Greene 1. Finishing of DEUG The entire genome of Drosophila eugracilis has recently been sequenced using Roche
|
|
|
- Camron Fletcher
- 5 years ago
- Views:
Transcription
1 Greene 1 Harley Greene Bio434W Elgin Finishing of DEUG Abstract The entire genome of Drosophila eugracilis has recently been sequenced using Roche 454 pyrosequencing and Illumina paired-end reads sequencing. In this project, the contig DEUG , a 100 kb genomic region of the D. eugracilis dot chromosome, was finished by confirming and correcting the consensus sequence. The original assembly of the region contained 1 gap, 138 highly discrepant regions, 35 low coverage regions, and 1 region of low consensus quality. These regions were inspected for mononucleotide runs (MNRs), which often misinform the consensus sequence due to 454 pyrosequencing errors and require correcting. Here, 31 MNRs were either corrected by sequencing read inspection or confirmed. PCR primers were created to cover the unresolved gap; Sanger sequencing data will be required for this purpose. No polymorphisms were identified in DEUG Besides the one gap, this genomic region is ready for annotating. Introduction DNA in eukaryotic cells is found in one of two formations: euchromatin and heterochromatin. Euchromatin encompasses transcriptionally active genes which are loosely packaged in the nucleus to allow for easy access of transcription factors and RNA polymerase. Heterochromatin is typically associated with transcriptionally inactive genes and these domains are relatively compact. Heterochromatin is most often found near the centromeres and telomeres of chromosomes. Heterochromatin can be modified to allow for transcription, but it is much less
2 Greene 2 accessible than euchromatin. Additionally, heterochromatin and euchromatin have unique histone and DNA modifications. For example, chromatin regions that are transcriptionally active and are associated with euchromatin often have the histone acetylation on H3K9. These marks provide a clear way to differentiate heterochromatin and euchromatin and provide insight into gene regulation mechanisms. The species Drosophila eugracilis has a small dot chromosome (the F element) containing approximately 80 actively transcribed genes, but by most measures is entirely heterochromatic. The dot chromosome illustrates an unusual scenario of heterochromatic genes being expressed at the same level as euchromatic genes from other chromosomes. D. eugracilis has recently been sequenced but still requires finishing and annotating. Finishing and annotating D. eugracilis will allow for genomic comparisons with Drosophila melanogaster, and other evolutionarily close neighbor species, to study the mechanisms of heterochromatic gene expression. The initial D. eugracilis sequenced genome was constructed using two types of sequencing data: Roche 454 pyrosequencing and Illumina paired end reads. 454 pyrosequencing, which produces long reads (~450 nts) compared to Illumina, has been known to drop off in sequencing quality at mononucleotide runs (MNRs). On the other hand, Illumina sequencing reads are more precise, but are shorter in comparison ( nts). Therefore, the short Illumina reads can be used to correct errors in incorrect MNRs reported in 454 reads. In this report, the 100 kb contig DEUG was finished by analyzing its consensus sequence and making changes to the consensus when necessary. All analysis and sequence changes were conducted on the program Consed.
3 Greene 3 Initial Assembly Contig DEUG maps to bases 90, ,000 of the D. eugracilis dot chromosome (Figure 1). The initial Assembly View (Figure 2) showed a single contig of 100 kb. The contig contained one gap, 138 highly discrepant regions, 35 low coverage regions, and 1 region of low consensus quality. Additionally, there were several regions of repetitious sequences and several incorrectly matched forward/reverse read pairs. The incorrectly matched read pairs were likely marked incorrect due to the highly repetitive nature of this contig s sequence. Figure 1: Position of DEUG in D. eugracilis dot chromosome: contig finished in this report, DEUG , is highlighted by red box. It represents region 90, ,000 in the D. eugracilis dot chromosome. Figure 2: Initial Assembly View of DEUG : 100,000 bp region visualized using assembly view on Consed. Green line represents depth of reads across the contig. Red lines indicate incorrect spacing or orientation between forward and reverse read pairs. Black and orange lines indicate repetitious sequences that map to multiple locations. High Quality Discrepancies The contig was first examined for high quality discrepancies. High quality discrepancies were identified as regions where at least three reads did not match the consensus sequence, ignoring bases with a Phred quality score below 30. In total, 138 highly discrepant regions were identified using Consed. These high quality discrepancies were then examined for MNRs. Of the
4 Greene highly discrepant regions, 73 were MNRs. For each MNR, the 454 and Illumina reads were inspected to ensure the consensus was correct. At each location, the number of bases in the MNR consensus were counted and compared to the high quality Illumina reads. The consensus was confirmed if the Illumina reads matched the consensus. The consensus required editing if the reads did not match. Thirty-one MNRs required editing to correct the consensus sequence. All MNRs identified were all mono-a or mono-t runs, which is unsurprising since heterochromatin is known to be A/T rich. Almost all of the changed bases were due to slightly misaligned reads which shortened the MNR in the consensus and were corrected by adding a single base to the beginning or end of the MNR. The addition of an A at position 42,613 illustrates this typical problem (Figure 3). The consensus sequence showed eight As with a pad (*) inserted after the fifth A. The first three Illumina reads have a ninth A at the end of the MNR, Figure 3: Common MNR correction: This is a common example of a misaligned MNR that was resolved by adding a single base. Here, a MNR of eight As was seen in the consensus but a MNR of nine As was seen in the Illumina reads (region within blue box). A single A was added at position 42,613 to resolve the area. Read names that start with USI refer to Illumina data and read names that start with a G refer to 454 data.
5 Greene 5 which is not represented in the consensus. Below the Illumina reads are 37 low quality 454 reads which provide little help for this situation. Below the 454 reads are 19 more Illumina reads that have a ninth A at the beginning of the mono-a run. Since all of the Illumina reads indicated that the MNR should have nine As instead of eight As, an A was inserted into the consensus. Besides these easily reconcilable errors, there were a few more interesting and difficult cases. One interesting case was a highly discrepant position at 38,334 (Figure 4). This region represented an overlap between two repeat elements present on the dot chromosome. The consensus showed six Ts, with the first and last position of the MNR marked as highly discrepant. The first 14 Illumina reads only had five Ts with a pad inserted in the first position of the MNR. Additionally, there were three Illumina reads, and many mid- and low-quality 454 reads (indicated by grey boxes around letters), that also had five Ts, but with a pad inserted in the last position of the MNR. Due to the misalignment of the reads, an extra T had been added to the Figure 4: Highly discrepant position at 38,334: 6Ts are seen in the consensus sequence but Illumina and 454 reads have only 5Ts with a pad on one side (green box). The blue tag on the consensus corresponds to a repeat element tag for likely transposable elements. The purple tag corresponds to two overlapping repeat tags.
6 Greene 6 consensus sequence. To remedy the error, the first T in the MNR was replaced with a pad to align with the Illumina reads (Figure 5). This instance was the only time an element besides an A or a T was added to correct the consensus sequence. Figure 5: Corrected consensus at position 38,334: A T at the beginning of the MNR was replaced with a pad (*) to align consensus with high quality Illumina reads (green box). MNR changed from 6Ts to 5Ts. Another difficult case was at position 6397, where two MNRs appeared in a row: seven Ts and eight As with a pad in between them. Five Illumina reads had an A instead of a T to the right of the pad and three Illumina reads which had an extra A at the end of the mono-a run. Additionally, 17 Illumina reads had an extra A at the beginning of the mono-a run, having nine As instead of the eight As in the consensus. The 454 reads were of very low quality and were not helpful in correcting the issue. An A was added to replace the pad between the two MNRs,
7 Greene 7 making the mono-a run nine As instead of eight As and confirming the mono-t run of seven Ts (Figure 6). Figure 6: Corrected highly discrepant position at 6397: Consensus changed from MNRs of seven Ts and eight As to seven Ts and nine As to align with high quality Illumina reads (green box). Lowercase a seen in consensus marks edit made to consensus sequence. Blue tag on T corresponds to comment tag added to consensus. In total, 73 (53%) of all identified high quality discrepancies were MNRs. Forty-two of the MNRs were confirmed and 31 bases were added to correct the remaining MNRs (Table 1). Except for the one mentioned position where a pad was added, all corrections involved the addition of an A or a T. Polymorphisms After all MNRs were identified, the remaining high quality discrepancies were analyzed for polymorphisms. A polymorphism corresponds to a position where the high quality reads indicate that two bases are equally likely. In other words, it is a position where there is an approximate 50/50 split among the high quality reads as to which base should be in the consensus. No polymorphisms were identified in the contig, although there was one interesting
8 Greene 8 case (Figure 7). At position 18,829, there was an MNR of 5As, followed by a T and another 3As. In 9 Illumina reads, the T is replaced with an A, resulting in a 9A MNR. In the remaining Illumina and 454 reads (over 20 reads), the T is kept in that position. Based on this data, the consensus was not changed and the T was kept within the MNR. This region is particularly interesting because the incorrect Illumina reads, except for one of them, do not show other signs of being incorrectly mapped. On the other hand, this region is in the middle of a repeat tag, where incorrect mapping and misalignment is very frequent. This region was not marked as a potential polymorphism and no polymorphisms were identified in the contig. Figure 7: Interesting high quality discrepancy at position 18,829: 9 high quality Illumina reads indicate an incorrect base (A) compared to the consensus (T) (region within green box). The consensus was not changed because other Illumina and 454 reads show inclusion of the T. Additionally, the region was not marked as polymorphism because there is not a 50/50 split between reads, and the region is in a repeat tag.
