteins from amino acids, using information stored in the genetic code. Assemble models of four very
Protein Synthesis Instructions The purpose of today s lab is to: Understand how a cell manufactures proteins from amino acids, using information stored in the genetic code. Assemble models of four very
Problem Set Unit The base ratios in the DNA and RNA for an onion (Allium cepa) are given below.
 Problem Set Unit 3 Name 1. Which molecule is found in both DNA and RNA? A. Ribose B. Uracil C. Phosphate D. Amino acid 2. Which molecules form the nucleotide marked in the diagram? A. phosphate, deoxyribose
Problem Set Unit 3 Name 1. Which molecule is found in both DNA and RNA? A. Ribose B. Uracil C. Phosphate D. Amino acid 2. Which molecules form the nucleotide marked in the diagram? A. phosphate, deoxyribose
Basic concepts of molecular biology
 Basic concepts of molecular biology Gabriella Trucco Email: gabriella.trucco@unimi.it What is life made of? 1665: Robert Hooke discovered that organisms are composed of individual compartments called cells
Basic concepts of molecular biology Gabriella Trucco Email: gabriella.trucco@unimi.it What is life made of? 1665: Robert Hooke discovered that organisms are composed of individual compartments called cells
Protein Synthesis. Application Based Questions
 Protein Synthesis Application Based Questions MRNA Triplet Codons Note: Logic behind the single letter abbreviations can be found at: http://www.biology.arizona.edu/biochemistry/problem_sets/aa/dayhoff.html
Protein Synthesis Application Based Questions MRNA Triplet Codons Note: Logic behind the single letter abbreviations can be found at: http://www.biology.arizona.edu/biochemistry/problem_sets/aa/dayhoff.html
Supplementary Figure 1. Antigens generated for mab development (a) K9M1P1-mIgG and hgh-k9m1p1 antigen (~37 kda) expression verified by western blot
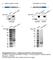 Supplementary Figure 1. Antigens generated for mab development (a) K9M1P1-mIgG and hgh-k9m1p1 antigen (~37 kda) expression verified by western blot (vector: ~25 kda). (b) Silver staining was used to assess
Supplementary Figure 1. Antigens generated for mab development (a) K9M1P1-mIgG and hgh-k9m1p1 antigen (~37 kda) expression verified by western blot (vector: ~25 kda). (b) Silver staining was used to assess
DNA.notebook March 08, DNA Overview
 DNA Overview Deoxyribonucleic Acid, or DNA, must be able to do 2 things: 1) give instructions for building and maintaining cells. 2) be copied each time a cell divides. DNA is made of subunits called nucleotides
DNA Overview Deoxyribonucleic Acid, or DNA, must be able to do 2 things: 1) give instructions for building and maintaining cells. 2) be copied each time a cell divides. DNA is made of subunits called nucleotides
Alpha-helices, beta-sheets and U-turns within a protein are stabilized by (hint: two words).
 1 Quiz1 Q1 2011 Alpha-helices, beta-sheets and U-turns within a protein are stabilized by (hint: two words) Value Correct Answer 1 noncovalent interactions 100% Equals hydrogen bonds (100%) Equals H-bonds
1 Quiz1 Q1 2011 Alpha-helices, beta-sheets and U-turns within a protein are stabilized by (hint: two words) Value Correct Answer 1 noncovalent interactions 100% Equals hydrogen bonds (100%) Equals H-bonds
Using DNA sequence, distinguish species in the same genus from one another.
 Species Identification: Penguins 7. It s Not All Black and White! Name: Objective Using DNA sequence, distinguish species in the same genus from one another. Background In this activity, we will observe
Species Identification: Penguins 7. It s Not All Black and White! Name: Objective Using DNA sequence, distinguish species in the same genus from one another. Background In this activity, we will observe
SUPPLEMENTARY INFORMATION FOR: Mycobacterium tuberculosis nitrogen assimilation and host colonization require aspartate
 SUPPLEMENTARY INFORMATION FOR: Mycobacterium tuberculosis nitrogen assimilation and host colonization require aspartate Alexandre Gouzy 1,2, Gérald Larrouy-Maumus 3, Ting-Di Wu 4,, Antonio Peixoto 1,2,
SUPPLEMENTARY INFORMATION FOR: Mycobacterium tuberculosis nitrogen assimilation and host colonization require aspartate Alexandre Gouzy 1,2, Gérald Larrouy-Maumus 3, Ting-Di Wu 4,, Antonio Peixoto 1,2,
NORWEGIAN UNIVERSITY OF SCIENCE AND TECHNOLOGY DEPARTMENT OF BIOTECHNOLOGY Professor Bjørn E. Christensen, Department of Biotechnology
 Page 1 NRWEGIAN UNIVERSITY F SCIENCE AND TECNLGY DEPARTMENT F BITECNLGY Professor Bjørn E. Christensen, Department of Biotechnology Contact during the exam: phone: 73593327, 92634016 EXAM TBT4135 BIPLYMERS
Page 1 NRWEGIAN UNIVERSITY F SCIENCE AND TECNLGY DEPARTMENT F BITECNLGY Professor Bjørn E. Christensen, Department of Biotechnology Contact during the exam: phone: 73593327, 92634016 EXAM TBT4135 BIPLYMERS
Bioinformatics. ONE Introduction to Biology. Sami Khuri Department of Computer Science San José State University Biology/CS 123A Fall 2012
 Bioinformatics ONE Introduction to Biology Sami Khuri Department of Computer Science San José State University Biology/CS 123A Fall 2012 Biology Review DNA RNA Proteins Central Dogma Transcription Translation
Bioinformatics ONE Introduction to Biology Sami Khuri Department of Computer Science San José State University Biology/CS 123A Fall 2012 Biology Review DNA RNA Proteins Central Dogma Transcription Translation
Nucleic acid and protein Flow of genetic information
 Nucleic acid and protein Flow of genetic information References: Glick, BR and JJ Pasternak, 2003, Molecular Biotechnology: Principles and Applications of Recombinant DNA, ASM Press, Washington DC, pages.
