Supplementary Fig. 1 Expression levels of 5 mirnas cycling in PDF cells under LD conditions.
|
|
|
- Ralph Morris
- 5 years ago
- Views:
Transcription
1 Supplementary Fig. 1 Expression levels of 5 mirnas cycling in PDF cells under LD conditions. RT-qPCR quantification of mirna levels in PDF cells entrained under LD cycles in WT flies (PDF-GAL4;UAS-mCD8::GFP). mirna expression levels are normalized to 2S rrna. 2 biological replicates are concatenated to show cycling. a.u. represents artificial units.
2 Supplementary Fig. 2 Expression levels of mir-92a under LD or DD conditions. The average expression levels of 3 biological replicates under either LD or DD conditions in PDF-GAL4;UAS-mCD8::GFP flies are double plotted and superimposed to show the phase difference.
3 Supplementary Fig. 3 Statistical analysis of maximum axonal crosses quantification. Axonal crosses were quantified with Sholl analysis (Fig. 2A). Maximum axonal crosses indicate the position where maximum amounts of intersections are found between axon branches of a projection and concentric circles in Sholl analysis. N = 14. Error bars represent ±SEM, n.s. indicates non-significant, *P < 0.05, **P < 0.01, one-way ANOVA.
4 Supplementary Fig. 4 Adult-specific manipulation of mir-92a levels changes PDF cell fasciculation status. Immunostaining of PDF cell projections with anti-pdf antibody at ZT2 or ZT14. mir-92aoe indicates PDF-GSG;UAS-mCD8::GFP;UAS-mir-92aOE flies and is compared to its corresponding control PDF-GSG;UAS-mCD8::GFP/+ (with w1118 background). mir-92asp
5 indicates PDF-GSG;UAS-mCD8::GFP;UAS-mir-92aSP flies and is compared to its control, PDF-GSG;UAS-mCD8::GFP;UAS-scramble. Flies were fed on food containing 0.2 mg/ml Mifepristone (Sigma-Aldrich) for 1 week and entrained for at least 3 days under LD cycles prior to the assay. (A) Representative images of PDF cell projections of the indicated genotype at the indicated time. Scale bar equals 25 µm. (B) Quantification with Sholl analysis. Statistics were done on the points with maximum axonal crosses. N = 14. Error bars represent ±SEM, n.s. indicates non-significant, **P < 0.01, one-way ANOVA.
6 Supplementary Fig. 5 GCaMP6 live imaging of PDF neurons may show an altered responsiveness to nicotine in mir-92aoe and mir-92asp flies. Flies expressing GCaMP6 in PDF cells (PDF-GSG;UAS-GCaMP6f) in addition to mir-92a manipulations (UAS-mir-92aOE or UAS-mir- 92aSP) in an adult-specific manner were imaged for fluorescence level changes with 3x10-6 M nicotine perfusion after 30 seconds of baseline recording and the washedout at 60 seconds. Flies were fed on food containing 0.2 mg/ml Mifepristone (Sigma-Aldrich) for 1 week and entrained for at least 3 LD cycles prior to the assay. Measurements for both scramble and mir-92asp were performed between ZT18 22 (when endogenous mir-92a levels in PDF cells are high), and w1118 and mir-92aoe between ZT6 10 (when endogenous mir-92a levels in PDF cells are low). Average fluorescence levels of PDF cell bodies (l-lnvs) are plotted (top panels) and quantified (bottom panel). Each dot represents one brain. Bars represent the mean. Changes are statistically insignificant by two-way ANOVA.
7 M e a n f l u o r e s c e n c e s i g n a l * * * s c r a m b l e m i r a S P w m i r a O E Z T Z T Supplementary Fig. 6 GCaMP6 baseline fluorescence levels in PDF cell projections are affected by manipulation of mir-92a in PDF cells. Flies expressing GCaMP6 in PDF cells (PDF-GAL4;UAS-GCaMP6f) in addition to mir-92a manipulation (UAS-mir-92aOE or UAS-mir-92aSP) were imaged for baseline fluorescence levels. Fluorescence levels in PDF cell projections (s-lnvs) are plotted. Each dot represents an individual measurement. Measurements for both scramble and mir-92asp were performed between ZT18 22 (when endogenous mir-92a levels in PDF cells are high), and w1118 and mir-92aoe between ZT6 10 (when endogenous mir-92a levels in PDF cells are low). Error bars represent ±SD, *P < 0.05, ** P < 0.01, two-tailed t-test.
8 Supplementary Fig. 7 Sleep profiles of mir-92a mutant and control flies. (A) Sleep profiles of female flies entrained under LD cycles. mir-92a levels were manipulated in wake-promoting neurons driven by Dvpdf-GAL4. (B) Sleep profiles from the control experiment with no GAL4 driver. Sleep duration is quantified to the right. N = 32. Error bars represent ±SEM, n.s. represents non-significant, **P < 0.01, **** P < , two-way ANOVA.
9 Supplementary Fig. 8 mir-92a expression levels in dopaminergic neurons of flies are not affected by light pulses. RT-qPCR quantification of mir-92a levels in dopaminergic neurons. Flies were entrained for at least 3 days under LD cycles before exposure to a 10-min light pulse at either ZT15 or ZT21. Dopaminergic neurons were sorted at either ZT16 or ZT22 (50 min after the light pulse). No LP indicates no exposure to light, and LP indicates light pulse exposure. N = 3. Error bars represent ±SEM, n.s. represents non-significant, two-tailed t-test.
10 Supplementary Fig. 9 Sleep and circadian effects from manipulation of mir-92a levels in PDF cells. (A) The left panel shows a scatter plot of rhythmicity index, and the right panel shows a box plot of period lengths. PDF-GAL4 was crossed to w1118, UAS-mir-92aOE, UASscramble and UAS-mir-92aSP for comparison. Each dot indicates one fly. N = 16. Error bars represent ±SD, n.s. represents non-significant, one-way ANOVA. (B) Sleep profiles of female flies of the same genotypes as in (A). Quantification of sleep duration is to the right. N = 16. Error bars represent ±SEM, n.s. represents nonsignificant, *P < 0.05, **P < 0.01, **** P < , two-way ANOVA.
11 Supplementary Fig. 10 Scheme of S2 cell reporter assay. (A) Three plasmids were co-transfected into S2 cells. Ub-GAL4 and UAS-mir-92a cotransfection enabled expression of mir-92a in the cells, and psicheck2-3 UTR served as a reporter for the suppression assay.
12 (B) Plasmid map of psicheck2. The 3 UTR of sirt2 was inserted downstream of renilla. WT sirt2 3 UTR shows base-paring with mir-92a, and the binding site in Mut. sirt2 is mutated (brown). The plasmid map is adapted from Promega psicheck -2 Vector.
13 Supplementary Fig. 11 Quantification of sirt2 RNAi efficiency. Control (#36303, Bloomington stock center, WT background line for transgene injection) and two RNAi lines (RNAi-1: #32482; RNAi-2: #31363) were crossed to Tubulin-GAL4 drivers for expression in whole flies. Total RNA was extracted from fly heads and sirt2 mrna levels were quantified with RT-qPCR with primers listed in Supplementary Table 2. N = 3. Error bars represent ±SEM, ***P < 0.001, one-way ANOVA.
14 Supplementary Fig. 12 Quantification of PDF cell projection axonal crosses of the indicated genotypes. Quantification with Sholl analysis was done as described above. All flies also express the PDF- GAL4 driver. N = 14. Error bars represent ±SEM. n.s. represents non-significant, *P < 0.05, **P < 0.01, ***P < 0.001, one-way ANOVA.
15 Supplementary Fig. 13 Adult-specific sirt2 knockdown alters the PDF cell fasciculation phenotype. Immunostaining of PDF cell projections with anti-pdf antibody at ZT2 or ZT14. The control is PDF-GSG;UAS-mCD8::GFP/+ (with #36303 genetic background). Sirt2 RNAi-1/2 is PDF- GSG;UAS-mCD8::GFP;UAS-sirt2 RNAi-1/2. Flies were fed on food containing 0.2 mg/ml Mifepristone (Sigma-Aldrich) for 1 week and entrained for at least 3 LD cycles prior to the assay. (A) Representative images of PDF cell projections of the indicated genotype at the indicated time. Scale bar equals 25 µm.