9 Greene 9 Low Coverage Regions After going through all high quality discrepancies, the next regions investigated were areas with low coverage. Areas of low coverage corresponded to regions with fewer than 40 reads covering the sequence. Thirty-five areas of low coverage were identified. One of these areas was the gap found in the contig and is discussed in the next section. In the remaining 34 low coverage regions, MNRs were identified, but no evidence was found to change the consensus at these locations. There were at least five Illumina reads in each region that matched the consensus. An example of an area of low coverage is show in Figure 8. In the end, no bases were changed in low coverage regions. Figure 8: Example of low coverage region: MNR of As in area of low coverage starts at position 37,044 and is highlighted by orange box. Even though this is an area of low coverage, there is no evidence that the consensus should be changed.
10 Greene 10 Low Consensus Quality Regions With all high quality discrepancies and areas of low coverage analyzed for MNRs and polymorphisms, the next step was to look at regions of low consensus quality. Regions with Phred quality scores less than or equal to 25 or 98 were considered low consensus quality regions. (Regions that are edited by the finisher are automatically given a quality score of 98.) In total, there were 32 low consensus quality regions, but 31 of those were edits made to the consensus sequence. Therefore, there was only one low consensus quality region to examine. This low consensus quality region was found at positions 90,392-90,415 and corresponded to a gap in the contig (Figure 9). To attempt to resolve the gap by a forced join, a unique sequence had to be found on either side of the gap. The sequence ATAAAGTGTATAAAATATATTA was identified to be slightly upstream of the gap and immediately following the gap (Figure 10). Searching for the sequence throughout the contig showed that it only appeared at these two locations. The contig-spanning read used to initially assemble the contig (KB464927: ) was removed to allow for tearing. The contig was torn at the first A of the overlapping sequence, creating two contigs (Figure 11). Next, the two contigs were compared using the overlapping sequence and were aligned to see if a forced join could be completed (Figure 12). Figure 9: Picture of gap: Gap found at positions 90,392-90,415 (region within red box).
11 Greene 11 Figure 10: Common sequence found on either side of gap: The sequence ATAAAGTGTATAAAATATATTA, highlighted by yellow box, was found on either side of the gap, indicating that the gap may be resolved by matching up the sequences. This unique sequence was only found at these two locations. Figure 11: Assembly View post-tear: The Assembly View of the contigs after the initial contig was torn shows two sepearte contigs ( and ). As with the initial Assembly View, the green line represents depth of reads across the contig. Red lines indicate incorrect spacing or orientation between forward and reverse read pairs. Black and orange lines indicate repetitious elements that map to multiple locations. The blue/green lines near the gap indicate paired reads from the same sequencing reaction that span the gap. Figure 12: Compare Contig View: Upon comparing the two contigs, the overlapping sequence matches up (red box), but unique sequences on either side prevent a forced join.
12 Greene 12 Unfortunately, even though the overlapping sequence was the same for both contigs, there were unique sequences on either side of the overlapping sequence that prevented a force join. Additionally, when looking at the post-tear Assembly View in Figure 11, Crossmatch results show that there are no repeat sequences found on both sides of the gap. Furthermore, there were many paired end reads spanning the gap, indicating that a forced join would not resolve the gap (estimated size of ~1200bp). Since the gap could not be resolved, more sequencing data is needed. PCR primers were created using Consed to cover the gap and the neighboring low quality areas (Table 2). Two primer pairs were chosen to order. Each pair met ideal PCR primer criteria, with the distance between them being less than 1000 bp and the primers having the same melting temperature. Two pairs were chosen to maximize the coverage of the area. Once sequencing data for this region is added, the contig will be finished and annotation may commence. Final Assembly/Conclusion Figure 13 shows the final assembly of contig DEUG It is not drastically different from the initial assembly of DEUG , but it shows the edited bases (31 total) and the PCR primers created to cover the gap. The red lines, representing misaligned reads, were not removed and realigned to the contig because they did not contaminate the consensus sequence. Although not visible in the photo, all MNRs associated with high quality discrepancies and regions of low coverage (73 total) were investigated and either confirmed or corrected. Upon receiving sequencing data for the gap, the contig will be ready for annotation.
13 Greene 13 Figure 13: Final Assembly View of DEUG : Green marks below contig represent edited bases and yellow marks below contig represent PCR primers constructed. Acknowledgments This research project would not have been possible without the help of the Bio434W teaching team. I would like to thank Dr. Elgin, Dr. Shaffer, Wilson Leung, Lee Trani, and Ryan Freidman for their overall guidance and wisdom, their expertise on Consed, and their assistance in my project, and Dr. Bednarski for improving my writing. I would also like to thank Washington University in St. Louis and the Genomics Education Partnership for making this research possible.
14 Greene 14 Table 1: List of MNRs: All MNRs identified in Contig DEUG ; under Evidence, number of reads used as evidence given if sequence was changed. Position Analysis Change to Evidence Consensus 3338 MNR of Ts +T Illumina (21 reads) 3950 MNR of As confirmed consensus Illumina 3976 MNR of As +A Illumina (20 reads) 6252 MNR of As confirmed consensus Illumina 6388 MNR of Ts confirmed consensus Illumina 6397 MNR of As +A Illumina (17 reads) 6864 MNR of As confirmed consensus Illumina 8016 MNR of Ts confirmed consensus Illumina 8111 MNR of Ts +T Illumina (15 reads) 9473 MNR of As +A Illumina (20 reads) 12,593 MNR of As confirmed consensus Illumina 14,047 MNR of As +A Illumina (18 reads) 14,780 MNR of As +A Illumina (9 reads) 15,954 MNR of As +A Illumina (15 reads) 16,977 MNR of As +A Illumina (10 reads) 17,428 MNR of Ts confirmed consensus Illumina 18,804 MNR of Ts confirmed consensus Illumina 18,822 2 MNRs of As with T in the middle confirmed consensus Illumina (9 Illumina reads incorrectly have single MNR of As) 19,979 MNR of As confirmed consensus Illumina 20,016 MNR of As confirmed consensus Illumina 20,958 MNR of As confirmed consensus Illumina 21,498 MNR of As confirmed consensus Illumina 21,567-21,568 MNR of As confirmed consensus Illumina 21,872 MNR of As confirmed consensus Illumina 22,275 MNR of As confirmed consensus Illumina 24,432 MNR of As confirmed consensus Illumina 27,832 MNR of As confirmed consensus Illumina 28,878 MNR of As +A Illumina (5 reads) 30,354 MNR of As confirmed consensus Illumina 31,249 MNR of Ts +T Illumina (10 reads) 31,559 MNR of Ts confirmed consensus Illumina 34,946 MNR of Ts confirmed consensus Illumina 36,261 MNR of As confirmed consensus Illumina 37,842 MNR of Ts confirmed consensus Illumina 38,334 MNR of Ts + pad (*) Ilumina (20 reads) 41,244 MNR of Ts +T Illumina (21 reads) 42,613 MNR of As +A Illumina (22 reads)
15 Greene 15 44,295 MNR of As confirmed consensus Illumina 46,084 MNR of Ts confirmed consensus Illumina 47,016 MNR of As confirmed consensus Illumina 52,185 MNR of Ts +T Illumina (12 reads) 52,486 MNR of Ts confirmed consensus Illumina 53,822 MNR of As confirmed consensus Illumina 55,540 MNR of Ts confirmed consensus Illumina 55,855 MNR of As +A Illumina (17 reads) 56,880 MNR of Ts +T Illumina (20 reads) 59,237 MNR of Ts confirmed consensus Illumina 59,270 MNR of As +A Illumina (15 reads) 60,396 MNR of As +A Illumina (15 reads) 61,252 MNR of As +A Illumina (19 reads) 61,426 MNR of As confirmed consensus Illumina 63,510 MNR of As confirmed consensus Illumina 68,006 MNR of As confirmed consensus Illumina 70,795 MNR of Ts confirmed consensus Illumina 70,841 MNR of Ts confirmed consensus Illumina 71,038 MNR of Ts +T Illumina (27 reads) 72,339 MNR of Ts confirmed consensus Illumina 73,391 MNR of Ts confirmed consensus Illumina 75,506 MNR of Ts confirmed consensus Illumina 76,163 MNR of As +A Illumina (19) 78, 926 MNR of Ts confirmed consensus Illumina 81,058 MNR of As +A Illumina (25) 81,609 MNR of Ts +T Illumina (20) 83,114 MNR of Ts +T Illumina (11) 86,256 MNR of As +A Illumina (17) 88,304 MNR of As +A Illumina (15) 90, 971 MNR of As +A Illumina (29) 93,092 MNR of Ts +T Illumina (6) 94,002 MNR of Ts confirmed consensus Illumina 94,433 MNR of Ts confirmed consensus Illumina 94,678-94,679 MNR of As confirmed consensus Illumina 94,861 MNR of As confirmed consensus Illumina 95,113 MNR of As +A Illumina (13 reads) 96,593 MNR of Ts +T Illumina (26 reads) 98,193 MNR of As +A Illumina (13 reads)
