Disengaging the Smc3/kleisin interface releases cohesin from Drosophila chromosomes during interphase and mitosis
|
|
|
- Della Lester
- 5 years ago
- Views:
Transcription
1 Manuscript EMBO Disengaging the Smc3/kleisin interface releases cohesin from Drosophila chromosomes during interphase and mitosis Christian S. Eichinger, Alexander Kurze, Raquel A. Oliveira, Kim A. Nasmyth Corresponding author: Kim A. Nasmyth, University of Oxford Review timeline: Submission date: 30 August 2012 Editorial Decision: 03 October 2012 Revision received: 05 December 2012 Acceptance letter: 12 December 2012 Accepted: 13 December 2012 Editor: Hartmut Vodermaier Transaction Report: (Note: With the exception of the correction of typographical or spelling errors that could be a source of ambiguity, letters and reports are not edited. The original formatting of letters and referee reports may not be reflected in this compilation.) 1st Editorial Decision 03 October 2012 After some delay associated with the evaluation of back-to-back submissions, we have now received the feedback of two referees, copied below for your information. As you will see, both referees consider the demonstration of a conserved cohesin exit gate and its role in prophase cohesin release in metazoans important and therefore in principle suited for publication in a broad general journal. Both of them nevertheless demand strengthening of certain aspects of this work, in order to provide the required strong support for the main conclusions. Most of these points seem to be well-taken and easily addressable, however the one major concern of referee 1 may require some more substantial follow-up work to rule out potential alternative explanations for the results in Figure 5. I would therefore like to invite you to respond to the referees' comments through the form of a revised version of the manuscript. Given your recent publication on the cohesin exit gate in yeast, it is my hope that you will be able to resubmit your manuscript in a timely manner, nevertheless I feel it will be primarily important to diligently and thoroughly address the raised concerns during this revision. As per our EMBO Journal editorial policies, related or competing manuscripts published during this revision period will have no negative impact on our final assessment of your revised study. Finally, when revising the manuscript text and organization, I feel it would be important to touch on the Chan et al yeast paper and its key findings already in the introduction section, while at the same time stressing the rationale and conceptual importance of the present, parallel in vivo efforts in a metazoan organism (as you have done in the cover letter). Thank you for the opportunity to consider this work for publication, and please do get back to me should you have any comments or require further clarifications regarding the referee reports and this decision. I look forward to your revision! European Molecular Biology Organization 1
2 REFEREE REPORTS: Referee #1 (Remarks to the Author): In the manuscript by Eichinger et al, the authors investigate whether disengagement of the Smc3- Rad21 interface is necessary for cohesin release during interphase and mitosis in Drosophila. The requirement of this process has been recently demonstrated for the turnover of cohesin at pericentic chromatin during mitosis in S. cerevisiae by the same group (Chan 2012). I believe that the current study provides further proof of the existence of an exit gate for cohesin in a metazoan organism and, moreover, shows its importance for cohesin dynamics during interphase and for the prophase pathway. I have enjoyed reading this paper and watching the very nice movies. Thus, if the authors can address the one major concern that I explain below, I would support its publication in Embo J. My major concern refers to the result in Figure 5. Cleavage of the peptide linking Smc3-Rad21-GFP leads to extensive release of cohesin complexes. This should not be the case if these complexes were behaving as the endogenous. My concern is then that the over expressed Smc3-Rad21-GFP heterodimer binds to chromatin through the Smc3 subunit but does not form a bonafide cohesin complex and thus does not turn over (figure 4). In this scenario, when TEV is injected, Rad21-GFP is released (figure 5). Similarly, the heterodimer would persist on chromatin though prophasemetaphase (figure 6D) and a Rad21-GFP fragment would be released upon separase cleavage in anaphase. To rule out this possibility, I would like to see: 1. Immunoprecipitation with anti-gfp from salivary gland extracts (after induction of Smc3-Rad21- GFP) brings down not only Rad21 and Smc3, but also Smc1, Scc3, Pds5 and Wapl. 2. Repeat the experiment with the GFP tag in Smc3 instead of Rad Demonstrate by an alternative method that cohesin complexes containing Smc3-Rad21-GFP remain on chromatin after TEV injection (isolation of salivary gland chromosomes after TEV injection and analysis of chromatin by western blot). 4. Show that dissociation of GFP labelled complexes from salivary gland chromosomes upon TEV injection depends on Wapl. Minor points: -page 7, "In the salivary glands, we found comparable levels of the fusion protein..." It is not clear what levels are being compared. Please clarify. In the salivary glands there seems to be a huge overexpression of the transgene with respect to endogenous protein. - In the last experiment regarding the prophase pathway (Figure 6D), the authors indicate "cohesin containing an intact Smc3-Rad21-GFP fusion protein persisted...until the onset of anaphase." Why is there not a movie showing this? European Molecular Biology Organization 2
3 Additional suggestions: -In the Introduction: -The prophase pathway was initially identified in Xenopus (Losada 1998) and the involvement of mitotic kinases was also first described by Sumara 2002 and Losada In page 3 of Introduction, we read "Wapl is recruited to cohesin by binding...pds5, which..." and Chan KL, personal communication is cited. I do not think this is adequate for the Introduction. Maybe for the first part of the sentence Chan et al (2012) should be cited instead, although it should be also mentioned that the requirement of Pds5 for Wapl recruitment does not exist in Xenopus (Shintomi 2009). - Results By the end of the first paragraph in page 9, Kueng 2006 should be cited: In that study it was already shown by live cell imaging in HeLa cells that despite failure of the prophase pathway in Wapl sirna cells, all cohesin was released in anaphase. -Figure Legends. Although I am no fan of long Figure legends, I would encourage the authors to provide some more info in Figures 5 and 6 to allow the reader to understand the figure independently of the main text. Also, magnification bars should be added. Referee #2 (Remarks to the Author): Experiments in yeast have suggested that cohesin undergoes DNA entrapment and release dynamically, through transient opening of Smc1/3 interface and Smc3/alpha-kleisin (Scc1) interface, respectively. It is well known that DNA is released from cohesin by the proteolytic cleavage of kleisin upon anaphase onset, but the proteolysis-independent dissociation of cohesin in prophase/prometaphase through what it is called prophase pathway is not well understood. This paper eloquently addresses the long-standing question of how cohesin is released nonproteolytically. The authors made use of live cell imaging analyses in Drosophila non-dividing salivary gland cells and showed that cohesin turnover in these interphase cells, which required proficient Wapl. Thus, in Wapl mutant cells, cohesin over-enriched at chromosomal loci revealed characteristic structures, but artificial cleavage of kleisin Rad21 caused immediate dissociation of cohesin from chromatin. This finding significantly implied that cohesins that are not conferring cohesion during interphase associates to chromosomes in a topological manner. To my knowledge this is the first demonstration that cohesin association is basically topological. The authors then tested the idea that DNA might escape from the cohesin ring through the transient disconnection of the Smc3-Rad21 interaction, in a manner depending on Wapl, the hypothesis based on yeast works. To address this the authors tethered Smc3 and Rad21 heads by short polypeptide links, which can be artificially cleaved by TEV protease. Cohesin ring with Smc3-Rad21 fusion caused over-enrichment of the complex on chromatin, as seen in Wapl mutant, but this was released by cleaving the tethering peptide. Crucially, the authors described that not only on polytene chromosomes in salivary gland cells but also in prophase in neuroblasts, Wapl-promoting cohesin release is blocked by fusing Smc3 and kleisin. Based on these results the authors concluded that proteolysis-independent release of cohesin from chromatin is universally mediated by escape of DNA through Smc3/Rad21 gate. I found the paper provide important results indicating how cohesin complexes are associated with chromatin and how most of them are released before anaphase. It is true that similar lines of conclusions are already drawn in yeast, nevertheless I do see that the current study goes beyond the yeast studies, as explicitly described in the discussion. It is a significant step toward our understanding of cohesin regulation and clearly deserves for the publication. That said, I have several concerns that might taken into consideration, which should be addressed without much difficulty. European Molecular Biology Organization 3
