Ahi1, whose human ortholog is mutated in Joubert syndrome, is required for Rab8a localization, ciliogenesis and vesicle trafficking
|
|
|
- Tyrone Whitehead
- 6 years ago
- Views:
Transcription
1 Human Molecular Genetics, 2009, Vol. 18, No doi: /hmg/ddp335 Advance Access published on July 22, 2009 Ahi1, whose human ortholog is mutated in Joubert syndrome, is required for Rab8a localization, ciliogenesis and vesicle trafficking Yi-Chun Hsiao 1, Zachary J. Tong 1, Jennifer E. Westfall 1, Jeffrey G. Ault 2, Patrick S. Page-McCaw 1 and Russell J. Ferland 1,2, 1 Department of Biology, Center for Biotechnology and Interdisciplinary Studies, Rensselaer Polytechnic Institute, Troy, NY 12180, USA and 2 The Wadsworth Center, New York State Department of Health, Albany, NY 12201, USA Received February 17, 2009; Revised and Accepted July 17, 2009 The primary non-motile cilium, a membrane-ensheathed, microtubule-bundled organelle, extends from virtually all cells and is important for development. Normal functioning of the cilium requires proper axoneme assembly, membrane biogenesis and ciliary protein localization, in tight coordination with the intraflagellar transport system and vesicular trafficking. Disruptions at any level can induce severe alterations in cell function, giving rise to a myriad of human genetic diseases known as ciliopathies. Here we show that the Abelson helper integration site 1 (Ahi1) gene, whose human ortholog is mutated in Joubert syndrome, regulates cilium formation via its interaction with Rab8a, a small GTPase critical for polarized membrane trafficking. We find that the Ahi1 protein localizes to a single centriole, the mother centriole, which becomes the basal body of the primary cilium. In order to determine whether Ahi1 functions in ciliogenesis, loss of function analysis of Ahi1 was performed in cell culture models of ciliogenesis. Knockdown of Ahi1 expression by shrnai in cells or targeted deletion of Ahi1 (Ahi1 knockout mouse) leads to impairments in ciliogenesis. In Ahi1-knockdown cells, Rab8a is destabilized and does not properly localize to the basal body. Since Rab8a is implicated in vesicular trafficking, we next examined this process in Ahi1-knockdown cells. Defects in the trafficking of endocytic vesicles from the plasma membrane to the Golgi and back to the plasma membrane were observed in Ahi1-knockdown cells. Overall, our data indicate that the distribution and functioning of Rab8a is regulated by Ahi1, not only affecting cilium formation, but also vesicle transport. INTRODUCTION The primary cilium is a conserved cell structure extending from the apical cell surface into the extracellular space (1). It extends from virtually all cells and is important for normal development (1 6). The cilium is thought to act as a sensory organelle for the cell (7). For instance, the formation and functioning of cilia have been implicated in the maintenance of the sonic hedgehog (Shh) and Wnt signalling pathways, leading to their critical roles in the patterning of the vertebrate body plan (2 5,8). An emerging body of evidence has implicated the cilium in a range of human developmental disorders. Such diseases are now referred to as ciliopathies (9), reflecting the fact that the proper assembly and maintenance of the cilium are critical for normal development and cell signalling (2 5,8). Ciliogenesis and cilium function require axoneme assembly, intraflagellar transport (IFT), membrane biogenesis and proper compartmentation of signalling proteins (1). One important cellular and ciliary process is the coordination of polarized vesicle trafficking, a process modulated by members of the Rab and Arf small GTPase protein families (10,11). Rab proteins are small, monomeric GTPases that comprise the largest family of proteins within the Ras superfamily. Like other GTPases, Rab activity is regulated by guanine nucleotide exchange and modulation of GTP hydrolysis (12). Inactive GDP-bound Rab is activated by the exchange of GDP for GTP. Rabs are tightly controlled by a host of effector proteins To whom correspondence should be addressed at: Biology Department, Rensselaer Polytechnic Institute, 110 8th Street, BT2227, Troy, NY 12180, USA. Tel: þ ; Fax: þ ; ferlar@rpi.edu # The Author Published by Oxford University Press. All rights reserved. For Permissions, please journals.permissions@oxfordjournals.org
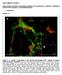
2 Human Molecular Genetics, 2009, Vol. 18, No including GDP/GTP exchange factors (GEFs), GTPase activating proteins (GAPs) and GDP dissociation inhibitors (GDIs) (12,13). Rabs are critical mediators of both exocytotic and endocytotic membrane trafficking, facilitating the formation of protein complexes that are involved in vesicle targeting, transport and fusion (12,14 16). Rab5, 8a and 11 are involved in vesicular trafficking between the Golgi network and the plasma membrane (17). Interestingly, Rab8a also has been found in the primary cilium, where it plays important roles in ciliogenesis and ciliary function (11). Alterations to the Rab8a pathway have a significant impact on ciliary membrane biogenesis (11,18 20); however, the mechanism by which Rab8a is recruited to the basal body, which is necessary for its effects on ciliogenesis, remains unclear. Joubert syndrome (JBTS) is an autosomal recessive neurodevelopmental disorder in humans characterized by retinal dystrophy and by defects in the development of the hindbrain (21). The Abelson helper integration site 1 (AHI1) gene is one of seven identified genes [NPHP1 (22), NPHP6/CEP290 (23,24), MKS3 (25), RPGRIP1L/NPHP8 (26,27), ARL13B (28) and CC2D2A (29)] implicated in JBTS, with 10 15% of all individuals with JBTS having mutations in AHI1 (30 32). AHI1 is a cytoplasmic multidomain protein composed of an N-terminal coiled-coil domain, WD40 repeats and a C- terminal SH3 domain (31,33). Given these domains, it is presumed that AHI1 acts as a scaffolding protein (31). AHI1 is expressed in the neurons of the hindbrain, midbrain and ventral forebrain; but it is also present in the pituitary, testis and kidney (34). Little is known about the function of AHI1/Ahi1 or how mutations in AHI1 lead to JBTS, but JBTS is considered to be a disease of the primary cilium, based on clinical-symptom similarities with other ciliopathies (9,35). Many of the proteins implicated in JBTS appear to be cytoplasmic proteins that are not membrane-associated, suggesting that these proteins may be involved in the signal transduction events that occur following receptor activation. Whether AHI1 shares similar properties and can be grouped with these proteins, as a cause of disorders now being termed ciliopathies, and/or whether AHI1 defines a separate pathway involved in manifesting this phenotype is currently unknown. Elucidation of the role of AHI1 in ciliary function is critical not only for a better understanding of JBTS, but also for identifying the role of the cilium in development and cell function. Given the overlap between JBTS and other ciliopathies, we postulated that Ahi1 may be necessary for proper primary cilium formation or function. We tested our hypothesis by examining whether inner medullary collecting duct cells (IMCD3) that had shrnai knockdown of Ahi1 had normal primary cilium formation, and whether any defects in ciliogenesis were caused by altered Rab8a function or vesicular trafficking defects. Here we show that Ahi1 is found at both the mother centriole and the basal body of the primary cilium. Knockdown of Ahi1 expression by shrnai leads to (i) loss of primary non-motile cilia, (ii) defects in vesicular trafficking and (iii) disruption of Rab8a function. In Ahi1-knockdown cells, endocytic vesicles are formed, but do not properly traffic. Vesicles, transporting the transferrin receptor, form and traffic centripetally, but the transferrin receptor does not recycle to the plasma membrane. In cells with knockdown of Ahi1, Rab8a is destabilized leading to decreases in its expression levels, and cannot properly localize to the basal body. Lastly, in further support for a role of Ahi1 in ciliogenesis, we found that mouse embryonic fibroblasts obtained from mice with a targeted deletion of Ahi1 have significantly decreased numbers of cells with primary non-motile cilia. Together, these data indicate a role for Ahi1 in the process of ciliogenesis as well as in the coordination of polarized vesicle trafficking. Our results begin to clarify the role of Ahi1 in JBTS in addition to further implicating the primary cilium in the pathogenesis observed in JBTS. RESULTS Ahi1 localizes to the mother centriole We find that Ahi1, the mouse ortholog of AHI1, is found at the centrosome as demonstrated by co-localization with the centrosomal marker, g-tubulin, in mouse renal inner medullary collecting duct (IMCD3) cells (Fig. 1A), NIH/3T3 cells (mouse fibroblast cell line; Supplementary Material, Fig. S1A) and spermatogonia cells (Supplementary Material, Fig. S1B); all cell lines containing a primary non-motile cilium. Analysis of the distribution of Ahi1 at each phase of the cell cycle demonstrated that Ahi1 is present in the centrioles at all stages (Supplementary Material, Fig. S2). We further noticed that only one of the centrioles shows Ahi1 immunostaining (Fig. 1A). To determine whether Ahi1 is present in the mother or the daughter centriole (36), we performed co-staining of g-tubulin with two markers of the mother centriole, ninein and Odf2 (37,38), in IMCD3 cells transfected with an enhanced green fluorescent protein tagged mouse Ahi1 plasmid (Ahi1-EGFP). In agreement with the results of the Ahi1 immunostaining experiments, Ahi1-EGFP co-localizes with g-tubulin (Fig. 1B). Ninein has been found to label both the mother and the daughter centriole, with ninein having greater localization at the mother centriole than the daughter centriole (38). We find that Ahi1-EGFP is found in association with the centriole having higher localization of ninein, the mother centriole (Fig. 1B). In further support, Ahi1-EGFP is also co-localized only at the Odf2- positive centriole (Fig. 1C). These data indicate that Ahi1 associates with a single centriole, the Odf2- and nineinpositive mother centriole, in cycling cells. Ahi1 localizes to the basal body of the primary cilium While Ahi1 was found throughout the cytoplasm, very little expression was observed in either the juxtamembrane domain or in the nucleus of confluent cells. Since the mother centriole is the origin of the basal body of the primary cilium (1,39), we sought to determine whether Ahi1 is directly associated with the basal body. We performed Ahi1 immunostaining on IMCD3 cells after serum deprivation, a treatment that induces the formation of primary nonmotile cilia (40,41). As expected, we observe a single elongated primary non-motile cilium, as determined by acetylated a-tubulin immunostaining, in the vast majority of the serum-starved cells (Fig. 2A). Ahi1 immunostaining mapped to the base of the primary non-motile cilium, specifically the
