Supporting Information
|
|
|
- Brendan Harvey
- 5 years ago
- Views:
Transcription
1 Supporting Information Chen et al /pnas Fig. S1. The sae2δ-suppressing mre11 alleles: protein level and location. (A) Protein level of Mre11 and Mre11 P11L expressed from the endogenous MRE11 locus in wild-type and sae2δ derivatives (Left) and Mre11, Mre11 H37Y, Mre11 Q7H, Mre11 T74A, and Mre11 E11G expressed from prs416 in mre11δ sae2δ cells (Right). (B) Sequence alignment of the N-terminal region of Mre11 from different species. Sequence conservation is indicated by colors, with red representing 1% and blue representing %. The Mre11 nuclease-related phosphoesterase motifs I III, the Nbs1 interacting loop, and two human disease-associated mre11 mutations are indicated. Residues that suppress sae2δ DNA damage sensitivity when mutated are marked by green circles. 1of6
2 Fig. S2. The sae2δ-suppressing mre11 alleles do not restore Mre11 endonuclease activity. (A) Indicated mre11 alleles were expressed from a plasmid in mre11δ or mre11δ sae2δ cells harboring the lys2-aluir and lys2-δ5 ectopic recombination reporter. Cells were patched as triplicates on SC-URA medium to maintain the plasmid and replicated to SC-URA-LYS medium to score for Lys + recombinants. (B) Sporulation efficiency from indicated diploid cells were determined by scoring number of tetrads/number of total cells by microscopy 2 d after growing cells on sporulation medium at 3 C. 2of6
3 Fig. S3. mre11-p11l does not restore end resection or affect HR. (A) Spores from tetrad dissection showing that mre11-p11l does not suppress the sae2δ sgs1δ synthetic lethality. (B) Southern blot analysis of KpnI-digested genomic DNA from indicated strains taken at different time points after HO induction in G 2 /M arrested cells. Uncut fragment (14.5 kb), cut fragments (12 and 2.5 kb), and the SSA product (8 kb) were detected by a leu2 probe. A ssdna-resection intermediate is marked by the asterisk. (C) A mating type switching assay was used to monitor HR efficiency on a HO-induced DSB at the MATα locus. Southern blot was performed and the ratio of MATa product among total DNA at 6-h after HO induction is indicated. Fig. S4. The mre11 alleles suppress the checkpoint recovery defect in sae2δ.(a) Western blot analysis showing Rad53 phosphorylation and dephosphorylation in response to MMS. Log-phase growing cells (t = ) from indicated strains were treated with.15% MMS for 1 h and released into fresh media (t = 1h). Protein samples from different time points before and after MMS treatment were analyzed by using anti-rad53 antibodies. (B) Tenfold serial dilutions of logphase cultures of the indicated strains were spotted onto YPD medium with no MMS or.1% MMS. 3of6
4 A B C % of cells with Mre11 foci time post IR 4 Gy (h) WT sae2 tel1 sae2 tel1 (MW) Marker M WT RX M PL RX Rad5 Xrs2 Mre11 Degraded, % Time, min Mn 2+ WT Mn 2+ PL Mg 2+ WT Mg 2+ PL D E 1mM ATP; 5mM Mg 2+ 1mM ATP; 2mM Mn 2+ M WT RX M PL RX M WT RX M PL RX DNA bound, % Mg 2+ ssdna,wt ssdna,pl dsdna,wt dsdna,pl ssdna (1nt) dsdna (1nt) 1mM ATP; 5mM Mg 2+ 1mM ATP; 2mM Mn 2+ M WT RX M PL RX M WT RX M PL RX DNA bound, % Mn Fig. S5. Nuclease and DNA binding activity of the MRX and M P11L RX complexes. (A) Quantification of Mre11-YFP foci from indicated strains. The percentage of cells with one or more YFP foci at different time points after IR was quantitated. The mean values from three independent trials are plotted, and error bars show SD. (B) Purified MRX and M P11L RX protein complexes were run on a 4 15% gel and stained with SYPRO-Orange. (C) The M P11L RX proteins have weaker 3-5 exonuclease activity than MRX. The mean values from two independent trials are plotted, and error bars show SE. (D) DNA binding activity of WT and mutant complexes in the presence of ATP and 5 mm Mg 2+ or 2 mm Mn 2+.(E) Quantification of the data shown in D. The mean values from two independent trials are plotted, and error bars show SE. 4of6
5 Table S1. Yeast strains Strain Genotype Source W1588-4C MATa 1 LSY191 MATa sae2::kanmx6 2 LSY2954-2B MATa mre11-p11l-natmx4 This study LSY2882-1A MATa mre11-p11l-natmx4 sae2::kanmx6 This study LSY2872-2A MATα rad5s::ura3 This study LSY2872-1A MATa mre11-p11l-natmx4 rad5s::ura3 This study LSY779 MATa mre11::leu2 3 LSY2719-2A MATα mre11::leu2 sae2::kanmx6 This study ALE94 MATα ade5-1 his7-2 leu2-3,112::p35l3 (LEU2) trp1-289 ura3 lys2::aluir 4 ALE18 MATα ade5-1 his7-2 leu2-3,112::p35l3 (LEU2) trp1-289 ura3 lys2::aluir sae2::hgrb 4 LSY293 MATα ade5-1 his7-2 leu2-3,112::p35l3 (LEU2) trp1-289 ura3 lys2::aluir mre11::trp1 This study LSY2931 MATα a ade5-1 his7-2 leu2-3,112::p35l3 (LEU2) trp1-289 ura3 lys2::aluir mre11::trp1 sae2::hgrb This study LSY2933-2B MATα sae2::kanmx6 exo1::ura3 This study mre11-p11l-natmx4 LSY2938 MATα sae2::kanmx6 exo1::ura3 This study LSY349-3B MATa sae2::kanmx6 yku7::his3 RAD5 This study LSY2954-1C MATa mre11-p11l-natmx4 yku7::his3 sae2::kanmx6 This study YMV45 MAT::hisG hml::ade1 hmr::ade1 ade1 lys5 ura3-52 trp1 ho ade3::pgal-ho leu2-cs leu2 fragment 5 inserted 4.6 kb upstream of leu2-cs LSY1951 (YMV45 sae2) MAT::hisG hml::ade1 hmr::ade1 ade1 lys5 ura3-52 trp1 ho ade3::pgal-ho leu2-cs leu2 fragment 6 inserted 4.6 kb upstream of leu2-cs sae2::kanmx6 LSY389 MAT::hisG hml::ade1 hmr::ade1 ade1 lys5 ura3-52 trp1 ho ade3::pgal-ho leu2-cs leu2 fragment This study inserted 4.6 kb upstream of leu2-cs mre11-p11l-natmx4 LSY386 MAT::hisG hml::ade1 hmr::ade1 ade1 lys5 ura3-52 trp1 ho ade3::pgal-ho leu2-cs leu2 fragment This study inserted 4.6 kb upstream of leu2-cs sae2::kanmx6 mre11-p11l-natmx4 LSY2917 MATa bar1::leu2 This study LSY2944-6A MATa bar1::leu2 mre11-p11l-natmx4 This study LSY2918 MATa bar1::leu2 sae2::kanmx6 This study LSY2919 MATa bar1::leu2 sae2::kanmx6 mre11-p11l-natmx4 This study LSY1996 MATa tel1::hphmx4 E. Mimitou LSY287-3A MATa sae2::kanmx6 tel1::hphmx4 This study LSY351-4B MATa sae2::kanmx6 tel1::hphmx4 mre11-p11l-natmx4 This study LSY351-3D MATα tel1::hphmx4 mre11-p11l-natmx4 LSY C MATa mec1::trp1 sml1::his3 This study LSY B MATα mec1::trp1 sml1::his3 sae2::kanmx6 This study LSY2988-1D MATa mec1::trp1 sml1::his3 sae2::kanmx6 This study mre11-p11l-natmx4 LSY2988-1C MATa