NIH Public Access Author Manuscript Nature. Author manuscript; available in PMC 2010 October 1.
|
|
|
- Marybeth Gilbert
- 6 years ago
- Views:
Transcription
1 NIH Public Access Author Manuscript Published in final edited form as: Nature April 15; 464(7291): doi: /nature Functional genomic screen for modulators of ciliogenesis and cilium length Joon Kim 1, Ji Eun Lee 1, Susanne Heynen 2, Eigo Suyama 3, Keiichiro Ono 4, KiYoung Lee 4,5, Trey Ideker 4, Pedro Aza-Blac 3, and Joseph G. Gleeson 1 1 Departments of Neurosciences and Pediatrics, Institute for Genomic Medicine, Howard Hughes Medical Institute, University of California San Diego, La Jolla, CA 92093, USA 2 High Content Screening Systems, Burnham Institute, La Jolla, CA 92037, USA 3 Functional Genomics Core, Burnham Institute, La Jolla, CA 92037, USA 4 Departments of Medicine and Bioengineering, University of California San Diego, La Jolla, CA 92093, USA 5 Department of Biomedical Informatics, Ajou University School of Medicine, Suwon , Korea Abstract Primary cilia are evolutionarily conserved cellular organelles that organize diverse signaling pathways 1,2. Defects in the formation or function of primary cilia are associated with a spectrum of human diseases and developmental abnormalities 3. Genetic screens in model organisms have discovered core machineries of cilium assembly and maintenance 4. However, regulatory molecules that coordinate the biogenesis of primary cilia with other cellular processes, including cytoskeletal organization, vesicle trafficking and cell-cell adhesion, remain to be identified. Here we report the results of a functional genomic screen using RNA interference (RNAi) to identify human genes involved in ciliogenesis control. The screen identified 36 positive and 13 negative ciliogenesis modulators, which include molecules involved in actin dynamics and vesicle trafficking. Further investigation demonstrated that blocking actin assembly facilitates ciliogenesis by stabilizing the pericentrosomal preciliary compartment (PPC), a previously uncharacterized compact vesiculotubular structure storing transmembrane proteins destined for cilia during the early phase of ciliogenesis. PPC was labeled by recycling endosome markers. Moreover, knockdown of modulators that are involved in the endocytic recycling pathway affected the formation of PPC as well as ciliogenesis. Our results uncover a critical regulatory step that couples actin dynamics and endocytic recycling with ciliogenesis, and also provide potential target molecules for future study. Users may view, print, copy, download and text and data- mine the content in such documents, for the purposes of academic research, subject always to the full Conditions of use: Correspondence and requests for materials should be addressed to Joseph G. Gleeson: 9500 Gilman Drive, M/C 0665, Leichtag Biomedical Research Building R482, University of California San Diego, La Jolla, CA 92093, Tel: , Fax: , jogleeson@ucsd.edu. Author Information Reprints and permissions information is available at Supplementary Information is linked to the online version of the paper a Author Contributions J.G.G. conceived and directed the project. J.K., S.H., and P.A. designed the screen and J.K., S.H. and E.S. performed the screen. J.K., K.O., K.L. and T.I. analyzed the screen data. J.K. designed the follow-up experiments and J.K. and J.L. performed the experiments. J.K. interpreted the results and J.K. and J.G.G. wrote the paper with contributions from S.H and P.A. The authors declare competing financial interests (patent application).
2 Kim et al. Page 2 Cilium assembly and disassembly are interconnected with several complex cellular processes such as cell cycle, cell polarization and cell migration, suggesting that there may be large numbers of ciliogenesis modulators 1,5. In accordance with this, recent proteomic analyses together with comparative genomics and bioinformatics studies have identified over a thousand cilia- or basal body-associated proteins, referred to as the ciliome 6,7. Although these approaches can provide a comprehensive list of candidates, the discovery of key regulators of ciliogenesis, which could reveal potential therapeutic targets, requires functional analysis. Thus we developed a high-throughput assay using small interfering RNA (sirna) to evaluate the functional impact of 7784 therapeutically relevant genes across the human genome (Supplementary Table 1). The screen was based on an in vitro ciliogenesis model: serum starvation-induced ciliogenesis in telomerase-immortalized human retinal pigmented epithelial (htrpe) cells. EGFP-tagged Smoothed (Smo), a transmembrane protein that accumulates in primary cilia 8, was used as a cilium marker for automated quantification of ciliated cells (Supplementary Fig. 1a,b,c). Sensitivity of the screen strategy was assessed by sirnas targeting KIF3A, a critical protein for cilium assembly (Supplementary Fig. 1d). We selected a screen condition suitable for identifying both positive and negative modulators: sirna transfection was performed at a reduced (suboptimal) cell density in order to detect these dynamic ranges of ciliogenesis activity, and ciliation was assessed after 48 hr serum starvation (Supplementary Fig. 1e,f). The primary screen identified 153 positive modulator hits and 79 negative modulator hits (Supplementary Table 1 and Supplementary Fig. 2a). We next performed a confirmation screen with the following modifications: (1) optimal cell density for ciliogenesis (confluent at the time of serum starvation) was used to minimize the selection of sirnas which indirectly influence ciliogenesis through an effect on cell density, (2) cilium length was measured for validating negative modulators because length increase was a highly specific indicator for enhanced ciliogenesis, where optimal ciliogenesis condition was used (Supplementary Fig. 2b). The confirmation screen identified 40 positive and 13 negative modulators, which are associated with various molecular processes (Supplementary Table 2 and Supplementary Fig. 3). Remarkably, the screen hits include INPP5E, a gene recently identified as mutated in a human ciliopathy, Joubert syndrome 9. A positive modulator Agtpbp1 (formerly Nna1) is mutated in Purkinje cell degeneration (pcd) mice, which exhibit ciliopathy phenotypes: retinal degeneration and defective spermatogenesis 10. Although the library was selected for therapeutically relevant genes and thus not highly representative of ciliome genes (tending to be structural), we did confirm 12 genes encoding ciliome proteins (Supplementary Table 3 and Supplementary Fig. 3). We rescreened the confirmed positive modulators using anti-acetylated tubulin immunofluorescence as a cilium marker, and found 4 genes that did not affect cilium assembly (Supplementary Table 3 and Supplementary Fig. 3). The confirmed negative modulators were further classified according to silencing phenotypes observed in the absence of serum starvation. Silencing of 5 genes facilitated ciliogenesis even without serum-starvation (Supplementary Table 3 and Supplementary Fig. 3). Among the ciliogenesis modulators with high confirmation screen scores (Supplementary Table 2) are two Gelsolin (GSN) family proteins GSN and AVIL, which regulate cytoskeletal actin organization by severing actin filaments 11. Depletion of GSN proteins by two independent sirnas significantly reduced ciliated cell numbers, suggesting that actin filament severing is involved in ciliogenesis (Fig. 1a,b,c). In contrast, silencing of actinrelated protein ACTR3 (also called ARP3), which is a major constituent of the ARP2/3 complex that is necessary for nucleating actin polymerization at filament branches 12, caused a significant increase in cilium length (Fig. 1a,d,e) and also facilitated ciliogenesis
3 Kim et al. Page 3 independent of serum starvation (Fig. 1f). These observations suggest an inhibitory role of branched actin network formation in ciliogenesis. To examine the link between actin dynamics and ciliogenesis, we treated htrpe cells with actin polymerization inhibitor cytochalasind, which facilitated ciliogenesis independent of serum starvation and promoted cilium elongation (Fig. 1g and Supplementary Fig. 4). Ciliogenic effect of cytochalasind was also observed in HEK293T cells (Supplementary Fig. 5a,b). Moreover, cytochalasind treatment significantly rescued ciliogenesis defect caused by GSN knockdown (Fig. 1h). CytochalasinD facilitated ciliogenesis at doses below that which affect stress fiber formation, excluding the possibility of global actin cytoskeleton rearrangement in ciliogenesis control (Supplementary Fig. 5c). Involvement of actin dynamics in ciliogenesis is further supported by the finding that α-parvin (PARVA), a component of the focal adhesion complex regulating actin cytoskeletal dynamics and cell signaling 13, exhibits knockdown phenotypes similar to that of ACTR3 (Supplementary Fig. 6). In order to identify the mechanism of altered ciliogenesis, we performed time-course live imaging. Remarkably, a number of non-ciliated htrpe cells transfected with ACTR3 sirnas (14/30 cells) developed pronounced primary cilia (2.9±0.9 μm in length) within 2 hrs of serum starvation, whereas none of the control cells (0/30 cells) displayed cilia longer than 1.5 μm (Fig. 1i and Supplementary Fig. 7a). Furthermore, faster cilium elongation after the initiation of ciliogenesis was observed in ACTR3 depleted cells [mean cilium extension rate for first 