Editorial Note: The figure on page 18 (Figure R3A) is sourced from Integr Biol (Camb). 4(11): , copyright 2012
|
|
|
- Elmer Dickerson
- 6 years ago
- Views:
Transcription
1 Editorial Note: The figure on page 7 is reprinted by permission from Macmillan Publishers Ltd: Scientific Reports 3 Article Number 1449, copyright 2013 Editorial Note: The figure on page 18 (Figure R3A) is sourced from Integr Biol (Camb). 4(11): , copyright 2012
2 Reviewers' comments: Reviewer #1 (Remarks to the Author): In this manuscript, Takaki et al. investigate how the actin cytoskeleton controls nuclear morphology and how this impacts on genomic integrity. It is not well understood how mechanical forces of cytoskeleton govern the shape of cell nuclei, and how misregulation of cytoskeletal contractility leads to aberrant nuclear morphogenesis that are typical for cancer cells. Hence, this study will be very interesting for cell biologists as well as cancer researchers. The authors identified by RNAi screening for nuclear shape regulators two subunits of the myosin phosphatase complex and show that they are important to maintain a round nuclear morphology, a sealed nuclear envelope, and a stable genome. Through a series of elegant and very rigorously controlled loss-of-function and gain-of-function experiments, the authors demonstrate that excessive actomyosin contractility deforms nuclei and damages DNA, in vitro as well as in a mouse tumor model. They further show that nuclear envelope stiffness, provided by the lamina, protects nuclei against deformation by excessive contractility of the actomyosin cytoskeleton. These findings are exciting and very well supported by the data. I am convinced that this study will be highly relevant for a broad readership and thus strongly recommend publication in Nature Communications. Reviewer #2 (Remarks to the Author): This manuscript by Petronczki and colleagues describes the control of actomyosin contractility as a critical modulator of nuclear morphology and genome integrity in cancer cells. Using a phosphatase-focused RNAi screen, the authors uncovered myosin phosphatase (PPP1R12A/PPP1CB) as a key factor in maintaining nuclear shape. The authors elegantly dissect the molecular pathway involved in this process as well as the impact of PPP1R12A/PPP1CB loss on nuclear integrity. The myosin regulatory light chain MRLC is identified as the responsible myosin phosphatase target, and inhibition of upstream Rho-ROCK signaling, phospho-mutant MRLC or myosin ATPase inhibition are able to revert the nuclear rupture phenotype. Finally, the authors demonstrate a role for Rho-ROCK signaling in altering nuclear shape in tumors in vivo and show that this pathway, when perturbed, may contribute to genomic instability. Together, this is a compelling study that significantly advances our understanding of nuclear dysmorphia and its potential adverse consequences in cancer. The following issues should, however, be clarified: 1) The cause for the observed increase in genomic instability is speculative at this point. A more thorough investigation of the types of DNA lesions (i.e. double-strand and/or single-strand breaks) as well as possible cell cycle dependence would significantly strengthen this aspect of the manuscript. Are lesions a result of replication or do they occur throughout interphase? The authors propose F-actin contraction as a cause for nuclear rupture, implying the generation of ruptured chromosomes and, hence, DSBs, which can be tested (e.g. using alkaline versus neutral comet assays). 2) On a related note, the authors should place their work in the context of recent findings from the Foiani lab, demonstrating that mechanical stress is necessary to activate ATR during S phase and thereby facilitate DNA replication. 3) The bursts of mcherry export (Fig. 1d) are striking and suggest that nuclear rupture(s) also occur in bursts. The authors should at a minimum comment on this observation and discuss possible causes.
3 4) Only ~ ½ of the tested cell lines appear to respond to myosin phosphatase depletion (Fig. S11a). No effect is seen in two htert-immortalized, non-transformed cell lines. Why do the authors think that is? And what is the effect in primary cells? 5) The authors should include at least on additional marker for apoptosis. Reviewer #3 (Remarks to the Author): Recent work on nucleus structure has shown that the nuclear envelope (NE) is much more dynamic than previously realized. Rupture and repair of the NE has been shown to occur in laminopathy and cancer cells in culture and migrating cancer and immune cells. NE rupture is speculated to contribute to genome instability in cancer cells by causing DNA damage. One initiator of NE structure defects leading to NE rupture is increased tension on the membrane from either mechanical nucleus compression or assembly of actin bundles. Here Takaki et al. identify a mechanism that drives accumulation of contractile actin bundles resulting in increased NE structure defects and NE rupture and reinforces the idea that the actin cytoskeleton is a major determinant of nucleus morphology and stability. Specifically, they identify a phosphatase complex (PPP1R12A/PPP1CB) that is required to maintain nucleus circularity and integrity by antagonizing Rho-ROCK kinase activation of a myosin complex protein. They show that loss of the phosphatase causes an increase in actin bundles in the cell that correspond with 1) increased lobulation of the nucleus, 2) blebbing of chromatin through disruptions in the nuclear lamina, and 3) rupture of the NE causing loss of nucleus compartmentalization. In addition, this paper shows that increased actomyosin activity is correlated with increased DNA damage. The authors connect these in vitro findings to in vivo tumor conditions by demonstrating that xenograft nuclei become rounder when treated with actomyosin inhibitors. However they do not convincingly show that the cells have defects in NE stability or DNA damage accumulation prior to drug treatment and thus it is unclear what the significance of this finding is. Overall this paper does an excellent job characterizing a new mechanism that inhibits actin accumulation to prevent NE instability, but some additional control experiments are required to solidify these findings. Major Issues: 1. The phrases describing the nucleus phenotypes are used inconsistently throughout the paper and it is unclear what the authors think is really happening. For instance, in an early description they say they observe DNA fragments outside the nuclear lamina yet there is no experiment demonstrating that these DNA structures represent a broken piece of a chromosome that is unconnected to the rest of the genome. In fact at least one of their images (Fig. 2f) has a thinner DNA band connecting these extended regions to the rest of the nucleus. Thus it is unclear whether the phenotype is fragmentation of a single nucleus into multiple nuclei or a single nucleus with multiple lobes or blebs. From their images it looks like they are seeing the formation of multiple nucleus blebs, some at the sites of lamina holes, as previously described (e.g. Sullivan et al., JCB, 1999, de Noronha et al., Science, 2001, Shimi et al, Genes Dev, 2008). A more careful description of the nucleus morphology and single plane images of where the lobes connect to the main nucleus would help to clarify this issue as well as a discussion of how similar these structures are to those previously described. In addition, the authors use different three phrases - nucleus rupture, NE rupture, and lamina rupture to describe defects in lamina structure. This becomes confusing since it is not clear when they are talking about lamina defects versus rupture of the nuclear membrane. The authors need to define these terms and stick with a single phrase to describe each event. Finally, using the phrase lamina rupture implies that new holes are forming in the nuclear lamina. Although this is likely, to distinguish the formation of new holes from the expansion of existing holes the authors need to expand their description of the nuclear lamina in untreated
4 cells, ideally using super-resolution microscopy. 2. The authors use cell lines overexpressing Lap2B to conclude that nuclear envelope reassembly at mitosis is normal in phosphatase depleted cells, that actin accumulates at chromatin necks, and to observe the in vivo dynamics of MDA-MB-231 cells. However, Lap2B is a structural component of the nuclear lamina that interacts with the lamins and chromatin regulatory proteins and is required for proper nuclear envelope assembly. Thus, the authors need to show additional controls demonstrating that Lap2B overexpression does not have additional affects on nucleus and lamina morphology, and on the kinetics of NE assembly. 3. The authors need an additional control to show that phosphatase disruption does not cause loss of lamin proteins from the NE, which can cause similar shape changes and increased NE rupture. This needs to be examined in the presence and absence of actomyosin inhibitors as well since actin structure has been shown to regulate lamin levels and phosphorylation status. 4. One of the major observations in the authors in vivo xenograft experiments is that, in the presence of actin, they see un-laminated chromatin, indicative of larger NE structural defects and similar to what they see in culture. However the MDA-MB-231 cells in culture shows little evidence of lamina disruption (Fig. S11), and they show no evidence of lamina rupturing in the xenograft. In addition, it is not clear in the movie that the red spot interpreted as un-laminated chromatin is part of the same nucleus or same cell as the laminated chromatin. Thus, their evidence seems to show only that MDA-MB-231 nuclei are soft and responsive to weak actin forces in xenografts, rather than that actin is driving significant changes in NE structure that could cause NE rupture or DNA damage in MDA-MB-231 cells. 5. The discussion of the result that increased actin contractility causes genome instability due to increased DNA damage is a little confusing. The karyotype images were taken from cells where the DNA damage checkpoint was inhibited by caffeine. Thus these images do not necessarily represent the real amount of chromatin fragmentation that would occur during mitosis as a result of increased damage. This should be clarified in the text. Minor issues 1. The student s T-test is not sufficient to determine the significance for data with more than two samples Graphs like Fig. 1b should be analyzed using a one-way ANOVA to determine the significance for the entire family in conjunction with a multiple comparisons post-test. In graphs such as Fig. 2c and 4a, where the interaction of multiple conditions are being tested (sirna + drug) the authors need to use a 2-way ANOVA plus a multiple comparisons post-test and always include the data for the ctl sirna + drug condition (Fig. 4a). In addition, the authors should use a chi-square analysis to assess the significance of the nominal data presented in graphs such as Fig 1a. 2. The authors rule out the possibility that defects in NE assembly could contribute to altered NE structure in their experiments. However, what they show is that mitosis is not necessary for these alterations. To rule out that NE assembly is not contributing they would have to do additional analysis of the timing of NE assembly, the extent of chromosome decompaction during reassembly, and the timing of protein recruitment during NE assembly in cells depleted of the phosphatase complex.
