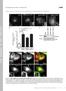
Fig. S1. Endocytosis of extracellular cargo does not depend on dia. (A) To visualize endocytosis, fluorescently labelled wheat germ agglutinin
|
|
|
- Juliet Atkins
- 6 years ago
- Views:
Transcription
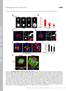
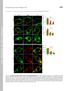
1 Fig. S1. Endocytosis of extracellular cargo does not depend on dia. (A) To visualize endocytosis, fluorescently labelled wheat germ agglutinin (WGA-Alexa555) was injected into the extracellular perivitteline space. (B) Time-lapse recording 20 min after onset of.cellularization with a frame rate of 1 in 3 s. Arrow heads indicate endocytic events in wild type and dia embryos. (C) Photographs of WGA555 injected wild type and dia[sy5] embryos (frame rate of 1 in 5 min). Fluorescent particles in basal cytoplasm were marked in red. (D) Quantification of basal particles in a given section in each three wild type and dia[sy5] embryos. (E) WGA555 (red) injected wild type and dia[sy5] embryos fixed and stained for the FC marker Slam (green). Section in large magnification. Scale bar 10 μm. (D). Fig. S2. Cip4-GFPΔSH3 does not induced cellularization defects. Embryos expressing Cip4-GFPΔSH3 were fixed and stained for (A) GFP (white/green), F-actin (white/red) and DAPI (blue) or (B) Dlg (white/green), Slam (white/red) and DAPI (blue). (A) Surface view and cross section of the same embryo. Fig. S3. Constructs used in this study. Tags are labelled in grey (ZZ, GST), blue (myc) and green (GFP, Cherry). DiaN and DiaC contained a His6 tag at the C-terminus. The domain structure of Dia (FH3+RBD, FH1, FH2, DAD) is indicated in red, of Cip4 (F-BAR, HR1, SH3), in yellow. Numbers in parentheses indicated the amino acid residues contained in the construct. The Delta sign indicates the amino acid residues lacking in the construct. Figure S4. Proteins used in this study. Samples of the purified proteins as designated were analysed by SDS polyacrylamide electrophoresis and stained by Coomassie. Figure S5. dia locus and alleles. Exons and coding sequence of dia are indicated by boxes and blue shading. Domain structure of Dia protein is indicated by boxes. dia[1] is a transposon insertion in the first exon (Castrillon1994). dia[5] is a derivative isolated after mobilisation of dia[1], which has not been molecularly characterised (Afshar2000). dia[syn] alleles were induced by chemical mutagenesis. Point mutations were determined by sequencing of the isogenic chromosomes. Resulting alteration in the sequence of amino acid residues are indicated. dia[sy4] and dia[sy6] are clonal. Figure S6. Actin filament elongation rates in the absence of presence of DiaC or Cip4. Analyses of actin assembly by single filament TIRF microscopy. 1.3 µm actin (23% Atto488 labelled) in the presence or absence of the Dia and Cip4 constructs at the concentrations indicated were used. Error bars indicate S.E.M. More than 20 filaments of at least three independent assays were analyzed for each condition.
2 Supplemental material. Figure S1: Yan et al A Injection of labelled WGA into the perivitteline space dia WT B dia WT C D No. of WGA vesicles WT dia t (min) E
3 Supplemental material. Figure S2: Yan et al GFP-Cip4-ΔSH3 GFP-Cip4 ΔSH3 A Dlg Slam GFP-Cip4 ΔSH3 B F-actin
4 Supplemental material. Figure S3: Yan et al ZZ-DiaN-His6 (1..511) ZZ-DiaC-His6 ( ) FH1 FH2 DAD Cip4 (1..631) Cip4ΔSH3 (1..565) GST-Cip4 (1..631) GST-Cip4ΔBAR ( ) GST-Cip4ΔBARΔSH3 ( ) GST-SH3 ( ) BAR SH3 myc-dia ( ) Dia-GFP/Cherry ( ) DiaFH1-GFP/Cherry ( ) DiaΔFH1-GFP/Cherry (Δ ) Cip4-GFP/Cherry (1..631) Cip4ΔSH3-GFP/Cherry (1..565)
5 Supplemental material. Figure S4: Yan et al MW (kd) ZZ-DiaC ZZ-DiaN Cip4ΔSH3 Cip4 GST-Cip4ΔBAR GST-Cip4ΔBARΔSH3 GST-SH3 Chic/profilin
6 Supplemental material. Figure S5. Yan et al dia1 dia5 10k SY5 SY3 S92L R146H SY2 G161R SY1 455stop SY4/6 682stop RBD DID cc FH1 FH2 DID
7 Supplemental material. Figure S Elongation rate [subunits/s] µm actin + 10 nm DiaC nm Cip nm DiaC nm Cip4
8 Movie 1. GFPslam in wild type embryo. GFPslam labels the basal domain of the metaphase furrow in mitosis and the furrow canal in interphase 14. Dynamical membrane extensions are observed in mitosis and initial interphase 14. In telophase 13 and progressively in interphase 14 the extensions disappear. Frame rate 1/5s, pixel size 130 nm, focal depth µm. The time lapse recording shows the dynamics of GFPslam from interphase 13 to interphase 14. Movie 2. GFPslam in dia[sy5] embryo. In dia embryos the metaphase furrows do not form properly and the furrow canals that form are often severely dilated. The dynamical membrane extensions remain visible throughout cellularization. GFPslam is expressed maternally by a tubulinvp16 GAL4 driver line. Frame rate 1/5s, pixel size 130 nm, focal depth µm. The time lapse recording shows the dynamics of GFPslam from interphase 13 to interphase 14. Movie 3. WGA endocytosis in wild type embryo. Frame rat 1/3s. The time lapse recordings show the incorporation of fluorescent protein (wheat germ agglutinin-alexa555) injected into the extracellular perivitteline space. Note that endocytic particles appear all along the furrow. Movie 4. WGA endocytosis in dia[sy5] embryo. Frame rat 1/3s. The time lapse recordings show the incorporation of fluorescent protein (wheat germ agglutinin-alexa555) injected into the extracellular perivitteline space. Note that endocytic particles appear all along the furrow.
9 Movie 5. Wild type wing imaginal disc, GFP-tubulin, Histone2Av-RFP. Movie 6. Wing imaginal disc expressing Cip4[myt], GFP-tubulin, Histone2Av-RFP. Movie 7. Wing imaginal disc expressing dia RNAi, GFP-tubulin, Histone2Av-RFP. The time lapse recordings show the mitotic cells in wing imaginal discs recognizable by the chromosome dynamics labelled by Histone2Av-RFP. During cytokinesis the separation of the daughter cells is marked by the labeling of microtubules. In cell lacking cytokinesis two nuclei share a common arrangement of microtubules.