16 (B) Quantification was done with Sholl analysis. Statistics were done at the points with maximum axonal crosses. N = 14. Error bars represent ±SEM, n.s. indicates nonsignificant, ***P < 0.001, one-way ANOVA.
17 F l u o r e s c e n c e l e v e l s * * * * * b a s e l i n e l e v e l s m a x i m u m l e v e l s c o n t r o l s i r t 2 R N A i - 1 s i r t 2 R N A i - 2 Supplementary Fig. 14 GCaMP6 live imaging of PDF neurons shows reduced responsiveness to nicotine with adult-specific sirt2 knockdown. Flies expressing GCaMP6 in PDF cells (PDF-GSG;UAS-GCaMP6f) in addition to RNAi against sirt2 were imaged for fluorescence level changes with 3x10-6 M nicotine perfusion after 30 seconds of baseline recording and then a wash-out at 60 seconds. Flies were fed on food containing 0.2 mg/ml Mifepristone (Sigma-Aldrich) for 1 week and entrained for at least 3 LD cycles prior to the assay. Measurements were performed between ZT2-6. Average fluorescence levels in PDF cell bodies (l-lnvs) are quantified. Each dot represents one brain. Bars represent mean. *P < 0.05, ****P < , two-way ANOVA.
18 Supplementary Fig. 15 sirt2 knockdown in dopaminergic neurons increases sleep duration. (A) sirt2 was knocked-down in dopaminergic neurons using sirt2 RNAi-2 in combination with TH-GAL4/UAS-dicer2 for higher knockdown efficiency. Sleep duration is quantified to the right. N = 16. Error bars represent ±SEM, n.s. represents non-significant, ****P < , two-way ANOVA. (B) A control experiment with no GAL4 driver. Sleep duration was quantified to the right. N = 16. Error bars represent ±SEM, n.s. represents non-significant, two-way ANOVA.
19 Supplementary Fig. 16 A sirt2 amorphic mutant strain shows PDF projection abnormalities. Immunostaining of PDF cell projections with anti-pdf antibody at ZT2 or ZT14. w1118 WT was used to compare with the sirt2 mutant flies. Flies were entrained for at least 3 LD cycles prior to the assay. (A) Representative images of PDF cell projections of the indicated genotypes at the indicated times. Scale bar equals 25 µm. (B) The defasciculation index (DI) was calculated according to 31. High DI indicates more defasciculation and lower DI indicates nire fasciculation. Each dot represents one projection. Bars indicate the mean. n.s. indicates non-significant, *P < 0.05, two-way ANOVA. (C) Axonal crosses were quantified with Sholl analysis as described above. (D) Statistics were done at the points with maximum axonal crosses. N = Error bars represent ±SEM, n.s. indicates non-significant, **P < 0.01, ****P < , two-way ANOVA.
20 Supplementary Fig. 17 sirt2 amorphic flies show increased sleep duration in LD as well as enhanced arrhythmicity in DD. (A) Sleep profiles of female sirt2 amorphic flies compared to w1118 flies under LD conditions. Quantification is to the right. N = 32. Error bars represent ±SEM, n.s. represents non-significant, ****P < , two-way ANOVA. (B) Actograms of male sirt2 amorphic flies compared to w1118 flies during 3 days of LD cycles followed by 8 days of DD cycles. The actograms show the average activity of 32 flies. The white background indicates lights-on, and the grey background indicates lightsoff. The rhythmicity index was calculated for individual flies. N = 32. Error bars represent ±SD, ****P < , two-tailed t-test.
21 Supplementary Fig. 18 Validation of mir-92a expression in PDF cells using traditional stem-loop RT-qPCR. Extracted RNA from sorted PDF cells was quantified. Shown to the left are PDF cell mir-92a levels at ZT6 and ZT18 from flies entrained under LD cycles. Shown to the right are PDF cell mir-92a levels from flies exposed to a 10-min light pulse at either ZT15 or ZT21 (same paradigm as Fig. 4A). ZT16/22 indicates mir-92a expression in controls with no exposure to light pulses. LP16/22 indicates mir-92a expression in flies with light pulse exposure. N = 2 biological replicates. Bar graphs show the average values of the replicates.
22 Supplementary Table 1 TRAP values of mir-92a target candidates with decreased IP/IN levels in mir-92a overexpression flies and increased levels in mir-92asp flies GENE NAMES MIR- 92AOE/W1 118 (IP/IN)/(IP/ IN)-1 MIR- 92AOE/W1 118 (IP/IN)/(IP/ IN)-2 AVERAGE MIR- 92AOE/W1 118 (IP/IN)/(IP /IN) MIR- 92ASP/SCRA MBLE (IP/IN)/(IP/IN )-1 MIR- 92ASP/SCRA MBLE (IP/IN)/(IP/IN )-2 AVERAGE MIR- 92ASP/SCRA MBLE (IP/IN)/(IP/IN ) CG CG CG CG CG REGUCAL CIN CG CG CG L(3)73AH KAT RAB ARR DER CG CG SIRT CG HIL CG CG ONECUT LOLA CG PDK ELAV CG POR
23 Supplementary Table 2 Primer list sirt2 3'UTR fwd sirt2 3'UTR rv sirt2 mut 3'UTR fwd sirt2 mut 3'UTR rv Forward primer: Small RNA PCR Primer 2 Reverse primer RT primer 3' linker (linker1) GCAGTAATTCTAGGCGATCGCACCCTAAGATTAGTTAGTAACATCCGT AGTTAATTTGTAGTTG AATGAAAATAAAGATATTTTATTGCGGCCAGCTTAGCCAGCAATGCC TGCGCTT CCGTAGTTAATTTGTAGTTGAATTcacgttaTTGTTTACATGTGGATTAC GTAATCCACATGTAAACAAtaacgtgAATTCAACTACAAATTAACTACGG AATGATACGGCGACCACCGACAGGTTCAGAGTTCTACAGTCCGA CAAGCAGAAGACGGCATACGAGATACATCGGTGACTGGAGTTATTGA TGGTGCCTACAG ATTGATGGTGCCTACAG rappctgtaggcaccatcaat/ddc/ 5 'linker GUUCAGAGUUCUACAGUCCGACGAUC dme-mir- 92a-3p fwd pr 2S fwd pr dme-mir- 184 fwd pr dme-mir- 999 fwd pr dme-mir- 981 fwd pr dme-mir- 210 fwd pr dme-mir- 276a fwd pr sirt2 fwd sirt2 rv ACACTCCAGCTGGGcattgcacttgtccc ACACTCCAGCTGGGtacaaccctcaacca ACACTCCAGCTGGGtggacggagaactga ACACTCCAGCTGGGtgttaactgtaagac ACACTCCAGCTGGGttcgttgtcgacgaa ACACTCCAGCTGGGttgtgcgtgtgacag ACACTCCAGCTGGGtaggaacttcatacc TTACCATGGGCGACATCGAA GTCGTCACTGGACGAGGAAT
Supplementary Figure1: ClustalW comparison between Tll, Dsf and NR2E1.
