Supplementary Figure 1. shrna screen against CSD-containing proteins in hescs. (a) Brightfield images of H9 hescs. Knockdown of YBX1, YBX2 and YBX3
|
|
|
- Gary Jordan
- 6 years ago
- Views:
Transcription
1 Supplementary Figure 1. shrna screen against CSD-containing proteins in hescs. (a) Brightfield images of H9 hescs. Knockdown of YBX1, YBX2 and YBX3 do not change hesc colony morphology. Scale bar represents 500 µm. (b) Data represent the mean ± s.e.m. of mrna relative expression levels to non-targeting (NT) shrna H9 hescs (n= 3 independent experiments). All the statistical comparisons were made by Student s t-test for unpaired samples. P-value: *(P<0.05), **(P<0.01), ***(P<0.001).
2 Supplementary Figure 2. Loss of CSDE1 promotes differentiation of hescs. Brightfield images of H9 (a) and H1 (b) hescs. Scale bars represent 500 µm.
3 Supplementary Figure 3. CSDE1 protein levels decrease during neural differentiation of H1 hescs. Western blot analysis with antibody to CSDE1. GAPDH is the loading control. The graph represents the CSDE1 relative percentage values (corrected for GAPDH loading control) to H1 hescs (mean ± s.e.m. of three independent experiments).
4 Supplementary Figure 4. CSDE1 KD hesc cultures are maintained by transferring typical undifferentiated colonies followed by daily monitoring to remove differentiated cells. Under this protocol, we did not observe differences in the percentage of OCT4 and PAX6-positive cells/total nuclei (mean ± s.e.m. of three independent experiments, 200 total cells per experiment). Statistical comparisons were made by Student s t-test for unpaired samples. On the contrary, when we grew hescs without removing differentiated colonies, flattened cells proliferated in CSDE1 KD lines resulting in decreased percentage of OCT4-positive cells as compared with control hesc (please see Fig. 2e-f)
5 Supplementary Figure 5. Bromodeoxyuridine (BrdU) proliferation assay. Loss of CSDE1 does not induce significant differences in the percentage of BrdU positive cells (H9 NT hescs vs CSDE1 shrna#1 hescs (P= 0.13), H9 NT hescs vs CSDE1 shrna #2 hescs (P= 0.08)). The results are represented as averages of at least 1400 cells scored in six different experiments. Statistical comparisons were made by Student s t-test for unpaired samples.
6 Supplementary Figure 6. Loss of CSDE1 does not trigger the expression of endoderm markers in hescs. Quantitative PCR (qpcr) analysis of hescs cultured for five days without removing differentiated cells. Graph (relative expression to non-targeting shrna) represents the mean ± s.e.m. of three independent experiments. Statistical comparisons were made by Student s t-test for unpaired samples (P-value: *(P<0.05)).
7 Supplementary Figure 7. Knockdown of CSDE1 with a shrna to the 3 UTR region of the CSDE1 transcript also induces spontaneous proliferation of PAX6-positive cells. (a) Western blot analysis with antibodies to CSDE1 in H9 hesc lysates. β-actin is the loading control. CSDE1 shrna #3 targets the 3 UTR region of CSDE1 transcript whereas the shrnas #1 and #2 used in Fig. 2e target the coding region. (b) Percentage of OCT4 and PAX6-positive cells/total nuclei after five days without removing differentiated cells (mean ± s.e.m. of three independent experiments, total cells per experiment). Statistical comparisons were made by Student s t-test for unpaired samples (P-value: *(P<0.05)).
8 Supplementary Figure 8. CSDE1 protein expression levels in distinct knockout and knockdown hesc lines. Western blot analysis with antibodies to CSDE1 in (a) H9, (b) H1, (c) HUES9 and (d) HUES6 hesc lysates. As a loading control, we used β-actin (a-c) or GAPDH (d).
9 Supplementary Figure 9. Knockdown of CSDE1 with a shrna to the 3 UTR region of the CSDE1 transcript also accelerates neural differentiation. Percentage of OCT4 and PAX6- positive cells/total nuclei at different days after neural induction treatment of undifferentiated H9 hescs (mean ± s.e.m. (n= 3, total cells per data point). Statistical comparisons were made by Student s t-test for unpaired samples. P-value: **(P<0.01).
10 Supplementary Figure 10. Knockout of CSDE1 accelerates neural differentiation. Percentage of OCT4 and PAX6-positive cells/total nuclei at different days after neural induction treatment of undifferentiated H9 hesc colonies (mean ± s.e.m. of three independent experiments, total cells per data point). Statistical comparisons were made by Student s t-test for unpaired samples. P-value: *(P<0.05), **(P<0.01). It is important to note that we observed a slower differentiation rate into PAX6-positive cells in these differentiation experiments compared to Figure 3a. Although neural induction was performed using H9 hescs in both set of experiments, the control non-targeting shrna as well as CSDE1 shrna H9 lines used in Figure 3a were treated with puromycin every 3-4 days to select shrna-expressing cells. On the contrary, both control wild-type (WT) and CSDE1 -/- H9 hescs were not maintained under regular puromycin treatments. This could contribute to the different rates observed in both neural induction experiments.
11 Supplementary Figure 11. Loss of CSDE1 promotes neural differentiation of distinct hesc lines and ipscs. (a) Percentage of OCT4 and PAX6-positive cells/total nuclei at different days after neural induction of undifferentiated H1 hescs (mean ± s.e.m. of three independent experiments, approximately 200 total cells per experiment). (b) Percentage of OCT4 and PAX6- positive cells/total nuclei at different days after neural induction of undifferentiated HUES6 hescs (mean ± s.e.m., n= 3, total cells per data point). (c) Percentage of OCT4 and PAX6-positive cells/total nuclei at different days after neural induction of undifferentiated ipscs (mean ± s.e.m. of three independent experiments, total cells per data point). Statistical comparisons were made by Student s t-test for unpaired samples. P-value: *(P<0.05), **(P<0.01), ***(P<0.001), **** (P<0.0001).
12 Supplementary Figure 12. CSDE1 KD cells exhibit increased protein levels of NPC markers. Western blot analysis of H9 cells after 10 days of neural induction treatment with antibodies to PAX6 and NES. β-actin is the loading control.
13 Supplementary Figure 13. CSDE1 KD hescs exhibit diminished induction of cardiac markers during their differentiation into cardiomyocytes. (a-d) qpcr analysis at distinct stages of cardiomyocyte differentiation from H1 hescs. Specific markers for each differentiation stage are shown in the corresponding graph. (a) Graph (relative expression to NT shrna mesoderm) represent the mean ± s.e.m. (n= 3). (b) Graph (relative expression to NT shrna cardiac mesoderm) represent the mean ± s.e.m. (n= 3). (c) Graph (relative expression to NT shrna cardiac progenitors) represent the mean ± s.e.m. (n= 3). (d) Graph (relative expression to NT shrna cardiomyocytes) represent the mean ± s.e.m. (n= 3). All the statistical comparisons were made by Student s t-test for unpaired samples. P-value: *(P<0.05), **(P<0.01), ***(P<0.001), **** (P<0.0001).
14 Supplementary Figure 14. CSDE1 binds to complexes involved in mrna translation and stability in hescs. (a) Co-immunoprecipitation with CSDE1 and FLAG antibodies followed by western blot with CSDE1 antibody in wild-type H9 hesc lysates. (b) Polysome profiling experiments followed by western blot with antibodies to CSDE1, RPS27 (40S ribosomal subunit) and RPL7 (60S ribosomal subunit). (c) Gene Ontology (GO) Cellular Component (CC) and GO Molecular Function (MF)-terms significantly enriched after co-ip experiments with CSDE1 antibody (as defined by cut-off t-test Difference >2, p-value <0.02).