10 Movie 8. S2 cells expressing Dia-GFP, DiaDFH1-GFP, or DiaFH1-GFP. Movie 9. S2 cells expressing full length Dia-GFP and Cip4-Cherry. Movie 10. S2 cells expressing full length dia-cherry (green) and Cip4DSH3-GFP (red). Movie 11. S2 cells expressing DiaDFH1-GFP and Cip4-Cherry. Movie 12. S2 cells expressing DiaFH1-GFP and Cip4-Cherry.
11 Movie 13. S2 cells expressing DiaDDAD-GFP and Cip4-Cherry. S2 cells expressing differentially tagged Dia and Cip4 proteins. Cip4 expression induces tubular invaginations at the plasma membrane and labels intracellular vesicles. The time lapse recordings show the dynamics of the Dia and Cip4 proteins in relation to each other in interphase cells. Movie 14. S2 cells expressing Cip4-Cherry treated with dia RNAi. S2 cells stably expressing Cip4-GFP. Expression of Cip4-GFP was induced 3 days after treatment with dia RNAi. Movie 15. Analysis of actin assembly by single filament TIRF microscopy. Dynamics of actin filaments visualized by fluorescently labelled actin monomers in the presence profilin, DiaC, Cip4DBAR, Cip4DBARDSH3 as indicated.
The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit
 Cell Reports, Volume 5 Supplemental Information The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit Andrey Poleshko, Katelyn M. Mansfield, Caroline
Cell Reports, Volume 5 Supplemental Information The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit Andrey Poleshko, Katelyn M. Mansfield, Caroline
(phosphatase tensin) domain is shown in dark gray, the FH1 domain in black, and the
 Supplemental Figure 1. Predicted Domain Organization of the AFH14 Protein. (A) Schematic representation of the predicted domain organization of AFH14. The PTEN (phosphatase tensin) domain is shown in dark
Supplemental Figure 1. Predicted Domain Organization of the AFH14 Protein. (A) Schematic representation of the predicted domain organization of AFH14. The PTEN (phosphatase tensin) domain is shown in dark
JCB. Supplemental material THE JOURNAL OF CELL BIOLOGY. Hong et al.,
 Supplemental material JCB Hong et al., http://www.jcb.org/cgi/content/full/jcb.201412127/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Analysis of purified proteins by SDS-PAGE and pull-down assays. (A) Coomassie-stained
Supplemental material JCB Hong et al., http://www.jcb.org/cgi/content/full/jcb.201412127/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Analysis of purified proteins by SDS-PAGE and pull-down assays. (A) Coomassie-stained
Figure S1 is related to Figure 1B, showing more details of outer segment of
 Supplemental Information Supplementary Figure legends and Figures Figure S1. Electron microscopic images in Sema4A +/+ and Sema4A / retinas Figure S1 is related to Figure 1B, showing more details of outer
Supplemental Information Supplementary Figure legends and Figures Figure S1. Electron microscopic images in Sema4A +/+ and Sema4A / retinas Figure S1 is related to Figure 1B, showing more details of outer
Organelle Atlas APPENDIX TO CHAPTER 4 4A.1
 APPENDIX TO CHAPTER 4 This atlas contains images of cellular organelles visualized using a wide variety of reagents and techniques and demonstrates the diversity of methods for exploring the cell. The
APPENDIX TO CHAPTER 4 This atlas contains images of cellular organelles visualized using a wide variety of reagents and techniques and demonstrates the diversity of methods for exploring the cell. The
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Wang et al., http://www.jcb.org/cgi/content/full/jcb.201405026/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Generation and characterization of unc-40 alleles. (A and
Supplemental material Wang et al., http://www.jcb.org/cgi/content/full/jcb.201405026/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Generation and characterization of unc-40 alleles. (A and
Figure S1. Verification of ihog Mutation by Protein Immunoblotting Figure S2. Verification of ihog and boi
 Figure S1. Verification of ihog Mutation by Protein Immunoblotting Extracts from S2R+ cells, embryos, and adults were analyzed by immunoprecipitation and immunoblotting with anti-ihog antibody. The Ihog
Figure S1. Verification of ihog Mutation by Protein Immunoblotting Extracts from S2R+ cells, embryos, and adults were analyzed by immunoprecipitation and immunoblotting with anti-ihog antibody. The Ihog
Supplemental Information. Kinesin 1 Drives Autolysosome Tubulation
 Developmental Cell, Volume 37 Supplemental Information Kinesin 1 Drives Autolysosome Tubulation Wanqing Du, Qian Peter Su, Yang Chen, Yueyao Zhu, Dong Jiang, Yueguang Rong, Senyan Zhang, Yixiao Zhang,
Developmental Cell, Volume 37 Supplemental Information Kinesin 1 Drives Autolysosome Tubulation Wanqing Du, Qian Peter Su, Yang Chen, Yueyao Zhu, Dong Jiang, Yueguang Rong, Senyan Zhang, Yixiao Zhang,
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Allison et al., http://www.jcb.org/cgi/content/full/jcb.201211045/dc1 Figure S1. Spastin depletion causes increased endosomal tubulation.
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Allison et al., http://www.jcb.org/cgi/content/full/jcb.201211045/dc1 Figure S1. Spastin depletion causes increased endosomal tubulation.
