Figure S1. USP-46 is expressed in several tissues including the nervous system
|
|
|
- Polly Young
- 5 years ago
- Views:
Transcription
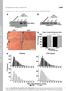
1 Supplemental Figure legends Figure S1. USP-46 is expressed in several tissues including the nervous system Transgenic animals expressing a transcriptional reporter (P::GFP) were imaged using epifluorescence (1) and Nomarski optics (2). P::GFP is expressed in the pharynx (A), in many neurons in the head (A), in the nerve ring (B), in body wall muscle (C), in the intestine (D), and in vulval muscles (E). The asterisks (*) in (D) and (E) mark the position of the vulva. (F-H) USP-46 is expressed in GLR-1 expressing head neurons. Transgenic animals co-expressing a transcriptional reporter (P::GFP)(F), and a glr-1 transcriptional reporter (Pglr-1::dsRED)(G) were imaged. (H) Merged image of (F) and (G). (F2-H2) White dotted lines outline the neuronal cell bodies for clarity. The neuron on the far left expresses (F2) but not glr-1 (G2). Neurons co-expressing and glr-1 are indicated by white asterisks (*) in (H2). Figure S2. Analysis of Multiple Synaptic Markers in mutant animals (A-B) Representative images of MAGI-1::YFP puncta in the anterior ventral nerve cords of wild type (A) and (ok2232)(b) L4 larvae expressing MAGI-1::YFP. The asterisk marks a neuronal cell body. (C) Quantification of MAGI-1::YFP puncta densities (puncta/10 m) for the strains pictured in (A-B). Shown are the means and standard errors (SEM) for n = 24 wild type and n = 24 (ok2232). (D-E) Representative images of LIN-10::GFP puncta in the anterior ventral nerve cords of wild type (D) and (ok2232)(e) L4 larvae expressing LIN-10::GFP. (F) Quantification of LIN-10::GFP puncta densities (puncta/10 m) for the strains pictured in (D-E). Shown are the means and standard errors (SEM) for n = 23 wild type and n = 18 (ok2232). 1
2 (G-H) Representative images of SOL-1::YFP puncta in the anterior ventral nerve cords of wild type (G) and (ok2232)(h) L4 larvae expressing SOL-1::YFP. (I) Quantification of SOL-1::YFP puncta densities (puncta/10 m) for the strains pictured in (G-H). Shown are the means and standard errors (SEM) for n = 13 wild type and n = 16 (ok2232). Figure S3. In vitro binding assay of GST-GLR-1 and USP-46 USP-46 or the last three PDZ domains of MAGI-1 were in vitro translated and labeled with 35 S-methionine. The labeled proteins were incubated with GST, lanes 2 and 5, or GST fused to the C-terminus of GLR-1 (GST-GLR-1C), lanes 3 and 6, and subsequently pulled down with glutathione sepharose beads. Lanes 1 and 4 show 10% of the input of 35 S-labeled protein. Reactions were analyzed by SDS-PAGE and Coomassie staining (lower panel) and fluorography (upper panel). Arrows indicate the position of GST alone or GST-GLR-1C (lower panel). Figure S4. Analysis of ubiquitinated GLR-1 (A) Control immunoprecipitation experiment detecting ubiquitinated GLR-1::GFP from membrane preparations of mixed stage wild type animals expressing GLR-1::GFP (nuis24). Total GLR-1::GFP was first immunoprecipitated using polyclonal anti-gfp antibodies (1 st IP). This was followed by a second, sequential immunoprecipitation of ubiquitin-glr-1 conjugates using polyclonal anti-ubiquitin antibodies (2 nd IP) (top panel, lanes 1 and 2). In this experiment, purified, free ubiquitin monomers (+Ub) or an equal concentration of bovine serum albumin (BSA) (-Ub) were preincubated with the antiubiquitin polyclonal antibodies prior to use in the 2 nd IP step. Excess free ubiquitin or excess BSA was also included in the corresponding 2 nd IP. Western blot analyses were used to detect immunoprecipitated proteins from each IP. Immunoprecipitated GLR- 2
3 1::GFP from the 1 st IP was detected with monoclonal anti-gfp antibodies (lower blot). Ubiquitin-GLR-1::GFP conjugates immunoprecipitated in the 2 nd IP were detected with monoclonal anti-gfp antibodies (upper blot). Note that the 3 rd lane contains Total membranes and indicates that non-ubiquitinated GLR-1::GFP runs below the apparent molecular weight of the Ub-GLR-1::GFP bands. (B) Control Immunoprecipitation experiment detecting ubiquitinated GLR-1::GFP in membranes prepared from mixed stage populations of wild type animals expressing GLR-1::GFP (nuis25). Total GLR-1::GFP was first immunoprecipitated using polyclonal anti-gfp antibodies (1 st IP). This was followed by a second, sequential immunoprecipitation of ubiquitin-glr-1 conjugates using polyclonal anti-ubiquitin antibodies (2 nd IP). Western blot analyses were used to detect immunoprecipitated proteins from each IP. Immunoprecipitated GLR-1::GFP from the 1 st IP and 2 nd IP were detected with monoclonal anti-gfp antibodies (left blot). Ubiquitin-GLR-1::GFP conjugates immunoprecipitated in the 2 nd IP were detected with monoclonal antiubiquitin antibodies (right blot). 120 times more material was loaded for the blots of the 2 nd IPs than for the anti-gfp blot of the 1 st IP. Figure S5. In vitro DUB activity of GST-USP-46 (A) Recombinant UCH-L3 (positive control) or GST-USP-46 were incubated with the inhibitor HA-tagged vinyl methyl ester (HA-Ub-VME) to covalently label active DUB species. Reaction with HA-Ub-VME causes an increase in the apparent molecular weight of the DUB (arrows). Reactions were analyzed by SDS-PAGE followed by either Coomassie staining (left panel) or Western blotting with anti-ha antibodies to detect HAtagged DUB species (right panel). About 50% of UCH-L3 is active and covalently labeled by HA-Ub-VME as detected by the anti-ha Western blot and the mobility shift in the coomaasie stained gel (lower arrows). In contrast, only a small percentage of GST- 3