5 Response to reviewers comments General We were very pleased at the overall positive nature of the comments and we are grateful for the constructive criticism and suggestions provided by the referees. In particular, that these findings are exciting and very well supported by the data (reviewer #1), the work is a compelling study that significantly advances our understanding of nuclear dysmorphia (reviewer #2), and that we do an excellent job characterizing a new mechanism that inhibits actin accumulation to prevent NE instability (reviewer #3). We also note that reviewers #2 and #3 make several pertinent suggestions to improve the manuscript. We have addressed the points raised by the reviewers by conducting a series of new experiments and analyses as well as by making changes to the text. We provide new evidence about the nature of DNA damage induced by actomyosin forces, we have performed analysis of primary patient cells both tumour and stroma, and have performed additional control experiments (impact of nuclear envelope markers on phenotype, lamin level analysis etc). In addition, we have expanded the discussion section and put our work in better context of the literature. Please see below for a point- by- point response to the reviewers comments. Response to Reviewer #1 In this manuscript, Takaki et al. investigate how the actin cytoskeleton controls nuclear morphology and how this impacts on genomic integrity. It is not well understood how mechanical forces of cytoskeleton govern the shape of cell nuclei, and how misregulation of cytoskeletal contractility leads to aberrant nuclear morphogenesis that are typical for cancer cells. Hence, this study will be very interesting for cell biologists as well as cancer researchers. The authors identified by RNAi screening for nuclear shape regulators two subunits of the myosin phosphatase complex and show that they are important to maintain a round nuclear morphology, a sealed nuclear envelope, and a stable genome. Through a series of elegant and very rigorously controlled loss- of- function and gain- of- function experiments, the authors demonstrate that excessive actomyosin contractility deforms nuclei and damages DNA, in vitro as well as in a mouse tumor model. They further show that nuclear envelope stiffness, provided by the lamina, protects nuclei against deformation by excessive contractility of the actomyosin cytoskeleton. These findings are exciting and very well supported by the data. I am convinced that this study will be highly relevant for a broad readership and thus strongly recommend publication in Nature Communications. Response: We are delighted that the reviewer expresses such strong support for the work and does not raise any issues that need addressing. Response to Reviewer #2
6 This manuscript by Petronczki and colleagues describes the control of actomyosin contractility as a critical modulator of nuclear morphology and genome integrity in cancer cells. Using a phosphatase- focused RNAi screen, the authors uncovered myosin phosphatase (PPP1R12A/PPP1CB) as a key factor in maintaining nuclear shape. The authors elegantly dissect the molecular pathway involved in this process as well as the impact of PPP1R12A/PPP1CB loss on nuclear integrity. The myosin regulatory light chain MRLC is identified as the responsible myosin phosphatase target, and inhibition of upstream Rho- ROCK signaling, phospho- mutant MRLC or myosin ATPase inhibition are able to revert the nuclear rupture phenotype. Finally, the authors demonstrate a role for Rho- ROCK signaling in altering nuclear shape in tumors in vivo and show that this pathway, when perturbed, may contribute to genomic instability. Together, this is a compelling study that significantly advances our understanding of nuclear dysmorphia and its potential adverse consequences in cancer. The following issues should, however, be clarified: Response: We are very pleased that the reviewer finds the work compelling and a significant advance. 1) The cause for the observed increase in genomic instability is speculative at this point. A more thorough investigation of the types of DNA lesions (i.e. double- strand and/or single- strand breaks) as well as possible cell cycle dependence would significantly strengthen this aspect of the manuscript. Are lesions a result of replication or do they occur throughout interphase? The authors propose F- actin contraction as a cause for nuclear rupture, implying the generation of ruptured chromosomes and, hence, DSBs, which can be tested (e.g. using alkaline versus neutral comet assays). Response: We agree that it is important to clarify the type of DNA damage in more detail. To this end, we have now performed staining for 53BP1, which is recruited to double strand breaks (new Figure 6b). This shows that PPP1R12A depletion leads to a clear increase in 53BP1 foci indicating double strand breaks. Comet assays were not possible because embedding in agar undermines the anchorage to a substrate required for the generation of large contractile actomyosin forces. Using nucleotide analog incorporation experiments we have addressed the relationship between DNA damage marker induction and DNA replication in PPP1R12A- depleted cells. Our findings suggest that DNA damage in PPP1R12A- depleted cells is not restricted to S phase but can occur throughout interphase (new Supplementary Figure 14). We also provide data indicating that DNA damage markers in PPP1R12A- depleted cells can be focused and concentrated in DNA segments that lack nuclear envelope markers (new Supplementary Figure 6c). This finding is discussed in the context of work from the Pellman lab describing genome instability in micronuclei. 2) On a related note, the authors should place their work in the context of recent findings from the Foiani lab, demonstrating that mechanical stress is necessary to activate ATR during S phase and thereby facilitate DNA replication.
7 Response: We thank the reviewer for this suggestion. We now refer to this study and put the implications of our work in the context of the findings of cited study in the discussion section of our manuscript (page 13). 3) The bursts of mcherry export (Fig. 1d) are striking and suggest that nuclear rupture(s) also occur in bursts. The authors should at a minimum comment on this observation and discuss possible causes. Response: In the revised version of the manuscript we discuss this phenomenon of nuclear ruptures and the breakdown of the nuclear compartment in more detail (see discussion section). Furthermore, we provide new data suggesting that the transient nature of these bursts could be explained by the association of extruded chromatin segments with SEC61- positive ER membranes (new Supplementary Figure 10b). 4) Only ~ ½ of the tested cell lines appear to respond to myosin phosphatase depletion (Fig. S11a). No effect is seen in two htert- immortalized, non- transformed cell lines. Why do the authors think that is? And what is the effect in primary cells? Response: We agree that this is an intriguing result. Following the reviewer s suggestion, we have now probed the effect of PPP1R12A depletion in primary patient cells. Specifically, we isolated both the transformed cancer cell compartment and non- transformed stromal fibroblasts from a squamous cell carcinoma patient. PPP1R12A depletion in the cancer cells (VSCC10) led to a clear increase in nuclear dysmorphia, whereas the effect in the stromal fibroblasts (VCAF10) was less pronounced (new Figure 4b). These new data exclude that the differences between cell lines are related to the primary vs established cell lines or the length of time in culture. We believe that the differences between cell lines relate to the intrinsic physical properties of the nucleus. Analysis by The Physical Sciences Oncology Centers Network ( Figure 2f(iii) shown below for the benefit of the reviewer) shows that the nuclei of transformed breast MDA- MB- 231 cells are much softer when deformed by >0.2μm than non- transformed MCF10A. This supports the emerging view that the nuclei of malignant cells are softer than that of normal cells. Consistent with this, we observe the greatest nuclear deformation in HeLa, MDA- MB- 231, A549, and VSCC10 (all cancer cells) and almost no PPP1R12A loss- mediated damage of nuclear morphology in non- transformed htert- RPE1 cell and primary BJ fibroblasts. This point is now included in the discussion section of our revised manuscript.
8 Figure reproduced from Scientific Reports showing the different Young s modulus of the nucleus of MDA- MB- 231 and MCF10A cells. Note that for deformation >0.1μm the MDAMB231 cell nucleus is softer. 5) The authors should include at least on additional marker for apoptosis. Response: We now show cleaved PARP as an apoptosis marker in addition to mitochondrial cytochrome c release (new Supplementary Figure 5b). This new dataset supports our conclusion that PPP1R12A depletion does not cause nuclear morphology changes through induction of apoptosis. Response to Reviewer #3 Recent work on nucleus structure has shown that the nuclear envelope (NE) is much more dynamic than previously realized. Rupture and repair of the NE has been shown to occur in laminopathy and cancer cells in culture and migrating cancer and immune cells. NE rupture is speculated to contribute to genome instability in cancer cells by causing DNA damage. One initiator of NE structure defects leading to NE rupture is increased tension on the membrane from either mechanical nucleus compression or assembly of actin bundles. Here Takaki et al. identify a mechanism that drives accumulation of contractile actin bundles resulting in increased NE structure defects and NE rupture and reinforces the idea that the actin cytoskeleton is a major determinant of nucleus morphology and stability. Specifically, they identify a phosphatase complex (PPP1R12A/PPP1CB) that is required to maintain nucleus circularity and integrity by antagonizing Rho- ROCK kinase activation of a myosin complex protein. They show that loss of the phosphatase causes an increase in actin bundles in the cell that correspond with 1) increased lobulation of the nucleus, 2) blebbing of chromatin through disruptions in the nuclear lamina, and 3) rupture of the NE causing loss of nucleus compartmentalization. In addition, this paper shows that increased actomyosin activity is correlated with increased DNA damage. The authors connect these in vitro findings to in vivo tumor conditions by demonstrating that xenograft nuclei become rounder when treated with actomyosin inhibitors. However they do not convincingly show that the cells have defects in NE stability or DNA damage accumulation prior to drug treatment and thus it is unclear what the significance of this finding is. Overall this paper does an excellent job characterizing a new mechanism that inhibits actin accumulation to prevent NE
9 instability, but some additional control experiments are required to solidify these findings. Response: We were pleased with the recognition that we have characterized a new mechanism that prevents NE instability. We are also grateful for the various technical comments that have prompted us to perform some further experimental controls and to clarify our language. 1. The phrases describing the nucleus phenotypes are used inconsistently throughout the paper and it is unclear what the authors think is really happening. For instance, in an early description they say they observe DNA fragments outside the nuclear lamina yet there is no experiment demonstrating that these DNA structures represent a broken piece of a chromosome that is unconnected to the rest of the genome. In fact at least one of their images (Fig. 2f) has a thinner DNA band connecting these extended regions to the rest of the nucleus. Thus it is unclear whether the phenotype is fragmentation of a single nucleus into multiple nuclei or a single nucleus with multiple lobes or blebs. From their images it looks like they are seeing the formation of multiple nucleus blebs, some at the sites of lamina holes, as previously described (e.g. Sullivan et al., JCB, 1999, de Noronha et al., Science, 2001, Shimi et al, Genes Dev, 2008). A more careful description of the nucleus morphology and single plane images of where the lobes connect to the main nucleus would help to clarify this issue as well as a discussion of how similar these structures are to those previously described. Response: The reviewer is correct that there is some ambiguity about whether DNA observed outside the nuclear envelope remains connected to the main body of the nucleus. We have now performed more careful analysis of confocal z- sections to address this. Supplementary Figure 3b confirms the reviewer s observation that thin DNA connections to the nucleus remain. However, in 18% of cases there was no DNA strand connecting the fragment to the nucleus. We therefore propose that nuclear envelope rupture typically leads to the extrusion of DNA into the cytoplasm, but that the extruded segments normally remain linked to the bulk of the nucleus. Only in more extreme circumstances, possibly when the forces involved are particularly high, does the extruded DNA become truly fragmented or is an entire chromosome extruded. We have adapted the text accordingly and have related the phenotype observed in myosin phosphatase depleted cells to previously described nuclear envelope aberrations in the suggested references. In addition, the authors use different three phrases - nucleus rupture, NE rupture, and lamina rupture to describe defects in lamina structure. This becomes confusing since it is not clear when they are talking about lamina defects versus rupture of the nuclear membrane. The authors need to define these terms and stick with a single phrase to describe each event. Response: We thank the reviewer for requesting that we standardize our terminology to improve the readability of the manuscript. Our data suggest persistent, frequent and widespread rupture of the nuclear lamina and transient breaches of the nuclear membrane. We now use the term NE rupture to cover both ongoing phenomena as this does not imply
10 particular defects in the lamina, nor does it imply complete destruction of the nucleus. Finally, using the phrase lamina rupture implies that new holes are forming in the nuclear lamina. Although this is likely, to distinguish the formation of new holes from the expansion of existing holes the authors need to expand their description of the nuclear lamina in untreated cells, ideally using super- resolution microscopy. Response: We agree with the reviewer regarding the possible implications of the term lamina rupture. As mentioned above, we now use the more neutral term of nuclear envelope rupture. It would be fascinating to observe the process with higher spatio- temporal resolution; however, to properly distinguish nuclear pores and lamina sub- structures a resolution in the tens of nm is required (a nuclear pore is ~120nm across) and this is only achievable by PALM/STORM type of super- resolution microscopy methods, but we know from previous studies that organelles within highly contractile cells are moving at speeds ~0.1μm/s (analysis conducted for Tozluoglu et al Nature Cell Biology 2013, although it was not included in the final manuscript). Unfortunately, PALM/STORM cannot operate at the necessary frame rates for such fast motion. Therefore, while dynamic super- resolution imaging of nuclear envelope sub- structures would be very interesting, it is not possible with current technology. 2. The authors use cell lines overexpressing Lap2B to conclude that nuclear envelope reassembly at mitosis is normal in phosphatase depleted cells, that actin accumulates at chromatin necks, and to observe the in vivo dynamics of MDA- MB- 231 cells. However, Lap2B is a structural component of the nuclear lamina that interacts with the lamins and chromatin regulatory proteins and is required for proper nuclear envelope assembly. Thus, the authors need to show additional controls demonstrating that Lap2B overexpression does not have additional affects on nucleus and lamina morphology, and on the kinetics of NE assembly. Response: We agree that this is a key control. We now show in new Supplementary Figure 4a&b that expression of the live cell markers AcGFP- LAP2ß and H2B- mcherry does neither change the baseline circularity of cells, nor does it alter the penetrance of nuclear morphology aberrations caused by depletion of PPP1R12A. This finding is in line with our observation that control cells stably expressing AcGFP- LAP2ß and H2B- mcherry do not show bursts of nuclear protein leakage (no exchange event recorded in 83 cells over 4 h; Supplementary Movie 8). Also we would like highlight the fact that most of our fixed cell analyses of nuclear envelope markers was performed in cells that do not overexpress nuclear envelope marker proteins (e.g. Fig. 1a and Supplementary Figure 3a,b). 3. The authors need an additional control to show that phosphatase disruption does not cause loss of lamin proteins from the NE, which can cause similar shape changes and increased NE rupture. This needs to be examined in the presence and absence of actomyosin inhibitors as well since actin structure has been shown to regulate lamin levels and phosphorylation status.
