A fail-safe mechanism in the septal ring assembly pathway generated by the sequential recruitment of cell separation amidases and their activators
|
|
|
- Eleanore Hall
- 5 years ago
- Views:
Transcription
1 Supplemental Information for: A fail-safe mechanism in the septal ring assembly pathway generated by the sequential recruitment of cell separation amidases and their activators Nick T. Peters, Thuy Dinh, and Thomas G. Bernhardt * Department of Microbiology and Molecular Genetics Harvard Medical School 200 Longwood Avenue Boston, MA *To whom correspondence should be addressed: Thomas G. Bernhardt, Ph.D. Department of Microbiology and Molecular Genetics Harvard Medical School Armenise Building, Room 302A 200 Longwood Avenue Boston, Massachusetts Phone: (617) Fax: (617) thomas_bernhardt@hms.harvard.edu Running title: Sequential recruitment of EnvC and AmiB to the division site
2 A1 A2 A3 DIC ZapA-mCherry FtsN-GFP Figure S1
3 DIC mcherry GFP A1 A2 A3 FtsN ZapA-mCherry FtsN-GFP B1 B2 B3 EnvC EnvC-mCherry ZapA-GFP C1 C2 C3 NlpD NlpD-mCherry ZapA-GFP D1 D2 D3 AmiC AmiB ZapA-mCherry AmiB- sf GFP E1 E2 E3 ZapA-mCherry AmiC-GFP Figure S2
4 Supplemental Figure Legends Figure S1. GFP-FtsN is recruited to septal rings following cephalexin treatment. Cells of HC284(attHKNP21) [zapa-mcherry (Plac::gfp-ftsN)] (A) were grown overnight in LB at 37 C. They were then diluted 1:100 in M9 maltose supplemented with 10 μm IPTG. Upon reaching an OD600 of 0.05, 10 μg/ml cephalexin was added. Cells were grown for 3-4 doublings and then imaged on 2% agarose pads made with 10 μg/ml cephalexin added to ensure consistent drug treatment. The GFP-FtsN fusion colocalized with ZapA-mCherry septal rings 61.6% (n=154) of the time. DIC is shown in panel 1, mcherry is shown in panel 2, and GFP is shown in panel 3. Bar equals 4 microns. Figure S2. FtsN, amidase, and LytM factor localization in the absence of cephalexin. Cells of HC284(attHKNP21) [zapa-mcherry (Plac::gfp-ftsN)] (A), NP32(attHKNP18) [ envc zapa-gfp (Plac::envC-mCherry)] (B), NP94(attHKTB314) [ΔnlpD zapa-gfp (Plac::nlpD-mCherry)] (C), NP90(attHKNP19) [ΔamiB zapa-mcherry (Plac:: ss dsba-amib-sfgfp)] (D), NP84 (atthknp16) [ΔamiC zapa-mcherry (Plac-m3::amiC-gfp)] (E), were grown overnight in LB at 37 C. Cultures were diluted 1:100 in M9 maltose plus 10 μm IPTG and grown at 30 C until they reached and OD600 between Cells were then imaged on 2% agarose pads with DIC (panels 1), mcherry (panels 2), and GFP/sfGFP (panels 3). Bar equals 4 microns.
5 Table S1. Localization statistics for NlpD and AmiC. Strain Genotype #cells %cells w/ rings TB28(attHKTB314) TB145(attHKTB314) TB143(attHKTB314) TU217(attHKTB314) TB28 (atthknp16) TB137(attHKNP16) TB139(attHKTB314) TU217(attHKTB314) WT (Plac::nlpD-mCherry) nlpd (Plac::nlpD-mCherry) amic (Plac::nlpD-mCherry) amic nlpd (Plac::nlpD-mCherry) WT (Plac::amiC-gfp) amic (Plac::amiC-gfp) nlpd (Plac::amiC-gfp) amic nlpd (Plac::amiC-gfp) * * * *AmiC rings were only observed in deeply constricted cells.
6 Plasmid construction In all cases, PCR was performed using KOD polymerase (Novagen) according to the instructions. Restriction sites for use in plasmid constructions are italicized and underlined in the primer sequences given below. Plasmid DNA and PCR fragments were purified using the Qiaprep spin miniprep kit (Qiagen) or the Qiaquick PCR purification kit (Qiagen), respectively. To construct ptd80 [attλ cat laci q Plac::envC-mCherry], the Plac::envC-mCherry containing EcoRI-HindIII fragment of ptb316 [atthk022 bla laci q Plac::envC-mCherry] (7) was used to replace the corresponding Para::envC fragment of ptd25 [attλ cat Para::envC] (7). For ptd82 [atthk022 bla laci q Plac:: ss dsba-amib(23-191)-sfgfp], amib(23-191) was amplified with the primers 5ʼ-GTCAGGATCCGCGACGCTCTCTGATATTCAGGTTTC-3ʼ and 5ʼ- GTCACTCGAGTTTATCGCCAGTGTTAGCCGTCG-3ʼ. The resulting fragment was digested with BamHI and XhoI and used to replace the corresponding amib(23-445) fragment of ptb311 [atthk022 bla laci q Plac:: ss dsba-amib(23-445)-sfgfp]. pnp3 [atthk022 bla laci q Plac:: ss dsba-amib (23-445)-mCherry] was constructed in several steps. First ptu148 [atthk022 bla laci q Plac:: ss dsba-mcherry] was constructed by replacing the sfgfp containing AvrII-HindIII fragment of ptb282 [atthk022 bla laci q Plac:: ss dsba-sfgfp] (6) with an appropriately digested mcherry fragment amplified from ptu136 [atthk022 bla laci q Plac:: ss dsba-mcherry] (7) using the primers 5ʼ- GTCACCTAGGCAAGATCCGGCTGGTCTGTCCAAGGGCG-3ʼ and 5ʼ- GTCAAAGCTTGTCGACTTATTTGTACAGCTCATCCATGCCACC-3ʼ. ptu136 and ptu148 are similar vectors but have different cloning sites for the construction of exported N- or C-terminal
7 mcherry fusions, respectively. ptu190 [atthk022 bla laci q Plac:: ss dsba-amib (23-445)- mcherry] was made by inserting the amib containing BamHI-XhoI fragment from ptb311 into ptu148 digested with the same enzymes. pnp3 was then constructed by replacing the mcherry containing XhoI-HindIII fragment of ptu190 with mcherry amplified from ptu136 with the primers 5ʼ- GTCACTCGAGGGTCCGGCTGGTCTGTCCAAGGGCGAGGAGGATAACC-3ʼ and 5ʼ- GTCAAAGCTTGTCGACTTATTTGTACAGCTCATCCATGCCACC-3ʼ. pnp3 and ptu190 are identical except that the former codes for a shorter linker sequence (LEGPAGL) between AmiB and mcherry. The shorter linker yielded superior localization results relative to the longer linker (LEPYASAQPRQDPAGL) (data not shown). To construct pdy71 [atthk022 teta tetr laci q Plac::], the base vector for Tet R containing integration plasmids needed for experiments with cephalexin, several rounds of mutagenesis were required to remove XbaI, HindIII, EcoRI and NdeI sites from the teta tetr genes. The teta tetr cassette was amplified from a Tn10 containing strain using the primers 5ʼ- ATTCTAGATCTCTAAAGGGTGGTTAACTCGACATCTTGG-3ʼ and 5ʼ- ATTCTGCGGCCGCAGGCCAATTTATTGCTATTTACCGCGG-3ʼ. Then single primer site directed mutagenesis was used to eliminate XbaI (AATTAGGAATTAATGATGTCCAGATTAGATAAAAGTAAAGT), HindIII (CGTAAACTCGCCCAGAAGCTCGGTGTAGAGCAGCCTACATT), EcoRI (AAATAAAGTGATGTATACCGAGTTCGATTGCGTCTCAACCC), and NdeI (TTCGGCCTTGAATTGATCATCTGCGGATTAGAAAAACAACT) resulting in pdy71.
