supplementary information
|
|
|
- Tracy Vincent Byrd
- 5 years ago
- Views:
Transcription
1 DOI: /ncb2007 Figure S1 Temperature sensitivity of DNA ligase I alleles. a, SSL204 (CDC9), SSL612 (cdc9-1) and SSL613 (cdc9-2) cells were spotted in ten-fold serial dilutions on YPD plates and incubated at the indicated temperatures for 2 days. b, Asynchronous cultures of SSL204 (CDC9), SSL612 (cdc9-1) and SSL613 (cdc9-2) were grown at 25 C and shifted to the restrictive temperature of 35 C for 90 or 180 min. Total protein was TCA precipitated 1. Cdc9 was detected with an anti-cdc9 antibody. Ponceau S staining of the membrane prior to blotting served as a loading control. The asterisk denotes a degradation product. 1
2 Figure S2 cdc9-td cells lack DNA ligase I at elevated temperatures. a, ABy021 (UBR1 CDC9), ABy010 (GAL-UBR1 CDC9), and ABy008 (GAL- UBR1 cdc9-td) cells were streaked onto complete medium containing either 2% glucose (Glu.) or 2% galactose (Gal.) as the sole carbon source. The plates were incubated for two days at 28 C or 37 C as noted. b, Asynchronous cultures of ABy010 (GAL-UBR1 CDC9) and ABy008 (GAL- UBR1 cdc9-td) were grown overnight at 28 C in YP plus raffinose medium supplemented with 10 µm CuSO 4. At OD 600 = 0.6, the culture was split, centrifuged and resuspended in either YP (Glu.) or YP plus galactose (Gal.) and incubated at 37 C. Samples were collected at 1, 2 or 3 hours. Cdc9- td-ha and histone H3 were detected by immunoblotting with anti-ha (16B12, Covance) and anti-histone H3 (Abcam) antibodies, respectively. Ponceau S staining served as an additional loading control. c, DNA was isolated from ABy317 cells (GAL-UBR1 cdc9-td) grown asynchronously at 28 C. In parallel, cells were arrested in G1 with alpha-factor. Following arrest, cultures were released and shifted to 37 C in the presence of galactose. RIP mapping was performed at the replication origin ARS1 using the primer ARS1/630 as described previously 2. The transition point (TP) maps between elements B1 and B2. Okazaki fragment initiation sites visible at 28 C but not at 37 C (in the absence of DNA ligase I) are indicated by O1, O2 and O3. Sequencing reactions were fractionated adjacent to the primer extension reactions on a 6% denaturing polyacrylamide gel. 2
3 Figure S3 Deletion of MRC1 or RAD9 abrogates the S phase arrest in cdc9 mutants. a, Asynchronous cultures of SSL204 (CDC9), SSL612 (cdc9-1), ABy296 (CDC9 rad9 ), ABy297 (cdc9-1 rad9 ), ABy287 (CDC9 mrc1 ) and ABy293 (cdc9-1 mrc1 ) were grown at 25 C and shifted to the restrictive temperature of 35 C for 1.5, 3 or 4.5 h. DNA was stained with Sytox Green and monitored using flow cytometry. b, Asynchronous cultures of ABy352 (CDC9 prnr1), ABy353 (cdc9-1 prnr1), ABy356 (CDC9 mrc1 rad9 prnr1) and ABy357 (cdc9-1 mrc1 rad9 prnr1) were grown at 25 C and shifted to the restrictive temperature of 35 C for the indicated times. DNA was stained with Sytox Green and monitored using flow cytometry. c, Asynchronous cultures of ABy343 (CDC9 mrc1 +vector), ABy344 (CDC9 mrc1 +mrc1 AQ ), ABy345 (CDC9 mrc1 +MRC1), ABy346 (cdc9-1 mrc1 + vector), ABy347 (cdc9-1 mrc1 +mrc1 AQ ), and ABy348 (cdc9-1 mrc1 +MRC1) were grown at 25 C and shifted to the restrictive temperature of 35 C for 3 h. DNA was stained with Sytox Green and monitored using flow cytometry. In a-c, the vertical line indicates a 2C DNA content. 3
4 Figure S4 Detection of robust PCNA signal in cdc9 mutants using a yeast specific PCNA antibody. Asynchronous cultures were grown at 25 C and shifted to 35 C for 3 h. Total protein was TCA-precipitated and PCNA was detected using a yeast specific PCNA antibody (S871) 3. α-tubulin served as a loading control. 4
5 Figure S5 PCNA ubiquitination in DNA ligase I mutants is dependent on Mms2, Rad5 and Ubc4 but not Rad6. a, SSL204 (CDC9), SSL612 (cdc9-1), ABy375 (cdc9-1 mms2δ), ABy388 (cdc9-1 ubc13δ), ABy410 (cdc9-1 rad5δ) were grown asynchronously and shifted to non-permissive temperature of 35 C for 3 h. Total protein was TCA-precipitated and undiluted samples run on SDS-PAGE. Poly-ubiquitinated PCNA was detected with anti-pcna antibody (S871) 3. The asterisks indicate nonspecific bands. Ponceau S staining served as a loading control. b, Asynchronous cultures of SSL612 (cdc9-1) and ABy520 (cdc9-1 rad6δ) were grown overnight at 25 C to a density of 0.6 at 600 nm. Cultures were shifted to the non-permissive temperature of 35 C for 3 h. c, Asynchronous cultures of SSL204 (CDC9), SSL612 (cdc9-1), ABy538 (cdc9-1 ubc4δ), and ABy539 (cdc9-1 ubc5δ) were shifted to 35 C for 3 h. In b-c, total protein was TCA-precipitated, diluted and PCNA was detected by anti-pcna antibody (S871) 3. In b-c, α-tubulin served as a loading control. 5
6 Figure S6 Mono-ubiquitination of PCNA in cdc9 mutants is sufficient for Rad53 activation. a, Asynchronous cultures of ABy350 (cdc9-1 Cu- Myc-Ub) and ABy485 (cdc9-1 Cu-Myc-Ub-K29R) were grown overnight at 25 C to a density of 0.6 at 600 nm. Cells were shifted to 35 C for 3 h in the presence of copper sulfate to induce expression of Myc-tagged ubiquitin. Samples were taken at the indicated times and total protein was TCA-precipitated.Rad53 was detected by Western blotting using an anti- Rad53 antibody. Ponceau S served as a loading control. b, Asynchronous cultures of ABy349 (CDC9 Cu-Myc-Ub), ABy350 (cdc9-1 Cu-Myc-Ub) and ABy485 (cdc9-1 Cu-Myc-Ub-K29R) were grown as described in a. Total protein was TCA-precipitated. Poly-ubiquitinated PCNA was detected by Western blot with a yeast specific anti-pcna antibody (S871) 3. The asterisks indicate nonspecific bands. Ponceau S staining served as a loading control. 6
7 Figure S7 A second site suppressor in ABy459 (cdc9-1 pol30 ppol30- K107R) results in elevated DNA ligase I levels and loss of temperature sensitivity. a, Asynchronous cultures of SSL204 (CDC9), SSL612 (cdc9-1), ABy459 (cdc9-1 pol30 ppol30-k107r) were shifted to 35 C for 90 and 180 min. DNA ligase I was detected in TCA-precipitated protein with an anti- Cdc9 antibody and α-tubulin served as a loading control. The asterisk denotes a degradation product. b, Cells were spotted in ten-fold serial dilutions on SCtrp plates and incubated at the indicated temperatures for 2 days. 7