11 Response: We now show in new Supplementary Figure 4c & d that depletion of PPP1R12A does not have a major impact on the levels of lamin A or lamin B as analysed by both western blot and immuno- fluorescence microscopy. 4. One of the major observations in the authors in vivo xenograft experiments is that, in the presence of actin, they see un- laminated chromatin, indicative of larger NE structural defects and similar to what they see in culture. However the MDA- MB- 231 cells in culture shows little evidence of lamina disruption (Fig. S11), and they show no evidence of lamina rupturing in the xenograft. In addition, it is not clear in the movie that the red spot interpreted as un- laminated chromatin is part of the same nucleus or same cell as the laminated chromatin. Thus, their evidence seems to show only that MDA- MB- 231 nuclei are soft and responsive to weak actin forces in xenografts, rather than that actin is driving significant changes in NE structure that could cause NE rupture or DNA damage in MDA- MB- 231 cells. Response: We now provide the individual confocal sections to unambiguously show that the un- laminated H2B signal is associated with the cell, and not an out of plain artefact (new Supplementary Figure 13b). 5. The discussion of the result that increased actin contractility causes genome instability due to increased DNA damage is a little confusing. The karyotype images were taken from cells where the DNA damage checkpoint was inhibited by caffeine. Thus these images do not necessarily represent the real amount of chromatin fragmentation that would occur during mitosis as a result of increased damage. This should be clarified in the text. Response: We thank the reviewer for pointing this out and now clarify the use of caffeine in the text (page 11). Caffeine was used to allow cells with damaged chromosomes to enter to mitosis. We propose that PPP1R12A suppresses actomyosin- driven chromatin fragmentation in interphase and the purpose of the experiment is to reveal the aberrations in chromosome structure accumulated prior to mitosis. Minor issues 1. The student s T- test is not sufficient to determine the significance for data with more than two samples Graphs like Fig. 1b should be analyzed using a one- way ANOVA to determine the significance for the entire family in conjunction with a multiple comparisons post- test. In graphs such as Fig. 2c and 4a, where the interaction of multiple conditions are being tested (sirna + drug) the authors need to use a 2- way ANOVA plus a multiple comparisons post- test and always include the data for the ctl sirna + drug condition (Fig. 4a). In addition, the authors should use a chi- square analysis to assess the significance of the nominal data presented in graphs such as Fig 1a. Response: We thank the reviewer for pointing out more appropriate statistical tests and have now performed ANOVA analysis in the Figures 1b, 2c, and 4a in line with his/her suggestions.
12 2. The authors rule out the possibility that defects in NE assembly could contribute to altered NE structure in their experiments. However, what they show is that mitosis is not necessary for these alterations. To rule out that NE assembly is not contributing they would have to do additional analysis of the timing of NE assembly, the extent of chromosome decompaction during reassembly, and the timing of protein recruitment during NE assembly in cells depleted of the phosphatase complex. Response: We have added new data (new Supplementary Figure 6c) in which we measure the length of time for nuclear envelope reformation following mitosis. This analysis shows that there is no change in nuclear envelope reformation during mitotic exit in PPP1R12A depleted cells.
13 Reviewers' comments: Reviewer #2 (Remarks to the Author): The authors have satisfactorily addressed my concerns. Reviewer #3 (Remarks to the Author): In the revised manuscript the authors have done an excellent job of addressing my concerns. I do have a comment on one of the new sections, the discussion of supplemental figure 10b, which shows that nucleus fragments associate with Sec61B-containing membranes. The discussion is a little confusing - the authors don't make clear that Sec61B labels the nuclear membranes as well as the ER, and thus all the fragments that have mcherry-nls should have a rim of Sec61B. There were also two papers (Denais et al and Raab et al, Science 2016) that described a role for the ESCRTIII complex in membrane resealing after nuclear envelope rupture that should be mentioned in this section. Reviewer #4 (Remarks to the Author): 1. This paper primarily identifies PPP1R12A and PPP1CB as 'proteins safeguarding nuclear integrity'. However, this discovery does not appear to be novel, as stated in the text: "Previous screens have also identified PPP1R12A and PPP1CB as factors controlling nuclear morphology (references 10, 11). That the lamina confers stiffness on the nucleus toward cytoplasmic forces is also well known. 2. The screen for nuclear shape was based on visual inspection, which makes it difficult to predict if others can reproduce this work. This is also a problem in other experiments (not the screen alone). 3. Circularity is quantified in later assays as a measure of irregularity. But circularity is low for elongated nuclei which are 'smooth' where no irregularities are present in the nuclear shape. Circularity is not necessarily a measure of irregularity, which makes it difficult to interpret the data and any differences. 4. Data in Figure 3B from different cancer cell lines show that nuclear circularity is higher in these cell lines when myosin is inhibited with blebbistatin. However,PPP1R12A and PPP1CB levels in these cell lines appear comparable (at least from a visual inspection of Fig. S12), therefore it is not clear how this data should be interpreted in the light of the mechanism that the paper proposes. 5. Relevant to point 4, treatment of cells with inhibitors, especially inhibitors like blebbistatin, change cell shape which can change nuclear shape, for example by rounding cells up, or causing a fragmented cytoplasm. It is not clear that the observed effects are due to actomyosin activity or indirect effects on the cell shape itself, or some other mechanism. For example, another possibility is that upon inhibition of myosin, cells with irregular nuclei detach preferentially, enriching the population in normal looking nuclei. 6. Figure 2f is very difficult to interpret in terms of F-actin filaments causing nuclear deformations given the low resolution. It is claimed that Figure 2e shows how actomyosin fibers 'bisect the nucleus and cause nuclear fragments'. But in Figure 2e, at least a few examples can be seen where actin fibers are not bisecting or touching the nucleus. Are all deformations of the nucleus due to impinging actomyosin fibers? The paper does not put forth a convincing mechanism for how the nucleus is fragmenting upon knockdown of PPP1R12A and PPP1CB.
14 7. The invivo measurements are unconvincing. Nuclear abnormalities are not clearly visible in the picture provided (Figure 3c). It is not clear that the decrease in variance of the measured parameters necessarily indicates a smoother nucleus upon treatment with Y Could the results in Figure 4 be interpreted in another way? Chromosomal spreads are done by colliding cells with a solid surface which forces the chromosomes apart. Knockdown of phosphatases makes the cytoplasm stiffer which might cause breaks in chromosomes. 9.A direct comparison of actomyosin forces generated by the different cancer cell lines, along with a comparison of mechanical properties of the nucleus would be required along with evidence that myosin inhibition dynamically causes the highly abnormal nucleus to relax back to its 'smooth' shape.
15 Response to Reviewers comments We would like to thank Reviewers #2 & #3 for their further comments, and to thank you Reviewer #4 for the criticism and comments on our work. We were very gratified that reviewer #2 stated that we had satisfactorily addressed [my] concerns, and that reviewer #3 felt that we have done an excellent job of addressing [my] concerns. We note that he/she had one remaining point around SEC61B localization requiring clarification. Unfortunately, the comments of reviewer #4 were not forwarded to us in October last year and we were only made aware of these comments recently, several days after the resubmission of our revised manuscript at the end of March. Nonetheless, we address numerous points raised by referee #4 by new data incorporated into the revised version, or by clarification of the text. We hope that our comments and the data below can clarify and address the points raised by referee #4 in this exceptional situation. This is in addition to the four months of new experimental work that has resolved the concerns of reviewers #2 & #3. Response to Reviewer #3 1. I do have a comment on one of the new sections, the discussion of supplemental figure 10b, which shows that nucleus fragments associate with Sec61B- containing membranes. The discussion is a little confusing - the authors don't make clear that Sec61B labels the nuclear membranes as well as the ER, and thus all the fragments that have mcherry- NLS should have a rim of Sec61B. There were also two papers (Denais et al and Raab et al, Science 2016) that described a role for the ESCRTIII complex in membrane resealing after nuclear envelope rupture that should be mentioned in this section. We thank the reviewer for raising this point. We have now modified the text in the discussion section to clarify that SEC61B- positive membranes can be derived from the rough ER or the nuclear membrane. Furthermore, we have cited the two papers demonstrating that ESCRT- III promotes the sealing of nuclear envelope rupture events upon nuclear confinement (page 14). Whether ESCRT- III is implicated in repairing actomyosin- inflicted nuclear rupture events is an interesting question for future research and now also mentioned in the discussion. Response to Reviewer #4 1. This paper primarily identifies PPP1R12A and PPP1CB as 'proteins safeguarding nuclear integrity'. However, this discovery does not appear to be novel, as stated in the text: "Previous screens have also identified PPP1R12A and PPP1CB as factors controlling nuclear morphology (references 10, 11). That the lamina confers stiffness on the nucleus toward cytoplasmic forces is also well known. The reviewer is correct that the finding that depletion of PPP1R12A and PPP1CB causes changes in nuclear morphology detected by DAPI staining was made previously. This fact is appropriately cited in our manuscript (middle of page 4). However, the type of nuclear alteration and the mechanism underlying it was unknown. Our work addresses these points. We for the first time describe the type of nuclear morphology and integrity defects occurring in myosin phosphatase- depleted ce lls an d, im portantly, we di scover th e mechanism underlying these nuclear aberrations. We show that loss of myosin phosphatase inflicts
16 nuclear damage, including nuclear envelope rupture, loss of the nuclear compartment, double- stranded DNA breaks, and genome instability, via unrestrained actomyosin activity. We further use intravital imaging to demonstrate nuclear rupture events in non- migrating cells in vivo, and provide evidence that ROCK- mediated actomyosin contractility regulates this process. 2. The screen for nuclear shape was based on visual inspection, which makes it difficult to predict if others can reproduce this work. This is also a problem in other experiments (not the screen alone). The original sirna screen was performed using visual inspection. The full data set derived from the screen is provided in Supplementary Table 1. sirna data in general require downstream validation using sirna pool deconvolution and transgenic rescue experiments, as we have performed successfully for PPP1R12A and PPP1CB. Thus, the primary screen result list can be utilized by other researchers to shortlist additional phosphatase genes as candidates for factors controlling cell division or nuclear integrity. In addition to PPP1R12A and PPP1CB, the catalytic subunit isoforms of PPP2A that are known to be required to preserve sister chromatid cohesion until metaphase, are highly scoring hits in our screen supporting the ability of our approach to identify relevant genes. We would respectfully disagree with the comment regarding the quality of analyses presented in the paper. Data for many key findings in the paper are collected using quantitative image analysis algorithms and are supported by multiple experiments and approaches. The quality of our analyses was also commended by other referees. 3. Circularity is quantified in later assays as a measure of irregularity. But circularity is low for elongated nuclei which are 'smooth' where no irregularities are present in the nuclear shape. Circularity is not necessarily a measure of irregularity, which makes it difficult to interpret the data and any differences. This is correct and the reason why we use additional classifiers in many of the figures. The normal/deformed/ruptured categories distinguish a smooth long nucleus from a rounder, more irregular one. In our manuscript, the changes in nuclear shape between control cells, myosin phosphatase- depleted cells and depleted cells treated with contractility inhibitors are striking, highly significant using circularity analysis and discernable by looking at the images. This differences in the circularity metric are primarily a reflection of perimeter of the nucleus becoming more tortuous upon PPP1R12A depletion, not a change in elongation, which would be most accurately scored by an aspect ratio metric. The circularity analysis presented in our paper is in strong agreement with the other visual qualifiers (e.g. ruptured nuclear envelope). Below we now provide an additional Figure (Figure R1) showing that treatment with the ROCK inhibitor restores nuclear shape and integrity in a panel of cancer cell lines depleted of PPP1R12A. Therefore, we believe that the use of a combination of classifiers including circularity is a suitable and fit- for- purpose approach to analyze nuclear phenotypes in this study.
