Supplemental data and methods Molecular transformation from committed progenitor to leukemia stem cell initiated by MLL-AF9
|
|
|
- Doris French
- 5 years ago
- Views:
Transcription
1 Supplemental data and methods Molecular transformation from committed progenitor to leukemia stem cell initiated by MLL-AF9 Andrei V. Krivtsov, David Twomey, Zhaohui Feng, Matthew C. Stubbs, Yingzi Wang, Joerg Faber, Jason E. Levine, Jing Wang, William C. Hahn, D. Gary Gilliland, Todd R. Golub, & Scott A. Armstrong 1
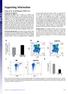
2 Supplementary Figure 1: Isolation of normal myeloid progenitor populations and transduction of GMP. Normal progenitors and HSC were sorted as described in supplementary methods, and GMP were transduced with either MSCV-GFP or MSCV- MLL-AF9-GFP retroviruses. Forty hours later the GFP positive cells were isolated for further experiments 2
3 a Bone Marrow (60X) Liver (10X) Spleen (60X) Supplementary Figure 2: Characterization of primary murine leukemias induced by transduction of GMP with an MLL-AF9 retrovirus. a) Histological evaluation of bone marrow, liver and spleen from a mouse with leukemia. The bone marrow is completely replaced by large cells, with large nuclei and prominent nucleoli. The spleen is enlarged and engorged with cells similar to those in the bone marrow. The liver shows infiltration of leukemic cells into the parenchyma. The experiment was repeated 3 times with similar results. 3
4 b c b) FACS analysis of bone marrow demonstrates 93% of bone marrow cells are GFP positive. Most leukemic cells are Mac1+/Gr1+. Leukemic cells do not express the lymphoid markers B220 and CD3. c) Southern blot analysis was performed on genomic DNA isolated from 4 MLL-AF9 induced leukemias and demonstrates the leukemias are oligoclonal. The southern blot was performed using a probe for GFP. 4
5 Supplementary Figure 3: Comparison of colony forming activity between L-GMP, Lin- Kit-, and lin+ cells. L-GMP were sorted from two mice that developed AML after transduction of normal GMP with MLL-AF cells were subsequently cultured in methylcellulose supplemented with SCF, IL3 and IL6. This experiment was repeated two times with cells isolated from 4 different mice with similar results. 5
6 a Bone Marrow (60X) Liver (10X) Spleen (60x) B. B. Supplementary Figure 4: Characterization of leukemias generated by introduction of 20 L-GMP into secondary recipients. a) Histological analysis shows infiltration of larger cells with large nuclei similar to the leukemias in primary recipients. 6
7 b c b) Immunophenotypic analysis of myeloid progenitors from a leukemia generated by injection of 20 L-GMP shows recapitulation of the initial disease shown in fig. 1 c c) FACS analysis demonstrates leukemia cells are GFP+, Mac1+, Gr1+, CD3-, B220- similar to primary leukemias. Progenitor analysis shown in fig. 1 of the manuscript further demonstrate the similarity between the primary and secondary leukemias. 7
8 Supplementary Figure 5: Immunophenotypic analysis of L-GMP propagated in liquid culture (AKLG cells) supplemented with IL3. 8
9 Supplementary Figure 6: The top 50 probe sets for genes that show decreased expression in the HSC and L-GMP as compared to normal progenitors are shown. Permutation testing demonstrates ~300 probe sets (262 genes) in this signature (p<0.001) 9
10 a b P < c P < Supplementary Figure 7: The self-renewal associated signature is a subset of the HSC signature-comparison with a previously published HSC signature. Due to the presence of MPP in our HSC population, it was possible that some portion of our HSCassociated signature is actually part of the MPP signature, and thus the true HSC signature might be identified only when we add the L-GMP to the analysis. a) To assess this, we first identified the genes highly expressed in our HSC population compared to other normal progenitors (approximately 1137 genes), and ranked them based on correlation with the profile of high expression in HSC and L-GMP and low level expression in progenitors a). Next, we divided our HSC signature into those genes that are also highly expressed in L-GMP and those that are not, and looked for enrichment (with GSEA) of these genes in a previously published dataset that compares highly purified long-term-hsc (LT-HSC) to total bone marrow 19. The genes highly expressed in HSC and L-GMP are enriched in the LT-HSC signature b), as is the remainder of the HSC signature c). Therefore the self-renewal associated signature we have identified is a subset of a larger HSC signature. 10
11 a b p=0.6 Supplementary Figure 8: The self-renewal associated signature is not merely a prominent portion of the HSC signature. As L-GMP contain leukemia stem cells with a calculated frequency of 1:6, it remained possible that the self-renewal associated signature was merely a prominent portion of the complete HSC signature expressed in only 1:6 L-GMP. If the self-renewal associated signature is only the visible portion of the full HSC signature, then the genes in this signature should be the most differentially expressed genes in a comparison between HSC and normal committed progenitors. a) To address this, we first ranked genes based on correlation with high-level expression in HSC compared to normal progenitors (leaving out L-GMP expression in determination of ranking) and assessed where genes in the self-renewal signature fell in the ranked list. Genes in the self-renewal associated signature are distributed throughout the list without obvious clustering toward the top. The red bars next to the gene expression data indicate when a member of the 420-gene self-renewal associated signature is found. b) Next, we performed a GSEA using the 420-gene self-renewal associated as a gene set against the HSC signature ranked as in a. As expected, there was no enrichment of this signature toward the top of the list (p=0.6). Thus, it is not the case that the self-renewal signature is a prominent portion of the full HSC signature that is identifiable above the background "GMP-like" signature. 11
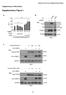
12 Supplementary Figure 9: L-GMP were transduced with lentiviruses expressing either a control shrna (luciferase) or either of two Mef2c directed shrna (Mef2c-F6, Mef2c- F7) and 1000 cells were plated in methylcellulose supplemented with cytokines and puromycin. After selection with puromycin, Mef2c RNA levels were assessed by realtime PCR 12
13 Supplementary Figure 10: Model for development of leukemia stem cells from committed progenitors. Our data supports a model where the MLL-AF9 fusion can induce serial replating activity and leukemia from GMP. The signature induced soon after MLL-AF9 expression includes a subset of genes that are part of a larger self-renewal associated signature that accumulates in the developing leukemia stem cell. However, the leukemia stem cell retains a global identity that is most similar to the GMP from which it arose. 13
14 Supplementary Methods: Mice, antibodies, and sorting of hematopoietic progenitors Bone marrow was collected from both femur and tibia of C57BL/6 donors by grinding the muscle free bones. Red blood cells were lysed on ice using red blood cells lysis buffer Puregene RBC Lysis Solution (cat# D-5001 Gentra Systems) of nucleated bone marrow cells were incubated 40 min on ice with 100 μl of each of the following lineage specific antibodies: Anti- mouse CD3 (Cat# RM3400, Caltag, CA), anti-mouse CD4 (Cat# MCD0400, Caltag, CA), anti-mouse CD8a (Cat# MCD0800, Caltag, CA), antimouse CD19 (Cat# RM7700, Caltag, CA), anti-mouse B220 (CD45R) (Cat# RM2600, Caltag, CA), anti-mouse Gr1 (Cat# RM3000, Caltag, CA), anti-mouse TER119 (Cat# MTER00, Caltag, CA) and anti-mouse CD127 (IL-7R) (Cat# , Bioscience). After double washing in PBS the cell suspension was incubated with 50 μl of secondary Goat-anti-rat F(ab)2 fragments labeled with Tri-Color (TC) (Cat# R40106, Caltag, CA). The cells were washed again and unbound Goat-anti-rat F(ab)2 fragments were blocked with 100 μg of rat-igg (Cat# I-8015, Sigma, MO) for 10 min on ice. The cells were labeled with 50 μl of each Sca1-bio (Cat# , BD, CA), Anti-mouse CD16/32 (Cat# , BD, CA); Anti-mouse CD117 (c-kit) (Cat# , BD, CA); Anti-mouse CD34 (Cat# , BD, CA) for 30 min on ice, washed in PBS and Sca1-bio antibody was then developed with 2 μl of Streptavidine-APC-Cy7 (Cat# SA1014, Caltag, CA), dead cells were labeled with 7-AAD (Cat# A-1310, Molecular Probes, OR) for 15 min before the sorting. Mouse population enriched in hematopoietic stem cells (Lin - ; IL-7R - ; Kit + ; Sca1 + ), and myeloid progenitors: CMP (IL-7R - Sca1 - Lin - Kit + CD34 + Fcγ RII/III lo ); GMP (IL-7R - Sca1 - Lin - Kit + CD34 + Fcγ RII/III hi ) and MEP (IL-7R - Sca1 - Lin - Kit + CD34 - Fcγ RII/III lo ) were sorted using FACSAria multicolor cell sorter equipper with 488 and 635 nm lasers (BD, San Diego, CA). RNA was isolated from multiple independently isolated samples containing normal HSC, CMP, GMP, MEP. Since L-GMP expressed GFP we included anti-mouse Sca1 (Caltag) in the lineage mix, and used anti-mouse CD34-bio (BD) and Streptavidin-APC-Cy7 (Caltag) when we sorted this population from mice with AML. The CD48 antibody # was obtained from ebioscience. L-GMP samples 1 and 2 were isolated from the bone marrow and spleen of a single mouse. Other L-GMP samples are from 4 separate mice. Retroviruses, Infections, Culture of hematopoietic progenitors The MLL-AF9 cdna was generously provided by Dr. Jay Hess and re-cloned into an MSCV based vector followed by IRES-GFP cassette (pmig). Mef2C, HoxA9, HoxA7, HoxA10, and HoxA5 were amplified from total RNA isolated from L-GMP using primers specific for each gene. The resultant PCR product was cloned into pmscv-puro (Clontech) and fully sequenced.the Mef2c shrna sequences were Mef2c-F6- GCCTCAGTGATACAGTATAAA and Mef2c-F7-CCATCAGTGAATCAAAGGATA. Ecotropic retroviral supernatants were produced by transient co-transfection of 293T cells as previously described 29. ShRNA in lentiviral vectors were obtained from the RNAi 14
