Structure, evolution and function of the bi-directionally transcribed iab4 microrna locus in insects
|
|
|
- Marilyn Armstrong
- 6 years ago
- Views:
Transcription
1 Supplementary data Structure, evolution and function of the bi-directionally transcribed iab4 microrna locus in insects Jerome H.L. Hui, Antonio Marco, Suzanne Hunt, Janet Melling, Sam Griffiths-Jones, Matthew Ronshaugen
2 Contents Table S1. Primers sequences for making different constructs as described in the main text. Table S2. Primers sequences for introducing mutagensis on the 3 UTRs. The residues highlighted in red are the sequences changed from the wild-type. Table S3. Statistical tests to analyse whether the overlapping of iab-4 and iab-8 binding sites is higher than expected by chance (for details, see Materials and Methods in the main text). Table S4. Statistical tests to analyse whether the overlapping of mir-307a and mir-307b binding sites is higher than expected by chance (for details, see Materials and Methods in the main text). Figure S1. Arm-specific mirna processing in Drosophila S2 cells. Tribolium iab4/iab-8 constructs were transfected into S2 cells and expressed under the control of the actin promoter. LNA mirna real-time PCR assays were used to estimate 5 and 3 mirna arm levels. Figure S2. Expression patterns and iab-4/8 targeted sites of A-C) tca-cx/scr, D-F) tcaptl/antp, and G-H) tca-abdb. Note that the expression domains of these genes follow the rules of spatial and temporal colinearity according their positions within the intact Tribolium Hox cluster. I) Relative luficerase activity of the other Hox 3 UTRs and mutagenated 3 UTRs (M) repressed by Tribolium/and Drosophila iab-4 and iab-8. Note the presence of repression is indicated in negative value. Figure S3. Putative 3'UTRs of Tribolium castaneum HOX genes.
3 Table S1. Primers sequences for making the different constructs. Target (vector) Primers tca-iab-4 (pac5.1) FP-KpnI;5'-TTTGGGTACCCATACTGCTCCCGAGGGAACGAGGGATTACTTTTAT-3' RP-EcoRI;5'-CAGGGAATTCCATTGTTAAAAAGCGTCAAGACTTAGCTGGAGT-3' tca-iab-8 (pac5.1) FP-KpnI;5'-TTTGGGTACCCGTTGAGTGTGTTTTACATCGATTAACTGATAT-3' RP-EcoRI;5'-CAGGGAATTCCATACTGCTCCCGAGGGAACGAGGGATTACTTTTAT-3' tca-ubx-3'utr (psicheck2) FP-XhoI;5'-CCCCTCGAGGGATTACTTCGGTGCTGTTGGCGGATCAGCATAA-3' RP-NotI;5'-GGGGCGGCCGCCCGAAAAACCATTAGCCACGTTACACTCCCAAA-3' tca-abda-3'utr (psicheck2) FP-XhoI;5'-TCCTCGAGCGAGTGACTTGACCTTTTGTTTTATAC-3' RP-NotI;5'-TAAGGCGGCCGCTGGCGTATACGTATTCATTTTCTTTAC-3' tca-scr-3'utr (psicheck2) FP-XhoI;5'-ACCTCGAGGACGTGTTTATGTGATTCCATTAGA-3' RP-NotI;5'-AAGCGGCCGCCTAAATTGTACGTCTTTCACACACAC-3' tca-abdb-3'utr (psicheck2) FP-XhoI;5'-ACCTCGAGCAGTGCCAAAGAGTTATTTAGGAAA-3' RP-NotI;5'-AAGCGGCCGCGTTAAAGACCGCGTTAACTATTCTGT-3' tca-antp/ptl-3'utr(psicheck2) FP-XhoI;5'-GTTCCTCGAGACTCCTCGTCACGATAACCATGTACCTAATT-3' RP-NotI;5'-TAAGGCGGCCGCGGTAATTTGTCCGAAATTAAGTGTCGGCCAAACT-3' dme-iab4-s (pac5.1) FP-KpnI;5'-TTGGTACCCATCTTTACATCTGGATATGGTTTCATCT-3' RP-EcoRI;5'-TGGAATTCAAGTGTAAGACCATATAACAAAGTGCTACGT-3' dme-iab4-as (iab-8)(pac5.1) FP-EcoRI;5'-TTGGAATTCCATCTTTACATCTGGATATGGTTTCATCT-3' RP-KpnI;5'-TTGGTACCAAGTGTAAGACCATATAACAAAGTGCTACGT-3' dme-ubx-3'utr (psicheck2) FP-XhoI;5 -ACCCTCGAGGTTGAATGCGAGCAAAATTCTGTTGATA-3' RP-NotI;5'-ATTTAAAAGGCGGCCGCTTCACTCTAGTCGTTTGGGCGAGAA-3 dme-abda-3'utr (psicheck2) FP-XhoI;5'-TCCTCGAGGATAGCAAGCAGAAGATAAACTCGAT-3' RP-NotI;5'-TTGCGGCCGCAACAGAAGTTTTAGCAGTAGGGTTCA-3' dme-antp-3'utr (psicheck2) FP-XhoI;5'-TGTCCTCGAGGATCGACGGAGTCTACCCACTTAAATGAA-3' RP-NotI;5'-ATAGGCGGCCGCTTCCGCTTTGCAGCCCTTTCACTGCATTCT-3' iab-4-5p perfect complementary (psicheck2) FP;5 -GGTTCCCTCGAGTCAGGATACATTCAGTATACGTCCCGGGAATTCGTTTAAACC -3 RP;5 -TATACGGTCCCCATGGAGC -3 iab-4-3p perfect complementary (psicheck2) FP;5 -GGTTCCCTCGAGTTACGTATACTGAAGGTATACCGCCCGGGAATTCGTTTAAACC-3 RP;5 -TATACGGTCCCCATGGAGC-3 iab-8-5p perfect complementary (psicheck2) FP;5 -GGTTCCCTCGAGGGTATACCTTCAGTATACGTAACCCGGGAATTCGTTTAAACC-3 RP;5 -TATACGGTCCCCATGGAGC-3 iab-8-3p perfect complementary (psicheck2) FP;5 -GGTTCCCTCGAGTAAACGTATACTGAATGTATCCCCCGGGAATTCGTTTAAACC-3 RP;5 -TATACGGTCCCCATGGAGC-3 FP: forward primer; RP: reverse primer. Added restriction site is also indicated.
4 Table S2. Primers used in mutagenesis of 3'UTRs. Target Primers (changes are shown in bold) tca-antp/ptl-3'utr tca-antp-m-fp;5'-cgtaacgctgctacaaatcgagtatcgctaactaattattttaat-3' tca-antp-m-rp; ATTAAAATAATTAGTTAGCGATACTCGATTTGTAGCAGCGTTACG-3' tca-ubx-3'utr tca-ubx-m1;5'-gcgatcgcaactatataaccaattgggtttatgtgcc-3' tca-ubx-mc;5'-ggcacataaacccaattggttatatagttgcgatcgc-3' tca-ubx-m2;5'-ggaaacttaacgatataaccaaattataatcctagta-3' tca-ubx-m2c;5'-tactaggattataatttggttatatcgttaagtttcc-3' tca-ubx-m3;5'-gtacaagaaatgtaataccaaatgtactaattata-3' tca-ubx-m3c;5'-tataattagtacatttggttatacatttcttgtac-3' tca-abd-a-3'utr tca-abda-m1;5'-ctcttagtactcttatcgctaatttggtgtcagggtag-3' tca-abda-m1c;5'-ctaccctgacaccaaattagcgataagagtactaagag-3' tca-abda-m2;5'-gccttctccacatttagatcgctaaaattgtacaatattttc-3' tca-abda-m2c;5'-gaaaatattgtacaattttagcgatctaaatgtggagaaggc-3' tca-abda-m3;5'-gtaaagaaaatgaatcgctatcgcccagcggccgctggccgc-3' tca-abda-m3c;5'-gcggccagcggccgctgggcgatagcgattcattttctttac-3'
5 Table S3. Target overlap between iab-4 and iab-8. P-values for randomization tests. See main text for details. RAND and SIMUL account for two different ways of calculating the null distribution as described in Marco et al. (2012) Silence 3:8 Canonical seeds RAND Canonical seeds SIMUL miranda RAND miranda SIMUL
6 Table S4. Target overlap between mir-307a and mir-307b. P-values for randomization tests. See main text for details. RAND and SIMUL account for two different ways of calculating the null distribution as described in Marco et al. (2012) Silence 3:8 Canonical seeds RAND Canonical seeds SIMUL miranda RAND miranda SIMUL