17 Figure R1: A panel of cell lines was transfected with the indicated sirna duplexes for 56 hrs and treated with 7.5 um Y for 24 hrs before fixation as indicated. Nuclear morphology was subsequently analyzed by Lamin B1 and DNA staining. (n > 200 cells each) 4. Data in Figure 3B from different cancer cell lines show that nuclear circularity is higher in these cell lines when myosin is inhibited with blebbistatin. However,PPP1R12A and PPP1CB levels in these cell lines appear comparable (at least from a visual inspection of Fig. S12), therefore it is not clear how this data should be interpreted in the light of the mechanism that the paper proposes. The experiment in the original Figure 3b demonstrates that actomyosin activity makes an important contribution to nuclear shape in a panel of cell lines even if not deregulated by depletion of myosin phosphatase. However, as the reviewer correctly points out the protein levels of myosin phosphatase subunits do not significantly differ between cell lines whose nuclei are more deformed by depletion of the phosphatase or more circularized by inhibition of contractility. The reason for this is that PPP1R12A is regulated by phosphorylation, and intersects with Rho- ROCK- MLC2 pathway activity at multiple levels (Kawano et al JCB 1999, Feng et al JBC 1999, Wilkinson et al NCB 2005). The difference between the cell lines is likely to relate to differences in activity that are related to post- translational modifications in PPP1R12A and additional differences, such as the expression of positive contractility regulators (such as RhoA, ROCK kinases, and the key RhoGEF Ect2) and of lamins. Below we provide an immunoblotting analysis of contractility regulators in a panel of cancer cell lines (Figure R2). The cell lines that show the most pronounced effect of PPP1R12A depletion have consistently high levels of Ect2, RhoA, ROCK1, and ROCK2. The lack of effect of PPP1R12A depletion in MCF- 7 and DLD- 1 cells is, quite possibly, due to the low levels of RhoA expression. This would be consistent with the data using C3 toxin to block RhoA in Figure 2b. Given the likely multifactorial nature of the differences in cellular responses in complex non- isogenic models, such as cancer cell lines, we decided not to speculate on this point in our manuscript. In isogenic systems we show the absence of lamin A sensitizes to the impact of actomyosin contractility (Figure 4a).
18 Figure R2: A panel of cell lines was transfected with the indicated sirna duplexes for 56 hrs and analyzed by immunofluorescence microscopy (n > 200 cells) (left panel). Protein extract of the indicated cell lines were analyzed by immunoblotting (right panel). 5. Relevant to point 4, treatment of cells with inhibitors, especially inhibitors like blebbistatin, change cell shape which can change nuclear shape, for example by rounding cells up, or causing a fragmented cytoplasm. It is not clear that the observed effects are due to actomyosin activity or indirect effects on the cell shape itself, or some other mechanism. For example, another possibility is that upon inhibition of myosin, cells with irregular nuclei detach preferentially, enriching the population in normal looking nuclei. We did not observe cell loss due to detachment or massive cell rounding following treatment with blebbistatin or Y A low concentration of blebbistatin (5 µm) that was insufficient to abrogate cytokinesis was able to restore nuclear architecture in PPP1R12A and PPP1CB- depleted cells. Cells treated with contractility inhibitors at the concentrations used in our work showed reduced actomyosin fibers but remained attached to the surface these new data are shown in Supplementary Figure 7b. Importantly, the Y washout experiment shown in Supplementary Figure 8 of the revised manuscript uses live cell imaging to show the alleviation of ROCK inhibition (by washout of a ROCK inhibitor) causes nuclear morphology changes and nuclear envelope rupture in PPP1R12A- depleted cells. A corollary of this result is the conclusion that the suppression of nuclear damage by contractility inhibitors cannot be explained by cell loss due to the detachment. 6. Figure 2f is very difficult to interpret in terms of F- actin filaments causing nuclear deformations given the low resolution. It is claimed that Figure 2e shows how actomyosin fibers 'bisect the nucleus and cause nuclear fragments'. But in Figure 2e, at least a few examples can be seen where actin fibers are not bisecting or touching the nucleus. Are all deformations of the nucleus due to impinging actomyosin fibers? The paper does not put forth a convincing mechanism for how the nucleus is fragmenting upon knockdown of PPP1R12A and PPP1CB.
19 We do not claim that all actomyosin filaments in PPP1R12A- depleted cells are engaged in nuclear compression and fragmentation. Our data demonstrate is that actomyosin activity is key to nuclear damage in myosin phosphatase- depleted cells. At nuclear compression sites proximal to nuclear envelope rupture sites, we do detect actomyosin filaments consistent with our hypothesis. These filaments are shown in Fig 3c and in the newly added Supplementary Fig. 10c of the revised manuscript. Despite the dynamic movement of nuclei in PPP1R12A- depleted cells, we find that 81.3% (165 out of 203) of nuclear compression and rupture sites were associated with actomyosin filaments in fixed cells these new data are now alsi included in Supplementary Fig. 10b. In addition, we have performed physical micromanipulation experiments in collaboration with the Piel lab that showed that nuclear envelope rupture and chromatin extrusion, akin to the phenotypes observed in PPP1R12A- depleted cells, can be recapitulated by compressing nuclei (Figure R3 below). A movie of the compression experiments is uploaded for the benefit of the reviewer (Nuclear compression.avi). This shows that compressive force alone can trigger nuclear envelope rupture (the catastrophic mitosis at the end of the movie is interesting, but not pertinent to this work). Thus, our data strongly demonstrate that actomyosin filaments are associated with nuclear compression and nuclear envelope rupture sites and suggest that these filaments are causally related to the nuclear alteration. The correlative light and electron microscopy image analysis (clem) in Figure S10b of the revised manuscript shows a high- resolution image of actin filaments at a compression and rupture site. + Figure R3: Experimental setup (left panel). Frame from live cell recordings of mechanical compression of cells using a PDMS piston (arrows) (H2B, green; LAP2, red) (right panel). The live- cell movie of the compression experiment will be attached to these comments. Arrows indicate nuclear lamina rupture and chromatin extrusion sites. 7. The in vivo measurements are unconvincing. Nuclear abnormalities are not clearly visible in the picture provided (Figure 3c). It is not clear that the decrease in variance of the measured parameters necessarily indicates a smoother nucleus upon treatment with Y Also at the request of reviewer #3 we have clarified the z- section issue to more convincingly make this point (Supplementary Figure S14a). We also provide higher magnification in vivo images taken with a 63x 1.2NA objective now included in Supplementary Figure 14b of the revised manuscript; and in greater detail in Figure R4 on the following page. Z- sections and separate channels are shown, with the arrow indicating a point of nuclear envelope rupture. These data can be included in the manuscript to address the reviewer s comment.
20 Figure R4: Intravital images of cells in a mammary tumors (MDA- MB- 231) with H2B shown in red and LAP2β in green. Left panel shows different confocal z- sections, right panel shows single channels, * indicates the z- section split into single channels in the right- hand panels. Scale bar is 10μm. Finally, the reviewer is correct that the variance measurements do not demonstrate smoothness, but this is not what we claim. We claim that the variance over time reflects the dynamic deformation of the nucleus. The overall changes in nuclear morphology are not as pronounced as in vitro; this, most likely, results from the constraints on shape conferred by the complex matrix geometry in vivo. This is not changed by short- term administration of Y Despite this, the metrics of nuclear morphology provided in Supp. Fig. 13b indicate an increase in circularity, with no significant change in aspect ratio, which is consistent with the nuclei becoming smoother even though we do not go as far as making this statement. 8. Could the results in Figure 4 be interpreted in another way? Chromosomal spreads are done by colliding cells with a solid surface which forces the chromosomes apart. Knockdown of phosphatases makes the cytoplasm stiffer which might cause breaks in chromosomes. The experimental protocol used for chromosome spreading including fixation and hypotonic swelling prior to spreading makes it very unlikely that chromosome breakage is caused by a
Description: Nuclear morphology and dynamics in nontargeting sirna transfected cells. HeLa Kyoto
 Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Tables Title of file for HTML: Supplementary Movie 1 Description: Nuclear morphology and dynamics
Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Tables Title of file for HTML: Supplementary Movie 1 Description: Nuclear morphology and dynamics
Section 10. Junaid Malek, M.D.
 Section 10 Junaid Malek, M.D. Cell Division Make sure you understand: How do cells know when to divide? (What drives the cell cycle? Why is it important to regulate this?) How is DNA replication regulated?
Section 10 Junaid Malek, M.D. Cell Division Make sure you understand: How do cells know when to divide? (What drives the cell cycle? Why is it important to regulate this?) How is DNA replication regulated?
The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit
 Cell Reports, Volume 5 Supplemental Information The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit Andrey Poleshko, Katelyn M. Mansfield, Caroline
Cell Reports, Volume 5 Supplemental Information The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit Andrey Poleshko, Katelyn M. Mansfield, Caroline
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
Immunofluorescence images of different core histones and different histone exchange assay.