3 3928 Human Molecular Genetics, 2009, Vol. 18, No. 20 Figure 1. Ahi1 localizes to the mother centriole. (A) IMCD3 cells were immunostained for the centrosomal marker g-tubulin (green), and for endogenous Ahi1 (red). Arrowheads indicate the co-localization of g-tubulin and Ahi1 to only one of the paired centrioles. (B) Ahi1-EGFP (green) transfected IMCD3 cells were stained with g-tubulin (violet) and with ninein (red), a marker for the mother centriole. Arrowheads indicate the co-localization of Ahi1, g-tubulin and ninein in the mother centriole. (C) Ahi1-EGFP (green) transfected IMCD3 cells were stained with g-tubulin (violet) and with Odf2 (red), a mother centriole marker. Arrowheads indicate the co-localization of Ahi1, g-tubulin, and Odf2 only in the mother centriole. DNA is visualized with Hoechst (blue). Scale bars ¼ 5 mm. basal body (Fig. 2A) (42). The mapping of Ahi1 to the basal body of the cilium was also observed in other mouse cell lines derived from brain, skin and gonadal tissue (Supplementary Material, Fig. S3). In some cilia, we observed staining for Ahi1, not only in the basal body, but also faintly in the cilium axoneme (Fig. 2A); nevertheless, Ahi1 staining was always most pronounced in the basal body. These results indicate that Ahi1 associates with the mother centriole-derived organelle, the basal body, in cells that have Ahi1 expression and a primary non-motile cilium (42). Ahi1 is required for primary cilium formation Given the mapping of Ahi1 to the ciliary basal body, we examined whether Ahi1 is necessary for cilium formation. Knockdown of Ahi1 in IMCD3 cells (shahi1), by either stable integration or transient transfection of three independent shrnai constructs targeted for Ahi1, produced a significant decrease in Ahi1 expression when compared with wild-type or scramble shrnai-transfected cells [Fig. 2B (RNAi sequence 1:.90% decrease in Ahi1 expression); data not shown (RNAi sequences 2 and 3)]. No Ahi1 expression to the centriole or basal body was seen in shahi1 cells [Fig. 2C (RNAi sequence 1); data not shown (RNAi sequences 2 and 3)]. Following serum deprivation, both scramble shrnai cells and wild-type cells have robust cilium outgrowth in nearly every cell, as assessed by acetylated a-tubulin (Fig. 3A) and Arl13b [an additional marker for the primary cilium (3); data not shown] immunostaining. However, in Ahi1-knockdown cells, we observed a dramatic reduction in cilium formation. Greater than 80% of Ahi1- knockdown cells lacked a primary cilium (Fig. 3A and B). These results suggest that Ahi1 is necessary for the formation of the primary non-motile cilium. In addition, we isolated mouse embryonic fibroblasts (MEFs) from mice with a targeted deletion of Ahi1 to determine whether cells from an Ahi1 knockout mouse lacked cilia (Supplementary Material, Fig. S4). Primary cilia were observed on 57% of all MEFs cultured from Ahi1 þ/þ embryos (Fig. 3C). However, there was a significant reduction in ciliated MEFs cultured from Ahi1 2/2 embryos with only 10% of the cells having a cilium (Fig. 3C). Overall, the lack of primary non-motile cilia in cells with either knockdown of Ahi1 or deletion of Ahi1 supports the involvement of Ahi1 in the process of ciliogenesis. Scanning electron microscopy (SEM) was used to visualize the cilium in Ahi1-knockdown and control cell lines. A primary cilium was easily observable on the surface of
4 Human Molecular Genetics, 2009, Vol. 18, No Figure 2. Ahi1 localizes to the basal body of the primary non-motile cilium and its expression is lowered in Ahi1-knockdown IMCD3 cells. (A) Serum-deprived IMCD3 cells have robust cilium formation (one/cell). Ciliated IMCD3 cells were stained with acetylated a-tubulin (green), a ciliary axoneme marker, and for Ahi1 (red). Arrowheads indicate the localization of Ahi1 to the base of a primary cilium. DNA is visualized with Hoechst (blue). For evaluation of knockdown efficiency of Ahi1, IMCD3 cells stably expressing shrnai against Ahi1 were established and analysed for Ahi1 protein expression by (B) western blotting and (C) immunostaining. (B) Protein levels of Ahi1 in four independent Ahi1-knockdown IMCD3 stable clones, two control shrnai scramble stable clones and wild-type cells were analysed by western blotting. Ahi1 (130 kda, lower band) can be detected in scramble control and wild-type cells, but not in cells expressing a shrnai against Ahi1. The top band is considered to be a non-specific band (34). (C) Immunostaining for g-tubulin (green), for Ahi1 (red) and for DNA (Hoechst (blue)) was performed in wild-type IMCD3 cells and in Ahi1-knockdown cell line 1 (shahi1-1). Scale bars ¼ 5 mm. the majority of wild-type and scramble shrnai cells, but that was not the case for the Ahi1-knockdown cell lines (Fig. 4A). The few putative primary cilia that we observed in the Ahi1- knockdown cells were shorter than normal (Fig. 4B). We also noticed that the acetylated a-tubulin immunostaining in Ahi1- knockdown cells was diffusely distributed throughout the cytoplasm, unlike the pattern in control cells where a-tubulin was restricted mostly to the primary cilium
5 3930 Human Molecular Genetics, 2009, Vol. 18, No. 20 Figure 3. Ahi1 is required for the normal formation of primary non-motile cilium. (A) Control cells (scramble shrnai), Ahi1-knockdown cell line 1 (shahi1-1) and Ahi1-knockdown cell line 2 (shahi1-2) (both shahi1-1 and shahi1-2 were RNAi sequence 1) were cultured under serum-deprived conditions, for initiation of robust cilium formation. Cells were immunostained for acetylated a-tubulin (green; marker of the primary cilium) and for Ahi1 (red). Note that the acetylated a-tubulin staining was actually acquired on a Cy5 channel and has been pseudocoloured green (the true FITC channel visualizes the shahi1 construct that is GFP tagged and is not shown in this image). Higher magnification images of the boxed regions are shown in the right column. The histogram shows the percentage of cells having a primary cilium based on counts of cells with acetylated a-tubulin-positive cilia, for Ahi1-knockdown cells (shahi1-1, -2 and -3), control-scramble cells and wild-type cells. Ten randomly acquired fields (a minimum of 500 cells were counted for each clone) were analysed. The error bars represent the standard error of the mean. Asterisks indicate significance from scramble and wild-type cells (P, ) using Chi-square analyses. (B) Ahi1-knockdown cells that had been transiently transfected with RNAi sequence 3 were cultured under serum-deprived conditions, for initiation of robust cilium formation. Cells were immunostained for acetylated a-tubulin (red) with the green channel highlighting cells that had been transfected with the shrnai construct. The histogram shows the percentage of cells having a primary cilium based on counts of cells with acetylated a-tubulin-positive cilia, for Ahi1-knockdown cells that were transfected with shrnai constructs against Ahi1 (RNAi sequences 2 and 3), a scramble shrnai construct or no construct (wild-type cells). Three randomly acquired fields (a minimum of 200 cells were counted for each clone) were analysed. The error bars represent the standard error of the mean. Asterisks indicate significance from scramble and wild-type cells (P, ) using Chi-square analyses. (C) Mouse embryonic fibroblasts (MEFs) cultured from wild-type (Ahi1 þ/þ ) and Ahi1 knockout (Ahi1 2/2 ) mouse embryos. MEFs were immunostained for acetylated a-tubulin (green; marker of the primary cilium). The histogram shows the percentage of Ahi1 þ/þ and Ahi1 2/2 MEFs having a primary cilium based on counts of cells with an acetylated a-tubulin-positive cilium. Three randomly acquired fields (a minimum of 100 cells were counted for each field) were analysed for each mouse. The error bars represent the standard error of the mean. Asterisk indicates significance from wild-type (Ahi1 þ/þ ) MEFs (P, ) using Chi-square analysis. Scale bars ¼ 5 mm.
6 Human Molecular Genetics, 2009, Vol. 18, No Figure 4. Ahi1 is required for the normal formation of primary cilia. (A) Low magnification scanning electron micrographic images of wild-type and Ahi1- knockdown (RNAi sequence 1) IMCD3 cells under conditions optimized for ciliogenesis. Wild-type cells (top) each clearly have an elongated and pronounced primary cilium (black arrowheads) on the cell surface. However, Ahi1-knockdown cells (bottom) have a striking lack of primary cilia. (B) Higher magnification scanning electron micrographs of the primary cilium (black arrowheads) and microvilli (black arrows) in wild-type (left) and Ahi1-knockdown IMCD3 (right) cells. Primary cilia were very difficult to find in Ahi1-knockdown IMCD3 cells; when putatively identified, they tended to be short and stumpy (right). (C) Apical and basal actin filament staining by phalloidin in wild-type, scramble and Ahi1-knockdown cells showing a disorganization and decrease in actin filaments in Ahi1-knockdown cells. Scale bars ¼ 2 mm in (A), 0.3 mm in (B) and 5 mm in (C). (Fig. 3A and B; data not shown). Given the observed defects in tubulin, we also explored whether there were abnormalities in the actin cytoskeleton. We examined actin organization in control, scramble and Ahi1-knockdown cells using phalloidin, a marker for actin filaments (43). Unlike control cells, which had an organized actin filament pattern, the actin filaments in Ahi1-knockdown cells had significant disorganization of apical actin and notably decreased actin stress fibres (basal) (Fig. 4C), with the shahi1-1 line being more severely affected than the shahi1-2 line. Overall, these results suggest that Ahi1 is a protein that is critical for ciliogenesis, and may also affect the organization and formation of cytoskeletal networks (microtubules and actin) necessary for cilia formation. Ahi1 is required for localization of Rab8a to the basal body of the primary cilium Rab proteins are generally thought to control polarized membrane trafficking in cells (16), and disruptions in Rab function result in significant alterations in protein homeostasis (44). Several members of the Rab GTPase family, particularly Rab8a, have been identified as critical modulators of ciliogenesis (18,45). Given the importance of Rab8a in ciliogenesis, we examined whether Ahi1 and Rab8a were found in similar cellular domains. We observed that both Ahi1 and Rab8a were seen to co-localize to the basal body of the developing and mature cilium (Fig. 5A, bottom). Next, we sought to determine whether Ahi1 interacts with Rab8a in regulating cilium formation. Since our mouse Ahi1 antibody cannot recognize human AHI1 in HEK293 cells, we used this cell line for testing whether Ahi1 and Rab8a form a complex. In HEK293 cells that were co-transfected with Ahi1-EGFP and wild-type HA-tagged Rab8a (HA-Rab8a) constructs, we observed an association of HA-Rab8a with Ahi1-EGFP by co-immunoprecipitation methods (Fig. 5A, top), but not in cells transfected with either Ahi1-EGFP or HA-Rab8a alone (Fig. 5A, top). Similar results were obtained using myc-tagged Ahi1 (myc-ahi1) and HA-Rab8a constructs (Supplementary Material, Fig. S5A) or IMCD3 cell lysates (Supplementary Material, Fig. S5B), but not using myc-ahi1 and HA-Rab11 constructs (control experiment; Supplementary Material, Fig. S5C). In addition, bead only controls were devoid of nonspecific binding (data not shown). Since these experiments were conducted in sub-confluent HEK293 cells, with relatively few cilia present, these data further suggest that this interaction is apparently not dependent on the presence of a cilium. In order to determine whether the association of Rab8a and Ahi1 was functionally important, we examined the expression and localization of Rab8a in Ahi1-knockdown cells. Significantly lower Rab8a expression was observed, by western blotting, in Ahi1-knockdown cells when compared with the expression in control cells (Fig. 5B) suggesting a role for Ahi1 in stabilizing Rab8a expression. We also observed a significant re-distribution of Rab8a in Ahi1-knockdown cells, relative to the pattern in control cells. In control cells, endogenous Rab8a immunostaining was found mainly at cell junctions and close to the basal body of primary cilium (Fig. 5C). In contrast, in Ahi1-knockdown cells, Rab8a
