Finishing of DFIC This project sought to finish DFIC , the terminal 45 kb of the Drosophila
|
|
|
- Solomon Stokes
- 6 years ago
- Views:
Transcription
1 Lin 1 Kevin Lin Bio 434W Dr. Elgin 26 February 2016 Finishing of DFIC Abstract This project sought to finish DFIC , the terminal 45 kb of the Drosophila ficusphila dot chromosome. The initial project contained 167 high quality discrepancies, 109 mononucleotide runs in low coverage regions, and thirteen low consensus quality regions, but contained no gaps. The primary challenge of this project was the large number of misaligned and mismapped reads due to the numerous low coverage and repeat regions present in the region. The final assembly has all mononucleotide runs resolved and is ready for annotation. Introduction Interphase DNA can be broadly categorized into euchromatin or heterochromatin based on its level of compaction. Euchromatin is less condensed chromatin typically found on chromosome arms and is generally accessible for transcription and gene rich. Heterochromatin is more condensed chromatin, typically found in centromeres and telomeres, and is associated with gene silencing. This semester we are studying the Drosophila dot chromosome (Muller F element) due to its unique feature of being almost entirely heterochromatic, yet still containing many actively transcribed genes. Investigating how genes in a heterochromatic environment are expressed may reveal new insights concerning the role sequence characteristics and organizational features play in gene expression. Our long term goal is to identify regulatory elements in the F element of Drosophila ficusphila, which we can compare with data from other
2 Lin 2 Drosophila species to look for conserved motifs. D. ficusphila was chosen to be analyzed because it has an ideal evolutionary distance from D. melanogaster for comparing regulatory motifs between species. The DNA sequence data we are using was initially generated using Roche 454 technology, which produced a version 1 assembly. However, this initial assembly has many consensus errors in regions with long mononucleotide runs (MNRs) due to a limitation of 454 sequencing technology, specifically its ability to accurately determine the length of long MNRs. To improve the quality of the sequencing data, the genome was subsequently sequenced using Illumina technology. Illumina sequencing does not have a problem with MNRs, but does suffer from shorter read lengths, which increases the probability of mismapping. The reads obtained from the Illumina sequencing were mapped back to the assembly generated by 454 technology and the consensus sequence was reconciled to be consistent with the new Illumina reads, generating a version 2 assembly. While the version 2 assembly fixed many of the MNR regions, there are still many errors present. The primary goal of this finishing project is to correct any errant MNR regions using the additional Illumina data and to call for additional sequencing reactions as needed. Initial Assembly The initial Assembly View (Figure 1) showed a single contig of approximately bases with a number of repetitious sequences and discrepant mate pairs present. All of the discrepant mate pairs fall in regions that are part of a repetitious sequence. Because our project focuses on the genes, this class of discrepant reads were not fixed. The initial assembly also contained thirteen low quality consensus regions, 167 highly discrepant regions, but no gaps.
3 Lin 3 Figure 1: Initial assembly view of DFIC Orange and black lines indicate the presence of repetitious sequences. Red lines indicate discrepant forward-reverse pairs. High Quality Discrepancies High quality discrepancies (HDQ) were found by searching the main contig for positions with at least three high quality (Phred >30) reads that differ from the main contig. The primary objective when searching highly discrepant positions was to find erroneous mononucleotide runs (MNR) in the consensus sequence that needed to be corrected, which could be done using high quality Illumina data. The initial search showed 167 HQDs. Of the 167 HQDs, 130 did not correspond to a mononucleotide run, but rather were the result of a small number of reads being discrepant, while an overwhelming majority of reads supported the consensus. These regions were not further analyzed as the data presented clearly supported the consensus sequence. The remaining 37 HQDs corresponded to MNRs or ambiguous bases and were analyzed further. Thirty-two of the HQDs corresponded to errors resulting from MNRs; analysis resulted in 22 corrections to the consensus sequence (Supplementary Table 1). Errors in MNR regions are common due to the limitations of the 454 sequencing used to generate the initial assembly; the Illumina data was needed to accurately determine the correct number of bases in a mononucleotide run. Many of the corrections to the consensus sequence involving
4 Lin 4 mononucleotide runs were straightforward adjustments, as all of the Illumina data was in agreement. Figure 2 shows a typical MNR where almost all of the high quality Illumina reads support changing the consensus sequence from 11 to 12 T s. While a few of the Illumina reads did not support changing the consensus to 12 T s, the contradictory bases that differed from the majority of Illumina reads were low quality due to being at the end of the reads. Therefore these reads were discounted. A common problem encountered when correcting HQDs associated with MNRs was misalignment of the Illumina reads as shown in Figure 3. This problem arose because the 454 sequencing technology used to generate the version 1 assembly often had errors in the MNR regions, which would cause the Illumina data that was later added to be misaligned by Consed. MNR regions associated with misaligned Illumina reads were resolved using a similar method of comparing the high quality Illumina reads to the consensus sequence, and determining whether the Illumina data supports the consensus. However, it was important to manually count the number of bases in a MNR in the misaligned Illumina reads, as reads that initially appeared to be different often supported the same consensus sequence. The region shown in Figure 3 illustrates this critical technique. Five of the Illumina reads are misaligned on the right end of the reads and three of the Illumina reads are misaligned on the left end of the reads. However, both sets of Illumina reads contain 9A s and 9T s, which supported the addition of an A and a T to the corresponding monoa and monot runs in the consensus.
5 Lin 5 Figure 2: Typical MNR region. The Illumina data indicates a T needs to be added to the consensus. The blue box contains Illumina reads that show the missing T at the end of the MNR and the orange box contains Illumina reads that show the missing T at the beginning of the MNR run. Note that the sequence represented is the same in both. The red boxes highlight Illumina reads that disagree with the consensus but are likely to incorrect due to the low quality bases at the end of each read.
6 Lin 6 Figure 3: MNR with misaligned sequences. The yellow box highlights the alignment of the TATT sequence prior to the monoa run and the blue box highlights the alignment of the TCT sequence that follows the monot run. There were five HQDs remaining that were not associated with a MNR, but required further analysis due to a high number of reads being discrepant with the consensus sequence. All five of these HQDs were located within repeat regions, which were tagged prior to receiving the project. Repeat regions are challenging to analyze because it is common for reads to be mismapped due to very similar repeats being present across the entire genome. Region (Figure 4) was a particularly difficult region that I was unable to resolve due to multiple discrepancies between reads. The consensus sequence immediately following a long monot and prior to the AGTAAAT sequence was CTAC. Illumina reads mapped to this region, however, mostly supported changing the consensus to TCAC or TTAC. However, the 454 data mapped to this region supported the consensus, CTAC, or the sequence CTACT. Due to the longer read length of 454 reads when compared to Illumina reads, it is more likely that the 454 reads are mapped to the correct location of the genome. Additionally, even
7 Lin 7 though the region immediately follows a monot run, the region of interest is not a part of the MNR and therefore the 454 reads are likely to be reliable. As a result, I decided not to change the consensus at this site based on the evidence that most of the 454 data supported the consensus sequence. A DataNeeded tag was not added to this location despite the ambiguity because this region is within a repeat, and determining the precise sequence of each repeat region is not essential for our research goal of searching for regulatory motifs. The high quality discrepancy at bases 14487, 41739, and were also in repeat regions and not associated with a MNR. All three of these regions contained reads supporting two different consensus sequences, but similar to region , the data did not support changing the consensus sequence. Figure 4: Ambiguous sequence following a long monot run in a repeat region. Reads boxed in blue show TCAT at the end of the monot run, reads boxed in yellow show TTAC at the end of the monot run, and reads boxed in red show CTAC and CTACT at the end of the monot run.
