Enhanced Chromatin Dynamics by FACT Promotes Transcriptional Restart after UV-Induced DNA Damage
|
|
|
- Lionel Stevenson
- 6 years ago
- Views:
Transcription
1 Article Enhanced Chromatin Dynamics by FACT Promotes Transcriptional Restart after UV-Induced DNA Damage Christoffel Dinant, 1,2,3 Giannis Ampatziadis-Michailidis, 4 Hannes Lans, 1 Maria Tresini, 1 Anna Lagarou, 4,7 Malgorzata Grosbart, 1 Arjan F. Theil, 1 Wiggert A. van Cappellen, 2 Hiroshi Kimura, 5 Jiri Bartek, 3,6 Maria Fousteri, 4 Adriaan B. Houtsmuller, 2, * Wim Vermeulen, 1, * and Jurgen A. Marteijn 1, * 1 Department of Genetics, Erasmus Medical Centre, Rotterdam 3015 GE, The Netherlands 2 Department of Pathology, Erasmus Medical Centre, Optical Imaging Centre (OIC), Rotterdam 3015 GE, The Netherlands 3 Genome Integrity Unit, Danish Cancer Society Research Centre, Strandboulevarden 49, DK-2100 Copenhagen, Denmark 4 Institute of Biomedical Sciences, BSRC Alexander Fleming, Vari, Athens, Greece 5 Graduate School of Frontier Biosciences, Osaka University, Osaka, Japan 6 Institute of Molecular and Translational Medicine, Palacky University, CZ Olomouc, Czech Republic 7 Present address: Institut de Pharmacologie et de Biologie Structurale (IPBS), CNRS/UPS, 205 Route de Narbonne, F Toulouse, France *Correspondence: a.houtsmuller@erasmusmc.nl (A.B.H.), w.vermeulen@erasmusmc.nl (W.V.), j.marteijn@erasmusmc.nl (J.A.M.) SUMMARY Chromatin remodeling is tightly linked to all DNAtransacting activities. To study chromatin remodeling during DNA repair, we established quantitative fluorescence imaging methods to measure the exchange of histones in chromatin in living cells. We show that particularly H2A and H2B are evicted and replaced at an accelerated pace at sites of UVinduced DNA damage. This accelerated exchange of H2A/H2B is facilitated by SPT16, one of the two subunits of the histone chaperone FACT (facilitates chromatin transcription) but largely independent of its partner SSRP1. Interestingly, SPT16 is targeted to sites of UV light-induced DNA damage-arrested transcription and is required for efficient restart of RNA synthesis upon damage removal. Together, our data uncover an important role for chromatin dynamics at the crossroads of transcription and the UV-induced DNA damage response. INTRODUCTION Cells are continuously exposed to endogenous metabolites and environmental agents that induce lesions in DNA. These lesions interfere with transcription and replication, resulting in cellular malfunction and DNA damage-induced mutagenesis of genetic information (Hoeijmakers, 2009). Helix-distorting DNA lesions, for instance, those induced by UV-C exposure, located anywhere in the genome are recognized by the concerted action of XPC- and DDB2-containing complexes to initiate global genome nucleotide excision repair (GG-NER) (Nouspikel, 2009). DNA lesions in the transcribed strand of active genes which interfere with RNA polymerase II (RNApolII) elongation activate a dedicated NER branch, termed transcription-coupled NER (TC-NER) (Hanawalt and Spivak, 2008). Both GG-NER and TC-NER funnel into a shared pathway that removes lesions by helix unwinding, excision of the damaged strand, DNA resynthesis over the single-stranded gap, and nick ligation (Nouspikel, 2009). A complex, as-yet poorly defined mechanism is coupled to TC-NER to allow transcription resumption after completion of TC-NER (Hanawalt and Spivak, 2008). Transcriptional control is largely dependent on the chromatin structure (Avvakumov et al., 2011). Similarly, efficient DNA repair requires plasticity of the chromatin structure for repair factors to gain access to the DNA template (Soria et al., 2012). Importantly, not only the access of repair factors to the DNA and the subsequent swift repair of DNA lesions is required for the recovery of transcription, but also proper restoration of the predamage chromatin structure (Green and Almouzni, 2003; Smerdon, 1991). Besides posttranslational histone modifications that increase access to DNA, nucleosome sliding and histone removal are thought to expose the DNA and thereby facilitate binding of, for example, DNA repair or transcription factors to damaged DNA (Avvakumov et al., 2011; Gong et al., 2005; Soria et al., 2012). Histone displacement through sliding, removal, and reinsertion into chromatin is achieved by two groups of proteins: ATP-dependent chromatin remodelers and histone chaperones. Remodelers use ATP hydrolysis to remove histones from DNA or slide whole nucleosomes along DNA fibers, a mechanism suggested to promote DNA accessibility (Lans et al., 2012). Histone chaperones primarily load histones onto DNA to restore chromatin after access is no longer required, and conversely they also catalyze histone removal (Avvakumov et al., 2011; De Koning et al., 2007). Accordingly, early steps of NER involve ATP-dependent relaxation of chromatin (Lans et al., 2012; Luijsterburg et al., 2012), whereas restoration of chromatin after NER completion depends on histone chaperones CAF1 and ASF1 (Polo et al., 2006). Direct measurement of histone removal and reinsertion kinetics in response to DNA damage in living cells, representing Molecular Cell 51, , August 22, 2013 ª2013 Elsevier Inc. 469
2 chromatin remodeling in vivo, has escaped scrutiny. We have investigated the dynamics of GFP-tagged histones in living cells in response to UV-C irradiation and discovered an important function for FACT-subunit SPT16 in accelerating the exchange of H2A and H2B and promoting transcription restart after DNA repair of the lesions blocking transcription. RESULTS Histone Exchange upon UV-Induced DNA Damage We designed three methods to determine the mobility of histones after DNA damage induction in living cells. First, we used a fluorescence recovery after photobleaching (FRAP) approach to compare histone mobility in UV-damaged versus unperturbed chromatin in living cells. In cells expressing GFP-tagged histones (Kimura and Cook, 2001), the fluorescence in half of the nucleus was photobleached, and local UV damage (LUD) was introduced in the photobleached part using a UV-C laser (Dinant et al., 2007) (Figure 1A). Incorporation of fluorescent histones in the bleached area will be faster in regions with a higher histone exchange rate, leading to increased local fluorescence. Interestingly, we observed increased fluorescence recovery corresponding to an 2-fold increased exchange rate of GFP-tagged histones H2A and H2B at the LUD, whereas no accelerated H3.1 or H4 exchange was detected (Figures 1B 1E). Local concentrations of histone proteins were not affected by UV damage (see Figures S1A and S1B online), indicating that the fluorescence increase at LUD after photobleaching directly reflects enhanced H2A/H2Bchromatin exchange rather than histone accumulation. Besides half-nucleus FRAP, we used two alternative approaches to study histone dynamics upon DNA damage. Induction of LUD in cells 6 8 hr after transfection with, before the incorporation of newly synthesized fluorescent H2A molecules had reached a steady state, resulted in faster increase of fluorescence at damaged areas (Figure S1C). And similarly, upon PEG-mediated cell fusion of cells expressing with cells expressing H2B-RFP, local UV irradiation caused accelerated incorporation of RFP signals in green nuclei and of GFP signals in red nuclei at LUD (Figure S1D). Both alternative methods confirmed our results obtained by FRAP experiments. DNA damage-induced H2A/H2B exchange was independent of DNA replication, as it occurred in both S phase and non-s phase cells, identified by the presence and absence of characteristic replication foci of mcherry-tagged proliferating cell nuclear antigen (PCNA), which itself is also recruited to LUD both in S phase and non-s phase cells (Essers et al., 2005) (Figure S1E). Together these data clearly show that UV-C irradiation induces accelerated removal from and reincorporation into chromatin of H2A/H2B dimers. Histone Exchange upon DNA Damage Requires FACT The key proteins involved in mediating nucleosome assembly are histone chaperones (Ransom et al., 2010). We tested the involvement of two well-studied H2A/H2B chaperones, NAP1L1 (Zlatanova et al., 2007) and FACT (Reinberg and Sims, 2006), in accelerated histone exchange at sites of DNA damage. While sirna-mediated knockdown of NAP1L1 resulted in a clear reduction of NAP1L1 protein levels, accelerated exchange of H2A at LUD was still observed (Figures S2A and S2B). In contrast, simultaneous sirna-mediated knockdown of both SSRP1 and SPT16 subunits of the FACT heterodimer resulted in the complete absence of accelerated H2A exchange at LUD, compared to control transfected cells (Figure 2A, Figures S2C and S2D). Importantly, only a small decrease of general H2A exchange was observed in unperturbed regions upon FACT depletion (Figure S2D). It has been described that FACT is modified by posttranslational modifications upon DNA damage, thereby regulating its interaction with chromatin. SSRP1 becomes phosphorylated by CK2 upon UV damage (Krohn et al., 2003), while the SPT16 subunit gets PARylated by PARP1 upon oxidative damage (Heo et al., 2008; Huang et al., 2006). To test whether these FACT modifications are involved in UV-induced accelerated histone exchange, we performed the live-cell histone exchange assay in the presence of PARP1 or CK2 inhibitors. No effect on the H2A exchange was observed upon CK2 inhibition (Pagano et al., 2004), while a small decrease in H2A exchange was found when PARP1 was inhibited (Menear et al., 2008) (Figures S2E S2H), indicating that PARylation events, possibly of SPT16 itself, have a minor stimulatory effect on histone mobility upon UV damage. The Role of SPT16 in the DNA Damage Response To investigate in more detail the in vivo role of FACT during the UV-induced DNA damage response (UV-DDR), we generated stable cell lines expressing full-length GFP-tagged SSRP1 or SPT16 (Figure S3A). Both SSRP1 and SPT16 accumulated at sites of LUD in living cells (Figure 2B). In addition, antibody staining of endogenous SPT16 confirmed colocalization both with CPD (cyclobutane pyrimidine dimer), a marker for UV damage, and with sites of increased exchange (Figure 2C). This recruitment to sites of DNA damage is a reversible event, as it was no longer detected 24 hr after DNA damage infliction, when most lesions are removed (Figure S3B). NER is the main DNA repair system that removes helix-distorting lesions such as CPDs induced by UV-C irradiation. We used the ChIP-on-western technique, a dedicated chromatin immunoprecipitation (ChIP) procedure based on classical ChIP, and optimized to reveal the composition of transient protein complexes at damaged DNA (Coin et al., 2008; Fousteri et al., 2006; Kannouche et al., 2004) to study the in vivo composition of FACT-associated complexes upon UV damage induction. This approach showed that SPT16 coimmunoprecipitated with the core NER factor XPB (p89 subunit of TFIIH) upon UV irradiation. Another core NER factor, XPA, used as positive control, also coprecipitated with p89 (Figure 2D). These data indicate the presence of SPT16 at chromatin sites undergoing NER. RNAimediated knockdown of both FACT subunits in NER-proficient HeLa cells led to increased sensitivity to UV-C irradiation (Figure 2E), indicative of a function in the UV-induced DDR. Strikingly, while expression of both FACT subunits was efficiently reduced (Figure S2C), knockdown of the SSRP1 subunit alone did not result in UV sensitivity, whereas depletion of only SPT16 resulted in similar UV hypersensitivity to simultaneous knockdown of both FACT subunits. Consistent with the effect on UV survival, knockdown of SPT16 strongly reduced the H2A exchange upon UV damage, while SSRP1 depletion did not have a clear effect on histone exchange (Figure S3C). These data suggest that the 470 Molecular Cell 51, , August 22, 2013 ª2013 Elsevier Inc.