4 USP-46 is active and covalently modified by HA-Ub-VME (top arrows). The asterisks indicate the apparent molecular weight of unreacted, inactive GST-USP-46. MW (KDa) markers are shown. (B) USP-46 deubiquitination assay. Recombinant GST-USP-46 or catalytically inactive GST-USP-46(C>A) were incubated with GLR-1::GFP isolated from membrane fractions of mixed stage wild type worms expressing GLR-1::GFP in DUB reaction buffer. The relative amounts of ubiquitinated GLR-1::GFP was subsequently determined. Total GLR-1::GFP was first immunoprecipitated using polyclonal anti-gfp antibodies (1 st IP), followed by a second, sequential immunoprecipitation of ubiquitin- GLR-1 conjugates using polyclonal anti-ubiquitin antibodies (2 nd IP). Western blot analyses with monoclonal anti-gfp antibodies were used to detect immunoprecipitated GLR-1::GFP from the 1 st IP (left panel) and Ubiquitin-GLR-1::GFP conjugates immunoprecipitated in the 2 nd IP (right panel). 120 times more material was loaded for the anti-gfp blot of the 2 nd IP than for the anti-gfp blot of the 1 st IP. Numbers listed below the right panel indicate the normalized ratios of the amount of ubiquitinated-glr-1 (right panel) to the amount of total immunoprecipitated GLR-1::GFP (left panel) (Ub- GLR-1/GLR-1). MW (kda) markers are indicated. (C) Graph representing the average ratio of Ub-GLR-1/GLR-1 normalized to GST-USP-46(C>A) treated samples (n=3). SEM shown. Figure S6. Quantification of Puncta Widths for experiments shown in Figures 5 and 6 (A) Quantification of GLR-1::GFP puncta widths for the strains pictured in Figure 5A-D. Shown are the means and standard errors (SEM) for n = 59 wild type, n = 41 (ok2232), n = 20 unc-11 AP180(e47), and n = 24 unc-11 AP180; animals. Values that differ significantly from wild type (Student s t test) are denoted by asterisks (*) above each bar. All significant differences are indicated as follows: * p 0.01, ** p
5 (B) Quantification of GLR-1::GFP puncta widths for the strains pictured in Figure 6A-D. Shown are the means and standard errors (SEM) for n = 26 wild type, n = 19 (ok2232), n = 22 vps-4(dn), and n = 26 vps-4(dn); animals. Values that differ significantly from wild type (Student s t test) are denoted by asterisks (*) above each bar. All significant differences are indicated as follows: # p 0.02, * p 0.01, ** p n.s. denotes no significant difference between the indicated strains (p > 0.05). Figure S7. USP-46 localization in the VNC (A-B) Representative images of mcherry::usp-46 (A) or GLR-1::GFP (B) expressed under the control of the glr-1 promoter in the anterior ventral nerve cords of wild type L4 larvae. (C) Merged image of (A) and (B). Arrowheads indicate mcherry::usp-46 puncta that do not co-localize with GLR-1::GFP punta. Arrows indicate mcherry::usp-46 puncta that exhibit some overlap. Figure S8. USP-46 localizes to an endosomal compartment but not the ER (A) Representative images of the ER marker KDEL::venus (A) and mcherry::usp-46 (B) expressed under the control of the glr-1 promoter in PVC interneuron cell bodies. (C) Merged image of (A) and (B). (D-E) Endo H assay. (D) GLR-1::GFP containing membranes from wild type or (ok2232) mutant animals were either left untreated (lanes 1 and 4), digested with Endo H (lanes 2 and 5) or digested with PNGase F (lanes 3 and 6). Western blot analyses with anti-gfp antibodies were used to detect GLR- 1::GFP. Endo H-sensitive (Endo H-S) and Endo H-resistant (Endo H-R) GLR-1::GFP bands are indicated. (E) Quantitation of the percentage of Endo H-sensitive GLR- 1::GFP averaged from three experiments. Errors bars show standard error of the mean (SEM). (F-G) Representative images of mcherry::usp-46 (F) or Venus::RAB-5 (G) puncta expressed under the control of the glr-1 promoter in the anterior ventral nerve 5
6 cords of wild type L4 larvae. (H) Merged image of (F) and (G). Arrows indicate mcherry::usp-46 puncta that are co-localized with Venus::RAB-5 puncta. (I-J) Representative images of mcherry::usp-46 (I-J) or Venus::RAB-5 (K-L) puncta expressed under the control of the glr-1 promoter in the PVC neuron cell bodies of wild type animals. (M) Merged image of (I) and (K). (N) Merged image of (J) and (L). (O) Analysis of ubiquitinated GLR-1 in animals expressing vps-4(dn). Representative immunoprecipitation experiments showing the relative amounts of ubiquitinated GLR- 1::GFP in membranes prepared from mixed stage populations of wild type or mutant animals expressing GLR-1::GFP and vps-4(dn). Total GLR-1::GFP was first immunoprecipitated using polyclonal anti-gfp antibodies (1 st IP), followed by a second, sequential immunoprecipitation of ubiquitin-glr-1 conjugates using polyclonal antiubiquitin antibodies (2 nd IP). Western blot analyses with monoclonal anti-gfp antibodies were used to detect immunoprecipitated GLR-1::GFP from the 1 st IP (left panel) and Ubiquitin-GLR-1::GFP conjugates immunoprecipitated in the 2 nd IP (right panel). 120 times more material was loaded for the anti-gfp blot of the 2 nd IP than for the anti-gfp blot of the 1 st IP. Numbers listed below the right panel indicate the normalized ratios of the amount of ubiquitinated-glr-1 (right panel) to the amount of total immunoprecipitated GLR-1::GFP (left panel) (Ub-GLR-1/GLR-1). MW (kda) markers are indicated. Similar results were observed in three independent immunoprecipitation experiments performed on a total of two membrane preparations per strain. 6
7 A1 pharynx A2 B1 B2 head neurons nerve ring C1 D1 intestine C2 body wall muscle D2 * * E1 vulva muscle E2 * * F1 G1 H1 F2 G2 H2 5 m * P::GFP Pglr-1::dsRED Merge * * *
8 MAGI-1::YFP A B * 10 m C D ensity (puncta/10 m) WT LIN-10::GFP D E 10 m F D ensity (puncta/10 m) WT G H SOL-1::YFP 10 m I D ensity (puncta/10 m) WT
9 MAGI-1 (3PDZ) USP-46 Input GST GST-GLR-1C Input GST GST-GLR-1C 35 S-MAGI-1 (3PDZ) 35 S-USP-46 MW MW Coomassie stain GST-GLR-1C GST
10 A -Ub +Ub Total GLR-1::GFP Ub-GLR-1 1st IP: -GFP 2nd IP: -Ub Blot: -GFP GLR-1::GFP 1st IP: -GFP Blot: -GFP B Total 1st IP: -GFP 2nd IP: -Ub 2nd IP: -Ub GLR-1::GFP Ub-GLR Blot: -GFP Blot: -Ub
11 A HA-Ub-VME HA-Ub-VME UCHL3 GST- USP46 UCHL3 GST- USP * * Labelled GST-USP Labelled UCHL Coomassie stained gel HA Western blot (HA-Ub conjugates) B C 216 GLR st IP: -GFP WB: -GFP GST-USP-46 C>A WT Ub-GLR-1 GLR-1 Ratio: 2nd IP: -Ub WB: -GFP GST-USP-46 C>A WT Ub-GLR-1 Ub-GLR-1/GLR-1 ratio GST-USP-46 (C>A) GST-USP-46 (WT)
12 A Puncta Width ( m) B Puncta Width ( m) ** ** ** ** # ** n.s. 0 unc-11 AP180 unc-11 AP180; 0 vps-4(dn) vps-4(dn);
13 A mcherry::usp-46 B GLR-1::GFP C Merge 10 m
14 A B C 5 m KDEL::venus mcherry::usp-46 Merged D 200 Untreated Endo H PNGase F Untreated Endo H PNGase F E Endo H-S (%) Endo H-R Endo H-S 0 F mcherry-usp-46 I J mcherry-usp-46 mcherry-usp-46 G Venus::RAB-5 K L Venus::RAB-5 Venus::RAB-5 H Merge M 5 m N 10 m Merge Merge O 1st IP: -GFP WB: -GFP 2nd IP: -Ub WB: -GFP DN-VPS-4 DN-VPS-4; DN-VPS-4 DN-VPS-4; 216 Ub-GLR-1 GLR Ub-GLR-1 GLR-1 Ratio:
Nature Structural & Molecular Biology: doi: /nsmb.3308
 Supplementary Figure 1 Analysis of CED-3 autoactivation and CED-4-induced CED-3 activation. (a) Repeat experiments of Fig. 1a. (b) Repeat experiments of Fig. 1b. (c) Quantitative analysis of three independent
Supplementary Figure 1 Analysis of CED-3 autoactivation and CED-4-induced CED-3 activation. (a) Repeat experiments of Fig. 1a. (b) Repeat experiments of Fig. 1b. (c) Quantitative analysis of three independent
Supplementary Figure S1 Purification of deubiquitinases HEK293 cells were transfected with the indicated DUB-expressing plasmids.