6 hrs after serum starvation: ACTR3 depleted cells, 0.75±0.27 μm/hr (n=17); control cells, 0.11±0.06 μm/hr (n=6)]. These observations suggest that slow progression of ciliogenesis in htrpe cells is not the result of slow transduction of serum starvationmediated ciliogenic signal or unavailability of core cilium assembly machineries, but is ascribed to the presence of an inhibitory regulation involving ACTR3. Earlier electron microscopic studies found small vesicles tightly associated with the centriole initiating cilium assembly in fibroblasts and smooth muscle cells 14,15. This suggests that ciliogenesis in non-polarized cells could be initiated by vesicle docking to the basal body, whereas basal body docking to the apical plasma membrane initiates ciliogenesis in polarized epithelial cells. More recent studies also have reported that the biogenesis of ciliary membrane that is coordinated with axoneme assembly may involve fusion of transport vesicles at the base of cilia 16,17. SmoEGFP allowed us to visualize transport vesicles targeted to the ciliary base. Interestingly, a subset of htrpe-smoegfp cells ( 10 %: 42/434 interphase cells) cultured in the presence of serum exhibited compact SmoEGFP positive vesiculotubular structures (termed here PPC) tightly associated with the centrosome (Fig. 1J). In these cells, ciliogenesis occurred at PPC during 4 hr live imaging (Fig. 1k). Notably, an attenuation of SmoEGFP positive PPC was observed after the initiation of ciliogenesis (Fig. 2k and Supplementary Fig. 7b, control cells). After 4 hr serum starvation (when cilia are detected in 20 % of cells), 3.4 % of total cilia (17/500 cilia) were accompanied by PPC, suggesting that the transition of PPC to cilia occurs over the course of hours. PPC positive cilia were rarely observed after 24 hr serum starvation (2/500 cilia), confirming that PPC is a transient structure (Supplementary Fig. 8a). SmoEGFP revealed similar transient structures in IMCD cells (Supplementary Fig. 8b). Remarkably, the majority of newly formed cilia (13/17 cilia) displaying mature lengths (4.5±1.9 μm) in ACTR3 depleted-live cells maintained pronounced PPC at the ciliary base (Fig. 1i and Supplementary Fig. 7b) although PPC eventually disappeared after 24 hr serum starvation as in control (Supplementary Fig. 8a). Moreover, facilitated ciliogenesis in cytochalasind treated cells was preceded by a promotion of PPC formation [16 PPC positive cells/30 live-imaged cells treated with cytochalasind for 2 hrs vs. 5 PPC positive cells/30
4 Kim et al. Page 4 control cells (Fig. 1l)]. Short-term live imaging showed that cytochalasind also affects the stability of the pre-existing PPC (Supplementary Fig. 9). These observations suggest that actin network formation may negatively modulate ciliogenesis by destabilizing the PPC, a potential temporary reservoir of lipid and membrane proteins for efficient ciliogenesis. Recycling endosomes are a dynamic vesiculotubular compartment, exporting endocytosed membrane proteins and lipids to the cell surface via vesicular intermediates 18. We found that PPC extensively overlaps with a subset of recycling endosomes concentrated around the centrosome: PPC was labeled by endocytosed transferrin and recycling endosome marker Rab11 (Fig. 2a and Supplementary Fig. 10). PTPN23, a nontransmembrane tyrosine phosphatase, has been implicated in cargo sorting at early endocytic compartments 19, and is one of the high-scored hits. Silencing of PTPN23 significantly reduced the number of ciliated cells (Fig. 2b and Supplementary Fig.11). In addition, PTPN23 knockdown caused an accumulation of SmoEGFP on the early endosomes marked by EEA1 (Fig. 2c). PPC was not observed from the cells exhibiting early endosomal SmoEGFP accumulation (0/80 cells; Supplementary Fig. 12). Supporting the link between recycling endosomes and ciliogenesis, knockdown of ASAP1 (not included in the initial screen library), a gene required for pericentrosomal enrichment of recycling endosomes 20, caused a decrease in the number of ciliated cells (Supplementary Fig. 13a,b,c). These observations suggest that a trafficking pathway connecting endocytic compartments to PPC is involved in cilium assembly. Pharmacological blocking of endocytic degradation pathway to lysosomes using concanamycina did not block PPC formation or ciliogenesis, indicating that endocytic recycling pathway is specifically involved in ciliogenesis (Supplementary Fig. 13d). Detection of PTPN23 immunoreactivity at the basal bodies further supported a direct role for PTPN23 in ciliary vesicle targeting (Fig. 2d). Among the unexpected screen hits was PLA2G3, a secreted phospholipase 21. Involvement of cytoplasmic Phospholipase A2 enzymes in intracellular vesicle trafficking has been suggested based on their potential for introducing membrane curvature 22. Depletion of PLA2G3 facilitated cilium extension and induced ciliogenesis independent of serum starvation, indicating that PLA2G3 is a negative ciliogenesis regulator (Fig. 2e,f and Supplementary Fig. 14). Confirming this result, application of oleyloxyethyl phosphorylcholine (OPC), an inhibitor for secreted phospholipase 23, increased the number of ciliated cells (Supplementary Fig. 15a). Notably, PLA2G3 immunofluorescence was detected at the centriole pair (Fig. 2g). In accord with its localization, knockdown of PLA2G3 significantly increased the number of cells displaying recycling endosomes concentrated at high levels around the centrosome [17.4±1.4 % in control cells and 48.2±2.7 % in PLA2G3 depleted cells examined after 56 hr transfection without serum starvation; Student's t-test, P=0.006 (Fig. 2h)]. The majority of pericentrosomal recycling endosomes in PLA2G3 depleted cells (122/153) were associated with PPC or cilia. Overexpressed PLA2G3 exhibited an extensive co-localization with endocytosed transferrin and inhibited ciliogenesis (Supplementary Fig. 14d,e,f). These results further demonstrate the link between endocytic recycling pathway and ciliogenesis. Moreover, higher intensities of SmoEGFP fluorescence was observed in cilia from cells depleted of PLA2G3, demonstrating an inhibitory role for PLA2G3 in ciliary membrane protein targeting (Supplementary Fig. 15b,c). The fact that both axoneme assembly and ciliary membrane biogenesis were facilitated by inhibition of actin polymerization (Supplementary Fig. 16) prompted us to test if cytochalasind treatment could rescue ciliogenesis defect caused by loss-of-function mutation in core ciliogenesis machinery. IFT88 transports molecules required for cilium assembly 4, and cells carrying homozygous IFT88 hypomorphic mutation (orpk/orpk) fail to grow cilia with normal length 24. Remarkably, 16 hr cytochalasind treatment partially, but
5 Kim et al. Page 5 Method Summary Cell culture sirna library screen significantly, rescued ciliogenesis defect of IFT88 mutant cells (Fig. 3). A recent study showed that IFT particles are associated with recycling endosomes and are implicated in the recycling of T-cell receptors in lymphocytes 25. Thus it is likely that cytochalasind facilitates ciliary recruitment of IFT particles through the endocytic recycling pathway. CytochalasinD can interfere with many cellular processes involving actin polymerization, and thus is unlikely to be useful for ciliopathy treatment. However, small molecules specifically targeting actin modulators implicated in ciliogenesis might be good candidates for ciliopathy treatment. In summary, our high-throughput functional screen identified modulators of ciliogenesis with diverse molecular functions, connecting ciliogenesis with other basic cellular processes. Furthermore, we discovered several proteins involved in the control of cilium length, a key determinant of normal cilium function. We anticipate that the development of small molecules that target these proteins may provide novel strategies for the treatment of the ciliopathies. htrpe cells were maintained in DMEM:F12 supplemented with 10 % fetal bovine serum. Plasmid DNA harboring mouse SmoEGFP fusion gene was transfected to htrpe cells and a stable cell line (htrpe-smoegfp) was established by G418 selection. For ciliogenesis induction, culture medium was replaced with DMEM (without supplement) when cells are 90 % confluent, and cultured for 48 hrs before fixation. Overexpressed SmoEGFP was targeted to ciliary membrane independent of Shh. An arrayed library containing unique sirnas targeting 7784 human genes (Ambion; human druggable genome sirna library V3.1; including sirnas targeting 665 Kinases, 101 Kinase modulators, 231 Phosphatases, 307 Ligases, 57 Lipases, 444 Signaling molecules and 426 Cytoskeleton interacting proteins) was screened in duplicates. A highthroughput imaging was done using IC100 automated microscope system (Beckman Coulter), and the images were analyzed by Cytoshop software. For a confirmation screen, re-arrayed 913 sirnas targeting 232 primary screen hits were tested in quadruplicates. Supplementary Material Acknowledgments References Refer to Web version on PubMed Central for supplementary material. The screen was performed at the Burnham Institute Functional Genomics Core (LJINCC grant P30 NS to S.Lipton) with technical assistance from A.Cortez and E.Sisman and with imaging support from the UCSD Microscopy Core (P30 NS and P30 CA23100). We thank P.A.Beachy for SmoEGFP plasmid and J.F.Reiter for IFT88 orpk/orpk MEFs. We thank C.Kintner, V.M.Fowler, S.J.Field and S.Ferro-Novick for discussion. This work was supported by grants from NARSAD (Young Investigator Award to J.K.), NINDS (RO1 NS to J.G.G) and the Howard Hughes Medical Institute. 1. Gerdes JM, Davis EE, Katsanis N. The vertebrate primary cilium in development, homeostasis, and disease. Cell. 2009; 137: [PubMed: ] 2. Eggenschwiler JT, Anderson KV. Cilia and developmental signaling. Annu Rev Cell Dev Biol. 2007; 23: [PubMed: ]