16 Greene 16 Table 2: PCR Primers: In table are two pairs of PCR primers chosen to cover gap from 90,392-90,415. P1 = primer 1; P2 = primer 2; Mp = melting temperature; Start-end = beginning and end position of primer in contig. Distance P1 Start-end Mp P2 Start-end mp 311 gtcgaaaatatcgtatgatatcaat tttatcaatttaaagaataaaattagacac gtcgaaaatatcgtatgatatcaat atgtgtaaacgctatacttagatgtc
The goal of this project was to prepare the DEUG contig which covers the
 Prakash 1 Jaya Prakash Dr. Elgin, Dr. Shaffer Biology 434W 10 February 2017 Finishing of DEUG4927010 Abstract The goal of this project was to prepare the DEUG4927010 contig which covers the terminal 99,279
Prakash 1 Jaya Prakash Dr. Elgin, Dr. Shaffer Biology 434W 10 February 2017 Finishing of DEUG4927010 Abstract The goal of this project was to prepare the DEUG4927010 contig which covers the terminal 99,279
Finishing of DFIC This project sought to finish DFIC , the terminal 45 kb of the Drosophila
 Lin 1 Kevin Lin Bio 434W Dr. Elgin 26 February 2016 Finishing of DFIC6622001 Abstract This project sought to finish DFIC6622001, the terminal 45 kb of the Drosophila ficusphila dot chromosome. The initial
Lin 1 Kevin Lin Bio 434W Dr. Elgin 26 February 2016 Finishing of DFIC6622001 Abstract This project sought to finish DFIC6622001, the terminal 45 kb of the Drosophila ficusphila dot chromosome. The initial
Finishing of DELE Drosophila elegans has been sequenced using Roche 454 pyrosequencing and Illumina
 Sarah Swiezy Dr. Elgin, Dr. Shaffer Bio 434W 27 February 2015 Finishing of DELE8596009 Abstract Drosophila elegans has been sequenced using Roche 454 pyrosequencing and Illumina technology. DELE8596009,
Sarah Swiezy Dr. Elgin, Dr. Shaffer Bio 434W 27 February 2015 Finishing of DELE8596009 Abstract Drosophila elegans has been sequenced using Roche 454 pyrosequencing and Illumina technology. DELE8596009,
Finishing of Fosmid 1042D14. Project 1042D14 is a roughly 40 kb segment of Drosophila ananassae
 Schefkind 1 Adam Schefkind Bio 434W 03/08/2014 Finishing of Fosmid 1042D14 Abstract Project 1042D14 is a roughly 40 kb segment of Drosophila ananassae genomic DNA. Through a comprehensive analysis of forward-
Schefkind 1 Adam Schefkind Bio 434W 03/08/2014 Finishing of Fosmid 1042D14 Abstract Project 1042D14 is a roughly 40 kb segment of Drosophila ananassae genomic DNA. Through a comprehensive analysis of forward-
Finishing Drosophila elegans Contig DELE This project aimed to finish the contig DELE from the F element (chromosome 6)
 Ji Han (Ricky) Lee 2015. 02. 27 Bio 434W Professor Elgin Finishing Drosophila elegans Contig DELE8596013 Abstract This project aimed to finish the contig DELE8596013 from the F element (chromosome 6) of
Ji Han (Ricky) Lee 2015. 02. 27 Bio 434W Professor Elgin Finishing Drosophila elegans Contig DELE8596013 Abstract This project aimed to finish the contig DELE8596013 from the F element (chromosome 6) of
Finishing Drosophila Ananassae Fosmid 2728G16
 Finishing Drosophila Ananassae Fosmid 2728G16 Kyle Jung March 8, 2013 Bio434W Professor Elgin Page 1 Abstract For my finishing project, I chose to finish fosmid 2728G16. This fosmid carries a segment of
Finishing Drosophila Ananassae Fosmid 2728G16 Kyle Jung March 8, 2013 Bio434W Professor Elgin Page 1 Abstract For my finishing project, I chose to finish fosmid 2728G16. This fosmid carries a segment of
Sundaram DGA43A19 Page 1. Finishing Drosophila grimshawi Fosmid: DGA43A19 Varun Sundaram 2/16/09
 Sundaram DGA43A19 Page 1 Finishing Drosophila grimshawi Fosmid: DGA43A19 Varun Sundaram 2/16/09 Sundaram DGA43A19 Page 2 Abstract My project focused on the fosmid clone DGA43A19. The main problems with
Sundaram DGA43A19 Page 1 Finishing Drosophila grimshawi Fosmid: DGA43A19 Varun Sundaram 2/16/09 Sundaram DGA43A19 Page 2 Abstract My project focused on the fosmid clone DGA43A19. The main problems with
Finishing Drosophila Grimshawi Fosmid Clone DGA19A15. Matthew Kwong Bio4342 Professor Elgin February 23, 2010
 Finishing Drosophila Grimshawi Fosmid Clone DGA19A15 Matthew Kwong Bio4342 Professor Elgin February 23, 2010 1 Abstract The primary aim of the Bio4342 course, Research Explorations and Genomics, is to
Finishing Drosophila Grimshawi Fosmid Clone DGA19A15 Matthew Kwong Bio4342 Professor Elgin February 23, 2010 1 Abstract The primary aim of the Bio4342 course, Research Explorations and Genomics, is to
Finishing Fosmid DMAC-27a of the Drosophila mojavensis third chromosome
 Finishing Fosmid DMAC-27a of the Drosophila mojavensis third chromosome Ruth Howe Bio 434W 27 February 2010 Abstract The fourth or dot chromosome of Drosophila species is composed primarily of highly condensed,
Finishing Fosmid DMAC-27a of the Drosophila mojavensis third chromosome Ruth Howe Bio 434W 27 February 2010 Abstract The fourth or dot chromosome of Drosophila species is composed primarily of highly condensed,
Finishing Drosophila grimshawi Fosmid Clone DGA23F17. Kenneth Smith Biology 434W Professor Elgin February 20, 2009
 Finishing Drosophila grimshawi Fosmid Clone DGA23F17 Kenneth Smith Biology 434W Professor Elgin February 20, 2009 Smith 2 Abstract: My fosmid, DGA23F17, did not present too many challenges when I finished
Finishing Drosophila grimshawi Fosmid Clone DGA23F17 Kenneth Smith Biology 434W Professor Elgin February 20, 2009 Smith 2 Abstract: My fosmid, DGA23F17, did not present too many challenges when I finished
A Guide to Consed Michelle Itano, Carolyn Cain, Tien Chusak, Justin Richner, and SCR Elgin.
 1 A Guide to Consed Michelle Itano, Carolyn Cain, Tien Chusak, Justin Richner, and SCR Elgin. Main Window Figure 1. The Main Window is the starting point when Consed is opened. From here, you can access
1 A Guide to Consed Michelle Itano, Carolyn Cain, Tien Chusak, Justin Richner, and SCR Elgin. Main Window Figure 1. The Main Window is the starting point when Consed is opened. From here, you can access
Finished (Almost) Sequence of Drosophila littoralis Chromosome 4 Fosmid Clone XAAA73. Seth Bloom Biology 4342 March 7, 2004
 Finished (Almost) Sequence of Drosophila littoralis Chromosome 4 Fosmid Clone XAAA73 Seth Bloom Biology 4342 March 7, 2004 Summary: I successfully sequenced Drosophila littoralis fosmid clone XAAA73. The
Finished (Almost) Sequence of Drosophila littoralis Chromosome 4 Fosmid Clone XAAA73 Seth Bloom Biology 4342 March 7, 2004 Summary: I successfully sequenced Drosophila littoralis fosmid clone XAAA73. The
Finishing Drosophila ananassae Fosmid 2410F24
 Nick Spies Research Explorations in Genomics Finishing Report Elgin, Shaffer and Leung 23 February 2013 Abstract: Finishing Drosophila ananassae Fosmid 2410F24 Finishing Drosophila ananassae fosmid clone
Nick Spies Research Explorations in Genomics Finishing Report Elgin, Shaffer and Leung 23 February 2013 Abstract: Finishing Drosophila ananassae Fosmid 2410F24 Finishing Drosophila ananassae fosmid clone
MODULE TSS1: TRANSCRIPTION START SITES INTRODUCTION (BASIC)
 MODULE TSS1: TRANSCRIPTION START SITES INTRODUCTION (BASIC) Lesson Plan: Title JAMIE SIDERS, MEG LAAKSO & WILSON LEUNG Identifying transcription start sites for Peaked promoters using chromatin landscape,
MODULE TSS1: TRANSCRIPTION START SITES INTRODUCTION (BASIC) Lesson Plan: Title JAMIE SIDERS, MEG LAAKSO & WILSON LEUNG Identifying transcription start sites for Peaked promoters using chromatin landscape,
Genome Projects. Part III. Assembly and sequencing of human genomes
 Genome Projects Part III Assembly and sequencing of human genomes All current genome sequencing strategies are clone-based. 1. ordered clone sequencing e.g., C. elegans well suited for repetitive sequences
Genome Projects Part III Assembly and sequencing of human genomes All current genome sequencing strategies are clone-based. 1. ordered clone sequencing e.g., C. elegans well suited for repetitive sequences
The fourth chromosome: targeting heterochromatin formation in Drosophila. Drosophila melanogaster chromosomes
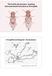 The fourth chromosome: targeting heterochromatin formation in Drosophila Drosophila melanogaster chromosomes modified from TS Painter, 1934, J. Hered 25: 465-476. 1 DNA packaging domains HATs Euchromatin
The fourth chromosome: targeting heterochromatin formation in Drosophila Drosophila melanogaster chromosomes modified from TS Painter, 1934, J. Hered 25: 465-476. 1 DNA packaging domains HATs Euchromatin
Annotation of contig27 in the Muller F Element of D. elegans. Contig27 is a 60,000 bp region located in the Muller F element of the D. elegans.
