Supplemental Data. Furlan et al. Plant Cell (2017) /tpc
|
|
|
- Sheryl Chapman
- 5 years ago
- Views:
Transcription
1 Supplemental Data. Furlan et al. Plant Cell (0) 0.0/tpc..00.
2 Supplemental Data. Furlan et al. Plant Cell (0) 0.0/tpc..00.
3 Supplemental Data. Furlan et al. Plant Cell (0) 0.0/tpc..00. Supplemental Figure. PUB displays autoubiquitination activity in vitro. (A) In vitro autoubiquitination assay with MBP-PUB and the inactive W0A mutant using His-UBA and His-UBC. MPB-PUB was incubated with the indicated components for h, resolved by PAGE and analysed by immunoblot (IB) with the indicated anti-bodies. (B) In vitro autoubiquitination sites identified on PUB. MS spectra of peptides with a diglycine footprint found on GST-PUB after in vitro autoubiquitination assay using His-UBA and His-UBC.
4 Supplemental Data. Furlan et al. Plant Cell (0) 0.0/tpc Supplemental Figure. PUBprom:GFP-PUB complements the pub pub pub phenotype. (A) Primary root lengths seedlings measured seven days after transplanting onto media containing µm flg or DMSO as control. A homozygous T line carrying a single copy of the PUBprom:GFP-PUB construct in the pub pub pub background was used. Data shown as mean of three independent experiments +/- SD (n 0). Asterisk indicates significantly different values between wild-type and pub pub pub with p<0.0 (one way ANOVA, Tukey post hoc test). (B) PUB accumulation is induced by flg in the PUBprom:GFP-PUB/pub pub pub line. Protein expression was induced by flg (µm) treatment in two week-old seedlings carrying a PUBprom:GFP-PUB construct. Total protein samples were analysed by IB using anti-gfp. Coomassie brilliant blue (CBB) is shown as loading control.
5 Supplemental Data. Furlan et al. Plant Cell (0) 0.0/tpc Supplemental Figure. Post-translational modification of PUB and PUB. (A) Protein expression from experiments in Figure a. cmyc-nyfp-mpk or cmyc-nyfp- MPK were coexpressed with HA-cYFP-PUB W0A as indicated in Figure A. Total proteins were analysed by IB with anti-cmyc and anti-ha. Equal loading shown by coomassie brilliant blue (CBB). (B) PUB does not ubiquitinate MPK or MPK in vitro. In vitro ubiquitination assay with MBP-PUB and GST-MPK or GST-MPK using His-UBA and His-UBC and incubated for h. Proteins were analysed by IB with anti-gst, anti-ubiquitin and anti-pub. (C) PUB is also phosphorylated by MPK in vitro. Recombinant MBP-PUB or MBP- PUB were incubated alone or with activated MPK, MPK and MPK and separated by PAGE (MAPKs indicated by asterisk). Autophosphorylation and trans-phosphorylation were visualized with radioactive ATP and autoradiography. The artificial kinase substrate, myelin basic protein (MBP), was used to adjust equal MPK activities (right panel). The protein loading control is shown by CBB staining.
6 Supplemental Data. Furlan et al. Plant Cell (0) 0.0/tpc..00. Supplemental Figure. In vitro and in vivo phosphorylation sites of PUB.
7 Supplemental Data. Furlan et al. Plant Cell (0) 0.0/tpc..00. (A) LC-MS/MS analysis spectra of PUB phosphorylated tryptic peptides from in vitro phosphorylation assay with MPK. To obtain phosphorylated PUB, GST-PUB was incubated alone or with activated GST-MPK, GST-MPK and untagged MPK. (B) LC-MS/MS analysis spectra of PUB phosphorylated tryptic peptides from in vivo samples. UBQ0prom:GFP-PUB transgenic seedlings grown for days in liquid media were treated with µm flg or water (mock) and harvested after 0min. GFP-PUB was immune-purified resolved by PAGE and gel bands were analysed by LC-MS/MS. Phosphorylated peptides were not detected in the control samples.
8 Supplemental Data. Furlan et al. Plant Cell (0) 0.0/tpc..00. Supplemental Figure. Phylogenetic relations between Arabidopsis PUB proteins class II and III.
9 Supplemental Data. Furlan et al. Plant Cell (0) 0.0/tpc..00. Alignment was performed using MAFFT and manually edited and used for phylogenetic analysis using maximum likelihood with 000 random drawn samples (bootstraps). Bootstrap values greater than 0. are shown at the corresponding node. The phylogenetic tree was rooted using AtCHIP as outgroup. Highlighted are PUB proteins with a conserved threonine followed by proline at position (orange circle) and (red arrowhead). Scale bar denotes 0. amino acid substitutions per site. 0 Supplemental Figure. PUB stabilization is MPK dependent. Luc-PUB fusion was coexpressed with MPK or MPK in mpk protoplasts. Protein levels of YFP-MPK and YFP-MPK of experiments shown in Figure B. Proteins were analyzed by IB using anti-gfp.
10 Supplemental Data. Furlan et al. Plant Cell (0) 0.0/tpc Supplemental Figure. Transgenic lines express comparable levels of transcript of PUB WT and mutant variants. (A) Quantitative real-time PCR of selected T homozygous transgenic lines carrying UBQ0prom:GFP-PUB WT and variants W0A, T/A and T/E. Samples were taken from adult plants and PPA (ATG0) was used as a reference gene. Data shown as mean +/- SD (n=). (B) Analysis of the relative amounts of PUB and mutant variants (line A). The band intensity of PUB WT was determined, set to 00% and the relative amounts for the other lines were calculated. Data shown as mean +/- SEM of four independent experiments. Letters indicate significantly different values at p<0.0 (one way ANOVA, Tukey post hoc test). (C) Representative immunoblot showing accumulation of PUB WT and W0A, T/A and T/E variants in transgenic lines A used to calculate relative protein amounts in (B). 0
11 Supplemental Data. Furlan et al. Plant Cell (0) 0.0/tpc Supplemental Figure. PUB homo- and hetero-oligomerizes. (Supports Figure.) (A) Protein levels of split luciferase experiments shown in Figure F. N- and C-terminal fragments of luciferase fused to PUB U-box WT, TA, TW and TI variants were analyzed by IB with anti-luc. (B) Protein levels of split luciferase experiments shown in Figure G. N- and C-terminal fragments of luciferase fused to PUB U-box WT, W0A, IR and F0E variants were analyzed by IB with anti-luc. (C) Model of the PUB U-box dimer highlighting exchanged residues. Front protomer shown in cartoon style (turquoise) and back protomer shows electrostatic surface charges. (D) Protein levels of split luciferase experiments shown in Figure H. N- and C-terminal luciferase fusion proteins of PUB U-box coexpressed with MKK KR (inactive) or MKK DD (constitutive active) were analyzed by IB with anti-luc. Activation of MPK and MPK was confirmed using anti-erk/.
