Supplementary Figure 1. Expressions of stem cell markers decreased in TRCs on 2D plastic. TRCs were cultured on plastic for 1, 3, 5, or 7 days,
|
|
|
- Junior Harvey
- 5 years ago
- Views:
Transcription
1 Supplementary Figure 1. Expressions of stem cell markers decreased in TRCs on 2D plastic. TRCs were cultured on plastic for 1, 3, 5, or 7 days, respectively, and their mrnas were quantified by real time RT-PCR for expressions of Bmi-1 (a), C-kit (b), Nestin (c), and Tert (d). Mean±s.e.m., n=3 independent experiments. * p<
2 Supplementary Figure 2. Low levels of apoptosis for cells in various conditions. The cells in Figure 1c were cultured in 90-Pa soft fibrin gels and live stained with 10 µg ml -1 propidium iodide (PI). The fluorescence images were taken from day 1 through day 5 for apoptosis analysis. The data represent the normalized fluorescence intensity (mean gray values divided by 255). The filled black circle or the filled triangle denotes a positive control, in which 2 µm doxorubicin was used to induce apoptosis 12 hr before day 5 or 100% methanol was used to fix the cells at day 5 when control B16-F1 cells were cultured in 90-Pa fibrin gels. Each value was averaged from > 20 colonies. The gray value of 0 means no fluorescence (0% apoptosis), while 1 means the highest fluorescence (100% apoptosis). * and # indicates significant differences (p<0.0001) between Control cells + Methanol or Control cells + Doxorubicin and all the others at day 5, respectively. 2
3 Supplementary Figure 3. Reduction of colony number in TRCs after culture on rigid plastic. TRC: B16-F1 cells were cultured in 90-Pa fibrin gels for 5 days. These TRCs were then re-plated on rigid plastic for 1, 3, 5, and 7 days, respectively, and re-seeded into 90-Pa fibrin gels. B16-F1 cells cultured on plastic were used as a control. There are significant differences between Control and all other conditions (all p<0.02) and between TRC and 1, or 3, or 5, or 7 days (all p<0.05, except 5 days at day 1, where p=0.055). Mean ± s.e.m.; n=2 independent experiments. * and # indicates significant differences (p<0.05) between Control cells or TRC and all the others, respectively. 3
4 Supplementary Figure 4. Schematic of the H3K9 FRET probe. (a) H3K9 FRET probe and its specific function. See Methods section for details. (b) Expression of H3K9 FRET probe fluorescence within nucleus (left) and the quantification of YPet/CFP ratio in the nucleus was always performed in the area within the dashed blue lines (~1.5 m from the nuclear periphery) (right). Scale Bars=5 m. 4
5 Supplementary Figure 5. Traction force of control B16-F1 cells increases with substrate stiffness, depends on the specific type of ECM protein, and is associated with H3K9 methylation. (a) Representative images of traction map of B16-F1 cells on polyacrylamide gels with various stiffnesses coated with fibrinogen or collagen 1. Scale bar=20 m for all columns. (b) Traction forces increase with substrate stiffness and are much larger on collagen coated substrates. Mean±s.e.m.; n=10 cells per condition. (c) Inhibition of traction forces decreases H3K9 methylation. Control B16-F1 cells were transfected with H3K9 FRET probe and plated on collagen-coated 8 kpa gels. Tractions and H3K9 methylation level were measure before (0 min) and 20 minutes after the addition of 25 ML7 (20 min+25 M ML7). Mean±s.e.m.; n=6 cells per condition. *p <
6 Supplementary Figure 6. H3K9 methylation in TRCs and control melanoma cells. (a) Western blotting analysis of H3K9 di- (me2) or tri-methylation (me3) in various conditions. 7 days: TRCs were cultured on rigid plastic for 7 days; Neg Ctr: control melanoma cells transfected with negative control sirna; sirna: control melanoma cells transfected with G9a (for H3K9 me2) or SUV39h1 (for H3K9 me3) sirna #1. (b) ChIP assays for H3K9 me2 (left panel) and me3 (right panel) in the promoter region of Sox2. The immunoprecipitated DNAs at the indicated conditions were used for RT-PCR. Mouse IgG was used as negative control. To exclude non-specific effect, the relative expression in the GAPDH promoter was also shown. 6
7 Supplementary Figure 7. Knockdown of G9a or SUV39h1 in control melanoma cells using another set of sirnas also promotes Sox2 expression and self-renewal capability. (a) Knockdown of G9a or SUV39h1 in control melanoma cells using another set of sirnas significantly increases their Sox2 expression. *p<0.05. (b) Knockdown of G9a or SUV39h1 greatly increases the growth of tumor spheroids; (c) Knockdown of G9a or SUV39h1 increases the number of tumor spheroids. (d) Knockdown of G9a or SUV39h1 does not increase the colony number. In (b), there are significant differences between Negative Control and G9a sirna #2 at day 5 (p=0.0015), or SUV39h1 sirna #2 from day 2 to day 5 (all p<0.001), and between TRC and Negative control at day 4 and 5 (both p<0.03), or SUV39h1 sirna #2 from day 3 to day 5 (all p<0.02), where * and # indicates differences comparing silenced cells with Negative control and TRC, respectively. In (c), there are significant differences between TRC and all the others from day 1 through day 5 (all p<0.001) and differences between Negative control and G9a sirna #2 at day 4 and day 5 (both p<0.02), or SUV39h1 sirna #2 at day 5 (p=0.03). # indicates significant differences between TRC and all the others. Mean±s.e.m.; n=3 independent experiments for all subfigures. In (d), there are differences between Negative Control and G9a sirna #1 or SUV39h1 sirna #1 from day 1 through day 5 (all p<0.05, except G9a sirna #1 at day 2 and SUV39h1 sirna #1 at day 1, where both p>0.05). * indicates significant difference between Negative Control and G9a sirna #1 or SUV39h1 sirna #1 and # indicates significant differences between TRC and all others. 7
8 Supplementary Figure 8. Silencing Sox2 in TRCs does not affect expression of G9a and SUV39h1. TRCs were transfected with Sox2 shrna for 13 hrs and the total proteins of these cells were extracted for western blot analysis on G9a and SUV39h1. No significant differences were found after Sox2 silencing. Control cells: control B16-F1 cells cultured on plastic. Mean±s.e.m.; n = 3 independent experiments. Note that TRCs express no G9a or SUV39h1 protein. 8
9 Supplementary Figure 9. Inhibiting Sox2 expression abrogates the increase in self-renewal capacity induced by H3K9 demethylation in control melanoma cells. Control melanoma cells were transfected with G9a or SUV39h1 sirna and Sox2 shrna concurrently and then seeded in soft fibrin gels. Their colony size (a) and number (b) were monitored from day 1 through day 5. Neg Ctr: negative control sirna. Mean±s.e.m.; two independent experiments. In (a), there are significant differences between Neg Ctr and all the others from day 2 to day 5 (all p<0.02); in (b), there are only differences between Neg Ctr and Neg Ctr+Sox2 shrna#3 at day 2 (p=0.026), or G9a sirna#1+sox2 shrna#3 at day 2 and 3 (both p<0.05). * indicates significant differences between Neg Ctr and all the others. 9
10 Supplementary Figure 10. Inhibiting H3K9 demethylase KDM4 dramatically prevents colony formation and growth. H3K9 demethylase KDM4 inhibitors JIB-04 (a, b) and DMOG (c, d) greatly suppress the colony size and number. Control melanoma cells were cultured in 90- Pa fibrin gels in the presence or absence of different doses of inhibitors. n=2 independent experiments. In (a) and (b), there are significant differences between Control and 0.2 µm or 1 µm JIB-04 from day 2 through day 5 (all p< in (a); all p<0.02 in (b)). In (c) and (d), there are differences between Control and 0.8 mm or 4 mm DMOG from day 2 through day 5 (all p< in (c); all p<0.05 in (d). * indicates significant differences between Control and all the others. 10
11 Supplementary Figure 11. Knockdown of Sox2 in TRCs greatly decreases the expressions of Sox2, Bmi-1, and C-kit. TRCs were transfected with Sox2 shrna and their mrnas were used to quantify the expressions of Bmi-1 and C-kit by real time RT-PCR. Mean ± s.e.m.; n=3 independent experiments; *p<
