OCTOBER 22, 2010 VOLUME 285 NUMBER 43 JOURNAL OF BIOLOGICAL CHEMISTRY 33281
|
|
|
- Priscilla Taylor
- 6 years ago
- Views:
Transcription
1 THE JOURNAL OF BIOLOGICAL CHEMISTRY VOL. 285, NO. 43, pp , October 22, by The American Society for Biochemistry and Molecular Biology, Inc. Printed in the U.S.A. Requirement for Lamin B Receptor and Its Regulation by Importin and Phosphorylation in Nuclear Envelope Assembly during Mitotic Exit * Received for publication, January 8, 2010, and in revised form, June 24, 2010 Published, JBC Papers in Press, June 24, 2010, DOI /jbc.M Xuelong Lu, Yang Shi, Quanlong Lu, Yan Ma, Jia Luo, Qingsong Wang, Jianguo Ji, Qing Jiang, and Chuanmao Zhang 1 From the The Key Laboratory of Cell Proliferation and Differentiation of the Ministry of Education and the State Key Laboratory of Bio-membrane and Membrane Bio-engineering, and the State Laboratory of Protein Engineering and Plant Genetic Engineering, College of Life Sciences, Peking University, Beijing , China Lamin B receptor (LBR), a chromatin and lamin B-binding protein in the inner nuclear membrane, has been proposed to target the membrane precursor vesicles to chromatin mediated by importin during the nuclear envelope (NE) assembly. However, the mechanisms for the binding of LBR with importin and the membrane targeting by LBR in NE assembly remain largely unknown. In this report, we show that the amino acids (aa) of LBR sequences are required to bind with importin at aa , and the binding is essential for the NE membrane precursor vesicle targeting to the chromatin during the NE assembly at the end of mitosis. We also show that this binding is cell cycle-regulated and dependent on the phosphorylation of LBR Ser-71 by p34 cdc2 kinase. RNAi knockdown of LBR causes the NE assembly failure and abnormal chromatin decondensation of the daughter cell nuclei, leading to the daughter cell death at early G 1 phase by apoptosis. Perturbation of the interaction of LBR with importin by deleting the LBR N-terminal spanning region or aa also induces the NE assembly failure, the abnormal chromatin decondensation, and the daughter cell death. The first transmembrane domain of LBR promotes the NE production and expansion, because overexpressing this domain is sufficient to induce membrane overproduction of the NE. Thus, these results demonstrate that LBR targets the membrane precursor vesicles to chromatin by interacting with importin in a LBR phosphorylation-dependent manner during the NE assembly at the end of mitosis and that the first transmembrane domain of LBR promotes the LBR-bearing membrane production and the NE expansion in interphase. * This work was supported by the National Natural Science Foundation of China (Grants , , , , , and ) and by the State Key Basic Research and Development Plan (Grants 2010CB833705, 2007CB914502, 2006CB910101, and 2010CB912203). 1 To whom correspondence should be addressed: Tel.: ; Fax: ; zhangcm@pku.edu.cn. 2 The abbreviations used are: NE, nuclear envelope; LBR, lamin B receptor; aa, amino acid(s). Nucleus, the largest organelle that contains the genome of the eukaryotic organisms, is surrounded by a continuous nuclear envelope (NE) 2 composed of a pair of inner and outer nuclear membranes, studded by numerous nuclear pore complexes. Through the nuclear pore complexes, the NE controls the flow of molecules between the nucleus and the cytoplasm in a tightly regulated manner (1 5). The NE is highly dynamic during the cell cycle. It disassembles into membranous vesicles or tubules and disperses into the cytoplasm at the onset of prometaphase and reassembles around the newly replicated chromosomes at the end of mitosis using their previously disassembled components (2, 4, 6). Recently, it was shown that Ran, a small Ras-like nuclear GTPase, and its binding proteins regulate the NE assembly process (7 13). Ran exists in GTP- and GDP-bound states that interact differently with effectors. Conversion between these two states and the assembly or disassembly of the effector complexes requires the interaction of regulator proteins, the nucleus-based guanine nucleotide exchange factor RCC1 and the cytoplasm-localized GTPase-activating protein RanGAP1 (6, 14). The compartmentalization of the regulators produces a high concentration gradient of Ran-GTP in the nucleus and Ran-GDP in the cytoplasm. In mitosis, after the NE breakdown, the nucleoplasm and the cytoplasm are mixed; Ran diffuses into the cytoplasm with many other soluble nuclear proteins; RCC1 dynamically binds with the mitotic chromosomes and promotes the establishment of a concentration gradient of Ran-GTP/GDP where it is found at highest near the chromosomes and lowest at the centrosomes. During the NE assembly at the end of mitosis, Ran rebinds the chromatin and regulates the NE assembly, which depends on its GTPase cycle and its regulators (6 11, 14). Although Ran and its regulators have been shown crucial for the NE assembly, the mechanism for the NE precursor vesicle recruitment to chromatin and the downstream effectors remain poorly understood (8 11, 15). Importin interacts with the FXFG domain of nucleoporins through its FXFG domainbinding sites (16, 17). Importin likely works with other Ranbinding proteins to regulate the nuclear pore complexes and NE formation through interacting with the FXFG domain nucleoporins and targeting them to the chromatin (8, 13). For example, importin depletion in Xenopus egg extracts blocks the NE assembly, whereas adding back exogenous importin rescues the NE formation in an appropriate concentration-dependent manner (8). Moreover, importin -coated beads alone could induce the NE assembly in Xenopus egg extracts under the regulation of Ran (8). In Caenorhabditis elegans embryos, importin knockdown by RNAi also blocks the NE assembly OCTOBER 22, 2010 VOLUME 285 NUMBER 43 JOURNAL OF BIOLOGICAL CHEMISTRY 33281
2 (18). We have recently reported that importin plays a crucial role in the recruitment of the lamin B receptor (LBR)-bearing NE precursor membrane vesicles to the chromatin surface and the NE assembly through a direct interaction with the N-terminal spanning domain of LBR at amino acids (aa) (15). Furthermore, these activities were carried out in a Ran-sensitive and importin -independent manner (15). LBR appears to be a central player in targeting nuclear membranes to the chromatin (4, 15). LBR is a polytopic inner nuclear envelope protein with eight predicted hydrophobic transmembrane domains (76). The hydrophilic N-terminal spanning region of LBR is composed of 200 aa and exposed to the nucleoplasm, where it interacts with chromosome/chromatin proteins such as B-type lamins, HA95, and heterochromatin protein 1 (HP1) (19 30). The interaction of LBR with HP1 may not be direct, but is mediated through the core histones H3/H4 (31). The N-terminal spanning region of LBR contains a conserved arginine/serine (RS)-rich region, which may be phosphorylated on some serine and threonine residues by multiple RS kinases in a cell-cycle-dependent manner (32, 33). LBR phosphorylation appears to regulate its binding to chromatin proteins (32 37). The C terminus of LBR is a short hydrophilic spanning region facing to the nucleoplasm with an unknown function (38). The large middle part from aa of the LBR houses the 8 transmembrane domains. The first transmembrane domain, but not the others, is critical for confining this protein to the inner nuclear membrane (39). During the NE assembly at the end of mitosis, it is found that LBR plays a central role in targeting the NE precursor membrane vesicles to the chromatin mediated by importins. Overexpression of LBR in yeast cells induces over-generation of membrane and formation of membrane stacks in the cytoplasm (40). In sea urchins, LBR is required for targeting the LBR-bearing membrane vesicles to the chromatin and the NE assembly during male pronuclear formation (41). In culture cells, LBR fused with the green fluorescent protein (GFP) has been used to follow the dynamics of the NE assembly and the mechanism investigation of the NE membrane precursor targeting to the chromatin (42, 43). In a more recent study we found that, during the NE reassembly, the targeting of the NE membrane precursor vesicles to the chromatin is mediated by the interaction of the N-terminal spanning region with importin (15). Once meeting with Ran- GTP on the surface of the chromatin generated by RCC1, importin preferentially binds with Ran-GTP and releases the LBR-bearing vesicles to assemble the NE. With the hydrolysis of the GTP molecule of Ran, the membrane vesicles fuse to form the bi-layered NE and importin leaves Ran for next cycle (6, 15). However, how the interaction between importin and LBR is regulated and why overexpression of LBR causes overgeneration of the membrane remain unclear. In this work, we report that the targeting of the LBR-bearing NE precursor membrane vesicles to the chromatin during the NE assembly is mediated by importin through interaction of its aa with the aa of LBR. LBR knockdown causes the NE assembly failure and the cell death in early G 1 phase. We also show that the phosphorylation status of LBR serine 71 regulates the interaction between LBR and importin. Deletion of the N-terminal spanning domain or aa of LBR reduces the binding strength of LBR to the chromatin in interphase, increases the lateral movement potential of LBR, prevents successful targeting of the LBR-bearing NE precursor membrane vesicles to the chromatin at the end of mitosis, and causes the cell death of the daughter cells in next early G 1 phase. We also find that the first transmembrane domain of LBR is responsible for the membrane production. Together, our results demonstrate that LBR performs a crucial role in recruiting the LBRbearing NE precursor membrane vesicles to the chromatin during the NE assembly mediated by importin in a phosphorylation-dependent manner in mitosis and that the first transmembrane domain of LBR is responsible for the membrane production and the targeting of this protein to the NE during the nuclear growth and the NE expansion in interphase. EXPERIMENTAL PROCEDURES Plasmid Construction Wild-type Xenopus LBR (NCBI Y17842) was cloned from a cdna library of Xenopus oocytes (Clontech) and inserted into pegfp-c2 vector (Invitrogen) at the EcoRI and SalI restriction sites. GFP-xLBR was constructed by inserting the cdna encoding aa into the EcoRI/SalI sites of pegfp-c2. GFP-xLBR 1 210, GFP-xLBR , GFP-xLBR , GFP-xLBR , GFP-xLBR , GFP-xLBR , and GFP-xLBR were subcloned by inserting the cdna sequence encoding the corresponding amino acids into the EcoRI/SalI sites of pegfp-c2. GFPxLBR 1 260, GFP-xLBR , GFP-xLBR , GFPxLBR 1 621, GFP-xLBR S71A, GFP-xLBR S71D, and GFP-xLBR were cloned into the PstI and SalI sites of pegfp-c1. Full-length hlbr and hlbr were cloned into the EcoRI and SalI restriction sites of pegfp-c2 by similar methods. GFP-xLBR S71A and GFP-xLBR S71D were constructed by a single site mutagenesis method. xlbr 1 210, xlbr S71A, and xlbr S71D were subcloned into the EcoRI/SalI sites of pet-28a vector. GST-importin 1 876, GSTimportin 1 462, GST-importin , GST-importin 1 380, GST-importin , GST-importin , GST-importin , and GST-importin were constructed by cloning into the BamHI site of pgex-4t-1 vector. Protein Expression and Purification Escherichia coli strain BL21 (plys) was transformed with His-xLBR 1 210, His-xLBR S71A, His-xLBR 1 210S71D, GST-importin 1 876, GST-importin 1 462, GST-importin , GST-importin 1 380, GST-importin , GST-importin , GSTimportin , and GST-importin To produce the recombinant His-tagged LBR proteins, the bacterial cells were grown to an A 600 of 1.0. Isopropyl- -D-thiogalactopyranoside was then added to a final concentration of 0.1 mm, and the cells were incubated at 30 C for more than 5htoinduce protein expression. To produce recombinant GST-tagged importin proteins, the bacterial cell cultures were grown to an A 600 of 0.5. Isopropyl-beta-D-thiogalactopyranoside was then added at a final concentration of 0.1 mm, and the cultures were incubated at 17 C for 6 h to induce the protein expression. The bacterial cells were pelleted by centrifugation at 6,000 g for 10 min at 4 C. Proteins were purified with either Talon-Resin (BD Bioscience) or Glutathione-Sepharose 4B (Pharmacia Biotech Inc.) following the manufacturer s instructions JOURNAL OF BIOLOGICAL CHEMISTRY VOLUME 285 NUMBER 43 OCTOBER 22, 2010
