Supplemental Data includes Supplemental Experimental Procedures, 4 Figures and 5 Movie Legends.
|
|
|
- Lorraine Eileen Dalton
- 5 years ago
- Views:
Transcription
1 SUPPLEMENTAL DATA Supplemental Data includes Supplemental Experimental Procedures, 4 Figures and 5 Movie Legends. Supplemental Experimental Procedures Dominant negative nesprin KASH (DN KASH) plasmid construction- To generate DN KASH fluorescently labeled with an amino-terminal mcherry domain, mcherry was amplified from pcdh-ef1- MCS1-puro-mCherry 2 (kindly provided by Dr. P. Patwari (1)) by PCR using the primers 5 - AATGGATCCTAACCATGGTGAGCAAGGGC-3 and 5 - CTTGTACAGCTCGTCCATGCCGCCGGT-3 and then subsequently subcloned into the BamHI and EcoRV (blunt end) sites of the pcdna4/his-max (Invitrogen) vector to create mcherry- pcdna4/his- Max. The KASH domain from GFP-mouse nesprin-1α (2) was amplified by PCR using primers 5 - ATAGCGGCCGCCGCGGCTTCCTGTTCAGAGTC-3 and 5 - TATTCTAGATCAGAGTGGAGGAGGGCCATT-3. The PCR product was digested with NotI and XbaI and inserted downstream of the mcherry sequence into the mcherry- pcdna4/his-max plasmid. mcherry-nesprin-1αkash was subcloned from pcdna4/his-max mcherry-nesprin-1αkash, into the BamHI and NotI (blunt end) sites of pcdh-ef1-mcs1-puro (System Biosciences) to create pcdh-ef1- MCS1-puro-mCherry-Nesprin-1αKASH. As a mock control (mcherry alone), pcdh-ef1-mcs1-puromcherry was used. All enzymes were obtained from New England Biolabs, Inc. The resulting plasmids are referred to as DN KASH (pcdh-ef1-mcs1-puro-mcherry-nesprin-1αkash) and mcherry control (pcdh-ef1-mcs1-puro-mcherry) in this manuscript. Dominant negative SUN1 luminal domain (DN SUN1L) plasmid construction- The 5 end of SS-HA- Sun1L-KDEL was amplified from pcdna3.1ss HA-Sun1L KDEL (3) by PCR using the pair of primers 5 - ATGCGGCCGCTTCGAACTATAGGGAGACCCAAGCTGG-3 and 3'- ATGAATTCATCGATAGTCGAGGCTGATCAGCGGTT-5. The SS-HA-Sun1L-KDEL sequence was then excised using NheI and EcoRI and the product was ligated into the NheI and EcoRI sites of the pcdh-cmv-mcs-ef1-copgfp-t2a-puro (System Biosciences) cut vector. The resulting plasmid was pcdh-cmv-mcs-ef1-copgfp-t2a-puross-ha-sun1l-kdel and is referred to as DN SUN1L in the manuscript. All enzymes were obtained from New England Biolabs, Inc. Primer sequences for strain-induced expression of mechanosensitive gene experiments- Gene expression of strain-induced cells was quantified by real-time PCR using probes for mechanosensitive genes Egr-1, Iex-1, Pai-I, tenascin C, talin and vinculin, using the following primers: mouse Egr CCTATGAGCACCTGACCACA-3 and 5 -TCGTTTGGCTGGGATAACTC-3 ; mouse Iex-1: 5 - GCCGAAGGGTGCTCTACC-3 and 5 -AAATCTGGCAGAAGATGATGG-3 ; mouse Pai-I 5 - GACTCTGGATGAGTGGAAAGC-3 and 5 -GGCGGCCTAAGTCTCCAAAAT-3 ; mouse Tenascin C 5 -ACGGCTACCACAGAAGCTG-3 and 5 -ATGGCTGTTGTTGCTATGGCA-3 ; mouse Talin 5 - CCTGCCGCATGATTCGTGA-3 and 5 - TCGGAGCATGTAGTAGTCCAAA-3 ; and mouse Vinculin 5 - GCACATCTGACCTACTGCTTAC-3 and 5 - TCTTAGTCATTCCTGGCCCAA-3. Expression was normalized to an endogenous control, TATA binding protein, using primers 5 - AGAACAATCCAGACTAGCAGCA-3 and 5 -GGGAACTTCACATCACAGCTC-3, and compared to unstrained controls and strained mcherry controls using the ΔΔC t method. 1
2 Supplemental Figures Figure S1. ER-targeted DN SUN1L localizes to the nuclear rim and ER. (A-B) Immunofluorescence images of fibroblasts transiently expressing DN SUN1L (containing a HA-tag; A) or GFP alone (B). Cells were stained for HA-tag (second panel) and DNA (Hoechst 33342, third panel). Fourth panel, merged image. The anti-ha-staining confirms localization of DN SUN1L to the nuclear envelope (perinuclear space) and endoplasmic reticulum. Scale bar, 10 µm. 2
3 Figure S2. Nesprin-2αext, mini-nesprin-2g, SS-GFP-KDEL and SUN1 control constructs do not displace endogenous nesprins. (A-D) Immunofluorescence images of fibroblasts transiently expressing EGFP-nesprin-2αext (A), GFP-mini-nesprin-2G (B), SS-GFP-KDEL (C) and GFP-SUN1 (D). Cells were stained for nesprin-2 (second panel) and DNA (Hoechst 33342, third panel). Fourth panel, merged image. The SS-GFP-KDEL construct is targeted to the perinuclear space and endoplasmic reticulum. The antinesprin-2 staining indicates that endogenous nesprin-2 remains at the nuclear envelope in cells expressing each of the control constructs. Scale bar, 10 µm. 3
4 Figure S3. LINC complex disruption does not alter mechanosensitive gene expression. (A) Mechanically-induced gene expression of Egr-1 and Iex-1 normalized to an endogenous control, TATA binding protein (TBP), and compared to unstrained controls and strained mcherry controls. Qualitative similar results were obtained for other mechanosensitive genes. Data are represented as mean ± s.e.m.; *, P <
5 Figure S4. Microtubule organization is normal in LINC disrupted fibroblasts. (A-B) Immunofluorescence analysis of fibroblasts expressing DN KASH domain fused to mcherry (A) or mcherry alone (B). First panel, mcherry signal. Cells were stained for β-tubulin (second panel) and DNA (third panel). Forth panel, merged image. Last panel, magnification of perinuclear area. Scale bar, 10 µm. 5
6 Movie Legends Movie 1. Microneedle manipulation assay in a mouse embryonic fibroblast. Phase contrast (left) and fluorescence (right) images of a cell labeled with Hoechst nuclear stain (blue) and MitroTracker Green mitochondrial stain (green). A microneedle was carefully inserted into the cytoskeleton 5 µm away from the nuclear periphery and moved 20 µm towards the cell periphery at 1 µm/second while collecting phase contrast and fluorescence images every 10 seconds, using a 60 objective (0.70 N.A.) on a inverted epifluorescence Olympus IX-70 microscope, with a digital CCDcamera (CoolSNAP EZ; Roper Scientific). Movie of the same cell shown in Fig. 2A-E. Movie 2. Induced nuclear and cytoskeletal deformation during microneedle manipulation in a mcherry expressing fibroblast. Phase contrast (left) and fluorescence (right) images of a cell expressing mcherry alone and labeled with nuclear stain (blue) and MitroTracker Green (green). A microneedle was carefully inserted into the cytoskeleton 5 µm away from the nuclear periphery and moved 10 µm towards the cell periphery at 1 µm/second while collecting phase contrast and fluorescence images every 10 seconds, using a 60 objective (0.70 N.A.) on a inverted epifluorescence Olympus IX-70 microscope, with a digital CCD-camera (CoolSNAP EZ; Roper Scientific). Movie 3. Induced nuclear and cytoskeletal deformation during microneedle