Applied Microbiology and Biotechnology. De-bugging and maximizing plant cytochrome P450 production in Escherichia coli with C-terminal GFP-fusions
|
|
|
- Zoe Lee
- 6 years ago
- Views:
Transcription
1 Applied Microbiology and Biotechnology De-bugging and maximizing plant cytochrome P450 production in Escherichia coli with C-terminal GFP-fusions Ulla Christensen 1, Dario V. Albacete 1, Karina M. Søgaard 1, Tonja Wolff 1, Morten T. Nielsen 1, Scott James Harrison 1, Anders Holmgaard Hansen 1, Birger Lindberg Møller 2,3, Susanna Seppälä 1,4 and Morten H. H. Nørholm 1,4 1 Novo Nordisk Foundation Center for Biosustainability, Technical University of Denmark, Denmark; 2 Plant Biochemistry Laboratory, Department of Plant and Environmental Sciences, University of Copenhagen, Thorvaldsensvej 40, Frederiksberg C, Copenhagen, Denmark; 3 Center for Synthetic Biology: biosynergy, University of Copenhagen, Thorvaldsensvej 40, Frederiksberg C, Copenhagen, Denmark. 4 Address correspondence to MHHN, morno@biosustain.dtu.dk. Phone: Fax: or SS, susse@biosustain.dtu.dk. 1
2 Supplementary figures Fig. S1 Mining of a library of membrane encoding genes. (A) Production levels of 520 different membrane proteins fused to GFP, based on whole cell fluorescence data previously published (Daley et al. 2005). SohB and YafU, selected based on their high expression levels, small size and topology, are highlighted in red color. (B) Production levels of 183 different membrane proteins fused to alkaline phosphate (PhoA), based on enzyme activity data previously published (Daley et al. 2005). LepB, selected based on the high expression level, small size and topology, is highlighted in red color. 2
3 Fig. S2 Expression of differently truncated variants of CYP79A1-TEV-GFP-his8 with different N-terminal modifications. (A) 3 h and (B) 22 h induction of expression of native CYP79A1, CYP79A1Δ1-35, 28-tag and SohB fused to CYP79A1: CYP79A1Δ1-69, CYP79A1Δ1-59, CYP79A1Δ1-43, CYP79A1Δ1-35 or CYP79A1 with an artificially extended linker region CYP79A1Δ Constructs were expressed in KRX and cell lysates analyzed by in-gel fluorescence. Quantification of full-length CYP79A1-TEV-GFP-his8 vs free GFP was performed using the Image J software. 3
4 Fig S3 Functional assaying of the highest expressing CYP71E1 and CYP79B2 constructs. (A) Microsomes from SohB 1-37-CYP71E1 and SohB 1-35-CYP79A1 expressing cell were mixed with purified SbCPR2b, NADPH and 14 C-tyrosine and the products were extracted by ethyl acetate and separated by thin layer chromatography (TLC). (B) Similarly, microsomes from SohB 1-45-CYP79B2 expressing cells were mixed with purified SbCPR2b, NADPH and 14 C-tryptophane and the product extracted by ethyl acetate and separated by TLC. 4
5 Fig. S4 Correlation between enzyme activity and fluorescence under short term (3h) expression conditions. Oxime production levels and whole-cell fluorescence was plotted from the different full-length CYP79A1 constructs (red squares) and the shortest (D1-35) truncations (green triangles). 5
6 Fig. S5 Effect of GFP fusions on the activity of different CYP79A1 variants. (A) Differently engineered versions of CYP79A1 with (green bars) or without (grey bars) fusions to GFP were co-produced with the compatible reductase electron donor CPR2b in the KRX strain and conversion of L-Tyrosine to (E)-p-hydroxyphenylacetaldoxime assayed by (A) adding the substrate to whole cells and analyzing the methanol-extracted product by HPLC or (B) by extracting the product without any externally added substrate and analysis by LC-MS. Cells were induced for 3h in A and 22 h in B. Empty vector control and 28-tag:native CYP79A1 expressed without CPR were included as references. Error bars indicate standard error of the mean (n=3). 6
7 Fig. S6. Effect of lysozyme on the production of CYP enzymes. BL21 (DE3) was transformed with the six different 28-tag:CYP:tev-gfp-his8 expression constructs in combination with plyssrare2, plyss or the corresponding plasmid after having deleted the lyss gene. Cells were grown in TB media and induced for 3 h, as cell cultures reached the exponential growing phase. Overall expression was monitored by whole cell fluorescence detection. Error bars indicate standard error of the mean (n=3). 7
8 Supplementary tables All pet28a(+)-based plasmids in this work had an AsiSI site removed like this Constructs Fwd oligonucleotide (5 - pet28a(+)-28tag-asisicastev-gfp-his8 3 ) ATCGCGUATTTCGTCT CGCTC ATCGCAAGCUTGCGAT CGCTGCTGAGGGGGTT ATATCTCCTTCTTGGA TCCAG Rev oligonucleotide (5-3 ) ACGCGAUCACTGTTAA AAGGACAATTACAAA CAGGAATC AGCTTGCGAUCGCCAG CTGAGGGAGAAAACC TGTACTTCCAGGGTC Template DNA See below pet28a(+)-28tag tev-gfp-his8 (Nørholm et al., 2013) ATCGCAAGCUTGCGAT AGCTTGCGAUCGCCAG pet28a(+)-malllavf- pet28a(+)-28tag-asisicas-tev- AsiSIcas-tev-gfp-his8 CGCTGCTGAGGGAAA CTGAGGGAGAAAACC gfp-his8 AACTGCTAATAACAG TGTACTTCCAGGGTC (this work) AGCCATATCATCCTCG AGTCTCCTTCTTAAAG pet28a(+)-sohb-asisicas-tev- ATCGCAAGCUTGCGAT AGCTTGCGAUCGCCAG pet28a(+)-sohb- -tev-gfp-his8 gfp-his8 CGCTGCTGAGGGGCTG CTGAGGGAGAAAACC (Daley et al., 2005) AGATTGTTGACCCGTA TGTACTTCCAGGGTC AC pet28a(+)-yafu-asisicas-tev- ATCGCAAGCUTGCGAT AGCTTGCGAUCGCCAG pet28a(+)-yafu-tev-gfp-his8 gfp-his8 CGCTGCTGAGGGGCTG CTGAGGGAGAAAACC (Daley et al., 2005) AGATTGTTGACCCGTA TGTACTTCCAGGGTC AC pet28a(+)-tev-gfp-his8 ATCCTCGAGUCTCCTT ACCTGGAUCCGAAAA pet28a(+)-28tag-asisicas-tev- CTTAAAG CCTGTACTTC gfp-his8 (this work) ACCAGTGCUTCATTGA AGCACTGGUCTAGCAT pet28a(+)-strep-hrv3c- pet28a(+)-strep-hrv3c- CPR2b-tev CCCTGGAAGTACAGGT AACCCCTTGGGGCCTC CPR2b-tev-gfp-his8 Table S1 DNA constructs made by single fragment PCR and uracil excision 8
