Inheritance, gene expression, and lignin characterization in a mutant pine deficient in cinnamyl alcohol dehydrogenase
|
|
|
- Nickolas Silvester Marsh
- 5 years ago
- Views:
Transcription
1 Proc. Natl. Acad. Sci. USA Vol. 94, pp , July 1997 Plant Biology Inheritance, gene expression, and lignin characterization in a mutant pine deficient in cinnamyl alcohol dehydrogenase JOHN J. MACKAY*, DAVID M. O MALLEY*, TIMOTHY PRESNELL, FITZGERALD L. BOOKER, MALCOLM M. CAMPBELL*,ROSS W. WHETTEN*, AND RONALD R. SEDEROFF* ** Departments of *Forestry, Genetics, Wood and Paper Science, and Crop Science, North Carolina State University, Raleigh, NC Contributed by Ronald R. Sederoff, May 21, 1997 ABSTRACT We have discovered a mutant loblolly pine (Pinus taeda L.) in which expression of the gene encoding cinnamyl alcohol dehydrogenase (CAD; EC ) is severely reduced. The products of CAD, cinnamyl alcohols, are the precursors of lignin, a major cell wall polymer of plant vascular tissues. Lignin composition in this mutant shows dramatic modifications, including increased incorporation of the substrate of CAD (coniferaldehyde), indicating that CAD may modulate lignin composition in pine. The recessive cad-n1 allele, which causes this phenotype, was discovered in a tree heterozygous for this mutant allele. It is inherited as a simple Mendelian locus that maps to the same genomic region as the cad locus. In mutant plants, CAD activity and abundance of cad RNA transcript are low, and free CAD substrate accumulates to a high level. The wood of the mutant is brown, whereas the wood in wild types is nearly white. The wood phenotype resembles that of brown midrib (bm) mutants and some transgenic plants in which xylem is red-brown due to a reduction in CAD activity. However, unlike transgenics with reduced CAD, the pine mutant has decreased lignin content. Wood in which the composition of lignin varies beyond previous expectations still provides vascular function and mechanical support. Lignin is a major component of the cell wall embedding matrix that reinforces the cellulose microfibrils of plant vascular tissues (1, 2). Thus, lignin confers compressive strength and bending stiffness necessary for mechanical support; it also provides a hydrophobic surface, essential for longitudinal water transport, and provides a barrier against pathogens. Lignin is a complex phenolic polymer, usually derived from various proportions of three cinnamyl alcohol precursors, p-coumaryl, coniferyl, and sinapyl alcohols (3). The biological role of this variation in composition and its significance in the structure function relationship of lignins remain poorly understood (4). The precursors of lignin (monolignols) are products of a specialized branch of phenylpropanoid metabolism (Fig. 1). Many enzymes implicated in monolignol biosynthesis have been characterized and the corresponding genes have been cloned (1, 2). The last step of the monolignol pathway, the reduction of cinnamaldehydes to cinnamyl alcohols, is catalyzed by cinnamyl alcohol dehydrogenase (CAD; EC ). Antisense suppression of CAD to as low as 7% of wild type in tobacco significantly increased the proportion of cinnamaldehyde subunits in lignin, but no effect was detected on the amount of lignin (5). The requirement for CAD in the synthesis of lignin precursors and the role of cinnamaldehydes as lignin precursors therefore need to be reassessed by more specific loss-of-function gain-of-function experiments. The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C solely to indicate this fact by The National Academy of Sciences $ PNAS is available online at http: FIG. 1. Schematic of monolignol biosynthesis. The general phenylpropanoid pathway synthesizes the p-hydroxycinnamic acids (p- HCA) via the enzymes PAL (phenylalanine ammonia-lyase), C4H (cinnamate 4-hydroxylase), C3H (p-coumarate 3-hydroxylase), COMT (caffeate 5-hydroxyferulate O-methyltransferase), and F5H (ferulate 5-hydroxylase). Hydroxycinnamoyl-CoA thioesters (p-hca-coa), formed by 4CL (4 coumarate:coa ligase) are reduced by CCR (cinnamoyl-coa reductase) and CAD (cinnamyl alcohol dehydrogenase) to give p-hydroxycinnamyl alcohols. In trees, lignin can account for 18 36% of the dry weight of wood (6). Therefore lignin mutants in tree species would be of particular interest to verify the biological role of lignin biosynthetic enzymes. However, lignin mutants have been reported only in herbaceous plants. Brown midrib (bm) mutants, found in monocots, have decreased lignin content and or modified lignin composition (7, 8). Here, we identify and describe a cad mutant in a woody gymnosperm, loblolly pine (Pinus taeda L.). A mutant cad allele was identified in a heterozygous parent and genetically mapped in a haploid mapping population. This allele results in severely decreased cad gene expression and, in homozygotes, modifies wood color and lignin composition. This mutant provides new insights into the role of CAD and the biosynthesis of lignin in woody plants. Abbreviations: AcBr, acetyl bromide; AF, alkaline fraction; CAD, cinnamyl alcohol dehydrogenase; COMT, caffeate 5-hydroxyferulate O-methyltransferase; CWR, cell wall residue; FTIR, Fouriertransform infrared; G6PDH, glucose-6-phosphate dehydrogenase; MWL, milled wood lignin; RAPD, random amplified polymorphic DNA. Present address: Westvaco, Charleston Research Center, Charleston, SC Present address: Department of Plant Sciences, Oxford University, Oxford, OX1 3RB, United Kingdom. **To whom reprint requests should be addressed at: Forest Biotechnology Group, Box 8008, Department of Forestry, North Carolina State University, Raleigh, NC volvo@unity.ncsu.edu. All precusors of lignin are p-hydroxycinnamyl alcohols. 8255
2 8256 Plant Biology: MacKay et al. Proc. Natl. Acad. Sci. USA 94 (1997) MATERIALS AND METHODS Plant Material and Wood Sample Preparation. Loblolly pine seeds, tissue, and wood samples were from the North Carolina State University Industry Cooperative Tree Improvement Program, Westvaco Corporation, and the Baruch Experimental Forest and were isolated as previously (9). Wood samples were debarked and dried at room temperature for 2 days [for milled wood lignin (MWL)] or at 65 C overnight and ground in a Wiley mill through a 60-mesh screen (10). Extractive free wood cell wall residue (CWR) was prepared according to Halpin et al. (5). Allozyme Gels and Enzyme Assays. Allozyme gel analyses used standard conditions for CAD activity (9, 11) and for glucose-6-phosphate dehydrogenase (G6PDH), alcohol dehydrogenase (ADH), and shikimic acid dehydrogenase (SDH) (12). The reverse reaction of CAD (the oxidation of coniferyl alcohol to coniferaldehyde) was assayed (11) in whole-cell extracts (13) from mg of fresh tissue per assay. Mean enzyme activities were obtained from six plants, each sampled in duplicate. The forward reaction (the reduction of coniferaldehyde to coniferyl alcohol) was assayed in partially purified protein extracts (11) from 150 mg of tissue, precipitated in the 30 70% range of saturated ammonium sulfate (14). DNA Isolation, Random Amplified Polymorphic DNA (RAPD) Marker Analysis, and Genetic Mapping. Genomic DNA was isolated from needles (15) and individual megagametophytes (Puregene DNA isolation kit, Gentra Systems). Approximately one-third of each haploid megagametophyte used in mapping experiments (5 to 6 mg) was used for allozyme gel analysis and the rest (9 12 mg) was stored at 80 C for DNA isolation. Methods for linkage analysis using RAPD markers and MAPMAKER for Macintosh version 2.0 (DuPont) were previously described (16, 17). For bulked segregant analysis (18), two pooled DNA samples each prepared from 12 megagametophytes and representing the two CAD allozymes of clone were screened with 170 RAPD primers (Operon Technologies, Alameda, CA) to identify linked markers. Cosegregation of RAPD markers and CAD allozymes was verified using 74 megagametophytes from clone and 108 megagametophytes from clone Isolation of RNA and Analysis of Transcript Levels. Total RNA (19) for Northern blots was separated in 1.2% agarose gels with 2.2 M formaldehyde and transferred to Zetaprobe nylon membranes (Bio-Rad). A full-length cad cdna (9) and a 170-bp fragment of a pine ribosomal protein cdna were 32 P radiolabeled with Ready-To-Go Labelling Beads (Pharmacia Biotech) and used as probes. The prehybridization, hybridization (in the presence of 50% deionized formamide), and membrane washes followed Brown and MacKey (20). Densitometric scanning of autoradiographs used a personal densitometer SI (Molecular Devices) and quantitations used the program IMAGEQUANT (Molecular Devices). Isolation and Analysis of Phenolic Extractives. Phenolic extractives were recovered by successive extraction of ground wood with 50% ethanol (once) and 50% methanol (five times) at 70 C, each for 2 hr. Total phenolic content was determined with the Folin and Ciocalteu reagent (21), using ferulic acid as a standard. Extracts were hydrolyzed in 1 M NaOH for 1 hr at room temperature and analyzed by reverse-phase HPLC using a Microsorb C 18 column (Rainin Instruments) with a 0% to 99% acetonitrile gradient in 1% acetic acid (21). Compounds were identified on the basis of coelution with standards and the UV spectrum of corresponding eluted fractions. Isolation of Lignin and Fourier-Transform Infrared (FTIR) Analysis of Lignin and CWR. MWL was prepared from one homozygous mutant and one wild-type 12-year old tree. Ground wood samples were ball milled (22) and extracted twice with 96:4 (vol vol) dioxane water (23). Extracts were freeze-dried, dissolved in 90% acetic acid, precipitated and washed with water and freeze-dried, dissolved in 2:1 (vol vol) 1,2-dichloroethane ethanol, precipitated with diethyl ether, washed with petroleum ether, and dried in vacuo. The yields of the MWL preparations were 19% of the total lignin for the mutant and 20% for the wild type. FTIR transmission spectra of MWL and CWR were acquired using a Perkin Elmer 16PC spectrophotometer. MWL (1 mg) or CWR (3 mg) was pressed into pellets with 300 mg of KBr. Spectral data were converted to absorbance units and vector-normalized using PECSS software (Perkin Elmer). Lignin Content Determinations. Lignin content of CWR was estimated by using both Klason and acetyl bromide (AcBr) methods. Klason acid-insoluble lignin was determined according to Effland (24), using samples of mg of CWR. The A 205 of the Klason acid filtrate was used to calculate the acid-soluble lignin (25). The AcBr method (26) used samples of 5 6 mg of CWR with or without a saponification pretreatment, consisting of three extractions with 1 M NaOH. RESULTS Detection and Inheritance of a Mutant cad Allele on Allozyme Gels. In loblolly pine, CAD is encoded by a single gene which has several alleles encoding proteins of different electrophoretic mobilities (9). In many conifers, including pines, allelic variation is easily analyzed in the megagametophyte, a haploid (1n) seed tissue derived from the megaspore (27). Megagametophytes from a single tree can be used as a segregating haploid population, in which alleles at each locus heterozygous in the parent will segregate 1:1. Furthermore, the allele present in the megagametophyte is identical to the maternally inherited allele of the embryo in the same seed. We uncovered a low-activity CAD allozyme segregating in megagametophytes from a heterozygous parent. In a genotype of loblolly pine (clone 7-56), megagametophytes from 50% of the seeds had a null allozyme phenotype for CAD (Fig. 2a), although all showed activity for G6PDH (Fig. 2a), shikimic acid dehydrogenase, and alcohol dehydrogenase (not shown). The segregation ratio of the low CAD activity phenotype was FIG. 2. Inheritance of the low-activity CAD allozyme. (a) Each lane contains megagametophyte extract from one seed of clone 7-56, costained for CAD and G6PDH. (b) Inheritance in megagametophytes (M) and embryos (E) within individual seeds, from a cross of two trees with four different cad alleles: the low-activity allozyme and a slow (S) mobility allozyme, clone 7-56, maternal parent; intermediate (I) and fast (F) alleles, from the pollen parent.