12 Supplementary Figure 12. Silencing Sox2 expression has no effect on the colony number and size in stiff (1050-Pa) fibrin gels. Control melanoma cells were transfected with Sox2 shrna or scrambled control and then cultured in 1050-Pa fibrin gels. Mean ± s.e.m.; n>80 colonies from 2 independent experiments; all p>0.05 except at day 2 in (a), where p< as indicated by *. 12
13 Supplementary Figure 13. Sox2 overexpression rescues the self-renewal capability of control melanoma cells. Control cells were transfected with Sox2 cdna and then cultured in soft fibrin gels. The colony size (a) and number (b) were measured from day 1 through day 5. There are no differences in (a) in colony size and significant differences between Control and Sox2 cdna from day 1 through day 5 in (b). Mean+/-s.e.m; *p<0.01 between Control and Sox2 cdna. 13
14 Supplementary Figure 14. Sox2 expression and H3K9 demethylation are critical in the selection and propagation of TRCs by soft fibrin gels. (a) Inhibition of Sox2 up-regulation almost completely suppresses spheroid growth. Control melanoma cells were transfected with Sox2 shrna and then cultured in soft fibrin gels. (b) Inhibition of Sox2 up-regulation significantly decreases spheroid formation. (c) Daminozide, inhibitor of H3K9 demethylase KDM7, significantly prevents colony growth but not spheroid formation (d). There are significant differences between Scrambled control and Sox2 shrna #3 in (a) (all p< ) and in (b) (all p<0.05) from day 1 through day 5. In (c), there are differences between Control and 2 µm or 20 µm Daminozide at day 4 and day 5 (all p<0.05). In (d), there are no differences between Control and 2 µm or 20 µm Daminozide. * indicates significant differences between Control and Sox2 shrna #3 in (a) and (b) or between Control and 2 µm or 20 µm Daminozide in (c). Statistics analysis was also conducted between Sox2 shrna #3 in (a) or (b) and 2 µm or 20 µm Daminozide in (c) or (d), where all the data were first normalized by the respective Control. There are significant differences between Sox2 shrna #3 and 2 µm or 20 µm Daminozide from day 1 through day 5 in colony size in (a, c) (all p< ) and in colony number in (b, d) (all p<0.04). 14
15 Supplementary Figure 15. Silencing Cdc42 decreases H3K9 methylation but not Sox2 expression. (a) Representative western blotting images of H3K9me2 and me3 after Cdc42 knockdown in control melanoma cells. (b) Efficiency of Cdc42 knockdown. Mean±s.e.m.; n=3 independent experiments; *p<0.05. (c) Silencing Cdc42 has no effect on Sox2 expression. Mean±s.e.m., n=7 independent experiments. 15
16 Supplementary Figure 16. Melanoma TRCs are the progenitors of control melanoma cells but more differentiated than melanoblasts. (a, b) Upregulation of self-renewal marker Sox2 and downregulation of melanocyte-specific gene Mitf in melanoma TRCs. (c, d) TRCs and control B16-F1 cells express similar levels of melanin-related genes. After 5-day culture in 90-Pa fibrin gels, total mrna of B16-F1 spheroid cells (TRCs) were extracted for detection of Sox2, Mitf (a, b), Pax3, DCT, Gpnmb, Tyrp1, and Sox10 (c, d) expression by RT-PCR (a, c) and real time RT-PCR (b, d). B16-F1 cells cultured on 2D rigid dish were used as a control. A representative of 3 independent experiments in (a, c). Mean±s.e.m.; n=3 independent experiments; *p<
17 Supplementary Figure 17. Regulation of Mitf by Sox2. (a) Sox2 silencing in TRCs increases expression of Mitf. (b) Efficiency of Mitf knockdown. (c) Silencing Mitf in control melanoma cells (Neg Ctr) does not affect Sox2 expression. TRCs (or control cells) were transfected with Sox2 shrna (or Mitf sirna) and total mrnas were extracted to quantify the expression of Mitf or Sox2 by real time RT-PCR. Mean±s.e.m., n=3 independent experiments. *, p<
18 Supplementary Figure 18. Expression of melanocyte-related gene Mitf increased in TRCs after culture on rigid plastic. (a) Representative image of expression of melanocyte-related genes Mitf. Control: B16-F1 cultured on rigid plastic. TRC: Control B16-F1 cells were cultured in 90-Pa fibrin gels for 5 days. 1 day, 3 days, 5 days, or 7 days: TRCs were seeded on 2D rigid dishes for 1, 3, 5, or 7days. (b) Quantification of Mitf expression by real time RT-PCR. Mean±s.e.m., n=3 independent experiments.*p<0.05. (c) Images with side markers show the gene sizes of GAPDH and Sox2 on the original gel in Fig. 1a. (d) Original Western blot images show the molecular weights of GAPDH and Sox2 in Fig.1a. 18
19 Supplementary Figure 19. Matrix stiffness but not porosity affects round colony formation. Control melanoma cells were cultured in soft fibrin gels for 5 days. In the same gel, those cells near the rigid bottom formed spread colonies (a, b), while the cells far away from the rigid bottom formed round colonies (c, d). z: distances from the rigid bottom. Note that in (b) a round shadow is the round colony in (c) that is ~170 m above (b). 19
20 Supplementary Figure 20. Additional soluble fibrinogen does not affect colony number and growth in soft fibrin gels. Control B16-F1 cells were cultured in soft fibrin gels (1 mg ml -1 ; 90- Pa) in the absence and presence of additional soluble fibrinogen (7 mg ml -1 soluble fibrinogen; total fibrinogen amount is equivalent to that of the 1050-Pa (8 mg ml -1 ) fibrin gel). Mean±s.e.m., n=3 independent experiments. 20
21 Supplementary Table 1. Sox2 is necessary for high efficiency of tumorigenicity in vivo. Sox2 expression in TRCs was silenced following the same protocol as in Figure 4d. These cells were then trypsinized and the cell number was counted. 100 of untreated TRCs, TRCs treated with Sox2 shrna, or scrambled control, or control melanoma cells (harvested from rigid plastic) were injected subcutaneously or intravenously via tail veins into wild-type syngeneic C57BL/6 mice, respectively. Tumor number and size measured at day 28 for subcutaneous injection (s.c.) are shown. The data are summarized from two independent experiments (each group 8 mice were used). Untreated TRCs + TRCs+ Plastic TRCs scrambled Sox2 shrna shrna # of Mice with 9/16 3/8 2/16 * 0/8 Tumor (s.c.) Tumor Size >3434 > (mm 3 ) (s.c.) s.c.: subcutaneous injection. *, p<0.024 (Fisher s exact test) compared with untreated TRCs. 21
22 Supplementary Table 2. Primers for reverse transcription PCR (RT-PCR). 22
23 Supplementary Table 3. Primers for real time RT-PCR. 23
24 Supplementary Table 4. Primers for ChIP assay. 24
Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured
 Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured in 90-Pa 3D fibrin gels for 5 days in the presence
Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured in 90-Pa 3D fibrin gels for 5 days in the presence
Supplementary Figure 1. IFN-γ induces TRC dormancy. a, IFN-γ induced dormancy
 Supplementary Figure 1. IFN-γ induces TRC dormancy. a, IFN-γ induced dormancy of various tumor type TRCs, including H22 (murine hepatocarcinoma) and CT26 (murine colon cancer). Bar, 50 µm. b, B16 cells
Supplementary Figure 1. IFN-γ induces TRC dormancy. a, IFN-γ induced dormancy of various tumor type TRCs, including H22 (murine hepatocarcinoma) and CT26 (murine colon cancer). Bar, 50 µm. b, B16 cells
SOFT 3D FIBRIN MATRICES DOWNREGULATE FAK EXPRESSION TO PROMOTE SELF-RENEWAL OF TUMOR-REPOPULATING CELLS ADAM RICHARD WOOD THESIS
 SOFT 3D FIBRIN MATRICES DOWNREGULATE FAK EXPRESSION TO PROMOTE SELF-RENEWAL OF TUMOR-REPOPULATING CELLS BY ADAM RICHARD WOOD THESIS Submitted in partial fulfillment of the requirements for the degree of
SOFT 3D FIBRIN MATRICES DOWNREGULATE FAK EXPRESSION TO PROMOTE SELF-RENEWAL OF TUMOR-REPOPULATING CELLS BY ADAM RICHARD WOOD THESIS Submitted in partial fulfillment of the requirements for the degree of
Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan).