9 Oligonucleotide name and use For cloning into AsiSI cassette CYP720B4_fwd CYP79A1 _fwd CYP79B2 _fwd CYP51G1 _fwd CYP71E1_fwd CYP51H10 _fwd CYP720B4_rev CYP79A1 _ rev CYP79B2 _ rev CYP51G1 _ rev CYP71E1_ rev CYP51H10 _rev For cloning truncated CYPs into AsiSI cassette tcyp720b4_fwd t1cyp79a1 _fwd t2cyp79a1 _fwd t3cyp79a1 _fwd t4cyp79a1 _fwd tcyp79b2_fwd tcyp51g1 _fwd tcyp71e1_fwd tcyp51h10 _fwd For cloning into PCR amplified pet28-variants ncyp720b4_fwd ncyp79a1 _fwd ncyp79b2 _fwd ncyp51g1 _fwd ncyp71e1_fwd ncyp51h10 _fwd ncyp720b4_rev ncyp79a1 _rev ncyp79b2 _rev CYP51G1 _rev ncyp71e1_rev ncyp51h10 _rev strepcpr2b_fwd CPR2b_rev Oligonucleotide sequence (5-3 ) GCAGCGAUGGCGCCCATGGCAGACCAAATAT GCAGCGAUGGCGACAATGGAGGTAGAGGCCG GCAGCGAUGAACACTTTTACCTCAAACTCTT GCAGCGAUGGATCTCGCCGACATCCCGCAGC GCAGCGAUGGCCACCACCGCCACCCCGCAGC GCAGCGAUGATGGACATGACAATTTGCGTCG GCTGGCGAUCCTTCATTCTCTACTCTACCATGA GCTGGCGAUCCGATGGAGATGGACGGGTAGAGG GCTGGCGAUCCCTTCACCGTCGGGTAGAGATGC GCTGGCGAUCCGTTGTCCACAACAAGCTTCCGC GCTGGCGAUCCGGCGGCGCGGCGGTTCTTGTAT GCTGGCGAUCCGTTTGCAGGCATACGACATCTC GCAGCGAUGCCTGGGTCGACTGGATGGCCGC GCAGCGAUGCCACGCAAAAGCACCACCAAGT GCAGCGAUGCCGGCCGGCGTTGGCAACCCGC GCAGCGAUGTACCTGGCCCGAGCCCTGAGGC GCAGCGAUGTACCTGGCCCGAGCCCTGAGGCGGCCATACCTGGCCCGAGCCCTGAGG GCAGCGAUGCCGGGTCCCACAGGATGGCCGA GCAGCGAUGCCGACGATCCCGGGGGCGCCGG GCAGCGAUGCCGGGCCCTGCGCAGCTGCCGA GCAGCGAUGCCTCCACCGGTGGTGACAGGAA ACTCGAGGAUGGCGGCGCCCATGGCAGACCAAATAT ACTCGAGGAUGGCGGCGACAATGGAGGTAGAGGCCG ACTCGAGGAUGGCGAACACTTTTACCTCAAACTCTT ACTCGAGGAUGGCGGATCTCGCCGACATCCCGCAGC ACTCGAGGAUGGCGGCCACCACCGCCACCCCGCAGC ACTCGAGGAUGGCGATGGACATGACAATTTGCGTCG ATCCAGGUACTTCATTCTCTACTCTACCATGA ATCCAGGUACGATGGAGATGGACGGGTAGAGG ATCCAGGUACCTTCACCGTCGGGTAGAGATGC ATCCAGGUACGTTGTCCACAACAAGCTTCCGC ATCCAGGUACGGCGGCGCGGCGGTTCTTGTAT ATCCAGGUACGTTTGCAGGCATACGACATCTC ACTCGAGGAUGGCGGCAAGCTGGAGCCACCCGCAGT ATCCAGGUACCCAGACATCACGCAGATAGCGG Table S2 Gene specific oligonucleotides used in this work 9
10 N-terminal tag Oligonucleotide sequence (5-3 ) ATGGAATTATCACAAGTTTGTACAAAAAAGGAGGCTGGCGCCGGAACCAATTCAGTCGACTGGA 28-tag TCCAAGAAGGAGATATAACC ATGGAATTGTTGTCTGAATATGGTTTGTTTTTGGCGAAAATCGTTACCGTTGTGCTAGCGATTGC SohB GGCGATTGCCGCCATTATTGTCAATGTTGCTCAACGTAATAAACGCCAGCGTGGCGAGTTACGG GTCAACAATCTCAGC MALLAVF ATGGCTCTGTTATTAGCAGTTTTT ATGAGTTCAGAGCGAGATCTGGTTAATTTTCTTGGCGATTTTTCAATGGATGTGGCCAAAGCAGT TATAGCCGGTGGTGTTGCAACCGCTATTGGAAGTCTGGCTTCTTTTGCCTGTGTTAGCTTTGGCTT YafU TCCAGTAATTCTTGTCGGAGGAGCAATTTTACTGACAGGGATAGTGTGTACAGTTGTTTTAAATG AAATCGATGCTCAATGCCATTTATCAGAAAAATTAAAATATGCAATTAGAGATGGACTAAAACG GCAA ATGGCGAATATGTTTGCCCTGATTCTGGTGATTGCCACACTGGTGACGGGCATTTTATGGTGCGT GGATAAATTCTTTTTCGCACCTAAACGGCGGGAACGTCAGGCAGCGGCGCAGGCGGCTGCCGGG GACTCACTGGATAAAGCAACGTTGAAAAAGGTTGCGCCGAAGCCTGGCTGGCTGGAAACCGGT GCTTCTGTTTTTCCGGTACTGGCTATCGTATTGATTGTGCGTTCGTTTATTTATGAACCGTTCCAG ATCCCGTCAGGTTCGATGATGCCGACTCTGAACTCTACTGATTTTATTCTGGTAGAGAAGTTTGC Lep TTATGGCATTAAAGATCCTATCTACCAGAAAACGCTGATCGAAACCGGTCATCCGAAACGCGGC GATATCGTGGTCTTTAAATATCCGGAAGATCCAAAGCTTGATTACATCAAGCGCGCGGTGGGTT TACCGGGCGATAAAGTCACTTACGATCCGGTCTCAAAAGAGCTGACGATTCAACCGGGATGCAG TTCCGGCCAGGCGTGTGAAAACGCGCTGCCGGTCACCTACTCAAACGTGGAACCGAGCGATTTC GTTCAGACCTTCTCACGCCGTAATGGTGGGGAAGCGACCAGCGGATTCTTTGAAGTGCCGAAAC AGGAAACCAAAGAAAATGGAATTCGTCTTTCCGAGACTAGTGGTGGTCCTGGA Table S3 Nucleotide sequences of N-terminal modifications used in this work 10
Design. Construction. Characterization
 Design Construction Characterization DNA mrna (messenger) A C C transcription translation C A C protein His A T G C T A C G Plasmids replicon copy number incompatibility selection marker origin of replication
Design Construction Characterization DNA mrna (messenger) A C C transcription translation C A C protein His A T G C T A C G Plasmids replicon copy number incompatibility selection marker origin of replication
Deep sequencing reveals global patterns of mrna recruitment
 Supplementary information for: Deep sequencing reveals global patterns of mrna recruitment during translation initiation Rong Gao 1#*, Kai Yu 1#, Ju-Kui Nie 1,Teng-Fei Lian 1, Jian-Shi Jin 1, Anders Liljas
Supplementary information for: Deep sequencing reveals global patterns of mrna recruitment during translation initiation Rong Gao 1#*, Kai Yu 1#, Ju-Kui Nie 1,Teng-Fei Lian 1, Jian-Shi Jin 1, Anders Liljas
Supplementary Information for: Engineered synthetic scaffolds for organising proteins within bacterial cytoplasms
 Lee et al. Supplementary Information 1 Supplementary Information for: Engineered synthetic scaffolds for organising proteins within bacterial cytoplasms Authors: Matthew J. Lee, 1 Judith Mantell, 2,3 Lorna
Lee et al. Supplementary Information 1 Supplementary Information for: Engineered synthetic scaffolds for organising proteins within bacterial cytoplasms Authors: Matthew J. Lee, 1 Judith Mantell, 2,3 Lorna
Supplementary Results Supplementary Table 1. P1 and P2 enrichment scores for wild-type subtiligase.
 Supplementary Results Supplementary Table 1. P1 and P2 enrichment scores for wild-type subtiligase. Supplementary Table 2. Masses of subtiligase mutants measured by LC-MS. Supplementary Table 2 (cont d).
Supplementary Results Supplementary Table 1. P1 and P2 enrichment scores for wild-type subtiligase. Supplementary Table 2. Masses of subtiligase mutants measured by LC-MS. Supplementary Table 2 (cont d).
