Quantitative comparison of different fluorescent protein couples for fast FRET-FLIM acquisition
|
|
|
- Gordon Stevens
- 6 years ago
- Views:
Transcription
1 Biophysical Journal, Volume 97 Supporting Material Quantitative comparison of different fluorescent protein couples for fast FRET-FLIM acquisition Sergi Padilla, Nicolas Audugé, Hervé Lalucque, Jean Claude Mevel, Maïté Coppey-Moisan, and Marc Tramier 1
2 SUPPLEMENTARY MATERIALS MATERIALS AN METHOS Plasmid constructs The plasmids coding the mcherry-egfp or EGFP-EGFP tandems have been described elsewhere (1). The plasmid coding the mstrawberry-egfp tandem was obtained thanks to the same strategy used for mcherry-egfp (1). In short, the EGFP fragment from pegfp-c1 (Clontech, city, USA) is replaced by the mstrawberry coding sequence from prsetmstrawberry (generous gift of r. R. Tsien, UCS, USA) using the BsrGI and NheI sites, leading to pmstrawberry-c1. The first EGFP sequence in pbunny (tandem EGFP-EGFP, (1)) is then replaced by the mstrawberry sequence from pmstrawberry-c1 using NheI and BspEI sites, giving the mstrawberry-egfp plasmid. The mtfp1-morange tandem, pmorange-n1 and pmtfp1-c1 plasmids are generous gifts from r O. Albagli (IGR, Villejuif, France). To obtain the mtfp1-egfp plasmid, the first EGFP sequence in pbunny (tandem EGFP-EGFP, (1)) is substituted by the mtfp1 coding sequence from pmtfp1-c1 using NheI and BspEI sites. The mtfp1-eyfp plasmid is constructed by replacing the EGFP fragment from mtfp1-egfp plasmid by the EYFP sequence from peyfp-n1 (Clontech) taking advantage of BamHI and NotI sites. To construct the plasmid coding the Halotag-EGFP tandem using PCR, the sequence containing the linker and EGFP are amplified by PCR from mcherry- EGFP with the primers CTGGCCGGCATGGACGAGCTGTACAAGTCC-3' (the underlined NaeI site is added to facilitate the cloning) and 5'-CTCTACAAATGTGGTATGGC-3'. The PCR product is then digested by NaeI and NotI and cloned into the Halotag pht vector (Promega) hydrolysed by the same enzymes, leading to Halotag-EGFP plasmid. The constructions coding for the two tandems between EGFP and tdimer(1) or mrfp1 (), named TdRed-EGFP or mrfp1-egfp respectively, are generous gifts from r S. Ahmed (Center for Molecular Medicine, Biopolis Singapore). The cloning strategy enables us to maintain the same peptide linker (SGLRSRGPPVAT) between the FRET partners for the mcherry-egfp, EGFP-EGFP, mstrawberry-egfp, mtfp1-egfp, mtfp1-eyfp and Halotag-EGFP tandems. The plasmids coding for EGFP-H4 and mtfp1-h4 were obtained by cloning histone H4 cna (IMAGE:130477) in pegfp-c1 (Clontech) and pmtfp1-c1 using EcoRI and KpnI sites of the MCS. Lifetime data analysis Using the time domain fluorescence lifetime microscope, the acquired fluorescence decays were deconvoluted with the instrument response function and fitted by a Marquardt nonlinear least-square algorithm using Globals Unlimited software (Laboratory for Fluorescence ynamics; University of California, Irvine). Three different approaches were considered when performing the fits: (i) A two species model in which two populations are taken into consideration (an interacting fraction corresponding to a population which relaxes through FRET (f ) and a non-interacting fraction in which the donor lifetime remains undisturbed (1-f )). In this case the donor lifetime obtained out of the single exponential fit from cells expressing the donor alone was
3 assigned and fixed into the non-interacting fraction of the double exponential model for cells expressing the corresponding tandem (3,4). i( t) = (1 f ) e + f e t /τ t /τ F (Eq.S1) In Eq.1 f stands for the fraction of interacting donor, τ is the fixed donor lifetime and τ F is the discrete FRET lifetime. (ii) Using the same formalism, it was also considered that instead of a discrete FRET lifetime, a distribution of lifetimes could arise from the different orientations between donor and acceptors. In this case we used a stretched exponential approach (5,6) for the FRET lifetime and discrete for the donor. β t / τ ( t / τ F ) ( t) = (1 f ) e + f e i (Eq.S) All parameters in Eq. are already defined in Eq.1 but the coefficient β, which is related to the heterogeneity parameter of the sample. (iii) The last considered approach was to leave both lifetimes free, still considering a discrete double exponential for the tandem. This system would account for the possibility of having two discrete FRET lifetimes out of the interaction between donor and acceptor (6). i( t) = A e + A e t / τ1 t / τ 1 (Eq.S3) In Eq.3 A 1 and A are the pre-exponential factors (which in turn are related to the fraction of each FRET population) and τ 1 and τ are the two FRET lifetimes. For conventional bi-exponential analysis using fixed donor lifetime, the FRET efficiency, E, was calculated as follows: E = 1 τ (Eq.S4) F τ where τ was the fixed fluorescence lifetime of the donor, and τ F the shortened FRET lifetime. The mathematical approach in order to recover the minimal fraction of interacting donor (mf ) out of the diminution of donor mean lifetime alone and in the presence of acceptor was described elsewhere (4). Briefly, considering a two species system with single exponential 3
4 donor and taking into account the definition of mean lifetime, the fraction of donor in interaction (f ) can be defined as f = [1 ( < τ > / τ )] [1 ( < τ > / τ ) ( τ / τ ) + ( < τ > / τ ) ( τ F / τ )] (Eq.S5) In which <τ> represents the mean lifetime, τ stands for the donor lifetime and τ F is the FRET lifetime. If we represent f as a function of τ F /τ and <τ>/τ, a minimal value of f described for each value of τ F /τ is revealed. The function which describes this minimum is the minimal fraction of donor undergoing FRET (mf ). This means that f can be minimized following τ F /τ. The partial derivative ( f / (τ F/ τ )) is zero for τ F = <τ>/. Replacing τ F by <τ>/ in Eq.5 gives mf = [ 1 ( < τ > / τ )] [( < τ > / τ ) 1] (Eq.S6) Analysis of the data was done using imagej (Rasband, W.S., ImageJ, U. S. National Institutes of Health, Bethesda, Maryland, USA, Raw images were first smoothened by a 3x3 mask to decrease the noise. Calculation of the mean lifetime (4) was applied on background subtracted stacks of time-gated images. Eq.6 was then applied on mean lifetime images using fixed value of τ to recover mf. F RESULTS f is influenced by the difference between donor and acceptor maturation rates In order to find a possible explanation for the low f values recovered for most of the tandems analyzed we hypothesized that differences in donor and acceptor FP maturation rates might play a role. ifferent maturation rates for different FP s would lead to different situations, incomplete maturation of the donor, incomplete maturation of the acceptor or incomplete maturation of both, donor and acceptor. We