15 Consortium, and viral particles generated by co-transfection of 293T cells with lentiviral packaging plasmids. For GMP transduction, 5x10 4 to 5x10 5 GMP were incubated with retroviral supernatant including 20% FCS; 20 ng/ml mscf (Peprotech) 10 ng/ml mil-3 (Peprotech), 10 ng/ml mil-6 (Peprotech), 1 penicillin-streptomycin (Invitrogen), 7 μg/ml polybrene (Sigma), and spun at 2000 rpm for 60 minutes at 37 o C. 40 hours after the infection GMP were resorted for GFP + PI- cells; then GFP + cells were either injected (tail vein) into sub-lethally (600 RAD) irradiated recipient mice, collected for RNA extraction, or plated in Methylcellulose media M3234 (Stem Cell Technologies) supplemented with 20 ng/ml mscf (Peprotech) 10 ng/ml IL-3 (Peprotech), 10 ng/ml IL-6 (Peprotech), 10 ng/ml GM- CSF (Peprotech) and 1 penicillin /streptomycin. For Mef2c shrna experiments, L-GMP sorted from leukemic mice were incubated with lentiviral vectors as described for retroviruses and plated in methylcellulose media M3234 or liquid culture (Stem Cell Technologies) supplemented with 10 ng/ml IL-3 (Peprotech), and 1 penicillin/streptomycin (Gibco) with 2.5 ug/ml puromycin (Sigma). Quantitative PCR RNA was isolated from colonies derived from L-GMP and cdna was generated using standard techniques. Real time PCR was performed using SYBR green detection reagents on a Sequence Detection System 7700 (Applied Biosystems) using primers for Mef2c and Gapdh. Ct values normalized against Gapdh as a housekeeping gene. Relative changes in concentrations were calculated according to the ΔΔCt method. Data analysis and statistical methods After hybridization, the raw expression data was normalized as previously described to account for differences in chip intensities 20. Gene expression was then analyzed using the GeneCluster 2 /GenePattern software or gene set enrichment analysis (GSEA) software (available at Hierarchical and K-means clustering were performed using the cluster software obtained from The data were preprocessed using minimum and maximum expression values, a max/min filter, and max-min filter. The filtering for hierarchical clustering was max/min>2 and max-min>80. The filtering for K-means clustering was max/min=3 and max-min=100. For comparisons of gene expression between two groups, the expression level correlated with a particular class was determined by comparing the means between the two groups using the signal-to-noise statistic 20. Stem cell frequency was calculated with the L-calc program from stem cell technologies. For murine/human comparisons, we first converted the murine probe set numbers to gene symbols using the latest Affymetrix annotation. Next we converted those gene symbols to 15
16 (HU133A) probes using Affymetrix annotation. This new list of human probe sets was then used to assess human gene expression data. Gene Set Enrichment Analysis (GSEA) GSEA provides a general statistical method to test for the enrichment of sets of genes in expression data, and has been particularly useful in identifying molecular pathways at play in complex gene expression signatures, as recently reported 23. GSEA considers a priori defined Gene Sets, for example, genes in a signature such as the self-renewal associated signature or members of a pathway. It then provides a method to determine whether the members of these sets are over-represented at the top (or bottom) of a Gene List of markers which have been ordered by their correlation with a specific phenotype or class distinction, and produces a Gene Set Gene List specific Enrichment Score (ES). The current implementation of GSEA is based on a Kolmogorov Smirnov (KS) score to estimate the difference between the empirical cumulative distribution functions P hit and P miss, representing the fraction of genes from the of Gene Set G that are present ( hits ) or absent ( misses ) in the Gene List up to a given position: P i hit () = () i M j, P j = 1 N miss i H () = i ( 1 M ( j ), j = 1 N M The membership function M() j takes the value 1 for a hit (i.e., the gene is in G) and 0 for a miss (i.e., the gene is not in G) at location j in the Gene List. NH (N M ) is the total number of genes from G that are found (not found) in the Gene List. The difference between the two distributions is a running enrichment score S( i)and the maximum is the Maximum Enrichment Score (ES) ES = max S()= i max P hit () i P miss i i=1,..,n i =1,..,N The significance of an observed ES( G) is obtained by permutation testing: reshuffling the phenotype labels and re-sorting the Gene List to determine how often an observed ES() G occurs by chance. Statistical significance is computed by comparing the observed ES() G with a histogram of ES( G,π ) values corresponding to the enrichment of the same Gene Set G but with reshuffled data according to a set of permutationsπ = ( 1,...,Π). The running enrichment score (red line in supplemental figure 7b.c. and 8b) is graphed vs. the gene # in a gene list ordered based on the correlation of interest. Simply, the higher the ES score and the earlier in the ordered gene list the max ES score is obtained, the greater the enrichment of the gene set. () 16
17 Cell population Transduction efficiency (%GFP + ) Transplanted cells (#) Estimated transduced cells (#) Transplanted mice (#) # AML Normal GMP /7 (100%) (MLL-AF9) /10 (100%) /5 (20%) /5(0%) /5 (0%) leukemic BM /6 (100%) /3 (100%) /7 (100%) /2 (100%) /14 (50%) /5 (0%) L-GMP /1 (100%) /11 (100%) /7 (100%) /11 (100%) /22 (100%) 4 6 1/6 (17%) Normal GMP (MSCV-GFP) AKLG Cells 100, /3 (100%) 5, /3 (0%) Supplemental Table 1: Mouse transplantation data 17
MicroRNAs Modulate Hematopoietic Lineage Differentiation
 Chen et al., page 1 MicroRNAs Modulate Hematopoietic Lineage Differentiation Chang-Zheng Chen, Ling Li, Harvey F. Lodish, David. Bartel Supplemental Online Material Methods Cell isolation Murine bone marrow
Chen et al., page 1 MicroRNAs Modulate Hematopoietic Lineage Differentiation Chang-Zheng Chen, Ling Li, Harvey F. Lodish, David. Bartel Supplemental Online Material Methods Cell isolation Murine bone marrow
Supplemental Information Inventory
 Cell Stem Cell, Volume 6 Supplemental Information Distinct Hematopoietic Stem Cell Subtypes Are Differentially Regulated by TGF-β1 Grant A. Challen, Nathan C. Boles, Stuart M. Chambers, and Margaret A.
Cell Stem Cell, Volume 6 Supplemental Information Distinct Hematopoietic Stem Cell Subtypes Are Differentially Regulated by TGF-β1 Grant A. Challen, Nathan C. Boles, Stuart M. Chambers, and Margaret A.
7-amino actinomycin D (7ADD) was added to all samples 10 minutes prior to analysis on the flow cytometer in order to gate 7AAD viable cells.
 Antibody staining for Ho uptake analyses For HSC staining, 10 7 BM cells from Ho perfused mice were stained with biotinylated lineage antibodies (CD3, CD5, B220, CD11b, Gr-1, CD41, Ter119), anti Sca-1-PECY7,
Antibody staining for Ho uptake analyses For HSC staining, 10 7 BM cells from Ho perfused mice were stained with biotinylated lineage antibodies (CD3, CD5, B220, CD11b, Gr-1, CD41, Ter119), anti Sca-1-PECY7,
The GOODELL laboratory
 The GOODELL laboratory Author Nathan Boles Feb.5, 2009 Title Hematopoietic Progenitor Staining Introduction This protocol describes the analysis or isolation of the early hematopoietic progenitors. HBSS+
The GOODELL laboratory Author Nathan Boles Feb.5, 2009 Title Hematopoietic Progenitor Staining Introduction This protocol describes the analysis or isolation of the early hematopoietic progenitors. HBSS+
Flow cytometry Stained cells were analyzed and sorted by SORP FACS Aria (BD Biosciences).