8 To construct pdy75 [atthk022 teta tetr arac Para:: ss dsba-amib], the Plac:: ss dsba-amib (23-445)-sfgfp containing BglII-HindIII fragment from ptb311[atthk022 bla laci q Plac:: ss dsbaamib(23-445)-sfgfp] (6) was used to replace the corresponding fragment of pdy71. To construct pnp16, the amic-gfp containing XbaI-HindIII fragment from ptb28 [bla laci q Plac::amiC-gfp] (2) replaced the corresponding fragment in phc583 [atthk022 teta tetr laci q Plac-m3::slmA]. The phc583 plasmid was generated by BgIII-HindIII digestion of the pdy75 backbone and replacing it with a corresponding fragment from phc531 [atthk022 bla laci q Placm3::slmA] (3). Plac-m3 is a synthetic lac promoter variant with a more consensus -10 element (TATGTT changed to TATATT). To construct pnp18 [atthk022 teta tetr laci q Plac::envC-mCherry], pnp19 [atthk022 teta tetr laci q Plac:: ss dsba-amib-sfgfp], and pnp20 [atthk022 teta tetr laci q Plac::nlpD-mCherry] the envc-mcherry, ss dsba-amib-sfgfp, or nlpd-mcherry containing XbaI-HindIII fragments from ptb316 [atthk022 bla laci q Plac::envC-mCherry] (7), ptb311 [atthk022 bla laci q Plac:: ss dsbaamib(23-445)-sfgfp] (6), or ptb314 [atthk022 bla laci q Plac::nlpD-mCherry] (7), respectively were used to replace the corresponding fragment of pmm60 [atthk022 teta tetr laci q Plac::ycfM-sfgfp]. To construct pmm60, the ycfm-sfgfp containing BgIII-HindIII fragment of pcb28 [atthk022 bla laci q Plac::ycfM-sfgfp] (5) was used to replace the corresponding fragment of pdy75. To construct pnp21 [atthk022 teta tetr laci q Plac::gfp-ftsN], the gfp-ftsn containing XbaI-HindIII fragment from pkp5 [attλ cat arac Para::gfp-ftsN] was used to replace the corresponding fragment of pmm60. The pkp5 plasmid was created by XbaI-HindIII digestion of ptb285 [attλ
9 cat arac Para::] (3) and inserting a corresponding fragment from ptu194 [atthk022 bla laci q Psyn135::mCherry-ZapA]. The ptu194 plasmid was generated by XbaI-HindIII digestion of ptu191 [atthk022 bla laci q Psyn135::gfp-ftsN] and inserting a corresponding fragment from pch201 [atthk022 bla laci q Plac::gfp-ftsN] (4). The ptu191 plasmid was created by EcoRI-SaII digestion of ptb265 [atthk022 bla laci q Para::] (7) and replaced with a corresponding fragment from pez4 [attλ cat arac Psyn135::gfp-zapA] (1).
10 REFERENCES 1. Bendezú F., C. Hale, T. Bernhardt, and P. de Boer RodZ (YfgA) is required for proper assembly of the MreB actin cytoskeleton and cell shape in E. coli. EMBO J 28: Bernhardt T. G., and P. A. J. de Boer The Escherichia coli amidase AmiC is a periplasmic septal ring component exported via the twin-arginine transport pathway. Mol Microbiol 48: Cho H., H. R. McManus, S. L. Dove, and T. G. Bernhardt Nucleoid occlusion factor SlmA is a DNA-activated FtsZ polymerization antagonist. Proc Nat Acad Sci USA 108: Hale C. A., and P. A. J. de Boer ZipA is required for recruitment of FtsK, FtsQ, FtsL, and FtsN to the septal ring in Escherichia coli. J Bacteriol 184: Paradis-Bleau C., M. Markovski, T. Uehara, T. J. Lupoli, S. Walker, D. E. Kahne, and T. G. Bernhardt Lipoprotein cofactors located in the outer membrane activate bacterial cell wall polymerases. Cell 143: Uehara T., K. R. Parzych, T. Dinh, and T. G. Bernhardt Daughter cell separation is controlled by cytokinetic ring-activated cell wall hydrolysis. EMBO J 29: Uehara T., T. Dinh, and T. G. Bernhardt LytM-domain factors are required for daughter cell separation and rapid ampicillin-induced lysis in Escherichia coli. J Bacteriol 191:
LytM-Domain Factors Are Required for Daughter Cell Separation and Rapid Ampicillin-Induced Lysis in Escherichia coli
 JOURNAL OF BACTERIOLOGY, Aug. 2009, p. 5094 5107 Vol. 191, No. 16 0021-9193/09/$08.00 0 doi:10.1128/jb.00505-09 Copyright 2009, American Society for Microbiology. All Rights Reserved. LytM-Domain Factors
JOURNAL OF BACTERIOLOGY, Aug. 2009, p. 5094 5107 Vol. 191, No. 16 0021-9193/09/$08.00 0 doi:10.1128/jb.00505-09 Copyright 2009, American Society for Microbiology. All Rights Reserved. LytM-Domain Factors
Supporting information
 Supporting information Construction of strains and plasmids To create ptc67, a PCR product obtained with primers cc2570-162f (gcatgggcaagcttgaggacggcgtcatgt) and cc2570+512f (gaggccgtggtaccatagaggcgggcg),
Supporting information Construction of strains and plasmids To create ptc67, a PCR product obtained with primers cc2570-162f (gcatgggcaagcttgaggacggcgtcatgt) and cc2570+512f (gaggccgtggtaccatagaggcgggcg),
FtsEX is required for CwlO peptidoglycan hydrolase activity during cell wall elongation in Bacillus subtilis
 FtsEX is required for CwlO peptidoglycan hydrolase activity during cell wall elongation in Bacillus subtilis Jeffrey Meisner, Paula Montero Llopis, Lok-To Sham, Ethan Garner, Thomas G. Bernhardt, David
FtsEX is required for CwlO peptidoglycan hydrolase activity during cell wall elongation in Bacillus subtilis Jeffrey Meisner, Paula Montero Llopis, Lok-To Sham, Ethan Garner, Thomas G. Bernhardt, David
Supplementary information. Interplay between Penicillin-binding proteins and SEDS proteins promotes. bacterial cell wall synthesis
 Supplementary information Interplay between Penicillin-binding proteins and SEDS proteins promotes bacterial cell wall synthesis Sophie Leclercq 1, Adeline Derouaux 1, Samir Olatunji 1, Claudine Fraipont
Supplementary information Interplay between Penicillin-binding proteins and SEDS proteins promotes bacterial cell wall synthesis Sophie Leclercq 1, Adeline Derouaux 1, Samir Olatunji 1, Claudine Fraipont
Certificate of Analysis
 Certificate of Analysis pet6xhn Expression Vector Set Contents Product Information... 1 pet6xhn-n, pet6xhn-c, and pet6xhn-gfpuv Vector Information... 2 Location of Features... 4 Additional Information...
Certificate of Analysis pet6xhn Expression Vector Set Contents Product Information... 1 pet6xhn-n, pet6xhn-c, and pet6xhn-gfpuv Vector Information... 2 Location of Features... 4 Additional Information...