 David Wang Bio 434W 4/27/15 Annotation of contig27 in the Muller F Element of D. elegans Abstract Contig27 is a 60,000 bp region located in the Muller F element of the D. elegans. Genscan predicted six
David Wang Bio 434W 4/27/15 Annotation of contig27 in the Muller F Element of D. elegans Abstract Contig27 is a 60,000 bp region located in the Muller F element of the D. elegans. Genscan predicted six
CHAPTERS , 17: Eukaryotic Genetics
 CHAPTERS 14.1 14.6, 17: Eukaryotic Genetics 1. Review the levels of DNA packing within the eukaryote nucleus. Label each level. (A similar diagram is on pg 188 of your textbook.) 2. How do the coding regions
CHAPTERS 14.1 14.6, 17: Eukaryotic Genetics 1. Review the levels of DNA packing within the eukaryote nucleus. Label each level. (A similar diagram is on pg 188 of your textbook.) 2. How do the coding regions
Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Supplementary Material
 Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions Joshua N. Burton 1, Andrew Adey 1, Rupali P. Patwardhan 1, Ruolan Qiu 1, Jacob O. Kitzman 1, Jay Shendure 1 1 Department
Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions Joshua N. Burton 1, Andrew Adey 1, Rupali P. Patwardhan 1, Ruolan Qiu 1, Jacob O. Kitzman 1, Jay Shendure 1 1 Department
Unfortunately, plate data was not available to generate an initial low coverage
 Finishing Report Carolyn Cain Fosmid: XBAA-30G19 Fosmid XBAA-30G19 was challenging to assemble, but ultimately came together in a satisfactory final assembly. The initial problems in my assembly were gaps,
Finishing Report Carolyn Cain Fosmid: XBAA-30G19 Fosmid XBAA-30G19 was challenging to assemble, but ultimately came together in a satisfactory final assembly. The initial problems in my assembly were gaps,
Draft 3 Annotation of DGA06H06, Contig 1 Jeannette Wong Bio4342W 27 April 2009
 Page 1 Draft 3 Annotation of DGA06H06, Contig 1 Jeannette Wong Bio4342W 27 April 2009 Page 2 Introduction: Annotation is the process of analyzing the genomic sequence of an organism. Besides identifying
Page 1 Draft 3 Annotation of DGA06H06, Contig 1 Jeannette Wong Bio4342W 27 April 2009 Page 2 Introduction: Annotation is the process of analyzing the genomic sequence of an organism. Besides identifying
Y1 Biology 131 Syllabus - Academic Year
 Y1 Biology 131 Syllabus - Academic Year 2016-2017 Monday 28/11/2016 DNA Packaging Week 11 Tuesday 29/11/2016 Regulation of gene expression Wednesday 22/9/2014 Cell cycle Sunday 4/12/2016 Tutorial Monday
Y1 Biology 131 Syllabus - Academic Year 2016-2017 Monday 28/11/2016 DNA Packaging Week 11 Tuesday 29/11/2016 Regulation of gene expression Wednesday 22/9/2014 Cell cycle Sunday 4/12/2016 Tutorial Monday
Mate-pair library data improves genome assembly
 De Novo Sequencing on the Ion Torrent PGM APPLICATION NOTE Mate-pair library data improves genome assembly Highly accurate PGM data allows for de Novo Sequencing and Assembly For a draft assembly, generate
De Novo Sequencing on the Ion Torrent PGM APPLICATION NOTE Mate-pair library data improves genome assembly Highly accurate PGM data allows for de Novo Sequencing and Assembly For a draft assembly, generate
B. Incorrect! Centromeric DNA is largely heterochromatin, which is inactive DNA.
 MCAT Biology - Problem Drill 06: Molecular Biology of Eukaryotes Question No. 1 of 10 1. Which type of DNA would have the highest level of expression? Question #01 (A) Heterochromatin. (B) Centromeric
MCAT Biology - Problem Drill 06: Molecular Biology of Eukaryotes Question No. 1 of 10 1. Which type of DNA would have the highest level of expression? Question #01 (A) Heterochromatin. (B) Centromeric
Annotation of Contig8 Sakura Oyama Dr. Elgin, Dr. Shaffer, Dr. Bednarski Bio 434W May 2, 2016
 Annotation of Contig8 Sakura Oyama Dr. Elgin, Dr. Shaffer, Dr. Bednarski Bio 434W May 2, 2016 Abstract Contig8, a 45 kb region of the fourth chromosome of Drosophila ficusphila, was annotated using the
Annotation of Contig8 Sakura Oyama Dr. Elgin, Dr. Shaffer, Dr. Bednarski Bio 434W May 2, 2016 Abstract Contig8, a 45 kb region of the fourth chromosome of Drosophila ficusphila, was annotated using the
MODULE 1: INTRODUCTION TO THE GENOME BROWSER: WHAT IS A GENE?
 MODULE 1: INTRODUCTION TO THE GENOME BROWSER: WHAT IS A GENE? Lesson Plan: Title Introduction to the Genome Browser: what is a gene? JOYCE STAMM Objectives Demonstrate basic skills in using the UCSC Genome
MODULE 1: INTRODUCTION TO THE GENOME BROWSER: WHAT IS A GENE? Lesson Plan: Title Introduction to the Genome Browser: what is a gene? JOYCE STAMM Objectives Demonstrate basic skills in using the UCSC Genome
Control of Eukaryotic Gene Expression (Learning Objectives)
 Control of Eukaryotic Gene Expression (Learning Objectives) 1. Compare and contrast chromatin and chromosome: composition, proteins involved and level of packing. Explain the structure and function of
Control of Eukaryotic Gene Expression (Learning Objectives) 1. Compare and contrast chromatin and chromosome: composition, proteins involved and level of packing. Explain the structure and function of
Molecular Cell Biology - Problem Drill 06: Genes and Chromosomes
 Molecular Cell Biology - Problem Drill 06: Genes and Chromosomes Question No. 1 of 10 1. Which of the following statements about genes is correct? Question #1 (A) Genes carry the information for protein
Molecular Cell Biology - Problem Drill 06: Genes and Chromosomes Question No. 1 of 10 1. Which of the following statements about genes is correct? Question #1 (A) Genes carry the information for protein
Sequence assembly. Jose Blanca COMAV institute bioinf.comav.upv.es
 Sequence assembly Jose Blanca COMAV institute bioinf.comav.upv.es Sequencing project Unknown sequence { experimental evidence result read 1 read 4 read 2 read 5 read 3 read 6 read 7 Computational requirements
Sequence assembly Jose Blanca COMAV institute bioinf.comav.upv.es Sequencing project Unknown sequence { experimental evidence result read 1 read 4 read 2 read 5 read 3 read 6 read 7 Computational requirements
3I03 - Eukaryotic Genetics Repetitive DNA
 Repetitive DNA Satellite DNA Minisatellite DNA Microsatellite DNA Transposable elements LINES, SINES and other retrosequences High copy number genes (e.g. ribosomal genes, histone genes) Multifamily member
Repetitive DNA Satellite DNA Minisatellite DNA Microsatellite DNA Transposable elements LINES, SINES and other retrosequences High copy number genes (e.g. ribosomal genes, histone genes) Multifamily member
Transcription Start Sites Project Report
 Transcription Start Sites Project Report Student name: Student email: Faculty advisor: College/university: Project details Project name: Project species: Date of submission: Number of genes in project:
Transcription Start Sites Project Report Student name: Student email: Faculty advisor: College/university: Project details Project name: Project species: Date of submission: Number of genes in project:
AP Biology. The BIG Questions. Chapter 19. Prokaryote vs. eukaryote genome. Prokaryote vs. eukaryote genome. Why turn genes on & off?