8 Figure S8 A PCNA mutation at K107 in DNA ligase I deficient cells inhibits cell proliferation. Wild-type (CDC9) and DNA ligase I deficient cells (cdc9-1) either with or without pol30-k107r integrated at the endogenous PCNA locus, all expressing POL30 from a low copy number plasmid, were grown on SC-ura and FOA plates for 2-3 days at 25 C. Wild-type (CDC9) and DNA ligase I deficient cells (cdc9-1) carrying a URA3 disruption of the BAR1 gene served as controls. 8
9 Figure S9 cdc9-1* pol30-k107r has slightly elevated DNA ligase I levels but still retains its temperature sensitivity. a, Cultures of SSL204 (CDC9), SSL612 (cdc9-1) and ABy782 (cdc9-1* pol30-k107r) were grown asynchronously at 25 C and total protein was TCA-precipitated. DNA ligase I was detected using anti-cdc9 antibody and α-tubulin served as a loading control. The asterisk denotes a degradation product. b, Cultures of SSL204 (CDC9), ABy685 (CDC9 pol30-k107r), SSL612 (cdc9-1), ABy782 (cdc9-1* pol30-k107r) were spotted in ten-fold serial dilutions on YPD plates and incubated at the indicated temperatures for 2-3 days. 9
10 Figure S10 Evolutionary conservation of lysines 107 and 110 in selected eukaryotes. Lysine 107 in PCNA is conserved in S. pombe and C. elegans but higher eukaryotes have a conserved lysine at position
11 Figure S11 Generation of LIG1 shrna stable cell lines. U20S cells stably expressing either control or DNA ligase I shrna were harvested and subjected to Western blot analysis with the indicated antibodies. DNA ligase I expression was determined in whole cell extracts, whereas PCNA expression was examined in chromatin-bound protein fractions. α-tubulin and histone H3 served as loading controls. 11
12 Figure S12 PCNA ubiquitination does not occur in rad27 and dna2-1 mutants. Asynchronous cultures were grown at 25 C and shifted to the restrictive temperature of 35 C for 3 h. Total protein was TCA-precipitated and PCNA was detected by a yeast specific anti-pcna antibody (S871) 3. α-tubulin served as a loading control. 12
13 Figure S13 Original Western blots for figures 2 to 5. The original, uncropped Western blots for figures 2a-d, 3a-b, 4b-c and 5a-g are shown. Molecular weight markers are indicated on the side. 13
14 Figure S13 continued 14
15 Supplementary Table 1. List of Yeast Strains Used in. Strain Name Relevant Genotype Source W303-derived strains ABy021 bar1::leu2 1 YKL83 GAL-UBR1 (HIS3) 4 ABy778 GAL-UBR1 (HIS3) pol30::leu2 [YCplac22-POL30-K107R] ABy010 GAL-UBR1 (HIS3) bar1::leu2 ABy317 GAL-UBR1 (HIS3) cdc9::cdc9-td (URA3) ABy780 GAL-UBR1 (HIS3) cdc9::cdc9-td (URA3) pol30::leu2 [YCplac22-POL30-K107R] ABy008 GAL-UBR1 (HIS3) bar1::leu2 cdc9::cdc9-td (URA3) SSL204-derived strains SSL204 MATa ade2 his3d200 trp1 leu2 ura3-52 ABy287 mrc1::trp1 ABy296 rad9::trp1 ABy343 mrc1::trp1 [prs316] ABy344 mrc1::trp1 [pao138] ABy345 mrc1::trp1 [pmrc1] ABy349 [YEp105] ABy352 [prnr1] ABy356 mrc1::trp1 rad9::ura3 [prnr1] ABy458 bar1::ura3 ABy517 pol30::ura3 [YCplac22-POL30] 5
16 ABy669 [prs316-pol30] ABy685 pol30::pol30-k107r (LEU2) ABy704 pol30::pol30-k107r (LEU2) [prs316-pol30] ABy366 dna2-1 [YEp105] ABy365 rad27δ [YEp105] SSL612 cdc9-1 ABy293 cdc9-1 mrc1::trp1 ABy297 cdc9-1 rad9::trp1 ABy346 cdc9-1 mrc1::trp1 [prs316] ABy347 cdc9-1 mrc1::trp1 [pao138] ABy348 cdc9-1 mrc1::trp1 [pmrc1] ABy350 cdc9-1 [YEp105] ABy353 cdc9-1 [prnr1] ABy357 cdc9-1 mrc1::trp1 rad9::ura3 [prnr1] ABy375 cdc9-1 mms2::ura3 ABy388 cdc9-1 ubc13::ura3 ABy392 cdc9-1 rad18::ura3 ABy410 cdc9-1 rad5::ura3 ABy520 cdc9-1 rad6::ura3 ABy538 cdc9-1 ubc4::ura3 ABy539 cdc9-1 ubc5::ura3 ABy579 cdc9-1 ubc5::ura3 ubc4::ubc4-nes-3ha (TRP1) ABy439 cdc9-1 pol30::ura3 [YCplac22-POL30] 6
17 ABy440 cdc9-1 pol30::ura3 [YCplac22-POL30-K164R] ABy441 cdc9-1 pol30::ura3 [YCplac22-POL30-K127R/164R] ABy442 cdc9-1 pol30::ura3 [YCplac22-POL30-K242R] ABy459 cdc9-1 pol30::ura3 [YCplac22-POL30-K107R] ABy461 cdc9-1 pol30::ura3 [YCplac22-POL30-K108R] ABy463 cdc9-1 pol30::ura3 [YCplac22-POL30-K183R] ABy465 cdc9-1 POL30::URA3 [YCplac22-POL30-K253R] ABy467 cdc9-1 POL30::URA3 [YCplac22-POL30-K117R] ABy469 cdc9-1 POL30::URA3 [YCplac22-POL30-K168R] ABy471 cdc9-1 POL30::URA3 [YCplac22-POL30-K127R] ABy479 cdc9-1 [YEp105-Ub-K48,63R] ABy485 cdc9-1 [YEp105-Ub-K29R] ABy489 cdc9-1 [YEp105-Ub-KO] ABy497 cdc9-1 [YEp105-Ub-G75,76A] ABy453 cdc9-1 bar1::ura3 ABy712 cdc9-1 [prs316-pol30] ABy777 cdc9-1 pol30::pol30-k107r (LEU2) [prs316-pol30] Aby782 cdc9-1* pol30::pol30-k107r from tetrad dissection SSL613 cdc9-2 ABy351 cdc9-2 [YEp105] 6
18 References 1. Ricke, R.M. & Bielinsky, A.K. A conserved Hsp10-like domain in Mcm10 is required to stabilize the catalytic subunit of DNA polymerase-alpha in budding yeast. J. Biol. Chem. 281, (2006). 2. Bielinsky, A.K. & Gerbi, S.A. Chromosomal ARS1 has a single leading strand start site. Mol. Cell 3, (1999). 3. Zhang, Z., Shibahara, K. & Stillman, B. PCNA connects DNA replication to epigenetic inheritance in yeast. Nature 408, (2000). 4. Labib, K., Tercero, J.A. & Diffley, J.F. Uninterrupted MCM2-7 function required for DNA replication fork progression. Science 288, (2000). 5. Dornfeld, K.J. & Livingston, D.M. Effects of controlled RAD52 expression on repair and recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 11, (1991). 6. Ireland, M.J., Reinke, S.S. & Livingston, D.M. The impact of lagging strand replication mutations on the stability of CAG repeat tracts in yeast. Genetics 155, (2000).
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Detection of MCM-subunit SUMOylation under normal growth conditions. a. Sumoylated forms of MCM subunits show differential shifts when SUMO is attached to differently sized tags.
Supplementary Figure 1 Detection of MCM-subunit SUMOylation under normal growth conditions. a. Sumoylated forms of MCM subunits show differential shifts when SUMO is attached to differently sized tags.