mec1::trp1 sml1::his3 mre11-p11l-natmx4 This study W3483-1A MATa ADE2 MRE11-YFP bar1::leu2 7 LSY336 MATa ADE2 MRE11-YFP bar1::leu2 sae2::kanmx6 This study LSY337 MATa ADE2 mre11-p11l-yfp bar1::leu2 sae2::kanmx6 This study LSY3128 MATa ADE2 mre11-h37y-yfp bar1::leu2 sae2::kanmx6 This study LSY329 MATa ADE2 mre11-p11l-yfp bar1::leu2 This study LSY385 MATa ADE2 mre11-h37y-yfp bar1::leu2 This study LSY A MATa ADE2 MRE11-YFP bar1::leu2 tel1::hphmx4 This study LSY3245-1C MATα ADE2 MRE11-YFP bar1::leu2 sae2::kanmx6 tel1::hphmx4 This study LSY379-1D MATa leu2::pgal-ho-leu2 hml::oriprs hmr::ampr This study LSY379-16A MATa leu2::pgal-ho-leu2 hml::oriprs hmr::ampr sae2::kanmx6 This study LSY379-1B MATa leu2::pgal-ho-leu2 hml::oriprs hmr::ampr This study mre11-p11l-natmx4 LSY379-13C MATa leu2::pgal-ho-leu2 hml::oriprs hmr::ampr This study mre11-p11l-natmx4 sae2::kanmx6 LSY388 MATα lys2::pgal-iscei ade2-isir-14mh This study LSY D MATα lys2::pgal-iscei ade2-isir-14mh sae2::kanmx6 This study LSY3173-4A MATα lys2::pgal-iscei ade2-isir-14mh mre11-p11l-natmx4 This study LSY3173-1C MATα lys2::pgal-iscei ade2-isir-14mh sae2::kanmx6 mre11-p11l-natmx4 This study LSY785 MATa yku7::his3 2 LSY3181-4B MATα sae2::kanmx6 yku7::his3 exo1::ura3 This study LSY318-2B MATα ade3::pgal-ho sae2::kanmx6 This study LSY318-3C MATα ade3::pgal-ho mre11-p11l-natmx4 This study LSY318-1A MATα ade3::pgal-ho mre11-p11l-natmx4 sae2::kanmx6 This study LSY19 MATα ade3::pgal-ho 2 LSY3184 MATa/MATα This study 5of6
6 Table S1. Cont. Strain Genotype Source LSY3185 MATa/MATα sae2::kanmx6/sae2::kanmx6 This study LSY3186 MATa/MATα mre11-p11l-natmx4/mre11-p11l-natmx4 This study LSY3187 MATa/MATα sae2::kanmx6/sae2::kanmx6 This study MRE11/mre11-P11L-NatMX4 LSY3188 MATa/MATα sae2::kanmx6/sae2::kanmx6 This study mre11-p11l-natmx4/mre11-p11l-natmx4 LSY3189 MATa/MATα sae2::kanmx6/ sae2::kanmx6 This study mre11- P11L-NatMX4/mre11::LEU2 LSY319 MATa/MATα sae2::kanmx6/sae2::kanmx6 MRE11/mre11::LEU2 This study Most strains are of the W33 background (trp1-1 his3-11,15 can1-1 ura3-1 leu2-3,112 ade2-1 RAD5), only the mating type and differences from this genotype are shown. The full genotypes are shown for all of the non-w33 strains (ALE94, ALE18, LSY293, LSY2931, YMV45, LSY1951, LSY389, and LSY386). 1. Zou H, Rothstein R (1997) Holliday junctions accumulate in replication mutants via a RecA homolog-independent mechanism. Cell 9(1): Mimitou EP, Symington LS (21) Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J 29(19): Moreau S, Ferguson JR, Symington LS (1999) The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol Cell Biol 19(1): Lobachev KS, Gordenin DA, Resnick MA (22) The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 18(2): Vaze MB, et al. (22) Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Mol Cell 1(2): Clerici M, Mantiero D, Lucchini G, Longhese MP (25) The Saccharomyces cerevisiae Sae2 protein promotes resection and bridging of double strand break ends. J Biol Chem 28(46): Lisby M, Barlow JH, Burgess RC, Rothstein R (24) Choreography of the DNA damage response: Spatiotemporal relationships among checkpoint and repair proteins. Cell 118(6): of6
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11355 Supplemental Table 1. Gene targeting efficiency in mutants of nonessential genes. This table is presented as a separate Excel file. Supplemental Table 2. Genes with altered expression
doi:10.1038/nature11355 Supplemental Table 1. Gene targeting efficiency in mutants of nonessential genes. This table is presented as a separate Excel file. Supplemental Table 2. Genes with altered expression
Supplementary Table 1a. Number of crossover (CO), noncrossover (NCO) and breakinduced replication (BIR) events in red/white sectored colonies
 Supplementary Table 1a. Number of crossover (CO), noncrossover (NCO) and breakinduced replication (BIR) events in red/white sectored colonies CO NCO BIR Total WT 90 939 15 144 yen1 114 34 4 44 114 55 15
Supplementary Table 1a. Number of crossover (CO), noncrossover (NCO) and breakinduced replication (BIR) events in red/white sectored colonies CO NCO BIR Total WT 90 939 15 144 yen1 114 34 4 44 114 55 15
SUPPLEMENTARY INFORMATION
 Supplementary Table 1 Components of the RAD6 damage tolerance pathway in yeast and humans S. cerevisiae Homo sapiens Functions Rad6 HHR6A and HHR6B Ubiquitin conjugating enzyme (E2) Rad18 RAD18 Ubiquitin
Supplementary Table 1 Components of the RAD6 damage tolerance pathway in yeast and humans S. cerevisiae Homo sapiens Functions Rad6 HHR6A and HHR6B Ubiquitin conjugating enzyme (E2) Rad18 RAD18 Ubiquitin
Supplemental Material for Xue et al. List. Supplemental Figure legends. Figure S1. Related to Figure 1. Figure S2. Related to Figure 3
 Supplemental Material for Xue et al List Supplemental Figure legends Figure S1. Related to Figure 1 Figure S. Related to Figure 3 Figure S3. Related to Figure 4 Figure S4. Related to Figure 4 Figure S5.
Supplemental Material for Xue et al List Supplemental Figure legends Figure S1. Related to Figure 1 Figure S. Related to Figure 3 Figure S3. Related to Figure 4 Figure S4. Related to Figure 4 Figure S5.
GGTCACTCTGCA!CAATGCG! ATCCC!TATTGGGAT! ACGT!CCAGTGAGACGT! CCCTAT! TGCA!GGTCACTCTGCA! GGGATA! Minor NHEJ Event (Ade- )
 a 12 bp I-ceI I-ceI 12 bp AACAAG! CCAGTGAGACGT! GTTACGC! TAGGGATAACAGGGTAAT!ATAT!ATTACCCTGTTATCCCTA!ACGT! CCAGTGAGACGT! CCCTAT! TTGTTC! GGTCACTCTGCA! CAATGCG!ATCCCTATTGTCCCATTA!TATA!TAATGGGACAATAGGGAT!TGCA!GGTCACTCTGCA!
a 12 bp I-ceI I-ceI 12 bp AACAAG! CCAGTGAGACGT! GTTACGC! TAGGGATAACAGGGTAAT!ATAT!ATTACCCTGTTATCCCTA!ACGT! CCAGTGAGACGT! CCCTAT! TTGTTC! GGTCACTCTGCA! CAATGCG!ATCCCTATTGTCCCATTA!TATA!TAATGGGACAATAGGGAT!TGCA!GGTCACTCTGCA!
Supplementary Figure 1. Nature Structural & Molecular Biology: doi: /nsmb.3494
 Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Detection of MCM-subunit SUMOylation under normal growth conditions. a. Sumoylated forms of MCM subunits show differential shifts when SUMO is attached to differently sized tags.
Supplementary Figure 1 Detection of MCM-subunit SUMOylation under normal growth conditions. a. Sumoylated forms of MCM subunits show differential shifts when SUMO is attached to differently sized tags.