7 3932 Human Molecular Genetics, 2009, Vol. 18, No. 20 Figure 5. Ahi1 is involved in ciliogenesis through its interactions with the small GTPase, Rab8a. (A, top) Co-immunoprecipitation of Ahi1-EGFP with HA-Rab8a in lysates from HEK293 cells transfected with Ahi1-EGFP and HA-Rab8a, but not in cells transfected with HA-Rab8a alone or Ahi1-EGFP alone. Resin only controls had no labelling (data not shown). (A, bottom) Co-localization of both Ahi1 (green) and Rab8a (red) at the basal body in wild-type cells (denoted by white arrowheads). (B) The protein level of endogenous Rab8a from Ahi1-knockdown (shahi1-1 and -2) and from scramble shrnai control cells was analysed by western blotting (top) and graphically displayed (bottom). The level of b-tubulin represents the loading control. The error bars represent the standard error of the mean. (C) Wild-type (top panel) and Ahi1-knockdown IMCD3 cells (lower panel) were stained with antibodies against Rab8a (green) and Arl13b (red; marker for the basal body and the primary cilium). Right panel represents the merged images of Rab8a, Arl13b and DNA (blue) staining. White arrowheads point to the basal body. With this Rab8a antibody, localization is only found at the basal body and does not show any apparent localization to the cilium. Scale bars ¼5 mm in (A and C). expression was significantly lower, and it could not be detected at the basal body (Fig. 5C). This result suggests that Ahi1 is required for Rab8a localization to the basal body and further indicates that failure of Ahi1 and Rab8a to associate leads to Rab8a destabilization. To test this model, HA-Rab8a was overexpressed in control, scramble and Ahi1-knockdown cells. Overexpression of HA-Rab8a was chosen instead of using a Rab8a antibody because our previous data demonstrated a significant reduction in Rab8a levels in Ahi1-knockdown cells to the point that it is not detectable by fluorescent microscopy. In control cells, HA-Rab8a was found mainly in the primary cilium, but variably in the basal body of wild-type cells under cilia growth-optimizing conditions (Fig. 6A, without serum); however, when cells were not in cilia growth-optimizing conditions, HA-Rab8a was found at the basal body (Fig. 6A, with serum). In addition, HA-Rab8a was also found localized throughout the cytoplasm, in the Golgi complex (using the Golgi marker, Golph4), and variably in cytoplasmic vesicles (Fig. 6A, bottom). Overall, these results indicate that Rab8a recruitment to the basal body of the primary cilium precedes ciliogenesis. During ciliary biogenesis, Rab8a transitions from the basal body to the primary cilium itself. Overexpression of HA-Rab8a in Ahi1-knockdown cells resulted in not only a failure to rescue cilium formation (Fig. 6B and C), but also an inability for Rab8a to localize to the basal body [Fig. 6D (using the basal body/cilia marker, Arl13b) and Supplementary Material, Fig. S6 (using the basal body marker, g-tubulin)]. Diffuse expression of HA-Rab8a was found throughout much of the cytoplasm in
8 Human Molecular Genetics, 2009, Vol. 18, No Figure 6. Overexpression of HA-Rab8a in Ahi1-knockdown cells results in an inability to rescue ciliogenesis and an inability for HA-Rab8a to localize to the basal body of the primary cilium. (A) Wild-type IMCD3 cells, cultured under non-cilia forming conditions (top) or under serum-deprived conditions so as to promote cilium formation (bottom), and transfected with an HA-Rab8a expression plasmid were immunostained for Ahi1 (green) and for HA (red). Right panel represents the merged images of Ahi1, HA-Rab8a and DNA (blue) staining. White arrowheads point to the basal body. The bottom panel represents HA-Rab8a localization to the Golgi complex (using the Golgi marker Golph4 (green)). (B) Wild-type, scramble and Ahi1-knockdown cells transfected with HA-Rab8a were immunostained with acetylated a-tubulin (green) for primary cilia and HA for Rab8a (red) distribution. Arrowheads indicate the co-localization of HA-Rab8a and acetylated a-tubulin staining. Right panel represents the merged images of HA-Rab8a, acetylated a-tubulin and DNA (blue) staining. (C) Quantification of cilium formation in HA-Rab8a expressing wild-type, scramble and Ahi1-knockdown cells. Cells were co-stained with HA and Arl13b. Fifty HA-positive cells were counted from each slide and the number of primary cilium was determined. Data were collected from three independent experiments. The error bars represent the standard error of the mean. Asterisks indicate significance from scramble and wild-type cells (P, ) using Chi-square analyses. (D) Wild-type and scramble cells [grown to sub-confluence (resulting in few cilia); left two panels] and Ahi1-knockdown cells (grown under cilia optimized conditions; right two panels) transfected with HA-Rab8a were stained with antibodies against HA (red, for HA-Rab8a) and Arl13b (green, for the basal body). Arrowheads point to Arl13b labelled basal bodies. The lower panel represents the merged images of HA-Rab8a, Arl13b and DNA (blue) staining. This experiment was designed to capture wild-type and scramble cells at a time when few cells had cilia, so as to easily observe the association of HA-Rab8a in the basal body (before the formation of cilia where HA-Rab8a would enter the cilium making it difficult to differentiate basal body from the primary cilium). Scale bars ¼5 mm in (A, B and D).
9 3934 Human Molecular Genetics, 2009, Vol. 18, No. 20 control, scramble and Ahi1-knockdown cells; however, unlike control and scramble cells, HA-Rab8a was never found to localize to the basal body in Ahi1-knockdown cells (Fig. 6B and D and Supplementary Material, Fig. S6). Since Rab8a is not found at the basal body in Ahi1-knockdown cells (Fig. 6B and D and Supplementary Material, Fig. S6), this indicates a fundamental role for Ahi1 in the proper targeting of Rab8a to the basal body. Furthermore, since Rab8a has been proposed to mark the cilium membrane domain (11), the proper distribution of Rab8a is likely critical for cilium formation. Since the normal distribution of Rab8a was disrupted in Ahi1-knockdown cells, particularly in the region of the ciliary basal body, a plausible physical explanation to account for the impairment in ciliogenesis seen in Ahi1- knockdown cells is that Ahi1 is required for Rab8a targeting to the appropriate membrane domain in Ahi1 expressing cells. Ahi1 is required for vesicular trafficking Rab8a regulates the transport of membrane proteins from the Golgi network to the base of the primary cilium and also to the surface of a polarized cell (11,17,44), suggesting a fundamental role of Rab8a in the directional transport of proteins and vesicles to plasma membrane domains. Since the Rab8a distribution was disrupted in Ahi1-knockdown cells, we next examined whether vesicular trafficking was disrupted in these cells. To begin to determine whether clathrinindependent endocytic transport is altered in Ahi1-knockdown cells, we used fluorescently labelled Cholera toxin B (CTxB) to follow endocytic vesicles (46). We found in control cells that CTxB-labelled vesicles were rapidly transported to the Golgi as evidenced by co-localization with the Golgi marker, GM130 (Fig. 7A). In contrast, CTxB-labelled vesicles were not transported to the Golgi in Ahi1-knockdown cells (Fig. 7A). Instead, CTxB vesicles were seen to be randomly distributed in the cytoplasm of these Ahi1-knockdown cells (Fig. 7A). Although longer CTxB exposure resulted in transport of some vesicles to the Golgi, most CTxB vesicles were unable to reach the Golgi. These results suggest that Ahi1 is necessary for trafficking of vesicles, but not for endocytosis itself. To determine whether this failure of endocytic vesicle trafficking was a general property of endocytic vesicles in Ahi1- knockdown cells, we examined whether clathrin-dependent endocytosis required Ahi1 by analysing the transferrin and transferrin receptor (Tf/TfR) endocytic pathway. In both control and Ahi1-knockdown cells, fluorescently labelled Tf was found to bind to the cell surface, and to then become incorporated into vesicles and transported toward the perinuclear region (Fig. 7B; 10 and 30 min pulse). This result demonstrates that Ahi1 is not required for general vesicular trafficking, as clathrin-dependent vesicle trafficking to the perinuclear region remains normal, while vesicles derived from clathrin-independent endocytosis do not traffic normally. The Tf/TfR system also allowed us to examine vesicle cycling from the Golgi to the plasma membrane. Whereas control cells recycled the Tf/TfR back to the plasma membrane after 60 min, Ahi1-knockdown cells accumulated Tf/TfR vesicles in the perinuclear region, indicating that vesicle recycling to the plasma membrane is disrupted (Fig. 7B; 60 min pulse). Our results demonstrate that trafficking of proteins/vesicles is altered in the absence of Ahi1, particularly from the Golgi to the plasma membrane, again suggesting that disruptions of the Rab8a-dependent membrane targeting pathway could be responsible for the failure of ciliogenesis and ciliary biogenesis. Since we observed an alteration in vesicular trafficking in Ahi1-knockdown cells, we hypothesized that Golgi organization was disrupted in these cells. Analysis of Golgi structure in Ahi1-knockdown cells showed abnormalities in Golgi structure, using both a cis-golgi (GM130) marker (Fig. 7C) and a trans-golgi (Golph4) marker (data not shown). The Golgi in Ahi1-knockdown cells appeared significantly less-extensive and fragmented in comparison to the perinuclear ribbon-like network observed in control cells (Fig. 7C). In addition, the Golgi in Ahi1-knockdown cells was abnormally positioned, appearing on the apical end of the nucleus, in contrast to the geometry in control cells where the Golgi was positioned laterally with respect to the nucleus (Fig. 7C). Golgi organization was thus apparently altered in Ahi1-knockdown cells, but whether the mechanism for this alteration relates directly to Ahi1 disruption, or whether it is an indirect result of vesicle accumulation remains to be established. Since proper Golgi position, vesicular trafficking and cilium formation are linked to cellular polarity, our results support a role for Ahi1 in establishing polarity or orienting the endocytic system to an existing cellular polarity. DISCUSSION Ahi1 is required for ciliogenesis Many genes have recently been identified in human genetic syndromes that affect the formation or function of the primary cilium. Two of these genetic syndromes, Bardet Biedl syndrome (BBS) and JBTS, present with different symptoms, but these multigenic syndromes are thought to affect the same organelle, the primary non-motile cilium (47). In this study, we have investigated the role of the JBTS gene, Ahi1, in the function of the primary non-motile cilium. Using welldeveloped cell culture models of ciliogenesis, we find that Ahi1 is expressed at the mother centriole and basal body in ciliated cells (42). Using multiple shrnai constructs and mouse embryonic fibroblasts derived from mice with a targeted deletion of Ahi1, we find that depletion of Ahi1 in these cells results in loss of the primary cilium. It still remains to be determined whether the loss of primary cilium in individuals with JBTS and AHI1 mutations is a significant factor in the pathogenesis of JBTS or whether the organ abnormalities in JBTS are related to other molecular mechanisms (i.e. defects in vesicular trafficking). However, our data clearly indicate that the loss of Ahi1 in cells results in defects in the primary non-motile cilium. Primary cilia have an important role in signalling in the cell, and molecules such as Shh and Wnt, which are critical for development, transduce their signals through cilia (2 5,8). Thus, embryonic defects including embryonic lethality would be expected if a gene involved in the formation of cilia is defective, but the severity of that defect may vary due to differing ciliary-dependent signalling requirements.