SUPPLEMENTARY INFORMATION
 Supplementary Table 1 Components of the RAD6 damage tolerance pathway in yeast and humans S. cerevisiae Homo sapiens Functions Rad6 HHR6A and HHR6B Ubiquitin conjugating enzyme (E2) Rad18 RAD18 Ubiquitin
Supplementary Table 1 Components of the RAD6 damage tolerance pathway in yeast and humans S. cerevisiae Homo sapiens Functions Rad6 HHR6A and HHR6B Ubiquitin conjugating enzyme (E2) Rad18 RAD18 Ubiquitin
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11355 Supplemental Table 1. Gene targeting efficiency in mutants of nonessential genes. This table is presented as a separate Excel file. Supplemental Table 2. Genes with altered expression
doi:10.1038/nature11355 Supplemental Table 1. Gene targeting efficiency in mutants of nonessential genes. This table is presented as a separate Excel file. Supplemental Table 2. Genes with altered expression
Supplemental Materials and Methods. Yeast strains. Strain genotypes are listed in Table S1. To generate MATainc::LEU2,
 Supplemental Materials and Methods Yeast strains. Strain genotypes are listed in Table S1. To generate MATainc::LEU2, the LEU2 marker was inserted by PCR-mediated recombination (BRACHMANN et al. 1998)
Supplemental Materials and Methods Yeast strains. Strain genotypes are listed in Table S1. To generate MATainc::LEU2, the LEU2 marker was inserted by PCR-mediated recombination (BRACHMANN et al. 1998)
Yeast strains used in this study Strain genotype source y2007 MATa sld3-600,609,622a-dpb11( )::kanmx, his3::pgal1-
 Supplementary Table Yeast strains used in this study Strain genotype source y2007 MATa sld3-600,609,622a-dpb11(253-764)::kanmx, his3::pgal1-1 4HA-sld2 T84D::HIS3, bar1δ::trp1 YB291 MATa cdc7-4 leu2-3,
Supplementary Table Yeast strains used in this study Strain genotype source y2007 MATa sld3-600,609,622a-dpb11(253-764)::kanmx, his3::pgal1-1 4HA-sld2 T84D::HIS3, bar1δ::trp1 YB291 MATa cdc7-4 leu2-3,
supplementary information
 supplementary information Figure S1 (a) Alignment of the Sgf11 amino acid sequences from S. cerevisiae, Yarrowia lipolytica and S. pombe with Sgf73 sequences from S. cerevisiae, S. pombe and Candida albicans.
supplementary information Figure S1 (a) Alignment of the Sgf11 amino acid sequences from S. cerevisiae, Yarrowia lipolytica and S. pombe with Sgf73 sequences from S. cerevisiae, S. pombe and Candida albicans.
Supporting Information
 Supporting Information Chen et al. 1.173/pnas.153331112 Fig. S1. The sae2δ-suppressing mre11 alleles: protein level and location. (A) Protein level of Mre11 and Mre11 P11L expressed from the endogenous
Supporting Information Chen et al. 1.173/pnas.153331112 Fig. S1. The sae2δ-suppressing mre11 alleles: protein level and location. (A) Protein level of Mre11 and Mre11 P11L expressed from the endogenous
SUPPLEMENTARY INFORMATION
 doi: 1.138/nature7652 Supplementary Figure legends Figure 1. Diagrammatic representation of the rdna locus on yeast chromosome XII and the processing pathway from 35S full length nascent rrna to mature
doi: 1.138/nature7652 Supplementary Figure legends Figure 1. Diagrammatic representation of the rdna locus on yeast chromosome XII and the processing pathway from 35S full length nascent rrna to mature
Coleman et al., Supplementary Figure 1
 Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Figure S1, related to Figure 1. Characterization of biosensor behavior in vivo.
 Developmental Cell, Volume 23 Supplemental Information Separase Biosensor Reveals that Cohesin Cleavage Timing Depends on Phosphatase PP2A Cdc55 Regulation Gilad Yaakov, Kurt Thorn, and David O. Morgan
Developmental Cell, Volume 23 Supplemental Information Separase Biosensor Reveals that Cohesin Cleavage Timing Depends on Phosphatase PP2A Cdc55 Regulation Gilad Yaakov, Kurt Thorn, and David O. Morgan
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Supplementary figures Supplementary Figure 1: Suv39h1, but not Suv39h2, promotes HP1α sumoylation in vivo. In vivo HP1α sumoylation assay. Top: experimental scheme. Middle: we
SUPPLEMENTARY INFORMATION Supplementary figures Supplementary Figure 1: Suv39h1, but not Suv39h2, promotes HP1α sumoylation in vivo. In vivo HP1α sumoylation assay. Top: experimental scheme. Middle: we
SUPPLEMENTARY MATERIALS
 SUPPLEMENTARY MATERIALS SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1 Condensin is required for condensation of rdna array during starvation (A) Loss of condensin blocks rdna condensation during
SUPPLEMENTARY MATERIALS SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1 Condensin is required for condensation of rdna array during starvation (A) Loss of condensin blocks rdna condensation during
supplementary information
 DOI: 10.1038/ncb2172 Figure S1 p53 regulates cellular NADPH and lipid levels via inhibition of G6PD. (a) U2OS cells stably expressing p53 shrna or a control shrna were transfected with control sirna or
DOI: 10.1038/ncb2172 Figure S1 p53 regulates cellular NADPH and lipid levels via inhibition of G6PD. (a) U2OS cells stably expressing p53 shrna or a control shrna were transfected with control sirna or
Materials and Methods: All strains were derivatives of SK1 and are listed in Supplemental Table 1.
 Supplemental Online Material: Materials and Methods: All strains were derivatives of SK1 and are listed in Supplemental Table 1. Constructs: pclb2-3ha-cdc5 and pclb2-3ha-cdc2 strains were constructed by
Supplemental Online Material: Materials and Methods: All strains were derivatives of SK1 and are listed in Supplemental Table 1. Constructs: pclb2-3ha-cdc5 and pclb2-3ha-cdc2 strains were constructed by
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11326 Supplementary Figure 1: Histone exchange increases over the ORF in a set2 mutant. (a) Gene average analysis. Schematic representation of the bin distribution over the coding and
doi:10.1038/nature11326 Supplementary Figure 1: Histone exchange increases over the ORF in a set2 mutant. (a) Gene average analysis. Schematic representation of the bin distribution over the coding and
indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) B. Male mice of
 SUPPLEMENTRY FIGURE LEGENDS Figure S1. USP44 loss leads to chromosome missegregation.. Genotypes obtained from the indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) 13.5.. Male mice
SUPPLEMENTRY FIGURE LEGENDS Figure S1. USP44 loss leads to chromosome missegregation.. Genotypes obtained from the indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) 13.5.. Male mice
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2579 Figure S1 Incorporation of heavy isotope-labeled amino acids and enrichment of di-glycine modified peptides. The incorporation of isotopelabeled amino acids in peptides was calculated
DOI: 10.1038/ncb2579 Figure S1 Incorporation of heavy isotope-labeled amino acids and enrichment of di-glycine modified peptides. The incorporation of isotopelabeled amino acids in peptides was calculated
Supplemental Data. Farmer et al. (2010) Plant Cell /tpc
 Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
Supplemental Data. Wu et al. (2). Plant Cell..5/tpc RGLG Hormonal treatment H2O B RGLG µm ABA µm ACC µm GA Time (hours) µm µm MJ µm IA
 Supplemental Data. Wu et al. (2). Plant Cell..5/tpc..4. A B Supplemental Figure. Immunoblot analysis verifies the expression of the AD-PP2C and BD-RGLG proteins in the Y2H assay. Total proteins were extracted
Supplemental Data. Wu et al. (2). Plant Cell..5/tpc..4. A B Supplemental Figure. Immunoblot analysis verifies the expression of the AD-PP2C and BD-RGLG proteins in the Y2H assay. Total proteins were extracted
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Han et al., http://www.jcb.org/cgi/content/full/jcb.201311007/dc1 Figure S1. SIVA1 interacts with PCNA. (A) HEK293T cells were transiently
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Han et al., http://www.jcb.org/cgi/content/full/jcb.201311007/dc1 Figure S1. SIVA1 interacts with PCNA. (A) HEK293T cells were transiently
Phosphorylation of RNA polymerase CTD dictates transcription
 Supplementary Information Phosphorylation of RNA polymerase CTD dictates transcription termination choice Rajani Kanth Gudipati 1,2,3, Tommaso Villa 1,2,3, Jocelyne Boulay 1,2 and Domenico Libri 1,2. 1
Supplementary Information Phosphorylation of RNA polymerase CTD dictates transcription termination choice Rajani Kanth Gudipati 1,2,3, Tommaso Villa 1,2,3, Jocelyne Boulay 1,2 and Domenico Libri 1,2. 1
SUPPLEMENTARY INFORMATION
 DOI: 1.1/ncb2918 Supplementary Figure 1 (a) Biotin-dUTP labelling does not affect S phase progression. Cells synchronized in mid-s phase were labelled with biotindutp during a 5 min hypotonic shift (red)
DOI: 1.1/ncb2918 Supplementary Figure 1 (a) Biotin-dUTP labelling does not affect S phase progression. Cells synchronized in mid-s phase were labelled with biotindutp during a 5 min hypotonic shift (red)
Supplementary Information for. Coordination of multiple enzyme activities by a single PCNA in archaeal Okazaki fragment.