Construct Design and Cloning Guide for Cas9-triggered homologous recombination
 Construct Design and Cloning Guide for Cas9-triggered homologous recombination Written by Dan Dickinson (ddickins@live.unc.edu) and last updated December 2013. Reference: Dickinson DJ, Ward JD, Reiner
Construct Design and Cloning Guide for Cas9-triggered homologous recombination Written by Dan Dickinson (ddickins@live.unc.edu) and last updated December 2013. Reference: Dickinson DJ, Ward JD, Reiner
Supplemental Information
 Supplemental Information ATP-dependent unwinding of U4/U6 snrnas by the Brr2 helicase requires the C-terminus of Prp8 Corina Maeder 1,3, Alan K. Kutach 1,2,3, and Christine Guthrie 1 1 Department of Biochemistry
Supplemental Information ATP-dependent unwinding of U4/U6 snrnas by the Brr2 helicase requires the C-terminus of Prp8 Corina Maeder 1,3, Alan K. Kutach 1,2,3, and Christine Guthrie 1 1 Department of Biochemistry
Supplementary Information
 Supplementary Information Supplementary Figures Figure S1. Study of mgtl translation in vitro. (A) Detection of 5 LR RNA using wild-type and anti-sd (91-95) substituted templates in a transcription-translation
Supplementary Information Supplementary Figures Figure S1. Study of mgtl translation in vitro. (A) Detection of 5 LR RNA using wild-type and anti-sd (91-95) substituted templates in a transcription-translation
Chapter 20: Biotechnology
 Name Period The AP Biology exam has reached into this chapter for essay questions on a regular basis over the past 15 years. Student responses show that biotechnology is a difficult topic. This chapter
Name Period The AP Biology exam has reached into this chapter for essay questions on a regular basis over the past 15 years. Student responses show that biotechnology is a difficult topic. This chapter
Protocol for cloning SEC-based repair templates using Gibson assembly and ccdb negative selection
 Protocol for cloning SEC-based repair templates using Gibson assembly and ccdb negative selection Written by Dan Dickinson (daniel.dickinson@austin.utexas.edu) and last updated January 2018. A version
Protocol for cloning SEC-based repair templates using Gibson assembly and ccdb negative selection Written by Dan Dickinson (daniel.dickinson@austin.utexas.edu) and last updated January 2018. A version
Contents... vii. List of Figures... xii. List of Tables... xiv. Abbreviatons... xv. Summary... xvii. 1. Introduction In vitro evolution...
 vii Contents Contents... vii List of Figures... xii List of Tables... xiv Abbreviatons... xv Summary... xvii 1. Introduction...1 1.1 In vitro evolution... 1 1.2 Phage Display Technology... 3 1.3 Cell surface
vii Contents Contents... vii List of Figures... xii List of Tables... xiv Abbreviatons... xv Summary... xvii 1. Introduction...1 1.1 In vitro evolution... 1 1.2 Phage Display Technology... 3 1.3 Cell surface
An expression tag toolbox for microbial production of membrane bound plant cytochromes P450
 Downloaded from orbit.dtu.dk on: Jul 08, 2018 An expression tag toolbox for microbial production of membrane bound plant cytochromes P450 Vazquez Albacete, Dario; Cavaleiro, Mafalda; Christensen, Ulla;
Downloaded from orbit.dtu.dk on: Jul 08, 2018 An expression tag toolbox for microbial production of membrane bound plant cytochromes P450 Vazquez Albacete, Dario; Cavaleiro, Mafalda; Christensen, Ulla;
Molecular Biology Techniques Supporting IBBE
 Molecular Biology Techniques Supporting IBBE Jared Cartwright Protein Production Lab Head Contact Details: email jared.cartwright@york.ac.uk Phone 01904 328797 Presentation Aims Gene synthesis Cloning
Molecular Biology Techniques Supporting IBBE Jared Cartwright Protein Production Lab Head Contact Details: email jared.cartwright@york.ac.uk Phone 01904 328797 Presentation Aims Gene synthesis Cloning
Molecular Genetics II - Genetic Engineering Course (Supplementary notes)
 1 von 12 21.02.2015 15:13 Molecular Genetics II - Genetic Engineering Course (Supplementary notes) Figures showing examples of cdna synthesis (currently 11 figures) cdna is a DNA copy synthesized from
1 von 12 21.02.2015 15:13 Molecular Genetics II - Genetic Engineering Course (Supplementary notes) Figures showing examples of cdna synthesis (currently 11 figures) cdna is a DNA copy synthesized from
Combinatorial Evolution of Enzymes and Synthetic pathways Using
 Supplementary data Combinatorial Evolution of Enzymes and Synthetic pathways Using One-Step PCR Peng Jin,, Zhen Kang,,, *, Junli Zhang,, Linpei Zhang,, Guocheng Du,, and Jian Chen, The Key Laboratory of
Supplementary data Combinatorial Evolution of Enzymes and Synthetic pathways Using One-Step PCR Peng Jin,, Zhen Kang,,, *, Junli Zhang,, Linpei Zhang,, Guocheng Du,, and Jian Chen, The Key Laboratory of
Genome Sequence Assembly
 Genome Sequence Assembly Learning Goals: Introduce the field of bioinformatics Familiarize the student with performing sequence alignments Understand the assembly process in genome sequencing Introduction:
Genome Sequence Assembly Learning Goals: Introduce the field of bioinformatics Familiarize the student with performing sequence alignments Understand the assembly process in genome sequencing Introduction:
7.1 Techniques for Producing and Analyzing DNA. SBI4U Ms. Ho-Lau
 7.1 Techniques for Producing and Analyzing DNA SBI4U Ms. Ho-Lau What is Biotechnology? From Merriam-Webster: the manipulation of living organisms or their components to produce useful usually commercial
7.1 Techniques for Producing and Analyzing DNA SBI4U Ms. Ho-Lau What is Biotechnology? From Merriam-Webster: the manipulation of living organisms or their components to produce useful usually commercial
Chapter 6 - Molecular Genetic Techniques
 Chapter 6 - Molecular Genetic Techniques Two objects of molecular & genetic technologies For analysis For generation Molecular genetic technologies! For analysis DNA gel electrophoresis Southern blotting
Chapter 6 - Molecular Genetic Techniques Two objects of molecular & genetic technologies For analysis For generation Molecular genetic technologies! For analysis DNA gel electrophoresis Southern blotting
Supplementary Information
 Single day construction of multi-gene circuits with 3G assembly Andrew D. Halleran 1, Anandh Swaminathan 2, and Richard M. Murray 1, 2 1. Bioengineering, California Institute of Technology, Pasadena, CA.
Single day construction of multi-gene circuits with 3G assembly Andrew D. Halleran 1, Anandh Swaminathan 2, and Richard M. Murray 1, 2 1. Bioengineering, California Institute of Technology, Pasadena, CA.
Supporting Information. Sequence Independent Cloning and Posttranslational. Enzymatic Ligation
 Supporting Information Sequence Independent Cloning and Posttranslational Modification of Repetitive Protein Polymers through Sortase and Sfp-mediated Enzymatic Ligation Wolfgang Ott,,, Thomas Nicolaus,,
Supporting Information Sequence Independent Cloning and Posttranslational Modification of Repetitive Protein Polymers through Sortase and Sfp-mediated Enzymatic Ligation Wolfgang Ott,,, Thomas Nicolaus,,
Ch.15 Section 4 Regulation of Gene Expression pgs Complete the attached Active Reading Guides for the above sections.