are only concerned for the second situation since FRET-FLIM focuses on the variation of the donor fluorescence decay profile. Since tandems are constantly produced in the cell, we designed an experiment in which protein production is stopped, so that all tandems have the time to maturate. We tested cells expressing mcherry-egfp tandem treated with cycloheximide (100 μg/ml) for 1hr, hr and 4hr, respectively. Cycloheximide is known to block protein synthesis (7). We first controlled that cycloheximide did not affect EGFP lifetime by treating cells expressing EGFP alone for 1h, h and 4h. For all these conditions, the lifetime remained always at.60+/-0.01 ns (n = 30). Fig.S1 shows a representative experiment in which the fluorescence decay profile of EGFP alone is compared to the tandem mcherry-egfp without drug and tandem mcherry-egfp treated for hours. It can be observed that FRET occurs for both conditions, as expected. But for the cell treated with cycloheximide the fluorescence decay is much faster than the cell without drug. This clearly shows the influence of cycloheximide upon f : 0.50+/-0.0 (n = 10) for 1hr treatment, 0.53+/-0.0 (n = 10) for hr treatment and 0.55+/-0.0 (n = 10) for 4hr treatment, to be compared to 0.45+/-0.0 (n = 10) without cycloheximide (Table 1). Slight increase in f is observed meaning that the difference in maturation rate plays a role in the fraction of unobserved acceptor. Note that E remained always constant for all experiments: E (mcherry-egfp) = 0.58+/-0.0, E (mcherry-egfp (1h treatment) = 0.56+/-0.08 (n = 10), E (mcherry-egfp (h treatment) = 0.58+/-0.05 (n = 10) and E (mcherry-egfp (4h treatment) = 0.58+/-0.04 (n = 10). 4
5 Figure S1. Effect of protein maturation on f determination. Middle panel: Fluorescence decay profiles for EGFP alone (green decays), mcherry-egfp tandem with (dark red decays) or without (red decay) cycloheximide, in live He-La cells using a TSCSPC system, are fitted (black lines) with a single exponential (for EGFP alone) and a discrete double exponential (for tandem). The residues of these fits are presented in the upper panel. The lower panel focuses the beginning of the fluorescence decays (black square in the middle panel). Confidence plot of the stretched exponential analysis Two confidence plots using a double exponential stretched fit for mcherry-egfp and mtfp1-eyfp are shown in Fig.S (discrete lifetime for the donor (τ ) and stretched lifetime for FRET (β, τ F )). For these two tandems only mtfp1-eyfp presents a minimal chi-square (χ ) as a function of β, which is not the case for mcherry-egfp. Interestingly, in the case of other tandems like mrfp1-egfp, mstrawberry-egfp, HaloTag(TMR)-EGFP and mtfp1- morange exhibiting also a low f values, it was not possible to find a minimum χ as a function of β. The use of a stretched exponential that describes and quantifies FRET is here 5
6 unclear since χ behaves linearly for values 0.7 < β < 1. The confidence plot for mtfp1- EYFP, however, presented a minimum (β = 0.7 +/-0.01, n = 6) suggesting that, in this case, the stretched exponential analysis is an alternative to the discrete double exponential model (Table ). Moreover, high f values (close to 1) are found using the stretched exponential approach opening the discussion of the occurrence and extent of a spectroscopic heterogeneity of the acceptor population and/or a certain distribution of the orientation between donors and acceptors. Figure S. Confidence plot of mtfp1-yfp and mcherry-egfp tandems using stretched exponential model. Confidence plot for mcherry-egfp (black circles) and mtfp1-eyfp (black squares) using a double exponential model, discrete for donor lifetime and stretched for the FRET lifetime. The confidence plot was obtained calculating the χ values fixing the stretched parameter (β) to 0.5, 0.6, 0.7, 0.8, 0.9 and 1. References 1. Tramier, M., M. Zahid, J.C. Mevel, M.J. Masse and M. Coppey-Moisan Sensitivity of CFP/YFP and GFP/mCherry pairs to donor photobleaching on FRET determination by fluorescence lifetime imaging microscopy in living cells. Microsc. Res. Tech. 11: Campbell R.E, O. Tour, A.E Palmer, P.A Steinbach, G.S. Baird,.A. Zacharias and R.Y. Tsien. 00. A monomeric red fluorescent protein. Proc Natl Acad Sci U S A. 99: Emiliani, V.,. Sanvitto, M. Tramier, T. Piolot, Z. Petrasek, K. Kemnitz, C. urieux, and M. Coppey-Moisan Low intensity two-dimensional imaging of fluorescence lifetimes in living cells. Appl. Phys. Lett. 83:
7 4. Padilla-Parra, S., N. Audugé, M. Coppey-Moisan and M. Tramier Quantitative FRET analysis by fast acquisition time domain FLIM at high spatial resolution in living cells. Biophys. J. 95: Lee, K.C., J. Siegel, S.E. Webb, S. Lévêque-Fort, M.J. Cole, R. Jones, K. owling, M.J. Lever and P.M. French Application of the stretched exponential function to fluorescence lifetime imaging. Biophys J. 81: Wu, B., Y. Chen and J.. Müller Fluorescence fluctuation spectroscopy of mcherry in living cells. Biophys. J. 96: Chu, C and A.J. Shatkin Apoptosis and autophagy induction in mammalian cells by small interfering RNA knockdown of mrna capping enzymes. Mol. Cell Biol. 8:
Quantitative Comparison of Different Fluorescent Protein Couples for Fast FRET-FLIM Acquisition
 2368 Biophysical Journal Volume 97 October 2009 2368 2376 Quantitative Comparison of Different Fluorescent Protein Couples for Fast FRET-FLIM Acquisition Sergi Padilla-Parra,* Nicolas Audugé, Hervé Lalucque,
2368 Biophysical Journal Volume 97 October 2009 2368 2376 Quantitative Comparison of Different Fluorescent Protein Couples for Fast FRET-FLIM Acquisition Sergi Padilla-Parra,* Nicolas Audugé, Hervé Lalucque,
Supplementary Figure S1. Np95 interacts with de novo methyltransferases Dnmt3a and 3b. (A) Co-immunoprecipitation of endogenous Dnmt3a2 (left and
 Supplementary Figure S1. Np95 interacts with de novo methyltransferases Dnmt3a and 3b. (A) Co-immunoprecipitation of endogenous Dnmt3a2 (left and right), Dnmt3b isoforms (left) and Dnmt1 (right) with GFP-Np95
Supplementary Figure S1. Np95 interacts with de novo methyltransferases Dnmt3a and 3b. (A) Co-immunoprecipitation of endogenous Dnmt3a2 (left and right), Dnmt3b isoforms (left) and Dnmt1 (right) with GFP-Np95
Creation of circularly permutated yellow fluorescent proteins using fluorescence screening and a tandem fusion template
 Biotechnology Letters (2006) 28: 471 475 Ó Springer 2006 DOI 10.1007/s10529-006-0007-6 Creation of circularly permutated yellow fluorescent proteins using fluorescence screening and a tandem fusion template
Biotechnology Letters (2006) 28: 471 475 Ó Springer 2006 DOI 10.1007/s10529-006-0007-6 Creation of circularly permutated yellow fluorescent proteins using fluorescence screening and a tandem fusion template
A quantitative protocol for intensity-based live cell FRET imaging.