 The BIG Questions Chapter 19. Control of Eukaryotic Genome How are genes turned on & off in eukaryotes? How do cells with the same genes differentiate to perform completely different, specialized functions?
The BIG Questions Chapter 19. Control of Eukaryotic Genome How are genes turned on & off in eukaryotes? How do cells with the same genes differentiate to perform completely different, specialized functions?
The wrong file for Lecture 8 was posted on the website. I ve sent the correct file and it should be posted by the time class is out.
 The wrong file for Lecture 8 was posted on the website. I ve sent the correct file and it should be posted by the time class is out. Grade Distribution for EXAM 1 45 40 38 No. Scores/Grade 35 30 25 20
The wrong file for Lecture 8 was posted on the website. I ve sent the correct file and it should be posted by the time class is out. Grade Distribution for EXAM 1 45 40 38 No. Scores/Grade 35 30 25 20
GEP Project Management System: Order Finishing Reactions
 GEP Project Management System: Order Finishing Reactions Author Wilson Leung wleung@wustl.edu Document History Initial Draft 06/04/2007 First Revision 01/10/2009 Second Revision 01/07/2012 Third Revision
GEP Project Management System: Order Finishing Reactions Author Wilson Leung wleung@wustl.edu Document History Initial Draft 06/04/2007 First Revision 01/10/2009 Second Revision 01/07/2012 Third Revision
Genome annotation & EST
 Genome annotation & EST What is genome annotation? The process of taking the raw DNA sequence produced by the genome sequence projects and adding the layers of analysis and interpretation necessary
Genome annotation & EST What is genome annotation? The process of taking the raw DNA sequence produced by the genome sequence projects and adding the layers of analysis and interpretation necessary
Annotating 7G24-63 Justin Richner May 4, Figure 1: Map of my sequence
 Annotating 7G24-63 Justin Richner May 4, 2005 Zfh2 exons Thd1 exons Pur-alpha exons 0 40 kb 8 = 1 kb = LINE, Penelope = DNA/Transib, Transib1 = DINE = Novel Repeat = LTR/PAO, Diver2 I = LTR/Gypsy, Invader
Annotating 7G24-63 Justin Richner May 4, 2005 Zfh2 exons Thd1 exons Pur-alpha exons 0 40 kb 8 = 1 kb = LINE, Penelope = DNA/Transib, Transib1 = DINE = Novel Repeat = LTR/PAO, Diver2 I = LTR/Gypsy, Invader
Course summary. Today. PCR Polymerase chain reaction. Obtaining molecular data. Sequencing. DNA sequencing. Genome Projects.
 Goals Organization Labs Project Reading Course summary DNA sequencing. Genome Projects. Today New DNA sequencing technologies. Obtaining molecular data PCR Typically used in empirical molecular evolution
Goals Organization Labs Project Reading Course summary DNA sequencing. Genome Projects. Today New DNA sequencing technologies. Obtaining molecular data PCR Typically used in empirical molecular evolution
Assemblytics: a web analytics tool for the detection of assembly-based variants Maria Nattestad and Michael C. Schatz
 Assemblytics: a web analytics tool for the detection of assembly-based variants Maria Nattestad and Michael C. Schatz Table of Contents Supplementary Note 1: Unique Anchor Filtering Supplementary Figure
Assemblytics: a web analytics tool for the detection of assembly-based variants Maria Nattestad and Michael C. Schatz Table of Contents Supplementary Note 1: Unique Anchor Filtering Supplementary Figure
Heterochromatin Silencing
 Heterochromatin Silencing Heterochromatin silencing Most DNA in eukaryotes consists of repetitive DNA, including retrotransposons, transposable elements. Packaged into a condensed form: Heterochromatin:
Heterochromatin Silencing Heterochromatin silencing Most DNA in eukaryotes consists of repetitive DNA, including retrotransposons, transposable elements. Packaged into a condensed form: Heterochromatin:
Rapid Transcriptome Characterization for a nonmodel organism using 454 pyrosequencing
 Rapid Transcriptome Characterization for a nonmodel organism using 454 pyrosequencing "#$%&'()*+,"(-*."#$%&/.,"*01*0.,(%-*.&0("2*01*3,$,45,"-*4#66&*71** 3"#)(82,"-*2&9:)($*)1*"(03&"2-*#)66(*.(8$6#*;
Rapid Transcriptome Characterization for a nonmodel organism using 454 pyrosequencing "#$%&'()*+,"(-*."#$%&/.,"*01*0.,(%-*.&0("2*01*3,$,45,"-*4#66&*71** 3"#)(82,"-*2&9:)($*)1*"(03&"2-*#)66(*.(8$6#*;
Introduction to metagenome assembly. Bas E. Dutilh Metagenomic Methods for Microbial Ecologists, NIOO September 18 th 2014
 Introduction to metagenome assembly Bas E. Dutilh Metagenomic Methods for Microbial Ecologists, NIOO September 18 th 2014 Sequencing specs* Method Read length Accuracy Million reads Time Cost per M 454
Introduction to metagenome assembly Bas E. Dutilh Metagenomic Methods for Microbial Ecologists, NIOO September 18 th 2014 Sequencing specs* Method Read length Accuracy Million reads Time Cost per M 454
The Journey of DNA Sequencing. Chromosomes. What is a genome? Genome size. H. Sunny Sun
 The Journey of DNA Sequencing H. Sunny Sun What is a genome? Genome is the total genetic complement of a living organism. The nuclear genome comprises approximately 3.2 * 10 9 nucleotides of DNA, divided
The Journey of DNA Sequencing H. Sunny Sun What is a genome? Genome is the total genetic complement of a living organism. The nuclear genome comprises approximately 3.2 * 10 9 nucleotides of DNA, divided
GENETICS - CLUTCH CH.15 GENOMES AND GENOMICS.
 !! www.clutchprep.com CONCEPT: OVERVIEW OF GENOMICS Genomics is the study of genomes in their entirety Bioinformatics is the analysis of the information content of genomes - Genes, regulatory sequences,
!! www.clutchprep.com CONCEPT: OVERVIEW OF GENOMICS Genomics is the study of genomes in their entirety Bioinformatics is the analysis of the information content of genomes - Genes, regulatory sequences,
We begin with a high-level overview of sequencing. There are three stages in this process.
 Lecture 11 Sequence Assembly February 10, 1998 Lecturer: Phil Green Notes: Kavita Garg 11.1. Introduction This is the first of two lectures by Phil Green on Sequence Assembly. Yeast and some of the bacterial
Lecture 11 Sequence Assembly February 10, 1998 Lecturer: Phil Green Notes: Kavita Garg 11.1. Introduction This is the first of two lectures by Phil Green on Sequence Assembly. Yeast and some of the bacterial
Introduction to Medical Genetics: Human Chromosome
 Introduction to Medical Genetics: Human Chromosome Ashley Soosay This OpenCourseWare@UNIMAS and its related course materials are licensed under a Creative Commons Attribution NonCommercial ShareAlike 4.0
Introduction to Medical Genetics: Human Chromosome Ashley Soosay This OpenCourseWare@UNIMAS and its related course materials are licensed under a Creative Commons Attribution NonCommercial ShareAlike 4.0
Aaditya Khatri. Abstract
 Abstract In this project, Chimp-chunk 2-7 was annotated. Chimp-chunk 2-7 is an 80 kb region on chromosome 5 of the chimpanzee genome. Analysis with the Mapviewer function using the NCBI non-redundant database
Abstract In this project, Chimp-chunk 2-7 was annotated. Chimp-chunk 2-7 is an 80 kb region on chromosome 5 of the chimpanzee genome. Analysis with the Mapviewer function using the NCBI non-redundant database
Biol 478/595 Intro to Bioinformatics
 Biol 478/595 Intro to Bioinformatics September M 1 Labor Day 4 W 3 MG Database Searching Ch. 6 5 F 5 MG Database Searching Hw1 6 M 8 MG Scoring Matrices Ch 3 and Ch 4 7 W 10 MG Pairwise Alignment 8 F 12
Biol 478/595 Intro to Bioinformatics September M 1 Labor Day 4 W 3 MG Database Searching Ch. 6 5 F 5 MG Database Searching Hw1 6 M 8 MG Scoring Matrices Ch 3 and Ch 4 7 W 10 MG Pairwise Alignment 8 F 12
Unit 6: Molecular Genetics & DNA Technology Guided Reading Questions (100 pts total)
 Name: AP Biology Biology, Campbell and Reece, 7th Edition Adapted from chapter reading guides originally created by Lynn Miriello Chapter 16 The Molecular Basis of Inheritance Unit 6: Molecular Genetics
Name: AP Biology Biology, Campbell and Reece, 7th Edition Adapted from chapter reading guides originally created by Lynn Miriello Chapter 16 The Molecular Basis of Inheritance Unit 6: Molecular Genetics
Chapter 19 Genetic Regulation of the Eukaryotic Genome. A. Bergeron AP Biology PCHS
 Chapter 19 Genetic Regulation of the Eukaryotic Genome A. Bergeron AP Biology PCHS 2 Do Now - Eukaryotic Transcription Regulation The diagram below shows five genes (with their enhancers) from the genome
Chapter 19 Genetic Regulation of the Eukaryotic Genome A. Bergeron AP Biology PCHS 2 Do Now - Eukaryotic Transcription Regulation The diagram below shows five genes (with their enhancers) from the genome
Supplementary Figures
 Supplementary Figures A B Supplementary Figure 1. Examples of discrepancies in predicted and validated breakpoint coordinates. A) Most frequently, predicted breakpoints were shifted relative to those derived
Supplementary Figures A B Supplementary Figure 1. Examples of discrepancies in predicted and validated breakpoint coordinates. A) Most frequently, predicted breakpoints were shifted relative to those derived
3. human genomics clone genes associated with genetic disorders. 4. many projects generate ordered clones that cover genome
 Lectures 30 and 31 Genome analysis I. Genome analysis A. two general areas 1. structural 2. functional B. genome projects a status report 1. 1 st sequenced: several viral genomes 2. mitochondria and chloroplasts
Lectures 30 and 31 Genome analysis I. Genome analysis A. two general areas 1. structural 2. functional B. genome projects a status report 1. 1 st sequenced: several viral genomes 2. mitochondria and chloroplasts
The Diploid Genome Sequence of an Individual Human
 The Diploid Genome Sequence of an Individual Human Maido Remm Journal Club 12.02.2008 Outline Background (history, assembling strategies) Who was sequenced in previous projects Genome variations in J.
