TraceTools: a new DNA basecaller
|
|
|
- Cordelia Miles
- 6 years ago
- Views:
Transcription
1 Full Length Paper TraceTools: a new DNA basecaller Mohamad T. Musavi *1, Cristian Domnisoru 2, Padma Natarajan 1, Rency Varghese 1, Mike Toothaker 1, Jehad Dawood 1, Habtom Ressom 1, Rebecca Van Beneden 3, Patty Singer 3 *corresponding author: musavi@eece.maine.edu Phone: (207) ; Fax: (207) Intelligent Systems Laboratory Department of Electrical and Computer Engineering University of Maine, 5708 Barrows Hall, Orono, ME 04473, USA 2 University of St. Thomas, St. Paul, MN 55105, USA 3 DNA Sequencing Facility, School of Marine Sciences University of Maine, 351 Hitchner Hall, Orono, ME 04473, USA Abstract This paper presents a novel basecalling software, TraceTools, with a unique user-friendly confidence value feature. Because of the differences between ABI data pre-processing and those of this software, TraceTools uses the raw data generated by ABI machines as opposed to the ABI processed data. For developing and testing TraceTools, a comprehensive database of correct DNA sequences corresponding to the ABI raw data was constructed. The ABI raw trace data was obtained from North Carolina State University. This comprehensive database was used for comparing the accuracy of TraceTools with other popular basecalling programs such as Phred and ABI. The results of this comparison on over 3000 data files with around 750 bases per file show that TraceTools performs better than ABI and Phred. The confidence values also provide a reliable measure of success in the bases called. Keywords ABI, basecalling accuracy, confidence values, DNA basecalling, Phred, TraceTools. 1
2 1. INTRODUCTION While the automated DNA basecalling is an established procedure for years, the need for increased accuracy, longer reads and increased confidence remains very important for any sequencing center. Current sequencing technology produces error rates in the order of 1.5% for a 550 base pairs read (ABI 3700 analyzer specifications). For longer reads, the error rate is higher, reaching 2.5% or more. Many sequencing projects are underway at a lower coverage rate due to the cost associated with sequencing. This implies acceptance of a large number of gaps and a significant total gap length. In particular, these limitations are severe for Bacterial Artificial Chromosomes (BAC) sequencing implying an increased cost for sequencing and less data output for future investigations. The technology for sequencing DNA has rapidly evolved from gel based to capillary electrophoresis (CE). Though the machines have been replaced there is hardly any change in the appearance of the data to be analyzed from a user s perspective. What a user sees is a succession of peaks of four different colors corresponding to the four bases: G, T, A, and C (Guanine-black, Thymine-red, Adenine-green, Cytosine-blue). If the peaks were clearly separated and big enough when compared to the noise at the baseline, making basecalls would be easier either through visual inspection or using automated software. The most used sequencing systems are the ABI sequencing machines. In general, DNA fragments are tagged with fluorescent dyes at lengths corresponding to the number of bases in the fragment. The strands are then separated by length using electrophoresis. Individual samples to be scanned are passed through separate capillaries. A laser beam scans the strands and the reflected intensities from each of the four bases are recorded. Though the output of this physical process is affected by noise, interference between the four filters and other phenomena is less understood. The ABI sequencing software for gel-based machines achieved an error rate of about 6.1% (Koonin, 1997). The ABI PRISM 3700 produces error rates of the order of 1.5% for a 550 base pairs 2
3 read (ABI 3700 analyzer specifications). This is very high considering the requirements of sequencing projects. Olson (Olson and Green, 1998), Richterich (Richterich, 1998), and Ewing (Ewing et. al, 1998), have indicated a commonly accepted base-pair accuracy goal of 99.99%. Sequences with an accuracy lower than 99.90% is considered of little help in identifying single nucleotide polymorphisms in humans; see Felsenfeld et. al., The primary alternative to ABI s software is Phred, developed over the years by a group of contributing researchers and supported by researchers at the Department of Molecular Biology, University of Washington. Phred uses an advanced heuristic algorithm based on Fourier analysis and dynamic programming for base selection. Phred constantly outperforms the ABI basecaller, even for the CE machines. An excellent additional feature of Phred is that for each basecall it provides a quality value. This established a de facto standard for sequencing centers. Although ABI didn t provide quality values in their earlier software, they have recently integrated a quality value similar to Phred s in their ABI 3730 machines. The data processing steps from the raw data to called bases can be divided in two parts: preprocessing and basecalling. Pre-processing step includes adjustment for mobility shift, removal of the interference between the channels (cross-talk removal), baseline adjustment, noise filtering, and finally preparation of a normalized set of four streams of peaks for identification of the bases. As this part changes with the technology, the maker of the sequencing machine is responsible to create the software for pre-processing. In the case of ABI, the change from gel based to CE has clearly changed the profile of the physical system output. The computational effort was directed to create preprocessing software that will have an output similar to the one provided by the old machines. This would provide the flexibility to use existing basecalling programs, with minor modifications if necessary, for the second part of data processing. 3
4 It is clear that the overall performance of DNA base identification would depend on the accuracy of each of the two data processing steps. No matter how well-designed a basecaller would be, a less accurate pre-processing will affect the overall performance. In our previous work (Domnisoru et. al, 2000; Domnisoru and Musavi, 2003), several techniques were provided as improvements to the current ABI pre-processing techniques. For example, it was shown that the cross-talk removal should be performed before the base-line adjustment in order to preserve the integrity of the original data. Furthermore, the cross-talk removal is more effective if performed on the difference signal instead of the signal itself. This paper describes and provides results on the software (TraceTools) that was built based on our pre-processing and basecalling algorithms. The paper is organized as follows. Section 2 describes materials and methods used for developing TraceTools. Section 2.1 describes the database preparation needed for obtaining the correct sequence of bases for each DNA file. Section 2.2 describes the data processing and the basecalling algorithm used by TraceTools. Section 2.3 provides information on assigning confidence values. Section 2.4 gives a brief description of the main features of TraceTools. Section 3 provides a discussion on the results obtained by testing, validation, and comparison of TraceTools with Phred and ABI basecallers. Section 4 provides conclusion. 2. MATERIALS AND METHODS 2.1 DATABASE PREPARATION The database for this study requires two sets of files: raw data files and the corresponding correct sequence files. The raw data file is used as the input to the basecalling system and the correct sequence is used as the ground truth for accuracy measurement. About 4000 raw data files from ABI 3700 machine were obtained from North Carolina State University Fungal Genomics Lab. This center was one of only a few places that kept the raw data when saving the ABI files. Most centers discard the raw data and only keep the sequence information to save memory. Availability of raw data is a 4
5 requirement because TraceTools pre-processing steps are performed independent of ABI preprocessing technique. Note that Phred uses the ABI pre-processed data. The corresponding correct sequences (ground truth) for the raw files were not readily available. Instead, a large contig of correct sequences, comprised of thousands of overlapping sequences, was available. The ground truth sequence for each raw data file had to be extracted from this large contig (McNally et. al, 2002). This contig contained the sequences of ABI raw files assembled within it and the information for making the correct basecalls for each sequence. The data quality of the obtained ABI files was predominantly good in the center portion, but poor at the beginning and end of the file. The purpose of this contig was to provide the correct basecalls for each sequence so that we could obtain a file containing the most accurate basecalls from start to end (with the exception of the primer). This correct file would enable making reliable evaluation of the basecalling program. Each base in the contig was formed from as many as five or six sequence files which had overlapped at that base position thereby providing concurrent information about the location of that base. This information from many overlapping files for a base increased the confidence of claiming that base as the correct base for that position in the sequence. With the availability of the large contig, the next step was to search the contig for the correct sequence corresponding to each raw file in the database. There were three steps involved in finding the correct sequences. First, Phred basecalling software was run on each raw data file in the database and a sequence was obtained. Phred was used because it has demonstrated very good accuracy when tested over wide range of sequencing methods; see Richterich (Richterich, 1998) and Ewing et al. (Ewing et. al., 1998). Furthermore, Phred has been shown to be more accurate than ABI basecalling software in terms of insertion, deletion, substitution and total error. Subsequently blast2seq (Tatusova and Madden, 1999) was run on each Phred generated sequence with the large contig providing information on the alignment location within the contig. If a match was found, the corresponding sequence from the contig was saved as the correct sequence for the file (except for the primer region). 5