 Supplementary Figure S1 Purification of deubiquitinases HEK293 cells were transfected with the indicated DUB-expressing plasmids. The cells were harvested 72 h after transfection. FLAG-tagged deubiquitinases
Supplementary Figure S1 Purification of deubiquitinases HEK293 cells were transfected with the indicated DUB-expressing plasmids. The cells were harvested 72 h after transfection. FLAG-tagged deubiquitinases
Nature Structural & Molecular Biology: doi: /nsmb.1583
 Acetylation by GCN5 regulates CDC6 phosphorylation in the S-phase of the cell cycle Roberta Paolinelli 1,2, Ramiro Mendoza-Maldonado 2, Anna Cereseto 1 and Mauro Giacca 2 1 Molecular Biology Laboratory,
Acetylation by GCN5 regulates CDC6 phosphorylation in the S-phase of the cell cycle Roberta Paolinelli 1,2, Ramiro Mendoza-Maldonado 2, Anna Cereseto 1 and Mauro Giacca 2 1 Molecular Biology Laboratory,
Supplemental Information. Pacer Mediates the Function of Class III PI3K. and HOPS Complexes in Autophagosome. Maturation by Engaging Stx17
 Molecular Cell, Volume 65 Supplemental Information Pacer Mediates the Function of Class III PI3K and HOPS Complexes in Autophagosome Maturation by Engaging Stx17 Xiawei Cheng, Xiuling Ma, Xianming Ding,
Molecular Cell, Volume 65 Supplemental Information Pacer Mediates the Function of Class III PI3K and HOPS Complexes in Autophagosome Maturation by Engaging Stx17 Xiawei Cheng, Xiuling Ma, Xianming Ding,
SUPPLEMENTAL MATERIAL. Supplemental Methods:
 SUPPLEMENTAL MATERIAL Supplemental Methods: Immunoprecipitation- As we described but with some modifications [22]. As part of another ongoing project, lysate from human umbilical vein endothelial cells
SUPPLEMENTAL MATERIAL Supplemental Methods: Immunoprecipitation- As we described but with some modifications [22]. As part of another ongoing project, lysate from human umbilical vein endothelial cells
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Supplementary figures Supplementary Figure 1: Suv39h1, but not Suv39h2, promotes HP1α sumoylation in vivo. In vivo HP1α sumoylation assay. Top: experimental scheme. Middle: we
SUPPLEMENTARY INFORMATION Supplementary figures Supplementary Figure 1: Suv39h1, but not Suv39h2, promotes HP1α sumoylation in vivo. In vivo HP1α sumoylation assay. Top: experimental scheme. Middle: we
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Nakajima and Tanoue, http://www.jcb.org/cgi/content/full/jcb.201104118/dc1 Figure S1. DLD-1 cells exhibit the characteristic morphology
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Nakajima and Tanoue, http://www.jcb.org/cgi/content/full/jcb.201104118/dc1 Figure S1. DLD-1 cells exhibit the characteristic morphology
Supplementary Figure 1. α-synuclein is truncated in PD and LBD brains. Nature Structural & Molecular Biology: doi: /nsmb.
 Supplementary Figure 1 α-synuclein is truncated in PD and LBD brains. (a) Specificity of anti-n103 antibody. Anti-N103 antibody was coated on an ELISA plate and different concentrations of full-length
Supplementary Figure 1 α-synuclein is truncated in PD and LBD brains. (a) Specificity of anti-n103 antibody. Anti-N103 antibody was coated on an ELISA plate and different concentrations of full-length
Stargazin regulates AMPA receptor trafficking through adaptor protein. complexes during long term depression
 Supplementary Information Stargazin regulates AMPA receptor trafficking through adaptor protein complexes during long term depression Shinji Matsuda, Wataru Kakegawa, Timotheus Budisantoso, Toshihiro Nomura,
Supplementary Information Stargazin regulates AMPA receptor trafficking through adaptor protein complexes during long term depression Shinji Matsuda, Wataru Kakegawa, Timotheus Budisantoso, Toshihiro Nomura,
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Han et al., http://www.jcb.org/cgi/content/full/jcb.201311007/dc1 Figure S1. SIVA1 interacts with PCNA. (A) HEK293T cells were transiently
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Han et al., http://www.jcb.org/cgi/content/full/jcb.201311007/dc1 Figure S1. SIVA1 interacts with PCNA. (A) HEK293T cells were transiently
Figure S1. Verification of ihog Mutation by Protein Immunoblotting Figure S2. Verification of ihog and boi
 Figure S1. Verification of ihog Mutation by Protein Immunoblotting Extracts from S2R+ cells, embryos, and adults were analyzed by immunoprecipitation and immunoblotting with anti-ihog antibody. The Ihog
Figure S1. Verification of ihog Mutation by Protein Immunoblotting Extracts from S2R+ cells, embryos, and adults were analyzed by immunoprecipitation and immunoblotting with anti-ihog antibody. The Ihog
Supplementary Figure 1 Collision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK2, PPK3 and PPK4 respectively.
 Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
Hossain_Supplemental Figure 1
 Hossain_Supplemental Figure 1 GFP-PACT GFP-PACT Motif I GFP-PACT Motif II A. MG132 (1µM) GFP Tubulin GFP-PACT Pericentrin GFP-PACT GFP-PACT Pericentrin Fig. S1. Expression and localization of Orc1 PACT
Hossain_Supplemental Figure 1 GFP-PACT GFP-PACT Motif I GFP-PACT Motif II A. MG132 (1µM) GFP Tubulin GFP-PACT Pericentrin GFP-PACT GFP-PACT Pericentrin Fig. S1. Expression and localization of Orc1 PACT
(A) Schematic depicting morphology of cholinergic DA and DB motor neurons in an L1
 SUPPLEMENTAL MATERIALS Supplemental Figure 1 Figure S1. Expression of the non-alpha nachr subunit ACR-2. (A) Schematic depicting morphology of cholinergic DA and DB motor neurons in an L1 animal. Wide-field
SUPPLEMENTAL MATERIALS Supplemental Figure 1 Figure S1. Expression of the non-alpha nachr subunit ACR-2. (A) Schematic depicting morphology of cholinergic DA and DB motor neurons in an L1 animal. Wide-field
Cellular/Molecular. The Journal of Neuroscience, January 26, (4):
 The Journal of Neuroscience, January 26, 2011 31(4):1341 1354 1341 Cellular/Molecular The Deubiquitinating Enzyme USP-46 Negatively Regulates the Degradation of Glutamate Receptors to Control Their Abundance
The Journal of Neuroscience, January 26, 2011 31(4):1341 1354 1341 Cellular/Molecular The Deubiquitinating Enzyme USP-46 Negatively Regulates the Degradation of Glutamate Receptors to Control Their Abundance
Supplemental Materials and Methods
 Supplemental Materials and Methods Co-immunoprecipitation (Co-IP) assay Cells were lysed with NETN buffer (20 mm Tris-HCl, ph 8.0, 0 mm NaCl, 1 mm EDT, 0.5% Nonidet P-40) containing 50 mm β-glycerophosphate,
Supplemental Materials and Methods Co-immunoprecipitation (Co-IP) assay Cells were lysed with NETN buffer (20 mm Tris-HCl, ph 8.0, 0 mm NaCl, 1 mm EDT, 0.5% Nonidet P-40) containing 50 mm β-glycerophosphate,
Coleman et al., Supplementary Figure 1
 Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Supplementary data. sienigma. F-Enigma F-EnigmaSM. a-p53
 Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
42 fl organelles = 34.5 fl (1) 3.5X X 0.93 = 78,000 (2)
 SUPPLEMENTAL DATA Supplementary Experimental Procedures Fluorescence Microscopy - A Zeiss Axiovert 200M microscope equipped with a Zeiss 100x Plan- Apochromat (1.40 NA) DIC objective and Hamamatsu Orca
SUPPLEMENTAL DATA Supplementary Experimental Procedures Fluorescence Microscopy - A Zeiss Axiovert 200M microscope equipped with a Zeiss 100x Plan- Apochromat (1.40 NA) DIC objective and Hamamatsu Orca
Supplementary Figure 1. APP cleavage assay. HEK293 cells were transfected with various
 Supplementary Figure 1. APP cleavage assay. HEK293 cells were transfected with various GST-tagged N-terminal truncated APP fragments including GST-APP full-length (FL), APP (123-695), APP (189-695), or