 Mice C57BL/6-Ly5.1 or -Ly5.2 congenic mice were used for LSK transduction and competitive repopulation assays. Animal care was in accordance with the guidelines of Keio University for animal and recombinant
Mice C57BL/6-Ly5.1 or -Ly5.2 congenic mice were used for LSK transduction and competitive repopulation assays. Animal care was in accordance with the guidelines of Keio University for animal and recombinant
Supplementary Fig. 1: Characterization of Asxl2 -/- mouse model. (a) HSCs and their
 Supplementary Fig. 1: Characterization of Asxl2 -/- mouse model. (a) HSCs and their differentiated cell populations were sorted from the BM of WT mice using respective surface markers, and Asxl2 mrna expression
Supplementary Fig. 1: Characterization of Asxl2 -/- mouse model. (a) HSCs and their differentiated cell populations were sorted from the BM of WT mice using respective surface markers, and Asxl2 mrna expression
The trithorax protein partner menin acts in tandem with EZH2 to suppress C/EBPα and differentiation in MLL-AF9 leukemia
 SUPPLEMENTARY APPENDIX The trithorax protein partner menin acts in tandem with EZH2 to suppress C/EBPα and differentiation in MLL-AF9 leukemia Austin T. Thiel, Zijie Feng, Dhruv K. Pant, Lewis A. Chodosh,
SUPPLEMENTARY APPENDIX The trithorax protein partner menin acts in tandem with EZH2 to suppress C/EBPα and differentiation in MLL-AF9 leukemia Austin T. Thiel, Zijie Feng, Dhruv K. Pant, Lewis A. Chodosh,
Endogenous MYC was detected by Western blotting using the N-262 polyclonal antibody (Santa
 SUPPLEMENTARY METHODS Antibodies Endogenous MYC was detected by Western blotting using the N-262 polyclonal antibody (Santa Cruz) or the 9E10 monoclonal antibody (Vanderbilt Antibody and Protein Resource).
SUPPLEMENTARY METHODS Antibodies Endogenous MYC was detected by Western blotting using the N-262 polyclonal antibody (Santa Cruz) or the 9E10 monoclonal antibody (Vanderbilt Antibody and Protein Resource).
SUPPLEMENTARY INFORMATION
 DOI: 1.138/ncb3342 EV O/E 13 4 B Relative expression 1..8.6.4.2 shctl Pcdh2_1 C Pcdh2_2 Number of shrns 12 8 4 293T mterc +/+ G3 mterc -/- D % of GFP + cells 1 8 6 4 2 Per2 p=.2 p>
DOI: 1.138/ncb3342 EV O/E 13 4 B Relative expression 1..8.6.4.2 shctl Pcdh2_1 C Pcdh2_2 Number of shrns 12 8 4 293T mterc +/+ G3 mterc -/- D % of GFP + cells 1 8 6 4 2 Per2 p=.2 p>
Table S1. Antibodies and recombinant proteins used in this study
 Table S1. Antibodies and recombinant proteins used in this study Labeled Antibody Clone Cat. no. Streptavidin-PerCP BD Biosciences 554064 Biotin anti-mouse CD25 7D4 BD Biosciences 553070 PE anti-mouse
Table S1. Antibodies and recombinant proteins used in this study Labeled Antibody Clone Cat. no. Streptavidin-PerCP BD Biosciences 554064 Biotin anti-mouse CD25 7D4 BD Biosciences 553070 PE anti-mouse
Supplemental Information. Targeting HIF1 Eliminates Cancer Stem Cells. in Hematological Malignancies. Inventory of Supplemental Information
 Cell Stem Cell, Volume 8 Supplemental Information Targeting HIF1 Eliminates Cancer Stem Cells in Hematological Malignancies Yin Wang, Yan Liu, Sami N. Malek, Pan Zheng, and Yang Liu Inventory of Supplemental
Cell Stem Cell, Volume 8 Supplemental Information Targeting HIF1 Eliminates Cancer Stem Cells in Hematological Malignancies Yin Wang, Yan Liu, Sami N. Malek, Pan Zheng, and Yang Liu Inventory of Supplemental
Nature Immunology: doi: /ni Supplementary Figure 1
 Supplementary Figure 1 Validation of the monoclonal antibody to mouse ACKR1 and expression of ACKR1 by BM hematopoietic cells. (a to d) Comparison of immunostaining of BM cells by anti-mouse ACKR1 antibodies:
Supplementary Figure 1 Validation of the monoclonal antibody to mouse ACKR1 and expression of ACKR1 by BM hematopoietic cells. (a to d) Comparison of immunostaining of BM cells by anti-mouse ACKR1 antibodies:
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES Supplementary Figure 1 Histogram displaying the distribution of DNA barcode copy numbers for each virus library. 100,000 BB88 cells were infected by lentivirus that carried the corresponding
SUPPLEMENTARY FIGURES Supplementary Figure 1 Histogram displaying the distribution of DNA barcode copy numbers for each virus library. 100,000 BB88 cells were infected by lentivirus that carried the corresponding
In vivo BrdU Incorporation Assay for Murine Hematopioetic Stem Cells Ningfei An, Yubin Kang *
 In vivo BrdU Incorporation Assay for Murine Hematopioetic Stem Cells Ningfei An, Yubin Kang * Division of Hematology-Oncology, Department of Medicine, Medical University of South Carolina, Charleston,
In vivo BrdU Incorporation Assay for Murine Hematopioetic Stem Cells Ningfei An, Yubin Kang * Division of Hematology-Oncology, Department of Medicine, Medical University of South Carolina, Charleston,
Multiple layers of B cell memory with different effector functions. Ismail Dogan, Barbara Bertocci, Valérie Vilmont, Frédéric Delbos,
 Multiple layers of B cell memory with different effector functions Ismail Dogan, Barbara Bertocci, Valérie Vilmont, Frédéric Delbos, Jérome Mégret, Sébastien Storck, Claude-Agnès Reynaud & Jean-Claude
Multiple layers of B cell memory with different effector functions Ismail Dogan, Barbara Bertocci, Valérie Vilmont, Frédéric Delbos, Jérome Mégret, Sébastien Storck, Claude-Agnès Reynaud & Jean-Claude
Mutagenesis and generation of expression vectors Gene expression profiling Lentiviral production and infection of TF1 cells
 Mutagenesis and generation of expression vectors Human c-kit wild-type cdna encoding the short c-kit isoform was excised from vector pbshkitwt (kind gift from Dr. L Gros, France) by Sal I-Acc65 I digestion.
Mutagenesis and generation of expression vectors Human c-kit wild-type cdna encoding the short c-kit isoform was excised from vector pbshkitwt (kind gift from Dr. L Gros, France) by Sal I-Acc65 I digestion.
FACS. Chromatin immunoprecipitation (ChIP) and ChIP-chip arrays 3 4
 FACS Stem progenitor sorting for progenitor analysis Adult bone marrow cells were depleted with Miltenyi lineage depletion column (Miltenyi). For identification of CMPs, GMPs, and MEPs the cells were incubated
FACS Stem progenitor sorting for progenitor analysis Adult bone marrow cells were depleted with Miltenyi lineage depletion column (Miltenyi). For identification of CMPs, GMPs, and MEPs the cells were incubated
For knockdown of individual genes, the following TRC library clones were used: Gene Species Library TRC number Designation in manuscript
 Supplemental Methods shrna constructs for validation experiments For knockdown of MLL-AF9, a custom shrna with the following stem sequence directed against the fusion breakpoint (underlined) was designed
Supplemental Methods shrna constructs for validation experiments For knockdown of MLL-AF9, a custom shrna with the following stem sequence directed against the fusion breakpoint (underlined) was designed
SANTA CRUZ BIOTECHNOLOGY, INC.
 TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
88.7 <GFP-A>: EGFP GFP. Null. % of Cells! <APC-A> B220!