3 Cell Culture, Transfection, Synchronization, and RNAi HeLa cells were grown in DMEM medium (Invitrogen), supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 g/ml streptomycin (Sigma), on 35-mm diameter Petri dishes at 37 C in a 5% CO 2 atmosphere and transfected using the calcium precipitation method as described previously (44). Briefly, the precipitated plasmid DNA was left with the cells in the Petri dishes for 6 h. Cells were then washed with PBS and grown for another h before direct observation and fixation for immunofluorescence microscopy. For cell synchronization, HeLa cells were grown in DMEM, treated in thymidine (2.5 mm) for 20 h, released in fresh medium for 4 h, and then treated with nocodazole (50 nm) for 12 h. Almost all the cells were blocked at metaphase. For RNAi, HeLa cells were transfected with LBR- HSS105976, LBR-HSS105977, and LBR-HSS by Lipofectamine Samples were assayed at 72 h after transfection. Co-immunoprecipitation and GST Fusion Protein Pulldown Assays HeLa cells were washed once with cold PBS and lysed in cell lysis buffer (20 mm Tris-HCl, ph 8.0, 150 mm NaCl, 2 mm EGTA, 0.5 mm EDTA, 0.5% Nonidet P-40, 5 mm NaF, 1 mm Na3VO 4,1mM PMSF, 10 g/ml aprotinin, and 5 g/ml Pepstatin A) for 20 min on ice and centrifuged at 13,000 rpm for 15 min to obtain clear cell lysates. For co-immunoprecipitation assays, the clarified cell lysates were incubated with indicated antibodies for 60 min on ice. Then 15 l of protein A- or G-Sepharose (75% slurry) was added, and the mixtures were rotated for 2 h at 4 C. The beads were washed three times with lysis buffer and harvested by brief centrifugation and finally suspended in gel sample buffer. For GST pulldown assay, 5 g of soluble GST or GST-fused proteins bound to 15 l of Glutathione-Sepharose beads (75% slurry) were incubated with lysates from HeLa cells for 3 h at 4 C. The beads were washed five times with lysis buffer and harvested by brief centrifugation and finally suspended in gel sample buffer. The beads-bound proteins were separated on 10% SDS-PAGE gels and analyzed by immunoblotting (IB) with the indicated antibodies. Gel Electrophoresis and IB After being separated on 10% SDS-PAGE gels, the protein samples were transferred onto nitrocellulose filters in the transfer buffer (25 mm Tris, 192 mm glycine, and 20% methanol) for 1 h at 100 V. The filters were blocked in TTBS (20 mm Tris-HCl (ph 7.4), 500 mm NaCl, and 0.3% Tween 20) containing 5% nonfat milk for 1 h at room temperature and probed with anti-gfp monoclonal antibody (Santa Cruz Biotechnology, diluted 1:1000 in TTBS with 5% nonfat milk) or probed with anti-importin monoclonal antibody (BD Transduction, diluted 1:1000 in TTBS with 5% nonfat milk) overnight at 4 C. The filters were then washed three times and blocked again for 30 min in TTBS containing 5% nonfat milk and incubated with horseradish peroxidase (HRP)- conjugated goat anti-mouse secondary antibody (Jackson, diluted 1:1000 in TTBS with 5% nonfat milk) for 1 h at room temperature. After a thorough wash in TTBS, the filters were developed for visualization by enhanced chemiluminescence (Sigma) and x-ray films. Immunofluorescence Microscopy Transfected cells were grown to 60% confluency on 35-mm diameter Petri dishes, washed three times with PBS, and fixed at room temperature in 4% paraformaldehyde for 15 min. The cells were then incubated with primary antibodies diluted in PBS containing 3% BSA for 1 h at room temperature or overnight at 4 C. The antibody against cleaved caspase 3 was from Cell Signaling Technology and diluted at 1:1000. The antibody against hlbr was generated by injecting rabbits and diluted 1:500. The cells were then washed five times in PBS and incubated with secondary antibodies diluted in PBS containing 3% BSA (TRITC goat anti-rabbit Ig (DAKO), diluted 1:200) at room temperature for 45 min to 1 h at room temperature. The cells were then washed five times in PBS and mounted in Mowiol (Sigma) containing DAPI. Samples were viewed under a Zeiss immunofluorescence microscope 200M equipped with a 63 objective. Images were captured using a cooled charged-coupled device AxioCamMRm camera. Live Imaging HeLa cells grown in a glass-bottomed Petri dish in a temperature-controlled chamber at 37 C with 5% CO 2 were transfected with GFP-xLBR , GFP-xLBR , or full-length GFP-xLBR (as control). The images of the cells expressing the GFP fusion protein were acquired every 5 min by a cooled charged-coupled device AxioCamMRm camera on an Axiovert 200M microscope. The Axiovert software was used to collect and process the data. Preparation of Xenopus Egg Extract and Demembraned Xenopus Sperm Chromatin and the NE Assembly in Vitro Low speed Xenopus egg extracts were prepared as described previously (45 47). The extracts were then frozen and stored in aliquots in liquid nitrogen until use. Sperms were released from testes by gently squeezing in nuclear isolation buffer (NIB, 15 mm NaCl, 60 mm KCl, 15 mm Tris, ph 7.5, 1 mm dithiothreitol, 0.5 mm spermine, and 0.25 M sucrose) and centrifuged at 100 g for 1 min at 4 C to remove somatic tissues. Sperm-containing supernatant was centrifuged at 1,500 g for 10 min at 4 C. The pellet was re-suspended in NIB and incubated in the presence of 0.05% lysolecithin for 8 10 min at 23 C. The reaction was stopped by adding 3 volumes of cold NIB plus 3% BSA. After washing for three times with NIB, the demembraned sperm chromatin were stored in aliquots in liquid nitrogen at 10 8 /ml. Low speed egg extract was mixed with an ATP-regenerating system and the demembraned sperm chromatin. The reaction system contained 20 l of low speed egg extract, 1 l of ATPregenerating system, and 2000/ l demembraned sperm chromatin. The proteins His-xLBR 1 210, His-xLBR S71A, His-xLBR S71D, or EB buffer (extract buffer) alone was added into the reaction system to a final concentration of 10 M. The samples were removed after incubation at 23 C for the time indicated and stained on a slide with 3,3-dihexyloxacarbocyanine without fixation. The samples were immediately viewed under a Zeiss 200M immunofluorescence microscope equipped with a 63 objective. Images were captured using a cooled charged-coupled device AxioCamMRm camera. FRAP Analysis Fluorescence recovery after photobleaching (FRAP) was performed with a Leica confocal microscope using a 488-nm line of a 400-milliwatt Kr/Ar laser in conjunction with a 100 objective for optimum resolution. For quantitative essay, the framed areas were photobleached at full laser power (100% power and 100% transmission) for 2 s, and recovery of OCTOBER 22, 2010 VOLUME 285 NUMBER 43 JOURNAL OF BIOLOGICAL CHEMISTRY 33283
4 the fluorescence was monitored by scanning the whole cell at low laser power (19% power and 19% transmission) in a 20-s intervals. The scanning laser intensity did not significantly photobleach the specimen over the time course of the experiment. MS Analysis of the Phosphorylated Proteins 2 g of HishLBR was incubated with cyclin B/Cdc2 (New England Biolabs) in 50 mm Tris, ph 7.5, 10 mm MgCl 2,2mM EGTA, 5 mm DTT, 100 mm ATP, for 30 min at 30 C. The reactions were subjected to SDS-PAGE, and the gels were processed for MS analysis. The targeted gel band was cut and destained by 25 mm NH 4 HCO 3 in 50% acetonitrile. The gel band was dried in 100% acetonitrile and digested in 25 mm NH 4 HCO 3 (ph 8.0) with modified porcine trypsin (Promega, Madison, WI) using a trypsin-to-protein ratio of 1:50 (w/w) overnight at 37 C. The peptide mixtures were extracted from the trypsin-digested gel band using 50% acetonitrile/1% trifluoroacetic acid, redissolved with 10 l of 0.2% formic acid, and analyzed by online nanolc (Microtech Scientific) connected to an LTQ Orbitrap mass spectrometer (ThermoFisher Scientific). Samples were eluted with a linear gradient from 2% to 35% buffer B (100% acetonitrile and 0.1% acetic acid) at a flow rate of 500 nl/min over 40 min using a 150- m inner diameter 150-mm reverse-phase C18 columns (Microtech Scientific). The MS was operated in data-dependent MS/MS mode in which each full MS scan was followed by five MS/MS scans where the five most abundant peptide ions were selected for collision-induced dissociation using a normalized collision energy of 35% in the ion trap. Survey MS scans were acquired in the Orbitrap analyzer with resolution of 60,000 (m/z 400) in the positive ion mode. Raw MS files were processed with Proteome Discoverer 1.0 (ThermoFisher Scientific), and searched against the IPI-human (version 3.55 European Bioinformatics Institute) containing both forward and reversed protein sequences. Initial maximum precursor and fragment mass deviations were set to 7 ppm and 0.5 Da, respectively. The search parameters included variable modifications for oxidation of methionine, and phosphorylation of serine, threonine, and tyrosine and variable modifications for cysteine carbamidomethylation. The maximum peptide false discovery rate was set to RESULTS LBR Interacts with Importin at Mitosis and They Become Disassociated at the End of Mitosis The NE assembly is one of the key steps in generating daughter nuclei during the cell division in eukaryotic cells. LBR has been shown to play a vital role in targeting the membrane vesicles to the chromatin during the NE assembly at the end of mitosis (15). Although it is known that the targeting of the membrane to the chromatin by the LBR is carried out through interaction of the aa of its N-terminal spanning region with importin, the regulation of the interaction remains unclear. To investigate how the interaction of LBR with importin is regulated, we cloned the N-terminal spanning region aa of Xenopus and human LBR genes and transiently expressed both GFP-xLBR and GFPhLBR in human HeLa cells (Fig. 1). By immunostaining with an anti-importin antibody, we observed that GFP-LBR co-localized with importin in the cytoplasm and spindle in mitosis and in the nucleus in interphase (Fig. 1A). We then selected the low-to-middle level GFP-LBR-expressing cells and FIGURE 1. LBR interacts with importin at mitosis and dissociates at the end of mitosis. A, HeLa cells, transiently transfected with GFP-xLBR 1 210, were co-stained with anti-importin and DAPI for DNA. Note that this truncate LBR localizes to the nucleus in interphase and moves to mitotic spindle and other cytoplasmic place where it co-localizes with importin in mitosis. B, co-immunoprecipitation of importin with GFP-xLBR. The clarified xlbr expressing cells in both interphase and mitosis were lysed and incubated with a rabbit anti-gfp or rabbit-igg (as control) antibody for 60 min on ice. Then 15 l of protein A- or G-Sepharose (75% slurry) were added, and the mixtures were rotated for 2 h at 4 C.The beads were washed three times with lysis buffer and harvested by brief centrifugation and finally suspended in gel sample buffer. The protein samples were separated by SDS-PAGE gel and processed for Western blot with anti-importin or anti-gfp antibody. Note that the binding of the truncate GFP-xLBR with importin occurred only in mitosis. C, HeLa cells were arrested by double thymidine block/release to various cell cycle phases and lysed to generate time-course samples, and the time at the metaphase onset is referred to as 0 min. The cell lysates were processed for GST pulldown assay between GFP-xLBR and GST-importin using an anti-gfp antibody, followed by Western blot. D, HeLa cells were arrested by thymidine/nocodazole block at metaphase and released. Cell lysates were prepared at the time indicated after release from metaphase and GST pulldown assay between GFP-xLBR and GST-importin was performed using anti-gfp antibody. The lysate GFP-LBR in GST pulldown assay was used as control. Note that the interaction of GFP-LBR became weaker along with the release from metaphase. synchronized them to interphase or mitosis and performed coimmunoprecipitation experiments using an anti-gfp antibody, followed by Western blots using the anti-importin antibody to reveal the interaction of GFP-LBR with importin. The results showed that the endogenous importin co-precipitated with both GFP-xLBR and GFP-hLBR from the mitotic but not the interphase cell lysates (Fig. 1B). This result indicates that LBR interacts with importin mostly during mitosis. We then set out to investigate if the interaction between importin and LBR is dynamic during mitosis. We synchronized GFP-xLBR expressing HeLa cells to various phases of the cell cycle by double thymidine block/release and performed pulldown assays with GST-importin as the bait JOURNAL OF BIOLOGICAL CHEMISTRY VOLUME 285 NUMBER 43 OCTOBER 22, 2010