manipulation in a dominant negative KASH expressing fibroblast. Phase contrast (left) and fluorescence (right) images of a cell expressing DN KASH and labeled with nuclear stain (blue) and MitroTracker Green (green). A microneedle was carefully inserted into the cytoskeleton 5 µm away from the nuclear periphery and moved 10 µm towards the cell periphery at 1 µm/second while collecting phase contrast and fluorescence images every 10 seconds, using a 60 objective (0.70 N.A.) on a inverted epifluorescence Olympus IX-70 microscope, with a digital CCD-camera (CoolSNAP EZ; Roper Scientific). Movie 4. Nuclear deformation in response to substrate strain application. Phase contrast images of mcherry (top) and DN KASH (bottom) expressing fibroblasts plated on fibronectin-coated silicone membranes and subjected to 20% uniaxial substrate strain. Membranes were placed on a custom-made strain device mounted on an inverted epifluorescence Olympus IX-70 microscope, using a 60 objective (0.70 N.A.) and a CoolSNAP EZ camera (Roper Scientific). One image was acquired before strain and a second image was acquired during strain. Movie of the same cells shown in Fig. 4C, D. Movie 5. Nuclear deformation in response to substrate strain application. Fluorescence images of the nuclei of mcherry (top) and DN KASH (bottom) expressing fibroblasts plated on fibronectin-coated silicone membranes and labeled with nuclear stain. The cells were subjected to 20% uniaxial substrate strain. Membranes were placed on a custom-made strain device mounted on an inverted epifluorescence Olympus IX-70 microscope, using a 60 objective (0.70 N.A.) and a CoolSNAP EZ camera (Roper Scientific). One image was acquired before strain and a second image was acquired during strain. Movies correspond to the same cells as shown in Movie 4. 6
7 References 1. Patwari, P., Chutkow, W. A., Cummings, K., Verstraeten, V. L., Lammerding, J., Schreiter, E. R., and Lee, R. T. (2009) J Biol Chem 284, Zhang, Q., Skepper, J. N., Yang, F., Davies, J. D., Hegyi, L., Roberts, R. G., Weissberg, P. L., Ellis, J. A., and Shanahan, C. M. (2001) J Cell Sci 114, Crisp, M., Liu, Q., Roux, K., Rattner, J. B., Shanahan, C., Burke, B., Stahl, P. D., and Hodzic, D. (2006) J Cell Biol 172,
Schematic representation of the endogenous PALB2 locus and gene-disruption constructs
 Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
SUPPLEMENTARY INFORMATION
 Clustering of a kinesin-14 motor enables processive retrograde microtubule-based transport in plants NATURE PLANTS www.nature.com/natureplants 1 2 NATURE PLANTS www.nature.com/natureplants NATURE PLANTS
Clustering of a kinesin-14 motor enables processive retrograde microtubule-based transport in plants NATURE PLANTS www.nature.com/natureplants 1 2 NATURE PLANTS www.nature.com/natureplants NATURE PLANTS
Figure S1: Insert sequences for GR1, GR2, Qc3c_AgeI and GR3 vectors
 Figure S1: Insert sequences for GR1, GR2, Qc3c_AgeI and GR3 vectors A Vector GR1 SacI BamHI CTCTGGCTAACTAGGC Insert 5/7nt - G TCGAGAGACCGATTGATCCG Insert 5/7nt - CCTAG 1 G CAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAAC
Figure S1: Insert sequences for GR1, GR2, Qc3c_AgeI and GR3 vectors A Vector GR1 SacI BamHI CTCTGGCTAACTAGGC Insert 5/7nt - G TCGAGAGACCGATTGATCCG Insert 5/7nt - CCTAG 1 G CAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAAC
Supplementary Methods
 Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
SAS6-like protein in Plasmodium indicates that conoid-associated apical complex proteins persist in invasive stages within the mosquito vector
 SAS6-like protein in Plasmodium indicates that conoid-associated apical complex proteins persist in invasive stages within the mosquito vector Richard J. Wall 1,#, Magali Roques 1,+, Nicholas J. Katris
SAS6-like protein in Plasmodium indicates that conoid-associated apical complex proteins persist in invasive stages within the mosquito vector Richard J. Wall 1,#, Magali Roques 1,+, Nicholas J. Katris
Actin cap associated focal adhesions and their distinct role in cellular mechanosensing
 Actin cap associated focal adhesions and their distinct role in cellular mechanosensing Dong-Hwee Kim 1,2, Shyam B. Khatau 1,2, Yunfeng Feng 1,3, Sam Walcott 1,4, Sean X. Sun 1,2,4, Gregory D. Longmore
Actin cap associated focal adhesions and their distinct role in cellular mechanosensing Dong-Hwee Kim 1,2, Shyam B. Khatau 1,2, Yunfeng Feng 1,3, Sam Walcott 1,4, Sean X. Sun 1,2,4, Gregory D. Longmore
Cytotoxicity of Botulinum Neurotoxins Reveals a Direct Role of
 Supplementary Information Cytotoxicity of Botulinum Neurotoxins Reveals a Direct Role of Syntaxin 1 and SNAP-25 in Neuron Survival Lisheng Peng, Huisheng Liu, Hongyu Ruan, William H. Tepp, William H. Stoothoff,
Supplementary Information Cytotoxicity of Botulinum Neurotoxins Reveals a Direct Role of Syntaxin 1 and SNAP-25 in Neuron Survival Lisheng Peng, Huisheng Liu, Hongyu Ruan, William H. Tepp, William H. Stoothoff,
Supporting information
 Supporting information Construction of strains and plasmids To create ptc67, a PCR product obtained with primers cc2570-162f (gcatgggcaagcttgaggacggcgtcatgt) and cc2570+512f (gaggccgtggtaccatagaggcgggcg),
Supporting information Construction of strains and plasmids To create ptc67, a PCR product obtained with primers cc2570-162f (gcatgggcaagcttgaggacggcgtcatgt) and cc2570+512f (gaggccgtggtaccatagaggcgggcg),
University of Dundee. Published in: Scientific Reports. DOI: /srep Publication date: 2016
 University of Dundee SAS6-like protein in Plasmodium indicates that conoid-associated apical complex proteins persist in invasive stages within the mosquito vector Wall, Richard J.; Roques, Magali; Katris,
University of Dundee SAS6-like protein in Plasmodium indicates that conoid-associated apical complex proteins persist in invasive stages within the mosquito vector Wall, Richard J.; Roques, Magali; Katris,
Programmable Sequence-Specific Transcriptional Regulation of Mammalian Genome Using Designer TAL Effectors
 Supplementary Information Programmable Sequence-Specific Transcriptional Regulation of Mammalian Genome Using Designer TAL Effectors Feng Zhang 1,2,3,5,7 *,±, Le Cong 2,3,4 *, Simona Lodato 5,6, Sriram
Supplementary Information Programmable Sequence-Specific Transcriptional Regulation of Mammalian Genome Using Designer TAL Effectors Feng Zhang 1,2,3,5,7 *,±, Le Cong 2,3,4 *, Simona Lodato 5,6, Sriram
Supplemental Information. Loss of MicroRNA-7 Regulation Leads. to a-synuclein Accumulation and. Dopaminergic Neuronal Loss In Vivo