 AP Biology 2018-2019 Summer Assignment Due Wednesday 9/5/2018 Text Book Reading Ch.13 The Molecular Basis of Life pgs. 245-267 Ch.15 Section 4 Regulation of Gene Expression pgs. 307-309 Active Reading
AP Biology 2018-2019 Summer Assignment Due Wednesday 9/5/2018 Text Book Reading Ch.13 The Molecular Basis of Life pgs. 245-267 Ch.15 Section 4 Regulation of Gene Expression pgs. 307-309 Active Reading
Molecular Biology Services. Make Research Easy
 Molecular Biology Services Make Research Easy Gene-on-Demand Technology Platform Any gene conceivable in any vector you desire GenScript s Gene-on-Demand technology platform combines patented OptimumGene
Molecular Biology Services Make Research Easy Gene-on-Demand Technology Platform Any gene conceivable in any vector you desire GenScript s Gene-on-Demand technology platform combines patented OptimumGene
Supplementary Fig. 1. Characteristics of transcription elongation by YonO. a. YonO forms a saltstable EC. Immobilized ECs were washed with
 Supplementary Fig. 1. Characteristics of transcription elongation by YonO. a. YonO forms a saltstable EC. Immobilized ECs were washed with transcription buffer with or without a high salt concentration
Supplementary Fig. 1. Characteristics of transcription elongation by YonO. a. YonO forms a saltstable EC. Immobilized ECs were washed with transcription buffer with or without a high salt concentration
Antisense RNA Targeting the First Periplasmic Domain of YidC in Escherichia coli Appears to Induce Filamentation but Does Not Affect Cell Viability
 Antisense RNA Targeting the First Periplasmic Domain of YidC in Escherichia coli Appears to Induce Filamentation but Does Not Affect Cell Viability Riaaz Lalani, Nathaniel Susilo, Elisa Xiao, Andrea Xu
Antisense RNA Targeting the First Periplasmic Domain of YidC in Escherichia coli Appears to Induce Filamentation but Does Not Affect Cell Viability Riaaz Lalani, Nathaniel Susilo, Elisa Xiao, Andrea Xu
SUPPLEMENTARY DATA: Supplementary Figures:
 SUPPLEMENTARY DATA: Supplementary Figures: Supplementary Fig. S1: In vivo recombinant Mtb reporter assay: Dependence of mcherry expression as a function of E. coli growth: A. Fluorescent intensity of mcherry
SUPPLEMENTARY DATA: Supplementary Figures: Supplementary Fig. S1: In vivo recombinant Mtb reporter assay: Dependence of mcherry expression as a function of E. coli growth: A. Fluorescent intensity of mcherry
Selected Techniques Part I
 1 Selected Techniques Part I Gel Electrophoresis Can be both qualitative and quantitative Qualitative About what size is the fragment? How many fragments are present? Is there in insert or not? Quantitative
1 Selected Techniques Part I Gel Electrophoresis Can be both qualitative and quantitative Qualitative About what size is the fragment? How many fragments are present? Is there in insert or not? Quantitative
Basics of Recombinant DNA Technology Biochemistry 302. March 5, 2004 Bob Kelm
 Basics of Recombinant DNA Technology Biochemistry 302 March 5, 2004 Bob Kelm Applications of recombinant DNA technology Mapping and identifying genes (DNA cloning) Propagating genes (DNA subcloning) Modifying
Basics of Recombinant DNA Technology Biochemistry 302 March 5, 2004 Bob Kelm Applications of recombinant DNA technology Mapping and identifying genes (DNA cloning) Propagating genes (DNA subcloning) Modifying
Supporting Information
 Supporting Information Gómez-Marín et al. 10.1073/pnas.1505463112 SI Materials and Methods Generation of BAC DK74B2-six2a::GFP-six3a::mCherry-iTol2. BAC clone (number 74B2) from DanioKey zebrafish BAC
Supporting Information Gómez-Marín et al. 10.1073/pnas.1505463112 SI Materials and Methods Generation of BAC DK74B2-six2a::GFP-six3a::mCherry-iTol2. BAC clone (number 74B2) from DanioKey zebrafish BAC
Transport of Potato Lipoxygenase into the Vacuole Larsen, Mia Kruse Guldstrand; Welinder, Karen Gjesing; Jørgensen, Malene
 Aalborg Universitet Transport of Potato Lipoxygenase into the Vacuole Larsen, Mia Kruse Guldstrand; Welinder, Karen Gjesing; Jørgensen, Malene Publication date: 2009 Document Version Publisher's PDF, also
Aalborg Universitet Transport of Potato Lipoxygenase into the Vacuole Larsen, Mia Kruse Guldstrand; Welinder, Karen Gjesing; Jørgensen, Malene Publication date: 2009 Document Version Publisher's PDF, also
Reading Lecture 8: Lecture 9: Lecture 8. DNA Libraries. Definition Types Construction
 Lecture 8 Reading Lecture 8: 96-110 Lecture 9: 111-120 DNA Libraries Definition Types Construction 142 DNA Libraries A DNA library is a collection of clones of genomic fragments or cdnas from a certain
Lecture 8 Reading Lecture 8: 96-110 Lecture 9: 111-120 DNA Libraries Definition Types Construction 142 DNA Libraries A DNA library is a collection of clones of genomic fragments or cdnas from a certain
Supplementary Methods
 Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
CHAPTER 9 DNA Technologies
 CHAPTER 9 DNA Technologies Recombinant DNA Artificially created DNA that combines sequences that do not occur together in the nature Basis of much of the modern molecular biology Molecular cloning of genes
CHAPTER 9 DNA Technologies Recombinant DNA Artificially created DNA that combines sequences that do not occur together in the nature Basis of much of the modern molecular biology Molecular cloning of genes
Recitation CHAPTER 9 DNA Technologies
 Recitation CHAPTER 9 DNA Technologies DNA Cloning: General Scheme A cloning vector and eukaryotic chromosomes are separately cleaved with the same restriction endonuclease. (A single chromosome is shown
Recitation CHAPTER 9 DNA Technologies DNA Cloning: General Scheme A cloning vector and eukaryotic chromosomes are separately cleaved with the same restriction endonuclease. (A single chromosome is shown
Biotechnology. Chapter 20. Biology Eighth Edition Neil Campbell and Jane Reece. PowerPoint Lecture Presentations for
 Chapter 20 Biotechnology PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece Lectures by Chris Romero, updated by Erin Barley with contributions from Joan Sharp Copyright
Chapter 20 Biotechnology PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece Lectures by Chris Romero, updated by Erin Barley with contributions from Joan Sharp Copyright
Bootcamp: Molecular Biology Techniques and Interpretation
 Bootcamp: Molecular Biology Techniques and Interpretation Bi8 Winter 2016 Today s outline Detecting and quantifying nucleic acids and proteins: Basic nucleic acid properties Hybridization PCR and Designing
Bootcamp: Molecular Biology Techniques and Interpretation Bi8 Winter 2016 Today s outline Detecting and quantifying nucleic acids and proteins: Basic nucleic acid properties Hybridization PCR and Designing
XXII DNA cloning and sequencing. Outline
 XXII DNA cloning and sequencing 1) Deriving DNA for cloning Outline 2) Vectors; forming recombinant DNA; cloning DNA; and screening for clones containing recombinant DNA [replica plating and autoradiography;
XXII DNA cloning and sequencing 1) Deriving DNA for cloning Outline 2) Vectors; forming recombinant DNA; cloning DNA; and screening for clones containing recombinant DNA [replica plating and autoradiography;
SUMOstar Gene Fusion Technology
 Gene Fusion Technology NEW METHODS FOR ENHANCING FUNCTIONAL PROTEIN EXPRESSION AND PURIFICATION IN INSECT CELLS White Paper June 2007 LifeSensors Inc. 271 Great Valley Parkway Malvern, PA 19355 www.lifesensors.com
Gene Fusion Technology NEW METHODS FOR ENHANCING FUNCTIONAL PROTEIN EXPRESSION AND PURIFICATION IN INSECT CELLS White Paper June 2007 LifeSensors Inc. 271 Great Valley Parkway Malvern, PA 19355 www.lifesensors.com
Molecular Cell Biology - Problem Drill 11: Recombinant DNA
 Molecular Cell Biology - Problem Drill 11: Recombinant DNA Question No. 1 of 10 1. Which of the following statements about the sources of DNA used for molecular cloning is correct? Question #1 (A) cdna
Molecular Cell Biology - Problem Drill 11: Recombinant DNA Question No. 1 of 10 1. Which of the following statements about the sources of DNA used for molecular cloning is correct? Question #1 (A) cdna
462 BCH. Biotechnology & Genetic engineering. (Practical)
 462 BCH Biotechnology & Genetic engineering (Practical) Nora Aljebrin Office: Building 5, 3 rd floor, 5T304 E.mail: naljebrin@ksu.edu.sa All lectures and lab sheets are available on my website: Fac.ksu.edu.sa\naljebrin
462 BCH Biotechnology & Genetic engineering (Practical) Nora Aljebrin Office: Building 5, 3 rd floor, 5T304 E.mail: naljebrin@ksu.edu.sa All lectures and lab sheets are available on my website: Fac.ksu.edu.sa\naljebrin
TECHNICAL BULLETIN. In Vitro Bacterial Split Fluorescent Protein Fold n Glow Solubility Assay Kits
 In Vitro Bacterial Split Fluorescent Protein Fold n Glow Solubility Assay Kits Catalog Numbers APPA001 In Vitro Bacterial Split GFP "Fold 'n' Glow" Solubility Assay Kit (Green) APPA008 In Vitro Bacterial
In Vitro Bacterial Split Fluorescent Protein Fold n Glow Solubility Assay Kits Catalog Numbers APPA001 In Vitro Bacterial Split GFP "Fold 'n' Glow" Solubility Assay Kit (Green) APPA008 In Vitro Bacterial
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling
 Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Fig. S1. CrgA intracellular levels in M. tuberculosis. Ten and twenty micrograms of
 Supplementary data Fig. S1. CrgA intracellular levels in M. tuberculosis. Ten and twenty micrograms of cell free protein lysates from WT M. tuberculosis (Rv) together with various known concentrations
Supplementary data Fig. S1. CrgA intracellular levels in M. tuberculosis. Ten and twenty micrograms of cell free protein lysates from WT M. tuberculosis (Rv) together with various known concentrations
Enhancers mutations that make the original mutant phenotype more extreme. Suppressors mutations that make the original mutant phenotype less extreme
 Interactomics and Proteomics 1. Interactomics The field of interactomics is concerned with interactions between genes or proteins. They can be genetic interactions, in which two genes are involved in the
Interactomics and Proteomics 1. Interactomics The field of interactomics is concerned with interactions between genes or proteins. They can be genetic interactions, in which two genes are involved in the
Molecular Cloning. Genomic DNA Library: Contains DNA fragments that represent an entire genome. cdna Library:
 Molecular Cloning Genomic DNA Library: Contains DNA fragments that represent an entire genome. cdna Library: Made from mrna, and represents only protein-coding genes expressed by a cell at a given time.