10 Human Molecular Genetics, 2009, Vol. 18, No Figure 7. Membrane trafficking is altered in Ahi1-knockdown cells. (A) Cholera toxin endocytic assay. Wild-type (top row) and Ahi1-knockdown cells (bottom row) were incubated with Alexa Fluor 546-conjugated Cholera toxin subunit B (CTxB; red) for 30 min after cold incubation. GM130 (green) staining delineates the Golgi. The plot shows the percentages of cells showing co-localization of CTxB and GM130 staining. In each case,.200 cells were scored in three independent tests [asterisk indicates a significant effect (P, ) using Chi-square analysis in comparing Ahi1-knockdown cells with scramble and wild-type cells]. (B) Transferrin endocytic assay. Wild-type (right) and Ahi1-knockdown (left) cells were incubated with Alexa Fluor 546-conjugated transferrin (Tf; red) for 10, 30 or 60 min after cold incubation. DNA is visualized with Hoechst (blue). Note that following a 60 min pulse, Tf is found throughout the cell membrane in wild-type cells, but in Ahi1-knockdown cells, Tf has accumulated in the perinuclear region of the cell (and not the membrane). The plot shows the percentages of cells with Tf localized to the perinuclear region of the cell after a 60 min pulse. In each case,.200 cells were scored with the asterisk indicating a significant effect (P, ) using Chi-square analysis in comparing Ahi1-knockdown cells with scramble and wild-type cells. (C) The Golgi structure and position in Ahi1-knockdown cells. Ahi1-knockdown (shahi1-1 and -2), control scramble and wild-type IMCD3 cells cultured under serum-deprived conditions were stained with the cis-golgi compartment marker, GM130 (red), to permit examination of the Golgi structure. Similar results were obtained with the trans-golgi compartment marker, Golph4 (data not shown). Scale bar¼5 mm in(a C). Genes known to be required for the formation of primary cilia have been found to be embryonic lethal in the mouse. For instance, cells with KIF3A defects (KIF3A is a component of kinesin II) cannot form primary cilium. Mice with targeted deletions of Kif3a show embryonic lethality, which is thought to be due to impairments in Shh signalling (48 50). However, abnormal ciliary function can result in a variety of phenotypes, not including embryonic lethality. For instance, the JBTS-causing genes NPHP1 and CEP290 encode centrosomal/basal body proteins (19,51). However, NPHP1 and CEP290 mutations, in both humans and mice, do not cause embryonic lethality, but manifest other ciliary defects, such as infertility and retinal degeneration (22 24,52 54). Conversely, other JBTS-causing genes, such as RPGRIP1L and ARL13B, which also encode centrosomal/basal body proteins, rarely cause embryonic lethality in humans, but almost always cause embryonic lethality in mice (3,27,28,55). However, little is known about the mechanisms behind these pleiotropic phenotypes. Our results indicate that significant ciliogenesis defects are observed with either Ahi1-knockdown by shrnai or in cells from Ahi1 knockout mice, but like mutations in other JS-related genes, no embryonic lethality is observed in human patients with AHI1 mutations (30,31). One possible
11 3936 Human Molecular Genetics, 2009, Vol. 18, No. 20 explanation for the differences in phenotypic expression is likely tissue specificity. Ahi1 is not expressed throughout the body, but is only expressed in a limited number of tissues (34). It is only in the cells in which Ahi1 is expressed, that there appears to be ciliary defects. Therefore, absence of Ahi1 may not impair global ciliogenesis, but only effect the formation of cilia in certain cells and tissues (i.e. those expressing Ahi1). Furthermore, as described earlier, mutations in several ciliopathy genes can result in different phenotype severities in humans. This widely varying phenotype appears to be common in many human ciliopathies, such as BBS, Meckel Gruber syndrome and JBTS (9). Ahi1 and Rab8a cooperation in ciliogenesis Multiple processes are required for the formation of the primary cilium: transport of the mother centriole to the cell membrane, axoneme assembly, IFT and polarized membrane transport to the ciliary membrane domain (1). Rab8a is known to target polarized membrane transport from the trans- Golgi complex to the basolateral membrane of cells (17). Interestingly, Rab8a has also been implicated as playing a crucial role in ciliogenesis (11,56). In BBS, a related ciliopathy, cilium formation is impaired due to improper Rab8a signalling at the cilia (11). Rab8a entry into the ciliary axoneme is regulated by the BBsome, a multiprotein complex located at the basal body (11). Also, CEP290, another gene mutated in JBTS has also been shown to be critical for normal ciliogenesis through a Rab8a mediated pathway (19,20). However, how Rab8a is targeted to the basal body is still largely unknown. We have utilized our Ahi1-knockdown cell lines to determine what role Ahi1 plays in this complex process. We find the localization of Rab8a to the basal body is disrupted in Ahi1-knockdown cells as are the cytosolic levels of Rab8a. Importantly, despite overexpression of Rab8a, proper localization to the basal body and primary cilium formation are not rescued. This indicates that Ahi1 is required for proper Rab8a targeting to the basal body, as well as having a role in the stabilization of the Rab8a protein. These data provide some insight on how Rab8a is localized to the basal body through an Ahi1-dependent mechanism. Whether Cep290 and Ahi1 work cooperatively in mediating the Rab8adependent role on ciliogenesis, or whether Cep290 and Ahi1 work through distinctive mechanisms remains unknown. However, if Cep290 and Ahi1 work cooperatively, then it reasons that the common phenotypes observed in JBTS may be through an effect on Rab8a trafficking and localization to the basal body. Ahi1 is required for proper membrane trafficking and Golgi organization Defects in vesicular trafficking are also apparent in Ahi1- knockdown cells. In cultured Ahi1-knockdown cells, Tf/TfR and CTxB endocytosis occurs, but only the Tf/TfR endocytic vesicles are able to traffic to the perinuclear region; trafficking of CTxB to the Golgi is retarded in Ahi1-knockdown cells. This suggests that the CTxB vesicles are unable to associate with the transport apparatus. Rab8a is required for the proper transport of CTxB to the Golgi and for transport of Tf/TfR to the perinuclear region of the cell (17,46). Defects in Rab8a function can lead to the disruption of these endocytic pathways (17,46). Furthermore, our data indicate that Ahi1 is likely regulating Rab8a in specific membrane trafficking routes, but not in a universal manner. Remarkably, the Tf/ TfR containing vesicles traffic correctly to the perinuclear region, but cannot return to the cellular membrane. Since the Tf/TfR recycling process is regulated by other Rab proteins that are concentrated in the pericentriolar region, and are required for Tf/TfR recycling through the endosomal recycling compartment (57,58), this suggests a role for Ahi1 in other Rab protein-mediated processes. Our data indicate that Ahi1 may be required for coupling of vesicles to the transport apparatus, but in its absence may not provide a specific directional cue. Taken together, Ahi1 affects the function of Rab8a in specific trafficking pathways, as well as potentially affecting other Rab proteins involved in membrane trafficking (58 60). The structure of the Golgi appears altered in Ahi1- knockdown cells; the Golgi is less-extensive, fragmented and has altered polarity with respect to the nucleus. These characteristics are often found in cells with abnormal membrane trafficking (61). However, it is clear that Ahi1 is not required for general Golgi function, since Ahi1 expression is not widespread in all cell types (34,62). This suggests that the defects in Golgi structure might possibly relate to the excess build-up of cytoplasmic vesicles either to or from the Golgi in Ahi1-knockdown cells (63,64). This build-up could itself result in abnormal Golgi. In addition, aberrant Golgi structure may also result from an excess of vesicles in the cytoplasm that then would provide a loss of membrane back to the Golgi or within the Golgi (63,64). Ahi1 and the formation of polarity Significant alterations in the cytoskeleton were found in Ahi1- knockdown cells suggesting a role of Ahi1 in cytoskeletal dynamics. However, given the lack of organized actin filaments and acetylated-tubulin in cells lacking Ahi1, the decreases in cilia may be accounted for by a more global effect of Ahi1 in regulating the actin/microtubule cytoskeleton. Microtubule involvement in the formation of cilia has long been recognized as an important contributor to this process, but the role of actin in ciliogenesis has only recently been appreciated through studies demonstrating that apical enrichments of actin are required for the proper docking of the basal body to the cell membrane to initiate axoneme outgrowth in ciliogenesis (65 67). Given the abnormal localization of the Golgi and the absence of primary cilium in Ahi1-knockdown cells, these findings suggest a potential role of Ahi1 in establishing or responding to cellular polarity, or in polarized membrane trafficking in other cell types. Since Ahi1 is highly expressed in neurons and the major anatomical defects in JBTS involve brain structures (21,30,31,34,68,69), whether Ahi1 is involved in neuronal polarity remains to be determined. Neurons are highly polarized cells, where specific membrane trafficking is required for neurite development (70,71). One of the major neuroanatomical defects in JBTS is abnormalities in axonal decussation in the cerebellar vermis and the brainstem;
12 Human Molecular Genetics, 2009, Vol. 18, No however, little is known about the mechanisms involved in these processes. The necessity of Ahi1 in regulating Rab8adependent polarized membrane trafficking may provide insight into the mechanisms involved in JBTS and in the establishment of polarity in developing neurons. Conclusion Overall, our data suggest Ahi1 is a crucial regulator of cilium formation through the targeted localization of Rab8a to the basal body and a regulator of both vesicular trafficking to and from the Golgi. Given the role of Rab8a in membrane trafficking in the primary cilium, our data provide strong evidence that Ahi1 plays an important role in targeting ciliary membrane and/or receptors to the growing cilium. In the absence of Ahi1, cells are incapable of targeting membrane to the ciliary membrane domain and are therefore unable to form a cilium. Lastly, our data also illustrate a role of Ahi1 in vesicle trafficking in the cell. It has been proposed that the primary cilium functions as a central sensory organelle for the cell (7). In order to serve this function, receptors must localize to this structure and it is likely that this occurs by transport of vesicles to the ciliary membrane domain and subsequent transport to the ciliary tip via IFT (2 5,8). However, very little is known about the regulation of these pathways and how this specificity in receptor recruitment to the primary cilium is achieved. Our present work adds to the growing list of genes involved in either the formation or function of the cilium (1,72 74). In stark contrast to previously identified genes, Ahi1 is expressed in only a subset of all ciliated cells (34). The unexpected role of Ahi1 in vesicular trafficking suggests a fundamental role of Ahi1 in the regulation of cilia bound vesicular traffic, possibly responsible for providing specificity to cilia. If true, this indicates that cells may have adopted differing mechanisms for ciliogenesis and for the specific targeting of only certain receptors to the tips of the primary cilium. A special subset of proteins, to which Ahi1 might belong, may act as adaptors that recruit proteins to the primary cilium in a tissue-specific manner. While there does not appear to be any Ahi1 family members in mammals, most analyses have only been conducted using bioinformatic approaches (31,33,42). Since these analyses rely mostly on protein sequence and structure, proteins that might not fit these criteria, but still serve a similar function as Ahi1, would be missed. Our data begin to suggest that Ahi1 may be a protein that is present only in a subset of ciliated cells that may be involved in the regulation or trafficking of proteins necessary for the formation and/or functioning of different subtypes of cilium with presumed independent functions. MATERIALS AND METHODS Cell culture and transfection Mouse inner medullary collecting duct cells (IMCD3; from ATCC, Manassas, VA, USA) were maintained in DMEM/ F12 medium (Cellgro, Manassas, VA, USA) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT, USA), non-essential amino acids (BioWhittaker, East Rutherford, NJ, USA) and penicillin (100 U/ml)/streptomycin (100 mg/ ml) (GIBCO, Carlsbad, CA, USA) at 378C in a humidified incubator under 5% CO 2. To induce cilium formation, we cultured IMCD3 cells to confluence, followed by an additional 2 days in serum-deprived medium. Stable and transiently transfected IMCD3 cells, containing either shrnai sequences directed against Ahi1 or a scramble sequence, were analysed. The RNAi targeting sequences for mouse Ahi1 (RNAi 1: 5 0 -GATTTCTCACCCAATGGTAAA- 3 0 ; RNAi 2: 5 0 -TGAAATTCCTTCTGGACGTTT-3 0 ; RNAi 3: 5 0 -GAAACTGTCACAGAGGTGATA-3 0 ) and for a control scramble sequence (5 0 -GAAACAAGGGTGCCAGTGTCT-3 0 ) were synthesized. The sense and antisense oligonucleotides were cloned into a psuper-neo/gfp plasmid according to the manufacturer s instructions (OligoEngine, Seattle, WA, USA). To establish the stable Ahi1-knockdown cells, we linearized the psuper-neo/gfp-shrna plasmids with SacI, and transfected them into IMCD3 cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). After 24 h, cells were cultured in G418 selecting medium (1 mg/ml G418; Sigma, St Louis, MO, USA) until cell colonies formed. For cells with Ahi1 RNAi knockdown, 10 EGFP-expressing/ G418-resistant clones were assayed for Ahi1 protein levels, by western blotting, resulting in four lines (shahi1-1, -2, -3, -4). Control scramble RNAi-expressing IMCD3 cells were established using the same procedure (scramble-1 and -2). Once the stable clones were established, cells were maintained in normal IMCD3 medium. For transient transfection, IMCD3 cells were plated on glass coverslips in 24-well culture dishes for 24 h before transfection with Lipofectamine Cells were fixed for immunostaining 24 h after transfection. Cell cycle synchronization Localization of Ahi1 at time points throughout the cell cycle was determined through synchronization of the cells at various stages of the cell cycle. To enrich the population of cells at specific stages of the cell cycle, we cultured IMCD3 cells under the following conditions: for G0/G1 phase (serumdepriving medium for 24 h); for G1 phase (100 mm mimosine (Sigma) for 16 h); for G1/S phase (1 mg/ml aphidicolin (Sigma) for 16 h); for M phase (100 nm paclitaxel (Sigma) for 16 h). Low cell-density IMCD3 cultures, in normal medium without any treatments, were also assessed for cell cycle stage on the basis of DNA and cell morphological-based criteria. Immunostaining and imaging Overexpressed hemagglutinin (HA)-tagged Rab8a was transfected into the stably integrated IMCD3 cells using Lipofectamine 2000, with fixation of cells 48 h later. In addition, untransfected cell lines were also analysed throughout our studies. Cells were either fixed with ice-cold methanol at 2208C for 15 min or with 4% paraformaldehyde, and were then permeabilized in 0.04% Triton X-100/PBS (PBSTx) for 10 min at room temperature. Fixed cells were incubated in 10% normal donkey serum (for Odf2 antibodies) or 10% normal
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or
 Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
SONOMA STATE UNIVERSITY DEPARTMENT OF BIOLOGY BIOLOGY 344: CELL BIOLOGY Fall 2013
 SONOMA STATE UNIVERSITY DEPARTMENT OF BIOLOGY BIOLOGY 344: CELL BIOLOGY Fall 2013 Instructor Murali C. Pillai, PhD Office 214 Darwin Hall Telephone (707) 664-2981 E-mail pillai@sonoma.edu Website www.sonoma.edu/users/p/pillai
SONOMA STATE UNIVERSITY DEPARTMENT OF BIOLOGY BIOLOGY 344: CELL BIOLOGY Fall 2013 Instructor Murali C. Pillai, PhD Office 214 Darwin Hall Telephone (707) 664-2981 E-mail pillai@sonoma.edu Website www.sonoma.edu/users/p/pillai
Beta3 integrin promotes long-lasting activation and polarization of Vascular Endothelial Growth Factor Receptor 2 by immobilized ligand
 SUPPLEMENTAL FIGURES Beta3 integrin promotes long-lasting activation and polarization of Vascular Endothelial Growth Factor Receptor 2 by immobilized ligand C. Ravelli et al. FIGURE S. I Figure S. I: Gremlin
SUPPLEMENTAL FIGURES Beta3 integrin promotes long-lasting activation and polarization of Vascular Endothelial Growth Factor Receptor 2 by immobilized ligand C. Ravelli et al. FIGURE S. I Figure S. I: Gremlin
The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit
 Cell Reports, Volume 5 Supplemental Information The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit Andrey Poleshko, Katelyn M. Mansfield, Caroline
Cell Reports, Volume 5 Supplemental Information The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit Andrey Poleshko, Katelyn M. Mansfield, Caroline
Lab Module 7: Cell Adhesion
 Lab Module 7: Cell Adhesion Tissues are made of cells and materials secreted by cells that occupy the spaces between the individual cells. This material outside of cells is called the Extracellular Matrix
Lab Module 7: Cell Adhesion Tissues are made of cells and materials secreted by cells that occupy the spaces between the individual cells. This material outside of cells is called the Extracellular Matrix
by Neurobasal medium (supplemented with B27, 0.5mM glutamine, and 100 U/mL
 Supplementary Materials and methods Neuronal cultures and transfection The hippocampus was dissected from E8 rat embryos, dissociated, and neurons plated onto glass coverslips coated with poly-ornithine
Supplementary Materials and methods Neuronal cultures and transfection The hippocampus was dissected from E8 rat embryos, dissociated, and neurons plated onto glass coverslips coated with poly-ornithine
Chapter 15. Cytoskeletal Systems. Lectures by Kathleen Fitzpatrick Simon Fraser University Pearson Education, Inc.