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION WWW.NATURE.COM/NATURECELLBIOLOGY 1 SUPPLEMENTARY INFORMATION 2 WWW.NATURE.COM/NATURECELLBIOLOGY SUPPLEMENTARY INFORMATION WWW.NATURE.COM/NATURECELLBIOLOGY 3 SUPPLEMENTARY INFORMATION
SUPPLEMENTARY INFORMATION WWW.NATURE.COM/NATURECELLBIOLOGY 1 SUPPLEMENTARY INFORMATION 2 WWW.NATURE.COM/NATURECELLBIOLOGY SUPPLEMENTARY INFORMATION WWW.NATURE.COM/NATURECELLBIOLOGY 3 SUPPLEMENTARY INFORMATION
Supplemental Figure 1 Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical
 Supplemental Figure Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical in all six REEPs are highlighted in green. Additional
Supplemental Figure Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical in all six REEPs are highlighted in green. Additional
JCB. Supplemental material THE JOURNAL OF CELL BIOLOGY. Prospéri et al.,
 Supplemental material JCB Prospéri et al., http://www.jcb.org/cgi/content/full/jcb.201501018/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Myo1b Tail interacts with YFP-EphB2 coated beads and genistein inhibits
Supplemental material JCB Prospéri et al., http://www.jcb.org/cgi/content/full/jcb.201501018/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Myo1b Tail interacts with YFP-EphB2 coated beads and genistein inhibits
Nature Structural & Molecular Biology: doi: /nsmb.3308
 Supplementary Figure 1 Analysis of CED-3 autoactivation and CED-4-induced CED-3 activation. (a) Repeat experiments of Fig. 1a. (b) Repeat experiments of Fig. 1b. (c) Quantitative analysis of three independent
Supplementary Figure 1 Analysis of CED-3 autoactivation and CED-4-induced CED-3 activation. (a) Repeat experiments of Fig. 1a. (b) Repeat experiments of Fig. 1b. (c) Quantitative analysis of three independent
System of protein filaments in the cytoplasm of. eukaryotic cell that gives the cell shape and capacity for
 Cytoskeleton System of protein filaments in the cytoplasm of eukaryotic cell that gives the cell shape and capacity for directed movement. The protein filaments are responsible for the shaping, moving,
Cytoskeleton System of protein filaments in the cytoplasm of eukaryotic cell that gives the cell shape and capacity for directed movement. The protein filaments are responsible for the shaping, moving,
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rodríguez-Fraticelli et al., http://www.jcb.org/cgi/content/full/jcb.201203075/dc1 Figure S1. Cell spreading and lumen formation in confined
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rodríguez-Fraticelli et al., http://www.jcb.org/cgi/content/full/jcb.201203075/dc1 Figure S1. Cell spreading and lumen formation in confined
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature06721 SUPPLEMENTARY INFORMATION. Supplemental Figure Legends Supplemental Figure 1 The distribution of hatx-1[82q] in Cos7 cells. Cos7 cells are co-transfected with hatx-1[82q]-gfp (green)
doi: 10.1038/nature06721 SUPPLEMENTARY INFORMATION. Supplemental Figure Legends Supplemental Figure 1 The distribution of hatx-1[82q] in Cos7 cells. Cos7 cells are co-transfected with hatx-1[82q]-gfp (green)
Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product.
 Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Biophysics of contractile ring assembly
 Biophysics of contractile ring assembly Dimitrios Vavylonis Department of Physics, Lehigh University October 1, 2007 Physical biology of the cell Physical processes in cell organization and function: Transport
Biophysics of contractile ring assembly Dimitrios Vavylonis Department of Physics, Lehigh University October 1, 2007 Physical biology of the cell Physical processes in cell organization and function: Transport
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Nakajima and Tanoue, http://www.jcb.org/cgi/content/full/jcb.201104118/dc1 Figure S1. DLD-1 cells exhibit the characteristic morphology
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Nakajima and Tanoue, http://www.jcb.org/cgi/content/full/jcb.201104118/dc1 Figure S1. DLD-1 cells exhibit the characteristic morphology
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
Sélection Internationale Ens Ulm 2012, Cell Biology
 Sélection Internationale Ens Ulm 2012, Cell Biology Please read the whole subject before starting. Questions 1-8 and 12-14 are general biology questions. In questions 9, 10, 11 and15, we ask you to interpret
Sélection Internationale Ens Ulm 2012, Cell Biology Please read the whole subject before starting. Questions 1-8 and 12-14 are general biology questions. In questions 9, 10, 11 and15, we ask you to interpret
Supplementary Information. Tau co-organizes dynamic microtubule and actin networks. and Isabelle Arnal 1,2,*
 Supplementary Information Tau co-organizes dynamic microtubule and actin networks Auréliane Elie 1,2, Elea Prezel 1,2, Christophe Guérin 3, Eric Denarier 1,2,4, Sacnicte Ramirez- Rios 1,2, Laurence Serre
Supplementary Information Tau co-organizes dynamic microtubule and actin networks Auréliane Elie 1,2, Elea Prezel 1,2, Christophe Guérin 3, Eric Denarier 1,2,4, Sacnicte Ramirez- Rios 1,2, Laurence Serre
indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) B. Male mice of
 SUPPLEMENTRY FIGURE LEGENDS Figure S1. USP44 loss leads to chromosome missegregation.. Genotypes obtained from the indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) 13.5.. Male mice
SUPPLEMENTRY FIGURE LEGENDS Figure S1. USP44 loss leads to chromosome missegregation.. Genotypes obtained from the indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) 13.5.. Male mice
(a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for
 Supplementary Figure 1 Generation of Diaph3 ko mice. (a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for different regions of Diaph3 mrna from WT, heterozygote
Supplementary Figure 1 Generation of Diaph3 ko mice. (a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for different regions of Diaph3 mrna from WT, heterozygote
Supplemental Data. Liu et al. (2013). Plant Cell /tpc
 Supplemental Figure 1. The GFP Tag Does Not Disturb the Physiological Functions of WDL3. (A) RT-PCR analysis of WDL3 expression in wild-type, WDL3-GFP, and WDL3 (without the GFP tag) transgenic seedlings.
Supplemental Figure 1. The GFP Tag Does Not Disturb the Physiological Functions of WDL3. (A) RT-PCR analysis of WDL3 expression in wild-type, WDL3-GFP, and WDL3 (without the GFP tag) transgenic seedlings.
Mitotic cell rounding and epithelial thinning regulate lumen
 Mitotic cell rounding and epithelial thinning regulate lumen growth and shape Esteban Hoijman, Davide Rubbini, Julien Colombelli and Berta Alsina SUPPLEMENTARY FIGURES Supplementary Figure 1 (a-c) Localization
Mitotic cell rounding and epithelial thinning regulate lumen growth and shape Esteban Hoijman, Davide Rubbini, Julien Colombelli and Berta Alsina SUPPLEMENTARY FIGURES Supplementary Figure 1 (a-c) Localization
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Prashar et al., http://www.jcb.org/cgi/content/full/jcb.201304095/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. FBT phagocytosis in cells expressing PM-GFP. (A) T-PC
Supplemental material Prashar et al., http://www.jcb.org/cgi/content/full/jcb.201304095/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. FBT phagocytosis in cells expressing PM-GFP. (A) T-PC
Xu et al., Supplementary Figures 1-7
 Xu et al., Supplementary Figures 1-7 Supplementary Figure 1. PIPKI is required for ciliogenesis. (a) PIPKI localizes at the basal body of primary cilium. RPE-1 cells treated with two sirnas targeting to
Xu et al., Supplementary Figures 1-7 Supplementary Figure 1. PIPKI is required for ciliogenesis. (a) PIPKI localizes at the basal body of primary cilium. RPE-1 cells treated with two sirnas targeting to
Supplementary Materials
 Supplementary Materials Supplementary Figure 1. PKM2 interacts with MLC2 in cytokinesis. a, U87, U87/EGFRvIII, and HeLa cells in cytokinesis were immunostained with DAPI and an anti-pkm2 antibody. Thirty
Supplementary Materials Supplementary Figure 1. PKM2 interacts with MLC2 in cytokinesis. a, U87, U87/EGFRvIII, and HeLa cells in cytokinesis were immunostained with DAPI and an anti-pkm2 antibody. Thirty
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2386 Figure 1 Src-containing puncta are not focal adhesions, podosomes or endosomes. (a) FAK-/- were stained with anti-py416 Src (green) and either (in red) the focal adhesion protein paxillin,
DOI: 10.1038/ncb2386 Figure 1 Src-containing puncta are not focal adhesions, podosomes or endosomes. (a) FAK-/- were stained with anti-py416 Src (green) and either (in red) the focal adhesion protein paxillin,
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table.
 Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
JCB. Supplemental material THE JOURNAL OF CELL BIOLOGY. Paul et al.,
 Supplemental material JCB Paul et al., http://www.jcb.org/cgi/content/full/jcb.201502040/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Mutant p53-expressing cells display limited retrograde actin flow at
Supplemental material JCB Paul et al., http://www.jcb.org/cgi/content/full/jcb.201502040/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Mutant p53-expressing cells display limited retrograde actin flow at
Supplementary Figure 1 Collision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK2, PPK3 and PPK4 respectively.
 Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
Supplemental Material
 Supplemental Material 1 Figure S1. Phylogenetic analysis of Cep72 and Lrrc36, comparative localization of Cep72 and Lrrc36 and Cep72 antibody characterization (A) Phylogenetic alignment of Cep72 and Lrrc36
Supplemental Material 1 Figure S1. Phylogenetic analysis of Cep72 and Lrrc36, comparative localization of Cep72 and Lrrc36 and Cep72 antibody characterization (A) Phylogenetic alignment of Cep72 and Lrrc36
Supplemental Fig. 1. Mcr alleles show defects in tracheal tube size and luminal protein accumulation. (A-F) Confocal projections of living stage 15
 Supplemental Fig. 1. Mcr alleles show defects in tracheal tube size and luminal protein accumulation. (A-F) Confocal projections of living stage 15 embryos expressing GFP and Verm-RFP in tracheal cells
Supplemental Fig. 1. Mcr alleles show defects in tracheal tube size and luminal protein accumulation. (A-F) Confocal projections of living stage 15 embryos expressing GFP and Verm-RFP in tracheal cells
Supplementary Figure S1. N-terminal fragments of LRRK1 bind to Grb2.
 Myc- HA-Grb2 Mr(K) 105 IP HA 75 25 105 1-1163 1-595 - + - + - + 1164-1989 Blot Myc HA total lysate 75 25 Myc HA Supplementary Figure S1. N-terminal fragments of bind to Grb2. COS7 cells were cotransfected
Myc- HA-Grb2 Mr(K) 105 IP HA 75 25 105 1-1163 1-595 - + - + - + 1164-1989 Blot Myc HA total lysate 75 25 Myc HA Supplementary Figure S1. N-terminal fragments of bind to Grb2. COS7 cells were cotransfected
Asymmetric endocytosis and remodeling of β1-integrin adhesions during growth cone chemorepulsion by MAG
 Asymmetric endocytosis and remodeling of β1-integrin adhesions during growth cone chemorepulsion by MAG Jacob H. Hines, Mohammad Abu-Rub and John R. Henley Supplementary Figure 1. Validation of FM 5-95
Asymmetric endocytosis and remodeling of β1-integrin adhesions during growth cone chemorepulsion by MAG Jacob H. Hines, Mohammad Abu-Rub and John R. Henley Supplementary Figure 1. Validation of FM 5-95
monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal antibody was examined in
 Supplementary information Supplementary figures Supplementary Figure 1 Determination of the s pecificity of in-house anti-rhbdd1 mouse monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal
Supplementary information Supplementary figures Supplementary Figure 1 Determination of the s pecificity of in-house anti-rhbdd1 mouse monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal
J. Cell Sci. 128: doi: /jcs : Supplementary Material. Supplemental Figures. Journal of Cell Science Supplementary Material
 Supplemental Figures Figure S1. Trio controls endothelial barrier function. (A) TagRFP-shTrio constructs were expressed in ECs. Western blot shows efficient Trio knockdown in TagRFP-expressing ECs. (B)
Supplemental Figures Figure S1. Trio controls endothelial barrier function. (A) TagRFP-shTrio constructs were expressed in ECs. Western blot shows efficient Trio knockdown in TagRFP-expressing ECs. (B)
Supplemental Data. Farmer et al. (2010) Plant Cell /tpc
 Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
Fluorescence Light Microscopy for Cell Biology
 Fluorescence Light Microscopy for Cell Biology Why use light microscopy? Traditional questions that light microscopy has addressed: Structure within a cell Locations of specific molecules within a cell
Fluorescence Light Microscopy for Cell Biology Why use light microscopy? Traditional questions that light microscopy has addressed: Structure within a cell Locations of specific molecules within a cell
λ N -GFP: an RNA reporter system for live-cell imaging
 λ N -GFP: an RNA reporter system for live-cell imaging Nathalie Daigle & Jan Ellenberg Supplementary Figures and Text: Supplementary Figure 1 localization in the cytoplasm. 4 λ N22-3 megfp-m9 serves as
λ N -GFP: an RNA reporter system for live-cell imaging Nathalie Daigle & Jan Ellenberg Supplementary Figures and Text: Supplementary Figure 1 localization in the cytoplasm. 4 λ N22-3 megfp-m9 serves as
