Revision Checklist for Science Signaling Research Manuscripts: Data Requirements and Style Guidelines
|
|
|
- Maximilian Johnson
- 6 years ago
- Views:
Transcription
1 Revision Checklist for Science Signaling Research Manuscripts: Data Requirements and Style Guidelines Further information can be found at: Please feel free to your editor with specific questions or if you have a question about manuscript preparation at initial submission. Scientific or stylistic problem Instructions for Revision Data and Analysis Standards - CRITICAL Data not shown All data must be shown. Please include data that is not shown in the Supplementary Materials, if the data are not critical to the interpretation of the findings, or remove any phrases or sentences that refer to data that is not in the manuscript. Be sure not to introduce references to data not in the manuscript or that have been previously published as you make revisions in response to the referees. Include all additional data either in the main figures and tables or in the Supplementary Materials. Be sure any added data is presented in order. Actions or notes to the Editor N values N values must be provided in the figure legends for every data panel (this includes the Supplementary Figures), and what N represents (for example, cells, blots, mice, or experiments) must be indicated. For data requiring quantification and statistical analysis, N must represent at least 3 independent biological replicates. For data that does not require quantification and statistical analysis, N=1 is not acceptable, and N of at least 3 is preferable. Representative data may be shown. Data added in response to the referee comments must be represent N of at least 2. If quantification and statistical analysis are required, N must represent at least 3 biological replicates. It is not scientifically valid to present standard error or deviation bars or perform statistical analysis on data from N < 3 biological replicates. N values and what they represent must be specified in the figure legend for the added data. Quantification and statistical analysis When making claims about changes (or lack of change), please show quantification of at least 3 independent assays and perform statistical analysis. Statistical analysis should be performed on amalgamated data from at least 3 independent experiments, not on the results of a single experiment. Western blots: Key statements about increases, decreases, or lack of changes in protein abundance, phosphorylation, association, and activation must be supported by quantification of data amalgamated from at least 3 Western blots (that represent independent biological replicates) and statistical analysis where appropriate. Showing the densitometry from a single Western blot is not acceptable; amalgamated data from at least 3 Western blots must be presented: The signal for the protein of interest must be normalized to that of a loading control when quantifying signals from lysates. The signal for a phosphorylated form of a protein must be normalized to that for the total abundance of that protein. When quantifying changes in protein-protein interactions, the signal for the immunoprecipitated protein must be normalized to that in the total lysate. Statistics tests Western blotting Please add statistical analysis to graphs wherever possible to support statements about increases, decreases, or lack of change in the variable being measured. After each P value, state the statistical method used. Please note that t-tests and ANOVAs may not be appropriate for data with a non-normal distribution (such as percentages or qrt-pcr data). t-tests may not be appropriate for multiple overlapping comparisons in an experiment. For multiple comparison tests, such as ANOVAs, please specify the post-test used to derive P values. For guidance on selecting an appropriate statistical test, please consult a statistician and include the correspondence with the statistician with the revision. Please do not cite previously published papers as evidence that the statistical tests used are appropriate for the analysis of your data. For all Western blotting data, including figure panels that do not require quantification: 1
2 Western blots for phosphorylated forms of a protein of interest must be accompanied by Western blots for the total protein, to show that changes or lack of changes in the phosphorylation of the protein of interest are not due to changes in the overall abundance of the protein. This requirement also applies to other posttranslational modifications. Each set of Western blotting data must include an input blot, such as for actin or tubulin. Input blots (not actin or tubulin) must be shown for all coimmunoprecipitation blots for both the protein being pulled down and the protein(s) of interest that coprecipitated. Non-contiguous lanes within or between blots should be made obvious (delineated with borders, if necessary) and stated as such in the legend. RNA interference Genotypes, genes, and mrnas Scale bars Graph units Availability of RNAi, microarray, mass spectrometry, or structure data Ideally, to control for off-target effects, knockdown experiments should be performed with more than one sirna or shrna OR rescue experiments should be performed in knockdown cells. As well, knockdown efficiency should be shown by Western blotting; this data can be presented in the Supplementary Materials. Please italicize the names of genotypes, genes, and mrnas, and use the proper gene names. Primary and precursor forms of mirnas are italicized (the functional, mature form is not). All microscopy images (including histology, light microscopy, and immunofluorescence) must have scale bars. A single bar is sufficient for a group of images in a panel if they are all the same scale; if not, please include the appropriate scale bar within each image. Note its size in the legend, not in the figure itself. Please include the units on every X and Y axis, as applicable. For quantifications based on densitometry, please use (a.u.) for arbitrary units. Critical: Per the Science journals policy on data availability, all RNAi screens, microarray, mass spectrometry, or structure data must be deposited in a publicly accessible repository prior to publication. Please provide the accession number in the paragraph after the last reference in the section called Data and materials availability. **Note: If your paper passes peer review, we will not edit the paper and schedule it for publication until we have this deposition information. Organization and Presentation Title and abstract Note that the title may not exceed 135 characters including spaces, and the abstract may not length exceed 250 words. Sections Please ensure the main text is organized under the following subheadings: Abstract Introduction Results Discussion Materials and Methods Supplementary Materials (see Supplementary Materials list further below) References and Notes Figure Legends For the Supplementary Materials, please provide a separate Word file that contains the figures and tables, each followed by its legend (including a title). The legends should be single-spaced and use Times New Roman as the font. Supplementary Materials list Each item in the Supplementary Materials must have a short title. Insert a list of these short titles between the Materials and Methods section and the References and Notes section. The list in the main text must match exactly to those in the Supplementary Materials, in the following order (not all elements may appear): Figures Tables 2