 1 2 3 4 5 6 7 8 Supplemental Materials and Methods Cell proliferation assay Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). GCs were plated at 96-well
1 2 3 4 5 6 7 8 Supplemental Materials and Methods Cell proliferation assay Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). GCs were plated at 96-well
Supplementary Table S1
 Primers used in RT-qPCR, ChIP and Bisulphite-Sequencing. Quantitative real-time RT-PCR primers Supplementary Table S1 gene Forward primer sequence Reverse primer sequence Product TRAIL CAACTCCGTCAGCTCGTTAGAAAG
Primers used in RT-qPCR, ChIP and Bisulphite-Sequencing. Quantitative real-time RT-PCR primers Supplementary Table S1 gene Forward primer sequence Reverse primer sequence Product TRAIL CAACTCCGTCAGCTCGTTAGAAAG
Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and
 Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
hnrnp D/AUF1 Rabbit IgG hnrnp M
 Mouse IgG Goat IgG Rabbit IgG Mouse IgG hnrnp F Goat IgG Mouse IgG Kb 6 4 3 2 15 5 Supplementary Figure S1. In vivo binding of TERRA-bound RBPs to target RNAs. Immunoprecipitation (IP) assay using 3 mg
Mouse IgG Goat IgG Rabbit IgG Mouse IgG hnrnp F Goat IgG Mouse IgG Kb 6 4 3 2 15 5 Supplementary Figure S1. In vivo binding of TERRA-bound RBPs to target RNAs. Immunoprecipitation (IP) assay using 3 mg
Supplementary Information. HBx-upregulated lncrna UCA1 promotes cell growth and tumorigenesis
 Supplementary Information HBx-upregulated lncrna UCA1 promotes cell growth and tumorigenesis by recruiting EZH2 and repressing p27kip1/cdk2 signaling Jiao-Jiao Hu 1, Wei Song 1, Shao-Dan Zhang 1, Xiao-Hui
Supplementary Information HBx-upregulated lncrna UCA1 promotes cell growth and tumorigenesis by recruiting EZH2 and repressing p27kip1/cdk2 signaling Jiao-Jiao Hu 1, Wei Song 1, Shao-Dan Zhang 1, Xiao-Hui
SUPPLEMENTARY INFORMATION
 Soft fibrin gels promote selection and growth of tumorigenic cells Jing Liu $, Youhua Tan $, Huafeng Zhang, Yi Zhang, Pingwei Xu, Junwei Chen, Yeh-Chuin Poh, Ke Tang, Ning Wang*, Bo Huang* $ These authors
Soft fibrin gels promote selection and growth of tumorigenic cells Jing Liu $, Youhua Tan $, Huafeng Zhang, Yi Zhang, Pingwei Xu, Junwei Chen, Yeh-Chuin Poh, Ke Tang, Ning Wang*, Bo Huang* $ These authors
Nature Biotechnology: doi: /nbt Supplementary Figure 1
 Supplementary Figure 1 Schematic and results of screening the combinatorial antibody library for Sox2 replacement activity. A single batch of MEFs were plated and transduced with doxycycline inducible
Supplementary Figure 1 Schematic and results of screening the combinatorial antibody library for Sox2 replacement activity. A single batch of MEFs were plated and transduced with doxycycline inducible
Supplementary Data. Supplementary Methods Three-step protocol for spontaneous differentiation of mouse induced pluripotent stem (embryonic stem) cells
 Supplementary Data Supplementary Methods Three-step protocol for spontaneous differentiation of mouse induced pluripotent stem (embryonic stem) cells Mouse induced pluripotent stem cells (ipscs) were cultured
Supplementary Data Supplementary Methods Three-step protocol for spontaneous differentiation of mouse induced pluripotent stem (embryonic stem) cells Mouse induced pluripotent stem cells (ipscs) were cultured
Supplementary Figure Legend
 Supplementary Figure Legend Supplementary Figure S1. Effects of MMP-1 silencing on HEp3-hi/diss cell proliferation in 2D and 3D culture conditions. (A) Downregulation of MMP-1 expression in HEp3-hi/diss
Supplementary Figure Legend Supplementary Figure S1. Effects of MMP-1 silencing on HEp3-hi/diss cell proliferation in 2D and 3D culture conditions. (A) Downregulation of MMP-1 expression in HEp3-hi/diss
SUPPLEMENTARY INFORMATION
 Transcription upregulation via force-induced direct stretching of chromatin Arash Tajik 1, 2#, Yuejin Zhang 1#, Fuxiang Wei 1#, Jian Sun 2#, Qiong Jia 1, Wenwen Zhou 1, Rishi Singh 2, Nimish Khanna 3,
Transcription upregulation via force-induced direct stretching of chromatin Arash Tajik 1, 2#, Yuejin Zhang 1#, Fuxiang Wei 1#, Jian Sun 2#, Qiong Jia 1, Wenwen Zhou 1, Rishi Singh 2, Nimish Khanna 3,
Supplementary Information
 Supplementary Information Supplementary Fig. 1 Morphology of NP69 and CNE1 cells. Phase contrast images of normal nasopharyngeal epithelial NP69 cells (left) and the well differentiated NPC CNE1 cells
Supplementary Information Supplementary Fig. 1 Morphology of NP69 and CNE1 cells. Phase contrast images of normal nasopharyngeal epithelial NP69 cells (left) and the well differentiated NPC CNE1 cells
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
SUPPLEMENTAL MATERIAL. Carvajal et al. Supplemental Table 1. List of quantitative PCR primers used for cdna analyses and
 UPPLEMEAL MATERIAL Carvajal et al. upplemental Table 1. List of quantitative PCR primers used for cdna analyses and chromatin immunoprecipitation assays. Figure 1. DNA damage-induced transcriptional repression
UPPLEMEAL MATERIAL Carvajal et al. upplemental Table 1. List of quantitative PCR primers used for cdna analyses and chromatin immunoprecipitation assays. Figure 1. DNA damage-induced transcriptional repression
MeCP2. MeCP2/α-tubulin. GFP mir1-1 mir132
 Conservation Figure S1. Schematic showing 3 UTR (top; thick black line), mir132 MRE (arrow) and nucleotide sequence conservation (vertical black lines; http://genome.ucsc.edu). a GFP mir1-1 mir132 b GFP
Conservation Figure S1. Schematic showing 3 UTR (top; thick black line), mir132 MRE (arrow) and nucleotide sequence conservation (vertical black lines; http://genome.ucsc.edu). a GFP mir1-1 mir132 b GFP
Nature Medicine: doi: /nm.4169
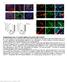 Supplementary Fig.1. EC-specific deletion of Ccm3 by Cdh5-CreERT2. a. mt/mg reporter mice were bred with Cdh5CreERT2 deleter mice followed by tamoxifen feeding from P1 to P3. mg expression was specifically
Supplementary Fig.1. EC-specific deletion of Ccm3 by Cdh5-CreERT2. a. mt/mg reporter mice were bred with Cdh5CreERT2 deleter mice followed by tamoxifen feeding from P1 to P3. mg expression was specifically
SUPPLEMENTARY INFORMATION
 (Supplementary Methods and Materials) GST pull-down assay GST-fusion proteins Fe65 365-533, and Fe65 538-700 were expressed in BL21 bacterial cells and purified with glutathione-agarose beads (Sigma).
