Supplementary Figure 1. Confirmation of sirna in PC3 and H1299 cells PC3 (a) and H1299 (b) cells were transfected with sirna oligonucleotides
|
|
|
- Lynn Stanley
- 5 years ago
- Views:
Transcription
1 Supplementary Figure 1. Confirmation of sirna in PC3 and H1299 cells PC3 (a) and H1299 (b) cells were transfected with sirna oligonucleotides targeting RCP (SMARTPool (RCP) or two individual oligos (RCP#1 & RCP#2), EphA2 (two individual oligos (EphA2#1 & EphA2#2), FIP2, FIP3, α5 integrin (α5), Rab14, Rab11, Rab6 or a non-targeting control (si-nt). 24 hr following transfection, cells were lysed and the levels of the indicated proteins determined by immunoblotting or by qpcr (for FIP2 in H1299 cells). 1
2 Supplementary Figure 2. RCP and EphA2 are not required for HGF-driven migration of non-contacting H1299 cells, and knockdown of Rab11 does not influence HGFdriven cellular accumulation of EphA2 (a) H1299 cells were transfected with sirnas targeting RCP (si-rcp), EphA2 (si-epha2) or a non-targeting control (si-nt). Transfected cells were seeded sparsely for sufficient time for them to attach, but not to divide and form colonies. Cells were then treated with HGF (10 ng ml -1 ) and cell migration was recorded using time-lapse video microscopy. Cell migration speed was quantified using ImageJ manual tracking and chemotaxis plugin. Cells were excluded from this analysis if they contacted another cell. Data are represented as box and whiskers plots (whiskers: percentile, + represents the mean). *** p<0.001; Mann- Whitney test. Data are from 3 independent experiments with >50 cells being tracked per condition. (b) H1299 cells were transfected with sirnas targeting Rab11a and Rab11b (si-rab11) or a non-targeting control (si-nt) and plated onto 10 cm plastic dishes. Cells were surfacelabelled with NHS-S-S-Biotin (0.13 mg ml -1 ) at 4ºC and internalization allowed to proceed at 37ºC for the indicated times with or without HGF (10 ng ml -1 ). Biotin remaining at the cell surface was removed by exposure to MesNa at 4 C and the quantity of biotinylated receptors within the cells determined by capture-elisa using microtitre wells coated with monoclonal antibodies recognising EphA2. Values are mean ± SEM, n=4 biological replicates from 2 independent experiments, *** p<0.001 (si-rab11 + HGF versus si-nt HGF); Two-way ANOVA, Bonferroni post-test. 2
3 Supplementary Figure 3. High resolution imaging of EphA2 and RCP localization. (a) Untransfected H1299 cells were incubated in the presence or absence of HGF (10 ng ml - 1 ) and then fixed in 4% paraformaldehyde. Endogenous RCP, EphA2 and nuclei were visualised by immunofluorescence followed by high resolution imaging using an Airy-scan microscope. Bar, 10 μm. These images are whole cell fluorescence micrographs and single channel fluorescence images corresponding to the merged and cropped images displayed in the right panels of Figure 3e. 3
4 (b) H1299 cells were transfected with EphA2-GFP or EphA2 897A -GFP in combination with mcherry-rcp or mcherry-rcp 435A, in the presence or absence of sirnas targeting LMTK3 (si-lmtk3) as indicated. Cells were incubated in the presence or absence of HGF (10 ng ml - 1 ), fixed in 4% paraformaldehyde and EphA2-GFP and mcherry-rcps visualised using a high resolution Airy-scan microscope. Bar, 10 μm. (c) H1299 cells were transfected with GFP-RCP or GFP-RCP 435A. Transfected cells were incubated in the presence or absence of HGF (10 ng ml -1 ) and then fixed in 4% paraformaldehyde. Endogenous phospho-ser 897 -EphA2 (EphA2 ps897 ) was stained with a phosphospecific antibody and visualised with respect to GFP-RCPs using an Airy-scan microscope. Bar, 10 μm. 4
5 Supplementary Figure 4. Validation of anti-phosphoser 435 RCP (a) Schematic representation of the domain structure of RCP indicating two phosphopeptides identified by mass spectrometry. (b) A2780 cells were transfected with GFP, GFP-RCP or GFP-RCP 435A and plated onto 15 cm plastic dishes. 48 hr following transfection, cells were lysed and analysed by immunoblotting using an antibody raised in rabbits against a synthetic peptide corresponding to RCP (pser 435 ), anti-gfp and anti-rcp as indicated. In the right panel, cells were treated with staurosporine or vehicle control 30 min prior to lysis. 5
6 Supplementary Figure 5. Ligand-dependent signalling of EphA2 (a) H1299 cells were transfected with a sirnas targeting ephrin A1 (si-ephrina1; a SMARTPool (SP) or two individual sirna oligonucleotides #1 & #2) or a non-targeting control (si-nt). Cell scattering in the presence and absence of HGF was then determined and quantified as for Figure 2d-e. Data are represented as box and whiskers plots (whiskers: percentile, + represents the mean). Data are from 3 independent experiments. The right panel displays the effectiveness of the ephrin A1 knockdown as determined by qpcr. (b) H1299 cells were transfected with a non-targeting sirna or with an sirna targeting EphA2 (si-epha2) in combination with sirna-resistant forms of EphA2-GFP or EphA2 588F - GFP. Cells were treated with recombinant ephrina1 (ephrina1-fc) for 10 min, and the expression levels and tyrosine phosphorylation of the sirna-resistant forms of EphA2-GFP 6
7 and EphA2 588F -GFP were determined in EphA2 knockdown cells by Western blotting with antibodies recognising EphA2 or ptyr 588 -EphA2. (c) H1299 cells expressing EphA2-GFP or EphA2 588F -GFP were treated with recombinant ephrin-a1 for 30 min or were left untreated. Cells were fixed and the distribution of the GFPtagged Ephs (green signal) determined using confocal microscopy. The cells were counterstained with DAPI (blue) to visualise nuclei. Bar, 20 μm. (d) H1299 cells were transfected with a sirna targeting EphA2 in combination with sirnaresistant forms of EphA2-GFP or EphA2 588F -GFP. Internalisation of EphA2-GFPs in the presence and absence of HGF was then determined as for Figure 3b, but with the ELISA plate coated with anti-gfp to specifically detect only the GFP-tagged receptor. Values are mean ± SEM n=4 biological replicates from 2 independent experiments. Black asterisks *** p<0.001, ** p<0.01 (EphA2 wt -GFP + HGF versus EphA2 wt -GFP - HGF); Orange asterisks ** p<0.01, * p<0.05 (EphA2 588F -GFP + HGF versus EphA2 588F -GFP - HGF); two-way ANOVA, Bonferroni post-test. (e) H1299 cells were transfected with EphA2-GFP or EphA2 588F -GFP in combination with mcherry-rcp. Trafficking of EphA2-GFP and mcherry-rcps in the presence and absence of HGF (added 30 min prior to collecting the movies) was visualised by fluorescence confocal time-lapse microscopy and colocalisation was quantified as for Figure 3e. Stills were extracted from movies at the indicated time points, and these display the region of interest indicated by the white box (only stills from the EphA2 588F -GFP movies are displayed). Bar, 10 µm. Fractions of co-localizing pixels are mean ± SEM from 3 separate experiments incorporating >10 cells/condition/experiment. ** p<0.01; 1 way ANOVA (Kruskal-Wallis test, Dunn's Multiple Comparison Test). (f) H1299 cells were transfected with a non-targeting sirna or with an sirna targeting EphA2 (si-epha2) in combination with a control vector (control) or an sirna-resistant form of EphA2 588F -GFP. Cell scattering in the presence and absence of HGF was then determined and quantified as for Figure 6c. Data are represented as box and whiskers plots (whiskers: percentile, + represents the mean). *** p<0.001; 1 way ANOVA (Kruskal-Wallis test, Dunn's Multiple Comparison Test). Data are from 3 independent experiments with >50 cells being tracked per condition. 7
8 Supplementary Figure 6. Ligand-independent signalling of EphA2 H1299 cells were transfected with sirnas targeting ephrin-a1 (si-ephrina1) (a), RCP (si- RCP), Rab14 (si-rab14), LMTK3 (si-lmtk3) (b), a non-targeting control (si-nt) or were left untransfected. Cells were treated with HGF (10 ng ml -1 ) for the indicated times and then lysed. Levels of phosphor-ser 897 -EphA2 (EphA2 ps897 ), phospho-tyr 588 -EphA2 (EphA2 py588 ), EphA2, RCP and Rab14 were determined by Western blotting as indicated. 8
9 Supplementary Figure 7. Generation and validation of mice with floxed RCP alleles (a) Schematic representation of the targeting construct used to generate RCP floxed mice. A conditional RCP allele was generated by flanking exon 2 of the RCP gene with LoxP sites in mouse ES cells. Mice were generated by injection of ES cells into blastocysts and implantation of these into pseudopregnant animals. The neomycin cassette was subsequently removed. (b) Effective RCP gene deletion was confirmed by adenoviral infection of Cre recombinase into MEFs derived from RCP fl/fl animals followed by immunoblotting using in-house antibodies recognising mouse RCP. (c) Primary mouse cell lines were derived from PDACs harvested from KPC (Pdx1-Cre, Kras G12D/+, p53 R172H/+ ), KPC:EPHA2 -/- and KPC:RCP fl/fl mice. At least 3 cell lines were derived per condition and the knockout of EphA2 and RCP was confirmed by immunoblotting. (d) Primary mouse cell lines were derived from PDACs harvested from KPC (Pdx1-Cre, Kras G12D/+, p53 R172H/+ ), KPC:EPHA2 -/- and KPC:RCP fl/fl mice. The EPHA2 #2 knockout PDAC 9
10 line was stably transfected with EphA2-GFP or EphA2 897A -GFP, and the RCP #2 knockout line was stably transfected with GFP-RCP or GFP-RCP 435A as indicated. The propensity of these cells to form tight or scattered colonies was determined using phase contrast microscopy (d), Bar, 100μm. Higher magnification images of these fields are displayed in Figure 7d 10
11 11
12 Supplementary figure 8. Uncropped blots for figure 1b-d The uncropped blots displayed in figures 1b-d are presented. In some cases, the total lane displays a different exposure of the same blot as the one presenting the immunoprecipitated (IP) samples, and this is indicated as appropriate. The cropped regions are denoted by red boxes. 12
13 Supplementary figure 9. Uncropped blots for figures 3c, 4c-d & 6d The uncropped blots displayed in figures 3, 4 & 6 are presented. The cropped regions are denoted by red boxes. 13
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rainero et al., http://www.jcb.org/cgi/content/full/jcb.201109112/dc1 Figure S1. The expression of DGK- is reduced upon transfection
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rainero et al., http://www.jcb.org/cgi/content/full/jcb.201109112/dc1 Figure S1. The expression of DGK- is reduced upon transfection
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3363 Supplementary Figure 1 Several WNTs bind to the extracellular domains of PKD1. (a) HEK293T cells were co-transfected with indicated plasmids. Flag-tagged proteins were immunoprecipiated
DOI: 10.1038/ncb3363 Supplementary Figure 1 Several WNTs bind to the extracellular domains of PKD1. (a) HEK293T cells were co-transfected with indicated plasmids. Flag-tagged proteins were immunoprecipiated
Supplementary Figure 1. APP cleavage assay. HEK293 cells were transfected with various
 Supplementary Figure 1. APP cleavage assay. HEK293 cells were transfected with various GST-tagged N-terminal truncated APP fragments including GST-APP full-length (FL), APP (123-695), APP (189-695), or
Supplementary Figure 1. APP cleavage assay. HEK293 cells were transfected with various GST-tagged N-terminal truncated APP fragments including GST-APP full-length (FL), APP (123-695), APP (189-695), or
monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal antibody was examined in
 Supplementary information Supplementary figures Supplementary Figure 1 Determination of the s pecificity of in-house anti-rhbdd1 mouse monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal
Supplementary information Supplementary figures Supplementary Figure 1 Determination of the s pecificity of in-house anti-rhbdd1 mouse monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal
JCB. Supplemental material THE JOURNAL OF CELL BIOLOGY. Paul et al.,
 Supplemental material JCB Paul et al., http://www.jcb.org/cgi/content/full/jcb.201502040/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Mutant p53-expressing cells display limited retrograde actin flow at
Supplemental material JCB Paul et al., http://www.jcb.org/cgi/content/full/jcb.201502040/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Mutant p53-expressing cells display limited retrograde actin flow at
Nature Neuroscience: doi: /nn Supplementary Figure 1
 Supplementary Figure 1 PCR-genotyping of the three mouse models used in this study and controls for behavioral experiments after semi-chronic Pten inhibition. a-c. DNA from App/Psen1 (a), Pten tg (b) and