4 1. Provided that Wapl-promoted release of chromatin involves DNA's escape through the Smc3- kleisin gate, I am curious to know if and to which extent the TEV-induced release of Smc3-Rad21- GFP from polytene chromosomes depends on Wapl in this experimental setting (Figure 5). If the release is primarily driven by Wapl, what one would expect to see in a Wapl mutant background is a marked enrichment of Smc3-Rad21-GFP on chromosomes, which do not grossly affected after microinjecting the active TEV. Is it the case? These results would be in sharp contrast to the experiment cleaving Rad21 (Figure 2E). 2. Smc3-Rad21-GFP fusion protein is found to accumulate at chromosomal loci that result in emergence of bands or circular structure appearance (Figure 3D). To estimate the net effect of the tethering of Smc3 and Rad21, it is informative to show side-by-side the picture of Smc3-Rad21-GFP when TEV was co-expressed. As for the Western blots in Figure 3, antibodies used for the analyses are missing. In the blots in Figure 3C,, is it possible to explain why endogenous Rad21 band is not detectable in Smc-Rad21-GFP lanes? 3. FRAP analysis in Figure 4 shows the turnover rate of Smc3-Rad21-GFP with our without the Smc3/Rad21 tethering. The authors' interpretation of the data for +TEV kinetics is "similar to wild type", but the fluorescence recovery seems to be consistently lower (60-70% of RFI) than that of wild type which is shown in Figure 2C. It seems to imply that there is more stably bound fraction of non-tethered Smc3-Rad21-GFP than Rad21-GFP. What could be a possible explanation for this? My suggestion is to compare the half-recovery time here instead, and say it is similar to wild-type, if that is the case. 4. The supplementary movie data in neuroblast provide a unique opportunity to show that the dissociation of cohesin from chromosome arms in prophase and from centromeres in anaphase, and that in Wapl mutants considerable amount of cohesin remains throughout the chromosome lengths until anaphase (Movie S10 and S11). It will be helpful to additionally provide the movie data for Smc3-Rad21-GFP with or without TEV, because it is difficult to tell that cohesin remains on arms solely from a still image (Figure 6D). For more comprehensive presentation for Figure 6D, drawings of schematic illustration may help. 5. The paper provides mechanistic explanation for the first time for how prophase pathway might work to release cohesin from chromosomes. It has long been known that the prophase pathway involves activity of mitotic kinases such as Plk1 and phosphorylation of Scc3/SA2. Therefore a perspective view for how these signals might contribute to promote Wapl-mediated cohesin release would be interesting to discuss. 1st Revision - authors' response 05 December 2012 Regarding editor s remarks: In the introduction of our new manuscript, we describe briefly the parallel work of Chan et al. and the rational and conceptual importance of the present manuscript. Also in the discussion, the conclusions of the parallel yeast study is put in relation to our work. Referee #1 (Remarks to the Author): In the manuscript by Eichinger et al, the authors investigate whether disengagement of the Smc3- Rad21 interface is necessary for cohesin release during interphase and mitosis in Drosophila. The requirement of this process has been recently demonstrated for the turnover of cohesin at pericentic chromatin during mitosis in S. cerevisiae by the same group (Chan 2012). I believe that the current study provides further proof of the existence of an exit gate for cohesin in a metazoan organism and, moreover, shows its importance for cohesin dynamics during interphase and for the prophase pathway. I have enjoyed reading this paper and watching the very nice movies. Thus, if the authors can address the one major concern that I explain below, I would support its publication in Embo J. European Molecular Biology Organization 4
5 My major concern refers to the result in Figure 5. Cleavage of the peptide linking Smc3-Rad21- GFP leads to extensive release of cohesin complexes. This should not be the case if these complexes were behaving as the endogenous. My concern is then that the over expressed Smc3-Rad21-GFP heterodimer binds to chromatin through the Smc3 subunit but does not form a bonafide cohesin complex and thus does not turn over (figure 4). In this scenario, when TEV is injected, Rad21-GFP is released (figure 5). Similarly, the heterodimer would persist on chromatin though prophasemetaphase (figure 6D) and a Rad21-GFP fragment would be released upon separase cleavage in anaphase. We agree that referee #1 s major concern whether or not the Smc3-Rad21-GFP fusion protein is incorporated into a bona fide cohesin ring needs to be clarified. For this, we carried out two experiments as suggested by referee 1: 1) First and foremost, we set up a fly cross that generates larvae, which express the Smc3-Rad21- GFP fusion protein in a wapl C204 mutant background and injected TEV protease after the fusion protein has been loaded onto chromatin. This experiment clearly demonstrated that Rad21-GFP is not released from Smc3 merely by cleaving the linker between the two proteins. Crucially, release also depends on the cohesin-associated protein Wapl. This experiment is now described by the revised version as Figure 5C and Supplementary Movie S10 and S11 (Movies S10 and S11 from our first submission are now named Movies S12 and S13, respectively). 2) Second, we carried out a Co-Immunoprecipitation experiment showing that the Smc3-Rad21- GFP fusion protein does interact with endogenous Smc1 but not with endogenous Rad21, thus showing that the Smc3-Rad21-GFP fusion protein is incorporated into a bona fide cohesin complex. The result of this experiment is included in the revised manuscript as a new Supplementary Figure S1. Further details on each of referee 1 s comments are described in the following: To rule out this possibility, I would like to see: 1. Immunoprecipitation with anti-gfp from salivary gland extracts (after induction of Smc3-Rad21- GFP) brings down not only Rad21 and Smc3, but also Smc1, Scc3, Pds5 and Wapl. We carried out an immunoprecipitation experiment (using anti-gfp beads) of chromatin-bound proteins from larval extracts after induction of Smc3-Rad21-GFP and tested binding for endogenous Smc1 and endogenous Rad21 using available antibodies against Drosophila Rad21 and Smc1. In our attempts to use salivary gland extracts we did not obtain enough material to purify chromatin-bound proteins and to carry out subsequent co-immunoprecipitation. However, using third instar larval extracts, we found that Smc3-Rad21-GFP interacts with endogenous Smc1, but not endogenous Rad21, indicating that chromatin-bound Smc3-Rad21-GFP forms an intact cohesin ring with endogenous Smc1 and does not incorporate another endogenous Rad21. The data of this experiment are shown in the new Supplementary Figure Repeat the experiment with the GFP tag in Smc3 instead of Rad21. Unfortunately, we do not have transgenic flies where an Smc3-Rad21 fusion protein is tagged at the N-terminus of Smc3 with GFP. Thus, in order to do this experiment, the generation of new transgenic flies would be necessary. This is unfortunately not doable in a reasonable amount of time. We believe however that our new experimental data described in referee 1 s points 1 and 4 show strong evidence against his/her major concern and indicates that Smc3-Rad21-GFP forms bona-fide cohesin complexes and that the release of Rad21-GFP from chromatin after cleaving the linkage between Smc3 and Rad21 depends on Wapl. 3. Demonstrate by an alternative method that cohesin complexes containing Smc3-Rad21-GFP remain on chromatin after TEV injection (isolation of salivary gland chromosomes after TEV injection and analysis of chromatin by western blot). Although we agree that this experiment would be very informative, it is unfortunately technically European Molecular Biology Organization 5
6 impossible to do for two reasons: 1) Proteins or mrnas are injected directly into the cytoplasm of only a single or of very few cells of a salivary gland in order to keep the tissue and its physiology intact. For this experiment however, one would need to inject every single cell in the tissue (one salivary gland contains around 120 cells). 2) Although live imaging after injection is possible without perturbing the physiology of the gland, it is technically very difficult to recover an injected intact gland reliably from the injection chamber for Western Blot analysis. 4. Show that dissociation of GFP labelled complexes from salivary gland chromosomes upon TEV injection depends on Wapl. We agree that this is a very important biological question that can be addressed using our expertise. We therefore set up a fly cross that generates progeny, which expresses the Smc3-Rad21-GFP protein upon heat-shock in a wapl C204 mutant background. We injected salivary gland cells with TEV protease in exactly the same manner as in a wildtype background. In a wildtype background, the fusion protein is released from chromatin within minutes (Figure 5A and B; Supplementary Movies S8 and S9). In sharp contrast, our new experiments clearly show that there is no release or turnover (measured by FRAP) of Smc3-Rad21-GFP seen in a wapl C204 mutant upon TEV protease injection (Figure 5C; Supplementary Movies S10 and S11). This indicates that separaseindependent cohesin release happens via opening of the Smc3/kleisin interface and that this process is mediated by the protein Wapl. This experiment also proves that Rad21-GFP is not released from Smc3 only by cleaving the linker between the two proteins, but that the process also depends on the cohesin-associated protein Wapl. Minor points: - page 7, "In the salivary glands, we found comparable levels of the fusion protein..." It is not clear what levels are being compared. Please clarify. In the salivary glands there seems to be a huge overexpression of the transgene with respect to endogenous protein. Yes, the Smc3-Rad21-GFP protein is indeed overexpressed in salivary glands as compared to the endogenous level at the time and condition at which the experiment was carried out and we therefore corrected the text accordingly. - In the last experiment regarding the prophase pathway (Figure 6D), the authors indicate "cohesin containing an intact Smc3-Rad21-GFP fusion protein persisted...until the onset of anaphase." Why is there not a movie showing this? We had many attempts to analyse the behavior of the fusion in live brains, however getting highquality time-lapse movies is technically very challenging due to low expression or incorporation of Smc3-Rad21-GFP in addition to the endogenous cohesin complex. This difficulty may result from the fact that there is not a constant turnover of cohesin in neuroblasts as compared to salivary glands where the Smc3-Rad21-GFP fusion protein is highly expressed, rapidly loaded onto chromatin and presumably replaces most of the dynamic endogenous cohesin. In order to get solid expression and chromatin binding of the fusion protein, we were therefore using conditions which do not allow time-lapse/live imaging analysis. Namely, we carried out heat-shock induction and recovery in live animals, thus ensuring best culture conditions. Afterwards, we dissected and analyzed brain tissues and imaged neuroblasts, which have undergone nuclear envelope breakdown but not metaphase-toanaphase transition. We also used poly-lysine-coated slides to improve the still imaging quality, which again is of disadvantage for live imaging as it interferes with proper cell divisions. Due to these experimental limitations, we changed the text accordingly to soften our conclusion to: This revealed that cohesin containing an intact Smc3-Rad21-GFP fusion protein persisted on chromosome arms after nuclear envelope breakdown. Additional suggestions: - In the Introduction: - The prophase pathway was initially identified in Xenopus (Losada 1998) and the involvement of mitotic kinases was also first described by Sumara 2002 and Losada The citations were added accordingly in the introduction of our revised manuscript. - In page 3 of Introduction, we read "Wapl is recruited to cohesin by binding...pds5, which..." and European Molecular Biology Organization 6