15 Supplementary Figure 15. Analysis of germ layer markers in CSDE1 KD H9 hesc cultures primarily formed by undifferentiated colonies. qpcr analysis of ectodermal (PAX6, NES, FGF5), mesodermal (MSX1) and endodermal (ALB, GATA4, GATA6) germ layer markers in hescs cultures daily monitored to remove differentiated cells. Graph (relative expression to NT shrna) represents the mean ± s.e.m. of four independent experiments with three biological replicates. Statistical comparisons were made by Student s t-test for unpaired samples. P-value: *(P<0.05), **(P<0.01), ***(P<0.001).
16 Supplementary Figure 16. FABP7 is highly expressed in NPCs derived from hescs. Western blot analysis of H9 hescs and their differentiated counterparts with antibody to FABP7. β-actin is the loading control.
17 Supplementary Figure 17. Loss of CSDE1 results in increased levels of FABP7. Western blot analysis in (a) H9 hesc, (b) H1 hesc, (c) HUES9 hesc, (d) HUES6 hesc and (e) ipsc lysates. As a loading control, we used β-actin (a, b, d, e) or GAPDH (c).
18 Supplementary Figure 18. Loss of CSDE1 does not alter the levels of radial glia markers such as GFAP, SLC1A3 or S100ß. (a) Western blot analysis of H9 hescs with antibodies to GFAP. β-actin is the loading control. (b) Graph (relative expression to NT shrna H9 hescs) represents the mean ± s.e.m. of two independent experiments with three biological replicates. All the statistical comparisons were made by Student s t-test for unpaired samples. P-value: ***(P<0.001).
19 Supplementary Figure 19. VIM and FABP7 RNA probes pull down CSDE1 protein from H9 hesc lysates. Pull-down assay using in vitro transcribed biotinylated VIM, FABP7 and 5 UTR msl2 RNA probes (negative control). RNA pull-down specificity was assessed by Western blotting using antibodies to CSDE1, unrelated RBPs (CIRBP and CELF1) and β-actin.
20 Supplementary Figure 20. Knockdown of CSDE1 results in decreased protein levels of SEMA4A. Western blot analysis of (a) H9 hescs and (b) HUES9 hescs with antibody to SEMA4A. GAPDH is the loading control.
21 Supplementary Figure 21. Uncropped images of most important western blots. Uncropped images are presented with molecular weight ladders.
22 Gene names Domain hesc vs neurons ratio q-value LIN28A CSD LIN28B CSD Not detected YBX1 CSD YBX2 CSD YBX3 CSD Not detected CSDE1 CSD DIS3 CSD and S EIF1AX S EIF2A S1 Not detected EIF5A S EXOSC3 S1 Not detected DHX8 S1 Not detected Supplementary Table 1. CSD and CSD-like proteins are increased in hescs compared with neurons. Tandem mass tag (TMT) quantitative proteomics comparing H9 hescs with their differentiated neuronal counterparts. CSD and CSD-like proteins are significantly increased in hescs. Statistical comparisons were made by Student s t-test (n= 3). False Discovery Rate (FDR) adjusted p-value (q-value) <0.05 was considered significant.
23 (a) Gene names Protein names RPS27;RPS27L 40S ribosomal protein S27 SMNDC1 Survival of motor neuron-related-splicing factor 30 FAM103A1 RNMT-activating mini protein SMC1A Structural maintenance of chromosomes protein 1A PES1 Pescadillo homolog PUM1 Pumilio homolog 1 CHCHD6 MICOS complex subunit MIC25 HIST1H2BH;HIST2H2BF Histone H2B type 1-H;Histone H2B type 2-F;Histone H2B SSSCA1 Sjoegren syndrome/scleroderma autoantigen 1 PSMC2 26S protease regulatory subunit 7 GDI2;GDI1 Rab GDP dissociation inhibitor beta;rab GDP dissociation inhibitor alpha NCAPG Condensin complex subunit 3 TAGLN2 Transgelin-2 NME1 Nucleoside diphosphate kinase A PTMS Parathymosin G6PD Glucose-6-phosphate 1-dehydrogenase AMPD2 AMP deaminase 2 SAMM50 Sorting and assembly machinery component 50 homolog MAP7 Ensconsin REPS1 Calcium binding ZNF512 TransriptionZinc finger protein 512 (b) Gene names Protein names T-test difference -log P-value CSDE1 Cold shock domain-containing protein E STRAP Serine-threonine kinase receptor-associated protein SUGP2 SURP and G-patch domain-containing protein IMMT MICOS complex subunit MIC Supplementary Table 2. Protein interactors of CSDE1 in H9 hescs. Protein label-free quantification (LFQ) values from co-immunoprecipitation (co-ip) experiments using CSDE1 antibody compared to control co-ip with FLAG antibody. (a) List of proteins only coimmunoprecipitated with CSDE1 antibody but not with FLAG antibody (n= 3). (b) List of proteins significantly increased after co-ip with CSDE1 compared with FLAG antibody (Student s t-test, n= 3, FDR< 0.05).
24 Gene names FABP7 Protein names Fatty acid-binding protein, brain T-test difference CSDE1 shrna 1 CSDE1 shrna 2 CSDE1 shrna 1 -log P-value CSDE1 shrna VIL1 Villin FGFBP3 PAIP2 Fibroblast growth factorbinding protein 3 Polyadenylate-binding protein-interacting protein FLNC Filamin-C S100A4 Protein S100-A VIM Vimentin CIZ1 Cip1-interacting zinc finger protein DNM1 Dynamin SMARCC2 CSDE1 SWI/SNF complex subunit SMARCC2 Cold shock domaincontaining protein E VSNL1 Visinin-like protein THUMPD3 EIF4A2 THUMP domain-containing protein 3 Eukaryotic initiation factor 4A-II ENPP1 Alkaline phosphodiesterase I PRPS1 DCLK1 Ribose-phosphate pyrophosphokinase 1 Serine/threonine-protein kinase DCLK SPG20 Spartin ANXA2 Annexin A TBC1D15 PTBP2 TBC1 domain family member 15 Polypyrimidine tract-binding protein Supplementary Table 3. List of significantly changed proteins in quantitative proteomic analysis of CSDE1 KD H9 hescs. Knockdown of CSDE1 induces an increase in the levels of FABP7 and VIM. Means are calculated from the log2 of LFQ values (LFQ CSDE1 shrna hescs/non-targeting shrna hescs). Statistical comparisons were made by Student s t-test (n= 3, FDR< 0.15).
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate
 Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
Figure S1: NUN preparation yields nascent, unadenylated RNA with a different profile from Total RNA.
 Summary of Supplemental Information Figure S1: NUN preparation yields nascent, unadenylated RNA with a different profile from Total RNA. Figure S2: rrna removal procedure is effective for clearing out
Summary of Supplemental Information Figure S1: NUN preparation yields nascent, unadenylated RNA with a different profile from Total RNA. Figure S2: rrna removal procedure is effective for clearing out
Int. J. Mol. Sci. 2016, 17, 1259; doi: /ijms
 S1 of S5 Supplementary Materials: Fibroblast-Derived Extracellular Matrix Induces Chondrogenic Differentiation in Human Adipose-Derived Mesenchymal Stromal/Stem Cells in Vitro Kevin Dzobo, Taegyn Turnley,
S1 of S5 Supplementary Materials: Fibroblast-Derived Extracellular Matrix Induces Chondrogenic Differentiation in Human Adipose-Derived Mesenchymal Stromal/Stem Cells in Vitro Kevin Dzobo, Taegyn Turnley,
GENESDEV/2007/ Supplementary Figure 1 Elkabetz et al.,
 GENESDEV/2007/089581 Supplementary Figure 1 Elkabetz et al., GENESDEV/2007/089581 Supplementary Figure 2 Elkabetz et al., GENESDEV/2007/089581 Supplementary Figure 3 Elkabetz et al., GENESDEV/2007/089581
GENESDEV/2007/089581 Supplementary Figure 1 Elkabetz et al., GENESDEV/2007/089581 Supplementary Figure 2 Elkabetz et al., GENESDEV/2007/089581 Supplementary Figure 3 Elkabetz et al., GENESDEV/2007/089581
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table.
 Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 CSRP2 PFKP ADFP ADM C10orf10 GPI LOX PLEKHA2 WIPF1
 Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 4.52E-18 PDK1 6.77E-18 CSRP2 4.42E-17 PFKP 1.23E-14 MSH2 3.79E-13 NARF_A 5.56E-13 ADFP 5.56E-13 FAM13A1 1.56E-12 FAM29A_A 1.22E-11 CA9 1.54E-11
Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 4.52E-18 PDK1 6.77E-18 CSRP2 4.42E-17 PFKP 1.23E-14 MSH2 3.79E-13 NARF_A 5.56E-13 ADFP 5.56E-13 FAM13A1 1.56E-12 FAM29A_A 1.22E-11 CA9 1.54E-11
ENCODE RBP Antibody Characterization Guidelines
 ENCODE RBP Antibody Characterization Guidelines Approved on November 18, 2016 Background An integral part of the ENCODE Project is to characterize the antibodies used in the experiments. This document
ENCODE RBP Antibody Characterization Guidelines Approved on November 18, 2016 Background An integral part of the ENCODE Project is to characterize the antibodies used in the experiments. This document
Human embryonic stem cells for in-vitro developmental toxicity testing
 Human embryonic stem cells for in-vitro developmental toxicity testing HESI workshop on alternatives assays for developmental toxicity Raimund Strehl Cellartis The company Founded in early 2001, University
Human embryonic stem cells for in-vitro developmental toxicity testing HESI workshop on alternatives assays for developmental toxicity Raimund Strehl Cellartis The company Founded in early 2001, University
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/3/6/ra27/dc Supplementary Materials for AAA+ Proteins and Coordinate PIKK Activity and Function in Nonsense-Mediated mrna Decay Natsuko Izumi, Akio Yamashita,*
www.sciencesignaling.org/cgi/content/full/3/6/ra27/dc Supplementary Materials for AAA+ Proteins and Coordinate PIKK Activity and Function in Nonsense-Mediated mrna Decay Natsuko Izumi, Akio Yamashita,*
Xeno-Free Systems for hesc & hipsc. Facilitating the shift from Stem Cell Research to Clinical Applications
 Xeno-Free Systems for hesc & hipsc Facilitating the shift from Stem Cell Research to Clinical Applications NutriStem Defined, xeno-free (XF), serum-free media (SFM) specially formulated for growth and
Xeno-Free Systems for hesc & hipsc Facilitating the shift from Stem Cell Research to Clinical Applications NutriStem Defined, xeno-free (XF), serum-free media (SFM) specially formulated for growth and
Supplementary Figures Montero et al._supplementary Figure 1
 Montero et al_suppl. Info 1 Supplementary Figures Montero et al._supplementary Figure 1 Montero et al_suppl. Info 2 Supplementary Figure 1. Transcripts arising from the structurally conserved subtelomeres
Montero et al_suppl. Info 1 Supplementary Figures Montero et al._supplementary Figure 1 Montero et al_suppl. Info 2 Supplementary Figure 1. Transcripts arising from the structurally conserved subtelomeres
Supplementary Figure 1: Derivation and characterization of RN ips cell lines. (a) RN ips cells maintain expression of pluripotency markers OCT4 and
 Supplementary Figure 1: Derivation and characterization of RN ips cell lines. (a) RN ips cells maintain expression of pluripotency markers OCT4 and SSEA4 after 10 passages in mtesr 1 medium. (b) Schematic
Supplementary Figure 1: Derivation and characterization of RN ips cell lines. (a) RN ips cells maintain expression of pluripotency markers OCT4 and SSEA4 after 10 passages in mtesr 1 medium. (b) Schematic
Isolation, culture, and transfection of primary mammary epithelial organoids
 Supplementary Experimental Procedures Isolation, culture, and transfection of primary mammary epithelial organoids Primary mammary epithelial organoids were prepared from 8-week-old CD1 mice (Charles River)
Supplementary Experimental Procedures Isolation, culture, and transfection of primary mammary epithelial organoids Primary mammary epithelial organoids were prepared from 8-week-old CD1 mice (Charles River)
Supplementary Figure 1. Characterization of EVs (a) Phase-contrast electron microscopy was used to visualize resuspended EV pellets.
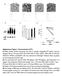 Supplementary Figure 1. Characterization of EVs (a) Phase-contrast electron microscopy was used to visualize resuspended EV pellets. Scale bar represent 100 nm. The sizes of EVs from MDA-MB-231-D3H1 (D3H1),
Supplementary Figure 1. Characterization of EVs (a) Phase-contrast electron microscopy was used to visualize resuspended EV pellets. Scale bar represent 100 nm. The sizes of EVs from MDA-MB-231-D3H1 (D3H1),
A simple choice. Essential 8 Media
 A simple choice Essential 8 Media Your stem cells thrive with the essentials Gibco Essential 8 Medium is a feeder-free, xeno-free medium originally developed in the laboratory of stem cell research pioneer
A simple choice Essential 8 Media Your stem cells thrive with the essentials Gibco Essential 8 Medium is a feeder-free, xeno-free medium originally developed in the laboratory of stem cell research pioneer
Fig Ch 17: From Gene to Protein
 Fig. 17-1 Ch 17: From Gene to Protein Basic Principles of Transcription and Translation RNA is the intermediate between genes and the proteins for which they code Transcription is the synthesis of RNA
Fig. 17-1 Ch 17: From Gene to Protein Basic Principles of Transcription and Translation RNA is the intermediate between genes and the proteins for which they code Transcription is the synthesis of RNA
MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1. Lecture 25: Mass Spectrometry Applications
![MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1. Lecture 25: Mass Spectrometry Applications MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1. Lecture 25: Mass Spectrometry Applications](/thumbs/74/71368870.jpg) MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1 Lecture 25: Mass Spectrometry Applications Measuring Protein Abundance o ICAT o DIGE Identifying Post-Translational Modifications Protein-protein
MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1 Lecture 25: Mass Spectrometry Applications Measuring Protein Abundance o ICAT o DIGE Identifying Post-Translational Modifications Protein-protein
HUMAN EMBRYONIC STEM CELL (hesc) ASSESSMENT BY REAL-TIME POLYMERASE CHAIN REACTION (PCR)
 HUMAN EMBRYONIC STEM CELL (hesc) ASSESSMENT BY REAL-TIME POLYMERASE CHAIN REACTION (PCR) OBJECTIVE: is performed to analyze gene expression of human Embryonic Stem Cells (hescs) in culture. Real-Time PCR
HUMAN EMBRYONIC STEM CELL (hesc) ASSESSMENT BY REAL-TIME POLYMERASE CHAIN REACTION (PCR) OBJECTIVE: is performed to analyze gene expression of human Embryonic Stem Cells (hescs) in culture. Real-Time PCR
Regulation of Synaptic Structure and Function by FMRP- Associated MicroRNAs mir-125b and mir-132
 Neuron, Volume 65 Regulation of Synaptic Structure and Function by FMRP- Associated MicroRNAs mir-125b and mir-132 Dieter Edbauer, Joel R. Neilson, Kelly A. Foster, Chi-Fong Wang, Daniel P. Seeburg, Matthew
Neuron, Volume 65 Regulation of Synaptic Structure and Function by FMRP- Associated MicroRNAs mir-125b and mir-132 Dieter Edbauer, Joel R. Neilson, Kelly A. Foster, Chi-Fong Wang, Daniel P. Seeburg, Matthew
Learning Objectives. Define RNA interference. Define basic terminology. Describe molecular mechanism. Define VSP and relevance