Supplemental Information
 Supplemental Information Supplemental material and methods ATPase assay Mph1 (35 nm) or mph1 D209N (100 nm) was incubated at 30 o C with 1.5 mm [γ- 32 P] ATP and φx174 circular (+) strand DNA (25 µm nucleotides)
Supplemental Information Supplemental material and methods ATPase assay Mph1 (35 nm) or mph1 D209N (100 nm) was incubated at 30 o C with 1.5 mm [γ- 32 P] ATP and φx174 circular (+) strand DNA (25 µm nucleotides)
supplementary information
 DOI: 10.1038/ncb2007 Figure S1 Temperature sensitivity of DNA ligase I alleles. a, SSL204 (CDC9), SSL612 (cdc9-1) and SSL613 (cdc9-2) cells were spotted in ten-fold serial dilutions on YPD plates and incubated
DOI: 10.1038/ncb2007 Figure S1 Temperature sensitivity of DNA ligase I alleles. a, SSL204 (CDC9), SSL612 (cdc9-1) and SSL613 (cdc9-2) cells were spotted in ten-fold serial dilutions on YPD plates and incubated
* + * RecA * + + for RecA-dependent strand + + MIT Department of Biology 7.28, Spring Molecular Biology
 MIT Department of Biology 7.28, Spring 2005 - Molecular Biology 1) Question 1. 1A. You are studying the function of in vitro along with a lab partner. You run several reactions to examine the requirements
MIT Department of Biology 7.28, Spring 2005 - Molecular Biology 1) Question 1. 1A. You are studying the function of in vitro along with a lab partner. You run several reactions to examine the requirements
SUPPLEMENTARY INFORMATION
 doi:1.138/nature11676 Replication template exchange Ori Rearrangement: acentric and/or dicentric Model for HR-dependent restart Restart on wrong template HJ resolution either restores original Restart
doi:1.138/nature11676 Replication template exchange Ori Rearrangement: acentric and/or dicentric Model for HR-dependent restart Restart on wrong template HJ resolution either restores original Restart
Recombination Models. A. The Holliday Junction Model
 A. The Holliday Junction Model Recombination Models Fig. 1 illustrates the formation of a Holliday junction (I) and its branch migration (II).The branch point (the blue X ) at the left in I has moved towards
A. The Holliday Junction Model Recombination Models Fig. 1 illustrates the formation of a Holliday junction (I) and its branch migration (II).The branch point (the blue X ) at the left in I has moved towards
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature10331 Supplemental Figures and Legends Supplementary Figure 1. Pch2-dependent suppression of DSBs at the rdna borders. a, Example of a CHEF gel/southern blot
SUPPLEMENTARY INFORMATION doi:10.1038/nature10331 Supplemental Figures and Legends Supplementary Figure 1. Pch2-dependent suppression of DSBs at the rdna borders. a, Example of a CHEF gel/southern blot
SUPPLEMENTARY MATERIALS
 SUPPLEMENTARY MATERIALS SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1 Condensin is required for condensation of rdna array during starvation (A) Loss of condensin blocks rdna condensation during
SUPPLEMENTARY MATERIALS SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1 Condensin is required for condensation of rdna array during starvation (A) Loss of condensin blocks rdna condensation during
Cover Page. The handle holds various files of this Leiden University dissertation.
 Cover Page The handle http://hdl.handle.net/1887/21009 holds various files of this Leiden University dissertation. Author: Guénolé, Aude Title: Dissection of DNA damage responses using multiconditional
Cover Page The handle http://hdl.handle.net/1887/21009 holds various files of this Leiden University dissertation. Author: Guénolé, Aude Title: Dissection of DNA damage responses using multiconditional
Chapter 11 Homologous Recombination at the Molecular Level. 吳彰哲 (Chang-Jer Wu) Department of Food Science National Taiwan Ocean University
 Chapter 11 Homologous Recombination at the Molecular Level 吳彰哲 (Chang-Jer Wu) Department of Food Science National Taiwan Ocean University 1 Introduction All DNA is recombinant DNA During meiosis homologous
Chapter 11 Homologous Recombination at the Molecular Level 吳彰哲 (Chang-Jer Wu) Department of Food Science National Taiwan Ocean University 1 Introduction All DNA is recombinant DNA During meiosis homologous
Western blot showing expression of mutant Sec63p. The strains used in Figure 7 were treated
 Supplemntary figure legend Western blot showing expression of mutant Sec63p. The strains used in Figure 7 were treated with methionine to repress transcription of genomic SEC63, and total protein extracts
Supplemntary figure legend Western blot showing expression of mutant Sec63p. The strains used in Figure 7 were treated with methionine to repress transcription of genomic SEC63, and total protein extracts
Supplemental Materials and Methods. Yeast strains. Strain genotypes are listed in Table S1. To generate MATainc::LEU2,
 Supplemental Materials and Methods Yeast strains. Strain genotypes are listed in Table S1. To generate MATainc::LEU2, the LEU2 marker was inserted by PCR-mediated recombination (BRACHMANN et al. 1998)
Supplemental Materials and Methods Yeast strains. Strain genotypes are listed in Table S1. To generate MATainc::LEU2, the LEU2 marker was inserted by PCR-mediated recombination (BRACHMANN et al. 1998)
A) (5 points) As the starting step isolate genomic DNA from
 GS Final Exam Spring 00 NAME. bub ts is a recessive temperature sensitive mutation in yeast. At º C bub ts cells grow normally, but at º C they die. Use the information below to clone the wild-type BUB
GS Final Exam Spring 00 NAME. bub ts is a recessive temperature sensitive mutation in yeast. At º C bub ts cells grow normally, but at º C they die. Use the information below to clone the wild-type BUB
NIH Public Access Author Manuscript Nat Struct Mol Biol. Author manuscript; available in PMC 2014 October 01.
 NIH Public Access Author Manuscript Published in final edited form as: Nat Struct Mol Biol. 2014 April ; 21(4): 405 412. doi:10.1038/nsmb.2786. RPA Antagonizes Microhomology-Mediated Repair of DNA Double-Strand
NIH Public Access Author Manuscript Published in final edited form as: Nat Struct Mol Biol. 2014 April ; 21(4): 405 412. doi:10.1038/nsmb.2786. RPA Antagonizes Microhomology-Mediated Repair of DNA Double-Strand
Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and
 Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
7.06 Problem Set Four, 2006
 7.06 Problem Set Four, 2006 1. Explain the molecular mechanism behind each of the following events that occur during the cell cycle, making sure to discuss specific proteins that are involved in making
7.06 Problem Set Four, 2006 1. Explain the molecular mechanism behind each of the following events that occur during the cell cycle, making sure to discuss specific proteins that are involved in making
A RRM1 H2AX DAPI. RRM1 H2AX DAPI Merge. Cont. sirna RRM1
 A H2AX DAPI H2AX DAPI Merge Cont sirna Figure S1: Accumulation of RRM1 at DNA damage sites (A) HeLa cells were subjected to in situ detergent extraction without IR irradiation, and immunostained with the
A H2AX DAPI H2AX DAPI Merge Cont sirna Figure S1: Accumulation of RRM1 at DNA damage sites (A) HeLa cells were subjected to in situ detergent extraction without IR irradiation, and immunostained with the
Yeast strains used in this study Strain genotype source y2007 MATa sld3-600,609,622a-dpb11( )::kanmx, his3::pgal1-
 Supplementary Table Yeast strains used in this study Strain genotype source y2007 MATa sld3-600,609,622a-dpb11(253-764)::kanmx, his3::pgal1-1 4HA-sld2 T84D::HIS3, bar1δ::trp1 YB291 MATa cdc7-4 leu2-3,
Supplementary Table Yeast strains used in this study Strain genotype source y2007 MATa sld3-600,609,622a-dpb11(253-764)::kanmx, his3::pgal1-1 4HA-sld2 T84D::HIS3, bar1δ::trp1 YB291 MATa cdc7-4 leu2-3,
 DOI: 10.1038/ncb3259 A Ismail et al. Supplementary Figure 1 B 60000 45000 SSC 30000 15000 Live cells 0 0 15000 30000 45000 60000 FSC- PARR 60000 45000 PARR Width 30000 FSC- 15000 Single cells 0 0 15000
DOI: 10.1038/ncb3259 A Ismail et al. Supplementary Figure 1 B 60000 45000 SSC 30000 15000 Live cells 0 0 15000 30000 45000 60000 FSC- PARR 60000 45000 PARR Width 30000 FSC- 15000 Single cells 0 0 15000
Schematic representation of the endogenous PALB2 locus and gene-disruption constructs
 Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supplementary Figure 1. Chromosome 3 is devoid of the telomere-proximal subtelomeric
 Supplementary Figure 1. Chromosome 3 is devoid of the telomere-proximal subtelomeric common sequences. (a) Schematic illustration of the telomere-proximal site of subtelomeres. The restriction map is based
Supplementary Figure 1. Chromosome 3 is devoid of the telomere-proximal subtelomeric common sequences. (a) Schematic illustration of the telomere-proximal site of subtelomeres. The restriction map is based
MRC Radiation Oncology and Biology Unit, Harwell, Didcot, Oxfordshire OX11 0RD, United Kingdom
 MOLECULAR AND CELLULAR BIOLOGY, Nov. 2007, p. 7745 7757 Vol. 27, No. 21 0270-7306/07/$08.00 0 doi:10.1128/mcb.00462-07 Copyright 2007, American Society for Microbiology. All Rights Reserved. Break-Induced
MOLECULAR AND CELLULAR BIOLOGY, Nov. 2007, p. 7745 7757 Vol. 27, No. 21 0270-7306/07/$08.00 0 doi:10.1128/mcb.00462-07 Copyright 2007, American Society for Microbiology. All Rights Reserved. Break-Induced
Figure S1, related to Figure 1. Characterization of biosensor behavior in vivo.