Supplementary Figure 1 PCR-genotyping of the three mouse models used in this study and controls for behavioral experiments after semi-chronic Pten inhibition. a-c. DNA from App/Psen1 (a), Pten tg (b) and
Journal of Cell Science Supplementary Material
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 SUPPLEMENTARY FIGURE LEGENDS Figure S1: Eps8 is localized at focal adhesions and binds directly to FAK (A) Focal
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 SUPPLEMENTARY FIGURE LEGENDS Figure S1: Eps8 is localized at focal adhesions and binds directly to FAK (A) Focal
Stargazin regulates AMPA receptor trafficking through adaptor protein. complexes during long term depression
 Supplementary Information Stargazin regulates AMPA receptor trafficking through adaptor protein complexes during long term depression Shinji Matsuda, Wataru Kakegawa, Timotheus Budisantoso, Toshihiro Nomura,
Supplementary Information Stargazin regulates AMPA receptor trafficking through adaptor protein complexes during long term depression Shinji Matsuda, Wataru Kakegawa, Timotheus Budisantoso, Toshihiro Nomura,
Supplementary Fig. 1. Multiple five micron sections of liver tissues of rats treated
 Supplementary Figure Legends Supplementary Fig. 1. Multiple five micron sections of liver tissues of rats treated with either vehicle (left; n=3) or CCl 4 (right; n=3) were co-immunostained for NRP-1 (green)
Supplementary Figure Legends Supplementary Fig. 1. Multiple five micron sections of liver tissues of rats treated with either vehicle (left; n=3) or CCl 4 (right; n=3) were co-immunostained for NRP-1 (green)
Beta3 integrin promotes long-lasting activation and polarization of Vascular Endothelial Growth Factor Receptor 2 by immobilized ligand
 SUPPLEMENTAL FIGURES Beta3 integrin promotes long-lasting activation and polarization of Vascular Endothelial Growth Factor Receptor 2 by immobilized ligand C. Ravelli et al. FIGURE S. I Figure S. I: Gremlin
SUPPLEMENTAL FIGURES Beta3 integrin promotes long-lasting activation and polarization of Vascular Endothelial Growth Factor Receptor 2 by immobilized ligand C. Ravelli et al. FIGURE S. I Figure S. I: Gremlin
Supplementary Figure 1. Additional RNAi screen data
 Supplementary Figure 1. Additional RNAi screen data A. Cisplatin induced ATR autophosphorylation. Western blot illustrating ATR and phospho-atr (T1989) in cells exposed to 1 µm cisplatin for 24 hours prior
Supplementary Figure 1. Additional RNAi screen data A. Cisplatin induced ATR autophosphorylation. Western blot illustrating ATR and phospho-atr (T1989) in cells exposed to 1 µm cisplatin for 24 hours prior
Supplementary Materials
 Supplementary Materials Supplementary Figure 1. PKM2 interacts with MLC2 in cytokinesis. a, U87, U87/EGFRvIII, and HeLa cells in cytokinesis were immunostained with DAPI and an anti-pkm2 antibody. Thirty
Supplementary Materials Supplementary Figure 1. PKM2 interacts with MLC2 in cytokinesis. a, U87, U87/EGFRvIII, and HeLa cells in cytokinesis were immunostained with DAPI and an anti-pkm2 antibody. Thirty
Supplementary information. Supplementary Figures
 Supplementary information Supplementary Figures Supplementary Figure 1. A. i. HA-JMY expressing U2OS cells were treated with SAHA (6h). DAPI was used to visualise nuclei. ii. U2OS cells stably expressing
Supplementary information Supplementary Figures Supplementary Figure 1. A. i. HA-JMY expressing U2OS cells were treated with SAHA (6h). DAPI was used to visualise nuclei. ii. U2OS cells stably expressing
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/10/496/eaam6291/dc1 Supplementary Materials for Regulation of autophagy, NF-κB signaling, and cell viability by mir-124 in KRAS mutant mesenchymal-like NSCLC cells
www.sciencesignaling.org/cgi/content/full/10/496/eaam6291/dc1 Supplementary Materials for Regulation of autophagy, NF-κB signaling, and cell viability by mir-124 in KRAS mutant mesenchymal-like NSCLC cells
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2271 Supplementary Figure a! WM266.4 mock WM266.4 #7 sirna WM266.4 #10 sirna SKMEL28 mock SKMEL28 #7 sirna SKMEL28 #10 sirna WM1361 mock WM1361 #7 sirna WM1361 #10 sirna 9 WM266. WM136
DOI: 10.1038/ncb2271 Supplementary Figure a! WM266.4 mock WM266.4 #7 sirna WM266.4 #10 sirna SKMEL28 mock SKMEL28 #7 sirna SKMEL28 #10 sirna WM1361 mock WM1361 #7 sirna WM1361 #10 sirna 9 WM266. WM136
Supporting Information
 Supporting Information Stavru et al. 0.073/pnas.357840 SI Materials and Methods Immunofluorescence. For immunofluorescence, cells were fixed for 0 min in 4% (wt/vol) paraformaldehyde (Electron Microscopy
Supporting Information Stavru et al. 0.073/pnas.357840 SI Materials and Methods Immunofluorescence. For immunofluorescence, cells were fixed for 0 min in 4% (wt/vol) paraformaldehyde (Electron Microscopy
Supplemental Fig. 1: PEA-15 knockdown efficiency assessed by immunohistochemistry and qpcr
 Supplemental figure legends Supplemental Fig. 1: PEA-15 knockdown efficiency assessed by immunohistochemistry and qpcr A, LβT2 cells were transfected with either scrambled or PEA-15 sirna. Cells were then
Supplemental figure legends Supplemental Fig. 1: PEA-15 knockdown efficiency assessed by immunohistochemistry and qpcr A, LβT2 cells were transfected with either scrambled or PEA-15 sirna. Cells were then
supplementary information
 DOI: 10.1038/ncb2172 Figure S1 p53 regulates cellular NADPH and lipid levels via inhibition of G6PD. (a) U2OS cells stably expressing p53 shrna or a control shrna were transfected with control sirna or
DOI: 10.1038/ncb2172 Figure S1 p53 regulates cellular NADPH and lipid levels via inhibition of G6PD. (a) U2OS cells stably expressing p53 shrna or a control shrna were transfected with control sirna or
SUPPLEMENTARY INFORMATION
 (Supplementary Methods and Materials) GST pull-down assay GST-fusion proteins Fe65 365-533, and Fe65 538-700 were expressed in BL21 bacterial cells and purified with glutathione-agarose beads (Sigma).
(Supplementary Methods and Materials) GST pull-down assay GST-fusion proteins Fe65 365-533, and Fe65 538-700 were expressed in BL21 bacterial cells and purified with glutathione-agarose beads (Sigma).