Molecular Cloning Genomic DNA Library: Contains DNA fragments that represent an entire genome. cdna Library: Made from mrna, and represents only protein-coding genes expressed by a cell at a given time.
oligonucleotide primers listed in Supplementary Table 2 and cloned into ppcr-script (Stratagene) before digestion
 Supplementary Methods. Construction of BiFC vectors The full-length ARC3 cdna, ARC3 1-1794 and ARC3 1-1083 were amplified from ppcr-script/arc3 using the oligonucleotide primers listed in Supplementary
Supplementary Methods. Construction of BiFC vectors The full-length ARC3 cdna, ARC3 1-1794 and ARC3 1-1083 were amplified from ppcr-script/arc3 using the oligonucleotide primers listed in Supplementary
Affibody scaffolds improve sesquiterpene production in. Saccharomyces cerevisiae
 Affibody scaffolds improve sesquiterpene production in Saccharomyces cerevisiae Stefan Tippmann 1,*, Josefine Anfelt 3*, Florian David 1,, Jacqueline M. Rand 1,, Verena Siewers 1,, Mathias Uhlén 3,5, Jens
Affibody scaffolds improve sesquiterpene production in Saccharomyces cerevisiae Stefan Tippmann 1,*, Josefine Anfelt 3*, Florian David 1,, Jacqueline M. Rand 1,, Verena Siewers 1,, Mathias Uhlén 3,5, Jens
Rotation Report Sample Version 3. Due Date: August 9, Analysis of the Guanine Nucleotide Exchange Activity of the S. cerevisiae Ats1 Protein
 Rotation Report Sample Version 3 Due Date: August 9, 1998 Analysis of the Guanine Nucleotide Exchange Activity of the S. cerevisiae Ats1 Protein Anita H. Corbett Advisor: Amy Jones Rotation 1 Abstract:
Rotation Report Sample Version 3 Due Date: August 9, 1998 Analysis of the Guanine Nucleotide Exchange Activity of the S. cerevisiae Ats1 Protein Anita H. Corbett Advisor: Amy Jones Rotation 1 Abstract:
Texas A&M University-Corpus Christi CHEM4402 Biochemistry II Laboratory Laboratory 4 - Polymerase Chain Reaction (PCR)
 Texas A&M University-Corpus Christi CHEM4402 Biochemistry II Laboratory Laboratory 4 - Polymerase Chain Reaction (PCR) Progressing with the sequence of experiments, we are now ready to amplify the green
Texas A&M University-Corpus Christi CHEM4402 Biochemistry II Laboratory Laboratory 4 - Polymerase Chain Reaction (PCR) Progressing with the sequence of experiments, we are now ready to amplify the green
Use of In-Fusion Cloning for Simple and Efficient Assembly of Gene Constructs No restriction enzymes or ligation reactions necessary
 No restriction enzymes or ligation reactions necessary Background The creation of genetic circuits and artificial biological systems typically involves the use of modular genetic components biological
No restriction enzymes or ligation reactions necessary Background The creation of genetic circuits and artificial biological systems typically involves the use of modular genetic components biological
The yeast two-hybrid assay
 Master program Molecular and Cellular Life Sciences Block 1: Intracellular membrane processes 11th October 2011 The yeast two-hybrid assay Fulvio Reggiori Department of Cell Biology, UMC Utrecht For what
Master program Molecular and Cellular Life Sciences Block 1: Intracellular membrane processes 11th October 2011 The yeast two-hybrid assay Fulvio Reggiori Department of Cell Biology, UMC Utrecht For what
STANDARD CLONING PROCEDURES. Shotgun cloning (using a plasmid vector and E coli as a host).
 STANDARD CLONING PROCEDURES Shotgun cloning (using a plasmid vector and E coli as a host). 1) Digest donor DNA and plasmid DNA with the same restriction endonuclease 2) Mix the fragments together and treat
STANDARD CLONING PROCEDURES Shotgun cloning (using a plasmid vector and E coli as a host). 1) Digest donor DNA and plasmid DNA with the same restriction endonuclease 2) Mix the fragments together and treat
Arsenite oxidation regulator AioR regulates bacterial chemotaxis towards. arsenite in Agrobacterium tumefaciens GW4
 1 2 Arsenite oxidation regulator AioR regulates bacterial chemotaxis towards arsenite in Agrobacterium tumefaciens GW4 3 4 5 Kaixiang Shi 1, Xia Fan 1, Zixu Qiao 1, Yushan Han 1, Timothy R. McDermott 2,
1 2 Arsenite oxidation regulator AioR regulates bacterial chemotaxis towards arsenite in Agrobacterium tumefaciens GW4 3 4 5 Kaixiang Shi 1, Xia Fan 1, Zixu Qiao 1, Yushan Han 1, Timothy R. McDermott 2,
Computational Biology I LSM5191
 Computational Biology I LSM5191 Lecture 5 Notes: Genetic manipulation & Molecular Biology techniques Broad Overview of: Enzymatic tools in Molecular Biology Gel electrophoresis Restriction mapping DNA
Computational Biology I LSM5191 Lecture 5 Notes: Genetic manipulation & Molecular Biology techniques Broad Overview of: Enzymatic tools in Molecular Biology Gel electrophoresis Restriction mapping DNA
Illumatool ΤΜ Tunable Light System: A Non-Destructive Light Source For Molecular And Cellular Biology Applications. John Fox, Lightools Research.