Supplementary Figure 1. APP cleavage assay. HEK293 cells were transfected with various GST-tagged N-terminal truncated APP fragments including GST-APP full-length (FL), APP (123-695), APP (189-695), or
Figure legends for supplement
 Figure legends for supplement Supplemental Figure 1 Characterization of purified and recombinant proteins Relevant fractions related the final stage of the purification protocol(bingham et al., 1998; Toba
Figure legends for supplement Supplemental Figure 1 Characterization of purified and recombinant proteins Relevant fractions related the final stage of the purification protocol(bingham et al., 1998; Toba
supplementary information
 DOI: 10.1038/ncb2172 Figure S1 p53 regulates cellular NADPH and lipid levels via inhibition of G6PD. (a) U2OS cells stably expressing p53 shrna or a control shrna were transfected with control sirna or
DOI: 10.1038/ncb2172 Figure S1 p53 regulates cellular NADPH and lipid levels via inhibition of G6PD. (a) U2OS cells stably expressing p53 shrna or a control shrna were transfected with control sirna or
Methanol fixation allows better visualization of Kal7. To compare methods for
 Supplementary Data Methanol fixation allows better visualization of Kal7. To compare methods for visualizing Kal7 in dendrites, mature cultures of dissociated hippocampal neurons (DIV21) were fixed with
Supplementary Data Methanol fixation allows better visualization of Kal7. To compare methods for visualizing Kal7 in dendrites, mature cultures of dissociated hippocampal neurons (DIV21) were fixed with
Supplementary Figure 1. Drawing of spinal cord open-book preparations and DiI tracing. Nature Neuroscience: doi: /nn.3893
 Supplementary Figure 1 Drawing of spinal cord open-book preparations and DiI tracing. Supplementary Figure 2 In ovo electroporation of dominant-negative PlexinA1 in commissural neurons induces midline
Supplementary Figure 1 Drawing of spinal cord open-book preparations and DiI tracing. Supplementary Figure 2 In ovo electroporation of dominant-negative PlexinA1 in commissural neurons induces midline
Supplementary information. Supplementary Figures
 Supplementary information Supplementary Figures Supplementary Figure 1. A. i. HA-JMY expressing U2OS cells were treated with SAHA (6h). DAPI was used to visualise nuclei. ii. U2OS cells stably expressing
Supplementary information Supplementary Figures Supplementary Figure 1. A. i. HA-JMY expressing U2OS cells were treated with SAHA (6h). DAPI was used to visualise nuclei. ii. U2OS cells stably expressing
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
Supplementary Fig. 1 Identification of Nedd4 as an IRS-2-associated protein in camp-treated FRTL-5 cells.
 Supplementary Fig. 1 Supplementary Fig. 1 Identification of Nedd4 as an IRS-2-associated protein in camp-treated FRTL-5 cells. (a) FRTL-5 cells were treated with 1 mm dibutyryl camp for 24 h, and the lysates
Supplementary Fig. 1 Supplementary Fig. 1 Identification of Nedd4 as an IRS-2-associated protein in camp-treated FRTL-5 cells. (a) FRTL-5 cells were treated with 1 mm dibutyryl camp for 24 h, and the lysates
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/9/429/ra54/dc1 Supplementary Materials for Dephosphorylation of the adaptor LAT and phospholipase C by SHP-1 inhibits natural killer cell cytotoxicity Omri Matalon,
www.sciencesignaling.org/cgi/content/full/9/429/ra54/dc1 Supplementary Materials for Dephosphorylation of the adaptor LAT and phospholipase C by SHP-1 inhibits natural killer cell cytotoxicity Omri Matalon,
SUPPLEMENTAL MATERIAL. Supplemental material contains Supplemental Figure Legends and Supplemental Figures 1 to
 SUPPLEMENTAL MATERIAL Supplemental material contains Supplemental Figure Legends and Supplemental Figures 1 to 6. SUPPLEMENTAL FIGURE LEGENDS Supplemental Figure 1. Overview of the mechanisms by which
SUPPLEMENTAL MATERIAL Supplemental material contains Supplemental Figure Legends and Supplemental Figures 1 to 6. SUPPLEMENTAL FIGURE LEGENDS Supplemental Figure 1. Overview of the mechanisms by which
Supporting Information
 Supporting Information Üstün and Börnke, 2015 Figure S1: XopJ does not display acetyltransferase activity. Autoacetylation activity in vitro. Acetylation reactions using MBP, MBP-XopJ, GST and GST-HopZ1a
Supporting Information Üstün and Börnke, 2015 Figure S1: XopJ does not display acetyltransferase activity. Autoacetylation activity in vitro. Acetylation reactions using MBP, MBP-XopJ, GST and GST-HopZ1a
Supplementary Figure 1. The Hsp70 acetylation level is related to the co-chaperone binding of Hsp70 under various stress conditions.
 Supplementary Figure 1. The Hsp70 acetylation level is related to the co-chaperone binding of Hsp70 under various stress conditions. 1 (a) Etoposide treatment gradually changes acetylation level and co-chaperone
Supplementary Figure 1. The Hsp70 acetylation level is related to the co-chaperone binding of Hsp70 under various stress conditions. 1 (a) Etoposide treatment gradually changes acetylation level and co-chaperone
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Moutin et al., http://www.jcb.org/cgi/content/full/jcb.201110101/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Tagged Homer1a and Homer are functional and display different
Supplemental material Moutin et al., http://www.jcb.org/cgi/content/full/jcb.201110101/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Tagged Homer1a and Homer are functional and display different
Supplemental Data. Zhang et al. Plant Cell (2014) /tpc
 Supplemental Data. Zhang et al. Plant Cell (214) 1.115/tpc.114.134163 55 - T C N SDIRIP1-GFP 35-25 - Psb 18 - Histone H3 Supplemental Figure 1. Detection of SDIRIP1-GFP in the nuclear fraction by Western
Supplemental Data. Zhang et al. Plant Cell (214) 1.115/tpc.114.134163 55 - T C N SDIRIP1-GFP 35-25 - Psb 18 - Histone H3 Supplemental Figure 1. Detection of SDIRIP1-GFP in the nuclear fraction by Western
Post-translational modification
 Protein expression Western blotting, is a widely used and accepted technique to detect levels of protein expression in a cell or tissue extract. This technique measures protein levels in a biological sample
Protein expression Western blotting, is a widely used and accepted technique to detect levels of protein expression in a cell or tissue extract. This technique measures protein levels in a biological sample
Supplemental Data. Sethi et al. (2014). Plant Cell /tpc
 Supplemental Data Supplemental Figure 1. MYC2 Binds to the E-box but not the E1-box of the MPK6 Promoter. (A) E1-box and E-box (wild type) containing MPK6 promoter fragment. The region shown in red denotes
Supplemental Data Supplemental Figure 1. MYC2 Binds to the E-box but not the E1-box of the MPK6 Promoter. (A) E1-box and E-box (wild type) containing MPK6 promoter fragment. The region shown in red denotes
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11070 Supplementary Figure 1 Purification of FLAG-tagged proteins. a, Purification of FLAG-RNF12 by FLAG-affinity from nuclear extracts of wild-type (WT) and two FLAG- RNF12 transgenic
doi:10.1038/nature11070 Supplementary Figure 1 Purification of FLAG-tagged proteins. a, Purification of FLAG-RNF12 by FLAG-affinity from nuclear extracts of wild-type (WT) and two FLAG- RNF12 transgenic
Supplemental Materials
 Supplemental Materials Flores-Pérez et al., Supplemental Materials, page 1 of 5 Supplemental Figure S1. Pull-down and BiFC controls, and quantitative analyses associated with the BiFC studies. (A) Controls
Supplemental Materials Flores-Pérez et al., Supplemental Materials, page 1 of 5 Supplemental Figure S1. Pull-down and BiFC controls, and quantitative analyses associated with the BiFC studies. (A) Controls
This is the author's accepted version of the manuscript.
