Figure 1-Figure Supplement 1 Validation of knockdown efficiency for trpa1 RNAi and RyR RNAi lines.
|
|
|
- Charles Kelly
- 6 years ago
- Views:
Transcription
1 Figure 1-Figure Supplement 1 Validation of knockdown efficiency for trpa1 RNAi and RyR RNAi lines. (A) Cartoon depicting the four characterized trpa1 isoforms. The target regions of trpa1 RNAi lines used in this study are shown in dashed rectangles. The regions amplified with isoform-specific primers are shown in square brackets. (B) Primers spanning exons for specific splicing events are used in RT-PCR to distinguish trpa1 isoform expression in fly midguts and heads. (C) RT-qPCR analysis of mrnas from midguts expressing different lines of trpa1 RNAi ubiquitously with inducible DaGal4ts for 5d, with primers designed for different types of trpa1 isoforms. GAPDH and rp49 are reference genes for normalization. To determine RNAi knockdown efficiency, the ratio of normalized trpa1 expression to corresponding control groups expressing either Luc RNAi (for BL lines) or carrying an empty insertional landing site (v60100 for v37249) is calculated. The data are presented as mean ± SEM for three technical replicates. We repeated the experiments (two biological replicates) and observed consistent results. (D) RT-qPCR measurement of midguts ubiquitously expressing LucRNAi or RyR RNAi for 5d, with two different sets of primers designed for all RyR isoforms. GAPDH and rp49 are used for normalization. The data are presented as mean ± SEM for three technical replicates. We repeated the experiments (two biological replicates) and observed consistent results. (E) Quantification of relative esg+ cell density in the posterior midgut region. esg+ cell number is divided by the imaged area size for density calculation. N > 5 midguts per genotype per time point are analyzed. Data are represented as mean ± SEM.
2 Figure 1-Figure Supplement 2 trpa1 RNAi and RyR RNAi do not cause ISC apoptosis. (A C) Immunostaining of midguts expressing Luc RNAi, trpa1 RNAi or RyR RNAi in ISCs for apoptosis marker cleaved-caspase 3. The channels of cleaved-caspase 3 signal are shown to the right of the merged images. No signal can be detected except some background staining in the trachea and muscle. (D F) Midguts expressing Luc RNAi, trpa1 RNAi or RyR RNAi in ISCs with the last 2d feeding 2 mm paraquat (D F ), are stained for
3 Figure 1-Figure Supplement 3 ISC depletion results in midgut shortening. (A) Length quantification of midguts with inducible expression of rpr in ISCs. In the rpr off group, t kept at 18 C to prevent Gal4 activity and rpr expression; in the rpr on group, rpr expression is ind for 4d and turned off at 18 C for 3d feeding normal food or bleomycin; in the post-rpr group, after expression of rpr, the flies are placed back at 18 C for specified days before bleomycin treatment. midguts are analyzed per genotype per treatment. Data are represented as mean ± SEM. (B) Mito quantification of midguts with inducible expression of rpr in ISCs. Mitosis activity in the midgut can after depletion of the esg+ cell population by 4d rpr expression, N > 7 midguts are analyzed per ge treatment. Data are represented as mean ± SEM.
4 Figure 2-Figure Supplement 1 Lineage-tracing experiments provide evidence that TRPA1 or RyR-deficient ISCs can differentiate. (A B) MARCM clones expressing Luc RNAi or trpa1 RNAi for 22d are examined for their survival and stained with anti-pdm1 antibody to examine EC differentiation. Arrowheads highlight examples of ECs generated from ISCs expressing LucRNAi or trpa1 RNAi. (C D) MARCM clones that are homozygous RyR mutant are stained with anti-pdm1 antibody. Arrowheads highlight examples of ECs in the MARCM clones that are Pdm1+. (E F) Immunostaining of midguts expressing Luc RNAi or trpa1 RNAi in the ISC lineage (labeled by GFP) for EE marker, Pros. Arrowheads highlight examples of EEs generated by ISCs during the 6d period of tracing. (G H) Immunostaining of midguts expressing Luc RNAi or trpa1 RNAi in the ISC lineage (labeled by GFP) for EC marker, Pdm1. Arrowheads highlight examples of ECs generated by ISCs during the 6d period of tracing.