 B. C. 1 Cell Number (X18) A. % of Cells % of Max 8 Con 6 4 88.7 1 13 14 : EGFP 4 6 4 1 WT 8 % of% ofcells Max 6 6 4 G. 1 13 B Hdac3-/- 14 15 1 13 14 15 Ter119 Ly6C Wild type WT 4
B. C. 1 Cell Number (X18) A. % of Cells % of Max 8 Con 6 4 88.7 1 13 14 : EGFP 4 6 4 1 WT 8 % of% ofcells Max 6 6 4 G. 1 13 B Hdac3-/- 14 15 1 13 14 15 Ter119 Ly6C Wild type WT 4
We designed the targeting vector in such a way that the first exon was flanked by two LoxP sites and
 Mice We designed the targeting vector in such a way that the first exon was flanked by two LoxP sites and an Frt flanked neo cassette (Fig. S1A). Targeted BA1 (C57BL/6 129/SvEv) hybrid embryonic stem cells
Mice We designed the targeting vector in such a way that the first exon was flanked by two LoxP sites and an Frt flanked neo cassette (Fig. S1A). Targeted BA1 (C57BL/6 129/SvEv) hybrid embryonic stem cells
Supplementary Figures
 Supplementary Figures Supplementary Figure 1: Phenotypically defined hematopoietic stem and progenitor cell populations show distinct mitochondrial activity and mass. (A) Isolation by FACS of commonly
Supplementary Figures Supplementary Figure 1: Phenotypically defined hematopoietic stem and progenitor cell populations show distinct mitochondrial activity and mass. (A) Isolation by FACS of commonly
Supplementary Figure 1. Effect of FRC-specific ablation of Myd88 on PP and mln organization.
 Supplementary Figure 1 Effect of FRC-specific ablation of Myd88 on PP and mln organization. (a) PP numbers in 8 10 week old Cre-negative littermate (Ctrl) and Myd88-cKO mice (n = 11 mice; each dot represents
Supplementary Figure 1 Effect of FRC-specific ablation of Myd88 on PP and mln organization. (a) PP numbers in 8 10 week old Cre-negative littermate (Ctrl) and Myd88-cKO mice (n = 11 mice; each dot represents
MLN8237 induces proliferation arrest, expression of differentiation markers and
 Supplementary Figure Legends Supplementary Figure 1 827 induces proliferation arrest, expression of differentiation markers and polyploidization of a human erythroleukemia cell line with the activating
Supplementary Figure Legends Supplementary Figure 1 827 induces proliferation arrest, expression of differentiation markers and polyploidization of a human erythroleukemia cell line with the activating
Supplemental Table S1. RT-PCR primers used in this study
 Supplemental Table S1. RT-PCR primers used in this study -----------------------------------------------------------------------------------------------------------------------------------------------
Supplemental Table S1. RT-PCR primers used in this study -----------------------------------------------------------------------------------------------------------------------------------------------
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1: Vector maps of TRMPV and TRMPVIR variants. Many derivatives of TRMPV have been generated and tested. Unless otherwise noted, experiments in this paper use
Supplementary Figure 1 Supplementary Figure 1: Vector maps of TRMPV and TRMPVIR variants. Many derivatives of TRMPV have been generated and tested. Unless otherwise noted, experiments in this paper use
Antibody used for FC Figure S1. Multimodal characterization of NIR dyes in vitro Figure S2. Ex vivo analysis of HL60 cells homing
 Antibody used for FC The following antibodies were used following manufacturer s instructions: anti-human CD4 (clone HI3, IgG1, k - Becton Dickinson), anti-human CD33 (clone WM3, IgG1, k- Becton Dickinson),
Antibody used for FC The following antibodies were used following manufacturer s instructions: anti-human CD4 (clone HI3, IgG1, k - Becton Dickinson), anti-human CD33 (clone WM3, IgG1, k- Becton Dickinson),
Figure S2. Response of mouse ES cells to GSK3 inhibition. Mentioned in discussion
 Stem Cell Reports, Volume 1 Supplemental Information Robust Self-Renewal of Rat Embryonic Stem Cells Requires Fine-Tuning of Glycogen Synthase Kinase-3 Inhibition Yaoyao Chen, Kathryn Blair, and Austin
Stem Cell Reports, Volume 1 Supplemental Information Robust Self-Renewal of Rat Embryonic Stem Cells Requires Fine-Tuning of Glycogen Synthase Kinase-3 Inhibition Yaoyao Chen, Kathryn Blair, and Austin
SUPPLEMENTAL METHODS. Chromatin immunoprecipitation (ChIP) assay. Cell culture. Non-ablated mouse transplantation. Hematology and flow cytometry
 SUPPLEMENTAL METHODS Cell culture EML C1 cells were cultured in IMDM 20% horse serum, 1% antibiotic-antimycotic (Gibco by Life Technologies) 15% SCF conditioned medium from BHK/ MKL cells or 50 100 ng/ml
SUPPLEMENTAL METHODS Cell culture EML C1 cells were cultured in IMDM 20% horse serum, 1% antibiotic-antimycotic (Gibco by Life Technologies) 15% SCF conditioned medium from BHK/ MKL cells or 50 100 ng/ml
Figure S1. Phenotypic characterization of AND-1_WASKO cell lines. AND- 1_WASKO_C1.1 (WASKO_C1.1) and AND-1_WASKO_C1.2 (WASKO_C1.
 LEGENDS TO SUPPLEMENTARY FIGURES Figure S1. Phenotypic characterization of AND-1_WASKO cell lines. AND- 1_WASKO_C1.1 (WASKO_C1.1) and AND-1_WASKO_C1.2 (WASKO_C1.2) were stained with the antibodies oct3/4
LEGENDS TO SUPPLEMENTARY FIGURES Figure S1. Phenotypic characterization of AND-1_WASKO cell lines. AND- 1_WASKO_C1.1 (WASKO_C1.1) and AND-1_WASKO_C1.2 (WASKO_C1.2) were stained with the antibodies oct3/4
Nature Immunology: doi: /ni.3694
 Supplementary Figure 1 Expression of Bhlhe41 and Bhlhe40 in B cell development and mature B cell subsets. (a) Scatter plot showing differential expression of genes between splenic B-1a cells and follicular
Supplementary Figure 1 Expression of Bhlhe41 and Bhlhe40 in B cell development and mature B cell subsets. (a) Scatter plot showing differential expression of genes between splenic B-1a cells and follicular
Application Note. Abstract. Reynolds S, 1 Edinger M, 1 Gray J, 1 Tanaka S, 2 Collins P 1 1
 Identification and Functionality of Adult Mouse Hematopoietic Stem Cell Side Populations after Enrichment on the BD FACSAria II Flow Cytometer Equipped with a 375-nm Near UV Laser Reynolds S, 1 Edinger
Identification and Functionality of Adult Mouse Hematopoietic Stem Cell Side Populations after Enrichment on the BD FACSAria II Flow Cytometer Equipped with a 375-nm Near UV Laser Reynolds S, 1 Edinger
SI Appendix. Tumor-specific CD8 + Tc9 cells are superior effector than Tc1 cells for. adoptive immunotherapy of cancers
 SI Appendix Tumor-specific CD8 + Tc9 cells are superior effector than Tc1 cells for adoptive immunotherapy of cancers Yong Lu, Bangxing Hong, Haiyan Li, Yuhuan Zheng, Mingjun Zhang, Siqing Wang, Jianfei
SI Appendix Tumor-specific CD8 + Tc9 cells are superior effector than Tc1 cells for adoptive immunotherapy of cancers Yong Lu, Bangxing Hong, Haiyan Li, Yuhuan Zheng, Mingjun Zhang, Siqing Wang, Jianfei
Supporting Information
 (xe number cells) LSK (% gated) Supporting Information Youm et al../pnas. SI Materials and Methods Quantification of sjtrecs. CD + T subsets were isolated from splenocytes using mouse CD + T cells positive
(xe number cells) LSK (% gated) Supporting Information Youm et al../pnas. SI Materials and Methods Quantification of sjtrecs. CD + T subsets were isolated from splenocytes using mouse CD + T cells positive
Supplemental methods Supplemental figure and legend...7. Supplemental table.. 8
 Supplemental Digital Content (SDC) Contents Supplemental methods..2-6 Supplemental figure and legend...7 Supplemental table.. 8 1 SDC, Supplemental Methods Flow cytometric analysis of intracellular phosphorylated
Supplemental Digital Content (SDC) Contents Supplemental methods..2-6 Supplemental figure and legend...7 Supplemental table.. 8 1 SDC, Supplemental Methods Flow cytometric analysis of intracellular phosphorylated
administration of tamoxifen. Bars show mean ± s.e.m (n=10-11). P-value was determined by
 Supplementary Figure 1. Chimerism of CD45.2 + GFP + cells at 1 month post transplantation No significant changes were detected in chimerism of CD45.2 + GFP + cells between recipient mice repopulated with
Supplementary Figure 1. Chimerism of CD45.2 + GFP + cells at 1 month post transplantation No significant changes were detected in chimerism of CD45.2 + GFP + cells between recipient mice repopulated with
Hematopoietic stem cell transplantation without irradiation
 nature methods Hematopoietic stem cell transplantation without irradiation Claudia Waskow, Vikas Madan, Susanne Bartels, Céline Costa, Rosel Blasig & Hans-Reimer Rodewald Supplementary figures and text:
nature methods Hematopoietic stem cell transplantation without irradiation Claudia Waskow, Vikas Madan, Susanne Bartels, Céline Costa, Rosel Blasig & Hans-Reimer Rodewald Supplementary figures and text:
Mayumi Egawa, Kaori Mukai, Soichiro Yoshikawa, Misako Iki, Naofumi Mukaida, Yohei Kawano, Yoshiyuki Minegishi, and Hajime Karasuyama
 Immunity, Volume 38 Supplemental Information Inflammatory Monocytes Recruited to Allergic Skin Acquire an Anti-inflammatory M2 Phenotype via Basophil-Derived Interleukin-4 Mayumi Egawa, Kaori Mukai, Soichiro
Immunity, Volume 38 Supplemental Information Inflammatory Monocytes Recruited to Allergic Skin Acquire an Anti-inflammatory M2 Phenotype via Basophil-Derived Interleukin-4 Mayumi Egawa, Kaori Mukai, Soichiro
Supplementary Figure 1. Reintroduction of HDAC1 in HDAC1-/- ES cells reverts the
 SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1. Reintroduction of HDAC1 in HDAC1-/- ES cells reverts the phenotype of HDAC1-/- teratomas. 3x10 6 HDAC1 reintroduced (HDAC1-/-re) and empty vector infected
SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1. Reintroduction of HDAC1 in HDAC1-/- ES cells reverts the phenotype of HDAC1-/- teratomas. 3x10 6 HDAC1 reintroduced (HDAC1-/-re) and empty vector infected
Supporting Information
 Supporting Information Narni-Mancinelli et al. 10.1073/pnas.1112064108 SI Materials and Methods Cell Preparation. Mice were anesthetized and immediately perfused with PBS before organs were collected.