1. The microtubule wall is composed of globular proteins arranged in longitudinal rows called.
 Name: Quiz name: Quiz 7 ate: 1. The microtubule wall is composed of globular proteins arranged in longitudinal rows called. microfilaments protofilaments prototubules microtubular subunits 2. Which of
Name: Quiz name: Quiz 7 ate: 1. The microtubule wall is composed of globular proteins arranged in longitudinal rows called. microfilaments protofilaments prototubules microtubular subunits 2. Which of
Stargazin regulates AMPA receptor trafficking through adaptor protein. complexes during long term depression
 Supplementary Information Stargazin regulates AMPA receptor trafficking through adaptor protein complexes during long term depression Shinji Matsuda, Wataru Kakegawa, Timotheus Budisantoso, Toshihiro Nomura,
Supplementary Information Stargazin regulates AMPA receptor trafficking through adaptor protein complexes during long term depression Shinji Matsuda, Wataru Kakegawa, Timotheus Budisantoso, Toshihiro Nomura,
Microtubules. Forms a sheet of protofilaments that folds to a tube Both tubulins bind GTP
 Microtubules Polymerize from a-tubulin/ß-tubulin dimers Hollow tube of 13 protofilaments In vitro- filament number varies In vivo- always 13 protofilaments Forms a sheet of protofilaments that folds to
Microtubules Polymerize from a-tubulin/ß-tubulin dimers Hollow tube of 13 protofilaments In vitro- filament number varies In vivo- always 13 protofilaments Forms a sheet of protofilaments that folds to
Supplemental figures (1-9) Gu et al. ADF/Cofilin-Mediated Actin Dynamics Regulate AMPA Receptor Trafficking during Synaptic Plasticity
 Supplemental figures (1-9) Gu et al. ADF/Cofilin-Mediated Actin Dynamics Regulate AMPA Receptor Trafficking during Synaptic Plasticity Supplemental figure 1. The quantification method to determine the
Supplemental figures (1-9) Gu et al. ADF/Cofilin-Mediated Actin Dynamics Regulate AMPA Receptor Trafficking during Synaptic Plasticity Supplemental figure 1. The quantification method to determine the
Supplemental Information. Plk1/Polo Phosphorylates Sas-4 at the Onset. of Mitosis for an Efficient Recruitment
 Cell Reports, Volume 25 Supplemental Information Plk1/Polo Phosphorylates Sas-4 at the Onset of Mitosis for an Efficient Recruitment of Pericentriolar Material to Centrosomes Anand Ramani, Aruljothi Mariappan,
Cell Reports, Volume 25 Supplemental Information Plk1/Polo Phosphorylates Sas-4 at the Onset of Mitosis for an Efficient Recruitment of Pericentriolar Material to Centrosomes Anand Ramani, Aruljothi Mariappan,
MT minus end catastrophe
 DOI: 1.138/ncb3241 MT minus end catastrophe frequency [s -1 ].2.15.1.5 control mgfp- TPX2 * * mgfp- TPX2 mini Supplementary Figure 1. TPX2 reduces catastrophes at the microtubule minus ends. Modified box-and-whiskers
DOI: 1.138/ncb3241 MT minus end catastrophe frequency [s -1 ].2.15.1.5 control mgfp- TPX2 * * mgfp- TPX2 mini Supplementary Figure 1. TPX2 reduces catastrophes at the microtubule minus ends. Modified box-and-whiskers
Supplementary Figure 1. Localization of MST1 in RPE cells. Proliferating or ciliated HA- MST1 expressing RPE cells (see Fig. 5b for establishment of
 Supplementary Figure 1. Localization of MST1 in RPE cells. Proliferating or ciliated HA- MST1 expressing RPE cells (see Fig. 5b for establishment of the cell line) were immunostained for HA, acetylated
Supplementary Figure 1. Localization of MST1 in RPE cells. Proliferating or ciliated HA- MST1 expressing RPE cells (see Fig. 5b for establishment of the cell line) were immunostained for HA, acetylated
Supplemental Information. Pacer Mediates the Function of Class III PI3K. and HOPS Complexes in Autophagosome. Maturation by Engaging Stx17
 Molecular Cell, Volume 65 Supplemental Information Pacer Mediates the Function of Class III PI3K and HOPS Complexes in Autophagosome Maturation by Engaging Stx17 Xiawei Cheng, Xiuling Ma, Xianming Ding,
Molecular Cell, Volume 65 Supplemental Information Pacer Mediates the Function of Class III PI3K and HOPS Complexes in Autophagosome Maturation by Engaging Stx17 Xiawei Cheng, Xiuling Ma, Xianming Ding,
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature06147 SUPPLEMENTARY INFORMATION Figure S1 The genomic and domain structure of Dscam. The Dscam gene comprises 24 exons, encoding a signal peptide (SP), 10 IgSF domains, 6 fibronectin
doi: 10.1038/nature06147 SUPPLEMENTARY INFORMATION Figure S1 The genomic and domain structure of Dscam. The Dscam gene comprises 24 exons, encoding a signal peptide (SP), 10 IgSF domains, 6 fibronectin
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/404/ra120/dc1 Supplementary Materials for The subcellular localization and activity of cortactin is regulated by acetylation and interaction with Keap1 Akihiro
www.sciencesignaling.org/cgi/content/full/8/404/ra120/dc1 Supplementary Materials for The subcellular localization and activity of cortactin is regulated by acetylation and interaction with Keap1 Akihiro
Nature Biotechnology: doi: /nbt.4166
 Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
Regulation of Acetylcholine Receptor Clustering by ADF/Cofilin- Directed Vesicular Trafficking
 Regulation of Acetylcholine Receptor Clustering by ADF/Cofilin- Directed Vesicular Trafficking Chi Wai Lee, Jianzhong Han, James R. Bamburg, Liang Han, Rachel Lynn, and James Q. Zheng Supplementary Figures
Regulation of Acetylcholine Receptor Clustering by ADF/Cofilin- Directed Vesicular Trafficking Chi Wai Lee, Jianzhong Han, James R. Bamburg, Liang Han, Rachel Lynn, and James Q. Zheng Supplementary Figures
Supplementary Fig. 1: Ultrastructural localization of Hh, Ihog and Disp in wing discs. A) Thawed 200 nm thick cryosections of wing disc expressing
 Supplementary Fig. 1: Ultrastructural localization of Hh, Ihog and Disp in wing discs. A) Thawed 200 nm thick cryosections of wing disc expressing Hh-GFP were immunostained with an anti-gfp antibody followed
Supplementary Fig. 1: Ultrastructural localization of Hh, Ihog and Disp in wing discs. A) Thawed 200 nm thick cryosections of wing disc expressing Hh-GFP were immunostained with an anti-gfp antibody followed
At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in
 Supplementary Materials and Methods Barrier function assays At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in acidic X-gal mix (100 mm phosphate buffer at ph4.3, 3 mm
Supplementary Materials and Methods Barrier function assays At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in acidic X-gal mix (100 mm phosphate buffer at ph4.3, 3 mm
Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity.
 Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity. (A) Amino acid alignment of HDA5, HDA15 and HDA18. The blue line
Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity. (A) Amino acid alignment of HDA5, HDA15 and HDA18. The blue line
1. I can describe the stages of the cell cycle.
 Unit 5 Study Guide Cell Cycle pg. 1 1. I can describe the stages of the cell cycle. Interphase = period in between division G1 = growth phase S = DNA replication G2 = Preparation for division (extra copies
Unit 5 Study Guide Cell Cycle pg. 1 1. I can describe the stages of the cell cycle. Interphase = period in between division G1 = growth phase S = DNA replication G2 = Preparation for division (extra copies
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Craft et al., http://www.jcb.org/cgi/content/full/jcb.201409036/dc1 Figure S1. GFP -tubulin interacts with endogenous -tubulin. Ciliary
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Craft et al., http://www.jcb.org/cgi/content/full/jcb.201409036/dc1 Figure S1. GFP -tubulin interacts with endogenous -tubulin. Ciliary
Supplementary Figure 1. α-synuclein is truncated in PD and LBD brains. Nature Structural & Molecular Biology: doi: /nsmb.