 A quantitative protocol for intensity-based live cell FRET imaging. Kaminski CF, Rees EJ, Schierle GS. Methods Mol Biol. 2014; 1076:445-454. Department of Chemical Engineering and Biotechnology, Pembroke
A quantitative protocol for intensity-based live cell FRET imaging. Kaminski CF, Rees EJ, Schierle GS. Methods Mol Biol. 2014; 1076:445-454. Department of Chemical Engineering and Biotechnology, Pembroke
λ N -GFP: an RNA reporter system for live-cell imaging
 λ N -GFP: an RNA reporter system for live-cell imaging Nathalie Daigle & Jan Ellenberg Supplementary Figures and Text: Supplementary Figure 1 localization in the cytoplasm. 4 λ N22-3 megfp-m9 serves as
λ N -GFP: an RNA reporter system for live-cell imaging Nathalie Daigle & Jan Ellenberg Supplementary Figures and Text: Supplementary Figure 1 localization in the cytoplasm. 4 λ N22-3 megfp-m9 serves as
FRET measurement between YFP and CFP
 FRET measurement between YFP and CFP EYFP and ECFP function as a donor-acceptor pair for fluorescence resonance energy transfer (FRET), in which excitation of the donor (cyan) molecule leads to emission
FRET measurement between YFP and CFP EYFP and ECFP function as a donor-acceptor pair for fluorescence resonance energy transfer (FRET), in which excitation of the donor (cyan) molecule leads to emission
Supplementary information. for the paper:
 Supplementary information for the paper: Screening for protein-protein interactions using Förster resonance energy transfer (FRET) and fluorescence lifetime imaging microscopy (FLIM) Anca Margineanu 1*,
Supplementary information for the paper: Screening for protein-protein interactions using Förster resonance energy transfer (FRET) and fluorescence lifetime imaging microscopy (FLIM) Anca Margineanu 1*,
Design. Construction. Characterization
 Design Construction Characterization DNA mrna (messenger) A C C transcription translation C A C protein His A T G C T A C G Plasmids replicon copy number incompatibility selection marker origin of replication
Design Construction Characterization DNA mrna (messenger) A C C transcription translation C A C protein His A T G C T A C G Plasmids replicon copy number incompatibility selection marker origin of replication
F* techniques: FRAP, FLIP, FRET, FLIM,
 F* techniques: FRAP, FLIP, FRET, FLIM, FCS Antonia Göhler March 2015 Fluorescence explained in the Bohr model Absorption of light (blue) causes an electron to move to a higher energy orbit. After a particular
F* techniques: FRAP, FLIP, FRET, FLIM, FCS Antonia Göhler March 2015 Fluorescence explained in the Bohr model Absorption of light (blue) causes an electron to move to a higher energy orbit. After a particular
Chapter 4. the biological community to assay for protein-protein interactions. FRET describes the
 31 Chapter 4 Determination of nachr stoichiometry using Normalized Försters Resonance Energy Transfer (NFRET) Försters resonance energy transfer (FRET) has become a technique widely used in the biological
31 Chapter 4 Determination of nachr stoichiometry using Normalized Försters Resonance Energy Transfer (NFRET) Försters resonance energy transfer (FRET) has become a technique widely used in the biological
Supplementary Information
 Supplementary Information Supplementary Figures Figure S1. Study of mgtl translation in vitro. (A) Detection of 5 LR RNA using wild-type and anti-sd (91-95) substituted templates in a transcription-translation
Supplementary Information Supplementary Figures Figure S1. Study of mgtl translation in vitro. (A) Detection of 5 LR RNA using wild-type and anti-sd (91-95) substituted templates in a transcription-translation
Fluorescent protein. Origin of fluorescent proteins. Structural and biophysical analysis of FPs. Application in molecular biology and biotechnology
 Origin of fluorescent proteins types of FPs mechanism of fluorescence Fluorescent protein Structural and biophysical analysis of FPs Application in molecular biology and biotechnology Variety of organisms
Origin of fluorescent proteins types of FPs mechanism of fluorescence Fluorescent protein Structural and biophysical analysis of FPs Application in molecular biology and biotechnology Variety of organisms
Welcome! openmicberkeley.wordpress.com. Open Berkeley
 Welcome! openmicberkeley.wordpress.com Agenda Jen Lee: Introduction to FRET Marla Feller: Using FRET sensors to look at time resolved measurements Becky Lamason: Using FRET to determine if a bacterial
Welcome! openmicberkeley.wordpress.com Agenda Jen Lee: Introduction to FRET Marla Feller: Using FRET sensors to look at time resolved measurements Becky Lamason: Using FRET to determine if a bacterial
Workshop advanced light microscopy
 Workshop advanced light microscopy Multi-mode confocal laser scanning microscope Jan Willem Borst Laboratory of Biochemistry Biomolecular Networks www.bic.wur.nl MicroSpectroscopy Centre Wageningen Microspectroscopy
Workshop advanced light microscopy Multi-mode confocal laser scanning microscope Jan Willem Borst Laboratory of Biochemistry Biomolecular Networks www.bic.wur.nl MicroSpectroscopy Centre Wageningen Microspectroscopy
Fluorescent Protein Vectors
 Fluorescent Protein Vectors Living Colors vectors for easy cloning, expression, and subcellular localization applications Visualize a variety of targets with our subcellular localization vectors Establish
Fluorescent Protein Vectors Living Colors vectors for easy cloning, expression, and subcellular localization applications Visualize a variety of targets with our subcellular localization vectors Establish
Live-cell visualization of excitation energy dynamics in chloroplast thylakoid structures
 Supplementary Information Live-cell visualization of excitation energy dynamics in chloroplast thylakoid structures Masakazu Iwai, Makio Yokono, Kazuo Kurokawa, Akira Ichihara & Akihiko Nakano Supplementary
Supplementary Information Live-cell visualization of excitation energy dynamics in chloroplast thylakoid structures Masakazu Iwai, Makio Yokono, Kazuo Kurokawa, Akira Ichihara & Akihiko Nakano Supplementary
FLIM Fluorescence Lifetime IMaging
 FLIM Fluorescence Lifetime IMaging Fluorescence lifetime t I(t) = F0 exp( ) τ 1 τ = k f + k nr k nr = k IC + k ISC + k bl Batiaens et al, Trends in Cell Biology, 1999 τ τ = fluorescence lifetime (~ns to
FLIM Fluorescence Lifetime IMaging Fluorescence lifetime t I(t) = F0 exp( ) τ 1 τ = k f + k nr k nr = k IC + k ISC + k bl Batiaens et al, Trends in Cell Biology, 1999 τ τ = fluorescence lifetime (~ns to
Combining Techniques to Answer Molecular Questions
 Combining Techniques to Answer Molecular Questions UNIT FM02 How to cite this article: Curr. Protoc. Essential Lab. Tech. 9:FM02.1-FM02.5. doi: 10.1002/9780470089941.etfm02s9 INTRODUCTION This manual is
Combining Techniques to Answer Molecular Questions UNIT FM02 How to cite this article: Curr. Protoc. Essential Lab. Tech. 9:FM02.1-FM02.5. doi: 10.1002/9780470089941.etfm02s9 INTRODUCTION This manual is
Figure S1: Insert sequences for GR1, GR2, Qc3c_AgeI and GR3 vectors
 Figure S1: Insert sequences for GR1, GR2, Qc3c_AgeI and GR3 vectors A Vector GR1 SacI BamHI CTCTGGCTAACTAGGC Insert 5/7nt - G TCGAGAGACCGATTGATCCG Insert 5/7nt - CCTAG 1 G CAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAAC
Figure S1: Insert sequences for GR1, GR2, Qc3c_AgeI and GR3 vectors A Vector GR1 SacI BamHI CTCTGGCTAACTAGGC Insert 5/7nt - G TCGAGAGACCGATTGATCCG Insert 5/7nt - CCTAG 1 G CAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAAC
Supporting Information
 Supporting Information Development of a 2,4-Dinitrotoluene-Responsive Synthetic Riboswitch in E. coli cells Molly E. Davidson, Svetlana V. Harbaugh, Yaroslav G. Chushak, Morley O. Stone, Nancy Kelley-
Supporting Information Development of a 2,4-Dinitrotoluene-Responsive Synthetic Riboswitch in E. coli cells Molly E. Davidson, Svetlana V. Harbaugh, Yaroslav G. Chushak, Morley O. Stone, Nancy Kelley-
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10016 Supplementary discussion on binding site density for protein complexes on the surface: The density of biotin sites on the chip is ~10 3 biotin-peg per µm 2. The biotin sites are
doi:10.1038/nature10016 Supplementary discussion on binding site density for protein complexes on the surface: The density of biotin sites on the chip is ~10 3 biotin-peg per µm 2. The biotin sites are
SUPPLEMENTARY INFORMATION
 Clustering of a kinesin-14 motor enables processive retrograde microtubule-based transport in plants NATURE PLANTS www.nature.com/natureplants 1 2 NATURE PLANTS www.nature.com/natureplants NATURE PLANTS
Clustering of a kinesin-14 motor enables processive retrograde microtubule-based transport in plants NATURE PLANTS www.nature.com/natureplants 1 2 NATURE PLANTS www.nature.com/natureplants NATURE PLANTS
Supplementary Fig.1. Over-expression of RNase L in stable polyclonal cell line
 Supplemental Data mrna Protein A kda 75 40 NEO/vector NEO/RNase L RNASE L β-actin RNASE L β-actin B % of control (neo vector), normalized 240 ** 200 160 120 80 40 0 Neo Neo/RNase L RNase L Protein Supplementary
Supplemental Data mrna Protein A kda 75 40 NEO/vector NEO/RNase L RNASE L β-actin RNASE L β-actin B % of control (neo vector), normalized 240 ** 200 160 120 80 40 0 Neo Neo/RNase L RNase L Protein Supplementary
Supplemental Data includes Supplemental Experimental Procedures, 4 Figures and 5 Movie Legends.