(Supplementary Methods and Materials) GST pull-down assay GST-fusion proteins Fe65 365-533, and Fe65 538-700 were expressed in BL21 bacterial cells and purified with glutathione-agarose beads (Sigma).
Regulation of transcription by the MLL2 complex and MLL complex-associated AKAP95
 Supplementary Information Regulation of transcription by the complex and MLL complex-associated Hao Jiang, Xiangdong Lu, Miho Shimada, Yali Dou, Zhanyun Tang, and Robert G. Roeder Input HeLa NE IP lot:
Supplementary Information Regulation of transcription by the complex and MLL complex-associated Hao Jiang, Xiangdong Lu, Miho Shimada, Yali Dou, Zhanyun Tang, and Robert G. Roeder Input HeLa NE IP lot:
T-iPSC. Gra-iPSC. B-iPSC. TTF-iPSC. Supplementary Figure 1. Nature Biotechnology: doi: /nbt.1667
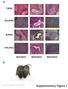 a T-iPSC Gra-iPSC B-iPSC TTF-iPSC Ectoderm Endoderm Mesoderm b Nature Biotechnology: doi:.38/nbt.667 Supplementary Figure Klf4 transgene expression Oct4 transgene expression.7 Fold GAPDH.6.5.4.3 Fold GAPDH
a T-iPSC Gra-iPSC B-iPSC TTF-iPSC Ectoderm Endoderm Mesoderm b Nature Biotechnology: doi:.38/nbt.667 Supplementary Figure Klf4 transgene expression Oct4 transgene expression.7 Fold GAPDH.6.5.4.3 Fold GAPDH
Supplementary Figure 1.
 Supplementary Figure 1. Quantification of western blot analysis of fibroblasts (related to Figure 1) (A-F) Quantification of western blot analysis for control and IR-Mut fibroblasts. Data are expressed
Supplementary Figure 1. Quantification of western blot analysis of fibroblasts (related to Figure 1) (A-F) Quantification of western blot analysis for control and IR-Mut fibroblasts. Data are expressed
14_integrins_EGFR
 α1 Integrin α1 -/- fibroblasts from integrin α1 knockout animals Figure S1. Serum-starved fibroblasts from α -/- 1 and +/+ mice were stimulated with 10% FBS for 30 min. (FBS) or plated on collagen I (CI)
α1 Integrin α1 -/- fibroblasts from integrin α1 knockout animals Figure S1. Serum-starved fibroblasts from α -/- 1 and +/+ mice were stimulated with 10% FBS for 30 min. (FBS) or plated on collagen I (CI)
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION DOI: 1.138/NMAT3777 Biophysical regulation of epigenetic state and cell reprogramming Authors: Timothy L. Downing 1,2, Jennifer Soto 1,2, Constant Morez 2,3,, Timothee Houssin
SUPPLEMENTARY INFORMATION DOI: 1.138/NMAT3777 Biophysical regulation of epigenetic state and cell reprogramming Authors: Timothy L. Downing 1,2, Jennifer Soto 1,2, Constant Morez 2,3,, Timothee Houssin
Supplementary Figure 1. TSA (10 nmol/l), non-class-selective HDAC inhibitor, potentiates
 Supplementary Figure 1. TSA (10 nmol/l), non-class-selective HDAC inhibitor, potentiates vascular calcification (VC). (a) Von Kossa staining shows that TSA potentiated the Pi-induced VC. Scale bar, 100
Supplementary Figure 1. TSA (10 nmol/l), non-class-selective HDAC inhibitor, potentiates vascular calcification (VC). (a) Von Kossa staining shows that TSA potentiated the Pi-induced VC. Scale bar, 100
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.
 Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
HCT116 SW48 Nutlin: p53
 Figure S HCT6 SW8 Nutlin: - + - + p GAPDH Figure S. Nutlin- treatment induces p protein. HCT6 and SW8 cells were left untreated or treated for 8 hr with Nutlin- ( µm) to up-regulate p. Whole cell lysates
Figure S HCT6 SW8 Nutlin: - + - + p GAPDH Figure S. Nutlin- treatment induces p protein. HCT6 and SW8 cells were left untreated or treated for 8 hr with Nutlin- ( µm) to up-regulate p. Whole cell lysates
Supplementary Figure 1. (A) Cell proliferative ability and (B) the invasiveness of
 LEGEND FOR SUPPLEMENTARY FIGURES Supplementary Figure 1. (A) Cell proliferative ability and (B) the invasiveness of LLC-1, LLC-3, and LLC-5 cell line series were quantified by BrdU assay after 72 h of
LEGEND FOR SUPPLEMENTARY FIGURES Supplementary Figure 1. (A) Cell proliferative ability and (B) the invasiveness of LLC-1, LLC-3, and LLC-5 cell line series were quantified by BrdU assay after 72 h of
Supplementary Figure 1. Reintroduction of HDAC1 in HDAC1-/- ES cells reverts the
 SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1. Reintroduction of HDAC1 in HDAC1-/- ES cells reverts the phenotype of HDAC1-/- teratomas. 3x10 6 HDAC1 reintroduced (HDAC1-/-re) and empty vector infected
SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1. Reintroduction of HDAC1 in HDAC1-/- ES cells reverts the phenotype of HDAC1-/- teratomas. 3x10 6 HDAC1 reintroduced (HDAC1-/-re) and empty vector infected
WT Day 90 after injections
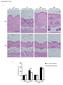 Supplementary Figure 1 a Day 1 after injections Day 9 after injections Klf5 +/- Day 1 after injections Klf5 +/- Day 9 after injections BLM PBS b Day 1 after injections Dermal thickness (μm) 3 1 Day 9 after
Supplementary Figure 1 a Day 1 after injections Day 9 after injections Klf5 +/- Day 1 after injections Klf5 +/- Day 9 after injections BLM PBS b Day 1 after injections Dermal thickness (μm) 3 1 Day 9 after
 Figure S1. Generation of HspA4 -/- mice. (A) Structures of the wild-type (Wt) HspA4 -/- allele, targeting vector and targeted allele are shown together with the relevant restriction sites. The filled rectangles
Figure S1. Generation of HspA4 -/- mice. (A) Structures of the wild-type (Wt) HspA4 -/- allele, targeting vector and targeted allele are shown together with the relevant restriction sites. The filled rectangles
Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate
 Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
SUPPLEMENTAL MATERIALS SIRTUIN 1 PROMOTES HYPEROXIA-INDUCED LUNG EPITHELIAL DEATH INDEPENDENT OF NRF2 ACTIVATION
 SUPPLEMENTAL MATERIALS SIRTUIN PROMOTES HYPEROXIA-INDUCED LUNG EPITHELIAL DEATH INDEPENDENT OF NRF ACTIVATION Haranatha R. Potteti*, Subbiah Rajasekaran*, Senthilkumar B. Rajamohan*, Chandramohan R. Tamatam,
SUPPLEMENTAL MATERIALS SIRTUIN PROMOTES HYPEROXIA-INDUCED LUNG EPITHELIAL DEATH INDEPENDENT OF NRF ACTIVATION Haranatha R. Potteti*, Subbiah Rajasekaran*, Senthilkumar B. Rajamohan*, Chandramohan R. Tamatam,
Electromagnetic Fields Mediate Efficient Cell Reprogramming Into a Pluripotent State
 Electromagnetic Fields Mediate Efficient Cell Reprogramming Into a Pluripotent State Soonbong Baek, 1 Xiaoyuan Quan, 1 Soochan Kim, 2 Christopher Lengner, 3 Jung- Keug Park, 4 and Jongpil Kim 1* 1 Lab
Electromagnetic Fields Mediate Efficient Cell Reprogramming Into a Pluripotent State Soonbong Baek, 1 Xiaoyuan Quan, 1 Soochan Kim, 2 Christopher Lengner, 3 Jung- Keug Park, 4 and Jongpil Kim 1* 1 Lab
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified
 Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Supplementary Fig. 1. (A) Working model. The pluripotency transcription factor OCT4
 SUPPLEMENTARY FIGURE LEGENDS Supplementary Fig. 1. (A) Working model. The pluripotency transcription factor OCT4 directly up-regulates the expression of NIPP1 and CCNF that together inhibit protein phosphatase
SUPPLEMENTARY FIGURE LEGENDS Supplementary Fig. 1. (A) Working model. The pluripotency transcription factor OCT4 directly up-regulates the expression of NIPP1 and CCNF that together inhibit protein phosphatase
Supplementary Table 1. Sequences for BTG2 and BRCA1 sirnas.