 Supplementary Information for Coordination of multiple enzyme activities by a single PCNA in archaeal Okazaki fragment maturation Thomas R. Beattie and Stephen D. Bell Sir William Dunn School of Pathology,
Supplementary Information for Coordination of multiple enzyme activities by a single PCNA in archaeal Okazaki fragment maturation Thomas R. Beattie and Stephen D. Bell Sir William Dunn School of Pathology,
Supplementary Online Material. An Expanded Eukaryotic Genetic Code 2QH, UK. * To whom correspondence should be addressed.
 Supplementary Online Material An Expanded Eukaryotic Genetic Code Jason W. Chin 1, T. Ashton Cropp, J. Christopher Anderson, Mridul Mukherji, Zhiwen Zhang and Peter G. Schultz* Department of Chemistry
Supplementary Online Material An Expanded Eukaryotic Genetic Code Jason W. Chin 1, T. Ashton Cropp, J. Christopher Anderson, Mridul Mukherji, Zhiwen Zhang and Peter G. Schultz* Department of Chemistry
plasmid shuffle strain
 Histone-Histone Interactions and Centromere Function LYNN GLOWCZEWSKI, PEIRONG YANG, TATYANA KALASHNIKOVA, MARIA SOLEDAD SANTISTEBAN, AND M. MITCHELL SMITH* Department of Microbiology and Cancer Center,
Histone-Histone Interactions and Centromere Function LYNN GLOWCZEWSKI, PEIRONG YANG, TATYANA KALASHNIKOVA, MARIA SOLEDAD SANTISTEBAN, AND M. MITCHELL SMITH* Department of Microbiology and Cancer Center,
THE JOURNAL OF CELL BIOLOGY
 Supplemental Material THE JOURNL OF CELL BIOLOGY Onischenko et al., http://www.jcb.org/cgi/content/full/jcb.200810030/dc1 Figure S1. Characterization of Ndc1 interactions. () Intracellular distribution
Supplemental Material THE JOURNL OF CELL BIOLOGY Onischenko et al., http://www.jcb.org/cgi/content/full/jcb.200810030/dc1 Figure S1. Characterization of Ndc1 interactions. () Intracellular distribution
SUPPLEMENTARY INFORMATION
 doi:1.138/nature1895 i Core histones CAF-1 v ii Reb1 CAF-1 Fen1 vi Mature Okazaki fragment iii PCNA Reb1 iv vii Mature Okazaki fragment Reb1 Mature Okazaki fragments Figure S1. Model for nucleosome-mediated
doi:1.138/nature1895 i Core histones CAF-1 v ii Reb1 CAF-1 Fen1 vi Mature Okazaki fragment iii PCNA Reb1 iv vii Mature Okazaki fragment Reb1 Mature Okazaki fragments Figure S1. Model for nucleosome-mediated
3.1 The Role of the Enzymatic Activity of Sir2 for Efficient. The Sir proteins are recruited to DNA by site-specific DNA-binding proteins and
 35 3 RESULTS 3.1 The Role of the Enzymatic Activity of Sir2 for Efficient Association of the SIR Complex with DNA The Sir proteins are recruited to DNA by site-specific DNA-binding proteins and subsequently
35 3 RESULTS 3.1 The Role of the Enzymatic Activity of Sir2 for Efficient Association of the SIR Complex with DNA The Sir proteins are recruited to DNA by site-specific DNA-binding proteins and subsequently
PDIP46 (DNA polymerase δ interacting protein 46) is an activating factor for human DNA polymerase δ
 PDIP46 (DNA polymerase δ interacting protein 46) is an activating factor for human DNA polymerase δ Supplementary Material Figure S1. PDIP46 is associated with Pol isolated by immunoaffinity chromatography.
PDIP46 (DNA polymerase δ interacting protein 46) is an activating factor for human DNA polymerase δ Supplementary Material Figure S1. PDIP46 is associated with Pol isolated by immunoaffinity chromatography.
An mrna decapping mutant deficient in P body assembly limits mrna stabilization in response to osmotic stress
 An mrna decapping mutant deficient in P body assembly limits mrna stabilization in response to osmotic stress SUPPLEMENTAL INFORMATION Susanne Huch and Tracy Nissan Supplementary Information Figure S1.
An mrna decapping mutant deficient in P body assembly limits mrna stabilization in response to osmotic stress SUPPLEMENTAL INFORMATION Susanne Huch and Tracy Nissan Supplementary Information Figure S1.
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature10331 Supplemental Figures and Legends Supplementary Figure 1. Pch2-dependent suppression of DSBs at the rdna borders. a, Example of a CHEF gel/southern blot
SUPPLEMENTARY INFORMATION doi:10.1038/nature10331 Supplemental Figures and Legends Supplementary Figure 1. Pch2-dependent suppression of DSBs at the rdna borders. a, Example of a CHEF gel/southern blot
Supplementary Material
 Supplementary Material Supplementary Methods Cell synchronization. For synchronized cell growth, thymidine was added to 30% confluent U2OS cells to a final concentration of 2.5mM. Cells were incubated
Supplementary Material Supplementary Methods Cell synchronization. For synchronized cell growth, thymidine was added to 30% confluent U2OS cells to a final concentration of 2.5mM. Cells were incubated
% Viability. isw2 ino isw2 ino isw2 ino isw2 ino mM HU 4-NQO CPT
 a Drug concentration b 1.3% MMS nhp1 nhp1 8 nhp1 mag1.5% MMS.3% MMS nhp1 nhp1 ino8 9 ino8 9 % Viability 4.5% MMS ino8 9 ino8 9 2.5.1.15 % MMS c d nhp1 nhp1 nhp1 nhp1 nhp1 nhp1 Control (YPD) γ IR (1 gy)
a Drug concentration b 1.3% MMS nhp1 nhp1 8 nhp1 mag1.5% MMS.3% MMS nhp1 nhp1 ino8 9 ino8 9 % Viability 4.5% MMS ino8 9 ino8 9 2.5.1.15 % MMS c d nhp1 nhp1 nhp1 nhp1 nhp1 nhp1 Control (YPD) γ IR (1 gy)
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or
 Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
Supplemental Material: Rev1 promotes replication through UV lesions in conjunction with DNA
 Supplemental Material: Rev1 promotes replication through UV lesions in conjunction with DNA polymerases,, and, but not with DNA polymerase Jung-Hoon Yoon, Jeseong Park, Juan Conde, Maki Wakamiya, Louise
Supplemental Material: Rev1 promotes replication through UV lesions in conjunction with DNA polymerases,, and, but not with DNA polymerase Jung-Hoon Yoon, Jeseong Park, Juan Conde, Maki Wakamiya, Louise
Supplementary Figure 1. Nature Structural & Molecular Biology: doi: /nsmb.3494
 Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Racine et al, Histone H2B ubiquitylation promotes activity of the intact Set1 histone methyltransferase complex in fission yeast
 Supplemental Materials Racine et al, Histone H2B ubiquitylation promotes activity of the intact Set histone methyltransferase complex in fission yeast Supplemental Experimental Procedures Table S Figures
Supplemental Materials Racine et al, Histone H2B ubiquitylation promotes activity of the intact Set histone methyltransferase complex in fission yeast Supplemental Experimental Procedures Table S Figures
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3209 Supplementary Figure 1 IR induces the association of FH with chromatin. a, U2OS cells synchronized by thymidine double block (2 mm) underwent no release (G1 phase) or release for 2
DOI: 10.1038/ncb3209 Supplementary Figure 1 IR induces the association of FH with chromatin. a, U2OS cells synchronized by thymidine double block (2 mm) underwent no release (G1 phase) or release for 2
supplementary information