6 The search between the contig and the sequence file was also performed for a possible reverse complement match. A program written in C++ automated the above search process. This program initially used loops to sequentially open each file in the database. The result of this search method produced 3362 correct sequence files within the contig corresponding to 3362 raw ABI files. These files were used to compare the accuracy of TraceTools basecalls with other currently popular basecalling programs such as ABI and Phred. Though the files found within the contig contained the correct bases for the raw data, they did not contain the primer. For an ABI file, a primer is used for every file: one for forward read and another for reverse. This primer does not appear in the contig as the contig was only formed as an assembled string of bases containing the basecalls of each sequence. A primer sticks to the DNA template, and its use initiates replication also necessary for DNA sequencing and Polymerase Chain Reaction (Lyons, 1998). The bases within the primer are not a part of the sequence. In order to incorporate the primer information in the correct sequence, the corresponding part from the original files is copied. However, while assessing the accuracy of the basecalls this part is not used. For accuracy calculations only the bases that are a part of the contig are considered. The overall process of database preparation for accuracy measurement and comparison proved very complex and time-consuming. Overall, it involved obtaining the ABI basecalls, Phred basecalls, searching the large contig for correct sequences, re-writing to a new file the information from the contig where the sequence overlaps, and finally adding the primer to the new file. These steps finally resulted in a five-part database of ABI raw data files, a large contig of DNA bases assembled from that raw data, the ABI basecalls, the Phred basecalls, and the correct basecalls. Using the prepared database and different comparative techniques, the percent accuracy for TraceTools was found and compared with those of ABI and Phred. The outcome is presented in the results section. 6
7 2.2 DATA PROCESSING AND BASECALLING The algorithm for basecalling used by TraceTools is based on processing the raw data contained in the ABI sequencing files. The general approach is oriented toward preserving the information contained in the raw data and avoiding the use of traditional filtering techniques. A detailed presentation of the proposed approach is presented in Domnisoru et. al., 2000 and Domnisoru and Musavi, The algorithm has two main steps: 1) data processing - where the raw data information is filtered and a model for the spacing between consecutive bases is constructed, and 2) basecalling - where the base spacing information is used to predict the location of the bases and make basecalls. Several pre-processing steps are employed to ensure the extraction of a model for the base spacing from the raw data file. A preliminary filtering is applied to smooth the signals. The cross talk parameters are detected automatically from the trace information and the cross talk removal itself is applied to the variation of the signals as opposed to the signals directly. The signals are reconstructed from their variation and aligned at a baseline. The next step is the detection of the peak candidates based on the local peakness and height of the signals. Using the peak candidates, a preliminary model for the base spacing is determined. The peak candidates not fitting the model are eliminated followed by a recalculation of the base spacing model. Note that the base spacing model is differentiated for each combination of possible two consecutive bases. There are 16 such combinations one for each pair (AA, AC, AG, AT, CA, CC, CG, ). Therefore, the base spacing model has 16 sub-models for each possibility. The final step, the basecalling, is a hybrid algorithm based on the prediction of the spacing between bases and a fuzzy logic basecall. At every base, say A, the base position is submitted to the four models (AA, AC, AG, AT) that start with the current base and the location of the next four possible base candidates are predicted. The algorithm then evaluates the peakness of the signals, the height, and the slope for all the four predicted locations on a local basis. A fuzzy system would then 7
8 use the peakness, the height, and the slope information to make a basecall. After the basecalling is performed once, the base spacing model is recalculated and the basecalling part is redone using the updated spacing information. 2.3 CONFIDENCE VALUES As the basecalling algorithm s error rates drop, the smaller basecall errors could be difficult to locate. Hence, assembling algorithms and human operators use a confidence value measure to determine how well the basecalling algorithm has performed for each basecall. This will clearly make it easier to uncover potential errors and correct them, thus increasing the throughput of genetic sequencing. So far, establishing such a confidence value for the basecalls was done primarily in support of the development of the Phred basecalling system (Ewing et. al, 1998). They provided a predictive measure, known as quality value, which would directly correlate to true trace error rates and help locate possible errors. This model utilized one static model for all sequences and it is based on sequencing error rate not the local information at each base, hence, resulting in a not so reliable measure of success for basecalls. Furthermore, their model would be inadequate with continuously evolving new sequencing techniques and process variability. ABI software previously didn t provide any confidence value for its basecalls. However, its latest version, ABI 3730, provides a measure that is similar to Phred quality value and in fact it uses the same technique. The only difference is that ABI 3730 has replaced Phred s numerical values with color coded bars for easy and quick recognition of error areas. In contrast, our model employs fuzzy logic, which provides flexibility, adaptability and intuition through the use of linguistic variables and fuzzy membership functions. More importantly, the measure is a true confidence values based on the information at the base. In the fuzzy model, three trace features from the basecalling algorithm are collected and used as inputs (French et. al, 2002). The first feature is the height, which is simply the height of a peak. The 8
9 second is the peakness, which is a measure related to the concavity at the top of a peak. The final feature is the base spacing, which is the difference in location from one peak to another. In addition to the first most likely candidate (the base called ), the peakness and height are also found for the second ( 2 nd ) likely candidate and used in the fuzzy model. As shown in Figure 1, the fuzzy system involves four subsystems that are designated as Fuzzy Peakness, Fuzzy Height, Fuzzy Spacing, and Fuzzy Confidence. The first three subsystems calculate C p, C H, and C S based on peaknesses (P called and P 2nd ), heights (H called and H 2nd ), and spacings ( S previous and S next ), respectively. The linguistic terms for C H are defined as very low (VL), low (L), medium (M), high (H), and very high (VH) and for C p and C S are defined as low (L), medium (M), and high (H), The Fuzzy Confidence system takes in the confidence values provided by the other three subsystems and computes the overall confidence value (C) of the base called. The fuzzy linguistic terms for C are very low (VL), low (L), medium (M), high (H), and very high (VH). Note that since there are 3 input variables for the fuzzy confidence subsystem, there could be as many as 45 (3x5x3) rules, of which some are unlikely to happen. The fuzzy operator AND is used for all fuzzy rule premises involved in the subsystems and the confidence value of height is given more weight in setting up the fuzzy rules. Figure TRACETOOLS SOFTWARE The above algorithms for data preprocessing, basecalling, and confidence value have been integrated in a software, called TraceTools. This software is designed to process ABI 3700 chromatograms. TraceTools, which is a Windows based software, can display both the raw and the processed data after making basecalls. The display of raw data allows the user to view the data as recorded by the sequencing machine. When the basecalls made are uncertain, this display feature would help the user make confident decisions after investigating the raw data. The basecalling is done 9
10 through a processing of the raw data rather than using the pre-processed results obtained by the ABI software. TraceTools also displays a confidence value associated with each basecall through a colorcoded rectangular bar. After calling bases, TraceTools can write the sequences to files in FASTA format. Below is a screenshot of TraceTools window showing both the raw data (upper window) and its processed version and called bases (lower window). Other features of TraceTools are: exporting raw and processed data into text files, printing data and display screen, searching for particular sequences by exact match, copying and pasting, and a help menu. More features are currently being added to the system. TraceTools confidence value is a continuous value between 0 and 1, with 1 indicating the highest confidence. For ease of recognitions of poor calls, the confidence values are however, indicated through rectangular bars in the display. Green indicates high confidence (50% or higher). The green box is further split into three parts to indicate confidence between 50 and 100%. If just the bottom 1/3 of the bar is colored green, the confidence value is 50% - 60%. If the bottom 2/3 are colored green, the confidence measure is 60% - 80%. A fully colored green bar indicates 80% - 100% confidence. Yellow colored bar indicates 25% to 50% confidence. Red indicates low confidence in the results obtained (25% or below). This makes it very easy for possible operator s intervention. Figure 2 shows bases with very high confidence (full green), medium confidence (yellow) and poor confidence (red). Figure 2 3. RESULTS AND DISCUSSION In this section, the results of TraceTools testing is presented and compared with the ABI and Phred software. The accuracy of TraceTool basecalling for a total of 3362 chromatogram files, as described in the data preparation section, was recorded. Several restrictions such as the trimmed ends of sequences and the error calculation for only the best matching part prevented the application of 10