3 Plant Biology: MacKay et al. Proc. Natl. Acad. Sci. USA 94 (1997) 8257 FIG. 3. Genetic maps of the cad locus in loblolly pine. Genetic maps of the region flanking the cad locus were generated for clones (a) and 7-56 (b) (on linkage group 5). RF, recombination fraction; Dist (cm), distance in centimorgans; RAPD markers names are Operon primer names followed by the estimated DNA fragment size in base pairs. determined from 121 megagametophytes and was 62:59, not significantly different from a 1:1 ratio ( ). These results are strong evidence that clone 7-56 is heterozygous for a mutation in a single genetic locus affecting CAD enzyme activity. This segregation ratio could result from a mutation with either a cis-acting or trans-acting effect on the cad gene product. To distinguish between these alternatives, the inheritance of the low-activity allozyme was analyzed in diploid tissue. Inheritance patterns in the megagametophyte and embryo of individual seeds showed that offspring that inherit the low-activity allozyme from one parent (heterozygous offspring) have one active Cad allele. CAD is a dimeric protein, therefore, heterozygotes for fast and slow electrophoretic alleles show three bands for CAD in diploid tissues, consisting of two homodimers and one heterodimer (9). In controlled crosses of clone 7-56, maternal inheritance of the low-activity allozyme was unambiguously determined in megagametophytes and gave a single band of CAD activity in the embryos, a homodimeric band of staining contributed by the other parent (Fig. 2b). This type of staining pattern was observed in heterozygotes that inherited this allozyme paternally or maternally and was consistent with a single active Cad allele. The simplest explanation for these results is the inheritance of a cis-acting mutation that results in a mutant cad allele, designated cad-n1. By contrast, a dominant mutation in a transacting gene would suppress both Cad alleles, and a recessive trans-acting mutation would result in two active Cad alleles. Genetic Mapping of the cad-n1 Allele by Using RAPD Markers. The cad locus and the locus responsible for the low-activity CAD allozyme were mapped independently in two unrelated pine genotypes, using megagametophytes, and both were located in the same genomic region (Fig. 3). We identified RAPD markers linked to the cad locus by bulk segregant analysis in a loblolly pine clone (6-1031) which contained two electrophoretically distinct wild-type CAD allozymes. On the genomic map of clone 7-56 (28), the low CAD activity phenotype associated with the cad-n1 allele mapped to linkage group 5. In clones and 7-56, the cad locus and the locus of the cad-n1 allele each cosegregated with genetic markers common to both individuals. The conserved map order and map distances identified a syntenic region that explains the Mendelian inheritance of these two types of allozyme variation. This result is consistent with genetic control of the low-activity phenotype by the single cad locus of pine (9) and its designation as a mutant cad allele. Segregation and Phenotype of the cad-n1 Mutant Plants. The cad genotype of plants obtained by selfing of clone 7-56 were determined using RAPD markers. cad genotypes could not be determined directly because there was no known DNA sequence polymorphism within the cad gene. Therefore RAPD markers were used to define a multilocus haplotype flanking the cad locus (Fig. 3; ref. 16). The inferred segregation ratio of cad genotypes was 1:2:1, following Mendelian segregation. Under greenhouse conditions, the morphology and growth of cad-n1 homozygotes could not be distinguished easily from that of other genotypes, although several mutants had slightly less straight stems (Fig. 4a). The wood of the cad-n1 homozygotes, however, was distinctly brown, whereas other genotypes had nearly white wood, typical for pine (Fig. 4b). The brown color was evenly distributed throughout the wood of cad-n1 homozygotes and persistent in several 2-yearold saplings (not shown) and even in a 12-year-old tree (Fig. 4d). It was darkest immediately after debarking but was stable in wood samples for several months (Fig. 4d). The color was observed only in tracheids and not in nonlignified ray parenchyma cells, which form lightly colored radial files in xylem cross sections (Fig. 4b). Weisner reagent (10) stained the wood of wild types pink as opposed to dark red for the mutants (Fig. 4c), indicative of an accumulation of cinnamaldehydes (10, 29). FIG. 4. Phenotype of the cad-n1 mutant. (a) Seedlings at 10 months, homozygous Cad (left) and cad-n1 (right). (b and c) Stem sections from 7-month-old Cad (lower) and cad-n1 (upper) homozygotes unstained (b) and stained with Weisner reagent (c). (d) Wood chips from 12-year-old trees (homozygous Cad, left; cad-n1, right).
4 8258 Plant Biology: MacKay et al. Proc. Natl. Acad. Sci. USA 94 (1997) FIG. 5. Northern analyses of steady-state cad message. (a) RNA blots probed with cad (Upper) and a pine ribosomal protein cdna (Lower). Samples are total RNA (shoot tips, 10 g; others, 5 g) from homozygous or haploid cad-n1 ( ) and wild-type ( ) genotypes. (b) Total RNAs (40 g, 15 g, or 3 g) from four cad-n1 (lanes 1 4, 8 11, and 15 18) and three wild-type homozygotes (lanes 5 7, 12 14, and 19 21). (c) Histogram of the cad hybridization signal normalized against the ribosomal protein signal for each lane of b. Together, the wood color, its cell type distribution, and the intense Weisner stain are consistent with a mutation in lignin biosynthesis that affects CAD activity, resulting in an increase in cinnamaldehyde (14, 30). Cad Gene Expression. CAD enzyme activity was assayed in differentiating xylem tissues from 10-month-old pine seedlings by monitoring the oxidation of coniferyl alcohol to coniferaldehyde (11). The wild-type homozygotes had nkat g of fresh weight tissue, whereas the cad-n1 homozygotes had nkat g, only 1% on average of wild-type CAD enzyme activity. The reduction of cinnamaldehydes to cinnamyl alcohols is the reaction carried out in vivo by CAD to synthesize monolignols. It was monitored in partially purified xylem protein extracts and, again, cad-n1 homozygotes had between 0% and 1% of the wild-type level of activity. Heterozygosity for the cad-n1 allele resulted in half the wild-type enzyme activity, consistent with heterozygotes having only one active Cad allele. The relative abundance of cad RNA transcript was decreased in tissue samples from mutant plants, including megagametophytes, elongating shoot tips, and differentiating xylem (Fig. 5a). Northern blots were probed with a full-length CAD cdna probe and a pine ribosomal protein cdna as an internal standard. The relative abundance of steady-state cad RNA was estimated in total RNA from elongating shoot tips, using 3, 15, and 40 g from each of several plants (Fig. 5b). It was, on average, 22 times higher in wild-type than in mutant plants, based upon hybridization intensities normalized against the hybridization to the ribosomal probe (Fig. 5c). RNase protection assays with xylem total RNA also showed that the cad message was at least 20 times more abundant in the wild types (not shown). The relative abundance of cad RNA message indicated that the cad-n1 allele is expressed at a very low level, sufficient to account for most of the decrease in CAD enzyme activity. CAD Substrate Accumulation in cad-n1 Plants. The total phenolics in ethanol and methanol extracts from finely ground wood were increased 2- to 3-fold in the cad-n1 homozygotes compared with wild-type plants. Extraction with ethanol (Table 1) and successive extractions with methanol (not shown) each gave increases in the relative abundance of phenolics, of similar magnitude, but methanol extracts contained lower concentrations of phenolics. Coniferaldehyde (1b; see Fig. 1), the substrate of CAD, and vanillin were the most abundant extractives, and both increased by at least one order of magnitude in the mutant. Ferulic acid and p-hydroxybenzaldehyde (1a) also increased significantly but were not as abundant. Under alkaline ph, coniferaldehyde reacts to give vanillin and acetaldehyde through a reversed aldol reaction (31). Alkaline hydrolysis of the ethanol extracts increased the ratio of vanillin to coniferaldehyde from 1:1 to 2:1; therefore at least part of the vanillin was derived from accumulated CAD substrate. Decreased Total Lignin Content in Homozygous cad-n1 Plants. Total lignin content estimated from Klason and acidsoluble lignin was decreased from 31.7% 0.8% of dry weight in the wild type to 28.9% 0.5% in the cad-n1 homozygotes (Table 2). In Klason determinations, CWR is treated with sulfuric acid and the residual acid-insoluble material represents most of the lignin (24). However, a small but variable fraction of the lignin is soluble in the acid. In samples from mutant plants, the acid-insoluble lignin was decreased to 91% of wild type, and the acid-soluble lignin remained unchanged. We confirmed the small decrease in lignin content by using the AcBr method, in which lignin is determined from the absorbance (A 280 ) of the CWR solubilized in AcBr (26). A saponification pretreatment was needed to completely solubilize the CWR from cad-n1 homozygotes and was used with Table 1. cad genotype Analysis of major ethanol-soluble phenolic extractives Total phenolics* Compound in extract, mg g dry weight Vanillin Coniferaldehyde p-hydroxybenzaldehyde Ferulic acid Cad Cad Cad cad-n ND ND ND ND cad-n1 cad-n Fold increase *Data are mg of ferulic acid equivalents per g dry weight; mean SEM from six plants. HPLC determination in extracts pooled from four seedlings of each genotype; results are from one typical HPLC analysis. ND, not determined. Fold increase: cad-n1 cad-n1 value divided by the Cad Cad value.