8 Lin 8 The large number of discrepant reads at HQD sites and prompted me to revisit these sites after finishing the remainder of the project. Closer analysis of this region in assembly view (Figure 5) revealed that the region from 14.5 to 15 kb has a low number of paired-end reads and a high number of discrepant mate pairs. In addition, CrossMatch indicates that this is a repetitious region. Thus, the data suggests that there may be a tandem array in this region. As discussed in the lab meeting, the cost of additional sequencing reactions in a region such as this one typically exceeds the potential benefits. Therefore, most researchers do not order additional sequencing reactions for this type of region. Moreover, our research question does not depend on accurate sequences of repeat regions. As a result, no additional primers were ordered. Figure 5: Assembly view of region from 14 to 15kb. Paired-end reads are shown in blue and discrepant mate pairs are shown in red. However, I did design a potential primer pair for additional sequencing of the kb region as practice because in other circumstances it may make sense to order additional sequencing. I used Consed to search for primer pairs that would cover the kb region which generated the following results (Figure 6). Finding an appropriate primer for this region
9 Lin 9 was difficult because almost the entire consensus sequence from kb consists of repeat regions. The only left primer not entirely located within a repeat region was the one from bases , which is seen in pairs 7,8,13, and 14. Although this left primer was not ideal due to the high AT content, it was the only primer not fully contained in a repeat region. Pair 14 was chosen and tagged in Consed via comment tags for the left and right primers but upon further thought pair 7 or 8 would be a better choice due to the right primer being closer to the region of interest. Figure 6: Potential PCR primer pairs generated by Consed for the region kb. Pair 14 (highlighted) was ultimately chosen for the submitted file but pair 7 or 8 may have been a better choice. One final region of interest is at base (Figure 7), located near the the end of the chromosome, a region which has low coverage and is associated with a repeat. This region is unique because none of the reads support the consensus sequence. Therefore, the consensus was changed from * to a T based on 6 of the 7 reads mapped to that site supporting a T at position
10 Lin 10 Figure 7: Base 42362, which is not supported by any of the 454 or Illumina reads. The read boxed in blue corresponds to a user-generated assembly piece generated to help break the original assembly up into 100 kb projects. Low Coverage Regions Only searching regions with 3 or more HQDs may potentially leave out MNRs that need to be corrected in regions that do not contain a high number of reads. Thus, regions of low coverage (<40 reads) were also searched for MNRs of five or more bases in order to screen for MNRs that need further analysis. MNRs found in regions of low coverage (109 instances) were closely inspected using the same process as described for MNRs found in HQDs (Supplementary Table 2), which resulted in 4 changes to the consensus sequence. One of the MNRs that was corrected, a monot run at (Figure 8), was notable because no Illumina reads spanned the MNR region. Therefore, the only option was to use the 454 data to determine the most likely consensus. Of the sixteen 454 reads covering the monot run, 10 supported changing the consensus from 5 T s to 7 T s. Moreover, 454 sequencing generally scores too few rather than too many nucleotides. As a result, the consensus was changed to include 7 T s in the consensus based on the 454 data because there were no high quality Illumina reads in this region. A DataNeeded tag was not added as this MNR is in a repeat region.
11 Lin 11 Figure 8: MNR at site This is a low coverage region, supported only by 454 reads. The search for MNRs in low consensus quality regions also yielded a particularly challenging region around (Figure 9). This was a repeat region with many potentially misaligned or mismapped Illumina reads. A number of discrepant Illumina reads were pulled out of the main contig either because of discrepancies at sites 5989 and 5992 or due to potential discrepancies to the right of the monoa run. Figure 9: Region The reads highlighted in purple on the left were removed from the contig. Reads boxed in red were removed due to discrepancies at bases 5989 and The removed reads formed three contigs with 1, 5, and 8 reads. I used CrossMatch to check if the contigs with 5 and 8 reads (contigs and respectively) mapped back to the main contig. Both were found to potentially map back to the region from which they
12 Lin 12 were pulled (Figure 10). These results were more closely examined by comparing each contig back to the consensus sequence. Figure 11 shows the Compare Contig window for the contig with five reads revealing zero discrepancies; therefore it was mapped back. In contrast, Figure 12 shows the Compare Contig window for the contig with eight reads which has six discrepancies, some of which do not correspond to mononucleotide runs. It is likely that the reads in this contig were mismapped; therefore this contig was not mapped back. After removal of mismapped reads, region (Figure 13) had a monoa run of seventeen bases in the consensus sequence. Six Illumina reads spanned the length of the monoa run, four supported a consensus of a twentyone base monoa run, and two support a consensus of a 20 base monoa run. In the absence of additional information, four A s were added to the consensus to reflect the majority of the Illumina data. While the region is still ambiguous, a DataNeeded tag was not added due to this region being in a repeat and thus being difficult to resolve, and of low interest. Figure 10: CrossMatch results for two of the contigs containing the reads pulled out from the region. Both were found to map back to the original region they were pulled from.
13 Lin 13 Figure 11: Compare contig window for the contig with 5 reads. This contig was mapped back. Figure 12: Compare contig window for the contig with 8 reads. This contig was not mapped back. Figure 13: Bases after mismapped reads were removed. Red boxes highlight Illumina reads that support a monoa run of 21 bases and blue boxes highlight Illumina reads that support a monoa run of 20 bases. Note that the first highlighted read appears to be discrepant to the consensus sequence to the left of the monoa run, but this is simply a misalignment.
14 Lin 14 Low Consensus Quality As a final check, the main contig was searched for bases with low consensus quality (Phred <30). Thirty two regions of low consensus quality were identified (Supplementary Table 3) but no changes were made to the consensus. Thirty of these regions were associated with reads not being properly aligned after the region was previously corrected. The remaining two regions of low consensus quality were in the first and last 2.5 kb of the contig and were simply the result of low coverage, which is expected to occur at either end of a contig. Final Assembly The Assembly View of the final contig (Figure 14) looks largely the same as the original contig because no gaps were joined, nor was the contig torn during this finishing project. However, the consensus sequence for all of the MNRs has been checked and adjusted as needed. A detailed summary of all changes to the consensus sequence can be found in Figure 15. While the final assembly still has some unresolved regions, notably the kb region (associated with a repeat and a large number of inconsistent mate pairs), these regions are unlikely to affect annotation. Therefore, the finishing project of DFIC is complete and DFIC is ready for annotation. Figure 14: Final assembly view
15 Lin 15 Changes to consensus sequence Contig Position Change Source of data T Illumina A Illumina AAAA Illumina A Illumina A Illumina T Illumina A Illumina T Illumina TT Illumina T Illumina A Illumina A Illumina T Illumina A Illumina T Illumina T Illumina TT 454 only A Illumina T Illumina T Illumina T Illumina T Illumina G Illumina T Illumina A Illumina T Illumina T Illumina Figure 15: Summary of all changes to consensus sequence.
16 Lin 16 Acknowledgements I wish to thank the Bio4342 course staff, Dr. Elgin, Dr. Shaffer, and Wilson Leung for organizing this project and providing guidance throughout the completion of the project. I would also like to thank professional finisher Lee Trani from the Genome Institute for his assistance with Consed and Dr. Bednarski for helping with our writing.
17 Lin 17 Supplemental Figures High Quality Discrepancies Contig Position Analysis Action Source of data monoa run -A Illumina monot run +T Illumina monoa run +A Illumina monoa run +A Illumina monot run +T Illumina monoa run NC Illumina monoa run NC Illumina monot run +T Illumina non-mnr discrepancies NC G/A discrepancy NC monoa run +A Illumina monot run +T Illumina monoa run NC Illumina monot run +T Illumina monoa run +A Illumina monoa run +A Illumina monot run +T Illumina monoa run +A Illumina monot run +T Illumina monot run +T Illumina monoa run +A Illumina monot run +T Illumina monot run +T Illumina monot run NC Illumina monot run +T Illumina monot run NC Illumina monot run +T Illumina monot run +T Illumina monot run NC Illumina monoa run NC Illumina monot run NC Illumina monoa run +A Illumina monot run NC Illumina missing base +T 454+Illumina monoa run NC Illumina A/G discrepancy NC 454+Illumina A/G discrepancy NC 454+Illumina Supplementary Table 1: Table of HQD regions analyzed. NC indicates no change.
18 Lin 18 Low Coverage Regions Analyzed Contig Position Analysis Action Source of data monoc run NC Illumina monoc run NC Illumina monot run NC Illumina monoa run +AAAA Illumina monot run NC Illumina monoa run PF n/a monot run NC Illumina monot run NC Illumina monoa run PF n/a monot run NC Illumina monot run NC Illumina monoa run NC Illumina monoa run NC Illumina monot run NC Illumina monot run NC Illumina monoa run NC Illumina monoa run NC Illumina monoa run PF n/a monoa run NC Illumina monot run NC Illumina monoa run NC Illumina monoa run NC Illumina monoa run PF n/a monot run NC Illumina monog run NC Illumina monot run NC Illumina monot run +TT Illumina monot run NC Illumina monot run PF n/a monoa run NC Illumina monoa run PF n/a monoa run NC Illumina monot run NC Illumina monot run NC Illumina monoa run PF n/a monot run PF n/a monoa run NC Illumina monot run NC Illumina monoa run NC Illumina monoa run NC Illumina monoa run NC Illumina monot run NC Illumina monoa run PF n/a monot run PF n/a monot run NC Illumina monot run NC Illumina monoa run NC Illumina monoa run NC Illumina monoa run NC Illumina monoa run NC Illumina monot run NC Illumina monot run NC Illumina monoa run NC Illumina monoa run NC Illumina monot run NC Illumina monoa run NC Illumina monot run NC Illumina monot run NC Illumina monoa run NC Illumina monoa run NC Illumina monoa run NC Illumina monot run PF n/a monot run NC Illumina monot run +TT monoa run NC Illumina monoa run NC Illumina monot run NC Illumina monot run NC Illumina monot run NC Illumina monot run PF n/a monoa run NC Illumina monot run +T Illumina monot run NC monoa run NC Illumina monog run NC monot run NC 454+Illumina monog run NC Illumina monoa run NC Illumina monot run NC Illumina monoa run NC 454+Illumina monog run +G Illumina monoa run NC Illumina monot run NC Illumina monoa run NC Illumina monog run NC Illumina monoc run NC Illumina monot run NC Illumina monoa run NC Illumina monot run NC Illumina monot run NC Illumina monot run NC Illumina monoa run NC Illumina monot run NC Illumina monot run NC Illumina monot run NC Illumina monot run NC Illumina monot run NC Illumina monoa run NC Illumina monot run NC Illumina monoa run NC Illumina monoa run NC Illumina monoa run NC Illumina monot run NC Illumina monoa run NC Illumina monoa run NC Illumina monoa run NC Illumina monot run NC Illumina monoa run NC 454 Supplementary Table 2: Table of MNRs in low coverage regions analyzed. NC indicates no change, PF indicates region previously fixed.