 Supplementary Figure 1 α-synuclein is truncated in PD and LBD brains. (a) Specificity of anti-n103 antibody. Anti-N103 antibody was coated on an ELISA plate and different concentrations of full-length
Supplementary Figure 1 α-synuclein is truncated in PD and LBD brains. (a) Specificity of anti-n103 antibody. Anti-N103 antibody was coated on an ELISA plate and different concentrations of full-length
Aminoacid change in chromophore. PIN3::PIN3-GFP GFP S65 (no change) (5.9) (Kneen et al., 1998)
 Supplemental Table. Table S1. Fluorophore characteristics of used fluorescent proteins, Related to Figure 2 and 3. Transgenic line Fluprescent marker Aminoacid change in chromophore pk(a) Ref. PIN3::PIN3-GFP
Supplemental Table. Table S1. Fluorophore characteristics of used fluorescent proteins, Related to Figure 2 and 3. Transgenic line Fluprescent marker Aminoacid change in chromophore pk(a) Ref. PIN3::PIN3-GFP
Verprolin function in endocytosis and actin organization
 Verprolin function in endocytosis and actin organization Roles of the Las17p (yeast WASP)-binding domain and a novel C-terminal actin-binding domain Thirumaran Thanabalu 1,2, Rajamuthiah Rajmohan 2, Lei
Verprolin function in endocytosis and actin organization Roles of the Las17p (yeast WASP)-binding domain and a novel C-terminal actin-binding domain Thirumaran Thanabalu 1,2, Rajamuthiah Rajmohan 2, Lei
PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF
 YFP-PHF1 CFP-PHT1;2 PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF + CFP-PHT1;2 Negative control!-gfp Supplemental Figure 1: PHT1;2 accumulation is PHF1 dependent. Immunoblot analysis on total protein extract
YFP-PHF1 CFP-PHT1;2 PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF + CFP-PHT1;2 Negative control!-gfp Supplemental Figure 1: PHT1;2 accumulation is PHF1 dependent. Immunoblot analysis on total protein extract
Ac(n cytoskeleton 12/7/11. The Cytoskeleton. Func0ons of the Cytoskeleton. B Visegrady. Elements of the Cytoskeleton
 The Cytoskeleton Ac(n cytoskeleton B Visegrady The cytoskeleton is an intricate network of protein filaments that extends throughout the cytoplasm. A skin cell (fibroblast) stained with Coomassie blue.
The Cytoskeleton Ac(n cytoskeleton B Visegrady The cytoskeleton is an intricate network of protein filaments that extends throughout the cytoplasm. A skin cell (fibroblast) stained with Coomassie blue.
To assess the localization of Citrine fusion proteins, we performed antibody staining to
 Trinh et al 1 SUPPLEMENTAL MATERIAL FlipTraps recapitulate endogenous protein localization To assess the localization of Citrine fusion proteins, we performed antibody staining to compare the expression
Trinh et al 1 SUPPLEMENTAL MATERIAL FlipTraps recapitulate endogenous protein localization To assess the localization of Citrine fusion proteins, we performed antibody staining to compare the expression
Supplemental material JCB. Pitaval et al., Cell shape and ciliogenesis Pitaval et al.
 Supplemental material THE JOURNAL OF CELL BIOLOGY Pitaval et al., http://www.jcb.org/cgi/content/full/jcb.201004003/dc1 S1 Figure S1. Cell cycle exit analysis. (A) Experimental procedure. RPE1 cells were
Supplemental material THE JOURNAL OF CELL BIOLOGY Pitaval et al., http://www.jcb.org/cgi/content/full/jcb.201004003/dc1 S1 Figure S1. Cell cycle exit analysis. (A) Experimental procedure. RPE1 cells were
Supplementary Information
 Supplementary Information Supplementary Figure 1: Over-expression of CD300f in NIH3T3 cells enhances their capacity to phagocytize AC. (a) NIH3T3 cells were stably transduced by EV, CD300f WT or CD300f
Supplementary Information Supplementary Figure 1: Over-expression of CD300f in NIH3T3 cells enhances their capacity to phagocytize AC. (a) NIH3T3 cells were stably transduced by EV, CD300f WT or CD300f
2. Know the parts of a light microscope and general rules for using and focusing a microscope, such as:
 SNC 2DI Exam Review: Biology Unit 1. Understand the meaning of the following terms. Be able to recognize their definitions: Biology Mounting medium Telophase Organelle Cell Theory Cell cycle Cytokinesis
SNC 2DI Exam Review: Biology Unit 1. Understand the meaning of the following terms. Be able to recognize their definitions: Biology Mounting medium Telophase Organelle Cell Theory Cell cycle Cytokinesis
Supplementary Figure 1. Confirmation of sirna in PC3 and H1299 cells PC3 (a) and H1299 (b) cells were transfected with sirna oligonucleotides
 Supplementary Figure 1. Confirmation of sirna in PC3 and H1299 cells PC3 (a) and H1299 (b) cells were transfected with sirna oligonucleotides targeting RCP (SMARTPool (RCP) or two individual oligos (RCP#1
Supplementary Figure 1. Confirmation of sirna in PC3 and H1299 cells PC3 (a) and H1299 (b) cells were transfected with sirna oligonucleotides targeting RCP (SMARTPool (RCP) or two individual oligos (RCP#1
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rainero et al., http://www.jcb.org/cgi/content/full/jcb.201109112/dc1 Figure S1. The expression of DGK- is reduced upon transfection
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rainero et al., http://www.jcb.org/cgi/content/full/jcb.201109112/dc1 Figure S1. The expression of DGK- is reduced upon transfection
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature12138 Supplementary Figure 1. Knockdown of KRAS leads to a reduction in macropinocytosis. (a) KRAS knockdown in MIA PaCa-2 cells expressing KRASspecific shrnas
SUPPLEMENTARY INFORMATION doi:10.1038/nature12138 Supplementary Figure 1. Knockdown of KRAS leads to a reduction in macropinocytosis. (a) KRAS knockdown in MIA PaCa-2 cells expressing KRASspecific shrnas
Aoki et al.,
 JCB: SUPPLEMENTAL MATERIAL Aoki et al., http://www.jcb.org/cgi/content/full/jcb.200609017/dc1 Supplemental materials and methods Calculation of the raw number of translocated proteins First, the average
JCB: SUPPLEMENTAL MATERIAL Aoki et al., http://www.jcb.org/cgi/content/full/jcb.200609017/dc1 Supplemental materials and methods Calculation of the raw number of translocated proteins First, the average
Supplemental Figure 1. Alignment of the NbGAPC amino acid sequences with their Arabidopsis homologues.