7 Chan KL, personal communication is cited. I do not think this is adequate for the Introduction. Maybe for the first part of the sentence Chan et al (2012) should be cited instead, although it should be also mentioned that the requirement of Pds5 for Wapl recruitment does not exist in Xenopus (Shintomi 2009). We changed the text accordingly in our revised manuscript. - Results By the end of the first paragraph in page 9, Kueng 2006 should be cited: In that study it was already shown by live cell imaging in HeLa cells that despite failure of the prophase pathway in Wapl sirna cells, all cohesin was released in anaphase. We changed the text accordingly in our revised manuscript. - Figure Legends. Although I am no fan of long Figure legends, I would encourage the authors to provide some more info in Figures 5 and 6 to allow the reader to understand the figure independently of the main text. Also, magnification bars should be added. We changed the figure legends accordingly in our revised manuscript. Referee #2 (Remarks to the Author): Experiments in yeast have suggested that cohesin undergoes DNA entrapment and release dynamically, through transient opening of Smc1/3 interface and Smc3/alpha-kleisin (Scc1) interface, respectively. It is well known that DNA is released from cohesin by the proteolytic cleavage of kleisin upon anaphase onset, but the proteolysis-independent dissociation of cohesin in prophase/prometaphase through what it is called prophase pathway is not well understood. This paper eloquently addresses the long-standing question of how cohesin is released nonproteolytically. The authors made use of live cell imaging analyses in Drosophila non-dividing salivary gland cells and showed that cohesin turnover in these interphase cells, which required proficient Wapl. Thus, in Wapl mutant cells, cohesin over-enriched at chromosomal loci revealed characteristic structures, but artificial cleavage of kleisin Rad21 caused immediate dissociation of cohesin from chromatin. This finding significantly implied that cohesins that are not conferring cohesion during interphase associates to chromosomes in a topological manner. To my knowledge this is the first demonstration that cohesin association is basically topological. The authors then tested the idea that DNA might escape from the cohesin ring through the transient disconnection of the Smc3-Rad21 interaction, in a manner depending on Wapl, the hypothesis based on yeast works. To address this the authors tethered Smc3 and Rad21 heads by short polypeptide links, which can be artificially cleaved by TEV protease. Cohesin ring with Smc3-Rad21 fusion caused over-enrichment of the complex on chromatin, as seen in Wapl mutant, but this was released by cleaving the tethering peptide. Crucially, the authors described that not only on polytene chromosomes in salivary gland cells but also in prophase in neuroblasts, Wapl-promoting cohesin release is blocked by fusing Smc3 and kleisin. Based on these results the authors concluded that proteolysis-independent release of cohesin from chromatin is universally mediated by escape of DNA through Smc3/Rad21 gate. I found the paper provide important results indicating how cohesin complexes are associated with chromatin and how most of them are released before anaphase. It is true that similar lines of conclusions are already drawn in yeast, nevertheless I do see that the current study goes beyond the yeast studies, as explicitly described in the discussion. It is a significant step toward our understanding of cohesin regulation and clearly deserves for the publication. That said, I have several concerns that might taken into consideration, which should be addressed without much difficulty. 1. Provided that Wapl-promoted release of chromatin involves DNA's escape through the Smc3- kleisin gate, I am curious to know if and to which extent the TEV-induced release of Smc3-Rad21- GFP from polytene chromosomes depends on Wapl in this experimental setting (Figure 5). If the release is primarily driven by Wapl, what one would expect to see in a Wapl mutant background is a marked enrichment of Smc3-Rad21-GFP on chromosomes, which do not grossly affected after European Molecular Biology Organization 7
8 microinjecting the active TEV. Is it the case? These results would be in sharp contrast to the experiment cleaving Rad21 (Figure 2E). This point raises the same question as referee 1 (see before) and we agree that this is a very important biological question that can be addressed using our expertise. We therefore set up a fly cross that generates progeny, which expresses the Smc3-Rad21-GFP protein upon heat-shock in a wapl C204 mutant background. We injected salivary gland cells with TEV protease in exactly the same manner as in a wildtype background. In a wildtype background, the fusion protein is released from chromatin within minutes (Figure 5A and B; Supplementary Movies S8 and S9). In sharp contrast, our new experiments clearly show that there is no release or turnover (measured by FRAP) of Smc3-Rad21-GFP seen in a wapl C204 mutant upon TEV protease injection (Figure 5C; Supplementary Movies S10 and S11). This indicates that separase-independent cohesin release happens via opening of the Smc3/kleisin interface and that this process is mediated by the protein Wapl. This experiment also proves that Rad21-GFP is not released from Smc3 only by cleaving the linker between the two proteins, but that the process also depends on the cohesin-associated protein Wapl. Moreover, it seem that Wapl-dependent release affects the entire cohesin population in Drosophila salivary glands. 2. Smc3-Rad21-GFP fusion protein is found to accumulate at chromosomal loci that result in emergence of bands or circular structure appearance (Figure 3D). To estimate the net effect of the tethering of Smc3 and Rad21, it is informative to show side-by-side the picture of Smc3-Rad21-GFP when TEV was co-expressed. As for the Western blots in Figure 3, antibodies used for the analyses are missing. In the blots in Figure 3C, is it possible to explain why endogenous Rad21 band is not detectable in Smc-Rad21-GFP lanes? The net effect of tethering Smc3 and Rad21 compared to its co-expression with TEV protease is basically shown in Figure 4A upper and lower panel. The key difference between the two conditions is the observation of less pronounced circular structures regarding the cohesin localization pattern. We added a description of antibodies in Figure legend 3 as well as in the methods part in our revised manuscript. We can only speculate on why the endogenous Rad21 levels may be lower in the Smc3-Rad21-GFP without TEV co-expression as compared to co-expressed TEV. Given our newly added experiments (Cleavage of the linker in the Smc3-Rad21-GFP fusion protein by TEV injection and Coimmunoprecipitation of Smc3-Rad21-GFP), we strongly believe that the Smc3-Rad21-GFP protein occupies most of the possible cohesin binding sites (without turning over) and that endogenous Rad21 remains mainly in the soluble pool where it could be more prone to degradation. 3. FRAP analysis in Figure 4 shows the turnover rate of Smc3-Rad21-GFP with our without the Smc3/Rad21 tethering. The authors' interpretation of the data for +TEV kinetics is "similar to wild type", but the fluorescence recovery seems to be consistently lower (60-70% of RFI) than that of wild type which is shown in Figure 2C. It seems to imply that there is more stably bound fraction of non-tethered Smc3-Rad21-GFP than Rad21-GFP. What could be a possible explanation for this? My suggestion is to compare the half-recovery time here instead, and say it is similar to wild-type, if that is the case. The expression similar to wild-type may indeed have been not accurate and therefore misleading. The only main point we wanted to make here is the observation that co-expression of TEV together with the Smc3-Rad21-GFP fusion leads to a very significant portion of fluorescence recovery (around 70% of RFI). The fact that recovery does not go back to the level seen in a wild-type (Figure 1C) can have several reasons: 1) TEV protease may need time to access and cleave the linker, 2) TEV protease concentration is lower compared to TEV injected cells, 3) Some fusion protein is present in a non-cleaved version which presumably represents a fraction of around 20 % as indicated by Western blot (Figure 3C). 4. The supplementary movie data in neuroblast provide a unique opportunity to show that the dissociation of cohesin from chromosome arms in prophase and from centromeres in anaphase, and that in Wapl mutants considerable amount of cohesin remains throughout the chromosome lengths European Molecular Biology Organization 8
9 until anaphase (Movie S10 and S11). It will be helpful to additionally provide the movie data for Smc3-Rad21-GFP with or without TEV, because it is difficult to tell that cohesin remains on arms solely from a still image (Figure 6D). For more comprehensive presentation for Figure 6D, drawings of schematic illustration may help. A similar point has also been raised by referee 1 (see before). We indeed had many attempts to analyse the behavior of the fusion in live brains, however getting high-quality time-lapse movies is technically very challenging due to low expression or incorporation of Smc3-Rad21-GFP in addition to the endogenous cohesin. This difficulty may result from the fact that there is not a constant turnover of cohesin in neuroblasts as compared to salivary glands where the Smc3-Rad21-GFP fusion protein is highly expressed, rapidly loaded onto chromatin and presumably replaces most of the dynamic endogenous cohesin. In order to get solid expression and chromatin binding of the fusion protein, we were therefore using conditions which do not allow time-lapse/live imaging analysis. Namely, we carried out heat-shock induction and recovery in live animals, thus ensuring best culture conditions. Afterwards, we dissected and analyzed brain tissues and imaged neuroblasts, which have undergone nuclear envelope breakdown but not metaphase-to-anaphase transition. We also used poly-lysine-coated slides to improve the still imaging quality, we again is of disadvantage for live imaging as it interferes with proper cell divisions. Due to these experimental limitations, we changed the text accordingly to soften our conclusion to: This revealed that cohesin containing an intact Smc3-Rad21-GFP fusion protein persisted on chromosome arms after nuclear envelope breakdown. 5. The paper provides mechanistic explanation for the first time for how prophase pathway might work to release cohesin from chromosomes. It has long been known that the prophase pathway involves activity of mitotic kinases such as Plk1 and phosphorylation of Scc3/SA2. Therefore a perspective view for how these signals might contribute to promote Wapl-mediated cohesin release would be interesting to discuss. We have added a comment in the discussion on how SA/Scc3-P and Plk1 may contribute to the Wapl-mediated cohesin release in prophase and changed text accordingly in our revised manuscript. Acceptance letter 12 December 2012 Thank you for submitting your revised manuscript for our consideration. I have now had a chance to look through it and your responses, and referee 1 has also taken another look at the study and found their concerns satisfactorily addressed. I am therefore happy to inform you that we have decided to accept your manuscript for publication in The EMBO Journal at this stage. Referee #1 (Remarks to the Author) I read the revised manuscript and the rebuttal letter. I am satisfied with the revisions made and the additional data included so from my part it is OK to publish it. European Molecular Biology Organization 9