 P-Box Dsf -----------------MG-TAG--DRLLD-IPCKVCGDRSSGKHYGIYSCDGCSGFFKR 39 NR2E1 -----------------MSKPAGSTSRILD-IPCKVCGDRSSGKHYGVYACDGCSGFFKR 42 Tll MQSSEGSPDMMDQKYNSVRLSPAASSRILYHVPCKVCRDHSSGKHYGIYACDGCAGFFKR
P-Box Dsf -----------------MG-TAG--DRLLD-IPCKVCGDRSSGKHYGIYSCDGCSGFFKR 39 NR2E1 -----------------MSKPAGSTSRILD-IPCKVCGDRSSGKHYGVYACDGCSGFFKR 42 Tll MQSSEGSPDMMDQKYNSVRLSPAASSRILYHVPCKVCRDHSSGKHYGIYACDGCAGFFKR
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10810 Supplementary Fig. 1: Mutation of the loqs gene leads to shortened lifespan and adult-onset brain degeneration. a. Northern blot of control and loqs f00791 mutant flies. loqs f00791
doi:10.1038/nature10810 Supplementary Fig. 1: Mutation of the loqs gene leads to shortened lifespan and adult-onset brain degeneration. a. Northern blot of control and loqs f00791 mutant flies. loqs f00791
Supporting Information
 Supporting Information Ho et al. 10.1073/pnas.0808899106 SI Materials and Methods In immunostaining, antibodies from Developmental Studies Hybridoma ank were -FasII (mouse, 1:200), - (mouse, 1:100), and
Supporting Information Ho et al. 10.1073/pnas.0808899106 SI Materials and Methods In immunostaining, antibodies from Developmental Studies Hybridoma ank were -FasII (mouse, 1:200), - (mouse, 1:100), and
Derepressing muscleblind expression by mirna sponges ameliorates myotonic dystrophy-like. Ruben Artero 1,2 *
 Derepressing muscleblind expression by mirna sponges ameliorates myotonic dystrophy-like phenotypes in Drosophila. Estefania Cerro-Herreros 1,2, Juan M. Fernandez-Costa 1,2, María Sabater-Arcis 1,2, Beatriz
Derepressing muscleblind expression by mirna sponges ameliorates myotonic dystrophy-like phenotypes in Drosophila. Estefania Cerro-Herreros 1,2, Juan M. Fernandez-Costa 1,2, María Sabater-Arcis 1,2, Beatriz
Supplementary Figure 1 (related to Figure 1, 2 and Table 1)
 Supplementary Figure 1 (related to Figure 1, 2 and Table 1) Role of E75 in regulating circadian rhythms. (A) E75 mrna expression in TG 27 controls, TG 27 >NE-30-49-10, TG 27 >UAS-E75 (II), TG 27 >UAS-E75
Supplementary Figure 1 (related to Figure 1, 2 and Table 1) Role of E75 in regulating circadian rhythms. (A) E75 mrna expression in TG 27 controls, TG 27 >NE-30-49-10, TG 27 >UAS-E75 (II), TG 27 >UAS-E75
Supplementary Figure 1 Validate the expression of mir-302b after bacterial infection by northern
 Supplementary Figure 1 Validate the expression of mir-302b after bacterial infection by northern blot. Northern blot analysis of mir-302b expression following infection with PAO1, PAK and Kp in (A) lung
Supplementary Figure 1 Validate the expression of mir-302b after bacterial infection by northern blot. Northern blot analysis of mir-302b expression following infection with PAO1, PAK and Kp in (A) lung
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Supplementary figure 1: List of primers/oligonucleotides used in this study. 1 Supplementary figure 2: Sequences and mirna-targets of i) mcherry expresses in transgenic fish used
SUPPLEMENTARY INFORMATION Supplementary figure 1: List of primers/oligonucleotides used in this study. 1 Supplementary figure 2: Sequences and mirna-targets of i) mcherry expresses in transgenic fish used
B. Transgenic plants with strong phenotype (%)
 A. TCTAGTTGTTGTTGTTATGGTCTAGTTGTTGTTGTTATGGTCTAATTT AAATATGGTCTAAAGAAGAAGAATATGGTCTAAAGAAGAAGAATATGG 2XP35S STTM165 5 GGGGGATGAAGctaCCTGGTCCGA3 3 CCCCCUACUUC---GGACCAGGCU5 mir165 HindIII mir165 96 nt GTTGTTGTTGTTATGGTCTAGTTGTTGTTGTTATGGTCTAATTT
A. TCTAGTTGTTGTTGTTATGGTCTAGTTGTTGTTGTTATGGTCTAATTT AAATATGGTCTAAAGAAGAAGAATATGGTCTAAAGAAGAAGAATATGG 2XP35S STTM165 5 GGGGGATGAAGctaCCTGGTCCGA3 3 CCCCCUACUUC---GGACCAGGCU5 mir165 HindIII mir165 96 nt GTTGTTGTTGTTATGGTCTAGTTGTTGTTGTTATGGTCTAATTT
Fig. S1. Negative controls for in situ hybridization results using bam antisense probe. Fig. S2. Overexpression of either mir-275 or mir306
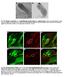 Fig. S1. Negative controls for in situ hybridization results using bam antisense probe. (A) In situ hybridization using bam sense probe in wild-type testis. (B) In situ hybridization using bam antisense
Fig. S1. Negative controls for in situ hybridization results using bam antisense probe. (A) In situ hybridization using bam sense probe in wild-type testis. (B) In situ hybridization using bam antisense
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Endogenous gene tagging to study subcellular localization and chromatin binding. a, b, Schematic of experimental set-up to endogenously tag RNAi factors using the CRISPR Cas9 technology,
Supplementary Figure 1 Endogenous gene tagging to study subcellular localization and chromatin binding. a, b, Schematic of experimental set-up to endogenously tag RNAi factors using the CRISPR Cas9 technology,
Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected
 Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected with the sirna against lnc-2, lnc-6, lnc-7, and the
Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected with the sirna against lnc-2, lnc-6, lnc-7, and the
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling
 Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary Figure 1. Nur77 and leptin-controlled obesity. (A) (B) (C)
 Supplementary Figure 1. Nur77 and leptin-controlled obesity. (A) Effect of leptin on body weight and food intake between WT and KO mice at the age of 12 weeks (n=7). Mice were i.c.v. injected with saline
Supplementary Figure 1. Nur77 and leptin-controlled obesity. (A) Effect of leptin on body weight and food intake between WT and KO mice at the age of 12 weeks (n=7). Mice were i.c.v. injected with saline
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified
 Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
SUPPLEMENTARY INFORMATION
 doi:.38/nature899 Supplementary Figure Suzuki et al. a c p7 -/- / WT ratio (+)/(-) p7 -/- / WT ratio Log X 3. Fold change by treatment ( (+)/(-)) Log X.5 3-3. -. b Fold change by treatment ( (+)/(-)) 8
doi:.38/nature899 Supplementary Figure Suzuki et al. a c p7 -/- / WT ratio (+)/(-) p7 -/- / WT ratio Log X 3. Fold change by treatment ( (+)/(-)) Log X.5 3-3. -. b Fold change by treatment ( (+)/(-)) 8
IGF-1 promotes the development and cytotoxic activity of human NK cells regulated
 Supplementary Information for IGF-1 promotes the development and cytotoxic activity of human NK cells regulated by mir-483-3p Fang Ni 1, 2, Rui Sun 1, Binqing Fu 1, Fuyan Wang 1, Chuang Guo 1, Zhigang
Supplementary Information for IGF-1 promotes the development and cytotoxic activity of human NK cells regulated by mir-483-3p Fang Ni 1, 2, Rui Sun 1, Binqing Fu 1, Fuyan Wang 1, Chuang Guo 1, Zhigang
embryos. Asterisk represents loss of or reduced expression. Brackets represent
 Supplemental Figures Supplemental Figure 1. tfec expression is highly enriched in tail endothelial cells (A- B) ISH of tfec at 15 and 16hpf in WT embryos. (C- D) ISH of tfec at 36 and 38hpf in WT embryos.
Supplemental Figures Supplemental Figure 1. tfec expression is highly enriched in tail endothelial cells (A- B) ISH of tfec at 15 and 16hpf in WT embryos. (C- D) ISH of tfec at 36 and 38hpf in WT embryos.
Document S1. Supplemental Experimental Procedures and Three Figures (see next page)
 Supplemental Data Document S1. Supplemental Experimental Procedures and Three Figures (see next page) Table S1. List of Candidate Genes Identified from the Screen. Candidate genes, corresponding dsrnas
Supplemental Data Document S1. Supplemental Experimental Procedures and Three Figures (see next page) Table S1. List of Candidate Genes Identified from the Screen. Candidate genes, corresponding dsrnas
TRIM31 is recruited to mitochondria after infection with SeV.
 Supplementary Figure 1 TRIM31 is recruited to mitochondria after infection with SeV. (a) Confocal microscopy of TRIM31-GFP transfected into HEK293T cells for 24 h followed with SeV infection for 6 h. MitoTracker
Supplementary Figure 1 TRIM31 is recruited to mitochondria after infection with SeV. (a) Confocal microscopy of TRIM31-GFP transfected into HEK293T cells for 24 h followed with SeV infection for 6 h. MitoTracker
Revision Checklist for Science Signaling Research Manuscripts: Data Requirements and Style Guidelines
 Revision Checklist for Science Signaling Research Manuscripts: Data Requirements and Style Guidelines Further information can be found at: http://stke.sciencemag.org/sites/default/files/researcharticlerevmsinstructions_0.pdf.