 YMTHE, Volume 25 Supplemental Information Loss of MicroRNA-7 Regulation Leads to a-synuclein Accumulation and Dopaminergic Neuronal Loss In Vivo Kirsty J. McMillan, Tracey K. Murray, Nora Bengoa-Vergniory,
YMTHE, Volume 25 Supplemental Information Loss of MicroRNA-7 Regulation Leads to a-synuclein Accumulation and Dopaminergic Neuronal Loss In Vivo Kirsty J. McMillan, Tracey K. Murray, Nora Bengoa-Vergniory,
Fluorescent labeling of the nuclear envelope by localizing GFP on the inner nuclear membrane
 Supporting Information Fluorescent labeling of the nuclear envelope by localizing GFP on the inner nuclear membrane Toshiyuki Taniyama 1, Natsumi Tsuda 1, and Shinji Sueda 1,2,* 1 Department of Bioscience
Supporting Information Fluorescent labeling of the nuclear envelope by localizing GFP on the inner nuclear membrane Toshiyuki Taniyama 1, Natsumi Tsuda 1, and Shinji Sueda 1,2,* 1 Department of Bioscience
Supporting Information
 Supporting Information Stavru et al. 0.073/pnas.357840 SI Materials and Methods Immunofluorescence. For immunofluorescence, cells were fixed for 0 min in 4% (wt/vol) paraformaldehyde (Electron Microscopy
Supporting Information Stavru et al. 0.073/pnas.357840 SI Materials and Methods Immunofluorescence. For immunofluorescence, cells were fixed for 0 min in 4% (wt/vol) paraformaldehyde (Electron Microscopy
Self-labelling enzymes as universal tags for fluorescence microscopy, superresolution microscopy and electron microscopy
 Supplementary Materials Self-labelling enzymes as universal tags for fluorescence microscopy, superresolution microscopy and electron microscopy Viktoria Liss, Britta Barlag, Monika Nietschke and Michael
Supplementary Materials Self-labelling enzymes as universal tags for fluorescence microscopy, superresolution microscopy and electron microscopy Viktoria Liss, Britta Barlag, Monika Nietschke and Michael
SUPPLEMENTARY INFORMATION
 a 14 12 Densitometry (AU) 1 8 6 4 2 t b 16 NMHC-IIA GAPDH NMHC-IIB Densitometry (AU) 14 12 1 8 6 4 2 1 nm 1 nm 1 nm 1 nm sirna 1 nm 1 nm Figure S1 S4 Quantification of protein levels. (a) The microtubule
a 14 12 Densitometry (AU) 1 8 6 4 2 t b 16 NMHC-IIA GAPDH NMHC-IIB Densitometry (AU) 14 12 1 8 6 4 2 1 nm 1 nm 1 nm 1 nm sirna 1 nm 1 nm Figure S1 S4 Quantification of protein levels. (a) The microtubule
Chapter One. Construction of a Fluorescent α5 Subunit. Elucidation of the unique contribution of the α5 subunit is complicated by several factors
 4 Chapter One Construction of a Fluorescent α5 Subunit The significance of the α5 containing nachr receptor (α5* receptor) has been a challenging question for researchers since its characterization by
4 Chapter One Construction of a Fluorescent α5 Subunit The significance of the α5 containing nachr receptor (α5* receptor) has been a challenging question for researchers since its characterization by
Supplementary Materials for
 www.advances.sciencemag.org/cgi/content/full/1/7/e1500454/dc1 Supplementary Materials for CRISPR-Cas9 delivery to hard-to-transfect cells via membrane deformation Xin Han, Zongbin Liu, Myeong chan Jo,
www.advances.sciencemag.org/cgi/content/full/1/7/e1500454/dc1 Supplementary Materials for CRISPR-Cas9 delivery to hard-to-transfect cells via membrane deformation Xin Han, Zongbin Liu, Myeong chan Jo,
Nature Medicine doi: /nm.2548
 Supplementary Table 1: Genotypes of offspring and embryos from matings of Pmm2 WT/F118L mice with Pmm2 WT/R137H mice total events Pmm2 WT/WT Pmm2 WT/R137H Pmm2 WT/F118L Pmm2 R137H/F118L offspring 117 (100%)
Supplementary Table 1: Genotypes of offspring and embryos from matings of Pmm2 WT/F118L mice with Pmm2 WT/R137H mice total events Pmm2 WT/WT Pmm2 WT/R137H Pmm2 WT/F118L Pmm2 R137H/F118L offspring 117 (100%)
Supporting Information
 Supporting Information Shao et al. 10.1073/pnas.1504837112 SI Materials and Methods Immunofluorescence and Immunoblotting. For immunofluorescence, cells were fixed with 4% paraformaldehyde and permeabilized
Supporting Information Shao et al. 10.1073/pnas.1504837112 SI Materials and Methods Immunofluorescence and Immunoblotting. For immunofluorescence, cells were fixed with 4% paraformaldehyde and permeabilized
Supplementary Figure S1. Np95 interacts with de novo methyltransferases Dnmt3a and 3b. (A) Co-immunoprecipitation of endogenous Dnmt3a2 (left and
 Supplementary Figure S1. Np95 interacts with de novo methyltransferases Dnmt3a and 3b. (A) Co-immunoprecipitation of endogenous Dnmt3a2 (left and right), Dnmt3b isoforms (left) and Dnmt1 (right) with GFP-Np95
Supplementary Figure S1. Np95 interacts with de novo methyltransferases Dnmt3a and 3b. (A) Co-immunoprecipitation of endogenous Dnmt3a2 (left and right), Dnmt3b isoforms (left) and Dnmt1 (right) with GFP-Np95
Engineering splicing factors with designed specificities
 nature methods Engineering splicing factors with designed specificities Yang Wang, Cheom-Gil Cheong, Traci M Tanaka Hall & Zefeng Wang Supplementary figures and text: Supplementary Figure 1 Supplementary
nature methods Engineering splicing factors with designed specificities Yang Wang, Cheom-Gil Cheong, Traci M Tanaka Hall & Zefeng Wang Supplementary figures and text: Supplementary Figure 1 Supplementary
Single-Molecule Imaging Reveals Dynamics of CREB Transcription Factor Bound to Its Target Sequence
 Supplementary Information Single-Molecule Imaging Reveals Dynamics of CREB Transcription Factor Bound to Its Target Sequence Noriyuki Sugo, Masatoshi Morimatsu, Yoshiyuki Arai, Yoshinori Kousoku, Aya Ohkuni,
Supplementary Information Single-Molecule Imaging Reveals Dynamics of CREB Transcription Factor Bound to Its Target Sequence Noriyuki Sugo, Masatoshi Morimatsu, Yoshiyuki Arai, Yoshinori Kousoku, Aya Ohkuni,
NAME TA SEC Problem Set 4 FRIDAY October 15, Answers to this problem set must be inserted into the box outside
 MIT Biology Department 7.012: Introductory Biology - Fall 2004 Instructors: Professor Eric Lander, Professor Robert A. Weinberg, Dr. Claudette Gardel NAME TA SEC 7.012 Problem Set 4 FRIDAY October 15,
MIT Biology Department 7.012: Introductory Biology - Fall 2004 Instructors: Professor Eric Lander, Professor Robert A. Weinberg, Dr. Claudette Gardel NAME TA SEC 7.012 Problem Set 4 FRIDAY October 15,
Organelle Atlas APPENDIX TO CHAPTER 4 4A.1
 APPENDIX TO CHAPTER 4 This atlas contains images of cellular organelles visualized using a wide variety of reagents and techniques and demonstrates the diversity of methods for exploring the cell. The