19 Lin 19 Low Consensus Quality Regions Contig Position Analysis Action Source of data isolated low consensus quality NC n/a isolated low consensus quality PF n/a , 6011 isolated low consensus quality PF n/a isolated low consensus quality PF n/a isolated low consensus quality PF n/a isolated low consensus quality PF n/a isolated low consensus quality PF n/a isolated low consensus quality NC isolated low consensus quality PF n/a isolated low consensus quality PF n/a isolated low consensus quality PF n/a isolated low consensus quality PF n/a isolated low consensus quality PF n/a isolated low consensus quality PF n/a isolated low consensus quality PF n/a isolated low consensus quality PF n/a isolated low consensus quality PF n/a isolated low consensus quality PF n/a isolated low consensus quality PF n/a isolated low consensus quality PF n/a isolated low consensus quality PF n/a isolated low consensus quality PF n/a isolated low consensus quality PF n/a isolated low consensus quality PF n/a isolated low consensus quality PF n/a isolated low consensus quality PF n/a isolated low consensus quality PF n/a isolated low consensus quality PF n/a isolated low consensus quality PF n/a isolated low consensus quality PF n/a isolated low consensus quality NC n/a Supplementary Table 3: Table of low consensus quality regions analyzed. NC indicates no change, PF indicates region previously fixed.
Finishing of DELE Drosophila elegans has been sequenced using Roche 454 pyrosequencing and Illumina
 Sarah Swiezy Dr. Elgin, Dr. Shaffer Bio 434W 27 February 2015 Finishing of DELE8596009 Abstract Drosophila elegans has been sequenced using Roche 454 pyrosequencing and Illumina technology. DELE8596009,
Sarah Swiezy Dr. Elgin, Dr. Shaffer Bio 434W 27 February 2015 Finishing of DELE8596009 Abstract Drosophila elegans has been sequenced using Roche 454 pyrosequencing and Illumina technology. DELE8596009,
Finishing Fosmid DMAC-27a of the Drosophila mojavensis third chromosome
 Finishing Fosmid DMAC-27a of the Drosophila mojavensis third chromosome Ruth Howe Bio 434W 27 February 2010 Abstract The fourth or dot chromosome of Drosophila species is composed primarily of highly condensed,
Finishing Fosmid DMAC-27a of the Drosophila mojavensis third chromosome Ruth Howe Bio 434W 27 February 2010 Abstract The fourth or dot chromosome of Drosophila species is composed primarily of highly condensed,
Finishing Drosophila Ananassae Fosmid 2728G16
 Finishing Drosophila Ananassae Fosmid 2728G16 Kyle Jung March 8, 2013 Bio434W Professor Elgin Page 1 Abstract For my finishing project, I chose to finish fosmid 2728G16. This fosmid carries a segment of
Finishing Drosophila Ananassae Fosmid 2728G16 Kyle Jung March 8, 2013 Bio434W Professor Elgin Page 1 Abstract For my finishing project, I chose to finish fosmid 2728G16. This fosmid carries a segment of
Finished (Almost) Sequence of Drosophila littoralis Chromosome 4 Fosmid Clone XAAA73. Seth Bloom Biology 4342 March 7, 2004
 Finished (Almost) Sequence of Drosophila littoralis Chromosome 4 Fosmid Clone XAAA73 Seth Bloom Biology 4342 March 7, 2004 Summary: I successfully sequenced Drosophila littoralis fosmid clone XAAA73. The
Finished (Almost) Sequence of Drosophila littoralis Chromosome 4 Fosmid Clone XAAA73 Seth Bloom Biology 4342 March 7, 2004 Summary: I successfully sequenced Drosophila littoralis fosmid clone XAAA73. The
Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Supplementary Material
 Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions Joshua N. Burton 1, Andrew Adey 1, Rupali P. Patwardhan 1, Ruolan Qiu 1, Jacob O. Kitzman 1, Jay Shendure 1 1 Department
Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions Joshua N. Burton 1, Andrew Adey 1, Rupali P. Patwardhan 1, Ruolan Qiu 1, Jacob O. Kitzman 1, Jay Shendure 1 1 Department
Chimp Chunk 3-14 Annotation by Matthew Kwong, Ruth Howe, and Hao Yang
 Chimp Chunk 3-14 Annotation by Matthew Kwong, Ruth Howe, and Hao Yang Ruth Howe Bio 434W April 1, 2010 INTRODUCTION De novo annotation is the process by which a finished genomic sequence is searched for
Chimp Chunk 3-14 Annotation by Matthew Kwong, Ruth Howe, and Hao Yang Ruth Howe Bio 434W April 1, 2010 INTRODUCTION De novo annotation is the process by which a finished genomic sequence is searched for
De Novo Assembly of High-throughput Short Read Sequences
 De Novo Assembly of High-throughput Short Read Sequences Chuming Chen Center for Bioinformatics and Computational Biology (CBCB) University of Delaware NECC Third Skate Genome Annotation Workshop May 23,
De Novo Assembly of High-throughput Short Read Sequences Chuming Chen Center for Bioinformatics and Computational Biology (CBCB) University of Delaware NECC Third Skate Genome Annotation Workshop May 23,
Annotating Fosmid 14p24 of D. Virilis chromosome 4
 Lo 1 Annotating Fosmid 14p24 of D. Virilis chromosome 4 Lo, Louis April 20, 2006 Annotation Report Introduction In the first half of Research Explorations in Genomics I finished a 38kb fragment of chromosome
Lo 1 Annotating Fosmid 14p24 of D. Virilis chromosome 4 Lo, Louis April 20, 2006 Annotation Report Introduction In the first half of Research Explorations in Genomics I finished a 38kb fragment of chromosome
GEP Project Management System: Order Finishing Reactions
 GEP Project Management System: Order Finishing Reactions Author Wilson Leung wleung@wustl.edu Document History Initial Draft 06/04/2007 First Revision 01/10/2009 Second Revision 01/07/2012 Third Revision
GEP Project Management System: Order Finishing Reactions Author Wilson Leung wleung@wustl.edu Document History Initial Draft 06/04/2007 First Revision 01/10/2009 Second Revision 01/07/2012 Third Revision
De novo Genome Assembly
 De novo Genome Assembly A/Prof Torsten Seemann Winter School in Mathematical & Computational Biology - Brisbane, AU - 3 July 2017 Introduction The human genome has 47 pieces MT (or XY) The shortest piece
De novo Genome Assembly A/Prof Torsten Seemann Winter School in Mathematical & Computational Biology - Brisbane, AU - 3 July 2017 Introduction The human genome has 47 pieces MT (or XY) The shortest piece
Mate-pair library data improves genome assembly
 De Novo Sequencing on the Ion Torrent PGM APPLICATION NOTE Mate-pair library data improves genome assembly Highly accurate PGM data allows for de Novo Sequencing and Assembly For a draft assembly, generate
De Novo Sequencing on the Ion Torrent PGM APPLICATION NOTE Mate-pair library data improves genome assembly Highly accurate PGM data allows for de Novo Sequencing and Assembly For a draft assembly, generate
Fig. 16-7a. 5 end Hydrogen bond 3 end. 1 nm. 3.4 nm nm
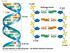 Fig. 16-7a end Hydrogen bond end 1 nm 3.4 nm 0.34 nm (a) Key features of DNA structure end (b) Partial chemical structure end Fig. 16-8 Adenine (A) Thymine (T) Guanine (G) Cytosine (C) Concept 16.2: Many
Fig. 16-7a end Hydrogen bond end 1 nm 3.4 nm 0.34 nm (a) Key features of DNA structure end (b) Partial chemical structure end Fig. 16-8 Adenine (A) Thymine (T) Guanine (G) Cytosine (C) Concept 16.2: Many
Outline. Annotation of Drosophila Primer. Gene structure nomenclature. Muller element nomenclature. GEP Drosophila annotation projects 01/04/2018
 Outline Overview of the GEP annotation projects Annotation of Drosophila Primer January 2018 GEP annotation workflow Practice applying the GEP annotation strategy Wilson Leung and Chris Shaffer AAACAACAATCATAAATAGAGGAAGTTTTCGGAATATACGATAAGTGAAATATCGTTCT
Outline Overview of the GEP annotation projects Annotation of Drosophila Primer January 2018 GEP annotation workflow Practice applying the GEP annotation strategy Wilson Leung and Chris Shaffer AAACAACAATCATAAATAGAGGAAGTTTTCGGAATATACGATAAGTGAAATATCGTTCT
Sequence assembly. Jose Blanca COMAV institute bioinf.comav.upv.es
 Sequence assembly Jose Blanca COMAV institute bioinf.comav.upv.es Sequencing project Unknown sequence { experimental evidence result read 1 read 4 read 2 read 5 read 3 read 6 read 7 Computational requirements
Sequence assembly Jose Blanca COMAV institute bioinf.comav.upv.es Sequencing project Unknown sequence { experimental evidence result read 1 read 4 read 2 read 5 read 3 read 6 read 7 Computational requirements
6.2 Chromatin is divided into euchromatin and heterochromatin
 6.2 Chromatin is divided into euchromatin and heterochromatin Individual chromosomes can be seen only during mitosis. During interphase, the general mass of chromatin is in the form of euchromatin. Euchromatin
6.2 Chromatin is divided into euchromatin and heterochromatin Individual chromosomes can be seen only during mitosis. During interphase, the general mass of chromatin is in the form of euchromatin. Euchromatin
Genes found in the genome include protein-coding genes and non-coding RNA genes. Which nucleotide is not normally found in non-coding RNA genes?