Disengaging the Smc3/kleisin interface releases cohesin. from Drosophilachromosomes during interphase and mitosis
 Disengaging the Smc3/kleisin interface releases cohesin from Drosophilachromosomes during interphase and mitosis Christian S Eichinger 1*, Alexandre Kurze 1*, Raquel A Oliveira 1 and Kim Nasmyth 1, a a
Disengaging the Smc3/kleisin interface releases cohesin from Drosophilachromosomes during interphase and mitosis Christian S Eichinger 1*, Alexandre Kurze 1*, Raquel A Oliveira 1 and Kim Nasmyth 1, a a
hsssu72 phosphatase is a cohesin-binding protein that regulates the resolution of sister chromatid arm cohesion
 Manuscript EMBO-2010-74043 hsssu72 phosphatase is a cohesin-binding protein that regulates the resolution of sister chromatid arm cohesion Hyun-Soo Kim, Kwan-Hyuck Baek, Geun-Hyoung Ha, Jae-Chul Lee, Yu-Na
Manuscript EMBO-2010-74043 hsssu72 phosphatase is a cohesin-binding protein that regulates the resolution of sister chromatid arm cohesion Hyun-Soo Kim, Kwan-Hyuck Baek, Geun-Hyoung Ha, Jae-Chul Lee, Yu-Na
Mammalian EGF receptor activation by the rhomboid protease RHBDL2
 Manuscript EMBOR-2011-34771 Mammalian EGF receptor activation by the rhomboid protease RHBDL2 Colin Adrain, Kvido Strisovsky, Markus Zettl, Landian Hu, Marius K. Lemberg, Matthew Freeman Corresponding
Manuscript EMBOR-2011-34771 Mammalian EGF receptor activation by the rhomboid protease RHBDL2 Colin Adrain, Kvido Strisovsky, Markus Zettl, Landian Hu, Marius K. Lemberg, Matthew Freeman Corresponding
7.06 Problem Set Four, 2006
 7.06 Problem Set Four, 2006 1. Explain the molecular mechanism behind each of the following events that occur during the cell cycle, making sure to discuss specific proteins that are involved in making
7.06 Problem Set Four, 2006 1. Explain the molecular mechanism behind each of the following events that occur during the cell cycle, making sure to discuss specific proteins that are involved in making
7.06 Cell Biology EXAM #2 March 20, 2003
 7.06 Cell Biology EXAM #2 March 20, 2003 This is an open book exam, and you are allowed access to books, a calculator, and notes but not computers or any other types of electronic devices. Please write
7.06 Cell Biology EXAM #2 March 20, 2003 This is an open book exam, and you are allowed access to books, a calculator, and notes but not computers or any other types of electronic devices. Please write
3'UTR elements inhibit Ras-induced C/EBPß posttranslational activation and senescence in tumor cells
 Manuscript EMBO-2011-77536 3'UTR elements inhibit Ras-induced C/EBPß posttranslational activation and senescence in tumor cells Sandip K. Basu, Radek Malik, Christopher J. Huggins, Sook Lee, Thomas Sebastian,
Manuscript EMBO-2011-77536 3'UTR elements inhibit Ras-induced C/EBPß posttranslational activation and senescence in tumor cells Sandip K. Basu, Radek Malik, Christopher J. Huggins, Sook Lee, Thomas Sebastian,
R-loops cause replication impairment and genome instability during meiosis
 Manuscript EMBOR-2012-35976 R-loops cause replication impairment and genome instability during meiosis Maikel Castellano-Pozo, Tatiana Garcia-Muse and Andres Aguilera Corresponding author: Andres Aguilera,
Manuscript EMBOR-2012-35976 R-loops cause replication impairment and genome instability during meiosis Maikel Castellano-Pozo, Tatiana Garcia-Muse and Andres Aguilera Corresponding author: Andres Aguilera,
NuMA phosphorylation by CDK1 couples mitotic progression with cortical dynein function
 Manuscript EMBO-2013-85125 NuMA phosphorylation by CDK1 couples mitotic progression with cortical dynein function Sachin Kotak, Coralie Busso, Pierre Gönczy Corresponding author: Pierre Gönczy, ISREC Review
Manuscript EMBO-2013-85125 NuMA phosphorylation by CDK1 couples mitotic progression with cortical dynein function Sachin Kotak, Coralie Busso, Pierre Gönczy Corresponding author: Pierre Gönczy, ISREC Review
Plant virus mediated induction of mir168 is associated with repression of ARGONAUTE1 accumulation
 Manuscript EMBO-2010-73919 Plant virus mediated induction of mir168 is associated with repression of ARGONAUTE1 accumulation Éva Várallyay, Anna Válóczi, Ákos Ágyi, József Burgyán and Zoltan Havelda Corresponding
Manuscript EMBO-2010-73919 Plant virus mediated induction of mir168 is associated with repression of ARGONAUTE1 accumulation Éva Várallyay, Anna Válóczi, Ákos Ágyi, József Burgyán and Zoltan Havelda Corresponding
Section 10. Junaid Malek, M.D.
 Section 10 Junaid Malek, M.D. Cell Division Make sure you understand: How do cells know when to divide? (What drives the cell cycle? Why is it important to regulate this?) How is DNA replication regulated?
Section 10 Junaid Malek, M.D. Cell Division Make sure you understand: How do cells know when to divide? (What drives the cell cycle? Why is it important to regulate this?) How is DNA replication regulated?
Supplemental Information. Plk1/Polo Phosphorylates Sas-4 at the Onset. of Mitosis for an Efficient Recruitment
 Cell Reports, Volume 25 Supplemental Information Plk1/Polo Phosphorylates Sas-4 at the Onset of Mitosis for an Efficient Recruitment of Pericentriolar Material to Centrosomes Anand Ramani, Aruljothi Mariappan,
Cell Reports, Volume 25 Supplemental Information Plk1/Polo Phosphorylates Sas-4 at the Onset of Mitosis for an Efficient Recruitment of Pericentriolar Material to Centrosomes Anand Ramani, Aruljothi Mariappan,
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
Supplementary Figure 1. α-synuclein is truncated in PD and LBD brains. Nature Structural & Molecular Biology: doi: /nsmb.
 Supplementary Figure 1 α-synuclein is truncated in PD and LBD brains. (a) Specificity of anti-n103 antibody. Anti-N103 antibody was coated on an ELISA plate and different concentrations of full-length
Supplementary Figure 1 α-synuclein is truncated in PD and LBD brains. (a) Specificity of anti-n103 antibody. Anti-N103 antibody was coated on an ELISA plate and different concentrations of full-length
Pretaporter, a Drosophila protein serving as a ligand for Draper in the phagocytosis of apoptotic cells
 Manuscript EMBO-2009-71518 Pretaporter, a Drosophila protein serving as a ligand for Draper in the phagocytosis of apoptotic cells Takayuki Kuraishi, Yukiko Nakagawa, Kaz Nagaosa, Yumi Hashimoto, Takashi
Manuscript EMBO-2009-71518 Pretaporter, a Drosophila protein serving as a ligand for Draper in the phagocytosis of apoptotic cells Takayuki Kuraishi, Yukiko Nakagawa, Kaz Nagaosa, Yumi Hashimoto, Takashi
Figure S1, related to Figure 1. Characterization of biosensor behavior in vivo.
 Developmental Cell, Volume 23 Supplemental Information Separase Biosensor Reveals that Cohesin Cleavage Timing Depends on Phosphatase PP2A Cdc55 Regulation Gilad Yaakov, Kurt Thorn, and David O. Morgan
Developmental Cell, Volume 23 Supplemental Information Separase Biosensor Reveals that Cohesin Cleavage Timing Depends on Phosphatase PP2A Cdc55 Regulation Gilad Yaakov, Kurt Thorn, and David O. Morgan
Nucleophosmin Deposition During mrna 3' End Processing Influences Poly(A) Tail Length
 Manuscript EMBO-2011-77995 Nucleophosmin Deposition During mrna 3' End Processing Influences Poly(A) Tail Length Fumihiko Sagawa, Hend Ibrahim, Angela L. Morrison, Carol J. Wilusz and Jeffrey Wilusz Corresponding
Manuscript EMBO-2011-77995 Nucleophosmin Deposition During mrna 3' End Processing Influences Poly(A) Tail Length Fumihiko Sagawa, Hend Ibrahim, Angela L. Morrison, Carol J. Wilusz and Jeffrey Wilusz Corresponding
Nature Structural & Molecular Biology: doi: /nsmb.1583
 Acetylation by GCN5 regulates CDC6 phosphorylation in the S-phase of the cell cycle Roberta Paolinelli 1,2, Ramiro Mendoza-Maldonado 2, Anna Cereseto 1 and Mauro Giacca 2 1 Molecular Biology Laboratory,
Acetylation by GCN5 regulates CDC6 phosphorylation in the S-phase of the cell cycle Roberta Paolinelli 1,2, Ramiro Mendoza-Maldonado 2, Anna Cereseto 1 and Mauro Giacca 2 1 Molecular Biology Laboratory,
PEER REVIEW FILE. Reviewers' Comments: Reviewer #2 (Remarks to the Author):
 Editorial Note: This manuscript has been previously reviewed at another journal that is not operating a transparent peer review scheme. This document only contains reviewer comments and rebuttal letters
Editorial Note: This manuscript has been previously reviewed at another journal that is not operating a transparent peer review scheme. This document only contains reviewer comments and rebuttal letters
Cell Cycle. Trends in Cell Biology
 Cell Cycle Trends in Cell Biology Cyclic proteolysis Two distinct enzyme complexes proteolysis of cyclin-cdk complexes SCF (Skp1-Cul1-F box) Ubiquitylation and destruction of G1/S-cyclins and CKI proteins
Cell Cycle Trends in Cell Biology Cyclic proteolysis Two distinct enzyme complexes proteolysis of cyclin-cdk complexes SCF (Skp1-Cul1-F box) Ubiquitylation and destruction of G1/S-cyclins and CKI proteins
The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit
 Cell Reports, Volume 5 Supplemental Information The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit Andrey Poleshko, Katelyn M. Mansfield, Caroline
Cell Reports, Volume 5 Supplemental Information The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit Andrey Poleshko, Katelyn M. Mansfield, Caroline
Centromere-Independent Accumulation of Cohesin at Ectopic Heterochromatin Sites Induces Chromosome Stretching during Anaphase
 Centromere-Independent Accumulation of Cohesin at Ectopic Heterochromatin Sites Induces Chromosome Stretching during Anaphase Raquel A. Oliveira 1,2. *, Shaila Kotadia 3., Alexandra Tavares 1, Mihailo
Centromere-Independent Accumulation of Cohesin at Ectopic Heterochromatin Sites Induces Chromosome Stretching during Anaphase Raquel A. Oliveira 1,2. *, Shaila Kotadia 3., Alexandra Tavares 1, Mihailo
Practice Problems 5. Location of LSA-GFP fluorescence
 Life Sciences 1a Practice Problems 5 1. Soluble proteins that are normally secreted from the cell into the extracellular environment must go through a series of steps referred to as the secretory pathway.