 This is the author's accepted version of the manuscript. The definitive version is published in Nature Communications Online Edition: 2015/4/16 (Japan time), doi:10.1038/ncomms7780. The final version published
This is the author's accepted version of the manuscript. The definitive version is published in Nature Communications Online Edition: 2015/4/16 (Japan time), doi:10.1038/ncomms7780. The final version published
Input. Pulldown GST. GST-Ahi1
 A kd 250 150 100 75 50 37 25 -GST-Ahi1 +GST-Ahi1 kd 148 98 64 50 37 22 GST GST-Ahi1 C kd 250 148 98 64 50 Control Input Hap1A Hap1 Pulldown GST GST-Ahi1 Hap1A Hap1 Hap1A Hap1 Figure S1. (A) Ahi1 western
A kd 250 150 100 75 50 37 25 -GST-Ahi1 +GST-Ahi1 kd 148 98 64 50 37 22 GST GST-Ahi1 C kd 250 148 98 64 50 Control Input Hap1A Hap1 Pulldown GST GST-Ahi1 Hap1A Hap1 Hap1A Hap1 Figure S1. (A) Ahi1 western
Supplementary methods Shoc2 In Vitro Ubiquitination Assay
 Supplementary methods Shoc2 In Vitro Ubiquitination Assay 35 S-labelled Shoc2 was prepared using a TNT quick Coupled transcription/ translation System (Promega) as recommended by manufacturer. For the
Supplementary methods Shoc2 In Vitro Ubiquitination Assay 35 S-labelled Shoc2 was prepared using a TNT quick Coupled transcription/ translation System (Promega) as recommended by manufacturer. For the
SUPPLEMENTARY INFORMATION
 Supplementary Figure 1 sirna and shrna mediated depletion of ATP7A results in loss of melanosomal ATP7A staining. a-h, sirna mediated ATP7A depletion. Immunofluorescence microscopy (IFM) analysis of ATP7A
Supplementary Figure 1 sirna and shrna mediated depletion of ATP7A results in loss of melanosomal ATP7A staining. a-h, sirna mediated ATP7A depletion. Immunofluorescence microscopy (IFM) analysis of ATP7A
JCB. Supplemental material THE JOURNAL OF CELL BIOLOGY. Hong et al.,
 Supplemental material JCB Hong et al., http://www.jcb.org/cgi/content/full/jcb.201412127/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Analysis of purified proteins by SDS-PAGE and pull-down assays. (A) Coomassie-stained
Supplemental material JCB Hong et al., http://www.jcb.org/cgi/content/full/jcb.201412127/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Analysis of purified proteins by SDS-PAGE and pull-down assays. (A) Coomassie-stained
A novel mechanism of post-translational modulation of HMGA functions by the histone chaperone nucleophosmin.
 A novel mechanism of post-translational modulation of HMGA functions by the histone chaperone nucleophosmin. Laura Arnoldo 1,#, Riccardo Sgarra 1,#, Eusebio Chiefari 2, Stefania Iiritano 2, Biagio Arcidiacono
A novel mechanism of post-translational modulation of HMGA functions by the histone chaperone nucleophosmin. Laura Arnoldo 1,#, Riccardo Sgarra 1,#, Eusebio Chiefari 2, Stefania Iiritano 2, Biagio Arcidiacono
J. Cell Sci. 128: doi: /jcs : Supplementary Material. Supplemental Figures. Journal of Cell Science Supplementary Material
 Supplemental Figures Figure S1. Trio controls endothelial barrier function. (A) TagRFP-shTrio constructs were expressed in ECs. Western blot shows efficient Trio knockdown in TagRFP-expressing ECs. (B)
Supplemental Figures Figure S1. Trio controls endothelial barrier function. (A) TagRFP-shTrio constructs were expressed in ECs. Western blot shows efficient Trio knockdown in TagRFP-expressing ECs. (B)
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3363 Supplementary Figure 1 Several WNTs bind to the extracellular domains of PKD1. (a) HEK293T cells were co-transfected with indicated plasmids. Flag-tagged proteins were immunoprecipiated
DOI: 10.1038/ncb3363 Supplementary Figure 1 Several WNTs bind to the extracellular domains of PKD1. (a) HEK293T cells were co-transfected with indicated plasmids. Flag-tagged proteins were immunoprecipiated
Supplemental material
 Supplemental material THE JOURNAL OF CELL BIOLOGY Gillespie et al., http://www.jcb.org/cgi/content/full/jcb.200907037/dc1 repressor complex induced by p38- Gillespie et al. Figure S1. Reduced fiber size
Supplemental material THE JOURNAL OF CELL BIOLOGY Gillespie et al., http://www.jcb.org/cgi/content/full/jcb.200907037/dc1 repressor complex induced by p38- Gillespie et al. Figure S1. Reduced fiber size
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Craft et al., http://www.jcb.org/cgi/content/full/jcb.201409036/dc1 Figure S1. GFP -tubulin interacts with endogenous -tubulin. Ciliary
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Craft et al., http://www.jcb.org/cgi/content/full/jcb.201409036/dc1 Figure S1. GFP -tubulin interacts with endogenous -tubulin. Ciliary
SUPPLEMENTARY INFORMATION
 (Supplementary Methods and Materials) GST pull-down assay GST-fusion proteins Fe65 365-533, and Fe65 538-700 were expressed in BL21 bacterial cells and purified with glutathione-agarose beads (Sigma).
(Supplementary Methods and Materials) GST pull-down assay GST-fusion proteins Fe65 365-533, and Fe65 538-700 were expressed in BL21 bacterial cells and purified with glutathione-agarose beads (Sigma).