 Developmental Cell, Volume 23 Supplemental Information Separase Biosensor Reveals that Cohesin Cleavage Timing Depends on Phosphatase PP2A Cdc55 Regulation Gilad Yaakov, Kurt Thorn, and David O. Morgan
Developmental Cell, Volume 23 Supplemental Information Separase Biosensor Reveals that Cohesin Cleavage Timing Depends on Phosphatase PP2A Cdc55 Regulation Gilad Yaakov, Kurt Thorn, and David O. Morgan
Gene Targeting and Hit-and-Run Transformation. Senior Thesis. Presented to. The Faculty of the School of Arts and Sciences Brandeis University
 Gene Targeting and Hit-and-Run Transformation Senior Thesis Presented to The Faculty of the School of Arts and Sciences Brandeis University Undergraduate Program in Biology James E. Haber, Advisor In partial
Gene Targeting and Hit-and-Run Transformation Senior Thesis Presented to The Faculty of the School of Arts and Sciences Brandeis University Undergraduate Program in Biology James E. Haber, Advisor In partial
Coleman et al., Supplementary Figure 1
 Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Supplemental Data Genetic and Epigenetic Regulation of the FLO Gene Family Generates Cell-Surface Variation in Yeast
 Supplemental Data Genetic and Epigenetic Regulation of the FLO Gene Family Generates Cell-Surface Variation in Yeast S1 Adrian Halme, Stacie Bumgarner, Cora Styles and Gerald R. Fink Yeast Strain Construction
Supplemental Data Genetic and Epigenetic Regulation of the FLO Gene Family Generates Cell-Surface Variation in Yeast S1 Adrian Halme, Stacie Bumgarner, Cora Styles and Gerald R. Fink Yeast Strain Construction
supplementary information
 supplementary information Figure S1 (a) Alignment of the Sgf11 amino acid sequences from S. cerevisiae, Yarrowia lipolytica and S. pombe with Sgf73 sequences from S. cerevisiae, S. pombe and Candida albicans.
supplementary information Figure S1 (a) Alignment of the Sgf11 amino acid sequences from S. cerevisiae, Yarrowia lipolytica and S. pombe with Sgf73 sequences from S. cerevisiae, S. pombe and Candida albicans.
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11326 Supplementary Figure 1: Histone exchange increases over the ORF in a set2 mutant. (a) Gene average analysis. Schematic representation of the bin distribution over the coding and
doi:10.1038/nature11326 Supplementary Figure 1: Histone exchange increases over the ORF in a set2 mutant. (a) Gene average analysis. Schematic representation of the bin distribution over the coding and
SUPPLEMENTARY INFORMATION
 Supplementary Table 1. Oligonucleotides used in this study. Name Description Size Structure from 5 Sequence a 80STUECOV.12D 80 [D 34]-ins::D 12-[D 34] 5'-TGTTAGGTGCTGTGGGTGGTCCTAAATGGGGTAC-AGGCCTGATATC-CGGTAGTGTTAGACCTGAACAAGGTTTACTAAAA
Supplementary Table 1. Oligonucleotides used in this study. Name Description Size Structure from 5 Sequence a 80STUECOV.12D 80 [D 34]-ins::D 12-[D 34] 5'-TGTTAGGTGCTGTGGGTGGTCCTAAATGGGGTAC-AGGCCTGATATC-CGGTAGTGTTAGACCTGAACAAGGTTTACTAAAA
An mrna decapping mutant deficient in P body assembly limits mrna stabilization in response to osmotic stress
 An mrna decapping mutant deficient in P body assembly limits mrna stabilization in response to osmotic stress SUPPLEMENTAL INFORMATION Susanne Huch and Tracy Nissan Supplementary Information Figure S1.
An mrna decapping mutant deficient in P body assembly limits mrna stabilization in response to osmotic stress SUPPLEMENTAL INFORMATION Susanne Huch and Tracy Nissan Supplementary Information Figure S1.
Tbf1 and Vid22 promote resection and nonhomologous end joining of DNA double-strand break ends
 The EMBO Journal (2013) 32, 275 289 www.embojournal.org Tbf1 and Vid22 promote resection and nonhomologous end joining of DNA double-strand break ends THE EMBO JOURNAL Diego Bonetti, Savani Anbalagan,
The EMBO Journal (2013) 32, 275 289 www.embojournal.org Tbf1 and Vid22 promote resection and nonhomologous end joining of DNA double-strand break ends THE EMBO JOURNAL Diego Bonetti, Savani Anbalagan,
Localized Histone Acetylation and Deacetylation Triggered by the Homologous Recombination Pathway of Double-Strand DNA Repair
 MOLECULAR AND CELLULAR BIOLOGY, June 2005, p. 4903 4913 Vol. 25, No. 12 0270-7306/05/$08.00 0 doi:10.1128/mcb.25.12.4903 4913.2005 Copyright 2005, American Society for Microbiology. All Rights Reserved.
MOLECULAR AND CELLULAR BIOLOGY, June 2005, p. 4903 4913 Vol. 25, No. 12 0270-7306/05/$08.00 0 doi:10.1128/mcb.25.12.4903 4913.2005 Copyright 2005, American Society for Microbiology. All Rights Reserved.
DNA polymerases δ and λ cooperate in repairing double-strand breaks by microhomology-mediated end-joining in Saccharomyces cerevisiae
 DNA polymerases δ and λ cooperate in repairing double-strand breaks by microhomology-mediated end-joining in Saccharomyces cerevisiae Damon Meyer a,1, Becky Xu Hua Fu a,2, and Wolf-Dietrich Heyer a,b,3
DNA polymerases δ and λ cooperate in repairing double-strand breaks by microhomology-mediated end-joining in Saccharomyces cerevisiae Damon Meyer a,1, Becky Xu Hua Fu a,2, and Wolf-Dietrich Heyer a,b,3
Supplemental Figure legends Figure S1. (A) (B) (C) (D) Figure S2. Figure S3. (A-E) Figure S4. Figure S5. (A, C, E, G, I) (B, D, F, H, Figure S6.