11 Blast for comparing the outcome of basecalling with the corresponding correct sequence. Therefore, an alignment program was developed that when provided with two sequences produced the alignment of those sequences and reported accuracy as well as a visual base-by-base comparison of the files. This visual comparison allows the user to view where the files do not match and investigate the reason for mismatch. The basecalls by ABI, Phred, and TraceTools were compared against the correct sequences. The overall accuracy for TraceTools was higher than ABI and Phred. Figure 3 shows the histogram of accuracy for ABI, Phred and TraceTools for the accuracy range %. Figure 4 shows the difference histogram between TraceTools and ABI and between TraceTools and Phred for the same accuracy range. As can be seen from the figures, TraceTools performs better than ABI and Phred for a majority of the files. From Figure 4, it can also be seen that TraceTools had 100 more files than Phred and 300 more files than ABI in the high accuracy range of %. In other words TraceTools had 3% more files than Phred and 9% more files than ABI that were highly accurate. Our experiments with TraceTools confidence values also provided more accurate representation of the basecall correctness as compared to Phred and ABI quality values. As an example, Figure 5 gives a screenshot of TraceTools where a section of a sequence is shown and the confidence values are also indicated by colored bars above the bases. Note that the numerical values for these confidence measures are available, as given in the 2 nd row of Table 1. However, color-coded bars are used in the display for ease of recognition. The 1 st row of Table 1 gives the corresponding Phred s quality values. By looking at Figure 5, it is obvious that the measure of correctness of any basecaller for calling the first T base should have the best confidence value among all the other bases. TraceTools has assigned a confidence value of 0.93 (out of 1), which is the highest among all other values. While Phred s quality value for the same base is 12 (out of 50), which is surprisingly the lowest. Note that in Phred, the higher the number is, the better the basecall should be. Similar observations can be made for other bases. For example, the trace data in Figure 5 clearly shows that the 2 nd T base should have a better confidence value than any of the two C bases around it. While 11
12 TraceTools clearly shows this distinction in its confidence value, Phred s quality value provides exactly the opposite. This shows an inconsistency in the assignment of confidence values or quality values by Phred. Figure 3 Figure 4 Figure 5 Table 1 There are a couple of points that one needs to consider regarding the accuracy results presented above. One point is the fact that the contig is not 100% full proof. In other words, the contig that was used for the extraction of correct sequences has shown to have some errors in it. And the other point is that the accuracy results may be biased in the favor of Phred because as it was explained before, Phred s generated sequences were used with Blast to extract the correct sequences from the contig. The errors in contig was observed in the accuracy analysis. In finding the accuracy, the traces were also analyzed and confirmed manually for a large number of files. In doing this, it was observed that in some cases, TraceTools made the right call, but the contig (ground truth) was incorrect. To give some examples, Figures 6 shows a screenshot of the TraceTools result for the chromatogram file tested. Shown to the right of the screenshot is the part of the sequence called by TraceTools (bottom) and indicated by contig (top). The same is also given for ABI and Phred. As seen, TraceTools calls the base at the vertical line a C and contig indicates it as a T. However, analyzing the results by comparing it with the raw data shows that TraceTools indeed made the correct basecall. This fact is also verified by ABI and Phred, as indicated on the figure. Even though TraceTools makes a correct basecall in cases such as the above, it is not accounted for in the calculation of its accuracy as the accuracy percentages are calculated with respect to the ground truth (contig). 12
13 Figure 7 shows an example when TraceTools was correct and the called base matched that of the contig while others failed. The figure shows the result of Phred, ABI and TraceTools for a test file (bottom row). The bases indicated by the contig are shown in the top row. The vertical lines show that a match exists between the basecaller results and the contig. As evident from the results TraceTools makes the basecall C similar to the contig, but ABI calls it a G and Phred calls it a T. Figure 6 Figure 7 4. CONCLUSION A new sequencing system, TraceTools, has been presented that has shown improved results over the ABI and Phred software on both the called bases and their confidence values. The increase in the accuracy can be attributed to TraceTools unique features of filtering, mobility shift correction, adaptive base space prediction, cross talk removal, and fuzzy confidence value calculation. The colorcoded confidence values assist the user in quick identification of bases with low confidence values. AVAILABILITY A copy of TraceTools trial version for sequencing ABI 3700 files can be downloaded from the project web site at: ACKNOWLEDGEMENTS This research was funded in part by NSF grant #DBI and #EEC The authors would like to acknowledge Dr. Ralph Dean of the Fungal Genomics Laboratory at North Carolina State University for generously providing the ABI raw data and corresponding contig used for this study. The authors also extend their appreciation to Ms. Ceara McNally and Mr. Brian French for software development and database preparation. 13
14 REFERENCES ABI PRISM 3700 DNA Analyzer Specifications, at: products/specifications.cfm?prod_id=40. Domnisoru, C., Zhan, X., and Musavi, M.T., (2000), Electrophoresis, 14, Domnisoru, C. and Musavi, M., (2003), Method for reducing cross talk within DNA data, United States Patent Office #6,598,013. Ewing, B., Hillier, L., Wendl, M. C., and Green, P., (1998), Genome Research, 8, Felsenfeld, A., Peterson, J., Schloss, J., and Guyer, M., (1999), Genome Research, 9, 1-4. French, B., Domnisoru, C., Ressom, H., and Musavi, M.T., (2002), Proceedings of the International Conference on Mathematics and Engineering Techniques in Medicine and Biological Sciences (METMBS), Las Vegas, Nevada, USA, June 27, 2002, Koonin,S., (1997), Genome Informatics, Human Genome Project, The MITRE Corporation, JASON Program Office. Lyons, R. H., (1998), A Molecular Biology Glossary McNally, C., Domnisoru, C., and Musavi, M.T., (2002), Proceedings of the International Conference on Mathematics and Engineering Techniques in Medicine and Biological Sciences (METMBS), Las Vegas, Nevada, June 27, 2002, Olson, M. and Green, P., (1998), Genome Research, 8, Richterich, P., (1998), Genome Research, 8, Tatusova, T.A. and Madden, T.L., (1999), FEMS Microbiol Lett., 174:
15 P called P 2nd Fuzzy Peakness C P Fuzzy Height Fuzzy H called C H C Confidence H 2nd S previous S next Fuzzy Spacing C S Figure 1: Block diagram of the overall fuzzy logic system for calculation of confidence value. Figure 2: TraceTools window showing a sequence with bases called and their confidence values indicated by color-coded bars; upper window shows unprocessed data. 15
16 Figure 3: Histogram of accuracy for ABI, Phred and TraceTools for range %. Figure 4: Difference histograms between TraceTools and ABI & TraceTools and Phred. 16
17 Figure 5: A screenshot of TraceTools for confidence value analysis. Contig ( top row) 1-TraceTools, 2- Phred and 3- ABI (bottom row) Figure 6: Screenshot of TraceTools results for a chromatogram file (upper window: raw data; lower window: base calls made by TraceTools) (Right) TraceTools, Phred and ABI base calls w.r.t. Contig; vertical lines show match between them. 17
18 Contig Contig Contig Phred ABI TraceTools Figure 7: Phred, ABI and TraceTools base calls w.r.t the contig for a chromatogram file. Phred (Max value50) TraceTools (Max value 1) Confidence values for the bases called A T C T C G Table 1: Confidence values for Phred and TraceTools for the sequence shown in Figure 5. 18
A Guide to Consed Michelle Itano, Carolyn Cain, Tien Chusak, Justin Richner, and SCR Elgin.
 1 A Guide to Consed Michelle Itano, Carolyn Cain, Tien Chusak, Justin Richner, and SCR Elgin. Main Window Figure 1. The Main Window is the starting point when Consed is opened. From here, you can access
1 A Guide to Consed Michelle Itano, Carolyn Cain, Tien Chusak, Justin Richner, and SCR Elgin. Main Window Figure 1. The Main Window is the starting point when Consed is opened. From here, you can access
We begin with a high-level overview of sequencing. There are three stages in this process.
 Lecture 11 Sequence Assembly February 10, 1998 Lecturer: Phil Green Notes: Kavita Garg 11.1. Introduction This is the first of two lectures by Phil Green on Sequence Assembly. Yeast and some of the bacterial