Revision Checklist for Science Signaling Research Manuscripts: Data Requirements and Style Guidelines Further information can be found at: http://stke.sciencemag.org/sites/default/files/researcharticlerevmsinstructions_0.pdf.
Regulation of Synaptic Structure and Function by FMRP- Associated MicroRNAs mir-125b and mir-132
 Neuron, Volume 65 Regulation of Synaptic Structure and Function by FMRP- Associated MicroRNAs mir-125b and mir-132 Dieter Edbauer, Joel R. Neilson, Kelly A. Foster, Chi-Fong Wang, Daniel P. Seeburg, Matthew
Neuron, Volume 65 Regulation of Synaptic Structure and Function by FMRP- Associated MicroRNAs mir-125b and mir-132 Dieter Edbauer, Joel R. Neilson, Kelly A. Foster, Chi-Fong Wang, Daniel P. Seeburg, Matthew
Supplementary Figure 1. The LIR motif in Kenny is necessary for its targeting to the lysosomes. (a-b) Confocal sections from fat body cells from
 Supplementary Figure 1. The LIR motif in Kenny is necessary for its targeting to the lysosomes. (a-b) Confocal sections from fat body cells from larvae expressing GFP-Kenny WT (a-a ) or GFP-Kenny F7A/L10A
Supplementary Figure 1. The LIR motif in Kenny is necessary for its targeting to the lysosomes. (a-b) Confocal sections from fat body cells from larvae expressing GFP-Kenny WT (a-a ) or GFP-Kenny F7A/L10A
SUPPLEMENTARY INFORMATION
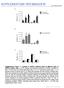 SUPPLEMENTARY INFORMATION doi:10.1038/nature12474 Supplementary Figure 1 Analysis of mtdna mutation loads in different types of mtdna mutator mice. a, PCR, cloning, and sequencing analysis of mtdna mutations
SUPPLEMENTARY INFORMATION doi:10.1038/nature12474 Supplementary Figure 1 Analysis of mtdna mutation loads in different types of mtdna mutator mice. a, PCR, cloning, and sequencing analysis of mtdna mutations
Supplemental Data. Na Xu et al. (2016). Plant Cell /tpc
 Supplemental Figure 1. The weak fluorescence phenotype is not caused by the mutation in At3g60240. (A) A mutation mapped to the gene At3g60240. Map-based cloning strategy was used to map the mutated site
Supplemental Figure 1. The weak fluorescence phenotype is not caused by the mutation in At3g60240. (A) A mutation mapped to the gene At3g60240. Map-based cloning strategy was used to map the mutated site
Supplementary Figure 1. ips_3y+mir-302b. ips_3y+mir-372. ips_3y+mir-302b + mir-372. ips_4y. ips_4y+mir-302b. ips_4y + mir-372
 Supplementary Figure 1 ips_3y+mir-32b ips_3y+ ips_3y+mir-32b + ips_4y ips_4y+mir-32b ips_4y + Nature Biotechnology: doi:1.138/nb.t.1862 TRA-1-6 DAPI Oct3/4 Supplementary Figure 1: ips cells derived from
Supplementary Figure 1 ips_3y+mir-32b ips_3y+ ips_3y+mir-32b + ips_4y ips_4y+mir-32b ips_4y + Nature Biotechnology: doi:1.138/nb.t.1862 TRA-1-6 DAPI Oct3/4 Supplementary Figure 1: ips cells derived from
Comparative Analysis of Argonaute-Dependent Small RNA Pathways in Drosophila
 Molecular Cell, Volume 32 Supplemental Data Comparative Analysis of Argonaute-Dependent Small RNA Pathways in Drosophila Rui Zhou, Ikuko Hotta, Ahmet M. Denli, Pengyu Hong, Norbert Perrimon, and Gregory
Molecular Cell, Volume 32 Supplemental Data Comparative Analysis of Argonaute-Dependent Small RNA Pathways in Drosophila Rui Zhou, Ikuko Hotta, Ahmet M. Denli, Pengyu Hong, Norbert Perrimon, and Gregory
Supplementary Figure 1 A
 Supplementary Figure A B M. mullata p53, 3 UTR Luciferase activity (%) mir-5b 8 Le et al. Supplementary Information NC-DP - + - - - - - NC-DP - - + - - - - NC-DP3 - - - + - - - 5b-DP - - - - + + + NC-AS
Supplementary Figure A B M. mullata p53, 3 UTR Luciferase activity (%) mir-5b 8 Le et al. Supplementary Information NC-DP - + - - - - - NC-DP - - + - - - - NC-DP3 - - - + - - - 5b-DP - - - - + + + NC-AS
(a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for
 Supplementary Figure 1 Generation of Diaph3 ko mice. (a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for different regions of Diaph3 mrna from WT, heterozygote
Supplementary Figure 1 Generation of Diaph3 ko mice. (a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for different regions of Diaph3 mrna from WT, heterozygote
Supporting Information
 Supporting Information SI Materials and Methods RT-qPCR The 25 µl qrt-pcr reaction mixture included 1 µl of cdna or DNA, 12.5 µl of 2X SYBER Green Master Mix (Applied Biosystems ), 5 µm of primers and
Supporting Information SI Materials and Methods RT-qPCR The 25 µl qrt-pcr reaction mixture included 1 µl of cdna or DNA, 12.5 µl of 2X SYBER Green Master Mix (Applied Biosystems ), 5 µm of primers and
Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan).
 1 2 3 4 5 6 7 8 Supplemental Materials and Methods Cell proliferation assay Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). GCs were plated at 96-well
1 2 3 4 5 6 7 8 Supplemental Materials and Methods Cell proliferation assay Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). GCs were plated at 96-well
SUPPLEMENTAL FIGURES AND TABLES
 SUPPLEMENTAL FIGURES AND TABLES A B Flag-ALDH1A1 IP: α-ac HEK293T WT 91R 128R 252Q 367R 41/ 419R 435R 495R 412R C Flag-ALDH1A1 NAM IP: HEK293T + + - + D NAM - + + E Relative ALDH1A1 activity 1..8.6.4.2
SUPPLEMENTAL FIGURES AND TABLES A B Flag-ALDH1A1 IP: α-ac HEK293T WT 91R 128R 252Q 367R 41/ 419R 435R 495R 412R C Flag-ALDH1A1 NAM IP: HEK293T + + - + D NAM - + + E Relative ALDH1A1 activity 1..8.6.4.2
File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description:
 File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description: Supplementary Figure 1. dcas9-mq1 fusion protein induces de novo
File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description: Supplementary Figure 1. dcas9-mq1 fusion protein induces de novo
Figure 1-Figure Supplement 1 Validation of knockdown efficiency for trpa1 RNAi and RyR RNAi lines.
 Figure 1-Figure Supplement 1 Validation of knockdown efficiency for trpa1 RNAi and RyR RNAi lines. (A) Cartoon depicting the four characterized trpa1 isoforms. The target regions of trpa1 RNAi lines used
Figure 1-Figure Supplement 1 Validation of knockdown efficiency for trpa1 RNAi and RyR RNAi lines. (A) Cartoon depicting the four characterized trpa1 isoforms. The target regions of trpa1 RNAi lines used
Supplementary
 Supplementary information Supplementary Material and Methods Plasmid construction The transposable element vectors for inducible expression of RFP-FUS wt and EGFP-FUS R521C and EGFP-FUS P525L were derived
Supplementary information Supplementary Material and Methods Plasmid construction The transposable element vectors for inducible expression of RFP-FUS wt and EGFP-FUS R521C and EGFP-FUS P525L were derived
MeCP2. MeCP2/α-tubulin. GFP mir1-1 mir132
 Conservation Figure S1. Schematic showing 3 UTR (top; thick black line), mir132 MRE (arrow) and nucleotide sequence conservation (vertical black lines; http://genome.ucsc.edu). a GFP mir1-1 mir132 b GFP
Conservation Figure S1. Schematic showing 3 UTR (top; thick black line), mir132 MRE (arrow) and nucleotide sequence conservation (vertical black lines; http://genome.ucsc.edu). a GFP mir1-1 mir132 b GFP
Supplementary Figure 1, related to Figure 1. GAS5 is highly expressed in the cytoplasm of hescs, and positively correlates with pluripotency.