 Midterm Q Genes found in the genome include protein-coding genes and non-coding RNA genes Which nucleotide is not normally found in non-coding RNA genes? G T 3 A 4 C 5 U 00% Midterm Q Which of the following
Midterm Q Genes found in the genome include protein-coding genes and non-coding RNA genes Which nucleotide is not normally found in non-coding RNA genes? G T 3 A 4 C 5 U 00% Midterm Q Which of the following
Annotating 7G24-63 Justin Richner May 4, Figure 1: Map of my sequence
 Annotating 7G24-63 Justin Richner May 4, 2005 Zfh2 exons Thd1 exons Pur-alpha exons 0 40 kb 8 = 1 kb = LINE, Penelope = DNA/Transib, Transib1 = DINE = Novel Repeat = LTR/PAO, Diver2 I = LTR/Gypsy, Invader
Annotating 7G24-63 Justin Richner May 4, 2005 Zfh2 exons Thd1 exons Pur-alpha exons 0 40 kb 8 = 1 kb = LINE, Penelope = DNA/Transib, Transib1 = DINE = Novel Repeat = LTR/PAO, Diver2 I = LTR/Gypsy, Invader
MODULE 1: INTRODUCTION TO THE GENOME BROWSER: WHAT IS A GENE?
 MODULE 1: INTRODUCTION TO THE GENOME BROWSER: WHAT IS A GENE? Lesson Plan: Title Introduction to the Genome Browser: what is a gene? JOYCE STAMM Objectives Demonstrate basic skills in using the UCSC Genome
MODULE 1: INTRODUCTION TO THE GENOME BROWSER: WHAT IS A GENE? Lesson Plan: Title Introduction to the Genome Browser: what is a gene? JOYCE STAMM Objectives Demonstrate basic skills in using the UCSC Genome
DNA Replication. The Organization of DNA. Recall:
 Recall: The Organization of DNA DNA Replication Chromosomal form appears only during mitosis, and is used in karyotypes. folded back upon itself (chromosomes) coiled around itself (chromatin) wrapped around
Recall: The Organization of DNA DNA Replication Chromosomal form appears only during mitosis, and is used in karyotypes. folded back upon itself (chromosomes) coiled around itself (chromatin) wrapped around
Connect-A-Contig Paper version
 Teacher Guide Connect-A-Contig Paper version Abstract Students align pieces of paper DNA strips based on the distance between markers to generate a DNA consensus sequence. The activity helps students see
Teacher Guide Connect-A-Contig Paper version Abstract Students align pieces of paper DNA strips based on the distance between markers to generate a DNA consensus sequence. The activity helps students see
Lack of Relationship Between Amount of DNA and Organism Complexity. In eukaryotes, genes are often much larger than the coding region
 Genomes, Heterochromatin, and the Dot Chromosome: Comparative Genomics in Drosophila Sarah CR Elgin Lab Genomics Education Partnership TA Workshop Washington University in St. Louis August, 2006 Minimum
Genomes, Heterochromatin, and the Dot Chromosome: Comparative Genomics in Drosophila Sarah CR Elgin Lab Genomics Education Partnership TA Workshop Washington University in St. Louis August, 2006 Minimum
3I03 - Eukaryotic Genetics Repetitive DNA
 Repetitive DNA Satellite DNA Minisatellite DNA Microsatellite DNA Transposable elements LINES, SINES and other retrosequences High copy number genes (e.g. ribosomal genes, histone genes) Multifamily member
Repetitive DNA Satellite DNA Minisatellite DNA Microsatellite DNA Transposable elements LINES, SINES and other retrosequences High copy number genes (e.g. ribosomal genes, histone genes) Multifamily member
user s guide Question 3
 Question 3 During a positional cloning project aimed at finding a human disease gene, linkage data have been obtained suggesting that the gene of interest lies between two sequence-tagged site markers.
Question 3 During a positional cloning project aimed at finding a human disease gene, linkage data have been obtained suggesting that the gene of interest lies between two sequence-tagged site markers.
601 CTGTCCACACAATCTGCCCTTTCGAAAGATCCCAACGAAAAGAGAGACCACATGGTCCTT GACAGGTGTGTTAGACGGGAAAGCTTTCTAGGGTTGCTTTTCTCTCTGGTGTACCAGGAA >>>>>>>>>>>>>>>>>>
 BIO450 Primer Design Tutorial The most critical step in your PCR experiment will be designing your oligonucleotide primers. Poor primers could result in little or even no PCR product. Alternatively, they
BIO450 Primer Design Tutorial The most critical step in your PCR experiment will be designing your oligonucleotide primers. Poor primers could result in little or even no PCR product. Alternatively, they
Chapter 5 DNA and Chromosomes
 Chapter 5 DNA and Chromosomes DNA as the genetic material Heat-killed bacteria can transform living cells S Smooth R Rough Fred Griffith, 1920 DNA is the genetic material Oswald Avery Colin MacLeod Maclyn
Chapter 5 DNA and Chromosomes DNA as the genetic material Heat-killed bacteria can transform living cells S Smooth R Rough Fred Griffith, 1920 DNA is the genetic material Oswald Avery Colin MacLeod Maclyn
DNA: The Genetic Material. Chapter 10
 DNA: The Genetic Material Chapter 10 DNA as the Genetic Material DNA was first extracted from nuclei in 1870 named nuclein after their source. Chemical analysis determined that DNA was a weak acid rich
DNA: The Genetic Material Chapter 10 DNA as the Genetic Material DNA was first extracted from nuclei in 1870 named nuclein after their source. Chemical analysis determined that DNA was a weak acid rich
Supplementary Figure 1
 Nucleotide Content E. coli End. Neb. Tr. End. Neb. Tr. Supplementary Figure 1 Fragmentation Site Profiles CRW1 End. Son. Tr. End. Son. Tr. Human PA1 Position Fragmentation site profiles. Nucleotide content
Nucleotide Content E. coli End. Neb. Tr. End. Neb. Tr. Supplementary Figure 1 Fragmentation Site Profiles CRW1 End. Son. Tr. End. Son. Tr. Human PA1 Position Fragmentation site profiles. Nucleotide content
Plant Molecular and Cellular Biology Lecture 9: Nuclear Genome Organization: Chromosome Structure, Chromatin, DNA Packaging, Mitosis Gary Peter
 Plant Molecular and Cellular Biology Lecture 9: Nuclear Genome Organization: Chromosome Structure, Chromatin, DNA Packaging, Mitosis Gary Peter 9/16/2008 1 Learning Objectives 1. List and explain how DNA
Plant Molecular and Cellular Biology Lecture 9: Nuclear Genome Organization: Chromosome Structure, Chromatin, DNA Packaging, Mitosis Gary Peter 9/16/2008 1 Learning Objectives 1. List and explain how DNA
Genome Sequence Assembly
 Genome Sequence Assembly Learning Goals: Introduce the field of bioinformatics Familiarize the student with performing sequence alignments Understand the assembly process in genome sequencing Introduction:
Genome Sequence Assembly Learning Goals: Introduce the field of bioinformatics Familiarize the student with performing sequence alignments Understand the assembly process in genome sequencing Introduction:
3. human genomics clone genes associated with genetic disorders. 4. many projects generate ordered clones that cover genome
 Lectures 30 and 31 Genome analysis I. Genome analysis A. two general areas 1. structural 2. functional B. genome projects a status report 1. 1 st sequenced: several viral genomes 2. mitochondria and chloroplasts
Lectures 30 and 31 Genome analysis I. Genome analysis A. two general areas 1. structural 2. functional B. genome projects a status report 1. 1 st sequenced: several viral genomes 2. mitochondria and chloroplasts
The Genome Analysis Centre. Building Excellence in Genomics and Computational Bioscience
 Building Excellence in Genomics and Computational Bioscience Wheat genome sequencing: an update from TGAC Sequencing Technology Development now Plant & Microbial Genomics Group Leader Matthew Clark matt.clark@tgac.ac.uk
Building Excellence in Genomics and Computational Bioscience Wheat genome sequencing: an update from TGAC Sequencing Technology Development now Plant & Microbial Genomics Group Leader Matthew Clark matt.clark@tgac.ac.uk
Y1 Biology 131 Syllabus - Academic Year
 Y1 Biology 131 Syllabus - Academic Year 2016-2017 Monday 28/11/2016 DNA Packaging Week 11 Tuesday 29/11/2016 Regulation of gene expression Wednesday 22/9/2014 Cell cycle Sunday 4/12/2016 Tutorial Monday
Y1 Biology 131 Syllabus - Academic Year 2016-2017 Monday 28/11/2016 DNA Packaging Week 11 Tuesday 29/11/2016 Regulation of gene expression Wednesday 22/9/2014 Cell cycle Sunday 4/12/2016 Tutorial Monday
TIGR THE INSTITUTE FOR GENOMIC RESEARCH
 Introduction to Genome Annotation: Overview of What You Will Learn This Week C. Robin Buell May 21, 2007 Types of Annotation Structural Annotation: Defining genes, boundaries, sequence motifs e.g. ORF,
Introduction to Genome Annotation: Overview of What You Will Learn This Week C. Robin Buell May 21, 2007 Types of Annotation Structural Annotation: Defining genes, boundaries, sequence motifs e.g. ORF,
user s guide Question 3
 Question 3 During a positional cloning project aimed at finding a human disease gene, linkage data have been obtained suggesting that the gene of interest lies between two sequence-tagged site markers.