 SUPPLEMENTAL DATA Supplemental Data includes Supplemental Experimental Procedures, 4 Figures and 5 Movie Legends. Supplemental Experimental Procedures Dominant negative nesprin KASH (DN KASH) plasmid construction-
SUPPLEMENTAL DATA Supplemental Data includes Supplemental Experimental Procedures, 4 Figures and 5 Movie Legends. Supplemental Experimental Procedures Dominant negative nesprin KASH (DN KASH) plasmid construction-
Engineering splicing factors with designed specificities
 nature methods Engineering splicing factors with designed specificities Yang Wang, Cheom-Gil Cheong, Traci M Tanaka Hall & Zefeng Wang Supplementary figures and text: Supplementary Figure 1 Supplementary
nature methods Engineering splicing factors with designed specificities Yang Wang, Cheom-Gil Cheong, Traci M Tanaka Hall & Zefeng Wang Supplementary figures and text: Supplementary Figure 1 Supplementary
Dual-Color Photon Counting Histogram Analysis of mrfp1 and EGFP in Living Cells
 Biophysical Journal Volume 91 December 2006 4273 4284 4273 Dual-Color Photon Counting Histogram Analysis of mrfp1 and EGFP in Living Cells Lindsey N. Hillesheim, Yan Chen, and Joachim D. Müller School
Biophysical Journal Volume 91 December 2006 4273 4284 4273 Dual-Color Photon Counting Histogram Analysis of mrfp1 and EGFP in Living Cells Lindsey N. Hillesheim, Yan Chen, and Joachim D. Müller School
A quantitative investigation of linker histone interactions with nucleosomes and chromatin
 White et al Supplementary figures and figure legends A quantitative investigation of linker histone interactions with nucleosomes and chromatin 1 Alison E. White, Aaron R. Hieb, and 1 Karolin Luger 1 White
White et al Supplementary figures and figure legends A quantitative investigation of linker histone interactions with nucleosomes and chromatin 1 Alison E. White, Aaron R. Hieb, and 1 Karolin Luger 1 White
Dynamic map of protein interactions in the Escherichia coli chemotaxis pathway. Attractant exchange profile during kinetics measurements.
 Supplementary data Dynamic map of protein interactions in the Escherichia coli chemotaxis pathway David Kentner, Victor Sourjik Table of content: Figure S1 Figure S2 Figure S3 Table SI Microscopy setups
Supplementary data Dynamic map of protein interactions in the Escherichia coli chemotaxis pathway David Kentner, Victor Sourjik Table of content: Figure S1 Figure S2 Figure S3 Table SI Microscopy setups
HeLa cells stably transfected with an empty pcdna3 vector (HeLa Neo) or with a plasmid
 SUPPLEMENTAL MATERIALS AND METHODS Cell culture, transfection and treatments. HeLa cells stably transfected with an empty pcdna3 vector (HeLa Neo) or with a plasmid encoding vmia (HeLa vmia) 1 were cultured
SUPPLEMENTAL MATERIALS AND METHODS Cell culture, transfection and treatments. HeLa cells stably transfected with an empty pcdna3 vector (HeLa Neo) or with a plasmid encoding vmia (HeLa vmia) 1 were cultured
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature08627 Supplementary Figure 1. DNA sequences used to construct nucleosomes in this work. a, DNA sequences containing the 601 positioning sequence (blue)24 with a PstI restriction site
doi: 10.1038/nature08627 Supplementary Figure 1. DNA sequences used to construct nucleosomes in this work. a, DNA sequences containing the 601 positioning sequence (blue)24 with a PstI restriction site
The Green Fluorescent Protein. w.chem.uwec.edu/chem412_s99/ppt/green.ppt
 The Green Fluorescent Protein w.chem.uwec.edu/chem412_s99/ppt/green.ppt www.chem.uwec.edu/chem412_s99/ppt/green.ppt Protein (gene) is from a jellyfish: Aequorea victoria www.chem.uwec.edu/chem412_s99/ppt/green.ppt
The Green Fluorescent Protein w.chem.uwec.edu/chem412_s99/ppt/green.ppt www.chem.uwec.edu/chem412_s99/ppt/green.ppt Protein (gene) is from a jellyfish: Aequorea victoria www.chem.uwec.edu/chem412_s99/ppt/green.ppt
Multiplexed 3D FRET imaging in deep tissue of live embryos Ming Zhao, Xiaoyang Wan, Yu Li, Weibin Zhou and Leilei Peng
 Scientific Reports Multiplexed 3D FRET imaging in deep tissue of live embryos Ming Zhao, Xiaoyang Wan, Yu Li, Weibin Zhou and Leilei Peng 1 Supplementary figures and notes Supplementary Figure S1 Volumetric
Scientific Reports Multiplexed 3D FRET imaging in deep tissue of live embryos Ming Zhao, Xiaoyang Wan, Yu Li, Weibin Zhou and Leilei Peng 1 Supplementary figures and notes Supplementary Figure S1 Volumetric
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Mass spectrometry characterization of Au 25, Au 38, Au 144, Au 333, Au ~520 and Au ~940 nanoclusters. (a) MALDI-mass spectra of Au 144, Au 333, Au ~520 and
Supplementary Figures Supplementary Figure 1. Mass spectrometry characterization of Au 25, Au 38, Au 144, Au 333, Au ~520 and Au ~940 nanoclusters. (a) MALDI-mass spectra of Au 144, Au 333, Au ~520 and
Self-labelling enzymes as universal tags for fluorescence microscopy, superresolution microscopy and electron microscopy
 Supplementary Materials Self-labelling enzymes as universal tags for fluorescence microscopy, superresolution microscopy and electron microscopy Viktoria Liss, Britta Barlag, Monika Nietschke and Michael
Supplementary Materials Self-labelling enzymes as universal tags for fluorescence microscopy, superresolution microscopy and electron microscopy Viktoria Liss, Britta Barlag, Monika Nietschke and Michael
FRET Biosensors and Live Cell Imaging
 08/3/2015 Summer Workshop FRET Imaging and Quantification FRET Biosensors and Live Cell Imaging Peter Yingxiao Wang Associate Professor Department of Bioengineering, Institute of Engineering in Medicine,
08/3/2015 Summer Workshop FRET Imaging and Quantification FRET Biosensors and Live Cell Imaging Peter Yingxiao Wang Associate Professor Department of Bioengineering, Institute of Engineering in Medicine,
Fluorescent labeling of the nuclear envelope by localizing GFP on the inner nuclear membrane
 Supporting Information Fluorescent labeling of the nuclear envelope by localizing GFP on the inner nuclear membrane Toshiyuki Taniyama 1, Natsumi Tsuda 1, and Shinji Sueda 1,2,* 1 Department of Bioscience
Supporting Information Fluorescent labeling of the nuclear envelope by localizing GFP on the inner nuclear membrane Toshiyuki Taniyama 1, Natsumi Tsuda 1, and Shinji Sueda 1,2,* 1 Department of Bioscience
D e c N o. 2 8
 D e c. 2 0 0 7 N o. 2 8 CONFOCAL APPLICATION LETTER resolution FRET Acceptor Photobleaching LAS AF Application Wizard FRET with Leica TCS SP5 LAS AF Version 1.7.0 Introduction Fluorescence Resonance Energy
D e c. 2 0 0 7 N o. 2 8 CONFOCAL APPLICATION LETTER resolution FRET Acceptor Photobleaching LAS AF Application Wizard FRET with Leica TCS SP5 LAS AF Version 1.7.0 Introduction Fluorescence Resonance Energy
Supplementary Methods. Plasmid construction. NS5A-fluorophore fusion Jc1 genomes were generated
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 Supplementary Methods Plasmid construction. NS5A-fluorophore fusion Jc1 genomes were generated by introducing a linker containing an XbaI and
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 Supplementary Methods Plasmid construction. NS5A-fluorophore fusion Jc1 genomes were generated by introducing a linker containing an XbaI and
Challenges to measuring intracellular Ca 2+ Calmodulin: nature s Ca 2+ sensor
 Calcium Signals in Biological Systems Lecture 3 (2/9/0) Measuring intracellular Ca 2+ signals II: Genetically encoded Ca 2+ sensors Henry M. Colecraft, Ph.D. Challenges to measuring intracellular Ca 2+
Calcium Signals in Biological Systems Lecture 3 (2/9/0) Measuring intracellular Ca 2+ signals II: Genetically encoded Ca 2+ sensors Henry M. Colecraft, Ph.D. Challenges to measuring intracellular Ca 2+
Chapter 20 Recombinant DNA Technology. Copyright 2009 Pearson Education, Inc.