3 A time UV-C laser (266nm) Photobleaching half of nucleus Local UV-damage B Pre-bleach Bleach and damage After damage C GFP α-cpd Merge H4-GFP H3.1-GFP H2B-GFP H4-GFP H3.1-GFP H2B-GFP D Relative Fluorescence UV H2B-GFP+UV H2B-GFP E Relative Fluorescence H3.1-GFP+UV H3.1-GFP H4-GFP+UV H4-GFP Time (s) Time (s) Figure 1. Increased Histone H2A and H2B Exchange at UV-Induced DNA Damage (A) Method for measuring histone exchange at local laser-directed UV-C DNA damage (LUD). Half of the nucleus of cells stably expressing GFP-tagged histones is photobleached. In the photobleached part, UV-C damage is inflicted with a UV-C (266 nm) laser. Recovery of fluorescence for both damaged and undamaged areas is measured. (B) and H2B-GFP exchange is accelerated at LUD, whereas H3.1-GFP and H4-GFP do not display DNA damage-induced accelerated exchange rates. (Left panel) Unbleached cells, (middle panel) images of half nucleus bleached immediately captured after local UV-C damage infliction, and (right panel) 15 min after UV damage. (C) Accelerated histone exchange localizes to damaged areas marked by CPD staining. (Left panel) GFP signal, (middle panel) anti-cpd (cyclobutane pyrimidine dimer) antibody staining used as UV-induced DNA damage marker, and (right panel) merge of GFP and anti-cpd signals. (D and E) Quantification of histone exchange in unperturbed or locally UV-C-exposed regions of the bleached part of the nucleus. Fluorescence recovery directly after LUD is plotted against time in seconds for H2A and H2B-GFP (D) and H3.1- and H4-GFP (E) (n > 12 cells, mean ± SEM). See also Figure S1. Molecular Cell 51, , August 22, 2013 ª2013 Elsevier Inc. 471
4 A sictrl sifact B Before Damage Directly after Damage 5 Sec after Damage 30 Sec after Damage After damage After bleach SSRP1-GFP SPT16-GFP C α-spt16 α-cpd D ChIP: α-p89 UV (20 J/m 2 ) - 1 hr p89 (XPB) Spt16 XPA E Relative Colony Number sictrl sixpf sifact sispt16 sissrp F After damage Before damage sictrl sissrp1 sictrl sispt16 UV dose J/m 2 SPT16 -GFP SSRP1 -GFP Figure 2. H2A Exchange Is Dependent on the SPT16 Subunit of FACT (A) histone exchange after sirna transfection of nontargeting control or sirna targeting both SSRP1 and SPT16 together (sifact). (Top row) Immediate after LUD induction, (bottom row) 30 min after LUD. (B) Live-cell imaging analysis after LUD infliction (arrow) in MRC5 cells stably expressing SPT16-GFP (top row) and SSRP1-GFP (bottom row). (C) Immunofluorescent analysis of SP16 accumulation on LUD. GFP-H2A-expressing cells with a bleached half nucleus were fixed 30 min after LUD infliction, GFP imaged (left panel), and immunostained for SPT16 (middle panel) and CPD (right panel). SPT16 accumulates at LUD and colocalizes with accelerated exchange of and CPD staining. (D) ChIP-on-western analysis of mock and UV irradiated (1 hr after 20 J/m 2 ) in vivo crosslinked VH10 (sv40) cells chromatin immunoprecipitated with p89 antibodies. Samples were normalized to p89 levels; immunoblot analysis of the coimmunoprecipitated proteins was performed for SPT16 and XPA. (E) UV-C sensitivity of HeLa cells with sirna-mediated knockdown for the indicated proteins, determined by colony-forming ability (mean ± SD). (F) SPT16-GFP is recruited to LUD in cells treated with control or SSRP1 sirna, but SSRP1-GFP does not accumulate at LUD in the presence of sirna against SPT16. See also Figure S Molecular Cell 51, , August 22, 2013 ª2013 Elsevier Inc.
5 A B CS-A XP-C XP-A CS-A H2A CS-B XP-E DAPI α -CPD α -Spt16 XP-C H2B XP-A Figure 3. NER-Independent FACT Implication in UV-DDR (A) Accelerated histone exchange in CS-A, CS-B, and XP-E patient cells expressing ; XP-C patient cells expressing H2B-GFP; and XP-A patient cells expressing H2B-YFP. Arrow indicates site of LUD. (B) Endogenous SPT16 localizes to LUD as indicated by staining with anti-cpd antibodiesin CS-A, XP-A, and XP-C cells. See also Figure S3. SPT16 function at damaged chromatin does not require SSRP1. In addition, accumulation of SSRP1-GFP at LUD required SPT16 expression, whereas SPT16 was recruited to LUD in the absence of SSRP1 (Figure 2F and Figure S3E). Together these results indicate that although SSRP1 is present at sites of DNA damage, only its partner in the FACT heterodimer, SPT16, plays an important role in the UV-DDR. Accelerated Histone Exchange Occurs Also in the Absence of NER NER is initiated by two damage recognition pathways. When lesions are present specifically in the transcribed strand of active genes and block RNApolII-mediated transcription elongation, they are targeted by TC-NER, whereas lesions located anywhere in the genome can trigger GG-NER (Nouspikel, 2009). Interestingly, neither accumulation of SPT16 at LUD nor the accelerated exchange of H2A/H2B required initiation of NER. Both still occurred in the absence of damage recognition factors for GG-NER (in patient cells lacking functional XPC or DDB2) or for TC-NER (in patient cells lacking functional CSA or CSB) (Figures 3A and 3B). Also in cells derived from an XP-A patient, in which both NER subpathways are defective due to the complete absence of the crucial NER factor XPA, H2A exchange and SPT16 accumulation still took place (Figures 3A and 3B). These data suggest that both targeting of Spt16 and damage-induced accelerated histone exchange occur independent of NER. SPT16 Is Involved in Transcription Restart after TC-NER Completion Because SPT16 is involved in the survival upon UV-C induced damage (Figure 2E), we determined whether SPT16 activity specifically affects TC-NER or GG-NER. RNAi-mediated knockdown of both FACT subunits did not interfere with the recruitment of early GG-NER factors XPC and DDB2, nor with TC-NER factor CSB (Figure S4A), suggesting that the role of SPT16 in the UV damage response is not needed for the recruitment of these factors that are involved in damage recognition. A ChIP-onwestern analysis after immunoprecipitation with an antibody against DDB1, which functions in both GG-NER and TC-NER via interactions with DDB2 and CSA, respectively (Fousteri et al., 2006; Groisman et al., 2003), revealed a clear enrichment of endogenous SPT16 and SSRP1 in active chromatin-bound NER complexes in NER-proficient cells upon UV-C damage induction (Figure 4A). However, in CS-B patient cells (GG-NER proficient), where DDB1 can only be recruited to DNA damage via the GG-NER factor DDB2 (Fousteri et al., 2006), SPT16 did not coimmunoprecipitate with DDB1, indicating that SPT16 specifically binds to chromatin sites undergoing TC-NER and not GG-NER. In line with this, ChIP-on-western for SPT16 revealed that in the absence of CSB, when no functional TC-NER complexes are formed (Fousteri et al., 2006), no UVinduced interaction with XPA was observed (Figure S4B). The specific presence of SPT16 in TC-NER complexes was further confirmed by ChIP-on-western experiments with the elongating form of RNApolII (RNApolIIo) and the TC-NER master organizer CSB (Figure 4B). Moreover, ChIP for p89 (XPB) showed an interaction with the FACT complex in XP-C patient cells (TC-NER proficient), but not in CS-B patient cells, indicating that FACT can only be found in the vicinity of active NER proteins at UV-damage-blocked transcription sites and not at UV lesions in the rest of the genome (Figure S4C). It is important to mention that the CSB-dependent interaction in these ChIP experiments only indicates that SPT16 specifically interacts with the TC-NER complex. It does not show that CSB is required for the SPT16 recruitment to DNA damage. On the contrary, the FACTfacilitated H2A/H2B exchange at the site of damage occurred independently of CS proteins (Figures 3A and 3B). Accordingly, in the absence of CSB, SPT16 coimmunoprecipitated with RNAPolIIo (Figure S4D), whereas binding of most of the other TC-NER-specific factors is dependent on the presence of CSB (Fousteri et al., 2006; Schwertman et al., 2012). Together these results show that while FACT is present at sites of active TC-NER, the recruitment to this repair complex is mediated via a different mechanism. A key event in the initiation of TC-NER is the stalling of RNApolII on DNA lesions, which results in the stable association of the TC-NER-specific factors CSA and CSB (Fousteri et al., 2006). We tested whether stalling of RNApolII at DNA lesions itself might be the trigger to recruit FACT as well, thereby initiating the H2A/H2B histone exchange at sites of UV damage. Inhibition of active transcription by RNAPolII using DRB or a-amanitin (Bensaude, 2011), which effectively blocks the accumulation of TC-NER proteins (Schwertman et al., 2012; van den Boom et al., 2004), did not interfere with exchange at LUD or SPT16 accumulation (Figures 4C and 4D and Figure S4E). In addition, FRAP experiments showed no difference in mobility of SPT16 upon RNApolII transcription inhibition, indicating that stalling of RNApolII, in the absence of UV-C damage, is not sufficient to immobilize SPT16 (Figures S4F and S4G). Together this Molecular Cell 51, , August 22, 2013 ª2013 Elsevier Inc. 473
6 A ChIP: α- DDB1 No ab - 1 hr - 1 hr - B α-rnapiio UV (20 J/m 2 ) - 1 hr DDB1 UV (20 J/m 2 ) RNAPIIo α-csb - 1 hr - 1 hr CSB Spt16 Spt16 Spt16 Wt CS-B SSRP1 RNAPIIo Wt SSRP1 CSA SSRP1 RNAPIIo P89 (XPB) Wt CS-B C Untreated α-amanitin DRB D Untreated α-amanitin DRB SPT16-GFP E Unscheduled Repair Synthesis (UDS) F 125 Recovery of RNA synthesis (RRS) Relative EDU incorporation Relative EU incorporation sictrl sixpa sifact sissrp1 sispt16 0 0hrs 2hrs 16hrs 0hrs 2hrs 16hrs 0hrs 2hrs 16hrs 0hrs 2hrs 16hrs 0hrs 2hrs 16hrs sictrl sixpa sifact sispt16 sissrp1 G 16 hrs after 12 J/m2 UV-C EU-Alexa 594 GFP-CLIP170 DAPI Merge sifact/sictrl Figure 4. SPT16 Is Specifically Involved in TC-NER (A) Mock and UV irradiated (1 hr after 20 J/m 2 ) VH10 and CS1AN (CS-B) cells were in vivo crosslinked and chromatin immunoprecipitated with DDB1 antibodies for ChIP-on-western analysis. RNApolIIo (elongating form) and both FACT subunits only coimmunoprecipitated with DDB1 in TC-NER-proficient cells, indicative for their TC-NER-specific role. (B) RNApolIIo and CSB ChIP with and without UV irradiation (1 hr after 20 J/m 2 ) in htert-immortalized VH10 cells and CS1AN (CS-B) cells, showing UV-induced association of FACT to TC-NER complexes. CSB ChIP in CS-B cells is shown as a control for antibody specificity. For (A) and (B), samples were normalized to equal levels of the chromatin immunoprecipitated protein. Immunoblot analysis of the coimmunoprecipitated proteins was performed with antibodies as indicated. (C and D) (C) Accelerated exchange of or (D) recruitment of SPT16-GFP was not affected by transcription inhibition by 16 hr of a-amanitin (25 mg/ml) or 2 hr of DRB (100 mm) before damage infliction. (E) Unscheduled DNA repair synthesis (UDS) determined by EdU incorporation over 3 hr immediately after UV-C exposure (16 J/m 2 ) in htert-immortalized C5RO cells after sirna-mediated knockdown of the indicated proteins. (n > 200 cells per sample; RFI, relative fluorescence intensity; mean ± SEM). (F) Recovery of RNA synthesis (RRS) after UV-induced inhibition, using 2 hr pulse labeling with EU without UV damage and 2 and 16 hr after UV-C exposure (8 J/m 2 ) in U2OS cells after sirna-mediated knockdown of the indicated proteins. Values were normalized to predamage values (n > 200 cells per sample; RFI, relative fluorescence intensity; mean ± SEM). (G) U2OS cells expressing GFP-CLIP170 (Dragestein et al., 2008) (for identification of sictrl-transfected cells) were transfected with sictrl and mixed with cells cotransfected with sissrp1 and sispt16. Transcription levels 16 hr after 12 J/m 2 as determined by 2 hr pulse labeling with EU were not recovered in SRRP1/ Spt16 knockdown cells. After fixation, EU is fluorescent labeled with Alexa 594. See also Figure S Molecular Cell 51, , August 22, 2013 ª2013 Elsevier Inc.