Lecture 11 Sequence Assembly February 10, 1998 Lecturer: Phil Green Notes: Kavita Garg 11.1. Introduction This is the first of two lectures by Phil Green on Sequence Assembly. Yeast and some of the bacterial
Fast, Accurate and Sensitive DNA Variant Detection from Sanger Sequencing:
 Fast, Accurate and Sensitive DNA Variant Detection from Sanger Sequencing: Patented, Anti-Correlation Technology Provides 99.5% Accuracy & Sensitivity to 5% Variant Knowledge Base and External Annotation
Fast, Accurate and Sensitive DNA Variant Detection from Sanger Sequencing: Patented, Anti-Correlation Technology Provides 99.5% Accuracy & Sensitivity to 5% Variant Knowledge Base and External Annotation
4.1. Genetics as a Tool in Anthropology
 4.1. Genetics as a Tool in Anthropology Each biological system and every human being is defined by its genetic material. The genetic material is stored in the cells of the body, mainly in the nucleus of
4.1. Genetics as a Tool in Anthropology Each biological system and every human being is defined by its genetic material. The genetic material is stored in the cells of the body, mainly in the nucleus of
Next Generation Sequencing. Dylan Young Biomedical Engineering
 Next Generation Sequencing Dylan Young Biomedical Engineering What is DNA? Molecule composed of Adenine (A) Guanine (G) Cytosine (C) Thymine (T) Paired as either AT or CG Provides genetic instructions
Next Generation Sequencing Dylan Young Biomedical Engineering What is DNA? Molecule composed of Adenine (A) Guanine (G) Cytosine (C) Thymine (T) Paired as either AT or CG Provides genetic instructions
Sanger Sequencing: Troubleshooting Guide
 If you need help analysing your Sanger sequencing output, this guide can help. CONTENTS 1 Introduction... 2 2 Sequence Data Evaluation... 2 3 Troubleshooting... 4 3.1 Reviewing the Sequence... 4 3.1.1
If you need help analysing your Sanger sequencing output, this guide can help. CONTENTS 1 Introduction... 2 2 Sequence Data Evaluation... 2 3 Troubleshooting... 4 3.1 Reviewing the Sequence... 4 3.1.1
MassArray Analytical Tools
 MassArray Analytical Tools Reid F. Thompson and John M. Greally April 30, 08 Contents Introduction Changes for MassArray in current BioC release 3 3 Optimal Amplicon Design 4 4 Conversion Controls 0 5
MassArray Analytical Tools Reid F. Thompson and John M. Greally April 30, 08 Contents Introduction Changes for MassArray in current BioC release 3 3 Optimal Amplicon Design 4 4 Conversion Controls 0 5
1. A brief overview of sequencing biochemistry
 Supplementary reading materials on Genome sequencing (optional) The materials are from Mark Blaxter s lecture notes on Sequencing strategies and Primary Analysis 1. A brief overview of sequencing biochemistry
Supplementary reading materials on Genome sequencing (optional) The materials are from Mark Blaxter s lecture notes on Sequencing strategies and Primary Analysis 1. A brief overview of sequencing biochemistry
Nature Biotechnology: doi: /nbt Supplementary Figure 1. Read Complexity
 Supplementary Figure 1 Read Complexity A) Density plot showing the percentage of read length masked by the dust program, which identifies low-complexity sequence (simple repeats). Scrappie outputs a significantly
Supplementary Figure 1 Read Complexity A) Density plot showing the percentage of read length masked by the dust program, which identifies low-complexity sequence (simple repeats). Scrappie outputs a significantly
PRESENTING SEQUENCES 5 GAATGCGGCTTAGACTGGTACGATGGAAC 3 3 CTTACGCCGAATCTGACCATGCTACCTTG 5
 Molecular Biology-2017 1 PRESENTING SEQUENCES As you know, sequences may either be double stranded or single stranded and have a polarity described as 5 and 3. The 5 end always contains a free phosphate
Molecular Biology-2017 1 PRESENTING SEQUENCES As you know, sequences may either be double stranded or single stranded and have a polarity described as 5 and 3. The 5 end always contains a free phosphate
Lecture Four. Molecular Approaches I: Nucleic Acids
 Lecture Four. Molecular Approaches I: Nucleic Acids I. Recombinant DNA and Gene Cloning Recombinant DNA is DNA that has been created artificially. DNA from two or more sources is incorporated into a single
Lecture Four. Molecular Approaches I: Nucleic Acids I. Recombinant DNA and Gene Cloning Recombinant DNA is DNA that has been created artificially. DNA from two or more sources is incorporated into a single
Genome 373: Mapping Short Sequence Reads II. Doug Fowler
 Genome 373: Mapping Short Sequence Reads II Doug Fowler The final Will be in this room on June 6 th at 8:30a Will be focused on the second half of the course, but will include material from the first half
Genome 373: Mapping Short Sequence Reads II Doug Fowler The final Will be in this room on June 6 th at 8:30a Will be focused on the second half of the course, but will include material from the first half
DNA Replication: Paper Clip Activity
 DNA Replication: Paper Clip Activity Name Hour: Date: Quick Review: Each DNA molecule has a unique structure that makes it different from other DNA molecules (Remember A chromosome is condensed DNA and
DNA Replication: Paper Clip Activity Name Hour: Date: Quick Review: Each DNA molecule has a unique structure that makes it different from other DNA molecules (Remember A chromosome is condensed DNA and
Finishing Fosmid DMAC-27a of the Drosophila mojavensis third chromosome
 Finishing Fosmid DMAC-27a of the Drosophila mojavensis third chromosome Ruth Howe Bio 434W 27 February 2010 Abstract The fourth or dot chromosome of Drosophila species is composed primarily of highly condensed,
Finishing Fosmid DMAC-27a of the Drosophila mojavensis third chromosome Ruth Howe Bio 434W 27 February 2010 Abstract The fourth or dot chromosome of Drosophila species is composed primarily of highly condensed,
Biochemistry. Dr. Shariq Syed. Shariq AIKC/FinalYB/2014
 Biochemistry Dr. Shariq Syed Shariq AIKC/FinalYB/2014 What is DNA Sequence?? Our Genome is made up of DNA Biological instructions are written in our DNA in chemical form The order (sequence) in which nucleotides
Biochemistry Dr. Shariq Syed Shariq AIKC/FinalYB/2014 What is DNA Sequence?? Our Genome is made up of DNA Biological instructions are written in our DNA in chemical form The order (sequence) in which nucleotides
Finishing of Fosmid 1042D14. Project 1042D14 is a roughly 40 kb segment of Drosophila ananassae
 Schefkind 1 Adam Schefkind Bio 434W 03/08/2014 Finishing of Fosmid 1042D14 Abstract Project 1042D14 is a roughly 40 kb segment of Drosophila ananassae genomic DNA. Through a comprehensive analysis of forward-
Schefkind 1 Adam Schefkind Bio 434W 03/08/2014 Finishing of Fosmid 1042D14 Abstract Project 1042D14 is a roughly 40 kb segment of Drosophila ananassae genomic DNA. Through a comprehensive analysis of forward-
ENGR 213 Bioengineering Fundamentals April 25, A very coarse introduction to bioinformatics
 A very coarse introduction to bioinformatics In this exercise, you will get a quick primer on how DNA is used to manufacture proteins. You will learn a little bit about how the building blocks of these
A very coarse introduction to bioinformatics In this exercise, you will get a quick primer on how DNA is used to manufacture proteins. You will learn a little bit about how the building blocks of these
Interest Grabber Notebook #1
 Chapter 13 Interest Grabber Notebook #1 A New Breed The tomatoes in your salad and the dog in your backyard are a result of selective breeding. Over thousands of years, humans have developed breeds of
Chapter 13 Interest Grabber Notebook #1 A New Breed The tomatoes in your salad and the dog in your backyard are a result of selective breeding. Over thousands of years, humans have developed breeds of
Sundaram DGA43A19 Page 1. Finishing Drosophila grimshawi Fosmid: DGA43A19 Varun Sundaram 2/16/09
 Sundaram DGA43A19 Page 1 Finishing Drosophila grimshawi Fosmid: DGA43A19 Varun Sundaram 2/16/09 Sundaram DGA43A19 Page 2 Abstract My project focused on the fosmid clone DGA43A19. The main problems with
Sundaram DGA43A19 Page 1 Finishing Drosophila grimshawi Fosmid: DGA43A19 Varun Sundaram 2/16/09 Sundaram DGA43A19 Page 2 Abstract My project focused on the fosmid clone DGA43A19. The main problems with
Interest Grabber Notebook #1
 Chapter 13 Interest Grabber Notebook #1 A New Breed The tomatoes in your salad and the dog in your backyard are a result of selective breeding. Over thousands of years, humans have developed breeds of
Chapter 13 Interest Grabber Notebook #1 A New Breed The tomatoes in your salad and the dog in your backyard are a result of selective breeding. Over thousands of years, humans have developed breeds of
Bio-inspired Computing for Network Modelling
 ISSN (online): 2289-7984 Vol. 6, No.. Pages -, 25 Bio-inspired Computing for Network Modelling N. Rajaee *,a, A. A. S. A Hussaini b, A. Zulkharnain c and S. M. W Masra d Faculty of Engineering, Universiti
ISSN (online): 2289-7984 Vol. 6, No.. Pages -, 25 Bio-inspired Computing for Network Modelling N. Rajaee *,a, A. A. S. A Hussaini b, A. Zulkharnain c and S. M. W Masra d Faculty of Engineering, Universiti
Interpretation of sequence results
 Interpretation of sequence results An overview on DNA sequencing: DNA sequencing involves the determination of the sequence of nucleotides in a sample of DNA. It use a modified PCR reaction where both
Interpretation of sequence results An overview on DNA sequencing: DNA sequencing involves the determination of the sequence of nucleotides in a sample of DNA. It use a modified PCR reaction where both
Assemblytics: a web analytics tool for the detection of assembly-based variants Maria Nattestad and Michael C. Schatz
 Assemblytics: a web analytics tool for the detection of assembly-based variants Maria Nattestad and Michael C. Schatz Table of Contents Supplementary Note 1: Unique Anchor Filtering Supplementary Figure
Assemblytics: a web analytics tool for the detection of assembly-based variants Maria Nattestad and Michael C. Schatz Table of Contents Supplementary Note 1: Unique Anchor Filtering Supplementary Figure