5 Plant Biology: MacKay et al. Proc. Natl. Acad. Sci. USA 94 (1997) 8259 Table 2. Effect of the cad-n1 allele on lignin content (% dry weight of CWR) Klason method AcBr method* cad genotype Acid-insoluble Acid-soluble Total AcBr AF Total Cad Cad Cad cad-n ND ND ND cad-n1 cad-n Relative content 91% 91% 86% *CWR were pre-extracted with 1 M NaOH (AF); phenolic content was determined in the AF and lignin content was determined in the AcBr soluble fraction (AcBr). ND, not determined. Relative content: relative lignin content of cad-n1 homozygotes compared to Cad homozygotes. all genotypes. The phenolics in the alkaline fraction (AF) from the pretreatment of cad-n1 homozygotes accounted for 8% (by weight) of the CWR as opposed to only 0.9% in the wild-type plants (Table 2). The total lignin of the mutant, the sum of the AF and the AcBr-solubilized lignin, was 86% of wild type, consistent with a small decrease in the cad-n1 mutant. Characterization of the Lignin: Evidence of Changes in Lignin Composition. FTIR difference spectra of MWL and of ground wood (CWR) showed an increase in functional groups consistent with an enrichment of coniferaldehyde (1b) subunits in the mutant lignin (Fig. 6, Table 3). MWL is most representative of lignin in plant tissues (22), and FTIR spectra of MWL have been extensively characterized (32). Changes in absorption at several wavelengths in the cm 1 range provide strong evidence that the MWL from the cad-n1 tree is enriched in carbonyl groups conjugated to aromatic rings, as expected for an increase in coniferaldehyde. The spectra of CWR from mutant plants gave similar results (Fig. 6), consistent with spectra from xylem strips of antisense cad tobacco (33). Conclusions from the MWL sample from the mutant were thus extended to the samples containing the entire cell wall lignin from several plants. FIG. 6. FTIR difference spectra for MWLs and CWRs. Difference spectra were obtained by subtracting the absorption spectrum of wild type from the spectrum of the mutant: peaks (above the origin) represent stronger absorption in the mutant, troughs are stronger in the wild type. Assignments of absorption bands (10, 32) are 1, cm 1 : carbonyl group (CAO) conjugated to aromatic ring; 2, 1595 cm 1 : aromatic ring with CAO stretching; 3, 1510 cm 1 : aromatic ring with COO stretching; 4, 1270 cm 1 :COO stretching, aromatic (methoxyl); 5, 1040 cm 1 : dialkyl ether linkages (between cinnamyl alcohol subunits). CWR spectrum is the average from four pairwise comparisons of cad-n1 and wild-type plants. FTIR absorption ratios A 1665 A 1510 and A 1595 A 1510 are diagnostic for cinnamaldehyde content (29, 33) and were indicative of their increase. MWL of the mutant had a 2- to 3-fold increase in these ratios and CWR of mutants had a smaller increase (Table 3). Dehydrogenation lignin polymers (DHPs) can be synthesized in vitro from cinnamyl alcohols (2a, 2b), and addition of cinnamaldehyde (1a, b) to the synthesis can increase the A 1665 A 1510 and A 1595 A 1510 ratios of such DHPs (29). A synthesis reaction mixture containing 50% cinnamaldehyde was required to obtain an increase of magnitude similar to that observed with the MWL of cad-n1 plants. Finally, using NaBH 4 reduction (34), we estimated that MWL of the cad-n1 mutant contained at least 25% aldehyde as opposed to 10% in the wild type. DISCUSSION We have characterized a mutant allele (cad-n1) ofthecad gene, discovered in a heterozygous loblolly pine tree through segregation analysis. Genetic mapping of the low-activity CAD allozyme phenotype to the same genomic region as the cad locus is consistent with a single genetic lesion specifically affecting CAD. The phenotype of the vascular tissue is similar to that of bm mutants in maize and related grasses (2, 4, 7, 8). Several bm mutants have been linked to changes in the lignin biosynthetic enzymes COMT (35), CAD (36), or both (14). A mutation in the enzyme ferulate 5-hydroxylase in Arabidopsis thaliana affects lignin composition by reducing the formation of sinapyl alcohol but does not result in a bm phenotype (37). Recent reports have described the modification of lignin composition using antisense cad genes (5, 14, 30, 33). No decrease in lignin content was detected, even when the residual CAD activity was as low as 7% of wild type (5). The cad-n1 allele has a small effect on lignin content and has lower residual CAD activity than antisense plants and bm mutants described so far. The cad-n1 allele was associated with very low steady-state cad RNA transcript and accumulation of CAD substrate to high levels in xylem. Thus, this allele causes a stronger block in CAD enzyme activity than in previously characterized plants (5, 14, 30). Reduction or suppression of CAD activity in these diverse plant species has either no detectable effect or a small effect on lignin content, indicating that CAD is normally not rate limiting and has a minor role in regulating metabolic flux through the lignin biosynthetic pathway. However, some minimal level of CAD enzyme is needed to achieve wild-type lignin content, suggesting that a small amount of CAD activity is required for normal flux. In contrast, reduction of phenylal- Table 3. Diagnostic FTIR absorption ratios of MWL and CWR A 1665 A 1510 A 1595 A 1510 Sample Cad cad-n1 Cad cad-n1 MWL CWR Materials in this table were Cad and cad-n1 homozygotes.
6 8260 Plant Biology: MacKay et al. Proc. Natl. Acad. Sci. USA 94 (1997) anine ammonia-lyase activity had a more dramatic effect on regulation of flux into this pathway and on lignin content (38). Mechanisms for the incorporation of cinnamaldehydes directly into the lignin polymer could account for such a small decrease in lignin resulting from decreased CAD activity. Cinnamaldehyde residues normally account for 3 5% of lignin subunits (10, 31), but cell walls of some bm mutants, the pine cad-n1 mutant, and some transgenic antisense cad plants are enriched in such cinnamaldehyde residues (5, 14, 30, 33). The isolated lignin of transgenic poplar (30) and of the pine cad-n1 mutant (ref. 39; this report) provide conclusive evidence of the incorporation of aromatic aldehyde subunits into the lignin polymer at significantly higher levels. Cinnamaldehydes are also efficiently incorporated into in vitro synthesized phenolic dehydrogenation polymers (DHPs), which are dark red (29). In this context, the accumulation of free coniferaldehyde to much higher levels in the cad-n1 mutant is paradoxical; however, incorporation of coniferaldehyde is usually minor in vivo and may become more significant only when its concentration is elevated. The efficiency of incorporation of coniferaldehyde into lignin may be sufficient to maintain flux through the pathway but not enough to reduce the high concentrations of free aldehyde. The extent to which cinnamaldehyde subunits may be increased in lignin and the enzymatic activities involved in their incorporation are unknown. Alternative routes or mechanisms for the formation of lignin precursors from coniferaldehyde could be utilized or upregulated in the cad-n1 mutant. It was recently shown by NMR that this mutant incorporates a major proportion of dihydroconiferyl alcohol as a novel lignin precursor (39). Thus, alternative reduction of coniferaldehyde is likely to be carried out by enzymes other than CAD in the formation of modified lignin. In view of the viability and lignin composition of this mutant, what is the role of CAD? We suggest that CAD may modulate the ratio of cinnamaldehydes to cinnamyl alcohols in lignin. The proportion of aldehyde and alcohol lignin precursors could affect lignin structure (33), properties (30), and ultimately lignin function. Under controlled conditions, cad-n1 mutants did not have aberrant development or growth, suggesting that vascular and mechanical support functions were not significantly impaired. However, it remains to be determined how these functions may be affected under conditions of stress, such as drought. Analysis of the biosynthesis, structure, and functional properties of genetically altered lignin should continue to provide new insights into the structure function relationship of lignin and its evolution in higher plants. We thank K. Hill, whose work suggested a null CAD allozyme in clone 7-56, and B. Crane, C.-L. Chen, H.-M. Chang, J. Ralph, W. Vermerris, and I. Allona for helpful advice or assistance. The 12-yearold trees were provided by L. Pearson (Westvaco Corporation) and G. Askew (Clemson University). This work was supported by the North Carolina State University Forest Biotechnology Industrial Associates, Grant GN from the National Institutes of Health, and Grant DE-FG592ER20085 from the Department of Energy. 1. Whetten, R. W. & Sederoff, R. R. (1995) Plant Cell 7, Boudet, A. M., Lapierre, C. & Grima-Pettenati, J. (1995) New Phytol. 129, Freudenberg, K. (1959) Nature (London) 183, Campbell, M. M. & Sederoff, R. R. (1995) Plant Physiol. 110, Halpin, C., Knight, M. E., Foxon, G. A., Campbell, M. M., Boudet, A. M., Boon, J. J., Chabbert, B., Tollier, M.-T. & Schuch, W. (1994) Plant J. 6, Sarkanen, K. V. & Hergert, H. L. (1971) in Lignins, Occurrence, Formation, Structure and Reactions, eds. Sarkanen, K. V. & Ludwig, C. H. (Wiley Interscience, New York), pp Kuc, J. & Nelson, O. E. (1964) Arch. Biochem. Biophys. 105, Muller, L. D., Barnes, R. F., Bauman, L. F. & Colenbrander, V. F. (1971) Crop Sci 11, MacKay, J. J., Liu, W., Whetten, R. W., Sederoff, R. R. & O Malley, D. M. (1995) Mol. Gen. Genet. 247, Browning B. L. (1967) Methods of Wood Chemistry (Inst. Paper Chem., Appleton, WI). 11. O Malley, D., Porter, S. & Sederoff, R. R. (1992) Plant Physiol. 98, Gabriel, O. & Gersten D. M. (1992) Anal. Biochem. 293, Campbell, M. M. & Ellis, B. E. (1992) Planta 486, Pillonel, C., Mulder, M. M., Boon, J. J., Forster, B. & Binder, A. (1991) Planta 185, Doyle, J. J. & Doyle, J. L. (1987) Focus 12, Plomion, C., Liu, B.-H. & O Malley, D. (1996) Theor. Appl. Genet. 93, Grattapaglia, D. & Sederoff, R. (1994) Genetics 137, Michelmore, R. W., Paran, I. & Kesseli, R. V. (1991) Proc. Natl. Acad. Sci. USA 88, Chang, S., Puryear, J. & Cairney, J. (1993) Plant Mol. Biol. Rep. 11, Brown, T. & MacKey, K. (1997) in Current Protocols in Molecular Biology, eds. Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Siedman, J. G., Smith, J. A. & Struchl, K. (Wiley, New York), pp Booker, F. L., Anttonen, S. & Heagle, A. S. (1996) New Phytol. 132, Lundquist, K. (1992) in Methods in Lignin Chemistry, eds. Lin, S. E. & Dence, C. W. (Springer, Berlin), pp Björkman, A. (1954) Nature (London) 174, Effland, M. J. (1977) Tappi 60, Dence, C. W. (1992) in Methods in Lignin Chemistry, eds. Lin, S. E. & Dence, C. W. (Springer, Berlin), pp Iiyama, K. & Wallis, A. F. A. (1988) Wood Sci. Technol. 22, Gifford, M. & Foster, A. S. (1989) Morphology and Evolution of Vascular Plants (Freeman, New York), 3rd Ed. 28. O Malley, D. M., Grattapaglia, D., Chaparro, J. X., Wilcox P. L., Amerson, H. V., Liu, B.-H., Whetten, R., McKeand, S. E., Kuhlman, E. G., McCord, S., Crane, B. & Sederoff, R. R. (1995) in Genomes of Plants and Animals: 21st Stadler Genetics Symposium, eds. Gustafson, J. P. & Flavell, R. B. (Plenum, New York), pp Higuchi, T., Ito T., Umezawa, T., Hibino, T. & Shibata, D. (1994) J. Biotechnol. 37, Baucher, M., Chabbert, B., Pilate, G., van Doorsselaere, J., Tollier, M.-T., Petit-Conil, M., Cornu, D., Monties, B., van Montagu, M., Inzé, D., Jouanin, L. & Boerjan, W. (1996) Plant Physiol. 112, Hrutfiord, B. F. (1971) in Lignins, Occurrence, Formation, Structure and Reactions, eds. Sarkanen, K. V. & Ludwig, C. H. (Wiley Interscience, New York), pp Faix, O. (1992) in Methods in Lignin Chemistry, eds. Lin, S. E. & Dence, C. W. (Springer, Berlin), pp Stewart, D., Yahiaoui, N., McDougall, G. J., Myton, K., Marque, C., Boudet, A. M. & Haigh, J. (1997) Planta 201, Chen, C.-L. (1992) in Methods in Lignin Chemistry, eds. Lin, S. E. & Dence, C. W. (Springer, Berlin), pp Vignols, F., Rigau, J., Torres, M. A., Capellades, M. & Puigdomènech, P. (1995) Plant Cell 7, Halpin, C., Foxon, G. A. & Fentem, P. A. (1995) in Biotechnology in Animal Feeds and Animal Feeding, eds. Wallace, R. J. & Chesson, A. (VCH, New York) pp Meyer, K., Cusumano, J. C., Somerville, C. & Chapple, C. C. S. (1996) Proc. Natl. Acad. Sci. USA 93, Bate, N. J., Orr, J., Ni, W., Meromi, A., Nadler-Hassar, T., Doerner, P. W., Dixon, R. A., Lamb, C. J. & Elkind, Y. (1994) Proc. Natl. Acad. Sci. USA 91, Ralph, J., MacKay, J. J., Hatfield, R. D., O Malley, D. M., Whetten, R. W. & Sederoff, R. R. (1997) Science, in press.