 Molecular Cell, Volume 51 Supplemental Information Enhanced Chromatin Dynamics by FACT Promotes Transcriptional Restart after UV-Induced DNA Damage Christoffel Dinant, Giannis Ampatziadis-Michailidis,
Molecular Cell, Volume 51 Supplemental Information Enhanced Chromatin Dynamics by FACT Promotes Transcriptional Restart after UV-Induced DNA Damage Christoffel Dinant, Giannis Ampatziadis-Michailidis,
Cell cycle oscillations
 Positive and negative feedback produce a cell cycle Fast Slow Cell cycle oscillations Active Cdk1-Cyclin Inactive Cdk1-Cyclin Active APC 1 Ubiquitin mediated proteolysis Glycine Isopeptide bond Lysine
Positive and negative feedback produce a cell cycle Fast Slow Cell cycle oscillations Active Cdk1-Cyclin Inactive Cdk1-Cyclin Active APC 1 Ubiquitin mediated proteolysis Glycine Isopeptide bond Lysine
Chapter 3. DNA Replication & The Cell Cycle
 Chapter 3 DNA Replication & The Cell Cycle DNA Replication and the Cell Cycle Before cells divide, they must duplicate their DNA // the genetic material DNA is organized into strands called chromosomes
Chapter 3 DNA Replication & The Cell Cycle DNA Replication and the Cell Cycle Before cells divide, they must duplicate their DNA // the genetic material DNA is organized into strands called chromosomes
Cell Nucleus. Chen Li. Department of Cellular and Genetic Medicine
 Cell Nucleus Chen Li Department of Cellular and Genetic Medicine 13 223 chenli2008@fudan.edu.cn Outline A. Historical background B. Structure of the nucleus: nuclear pore complex (NPC), lamina, nucleolus,
Cell Nucleus Chen Li Department of Cellular and Genetic Medicine 13 223 chenli2008@fudan.edu.cn Outline A. Historical background B. Structure of the nucleus: nuclear pore complex (NPC), lamina, nucleolus,
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
Learning Objectives. Define RNA interference. Define basic terminology. Describe molecular mechanism. Define VSP and relevance
 Learning Objectives Define RNA interference Define basic terminology Describe molecular mechanism Define VSP and relevance Describe role of RNAi in antigenic variation A Nobel Way to Regulate Gene Expression
Learning Objectives Define RNA interference Define basic terminology Describe molecular mechanism Define VSP and relevance Describe role of RNAi in antigenic variation A Nobel Way to Regulate Gene Expression
Tumor Growth Suppression Through the Activation of p21, a Cyclin-Dependent Kinase Inhibitor
 Tumor Growth Suppression Through the Activation of p21, a Cyclin-Dependent Kinase Inhibitor Nicholas Love 11/28/01 A. What is p21? Introduction - p21 is a gene found on chromosome 6 at 6p21.2 - this gene
Tumor Growth Suppression Through the Activation of p21, a Cyclin-Dependent Kinase Inhibitor Nicholas Love 11/28/01 A. What is p21? Introduction - p21 is a gene found on chromosome 6 at 6p21.2 - this gene
CELLULAR PROCESSES; REPRODUCTION. Unit 5
 CELLULAR PROCESSES; REPRODUCTION Unit 5 Cell Cycle Chromosomes and their make up Crossover Cytokines Diploid (haploid diploid and karyotypes) Mitosis Meiosis What is Cancer? Somatic Cells THE CELL CYCLE
CELLULAR PROCESSES; REPRODUCTION Unit 5 Cell Cycle Chromosomes and their make up Crossover Cytokines Diploid (haploid diploid and karyotypes) Mitosis Meiosis What is Cancer? Somatic Cells THE CELL CYCLE
Use of Phase Contrast Imaging to Track Morphological Cellular Changes due to Apoptotic Activity
 A p p l i c a t i o n N o t e Use of Phase Contrast Imaging to Track Morphological Cellular Changes due to Apoptotic Activity Brad Larson and Peter Banks, Applications Department, BioTek Instruments, Inc.,
A p p l i c a t i o n N o t e Use of Phase Contrast Imaging to Track Morphological Cellular Changes due to Apoptotic Activity Brad Larson and Peter Banks, Applications Department, BioTek Instruments, Inc.,
NUCLEUS. Fig. 2. Various stages in the condensation of chromatin
 NUCLEUS Animal cells contain DNA in nucleus (contains ~ 98% of cell DNA) and mitochondrion. Both compartments are surrounded by an envelope (double membrane). Nuclear DNA represents some linear molecules
NUCLEUS Animal cells contain DNA in nucleus (contains ~ 98% of cell DNA) and mitochondrion. Both compartments are surrounded by an envelope (double membrane). Nuclear DNA represents some linear molecules
Materials and Methods: All strains were derivatives of SK1 and are listed in Supplemental Table 1.
 Supplemental Online Material: Materials and Methods: All strains were derivatives of SK1 and are listed in Supplemental Table 1. Constructs: pclb2-3ha-cdc5 and pclb2-3ha-cdc2 strains were constructed by
Supplemental Online Material: Materials and Methods: All strains were derivatives of SK1 and are listed in Supplemental Table 1. Constructs: pclb2-3ha-cdc5 and pclb2-3ha-cdc2 strains were constructed by
Cell Division. Use Target Reading Skills. This section explains how cells grow and divide.
 Name Date Class Cell Processes Guided Reading and Study Cell Division This section explains how cells grow and divide. Use Target Reading Skills As you read, make a cycle diagram that shows the events
Name Date Class Cell Processes Guided Reading and Study Cell Division This section explains how cells grow and divide. Use Target Reading Skills As you read, make a cycle diagram that shows the events
SureSilencing sirna Array Technology Overview
 SureSilencing sirna Array Technology Overview Pathway-Focused sirna-based RNA Interference Topics to be Covered Who is SuperArray? Brief Introduction to RNA Interference Challenges Facing RNA Interference
SureSilencing sirna Array Technology Overview Pathway-Focused sirna-based RNA Interference Topics to be Covered Who is SuperArray? Brief Introduction to RNA Interference Challenges Facing RNA Interference
SONOMA STATE UNIVERSITY DEPARTMENT OF BIOLOGY BIOLOGY 344: CELL BIOLOGY Fall 2013
 SONOMA STATE UNIVERSITY DEPARTMENT OF BIOLOGY BIOLOGY 344: CELL BIOLOGY Fall 2013 Instructor Murali C. Pillai, PhD Office 214 Darwin Hall Telephone (707) 664-2981 E-mail pillai@sonoma.edu Website www.sonoma.edu/users/p/pillai
SONOMA STATE UNIVERSITY DEPARTMENT OF BIOLOGY BIOLOGY 344: CELL BIOLOGY Fall 2013 Instructor Murali C. Pillai, PhD Office 214 Darwin Hall Telephone (707) 664-2981 E-mail pillai@sonoma.edu Website www.sonoma.edu/users/p/pillai
Three major types of cytoskeleton
 The Cytoskeleton Organizes and stabilizes cells Pulls chromosomes apart Drives intracellular traffic Supports plasma membrane and nuclear envelope Enables cellular movement Guides growth of the plant cell
The Cytoskeleton Organizes and stabilizes cells Pulls chromosomes apart Drives intracellular traffic Supports plasma membrane and nuclear envelope Enables cellular movement Guides growth of the plant cell
Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product.
 Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplementary Figure 1. RAD51 and RAD51 paralogs are enriched spontaneously onto
 Supplementary Figure legends Supplementary Figure 1. and paralogs are enriched spontaneously onto the S-phase chromatin during DN replication. () Chromatin fractionation was carried out as described in
Supplementary Figure legends Supplementary Figure 1. and paralogs are enriched spontaneously onto the S-phase chromatin during DN replication. () Chromatin fractionation was carried out as described in
General Education Learning Outcomes
 BOROUGH OF MANHATTAN COMMUNITY COLLEGE City University of New York Department of Science Title of Course: Cell Biology Class hours 3 BIO Section: 260 Lab hours 3 Semester Spring 2018 Credits 4 Schedule:
BOROUGH OF MANHATTAN COMMUNITY COLLEGE City University of New York Department of Science Title of Course: Cell Biology Class hours 3 BIO Section: 260 Lab hours 3 Semester Spring 2018 Credits 4 Schedule:
Supporting information. Single-cell and subcellular pharmacokinetic imaging allows insight into drug action in vivo
 Supporting information Single-cell and subcellular pharmacokinetic imaging allows insight into drug action in vivo Greg Thurber 1, Katy Yang 1, Thomas Reiner 1, Rainer Kohler 1, Peter Sorger 2, Tim Mitchison
Supporting information Single-cell and subcellular pharmacokinetic imaging allows insight into drug action in vivo Greg Thurber 1, Katy Yang 1, Thomas Reiner 1, Rainer Kohler 1, Peter Sorger 2, Tim Mitchison
BME Engineering Molecular Cell Biology. The Cytoskeleton (I): Actin The Cytoskeleton (II): Microtubule & Intermediate Filament
 BME 42-620 Engineering Molecular Cell Biology Lecture 09: The Cytoskeleton (I): Actin The Cytoskeleton (II): Microtubule & Intermediate Filament BME42-620 Lecture 09, September 27, 2011 1 Outline Overviewofcytoskeletal
BME 42-620 Engineering Molecular Cell Biology Lecture 09: The Cytoskeleton (I): Actin The Cytoskeleton (II): Microtubule & Intermediate Filament BME42-620 Lecture 09, September 27, 2011 1 Outline Overviewofcytoskeletal
Cell cycle. Chen Li. Department of cellular and genetic medicine
 Cell cycle Chen Li Department of cellular and genetic medicine 13 223 chenli2008@fudan.edu.cn Outline A. Historical background B. Phases of cell cycle C. DNA replication D. Telomere & telomerase E. DNA
Cell cycle Chen Li Department of cellular and genetic medicine 13 223 chenli2008@fudan.edu.cn Outline A. Historical background B. Phases of cell cycle C. DNA replication D. Telomere & telomerase E. DNA
Cell Proliferation and Death
 Cell Proliferation and Death Derek Davies, Cancer Research UK http://www.london-research-institute.org.uk/technologies/120 Proliferation A cell Apoptosis Cell death Proliferation signals Senescence DNA
Cell Proliferation and Death Derek Davies, Cancer Research UK http://www.london-research-institute.org.uk/technologies/120 Proliferation A cell Apoptosis Cell death Proliferation signals Senescence DNA
Fig. S1. Endocytosis of extracellular cargo does not depend on dia. (A) To visualize endocytosis, fluorescently labelled wheat germ agglutinin
 Fig. S1. Endocytosis of extracellular cargo does not depend on dia. (A) To visualize endocytosis, fluorescently labelled wheat germ agglutinin (WGA-Alexa555) was injected into the extracellular perivitteline
Fig. S1. Endocytosis of extracellular cargo does not depend on dia. (A) To visualize endocytosis, fluorescently labelled wheat germ agglutinin (WGA-Alexa555) was injected into the extracellular perivitteline
Plant Molecular and Cellular Biology Lecture 9: Nuclear Genome Organization: Chromosome Structure, Chromatin, DNA Packaging, Mitosis Gary Peter
 Plant Molecular and Cellular Biology Lecture 9: Nuclear Genome Organization: Chromosome Structure, Chromatin, DNA Packaging, Mitosis Gary Peter 9/16/2008 1 Learning Objectives 1. List and explain how DNA
Plant Molecular and Cellular Biology Lecture 9: Nuclear Genome Organization: Chromosome Structure, Chromatin, DNA Packaging, Mitosis Gary Peter 9/16/2008 1 Learning Objectives 1. List and explain how DNA
Automated Imaging and Dual-Mask Analysis of γh2ax Foci to Determine DNA Damage on an Individual Cell Basis
 A p p l i c a t i o n N o t e Automated Imaging and Dual-Mask Analysis of γh2ax Foci to Determine DNA Damage on an Individual Cell Basis Brad Larson, BioTek Instruments, Inc., Winooski, VT USA Asha Sinha
A p p l i c a t i o n N o t e Automated Imaging and Dual-Mask Analysis of γh2ax Foci to Determine DNA Damage on an Individual Cell Basis Brad Larson, BioTek Instruments, Inc., Winooski, VT USA Asha Sinha
Chapter 8 DNA STRUCTURE AND CHROMOSOMAL ORGANIZATION
 Chapter 8 DNA STRUCTURE AND CHROMOSOMAL ORGANIZATION Chapter Summary Even though DNA has been known as a biochemical compound for over 100 years, it was not implicated as the carrier of hereditary information
Chapter 8 DNA STRUCTURE AND CHROMOSOMAL ORGANIZATION Chapter Summary Even though DNA has been known as a biochemical compound for over 100 years, it was not implicated as the carrier of hereditary information
FRAUNHOFER IME SCREENINGPORT
 FRAUNHOFER IME SCREENINGPORT Detection technologies used in drug discovery Introduction Detection technologies in drug discovery is unlimited only a biased snapshot can be presented Differences can presented
FRAUNHOFER IME SCREENINGPORT Detection technologies used in drug discovery Introduction Detection technologies in drug discovery is unlimited only a biased snapshot can be presented Differences can presented
BIO 315 Lab Exam I. Section #: Name:
 Section #: Name: Also provide this information on the computer grid sheet given to you. (Section # in special code box) BIO 315 Lab Exam I 1. In labeling the parts of a standard compound light microscope
Section #: Name: Also provide this information on the computer grid sheet given to you. (Section # in special code box) BIO 315 Lab Exam I 1. In labeling the parts of a standard compound light microscope
CHAPTER 17 FROM GENE TO PROTEIN. Section C: The Synthesis of Protein
 CHAPTER 17 FROM GENE TO PROTEIN Section C: The Synthesis of Protein 1. Translation is the RNA-directed synthesis of a polypeptide: a closer look 2. Signal peptides target some eukaryotic polypeptides to
CHAPTER 17 FROM GENE TO PROTEIN Section C: The Synthesis of Protein 1. Translation is the RNA-directed synthesis of a polypeptide: a closer look 2. Signal peptides target some eukaryotic polypeptides to
Chapter 5 DNA and Chromosomes
 Chapter 5 DNA and Chromosomes DNA as the genetic material Heat-killed bacteria can transform living cells S Smooth R Rough Fred Griffith, 1920 DNA is the genetic material Oswald Avery Colin MacLeod Maclyn
Chapter 5 DNA and Chromosomes DNA as the genetic material Heat-killed bacteria can transform living cells S Smooth R Rough Fred Griffith, 1920 DNA is the genetic material Oswald Avery Colin MacLeod Maclyn
Supporting Information
 Supporting Information Chakrabarty et al. 10.1073/pnas.1018001108 SI Materials and Methods Cell Lines. All cell lines were purchased from the American Type Culture Collection. Media and FBS were purchased
Supporting Information Chakrabarty et al. 10.1073/pnas.1018001108 SI Materials and Methods Cell Lines. All cell lines were purchased from the American Type Culture Collection. Media and FBS were purchased