 Chapter 20 Recombinant DNA Technology Copyright 2009 Pearson Education, Inc. 20.1 Recombinant DNA Technology Began with Two Key Tools: Restriction Enzymes and DNA Cloning Vectors Recombinant DNA refers
Chapter 20 Recombinant DNA Technology Copyright 2009 Pearson Education, Inc. 20.1 Recombinant DNA Technology Began with Two Key Tools: Restriction Enzymes and DNA Cloning Vectors Recombinant DNA refers
Supplementary Figure 1. Diagram for CATCHA construct. Nature Biotechnology doi: /nbt.3444
 Supplementary Figure 1 Diagram for CATCHA construct. Supplementary Figure 2 Representative view of ebony (left) and non-ebony (right) F2 flies from experiments described in Fig. 1c. F0 #1 F0 #2 F0 #3 F0
Supplementary Figure 1 Diagram for CATCHA construct. Supplementary Figure 2 Representative view of ebony (left) and non-ebony (right) F2 flies from experiments described in Fig. 1c. F0 #1 F0 #2 F0 #3 F0
CoralHue Fluo-chase Kit
 Amalgaam For Research Use Only. Not for use in diagnostic procedures. Protein-Protein Interaction Analysis CoralHue Fluo-chase Kit Code No. AM-1100 15 B Constitution Way Woburn, MA 01801 Phone: 1.800.200.5459
Amalgaam For Research Use Only. Not for use in diagnostic procedures. Protein-Protein Interaction Analysis CoralHue Fluo-chase Kit Code No. AM-1100 15 B Constitution Way Woburn, MA 01801 Phone: 1.800.200.5459
Cre Stoplight with Living Colors is a faster, brighter
 Cre Stoplight with Living Colors is a faster, brighter reporter for Cre recombinase. Drago A Guggiana-Nilo 1, Anne Marie Quinn 2,Thomas E. Hughes 1 1 Department of Cell Biology and Neuroscience, Montana
Cre Stoplight with Living Colors is a faster, brighter reporter for Cre recombinase. Drago A Guggiana-Nilo 1, Anne Marie Quinn 2,Thomas E. Hughes 1 1 Department of Cell Biology and Neuroscience, Montana
Supplementary Information
 Supplementary Information STED nanoscopy combined with optical tweezers reveals protein dynamics on densely covered DNA Iddo Heller, Gerrit Sitters, Onno D. Broekmans, Géraldine Farge, Carolin Menges,
Supplementary Information STED nanoscopy combined with optical tweezers reveals protein dynamics on densely covered DNA Iddo Heller, Gerrit Sitters, Onno D. Broekmans, Géraldine Farge, Carolin Menges,
Donor DNA Utilization during Gene Targeting with Zinc- finger Nucleases
 Donor DNA Utilization during Gene Targeting with Zinc- finger Nucleases Kelly J. Beumer, Jonathan K. Trautman, Kusumika Mukherjee and Dana Carroll Department of Biochemistry University of Utah School of
Donor DNA Utilization during Gene Targeting with Zinc- finger Nucleases Kelly J. Beumer, Jonathan K. Trautman, Kusumika Mukherjee and Dana Carroll Department of Biochemistry University of Utah School of
High-resolution Identification and Separation of Living Cell Types by Multiple microrna-responsive Synthetic mrnas
 Supplementary information High-resolution Identification and Separation of Living Cell Types by Multiple microrna-responsive Synthetic mrnas Kei Endo *, Karin Hayashi, Hirohide Saito * Contents: 1. Supplementary
Supplementary information High-resolution Identification and Separation of Living Cell Types by Multiple microrna-responsive Synthetic mrnas Kei Endo *, Karin Hayashi, Hirohide Saito * Contents: 1. Supplementary
Supporting information
 Supporting information Construction of strains and plasmids To create ptc67, a PCR product obtained with primers cc2570-162f (gcatgggcaagcttgaggacggcgtcatgt) and cc2570+512f (gaggccgtggtaccatagaggcgggcg),
Supporting information Construction of strains and plasmids To create ptc67, a PCR product obtained with primers cc2570-162f (gcatgggcaagcttgaggacggcgtcatgt) and cc2570+512f (gaggccgtggtaccatagaggcgggcg),
Table S1. List of DNA constructs and primers, part 1 Construct
 SUPPLEMENTARY TABLES: Table S1. List of DNA constructs and primers, part 1 Construct Comment (Expressed protein) Vector, PCR primers and template or source of Restriction Sites name Resistance construct
SUPPLEMENTARY TABLES: Table S1. List of DNA constructs and primers, part 1 Construct Comment (Expressed protein) Vector, PCR primers and template or source of Restriction Sites name Resistance construct
To clone eif4a fragments into SalI-NotI sites of pacycduet-1 (pmb09); and petduet-1
 Supplementary Methods DNA constructs and strains To clone eif4a fragments into SalI-NotI sites of pacycduet-1 (pmb09); and petduet-1 (Novagen) (pmb11), we PCR-amplified each domain from the mouse eif4a
Supplementary Methods DNA constructs and strains To clone eif4a fragments into SalI-NotI sites of pacycduet-1 (pmb09); and petduet-1 (Novagen) (pmb11), we PCR-amplified each domain from the mouse eif4a
Supplementary Figure 1. FRET probe labeling locations in the Cas9-RNA-DNA complex.
 Supplementary Figure 1. FRET probe labeling locations in the Cas9-RNA-DNA complex. (a) Cy3 and Cy5 labeling locations shown in the crystal structure of Cas9-RNA bound to a cognate DNA target (PDB ID: 4UN3)
Supplementary Figure 1. FRET probe labeling locations in the Cas9-RNA-DNA complex. (a) Cy3 and Cy5 labeling locations shown in the crystal structure of Cas9-RNA bound to a cognate DNA target (PDB ID: 4UN3)
Optimization of fluorescent proteins for novel quantitative multiparameter microscopy approaches Kremers, G.J.
 UvA-DARE (Digital Academic Repository) Optimization of fluorescent proteins for novel quantitative multiparameter microscopy approaches Kremers, G.J. Link to publication Citation for published version
UvA-DARE (Digital Academic Repository) Optimization of fluorescent proteins for novel quantitative multiparameter microscopy approaches Kremers, G.J. Link to publication Citation for published version
Functional activity of fluorescence-labeled ribosome complexes used in this study, as determined by the time-resolved puromycin assay.