APPENDIX TO CHAPTER 4 This atlas contains images of cellular organelles visualized using a wide variety of reagents and techniques and demonstrates the diversity of methods for exploring the cell. The
Table S1. List of DNA constructs and primers, part 1 Construct
 SUPPLEMENTARY TABLES: Table S1. List of DNA constructs and primers, part 1 Construct Comment (Expressed protein) Vector, PCR primers and template or source of Restriction Sites name Resistance construct
SUPPLEMENTARY TABLES: Table S1. List of DNA constructs and primers, part 1 Construct Comment (Expressed protein) Vector, PCR primers and template or source of Restriction Sites name Resistance construct
Supplemental Materials and Methods. Yeast strains. Strain genotypes are listed in Table S1. To generate MATainc::LEU2,
 Supplemental Materials and Methods Yeast strains. Strain genotypes are listed in Table S1. To generate MATainc::LEU2, the LEU2 marker was inserted by PCR-mediated recombination (BRACHMANN et al. 1998)
Supplemental Materials and Methods Yeast strains. Strain genotypes are listed in Table S1. To generate MATainc::LEU2, the LEU2 marker was inserted by PCR-mediated recombination (BRACHMANN et al. 1998)
BIO 315 Lab Exam I. Section #: Name:
 Section #: Name: Also provide this information on the computer grid sheet given to you. (Section # in special code box) BIO 315 Lab Exam I 1. In labeling the parts of a standard compound light microscope
Section #: Name: Also provide this information on the computer grid sheet given to you. (Section # in special code box) BIO 315 Lab Exam I 1. In labeling the parts of a standard compound light microscope
T H E J O U R N A L O F C E L L B I O L O G Y
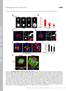 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rodríguez-Fraticelli et al., http://www.jcb.org/cgi/content/full/jcb.201203075/dc1 Figure S1. Cell spreading and lumen formation in confined
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rodríguez-Fraticelli et al., http://www.jcb.org/cgi/content/full/jcb.201203075/dc1 Figure S1. Cell spreading and lumen formation in confined
Design. Construction. Characterization
 Design Construction Characterization DNA mrna (messenger) A C C transcription translation C A C protein His A T G C T A C G Plasmids replicon copy number incompatibility selection marker origin of replication
Design Construction Characterization DNA mrna (messenger) A C C transcription translation C A C protein His A T G C T A C G Plasmids replicon copy number incompatibility selection marker origin of replication
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling
 Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
The fusion used in most of these studies (MinC4-GFP) contains GFP and
 Gregory, Becker and Pogliano, Supplemental Materials page 1 Supplemental Methods Description of the MinC and MinD GFP insertions The fusion used in most of these studies (MinC4-GFP) contains GFP and flanking
Gregory, Becker and Pogliano, Supplemental Materials page 1 Supplemental Methods Description of the MinC and MinD GFP insertions The fusion used in most of these studies (MinC4-GFP) contains GFP and flanking
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table.
 Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) B. Male mice of
 SUPPLEMENTRY FIGURE LEGENDS Figure S1. USP44 loss leads to chromosome missegregation.. Genotypes obtained from the indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) 13.5.. Male mice
SUPPLEMENTRY FIGURE LEGENDS Figure S1. USP44 loss leads to chromosome missegregation.. Genotypes obtained from the indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) 13.5.. Male mice
transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected with the wwp-luc reporter, and FLAG-tagged FHL1,
 Supplementary Data Supplementary Figure Legends Supplementary Figure 1 FHL-mediated TGFβ-responsive reporter transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected
Supplementary Data Supplementary Figure Legends Supplementary Figure 1 FHL-mediated TGFβ-responsive reporter transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected
SUPPLEMENTARY INFORMATION
 doi:.38/nature899 Supplementary Figure Suzuki et al. a c p7 -/- / WT ratio (+)/(-) p7 -/- / WT ratio Log X 3. Fold change by treatment ( (+)/(-)) Log X.5 3-3. -. b Fold change by treatment ( (+)/(-)) 8
doi:.38/nature899 Supplementary Figure Suzuki et al. a c p7 -/- / WT ratio (+)/(-) p7 -/- / WT ratio Log X 3. Fold change by treatment ( (+)/(-)) Log X.5 3-3. -. b Fold change by treatment ( (+)/(-)) 8
Supplementary
 Supplementary information Supplementary Material and Methods Plasmid construction The transposable element vectors for inducible expression of RFP-FUS wt and EGFP-FUS R521C and EGFP-FUS P525L were derived
Supplementary information Supplementary Material and Methods Plasmid construction The transposable element vectors for inducible expression of RFP-FUS wt and EGFP-FUS R521C and EGFP-FUS P525L were derived
Myofibroblasts in kidney fibrosis. LeBleu et al.
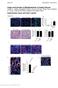 Origin and Function of Myofibroblasts in Kidney Fibrosis Valerie S. LeBleu, Gangadhar Taduri, Joyce O Connell, Yingqi Teng, Vesselina G. Cooke, Craig Woda, Hikaru Sugimoto and Raghu Kalluri Supplementary
Origin and Function of Myofibroblasts in Kidney Fibrosis Valerie S. LeBleu, Gangadhar Taduri, Joyce O Connell, Yingqi Teng, Vesselina G. Cooke, Craig Woda, Hikaru Sugimoto and Raghu Kalluri Supplementary
Supporting Information
 Supporting Information Development of a 2,4-Dinitrotoluene-Responsive Synthetic Riboswitch in E. coli cells Molly E. Davidson, Svetlana V. Harbaugh, Yaroslav G. Chushak, Morley O. Stone, Nancy Kelley-
Supporting Information Development of a 2,4-Dinitrotoluene-Responsive Synthetic Riboswitch in E. coli cells Molly E. Davidson, Svetlana V. Harbaugh, Yaroslav G. Chushak, Morley O. Stone, Nancy Kelley-
Cancer cells that survive radiation therapy acquire HIF-1 activity and translocate toward tumor blood vessels Supplementary Information
 Cancer cells that survive radiation therapy acquire HIF-1 activity and translocate toward tumor blood vessels Supplementary Information 1. Supplementary Figure S1-S10: Pages 2-11 2. Supplementary References:
Cancer cells that survive radiation therapy acquire HIF-1 activity and translocate toward tumor blood vessels Supplementary Information 1. Supplementary Figure S1-S10: Pages 2-11 2. Supplementary References:
Nature Methods: doi: /nmeth Supplementary Figure 1. Schematics of SAM, dcas9-suntag and dcas9-vpr.