 DOI: 10.1038/ncb2116 Figure S1 CDK phosphorylation of EZH2 in cells. (a) Comparison of candidate CDK phosphorylation sites on EZH2 with known CDK substrates by multiple sequence alignments. (b) CDK1 and
DOI: 10.1038/ncb2116 Figure S1 CDK phosphorylation of EZH2 in cells. (a) Comparison of candidate CDK phosphorylation sites on EZH2 with known CDK substrates by multiple sequence alignments. (b) CDK1 and
 DOI: 10.1038/ncb3259 A Ismail et al. Supplementary Figure 1 B 60000 45000 SSC 30000 15000 Live cells 0 0 15000 30000 45000 60000 FSC- PARR 60000 45000 PARR Width 30000 FSC- 15000 Single cells 0 0 15000
DOI: 10.1038/ncb3259 A Ismail et al. Supplementary Figure 1 B 60000 45000 SSC 30000 15000 Live cells 0 0 15000 30000 45000 60000 FSC- PARR 60000 45000 PARR Width 30000 FSC- 15000 Single cells 0 0 15000
MATERIALS AND METHODS
 MATERIALS AND METHODS Strains Strains used in this study are shown in Table 1. The yap1 deletion strain was constructed by homologous recombination using G418 resistance as a selectable marker. The KanMX4
MATERIALS AND METHODS Strains Strains used in this study are shown in Table 1. The yap1 deletion strain was constructed by homologous recombination using G418 resistance as a selectable marker. The KanMX4
Supplementary Figures 1-12
 Supplementary Figures 1-12 Supplementary Figure 1. The specificity of anti-abi1 antibody. Total Proteins extracted from the wild type seedlings or abi1-3 null mutant seedlings were used for immunoblotting
Supplementary Figures 1-12 Supplementary Figure 1. The specificity of anti-abi1 antibody. Total Proteins extracted from the wild type seedlings or abi1-3 null mutant seedlings were used for immunoblotting
Supplementary Information for. Regulation of Rev1 by the Fanconi Anemia Core Complex
 Supplementary Information for Regulation of Rev1 by the Fanconi Anemia Core Complex Hyungjin Kim, Kailin Yang, Donniphat Dejsuphong, Alan D. D Andrea* *Corresponding Author: Alan D. D Andrea, M.D. Alan_dandrea@dfci.harvard.edu
Supplementary Information for Regulation of Rev1 by the Fanconi Anemia Core Complex Hyungjin Kim, Kailin Yang, Donniphat Dejsuphong, Alan D. D Andrea* *Corresponding Author: Alan D. D Andrea, M.D. Alan_dandrea@dfci.harvard.edu
Name Genotype Reference
 Supplemental Data Supplemental Table 1 S. cerevisiae strains. Name Genotype Reference RMY200 UKY403 W303-1a MATa ade2-101 (och) his3 200 lys2-801 (amp) trp1 901 ura3-52 hht1,hhf1::leu2 hht2,hhf2::his3
Supplemental Data Supplemental Table 1 S. cerevisiae strains. Name Genotype Reference RMY200 UKY403 W303-1a MATa ade2-101 (och) his3 200 lys2-801 (amp) trp1 901 ura3-52 hht1,hhf1::leu2 hht2,hhf2::his3
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Impaired ITS2 processing results in mislocalization of foot-factors to the cytoplasm. (a) Schematic representation of ITS2 processing. The endonuclease Las1 initiates ITS2 processing
Supplementary Figure 1 Impaired ITS2 processing results in mislocalization of foot-factors to the cytoplasm. (a) Schematic representation of ITS2 processing. The endonuclease Las1 initiates ITS2 processing
Nature Structural & Molecular Biology: doi: /nsmb.1583
 Acetylation by GCN5 regulates CDC6 phosphorylation in the S-phase of the cell cycle Roberta Paolinelli 1,2, Ramiro Mendoza-Maldonado 2, Anna Cereseto 1 and Mauro Giacca 2 1 Molecular Biology Laboratory,
Acetylation by GCN5 regulates CDC6 phosphorylation in the S-phase of the cell cycle Roberta Paolinelli 1,2, Ramiro Mendoza-Maldonado 2, Anna Cereseto 1 and Mauro Giacca 2 1 Molecular Biology Laboratory,
Notes of Protein Engineering
 Notes of Protein Engineering Week 2 (01/ 08/ 2016-07/ 08/ 2016) Aug 5th 1. PCR Verify that the E.coli transformed with prs416 linked with parts is well constructed. Fast Taq Polymerase 50μl Protocol; Strategy:
Notes of Protein Engineering Week 2 (01/ 08/ 2016-07/ 08/ 2016) Aug 5th 1. PCR Verify that the E.coli transformed with prs416 linked with parts is well constructed. Fast Taq Polymerase 50μl Protocol; Strategy:
S156AT168AY175A (AAA) were purified as GST-fusion proteins and incubated with GSTfused
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 Supplemental Materials Supplemental Figure S1 (a) Phenotype of the wild type and grik1-2 grik2-1 plants after 8 days in darkness.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 Supplemental Materials Supplemental Figure S1 (a) Phenotype of the wild type and grik1-2 grik2-1 plants after 8 days in darkness.
human Cdc45 Figure 1c. (c)
 1 Details of the refined crystallographic model of human Cdc45 and comparison of its active-site region with that of bacterial RecJ. (a) Stereo view of a representative example of the final 2F o -F c electron
1 Details of the refined crystallographic model of human Cdc45 and comparison of its active-site region with that of bacterial RecJ. (a) Stereo view of a representative example of the final 2F o -F c electron
c Nup82-TAP purification
 a Dbp5-TAP Nup159-TAP b Dbp5-TAP Dbp5-TAP/dyn2Δ Nup116 Dbp5 Nup159 Nsp1 Nup82 Nup159 Nup116 Nsp1 Nup82 Dbp5 # # 1 2 1 2 Western c Nup82-TAP purification input flow through pull-down 1 2 3 α-nup82 1 2 3
a Dbp5-TAP Nup159-TAP b Dbp5-TAP Dbp5-TAP/dyn2Δ Nup116 Dbp5 Nup159 Nsp1 Nup82 Nup159 Nup116 Nsp1 Nup82 Dbp5 # # 1 2 1 2 Western c Nup82-TAP purification input flow through pull-down 1 2 3 α-nup82 1 2 3
Supplemental Data. Sethi et al. (2014). Plant Cell /tpc
 Supplemental Data Supplemental Figure 1. MYC2 Binds to the E-box but not the E1-box of the MPK6 Promoter. (A) E1-box and E-box (wild type) containing MPK6 promoter fragment. The region shown in red denotes
Supplemental Data Supplemental Figure 1. MYC2 Binds to the E-box but not the E1-box of the MPK6 Promoter. (A) E1-box and E-box (wild type) containing MPK6 promoter fragment. The region shown in red denotes
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3562 In the format provided by the authors and unedited. Supplementary Figure 1 Glucose deficiency induced FH-ATF2 interaction. In b-m, immunoblotting or immunoprecipitation analyses were
DOI: 10.1038/ncb3562 In the format provided by the authors and unedited. Supplementary Figure 1 Glucose deficiency induced FH-ATF2 interaction. In b-m, immunoblotting or immunoprecipitation analyses were
Hoffmann et al., http ://www.jcb.org /cgi /content /full /jcb /DC1
 Supplemental material JCB Hoffmann et al., http ://www.jcb.org /cgi /content /full /jcb.201506071 /DC1 THE JOU RNAL OF CELL BIO LOGY Figure S1. TRA IP harbors a PCNA-binding PIP box required for its recruitment
Supplemental material JCB Hoffmann et al., http ://www.jcb.org /cgi /content /full /jcb.201506071 /DC1 THE JOU RNAL OF CELL BIO LOGY Figure S1. TRA IP harbors a PCNA-binding PIP box required for its recruitment
Localized Histone Acetylation and Deacetylation Triggered by the Homologous Recombination Pathway of Double-Strand DNA Repair
 MOLECULAR AND CELLULAR BIOLOGY, June 2005, p. 4903 4913 Vol. 25, No. 12 0270-7306/05/$08.00 0 doi:10.1128/mcb.25.12.4903 4913.2005 Copyright 2005, American Society for Microbiology. All Rights Reserved.