13.1 RNA Lesson Objectives Contrast RNA and DNA. Explain the process of transcription.
 13.1 RNA Lesson Objectives Contrast RNA and DNA. Explain the process of transcription. The Role of RNA 1. Complete the table to contrast the structures of DNA and RNA. DNA Sugar Number of Strands Bases
13.1 RNA Lesson Objectives Contrast RNA and DNA. Explain the process of transcription. The Role of RNA 1. Complete the table to contrast the structures of DNA and RNA. DNA Sugar Number of Strands Bases
Pre-Lab: Molecular Biology
 Pre-Lab: Molecular Biology Name 1. What are the three chemical parts of a nucleotide. Draw a simple sketch to show how the three parts are arranged. 2. What are the rules of base pairing? 3. In double
Pre-Lab: Molecular Biology Name 1. What are the three chemical parts of a nucleotide. Draw a simple sketch to show how the three parts are arranged. 2. What are the rules of base pairing? 3. In double
COS 597c: Topics in Computational Molecular Biology. DNA arrays. Background
 COS 597c: Topics in Computational Molecular Biology Lecture 19a: December 1, 1999 Lecturer: Robert Phillips Scribe: Robert Osada DNA arrays Before exploring the details of DNA chips, let s take a step
COS 597c: Topics in Computational Molecular Biology Lecture 19a: December 1, 1999 Lecturer: Robert Phillips Scribe: Robert Osada DNA arrays Before exploring the details of DNA chips, let s take a step
Finishing Drosophila Ananassae Fosmid 2728G16
 Finishing Drosophila Ananassae Fosmid 2728G16 Kyle Jung March 8, 2013 Bio434W Professor Elgin Page 1 Abstract For my finishing project, I chose to finish fosmid 2728G16. This fosmid carries a segment of
Finishing Drosophila Ananassae Fosmid 2728G16 Kyle Jung March 8, 2013 Bio434W Professor Elgin Page 1 Abstract For my finishing project, I chose to finish fosmid 2728G16. This fosmid carries a segment of
Next Gen Sequencing. Expansion of sequencing technology. Contents
 Next Gen Sequencing Contents 1 Expansion of sequencing technology 2 The Next Generation of Sequencing: High-Throughput Technologies 3 High Throughput Sequencing Applied to Genome Sequencing (TEDed CC BY-NC-ND
Next Gen Sequencing Contents 1 Expansion of sequencing technology 2 The Next Generation of Sequencing: High-Throughput Technologies 3 High Throughput Sequencing Applied to Genome Sequencing (TEDed CC BY-NC-ND
Finished (Almost) Sequence of Drosophila littoralis Chromosome 4 Fosmid Clone XAAA73. Seth Bloom Biology 4342 March 7, 2004
 Finished (Almost) Sequence of Drosophila littoralis Chromosome 4 Fosmid Clone XAAA73 Seth Bloom Biology 4342 March 7, 2004 Summary: I successfully sequenced Drosophila littoralis fosmid clone XAAA73. The
Finished (Almost) Sequence of Drosophila littoralis Chromosome 4 Fosmid Clone XAAA73 Seth Bloom Biology 4342 March 7, 2004 Summary: I successfully sequenced Drosophila littoralis fosmid clone XAAA73. The
Basic Bioinformatics: Homology, Sequence Alignment,
 Basic Bioinformatics: Homology, Sequence Alignment, and BLAST William S. Sanders Institute for Genomics, Biocomputing, and Biotechnology (IGBB) High Performance Computing Collaboratory (HPC 2 ) Mississippi
Basic Bioinformatics: Homology, Sequence Alignment, and BLAST William S. Sanders Institute for Genomics, Biocomputing, and Biotechnology (IGBB) High Performance Computing Collaboratory (HPC 2 ) Mississippi
7.1 Techniques for Producing and Analyzing DNA. SBI4U Ms. Ho-Lau
 7.1 Techniques for Producing and Analyzing DNA SBI4U Ms. Ho-Lau What is Biotechnology? From Merriam-Webster: the manipulation of living organisms or their components to produce useful usually commercial
7.1 Techniques for Producing and Analyzing DNA SBI4U Ms. Ho-Lau What is Biotechnology? From Merriam-Webster: the manipulation of living organisms or their components to produce useful usually commercial
Introduction to some aspects of molecular genetics
 Introduction to some aspects of molecular genetics Julius van der Werf (partly based on notes from Margaret Katz) University of New England, Armidale, Australia Genetic and Physical maps of the genome...
Introduction to some aspects of molecular genetics Julius van der Werf (partly based on notes from Margaret Katz) University of New England, Armidale, Australia Genetic and Physical maps of the genome...
10/20/2009 Comp 590/Comp Fall
 Lecture 14: DNA Sequencing Study Chapter 8.9 10/20/2009 Comp 590/Comp 790-90 Fall 2009 1 DNA Sequencing Shear DNA into millions of small fragments Read 500 700 nucleotides at a time from the small fragments
Lecture 14: DNA Sequencing Study Chapter 8.9 10/20/2009 Comp 590/Comp 790-90 Fall 2009 1 DNA Sequencing Shear DNA into millions of small fragments Read 500 700 nucleotides at a time from the small fragments
DNA Sequencing and Assembly
 DNA Sequencing and Assembly CS 262 Lecture Notes, Winter 2016 February 2nd, 2016 Scribe: Mark Berger Abstract In this lecture, we survey a variety of different sequencing technologies, including their
DNA Sequencing and Assembly CS 262 Lecture Notes, Winter 2016 February 2nd, 2016 Scribe: Mark Berger Abstract In this lecture, we survey a variety of different sequencing technologies, including their
Artisan Technology Group is your source for quality new and certified-used/pre-owned equipment
 Artisan Technology Group is your source for quality new and certified-used/pre-owned equipment FAST SHIPPING AND DELIVERY TENS OF THOUSANDS OF IN-STOCK ITEMS EQUIPMENT DEMOS HUNDREDS OF MANUFACTURERS SUPPORTED
Artisan Technology Group is your source for quality new and certified-used/pre-owned equipment FAST SHIPPING AND DELIVERY TENS OF THOUSANDS OF IN-STOCK ITEMS EQUIPMENT DEMOS HUNDREDS OF MANUFACTURERS SUPPORTED
Unfortunately, plate data was not available to generate an initial low coverage
 Finishing Report Carolyn Cain Fosmid: XBAA-30G19 Fosmid XBAA-30G19 was challenging to assemble, but ultimately came together in a satisfactory final assembly. The initial problems in my assembly were gaps,
Finishing Report Carolyn Cain Fosmid: XBAA-30G19 Fosmid XBAA-30G19 was challenging to assemble, but ultimately came together in a satisfactory final assembly. The initial problems in my assembly were gaps,
The most popular method for doing this is called the dideoxy method or Sanger method (named after its inventor, Frederick Sanger, who was awarded the
 DNA Sequencing DNA sequencing is the determination of the precise sequence of nucleotides in a sample of DNA. The most popular method for doing this is called the dideoxy method or Sanger method (named
DNA Sequencing DNA sequencing is the determination of the precise sequence of nucleotides in a sample of DNA. The most popular method for doing this is called the dideoxy method or Sanger method (named
Session 7 Glycerol Stocks & Sequencing Clones
 Session 7 Glycerol Stocks & Sequencing Clones Learning Objective: In this lab you will prepare several of your clones for DNA sequencing and make glycerol stock cultures as a stable and uniform starting
Session 7 Glycerol Stocks & Sequencing Clones Learning Objective: In this lab you will prepare several of your clones for DNA sequencing and make glycerol stock cultures as a stable and uniform starting
13-2 Manipulating DNA Slide 1 of 32
 1 of 32 The Tools of Molecular Biology The Tools of Molecular Biology How do scientists make changes to DNA? Scientists use their knowledge of the structure of DNA and its chemical properties to study
1 of 32 The Tools of Molecular Biology The Tools of Molecular Biology How do scientists make changes to DNA? Scientists use their knowledge of the structure of DNA and its chemical properties to study
Outline. Structure of DNA DNA Functions Transcription Translation Mutation Cytogenetics Mendelian Genetics Quantitative Traits Linkage
 Genetics Outline Structure of DNA DNA Functions Transcription Translation Mutation Cytogenetics Mendelian Genetics Quantitative Traits Linkage Chromosomes are composed of chromatin, which is DNA and associated
Genetics Outline Structure of DNA DNA Functions Transcription Translation Mutation Cytogenetics Mendelian Genetics Quantitative Traits Linkage Chromosomes are composed of chromatin, which is DNA and associated
Computational Biology 2. Pawan Dhar BII
 Computational Biology 2 Pawan Dhar BII Lecture 1 Introduction to terms, techniques and concepts in molecular biology Molecular biology - a primer Human body has 100 trillion cells each containing 3 billion
Computational Biology 2 Pawan Dhar BII Lecture 1 Introduction to terms, techniques and concepts in molecular biology Molecular biology - a primer Human body has 100 trillion cells each containing 3 billion
Chapter 8: Recombinant DNA. Ways this technology touches us. Overview. Genetic Engineering
 Chapter 8 Recombinant DNA and Genetic Engineering Genetic manipulation Ways this technology touches us Criminal justice The Justice Project, started by law students to advocate for DNA testing of Death
Chapter 8 Recombinant DNA and Genetic Engineering Genetic manipulation Ways this technology touches us Criminal justice The Justice Project, started by law students to advocate for DNA testing of Death
BST227 Introduction to Statistical Genetics. Lecture 8: Variant calling from high-throughput sequencing data
 BST227 Introduction to Statistical Genetics Lecture 8: Variant calling from high-throughput sequencing data 1 PC recap typical genome Differs from the reference genome at 4-5 million sites ~85% SNPs ~15%
BST227 Introduction to Statistical Genetics Lecture 8: Variant calling from high-throughput sequencing data 1 PC recap typical genome Differs from the reference genome at 4-5 million sites ~85% SNPs ~15%
Chapter 17. PCR the polymerase chain reaction and its many uses. Prepared by Woojoo Choi
 Chapter 17. PCR the polymerase chain reaction and its many uses Prepared by Woojoo Choi Polymerase chain reaction 1) Polymerase chain reaction (PCR): artificial amplification of a DNA sequence by repeated
Chapter 17. PCR the polymerase chain reaction and its many uses Prepared by Woojoo Choi Polymerase chain reaction 1) Polymerase chain reaction (PCR): artificial amplification of a DNA sequence by repeated
Studying the Human Genome. Lesson Overview. Lesson Overview Studying the Human Genome
 Lesson Overview 14.3 Studying the Human Genome THINK ABOUT IT Just a few decades ago, computers were gigantic machines found only in laboratories and universities. Today, many of us carry small, powerful
Lesson Overview 14.3 Studying the Human Genome THINK ABOUT IT Just a few decades ago, computers were gigantic machines found only in laboratories and universities. Today, many of us carry small, powerful
What is Bioinformatics? Bioinformatics is the application of computational techniques to the discovery of knowledge from biological databases.