 Supplementary Figure 1 Schematics of SAM, dcas9-suntag and dcas9-vpr. (a) Schematics of SAM. SAM consists of two chimeric proteins, dcas9 fused with VP64 and MS2-coat protein fused with p65 and HSF1 activator
Supplementary Figure 1 Schematics of SAM, dcas9-suntag and dcas9-vpr. (a) Schematics of SAM. SAM consists of two chimeric proteins, dcas9 fused with VP64 and MS2-coat protein fused with p65 and HSF1 activator
SUPPLEMENTARY INFORMATION FILE
 SUPPLEMENTARY INFORMATION FILE Existence of a microrna pathway in anucleate platelets Patricia Landry, Isabelle Plante, Dominique L. Ouellet, Marjorie P. Perron, Guy Rousseau & Patrick Provost 1. SUPPLEMENTARY
SUPPLEMENTARY INFORMATION FILE Existence of a microrna pathway in anucleate platelets Patricia Landry, Isabelle Plante, Dominique L. Ouellet, Marjorie P. Perron, Guy Rousseau & Patrick Provost 1. SUPPLEMENTARY
Gene Expression Technology
 Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
No wash 2 Washes 2 Days ** ** IgG-bead phagocytosis (%)
 Supplementary Figures Supplementary Figure 1. No wash 2 Washes 2 Days Tat Control ** ** 2 4 6 8 IgG-bead phagocytosis (%) Supplementary Figure 1. Reversibility of phagocytosis inhibition by Tat. Human
Supplementary Figures Supplementary Figure 1. No wash 2 Washes 2 Days Tat Control ** ** 2 4 6 8 IgG-bead phagocytosis (%) Supplementary Figure 1. Reversibility of phagocytosis inhibition by Tat. Human
At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in
 Supplementary Materials and Methods Barrier function assays At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in acidic X-gal mix (100 mm phosphate buffer at ph4.3, 3 mm
Supplementary Materials and Methods Barrier function assays At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in acidic X-gal mix (100 mm phosphate buffer at ph4.3, 3 mm
BIO 315 Lab Exam I. Section #: Name:
 Section #: Name: Also provide this information on the computer grid sheet given to you. (Section # in special code box) BIO 315 Lab Exam I 1. In labeling the parts of a standard compound light microscope
Section #: Name: Also provide this information on the computer grid sheet given to you. (Section # in special code box) BIO 315 Lab Exam I 1. In labeling the parts of a standard compound light microscope
JCB Online Supplemental Material
 JCB Online Supplemental Material Schinzel et al. Results At least one positive charge at the very COOH terminus is needed for mitochondrial sorting It has been reported that two consecutive positive charges
JCB Online Supplemental Material Schinzel et al. Results At least one positive charge at the very COOH terminus is needed for mitochondrial sorting It has been reported that two consecutive positive charges
Nature Biotechnology: doi: /nbt.4166
 Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
Yeast 2-Hybrid Kayla Nygaard
 Yeast 2-Hybrid 2.26.18 Kayla Nygaard Y2H - What is it? A method to screen for protein-protein interactions in yeast Capitalizes on GAL4 system in yeast. GAL4 has 2 domains DNA-Binding Domain (DB) Transcriptional
Yeast 2-Hybrid 2.26.18 Kayla Nygaard Y2H - What is it? A method to screen for protein-protein interactions in yeast Capitalizes on GAL4 system in yeast. GAL4 has 2 domains DNA-Binding Domain (DB) Transcriptional
Supplementary Figures
 Supplementary Figures Wossidlo et al. S1 Supplementary Figure S1 5hmC (clone 63.3) immunostaining on mouse zygotes. a, Representative images of indirect immunostainings using antibodies against 5hmC (clone
Supplementary Figures Wossidlo et al. S1 Supplementary Figure S1 5hmC (clone 63.3) immunostaining on mouse zygotes. a, Representative images of indirect immunostainings using antibodies against 5hmC (clone
pbroad3-lacz An optimized vector for mouse and rat transgenesis Catalog # pbroad3-lacz
 pbroad3-lacz An optimized vector for mouse and rat transgenesis Catalog # pbroad3-lacz For research use only Version # 03B04-MT PRODUCT INFORMATION Content: - 20 µg of pbroad3-lacz provided as lyophilized
pbroad3-lacz An optimized vector for mouse and rat transgenesis Catalog # pbroad3-lacz For research use only Version # 03B04-MT PRODUCT INFORMATION Content: - 20 µg of pbroad3-lacz provided as lyophilized
Sarker et al. Supplementary Material. Subcellular Fractionation
 Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
Supplemental Information. Pacer Mediates the Function of Class III PI3K. and HOPS Complexes in Autophagosome. Maturation by Engaging Stx17
 Molecular Cell, Volume 65 Supplemental Information Pacer Mediates the Function of Class III PI3K and HOPS Complexes in Autophagosome Maturation by Engaging Stx17 Xiawei Cheng, Xiuling Ma, Xianming Ding,
Molecular Cell, Volume 65 Supplemental Information Pacer Mediates the Function of Class III PI3K and HOPS Complexes in Autophagosome Maturation by Engaging Stx17 Xiawei Cheng, Xiuling Ma, Xianming Ding,
Supplement Figure 1. Plin5 Plin2 Plin1. KDEL-DSRed. Plin-YFP. Merge
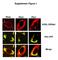 Supplement Figure 1 Plin5 Plin2 Plin1 KDEL-DSRed Plin-YFP Merge Supplement Figure 2 A. Plin5-Ab MitoTracker Merge AML12 B. Plin5-YFP Cytochrome c-cfp merge Supplement Figure 3 Ad.GFP Ad.Plin5 Supplement
Supplement Figure 1 Plin5 Plin2 Plin1 KDEL-DSRed Plin-YFP Merge Supplement Figure 2 A. Plin5-Ab MitoTracker Merge AML12 B. Plin5-YFP Cytochrome c-cfp merge Supplement Figure 3 Ad.GFP Ad.Plin5 Supplement
Table 1. Primers, annealing temperatures, and product sizes for PCR amplification.
 Table 1. Primers, annealing temperatures, and product sizes for PCR amplification. Gene Direction Primer sequence (5 3 ) Annealing Temperature Size (bp) BRCA1 Forward TTGCGGGAGGAAAATGGGTAGTTA 50 o C 292
Table 1. Primers, annealing temperatures, and product sizes for PCR amplification. Gene Direction Primer sequence (5 3 ) Annealing Temperature Size (bp) BRCA1 Forward TTGCGGGAGGAAAATGGGTAGTTA 50 o C 292
Partial list of differentially expressed genes from cdna microarray, comparing MUC18-
 Supplemental Figure legends Table-1 Partial list of differentially expressed genes from cdna microarray, comparing MUC18- silenced and NT-transduced A375SM cells. Supplemental Figure 1 Effect of MUC-18
Supplemental Figure legends Table-1 Partial list of differentially expressed genes from cdna microarray, comparing MUC18- silenced and NT-transduced A375SM cells. Supplemental Figure 1 Effect of MUC-18
Nature Structural & Molecular Biology: doi: /nsmb.3308
 Supplementary Figure 1 Analysis of CED-3 autoactivation and CED-4-induced CED-3 activation. (a) Repeat experiments of Fig. 1a. (b) Repeat experiments of Fig. 1b. (c) Quantitative analysis of three independent
Supplementary Figure 1 Analysis of CED-3 autoactivation and CED-4-induced CED-3 activation. (a) Repeat experiments of Fig. 1a. (b) Repeat experiments of Fig. 1b. (c) Quantitative analysis of three independent
mod-1::mcherry unc-47::gfp RME ser-4::gfp vm2 vm2 vm2 vm2 VNC
 A mod-1::mcherry unc-47::gfp RME B ser-4::gfp vm2 vm2 VNC vm2 vm2 VN Figure S1 Further details of mod- 1 and ser- 4 reporter expression patterns. (A) Adult head region showing mod- 1::mCherry and unc-
A mod-1::mcherry unc-47::gfp RME B ser-4::gfp vm2 vm2 VNC vm2 vm2 VN Figure S1 Further details of mod- 1 and ser- 4 reporter expression patterns. (A) Adult head region showing mod- 1::mCherry and unc-
(Toll Free) (outside US) Page 1
 888-266-5066 (Toll Free) 650-968-2200 (outside US) Page 1 System Biosciences (SBI) Contents User Manual I. Introduction and Background... 3 II. Available CytoTracers... 5 III. Protocol... 5 A. Transfection
888-266-5066 (Toll Free) 650-968-2200 (outside US) Page 1 System Biosciences (SBI) Contents User Manual I. Introduction and Background... 3 II. Available CytoTracers... 5 III. Protocol... 5 A. Transfection