Supplementary figures
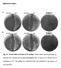 Supplementary figures A ΔrelAΔlon ΔrelA ΔrelAΔlon Δlon ppgpp 0 Δlon B ΔrelAΔlon ΔrelA ΔrelAΔlon Δlon ppgpp 0 Δlon Fig. S1. Growth defect of strains in LB medium. Strains whose relevant genotypes are indicated
Supplementary figures A ΔrelAΔlon ΔrelA ΔrelAΔlon Δlon ppgpp 0 Δlon B ΔrelAΔlon ΔrelA ΔrelAΔlon Δlon ppgpp 0 Δlon Fig. S1. Growth defect of strains in LB medium. Strains whose relevant genotypes are indicated
igem2013 Microbiology BMB SDU
 igem2013 Microbiology BMB SDU Project type: USER cloning Project title: USER cloning of DXS GFP reporter system Sub project: Creation date: 13.07.24 Written by: PRA, MHK Performed by: PRA, MHK 1. SOPs
igem2013 Microbiology BMB SDU Project type: USER cloning Project title: USER cloning of DXS GFP reporter system Sub project: Creation date: 13.07.24 Written by: PRA, MHK Performed by: PRA, MHK 1. SOPs
Differences in MinC/MinD Sensitivity between Polar and Internal Z Rings in Escherichia coli
 JOURNAL OF BACTERIOLOGY, Jan. 2011, p. 367 376 Vol. 193, No. 2 0021-9193/11/$12.00 doi:10.1128/jb.01095-10 Copyright 2011, American Society for Microbiology. All Rights Reserved. Differences in MinC/MinD
JOURNAL OF BACTERIOLOGY, Jan. 2011, p. 367 376 Vol. 193, No. 2 0021-9193/11/$12.00 doi:10.1128/jb.01095-10 Copyright 2011, American Society for Microbiology. All Rights Reserved. Differences in MinC/MinD
Supplementary Information
 Supplementary Information Supplementary Figures Figure S1. Study of mgtl translation in vitro. (A) Detection of 5 LR RNA using wild-type and anti-sd (91-95) substituted templates in a transcription-translation
Supplementary Information Supplementary Figures Figure S1. Study of mgtl translation in vitro. (A) Detection of 5 LR RNA using wild-type and anti-sd (91-95) substituted templates in a transcription-translation
Supporting Information
 Supporting Information Development of a 2,4-Dinitrotoluene-Responsive Synthetic Riboswitch in E. coli cells Molly E. Davidson, Svetlana V. Harbaugh, Yaroslav G. Chushak, Morley O. Stone, Nancy Kelley-
Supporting Information Development of a 2,4-Dinitrotoluene-Responsive Synthetic Riboswitch in E. coli cells Molly E. Davidson, Svetlana V. Harbaugh, Yaroslav G. Chushak, Morley O. Stone, Nancy Kelley-
Fig. S1. CrgA intracellular levels in M. tuberculosis. Ten and twenty micrograms of
 Supplementary data Fig. S1. CrgA intracellular levels in M. tuberculosis. Ten and twenty micrograms of cell free protein lysates from WT M. tuberculosis (Rv) together with various known concentrations
Supplementary data Fig. S1. CrgA intracellular levels in M. tuberculosis. Ten and twenty micrograms of cell free protein lysates from WT M. tuberculosis (Rv) together with various known concentrations
The fusion used in most of these studies (MinC4-GFP) contains GFP and
 Gregory, Becker and Pogliano, Supplemental Materials page 1 Supplemental Methods Description of the MinC and MinD GFP insertions The fusion used in most of these studies (MinC4-GFP) contains GFP and flanking
Gregory, Becker and Pogliano, Supplemental Materials page 1 Supplemental Methods Description of the MinC and MinD GFP insertions The fusion used in most of these studies (MinC4-GFP) contains GFP and flanking
Divisome under Construction: Distinct Domains of the Small Membrane Protein FtsB Are Necessary for Interaction with Multiple Cell Division Proteins
 JOURNAL OF BACTERIOLOGY, Apr. 2009, p. 2815 2825 Vol. 191, No. 8 0021-9193/09/$08.00 0 doi:10.1128/jb.01597-08 Copyright 2009, American Society for Microbiology. All Rights Reserved. Divisome under Construction:
JOURNAL OF BACTERIOLOGY, Apr. 2009, p. 2815 2825 Vol. 191, No. 8 0021-9193/09/$08.00 0 doi:10.1128/jb.01597-08 Copyright 2009, American Society for Microbiology. All Rights Reserved. Divisome under Construction:
Antisense RNA Targeting the First Periplasmic Domain of YidC in Escherichia coli Appears to Induce Filamentation but Does Not Affect Cell Viability
 Antisense RNA Targeting the First Periplasmic Domain of YidC in Escherichia coli Appears to Induce Filamentation but Does Not Affect Cell Viability Riaaz Lalani, Nathaniel Susilo, Elisa Xiao, Andrea Xu
Antisense RNA Targeting the First Periplasmic Domain of YidC in Escherichia coli Appears to Induce Filamentation but Does Not Affect Cell Viability Riaaz Lalani, Nathaniel Susilo, Elisa Xiao, Andrea Xu
Figure S1. gfp tola and pal mcherry can complement deletion mutants of tola and pal respectively. (A)When strain LS4522 was grown in the presence of
 Figure S1. gfp tola and pal mcherry can complement deletion mutants of tola and pal respectively. (A)When strain LS4522 was grown in the presence of xylose, inducing expression of gfp-tola, localization
Figure S1. gfp tola and pal mcherry can complement deletion mutants of tola and pal respectively. (A)When strain LS4522 was grown in the presence of xylose, inducing expression of gfp-tola, localization
Molecular Biology Techniques Supporting IBBE
 Molecular Biology Techniques Supporting IBBE Jared Cartwright Protein Production Lab Head Contact Details: email jared.cartwright@york.ac.uk Phone 01904 328797 Presentation Aims Gene synthesis Cloning
Molecular Biology Techniques Supporting IBBE Jared Cartwright Protein Production Lab Head Contact Details: email jared.cartwright@york.ac.uk Phone 01904 328797 Presentation Aims Gene synthesis Cloning
Cold Fusion Cloning Kit. Cat. #s MC100A-1, MC101A-1. User Manual
 Fusion Cloning technology Cold Fusion Cloning Kit Store the master mixture and positive controls at -20 C Store the competent cells at -80 C. (ver. 120909) A limited-use label license covers this product.
Fusion Cloning technology Cold Fusion Cloning Kit Store the master mixture and positive controls at -20 C Store the competent cells at -80 C. (ver. 120909) A limited-use label license covers this product.
NZYGene Synthesis kit
 Kit components Component Concentration Amount NZYGene Synthesis kit Catalogue number: MB33901, 10 reactions GS DNA Polymerase 1U/ μl 30 μl Reaction Buffer for GS DNA Polymerase 10 150 μl dntp mix 2 mm
Kit components Component Concentration Amount NZYGene Synthesis kit Catalogue number: MB33901, 10 reactions GS DNA Polymerase 1U/ μl 30 μl Reaction Buffer for GS DNA Polymerase 10 150 μl dntp mix 2 mm
Data Sheet Quick PCR Cloning Kit
 Data Sheet Quick PCR Cloning Kit 6044 Cornerstone Ct. West, Ste. E DESCRIPTION: The Quick PCR Cloning Kit is a simple and highly efficient method to insert any gene or DNA fragment into a vector, without
Data Sheet Quick PCR Cloning Kit 6044 Cornerstone Ct. West, Ste. E DESCRIPTION: The Quick PCR Cloning Kit is a simple and highly efficient method to insert any gene or DNA fragment into a vector, without
igem2013 Microbiology BMB SDU
 igem2013 Microbiology BMB SDU Project type: USER cloning Project title: USER cloning of psb1c3, lac promoter and DXS (B. sub) Sub project: Creation date: 13.08.03 Written by: PRA Performed by: PRA 1. SOPs
igem2013 Microbiology BMB SDU Project type: USER cloning Project title: USER cloning of psb1c3, lac promoter and DXS (B. sub) Sub project: Creation date: 13.08.03 Written by: PRA Performed by: PRA 1. SOPs
7.02 Recombinant DNA Methods Spring 2005 Exam Study Questions Answer Key
 MIT Department of Biology 7.02 Experimental Biology & Communication, Spring 2005 7.02/10.702 Spring 2005 RDM Exam Study Questions 7.02 Recombinant DNA Methods Spring 2005 Exam Study Questions Answer Key
MIT Department of Biology 7.02 Experimental Biology & Communication, Spring 2005 7.02/10.702 Spring 2005 RDM Exam Study Questions 7.02 Recombinant DNA Methods Spring 2005 Exam Study Questions Answer Key
Lab Book igem Stockholm Lysostaphin. Week 6
 Lysostaphin Week 6 Summarized below are the experiments conducted this week in chronological order. Click on the experiment name to view it. To go back to this summary, click Summary in the footer. Summary
Lysostaphin Week 6 Summarized below are the experiments conducted this week in chronological order. Click on the experiment name to view it. To go back to this summary, click Summary in the footer. Summary
Materials and methods. by University of Washington Yeast Resource Center) from several promoters, including
 Supporting online material for Elowitz et al. report Materials and methods Strains and plasmids. Plasmids expressing CFP or YFP (wild-type codons, developed by University of Washington Yeast Resource Center)
Supporting online material for Elowitz et al. report Materials and methods Strains and plasmids. Plasmids expressing CFP or YFP (wild-type codons, developed by University of Washington Yeast Resource Center)
Supplemental Information: Table S1, Plasmid and Strain Construction, Supplemental Figures, and Movie Legends
 Supplemental Information: Table S1, Plasmid and Strain Construction, Supplemental Figures, and Movie Legends Supplemental Table Table S1: Strains and plasmids Na me Genotype/description Reference/source
Supplemental Information: Table S1, Plasmid and Strain Construction, Supplemental Figures, and Movie Legends Supplemental Table Table S1: Strains and plasmids Na me Genotype/description Reference/source
igem2013 Microbiology BMB SDU
 igem2013 Microbiology BMB SDU Project type: Biobrick Project title: DXS biobrick from B. subtilis Sub project: Creation date: 18.06.13 Written by: HWJ, PRA & SF Performed by: HWJ, ASF, SIS, MHK, SF & PRA
igem2013 Microbiology BMB SDU Project type: Biobrick Project title: DXS biobrick from B. subtilis Sub project: Creation date: 18.06.13 Written by: HWJ, PRA & SF Performed by: HWJ, ASF, SIS, MHK, SF & PRA
Programmable Sequence-Specific Transcriptional Regulation of Mammalian Genome Using Designer TAL Effectors
 Supplementary Information Programmable Sequence-Specific Transcriptional Regulation of Mammalian Genome Using Designer TAL Effectors Feng Zhang 1,2,3,5,7 *,±, Le Cong 2,3,4 *, Simona Lodato 5,6, Sriram
Supplementary Information Programmable Sequence-Specific Transcriptional Regulation of Mammalian Genome Using Designer TAL Effectors Feng Zhang 1,2,3,5,7 *,±, Le Cong 2,3,4 *, Simona Lodato 5,6, Sriram
Artificial Repressors for Controlling Gene Expression in Bacteria
 Artificial Repressors for Controlling Gene Expression in Bacteria Electronic Supplementary Information Table of Contents Methods...2 SI Table 1: List of Oligonucleotides Used for Plasmid Construction...6
Artificial Repressors for Controlling Gene Expression in Bacteria Electronic Supplementary Information Table of Contents Methods...2 SI Table 1: List of Oligonucleotides Used for Plasmid Construction...6
Supplementary Information
 Single day construction of multi-gene circuits with 3G assembly Andrew D. Halleran 1, Anandh Swaminathan 2, and Richard M. Murray 1, 2 1. Bioengineering, California Institute of Technology, Pasadena, CA.