 What is Bioinformatics? Bioinformatics is the application of computational techniques to the discovery of knowledge from biological databases. Bioinformatics is the marriage of molecular biology with computer
What is Bioinformatics? Bioinformatics is the application of computational techniques to the discovery of knowledge from biological databases. Bioinformatics is the marriage of molecular biology with computer
Introduction to Sequencher. Tom Randall Center for Bioinformatics
 Introduction to Sequencher Tom Randall Center for Bioinformatics tarandal@email.unc.edu Introduction Importing, viewing and manipulating chromatographs Trimming chromatographs Assembly into contigs Editing
Introduction to Sequencher Tom Randall Center for Bioinformatics tarandal@email.unc.edu Introduction Importing, viewing and manipulating chromatographs Trimming chromatographs Assembly into contigs Editing
R = G A (purine) Y = T C (pyrimidine) K = G T (Keto) M = A C (amino) S = G C (Strong bonds) W = A T (Weak bonds)
 1 Nucleic acids can form heterocopolymers as DNA or RNA, thus their structural formula can be described (to a close approximation) simply by listing the nucleotide bases in a defined order. A oneletter
1 Nucleic acids can form heterocopolymers as DNA or RNA, thus their structural formula can be described (to a close approximation) simply by listing the nucleotide bases in a defined order. A oneletter
4. Analysing genes II Isolate mutants*
 .. 4. Analysing s II Isolate mutants* Using the mutant to isolate the classify mutants by complementation analysis wild type study phenotype of mutants mutant 1 - use mutant to isolate sequence put individual
.. 4. Analysing s II Isolate mutants* Using the mutant to isolate the classify mutants by complementation analysis wild type study phenotype of mutants mutant 1 - use mutant to isolate sequence put individual
Comparing a few SNP calling algorithms using low-coverage sequencing data
 Yu and Sun BMC Bioinformatics 2013, 14:274 RESEARCH ARTICLE Open Access Comparing a few SNP calling algorithms using low-coverage sequencing data Xiaoqing Yu 1 and Shuying Sun 1,2* Abstract Background:
Yu and Sun BMC Bioinformatics 2013, 14:274 RESEARCH ARTICLE Open Access Comparing a few SNP calling algorithms using low-coverage sequencing data Xiaoqing Yu 1 and Shuying Sun 1,2* Abstract Background:
Genome Sequence Assembly
 Genome Sequence Assembly Learning Goals: Introduce the field of bioinformatics Familiarize the student with performing sequence alignments Understand the assembly process in genome sequencing Introduction:
Genome Sequence Assembly Learning Goals: Introduce the field of bioinformatics Familiarize the student with performing sequence alignments Understand the assembly process in genome sequencing Introduction:
7. Troubleshooting Guidelines
 7. Troubleshooting Guidelines The Pyrosequencing TM technique provides the user with sequence information for several nucleotides 3 of the sequencing primer apart from the polymorphic site. This gives
7. Troubleshooting Guidelines The Pyrosequencing TM technique provides the user with sequence information for several nucleotides 3 of the sequencing primer apart from the polymorphic site. This gives
NUCLEIC ACIDS. DNA (Deoxyribonucleic Acid) and RNA (Ribonucleic Acid): information storage molecules made up of nucleotides.
 NUCLEIC ACIDS DNA (Deoxyribonucleic Acid) and RNA (Ribonucleic Acid): information storage molecules made up of nucleotides. Base Adenine Guanine Cytosine Uracil Thymine Abbreviation A G C U T DNA RNA 2
NUCLEIC ACIDS DNA (Deoxyribonucleic Acid) and RNA (Ribonucleic Acid): information storage molecules made up of nucleotides. Base Adenine Guanine Cytosine Uracil Thymine Abbreviation A G C U T DNA RNA 2
3.A.1 DNA and RNA: Structure and Replication
 3.A.1 DNA and RNA: Structure and Replication Each DNA polymer is made of Nucleotides (monomer) which are made of: a) Phosphate group: Negatively charged and polar b) Sugar: deoxyribose- a 5 carbon sugar
3.A.1 DNA and RNA: Structure and Replication Each DNA polymer is made of Nucleotides (monomer) which are made of: a) Phosphate group: Negatively charged and polar b) Sugar: deoxyribose- a 5 carbon sugar
DNA REPLICATION & BIOTECHNOLOGY Biology Study Review
 DNA REPLICATION & BIOTECHNOLOGY Biology Study Review DNA DNA is found in, in the nucleus. It controls cellular activity by regulating the production of, which includes It is a very long molecule made up
DNA REPLICATION & BIOTECHNOLOGY Biology Study Review DNA DNA is found in, in the nucleus. It controls cellular activity by regulating the production of, which includes It is a very long molecule made up
Biology (2017) INTERNATIONAL GCSE. TOPIC GUIDE: Protein synthesis: transcription and translation. Pearson Edexcel International GCSE in Science
 INTERNATIONAL GCSE Biology (2017) TOPIC GUIDE: Protein synthesis: transcription and translation Pearson Edexcel International GCSE in Science Introduction to the teaching of protein synthesis Specification
INTERNATIONAL GCSE Biology (2017) TOPIC GUIDE: Protein synthesis: transcription and translation Pearson Edexcel International GCSE in Science Introduction to the teaching of protein synthesis Specification
THE STRUCTURE AND FUNCTION OF DNA
 THE STRUCTURE AND FUNCTION OF DNA 1. DNA is our genetic code!!! It is passed from generation to generation. It carries information that controls the functions of our cells. DNA stands for deoxyribonucleic
THE STRUCTURE AND FUNCTION OF DNA 1. DNA is our genetic code!!! It is passed from generation to generation. It carries information that controls the functions of our cells. DNA stands for deoxyribonucleic
A DNA Computing Model to Solve 0-1 Integer. Programming Problem
 Applied Mathematical Sciences, Vol. 2, 2008, no. 59, 2921-2929 A DNA Computing Model to Solve 0-1 Integer Programming Problem Sanchita Paul Lecturer, Department of Computer Science & Engineering Birla
Applied Mathematical Sciences, Vol. 2, 2008, no. 59, 2921-2929 A DNA Computing Model to Solve 0-1 Integer Programming Problem Sanchita Paul Lecturer, Department of Computer Science & Engineering Birla
High Throughput Sequencing Technologies. J Fass UCD Genome Center Bioinformatics Core Tuesday December 16, 2014
 High Throughput Sequencing Technologies J Fass UCD Genome Center Bioinformatics Core Tuesday December 16, 2014 Sequencing Explosion www.genome.gov/sequencingcosts http://t.co/ka5cvghdqo Sequencing Explosion
High Throughput Sequencing Technologies J Fass UCD Genome Center Bioinformatics Core Tuesday December 16, 2014 Sequencing Explosion www.genome.gov/sequencingcosts http://t.co/ka5cvghdqo Sequencing Explosion
Course summary. Today. PCR Polymerase chain reaction. Obtaining molecular data. Sequencing. DNA sequencing. Genome Projects.
 Goals Organization Labs Project Reading Course summary DNA sequencing. Genome Projects. Today New DNA sequencing technologies. Obtaining molecular data PCR Typically used in empirical molecular evolution
Goals Organization Labs Project Reading Course summary DNA sequencing. Genome Projects. Today New DNA sequencing technologies. Obtaining molecular data PCR Typically used in empirical molecular evolution
SAMPLE LITERATURE Please refer to included weblink for correct version.
 Edvo-Kit #340 DNA Informatics Experiment Objective: In this experiment, students will explore the popular bioninformatics tool BLAST. First they will read sequences from autoradiographs of automated gel
Edvo-Kit #340 DNA Informatics Experiment Objective: In this experiment, students will explore the popular bioninformatics tool BLAST. First they will read sequences from autoradiographs of automated gel
Finishing Drosophila Grimshawi Fosmid Clone DGA19A15. Matthew Kwong Bio4342 Professor Elgin February 23, 2010
 Finishing Drosophila Grimshawi Fosmid Clone DGA19A15 Matthew Kwong Bio4342 Professor Elgin February 23, 2010 1 Abstract The primary aim of the Bio4342 course, Research Explorations and Genomics, is to
Finishing Drosophila Grimshawi Fosmid Clone DGA19A15 Matthew Kwong Bio4342 Professor Elgin February 23, 2010 1 Abstract The primary aim of the Bio4342 course, Research Explorations and Genomics, is to
Decoding of Superimposed Traces Produced by Direct Sequencing of Heterozygous Indels Dmitriev, D.A. & Rakitov, R.A.