B. ADM: C. D. Apoptosis: 1.68% 2.99% 1.31% Figure.S1,Li et al. number. invaded cells. HuH7 BxPC-3 DLD-1.
 A. - Figure.S1,Li et al. B. : - + - + - + E-cadherin CK19 α-sma vimentin β -actin C. D. Apoptosis: 1.68% 2.99% 1.31% - : - + - + - + Apoptosis: 48.33% 45.32% 44.59% E. invaded cells number 400 300 200
A. - Figure.S1,Li et al. B. : - + - + - + E-cadherin CK19 α-sma vimentin β -actin C. D. Apoptosis: 1.68% 2.99% 1.31% - : - + - + - + Apoptosis: 48.33% 45.32% 44.59% E. invaded cells number 400 300 200
Supplemental Figure 1 Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical
 Supplemental Figure Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical in all six REEPs are highlighted in green. Additional
Supplemental Figure Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical in all six REEPs are highlighted in green. Additional
Lecture 22: Molecular techniques DNA cloning and DNA libraries
 Lecture 22: Molecular techniques DNA cloning and DNA libraries DNA cloning: general strategy -> to prepare large quantities of identical DNA Vector + DNA fragment Recombinant DNA (any piece of DNA derived
Lecture 22: Molecular techniques DNA cloning and DNA libraries DNA cloning: general strategy -> to prepare large quantities of identical DNA Vector + DNA fragment Recombinant DNA (any piece of DNA derived
Revised: RG-RV2 by Fukuhara et al.
 Supplemental Figure 1 The generation of Spns2 conditional knockout mice. (A) Schematic representation of the wild type Spns2 locus (Spns2 + ), the targeted allele, the floxed allele (Spns2 f ) and the
Supplemental Figure 1 The generation of Spns2 conditional knockout mice. (A) Schematic representation of the wild type Spns2 locus (Spns2 + ), the targeted allele, the floxed allele (Spns2 f ) and the
MIT Department of Biology 7.013: Introductory Biology - Spring 2005 Instructors: Professor Hazel Sive, Professor Tyler Jacks, Dr.
 MIT Department of Biology 7.01: Introductory Biology - Spring 2005 Instructors: Professor Hazel Sive, Professor Tyler Jacks, Dr. Claudette Gardel iv) Would Xba I be useful for cloning? Why or why not?
MIT Department of Biology 7.01: Introductory Biology - Spring 2005 Instructors: Professor Hazel Sive, Professor Tyler Jacks, Dr. Claudette Gardel iv) Would Xba I be useful for cloning? Why or why not?
The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit
 Cell Reports, Volume 5 Supplemental Information The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit Andrey Poleshko, Katelyn M. Mansfield, Caroline
Cell Reports, Volume 5 Supplemental Information The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit Andrey Poleshko, Katelyn M. Mansfield, Caroline
Selected Techniques Part I
 1 Selected Techniques Part I Gel Electrophoresis Can be both qualitative and quantitative Qualitative About what size is the fragment? How many fragments are present? Is there in insert or not? Quantitative
1 Selected Techniques Part I Gel Electrophoresis Can be both qualitative and quantitative Qualitative About what size is the fragment? How many fragments are present? Is there in insert or not? Quantitative
Description: Nuclear morphology and dynamics in nontargeting sirna transfected cells. HeLa Kyoto
 Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Tables Title of file for HTML: Supplementary Movie 1 Description: Nuclear morphology and dynamics
Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Tables Title of file for HTML: Supplementary Movie 1 Description: Nuclear morphology and dynamics
SUPPLEMENTAL MATERIAL
 SUPPLEMENTAL MATERIAL MATERIALS AND METHODS Generation of TSPOΔ/Δ murine embryonic fibroblasts Embryos were harvested from 13.5-day pregnant TSPOfl/fl mice. After dissection to eviscerate and remove the
SUPPLEMENTAL MATERIAL MATERIALS AND METHODS Generation of TSPOΔ/Δ murine embryonic fibroblasts Embryos were harvested from 13.5-day pregnant TSPOfl/fl mice. After dissection to eviscerate and remove the
Shah, N.R., et al., Supplemental Data SUPPLEMENTAL TEXT. Cloning:
 Shah, N.R., et al., Supplemental Data SUPPLEMENTAL TEXT Cloning: pnmlgmab: the region containing lgma,lgmb, and the intergenic region upstream of lgma was amplified by PCR using genomic DNA of B. pertussis
Shah, N.R., et al., Supplemental Data SUPPLEMENTAL TEXT Cloning: pnmlgmab: the region containing lgma,lgmb, and the intergenic region upstream of lgma was amplified by PCR using genomic DNA of B. pertussis
Supplemental Information. Autoregulatory Feedback Controls. Sequential Action of cis-regulatory Modules. at the brinker Locus
 Developmental Cell, Volume 26 Supplemental Information Autoregulatory Feedback Controls Sequential Action of cis-regulatory Modules at the brinker Locus Leslie Dunipace, Abbie Saunders, Hilary L. Ashe,
Developmental Cell, Volume 26 Supplemental Information Autoregulatory Feedback Controls Sequential Action of cis-regulatory Modules at the brinker Locus Leslie Dunipace, Abbie Saunders, Hilary L. Ashe,
SONOMA STATE UNIVERSITY DEPARTMENT OF BIOLOGY BIOLOGY 344: CELL BIOLOGY Fall 2013
 SONOMA STATE UNIVERSITY DEPARTMENT OF BIOLOGY BIOLOGY 344: CELL BIOLOGY Fall 2013 Instructor Murali C. Pillai, PhD Office 214 Darwin Hall Telephone (707) 664-2981 E-mail pillai@sonoma.edu Website www.sonoma.edu/users/p/pillai
SONOMA STATE UNIVERSITY DEPARTMENT OF BIOLOGY BIOLOGY 344: CELL BIOLOGY Fall 2013 Instructor Murali C. Pillai, PhD Office 214 Darwin Hall Telephone (707) 664-2981 E-mail pillai@sonoma.edu Website www.sonoma.edu/users/p/pillai
Red Type Indicates Unique Site
 3600 G0605 pscaavmcmvmcsbghpa Plasmid Features: Coordinates Feature 980-1084 AAV2 5 ITR 1144-1666 modified CMV 1667-1761 MCS 1762-1975 BgHpA 1987-2114 AAV2 3 ITR 3031-3891 B-lactamase (Ampicillin) Antibiotic
3600 G0605 pscaavmcmvmcsbghpa Plasmid Features: Coordinates Feature 980-1084 AAV2 5 ITR 1144-1666 modified CMV 1667-1761 MCS 1762-1975 BgHpA 1987-2114 AAV2 3 ITR 3031-3891 B-lactamase (Ampicillin) Antibiotic
Supplemental Information
 Supplemental Information ATP-dependent unwinding of U4/U6 snrnas by the Brr2 helicase requires the C-terminus of Prp8 Corina Maeder 1,3, Alan K. Kutach 1,2,3, and Christine Guthrie 1 1 Department of Biochemistry
Supplemental Information ATP-dependent unwinding of U4/U6 snrnas by the Brr2 helicase requires the C-terminus of Prp8 Corina Maeder 1,3, Alan K. Kutach 1,2,3, and Christine Guthrie 1 1 Department of Biochemistry
Supplementary Figure 1 Collision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK2, PPK3 and PPK4 respectively.
 Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
CoralHue Fluo-chase Kit
 Amalgaam For Research Use Only. Not for use in diagnostic procedures. Protein-Protein Interaction Analysis CoralHue Fluo-chase Kit Code No. AM-1100 15 B Constitution Way Woburn, MA 01801 Phone: 1.800.200.5459
Amalgaam For Research Use Only. Not for use in diagnostic procedures. Protein-Protein Interaction Analysis CoralHue Fluo-chase Kit Code No. AM-1100 15 B Constitution Way Woburn, MA 01801 Phone: 1.800.200.5459
pwhere An optimized vector for mouse and rat transgenesis Catalog # pwhere For research use only Version # 05B11SV
 pwhere An optimized vector for mouse and rat transgenesis Catalog # pwhere For research use only Version # 05B11SV PRODUCT INFORMATION Content: pwhere is provided as 20 µg of lyophilized DNA. Storage and
pwhere An optimized vector for mouse and rat transgenesis Catalog # pwhere For research use only Version # 05B11SV PRODUCT INFORMATION Content: pwhere is provided as 20 µg of lyophilized DNA. Storage and
Supplemental Table 1 Primers used in study. Human. Mouse
 Supplemental Table 1 Primers used in study Human Forward primer region(5-3 ) Reverse primer region(5-3 ) RT-PCR GAPDH gagtcaacggatttggtcgt ttgattttggagggatctcg Raftlin atgggttgcggattgaacaagttaga ctgaggtataacaccaacgaatttcaggc
Supplemental Table 1 Primers used in study Human Forward primer region(5-3 ) Reverse primer region(5-3 ) RT-PCR GAPDH gagtcaacggatttggtcgt ttgattttggagggatctcg Raftlin atgggttgcggattgaacaagttaga ctgaggtataacaccaacgaatttcaggc
Supplemental Data. Na Xu et al. (2016). Plant Cell /tpc
 Supplemental Figure 1. The weak fluorescence phenotype is not caused by the mutation in At3g60240. (A) A mutation mapped to the gene At3g60240. Map-based cloning strategy was used to map the mutated site
Supplemental Figure 1. The weak fluorescence phenotype is not caused by the mutation in At3g60240. (A) A mutation mapped to the gene At3g60240. Map-based cloning strategy was used to map the mutated site
Different Upstream Activators Stimulate Transcription Through Distinct Coactivator Complexes
 Supplemental Material for Different Upstream Activators Stimulate Transcription Through Distinct Coactivator Complexes Dong-ki Lee, Soyoun Kim, and John T. Lis Due to be published as a Research Communication
Supplemental Material for Different Upstream Activators Stimulate Transcription Through Distinct Coactivator Complexes Dong-ki Lee, Soyoun Kim, and John T. Lis Due to be published as a Research Communication
Supplementary Information. A superfolding Spinach2 reveals the dynamic nature of. trinucleotide repeat RNA
 Supplementary Information A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat RNA Rita L. Strack 1, Matthew D. Disney 2 & Samie R. Jaffrey 1 1 Department of Pharmacology, Weill Medical
Supplementary Information A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat RNA Rita L. Strack 1, Matthew D. Disney 2 & Samie R. Jaffrey 1 1 Department of Pharmacology, Weill Medical
LINGO-1, A TRANSMEMBRANE SIGNALING PROTEIN, INHIBITS OLIGODENDROCYTE DIFFERENTIATION AND MYELINATION THROUGH INTERCELLULAR SELF- INTERACTIONS.
 Supplemental Data: LINGO-1, A TRANSMEMBRANE SIGNALING PROTEIN, INHIBITS OLIGODENDROCYTE DIFFERENTIATION AND MYELINATION THROUGH INTERCELLULAR SELF- INTERACTIONS. Scott Jepson, Bryan Vought, Christian H.
Supplemental Data: LINGO-1, A TRANSMEMBRANE SIGNALING PROTEIN, INHIBITS OLIGODENDROCYTE DIFFERENTIATION AND MYELINATION THROUGH INTERCELLULAR SELF- INTERACTIONS. Scott Jepson, Bryan Vought, Christian H.
Integrated Omics Study Delineates the Dynamics of Lipid Droplets in Rhodococcus Opacus PD630
 School of Natural Sciences and Mathematics 2013-10-22 Integrated Omics Study Delineates the Dynamics of Lipid Droplets in Rhodococcus Opacus PD630 UTD AUTHOR(S): Michael Qiwei Zhang 2013 The Authors This
School of Natural Sciences and Mathematics 2013-10-22 Integrated Omics Study Delineates the Dynamics of Lipid Droplets in Rhodococcus Opacus PD630 UTD AUTHOR(S): Michael Qiwei Zhang 2013 The Authors This
SUPPLEMENTARY INFORMATION. Tolerance of a knotted near infrared fluorescent protein to random circular permutation
 SUPPLEMENTARY INFORMATION Tolerance of a knotted near infrared fluorescent protein to random circular permutation Naresh Pandey 1,3, Brianna E. Kuypers 2,4, Barbara Nassif 1, Emily E. Thomas 1,3, Razan
SUPPLEMENTARY INFORMATION Tolerance of a knotted near infrared fluorescent protein to random circular permutation Naresh Pandey 1,3, Brianna E. Kuypers 2,4, Barbara Nassif 1, Emily E. Thomas 1,3, Razan
Supplementary Information
 Supplementary Information Supplementary Figures Figure S1. Study of mgtl translation in vitro. (A) Detection of 5 LR RNA using wild-type and anti-sd (91-95) substituted templates in a transcription-translation
Supplementary Information Supplementary Figures Figure S1. Study of mgtl translation in vitro. (A) Detection of 5 LR RNA using wild-type and anti-sd (91-95) substituted templates in a transcription-translation
Figure legends for supplement
 Figure legends for supplement Supplemental Figure 1 Characterization of purified and recombinant proteins Relevant fractions related the final stage of the purification protocol(bingham et al., 1998; Toba
Figure legends for supplement Supplemental Figure 1 Characterization of purified and recombinant proteins Relevant fractions related the final stage of the purification protocol(bingham et al., 1998; Toba
Somatic Primary pirna Biogenesis Driven by cis-acting RNA Elements and Trans-Acting Yb
 Cell Reports Supplemental Information Somatic Primary pirna Biogenesis Driven by cis-acting RNA Elements and Trans-Acting Yb Hirotsugu Ishizu, Yuka W. Iwasaki, Shigeki Hirakata, Haruka Ozaki, Wataru Iwasaki,
Cell Reports Supplemental Information Somatic Primary pirna Biogenesis Driven by cis-acting RNA Elements and Trans-Acting Yb Hirotsugu Ishizu, Yuka W. Iwasaki, Shigeki Hirakata, Haruka Ozaki, Wataru Iwasaki,
BIOTECHNOLOGY. Sticky & blunt ends. Restriction endonucleases. Gene cloning an overview. DNA isolation & restriction
 BIOTECHNOLOGY RECOMBINANT DNA TECHNOLOGY Recombinant DNA technology involves sticking together bits of DNA from different sources. Made possible because DNA & the genetic code are universal. 2004 Biology