7 Figure S1.
8 Figure S2.
9 Figure S3. Abd-B abd-a Ptl/Antp Scr Ubx Ubx 3'UTR
SUPPLEMENTARY INFORMATION
 doi:.38/nature899 Supplementary Figure Suzuki et al. a c p7 -/- / WT ratio (+)/(-) p7 -/- / WT ratio Log X 3. Fold change by treatment ( (+)/(-)) Log X.5 3-3. -. b Fold change by treatment ( (+)/(-)) 8
doi:.38/nature899 Supplementary Figure Suzuki et al. a c p7 -/- / WT ratio (+)/(-) p7 -/- / WT ratio Log X 3. Fold change by treatment ( (+)/(-)) Log X.5 3-3. -. b Fold change by treatment ( (+)/(-)) 8
1. Primers for PCR to amplify hairpin stem-loop precursor mir-145 plus different flanking sequence from human genomic DNA.
 Supplemental data: 1. Primers for PCR to amplify hairpin stem-loop precursor mir-145 plus different flanking sequence from human genomic DNA. Strategy#1: 20nt at both sides: #1_BglII-Fd primer : 5 -gga
Supplemental data: 1. Primers for PCR to amplify hairpin stem-loop precursor mir-145 plus different flanking sequence from human genomic DNA. Strategy#1: 20nt at both sides: #1_BglII-Fd primer : 5 -gga
mirnaselect pmir-gfp Reporter System
 Product Data Sheet mirnaselect pmir-gfp Reporter System CATALOG NUMBER: MIR-GFP STORAGE: -80ºC QUANTITY: 100 µl of bacterial glycerol stock Components 1. mirnaselect pmir-gfp Reporter Vector (Part No.
Product Data Sheet mirnaselect pmir-gfp Reporter System CATALOG NUMBER: MIR-GFP STORAGE: -80ºC QUANTITY: 100 µl of bacterial glycerol stock Components 1. mirnaselect pmir-gfp Reporter Vector (Part No.
RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by. 5 AGACACAAACACCAUUGUCACACUCCACAGC; Rand-2 OMe,
 Materials and methods Oligonucleotides and DNA constructs RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by Dharmacon Inc. (Lafayette, CO). The sequences were: 122-2 OMe, 5
Materials and methods Oligonucleotides and DNA constructs RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by Dharmacon Inc. (Lafayette, CO). The sequences were: 122-2 OMe, 5
Comparative Analysis of Argonaute-Dependent Small RNA Pathways in Drosophila
 Molecular Cell, Volume 32 Supplemental Data Comparative Analysis of Argonaute-Dependent Small RNA Pathways in Drosophila Rui Zhou, Ikuko Hotta, Ahmet M. Denli, Pengyu Hong, Norbert Perrimon, and Gregory
Molecular Cell, Volume 32 Supplemental Data Comparative Analysis of Argonaute-Dependent Small RNA Pathways in Drosophila Rui Zhou, Ikuko Hotta, Ahmet M. Denli, Pengyu Hong, Norbert Perrimon, and Gregory
Blimp-1/PRDM1 rabbit monoclonal antibody (C14A4) was purchased from Cell Signaling
 1 2 3 4 5 6 7 8 9 10 11 Supplementary Methods Antibodies Blimp-1/PRDM1 rabbit monoclonal antibody (C14A4) was purchased from Cell Signaling (Danvers, MA) and used at 1:1000 to detect the total PRDM1 protein
1 2 3 4 5 6 7 8 9 10 11 Supplementary Methods Antibodies Blimp-1/PRDM1 rabbit monoclonal antibody (C14A4) was purchased from Cell Signaling (Danvers, MA) and used at 1:1000 to detect the total PRDM1 protein
SUPPLEMENTARY INFORMATION
 Supplementary Figure a T m ( C) Seq. '-3' uguuugugguaacagugugaggu L 62 AttGtcAcaCtcC L2 6 ccattgtcacactcc L3 66 attgtcacactcc 7 ccattgtcacactcca L 73 ccattgtcacactcc L6 74 AttGTcaCaCtCC L7 7 attgtcacactcc
Supplementary Figure a T m ( C) Seq. '-3' uguuugugguaacagugugaggu L 62 AttGtcAcaCtcC L2 6 ccattgtcacactcc L3 66 attgtcacactcc 7 ccattgtcacactcca L 73 ccattgtcacactcc L6 74 AttGTcaCaCtCC L7 7 attgtcacactcc
Functional microrna targets in protein coding sequences. Merve Çakır
 Functional microrna targets in protein coding sequences Martin Reczko, Manolis Maragkakis, Panagiotis Alexiou, Ivo Grosse, Artemis G. Hatzigeorgiou Merve Çakır 27.04.2012 microrna * micrornas are small
Functional microrna targets in protein coding sequences Martin Reczko, Manolis Maragkakis, Panagiotis Alexiou, Ivo Grosse, Artemis G. Hatzigeorgiou Merve Çakır 27.04.2012 microrna * micrornas are small
Structure, evolution and function of the bi-directionally transcribed iab-4/iab-8 microrna locus in arthropods
 3352 3361 Nucleic Acids Research, 2013, Vol. 41, No. 5 Published online 17 January 2013 doi:10.1093/nar/gks1445 Structure, evolution and function of the bi-directionally transcribed iab-4/iab-8 microrna
3352 3361 Nucleic Acids Research, 2013, Vol. 41, No. 5 Published online 17 January 2013 doi:10.1093/nar/gks1445 Structure, evolution and function of the bi-directionally transcribed iab-4/iab-8 microrna
Supplementary Figure 1 A
 Supplementary Figure A B M. mullata p53, 3 UTR Luciferase activity (%) mir-5b 8 Le et al. Supplementary Information NC-DP - + - - - - - NC-DP - - + - - - - NC-DP3 - - - + - - - 5b-DP - - - - + + + NC-AS
Supplementary Figure A B M. mullata p53, 3 UTR Luciferase activity (%) mir-5b 8 Le et al. Supplementary Information NC-DP - + - - - - - NC-DP - - + - - - - NC-DP3 - - - + - - - 5b-DP - - - - + + + NC-AS
Nature Methods: doi: /nmeth Supplementary Figure 1. Construction of a sensitive TetR mediated auxotrophic off-switch.
 Supplementary Figure 1 Construction of a sensitive TetR mediated auxotrophic off-switch. A Production of the Tet repressor in yeast when conjugated to either the LexA4 or LexA8 promoter DNA binding sequences.