Figure S1 is related to Figure 1B, showing more details of outer segment of
 Supplemental Information Supplementary Figure legends and Figures Figure S1. Electron microscopic images in Sema4A +/+ and Sema4A / retinas Figure S1 is related to Figure 1B, showing more details of outer
Supplemental Information Supplementary Figure legends and Figures Figure S1. Electron microscopic images in Sema4A +/+ and Sema4A / retinas Figure S1 is related to Figure 1B, showing more details of outer
JCB. Supplemental material THE JOURNAL OF CELL BIOLOGY. Prospéri et al.,
 Supplemental material JCB Prospéri et al., http://www.jcb.org/cgi/content/full/jcb.201501018/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Myo1b Tail interacts with YFP-EphB2 coated beads and genistein inhibits
Supplemental material JCB Prospéri et al., http://www.jcb.org/cgi/content/full/jcb.201501018/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Myo1b Tail interacts with YFP-EphB2 coated beads and genistein inhibits
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2386 Figure 1 Src-containing puncta are not focal adhesions, podosomes or endosomes. (a) FAK-/- were stained with anti-py416 Src (green) and either (in red) the focal adhesion protein paxillin,
DOI: 10.1038/ncb2386 Figure 1 Src-containing puncta are not focal adhesions, podosomes or endosomes. (a) FAK-/- were stained with anti-py416 Src (green) and either (in red) the focal adhesion protein paxillin,
Nature Medicine doi: /nm.2548
 Supplementary Table 1: Genotypes of offspring and embryos from matings of Pmm2 WT/F118L mice with Pmm2 WT/R137H mice total events Pmm2 WT/WT Pmm2 WT/R137H Pmm2 WT/F118L Pmm2 R137H/F118L offspring 117 (100%)
Supplementary Table 1: Genotypes of offspring and embryos from matings of Pmm2 WT/F118L mice with Pmm2 WT/R137H mice total events Pmm2 WT/WT Pmm2 WT/R137H Pmm2 WT/F118L Pmm2 R137H/F118L offspring 117 (100%)
Supplementary Figure 1. Immunoprecipitation of synthetic SUMOm-remnant peptides using UMO monoclonal antibody. (a) LC-MS analyses of tryptic
 Supplementary Figure 1. Immunoprecipitation of synthetic SUMOm-remnant peptides using UMO 1-7-7 monoclonal antibody. (a) LC-MS analyses of tryptic digest from HEK293 cells spiked with 6 SUMOmremnant peptides
Supplementary Figure 1. Immunoprecipitation of synthetic SUMOm-remnant peptides using UMO 1-7-7 monoclonal antibody. (a) LC-MS analyses of tryptic digest from HEK293 cells spiked with 6 SUMOmremnant peptides
JCB. Supplemental material THE JOURNAL OF CELL BIOLOGY. Hong et al.,
 Supplemental material JCB Hong et al., http://www.jcb.org/cgi/content/full/jcb.201412127/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Analysis of purified proteins by SDS-PAGE and pull-down assays. (A) Coomassie-stained
Supplemental material JCB Hong et al., http://www.jcb.org/cgi/content/full/jcb.201412127/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Analysis of purified proteins by SDS-PAGE and pull-down assays. (A) Coomassie-stained
(a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for
 Supplementary Figure 1 Generation of Diaph3 ko mice. (a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for different regions of Diaph3 mrna from WT, heterozygote
Supplementary Figure 1 Generation of Diaph3 ko mice. (a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for different regions of Diaph3 mrna from WT, heterozygote
Plutoni et al., http :// /cgi /content /full /jcb /DC1
 Supplemental material JCB Plutoni et al., http ://www.jcb.org /cgi /content /full /jcb.201505105 /DC1 THE JOU RNAL OF CELL BIO LOGY Figure S1. Cell morphology during migration. (a) Protein extracts (20
Supplemental material JCB Plutoni et al., http ://www.jcb.org /cgi /content /full /jcb.201505105 /DC1 THE JOU RNAL OF CELL BIO LOGY Figure S1. Cell morphology during migration. (a) Protein extracts (20
TRANSGENIC ANIMALS. -transient transfection of cells -stable transfection of cells. - Two methods to produce transgenic animals:
 TRANSGENIC ANIMALS -transient transfection of cells -stable transfection of cells - Two methods to produce transgenic animals: 1- DNA microinjection - random insertion 2- embryonic stem cell-mediated gene
TRANSGENIC ANIMALS -transient transfection of cells -stable transfection of cells - Two methods to produce transgenic animals: 1- DNA microinjection - random insertion 2- embryonic stem cell-mediated gene
Supplement Figure 1. Plin5 Plin2 Plin1. KDEL-DSRed. Plin-YFP. Merge
 Supplement Figure 1 Plin5 Plin2 Plin1 KDEL-DSRed Plin-YFP Merge Supplement Figure 2 A. Plin5-Ab MitoTracker Merge AML12 B. Plin5-YFP Cytochrome c-cfp merge Supplement Figure 3 Ad.GFP Ad.Plin5 Supplement
Supplement Figure 1 Plin5 Plin2 Plin1 KDEL-DSRed Plin-YFP Merge Supplement Figure 2 A. Plin5-Ab MitoTracker Merge AML12 B. Plin5-YFP Cytochrome c-cfp merge Supplement Figure 3 Ad.GFP Ad.Plin5 Supplement
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/282/ra53/dc1 Supplementary Materials for Estrogen Alters the Splicing of Type 1 Corticotropin-Releasing Hormone Receptor in Breast Cancer Cells Suchita Lal,
www.sciencesignaling.org/cgi/content/full/6/282/ra53/dc1 Supplementary Materials for Estrogen Alters the Splicing of Type 1 Corticotropin-Releasing Hormone Receptor in Breast Cancer Cells Suchita Lal,
14_integrins_EGFR
 α1 Integrin α1 -/- fibroblasts from integrin α1 knockout animals Figure S1. Serum-starved fibroblasts from α -/- 1 and +/+ mice were stimulated with 10% FBS for 30 min. (FBS) or plated on collagen I (CI)
α1 Integrin α1 -/- fibroblasts from integrin α1 knockout animals Figure S1. Serum-starved fibroblasts from α -/- 1 and +/+ mice were stimulated with 10% FBS for 30 min. (FBS) or plated on collagen I (CI)
indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) B. Male mice of
 SUPPLEMENTRY FIGURE LEGENDS Figure S1. USP44 loss leads to chromosome missegregation.. Genotypes obtained from the indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) 13.5.. Male mice
SUPPLEMENTRY FIGURE LEGENDS Figure S1. USP44 loss leads to chromosome missegregation.. Genotypes obtained from the indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) 13.5.. Male mice
Supplementary Information. A novel human endogenous retroviral protein inhibits cell-cell fusion. Supplementary Figures:
 Supplementary Information A novel human endogenous retroviral protein inhibits cell-cell fusion Jun Sugimoto, Makiko Sugimoto, Helene Bernstein, Yoshihiro Jinno and Danny J. Schust Supplementary Figures:
Supplementary Information A novel human endogenous retroviral protein inhibits cell-cell fusion Jun Sugimoto, Makiko Sugimoto, Helene Bernstein, Yoshihiro Jinno and Danny J. Schust Supplementary Figures:
SUPPLEMENTARY INFORMATION
 The Supplementary Information (SI) Methods Cell culture and transfections H1299, U2OS, 293, HeLa cells were maintained in DMEM medium supplemented with 10% fetal bovine serum. H1299 and 293 cells were
The Supplementary Information (SI) Methods Cell culture and transfections H1299, U2OS, 293, HeLa cells were maintained in DMEM medium supplemented with 10% fetal bovine serum. H1299 and 293 cells were
Supplemental Information. Pacer Mediates the Function of Class III PI3K. and HOPS Complexes in Autophagosome. Maturation by Engaging Stx17
 Molecular Cell, Volume 65 Supplemental Information Pacer Mediates the Function of Class III PI3K and HOPS Complexes in Autophagosome Maturation by Engaging Stx17 Xiawei Cheng, Xiuling Ma, Xianming Ding,
Molecular Cell, Volume 65 Supplemental Information Pacer Mediates the Function of Class III PI3K and HOPS Complexes in Autophagosome Maturation by Engaging Stx17 Xiawei Cheng, Xiuling Ma, Xianming Ding,
Supplementary Figure 1. α-synuclein is truncated in PD and LBD brains. Nature Structural & Molecular Biology: doi: /nsmb.