 Supplementary Table 1. Sequences for BTG2 and BRCA1 sirnas. Target Gene Non-target / Control BTG2 BRCA1 NFE2L2 Target Sequence ON-TARGET plus Non-targeting sirna # 1 (Cat# D-001810-01-05) sirna1: GAACCGACAUGCUCCCGGA
Supplementary Table 1. Sequences for BTG2 and BRCA1 sirnas. Target Gene Non-target / Control BTG2 BRCA1 NFE2L2 Target Sequence ON-TARGET plus Non-targeting sirna # 1 (Cat# D-001810-01-05) sirna1: GAACCGACAUGCUCCCGGA
Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 CSRP2 PFKP ADFP ADM C10orf10 GPI LOX PLEKHA2 WIPF1
 Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 4.52E-18 PDK1 6.77E-18 CSRP2 4.42E-17 PFKP 1.23E-14 MSH2 3.79E-13 NARF_A 5.56E-13 ADFP 5.56E-13 FAM13A1 1.56E-12 FAM29A_A 1.22E-11 CA9 1.54E-11
Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 4.52E-18 PDK1 6.77E-18 CSRP2 4.42E-17 PFKP 1.23E-14 MSH2 3.79E-13 NARF_A 5.56E-13 ADFP 5.56E-13 FAM13A1 1.56E-12 FAM29A_A 1.22E-11 CA9 1.54E-11
Supplementary Figure 1. Co-localization of GLUT1 and DNAL4 in BeWo cells cultured
 Supplementary Figure 1. Co-localization of GLUT1 and DNAL4 in BeWo cells cultured under static conditions. Cells were seeded in the chamber area of the device and cultured overnight without medium perfusion.
Supplementary Figure 1. Co-localization of GLUT1 and DNAL4 in BeWo cells cultured under static conditions. Cells were seeded in the chamber area of the device and cultured overnight without medium perfusion.
Supplementary Information
 Supplementary Information Supplementary Figures Supplementary Figure 1. MLK1-4 phosphorylate MEK in the presence of RAF inhibitors. (a) H157 cells were transiently transfected with Flag- or HA-tagged MLK1-4
Supplementary Information Supplementary Figures Supplementary Figure 1. MLK1-4 phosphorylate MEK in the presence of RAF inhibitors. (a) H157 cells were transiently transfected with Flag- or HA-tagged MLK1-4
Sélection Internationale Ens Ulm 2012, Cell Biology
 Sélection Internationale Ens Ulm 2012, Cell Biology Please read the whole subject before starting. Questions 1-8 and 12-14 are general biology questions. In questions 9, 10, 11 and15, we ask you to interpret
Sélection Internationale Ens Ulm 2012, Cell Biology Please read the whole subject before starting. Questions 1-8 and 12-14 are general biology questions. In questions 9, 10, 11 and15, we ask you to interpret
(A-B) P2ry14 expression was assessed by (A) genotyping (upper arrow: WT; lower
 Supplementary Figures S1. (A-B) P2ry14 expression was assessed by (A) genotyping (upper arrow: ; lower arrow: KO) and (B) q-pcr analysis with Lin- cells, The white vertical line in panel A indicates that
Supplementary Figures S1. (A-B) P2ry14 expression was assessed by (A) genotyping (upper arrow: ; lower arrow: KO) and (B) q-pcr analysis with Lin- cells, The white vertical line in panel A indicates that
monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal antibody was examined in
 Supplementary information Supplementary figures Supplementary Figure 1 Determination of the s pecificity of in-house anti-rhbdd1 mouse monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal
Supplementary information Supplementary figures Supplementary Figure 1 Determination of the s pecificity of in-house anti-rhbdd1 mouse monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal
supplementary information
 DOI: 10.1038/ncb2116 Figure S1 CDK phosphorylation of EZH2 in cells. (a) Comparison of candidate CDK phosphorylation sites on EZH2 with known CDK substrates by multiple sequence alignments. (b) CDK1 and
DOI: 10.1038/ncb2116 Figure S1 CDK phosphorylation of EZH2 in cells. (a) Comparison of candidate CDK phosphorylation sites on EZH2 with known CDK substrates by multiple sequence alignments. (b) CDK1 and
TE5 KYSE510 TE7 KYSE70 KYSE140
 TE5 KYSE5 TT KYSE7 KYSE4 Supplementary Figure. Hockey stick plots showing input normalized, rank ordered H3K7ac signals for the candidate SE-associated lncrnas in this study. Rpm Rpm Rpm Chip-seq H3K7ac
TE5 KYSE5 TT KYSE7 KYSE4 Supplementary Figure. Hockey stick plots showing input normalized, rank ordered H3K7ac signals for the candidate SE-associated lncrnas in this study. Rpm Rpm Rpm Chip-seq H3K7ac
Spironolactone ameliorates PIT1-dependent vascular osteoinduction in klotho-hypomorphic mice
 Spironolactone ameliorates PIT1-dependent vascular osteoinduction in klotho-hypomorphic mice Supplementary Material Supplementary Methods Materials Spironolactone, aldosterone and β-glycerophosphate were
Spironolactone ameliorates PIT1-dependent vascular osteoinduction in klotho-hypomorphic mice Supplementary Material Supplementary Methods Materials Spironolactone, aldosterone and β-glycerophosphate were
Emanuela Tumini, Sonia Barroso, Carmen Pérez Calero and Andrés Aguilera
 SUPPLEMENTARY INFORMATION Roles of human POLD1 and POLD3 in genome stability Emanuela Tumini, Sonia Barroso, Carmen Pérez Calero and Andrés Aguilera SUPPLEMENTARY METHODS Cell proliferation After sirna
SUPPLEMENTARY INFORMATION Roles of human POLD1 and POLD3 in genome stability Emanuela Tumini, Sonia Barroso, Carmen Pérez Calero and Andrés Aguilera SUPPLEMENTARY METHODS Cell proliferation After sirna
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the
 Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
Supplementary Information
 Supplementary Information MLL histone methylases regulate expression of HDLR- in presence of estrogen and control plasma cholesterol in vivo Khairul I. Ansari 1, Sahba Kasiri 1, Imran Hussain 1, Samara
Supplementary Information MLL histone methylases regulate expression of HDLR- in presence of estrogen and control plasma cholesterol in vivo Khairul I. Ansari 1, Sahba Kasiri 1, Imran Hussain 1, Samara
Figure S1. Specificity of polyclonal anti stabilin-1 and anti stabilin-2 antibodies Lysates of 293T cells transfected with empty vector, mouse
 Figure S1. Specificity of polyclonal anti stabilin-1 and anti stabilin-2 antibodies Lysates of 293T cells transfected with empty vector, mouse stabilin-1, or mouse stabilin-2 were immunoblotted using anti
Figure S1. Specificity of polyclonal anti stabilin-1 and anti stabilin-2 antibodies Lysates of 293T cells transfected with empty vector, mouse stabilin-1, or mouse stabilin-2 were immunoblotted using anti
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Fig. 1 shrna mediated knockdown of ZRSR2 in K562 and 293T cells. (a) ZRSR2 transcript levels in stably transduced K562 cells were determined using qrt-pcr. GAPDH was
Supplementary Figure 1 Supplementary Fig. 1 shrna mediated knockdown of ZRSR2 in K562 and 293T cells. (a) ZRSR2 transcript levels in stably transduced K562 cells were determined using qrt-pcr. GAPDH was
We performed RT-PCR, cloning, sequencing and qrt-pcr in murine melanoma. cell lines and melanocytic tumors from RET-mice in accordance with the method
 Supplementary Material and Methods Quantitative RT-PCR (qrt-pcr) We performed RT-PCR, cloning, sequencing and qrt-pcr in murine melanoma cell lines and melanocytic tumors from RET-mice in accordance with
Supplementary Material and Methods Quantitative RT-PCR (qrt-pcr) We performed RT-PCR, cloning, sequencing and qrt-pcr in murine melanoma cell lines and melanocytic tumors from RET-mice in accordance with
EpiQuik Methyl-CpG Binding Domain Protein 2 ChIP Kit
 EpiQuik Methyl-CpG Binding Domain Protein 2 ChIP Kit Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Methyl-CpG Binding Domain Protein 2 ChIP Kit is suitable for combining the
EpiQuik Methyl-CpG Binding Domain Protein 2 ChIP Kit Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Methyl-CpG Binding Domain Protein 2 ChIP Kit is suitable for combining the
TOOLS sirna and mirna. User guide