 Supplemental Figure legends Figure S1. Map-based cloning and complementation testing for ZOP1. (A) ZOP1 was mapped to a ~273-kb interval on Chromosome 1. In the interval, a single-nucleotide G to A substitution
Supplemental Figure legends Figure S1. Map-based cloning and complementation testing for ZOP1. (A) ZOP1 was mapped to a ~273-kb interval on Chromosome 1. In the interval, a single-nucleotide G to A substitution
Role of RAD52 Epistasis Group Genes in Homologous Recombination and Double-Strand Break Repair
 MICROBIOLOGY AND MOLECULAR BIOLOGY REVIEWS, Dec. 2002, p. 630 670 Vol. 66, No. 4 1092-2172/02/$04.00 0 DOI: 10.1128/MMBR.66.4.630 670.2002 Copyright 2002, American Society for Microbiology. All Rights
MICROBIOLOGY AND MOLECULAR BIOLOGY REVIEWS, Dec. 2002, p. 630 670 Vol. 66, No. 4 1092-2172/02/$04.00 0 DOI: 10.1128/MMBR.66.4.630 670.2002 Copyright 2002, American Society for Microbiology. All Rights
Supplementary Figure Legends
 Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
Materials and Methods: All strains were derivatives of SK1 and are listed in Supplemental Table 1.
 Supplemental Online Material: Materials and Methods: All strains were derivatives of SK1 and are listed in Supplemental Table 1. Constructs: pclb2-3ha-cdc5 and pclb2-3ha-cdc2 strains were constructed by
Supplemental Online Material: Materials and Methods: All strains were derivatives of SK1 and are listed in Supplemental Table 1. Constructs: pclb2-3ha-cdc5 and pclb2-3ha-cdc2 strains were constructed by
SUPPLEMENTARY INFORMATION
 Figure S1: Activation of the ATM pathway by I-PpoI. A. HEK293T cells were either untransfected, vector transfected, transfected with an I-PpoI expression vector, or subjected to 2Gy γ-irradiation. 24 hrs
Figure S1: Activation of the ATM pathway by I-PpoI. A. HEK293T cells were either untransfected, vector transfected, transfected with an I-PpoI expression vector, or subjected to 2Gy γ-irradiation. 24 hrs
The Mre11-Rad50-Xrs2 Protein Complex Facilitates Homologous Recombination-Based Double-Strand Break Repair in Saccharomyces cerevisiae
 MOLECULAR AND CELLULAR BIOLOGY, Nov. 1999, p. 7681 7687 Vol. 19, No. 11 0270-7306/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. The Mre11-Rad50-Xrs2 Protein Complex
MOLECULAR AND CELLULAR BIOLOGY, Nov. 1999, p. 7681 7687 Vol. 19, No. 11 0270-7306/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. The Mre11-Rad50-Xrs2 Protein Complex
SUPPLEMENTAL MATERIAL. Supplemental material contains Supplemental Figure Legends and Supplemental Figures 1 to
 SUPPLEMENTAL MATERIAL Supplemental material contains Supplemental Figure Legends and Supplemental Figures 1 to 6. SUPPLEMENTAL FIGURE LEGENDS Supplemental Figure 1. Overview of the mechanisms by which
SUPPLEMENTAL MATERIAL Supplemental material contains Supplemental Figure Legends and Supplemental Figures 1 to 6. SUPPLEMENTAL FIGURE LEGENDS Supplemental Figure 1. Overview of the mechanisms by which
% Viability. isw2 ino isw2 ino isw2 ino isw2 ino mM HU 4-NQO CPT
 a Drug concentration b 1.3% MMS nhp1 nhp1 8 nhp1 mag1.5% MMS.3% MMS nhp1 nhp1 ino8 9 ino8 9 % Viability 4.5% MMS ino8 9 ino8 9 2.5.1.15 % MMS c d nhp1 nhp1 nhp1 nhp1 nhp1 nhp1 Control (YPD) γ IR (1 gy)
a Drug concentration b 1.3% MMS nhp1 nhp1 8 nhp1 mag1.5% MMS.3% MMS nhp1 nhp1 ino8 9 ino8 9 % Viability 4.5% MMS ino8 9 ino8 9 2.5.1.15 % MMS c d nhp1 nhp1 nhp1 nhp1 nhp1 nhp1 Control (YPD) γ IR (1 gy)
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3209 Supplementary Figure 1 IR induces the association of FH with chromatin. a, U2OS cells synchronized by thymidine double block (2 mm) underwent no release (G1 phase) or release for 2
DOI: 10.1038/ncb3209 Supplementary Figure 1 IR induces the association of FH with chromatin. a, U2OS cells synchronized by thymidine double block (2 mm) underwent no release (G1 phase) or release for 2
Supplementary Figure legends
 Supplementary Figure legends Figure S. Histone shuffle strain construction and characterization. A. Schematic representing the histone shuffle strain. Both genomic copies of the and genes are deleted (genes
Supplementary Figure legends Figure S. Histone shuffle strain construction and characterization. A. Schematic representing the histone shuffle strain. Both genomic copies of the and genes are deleted (genes
Single Cell Visualization of the DNA repair mechanism in vivo
 Single Cell Visualization of the DNA repair mechanism in vivo Jen Makridakis, Jane kondev, Jim Haber Azadeh Samadani Department of Physics Brandeis University Objectives DNA double-strand break constitutes
Single Cell Visualization of the DNA repair mechanism in vivo Jen Makridakis, Jane kondev, Jim Haber Azadeh Samadani Department of Physics Brandeis University Objectives DNA double-strand break constitutes
HEK293T. Fig. 1 in the
 Supplementary Information Supplementary Figure 1 Zinc uptake assay of hzip4 and hzip4-δecd transiently expressed in HEK293T cells. The results of one representative e experiment are shown in Fig. 1 in
Supplementary Information Supplementary Figure 1 Zinc uptake assay of hzip4 and hzip4-δecd transiently expressed in HEK293T cells. The results of one representative e experiment are shown in Fig. 1 in
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the
 Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
3.1 The Role of the Enzymatic Activity of Sir2 for Efficient. The Sir proteins are recruited to DNA by site-specific DNA-binding proteins and
 35 3 RESULTS 3.1 The Role of the Enzymatic Activity of Sir2 for Efficient Association of the SIR Complex with DNA The Sir proteins are recruited to DNA by site-specific DNA-binding proteins and subsequently
35 3 RESULTS 3.1 The Role of the Enzymatic Activity of Sir2 for Efficient Association of the SIR Complex with DNA The Sir proteins are recruited to DNA by site-specific DNA-binding proteins and subsequently
SUPPLEMENTARY INFORMATION
 Ca 2+ /calmodulin Regulates Salicylic Acid-mediated Plant Immunity Liqun Du, Gul S. Ali, Kayla A. Simons, Jingguo Hou, Tianbao Yang, A.S.N. Reddy and B. W. Poovaiah * *To whom correspondence should be
Ca 2+ /calmodulin Regulates Salicylic Acid-mediated Plant Immunity Liqun Du, Gul S. Ali, Kayla A. Simons, Jingguo Hou, Tianbao Yang, A.S.N. Reddy and B. W. Poovaiah * *To whom correspondence should be
Supplementary Figures 1-12
 Supplementary Figures 1-12 Supplementary Figure 1. The specificity of anti-abi1 antibody. Total Proteins extracted from the wild type seedlings or abi1-3 null mutant seedlings were used for immunoblotting
Supplementary Figures 1-12 Supplementary Figure 1. The specificity of anti-abi1 antibody. Total Proteins extracted from the wild type seedlings or abi1-3 null mutant seedlings were used for immunoblotting
Supplementary Methods
 Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Supplemental Materials and Methods
 Supplemental Materials and Methods Proteins and reagents Proteins were purified as described previously: RecA, RecQ, and SSB proteins (Harmon and Kowalczykowski 1998); RecF protein (Morimatsu and Kowalczykowski
Supplemental Materials and Methods Proteins and reagents Proteins were purified as described previously: RecA, RecQ, and SSB proteins (Harmon and Kowalczykowski 1998); RecF protein (Morimatsu and Kowalczykowski
Supporting Information
 Supporting Information Kilian et al. 10.1073/pnas.1105861108 SI Materials and Methods Determination of the Electric Field Strength Required for Successful Electroporation. The transformation construct
Supporting Information Kilian et al. 10.1073/pnas.1105861108 SI Materials and Methods Determination of the Electric Field Strength Required for Successful Electroporation. The transformation construct