Question 3 During a positional cloning project aimed at finding a human disease gene, linkage data have been obtained suggesting that the gene of interest lies between two sequence-tagged site markers.
Next Gen Sequencing. Expansion of sequencing technology. Contents
 Next Gen Sequencing Contents 1 Expansion of sequencing technology 2 The Next Generation of Sequencing: High-Throughput Technologies 3 High Throughput Sequencing Applied to Genome Sequencing (TEDed CC BY-NC-ND
Next Gen Sequencing Contents 1 Expansion of sequencing technology 2 The Next Generation of Sequencing: High-Throughput Technologies 3 High Throughput Sequencing Applied to Genome Sequencing (TEDed CC BY-NC-ND
Lecture 21: Epigenetics Nurture or Nature? Chromatin DNA methylation Histone Code Twin study X-chromosome inactivation Environemnt and epigenetics
 Lecture 21: Epigenetics Nurture or Nature? Chromatin DNA methylation Histone Code Twin study X-chromosome inactivation Environemnt and epigenetics Epigenetics represents the science for the studying heritable
Lecture 21: Epigenetics Nurture or Nature? Chromatin DNA methylation Histone Code Twin study X-chromosome inactivation Environemnt and epigenetics Epigenetics represents the science for the studying heritable
Sequence Assembly and Alignment. Jim Noonan Department of Genetics
 Sequence Assembly and Alignment Jim Noonan Department of Genetics james.noonan@yale.edu www.yale.edu/noonanlab The assembly problem >>10 9 sequencing reads 36 bp - 1 kb 3 Gb Outline Basic concepts in genome
Sequence Assembly and Alignment Jim Noonan Department of Genetics james.noonan@yale.edu www.yale.edu/noonanlab The assembly problem >>10 9 sequencing reads 36 bp - 1 kb 3 Gb Outline Basic concepts in genome
COMPUTER RESOURCES II:
 COMPUTER RESOURCES II: Using the computer to analyze data, using the internet, and accessing online databases Bio 210, Fall 2006 Linda S. Huang, Ph.D. University of Massachusetts Boston In the first computer
COMPUTER RESOURCES II: Using the computer to analyze data, using the internet, and accessing online databases Bio 210, Fall 2006 Linda S. Huang, Ph.D. University of Massachusetts Boston In the first computer
Chapter 13. The Nucleus. The nucleus is the hallmark of eukaryotic cells; the very term eukaryotic means having a "true nucleus".
 Chapter 13 The Nucleus The nucleus is the hallmark of eukaryotic cells; the very term eukaryotic means having a "true nucleus". Fig.13.1. The EM of the Nucleus of a Eukaryotic Cell 13.1. The Nuclear Envelope
Chapter 13 The Nucleus The nucleus is the hallmark of eukaryotic cells; the very term eukaryotic means having a "true nucleus". Fig.13.1. The EM of the Nucleus of a Eukaryotic Cell 13.1. The Nuclear Envelope
1. A brief overview of sequencing biochemistry
 Supplementary reading materials on Genome sequencing (optional) The materials are from Mark Blaxter s lecture notes on Sequencing strategies and Primary Analysis 1. A brief overview of sequencing biochemistry
Supplementary reading materials on Genome sequencing (optional) The materials are from Mark Blaxter s lecture notes on Sequencing strategies and Primary Analysis 1. A brief overview of sequencing biochemistry
Last Update: 12/31/2017. Recommended Background Tutorial: An Introduction to NCBI BLAST
 BLAST Exercise: Detecting and Interpreting Genetic Homology Adapted by T. Cordonnier, C. Shaffer, W. Leung and SCR Elgin from Detecting and Interpreting Genetic Homology by Dr. J. Buhler Recommended Background
BLAST Exercise: Detecting and Interpreting Genetic Homology Adapted by T. Cordonnier, C. Shaffer, W. Leung and SCR Elgin from Detecting and Interpreting Genetic Homology by Dr. J. Buhler Recommended Background
Interpreting RNA-seq data (Browser Exercise II)
 Interpreting RNA-seq data (Browser Exercise II) In previous exercises, you spent some time learning about gene pages and examining genes in the context of the GBrowse genome browser. It is important to
Interpreting RNA-seq data (Browser Exercise II) In previous exercises, you spent some time learning about gene pages and examining genes in the context of the GBrowse genome browser. It is important to
DNA Structure & the Genome. Bio160 General Biology
 DNA Structure & the Genome Bio160 General Biology Lecture Outline I. DNA A nucleic acid II. Chromosome Structure III. Chromosomes and Genes IV. DNA vs. RNA I. DNA A Nucleic Acid Structure of DNA: Remember:
DNA Structure & the Genome Bio160 General Biology Lecture Outline I. DNA A nucleic acid II. Chromosome Structure III. Chromosomes and Genes IV. DNA vs. RNA I. DNA A Nucleic Acid Structure of DNA: Remember:
Contact us for more information and a quotation
 GenePool Information Sheet #1 Installed Sequencing Technologies in the GenePool The GenePool offers sequencing service on three platforms: Sanger (dideoxy) sequencing on ABI 3730 instruments Illumina SOLEXA
GenePool Information Sheet #1 Installed Sequencing Technologies in the GenePool The GenePool offers sequencing service on three platforms: Sanger (dideoxy) sequencing on ABI 3730 instruments Illumina SOLEXA
Multiple choice questions (numbers in brackets indicate the number of correct answers)
 1 Multiple choice questions (numbers in brackets indicate the number of correct answers) February 1, 2013 1. Ribose is found in Nucleic acids Proteins Lipids RNA DNA (2) 2. Most RNA in cells is transfer
1 Multiple choice questions (numbers in brackets indicate the number of correct answers) February 1, 2013 1. Ribose is found in Nucleic acids Proteins Lipids RNA DNA (2) 2. Most RNA in cells is transfer
Figure S1. Unrearranged locus. Rearranged locus. Concordant read pairs. Region1. Region2. Cluster of discordant read pairs, bundle
 Figure S1 a Unrearranged locus Rearranged locus Concordant read pairs Region1 Concordant read pairs Cluster of discordant read pairs, bundle Region2 Concordant read pairs b Physical coverage 5 4 3 2 1
Figure S1 a Unrearranged locus Rearranged locus Concordant read pairs Region1 Concordant read pairs Cluster of discordant read pairs, bundle Region2 Concordant read pairs b Physical coverage 5 4 3 2 1
High throughput omics and BIOINFORMATICS
 High throughput omics and BIOINFORMATICS Giuseppe D'Auria Seville, February 2009 Genomes from isolated bacteria $ $ $ $ $ $ $ $ $$ $ $ $ $ $ $ $ se q se uen q c se uen ing q c se uen ing qu c en ing c
High throughput omics and BIOINFORMATICS Giuseppe D'Auria Seville, February 2009 Genomes from isolated bacteria $ $ $ $ $ $ $ $ $$ $ $ $ $ $ $ $ se q se uen q c se uen ing q c se uen ing qu c en ing c
7 Gene Isolation and Analysis of Multiple
 Genetic Techniques for Biological Research Corinne A. Michels Copyright q 2002 John Wiley & Sons, Ltd ISBNs: 0-471-89921-6 (Hardback); 0-470-84662-3 (Electronic) 7 Gene Isolation and Analysis of Multiple
Genetic Techniques for Biological Research Corinne A. Michels Copyright q 2002 John Wiley & Sons, Ltd ISBNs: 0-471-89921-6 (Hardback); 0-470-84662-3 (Electronic) 7 Gene Isolation and Analysis of Multiple
Genomic region (ENCODE) Gene definitions
 DNA From genes to proteins Bioinformatics Methods RNA PROMOTER ELEMENTS TRANSCRIPTION Iosif Vaisman mrna SPLICE SITES SPLICING Email: ivaisman@gmu.edu START CODON STOP CODON TRANSLATION PROTEIN From genes
DNA From genes to proteins Bioinformatics Methods RNA PROMOTER ELEMENTS TRANSCRIPTION Iosif Vaisman mrna SPLICE SITES SPLICING Email: ivaisman@gmu.edu START CODON STOP CODON TRANSLATION PROTEIN From genes
Thr Gly Tyr. Gly Lys Asn