Supplementary Figure 1 Construction of a sensitive TetR mediated auxotrophic off-switch. A Production of the Tet repressor in yeast when conjugated to either the LexA4 or LexA8 promoter DNA binding sequences.
B. Transgenic plants with strong phenotype (%)
 A. TCTAGTTGTTGTTGTTATGGTCTAGTTGTTGTTGTTATGGTCTAATTT AAATATGGTCTAAAGAAGAAGAATATGGTCTAAAGAAGAAGAATATGG 2XP35S STTM165 5 GGGGGATGAAGctaCCTGGTCCGA3 3 CCCCCUACUUC---GGACCAGGCU5 mir165 HindIII mir165 96 nt GTTGTTGTTGTTATGGTCTAGTTGTTGTTGTTATGGTCTAATTT
A. TCTAGTTGTTGTTGTTATGGTCTAGTTGTTGTTGTTATGGTCTAATTT AAATATGGTCTAAAGAAGAAGAATATGGTCTAAAGAAGAAGAATATGG 2XP35S STTM165 5 GGGGGATGAAGctaCCTGGTCCGA3 3 CCCCCUACUUC---GGACCAGGCU5 mir165 HindIII mir165 96 nt GTTGTTGTTGTTATGGTCTAGTTGTTGTTGTTATGGTCTAATTT
Schematic representation of the endogenous PALB2 locus and gene-disruption constructs
 Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Fig. S1. Negative controls for in situ hybridization results using bam antisense probe. Fig. S2. Overexpression of either mir-275 or mir306
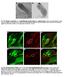 Fig. S1. Negative controls for in situ hybridization results using bam antisense probe. (A) In situ hybridization using bam sense probe in wild-type testis. (B) In situ hybridization using bam antisense
Fig. S1. Negative controls for in situ hybridization results using bam antisense probe. (A) In situ hybridization using bam sense probe in wild-type testis. (B) In situ hybridization using bam antisense
Document S1. Supplemental Experimental Procedures and Three Figures (see next page)
 Supplemental Data Document S1. Supplemental Experimental Procedures and Three Figures (see next page) Table S1. List of Candidate Genes Identified from the Screen. Candidate genes, corresponding dsrnas
Supplemental Data Document S1. Supplemental Experimental Procedures and Three Figures (see next page) Table S1. List of Candidate Genes Identified from the Screen. Candidate genes, corresponding dsrnas
Therapeutic & Prevention Application of Nucleic Acids
 Therapeutic & Prevention Application of Nucleic Acids Seyed Amir Hossein Jalali Institute of Biotechnology and Bioengineering, Isfahan University Of Technology (IUT). 30.7.2015 * Plasmids * DNA Aptamers
Therapeutic & Prevention Application of Nucleic Acids Seyed Amir Hossein Jalali Institute of Biotechnology and Bioengineering, Isfahan University Of Technology (IUT). 30.7.2015 * Plasmids * DNA Aptamers
supplementary information
 Figure S1 ZEB1 full length mrna. (a) Analysis of the ZEB1 mrna using the UCSC genome browser (http://genome.ucsc.edu) revealed truncation of the annotated Refseq sequence (NM_030751). The probable terminus
Figure S1 ZEB1 full length mrna. (a) Analysis of the ZEB1 mrna using the UCSC genome browser (http://genome.ucsc.edu) revealed truncation of the annotated Refseq sequence (NM_030751). The probable terminus
MicroRNA Destabilization Enables. Dynamic Regulation of the mir-16 Family. In Response to Cell-Cycle Changes SUPPLEMENTAL INFORMATION
 Molecular Cell, Volume 43 SUPPLEMENTAL INFORMATION MicroRNA Destabilization Enables Dynamic Regulation of the mir-16 Family In Response to Cell-Cycle Changes Olivia S. Risland, Sue-Jean Hong, and David
Molecular Cell, Volume 43 SUPPLEMENTAL INFORMATION MicroRNA Destabilization Enables Dynamic Regulation of the mir-16 Family In Response to Cell-Cycle Changes Olivia S. Risland, Sue-Jean Hong, and David
UTR Reporter Vectors and Viruses
 UTR Reporter Vectors and Viruses 3 UTR, 5 UTR, Promoter Reporter (Version 1) Applied Biological Materials Inc. #1-3671 Viking Way Richmond, BC V6V 2J5 Canada Notice to Purchaser All abm products are for
UTR Reporter Vectors and Viruses 3 UTR, 5 UTR, Promoter Reporter (Version 1) Applied Biological Materials Inc. #1-3671 Viking Way Richmond, BC V6V 2J5 Canada Notice to Purchaser All abm products are for
mir-24-mediated down-regulation of H2AX suppresses DNA repair
 Supplemental Online Material mir-24-mediated down-regulation of H2AX suppresses DNA repair in terminally differentiated blood cells Ashish Lal 1,4, Yunfeng Pan 2,4, Francisco Navarro 1,4, Derek M. Dykxhoorn
Supplemental Online Material mir-24-mediated down-regulation of H2AX suppresses DNA repair in terminally differentiated blood cells Ashish Lal 1,4, Yunfeng Pan 2,4, Francisco Navarro 1,4, Derek M. Dykxhoorn
Supplementary Online Material. the flowchart of Supplemental Figure 1, with the fraction of known human loci retained
 SOM, page 1 Supplementary Online Material Materials and Methods Identification of vertebrate mirna gene candidates The computational procedure used to identify vertebrate mirna genes is summarized in the
SOM, page 1 Supplementary Online Material Materials and Methods Identification of vertebrate mirna gene candidates The computational procedure used to identify vertebrate mirna genes is summarized in the
Supplementary Information Design of small molecule-responsive micrornas based on structural requirements for Drosha processing
 Supplementary Information Design of small molecule-responsive micrornas based on structural requirements for Drosha processing Chase L. Beisel, Yvonne Y. Chen, Stephanie J. Culler, Kevin G. Hoff, & Christina
Supplementary Information Design of small molecule-responsive micrornas based on structural requirements for Drosha processing Chase L. Beisel, Yvonne Y. Chen, Stephanie J. Culler, Kevin G. Hoff, & Christina
Supplementary Information
 Supplementary Information Super-resolution imaging of fluorescently labeled, endogenous RNA Polymerase II in living cells with CRISPR/Cas9-mediated gene editing Won-Ki Cho 1, Namrata Jayanth 1, Susan Mullen
Supplementary Information Super-resolution imaging of fluorescently labeled, endogenous RNA Polymerase II in living cells with CRISPR/Cas9-mediated gene editing Won-Ki Cho 1, Namrata Jayanth 1, Susan Mullen
Alteration of microrna expression correlates to fatty acidmediated. insulin resistance in mouse myoblasts
 Alteration of microrna expression correlates to fatty acidmediated insulin resistance in mouse myoblasts Supplemental Data Correlation coefficient matrix con PA PA 0.9425 1.0000 PA+OA 0.9407 0.9626 con
Alteration of microrna expression correlates to fatty acidmediated insulin resistance in mouse myoblasts Supplemental Data Correlation coefficient matrix con PA PA 0.9425 1.0000 PA+OA 0.9407 0.9626 con
SUPPLEMENTARY INFORMATION
 Gene replacements and insertions in rice by intron targeting using CRISPR Cas9 Table of Contents Supplementary Figure 1. sgrna-induced targeted mutations in the OsEPSPS gene in rice protoplasts. Supplementary
Gene replacements and insertions in rice by intron targeting using CRISPR Cas9 Table of Contents Supplementary Figure 1. sgrna-induced targeted mutations in the OsEPSPS gene in rice protoplasts. Supplementary
Analyze microrna Activity in Cells Using Luciferase Reporters. Trista Schagat, Ph.D. December 2013
 Analyze microrna Activity in Cells Using Luciferase Reporters Trista Schagat, Ph.D. December 2013 agenda microrna Overview 3 5 3 5 3 5 How to and Tips Design Transfect Assay Analyze Case Studies 2 micrornas
Analyze microrna Activity in Cells Using Luciferase Reporters Trista Schagat, Ph.D. December 2013 agenda microrna Overview 3 5 3 5 3 5 How to and Tips Design Transfect Assay Analyze Case Studies 2 micrornas
Identification of individual motifs on the genome scale. Some slides are from Mayukh Bhaowal
 Identification of individual motifs on the genome scale Some slides are from Mayukh Bhaowal Two papers Nature 423, 241-254 (15 May 2003) Sequencing and comparison of yeast species to identify genes and
Identification of individual motifs on the genome scale Some slides are from Mayukh Bhaowal Two papers Nature 423, 241-254 (15 May 2003) Sequencing and comparison of yeast species to identify genes and
Supporting Information
 Supporting Information Schnall-Levin et al. 10.1073/pnas.1006172107 SI Text Cell Transfections. S2R þ cells were maintained in Schneider s medium (Invitrogen), supplemented with 10% FBS and 1% pen-strep.