Understanding Gene Function and Control in Lignin Formation In Wood
 Understanding Gene Function and Control in Lignin Formation In Wood Vincent L. Chiang North Carolina State University Raleigh, NC Tremendous effort has been devoted to developing genetically engineered
Understanding Gene Function and Control in Lignin Formation In Wood Vincent L. Chiang North Carolina State University Raleigh, NC Tremendous effort has been devoted to developing genetically engineered
Population Genetics. If we closely examine the individuals of a population, there is almost always PHENOTYPIC
 1 Population Genetics How Much Genetic Variation exists in Natural Populations? Phenotypic Variation If we closely examine the individuals of a population, there is almost always PHENOTYPIC VARIATION -
1 Population Genetics How Much Genetic Variation exists in Natural Populations? Phenotypic Variation If we closely examine the individuals of a population, there is almost always PHENOTYPIC VARIATION -
Mapping and Mapping Populations
 Mapping and Mapping Populations Types of mapping populations F 2 o Two F 1 individuals are intermated Backcross o Cross of a recurrent parent to a F 1 Recombinant Inbred Lines (RILs; F 2 -derived lines)
Mapping and Mapping Populations Types of mapping populations F 2 o Two F 1 individuals are intermated Backcross o Cross of a recurrent parent to a F 1 Recombinant Inbred Lines (RILs; F 2 -derived lines)
Assembly of a lignin modification toolbox
 Assembly of a lignin modification toolbox Investigators Clint Chapple, Distinguished Professor and Head, Department of Biochemistry; Alan Friedman, Associate Professor, Department of Biological Sciences;
Assembly of a lignin modification toolbox Investigators Clint Chapple, Distinguished Professor and Head, Department of Biochemistry; Alan Friedman, Associate Professor, Department of Biological Sciences;
Enzyme that uses RNA as a template to synthesize a complementary DNA
 Biology 105: Introduction to Genetics PRACTICE FINAL EXAM 2006 Part I: Definitions Homology: Comparison of two or more protein or DNA sequence to ascertain similarities in sequences. If two genes have
Biology 105: Introduction to Genetics PRACTICE FINAL EXAM 2006 Part I: Definitions Homology: Comparison of two or more protein or DNA sequence to ascertain similarities in sequences. If two genes have
Molecular Cell Biology - Problem Drill 11: Recombinant DNA
 Molecular Cell Biology - Problem Drill 11: Recombinant DNA Question No. 1 of 10 1. Which of the following statements about the sources of DNA used for molecular cloning is correct? Question #1 (A) cdna
Molecular Cell Biology - Problem Drill 11: Recombinant DNA Question No. 1 of 10 1. Which of the following statements about the sources of DNA used for molecular cloning is correct? Question #1 (A) cdna
Efficient Method for Isolation of High Quality Concentrated Cellular RNA with Extremely Low Levels of Genomic DNA Contamination Application
 Efficient Method for Isolation of High Quality Concentrated Cellular RNA with Extremely Low Levels of Genomic DNA Contamination Application Gene Expression Authors Ilgar Abbaszade, Claudia Robbins, John
Efficient Method for Isolation of High Quality Concentrated Cellular RNA with Extremely Low Levels of Genomic DNA Contamination Application Gene Expression Authors Ilgar Abbaszade, Claudia Robbins, John
Biology 105: Introduction to Genetics PRACTICE FINAL EXAM Part I: Definitions. Homology: Reverse transcriptase. Allostery: cdna library
 Biology 105: Introduction to Genetics PRACTICE FINAL EXAM 2006 Part I: Definitions Homology: Reverse transcriptase Allostery: cdna library Transformation Part II Short Answer 1. Describe the reasons for
Biology 105: Introduction to Genetics PRACTICE FINAL EXAM 2006 Part I: Definitions Homology: Reverse transcriptase Allostery: cdna library Transformation Part II Short Answer 1. Describe the reasons for
Applicazioni biotecnologiche
 Applicazioni biotecnologiche Analisi forense Sintesi di proteine ricombinanti Restriction Fragment Length Polymorphism (RFLP) Polymorphism (more fully genetic polymorphism) refers to the simultaneous occurrence
Applicazioni biotecnologiche Analisi forense Sintesi di proteine ricombinanti Restriction Fragment Length Polymorphism (RFLP) Polymorphism (more fully genetic polymorphism) refers to the simultaneous occurrence
Eucalyptus pulping efficiency
 Biotech enhanced levels of syringyl lignin improves Eucalyptus pulping efficiency Maud Hinchee, Ph.D. : Chief Science Officer, ArborGen, USA, mahinch@arborgen.com Ana Gabriela M. C. Bassa,MSc : R&D Manager,
Biotech enhanced levels of syringyl lignin improves Eucalyptus pulping efficiency Maud Hinchee, Ph.D. : Chief Science Officer, ArborGen, USA, mahinch@arborgen.com Ana Gabriela M. C. Bassa,MSc : R&D Manager,
Conifer Translational Genomics Network Coordinated Agricultural Project
 Conifer Translational Genomics Network Coordinated Agricultural Project Genomics in Tree Breeding and Forest Ecosystem Management ----- Module 2 Genes, Genomes, and Mendel Nicholas Wheeler & David Harry
Conifer Translational Genomics Network Coordinated Agricultural Project Genomics in Tree Breeding and Forest Ecosystem Management ----- Module 2 Genes, Genomes, and Mendel Nicholas Wheeler & David Harry
Enhancers mutations that make the original mutant phenotype more extreme. Suppressors mutations that make the original mutant phenotype less extreme
 Interactomics and Proteomics 1. Interactomics The field of interactomics is concerned with interactions between genes or proteins. They can be genetic interactions, in which two genes are involved in the
Interactomics and Proteomics 1. Interactomics The field of interactomics is concerned with interactions between genes or proteins. They can be genetic interactions, in which two genes are involved in the
Erhard et al. (2013). Plant Cell /tpc
 Supplemental Figure 1. c1-hbr allele structure. Diagram of the c1-hbr allele found in stocks segregating 1:1 for rpd1-1 and rpd1-2 homozygous mutants showing the presence of a 363 base pair (bp) Heartbreaker
Supplemental Figure 1. c1-hbr allele structure. Diagram of the c1-hbr allele found in stocks segregating 1:1 for rpd1-1 and rpd1-2 homozygous mutants showing the presence of a 363 base pair (bp) Heartbreaker
Biology Evolution Dr. Kilburn, page 1 Mutation and genetic variation
 Biology 203 - Evolution Dr. Kilburn, page 1 In this unit, we will look at the mechanisms of evolution, largely at the population scale. Our primary focus will be on natural selection, but we will also
Biology 203 - Evolution Dr. Kilburn, page 1 In this unit, we will look at the mechanisms of evolution, largely at the population scale. Our primary focus will be on natural selection, but we will also
Supplementary Information. The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato
 Supplementary Information The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato Uri Krieger 1, Zachary B. Lippman 2 *, and Dani Zamir 1 * 1. The Hebrew University of Jerusalem Faculty
Supplementary Information The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato Uri Krieger 1, Zachary B. Lippman 2 *, and Dani Zamir 1 * 1. The Hebrew University of Jerusalem Faculty
Solutions to 7.02 Quiz II 10/27/05
 Solutions to 7.02 Quiz II 10/27/05 Class Average = 83 Standard Deviation = 9 Range Grade % 87-100 A 43 74-86 B 39 55-73 C 17 > 54 D 1 Question 1 (56 points) While studying deep sea bacteria, you discover
Solutions to 7.02 Quiz II 10/27/05 Class Average = 83 Standard Deviation = 9 Range Grade % 87-100 A 43 74-86 B 39 55-73 C 17 > 54 D 1 Question 1 (56 points) While studying deep sea bacteria, you discover
Lecture Four. Molecular Approaches I: Nucleic Acids
 Lecture Four. Molecular Approaches I: Nucleic Acids I. Recombinant DNA and Gene Cloning Recombinant DNA is DNA that has been created artificially. DNA from two or more sources is incorporated into a single
Lecture Four. Molecular Approaches I: Nucleic Acids I. Recombinant DNA and Gene Cloning Recombinant DNA is DNA that has been created artificially. DNA from two or more sources is incorporated into a single
Using mutants to clone genes
 Using mutants to clone genes Objectives: 1. What is positional cloning? 2. What is insertional tagging? 3. How can one confirm that the gene cloned is the same one that is mutated to give the phenotype
Using mutants to clone genes Objectives: 1. What is positional cloning? 2. What is insertional tagging? 3. How can one confirm that the gene cloned is the same one that is mutated to give the phenotype
1) (15 points) Next to each term in the left-hand column place the number from the right-hand column that best corresponds:
 1) (15 points) Next to each term in the left-hand column place the number from the right-hand column that best corresponds: natural selection 21 1) the component of phenotypic variance not explained by
1) (15 points) Next to each term in the left-hand column place the number from the right-hand column that best corresponds: natural selection 21 1) the component of phenotypic variance not explained by
Simultaneous Suppression of Multiple Genes by Single Transgenes. Down-Regulation of Three Unrelated Lignin Biosynthetic Genes in Tobacco 1
 Simultaneous Suppression of Multiple Genes by Single Transgenes. Down-Regulation of Three Unrelated Lignin Biosynthetic Genes in Tobacco 1 James C. Abbott 2, Abdellah Barakate, Gaelle Pinçon, Michel Legrand,