Nucleic acid and protein Flow of genetic information References: Glick, BR and JJ Pasternak, 2003, Molecular Biotechnology: Principles and Applications of Recombinant DNA, ASM Press, Washington DC, pages.
Automated bioreactor sampling for on-line analysis of amino acids using two approaches: HPLC and Cedex
 Application note #004/Nov 2018 Automated bioreactor sampling for on-line analysis of amino acids using two approaches: HPLC and Cedex Bio HT Alexandra Hofer Securecell AG, Zürcherstrasse 137, 8952 Schlieren,
Application note #004/Nov 2018 Automated bioreactor sampling for on-line analysis of amino acids using two approaches: HPLC and Cedex Bio HT Alexandra Hofer Securecell AG, Zürcherstrasse 137, 8952 Schlieren,
First&year&tutorial&in&Chemical&Biology&(amino&acids,&peptide&and&proteins)&! 1.&!
 First&year&tutorial&in&Chemical&Biology&(amino&acids,&peptide&and&proteins& 1.& a. b. c. d. e. 2.& a. b. c. d. e. f. & UsingtheCahn Ingold Prelogsystem,assignstereochemicaldescriptorstothe threeaminoacidsshownbelow.
First&year&tutorial&in&Chemical&Biology&(amino&acids,&peptide&and&proteins& 1.& a. b. c. d. e. 2.& a. b. c. d. e. f. & UsingtheCahn Ingold Prelogsystem,assignstereochemicaldescriptorstothe threeaminoacidsshownbelow.
1/4/18 NUCLEIC ACIDS. Nucleic Acids. Nucleic Acids. ECS129 Instructor: Patrice Koehl
 NUCLEIC ACIDS ECS129 Instructor: Patrice Koehl Nucleic Acids Nucleotides DNA Structure RNA Synthesis Function Secondary structure Tertiary interactions Wobble hypothesis DNA RNA Replication Transcription
NUCLEIC ACIDS ECS129 Instructor: Patrice Koehl Nucleic Acids Nucleotides DNA Structure RNA Synthesis Function Secondary structure Tertiary interactions Wobble hypothesis DNA RNA Replication Transcription
NUCLEIC ACIDS. ECS129 Instructor: Patrice Koehl
 NUCLEIC ACIDS ECS129 Instructor: Patrice Koehl Nucleic Acids Nucleotides DNA Structure RNA Synthesis Function Secondary structure Tertiary interactions Wobble hypothesis DNA RNA Replication Transcription
NUCLEIC ACIDS ECS129 Instructor: Patrice Koehl Nucleic Acids Nucleotides DNA Structure RNA Synthesis Function Secondary structure Tertiary interactions Wobble hypothesis DNA RNA Replication Transcription
03-511/711 Computational Genomics and Molecular Biology, Fall
 03-511/711 Computational Genomics and Molecular Biology, Fall 2011 1 Problem Set 0 Due Tuesday, September 6th This homework is intended to be a self-administered placement quiz, to help you (and me) determine
03-511/711 Computational Genomics and Molecular Biology, Fall 2011 1 Problem Set 0 Due Tuesday, September 6th This homework is intended to be a self-administered placement quiz, to help you (and me) determine
Dr. R. Sankar, BSE 631 (2018)
 Pauling, Corey and Branson Diffraction of DNA http://www.nature.com/scitable/topicpage/dna-is-a-structure-that-encodes-biological-6493050 In short, stereochemistry is important in determining which helices
Pauling, Corey and Branson Diffraction of DNA http://www.nature.com/scitable/topicpage/dna-is-a-structure-that-encodes-biological-6493050 In short, stereochemistry is important in determining which helices
Name. Student ID. Midterm 2, Biology 2020, Kropf 2004
 Midterm 2, Biology 2020, Kropf 2004 1 1. RNA vs DNA (5 pts) The table below compares DNA and RNA. Fill in the open boxes, being complete and specific Compare: DNA RNA Pyrimidines C,T C,U Purines 3-D structure
Midterm 2, Biology 2020, Kropf 2004 1 1. RNA vs DNA (5 pts) The table below compares DNA and RNA. Fill in the open boxes, being complete and specific Compare: DNA RNA Pyrimidines C,T C,U Purines 3-D structure
Case 7 A Storage Protein From Seeds of Brassica nigra is a Serine Protease Inhibitor
 Case 7 A Storage Protein From Seeds of Brassica nigra is a Serine Protease Inhibitor Focus concept Purification of a novel seed storage protein allows sequence analysis and determination of the protein
Case 7 A Storage Protein From Seeds of Brassica nigra is a Serine Protease Inhibitor Focus concept Purification of a novel seed storage protein allows sequence analysis and determination of the protein
In silico measurements of twist and bend. moduli for beta solenoid protein self-
 In silico measurements of twist and bend moduli for beta solenoid protein self- assembly units Leonard P. Heinz, Krishnakumar M. Ravikumar, and Daniel L. Cox Department of Physics and Institute for Complex
In silico measurements of twist and bend moduli for beta solenoid protein self- assembly units Leonard P. Heinz, Krishnakumar M. Ravikumar, and Daniel L. Cox Department of Physics and Institute for Complex
C57BL/6N Female. C57BL/6N Male. Prl2 WT Allele. Prl2 Targeted Allele **** **** WT Het KO PRL bp. 230 bp CNX. C57BL/6N Male 30.