Life Sciences 1a Practice Problems 5 1. Soluble proteins that are normally secreted from the cell into the extracellular environment must go through a series of steps referred to as the secretory pathway.
7.06 Problem Set #3, 2006
 7.06 Problem Set #3, 2006 1. You are studying the EGF/Ras/MAPK pathway in cultured cells. When the pathway is activated, cells are signaled to proliferate. You generate various mutants described below.
7.06 Problem Set #3, 2006 1. You are studying the EGF/Ras/MAPK pathway in cultured cells. When the pathway is activated, cells are signaled to proliferate. You generate various mutants described below.
Supplementary Figure Legends
 Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Ono et al., http://www.jcb.org/cgi/content/full/jcb.201208008/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Chromatin-binding properties of MCM2 and condensin II during
Supplemental material Ono et al., http://www.jcb.org/cgi/content/full/jcb.201208008/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Chromatin-binding properties of MCM2 and condensin II during
Efficient translation initiation dictates codon usage at gene start in bacteria
 Molecular Systems Biology Peer Review Process File Efficient translation initiation dictates codon usage at gene start in bacteria Kajetan Bentele, Paul Saffert, Robert Rauscher, Zoya Ignatova, Nils Blüthgen
Molecular Systems Biology Peer Review Process File Efficient translation initiation dictates codon usage at gene start in bacteria Kajetan Bentele, Paul Saffert, Robert Rauscher, Zoya Ignatova, Nils Blüthgen
3.1 The Role of the Enzymatic Activity of Sir2 for Efficient. The Sir proteins are recruited to DNA by site-specific DNA-binding proteins and
 35 3 RESULTS 3.1 The Role of the Enzymatic Activity of Sir2 for Efficient Association of the SIR Complex with DNA The Sir proteins are recruited to DNA by site-specific DNA-binding proteins and subsequently
35 3 RESULTS 3.1 The Role of the Enzymatic Activity of Sir2 for Efficient Association of the SIR Complex with DNA The Sir proteins are recruited to DNA by site-specific DNA-binding proteins and subsequently
Genetics Lecture Notes Lectures 6 9
 Genetics Lecture Notes 7.03 2005 Lectures 6 9 Lecture 6 Until now our analysis of genes has focused on gene function as determined by phenotype differences brought about by different alleles or by a direct
Genetics Lecture Notes 7.03 2005 Lectures 6 9 Lecture 6 Until now our analysis of genes has focused on gene function as determined by phenotype differences brought about by different alleles or by a direct
6.2 Chromatin is divided into euchromatin and heterochromatin
 6.2 Chromatin is divided into euchromatin and heterochromatin Individual chromosomes can be seen only during mitosis. During interphase, the general mass of chromatin is in the form of euchromatin. Euchromatin
6.2 Chromatin is divided into euchromatin and heterochromatin Individual chromosomes can be seen only during mitosis. During interphase, the general mass of chromatin is in the form of euchromatin. Euchromatin
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the
 Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
Revision Checklist for Science Signaling Research Manuscripts: Data Requirements and Style Guidelines
 Revision Checklist for Science Signaling Research Manuscripts: Data Requirements and Style Guidelines Further information can be found at: http://stke.sciencemag.org/sites/default/files/researcharticlerevmsinstructions_0.pdf.
Revision Checklist for Science Signaling Research Manuscripts: Data Requirements and Style Guidelines Further information can be found at: http://stke.sciencemag.org/sites/default/files/researcharticlerevmsinstructions_0.pdf.
Materials and Methods: All strains were derivatives of SK1 and are listed in Supplemental Table 1.
 Supplemental Online Material: Materials and Methods: All strains were derivatives of SK1 and are listed in Supplemental Table 1. Constructs: pclb2-3ha-cdc5 and pclb2-3ha-cdc2 strains were constructed by
Supplemental Online Material: Materials and Methods: All strains were derivatives of SK1 and are listed in Supplemental Table 1. Constructs: pclb2-3ha-cdc5 and pclb2-3ha-cdc2 strains were constructed by
The mechanism of translation initiation on Aichi Virus RNA mediated by a novel type of picornavirus IRES
 Manuscript EMBO-2011-78346 The mechanism of translation initiation on Aichi Virus RNA mediated by a novel type of picornavirus IRES Yingpu Yu, Trevor R. Sweeney, Panagiota Kafasla, Richard J. Jackson,
Manuscript EMBO-2011-78346 The mechanism of translation initiation on Aichi Virus RNA mediated by a novel type of picornavirus IRES Yingpu Yu, Trevor R. Sweeney, Panagiota Kafasla, Richard J. Jackson,
Coleman et al., Supplementary Figure 1
 Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Post-translational modification
 Protein expression Western blotting, is a widely used and accepted technique to detect levels of protein expression in a cell or tissue extract. This technique measures protein levels in a biological sample
Protein expression Western blotting, is a widely used and accepted technique to detect levels of protein expression in a cell or tissue extract. This technique measures protein levels in a biological sample
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11070 Supplementary Figure 1 Purification of FLAG-tagged proteins. a, Purification of FLAG-RNF12 by FLAG-affinity from nuclear extracts of wild-type (WT) and two FLAG- RNF12 transgenic
doi:10.1038/nature11070 Supplementary Figure 1 Purification of FLAG-tagged proteins. a, Purification of FLAG-RNF12 by FLAG-affinity from nuclear extracts of wild-type (WT) and two FLAG- RNF12 transgenic
Structure of a novel phosphotyrosine-binding domain in Hakai that targets E-cadherin
 Manuscript EMBO-2011-79235 Structure of a novel phosphotyrosine-binding domain in Hakai that targets E-cadherin Manjeet Mukherjee, Soah Yee Chow, Permeen Yusoff, Jayaraman Seetharaman, Cherlyn Ng, Saravanan
Manuscript EMBO-2011-79235 Structure of a novel phosphotyrosine-binding domain in Hakai that targets E-cadherin Manjeet Mukherjee, Soah Yee Chow, Permeen Yusoff, Jayaraman Seetharaman, Cherlyn Ng, Saravanan
Jiali Li, Sukhvir P Mahal, Cheryl A Demczyk and Charles Weissmann
 Manuscript EMBOR-2011-35144 Mutability of prions Jiali Li, Sukhvir P Mahal, Cheryl A Demczyk and Charles Weissmann Corresponding author: Charles Weissmann, Scripps Florida Review timeline: Submission date:
Manuscript EMBOR-2011-35144 Mutability of prions Jiali Li, Sukhvir P Mahal, Cheryl A Demczyk and Charles Weissmann Corresponding author: Charles Weissmann, Scripps Florida Review timeline: Submission date:
Supplementary Information for. Regulation of Rev1 by the Fanconi Anemia Core Complex
 Supplementary Information for Regulation of Rev1 by the Fanconi Anemia Core Complex Hyungjin Kim, Kailin Yang, Donniphat Dejsuphong, Alan D. D Andrea* *Corresponding Author: Alan D. D Andrea, M.D. Alan_dandrea@dfci.harvard.edu
Supplementary Information for Regulation of Rev1 by the Fanconi Anemia Core Complex Hyungjin Kim, Kailin Yang, Donniphat Dejsuphong, Alan D. D Andrea* *Corresponding Author: Alan D. D Andrea, M.D. Alan_dandrea@dfci.harvard.edu
Learning Objectives. Define RNA interference. Define basic terminology. Describe molecular mechanism. Define VSP and relevance
 Learning Objectives Define RNA interference Define basic terminology Describe molecular mechanism Define VSP and relevance Describe role of RNAi in antigenic variation A Nobel Way to Regulate Gene Expression
Learning Objectives Define RNA interference Define basic terminology Describe molecular mechanism Define VSP and relevance Describe role of RNAi in antigenic variation A Nobel Way to Regulate Gene Expression
Chromosome Structural Analysis
 Chromosome Structural Analysis A Practical Approach Edited by WENDY A. BICKMORE Cell Genetics Section, MRC Human Genetics Unit, Western General Hospital, Edinburgh OXJORD UNIVERSITY PRESS 1999 List of
Chromosome Structural Analysis A Practical Approach Edited by WENDY A. BICKMORE Cell Genetics Section, MRC Human Genetics Unit, Western General Hospital, Edinburgh OXJORD UNIVERSITY PRESS 1999 List of
Sororin Mediates Sister Chromatid Cohesion by Antagonizing Wapl
 Sororin Mediates Sister Chromatid Cohesion by Antagonizing Wapl Tomoko Nishiyama, 1 Rene Ladurner, 1 Julia Schmitz, 1,4 Emanuel Kreidl, 1 Alexander Schleiffer, 1 Venugopal Bhaskara, 1 Masashige Bando,
Sororin Mediates Sister Chromatid Cohesion by Antagonizing Wapl Tomoko Nishiyama, 1 Rene Ladurner, 1 Julia Schmitz, 1,4 Emanuel Kreidl, 1 Alexander Schleiffer, 1 Venugopal Bhaskara, 1 Masashige Bando,
Structural diversity and dynamics of genomic replication origins in Schizosaccharomyces pombe
 Manuscript EMBO-2009-71947 Structural diversity and dynamics of genomic replication origins in Schizosaccharomyces pombe Cristina Cotobal, Monica Segurado and Francisco Antequera Corresponding author:
Manuscript EMBO-2009-71947 Structural diversity and dynamics of genomic replication origins in Schizosaccharomyces pombe Cristina Cotobal, Monica Segurado and Francisco Antequera Corresponding author:
Mechanism of transcriptional repression at a bacterial promoter by analysis of single molecules
 Manuscript EMBO-2011-77282 Mechanism of transcriptional repression at a bacterial promoter by analysis of single molecules Alvaro Sanchez, Melisa L. Osborne, Larry J. Friedman, Jane Kondev and Jeff Gelles
Manuscript EMBO-2011-77282 Mechanism of transcriptional repression at a bacterial promoter by analysis of single molecules Alvaro Sanchez, Melisa L. Osborne, Larry J. Friedman, Jane Kondev and Jeff Gelles
Cell Division. Use Target Reading Skills. This section explains how cells grow and divide.