BIOTECHNOLOGY RECOMBINANT DNA TECHNOLOGY Recombinant DNA technology involves sticking together bits of DNA from different sources. Made possible because DNA & the genetic code are universal. 2004 Biology
HPV E6 oncoprotein targets histone methyltransferases for modulating specific. Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu,
 1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
SUPPLEMENTARY EXPEMENTAL PROCEDURES
 SUPPLEMENTARY EXPEMENTAL PROCEDURES Plasmids- Total RNAs were extracted from HeLaS3 cells and reverse-transcribed using Superscript III Reverse Transcriptase (Invitrogen) to obtain DNA template for the
SUPPLEMENTARY EXPEMENTAL PROCEDURES Plasmids- Total RNAs were extracted from HeLaS3 cells and reverse-transcribed using Superscript III Reverse Transcriptase (Invitrogen) to obtain DNA template for the
CONTENTS. LIST OF FIGURES... i. LIST OF TABLES... iii. SUMMARY OF THE WORK DONE... vi
 CONTENTS SECTION ~ LIST OF FIGURES... i LIST OF TABLES... iii LIST OF ABBREVIATIONS USED... ~... iv SUMMARY OF THE WORK DONE... vi 1. INTROD UCTION... :.................................... 1 1.1. AN OVERVIEW
CONTENTS SECTION ~ LIST OF FIGURES... i LIST OF TABLES... iii LIST OF ABBREVIATIONS USED... ~... iv SUMMARY OF THE WORK DONE... vi 1. INTROD UCTION... :.................................... 1 1.1. AN OVERVIEW
Supporting Information
 Supporting Information Gómez-Marín et al. 10.1073/pnas.1505463112 SI Materials and Methods Generation of BAC DK74B2-six2a::GFP-six3a::mCherry-iTol2. BAC clone (number 74B2) from DanioKey zebrafish BAC
Supporting Information Gómez-Marín et al. 10.1073/pnas.1505463112 SI Materials and Methods Generation of BAC DK74B2-six2a::GFP-six3a::mCherry-iTol2. BAC clone (number 74B2) from DanioKey zebrafish BAC
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature06721 SUPPLEMENTARY INFORMATION. Supplemental Figure Legends Supplemental Figure 1 The distribution of hatx-1[82q] in Cos7 cells. Cos7 cells are co-transfected with hatx-1[82q]-gfp (green)
doi: 10.1038/nature06721 SUPPLEMENTARY INFORMATION. Supplemental Figure Legends Supplemental Figure 1 The distribution of hatx-1[82q] in Cos7 cells. Cos7 cells are co-transfected with hatx-1[82q]-gfp (green)
SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURES
 SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURES Supplementary Figure 1. Generation of inducible BICD2 knock-out mice. A) The mouse BICD2 locus and gene targeting constructs. To generate an inducible Bicd2
SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURES Supplementary Figure 1. Generation of inducible BICD2 knock-out mice. A) The mouse BICD2 locus and gene targeting constructs. To generate an inducible Bicd2
Hyper sensitive protein detection by Tandem-HTRF reveals Cyclin D1
 Hyper sensitive protein detection by Tandem-HTRF reveals Cyclin D1 dynamics in adult mouse Alexandre Zampieri, Julien Champagne, Baptiste Auzemery, Ivanna Fuentes, Benjamin Maurel and Frédéric Bienvenu
Hyper sensitive protein detection by Tandem-HTRF reveals Cyclin D1 dynamics in adult mouse Alexandre Zampieri, Julien Champagne, Baptiste Auzemery, Ivanna Fuentes, Benjamin Maurel and Frédéric Bienvenu
Chapter 20 Recombinant DNA Technology. Copyright 2009 Pearson Education, Inc.
 Chapter 20 Recombinant DNA Technology Copyright 2009 Pearson Education, Inc. 20.1 Recombinant DNA Technology Began with Two Key Tools: Restriction Enzymes and DNA Cloning Vectors Recombinant DNA refers
Chapter 20 Recombinant DNA Technology Copyright 2009 Pearson Education, Inc. 20.1 Recombinant DNA Technology Began with Two Key Tools: Restriction Enzymes and DNA Cloning Vectors Recombinant DNA refers
J. Cell Sci. 128: doi: /jcs : Supplementary Material. Supplemental Figures. Journal of Cell Science Supplementary Material
 Supplemental Figures Figure S1. Trio controls endothelial barrier function. (A) TagRFP-shTrio constructs were expressed in ECs. Western blot shows efficient Trio knockdown in TagRFP-expressing ECs. (B)
Supplemental Figures Figure S1. Trio controls endothelial barrier function. (A) TagRFP-shTrio constructs were expressed in ECs. Western blot shows efficient Trio knockdown in TagRFP-expressing ECs. (B)
Rotation Report Sample Version 3. Due Date: August 9, Analysis of the Guanine Nucleotide Exchange Activity of the S. cerevisiae Ats1 Protein
 Rotation Report Sample Version 3 Due Date: August 9, 1998 Analysis of the Guanine Nucleotide Exchange Activity of the S. cerevisiae Ats1 Protein Anita H. Corbett Advisor: Amy Jones Rotation 1 Abstract:
Rotation Report Sample Version 3 Due Date: August 9, 1998 Analysis of the Guanine Nucleotide Exchange Activity of the S. cerevisiae Ats1 Protein Anita H. Corbett Advisor: Amy Jones Rotation 1 Abstract:
To assess the localization of Citrine fusion proteins, we performed antibody staining to
 Trinh et al 1 SUPPLEMENTAL MATERIAL FlipTraps recapitulate endogenous protein localization To assess the localization of Citrine fusion proteins, we performed antibody staining to compare the expression
Trinh et al 1 SUPPLEMENTAL MATERIAL FlipTraps recapitulate endogenous protein localization To assess the localization of Citrine fusion proteins, we performed antibody staining to compare the expression
oligonucleotide primers listed in Supplementary Table 2 and cloned into ppcr-script (Stratagene) before digestion
 Supplementary Methods. Construction of BiFC vectors The full-length ARC3 cdna, ARC3 1-1794 and ARC3 1-1083 were amplified from ppcr-script/arc3 using the oligonucleotide primers listed in Supplementary
Supplementary Methods. Construction of BiFC vectors The full-length ARC3 cdna, ARC3 1-1794 and ARC3 1-1083 were amplified from ppcr-script/arc3 using the oligonucleotide primers listed in Supplementary
Gene disruption and construction of C-terminally tagged strains
 Supplementary information Supplementary Methods Gene disruption and construction of C-terminally tagged strains A PCR-based gene targeting method (Bähler et al., 1998; Sato et al., 2005) was used for constructing
Supplementary information Supplementary Methods Gene disruption and construction of C-terminally tagged strains A PCR-based gene targeting method (Bähler et al., 1998; Sato et al., 2005) was used for constructing
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.
 Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