 Learning Objectives Define RNA interference Define basic terminology Describe molecular mechanism Define VSP and relevance Describe role of RNAi in antigenic variation A Nobel Way to Regulate Gene Expression
Learning Objectives Define RNA interference Define basic terminology Describe molecular mechanism Define VSP and relevance Describe role of RNAi in antigenic variation A Nobel Way to Regulate Gene Expression
Gene Expression: Transcription
 Gene Expression: Transcription The majority of genes are expressed as the proteins they encode. The process occurs in two steps: Transcription = DNA RNA Translation = RNA protein Taken together, they make
Gene Expression: Transcription The majority of genes are expressed as the proteins they encode. The process occurs in two steps: Transcription = DNA RNA Translation = RNA protein Taken together, they make
Real-time PCR. TaqMan Protein Assays. Unlock the power of real-time PCR for protein analysis
 Real-time PCR TaqMan Protein Assays Unlock the power of real-time PCR for protein analysis I can use my real-time PCR instrument to quantitate protein? Do protein levels correlate with related mrna levels
Real-time PCR TaqMan Protein Assays Unlock the power of real-time PCR for protein analysis I can use my real-time PCR instrument to quantitate protein? Do protein levels correlate with related mrna levels
HCT116 SW48 Nutlin: p53
 Figure S HCT6 SW8 Nutlin: - + - + p GAPDH Figure S. Nutlin- treatment induces p protein. HCT6 and SW8 cells were left untreated or treated for 8 hr with Nutlin- ( µm) to up-regulate p. Whole cell lysates
Figure S HCT6 SW8 Nutlin: - + - + p GAPDH Figure S. Nutlin- treatment induces p protein. HCT6 and SW8 cells were left untreated or treated for 8 hr with Nutlin- ( µm) to up-regulate p. Whole cell lysates
Directed Differentiation of Human Induced Pluripotent Stem Cells. into Fallopian Tube Epithelium
 Directed Differentiation of Human Induced Pluripotent Stem Cells into Fallopian Tube Epithelium Nur Yucer 1,2, Marie Holzapfel 2, Tilley Jenkins Vogel 2, Lindsay Lenaeus 1, Loren Ornelas 1, Anna Laury
Directed Differentiation of Human Induced Pluripotent Stem Cells into Fallopian Tube Epithelium Nur Yucer 1,2, Marie Holzapfel 2, Tilley Jenkins Vogel 2, Lindsay Lenaeus 1, Loren Ornelas 1, Anna Laury
Differential Association of Chromatin Proteins Identifies BAF60a/SMARCD1 as a Regulator of Embryonic Stem Cell Differentiation
 Article Differential Association of Chromatin Proteins Identifies BAF60a/SMARCD1 as a Regulator of Embryonic Stem Cell Differentiation Graphical Abstract Authors Adi Alajem, Alva Biran,..., Siu Kwan Sze,
Article Differential Association of Chromatin Proteins Identifies BAF60a/SMARCD1 as a Regulator of Embryonic Stem Cell Differentiation Graphical Abstract Authors Adi Alajem, Alva Biran,..., Siu Kwan Sze,
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling
 Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
The microtubule-associated tau protein has intrinsic acetyltransferase activity. Todd J. Cohen, Dave Friedmann, Andrew W. Hwang, Ronen Marmorstein and
 SUPPLEMENTARY INFORMATION: The microtubule-associated tau protein has intrinsic acetyltransferase activity Todd J. Cohen, Dave Friedmann, Andrew W. Hwang, Ronen Marmorstein and Virginia M.Y. Lee Cohen
SUPPLEMENTARY INFORMATION: The microtubule-associated tau protein has intrinsic acetyltransferase activity Todd J. Cohen, Dave Friedmann, Andrew W. Hwang, Ronen Marmorstein and Virginia M.Y. Lee Cohen
Chapter 13. The Nucleus. The nucleus is the hallmark of eukaryotic cells; the very term eukaryotic means having a "true nucleus".
 Chapter 13 The Nucleus The nucleus is the hallmark of eukaryotic cells; the very term eukaryotic means having a "true nucleus". Fig.13.1. The EM of the Nucleus of a Eukaryotic Cell 13.1. The Nuclear Envelope
Chapter 13 The Nucleus The nucleus is the hallmark of eukaryotic cells; the very term eukaryotic means having a "true nucleus". Fig.13.1. The EM of the Nucleus of a Eukaryotic Cell 13.1. The Nuclear Envelope
Supporting Information. SI Material and Methods
 Supporting Information SI Material and Methods RNA extraction and qrt-pcr RNA extractions were done using the classical phenol-chloroform method. Total RNA samples were treated with Turbo DNA-free (Ambion)
Supporting Information SI Material and Methods RNA extraction and qrt-pcr RNA extractions were done using the classical phenol-chloroform method. Total RNA samples were treated with Turbo DNA-free (Ambion)
An investigation of the role of integrin alpha-6 in human induced pluripotent stem cell development and pluripotency
 An investigation of the role of integrin alpha-6 in human induced pluripotent stem cell development and pluripotency Submitted by Genna Wilber Biology and English To The Honors College Oakland University
An investigation of the role of integrin alpha-6 in human induced pluripotent stem cell development and pluripotency Submitted by Genna Wilber Biology and English To The Honors College Oakland University
Stem Cell Services. Driving Innovation for Stem Cell Researchers
 Driving Innovation for Stem Cell Researchers Stem Cell Services Partner with us and have access to the most advanced and comprehensive stem cell services available today. 675 W. Kendall St. Cambridge,
Driving Innovation for Stem Cell Researchers Stem Cell Services Partner with us and have access to the most advanced and comprehensive stem cell services available today. 675 W. Kendall St. Cambridge,
7.06 Problem Set #3, Spring 2005
 7.06 Problem Set #3, Spring 2005 1. The Drosophila compound eye is composed of about 800 units called ommatidia. Each ommatidium contains eight photoreceptor neurons (R1 through R8), which develop in a
7.06 Problem Set #3, Spring 2005 1. The Drosophila compound eye is composed of about 800 units called ommatidia. Each ommatidium contains eight photoreceptor neurons (R1 through R8), which develop in a
To isolate single GNS 144 cell clones, cells were plated at a density of 1cell/well
 Supplemental Information: Supplemental Methods: Cell culture To isolate single GNS 144 cell clones, cells were plated at a density of 1cell/well in 96 well Primaria plates in GNS media and incubated at
Supplemental Information: Supplemental Methods: Cell culture To isolate single GNS 144 cell clones, cells were plated at a density of 1cell/well in 96 well Primaria plates in GNS media and incubated at
Nucleic acids deoxyribonucleic acid (DNA) ribonucleic acid (RNA) nucleotide
 Nucleic Acids Nucleic acids are molecules that store information for cellular growth and reproduction There are two types of nucleic acids: - deoxyribonucleic acid (DNA) and ribonucleic acid (RNA) These
Nucleic Acids Nucleic acids are molecules that store information for cellular growth and reproduction There are two types of nucleic acids: - deoxyribonucleic acid (DNA) and ribonucleic acid (RNA) These
Microarrays and Stem Cells
 Microarray Background Information Microarrays and Stem Cells Stem cells are the building blocks that allow the body to produce new cells and repair tissues. Scientists are actively investigating the potential
Microarray Background Information Microarrays and Stem Cells Stem cells are the building blocks that allow the body to produce new cells and repair tissues. Scientists are actively investigating the potential