5 in corresponding cell lysates, followed by Western blot using an anti-gfp antibody. The result showed that a weak interaction between importin and the LBR started during the S and G 2 phases, then became stronger as the cells entered mitosis, reached peak at 90 min after entry into mitosis, and declined until it disappeared during the G 1 phase (Fig. 1C). We also performed similar pulldown assays using metaphase-arrested and -released cells expressing GFP-xLBR and GST-importin Consistent with the previous observations, we found that a strong interaction between LBR and importin occurred within 30 min after metaphase onset. Following that, the interaction attenuated till it became undetectable after 50 min when most of the cells entered the G 1 phase (Fig. 1D). Together, these results demonstrate that the strong binding of LBR with importin is mitosis-specific and that they dissociate at the end of mitosis. aa Are Required for the Binding of LBR with aa of Importin and the Binding Is Regulated by Phosphorylation of LBR at Ser-71 Next, we decided to investigate the binding sites on both LBR and importin. First, we constructed and purified a series of truncated importin proteins fused with GST: GST-importin 1 876, GST-importin 1 462, GSTimportin 1 380, GST-importin , GST-importin , GST-importin , GST-importin , and GST-importin (Fig. 2A). Then we performed pulldown assays using these GST-importin truncate proteins and the lysate of mitotic HeLa cells transfected with GFP-xLBR to determine the interaction between importin and LBR. Our results showed that the binding affinity of GST-importin 1 462, GSTimportin , or GST-importin with LBR was as the same as that of full-length importin, whereas the binding affinity of other truncated importin proteins with LBR was not detectable (Fig. 2B). These results indicate that the aa region of importin is the minimal length required for binding with LBR (Fig. 2, A and B). It has been reported that there is a conserved serine and arginine (SR)-rich sequence in the LBR N-terminal spanning region, which contains a number of potential phosphorylation sites (Fig. 2C) (25). To reveal the binding sites on LBR for importin, we constructed GFP-xLBR , GFP-xLBR , GFP-xLBR , GFP-xLBR , GFP-xLBR (aa deleted GFP-xLBR ), and GFP-xLBR (aa deleted GFP-xLBR ) based on SR-rich sequence features. Then we expressed these truncated proteins in HeLa cells and carried out pulldown assays to detect the interaction between importin and these truncate proteins. The result showed that both GFP-xLBR and xlbr bound with importin efficiently, whereas GFP-xLBR showed relatively weak affinity with importin. When the SR-rich sequence was disrupted or deleted, as in truncates GFPxLBR , GFP-xLBR , GFP-xLBR ,or GFP-xLBR , these truncate proteins completely lost their binding ability with importin (Fig. 2D). This result indicates that the SR-rich sequence is crucial for the interaction of LBR with importin. As it has been reported that serine 71 of LBR is phosphorylated by the mitotic kinase CDK1 at mitosis (32, 33), we mutated this amino acid to alanine (S71A) or aspartic acid (S71D) to mimic its unphosphorylated or phosphorylated state to determine if this site is important for the binding of LBR with importin. Then we transiently expressed these two mutants in HeLa cells. The cells expressing GFP-xLBR S71A and GFP-xLBR S71D were synchronized, lysated, and subjected to pulldown assays as aforementioned. The result showed that unphosphorylation-mimicking mutant GFP-xLBR S71A could not bind with importin, whereas the phosphorylationmimicking mutant GFP-xLBR S71D bound importin like the wild-type xlbr (control) (Fig. 2, D and E). Taken together, these results demonstrate that phosphorylation of xlbr at serine 71 regulates the interaction between LBR and importin in mitosis. We then asked whether disrupting the interaction between LBR and importin interferes the NE assembly. We transiently expressed the full-length mutants GFP-LBR S71A, GFP- LBR S71D, and the wild-type GFP-LBR in HeLa cells followed by fluorescence microscopy. The result showed that, like the wild-type, both GFP-LBR S71D and GFP-LBR S71A caused no significant defects to the cells at interphase (data not shown). However, once the cell entered mitosis, the unphosphorylation-mimicking mutant GFP-LBR S71A showed a perturbed distribution and often aggregated into membrane stacks in the cytoplasm. At the end of mitosis (telophase/g 1 ), this mutant protein did not become associated with the NE but remained in the membrane stacks (Fig. 2F). In contrast, the phosphorylation-mimicking mutant GFP-LBR S71D showed a similar NE distribution to the wild type at the end of mitosis (telophase/g 1 ), except that the GFP-LBR S71D -expressing metaphase cells showed a likely continuous membrane around the aligned metaphase chromosomes (Fig. 2F). To explore and confirm whether serine 71 of LBR is phosphorylated during mitosis in HeLa cells, we performed phosphorylation reactions in vitro using his-hlbr and Cdc2-cyclin B. The sample was separated by SDS-PAGE, and the targeted gel band was cut, destained, and trypsin-digested. Then we analyzed the tryptic peptide mixtures using high resolution LTQ Orbitrap mass spectrometry and found that serine 71 was indeed phosphorylated (Fig. 2G). Together, these results indicate that the interaction of LBR with importin regulated by phosphorylation at LBR Ser-71 is required for the NE membrane precursor vesicle targeting to the chromatin and the NE assembly. Deletion of the N-terminal Spanning Region Causes LBR Lateral Mobile and the Cell Death in Early G 1 Due to the Failure of the NE Assembly Knowing that the N-terminal spanning region of LBR is critical for its interaction with importin and the NE assembly, we set out to examine the effects to the cells when deleting this N-terminal spanning region. We transfected the cells with the truncate GFP-xLBR and the full-length GFP-xLBR as control. We observed that, similar to the full-length GFP-xLBR 1 621, the truncate GFP-xLBR caused no visible defects to the interphase cells at current cell cycle, no matter at low or high level expression (Fig. 3A). We immunostained the cells with a human LBR-specific antibody (Fig. 3B) and found that the endogenous LBR (in red) was interweaved with the truncate exogenous xlbr or the fulllength foreign xlbr in the nuclear membrane (Fig. 3A, arrows). It is noteworthy that, similar to the full-length GFP- OCTOBER 22, 2010 VOLUME 285 NUMBER 43 JOURNAL OF BIOLOGICAL CHEMISTRY 33285
6 FIGURE 2. Phosphorylation of LBR Ser-71 is required for the interaction of LBR with importin, the NE precursor membrane vesicle targeting to the chromatin, and the NE assembly. A, schematic representation of the importin truncates and their respective binding ability to GFP-xLBR The gray zone indicates the minimal length (aa ) of the truncated importin proteins necessary for binding with LBR. B, GST pulldown assay between the GSTimportin proteins and GFP-xLBR g of soluble GST or GST-importin proteins bound to 15 l of glutathione-sepharose beads (75% slurry) was incubated with lysate of HeLa cells expressing GFP-xLBR for3hat4 C.Thebound proteins of the beads were processed for Western blot using an anti-gfp antibody. Note that importin was the minimum size to bind with GFP-xLBR 1 210, and any truncate containing this fragment such as importin and importin as well as the full-length importin could bind with GFP-xLBR equally. C, part of xlbr amino acid sequence, showing the serine-rich conserved region (SR-rich region). D, pulldown assay using GST-importin coated beads and mitotic HeLa cell lysates expressing different GFP-xLBR truncated proteins or GFP as control, followed by a Western blot analysis with an anti-gfp antibody. Note that the aa region of xlbr is required for its binding with importin and its serine 71 is critical for this binding. E, pulldown assay using GST-importin coated beads and the lysate of mitotic HeLa cells expressing the wild-type truncate GFP-xLBR 1 210, the unphosphorylation-mimicking mutant GFP-xLBR S71A or the phosphorylationmimicking mutant GFP-xLBR S71D, or GFP alone as control, followed by a Western blot analysis with an anti-gfp antibody. Note that importin only bound with wild-type GFP-xLBR and the serine 71 phosphorylation-mimicking mutant GFP-xLBR S71D. F, HeLa cells expressing the wild-type GFP-LBR, GFP-LBR S71D, and GFP-xLBR S71A were fixed and count-stained with DAPI. Images were collected using a Zeiss immunofluorescence microscope 200M equipped with a 63 objective and a cooled charged-coupled device AxioCamMRm camera. Bars,10 m. G, phosphorylation site identification of the peptide ( 64 KGGSTSSSPSR) by nanolc-ms/ms from in-gel tryptic digest of hlbr. MS/MS fragmentation of the doubly charged ion m/z gave a sequential b and y ion sequence to match the sequence of one peptide from hlbr ( 64 KGGSTSSSPSR, [M H] Da) with a phosphate modification. The b and y ions showed that the phosphorylation site was located at the serine 71 position. xlbr 1 621, the truncate GFP-xLBR also induced the membrane stack formation in the cytoplasm and interweaved with the endogenous human LBR (red in Fig. 3A, arrowheads). It has been reported that one role of the N-terminal region of LBR is to anchor this protein and the NE to the chromatin (39, 48, 49). In this work, we set out to determine whether the protein is mobile when the function of N-terminal region of LBR is disturbed. We carried out a FRAP experiment using the cells with low-to-middle level expression of the full-length GFPxLBR as a control and the truncate GFP-xLBR (Fig JOURNAL OF BIOLOGICAL CHEMISTRY VOLUME 285 NUMBER 43 OCTOBER 22, 2010
7 FIGURE 3. GFP-xLBR could locate in the NE and induce endogenous LBR-containing membrane overproduction and stack formation in cytoplasm and these truncated proteins are laterally mobile. A, HeLa cells overexpressing full-length GFP-xLBR or the N-terminal spanning region-deleted truncate GFPxLBR were fixed and immunostained with hlbr-specific antibody, and counterstained with DAPI for DNA. Note that both the full-length and the truncated LBR localized on the NE (arrows) and induced membrane overproduction and stack formation containing endogenous LBR in the cytoplasm (arrowheads). B, characterization of anti-human hlbr antibody used in A. This antibody specifically recognizes endogenous hlbr but not the exogenous GFP-xLBR. C, FRAP assay for the full-length GFP-xLBR on the NE. Note that the fluorescence in the photo-bleached regions could not recover within as long as 300 s, indicating that the full-length xlbr was static on the NE. D, FRAP assay for the GFP-xLBR on the NE. Note that the fluorescence in the photo-bleached regions recovered rapidly, indicating that GFP-xLBR was mobile on the NE. Bars,10 m. LBR Depletion Causes NE Assembly Failure and Apoptosis 3, C and D). As expected, the fluorescence in the bleached NE regions of the full-length GFP-xLBR expressing cells could not recover within 300 s after the photo-bleaching, indicating that the full-length GFP-xLBR in the NE was totally immobilized as previously reported (48). In contrast, the majority of the truncate GFP-xLBR in the NE was mobile, as indicated by the fast recovery of the fluorescence of the photobleached region (Fig. 3D). This result suggests that the truncate GFP-xLBR is laterally mobile and that it moves rapidly from ER to the NE, although this truncate protein has no root to anchor itself to the chromatin and/or the nuclear lamina. Because the truncate GFP-xLBR was mobile within the NE and showed no visible harm to the cell in current cell cycle, we wished to investigate if it is harmful to the cell in the next cell cycle. We transiently expressed the truncate GFPxLBR , full-length xlbr in HeLa cells and followed the behavior of the dividing cells. We found that, although GFPxLBR is not significantly toxic to the cells in current cell cycle as aforementioned (Fig. 4A), the daughter cells overexpressing the truncate GFP-xLBR could not spread well after division (Fig. 4B), compared with the daughter cells overexpressing the full-length xlbr (data not shown). Furthermore, the truncate GFP-xLBR protein seemed to form vesicle-like or aggregate structures (Fig. 4B). By DAPI staining to visualize DNA, we found that the chromatin in these daughter cells could not decondense well, the NE around the chromatin could not be well assembled and the cells were likely dying (Fig. 4C). We repeated these experiments three times and found that the likely dying cells stood for 70% of the paired G 1 cells overexpressing the truncate GFP-xLBR , whereas only 3% were dying among the paired cells expressing full-length xlbr (Fig. 4D). By time-lapse microscopy, we examined the behavior of the paired daughter cells during division (Fig. 4, E G). We observed that the truncate GFP-xLBR and GFP-xLBR did not take part in the NE assembly around the daughter nuclei, and the nuclei could not be well assembled, compared with those in the cells overexpressing full-length GFP-xLBR. Furthermore, the daughter cells overexpressing GFP-xLBR and GFP-xLBR could not spread well and finally showed apoptotic characteristics such as bubbling, chromatin condensing, and apoptotic body-like structure formation (Fig. 4, E and G). To confirm if the dying paired G 1 cells were apoptotic, we immunostained these cells with an antibody against the cleaved caspase 3, a marker for apoptotic cells (Fig. 5, A and B). We observed that the cleaved caspase 3 staining was totally negative in the mother cells (in the current cell cycle) and clearly positive in the paired cells (G 1 phase of the next cell cycle). These results indicate that N terminus-deleted LBR and GFPxLBR cannot target the NE precursor membrane vesicles to the chromatin to form the NE, and the daughter cells without a functional NE would eventually die in G 1 phase through apoptosis. Then we performed LBR RNAi and rescue experiment in HeLa cells to see if the function of LBR is critical for the NE assembly. At 72 h after sirna transfection, the LBR protein level was decreased substantially detected by immuno- OCTOBER 22, 2010 VOLUME 285 NUMBER 43 JOURNAL OF BIOLOGICAL CHEMISTRY 33287