Supporting Information Schnall-Levin et al. 10.1073/pnas.1006172107 SI Text Cell Transfections. S2R þ cells were maintained in Schneider s medium (Invitrogen), supplemented with 10% FBS and 1% pen-strep.
High-resolution Identification and Separation of Living Cell Types by Multiple microrna-responsive Synthetic mrnas
 Supplementary information High-resolution Identification and Separation of Living Cell Types by Multiple microrna-responsive Synthetic mrnas Kei Endo *, Karin Hayashi, Hirohide Saito * Contents: 1. Supplementary
Supplementary information High-resolution Identification and Separation of Living Cell Types by Multiple microrna-responsive Synthetic mrnas Kei Endo *, Karin Hayashi, Hirohide Saito * Contents: 1. Supplementary
Endo et al., supplementary Fig S1
 Endo et al., supplementary Fig S1 supplementary Fig S1. AGO2 does not recognize the central region of mir390/mir390* duplex for RISC assembly. (A) The structure of wild-type mir390/mir390* and its variants
Endo et al., supplementary Fig S1 supplementary Fig S1. AGO2 does not recognize the central region of mir390/mir390* duplex for RISC assembly. (A) The structure of wild-type mir390/mir390* and its variants
An antibiotic selection marker for nematode transgenesis
 nature methods An antibiotic selection marker for nematode transgenesis Rosina Giordano-Santini, Stuart Milstein, Nenad Svrzikapa, Domena Tu, Robert Johnsen, David Baillie, Marc Vidal & Denis Dupuy Supplementary
nature methods An antibiotic selection marker for nematode transgenesis Rosina Giordano-Santini, Stuart Milstein, Nenad Svrzikapa, Domena Tu, Robert Johnsen, David Baillie, Marc Vidal & Denis Dupuy Supplementary
Total RNA was isolated using Trizol reagent (Invitrogen) and reverse transcribed using
 Supplementary Methods RNA Isolation and Quantitative RT-PCR Total RNA was isolated using Trizol reagent (Invitrogen) and reverse transcribed using random hexamers and superscript II reverse transcriptase
Supplementary Methods RNA Isolation and Quantitative RT-PCR Total RNA was isolated using Trizol reagent (Invitrogen) and reverse transcribed using random hexamers and superscript II reverse transcriptase
Cell line CDH1 mrna AS-paRNA S-paRNA
 a b Cell line mrna AS-paRNA S-paRNA HCT116 P53 +/+ 39.47 + + HCT116 P53 -/- 35.36 + + LNCAP 1562.4 + - MCF7 C1 1864.76 + - MCF7 C2 1812.81 + - Supplementary Figure 1. Promoter-associated transcripts at
a b Cell line mrna AS-paRNA S-paRNA HCT116 P53 +/+ 39.47 + + HCT116 P53 -/- 35.36 + + LNCAP 1562.4 + - MCF7 C1 1864.76 + - MCF7 C2 1812.81 + - Supplementary Figure 1. Promoter-associated transcripts at
MicroRNA Expression Plasmids
 MicroRNA Expression Plasmids Application Guide Table of Contents Package Contents and Related Products... 2 Related, Optional Reagents... 2 Related OriGene Products... 2 Cloning vector:... 3 Vector map
MicroRNA Expression Plasmids Application Guide Table of Contents Package Contents and Related Products... 2 Related, Optional Reagents... 2 Related OriGene Products... 2 Cloning vector:... 3 Vector map
Reviewers' Comments: Reviewer #1 (Remarks to the Author)
 Reviewers' Comments: Reviewer #1 (Remarks to the Author) The manuscript reports a novel form of microrna sponge, designated a "zipper" an LNA antisense sequence that bridges from the 3' end of one microrna
Reviewers' Comments: Reviewer #1 (Remarks to the Author) The manuscript reports a novel form of microrna sponge, designated a "zipper" an LNA antisense sequence that bridges from the 3' end of one microrna
Candidate region (0.74 Mb) ATC TCT GGG ACT CAT GAG CAG GAG GCT AGC ATC TCT GGG ACT CAT TAG CAG GAG GCT AGC
 A idm-3 idm-3 B Physical distance (Mb) 4.6 4.86.6 8.4 C Chr.3 Recom. Rate (%) ATG 3.9.9.9 9.74 Candidate region (.74 Mb) n=4 TAA D idm-3 G3T(E4) G4A(W988) WT idm-3 ATC TCT GGG ACT CAT GAG CAG GAG GCT AGC
A idm-3 idm-3 B Physical distance (Mb) 4.6 4.86.6 8.4 C Chr.3 Recom. Rate (%) ATG 3.9.9.9 9.74 Candidate region (.74 Mb) n=4 TAA D idm-3 G3T(E4) G4A(W988) WT idm-3 ATC TCT GGG ACT CAT GAG CAG GAG GCT AGC
SUPPLEMENTARY FIGURES AND TABLES
 SUPPLEMENTARY FIGURES AND TABLES Suppl. Table 1 Oligonucleotides used in the study The table shows the structure and nucleotide position of the oligonucleotides used in the study along with their application
SUPPLEMENTARY FIGURES AND TABLES Suppl. Table 1 Oligonucleotides used in the study The table shows the structure and nucleotide position of the oligonucleotides used in the study along with their application
Somatic Primary pirna Biogenesis Driven by cis-acting RNA Elements and Trans-Acting Yb
 Cell Reports Supplemental Information Somatic Primary pirna Biogenesis Driven by cis-acting RNA Elements and Trans-Acting Yb Hirotsugu Ishizu, Yuka W. Iwasaki, Shigeki Hirakata, Haruka Ozaki, Wataru Iwasaki,
Cell Reports Supplemental Information Somatic Primary pirna Biogenesis Driven by cis-acting RNA Elements and Trans-Acting Yb Hirotsugu Ishizu, Yuka W. Iwasaki, Shigeki Hirakata, Haruka Ozaki, Wataru Iwasaki,
MicroRNA Expression Plasmids
 MicroRNA Expression Plasmids Application Guide Table of Contents Package Contents and Related Products... 2 Related, Optional Reagents... 2 Related OriGene Products... 2 Cloning vector:... 3 Vector map
MicroRNA Expression Plasmids Application Guide Table of Contents Package Contents and Related Products... 2 Related, Optional Reagents... 2 Related OriGene Products... 2 Cloning vector:... 3 Vector map
3 Designing Primers for Site-Directed Mutagenesis
 3 Designing Primers for Site-Directed Mutagenesis 3.1 Learning Objectives During the next two labs you will learn the basics of site-directed mutagenesis: you will design primers for the mutants you designed
3 Designing Primers for Site-Directed Mutagenesis 3.1 Learning Objectives During the next two labs you will learn the basics of site-directed mutagenesis: you will design primers for the mutants you designed
Investigating the regulation of mirna biogenesis and Argonaute2 by RNA binding proteins
 Investigating the regulation of mirna biogenesis and Argonaute2 by RNA binding proteins Patrick Peter Connerty Supervisor: Gyorgy Hutvagner Thesis Submitted for the Degree of Doctor of Philosophy (Science)
Investigating the regulation of mirna biogenesis and Argonaute2 by RNA binding proteins Patrick Peter Connerty Supervisor: Gyorgy Hutvagner Thesis Submitted for the Degree of Doctor of Philosophy (Science)
Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan).