12 Supplemental Data. Furlan et al. Plant Cell (0) 0.0/tpc..00. Supplemental Tables Table : Primers for genotyping Gene Locus `-Forward-` `-Reverse-` PUB AtG0 ATGTCCATGGGGAAGGAATAG AATGCCCGTCGTGGATATC PUB AtG0 CAATCTTGGTGCACCCTAAAC TTTTCATCAGCAGGGATATGC PUB AtG0 GACGACGTCGTATCAAAGGAC TCGATTGAGGATTGATCGATC MPK AtG0 ATTTTTGTCAACAATGGCCTG TCTGCCTTTTCACGGAATATG Table : Primers used for SDM on PUB and PUB U-box in entry vectors Mutation `-Forward-` `-Reverse-` Type II enzyme F0E ATATATGAAGACGAGTTCC ATATATGAAGACGAACTC BpiI TTTGTCCAATCTCTCT GGAAGGAATCTCTATCT IR ATATATGAAGACCGAGTTT ATATATGAAGACGAAACT BpiI CCACCGGAATAAC CGCACCGGATCT W0A ATATATGAAGACGCGCTCT ATATATGAAGACAAGAG BpiI TTTCCGGTAAGA CGCCTTCTCGATG TI ATATATGAAGACATTCCAA ATATATGAAGACTTTGGA BpiI ACCACAC ATAAGATCAGTT TA ATATATGGTCTCGGCGCC ATATATGGTCTCGGCGC BsaI GAACCACACTCTTCGCC CAGATCAGTTTCGGTTAT G TE ATATATGGTCTCGAGCCC ATATATGGTCTCGGGCT BsaI AACCACACTCTTC CAAGATCAGTTTCG TA ATATATGAAGACAGCTCCA ATATATGAAGACTTGGAG BpiI TE AAACCTCCGATC ATATATGAAGACGAGCCC AAACCTCCGATCTG Table : Primers for cloning CTGGGATCCTCTC ATATATGAAGACTTGGG CTCTGGGATCCTCTCT BpiI Diagnostic none none HaeII none HaeII BanII none BanII Gene Plasmid `-Forward-` `-Reverse-` PUB Ubox pentrc GGCCATTUATGGATCAAGAGAT AGAGA GGTGATTUCTAGATGCGAAGA TGAC no stop PUB pmal CATGTCGACATGGATCAAGAGA TAGAGA CATCTGCAGTCAAGCAGGATA CGAAT PUB pmal CATGAATTCATGAATATATATAC CATCTGCAGTTAGATCTTTGGC GTACA Exo0B petb GAATTCGATGGCTGAAGCCGGT GACGA Table : qrt-pcr CCTTTG CTCGAGTCAACTTGAGCTTTCC TTGA Gene `-Forward-` `-Reverse-` PPA TAACGTGGCCAAAATGATGC GTTCTCCACAACCGCTTGGT PUB TGCAATTGGGAGTTGTAGCA GATTCCCTCCAAACCCTAGC 0
Supplemental Data. Sethi et al. (2014). Plant Cell /tpc
 Supplemental Data Supplemental Figure 1. MYC2 Binds to the E-box but not the E1-box of the MPK6 Promoter. (A) E1-box and E-box (wild type) containing MPK6 promoter fragment. The region shown in red denotes
Supplemental Data Supplemental Figure 1. MYC2 Binds to the E-box but not the E1-box of the MPK6 Promoter. (A) E1-box and E-box (wild type) containing MPK6 promoter fragment. The region shown in red denotes
Supplemental Data. Xie et al. (2014). Plant Cell /tpc
 Supplemental Figure 1. The phylogenetic tree of rice and Arabidopsis Group A/B/C MAPKs that contain the TEY motif in their activation loop. Tobacco SIPK and WIPK, human ERK2 and P38 were also included
Supplemental Figure 1. The phylogenetic tree of rice and Arabidopsis Group A/B/C MAPKs that contain the TEY motif in their activation loop. Tobacco SIPK and WIPK, human ERK2 and P38 were also included
Figure S1. DELLA Proteins Act as Positive Regulators to Mediate GA-Regulated Anthocyanin
 Supplemental Information Figure S1. DELLA Proteins Act as Positive Regulators to Mediate GA-Regulated Anthocyanin Biosynthesis. (A) Effect of GA on anthocyanin content in WT and ga1-3 seedlings. Mock,
Supplemental Information Figure S1. DELLA Proteins Act as Positive Regulators to Mediate GA-Regulated Anthocyanin Biosynthesis. (A) Effect of GA on anthocyanin content in WT and ga1-3 seedlings. Mock,
S156AT168AY175A (AAA) were purified as GST-fusion proteins and incubated with GSTfused
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 Supplemental Materials Supplemental Figure S1 (a) Phenotype of the wild type and grik1-2 grik2-1 plants after 8 days in darkness.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 Supplemental Materials Supplemental Figure S1 (a) Phenotype of the wild type and grik1-2 grik2-1 plants after 8 days in darkness.
Supplemental Data. Cui et al. (2012). Plant Cell /tpc a b c d. Stem UBC32 ACTIN
 A Root Stem Leaf Flower Silique Senescence leaf B a b c d UBC32 ACTIN C * Supplemental Figure 1. Expression Pattern and Protein Sequence of UBC32 Homologues in Yeast, Human, and Arabidopsis. (A) Expression
A Root Stem Leaf Flower Silique Senescence leaf B a b c d UBC32 ACTIN C * Supplemental Figure 1. Expression Pattern and Protein Sequence of UBC32 Homologues in Yeast, Human, and Arabidopsis. (A) Expression
Supplemental Materials
 Supplemental Materials Flores-Pérez et al., Supplemental Materials, page 1 of 5 Supplemental Figure S1. Pull-down and BiFC controls, and quantitative analyses associated with the BiFC studies. (A) Controls
Supplemental Materials Flores-Pérez et al., Supplemental Materials, page 1 of 5 Supplemental Figure S1. Pull-down and BiFC controls, and quantitative analyses associated with the BiFC studies. (A) Controls
Supplemental Data. Li et al. (2015). Plant Cell /tpc
 Supplemental Data Supplemental Figure 1: Characterization of asr3 T-DNA knockout lines and complementation transgenic lines. (A) The scheme of At2G33550 (ASR3) with gray boxes indicating exons and dash
Supplemental Data Supplemental Figure 1: Characterization of asr3 T-DNA knockout lines and complementation transgenic lines. (A) The scheme of At2G33550 (ASR3) with gray boxes indicating exons and dash
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/5/244/ra72/dc1 Supplementary Materials for An Interaction Between BZR1 and DELLAs Mediates Direct Signaling Crosstalk Between Brassinosteroids and Gibberellins
www.sciencesignaling.org/cgi/content/full/5/244/ra72/dc1 Supplementary Materials for An Interaction Between BZR1 and DELLAs Mediates Direct Signaling Crosstalk Between Brassinosteroids and Gibberellins
The Arabidopsis Transcription Factor BES1 Is a Direct Substrate of MPK6 and Regulates Immunity
 Supplemental Data The Arabidopsis Transcription Factor BES1 Is a Direct Substrate of MPK6 and Regulates Immunity Authors: Sining Kang, Fan Yang, Lin Li, Huamin Chen, She Chen and Jie Zhang The following
Supplemental Data The Arabidopsis Transcription Factor BES1 Is a Direct Substrate of MPK6 and Regulates Immunity Authors: Sining Kang, Fan Yang, Lin Li, Huamin Chen, She Chen and Jie Zhang The following
Supplemental Data. Farmer et al. (2010) Plant Cell /tpc
 Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
Supplemental Information. A Tyrosine Phosphorylation Cycle. Regulates Fungal Activation of a Plant. Receptor Ser/Thr Kinase
 Cell Host & Microbe, Volume 23 Supplemental Information A Tyrosine Phosphorylation Cycle Regulates Fungal Activation of a Plant Receptor Ser/Thr Kinase Jun Liu, Bing Liu, Sufen Chen, Ben-Qiang Gong, Lijuan
Cell Host & Microbe, Volume 23 Supplemental Information A Tyrosine Phosphorylation Cycle Regulates Fungal Activation of a Plant Receptor Ser/Thr Kinase Jun Liu, Bing Liu, Sufen Chen, Ben-Qiang Gong, Lijuan
Supplemental Figure 1
 Supplemental Figure 1 A gta2-1 gta2-2 1kb AT4G08350 B Col-0 gta2-1 LP+RP+LBb1.3 C Col-0 gta2-2 LP+RP+LB1 D Col-0 gta2-2 gta2-1 GTA2 TUBLIN E Col0 gta2-1 gta2-2 Supplemental Figure 1. Phenotypic analysis
Supplemental Figure 1 A gta2-1 gta2-2 1kb AT4G08350 B Col-0 gta2-1 LP+RP+LBb1.3 C Col-0 gta2-2 LP+RP+LB1 D Col-0 gta2-2 gta2-1 GTA2 TUBLIN E Col0 gta2-1 gta2-2 Supplemental Figure 1. Phenotypic analysis
Supplemental Data. Hu et al. Plant Cell (2017) /tpc
 1 2 3 4 Supplemental Figure 1. DNA gel blot analysis of homozygous transgenic plants. (Supports Figure 1.) 5 6 7 8 Rice genomic DNA was digested with the restriction enzymes EcoRⅠ and BamHⅠ. Lanes in the
1 2 3 4 Supplemental Figure 1. DNA gel blot analysis of homozygous transgenic plants. (Supports Figure 1.) 5 6 7 8 Rice genomic DNA was digested with the restriction enzymes EcoRⅠ and BamHⅠ. Lanes in the
Supplementary Figure 1 Collision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK2, PPK3 and PPK4 respectively.
 Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
Supplemental Figure 1. Alignment of the NbGAPC amino acid sequences with their Arabidopsis homologues.
 Supplemental Figure 1. Alignment of the NbGAPC amino acid sequences with their Arabidopsis homologues. Homologs from N. benthamiana (NbGAPC1, NbGAPC2, NbGAPC3), Arabidopsis (AtGAPC1, AT3G04120; AtGAPC2,
Supplemental Figure 1. Alignment of the NbGAPC amino acid sequences with their Arabidopsis homologues. Homologs from N. benthamiana (NbGAPC1, NbGAPC2, NbGAPC3), Arabidopsis (AtGAPC1, AT3G04120; AtGAPC2,
Supplemental Data. Huo et al. (2013). Plant Cell /tpc
 Supplemental Data. Huo et al. (2013). Plant Cell 10.1105/tpc.112.108902 1 Supplemental Figure 1. Alignment of NCED4 Amino Acid Sequences from Different Lettuce Varieties. NCED4 amino acid sequences are
Supplemental Data. Huo et al. (2013). Plant Cell 10.1105/tpc.112.108902 1 Supplemental Figure 1. Alignment of NCED4 Amino Acid Sequences from Different Lettuce Varieties. NCED4 amino acid sequences are
Supplementary Data Supplementary Figures
 Supplementary Data Supplementary Figures Supplementary Figure 1. Pi04314 is expressed during infection, each GFP-Pi04314 fusion is stable and myr GFP-Pi04314 is removed from the nucleus while NLS GFP-Pi04314
Supplementary Data Supplementary Figures Supplementary Figure 1. Pi04314 is expressed during infection, each GFP-Pi04314 fusion is stable and myr GFP-Pi04314 is removed from the nucleus while NLS GFP-Pi04314
Supplemental Figure 1. Immunoblot analysis of wild-type and mutant forms of JAZ3
 Supplemental Data. Chung and Howe (2009). A Critical Role for the TIFY Motif in Repression of Jasmonate Signaling by a Stabilized Splice Variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis.
Supplemental Data. Chung and Howe (2009). A Critical Role for the TIFY Motif in Repression of Jasmonate Signaling by a Stabilized Splice Variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis.
Figure S1. Sequence alignments of ATRIP and ATR TopBP1 interacting regions.
 A H. sapiens 204 TKLQTS--ERANKLAAPSVSH VSPRKNPSVVIKPEACS-PQFGKTSFPTKESFSANMS LP 259 B. taurus 201 TKLQSS--ERANKLAVPTVSH VSPRKSPSVVIKPEACS-PQFGKPSFPTKESFSANKS LP 257 M. musculus 204 TKSQSN--GRTNKPAAPSVSH
A H. sapiens 204 TKLQTS--ERANKLAAPSVSH VSPRKNPSVVIKPEACS-PQFGKTSFPTKESFSANMS LP 259 B. taurus 201 TKLQSS--ERANKLAVPTVSH VSPRKSPSVVIKPEACS-PQFGKPSFPTKESFSANKS LP 257 M. musculus 204 TKSQSN--GRTNKPAAPSVSH
Supplemental Data. Hachez et al. Plant Cell (2014) /tpc Suppl. Figure 1A
 Suppl. Figure 1A Suppl. Figure 1B Supplemental Figure 1: Results of the commercial screening of interactants using split ubiquitin technique. (A) Isolated preys (192) using the bait construct pbt3-n- as
Suppl. Figure 1A Suppl. Figure 1B Supplemental Figure 1: Results of the commercial screening of interactants using split ubiquitin technique. (A) Isolated preys (192) using the bait construct pbt3-n- as
Supplementary Figures 1-12
 Supplementary Figures 1-12 Supplementary Figure 1. The specificity of anti-abi1 antibody. Total Proteins extracted from the wild type seedlings or abi1-3 null mutant seedlings were used for immunoblotting
Supplementary Figures 1-12 Supplementary Figure 1. The specificity of anti-abi1 antibody. Total Proteins extracted from the wild type seedlings or abi1-3 null mutant seedlings were used for immunoblotting
Supplementary information
 Supplementary information Supplementary figures Figure S1 Level of mycdet1 protein in DET1 OE-1, OE-2 and OE-3 transgenic lines. Total protein extract from wild type Col0, det1-1 mutant and DET1 OE lines
Supplementary information Supplementary figures Figure S1 Level of mycdet1 protein in DET1 OE-1, OE-2 and OE-3 transgenic lines. Total protein extract from wild type Col0, det1-1 mutant and DET1 OE lines
Supplemental Data. Wu et al. (2). Plant Cell..5/tpc RGLG Hormonal treatment H2O B RGLG µm ABA µm ACC µm GA Time (hours) µm µm MJ µm IA
 Supplemental Data. Wu et al. (2). Plant Cell..5/tpc..4. A B Supplemental Figure. Immunoblot analysis verifies the expression of the AD-PP2C and BD-RGLG proteins in the Y2H assay. Total proteins were extracted
Supplemental Data. Wu et al. (2). Plant Cell..5/tpc..4. A B Supplemental Figure. Immunoblot analysis verifies the expression of the AD-PP2C and BD-RGLG proteins in the Y2H assay. Total proteins were extracted
Supplemental Data. Lee et al. Plant Cell. (2010) /tpc Supplemental Figure 1. Protein and Gene Structures of DWA1 and DWA2.