Simultaneous Suppression of Multiple Genes by Single Transgenes. Down-Regulation of Three Unrelated Lignin Biosynthetic Genes in Tobacco 1 James C. Abbott 2, Abdellah Barakate, Gaelle Pinçon, Michel Legrand,
Midterm 1 Results. Midterm 1 Akey/ Fields Median Number of Students. Exam Score
 Midterm 1 Results 10 Midterm 1 Akey/ Fields Median - 69 8 Number of Students 6 4 2 0 21 26 31 36 41 46 51 56 61 66 71 76 81 86 91 96 101 Exam Score Quick review of where we left off Parental type: the
Midterm 1 Results 10 Midterm 1 Akey/ Fields Median - 69 8 Number of Students 6 4 2 0 21 26 31 36 41 46 51 56 61 66 71 76 81 86 91 96 101 Exam Score Quick review of where we left off Parental type: the
Chapter 5: Proteins: Primary Structure
 Instant download and all chapters Test Bank Fundamentals of Biochemistry Life at the Molecular Level 4th Edition Donald Voet https://testbanklab.com/download/test-bank-fundamentals-biochemistry-life-molecular-level-
Instant download and all chapters Test Bank Fundamentals of Biochemistry Life at the Molecular Level 4th Edition Donald Voet https://testbanklab.com/download/test-bank-fundamentals-biochemistry-life-molecular-level-
Saccharomyces cerevisiae. haploid =
 In this lecture we are going to consider experiments on yeast, a very useful organism for genetic study. Yeast is more properly known as Saccharomyces cerevisiae, which is the single-celled microbe used
In this lecture we are going to consider experiments on yeast, a very useful organism for genetic study. Yeast is more properly known as Saccharomyces cerevisiae, which is the single-celled microbe used
Supplemental Data. Benstein et al. (2013). Plant Cell /tpc
 Supplemental Figure 1. Purification of the heterologously expressed PGDH1, PGDH2 and PGDH3 enzymes by Ni-NTA affinity chromatography. Protein extracts (2 µl) of different fractions (lane 1 = total extract,
Supplemental Figure 1. Purification of the heterologously expressed PGDH1, PGDH2 and PGDH3 enzymes by Ni-NTA affinity chromatography. Protein extracts (2 µl) of different fractions (lane 1 = total extract,
Authors: Vivek Sharma and Ram Kunwar
 Molecular markers types and applications A genetic marker is a gene or known DNA sequence on a chromosome that can be used to identify individuals or species. Why we need Molecular Markers There will be
Molecular markers types and applications A genetic marker is a gene or known DNA sequence on a chromosome that can be used to identify individuals or species. Why we need Molecular Markers There will be
HLA-DR TYPING OF GENOMIC DNA
 HLA-DR TYPING OF GENOMIC DNA Zofia SZCZERKOWSKA, Joanna WYSOCKA Institute of Forensic Medicine, Medical University, Gdañsk, Poland ABSTRACT: Advances in molecular biology techniques allowed for introduction
HLA-DR TYPING OF GENOMIC DNA Zofia SZCZERKOWSKA, Joanna WYSOCKA Institute of Forensic Medicine, Medical University, Gdañsk, Poland ABSTRACT: Advances in molecular biology techniques allowed for introduction
2014 Pearson Education, Inc. CH 8: Recombinant DNA Technology
 CH 8: Recombinant DNA Technology Biotechnology the use of microorganisms to make practical products Recombinant DNA = DNA from 2 different sources What is Recombinant DNA Technology? modifying genomes
CH 8: Recombinant DNA Technology Biotechnology the use of microorganisms to make practical products Recombinant DNA = DNA from 2 different sources What is Recombinant DNA Technology? modifying genomes
SUBUNIT INTERACTION OF A TEMPERATURE-SENSITIVE ALCOHOL DEHYDROGENASE MUTANT IN MAIZE1
 SUBUNIT INTERACTION OF A TEMPERATURE-SENSITIVE ALCOHOL DEHYDROGENASE MUTANT IN MAIZE1 DREW SCHWARTZ Department of Botany, Indiana Uniuersity, Bloomington, Indiana 47401 Received November 11, 1970 EMPERATURE-sensitive
SUBUNIT INTERACTION OF A TEMPERATURE-SENSITIVE ALCOHOL DEHYDROGENASE MUTANT IN MAIZE1 DREW SCHWARTZ Department of Botany, Indiana Uniuersity, Bloomington, Indiana 47401 Received November 11, 1970 EMPERATURE-sensitive
Before starting, write your name on the top of each page Make sure you have all pages
 Biology 105: Introduction to Genetics Name Student ID Before starting, write your name on the top of each page Make sure you have all pages You can use the back-side of the pages for scratch, but we will
Biology 105: Introduction to Genetics Name Student ID Before starting, write your name on the top of each page Make sure you have all pages You can use the back-side of the pages for scratch, but we will
Week 7 - Natural Selection and Genetic Variation for Allozymes
 Week 7 - Natural Selection and Genetic Variation for Allozymes Introduction In today's laboratory exercise, we will explore the potential for natural selection to cause evolutionary change, and we will
Week 7 - Natural Selection and Genetic Variation for Allozymes Introduction In today's laboratory exercise, we will explore the potential for natural selection to cause evolutionary change, and we will
Supporting Information
 Supporting Information Yuan et al. 10.1073/pnas.0906869106 Fig. S1. Heat map showing that Populus ICS is coregulated with orthologs of Arabidopsis genes involved in PhQ biosynthesis and PSI function, but
Supporting Information Yuan et al. 10.1073/pnas.0906869106 Fig. S1. Heat map showing that Populus ICS is coregulated with orthologs of Arabidopsis genes involved in PhQ biosynthesis and PSI function, but
1. (a) Define sex linkage... State one example of sex linkage... Key. 1st generation. Male. Female
 1. Define sex linkage. State one example of sex linkage. Draw a simple pedigree chart that clearly shows sex linkage in humans. Use conventional symbols. Start with an affected woman and an unaffected
1. Define sex linkage. State one example of sex linkage. Draw a simple pedigree chart that clearly shows sex linkage in humans. Use conventional symbols. Start with an affected woman and an unaffected
I (ALLEN 1965; GIBSON 1970). In Tetrahymena pyriformis, syngen 1, a genetically
 SHORT PAPERS MUTANT SELECTION IN TETRAHYMENA PYRIFORMIS PETER S. CARLSON' Department of Biology, Wesleyan University, Middletown, Connecticut 06457 Received June 18, 1971 NTRACLONAL variation is a well
SHORT PAPERS MUTANT SELECTION IN TETRAHYMENA PYRIFORMIS PETER S. CARLSON' Department of Biology, Wesleyan University, Middletown, Connecticut 06457 Received June 18, 1971 NTRACLONAL variation is a well
CH 8: Recombinant DNA Technology
 CH 8: Recombinant DNA Technology Biotechnology the use of microorganisms to make practical products Recombinant DNA = DNA from 2 different sources What is Recombinant DNA Technology? modifying genomes
CH 8: Recombinant DNA Technology Biotechnology the use of microorganisms to make practical products Recombinant DNA = DNA from 2 different sources What is Recombinant DNA Technology? modifying genomes
Gene expression in the lignin biosynthesis pathway during soybean seed development
 Gene expression in the lignin biosynthesis pathway during soybean seed development A. Baldoni, E.V.R. Von Pinho, J.S. Fernandes, V.M. Abreu and M.L.M. Carvalho Laboratório Central de Sementes, Departamento
Gene expression in the lignin biosynthesis pathway during soybean seed development A. Baldoni, E.V.R. Von Pinho, J.S. Fernandes, V.M. Abreu and M.L.M. Carvalho Laboratório Central de Sementes, Departamento
A) (5 points) As the starting step isolate genomic DNA from
 GS Final Exam Spring 00 NAME. bub ts is a recessive temperature sensitive mutation in yeast. At º C bub ts cells grow normally, but at º C they die. Use the information below to clone the wild-type BUB
GS Final Exam Spring 00 NAME. bub ts is a recessive temperature sensitive mutation in yeast. At º C bub ts cells grow normally, but at º C they die. Use the information below to clone the wild-type BUB
1. Introduction Drought stress and climate change Three strategies of plants in response to water stress 3
 Contents 1. Introduction 1 1.1 Drought stress and climate change 3 1.2 Three strategies of plants in response to water stress 3 1.3 Three closely related species of Linderniaceae family are experimental
Contents 1. Introduction 1 1.1 Drought stress and climate change 3 1.2 Three strategies of plants in response to water stress 3 1.3 Three closely related species of Linderniaceae family are experimental
TriPure Isolation Reagent Clear, red solution; ready-to-use Cat. No (50 ml) Cat. No (250 ml)
 Clear, red solution; ready-to-use Cat. No. 1 667 157 (50 ml) Cat. No. 1 667 165 (250 ml) Principle Starting material Application Time required Results Key advantages During a one-step sample homogenization/lysis
Clear, red solution; ready-to-use Cat. No. 1 667 157 (50 ml) Cat. No. 1 667 165 (250 ml) Principle Starting material Application Time required Results Key advantages During a one-step sample homogenization/lysis
Test Bank for Molecular Cell Biology 7th Edition by Lodish
 Test Bank for Molecular Cell Biology 7th Edition by Lodish Link download full: http://testbankair.com/download/test-bank-formolecular-cell-biology-7th-edition-by-lodish/ Chapter 5 Molecular Genetic Techniques
Test Bank for Molecular Cell Biology 7th Edition by Lodish Link download full: http://testbankair.com/download/test-bank-formolecular-cell-biology-7th-edition-by-lodish/ Chapter 5 Molecular Genetic Techniques
Concepts: What are RFLPs and how do they act like genetic marker loci?