6 Kim et al. Page 6 3. Fliegauf M, Benzing T, Omran H. When cilia go bad: cilia defects and ciliopathies. Nat Rev Mol Cell Biol. 2007; 8: [PubMed: ] 4. Rosenbaum JL, Witman GB. Intraflagellar transport. Nature Rev Mol Cell Biol. 2002; 3: [PubMed: ] 5. Christensen ST, et al. The primary cilium coordinates signaling pathways in cell cycle control and migration during development and tissue repair. Curr Top Dev Biol. 2008; 85: [PubMed: ] 6. Inglis PN, Boroevich KA, Leroux MR. Piecing together a ciliome. Trends Genet. 2006; 22: [PubMed: ] 7. Gherman A, Davis EE, Katsanis N. The ciliary proteome database: an integrated community resource for the genetic and functional dissection of cilia. Nature Genet. 2006; 38: [PubMed: ] 8. Corbit KC, et al. Vertebrate Smoothened functions at the primary cilium. Nature. 2005; 437: [PubMed: ] 9. Bielas SL, et al. Mutations in INPP5E, encoding inositol polyphosphate-5-phosphatase E, link phosphatidyl inositol signaling to the ciliopathies. Nature Genet. 2009; 41: [PubMed: ] 10. Fernandez-Gonzalez A, et al. Purkinje cell degeneration (pcd) phenotypes caused by mutations in the axotomy-induced gene, Nna1. Science. 2002; 295: [PubMed: ] 11. Silacci P, et al. Gelsolin superfamily proteins: key regulators of cellular functions. Cell Mol Life Sci. 2004; 61: [PubMed: ] 12. Cooper JA, Schafer DA. Control of actin assembly and disassembly at filament ends. Curr Opin Cell Biol. 2000; 12: [PubMed: ] 13. Sepulveda JL, Wu C. The parvins. Cell Mol Life Sci. 2006; 63: [PubMed: ] 14. Sorokin S. Centrioles and the formation of rudimentary cilia by fibroblasts and smooth muscle cells. J Cell Biol. 1962; 15: [PubMed: ] 15. Archer FL, Wheatley DN. Cilia in cell-cultured fibroblasts. II. Incidence in mitotic and postmitotic BHK 21-C13 fibroblasts. J Anat. 1971; 109: [PubMed: ] 16. Nachury MV, et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007; 129: [PubMed: ] 17. Yoshimura S, et al. Functional dissection of Rab GTPases involved in primary cilium formation. J Cell Biol. 2007; 178: [PubMed: ] 18. Maxfield FR, McGraw TE. Endocytic recycling. Nature Rev Mol Cell Biol. 2004; 5: [PubMed: ] 19. Doyotte A, Mironov A, McKenzie E, Woodman P. The Bro1-related protein HD-PTP/PTPN23 is required for endosomal cargo sorting and multivesicular body morphogenesis. Proc Natl Acad Sci U S A. 2008; 105: [PubMed: ] 20. Inoue H, Ha VL, Prekeris R, Randazzo PA. Arf GTPase-activating protein ASAP1 interacts with Rab11 effector FIP3 and regulates pericentrosomal localization of transferrin receptor-positive recycling endosome. Mol Biol Cell. 2008; 19: [PubMed: ] 21. Valentin E, et al. Novel human secreted phospholipase A(2) with homology to the group III bee venom enzyme. J Biol Chem. 2000; 275: [PubMed: ] 22. Brown WJ, Chambers K, Doody A. Phospholipase A2 (PLA2) enzymes in membrane trafficking: mediators of membrane shape and function. Traffic. 2003; 4: [PubMed: ] 23. Hope WC, Chen T, Morgan DW. Secretory phospholipase A2 inhibitors and calmodulin antagonists as inhibitors of cytosolic phospholipase A2. Agents Actions. 1993; 39:C Spec No. [PubMed: ] 24. Yoder BK, et al. Polaris, a protein disrupted in orpk mutant mice, is required for assembly of renal cilium. Am J Physiol Renal Physiol. 2002; 282:F [PubMed: ] 25. Finetti F, et al. Intraflagellar transport is required for polarized recycling of the TCR/CD3 complex to the immune synapse. Nature Cell Biol. 2009; 11: [PubMed: ]
7 Kim et al. Page 7 Figure 1. Regulators of actin dynamics modulate cilium assembly a, b, GSN knockdown reduced ciliated cell numbers. a, d, ACTR3 knockdown increased the number of cells with longer cilium (> 6 μm). c, e, Western blots showing protein levels ( GSN sirna#1 was cytotoxic). f, ACTR3 knockdown induced ciliogenesis without serum starvation. g, CytochalasinD treatment for 8 hrs increased ciliated cell numbers without serum starvation. h, Ciliogenesis defect by GSN knockdown was rescued by 8 hr cytochalasind treatment. i, Live imaging of htrpe-smoegfp cells after 60 hr sirna transfection. Arrowheads, the pericentrosomal preciliary compartment (PPC) at the ciliary base. j, Morphology of PPC. Gamma-tubulin labels the centrosome. k, Live imaging of a PPC positive cell in serum-free medium. l, Live cell imaging in serum-free medium containing DMSO or cytochalasind. Values, mean ± SD [n = 4 (b,d), 2 (f) and 3 (g,h)]. Student's t-test: *P < 0.05, **P < Scale bars, 10 μm (a); 5 μm (i,l); 2.5 μm (j,k).
8 Kim et al. Page 8 Figure 2. Endocytic recycling pathway is linked to ciliogenesis a, PPC overlapping with endocytosed transferrin-alexa594 and Rab11 (arrowhead). b, PTPN23 knockdown decreased the number of ciliated cells. c, PTPN23 knockdown caused an accumulation of SmoEGFP on EEA1-positive early endosomes. d, PTPN23 immunofluorescence was detected at the ciliary base. e, PLA2G3 knockdown increased the number of cells with longer cilia (> 6 μm). f, PLA2G3 knockdown induced ciliogenesis without serum starvation. g, Pericentriolar localization of PLA2G3. h, PLA2G3 knockdown increased the number of cells exhibiting recycling endosomes concentrated at high levels around the centrosome (arrowheads). Values, mean ± SD [n = 4 (b,e) and 3 (f)]. Student's t- test: *P < 0.05, **P < Scale bars, 2.5 μm (a,d,g); 5 μm (c); 20 μm (h).
9 Kim et al. Page 9 Figure 3. Pharmacological rescue of ciliary defect on IFT88 mutant cells a, Cytochalasin D treatment for 16 hrs partially rescued cilium elongation defect caused by hypormorphic IFT88 mutation. b, Percentage of cells with the primary cilium longer than 1.5 μm. Values, mean ± SD (n = 3). Student's t-test: *P < Scale bar, 5 μm.
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
Xu et al., Supplementary Figures 1-7
 Xu et al., Supplementary Figures 1-7 Supplementary Figure 1. PIPKI is required for ciliogenesis. (a) PIPKI localizes at the basal body of primary cilium. RPE-1 cells treated with two sirnas targeting to
Xu et al., Supplementary Figures 1-7 Supplementary Figure 1. PIPKI is required for ciliogenesis. (a) PIPKI localizes at the basal body of primary cilium. RPE-1 cells treated with two sirnas targeting to
Ahi1, whose human ortholog is mutated in Joubert syndrome, is required for Rab8a localization, ciliogenesis and vesicle trafficking
 Human Molecular Genetics, 2009, Vol. 18, No. 20 3926 3941 doi:10.1093/hmg/ddp335 Advance Access published on July 22, 2009 Ahi1, whose human ortholog is mutated in Joubert syndrome, is required for Rab8a
Human Molecular Genetics, 2009, Vol. 18, No. 20 3926 3941 doi:10.1093/hmg/ddp335 Advance Access published on July 22, 2009 Ahi1, whose human ortholog is mutated in Joubert syndrome, is required for Rab8a
Supplemental Material
 Supplemental Material 1 Figure S1. Phylogenetic analysis of Cep72 and Lrrc36, comparative localization of Cep72 and Lrrc36 and Cep72 antibody characterization (A) Phylogenetic alignment of Cep72 and Lrrc36
Supplemental Material 1 Figure S1. Phylogenetic analysis of Cep72 and Lrrc36, comparative localization of Cep72 and Lrrc36 and Cep72 antibody characterization (A) Phylogenetic alignment of Cep72 and Lrrc36
β-arrestin mediated localization of Smoothened to the primary cilium
 Published as: Science. 2008 June 27; 320(5884): 1777 1781. β-arrestin mediated localization of Smoothened to the primary cilium Jeffrey J. Kovacs 1,2, Erin J. Whalen 1, Renshui Liu 1, Kunhong Xiao 3, Jihee
Published as: Science. 2008 June 27; 320(5884): 1777 1781. β-arrestin mediated localization of Smoothened to the primary cilium Jeffrey J. Kovacs 1,2, Erin J. Whalen 1, Renshui Liu 1, Kunhong Xiao 3, Jihee
OmicsLink shrna Clones guaranteed knockdown even in difficult-to-transfect cells
 OmicsLink shrna Clones guaranteed knockdown even in difficult-to-transfect cells OmicsLink shrna clone collections consist of lentiviral, and other mammalian expression vector based small hairpin RNA (shrna)
OmicsLink shrna Clones guaranteed knockdown even in difficult-to-transfect cells OmicsLink shrna clone collections consist of lentiviral, and other mammalian expression vector based small hairpin RNA (shrna)
OriGene GFC-Arrays for High-throughput Overexpression Screening of Human Gene Phenotypes
 OriGene GFC-Arrays for High-throughput Overexpression Screening of Human Gene Phenotypes High-throughput Gene Function Validation Tool Introduction sirna screening libraries enable scientists to identify
OriGene GFC-Arrays for High-throughput Overexpression Screening of Human Gene Phenotypes High-throughput Gene Function Validation Tool Introduction sirna screening libraries enable scientists to identify
Supplemental Information
 Supplemental Information Intrinsic protein-protein interaction mediated and chaperonin assisted sequential assembly of a stable Bardet Biedl syndome protein complex, the BBSome * Qihong Zhang 1#, Dahai
Supplemental Information Intrinsic protein-protein interaction mediated and chaperonin assisted sequential assembly of a stable Bardet Biedl syndome protein complex, the BBSome * Qihong Zhang 1#, Dahai
Supplementary Figure 1, related to Figure 1. GAS5 is highly expressed in the cytoplasm of hescs, and positively correlates with pluripotency.