 Decoding of Superimposed Traces Produced by Direct Sequencing of Heterozygous Indels Dmitriev, D.A. & Rakitov, R.A. Illinois Natural History Survey, Institute of Natural Resource Sustainability, University
Decoding of Superimposed Traces Produced by Direct Sequencing of Heterozygous Indels Dmitriev, D.A. & Rakitov, R.A. Illinois Natural History Survey, Institute of Natural Resource Sustainability, University
Genome Resources. Genome Resources. Maj Gen (R) Suhaib Ahmed, HI (M)
 Maj Gen (R) Suhaib Ahmed, I (M) The human genome comprises DNA sequences mostly contained in the nucleus. A small portion is also present in the mitochondria. The nuclear DNA is present in chromosomes.
Maj Gen (R) Suhaib Ahmed, I (M) The human genome comprises DNA sequences mostly contained in the nucleus. A small portion is also present in the mitochondria. The nuclear DNA is present in chromosomes.
Tutorial for Stop codon reassignment in the wild
 Tutorial for Stop codon reassignment in the wild Learning Objectives This tutorial has two learning objectives: 1. Finding evidence of stop codon reassignment on DNA fragments. 2. Detecting and confirming
Tutorial for Stop codon reassignment in the wild Learning Objectives This tutorial has two learning objectives: 1. Finding evidence of stop codon reassignment on DNA fragments. 2. Detecting and confirming
DNA. Shape = Double Helix (twisted ladder) The purpose of each cell having DNA is to have directions for the cell to make proteins
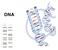 DNA DNA Deoxyribo- Nucleic Acid Shape = Double Helix (twisted ladder) The purpose of each cell having DNA is to have directions for the cell to make proteins Parts = nucleotide 1. Sugar (deoxyribose) 2.
DNA DNA Deoxyribo- Nucleic Acid Shape = Double Helix (twisted ladder) The purpose of each cell having DNA is to have directions for the cell to make proteins Parts = nucleotide 1. Sugar (deoxyribose) 2.
Molecular Biology: DNA sequencing
 Molecular Biology: DNA sequencing Author: Prof Marinda Oosthuizen Licensed under a Creative Commons Attribution license. SEQUENCING OF LARGE TEMPLATES As we have seen, we can obtain up to 800 nucleotides
Molecular Biology: DNA sequencing Author: Prof Marinda Oosthuizen Licensed under a Creative Commons Attribution license. SEQUENCING OF LARGE TEMPLATES As we have seen, we can obtain up to 800 nucleotides
User Bulletin. ABI PRISM 373 DNA Sequencer. Using the ABI PRISM 373 BigDye Filter Wheel. Contents. Introduction. February 16, 1998 (updated 06/2001)
 User Bulletin ABI PRISM 373 DNA Sequencer February 16, 1998 (updated 06/2001) SUBJECT: Using the ABI PRISM 373 BigDye Filter Wheel Contents The following topics are discussed in this bulletin: Topic See
User Bulletin ABI PRISM 373 DNA Sequencer February 16, 1998 (updated 06/2001) SUBJECT: Using the ABI PRISM 373 BigDye Filter Wheel Contents The following topics are discussed in this bulletin: Topic See
Auto Sequencer: A DNA Sequence Alignment and Assembly Tool
 Auto Sequencer: A DNA Sequence Alignment and Assembly Tool Abhi Aggarwal, Landon Zarowny, Robert E. Campbell Department of Chemistry, University of Alberta, Edmonton, Alberta Corresponding author: abhi2@ualberta.ca
Auto Sequencer: A DNA Sequence Alignment and Assembly Tool Abhi Aggarwal, Landon Zarowny, Robert E. Campbell Department of Chemistry, University of Alberta, Edmonton, Alberta Corresponding author: abhi2@ualberta.ca
Ah, Lou! There really are differences between us!
 Name Per Ah, Lou! There really are differences between us! Introduction The human genome (the total sum of our genetic makeup) is made up of approximately 6 billion base pairs distributed on 46 chromosomes.
Name Per Ah, Lou! There really are differences between us! Introduction The human genome (the total sum of our genetic makeup) is made up of approximately 6 billion base pairs distributed on 46 chromosomes.
MICROARRAYS: CHIPPING AWAY AT THE MYSTERIES OF SCIENCE AND MEDICINE
 MICROARRAYS: CHIPPING AWAY AT THE MYSTERIES OF SCIENCE AND MEDICINE National Center for Biotechnology Information With only a few exceptions, every
MICROARRAYS: CHIPPING AWAY AT THE MYSTERIES OF SCIENCE AND MEDICINE National Center for Biotechnology Information With only a few exceptions, every
Variant calling workflow for the Oncomine Comprehensive Assay using Ion Reporter Software v4.4
 WHITE PAPER Oncomine Comprehensive Assay Variant calling workflow for the Oncomine Comprehensive Assay using Ion Reporter Software v4.4 Contents Scope and purpose of document...2 Content...2 How Torrent
WHITE PAPER Oncomine Comprehensive Assay Variant calling workflow for the Oncomine Comprehensive Assay using Ion Reporter Software v4.4 Contents Scope and purpose of document...2 Content...2 How Torrent
DNA Fingerprinting. The DNA fingerprinting technique is summarized as follows:
 DNA Fingerprinting Introduction Laboratory techniques called DNA fingerprinting have been developed to identify or type an individual's DNA. One application of these techniques has been in solving crimes.
DNA Fingerprinting Introduction Laboratory techniques called DNA fingerprinting have been developed to identify or type an individual's DNA. One application of these techniques has been in solving crimes.
Era with Computational Biology/Toxicology
 USM Seminar 1/22/2010 Embracing the Post-Omics Era with Computational Biology/Toxicology Ping Gong Environmental Genomics and Genetics (EGG) Team @ Environmental Laboratory Outline Introduction Bioinformatics
USM Seminar 1/22/2010 Embracing the Post-Omics Era with Computational Biology/Toxicology Ping Gong Environmental Genomics and Genetics (EGG) Team @ Environmental Laboratory Outline Introduction Bioinformatics
The goal of this project was to prepare the DEUG contig which covers the
 Prakash 1 Jaya Prakash Dr. Elgin, Dr. Shaffer Biology 434W 10 February 2017 Finishing of DEUG4927010 Abstract The goal of this project was to prepare the DEUG4927010 contig which covers the terminal 99,279
Prakash 1 Jaya Prakash Dr. Elgin, Dr. Shaffer Biology 434W 10 February 2017 Finishing of DEUG4927010 Abstract The goal of this project was to prepare the DEUG4927010 contig which covers the terminal 99,279
Data Mining for Biological Data Analysis
 Data Mining for Biological Data Analysis Data Mining and Text Mining (UIC 583 @ Politecnico di Milano) References Data Mining Course by Gregory-Platesky Shapiro available at www.kdnuggets.com Jiawei Han
Data Mining for Biological Data Analysis Data Mining and Text Mining (UIC 583 @ Politecnico di Milano) References Data Mining Course by Gregory-Platesky Shapiro available at www.kdnuggets.com Jiawei Han
Molecular Biology. IMBB 2017 RAB, Kigali - Rwanda May 02 13, Francesca Stomeo
 Molecular Biology IMBB 2017 RAB, Kigali - Rwanda May 02 13, 2017 Francesca Stomeo Molecular biology is the study of biology at a molecular level, especially DNA and RNA - replication, transcription, translation,
Molecular Biology IMBB 2017 RAB, Kigali - Rwanda May 02 13, 2017 Francesca Stomeo Molecular biology is the study of biology at a molecular level, especially DNA and RNA - replication, transcription, translation,
How to view Results with Scaffold. Proteomics Shared Resource
 How to view Results with Scaffold Proteomics Shared Resource Starting out Download Scaffold from http://www.proteomes oftware.com/proteom e_software_prod_sca ffold_download.html Follow installation instructions
How to view Results with Scaffold Proteomics Shared Resource Starting out Download Scaffold from http://www.proteomes oftware.com/proteom e_software_prod_sca ffold_download.html Follow installation instructions
Alignment methods. Martijn Vermaat Department of Human Genetics Center for Human and Clinical Genetics
 Alignment methods Martijn Vermaat Department of Human Genetics Center for Human and Clinical Genetics Alignment methods Sequence alignment Assembly vs alignment Alignment methods Common issues Platform
Alignment methods Martijn Vermaat Department of Human Genetics Center for Human and Clinical Genetics Alignment methods Sequence alignment Assembly vs alignment Alignment methods Common issues Platform
Genome Assembly Using de Bruijn Graphs. Biostatistics 666
 Genome Assembly Using de Bruijn Graphs Biostatistics 666 Previously: Reference Based Analyses Individual short reads are aligned to reference Genotypes generated by examining reads overlapping each position
Genome Assembly Using de Bruijn Graphs Biostatistics 666 Previously: Reference Based Analyses Individual short reads are aligned to reference Genotypes generated by examining reads overlapping each position
2 Gene Technologies in Our Lives
 CHAPTER 15 2 Gene Technologies in Our Lives SECTION Gene Technologies and Human Applications KEY IDEAS As you read this section, keep these questions in mind: For what purposes are genes and proteins manipulated?