1. I can describe the stages of the cell cycle.
 Unit 5 Study Guide Cell Cycle pg. 1 1. I can describe the stages of the cell cycle. Interphase = period in between division G1 = growth phase S = DNA replication G2 = Preparation for division (extra copies
Unit 5 Study Guide Cell Cycle pg. 1 1. I can describe the stages of the cell cycle. Interphase = period in between division G1 = growth phase S = DNA replication G2 = Preparation for division (extra copies
PROTEOMICS AND FUNCTIONAL GENOMICS PRESENTATION KIMBERLY DONG
 PROTEOMICS AND FUNCTIONAL GENOMICS PRESENTATION KIMBERLY DONG A Lentiviral RNAi Library for Human and Mouse Genes Applied to an Arrayed Viral High-Content Screen Jason Moffat,1,2,4,10 Dorre A. Grueneberg,1,10
PROTEOMICS AND FUNCTIONAL GENOMICS PRESENTATION KIMBERLY DONG A Lentiviral RNAi Library for Human and Mouse Genes Applied to an Arrayed Viral High-Content Screen Jason Moffat,1,2,4,10 Dorre A. Grueneberg,1,10
7.06 Cell Biology EXAM #2 March 20, 2003
 7.06 Cell Biology EXAM #2 March 20, 2003 This is an open book exam, and you are allowed access to books, a calculator, and notes but not computers or any other types of electronic devices. Please write
7.06 Cell Biology EXAM #2 March 20, 2003 This is an open book exam, and you are allowed access to books, a calculator, and notes but not computers or any other types of electronic devices. Please write
supplementary information
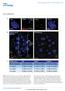 DOI: 10.1038/ncb2017 Figure S1 53BP1 sirna results in a heterochromatic DSB repair defect. Panel A: NIH3T3 cells were transfected with murine 53BP1 sirna. 48 hrs later, cells were irradiated with 3 Gy
DOI: 10.1038/ncb2017 Figure S1 53BP1 sirna results in a heterochromatic DSB repair defect. Panel A: NIH3T3 cells were transfected with murine 53BP1 sirna. 48 hrs later, cells were irradiated with 3 Gy
Chapter 18: Regulation of Gene Expression. 1. Gene Regulation in Bacteria 2. Gene Regulation in Eukaryotes 3. Gene Regulation & Cancer
 Chapter 18: Regulation of Gene Expression 1. Gene Regulation in Bacteria 2. Gene Regulation in Eukaryotes 3. Gene Regulation & Cancer Gene Regulation Gene regulation refers to all aspects of controlling
Chapter 18: Regulation of Gene Expression 1. Gene Regulation in Bacteria 2. Gene Regulation in Eukaryotes 3. Gene Regulation & Cancer Gene Regulation Gene regulation refers to all aspects of controlling
Genetic Basis of Development & Biotechnologies
 Genetic Basis of Development & Biotechnologies 1. Steps of embryonic development: cell division, morphogenesis, differentiation Totipotency and pluripotency 2. Plant cloning 3. Animal cloning Reproductive
Genetic Basis of Development & Biotechnologies 1. Steps of embryonic development: cell division, morphogenesis, differentiation Totipotency and pluripotency 2. Plant cloning 3. Animal cloning Reproductive
Molecular Cell Biology - Problem Drill 01: Introduction to Molecular Cell Biology
 Molecular Cell Biology - Problem Drill 01: Introduction to Molecular Cell Biology Question No. 1 of 10 1. Which statement describes how an organism is organized from most simple to most complex? Question
Molecular Cell Biology - Problem Drill 01: Introduction to Molecular Cell Biology Question No. 1 of 10 1. Which statement describes how an organism is organized from most simple to most complex? Question
MitoBiogenesis In-Cell ELISA Kit (Colorimetric)
 PROTOCOL MitoBiogenesis In-Cell ELISA Kit (Colorimetric) DESCRIPTION 1850 Millrace Drive, Suite 3A Eugene, Oregon 97403 MS643 Rev.2 For identifying inhibitors and activators of mitochondrial biogenesis
PROTOCOL MitoBiogenesis In-Cell ELISA Kit (Colorimetric) DESCRIPTION 1850 Millrace Drive, Suite 3A Eugene, Oregon 97403 MS643 Rev.2 For identifying inhibitors and activators of mitochondrial biogenesis
BIOLOGY. Chapter 16 GenesExpression
 BIOLOGY Chapter 16 GenesExpression CAMPBELL BIOLOGY TENTH EDITION Reece Urry Cain Wasserman Minorsky Jackson 18 Gene Expression 2014 Pearson Education, Inc. Figure 16.1 Differential Gene Expression results
BIOLOGY Chapter 16 GenesExpression CAMPBELL BIOLOGY TENTH EDITION Reece Urry Cain Wasserman Minorsky Jackson 18 Gene Expression 2014 Pearson Education, Inc. Figure 16.1 Differential Gene Expression results
NIH Public Access Author Manuscript Cell Cycle. Author manuscript; available in PMC 2010 March 1.
 NIH Public Access Author Manuscript Published in final edited form as: Cell Cycle. 2009 October 1; 8(19): 3133 3148. Temporal differences in DNA replication during the S phase using single fiber analysis
NIH Public Access Author Manuscript Published in final edited form as: Cell Cycle. 2009 October 1; 8(19): 3133 3148. Temporal differences in DNA replication during the S phase using single fiber analysis
To assess the localization of Citrine fusion proteins, we performed antibody staining to
 Trinh et al 1 SUPPLEMENTAL MATERIAL FlipTraps recapitulate endogenous protein localization To assess the localization of Citrine fusion proteins, we performed antibody staining to compare the expression
Trinh et al 1 SUPPLEMENTAL MATERIAL FlipTraps recapitulate endogenous protein localization To assess the localization of Citrine fusion proteins, we performed antibody staining to compare the expression
Small-Molecule Drug Target Identification/Deconvolution Technologies
 Small-Molecule Drug Target Identification/Deconvolution Technologies Case-Studies Shantani Target ID Technology Tool Box Target Deconvolution is not Trivial = A single Tool / Technology May Not necessarily
Small-Molecule Drug Target Identification/Deconvolution Technologies Case-Studies Shantani Target ID Technology Tool Box Target Deconvolution is not Trivial = A single Tool / Technology May Not necessarily
8/21/2014. From Gene to Protein
 From Gene to Protein Chapter 17 Objectives Describe the contributions made by Garrod, Beadle, and Tatum to our understanding of the relationship between genes and enzymes Briefly explain how information
From Gene to Protein Chapter 17 Objectives Describe the contributions made by Garrod, Beadle, and Tatum to our understanding of the relationship between genes and enzymes Briefly explain how information
Supplementary Figure 1. Characterization of EVs (a) Phase-contrast electron microscopy was used to visualize resuspended EV pellets.
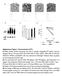 Supplementary Figure 1. Characterization of EVs (a) Phase-contrast electron microscopy was used to visualize resuspended EV pellets. Scale bar represent 100 nm. The sizes of EVs from MDA-MB-231-D3H1 (D3H1),
Supplementary Figure 1. Characterization of EVs (a) Phase-contrast electron microscopy was used to visualize resuspended EV pellets. Scale bar represent 100 nm. The sizes of EVs from MDA-MB-231-D3H1 (D3H1),
Cells and Tissues. Overview CELLS
 Cells and Tissues WIll The basic unit of structure and function in the human body is the cell. Each of a cell's parts, or organelles, as well as the entire cell, is organized to perform a specific function.
Cells and Tissues WIll The basic unit of structure and function in the human body is the cell. Each of a cell's parts, or organelles, as well as the entire cell, is organized to perform a specific function.
Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected
 Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected with the sirna against lnc-2, lnc-6, lnc-7, and the
Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected with the sirna against lnc-2, lnc-6, lnc-7, and the
PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF
 YFP-PHF1 CFP-PHT1;2 PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF + CFP-PHT1;2 Negative control!-gfp Supplemental Figure 1: PHT1;2 accumulation is PHF1 dependent. Immunoblot analysis on total protein extract
YFP-PHF1 CFP-PHT1;2 PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF + CFP-PHT1;2 Negative control!-gfp Supplemental Figure 1: PHT1;2 accumulation is PHF1 dependent. Immunoblot analysis on total protein extract
REGULATION OF PROTEIN SYNTHESIS. II. Eukaryotes
 REGULATION OF PROTEIN SYNTHESIS II. Eukaryotes Complexities of eukaryotic gene expression! Several steps needed for synthesis of mrna! Separation in space of transcription and translation! Compartmentation
REGULATION OF PROTEIN SYNTHESIS II. Eukaryotes Complexities of eukaryotic gene expression! Several steps needed for synthesis of mrna! Separation in space of transcription and translation! Compartmentation
Label-free, real-time live-cell assays for spheroids: IncuCyte bright-field analysis
 Introduction APPLICATION NOTE IncuCyte Live-Cell Analysis System Label-free, real-time live-cell assays for spheroids: IncuCyte bright-field analysis Susana L. Alcantara, Miniver Oliver, Kalpana Patel,
Introduction APPLICATION NOTE IncuCyte Live-Cell Analysis System Label-free, real-time live-cell assays for spheroids: IncuCyte bright-field analysis Susana L. Alcantara, Miniver Oliver, Kalpana Patel,
a Award Number: W81XWH TITLE:
 AD Award Number: W81XWH-04-1-0138 TITLE: Imaging Metastatic Prostate Cancer After Genetic Manipulation of Transcriptional Memory Regulators EZH2 and EED PRINCIPAL INVESTIGATOR: Lily Wu, M.D., Ph.D. CONTRACTING
AD Award Number: W81XWH-04-1-0138 TITLE: Imaging Metastatic Prostate Cancer After Genetic Manipulation of Transcriptional Memory Regulators EZH2 and EED PRINCIPAL INVESTIGATOR: Lily Wu, M.D., Ph.D. CONTRACTING
1. The microtubule wall is composed of globular proteins arranged in longitudinal rows called.
 Name: Quiz name: Quiz 7 ate: 1. The microtubule wall is composed of globular proteins arranged in longitudinal rows called. microfilaments protofilaments prototubules microtubular subunits 2. Which of
Name: Quiz name: Quiz 7 ate: 1. The microtubule wall is composed of globular proteins arranged in longitudinal rows called. microfilaments protofilaments prototubules microtubular subunits 2. Which of
The cohesin acetyltransferase Eco1 coordinates rdna replication and transcription
 Manuscript EMBOR-2013-37974 The cohesin acetyltransferase Eco1 coordinates rdna replication and transcription Shuai Lu, Kenneth K. Lee, Bethany Harris, Bo Xiong, Tania Bose, Anita Saraf, Gaye Hattem, Laurence
Manuscript EMBOR-2013-37974 The cohesin acetyltransferase Eco1 coordinates rdna replication and transcription Shuai Lu, Kenneth K. Lee, Bethany Harris, Bo Xiong, Tania Bose, Anita Saraf, Gaye Hattem, Laurence
Title: Combination of a third generation bisphosphonate and replication-competent adenoviruses augments the cytotoxicity on mesothelioma
 Author s response to reviews Title: Combination of a third generation bisphosphonate and replication-competent adenoviruses augments the cytotoxicity on mesothelioma Authors: Masatoshi Tagawa (mtagawa@chiba-cc.jp)
Author s response to reviews Title: Combination of a third generation bisphosphonate and replication-competent adenoviruses augments the cytotoxicity on mesothelioma Authors: Masatoshi Tagawa (mtagawa@chiba-cc.jp)
Introduction to Assay Development
 Introduction to Assay Development A poorly designed assay can derail a drug discovery program before it gets off the ground. While traditional bench-top assays are suitable for basic research and target
Introduction to Assay Development A poorly designed assay can derail a drug discovery program before it gets off the ground. While traditional bench-top assays are suitable for basic research and target
7.06 Problem Set #3, Spring 2005
 7.06 Problem Set #3, Spring 2005 1. The Drosophila compound eye is composed of about 800 units called ommatidia. Each ommatidium contains eight photoreceptor neurons (R1 through R8), which develop in a
7.06 Problem Set #3, Spring 2005 1. The Drosophila compound eye is composed of about 800 units called ommatidia. Each ommatidium contains eight photoreceptor neurons (R1 through R8), which develop in a
leading the way in research & development
 leading the way in research & development people. passion. possibilities. ABBVIE 2 immunology AbbVie Immunology has a demonstrated record of success in identifying and developing both small molecule and
leading the way in research & development people. passion. possibilities. ABBVIE 2 immunology AbbVie Immunology has a demonstrated record of success in identifying and developing both small molecule and
Supplementary Materials
 Supplementary Materials Construction of Synthetic Nucleoli in Human Cells Reveals How a Major Functional Nuclear Domain is Formed and Propagated Through Cell Divisision Authors: Alice Grob, Christine Colleran
Supplementary Materials Construction of Synthetic Nucleoli in Human Cells Reveals How a Major Functional Nuclear Domain is Formed and Propagated Through Cell Divisision Authors: Alice Grob, Christine Colleran
Supplementary Figure 1: MYCER protein expressed from the transgene can enhance
 Relative luciferase activity Relative luciferase activity MYC is a critical target FBXW7 MYC Supplementary is a critical Figures target 1-7. FBXW7 Supplementary Material A E-box sequences 1 2 3 4 5 6 HSV-TK
Relative luciferase activity Relative luciferase activity MYC is a critical target FBXW7 MYC Supplementary is a critical Figures target 1-7. FBXW7 Supplementary Material A E-box sequences 1 2 3 4 5 6 HSV-TK
Eukaryotic Transcription
 Eukaryotic Transcription I. Differences between eukaryotic versus prokaryotic transcription. II. (core vs holoenzyme): RNA polymerase II - Promotor elements. - General Pol II transcription factors (GTF).
Eukaryotic Transcription I. Differences between eukaryotic versus prokaryotic transcription. II. (core vs holoenzyme): RNA polymerase II - Promotor elements. - General Pol II transcription factors (GTF).
Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate
 Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
DNA STRUCTURE. Nucleotides: Nitrogenous Bases (Carry the Genetic Code) Expectation Sheet: DNA & Cell Cycle. I can statements: Basic Information:
 Expectation Sheet: DNA & Cell Cycle NAME: Test is 11/8/17 I can statements: I can discuss how DNA is found in all organisms and that the structure is common to all living things. I can diagram and label
Expectation Sheet: DNA & Cell Cycle NAME: Test is 11/8/17 I can statements: I can discuss how DNA is found in all organisms and that the structure is common to all living things. I can diagram and label
TheraLin. Universal Tissue Fixative Enabling Molecular Pathology
 TheraLin Universal Tissue Fixative Enabling Molecular Pathology TheraLin Universal Tissue Fixative Enabling Molecular Pathology Contents Page # TheraLin Universal Tissue Fixative 3 Introduction 5 Easy
TheraLin Universal Tissue Fixative Enabling Molecular Pathology TheraLin Universal Tissue Fixative Enabling Molecular Pathology Contents Page # TheraLin Universal Tissue Fixative 3 Introduction 5 Easy
Cell Growth and Reproduction
 Cell Growth and Reproduction Robert Hooke was the first person to describe cells, in the year 1665. He was looking through his microscope at a piece of cork when he noticed a lot of repeating honeycomb
Cell Growth and Reproduction Robert Hooke was the first person to describe cells, in the year 1665. He was looking through his microscope at a piece of cork when he noticed a lot of repeating honeycomb
Genomic Instability And Chromosome Architecture. Kevin Mills, Ph.D. Associate Professor, The Jackson Laboratory
 Genomic Instability And Chromosome Architecture Kevin Mills, Ph.D. Associate Professor, The Jackson Laboratory Genomic Instability is a Hallmark of Cancer Angiogenesis Invasion and Metastasis Metabolism
Genomic Instability And Chromosome Architecture Kevin Mills, Ph.D. Associate Professor, The Jackson Laboratory Genomic Instability is a Hallmark of Cancer Angiogenesis Invasion and Metastasis Metabolism
Answers to Module 1. An obligate aerobe is an organism that has an absolute requirement of oxygen for growth.
 Answers to Module 1 Short Answers 1) What is an obligate aerobe? An obligate aerobe is an organism that has an absolute requirement of oxygen for growth. What about facultative anaerobe? 2) Distinguish
Answers to Module 1 Short Answers 1) What is an obligate aerobe? An obligate aerobe is an organism that has an absolute requirement of oxygen for growth. What about facultative anaerobe? 2) Distinguish
S-sulfhydration of MEK1 leads to PARP-1 activation and DNA damage repair
 Manuscript EMBOR-2013-38213 S-sulfhydration of MEK1 leads to PARP-1 activation and DNA damage repair Kexin Zhao, YoungJun Ju, Shuangshuang Li, Zaid Al Taany, Rui Wang and Guangdong Yang Corresponding author:
Manuscript EMBOR-2013-38213 S-sulfhydration of MEK1 leads to PARP-1 activation and DNA damage repair Kexin Zhao, YoungJun Ju, Shuangshuang Li, Zaid Al Taany, Rui Wang and Guangdong Yang Corresponding author:
Chapter 15. Cytoskeletal Systems. Lectures by Kathleen Fitzpatrick Simon Fraser University Pearson Education, Inc.
 Chapter 15 Cytoskeletal Systems Lectures by Kathleen Fitzpatrick Simon Fraser University Table 15-1 - Microtubules Table 15-1 - Microfilaments Table 15-1 Intermediate Filaments Table 15-3 Microtubules
Chapter 15 Cytoskeletal Systems Lectures by Kathleen Fitzpatrick Simon Fraser University Table 15-1 - Microtubules Table 15-1 - Microfilaments Table 15-1 Intermediate Filaments Table 15-3 Microtubules
Thermo Scientific Dharmacon SMARTvector 2.0 Lentiviral shrna Particles
 Thermo Scientific Dharmacon SMARTvector 2.0 Lentiviral shrna Particles Long-term gene silencing shrna-specific design algorithm High titer, purified particles Thermo Scientific Dharmacon SMARTvector shrna
Thermo Scientific Dharmacon SMARTvector 2.0 Lentiviral shrna Particles Long-term gene silencing shrna-specific design algorithm High titer, purified particles Thermo Scientific Dharmacon SMARTvector shrna
Focus Application. Cell Migration. Featured Study: Inhibition of Cell Migration by Gene Silencing. xcelligence System Real-Time Cell Analyzer
 xcelligence System Real-Time Cell Analyzer Focus Application Cell Migration Featured Study: Inhibition of Cell Migration by Gene Silencing Markus Greiner and Richard Zimmermann Department of Medical Biochemistry
xcelligence System Real-Time Cell Analyzer Focus Application Cell Migration Featured Study: Inhibition of Cell Migration by Gene Silencing Markus Greiner and Richard Zimmermann Department of Medical Biochemistry
MOLECULAR BIOLOGY OF EUKARYOTES 2016 SYLLABUS
 03-442 Lectures: MWF 9:30-10:20 a.m. Doherty Hall 2105 03-742 Advanced Discussion Section: Time and place to be announced Probably Mon 4-6 p.m. or 6-8p.m.? Once we establish who is taking the advanced
03-442 Lectures: MWF 9:30-10:20 a.m. Doherty Hall 2105 03-742 Advanced Discussion Section: Time and place to be announced Probably Mon 4-6 p.m. or 6-8p.m.? Once we establish who is taking the advanced
If Dna Has The Instructions For Building Proteins Why Is Mrna Needed
 If Dna Has The Instructions For Building Proteins Why Is Mrna Needed if a strand of DNA has the sequence CGGTATATC, then the complementary each strand of DNA contains the info needed to produce the complementary
If Dna Has The Instructions For Building Proteins Why Is Mrna Needed if a strand of DNA has the sequence CGGTATATC, then the complementary each strand of DNA contains the info needed to produce the complementary
Molecular Cell Biology - Problem Drill 11: Recombinant DNA
 Molecular Cell Biology - Problem Drill 11: Recombinant DNA Question No. 1 of 10 1. Which of the following statements about the sources of DNA used for molecular cloning is correct? Question #1 (A) cdna
Molecular Cell Biology - Problem Drill 11: Recombinant DNA Question No. 1 of 10 1. Which of the following statements about the sources of DNA used for molecular cloning is correct? Question #1 (A) cdna
Section 10.3 Outline 10.3 How Is the Base Sequence of a Messenger RNA Molecule Translated into Protein?