MOLECULAR AND CELLULAR BIOLOGY, June 2005, p. 4903 4913 Vol. 25, No. 12 0270-7306/05/$08.00 0 doi:10.1128/mcb.25.12.4903 4913.2005 Copyright 2005, American Society for Microbiology. All Rights Reserved.
Supplementary Table 1a. Number of crossover (CO), noncrossover (NCO) and breakinduced replication (BIR) events in red/white sectored colonies
 Supplementary Table 1a. Number of crossover (CO), noncrossover (NCO) and breakinduced replication (BIR) events in red/white sectored colonies CO NCO BIR Total WT 90 939 15 144 yen1 114 34 4 44 114 55 15
Supplementary Table 1a. Number of crossover (CO), noncrossover (NCO) and breakinduced replication (BIR) events in red/white sectored colonies CO NCO BIR Total WT 90 939 15 144 yen1 114 34 4 44 114 55 15
Supplemental Information. The E2F functional analog SBF recruits the Rpd3(L) HDAC, via Whi5 and Stb1,
 The E2F functional analog SBF recruits the Rpd3(L) HDAC, via Whi5 and Stb1, and the FACT chromatin reorganizer, to yeast G1 cyclin promoters Shinya Takahata, Yaxin Yu, and David J. Stillman Supplemental
The E2F functional analog SBF recruits the Rpd3(L) HDAC, via Whi5 and Stb1, and the FACT chromatin reorganizer, to yeast G1 cyclin promoters Shinya Takahata, Yaxin Yu, and David J. Stillman Supplemental
Cover Page. The handle holds various files of this Leiden University dissertation.
 Cover Page The handle http://hdl.handle.net/1887/20360 holds various files of this Leiden University dissertation. Author: Eijk, Patrick van Title: Nucleotide excision repair in yeast Date: 2012-12-13
Cover Page The handle http://hdl.handle.net/1887/20360 holds various files of this Leiden University dissertation. Author: Eijk, Patrick van Title: Nucleotide excision repair in yeast Date: 2012-12-13
Supplementary data. sienigma. F-Enigma F-EnigmaSM. a-p53
 Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Sulfur transfer and activation by ubiquitin-like modifier system Uba4 Urm1 link protein urmylation and trna thiolation in yeast
 Supplemental Data Sulfur transfer and activation by ubiquitin-like modifier system Uba4 Urm1 link protein urmylation and trna thiolation in yeast André Jüdes 1, Alexander Bruch 1, Roland Klassen 1, Mark
Supplemental Data Sulfur transfer and activation by ubiquitin-like modifier system Uba4 Urm1 link protein urmylation and trna thiolation in yeast André Jüdes 1, Alexander Bruch 1, Roland Klassen 1, Mark
7.06 Problem Set Four, 2006
 7.06 Problem Set Four, 2006 1. Explain the molecular mechanism behind each of the following events that occur during the cell cycle, making sure to discuss specific proteins that are involved in making
7.06 Problem Set Four, 2006 1. Explain the molecular mechanism behind each of the following events that occur during the cell cycle, making sure to discuss specific proteins that are involved in making
Supplementary Figure 1. RAD51 and RAD51 paralogs are enriched spontaneously onto
 Supplementary Figure legends Supplementary Figure 1. and paralogs are enriched spontaneously onto the S-phase chromatin during DN replication. () Chromatin fractionation was carried out as described in
Supplementary Figure legends Supplementary Figure 1. and paralogs are enriched spontaneously onto the S-phase chromatin during DN replication. () Chromatin fractionation was carried out as described in
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the
 Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/291/ra78/dc1 Supplementary Materials for Regulation of Yeast G Protein Signaling by the Kinases That Activate the AMPK Homolog Snf1 Sarah T. Clement, Gauri Dixit,
www.sciencesignaling.org/cgi/content/full/6/291/ra78/dc1 Supplementary Materials for Regulation of Yeast G Protein Signaling by the Kinases That Activate the AMPK Homolog Snf1 Sarah T. Clement, Gauri Dixit,
Supplementary information
 Supplementary information The E3 ligase RNF8 regulates KU80 removal and NHEJ repair Lin Feng 1, Junjie Chen 1 1 Department of Experimental Radiation Oncology, The University of Texas M. D. Anderson Cancer
Supplementary information The E3 ligase RNF8 regulates KU80 removal and NHEJ repair Lin Feng 1, Junjie Chen 1 1 Department of Experimental Radiation Oncology, The University of Texas M. D. Anderson Cancer
7.03, 2006, Lecture 23 Eukaryotic Genes and Genomes IV
 1 Fall 2006 7.03 7.03, 2006, Lecture 23 Eukaryotic Genes and Genomes IV In the last three lectures we have thought a lot about analyzing a regulatory system in S. cerevisiae, namely Gal regulation that
1 Fall 2006 7.03 7.03, 2006, Lecture 23 Eukaryotic Genes and Genomes IV In the last three lectures we have thought a lot about analyzing a regulatory system in S. cerevisiae, namely Gal regulation that
7.05, 2005, Lecture 23 Eukaryotic Genes and Genomes IV
 7.05, 2005, Lecture 23 Eukaryotic Genes and Genomes IV In the last three lectures we have thought a lot about analyzing a regulatory system in S. cerevisiae, namely Gal regulation that involved a hand
7.05, 2005, Lecture 23 Eukaryotic Genes and Genomes IV In the last three lectures we have thought a lot about analyzing a regulatory system in S. cerevisiae, namely Gal regulation that involved a hand
Schematic representation of the endogenous PALB2 locus and gene-disruption constructs
 Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Schizosaccharomyces pombe Hst4 Functions in DNA Damage Response by Regulating Histone H3 K56 Acetylation
 EUKARYOTIC CELL, May 2008, p. 800 813 Vol. 7, No. 5 1535-9778/08/$08.00 0 doi:10.1128/ec.00379-07 Copyright 2008, American Society for Microbiology. All Rights Reserved. Schizosaccharomyces pombe Hst4
EUKARYOTIC CELL, May 2008, p. 800 813 Vol. 7, No. 5 1535-9778/08/$08.00 0 doi:10.1128/ec.00379-07 Copyright 2008, American Society for Microbiology. All Rights Reserved. Schizosaccharomyces pombe Hst4
Schizosaccharomyces pombe Hst4 Functions in DNA Damage Response by Regulating Histone H3 K56 Acetylation
 EUKARYOTIC CELL, May 2008, p. 800 813 Vol. 7, No. 5 1535-9778/08/$08.00 0 doi:10.1128/ec.00379-07 Copyright 2008, American Society for Microbiology. All Rights Reserved. Schizosaccharomyces pombe Hst4
EUKARYOTIC CELL, May 2008, p. 800 813 Vol. 7, No. 5 1535-9778/08/$08.00 0 doi:10.1128/ec.00379-07 Copyright 2008, American Society for Microbiology. All Rights Reserved. Schizosaccharomyces pombe Hst4
Lecture 22 Eukaryotic Genes and Genomes III
 Lecture 22 Eukaryotic Genes and Genomes III In the last three lectures we have thought a lot about analyzing a regulatory system in S. cerevisiae, namely Gal regulation that involved a hand full of genes.