 Supplementary Figure 1 α-synuclein is truncated in PD and LBD brains. (a) Specificity of anti-n103 antibody. Anti-N103 antibody was coated on an ELISA plate and different concentrations of full-length
Supplementary Figure 1 α-synuclein is truncated in PD and LBD brains. (a) Specificity of anti-n103 antibody. Anti-N103 antibody was coated on an ELISA plate and different concentrations of full-length
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/9/429/ra54/dc1 Supplementary Materials for Dephosphorylation of the adaptor LAT and phospholipase C by SHP-1 inhibits natural killer cell cytotoxicity Omri Matalon,
www.sciencesignaling.org/cgi/content/full/9/429/ra54/dc1 Supplementary Materials for Dephosphorylation of the adaptor LAT and phospholipase C by SHP-1 inhibits natural killer cell cytotoxicity Omri Matalon,
supplementary information
 DOI: 10.1038/ncb2116 Figure S1 CDK phosphorylation of EZH2 in cells. (a) Comparison of candidate CDK phosphorylation sites on EZH2 with known CDK substrates by multiple sequence alignments. (b) CDK1 and
DOI: 10.1038/ncb2116 Figure S1 CDK phosphorylation of EZH2 in cells. (a) Comparison of candidate CDK phosphorylation sites on EZH2 with known CDK substrates by multiple sequence alignments. (b) CDK1 and
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or
 Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
Supplemental Materials and Methods
 Supplemental Materials and Methods Co-immunoprecipitation (Co-IP) assay Cells were lysed with NETN buffer (20 mm Tris-HCl, ph 8.0, 0 mm NaCl, 1 mm EDT, 0.5% Nonidet P-40) containing 50 mm β-glycerophosphate,
Supplemental Materials and Methods Co-immunoprecipitation (Co-IP) assay Cells were lysed with NETN buffer (20 mm Tris-HCl, ph 8.0, 0 mm NaCl, 1 mm EDT, 0.5% Nonidet P-40) containing 50 mm β-glycerophosphate,
Single cell resolution in vivo imaging of DNA damage following PARP inhibition. Supplementary Data
 Single cell resolution in vivo imaging of DNA damage following PARP inhibition Katherine S. Yang, Rainer H. Kohler, Matthieu Landon, Randy Giedt, and Ralph Weissleder Supplementary Data Supplementary Figures
Single cell resolution in vivo imaging of DNA damage following PARP inhibition Katherine S. Yang, Rainer H. Kohler, Matthieu Landon, Randy Giedt, and Ralph Weissleder Supplementary Data Supplementary Figures
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Nakajima and Tanoue, http://www.jcb.org/cgi/content/full/jcb.201104118/dc1 Figure S1. DLD-1 cells exhibit the characteristic morphology
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Nakajima and Tanoue, http://www.jcb.org/cgi/content/full/jcb.201104118/dc1 Figure S1. DLD-1 cells exhibit the characteristic morphology
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Allison et al., http://www.jcb.org/cgi/content/full/jcb.201211045/dc1 Figure S1. Spastin depletion causes increased endosomal tubulation.
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Allison et al., http://www.jcb.org/cgi/content/full/jcb.201211045/dc1 Figure S1. Spastin depletion causes increased endosomal tubulation.
Nature Medicine: doi: /nm.4169
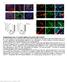 Supplementary Fig.1. EC-specific deletion of Ccm3 by Cdh5-CreERT2. a. mt/mg reporter mice were bred with Cdh5CreERT2 deleter mice followed by tamoxifen feeding from P1 to P3. mg expression was specifically
Supplementary Fig.1. EC-specific deletion of Ccm3 by Cdh5-CreERT2. a. mt/mg reporter mice were bred with Cdh5CreERT2 deleter mice followed by tamoxifen feeding from P1 to P3. mg expression was specifically
Supplementary Information
 Supplementary Information Supplementary Figures Supplementary Figure 1. MLK1-4 phosphorylate MEK in the presence of RAF inhibitors. (a) H157 cells were transiently transfected with Flag- or HA-tagged MLK1-4
Supplementary Information Supplementary Figures Supplementary Figure 1. MLK1-4 phosphorylate MEK in the presence of RAF inhibitors. (a) H157 cells were transiently transfected with Flag- or HA-tagged MLK1-4
Sarker et al. Supplementary Material. Subcellular Fractionation
 Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Han et al., http://www.jcb.org/cgi/content/full/jcb.201311007/dc1 Figure S1. SIVA1 interacts with PCNA. (A) HEK293T cells were transiently
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Han et al., http://www.jcb.org/cgi/content/full/jcb.201311007/dc1 Figure S1. SIVA1 interacts with PCNA. (A) HEK293T cells were transiently
Supplementary Figure 1. Localization of MST1 in RPE cells. Proliferating or ciliated HA- MST1 expressing RPE cells (see Fig. 5b for establishment of
 Supplementary Figure 1. Localization of MST1 in RPE cells. Proliferating or ciliated HA- MST1 expressing RPE cells (see Fig. 5b for establishment of the cell line) were immunostained for HA, acetylated
Supplementary Figure 1. Localization of MST1 in RPE cells. Proliferating or ciliated HA- MST1 expressing RPE cells (see Fig. 5b for establishment of the cell line) were immunostained for HA, acetylated
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
Supplementary Material
 Supplementary Material Supplementary Methods Cell synchronization. For synchronized cell growth, thymidine was added to 30% confluent U2OS cells to a final concentration of 2.5mM. Cells were incubated
Supplementary Material Supplementary Methods Cell synchronization. For synchronized cell growth, thymidine was added to 30% confluent U2OS cells to a final concentration of 2.5mM. Cells were incubated
Figure S1. Specificity of polyclonal anti stabilin-1 and anti stabilin-2 antibodies Lysates of 293T cells transfected with empty vector, mouse
 Figure S1. Specificity of polyclonal anti stabilin-1 and anti stabilin-2 antibodies Lysates of 293T cells transfected with empty vector, mouse stabilin-1, or mouse stabilin-2 were immunoblotted using anti
Figure S1. Specificity of polyclonal anti stabilin-1 and anti stabilin-2 antibodies Lysates of 293T cells transfected with empty vector, mouse stabilin-1, or mouse stabilin-2 were immunoblotted using anti
Nature Biotechnology: doi: /nbt Supplementary Figure 1
 Supplementary Figure 1 Schematic and results of screening the combinatorial antibody library for Sox2 replacement activity. A single batch of MEFs were plated and transduced with doxycycline inducible
Supplementary Figure 1 Schematic and results of screening the combinatorial antibody library for Sox2 replacement activity. A single batch of MEFs were plated and transduced with doxycycline inducible
Nanog-Luc. R-Luc. 40 HA-Klf Relative protein level of Klf USP21. Vector USP2 USP21 *** *** *** ***
 :Flag Relative protein level of Relative protein level of : Flag Vec 21LV 21SV 2 : HA HA- HA- HA- HA-Klf4 Relative mrna expression Relative protein level of Relative protein level of Relative protein level
:Flag Relative protein level of Relative protein level of : Flag Vec 21LV 21SV 2 : HA HA- HA- HA- HA-Klf4 Relative mrna expression Relative protein level of Relative protein level of Relative protein level
Post-translational modification
 Protein expression Western blotting, is a widely used and accepted technique to detect levels of protein expression in a cell or tissue extract. This technique measures protein levels in a biological sample