Yeast Mating Type. DefinitionandMutantPhenotype. Secondary article. Beth A Montelone, Kansas State University, Manhattan, Kansas, USA
 Beth A Montelone, Kansas State University, Manhattan, Kansas, USA Yeast mating type is determined by alleles of a single genetic locus that encodes regulatory proteins governing genes encoding mating pheromones,
Beth A Montelone, Kansas State University, Manhattan, Kansas, USA Yeast mating type is determined by alleles of a single genetic locus that encodes regulatory proteins governing genes encoding mating pheromones,
The RNA binding protein Npl3 promotes resection of DNA double-strand breaks by regulating the levels of Exo1
 6530 6545 Nucleic Acids Research, 2017, Vol. 45, No. 11 Published online 2 May 2017 doi: 10.1093/nar/gkx347 The RNA binding protein Npl3 promotes resection of DNA double-strand breaks by regulating the
6530 6545 Nucleic Acids Research, 2017, Vol. 45, No. 11 Published online 2 May 2017 doi: 10.1093/nar/gkx347 The RNA binding protein Npl3 promotes resection of DNA double-strand breaks by regulating the
Name Genotype Reference
 Supplemental Data Supplemental Table 1 S. cerevisiae strains. Name Genotype Reference RMY200 UKY403 W303-1a MATa ade2-101 (och) his3 200 lys2-801 (amp) trp1 901 ura3-52 hht1,hhf1::leu2 hht2,hhf2::his3
Supplemental Data Supplemental Table 1 S. cerevisiae strains. Name Genotype Reference RMY200 UKY403 W303-1a MATa ade2-101 (och) his3 200 lys2-801 (amp) trp1 901 ura3-52 hht1,hhf1::leu2 hht2,hhf2::his3
Chapter 11 DNA Replication and Recombination
 Chapter 11 DNA Replication and Recombination Copyright Copyright 2009 Pearson 2009 Pearson Education, Education, Inc. Inc. 11.1 DNA is reproduced by Semiconservative Replication The complementarity of
Chapter 11 DNA Replication and Recombination Copyright Copyright 2009 Pearson 2009 Pearson Education, Education, Inc. Inc. 11.1 DNA is reproduced by Semiconservative Replication The complementarity of
C24. ros1-1. ros1-1 rdm18-1. ros1-1 rdm18-2. ros1-1 nrpe1
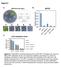 Figure S1 Methylation level 1 0.8 0.6 0.4 0.2 0 p35s methylation levels 24 ros1-1 ros1-1 rdm18-1 ros1-1 rdm18-2 ros1-1 nrpe1 mg mhg mhh Figure S1. RM18/PKL promotes silencing at the p35s-npt II transgene
Figure S1 Methylation level 1 0.8 0.6 0.4 0.2 0 p35s methylation levels 24 ros1-1 ros1-1 rdm18-1 ros1-1 rdm18-2 ros1-1 nrpe1 mg mhg mhh Figure S1. RM18/PKL promotes silencing at the p35s-npt II transgene
The Biotechnology Toolbox
 Chapter 15 The Biotechnology Toolbox Cutting and Pasting DNA Cutting DNA Restriction endonuclease or restriction enzymes Cellular protection mechanism for infected foreign DNA Recognition and cutting specific
Chapter 15 The Biotechnology Toolbox Cutting and Pasting DNA Cutting DNA Restriction endonuclease or restriction enzymes Cellular protection mechanism for infected foreign DNA Recognition and cutting specific
PLASMID INTEGRATION IN YEAST: CONCEPTIONS AND MISSCONCEPTIONS
 Current Studies of Biotechnology Volume II. - Environment PLASMID INTEGRATION IN YEAST: CONCEPTIONS AND MISSCONCEPTIONS ZORAN ZGAGA, KREŠIMIR GJURAČIĆ 1, IVAN-KREŠIMIR SVETEC, PETAR T. MITRIKESKI AND SANDRA
Current Studies of Biotechnology Volume II. - Environment PLASMID INTEGRATION IN YEAST: CONCEPTIONS AND MISSCONCEPTIONS ZORAN ZGAGA, KREŠIMIR GJURAČIĆ 1, IVAN-KREŠIMIR SVETEC, PETAR T. MITRIKESKI AND SANDRA
DNA Recombination II Biochemistry 302. Bob Kelm February 4, 2004
 DNA Recombination II Biochemistry 302 Bob Kelm February 4, 2004 Core interactions at junction crossover (A 6 C 7 C 8 or R 6 -C 7 -Y 8 motif) parallel stacked-x WC H-bonds red dots ACC H-bonds blue dots
DNA Recombination II Biochemistry 302 Bob Kelm February 4, 2004 Core interactions at junction crossover (A 6 C 7 C 8 or R 6 -C 7 -Y 8 motif) parallel stacked-x WC H-bonds red dots ACC H-bonds blue dots
SUPPLEMENTARY INFORMATION
 doi: 1.138/nature7652 Supplementary Figure legends Figure 1. Diagrammatic representation of the rdna locus on yeast chromosome XII and the processing pathway from 35S full length nascent rrna to mature
doi: 1.138/nature7652 Supplementary Figure legends Figure 1. Diagrammatic representation of the rdna locus on yeast chromosome XII and the processing pathway from 35S full length nascent rrna to mature
Analysis of gene function
 Genome 371, 22 February 2010, Lecture 12 Analysis of gene function Gene knockouts PHASE TWO: INTERPRETATION I THINK I FOUND A CORNER PIECE. 3 BILLION PIECES Analysis of a disease gene Gene knockout or
Genome 371, 22 February 2010, Lecture 12 Analysis of gene function Gene knockouts PHASE TWO: INTERPRETATION I THINK I FOUND A CORNER PIECE. 3 BILLION PIECES Analysis of a disease gene Gene knockout or
Autophagy induction [h]
![Autophagy induction [h] Autophagy induction [h]](/thumbs/80/80564099.jpg) A SD-N [h]: SD-N [h]: 1 2 4 1 2 4 1 2 4 -GFP 1 2 4 percentage GFP-Atg8 [%] 1 9 8 7 6 5 4 3 2 1 1 2 4 Autophagy induction [h] -13xmyc SD-N [h]: 1 2 4 -GFP -13xmyc 1 prape1 mape1 - - GFP - 13xmyc mape1 [%]
A SD-N [h]: SD-N [h]: 1 2 4 1 2 4 1 2 4 -GFP 1 2 4 percentage GFP-Atg8 [%] 1 9 8 7 6 5 4 3 2 1 1 2 4 Autophagy induction [h] -13xmyc SD-N [h]: 1 2 4 -GFP -13xmyc 1 prape1 mape1 - - GFP - 13xmyc mape1 [%]
SUPPLEMENTARY INFORMATION FIGURE LEGENDS
 SUPPLEMENTARY INFORMATION FIGURE LEGENDS Fig. S1. Radiation-induced phosphorylation of Rad50 at a specific site. A. Rad50 is an in vitro substrate for ATM. A series of Rad50-GSTs covering the entire molecule
SUPPLEMENTARY INFORMATION FIGURE LEGENDS Fig. S1. Radiation-induced phosphorylation of Rad50 at a specific site. A. Rad50 is an in vitro substrate for ATM. A series of Rad50-GSTs covering the entire molecule
Supporting information
 Supporting information Construction of strains and plasmids To create ptc67, a PCR product obtained with primers cc2570-162f (gcatgggcaagcttgaggacggcgtcatgt) and cc2570+512f (gaggccgtggtaccatagaggcgggcg),
Supporting information Construction of strains and plasmids To create ptc67, a PCR product obtained with primers cc2570-162f (gcatgggcaagcttgaggacggcgtcatgt) and cc2570+512f (gaggccgtggtaccatagaggcgggcg),
Supplemental Data. Hu et al. Plant Cell (2017) /tpc
 1 2 3 4 Supplemental Figure 1. DNA gel blot analysis of homozygous transgenic plants. (Supports Figure 1.) 5 6 7 8 Rice genomic DNA was digested with the restriction enzymes EcoRⅠ and BamHⅠ. Lanes in the
1 2 3 4 Supplemental Figure 1. DNA gel blot analysis of homozygous transgenic plants. (Supports Figure 1.) 5 6 7 8 Rice genomic DNA was digested with the restriction enzymes EcoRⅠ and BamHⅠ. Lanes in the
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/291/ra78/dc1 Supplementary Materials for Regulation of Yeast G Protein Signaling by the Kinases That Activate the AMPK Homolog Snf1 Sarah T. Clement, Gauri Dixit,
www.sciencesignaling.org/cgi/content/full/6/291/ra78/dc1 Supplementary Materials for Regulation of Yeast G Protein Signaling by the Kinases That Activate the AMPK Homolog Snf1 Sarah T. Clement, Gauri Dixit,
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Impaired ITS2 processing results in mislocalization of foot-factors to the cytoplasm. (a) Schematic representation of ITS2 processing. The endonuclease Las1 initiates ITS2 processing
Supplementary Figure 1 Impaired ITS2 processing results in mislocalization of foot-factors to the cytoplasm. (a) Schematic representation of ITS2 processing. The endonuclease Las1 initiates ITS2 processing
Your first goal is to determine the order of assembly of the 3 DNA polymerases during initiation.