 Supplemental Figure 1. Protein and Gene Structures of DWA1 and DWA2. (A) Protein structures of DWA1 and DWA2. WD40 region was determined based on the NCBI conserved domain databases (B, C) Schematic representation
Supplemental Figure 1. Protein and Gene Structures of DWA1 and DWA2. (A) Protein structures of DWA1 and DWA2. WD40 region was determined based on the NCBI conserved domain databases (B, C) Schematic representation
Supplemental material
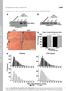 Supplemental material THE JOURNAL OF CELL BIOLOGY Gillespie et al., http://www.jcb.org/cgi/content/full/jcb.200907037/dc1 repressor complex induced by p38- Gillespie et al. Figure S1. Reduced fiber size
Supplemental material THE JOURNAL OF CELL BIOLOGY Gillespie et al., http://www.jcb.org/cgi/content/full/jcb.200907037/dc1 repressor complex induced by p38- Gillespie et al. Figure S1. Reduced fiber size
Supplemental Data. Na Xu et al. (2016). Plant Cell /tpc
 Supplemental Figure 1. The weak fluorescence phenotype is not caused by the mutation in At3g60240. (A) A mutation mapped to the gene At3g60240. Map-based cloning strategy was used to map the mutated site
Supplemental Figure 1. The weak fluorescence phenotype is not caused by the mutation in At3g60240. (A) A mutation mapped to the gene At3g60240. Map-based cloning strategy was used to map the mutated site
AD BD TOC1. Supplementary Figure 1: Yeast two-hybrid assays showing the interaction between
 AD X BD TOC1 AD BD X PIFΔAD PIF TOC1 TOC1 PIFΔAD PIF N TOC1 TOC1 C1 PIFΔAD PIF C1 TOC1 TOC1 C PIFΔAD PIF C TOC1 Supplementary Figure 1: Yeast two-hybrid assays showing the interaction between PIF and TOC1
AD X BD TOC1 AD BD X PIFΔAD PIF TOC1 TOC1 PIFΔAD PIF N TOC1 TOC1 C1 PIFΔAD PIF C1 TOC1 TOC1 C PIFΔAD PIF C TOC1 Supplementary Figure 1: Yeast two-hybrid assays showing the interaction between PIF and TOC1
Supplemental Figure 1. Conserved regions of the kinase domain of PEPR1, PEPR2, CLV1 and BRI1.
 Supplemental Figure 1. Conserved regions of the kinase domain of PEPR1, PEPR2, CLV1 and BRI1. The asterisks and colons indicate the important residues for ATP-binding pocket and substrate binding pocket,
Supplemental Figure 1. Conserved regions of the kinase domain of PEPR1, PEPR2, CLV1 and BRI1. The asterisks and colons indicate the important residues for ATP-binding pocket and substrate binding pocket,
pgbkt7 Anti- Myc AH109 strain (KDa) 50
 pgbkt7 (KDa) 50 37 Anti- Myc AH109 strain Supplementary Figure 1. Protein expression of CRN and TDR in yeast. To analyse the protein expression of CRNKD and TDRKD, total proteins extracted from yeast culture
pgbkt7 (KDa) 50 37 Anti- Myc AH109 strain Supplementary Figure 1. Protein expression of CRN and TDR in yeast. To analyse the protein expression of CRNKD and TDRKD, total proteins extracted from yeast culture
Supplemental Data. Steiner et al. Plant Cell. (2012) /tpc
 Supplemental Figure 1. SPY does not interact with free GST. Invitro pull-down assay using E. coli-expressed MBP-SPY and GST, GST-TCP14 and GST-TCP15. MBP-SPY was used as bait and incubated with equal amount
Supplemental Figure 1. SPY does not interact with free GST. Invitro pull-down assay using E. coli-expressed MBP-SPY and GST, GST-TCP14 and GST-TCP15. MBP-SPY was used as bait and incubated with equal amount
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified
 Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Supplemental Data. Guo et al. (2015). Plant Cell /tpc
 Supplemental Figure 1. The Mutant exb1-d Displayed Pleiotropic Phenotypes and Produced Branches in the Axils of Cotyledons. (A) Branches were developed in exb1-d but not in wild-type plants. (B) and (C)
Supplemental Figure 1. The Mutant exb1-d Displayed Pleiotropic Phenotypes and Produced Branches in the Axils of Cotyledons. (A) Branches were developed in exb1-d but not in wild-type plants. (B) and (C)
Supplemental Figure 1. Mutation in NLA Causes Increased Pi Uptake Activity and
 Supplemental Figure 1. Mutation in NLA Causes Increased Pi Uptake Activity and PHT1 Protein Amounts. (A) Shoot morphology of 19-day-old nla mutants under Pi-sufficient conditions. (B) [ 33 P]Pi uptake
Supplemental Figure 1. Mutation in NLA Causes Increased Pi Uptake Activity and PHT1 Protein Amounts. (A) Shoot morphology of 19-day-old nla mutants under Pi-sufficient conditions. (B) [ 33 P]Pi uptake
At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in
 Supplementary Materials and Methods Barrier function assays At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in acidic X-gal mix (100 mm phosphate buffer at ph4.3, 3 mm
Supplementary Materials and Methods Barrier function assays At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in acidic X-gal mix (100 mm phosphate buffer at ph4.3, 3 mm
supplementary information
 DOI: 10.1038/ncb2116 Figure S1 CDK phosphorylation of EZH2 in cells. (a) Comparison of candidate CDK phosphorylation sites on EZH2 with known CDK substrates by multiple sequence alignments. (b) CDK1 and
DOI: 10.1038/ncb2116 Figure S1 CDK phosphorylation of EZH2 in cells. (a) Comparison of candidate CDK phosphorylation sites on EZH2 with known CDK substrates by multiple sequence alignments. (b) CDK1 and
Supplemental Information. Reciprocal Regulation of the TOR Kinase and ABA. Receptor Balances Plant Growth and Stress Response
 Molecular Cell, Volume 69 Supplemental Information Reciprocal Regulation of the TOR Kinase and ABA Receptor Balances Plant Growth and Stress Response Pengcheng Wang, Yang Zhao, Zhongpeng Li, Chuan-Chih
Molecular Cell, Volume 69 Supplemental Information Reciprocal Regulation of the TOR Kinase and ABA Receptor Balances Plant Growth and Stress Response Pengcheng Wang, Yang Zhao, Zhongpeng Li, Chuan-Chih
Chemical hijacking of auxin signaling with an engineered auxin-tir1
 1 SUPPLEMENTARY INFORMATION Chemical hijacking of auxin signaling with an engineered auxin-tir1 pair Naoyuki Uchida 1,2*, Koji Takahashi 2*, Rie Iwasaki 1, Ryotaro Yamada 2, Masahiko Yoshimura 2, Takaho
1 SUPPLEMENTARY INFORMATION Chemical hijacking of auxin signaling with an engineered auxin-tir1 pair Naoyuki Uchida 1,2*, Koji Takahashi 2*, Rie Iwasaki 1, Ryotaro Yamada 2, Masahiko Yoshimura 2, Takaho
Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product.
 Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF
 YFP-PHF1 CFP-PHT1;2 PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF + CFP-PHT1;2 Negative control!-gfp Supplemental Figure 1: PHT1;2 accumulation is PHF1 dependent. Immunoblot analysis on total protein extract
YFP-PHF1 CFP-PHT1;2 PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF + CFP-PHT1;2 Negative control!-gfp Supplemental Figure 1: PHT1;2 accumulation is PHF1 dependent. Immunoblot analysis on total protein extract
Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity.
 Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity. (A) Amino acid alignment of HDA5, HDA15 and HDA18. The blue line
Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity. (A) Amino acid alignment of HDA5, HDA15 and HDA18. The blue line
Supplemental Figure 1. Transcript profiles of Arabidopsis IPMS1 and IPMS2 in different tissues and developmental stages.
 Transcript level (intensity) Supplemental Data. Xing et al. Plant Cell (2017) 10.1105/tpc.17.00186 1600 1400 1200 1000 800 600 400 200 0 IPMS1 IPMS2 Supplemental Figure 1. Transcript profiles of Arabidopsis
Transcript level (intensity) Supplemental Data. Xing et al. Plant Cell (2017) 10.1105/tpc.17.00186 1600 1400 1200 1000 800 600 400 200 0 IPMS1 IPMS2 Supplemental Figure 1. Transcript profiles of Arabidopsis
Supplemental Data. Liu et al. (2013). Plant Cell /tpc
 Supplemental Figure 1. The GFP Tag Does Not Disturb the Physiological Functions of WDL3. (A) RT-PCR analysis of WDL3 expression in wild-type, WDL3-GFP, and WDL3 (without the GFP tag) transgenic seedlings.