5 Figure 3-Figure Supplement 1 The knock-in design of trpa1gal4 CP2A. By CRISPR/Cas9-induced homologous recombination, Gal4 is inserted into the shared C terminal of trpa1 isoforms right before the stop codon. The knock-in could result in bi-cistronic transcripts expressing both TRPA1 and Gal4.
6 Figure 3-Figure Supplement 2 Additional evidence for trpa1 expression and function in ISCs. (A) RT-qPCR measurement of midguts expressing the cell death gene rpr in ISCs for 4d for the expression of esg, trpa1(using primers for CD isoforms), EC marker ε-trpsin (εtrp), Pdm1, or pros. GAPDH and rp49 are used for normalization. The data are presented as mean ± SEM for three technical replicates. We repeated the experiments (two biological replicates) and observed consistent results. (B) Mitosis quantification of midguts expressing Luc RNAi or trpa1 RNAi in self-renewing ISCs (DlGal4ts), differentiating ISCs or enteroblasts (Su(H)Gal4ts), muscles (24BGal4ts), or ubiquitously (tubgal4ts) for 9d, with last 2d feeding bleomycin. DlGal4ts and Su(H)Gal4ts are used to drive gene expression in ISCs and enteroblasts, respectively (Zeng et al., 2010). Data are represented as mean ± SEM.
7 Figure 4-Figure Supplement 1 Elevated ROS is a common stress signal in various midgut damage conditions. (A E) Dihydro-ethidium (DHE) stainings of live midguts expressing GFP in ISCs for 3d from flies fed with normal food, food containing 2 mm paraquat, 25 µg/ml bleomycin for 6 hr, pathogens Pseudomonas entomophila (P.e.), or Pseudomonas aeruginosa (PA14) for 18 hr. DHE signal channel is shown below the merged image. The control group for Pseudomonasinfection, using Bacteria-free LB media, is not shown in the figure because the midgut DHE staining looks the same as flies fed with normal food. Arrowheads highlight examples of stem cells that have high ROS concentration. (F G) DHE staining of midguts overexpressing active JNKK (Drosophila Hep ca ) for 1d in ISCs with EGT. (H J) DHE staining of midguts overexpressing Hep ca or cell death gene reaper (rpr) for 2d in ECs with MyO1AGal4ts. esggfp labels the ISCs.
8 Figure 5-Figure Supplement 1 The total number of ISCs expressing esggal4>gcamp6s reporter is not significantly reduced by trpa1 RNAi or RyR RNAi. (A C) Anti-GFP staining of midguts expressing GCamP6s alone, or together with trpa1 RNAi or RyR RNAi in ISCs detects GCamP6s expression. The images are acquired on a confocal microscope. The relative density of GCamP6s+ (driven by esggal4) cells, imaged on a Keyence microscope covering most of the posterior midgut region, is quantified in (D) while the total number of GCamP6s+ cells in the posterior midgut region is quantified in (E). N > 8 midguts are analyzed for each genotype. Data are represented as mean ± SEM.
9 Figure 5-Figure Supplement 2 Additional reporters showing that TRPA1 and RyR are required for ROS-mediated Ca 2+ increases in ISCs. (A B) Confocal calcium imaging of midguts that express bi-cistronic UAS-tdTomato-P2A-GCaMP5G reporter alone or together with trpa1 RNAi in ISCs with exposure to 2 mm paraquat for 5 min (A B ). The bi-cistronic reporter consists of tdtomato that labels all cells expressing the reporter, and GCaMP5G whose GFP signal intensity reflects intracellular calcium concentration (Daniels et al., 2014). (C E) Midguts expressing the TRIC lexgfp calcium reporter alone or together with trpa1 RNAi or RyR RNAi in ISCs for 6d, with the last 1d feeding 2 mm paraquat (C E ) are stained to examine stem cell Ca 2+ concentration in vivo. The conventional UAS- RFP reporter is used to label all stem cells. (F) Quantification of high Ca 2+ stem cells (GFP+) ratio to all stem cells (RFP+) for midguts in experiments (C E). N = 16, 10, six images are analyzed for the genotypes of control, trpa1 RNAi, and RyR RNAi (half feeding normally, half feeding paraquat for 1d). Data are represented as mean ± SEM.