 1 2 3 4 5 6 7 8 Supplemental Materials and Methods Cell proliferation assay Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). GCs were plated at 96-well
1 2 3 4 5 6 7 8 Supplemental Materials and Methods Cell proliferation assay Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). GCs were plated at 96-well
Regulation of transcription by the MLL2 complex and MLL complex-associated AKAP95
 Supplementary Information Regulation of transcription by the complex and MLL complex-associated Hao Jiang, Xiangdong Lu, Miho Shimada, Yali Dou, Zhanyun Tang, and Robert G. Roeder Input HeLa NE IP lot:
Supplementary Information Regulation of transcription by the complex and MLL complex-associated Hao Jiang, Xiangdong Lu, Miho Shimada, Yali Dou, Zhanyun Tang, and Robert G. Roeder Input HeLa NE IP lot:
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10810 Supplementary Fig. 1: Mutation of the loqs gene leads to shortened lifespan and adult-onset brain degeneration. a. Northern blot of control and loqs f00791 mutant flies. loqs f00791
doi:10.1038/nature10810 Supplementary Fig. 1: Mutation of the loqs gene leads to shortened lifespan and adult-onset brain degeneration. a. Northern blot of control and loqs f00791 mutant flies. loqs f00791
NIH Public Access Author Manuscript Fly (Austin). Author manuscript; available in PMC 2008 May 23.
 NIH Public Access Author Manuscript Published in final edited form as: Fly (Austin). 2007 December ; 1(6): 337 339. The Abdominal-B Promoter Tethering Element Mediates Promoter-Enhancer Specificity at
NIH Public Access Author Manuscript Published in final edited form as: Fly (Austin). 2007 December ; 1(6): 337 339. The Abdominal-B Promoter Tethering Element Mediates Promoter-Enhancer Specificity at
3 UTR (untranslated region) Reporter Clone and its vector, pmirtarget. Application Guide. OriGene Technologies, Inc
 3 UTR (untranslated region) Reporter Clone and its vector, pmirtarget Application Guide OriGene Technologies, Inc Package Contents and Storage Conditions 3 UTR reporter clone as 10ug lyophilized plasmid
3 UTR (untranslated region) Reporter Clone and its vector, pmirtarget Application Guide OriGene Technologies, Inc Package Contents and Storage Conditions 3 UTR reporter clone as 10ug lyophilized plasmid
Derepressing muscleblind expression by mirna sponges ameliorates myotonic dystrophy-like. Ruben Artero 1,2 *
 Derepressing muscleblind expression by mirna sponges ameliorates myotonic dystrophy-like phenotypes in Drosophila. Estefania Cerro-Herreros 1,2, Juan M. Fernandez-Costa 1,2, María Sabater-Arcis 1,2, Beatriz
Derepressing muscleblind expression by mirna sponges ameliorates myotonic dystrophy-like phenotypes in Drosophila. Estefania Cerro-Herreros 1,2, Juan M. Fernandez-Costa 1,2, María Sabater-Arcis 1,2, Beatriz
MicroRNA Expression Plasmids
 MicroRNA Expression Plasmids Application Guide Table of Contents Package Contents and Related Products... 2 Related, Optional Reagents... 2 Related OriGene Products... 2 Cloning vector:... 3 Vector map
MicroRNA Expression Plasmids Application Guide Table of Contents Package Contents and Related Products... 2 Related, Optional Reagents... 2 Related OriGene Products... 2 Cloning vector:... 3 Vector map
SUPPLEMENTARY FIGURES AND TABLES
 SUPPLEMENTARY FIGURES AND TABLES Supplementary Figure S1: Plot shows the percentage of initially identified backsplice junctions against the Read depth value. Junctional circrna candidate have one mate
SUPPLEMENTARY FIGURES AND TABLES Supplementary Figure S1: Plot shows the percentage of initially identified backsplice junctions against the Read depth value. Junctional circrna candidate have one mate
Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question in Section B and ONE question from Section C.
 UNIVERSITY OF EAST ANGLIA School of Biological Sciences Main Series UG Examination 2013-2014 MOLECULAR BIOLOGY BIO-2B02 Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question
UNIVERSITY OF EAST ANGLIA School of Biological Sciences Main Series UG Examination 2013-2014 MOLECULAR BIOLOGY BIO-2B02 Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question
Galina Gabriely, Ph.D. BWH/HMS
 Galina Gabriely, Ph.D. BWH/HMS Email: ggabriely@rics.bwh.harvard.edu Outline: microrna overview microrna expression analysis microrna functional analysis microrna (mirna) Characteristics mirnas discovered
Galina Gabriely, Ph.D. BWH/HMS Email: ggabriely@rics.bwh.harvard.edu Outline: microrna overview microrna expression analysis microrna functional analysis microrna (mirna) Characteristics mirnas discovered
Supplementary Materials
 Supplementary Materials Table S1. Oligonucleotide sequences and PCR conditions used to amplify the indicated genes. TA = annealing temperature; gdna = genomic DNA; cdna = complementary DNA; c = concentration.
Supplementary Materials Table S1. Oligonucleotide sequences and PCR conditions used to amplify the indicated genes. TA = annealing temperature; gdna = genomic DNA; cdna = complementary DNA; c = concentration.
Supplementary Fig. 1 Expression levels of 5 mirnas cycling in PDF cells under LD conditions.
 Supplementary Fig. 1 Expression levels of 5 mirnas cycling in PDF cells under LD conditions. RT-qPCR quantification of mirna levels in PDF cells entrained under LD cycles in WT flies (PDF-GAL4;UAS-mCD8::GFP).