 Supplementary Figure 1 Functional activity of fluorescence-labeled ribosome complexes used in this study, as determined by the time-resolved puromycin assay. Closed circles, unlabeled PRE complex; open
Supplementary Figure 1 Functional activity of fluorescence-labeled ribosome complexes used in this study, as determined by the time-resolved puromycin assay. Closed circles, unlabeled PRE complex; open
Chapter One. Construction of a Fluorescent α5 Subunit. Elucidation of the unique contribution of the α5 subunit is complicated by several factors
 4 Chapter One Construction of a Fluorescent α5 Subunit The significance of the α5 containing nachr receptor (α5* receptor) has been a challenging question for researchers since its characterization by
4 Chapter One Construction of a Fluorescent α5 Subunit The significance of the α5 containing nachr receptor (α5* receptor) has been a challenging question for researchers since its characterization by
Molecular Biology Techniques Supporting IBBE
 Molecular Biology Techniques Supporting IBBE Jared Cartwright Protein Production Lab Head Contact Details: email jared.cartwright@york.ac.uk Phone 01904 328797 Presentation Aims Gene synthesis Cloning
Molecular Biology Techniques Supporting IBBE Jared Cartwright Protein Production Lab Head Contact Details: email jared.cartwright@york.ac.uk Phone 01904 328797 Presentation Aims Gene synthesis Cloning
Supplementary Information
 Supplementary Information Fluorescent protein choice The dual fluorescence channel ratiometric promoter characterization plasmid we designed for this study required a pair of fluorescent proteins with
Supplementary Information Fluorescent protein choice The dual fluorescence channel ratiometric promoter characterization plasmid we designed for this study required a pair of fluorescent proteins with
High-precision FLIM FRET in fixed and living cells reveals heterogeneity in a simple CFP YFP fusion protein
 Biophysical Chemistry 127 (2007) 155 164 http://www.elsevier.com/locate/biophyschem High-precision FLIM FRET in fixed and living cells reveals heterogeneity in a simple CFP YFP fusion protein Michael Millington
Biophysical Chemistry 127 (2007) 155 164 http://www.elsevier.com/locate/biophyschem High-precision FLIM FRET in fixed and living cells reveals heterogeneity in a simple CFP YFP fusion protein Michael Millington
SUPPLEMENTARY NOTE 2. Supplememtary Note 2, Wehr et al., Monitoring Regulated Protein-Protein Interactions Using Split-TEV 1
 SUPPLEMENTARY NOTE 2 A recombinase reporter system for permanent reporter activation We made use of the Cre-loxP recombinase system for a maximal amplification and complete kinetic uncoupling within single
SUPPLEMENTARY NOTE 2 A recombinase reporter system for permanent reporter activation We made use of the Cre-loxP recombinase system for a maximal amplification and complete kinetic uncoupling within single
Supplementary information. Interplay between Penicillin-binding proteins and SEDS proteins promotes. bacterial cell wall synthesis
 Supplementary information Interplay between Penicillin-binding proteins and SEDS proteins promotes bacterial cell wall synthesis Sophie Leclercq 1, Adeline Derouaux 1, Samir Olatunji 1, Claudine Fraipont
Supplementary information Interplay between Penicillin-binding proteins and SEDS proteins promotes bacterial cell wall synthesis Sophie Leclercq 1, Adeline Derouaux 1, Samir Olatunji 1, Claudine Fraipont
Supporting Information
 Supporting Information Dirk H. Ortgies ƥδ, Meiling Tan Δ, Erving C. Ximendes ᵻΔ, Blanca del Rosal ƍ, Jie Hu, Lei Xu, Xindong Wang, Emma Martín Rodríguez &ƥ, Carlos Jacinto ᵻ, Nuria Fernandez ᴝƥ, Guanying
Supporting Information Dirk H. Ortgies ƥδ, Meiling Tan Δ, Erving C. Ximendes ᵻΔ, Blanca del Rosal ƍ, Jie Hu, Lei Xu, Xindong Wang, Emma Martín Rodríguez &ƥ, Carlos Jacinto ᵻ, Nuria Fernandez ᴝƥ, Guanying
Imaging Protein-Protein Interactions By Multiphoton FLIM S. M. Ameer-Beg 1, N. Edme 2, M. Peter 2, P. R. Barber 1, T. Ng 2, B. Vojnovic 1.
 Imaging Protein-Protein Interactions By Multiphoton FLIM S. M. Ameer-Beg 1, N. Edme 2, M. Peter 2, P. R. Barber 1, T. Ng 2, B. Vojnovic 1. 1. Advanced Technology Development Group, Gray Cancer Institute,
Imaging Protein-Protein Interactions By Multiphoton FLIM S. M. Ameer-Beg 1, N. Edme 2, M. Peter 2, P. R. Barber 1, T. Ng 2, B. Vojnovic 1. 1. Advanced Technology Development Group, Gray Cancer Institute,
Product Manual. RFP ELISA Kit. Catalog Number. FOR RESEARCH USE ONLY Not for use in diagnostic procedures
 Product Manual RFP ELISA Kit Catalog Number AKR-122 96 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Red fluorescent protein (DsRed) is a spontaneously fluorescent protein
Product Manual RFP ELISA Kit Catalog Number AKR-122 96 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Red fluorescent protein (DsRed) is a spontaneously fluorescent protein
Materials and methods. by University of Washington Yeast Resource Center) from several promoters, including
 Supporting online material for Elowitz et al. report Materials and methods Strains and plasmids. Plasmids expressing CFP or YFP (wild-type codons, developed by University of Washington Yeast Resource Center)
Supporting online material for Elowitz et al. report Materials and methods Strains and plasmids. Plasmids expressing CFP or YFP (wild-type codons, developed by University of Washington Yeast Resource Center)
Aminoacid change in chromophore. PIN3::PIN3-GFP GFP S65 (no change) (5.9) (Kneen et al., 1998)
 Supplemental Table. Table S1. Fluorophore characteristics of used fluorescent proteins, Related to Figure 2 and 3. Transgenic line Fluprescent marker Aminoacid change in chromophore pk(a) Ref. PIN3::PIN3-GFP
Supplemental Table. Table S1. Fluorophore characteristics of used fluorescent proteins, Related to Figure 2 and 3. Transgenic line Fluprescent marker Aminoacid change in chromophore pk(a) Ref. PIN3::PIN3-GFP
Programmable Sequence-Specific Transcriptional Regulation of Mammalian Genome Using Designer TAL Effectors
 Supplementary Information Programmable Sequence-Specific Transcriptional Regulation of Mammalian Genome Using Designer TAL Effectors Feng Zhang 1,2,3,5,7 *,±, Le Cong 2,3,4 *, Simona Lodato 5,6, Sriram
Supplementary Information Programmable Sequence-Specific Transcriptional Regulation of Mammalian Genome Using Designer TAL Effectors Feng Zhang 1,2,3,5,7 *,±, Le Cong 2,3,4 *, Simona Lodato 5,6, Sriram
NAME TA SEC Problem Set 4 FRIDAY October 15, Answers to this problem set must be inserted into the box outside
 MIT Biology Department 7.012: Introductory Biology - Fall 2004 Instructors: Professor Eric Lander, Professor Robert A. Weinberg, Dr. Claudette Gardel NAME TA SEC 7.012 Problem Set 4 FRIDAY October 15,
MIT Biology Department 7.012: Introductory Biology - Fall 2004 Instructors: Professor Eric Lander, Professor Robert A. Weinberg, Dr. Claudette Gardel NAME TA SEC 7.012 Problem Set 4 FRIDAY October 15,
Intercalation-based single-molecule fluorescence assay to study DNA supercoil
 Intercalation-based single-molecule fluorescence assay to study DNA supercoil dynamics Mahipal Ganji, Sung Hyun Kim, Jaco van der Torre, Elio Abbondanzieri*, Cees Dekker* Supplementary information Supplementary
Intercalation-based single-molecule fluorescence assay to study DNA supercoil dynamics Mahipal Ganji, Sung Hyun Kim, Jaco van der Torre, Elio Abbondanzieri*, Cees Dekker* Supplementary information Supplementary
Certificate of Analysis
 Certificate of Analysis Catalog No. Amount Lot Number 631986 10 μg Specified on product label. Product Information plvx-ef1α-mcherry-n1 is a lentiviral expression vector that can be used to generate high-titer
Certificate of Analysis Catalog No. Amount Lot Number 631986 10 μg Specified on product label. Product Information plvx-ef1α-mcherry-n1 is a lentiviral expression vector that can be used to generate high-titer
BBF RFC 25: Fusion Protein (Freiburg) Biobrick assembly standard
 BBF RFC 25 Fusionprotein Assembly BBF RFC 25: Fusion Protein (Freiburg) Biobrick assembly standard Kristian M. Müller, Katja M. Arndt, igem 2007 Team Freiburg, Raik Grünberg 1. Purpose 31.3.2009 This Request
BBF RFC 25 Fusionprotein Assembly BBF RFC 25: Fusion Protein (Freiburg) Biobrick assembly standard Kristian M. Müller, Katja M. Arndt, igem 2007 Team Freiburg, Raik Grünberg 1. Purpose 31.3.2009 This Request
Supplementary Methods Plasmid constructs
 Supplementary Methods Plasmid constructs. Mouse cdna encoding SHP-1, amplified from mrna of RAW264.7 macrophages with primer 5'cgtgcctgcccagacaaactgt3' and 5'cggaattcagacgaatgcccagatcacttcc3', was cloned
Supplementary Methods Plasmid constructs. Mouse cdna encoding SHP-1, amplified from mrna of RAW264.7 macrophages with primer 5'cgtgcctgcccagacaaactgt3' and 5'cggaattcagacgaatgcccagatcacttcc3', was cloned
Hyper sensitive protein detection by Tandem-HTRF reveals Cyclin D1
 Hyper sensitive protein detection by Tandem-HTRF reveals Cyclin D1 dynamics in adult mouse Alexandre Zampieri, Julien Champagne, Baptiste Auzemery, Ivanna Fuentes, Benjamin Maurel and Frédéric Bienvenu
Hyper sensitive protein detection by Tandem-HTRF reveals Cyclin D1 dynamics in adult mouse Alexandre Zampieri, Julien Champagne, Baptiste Auzemery, Ivanna Fuentes, Benjamin Maurel and Frédéric Bienvenu
Introduction. BRET Principle. K. M. Kroeger 1, K. A. Eidne 1, Bernd Hutter 2
 Bioluminescence Resonance Energy Transfer (BRET) as a means of monitoring dynamic receptor-protein interactions in living cells measured on LB 940 Mithras K. M. Kroeger 1, K. A. Eidne 1, Bernd Hutter 2
Bioluminescence Resonance Energy Transfer (BRET) as a means of monitoring dynamic receptor-protein interactions in living cells measured on LB 940 Mithras K. M. Kroeger 1, K. A. Eidne 1, Bernd Hutter 2
DNA RNA protein (which performs different functions in the cell, i.e. a catalytic reaction on a small molecule substrate)
 Synthetic Biology Is an emerging discipline that emphasizes the application of engineering principles to the design and construction of synthetic biological systems that exhibit dynamic or logical behavior.