Single day construction of multi-gene circuits with 3G assembly Andrew D. Halleran 1, Anandh Swaminathan 2, and Richard M. Murray 1, 2 1. Bioengineering, California Institute of Technology, Pasadena, CA.
Construction of a Mutant pbr322 Using Site-Directed Mutagenesis to Investigate the Exclusion Effects of pbr322 During Co-transformation with puc19
 Construction of a Mutant pbr322 Using Site-Directed Mutagenesis to Investigate the Exclusion Effects of pbr322 During Co-transformation with puc19 IVANA KOMLJENOVIC Department of Microbiology and Immunology,
Construction of a Mutant pbr322 Using Site-Directed Mutagenesis to Investigate the Exclusion Effects of pbr322 During Co-transformation with puc19 IVANA KOMLJENOVIC Department of Microbiology and Immunology,
Protocol for cloning SEC-based repair templates using Gibson assembly and ccdb negative selection
 Protocol for cloning SEC-based repair templates using Gibson assembly and ccdb negative selection Written by Dan Dickinson (daniel.dickinson@austin.utexas.edu) and last updated January 2018. A version
Protocol for cloning SEC-based repair templates using Gibson assembly and ccdb negative selection Written by Dan Dickinson (daniel.dickinson@austin.utexas.edu) and last updated January 2018. A version
Supplementary Information
 Supplementary Information RapGene: a fast and accurate strategy for synthetic gene assembly in Escherichia coli Massimiliano Zampini, Pauline Rees Stevens, Justin A. Pachebat, Alison Kingston-Smith, Luis
Supplementary Information RapGene: a fast and accurate strategy for synthetic gene assembly in Escherichia coli Massimiliano Zampini, Pauline Rees Stevens, Justin A. Pachebat, Alison Kingston-Smith, Luis
Human Viperin Causes Radical SAM Dependent Elongation of E. coli Hinting at its Physiological Role
 Supporting Information Human Viperin Causes Radical SAM Dependent Elongation of E. coli Hinting at its Physiological Role Micah T. Nelp, Anthony P. Young, Branden M. Stepanski, Vahe Bandarian* Department
Supporting Information Human Viperin Causes Radical SAM Dependent Elongation of E. coli Hinting at its Physiological Role Micah T. Nelp, Anthony P. Young, Branden M. Stepanski, Vahe Bandarian* Department
Creating pentr vectors by BP reaction
 Creating pentr vectors by BP reaction Tanya Lepikhova and Rafael Martinez 15072011 Overview: 1. Design primers to add the attb sites to gene of interest 2. Perform PCR with a high fidelity DNA polymerase
Creating pentr vectors by BP reaction Tanya Lepikhova and Rafael Martinez 15072011 Overview: 1. Design primers to add the attb sites to gene of interest 2. Perform PCR with a high fidelity DNA polymerase
Journal of Experimental Microbiology and Immunology (JEMI) Vol. 6:20-25 Copyright December 2004, M&I UBC
 Preparing Plasmid Constructs to Investigate the Characteristics of Thiol Reductase and Flavin Reductase With Regard to Solubilizing Insoluble Proteinase Inhibitor 2 in Bacterial Protein Overexpression
Preparing Plasmid Constructs to Investigate the Characteristics of Thiol Reductase and Flavin Reductase With Regard to Solubilizing Insoluble Proteinase Inhibitor 2 in Bacterial Protein Overexpression
Conversion of plasmids into Gateway compatible cloning
 Conversion of plasmids into Gateway compatible cloning Rafael Martinez 14072011 Overview: 1. Select the right Gateway cassette (A, B or C). 2. Design primers to amplify the right Gateway cassette from
Conversion of plasmids into Gateway compatible cloning Rafael Martinez 14072011 Overview: 1. Select the right Gateway cassette (A, B or C). 2. Design primers to amplify the right Gateway cassette from
Supplementary Information: The timing of transcriptional regulation in synthetic gene circuits. Corresponding author.
 Supplementary Information: The timing of transcriptional regulation in synthetic gene circuits Yu-Yu Cheng 1, Andrew J. Hirning 1, Krešimir Josić 1,2,3, and Matthew R. Bennett 1,4, 1 Department of Biosciences,
Supplementary Information: The timing of transcriptional regulation in synthetic gene circuits Yu-Yu Cheng 1, Andrew J. Hirning 1, Krešimir Josić 1,2,3, and Matthew R. Bennett 1,4, 1 Department of Biosciences,
Lab note. Hu Yang. 1. PCR reaction for mbp fragments using dissolved primers. 5x PHF buffer II. ddntp Mixture 2uL. Template plasmid 0.