 Name Date Class Cell Processes Guided Reading and Study Cell Division This section explains how cells grow and divide. Use Target Reading Skills As you read, make a cycle diagram that shows the events
Name Date Class Cell Processes Guided Reading and Study Cell Division This section explains how cells grow and divide. Use Target Reading Skills As you read, make a cycle diagram that shows the events
ksierzputowska.com Research Title: Using novel TALEN technology to engineer precise mutations in the genome of D. melanogaster
 Research Title: Using novel TALEN technology to engineer precise mutations in the genome of D. melanogaster Research plan: Specific aims: 1. To successfully engineer transgenic Drosophila expressing TALENs
Research Title: Using novel TALEN technology to engineer precise mutations in the genome of D. melanogaster Research plan: Specific aims: 1. To successfully engineer transgenic Drosophila expressing TALENs
transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected with the wwp-luc reporter, and FLAG-tagged FHL1,
 Supplementary Data Supplementary Figure Legends Supplementary Figure 1 FHL-mediated TGFβ-responsive reporter transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected
Supplementary Data Supplementary Figure Legends Supplementary Figure 1 FHL-mediated TGFβ-responsive reporter transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected
I have carefully read all reviewers' comments and believe that the authors have addressed most of them adequately.
 Response to reviewers Reviewer #1 (Remarks to the Author): I have carefully read all reviewers' comments and believe that the authors have addressed most of them adequately. We thank reviewer 1 for this
Response to reviewers Reviewer #1 (Remarks to the Author): I have carefully read all reviewers' comments and believe that the authors have addressed most of them adequately. We thank reviewer 1 for this
Time-scales and bottlenecks in mirna-dependent gene regulation.
 Time-scales and bottlenecks in mirna-dependent gene regulation. Jean Hausser, Afzal Pasha Syed, Nathalie Selevsek, Erik van Nimwegen, Lukasz Jaskiewicz, Ruedi Aebersold and Mihaela Zavolan Corresponding
Time-scales and bottlenecks in mirna-dependent gene regulation. Jean Hausser, Afzal Pasha Syed, Nathalie Selevsek, Erik van Nimwegen, Lukasz Jaskiewicz, Ruedi Aebersold and Mihaela Zavolan Corresponding
Stargazin regulates AMPA receptor trafficking through adaptor protein. complexes during long term depression
 Supplementary Information Stargazin regulates AMPA receptor trafficking through adaptor protein complexes during long term depression Shinji Matsuda, Wataru Kakegawa, Timotheus Budisantoso, Toshihiro Nomura,
Supplementary Information Stargazin regulates AMPA receptor trafficking through adaptor protein complexes during long term depression Shinji Matsuda, Wataru Kakegawa, Timotheus Budisantoso, Toshihiro Nomura,
This is the author's accepted version of the manuscript.
 This is the author's accepted version of the manuscript. The definitive version is published in Nature Communications Online Edition: 2015/4/16 (Japan time), doi:10.1038/ncomms7780. The final version published
This is the author's accepted version of the manuscript. The definitive version is published in Nature Communications Online Edition: 2015/4/16 (Japan time), doi:10.1038/ncomms7780. The final version published
IKK and NF- B-mediated regulation of Claspin impacts on ATR checkpoint function
 Manuscript EMBO-2010-73914 IKK and NF- B-mediated regulation of Claspin impacts on ATR checkpoint function Kenneth NS, Mudie S and Rocha S Corresponding author: Sonia Rocha, Dundee, University of Review
Manuscript EMBO-2010-73914 IKK and NF- B-mediated regulation of Claspin impacts on ATR checkpoint function Kenneth NS, Mudie S and Rocha S Corresponding author: Sonia Rocha, Dundee, University of Review
A mitochondrial phosphatase required for cardiolipin biosynthesis: the PGP phosphatase Gep4
 Manuscript EMBO-2010-74019 A mitochondrial phosphatase required for cardiolipin biosynthesis: the PGP phosphatase Gep4 Christof Osman, Mathias Haag, Britta Brügger, Felix Wieland, Thomas Langer Corresponding
Manuscript EMBO-2010-74019 A mitochondrial phosphatase required for cardiolipin biosynthesis: the PGP phosphatase Gep4 Christof Osman, Mathias Haag, Britta Brügger, Felix Wieland, Thomas Langer Corresponding
fga phenotypes were also analyzed in the tracheal system, the organ responsible for
 SUPPLEMENTARY TEXT Tracheal defects fga phenotypes were also analyzed in the tracheal system, the organ responsible for oxygen delivery in the fly. In fga homozygous mutant alleles, we observed defects
SUPPLEMENTARY TEXT Tracheal defects fga phenotypes were also analyzed in the tracheal system, the organ responsible for oxygen delivery in the fly. In fga homozygous mutant alleles, we observed defects
Figure legends for supplement
 Figure legends for supplement Supplemental Figure 1 Characterization of purified and recombinant proteins Relevant fractions related the final stage of the purification protocol(bingham et al., 1998; Toba
Figure legends for supplement Supplemental Figure 1 Characterization of purified and recombinant proteins Relevant fractions related the final stage of the purification protocol(bingham et al., 1998; Toba
An in silico mathematical model of the initiation of DNA replication
 An in silico mathematical model of the initiation of DNA replication Rohan D. Gidvani a1, Brendan J. McConkey a, Bernard P. Duncker a and Brian P. Ingalls a,b a Department of Biology, University of Waterloo,
An in silico mathematical model of the initiation of DNA replication Rohan D. Gidvani a1, Brendan J. McConkey a, Bernard P. Duncker a and Brian P. Ingalls a,b a Department of Biology, University of Waterloo,
A RRM1 H2AX DAPI. RRM1 H2AX DAPI Merge. Cont. sirna RRM1
 A H2AX DAPI H2AX DAPI Merge Cont sirna Figure S1: Accumulation of RRM1 at DNA damage sites (A) HeLa cells were subjected to in situ detergent extraction without IR irradiation, and immunostained with the
A H2AX DAPI H2AX DAPI Merge Cont sirna Figure S1: Accumulation of RRM1 at DNA damage sites (A) HeLa cells were subjected to in situ detergent extraction without IR irradiation, and immunostained with the
The E3 ubiquitin ligase RNF114 and TAB1 degradation are required for maternal-to-zygotic transition
 Manuscript EMBO-2016-42573 The E3 ubiquitin ligase RNF114 and TAB1 degradation are required for maternal-to-zygotic transition Ye Yang, Cheng Zhou, Ying Wang, Weixiao Liu, Chao Liu, Liying Wang,Yujiao
Manuscript EMBO-2016-42573 The E3 ubiquitin ligase RNF114 and TAB1 degradation are required for maternal-to-zygotic transition Ye Yang, Cheng Zhou, Ying Wang, Weixiao Liu, Chao Liu, Liying Wang,Yujiao
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Detection of MCM-subunit SUMOylation under normal growth conditions. a. Sumoylated forms of MCM subunits show differential shifts when SUMO is attached to differently sized tags.
Supplementary Figure 1 Detection of MCM-subunit SUMOylation under normal growth conditions. a. Sumoylated forms of MCM subunits show differential shifts when SUMO is attached to differently sized tags.
Lecture 22 Eukaryotic Genes and Genomes III
 Lecture 22 Eukaryotic Genes and Genomes III In the last three lectures we have thought a lot about analyzing a regulatory system in S. cerevisiae, namely Gal regulation that involved a hand full of genes.
Lecture 22 Eukaryotic Genes and Genomes III In the last three lectures we have thought a lot about analyzing a regulatory system in S. cerevisiae, namely Gal regulation that involved a hand full of genes.