 Supplementary Figure 1, related to Figure 1. GAS5 is highly expressed in the cytoplasm of hescs, and positively correlates with pluripotency. (a) Transfection of different concentration of GAS5-overexpressing
Supplementary Figure 1, related to Figure 1. GAS5 is highly expressed in the cytoplasm of hescs, and positively correlates with pluripotency. (a) Transfection of different concentration of GAS5-overexpressing
Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question in Section B and ONE question from Section C.
 UNIVERSITY OF EAST ANGLIA School of Biological Sciences Main Series UG Examination 2017-18 CELL BIOLOGY BIO-5005B Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question in Section
UNIVERSITY OF EAST ANGLIA School of Biological Sciences Main Series UG Examination 2017-18 CELL BIOLOGY BIO-5005B Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question in Section
Microtubules. Forms a sheet of protofilaments that folds to a tube Both tubulins bind GTP
 Microtubules Polymerize from a-tubulin/ß-tubulin dimers Hollow tube of 13 protofilaments In vitro- filament number varies In vivo- always 13 protofilaments Forms a sheet of protofilaments that folds to
Microtubules Polymerize from a-tubulin/ß-tubulin dimers Hollow tube of 13 protofilaments In vitro- filament number varies In vivo- always 13 protofilaments Forms a sheet of protofilaments that folds to
General Education Learning Outcomes
 BOROUGH OF MANHATTAN COMMUNITY COLLEGE City University of New York Department of Science Title of Course: Cell Biology Class hours 3 BIO Section: 260 Lab hours 3 Semester Spring 2018 Credits 4 Schedule:
BOROUGH OF MANHATTAN COMMUNITY COLLEGE City University of New York Department of Science Title of Course: Cell Biology Class hours 3 BIO Section: 260 Lab hours 3 Semester Spring 2018 Credits 4 Schedule:
Learning Objectives. Define RNA interference. Define basic terminology. Describe molecular mechanism. Define VSP and relevance
 Learning Objectives Define RNA interference Define basic terminology Describe molecular mechanism Define VSP and relevance Describe role of RNAi in antigenic variation A Nobel Way to Regulate Gene Expression
Learning Objectives Define RNA interference Define basic terminology Describe molecular mechanism Define VSP and relevance Describe role of RNAi in antigenic variation A Nobel Way to Regulate Gene Expression
Stargazin regulates AMPA receptor trafficking through adaptor protein. complexes during long term depression
 Supplementary Information Stargazin regulates AMPA receptor trafficking through adaptor protein complexes during long term depression Shinji Matsuda, Wataru Kakegawa, Timotheus Budisantoso, Toshihiro Nomura,
Supplementary Information Stargazin regulates AMPA receptor trafficking through adaptor protein complexes during long term depression Shinji Matsuda, Wataru Kakegawa, Timotheus Budisantoso, Toshihiro Nomura,
Chapter 15. Cytoskeletal Systems. Lectures by Kathleen Fitzpatrick Simon Fraser University Pearson Education, Inc.
 Chapter 15 Cytoskeletal Systems Lectures by Kathleen Fitzpatrick Simon Fraser University Table 15-1 - Microtubules Table 15-1 - Microfilaments Table 15-1 Intermediate Filaments Table 15-3 Microtubules
Chapter 15 Cytoskeletal Systems Lectures by Kathleen Fitzpatrick Simon Fraser University Table 15-1 - Microtubules Table 15-1 - Microfilaments Table 15-1 Intermediate Filaments Table 15-3 Microtubules
BME Engineering Molecular Cell Biology. The Cytoskeleton (I): Actin The Cytoskeleton (II): Microtubule & Intermediate Filament
 BME 42-620 Engineering Molecular Cell Biology Lecture 09: The Cytoskeleton (I): Actin The Cytoskeleton (II): Microtubule & Intermediate Filament BME42-620 Lecture 09, September 27, 2011 1 Outline Overviewofcytoskeletal
BME 42-620 Engineering Molecular Cell Biology Lecture 09: The Cytoskeleton (I): Actin The Cytoskeleton (II): Microtubule & Intermediate Filament BME42-620 Lecture 09, September 27, 2011 1 Outline Overviewofcytoskeletal
supplementary information
 DOI: 10.1038/ncb2156 Figure S1 Depletion of p114rhogef with different sirnas. Caco-2 (a) and HCE (b) cells were transfected with individual sirnas, pools of the two sirnas or the On Target (OnT) sirna
DOI: 10.1038/ncb2156 Figure S1 Depletion of p114rhogef with different sirnas. Caco-2 (a) and HCE (b) cells were transfected with individual sirnas, pools of the two sirnas or the On Target (OnT) sirna
CHAPTER 16 THE CYTOSKELETON
 CHAPTER 16 THE CYTOSKELETON THE SELF-ASSEMBLY AND DYNAMIC STRUCTURE OF CYTOSKELETAL FILAMENTS HOW CELLS REGULATE THEIR CYTOSKELETAL FILAMENTS MOLECULAR MOTORS THE CYTOSKELETON AND CELL BEHAVIOR THE SELF-ASSEMBLY
CHAPTER 16 THE CYTOSKELETON THE SELF-ASSEMBLY AND DYNAMIC STRUCTURE OF CYTOSKELETAL FILAMENTS HOW CELLS REGULATE THEIR CYTOSKELETAL FILAMENTS MOLECULAR MOTORS THE CYTOSKELETON AND CELL BEHAVIOR THE SELF-ASSEMBLY
Regulation of Acetylcholine Receptor Clustering by ADF/Cofilin- Directed Vesicular Trafficking
 Regulation of Acetylcholine Receptor Clustering by ADF/Cofilin- Directed Vesicular Trafficking Chi Wai Lee, Jianzhong Han, James R. Bamburg, Liang Han, Rachel Lynn, and James Q. Zheng Supplementary Figures
Regulation of Acetylcholine Receptor Clustering by ADF/Cofilin- Directed Vesicular Trafficking Chi Wai Lee, Jianzhong Han, James R. Bamburg, Liang Han, Rachel Lynn, and James Q. Zheng Supplementary Figures
Figure S1 is related to Figure 1B, showing more details of outer segment of
 Supplemental Information Supplementary Figure legends and Figures Figure S1. Electron microscopic images in Sema4A +/+ and Sema4A / retinas Figure S1 is related to Figure 1B, showing more details of outer
Supplemental Information Supplementary Figure legends and Figures Figure S1. Electron microscopic images in Sema4A +/+ and Sema4A / retinas Figure S1 is related to Figure 1B, showing more details of outer
The following antibodies were used in this study: NuMA - rabbit-anti NuMA (gift from
 Materials and Methods Antibodies: The following antibodies were used in this study: NuMA - rabbit-anti NuMA (gift from D. A. Compton); Dynein Intermediate Chain - 74.1 (Chemicon, Temecula, CA); Dynein
Materials and Methods Antibodies: The following antibodies were used in this study: NuMA - rabbit-anti NuMA (gift from D. A. Compton); Dynein Intermediate Chain - 74.1 (Chemicon, Temecula, CA); Dynein
A Tumor Necrosis Factor-Alpha-Mediated Pathway Promoting Autosomal Dominant Polycystic Kidney Disease.