 Lab note Hu Yang 6.30 1. Dissolve primers: primers for mbp fragments (standardization): SD_A, SD_C, SD_. primers for mbp fragments (commercialization): NdeI_A(), NdeI_C(), XhoI(). 2. Miniprep: plasmids
Lab note Hu Yang 6.30 1. Dissolve primers: primers for mbp fragments (standardization): SD_A, SD_C, SD_. primers for mbp fragments (commercialization): NdeI_A(), NdeI_C(), XhoI(). 2. Miniprep: plasmids
Transcriptional Interference in Convergent Promoters as a Means for Tunable Gene
 Supporting material for: Transcriptional Interference in Convergent Promoters as a Means for Tunable Gene Expression Antoni E. Bordoy, Usha S. Varanasi, Colleen M. Courtney, and Anushree Chatterjee Department
Supporting material for: Transcriptional Interference in Convergent Promoters as a Means for Tunable Gene Expression Antoni E. Bordoy, Usha S. Varanasi, Colleen M. Courtney, and Anushree Chatterjee Department
Molecular cloning: 6 RBS+ arac/ laci
 Molecular cloning: 6 RBS+ arac/ laci Resource: 6 RBS: from the parts: B0030, B0032, B0034, J61100, J61107, J61127; renamed as RBS1 RBS2 RBS6; arac: C0080 laci: C0012 July 6 th Plasmid mini prep: 6 RBS;
Molecular cloning: 6 RBS+ arac/ laci Resource: 6 RBS: from the parts: B0030, B0032, B0034, J61100, J61107, J61127; renamed as RBS1 RBS2 RBS6; arac: C0080 laci: C0012 July 6 th Plasmid mini prep: 6 RBS;
peco TM -T7-nGST, Eco cloning Kit User Manual (Patent pending)
 peco TM -T7-nGST, Eco cloning Kit User Manual (Patent pending) Cloning PCR products for E Coli expression of N-term GST-tagged protein Cat# Contents Amounts Application IC-1004 peco-t7-ngst vector built-in
peco TM -T7-nGST, Eco cloning Kit User Manual (Patent pending) Cloning PCR products for E Coli expression of N-term GST-tagged protein Cat# Contents Amounts Application IC-1004 peco-t7-ngst vector built-in
Polymerase Chain Reaction
 Polymerase Chain Reaction Amplify your insert or verify its presence 3H Taq platinum PCR mix primers Ultrapure Water PCR tubes PCR machine A. Insert amplification For insert amplification, use the Taq
Polymerase Chain Reaction Amplify your insert or verify its presence 3H Taq platinum PCR mix primers Ultrapure Water PCR tubes PCR machine A. Insert amplification For insert amplification, use the Taq
YG1 Control. YG1 PstI XbaI. YG5 PstI XbaI Water Buffer DNA Enzyme1 Enzyme2
 9/9/03 Aim: Digestion and gel extraction of YG, YG3, YG5 and 8/C. Strain: E. coli DH5α Plasmid: Bba_J600, psbc3 4,, 3 6 7, 8 9 0, 3, 4 8/C 8/C SpeI PstI 3 3 x3 YG YG PstI XbaI.5 3.5 x YG3 YG3 PstI XbaI
9/9/03 Aim: Digestion and gel extraction of YG, YG3, YG5 and 8/C. Strain: E. coli DH5α Plasmid: Bba_J600, psbc3 4,, 3 6 7, 8 9 0, 3, 4 8/C 8/C SpeI PstI 3 3 x3 YG YG PstI XbaI.5 3.5 x YG3 YG3 PstI XbaI
TECHNICAL BULLETIN. In Vitro Bacterial Split Fluorescent Protein Fold n Glow Solubility Assay Kits
 In Vitro Bacterial Split Fluorescent Protein Fold n Glow Solubility Assay Kits Catalog Numbers APPA001 In Vitro Bacterial Split GFP "Fold 'n' Glow" Solubility Assay Kit (Green) APPA008 In Vitro Bacterial
In Vitro Bacterial Split Fluorescent Protein Fold n Glow Solubility Assay Kits Catalog Numbers APPA001 In Vitro Bacterial Split GFP "Fold 'n' Glow" Solubility Assay Kit (Green) APPA008 In Vitro Bacterial
Synthetic Biology for
 Synthetic Biology for Plasmids and DNA Digestion Plasmids Plasmids are small DNA molecules that are separate from chromosomal DNA They are most commonly found as double stranded, circular DNA Typical plasmids
Synthetic Biology for Plasmids and DNA Digestion Plasmids Plasmids are small DNA molecules that are separate from chromosomal DNA They are most commonly found as double stranded, circular DNA Typical plasmids
Lecture 25 (11/15/17)
 Lecture 25 (11/15/17) Reading: Ch9; 328-332 Ch25; 990-995, 1005-1012 Problems: Ch9 (study-guide: applying); 1,2 Ch9 (study-guide: facts); 7,8 Ch25 (text); 1-3,5-7,9,10,13-15 Ch25 (study-guide: applying);
Lecture 25 (11/15/17) Reading: Ch9; 328-332 Ch25; 990-995, 1005-1012 Problems: Ch9 (study-guide: applying); 1,2 Ch9 (study-guide: facts); 7,8 Ch25 (text); 1-3,5-7,9,10,13-15 Ch25 (study-guide: applying);
Biosolar Conversion of N2 and H2O to Ammonia by Engineered N2-fixing Cyanobacteria
 The Journal of Undergraduate Research Volume 9 Journal of Undergraduate Research, Volume 9: 2011 Article 16 2011 Biosolar Conversion of N2 and H2O to Ammonia by Engineered N2-fixing Cyanobacteria Seth
The Journal of Undergraduate Research Volume 9 Journal of Undergraduate Research, Volume 9: 2011 Article 16 2011 Biosolar Conversion of N2 and H2O to Ammonia by Engineered N2-fixing Cyanobacteria Seth
Schematic representation of the endogenous PALB2 locus and gene-disruption constructs
 Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Red Type Indicates Unique Site
 3600 G0605 pscaavmcmvmcsbghpa Plasmid Features: Coordinates Feature 980-1084 AAV2 5 ITR 1144-1666 modified CMV 1667-1761 MCS 1762-1975 BgHpA 1987-2114 AAV2 3 ITR 3031-3891 B-lactamase (Ampicillin) Antibiotic
3600 G0605 pscaavmcmvmcsbghpa Plasmid Features: Coordinates Feature 980-1084 AAV2 5 ITR 1144-1666 modified CMV 1667-1761 MCS 1762-1975 BgHpA 1987-2114 AAV2 3 ITR 3031-3891 B-lactamase (Ampicillin) Antibiotic
Reading Lecture 3: 24-25, 45, Lecture 4: 66-71, Lecture 3. Vectors. Definition Properties Types. Transformation
 Lecture 3 Reading Lecture 3: 24-25, 45, 55-66 Lecture 4: 66-71, 75-79 Vectors Definition Properties Types Transformation 56 VECTORS- Definition Vectors are carriers of a DNA fragment of interest Insert
Lecture 3 Reading Lecture 3: 24-25, 45, 55-66 Lecture 4: 66-71, 75-79 Vectors Definition Properties Types Transformation 56 VECTORS- Definition Vectors are carriers of a DNA fragment of interest Insert
Supplementary Methods pcfd5 cloning protocol
 Supplementary Methods cloning protocol vermilion trna grna trna grna U6:3 Terminator AmpR attb is a vector for expressing one or multiple trna-flanked Cas9 grnas under the control of the strong, ubiquitous
Supplementary Methods cloning protocol vermilion trna grna trna grna U6:3 Terminator AmpR attb is a vector for expressing one or multiple trna-flanked Cas9 grnas under the control of the strong, ubiquitous
Plasmid construction SUPPLEMENTAL EXPERIMENTAL PROCEDURES
 SUPPLEMENTAL EXPERIMENTAL PROCEDURES Plasmid construction pjk005 [amye::phy-refz-gfp (spec)(amp)] was generated in a two-way ligation with a PCR product containing refz-gfp (oligonucleotide primers ojw020
SUPPLEMENTAL EXPERIMENTAL PROCEDURES Plasmid construction pjk005 [amye::phy-refz-gfp (spec)(amp)] was generated in a two-way ligation with a PCR product containing refz-gfp (oligonucleotide primers ojw020
Orthogonal Ribosome Biofirewall
 1 2 3 4 5 Supporting Information Orthogonal Ribosome Biofirewall Bin Jia,, Hao Qi,, Bing-Zhi Li,, Shuo pan,, Duo Liu,, Hong Liu,, Yizhi Cai, and Ying-Jin Yuan*, 6 7 8 9 10 11 12 Key Laboratory of Systems
1 2 3 4 5 Supporting Information Orthogonal Ribosome Biofirewall Bin Jia,, Hao Qi,, Bing-Zhi Li,, Shuo pan,, Duo Liu,, Hong Liu,, Yizhi Cai, and Ying-Jin Yuan*, 6 7 8 9 10 11 12 Key Laboratory of Systems
Cytochrome c heme lyase can mature a fusion peptide composed of the amino-terminal residues of horse cytochrome c
 Cytochrome c heme lyase can mature a fusion peptide composed of the amino-terminal residues of horse cytochrome c Wesley B. Asher a and Kara L. Bren a * a Department of Chemistry, University of Rochester,
Cytochrome c heme lyase can mature a fusion peptide composed of the amino-terminal residues of horse cytochrome c Wesley B. Asher a and Kara L. Bren a * a Department of Chemistry, University of Rochester,
Inefficient Tat-Dependent Export of Periplasmic Amidases in an Escherichia coli Strain with Mutations in Two DedA Family Genes
 JOURNAL OF BACTERIOLOGY, Feb. 2010, p. 807 818 Vol. 192, No. 3 0021-9193/10/$12.00 doi:10.1128/jb.00716-09 Copyright 2010, American Society for Microbiology. All Rights Reserved. Inefficient Tat-Dependent
JOURNAL OF BACTERIOLOGY, Feb. 2010, p. 807 818 Vol. 192, No. 3 0021-9193/10/$12.00 doi:10.1128/jb.00716-09 Copyright 2010, American Society for Microbiology. All Rights Reserved. Inefficient Tat-Dependent
5 µl 10X NEB µl ddh 2 O 5 µl StuI (10 units/µl) 2.5 µl XhoI (20 units/µl) *********************************** Incubate at 37 C overnight.