 TOOLS sirna and mirna User guide Introduction RNA interference (RNAi) is a powerful tool for suppression gene expression by causing the destruction of specific mrna molecules. Small Interfering RNAs (sirnas)
TOOLS sirna and mirna User guide Introduction RNA interference (RNAi) is a powerful tool for suppression gene expression by causing the destruction of specific mrna molecules. Small Interfering RNAs (sirnas)
Fig. S1 TGF RI inhibitor SB effectively blocks phosphorylation of Smad2 induced by TGF. FET cells were treated with TGF in the presence of
 Fig. S1 TGF RI inhibitor SB525334 effectively blocks phosphorylation of Smad2 induced by TGF. FET cells were treated with TGF in the presence of different concentrations of SB525334. Cells were lysed and
Fig. S1 TGF RI inhibitor SB525334 effectively blocks phosphorylation of Smad2 induced by TGF. FET cells were treated with TGF in the presence of different concentrations of SB525334. Cells were lysed and
Sarker et al. Supplementary Material. Subcellular Fractionation
 Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
Supporting Information
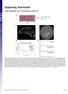 Supporting Information Zorde Khvalevsky et al..73/pnas.337.9 mm sikrsg Molecule PG Matrix ± mm right sc left retained in OER E T= days T= days (iver) T= days (PS) Fig. S. OER characteristics. () iagram
Supporting Information Zorde Khvalevsky et al..73/pnas.337.9 mm sikrsg Molecule PG Matrix ± mm right sc left retained in OER E T= days T= days (iver) T= days (PS) Fig. S. OER characteristics. () iagram
Flow cytometric determination of apoptosis by annexin V/propidium iodide double staining.
 Supplementary materials and methods Flow cytometric determination of apoptosis by annexin V/propidium iodide double staining. Cells were analyzed for phosphatidylserine exposure by an annexin-v FITC/propidium
Supplementary materials and methods Flow cytometric determination of apoptosis by annexin V/propidium iodide double staining. Cells were analyzed for phosphatidylserine exposure by an annexin-v FITC/propidium
SANTA CRUZ BIOTECHNOLOGY, INC.
 TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
(a) Immunoblotting to show the migration position of Flag-tagged MAVS
 Supplementary Figure 1 Characterization of six MAVS isoforms. (a) Immunoblotting to show the migration position of Flag-tagged MAVS isoforms. HEK293T Mavs -/- cells were transfected with constructs expressing
Supplementary Figure 1 Characterization of six MAVS isoforms. (a) Immunoblotting to show the migration position of Flag-tagged MAVS isoforms. HEK293T Mavs -/- cells were transfected with constructs expressing
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2743 Figure S1 stabilizes cellular protein level, post-transcriptionally. (a, b) and DDR1 were RNAi-depleted from HEK.293.-CBG cells. Western blots with indicated antibodies (a). RT-PCRs
DOI: 10.1038/ncb2743 Figure S1 stabilizes cellular protein level, post-transcriptionally. (a, b) and DDR1 were RNAi-depleted from HEK.293.-CBG cells. Western blots with indicated antibodies (a). RT-PCRs
(a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for
 Supplementary Figure 1 Generation of Diaph3 ko mice. (a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for different regions of Diaph3 mrna from WT, heterozygote
Supplementary Figure 1 Generation of Diaph3 ko mice. (a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for different regions of Diaph3 mrna from WT, heterozygote
- Supplementary Information -
 TRIM32 dependent transcription in adult neural progenitor cells regulates neuronal differentiation. - Supplementary Information - Anna-Lena Hillje 1, 2#, Maria Angeliki S. Pavlou 1, 2#, Elisabeth Beckmann
TRIM32 dependent transcription in adult neural progenitor cells regulates neuronal differentiation. - Supplementary Information - Anna-Lena Hillje 1, 2#, Maria Angeliki S. Pavlou 1, 2#, Elisabeth Beckmann
Supplementary Figure S1 Evi/Wls and Wnt3 are overexpressed in epithelial cells in colon cancers. (a) Validation of the specificity of the Wnt3
 Supplementary Figure S1 Evi/Wls and Wnt3 are overexpressed in epithelial cells in colon cancers. (a) Validation of the specificity of the Wnt3 antibody. HCT116 cells were reverse transfected with sicontrol
Supplementary Figure S1 Evi/Wls and Wnt3 are overexpressed in epithelial cells in colon cancers. (a) Validation of the specificity of the Wnt3 antibody. HCT116 cells were reverse transfected with sicontrol
Isolation, culture, and transfection of primary mammary epithelial organoids
 Supplementary Experimental Procedures Isolation, culture, and transfection of primary mammary epithelial organoids Primary mammary epithelial organoids were prepared from 8-week-old CD1 mice (Charles River)
Supplementary Experimental Procedures Isolation, culture, and transfection of primary mammary epithelial organoids Primary mammary epithelial organoids were prepared from 8-week-old CD1 mice (Charles River)
HPV E6 oncoprotein targets histone methyltransferases for modulating specific. Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu,
 1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
Supplementary Figure 1 PARP1 is involved in regulating the stability of mrnas from pro-inflammatory cytokine/chemokine mediators.
 Supplementary Figure 1 PARP1 is involved in regulating the stability of mrnas from pro-inflammatory cytokine/chemokine mediators. (a) A graphic depiction of the approach to determining the stability of
Supplementary Figure 1 PARP1 is involved in regulating the stability of mrnas from pro-inflammatory cytokine/chemokine mediators. (a) A graphic depiction of the approach to determining the stability of
 DOI: 10.1038/ncb3259 A Ismail et al. Supplementary Figure 1 B 60000 45000 SSC 30000 15000 Live cells 0 0 15000 30000 45000 60000 FSC- PARR 60000 45000 PARR Width 30000 FSC- 15000 Single cells 0 0 15000
DOI: 10.1038/ncb3259 A Ismail et al. Supplementary Figure 1 B 60000 45000 SSC 30000 15000 Live cells 0 0 15000 30000 45000 60000 FSC- PARR 60000 45000 PARR Width 30000 FSC- 15000 Single cells 0 0 15000
a. Increased active HPSE (50 kda) protein expression upon infection with multiple herpes viruses: bovine
 Fold change vs uninfeced MOI 0.1 MOI 1 MOI 0.1 MOI 1 MOI 0.001 MOI 0.01 Fold change vs uninfected a 6 4 2 50 kda HPSE * ** * * BHV PRV HSV-2 333 ** * 0 - + + - + + - + + b 50 kda HPSE 3 2 ** *** mock inf
Fold change vs uninfeced MOI 0.1 MOI 1 MOI 0.1 MOI 1 MOI 0.001 MOI 0.01 Fold change vs uninfected a 6 4 2 50 kda HPSE * ** * * BHV PRV HSV-2 333 ** * 0 - + + - + + - + + b 50 kda HPSE 3 2 ** *** mock inf
STAT3 signaling controls satellite cell expansion and skeletal muscle repair
 SUPPLEMENTARY INFORMATION STAT3 signaling controls satellite cell expansion and skeletal muscle repair Matthew Timothy Tierney 1 *, Tufan Aydogdu 2,3 *, David Sala 2, Barbora Malecova 2, Sole Gatto 2,
SUPPLEMENTARY INFORMATION STAT3 signaling controls satellite cell expansion and skeletal muscle repair Matthew Timothy Tierney 1 *, Tufan Aydogdu 2,3 *, David Sala 2, Barbora Malecova 2, Sole Gatto 2,
Supplementary data. sienigma. F-Enigma F-EnigmaSM. a-p53
 Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
cells (MLEC) that produce luciferase under the control of the PAI-1 promoter in response to
 Supplemental Materials and Methods TGF bioassay. To quantify the levels of active and total TGF, we used mink lung epithelial cells (MLEC) that produce luciferase under the control of the PAI-1 promoter
Supplemental Materials and Methods TGF bioassay. To quantify the levels of active and total TGF, we used mink lung epithelial cells (MLEC) that produce luciferase under the control of the PAI-1 promoter
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1.