7 shows that histone exchange facilitated by FACT at sites of UV-C damage-stalled transcription is not dependent on RNApolII stalling, nor does it require the presence of NER factors. This indicates that FACT recruitment to DNA damage is either a very early step in the NER pathway or occurs in parallel to, and independently of, NER. We found that RNAi-mediated knockdown of SPT16 had no effect on the amount of unscheduled DNA synthesis (UDS) after UV-C damage, which is a measure for GG-NER activity (Stefanini et al., 1993)(Figure 4E). Together with the absence of FACT near DNA lesions recognized by GG-NER (Figure 4A, Figure S4C), these data confirm that SPT16 has no essential function in GG-NER. Stalling of RNApolII at UV lesions causes inhibition of RNA synthesis. In TC-NER-proficient cells, recovery of RNA synthesis (RRS) after UV-induced inhibition usually starts several hours after damage infliction, when transcription-blocking lesions have been removed (Mayne and Lehmann, 1982). Interestingly, similar to depletion of the essential NER gene XPA, knockdown of both FACT subunits or of SPT16 alone resulted in a strong decrease in RRS (Figures 4F and 4G). Together these data indicate that, by promoting accelerated histone exchange, SPT16 plays an important role in the completion of TC-NER, allowing efficient restart of transcription after repair of the blocking lesions. DISCUSSION Here, we provide evidence for a role of the histone chaperone FACT in promoting transcription resumption after DNA damage-induced inhibition. FACT aids passage of RNApolII through chromatin templates by destabilizing the nucleosomes through the exchange of a histone H2A/H2B dimer (Hsieh et al., 2013; Reinberg and Sims, 2006). In addition, FACT is known to deposit the H2A/H2B dimer immediately behind the elongating transcription complex, likely to restore a more closed chromatin structure, thereby repressing intragenic transcription initiation from cryptic sites (Carvalho et al., 2013; Reinberg and Sims, 2006). The combined action of both removal and deposition of H2A/H2B during and immediately following transcription, respectively, also explains the observed transcription-dependent increased exchange of these dimers (Kimura and Cook, 2001). The ability of FACT to remove and reinsert H2A/H2B histones into chromatin in vitro has implicated it in all major DNA-associated processes, i.e., replication, transcription, and repair (Heo et al., 2008; Reinberg and Sims, 2006; Winkler and Luger, 2011). In addition, FACT has been shown to act during the UV-induced DDR in association with Casein Kinase II (CK2), in a role unrelated to chromatin remodeling (Keller and Lu, 2002; Krohn et al., 2003). We found that the SPT16-dependent accelerated H2A/H2B exchange occurs specifically at sites of damage-arrested transcription and is important for efficient recovery of UVinhibited RNA synthesis. Surprisingly, lesion recognition by stalled RNApolII is not required for SPT16 recruitment or accelerated H2A/H2B exchange at damaged sites, suggesting that this action occurs prior to or in parallel to TC-NER activation. Interestingly, these data show that the SPT16 histone chaperone activity is recruited independently from the ATP-dependent chromatin remodeler CSB (Citterio et al., 2000; Lans et al., 2012); however, both chromatin-remodeling activities are needed for efficient transcription-coupled repair. Importantly, the fact that H2A/H2B exchange still takes places at damaged chromatin in the absence of functional TC-NER and that recruitment of TC-NER factors occurs in the absence of FACT or enhanced H2A/H2B exchange shows that this exchange is not required to trigger TC-NER, nor is it a consequence of DNA repair itself. The FACT heterodimer can be posttranslationally modified by phosphorylation of SSRP1 by CK2 and by PARylation of SPT16 by PARP1 (Huang et al., 2006; Krohn et al., 2003). In our assay, CK2 inhibition had no effect on FACT activity, but PARP inhibition slightly decreased histone exchange at UV damage. This suggests that FACT recruitment or activity is stimulated by PARylation, a finding that conflicts with earlier studies showing that PARP1 activity inhibits FACT-directed H2AX exchange and promotes FACT dissociation from chromatin in vitro (Heo et al., 2008; Huang et al., 2006). This discrepancy might be explained by assuming that PARylation of factors that were not present in these in vitro experiments contributes to nucleosome plasticity in living cells. More experiments are needed to determine the precise role of PARP1 with regards to histone exchange in response to DNA damage. We have shown that H2A/H2B exchange is essential to promote transcription restart by RNApolII. There are multiple points during TC-NER where FACT activity might be required. Figure 5 describes three possible scenarios for FACT functions at sites of UV-damage-induced stalled transcription. In scenario 1, nucleosome deassembly behind the transcribing RNApolII is required for lesion-blocked RNApolII to reverse translocate (backtrack) (Cheung and Cramer, 2011). As removal of the blocking lesion is shown to occur via a nonallosteric recruitment of repair factors in the presence of RNAPII (Brueckner et al., 2007), remodeling of the chromatin in the vicinity of lesion-stalled RNAPolII is necessary to provide access of repair factors to the otherwise hidden blocking lesion. In this situation, the action of SPT16 will provide sufficient chromatin plasticity to remodel the arrested transcription complex to ensure efficient recruitment and activity of TC-NER proteins. Increased histone turnover might also promote the recruitment of a thus-far-unknown protein that is required for efficient repair and/or transcription restart after repair (scenario 2, Figure 5). The recruitment of such a protein could also occur via a direct interaction with SPT16, rather than indirectly through histone exchange. In scenario 3, accelerated exchange of H2A and H2B is required specifically for the restart of the stalled polymerase, or even damage bypass (Walmacq et al., 2012). In both situations prolonged enhanced plasticity of chromatin would allow a repeated cycle of failed transcription reinitiation events to take place, until a successful attempt leads to normal levels of histone exchange again. These three scenarios are by no means mutually exclusive and may even take place in a specific temporal order. These scenarios are likely not restricted to blocked transcription at NER-initiating lesions but may occur at any other type of damage that interferes with RNA polymerase elongation, such as collisions between RNA and DNA polymerases (Bermejo et al., 2012) or other lesions. Molecular Cell 51, , August 22, 2013 ª2013 Elsevier Inc. 475
8 H3-H4 H2A-H2B DNA damage FACT RNApolII and associated TC-NER factors Repaired DNA damage FACT needed for: 1. RNApolII backtracking 2. Recruitment of an unknown protein? 3. Transcription restart Figure 5. Three Scenarios for the Function of FACT-Induced Accelerated Histone Exchange at UV-Induced DNA Damage Upon stalling of RNAPolII on a UV lesion, the following three events may take place. In scenario 1, removal of an H2A-H2B dimer provides space for RNApolII to reverse translocate, which in turn allows efficient recruitment of downstream TC-NER proteins. In scenario 2, an unidentified TC-NER protein needs to be recruited to the DNA damage. This recruitment can only take place when H2A and H2B are removed via FACT, or via a direct interaction with FACT that is present near the damage site. In scenario 3, restart of transcription after TC-NER completion requires elevated levels of histone exchange mediated by FACT. The finding that specifically the SPT16 subunit of FACT seems required, rather than the complete heterodimeric complex, suggests a damage-specific chromatin-remodeling activity separating it from the canonical FACT function in transcription regulation in which both subunits are involved. It should be noted that, as is often the case for heterodimeric protein complexes, sirna-mediated knockdown of SSRP1 also results in the reduction of SPT16 protein levels, but not to the same extent as SPT16 knockdown itself (Figure S2C). The remaining SPT16 protein levels upon SSRP1 sirna depletion are apparently sufficient to perform its function in the UV-DDR. The separation of function between the SPT16 and SSRP1 subunits was unexpected because SSRP1 has been shown to bind to both cisplatin- and UV-C-induced DNA lesions via its HMG1 domain (Krohn et al., 2003; Yarnell et al., 2001). There are only limited additional data available for a separation of function between SPT16 and SSRP1. Thus far only SSRP1 has been found to have an independent-of-spt16 role in the transcription of a small subset of genes (Li et al., 2007). In spite of that, deletion of the SSRP1 homolog Pob3 in S. pombe is viable, but deletion of Spt16 is not (Lejeune et al., 2007). Other studies showing a function for only SSRP1 or SPT16 did not directly compare the activities of both FACT subunits. Recent mechanistic studies on FACT revealed that a C-terminal domain on SPT16, SPT16M, is mainly responsible for the interaction with nucleosomes and can bind to multiple sites on both H3-H4 and H2A-H2B histones, thereby shielding H2A-H2B from interactions with DNA, making space for the HMG1 domain of SSRP1 to bind DNA (Hondele et al., 2013; Kemble et al., 2013; Winkler et al., 2011). Other domains on SPT16 and SSRP1 bind nucleosomes cooperatively with SPT16M (Stuwe et al., 2008; Winkler et al., 2011). This suggests that FACT recruitment to nucleosomes is initiated by SPT16 and is in line with our observations that SSRP1 recruitment to DNA damage is dependent on SPT16. The recruitment of SPT16 to chromatin occurs partly via histone tail interactions (Winkler et al., 2011), which in turn suggests that SPT16 might be targeted to damaged chromatin via a histone tail modification that may occur quickly in response to UV-C irradiation. The method we have developed and applied here allows direct determination of in vivo histone dynamics at subnuclear regions in living cells. Using this technique we have uncovered a link between UV damage-induced chromatin remodeling and the restart of RNA synthesis. The regulation of the resumption of transcription after repair is highly important, given that improper restart leads to cellular malfunction and apoptosis, and that DNA damage-induced derailment of transcription is an important contributor to aging (Hanawalt and Spivak, 2008; Hoeijmakers, 2009). EXPERIMENTAL PROCEDURES Cell Lines and Constructs HeLa, U2OS, MRC5, XP4PA (XP-C), XP4PA (+GFP-XPC), CS3BE (CS-A), CS1AN (CS-B), CS1AN (+GFP-CSB), and VH10 (DDB2-GFP) (both sv40 and htert immortalized) cells were cultured under standard conditions in DMEM/ F10 medium supplemented with 10% fetal calf serum and antibiotics at 37 C and 5% CO 2. The H2B-GFP-, H3.1-GFP-, and H4-GFP-expressing cells have been previously published (Kimura and Cook, 2001). GFP-H2A (kind gift from Dr. M. Luijsterburg) and H2B-RFP were stably expressed in HeLa cells using neomycin selection. U2OS expressing GFP-tagged SPT16 and SSRP1 were, respectively, hygromycin and neomycin selected. The observed nuclear localization with enriched signal in nucleoli (Birch et al., 2009) is indicative of proper biological activity of these tagged proteins. For cell fusion experiments, cells were fused with 50% polyethylenegycol 1500 in PBS for 1.5 min as described in Schmidt-Zachmann et al. (1993). DNA constructs were transfected with FuGENE HD (Roche Applied Science), and sirna transfections were performed using RNAiMax (Invitrogen) or Hiperfect (QIAGEN) according to the manufacturer s instructions. Small interfering RNAs (smartpool) against SSRP1, SPT16, and NAP1L1 were supplied by Dharmacon. PARP1 inhibitor (AZD2281) and CK2 inhibitor (2-dimethylamino-4,5,6,7-tetrabromo-1H-benzimidazole [DMAT]) were used 1 hr before damage infliction at a final concentration of 10 mm. Transcription was inhibited with a-amanitin (25 mg/ml) or DRB (100 mm) for the indicated times. Microscopy For local UV-C irradiation experiments, a 2 mw pulsed (7.8 khz) diodepumped solid-state laser emitting at 266 nm (Rapp OptoElectronic, Hamburg GmbH) was connected to a Zeiss LSM 510 confocal microscope with an axiovert 200 M housing adapted for UV by all-quartz optics or to a Leica SP5 confocal microscope as described (Dinant et al., 2007; Schwertman et al., 476 Molecular Cell 51, , August 22, 2013 ª2013 Elsevier Inc.