Protein expression Western blotting, is a widely used and accepted technique to detect levels of protein expression in a cell or tissue extract. This technique measures protein levels in a biological sample
Supplementary Information (Ha, et. al) Supplementary Figures Supplementary Fig. S1
 Supplementary Information (Ha, et. al) Supplementary Figures Supplementary Fig. S1 a His-ORMDL3 ~ 17 His-ORMDL3 GST-ORMDL3 - + - + IPTG GST-ORMDL3 ~ b Integrated Density (ORMDL3/ -actin) 0.4 0.3 0.2 0.1
Supplementary Information (Ha, et. al) Supplementary Figures Supplementary Fig. S1 a His-ORMDL3 ~ 17 His-ORMDL3 GST-ORMDL3 - + - + IPTG GST-ORMDL3 ~ b Integrated Density (ORMDL3/ -actin) 0.4 0.3 0.2 0.1
Supplementary Figure 1
 Supplementary Figure 1 PTEN promotes virus-induced expression of IFNB1 and its downstream genes. (a) Quantitative RT-PCR analysis of IFNB1 mrna (left) and ELISA of IFN-β (right) in HEK 293 cells (2 10
Supplementary Figure 1 PTEN promotes virus-induced expression of IFNB1 and its downstream genes. (a) Quantitative RT-PCR analysis of IFNB1 mrna (left) and ELISA of IFN-β (right) in HEK 293 cells (2 10
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
T H E J O U R N A L O F C E L L B I O L O G Y
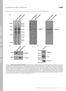 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Bays et al., http://www.jcb.org/cgi/content/full/jcb.201309092/dc1 Figure S1. Specificity of the phospho-y822 antibody. (A) Total cell
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Bays et al., http://www.jcb.org/cgi/content/full/jcb.201309092/dc1 Figure S1. Specificity of the phospho-y822 antibody. (A) Total cell
Supplementary Information
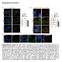 Supplementary Information Supplementary Figure S1 (a) P-cRAF colocalizes with LC3 puncta. Immunofluorescence (IF) depicting colocalization of P-cRAF (green) and LC3 puncta (red) in NIH/3T3 cells treated
Supplementary Information Supplementary Figure S1 (a) P-cRAF colocalizes with LC3 puncta. Immunofluorescence (IF) depicting colocalization of P-cRAF (green) and LC3 puncta (red) in NIH/3T3 cells treated
Supplementary material for: Materials and Methods:
 Supplementary material for: Iron-responsive degradation of iron regulatory protein 1 does not require the Fe-S cluster: S.L. Clarke, et al. Materials and Methods: Fe-S Cluster Reconstitution: Cells treated
Supplementary material for: Iron-responsive degradation of iron regulatory protein 1 does not require the Fe-S cluster: S.L. Clarke, et al. Materials and Methods: Fe-S Cluster Reconstitution: Cells treated
Supplementary Figure 1. Co-localization of GLUT1 and DNAL4 in BeWo cells cultured
 Supplementary Figure 1. Co-localization of GLUT1 and DNAL4 in BeWo cells cultured under static conditions. Cells were seeded in the chamber area of the device and cultured overnight without medium perfusion.
Supplementary Figure 1. Co-localization of GLUT1 and DNAL4 in BeWo cells cultured under static conditions. Cells were seeded in the chamber area of the device and cultured overnight without medium perfusion.
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Ricke et al., http://www.jcb.org/cgi/content/full/jcb.201205115/dc1 Figure S1. Kinetochore localization of mitotic regulators in wild-type
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Ricke et al., http://www.jcb.org/cgi/content/full/jcb.201205115/dc1 Figure S1. Kinetochore localization of mitotic regulators in wild-type
Figure legends for supplement
 Figure legends for supplement Supplemental Figure 1 Characterization of purified and recombinant proteins Relevant fractions related the final stage of the purification protocol(bingham et al., 1998; Toba
Figure legends for supplement Supplemental Figure 1 Characterization of purified and recombinant proteins Relevant fractions related the final stage of the purification protocol(bingham et al., 1998; Toba
Supplementary Figure 1. Differential contribution α5β1 and αv-class integrins to the early adhesion of mouse kidney fibroblasts.
 Supplementary Figure 1. Differential contribution α5β1 and αv-class integrins to the early adhesion of mouse kidney fibroblasts. (a) AFM-based single-cell force spectroscopy (SCFS) to characterize the
Supplementary Figure 1. Differential contribution α5β1 and αv-class integrins to the early adhesion of mouse kidney fibroblasts. (a) AFM-based single-cell force spectroscopy (SCFS) to characterize the
FIG S1: Calibration curves of standards for HPLC detection of Dopachrome (Dopac), Dopamine (DA) and Homovanillic acid (HVA), showing area of peak vs
 FIG S1: Calibration curves of standards for HPLC detection of Dopachrome (Dopac), Dopamine (DA) and Homovanillic acid (HVA), showing area of peak vs amount of standard analysed. Each point represents the
FIG S1: Calibration curves of standards for HPLC detection of Dopachrome (Dopac), Dopamine (DA) and Homovanillic acid (HVA), showing area of peak vs amount of standard analysed. Each point represents the
Quantitative analysis of Bidirectional Signaling (qbids)
 Quantitative analysis of Bidirectional Signaling (qbids) EphB2 + cells were labeled independently with light (C 12 N 14 ) arginine and lysine or heavy (C 13 N 15 ) arginine and lysine ephrin-b1 + cells
Quantitative analysis of Bidirectional Signaling (qbids) EphB2 + cells were labeled independently with light (C 12 N 14 ) arginine and lysine or heavy (C 13 N 15 ) arginine and lysine ephrin-b1 + cells
Supplementary Figure 1 Pfn1, but not other Pfn isoforms are expressed in
 Supplementary Figure 1 Pfn1, but not other Pfn isoforms are expressed in platelets. (a) RT-PCR of Pfn isoforms in control mouse platelets, Pfn1 -/- platelets and control heart. Expected band size for Pfn1
Supplementary Figure 1 Pfn1, but not other Pfn isoforms are expressed in platelets. (a) RT-PCR of Pfn isoforms in control mouse platelets, Pfn1 -/- platelets and control heart. Expected band size for Pfn1
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3209 Supplementary Figure 1 IR induces the association of FH with chromatin. a, U2OS cells synchronized by thymidine double block (2 mm) underwent no release (G1 phase) or release for 2
DOI: 10.1038/ncb3209 Supplementary Figure 1 IR induces the association of FH with chromatin. a, U2OS cells synchronized by thymidine double block (2 mm) underwent no release (G1 phase) or release for 2
Electronic Supplementary Information
 Electronic Supplementary Material (ESI) for Integrative Biology. This journal is The Royal Society of Chemistry 2015 Electronic Supplementary Information Table S1. Definition of quantitative cellular features
Electronic Supplementary Material (ESI) for Integrative Biology. This journal is The Royal Society of Chemistry 2015 Electronic Supplementary Information Table S1. Definition of quantitative cellular features
SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURES
 SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURES Supplementary Figure 1. Generation of inducible BICD2 knock-out mice. A) The mouse BICD2 locus and gene targeting constructs. To generate an inducible Bicd2
SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURES Supplementary Figure 1. Generation of inducible BICD2 knock-out mice. A) The mouse BICD2 locus and gene targeting constructs. To generate an inducible Bicd2
Figure S1. Figure S2. Figure S3 HB Anti-FSP27 (COOH-terminal peptide) Ab. Anti-GST-FSP27(45-127) Ab.