Lecture 22 Eukaryotic Genes and Genomes III In the last three lectures we have thought a lot about analyzing a regulatory system in S. cerevisiae, namely Gal regulation that involved a hand full of genes.
2/6/18. file:///users/jlc/ Downloads/ DNAi_replicatio n_vo2-lg.mov. 3x10 13 cells (Bianconi et al. Ann Hum Biol, 2013) DNA Replication in vivo
 file:///users/jlc/ Downloads/ DNAi_replicatio n_vo2-lg.mov 3x10 13 cells (Bianconi et al. Ann Hum Biol, 2013) 10 12 replicating cells (Cairns: Genetics (2006)174,1069). Lose about 1/3 of these per day
file:///users/jlc/ Downloads/ DNAi_replicatio n_vo2-lg.mov 3x10 13 cells (Bianconi et al. Ann Hum Biol, 2013) 10 12 replicating cells (Cairns: Genetics (2006)174,1069). Lose about 1/3 of these per day
Test Bank for Molecular Cell Biology 7th Edition by Lodish
 Test Bank for Molecular Cell Biology 7th Edition by Lodish Link download full: http://testbankair.com/download/test-bank-formolecular-cell-biology-7th-edition-by-lodish/ Chapter 5 Molecular Genetic Techniques
Test Bank for Molecular Cell Biology 7th Edition by Lodish Link download full: http://testbankair.com/download/test-bank-formolecular-cell-biology-7th-edition-by-lodish/ Chapter 5 Molecular Genetic Techniques
At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in
 Supplementary Materials and Methods Barrier function assays At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in acidic X-gal mix (100 mm phosphate buffer at ph4.3, 3 mm
Supplementary Materials and Methods Barrier function assays At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in acidic X-gal mix (100 mm phosphate buffer at ph4.3, 3 mm
Aspergillus fumigatus CalA binds to integrin α 5 β 1 and mediates host cell invasion
 In the format provided by the authors and unedited. SUPPLEMENTARY INFORMATION ARTICLE NUMBER: 16211 DOI: 10.1038/NMICROBIOL.2016.211 Aspergillus fumigatus CalA binds to integrin α 5 β 1 and mediates host
In the format provided by the authors and unedited. SUPPLEMENTARY INFORMATION ARTICLE NUMBER: 16211 DOI: 10.1038/NMICROBIOL.2016.211 Aspergillus fumigatus CalA binds to integrin α 5 β 1 and mediates host
indicated) in rich, 5-fluoroorotic acid (5FOA) or drop-out media containing 2% glucose,
 SUPPLEMENTARY MATERIAL MATERIALS AND METHODS Media and growth conditions. All strains were grown at 30 C (unless otherwise indicated) in rich, 5-fluoroorotic acid (5FOA) or drop-out media containing 2%
SUPPLEMENTARY MATERIAL MATERIALS AND METHODS Media and growth conditions. All strains were grown at 30 C (unless otherwise indicated) in rich, 5-fluoroorotic acid (5FOA) or drop-out media containing 2%
Separation of a functional deubiquitylating module from the SAGA complex by the proteasome regulatory particle
 Supplementary information for Separation of a functional deubiquitylating module from the SAGA complex by the proteasome regulatory particle Sungsu Lim 1, Jaechan Kwak 1, Minhoo Kim 1 and Daeyoup Lee 1,*
Supplementary information for Separation of a functional deubiquitylating module from the SAGA complex by the proteasome regulatory particle Sungsu Lim 1, Jaechan Kwak 1, Minhoo Kim 1 and Daeyoup Lee 1,*
A) (5 points) As the starting step isolate genomic DNA from
 GS Final Exam Spring 00 NAME. bub ts is a recessive temperature sensitive mutation in yeast. At º C bub ts cells grow normally, but at º C they die. Use the information below to clone the wild-type BUB
GS Final Exam Spring 00 NAME. bub ts is a recessive temperature sensitive mutation in yeast. At º C bub ts cells grow normally, but at º C they die. Use the information below to clone the wild-type BUB
Figure S1. Immunoblotting showing proper expression of tagged R and AVR proteins in N. benthamiana.
 SUPPLEMENTARY FIGURES Figure S1. Immunoblotting showing proper expression of tagged R and AVR proteins in N. benthamiana. YFP:, :CFP, HA: and (A) and HA, CFP and YFPtagged and AVR1-CO39 (B) were expressed
SUPPLEMENTARY FIGURES Figure S1. Immunoblotting showing proper expression of tagged R and AVR proteins in N. benthamiana. YFP:, :CFP, HA: and (A) and HA, CFP and YFPtagged and AVR1-CO39 (B) were expressed
Antibodies against PCNA were previously described [1]. To deplete PCNA from Xenopus egg
![Antibodies against PCNA were previously described [1]. To deplete PCNA from Xenopus egg Antibodies against PCNA were previously described [1]. To deplete PCNA from Xenopus egg](/thumbs/84/90687567.jpg) Supplementary information Supplementary methods PCNA antibody and immunodepletion Antibodies against PCNA were previously described [1]. To deplete PCNA from Xenopus egg extracts, one volume of protein
Supplementary information Supplementary methods PCNA antibody and immunodepletion Antibodies against PCNA were previously described [1]. To deplete PCNA from Xenopus egg extracts, one volume of protein
Supplementary Figure 1. Chromosome 3 is devoid of the telomere-proximal subtelomeric
 Supplementary Figure 1. Chromosome 3 is devoid of the telomere-proximal subtelomeric common sequences. (a) Schematic illustration of the telomere-proximal site of subtelomeres. The restriction map is based
Supplementary Figure 1. Chromosome 3 is devoid of the telomere-proximal subtelomeric common sequences. (a) Schematic illustration of the telomere-proximal site of subtelomeres. The restriction map is based
sicker; i.e. to test whether we can reverse the mutant phenotype of mec1-100 when the suppressor gene is ectopically over-expressed in mec1-100.
 Internship Report ~FMI (Basel, Switzerland)~ Kyoto University Faculty of Medicine 1 st grade Yuiko Hirata This summer I went to the FMI (Friedrich Miescher Institute for Biomedical Research) in Basel.
Internship Report ~FMI (Basel, Switzerland)~ Kyoto University Faculty of Medicine 1 st grade Yuiko Hirata This summer I went to the FMI (Friedrich Miescher Institute for Biomedical Research) in Basel.
Supplementary information
 Supplementary information Supplementary figures Figure S1 Level of mycdet1 protein in DET1 OE-1, OE-2 and OE-3 transgenic lines. Total protein extract from wild type Col0, det1-1 mutant and DET1 OE lines
Supplementary information Supplementary figures Figure S1 Level of mycdet1 protein in DET1 OE-1, OE-2 and OE-3 transgenic lines. Total protein extract from wild type Col0, det1-1 mutant and DET1 OE lines
MCB 110L EXAMINATION FOR SECTION #1 Spring, 2008 QUESTION YOUR SCORE FULL SCORE I 40 II 40 III 40 IV 40 V 40 TOTAL 200
 MCB 110L EAMINATION FOR SECTION #1 Spring, 2008 QUESTION YOUR SCORE FULL SCORE I 40 II 40 III 40 IV 40 V 40 TOTAL 200 YOUR NAME: (Please PRINT your name in ink on this line as legibly as possible.) YOUR
MCB 110L EAMINATION FOR SECTION #1 Spring, 2008 QUESTION YOUR SCORE FULL SCORE I 40 II 40 III 40 IV 40 V 40 TOTAL 200 YOUR NAME: (Please PRINT your name in ink on this line as legibly as possible.) YOUR
SUPPLEMENTARY INFORMATION
 doi:.38/nature46 SUPPLEMENTARY INFORMATION Supplementary Information for: CDK-dependent phosphorylation of Sld and Sld3 initiates DNA replication in budding yeast Seiji Tanaka, Toshiko Umemori, Kazuyuki
doi:.38/nature46 SUPPLEMENTARY INFORMATION Supplementary Information for: CDK-dependent phosphorylation of Sld and Sld3 initiates DNA replication in budding yeast Seiji Tanaka, Toshiko Umemori, Kazuyuki
Received 29 September 2005/Returned for modification 9 November 2005/Accepted 21 November 2005
 MOLECULAR AND CELLULAR BIOLOGY, Feb. 2006, p. 1496 1509 Vol. 26, No. 4 0270-7306/06/$08.00 0 doi:10.1128/mcb.26.4.1496 1509.2006 Copyright 2006, American Society for Microbiology. All Rights Reserved.