 Supplementary Figure 1. Characterization and high expression of Lnc-β-Catm in liver CSCs. (a) Heatmap of differently expressed lncrnas in Liver CSCs (CD13 + CD133 + ) and non-cscs (CD13 - CD133 - ) according
Supplementary Figure 1. Characterization and high expression of Lnc-β-Catm in liver CSCs. (a) Heatmap of differently expressed lncrnas in Liver CSCs (CD13 + CD133 + ) and non-cscs (CD13 - CD133 - ) according
Transcriptional regulation of BRCA1 expression by a metabolic switch: Di, Fernandez, De Siervi, Longo, and Gardner. H3K4Me3
 ChIP H3K4Me3 enrichment.25.2.15.1.5 H3K4Me3 H3K4Me3 ctrl H3K4Me3 + E2 NS + E2 1. kb kb +82 kb Figure S1. Estrogen promotes entry of MCF-7 into the cell cycle but does not significantly change activation-associated
ChIP H3K4Me3 enrichment.25.2.15.1.5 H3K4Me3 H3K4Me3 ctrl H3K4Me3 + E2 NS + E2 1. kb kb +82 kb Figure S1. Estrogen promotes entry of MCF-7 into the cell cycle but does not significantly change activation-associated
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2271 Supplementary Figure a! WM266.4 mock WM266.4 #7 sirna WM266.4 #10 sirna SKMEL28 mock SKMEL28 #7 sirna SKMEL28 #10 sirna WM1361 mock WM1361 #7 sirna WM1361 #10 sirna 9 WM266. WM136
DOI: 10.1038/ncb2271 Supplementary Figure a! WM266.4 mock WM266.4 #7 sirna WM266.4 #10 sirna SKMEL28 mock SKMEL28 #7 sirna SKMEL28 #10 sirna WM1361 mock WM1361 #7 sirna WM1361 #10 sirna 9 WM266. WM136
SUPPLEMENTAL FIGURE LEGENDS
 SUPPLEMENTAL FIGURE LEGENDS Suppl. Fig. 1. Notch and myostatin expression is unaffected in the absence of p65 during postnatal development. A & B. Myostatin and Notch-1 expression levels were determined
SUPPLEMENTAL FIGURE LEGENDS Suppl. Fig. 1. Notch and myostatin expression is unaffected in the absence of p65 during postnatal development. A & B. Myostatin and Notch-1 expression levels were determined
Supplementary Figure 1. Confirmation of sirna in PC3 and H1299 cells PC3 (a) and H1299 (b) cells were transfected with sirna oligonucleotides
 Supplementary Figure 1. Confirmation of sirna in PC3 and H1299 cells PC3 (a) and H1299 (b) cells were transfected with sirna oligonucleotides targeting RCP (SMARTPool (RCP) or two individual oligos (RCP#1
Supplementary Figure 1. Confirmation of sirna in PC3 and H1299 cells PC3 (a) and H1299 (b) cells were transfected with sirna oligonucleotides targeting RCP (SMARTPool (RCP) or two individual oligos (RCP#1
Supplementary Figure 1: Overexpression of EBV-encoded proteins Western blot analysis of the expression levels of EBV-encoded latency III proteins in
 Supplementary Figure 1: Overexpression of EBV-encoded proteins Western blot analysis of the expression levels of EBV-encoded latency III proteins in BL2 cells. The Ponceau S staining of the membranes or
Supplementary Figure 1: Overexpression of EBV-encoded proteins Western blot analysis of the expression levels of EBV-encoded latency III proteins in BL2 cells. The Ponceau S staining of the membranes or
Supplementary Figure 1. NORAD expression in mouse (A) and dog (B). The black boxes indicate the position of the regions alignable to the 12 repeat
 Supplementary Figure 1. NORAD expression in mouse (A) and dog (B). The black boxes indicate the position of the regions alignable to the 12 repeat units in the human genome. Annotated transposable elements
Supplementary Figure 1. NORAD expression in mouse (A) and dog (B). The black boxes indicate the position of the regions alignable to the 12 repeat units in the human genome. Annotated transposable elements
Supplementary
 Supplementary information Supplementary Material and Methods Plasmid construction The transposable element vectors for inducible expression of RFP-FUS wt and EGFP-FUS R521C and EGFP-FUS P525L were derived
Supplementary information Supplementary Material and Methods Plasmid construction The transposable element vectors for inducible expression of RFP-FUS wt and EGFP-FUS R521C and EGFP-FUS P525L were derived
T H E J O U R N A L O F C E L L B I O L O G Y
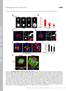 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rodríguez-Fraticelli et al., http://www.jcb.org/cgi/content/full/jcb.201203075/dc1 Figure S1. Cell spreading and lumen formation in confined
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rodríguez-Fraticelli et al., http://www.jcb.org/cgi/content/full/jcb.201203075/dc1 Figure S1. Cell spreading and lumen formation in confined
Supplementary Information Alternative splicing of CD44 mrna by ESRP1 enhances lung colonization of metastatic cancer cell
 Supplementary Information Alternative splicing of CD44 mrna by ESRP1 enhances lung colonization of metastatic cancer cell Supplementary Figures S1-S3 Supplementary Methods Supplementary Figure S1. Identification
Supplementary Information Alternative splicing of CD44 mrna by ESRP1 enhances lung colonization of metastatic cancer cell Supplementary Figures S1-S3 Supplementary Methods Supplementary Figure S1. Identification
CD93 and dystroglycan cooperation in human endothelial cell adhesion and migration
 /, Supplementary Advance Publications Materials 2016 CD93 and dystroglycan cooperation in human endothelial cell adhesion and migration Supplementary Materials Supplementary Figure S1: In ECs CD93 silencing
/, Supplementary Advance Publications Materials 2016 CD93 and dystroglycan cooperation in human endothelial cell adhesion and migration Supplementary Materials Supplementary Figure S1: In ECs CD93 silencing
Establishing sirna assays in primary human peripheral blood lymphocytes
 page 1 of 7 Establishing sirna assays in primary human peripheral blood lymphocytes This protocol is designed to establish sirna assays in primary human peripheral blood lymphocytes using the Nucleofector
page 1 of 7 Establishing sirna assays in primary human peripheral blood lymphocytes This protocol is designed to establish sirna assays in primary human peripheral blood lymphocytes using the Nucleofector
Cytotoxicity of Botulinum Neurotoxins Reveals a Direct Role of
 Supplementary Information Cytotoxicity of Botulinum Neurotoxins Reveals a Direct Role of Syntaxin 1 and SNAP-25 in Neuron Survival Lisheng Peng, Huisheng Liu, Hongyu Ruan, William H. Tepp, William H. Stoothoff,
Supplementary Information Cytotoxicity of Botulinum Neurotoxins Reveals a Direct Role of Syntaxin 1 and SNAP-25 in Neuron Survival Lisheng Peng, Huisheng Liu, Hongyu Ruan, William H. Tepp, William H. Stoothoff,
Supporting Information
 Supporting Information Su et al. 10.1073/pnas.1211604110 SI Materials and Methods Cell Culture and Plasmids. Tera-1 and Tera-2 cells (ATCC: HTB- 105/106) were maintained in McCoy s 5A medium with 15% FBS
Supporting Information Su et al. 10.1073/pnas.1211604110 SI Materials and Methods Cell Culture and Plasmids. Tera-1 and Tera-2 cells (ATCC: HTB- 105/106) were maintained in McCoy s 5A medium with 15% FBS
Supplementary Figure 1 (A), (B), and (C) Docking of a physiologic ligand of integrin αvβ3, the tenth type III RGD domain of wild-type fibronectin
 Supplementary Figure 1 (A), (B), and (C) Docking of a physiologic ligand of integrin αvβ3, the tenth type III RGD domain of wild-type fibronectin (A), D1-CD2 (B), and the D1-CD2 variant 3 (C) to Adomain
Supplementary Figure 1 (A), (B), and (C) Docking of a physiologic ligand of integrin αvβ3, the tenth type III RGD domain of wild-type fibronectin (A), D1-CD2 (B), and the D1-CD2 variant 3 (C) to Adomain
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1. Analyses of ECTRs by C-circle and T-circle assays.