 A Prl Allele Prl Targeted Allele 631 bp 1 3 4 5 6 bp 1 SA β-geo pa 3 4 5 6 Het B Het 631 bp bp PRL CNX C 1 C57BL/6N Male (14) Het (7) (1) 1 C57BL/6N Female (19) Het (31) (1) % survival 5 % survival 5 ****
A Prl Allele Prl Targeted Allele 631 bp 1 3 4 5 6 bp 1 SA β-geo pa 3 4 5 6 Het B Het 631 bp bp PRL CNX C 1 C57BL/6N Male (14) Het (7) (1) 1 C57BL/6N Female (19) Het (31) (1) % survival 5 % survival 5 ****
Nature Medicine: doi: /nm.4464
 Supplementary Fig. 1. Amino acid transporters and substrates used for selectivity screening. (A) Common transporters and amino acid substrates shown. Amino acids designated by one-letter codes. Transporters
Supplementary Fig. 1. Amino acid transporters and substrates used for selectivity screening. (A) Common transporters and amino acid substrates shown. Amino acids designated by one-letter codes. Transporters
Thr Gly Tyr. Gly Lys Asn
 Your unique body characteristics (traits), such as hair color or blood type, are determined by the proteins your body produces. Proteins are the building blocks of life - in fact, about 45% of the human
Your unique body characteristics (traits), such as hair color or blood type, are determined by the proteins your body produces. Proteins are the building blocks of life - in fact, about 45% of the human
Automated bioreactor sampling for on-line analysis of amino acids using two approaches: HPLC and Cedex Bio HT
 Application note #004/Nov 2018 Automated bioreactor sampling for on-line analysis of amino acids using two approaches: HPLC and Cedex Bio HT Alexandra Hofer Securecell AG, Zürcherstrasse 137, 8952 Schlieren,
Application note #004/Nov 2018 Automated bioreactor sampling for on-line analysis of amino acids using two approaches: HPLC and Cedex Bio HT Alexandra Hofer Securecell AG, Zürcherstrasse 137, 8952 Schlieren,
ENZYMES AND METABOLIC PATHWAYS
 ENZYMES AND METABOLIC PATHWAYS This document is licensed under the Attribution-NonCommercial-ShareAlike 2.5 Italy license, available at http://creativecommons.org/licenses/by-nc-sa/2.5/it/ 1. Enzymes build
ENZYMES AND METABOLIC PATHWAYS This document is licensed under the Attribution-NonCommercial-ShareAlike 2.5 Italy license, available at http://creativecommons.org/licenses/by-nc-sa/2.5/it/ 1. Enzymes build
Bi Lecture 3 Loss-of-function (Ch. 4A) Monday, April 8, 13
 Bi190-2013 Lecture 3 Loss-of-function (Ch. 4A) Infer Gene activity from type of allele Loss-of-Function alleles are Gold Standard If organism deficient in gene A fails to accomplish process B, then gene
Bi190-2013 Lecture 3 Loss-of-function (Ch. 4A) Infer Gene activity from type of allele Loss-of-Function alleles are Gold Standard If organism deficient in gene A fails to accomplish process B, then gene
BIOSTAT516 Statistical Methods in Genetic Epidemiology Autumn 2005 Handout1, prepared by Kathleen Kerr and Stephanie Monks
 Rationale of Genetic Studies Some goals of genetic studies include: to identify the genetic causes of phenotypic variation develop genetic tests o benefits to individuals and to society are still uncertain
Rationale of Genetic Studies Some goals of genetic studies include: to identify the genetic causes of phenotypic variation develop genetic tests o benefits to individuals and to society are still uncertain
Proteins: Wide range of func2ons. Polypep2des. Amino Acid Monomers
 Proteins: Wide range of func2ons Proteins coded in DNA account for more than 50% of the dry mass of most cells Protein func9ons structural support storage transport cellular communica9ons movement defense
Proteins: Wide range of func2ons Proteins coded in DNA account for more than 50% of the dry mass of most cells Protein func9ons structural support storage transport cellular communica9ons movement defense
The Stringent Response
 The Stringent Response When amino acids are limiting a response is triggered to shut down a wide range of biosynthetic processes This process is called the Stringent Response It results in the synthesis
The Stringent Response When amino acids are limiting a response is triggered to shut down a wide range of biosynthetic processes This process is called the Stringent Response It results in the synthesis
MBMB451A Section1 Fall 2008 KEY These questions may have more than one correct answer
 MBMB451A Section1 Fall 2008 KEY These questions may have more than one correct answer 1. In a double stranded molecule of DNA, the ratio of purines : pyrimidines is (a) variable (b) determined by the base
MBMB451A Section1 Fall 2008 KEY These questions may have more than one correct answer 1. In a double stranded molecule of DNA, the ratio of purines : pyrimidines is (a) variable (b) determined by the base
(A-B) P2ry14 expression was assessed by (A) genotyping (upper arrow: WT; lower
 Supplementary Figures S1. (A-B) P2ry14 expression was assessed by (A) genotyping (upper arrow: ; lower arrow: KO) and (B) q-pcr analysis with Lin- cells, The white vertical line in panel A indicates that
Supplementary Figures S1. (A-B) P2ry14 expression was assessed by (A) genotyping (upper arrow: ; lower arrow: KO) and (B) q-pcr analysis with Lin- cells, The white vertical line in panel A indicates that
Supplementary Figure 1.