 Section 10.3 Outline 10.3 How Is the Base Sequence of a Messenger RNA Molecule Translated into Protein? Messenger RNA Carries Information for Protein Synthesis from the DNA to Ribosomes Ribosomes Consist
Section 10.3 Outline 10.3 How Is the Base Sequence of a Messenger RNA Molecule Translated into Protein? Messenger RNA Carries Information for Protein Synthesis from the DNA to Ribosomes Ribosomes Consist
Daughter-cell-specific modulation of nuclear pore complexes controls cell cycle entry during asymmetric division
 SUPPLEMENTARY INFORMATION Articles https://doi.org/10.1038/s41556-018-0056-9 In the format provided by the authors and unedited. Daughter-cell-specific modulation of nuclear pore complexes controls cell
SUPPLEMENTARY INFORMATION Articles https://doi.org/10.1038/s41556-018-0056-9 In the format provided by the authors and unedited. Daughter-cell-specific modulation of nuclear pore complexes controls cell
Evaluate Compound Toxicity Early in the Drug Discovery Process Using High Content Analysis in Cell-Based Assays
 Application Note Evaluate Compound Toxicity Early in the Drug Discovery Process Using High Content Analysis in Cell-Based Assays Jacob Fog, Louise Gjelstrup, Frosty Loechel and Morten Præstegaard, Thermo
Application Note Evaluate Compound Toxicity Early in the Drug Discovery Process Using High Content Analysis in Cell-Based Assays Jacob Fog, Louise Gjelstrup, Frosty Loechel and Morten Præstegaard, Thermo
Whole Transcriptome Analysis of Illumina RNA- Seq Data. Ryan Peters Field Application Specialist
 Whole Transcriptome Analysis of Illumina RNA- Seq Data Ryan Peters Field Application Specialist Partek GS in your NGS Pipeline Your Start-to-Finish Solution for Analysis of Next Generation Sequencing Data
Whole Transcriptome Analysis of Illumina RNA- Seq Data Ryan Peters Field Application Specialist Partek GS in your NGS Pipeline Your Start-to-Finish Solution for Analysis of Next Generation Sequencing Data
Mammalian DNA2 helicase/nuclease cleaves G-quadruplex DNA and is required for telomere integrity
 Manuscript EMBO-2012-83769 Mammalian DNA2 helicase/nuclease cleaves G-quadruplex DNA and is required for telomere integrity Weiqiang Lin, Shilpa Sampathi, Huifang Dai, Changwei Liu, Mian Zhou, Jenny Hu,
Manuscript EMBO-2012-83769 Mammalian DNA2 helicase/nuclease cleaves G-quadruplex DNA and is required for telomere integrity Weiqiang Lin, Shilpa Sampathi, Huifang Dai, Changwei Liu, Mian Zhou, Jenny Hu,
Gene Expression Technology
 Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
Mystery microscope images
 Mystery microscope images Equipment: 6X framed microscopy images 6X cards saying what each microscope image shows 6X cards with the different microscopy techniques written on them 6X cards with the different
Mystery microscope images Equipment: 6X framed microscopy images 6X cards saying what each microscope image shows 6X cards with the different microscopy techniques written on them 6X cards with the different
Module1TheBasicsofRealTimePCR Monday, March 19, 2007
 Objectives Slide notes: Page 1 of 41 Module 1: The Basics Of Real Time PCR Slide notes: Module 1: The Basics of real time PCR Page 2 of 41 Polymerase Chain Reaction Slide notes: Here is a review of PCR,
Objectives Slide notes: Page 1 of 41 Module 1: The Basics Of Real Time PCR Slide notes: Module 1: The Basics of real time PCR Page 2 of 41 Polymerase Chain Reaction Slide notes: Here is a review of PCR,
7.17: Writing Up Results and Creating Illustrations
 7.17: Writing Up Results and Creating Illustrations A Results Exercise: Kansas and Pancakes Write a 5-sentence paragraph describing the results illustrated in this figure: - Describe the figure: highlights?
7.17: Writing Up Results and Creating Illustrations A Results Exercise: Kansas and Pancakes Write a 5-sentence paragraph describing the results illustrated in this figure: - Describe the figure: highlights?
Supporting Information
 Supporting Information Signal mingle: Micropatterns of BMP-2 and fibronectin on soft biopolymeric films regulate myoblast shape and SMAD signaling. Vincent Fitzpatrick, Laure Fourel, Olivier Destaing,
Supporting Information Signal mingle: Micropatterns of BMP-2 and fibronectin on soft biopolymeric films regulate myoblast shape and SMAD signaling. Vincent Fitzpatrick, Laure Fourel, Olivier Destaing,
of Medicine, Zhejiang University, Hangzhou, Zhejiang , China Michigan, 4424B MS-1, 1301 Catherine Street, Ann Arbor, MI 48109, USA.
 Supplemental figure legends: Neddylation inhibitor MLN4924 suppresses growth and migration of human gastric cancer cells Huiyin Lan 1,2#, Zaiming Tang 1#, Hongchuan Jin 2, and Yi Sun 1,3,4* 1 Institute
Supplemental figure legends: Neddylation inhibitor MLN4924 suppresses growth and migration of human gastric cancer cells Huiyin Lan 1,2#, Zaiming Tang 1#, Hongchuan Jin 2, and Yi Sun 1,3,4* 1 Institute
Improving CRISPR-Cas9 Gene Knockout with a Validated Guide RNA Algorithm
 Improving CRISPR-Cas9 Gene Knockout with a Validated Guide RNA Algorithm Anja Smith Director R&D Dharmacon, part of GE Healthcare Imagination at work crrna:tracrrna program Cas9 nuclease Active crrna is
Improving CRISPR-Cas9 Gene Knockout with a Validated Guide RNA Algorithm Anja Smith Director R&D Dharmacon, part of GE Healthcare Imagination at work crrna:tracrrna program Cas9 nuclease Active crrna is
6.2 Chromatin is divided into euchromatin and heterochromatin
 6.2 Chromatin is divided into euchromatin and heterochromatin Individual chromosomes can be seen only during mitosis. During interphase, the general mass of chromatin is in the form of euchromatin. Euchromatin
6.2 Chromatin is divided into euchromatin and heterochromatin Individual chromosomes can be seen only during mitosis. During interphase, the general mass of chromatin is in the form of euchromatin. Euchromatin
Review of Biomedical Image Processing
 BOOK REVIEW Open Access Review of Biomedical Image Processing Edward J Ciaccio Correspondence: ciaccio@columbia. edu Department of Medicine, Columbia University, New York, USA Abstract This article is
BOOK REVIEW Open Access Review of Biomedical Image Processing Edward J Ciaccio Correspondence: ciaccio@columbia. edu Department of Medicine, Columbia University, New York, USA Abstract This article is
supplementary information
 DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
The Nuclear Area Factor (NAF): a measure for cell apoptosis using microscopy and image analysis
 The Nuclear Area Factor (NAF): a measure for cell apoptosis using microscopy and image analysis Mark A. DeCoster Department of Biomedical Engineering and Institute for Micromanufacturing, Louisiana Tech
The Nuclear Area Factor (NAF): a measure for cell apoptosis using microscopy and image analysis Mark A. DeCoster Department of Biomedical Engineering and Institute for Micromanufacturing, Louisiana Tech
INVESTIGATION OF MSC DIFFERENTIATION ON ELECTROSPUN NANOFIBROUS SCAFFOLDS
 With support of NSF Award no. EEC-0754741 INVESTIGATION OF MSC DIFFERENTIATION ON ELECTROSPUN NANOFIBROUS SCAFFOLDS NSF Summer Undergraduate Fellowship in Sensor Technologies Emily Wible (Bioengineering)
With support of NSF Award no. EEC-0754741 INVESTIGATION OF MSC DIFFERENTIATION ON ELECTROSPUN NANOFIBROUS SCAFFOLDS NSF Summer Undergraduate Fellowship in Sensor Technologies Emily Wible (Bioengineering)
Higher Human Biology Unit 1: Human Cells Pupils Learning Outcomes
 Higher Human Biology Unit 1: Human Cells Pupils Learning Outcomes 1.1 Division and Differentiation in Human Cells I can state that cellular differentiation is the process by which a cell develops more
Higher Human Biology Unit 1: Human Cells Pupils Learning Outcomes 1.1 Division and Differentiation in Human Cells I can state that cellular differentiation is the process by which a cell develops more
Adenine % Guanine % Thymine % Cytosine %
 1. Explain each of the following statements in terms of your knowledge of the structure and function of DNA. (i) In all living organisms the ratio species to another. A C T G is constant but the ratio
1. Explain each of the following statements in terms of your knowledge of the structure and function of DNA. (i) In all living organisms the ratio species to another. A C T G is constant but the ratio
SUPPLEMENTAL FIGURE LEGENDS. Figure S1: Homology alignment of DDR2 amino acid sequence. Shown are
 SUPPLEMENTAL FIGURE LEGENDS Figure S1: Homology alignment of DDR2 amino acid sequence. Shown are the amino acid sequences of human DDR2, mouse DDR2 and the closest homologs in zebrafish and C. Elegans.
SUPPLEMENTAL FIGURE LEGENDS Figure S1: Homology alignment of DDR2 amino acid sequence. Shown are the amino acid sequences of human DDR2, mouse DDR2 and the closest homologs in zebrafish and C. Elegans.
Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna
 Supplemental Materials and Methods Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna (Mission shrna, Sigma) against mouse MMP14 were transfected into HEK293 cells using FuGene6
Supplemental Materials and Methods Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna (Mission shrna, Sigma) against mouse MMP14 were transfected into HEK293 cells using FuGene6
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1: Vector maps of TRMPV and TRMPVIR variants. Many derivatives of TRMPV have been generated and tested. Unless otherwise noted, experiments in this paper use
Supplementary Figure 1 Supplementary Figure 1: Vector maps of TRMPV and TRMPVIR variants. Many derivatives of TRMPV have been generated and tested. Unless otherwise noted, experiments in this paper use
Y1 Biology 131 Syllabus - Academic Year
 Y1 Biology 131 Syllabus - Academic Year 2016-2017 Monday 28/11/2016 DNA Packaging Week 11 Tuesday 29/11/2016 Regulation of gene expression Wednesday 22/9/2014 Cell cycle Sunday 4/12/2016 Tutorial Monday
Y1 Biology 131 Syllabus - Academic Year 2016-2017 Monday 28/11/2016 DNA Packaging Week 11 Tuesday 29/11/2016 Regulation of gene expression Wednesday 22/9/2014 Cell cycle Sunday 4/12/2016 Tutorial Monday