Supplementary Fig. 1 Expression levels of 5 mirnas cycling in PDF cells under LD conditions. RT-qPCR quantification of mirna levels in PDF cells entrained under LD cycles in WT flies (PDF-GAL4;UAS-mCD8::GFP).
mirwip: microrna target prediction based on micrornacontaining ribonucleoprotein enriched transcripts
 nature methods mirwip: microrna target prediction based on micrornacontaining ribonucleoprotein enriched transcripts Molly Hammell, Dang Long, Liang Zhang, Andrew Lee, C Steven Carmack, Min Han, Ye Ding
nature methods mirwip: microrna target prediction based on micrornacontaining ribonucleoprotein enriched transcripts Molly Hammell, Dang Long, Liang Zhang, Andrew Lee, C Steven Carmack, Min Han, Ye Ding
Summary MicroRNAs (mirnas) are genomically encoded small RNAs used by organisms to regulate the expression of proteins generated from messenger RNA
 Summary MicroRNAs (mirnas) are genomically encoded small RNAs used by organisms to regulate the expression of proteins generated from messenger RNA transcripts. The in vivo requirement of specific mirnas
Summary MicroRNAs (mirnas) are genomically encoded small RNAs used by organisms to regulate the expression of proteins generated from messenger RNA transcripts. The in vivo requirement of specific mirnas
Experimental genetics - I
 Experimental genetics - I Examples of diseases with genetic-links Hemophilia (complete loss or altered form of factor VIII): bleeding disorder Duchenne muscular dystrophy (altered form of dystrophin) muscle
Experimental genetics - I Examples of diseases with genetic-links Hemophilia (complete loss or altered form of factor VIII): bleeding disorder Duchenne muscular dystrophy (altered form of dystrophin) muscle
Construction of plant complementation vector and generation of transgenic plants
 MATERIAL S AND METHODS Plant materials and growth conditions Arabidopsis ecotype Columbia (Col0) was used for this study. SALK_072009, SALK_076309, and SALK_027645 were obtained from the Arabidopsis Biological
MATERIAL S AND METHODS Plant materials and growth conditions Arabidopsis ecotype Columbia (Col0) was used for this study. SALK_072009, SALK_076309, and SALK_027645 were obtained from the Arabidopsis Biological
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/10/496/eaam6291/dc1 Supplementary Materials for Regulation of autophagy, NF-κB signaling, and cell viability by mir-124 in KRAS mutant mesenchymal-like NSCLC cells
www.sciencesignaling.org/cgi/content/full/10/496/eaam6291/dc1 Supplementary Materials for Regulation of autophagy, NF-κB signaling, and cell viability by mir-124 in KRAS mutant mesenchymal-like NSCLC cells
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1137999/dc1 Supporting Online Material for Disrupting the Pairing Between let-7 and Enhances Oncogenic Transformation Christine Mayr, Michael T. Hemann, David P. Bartel*
www.sciencemag.org/cgi/content/full/1137999/dc1 Supporting Online Material for Disrupting the Pairing Between let-7 and Enhances Oncogenic Transformation Christine Mayr, Michael T. Hemann, David P. Bartel*
Fig. S1. eif6 expression in HEK293 transfected with shrna against eif6 or pcmv-eif6 vector.
 Fig. S1. eif6 expression in HEK293 transfected with shrna against eif6 or pcmv-eif6 vector. (a) Western blotting analysis and (b) qpcr analysis of eif6 expression in HEK293 T cells transfected with either
Fig. S1. eif6 expression in HEK293 transfected with shrna against eif6 or pcmv-eif6 vector. (a) Western blotting analysis and (b) qpcr analysis of eif6 expression in HEK293 T cells transfected with either
Supplementary Figures and Figure legends
 Supplementary Figures and Figure legends 3 Supplementary Figure 1. Conditional targeting construct for the murine Satb1 locus with a modified FLEX switch. Schematic of the wild type Satb1 locus; the conditional
Supplementary Figures and Figure legends 3 Supplementary Figure 1. Conditional targeting construct for the murine Satb1 locus with a modified FLEX switch. Schematic of the wild type Satb1 locus; the conditional
TABLE S1. Sequences of the primers used for RT-PCR and PCR
 TABLE S1. Sequences of the primers used for RT-PCR and PCR Primer Sequence (5 to 3 ) Purpose Cloning: posuprtid-1 & posuprtid-2 deletion plasmids DM01 AGCTCCAGCGGGTCGCCGGGGTTGGCC DM02 CCAGCGGGTCGCCGGGGTTGGCC
TABLE S1. Sequences of the primers used for RT-PCR and PCR Primer Sequence (5 to 3 ) Purpose Cloning: posuprtid-1 & posuprtid-2 deletion plasmids DM01 AGCTCCAGCGGGTCGCCGGGGTTGGCC DM02 CCAGCGGGTCGCCGGGGTTGGCC
Non-coding Function & Variation, MPRAs II. Mike White Bio /5/18
 Non-coding Function & Variation, MPRAs II Mike White Bio 5488 3/5/18 MPRA Review Problem 1: Where does your CRE DNA come from? DNA synthesis Genomic fragments Targeted regulome capture Problem 2: How do
Non-coding Function & Variation, MPRAs II Mike White Bio 5488 3/5/18 MPRA Review Problem 1: Where does your CRE DNA come from? DNA synthesis Genomic fragments Targeted regulome capture Problem 2: How do
Supplementary Information
 Supplementary Information Supplemental Material and Methods sirna, plasmids and antibodies Cell were transfected with 10 nm Hs_WIG1_1 sirna (siw1), Hs_WIG1_2 sirna (siw2), Hs_XRN1_1 (sixrn1), Hs_DCP1A_6
Supplementary Information Supplemental Material and Methods sirna, plasmids and antibodies Cell were transfected with 10 nm Hs_WIG1_1 sirna (siw1), Hs_WIG1_2 sirna (siw2), Hs_XRN1_1 (sixrn1), Hs_DCP1A_6
nature methods A paired-end sequencing strategy to map the complex landscape of transcription initiation
 nature methods A paired-end sequencing strategy to map the complex landscape of transcription initiation Ting Ni, David L Corcoran, Elizabeth A Rach, Shen Song, Eric P Spana, Yuan Gao, Uwe Ohler & Jun
nature methods A paired-end sequencing strategy to map the complex landscape of transcription initiation Ting Ni, David L Corcoran, Elizabeth A Rach, Shen Song, Eric P Spana, Yuan Gao, Uwe Ohler & Jun
MeCP2. MeCP2/α-tubulin. GFP mir1-1 mir132
 Conservation Figure S1. Schematic showing 3 UTR (top; thick black line), mir132 MRE (arrow) and nucleotide sequence conservation (vertical black lines; http://genome.ucsc.edu). a GFP mir1-1 mir132 b GFP
Conservation Figure S1. Schematic showing 3 UTR (top; thick black line), mir132 MRE (arrow) and nucleotide sequence conservation (vertical black lines; http://genome.ucsc.edu). a GFP mir1-1 mir132 b GFP
Supplemental Information. Autoregulatory Feedback Controls. Sequential Action of cis-regulatory Modules. at the brinker Locus
 Developmental Cell, Volume 26 Supplemental Information Autoregulatory Feedback Controls Sequential Action of cis-regulatory Modules at the brinker Locus Leslie Dunipace, Abbie Saunders, Hilary L. Ashe,
Developmental Cell, Volume 26 Supplemental Information Autoregulatory Feedback Controls Sequential Action of cis-regulatory Modules at the brinker Locus Leslie Dunipace, Abbie Saunders, Hilary L. Ashe,
Supplementary Figures 1-12
 Supplementary Figures 1-12 Supplementary Figure 1. The specificity of anti-abi1 antibody. Total Proteins extracted from the wild type seedlings or abi1-3 null mutant seedlings were used for immunoblotting