 / 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
/ 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
MT minus end catastrophe
 DOI: 1.138/ncb3241 MT minus end catastrophe frequency [s -1 ].2.15.1.5 control mgfp- TPX2 * * mgfp- TPX2 mini Supplementary Figure 1. TPX2 reduces catastrophes at the microtubule minus ends. Modified box-and-whiskers
DOI: 1.138/ncb3241 MT minus end catastrophe frequency [s -1 ].2.15.1.5 control mgfp- TPX2 * * mgfp- TPX2 mini Supplementary Figure 1. TPX2 reduces catastrophes at the microtubule minus ends. Modified box-and-whiskers
Supplementary Figure 1. IFN-γ induces TRC dormancy. a, IFN-γ induced dormancy
 Supplementary Figure 1. IFN-γ induces TRC dormancy. a, IFN-γ induced dormancy of various tumor type TRCs, including H22 (murine hepatocarcinoma) and CT26 (murine colon cancer). Bar, 50 µm. b, B16 cells
Supplementary Figure 1. IFN-γ induces TRC dormancy. a, IFN-γ induced dormancy of various tumor type TRCs, including H22 (murine hepatocarcinoma) and CT26 (murine colon cancer). Bar, 50 µm. b, B16 cells
Supplementary Information
 Supplementary Information Supplementary Figure 1: Over-expression of CD300f in NIH3T3 cells enhances their capacity to phagocytize AC. (a) NIH3T3 cells were stably transduced by EV, CD300f WT or CD300f
Supplementary Information Supplementary Figure 1: Over-expression of CD300f in NIH3T3 cells enhances their capacity to phagocytize AC. (a) NIH3T3 cells were stably transduced by EV, CD300f WT or CD300f
Artificial targeting of misfolded cytosolic proteins to endoplasmic reticulum as a mechanism for clearance
 Supplementary Information Artificial targeting of misfolded cytosolic proteins to endoplasmic reticulum as a mechanism for clearance Fen Liu 1, Deanna M. Koepp 2, and Kylie J. Walters 1* 1 Protein Processing
Supplementary Information Artificial targeting of misfolded cytosolic proteins to endoplasmic reticulum as a mechanism for clearance Fen Liu 1, Deanna M. Koepp 2, and Kylie J. Walters 1* 1 Protein Processing
Supplementary Figure 1. Adipogenic protein expression in WT and KO MEFs after 7 days of adipogenic differentiation.
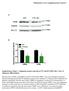 Merkestein et al. Supplementary Figure 1 A PLIN WT FTO KO 72 kda FABP4 17 kda HSC70 72 kda B Supplementary Figure 1. Adipogenic protein expression in WT and KO MEFs after 7 days of adipogenic differentiation.
Merkestein et al. Supplementary Figure 1 A PLIN WT FTO KO 72 kda FABP4 17 kda HSC70 72 kda B Supplementary Figure 1. Adipogenic protein expression in WT and KO MEFs after 7 days of adipogenic differentiation.
Specific threonine-4 phosphorylation and function of RNA polymerase II CTD during M phase progression
 Supplementary Figures Specific threonine- phosphorylation and function of RNA polymerase II CTD during M phase progression Corinna Hintermair 1, Kirsten Voß 1, Ignasi Forné 2, Martin Heidemann 1, Andrew
Supplementary Figures Specific threonine- phosphorylation and function of RNA polymerase II CTD during M phase progression Corinna Hintermair 1, Kirsten Voß 1, Ignasi Forné 2, Martin Heidemann 1, Andrew
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3164 Supplementary Figure 1 Validation of effective Gnas deletion and epithelial thickness. a, Representative genotyping in mice treated or not with tamoxifen to show Gnas deletion. To
DOI: 10.1038/ncb3164 Supplementary Figure 1 Validation of effective Gnas deletion and epithelial thickness. a, Representative genotyping in mice treated or not with tamoxifen to show Gnas deletion. To
Hyper sensitive protein detection by Tandem-HTRF reveals Cyclin D1
 Hyper sensitive protein detection by Tandem-HTRF reveals Cyclin D1 dynamics in adult mouse Alexandre Zampieri, Julien Champagne, Baptiste Auzemery, Ivanna Fuentes, Benjamin Maurel and Frédéric Bienvenu
Hyper sensitive protein detection by Tandem-HTRF reveals Cyclin D1 dynamics in adult mouse Alexandre Zampieri, Julien Champagne, Baptiste Auzemery, Ivanna Fuentes, Benjamin Maurel and Frédéric Bienvenu
Supplementary Figure 1. Expressions of stem cell markers decreased in TRCs on 2D plastic. TRCs were cultured on plastic for 1, 3, 5, or 7 days,
 Supplementary Figure 1. Expressions of stem cell markers decreased in TRCs on 2D plastic. TRCs were cultured on plastic for 1, 3, 5, or 7 days, respectively, and their mrnas were quantified by real time
Supplementary Figure 1. Expressions of stem cell markers decreased in TRCs on 2D plastic. TRCs were cultured on plastic for 1, 3, 5, or 7 days, respectively, and their mrnas were quantified by real time
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Supplementary Table 1 Supplementary Figures 1, 2, 3, 4, 5, 6, 7, 8 and legends Supplementary Video 1, 2 legends AON Sequence Position Length MW tcdna 5 -AACCTCGGCTTACCT-3 +2-13
SUPPLEMENTARY INFORMATION Supplementary Table 1 Supplementary Figures 1, 2, 3, 4, 5, 6, 7, 8 and legends Supplementary Video 1, 2 legends AON Sequence Position Length MW tcdna 5 -AACCTCGGCTTACCT-3 +2-13
GFP CCD2 GFP IP:GFP
 D1 D2 1 75 95 148 178 492 GFP CCD1 CCD2 CCD2 GFP D1 D2 GFP D1 D2 Beclin 1 IB:GFP IP:GFP Supplementary Figure 1: Mapping domains required for binding to HEK293T cells are transfected with EGFP-tagged mutant
D1 D2 1 75 95 148 178 492 GFP CCD1 CCD2 CCD2 GFP D1 D2 GFP D1 D2 Beclin 1 IB:GFP IP:GFP Supplementary Figure 1: Mapping domains required for binding to HEK293T cells are transfected with EGFP-tagged mutant
Rer1 and calnexin regulate endoplasmic reticulum retention of a peripheral myelin protein 22 mutant that causes type 1A Charcot-Marie-Tooth disease
 Rer1 and calnexin regulate endoplasmic reticulum retention of a peripheral myelin protein mutant that causes type 1A Charcot-Marie-Tooth disease Taichi Hara, Yukiko Hashimoto, Tomoko Akuzawa, Rika Hirai,
Rer1 and calnexin regulate endoplasmic reticulum retention of a peripheral myelin protein mutant that causes type 1A Charcot-Marie-Tooth disease Taichi Hara, Yukiko Hashimoto, Tomoko Akuzawa, Rika Hirai,
SUPPLEMENTARY INFORMATION
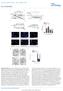 DOI: 10.1038/ncb3230 a GM13267(ZW) WCE C N M H 2 O 2 : p(s1981) 2 2 LDH Lamin A/C β-integrin c b d AT Flag- WT Flag- RQ H 2 O 2 IgG p (S1981) p (S1981) 2 IP: Flag N.S. 2 e f A: Untreated AT5 cells B: AT5
DOI: 10.1038/ncb3230 a GM13267(ZW) WCE C N M H 2 O 2 : p(s1981) 2 2 LDH Lamin A/C β-integrin c b d AT Flag- WT Flag- RQ H 2 O 2 IgG p (S1981) p (S1981) 2 IP: Flag N.S. 2 e f A: Untreated AT5 cells B: AT5
hnrnp D/AUF1 Rabbit IgG hnrnp M
 Mouse IgG Goat IgG Rabbit IgG Mouse IgG hnrnp F Goat IgG Mouse IgG Kb 6 4 3 2 15 5 Supplementary Figure S1. In vivo binding of TERRA-bound RBPs to target RNAs. Immunoprecipitation (IP) assay using 3 mg
Mouse IgG Goat IgG Rabbit IgG Mouse IgG hnrnp F Goat IgG Mouse IgG Kb 6 4 3 2 15 5 Supplementary Figure S1. In vivo binding of TERRA-bound RBPs to target RNAs. Immunoprecipitation (IP) assay using 3 mg
Revised: RG-RV2 by Fukuhara et al.