 Illumatool ΤΜ Tunable Light System: A Non-Destructive Light Source For Molecular And Cellular Biology Applications. John Fox, Lightools Research. Fluorescent dyes and proteins are basic analytical tools
Illumatool ΤΜ Tunable Light System: A Non-Destructive Light Source For Molecular And Cellular Biology Applications. John Fox, Lightools Research. Fluorescent dyes and proteins are basic analytical tools
Bi 8 Lecture 4. Ellen Rothenberg 14 January Reading: from Alberts Ch. 8
 Bi 8 Lecture 4 DNA approaches: How we know what we know Ellen Rothenberg 14 January 2016 Reading: from Alberts Ch. 8 Central concept: DNA or RNA polymer length as an identifying feature RNA has intrinsically
Bi 8 Lecture 4 DNA approaches: How we know what we know Ellen Rothenberg 14 January 2016 Reading: from Alberts Ch. 8 Central concept: DNA or RNA polymer length as an identifying feature RNA has intrinsically
Molecular Cloning. Joseph Sambrook. David W. Russell A LABORATORY MANUAL COLD SPRING HARBOR LABORATORY PRESS VOLUME.
 VOLUME Molecular Cloning A LABORATORY MANUAL THIRD EDITION www.molecularcloning.com Joseph Sambrook PETER MACCALLUM CANCER INSTITUTE AND THE UNIVERSITY OF MELBOURNE, AUSTRALIA David W. Russell UNIVERSITY
VOLUME Molecular Cloning A LABORATORY MANUAL THIRD EDITION www.molecularcloning.com Joseph Sambrook PETER MACCALLUM CANCER INSTITUTE AND THE UNIVERSITY OF MELBOURNE, AUSTRALIA David W. Russell UNIVERSITY
Supplementary information. Interplay between Penicillin-binding proteins and SEDS proteins promotes. bacterial cell wall synthesis
 Supplementary information Interplay between Penicillin-binding proteins and SEDS proteins promotes bacterial cell wall synthesis Sophie Leclercq 1, Adeline Derouaux 1, Samir Olatunji 1, Claudine Fraipont
Supplementary information Interplay between Penicillin-binding proteins and SEDS proteins promotes bacterial cell wall synthesis Sophie Leclercq 1, Adeline Derouaux 1, Samir Olatunji 1, Claudine Fraipont
3 -end. Sau3A. 3 -end TAA TAA. ~9.1kb SalI TAA. ~12.6kb. HindШ ATG 5,860bp TAA. ~12.7kb
 Supplemental Data. Chen et al. (2008). Badh2, encoding betaine aldehyde dehydrogenase, inhibits the biosynthesis of 2-acetyl-1-pyrroline, a major component in rice fragrance. A pcam-cah/cah Sau3A Promoter
Supplemental Data. Chen et al. (2008). Badh2, encoding betaine aldehyde dehydrogenase, inhibits the biosynthesis of 2-acetyl-1-pyrroline, a major component in rice fragrance. A pcam-cah/cah Sau3A Promoter
Human Viperin Causes Radical SAM Dependent Elongation of E. coli Hinting at its Physiological Role
 Supporting Information Human Viperin Causes Radical SAM Dependent Elongation of E. coli Hinting at its Physiological Role Micah T. Nelp, Anthony P. Young, Branden M. Stepanski, Vahe Bandarian* Department
Supporting Information Human Viperin Causes Radical SAM Dependent Elongation of E. coli Hinting at its Physiological Role Micah T. Nelp, Anthony P. Young, Branden M. Stepanski, Vahe Bandarian* Department
CHAPTER 20 DNA TECHNOLOGY AND GENOMICS. Section A: DNA Cloning
 Section A: DNA Cloning 1. DNA technology makes it possible to clone genes for basic research and commercial applications: an overview 2. Restriction enzymes are used to make recombinant DNA 3. Genes can
Section A: DNA Cloning 1. DNA technology makes it possible to clone genes for basic research and commercial applications: an overview 2. Restriction enzymes are used to make recombinant DNA 3. Genes can
Genetics and Genomics in Medicine Chapter 3. Questions & Answers
 Genetics and Genomics in Medicine Chapter 3 Multiple Choice Questions Questions & Answers Question 3.1 Which of the following statements, if any, is false? a) Amplifying DNA means making many identical
Genetics and Genomics in Medicine Chapter 3 Multiple Choice Questions Questions & Answers Question 3.1 Which of the following statements, if any, is false? a) Amplifying DNA means making many identical
Roche Molecular Biochemicals Technical Note No. LC 12/2000
 Roche Molecular Biochemicals Technical Note No. LC 12/2000 LightCycler Absolute Quantification with External Standards and an Internal Control 1. General Introduction Purpose of this Note Overview of Method
Roche Molecular Biochemicals Technical Note No. LC 12/2000 LightCycler Absolute Quantification with External Standards and an Internal Control 1. General Introduction Purpose of this Note Overview of Method
transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected with the wwp-luc reporter, and FLAG-tagged FHL1,
 Supplementary Data Supplementary Figure Legends Supplementary Figure 1 FHL-mediated TGFβ-responsive reporter transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected
Supplementary Data Supplementary Figure Legends Supplementary Figure 1 FHL-mediated TGFβ-responsive reporter transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected
MIT Department of Biology 7.013: Introductory Biology - Spring 2005 Instructors: Professor Hazel Sive, Professor Tyler Jacks, Dr.
 MIT Department of Biology 7.01: Introductory Biology - Spring 2005 Instructors: Professor Hazel Sive, Professor Tyler Jacks, Dr. Claudette Gardel iv) Would Xba I be useful for cloning? Why or why not?
MIT Department of Biology 7.01: Introductory Biology - Spring 2005 Instructors: Professor Hazel Sive, Professor Tyler Jacks, Dr. Claudette Gardel iv) Would Xba I be useful for cloning? Why or why not?
Lecture Four. Molecular Approaches I: Nucleic Acids
 Lecture Four. Molecular Approaches I: Nucleic Acids I. Recombinant DNA and Gene Cloning Recombinant DNA is DNA that has been created artificially. DNA from two or more sources is incorporated into a single
Lecture Four. Molecular Approaches I: Nucleic Acids I. Recombinant DNA and Gene Cloning Recombinant DNA is DNA that has been created artificially. DNA from two or more sources is incorporated into a single
Supplementary Figure 1. Nature Structural & Molecular Biology: doi: /nsmb.3494
 Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Integrated Omics Study Delineates the Dynamics of Lipid Droplets in Rhodococcus Opacus PD630
 School of Natural Sciences and Mathematics 2013-10-22 Integrated Omics Study Delineates the Dynamics of Lipid Droplets in Rhodococcus Opacus PD630 UTD AUTHOR(S): Michael Qiwei Zhang 2013 The Authors This
School of Natural Sciences and Mathematics 2013-10-22 Integrated Omics Study Delineates the Dynamics of Lipid Droplets in Rhodococcus Opacus PD630 UTD AUTHOR(S): Michael Qiwei Zhang 2013 The Authors This
Schematic representation of the endogenous PALB2 locus and gene-disruption constructs
 Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Lab Book igem Stockholm Lysostaphin. Week 8
 Lysostaphin Week 8 Summarized below are the experiments conducted this week in chronological order. Click on the experiment name to view it. To go back to this summary, click Summary in the footer. Summary
Lysostaphin Week 8 Summarized below are the experiments conducted this week in chronological order. Click on the experiment name to view it. To go back to this summary, click Summary in the footer. Summary
1. A brief overview of sequencing biochemistry
 Supplementary reading materials on Genome sequencing (optional) The materials are from Mark Blaxter s lecture notes on Sequencing strategies and Primary Analysis 1. A brief overview of sequencing biochemistry
Supplementary reading materials on Genome sequencing (optional) The materials are from Mark Blaxter s lecture notes on Sequencing strategies and Primary Analysis 1. A brief overview of sequencing biochemistry
Chapter 20 Recombinant DNA Technology. Copyright 2009 Pearson Education, Inc.