 Supplementary Figure 1. Quantification of western blot analysis of fibroblasts (related to Figure 1) (A-F) Quantification of western blot analysis for control and IR-Mut fibroblasts. Data are expressed
Supplementary Figure 1. Quantification of western blot analysis of fibroblasts (related to Figure 1) (A-F) Quantification of western blot analysis for control and IR-Mut fibroblasts. Data are expressed
Amino Acid Sequences and Evolutionary Relationships
 Amino Acid Sequences and Evolutionary Relationships One technique used to determine evolutionary relationships is to study the biochemical similarity of organisms. Though molds, aardvarks, and humans appear
Amino Acid Sequences and Evolutionary Relationships One technique used to determine evolutionary relationships is to study the biochemical similarity of organisms. Though molds, aardvarks, and humans appear
Chapter Twelve Protein Synthesis: Translation of the Genetic Message
 Mary K. Campbell Shawn O. Farrell international.cengage.com/ Chapter Twelve Protein Synthesis: Translation of the Genetic Message Paul D. Adams University of Arkansas 1 Translating the Genetic Message
Mary K. Campbell Shawn O. Farrell international.cengage.com/ Chapter Twelve Protein Synthesis: Translation of the Genetic Message Paul D. Adams University of Arkansas 1 Translating the Genetic Message
Steroids. Steroids. Proteins: Wide range of func6ons. lipids characterized by a carbon skeleton consis3ng of four fused rings
 Steroids Steroids lipids characterized by a carbon skeleton consis3ng of four fused rings 3 six sided, and 1 five sided Cholesterol important steroid precursor component in animal cell membranes Although
Steroids Steroids lipids characterized by a carbon skeleton consis3ng of four fused rings 3 six sided, and 1 five sided Cholesterol important steroid precursor component in animal cell membranes Although
Name Section Problem Set 3
 Name Section 7.013 Problem Set 3 The completed problem must be turned into the wood box outside 68120 by 4:40 pm, Thursday, March 13. Problem sets will not be accepted late. Question 1 Based upon your
Name Section 7.013 Problem Set 3 The completed problem must be turned into the wood box outside 68120 by 4:40 pm, Thursday, March 13. Problem sets will not be accepted late. Question 1 Based upon your
BIOLOGY Component 1 Basic Biochemistry and Cell Organisation
 Surname Centre Number Candidate Number Other Names 2 GCE AS NEW AS B400U10-1 S16-B400U10-1 BIOLOGY Component 1 Basic Biochemistry and Cell Organisation P.M. THURSDAY, 26 May 2016 1 hour 30 minutes For
Surname Centre Number Candidate Number Other Names 2 GCE AS NEW AS B400U10-1 S16-B400U10-1 BIOLOGY Component 1 Basic Biochemistry and Cell Organisation P.M. THURSDAY, 26 May 2016 1 hour 30 minutes For
Table 1. Primers, annealing temperatures, and product sizes for PCR amplification.
 Table 1. Primers, annealing temperatures, and product sizes for PCR amplification. Gene Direction Primer sequence (5 3 ) Annealing Temperature Size (bp) BRCA1 Forward TTGCGGGAGGAAAATGGGTAGTTA 50 o C 292
Table 1. Primers, annealing temperatures, and product sizes for PCR amplification. Gene Direction Primer sequence (5 3 ) Annealing Temperature Size (bp) BRCA1 Forward TTGCGGGAGGAAAATGGGTAGTTA 50 o C 292
Homework. A bit about the nature of the atoms of interest. Project. The role of electronega<vity
 Homework Why cited articles are especially useful. citeulike science citation index When cutting and pasting less is more. Project Your protein: I will mail these out this weekend If you haven t gotten
Homework Why cited articles are especially useful. citeulike science citation index When cutting and pasting less is more. Project Your protein: I will mail these out this weekend If you haven t gotten
Project 07/111 Final Report October 31, Project Title: Cloning and expression of porcine complement C3d for enhanced vaccines
 Project 07/111 Final Report October 31, 2007. Project Title: Cloning and expression of porcine complement C3d for enhanced vaccines Project Leader: Dr Douglas C. Hodgins (519-824-4120 Ex 54758, fax 519-824-5930)
Project 07/111 Final Report October 31, 2007. Project Title: Cloning and expression of porcine complement C3d for enhanced vaccines Project Leader: Dr Douglas C. Hodgins (519-824-4120 Ex 54758, fax 519-824-5930)
7.014 Problem Set 4 Answers to this problem set are to be turned in. Problem sets will not be accepted late. Solutions will be posted on the web.