Supplemental Figure 1. The GFP Tag Does Not Disturb the Physiological Functions of WDL3. (A) RT-PCR analysis of WDL3 expression in wild-type, WDL3-GFP, and WDL3 (without the GFP tag) transgenic seedlings.
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb54 sensitivity (+/-) 3 det- WS bri-5 BL (nm) bzr-dbri- bzr-d/ bri- g h i a b c d e f Hypcocotyl length(cm) 5 5 PAC Hypocotyl length (cm)..8..4. 5 5 PPZ - - + + + + - + - + - + PPZ - - + + +
DOI:.38/ncb54 sensitivity (+/-) 3 det- WS bri-5 BL (nm) bzr-dbri- bzr-d/ bri- g h i a b c d e f Hypcocotyl length(cm) 5 5 PAC Hypocotyl length (cm)..8..4. 5 5 PPZ - - + + + + - + - + - + PPZ - - + + +
A Nucleus-Encoded Chloroplast Protein YL1 Is Involved in Chloroplast. Development and Efficient Biogenesis of Chloroplast ATP Synthase in Rice
 A Nucleus-Encoded Chloroplast Protein YL1 Is Involved in Chloroplast Development and Efficient Biogenesis of Chloroplast ATP Synthase in Rice Fei Chen 1,*, Guojun Dong 2,*, Limin Wu 1, Fang Wang 3, Xingzheng
A Nucleus-Encoded Chloroplast Protein YL1 Is Involved in Chloroplast Development and Efficient Biogenesis of Chloroplast ATP Synthase in Rice Fei Chen 1,*, Guojun Dong 2,*, Limin Wu 1, Fang Wang 3, Xingzheng
Figure S1. USP-46 is expressed in several tissues including the nervous system
 Supplemental Figure legends Figure S1. USP-46 is expressed in several tissues including the nervous system Transgenic animals expressing a transcriptional reporter (P::GFP) were imaged using epifluorescence
Supplemental Figure legends Figure S1. USP-46 is expressed in several tissues including the nervous system Transgenic animals expressing a transcriptional reporter (P::GFP) were imaged using epifluorescence
- 1 - Supplemental Data
 - 1-1 Supplemental Data 2 3 4 5 6 7 8 9 Supplemental Figure S1. Differential expression of AtPIP Genes in DC3000-inoculated plants. Gene expression in leaves was analyzed by real-time RT-PCR and expression
- 1-1 Supplemental Data 2 3 4 5 6 7 8 9 Supplemental Figure S1. Differential expression of AtPIP Genes in DC3000-inoculated plants. Gene expression in leaves was analyzed by real-time RT-PCR and expression
Supplementary Fig. 1 Identification of Nedd4 as an IRS-2-associated protein in camp-treated FRTL-5 cells.
 Supplementary Fig. 1 Supplementary Fig. 1 Identification of Nedd4 as an IRS-2-associated protein in camp-treated FRTL-5 cells. (a) FRTL-5 cells were treated with 1 mm dibutyryl camp for 24 h, and the lysates
Supplementary Fig. 1 Supplementary Fig. 1 Identification of Nedd4 as an IRS-2-associated protein in camp-treated FRTL-5 cells. (a) FRTL-5 cells were treated with 1 mm dibutyryl camp for 24 h, and the lysates
(phosphatase tensin) domain is shown in dark gray, the FH1 domain in black, and the
 Supplemental Figure 1. Predicted Domain Organization of the AFH14 Protein. (A) Schematic representation of the predicted domain organization of AFH14. The PTEN (phosphatase tensin) domain is shown in dark
Supplemental Figure 1. Predicted Domain Organization of the AFH14 Protein. (A) Schematic representation of the predicted domain organization of AFH14. The PTEN (phosphatase tensin) domain is shown in dark
pmyrsaur78 psosein4+ psosetr1+ psosetr2+
 SAUR77 SAUR78 SAUR76 Figure S1 Phylogenetic analysis of Arabidopsis SAUR proteins. The analyses were conducted in MEGA5, using the Neighbor-Joining method. The percentage of replicate trees in which the
SAUR77 SAUR78 SAUR76 Figure S1 Phylogenetic analysis of Arabidopsis SAUR proteins. The analyses were conducted in MEGA5, using the Neighbor-Joining method. The percentage of replicate trees in which the
Supporting Information
 Supporting Information Deng et al. 10.1073/pnas.1102117108 Fig. S1. Predicted structure of Arabidopsis bzip60 RNA. Lowest free energy form (ΔG = 309.72 (initially 343.10) of bzip60 mrna folded by M-Fold
Supporting Information Deng et al. 10.1073/pnas.1102117108 Fig. S1. Predicted structure of Arabidopsis bzip60 RNA. Lowest free energy form (ΔG = 309.72 (initially 343.10) of bzip60 mrna folded by M-Fold
Nature Genetics: doi: /ng Supplementary Figure 1. ChIP-seq genome browser views of BRM occupancy at previously identified BRM targets.
 Supplementary Figure 1 ChIP-seq genome browser views of BRM occupancy at previously identified BRM targets. Gene structures are shown underneath each panel. Supplementary Figure 2 pref6::ref6-gfp complements
Supplementary Figure 1 ChIP-seq genome browser views of BRM occupancy at previously identified BRM targets. Gene structures are shown underneath each panel. Supplementary Figure 2 pref6::ref6-gfp complements
Figure S1. Immunoblotting showing proper expression of tagged R and AVR proteins in N. benthamiana.
 SUPPLEMENTARY FIGURES Figure S1. Immunoblotting showing proper expression of tagged R and AVR proteins in N. benthamiana. YFP:, :CFP, HA: and (A) and HA, CFP and YFPtagged and AVR1-CO39 (B) were expressed
SUPPLEMENTARY FIGURES Figure S1. Immunoblotting showing proper expression of tagged R and AVR proteins in N. benthamiana. YFP:, :CFP, HA: and (A) and HA, CFP and YFPtagged and AVR1-CO39 (B) were expressed
This is the author's accepted version of the manuscript.