The Diploid Genome Sequence of an Individual Human Maido Remm Journal Club 12.02.2008 Outline Background (history, assembling strategies) Who was sequenced in previous projects Genome variations in J.
Sequence Assembly and Alignment. Jim Noonan Department of Genetics
 Sequence Assembly and Alignment Jim Noonan Department of Genetics james.noonan@yale.edu www.yale.edu/noonanlab The assembly problem >>10 9 sequencing reads 36 bp - 1 kb 3 Gb Outline Basic concepts in genome
Sequence Assembly and Alignment Jim Noonan Department of Genetics james.noonan@yale.edu www.yale.edu/noonanlab The assembly problem >>10 9 sequencing reads 36 bp - 1 kb 3 Gb Outline Basic concepts in genome
Chapter 13. The Nucleus. The nucleus is the hallmark of eukaryotic cells; the very term eukaryotic means having a "true nucleus".
 Chapter 13 The Nucleus The nucleus is the hallmark of eukaryotic cells; the very term eukaryotic means having a "true nucleus". Fig.13.1. The EM of the Nucleus of a Eukaryotic Cell 13.1. The Nuclear Envelope
Chapter 13 The Nucleus The nucleus is the hallmark of eukaryotic cells; the very term eukaryotic means having a "true nucleus". Fig.13.1. The EM of the Nucleus of a Eukaryotic Cell 13.1. The Nuclear Envelope
DNA Replication. The Organization of DNA. Recall:
 Recall: The Organization of DNA DNA Replication Chromosomal form appears only during mitosis, and is used in karyotypes. folded back upon itself (chromosomes) coiled around itself (chromatin) wrapped around
Recall: The Organization of DNA DNA Replication Chromosomal form appears only during mitosis, and is used in karyotypes. folded back upon itself (chromosomes) coiled around itself (chromatin) wrapped around
Fig. 16-7a. 5 end Hydrogen bond 3 end. 1 nm. 3.4 nm nm
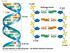 Fig. 16-7a end Hydrogen bond end 1 nm 3.4 nm 0.34 nm (a) Key features of DNA structure end (b) Partial chemical structure end Fig. 16-8 Adenine (A) Thymine (T) Guanine (G) Cytosine (C) Concept 16.2: Many
Fig. 16-7a end Hydrogen bond end 1 nm 3.4 nm 0.34 nm (a) Key features of DNA structure end (b) Partial chemical structure end Fig. 16-8 Adenine (A) Thymine (T) Guanine (G) Cytosine (C) Concept 16.2: Many
Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C
 CORRECTION NOTICE Nat. Genet. 47, 598 606 (2015) Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C Borbala Mifsud, Filipe Tavares-Cadete, Alice N Young, Robert Sugar,
CORRECTION NOTICE Nat. Genet. 47, 598 606 (2015) Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C Borbala Mifsud, Filipe Tavares-Cadete, Alice N Young, Robert Sugar,
Annotation of Drosophila erecta Contig 14. Kimberly Chau Dr. Laura Hoopes. Pomona College 24 February 2009
 Annotation of Drosophila erecta Contig 14 Kimberly Chau Dr. Laura Hoopes Pomona College 24 February 2009 1 Table of Contents I. Overview A. Introduction..1 B. Final Gene Model.....1 II. Genes A. Initial
Annotation of Drosophila erecta Contig 14 Kimberly Chau Dr. Laura Hoopes Pomona College 24 February 2009 1 Table of Contents I. Overview A. Introduction..1 B. Final Gene Model.....1 II. Genes A. Initial
1. The AGI (Arabidospis Genome Initiative) convention gene names or AtRTPrimer ID should
 We will show how users can select their desired types of primer-pairs, as we explain each of forms indicated by the blue-filled rectangles of Figure 1. Figure 1 Front-end webpage for searching desired
We will show how users can select their desired types of primer-pairs, as we explain each of forms indicated by the blue-filled rectangles of Figure 1. Figure 1 Front-end webpage for searching desired
Chimp Chunk 3-14 Annotation by Matthew Kwong, Ruth Howe, and Hao Yang
 Chimp Chunk 3-14 Annotation by Matthew Kwong, Ruth Howe, and Hao Yang Ruth Howe Bio 434W April 1, 2010 INTRODUCTION De novo annotation is the process by which a finished genomic sequence is searched for
Chimp Chunk 3-14 Annotation by Matthew Kwong, Ruth Howe, and Hao Yang Ruth Howe Bio 434W April 1, 2010 INTRODUCTION De novo annotation is the process by which a finished genomic sequence is searched for
Need a little extra help?