8 blotting (Fig. 5C). We also co-transfected the cells with sirna and RFP-H2B as a transfection reporter for microscopy. After 72 h, the cells were stained with an antibody against cleaved caspase 3. We found that LBR RNAi caused apoptosis of the daughter cells (Fig. 5D) and that the cells could be rescued by expressing RNAi-resistant xlbr (Fig. 5E). Taken together, these results demonstrate that the function of LBR is crucial for the NE assembly. The First Transmembrane Domain of LBR Induces the NE Membrane Production and Targeting to the Chromatin It has been previously shown that overexpression of exogenous full-length or N-terminal spanning region-deleted LBR in HeLa cells results in membrane overproduction and stack formation in the cytoplasm (15). To reveal how the membrane overproduction and stack formation were induced, we constructed GFP-fused fulllength xlbr and a number of truncated proteins: GFP-xLBR (the N-terminal spanning region), GFP-xLBR (the N-terminal spanning region plus the first transmembrane domain), GFPxLBR (xlbr without the N-terminal spanning region), GFPxLBR (xlbr without the N-terminal spanning region and the first transmembrane domain), GFPxLBR (xlbr without the first transmembrane domain), and GFP-xLBR (xlbr without the first and the second transmembrane domains) (Fig. 6A). We transiently expressed these proteins in HeLa cells and observed their distributions in the cells. The results showed that overexpressing fulllength GFP-xLBR and GFPxLBR efficiently induced membrane over-generation and stack formation and that the N-terminal spanning region was situated in the nucleus when expressed (Fig. 6B). Furthermore, we observed that the truncate xlbr 1 260, which only contains the N-terminal spanning region and the first transmembrane domain, not only localized to the JOURNAL OF BIOLOGICAL CHEMISTRY VOLUME 285 NUMBER 43 OCTOBER 22, 2010
9 FIGURE 5. Dysfunction or knockdown of LBR causes the cell death in early G 1 phase via apoptosis. A, the upper panel shows a mother cell, and the lower panels show three paired daughter cells. Note that overexpression of GFP-xLBR does not significantly harm the mother cell but causes the daughter cell death during the next G 1 phase through apoptosis, judging by positive staining of the cleaved caspase 3, an apoptosis marker. Bars, 10 m. B, the upper panel shows a mother cell, and the lower panels show three paired daughter cells. Note that Overexpression of GFP-xLBR does not significantly harm the mother cell but causes the daughter cell death during next G 1 phase through apoptosis. Bars,10 m. C, HeLa cells transfected with control or sirna were collected at 72 h after transfection and analyzed by Western blot with antibodies against human LBR and -tubulin serving as a loading control. D, HeLa cells co-transfected with control, sirna, and RFP-H2B (1:20) were immunostained with anti-cleaved caspase 3 antibody (green) at 72 h after transfection. The cells deleted endogenous LBR died in early G 1 phase, judging by cleaved caspase 3-positive staining. E, HeLa cells co-transfected with sirna, RFP-H2B, and sirna-resistant GFP-xLBR. The cells co-transfected with sirna, RFP-H2B, and GFP-xLBR showed a normal cell division, indicating that expression of the sirna-resistant xlbr rescued the cell death caused by hlbr knockdown. NE, but also efficiently induced the membrane stack formation, and that the truncate GFP-xLBR , which lacks the first transmembrane domain (aa ), distributed throughout the whole cell and caused no any membrane stack formation (Fig. 6B). Consistently, we also observed that both the truncate proteins GFP-xLBR (the first transmembrane domain deletion truncate mutant) and GFPxLBR (both the first and the second transmembrane domain deletion truncate mutant) localized throughout the cell but not the NE and did not induce membrane stack formation (Fig. 6B). These results not only confirm that the first transmembrane domain of LBR is essential for the NE localization of this protein as reported previously (39, 50), but also demonstrate that the transmembrane domain induces the membrane overproduction and the stack formation. Phosphorylation of LBR Ser-71 Regulates the NE Assembly around the Sperm Chromatin in Xenopus Egg Extracts To further study the role of the N-terminal spanning region of LBR in the NE assembly, we decided to take advantage of the Xenopus egg extracts system to examine the NE assembly (14). We prepared the His-tagged truncate LBR proteins His-xLBR 1 210, His-xLBR S71A, and HisxLBR S71D and the interphase Xenopus egg extract and the sperm chromatin for the nuclear assembly (46, 47). When the sperm chromatin were added into the extract, a complete NE assembled around the sperm chromatin within 150 min, visualized by lipid staining with a lipophilic fluorescent dye 3,3-dihexyloxacarbocyanine (Fig. 7A). When the wild-type N-terminal FIGURE 4. Overexpression of GFP-xLBR and GFP-xLBR results in the daughter cell death at early G 1 phase. A, HeLa cell transiently expressed GFP-xLBR in the presence of 2.5 mm thymidine in the medium, which was used to arrest the cells at interphase, were fixed and viewed directly. Note that the GFP-xLBR expressing cells were alive in interphase in current cell cycle. Bar,20 m. B, four paired representative likely dying early G 1 cells overexpressing the truncate GFP-xLBR Bar, 10 m. C, DAPI staining for DNA showing that the paired cells likely died by apoptosis. Bar, 10 m. D, statistical analyses showing that the majority of paired cells overexpressing xlbr likely died in early G 1 phase. The data were from three independent experiments. E, time-lapse images showing that the paired daughter cells overexpressing GFP-xLBR could not assemble their NE and finally died in early G 1 phase, judging by their morphology and Hoechst staining for DNA. F, time-lapse images showing that the daughter cells overexpressing full-length wild-type GFP-xLBR could assemble normal NE and nuclei, judging by their morphology and Hoechst staining. G, time-lapse images showing that the paired daughter cells overexpressing GFP-xLBR could not assemble their NE and finally died in early G 1 phase, judging by their morphology and Hoechst staining for DNA. Bars,20 m. OCTOBER 22, 2010 VOLUME 285 NUMBER 43 JOURNAL OF BIOLOGICAL CHEMISTRY 33289
10 it may not block the targeting of LBR-containing membrane vesicles to the chromatin for the NE assembly by importin. Because S71D could bind with importin (see Fig. 2E), it may block the targeting of LBR-containing membrane vesicles to the chromatin by importin for the NE assembly. Therefore, these results demonstrate that, even in the in vitro NE assembly system based on Xenopus egg extracts, phosphorylation of Ser-71 of LBR also regulates the binding of LBR and importin for the LBR-bearing membrane targeting to the chromatin and the NE assembly. FIGURE 6. The first transmembrane domain is essential for LBR to localize to the NE and for the membrane stack formation. A, schematic description of the GFP-fused xlbr constructs used to transfect the cells. M1 to M8 indicate the eight putative transmembrane domain positions. B, the cells expressing full-length or truncate GFP-xLBR proteins were counterstained with DAPI for DNA. Note that only the wild-type and the first transmembrane domain-containing truncate proteins GFP-xLBR and GFP-xLBR localize to the NE and induce membrane stack formation in the cytoplasm, whereas the other truncate proteins did not localize to the NE and induce the membrane stack formation. When the first transmembrane domain was deleted or codeleted with the second transmembrane domain, the deletion mutants showed no specific membrane distribution in the cells. Together, these results demonstrate that the first transmembrane domain is sufficient to induce the membrane formation and targets the membrane to the NE. Bars, 20 m. spanning region His-xLBR and its mutants were added, we found that the wild-type and the mutant S71D, but not S71A, blocked the NE assembly around the sperm chromatin (Fig. 7, B D). To understand these results, we supposed that, because S71A mutant did not bind with importin (see Fig. 2, D and E), DISCUSSION Importin is an evolutionarily conserved protein that serves as a main nucleocytoplasmic transport receptor to regulate the nuclear import together with importin regulated by Ran (51 56). Importin can also mediate the nuclear import of some proteins independently of importin (57 63). It is also found that, in early mitosis, importin plays an inhibitory role for mitotic spindle assembly by interacting with and suppressing the spindle assembly factors (64), and this inhibitory role can be eliminated by Ran-GTP, which interacts with importin and releases the spindle assembly factors (6). At the end of mitosis, importin takes part in the NE assembly (9, 12, 15, 65). Importin was first reported to take a role in the NE assembly in Xenopus egg extracts in an appropriate dosage-dependent manner, and if its concentration was increased in the extract by adding excess importin protein, it inhibits the NE assembly (9, 12, 13). In our previous report, we found that one role of importin in the NE assembly is to target the NE membrane precursor vesicles to the chromatin through interaction with the membrane-born LBR (15). We found that depletion of LBR causes the cell death via apoptosis at early G 1 phase due to the NE assembly failure. We have pinpointed the binding regions, which are aa in LBR and aa in importin. We also demonstrate that the interaction of both proteins is regulated by phosphorylation of LBR at Ser-71 and that the interaction plays a crucial role in mediating the LBR-containing membrane vesicle targeting to the chromatin for the NE assembly. LBR has been implicated in a number of important functions. It plays roles in sustaining the NE architecture, in NE assembly, in the lobulated (segmented) nuclear shape formation in the development of the neutrophiles, and in the regulation of proliferative and functional responses for innate immunity (66 69). It is also involved in some diseases, such as Greenberg skeletal dysplasia and Pelger-Huet anomaly (38, 70 72). However, the detailed analyses of LBR have been performed using its soluble and functional N-terminal spanning region, which provides essential chromatin docking sites at the NE (22, 24, 30, 73). During NE assembly, the N-terminal spanning region of LBR targets its associated membrane vesicles to the chromatin by interacting with importin (15). In this work, we have identified the interacting sites in both importin and LBR N- terminal spanning region. We observed that, without the N-terminal spanning region, LBR is mobile compared with the stock-still full-length LBR at the NE. Even so, the N-terminal spanning region-deleted LBR is not immediately lethal to the cell in the current cell cycle, and the cell could go to mitosis. However, when the cell divides, its daughter cells could not JOURNAL OF BIOLOGICAL CHEMISTRY VOLUME 285 NUMBER 43 OCTOBER 22, 2010