Dead end1 is an essential partner of NANOS2 for selective binding of target RNAs in male germ cell development
 Manuscript EMBO-2015-40828 Dead end1 is an essential partner of NANOS2 for selective binding of target RNAs in male germ cell development Atsushi Suzuki, Yuki Niimi, Kaori Shinmyozu, Zhi Zhou, Makoto Kiso,
Manuscript EMBO-2015-40828 Dead end1 is an essential partner of NANOS2 for selective binding of target RNAs in male germ cell development Atsushi Suzuki, Yuki Niimi, Kaori Shinmyozu, Zhi Zhou, Makoto Kiso,
Lecture 6. Chromosome Structure and Function in Mitosis
 Lecture 6 Chromosome Structure and Function in Mitosis Outline: Chromosome Organization and Function in Interphase Chromatin effects on replication Effects of nuclear organization on gene expression Chromosomes
Lecture 6 Chromosome Structure and Function in Mitosis Outline: Chromosome Organization and Function in Interphase Chromatin effects on replication Effects of nuclear organization on gene expression Chromosomes
Potent degradation of neuronal mirnas induced by highly complementary targets
 EMBO reports - Peer Review Process File - EMBOR-2015-40078 Manuscript EMBOR-2015-40078 Potent degradation of neuronal mirnas induced by highly complementary targets Manuel de la Mata, Dimos Gaidatzis,
EMBO reports - Peer Review Process File - EMBOR-2015-40078 Manuscript EMBOR-2015-40078 Potent degradation of neuronal mirnas induced by highly complementary targets Manuel de la Mata, Dimos Gaidatzis,
Immature small ribosomal subunits can engage in translation initiation in S. cerevisiae
 Manuscript EMBO-2009-70906 Immature small ribosomal subunits can engage in translation initiation in S. cerevisiae Julien Soudet, Kamila Belhabich-Baumas, Michèle Caizergues-Ferrer, Annie Mougin Corresponding
Manuscript EMBO-2009-70906 Immature small ribosomal subunits can engage in translation initiation in S. cerevisiae Julien Soudet, Kamila Belhabich-Baumas, Michèle Caizergues-Ferrer, Annie Mougin Corresponding
FANCJ promotes replication of G-quadruplex DNA structures
 Manuscript EMBO-2014-88663 FANCJ promotes replication of G-quadruplex DNA structures Pau Castillo Bosch, Sandra Segura-Bayona, Wouter Koole, Jane van Heteren, James Dewar, Marcel Tijsterman, Puck Knipscheer
Manuscript EMBO-2014-88663 FANCJ promotes replication of G-quadruplex DNA structures Pau Castillo Bosch, Sandra Segura-Bayona, Wouter Koole, Jane van Heteren, James Dewar, Marcel Tijsterman, Puck Knipscheer
ENCODE RBP Antibody Characterization Guidelines
 ENCODE RBP Antibody Characterization Guidelines Approved on November 18, 2016 Background An integral part of the ENCODE Project is to characterize the antibodies used in the experiments. This document
ENCODE RBP Antibody Characterization Guidelines Approved on November 18, 2016 Background An integral part of the ENCODE Project is to characterize the antibodies used in the experiments. This document
Phosphoproteome dynamics reveal novel ERK1/2 MAP kinase substrates with broad spectrum of functions
 Molecular Systems Biology Peer Review Process File Phosphoproteome dynamics reveal novel ERK1/2 MAP kinase substrates with broad spectrum of functions Mathieu Courcelles, Christophe Frémin, Laure Voisin,
Molecular Systems Biology Peer Review Process File Phosphoproteome dynamics reveal novel ERK1/2 MAP kinase substrates with broad spectrum of functions Mathieu Courcelles, Christophe Frémin, Laure Voisin,
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Craft et al., http://www.jcb.org/cgi/content/full/jcb.201409036/dc1 Figure S1. GFP -tubulin interacts with endogenous -tubulin. Ciliary
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Craft et al., http://www.jcb.org/cgi/content/full/jcb.201409036/dc1 Figure S1. GFP -tubulin interacts with endogenous -tubulin. Ciliary
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Han et al., http://www.jcb.org/cgi/content/full/jcb.201311007/dc1 Figure S1. SIVA1 interacts with PCNA. (A) HEK293T cells were transiently
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Han et al., http://www.jcb.org/cgi/content/full/jcb.201311007/dc1 Figure S1. SIVA1 interacts with PCNA. (A) HEK293T cells were transiently
Biology 105 First Exam
 September 26, 2003 Biology 105 First Exam 1. 2. 3. 4. 5. 6. 7. Total 1 1. (15 points) a. Draw the structure of the nucleotide monophosphate 5-Me cytosine. b. Draw the structure of the base analog azacytidine..
September 26, 2003 Biology 105 First Exam 1. 2. 3. 4. 5. 6. 7. Total 1 1. (15 points) a. Draw the structure of the nucleotide monophosphate 5-Me cytosine. b. Draw the structure of the base analog azacytidine..
Molecular Architecture of SMC proteins and the Yeast Cohesin Complex
 Molecular Architecture of SMC proteins and the Yeast Cohesin Complex Christian H. Haering 1,5, Jan Löwe 2,5, Andreas Hochwagen 1,3, and Kim Nasmyth 1,4 1 Research Institute of Molecular Pathology, Dr.
Molecular Architecture of SMC proteins and the Yeast Cohesin Complex Christian H. Haering 1,5, Jan Löwe 2,5, Andreas Hochwagen 1,3, and Kim Nasmyth 1,4 1 Research Institute of Molecular Pathology, Dr.
Epigenetics in. Saccharomyces cerevisiae. Chapter 4 2/4/14
 Epigenetics in Saccharomyces cerevisiae Chapter 4 2/4/14 The budding yeast - Saccharomyces cerevisiae The fission yeast - Schizosaccharomyces pombe The budding yeast, Saccharomyces cerevisiae and the fission
Epigenetics in Saccharomyces cerevisiae Chapter 4 2/4/14 The budding yeast - Saccharomyces cerevisiae The fission yeast - Schizosaccharomyces pombe The budding yeast, Saccharomyces cerevisiae and the fission
Metabolism and Chromosome Copy Number Control Mutually Exclusive Cell Fates in Bacillus subtilis
 Manuscript EMBO-2010-76658 Metabolism and Chromosome Copy Number Control Mutually Exclusive Cell Fates in Bacillus subtilis Yunrong Chai, Thomas Norman, Roberto Kolter, Richard M. Losick Corresponding
Manuscript EMBO-2010-76658 Metabolism and Chromosome Copy Number Control Mutually Exclusive Cell Fates in Bacillus subtilis Yunrong Chai, Thomas Norman, Roberto Kolter, Richard M. Losick Corresponding
7.03, 2006, Lecture 23 Eukaryotic Genes and Genomes IV
 1 Fall 2006 7.03 7.03, 2006, Lecture 23 Eukaryotic Genes and Genomes IV In the last three lectures we have thought a lot about analyzing a regulatory system in S. cerevisiae, namely Gal regulation that
1 Fall 2006 7.03 7.03, 2006, Lecture 23 Eukaryotic Genes and Genomes IV In the last three lectures we have thought a lot about analyzing a regulatory system in S. cerevisiae, namely Gal regulation that
7.05, 2005, Lecture 23 Eukaryotic Genes and Genomes IV
 7.05, 2005, Lecture 23 Eukaryotic Genes and Genomes IV In the last three lectures we have thought a lot about analyzing a regulatory system in S. cerevisiae, namely Gal regulation that involved a hand
7.05, 2005, Lecture 23 Eukaryotic Genes and Genomes IV In the last three lectures we have thought a lot about analyzing a regulatory system in S. cerevisiae, namely Gal regulation that involved a hand
MOLECULAR CELL BIOLOGY
 Lodish Berk Kaiser Krieger scott Bretscher Ploegh Matsudaira MOLECULAR CELL BIOLOGY SEVENTH EDITION CHAPTER 19 The Eukaryotic Cell Cycle Copyright 2013 by W. H. Freeman and Company Figure 19.1 The fate
Lodish Berk Kaiser Krieger scott Bretscher Ploegh Matsudaira MOLECULAR CELL BIOLOGY SEVENTH EDITION CHAPTER 19 The Eukaryotic Cell Cycle Copyright 2013 by W. H. Freeman and Company Figure 19.1 The fate
Supplemental Fig. 1: PEA-15 knockdown efficiency assessed by immunohistochemistry and qpcr
 Supplemental figure legends Supplemental Fig. 1: PEA-15 knockdown efficiency assessed by immunohistochemistry and qpcr A, LβT2 cells were transfected with either scrambled or PEA-15 sirna. Cells were then
Supplemental figure legends Supplemental Fig. 1: PEA-15 knockdown efficiency assessed by immunohistochemistry and qpcr A, LβT2 cells were transfected with either scrambled or PEA-15 sirna. Cells were then
Supplementary Materials
 Supplementary Materials Supplementary Figure 1. PKM2 interacts with MLC2 in cytokinesis. a, U87, U87/EGFRvIII, and HeLa cells in cytokinesis were immunostained with DAPI and an anti-pkm2 antibody. Thirty
Supplementary Materials Supplementary Figure 1. PKM2 interacts with MLC2 in cytokinesis. a, U87, U87/EGFRvIII, and HeLa cells in cytokinesis were immunostained with DAPI and an anti-pkm2 antibody. Thirty
Grb2 regulates B cell maturation, B cell memory responses and inhibits B cell Ca2+ signalling
 Manuscript EMBO-2010-76615 Grb2 regulates B cell maturation, B cell memory responses and inhibits B cell Ca2+ signalling Jochen Ackermann, Daniel Radtke, Thomas Winkler, Lars Nitschke Corresponding author:
Manuscript EMBO-2010-76615 Grb2 regulates B cell maturation, B cell memory responses and inhibits B cell Ca2+ signalling Jochen Ackermann, Daniel Radtke, Thomas Winkler, Lars Nitschke Corresponding author:
MCB130 midterm exam. 1) The exam has 6 pages (not including this page). Check that you have all 6 pages
 MCB130 midterm exam 1) The exam has 6 pages (not including this page). Check that you have all 6 pages 2) Please write your name and SID on the top of each page. The pages will be separated for grading
MCB130 midterm exam 1) The exam has 6 pages (not including this page). Check that you have all 6 pages 2) Please write your name and SID on the top of each page. The pages will be separated for grading
The RSC chromatin remodeling enzyme plays a unique role in directing the accurate positioning of nucleosomes
 Manuscript EMBO-2010-75466 The RSC chromatin remodeling enzyme plays a unique role in directing the accurate positioning of nucleosomes Christian J. Wippo, Lars Israel, Shinya Watanabe, Andreas Hochheimer,
Manuscript EMBO-2010-75466 The RSC chromatin remodeling enzyme plays a unique role in directing the accurate positioning of nucleosomes Christian J. Wippo, Lars Israel, Shinya Watanabe, Andreas Hochheimer,
Phosphorylation of CLIP-170 by Plk1 and CK2 promotes timely formation of kinetochore microtubule attachments
 The EMBO Journal (2010) 29, 2953 2965 & 2010 European Molecular Biology Organization All Rights Reserved 0261-4189/10 www.embojournal.org Phosphorylation of CLIP-170 by Plk1 and CK2 promotes timely formation
The EMBO Journal (2010) 29, 2953 2965 & 2010 European Molecular Biology Organization All Rights Reserved 0261-4189/10 www.embojournal.org Phosphorylation of CLIP-170 by Plk1 and CK2 promotes timely formation
Hossain_Supplemental Figure 1
 Hossain_Supplemental Figure 1 GFP-PACT GFP-PACT Motif I GFP-PACT Motif II A. MG132 (1µM) GFP Tubulin GFP-PACT Pericentrin GFP-PACT GFP-PACT Pericentrin Fig. S1. Expression and localization of Orc1 PACT
Hossain_Supplemental Figure 1 GFP-PACT GFP-PACT Motif I GFP-PACT Motif II A. MG132 (1µM) GFP Tubulin GFP-PACT Pericentrin GFP-PACT GFP-PACT Pericentrin Fig. S1. Expression and localization of Orc1 PACT
Lab 5 Cell Biology II
 Lab 5 Cell Biology II Learning Objectives Describe the structures and processes required for protein synthesis. Define a gene. Compare chromosomes and chromatin. Describe the process of DNA replication.