MOLECULAR AND CELLULAR BIOLOGY, Feb. 2006, p. 1496 1509 Vol. 26, No. 4 0270-7306/06/$08.00 0 doi:10.1128/mcb.26.4.1496 1509.2006 Copyright 2006, American Society for Microbiology. All Rights Reserved.
Supplemental Data. Cui et al. (2012). Plant Cell /tpc a b c d. Stem UBC32 ACTIN
 A Root Stem Leaf Flower Silique Senescence leaf B a b c d UBC32 ACTIN C * Supplemental Figure 1. Expression Pattern and Protein Sequence of UBC32 Homologues in Yeast, Human, and Arabidopsis. (A) Expression
A Root Stem Leaf Flower Silique Senescence leaf B a b c d UBC32 ACTIN C * Supplemental Figure 1. Expression Pattern and Protein Sequence of UBC32 Homologues in Yeast, Human, and Arabidopsis. (A) Expression
MID-TERM EXAMINATION
 PLNT3140 INTRODUCTORY CYTOGENETICS MID-TERM EXAMINATION 1 p.m. to 2:15 p.m. Thursday, October 18, 2012 Answer any combination of questions totalling to exactly 100 points. If you answer questions totalling
PLNT3140 INTRODUCTORY CYTOGENETICS MID-TERM EXAMINATION 1 p.m. to 2:15 p.m. Thursday, October 18, 2012 Answer any combination of questions totalling to exactly 100 points. If you answer questions totalling
Fig. S1. CrgA intracellular levels in M. tuberculosis. Ten and twenty micrograms of
 Supplementary data Fig. S1. CrgA intracellular levels in M. tuberculosis. Ten and twenty micrograms of cell free protein lysates from WT M. tuberculosis (Rv) together with various known concentrations
Supplementary data Fig. S1. CrgA intracellular levels in M. tuberculosis. Ten and twenty micrograms of cell free protein lysates from WT M. tuberculosis (Rv) together with various known concentrations
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified
 Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
PRODUCT INFORMATION. Composition of SOC medium supplied :
 Product Name : Competent Cell BL21(DE3)pLysS Code No. : DS260 Size : 100 μl 10 Competency : > 5 10 7 cfu/μg (puc19) Supplied product : SOC medium, 1 ml 10 This product is for research use only Description
Product Name : Competent Cell BL21(DE3)pLysS Code No. : DS260 Size : 100 μl 10 Competency : > 5 10 7 cfu/μg (puc19) Supplied product : SOC medium, 1 ml 10 This product is for research use only Description
Supplemental material
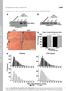 Supplemental material THE JOURNAL OF CELL BIOLOGY Gillespie et al., http://www.jcb.org/cgi/content/full/jcb.200907037/dc1 repressor complex induced by p38- Gillespie et al. Figure S1. Reduced fiber size
Supplemental material THE JOURNAL OF CELL BIOLOGY Gillespie et al., http://www.jcb.org/cgi/content/full/jcb.200907037/dc1 repressor complex induced by p38- Gillespie et al. Figure S1. Reduced fiber size
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb54 sensitivity (+/-) 3 det- WS bri-5 BL (nm) bzr-dbri- bzr-d/ bri- g h i a b c d e f Hypcocotyl length(cm) 5 5 PAC Hypocotyl length (cm)..8..4. 5 5 PPZ - - + + + + - + - + - + PPZ - - + + +
DOI:.38/ncb54 sensitivity (+/-) 3 det- WS bri-5 BL (nm) bzr-dbri- bzr-d/ bri- g h i a b c d e f Hypcocotyl length(cm) 5 5 PAC Hypocotyl length (cm)..8..4. 5 5 PPZ - - + + + + - + - + - + PPZ - - + + +
Chapter 11 DNA Replication and Recombination
 Chapter 11 DNA Replication and Recombination Copyright Copyright 2009 Pearson 2009 Pearson Education, Education, Inc. Inc. 11.1 DNA is reproduced by Semiconservative Replication The complementarity of
Chapter 11 DNA Replication and Recombination Copyright Copyright 2009 Pearson 2009 Pearson Education, Education, Inc. Inc. 11.1 DNA is reproduced by Semiconservative Replication The complementarity of
An in silico mathematical model of the initiation of DNA replication
 An in silico mathematical model of the initiation of DNA replication Rohan D. Gidvani a1, Brendan J. McConkey a, Bernard P. Duncker a and Brian P. Ingalls a,b a Department of Biology, University of Waterloo,
An in silico mathematical model of the initiation of DNA replication Rohan D. Gidvani a1, Brendan J. McConkey a, Bernard P. Duncker a and Brian P. Ingalls a,b a Department of Biology, University of Waterloo,
Your first goal is to determine the order of assembly of the 3 DNA polymerases during initiation.
 MIT Department of Biology 7.28, Spring 2005 - Molecular Biology 1 Question 1 You are studying the mechanism of DNA polymerase loading at eukaryotic DNA replication origins. Unlike the situation in bacteria,
MIT Department of Biology 7.28, Spring 2005 - Molecular Biology 1 Question 1 You are studying the mechanism of DNA polymerase loading at eukaryotic DNA replication origins. Unlike the situation in bacteria,
Supplemental Information
 Supplemental Information Supplemental material and methods ATPase assay Mph1 (35 nm) or mph1 D209N (100 nm) was incubated at 30 o C with 1.5 mm [γ- 32 P] ATP and φx174 circular (+) strand DNA (25 µm nucleotides)
Supplemental Information Supplemental material and methods ATPase assay Mph1 (35 nm) or mph1 D209N (100 nm) was incubated at 30 o C with 1.5 mm [γ- 32 P] ATP and φx174 circular (+) strand DNA (25 µm nucleotides)
Supplemental Data. Zhang et al. Plant Cell (2014) /tpc
 Supplemental Data. Zhang et al. Plant Cell (214) 1.115/tpc.114.134163 55 - T C N SDIRIP1-GFP 35-25 - Psb 18 - Histone H3 Supplemental Figure 1. Detection of SDIRIP1-GFP in the nuclear fraction by Western
Supplemental Data. Zhang et al. Plant Cell (214) 1.115/tpc.114.134163 55 - T C N SDIRIP1-GFP 35-25 - Psb 18 - Histone H3 Supplemental Figure 1. Detection of SDIRIP1-GFP in the nuclear fraction by Western
Nature Structural & Molecular Biology: doi: /nsmb.3308
 Supplementary Figure 1 Analysis of CED-3 autoactivation and CED-4-induced CED-3 activation. (a) Repeat experiments of Fig. 1a. (b) Repeat experiments of Fig. 1b. (c) Quantitative analysis of three independent
Supplementary Figure 1 Analysis of CED-3 autoactivation and CED-4-induced CED-3 activation. (a) Repeat experiments of Fig. 1a. (b) Repeat experiments of Fig. 1b. (c) Quantitative analysis of three independent
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2271 Supplementary Figure a! WM266.4 mock WM266.4 #7 sirna WM266.4 #10 sirna SKMEL28 mock SKMEL28 #7 sirna SKMEL28 #10 sirna WM1361 mock WM1361 #7 sirna WM1361 #10 sirna 9 WM266. WM136
DOI: 10.1038/ncb2271 Supplementary Figure a! WM266.4 mock WM266.4 #7 sirna WM266.4 #10 sirna SKMEL28 mock SKMEL28 #7 sirna SKMEL28 #10 sirna WM1361 mock WM1361 #7 sirna WM1361 #10 sirna 9 WM266. WM136