 Flowchart_Cassette Mutagenesis (Steps 0. A-L) 0. Original plasmid pt7-αhl-rl3-d8 In our strategy, for double-mutant E111C-K147C, we will make first K147C by a couple of oligo-pairs shorter than 40 bp (so
Flowchart_Cassette Mutagenesis (Steps 0. A-L) 0. Original plasmid pt7-αhl-rl3-d8 In our strategy, for double-mutant E111C-K147C, we will make first K147C by a couple of oligo-pairs shorter than 40 bp (so
Use of In-Fusion Cloning for Simple and Efficient Assembly of Gene Constructs No restriction enzymes or ligation reactions necessary
 No restriction enzymes or ligation reactions necessary Background The creation of genetic circuits and artificial biological systems typically involves the use of modular genetic components biological
No restriction enzymes or ligation reactions necessary Background The creation of genetic circuits and artificial biological systems typically involves the use of modular genetic components biological
GenBuilder TM Plus Cloning Kit User Manual
 GenBuilder TM Plus Cloning Kit User Manual Cat. No. L00744 Version 11242017 Ⅰ. Introduction... 2 I.1 Product Information... 2 I.2 Kit Contents and Storage... 2 I.3 GenBuilder Cloning Kit Workflow... 2
GenBuilder TM Plus Cloning Kit User Manual Cat. No. L00744 Version 11242017 Ⅰ. Introduction... 2 I.1 Product Information... 2 I.2 Kit Contents and Storage... 2 I.3 GenBuilder Cloning Kit Workflow... 2
Figure S1. Biosynthetic pathway of GDP-PerNAc. Mass spectrum of purified GDP-PerNAc. Nature Protocols: doi: /nprot
 Synthesis of GDP-PerNAc In E. coli O157, the biosynthesis of GDP- -N-acetyl-D-perosamine (GDP-PerNAc) involves three enzymes (Fig. S1). GDP-D-Mannose is converted by GDP-mannose-4,6-dehydratase (GMD) into
Synthesis of GDP-PerNAc In E. coli O157, the biosynthesis of GDP- -N-acetyl-D-perosamine (GDP-PerNAc) involves three enzymes (Fig. S1). GDP-D-Mannose is converted by GDP-mannose-4,6-dehydratase (GMD) into
Tear-Inducing Bacteria. Holly Milbach, Jenna Fox, Neha Yadav, Heather Goodrich
 Tear-Inducing Bacteria Holly Milbach, Jenna Fox, Neha Yadav, Heather Goodrich Goals The goal of our project was to isolate the Alliinase gene from an onion, which is the gene responsible for producing
Tear-Inducing Bacteria Holly Milbach, Jenna Fox, Neha Yadav, Heather Goodrich Goals The goal of our project was to isolate the Alliinase gene from an onion, which is the gene responsible for producing
Supplementary Material. Increased heterocyst frequency by patn disruption in Anabaena leads to enhanced photobiological
 Supplementary Material Increased heterocyst frequency by patn disruption in Anabaena leads to enhanced photobiological hydrogen production at high light intensity and high cell density Applied Microbiology
Supplementary Material Increased heterocyst frequency by patn disruption in Anabaena leads to enhanced photobiological hydrogen production at high light intensity and high cell density Applied Microbiology
An estimate of the physical distance between two linked markers in Haemophilus influenzae
 J. Biosci., Vol. 13, No. 3, September 1988, pp. 223 228. Printed in India. An estimate of the physical distance between two linked markers in Haemophilus influenzae Ε. Β. SAMIWALA, VASUDHA P. JOSHI and
J. Biosci., Vol. 13, No. 3, September 1988, pp. 223 228. Printed in India. An estimate of the physical distance between two linked markers in Haemophilus influenzae Ε. Β. SAMIWALA, VASUDHA P. JOSHI and
Manipulation of Purified DNA
 Manipulation of Purified DNA To produce the recombinant DNA molecule, the vector, as well as the DNA to be cloned, must be cut at specific points and then joined together in a controlled manner by DNA
Manipulation of Purified DNA To produce the recombinant DNA molecule, the vector, as well as the DNA to be cloned, must be cut at specific points and then joined together in a controlled manner by DNA
Highly efficient one-step PCR-based mutagenesis technique for large plasmids using high-fidelity DNA polymerase
 Highly efficient one-step PCR-based mutagenesis technique for large plasmids using high-fidelity DNA polymerase H. Liu, R. Ye and Y.Y. Wang Department of Medical Microbiology and Parasitology, School of
Highly efficient one-step PCR-based mutagenesis technique for large plasmids using high-fidelity DNA polymerase H. Liu, R. Ye and Y.Y. Wang Department of Medical Microbiology and Parasitology, School of
Rotation Report Sample Version 3. Due Date: August 9, Analysis of the Guanine Nucleotide Exchange Activity of the S. cerevisiae Ats1 Protein
 Rotation Report Sample Version 3 Due Date: August 9, 1998 Analysis of the Guanine Nucleotide Exchange Activity of the S. cerevisiae Ats1 Protein Anita H. Corbett Advisor: Amy Jones Rotation 1 Abstract:
Rotation Report Sample Version 3 Due Date: August 9, 1998 Analysis of the Guanine Nucleotide Exchange Activity of the S. cerevisiae Ats1 Protein Anita H. Corbett Advisor: Amy Jones Rotation 1 Abstract:
Ligase Independent Cloning (LIC) using petm-13/lic
 Ligase Independent Cloning (LIC) using petm-13/lic Creating a construct with a C-terminal His 6 -tag Ligase independent cloning (LIC) is a simple, fast and relatively cheap method to produce expression
Ligase Independent Cloning (LIC) using petm-13/lic Creating a construct with a C-terminal His 6 -tag Ligase independent cloning (LIC) is a simple, fast and relatively cheap method to produce expression
Supplementary Figure 1 Performance analysis of RedLibs (version 0.2.3). Supplementary Figure 2
 Supplementary Figure 1 Performance analysis of RedLibs (version 0.2.3). The required calculation time for the evaluation of all sub-libraries with a given target size are plotted against the combinatorial
Supplementary Figure 1 Performance analysis of RedLibs (version 0.2.3). The required calculation time for the evaluation of all sub-libraries with a given target size are plotted against the combinatorial
BS 50 Genetics and Genomics Week of Nov 29
 BS 50 Genetics and Genomics Week of Nov 29 Additional Practice Problems for Section Problem 1. A linear piece of DNA is digested with restriction enzymes EcoRI and HinDIII, and the products are separated
BS 50 Genetics and Genomics Week of Nov 29 Additional Practice Problems for Section Problem 1. A linear piece of DNA is digested with restriction enzymes EcoRI and HinDIII, and the products are separated
igem2013 Microbiology BMB SDU
 igem2013 Microbiology BMB SDU Project type: USER PCR Project title: USER PCR of Lac promoter-dxs (E. coli) without reporter gene Sub project: Creation date: 23.07.13 Written by: SF Performed by: SIS, SF,
igem2013 Microbiology BMB SDU Project type: USER PCR Project title: USER PCR of Lac promoter-dxs (E. coli) without reporter gene Sub project: Creation date: 23.07.13 Written by: SF Performed by: SIS, SF,
Hetero-Stagger PCR Cloning Kit
 Product Name: Code No: Size: DynaExpress Hetero-Stagger PCR Cloning Kit DS150 20 reactions Kit Components: Box 1 (-20 ) phst-1 Vector, linearized Annealing Buffer Ligase Mixture phst Forward Sequence Primer
Product Name: Code No: Size: DynaExpress Hetero-Stagger PCR Cloning Kit DS150 20 reactions Kit Components: Box 1 (-20 ) phst-1 Vector, linearized Annealing Buffer Ligase Mixture phst Forward Sequence Primer
ROLES OF FTSN AND DEDD IN INITIATING E. COLI CELL CONSTRICTION BING LIU
 ROLES OF FTSN AND DEDD IN INITIATING E. COLI CELL CONSTRICTION by BING LIU Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy Molecular Biology and Microbiology