9 2012). By focusing the UV C laser inside cell nuclei without scanning, only a limited area within the nucleus (diffraction limited spot) was irradiated. Cells for these experiments cells were grown on 25 or 24 mm diameter quartz coverslips to allow deep UV light to pass through the coverslip. Cells were imaged and irradiated through a NA Ultrafluar quartz objective. Immunofluorescence experiments were executed as described (Marteijn et al., 2009). FRAP experiments were performed as described previously (Hoogstraten et al., 2002). All FRAP data were normalized to the average prebleached fluorescence after removal of the background signal. All FRAP curves represent an average of at least 12 measured cells. Plasmid Constructs Murine pcmv-myc-ssrp1 and pcmv4a-spt16 were a kind gift of Professor Masayuki (Kihara et al., 2008). SSRP1 was amplified by PCR; during this reaction, SalI and XhoI were added to the cdna and cloned in PCR2.1 (Invitrogen). From this vector, SSRP1 was cloned into pegfp-n1 using Sal1/Xho1. To generate SPT16:GFP:HA the open reading frame (minus the STOP codon) of SPT16 was PCR amplified using primers that contained restriction sites for Hind III (forward) and BamHI (reverse). The amplified fragment was subcloned in a plhcx retroviral expression vector (Clontech Laboratories) which was previously modified to contain, following a BamHI restriction site, egfp (minus the initiation and stop codons), followed by the hemagglutinin (HA) tag and a STOP codon. PCR amplifications were performed on a MJ Scientific, Inc., PTC-100 Thermocycler using Phusion polymerase (Bioke). Amplified DNAs were purified using the Promega Wizard PCR purification kit. Following restriction digestion of the inserts and vectors, shrimp alkaline phosphatase treatment of the vectors, and agarose gel electrophoresis, the gel-excised DNAs were purified using the Promega Wizard gel extraction kit. DNA concentrations were calculated by spectrophotometry, and inserts were ligated into vectors at a 3:1 molar ratio. Plasmid DNAs were analyzed by restriction digestion and sequencing. Antibodies For ChIP rabbit polyclonal RNApolII (Abcam), a-p89/xpb (S-19, Santa Cruz), a-ddb1 (Abcam), a-spt16 (clone H-300), and a-ercc6 antibodies (Santa Cruz) were used. For western blot, a-rnapolii (H5, Babco; and Ser2, Active Motif), a-xpa (Abcam), a-gfp (Roche and Santa Cruz), a-csb (E-18, Santa- Cruz), a-csa, a-p89/xpb (S-19, Santa Cruz), a-ssrp1 (clone 10D7, Abcam and clone D-7 Santa Cruz), and a-spt16 (clone H-300, Santa Cruz) were used. Odyssey compatible secondary antibodies were from LI-COR, and horseradish peroxidase (HRP)-conjugated secondary antibodies were from Dako (a-mouse and a-goat), Southern Biotech (a-rabbit), and Sigma (a-igm). For immunofluorescence, the following antibodies were used: a-npm/b23 (Abcam), a-par (Alexis, Clone H10), a-spt16 (clone H-300, Santa Cruz), and a-cpd (TDM-2, MBL International) in combination with the corresponding secondary antibodies labeled with Alexa Fluor 488 or 594 as indicated (Invitrogen; The Jackson Laboratory). Immunofluorescence experiments were executed as described (Marteijn et al., 2009). Clonogenic Survival Assay Cells from indicated cell lines seeded in triplicate in 6-well plates (300 cells/ well) were treated with UV-C 1 day after seeding and recovered for 1 week before colonies were fixed and stained in 50% methanol, 7% acetic acid, and 0.1% Coomassie blue. Recovery of RNA Synthesis and Unscheduled DNA Synthesis after UV Irradiation Fluorescence-based RRS and UDS were performed as described (Nakazawa et al., 2010; Schwertman et al., 2012). In short, for RRS, the indicated cell lines were UV rradiated with 12 J/m 2. Transcription levels were determined 16 hr after UV by 2 hr ethynyluridine (EU)-incorporation. For UDS, VH10 cells were UV irradiated with 16 J/m 2 48 hr after sirna, and UDS was determined after 3 hr of 5-ethynyl-2 0 -deoxyuridine (EdU) incorporation. RRS and UDS were quantified by determining fluorescent intensities of >200 cells with ImageJ software of images obtained with a Zeiss LSM700. The cells displayed in Figure 4E were stained with a-gfp (Abcam) after the RRS procedure to visualize the GFP signal. In Vivo Crosslinking and ChIP-on-Western The procedure for ChIP in combination with western blot (ChIP-on-Western) has been described previously (Fousteri et al., 2006). Briefly, cells were mock treated or exposed to UV-C light (20 J/m 2 ) and left to recover for 1 hr at 37 C prior to in vivo crosslinking by 1% formaldehyde at 4 C. Lysis of the crosslinked cells and chromatin purification was performed as described previously (Fousteri et al., 2006). Chromatin shearing was achieved on the Bioruptor Sonicator (Diagenode) using cycles of ON, OFF. ChIP was performed on equal amounts of bp fragments of chromatin from treated and nontreated cells. Reversal of the crosslinks and elution of the precipitated proteins was performed by extended boiling in Laemmli SDSsample buffer and analyzed by western blotting (Fousteri et al., 2006). SUPPLEMENTAL INFORMATION Supplemental Information includes four figures and Supplemental References and can be found with this article online at ACKNOWLEDGMENTS We would like to thank Florian Lust and Sander Tuit for their assistance, and the Optical Imaging Centre (OIC) of the Erasmus MC for support with microscopes. We thank Niels Galjart for providing the stable GFP-CLIP170-expressing cell line. We also thank Loes van Cuijk for her help testing the activity of the used transcription inhibitors. This work was funded by the Lundbeck Foundation, the Novo Nordisk Foundation, and the Danish Cancer Society (C.D. and J.B.); the European Commission (projects DDResponse and Biomedreg) (J.B.); TOP ALW grant (H.L.); the Association for International Cancer Research (M.T. and W.V.); NSRF , Aristeia-2429, KRHPIS, and ERC-2012-StG (M.F.); ZonMW TOP grant , Horizon Zenith , and FP7 Marie Curie International Training Network address (W.V.); and Erasmus MC fellowship and Dutch Organization for Scientific Research ZonMW Veni Grant (J.A.M). Received: May 2, 2013 Revised: June 7, 2013 Accepted: July 17, 2013 Published: August 22, 2013 REFERENCES Avvakumov, N., Nourani, A., and Côté, J. (2011). Histone chaperones: modulators of chromatin marks. Mol. Cell 41, Bensaude, O. (2011). Inhibiting eukaryotic transcription: Which compound to choose? How to evaluate its activity? Transcription 2, Bermejo, R., Lai, M.S., and Foiani, M. (2012). Preventing replication stress to maintain genome stability: resolving conflicts between replication and transcription. Mol. Cell 45, Birch, J.L., Tan, B.C., Panov, K.I., Panova, T.B., Andersen, J.S., Owen- Hughes, T.A., Russell, J., Lee, S.C., and Zomerdijk, J.C. (2009). FACT facilitates chromatin transcription by RNA polymerases I and III. EMBO J. 28, Brueckner, F., Hennecke, U., Carell, T., and Cramer, P. (2007). CPD damage recognition by transcribing RNA polymerase II. Science 315, Carvalho, S., Raposo, A.C., Martins, F.B., Grosso, A.R., Sridhara, S.C., Rino, J., Carmo-Fonseca, M., and de Almeida, S.F. (2013). Histone methyltransferase SETD2 coordinates FACT recruitment with nucleosome dynamics during transcription. Nucleic Acids Res. 41, Cheung, A.C., and Cramer, P. (2011). Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature 471, Citterio, E., Van Den Boom, V., Schnitzler, G., Kanaar, R., Bonte, E., Kingston, R.E., Hoeijmakers, J.H., and Vermeulen, W. (2000). ATP-dependent chromatin remodeling by the Cockayne syndrome B DNA repair-transcription-coupling factor. Mol. Cell. Biol. 20, Molecular Cell 51, , August 22, 2013 ª2013 Elsevier Inc. 477
10 Coin, F., Oksenych, V., Mocquet, V., Groh, S., Blattner, C., and Egly, J.M. (2008). Nucleotide excision repair driven by the dissociation of CAK from TFIIH. Mol. Cell 31, De Koning, L., Corpet, A., Haber, J.E., and Almouzni, G. (2007). Histone chaperones: an escort network regulating histone traffic. Nat. Struct. Mol. Biol. 14, Dinant, C., de Jager, M., Essers, J., van Cappellen, W.A., Kanaar, R., Houtsmuller, A.B., and Vermeulen, W. (2007). Activation of multiple DNA repair pathways by sub-nuclear damage induction methods. J. Cell Sci. 120, Dragestein, K.A., van Cappellen, W.A., van Haren, J., Tsibidis, G.D., Akhmanova, A., Knoch, T.A., Grosveld, F., and Galjart, N. (2008). Dynamic behavior of GFP-CLIP-170 reveals fast protein turnover on microtubule plus ends. J. Cell Biol. 180, Essers, J., Theil, A.F., Baldeyron, C., van Cappellen, W.A., Houtsmuller, A.B., Kanaar, R., and Vermeulen, W. (2005). Nuclear dynamics of PCNA in DNA replication and repair. Mol. Cell. Biol. 25, Fousteri, M., Vermeulen, W., van Zeeland, A.A., and Mullenders, L.H. (2006). Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Mol. Cell 23, Gong, F., Kwon, Y., and Smerdon, M.J. (2005). Nucleotide excision repair in chromatin and the right of entry. DNA Repair (Amst.) 4, Green, C.M., and Almouzni, G. (2003). Local action of the chromatin assembly factor CAF-1 at sites of nucleotide excision repair in vivo. EMBO J. 22, Groisman, R., Polanowska, J., Kuraoka, I., Sawada, J., Saijo, M., Drapkin, R., Kisselev, A.F., Tanaka, K., and Nakatani, Y. (2003). The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell 113, Hanawalt, P.C., and Spivak, G. (2008). Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 9, Heo, K., Kim, H., Choi, S.H., Choi, J., Kim, K., Gu, J., Lieber, M.R., Yang, A.S., and An, W. (2008). FACT-mediated exchange of histone variant H2AX regulated by phosphorylation of H2AX and ADP-ribosylation of Spt16. Mol. Cell 30, Hoeijmakers, J.H. (2009). DNA damage, aging, and cancer. N. Engl. J. Med. 361, Hondele, M., Stuwe, T., Hassler, M., Halbach, F., Bowman, A., Zhang, E.T., Nijmeijer, B., Kotthoff, C., Rybin, V., Amlacher, S., et al. (2013). Structural basis of histone H2A-H2B recognition by the essential chaperone FACT. Nature 499, Hoogstraten, D., Nigg, A.L., Heath, H., Mullenders, L.H., van Driel, R., Hoeijmakers, J.H., Vermeulen, W., and Houtsmuller, A.B. (2002). Rapid switching of TFIIH between RNA polymerase I and II transcription and DNA repair in vivo. Mol. Cell 10, Hsieh, F.K., Kulaeva, O.I., Patel, S.S., Dyer, P.N., Luger, K., Reinberg, D., and Studitsky, V.M. (2013). Histone chaperone FACT action during transcription through chromatin by RNA polymerase II. Proc. Natl. Acad. Sci. USA 110, Huang, J.Y., Chen, W.H., Chang, Y.L., Wang, H.T., Chuang, W.T., and Lee, S.C. (2006). Modulation of nucleosome-binding activity of FACT by poly(adp-ribosyl)ation. Nucleic Acids Res. 34, Kannouche, P.L., Wing, J., and Lehmann, A.R. (2004). Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell 14, Keller, D.M., and Lu, H. (2002). p53 serine 392 phosphorylation increases after UV through induction of the assembly of the CK2.hSPT16.SSRP1 complex. J. Biol. Chem. 277, Kemble, D.J., Whitby, F.G., Robinson, H., McCullough, L.L., Formosa, T., and Hill, C.P. (2013). Structure of the Spt16 middle domain reveals functional features of the histone chaperone FACT. J. Biol. Chem. 288, Kihara, T., Kano, F., and Murata, M. (2008). Modulation of SRF-dependent gene expression by association of SPT16 with MKL1. Exp. Cell Res. 314, Kimura, H., and Cook, P.R. (2001). Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell Biol. 153, Krohn, N.M., Stemmer, C., Fojan, P., Grimm, R., and Grasser, K.D. (2003). Protein kinase CK2 phosphorylates the high mobility group domain protein SSRP1, inducing the recognition of UV-damaged DNA. J. Biol. Chem. 278, Lans, H., Marteijn, J.A., and Vermeulen, W. (2012). ATP-dependent chromatin remodeling in the DNA-damage response. Epigenetics Chromatin 5, 4. Lejeune, E., Bortfeld, M., White, S.A., Pidoux, A.L., Ekwall, K., Allshire, R.C., and Ladurner, A.G. (2007). The chromatin-remodeling factor FACT contributes to centromeric heterochromatin independently of RNAi. Curr. Biol. 17, Li, Y., Zeng, S.X., Landais, I., and Lu, H. (2007). Human SSRP1 has Spt16- dependent and -independent roles in gene transcription. J. Biol. Chem. 282, Luijsterburg, M.S., Lindh, M., Acs, K., Vrouwe, M.G., Pines, A., van Attikum, H., Mullenders, L.H., and Dantuma, N.P. (2012). DDB2 promotes chromatin decondensation at UV-induced DNA damage. J. Cell Biol. 197, Marteijn, J.A., Bekker-Jensen, S., Mailand, N., Lans, H., Schwertman, P., Gourdin, A.M., Dantuma, N.P., Lukas, J., and Vermeulen, W. (2009). Nucleotide excision repair-induced H2A ubiquitination is dependent on MDC1 and RNF8 and reveals a universal DNA damage response. J. Cell Biol. 186, Mayne, L.V., and Lehmann, A.R. (1982). Failure of RNA synthesis to recover after UV irradiation: an early defect in cells from individuals with Cockayne s syndrome and xeroderma pigmentosum. Cancer Res. 42, Menear, K.A., Adcock, C., Boulter, R., Cockcroft, X.L., Copsey, L., Cranston, A., Dillon, K.J., Drzewiecki, J., Garman, S., Gomez, S., et al. (2008). 4-[3-(4- cyclopropanecarbonylpiperazine-1-carbonyl)-4-fluorobenzyl]-2h-phthalazin- 1-one: a novel bioavailable inhibitor of poly(adp-ribose) polymerase-1. J. Med. Chem. 51, Nakazawa, Y., Yamashita, S., Lehmann, A.R., and Ogi, T. (2010). A semi-automated non-radioactive system for measuring recovery of RNA synthesis and unscheduled DNA synthesis using ethynyluracil derivatives. DNA Repair (Amst.) 9, Nouspikel, T. (2009). DNA repair in mammalian cells : Nucleotide excision repair: variations on versatility. Cell. Mol. Life Sci. 66, Pagano, M.A., Meggio, F., Ruzzene, M., Andrzejewska, M., Kazimierczuk, Z., and Pinna, L.A. (2004). 2-Dimethylamino-4,5,6,7-tetrabromo-1H-benzimidazole: a novel powerful and selective inhibitor of protein kinase CK2. Biochem. Biophys. Res. Commun. 321, Polo, S.E., Roche, D., and Almouzni, G. (2006). New histone incorporation marks sites of UV repair in human cells. Cell 127, Ransom, M., Dennehey, B.K., and Tyler, J.K. (2010). Chaperoning histones during DNA replication and repair. Cell 140, Reinberg, D., and Sims, R.J., 3rd. (2006). de FACTo nucleosome dynamics. J. Biol. Chem. 281, Schmidt-Zachmann, M.S., Dargemont, C., Kühn, L.C., and Nigg, E.A. (1993). Nuclear export of proteins: the role of nuclear retention. Cell 74, Schwertman, P., Lagarou, A., Dekkers, D.H., Raams, A., van der Hoek, A.C., Laffeber, C., Hoeijmakers, J.H., Demmers, J.A., Fousteri, M., Vermeulen, W., and Marteijn, J.A. (2012). UV-sensitive syndrome protein UVSSA recruits USP7 to regulate transcription-coupled repair. Nat. Genet. 44, Smerdon, M.J. (1991). DNA repair and the role of chromatin structure. Curr. Opin. Cell Biol. 3, Soria, G., Polo, S.E., and Almouzni, G. (2012). Prime, repair, restore: the active role of chromatin in the DNA damage response. Mol. Cell 46, Molecular Cell 51, , August 22, 2013 ª2013 Elsevier Inc.