11 FIGURE 7. Phosphorylation of LBR Ser-71 regulates the NE assembly around the sperm chromatin in Xenopus egg extract. When Xenopus egg extract was added with the sperm chromatin and the ATP-regenerating system and incubated at 23 C for a certain period, a complete NE could be assembled around the sperm chromatin. A, a time course of the NE assembly in the egg extract. The EB buffer was added into the extract as control. Note that a complete NE assembled around the well decondensed sperm chromatin, shown by fluorescent lipophilic dye 3,3-dihexyloxacarbocyanine staining for the NE and DAPI for DNA. B, when 10 M truncated wild-type protein His-xLBR was added into the extract, the NE assembly was significantly blocked and the chromatin decondensation was not completed. C, when 10 M truncated unphosphorylation-mimicking mutant protein His-xLBR S71A was added to the extract, the NE assembly and the chromatin decondensation were not seriously affected. D, when 10 M truncated phosphorylation-mimicking mutant protein His-xLBR S71D was added to the extract, the NE assembly and the chromatin decondensation were almost totally blocked. Together, these data indicate that phosphorylation of LBR Ser-71 regulates the NE assembly around the sperm chromatin in Xenopus egg extract. Bar,20 m. organize their complete and functional NE due to the LBRbearing NE membrane precursor vesicles not being targeted onto the chromatin, and their chromatin could not be fully decondensed. Finally, these daughter cells will die in early G 1 phase by apoptosis. This result demonstrates that LBR does play a role in the NE assembly. Besides, LBR may play a role in chromosome decondensation and nuclear structure organization through anchoring the chromatin to the NE and organizing local domains through binding some factors like lamin B, HP1, and chromatin/dna for some special nuclear functions (25, 28, 30). To perform these functions, LBR may be regulated through some modifications. During interphase, the RS region of LBR is mainly phosphorylated by NE-bound kinase-rs kinases; whereas in mitosis, the RS region is phosphorylated by p34 cdc2 at the site serine 71. RS kinases also phosphorylate other sites of LBR in interphase, and this process may be antagonized by phosphatase PP1 (34, 36, 74). The modification of the RS region is believed to modulate interaction between LBR and other nuclear proteins. Further phosphorylation of LBR by RS kinase completely abolishes the binding of p34/p32, indicating a distinct regulation mechanism between interphase and mitosis (32 35, 75). The function of these distinct kinases in regulating LBR activity so far remains unclear. In our present study, we found that Ser-71 phosphorylation of LBR by p34 cdc2 enhances the binding of LBR and importin during mitosis. As it has been reported that the PP1 stimulate the binding of LBR LBR Depletion Causes NE Assembly Failure and Apoptosis to the chromatin (74), here we propose that, during the NE assembly at the end of mitosis, once importin /LBR-bearing membrane vesicle complex approaches the surface of the chromatin, importin preferentially binds with Ran-GTP on the surface of the chromatin generated by RCC1 and releases the LBR-bearing membrane vesicle. Simultaneously, PP1 dephosphorylates LBR at Ser-71 and prevents it from rebinding with importin but to the chromatin to form the NE. Once the NE is assembled around the daughter nuclei, it grows to meet the requirement of the chromatin decondensation to a regular size and the function of the interphase nucleus. It has been shown that overexpression of LBR in yeast cells induced membrane overproduction and stack formation (40). We have previously reported that, in mammalian cells, overexpression of LBR resulted in overproduction of the NE, which either invaginated into the nucleus or budded off from the NE into the cytoplasm in an expression quantity-dependent manner (15). In this work, we found that the first transmembrane domain is an essential part for the NE growth and the membrane stack formation. With the N-terminal spanning region, the first transmembrane domain could induce the membrane stack formation when it is overexpressed. If the first transmembrane domain is deleted, the deletion mutant will not be able to localize to the NE, and its overexpression will not cause any membrane overproduction and membrane stack formation. Therefore, we propose that the first transmembrane domain of LBR is responsible for the NE generation and localization to the NE. In summary, we found that the interaction between importin and the N-terminal spanning region of LBR occurs between aa of importin and aa of LBR and that the interaction is regulated by the phosphorylation at the amino acid serine 71 of LBR. Disruption of this interaction by deletion of the N-terminal spanning region or aa of LBR inhibits the NE assembly. We also found that the first transmembrane domain of LBR is responsible for the NE generation. We propose that, during the NE assembly at the end of mitosis, LBRcontaining NE precursor membrane vesicles are targeted to the chromatin by the N-terminal spanning region of LBR through binding with importin in a LBR Ser-71 phosphorylation-dependent manner. Once it approaches the surface of the chromatin at the end of mitosis, the complex of importin /LBR disassociates through the preferential binding of importin with Ran-GTP and the dephosphorylation of the released LBR OCTOBER 22, 2010 VOLUME 285 NUMBER 43 JOURNAL OF BIOLOGICAL CHEMISTRY 33291
SUPPLEMENTARY INFORMATION
 The Supplementary Information (SI) Methods Cell culture and transfections H1299, U2OS, 293, HeLa cells were maintained in DMEM medium supplemented with 10% fetal bovine serum. H1299 and 293 cells were
The Supplementary Information (SI) Methods Cell culture and transfections H1299, U2OS, 293, HeLa cells were maintained in DMEM medium supplemented with 10% fetal bovine serum. H1299 and 293 cells were
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Nakajima and Tanoue, http://www.jcb.org/cgi/content/full/jcb.201104118/dc1 Figure S1. DLD-1 cells exhibit the characteristic morphology
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Nakajima and Tanoue, http://www.jcb.org/cgi/content/full/jcb.201104118/dc1 Figure S1. DLD-1 cells exhibit the characteristic morphology
Supplementary Material
 Supplementary Material Supplementary Methods Cell synchronization. For synchronized cell growth, thymidine was added to 30% confluent U2OS cells to a final concentration of 2.5mM. Cells were incubated
Supplementary Material Supplementary Methods Cell synchronization. For synchronized cell growth, thymidine was added to 30% confluent U2OS cells to a final concentration of 2.5mM. Cells were incubated
Sarker et al. Supplementary Material. Subcellular Fractionation
 Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit
 Cell Reports, Volume 5 Supplemental Information The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit Andrey Poleshko, Katelyn M. Mansfield, Caroline
Cell Reports, Volume 5 Supplemental Information The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit Andrey Poleshko, Katelyn M. Mansfield, Caroline
Gα 13 Activation Assay Kit
 A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Gα 13 Activation Assay Kit Catalog Number: 80401 20 assays NewEast Biosciences 1 Table of Content Product
A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Gα 13 Activation Assay Kit Catalog Number: 80401 20 assays NewEast Biosciences 1 Table of Content Product
Supplementary data. sienigma. F-Enigma F-EnigmaSM. a-p53
 Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Supplementary Materials and Methods
 Supplementary Materials and Methods sirna sequences used in this study The sequences of Stealth Select RNAi for ALK and FLOT-1 were as follows: ALK sense no.1 (ALK): 5 -AAUACUGACAGCCACAGGCAAUGUC-3 ; ALK
Supplementary Materials and Methods sirna sequences used in this study The sequences of Stealth Select RNAi for ALK and FLOT-1 were as follows: ALK sense no.1 (ALK): 5 -AAUACUGACAGCCACAGGCAAUGUC-3 ; ALK
Polyclonal ARHGAP25 antibody was prepared from rabbit serum after intracutaneous
 Preparation and purification of polyclonal antibodies Polyclonal ARHGAP25 antibody was prepared from rabbit serum after intracutaneous injections of glutathione S-transferase-ARHGAP25-(509-619) (GST-coiled
Preparation and purification of polyclonal antibodies Polyclonal ARHGAP25 antibody was prepared from rabbit serum after intracutaneous injections of glutathione S-transferase-ARHGAP25-(509-619) (GST-coiled
Gα i Activation Assay Kit
 A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Gα i Activation Assay Kit Catalog Number 80301 20 assays NewEast Biosciences, Inc 1 Table of Content Product
A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Gα i Activation Assay Kit Catalog Number 80301 20 assays NewEast Biosciences, Inc 1 Table of Content Product
Cdc42 Activation Assay Kit
 A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Cdc42 Activation Assay Kit Catalog Number: 80701 20 assays 1 Table of Content Product Description 3 Assay
A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Cdc42 Activation Assay Kit Catalog Number: 80701 20 assays 1 Table of Content Product Description 3 Assay
transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected with the wwp-luc reporter, and FLAG-tagged FHL1,
 Supplementary Data Supplementary Figure Legends Supplementary Figure 1 FHL-mediated TGFβ-responsive reporter transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected
Supplementary Data Supplementary Figure Legends Supplementary Figure 1 FHL-mediated TGFβ-responsive reporter transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected
Supplemental Information: Phosphorylation of CLIP-170 by Both Plk1 and CK2 Is Involved in the Timely Formation of Kinetochore-microtubule Attachments
 Supplemental Information: Phosphorylation of CLIP-170 by Both Plk1 and CK2 Is Involved in the Timely Formation of Kinetochore-microtubule Attachments Hongchang Li, X. Shawn Liu, Xiaoming Yang, Yingmin
Supplemental Information: Phosphorylation of CLIP-170 by Both Plk1 and CK2 Is Involved in the Timely Formation of Kinetochore-microtubule Attachments Hongchang Li, X. Shawn Liu, Xiaoming Yang, Yingmin
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Thompson et al., http://www.jcb.org/cgi/content/full/jcb.200909067/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Modification-specific antibodies do not detect unmodified
Supplemental material Thompson et al., http://www.jcb.org/cgi/content/full/jcb.200909067/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Modification-specific antibodies do not detect unmodified
Figure legends for supplement
 Figure legends for supplement Supplemental Figure 1 Characterization of purified and recombinant proteins Relevant fractions related the final stage of the purification protocol(bingham et al., 1998; Toba
Figure legends for supplement Supplemental Figure 1 Characterization of purified and recombinant proteins Relevant fractions related the final stage of the purification protocol(bingham et al., 1998; Toba
SUPPLEMENTARY INFORMATION
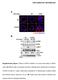 SUPPLEMENTARY INFORMATION Supplementary Figure 1 Effect of ROCK inhibition on lumen abnormality in MDCK cysts. (A) MDCK cells as indicated cultured in Matrigel were treated with and without Y27632 (10
SUPPLEMENTARY INFORMATION Supplementary Figure 1 Effect of ROCK inhibition on lumen abnormality in MDCK cysts. (A) MDCK cells as indicated cultured in Matrigel were treated with and without Y27632 (10
Analysing protein protein interactions using a GST-fusion protein to pull down the interacting target from the cell lysate Hong Wang and Xin Zeng
 Analysing protein protein interactions using a GST-fusion protein to pull down the interacting target from the cell lysate Hong Wang and Xin Zeng Department of Molecular Genetics, Biochemistry and Microbiology,
Analysing protein protein interactions using a GST-fusion protein to pull down the interacting target from the cell lysate Hong Wang and Xin Zeng Department of Molecular Genetics, Biochemistry and Microbiology,
B. ADM: C. D. Apoptosis: 1.68% 2.99% 1.31% Figure.S1,Li et al. number. invaded cells. HuH7 BxPC-3 DLD-1.
 A. - Figure.S1,Li et al. B. : - + - + - + E-cadherin CK19 α-sma vimentin β -actin C. D. Apoptosis: 1.68% 2.99% 1.31% - : - + - + - + Apoptosis: 48.33% 45.32% 44.59% E. invaded cells number 400 300 200
A. - Figure.S1,Li et al. B. : - + - + - + E-cadherin CK19 α-sma vimentin β -actin C. D. Apoptosis: 1.68% 2.99% 1.31% - : - + - + - + Apoptosis: 48.33% 45.32% 44.59% E. invaded cells number 400 300 200
Supplementary Information: Materials and Methods. GST and GST-p53 were purified according to standard protocol after
 Supplementary Information: Materials and Methods Recombinant protein expression and in vitro kinase assay. GST and GST-p53 were purified according to standard protocol after induction with.5mm IPTG for
Supplementary Information: Materials and Methods Recombinant protein expression and in vitro kinase assay. GST and GST-p53 were purified according to standard protocol after induction with.5mm IPTG for
Arf6 Activation Assay Kit
 A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Arf6 Activation Assay Kit Catalog Number: 82401 20 assays NewEast Biosciences 1 Table of Content Product
A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Arf6 Activation Assay Kit Catalog Number: 82401 20 assays NewEast Biosciences 1 Table of Content Product
Segments of the obstructed intestinal loops were fixed in 4% paraformaldehyde
 Supplementary text Supplementary materials and methods Histopathological examination Segments of the obstructed intestinal loops were fixed in 4% paraformaldehyde (PFA) and embedded in paraffin wax with
Supplementary text Supplementary materials and methods Histopathological examination Segments of the obstructed intestinal loops were fixed in 4% paraformaldehyde (PFA) and embedded in paraffin wax with
Supplemental Information
 Supplemental Information Intrinsic protein-protein interaction mediated and chaperonin assisted sequential assembly of a stable Bardet Biedl syndome protein complex, the BBSome * Qihong Zhang 1#, Dahai
Supplemental Information Intrinsic protein-protein interaction mediated and chaperonin assisted sequential assembly of a stable Bardet Biedl syndome protein complex, the BBSome * Qihong Zhang 1#, Dahai
SUPPLEMENTARY INFORMATION
 (Supplementary Methods and Materials) GST pull-down assay GST-fusion proteins Fe65 365-533, and Fe65 538-700 were expressed in BL21 bacterial cells and purified with glutathione-agarose beads (Sigma).