Supplementary Figures 1-12 Supplementary Figure 1. The specificity of anti-abi1 antibody. Total Proteins extracted from the wild type seedlings or abi1-3 null mutant seedlings were used for immunoblotting
Supplemental Figure S1 (related to Figure 1). The LOTUS domain of Oskar, Tejas, and Tapas is required for co-localization with Vasa Plasmids encoding
 Supplemental Figure S1 (related to Figure 1). The LOTUS domain of Oskar, Tejas, and Tapas is required for co-localization with Vasa Plasmids encoding N-terminal EGFP- or mcherry-protein fusions under the
Supplemental Figure S1 (related to Figure 1). The LOTUS domain of Oskar, Tejas, and Tapas is required for co-localization with Vasa Plasmids encoding N-terminal EGFP- or mcherry-protein fusions under the
Genome-Wide Survey of MicroRNA - Transcription Factor Feed-Forward Regulatory Circuits in Human. Supporting Information
 Genome-Wide Survey of MicroRNA - Transcription Factor Feed-Forward Regulatory Circuits in Human Angela Re #, Davide Corá #, Daniela Taverna and Michele Caselle # equal contribution * corresponding author,
Genome-Wide Survey of MicroRNA - Transcription Factor Feed-Forward Regulatory Circuits in Human Angela Re #, Davide Corá #, Daniela Taverna and Michele Caselle # equal contribution * corresponding author,
Supplementary Figure 1 Validate the expression of mir-302b after bacterial infection by northern
 Supplementary Figure 1 Validate the expression of mir-302b after bacterial infection by northern blot. Northern blot analysis of mir-302b expression following infection with PAO1, PAK and Kp in (A) lung
Supplementary Figure 1 Validate the expression of mir-302b after bacterial infection by northern blot. Northern blot analysis of mir-302b expression following infection with PAO1, PAK and Kp in (A) lung
QPCR ASSAYS FOR MIRNA EXPRESSION PROFILING
 TECH NOTE 4320 Forest Park Ave Suite 303 Saint Louis, MO 63108 +1 (314) 833-9764 mirna qpcr ASSAYS - powered by NAWGEN Our mirna qpcr Assays were developed by mirna experts at Nawgen to improve upon previously
TECH NOTE 4320 Forest Park Ave Suite 303 Saint Louis, MO 63108 +1 (314) 833-9764 mirna qpcr ASSAYS - powered by NAWGEN Our mirna qpcr Assays were developed by mirna experts at Nawgen to improve upon previously
Design. Construction. Characterization
 Design Construction Characterization DNA mrna (messenger) A C C transcription translation C A C protein His A T G C T A C G Plasmids replicon copy number incompatibility selection marker origin of replication
Design Construction Characterization DNA mrna (messenger) A C C transcription translation C A C protein His A T G C T A C G Plasmids replicon copy number incompatibility selection marker origin of replication
Long Noncoding RNA LOC Suppresses Apoptosis by. Targeting mir p and mir-4767 in Vascular Endothelial Cells
 Long Noncoding RNA LOC100129973 Suppresses Apoptosis by Targeting mir-4707-5p and mir-4767 in Vascular Endothelial Cells Wei Lu 1, ShuYa Huang 1, Le Su 1, BaoXiang Zhao 2, *, JunYing Miao 1, 3, * 1 Shandong
Long Noncoding RNA LOC100129973 Suppresses Apoptosis by Targeting mir-4707-5p and mir-4767 in Vascular Endothelial Cells Wei Lu 1, ShuYa Huang 1, Le Su 1, BaoXiang Zhao 2, *, JunYing Miao 1, 3, * 1 Shandong
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table.
 Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Kawai and Amano, http://www.jcb.org/cgi/content/full/jcb.201110008/dc1 Figure S1. Regulation of mirna expression by BRCA1. (A) Confirmation
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Kawai and Amano, http://www.jcb.org/cgi/content/full/jcb.201110008/dc1 Figure S1. Regulation of mirna expression by BRCA1. (A) Confirmation
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Supplementary figure 1: List of primers/oligonucleotides used in this study. 1 Supplementary figure 2: Sequences and mirna-targets of i) mcherry expresses in transgenic fish used
SUPPLEMENTARY INFORMATION Supplementary figure 1: List of primers/oligonucleotides used in this study. 1 Supplementary figure 2: Sequences and mirna-targets of i) mcherry expresses in transgenic fish used
Supplemental Figure 1. Immunoblot analysis of wild-type and mutant forms of JAZ3
 Supplemental Data. Chung and Howe (2009). A Critical Role for the TIFY Motif in Repression of Jasmonate Signaling by a Stabilized Splice Variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis.
Supplemental Data. Chung and Howe (2009). A Critical Role for the TIFY Motif in Repression of Jasmonate Signaling by a Stabilized Splice Variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis.
Supplementary Fig. 1 Generation and genotyping of ThPOK / mice
 Supplementary Fig. 1 Generation and genotyping of ThPOK / mice (a). The strategy used to target the ThPOK locus is schematically depicted. A targeting construct was generated by inserting a loxp site (filled
Supplementary Fig. 1 Generation and genotyping of ThPOK / mice (a). The strategy used to target the ThPOK locus is schematically depicted. A targeting construct was generated by inserting a loxp site (filled
Supplementary Figure 1. ips_3y+mir-302b. ips_3y+mir-372. ips_3y+mir-302b + mir-372. ips_4y. ips_4y+mir-302b. ips_4y + mir-372
 Supplementary Figure 1 ips_3y+mir-32b ips_3y+ ips_3y+mir-32b + ips_4y ips_4y+mir-32b ips_4y + Nature Biotechnology: doi:1.138/nb.t.1862 TRA-1-6 DAPI Oct3/4 Supplementary Figure 1: ips cells derived from
Supplementary Figure 1 ips_3y+mir-32b ips_3y+ ips_3y+mir-32b + ips_4y ips_4y+mir-32b ips_4y + Nature Biotechnology: doi:1.138/nb.t.1862 TRA-1-6 DAPI Oct3/4 Supplementary Figure 1: ips cells derived from
Supplementary information
 Supplementary information Plant phosphomannose isomerase as a selectable marker for rice transformation Lei Hu, Hao Li, Ruiying Qin, Rongfang Xu, Juan Li, Li Li, Pengcheng Wei & Jianbo Yang Figure S1.
Supplementary information Plant phosphomannose isomerase as a selectable marker for rice transformation Lei Hu, Hao Li, Ruiying Qin, Rongfang Xu, Juan Li, Li Li, Pengcheng Wei & Jianbo Yang Figure S1.
Supplementary information, Figure S1
 Supplementary information, Figure S1 (A) Schematic diagram of the sgrna and hspcas9 expression cassettes in a single binary vector designed for Agrobacterium-mediated stable transformation of Arabidopsis
Supplementary information, Figure S1 (A) Schematic diagram of the sgrna and hspcas9 expression cassettes in a single binary vector designed for Agrobacterium-mediated stable transformation of Arabidopsis
3. human genomics clone genes associated with genetic disorders. 4. many projects generate ordered clones that cover genome
 Lectures 30 and 31 Genome analysis I. Genome analysis A. two general areas 1. structural 2. functional B. genome projects a status report 1. 1 st sequenced: several viral genomes 2. mitochondria and chloroplasts
Lectures 30 and 31 Genome analysis I. Genome analysis A. two general areas 1. structural 2. functional B. genome projects a status report 1. 1 st sequenced: several viral genomes 2. mitochondria and chloroplasts
Expression constructs used in this study: Rtn1pQM-Ntag/A. The expression vector was generated as described previously (Mannan et al., 2006).