 Supplementary Figure 1, related to Figure 1. GAS5 is highly expressed in the cytoplasm of hescs, and positively correlates with pluripotency. (a) Transfection of different concentration of GAS5-overexpressing
Supplementary Figure 1, related to Figure 1. GAS5 is highly expressed in the cytoplasm of hescs, and positively correlates with pluripotency. (a) Transfection of different concentration of GAS5-overexpressing
Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon Song, Richard M. Amasino, Bosl Noh, and Yoo-Sun Noh
 Developmental Cell, Volume 22 Supplemental Information Control of Seed Germination by Light-Induced Histone Arginine Demethylation Activity Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon
Developmental Cell, Volume 22 Supplemental Information Control of Seed Germination by Light-Induced Histone Arginine Demethylation Activity Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon
Hoopfer et al., Supplemental Data Supplemental Figure S1. Quantification of ORN axon degeneration
 Hoopfer et al., Supplemental Data Supplemental Figure S1. Quantification of ORN axon degeneration (A) Mean GFP intensity of commissural ORN axons 1 day after antennae removal. 1 day after cutting, wt ORNs
Hoopfer et al., Supplemental Data Supplemental Figure S1. Quantification of ORN axon degeneration (A) Mean GFP intensity of commissural ORN axons 1 day after antennae removal. 1 day after cutting, wt ORNs
(a) Immunoblotting to show the migration position of Flag-tagged MAVS
 Supplementary Figure 1 Characterization of six MAVS isoforms. (a) Immunoblotting to show the migration position of Flag-tagged MAVS isoforms. HEK293T Mavs -/- cells were transfected with constructs expressing
Supplementary Figure 1 Characterization of six MAVS isoforms. (a) Immunoblotting to show the migration position of Flag-tagged MAVS isoforms. HEK293T Mavs -/- cells were transfected with constructs expressing
Sarker et al. Supplementary Material. Subcellular Fractionation
 Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
ASPP1 Fw GGTTGGGAATCCACGTGTTG ASPP1 Rv GCCATATCTTGGAGCTCTGAGAG
 Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
Nature Biotechnology: doi: /nbt Supplementary Figure 1
 Supplementary Figure 1 Schematic and results of screening the combinatorial antibody library for Sox2 replacement activity. A single batch of MEFs were plated and transduced with doxycycline inducible
Supplementary Figure 1 Schematic and results of screening the combinatorial antibody library for Sox2 replacement activity. A single batch of MEFs were plated and transduced with doxycycline inducible
a KYSE270-CON KYSE270-Id1
 a KYSE27-CON KYSE27- shcon shcon sh b Human Mouse CD31 Relative MVD 3.5 3 2.5 2 1.5 1.5 *** *** c KYSE15 KYSE27 sirna (nm) 5 1 Id2 Id2 sirna 5 1 sirna (nm) 5 1 Id2 sirna 5 1 Id2 [h] (pg per ml) d 3 2 1
a KYSE27-CON KYSE27- shcon shcon sh b Human Mouse CD31 Relative MVD 3.5 3 2.5 2 1.5 1.5 *** *** c KYSE15 KYSE27 sirna (nm) 5 1 Id2 Id2 sirna 5 1 sirna (nm) 5 1 Id2 sirna 5 1 Id2 [h] (pg per ml) d 3 2 1
Stargazin regulates AMPA receptor trafficking through adaptor protein. complexes during long term depression
 Supplementary Information Stargazin regulates AMPA receptor trafficking through adaptor protein complexes during long term depression Shinji Matsuda, Wataru Kakegawa, Timotheus Budisantoso, Toshihiro Nomura,
Supplementary Information Stargazin regulates AMPA receptor trafficking through adaptor protein complexes during long term depression Shinji Matsuda, Wataru Kakegawa, Timotheus Budisantoso, Toshihiro Nomura,
Functional analysis reveals that RBM10 mutations. contribute to lung adenocarcinoma pathogenesis by. deregulating splicing
 Supplementary Information Functional analysis reveals that RBM10 mutations contribute to lung adenocarcinoma pathogenesis by deregulating splicing Jiawei Zhao 1,+, Yue Sun 2,3,+, Yin Huang 6, Fan Song
Supplementary Information Functional analysis reveals that RBM10 mutations contribute to lung adenocarcinoma pathogenesis by deregulating splicing Jiawei Zhao 1,+, Yue Sun 2,3,+, Yin Huang 6, Fan Song
Xu et al., Supplementary Figures 1-7
 Xu et al., Supplementary Figures 1-7 Supplementary Figure 1. PIPKI is required for ciliogenesis. (a) PIPKI localizes at the basal body of primary cilium. RPE-1 cells treated with two sirnas targeting to
Xu et al., Supplementary Figures 1-7 Supplementary Figure 1. PIPKI is required for ciliogenesis. (a) PIPKI localizes at the basal body of primary cilium. RPE-1 cells treated with two sirnas targeting to
AD BD TOC1. Supplementary Figure 1: Yeast two-hybrid assays showing the interaction between
 AD X BD TOC1 AD BD X PIFΔAD PIF TOC1 TOC1 PIFΔAD PIF N TOC1 TOC1 C1 PIFΔAD PIF C1 TOC1 TOC1 C PIFΔAD PIF C TOC1 Supplementary Figure 1: Yeast two-hybrid assays showing the interaction between PIF and TOC1
AD X BD TOC1 AD BD X PIFΔAD PIF TOC1 TOC1 PIFΔAD PIF N TOC1 TOC1 C1 PIFΔAD PIF C1 TOC1 TOC1 C PIFΔAD PIF C TOC1 Supplementary Figure 1: Yeast two-hybrid assays showing the interaction between PIF and TOC1
Two classes of silencing RNAs move between Caenorhabditis elegans tissues.