Supplementary Information
 Supplementary Information Supplementary Figure 1. ZBTB20 expression in the developing DRG. ZBTB20 expression in the developing DRG was detected by immunohistochemistry using anti-zbtb20 antibody 9A10 on
Supplementary Information Supplementary Figure 1. ZBTB20 expression in the developing DRG. ZBTB20 expression in the developing DRG was detected by immunohistochemistry using anti-zbtb20 antibody 9A10 on
Thyroid peroxidase gene expression is induced by lipopolysaccharide involving Nuclear Factor (NF)-κB p65 subunit phosphorylation
 1 2 3 4 5 SUPPLEMENTAL DATA Thyroid peroxidase gene expression is induced by lipopolysaccharide involving Nuclear Factor (NF)-κB p65 subunit phosphorylation Magalí Nazar, Juan Pablo Nicola, María Laura
1 2 3 4 5 SUPPLEMENTAL DATA Thyroid peroxidase gene expression is induced by lipopolysaccharide involving Nuclear Factor (NF)-κB p65 subunit phosphorylation Magalí Nazar, Juan Pablo Nicola, María Laura
Roche Molecular Biochemicals Technical Note No. LC 10/2000
 Roche Molecular Biochemicals Technical Note No. LC 10/2000 LightCycler Overview of LightCycler Quantification Methods 1. General Introduction Introduction Content Definitions This Technical Note will introduce
Roche Molecular Biochemicals Technical Note No. LC 10/2000 LightCycler Overview of LightCycler Quantification Methods 1. General Introduction Introduction Content Definitions This Technical Note will introduce
Supplemental Information. PARP1 Represses PAP and Inhibits Polyadenylation during Heat Shock
 Molecular Cell, Volume 49 Supplemental Information PARP1 Represses PAP and Inhibits Polyadenylation during Heat Shock Dafne Campigli Di Giammartino, Yongsheng Shi, and James L. Manley Supplemental Information
Molecular Cell, Volume 49 Supplemental Information PARP1 Represses PAP and Inhibits Polyadenylation during Heat Shock Dafne Campigli Di Giammartino, Yongsheng Shi, and James L. Manley Supplemental Information
sirna Transfection Into Primary Neurons Using Fuse-It-siRNA
 sirna Transfection Into Primary Neurons Using Fuse-It-siRNA This Application Note describes a protocol for sirna transfection into sensitive, primary cortical neurons using Fuse-It-siRNA. This innovative
sirna Transfection Into Primary Neurons Using Fuse-It-siRNA This Application Note describes a protocol for sirna transfection into sensitive, primary cortical neurons using Fuse-It-siRNA. This innovative
Chapter 8 Lecture Outline. Transcription, Translation, and Bioinformatics
 Chapter 8 Lecture Outline Transcription, Translation, and Bioinformatics Replication, Transcription, Translation n Repetitive processes Build polymers of nucleotides or amino acids n All have 3 major steps
Chapter 8 Lecture Outline Transcription, Translation, and Bioinformatics Replication, Transcription, Translation n Repetitive processes Build polymers of nucleotides or amino acids n All have 3 major steps
SANTA CRUZ BIOTECHNOLOGY, INC.
 TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
8/21/2014. From Gene to Protein
 From Gene to Protein Chapter 17 Objectives Describe the contributions made by Garrod, Beadle, and Tatum to our understanding of the relationship between genes and enzymes Briefly explain how information
From Gene to Protein Chapter 17 Objectives Describe the contributions made by Garrod, Beadle, and Tatum to our understanding of the relationship between genes and enzymes Briefly explain how information
Supplementary Figure 1: MYCER protein expressed from the transgene can enhance
 Relative luciferase activity Relative luciferase activity MYC is a critical target FBXW7 MYC Supplementary is a critical Figures target 1-7. FBXW7 Supplementary Material A E-box sequences 1 2 3 4 5 6 HSV-TK
Relative luciferase activity Relative luciferase activity MYC is a critical target FBXW7 MYC Supplementary is a critical Figures target 1-7. FBXW7 Supplementary Material A E-box sequences 1 2 3 4 5 6 HSV-TK
Bio11 Announcements. Ch 21: DNA Biology and Technology. DNA Functions. DNA and RNA Structure. How do DNA and RNA differ? What are genes?
 Bio11 Announcements TODAY Genetics (review) and quiz (CP #4) Structure and function of DNA Extra credit due today Next week in lab: Case study presentations Following week: Lab Quiz 2 Ch 21: DNA Biology
Bio11 Announcements TODAY Genetics (review) and quiz (CP #4) Structure and function of DNA Extra credit due today Next week in lab: Case study presentations Following week: Lab Quiz 2 Ch 21: DNA Biology
MOLECULAR BIOLOGY OF EUKARYOTES 2016 SYLLABUS
 03-442 Lectures: MWF 9:30-10:20 a.m. Doherty Hall 2105 03-742 Advanced Discussion Section: Time and place to be announced Probably Mon 4-6 p.m. or 6-8p.m.? Once we establish who is taking the advanced
03-442 Lectures: MWF 9:30-10:20 a.m. Doherty Hall 2105 03-742 Advanced Discussion Section: Time and place to be announced Probably Mon 4-6 p.m. or 6-8p.m.? Once we establish who is taking the advanced
Supplementary Figure 1 Activated B cells are subdivided into three groups
 Supplementary Figure 1 Activated B cells are subdivided into three groups according to mitochondrial status (a) Flow cytometric analysis of mitochondrial status monitored by MitoTracker staining or differentiation
Supplementary Figure 1 Activated B cells are subdivided into three groups according to mitochondrial status (a) Flow cytometric analysis of mitochondrial status monitored by MitoTracker staining or differentiation
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1: Vector maps of TRMPV and TRMPVIR variants. Many derivatives of TRMPV have been generated and tested. Unless otherwise noted, experiments in this paper use
Supplementary Figure 1 Supplementary Figure 1: Vector maps of TRMPV and TRMPVIR variants. Many derivatives of TRMPV have been generated and tested. Unless otherwise noted, experiments in this paper use
RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by. 5 AGACACAAACACCAUUGUCACACUCCACAGC; Rand-2 OMe,
 Materials and methods Oligonucleotides and DNA constructs RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by Dharmacon Inc. (Lafayette, CO). The sequences were: 122-2 OMe, 5
Materials and methods Oligonucleotides and DNA constructs RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by Dharmacon Inc. (Lafayette, CO). The sequences were: 122-2 OMe, 5
Nature Structural and Molecular Biology: doi: /nsmb.2959
 Supplementary Figure 1 EIciNAs were pulled down with an antibody to Pol II. (a) Western blot showing that pol II was efficiently pulled down with a pol II antibody in HeLa cell lysates. (b) The enrichment
Supplementary Figure 1 EIciNAs were pulled down with an antibody to Pol II. (a) Western blot showing that pol II was efficiently pulled down with a pol II antibody in HeLa cell lysates. (b) The enrichment
Regulation of cellular differentiation by tissue specific transcription factor
 Regulation of cellular differentiation by tissue specific transcription factor T. Endoh, M. Mie, Y. Yanagida, M. Aizawa, E. Kobatake Department of Biological Information, Graduate School of Bioscience
Regulation of cellular differentiation by tissue specific transcription factor T. Endoh, M. Mie, Y. Yanagida, M. Aizawa, E. Kobatake Department of Biological Information, Graduate School of Bioscience