Supporting Information Narni-Mancinelli et al. 10.1073/pnas.1112064108 SI Materials and Methods Cell Preparation. Mice were anesthetized and immediately perfused with PBS before organs were collected.
BD IMag. Streptavidin Particles Plus - DM. Technical Data Sheet. Product Information
 Technical Data Sheet Streptavidin Particles Plus - DM Product Information Material Number: Size: Storage Buffer: 557812 5 ml Aqueous buffered solution containing BSA and 0.09% sodium azide. Description
Technical Data Sheet Streptavidin Particles Plus - DM Product Information Material Number: Size: Storage Buffer: 557812 5 ml Aqueous buffered solution containing BSA and 0.09% sodium azide. Description
TIB6 B16F1 LLC EL4. n=2 n=7 n=9 n=6
 Shojaei et al., Supplemental Fig. 1 9 Mean Inhibition by anti-vegf (%) 7 5 3 1 TIB6 n=2 n=7 n=9 n=6 Supplemental Figure 1 Differential growth inhibition by anti-vegf treatment. The percent of tumor growth
Shojaei et al., Supplemental Fig. 1 9 Mean Inhibition by anti-vegf (%) 7 5 3 1 TIB6 n=2 n=7 n=9 n=6 Supplemental Figure 1 Differential growth inhibition by anti-vegf treatment. The percent of tumor growth
Supporting Information
 Supporting Information Tal et al. 10.1073/pnas.0807694106 SI Materials and Methods VSV Infection and Quantification. Infection was carried out by seeding 5 10 5 MEF cells per well in a 6-well plate and
Supporting Information Tal et al. 10.1073/pnas.0807694106 SI Materials and Methods VSV Infection and Quantification. Infection was carried out by seeding 5 10 5 MEF cells per well in a 6-well plate and
FLOW CYTOMETRY METHODS FOR IDENTIFYING MOUSE HEMATOPOIETIC STEM AND PROGENITOR CELLS
 TECHNICAL BULLETIN FLOW CYTOMETRY METHODS FOR IDENTIFYING MOUSE HEMATOPOIETIC STEM AND PROGENITOR CELLS Background The phenotypic characterization of mouse hematopoietic stem and progenitor cells (HSPCs),
TECHNICAL BULLETIN FLOW CYTOMETRY METHODS FOR IDENTIFYING MOUSE HEMATOPOIETIC STEM AND PROGENITOR CELLS Background The phenotypic characterization of mouse hematopoietic stem and progenitor cells (HSPCs),
To determine the effect of N1IC in the susceptibility of T cells to the tolerogenic effect of tumorassociated
 Supplementary Methods Tolerogenic effect of MDSC To determine the effect of N1IC in the susceptibility of T cells to the tolerogenic effect of tumorassociated MDSC in vivo, we used a model described previously
Supplementary Methods Tolerogenic effect of MDSC To determine the effect of N1IC in the susceptibility of T cells to the tolerogenic effect of tumorassociated MDSC in vivo, we used a model described previously
Supplementary Information Supplementary Figure 1
 Bararia, Kwok et al, Supplementary Page 1 Supplementary Information Supplementary Figure 1 S1 Bararia, Kwok et al, Supplementary Page 2 Supplementary Figure 1 (cont.) S2 Bararia, Kwok et al, Supplementary
Bararia, Kwok et al, Supplementary Page 1 Supplementary Information Supplementary Figure 1 S1 Bararia, Kwok et al, Supplementary Page 2 Supplementary Figure 1 (cont.) S2 Bararia, Kwok et al, Supplementary
Mouse expression data were normalized using the robust multiarray algorithm (1) using
 Supplementary Information Bioinformatics statistical analysis of microarray data Mouse expression data were normalized using the robust multiarray algorithm (1) using a custom probe set definition that
Supplementary Information Bioinformatics statistical analysis of microarray data Mouse expression data were normalized using the robust multiarray algorithm (1) using a custom probe set definition that
F4/80, CD11b, Gr-1, NK1.1, CD3, CD4, CD8 and CD19. A-antigen was detected with FITCconjugated
 SDC MATERIALS AND METHODS Flow Cytometric Detection of A-Antigen Expression Single cell suspensions were prepared from bone marrow, lymph node and spleen. Peripheral blood was obtained and erythrocytes
SDC MATERIALS AND METHODS Flow Cytometric Detection of A-Antigen Expression Single cell suspensions were prepared from bone marrow, lymph node and spleen. Peripheral blood was obtained and erythrocytes
Best practices in panel design to optimize the isolation of cells of interest
 Sort Best practices in panel design to optimize the isolation of cells of interest For Research Use Only. Not for use in diagnostic or therapeutic procedures. Alexa Fluor is a registered trademark of Life
Sort Best practices in panel design to optimize the isolation of cells of interest For Research Use Only. Not for use in diagnostic or therapeutic procedures. Alexa Fluor is a registered trademark of Life
Fundamental properties of Stem Cells
 Stem cells Learning Goals: Define what a stem cell is and describe its general properties, using hematopoietic stem cells as an example. Describe to a non-scientist the current progress of human stem cell
Stem cells Learning Goals: Define what a stem cell is and describe its general properties, using hematopoietic stem cells as an example. Describe to a non-scientist the current progress of human stem cell
LINGO-1, A TRANSMEMBRANE SIGNALING PROTEIN, INHIBITS OLIGODENDROCYTE DIFFERENTIATION AND MYELINATION THROUGH INTERCELLULAR SELF- INTERACTIONS.
 Supplemental Data: LINGO-1, A TRANSMEMBRANE SIGNALING PROTEIN, INHIBITS OLIGODENDROCYTE DIFFERENTIATION AND MYELINATION THROUGH INTERCELLULAR SELF- INTERACTIONS. Scott Jepson, Bryan Vought, Christian H.
Supplemental Data: LINGO-1, A TRANSMEMBRANE SIGNALING PROTEIN, INHIBITS OLIGODENDROCYTE DIFFERENTIATION AND MYELINATION THROUGH INTERCELLULAR SELF- INTERACTIONS. Scott Jepson, Bryan Vought, Christian H.