 Two classes of silencing RNAs move between Caenorhabditis elegans tissues. Antony M Jose, Giancarlo A Garcia, and Craig P Hunter. Supplementary Figures, Figure Legends, and Tables. Supplementary Figure
Two classes of silencing RNAs move between Caenorhabditis elegans tissues. Antony M Jose, Giancarlo A Garcia, and Craig P Hunter. Supplementary Figures, Figure Legends, and Tables. Supplementary Figure
Supplemental Material: Rev1 promotes replication through UV lesions in conjunction with DNA
 Supplemental Material: Rev1 promotes replication through UV lesions in conjunction with DNA polymerases,, and, but not with DNA polymerase Jung-Hoon Yoon, Jeseong Park, Juan Conde, Maki Wakamiya, Louise
Supplemental Material: Rev1 promotes replication through UV lesions in conjunction with DNA polymerases,, and, but not with DNA polymerase Jung-Hoon Yoon, Jeseong Park, Juan Conde, Maki Wakamiya, Louise
Long-term, efficient inhibition of microrna function in mice using raav vectors
 Nature Methods Long-term, efficient inhibition of microrna function in mice using raav vectors Jun Xie, Stefan L Ameres, Randall Friedline, Jui-Hung Hung, Yu Zhang, Qing Xie, Li Zhong, Qin Su, Ran He,
Nature Methods Long-term, efficient inhibition of microrna function in mice using raav vectors Jun Xie, Stefan L Ameres, Randall Friedline, Jui-Hung Hung, Yu Zhang, Qing Xie, Li Zhong, Qin Su, Ran He,
supplementary information
 Figure S1 ZEB1 full length mrna. (a) Analysis of the ZEB1 mrna using the UCSC genome browser (http://genome.ucsc.edu) revealed truncation of the annotated Refseq sequence (NM_030751). The probable terminus
Figure S1 ZEB1 full length mrna. (a) Analysis of the ZEB1 mrna using the UCSC genome browser (http://genome.ucsc.edu) revealed truncation of the annotated Refseq sequence (NM_030751). The probable terminus
SUPPLEMENTARY INFORMATION
 Ca 2+ /calmodulin Regulates Salicylic Acid-mediated Plant Immunity Liqun Du, Gul S. Ali, Kayla A. Simons, Jingguo Hou, Tianbao Yang, A.S.N. Reddy and B. W. Poovaiah * *To whom correspondence should be
Ca 2+ /calmodulin Regulates Salicylic Acid-mediated Plant Immunity Liqun Du, Gul S. Ali, Kayla A. Simons, Jingguo Hou, Tianbao Yang, A.S.N. Reddy and B. W. Poovaiah * *To whom correspondence should be
OmicsLink shrna Clones guaranteed knockdown even in difficult-to-transfect cells
 OmicsLink shrna Clones guaranteed knockdown even in difficult-to-transfect cells OmicsLink shrna clone collections consist of lentiviral, and other mammalian expression vector based small hairpin RNA (shrna)
OmicsLink shrna Clones guaranteed knockdown even in difficult-to-transfect cells OmicsLink shrna clone collections consist of lentiviral, and other mammalian expression vector based small hairpin RNA (shrna)
Supplemental Information
 Supplemental Information Itemized List Materials and Methods, Related to Supplemental Figures S5A-C and S6. Supplemental Figure S1, Related to Figures 1 and 2. Supplemental Figure S2, Related to Figure
Supplemental Information Itemized List Materials and Methods, Related to Supplemental Figures S5A-C and S6. Supplemental Figure S1, Related to Figures 1 and 2. Supplemental Figure S2, Related to Figure
Supplementary information for: Mutant p53 gain-of-function induces epithelial-mesenchymal transition. through modulation of the mir-130b-zeb1 axis
 Supplementary information for: Mutant p53 gain-of-function induces epithelial-mesenchymal transition through modulation of the mir-3b-zeb axis AUTHORS: Peixin Dong, Mihriban Karaayvaz, Nan Jia, Masanori
Supplementary information for: Mutant p53 gain-of-function induces epithelial-mesenchymal transition through modulation of the mir-3b-zeb axis AUTHORS: Peixin Dong, Mihriban Karaayvaz, Nan Jia, Masanori
(A-B) P2ry14 expression was assessed by (A) genotyping (upper arrow: WT; lower
 Supplementary Figures S1. (A-B) P2ry14 expression was assessed by (A) genotyping (upper arrow: ; lower arrow: KO) and (B) q-pcr analysis with Lin- cells, The white vertical line in panel A indicates that
Supplementary Figures S1. (A-B) P2ry14 expression was assessed by (A) genotyping (upper arrow: ; lower arrow: KO) and (B) q-pcr analysis with Lin- cells, The white vertical line in panel A indicates that
SUPPLEMENTARY INFORMATION
 AS-NMD modulates FLM-dependent thermosensory flowering response in Arabidopsis NATURE PLANTS www.nature.com/natureplants 1 Supplementary Figure 1. Genomic sequence of FLM along with the splice sites. Sequencing
AS-NMD modulates FLM-dependent thermosensory flowering response in Arabidopsis NATURE PLANTS www.nature.com/natureplants 1 Supplementary Figure 1. Genomic sequence of FLM along with the splice sites. Sequencing
Supplementary Materials: 1. Supplementary Figures S1-S9. 2. Supplementary Tables S1-S2
 Supplementary Materials: 1. Supplementary Figures S1-S9 2. Supplementary Tables S1-S2 S1 1. Supplementary Figures Fig. S1. Genotypes of tcp20 mutants. Fig. S2. Root phenotypes of tcp20 mutants. Fig. S3.
Supplementary Materials: 1. Supplementary Figures S1-S9 2. Supplementary Tables S1-S2 S1 1. Supplementary Figures Fig. S1. Genotypes of tcp20 mutants. Fig. S2. Root phenotypes of tcp20 mutants. Fig. S3.
supplementary information
 DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
High-resolution Identification and Separation of Living Cell Types by Multiple microrna-responsive Synthetic mrnas
 Supplementary information High-resolution Identification and Separation of Living Cell Types by Multiple microrna-responsive Synthetic mrnas Kei Endo *, Karin Hayashi, Hirohide Saito * Contents: 1. Supplementary
Supplementary information High-resolution Identification and Separation of Living Cell Types by Multiple microrna-responsive Synthetic mrnas Kei Endo *, Karin Hayashi, Hirohide Saito * Contents: 1. Supplementary
Supplemental Data. Liu et al. (2013). Plant Cell /tpc
 Supplemental Figure 1. The GFP Tag Does Not Disturb the Physiological Functions of WDL3. (A) RT-PCR analysis of WDL3 expression in wild-type, WDL3-GFP, and WDL3 (without the GFP tag) transgenic seedlings.
Supplemental Figure 1. The GFP Tag Does Not Disturb the Physiological Functions of WDL3. (A) RT-PCR analysis of WDL3 expression in wild-type, WDL3-GFP, and WDL3 (without the GFP tag) transgenic seedlings.
Supplementary Table 1. Primers used to construct full-length or various truncated mutants of ISG12b2.
 Supplementary Table 1. Primers used to construct full-length or various truncated mutants of ISG12b2. Construct name ISG12b2 (No tag) HA-ISG12b2 (N-HA) ISG12b2-HA (C-HA; FL-HA) 94-283-HA (FL-GFP) 93-GFP
Supplementary Table 1. Primers used to construct full-length or various truncated mutants of ISG12b2. Construct name ISG12b2 (No tag) HA-ISG12b2 (N-HA) ISG12b2-HA (C-HA; FL-HA) 94-283-HA (FL-GFP) 93-GFP
Regulation of axonal and dendritic growth by the extracellular calcium-sensing
 Regulation of axonal and dendritic growth by the extracellular calcium-sensing receptor (CaSR). Thomas N. Vizard, Gerard W. O Keeffe, Humberto Gutierrez, Claudine H. Kos, Daniela Riccardi, Alun M. Davies
Regulation of axonal and dendritic growth by the extracellular calcium-sensing receptor (CaSR). Thomas N. Vizard, Gerard W. O Keeffe, Humberto Gutierrez, Claudine H. Kos, Daniela Riccardi, Alun M. Davies
Regulation of transcription by the MLL2 complex and MLL complex-associated AKAP95
 Supplementary Information Regulation of transcription by the complex and MLL complex-associated Hao Jiang, Xiangdong Lu, Miho Shimada, Yali Dou, Zhanyun Tang, and Robert G. Roeder Input HeLa NE IP lot:
Supplementary Information Regulation of transcription by the complex and MLL complex-associated Hao Jiang, Xiangdong Lu, Miho Shimada, Yali Dou, Zhanyun Tang, and Robert G. Roeder Input HeLa NE IP lot:
Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) (b)
 Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) A schematic overview of the production and amplification of a single pirna from a transposon transcript. The
Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) A schematic overview of the production and amplification of a single pirna from a transposon transcript. The
DCLK-immunopositive. Bars, 100 µm for B, 50 µm for C.
 Supplementary Figure S1. Characterization of rabbit polyclonal anti-dclk antibody. (A) Immunoblotting of COS7 cells transfected with DCLK1-GFP and DCLK2-GFP expression plasmids probed with anti-dclk antibody
Supplementary Figure S1. Characterization of rabbit polyclonal anti-dclk antibody. (A) Immunoblotting of COS7 cells transfected with DCLK1-GFP and DCLK2-GFP expression plasmids probed with anti-dclk antibody
Supplementary Information Cyclin Y inhibits plasticity-induced AMPA receptor exocytosis and LTP
 Supplementary Information Cyclin Y inhibits plasticity-induced AMPA receptor exocytosis and LTP Eunsil Cho, Dong-Hyun Kim, Young-Na Hur, Daniel J. Whitcomb, Philip Regan, Jung-Hwa Hong, Hanna Kim, Young
Supplementary Information Cyclin Y inhibits plasticity-induced AMPA receptor exocytosis and LTP Eunsil Cho, Dong-Hyun Kim, Young-Na Hur, Daniel J. Whitcomb, Philip Regan, Jung-Hwa Hong, Hanna Kim, Young
Supplemental Figure 1. HepG2 cells were transfected with GLI luciferase reporter construct
 Supplemental Figure 1. HepG2 cells were transfected with GLI luciferase reporter construct (pgl38xgli), EWS-FLI1 luciferase reporter construct (NROB1-Luc) with or without GLI1, EWS- FLI1 and cdnas respectively.