ROLES OF FTSN AND DEDD IN INITIATING E. COLI CELL CONSTRICTION by BING LIU Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy Molecular Biology and Microbiology
7.1 Techniques for Producing and Analyzing DNA. SBI4U Ms. Ho-Lau
 7.1 Techniques for Producing and Analyzing DNA SBI4U Ms. Ho-Lau What is Biotechnology? From Merriam-Webster: the manipulation of living organisms or their components to produce useful usually commercial
7.1 Techniques for Producing and Analyzing DNA SBI4U Ms. Ho-Lau What is Biotechnology? From Merriam-Webster: the manipulation of living organisms or their components to produce useful usually commercial
DNA Technology. Asilomar Singer, Zinder, Brenner, Berg
 DNA Technology Asilomar 1973. Singer, Zinder, Brenner, Berg DNA Technology The following are some of the most important molecular methods we will be using in this course. They will be used, among other
DNA Technology Asilomar 1973. Singer, Zinder, Brenner, Berg DNA Technology The following are some of the most important molecular methods we will be using in this course. They will be used, among other
Mutating Asn-666 to Glu in the O-helix region of the taq DNA polymerase gene
 Research in Pharmaceutical Sciences, April 2010; 5(1): 15-19 Received: Oct 2009 Accepted: Jan 2010 School of Pharmacy & Pharmaceutical Sciences 15 Isfahan University of Medical Sciences Original Article
Research in Pharmaceutical Sciences, April 2010; 5(1): 15-19 Received: Oct 2009 Accepted: Jan 2010 School of Pharmacy & Pharmaceutical Sciences 15 Isfahan University of Medical Sciences Original Article
Lab Notes: igem 2010
 Lab Notes: igem 2010 By Zairan Liu 7.5 Purification of digested PCR product: merp, mert, and merc Insert (merp / mert / merc digested with EcoRI and PstI): 7μL Vector (psb1a2 backbone digested with EcoRI
Lab Notes: igem 2010 By Zairan Liu 7.5 Purification of digested PCR product: merp, mert, and merc Insert (merp / mert / merc digested with EcoRI and PstI): 7μL Vector (psb1a2 backbone digested with EcoRI
SUPPLEMENTARY DATA: Supplementary Figures:
 SUPPLEMENTARY DATA: Supplementary Figures: Supplementary Fig. S1: In vivo recombinant Mtb reporter assay: Dependence of mcherry expression as a function of E. coli growth: A. Fluorescent intensity of mcherry
SUPPLEMENTARY DATA: Supplementary Figures: Supplementary Fig. S1: In vivo recombinant Mtb reporter assay: Dependence of mcherry expression as a function of E. coli growth: A. Fluorescent intensity of mcherry
pgm-t Cloning Kit Cat. # : GVT202 Size : 20 Reactions Store at -20
 pgm-t Cloning Kit Cat. # : GVT202 Size : 20 Reactions Store at -20 1 Kit Contents Contents pgm-t Cloning Kit pgm-t Vector (50 ng/μl) 20 μl T4 DNA Ligase (3 U/μl) 20 μl 10X T4 DNA Ligation Buffer 30 μl
pgm-t Cloning Kit Cat. # : GVT202 Size : 20 Reactions Store at -20 1 Kit Contents Contents pgm-t Cloning Kit pgm-t Vector (50 ng/μl) 20 μl T4 DNA Ligase (3 U/μl) 20 μl 10X T4 DNA Ligation Buffer 30 μl
SUPPLEMENTARY INFORMATION
 Gene replacements and insertions in rice by intron targeting using CRISPR Cas9 Table of Contents Supplementary Figure 1. sgrna-induced targeted mutations in the OsEPSPS gene in rice protoplasts. Supplementary
Gene replacements and insertions in rice by intron targeting using CRISPR Cas9 Table of Contents Supplementary Figure 1. sgrna-induced targeted mutations in the OsEPSPS gene in rice protoplasts. Supplementary
Cat # Box1 Box2. DH5a Competent E. coli cells CCK-20 (20 rxns) 40 µl 40 µl 50 µl x 20 tubes. Choo-Choo Cloning TM Enzyme Mix
 Molecular Cloning Laboratories User Manual Version 3.3 Product name: Choo-Choo Cloning Kits Cat #: CCK-10, CCK-20, CCK-096, CCK-384 Description: Choo-Choo Cloning is a highly efficient directional PCR
Molecular Cloning Laboratories User Manual Version 3.3 Product name: Choo-Choo Cloning Kits Cat #: CCK-10, CCK-20, CCK-096, CCK-384 Description: Choo-Choo Cloning is a highly efficient directional PCR
BBF RFC 98: GenBrick A Rapid Multi-Part Assembly Method for BioBricks
 BBF RFC 98 GenBrick Assembly BBF RFC 98: GenBrick A Rapid Multi-Part Assembly Method for BioBricks Lina Gasiūnaitė, Aleksandra Lewicka, Hristiana Pashkuleva, Hugo Villanueva, Harry Thornton, Maryia Trubitsyna,
BBF RFC 98 GenBrick Assembly BBF RFC 98: GenBrick A Rapid Multi-Part Assembly Method for BioBricks Lina Gasiūnaitė, Aleksandra Lewicka, Hristiana Pashkuleva, Hugo Villanueva, Harry Thornton, Maryia Trubitsyna,
Microbiology 微生物学 Spring-Summer
 Microbiology 微生物学 2017 Spring-Summer Relevant Information and Resources Course slides can be found at http://mypage.zju.edu.cn/haichun 教学工作 Course-related questions will be answered through emails. Textbook:
Microbiology 微生物学 2017 Spring-Summer Relevant Information and Resources Course slides can be found at http://mypage.zju.edu.cn/haichun 教学工作 Course-related questions will be answered through emails. Textbook:
Supporting Information
 Supporting Information Massie et al. 10.1073/pnas.1115663109 SI Materials and Methods Construction of DGC Overexpression Plasmids. The overexpression plasmids for each V. cholerae DGC were constructed
Supporting Information Massie et al. 10.1073/pnas.1115663109 SI Materials and Methods Construction of DGC Overexpression Plasmids. The overexpression plasmids for each V. cholerae DGC were constructed
Site-directed mutagenesis of proteins
 IFM/Kemi Linköpings Universitet August 2013/LGM Labmanual Site-directed mutagenesis of proteins Figur 1: Flow-chart of the site-directed mutagenesis lab exercise 2 Site-specific mutagenesis Introduction
IFM/Kemi Linköpings Universitet August 2013/LGM Labmanual Site-directed mutagenesis of proteins Figur 1: Flow-chart of the site-directed mutagenesis lab exercise 2 Site-specific mutagenesis Introduction
Discovery and Characterization of Three New Escherichia coli Septal Ring Proteins That Contain a SPOR Domain: DamX, DedD, and RlpA
 JOURNAL OF BACTERIOLOGY, Jan. 2010, p. 242 255 Vol. 192, No. 1 0021-9193/10/$12.00 doi:10.1128/jb.01244-09 Copyright 2010, American Society for Microbiology. All Rights Reserved. Discovery and Characterization
JOURNAL OF BACTERIOLOGY, Jan. 2010, p. 242 255 Vol. 192, No. 1 0021-9193/10/$12.00 doi:10.1128/jb.01244-09 Copyright 2010, American Society for Microbiology. All Rights Reserved. Discovery and Characterization
Supplementary Information
 Supplementary Information Experimental procedures Identifying the viable circular permutation sites of E. coli EPSPS Bioinformatics prediction and E. coli complementation assay were performed together
Supplementary Information Experimental procedures Identifying the viable circular permutation sites of E. coli EPSPS Bioinformatics prediction and E. coli complementation assay were performed together
TABLE S1. Sequences of the primers used for RT-PCR and PCR
 TABLE S1. Sequences of the primers used for RT-PCR and PCR Primer Sequence (5 to 3 ) Purpose Cloning: posuprtid-1 & posuprtid-2 deletion plasmids DM01 AGCTCCAGCGGGTCGCCGGGGTTGGCC DM02 CCAGCGGGTCGCCGGGGTTGGCC
TABLE S1. Sequences of the primers used for RT-PCR and PCR Primer Sequence (5 to 3 ) Purpose Cloning: posuprtid-1 & posuprtid-2 deletion plasmids DM01 AGCTCCAGCGGGTCGCCGGGGTTGGCC DM02 CCAGCGGGTCGCCGGGGTTGGCC
GenBuilder TM Plus Cloning Kit User Manual
 GenBuilder TM Plus Cloning Kit User Manual Cat.no L00744 Version 11242017 Ⅰ. Introduction... 2 I.1 Product Information... 2 I.2 Kit Contents and Storage... 2 I.3 GenBuilder Cloning Kit Workflow... 2 Ⅱ.