 MIT Department of Biology 7.28, Spring 2005 - Molecular Biology 1 Question 1 You are studying the mechanism of DNA polymerase loading at eukaryotic DNA replication origins. Unlike the situation in bacteria,
MIT Department of Biology 7.28, Spring 2005 - Molecular Biology 1 Question 1 You are studying the mechanism of DNA polymerase loading at eukaryotic DNA replication origins. Unlike the situation in bacteria,
sicker; i.e. to test whether we can reverse the mutant phenotype of mec1-100 when the suppressor gene is ectopically over-expressed in mec1-100.
 Internship Report ~FMI (Basel, Switzerland)~ Kyoto University Faculty of Medicine 1 st grade Yuiko Hirata This summer I went to the FMI (Friedrich Miescher Institute for Biomedical Research) in Basel.
Internship Report ~FMI (Basel, Switzerland)~ Kyoto University Faculty of Medicine 1 st grade Yuiko Hirata This summer I went to the FMI (Friedrich Miescher Institute for Biomedical Research) in Basel.
7.03 Problem Set 2 Due before 5 PM on Friday, September 29 Hand in answers in recitation section or in the box outside of
 7.03 Problem Set 2 Due before 5 PM on Friday, September 29 Hand in answers in recitation section or in the box outside of 68-120 1. Hemophilia A is a X-linked recessive disorder characterized by dysfunctional
7.03 Problem Set 2 Due before 5 PM on Friday, September 29 Hand in answers in recitation section or in the box outside of 68-120 1. Hemophilia A is a X-linked recessive disorder characterized by dysfunctional
Biology 105: Introduction to Genetics PRACTICE FINAL EXAM Part I: Definitions. Homology: Reverse transcriptase. Allostery: cdna library
 Biology 105: Introduction to Genetics PRACTICE FINAL EXAM 2006 Part I: Definitions Homology: Reverse transcriptase Allostery: cdna library Transformation Part II Short Answer 1. Describe the reasons for
Biology 105: Introduction to Genetics PRACTICE FINAL EXAM 2006 Part I: Definitions Homology: Reverse transcriptase Allostery: cdna library Transformation Part II Short Answer 1. Describe the reasons for
R1 12 kb R1 4 kb R1. R1 10 kb R1 2 kb R1 4 kb R1
 Bcor101 Sample questions Midterm 3 1. The maps of the sites for restriction enzyme EcoR1 (R1) in the wild type and mutated cystic fibrosis genes are shown below: Wild Type R1 12 kb R1 4 kb R1 _ _ CF probe
Bcor101 Sample questions Midterm 3 1. The maps of the sites for restriction enzyme EcoR1 (R1) in the wild type and mutated cystic fibrosis genes are shown below: Wild Type R1 12 kb R1 4 kb R1 _ _ CF probe
A Modified Digestion-Circularization PCR (DC-PCR) Approach to Detect Hypermutation- Associated DNA Double-Strand Breaks
 A Modified Digestion-Circularization PCR (DC-PCR) Approach to Detect Hypermutation- Associated DNA Double-Strand Breaks SARAH K. DICKERSON AND F. NINA PAPAVASILIOU Laboratory of Lymphocyte Biology, The
A Modified Digestion-Circularization PCR (DC-PCR) Approach to Detect Hypermutation- Associated DNA Double-Strand Breaks SARAH K. DICKERSON AND F. NINA PAPAVASILIOU Laboratory of Lymphocyte Biology, The
Defective break-induced replication leads to half-crossovers in Saccharomyces cerevisiae.
 Genetics: Published Articles Ahead of Print, published on August 9, 2008 as 10.1534/genetics.108.087940 Defective break-induced replication leads to half-crossovers in Saccharomyces cerevisiae. Angela
Genetics: Published Articles Ahead of Print, published on August 9, 2008 as 10.1534/genetics.108.087940 Defective break-induced replication leads to half-crossovers in Saccharomyces cerevisiae. Angela
1A) What is different about the resulting resection of ends by these two complexes?
 MIT Department of Biology 7.28, Spring 2005 - Molecular Biology Question 1. Your lab is studying the MRX complex in S. pombe, otherwise known as fission yeast. For your dissertation work, you are studying
MIT Department of Biology 7.28, Spring 2005 - Molecular Biology Question 1. Your lab is studying the MRX complex in S. pombe, otherwise known as fission yeast. For your dissertation work, you are studying
Regulation of ARE transcript 3 end processing by the. yeast Cth2 mrna decay factor
 Regulation of ARE transcript 3 end processing by the yeast Cth2 mrna decay factor Manoël Prouteau, Marie-Claire Daugeron and Bertrand Séraphin Supplementary Information Material and Methods Plasmid construction
Regulation of ARE transcript 3 end processing by the yeast Cth2 mrna decay factor Manoël Prouteau, Marie-Claire Daugeron and Bertrand Séraphin Supplementary Information Material and Methods Plasmid construction
_ DNA absorbs light at 260 wave length and it s a UV range so we cant see DNA, we can see DNA only by staining it.
 * GEL ELECTROPHORESIS : its a technique aim to separate DNA in agel based on size, in this technique we add a sample of DNA in a wells in the gel, then we turn on the electricity, the DNA will travel in
* GEL ELECTROPHORESIS : its a technique aim to separate DNA in agel based on size, in this technique we add a sample of DNA in a wells in the gel, then we turn on the electricity, the DNA will travel in
SUPPLEMENTARY INFORMATION
 doi:.8/nature979 Financial support: CS is a Royal Swedish Academy of Sciences Research Fellow supported by Knut and Alice Wallenbergs Foundation. This work was financed European research council (ERC starting
doi:.8/nature979 Financial support: CS is a Royal Swedish Academy of Sciences Research Fellow supported by Knut and Alice Wallenbergs Foundation. This work was financed European research council (ERC starting
Supplemental Data. Cui et al. (2012). Plant Cell /tpc a b c d. Stem UBC32 ACTIN
 A Root Stem Leaf Flower Silique Senescence leaf B a b c d UBC32 ACTIN C * Supplemental Figure 1. Expression Pattern and Protein Sequence of UBC32 Homologues in Yeast, Human, and Arabidopsis. (A) Expression
A Root Stem Leaf Flower Silique Senescence leaf B a b c d UBC32 ACTIN C * Supplemental Figure 1. Expression Pattern and Protein Sequence of UBC32 Homologues in Yeast, Human, and Arabidopsis. (A) Expression
Supplemental Data. Mec1/Tel1 Phosphorylation of the INO80. Chromatin Remodeling Complex Influences. DNA Damage Checkpoint Responses
 Cell, Volume 130 Supplemental Data Mec1/Tel1 Phosphorylation of the INO80 Chromatin Remodeling Complex Influences DNA Damage Checkpoint Responses Ashby J. Morrison, Jung-Ae Kim, Maria D. Person, Jessica
Cell, Volume 130 Supplemental Data Mec1/Tel1 Phosphorylation of the INO80 Chromatin Remodeling Complex Influences DNA Damage Checkpoint Responses Ashby J. Morrison, Jung-Ae Kim, Maria D. Person, Jessica
Can a cross-over event occur between genes A and D? Briefly support your answer.