 Restriction Fragment Length Polymorphisms (RFLPs) -1 Readings: Griffiths et al: 7th Edition: Ch. 12 pp. 384-386; Ch.13 pp404-407 8th Edition: pp. 364-366 Assigned Problems: 8th Ch. 11: 32, 34, 38-39 7th
Restriction Fragment Length Polymorphisms (RFLPs) -1 Readings: Griffiths et al: 7th Edition: Ch. 12 pp. 384-386; Ch.13 pp404-407 8th Edition: pp. 364-366 Assigned Problems: 8th Ch. 11: 32, 34, 38-39 7th
ksierzputowska.com Research Title: Using novel TALEN technology to engineer precise mutations in the genome of D. melanogaster
 Research Title: Using novel TALEN technology to engineer precise mutations in the genome of D. melanogaster Research plan: Specific aims: 1. To successfully engineer transgenic Drosophila expressing TALENs
Research Title: Using novel TALEN technology to engineer precise mutations in the genome of D. melanogaster Research plan: Specific aims: 1. To successfully engineer transgenic Drosophila expressing TALENs
9. What proteins will be affected by mutations in the trans-acting elements? Cis-acting elements?
 6. What regulates the expression of a gene? 7. What are the cis- and trans-acting elements? 8. Can a deficiency in a trans-acting element be overcome by the addition of another copy of the gene to a cell?
6. What regulates the expression of a gene? 7. What are the cis- and trans-acting elements? 8. Can a deficiency in a trans-acting element be overcome by the addition of another copy of the gene to a cell?
Concepts of Genetics, 10e (Klug/Cummings/Spencer/Palladino) Chapter 1 Introduction to Genetics
 1 Concepts of Genetics, 10e (Klug/Cummings/Spencer/Palladino) Chapter 1 Introduction to Genetics 1) What is the name of the company or institution that has access to the health, genealogical, and genetic
1 Concepts of Genetics, 10e (Klug/Cummings/Spencer/Palladino) Chapter 1 Introduction to Genetics 1) What is the name of the company or institution that has access to the health, genealogical, and genetic
Identification of a RAPD marker linked to a male fertility restoration gene in cotton
 TITLE Identification of a RAPD marker linked to a male fertility restoration gene in cotton (Gossypium hirsutum L.) Tien-Hung Lan 1, Charles G. Cook 2 & Andrew H. Paterson 1, * 1 Department of Soil and
TITLE Identification of a RAPD marker linked to a male fertility restoration gene in cotton (Gossypium hirsutum L.) Tien-Hung Lan 1, Charles G. Cook 2 & Andrew H. Paterson 1, * 1 Department of Soil and
Recombinant DNA Technology. The Role of Recombinant DNA Technology in Biotechnology. yeast. Biotechnology. Recombinant DNA technology.
 PowerPoint Lecture Presentations prepared by Mindy Miller-Kittrell, North Carolina State University C H A P T E R 8 Recombinant DNA Technology The Role of Recombinant DNA Technology in Biotechnology Biotechnology?
PowerPoint Lecture Presentations prepared by Mindy Miller-Kittrell, North Carolina State University C H A P T E R 8 Recombinant DNA Technology The Role of Recombinant DNA Technology in Biotechnology Biotechnology?
WesternMAX Alkaline Phosphatase Chemiluminescent Detection Kits
 WesternMAX Alkaline Phosphatase Chemiluminescent Detection Kits Code N221-KIT N220-KIT Description WesternMAX Chemiluminescent AP Kit, Anti-Mouse Includes: Alkaline Phosphatase (AP) Conjugated Anti-Mouse
WesternMAX Alkaline Phosphatase Chemiluminescent Detection Kits Code N221-KIT N220-KIT Description WesternMAX Chemiluminescent AP Kit, Anti-Mouse Includes: Alkaline Phosphatase (AP) Conjugated Anti-Mouse
High Pure RNA Isolation Kit for isolation of total RNA from 50 samples Cat. No
 for isolation of total RNA from 50 samples Cat. No. 1 88 665 Principle A single reagent lyses the sample lysis and inactivates RNase. In the presence of a chaotropic salt (guanidine HCl), the released
for isolation of total RNA from 50 samples Cat. No. 1 88 665 Principle A single reagent lyses the sample lysis and inactivates RNase. In the presence of a chaotropic salt (guanidine HCl), the released
Design. Construction. Characterization
 Design Construction Characterization DNA mrna (messenger) A C C transcription translation C A C protein His A T G C T A C G Plasmids replicon copy number incompatibility selection marker origin of replication
Design Construction Characterization DNA mrna (messenger) A C C transcription translation C A C protein His A T G C T A C G Plasmids replicon copy number incompatibility selection marker origin of replication
MUTANT: A mutant is a strain that has suffered a mutation and exhibits a different phenotype from the parental strain.
 OUTLINE OF GENETICS LECTURE #1 A. TERMS PHENOTYPE: Phenotype refers to the observable properties of an organism, such as morphology, growth rate, ability to grow under different conditions or media. For
OUTLINE OF GENETICS LECTURE #1 A. TERMS PHENOTYPE: Phenotype refers to the observable properties of an organism, such as morphology, growth rate, ability to grow under different conditions or media. For
Purification and characterization of cinnamyl alcohol-nadph-dehydrogenase from the leaf tissues of a basin mangrove Lumnitzera racemosa Willd.
 Indian Journal of Biochemistry & Biophysics Vol. 41, April-June 2004, pp. 96-101 Purification and characterization of cinnamyl alcohol-nadph-dehydrogenase from the leaf tissues of a basin mangrove Lumnitzera
Indian Journal of Biochemistry & Biophysics Vol. 41, April-June 2004, pp. 96-101 Purification and characterization of cinnamyl alcohol-nadph-dehydrogenase from the leaf tissues of a basin mangrove Lumnitzera
SCREENING AND PRESERVATION OF DNA LIBRARIES
 MODULE 4 LECTURE 5 SCREENING AND PRESERVATION OF DNA LIBRARIES 4-5.1. Introduction Library screening is the process of identification of the clones carrying the gene of interest. Screening relies on a
MODULE 4 LECTURE 5 SCREENING AND PRESERVATION OF DNA LIBRARIES 4-5.1. Introduction Library screening is the process of identification of the clones carrying the gene of interest. Screening relies on a
Introduction to some aspects of molecular genetics
 Introduction to some aspects of molecular genetics Julius van der Werf (partly based on notes from Margaret Katz) University of New England, Armidale, Australia Genetic and Physical maps of the genome...
Introduction to some aspects of molecular genetics Julius van der Werf (partly based on notes from Margaret Katz) University of New England, Armidale, Australia Genetic and Physical maps of the genome...
Phenotype analysis: biological-biochemical analysis. Genotype analysis: molecular and physical analysis
 1 Genetic Analysis Phenotype analysis: biological-biochemical analysis Behaviour under specific environmental conditions Behaviour of specific genetic configurations Behaviour of progeny in crosses - Genotype
1 Genetic Analysis Phenotype analysis: biological-biochemical analysis Behaviour under specific environmental conditions Behaviour of specific genetic configurations Behaviour of progeny in crosses - Genotype
Assembly of a lignin modification toolbox
 Assembly of a lignin modification toolbox Investigators Clint Chapple, Distinguished Professor and Head, Department of Biochemistry; Alan Friedman, Associate Professor, Department of Biological Sciences;
Assembly of a lignin modification toolbox Investigators Clint Chapple, Distinguished Professor and Head, Department of Biochemistry; Alan Friedman, Associate Professor, Department of Biological Sciences;
ACETYLATION AND THE DEVELOPMENT OF NEW PRODUCTS FROM RADIATA PINE
 ACETYLATION AND THE DEVELOPMENT OF NEW PRODUCTS FROM RADIATA PINE D.V. Plackett, R.M. Rowell*, and E.A. Close, Forest Research Institute (*US Forest Products Laboratory, Madison, Wisconsin) ABSTRACT Wood
ACETYLATION AND THE DEVELOPMENT OF NEW PRODUCTS FROM RADIATA PINE D.V. Plackett, R.M. Rowell*, and E.A. Close, Forest Research Institute (*US Forest Products Laboratory, Madison, Wisconsin) ABSTRACT Wood
7 Gene Isolation and Analysis of Multiple
 Genetic Techniques for Biological Research Corinne A. Michels Copyright q 2002 John Wiley & Sons, Ltd ISBNs: 0-471-89921-6 (Hardback); 0-470-84662-3 (Electronic) 7 Gene Isolation and Analysis of Multiple
Genetic Techniques for Biological Research Corinne A. Michels Copyright q 2002 John Wiley & Sons, Ltd ISBNs: 0-471-89921-6 (Hardback); 0-470-84662-3 (Electronic) 7 Gene Isolation and Analysis of Multiple
Non-Organic-Based Isolation of Mammalian microrna using Norgen s microrna Purification Kit
 Application Note 13 RNA Sample Preparation Non-Organic-Based Isolation of Mammalian microrna using Norgen s microrna Purification Kit B. Lam, PhD 1, P. Roberts, MSc 1 Y. Haj-Ahmad, M.Sc., Ph.D 1,2 1 Norgen
Application Note 13 RNA Sample Preparation Non-Organic-Based Isolation of Mammalian microrna using Norgen s microrna Purification Kit B. Lam, PhD 1, P. Roberts, MSc 1 Y. Haj-Ahmad, M.Sc., Ph.D 1,2 1 Norgen
BIO 304 Fall 2000 Exam II Name: ID #: 1. Fill in the blank with the best answer from the provided word bank. (2 pts each)
 1. Fill in the blank with the best answer from the provided word bank. (2 pts each) incomplete dominance conditional mutation penetrance expressivity pleiotropy Southern blotting hybridization epistasis