Supplemental Figure 1. HepG2 cells were transfected with GLI luciferase reporter construct (pgl38xgli), EWS-FLI1 luciferase reporter construct (NROB1-Luc) with or without GLI1, EWS- FLI1 and cdnas respectively.
 Figure S1. Generation of HspA4 -/- mice. (A) Structures of the wild-type (Wt) HspA4 -/- allele, targeting vector and targeted allele are shown together with the relevant restriction sites. The filled rectangles
Figure S1. Generation of HspA4 -/- mice. (A) Structures of the wild-type (Wt) HspA4 -/- allele, targeting vector and targeted allele are shown together with the relevant restriction sites. The filled rectangles
SUPPLEMENTARY INFORMATION
 Supplementary Figure a T m ( C) Seq. '-3' uguuugugguaacagugugaggu L 62 AttGtcAcaCtcC L2 6 ccattgtcacactcc L3 66 attgtcacactcc 7 ccattgtcacactcca L 73 ccattgtcacactcc L6 74 AttGTcaCaCtCC L7 7 attgtcacactcc
Supplementary Figure a T m ( C) Seq. '-3' uguuugugguaacagugugaggu L 62 AttGtcAcaCtcC L2 6 ccattgtcacactcc L3 66 attgtcacactcc 7 ccattgtcacactcca L 73 ccattgtcacactcc L6 74 AttGTcaCaCtCC L7 7 attgtcacactcc
sirna Transfection Into Primary Neurons Using Fuse-It-siRNA
 sirna Transfection Into Primary Neurons Using Fuse-It-siRNA This Application Note describes a protocol for sirna transfection into sensitive, primary cortical neurons using Fuse-It-siRNA. This innovative
sirna Transfection Into Primary Neurons Using Fuse-It-siRNA This Application Note describes a protocol for sirna transfection into sensitive, primary cortical neurons using Fuse-It-siRNA. This innovative
Supplemental Methods Cell lines and culture
 Supplemental Methods Cell lines and culture AGS, CL5, BT549, and SKBR were propagated in RPMI 64 medium (Mediatech Inc., Manassas, VA) supplemented with % fetal bovine serum (FBS, Atlanta Biologicals,
Supplemental Methods Cell lines and culture AGS, CL5, BT549, and SKBR were propagated in RPMI 64 medium (Mediatech Inc., Manassas, VA) supplemented with % fetal bovine serum (FBS, Atlanta Biologicals,
Engineering splicing factors with designed specificities
 nature methods Engineering splicing factors with designed specificities Yang Wang, Cheom-Gil Cheong, Traci M Tanaka Hall & Zefeng Wang Supplementary figures and text: Supplementary Figure 1 Supplementary
nature methods Engineering splicing factors with designed specificities Yang Wang, Cheom-Gil Cheong, Traci M Tanaka Hall & Zefeng Wang Supplementary figures and text: Supplementary Figure 1 Supplementary
0.9 5 H M L E R -C tr l in T w is t1 C M
 a. b. c. d. e. f. g. h. 2.5 C elltiter-g lo A ssay 1.1 5 M T S a s s a y Lum inescence (A.U.) 2.0 1.5 1.0 0.5 n s H M L E R -C tr l in C tr l C M H M L E R -C tr l in S n a il1 C M A bsorbance (@ 490nm
a. b. c. d. e. f. g. h. 2.5 C elltiter-g lo A ssay 1.1 5 M T S a s s a y Lum inescence (A.U.) 2.0 1.5 1.0 0.5 n s H M L E R -C tr l in C tr l C M H M L E R -C tr l in S n a il1 C M A bsorbance (@ 490nm
Chemical hijacking of auxin signaling with an engineered auxin-tir1
 1 SUPPLEMENTARY INFORMATION Chemical hijacking of auxin signaling with an engineered auxin-tir1 pair Naoyuki Uchida 1,2*, Koji Takahashi 2*, Rie Iwasaki 1, Ryotaro Yamada 2, Masahiko Yoshimura 2, Takaho
1 SUPPLEMENTARY INFORMATION Chemical hijacking of auxin signaling with an engineered auxin-tir1 pair Naoyuki Uchida 1,2*, Koji Takahashi 2*, Rie Iwasaki 1, Ryotaro Yamada 2, Masahiko Yoshimura 2, Takaho
Supplementary Methods
 Supplementary Methods MARCM-based forward genetic screen We used ethylmethane sulfonate (EMS) to mutagenize flies carrying FRT 2A and FRT 82B transgenes, which are sites for the FLP-mediated recombination
Supplementary Methods MARCM-based forward genetic screen We used ethylmethane sulfonate (EMS) to mutagenize flies carrying FRT 2A and FRT 82B transgenes, which are sites for the FLP-mediated recombination
Nature Methods: doi: /nmeth Supplementary Figure 1. Neither driver nor effector alone displays expression of GFP
 Supplementary Figure 1 Neither driver nor effector alone displays expression of GFP Comparison of GFP fluorescence in a driver- only strain (a) or an effector-only strain (b) to their driver + effector
Supplementary Figure 1 Neither driver nor effector alone displays expression of GFP Comparison of GFP fluorescence in a driver- only strain (a) or an effector-only strain (b) to their driver + effector
RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by. 5 AGACACAAACACCAUUGUCACACUCCACAGC; Rand-2 OMe,
 Materials and methods Oligonucleotides and DNA constructs RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by Dharmacon Inc. (Lafayette, CO). The sequences were: 122-2 OMe, 5
Materials and methods Oligonucleotides and DNA constructs RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by Dharmacon Inc. (Lafayette, CO). The sequences were: 122-2 OMe, 5
Competition for XPO5 binding between Dicer mrna, pre-mirna and viral RNA regulates human Dicer levels
 CORRECTION NOTICE Nat. Struct. Mol. Biol. 18, 323 327 (211) Competition for binding between mrna, pre-mirna and viral RNA regulates human levels Yamina Bennasser, Christine Chable-Bessia, Robinson Triboulet,
CORRECTION NOTICE Nat. Struct. Mol. Biol. 18, 323 327 (211) Competition for binding between mrna, pre-mirna and viral RNA regulates human levels Yamina Bennasser, Christine Chable-Bessia, Robinson Triboulet,
Supplementary Figure 1
 Supplementary Figure 1 Virus infection induces RNF128 expression. (a,b) RT-PCR analysis of Rnf128 (RNF128) mrna expression in mouse peritoneal macrophages (a) and THP-1 cells (b) upon stimulation with
Supplementary Figure 1 Virus infection induces RNF128 expression. (a,b) RT-PCR analysis of Rnf128 (RNF128) mrna expression in mouse peritoneal macrophages (a) and THP-1 cells (b) upon stimulation with
Supplementary Figure 1. TRIM9 does not affect AP-1, NF-AT or ISRE activity. (a,b) At 24h post-transfection with TRIM9 or vector and indicated
 Supplementary Figure 1. TRIM9 does not affect AP-1, NF-AT or ISRE activity. (a,b) At 24h post-transfection with TRIM9 or vector and indicated reporter luciferase constructs, HEK293T cells were stimulated
Supplementary Figure 1. TRIM9 does not affect AP-1, NF-AT or ISRE activity. (a,b) At 24h post-transfection with TRIM9 or vector and indicated reporter luciferase constructs, HEK293T cells were stimulated
Supplementary Information Alternative splicing of CD44 mrna by ESRP1 enhances lung colonization of metastatic cancer cell
 Supplementary Information Alternative splicing of CD44 mrna by ESRP1 enhances lung colonization of metastatic cancer cell Supplementary Figures S1-S3 Supplementary Methods Supplementary Figure S1. Identification
Supplementary Information Alternative splicing of CD44 mrna by ESRP1 enhances lung colonization of metastatic cancer cell Supplementary Figures S1-S3 Supplementary Methods Supplementary Figure S1. Identification
Supplemental Material Igreja and Izaurralde 1. CUP promotes deadenylation and inhibits decapping of mrna targets. Catia Igreja and Elisa Izaurralde
 Supplemental Material Igreja and Izaurralde 1 CUP promotes deadenylation and inhibits decapping of mrna targets Catia Igreja and Elisa Izaurralde Supplemental Materials and methods Functional assays and
Supplemental Material Igreja and Izaurralde 1 CUP promotes deadenylation and inhibits decapping of mrna targets Catia Igreja and Elisa Izaurralde Supplemental Materials and methods Functional assays and
Supplementary Figure 1. Reintroduction of HDAC1 in HDAC1-/- ES cells reverts the
 SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1. Reintroduction of HDAC1 in HDAC1-/- ES cells reverts the phenotype of HDAC1-/- teratomas. 3x10 6 HDAC1 reintroduced (HDAC1-/-re) and empty vector infected
SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1. Reintroduction of HDAC1 in HDAC1-/- ES cells reverts the phenotype of HDAC1-/- teratomas. 3x10 6 HDAC1 reintroduced (HDAC1-/-re) and empty vector infected
Nature Biotechnology: doi: /nbt Supplementary Figure 1. Design and sequence of system 1 for targeted demethylation.