Fig. S1. Phosphorylation of eif2 by PEK-1 is increased in ire-1 mutants. Fig. S2. ire-1
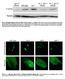 Fig. S1. Phosphorylation of eif2a by PEK-1 is increased in ire-1 mutants. Representative western blot of phosphorylated eif2a and tubulin of day-0 wild-type animals and ire-1 mutants treated with control,
Fig. S1. Phosphorylation of eif2a by PEK-1 is increased in ire-1 mutants. Representative western blot of phosphorylated eif2a and tubulin of day-0 wild-type animals and ire-1 mutants treated with control,
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full//59/ra7/dc1 Supplementary Materials for Dok-7 ctivates the Muscle Receptor Kinase MuSK and Shapes Synapse Formation kane Inoue, Kiyoko Setoguchi, Yosuke Matsubara,
www.sciencesignaling.org/cgi/content/full//59/ra7/dc1 Supplementary Materials for Dok-7 ctivates the Muscle Receptor Kinase MuSK and Shapes Synapse Formation kane Inoue, Kiyoko Setoguchi, Yosuke Matsubara,
Supplemental Figure 1 Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical
 Supplemental Figure Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical in all six REEPs are highlighted in green. Additional
Supplemental Figure Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical in all six REEPs are highlighted in green. Additional
MEFs were treated with the indicated concentrations of LLOMe for three hours, washed
 Supplementary Materials and Methods Cell Fractionation MEFs were treated with the indicated concentrations of LLOMe for three hours, washed with ice-cold PBS, collected by centrifugation, and then homogenized
Supplementary Materials and Methods Cell Fractionation MEFs were treated with the indicated concentrations of LLOMe for three hours, washed with ice-cold PBS, collected by centrifugation, and then homogenized
Viral RNAi suppressor reversibly binds sirna to. outcompete Dicer and RISC via multiple-turnover
 Supplementary Data Viral RNAi suppressor reversibly binds sirna to outcompete Dicer and RISC via multiple-turnover Renata A. Rawlings 1,2, Vishalakshi Krishnan 2 and Nils G. Walter 2 * 1 Biophysics and
Supplementary Data Viral RNAi suppressor reversibly binds sirna to outcompete Dicer and RISC via multiple-turnover Renata A. Rawlings 1,2, Vishalakshi Krishnan 2 and Nils G. Walter 2 * 1 Biophysics and
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Thompson et al., http://www.jcb.org/cgi/content/full/jcb.200909067/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Modification-specific antibodies do not detect unmodified
Supplemental material Thompson et al., http://www.jcb.org/cgi/content/full/jcb.200909067/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Modification-specific antibodies do not detect unmodified
indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) B. Male mice of
 SUPPLEMENTRY FIGURE LEGENDS Figure S1. USP44 loss leads to chromosome missegregation.. Genotypes obtained from the indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) 13.5.. Male mice
SUPPLEMENTRY FIGURE LEGENDS Figure S1. USP44 loss leads to chromosome missegregation.. Genotypes obtained from the indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) 13.5.. Male mice
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table.
 Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or
 Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
Supplementary Figure 1. GST pull-down analysis of the interaction of GST-cIAP1 (A, B), GSTcIAP1
 Legends Supplementary Figure 1. GST pull-down analysis of the interaction of GST- (A, B), GST mutants (B) or GST- (C) with indicated proteins. A, B, Cell lysate from untransfected HeLa cells were loaded
Legends Supplementary Figure 1. GST pull-down analysis of the interaction of GST- (A, B), GST mutants (B) or GST- (C) with indicated proteins. A, B, Cell lysate from untransfected HeLa cells were loaded
Supplemental Data. Wu et al. (2). Plant Cell..5/tpc RGLG Hormonal treatment H2O B RGLG µm ABA µm ACC µm GA Time (hours) µm µm MJ µm IA
 Supplemental Data. Wu et al. (2). Plant Cell..5/tpc..4. A B Supplemental Figure. Immunoblot analysis verifies the expression of the AD-PP2C and BD-RGLG proteins in the Y2H assay. Total proteins were extracted
Supplemental Data. Wu et al. (2). Plant Cell..5/tpc..4. A B Supplemental Figure. Immunoblot analysis verifies the expression of the AD-PP2C and BD-RGLG proteins in the Y2H assay. Total proteins were extracted
Polyclonal ARHGAP25 antibody was prepared from rabbit serum after intracutaneous
 Preparation and purification of polyclonal antibodies Polyclonal ARHGAP25 antibody was prepared from rabbit serum after intracutaneous injections of glutathione S-transferase-ARHGAP25-(509-619) (GST-coiled
Preparation and purification of polyclonal antibodies Polyclonal ARHGAP25 antibody was prepared from rabbit serum after intracutaneous injections of glutathione S-transferase-ARHGAP25-(509-619) (GST-coiled
Supplementary Fig.1 Luton
 Supplementary Fig.1 Luton a 175 Brain Thymus Spleen Small Intestine Kidney Testis HeLa b 250 Lung Kidney MDCK c EFA6B si Control si Mismatch #637 #1564 #1770 83 62 47.5 175 IB: anti-efa6b #B1 130 66 Lysates
Supplementary Fig.1 Luton a 175 Brain Thymus Spleen Small Intestine Kidney Testis HeLa b 250 Lung Kidney MDCK c EFA6B si Control si Mismatch #637 #1564 #1770 83 62 47.5 175 IB: anti-efa6b #B1 130 66 Lysates
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/404/ra120/dc1 Supplementary Materials for The subcellular localization and activity of cortactin is regulated by acetylation and interaction with Keap1 Akihiro
www.sciencesignaling.org/cgi/content/full/8/404/ra120/dc1 Supplementary Materials for The subcellular localization and activity of cortactin is regulated by acetylation and interaction with Keap1 Akihiro
Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity.
 Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity. (A) Amino acid alignment of HDA5, HDA15 and HDA18. The blue line
Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity. (A) Amino acid alignment of HDA5, HDA15 and HDA18. The blue line
Supplemental Data. Furlan et al. Plant Cell (2017) /tpc
 Supplemental Data. Furlan et al. Plant Cell (0) 0.0/tpc..00. Supplemental Data. Furlan et al. Plant Cell (0) 0.0/tpc..00. Supplemental Data. Furlan et al. Plant Cell (0) 0.0/tpc..00. Supplemental Figure.
Supplemental Data. Furlan et al. Plant Cell (0) 0.0/tpc..00. Supplemental Data. Furlan et al. Plant Cell (0) 0.0/tpc..00. Supplemental Data. Furlan et al. Plant Cell (0) 0.0/tpc..00. Supplemental Figure.