Supplemental Information. Myeloid-Derived Suppressor Cells Are. Controlled by Regulatory T Cells via TGF-b. during Murine Colitis
 Cell Reports, Volume 17 Supplemental Information Myeloid-Derived Suppressor Cells Are Controlled by Regulatory T Cells via TGF-b during Murine Colitis Cho-Rong Lee, Yewon Kwak, Taewoo Yang, Jung Hyun Han,
Cell Reports, Volume 17 Supplemental Information Myeloid-Derived Suppressor Cells Are Controlled by Regulatory T Cells via TGF-b during Murine Colitis Cho-Rong Lee, Yewon Kwak, Taewoo Yang, Jung Hyun Han,
Insulin-like growth factor 2 expressed in a novel fetal liver cell population is a growth factor for hematopoietic stem cells
 HEMATOPOIESIS Insulin-like growth factor 2 expressed in a novel fetal liver cell population is a growth factor for hematopoietic stem cells Cheng Cheng Zhang and Harvey F. Lodish Hematopoietic stem cells
HEMATOPOIESIS Insulin-like growth factor 2 expressed in a novel fetal liver cell population is a growth factor for hematopoietic stem cells Cheng Cheng Zhang and Harvey F. Lodish Hematopoietic stem cells
show PB hemoglobin concentration (Hgb) and platelet counts (Plt) in Kras ex3op/ex3op mice
 Supplemental Data Supplemental Figure Legends Supplemental Figure 1. Hematologic profile of Kras / mice. (A) Scatter plots show PB hemoglobin concentration (Hgb) and platelet counts (Plt) in Kras / mice
Supplemental Data Supplemental Figure Legends Supplemental Figure 1. Hematologic profile of Kras / mice. (A) Scatter plots show PB hemoglobin concentration (Hgb) and platelet counts (Plt) in Kras / mice
We used HEK293T cells for lentiviral supernatant production. HEK293T cells were
 Lentiviral transduction of CD34 + cells and cell lines We used HEK293T cells for lentiviral supernatant production. HEK293T cells were maintained in DMEM supplemented with 1% FCS, 1 U/ml penicillin-streptomycin,
Lentiviral transduction of CD34 + cells and cell lines We used HEK293T cells for lentiviral supernatant production. HEK293T cells were maintained in DMEM supplemented with 1% FCS, 1 U/ml penicillin-streptomycin,
Supplemental Methods Cell lines and culture
 Supplemental Methods Cell lines and culture AGS, CL5, BT549, and SKBR were propagated in RPMI 64 medium (Mediatech Inc., Manassas, VA) supplemented with % fetal bovine serum (FBS, Atlanta Biologicals,
Supplemental Methods Cell lines and culture AGS, CL5, BT549, and SKBR were propagated in RPMI 64 medium (Mediatech Inc., Manassas, VA) supplemented with % fetal bovine serum (FBS, Atlanta Biologicals,
Supplementary Methods
 Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Supplementary Figure 1
 DOI: 1.138/ncb273 Supplementary Figure 1 a 1x DAPI field images 4x contoured cells 2µm 5µm b Tissue map Dot plot Histogram Y X antigen X DAPI antigen X Figure S1 Laser Scanning Cytometry of bone marrow
DOI: 1.138/ncb273 Supplementary Figure 1 a 1x DAPI field images 4x contoured cells 2µm 5µm b Tissue map Dot plot Histogram Y X antigen X DAPI antigen X Figure S1 Laser Scanning Cytometry of bone marrow
Arrayed shrna screening 11,415 hairpins representing 1383 unique genes. encompassing the murine protein kinome, GTPases, and components of the vesicle
 Supplemental Material for Strohecker et al. Identification of 6-phosphofructo-2- kinase/fructose-2,6-bisphosphatase (PFKFB4) as a Novel Autophagy Regulator by High Content shrna Screening Supplemental
Supplemental Material for Strohecker et al. Identification of 6-phosphofructo-2- kinase/fructose-2,6-bisphosphatase (PFKFB4) as a Novel Autophagy Regulator by High Content shrna Screening Supplemental
TITLE: Identification of Prostate Cancer Metastasis-Suppressor Genes Using Genomic shrna Libraries
 AD Award Number: W81XWH-07-1-0184 TITLE: Identification of Prostate Cancer Metastasis-Suppressor Genes Using Genomic shrna Libraries PRINCIPAL INVESTIGATOR: Irwin H. Gelman, Ph.D. CONTRACTING ORGANIZATION:
AD Award Number: W81XWH-07-1-0184 TITLE: Identification of Prostate Cancer Metastasis-Suppressor Genes Using Genomic shrna Libraries PRINCIPAL INVESTIGATOR: Irwin H. Gelman, Ph.D. CONTRACTING ORGANIZATION:
Platinum HSC Retrovirus Expression System, Amphotropic
 Product Manual Platinum HSC Retrovirus Expression System, Amphotropic Catalog Number VPK-307 1 Kit FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Retroviral gene transfer is a
Product Manual Platinum HSC Retrovirus Expression System, Amphotropic Catalog Number VPK-307 1 Kit FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Retroviral gene transfer is a
Supplementary Information Alternative splicing of CD44 mrna by ESRP1 enhances lung colonization of metastatic cancer cell
 Supplementary Information Alternative splicing of CD44 mrna by ESRP1 enhances lung colonization of metastatic cancer cell Supplementary Figures S1-S3 Supplementary Methods Supplementary Figure S1. Identification
Supplementary Information Alternative splicing of CD44 mrna by ESRP1 enhances lung colonization of metastatic cancer cell Supplementary Figures S1-S3 Supplementary Methods Supplementary Figure S1. Identification
Nature Genetics: doi: /ng.2949
 Supplementary Figure 1. Abnormal differentiation in vivo and colony growth in vitro of B cells with triplication of chr.21 orthologs. (a) (Top) B220 and CD43 staining of bone marrow from Ts1Rhr and wild-type
Supplementary Figure 1. Abnormal differentiation in vivo and colony growth in vitro of B cells with triplication of chr.21 orthologs. (a) (Top) B220 and CD43 staining of bone marrow from Ts1Rhr and wild-type
Supplementary Figure 1. Characterization of the POP2 transcriptional and post-transcriptional regulatory elements. (A) POP2 nucleotide sequence
 1 5 6 7 8 9 10 11 1 1 1 Supplementary Figure 1. Characterization of the POP transcriptional and post-transcriptional regulatory elements. (A) POP nucleotide sequence depicting the consensus sequence for
1 5 6 7 8 9 10 11 1 1 1 Supplementary Figure 1. Characterization of the POP transcriptional and post-transcriptional regulatory elements. (A) POP nucleotide sequence depicting the consensus sequence for
MagniSort Mouse Hematopoietic Lineage Depletion Kit Catalog Number: RUO: For Research Use Only. Not for use in diagnostic procedures.
 Page 1 of 2 MagniSort Mouse Hematopoietic Lineage Depletion Kit RUO: For Research Use Only. Not for use in diagnostic procedures. Mouse bone marrow cells were unsorted (top row) or sorted with the MagniSort
Page 1 of 2 MagniSort Mouse Hematopoietic Lineage Depletion Kit RUO: For Research Use Only. Not for use in diagnostic procedures. Mouse bone marrow cells were unsorted (top row) or sorted with the MagniSort
CRISPR-based gene disruption in murine HSPCs. Ayumi Kitano and Daisuke Nakada
 CRISPR-based gene disruption in murine HSPCs Ayumi Kitano and Daisuke Nakada Nakada Lab Protocol INTRODUCTION We recently described fast, efficient, and cost-effective methods to directly modify the genomes
CRISPR-based gene disruption in murine HSPCs Ayumi Kitano and Daisuke Nakada Nakada Lab Protocol INTRODUCTION We recently described fast, efficient, and cost-effective methods to directly modify the genomes
isolated from ctr and pictreated mice. Activation of effector CD4 +
 Supplementary Figure 1 Bystander inflammation conditioned T reg cells have normal functional suppressive activity and ex vivo phenotype. WT Balb/c mice were treated with polyi:c (pic) or PBS (ctr) via
Supplementary Figure 1 Bystander inflammation conditioned T reg cells have normal functional suppressive activity and ex vivo phenotype. WT Balb/c mice were treated with polyi:c (pic) or PBS (ctr) via
Adenoviral Expression Systems. Lentivirus is not the only choice for gene delivery. Adeno-X
 Adenoviral Expression Systems Lentivirus is not the only choice for gene delivery 3 Adeno-X Why choose adenoviral gene delivery? Table I: Adenoviral vs. Lentiviral Gene Delivery Lentivirus Adenovirus Infects
Adenoviral Expression Systems Lentivirus is not the only choice for gene delivery 3 Adeno-X Why choose adenoviral gene delivery? Table I: Adenoviral vs. Lentiviral Gene Delivery Lentivirus Adenovirus Infects
Competitive Bone-marrow Transplantations Maria Maryanovich and Atan Gross *
 Competitive Bone-marrow Transplantations Maria Maryanovich and Atan Gross * Biological Regulation, Weizmann Institute of Science, Rehovot, Israel *For correspondence: atan.gross@weizmann.ac.il [Abstract]