3 References (range of the reference numbers that are only cited in the Supplementary Materials) Excel tables Movies Software Data files Refer to any recently published Science Signaling Research Article or Resource for example lists or see the online revision instructions for authors. Subheadings Figure order Figure legend length and content Figure size & formatting Please ensure that all subheadings in the Results and Discussion sections are the same style: either phrases or full sentences. Each section (A, B, etc) of the main figures must be called out in the text in order. Please ensure these are in order, and if not, rearrange the figures and their legends or rephrase the text as needed. Be sure that any data added to the manuscript in response to reviewers comments is also presented in order in the Results before being mentioned in the Discussion. Figure and table legends should not exceed 200 words if possible. Legends should describe the figure or table, not recapitulate the Materials and Methods or include interpretation that belongs in the Results. Example: (A) Western blotting analysis for x, y, and z performed at the indicated time points on lysates from xx cells treated with xx for x hours [or simply treated as indicated if the figure labels are explanatory]. Blots are representative of x experiments. (B) qrt-pcr for the expression of x, y, and z relative to w in xx cells transfected with xx sirna. (C) Apoptosis assessed by [insert assay here] was quantified in xx cells treated with xx. Data in (B) and (C) are means ± [SD or SEM] from [3 or more] independent experiments. We recommend using Adobe Illustrator or a program that can generate vector-based images to prepare your figures. We prefer that figures be provided as PDFs with editable text. Please prepare your main figures at high resolution (300 dpi minimum for gray scale and color; 1200 for line art), at 3.5, 5.0, 6.125, or 7.3 inches wide. Use the smallest of these sizes that retains legibility. For figure labels, please use Helvetica between 6 and 9 point in size; do not use bold for any labels within the figures. For panel labels, please use 10 point bold. Please try to leave some room at least in which to start the legend on the same page, and please left-align the panels; the legend may start in any corner white space. Across the figures: Bands in Western blots should be similar in width. Bars in all bar graphs should be a similar, narrow width, and graphs should occupy a similar sized area. Images should be a similar size (or as appropriate to see the necessary detail). Please make graphs and images small but retain label legibility. For an example of a paper with wellprepared figures, please see Figure labels Paragraph after last All language guidelines further detailed below apply to the figure labels as well. Most commonly: Do not use the word level (or levels), and do not use expression unless referring to genes or mrnas. If referring to mrnas, genes, or genotypes, use the proper gene name and italicize it. Ensure that gene and protein names conform to the appropriate species nomenclature. Avoid noting N.S. for non-significance within the figure unless the complexity warrants it; instead, indicate this and the test used to assess it in the legend. Avoid using anti-xx or α-xx to denote antibodies in the labels of Western blots (just label it with the protein name). The paragraph after the last reference should contain the following sections: 3
4 reference Acknowledgments, Funding Sources, Author Contributions (all authors must be mentioned), Competing Interests, and Data and Materials Availability (such as MTAs and accession numbers for data in public repositories). For example: Acknowledgments: We thank K.W. (AAAS) for providing the plasmids. We thank A.S. and L.T.M.F. for expert technical assistance. Funding: This work was supported by grants from AAAS (#88888 to N.R.G. and #77777 to J.F.F.). Author contributions: N.R.G. designed the project; A.V. and W.W. performed experiments; S.P. analyzed the mass spectrometry data; and L.K.F. and J.F.F. wrote the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium through the PRIDE partner repository with the dataset identifier PXD The plasmids require a material transfer agreement from AAAS, U.S.A. References and methods in the Supplementary Materials There must be a single reference list and Materials and Methods for the entire manuscript. (Exceptions for Materials and Methods in the Supplementary Materials may include detailed computational methodology.) Any reference not cited in the main text should be added to the end of the main text references and numbered sequentially in the main text reference list and then referred by those numbers in the Supplementary Materials. Language and Stylistic Guidelines Use of tenses Please use the past tense to describe the results in your manuscript and the present tense to describe previously published results. Significant Slashes Amino acid callouts We reserve the use of significant for describing results of statistical analysis. Do not use it as a replacement for important or substantial. If you have used it to describe results that have not been analyzed by statistical tests, please use an appropriate statistical test to support claims of significant differences. Per style, we try to avoid the use of the slash due to its ambiguity. Please use and or or instead of a slash where appropriate, or define terms that will include a slash, such as protein names or complexes. For example, ERK1/2 should be defined as extracellular signalregulated kinase 1 and 2 (ERK1/2). To refer to single amino acid residues, please use the 3 letter abbreviation and superscript the residue number (for example, Ser 25 ); single letter abbreviations book-ending a normalscripted residue are used when describing amino acid mutants (for example, S25A). Express We use express and its related words to refer to (1) gene transcriptional events or (2) transgenic plants or animals or to transfected cells. It should not be used to describe the presence or absence, abundance, distribution, or location of a signaling molecule. Levels Up and down regulate Acronyms Antibodies In vivo, in vitro, in We use levels to indicate hierarchy (such as at the transcriptional level ) and not to indicate amount, abundance, or concentration. Per style, we avoid using imprecise terms, such as up-regulated, down-regulated, controlled, regulated, and modulated where possible. Please change up-regulated to increased abundance, increased activity, or whatever is appropriate and use similar language for down-regulated. Please change regulates to inhibits, attenuates, or reduces or stimulates, enhances, or promotes, as appropriate. Please define all acronyms the first time that they are used in the Abstract and the first time that they are used in the main text. Per style we do not refer to antibodies as anti-xxx. Please change in the text to an antibody recognizing or an antibody against or an antibody specific for. If anti-xxx is used in the figure labels, please define it in the legend. Per style, we do not italicize in vivo, in vitro, ex vivo, in silico, or in situ, but we do italicize 4
5 situ Other style guidelines et al. Please avoid claims of novelty or being first. Please do not start sentences with Importantly or Interestingly. Please avoid hyperbole and delete dramatically wherever you have used it. Our style is to use wild-type in the main text and WT in the figures. Note that it is not our style to use via, e.g., or i.e.. Please delete and rephrase wherever you have used these terms. Note that it is not our style to use elevated as a synonym for increased. Please rephrase. We avoid the use of recent and similar words and presenting previously published results as a historical perspective. Note that it is not our style to start sentences with As shown in Fig. 1B. Please revise throughout the manuscript. 5
Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected
 Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected with the sirna against lnc-2, lnc-6, lnc-7, and the
Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected with the sirna against lnc-2, lnc-6, lnc-7, and the
SUPPLEMENTARY INFORMATION
 doi:.38/nature899 Supplementary Figure Suzuki et al. a c p7 -/- / WT ratio (+)/(-) p7 -/- / WT ratio Log X 3. Fold change by treatment ( (+)/(-)) Log X.5 3-3. -. b Fold change by treatment ( (+)/(-)) 8
doi:.38/nature899 Supplementary Figure Suzuki et al. a c p7 -/- / WT ratio (+)/(-) p7 -/- / WT ratio Log X 3. Fold change by treatment ( (+)/(-)) Log X.5 3-3. -. b Fold change by treatment ( (+)/(-)) 8
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table.
 Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or
 Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
* ** ** * IB: p-p90rsk. p90rsk (Ser380) (arbitrary units) (Ser380) p90rsk. IB: p90rsk. Tubulin. IB: Tubulin. Ang II (200 nm) Ang II (200 nm)
 I: p-p9rsk I: p9rsk I: C I: p-p9rsk I: p9rsk 5 (ka) 5 5 (min) Ang II ( nm) p-p9rsk (Ser8) p9rsk p-p9rsk (Ser8) p9rsk (h) Mannitol 5 mm -Glucose 5 mm p9rsk (Ser8) (arbitrary units) p-p9rsk (Ser8) (arbitrary
I: p-p9rsk I: p9rsk I: C I: p-p9rsk I: p9rsk 5 (ka) 5 5 (min) Ang II ( nm) p-p9rsk (Ser8) p9rsk p-p9rsk (Ser8) p9rsk (h) Mannitol 5 mm -Glucose 5 mm p9rsk (Ser8) (arbitrary units) p-p9rsk (Ser8) (arbitrary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling
 Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
TE5 KYSE510 TE7 KYSE70 KYSE140
 TE5 KYSE5 TT KYSE7 KYSE4 Supplementary Figure. Hockey stick plots showing input normalized, rank ordered H3K7ac signals for the candidate SE-associated lncrnas in this study. Rpm Rpm Rpm Chip-seq H3K7ac
TE5 KYSE5 TT KYSE7 KYSE4 Supplementary Figure. Hockey stick plots showing input normalized, rank ordered H3K7ac signals for the candidate SE-associated lncrnas in this study. Rpm Rpm Rpm Chip-seq H3K7ac
Supplementary Figure 1. Bone density was decreased in osteoclast-lineage cell specific Gna13 deficient mice. (a-c) PCR genotyping of mice by mouse
 Supplementary Figure 1. Bone density was decreased in osteoclast-lineage cell specific Gna13 deficient mice. (a-c) PCR genotyping of mice by mouse tail DNA. Primers were designed to detect Gna13-WT/f (~400bp/470bp)
Supplementary Figure 1. Bone density was decreased in osteoclast-lineage cell specific Gna13 deficient mice. (a-c) PCR genotyping of mice by mouse tail DNA. Primers were designed to detect Gna13-WT/f (~400bp/470bp)
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature09732 Supplementary Figure 1: Depletion of Fbw7 results in elevated Mcl-1 abundance. a, Total thymocytes from 8-wk-old Lck-Cre/Fbw7 +/fl (Control) or Lck-Cre/Fbw7 fl/fl (Fbw7 KO) mice
doi:10.1038/nature09732 Supplementary Figure 1: Depletion of Fbw7 results in elevated Mcl-1 abundance. a, Total thymocytes from 8-wk-old Lck-Cre/Fbw7 +/fl (Control) or Lck-Cre/Fbw7 fl/fl (Fbw7 KO) mice
SureSilencing sirna Array Technology Overview
 SureSilencing sirna Array Technology Overview Pathway-Focused sirna-based RNA Interference Topics to be Covered Who is SuperArray? Brief Introduction to RNA Interference Challenges Facing RNA Interference
SureSilencing sirna Array Technology Overview Pathway-Focused sirna-based RNA Interference Topics to be Covered Who is SuperArray? Brief Introduction to RNA Interference Challenges Facing RNA Interference
Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna
 Supplemental Materials and Methods Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna (Mission shrna, Sigma) against mouse MMP14 were transfected into HEK293 cells using FuGene6
Supplemental Materials and Methods Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna (Mission shrna, Sigma) against mouse MMP14 were transfected into HEK293 cells using FuGene6
Supplemental Data. Wu et al. (2). Plant Cell..5/tpc RGLG Hormonal treatment H2O B RGLG µm ABA µm ACC µm GA Time (hours) µm µm MJ µm IA
 Supplemental Data. Wu et al. (2). Plant Cell..5/tpc..4. A B Supplemental Figure. Immunoblot analysis verifies the expression of the AD-PP2C and BD-RGLG proteins in the Y2H assay. Total proteins were extracted
Supplemental Data. Wu et al. (2). Plant Cell..5/tpc..4. A B Supplemental Figure. Immunoblot analysis verifies the expression of the AD-PP2C and BD-RGLG proteins in the Y2H assay. Total proteins were extracted
ENCODE RBP Antibody Characterization Guidelines
 ENCODE RBP Antibody Characterization Guidelines Approved on November 18, 2016 Background An integral part of the ENCODE Project is to characterize the antibodies used in the experiments. This document
ENCODE RBP Antibody Characterization Guidelines Approved on November 18, 2016 Background An integral part of the ENCODE Project is to characterize the antibodies used in the experiments. This document
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than. SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with
 Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
7.17: Writing Up Results and Creating Illustrations
 7.17: Writing Up Results and Creating Illustrations A Results Exercise: Kansas and Pancakes Write a 5-sentence paragraph describing the results illustrated in this figure: - Describe the figure: highlights?
7.17: Writing Up Results and Creating Illustrations A Results Exercise: Kansas and Pancakes Write a 5-sentence paragraph describing the results illustrated in this figure: - Describe the figure: highlights?
Supplementary Figure 1. Characterization of EVs (a) Phase-contrast electron microscopy was used to visualize resuspended EV pellets.
 Supplementary Figure 1. Characterization of EVs (a) Phase-contrast electron microscopy was used to visualize resuspended EV pellets. Scale bar represent 100 nm. The sizes of EVs from MDA-MB-231-D3H1 (D3H1),
Supplementary Figure 1. Characterization of EVs (a) Phase-contrast electron microscopy was used to visualize resuspended EV pellets. Scale bar represent 100 nm. The sizes of EVs from MDA-MB-231-D3H1 (D3H1),
Chemical hijacking of auxin signaling with an engineered auxin-tir1
 1 SUPPLEMENTARY INFORMATION Chemical hijacking of auxin signaling with an engineered auxin-tir1 pair Naoyuki Uchida 1,2*, Koji Takahashi 2*, Rie Iwasaki 1, Ryotaro Yamada 2, Masahiko Yoshimura 2, Takaho
1 SUPPLEMENTARY INFORMATION Chemical hijacking of auxin signaling with an engineered auxin-tir1 pair Naoyuki Uchida 1,2*, Koji Takahashi 2*, Rie Iwasaki 1, Ryotaro Yamada 2, Masahiko Yoshimura 2, Takaho
Regulation of Synaptic Structure and Function by FMRP- Associated MicroRNAs mir-125b and mir-132
 Neuron, Volume 65 Regulation of Synaptic Structure and Function by FMRP- Associated MicroRNAs mir-125b and mir-132 Dieter Edbauer, Joel R. Neilson, Kelly A. Foster, Chi-Fong Wang, Daniel P. Seeburg, Matthew
Neuron, Volume 65 Regulation of Synaptic Structure and Function by FMRP- Associated MicroRNAs mir-125b and mir-132 Dieter Edbauer, Joel R. Neilson, Kelly A. Foster, Chi-Fong Wang, Daniel P. Seeburg, Matthew
Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product.
 Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Online Supplementary Information
 Online Supplementary Information NLRP4 negatively regulates type I interferon signaling by targeting TBK1 for degradation via E3 ubiquitin ligase DTX4 Jun Cui 1,4,6,7, Yinyin Li 1,5,6,7, Liang Zhu 1, Dan
Online Supplementary Information NLRP4 negatively regulates type I interferon signaling by targeting TBK1 for degradation via E3 ubiquitin ligase DTX4 Jun Cui 1,4,6,7, Yinyin Li 1,5,6,7, Liang Zhu 1, Dan
SUPPLEMENTARY INFORMATION. Small molecule activation of the TRAIL receptor DR5 in human cancer cells
 SUPPLEMENTARY INFORMATION Small molecule activation of the TRAIL receptor DR5 in human cancer cells Gelin Wang 1*, Xiaoming Wang 2, Hong Yu 1, Shuguang Wei 1, Noelle Williams 1, Daniel L. Holmes 1, Randal
SUPPLEMENTARY INFORMATION Small molecule activation of the TRAIL receptor DR5 in human cancer cells Gelin Wang 1*, Xiaoming Wang 2, Hong Yu 1, Shuguang Wei 1, Noelle Williams 1, Daniel L. Holmes 1, Randal
Journal of Cell Science Supplementary Material
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 SUPPLEMENTARY FIGURE LEGENDS Figure S1: Eps8 is localized at focal adhesions and binds directly to FAK (A) Focal