 Supplemental Figure 1. Alignment of the NbGAPC amino acid sequences with their Arabidopsis homologues. Homologs from N. benthamiana (NbGAPC1, NbGAPC2, NbGAPC3), Arabidopsis (AtGAPC1, AT3G04120; AtGAPC2,
Supplemental Figure 1. Alignment of the NbGAPC amino acid sequences with their Arabidopsis homologues. Homologs from N. benthamiana (NbGAPC1, NbGAPC2, NbGAPC3), Arabidopsis (AtGAPC1, AT3G04120; AtGAPC2,
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than. SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with
 Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
Sequence of C-terminal tail regions of myosin heavy chains in class XI of Nicotiana benthamiana (Nb
 Fig. S1 Sequence of C-terminal tail regions of myosin heavy chains in class XI of Nicotiana benthamiana (Nb myosin XI-2, -F and K) and BY-2 cell (Nt 170-kD myosin and Nt 175-kD myosin). Amino acids identical
Fig. S1 Sequence of C-terminal tail regions of myosin heavy chains in class XI of Nicotiana benthamiana (Nb myosin XI-2, -F and K) and BY-2 cell (Nt 170-kD myosin and Nt 175-kD myosin). Amino acids identical
2. Know the parts of a light microscope and general rules for using and focusing a microscope, such as:
 SNC 2DI Exam Review: Biology Unit 1. Understand the meaning of the following terms. Be able to recognize their definitions: Biology Mounting medium Telophase Organelle Cell Theory Cell cycle Cytokinesis
SNC 2DI Exam Review: Biology Unit 1. Understand the meaning of the following terms. Be able to recognize their definitions: Biology Mounting medium Telophase Organelle Cell Theory Cell cycle Cytokinesis
(A) Schematic depicting morphology of cholinergic DA and DB motor neurons in an L1
 SUPPLEMENTAL MATERIALS Supplemental Figure 1 Figure S1. Expression of the non-alpha nachr subunit ACR-2. (A) Schematic depicting morphology of cholinergic DA and DB motor neurons in an L1 animal. Wide-field
SUPPLEMENTAL MATERIALS Supplemental Figure 1 Figure S1. Expression of the non-alpha nachr subunit ACR-2. (A) Schematic depicting morphology of cholinergic DA and DB motor neurons in an L1 animal. Wide-field
1. I can describe the stages of the cell cycle.
 Unit 5 Study Guide Cell Cycle pg. 1 1. I can describe the stages of the cell cycle. Interphase = period in between division G1 = growth phase S = DNA replication G2 = Preparation for division (extra copies
Unit 5 Study Guide Cell Cycle pg. 1 1. I can describe the stages of the cell cycle. Interphase = period in between division G1 = growth phase S = DNA replication G2 = Preparation for division (extra copies
Three major types of cytoskeleton
 The Cytoskeleton Organizes and stabilizes cells Pulls chromosomes apart Drives intracellular traffic Supports plasma membrane and nuclear envelope Enables cellular movement Guides growth of the plant cell
The Cytoskeleton Organizes and stabilizes cells Pulls chromosomes apart Drives intracellular traffic Supports plasma membrane and nuclear envelope Enables cellular movement Guides growth of the plant cell
THE JOURNAL OF CELL BIOLOGY
 Supplemental Material THE JOURNAL OF CELL BIOLOGY Toso et al., http://www.jcb.org/cgi/content/full/jcb.200809055/dc1 Figure S1. Control experiments for sirna depletions used in Figs. 1 3. The phenotype
Supplemental Material THE JOURNAL OF CELL BIOLOGY Toso et al., http://www.jcb.org/cgi/content/full/jcb.200809055/dc1 Figure S1. Control experiments for sirna depletions used in Figs. 1 3. The phenotype
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
supplementary information
 DOI: 10.1038/ncb1979 Figure S1 Alignment and domain composition of HsTSN and PaTSN. Since both HsTSN (accession number AAA80488) and PaTSN (accession number CAL38976) have five staphylococcal nuclease
DOI: 10.1038/ncb1979 Figure S1 Alignment and domain composition of HsTSN and PaTSN. Since both HsTSN (accession number AAA80488) and PaTSN (accession number CAL38976) have five staphylococcal nuclease
Supplemental Fig. 1: PEA-15 knockdown efficiency assessed by immunohistochemistry and qpcr
 Supplemental figure legends Supplemental Fig. 1: PEA-15 knockdown efficiency assessed by immunohistochemistry and qpcr A, LβT2 cells were transfected with either scrambled or PEA-15 sirna. Cells were then
Supplemental figure legends Supplemental Fig. 1: PEA-15 knockdown efficiency assessed by immunohistochemistry and qpcr A, LβT2 cells were transfected with either scrambled or PEA-15 sirna. Cells were then
Actin cap associated focal adhesions and their distinct role in cellular mechanosensing
 Actin cap associated focal adhesions and their distinct role in cellular mechanosensing Dong-Hwee Kim 1,2, Shyam B. Khatau 1,2, Yunfeng Feng 1,3, Sam Walcott 1,4, Sean X. Sun 1,2,4, Gregory D. Longmore
Actin cap associated focal adhesions and their distinct role in cellular mechanosensing Dong-Hwee Kim 1,2, Shyam B. Khatau 1,2, Yunfeng Feng 1,3, Sam Walcott 1,4, Sean X. Sun 1,2,4, Gregory D. Longmore
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2271 Supplementary Figure a! WM266.4 mock WM266.4 #7 sirna WM266.4 #10 sirna SKMEL28 mock SKMEL28 #7 sirna SKMEL28 #10 sirna WM1361 mock WM1361 #7 sirna WM1361 #10 sirna 9 WM266. WM136
DOI: 10.1038/ncb2271 Supplementary Figure a! WM266.4 mock WM266.4 #7 sirna WM266.4 #10 sirna SKMEL28 mock SKMEL28 #7 sirna SKMEL28 #10 sirna WM1361 mock WM1361 #7 sirna WM1361 #10 sirna 9 WM266. WM136
Supplementary
 Supplementary information Supplementary Material and Methods Plasmid construction The transposable element vectors for inducible expression of RFP-FUS wt and EGFP-FUS R521C and EGFP-FUS P525L were derived
Supplementary information Supplementary Material and Methods Plasmid construction The transposable element vectors for inducible expression of RFP-FUS wt and EGFP-FUS R521C and EGFP-FUS P525L were derived
CHAPTER 16 THE CYTOSKELETON
 CHAPTER 16 THE CYTOSKELETON THE SELF-ASSEMBLY AND DYNAMIC STRUCTURE OF CYTOSKELETAL FILAMENTS HOW CELLS REGULATE THEIR CYTOSKELETAL FILAMENTS MOLECULAR MOTORS THE CYTOSKELETON AND CELL BEHAVIOR THE SELF-ASSEMBLY
CHAPTER 16 THE CYTOSKELETON THE SELF-ASSEMBLY AND DYNAMIC STRUCTURE OF CYTOSKELETAL FILAMENTS HOW CELLS REGULATE THEIR CYTOSKELETAL FILAMENTS MOLECULAR MOTORS THE CYTOSKELETON AND CELL BEHAVIOR THE SELF-ASSEMBLY
4.1 CELL DIVISION AND GENETIC MATERIAL
 4.1 CELL DIVISION AND GENETIC MATERIAL GENETICS Field of biology Study how genetic information is passed from one generation of organism/cells to the next THE CELL THEORY developed in mid-1800s 1. All
4.1 CELL DIVISION AND GENETIC MATERIAL GENETICS Field of biology Study how genetic information is passed from one generation of organism/cells to the next THE CELL THEORY developed in mid-1800s 1. All
supplementary information
 DOI: 10.1038/ncb2172 Figure S1 p53 regulates cellular NADPH and lipid levels via inhibition of G6PD. (a) U2OS cells stably expressing p53 shrna or a control shrna were transfected with control sirna or
DOI: 10.1038/ncb2172 Figure S1 p53 regulates cellular NADPH and lipid levels via inhibition of G6PD. (a) U2OS cells stably expressing p53 shrna or a control shrna were transfected with control sirna or
Materials and Methods: All strains were derivatives of SK1 and are listed in Supplemental Table 1.