 A Tumor Necrosis Factor-Alpha-Mediated Pathway Promoting Autosomal Dominant Polycystic Kidney Disease. Xiaogang Li, Brenda S. Magenheimer, Sheng Xia, Teri Johnson, Darren P. Wallace, James P. Calvet and
A Tumor Necrosis Factor-Alpha-Mediated Pathway Promoting Autosomal Dominant Polycystic Kidney Disease. Xiaogang Li, Brenda S. Magenheimer, Sheng Xia, Teri Johnson, Darren P. Wallace, James P. Calvet and
Supplementary Figure 1. Localization of MST1 in RPE cells. Proliferating or ciliated HA- MST1 expressing RPE cells (see Fig. 5b for establishment of
 Supplementary Figure 1. Localization of MST1 in RPE cells. Proliferating or ciliated HA- MST1 expressing RPE cells (see Fig. 5b for establishment of the cell line) were immunostained for HA, acetylated
Supplementary Figure 1. Localization of MST1 in RPE cells. Proliferating or ciliated HA- MST1 expressing RPE cells (see Fig. 5b for establishment of the cell line) were immunostained for HA, acetylated
Revision Checklist for Science Signaling Research Manuscripts: Data Requirements and Style Guidelines
 Revision Checklist for Science Signaling Research Manuscripts: Data Requirements and Style Guidelines Further information can be found at: http://stke.sciencemag.org/sites/default/files/researcharticlerevmsinstructions_0.pdf.
Revision Checklist for Science Signaling Research Manuscripts: Data Requirements and Style Guidelines Further information can be found at: http://stke.sciencemag.org/sites/default/files/researcharticlerevmsinstructions_0.pdf.
3'UTR elements inhibit Ras-induced C/EBPß posttranslational activation and senescence in tumor cells
 Manuscript EMBO-2011-77536 3'UTR elements inhibit Ras-induced C/EBPß posttranslational activation and senescence in tumor cells Sandip K. Basu, Radek Malik, Christopher J. Huggins, Sook Lee, Thomas Sebastian,
Manuscript EMBO-2011-77536 3'UTR elements inhibit Ras-induced C/EBPß posttranslational activation and senescence in tumor cells Sandip K. Basu, Radek Malik, Christopher J. Huggins, Sook Lee, Thomas Sebastian,
System. Dynamic Monitoring of Receptor Tyrosine Kinase Activation in Living Cells. Application Note No. 4 / March
 System Application Note No. 4 / March 2008 Dynamic Monitoring of Receptor Tyrosine Kinase Activation in Living Cells www.roche-applied-science.com Dynamic Monitoring of Receptor Tyrosine Kinase Activation
System Application Note No. 4 / March 2008 Dynamic Monitoring of Receptor Tyrosine Kinase Activation in Living Cells www.roche-applied-science.com Dynamic Monitoring of Receptor Tyrosine Kinase Activation
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2386 Figure 1 Src-containing puncta are not focal adhesions, podosomes or endosomes. (a) FAK-/- were stained with anti-py416 Src (green) and either (in red) the focal adhesion protein paxillin,
DOI: 10.1038/ncb2386 Figure 1 Src-containing puncta are not focal adhesions, podosomes or endosomes. (a) FAK-/- were stained with anti-py416 Src (green) and either (in red) the focal adhesion protein paxillin,
1/24/2012. Cell. Plasma Membrane
 Chapter 3 Outline Plasma Membrane Cytoplasm and Its Organelles Cell and Gene Expression Protein Synthesis and Secretion DNA Synthesis and Cell Division Cell Basic unit of structure and function in body
Chapter 3 Outline Plasma Membrane Cytoplasm and Its Organelles Cell and Gene Expression Protein Synthesis and Secretion DNA Synthesis and Cell Division Cell Basic unit of structure and function in body
SureSilencing sirna Array Technology Overview
 SureSilencing sirna Array Technology Overview Pathway-Focused sirna-based RNA Interference Topics to be Covered Who is SuperArray? Brief Introduction to RNA Interference Challenges Facing RNA Interference
SureSilencing sirna Array Technology Overview Pathway-Focused sirna-based RNA Interference Topics to be Covered Who is SuperArray? Brief Introduction to RNA Interference Challenges Facing RNA Interference
Supplementary Figure S1. N-terminal fragments of LRRK1 bind to Grb2.
 Myc- HA-Grb2 Mr(K) 105 IP HA 75 25 105 1-1163 1-595 - + - + - + 1164-1989 Blot Myc HA total lysate 75 25 Myc HA Supplementary Figure S1. N-terminal fragments of bind to Grb2. COS7 cells were cotransfected
Myc- HA-Grb2 Mr(K) 105 IP HA 75 25 105 1-1163 1-595 - + - + - + 1164-1989 Blot Myc HA total lysate 75 25 Myc HA Supplementary Figure S1. N-terminal fragments of bind to Grb2. COS7 cells were cotransfected
Fuse-It Membrane Fusion
 Fuse-It Membrane Fusion Next Generation Transfection Including: LifeAct Actin Visualization in Living Cells www..com Mechanisms and Methods of Molecular Transfer into Living Cells Fuse-It- Fuse-It-siRNA
Fuse-It Membrane Fusion Next Generation Transfection Including: LifeAct Actin Visualization in Living Cells www..com Mechanisms and Methods of Molecular Transfer into Living Cells Fuse-It- Fuse-It-siRNA
Supplemental Material: Rev1 promotes replication through UV lesions in conjunction with DNA
 Supplemental Material: Rev1 promotes replication through UV lesions in conjunction with DNA polymerases,, and, but not with DNA polymerase Jung-Hoon Yoon, Jeseong Park, Juan Conde, Maki Wakamiya, Louise
Supplemental Material: Rev1 promotes replication through UV lesions in conjunction with DNA polymerases,, and, but not with DNA polymerase Jung-Hoon Yoon, Jeseong Park, Juan Conde, Maki Wakamiya, Louise
SONOMA STATE UNIVERSITY DEPARTMENT OF BIOLOGY BIOLOGY 344: CELL BIOLOGY Fall 2013
 SONOMA STATE UNIVERSITY DEPARTMENT OF BIOLOGY BIOLOGY 344: CELL BIOLOGY Fall 2013 Instructor Murali C. Pillai, PhD Office 214 Darwin Hall Telephone (707) 664-2981 E-mail pillai@sonoma.edu Website www.sonoma.edu/users/p/pillai
SONOMA STATE UNIVERSITY DEPARTMENT OF BIOLOGY BIOLOGY 344: CELL BIOLOGY Fall 2013 Instructor Murali C. Pillai, PhD Office 214 Darwin Hall Telephone (707) 664-2981 E-mail pillai@sonoma.edu Website www.sonoma.edu/users/p/pillai
New microribbon production
 New microribbon production In vitro transformation Excystation brief exposure to acidic ph (~2) flagellar activity within 5-10 min after return to neutral ph breakdown of cyst wall (proteases) trophozoite
New microribbon production In vitro transformation Excystation brief exposure to acidic ph (~2) flagellar activity within 5-10 min after return to neutral ph breakdown of cyst wall (proteases) trophozoite
cell fusion live and fixed imaging genetics biochemistry in vitro systems inhibitors of cellular processes (transcription, replication, microtubules)
 DISCUSSION SECTIONS BY STUDENT NUMBER ENDING IN ODD NUMBERS 2-3, EVEN 3-4 Methods for Studying the Cell Cycle cell fusion live and fixed imaging genetics biochemistry in vitro systems inhibitors of cellular
DISCUSSION SECTIONS BY STUDENT NUMBER ENDING IN ODD NUMBERS 2-3, EVEN 3-4 Methods for Studying the Cell Cycle cell fusion live and fixed imaging genetics biochemistry in vitro systems inhibitors of cellular
Cytomics in Action: Cytokine Network Cytometry
 Cytomics in Action: Cytokine Network Cytometry Jonni S. Moore, Ph.D. Director, Clinical and Research Flow Cytometry and PathBioResource Associate Professor of Pathology & Laboratory Medicine University
Cytomics in Action: Cytokine Network Cytometry Jonni S. Moore, Ph.D. Director, Clinical and Research Flow Cytometry and PathBioResource Associate Professor of Pathology & Laboratory Medicine University
Technical tips Session 4
 Technical tips Session 4 Biotinylation assay: Streptavidin is a small bacterial protein that binds with high affinity to the vitamin biotin. This streptavidin-biotin combination can be used to link molecules
Technical tips Session 4 Biotinylation assay: Streptavidin is a small bacterial protein that binds with high affinity to the vitamin biotin. This streptavidin-biotin combination can be used to link molecules
Flag-Rac Vector V12 V12 N17 C40. Vector C40 pakt (T308) Akt1. Myc-DN-PAK1 (N-SP)
 a b FlagRac FlagRac V2 V2 N7 C4 V2 V2 N7 C4 p (T38) p (S99, S24) p Flag (Rac) NIH 3T3 COS c +Serum p (T38) MycDN (NSP) Mycp27 3 6 2 3 6 2 3 6 2 min p Myc ( or p27) Figure S (a) Effects of Rac mutants on
a b FlagRac FlagRac V2 V2 N7 C4 V2 V2 N7 C4 p (T38) p (S99, S24) p Flag (Rac) NIH 3T3 COS c +Serum p (T38) MycDN (NSP) Mycp27 3 6 2 3 6 2 3 6 2 min p Myc ( or p27) Figure S (a) Effects of Rac mutants on
Switch On Protein-Protein Interactions
 Switch On Protein-Protein Interactions Rapid and specific control of signal transduction pathways, protein activity, protein localization, transcription, and more Dimerizer protein 1 protein 2 Clontech
Switch On Protein-Protein Interactions Rapid and specific control of signal transduction pathways, protein activity, protein localization, transcription, and more Dimerizer protein 1 protein 2 Clontech
Smith Family Awards Program for Excellence in Biomedical Research 2018 Award Recipients
 Smith Family Awards Program for Excellence in Biomedical Research 2018 Award Recipients David Breslow, Ph.D. Assistant Professor of Molecular, Cellular & Developmental Biology Yale University Deconstructing
Smith Family Awards Program for Excellence in Biomedical Research 2018 Award Recipients David Breslow, Ph.D. Assistant Professor of Molecular, Cellular & Developmental Biology Yale University Deconstructing
JCB. Supplemental material THE JOURNAL OF CELL BIOLOGY. Paul et al.,
 Supplemental material JCB Paul et al., http://www.jcb.org/cgi/content/full/jcb.201502040/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Mutant p53-expressing cells display limited retrograde actin flow at
Supplemental material JCB Paul et al., http://www.jcb.org/cgi/content/full/jcb.201502040/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Mutant p53-expressing cells display limited retrograde actin flow at
Section 10. Junaid Malek, M.D.