Immunofluorescence images of different core histones and different histone exchange assay.
 Molecular Cell, Volume 51 Supplemental Information Enhanced Chromatin Dynamics by FACT Promotes Transcriptional Restart after UV-Induced DNA Damage Christoffel Dinant, Giannis Ampatziadis-Michailidis,
Molecular Cell, Volume 51 Supplemental Information Enhanced Chromatin Dynamics by FACT Promotes Transcriptional Restart after UV-Induced DNA Damage Christoffel Dinant, Giannis Ampatziadis-Michailidis,
Four different active promoter genes were chosen, ATXN7L2, PSRC1, CELSR2 and
 SUPPLEMENTARY MATERIALS AND METHODS Chromatin Immunoprecipitation for qpcr analysis Four different active promoter genes were chosen, ATXN7L2, PSRC1, CELSR2 and IL24, all located on chromosome 1. Primer
SUPPLEMENTARY MATERIALS AND METHODS Chromatin Immunoprecipitation for qpcr analysis Four different active promoter genes were chosen, ATXN7L2, PSRC1, CELSR2 and IL24, all located on chromosome 1. Primer
UV-sensitive syndrome protein UVSSA recruits USP7 to regulate transcription-coupled repair
 Supplementary Information UVsensitive syndrome protein UVSSA recruits to regulate transcriptioncoupled repair Petra Schwertman 1, Anna Lagarou 2, Dick H.W. Dekkers 3, Anja Raams 1, Adriana C. van der Hoek
Supplementary Information UVsensitive syndrome protein UVSSA recruits to regulate transcriptioncoupled repair Petra Schwertman 1, Anna Lagarou 2, Dick H.W. Dekkers 3, Anja Raams 1, Adriana C. van der Hoek
HPV E6 oncoprotein targets histone methyltransferases for modulating specific. Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu,
 1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
Molecular mechanisms of DNA repair and transcription studied by FRAP
 Molecular mechanisms of DNA repair and transcription studied by FRAP Adriaan Houtsmuller Josephine Nefkens Institute Erasmus Medical Centre Rotterdam The Netherlands Nucleus GFP-tagged proteins Confocal
Molecular mechanisms of DNA repair and transcription studied by FRAP Adriaan Houtsmuller Josephine Nefkens Institute Erasmus Medical Centre Rotterdam The Netherlands Nucleus GFP-tagged proteins Confocal
Supplementary Figure Legends
 Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
SUPPLEMENTAL MATERIAL. Supplemental material contains Supplemental Figure Legends and Supplemental Figures 1 to
 SUPPLEMENTAL MATERIAL Supplemental material contains Supplemental Figure Legends and Supplemental Figures 1 to 6. SUPPLEMENTAL FIGURE LEGENDS Supplemental Figure 1. Overview of the mechanisms by which
SUPPLEMENTAL MATERIAL Supplemental material contains Supplemental Figure Legends and Supplemental Figures 1 to 6. SUPPLEMENTAL FIGURE LEGENDS Supplemental Figure 1. Overview of the mechanisms by which
Supplementary Material
 Supplementary Material Supplementary Methods Cell synchronization. For synchronized cell growth, thymidine was added to 30% confluent U2OS cells to a final concentration of 2.5mM. Cells were incubated
Supplementary Material Supplementary Methods Cell synchronization. For synchronized cell growth, thymidine was added to 30% confluent U2OS cells to a final concentration of 2.5mM. Cells were incubated
Supplementary data. sienigma. F-Enigma F-EnigmaSM. a-p53
 Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the
 Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
The role of erna in chromatin looping
 The role of erna in chromatin looping Introduction Enhancers are cis-acting DNA regulatory elements that activate transcription of target genes 1,2. It is estimated that 400 000 to >1 million alleged enhancers
The role of erna in chromatin looping Introduction Enhancers are cis-acting DNA regulatory elements that activate transcription of target genes 1,2. It is estimated that 400 000 to >1 million alleged enhancers
Supplemental Material: Rev1 promotes replication through UV lesions in conjunction with DNA
 Supplemental Material: Rev1 promotes replication through UV lesions in conjunction with DNA polymerases,, and, but not with DNA polymerase Jung-Hoon Yoon, Jeseong Park, Juan Conde, Maki Wakamiya, Louise
Supplemental Material: Rev1 promotes replication through UV lesions in conjunction with DNA polymerases,, and, but not with DNA polymerase Jung-Hoon Yoon, Jeseong Park, Juan Conde, Maki Wakamiya, Louise
Sarker et al. Supplementary Material. Subcellular Fractionation
 Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
Design. Construction. Characterization
 Design Construction Characterization DNA mrna (messenger) A C C transcription translation C A C protein His A T G C T A C G Plasmids replicon copy number incompatibility selection marker origin of replication
Design Construction Characterization DNA mrna (messenger) A C C transcription translation C A C protein His A T G C T A C G Plasmids replicon copy number incompatibility selection marker origin of replication
Supplementary Figure 1. RAD51 and RAD51 paralogs are enriched spontaneously onto
 Supplementary Figure legends Supplementary Figure 1. and paralogs are enriched spontaneously onto the S-phase chromatin during DN replication. () Chromatin fractionation was carried out as described in
Supplementary Figure legends Supplementary Figure 1. and paralogs are enriched spontaneously onto the S-phase chromatin during DN replication. () Chromatin fractionation was carried out as described in
Non-Organic-Based Isolation of Mammalian microrna using Norgen s microrna Purification Kit
 Application Note 13 RNA Sample Preparation Non-Organic-Based Isolation of Mammalian microrna using Norgen s microrna Purification Kit B. Lam, PhD 1, P. Roberts, MSc 1 Y. Haj-Ahmad, M.Sc., Ph.D 1,2 1 Norgen
Application Note 13 RNA Sample Preparation Non-Organic-Based Isolation of Mammalian microrna using Norgen s microrna Purification Kit B. Lam, PhD 1, P. Roberts, MSc 1 Y. Haj-Ahmad, M.Sc., Ph.D 1,2 1 Norgen
Histone H3 Acetylation Antibody Panel Pack II Base Catalog # C K23A Histone H3K23ac (Acetyl H3K23) Polyclonal Antibody 25 µg 4 C 20 C
 Histone H3 Acetylation Antibody Panel Pack II Base Catalog # PACK CONTENTS Component Size Shipping Temperature Upon Receipt Checklist 3K23A Histone H3K23ac (Acetyl H3K23) Polyclonal Antibody 25 µg 4 C
Histone H3 Acetylation Antibody Panel Pack II Base Catalog # PACK CONTENTS Component Size Shipping Temperature Upon Receipt Checklist 3K23A Histone H3K23ac (Acetyl H3K23) Polyclonal Antibody 25 µg 4 C
Supplementary information
 Supplementary information The E3 ligase RNF8 regulates KU80 removal and NHEJ repair Lin Feng 1, Junjie Chen 1 1 Department of Experimental Radiation Oncology, The University of Texas M. D. Anderson Cancer
Supplementary information The E3 ligase RNF8 regulates KU80 removal and NHEJ repair Lin Feng 1, Junjie Chen 1 1 Department of Experimental Radiation Oncology, The University of Texas M. D. Anderson Cancer
Supplementary Figure S1. Np95 interacts with de novo methyltransferases Dnmt3a and 3b. (A) Co-immunoprecipitation of endogenous Dnmt3a2 (left and
 Supplementary Figure S1. Np95 interacts with de novo methyltransferases Dnmt3a and 3b. (A) Co-immunoprecipitation of endogenous Dnmt3a2 (left and right), Dnmt3b isoforms (left) and Dnmt1 (right) with GFP-Np95
Supplementary Figure S1. Np95 interacts with de novo methyltransferases Dnmt3a and 3b. (A) Co-immunoprecipitation of endogenous Dnmt3a2 (left and right), Dnmt3b isoforms (left) and Dnmt1 (right) with GFP-Np95
Learning Objectives. Define RNA interference. Define basic terminology. Describe molecular mechanism. Define VSP and relevance
 Learning Objectives Define RNA interference Define basic terminology Describe molecular mechanism Define VSP and relevance Describe role of RNAi in antigenic variation A Nobel Way to Regulate Gene Expression
Learning Objectives Define RNA interference Define basic terminology Describe molecular mechanism Define VSP and relevance Describe role of RNAi in antigenic variation A Nobel Way to Regulate Gene Expression
SUPPLEMENTARY INFORMATION
 (Supplementary Methods and Materials) GST pull-down assay GST-fusion proteins Fe65 365-533, and Fe65 538-700 were expressed in BL21 bacterial cells and purified with glutathione-agarose beads (Sigma).
(Supplementary Methods and Materials) GST pull-down assay GST-fusion proteins Fe65 365-533, and Fe65 538-700 were expressed in BL21 bacterial cells and purified with glutathione-agarose beads (Sigma).