 Chapter 15 Cytoskeletal Systems Lectures by Kathleen Fitzpatrick Simon Fraser University Table 15-1 - Microtubules Table 15-1 - Microfilaments Table 15-1 Intermediate Filaments Table 15-3 Microtubules
Chapter 15 Cytoskeletal Systems Lectures by Kathleen Fitzpatrick Simon Fraser University Table 15-1 - Microtubules Table 15-1 - Microfilaments Table 15-1 Intermediate Filaments Table 15-3 Microtubules
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table.
 Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Fig. S1. Endocytosis of extracellular cargo does not depend on dia. (A) To visualize endocytosis, fluorescently labelled wheat germ agglutinin
 Fig. S1. Endocytosis of extracellular cargo does not depend on dia. (A) To visualize endocytosis, fluorescently labelled wheat germ agglutinin (WGA-Alexa555) was injected into the extracellular perivitteline
Fig. S1. Endocytosis of extracellular cargo does not depend on dia. (A) To visualize endocytosis, fluorescently labelled wheat germ agglutinin (WGA-Alexa555) was injected into the extracellular perivitteline
Hossain_Supplemental Figure 1
 Hossain_Supplemental Figure 1 GFP-PACT GFP-PACT Motif I GFP-PACT Motif II A. MG132 (1µM) GFP Tubulin GFP-PACT Pericentrin GFP-PACT GFP-PACT Pericentrin Fig. S1. Expression and localization of Orc1 PACT
Hossain_Supplemental Figure 1 GFP-PACT GFP-PACT Motif I GFP-PACT Motif II A. MG132 (1µM) GFP Tubulin GFP-PACT Pericentrin GFP-PACT GFP-PACT Pericentrin Fig. S1. Expression and localization of Orc1 PACT
Microtubules. Forms a sheet of protofilaments that folds to a tube Both tubulins bind GTP
 Microtubules Polymerize from a-tubulin/ß-tubulin dimers Hollow tube of 13 protofilaments In vitro- filament number varies In vivo- always 13 protofilaments Forms a sheet of protofilaments that folds to
Microtubules Polymerize from a-tubulin/ß-tubulin dimers Hollow tube of 13 protofilaments In vitro- filament number varies In vivo- always 13 protofilaments Forms a sheet of protofilaments that folds to
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
GM130 Is Required for Compartmental Organization of Dendritic Golgi Outposts
 Current Biology, Volume 24 Supplemental Information GM130 Is Required for Compartmental Organization of Dendritic Golgi Outposts Wei Zhou, Jin Chang, Xin Wang, Masha G. Savelieff, Yinyin Zhao, Shanshan
Current Biology, Volume 24 Supplemental Information GM130 Is Required for Compartmental Organization of Dendritic Golgi Outposts Wei Zhou, Jin Chang, Xin Wang, Masha G. Savelieff, Yinyin Zhao, Shanshan
SUPPLEMENTARY INFORMATION FIGURE 1 - 1
 SUPPLEMENTARY INFORMATION FIGURE 1-1 SUPPLEMENTARY INFORMATION FIGURE 2-2 SUPPLEMENTARY INFORMATION METHODS GST-Pull-Down. Cultures of E. Coli (BL21) were transformed with pgex (Clontech) and pgex recombinant
SUPPLEMENTARY INFORMATION FIGURE 1-1 SUPPLEMENTARY INFORMATION FIGURE 2-2 SUPPLEMENTARY INFORMATION METHODS GST-Pull-Down. Cultures of E. Coli (BL21) were transformed with pgex (Clontech) and pgex recombinant
GFP CCD2 GFP IP:GFP
 D1 D2 1 75 95 148 178 492 GFP CCD1 CCD2 CCD2 GFP D1 D2 GFP D1 D2 Beclin 1 IB:GFP IP:GFP Supplementary Figure 1: Mapping domains required for binding to HEK293T cells are transfected with EGFP-tagged mutant
D1 D2 1 75 95 148 178 492 GFP CCD1 CCD2 CCD2 GFP D1 D2 GFP D1 D2 Beclin 1 IB:GFP IP:GFP Supplementary Figure 1: Mapping domains required for binding to HEK293T cells are transfected with EGFP-tagged mutant
Golgi are located near the MTOC while the ER is spread throughout the cytoplasm, so which of the following is probably true?
 Golgi are located near the MTOC while the ER is spread throughout the cytoplasm, so which of the following is probably true? A. COPII and COPI vesicles are transported with dynein. B. COPII and COPI vesicles
Golgi are located near the MTOC while the ER is spread throughout the cytoplasm, so which of the following is probably true? A. COPII and COPI vesicles are transported with dynein. B. COPII and COPI vesicles
Polyclonal ARHGAP25 antibody was prepared from rabbit serum after intracutaneous
 Preparation and purification of polyclonal antibodies Polyclonal ARHGAP25 antibody was prepared from rabbit serum after intracutaneous injections of glutathione S-transferase-ARHGAP25-(509-619) (GST-coiled
Preparation and purification of polyclonal antibodies Polyclonal ARHGAP25 antibody was prepared from rabbit serum after intracutaneous injections of glutathione S-transferase-ARHGAP25-(509-619) (GST-coiled
Nature Neuroscience: doi: /nn Supplementary Figure 1
 Supplementary Figure 1 PCR-genotyping of the three mouse models used in this study and controls for behavioral experiments after semi-chronic Pten inhibition. a-c. DNA from App/Psen1 (a), Pten tg (b) and
Supplementary Figure 1 PCR-genotyping of the three mouse models used in this study and controls for behavioral experiments after semi-chronic Pten inhibition. a-c. DNA from App/Psen1 (a), Pten tg (b) and
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling
 Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
ApoTrack Cytochrome c Apoptosis ICC Antibody
 ab110417 ApoTrack Cytochrome c Apoptosis ICC Antibody Instructions for Use For the Immunocytochemistry analysis of cytochrome c and a mitochondrial marker (Complex Vα) in apoptotic cells and nonapoptotic
ab110417 ApoTrack Cytochrome c Apoptosis ICC Antibody Instructions for Use For the Immunocytochemistry analysis of cytochrome c and a mitochondrial marker (Complex Vα) in apoptotic cells and nonapoptotic
Molecular Cell Biology - Problem Drill 01: Introduction to Molecular Cell Biology
 Molecular Cell Biology - Problem Drill 01: Introduction to Molecular Cell Biology Question No. 1 of 10 1. Which statement describes how an organism is organized from most simple to most complex? Question
Molecular Cell Biology - Problem Drill 01: Introduction to Molecular Cell Biology Question No. 1 of 10 1. Which statement describes how an organism is organized from most simple to most complex? Question
Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna
 Supplemental Materials and Methods Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna (Mission shrna, Sigma) against mouse MMP14 were transfected into HEK293 cells using FuGene6
Supplemental Materials and Methods Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna (Mission shrna, Sigma) against mouse MMP14 were transfected into HEK293 cells using FuGene6
Lab module 6b Receptor-mediated endocytosis
 Goal for the module Lab module 6b Receptor-mediated endocytosis To follow the movement of a degraded ligand, LDL, and a recycled ligand, transferrin, as they undergo endocytic processing. Pre-lab homework
Goal for the module Lab module 6b Receptor-mediated endocytosis To follow the movement of a degraded ligand, LDL, and a recycled ligand, transferrin, as they undergo endocytic processing. Pre-lab homework
NIH Public Access Author Manuscript Nature. Author manuscript; available in PMC 2010 October 1.