 Chapter 20 Recombinant DNA Technology Copyright 2009 Pearson Education, Inc. 20.1 Recombinant DNA Technology Began with Two Key Tools: Restriction Enzymes and DNA Cloning Vectors Recombinant DNA refers
Chapter 20 Recombinant DNA Technology Copyright 2009 Pearson Education, Inc. 20.1 Recombinant DNA Technology Began with Two Key Tools: Restriction Enzymes and DNA Cloning Vectors Recombinant DNA refers
SUPPLEMENTARY INFORMATION. Tolerance of a knotted near infrared fluorescent protein to random circular permutation
 SUPPLEMENTARY INFORMATION Tolerance of a knotted near infrared fluorescent protein to random circular permutation Naresh Pandey 1,3, Brianna E. Kuypers 2,4, Barbara Nassif 1, Emily E. Thomas 1,3, Razan
SUPPLEMENTARY INFORMATION Tolerance of a knotted near infrared fluorescent protein to random circular permutation Naresh Pandey 1,3, Brianna E. Kuypers 2,4, Barbara Nassif 1, Emily E. Thomas 1,3, Razan
Some types of Mutagenesis
 Mutagenesis What Is a Mutation? Genetic information is encoded by the sequence of the nucleotide bases in DNA of the gene. The four nucleotides are: adenine (A), thymine (T), guanine (G), and cytosine
Mutagenesis What Is a Mutation? Genetic information is encoded by the sequence of the nucleotide bases in DNA of the gene. The four nucleotides are: adenine (A), thymine (T), guanine (G), and cytosine
DNA RNA protein (which performs different functions in the cell, i.e. a catalytic reaction on a small molecule substrate)
 Synthetic Biology Is an emerging discipline that emphasizes the application of engineering principles to the design and construction of synthetic biological systems that exhibit dynamic or logical behavior.
Synthetic Biology Is an emerging discipline that emphasizes the application of engineering principles to the design and construction of synthetic biological systems that exhibit dynamic or logical behavior.
Chapter 9 Genetic Engineering
 Chapter 9 Genetic Engineering Biotechnology: use of microbes to make a protein product Recombinant DNA Technology: Insertion or modification of genes to produce desired proteins Genetic engineering: manipulation
Chapter 9 Genetic Engineering Biotechnology: use of microbes to make a protein product Recombinant DNA Technology: Insertion or modification of genes to produce desired proteins Genetic engineering: manipulation
SUPPLEMENTARY MATERIAL
 SUPPLEMENTARY MATERIAL Engineering a pyridoxal '-phosphate supply for cadaverine production by using Escherichia coli whole-cell biocatalysis Weichao Ma 1,,, Weijia Cao 1,, Bowen Zhang 1,, Kequan Chen
SUPPLEMENTARY MATERIAL Engineering a pyridoxal '-phosphate supply for cadaverine production by using Escherichia coli whole-cell biocatalysis Weichao Ma 1,,, Weijia Cao 1,, Bowen Zhang 1,, Kequan Chen
GENETIC ENGINEERING worksheet
 Section A: Genetic Engineering Overview 1. What is genetic engineering? 2. Put the steps of genetic engineering in order. Recombinant product is isolated, purified and analyzed before marketing. The DNA
Section A: Genetic Engineering Overview 1. What is genetic engineering? 2. Put the steps of genetic engineering in order. Recombinant product is isolated, purified and analyzed before marketing. The DNA
Protein isotopic enrichment for NMR studies
 Protein isotopic enrichment for NMR studies Protein NMR studies ARTGKYVDES sequence structure Structure of protein- ligand, protein-protein complexes Binding of molecules (perturbation mapping) Dynamic
Protein isotopic enrichment for NMR studies Protein NMR studies ARTGKYVDES sequence structure Structure of protein- ligand, protein-protein complexes Binding of molecules (perturbation mapping) Dynamic
Supporting Information
 Supporting Information Development of a 2,4-Dinitrotoluene-Responsive Synthetic Riboswitch in E. coli cells Molly E. Davidson, Svetlana V. Harbaugh, Yaroslav G. Chushak, Morley O. Stone, Nancy Kelley-
Supporting Information Development of a 2,4-Dinitrotoluene-Responsive Synthetic Riboswitch in E. coli cells Molly E. Davidson, Svetlana V. Harbaugh, Yaroslav G. Chushak, Morley O. Stone, Nancy Kelley-
VOLUME 2. Molecular Clonin g A LABORATORY MANUA L THIRD EDITIO N. Joseph Sambrook. David W. Russell
 VOLUME 2 Molecular Clonin g A LABORATORY MANUA L THIRD EDITIO N Joseph Sambrook David W. Russell Chapter 8 In Vitro Amplification of DNA by the Polymerase 8. 1 Chain Reaction 1 The Basic Polymerase Chain
VOLUME 2 Molecular Clonin g A LABORATORY MANUA L THIRD EDITIO N Joseph Sambrook David W. Russell Chapter 8 In Vitro Amplification of DNA by the Polymerase 8. 1 Chain Reaction 1 The Basic Polymerase Chain
Figure S1. Biosynthetic pathway of GDP-PerNAc. Mass spectrum of purified GDP-PerNAc. Nature Protocols: doi: /nprot
 Synthesis of GDP-PerNAc In E. coli O157, the biosynthesis of GDP- -N-acetyl-D-perosamine (GDP-PerNAc) involves three enzymes (Fig. S1). GDP-D-Mannose is converted by GDP-mannose-4,6-dehydratase (GMD) into
Synthesis of GDP-PerNAc In E. coli O157, the biosynthesis of GDP- -N-acetyl-D-perosamine (GDP-PerNAc) involves three enzymes (Fig. S1). GDP-D-Mannose is converted by GDP-mannose-4,6-dehydratase (GMD) into
Module 1 overview PRESIDENTʼS DAY
 1 Module 1 overview lecture lab 1. Introduction to the module 1. Start-up protein eng. 2. Rational protein design 2. Site-directed mutagenesis 3. Fluorescence and sensors 3. DA amplification PRESIDETʼS
1 Module 1 overview lecture lab 1. Introduction to the module 1. Start-up protein eng. 2. Rational protein design 2. Site-directed mutagenesis 3. Fluorescence and sensors 3. DA amplification PRESIDETʼS
BS1940 Course Topics Fall 2001 Drs. Hatfull and Arndt
 BS1940 Course Topics Fall 2001 Drs. Hatfull and Arndt Introduction to molecular biology Combining genetics, biochemistry, structural chemistry Information flow in biological systems: The Central Dogma
BS1940 Course Topics Fall 2001 Drs. Hatfull and Arndt Introduction to molecular biology Combining genetics, biochemistry, structural chemistry Information flow in biological systems: The Central Dogma
Supporting information
 Supporting information Construction of strains and plasmids To create ptc67, a PCR product obtained with primers cc2570-162f (gcatgggcaagcttgaggacggcgtcatgt) and cc2570+512f (gaggccgtggtaccatagaggcgggcg),
Supporting information Construction of strains and plasmids To create ptc67, a PCR product obtained with primers cc2570-162f (gcatgggcaagcttgaggacggcgtcatgt) and cc2570+512f (gaggccgtggtaccatagaggcgggcg),
Directe d Mutagenesis
 Directe d Mutagenesis A Practical Approac h M. J. McPHERSON 1. Mutagenesis facilitated by the removal or introduction of unique restriction sites 1 P. Carte r 1. Introduction to site-directed mutagenesis
Directe d Mutagenesis A Practical Approac h M. J. McPHERSON 1. Mutagenesis facilitated by the removal or introduction of unique restriction sites 1 P. Carte r 1. Introduction to site-directed mutagenesis
DNA amplification and analysis: minipcr TM Food Safety Lab
 Science for everyone, everywhere DNA amplification and analysis: minipcr TM Food Safety Lab Release date: 09 September 2014 Welcome Our goals for today: Review DNA amplification theory Solve a public health
Science for everyone, everywhere DNA amplification and analysis: minipcr TM Food Safety Lab Release date: 09 September 2014 Welcome Our goals for today: Review DNA amplification theory Solve a public health
Comparison of different methods for purification analysis of a green fluorescent Strep-tag fusion protein. Application