 Supplementary Figure 1 Analyses of ECTRs by C-circle and T-circle assays. (a) C-circle and (b) T-circle amplification reactions using genomic DNA from different cell lines in the presence (+) or absence
Supplementary Figure 1 Analyses of ECTRs by C-circle and T-circle assays. (a) C-circle and (b) T-circle amplification reactions using genomic DNA from different cell lines in the presence (+) or absence
EPIGENTEK. EpiQuik Chromatin Immunoprecipitation Kit. Base Catalog # P-2002 PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE
 EpiQuik Chromatin Immunoprecipitation Kit Base Catalog # P-2002 PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Chromatin Immunoprecipitation Kit is suitable for combining the specificity of
EpiQuik Chromatin Immunoprecipitation Kit Base Catalog # P-2002 PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Chromatin Immunoprecipitation Kit is suitable for combining the specificity of
Measuring gene expression and protein production
 Measuring gene expression and protein production Module 3, Lecture 5! 20.109 Spring 2013! Lessons from Lecture 4 Always look for the recovery file! The myth of multitasking! task-switching taps processing
Measuring gene expression and protein production Module 3, Lecture 5! 20.109 Spring 2013! Lessons from Lecture 4 Always look for the recovery file! The myth of multitasking! task-switching taps processing
Supplemental Information. DNp63 Inhibits Oxidative Stress-Induced Cell. Death, Including Ferroptosis, and Cooperates with
 Cell Reports, Volume 21 Supplemental Information DNp63 Inhibits Oxidative Stress-Induced Cell Death, Including Ferroptosis, and Cooperates with the BCL-2 Family to Promote Clonogenic Survival Gary X. Wang,
Cell Reports, Volume 21 Supplemental Information DNp63 Inhibits Oxidative Stress-Induced Cell Death, Including Ferroptosis, and Cooperates with the BCL-2 Family to Promote Clonogenic Survival Gary X. Wang,
Supplementary Information
 Supplementary Information Supplementary Figures Supplementary Figure 1. qrt-pcr analysis of the expression of candidate factors including SOX2, MITF, PAX3, DLX5, FOXD3, LEF1, MSX1, PAX6, SOX2 and SOX9
Supplementary Information Supplementary Figures Supplementary Figure 1. qrt-pcr analysis of the expression of candidate factors including SOX2, MITF, PAX3, DLX5, FOXD3, LEF1, MSX1, PAX6, SOX2 and SOX9
Primers used for PCR of conductin, SGK1 and GAPDH have been described in (Dehner et al,
 Supplementary METHODS Flow Cytometry (FACS) For FACS analysis, trypsinized cells were fixed in ethanol, rehydrated in PBS and treated with 40μg/ml propidium iodide and 10μ/ml RNase for 30 min at room temperature.
Supplementary METHODS Flow Cytometry (FACS) For FACS analysis, trypsinized cells were fixed in ethanol, rehydrated in PBS and treated with 40μg/ml propidium iodide and 10μ/ml RNase for 30 min at room temperature.
Supporting Information
 Supporting Information Chakrabarty et al. 10.1073/pnas.1018001108 SI Materials and Methods Cell Lines. All cell lines were purchased from the American Type Culture Collection. Media and FBS were purchased
Supporting Information Chakrabarty et al. 10.1073/pnas.1018001108 SI Materials and Methods Cell Lines. All cell lines were purchased from the American Type Culture Collection. Media and FBS were purchased
M X 500 µl. M X 1000 µl
 GeneGlide TM sirna Transfection Reagent (Catalog # M1081-300, -500, -1000; Store at 4 C) I. Introduction: BioVision s GeneGlide TM sirna Transfection reagent is a cationic proprietary polymer/lipid formulation,
GeneGlide TM sirna Transfection Reagent (Catalog # M1081-300, -500, -1000; Store at 4 C) I. Introduction: BioVision s GeneGlide TM sirna Transfection reagent is a cationic proprietary polymer/lipid formulation,
Supplementary Figure 1: Expression of RNF8, HERC2 and NEURL4 in the cerebellum and knockdown of RNF8 by RNAi (a) Lysates of the cerebellum from rat
 Supplementary Figure 1: Expression of RNF8, HERC2 and NEURL4 in the cerebellum and knockdown of RNF8 by RNAi (a) Lysates of the cerebellum from rat pups at P6, P14, P22, P30 and adult (A) rats were subjected
Supplementary Figure 1: Expression of RNF8, HERC2 and NEURL4 in the cerebellum and knockdown of RNF8 by RNAi (a) Lysates of the cerebellum from rat pups at P6, P14, P22, P30 and adult (A) rats were subjected
Transfection Reagent SPECIFICATIONS MATERIALS. For Research Use Only. Materials Supplied. Materials required, but not supplied
 TransIT-siQUEST Transfection Reagent Quick Reference Protocol, MSDS and Certificate of Analysis available at mirusbio.com/2110 INTRODUCTION TransIT-siQUEST is a broad spectrum sirna transfection reagent
TransIT-siQUEST Transfection Reagent Quick Reference Protocol, MSDS and Certificate of Analysis available at mirusbio.com/2110 INTRODUCTION TransIT-siQUEST is a broad spectrum sirna transfection reagent
Khaled_Fig. S MITF TYROSINASE R²= MITF PDE4D R²= Variance from mean mrna expression
 Khaled_Fig. S Variance from mean mrna expression.8 2 MITF.6 TYROSINASE.4 R²=.73.2.8.6.4.2.8.6.4.2.8.6.4.2 MALME3M SKMEL28 UACC257 MITF PDE4D R²=.63 4.5 5 MITF 3.5 4 PDE4B 2.5 3 R²=.2.5 2.5.8.6.4.2.8.6.4.2
Khaled_Fig. S Variance from mean mrna expression.8 2 MITF.6 TYROSINASE.4 R²=.73.2.8.6.4.2.8.6.4.2.8.6.4.2 MALME3M SKMEL28 UACC257 MITF PDE4D R²=.63 4.5 5 MITF 3.5 4 PDE4B 2.5 3 R²=.2.5 2.5.8.6.4.2.8.6.4.2
Supplementary Figure 1. Intracellular distribution of the EPE peptide. HeLa cells were serum-starved (16 h, 0.1%), and treated with EPE peptide,
 Supplementary Figure 1. Intracellular distribution of the EPE peptide. HeLa cells were serum-starved (16 h, 0.1%), and treated with EPE peptide, conjugated with either TAT or Myristic acid and biotin for
Supplementary Figure 1. Intracellular distribution of the EPE peptide. HeLa cells were serum-starved (16 h, 0.1%), and treated with EPE peptide, conjugated with either TAT or Myristic acid and biotin for
supplementary information
 DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