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 SUPPLEMENTARY FIGURE LEGENDS Figure S1: Eps8 is localized at focal adhesions and binds directly to FAK (A) Focal
sirna Transfection Into Primary Neurons Using Fuse-It-siRNA
 sirna Transfection Into Primary Neurons Using Fuse-It-siRNA This Application Note describes a protocol for sirna transfection into sensitive, primary cortical neurons using Fuse-It-siRNA. This innovative
sirna Transfection Into Primary Neurons Using Fuse-It-siRNA This Application Note describes a protocol for sirna transfection into sensitive, primary cortical neurons using Fuse-It-siRNA. This innovative
RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by. 5 AGACACAAACACCAUUGUCACACUCCACAGC; Rand-2 OMe,
 Materials and methods Oligonucleotides and DNA constructs RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by Dharmacon Inc. (Lafayette, CO). The sequences were: 122-2 OMe, 5
Materials and methods Oligonucleotides and DNA constructs RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by Dharmacon Inc. (Lafayette, CO). The sequences were: 122-2 OMe, 5
Gene Sequence Fragment size ΔEGFR F 5' GGGCTCTGGAGGAAAAGAAAG GT 3' 116 bp R 5' CTTCTTACACTTGCGGACGC 3'
 Supplementary Table 1: Real-time PCR primer sequences for ΔEGFR, wtegfr, IL-6, LIF and GAPDH. Gene Sequence Fragment size ΔEGFR F 5' GGGCTCTGGAGGAAAAGAAAG GT 3' 116 bp R 5' CTTCTTACACTTGCGGACGC 3' wtegfr
Supplementary Table 1: Real-time PCR primer sequences for ΔEGFR, wtegfr, IL-6, LIF and GAPDH. Gene Sequence Fragment size ΔEGFR F 5' GGGCTCTGGAGGAAAAGAAAG GT 3' 116 bp R 5' CTTCTTACACTTGCGGACGC 3' wtegfr
Supplementary Information Design of small molecule-responsive micrornas based on structural requirements for Drosha processing
 Supplementary Information Design of small molecule-responsive micrornas based on structural requirements for Drosha processing Chase L. Beisel, Yvonne Y. Chen, Stephanie J. Culler, Kevin G. Hoff, & Christina
Supplementary Information Design of small molecule-responsive micrornas based on structural requirements for Drosha processing Chase L. Beisel, Yvonne Y. Chen, Stephanie J. Culler, Kevin G. Hoff, & Christina
Isolation, culture, and transfection of primary mammary epithelial organoids
 Supplementary Experimental Procedures Isolation, culture, and transfection of primary mammary epithelial organoids Primary mammary epithelial organoids were prepared from 8-week-old CD1 mice (Charles River)
Supplementary Experimental Procedures Isolation, culture, and transfection of primary mammary epithelial organoids Primary mammary epithelial organoids were prepared from 8-week-old CD1 mice (Charles River)
ASPP1 Fw GGTTGGGAATCCACGTGTTG ASPP1 Rv GCCATATCTTGGAGCTCTGAGAG
 Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
Wnt16 smact merge VK/AB
 A WT Wnt6 smact merge VK/A KO ctrl IgG WT KO Wnt6 smact DAPI SUPPLEMENTAL FIGURE I: Wnt6 expression in MGP-deficient aortae. Immunostaining for Wnt6 and smooth muscle actin (smact) in aortae from 7 day
A WT Wnt6 smact merge VK/A KO ctrl IgG WT KO Wnt6 smact DAPI SUPPLEMENTAL FIGURE I: Wnt6 expression in MGP-deficient aortae. Immunostaining for Wnt6 and smooth muscle actin (smact) in aortae from 7 day
Figure S1. Figure S2. Figure S3 HB Anti-FSP27 (COOH-terminal peptide) Ab. Anti-GST-FSP27(45-127) Ab.
 / 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
/ 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate
 Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
Parthanatos mediates AIMP2-activated age-dependent dopaminergic neuronal loss
 SUPPLEMENTARY INFORMATION Parthanatos mediates AIMP2-activated age-dependent dopaminergic neuronal loss Yunjong Lee, Senthilkumar S. Karuppagounder, Joo-Ho Shin, Yun-Il Lee, Han Seok Ko, Debbie Swing,
SUPPLEMENTARY INFORMATION Parthanatos mediates AIMP2-activated age-dependent dopaminergic neuronal loss Yunjong Lee, Senthilkumar S. Karuppagounder, Joo-Ho Shin, Yun-Il Lee, Han Seok Ko, Debbie Swing,
Supplementary Figure 1: Expression of RNF8, HERC2 and NEURL4 in the cerebellum and knockdown of RNF8 by RNAi (a) Lysates of the cerebellum from rat
 Supplementary Figure 1: Expression of RNF8, HERC2 and NEURL4 in the cerebellum and knockdown of RNF8 by RNAi (a) Lysates of the cerebellum from rat pups at P6, P14, P22, P30 and adult (A) rats were subjected
Supplementary Figure 1: Expression of RNF8, HERC2 and NEURL4 in the cerebellum and knockdown of RNF8 by RNAi (a) Lysates of the cerebellum from rat pups at P6, P14, P22, P30 and adult (A) rats were subjected
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
Nature Neuroscience: doi: /nn Supplementary Figure 1
 Supplementary Figure 1 PCR-genotyping of the three mouse models used in this study and controls for behavioral experiments after semi-chronic Pten inhibition. a-c. DNA from App/Psen1 (a), Pten tg (b) and
Supplementary Figure 1 PCR-genotyping of the three mouse models used in this study and controls for behavioral experiments after semi-chronic Pten inhibition. a-c. DNA from App/Psen1 (a), Pten tg (b) and
Nature Immunology: doi: /ni Supplementary Figure 1. Zranb1 gene targeting.
 Supplementary Figure 1 Zranb1 gene targeting. (a) Schematic picture of Zranb1 gene targeting using an FRT-LoxP vector, showing the first 6 exons of Zranb1 gene (exons 7-9 are not shown). Targeted mice
Supplementary Figure 1 Zranb1 gene targeting. (a) Schematic picture of Zranb1 gene targeting using an FRT-LoxP vector, showing the first 6 exons of Zranb1 gene (exons 7-9 are not shown). Targeted mice
Supplementary Information
 Supplementary Information Live imaging reveals the dynamics and regulation of mitochondrial nucleoids during the cell cycle in Fucci2-HeLa cells Taeko Sasaki 1, Yoshikatsu Sato 2, Tetsuya Higashiyama 1,2,
Supplementary Information Live imaging reveals the dynamics and regulation of mitochondrial nucleoids during the cell cycle in Fucci2-HeLa cells Taeko Sasaki 1, Yoshikatsu Sato 2, Tetsuya Higashiyama 1,2,
Comparison of Commercial Transfection Reagents: Cell line optimized transfection kits for in vitro cancer research.
 Comparison of Commercial Transfection Reagents: Cell line optimized transfection kits for in vitro cancer research. by Altogen Labs, 11200 Manchaca Road, Suite 203 Austin TX 78748 USA Tel. (512) 433-6177
Comparison of Commercial Transfection Reagents: Cell line optimized transfection kits for in vitro cancer research. by Altogen Labs, 11200 Manchaca Road, Suite 203 Austin TX 78748 USA Tel. (512) 433-6177
S-sulfhydration of MEK1 leads to PARP-1 activation and DNA damage repair
 Manuscript EMBOR-2013-38213 S-sulfhydration of MEK1 leads to PARP-1 activation and DNA damage repair Kexin Zhao, YoungJun Ju, Shuangshuang Li, Zaid Al Taany, Rui Wang and Guangdong Yang Corresponding author:
Manuscript EMBOR-2013-38213 S-sulfhydration of MEK1 leads to PARP-1 activation and DNA damage repair Kexin Zhao, YoungJun Ju, Shuangshuang Li, Zaid Al Taany, Rui Wang and Guangdong Yang Corresponding author:
Learning Objectives. Define RNA interference. Define basic terminology. Describe molecular mechanism. Define VSP and relevance
 Learning Objectives Define RNA interference Define basic terminology Describe molecular mechanism Define VSP and relevance Describe role of RNAi in antigenic variation A Nobel Way to Regulate Gene Expression
Learning Objectives Define RNA interference Define basic terminology Describe molecular mechanism Define VSP and relevance Describe role of RNAi in antigenic variation A Nobel Way to Regulate Gene Expression
Coleman et al., Supplementary Figure 1
 Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Supplementary Figure 1. Utilization of publicly available antibodies in different applications.