 Section 10 Junaid Malek, M.D. Cell Division Make sure you understand: How do cells know when to divide? (What drives the cell cycle? Why is it important to regulate this?) How is DNA replication regulated?
Section 10 Junaid Malek, M.D. Cell Division Make sure you understand: How do cells know when to divide? (What drives the cell cycle? Why is it important to regulate this?) How is DNA replication regulated?
sirna Overview and Technical Tips
 1 sirna Overview and Technical Tips 2 CONTENTS 3 4 5 7 8 10 11 13 14 18 19 20 21 Introduction Applications How Does It Work? Handy Tips Troubleshooting Conclusions Further References Contact Us 3 INTRODUCTION
1 sirna Overview and Technical Tips 2 CONTENTS 3 4 5 7 8 10 11 13 14 18 19 20 21 Introduction Applications How Does It Work? Handy Tips Troubleshooting Conclusions Further References Contact Us 3 INTRODUCTION
1 Supplementary Figure 1. Kif13b expression and localization in serum starved
 1 2 3 4 5 6 Supplementary Figure 1. Kif13b expression and localization in serum starved cells. (a) Immunoblot of NIH3T3 cell lysates after 24 hrs of serum deprivation, using rabbit polyclonal KIF13B antiserum.
1 2 3 4 5 6 Supplementary Figure 1. Kif13b expression and localization in serum starved cells. (a) Immunoblot of NIH3T3 cell lysates after 24 hrs of serum deprivation, using rabbit polyclonal KIF13B antiserum.
MICB688L/MOCB639 Advanced Cell Biology Exam II
 MICB688L/MOCB639 Advanced Cell Biology Exam II May 10, 2001 Name: 1. Briefly describe the four major classes of cell surface receptors and their modes of action (immediate downstream only) (8) 2. Please
MICB688L/MOCB639 Advanced Cell Biology Exam II May 10, 2001 Name: 1. Briefly describe the four major classes of cell surface receptors and their modes of action (immediate downstream only) (8) 2. Please
Fig. S1. Endocytosis of extracellular cargo does not depend on dia. (A) To visualize endocytosis, fluorescently labelled wheat germ agglutinin
 Fig. S1. Endocytosis of extracellular cargo does not depend on dia. (A) To visualize endocytosis, fluorescently labelled wheat germ agglutinin (WGA-Alexa555) was injected into the extracellular perivitteline
Fig. S1. Endocytosis of extracellular cargo does not depend on dia. (A) To visualize endocytosis, fluorescently labelled wheat germ agglutinin (WGA-Alexa555) was injected into the extracellular perivitteline
Boston, MA PREPARED FOR: U.S. Army Medical Research and Materiel Command Fort Detrick, Maryland
 AWARD NUMBER: W81XWH-15-1-0139 TITLE: Polymeric RNAi Microsponge Delivery Simultaneously Targeting Multiple Genes for Novel Pathway Inhibition of Ovarian Cancer PRINCIPAL INVESTIGATOR: Michael Birrer CONTRACTING
AWARD NUMBER: W81XWH-15-1-0139 TITLE: Polymeric RNAi Microsponge Delivery Simultaneously Targeting Multiple Genes for Novel Pathway Inhibition of Ovarian Cancer PRINCIPAL INVESTIGATOR: Michael Birrer CONTRACTING
Antibody Structure. Antibodies
 Antibodies Secreted by B lymphocytes Great diversity and specificity: >10 9 different antibodies; can distinguish between very similar molecules Tag particles for clearance/destruction Protect against
Antibodies Secreted by B lymphocytes Great diversity and specificity: >10 9 different antibodies; can distinguish between very similar molecules Tag particles for clearance/destruction Protect against
Antibody Structure supports Function
 Antibodies Secreted by B lymphocytes Great diversity and specificity: >10 9 different antibodies; can distinguish between very similar molecules Tag particles for clearance/destruction Protect against
Antibodies Secreted by B lymphocytes Great diversity and specificity: >10 9 different antibodies; can distinguish between very similar molecules Tag particles for clearance/destruction Protect against
7.012 Problem Set 5. Question 1
 Name Section 7.012 Problem Set 5 Question 1 While studying the problem of infertility, you attempt to isolate a hypothetical rabbit gene that accounts for the prolific reproduction of rabbits. After much
Name Section 7.012 Problem Set 5 Question 1 While studying the problem of infertility, you attempt to isolate a hypothetical rabbit gene that accounts for the prolific reproduction of rabbits. After much
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or
 Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
FRAUNHOFER IME SCREENINGPORT
 FRAUNHOFER IME SCREENINGPORT Detection technologies used in drug discovery Introduction Detection technologies in drug discovery is unlimited only a biased snapshot can be presented Differences can presented
FRAUNHOFER IME SCREENINGPORT Detection technologies used in drug discovery Introduction Detection technologies in drug discovery is unlimited only a biased snapshot can be presented Differences can presented
Aminoacid change in chromophore. PIN3::PIN3-GFP GFP S65 (no change) (5.9) (Kneen et al., 1998)
 Supplemental Table. Table S1. Fluorophore characteristics of used fluorescent proteins, Related to Figure 2 and 3. Transgenic line Fluprescent marker Aminoacid change in chromophore pk(a) Ref. PIN3::PIN3-GFP
Supplemental Table. Table S1. Fluorophore characteristics of used fluorescent proteins, Related to Figure 2 and 3. Transgenic line Fluprescent marker Aminoacid change in chromophore pk(a) Ref. PIN3::PIN3-GFP
CRISPR GENOMIC SERVICES PRODUCT CATALOG
 CRISPR GENOMIC SERVICES PRODUCT CATALOG DESIGN BUILD ANALYZE The experts at Desktop Genetics can help you design, prepare and manufacture all of the components needed for your CRISPR screen. We provide
CRISPR GENOMIC SERVICES PRODUCT CATALOG DESIGN BUILD ANALYZE The experts at Desktop Genetics can help you design, prepare and manufacture all of the components needed for your CRISPR screen. We provide
Three major types of cytoskeleton
 The Cytoskeleton Organizes and stabilizes cells Pulls chromosomes apart Drives intracellular traffic Supports plasma membrane and nuclear envelope Enables cellular movement Guides growth of the plant cell
The Cytoskeleton Organizes and stabilizes cells Pulls chromosomes apart Drives intracellular traffic Supports plasma membrane and nuclear envelope Enables cellular movement Guides growth of the plant cell
University of Pennsylvania Perelman School of Medicine High Throughput Screening Core
 University of Pennsylvania Perelman School of Medicine High Throughput Screening Core Sara Cherry, Ph. D. David C. Schultz, Ph. D. dschultz@mail.med.upenn.edu (215) 573 9641 67 John Morgan Building Mission
University of Pennsylvania Perelman School of Medicine High Throughput Screening Core Sara Cherry, Ph. D. David C. Schultz, Ph. D. dschultz@mail.med.upenn.edu (215) 573 9641 67 John Morgan Building Mission
Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and
 Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
Chapter 3. DNA Replication & The Cell Cycle
 Chapter 3 DNA Replication & The Cell Cycle DNA Replication and the Cell Cycle Before cells divide, they must duplicate their DNA // the genetic material DNA is organized into strands called chromosomes
Chapter 3 DNA Replication & The Cell Cycle DNA Replication and the Cell Cycle Before cells divide, they must duplicate their DNA // the genetic material DNA is organized into strands called chromosomes
7.06 Cell Biology QUIZ #3
 Recitation Section: 7.06 Cell Biology QUIZ #3 This is an open book exam, and you are allowed access to books and notes, but not computers or any other types of electronic devices. Please write your answers
Recitation Section: 7.06 Cell Biology QUIZ #3 This is an open book exam, and you are allowed access to books and notes, but not computers or any other types of electronic devices. Please write your answers
Lullaby sirna Transfection Reagent - Results
 sirna Transfection Reagent - Results OZ Biosciences is delighted to announce the launching of a new sirna transfection reagent: -sirna. This lipid based transfection reagent is specifically designed for
sirna Transfection Reagent - Results OZ Biosciences is delighted to announce the launching of a new sirna transfection reagent: -sirna. This lipid based transfection reagent is specifically designed for
me239 mechanics of the cell homework biopolymers - motivation 3.1 biopolymers - motivation 3. biopolymers biopolymers biopolymers
 3. biopolymers the inner life of a cell, viel & lue, harvard [2006] me239 mechanics of the cell 1 2 biopolymers Figure 3.1. Biopolymers. Characteristic length scales on the cellular and sucellular level..