 This is the author's accepted version of the manuscript. The definitive version is published in Nature Communications Online Edition: 2015/4/16 (Japan time), doi:10.1038/ncomms7780. The final version published
This is the author's accepted version of the manuscript. The definitive version is published in Nature Communications Online Edition: 2015/4/16 (Japan time), doi:10.1038/ncomms7780. The final version published
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
Supplemental Data. Sun et al. Plant Cell. (2012) /tpc FLS2 -cmyc -GFP FLS2 -HA. FLS2-FLAG FLS2 -HA BAK1-HA flg22 (10mM)
 Supplemental Data. Sun et al. Plant ell. ()..5/tpc..9599 A FLS-FLA FLS -HA BAK-HA flg (mm) - B FLS -cmyc -FP FLS -HA IP antibody cmyc FLA FP cmyc IP ; WB: α-ha IP; WB: α-ha IP: α-fla; WB: α-ha IP ; WB:
Supplemental Data. Sun et al. Plant ell. ()..5/tpc..9599 A FLS-FLA FLS -HA BAK-HA flg (mm) - B FLS -cmyc -FP FLS -HA IP antibody cmyc FLA FP cmyc IP ; WB: α-ha IP; WB: α-ha IP: α-fla; WB: α-ha IP ; WB:
Supplemental Materials
 Supplemental Materials Supplemental Figure S. Phenotypic assessment of alb4 mutant plants under different stress conditions. (A) High-light stress and drought stress. Wild-type (WT) and alb4 mutant plants
Supplemental Materials Supplemental Figure S. Phenotypic assessment of alb4 mutant plants under different stress conditions. (A) High-light stress and drought stress. Wild-type (WT) and alb4 mutant plants
SUPPLEMENTARY INFORMATION
 Gene replacements and insertions in rice by intron targeting using CRISPR Cas9 Table of Contents Supplementary Figure 1. sgrna-induced targeted mutations in the OsEPSPS gene in rice protoplasts. Supplementary
Gene replacements and insertions in rice by intron targeting using CRISPR Cas9 Table of Contents Supplementary Figure 1. sgrna-induced targeted mutations in the OsEPSPS gene in rice protoplasts. Supplementary
Supplemental Data. Jing et al. (2013). Plant Cell /tpc
 Supplemental Figure 1. Characterization of epp1 Mutants. (A) Cotyledon angles of 5-d-old Col wild-type (gray bars) and epp1-1 (black bars) seedlings under red (R), far-red (FR) and blue (BL) light conditions,
Supplemental Figure 1. Characterization of epp1 Mutants. (A) Cotyledon angles of 5-d-old Col wild-type (gray bars) and epp1-1 (black bars) seedlings under red (R), far-red (FR) and blue (BL) light conditions,
Supplementary methods Shoc2 In Vitro Ubiquitination Assay
 Supplementary methods Shoc2 In Vitro Ubiquitination Assay 35 S-labelled Shoc2 was prepared using a TNT quick Coupled transcription/ translation System (Promega) as recommended by manufacturer. For the
Supplementary methods Shoc2 In Vitro Ubiquitination Assay 35 S-labelled Shoc2 was prepared using a TNT quick Coupled transcription/ translation System (Promega) as recommended by manufacturer. For the
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
Nature Structural & Molecular Biology: doi: /nsmb.3308
 Supplementary Figure 1 Analysis of CED-3 autoactivation and CED-4-induced CED-3 activation. (a) Repeat experiments of Fig. 1a. (b) Repeat experiments of Fig. 1b. (c) Quantitative analysis of three independent
Supplementary Figure 1 Analysis of CED-3 autoactivation and CED-4-induced CED-3 activation. (a) Repeat experiments of Fig. 1a. (b) Repeat experiments of Fig. 1b. (c) Quantitative analysis of three independent
 Supplemental Table 1. The progeny of crosses between MPK4/mpk4-2 and ANQ/anq-2. Genotype Number Number Number observed expected-1 expected-2 (1) MPK4/MPK4 ANQ/ANQ 32 12 21 (2) MPK4/MPK4 ANQ/anq-2 28 25
Supplemental Table 1. The progeny of crosses between MPK4/mpk4-2 and ANQ/anq-2. Genotype Number Number Number observed expected-1 expected-2 (1) MPK4/MPK4 ANQ/ANQ 32 12 21 (2) MPK4/MPK4 ANQ/anq-2 28 25
Supplementary Figure 1 qrt-pcr expression analysis of NLP8 with and without KNO 3 during germination.
 Supplementary Figure 1 qrt-pcr expression analysis of NLP8 with and without KNO 3 during germination. Seeds of Col-0 were harvested from plants grown at 16 C, stored for 2 months, imbibed for indicated
Supplementary Figure 1 qrt-pcr expression analysis of NLP8 with and without KNO 3 during germination. Seeds of Col-0 were harvested from plants grown at 16 C, stored for 2 months, imbibed for indicated
Supplementary Figure S1 Purification of deubiquitinases HEK293 cells were transfected with the indicated DUB-expressing plasmids.
 Supplementary Figure S1 Purification of deubiquitinases HEK293 cells were transfected with the indicated DUB-expressing plasmids. The cells were harvested 72 h after transfection. FLAG-tagged deubiquitinases
Supplementary Figure S1 Purification of deubiquitinases HEK293 cells were transfected with the indicated DUB-expressing plasmids. The cells were harvested 72 h after transfection. FLAG-tagged deubiquitinases
Supporting Information
 Supporting Information Üstün and Börnke, 2015 Figure S1: XopJ does not display acetyltransferase activity. Autoacetylation activity in vitro. Acetylation reactions using MBP, MBP-XopJ, GST and GST-HopZ1a
Supporting Information Üstün and Börnke, 2015 Figure S1: XopJ does not display acetyltransferase activity. Autoacetylation activity in vitro. Acetylation reactions using MBP, MBP-XopJ, GST and GST-HopZ1a
Supplementary information
 Supplementary information The E3 ligase RNF8 regulates KU80 removal and NHEJ repair Lin Feng 1, Junjie Chen 1 1 Department of Experimental Radiation Oncology, The University of Texas M. D. Anderson Cancer
Supplementary information The E3 ligase RNF8 regulates KU80 removal and NHEJ repair Lin Feng 1, Junjie Chen 1 1 Department of Experimental Radiation Oncology, The University of Texas M. D. Anderson Cancer
Supplementary data. sienigma. F-Enigma F-EnigmaSM. a-p53
 Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Supplemental Figure 1 Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical
 Supplemental Figure Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical in all six REEPs are highlighted in green. Additional
Supplemental Figure Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical in all six REEPs are highlighted in green. Additional
Supplemental Data. Wang et al. (2017). Plant Cell /tpc NIP1;2 NIP7;1
 Supplemental Data. Wang et al. (217). Plant Cell 1.115/tpc.16.825 NIP1;1 NIP1;2 NIP2;1 NIP3;1 NIP4;1 NIP5;1 NIP6;1 NIP7;1 Supplemental Figure 1. Distinct Localization of NIPs in Root Epidermal Cells. (Supports
Supplemental Data. Wang et al. (217). Plant Cell 1.115/tpc.16.825 NIP1;1 NIP1;2 NIP2;1 NIP3;1 NIP4;1 NIP5;1 NIP6;1 NIP7;1 Supplemental Figure 1. Distinct Localization of NIPs in Root Epidermal Cells. (Supports
Supplemental Data. Dai et al. (2013). Plant Cell /tpc Absolute FyPP3. Absolute
 A FyPP1 Absolute B FyPP3 Absolute Dry seeds Imbibed 24 hours Dry seeds Imbibed 24 hours C ABI5 Absolute Dry seeds Imbibed 24 hours Supplemental Figure 1. Expression of FyPP1, FyPP3 and ABI5 during seed
A FyPP1 Absolute B FyPP3 Absolute Dry seeds Imbibed 24 hours Dry seeds Imbibed 24 hours C ABI5 Absolute Dry seeds Imbibed 24 hours Supplemental Figure 1. Expression of FyPP1, FyPP3 and ABI5 during seed
Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and
 Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
Supplemental Data. Shimoda et al. (2011). Plant Cell /tpc B C D E
 (kda) 80 58 46 30 25 B C D E (0/15) (29/29) (0/29) (0/32) Supplemental Figure 1. Immunodetection of Myc-tagged CCaMK Protein Variants on ccamk-3 Roots. () Immunodetection of myc-tag fusion CCaMK WT, 1-340
(kda) 80 58 46 30 25 B C D E (0/15) (29/29) (0/29) (0/32) Supplemental Figure 1. Immunodetection of Myc-tagged CCaMK Protein Variants on ccamk-3 Roots. () Immunodetection of myc-tag fusion CCaMK WT, 1-340
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11070 Supplementary Figure 1 Purification of FLAG-tagged proteins. a, Purification of FLAG-RNF12 by FLAG-affinity from nuclear extracts of wild-type (WT) and two FLAG- RNF12 transgenic
doi:10.1038/nature11070 Supplementary Figure 1 Purification of FLAG-tagged proteins. a, Purification of FLAG-RNF12 by FLAG-affinity from nuclear extracts of wild-type (WT) and two FLAG- RNF12 transgenic
Supplemental Data. Seo et al. (2014). Plant Cell /tpc
 Supplemental Figure 1. Protein alignment of ABD1 from other model organisms. The alignment was performed with H. sapiens DCAF8, M. musculus DCAF8 and O. sativa Os10g0544500. The WD40 domains are underlined.