 Your unique body characteristics (traits), such as hair color or blood type, are determined by the proteins your body produces. Proteins are the building blocks of life - in fact, about 45% of the human
Your unique body characteristics (traits), such as hair color or blood type, are determined by the proteins your body produces. Proteins are the building blocks of life - in fact, about 45% of the human
The Molecular Basis of Inheritance
 The Molecular Basis of Inheritance Chapter 16 Objectives Describe the contributions of the following people: Griffith; Avery, McCary, and MacLeod; Hershey and Chase; Chargaff; Watson and Crick; Franklin;
The Molecular Basis of Inheritance Chapter 16 Objectives Describe the contributions of the following people: Griffith; Avery, McCary, and MacLeod; Hershey and Chase; Chargaff; Watson and Crick; Franklin;
Value Correct Answer Feedback. Student Response. A. Dicer enzyme. complex. C. the Dicer-RISC complex D. none of the above
 1 RNA mediated interference is a post-transcriptional gene silencing mechanism Which component of the RNAi pathway have been implicated in cleavage of the target mrna? A Dicer enzyme B the RISC-siRNA complex
1 RNA mediated interference is a post-transcriptional gene silencing mechanism Which component of the RNAi pathway have been implicated in cleavage of the target mrna? A Dicer enzyme B the RISC-siRNA complex
CSE182-L16. LW statistics/assembly
 CSE182-L16 LW statistics/assembly Silly Quiz Who are these people, and what is the occasion? Genome Sequencing and Assembly Sequencing A break at T is shown here. Measuring the lengths using electrophoresis
CSE182-L16 LW statistics/assembly Silly Quiz Who are these people, and what is the occasion? Genome Sequencing and Assembly Sequencing A break at T is shown here. Measuring the lengths using electrophoresis
Host : Dr. Nobuyuki Nukina Tutor : Dr. Fumitaka Oyama
 Method to assign the coding regions of ESTs Céline Becquet Summer Program 2002 Structural Neuropathology Lab Molecular Neuropathology Group RIKEN Brain Science Institute Host : Dr. Nobuyuki Nukina Tutor
Method to assign the coding regions of ESTs Céline Becquet Summer Program 2002 Structural Neuropathology Lab Molecular Neuropathology Group RIKEN Brain Science Institute Host : Dr. Nobuyuki Nukina Tutor
SOURCES AND RESOURCES:
 DNA Mass Production Lesson plan for grades 6-12 Length of lesson: 60-85 minutes (1.5 class periods) Authored by: Brotee Rahman, Environmental Science Institute, 5/17/2013 SOURCES AND RESOURCES: Andrew
DNA Mass Production Lesson plan for grades 6-12 Length of lesson: 60-85 minutes (1.5 class periods) Authored by: Brotee Rahman, Environmental Science Institute, 5/17/2013 SOURCES AND RESOURCES: Andrew
Figure 1: Testing the CIT: T.H. Morgan s Fruit Fly Mating Experiments
 I. Chromosomal Theory of Inheritance As early cytologists worked out the mechanism of cell division in the late 1800 s, they began to notice similarities in the behavior of BOTH chromosomes & Mendel s
I. Chromosomal Theory of Inheritance As early cytologists worked out the mechanism of cell division in the late 1800 s, they began to notice similarities in the behavior of BOTH chromosomes & Mendel s
Agenda. Web Databases for Drosophila. Gene annotation workflow. GEP Drosophila annotation projects 01/01/2018. Annotation adding labels to a sequence
 Agenda GEP annotation project overview Web Databases for Drosophila An introduction to web tools, databases and NCBI BLAST Web databases for Drosophila annotation UCSC Genome Browser NCBI / BLAST FlyBase
Agenda GEP annotation project overview Web Databases for Drosophila An introduction to web tools, databases and NCBI BLAST Web databases for Drosophila annotation UCSC Genome Browser NCBI / BLAST FlyBase
Introduction to Molecular Biology
 Introduction to Molecular Biology Bioinformatics: Issues and Algorithms CSE 308-408 Fall 2007 Lecture 2-1- Important points to remember We will study: Problems from bioinformatics. Algorithms used to solve
Introduction to Molecular Biology Bioinformatics: Issues and Algorithms CSE 308-408 Fall 2007 Lecture 2-1- Important points to remember We will study: Problems from bioinformatics. Algorithms used to solve
DNA AND CHROMOSOMES. Genetica per Scienze Naturali a.a prof S. Presciuttini
 DNA AND CHROMOSOMES This document is licensed under the Attribution-NonCommercial-ShareAlike 2.5 Italy license, available at http://creativecommons.org/licenses/by-nc-sa/2.5/it/ 1. The Building Blocks
DNA AND CHROMOSOMES This document is licensed under the Attribution-NonCommercial-ShareAlike 2.5 Italy license, available at http://creativecommons.org/licenses/by-nc-sa/2.5/it/ 1. The Building Blocks
PrimePCR Assay Validation Report
 Gene Information Gene Name collagen, type IV, alpha 1 Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID COL4A1 Human This gene encodes the major type IV alpha
Gene Information Gene Name collagen, type IV, alpha 1 Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID COL4A1 Human This gene encodes the major type IV alpha
NUCLEUS. Fig. 2. Various stages in the condensation of chromatin
 NUCLEUS Animal cells contain DNA in nucleus (contains ~ 98% of cell DNA) and mitochondrion. Both compartments are surrounded by an envelope (double membrane). Nuclear DNA represents some linear molecules
NUCLEUS Animal cells contain DNA in nucleus (contains ~ 98% of cell DNA) and mitochondrion. Both compartments are surrounded by an envelope (double membrane). Nuclear DNA represents some linear molecules
Chapter Eleven: Chromosome Structure and Transposable Elements
 Chapter Eleven: Chromosome Structure and Transposable Elements COMPREHENSION QUESTIONS Section 11.1 *1. How does supercoiling arise? What is the difference between positive and negative supercoiling? Supercoiling
Chapter Eleven: Chromosome Structure and Transposable Elements COMPREHENSION QUESTIONS Section 11.1 *1. How does supercoiling arise? What is the difference between positive and negative supercoiling? Supercoiling
Chapter 11. Gene Expression and Regulation. Lectures by Gregory Ahearn. University of North Florida. Copyright 2009 Pearson Education, Inc..
 Chapter 11 Gene Expression and Regulation Lectures by Gregory Ahearn University of North Florida Copyright 2009 Pearson Education, Inc.. 11.1 How Is The Information In DNA Used In A Cell? Most genes contain
Chapter 11 Gene Expression and Regulation Lectures by Gregory Ahearn University of North Florida Copyright 2009 Pearson Education, Inc.. 11.1 How Is The Information In DNA Used In A Cell? Most genes contain
Biology 201 (Genetics) Exam #3 120 points 20 November Read the question carefully before answering. Think before you write.
 Name KEY Section Biology 201 (Genetics) Exam #3 120 points 20 November 2006 Read the question carefully before answering. Think before you write. You will have up to 50 minutes to take this exam. After
Name KEY Section Biology 201 (Genetics) Exam #3 120 points 20 November 2006 Read the question carefully before answering. Think before you write. You will have up to 50 minutes to take this exam. After
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Origin use and efficiency are similar among WT, rrm3, pif1-m2, and pif1-m2; rrm3 strains. A. Analysis of fork progression around confirmed and likely origins (from cerevisiae.oridb.org).
Supplementary Figure 1 Origin use and efficiency are similar among WT, rrm3, pif1-m2, and pif1-m2; rrm3 strains. A. Analysis of fork progression around confirmed and likely origins (from cerevisiae.oridb.org).