 MIT Department of Biology 7.014 Introductory Biology, Spring 2005 Name: Section : 7.014 Problem Set 4 Answers to this problem set are to be turned in. Problem sets will not be accepted late. Solutions
MIT Department of Biology 7.014 Introductory Biology, Spring 2005 Name: Section : 7.014 Problem Set 4 Answers to this problem set are to be turned in. Problem sets will not be accepted late. Solutions
Cancer cells that survive radiation therapy acquire HIF-1 activity and translocate toward tumor blood vessels Supplementary Information
 Cancer cells that survive radiation therapy acquire HIF-1 activity and translocate toward tumor blood vessels Supplementary Information 1. Supplementary Figure S1-S10: Pages 2-11 2. Supplementary References:
Cancer cells that survive radiation therapy acquire HIF-1 activity and translocate toward tumor blood vessels Supplementary Information 1. Supplementary Figure S1-S10: Pages 2-11 2. Supplementary References:
Unit 1. DNA and the Genome
 Unit 1 DNA and the Genome Gene Expression Key Area 3 Vocabulary 1: Transcription Translation Phenotype RNA (mrna, trna, rrna) Codon Anticodon Ribosome RNA polymerase RNA splicing Introns Extrons Gene Expression
Unit 1 DNA and the Genome Gene Expression Key Area 3 Vocabulary 1: Transcription Translation Phenotype RNA (mrna, trna, rrna) Codon Anticodon Ribosome RNA polymerase RNA splicing Introns Extrons Gene Expression
ATP GTP UTP CTP
 Table S1. ATP (GTP) utilization for de novo synthesis of rtps ucleotide 5 phosphoribosyl ucleobase rmp rtp (total) pyrophosphate from glucose ATP 3 5 8 10 GTP 3 5 8 10 UTP 3 2 5 7 CTP 3 3 6 8 The numbers
Table S1. ATP (GTP) utilization for de novo synthesis of rtps ucleotide 5 phosphoribosyl ucleobase rmp rtp (total) pyrophosphate from glucose ATP 3 5 8 10 GTP 3 5 8 10 UTP 3 2 5 7 CTP 3 3 6 8 The numbers
EE550 Computational Biology
 EE550 Computational Biology Week 1 Course Notes Instructor: Bilge Karaçalı, PhD Syllabus Schedule : Thursday 13:30, 14:30, 15:30 Text : Paul G. Higgs, Teresa K. Attwood, Bioinformatics and Molecular Evolution,
EE550 Computational Biology Week 1 Course Notes Instructor: Bilge Karaçalı, PhD Syllabus Schedule : Thursday 13:30, 14:30, 15:30 Text : Paul G. Higgs, Teresa K. Attwood, Bioinformatics and Molecular Evolution,
Supplemental Material S1, Zhang et al. (2009)
 Supplemental Material S1, Zhang et al. (2009) Cloning of PS small RNA fragments. PS RNA labeled with [γ 32 P]-ATP (NEN) of a length from 30 to 90 nucleotides were isolated from the gel after denaturing
Supplemental Material S1, Zhang et al. (2009) Cloning of PS small RNA fragments. PS RNA labeled with [γ 32 P]-ATP (NEN) of a length from 30 to 90 nucleotides were isolated from the gel after denaturing
Amino Acid Sequences and Evolutionary Relationships. How do similarities in amino acid sequences of various species provide evidence for evolution?
 Amino Acid Sequences and Evolutionary Relationships Name: How do similarities in amino acid sequences of various species provide evidence for evolution? An important technique used in determining evolutionary
Amino Acid Sequences and Evolutionary Relationships Name: How do similarities in amino acid sequences of various species provide evidence for evolution? An important technique used in determining evolutionary
Chapter 13 From Genes to Proteins
 Chapter 13 From Genes to Proteins True/False Indicate whether the sentence or statement is true(a) or false(b). 1. RNA nucleotides contain the sugar ribose. 2. Only DNA molecules contain the nitrogen base
Chapter 13 From Genes to Proteins True/False Indicate whether the sentence or statement is true(a) or false(b). 1. RNA nucleotides contain the sugar ribose. 2. Only DNA molecules contain the nitrogen base
Supplemental Table 1. Amino acid sequences of synthetic kisspeptins
 Supplemental Data Supplemental Table 1. Amino acid sequences of synthetic kisspeptins Kisspeptins Symbol Sequence Human kisspeptin-10 H-10 Tyr-Asn-Trp-Asn-Ser-Phe-Gly-Leu-Arg-Phe-NH 2 Rodent/Xenopus 1a
Supplemental Data Supplemental Table 1. Amino acid sequences of synthetic kisspeptins Kisspeptins Symbol Sequence Human kisspeptin-10 H-10 Tyr-Asn-Trp-Asn-Ser-Phe-Gly-Leu-Arg-Phe-NH 2 Rodent/Xenopus 1a
Lac Operon contains three structural genes and is controlled by the lac repressor: (1) LacY protein transports lactose into the cell.
 Regulation of gene expression a. Expression of most genes can be turned off and on, usually by controlling the initiation of transcription. b. Lactose degradation in E. coli (Negative Control) Lac Operon
Regulation of gene expression a. Expression of most genes can be turned off and on, usually by controlling the initiation of transcription. b. Lactose degradation in E. coli (Negative Control) Lac Operon
Lecture 19A. DNA computing
 Lecture 19A. DNA computing What exactly is DNA (deoxyribonucleic acid)? DNA is the material that contains codes for the many physical characteristics of every living creature. Your cells use different
Lecture 19A. DNA computing What exactly is DNA (deoxyribonucleic acid)? DNA is the material that contains codes for the many physical characteristics of every living creature. Your cells use different
Accelerating skin wound healing by M-CSF through generating SSEA-1 and -3 stem cells. in the injured sites
 Accelerating skin wound healing by M-CSF through generating SSEA-1 and -3 stem cells in the injured sites Yunyuan Li, Reza Baradar Jalili, Aziz Ghahary Department of Surgery, University of British Columbia,
Accelerating skin wound healing by M-CSF through generating SSEA-1 and -3 stem cells in the injured sites Yunyuan Li, Reza Baradar Jalili, Aziz Ghahary Department of Surgery, University of British Columbia,
BS 50 Genetics and Genomics Week of Oct 24
 BS 50 Genetics and Genomics Week of Oct 24 Additional Practice Problems for Section Question 1: The following table contains a list of statements that apply to replication, transcription, both, or neither.
BS 50 Genetics and Genomics Week of Oct 24 Additional Practice Problems for Section Question 1: The following table contains a list of statements that apply to replication, transcription, both, or neither.