Supplemental Figure 1. Protein alignment of ABD1 from other model organisms. The alignment was performed with H. sapiens DCAF8, M. musculus DCAF8 and O. sativa Os10g0544500. The WD40 domains are underlined.
Fig. S1. CrgA intracellular levels in M. tuberculosis. Ten and twenty micrograms of
 Supplementary data Fig. S1. CrgA intracellular levels in M. tuberculosis. Ten and twenty micrograms of cell free protein lysates from WT M. tuberculosis (Rv) together with various known concentrations
Supplementary data Fig. S1. CrgA intracellular levels in M. tuberculosis. Ten and twenty micrograms of cell free protein lysates from WT M. tuberculosis (Rv) together with various known concentrations
AD-FIL AD-YAB2 -2 BD-JAZ3 AD-YAB3 AD-YAB5 AD-FIL -4 3AT BD-JAZ3 AD-YAB3
 3 4 9 10 11 12 AD-FIL AD-YAB5 2 B AD-YAB3 1 BD-JAZ 5 6 7 8 AD-FIL A AD-YAB2 Supplemental Data. Boter et al. (2015). Plant Cell 10.1105/tpc.15.00220 BD AD-YAB2-2 -2 BD-JAZ3 AD-YAB3 BD AD-YAB5-4 AD-FIL -4
3 4 9 10 11 12 AD-FIL AD-YAB5 2 B AD-YAB3 1 BD-JAZ 5 6 7 8 AD-FIL A AD-YAB2 Supplemental Data. Boter et al. (2015). Plant Cell 10.1105/tpc.15.00220 BD AD-YAB2-2 -2 BD-JAZ3 AD-YAB3 BD AD-YAB5-4 AD-FIL -4
Supplementary Figure S1. N-terminal fragments of LRRK1 bind to Grb2.
 Myc- HA-Grb2 Mr(K) 105 IP HA 75 25 105 1-1163 1-595 - + - + - + 1164-1989 Blot Myc HA total lysate 75 25 Myc HA Supplementary Figure S1. N-terminal fragments of bind to Grb2. COS7 cells were cotransfected
Myc- HA-Grb2 Mr(K) 105 IP HA 75 25 105 1-1163 1-595 - + - + - + 1164-1989 Blot Myc HA total lysate 75 25 Myc HA Supplementary Figure S1. N-terminal fragments of bind to Grb2. COS7 cells were cotransfected
Figure S1. Verification of ihog Mutation by Protein Immunoblotting Figure S2. Verification of ihog and boi
 Figure S1. Verification of ihog Mutation by Protein Immunoblotting Extracts from S2R+ cells, embryos, and adults were analyzed by immunoprecipitation and immunoblotting with anti-ihog antibody. The Ihog
Figure S1. Verification of ihog Mutation by Protein Immunoblotting Extracts from S2R+ cells, embryos, and adults were analyzed by immunoprecipitation and immunoblotting with anti-ihog antibody. The Ihog
GTTCGGGTTCC TTTTGAGCAG
 Supplementary Figures Splice variants of the SIP1 transcripts play a role in nodule organogenesis in Lotus japonicus. Wang C, Zhu H, Jin L, Chen T, Wang L, Kang H, Hong Z, Zhang Z. 5 UTR CDS 3 UTR TCTCAACCATCCTTTGTCTGCTTCCGCCGCATGGGTGAGGTCATTTTGTCTAGATGACGTGCAATTTACAATGA
Supplementary Figures Splice variants of the SIP1 transcripts play a role in nodule organogenesis in Lotus japonicus. Wang C, Zhu H, Jin L, Chen T, Wang L, Kang H, Hong Z, Zhang Z. 5 UTR CDS 3 UTR TCTCAACCATCCTTTGTCTGCTTCCGCCGCATGGGTGAGGTCATTTTGTCTAGATGACGTGCAATTTACAATGA
Nature Structural & Molecular Biology: doi: /nsmb.1583
 Acetylation by GCN5 regulates CDC6 phosphorylation in the S-phase of the cell cycle Roberta Paolinelli 1,2, Ramiro Mendoza-Maldonado 2, Anna Cereseto 1 and Mauro Giacca 2 1 Molecular Biology Laboratory,
Acetylation by GCN5 regulates CDC6 phosphorylation in the S-phase of the cell cycle Roberta Paolinelli 1,2, Ramiro Mendoza-Maldonado 2, Anna Cereseto 1 and Mauro Giacca 2 1 Molecular Biology Laboratory,
Supplemental Data. Fan et al. (2014). Plant Cell /tpc
 Supplemental Data. Fan et al. (). Plant Cell./tpc.. Cell wall EXP EXP EXP9 HLH/bHLH AIF HFR HBI PAR ABAR Atg778 At3g39 Atg38 Atg3 EXP EXP6 EXP8 Photosynthesis PSAD- PSAD- PSBY PSBO- PSAN LHCB6 PSBS LHCB
Supplemental Data. Fan et al. (). Plant Cell./tpc.. Cell wall EXP EXP EXP9 HLH/bHLH AIF HFR HBI PAR ABAR Atg778 At3g39 Atg38 Atg3 EXP EXP6 EXP8 Photosynthesis PSAD- PSAD- PSBY PSBO- PSAN LHCB6 PSBS LHCB
Supplementary Information: Materials and Methods. GST and GST-p53 were purified according to standard protocol after
 Supplementary Information: Materials and Methods Recombinant protein expression and in vitro kinase assay. GST and GST-p53 were purified according to standard protocol after induction with.5mm IPTG for
Supplementary Information: Materials and Methods Recombinant protein expression and in vitro kinase assay. GST and GST-p53 were purified according to standard protocol after induction with.5mm IPTG for
PIE1 ARP6 SWC6 KU70 ARP6 PIE1. HSA SNF2_N HELICc SANT. pie1-3 A1,A2 K1,K2 K1,K3 K3,LB2 A3, A4 A3,LB1 A1,A2 K1,K2 K1,K3. swc6-1 A3,A4.
 A B N-terminal SWC2 H2A.Z SWC6 ARP6 PIE1 HSA SNF2_N HELICc SANT C pie1-3 D PIE1 ARP6 5 Kb A1 200 bp A3 A2 LB1 arp6-3 A4 E A1,A2 A3, A4 A3,LB1 K1,K2 K1,K3 K3,LB2 SWC6 swc6-1 A1,A2 A3,A4 K1,K2 K1,K3 100
A B N-terminal SWC2 H2A.Z SWC6 ARP6 PIE1 HSA SNF2_N HELICc SANT C pie1-3 D PIE1 ARP6 5 Kb A1 200 bp A3 A2 LB1 arp6-3 A4 E A1,A2 A3, A4 A3,LB1 K1,K2 K1,K3 K3,LB2 SWC6 swc6-1 A1,A2 A3,A4 K1,K2 K1,K3 100
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Nakajima and Tanoue, http://www.jcb.org/cgi/content/full/jcb.201104118/dc1 Figure S1. DLD-1 cells exhibit the characteristic morphology
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Nakajima and Tanoue, http://www.jcb.org/cgi/content/full/jcb.201104118/dc1 Figure S1. DLD-1 cells exhibit the characteristic morphology
SUPPLEMENTARY INFORMATION FIGURE LEGENDS
 SUPPLEMENTARY INFORMATION FIGURE LEGENDS Fig. S1. Radiation-induced phosphorylation of Rad50 at a specific site. A. Rad50 is an in vitro substrate for ATM. A series of Rad50-GSTs covering the entire molecule
SUPPLEMENTARY INFORMATION FIGURE LEGENDS Fig. S1. Radiation-induced phosphorylation of Rad50 at a specific site. A. Rad50 is an in vitro substrate for ATM. A series of Rad50-GSTs covering the entire molecule
Supplementary Figure 1. α-synuclein is truncated in PD and LBD brains. Nature Structural & Molecular Biology: doi: /nsmb.