3. biopolymers the inner life of a cell, viel & lue, harvard [2006] me239 mechanics of the cell 1 2 biopolymers Figure 3.1. Biopolymers. Characteristic length scales on the cellular and sucellular level..
Electronic Supplementary Information
 Electronic Supplementary Material (ESI) for Integrative Biology. This journal is The Royal Society of Chemistry 2015 Electronic Supplementary Information Table S1. Definition of quantitative cellular features
Electronic Supplementary Material (ESI) for Integrative Biology. This journal is The Royal Society of Chemistry 2015 Electronic Supplementary Information Table S1. Definition of quantitative cellular features
Building Better Algae
 Building Better Algae Craig Marcus, Ph.D. Dept. of Environmental & Molecular Toxicology Domestication of Algae as a New Crop Must develop a rapid process (corn first domesticated ~4000 B.C.) Requires a
Building Better Algae Craig Marcus, Ph.D. Dept. of Environmental & Molecular Toxicology Domestication of Algae as a New Crop Must develop a rapid process (corn first domesticated ~4000 B.C.) Requires a
MCBII. Points this page
 1. What makes intermediate filaments (IFs) an inefficient track for motor proteins (4 pts)? A. The outer surface of IFs is hydrophobic. B. IFs are nonpolar structures. C. IFs contain coiled coil domains.
1. What makes intermediate filaments (IFs) an inefficient track for motor proteins (4 pts)? A. The outer surface of IFs is hydrophobic. B. IFs are nonpolar structures. C. IFs contain coiled coil domains.
HPV E6 oncoprotein targets histone methyltransferases for modulating specific. Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu,
 1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
Post-translational modification
 Protein expression Western blotting, is a widely used and accepted technique to detect levels of protein expression in a cell or tissue extract. This technique measures protein levels in a biological sample
Protein expression Western blotting, is a widely used and accepted technique to detect levels of protein expression in a cell or tissue extract. This technique measures protein levels in a biological sample
CELL BIOLOGY BIOL3030, 3 credits Fall 2012, Aug 20, Dec 14, 2012 Tuesday/Thursday 8:00 am-9:15 am Bowman-Oddy Laboratories 1049
 INSTRUCTOR: Dr. Song-Tao Liu WO3254B Tel: 419-530-7853 Email: Song-Tao.Liu@utoledo.edu OFFICE HOURS CELL BIOLOGY BIOL3030, 3 credits Fall 2012, Aug 20, 2012 - Dec 14, 2012 Tuesday/Thursday 8:00 am-9:15
INSTRUCTOR: Dr. Song-Tao Liu WO3254B Tel: 419-530-7853 Email: Song-Tao.Liu@utoledo.edu OFFICE HOURS CELL BIOLOGY BIOL3030, 3 credits Fall 2012, Aug 20, 2012 - Dec 14, 2012 Tuesday/Thursday 8:00 am-9:15
Supplementary Figure 1. Co-localization of GLUT1 and DNAL4 in BeWo cells cultured
 Supplementary Figure 1. Co-localization of GLUT1 and DNAL4 in BeWo cells cultured under static conditions. Cells were seeded in the chamber area of the device and cultured overnight without medium perfusion.
Supplementary Figure 1. Co-localization of GLUT1 and DNAL4 in BeWo cells cultured under static conditions. Cells were seeded in the chamber area of the device and cultured overnight without medium perfusion.
RXi Pharmaceuticals. BioPharm America September 26, 2017 NASDAQ: RXII. Property of RXi Pharmaceuticals
 RXi Pharmaceuticals BioPharm America September 26, 2017 NASDAQ: RXII Forward Looking Statements This presentation contains forward-looking statements within the meaning of the Private Securities Litigation
RXi Pharmaceuticals BioPharm America September 26, 2017 NASDAQ: RXII Forward Looking Statements This presentation contains forward-looking statements within the meaning of the Private Securities Litigation
Focus Application. Cell Migration. Featured Study: Inhibition of Cell Migration by Gene Silencing. xcelligence System Real-Time Cell Analyzer
 xcelligence System Real-Time Cell Analyzer Focus Application Cell Migration Featured Study: Inhibition of Cell Migration by Gene Silencing Markus Greiner and Richard Zimmermann Department of Medical Biochemistry
xcelligence System Real-Time Cell Analyzer Focus Application Cell Migration Featured Study: Inhibition of Cell Migration by Gene Silencing Markus Greiner and Richard Zimmermann Department of Medical Biochemistry
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling
 Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
PLEASE MAKE SURE THAT ALL PAGES ARE NUMBERED and IDENTIFIED!!!!!.
 Instructions: The following pages contain twelve questions. You must answer ten out of the twelve questions. Any one question may ask for more than one response so please read each question carefully for
Instructions: The following pages contain twelve questions. You must answer ten out of the twelve questions. Any one question may ask for more than one response so please read each question carefully for
Lab Module 7: Cell Adhesion
 Lab Module 7: Cell Adhesion Tissues are made of cells and materials secreted by cells that occupy the spaces between the individual cells. This material outside of cells is called the Extracellular Matrix
Lab Module 7: Cell Adhesion Tissues are made of cells and materials secreted by cells that occupy the spaces between the individual cells. This material outside of cells is called the Extracellular Matrix
Tracking Cellular Protein Localization and Movement in Cells with a Flexible Fluorescent Labeling Technology. Chad Zimprich January 2015
 Tracking Cellular Protein Localization and Movement in Cells with a Flexible Fluorescent Labeling Technology Chad Zimprich January 2015 Presentation verview HaloTag Fusion Technology Design Functionality
Tracking Cellular Protein Localization and Movement in Cells with a Flexible Fluorescent Labeling Technology Chad Zimprich January 2015 Presentation verview HaloTag Fusion Technology Design Functionality
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Thompson et al., http://www.jcb.org/cgi/content/full/jcb.200909067/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Modification-specific antibodies do not detect unmodified
Supplemental material Thompson et al., http://www.jcb.org/cgi/content/full/jcb.200909067/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Modification-specific antibodies do not detect unmodified
In-Cell Western QUANTITATIVE CELL-BASED ASSAYS ACCURATE QUANTIFICATION MULTIPLEX DETECTION HIGH SENSITIVITY DIRECT DETECTION
 In-Cell Western QUANTITATIVE CELL-BASED ASSAYS ACCURATE QUANTIFICATION MULTIPLEX DETECTION HIGH SENSITIVITY DIRECT DETECTION IMMUNOFLUORESCENT-BASED ASSAY MULTIPLE TARGETS In-Cell Western QUANTITATIVE
In-Cell Western QUANTITATIVE CELL-BASED ASSAYS ACCURATE QUANTIFICATION MULTIPLEX DETECTION HIGH SENSITIVITY DIRECT DETECTION IMMUNOFLUORESCENT-BASED ASSAY MULTIPLE TARGETS In-Cell Western QUANTITATIVE
Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product.
 Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
PLA2 domain in parvoviruses The enzymatic features of viral PLA2
 PLA2 domain in parvoviruses Sequence analysis of 21 Pavovirinae members revealed that they contain an 80 amino acid conserved region in their VP1up. 20 amino acids out of the 80 were fully conserved and
PLA2 domain in parvoviruses Sequence analysis of 21 Pavovirinae members revealed that they contain an 80 amino acid conserved region in their VP1up. 20 amino acids out of the 80 were fully conserved and
sirna Transfection Into Primary Neurons Using Fuse-It-siRNA
 sirna Transfection Into Primary Neurons Using Fuse-It-siRNA This Application Note describes a protocol for sirna transfection into sensitive, primary cortical neurons using Fuse-It-siRNA. This innovative
sirna Transfection Into Primary Neurons Using Fuse-It-siRNA This Application Note describes a protocol for sirna transfection into sensitive, primary cortical neurons using Fuse-It-siRNA. This innovative
Therapeutic & Prevention Application of Nucleic Acids
 Therapeutic & Prevention Application of Nucleic Acids Seyed Amir Hossein Jalali Institute of Biotechnology and Bioengineering, Isfahan University Of Technology (IUT). 30.7.2015 * Plasmids * DNA Aptamers
Therapeutic & Prevention Application of Nucleic Acids Seyed Amir Hossein Jalali Institute of Biotechnology and Bioengineering, Isfahan University Of Technology (IUT). 30.7.2015 * Plasmids * DNA Aptamers
Molecular Cell Biology - Problem Drill 01: Introduction to Molecular Cell Biology
 Molecular Cell Biology - Problem Drill 01: Introduction to Molecular Cell Biology Question No. 1 of 10 1. Which statement describes how an organism is organized from most simple to most complex? Question
Molecular Cell Biology - Problem Drill 01: Introduction to Molecular Cell Biology Question No. 1 of 10 1. Which statement describes how an organism is organized from most simple to most complex? Question
Lecture 13. Motor Proteins I
 Lecture 13 Motor Proteins I Introduction: The study of motor proteins has become a major focus in cell and molecular biology. Motor proteins are very interesting because they do what no man-made engines
Lecture 13 Motor Proteins I Introduction: The study of motor proteins has become a major focus in cell and molecular biology. Motor proteins are very interesting because they do what no man-made engines
Cancer cells that survive radiation therapy acquire HIF-1 activity and translocate toward tumor blood vessels Supplementary Information
 Cancer cells that survive radiation therapy acquire HIF-1 activity and translocate toward tumor blood vessels Supplementary Information 1. Supplementary Figure S1-S10: Pages 2-11 2. Supplementary References:
Cancer cells that survive radiation therapy acquire HIF-1 activity and translocate toward tumor blood vessels Supplementary Information 1. Supplementary Figure S1-S10: Pages 2-11 2. Supplementary References:
Supplementary Figure 1. RAD51 and RAD51 paralogs are enriched spontaneously onto
 Supplementary Figure legends Supplementary Figure 1. and paralogs are enriched spontaneously onto the S-phase chromatin during DN replication. () Chromatin fractionation was carried out as described in
Supplementary Figure legends Supplementary Figure 1. and paralogs are enriched spontaneously onto the S-phase chromatin during DN replication. () Chromatin fractionation was carried out as described in
Supplementary Figure 1. α-synuclein is truncated in PD and LBD brains. Nature Structural & Molecular Biology: doi: /nsmb.
 Supplementary Figure 1 α-synuclein is truncated in PD and LBD brains. (a) Specificity of anti-n103 antibody. Anti-N103 antibody was coated on an ELISA plate and different concentrations of full-length
Supplementary Figure 1 α-synuclein is truncated in PD and LBD brains. (a) Specificity of anti-n103 antibody. Anti-N103 antibody was coated on an ELISA plate and different concentrations of full-length
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature12138 Supplementary Figure 1. Knockdown of KRAS leads to a reduction in macropinocytosis. (a) KRAS knockdown in MIA PaCa-2 cells expressing KRASspecific shrnas
SUPPLEMENTARY INFORMATION doi:10.1038/nature12138 Supplementary Figure 1. Knockdown of KRAS leads to a reduction in macropinocytosis. (a) KRAS knockdown in MIA PaCa-2 cells expressing KRASspecific shrnas
Supplemental Movie Legend.
 Supplemental Movie Legend. Transfected T cells were dropped onto SEE superantigen-pulsed Raji B cells (approximate location indicated by circle). Maximum-intensity projections from Z-stacks (17 slices,
Supplemental Movie Legend. Transfected T cells were dropped onto SEE superantigen-pulsed Raji B cells (approximate location indicated by circle). Maximum-intensity projections from Z-stacks (17 slices,
How Targets Are Chosen. Chris Wayman 12 th April 2012
 How Targets Are Chosen Chris Wayman 12 th April 2012 A few questions How many ideas does it take to make a medicine? 10 20 20-50 50-100 A few questions How long does it take to bring a product from bench
How Targets Are Chosen Chris Wayman 12 th April 2012 A few questions How many ideas does it take to make a medicine? 10 20 20-50 50-100 A few questions How long does it take to bring a product from bench
C) Based on your knowledge of transcription, what factors do you expect to ultimately identify? (1 point)
 Question 1 Unlike bacterial RNA polymerase, eukaryotic RNA polymerases need additional assistance in finding a promoter and transcription start site. You wish to reconstitute accurate transcription by
Question 1 Unlike bacterial RNA polymerase, eukaryotic RNA polymerases need additional assistance in finding a promoter and transcription start site. You wish to reconstitute accurate transcription by
Functional characterisation
 Me Me Ac Ac Functional characterisation How can we know if measured changes in DNA methylation and function (phenotype) and linked, and in what way? DNMT Dianne Ford Professor of Molecular Nutritional
Me Me Ac Ac Functional characterisation How can we know if measured changes in DNA methylation and function (phenotype) and linked, and in what way? DNMT Dianne Ford Professor of Molecular Nutritional
The microtubule-associated tau protein has intrinsic acetyltransferase activity. Todd J. Cohen, Dave Friedmann, Andrew W. Hwang, Ronen Marmorstein and
 SUPPLEMENTARY INFORMATION: The microtubule-associated tau protein has intrinsic acetyltransferase activity Todd J. Cohen, Dave Friedmann, Andrew W. Hwang, Ronen Marmorstein and Virginia M.Y. Lee Cohen
SUPPLEMENTARY INFORMATION: The microtubule-associated tau protein has intrinsic acetyltransferase activity Todd J. Cohen, Dave Friedmann, Andrew W. Hwang, Ronen Marmorstein and Virginia M.Y. Lee Cohen
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10163 Supplementary Table 1 Efficiency of vector construction. Process wells recovered efficiency (%) Recombineering* 480 461 96 Intermediate plasmids 461 381 83 Recombineering efficiency
doi:10.1038/nature10163 Supplementary Table 1 Efficiency of vector construction. Process wells recovered efficiency (%) Recombineering* 480 461 96 Intermediate plasmids 461 381 83 Recombineering efficiency
Hector R. Wong, MD Division of Critical Care Medicine Cincinnati Children s Hospital Medical Center Cincinnati Children s Research Foundation
 Mechanisms of Sepsis Exploiting Pathogenic Genes Hector R. Wong, MD Division of Critical Care Medicine Cincinnati Children s Hospital Medical Center Cincinnati Children s Research Foundation Critical Care
Mechanisms of Sepsis Exploiting Pathogenic Genes Hector R. Wong, MD Division of Critical Care Medicine Cincinnati Children s Hospital Medical Center Cincinnati Children s Research Foundation Critical Care
The Ups & Downs of Gene Expression:
 The Ups & Downs of Gene Expression: Using Lipid-Based Transfection and RT-qPCR to Deliver Perfect Knockdown and Achieve Optimal Expression Results Hilary Srere, Ph.D. Topics What is RNAi? Methods of Delivery
The Ups & Downs of Gene Expression: Using Lipid-Based Transfection and RT-qPCR to Deliver Perfect Knockdown and Achieve Optimal Expression Results Hilary Srere, Ph.D. Topics What is RNAi? Methods of Delivery
Figure legends for supplement
 Figure legends for supplement Supplemental Figure 1 Characterization of purified and recombinant proteins Relevant fractions related the final stage of the purification protocol(bingham et al., 1998; Toba
Figure legends for supplement Supplemental Figure 1 Characterization of purified and recombinant proteins Relevant fractions related the final stage of the purification protocol(bingham et al., 1998; Toba
Practice Problems 5. Location of LSA-GFP fluorescence
 Life Sciences 1a Practice Problems 5 1. Soluble proteins that are normally secreted from the cell into the extracellular environment must go through a series of steps referred to as the secretory pathway.
Life Sciences 1a Practice Problems 5 1. Soluble proteins that are normally secreted from the cell into the extracellular environment must go through a series of steps referred to as the secretory pathway.
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/9/429/ra54/dc1 Supplementary Materials for Dephosphorylation of the adaptor LAT and phospholipase C by SHP-1 inhibits natural killer cell cytotoxicity Omri Matalon,
www.sciencesignaling.org/cgi/content/full/9/429/ra54/dc1 Supplementary Materials for Dephosphorylation of the adaptor LAT and phospholipase C by SHP-1 inhibits natural killer cell cytotoxicity Omri Matalon,
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the
 Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question in Section B and ONE question from Section C.
 UNIVERSITY OF EAST ANGLIA School of Biological Sciences Main Series UG Examination 2013-2014 CELL BIOLOGY BIO-2B06 Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question in
UNIVERSITY OF EAST ANGLIA School of Biological Sciences Main Series UG Examination 2013-2014 CELL BIOLOGY BIO-2B06 Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question in
T H E J O U R N A L O F C E L L B I O L O G Y
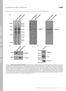 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Bays et al., http://www.jcb.org/cgi/content/full/jcb.201309092/dc1 Figure S1. Specificity of the phospho-y822 antibody. (A) Total cell
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Bays et al., http://www.jcb.org/cgi/content/full/jcb.201309092/dc1 Figure S1. Specificity of the phospho-y822 antibody. (A) Total cell