Amino Acid Sequences and Evolutionary Relationships
 Amino Acid Sequences and Evolutionary Relationships Pre-Lab Discussion Homologous structures -- those structures believed to have a common origin but not necessarily a common function -- provide some of
Amino Acid Sequences and Evolutionary Relationships Pre-Lab Discussion Homologous structures -- those structures believed to have a common origin but not necessarily a common function -- provide some of
6-Foot Mini Toober Activity
 Big Idea The interaction between the substrate and enzyme is highly specific. Even a slight change in shape of either the substrate or the enzyme may alter the efficient and selective ability of the enzyme
Big Idea The interaction between the substrate and enzyme is highly specific. Even a slight change in shape of either the substrate or the enzyme may alter the efficient and selective ability of the enzyme
Module 3. Lecture 5. Regulation of Gene Expression in Prokaryotes
 Module 3 Lecture 5 Regulation of Gene Expression in Prokaryotes Recap So far, we have looked at prokaryotic gene regulation using 3 operon models. lac: a catabolic operon which displays induction via negative
Module 3 Lecture 5 Regulation of Gene Expression in Prokaryotes Recap So far, we have looked at prokaryotic gene regulation using 3 operon models. lac: a catabolic operon which displays induction via negative
i-stop codon positions in the mcherry gene
 Supplementary Figure 1 i-stop codon positions in the mcherry gene The grnas (green) that can potentially generate stop codons from Trp (63 th and 98 th aa, upper panel) and Gln (47 th and 114 th aa, bottom
Supplementary Figure 1 i-stop codon positions in the mcherry gene The grnas (green) that can potentially generate stop codons from Trp (63 th and 98 th aa, upper panel) and Gln (47 th and 114 th aa, bottom
Case 7 A Storage Protein From Seeds of Brassica nigra is a Serine Protease Inhibitor Last modified 29 September 2005
 Case 7 A Storage Protein From Seeds of Brassica nigra is a Serine Protease Inhibitor Last modified 9 September 005 Focus concept Purification of a novel seed storage protein allows sequence analysis and
Case 7 A Storage Protein From Seeds of Brassica nigra is a Serine Protease Inhibitor Last modified 9 September 005 Focus concept Purification of a novel seed storage protein allows sequence analysis and
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3562 In the format provided by the authors and unedited. Supplementary Figure 1 Glucose deficiency induced FH-ATF2 interaction. In b-m, immunoblotting or immunoprecipitation analyses were
DOI: 10.1038/ncb3562 In the format provided by the authors and unedited. Supplementary Figure 1 Glucose deficiency induced FH-ATF2 interaction. In b-m, immunoblotting or immunoprecipitation analyses were
Solutions to 7.02 Quiz II 10/27/05
 Solutions to 7.02 Quiz II 10/27/05 Class Average = 83 Standard Deviation = 9 Range Grade % 87-100 A 43 74-86 B 39 55-73 C 17 > 54 D 1 Question 1 (56 points) While studying deep sea bacteria, you discover
Solutions to 7.02 Quiz II 10/27/05 Class Average = 83 Standard Deviation = 9 Range Grade % 87-100 A 43 74-86 B 39 55-73 C 17 > 54 D 1 Question 1 (56 points) While studying deep sea bacteria, you discover
Ali Yaghi. Tamara Wahbeh. Mamoun Ahram
 28 Ali Yaghi Tamara Wahbeh Mamoun Ahram This sheet is a continuation of protein purification methods. Isoelectric focusing Separation of proteins based on Isoelectric points(charge),and it is a horizontal
28 Ali Yaghi Tamara Wahbeh Mamoun Ahram This sheet is a continuation of protein purification methods. Isoelectric focusing Separation of proteins based on Isoelectric points(charge),and it is a horizontal
Assignment 13. In the Griffith experiment, why did mice die when injected with live R bacteria plus heatkilled
 Assignment 13 1. Multiple-choice (1 point) In the Griffith experiment, why did mice die when injected with live R bacteria plus heatkilled S bacteria? Some of the S bacteria were still alive. The R bacteria
Assignment 13 1. Multiple-choice (1 point) In the Griffith experiment, why did mice die when injected with live R bacteria plus heatkilled S bacteria? Some of the S bacteria were still alive. The R bacteria
7.014 Quiz II Handout
 7.014 Quiz II Handout Quiz II: Wednesday, March 17 12:05-12:55 54-100 **This will be a closed book exam** Quiz Review Session: Friday, March 12 7:00-9:00 pm room 54-100 Open Tutoring Session: Tuesday,
7.014 Quiz II Handout Quiz II: Wednesday, March 17 12:05-12:55 54-100 **This will be a closed book exam** Quiz Review Session: Friday, March 12 7:00-9:00 pm room 54-100 Open Tutoring Session: Tuesday,
Proteomics Informatics (BMSC-GA 4437)
 Proteomics Informatics (BMSC-GA 4437) Instructor David Fenyö Contact information David@FenyoLab.org htt://fenyolab.org/resentations/proteomics_informatics_2013/ Learning Objectives Be able analyze a roteomics
Proteomics Informatics (BMSC-GA 4437) Instructor David Fenyö Contact information David@FenyoLab.org htt://fenyolab.org/resentations/proteomics_informatics_2013/ Learning Objectives Be able analyze a roteomics
Molecular Cell Biology - Problem Drill 11: Recombinant DNA
 Molecular Cell Biology - Problem Drill 11: Recombinant DNA Question No. 1 of 10 1. Which of the following statements about the sources of DNA used for molecular cloning is correct? Question #1 (A) cdna
Molecular Cell Biology - Problem Drill 11: Recombinant DNA Question No. 1 of 10 1. Which of the following statements about the sources of DNA used for molecular cloning is correct? Question #1 (A) cdna