Lab 5 Cell Biology II Learning Objectives Describe the structures and processes required for protein synthesis. Define a gene. Compare chromosomes and chromatin. Describe the process of DNA replication.
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/310/5753/1487/dc1 Supporting Online Material for Stem Cell Self-Renewal Controlled by Chromatin Remodeling Factors Rongwen Xi and Ting Xie* *To whom correspondence should
www.sciencemag.org/cgi/content/full/310/5753/1487/dc1 Supporting Online Material for Stem Cell Self-Renewal Controlled by Chromatin Remodeling Factors Rongwen Xi and Ting Xie* *To whom correspondence should
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2386 Figure 1 Src-containing puncta are not focal adhesions, podosomes or endosomes. (a) FAK-/- were stained with anti-py416 Src (green) and either (in red) the focal adhesion protein paxillin,
DOI: 10.1038/ncb2386 Figure 1 Src-containing puncta are not focal adhesions, podosomes or endosomes. (a) FAK-/- were stained with anti-py416 Src (green) and either (in red) the focal adhesion protein paxillin,
Supplementary Figure 1. The Hsp70 acetylation level is related to the co-chaperone binding of Hsp70 under various stress conditions.
 Supplementary Figure 1. The Hsp70 acetylation level is related to the co-chaperone binding of Hsp70 under various stress conditions. 1 (a) Etoposide treatment gradually changes acetylation level and co-chaperone
Supplementary Figure 1. The Hsp70 acetylation level is related to the co-chaperone binding of Hsp70 under various stress conditions. 1 (a) Etoposide treatment gradually changes acetylation level and co-chaperone
Chapter 19. Control of Eukaryotic Genome. AP Biology
 Chapter 19. Control of Eukaryotic Genome The BIG Questions How are genes turned on & off in eukaryotes? How do cells with the same genes differentiate to perform completely different, specialized functions?
Chapter 19. Control of Eukaryotic Genome The BIG Questions How are genes turned on & off in eukaryotes? How do cells with the same genes differentiate to perform completely different, specialized functions?
Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and
 Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
Giardia RNAi Paper Discussion. Supplemental - VSG clonality. Variant-specific surface protein VSP9B10
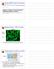 Giardia RNAi Paper Discussion Supplemental - VSG clonality VSP9B10 Green - VSP9B10 antibody Blue - DAPI Demonstration that cell line expresses a single VSP Variant-specific surface protein Outside Inside
Giardia RNAi Paper Discussion Supplemental - VSG clonality VSP9B10 Green - VSP9B10 antibody Blue - DAPI Demonstration that cell line expresses a single VSP Variant-specific surface protein Outside Inside
Plant Molecular and Cellular Biology Lecture 9: Nuclear Genome Organization: Chromosome Structure, Chromatin, DNA Packaging, Mitosis Gary Peter
 Plant Molecular and Cellular Biology Lecture 9: Nuclear Genome Organization: Chromosome Structure, Chromatin, DNA Packaging, Mitosis Gary Peter 9/16/2008 1 Learning Objectives 1. List and explain how DNA
Plant Molecular and Cellular Biology Lecture 9: Nuclear Genome Organization: Chromosome Structure, Chromatin, DNA Packaging, Mitosis Gary Peter 9/16/2008 1 Learning Objectives 1. List and explain how DNA
Rad51 replication fork recruitment is required for DNA damage tolerance
 Manuscript EMBO-2012-83909 Rad51 replication fork recruitment is required for DNA damage tolerance Rom n Gonzalez-Prieto, Ana M. Muñoz-Cabello, María J. Cabello-Lobato, Félix Prado Corresponding author:
Manuscript EMBO-2012-83909 Rad51 replication fork recruitment is required for DNA damage tolerance Rom n Gonzalez-Prieto, Ana M. Muñoz-Cabello, María J. Cabello-Lobato, Félix Prado Corresponding author:
RNA polymerase II kinetics in polo polyadenylation signal selection
 Manuscript EMBO-2010-75451 RNA polymerase II kinetics in polo polyadenylation signal selection Pedro A. B. Pinto, Telmo Henriques, Marta O. Freitas, Torcato Martins, Rita G. Domingues, Paulina S. Wyrzykowska,
Manuscript EMBO-2010-75451 RNA polymerase II kinetics in polo polyadenylation signal selection Pedro A. B. Pinto, Telmo Henriques, Marta O. Freitas, Torcato Martins, Rita G. Domingues, Paulina S. Wyrzykowska,
MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1. Lecture 25: Mass Spectrometry Applications
![MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1. Lecture 25: Mass Spectrometry Applications MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1. Lecture 25: Mass Spectrometry Applications](/thumbs/74/71368870.jpg) MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1 Lecture 25: Mass Spectrometry Applications Measuring Protein Abundance o ICAT o DIGE Identifying Post-Translational Modifications Protein-protein
MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1 Lecture 25: Mass Spectrometry Applications Measuring Protein Abundance o ICAT o DIGE Identifying Post-Translational Modifications Protein-protein
supplementary information
 DOI: 10.1038/ncb2172 Figure S1 p53 regulates cellular NADPH and lipid levels via inhibition of G6PD. (a) U2OS cells stably expressing p53 shrna or a control shrna were transfected with control sirna or
DOI: 10.1038/ncb2172 Figure S1 p53 regulates cellular NADPH and lipid levels via inhibition of G6PD. (a) U2OS cells stably expressing p53 shrna or a control shrna were transfected with control sirna or
Histone H3 Methylation Antibody Panel Pack I - Active Genes Base Catalog # C Component Size Shipping Temperature
 Histone H3 Methylation Antibody Panel Pack I - Active Genes Base Catalog # PACK CONTENTS Component Size Shipping Temperature Upon Receipt Checklist 3K4D Histone H3K4me2 (H3K4 Dimethyl) Polyclonal Antibody
Histone H3 Methylation Antibody Panel Pack I - Active Genes Base Catalog # PACK CONTENTS Component Size Shipping Temperature Upon Receipt Checklist 3K4D Histone H3K4me2 (H3K4 Dimethyl) Polyclonal Antibody
The role of erna in chromatin looping
 The role of erna in chromatin looping Introduction Enhancers are cis-acting DNA regulatory elements that activate transcription of target genes 1,2. It is estimated that 400 000 to >1 million alleged enhancers
The role of erna in chromatin looping Introduction Enhancers are cis-acting DNA regulatory elements that activate transcription of target genes 1,2. It is estimated that 400 000 to >1 million alleged enhancers
Sol narae (Sona) is a Drosophila ADAMTS involved in Wg signaling
 Sol narae (Sona) is a Drosophila ADAMTS involved in Wg signaling Go-Woon Kim, Jong-Hoon Won, Ok-Kyung Lee, Sang-Soo Lee, Jeong-Hoon Han, Orkhon Tsogtbaatar, Sujin Nam, Yeon Kim, and Kyung-Ok Cho* Supplementary
Sol narae (Sona) is a Drosophila ADAMTS involved in Wg signaling Go-Woon Kim, Jong-Hoon Won, Ok-Kyung Lee, Sang-Soo Lee, Jeong-Hoon Han, Orkhon Tsogtbaatar, Sujin Nam, Yeon Kim, and Kyung-Ok Cho* Supplementary
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION WWW.NATURE.COM/NATURECELLBIOLOGY 1 SUPPLEMENTARY INFORMATION 2 WWW.NATURE.COM/NATURECELLBIOLOGY SUPPLEMENTARY INFORMATION WWW.NATURE.COM/NATURECELLBIOLOGY 3 SUPPLEMENTARY INFORMATION
SUPPLEMENTARY INFORMATION WWW.NATURE.COM/NATURECELLBIOLOGY 1 SUPPLEMENTARY INFORMATION 2 WWW.NATURE.COM/NATURECELLBIOLOGY SUPPLEMENTARY INFORMATION WWW.NATURE.COM/NATURECELLBIOLOGY 3 SUPPLEMENTARY INFORMATION
At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in
 Supplementary Materials and Methods Barrier function assays At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in acidic X-gal mix (100 mm phosphate buffer at ph4.3, 3 mm
Supplementary Materials and Methods Barrier function assays At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in acidic X-gal mix (100 mm phosphate buffer at ph4.3, 3 mm