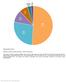 Supplementary Figure 1 Utilization of publicly available antibodies in different applications. The fraction of publicly available antibodies toward human protein targets is shown with data for the following
Supplementary Figure 1 Utilization of publicly available antibodies in different applications. The fraction of publicly available antibodies toward human protein targets is shown with data for the following
Confocal immunofluorescence microscopy
 Confocal immunofluorescence microscopy HL-6 and cells were cultured and cytospun onto glass slides. The cells were double immunofluorescence stained for Mt NPM1 and fibrillarin (nucleolar marker). Briefly,
Confocal immunofluorescence microscopy HL-6 and cells were cultured and cytospun onto glass slides. The cells were double immunofluorescence stained for Mt NPM1 and fibrillarin (nucleolar marker). Briefly,
A) B) Ladder. Supplementary Figure 1. Recombinant calpain 14 purification analysis. A) The general domain structure of classical
 A) B) Ladder C) r4 r4 Nt- -Ct 78 kda Supplementary Figure 1. Recombinant calpain 14 purification analysis. A) The general domain structure of classical calpains is shown. The protease core consists of
A) B) Ladder C) r4 r4 Nt- -Ct 78 kda Supplementary Figure 1. Recombinant calpain 14 purification analysis. A) The general domain structure of classical calpains is shown. The protease core consists of
Hossain_Supplemental Figure 1
 Hossain_Supplemental Figure 1 GFP-PACT GFP-PACT Motif I GFP-PACT Motif II A. MG132 (1µM) GFP Tubulin GFP-PACT Pericentrin GFP-PACT GFP-PACT Pericentrin Fig. S1. Expression and localization of Orc1 PACT
Hossain_Supplemental Figure 1 GFP-PACT GFP-PACT Motif I GFP-PACT Motif II A. MG132 (1µM) GFP Tubulin GFP-PACT Pericentrin GFP-PACT GFP-PACT Pericentrin Fig. S1. Expression and localization of Orc1 PACT
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1.
 Supplementary Figure 1. Characterization and high expression of Lnc-β-Catm in liver CSCs. (a) Heatmap of differently expressed lncrnas in Liver CSCs (CD13 + CD133 + ) and non-cscs (CD13 - CD133 - ) according
Supplementary Figure 1. Characterization and high expression of Lnc-β-Catm in liver CSCs. (a) Heatmap of differently expressed lncrnas in Liver CSCs (CD13 + CD133 + ) and non-cscs (CD13 - CD133 - ) according
Supplementary Information
 Supplementary Information MLL histone methylases regulate expression of HDLR- in presence of estrogen and control plasma cholesterol in vivo Khairul I. Ansari 1, Sahba Kasiri 1, Imran Hussain 1, Samara
Supplementary Information MLL histone methylases regulate expression of HDLR- in presence of estrogen and control plasma cholesterol in vivo Khairul I. Ansari 1, Sahba Kasiri 1, Imran Hussain 1, Samara
SUPPLEMENTAL FIGURE LEGENDS. Figure S1: Homology alignment of DDR2 amino acid sequence. Shown are
 SUPPLEMENTAL FIGURE LEGENDS Figure S1: Homology alignment of DDR2 amino acid sequence. Shown are the amino acid sequences of human DDR2, mouse DDR2 and the closest homologs in zebrafish and C. Elegans.
SUPPLEMENTAL FIGURE LEGENDS Figure S1: Homology alignment of DDR2 amino acid sequence. Shown are the amino acid sequences of human DDR2, mouse DDR2 and the closest homologs in zebrafish and C. Elegans.
Fatchiyah
 Fatchiyah Email: fatchiya@yahoo.co.id RNAs: mrna trna rrna RNAi DNAs: Protein: genome DNA cdna mikro-makro mono-poly single-multi Analysis: Identification human and animal disease Finger printing Sexing
Fatchiyah Email: fatchiya@yahoo.co.id RNAs: mrna trna rrna RNAi DNAs: Protein: genome DNA cdna mikro-makro mono-poly single-multi Analysis: Identification human and animal disease Finger printing Sexing
High Profile Publishing in Molecular Biology. Hélène Hodak, Marina Ostankovitch,
 High Profile Publishing in Molecular Biology Hélène Hodak, h.hodak@elsevier.com Marina Ostankovitch, m.ostankovitch@elsevier.com The field of Molecular Biology, inception to current trends Preparing a
High Profile Publishing in Molecular Biology Hélène Hodak, h.hodak@elsevier.com Marina Ostankovitch, m.ostankovitch@elsevier.com The field of Molecular Biology, inception to current trends Preparing a
Table I. Primers used in quantitative real-time PCR for detecting gene expressions in mouse liver tissues
 Table I. Primers used in quantitative real-time PCR for detecting gene expressions in mouse liver tissues Gene name Forward primer 5' to 3' Gene name Reverse primer 5' to 3' CC_F TGCCCTGTTGGGGTTG CC_R
Table I. Primers used in quantitative real-time PCR for detecting gene expressions in mouse liver tissues Gene name Forward primer 5' to 3' Gene name Reverse primer 5' to 3' CC_F TGCCCTGTTGGGGTTG CC_R
FCN1 (M-ficolin), which directly associates with immunoglobulin G1, is a molecular target of intravenous immunoglobulin therapy for Kawasaki disease
 FCN1 (M-ficolin), which directly associates with immunoglobulin G1, is a molecular target of intravenous immunoglobulin therapy for Kawasaki disease Daisuke Okuzaki, Kaori Ota, Shin-ichi Takatsuki, Yukari
FCN1 (M-ficolin), which directly associates with immunoglobulin G1, is a molecular target of intravenous immunoglobulin therapy for Kawasaki disease Daisuke Okuzaki, Kaori Ota, Shin-ichi Takatsuki, Yukari
Supplemental Information. Mitophagy Controls the Activities. of Tumor Suppressor p53. to Regulate Hepatic Cancer Stem Cells
 Molecular Cell, Volume 68 Supplemental Information Mitophagy Controls the Activities of Tumor Suppressor to Regulate Hepatic Cancer Stem Cells Kai Liu, Jiyoung Lee, Ja Yeon Kim, Linya Wang, Yongjun Tian,
Molecular Cell, Volume 68 Supplemental Information Mitophagy Controls the Activities of Tumor Suppressor to Regulate Hepatic Cancer Stem Cells Kai Liu, Jiyoung Lee, Ja Yeon Kim, Linya Wang, Yongjun Tian,
The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit
 Cell Reports, Volume 5 Supplemental Information The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit Andrey Poleshko, Katelyn M. Mansfield, Caroline
Cell Reports, Volume 5 Supplemental Information The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit Andrey Poleshko, Katelyn M. Mansfield, Caroline
Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon Song, Richard M. Amasino, Bosl Noh, and Yoo-Sun Noh
 Developmental Cell, Volume 22 Supplemental Information Control of Seed Germination by Light-Induced Histone Arginine Demethylation Activity Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon
Developmental Cell, Volume 22 Supplemental Information Control of Seed Germination by Light-Induced Histone Arginine Demethylation Activity Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon
HCT116 SW48 Nutlin: p53
 Figure S HCT6 SW8 Nutlin: - + - + p GAPDH Figure S. Nutlin- treatment induces p protein. HCT6 and SW8 cells were left untreated or treated for 8 hr with Nutlin- ( µm) to up-regulate p. Whole cell lysates
Figure S HCT6 SW8 Nutlin: - + - + p GAPDH Figure S. Nutlin- treatment induces p protein. HCT6 and SW8 cells were left untreated or treated for 8 hr with Nutlin- ( µm) to up-regulate p. Whole cell lysates
SANTA CRUZ BIOTECHNOLOGY, INC.
 TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
MISSION shrna Library: Next Generation RNA Interference
 Page 1 of 6 Page 1 of 6 Return to Web Version MISSION shrna Library: Next Generation RNA Interference By: Stephanie Uder, Henry George, Betsy Boedeker, LSI Volume 6 Article 2 Introduction The technology
Page 1 of 6 Page 1 of 6 Return to Web Version MISSION shrna Library: Next Generation RNA Interference By: Stephanie Uder, Henry George, Betsy Boedeker, LSI Volume 6 Article 2 Introduction The technology
Concepts and Methods in Developmental Biology
 Biology 4361 Developmental Biology Concepts and Methods in Developmental Biology June 16, 2009 Conceptual and Methodological Tools Concepts Genomic equivalence Differential gene expression Differentiation/de-differentiation
Biology 4361 Developmental Biology Concepts and Methods in Developmental Biology June 16, 2009 Conceptual and Methodological Tools Concepts Genomic equivalence Differential gene expression Differentiation/de-differentiation
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/10/494/eaan6284/dc1 Supplementary Materials for Activation of master virulence regulator PhoP in acidic ph requires the Salmonella-specific protein UgtL Jeongjoon
www.sciencesignaling.org/cgi/content/full/10/494/eaan6284/dc1 Supplementary Materials for Activation of master virulence regulator PhoP in acidic ph requires the Salmonella-specific protein UgtL Jeongjoon
Supplementary Figure Legend
 Supplementary Figure Legend Supplementary Figure S1. Effects of MMP-1 silencing on HEp3-hi/diss cell proliferation in 2D and 3D culture conditions. (A) Downregulation of MMP-1 expression in HEp3-hi/diss
Supplementary Figure Legend Supplementary Figure S1. Effects of MMP-1 silencing on HEp3-hi/diss cell proliferation in 2D and 3D culture conditions. (A) Downregulation of MMP-1 expression in HEp3-hi/diss
Gene Expression Technology
 Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
a Award Number: W81XWH TITLE:
 AD Award Number: W81XWH-04-1-0138 TITLE: Imaging Metastatic Prostate Cancer After Genetic Manipulation of Transcriptional Memory Regulators EZH2 and EED PRINCIPAL INVESTIGATOR: Lily Wu, M.D., Ph.D. CONTRACTING
AD Award Number: W81XWH-04-1-0138 TITLE: Imaging Metastatic Prostate Cancer After Genetic Manipulation of Transcriptional Memory Regulators EZH2 and EED PRINCIPAL INVESTIGATOR: Lily Wu, M.D., Ph.D. CONTRACTING
Table S1. Primers used in RT-PCR studies (all in 5 to 3 direction)
 Table S1. Primers used in RT-PCR studies (all in 5 to 3 direction) Epo Fw CTGTATCATGGACCACCTCGG Epo Rw TGAAGCACAGAAGCTCTTCGG Jak2 Fw ATCTGACCTTTCCATCTGGGG Jak2 Rw TGGTTGGGTGGATACCAGATC Stat5A Fw TTACTGAAGATCAAGCTGGGG
Table S1. Primers used in RT-PCR studies (all in 5 to 3 direction) Epo Fw CTGTATCATGGACCACCTCGG Epo Rw TGAAGCACAGAAGCTCTTCGG Jak2 Fw ATCTGACCTTTCCATCTGGGG Jak2 Rw TGGTTGGGTGGATACCAGATC Stat5A Fw TTACTGAAGATCAAGCTGGGG
Real-time PCR. TaqMan Protein Assays. Unlock the power of real-time PCR for protein analysis
 Real-time PCR TaqMan Protein Assays Unlock the power of real-time PCR for protein analysis I can use my real-time PCR instrument to quantitate protein? Do protein levels correlate with related mrna levels
Real-time PCR TaqMan Protein Assays Unlock the power of real-time PCR for protein analysis I can use my real-time PCR instrument to quantitate protein? Do protein levels correlate with related mrna levels
Supplementary Material. TRIB3 inhibits proliferation and promotes osteogenesis in hbmscs by regulating the. ERK1/2 signaling pathway
 Supplementary Material TRIB3 inhibits proliferation and promotes osteogenesis in hbmscs by regulating the ERK1/2 signaling pathway Cui Zhang 1, Fan-Fan Hong 1, Cui-Cui Wang 1, Liang Li 1, Jian-Ling Chen
Supplementary Material TRIB3 inhibits proliferation and promotes osteogenesis in hbmscs by regulating the ERK1/2 signaling pathway Cui Zhang 1, Fan-Fan Hong 1, Cui-Cui Wang 1, Liang Li 1, Jian-Ling Chen
SUPPLEMENTAL MATERIAL. Supplemental Methods. Cumate solution was from System Biosciences. Human complement C1q and complement
 SUPPLEMENTAL MATERIAL Supplemental Methods Reagents Cumate solution was from System Biosciences. Human complement Cq and complement C-esterase inhibitor (C-INH) were from Calbiochem. C-INH (Berinert) for
SUPPLEMENTAL MATERIAL Supplemental Methods Reagents Cumate solution was from System Biosciences. Human complement Cq and complement C-esterase inhibitor (C-INH) were from Calbiochem. C-INH (Berinert) for
Supplemental Information
 Supplemental Information Itemized List Materials and Methods, Related to Supplemental Figures S5A-C and S6. Supplemental Figure S1, Related to Figures 1 and 2. Supplemental Figure S2, Related to Figure
Supplemental Information Itemized List Materials and Methods, Related to Supplemental Figures S5A-C and S6. Supplemental Figure S1, Related to Figures 1 and 2. Supplemental Figure S2, Related to Figure
Introduction State the objectives of the work and provide adequate background, avoiding a detailed literature survey or a summary of the results.
 Format requirements for IJBCB articles For each article type, IJBCB has its own requirements regarding the article format. Please ensure you follow the exact guidelines (included below) for the specific
Format requirements for IJBCB articles For each article type, IJBCB has its own requirements regarding the article format. Please ensure you follow the exact guidelines (included below) for the specific
Figure S2. Response of mouse ES cells to GSK3 inhibition. Mentioned in discussion
 Stem Cell Reports, Volume 1 Supplemental Information Robust Self-Renewal of Rat Embryonic Stem Cells Requires Fine-Tuning of Glycogen Synthase Kinase-3 Inhibition Yaoyao Chen, Kathryn Blair, and Austin
Stem Cell Reports, Volume 1 Supplemental Information Robust Self-Renewal of Rat Embryonic Stem Cells Requires Fine-Tuning of Glycogen Synthase Kinase-3 Inhibition Yaoyao Chen, Kathryn Blair, and Austin
(A) Schematic illustration of sciatic nerve ligation. P, proximal; D, distal to the ligation site.