 Comparison of different methods for purification analysis of a green fluorescent Strep-tag fusion protein Application Petra Sebastian Meike Kuschel Stefan Schmidt Abstract This Application Note describes
Comparison of different methods for purification analysis of a green fluorescent Strep-tag fusion protein Application Petra Sebastian Meike Kuschel Stefan Schmidt Abstract This Application Note describes
Supplementary Figure 1 Abortive ligation (adenylation) of the gapped 5'-dRP-containing BER
 Supplementary Figure 1 Abortive ligation (adenylation) of the gapped 5'-dRP-containing BER intermediate by human BER ligases. The ligation efficiency of DNA ligase I (a) and XRCC1/DNA ligase III complex
Supplementary Figure 1 Abortive ligation (adenylation) of the gapped 5'-dRP-containing BER intermediate by human BER ligases. The ligation efficiency of DNA ligase I (a) and XRCC1/DNA ligase III complex
Box 7905, Engineering Building I, North Carolina State University, Raleigh, NC 27695, Phone: Fax:
 Inefficient ribosomal skipping enables simultaneous secretion and display of proteins in Saccharomyces cerevisiae Carlos A. Cruz-Teran 1, Karthik Tiruthani 1, Adam Mischler, and Balaji M. Rao * Department
Inefficient ribosomal skipping enables simultaneous secretion and display of proteins in Saccharomyces cerevisiae Carlos A. Cruz-Teran 1, Karthik Tiruthani 1, Adam Mischler, and Balaji M. Rao * Department
Understanding the Cellular Mechanism of the Excess Microsporocytes I (EMSI) Gene. Andrew ElBardissi, The Pennsylvania State University
 Understanding the Cellular Mechanism of the Excess Microsporocytes I (EMSI) Gene Andrew ElBardissi, The Pennsylvania State University Abstract: Hong Ma, The Pennsylvania State University The Excess Microsporocytes
Understanding the Cellular Mechanism of the Excess Microsporocytes I (EMSI) Gene Andrew ElBardissi, The Pennsylvania State University Abstract: Hong Ma, The Pennsylvania State University The Excess Microsporocytes
Biosynthesis of 3-Methoxy-5-Methyl Naphthoic Acid and Its Incorporation into the Antitumor Antibiotic Azinomycin B
 Biosynthesis of 3-Methoxy-5-Methyl Naphthoic Acid and Its Incorporation into the Antitumor Antibiotic Azinomycin B Wei Ding, a,b Wei Deng, b Mancheng Tang, b Qi Zhang, b Gongli Tang, b Yurong Bi, a and
Biosynthesis of 3-Methoxy-5-Methyl Naphthoic Acid and Its Incorporation into the Antitumor Antibiotic Azinomycin B Wei Ding, a,b Wei Deng, b Mancheng Tang, b Qi Zhang, b Gongli Tang, b Yurong Bi, a and
A fail-safe mechanism in the septal ring assembly pathway generated by the sequential recruitment of cell separation amidases and their activators
 Supplemental Information for: A fail-safe mechanism in the septal ring assembly pathway generated by the sequential recruitment of cell separation amidases and their activators Nick T. Peters, Thuy Dinh,
Supplemental Information for: A fail-safe mechanism in the septal ring assembly pathway generated by the sequential recruitment of cell separation amidases and their activators Nick T. Peters, Thuy Dinh,
NCERT. 2. An enzyme catalysing the removal of nucleotides from the ends of DNA is: a. endonuclease b. exonuclease c. DNA ligase d.
 BIOTECHNOLOGY PRINCIPLES AND PROCESSES 75 CHAPTER 11 BIOTECHNOLOGY: PRINCIPLES AND PROCESSES 1. Rising of dough is due to: MULTIPLE-CHOICE QUESTIONS a. Multiplication of yeast b. Production of CO 2 c.
BIOTECHNOLOGY PRINCIPLES AND PROCESSES 75 CHAPTER 11 BIOTECHNOLOGY: PRINCIPLES AND PROCESSES 1. Rising of dough is due to: MULTIPLE-CHOICE QUESTIONS a. Multiplication of yeast b. Production of CO 2 c.
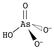 ph and redox potential (pe) are the most important factors controlling arsenic speciation. Phosphate (A) and arsenate (B) speciation are shown as a function of ph for the (V) oxidation states. H 3 PO 4
ph and redox potential (pe) are the most important factors controlling arsenic speciation. Phosphate (A) and arsenate (B) speciation are shown as a function of ph for the (V) oxidation states. H 3 PO 4
Mcbio 316: Exam 3. (10) 2. Compare and contrast operon vs gene fusions.
 Mcbio 316: Exam 3 Name (15) 1. Transposons provide useful tools for genetic analysis. List 5 different uses of transposon insertions. ANSWER: Many answers are possible, however, if multiple items on the
Mcbio 316: Exam 3 Name (15) 1. Transposons provide useful tools for genetic analysis. List 5 different uses of transposon insertions. ANSWER: Many answers are possible, however, if multiple items on the
Supplementary Online Material. An Expanded Eukaryotic Genetic Code 2QH, UK. * To whom correspondence should be addressed.
 Supplementary Online Material An Expanded Eukaryotic Genetic Code Jason W. Chin 1, T. Ashton Cropp, J. Christopher Anderson, Mridul Mukherji, Zhiwen Zhang and Peter G. Schultz* Department of Chemistry
Supplementary Online Material An Expanded Eukaryotic Genetic Code Jason W. Chin 1, T. Ashton Cropp, J. Christopher Anderson, Mridul Mukherji, Zhiwen Zhang and Peter G. Schultz* Department of Chemistry
Supplementary
 Supplementary information Supplementary Material and Methods Plasmid construction The transposable element vectors for inducible expression of RFP-FUS wt and EGFP-FUS R521C and EGFP-FUS P525L were derived
Supplementary information Supplementary Material and Methods Plasmid construction The transposable element vectors for inducible expression of RFP-FUS wt and EGFP-FUS R521C and EGFP-FUS P525L were derived
CLONING AND EXPRESSION OF PSEUDOMONAS AERUGINOSA PA01 GENES IN E.COLI DH5Α FOR THE DETECTION OF ARSENIC IN CONTAMINATED WATER SAMPLE
 CLONING AND EXPRESSION OF PSEUDOMONAS AERUGINOSA PA01 GENES IN E.COLI DH5Α FOR THE DETECTION OF ARSENIC IN CONTAMINATED WATER SAMPLE A report Submitted to KSCST for 40 th series of SPP 2017 Project Proposal
CLONING AND EXPRESSION OF PSEUDOMONAS AERUGINOSA PA01 GENES IN E.COLI DH5Α FOR THE DETECTION OF ARSENIC IN CONTAMINATED WATER SAMPLE A report Submitted to KSCST for 40 th series of SPP 2017 Project Proposal
Phosphorothioate-based BioBrick cloning (Potsdam) Standard
 BBF RFC 91: phosphorothioate-based BioBrick cloning 1 Document Version 1.0, October 26, 2012 Phosphorothioate-based BioBrick cloning (Potsdam) Standard igem Team Potsdam 2012 1, Tobias Baumann 1, Kristian
BBF RFC 91: phosphorothioate-based BioBrick cloning 1 Document Version 1.0, October 26, 2012 Phosphorothioate-based BioBrick cloning (Potsdam) Standard igem Team Potsdam 2012 1, Tobias Baumann 1, Kristian
Lecture 8: Affinity Chromatography-III
 Lecture 8: Affinity Chromatography-III Key words: Chromatography; Affinity chromatography; Protein Purification During this lecture, we shall be studying few more examples of affinity chromatography. The
Lecture 8: Affinity Chromatography-III Key words: Chromatography; Affinity chromatography; Protein Purification During this lecture, we shall be studying few more examples of affinity chromatography. The
Synthetic Biology for
 Synthetic Biology for Plasmids and DNA Digestion Plasmids Plasmids are small DNA molecules that are separate from chromosomal DNA They are most commonly found as double stranded, circular DNA Typical plasmids
Synthetic Biology for Plasmids and DNA Digestion Plasmids Plasmids are small DNA molecules that are separate from chromosomal DNA They are most commonly found as double stranded, circular DNA Typical plasmids