1. Fill in the blank with the best answer from the provided word bank. (2 pts each) incomplete dominance conditional mutation penetrance expressivity pleiotropy Southern blotting hybridization epistasis
Lecture 12. Genomics. Mapping. Definition Species sequencing ESTs. Why? Types of mapping Markers p & Types
 Lecture 12 Reading Lecture 12: p. 335-338, 346-353 Lecture 13: p. 358-371 Genomics Definition Species sequencing ESTs Mapping Why? Types of mapping Markers p.335-338 & 346-353 Types 222 omics Interpreting
Lecture 12 Reading Lecture 12: p. 335-338, 346-353 Lecture 13: p. 358-371 Genomics Definition Species sequencing ESTs Mapping Why? Types of mapping Markers p.335-338 & 346-353 Types 222 omics Interpreting
Site directed mutagenesis, Insertional and Deletion Mutagenesis. Mitesh Shrestha
 Site directed mutagenesis, Insertional and Deletion Mutagenesis Mitesh Shrestha Mutagenesis Mutagenesis (the creation or formation of a mutation) can be used as a powerful genetic tool. By inducing mutations
Site directed mutagenesis, Insertional and Deletion Mutagenesis Mitesh Shrestha Mutagenesis Mutagenesis (the creation or formation of a mutation) can be used as a powerful genetic tool. By inducing mutations
THE INSTITUTE OF PAPER CHEMISTRY, APPLETON, WISCONSIN IPC TECHNICAL PAPER SERIES NUMBER 256 PLANT GROWTH: THE ROLE OF POLYAMINES RUSSELL FEIRER
 THE INSTITUTE OF PAPER CHEMISTRY, APPLETON, WISCONSIN IPC TECHNICAL PAPER SERIES NUMBER 256 PLANT GROWTH: THE ROLE OF POLYAMINES RUSSELL FEIRER AUGUST, 1987 Plant Growth: The Role of Polyamines Russell
THE INSTITUTE OF PAPER CHEMISTRY, APPLETON, WISCONSIN IPC TECHNICAL PAPER SERIES NUMBER 256 PLANT GROWTH: THE ROLE OF POLYAMINES RUSSELL FEIRER AUGUST, 1987 Plant Growth: The Role of Polyamines Russell
Lecture 2-3: Using Mutants to study Biological processes
 Lecture 2-3: Using Mutants to study Biological processes Objectives: 1. Why use mutants? 2. How are mutants isolated? 3. What important genetic analyses must be done immediately after a genetic screen
Lecture 2-3: Using Mutants to study Biological processes Objectives: 1. Why use mutants? 2. How are mutants isolated? 3. What important genetic analyses must be done immediately after a genetic screen
Marker types. Potato Association of America Frederiction August 9, Allen Van Deynze
 Marker types Potato Association of America Frederiction August 9, 2009 Allen Van Deynze Use of DNA Markers in Breeding Germplasm Analysis Fingerprinting of germplasm Arrangement of diversity (clustering,
Marker types Potato Association of America Frederiction August 9, 2009 Allen Van Deynze Use of DNA Markers in Breeding Germplasm Analysis Fingerprinting of germplasm Arrangement of diversity (clustering,
Gene Expression Technology
 Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
Molecular studies (SSR) for screening of genetic variability among direct regenerants of sugarcane clone NIA-98
 Molecular studies (R) for screening of genetic variability among direct regenerants of sugarcane clone NIA-98 Dr. Imtiaz A. Khan Pr. cientist / PI sugarcane and molecular marker group NIA-2012 NIA-2010
Molecular studies (R) for screening of genetic variability among direct regenerants of sugarcane clone NIA-98 Dr. Imtiaz A. Khan Pr. cientist / PI sugarcane and molecular marker group NIA-2012 NIA-2010
Optimizing Saccharificationand Yield in Lignin-Modified plants
 Optimizing Saccharificationand Yield in Lignin-Modified plants Stanford, November 2-3, 2016 Wout Boerjan Department of Plant Systems Biology VIB-UGent Technologiepark 927 9052 Gent, Belgium woboe@psb.vib-ugent.be
Optimizing Saccharificationand Yield in Lignin-Modified plants Stanford, November 2-3, 2016 Wout Boerjan Department of Plant Systems Biology VIB-UGent Technologiepark 927 9052 Gent, Belgium woboe@psb.vib-ugent.be
Mathematical Biosciences
 Mathematical Biosciences 228 (2010) 78 89 Contents lists available at ScienceDirect Mathematical Biosciences journal homepage: www.elsevier.com/locate/mbs Mathematical modeling of monolignol biosynthesis
Mathematical Biosciences 228 (2010) 78 89 Contents lists available at ScienceDirect Mathematical Biosciences journal homepage: www.elsevier.com/locate/mbs Mathematical modeling of monolignol biosynthesis
Combining Techniques to Answer Molecular Questions
 Combining Techniques to Answer Molecular Questions UNIT FM02 How to cite this article: Curr. Protoc. Essential Lab. Tech. 9:FM02.1-FM02.5. doi: 10.1002/9780470089941.etfm02s9 INTRODUCTION This manual is
Combining Techniques to Answer Molecular Questions UNIT FM02 How to cite this article: Curr. Protoc. Essential Lab. Tech. 9:FM02.1-FM02.5. doi: 10.1002/9780470089941.etfm02s9 INTRODUCTION This manual is
Map-Based Cloning of Qualitative Plant Genes
 Map-Based Cloning of Qualitative Plant Genes Map-based cloning using the genetic relationship between a gene and a marker as the basis for beginning a search for a gene Chromosome walking moving toward
Map-Based Cloning of Qualitative Plant Genes Map-based cloning using the genetic relationship between a gene and a marker as the basis for beginning a search for a gene Chromosome walking moving toward
Unit 8: Genomics Guided Reading Questions (150 pts total)
 Name: AP Biology Biology, Campbell and Reece, 7th Edition Adapted from chapter reading guides originally created by Lynn Miriello Chapter 18 The Genetics of Viruses and Bacteria Unit 8: Genomics Guided
Name: AP Biology Biology, Campbell and Reece, 7th Edition Adapted from chapter reading guides originally created by Lynn Miriello Chapter 18 The Genetics of Viruses and Bacteria Unit 8: Genomics Guided
IN VIVO AND IN VITRO COMPLEMENTATION BETWEEN DHQ
 IN VIVO AND IN VITRO COMPLEMENTATION BETWEEN DHQ SYNTHETASE MUTANTS IN THE AROM GENE CLUSTER OF NEUROSPORA CRASSA MARY E. CASE, LEIGH BURGOYNE,' AND NORMAN H. GILES Department of Biology, Kline Biology
IN VIVO AND IN VITRO COMPLEMENTATION BETWEEN DHQ SYNTHETASE MUTANTS IN THE AROM GENE CLUSTER OF NEUROSPORA CRASSA MARY E. CASE, LEIGH BURGOYNE,' AND NORMAN H. GILES Department of Biology, Kline Biology
Q1 (1 point): Explain why a lettuce leaf wilts when it is placed in a concentrated salt solution.
 Short questions 1 point per question. Q1 (1 point): Explain why a lettuce leaf wilts when it is placed in a concentrated salt solution. Q2 (1 point): Put a cross by the correct answer(s) below. The Na
Short questions 1 point per question. Q1 (1 point): Explain why a lettuce leaf wilts when it is placed in a concentrated salt solution. Q2 (1 point): Put a cross by the correct answer(s) below. The Na
Chapter 9. Gene Interactions. As we learned in Chapter 3, Mendel reported that the pairs of loci he observed segregated independently
 Chapter 9 Gene Interactions Figure 9-1 The coat colour on this juvenile horse is called Bay Roan Tobiano. Bay is the brown base coat colour; Roan is the mixture of white hairs with the base coat, making
Chapter 9 Gene Interactions Figure 9-1 The coat colour on this juvenile horse is called Bay Roan Tobiano. Bay is the brown base coat colour; Roan is the mixture of white hairs with the base coat, making
BSCI410-Liu/Spring 06 Exam #1 Feb. 23, 06
 Your Name: Your UID# 1. (20 points) Match following mutations with corresponding mutagens (X-RAY, Ds transposon excision, UV, EMS, Proflavin) a) Thymidine dimmers b) Breakage of DNA backbone c) Frameshift
Your Name: Your UID# 1. (20 points) Match following mutations with corresponding mutagens (X-RAY, Ds transposon excision, UV, EMS, Proflavin) a) Thymidine dimmers b) Breakage of DNA backbone c) Frameshift
Exam Plant Genetics CSS/Hort 430/530
 Exam 1 2010 Plant Genetics CSS/Hort 430/530 Barley can produce seed which is hulled or hull-less. This trait is determined by one gene, hulled being the dominant phenotype and hull-less being the recessive.
Exam 1 2010 Plant Genetics CSS/Hort 430/530 Barley can produce seed which is hulled or hull-less. This trait is determined by one gene, hulled being the dominant phenotype and hull-less being the recessive.
Purification and Characterization of Cinnamyl Alcohol Dehydrogenase from Tobacco Stems1
 Plant Physiol. (1992) 98, 12-16 0032-0889/92/98/001 2/05/$01.00/0 Received for publication May 15, 1991 Accepted September 10, 1991 Purification and Characterization of Cinnamyl Alcohol Dehydrogenase from
Plant Physiol. (1992) 98, 12-16 0032-0889/92/98/001 2/05/$01.00/0 Received for publication May 15, 1991 Accepted September 10, 1991 Purification and Characterization of Cinnamyl Alcohol Dehydrogenase from
CONSTRUCTION OF GENOMIC LIBRARY
 MODULE 4-LECTURE 4 CONSTRUCTION OF GENOMIC LIBRARY 4-4.1. Introduction A genomic library is an organism specific collection of DNA covering the entire genome of an organism. It contains all DNA sequences
MODULE 4-LECTURE 4 CONSTRUCTION OF GENOMIC LIBRARY 4-4.1. Introduction A genomic library is an organism specific collection of DNA covering the entire genome of an organism. It contains all DNA sequences
-Genes on the same chromosome are called linked. Human -23 pairs of chromosomes, ~35,000 different genes expressed.