7.22 Example Problems for Exam 1 The exam will be of this format. It will consist of 2-3 sets scenarios.
 Massachusetts Institute of Technology Department of Biology 7.22, Fall 2005 - Developmental Biology Instructors: Professor Hazel Sive, Professor Martha Constantine-Paton 1 of 10 7.22 Fall 2005 sample exam
Massachusetts Institute of Technology Department of Biology 7.22, Fall 2005 - Developmental Biology Instructors: Professor Hazel Sive, Professor Martha Constantine-Paton 1 of 10 7.22 Fall 2005 sample exam
CHAPTERS , 17: Eukaryotic Genetics
 CHAPTERS 14.1 14.6, 17: Eukaryotic Genetics 1. Review the levels of DNA packing within the eukaryote nucleus. Label each level. (A similar diagram is on pg 188 of your textbook.) 2. How do the coding regions
CHAPTERS 14.1 14.6, 17: Eukaryotic Genetics 1. Review the levels of DNA packing within the eukaryote nucleus. Label each level. (A similar diagram is on pg 188 of your textbook.) 2. How do the coding regions
Multiple choice questions (numbers in brackets indicate the number of correct answers)
 1 Multiple choice questions (numbers in brackets indicate the number of correct answers) February 1, 2013 1. Ribose is found in Nucleic acids Proteins Lipids RNA DNA (2) 2. Most RNA in cells is transfer
1 Multiple choice questions (numbers in brackets indicate the number of correct answers) February 1, 2013 1. Ribose is found in Nucleic acids Proteins Lipids RNA DNA (2) 2. Most RNA in cells is transfer
Confocal immunofluorescence microscopy
 Confocal immunofluorescence microscopy HL-6 and cells were cultured and cytospun onto glass slides. The cells were double immunofluorescence stained for Mt NPM1 and fibrillarin (nucleolar marker). Briefly,
Confocal immunofluorescence microscopy HL-6 and cells were cultured and cytospun onto glass slides. The cells were double immunofluorescence stained for Mt NPM1 and fibrillarin (nucleolar marker). Briefly,
Solution Key Problem Set
 Solution Key- 7.013 Problem Set 5-2013 Question 1 During a summer hike you suddenly spot a huge grizzly bear. This emergency situation triggers a fight or flight response through a signaling pathway as
Solution Key- 7.013 Problem Set 5-2013 Question 1 During a summer hike you suddenly spot a huge grizzly bear. This emergency situation triggers a fight or flight response through a signaling pathway as
Modeling Cardiac Hypertrophy: Endothelin-1 Induction with qrt-pcr Analysis
 icell Cardiomyocytes Application Protocol Modeling Cardiac Hypertrophy: Endothelin-1 Induction with qrt-pcr Analysis Introduction Cardiac hypertrophy is characterized by several different cellular changes,
icell Cardiomyocytes Application Protocol Modeling Cardiac Hypertrophy: Endothelin-1 Induction with qrt-pcr Analysis Introduction Cardiac hypertrophy is characterized by several different cellular changes,
BEADLE & TATUM EXPERIMENT
 FROM DNA TO PROTEINS: gene expression Chapter 14 LECTURE OBJECTIVES What Is the Evidence that Genes Code for Proteins? How Does Information Flow from Genes to Proteins? How Is the Information Content in
FROM DNA TO PROTEINS: gene expression Chapter 14 LECTURE OBJECTIVES What Is the Evidence that Genes Code for Proteins? How Does Information Flow from Genes to Proteins? How Is the Information Content in
supplementary information
 DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
RNA-Guided Gene Activation by CRISPR-Cas9-Based Transcription Factors
 Supplementary Information RNA-Guided Gene Activation by CRISPR-Cas9-Based Transcription Factors Pablo Perez-Pinera 1, Daniel D. Kocak 1, Christopher M. Vockley 2,3, Andrew F. Adler 1, Ami M. Kabadi 1,
Supplementary Information RNA-Guided Gene Activation by CRISPR-Cas9-Based Transcription Factors Pablo Perez-Pinera 1, Daniel D. Kocak 1, Christopher M. Vockley 2,3, Andrew F. Adler 1, Ami M. Kabadi 1,
Nature Neuroscience: doi: /nn Supplementary Figure 1
 Supplementary Figure 1 PCR-genotyping of the three mouse models used in this study and controls for behavioral experiments after semi-chronic Pten inhibition. a-c. DNA from App/Psen1 (a), Pten tg (b) and
Supplementary Figure 1 PCR-genotyping of the three mouse models used in this study and controls for behavioral experiments after semi-chronic Pten inhibition. a-c. DNA from App/Psen1 (a), Pten tg (b) and
Figure S1. Figure S2. Figure S3 HB Anti-FSP27 (COOH-terminal peptide) Ab. Anti-GST-FSP27(45-127) Ab.
 / 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
/ 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
Nature Immunology: doi: /ni Supplementary Figure 1. Zranb1 gene targeting.
 Supplementary Figure 1 Zranb1 gene targeting. (a) Schematic picture of Zranb1 gene targeting using an FRT-LoxP vector, showing the first 6 exons of Zranb1 gene (exons 7-9 are not shown). Targeted mice
Supplementary Figure 1 Zranb1 gene targeting. (a) Schematic picture of Zranb1 gene targeting using an FRT-LoxP vector, showing the first 6 exons of Zranb1 gene (exons 7-9 are not shown). Targeted mice
AP Biology Gene Expression/Biotechnology REVIEW
 AP Biology Gene Expression/Biotechnology REVIEW Multiple Choice Identify the choice that best completes the statement or answers the question. 1. Gene expression can be a. regulated before transcription.
AP Biology Gene Expression/Biotechnology REVIEW Multiple Choice Identify the choice that best completes the statement or answers the question. 1. Gene expression can be a. regulated before transcription.
A15871 A15872 A15876 A15870
 TaqMan hpsc Scorecard Kits TaqMan hpsc Scorecard Panels A15871 A15872 A15876 A15870 Product Qty Cat. No. TaqMan hpsc Scorecard Kit 2 x 96w FAST 1 kit A15871 TaqMan hpsc Scorecard Kit 384w 1 kit A15872
TaqMan hpsc Scorecard Kits TaqMan hpsc Scorecard Panels A15871 A15872 A15876 A15870 Product Qty Cat. No. TaqMan hpsc Scorecard Kit 2 x 96w FAST 1 kit A15871 TaqMan hpsc Scorecard Kit 384w 1 kit A15872
Chapter 18: Regulation of Gene Expression. 1. Gene Regulation in Bacteria 2. Gene Regulation in Eukaryotes 3. Gene Regulation & Cancer
 Chapter 18: Regulation of Gene Expression 1. Gene Regulation in Bacteria 2. Gene Regulation in Eukaryotes 3. Gene Regulation & Cancer Gene Regulation Gene regulation refers to all aspects of controlling
Chapter 18: Regulation of Gene Expression 1. Gene Regulation in Bacteria 2. Gene Regulation in Eukaryotes 3. Gene Regulation & Cancer Gene Regulation Gene regulation refers to all aspects of controlling
Transcriptional Regulation in Eukaryotes
 Transcriptional Regulation in Eukaryotes Concepts, Strategies, and Techniques Michael Carey Stephen T. Smale COLD SPRING HARBOR LABORATORY PRESS NEW YORK 2000 Cold Spring Harbor Laboratory Press, 0-87969-537-4
Transcriptional Regulation in Eukaryotes Concepts, Strategies, and Techniques Michael Carey Stephen T. Smale COLD SPRING HARBOR LABORATORY PRESS NEW YORK 2000 Cold Spring Harbor Laboratory Press, 0-87969-537-4
RNA Polymerase III Subunit POLR3G Regulates Specific Subsets of
 Stem Cell Reports, Volume 8 Supplemental Information RNA Polymerase III Subunit POLR3G Regulates Specific Subsets of PolyA + and SmallRNA Transcriptomes and Splicing in Human Pluripotent Stem Cells Riikka
Stem Cell Reports, Volume 8 Supplemental Information RNA Polymerase III Subunit POLR3G Regulates Specific Subsets of PolyA + and SmallRNA Transcriptomes and Splicing in Human Pluripotent Stem Cells Riikka
36. The double bonds in naturally-occuring fatty acids are usually isomers. A. cis B. trans C. both cis and trans D. D- E. L-
 36. The double bonds in naturally-occuring fatty acids are usually isomers. A. cis B. trans C. both cis and trans D. D- E. L- 37. The essential fatty acids are A. palmitic acid B. linoleic acid C. linolenic
36. The double bonds in naturally-occuring fatty acids are usually isomers. A. cis B. trans C. both cis and trans D. D- E. L- 37. The essential fatty acids are A. palmitic acid B. linoleic acid C. linolenic
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature11112 Figure 1 Salient features of the m 6 A methylome. a, m 6 A punctuates RNA molecules mainly around the stop codon and in unusually long internal exons.