pgbkt7 Anti- Myc AH109 strain (KDa) 50
 pgbkt7 (KDa) 50 37 Anti- Myc AH109 strain Supplementary Figure 1. Protein expression of CRN and TDR in yeast. To analyse the protein expression of CRNKD and TDRKD, total proteins extracted from yeast culture
pgbkt7 (KDa) 50 37 Anti- Myc AH109 strain Supplementary Figure 1. Protein expression of CRN and TDR in yeast. To analyse the protein expression of CRNKD and TDRKD, total proteins extracted from yeast culture
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
HPV E6 oncoprotein targets histone methyltransferases for modulating specific. Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu,
 1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
SUPPLEMENTARY INFORMATION
 The Supplementary Information (SI) Methods Cell culture and transfections H1299, U2OS, 293, HeLa cells were maintained in DMEM medium supplemented with 10% fetal bovine serum. H1299 and 293 cells were
The Supplementary Information (SI) Methods Cell culture and transfections H1299, U2OS, 293, HeLa cells were maintained in DMEM medium supplemented with 10% fetal bovine serum. H1299 and 293 cells were
Recruitment of Grb2 to surface IgG and IgE provides antigen receptor-intrinsic costimulation to class-switched B cells
 SUPPLEMENTARY FIGURES Recruitment of Grb2 to surface IgG and IgE provides antigen receptor-intrinsic costimulation to class-switched B cells Niklas Engels, Lars Morten König, Christina Heemann, Johannes
SUPPLEMENTARY FIGURES Recruitment of Grb2 to surface IgG and IgE provides antigen receptor-intrinsic costimulation to class-switched B cells Niklas Engels, Lars Morten König, Christina Heemann, Johannes
CD93 and dystroglycan cooperation in human endothelial cell adhesion and migration
 /, Supplementary Advance Publications Materials 2016 CD93 and dystroglycan cooperation in human endothelial cell adhesion and migration Supplementary Materials Supplementary Figure S1: In ECs CD93 silencing
/, Supplementary Advance Publications Materials 2016 CD93 and dystroglycan cooperation in human endothelial cell adhesion and migration Supplementary Materials Supplementary Figure S1: In ECs CD93 silencing
GFP CCD2 GFP IP:GFP
 D1 D2 1 75 95 148 178 492 GFP CCD1 CCD2 CCD2 GFP D1 D2 GFP D1 D2 Beclin 1 IB:GFP IP:GFP Supplementary Figure 1: Mapping domains required for binding to HEK293T cells are transfected with EGFP-tagged mutant
D1 D2 1 75 95 148 178 492 GFP CCD1 CCD2 CCD2 GFP D1 D2 GFP D1 D2 Beclin 1 IB:GFP IP:GFP Supplementary Figure 1: Mapping domains required for binding to HEK293T cells are transfected with EGFP-tagged mutant
Supplemental Data. Farmer et al. (2010) Plant Cell /tpc
 Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
Supplementary Figure S1. N-terminal fragments of LRRK1 bind to Grb2.
 Myc- HA-Grb2 Mr(K) 105 IP HA 75 25 105 1-1163 1-595 - + - + - + 1164-1989 Blot Myc HA total lysate 75 25 Myc HA Supplementary Figure S1. N-terminal fragments of bind to Grb2. COS7 cells were cotransfected
Myc- HA-Grb2 Mr(K) 105 IP HA 75 25 105 1-1163 1-595 - + - + - + 1164-1989 Blot Myc HA total lysate 75 25 Myc HA Supplementary Figure S1. N-terminal fragments of bind to Grb2. COS7 cells were cotransfected
Sarker et al. Supplementary Material. Subcellular Fractionation
 Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
Figure S1 is related to Figure 1B, showing more details of outer segment of
 Supplemental Information Supplementary Figure legends and Figures Figure S1. Electron microscopic images in Sema4A +/+ and Sema4A / retinas Figure S1 is related to Figure 1B, showing more details of outer
Supplemental Information Supplementary Figure legends and Figures Figure S1. Electron microscopic images in Sema4A +/+ and Sema4A / retinas Figure S1 is related to Figure 1B, showing more details of outer
Conservation of oncofetal antigens on human embryonic stem cells enables discovery of monoclonal antibodies against cancer.
 Title Conservation of oncofetal antigens on human embryonic stem cells enables discovery of monoclonal antibodies against cancer. Authors H.L. Tan 1, *, C. Yong 1, B.Z. Tan 1, W.J. Fong 1, J. Padmanabhan
Title Conservation of oncofetal antigens on human embryonic stem cells enables discovery of monoclonal antibodies against cancer. Authors H.L. Tan 1, *, C. Yong 1, B.Z. Tan 1, W.J. Fong 1, J. Padmanabhan
VCP adaptor interactions are exceptionally dynamic and subject to differential modulation by a VCP inhibitor
 VCP adaptor interactions are exceptionally dynamic and subject to differential modulation by a VCP inhibitor Liang Xue 1, Emily E. Blythe 1, Elyse C. Freiberger 2, Jennifer Mamrosh 1, Alexander S. Hebert
VCP adaptor interactions are exceptionally dynamic and subject to differential modulation by a VCP inhibitor Liang Xue 1, Emily E. Blythe 1, Elyse C. Freiberger 2, Jennifer Mamrosh 1, Alexander S. Hebert
Tohru; Kinjo, Masataka; Taru, Hidenori; Suzuki, Tosh. WoS_67871_Suzuki_Supplemental_Materials.pdf (Supplem. Instructions for use
 Title Quantitative analysis of APP axonal transport in neu Chiba, Kyoko; Araseki, Masahiko; Nozawa, Keisuke; Fu Author(s) Tadashi; Hata, Saori; Saito, Yuhki; Uchida, Seiichi; Tohru; Kinjo, Masataka; Taru,
Title Quantitative analysis of APP axonal transport in neu Chiba, Kyoko; Araseki, Masahiko; Nozawa, Keisuke; Fu Author(s) Tadashi; Hata, Saori; Saito, Yuhki; Uchida, Seiichi; Tohru; Kinjo, Masataka; Taru,
JCB. Supplemental material THE JOURNAL OF CELL BIOLOGY. Kimura et al.,
 Supplemental material JCB Kimura et al., http://www.jcb.org/cgi/content/full/jcb.201503023/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. TRIMs regulate IFN-γ induced autophagy. (A and B) HC image analysis
Supplemental material JCB Kimura et al., http://www.jcb.org/cgi/content/full/jcb.201503023/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. TRIMs regulate IFN-γ induced autophagy. (A and B) HC image analysis
Supplementary Information for. Regulation of Rev1 by the Fanconi Anemia Core Complex
 Supplementary Information for Regulation of Rev1 by the Fanconi Anemia Core Complex Hyungjin Kim, Kailin Yang, Donniphat Dejsuphong, Alan D. D Andrea* *Corresponding Author: Alan D. D Andrea, M.D. Alan_dandrea@dfci.harvard.edu
Supplementary Information for Regulation of Rev1 by the Fanconi Anemia Core Complex Hyungjin Kim, Kailin Yang, Donniphat Dejsuphong, Alan D. D Andrea* *Corresponding Author: Alan D. D Andrea, M.D. Alan_dandrea@dfci.harvard.edu
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature06721 SUPPLEMENTARY INFORMATION. Supplemental Figure Legends Supplemental Figure 1 The distribution of hatx-1[82q] in Cos7 cells. Cos7 cells are co-transfected with hatx-1[82q]-gfp (green)
doi: 10.1038/nature06721 SUPPLEMENTARY INFORMATION. Supplemental Figure Legends Supplemental Figure 1 The distribution of hatx-1[82q] in Cos7 cells. Cos7 cells are co-transfected with hatx-1[82q]-gfp (green)
Supplementary Figure 1 Structural modeling and purification of V. cholerae ABH. (a) The migration of the purified rabh and catalytically inactive
 Supplementary Figure 1 Structural modeling and purification of V. cholerae ABH. (a) The migration of the purified rabh and catalytically inactive variants rabhs, rabhd, and rabhh are shown on 12% SDS-PAGE
Supplementary Figure 1 Structural modeling and purification of V. cholerae ABH. (a) The migration of the purified rabh and catalytically inactive variants rabhs, rabhd, and rabhh are shown on 12% SDS-PAGE
IMMUNOPRECIPITATION TROUBLESHOOTING TIPS
 IMMUNOPRECIPITATION TROUBLESHOOTING TIPS Creative Diagnostics Abstract Immunoprecipitation (IP) is the technique of precipitating a protein antigen out of solution using an antibody that specifically binds
IMMUNOPRECIPITATION TROUBLESHOOTING TIPS Creative Diagnostics Abstract Immunoprecipitation (IP) is the technique of precipitating a protein antigen out of solution using an antibody that specifically binds
Flag-Rac Vector V12 V12 N17 C40. Vector C40 pakt (T308) Akt1. Myc-DN-PAK1 (N-SP)
 a b FlagRac FlagRac V2 V2 N7 C4 V2 V2 N7 C4 p (T38) p (S99, S24) p Flag (Rac) NIH 3T3 COS c +Serum p (T38) MycDN (NSP) Mycp27 3 6 2 3 6 2 3 6 2 min p Myc ( or p27) Figure S (a) Effects of Rac mutants on
a b FlagRac FlagRac V2 V2 N7 C4 V2 V2 N7 C4 p (T38) p (S99, S24) p Flag (Rac) NIH 3T3 COS c +Serum p (T38) MycDN (NSP) Mycp27 3 6 2 3 6 2 3 6 2 min p Myc ( or p27) Figure S (a) Effects of Rac mutants on
UbiCREST Deubiquitinase Enzyme Set
 UbiCREST Deubiquitinase Enzyme Set Catalog Number K-400 This kit contains reagents sufficient for 5 UbiCREST analysis experiments. This package insert must be read in its entirety before using this product.