 Supplementary file 1 Sequences of primers used: C-mycTag-new-F: 5 CCGAATTCCGTCGGAGCAAGCTTGATTTA3 Ad.C-mycTag-R: 5 TTGCGGCCGCCTTTTGCTCCATGGTGAGGTC3 adpxt1-orf-f: 5 GGTGCGGCCGCTGGTGGCATGCAGCTTAGACACATTGG3
Supplementary file 1 Sequences of primers used: C-mycTag-new-F: 5 CCGAATTCCGTCGGAGCAAGCTTGATTTA3 Ad.C-mycTag-R: 5 TTGCGGCCGCCTTTTGCTCCATGGTGAGGTC3 adpxt1-orf-f: 5 GGTGCGGCCGCTGGTGGCATGCAGCTTAGACACATTGG3
Supplementary Materials
 Supplementary Materials Supplementary Methods Supplementary Discussion Supplementary Figure 1 Calculated frequencies of embryo cells bearing bi-allelic alterations. Targeted indel mutations induced by
Supplementary Materials Supplementary Methods Supplementary Discussion Supplementary Figure 1 Calculated frequencies of embryo cells bearing bi-allelic alterations. Targeted indel mutations induced by
Antisense RNA Insert Design for Plasmid Construction to Knockdown Target Gene Expression
 Vol. 1:7-15 Antisense RNA Insert Design for Plasmid Construction to Knockdown Target Gene Expression Ji, Tom, Lu, Aneka, Wu, Kaylee Department of Microbiology and Immunology, University of British Columbia
Vol. 1:7-15 Antisense RNA Insert Design for Plasmid Construction to Knockdown Target Gene Expression Ji, Tom, Lu, Aneka, Wu, Kaylee Department of Microbiology and Immunology, University of British Columbia
Figure S1: Insert sequences for GR1, GR2, Qc3c_AgeI and GR3 vectors
 Figure S1: Insert sequences for GR1, GR2, Qc3c_AgeI and GR3 vectors A Vector GR1 SacI BamHI CTCTGGCTAACTAGGC Insert 5/7nt - G TCGAGAGACCGATTGATCCG Insert 5/7nt - CCTAG 1 G CAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAAC
Figure S1: Insert sequences for GR1, GR2, Qc3c_AgeI and GR3 vectors A Vector GR1 SacI BamHI CTCTGGCTAACTAGGC Insert 5/7nt - G TCGAGAGACCGATTGATCCG Insert 5/7nt - CCTAG 1 G CAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAAC
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature05841 SUPPLEMENTARY INFORMATION www.nature.com/nature 1 www.nature.com/nature 2 www.nature.com/nature 3 www.nature.com/nature 4 a Hours of Development: eif-6(rnai): 4 wildtype 30 4 lin-4
doi: 10.1038/nature05841 SUPPLEMENTARY INFORMATION www.nature.com/nature 1 www.nature.com/nature 2 www.nature.com/nature 3 www.nature.com/nature 4 a Hours of Development: eif-6(rnai): 4 wildtype 30 4 lin-4
Long-term, efficient inhibition of microrna function in mice using raav vectors
 Nature Methods Long-term, efficient inhibition of microrna function in mice using raav vectors Jun Xie, Stefan L Ameres, Randall Friedline, Jui-Hung Hung, Yu Zhang, Qing Xie, Li Zhong, Qin Su, Ran He,
Nature Methods Long-term, efficient inhibition of microrna function in mice using raav vectors Jun Xie, Stefan L Ameres, Randall Friedline, Jui-Hung Hung, Yu Zhang, Qing Xie, Li Zhong, Qin Su, Ran He,
Enhancers mutations that make the original mutant phenotype more extreme. Suppressors mutations that make the original mutant phenotype less extreme
 Interactomics and Proteomics 1. Interactomics The field of interactomics is concerned with interactions between genes or proteins. They can be genetic interactions, in which two genes are involved in the
Interactomics and Proteomics 1. Interactomics The field of interactomics is concerned with interactions between genes or proteins. They can be genetic interactions, in which two genes are involved in the
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
Structural and functional analysis of the mir-282 microrna gene of Drosophila melanogaster. Ágnes Bujna
 Structural and functional analysis of the mir-282 microrna gene of Drosophila melanogaster Ph.D. thesis Ágnes Bujna Supervisor: Dr. Miklós Erdélyi University of Szeged, PhD School of Biological Sciences
Structural and functional analysis of the mir-282 microrna gene of Drosophila melanogaster Ph.D. thesis Ágnes Bujna Supervisor: Dr. Miklós Erdélyi University of Szeged, PhD School of Biological Sciences
Percent survival. Supplementary fig. S3 A.
 Supplementary fig. S3 A. B. 100 Percent survival 80 60 40 20 Ml 0 0 100 C. Fig. S3 Comparison of leukaemia incidence rate in the triple targeted chimaeric mice and germline-transmission translocator mice
Supplementary fig. S3 A. B. 100 Percent survival 80 60 40 20 Ml 0 0 100 C. Fig. S3 Comparison of leukaemia incidence rate in the triple targeted chimaeric mice and germline-transmission translocator mice
Predicting microrna targeting efficacy in Drosophila
 Agarwal et al. Genome Biology (2018) 19:152 https://doi.org/10.1186/s13059-018-1504-3 RESEARCH Predicting microrna targeting efficacy in Drosophila Vikram Agarwal 1,2,3,4, Alexander O. Subtelny 1,2,6,
Agarwal et al. Genome Biology (2018) 19:152 https://doi.org/10.1186/s13059-018-1504-3 RESEARCH Predicting microrna targeting efficacy in Drosophila Vikram Agarwal 1,2,3,4, Alexander O. Subtelny 1,2,6,
Rajesh Pandey et al. Alu-miRNA interactions modulate transcript isoform diversity in stress response and reveal signatures of positive selection
 Rajesh Pandey et al. Alu-miRNA interactions modulate transcript isoform diversity in stress response and reveal signatures of positive selection Rajesh Pandey 1, Aniket Bhattacharya 2,3#, Vivek Bhardwaj
Rajesh Pandey et al. Alu-miRNA interactions modulate transcript isoform diversity in stress response and reveal signatures of positive selection Rajesh Pandey 1, Aniket Bhattacharya 2,3#, Vivek Bhardwaj
Construct Design and Cloning Guide for Cas9-triggered homologous recombination
 Construct Design and Cloning Guide for Cas9-triggered homologous recombination Written by Dan Dickinson (ddickins@live.unc.edu) and last updated December 2013. Reference: Dickinson DJ, Ward JD, Reiner
Construct Design and Cloning Guide for Cas9-triggered homologous recombination Written by Dan Dickinson (ddickins@live.unc.edu) and last updated December 2013. Reference: Dickinson DJ, Ward JD, Reiner
Lecture 20: Drosophila melanogaster
 Lecture 20: Drosophila melanogaster Model organisms Polytene chromosome Life cycle P elements and transformation Embryogenesis Read textbook: 732-744; Fig. 20.4; 20.10; 20.15-26 www.mhhe.com/hartwell3
Lecture 20: Drosophila melanogaster Model organisms Polytene chromosome Life cycle P elements and transformation Embryogenesis Read textbook: 732-744; Fig. 20.4; 20.10; 20.15-26 www.mhhe.com/hartwell3
Name Class Date. Practice Test
 Name Class Date 12 DNA Practice Test Multiple Choice Write the letter that best answers the question or completes the statement on the line provided. 1. What do bacteriophages infect? a. mice. c. viruses.
Name Class Date 12 DNA Practice Test Multiple Choice Write the letter that best answers the question or completes the statement on the line provided. 1. What do bacteriophages infect? a. mice. c. viruses.
Supplementary Methods
 Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
SUPPLEMENTARY INFORMATION
 Materials and Methods Transgenic Plant Materials and DNA Constructs. VEX1::H2B-GFP, ACA3::H2B-GFP, KRP6::H2B-GFP, KRP6::mock21ts-GFP, KRP6::TE21ts- GFP and KRP6::miR161ts-GFP constructs were generated
Materials and Methods Transgenic Plant Materials and DNA Constructs. VEX1::H2B-GFP, ACA3::H2B-GFP, KRP6::H2B-GFP, KRP6::mock21ts-GFP, KRP6::TE21ts- GFP and KRP6::miR161ts-GFP constructs were generated