Protein analysis. Dr. Mamoun Ahram Summer semester, Resources This lecture Campbell and Farrell s Biochemistry, Chapters 5
 Protein analysis Dr. Mamoun Ahram Summer semester, 2015-2016 Resources This lecture Campbell and Farrell s Biochemistry, Chapters 5 Bases of protein separation Proteins can be purified on the basis Solubility
Protein analysis Dr. Mamoun Ahram Summer semester, 2015-2016 Resources This lecture Campbell and Farrell s Biochemistry, Chapters 5 Bases of protein separation Proteins can be purified on the basis Solubility
T H E J O U R N A L O F C E L L B I O L O G Y
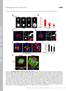 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rodríguez-Fraticelli et al., http://www.jcb.org/cgi/content/full/jcb.201203075/dc1 Figure S1. Cell spreading and lumen formation in confined
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rodríguez-Fraticelli et al., http://www.jcb.org/cgi/content/full/jcb.201203075/dc1 Figure S1. Cell spreading and lumen formation in confined
Supplementary Figure 1. Confirmation of sirna in PC3 and H1299 cells PC3 (a) and H1299 (b) cells were transfected with sirna oligonucleotides
 Supplementary Figure 1. Confirmation of sirna in PC3 and H1299 cells PC3 (a) and H1299 (b) cells were transfected with sirna oligonucleotides targeting RCP (SMARTPool (RCP) or two individual oligos (RCP#1
Supplementary Figure 1. Confirmation of sirna in PC3 and H1299 cells PC3 (a) and H1299 (b) cells were transfected with sirna oligonucleotides targeting RCP (SMARTPool (RCP) or two individual oligos (RCP#1
7.014 Quiz II 3/18/05. Write your name on this page and your initials on all the other pages in the space provided.
 7.014 Quiz II 3/18/05 Your Name: TA's Name: Write your name on this page and your initials on all the other pages in the space provided. This exam has 10 pages including this coversheet. heck that you
7.014 Quiz II 3/18/05 Your Name: TA's Name: Write your name on this page and your initials on all the other pages in the space provided. This exam has 10 pages including this coversheet. heck that you
蛋白質體學. Proteomics Amino acids, Peptides and Proteins 陳威戎 & 21
 蛋白質體學 Proteomics 2015 Amino acids, Peptides and Proteins 陳威戎 2015. 09. 14 & 21 Outline 1. Amino Acids 2. Peptides and Proteins 3. Covalent Structure of Proteins Amino Acids Proteins are polymers of amino
蛋白質體學 Proteomics 2015 Amino acids, Peptides and Proteins 陳威戎 2015. 09. 14 & 21 Outline 1. Amino Acids 2. Peptides and Proteins 3. Covalent Structure of Proteins Amino Acids Proteins are polymers of amino
Supplementary information Activation of AMP-activated protein kinase
 Supplementary information Activation of AMP-activated protein kinase 2 by nicotine instigates formation of abdominal aortic aneurysms in mice in vivo Shuangxi Wang 1,2,5, Cheng Zhang 1,2,5, Miao Zhang
Supplementary information Activation of AMP-activated protein kinase 2 by nicotine instigates formation of abdominal aortic aneurysms in mice in vivo Shuangxi Wang 1,2,5, Cheng Zhang 1,2,5, Miao Zhang
Supplementary Figure 1. Co-localization of GLUT1 and DNAL4 in BeWo cells cultured
 Supplementary Figure 1. Co-localization of GLUT1 and DNAL4 in BeWo cells cultured under static conditions. Cells were seeded in the chamber area of the device and cultured overnight without medium perfusion.
Supplementary Figure 1. Co-localization of GLUT1 and DNAL4 in BeWo cells cultured under static conditions. Cells were seeded in the chamber area of the device and cultured overnight without medium perfusion.
Student Number: a) Indicate which, if any, of the six proteins would be found in these three fractions.
 Name: Student Number: April 23, 2009, 9:00 AM -12:00 PM Page 1 (of 4) Biochemistry II Laboratory Section Final Examination Examiner: Dr. J. Galka 1. Answer ALL questions in the space provided. 2. The back
Name: Student Number: April 23, 2009, 9:00 AM -12:00 PM Page 1 (of 4) Biochemistry II Laboratory Section Final Examination Examiner: Dr. J. Galka 1. Answer ALL questions in the space provided. 2. The back
Supplementary Figure 1. Generation of B2M -/- ESCs. Nature Biotechnology: doi: /nbt.3860
 Supplementary Figure 1 Generation of B2M -/- ESCs. (a) Maps of the B2M alleles in cells with the indicated B2M genotypes. Probes and restriction enzymes used in Southern blots are indicated (H, Hind III;
Supplementary Figure 1 Generation of B2M -/- ESCs. (a) Maps of the B2M alleles in cells with the indicated B2M genotypes. Probes and restriction enzymes used in Southern blots are indicated (H, Hind III;
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
Daily Agenda. Warm Up: Review. Translation Notes Protein Synthesis Practice. Redos
 Daily Agenda Warm Up: Review Translation Notes Protein Synthesis Practice Redos 1. What is DNA Replication? 2. Where does DNA Replication take place? 3. Replicate this strand of DNA into complimentary
Daily Agenda Warm Up: Review Translation Notes Protein Synthesis Practice Redos 1. What is DNA Replication? 2. Where does DNA Replication take place? 3. Replicate this strand of DNA into complimentary
Supplementary Information (Ha, et. al) Supplementary Figures Supplementary Fig. S1
 Supplementary Information (Ha, et. al) Supplementary Figures Supplementary Fig. S1 a His-ORMDL3 ~ 17 His-ORMDL3 GST-ORMDL3 - + - + IPTG GST-ORMDL3 ~ b Integrated Density (ORMDL3/ -actin) 0.4 0.3 0.2 0.1
Supplementary Information (Ha, et. al) Supplementary Figures Supplementary Fig. S1 a His-ORMDL3 ~ 17 His-ORMDL3 GST-ORMDL3 - + - + IPTG GST-ORMDL3 ~ b Integrated Density (ORMDL3/ -actin) 0.4 0.3 0.2 0.1
The combination of a phosphate, sugar and a base forms a compound called a nucleotide.