Synthetic Biology Is an emerging discipline that emphasizes the application of engineering principles to the design and construction of synthetic biological systems that exhibit dynamic or logical behavior.
Supplemental figures Supplemental Figure 1: Fluorescence recovery for FRAP experiments depicted in Figure 1.
 Supplemental figures Supplemental Figure 1: Fluorescence recovery for FRAP experiments depicted in Figure 1. Percent of original fluorescence was plotted as a function of time following photobleaching
Supplemental figures Supplemental Figure 1: Fluorescence recovery for FRAP experiments depicted in Figure 1. Percent of original fluorescence was plotted as a function of time following photobleaching
Knockdown of chimeric glucocerebrosidase by green fluorescent protein-directed small interfering RNA
 T.N. Campbell and F.Y.M. Choy 282 Knockdown of chimeric glucocerebrosidase by green fluorescent protein-directed small interfering RNA Tessa N. Campbell and Francis Y.M. Choy Centre for Biomedical Research
T.N. Campbell and F.Y.M. Choy 282 Knockdown of chimeric glucocerebrosidase by green fluorescent protein-directed small interfering RNA Tessa N. Campbell and Francis Y.M. Choy Centre for Biomedical Research
Synonymous Modification Results in High Fidelity Gene Expression of Repetitive Protein and Nucleotide Sequences
 Synonymous Modification Results in High Fidelity Gene Expression of Repetitive Protein and Nucleotide Sequences Bin Wu *,1,2, Veronika Miskolci *,1, Hanae Sato 1, Evelina Tutucci 1, Charles A. Kenworthy
Synonymous Modification Results in High Fidelity Gene Expression of Repetitive Protein and Nucleotide Sequences Bin Wu *,1,2, Veronika Miskolci *,1, Hanae Sato 1, Evelina Tutucci 1, Charles A. Kenworthy
Molecular Genetics of Disease and the Human Genome Project
 9 Molecular Genetics of Disease and the Human Genome Project Fig. 1. The 23 chromosomes in the human genome. There are 22 autosomes (chromosomes 1 to 22) and two sex chromosomes (X and Y). Females inherit
9 Molecular Genetics of Disease and the Human Genome Project Fig. 1. The 23 chromosomes in the human genome. There are 22 autosomes (chromosomes 1 to 22) and two sex chromosomes (X and Y). Females inherit
Lecture Four. Molecular Approaches I: Nucleic Acids
 Lecture Four. Molecular Approaches I: Nucleic Acids I. Recombinant DNA and Gene Cloning Recombinant DNA is DNA that has been created artificially. DNA from two or more sources is incorporated into a single
Lecture Four. Molecular Approaches I: Nucleic Acids I. Recombinant DNA and Gene Cloning Recombinant DNA is DNA that has been created artificially. DNA from two or more sources is incorporated into a single
Certificate of Analysis
 Certificate of Analysis pet6xhn Expression Vector Set Contents Product Information... 1 pet6xhn-n, pet6xhn-c, and pet6xhn-gfpuv Vector Information... 2 Location of Features... 4 Additional Information...
Certificate of Analysis pet6xhn Expression Vector Set Contents Product Information... 1 pet6xhn-n, pet6xhn-c, and pet6xhn-gfpuv Vector Information... 2 Location of Features... 4 Additional Information...
Quantitative Real time PCR. Only for teaching purposes - not for reproduction or sale
 Quantitative Real time PCR PCR reaction conventional versus real time PCR real time PCR principles threshold cycle C T efficiency relative quantification reference genes primers detection chemistry GLP
Quantitative Real time PCR PCR reaction conventional versus real time PCR real time PCR principles threshold cycle C T efficiency relative quantification reference genes primers detection chemistry GLP
Chapter 8: Recombinant DNA. Ways this technology touches us. Overview. Genetic Engineering
 Chapter 8 Recombinant DNA and Genetic Engineering Genetic manipulation Ways this technology touches us Criminal justice The Justice Project, started by law students to advocate for DNA testing of Death
Chapter 8 Recombinant DNA and Genetic Engineering Genetic manipulation Ways this technology touches us Criminal justice The Justice Project, started by law students to advocate for DNA testing of Death
Genetic Engineering & Recombinant DNA
 Genetic Engineering & Recombinant DNA Chapter 10 Copyright The McGraw-Hill Companies, Inc) Permission required for reproduction or display. Applications of Genetic Engineering Basic science vs. Applied
Genetic Engineering & Recombinant DNA Chapter 10 Copyright The McGraw-Hill Companies, Inc) Permission required for reproduction or display. Applications of Genetic Engineering Basic science vs. Applied
CONFOCAL APPLICATION LETTER. June 2009 No. 35. FRET with FLIM Quantitative In-vivo Biochemistry
 CONFOCAL APPLICATION LETTER June 2009 No. 35 resolution FRET with FLIM Quantitative In-vivo Biochemistry Fig. 1 Emission spectrum of donor (here ECFP, blue line) must overlap with excitation spectrum of
CONFOCAL APPLICATION LETTER June 2009 No. 35 resolution FRET with FLIM Quantitative In-vivo Biochemistry Fig. 1 Emission spectrum of donor (here ECFP, blue line) must overlap with excitation spectrum of
OmicsLink shrna Clones guaranteed knockdown even in difficult-to-transfect cells
 OmicsLink shrna Clones guaranteed knockdown even in difficult-to-transfect cells OmicsLink shrna clone collections consist of lentiviral, and other mammalian expression vector based small hairpin RNA (shrna)
OmicsLink shrna Clones guaranteed knockdown even in difficult-to-transfect cells OmicsLink shrna clone collections consist of lentiviral, and other mammalian expression vector based small hairpin RNA (shrna)
CHAPTER 9 DNA Technologies
 CHAPTER 9 DNA Technologies Recombinant DNA Artificially created DNA that combines sequences that do not occur together in the nature Basis of much of the modern molecular biology Molecular cloning of genes
CHAPTER 9 DNA Technologies Recombinant DNA Artificially created DNA that combines sequences that do not occur together in the nature Basis of much of the modern molecular biology Molecular cloning of genes
Supplementary Figure 1 Telomerase RNA fragments used in single-molecule FRET experiments. A pseudoknot fragment (nts ) labeled at position U42
 Supplementary Figure 1 Telomerase RNA fragments used in single-molecule FRET experiments. A pseudoknot fragment (nts 32-195) labeled at position U42 with Cy3 (green circle) was constructed by a two piece
Supplementary Figure 1 Telomerase RNA fragments used in single-molecule FRET experiments. A pseudoknot fragment (nts 32-195) labeled at position U42 with Cy3 (green circle) was constructed by a two piece
3/11/10. Genetic Circuits. Edge Detection. Genetic Edge Detection Algorithm. Goal. What I cannot create I do not understand.