SUPPLEMENTARY INFORMATION
 Figure S1: Activation of the ATM pathway by I-PpoI. A. HEK293T cells were either untransfected, vector transfected, transfected with an I-PpoI expression vector, or subjected to 2Gy γ-irradiation. 24 hrs
Figure S1: Activation of the ATM pathway by I-PpoI. A. HEK293T cells were either untransfected, vector transfected, transfected with an I-PpoI expression vector, or subjected to 2Gy γ-irradiation. 24 hrs
Supplementary Information
 Supplementary Information stability is regulated by CK2-dependent interaction with R2TP complex Patrick von Morgen 1,2, Kamila Burdova 1, Thomas G. Flower 3, Nicola J. O'Reilly 4, Simon J. Boulton 5, Stephen
Supplementary Information stability is regulated by CK2-dependent interaction with R2TP complex Patrick von Morgen 1,2, Kamila Burdova 1, Thomas G. Flower 3, Nicola J. O'Reilly 4, Simon J. Boulton 5, Stephen
Supplementary Figure 1 Collision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK2, PPK3 and PPK4 respectively.
 Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
SUPPLEMENTARY INFORMATION
 The Supplementary Information (SI) Methods Cell culture and transfections H1299, U2OS, 293, HeLa cells were maintained in DMEM medium supplemented with 10% fetal bovine serum. H1299 and 293 cells were
The Supplementary Information (SI) Methods Cell culture and transfections H1299, U2OS, 293, HeLa cells were maintained in DMEM medium supplemented with 10% fetal bovine serum. H1299 and 293 cells were
 DOI: 10.1038/ncb3259 A Ismail et al. Supplementary Figure 1 B 60000 45000 SSC 30000 15000 Live cells 0 0 15000 30000 45000 60000 FSC- PARR 60000 45000 PARR Width 30000 FSC- 15000 Single cells 0 0 15000
DOI: 10.1038/ncb3259 A Ismail et al. Supplementary Figure 1 B 60000 45000 SSC 30000 15000 Live cells 0 0 15000 30000 45000 60000 FSC- PARR 60000 45000 PARR Width 30000 FSC- 15000 Single cells 0 0 15000
Histone H3K27 Methylation Antibody Panel Pack Base Catalog # C Component Size Shipping Temperature
 Histone H3K27 Methylation Antibody Panel Pack Base Catalog # PACK CONTENTS Component Size Shipping Temperature Upon Receipt Checklist 3K27M Histone H3K27me1 (H3K27 Monomethyl) Polyclonal Antibody 25 µg
Histone H3K27 Methylation Antibody Panel Pack Base Catalog # PACK CONTENTS Component Size Shipping Temperature Upon Receipt Checklist 3K27M Histone H3K27me1 (H3K27 Monomethyl) Polyclonal Antibody 25 µg
Oligonucleotides containing either a single cyclo-da lesion or a single CPD in an
 Materials and Methods Preparation of Constructs Oligonucleotides containing either a single cyclo-da lesion or a single CPD in an unique MfeI site were prepared as described previously (Brooks et al.,
Materials and Methods Preparation of Constructs Oligonucleotides containing either a single cyclo-da lesion or a single CPD in an unique MfeI site were prepared as described previously (Brooks et al.,
Transcriptional regulation of BRCA1 expression by a metabolic switch: Di, Fernandez, De Siervi, Longo, and Gardner. H3K4Me3
 ChIP H3K4Me3 enrichment.25.2.15.1.5 H3K4Me3 H3K4Me3 ctrl H3K4Me3 + E2 NS + E2 1. kb kb +82 kb Figure S1. Estrogen promotes entry of MCF-7 into the cell cycle but does not significantly change activation-associated
ChIP H3K4Me3 enrichment.25.2.15.1.5 H3K4Me3 H3K4Me3 ctrl H3K4Me3 + E2 NS + E2 1. kb kb +82 kb Figure S1. Estrogen promotes entry of MCF-7 into the cell cycle but does not significantly change activation-associated
Supplementary Discussion and Figures
 Supplementary Discussion and Figures Differences in chromatin retention between the two CHD3 isoforms The pan-nuclear loss of the KAP-1-interacting CHD3 isoform from chromatin (observed in Fig. 2A) is
Supplementary Discussion and Figures Differences in chromatin retention between the two CHD3 isoforms The pan-nuclear loss of the KAP-1-interacting CHD3 isoform from chromatin (observed in Fig. 2A) is
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Nakajima and Tanoue, http://www.jcb.org/cgi/content/full/jcb.201104118/dc1 Figure S1. DLD-1 cells exhibit the characteristic morphology
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Nakajima and Tanoue, http://www.jcb.org/cgi/content/full/jcb.201104118/dc1 Figure S1. DLD-1 cells exhibit the characteristic morphology
Supplementary Information for. Regulation of Rev1 by the Fanconi Anemia Core Complex
 Supplementary Information for Regulation of Rev1 by the Fanconi Anemia Core Complex Hyungjin Kim, Kailin Yang, Donniphat Dejsuphong, Alan D. D Andrea* *Corresponding Author: Alan D. D Andrea, M.D. Alan_dandrea@dfci.harvard.edu
Supplementary Information for Regulation of Rev1 by the Fanconi Anemia Core Complex Hyungjin Kim, Kailin Yang, Donniphat Dejsuphong, Alan D. D Andrea* *Corresponding Author: Alan D. D Andrea, M.D. Alan_dandrea@dfci.harvard.edu
Otefin, a Nuclear Membrane Protein, Determines
 Developmental Cell 14 Supplemental Data Otefin, a Nuclear Membrane Protein, Determines the Fate of Germline Stem Cells in Drosophila via Interaction with Smad Complexes Xiaoyong Jiang, Laixin Xia, Dongsheng
Developmental Cell 14 Supplemental Data Otefin, a Nuclear Membrane Protein, Determines the Fate of Germline Stem Cells in Drosophila via Interaction with Smad Complexes Xiaoyong Jiang, Laixin Xia, Dongsheng
Supplementary Information. Synergistic action of RNA polymerases in overcoming the nucleosomal barrier
 Supplementary Information Synergistic action of RNA polymerases in overcoming the nucleosomal barrier Jing Jin, Lu Bai, Daniel S. Johnson, Robert M. Fulbright, Maria L. Kireeva, Mikhail Kashlev, Michelle
Supplementary Information Synergistic action of RNA polymerases in overcoming the nucleosomal barrier Jing Jin, Lu Bai, Daniel S. Johnson, Robert M. Fulbright, Maria L. Kireeva, Mikhail Kashlev, Michelle
Schematic representation of the endogenous PALB2 locus and gene-disruption constructs
 Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or
 Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
Supplementary methods
 Supplementary methods Cell culture, infection, transfection, and RNA interference HEK293 cells and its derivatives were grown in DMEM supplemented with 10% FBS. Various constructs were introduced into
Supplementary methods Cell culture, infection, transfection, and RNA interference HEK293 cells and its derivatives were grown in DMEM supplemented with 10% FBS. Various constructs were introduced into
Histone H3 Methylation Antibody Panel Pack I - Active Genes Base Catalog # C Component Size Shipping Temperature
 Histone H3 Methylation Antibody Panel Pack I - Active Genes Base Catalog # PACK CONTENTS Component Size Shipping Temperature Upon Receipt Checklist 3K4D Histone H3K4me2 (H3K4 Dimethyl) Polyclonal Antibody
Histone H3 Methylation Antibody Panel Pack I - Active Genes Base Catalog # PACK CONTENTS Component Size Shipping Temperature Upon Receipt Checklist 3K4D Histone H3K4me2 (H3K4 Dimethyl) Polyclonal Antibody
Supplementary Fig S1 Nutlin-3a treatment does not affect cell cycle progression in the absence
 Supplementary Figure Legends Supplementary Fig S1 Nutlin-3a treatment does not affect cell cycle progression in the absence of p53 or p21. HCT116 cells which were null for either p53 (A) or p21 (B) were
Supplementary Figure Legends Supplementary Fig S1 Nutlin-3a treatment does not affect cell cycle progression in the absence of p53 or p21. HCT116 cells which were null for either p53 (A) or p21 (B) were
Dual Roles for PARP1 during Heat Shock: Transcriptional Activator and Posttranscriptional Inhibitor of Gene Expression
 Dual Roles for PARP1 during Heat Shock: Transcriptional Activator and Posttranscriptional Inhibitor of Gene Expression The MIT Faculty has made this article openly available. Please share how this access
Dual Roles for PARP1 during Heat Shock: Transcriptional Activator and Posttranscriptional Inhibitor of Gene Expression The MIT Faculty has made this article openly available. Please share how this access
Exam 2 BIO200, Winter 2012
 Exam 2 BIO200, Winter 2012 Name: Multiple Choice Questions: Circle the one best answer for each question. (2 points each) 1. The 5 cap structure is often described as a backwards G. What makes this nucleotide
Exam 2 BIO200, Winter 2012 Name: Multiple Choice Questions: Circle the one best answer for each question. (2 points each) 1. The 5 cap structure is often described as a backwards G. What makes this nucleotide
Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity.
 Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity. (A) Amino acid alignment of HDA5, HDA15 and HDA18. The blue line
Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity. (A) Amino acid alignment of HDA5, HDA15 and HDA18. The blue line
HeLa cells stably transfected with an empty pcdna3 vector (HeLa Neo) or with a plasmid
 SUPPLEMENTAL MATERIALS AND METHODS Cell culture, transfection and treatments. HeLa cells stably transfected with an empty pcdna3 vector (HeLa Neo) or with a plasmid encoding vmia (HeLa vmia) 1 were cultured
SUPPLEMENTAL MATERIALS AND METHODS Cell culture, transfection and treatments. HeLa cells stably transfected with an empty pcdna3 vector (HeLa Neo) or with a plasmid encoding vmia (HeLa vmia) 1 were cultured
Evaluation of Metafectene-Pro mediated transfection of sirna and/or plasmids into human and murine cells
 Evaluation of Metafectene-Pro mediated transfection of sirna and/or plasmids into human and murine cells Aaron A. Goodarzi and Penny A. Jeggo Genome Damage and Stability Centre, University of Sussex, East
Evaluation of Metafectene-Pro mediated transfection of sirna and/or plasmids into human and murine cells Aaron A. Goodarzi and Penny A. Jeggo Genome Damage and Stability Centre, University of Sussex, East
Evaluation of Metafectene-Pro mediated transfection of sirna and/or plasmids into human and murine cells
 Evaluation of Metafectene-Pro mediated transfection of sirna and/or plasmids into human and murine cells Aaron A. Goodarzi and Penny A. Jeggo Genome Damage and Stability Centre, University of Sussex, East
Evaluation of Metafectene-Pro mediated transfection of sirna and/or plasmids into human and murine cells Aaron A. Goodarzi and Penny A. Jeggo Genome Damage and Stability Centre, University of Sussex, East
transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected with the wwp-luc reporter, and FLAG-tagged FHL1,
 Supplementary Data Supplementary Figure Legends Supplementary Figure 1 FHL-mediated TGFβ-responsive reporter transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected
Supplementary Data Supplementary Figure Legends Supplementary Figure 1 FHL-mediated TGFβ-responsive reporter transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected
Supplementary Information: Materials and Methods. GST and GST-p53 were purified according to standard protocol after
 Supplementary Information: Materials and Methods Recombinant protein expression and in vitro kinase assay. GST and GST-p53 were purified according to standard protocol after induction with.5mm IPTG for
Supplementary Information: Materials and Methods Recombinant protein expression and in vitro kinase assay. GST and GST-p53 were purified according to standard protocol after induction with.5mm IPTG for
Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and
 Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Thompson et al., http://www.jcb.org/cgi/content/full/jcb.200909067/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Modification-specific antibodies do not detect unmodified
Supplemental material Thompson et al., http://www.jcb.org/cgi/content/full/jcb.200909067/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Modification-specific antibodies do not detect unmodified
The Need for a PARP Pharmacodynamic Assay
 The Need for a PARP Pharmacodynamic Assay Version 05/07/09 amsbiotechnology(europe)ltd info@amsbio.com www.amsbio.com (UK)+44(0)1235828200 (CH)+41(0)916045522 (DE)+49(0)69779099 DNA Repair Pathways Base
The Need for a PARP Pharmacodynamic Assay Version 05/07/09 amsbiotechnology(europe)ltd info@amsbio.com www.amsbio.com (UK)+44(0)1235828200 (CH)+41(0)916045522 (DE)+49(0)69779099 DNA Repair Pathways Base
Emanuela Tumini, Sonia Barroso, Carmen Pérez Calero and Andrés Aguilera
 SUPPLEMENTARY INFORMATION Roles of human POLD1 and POLD3 in genome stability Emanuela Tumini, Sonia Barroso, Carmen Pérez Calero and Andrés Aguilera SUPPLEMENTARY METHODS Cell proliferation After sirna
SUPPLEMENTARY INFORMATION Roles of human POLD1 and POLD3 in genome stability Emanuela Tumini, Sonia Barroso, Carmen Pérez Calero and Andrés Aguilera SUPPLEMENTARY METHODS Cell proliferation After sirna
RNA was isolated using NucleoSpin RNA II (Macherey-Nagel, Bethlehem, PA) according to the
 Supplementary Methods RT-PCR and real-time PCR analysis RNA was isolated using NucleoSpin RNA II (Macherey-Nagel, Bethlehem, PA) according to the manufacturer s protocol and quantified by measuring the
Supplementary Methods RT-PCR and real-time PCR analysis RNA was isolated using NucleoSpin RNA II (Macherey-Nagel, Bethlehem, PA) according to the manufacturer s protocol and quantified by measuring the
Supplemental Information. Pacer Mediates the Function of Class III PI3K. and HOPS Complexes in Autophagosome. Maturation by Engaging Stx17
 Molecular Cell, Volume 65 Supplemental Information Pacer Mediates the Function of Class III PI3K and HOPS Complexes in Autophagosome Maturation by Engaging Stx17 Xiawei Cheng, Xiuling Ma, Xianming Ding,
Molecular Cell, Volume 65 Supplemental Information Pacer Mediates the Function of Class III PI3K and HOPS Complexes in Autophagosome Maturation by Engaging Stx17 Xiawei Cheng, Xiuling Ma, Xianming Ding,
Histone H3 Methylation Antibody Panel Pack II - Repression Genes Base Catalog # C10003
 Histone H3 Methylation Antibody Panel Pack II - Repression Genes Base Catalog # PACK CONTENTS Component Size Shipping Temperature Upon Receipt Checklist 3R2DA Histone H3R2 Dimethyl Asymmetric (H3R2me2a)
Histone H3 Methylation Antibody Panel Pack II - Repression Genes Base Catalog # PACK CONTENTS Component Size Shipping Temperature Upon Receipt Checklist 3R2DA Histone H3R2 Dimethyl Asymmetric (H3R2me2a)
H3K36me3 polyclonal antibody
 H3K36me3 polyclonal antibody Cat. No. C15410192 Type: Polyclonal ChIP-grade/ChIP-seq grade Source: Rabbit Lot #: A1845P Size: 50 µg/32 µl Concentration: 1.6 μg/μl Specificity: Human, mouse, Arabidopsis,
H3K36me3 polyclonal antibody Cat. No. C15410192 Type: Polyclonal ChIP-grade/ChIP-seq grade Source: Rabbit Lot #: A1845P Size: 50 µg/32 µl Concentration: 1.6 μg/μl Specificity: Human, mouse, Arabidopsis,
OmicsLink shrna Clones guaranteed knockdown even in difficult-to-transfect cells
 OmicsLink shrna Clones guaranteed knockdown even in difficult-to-transfect cells OmicsLink shrna clone collections consist of lentiviral, and other mammalian expression vector based small hairpin RNA (shrna)
OmicsLink shrna Clones guaranteed knockdown even in difficult-to-transfect cells OmicsLink shrna clone collections consist of lentiviral, and other mammalian expression vector based small hairpin RNA (shrna)
Molecular Cell Biology - Problem Drill 11: Recombinant DNA
 Molecular Cell Biology - Problem Drill 11: Recombinant DNA Question No. 1 of 10 1. Which of the following statements about the sources of DNA used for molecular cloning is correct? Question #1 (A) cdna
Molecular Cell Biology - Problem Drill 11: Recombinant DNA Question No. 1 of 10 1. Which of the following statements about the sources of DNA used for molecular cloning is correct? Question #1 (A) cdna
Supplementary
 Supplementary information Supplementary Material and Methods Plasmid construction The transposable element vectors for inducible expression of RFP-FUS wt and EGFP-FUS R521C and EGFP-FUS P525L were derived
Supplementary information Supplementary Material and Methods Plasmid construction The transposable element vectors for inducible expression of RFP-FUS wt and EGFP-FUS R521C and EGFP-FUS P525L were derived
Replication Factor C Recruits DNA Polymerase to Sites of Nucleotide Excision Repair but Is Not Required for PCNA Recruitment
 MOLECULAR AND CELLULAR BIOLOGY, Oct. 2010, p. 4828 4839 Vol. 30, No. 20 0270-7306/10/$12.00 doi:10.1128/mcb.00285-10 Copyright 2010, American Society for Microbiology. All Rights Reserved. Replication
MOLECULAR AND CELLULAR BIOLOGY, Oct. 2010, p. 4828 4839 Vol. 30, No. 20 0270-7306/10/$12.00 doi:10.1128/mcb.00285-10 Copyright 2010, American Society for Microbiology. All Rights Reserved. Replication
Confocal immunofluorescence microscopy
 Confocal immunofluorescence microscopy HL-6 and cells were cultured and cytospun onto glass slides. The cells were double immunofluorescence stained for Mt NPM1 and fibrillarin (nucleolar marker). Briefly,
Confocal immunofluorescence microscopy HL-6 and cells were cultured and cytospun onto glass slides. The cells were double immunofluorescence stained for Mt NPM1 and fibrillarin (nucleolar marker). Briefly,
Giardia RNAi Paper Discussion. Supplemental - VSG clonality. Variant-specific surface protein VSP9B10
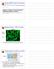 Giardia RNAi Paper Discussion Supplemental - VSG clonality VSP9B10 Green - VSP9B10 antibody Blue - DAPI Demonstration that cell line expresses a single VSP Variant-specific surface protein Outside Inside
Giardia RNAi Paper Discussion Supplemental - VSG clonality VSP9B10 Green - VSP9B10 antibody Blue - DAPI Demonstration that cell line expresses a single VSP Variant-specific surface protein Outside Inside
A RRM1 H2AX DAPI. RRM1 H2AX DAPI Merge. Cont. sirna RRM1
 A H2AX DAPI H2AX DAPI Merge Cont sirna Figure S1: Accumulation of RRM1 at DNA damage sites (A) HeLa cells were subjected to in situ detergent extraction without IR irradiation, and immunostained with the
A H2AX DAPI H2AX DAPI Merge Cont sirna Figure S1: Accumulation of RRM1 at DNA damage sites (A) HeLa cells were subjected to in situ detergent extraction without IR irradiation, and immunostained with the
Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question in Section B and ONE question from Section C.
 UNIVERSITY OF EAST ANGLIA School of Biological Sciences Main Series UG Examination 2013-2014 MOLECULAR BIOLOGY BIO-2B02 Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question
UNIVERSITY OF EAST ANGLIA School of Biological Sciences Main Series UG Examination 2013-2014 MOLECULAR BIOLOGY BIO-2B02 Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling
 Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary Figure 1
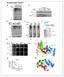 Supplementary Figure 1 A basic residue of BARD1 promotes Ub-transfer from BRCA1-E2~Ub. A. Scan of external facing BARD1 residues 91-99 for impact on Ub chain formation catalysed by the BRCA1- BARD1 ligase.
Supplementary Figure 1 A basic residue of BARD1 promotes Ub-transfer from BRCA1-E2~Ub. A. Scan of external facing BARD1 residues 91-99 for impact on Ub chain formation catalysed by the BRCA1- BARD1 ligase.
Tissue Acetyl-Histone H4 ChIP Kit
 Tissue Acetyl-Histone H4 ChIP Kit Catalog Number KA0672 48 assays Version: 03 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Background... 3 Principle
Tissue Acetyl-Histone H4 ChIP Kit Catalog Number KA0672 48 assays Version: 03 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Background... 3 Principle
Roche Molecular Biochemicals Technical Note No. LC 10/2000
 Roche Molecular Biochemicals Technical Note No. LC 10/2000 LightCycler Overview of LightCycler Quantification Methods 1. General Introduction Introduction Content Definitions This Technical Note will introduce
Roche Molecular Biochemicals Technical Note No. LC 10/2000 LightCycler Overview of LightCycler Quantification Methods 1. General Introduction Introduction Content Definitions This Technical Note will introduce
Antibodies against PCNA were previously described [1]. To deplete PCNA from Xenopus egg
![Antibodies against PCNA were previously described [1]. To deplete PCNA from Xenopus egg Antibodies against PCNA were previously described [1]. To deplete PCNA from Xenopus egg](/thumbs/84/90687567.jpg) Supplementary information Supplementary methods PCNA antibody and immunodepletion Antibodies against PCNA were previously described [1]. To deplete PCNA from Xenopus egg extracts, one volume of protein
Supplementary information Supplementary methods PCNA antibody and immunodepletion Antibodies against PCNA were previously described [1]. To deplete PCNA from Xenopus egg extracts, one volume of protein
Supplemental Information: Phosphorylation of CLIP-170 by Both Plk1 and CK2 Is Involved in the Timely Formation of Kinetochore-microtubule Attachments
 Supplemental Information: Phosphorylation of CLIP-170 by Both Plk1 and CK2 Is Involved in the Timely Formation of Kinetochore-microtubule Attachments Hongchang Li, X. Shawn Liu, Xiaoming Yang, Yingmin
Supplemental Information: Phosphorylation of CLIP-170 by Both Plk1 and CK2 Is Involved in the Timely Formation of Kinetochore-microtubule Attachments Hongchang Li, X. Shawn Liu, Xiaoming Yang, Yingmin
CONTENTS. LIST OF FIGURES... i. LIST OF TABLES... iii. SUMMARY OF THE WORK DONE... vi
 CONTENTS SECTION ~ LIST OF FIGURES... i LIST OF TABLES... iii LIST OF ABBREVIATIONS USED... ~... iv SUMMARY OF THE WORK DONE... vi 1. INTROD UCTION... :.................................... 1 1.1. AN OVERVIEW
CONTENTS SECTION ~ LIST OF FIGURES... i LIST OF TABLES... iii LIST OF ABBREVIATIONS USED... ~... iv SUMMARY OF THE WORK DONE... vi 1. INTROD UCTION... :.................................... 1 1.1. AN OVERVIEW
Mechanisms of Transcription. School of Life Science Shandong University
 Mechanisms of Transcription School of Life Science Shandong University Ch 12: Mechanisms of Transcription 1. RNA polymerase and the transcription cycle 2. The transcription cycle in bacteria 3. Transcription
Mechanisms of Transcription School of Life Science Shandong University Ch 12: Mechanisms of Transcription 1. RNA polymerase and the transcription cycle 2. The transcription cycle in bacteria 3. Transcription
Supplementary Figure 1 Phosphorylated tau accumulates in Nrf2 (-/-) mice. Hippocampal tissues obtained from Nrf2 (-/-) (10 months old, 4 male; 2
 Supplementary Figure 1 Phosphorylated tau accumulates in Nrf2 (-/-) mice. Hippocampal tissues obtained from Nrf2 (-/-) (10 months old, 4 male; 2 female) or wild-type (5 months old, 1 male; 11 months old,
Supplementary Figure 1 Phosphorylated tau accumulates in Nrf2 (-/-) mice. Hippocampal tissues obtained from Nrf2 (-/-) (10 months old, 4 male; 2 female) or wild-type (5 months old, 1 male; 11 months old,
CD93 and dystroglycan cooperation in human endothelial cell adhesion and migration
 /, Supplementary Advance Publications Materials 2016 CD93 and dystroglycan cooperation in human endothelial cell adhesion and migration Supplementary Materials Supplementary Figure S1: In ECs CD93 silencing
/, Supplementary Advance Publications Materials 2016 CD93 and dystroglycan cooperation in human endothelial cell adhesion and migration Supplementary Materials Supplementary Figure S1: In ECs CD93 silencing
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rainero et al., http://www.jcb.org/cgi/content/full/jcb.201109112/dc1 Figure S1. The expression of DGK- is reduced upon transfection
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rainero et al., http://www.jcb.org/cgi/content/full/jcb.201109112/dc1 Figure S1. The expression of DGK- is reduced upon transfection
Poly(ADP-ribose) polymerase 1 escorts XPC to UVinduced DNA lesions during nucleotide excision repair
 Poly(ADP-ribose) polymerase 1 escorts XPC to UVinduced DNA lesions during nucleotide excision repair Mihaela Robu a,1, Rashmi G. Shah a,1, Nupur K. Purohit a, Pengbo Zhou b, Hanspeter Naegeli c, and Girish
Poly(ADP-ribose) polymerase 1 escorts XPC to UVinduced DNA lesions during nucleotide excision repair Mihaela Robu a,1, Rashmi G. Shah a,1, Nupur K. Purohit a, Pengbo Zhou b, Hanspeter Naegeli c, and Girish
λ N -GFP: an RNA reporter system for live-cell imaging
 λ N -GFP: an RNA reporter system for live-cell imaging Nathalie Daigle & Jan Ellenberg Supplementary Figures and Text: Supplementary Figure 1 localization in the cytoplasm. 4 λ N22-3 megfp-m9 serves as
λ N -GFP: an RNA reporter system for live-cell imaging Nathalie Daigle & Jan Ellenberg Supplementary Figures and Text: Supplementary Figure 1 localization in the cytoplasm. 4 λ N22-3 megfp-m9 serves as
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Supplementary figures Supplementary Figure 1: Suv39h1, but not Suv39h2, promotes HP1α sumoylation in vivo. In vivo HP1α sumoylation assay. Top: experimental scheme. Middle: we
SUPPLEMENTARY INFORMATION Supplementary figures Supplementary Figure 1: Suv39h1, but not Suv39h2, promotes HP1α sumoylation in vivo. In vivo HP1α sumoylation assay. Top: experimental scheme. Middle: we
Impact of Nutraceuticals on TERT gene encoded protein
 Impact of Nutraceuticals on TERT gene encoded protein Xu Liu Department of Biological Sciences Fordham University, Bronx, New York, 10458 Abstract Telomerase is a Ribonucleo-protein polymerase that plays
Impact of Nutraceuticals on TERT gene encoded protein Xu Liu Department of Biological Sciences Fordham University, Bronx, New York, 10458 Abstract Telomerase is a Ribonucleo-protein polymerase that plays
Supplementary Figure S1 Purification of deubiquitinases HEK293 cells were transfected with the indicated DUB-expressing plasmids.