 NIH Public Access Author Manuscript Published in final edited form as: Nature. 2010 April 15; 464(7291): 1048 1051. doi:10.1038/nature08895. Functional genomic screen for modulators of ciliogenesis and
NIH Public Access Author Manuscript Published in final edited form as: Nature. 2010 April 15; 464(7291): 1048 1051. doi:10.1038/nature08895. Functional genomic screen for modulators of ciliogenesis and
Parthanatos mediates AIMP2-activated age-dependent dopaminergic neuronal loss
 SUPPLEMENTARY INFORMATION Parthanatos mediates AIMP2-activated age-dependent dopaminergic neuronal loss Yunjong Lee, Senthilkumar S. Karuppagounder, Joo-Ho Shin, Yun-Il Lee, Han Seok Ko, Debbie Swing,
SUPPLEMENTARY INFORMATION Parthanatos mediates AIMP2-activated age-dependent dopaminergic neuronal loss Yunjong Lee, Senthilkumar S. Karuppagounder, Joo-Ho Shin, Yun-Il Lee, Han Seok Ko, Debbie Swing,
MICB688L/MOCB639 Advanced Cell Biology Exam II
 MICB688L/MOCB639 Advanced Cell Biology Exam II May 10, 2001 Name: 1. Briefly describe the four major classes of cell surface receptors and their modes of action (immediate downstream only) (8) 2. Please
MICB688L/MOCB639 Advanced Cell Biology Exam II May 10, 2001 Name: 1. Briefly describe the four major classes of cell surface receptors and their modes of action (immediate downstream only) (8) 2. Please
7.06 Problem Set #3, Spring 2005
 7.06 Problem Set #3, Spring 2005 1. The Drosophila compound eye is composed of about 800 units called ommatidia. Each ommatidium contains eight photoreceptor neurons (R1 through R8), which develop in a
7.06 Problem Set #3, Spring 2005 1. The Drosophila compound eye is composed of about 800 units called ommatidia. Each ommatidium contains eight photoreceptor neurons (R1 through R8), which develop in a
BME Engineering Molecular Cell Biology. The Cytoskeleton (I): Actin The Cytoskeleton (II): Microtubule & Intermediate Filament
 BME 42-620 Engineering Molecular Cell Biology Lecture 09: The Cytoskeleton (I): Actin The Cytoskeleton (II): Microtubule & Intermediate Filament BME42-620 Lecture 09, September 27, 2011 1 Outline Overviewofcytoskeletal
BME 42-620 Engineering Molecular Cell Biology Lecture 09: The Cytoskeleton (I): Actin The Cytoskeleton (II): Microtubule & Intermediate Filament BME42-620 Lecture 09, September 27, 2011 1 Outline Overviewofcytoskeletal
Convoy TM Transfection Reagent
 Convoy TM Transfection Reagent Catalog No.11103 0.25ml (40-80 transfections in 35mm dishes) Catalog No.11105 0.5 ml (80-165 transfections in 35mm dishes) Catalog No.11110 1.0 ml (165-330 transfections
Convoy TM Transfection Reagent Catalog No.11103 0.25ml (40-80 transfections in 35mm dishes) Catalog No.11105 0.5 ml (80-165 transfections in 35mm dishes) Catalog No.11110 1.0 ml (165-330 transfections
Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product.
 Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
7.22 Example Problems for Exam 1 The exam will be of this format. It will consist of 2-3 sets scenarios.
 Massachusetts Institute of Technology Department of Biology 7.22, Fall 2005 - Developmental Biology Instructors: Professor Hazel Sive, Professor Martha Constantine-Paton 1 of 10 7.22 Fall 2005 sample exam
Massachusetts Institute of Technology Department of Biology 7.22, Fall 2005 - Developmental Biology Instructors: Professor Hazel Sive, Professor Martha Constantine-Paton 1 of 10 7.22 Fall 2005 sample exam
Supplemental Data Supplementary Figure Legends and Scheme Figure S1.
 Supplemental Data Supplementary Figure Legends and Scheme Figure S1. UTK1 inhibits the second EGF-induced wave of lamellipodia formation in TT cells. A and B, EGF-induced lamellipodia formation in TT cells,
Supplemental Data Supplementary Figure Legends and Scheme Figure S1. UTK1 inhibits the second EGF-induced wave of lamellipodia formation in TT cells. A and B, EGF-induced lamellipodia formation in TT cells,
Enhancers mutations that make the original mutant phenotype more extreme. Suppressors mutations that make the original mutant phenotype less extreme
 Interactomics and Proteomics 1. Interactomics The field of interactomics is concerned with interactions between genes or proteins. They can be genetic interactions, in which two genes are involved in the
Interactomics and Proteomics 1. Interactomics The field of interactomics is concerned with interactions between genes or proteins. They can be genetic interactions, in which two genes are involved in the
Thermo Scientific Dharmacon SMARTvector 2.0 Lentiviral shrna Particles
 Thermo Scientific Dharmacon SMARTvector 2.0 Lentiviral shrna Particles Long-term gene silencing shrna-specific design algorithm High titer, purified particles Thermo Scientific Dharmacon SMARTvector shrna
Thermo Scientific Dharmacon SMARTvector 2.0 Lentiviral shrna Particles Long-term gene silencing shrna-specific design algorithm High titer, purified particles Thermo Scientific Dharmacon SMARTvector shrna
General Education Learning Outcomes
 BOROUGH OF MANHATTAN COMMUNITY COLLEGE City University of New York Department of Science Title of Course: Cell Biology Class hours 3 BIO Section: 260 Lab hours 3 Semester Spring 2018 Credits 4 Schedule:
BOROUGH OF MANHATTAN COMMUNITY COLLEGE City University of New York Department of Science Title of Course: Cell Biology Class hours 3 BIO Section: 260 Lab hours 3 Semester Spring 2018 Credits 4 Schedule:
Asymmetric endocytosis and remodeling of β1-integrin adhesions during growth cone chemorepulsion by MAG
 Asymmetric endocytosis and remodeling of β1-integrin adhesions during growth cone chemorepulsion by MAG Jacob H. Hines, Mohammad Abu-Rub and John R. Henley Supplementary Figure 1. Validation of FM 5-95
Asymmetric endocytosis and remodeling of β1-integrin adhesions during growth cone chemorepulsion by MAG Jacob H. Hines, Mohammad Abu-Rub and John R. Henley Supplementary Figure 1. Validation of FM 5-95
Supplementary Figure 1: Expression of RNF8, HERC2 and NEURL4 in the cerebellum and knockdown of RNF8 by RNAi (a) Lysates of the cerebellum from rat
 Supplementary Figure 1: Expression of RNF8, HERC2 and NEURL4 in the cerebellum and knockdown of RNF8 by RNAi (a) Lysates of the cerebellum from rat pups at P6, P14, P22, P30 and adult (A) rats were subjected
Supplementary Figure 1: Expression of RNF8, HERC2 and NEURL4 in the cerebellum and knockdown of RNF8 by RNAi (a) Lysates of the cerebellum from rat pups at P6, P14, P22, P30 and adult (A) rats were subjected
The Two-Hybrid System
 Encyclopedic Reference of Genomics and Proteomics in Molecular Medicine The Two-Hybrid System Carolina Vollert & Peter Uetz Institut für Genetik Forschungszentrum Karlsruhe PO Box 3640 D-76021 Karlsruhe
Encyclopedic Reference of Genomics and Proteomics in Molecular Medicine The Two-Hybrid System Carolina Vollert & Peter Uetz Institut für Genetik Forschungszentrum Karlsruhe PO Box 3640 D-76021 Karlsruhe
Journal of Cell Science Supplementary Material
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 SUPPLEMENTARY FIGURE LEGENDS Figure S1: Eps8 is localized at focal adhesions and binds directly to FAK (A) Focal
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 SUPPLEMENTARY FIGURE LEGENDS Figure S1: Eps8 is localized at focal adhesions and binds directly to FAK (A) Focal
The Use of Genetically-Modified Mouse Models to Study the Actin Cytoskeleton
 The Use of Genetically-Modified Mouse Models to Study the Actin Cytoskeleton Anthony Kee (PhD) Cellular and Genetic Medicine Unit School of Medical Sciences (a.kee@unsw.edu.au) 2017 Structure of the Prac
The Use of Genetically-Modified Mouse Models to Study the Actin Cytoskeleton Anthony Kee (PhD) Cellular and Genetic Medicine Unit School of Medical Sciences (a.kee@unsw.edu.au) 2017 Structure of the Prac
Supplemental Information. Glutamylation Regulates Transport, Specializes. Function, and Sculpts the Structure of Cilia
 Current Biology, Volume 27 Supplemental Information Glutamylation Regulates Transport, Specializes Function, and Sculpts the Structure of Cilia Robert O'Hagan, Malan Silva, Ken C.Q. Nguyen, Winnie Zhang,
Current Biology, Volume 27 Supplemental Information Glutamylation Regulates Transport, Specializes Function, and Sculpts the Structure of Cilia Robert O'Hagan, Malan Silva, Ken C.Q. Nguyen, Winnie Zhang,
Regulation of Synaptic Structure and Function by FMRP- Associated MicroRNAs mir-125b and mir-132
 Neuron, Volume 65 Regulation of Synaptic Structure and Function by FMRP- Associated MicroRNAs mir-125b and mir-132 Dieter Edbauer, Joel R. Neilson, Kelly A. Foster, Chi-Fong Wang, Daniel P. Seeburg, Matthew
Neuron, Volume 65 Regulation of Synaptic Structure and Function by FMRP- Associated MicroRNAs mir-125b and mir-132 Dieter Edbauer, Joel R. Neilson, Kelly A. Foster, Chi-Fong Wang, Daniel P. Seeburg, Matthew
This Document Contains:
 This Document Contains: 1. In-Cell Western Protocol II. Cell Seeding and Stimulation Supplemental Protocol III. Complete Assay Example: Detailing the Seeding, Stimulation and Detection of the A431 Cellular
This Document Contains: 1. In-Cell Western Protocol II. Cell Seeding and Stimulation Supplemental Protocol III. Complete Assay Example: Detailing the Seeding, Stimulation and Detection of the A431 Cellular
β-arrestin mediated localization of Smoothened to the primary cilium
 Published as: Science. 2008 June 27; 320(5884): 1777 1781. β-arrestin mediated localization of Smoothened to the primary cilium Jeffrey J. Kovacs 1,2, Erin J. Whalen 1, Renshui Liu 1, Kunhong Xiao 3, Jihee
Published as: Science. 2008 June 27; 320(5884): 1777 1781. β-arrestin mediated localization of Smoothened to the primary cilium Jeffrey J. Kovacs 1,2, Erin J. Whalen 1, Renshui Liu 1, Kunhong Xiao 3, Jihee
ASPP1 Fw GGTTGGGAATCCACGTGTTG ASPP1 Rv GCCATATCTTGGAGCTCTGAGAG
 Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
CytoPainter Golgi Staining Kit Green Fluorescence
 ab139483 CytoPainter Golgi Staining Kit Green Fluorescence Instructions for Use Designed for the detection of Golgi bodies by microscopy This product is for research use only and is not intended for diagnostic
ab139483 CytoPainter Golgi Staining Kit Green Fluorescence Instructions for Use Designed for the detection of Golgi bodies by microscopy This product is for research use only and is not intended for diagnostic
affects the development of newborn neurons Brain Mind Institute and School of Life Sciences, Ecole Polytechnique Fédérale de
 Shedding of neurexin 3β ectodomain by ADAM10 releases a soluble fragment that affects the development of newborn neurons Erika Borcel* a, Magda Palczynska* a, Marine Krzisch b, Mitko Dimitrov a, Giorgio
Shedding of neurexin 3β ectodomain by ADAM10 releases a soluble fragment that affects the development of newborn neurons Erika Borcel* a, Magda Palczynska* a, Marine Krzisch b, Mitko Dimitrov a, Giorgio
Transfection of CRISPR/Cas9 Nuclease NLS ribonucleoprotein (RNP) into adherent mammalian cells using Lipofectamine RNAiMAX
 Transfection of CRISPR/Cas9 Nuclease NLS ribonucleoprotein (RNP) into adherent mammalian cells using Lipofectamine RNAiMAX INTRODUCTION The CRISPR/Cas genome editing system consists of a single guide RNA
Transfection of CRISPR/Cas9 Nuclease NLS ribonucleoprotein (RNP) into adherent mammalian cells using Lipofectamine RNAiMAX INTRODUCTION The CRISPR/Cas genome editing system consists of a single guide RNA
Online Supplementary Information
 Online Supplementary Information NLRP4 negatively regulates type I interferon signaling by targeting TBK1 for degradation via E3 ubiquitin ligase DTX4 Jun Cui 1,4,6,7, Yinyin Li 1,5,6,7, Liang Zhu 1, Dan
Online Supplementary Information NLRP4 negatively regulates type I interferon signaling by targeting TBK1 for degradation via E3 ubiquitin ligase DTX4 Jun Cui 1,4,6,7, Yinyin Li 1,5,6,7, Liang Zhu 1, Dan
SUPPLEMENTARY INFORMATION
 doi:.38/nature899 Supplementary Figure Suzuki et al. a c p7 -/- / WT ratio (+)/(-) p7 -/- / WT ratio Log X 3. Fold change by treatment ( (+)/(-)) Log X.5 3-3. -. b Fold change by treatment ( (+)/(-)) 8
doi:.38/nature899 Supplementary Figure Suzuki et al. a c p7 -/- / WT ratio (+)/(-) p7 -/- / WT ratio Log X 3. Fold change by treatment ( (+)/(-)) Log X.5 3-3. -. b Fold change by treatment ( (+)/(-)) 8
Supporting Information
 Supporting Information Su et al. 10.1073/pnas.1211604110 SI Materials and Methods Cell Culture and Plasmids. Tera-1 and Tera-2 cells (ATCC: HTB- 105/106) were maintained in McCoy s 5A medium with 15% FBS
Supporting Information Su et al. 10.1073/pnas.1211604110 SI Materials and Methods Cell Culture and Plasmids. Tera-1 and Tera-2 cells (ATCC: HTB- 105/106) were maintained in McCoy s 5A medium with 15% FBS
Description: Nuclear morphology and dynamics in nontargeting sirna transfected cells. HeLa Kyoto
 Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Tables Title of file for HTML: Supplementary Movie 1 Description: Nuclear morphology and dynamics
Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Tables Title of file for HTML: Supplementary Movie 1 Description: Nuclear morphology and dynamics
Supplemental Online Material. The mouse embryonic fibroblast cell line #10 derived from β-arrestin1 -/- -β-arrestin2 -/-
 #1074683s 1 Supplemental Online Material Materials and Methods Cell lines and tissue culture The mouse embryonic fibroblast cell line #10 derived from β-arrestin1 -/- -β-arrestin2 -/- knock-out animals
#1074683s 1 Supplemental Online Material Materials and Methods Cell lines and tissue culture The mouse embryonic fibroblast cell line #10 derived from β-arrestin1 -/- -β-arrestin2 -/- knock-out animals
Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question in Section B and ONE question from Section C.