 Name Section 7.014 Problem Set 5 Please print out this problem set and record your answers on the printed copy. Answers to this problem set are to be turned in to the box outside 68-120 by 5:00pm on Friday
Name Section 7.014 Problem Set 5 Please print out this problem set and record your answers on the printed copy. Answers to this problem set are to be turned in to the box outside 68-120 by 5:00pm on Friday
Juan Lucas Argueso, Amanda Wraith Kijas, Sumeet Sarin, Julie Heck, Marc Waase, and Eric Alani*
 MOLECULAR AND CELLULAR BIOLOGY, Feb. 2003, p. 873 886 Vol. 23, No. 3 0270-7306/03/$08.00 0 DOI: 10.1128/MCB.23.3.873 886.2003 Copyright 2003, American Society for Microbiology. All Rights Reserved. Systematic
MOLECULAR AND CELLULAR BIOLOGY, Feb. 2003, p. 873 886 Vol. 23, No. 3 0270-7306/03/$08.00 0 DOI: 10.1128/MCB.23.3.873 886.2003 Copyright 2003, American Society for Microbiology. All Rights Reserved. Systematic
AGATCCACTAGTTCTAGAGCTCAACTTGTTGGTTTGTCTTT TGTCCAAAAACGGTTTCCAG TGCCTTTCTGTTCGATTGAT
 1 Table S1. PCR primers Primer Use Sequence (5 3 ) JC17 SAT1 flipper GGCCCCCCCTCGAGGAAGTT JC18 SAT1 flipper GCTCTAGAACTAGTGGATCT JC57 5 NCR of CNA1 AATGATGGAGTTTATGGGAA JC58 5 NCR of CNA1 AACTTCCTCGAGGGGGGGCCGAAAAAAAAAAAGGGGGGGG
1 Table S1. PCR primers Primer Use Sequence (5 3 ) JC17 SAT1 flipper GGCCCCCCCTCGAGGAAGTT JC18 SAT1 flipper GCTCTAGAACTAGTGGATCT JC57 5 NCR of CNA1 AATGATGGAGTTTATGGGAA JC58 5 NCR of CNA1 AACTTCCTCGAGGGGGGGCCGAAAAAAAAAAAGGGGGGGG
Accuracy of DNA Repair During Replication in Saccharomyces Cerevisiae
 McNair Scholars Research Journal Volume 9 Issue 1 Article 6 Accuracy of DNA Repair During Replication in Saccharomyces Cerevisiae Eastern Michigan University, mdunn13@emich.edu Follow this and additional
McNair Scholars Research Journal Volume 9 Issue 1 Article 6 Accuracy of DNA Repair During Replication in Saccharomyces Cerevisiae Eastern Michigan University, mdunn13@emich.edu Follow this and additional
Enzyme that uses RNA as a template to synthesize a complementary DNA
 Biology 105: Introduction to Genetics PRACTICE FINAL EXAM 2006 Part I: Definitions Homology: Comparison of two or more protein or DNA sequence to ascertain similarities in sequences. If two genes have
Biology 105: Introduction to Genetics PRACTICE FINAL EXAM 2006 Part I: Definitions Homology: Comparison of two or more protein or DNA sequence to ascertain similarities in sequences. If two genes have
Bacterial DNA replication
 Bacterial DNA replication Summary: What problems do these proteins solve? Tyr OH attacks PO4 and forms a covalent intermediate Structural changes in the protein open the gap by 20 Å! 1 Summary: What problems
Bacterial DNA replication Summary: What problems do these proteins solve? Tyr OH attacks PO4 and forms a covalent intermediate Structural changes in the protein open the gap by 20 Å! 1 Summary: What problems
MID-TERM EXAMINATION
 PLNT3140 INTRODUCTORY CYTOGENETICS MID-TERM EXAMINATION 1 p.m. to 2:15 p.m. Thursday, October 18, 2012 Answer any combination of questions totalling to exactly 100 points. If you answer questions totalling
PLNT3140 INTRODUCTORY CYTOGENETICS MID-TERM EXAMINATION 1 p.m. to 2:15 p.m. Thursday, October 18, 2012 Answer any combination of questions totalling to exactly 100 points. If you answer questions totalling
BurrH: a new modular DNA binding protein for genome engineering
 Supplementary information for: BurrH: a new modular protein for genome engineering Alexandre Juillerat, Claudia Bertonati, Gwendoline Dubois, Valérie Guyot, Séverine Thomas, Julien Valton, Marine Beurdeley,
Supplementary information for: BurrH: a new modular protein for genome engineering Alexandre Juillerat, Claudia Bertonati, Gwendoline Dubois, Valérie Guyot, Séverine Thomas, Julien Valton, Marine Beurdeley,
ET - Recombination. Introduction
 GENERAL & APPLIED GENETICS Geert Van Haute August 2003 ET - Recombination Introduction Homologous recombination is of importance to a variety of cellular processes, including the maintenance of genomic
GENERAL & APPLIED GENETICS Geert Van Haute August 2003 ET - Recombination Introduction Homologous recombination is of importance to a variety of cellular processes, including the maintenance of genomic
Supplementary Figures and Figure legends
 Supplementary Figures and Figure legends 3 Supplementary Figure 1. Conditional targeting construct for the murine Satb1 locus with a modified FLEX switch. Schematic of the wild type Satb1 locus; the conditional
Supplementary Figures and Figure legends 3 Supplementary Figure 1. Conditional targeting construct for the murine Satb1 locus with a modified FLEX switch. Schematic of the wild type Satb1 locus; the conditional
7.014 Quiz II Handout
 7.014 Quiz II Handout Quiz II: Wednesday, March 17 12:05-12:55 54-100 **This will be a closed book exam** Quiz Review Session: Friday, March 12 7:00-9:00 pm room 54-100 Open Tutoring Session: Tuesday,
7.014 Quiz II Handout Quiz II: Wednesday, March 17 12:05-12:55 54-100 **This will be a closed book exam** Quiz Review Session: Friday, March 12 7:00-9:00 pm room 54-100 Open Tutoring Session: Tuesday,
DNA Metabolism. I. DNA Replication. A. Template concept: 1. How can you make a copy of a molecule? 2. Complementary Hydrogen bonding
 DNA Metabolism I. DNA Replication A. Template concept: 1. How can you make a copy of a molecule? 2. Complementary Hydrogen bonding B. DNA replication follows a set of fundamental rules 1. Semiconservative
DNA Metabolism I. DNA Replication A. Template concept: 1. How can you make a copy of a molecule? 2. Complementary Hydrogen bonding B. DNA replication follows a set of fundamental rules 1. Semiconservative
Received 29 September 2005/Returned for modification 9 November 2005/Accepted 21 November 2005
 MOLECULAR AND CELLULAR BIOLOGY, Feb. 2006, p. 1496 1509 Vol. 26, No. 4 0270-7306/06/$08.00 0 doi:10.1128/mcb.26.4.1496 1509.2006 Copyright 2006, American Society for Microbiology. All Rights Reserved.
MOLECULAR AND CELLULAR BIOLOGY, Feb. 2006, p. 1496 1509 Vol. 26, No. 4 0270-7306/06/$08.00 0 doi:10.1128/mcb.26.4.1496 1509.2006 Copyright 2006, American Society for Microbiology. All Rights Reserved.