 History Rosalin Franklin: Female scientist (x-ray crystallographer) who took the picture of DNA James Watson and Francis Crick: Solved the structure of DNA from information obtained by other scientist.
History Rosalin Franklin: Female scientist (x-ray crystallographer) who took the picture of DNA James Watson and Francis Crick: Solved the structure of DNA from information obtained by other scientist.
Supplementary Figure 1, related to Figure 1. GAS5 is highly expressed in the cytoplasm of hescs, and positively correlates with pluripotency.
 Supplementary Figure 1, related to Figure 1. GAS5 is highly expressed in the cytoplasm of hescs, and positively correlates with pluripotency. (a) Transfection of different concentration of GAS5-overexpressing
Supplementary Figure 1, related to Figure 1. GAS5 is highly expressed in the cytoplasm of hescs, and positively correlates with pluripotency. (a) Transfection of different concentration of GAS5-overexpressing
CHAPTER 1. DNA: The Hereditary Molecule SECTION D. What Does DNA Do? Chapter 1 Modern Genetics for All Students S 33
 HPER 1 DN: he Hereditary Molecule SEION D What Does DN Do? hapter 1 Modern enetics for ll Students S 33 D.1 DN odes For Proteins PROEINS DO HE nitty-gritty jobs of every living cell. Proteins are the molecules
HPER 1 DN: he Hereditary Molecule SEION D What Does DN Do? hapter 1 Modern enetics for ll Students S 33 D.1 DN odes For Proteins PROEINS DO HE nitty-gritty jobs of every living cell. Proteins are the molecules
Granby Transcription and Translation Services plc
 ompany Resources ranby Transcription and Translation Services plc has invested heavily in the Protein Synthesis business. mongst the resources available to new recruits are: the latest cellphones which
ompany Resources ranby Transcription and Translation Services plc has invested heavily in the Protein Synthesis business. mongst the resources available to new recruits are: the latest cellphones which
CFSSP: Chou and Fasman Secondary Structure Prediction server
 Wide Spectrum, Vol. 1, No. 9, (2013) pp 15-19 CFSSP: Chou and Fasman Secondary Structure Prediction server T. Ashok Kumar Department of Bioinformatics, Noorul Islam College of Arts and Science, Kumaracoil
Wide Spectrum, Vol. 1, No. 9, (2013) pp 15-19 CFSSP: Chou and Fasman Secondary Structure Prediction server T. Ashok Kumar Department of Bioinformatics, Noorul Islam College of Arts and Science, Kumaracoil
DLD1 * cl OD 570nm. OD 570nm NEAA
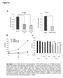 Figure S A. ATF4 mrna levels.2.8.6.4.2 HT8 shatf4- cl3 shatf4- cl4 ATF4 mrna levels.2.8.6.4.2 DLD shatf4-cl3 B. C. OD 57nm.7.6.5.4.3 shatf4 cl3 shatf4 cl4 OD 57nm.6.5.4.3.2.2. 2 4 6 day.. NEAA - + - +
Figure S A. ATF4 mrna levels.2.8.6.4.2 HT8 shatf4- cl3 shatf4- cl4 ATF4 mrna levels.2.8.6.4.2 DLD shatf4-cl3 B. C. OD 57nm.7.6.5.4.3 shatf4 cl3 shatf4 cl4 OD 57nm.6.5.4.3.2.2. 2 4 6 day.. NEAA - + - +
AOCS/SQT Amino Acid Round Robin Study
 AOCS/SQT Amino Acid Round Robin Study J. V. Simpson and Y. Zhang National Corn-to-Ethanol Research Center Edwardsville, IL Richard Cantrill Technical Director AOCS, Urbana, IL Amy Johnson Soybean Quality
AOCS/SQT Amino Acid Round Robin Study J. V. Simpson and Y. Zhang National Corn-to-Ethanol Research Center Edwardsville, IL Richard Cantrill Technical Director AOCS, Urbana, IL Amy Johnson Soybean Quality
OMICS International Conferences
 About OMICS Group OMICS Group is an amalgamation of Open Access Publications and worldwide international science conferences and events. Established in the year 2007 with the sole aim of making the information
About OMICS Group OMICS Group is an amalgamation of Open Access Publications and worldwide international science conferences and events. Established in the year 2007 with the sole aim of making the information
DNA RNA Protein Trait Protein Synthesis (Gene Expression) Notes Proteins (Review) Proteins make up all living materials
 DNA RNA Protein Trait Protein Synthesis (Gene Expression) Notes Proteins (Review) Proteins make up all living materials Proteins are composed of amino acids there are 20 different amino acids Different
DNA RNA Protein Trait Protein Synthesis (Gene Expression) Notes Proteins (Review) Proteins make up all living materials Proteins are composed of amino acids there are 20 different amino acids Different