 SUPPLEMENTRY INFORMTION SUPPLEMENTL FIGURES Figure S1. () Schematic illustration of sciatic nerve ligation. P, proximal; D, distal to the ligation site. () Western blot of ligated and unligated sciatic
SUPPLEMENTRY INFORMTION SUPPLEMENTL FIGURES Figure S1. () Schematic illustration of sciatic nerve ligation. P, proximal; D, distal to the ligation site. () Western blot of ligated and unligated sciatic
TOOLS sirna and mirna. User guide
 TOOLS sirna and mirna User guide Introduction RNA interference (RNAi) is a powerful tool for suppression gene expression by causing the destruction of specific mrna molecules. Small Interfering RNAs (sirnas)
TOOLS sirna and mirna User guide Introduction RNA interference (RNAi) is a powerful tool for suppression gene expression by causing the destruction of specific mrna molecules. Small Interfering RNAs (sirnas)
Supplemental Information. Boundary Formation through a Direct. Threshold-Based Readout. of Mobile Small RNA Gradients
 Developmental Cell, Volume 43 Supplemental Information Boundary Formation through a Direct Threshold-Based Readout of Mobile Small RNA Gradients Damianos S. Skopelitis, Anna H. Benkovics, Aman Y. Husbands,
Developmental Cell, Volume 43 Supplemental Information Boundary Formation through a Direct Threshold-Based Readout of Mobile Small RNA Gradients Damianos S. Skopelitis, Anna H. Benkovics, Aman Y. Husbands,
The mechanism of translation initiation on Aichi Virus RNA mediated by a novel type of picornavirus IRES
 Manuscript EMBO-2011-78346 The mechanism of translation initiation on Aichi Virus RNA mediated by a novel type of picornavirus IRES Yingpu Yu, Trevor R. Sweeney, Panagiota Kafasla, Richard J. Jackson,
Manuscript EMBO-2011-78346 The mechanism of translation initiation on Aichi Virus RNA mediated by a novel type of picornavirus IRES Yingpu Yu, Trevor R. Sweeney, Panagiota Kafasla, Richard J. Jackson,
Impact of Retinoic acid induced-1 (Rai1) on Regulators of Metabolism and Adipogenesis
 Impact of Retinoic acid induced-1 (Rai1) on Regulators of Metabolism and Adipogenesis The mammalian system undergoes ~24 hour cycles known as circadian rhythms that temporally orchestrate metabolism, behavior,
Impact of Retinoic acid induced-1 (Rai1) on Regulators of Metabolism and Adipogenesis The mammalian system undergoes ~24 hour cycles known as circadian rhythms that temporally orchestrate metabolism, behavior,
Tumor Growth Suppression Through the Activation of p21, a Cyclin-Dependent Kinase Inhibitor
 Tumor Growth Suppression Through the Activation of p21, a Cyclin-Dependent Kinase Inhibitor Nicholas Love 11/28/01 A. What is p21? Introduction - p21 is a gene found on chromosome 6 at 6p21.2 - this gene
Tumor Growth Suppression Through the Activation of p21, a Cyclin-Dependent Kinase Inhibitor Nicholas Love 11/28/01 A. What is p21? Introduction - p21 is a gene found on chromosome 6 at 6p21.2 - this gene
Galina Gabriely, Ph.D. BWH/HMS
 Galina Gabriely, Ph.D. BWH/HMS Email: ggabriely@rics.bwh.harvard.edu Outline: microrna overview microrna expression analysis microrna functional analysis microrna (mirna) Characteristics mirnas discovered
Galina Gabriely, Ph.D. BWH/HMS Email: ggabriely@rics.bwh.harvard.edu Outline: microrna overview microrna expression analysis microrna functional analysis microrna (mirna) Characteristics mirnas discovered
supplementary information
 DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
Supplemental Data. Steiner et al. Plant Cell. (2012) /tpc
 Supplemental Figure 1. SPY does not interact with free GST. Invitro pull-down assay using E. coli-expressed MBP-SPY and GST, GST-TCP14 and GST-TCP15. MBP-SPY was used as bait and incubated with equal amount
Supplemental Figure 1. SPY does not interact with free GST. Invitro pull-down assay using E. coli-expressed MBP-SPY and GST, GST-TCP14 and GST-TCP15. MBP-SPY was used as bait and incubated with equal amount
MSD Immuno-Dot-Blot Assays. A division of Meso Scale Diagnostics, LLC.
 MSD Immuno-Dot-Blot Assays Example: High Throughput Western Blots Replacements Traditional Western Blots High content Molecular weight and immunoreactivity Labor and protein intensive Inherently low throughput
MSD Immuno-Dot-Blot Assays Example: High Throughput Western Blots Replacements Traditional Western Blots High content Molecular weight and immunoreactivity Labor and protein intensive Inherently low throughput
Supplementary Figures Montero et al._supplementary Figure 1
 Montero et al_suppl. Info 1 Supplementary Figures Montero et al._supplementary Figure 1 Montero et al_suppl. Info 2 Supplementary Figure 1. Transcripts arising from the structurally conserved subtelomeres
Montero et al_suppl. Info 1 Supplementary Figures Montero et al._supplementary Figure 1 Montero et al_suppl. Info 2 Supplementary Figure 1. Transcripts arising from the structurally conserved subtelomeres
Supplementary Figure 1: MYCER protein expressed from the transgene can enhance
 Relative luciferase activity Relative luciferase activity MYC is a critical target FBXW7 MYC Supplementary is a critical Figures target 1-7. FBXW7 Supplementary Material A E-box sequences 1 2 3 4 5 6 HSV-TK
Relative luciferase activity Relative luciferase activity MYC is a critical target FBXW7 MYC Supplementary is a critical Figures target 1-7. FBXW7 Supplementary Material A E-box sequences 1 2 3 4 5 6 HSV-TK
Supplementary Material
 Fig S1. Interactions of OsDof24 with the OsC4PPDK promoter in rice protoplasts (a) Loss-of-function analysis of the OsC4PPDK promoter Effects of OsDof24 overexpression and the mapping of OsDof24-binding
Fig S1. Interactions of OsDof24 with the OsC4PPDK promoter in rice protoplasts (a) Loss-of-function analysis of the OsC4PPDK promoter Effects of OsDof24 overexpression and the mapping of OsDof24-binding
Title: Production and characterisation of monoclonal antibodies against RAI3 and its expression in human breast cancer
 Author's response to reviews Title: Production and characterisation of monoclonal antibodies against RAI3 and its expression in human breast cancer Authors: Hannah Jörißen (hannah.joerissen@molbiotech.rwth-aachen.de)
Author's response to reviews Title: Production and characterisation of monoclonal antibodies against RAI3 and its expression in human breast cancer Authors: Hannah Jörißen (hannah.joerissen@molbiotech.rwth-aachen.de)
RNA Interference (RNAi) (see also mirna, sirna, micrna, shrna, etc.)