 What I cannot create I do not understand. ~Richard Feynman A Synthetic Genetic Edge Detection Program Tabor JJ, Salis HM, Simpson ZB, Chevalier AA, Levskaya A, Marcotte EM, Voigt CA, Ellington AD Cell.
What I cannot create I do not understand. ~Richard Feynman A Synthetic Genetic Edge Detection Program Tabor JJ, Salis HM, Simpson ZB, Chevalier AA, Levskaya A, Marcotte EM, Voigt CA, Ellington AD Cell.
The dual-promoter lentiviral vector system was described elsewhere 1. Destabilized ZsGreen
 Supplementary materials and methods Vector construction and lentiviral vector preparation and infection The dual-promoter lentiviral vector system was described elsewhere 1. Destabilized ZsGreen (t 1/2
Supplementary materials and methods Vector construction and lentiviral vector preparation and infection The dual-promoter lentiviral vector system was described elsewhere 1. Destabilized ZsGreen (t 1/2
The role of erna in chromatin looping
 The role of erna in chromatin looping Introduction Enhancers are cis-acting DNA regulatory elements that activate transcription of target genes 1,2. It is estimated that 400 000 to >1 million alleged enhancers
The role of erna in chromatin looping Introduction Enhancers are cis-acting DNA regulatory elements that activate transcription of target genes 1,2. It is estimated that 400 000 to >1 million alleged enhancers
CRISPR-dCas9 mediated TET1 targeting for selective DNA demethylation at BRCA1 promoter
 CRISPR-dCas9 mediated TET1 targeting for selective DNA demethylation at BRCA1 promoter SUPPLEMENTARY DATA See Supplementary Sequence File: 1 Supplementary Figure S1: The total protein was extracted from
CRISPR-dCas9 mediated TET1 targeting for selective DNA demethylation at BRCA1 promoter SUPPLEMENTARY DATA See Supplementary Sequence File: 1 Supplementary Figure S1: The total protein was extracted from
SUPPLEMENTARY INFORMATION
 Materials and Methods Transgenic Plant Materials and DNA Constructs. VEX1::H2B-GFP, ACA3::H2B-GFP, KRP6::H2B-GFP, KRP6::mock21ts-GFP, KRP6::TE21ts- GFP and KRP6::miR161ts-GFP constructs were generated
Materials and Methods Transgenic Plant Materials and DNA Constructs. VEX1::H2B-GFP, ACA3::H2B-GFP, KRP6::H2B-GFP, KRP6::mock21ts-GFP, KRP6::TE21ts- GFP and KRP6::miR161ts-GFP constructs were generated
pri 201 DNA Series (High-Expression Vectors for Plant Transformation)
 For Research Use Cat #. 3264 3265 3266 3267 pri 201 DNA Series (High-Expression Vectors for Plant Transformation) Product Manual Table of Contents I. Description... 3 II. Product Information... 3 III.
For Research Use Cat #. 3264 3265 3266 3267 pri 201 DNA Series (High-Expression Vectors for Plant Transformation) Product Manual Table of Contents I. Description... 3 II. Product Information... 3 III.
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION WWW.NATURE.COM/NATURECELLBIOLOGY 1 SUPPLEMENTARY INFORMATION 2 WWW.NATURE.COM/NATURECELLBIOLOGY SUPPLEMENTARY INFORMATION WWW.NATURE.COM/NATURECELLBIOLOGY 3 SUPPLEMENTARY INFORMATION
SUPPLEMENTARY INFORMATION WWW.NATURE.COM/NATURECELLBIOLOGY 1 SUPPLEMENTARY INFORMATION 2 WWW.NATURE.COM/NATURECELLBIOLOGY SUPPLEMENTARY INFORMATION WWW.NATURE.COM/NATURECELLBIOLOGY 3 SUPPLEMENTARY INFORMATION
Fibrin Clots Are Equilibrium Polymers That Can Be Remodeled Without Proteolytic Digestion
 Supplementary Information Fibrin Clots Are Equilibrium Polymers That Can Be Remodeled Without Proteolytic Digestion Irina N. Chernysh 1, Chandrasekaran Nagaswami 1, Prashant K. Purohit 2 and John W. Weisel
Supplementary Information Fibrin Clots Are Equilibrium Polymers That Can Be Remodeled Without Proteolytic Digestion Irina N. Chernysh 1, Chandrasekaran Nagaswami 1, Prashant K. Purohit 2 and John W. Weisel
from chloroplast DNA were purified, radioactively labeled to the protocol for obtaining greening cultures in a photoautotrophic
 Proc. Nati. Acad. Sci. USA Vol. 76, No. 5, pp. 2258-2262, May 1979 Biochemistry Transcription program of the chloroplast genome of Euglena gracilis during chloroplast development (restriction endonuclease
Proc. Nati. Acad. Sci. USA Vol. 76, No. 5, pp. 2258-2262, May 1979 Biochemistry Transcription program of the chloroplast genome of Euglena gracilis during chloroplast development (restriction endonuclease
A photoprotection strategy for microsecond-resolution single-molecule fluorescence spectroscopy
 Nature Methods A photoprotection strategy for microsecond-resolution single-molecule fluorescence spectroscopy Luis A Campos, Jianwei Liu, Xiang Wang, Ravishankar Ramanathan, Douglas S English & Victor
Nature Methods A photoprotection strategy for microsecond-resolution single-molecule fluorescence spectroscopy Luis A Campos, Jianwei Liu, Xiang Wang, Ravishankar Ramanathan, Douglas S English & Victor
Certificate of Analysis
 Certificate of Analysis Table of Contents Product Information... 1 Description... 2 Location of Features... 3 Additional Information... 3 Quality Control Data... 4 Catalog No. Amount Lot Number 631971
Certificate of Analysis Table of Contents Product Information... 1 Description... 2 Location of Features... 3 Additional Information... 3 Quality Control Data... 4 Catalog No. Amount Lot Number 631971
Recombinant DNA recombinant DNA DNA cloning gene cloning
 DNA Technology Recombinant DNA In recombinant DNA, DNA from two different sources, often two species, are combined into the same DNA molecule. DNA cloning permits production of multiple copies of a specific
DNA Technology Recombinant DNA In recombinant DNA, DNA from two different sources, often two species, are combined into the same DNA molecule. DNA cloning permits production of multiple copies of a specific
Supplementary Information: The timing of transcriptional regulation in synthetic gene circuits. Corresponding author.
 Supplementary Information: The timing of transcriptional regulation in synthetic gene circuits Yu-Yu Cheng 1, Andrew J. Hirning 1, Krešimir Josić 1,2,3, and Matthew R. Bennett 1,4, 1 Department of Biosciences,
Supplementary Information: The timing of transcriptional regulation in synthetic gene circuits Yu-Yu Cheng 1, Andrew J. Hirning 1, Krešimir Josić 1,2,3, and Matthew R. Bennett 1,4, 1 Department of Biosciences,
Certificate of Analysis
 Certificate of Analysis Table of Contents Product Information... 1 Description... 2 Location of Features... 3 Additional Information... 3 Quality Control Data... 4 Catalog No. Amount Lot Number 631972
Certificate of Analysis Table of Contents Product Information... 1 Description... 2 Location of Features... 3 Additional Information... 3 Quality Control Data... 4 Catalog No. Amount Lot Number 631972
Supplementary Figures
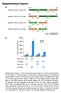 Supplementary Figures a HindIII XbaI pcdna3.1/v5-his A_Nfluc-VVD 1-398 37-186 Nfluc Linker 1 VVD HindIII XhoI pcdna3.1/v5-his A_VVD-Nfluc 37-186 1-398 VVD Linker 2 Nfluc HindIII XbaI pcdna3.1/v5-his A_Cfluc-VVD
Supplementary Figures a HindIII XbaI pcdna3.1/v5-his A_Nfluc-VVD 1-398 37-186 Nfluc Linker 1 VVD HindIII XhoI pcdna3.1/v5-his A_VVD-Nfluc 37-186 1-398 VVD Linker 2 Nfluc HindIII XbaI pcdna3.1/v5-his A_Cfluc-VVD