 Supplemental Figure 1 The generation of Spns2 conditional knockout mice. (A) Schematic representation of the wild type Spns2 locus (Spns2 + ), the targeted allele, the floxed allele (Spns2 f ) and the
Supplemental Figure 1 The generation of Spns2 conditional knockout mice. (A) Schematic representation of the wild type Spns2 locus (Spns2 + ), the targeted allele, the floxed allele (Spns2 f ) and the
Nature Biotechnology: doi: /nbt.4166
 Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
Parthanatos mediates AIMP2-activated age-dependent dopaminergic neuronal loss
 SUPPLEMENTARY INFORMATION Parthanatos mediates AIMP2-activated age-dependent dopaminergic neuronal loss Yunjong Lee, Senthilkumar S. Karuppagounder, Joo-Ho Shin, Yun-Il Lee, Han Seok Ko, Debbie Swing,
SUPPLEMENTARY INFORMATION Parthanatos mediates AIMP2-activated age-dependent dopaminergic neuronal loss Yunjong Lee, Senthilkumar S. Karuppagounder, Joo-Ho Shin, Yun-Il Lee, Han Seok Ko, Debbie Swing,
TRIM31 is recruited to mitochondria after infection with SeV.
 Supplementary Figure 1 TRIM31 is recruited to mitochondria after infection with SeV. (a) Confocal microscopy of TRIM31-GFP transfected into HEK293T cells for 24 h followed with SeV infection for 6 h. MitoTracker
Supplementary Figure 1 TRIM31 is recruited to mitochondria after infection with SeV. (a) Confocal microscopy of TRIM31-GFP transfected into HEK293T cells for 24 h followed with SeV infection for 6 h. MitoTracker
Supplementary Figure 1. TSA (10 nmol/l), non-class-selective HDAC inhibitor, potentiates
 Supplementary Figure 1. TSA (10 nmol/l), non-class-selective HDAC inhibitor, potentiates vascular calcification (VC). (a) Von Kossa staining shows that TSA potentiated the Pi-induced VC. Scale bar, 100
Supplementary Figure 1. TSA (10 nmol/l), non-class-selective HDAC inhibitor, potentiates vascular calcification (VC). (a) Von Kossa staining shows that TSA potentiated the Pi-induced VC. Scale bar, 100
Supplementary Fig. 1 Identification of Nedd4 as an IRS-2-associated protein in camp-treated FRTL-5 cells.
 Supplementary Fig. 1 Supplementary Fig. 1 Identification of Nedd4 as an IRS-2-associated protein in camp-treated FRTL-5 cells. (a) FRTL-5 cells were treated with 1 mm dibutyryl camp for 24 h, and the lysates
Supplementary Fig. 1 Supplementary Fig. 1 Identification of Nedd4 as an IRS-2-associated protein in camp-treated FRTL-5 cells. (a) FRTL-5 cells were treated with 1 mm dibutyryl camp for 24 h, and the lysates
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/5/233/ra50/dc1 Supplementary Materials for Epidermal Growth Factor Receptor Is Essential for Toll-Like Receptor 3 Signaling Michifumi Yamashita, Saurabh Chattopadhyay,
www.sciencesignaling.org/cgi/content/full/5/233/ra50/dc1 Supplementary Materials for Epidermal Growth Factor Receptor Is Essential for Toll-Like Receptor 3 Signaling Michifumi Yamashita, Saurabh Chattopadhyay,
JCB. Supplemental material THE JOURNAL OF CELL BIOLOGY. Kimura et al.,
 Supplemental material JCB Kimura et al., http://www.jcb.org/cgi/content/full/jcb.201503023/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. TRIMs regulate IFN-γ induced autophagy. (A and B) HC image analysis
Supplemental material JCB Kimura et al., http://www.jcb.org/cgi/content/full/jcb.201503023/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. TRIMs regulate IFN-γ induced autophagy. (A and B) HC image analysis
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
Supplementary Figure 1 (A), (B), and (C) Docking of a physiologic ligand of integrin αvβ3, the tenth type III RGD domain of wild-type fibronectin
 Supplementary Figure 1 (A), (B), and (C) Docking of a physiologic ligand of integrin αvβ3, the tenth type III RGD domain of wild-type fibronectin (A), D1-CD2 (B), and the D1-CD2 variant 3 (C) to Adomain
Supplementary Figure 1 (A), (B), and (C) Docking of a physiologic ligand of integrin αvβ3, the tenth type III RGD domain of wild-type fibronectin (A), D1-CD2 (B), and the D1-CD2 variant 3 (C) to Adomain
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than. SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with
 Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and
 Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
i-stop codon positions in the mcherry gene
 Supplementary Figure 1 i-stop codon positions in the mcherry gene The grnas (green) that can potentially generate stop codons from Trp (63 th and 98 th aa, upper panel) and Gln (47 th and 114 th aa, bottom
Supplementary Figure 1 i-stop codon positions in the mcherry gene The grnas (green) that can potentially generate stop codons from Trp (63 th and 98 th aa, upper panel) and Gln (47 th and 114 th aa, bottom
(a) Immunoblotting to show the migration position of Flag-tagged MAVS
 Supplementary Figure 1 Characterization of six MAVS isoforms. (a) Immunoblotting to show the migration position of Flag-tagged MAVS isoforms. HEK293T Mavs -/- cells were transfected with constructs expressing
Supplementary Figure 1 Characterization of six MAVS isoforms. (a) Immunoblotting to show the migration position of Flag-tagged MAVS isoforms. HEK293T Mavs -/- cells were transfected with constructs expressing
Supplementary Table 1. Sequences for BTG2 and BRCA1 sirnas.
 Supplementary Table 1. Sequences for BTG2 and BRCA1 sirnas. Target Gene Non-target / Control BTG2 BRCA1 NFE2L2 Target Sequence ON-TARGET plus Non-targeting sirna # 1 (Cat# D-001810-01-05) sirna1: GAACCGACAUGCUCCCGGA
Supplementary Table 1. Sequences for BTG2 and BRCA1 sirnas. Target Gene Non-target / Control BTG2 BRCA1 NFE2L2 Target Sequence ON-TARGET plus Non-targeting sirna # 1 (Cat# D-001810-01-05) sirna1: GAACCGACAUGCUCCCGGA