 Need a little extra help? Extra office hours! MWRF from 11:00 till 1:00 or by appointment: Marion.Brodhagen@wwu.edu. edu Tutoring center in Old Main 387: BIOL 205 drop-in tutoring at the following times:
Need a little extra help? Extra office hours! MWRF from 11:00 till 1:00 or by appointment: Marion.Brodhagen@wwu.edu. edu Tutoring center in Old Main 387: BIOL 205 drop-in tutoring at the following times:
DNA: The Genetic Material. Chapter 10
 DNA: The Genetic Material Chapter 10 DNA as the Genetic Material DNA was first extracted from nuclei in 1870 named nuclein after their source. Chemical analysis determined that DNA was a weak acid rich
DNA: The Genetic Material Chapter 10 DNA as the Genetic Material DNA was first extracted from nuclei in 1870 named nuclein after their source. Chemical analysis determined that DNA was a weak acid rich
NGS developments in tomato genome sequencing
 NGS developments in tomato genome sequencing 16-02-2012, Sandra Smit TATGTTTTGGAAAACATTGCATGCGGAATTGGGTACTAGGTTGGACCTTAGTACC GCGTTCCATCCTCAGACCGATGGTCAGTCTGAGAGAACGATTCAAGTGTTGGAAG ATATGCTTCGTGCATGTGTGATAGAGTTTGGTGGCCATTGGGATAGCTTCTTACC
NGS developments in tomato genome sequencing 16-02-2012, Sandra Smit TATGTTTTGGAAAACATTGCATGCGGAATTGGGTACTAGGTTGGACCTTAGTACC GCGTTCCATCCTCAGACCGATGGTCAGTCTGAGAGAACGATTCAAGTGTTGGAAG ATATGCTTCGTGCATGTGTGATAGAGTTTGGTGGCCATTGGGATAGCTTCTTACC
MODULE 5: TRANSLATION
 MODULE 5: TRANSLATION Lesson Plan: CARINA ENDRES HOWELL, LEOCADIA PALIULIS Title Translation Objectives Determine the codons for specific amino acids and identify reading frames by looking at the Base
MODULE 5: TRANSLATION Lesson Plan: CARINA ENDRES HOWELL, LEOCADIA PALIULIS Title Translation Objectives Determine the codons for specific amino acids and identify reading frames by looking at the Base
Lecture 21: Epigenetics Nurture or Nature? Chromatin DNA methylation Histone Code Twin study X-chromosome inactivation Environemnt and epigenetics
 Lecture 21: Epigenetics Nurture or Nature? Chromatin DNA methylation Histone Code Twin study X-chromosome inactivation Environemnt and epigenetics Epigenetics represents the science for the studying heritable
Lecture 21: Epigenetics Nurture or Nature? Chromatin DNA methylation Histone Code Twin study X-chromosome inactivation Environemnt and epigenetics Epigenetics represents the science for the studying heritable
ChIP-seq and RNA-seq
 ChIP-seq and RNA-seq Biological Goals Learn how genomes encode the diverse patterns of gene expression that define each cell type and state. Protein-DNA interactions (ChIPchromatin immunoprecipitation)
ChIP-seq and RNA-seq Biological Goals Learn how genomes encode the diverse patterns of gene expression that define each cell type and state. Protein-DNA interactions (ChIPchromatin immunoprecipitation)
Tutorial. In Silico Cloning. Sample to Insight. March 31, 2016
 In Silico Cloning March 31, 2016 Sample to Insight CLC bio, a QIAGEN Company Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.clcbio.com support-clcbio@qiagen.com In Silico Cloning
In Silico Cloning March 31, 2016 Sample to Insight CLC bio, a QIAGEN Company Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.clcbio.com support-clcbio@qiagen.com In Silico Cloning
Section C: The Control of Gene Expression
 Section C: The Control of Gene Expression 1. Each cell of a multicellular eukaryote expresses only a small fraction of its genes 2. The control of gene expression can occur at any step in the pathway from
Section C: The Control of Gene Expression 1. Each cell of a multicellular eukaryote expresses only a small fraction of its genes 2. The control of gene expression can occur at any step in the pathway from
Bioinformatics Course AA 2017/2018 Tutorial 2
 UNIVERSITÀ DEGLI STUDI DI PAVIA - FACOLTÀ DI SCIENZE MM.FF.NN. - LM MOLECULAR BIOLOGY AND GENETICS Bioinformatics Course AA 2017/2018 Tutorial 2 Anna Maria Floriano annamaria.floriano01@universitadipavia.it
UNIVERSITÀ DEGLI STUDI DI PAVIA - FACOLTÀ DI SCIENZE MM.FF.NN. - LM MOLECULAR BIOLOGY AND GENETICS Bioinformatics Course AA 2017/2018 Tutorial 2 Anna Maria Floriano annamaria.floriano01@universitadipavia.it
Annotating Fosmid 14p24 of D. Virilis chromosome 4
 Lo 1 Annotating Fosmid 14p24 of D. Virilis chromosome 4 Lo, Louis April 20, 2006 Annotation Report Introduction In the first half of Research Explorations in Genomics I finished a 38kb fragment of chromosome
Lo 1 Annotating Fosmid 14p24 of D. Virilis chromosome 4 Lo, Louis April 20, 2006 Annotation Report Introduction In the first half of Research Explorations in Genomics I finished a 38kb fragment of chromosome
Data Basics. Josef K Vogt Slides by: Simon Rasmussen Next Generation Sequencing Analysis
 Data Basics Josef K Vogt Slides by: Simon Rasmussen 2017 Generalized NGS analysis Sample prep & Sequencing Data size Main data reductive steps SNPs, genes, regions Application Assembly: Compare Raw Pre-
Data Basics Josef K Vogt Slides by: Simon Rasmussen 2017 Generalized NGS analysis Sample prep & Sequencing Data size Main data reductive steps SNPs, genes, regions Application Assembly: Compare Raw Pre-
RNA-Sequencing analysis
 RNA-Sequencing analysis Markus Kreuz 25. 04. 2012 Institut für Medizinische Informatik, Statistik und Epidemiologie Content: Biological background Overview transcriptomics RNA-Seq RNA-Seq technology Challenges
RNA-Sequencing analysis Markus Kreuz 25. 04. 2012 Institut für Medizinische Informatik, Statistik und Epidemiologie Content: Biological background Overview transcriptomics RNA-Seq RNA-Seq technology Challenges
ChIP-seq and RNA-seq. Farhat Habib
 ChIP-seq and RNA-seq Farhat Habib fhabib@iiserpune.ac.in Biological Goals Learn how genomes encode the diverse patterns of gene expression that define each cell type and state. Protein-DNA interactions
ChIP-seq and RNA-seq Farhat Habib fhabib@iiserpune.ac.in Biological Goals Learn how genomes encode the diverse patterns of gene expression that define each cell type and state. Protein-DNA interactions
NUCLEUS. Fig. 2. Various stages in the condensation of chromatin
 NUCLEUS Animal cells contain DNA in nucleus (contains ~ 98% of cell DNA) and mitochondrion. Both compartments are surrounded by an envelope (double membrane). Nuclear DNA represents some linear molecules
NUCLEUS Animal cells contain DNA in nucleus (contains ~ 98% of cell DNA) and mitochondrion. Both compartments are surrounded by an envelope (double membrane). Nuclear DNA represents some linear molecules
Chromosomes. M.Sc. Biotechnology. Hawler Medical University, Iraq
 Chromosomes Bashdar Mahmud Hussen M.Sc. Biotechnology Hawler Medical University, Iraq bashdar@res.hmu.edu.iq bmhscience@yahoo.com History of Chromosome Karl Nagali (1842) E. Russow (1872) first description
Chromosomes Bashdar Mahmud Hussen M.Sc. Biotechnology Hawler Medical University, Iraq bashdar@res.hmu.edu.iq bmhscience@yahoo.com History of Chromosome Karl Nagali (1842) E. Russow (1872) first description
Question 2: There are 5 retroelements (2 LINEs and 3 LTRs), 6 unclassified elements (XDMR and XDMR_DM), and 7 satellite sequences.
 Bio4342 Exercise 1 Answers: Detecting and Interpreting Genetic Homology (Answers prepared by Wilson Leung) Question 1: Low complexity DNA can be described as sequences that consist primarily of one or
Bio4342 Exercise 1 Answers: Detecting and Interpreting Genetic Homology (Answers prepared by Wilson Leung) Question 1: Low complexity DNA can be described as sequences that consist primarily of one or
Control of Eukaryotic Genes. AP Biology
 Control of Eukaryotic Genes The BIG Questions How are genes turned on & off in eukaryotes? How do cells with the same genes differentiate to perform completely different, specialized functions? Evolution
Control of Eukaryotic Genes The BIG Questions How are genes turned on & off in eukaryotes? How do cells with the same genes differentiate to perform completely different, specialized functions? Evolution
Factors affecting PCR
 Lec. 11 Dr. Ahmed K. Ali Factors affecting PCR The sequences of the primers are critical to the success of the experiment, as are the precise temperatures used in the heating and cooling stages of the
Lec. 11 Dr. Ahmed K. Ali Factors affecting PCR The sequences of the primers are critical to the success of the experiment, as are the precise temperatures used in the heating and cooling stages of the
Differences between prokaryotes & eukaryotes. Gene function
 GENE REGULATION Differences between prokaryotes & eukaryotes Gene function Description of Prokaryotic Chromosome and E.coli Review Differences between Prokaryotic & Eukaryotic Chromosomes Four differences
GENE REGULATION Differences between prokaryotes & eukaryotes Gene function Description of Prokaryotic Chromosome and E.coli Review Differences between Prokaryotic & Eukaryotic Chromosomes Four differences
Annotation Practice Activity [Based on materials from the GEP Summer 2010 Workshop] Special thanks to Chris Shaffer for document review Parts A-G
![Annotation Practice Activity [Based on materials from the GEP Summer 2010 Workshop] Special thanks to Chris Shaffer for document review Parts A-G Annotation Practice Activity [Based on materials from the GEP Summer 2010 Workshop] Special thanks to Chris Shaffer for document review Parts A-G](/thumbs/95/123686269.jpg) Annotation Practice Activity [Based on materials from the GEP Summer 2010 Workshop] Special thanks to Chris Shaffer for document review Parts A-G Introduction: A genome is the total genetic content of
Annotation Practice Activity [Based on materials from the GEP Summer 2010 Workshop] Special thanks to Chris Shaffer for document review Parts A-G Introduction: A genome is the total genetic content of
Wednesday, November 22, 17. Exons and Introns
 Exons and Introns Introns and Exons Exons: coded regions of DNA that get transcribed and translated into proteins make up 5% of the genome Introns and Exons Introns: non-coded regions of DNA Must be removed
Exons and Introns Introns and Exons Exons: coded regions of DNA that get transcribed and translated into proteins make up 5% of the genome Introns and Exons Introns: non-coded regions of DNA Must be removed
How does the human genome stack up? Genomic Size. Genome Size. Number of Genes. Eukaryotic genomes are generally larger.