Competitive Bone-marrow Transplantations Maria Maryanovich and Atan Gross * Biological Regulation, Weizmann Institute of Science, Rehovot, Israel *For correspondence: atan.gross@weizmann.ac.il [Abstract]
Supplementary Figure 1. Phenotype, morphology and distribution of embryonic GFP +
 Supplementary Figure 1 Supplementary Figure 1. Phenotype, morphology and distribution of embryonic GFP + haematopoietic cells in the intestine. GFP + cells from E15.5 intestines were obtained after collagenase
Supplementary Figure 1 Supplementary Figure 1. Phenotype, morphology and distribution of embryonic GFP + haematopoietic cells in the intestine. GFP + cells from E15.5 intestines were obtained after collagenase
Dnmt3a is Essential for Hematopoietic Stem Cell Differentiation
 Supplementary Information Dnmt3a is Essential for Hematopoietic Stem Cell Differentiation Grant A. Challen 1,2,3, Deqiang Sun 9*, Mira Jeong 1,2,5*, Min Luo 5*, Jaroslav Jelinek 8*, Jonathan S. Berg 10,12*,
Supplementary Information Dnmt3a is Essential for Hematopoietic Stem Cell Differentiation Grant A. Challen 1,2,3, Deqiang Sun 9*, Mira Jeong 1,2,5*, Min Luo 5*, Jaroslav Jelinek 8*, Jonathan S. Berg 10,12*,
SUPPLEMENTAL METHODS Animals Luciferase reporter assays Labeling of bone marrow sinusoidal and parenchymal B cells Epigenetic Analysis
 SUPPLEMENTAL METHODS Animals C57BL/6, Eµ/miR-125b, Eµ/miR-125b-Cas9, CD19cre IRF4 flox/flox, Eµ/miR-125b-CD19cre IRF4 flox/flox Tg mice were maintained in the Caltech Office of Laboratory Animal Resources
SUPPLEMENTAL METHODS Animals C57BL/6, Eµ/miR-125b, Eµ/miR-125b-Cas9, CD19cre IRF4 flox/flox, Eµ/miR-125b-CD19cre IRF4 flox/flox Tg mice were maintained in the Caltech Office of Laboratory Animal Resources
Nature Biotechnology: doi: /nbt Supplementary Figure 1
 Supplementary Figure 1 Schematic and results of screening the combinatorial antibody library for Sox2 replacement activity. A single batch of MEFs were plated and transduced with doxycycline inducible
Supplementary Figure 1 Schematic and results of screening the combinatorial antibody library for Sox2 replacement activity. A single batch of MEFs were plated and transduced with doxycycline inducible
Total RNA was isolated by Trizol Dnase-I treated and cdna synthesised using MMLV-RT
 Supplemental Material Methods Q-RT-PCR Total RNA was isolated by Trizol Dnase-I treated and cdna synthesised using MMLV-RT and random hexamer primers according to the manufacturer s protocols (Invitrogen,
Supplemental Material Methods Q-RT-PCR Total RNA was isolated by Trizol Dnase-I treated and cdna synthesised using MMLV-RT and random hexamer primers according to the manufacturer s protocols (Invitrogen,
LoxP. SFFV mcherry wpre SIN-LTR. shrna-p53. LoxP. shrna-scr
 LeGO-C-p53-shRNA SIN-LTR? RRE cppt H1/TO shrna-p53 SFFV mcherry wpre SIN-LTR LeGO-C-scr-shRNA SIN-LTR? RRE cppt H1/TO shrna-scr SFFV mcherry wpre SIN-LTR Supplementary Figure 3A: Scheme of the lentiviral
LeGO-C-p53-shRNA SIN-LTR? RRE cppt H1/TO shrna-p53 SFFV mcherry wpre SIN-LTR LeGO-C-scr-shRNA SIN-LTR? RRE cppt H1/TO shrna-scr SFFV mcherry wpre SIN-LTR Supplementary Figure 3A: Scheme of the lentiviral
Mature cells were first removed from the bone marrow by immunomagnetic cell separation. After
 Immunomagnetic cell separation and flow cytometry analysis Mature cells were first removed from the bone marrow by immunomagnetic cell separation. After incubation with blocking solution (IMDM, 2% FCS,
Immunomagnetic cell separation and flow cytometry analysis Mature cells were first removed from the bone marrow by immunomagnetic cell separation. After incubation with blocking solution (IMDM, 2% FCS,
SUPPLEMENTAL MATERIAL. Supplemental Methods. Cumate solution was from System Biosciences. Human complement C1q and complement
 SUPPLEMENTAL MATERIAL Supplemental Methods Reagents Cumate solution was from System Biosciences. Human complement Cq and complement C-esterase inhibitor (C-INH) were from Calbiochem. C-INH (Berinert) for
SUPPLEMENTAL MATERIAL Supplemental Methods Reagents Cumate solution was from System Biosciences. Human complement Cq and complement C-esterase inhibitor (C-INH) were from Calbiochem. C-INH (Berinert) for
Single-cell suspensions of C57BL/6J splenocytes were incubated with CD5 beads
 S S Single-cell suspensions of C57BL/6J splenocytes were incubated with CD5 beads (Miltenyi Biotech) and T cells were positively selected by magnetically activated cell sorting (MACS). 50x10 6 or greater
S S Single-cell suspensions of C57BL/6J splenocytes were incubated with CD5 beads (Miltenyi Biotech) and T cells were positively selected by magnetically activated cell sorting (MACS). 50x10 6 or greater
Supplementary Figure 1
 Supplementary Figure 1 Ex2 promotor region Cre IRES cherry pa Ex4 Ex5 Ex1 untranslated Ex3 Ex5 untranslated EYFP pa Rosa26 STOP loxp loxp Cre recombinase EYFP pa Rosa26 loxp 1 kb Interleukin-9 fate reporter
Supplementary Figure 1 Ex2 promotor region Cre IRES cherry pa Ex4 Ex5 Ex1 untranslated Ex3 Ex5 untranslated EYFP pa Rosa26 STOP loxp loxp Cre recombinase EYFP pa Rosa26 loxp 1 kb Interleukin-9 fate reporter
Isolation of ILC2 from Mouse Liver Tamar Mchedlidze and Stefan Wirtz *
 Isolation of ILC2 from Mouse Liver Tamar Mchedlidze and Stefan Wirtz * Medical Department 1, FAU Erlangen-Nuremberg, Erlangen, Germany *For correspondence: stefan.wirtz@uk-erlangen.de [Abstract] Group
Isolation of ILC2 from Mouse Liver Tamar Mchedlidze and Stefan Wirtz * Medical Department 1, FAU Erlangen-Nuremberg, Erlangen, Germany *For correspondence: stefan.wirtz@uk-erlangen.de [Abstract] Group
Nature Medicine doi: /nm.2548
 Supplementary Table 1: Genotypes of offspring and embryos from matings of Pmm2 WT/F118L mice with Pmm2 WT/R137H mice total events Pmm2 WT/WT Pmm2 WT/R137H Pmm2 WT/F118L Pmm2 R137H/F118L offspring 117 (100%)
Supplementary Table 1: Genotypes of offspring and embryos from matings of Pmm2 WT/F118L mice with Pmm2 WT/R137H mice total events Pmm2 WT/WT Pmm2 WT/R137H Pmm2 WT/F118L Pmm2 R137H/F118L offspring 117 (100%)
Protein and transcriptome quantitation using BD AbSeq Antibody-Oligonucleotide
 Protein and transcriptome quantitation using BD AbSeq Antibody-Oligonucleotide technology and the 10X Genomics Chromium Single Cell Gene Expression Solution Jocelyn G. Olvera, Brigid S. Boland, John T.
Protein and transcriptome quantitation using BD AbSeq Antibody-Oligonucleotide technology and the 10X Genomics Chromium Single Cell Gene Expression Solution Jocelyn G. Olvera, Brigid S. Boland, John T.
Supplementary Methods
 Supplementary Methods Generation of CX3CR1-GFP KI Mice Briefly, four individual DNA segments, including murine 3 -flanking region, neomycin resistance gene flanked by loxp sites, hcx 3 CR1-GFP, and murine
Supplementary Methods Generation of CX3CR1-GFP KI Mice Briefly, four individual DNA segments, including murine 3 -flanking region, neomycin resistance gene flanked by loxp sites, hcx 3 CR1-GFP, and murine
Supplementary Figure 1: Analysis of monocyte subsets and lineage relationships. (a) Gating strategy for definition of MDP and cmop populations in BM
 Supplementary Figure 1: Analysis of monocyte subsets and lineage relationships. (a) Gating strategy for definition of MDP and cmop populations in BM of Cx3cr1 GFP/+ mice related to Fig. 1a. MDP was defined
Supplementary Figure 1: Analysis of monocyte subsets and lineage relationships. (a) Gating strategy for definition of MDP and cmop populations in BM of Cx3cr1 GFP/+ mice related to Fig. 1a. MDP was defined
Nature Immunology: doi: /ni Supplementary Figure 1
 Supplementary Figure 1 Sox2 localizes in neutrophils of mouse spleen. (a) Microscopy analysis of Sox2 and MPO in wild-type mouse spleen. Nucl, nucleus. (b) Immunohistochemistry staining of Sox2 and MPO
Supplementary Figure 1 Sox2 localizes in neutrophils of mouse spleen. (a) Microscopy analysis of Sox2 and MPO in wild-type mouse spleen. Nucl, nucleus. (b) Immunohistochemistry staining of Sox2 and MPO
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Diffusion of MCF-7 and 3T3 cells. (a) High-resolution image (40x): DAPI and Cy5-CellTracker TM stained MCF-7 cells. (b) Corresponding high-resolution (40x)
Supplementary Figures Supplementary Figure 1. Diffusion of MCF-7 and 3T3 cells. (a) High-resolution image (40x): DAPI and Cy5-CellTracker TM stained MCF-7 cells. (b) Corresponding high-resolution (40x)
Yalin Emre, Magali Irla, Isabelle Dunand- Sauthier, Romain Ballet, Mehdi Meguenani, Stephane Jemelin, Christian Vesin, Walter Reith and Beat A.