(Supplementary Methods and Materials) GST pull-down assay GST-fusion proteins Fe65 365-533, and Fe65 538-700 were expressed in BL21 bacterial cells and purified with glutathione-agarose beads (Sigma).
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
Journal of Cell Science. 520 Research Article
 520 Research Article Lamin B receptor plays a role in stimulating nuclear envelope production and targeting membrane vesicles to chromatin during nuclear envelope assembly through direct interaction with
520 Research Article Lamin B receptor plays a role in stimulating nuclear envelope production and targeting membrane vesicles to chromatin during nuclear envelope assembly through direct interaction with
Supporting Information
 Supporting Information Deng et al. 10.1073/pnas.1515692112 SI Materials and Methods FPLC. All fusion proteins were expressed and purified through a three-step FPLC purification protocol, as described (20),
Supporting Information Deng et al. 10.1073/pnas.1515692112 SI Materials and Methods FPLC. All fusion proteins were expressed and purified through a three-step FPLC purification protocol, as described (20),
Section 10. Junaid Malek, M.D.
 Section 10 Junaid Malek, M.D. Cell Division Make sure you understand: How do cells know when to divide? (What drives the cell cycle? Why is it important to regulate this?) How is DNA replication regulated?
Section 10 Junaid Malek, M.D. Cell Division Make sure you understand: How do cells know when to divide? (What drives the cell cycle? Why is it important to regulate this?) How is DNA replication regulated?
RheB Activation Assay Kit
 A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based RheB Activation Assay Kit Catalog Number: 81201 20 assays NewEast Biosciences 1 FAX: 610-945-2008 Table
A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based RheB Activation Assay Kit Catalog Number: 81201 20 assays NewEast Biosciences 1 FAX: 610-945-2008 Table
At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in
 Supplementary Materials and Methods Barrier function assays At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in acidic X-gal mix (100 mm phosphate buffer at ph4.3, 3 mm
Supplementary Materials and Methods Barrier function assays At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in acidic X-gal mix (100 mm phosphate buffer at ph4.3, 3 mm
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Ono et al., http://www.jcb.org/cgi/content/full/jcb.201208008/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Chromatin-binding properties of MCM2 and condensin II during
Supplemental material Ono et al., http://www.jcb.org/cgi/content/full/jcb.201208008/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Chromatin-binding properties of MCM2 and condensin II during
Nascent Chromatin Capture. The NCC protocol is designed for SILAC-based massspectrometry
 Extended experimental procedure Nascent Chromatin Capture. The NCC protocol is designed for SILAC-based massspectrometry analysis of nascent versus mature chromatin composition (1). It requires a large
Extended experimental procedure Nascent Chromatin Capture. The NCC protocol is designed for SILAC-based massspectrometry analysis of nascent versus mature chromatin composition (1). It requires a large
Coleman et al., Supplementary Figure 1
 Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
ab Ran Activation Assay Kit
 ab173247 Ran Activation Assay Kit Instructions for Use For the simple and fast measurement of Ran activation. This product is for research use only and is not intended for diagnostic use. Version 1 Last
ab173247 Ran Activation Assay Kit Instructions for Use For the simple and fast measurement of Ran activation. This product is for research use only and is not intended for diagnostic use. Version 1 Last
IgG TrueBlot Protocol for Mouse, Rabbit or Goatderived Antibodies - For Research Use Only
 IgG TrueBlot Protocol for Mouse, Rabbit or Goatderived Antibodies - For Research Use Only Introduction The IgG TrueBlot for mouse, rabbit, or goat-derived antibodies represents unique series of respective
IgG TrueBlot Protocol for Mouse, Rabbit or Goatderived Antibodies - For Research Use Only Introduction The IgG TrueBlot for mouse, rabbit, or goat-derived antibodies represents unique series of respective
Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity.
 Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity. (A) Amino acid alignment of HDA5, HDA15 and HDA18. The blue line
Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity. (A) Amino acid alignment of HDA5, HDA15 and HDA18. The blue line
Rab5 Activation Assay Kit
 A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Rab5 Activation Assay Kit Catalog Number: 83701 20 assays 24 Whitewoods Lane 1 Table of Content Product
A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Rab5 Activation Assay Kit Catalog Number: 83701 20 assays 24 Whitewoods Lane 1 Table of Content Product
Antibodies against PCNA were previously described [1]. To deplete PCNA from Xenopus egg
![Antibodies against PCNA were previously described [1]. To deplete PCNA from Xenopus egg Antibodies against PCNA were previously described [1]. To deplete PCNA from Xenopus egg](/thumbs/84/90687567.jpg) Supplementary information Supplementary methods PCNA antibody and immunodepletion Antibodies against PCNA were previously described [1]. To deplete PCNA from Xenopus egg extracts, one volume of protein
Supplementary information Supplementary methods PCNA antibody and immunodepletion Antibodies against PCNA were previously described [1]. To deplete PCNA from Xenopus egg extracts, one volume of protein
Supplementary information
 Supplementary information The E3 ligase RNF8 regulates KU80 removal and NHEJ repair Lin Feng 1, Junjie Chen 1 1 Department of Experimental Radiation Oncology, The University of Texas M. D. Anderson Cancer
Supplementary information The E3 ligase RNF8 regulates KU80 removal and NHEJ repair Lin Feng 1, Junjie Chen 1 1 Department of Experimental Radiation Oncology, The University of Texas M. D. Anderson Cancer
Fig. S1. CrgA intracellular levels in M. tuberculosis. Ten and twenty micrograms of
 Supplementary data Fig. S1. CrgA intracellular levels in M. tuberculosis. Ten and twenty micrograms of cell free protein lysates from WT M. tuberculosis (Rv) together with various known concentrations
Supplementary data Fig. S1. CrgA intracellular levels in M. tuberculosis. Ten and twenty micrograms of cell free protein lysates from WT M. tuberculosis (Rv) together with various known concentrations
Appendix. Table of contents
 Appendix Table of contents 1. Appendix figures 2. Legends of Appendix figures 3. References Appendix Figure S1 -Tub STIL (41) DVFFYQADDEHYIPR (55) (64) AVLLDLEPR (72) 1 451aa (1071) YLNENQLSQLSVTR (1084)
Appendix Table of contents 1. Appendix figures 2. Legends of Appendix figures 3. References Appendix Figure S1 -Tub STIL (41) DVFFYQADDEHYIPR (55) (64) AVLLDLEPR (72) 1 451aa (1071) YLNENQLSQLSVTR (1084)
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. FANCD2 Western blot. Lane 1 marker, lane 2 LN9SV, lane 3 LN9SV treated with hydroxyurea (HU), lane 4 GM6914, lane 5 GM6914 treated with HU, lane 6 VU697L,
Supplementary Figures Supplementary Figure 1. FANCD2 Western blot. Lane 1 marker, lane 2 LN9SV, lane 3 LN9SV treated with hydroxyurea (HU), lane 4 GM6914, lane 5 GM6914 treated with HU, lane 6 VU697L,
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or
 Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
1. Cross-linking and cell harvesting
 ChIP is a powerful tool that allows the specific matching of proteins or histone modifications to regions of the genome. Chromatin is isolated and antibodies to the antigen of interest are used to determine
ChIP is a powerful tool that allows the specific matching of proteins or histone modifications to regions of the genome. Chromatin is isolated and antibodies to the antigen of interest are used to determine
Supporting Information
 Supporting Information Su et al. 10.1073/pnas.1211604110 SI Materials and Methods Cell Culture and Plasmids. Tera-1 and Tera-2 cells (ATCC: HTB- 105/106) were maintained in McCoy s 5A medium with 15% FBS
Supporting Information Su et al. 10.1073/pnas.1211604110 SI Materials and Methods Cell Culture and Plasmids. Tera-1 and Tera-2 cells (ATCC: HTB- 105/106) were maintained in McCoy s 5A medium with 15% FBS
Nature Structural & Molecular Biology: doi: /nsmb.1583
 Acetylation by GCN5 regulates CDC6 phosphorylation in the S-phase of the cell cycle Roberta Paolinelli 1,2, Ramiro Mendoza-Maldonado 2, Anna Cereseto 1 and Mauro Giacca 2 1 Molecular Biology Laboratory,
Acetylation by GCN5 regulates CDC6 phosphorylation in the S-phase of the cell cycle Roberta Paolinelli 1,2, Ramiro Mendoza-Maldonado 2, Anna Cereseto 1 and Mauro Giacca 2 1 Molecular Biology Laboratory,
Culture media, trypsin, penicillin and streptomycin were from Invitrogen (Breda, the Netherlands).
 Methods Materials Culture media, trypsin, penicillin and streptomycin were from Invitrogen (Breda, the Netherlands). Bovine fibroblast growth factor (BFGF), thrombin, forskolin, IBMX, H-89, BAPTA-AM and
Methods Materials Culture media, trypsin, penicillin and streptomycin were from Invitrogen (Breda, the Netherlands). Bovine fibroblast growth factor (BFGF), thrombin, forskolin, IBMX, H-89, BAPTA-AM and
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table.
 Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Technical tips Session 4
 Technical tips Session 4 Biotinylation assay: Streptavidin is a small bacterial protein that binds with high affinity to the vitamin biotin. This streptavidin-biotin combination can be used to link molecules
Technical tips Session 4 Biotinylation assay: Streptavidin is a small bacterial protein that binds with high affinity to the vitamin biotin. This streptavidin-biotin combination can be used to link molecules
Supplemental Data. LMO4 Controls the Balance between Excitatory. and Inhibitory Spinal V2 Interneurons
 Neuron, Volume 61 Supplemental Data LMO4 Controls the Balance between Excitatory and Inhibitory Spinal V2 Interneurons Kaumudi Joshi, Seunghee Lee, Bora Lee, Jae W. Lee, and Soo-Kyung Lee Supplemental
Neuron, Volume 61 Supplemental Data LMO4 Controls the Balance between Excitatory and Inhibitory Spinal V2 Interneurons Kaumudi Joshi, Seunghee Lee, Bora Lee, Jae W. Lee, and Soo-Kyung Lee Supplemental
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.
 Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
LINGO-1, A TRANSMEMBRANE SIGNALING PROTEIN, INHIBITS OLIGODENDROCYTE DIFFERENTIATION AND MYELINATION THROUGH INTERCELLULAR SELF- INTERACTIONS.
 Supplemental Data: LINGO-1, A TRANSMEMBRANE SIGNALING PROTEIN, INHIBITS OLIGODENDROCYTE DIFFERENTIATION AND MYELINATION THROUGH INTERCELLULAR SELF- INTERACTIONS. Scott Jepson, Bryan Vought, Christian H.