 Biochemistry 412 RNA Interference (RNAi) (see also mirna, sirna, micrna, shrna, etc.) April 8, 2008 The Discovery of the RNA Interference (RNAi) Phenomenon 1. Gene-specific inhibition of expression by
Biochemistry 412 RNA Interference (RNAi) (see also mirna, sirna, micrna, shrna, etc.) April 8, 2008 The Discovery of the RNA Interference (RNAi) Phenomenon 1. Gene-specific inhibition of expression by
GFP CCD2 GFP IP:GFP
 D1 D2 1 75 95 148 178 492 GFP CCD1 CCD2 CCD2 GFP D1 D2 GFP D1 D2 Beclin 1 IB:GFP IP:GFP Supplementary Figure 1: Mapping domains required for binding to HEK293T cells are transfected with EGFP-tagged mutant
D1 D2 1 75 95 148 178 492 GFP CCD1 CCD2 CCD2 GFP D1 D2 GFP D1 D2 Beclin 1 IB:GFP IP:GFP Supplementary Figure 1: Mapping domains required for binding to HEK293T cells are transfected with EGFP-tagged mutant
MOLECULAR BIOLOGY OF EUKARYOTES 2016 SYLLABUS
 03-442 Lectures: MWF 9:30-10:20 a.m. Doherty Hall 2105 03-742 Advanced Discussion Section: Time and place to be announced Probably Mon 4-6 p.m. or 6-8p.m.? Once we establish who is taking the advanced
03-442 Lectures: MWF 9:30-10:20 a.m. Doherty Hall 2105 03-742 Advanced Discussion Section: Time and place to be announced Probably Mon 4-6 p.m. or 6-8p.m.? Once we establish who is taking the advanced
Supporting Information
 Supporting Information Su et al. 10.1073/pnas.1211604110 SI Materials and Methods Cell Culture and Plasmids. Tera-1 and Tera-2 cells (ATCC: HTB- 105/106) were maintained in McCoy s 5A medium with 15% FBS
Supporting Information Su et al. 10.1073/pnas.1211604110 SI Materials and Methods Cell Culture and Plasmids. Tera-1 and Tera-2 cells (ATCC: HTB- 105/106) were maintained in McCoy s 5A medium with 15% FBS
Description: Nuclear morphology and dynamics in nontargeting sirna transfected cells. HeLa Kyoto
 Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Tables Title of file for HTML: Supplementary Movie 1 Description: Nuclear morphology and dynamics
Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Tables Title of file for HTML: Supplementary Movie 1 Description: Nuclear morphology and dynamics
ARQUIVOS BRASILEIROS DE CARDIOLOGIA (BRAZILIAN ARCHIVES OF CARDIOLOGY) GUIDELINES FOR PUBLICATION
 ARQUIVOS BRASILEIROS DE CARDIOLOGIA (BRAZILIAN ARCHIVES OF CARDIOLOGY) GUIDELINES FOR PUBLICATION 1. The Brazilian Archives of Cardiology (Arq Bras Cardiol) is a monthly publication of the Brazilian Society
ARQUIVOS BRASILEIROS DE CARDIOLOGIA (BRAZILIAN ARCHIVES OF CARDIOLOGY) GUIDELINES FOR PUBLICATION 1. The Brazilian Archives of Cardiology (Arq Bras Cardiol) is a monthly publication of the Brazilian Society
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/3/6/ra27/dc Supplementary Materials for AAA+ Proteins and Coordinate PIKK Activity and Function in Nonsense-Mediated mrna Decay Natsuko Izumi, Akio Yamashita,*
www.sciencesignaling.org/cgi/content/full/3/6/ra27/dc Supplementary Materials for AAA+ Proteins and Coordinate PIKK Activity and Function in Nonsense-Mediated mrna Decay Natsuko Izumi, Akio Yamashita,*
Supplementary Figure 1. RAD51 and RAD51 paralogs are enriched spontaneously onto
 Supplementary Figure legends Supplementary Figure 1. and paralogs are enriched spontaneously onto the S-phase chromatin during DN replication. () Chromatin fractionation was carried out as described in
Supplementary Figure legends Supplementary Figure 1. and paralogs are enriched spontaneously onto the S-phase chromatin during DN replication. () Chromatin fractionation was carried out as described in
Alternative Cleavage and Polyadenylation of RNA
 Developmental Cell 18 Supplemental Information The Spen Family Protein FPA Controls Alternative Cleavage and Polyadenylation of RNA Csaba Hornyik, Lionel C. Terzi, and Gordon G. Simpson Figure S1, related
Developmental Cell 18 Supplemental Information The Spen Family Protein FPA Controls Alternative Cleavage and Polyadenylation of RNA Csaba Hornyik, Lionel C. Terzi, and Gordon G. Simpson Figure S1, related
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION DOI: 10.1038/ncb3575 In the format provided by the authors and unedited. Supplementary Figure 1 Validation of key reagents and assays a, top, IHC with antibody recognizing specifically
SUPPLEMENTARY INFORMATION DOI: 10.1038/ncb3575 In the format provided by the authors and unedited. Supplementary Figure 1 Validation of key reagents and assays a, top, IHC with antibody recognizing specifically
Quantitative Real Time PCR USING SYBR GREEN
 Quantitative Real Time PCR USING SYBR GREEN SYBR Green SYBR Green is a cyanine dye that binds to double stranded DNA. When it is bound to D.S. DNA it has a much greater fluorescence than when bound to
Quantitative Real Time PCR USING SYBR GREEN SYBR Green SYBR Green is a cyanine dye that binds to double stranded DNA. When it is bound to D.S. DNA it has a much greater fluorescence than when bound to
Table S1. Primer sequences
 Table S1. Primer sequences Primers for quantitative PCR Tgf 1 Forward Tgf 1 Reverse Tgf 2Forward Tgf 2Reverse Tgf 3 Forward Tgf 3 Reverse Tgf r1 Forward Tgf r1 Reverse Tgf r2 Forward Tgf r2 Reverse Thbs1
Table S1. Primer sequences Primers for quantitative PCR Tgf 1 Forward Tgf 1 Reverse Tgf 2Forward Tgf 2Reverse Tgf 3 Forward Tgf 3 Reverse Tgf r1 Forward Tgf r1 Reverse Tgf r2 Forward Tgf r2 Reverse Thbs1
Charles Shuler, Ph.D. University of Southern California Los Angeles, California Approved for Public Release; Distribution Unlimited
 AD Award Number: DAMD17-01-1-0100 TITLE: Smad-Mediated Signaling During Prostate Growth and Development PRINCIPAL INVESTIGATOR: Charles Shuler, Ph.D. CONTRACTING ORGANIZATION: University of Southern California
AD Award Number: DAMD17-01-1-0100 TITLE: Smad-Mediated Signaling During Prostate Growth and Development PRINCIPAL INVESTIGATOR: Charles Shuler, Ph.D. CONTRACTING ORGANIZATION: University of Southern California
Corporate Medical Policy
 Corporate Medical Policy Proteogenomic Testing for Patients with Cancer (GPS Cancer Test) File Name: Origination: Last CAP Review: Next CAP Review: Last Review: proteogenomic_testing_for_patients_with_cancer_gps_cancer_test
Corporate Medical Policy Proteogenomic Testing for Patients with Cancer (GPS Cancer Test) File Name: Origination: Last CAP Review: Next CAP Review: Last Review: proteogenomic_testing_for_patients_with_cancer_gps_cancer_test
466 Asn (N) to Ala (A) Generate beta dimer Interface
 Table S1: Amino acid changes to the HexA α-subunit to convert the dimer interface from α to β and to introduce the putative GM2A binding surface from β- onto the α- subunit Residue position (α-numbering)
Table S1: Amino acid changes to the HexA α-subunit to convert the dimer interface from α to β and to introduce the putative GM2A binding surface from β- onto the α- subunit Residue position (α-numbering)