GenBuilder TM Plus Cloning Kit User Manual Cat.no L00744 Version 11242017 Ⅰ. Introduction... 2 I.1 Product Information... 2 I.2 Kit Contents and Storage... 2 I.3 GenBuilder Cloning Kit Workflow... 2 Ⅱ.
Cat. # Product Size DS130 DynaExpress TA PCR Cloning Kit (ptakn-2) 20 reactions Box 1 (-20 ) ptakn-2 Vector, linearized 20 µl (50 ng/µl) 1
 Product Name: Kit Component TA PCR Cloning Kit (ptakn-2) Cat. # Product Size DS130 TA PCR Cloning Kit (ptakn-2) 20 reactions Box 1 (-20 ) ptakn-2 Vector, linearized 20 µl (50 ng/µl) 1 2 Ligation Buffer
Product Name: Kit Component TA PCR Cloning Kit (ptakn-2) Cat. # Product Size DS130 TA PCR Cloning Kit (ptakn-2) 20 reactions Box 1 (-20 ) ptakn-2 Vector, linearized 20 µl (50 ng/µl) 1 2 Ligation Buffer
HE Swift Cloning Kit
 HE Swift Cloning Kit For high-efficient cloning of PCR products either blunt or sticky-end Kit Contents Contents VTT-BB05 phe Vector (35 ng/µl) 20 µl T4 DNA Ligase (3 U/µl) 20 µl 2 Reaction Buffer 100
HE Swift Cloning Kit For high-efficient cloning of PCR products either blunt or sticky-end Kit Contents Contents VTT-BB05 phe Vector (35 ng/µl) 20 µl T4 DNA Ligase (3 U/µl) 20 µl 2 Reaction Buffer 100
Δsig. ywa. yjbm. rela -re. rela
 A wt A. A, P rela -re la A/Δ yjbm A/Δ ywa C A, P hy-s igd, D (A, P IPTG hy-s igd, ) D D (+ IPTG ) D/Δ rela Figure S1 SigD B. Figure S1. SigD levels and swimming motility in (p)ppgpp synthetase mutants.
A wt A. A, P rela -re la A/Δ yjbm A/Δ ywa C A, P hy-s igd, D (A, P IPTG hy-s igd, ) D D (+ IPTG ) D/Δ rela Figure S1 SigD B. Figure S1. SigD levels and swimming motility in (p)ppgpp synthetase mutants.
igem2013 Microbiology BMB SDU
 igem2013 Microbiology BMB SDU Project type: Conventional cloning of IspG together with LacI-PLac-dxs(B.sub) in psb1c3. Project title: LacI-PLac-dxs(B.sub) in psb1c3 Sub project: Creation date: 26.08.2013
igem2013 Microbiology BMB SDU Project type: Conventional cloning of IspG together with LacI-PLac-dxs(B.sub) in psb1c3. Project title: LacI-PLac-dxs(B.sub) in psb1c3 Sub project: Creation date: 26.08.2013
Supporting Information-Tables
 Supporting Information-Tables Table S1. Bacterial strains and plasmids used in this work Bacterial strains Description Source of reference Streptococcus pneumoniae 1 Cp1015 non-capsulated and βl susceptible
Supporting Information-Tables Table S1. Bacterial strains and plasmids used in this work Bacterial strains Description Source of reference Streptococcus pneumoniae 1 Cp1015 non-capsulated and βl susceptible
XXII DNA cloning and sequencing. Outline
 XXII DNA cloning and sequencing 1) Deriving DNA for cloning Outline 2) Vectors; forming recombinant DNA; cloning DNA; and screening for clones containing recombinant DNA [replica plating and autoradiography;
XXII DNA cloning and sequencing 1) Deriving DNA for cloning Outline 2) Vectors; forming recombinant DNA; cloning DNA; and screening for clones containing recombinant DNA [replica plating and autoradiography;
Different Upstream Activators Stimulate Transcription Through Distinct Coactivator Complexes
 Supplemental Material for Different Upstream Activators Stimulate Transcription Through Distinct Coactivator Complexes Dong-ki Lee, Soyoun Kim, and John T. Lis Due to be published as a Research Communication
Supplemental Material for Different Upstream Activators Stimulate Transcription Through Distinct Coactivator Complexes Dong-ki Lee, Soyoun Kim, and John T. Lis Due to be published as a Research Communication
To clone eif4a fragments into SalI-NotI sites of pacycduet-1 (pmb09); and petduet-1
 Supplementary Methods DNA constructs and strains To clone eif4a fragments into SalI-NotI sites of pacycduet-1 (pmb09); and petduet-1 (Novagen) (pmb11), we PCR-amplified each domain from the mouse eif4a
Supplementary Methods DNA constructs and strains To clone eif4a fragments into SalI-NotI sites of pacycduet-1 (pmb09); and petduet-1 (Novagen) (pmb11), we PCR-amplified each domain from the mouse eif4a
0730 Measure the concentrations of Ligation products of reverse laci + promoter
 0730 Measure the concentrations of Ligation products of reverse laci + promoter 1 1.6536 2 2.4225 3 2.7404 4 2.8690 5 2.4858 6 2.5618 Enzyme digestion to test the ligation product Plasmid 8µl EcoRI 1.5
0730 Measure the concentrations of Ligation products of reverse laci + promoter 1 1.6536 2 2.4225 3 2.7404 4 2.8690 5 2.4858 6 2.5618 Enzyme digestion to test the ligation product Plasmid 8µl EcoRI 1.5
Supporting Information
 Supporting Information Risser and Callahan 10.1073/pnas.0909152106 SI Text Plasmid Construction. Plasmid pdr211 is a mobilizable shuttle vector containing P pete -pats. A fragment containing P pete was
Supporting Information Risser and Callahan 10.1073/pnas.0909152106 SI Text Plasmid Construction. Plasmid pdr211 is a mobilizable shuttle vector containing P pete -pats. A fragment containing P pete was
Classic cloning with pask-iba and pexpr-iba vectors
 Classic cloning with pask-iba and pexpr-iba vectors General protocol Last date of revision March 2017 Version PR08-0008 For research use only Important licensing information Products featuring CMV promoter,
Classic cloning with pask-iba and pexpr-iba vectors General protocol Last date of revision March 2017 Version PR08-0008 For research use only Important licensing information Products featuring CMV promoter,
Amgen Laboratory Series. Tabs C and E
 Amgen Laboratory Series Tabs C and E Chapter 2A Goals Describe the characteristics of plasmids Explain how plasmids are used in cloning a gene Describe the function of restriction enzymes Explain how to
Amgen Laboratory Series Tabs C and E Chapter 2A Goals Describe the characteristics of plasmids Explain how plasmids are used in cloning a gene Describe the function of restriction enzymes Explain how to
Team: The Students. Maria K. Andersen. Shiva Moghaddam. Ingrid Fadum Kjønstad. Patricia Adl. Ellen Stormo
 Team: The Students Maria K. Andersen Shiva Moghaddam Ellen Stormo Patricia Adl Ingrid Fadum Kjønstad Biotechnology/ Molecular medicine Master student Molecular medicine Master student Biotechnology Master
Team: The Students Maria K. Andersen Shiva Moghaddam Ellen Stormo Patricia Adl Ingrid Fadum Kjønstad Biotechnology/ Molecular medicine Master student Molecular medicine Master student Biotechnology Master
Biol/Chem 475 Spring 2007
 Biol/Chem 475 Spring 2007 Goal of lab: For most of the quarter, we will be exploring a gene family that was first discovered in fruitlfies and then found to be present in humans and worms and fish and
Biol/Chem 475 Spring 2007 Goal of lab: For most of the quarter, we will be exploring a gene family that was first discovered in fruitlfies and then found to be present in humans and worms and fish and