Supplemental Data: LINGO-1, A TRANSMEMBRANE SIGNALING PROTEIN, INHIBITS OLIGODENDROCYTE DIFFERENTIATION AND MYELINATION THROUGH INTERCELLULAR SELF- INTERACTIONS. Scott Jepson, Bryan Vought, Christian H.
SUPPLEMENTAL MATERIAL. Supplemental Methods:
 SUPPLEMENTAL MATERIAL Supplemental Methods: Immunoprecipitation- As we described but with some modifications [22]. As part of another ongoing project, lysate from human umbilical vein endothelial cells
SUPPLEMENTAL MATERIAL Supplemental Methods: Immunoprecipitation- As we described but with some modifications [22]. As part of another ongoing project, lysate from human umbilical vein endothelial cells
Sequence of C-terminal tail regions of myosin heavy chains in class XI of Nicotiana benthamiana (Nb
 Fig. S1 Sequence of C-terminal tail regions of myosin heavy chains in class XI of Nicotiana benthamiana (Nb myosin XI-2, -F and K) and BY-2 cell (Nt 170-kD myosin and Nt 175-kD myosin). Amino acids identical
Fig. S1 Sequence of C-terminal tail regions of myosin heavy chains in class XI of Nicotiana benthamiana (Nb myosin XI-2, -F and K) and BY-2 cell (Nt 170-kD myosin and Nt 175-kD myosin). Amino acids identical
Supplementary Material - Methods
 Novel Protein-Protein Interactions in the Schizophrenia interactome Supplementary Material - Methods Experimental validations of predicted interactions Table S1-1: Protein pairs that were validated by
Novel Protein-Protein Interactions in the Schizophrenia interactome Supplementary Material - Methods Experimental validations of predicted interactions Table S1-1: Protein pairs that were validated by
Supporting Online Material for
 Supporting Online Material for Spatiotemporal dynamics of Aurora B-PLK1-MCAK signaling axis orchestrates kinetochore bi-orientation and faithful chromosome segregation Hengyi Shao, Yuejia Huang, Liangyu
Supporting Online Material for Spatiotemporal dynamics of Aurora B-PLK1-MCAK signaling axis orchestrates kinetochore bi-orientation and faithful chromosome segregation Hengyi Shao, Yuejia Huang, Liangyu
Supplementary Protocol. sirna transfection methodology and performance
 Supplementary Protocol sirna transfection methodology and performance sirna oligonucleotides, DNA construct and cell line. Chemically synthesized 21 nt RNA duplexes were obtained from Ambion Europe, Ltd.
Supplementary Protocol sirna transfection methodology and performance sirna oligonucleotides, DNA construct and cell line. Chemically synthesized 21 nt RNA duplexes were obtained from Ambion Europe, Ltd.
Supplementary Information: Materials and Methods. Immunoblot and immunoprecipitation. Cells were washed in phosphate buffered
 Supplementary Information: Materials and Methods Immunoblot and immunoprecipitation. Cells were washed in phosphate buffered saline (PBS) and lysed in TNN lysis buffer (50mM Tris at ph 8.0, 120mM NaCl
Supplementary Information: Materials and Methods Immunoblot and immunoprecipitation. Cells were washed in phosphate buffered saline (PBS) and lysed in TNN lysis buffer (50mM Tris at ph 8.0, 120mM NaCl
Anti-HB-EGF (Human) mab
 Page 1 For Research Use Only. Not for use in diagnostic procedures. CODE No. D308-3 Anti-HB-EGF (Human) mab CLONALITY CLONE ISOTYPE QUANTITY SOURCE IMMUNOGEN FORMURATION STORAGE Monoclonal 3H4 Mouse IgG1
Page 1 For Research Use Only. Not for use in diagnostic procedures. CODE No. D308-3 Anti-HB-EGF (Human) mab CLONALITY CLONE ISOTYPE QUANTITY SOURCE IMMUNOGEN FORMURATION STORAGE Monoclonal 3H4 Mouse IgG1
Supplementary information, Figure S1A ShHTL7 interacted with MAX2 but not another F-box protein COI1.
 GR24 (μm) 0 20 0 20 GST-ShHTL7 anti-gst His-MAX2 His-COI1 PVDF staining Supplementary information, Figure S1A ShHTL7 interacted with MAX2 but not another F-box protein COI1. Pull-down assays using GST-ShHTL7
GR24 (μm) 0 20 0 20 GST-ShHTL7 anti-gst His-MAX2 His-COI1 PVDF staining Supplementary information, Figure S1A ShHTL7 interacted with MAX2 but not another F-box protein COI1. Pull-down assays using GST-ShHTL7
Supplemental Material
 Supplemental Material 1 Figure S1. Phylogenetic analysis of Cep72 and Lrrc36, comparative localization of Cep72 and Lrrc36 and Cep72 antibody characterization (A) Phylogenetic alignment of Cep72 and Lrrc36
Supplemental Material 1 Figure S1. Phylogenetic analysis of Cep72 and Lrrc36, comparative localization of Cep72 and Lrrc36 and Cep72 antibody characterization (A) Phylogenetic alignment of Cep72 and Lrrc36
Supporting Information
 Supporting Information Krieg et al. 10.1073/pnas.0907131106 SI Text Reagents. Recombinant human TNF- was from Peprotech. Monoclonal rat anti-rip2 was purchased from Alexis, whereas monoclonal mouse anti-xiap
Supporting Information Krieg et al. 10.1073/pnas.0907131106 SI Text Reagents. Recombinant human TNF- was from Peprotech. Monoclonal rat anti-rip2 was purchased from Alexis, whereas monoclonal mouse anti-xiap
Otefin, a Nuclear Membrane Protein, Determines
 Developmental Cell 14 Supplemental Data Otefin, a Nuclear Membrane Protein, Determines the Fate of Germline Stem Cells in Drosophila via Interaction with Smad Complexes Xiaoyong Jiang, Laixin Xia, Dongsheng
Developmental Cell 14 Supplemental Data Otefin, a Nuclear Membrane Protein, Determines the Fate of Germline Stem Cells in Drosophila via Interaction with Smad Complexes Xiaoyong Jiang, Laixin Xia, Dongsheng
The preparation of native chromatin from cultured human cells.
 Native chromatin immunoprecipitation protocol The preparation of native chromatin from cultured human cells. All solutions need to be ice cold. Sucrose containing solutions must be made up fresh on the
Native chromatin immunoprecipitation protocol The preparation of native chromatin from cultured human cells. All solutions need to be ice cold. Sucrose containing solutions must be made up fresh on the
Figure S1. Verification of ihog Mutation by Protein Immunoblotting Figure S2. Verification of ihog and boi
 Figure S1. Verification of ihog Mutation by Protein Immunoblotting Extracts from S2R+ cells, embryos, and adults were analyzed by immunoprecipitation and immunoblotting with anti-ihog antibody. The Ihog
Figure S1. Verification of ihog Mutation by Protein Immunoblotting Extracts from S2R+ cells, embryos, and adults were analyzed by immunoprecipitation and immunoblotting with anti-ihog antibody. The Ihog
Supplementary information
 Supplementary information Table of Content: Supplementary Results... 2 Supplementary Figure S1: Experimental validation of AP-MS results by coimmunprecipitation Western blot analysis.... 3 Supplementary
Supplementary information Table of Content: Supplementary Results... 2 Supplementary Figure S1: Experimental validation of AP-MS results by coimmunprecipitation Western blot analysis.... 3 Supplementary
Supplementary Fig.1 Luton
 Supplementary Fig.1 Luton a 175 Brain Thymus Spleen Small Intestine Kidney Testis HeLa b 250 Lung Kidney MDCK c EFA6B si Control si Mismatch #637 #1564 #1770 83 62 47.5 175 IB: anti-efa6b #B1 130 66 Lysates
Supplementary Fig.1 Luton a 175 Brain Thymus Spleen Small Intestine Kidney Testis HeLa b 250 Lung Kidney MDCK c EFA6B si Control si Mismatch #637 #1564 #1770 83 62 47.5 175 IB: anti-efa6b #B1 130 66 Lysates
Motivation From Protein to Gene
 MOLECULAR BIOLOGY 2003-4 Topic B Recombinant DNA -principles and tools Construct a library - what for, how Major techniques +principles Bioinformatics - in brief Chapter 7 (MCB) 1 Motivation From Protein
MOLECULAR BIOLOGY 2003-4 Topic B Recombinant DNA -principles and tools Construct a library - what for, how Major techniques +principles Bioinformatics - in brief Chapter 7 (MCB) 1 Motivation From Protein
Histone H3 Methylation Antibody Panel Pack I - Active Genes Base Catalog # C Component Size Shipping Temperature
 Histone H3 Methylation Antibody Panel Pack I - Active Genes Base Catalog # PACK CONTENTS Component Size Shipping Temperature Upon Receipt Checklist 3K4D Histone H3K4me2 (H3K4 Dimethyl) Polyclonal Antibody
Histone H3 Methylation Antibody Panel Pack I - Active Genes Base Catalog # PACK CONTENTS Component Size Shipping Temperature Upon Receipt Checklist 3K4D Histone H3K4me2 (H3K4 Dimethyl) Polyclonal Antibody
Four different active promoter genes were chosen, ATXN7L2, PSRC1, CELSR2 and
 SUPPLEMENTARY MATERIALS AND METHODS Chromatin Immunoprecipitation for qpcr analysis Four different active promoter genes were chosen, ATXN7L2, PSRC1, CELSR2 and IL24, all located on chromosome 1. Primer
SUPPLEMENTARY MATERIALS AND METHODS Chromatin Immunoprecipitation for qpcr analysis Four different active promoter genes were chosen, ATXN7L2, PSRC1, CELSR2 and IL24, all located on chromosome 1. Primer
Supplementary Figure 1. Immunoprecipitation of synthetic SUMOm-remnant peptides using UMO monoclonal antibody. (a) LC-MS analyses of tryptic
 Supplementary Figure 1. Immunoprecipitation of synthetic SUMOm-remnant peptides using UMO 1-7-7 monoclonal antibody. (a) LC-MS analyses of tryptic digest from HEK293 cells spiked with 6 SUMOmremnant peptides
Supplementary Figure 1. Immunoprecipitation of synthetic SUMOm-remnant peptides using UMO 1-7-7 monoclonal antibody. (a) LC-MS analyses of tryptic digest from HEK293 cells spiked with 6 SUMOmremnant peptides
Supplementary Materials
 Supplementary Materials Supplementary Figure 1. PKM2 interacts with MLC2 in cytokinesis. a, U87, U87/EGFRvIII, and HeLa cells in cytokinesis were immunostained with DAPI and an anti-pkm2 antibody. Thirty
Supplementary Materials Supplementary Figure 1. PKM2 interacts with MLC2 in cytokinesis. a, U87, U87/EGFRvIII, and HeLa cells in cytokinesis were immunostained with DAPI and an anti-pkm2 antibody. Thirty
RhoC Activation Assay Kit
 Product Manual RhoC Activation Assay Kit Catalog Number STA-403-C 20 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Small GTP-binding proteins (or GTPases) are a family
Product Manual RhoC Activation Assay Kit Catalog Number STA-403-C 20 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Small GTP-binding proteins (or GTPases) are a family
indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) B. Male mice of
 SUPPLEMENTRY FIGURE LEGENDS Figure S1. USP44 loss leads to chromosome missegregation.. Genotypes obtained from the indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) 13.5.. Male mice
SUPPLEMENTRY FIGURE LEGENDS Figure S1. USP44 loss leads to chromosome missegregation.. Genotypes obtained from the indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) 13.5.. Male mice
7.06 Problem Set #3, 2006
 7.06 Problem Set #3, 2006 1. You are studying the EGF/Ras/MAPK pathway in cultured cells. When the pathway is activated, cells are signaled to proliferate. You generate various mutants described below.
7.06 Problem Set #3, 2006 1. You are studying the EGF/Ras/MAPK pathway in cultured cells. When the pathway is activated, cells are signaled to proliferate. You generate various mutants described below.