CHAPTER 15 2 Gene Technologies in Our Lives SECTION Gene Technologies and Human Applications KEY IDEAS As you read this section, keep these questions in mind: For what purposes are genes and proteins manipulated?
CISC 889 Bioinformatics (Spring 2004) Lecture 3
 CISC 889 Bioinformatics (Spring 004) Lecture Genome Sequencing Li Liao Computer and Information Sciences University of Delaware Administrative Have you visited The NCBI website? Have you read Hunter s
CISC 889 Bioinformatics (Spring 004) Lecture Genome Sequencing Li Liao Computer and Information Sciences University of Delaware Administrative Have you visited The NCBI website? Have you read Hunter s
DNA, or Deoxyribonucleic Acid, is the genetic material in our cells. No two people (except identical twins) have the
 DNA, or Deoxyribonucleic Acid, is the genetic material in our cells. No two people (except identical twins) have the exact same DNA. DNA patterns from four sets of twins which are identical? DNA fingerprinting
DNA, or Deoxyribonucleic Acid, is the genetic material in our cells. No two people (except identical twins) have the exact same DNA. DNA patterns from four sets of twins which are identical? DNA fingerprinting
Identification of Putative Coding Regions in a 10kB Sequence of the E. coli 536 Genome. By Someone. Partner: Someone else
 1 Identification of Putative Coding Regions in a 10kB Sequence of the E. coli 536 Genome Introduction: By Someone Partner: Someone else The instructions required for the maintenance of life are encoded
1 Identification of Putative Coding Regions in a 10kB Sequence of the E. coli 536 Genome Introduction: By Someone Partner: Someone else The instructions required for the maintenance of life are encoded
DNA SEQUENCING BY SANGER METHOD
 DNA SEQUENCING BY SANGER METHOD First method described by Sanger and Coulson,1975 for DNA sequencing was called plus and minus. This method used E.coli DNA polymerase I and DNA ploymerase from bacteriophage
DNA SEQUENCING BY SANGER METHOD First method described by Sanger and Coulson,1975 for DNA sequencing was called plus and minus. This method used E.coli DNA polymerase I and DNA ploymerase from bacteriophage
ChIP-seq and RNA-seq. Farhat Habib
 ChIP-seq and RNA-seq Farhat Habib fhabib@iiserpune.ac.in Biological Goals Learn how genomes encode the diverse patterns of gene expression that define each cell type and state. Protein-DNA interactions
ChIP-seq and RNA-seq Farhat Habib fhabib@iiserpune.ac.in Biological Goals Learn how genomes encode the diverse patterns of gene expression that define each cell type and state. Protein-DNA interactions
Some types of Mutagenesis
 Mutagenesis What Is a Mutation? Genetic information is encoded by the sequence of the nucleotide bases in DNA of the gene. The four nucleotides are: adenine (A), thymine (T), guanine (G), and cytosine
Mutagenesis What Is a Mutation? Genetic information is encoded by the sequence of the nucleotide bases in DNA of the gene. The four nucleotides are: adenine (A), thymine (T), guanine (G), and cytosine
UltraFast Molecular Diagnostic System
 UltraFast Molecular Diagnostic System CONTENTS 01 PCR vs Real-time PCR 02 NANOBIOSYS Sample Prep G2-16TU 03 NANOBIOSYS Real-time PCR G2-4 01 PCR vs Real-time PCR What is DNA & What is PCR? NANOBIOSYS 4
UltraFast Molecular Diagnostic System CONTENTS 01 PCR vs Real-time PCR 02 NANOBIOSYS Sample Prep G2-16TU 03 NANOBIOSYS Real-time PCR G2-4 01 PCR vs Real-time PCR What is DNA & What is PCR? NANOBIOSYS 4
Representing Errors and Uncertainty in Plasma Proteomics
 Representing Errors and Uncertainty in Plasma Proteomics David J. States, M.D., Ph.D. University of Michigan Bioinformatics Program Proteomics Alliance for Cancer Genomics vs. Proteomics Genome sequence
Representing Errors and Uncertainty in Plasma Proteomics David J. States, M.D., Ph.D. University of Michigan Bioinformatics Program Proteomics Alliance for Cancer Genomics vs. Proteomics Genome sequence
CSCI 2570 Introduction to Nanocomputing
 CSCI 2570 Introduction to Nanocomputing DNA Computing John E Savage DNA (Deoxyribonucleic Acid) DNA is double-stranded helix of nucleotides, nitrogen-containing molecules. It carries genetic information
CSCI 2570 Introduction to Nanocomputing DNA Computing John E Savage DNA (Deoxyribonucleic Acid) DNA is double-stranded helix of nucleotides, nitrogen-containing molecules. It carries genetic information
Greene 1. Finishing of DEUG The entire genome of Drosophila eugracilis has recently been sequenced using Roche
 Greene 1 Harley Greene Bio434W Elgin Finishing of DEUG4927002 Abstract The entire genome of Drosophila eugracilis has recently been sequenced using Roche 454 pyrosequencing and Illumina paired-end reads
Greene 1 Harley Greene Bio434W Elgin Finishing of DEUG4927002 Abstract The entire genome of Drosophila eugracilis has recently been sequenced using Roche 454 pyrosequencing and Illumina paired-end reads
Supplementary Figures
 Supplementary Figures A B Supplementary Figure 1. Examples of discrepancies in predicted and validated breakpoint coordinates. A) Most frequently, predicted breakpoints were shifted relative to those derived
Supplementary Figures A B Supplementary Figure 1. Examples of discrepancies in predicted and validated breakpoint coordinates. A) Most frequently, predicted breakpoints were shifted relative to those derived
Back end. Sequencing of DNA clones. Lane finding and base calling - steps. Lane finding and base calling - software. Base calling - steps
 Sequencing of DNA clones Back end Front end - sample preparation Separation and detection - gel electrophoresis Lane finding Base calling Assembly Finishing Back end - determine input DNA Lane finding
Sequencing of DNA clones Back end Front end - sample preparation Separation and detection - gel electrophoresis Lane finding Base calling Assembly Finishing Back end - determine input DNA Lane finding
High Throughput Sequencing Technologies. UCD Genome Center Bioinformatics Core Monday 15 June 2015
 High Throughput Sequencing Technologies UCD Genome Center Bioinformatics Core Monday 15 June 2015 Sequencing Explosion www.genome.gov/sequencingcosts http://t.co/ka5cvghdqo Sequencing Explosion 2011 PacBio
High Throughput Sequencing Technologies UCD Genome Center Bioinformatics Core Monday 15 June 2015 Sequencing Explosion www.genome.gov/sequencingcosts http://t.co/ka5cvghdqo Sequencing Explosion 2011 PacBio
3 Designing Primers for Site-Directed Mutagenesis
 3 Designing Primers for Site-Directed Mutagenesis 3.1 Learning Objectives During the next two labs you will learn the basics of site-directed mutagenesis: you will design primers for the mutants you designed
3 Designing Primers for Site-Directed Mutagenesis 3.1 Learning Objectives During the next two labs you will learn the basics of site-directed mutagenesis: you will design primers for the mutants you designed
COMPUTER RESOURCES II:
 COMPUTER RESOURCES II: Using the computer to analyze data, using the internet, and accessing online databases Bio 210, Fall 2006 Linda S. Huang, Ph.D. University of Massachusetts Boston In the first computer
COMPUTER RESOURCES II: Using the computer to analyze data, using the internet, and accessing online databases Bio 210, Fall 2006 Linda S. Huang, Ph.D. University of Massachusetts Boston In the first computer
Sequencing the Human Genome
 The Biotechnology 339 EDVO-Kit # Sequencing the Human Genome Experiment Objective: In this experiment, DNA sequences obtained from automated sequencers will be submitted to Data bank searches using the
The Biotechnology 339 EDVO-Kit # Sequencing the Human Genome Experiment Objective: In this experiment, DNA sequences obtained from automated sequencers will be submitted to Data bank searches using the
Lesson Overview. Studying the Human Genome. Lesson Overview Studying the Human Genome
 Lesson Overview 14.3 Studying the Human Genome THINK ABOUT IT Just a few decades ago, computers were gigantic machines found only in laboratories and universities. Today, many of us carry small, powerful
Lesson Overview 14.3 Studying the Human Genome THINK ABOUT IT Just a few decades ago, computers were gigantic machines found only in laboratories and universities. Today, many of us carry small, powerful
Sequence Analysis Lab Protocol
 Sequence Analysis Lab Protocol You will need this handout of instructions The sequence of your plasmid from the ABI The Accession number for Lambda DNA J02459 The Accession number for puc 18 is L09136
Sequence Analysis Lab Protocol You will need this handout of instructions The sequence of your plasmid from the ABI The Accession number for Lambda DNA J02459 The Accession number for puc 18 is L09136
Lecture 2: Biology Basics Continued
 Lecture 2: Biology Basics Continued Central Dogma DNA: The Code of Life The structure and the four genomic letters code for all living organisms Adenine, Guanine, Thymine, and Cytosine which pair A-T and
Lecture 2: Biology Basics Continued Central Dogma DNA: The Code of Life The structure and the four genomic letters code for all living organisms Adenine, Guanine, Thymine, and Cytosine which pair A-T and