 Supplementary Figure 1 Design and sequence of system 1 for targeted demethylation. (a) Design of system 1 for targeted demethylation. TET1CD was fused to a catalytic inactive Cas9 nuclease (dcas9) and
Supplementary Figure 1 Design and sequence of system 1 for targeted demethylation. (a) Design of system 1 for targeted demethylation. TET1CD was fused to a catalytic inactive Cas9 nuclease (dcas9) and
p53 increases MHC class I expression by upregulating the endoplasmic reticulum aminopeptidase ERAP1
 Supplementary Information p53 increases MHC class I expression by upregulating the endoplasmic reticulum aminopeptidase ERAP1 Bei Wang 1, Dandan Niu 1, Liyun Lai 1, & Ee Chee Ren 1,2 1 Singapore Immunology
Supplementary Information p53 increases MHC class I expression by upregulating the endoplasmic reticulum aminopeptidase ERAP1 Bei Wang 1, Dandan Niu 1, Liyun Lai 1, & Ee Chee Ren 1,2 1 Singapore Immunology
Structural and functional analysis of the mir-282 microrna gene of Drosophila melanogaster. Ágnes Bujna
 Structural and functional analysis of the mir-282 microrna gene of Drosophila melanogaster Ph.D. thesis Ágnes Bujna Supervisor: Dr. Miklós Erdélyi University of Szeged, PhD School of Biological Sciences
Structural and functional analysis of the mir-282 microrna gene of Drosophila melanogaster Ph.D. thesis Ágnes Bujna Supervisor: Dr. Miklós Erdélyi University of Szeged, PhD School of Biological Sciences
Supplementary Figure 1. Co-localization of GLUT1 and DNAL4 in BeWo cells cultured
 Supplementary Figure 1. Co-localization of GLUT1 and DNAL4 in BeWo cells cultured under static conditions. Cells were seeded in the chamber area of the device and cultured overnight without medium perfusion.
Supplementary Figure 1. Co-localization of GLUT1 and DNAL4 in BeWo cells cultured under static conditions. Cells were seeded in the chamber area of the device and cultured overnight without medium perfusion.
Supplemental Data. Cui et al. (2012). Plant Cell /tpc a b c d. Stem UBC32 ACTIN
 A Root Stem Leaf Flower Silique Senescence leaf B a b c d UBC32 ACTIN C * Supplemental Figure 1. Expression Pattern and Protein Sequence of UBC32 Homologues in Yeast, Human, and Arabidopsis. (A) Expression
A Root Stem Leaf Flower Silique Senescence leaf B a b c d UBC32 ACTIN C * Supplemental Figure 1. Expression Pattern and Protein Sequence of UBC32 Homologues in Yeast, Human, and Arabidopsis. (A) Expression
Supplementary Figures
 Supplementary Figures Supplementary Figure 1 Experimental schema for the identification of circular RNAs in six normal tissues and seven cancerous tissues. Supplementary Fiure 2 Comparison of human circrnas
Supplementary Figures Supplementary Figure 1 Experimental schema for the identification of circular RNAs in six normal tissues and seven cancerous tissues. Supplementary Fiure 2 Comparison of human circrnas
Supplementary Figure 1 Collision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK2, PPK3 and PPK4 respectively.
 Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
Blimp-1/PRDM1 rabbit monoclonal antibody (C14A4) was purchased from Cell Signaling
 1 2 3 4 5 6 7 8 9 10 11 Supplementary Methods Antibodies Blimp-1/PRDM1 rabbit monoclonal antibody (C14A4) was purchased from Cell Signaling (Danvers, MA) and used at 1:1000 to detect the total PRDM1 protein
1 2 3 4 5 6 7 8 9 10 11 Supplementary Methods Antibodies Blimp-1/PRDM1 rabbit monoclonal antibody (C14A4) was purchased from Cell Signaling (Danvers, MA) and used at 1:1000 to detect the total PRDM1 protein
Trasposable elements: Uses of P elements Problem set B at the end
 Trasposable elements: Uses of P elements Problem set B at the end P-elements have revolutionized the way Drosophila geneticists conduct their research. Here, we will discuss just a few of the approaches
Trasposable elements: Uses of P elements Problem set B at the end P-elements have revolutionized the way Drosophila geneticists conduct their research. Here, we will discuss just a few of the approaches
Supplementary Figure 1. Characterization of the POP2 transcriptional and post-transcriptional regulatory elements. (A) POP2 nucleotide sequence
 1 5 6 7 8 9 10 11 1 1 1 Supplementary Figure 1. Characterization of the POP transcriptional and post-transcriptional regulatory elements. (A) POP nucleotide sequence depicting the consensus sequence for
1 5 6 7 8 9 10 11 1 1 1 Supplementary Figure 1. Characterization of the POP transcriptional and post-transcriptional regulatory elements. (A) POP nucleotide sequence depicting the consensus sequence for
Nature Biotechnology: doi: /nbt Supplementary Figure 1. In vitro validation of OTC sgrnas and donor template.
 Supplementary Figure 1 In vitro validation of OTC sgrnas and donor template. (a) In vitro validation of sgrnas targeted to OTC in the MC57G mouse cell line by transient transfection followed by 4-day puromycin
Supplementary Figure 1 In vitro validation of OTC sgrnas and donor template. (a) In vitro validation of sgrnas targeted to OTC in the MC57G mouse cell line by transient transfection followed by 4-day puromycin
Supplementary Figure 1. Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings.
 Supplementary Figure 1 Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings. (left) Representative bright-field images of wild type (wt), heterozygous (het)
Supplementary Figure 1 Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings. (left) Representative bright-field images of wild type (wt), heterozygous (het)
affects the development of newborn neurons Brain Mind Institute and School of Life Sciences, Ecole Polytechnique Fédérale de
 Shedding of neurexin 3β ectodomain by ADAM10 releases a soluble fragment that affects the development of newborn neurons Erika Borcel* a, Magda Palczynska* a, Marine Krzisch b, Mitko Dimitrov a, Giorgio
Shedding of neurexin 3β ectodomain by ADAM10 releases a soluble fragment that affects the development of newborn neurons Erika Borcel* a, Magda Palczynska* a, Marine Krzisch b, Mitko Dimitrov a, Giorgio
Supplementary Information
 Supplementary Information MLL histone methylases regulate expression of HDLR- in presence of estrogen and control plasma cholesterol in vivo Khairul I. Ansari 1, Sahba Kasiri 1, Imran Hussain 1, Samara
Supplementary Information MLL histone methylases regulate expression of HDLR- in presence of estrogen and control plasma cholesterol in vivo Khairul I. Ansari 1, Sahba Kasiri 1, Imran Hussain 1, Samara
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Fig. 1 shrna mediated knockdown of ZRSR2 in K562 and 293T cells. (a) ZRSR2 transcript levels in stably transduced K562 cells were determined using qrt-pcr. GAPDH was
Supplementary Figure 1 Supplementary Fig. 1 shrna mediated knockdown of ZRSR2 in K562 and 293T cells. (a) ZRSR2 transcript levels in stably transduced K562 cells were determined using qrt-pcr. GAPDH was
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3363 Supplementary Figure 1 Several WNTs bind to the extracellular domains of PKD1. (a) HEK293T cells were co-transfected with indicated plasmids. Flag-tagged proteins were immunoprecipiated
DOI: 10.1038/ncb3363 Supplementary Figure 1 Several WNTs bind to the extracellular domains of PKD1. (a) HEK293T cells were co-transfected with indicated plasmids. Flag-tagged proteins were immunoprecipiated