Supplemental Information
 Supplemental Information ATP-dependent unwinding of U4/U6 snrnas by the Brr2 helicase requires the C-terminus of Prp8 Corina Maeder 1,3, Alan K. Kutach 1,2,3, and Christine Guthrie 1 1 Department of Biochemistry
Supplemental Information ATP-dependent unwinding of U4/U6 snrnas by the Brr2 helicase requires the C-terminus of Prp8 Corina Maeder 1,3, Alan K. Kutach 1,2,3, and Christine Guthrie 1 1 Department of Biochemistry
SANTA CRUZ BIOTECHNOLOGY, INC.
 TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
Histone H3K27 Methylation Antibody Panel Pack Base Catalog # C Component Size Shipping Temperature
 Histone H3K27 Methylation Antibody Panel Pack Base Catalog # PACK CONTENTS Component Size Shipping Temperature Upon Receipt Checklist 3K27M Histone H3K27me1 (H3K27 Monomethyl) Polyclonal Antibody 25 µg
Histone H3K27 Methylation Antibody Panel Pack Base Catalog # PACK CONTENTS Component Size Shipping Temperature Upon Receipt Checklist 3K27M Histone H3K27me1 (H3K27 Monomethyl) Polyclonal Antibody 25 µg
CELL BIOLOGY - CLUTCH CH TECHNIQUES IN CELL BIOLOGY.
 !! www.clutchprep.com CONCEPT: LIGHT MICROSCOPE A light microscope uses light waves and magnification to view specimens Can be used to visualize transparent, and some cellular components - Can visualize
!! www.clutchprep.com CONCEPT: LIGHT MICROSCOPE A light microscope uses light waves and magnification to view specimens Can be used to visualize transparent, and some cellular components - Can visualize
Transcriptional regulation of BRCA1 expression by a metabolic switch: Di, Fernandez, De Siervi, Longo, and Gardner. H3K4Me3
 ChIP H3K4Me3 enrichment.25.2.15.1.5 H3K4Me3 H3K4Me3 ctrl H3K4Me3 + E2 NS + E2 1. kb kb +82 kb Figure S1. Estrogen promotes entry of MCF-7 into the cell cycle but does not significantly change activation-associated
ChIP H3K4Me3 enrichment.25.2.15.1.5 H3K4Me3 H3K4Me3 ctrl H3K4Me3 + E2 NS + E2 1. kb kb +82 kb Figure S1. Estrogen promotes entry of MCF-7 into the cell cycle but does not significantly change activation-associated
Comparison of different methods for purification analysis of a green fluorescent Strep-tag fusion protein. Application
 Comparison of different methods for purification analysis of a green fluorescent Strep-tag fusion protein Application Petra Sebastian Meike Kuschel Stefan Schmidt Abstract This Application Note describes
Comparison of different methods for purification analysis of a green fluorescent Strep-tag fusion protein Application Petra Sebastian Meike Kuschel Stefan Schmidt Abstract This Application Note describes
Immunofluorescence Confocal Microscopy of 3D Cultures Grown on Alvetex
 Immunofluorescence Confocal Microscopy of 3D Cultures Grown on Alvetex 1.0. Introduction Immunofluorescence uses the recognition of cellular targets by fluorescent dyes or antigen-specific antibodies coupled
Immunofluorescence Confocal Microscopy of 3D Cultures Grown on Alvetex 1.0. Introduction Immunofluorescence uses the recognition of cellular targets by fluorescent dyes or antigen-specific antibodies coupled
Mechanism of Induction and Suppression of Antiviral Immunity Directed by Virus-Derived Small RNAs in Drosophila
 Cell Host & Microbe, Volume 4 Supplemental Data Mechanism of Induction and Suppression of Antiviral Immunity Directed by Virus-Derived Small RNAs in Drosophila Roghiyh Aliyari, Qingfa Wu, Hong-Wei Li,
Cell Host & Microbe, Volume 4 Supplemental Data Mechanism of Induction and Suppression of Antiviral Immunity Directed by Virus-Derived Small RNAs in Drosophila Roghiyh Aliyari, Qingfa Wu, Hong-Wei Li,
Separose 4 Fast Flow beads stored in 100% isopropanol (Amersham. Bioscience) were washed four times with 1 mm HCl and twice with coupling buffer
 Supporting Information Materials and Methods Affinity chromatography. Three-hundred microliters of N-hydroxysuccinimideactivated Separose 4 Fast Flow beads stored in 100% isopropanol (Amersham Bioscience)
Supporting Information Materials and Methods Affinity chromatography. Three-hundred microliters of N-hydroxysuccinimideactivated Separose 4 Fast Flow beads stored in 100% isopropanol (Amersham Bioscience)
supplementary information
 DOI: 10.1038/ncb2172 Figure S1 p53 regulates cellular NADPH and lipid levels via inhibition of G6PD. (a) U2OS cells stably expressing p53 shrna or a control shrna were transfected with control sirna or
DOI: 10.1038/ncb2172 Figure S1 p53 regulates cellular NADPH and lipid levels via inhibition of G6PD. (a) U2OS cells stably expressing p53 shrna or a control shrna were transfected with control sirna or
RNA was isolated using NucleoSpin RNA II (Macherey-Nagel, Bethlehem, PA) according to the
 Supplementary Methods RT-PCR and real-time PCR analysis RNA was isolated using NucleoSpin RNA II (Macherey-Nagel, Bethlehem, PA) according to the manufacturer s protocol and quantified by measuring the
Supplementary Methods RT-PCR and real-time PCR analysis RNA was isolated using NucleoSpin RNA II (Macherey-Nagel, Bethlehem, PA) according to the manufacturer s protocol and quantified by measuring the
Replication-independent chromatin loading of Dnmt1 during G2 and M phases
 Replication-independent chromatin loading of Dnmt1 during G2 and M phases Hariharan P. Easwaran 1, Lothar Schermelleh 2, Heinrich Leonhardt 1,2,* and M. Cristina Cardoso 1,* 1 Max Delbrück Center for Molecular
Replication-independent chromatin loading of Dnmt1 during G2 and M phases Hariharan P. Easwaran 1, Lothar Schermelleh 2, Heinrich Leonhardt 1,2,* and M. Cristina Cardoso 1,* 1 Max Delbrück Center for Molecular
ab G alpha i Activation Assay Kit
 ab173234 G alpha i Activation Assay Kit Instructions for Use For the simple and fast measurement of G alpha i activation. This product is for research use only and is not intended for diagnostic use. Version
ab173234 G alpha i Activation Assay Kit Instructions for Use For the simple and fast measurement of G alpha i activation. This product is for research use only and is not intended for diagnostic use. Version
Lecture 7: Affinity Chromatography-II
 Lecture 7: Affinity Chromatography-II We have studied basics of affinity purification during last lecture. The current lecture is continuation of last lecture and we will cover following: 1. Few specific
Lecture 7: Affinity Chromatography-II We have studied basics of affinity purification during last lecture. The current lecture is continuation of last lecture and we will cover following: 1. Few specific
Identification of Microprotein-Protein Interactions via APEX Tagging
 Supporting Information Identification of Microprotein-Protein Interactions via APEX Tagging Qian Chu, Annie Rathore,, Jolene K. Diedrich,, Cynthia J. Donaldson, John R. Yates III, and Alan Saghatelian
Supporting Information Identification of Microprotein-Protein Interactions via APEX Tagging Qian Chu, Annie Rathore,, Jolene K. Diedrich,, Cynthia J. Donaldson, John R. Yates III, and Alan Saghatelian
Supplementary methods Shoc2 In Vitro Ubiquitination Assay
 Supplementary methods Shoc2 In Vitro Ubiquitination Assay 35 S-labelled Shoc2 was prepared using a TNT quick Coupled transcription/ translation System (Promega) as recommended by manufacturer. For the
Supplementary methods Shoc2 In Vitro Ubiquitination Assay 35 S-labelled Shoc2 was prepared using a TNT quick Coupled transcription/ translation System (Promega) as recommended by manufacturer. For the
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the
 Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
Supplementary methods
 Supplementary methods Cell culture, infection, transfection, and RNA interference HEK293 cells and its derivatives were grown in DMEM supplemented with 10% FBS. Various constructs were introduced into
Supplementary methods Cell culture, infection, transfection, and RNA interference HEK293 cells and its derivatives were grown in DMEM supplemented with 10% FBS. Various constructs were introduced into
DDA3 associates with microtubule plus ends and orchestrates microtubule dynamics and directional cell migration
 DDA3 associates with microtubule plus ends and orchestrates microtubule dynamics and directional cell migration Liangyu Zhang 1,2,4, Hengyi Shao 1,4, Tongge Zhu, Peng Xia 1, Zhikai Wang 1,2, Lifang Liu
DDA3 associates with microtubule plus ends and orchestrates microtubule dynamics and directional cell migration Liangyu Zhang 1,2,4, Hengyi Shao 1,4, Tongge Zhu, Peng Xia 1, Zhikai Wang 1,2, Lifang Liu
Schematic representation of the endogenous PALB2 locus and gene-disruption constructs
 Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
pt7ht vector and over-expressed in E. coli as inclusion bodies. Cells were lysed in 6 M
 Supplementary Methods MIG6 production, purification, inhibition, and kinase assays MIG6 segment 1 (30mer, residues 334 364) peptide was synthesized using standard solid-phase peptide synthesis as described
Supplementary Methods MIG6 production, purification, inhibition, and kinase assays MIG6 segment 1 (30mer, residues 334 364) peptide was synthesized using standard solid-phase peptide synthesis as described
ENCODE DCC Antibody Validation Document
 ENCODE DCC Antibody Validation Document Date of Submission 09/12/12 Name: Trupti Kawli Email: trupti@stanford.edu Lab Snyder Antibody Name: SREBP1 (sc-8984) Target: SREBP1 Company/ Source: Santa Cruz Biotechnology
ENCODE DCC Antibody Validation Document Date of Submission 09/12/12 Name: Trupti Kawli Email: trupti@stanford.edu Lab Snyder Antibody Name: SREBP1 (sc-8984) Target: SREBP1 Company/ Source: Santa Cruz Biotechnology
Protocol for in vitro transcription
 Protocol for in vitro transcription Assemble the reaction at room temperature in the following order: Component 10xTranscription Buffer rntp T7 RNA Polymerase Mix grna PCR DEPC H 2 O volume 2μl 2μl 2μl
Protocol for in vitro transcription Assemble the reaction at room temperature in the following order: Component 10xTranscription Buffer rntp T7 RNA Polymerase Mix grna PCR DEPC H 2 O volume 2μl 2μl 2μl
Protocol(Research use only)
 Immunohistochemistry (without pretreatment) p2 Immunohistochemistry (Microwave pretreatment) p3 Immunohistochemistry (Autoclave pretreatment) p4 Immunohistochemistry (Trypsin pretreatment) p5 Immunohistochemistry
Immunohistochemistry (without pretreatment) p2 Immunohistochemistry (Microwave pretreatment) p3 Immunohistochemistry (Autoclave pretreatment) p4 Immunohistochemistry (Trypsin pretreatment) p5 Immunohistochemistry
SUPPLEMENTARY INFORMATION FIGURE 1 - 1
 SUPPLEMENTARY INFORMATION FIGURE 1-1 SUPPLEMENTARY INFORMATION FIGURE 2-2 SUPPLEMENTARY INFORMATION METHODS GST-Pull-Down. Cultures of E. Coli (BL21) were transformed with pgex (Clontech) and pgex recombinant
SUPPLEMENTARY INFORMATION FIGURE 1-1 SUPPLEMENTARY INFORMATION FIGURE 2-2 SUPPLEMENTARY INFORMATION METHODS GST-Pull-Down. Cultures of E. Coli (BL21) were transformed with pgex (Clontech) and pgex recombinant
Checkpoint Kinase Activity Immunoblot Kit
 Product Manual Checkpoint Kinase Activity Immunoblot Kit Catalog Number STA- 413 20 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Cdc25C is a protein phosphatase responsible
Product Manual Checkpoint Kinase Activity Immunoblot Kit Catalog Number STA- 413 20 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Cdc25C is a protein phosphatase responsible