 UNIVERSITY OF EAST ANGLIA School of Biological Sciences Main Series UG Examination 2013-2014 CELL BIOLOGY BIO-2B06 Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question in
UNIVERSITY OF EAST ANGLIA School of Biological Sciences Main Series UG Examination 2013-2014 CELL BIOLOGY BIO-2B06 Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question in
RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by. 5 AGACACAAACACCAUUGUCACACUCCACAGC; Rand-2 OMe,
 Materials and methods Oligonucleotides and DNA constructs RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by Dharmacon Inc. (Lafayette, CO). The sequences were: 122-2 OMe, 5
Materials and methods Oligonucleotides and DNA constructs RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by Dharmacon Inc. (Lafayette, CO). The sequences were: 122-2 OMe, 5
SANTA CRUZ BIOTECHNOLOGY, INC.
 TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
The Conserved Isoleucine Valine Phenylalanine Motif Couples Activation State and Endocytic Functions of b-arrestins
 Traffic 2007 Blackwell Munksgaard # 2007 The Authors doi: 10.1111/j.1600-0854.2007.00578.x The Conserved Isoleucine Valine Phenylalanine Motif Couples Activation State and Endocytic Functions of b-arrestins
Traffic 2007 Blackwell Munksgaard # 2007 The Authors doi: 10.1111/j.1600-0854.2007.00578.x The Conserved Isoleucine Valine Phenylalanine Motif Couples Activation State and Endocytic Functions of b-arrestins
Epigenetics, Environment and Human Health
 Epigenetics, Environment and Human Health A. Karim Ahmed National Council for Science and the Environment Washington, DC May, 2015 Epigenetics A New Biological Paradigm A Question about Cells: All cells
Epigenetics, Environment and Human Health A. Karim Ahmed National Council for Science and the Environment Washington, DC May, 2015 Epigenetics A New Biological Paradigm A Question about Cells: All cells
Three major types of cytoskeleton
 The Cytoskeleton Organizes and stabilizes cells Pulls chromosomes apart Drives intracellular traffic Supports plasma membrane and nuclear envelope Enables cellular movement Guides growth of the plant cell
The Cytoskeleton Organizes and stabilizes cells Pulls chromosomes apart Drives intracellular traffic Supports plasma membrane and nuclear envelope Enables cellular movement Guides growth of the plant cell
Supplementary Materials for. Combinatory screening of DNA aptamers for molecular imaging of HER2 in cancer
 Supplementary Materials for Combinatory screening of DNA aptamers for molecular imaging of HER2 in cancer Guizhi Zhu, Huimin Zhang,, Orit Jacobson, Zhantong Wang, Haojun Chen,, Xiangyu Yang, ǁ, Gang Niu,
Supplementary Materials for Combinatory screening of DNA aptamers for molecular imaging of HER2 in cancer Guizhi Zhu, Huimin Zhang,, Orit Jacobson, Zhantong Wang, Haojun Chen,, Xiangyu Yang, ǁ, Gang Niu,
FRAUNHOFER IME SCREENINGPORT
 FRAUNHOFER IME SCREENINGPORT Detection technologies used in drug discovery Introduction Detection technologies in drug discovery is unlimited only a biased snapshot can be presented Differences can presented
FRAUNHOFER IME SCREENINGPORT Detection technologies used in drug discovery Introduction Detection technologies in drug discovery is unlimited only a biased snapshot can be presented Differences can presented
QUESTIONS 16 THROUGH 30 FROM EXAM 3 OF FALL, 2010
 BISC403 Genetic and Evolutionary Biology Spring, 2011 April 19, 2011 Summary of requirements for Exam 3 (to be given on April 26 plus third exam from fall, 2010) The primary responsibility is for any topic
BISC403 Genetic and Evolutionary Biology Spring, 2011 April 19, 2011 Summary of requirements for Exam 3 (to be given on April 26 plus third exam from fall, 2010) The primary responsibility is for any topic
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than. SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with
 Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
Mapping the NPHP-JBTS-MKS Protein Network Reveals Ciliopathy Disease Genes and Pathways
 Mapping the NPHP-JBTS-MKS Protein Network Reveals Ciliopathy Disease Genes and Pathways Liyun Sang, 1,16 Julie J. Miller, 2,16 Kevin C. Corbit, 3 Rachel H. Giles, 4 Matthew J. Brauer, 1 Edgar A. Otto,
Mapping the NPHP-JBTS-MKS Protein Network Reveals Ciliopathy Disease Genes and Pathways Liyun Sang, 1,16 Julie J. Miller, 2,16 Kevin C. Corbit, 3 Rachel H. Giles, 4 Matthew J. Brauer, 1 Edgar A. Otto,
Additional Practice Problems for Reading Period
 BS 50 Genetics and Genomics Reading Period Additional Practice Problems for Reading Period Question 1. In patients with a particular type of leukemia, their leukemic B lymphocytes display a translocation
BS 50 Genetics and Genomics Reading Period Additional Practice Problems for Reading Period Question 1. In patients with a particular type of leukemia, their leukemic B lymphocytes display a translocation
Endocytosis of Hedgehog through Dispatched Regulates Long-Range Signaling
 evelopmental Cell Supplemental Information Endocytosis of Hedgehog through ispatched Regulates Long-Range Signaling Gisela Angelo, Tamás Matusek, S. Pizette, and Pascal P. Thérond Ratio Hh-Np/Hh-FL SUPPLEMENTAL
evelopmental Cell Supplemental Information Endocytosis of Hedgehog through ispatched Regulates Long-Range Signaling Gisela Angelo, Tamás Matusek, S. Pizette, and Pascal P. Thérond Ratio Hh-Np/Hh-FL SUPPLEMENTAL
MIT Department of Biology 7.013: Introductory Biology - Spring 2005 Instructors: Professor Hazel Sive, Professor Tyler Jacks, Dr.
 MIT Department of Biology 7.01: Introductory Biology - Spring 2005 Instructors: Professor Hazel Sive, Professor Tyler Jacks, Dr. Claudette Gardel iv) Would Xba I be useful for cloning? Why or why not?
MIT Department of Biology 7.01: Introductory Biology - Spring 2005 Instructors: Professor Hazel Sive, Professor Tyler Jacks, Dr. Claudette Gardel iv) Would Xba I be useful for cloning? Why or why not?
SUPPLEMENTARY INFORMATION. Small molecule activation of the TRAIL receptor DR5 in human cancer cells
 SUPPLEMENTARY INFORMATION Small molecule activation of the TRAIL receptor DR5 in human cancer cells Gelin Wang 1*, Xiaoming Wang 2, Hong Yu 1, Shuguang Wei 1, Noelle Williams 1, Daniel L. Holmes 1, Randal
SUPPLEMENTARY INFORMATION Small molecule activation of the TRAIL receptor DR5 in human cancer cells Gelin Wang 1*, Xiaoming Wang 2, Hong Yu 1, Shuguang Wei 1, Noelle Williams 1, Daniel L. Holmes 1, Randal
Using Sapphire700 Stain and DRAQ5 Stain for Cell Number Normalization
 Using Sapphire700 Stain and DRAQ5 Stain for Cell Number Normalization Developed for: Aerius, Odyssey Classic, Odyssey CLx, and Odyssey Sa Infrared Imaging Systems Please refer to your manual to confirm
Using Sapphire700 Stain and DRAQ5 Stain for Cell Number Normalization Developed for: Aerius, Odyssey Classic, Odyssey CLx, and Odyssey Sa Infrared Imaging Systems Please refer to your manual to confirm
3. Results. 3.1 Generation of HEK293 cell clones stably expressing ETA and ETB receptors
 3. Results 3.1 Generation of HEK293 cell clones stably expressing ETA and ETB receptors To investigate the dimerisation of the endothelin receptor subtypes HEK293 cells were stably transfected with plasmids
3. Results 3.1 Generation of HEK293 cell clones stably expressing ETA and ETB receptors To investigate the dimerisation of the endothelin receptor subtypes HEK293 cells were stably transfected with plasmids
The MAP Kinase Family
 The MAP Kinase Family Extracellular stimuli Classical MAP kinases Atypical MAP kinases MAPKKK MLK1/2/3/7; LZK RAF-1/A/B TAK1; TPL2 c-mos MEKK1-4; DLK ASK1/2; MLTK TAO1/2 ASK1 TAK1 MEKK1-4 MEKK2/3 TPL2???
The MAP Kinase Family Extracellular stimuli Classical MAP kinases Atypical MAP kinases MAPKKK MLK1/2/3/7; LZK RAF-1/A/B TAK1; TPL2 c-mos MEKK1-4; DLK ASK1/2; MLTK TAO1/2 ASK1 TAK1 MEKK1-4 MEKK2/3 TPL2???