 Linkage -Genes on the same chromosome are called linked Human -23 pairs of chromosomes, ~35,000 different genes expressed. - average of 1,500 genes/chromosome Following Meiosis Parental chromosomal types
Linkage -Genes on the same chromosome are called linked Human -23 pairs of chromosomes, ~35,000 different genes expressed. - average of 1,500 genes/chromosome Following Meiosis Parental chromosomal types
Motivation From Protein to Gene
 MOLECULAR BIOLOGY 2003-4 Topic B Recombinant DNA -principles and tools Construct a library - what for, how Major techniques +principles Bioinformatics - in brief Chapter 7 (MCB) 1 Motivation From Protein
MOLECULAR BIOLOGY 2003-4 Topic B Recombinant DNA -principles and tools Construct a library - what for, how Major techniques +principles Bioinformatics - in brief Chapter 7 (MCB) 1 Motivation From Protein
Two possible explanations for dominant mutations Haploinsufficiency vs Dominant negative
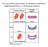 Two possible explanations for dominant mutations Haploinsufficiency vs Dominant negative Incomplete dominance for flower colour in Mirabilis jalapa R 1 R 1 R 2 R 2 R 1 R 2 R 1 Red pigment R 2 No pigment
Two possible explanations for dominant mutations Haploinsufficiency vs Dominant negative Incomplete dominance for flower colour in Mirabilis jalapa R 1 R 1 R 2 R 2 R 1 R 2 R 1 Red pigment R 2 No pigment
Lecture 5: Inbreeding and Allozymes. Sept 1, 2006
 Lecture 5: Inbreeding and Allozymes Sept 1, 2006 Last Time Tandem repeats and recombination Organellar DNA Introduction to DNA in populations Organelle Inheritance Organelles can be excluded from one parent's
Lecture 5: Inbreeding and Allozymes Sept 1, 2006 Last Time Tandem repeats and recombination Organellar DNA Introduction to DNA in populations Organelle Inheritance Organelles can be excluded from one parent's
Coniferyl aldehyde 5-hydroxylation and methylation direct syringyl lignin biosynthesis in angiosperms
 Proc. Natl. Acad. Sci. USA Vol. 96, pp. 8955 8960, August 1999 Biochemistry Coniferyl aldehyde 5-hydroxylation and methylation direct syringyl lignin biosynthesis in angiosperms KEISHI OSAKABE*, CHENG
Proc. Natl. Acad. Sci. USA Vol. 96, pp. 8955 8960, August 1999 Biochemistry Coniferyl aldehyde 5-hydroxylation and methylation direct syringyl lignin biosynthesis in angiosperms KEISHI OSAKABE*, CHENG
Part I: Predicting Genetic Outcomes
 Part I: Predicting Genetic Outcomes Deoxyribonucleic acid (DNA) is found in every cell of living organisms, and all of the cells in each organism contain the exact same copy of that organism s DNA. Because
Part I: Predicting Genetic Outcomes Deoxyribonucleic acid (DNA) is found in every cell of living organisms, and all of the cells in each organism contain the exact same copy of that organism s DNA. Because
Using mutants to clone genes
 Using mutants to clone genes Objectives 1. What is positional cloning? 2. What is insertional tagging? 3. How can one confirm that the gene cloned is the same one that is mutated to give the phenotype
Using mutants to clone genes Objectives 1. What is positional cloning? 2. What is insertional tagging? 3. How can one confirm that the gene cloned is the same one that is mutated to give the phenotype
RNA Blue REAGENT FOR RAPID ISOLATION OF PURE AND INTACT RNA (Cat. No. R011, R012, R013)
 RNA Blue REAGENT FOR RAPID ISOLATION OF PURE AND INTACT RNA (Cat. No. R011, R012, R013) WARNING: RNA Blue contains phenol and some other toxic components. After contact with skin, wash immediately with
RNA Blue REAGENT FOR RAPID ISOLATION OF PURE AND INTACT RNA (Cat. No. R011, R012, R013) WARNING: RNA Blue contains phenol and some other toxic components. After contact with skin, wash immediately with
Genomic Sequencing. Genomic Sequencing. Maj Gen (R) Suhaib Ahmed, HI (M)
 Maj Gen (R) Suhaib Ahmed, HI (M) The process of determining the sequence of an unknown DNA is called sequencing. There are many approaches for DNA sequencing. In the last couple of decades automated Sanger
Maj Gen (R) Suhaib Ahmed, HI (M) The process of determining the sequence of an unknown DNA is called sequencing. There are many approaches for DNA sequencing. In the last couple of decades automated Sanger
The AmMYB308 and AmMYB330 Transcription Factors from Antirrhinum Regulate Phenylpropanoid and Lignin Biosynthesis in Transgenic Tobacco
 The Plant Cell, Vol. 10, 135 154, February 1998 1998 American Society of Plant Physiologists RESEARCH ARTICLE The AmMYB308 and AmMYB330 Transcription Factors from Antirrhinum Regulate Phenylpropanoid and
The Plant Cell, Vol. 10, 135 154, February 1998 1998 American Society of Plant Physiologists RESEARCH ARTICLE The AmMYB308 and AmMYB330 Transcription Factors from Antirrhinum Regulate Phenylpropanoid and
Gene Tagging with Random Amplified Polymorphic DNA (RAPD) Markers for Molecular Breeding in Plants
 Critical Reviews in Plant Sciences, 20(3):251 275 (2001) Gene Tagging with Random Amplified Polymorphic DNA (RAPD) Markers for Molecular Breeding in Plants S. A. Ranade, * Nuzhat Farooqui, Esha Bhattacharya,
Critical Reviews in Plant Sciences, 20(3):251 275 (2001) Gene Tagging with Random Amplified Polymorphic DNA (RAPD) Markers for Molecular Breeding in Plants S. A. Ranade, * Nuzhat Farooqui, Esha Bhattacharya,
A facile and fast method for quantitating lignin in lignocellulosic biomass using acidic
 Electronic Supplementary Material (ESI) for Green Chemistry. This journal is The Royal Society of Chemistry 2016 Electronic Supporting Information (ESI) A facile and fast method for quantitating lignin
Electronic Supplementary Material (ESI) for Green Chemistry. This journal is The Royal Society of Chemistry 2016 Electronic Supporting Information (ESI) A facile and fast method for quantitating lignin
3. A form of a gene that is only expressed in the absence of a dominant alternative is:
 Student Name: Teacher: Date: District: Robeson Assessment: 9_12 Agriculture AU71 - Biotech and Agrisci Rsch I Test 3 Description: Obj 12 - Simple Mendelian Genetics Form: 501 1. The genotype of an organism
Student Name: Teacher: Date: District: Robeson Assessment: 9_12 Agriculture AU71 - Biotech and Agrisci Rsch I Test 3 Description: Obj 12 - Simple Mendelian Genetics Form: 501 1. The genotype of an organism
Biomass conversion: opportunities in lignin management. Clint Chapple Department of Biochemistry Purdue University
 Biomass conversion: opportunities in lignin management Clint Chapple Department of Biochemistry Purdue University Targets for biomass improvement Yield Water and nutrient use efficiency Pest and pathogen
Biomass conversion: opportunities in lignin management Clint Chapple Department of Biochemistry Purdue University Targets for biomass improvement Yield Water and nutrient use efficiency Pest and pathogen
Phenotype analysis: biological-biochemical analysis. Genotype analysis: molecular and physical analysis
 1 Genetic Analysis Phenotype analysis: biological-biochemical analysis Behaviour under specific environmental conditions Behaviour of specific genetic configurations Behaviour of progeny in crosses - Genotype
1 Genetic Analysis Phenotype analysis: biological-biochemical analysis Behaviour under specific environmental conditions Behaviour of specific genetic configurations Behaviour of progeny in crosses - Genotype
DNA Damage Quantification Kit
 Revised February 27, 2009 DNA Damage Quantification Kit - AP Site Counting - 5 Samples Technical Manual Product Code: DK02-10 Contents General Information...1 Description of the Kit...1 Contents...1 How
Revised February 27, 2009 DNA Damage Quantification Kit - AP Site Counting - 5 Samples Technical Manual Product Code: DK02-10 Contents General Information...1 Description of the Kit...1 Contents...1 How
Variation Chapter 9 10/6/2014. Some terms. Variation in phenotype can be due to genes AND environment: Is variation genetic, environmental, or both?
 Frequency 10/6/2014 Variation Chapter 9 Some terms Genotype Allele form of a gene, distinguished by effect on phenotype Haplotype form of a gene, distinguished by DNA sequence Gene copy number of copies
Frequency 10/6/2014 Variation Chapter 9 Some terms Genotype Allele form of a gene, distinguished by effect on phenotype Haplotype form of a gene, distinguished by DNA sequence Gene copy number of copies
4 Mutant Hunts - To Select or to Screen (Perhaps Even by Brute Force)
 Genetic Techniques for Biological Research Corinne A. Michels Copyright q 2002 John Wiley & Sons, Ltd ISBNs: 0-471-89921-6 (Hardback); 0-470-84662-3 (Electronic) 4 Mutant Hunts - To Select or to Screen
Genetic Techniques for Biological Research Corinne A. Michels Copyright q 2002 John Wiley & Sons, Ltd ISBNs: 0-471-89921-6 (Hardback); 0-470-84662-3 (Electronic) 4 Mutant Hunts - To Select or to Screen
Construction of plant complementation vector and generation of transgenic plants
 MATERIAL S AND METHODS Plant materials and growth conditions Arabidopsis ecotype Columbia (Col0) was used for this study. SALK_072009, SALK_076309, and SALK_027645 were obtained from the Arabidopsis Biological
MATERIAL S AND METHODS Plant materials and growth conditions Arabidopsis ecotype Columbia (Col0) was used for this study. SALK_072009, SALK_076309, and SALK_027645 were obtained from the Arabidopsis Biological
Biology 163 Laboratory in Genetics, Final Exam, Dec. 10, 2005
 1 Biology 163 Laboratory in Genetics, Final Exam, Dec. 10, 2005 Honor Pledge: I have neither given nor received any unauthorized help on this exam: Name Printed: Signature: 1. (2 pts) If you see the following
1 Biology 163 Laboratory in Genetics, Final Exam, Dec. 10, 2005 Honor Pledge: I have neither given nor received any unauthorized help on this exam: Name Printed: Signature: 1. (2 pts) If you see the following
Marathon TM cdna Amplification Kit Protocol-at-a-Glance
 (PT1115-2) Marathon cdna amplification is a fairly complex, multiday procedure. Please read the User Manual before using this abbreviated protocol, and refer to it often for interpretation of results during
(PT1115-2) Marathon cdna amplification is a fairly complex, multiday procedure. Please read the User Manual before using this abbreviated protocol, and refer to it often for interpretation of results during
Genetic Engineering & Recombinant DNA
 Genetic Engineering & Recombinant DNA Chapter 10 Copyright The McGraw-Hill Companies, Inc) Permission required for reproduction or display. Applications of Genetic Engineering Basic science vs. Applied
Genetic Engineering & Recombinant DNA Chapter 10 Copyright The McGraw-Hill Companies, Inc) Permission required for reproduction or display. Applications of Genetic Engineering Basic science vs. Applied
Ribonucleic acid (RNA)
 Ribonucleic acid (RNA) Ribonucleic acid (RNA) is more often found in nature as a singlestrand folded onto itself. Cellular organisms use messenger RNA (mrna) to convey genetic information (using the nitrogenous
Ribonucleic acid (RNA) Ribonucleic acid (RNA) is more often found in nature as a singlestrand folded onto itself. Cellular organisms use messenger RNA (mrna) to convey genetic information (using the nitrogenous