UbiCREST Deubiquitinase Enzyme Set Catalog Number K-400 This kit contains reagents sufficient for 5 UbiCREST analysis experiments. This package insert must be read in its entirety before using this product.
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the
 Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
SANTA CRUZ BIOTECHNOLOGY, INC.
 TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
unc-33/crmp and ankyrin organize microtubules and localize kinesin to polarize
 Supplementary Information unc-33/crmp and ankyrin organize microtubules and localize kinesin to polarize axon-dendrite sorting Tapan A. Maniar 1, Miriam Kaplan 1, George J. Wang 2, Kang Shen 2, Li Wei
Supplementary Information unc-33/crmp and ankyrin organize microtubules and localize kinesin to polarize axon-dendrite sorting Tapan A. Maniar 1, Miriam Kaplan 1, George J. Wang 2, Kang Shen 2, Li Wei
S156AT168AY175A (AAA) were purified as GST-fusion proteins and incubated with GSTfused
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 Supplemental Materials Supplemental Figure S1 (a) Phenotype of the wild type and grik1-2 grik2-1 plants after 8 days in darkness.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 Supplemental Materials Supplemental Figure S1 (a) Phenotype of the wild type and grik1-2 grik2-1 plants after 8 days in darkness.
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2386 Figure 1 Src-containing puncta are not focal adhesions, podosomes or endosomes. (a) FAK-/- were stained with anti-py416 Src (green) and either (in red) the focal adhesion protein paxillin,
DOI: 10.1038/ncb2386 Figure 1 Src-containing puncta are not focal adhesions, podosomes or endosomes. (a) FAK-/- were stained with anti-py416 Src (green) and either (in red) the focal adhesion protein paxillin,
Supplementary Figure 1 Phosphorylated tau accumulates in Nrf2 (-/-) mice. Hippocampal tissues obtained from Nrf2 (-/-) (10 months old, 4 male; 2
 Supplementary Figure 1 Phosphorylated tau accumulates in Nrf2 (-/-) mice. Hippocampal tissues obtained from Nrf2 (-/-) (10 months old, 4 male; 2 female) or wild-type (5 months old, 1 male; 11 months old,
Supplementary Figure 1 Phosphorylated tau accumulates in Nrf2 (-/-) mice. Hippocampal tissues obtained from Nrf2 (-/-) (10 months old, 4 male; 2 female) or wild-type (5 months old, 1 male; 11 months old,
Supplementary material for: Materials and Methods:
 Supplementary material for: Iron-responsive degradation of iron regulatory protein 1 does not require the Fe-S cluster: S.L. Clarke, et al. Materials and Methods: Fe-S Cluster Reconstitution: Cells treated
Supplementary material for: Iron-responsive degradation of iron regulatory protein 1 does not require the Fe-S cluster: S.L. Clarke, et al. Materials and Methods: Fe-S Cluster Reconstitution: Cells treated
(A) Schematic illustration of sciatic nerve ligation. P, proximal; D, distal to the ligation site.
 SUPPLEMENTRY INFORMTION SUPPLEMENTL FIGURES Figure S1. () Schematic illustration of sciatic nerve ligation. P, proximal; D, distal to the ligation site. () Western blot of ligated and unligated sciatic
SUPPLEMENTRY INFORMTION SUPPLEMENTL FIGURES Figure S1. () Schematic illustration of sciatic nerve ligation. P, proximal; D, distal to the ligation site. () Western blot of ligated and unligated sciatic
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rainero et al., http://www.jcb.org/cgi/content/full/jcb.201109112/dc1 Figure S1. The expression of DGK- is reduced upon transfection
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rainero et al., http://www.jcb.org/cgi/content/full/jcb.201109112/dc1 Figure S1. The expression of DGK- is reduced upon transfection
Supplemental Information
 Supplemental Information Intrinsic protein-protein interaction mediated and chaperonin assisted sequential assembly of a stable Bardet Biedl syndome protein complex, the BBSome * Qihong Zhang 1#, Dahai
Supplemental Information Intrinsic protein-protein interaction mediated and chaperonin assisted sequential assembly of a stable Bardet Biedl syndome protein complex, the BBSome * Qihong Zhang 1#, Dahai
Supplemental Figure S1. PGRN Binding to Sortilin.
 1 Neuron, volume 68 Supplemental Data Sortilin-Mediated Endocytosis Determines Levels of the Frontotemporal Dementia Protein, Progranulin Fenghua Hu, Thihan Padukkavidana, Christian B. Vægter, Owen A.
1 Neuron, volume 68 Supplemental Data Sortilin-Mediated Endocytosis Determines Levels of the Frontotemporal Dementia Protein, Progranulin Fenghua Hu, Thihan Padukkavidana, Christian B. Vægter, Owen A.
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/3/146/ra80/dc1 Supplementary Materials for DNMT1 Stability Is Regulated by Proteins Coordinating Deubiquitination and Acetylation-Driven Ubiquitination Zhanwen
www.sciencesignaling.org/cgi/content/full/3/146/ra80/dc1 Supplementary Materials for DNMT1 Stability Is Regulated by Proteins Coordinating Deubiquitination and Acetylation-Driven Ubiquitination Zhanwen
seminal vesicle thymus kidney lung liver
 a 1 HEK293 1 B 3 3 T GFPSMAD TGFβ (T)/ BMP (B) IB: SMAD1/3TP IB: GFP b wild type mouse tissue kidney thymus seminal vesicle liver lung brain adipose tissue muscle pancreas heart uterus spleen testis IB:
a 1 HEK293 1 B 3 3 T GFPSMAD TGFβ (T)/ BMP (B) IB: SMAD1/3TP IB: GFP b wild type mouse tissue kidney thymus seminal vesicle liver lung brain adipose tissue muscle pancreas heart uterus spleen testis IB:
Future of Novel Drugs: DUBs Drug Discovery Platform LifeSensors Inc.
 Future of Novel Drugs: DUBs Drug Discovery Platform LifeSensors Inc. 271 Great Valley Parkway Malvern PA 19355 Phone: 610-644-8845 Fax: 610-644-8616 www.lifesensors.com LifeSensors Capabilities ~35 active
Future of Novel Drugs: DUBs Drug Discovery Platform LifeSensors Inc. 271 Great Valley Parkway Malvern PA 19355 Phone: 610-644-8845 Fax: 610-644-8616 www.lifesensors.com LifeSensors Capabilities ~35 active