 How does the human genome stack up? Organism Human (Homo sapiens) Laboratory mouse (M. musculus) Mustard weed (A. thaliana) Roundworm (C. elegans) Fruit fly (D. melanogaster) Yeast (S. cerevisiae) Bacterium
How does the human genome stack up? Organism Human (Homo sapiens) Laboratory mouse (M. musculus) Mustard weed (A. thaliana) Roundworm (C. elegans) Fruit fly (D. melanogaster) Yeast (S. cerevisiae) Bacterium
Plant Molecular and Cellular Biology Lecture 9: Nuclear Genome Organization: Chromosome Structure, Chromatin, DNA Packaging, Mitosis Gary Peter
 Plant Molecular and Cellular Biology Lecture 9: Nuclear Genome Organization: Chromosome Structure, Chromatin, DNA Packaging, Mitosis Gary Peter 9/16/2008 1 Learning Objectives 1. List and explain how DNA
Plant Molecular and Cellular Biology Lecture 9: Nuclear Genome Organization: Chromosome Structure, Chromatin, DNA Packaging, Mitosis Gary Peter 9/16/2008 1 Learning Objectives 1. List and explain how DNA
Annotation of contig62 from Drosophila elegans Dot Chromosome
 Abstract: Annotation of contig62 from Drosophila elegans Dot Chromosome 1 Maxwell Wang The goal of this project is to annotate the Drosophila elegans Dot chromosome contig62. Contig62 is a 32,259 bp contig
Abstract: Annotation of contig62 from Drosophila elegans Dot Chromosome 1 Maxwell Wang The goal of this project is to annotate the Drosophila elegans Dot chromosome contig62. Contig62 is a 32,259 bp contig
H3K36me3 polyclonal antibody
 H3K36me3 polyclonal antibody Cat. No. C15410192 Type: Polyclonal ChIP-grade/ChIP-seq grade Source: Rabbit Lot #: A1845P Size: 50 µg/32 µl Concentration: 1.6 μg/μl Specificity: Human, mouse, Arabidopsis,
H3K36me3 polyclonal antibody Cat. No. C15410192 Type: Polyclonal ChIP-grade/ChIP-seq grade Source: Rabbit Lot #: A1845P Size: 50 µg/32 µl Concentration: 1.6 μg/μl Specificity: Human, mouse, Arabidopsis,
Lack of Relationship Between Amount of DNA and Organism Complexity. In eukaryotes, genes are often much larger than the coding region
 Genomes, Heterochromatin, and the Dot Chromosome: Comparative Genomics in Drosophila Sarah CR Elgin Lab Genomics Education Partnership TA Workshop Washington University in St. Louis August, 2006 Minimum
Genomes, Heterochromatin, and the Dot Chromosome: Comparative Genomics in Drosophila Sarah CR Elgin Lab Genomics Education Partnership TA Workshop Washington University in St. Louis August, 2006 Minimum
MCDB 1041 Class 21 Splicing and Gene Expression
 MCDB 1041 Class 21 Splicing and Gene Expression Learning Goals Describe the role of introns and exons Interpret the possible outcomes of alternative splicing Relate the generation of protein from DNA to
MCDB 1041 Class 21 Splicing and Gene Expression Learning Goals Describe the role of introns and exons Interpret the possible outcomes of alternative splicing Relate the generation of protein from DNA to
Molecular Biology (2)
 Molecular Biology (2) DNA replication Mamoun Ahram, PhD Second semester, 2018-2019 Resources This lecture Cooper, pp. 191-207 2 Some basic information The entire DNA content of the cell is known as genome.
Molecular Biology (2) DNA replication Mamoun Ahram, PhD Second semester, 2018-2019 Resources This lecture Cooper, pp. 191-207 2 Some basic information The entire DNA content of the cell is known as genome.
Outline. Annotation of Drosophila Primer. Gene structure nomenclature. Muller element nomenclature. GEP Drosophila annotation projects 01/04/2018
 Outline Overview of the GEP annotation projects Annotation of Drosophila Primer January 2018 GEP annotation workflow Practice applying the GEP annotation strategy Wilson Leung and Chris Shaffer AAACAACAATCATAAATAGAGGAAGTTTTCGGAATATACGATAAGTGAAATATCGTTCT
Outline Overview of the GEP annotation projects Annotation of Drosophila Primer January 2018 GEP annotation workflow Practice applying the GEP annotation strategy Wilson Leung and Chris Shaffer AAACAACAATCATAAATAGAGGAAGTTTTCGGAATATACGATAAGTGAAATATCGTTCT
Designing TaqMan MGB Probe and Primer Sets for Gene Expression Using Primer Express Software Version 2.0
 Designing TaqMan MGB Probe and Primer Sets for Gene Expression Using Primer Express Software Version 2.0 Overview This tutorial details how a TaqMan MGB Probe can be designed over a specific region of
Designing TaqMan MGB Probe and Primer Sets for Gene Expression Using Primer Express Software Version 2.0 Overview This tutorial details how a TaqMan MGB Probe can be designed over a specific region of
NEXT GENERATION SEQUENCING. Farhat Habib
 NEXT GENERATION SEQUENCING HISTORY HISTORY Sanger Dominant for last ~30 years 1000bp longest read Based on primers so not good for repetitive or SNPs sites HISTORY Sanger Dominant for last ~30 years 1000bp
NEXT GENERATION SEQUENCING HISTORY HISTORY Sanger Dominant for last ~30 years 1000bp longest read Based on primers so not good for repetitive or SNPs sites HISTORY Sanger Dominant for last ~30 years 1000bp
Delve AP Biology Lecture 7: 10/30/11 Melissa Ko and Anne Huang
 Today s Agenda: I. DNA Structure II. DNA Replication III. DNA Proofreading and Repair IV. The Central Dogma V. Transcription VI. Post-transcriptional Modifications Delve AP Biology Lecture 7: 10/30/11
Today s Agenda: I. DNA Structure II. DNA Replication III. DNA Proofreading and Repair IV. The Central Dogma V. Transcription VI. Post-transcriptional Modifications Delve AP Biology Lecture 7: 10/30/11
A) The constituent monomer of DNA and RNA. C) The basic structural unit of chromatin with "bead-on-a-string" morphology
 MATCHING. Choose the item in column 2 that best matches each item in column 1. Please select the best match for each term. 1) Centromere 2) Nucleoid 3) Nucleotide 4) Chromosome 5) Nucleosome A) The constituent
MATCHING. Choose the item in column 2 that best matches each item in column 1. Please select the best match for each term. 1) Centromere 2) Nucleoid 3) Nucleotide 4) Chromosome 5) Nucleosome A) The constituent
BIO 4342 Lecture on Repeats
 BIO 4342 Lecture on Repeats Jeremy Buhler June 14, 2006 1 How RepeatMasker Works Running RepeatMasker is the most common first step in annotating genomic DNA sequences. What exactly does it do? Given a
BIO 4342 Lecture on Repeats Jeremy Buhler June 14, 2006 1 How RepeatMasker Works Running RepeatMasker is the most common first step in annotating genomic DNA sequences. What exactly does it do? Given a
C3BI. VARIANTS CALLING November Pierre Lechat Stéphane Descorps-Declère
 C3BI VARIANTS CALLING November 2016 Pierre Lechat Stéphane Descorps-Declère General Workflow (GATK) software websites software bwa picard samtools GATK IGV tablet vcftools website http://bio-bwa.sourceforge.net/
C3BI VARIANTS CALLING November 2016 Pierre Lechat Stéphane Descorps-Declère General Workflow (GATK) software websites software bwa picard samtools GATK IGV tablet vcftools website http://bio-bwa.sourceforge.net/
Genomic resources. for non-model systems
 Genomic resources for non-model systems 1 Genomic resources Whole genome sequencing reference genome sequence comparisons across species identify signatures of natural selection population-level resequencing
Genomic resources for non-model systems 1 Genomic resources Whole genome sequencing reference genome sequence comparisons across species identify signatures of natural selection population-level resequencing
Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) (b)
 Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) A schematic overview of the production and amplification of a single pirna from a transposon transcript. The
Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) A schematic overview of the production and amplification of a single pirna from a transposon transcript. The
Chapter 5 DNA and Chromosomes
 Chapter 5 DNA and Chromosomes DNA as the genetic material Heat-killed bacteria can transform living cells S Smooth R Rough Fred Griffith, 1920 DNA is the genetic material Oswald Avery Colin MacLeod Maclyn
Chapter 5 DNA and Chromosomes DNA as the genetic material Heat-killed bacteria can transform living cells S Smooth R Rough Fred Griffith, 1920 DNA is the genetic material Oswald Avery Colin MacLeod Maclyn
Chapter 6: Transcription and RNA Processing in Eukaryotes
 3. Basic Genetics Plant Molecular Biology Chapter 6: Transcription and RNA Processing in Eukaryotes - Genetic organization in eukaryote - Transcription in eukaryote - - RNA processing in eukaryote - Translation
3. Basic Genetics Plant Molecular Biology Chapter 6: Transcription and RNA Processing in Eukaryotes - Genetic organization in eukaryote - Transcription in eukaryote - - RNA processing in eukaryote - Translation
PrimePCR Assay Validation Report
 Gene Information Gene Name keratin 78 Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID KRT78 Human This gene is a member of the type II keratin gene family
Gene Information Gene Name keratin 78 Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID KRT78 Human This gene is a member of the type II keratin gene family