SUPPLEMENTARY INFORMATION doi:10.1038/nature11112 Figure 1 Salient features of the m 6 A methylome. a, m 6 A punctuates RNA molecules mainly around the stop codon and in unusually long internal exons.
SUPPLEMENTAL MATERIALS
 SUPPLEMENL MERILS Eh-seq: RISPR epitope tagging hip-seq of DN-binding proteins Daniel Savic, E. hristopher Partridge, Kimberly M. Newberry, Sophia. Smith, Sarah K. Meadows, rian S. Roberts, Mark Mackiewicz,
SUPPLEMENL MERILS Eh-seq: RISPR epitope tagging hip-seq of DN-binding proteins Daniel Savic, E. hristopher Partridge, Kimberly M. Newberry, Sophia. Smith, Sarah K. Meadows, rian S. Roberts, Mark Mackiewicz,
Cytomics in Action: Cytokine Network Cytometry
 Cytomics in Action: Cytokine Network Cytometry Jonni S. Moore, Ph.D. Director, Clinical and Research Flow Cytometry and PathBioResource Associate Professor of Pathology & Laboratory Medicine University
Cytomics in Action: Cytokine Network Cytometry Jonni S. Moore, Ph.D. Director, Clinical and Research Flow Cytometry and PathBioResource Associate Professor of Pathology & Laboratory Medicine University
A novel two-step genome editing strategy with CRISPR-Cas9 provides new insights into telomerase action and TERT gene expression
 Xi et al. Genome Biology (2015) 16:231 DOI 10.1186/s13059-015-0791-1 RESEARCH A novel two-step genome editing strategy with CRISPR-Cas9 provides new insights into telomerase action and TERT gene expression
Xi et al. Genome Biology (2015) 16:231 DOI 10.1186/s13059-015-0791-1 RESEARCH A novel two-step genome editing strategy with CRISPR-Cas9 provides new insights into telomerase action and TERT gene expression
ProtoScript. First Strand cdna Synthesis Kit DNA AMPLIFICATION & PCR. Instruction Manual. NEB #E6300S/L 30/150 reactions Version 2.
 DNA AMPLIFICATION & PCR ProtoScript First Strand cdna Synthesis Kit Instruction Manual NEB #E6300S/L 30/150 reactions Version 2.2 11/16 be INSPIRED drive DISCOVERY stay GENUINE This product is intended
DNA AMPLIFICATION & PCR ProtoScript First Strand cdna Synthesis Kit Instruction Manual NEB #E6300S/L 30/150 reactions Version 2.2 11/16 be INSPIRED drive DISCOVERY stay GENUINE This product is intended
Gene Expression and Heritable Phenotype. CBS520 Eric Nabity
 Gene Expression and Heritable Phenotype CBS520 Eric Nabity DNA is Just the Beginning DNA was determined to be the genetic material, and the structure was identified as a (double stranded) double helix.
Gene Expression and Heritable Phenotype CBS520 Eric Nabity DNA is Just the Beginning DNA was determined to be the genetic material, and the structure was identified as a (double stranded) double helix.
Chapter 15 Gene Technologies and Human Applications
 Chapter Outline Chapter 15 Gene Technologies and Human Applications Section 1: The Human Genome KEY IDEAS > Why is the Human Genome Project so important? > How do genomics and gene technologies affect
Chapter Outline Chapter 15 Gene Technologies and Human Applications Section 1: The Human Genome KEY IDEAS > Why is the Human Genome Project so important? > How do genomics and gene technologies affect
Division Ave. High School AP Biology
 Control of Eukaryotic Genes 2007-2008 The BIG Questions n How are genes turned on & off in eukaryotes? n How do cells with the same genes differentiate to perform completely different, specialized functions?
Control of Eukaryotic Genes 2007-2008 The BIG Questions n How are genes turned on & off in eukaryotes? n How do cells with the same genes differentiate to perform completely different, specialized functions?
Replication Review. 1. What is DNA Replication? 2. Where does DNA Replication take place in eukaryotic cells?
 Replication Review 1. What is DNA Replication? 2. Where does DNA Replication take place in eukaryotic cells? 3. Where does DNA Replication take place in the cell cycle? 4. 4. What guides DNA Replication?
Replication Review 1. What is DNA Replication? 2. Where does DNA Replication take place in eukaryotic cells? 3. Where does DNA Replication take place in the cell cycle? 4. 4. What guides DNA Replication?
Concepts and Methods in Developmental Biology
 Biology 4361 Developmental Biology Concepts and Methods in Developmental Biology June 16, 2009 Conceptual and Methodological Tools Concepts Genomic equivalence Differential gene expression Differentiation/de-differentiation
Biology 4361 Developmental Biology Concepts and Methods in Developmental Biology June 16, 2009 Conceptual and Methodological Tools Concepts Genomic equivalence Differential gene expression Differentiation/de-differentiation
Contents... vii. List of Figures... xii. List of Tables... xiv. Abbreviatons... xv. Summary... xvii. 1. Introduction In vitro evolution...
 vii Contents Contents... vii List of Figures... xii List of Tables... xiv Abbreviatons... xv Summary... xvii 1. Introduction...1 1.1 In vitro evolution... 1 1.2 Phage Display Technology... 3 1.3 Cell surface
vii Contents Contents... vii List of Figures... xii List of Tables... xiv Abbreviatons... xv Summary... xvii 1. Introduction...1 1.1 In vitro evolution... 1 1.2 Phage Display Technology... 3 1.3 Cell surface
TRANSGENIC TECHNOLOGIES: Gene-targeting
 TRANSGENIC TECHNOLOGIES: Gene-targeting Reverse Genetics Wild-type Bmp7 -/- Forward Genetics Phenotype Gene or Mutations First Molecular Analysis Second Reverse Genetics Gene Phenotype or Molecular Analysis
TRANSGENIC TECHNOLOGIES: Gene-targeting Reverse Genetics Wild-type Bmp7 -/- Forward Genetics Phenotype Gene or Mutations First Molecular Analysis Second Reverse Genetics Gene Phenotype or Molecular Analysis
Do you remember. What is a gene? What is RNA? How does it differ from DNA? What is protein?
 Lesson 1 - RNA Do you remember What is a gene? What is RNA? How does it differ from DNA? What is protein? Gene Segment of DNA that codes for building a protein DNA code is copied into RNA form, and RNA
Lesson 1 - RNA Do you remember What is a gene? What is RNA? How does it differ from DNA? What is protein? Gene Segment of DNA that codes for building a protein DNA code is copied into RNA form, and RNA
GENE REGULATION slide shows by Kim Foglia modified Slides with blue edges are Kim s
 GENE REGULATION slide shows by Kim Foglia modified Slides with blue edges are Kim s 2007-2008 Bacterial metabolism Bacteria need to respond quickly to changes in their environment STOP GO if they have
GENE REGULATION slide shows by Kim Foglia modified Slides with blue edges are Kim s 2007-2008 Bacterial metabolism Bacteria need to respond quickly to changes in their environment STOP GO if they have
Figure S1. Purity of primary cultures of renal proximal tubular epithelial culture ascertained by cytokeratin staining.