 Supplementary Figure 1 α-synuclein is truncated in PD and LBD brains. (a) Specificity of anti-n103 antibody. Anti-N103 antibody was coated on an ELISA plate and different concentrations of full-length
Supplementary Figure 1 α-synuclein is truncated in PD and LBD brains. (a) Specificity of anti-n103 antibody. Anti-N103 antibody was coated on an ELISA plate and different concentrations of full-length
7.012 Problem Set 5. Question 1
 Name Section 7.012 Problem Set 5 Question 1 While studying the problem of infertility, you attempt to isolate a hypothetical rabbit gene that accounts for the prolific reproduction of rabbits. After much
Name Section 7.012 Problem Set 5 Question 1 While studying the problem of infertility, you attempt to isolate a hypothetical rabbit gene that accounts for the prolific reproduction of rabbits. After much
Supplemental Data. Wang et al. Plant Cell. (2013) /tpc
 SNL1 SNL Supplemental Figure 1. The Expression Patterns of SNL1 and SNL in Different Tissues of Arabidopsis from Genevestigator Web Site (https://www.genevestigator.com/gv/index.jsp). 1 A 1. 1..8.6.4..
SNL1 SNL Supplemental Figure 1. The Expression Patterns of SNL1 and SNL in Different Tissues of Arabidopsis from Genevestigator Web Site (https://www.genevestigator.com/gv/index.jsp). 1 A 1. 1..8.6.4..
Supplemental Figure 1. Phos-Tag mobility shift detection of in vivo phosphorylated ERF6 in 35S:ERF6 WT plants after B. cinerea inoculation.
 Supplemental Figure 1. Phos-Tag mobility shift detection of in vivo phosphorylated ERF6 in 35S:ERF6 WT plants after B. cinerea inoculation. Protein extracts from 35S:ERF6 WT seedlings treated with B. cinerea
Supplemental Figure 1. Phos-Tag mobility shift detection of in vivo phosphorylated ERF6 in 35S:ERF6 WT plants after B. cinerea inoculation. Protein extracts from 35S:ERF6 WT seedlings treated with B. cinerea
Supplementary Figure1: ClustalW comparison between Tll, Dsf and NR2E1.
 P-Box Dsf -----------------MG-TAG--DRLLD-IPCKVCGDRSSGKHYGIYSCDGCSGFFKR 39 NR2E1 -----------------MSKPAGSTSRILD-IPCKVCGDRSSGKHYGVYACDGCSGFFKR 42 Tll MQSSEGSPDMMDQKYNSVRLSPAASSRILYHVPCKVCRDHSSGKHYGIYACDGCAGFFKR
P-Box Dsf -----------------MG-TAG--DRLLD-IPCKVCGDRSSGKHYGIYSCDGCSGFFKR 39 NR2E1 -----------------MSKPAGSTSRILD-IPCKVCGDRSSGKHYGVYACDGCSGFFKR 42 Tll MQSSEGSPDMMDQKYNSVRLSPAASSRILYHVPCKVCRDHSSGKHYGIYACDGCAGFFKR
Supplementary Figure 1
 Supplementary Figure 1 PTEN promotes virus-induced expression of IFNB1 and its downstream genes. (a) Quantitative RT-PCR analysis of IFNB1 mrna (left) and ELISA of IFN-β (right) in HEK 293 cells (2 10
Supplementary Figure 1 PTEN promotes virus-induced expression of IFNB1 and its downstream genes. (a) Quantitative RT-PCR analysis of IFNB1 mrna (left) and ELISA of IFN-β (right) in HEK 293 cells (2 10
Supplementary Figure 1. Nature Structural & Molecular Biology: doi: /nsmb.3494
 Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Figure S1, related to Figure 1. Characterization of biosensor behavior in vivo.
 Developmental Cell, Volume 23 Supplemental Information Separase Biosensor Reveals that Cohesin Cleavage Timing Depends on Phosphatase PP2A Cdc55 Regulation Gilad Yaakov, Kurt Thorn, and David O. Morgan
Developmental Cell, Volume 23 Supplemental Information Separase Biosensor Reveals that Cohesin Cleavage Timing Depends on Phosphatase PP2A Cdc55 Regulation Gilad Yaakov, Kurt Thorn, and David O. Morgan
A Repressor Complex Governs the Integration of
 Developmental Cell 15 Supplemental Data A Repressor Complex Governs the Integration of Flowering Signals in Arabidopsis Dan Li, Chang Liu, Lisha Shen, Yang Wu, Hongyan Chen, Masumi Robertson, Chris A.
Developmental Cell 15 Supplemental Data A Repressor Complex Governs the Integration of Flowering Signals in Arabidopsis Dan Li, Chang Liu, Lisha Shen, Yang Wu, Hongyan Chen, Masumi Robertson, Chris A.
Hossain_Supplemental Figure 1
 Hossain_Supplemental Figure 1 GFP-PACT GFP-PACT Motif I GFP-PACT Motif II A. MG132 (1µM) GFP Tubulin GFP-PACT Pericentrin GFP-PACT GFP-PACT Pericentrin Fig. S1. Expression and localization of Orc1 PACT
Hossain_Supplemental Figure 1 GFP-PACT GFP-PACT Motif I GFP-PACT Motif II A. MG132 (1µM) GFP Tubulin GFP-PACT Pericentrin GFP-PACT GFP-PACT Pericentrin Fig. S1. Expression and localization of Orc1 PACT
Supplemental Data. Tang et al. Plant Cell. (2012) /tpc
 Supplemental Figure 1. Relative Pchlide Fluorescence of Various Mutants and Wild Type. Seedlings were grown in darkness for 5 d. Experiments were repeated 3 times with same results. Supplemental Figure
Supplemental Figure 1. Relative Pchlide Fluorescence of Various Mutants and Wild Type. Seedlings were grown in darkness for 5 d. Experiments were repeated 3 times with same results. Supplemental Figure
Supplemental Figure 1.
 Supplemental Data. Charron et al. Dynamic landscapes of four histone modifications during de-etiolation in Arabidopsis. Plant Cell (2009). 10.1105/tpc.109.066845 Supplemental Figure 1. Immunodetection
Supplemental Data. Charron et al. Dynamic landscapes of four histone modifications during de-etiolation in Arabidopsis. Plant Cell (2009). 10.1105/tpc.109.066845 Supplemental Figure 1. Immunodetection
Supplementary Figure 1. BES1 specifically inhibits ABA responses in early seedling
 Supplementary Figure 1. BES1 specifically inhibits ABA responses in early seedling development. a. Exogenous BR application overcomes the hypersensitivity of bzr1-1d seedlings to ABA. Seed germination
Supplementary Figure 1. BES1 specifically inhibits ABA responses in early seedling development. a. Exogenous BR application overcomes the hypersensitivity of bzr1-1d seedlings to ABA. Seed germination
Design. Construction. Characterization
 Design Construction Characterization DNA mrna (messenger) A C C transcription translation C A C protein His A T G C T A C G Plasmids replicon copy number incompatibility selection marker origin of replication
Design Construction Characterization DNA mrna (messenger) A C C transcription translation C A C protein His A T G C T A C G Plasmids replicon copy number incompatibility selection marker origin of replication