Nature Biotechnology: doi: /nbt Supplementary Figure 1. Number and length distributions of the inferred fosmids.
 Supplementary Figure 1 Number and length distributions of the inferred fosmids. Fosmid were inferred by mapping each pool s sequence reads to hg19. We retained only those reads that mapped to within a
Supplementary Figure 1 Number and length distributions of the inferred fosmids. Fosmid were inferred by mapping each pool s sequence reads to hg19. We retained only those reads that mapped to within a
Chimp Sequence Annotation: Region 2_3
 Chimp Sequence Annotation: Region 2_3 Jeff Howenstein March 30, 2007 BIO434W Genomics 1 Introduction We received region 2_3 of the ChimpChunk sequence, and the first step we performed was to run RepeatMasker
Chimp Sequence Annotation: Region 2_3 Jeff Howenstein March 30, 2007 BIO434W Genomics 1 Introduction We received region 2_3 of the ChimpChunk sequence, and the first step we performed was to run RepeatMasker
Chapter 2 The Structure of Genes and Genomes. Electron micrograph of a metaphase chromosome
 Chapter 2 The Structure of Genes and Genomes Electron micrograph of a metaphase chromosome Genetic information is stored in double stranded DNA DNA structure, building blocks: (A) Bases DNA structure,
Chapter 2 The Structure of Genes and Genomes Electron micrograph of a metaphase chromosome Genetic information is stored in double stranded DNA DNA structure, building blocks: (A) Bases DNA structure,
Research Finishing a whole-genome shotgun: Release 3 of the Drosophila melanogaster euchromatic genome sequence
 http://genomebiology.com/2002/3/12/research/0079.1 Research Finishing a whole-genome shotgun: Release 3 of the Drosophila melanogaster euchromatic genome sequence Susan E Celniker*, David A Wheeler, Brent
http://genomebiology.com/2002/3/12/research/0079.1 Research Finishing a whole-genome shotgun: Release 3 of the Drosophila melanogaster euchromatic genome sequence Susan E Celniker*, David A Wheeler, Brent
BENG 183 Trey Ideker. Genome Assembly and Physical Mapping
 BENG 183 Trey Ideker Genome Assembly and Physical Mapping Reasons for sequencing Complete genome sequencing!!! Resequencing (Confirmatory) E.g., short regions containing single nucleotide polymorphisms
BENG 183 Trey Ideker Genome Assembly and Physical Mapping Reasons for sequencing Complete genome sequencing!!! Resequencing (Confirmatory) E.g., short regions containing single nucleotide polymorphisms
Wednesday, November 22, 17. Exons and Introns
 Exons and Introns Introns and Exons Exons: coded regions of DNA that get transcribed and translated into proteins make up 5% of the genome Introns and Exons Introns: non-coded regions of DNA Must be removed
Exons and Introns Introns and Exons Exons: coded regions of DNA that get transcribed and translated into proteins make up 5% of the genome Introns and Exons Introns: non-coded regions of DNA Must be removed
Recombination. The kinetochore ("spindle attachment ) always separates reductionally at anaphase I and equationally at anaphase II.
 Recombination Chromosome Separations At Anaphase I And II Mather (1935 pp. 53-62). Reductional vs. equational separations Reductional Division: Sister chromatids go to same pole at anaphase I Equational
Recombination Chromosome Separations At Anaphase I And II Mather (1935 pp. 53-62). Reductional vs. equational separations Reductional Division: Sister chromatids go to same pole at anaphase I Equational
PIP-seq. Cells. Permanganate ChIP-Seq
 PIP-seq ells Formaldehyde Permanganate 5 Harvest Lyse Sonicate First dapter Ligation 3 3 5 hip Elute Reverse rosslinks Piperidine cleavage 5 3 3 5 Primer Extension Second dapter Ligation 5 3 3 5 Deep Sequencing
PIP-seq ells Formaldehyde Permanganate 5 Harvest Lyse Sonicate First dapter Ligation 3 3 5 hip Elute Reverse rosslinks Piperidine cleavage 5 3 3 5 Primer Extension Second dapter Ligation 5 3 3 5 Deep Sequencing
CHAPTER 4 STURTEVANT: THE FIRST GENETIC MAP: DROSOPHILA X CHROMOSOME LINKED GENES MAY BE MAPPED BY THREE-FACTOR TEST CROSSES STURTEVANT S EXPERIMENT
 CHAPTER 4 STURTEVANT: THE FIRST GENETIC MAP: DROSOPHILA X CHROMOSOME In 1913, Alfred Sturtevant drew a logical conclusion from Morgan s theories of crossing-over, suggesting that the information gained
CHAPTER 4 STURTEVANT: THE FIRST GENETIC MAP: DROSOPHILA X CHROMOSOME In 1913, Alfred Sturtevant drew a logical conclusion from Morgan s theories of crossing-over, suggesting that the information gained
Genome Sequencing-- Strategies
 Genome Sequencing-- Strategies Bio 4342 Spring 04 What is a genome? A genome can be defined as the entire DNA content of each nucleated cell in an organism Each organism has one or more chromosomes that
Genome Sequencing-- Strategies Bio 4342 Spring 04 What is a genome? A genome can be defined as the entire DNA content of each nucleated cell in an organism Each organism has one or more chromosomes that
Agenda. Annotation of Drosophila. Muller element nomenclature. Annotation: Adding labels to a sequence. GEP Drosophila annotation projects 01/03/2018
 Agenda Annotation of Drosophila January 2018 Overview of the GEP annotation project GEP annotation strategy Types of evidence Analysis tools Web databases Annotation of a single isoform (walkthrough) Wilson
Agenda Annotation of Drosophila January 2018 Overview of the GEP annotation project GEP annotation strategy Types of evidence Analysis tools Web databases Annotation of a single isoform (walkthrough) Wilson
Mapping strategies for sequence reads
 Mapping strategies for sequence reads Ernest Turro University of Cambridge 21 Oct 2013 Quantification A basic aim in genomics is working out the contents of a biological sample. 1. What distinct elements
Mapping strategies for sequence reads Ernest Turro University of Cambridge 21 Oct 2013 Quantification A basic aim in genomics is working out the contents of a biological sample. 1. What distinct elements
Chapter 20: Biotechnology
 Name Period The AP Biology exam has reached into this chapter for essay questions on a regular basis over the past 15 years. Student responses show that biotechnology is a difficult topic. This chapter
Name Period The AP Biology exam has reached into this chapter for essay questions on a regular basis over the past 15 years. Student responses show that biotechnology is a difficult topic. This chapter
Whole Transcriptome Analysis of Illumina RNA- Seq Data. Ryan Peters Field Application Specialist
 Whole Transcriptome Analysis of Illumina RNA- Seq Data Ryan Peters Field Application Specialist Partek GS in your NGS Pipeline Your Start-to-Finish Solution for Analysis of Next Generation Sequencing Data
Whole Transcriptome Analysis of Illumina RNA- Seq Data Ryan Peters Field Application Specialist Partek GS in your NGS Pipeline Your Start-to-Finish Solution for Analysis of Next Generation Sequencing Data
PrimePCR Assay Validation Report
 Gene Information Gene Name Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin)
Gene Information Gene Name Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin)
Human SNP haplotypes. Statistics 246, Spring 2002 Week 15, Lecture 1
 Human SNP haplotypes Statistics 246, Spring 2002 Week 15, Lecture 1 Human single nucleotide polymorphisms The majority of human sequence variation is due to substitutions that have occurred once in the
Human SNP haplotypes Statistics 246, Spring 2002 Week 15, Lecture 1 Human single nucleotide polymorphisms The majority of human sequence variation is due to substitutions that have occurred once in the
Chapter 15 Gene Technologies and Human Applications
 Chapter Outline Chapter 15 Gene Technologies and Human Applications Section 1: The Human Genome KEY IDEAS > Why is the Human Genome Project so important? > How do genomics and gene technologies affect
Chapter Outline Chapter 15 Gene Technologies and Human Applications Section 1: The Human Genome KEY IDEAS > Why is the Human Genome Project so important? > How do genomics and gene technologies affect
TECH NOTE SMARTer T-cell receptor profiling in single cells
 TECH NOTE SMARTer T-cell receptor profiling in single cells Flexible workflow: Illumina-ready libraries from FACS or manually sorted single cells Ease of use: Optimized indexing allows for pooling 96 cells
TECH NOTE SMARTer T-cell receptor profiling in single cells Flexible workflow: Illumina-ready libraries from FACS or manually sorted single cells Ease of use: Optimized indexing allows for pooling 96 cells