 Supplemental Online Material: Materials and Methods: All strains were derivatives of SK1 and are listed in Supplemental Table 1. Constructs: pclb2-3ha-cdc5 and pclb2-3ha-cdc2 strains were constructed by
Supplemental Online Material: Materials and Methods: All strains were derivatives of SK1 and are listed in Supplemental Table 1. Constructs: pclb2-3ha-cdc5 and pclb2-3ha-cdc2 strains were constructed by
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2880 Supplementary Figure 1 Sequence alignment of Deup1 and Cep63. The protein sequence alignment was generated by the Clustal X 2.0 multiple sequence alignment program using default parameters.
DOI: 10.1038/ncb2880 Supplementary Figure 1 Sequence alignment of Deup1 and Cep63. The protein sequence alignment was generated by the Clustal X 2.0 multiple sequence alignment program using default parameters.
These molecules make up the ladder of the DNA Bound by weak hydrogen bonds. 4 Different Types (2 specific matches) look at the
 B.A.T. Review DNA & Cell Cycle Test is 11/3/16 NAME: PERIOD DNA STRUCTURE VOCABULARY Mitosis Chromosomes Interphase G1,S,G2,M Prophase Metaphase Anaphase Telophase Cytokinesis Cleavage Furrow Spindle Fibers
B.A.T. Review DNA & Cell Cycle Test is 11/3/16 NAME: PERIOD DNA STRUCTURE VOCABULARY Mitosis Chromosomes Interphase G1,S,G2,M Prophase Metaphase Anaphase Telophase Cytokinesis Cleavage Furrow Spindle Fibers
Supplemental Figure 1. VLN5 retains conserved residues at both type 1 and type 2 Ca 2+ -binding
 Supplemental Figure 1. VLN5 retains conserved residues at both type 1 and type 2 Ca 2+ -binding sites in the G1 domain. Multiple sequence alignment was performed with DNAMAN6.0.40. Secondary structural
Supplemental Figure 1. VLN5 retains conserved residues at both type 1 and type 2 Ca 2+ -binding sites in the G1 domain. Multiple sequence alignment was performed with DNAMAN6.0.40. Secondary structural
THE JOURNAL OF CELL BIOLOGY
 Supplemental Material THE JOURNAL OF CELL BIOLOGY Yeung et al., http://www.jcb.org/cgi/content/full/jcb.200903020/dc1 Figure S1. Assessment of the surface charge of maturing phagosomes. (A C and F H) RAW
Supplemental Material THE JOURNAL OF CELL BIOLOGY Yeung et al., http://www.jcb.org/cgi/content/full/jcb.200903020/dc1 Figure S1. Assessment of the surface charge of maturing phagosomes. (A C and F H) RAW
42 fl organelles = 34.5 fl (1) 3.5X X 0.93 = 78,000 (2)
 SUPPLEMENTAL DATA Supplementary Experimental Procedures Fluorescence Microscopy - A Zeiss Axiovert 200M microscope equipped with a Zeiss 100x Plan- Apochromat (1.40 NA) DIC objective and Hamamatsu Orca
SUPPLEMENTAL DATA Supplementary Experimental Procedures Fluorescence Microscopy - A Zeiss Axiovert 200M microscope equipped with a Zeiss 100x Plan- Apochromat (1.40 NA) DIC objective and Hamamatsu Orca
Supplementary Figure 1. Serial deletion mutants of BLITz.
 Supplementary Figure 1 Serial deletion mutants of BLITz. (a) Design of TEVseq insertion into J -helix. C-terminal end of J -helix was serially deleted and replaced by TEVseq. TEV cleavage site is labeled
Supplementary Figure 1 Serial deletion mutants of BLITz. (a) Design of TEVseq insertion into J -helix. C-terminal end of J -helix was serially deleted and replaced by TEVseq. TEV cleavage site is labeled
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Supplementary figures Supplementary Figure 1: Suv39h1, but not Suv39h2, promotes HP1α sumoylation in vivo. In vivo HP1α sumoylation assay. Top: experimental scheme. Middle: we
SUPPLEMENTARY INFORMATION Supplementary figures Supplementary Figure 1: Suv39h1, but not Suv39h2, promotes HP1α sumoylation in vivo. In vivo HP1α sumoylation assay. Top: experimental scheme. Middle: we
Human Anatomy & Physiology I Dr. Sullivan Unit IV Cellular Function Chapter 4, Chapter 27 (meiosis only)
 Human Anatomy & Physiology I Dr. Sullivan Unit IV Cellular Function Chapter 4, Chapter 27 (meiosis only) I. Protein Synthesis: creation of new proteins a. Much of the cellular machinery is devoted to synthesizing
Human Anatomy & Physiology I Dr. Sullivan Unit IV Cellular Function Chapter 4, Chapter 27 (meiosis only) I. Protein Synthesis: creation of new proteins a. Much of the cellular machinery is devoted to synthesizing
Supplementary Online Material. Structural mimicry in transcription regulation of human RNA polymerase II by the. DNA helicase RECQL5
 Supplementary Online Material Structural mimicry in transcription regulation of human RNA polymerase II by the DNA helicase RECQL5 Susanne A. Kassube, Martin Jinek, Jie Fang, Susan Tsutakawa and Eva Nogales
Supplementary Online Material Structural mimicry in transcription regulation of human RNA polymerase II by the DNA helicase RECQL5 Susanne A. Kassube, Martin Jinek, Jie Fang, Susan Tsutakawa and Eva Nogales