 Supplementary Figure S1 Purification of deubiquitinases HEK293 cells were transfected with the indicated DUB-expressing plasmids. The cells were harvested 72 h after transfection. FLAG-tagged deubiquitinases
Supplementary Figure S1 Purification of deubiquitinases HEK293 cells were transfected with the indicated DUB-expressing plasmids. The cells were harvested 72 h after transfection. FLAG-tagged deubiquitinases
Figure S1, related to Figure 1. Characterization of biosensor behavior in vivo.
 Developmental Cell, Volume 23 Supplemental Information Separase Biosensor Reveals that Cohesin Cleavage Timing Depends on Phosphatase PP2A Cdc55 Regulation Gilad Yaakov, Kurt Thorn, and David O. Morgan
Developmental Cell, Volume 23 Supplemental Information Separase Biosensor Reveals that Cohesin Cleavage Timing Depends on Phosphatase PP2A Cdc55 Regulation Gilad Yaakov, Kurt Thorn, and David O. Morgan
Multiple choice questions (numbers in brackets indicate the number of correct answers)
 1 Multiple choice questions (numbers in brackets indicate the number of correct answers) February 1, 2013 1. Ribose is found in Nucleic acids Proteins Lipids RNA DNA (2) 2. Most RNA in cells is transfer
1 Multiple choice questions (numbers in brackets indicate the number of correct answers) February 1, 2013 1. Ribose is found in Nucleic acids Proteins Lipids RNA DNA (2) 2. Most RNA in cells is transfer
Histone H3 Methylation Antibody Panel Pack III - Active Genes Base Catalog # C Component Size Shipping Temperature
 Histone H3 Methylation Antibody Panel Pack III - Active Genes Base Catalog # PACK CONTENTS Component Size Shipping Temperature Upon Receipt Checklist 3R17A Histone H3R17 Dimethyl Asymmetric (H3R17me2a)
Histone H3 Methylation Antibody Panel Pack III - Active Genes Base Catalog # PACK CONTENTS Component Size Shipping Temperature Upon Receipt Checklist 3R17A Histone H3R17 Dimethyl Asymmetric (H3R17me2a)
Supplemental Materials and Methods. Yeast strains. Strain genotypes are listed in Table S1. To generate MATainc::LEU2,
 Supplemental Materials and Methods Yeast strains. Strain genotypes are listed in Table S1. To generate MATainc::LEU2, the LEU2 marker was inserted by PCR-mediated recombination (BRACHMANN et al. 1998)
Supplemental Materials and Methods Yeast strains. Strain genotypes are listed in Table S1. To generate MATainc::LEU2, the LEU2 marker was inserted by PCR-mediated recombination (BRACHMANN et al. 1998)
Technical tips Session 5
 Technical tips Session 5 Chromatine Immunoprecipitation (ChIP): This is a powerful in vivo method to quantitate interaction of proteins associated with specific regions of the genome. It involves the immunoprecipitation
Technical tips Session 5 Chromatine Immunoprecipitation (ChIP): This is a powerful in vivo method to quantitate interaction of proteins associated with specific regions of the genome. It involves the immunoprecipitation
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table.
 Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Plant Molecular and Cellular Biology Lecture 9: Nuclear Genome Organization: Chromosome Structure, Chromatin, DNA Packaging, Mitosis Gary Peter
 Plant Molecular and Cellular Biology Lecture 9: Nuclear Genome Organization: Chromosome Structure, Chromatin, DNA Packaging, Mitosis Gary Peter 9/16/2008 1 Learning Objectives 1. List and explain how DNA
Plant Molecular and Cellular Biology Lecture 9: Nuclear Genome Organization: Chromosome Structure, Chromatin, DNA Packaging, Mitosis Gary Peter 9/16/2008 1 Learning Objectives 1. List and explain how DNA
supplementary information
 DOI: 10.1038/ncb1864 Figure S1 Apak specifically inhibits p53 transcriptional activity. Transcription activity of p53 was measured in U2OS (p53 wild-type) and H1299 (p53 deficient) cells which were transfected
DOI: 10.1038/ncb1864 Figure S1 Apak specifically inhibits p53 transcriptional activity. Transcription activity of p53 was measured in U2OS (p53 wild-type) and H1299 (p53 deficient) cells which were transfected
Lecture 22 Eukaryotic Genes and Genomes III
 Lecture 22 Eukaryotic Genes and Genomes III In the last three lectures we have thought a lot about analyzing a regulatory system in S. cerevisiae, namely Gal regulation that involved a hand full of genes.
Lecture 22 Eukaryotic Genes and Genomes III In the last three lectures we have thought a lot about analyzing a regulatory system in S. cerevisiae, namely Gal regulation that involved a hand full of genes.
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Han et al., http://www.jcb.org/cgi/content/full/jcb.201311007/dc1 Figure S1. SIVA1 interacts with PCNA. (A) HEK293T cells were transiently
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Han et al., http://www.jcb.org/cgi/content/full/jcb.201311007/dc1 Figure S1. SIVA1 interacts with PCNA. (A) HEK293T cells were transiently
SUPPLEMENTAL MATERIALS. Chromatin immunoprecipitation assays. Single-cell suspensions of pituitary
 SUPPLEMENTAL MATERIALS METHODS Chromatin immunoprecipitation assays. Single-cell suspensions of pituitary and liver cells were prepared from the indicated transgenic mice, as described (Ho et al, 2002).
SUPPLEMENTAL MATERIALS METHODS Chromatin immunoprecipitation assays. Single-cell suspensions of pituitary and liver cells were prepared from the indicated transgenic mice, as described (Ho et al, 2002).
Supplementary Information
 Journal : Nature Biotechnology Supplementary Information Targeted genome engineering in human cells with RNA-guided endonucleases Seung Woo Cho, Sojung Kim, Jong Min Kim, and Jin-Soo Kim* National Creative
Journal : Nature Biotechnology Supplementary Information Targeted genome engineering in human cells with RNA-guided endonucleases Seung Woo Cho, Sojung Kim, Jong Min Kim, and Jin-Soo Kim* National Creative
Cytotoxicity of Botulinum Neurotoxins Reveals a Direct Role of
 Supplementary Information Cytotoxicity of Botulinum Neurotoxins Reveals a Direct Role of Syntaxin 1 and SNAP-25 in Neuron Survival Lisheng Peng, Huisheng Liu, Hongyu Ruan, William H. Tepp, William H. Stoothoff,
Supplementary Information Cytotoxicity of Botulinum Neurotoxins Reveals a Direct Role of Syntaxin 1 and SNAP-25 in Neuron Survival Lisheng Peng, Huisheng Liu, Hongyu Ruan, William H. Tepp, William H. Stoothoff,
Supplementary Figure 1. Nature Structural & Molecular Biology: doi: /nsmb.3494
 Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Technical tips Session 4
 Technical tips Session 4 Biotinylation assay: Streptavidin is a small bacterial protein that binds with high affinity to the vitamin biotin. This streptavidin-biotin combination can be used to link molecules
Technical tips Session 4 Biotinylation assay: Streptavidin is a small bacterial protein that binds with high affinity to the vitamin biotin. This streptavidin-biotin combination can be used to link molecules
SUPPLEMENTARY INFORMATION
 Local auxin metabolism regulates environment-induced hypocotyl elongation Zuyu Zheng 1,2, Yongxia Guo 3, Ondřej Novák 4,5, William Chen 2, Karin Ljung 4, Joseph P. Noel 1,3, *, and Joanne Chory 1,2, *
Local auxin metabolism regulates environment-induced hypocotyl elongation Zuyu Zheng 1,2, Yongxia Guo 3, Ondřej Novák 4,5, William Chen 2, Karin Ljung 4, Joseph P. Noel 1,3, *, and Joanne Chory 1,2, *
SUPPLEMENTARY INFORMATION
 DOI: 1.138/ncb37 a Supplementary Figure 1 Dapi EU γh2ax Overlay b Dapi NBS1-GFP Firbillarin Overlay c Pre-laser 2 min 5 min 1 min 2 min 3 min 4 min 5 min 6 min 7 min d Dapi GFP-MRE11 γh2ax Overlay Supplementary
DOI: 1.138/ncb37 a Supplementary Figure 1 Dapi EU γh2ax Overlay b Dapi NBS1-GFP Firbillarin Overlay c Pre-laser 2 min 5 min 1 min 2 min 3 min 4 min 5 min 6 min 7 min d Dapi GFP-MRE11 γh2ax Overlay Supplementary
The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit
 Cell Reports, Volume 5 Supplemental Information The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit Andrey Poleshko, Katelyn M. Mansfield, Caroline
Cell Reports, Volume 5 Supplemental Information The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit Andrey Poleshko, Katelyn M. Mansfield, Caroline
Xeroderma Pigmentosum Group A Protein Loads as a Separate Factor onto DNA Lesions
 MOLECULAR AND CELLULAR BIOLOGY, Aug. 2003, p. 5755 5767 Vol. 23, No. 16 0270-7306/03/$08.00 0 DOI: 10.1128/MCB.23.16.5755 5767.2003 Copyright 2003, American Society for Microbiology. All Rights Reserved.
MOLECULAR AND CELLULAR BIOLOGY, Aug. 2003, p. 5755 5767 Vol. 23, No. 16 0270-7306/03/$08.00 0 DOI: 10.1128/MCB.23.16.5755 5767.2003 Copyright 2003, American Society for Microbiology. All Rights Reserved.
Chapter 20 Recombinant DNA Technology. Copyright 2009 Pearson Education, Inc.
 Chapter 20 Recombinant DNA Technology Copyright 2009 Pearson Education, Inc. 20.1 Recombinant DNA Technology Began with Two Key Tools: Restriction Enzymes and DNA Cloning Vectors Recombinant DNA refers
Chapter 20 Recombinant DNA Technology Copyright 2009 Pearson Education, Inc. 20.1 Recombinant DNA Technology Began with Two Key Tools: Restriction Enzymes and DNA Cloning Vectors Recombinant DNA refers
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