Genetic Identity. Steve Harris SPASH - Biotechnology
 Genetic Identity Steve Harris SPASH - Biotechnology Comparison of Organisms ORGANISM GENES BASE PAIRS Lambda Phage 40 50,000 E.coli 400 5,000,000 Yeast 13,000 15,000,000 Human 20,000 3,000,000,000 (3 billion)
Genetic Identity Steve Harris SPASH - Biotechnology Comparison of Organisms ORGANISM GENES BASE PAIRS Lambda Phage 40 50,000 E.coli 400 5,000,000 Yeast 13,000 15,000,000 Human 20,000 3,000,000,000 (3 billion)
Chapter 3. DNA Replication & The Cell Cycle
 Chapter 3 DNA Replication & The Cell Cycle DNA Replication and the Cell Cycle Before cells divide, they must duplicate their DNA // the genetic material DNA is organized into strands called chromosomes
Chapter 3 DNA Replication & The Cell Cycle DNA Replication and the Cell Cycle Before cells divide, they must duplicate their DNA // the genetic material DNA is organized into strands called chromosomes
ALSO: look at figure 5-11 showing exonintron structure of the beta globin gene
 S08 Biology 205 6/4/08 Reading Assignment Chapter 7: From DNA to Protein: How cells read the genome pg 237-243 on exons and introns (you are not responsible for the biochemistry of splicing: figures 7-15,16
S08 Biology 205 6/4/08 Reading Assignment Chapter 7: From DNA to Protein: How cells read the genome pg 237-243 on exons and introns (you are not responsible for the biochemistry of splicing: figures 7-15,16
Bioinformatics in next generation sequencing projects
 Bioinformatics in next generation sequencing projects Rickard Sandberg Assistant Professor Department of Cell and Molecular Biology Karolinska Institutet May 2013 Standard sequence library generation Illumina
Bioinformatics in next generation sequencing projects Rickard Sandberg Assistant Professor Department of Cell and Molecular Biology Karolinska Institutet May 2013 Standard sequence library generation Illumina
Sequencing technologies. Jose Blanca COMAV institute bioinf.comav.upv.es
 Sequencing technologies Jose Blanca COMAV institute bioinf.comav.upv.es Outline Sequencing technologies: Sanger 2nd generation sequencing: 3er generation sequencing: 454 Illumina SOLiD Ion Torrent PacBio
Sequencing technologies Jose Blanca COMAV institute bioinf.comav.upv.es Outline Sequencing technologies: Sanger 2nd generation sequencing: 3er generation sequencing: 454 Illumina SOLiD Ion Torrent PacBio
132 Grundlagen der Bioinformatik, SoSe 14, D. Huson, June 22, This exposition is based on the following source, which is recommended reading:
 132 Grundlagen der Bioinformatik, SoSe 14, D. Huson, June 22, 214 1 Gene Prediction Using HMMs This exposition is based on the following source, which is recommended reading: 1. Chris Burge and Samuel
132 Grundlagen der Bioinformatik, SoSe 14, D. Huson, June 22, 214 1 Gene Prediction Using HMMs This exposition is based on the following source, which is recommended reading: 1. Chris Burge and Samuel
Next Generation Sequencing Lecture Saarbrücken, 19. March Sequencing Platforms
 Next Generation Sequencing Lecture Saarbrücken, 19. March 2012 Sequencing Platforms Contents Introduction Sequencing Workflow Platforms Roche 454 ABI SOLiD Illumina Genome Anlayzer / HiSeq Problems Quality
Next Generation Sequencing Lecture Saarbrücken, 19. March 2012 Sequencing Platforms Contents Introduction Sequencing Workflow Platforms Roche 454 ABI SOLiD Illumina Genome Anlayzer / HiSeq Problems Quality
Grundlagen der Bioinformatik, SoSe 11, D. Huson, July 4, This exposition is based on the following source, which is recommended reading:
 Grundlagen der Bioinformatik, SoSe 11, D. Huson, July 4, 211 155 12 Gene Prediction Using HMMs This exposition is based on the following source, which is recommended reading: 1. Chris Burge and Samuel
Grundlagen der Bioinformatik, SoSe 11, D. Huson, July 4, 211 155 12 Gene Prediction Using HMMs This exposition is based on the following source, which is recommended reading: 1. Chris Burge and Samuel
Certificate of Analysis of the Holotype HLA 24/7 Configuration A1 & CE
 Certificate of Analysis of the Holotype HLA 24/7 Configuration A1 & CE Product name Holotype HLA 24/7 Configuration A1 & CE Reference number H52 LOT number 00027 (N4/002-P5/006-E1/004-R2/006) Expiration
Certificate of Analysis of the Holotype HLA 24/7 Configuration A1 & CE Product name Holotype HLA 24/7 Configuration A1 & CE Reference number H52 LOT number 00027 (N4/002-P5/006-E1/004-R2/006) Expiration
Files for this Tutorial: All files needed for this tutorial are compressed into a single archive: [BLAST_Intro.tar.gz]
![Files for this Tutorial: All files needed for this tutorial are compressed into a single archive: [BLAST_Intro.tar.gz] Files for this Tutorial: All files needed for this tutorial are compressed into a single archive: [BLAST_Intro.tar.gz]](/thumbs/75/71877651.jpg) BLAST Exercise: Detecting and Interpreting Genetic Homology Adapted by W. Leung and SCR Elgin from Detecting and Interpreting Genetic Homology by Dr. J. Buhler Prequisites: None Resources: The BLAST web
BLAST Exercise: Detecting and Interpreting Genetic Homology Adapted by W. Leung and SCR Elgin from Detecting and Interpreting Genetic Homology by Dr. J. Buhler Prequisites: None Resources: The BLAST web
Differential Gene Expression
 Biology 4361 Developmental Biology Differential Gene Expression September 28, 2006 Chromatin Structure ~140 bp ~60 bp Transcriptional Regulation: 1. Packing prevents access CH 3 2. Acetylation ( C O )
Biology 4361 Developmental Biology Differential Gene Expression September 28, 2006 Chromatin Structure ~140 bp ~60 bp Transcriptional Regulation: 1. Packing prevents access CH 3 2. Acetylation ( C O )
Tutorial. In Silico Cloning. Sample to Insight. March 31, 2016
 In Silico Cloning March 31, 2016 Sample to Insight CLC bio, a QIAGEN Company Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.clcbio.com support-clcbio@qiagen.com In Silico Cloning
In Silico Cloning March 31, 2016 Sample to Insight CLC bio, a QIAGEN Company Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.clcbio.com support-clcbio@qiagen.com In Silico Cloning
ChampionChIP Quick, High Throughput Chromatin Immunoprecipitation Assay System
 ChampionChIP Quick, High Throughput Chromatin Immunoprecipitation Assay System Liyan Pang, Ph.D. Application Scientist 1 Topics to be Covered Introduction What is ChIP-qPCR? Challenges Facing Biological
ChampionChIP Quick, High Throughput Chromatin Immunoprecipitation Assay System Liyan Pang, Ph.D. Application Scientist 1 Topics to be Covered Introduction What is ChIP-qPCR? Challenges Facing Biological
LINKAGE AND CHROMOSOME MAPPING IN EUKARYOTES
 LINKAGE AND CHROMOSOME MAPPING IN EUKARYOTES Objectives: Upon completion of this lab, the students should be able to: Understand the different stages of meiosis. Describe the events during each phase of
LINKAGE AND CHROMOSOME MAPPING IN EUKARYOTES Objectives: Upon completion of this lab, the students should be able to: Understand the different stages of meiosis. Describe the events during each phase of
Protocol for cloning SEC-based repair templates using Gibson assembly and ccdb negative selection
 Protocol for cloning SEC-based repair templates using Gibson assembly and ccdb negative selection Written by Dan Dickinson (daniel.dickinson@austin.utexas.edu) and last updated January 2018. A version
Protocol for cloning SEC-based repair templates using Gibson assembly and ccdb negative selection Written by Dan Dickinson (daniel.dickinson@austin.utexas.edu) and last updated January 2018. A version
Single Cell Genomics
 Single Cell Genomics Application Cost Platform/Protoc ol Note Single cell 3 mrna-seq cell lysis/rt/library prep $2460/Sample 10X Genomics Chromium 500-10,000 cells/sample Single cell 5 V(D)J mrna-seq cell
Single Cell Genomics Application Cost Platform/Protoc ol Note Single cell 3 mrna-seq cell lysis/rt/library prep $2460/Sample 10X Genomics Chromium 500-10,000 cells/sample Single cell 5 V(D)J mrna-seq cell
Sequence Variations. Baxevanis and Ouellette, Chapter 7 - Sequence Polymorphisms. NCBI SNP Primer:
 Sequence Variations Baxevanis and Ouellette, Chapter 7 - Sequence Polymorphisms NCBI SNP Primer: http://www.ncbi.nlm.nih.gov/about/primer/snps.html Overview Mutation and Alleles Linkage Genetic variation
Sequence Variations Baxevanis and Ouellette, Chapter 7 - Sequence Polymorphisms NCBI SNP Primer: http://www.ncbi.nlm.nih.gov/about/primer/snps.html Overview Mutation and Alleles Linkage Genetic variation