 Supplementary information Supplementary figures Figure S1. Purity of primary cultures of renal proximal tubular epithelial culture ascertained by cytokeratin staining. Figure S2. Induction of Nur77 in
Supplementary information Supplementary figures Figure S1. Purity of primary cultures of renal proximal tubular epithelial culture ascertained by cytokeratin staining. Figure S2. Induction of Nur77 in
Isolation and Analysis of Pluripotent, Neural, and Hematopoietic Stem Cells
 Isolation and Analysis of Pluripotent, Neural, and Hematopoietic Stem Cells Christian Carson BD Biosciences R&D Scientist Stem Cell 23-10679-00 Overview Introduction Challenges in stem cell research Antibody
Isolation and Analysis of Pluripotent, Neural, and Hematopoietic Stem Cells Christian Carson BD Biosciences R&D Scientist Stem Cell 23-10679-00 Overview Introduction Challenges in stem cell research Antibody
Transcription Eukaryotic Cells
 Transcription Eukaryotic Cells Packet #20 1 Introduction Transcription is the process in which genetic information, stored in a strand of DNA (gene), is copied into a strand of RNA. Protein-encoding genes
Transcription Eukaryotic Cells Packet #20 1 Introduction Transcription is the process in which genetic information, stored in a strand of DNA (gene), is copied into a strand of RNA. Protein-encoding genes
Cortical Neural Induction Kit. Protocol version 1.0
 Cortical Neural Induction Kit Protocol version 1.0 Protocol version 1.0 Table of Contents Product Information 2 Preparation of Reagents 3 Protocol Overview 4 Seeding ipscs 4 Cortical Neural Induction
Cortical Neural Induction Kit Protocol version 1.0 Protocol version 1.0 Table of Contents Product Information 2 Preparation of Reagents 3 Protocol Overview 4 Seeding ipscs 4 Cortical Neural Induction
ADVANCED MEDIA TECHNOLOGY
 ADVANCED MEDIA TECHNOLOGY For Human ES and ips Cell Research The right medium makes all the difference in ensuring successful ES and ips cell culture. Stemgent offers high-quality, chemically-defined,
ADVANCED MEDIA TECHNOLOGY For Human ES and ips Cell Research The right medium makes all the difference in ensuring successful ES and ips cell culture. Stemgent offers high-quality, chemically-defined,
Recapitulation of spinal motor neuron-specific disease phenotypes in a human cell model of spinal muscular atrophy
 npg Modeling spinal muscular atrophy in human neurons 378 ORIGINAL ARTICLE Cell Research (2013) 23:378-393. 2013 IBCB, SIBS, CAS All rights reserved 1001-0602/13 $ 32.00 www.nature.com/cr npg Recapitulation
npg Modeling spinal muscular atrophy in human neurons 378 ORIGINAL ARTICLE Cell Research (2013) 23:378-393. 2013 IBCB, SIBS, CAS All rights reserved 1001-0602/13 $ 32.00 www.nature.com/cr npg Recapitulation
Contents. The Right Surface for Every Cell Extracellular Matrices and Biologically Coated Surfaces ECM Mimetic and Advanced Surfaces...
 Contents The Right Surface for Every Cell... 1 Extracellular Matrices and Biologically Coated Surfaces... 2 Corning Matrigel Matrix... 2 Corning BioCoat Cultureware... 3 ECM Mimetic and Advanced Surfaces...
Contents The Right Surface for Every Cell... 1 Extracellular Matrices and Biologically Coated Surfaces... 2 Corning Matrigel Matrix... 2 Corning BioCoat Cultureware... 3 ECM Mimetic and Advanced Surfaces...
SureSilencing sirna Array Technology Overview
 SureSilencing sirna Array Technology Overview Pathway-Focused sirna-based RNA Interference Topics to be Covered Who is SuperArray? Brief Introduction to RNA Interference Challenges Facing RNA Interference
SureSilencing sirna Array Technology Overview Pathway-Focused sirna-based RNA Interference Topics to be Covered Who is SuperArray? Brief Introduction to RNA Interference Challenges Facing RNA Interference
Tumor Growth Suppression Through the Activation of p21, a Cyclin-Dependent Kinase Inhibitor
 Tumor Growth Suppression Through the Activation of p21, a Cyclin-Dependent Kinase Inhibitor Nicholas Love 11/28/01 A. What is p21? Introduction - p21 is a gene found on chromosome 6 at 6p21.2 - this gene
Tumor Growth Suppression Through the Activation of p21, a Cyclin-Dependent Kinase Inhibitor Nicholas Love 11/28/01 A. What is p21? Introduction - p21 is a gene found on chromosome 6 at 6p21.2 - this gene
RNA synthesis/transcription I Biochemistry 302. February 6, 2004 Bob Kelm
 RNA synthesis/transcription I Biochemistry 302 February 6, 2004 Bob Kelm Overview of RNA classes Messenger RNA (mrna) Encodes protein Relatively short half-life ( 3 min in E. coli, 30 min in eukaryotic
RNA synthesis/transcription I Biochemistry 302 February 6, 2004 Bob Kelm Overview of RNA classes Messenger RNA (mrna) Encodes protein Relatively short half-life ( 3 min in E. coli, 30 min in eukaryotic
IMMUNOGLOBULIN GENES UNDERGO TWO DNA REARRANGEMENTS
 A Prototype Ig Gene: Murine Kappa About 10 0 V κ gene segments 4 J Gene Segment s 1 C κ Gene Segmen t Multiple V gene segments, distant from J and C A few J gene segments One C gene segment GERMLINE Ig
A Prototype Ig Gene: Murine Kappa About 10 0 V κ gene segments 4 J Gene Segment s 1 C κ Gene Segmen t Multiple V gene segments, distant from J and C A few J gene segments One C gene segment GERMLINE Ig
ENCODE DCC Antibody Validation Document
 ENCODE DCC Antibody Validation Document Date of Submission 09/12/12 Name: Trupti Kawli Email: trupti@stanford.edu Lab Snyder Antibody Name: SREBP1 (sc-8984) Target: SREBP1 Company/ Source: Santa Cruz Biotechnology
ENCODE DCC Antibody Validation Document Date of Submission 09/12/12 Name: Trupti Kawli Email: trupti@stanford.edu Lab Snyder Antibody Name: SREBP1 (sc-8984) Target: SREBP1 Company/ Source: Santa Cruz Biotechnology
DNA is the genetic material. DNA structure. Chapter 7: DNA Replication, Transcription & Translation; Mutations & Ames test
 DNA is the genetic material Chapter 7: DNA Replication, Transcription & Translation; Mutations & Ames test Dr. Amy Rogers Bio 139 General Microbiology Hereditary information is carried by DNA Griffith/Avery
DNA is the genetic material Chapter 7: DNA Replication, Transcription & Translation; Mutations & Ames test Dr. Amy Rogers Bio 139 General Microbiology Hereditary information is carried by DNA Griffith/Avery
Neural Induction. Chapter One
 Neural Induction Chapter One Fertilization Development of the Nervous System Cleavage (Blastula, Gastrula) Neuronal Induction- Neuroblast Formation Cell Migration Mesodermal Induction Lateral Inhibition
Neural Induction Chapter One Fertilization Development of the Nervous System Cleavage (Blastula, Gastrula) Neuronal Induction- Neuroblast Formation Cell Migration Mesodermal Induction Lateral Inhibition
Supplemental Information. Antagonistic Activities of Sox2. and Brachyury Control the Fate Choice. of Neuro-Mesodermal Progenitors
 Developmental Cell, Volume 42 Supplemental Information Antagonistic Activities of Sox2 and Brachyury Control the Fate Choice of Neuro-Mesodermal Progenitors Frederic Koch, Manuela Scholze, Lars Wittler,
Developmental Cell, Volume 42 Supplemental Information Antagonistic Activities of Sox2 and Brachyury Control the Fate Choice of Neuro-Mesodermal Progenitors Frederic Koch, Manuela Scholze, Lars Wittler,