12 Interaction of Genes
 12 Interaction of Genes Yeast genetics has been particularly amenable for identifying and characterizing gene products that directly or indirectly interact with each other, especially when two mutations
12 Interaction of Genes Yeast genetics has been particularly amenable for identifying and characterizing gene products that directly or indirectly interact with each other, especially when two mutations
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/4/157/ra4/dc1 Supplementary Materials for Genome-Wide RNAi Screen Reveals Disease-Associated Genes That Are Common to Hedgehog and Wnt Signaling Leni S. Jacob,
www.sciencesignaling.org/cgi/content/full/4/157/ra4/dc1 Supplementary Materials for Genome-Wide RNAi Screen Reveals Disease-Associated Genes That Are Common to Hedgehog and Wnt Signaling Leni S. Jacob,
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1: Vector maps of TRMPV and TRMPVIR variants. Many derivatives of TRMPV have been generated and tested. Unless otherwise noted, experiments in this paper use
Supplementary Figure 1 Supplementary Figure 1: Vector maps of TRMPV and TRMPVIR variants. Many derivatives of TRMPV have been generated and tested. Unless otherwise noted, experiments in this paper use
Problem Set 4
 7.016- Problem Set 4 Question 1 Arginine biosynthesis is an example of multi-step biochemical pathway where each step is catalyzed by a specific enzyme (E1, E2 and E3) as is outlined below. E1 E2 E3 A
7.016- Problem Set 4 Question 1 Arginine biosynthesis is an example of multi-step biochemical pathway where each step is catalyzed by a specific enzyme (E1, E2 and E3) as is outlined below. E1 E2 E3 A
T H E J O U R N A L O F C E L L B I O L O G Y
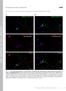 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Kanaani et al., http://www.jcb.org/cgi/content/full/jcb.200912101/dc1 Figure S1. The K2 rabbit polyclonal antibody is specific for GAD67,
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Kanaani et al., http://www.jcb.org/cgi/content/full/jcb.200912101/dc1 Figure S1. The K2 rabbit polyclonal antibody is specific for GAD67,
Supplementary Figure 1. RAD51 and RAD51 paralogs are enriched spontaneously onto
 Supplementary Figure legends Supplementary Figure 1. and paralogs are enriched spontaneously onto the S-phase chromatin during DN replication. () Chromatin fractionation was carried out as described in
Supplementary Figure legends Supplementary Figure 1. and paralogs are enriched spontaneously onto the S-phase chromatin during DN replication. () Chromatin fractionation was carried out as described in
Coleman et al., Supplementary Figure 1
 Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Zool 3200: Cell Biology Exam 2 2/20/15
 Name: TRASK Zool 3200: Cell Biology Exam 2 2/20/15 Answer each of the following short and longer answer questions in the space provided; circle the BEST answer or answers for each multiple choice question
Name: TRASK Zool 3200: Cell Biology Exam 2 2/20/15 Answer each of the following short and longer answer questions in the space provided; circle the BEST answer or answers for each multiple choice question
Section 10. Junaid Malek, M.D.
 Section 10 Junaid Malek, M.D. Cell Division Make sure you understand: How do cells know when to divide? (What drives the cell cycle? Why is it important to regulate this?) How is DNA replication regulated?
Section 10 Junaid Malek, M.D. Cell Division Make sure you understand: How do cells know when to divide? (What drives the cell cycle? Why is it important to regulate this?) How is DNA replication regulated?
X2-C/X1-Y X2-C/VCAM-Y. FRET efficiency. Ratio YFP/CFP
 FRET efficiency.7.6..4.3.2 X2-C/X1-Y X2-C/VCAM-Y.1 1 2 3 Ratio YFP/CFP Supplemental Data 1. Analysis of / heterodimers in live cells using FRET. FRET saturation curves were obtained using cells transiently
FRET efficiency.7.6..4.3.2 X2-C/X1-Y X2-C/VCAM-Y.1 1 2 3 Ratio YFP/CFP Supplemental Data 1. Analysis of / heterodimers in live cells using FRET. FRET saturation curves were obtained using cells transiently
To isolate single GNS 144 cell clones, cells were plated at a density of 1cell/well
 Supplemental Information: Supplemental Methods: Cell culture To isolate single GNS 144 cell clones, cells were plated at a density of 1cell/well in 96 well Primaria plates in GNS media and incubated at
Supplemental Information: Supplemental Methods: Cell culture To isolate single GNS 144 cell clones, cells were plated at a density of 1cell/well in 96 well Primaria plates in GNS media and incubated at
Supplementary Figures
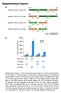 Supplementary Figures a HindIII XbaI pcdna3.1/v5-his A_Nfluc-VVD 1-398 37-186 Nfluc Linker 1 VVD HindIII XhoI pcdna3.1/v5-his A_VVD-Nfluc 37-186 1-398 VVD Linker 2 Nfluc HindIII XbaI pcdna3.1/v5-his A_Cfluc-VVD
Supplementary Figures a HindIII XbaI pcdna3.1/v5-his A_Nfluc-VVD 1-398 37-186 Nfluc Linker 1 VVD HindIII XhoI pcdna3.1/v5-his A_VVD-Nfluc 37-186 1-398 VVD Linker 2 Nfluc HindIII XbaI pcdna3.1/v5-his A_Cfluc-VVD
HeLa cells stably transfected with an empty pcdna3 vector (HeLa Neo) or with a plasmid
 SUPPLEMENTAL MATERIALS AND METHODS Cell culture, transfection and treatments. HeLa cells stably transfected with an empty pcdna3 vector (HeLa Neo) or with a plasmid encoding vmia (HeLa vmia) 1 were cultured
SUPPLEMENTAL MATERIALS AND METHODS Cell culture, transfection and treatments. HeLa cells stably transfected with an empty pcdna3 vector (HeLa Neo) or with a plasmid encoding vmia (HeLa vmia) 1 were cultured
Thermo Scientific GTPase Research Tools
 Thermo Scientific Research Tools Active Pull-Down Assays We offer two different tools to study biology, one for active monitoring and one for global profiling. The Thermo Scientific Pierce Active Pull-Down
Thermo Scientific Research Tools Active Pull-Down Assays We offer two different tools to study biology, one for active monitoring and one for global profiling. The Thermo Scientific Pierce Active Pull-Down
NTM486-04, NTM174-04,
 Transfection of transformed human trabecular meshwork TM5, and primary human NTM210-05, NTM486-04, NTM174-04, and NTM153-00 cells with Metafectene Easy Adnan Dibas1A,C, Ming Jiang1A,C, Thomas Yorio1A,C.
Transfection of transformed human trabecular meshwork TM5, and primary human NTM210-05, NTM486-04, NTM174-04, and NTM153-00 cells with Metafectene Easy Adnan Dibas1A,C, Ming Jiang1A,C, Thomas Yorio1A,C.
PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF
 YFP-PHF1 CFP-PHT1;2 PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF + CFP-PHT1;2 Negative control!-gfp Supplemental Figure 1: PHT1;2 accumulation is PHF1 dependent. Immunoblot analysis on total protein extract
YFP-PHF1 CFP-PHT1;2 PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF + CFP-PHT1;2 Negative control!-gfp Supplemental Figure 1: PHT1;2 accumulation is PHF1 dependent. Immunoblot analysis on total protein extract
ab G alpha i Activation Assay Kit
 ab173234 G alpha i Activation Assay Kit Instructions for Use For the simple and fast measurement of G alpha i activation. This product is for research use only and is not intended for diagnostic use. Version
ab173234 G alpha i Activation Assay Kit Instructions for Use For the simple and fast measurement of G alpha i activation. This product is for research use only and is not intended for diagnostic use. Version
M X 500 µl. M X 1000 µl
 GeneGlide TM sirna Transfection Reagent (Catalog # M1081-300, -500, -1000; Store at 4 C) I. Introduction: BioVision s GeneGlide TM sirna Transfection reagent is a cationic proprietary polymer/lipid formulation,
GeneGlide TM sirna Transfection Reagent (Catalog # M1081-300, -500, -1000; Store at 4 C) I. Introduction: BioVision s GeneGlide TM sirna Transfection reagent is a cationic proprietary polymer/lipid formulation,
cell and tissue imaging by fluorescence microscopy
 cell and tissue imaging by fluorescence microscopy Steven NEDELLEC Plateforme Micropicell SFR Santé François Bonamy Nantes 1 A matter of size Limit of resolution 0.15mm aims: building the image of an object
cell and tissue imaging by fluorescence microscopy Steven NEDELLEC Plateforme Micropicell SFR Santé François Bonamy Nantes 1 A matter of size Limit of resolution 0.15mm aims: building the image of an object
Xfect Protein Transfection Reagent
 Xfect Protein Transfection Reagent Mammalian Expression Systems Rapid, high-efficiency, low-toxicity protein transfection Transfect a large amount of active protein Virtually no cytotoxicity, unlike lipofection
Xfect Protein Transfection Reagent Mammalian Expression Systems Rapid, high-efficiency, low-toxicity protein transfection Transfect a large amount of active protein Virtually no cytotoxicity, unlike lipofection
pdsipher and pdsipher -GFP shrna Vector User s Guide
 pdsipher and pdsipher -GFP shrna Vector User s Guide NOTE: PLEASE READ THE ENTIRE PROTOCOL CAREFULLY BEFORE USE Page 1. Introduction... 1 2. Vector Overview... 1 3. Vector Maps 2 4. Materials Provided...
pdsipher and pdsipher -GFP shrna Vector User s Guide NOTE: PLEASE READ THE ENTIRE PROTOCOL CAREFULLY BEFORE USE Page 1. Introduction... 1 2. Vector Overview... 1 3. Vector Maps 2 4. Materials Provided...
Use of Phase Contrast Imaging to Track Morphological Cellular Changes due to Apoptotic Activity
 A p p l i c a t i o n N o t e Use of Phase Contrast Imaging to Track Morphological Cellular Changes due to Apoptotic Activity Brad Larson and Peter Banks, Applications Department, BioTek Instruments, Inc.,
A p p l i c a t i o n N o t e Use of Phase Contrast Imaging to Track Morphological Cellular Changes due to Apoptotic Activity Brad Larson and Peter Banks, Applications Department, BioTek Instruments, Inc.,
Rapid Learning Center Presents. Teach Yourself AP Biology in 24 Hours
 Rapid Learning Center Chemistry :: Biology :: Physics :: Math Rapid Learning Center Presents Teach Yourself AP Biology in 24 Hours 1/35 *AP is a registered trademark of the College Board, which does not
Rapid Learning Center Chemistry :: Biology :: Physics :: Math Rapid Learning Center Presents Teach Yourself AP Biology in 24 Hours 1/35 *AP is a registered trademark of the College Board, which does not
Product: Arrest-In TM Transfection Reagent for RNAi
 Product: Arrest-In TM Transfection Reagent for RNAi Catalog #: ATR1740, ATR1741, ATR1742, ATR1743 Product Description Arrest-In transfection reagent is a proprietary polymeric formulation, developed and
Product: Arrest-In TM Transfection Reagent for RNAi Catalog #: ATR1740, ATR1741, ATR1742, ATR1743 Product Description Arrest-In transfection reagent is a proprietary polymeric formulation, developed and
Introduction State the objectives of the work and provide adequate background, avoiding a detailed literature survey or a summary of the results.
 Format requirements for IJBCB articles For each article type, IJBCB has its own requirements regarding the article format. Please ensure you follow the exact guidelines (included below) for the specific
Format requirements for IJBCB articles For each article type, IJBCB has its own requirements regarding the article format. Please ensure you follow the exact guidelines (included below) for the specific
Revision Checklist for Science Signaling Research Manuscripts: Data Requirements and Style Guidelines
 Revision Checklist for Science Signaling Research Manuscripts: Data Requirements and Style Guidelines Further information can be found at: http://stke.sciencemag.org/sites/default/files/researcharticlerevmsinstructions_0.pdf.
Revision Checklist for Science Signaling Research Manuscripts: Data Requirements and Style Guidelines Further information can be found at: http://stke.sciencemag.org/sites/default/files/researcharticlerevmsinstructions_0.pdf.
E. Incorrect! The four different DNA nucleotides follow a strict base pairing arrangement:
 AP Biology - Problem Drill 10: Molecular and Human Genetics Question No. 1 of 10 Instructions: (1) Read the problem and answer choices carefully, (2) Work the problems on paper as 1. Which of the following
AP Biology - Problem Drill 10: Molecular and Human Genetics Question No. 1 of 10 Instructions: (1) Read the problem and answer choices carefully, (2) Work the problems on paper as 1. Which of the following
This is a closed book, closed note exam. No calculators, phones or any electronic device are allowed.
 MCB 104 MIDTERM #2 October 23, 2013 ***IMPORTANT REMINDERS*** Print your name and ID# on every page of the exam. You will lose 0.5 point/page if you forget to do this. Name KEY If you need more space than
MCB 104 MIDTERM #2 October 23, 2013 ***IMPORTANT REMINDERS*** Print your name and ID# on every page of the exam. You will lose 0.5 point/page if you forget to do this. Name KEY If you need more space than