 Supplementary information Thymic epithelial cell expansion through matricellular protein CYR61 boosts progenitor homing and T- cell output Yalin Emre, Magali Irla, Isabelle Dunand- Sauthier, Romain Ballet,
Supplementary information Thymic epithelial cell expansion through matricellular protein CYR61 boosts progenitor homing and T- cell output Yalin Emre, Magali Irla, Isabelle Dunand- Sauthier, Romain Ballet,
The transcrip-on factor NR4A1 (Nur77) controls bone marrow differen-a-on and survival of Ly6C monocytes
 Supplementary Informa0on The transcrip-on factor NR4A1 (Nur77) controls bone marrow differen-a-on and survival of Ly6C monocytes Richard N. Hanna 1, Leo M. Carlin 2, Harper G. Hubbeling 3, Dominika Nackiewicz
Supplementary Informa0on The transcrip-on factor NR4A1 (Nur77) controls bone marrow differen-a-on and survival of Ly6C monocytes Richard N. Hanna 1, Leo M. Carlin 2, Harper G. Hubbeling 3, Dominika Nackiewicz
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION ARTICLE NUMBER: 16108 DOI: 10.1038/NMICROBIOL.2016.108 The binary toxin CDT enhances Clostridium difficile virulence by suppressing protective colonic eosinophilia Carrie A. Cowardin,
SUPPLEMENTARY INFORMATION ARTICLE NUMBER: 16108 DOI: 10.1038/NMICROBIOL.2016.108 The binary toxin CDT enhances Clostridium difficile virulence by suppressing protective colonic eosinophilia Carrie A. Cowardin,
Data Sheet GITR / NF-ĸB-Luciferase Reporter (Luc) - Jurkat Cell Line Catalog #60546
 Data Sheet GITR / NF-ĸB-Luciferase Reporter (Luc) - Jurkat Cell Line Catalog #60546 Description This cell line expresses a surface human GITR (glucocorticoid-induced TNFR family related gene; TNFRSF18;
Data Sheet GITR / NF-ĸB-Luciferase Reporter (Luc) - Jurkat Cell Line Catalog #60546 Description This cell line expresses a surface human GITR (glucocorticoid-induced TNFR family related gene; TNFRSF18;
Supplementary Data. Supplementary Materials and Methods Twenty-cell gene expression profiling. Stem cell transcriptome analysis
 Supplementary Data The Supplementary Data includes Supplementary Materials and Methods, and Figures (Supplementary Figs. S1 S10). Supplementary Materials and Methods Twenty-cell gene expression profiling
Supplementary Data The Supplementary Data includes Supplementary Materials and Methods, and Figures (Supplementary Figs. S1 S10). Supplementary Materials and Methods Twenty-cell gene expression profiling
(A-B) P2ry14 expression was assessed by (A) genotyping (upper arrow: WT; lower
 Supplementary Figures S1. (A-B) P2ry14 expression was assessed by (A) genotyping (upper arrow: ; lower arrow: KO) and (B) q-pcr analysis with Lin- cells, The white vertical line in panel A indicates that
Supplementary Figures S1. (A-B) P2ry14 expression was assessed by (A) genotyping (upper arrow: ; lower arrow: KO) and (B) q-pcr analysis with Lin- cells, The white vertical line in panel A indicates that
Table S1. Nucleotide sequences of synthesized oligonucleotides for quantitative
 Table S1. Nucleotide sequences of synthesized oligonucleotides for quantitative RT-PCR For CD8-1 transcript, forward primer CD8-1F 5 -TAGTAACCAGAGGCCGCAAGA-3 reverse primer CD8-1R 5 -TCTACTAAGGTGTCCCATAGCATGAT-3
Table S1. Nucleotide sequences of synthesized oligonucleotides for quantitative RT-PCR For CD8-1 transcript, forward primer CD8-1F 5 -TAGTAACCAGAGGCCGCAAGA-3 reverse primer CD8-1R 5 -TCTACTAAGGTGTCCCATAGCATGAT-3
Supplementary methods. RNA isolation, cdna synthesis, and quantitative real-time PCR. Total RNAs were
 Supplementary methods RNA isolation, cdna synthesis, and quantitative real-time PCR. Total RNAs were extracted from cells by using an RNeasy Mini Kit (QIAGEN) and then reverse-transcribed using the SuperScript
Supplementary methods RNA isolation, cdna synthesis, and quantitative real-time PCR. Total RNAs were extracted from cells by using an RNeasy Mini Kit (QIAGEN) and then reverse-transcribed using the SuperScript
Mammosphere formation assay. Mammosphere culture was performed as previously described (13,
 Supplemental Text Materials and Methods Mammosphere formation assay. Mammosphere culture was performed as previously described (13, 17). For co-culture with fibroblasts and treatment with CM or CCL2, fibroblasts
Supplemental Text Materials and Methods Mammosphere formation assay. Mammosphere culture was performed as previously described (13, 17). For co-culture with fibroblasts and treatment with CM or CCL2, fibroblasts
Flow-cytometry assessment of parasitemia in cell parasite co-culture assay using hydroethidine staining Western blotting RT-PCR
 Flow-cytometry assessment of parasitemia in cell parasite co-culture assay using hydroethidine staining The capability of cells, either PBMC or purified T- cell lines, to inhibit parasite growth or merozoite
Flow-cytometry assessment of parasitemia in cell parasite co-culture assay using hydroethidine staining The capability of cells, either PBMC or purified T- cell lines, to inhibit parasite growth or merozoite
Supplementary Figure 1 Phosphorylated tau accumulates in Nrf2 (-/-) mice. Hippocampal tissues obtained from Nrf2 (-/-) (10 months old, 4 male; 2
 Supplementary Figure 1 Phosphorylated tau accumulates in Nrf2 (-/-) mice. Hippocampal tissues obtained from Nrf2 (-/-) (10 months old, 4 male; 2 female) or wild-type (5 months old, 1 male; 11 months old,
Supplementary Figure 1 Phosphorylated tau accumulates in Nrf2 (-/-) mice. Hippocampal tissues obtained from Nrf2 (-/-) (10 months old, 4 male; 2 female) or wild-type (5 months old, 1 male; 11 months old,
Interferon- -producing immature myeloid cells confer protection. against severe invasive group A Streptococcus infections. Supplementary Information
 Supplementary Information Interferon- -producing immature myeloid cells confer protection against severe invasive group A Streptococcus infections Takayuki Matsumura, Manabu Ato, Tadayoshi Ikebe, Makoto
Supplementary Information Interferon- -producing immature myeloid cells confer protection against severe invasive group A Streptococcus infections Takayuki Matsumura, Manabu Ato, Tadayoshi Ikebe, Makoto
Supporting Information
 CD38 CD38 CD43 CD5 CD43 CD11b CD3 Supporting Information Chu et al. 10.1073/pnas.1613884113 ntibodies The following anti-mouse antibodies were purchased from iolegend: anti-220 (R3-62), anti- (6D5), anti-cd138
CD38 CD38 CD43 CD5 CD43 CD11b CD3 Supporting Information Chu et al. 10.1073/pnas.1613884113 ntibodies The following anti-mouse antibodies were purchased from iolegend: anti-220 (R3-62), anti- (6D5), anti-cd138
RPL27A is a target of mir-595 and may contribute to the myelodysplastic phenotype through ribosomal dysgenesis
 www.impactjournals.com/oncotarget/ Oncotarget, Supplementary Materials 2016 RPL27A is a target of mir-595 and may contribute to the myelodysplastic phenotype through ribosomal dysgenesis Supplementary
www.impactjournals.com/oncotarget/ Oncotarget, Supplementary Materials 2016 RPL27A is a target of mir-595 and may contribute to the myelodysplastic phenotype through ribosomal dysgenesis Supplementary