10 Figure 6-Figure Supplement 1 Prolonged induction of high cytosolic Ca 2+ in ISCs results in a nonspecific and variable pattern of Ras/MAPK activation. (A C) Midguts expressing SERCA RNAi together with Luc RNAi, Ras1 RNAi, or Yki RNAi in ISCs for 5d are stained for dperk. The channel of dperk signal is shown below the merged image. A and A are biological replicates of the same genotype showing examples of variation. Although perk induction is still detectable in some ISCs, the signal intensity varies among different ISCs and different midguts.
11 Figure 6-Figure Supplement 2 Src might be a mechanism by which cytosolic Ca 2+ can activate Ras/MAPK in ISCs. (A D) Midguts expressing the constitutive active form of Src42A (Src42A ca ), Src64B, or Ras1 A in ISCs for 2d are stained for the Ras/MAPK activity marker dperk. The channel of dperk signal is shown below the merged image. w- genetic background is used as the control for transgenic expression. (E G) Midguts expressing trpa1 RNAi alone or together with Src42A ca / Src64B in ISCs for 2d are stained for dperk. The channel of dperk signal is shown below the merged image. (H) Mitosis quantification of midguts expressing trpa1 RNAi with GFP, Src42A ca, or Src64B in ISCs for 3d. N > 5 midguts are analyzed for each genotype. Data are represented as mean ± SEM. (I) Mitosis quantification of midguts expressing SERCARNAi with Luc RNAi, Src42A RNAi, or Src64B RNAi in ISCs for 4d. N > 4 midguts are analyzed for each genotype. Data are represented as mean ± SEM.
12 Figure 7-Figure Supplement 1 Ras/MAPK activity, but not CanA1/CrebB, is required for ISC proliferation induced by calcium influx. (A E) Midguts expressing SERCA RNAi together with Luc RNAi, EGFR RNAi, CanA1 RNAi, CrebB RNAi, or CRTC RNAi in ISCs for 3d are stained for mitosis marker ph3. (F) Mitosis quantification of midguts expressing SERCA RNAi together with Luc RNAi, EGFR RNAi (three different lines), Ras1 RNAi, Yki RNAi, CanA1 RNAi, CrebB RNAi (two different lines), or CRTCRNAi in ISCs for 5d. N > 7 midguts are analyzed for each genotype. Data are represented as mean ± SEM. (G) Mitosis quantification of midguts expressing SERCA RNAi 2 alone, or together with CanA1 RNAi fb5, or Yki RNAi v in ISCs for 5d. N > 5 midguts are analyzed for each genotype. Data are represented as mean ± SEM. For CanA1 RNAi line fb5, flies with a similar w- genetic background carrying an empty insertional landing site (v60100) are used as the control (Ctrl v ). (H) RTqPCR measurement of CRTC expression in midguts ubiquitously expressing Luc RNAi or CRTC RNAi for 5d. GAPDH and rp49 are used for normalization. The data are presented as mean ± SEM for three technical replicates. (I) RT-qPCR measurement of CanA1 RNAi fb5 knockdown efficiency. L3 larvae expressing RNAi for 2 days are used for better quantification because midgut CanA1 expression is barely detectable. GAPDH and rp49 are used for normalization. The data are presented as mean ± SEM for three technical replicates.
13 Figure 7-Figure Supplement 2 Ligands for receptor tyrosine kinases (RTKs) are affected by cytosolic Ca 2+ signaling. (A) RT-qPCR measurement of midguts expressing Luc RNAi or trpa1 RNAi in ISCs for 6d, with the last day feeding on normal food or paraquat. rp49 is used for normalization. The data are presented as mean ± SEM for three technical replicates. While expression of JAK/ Stat pathway ligands upd1 and upd2 remains unaffected, multiple RTK ligands (spi, krn, vn, pvf1, pvf3, dilp3, highlighted by gray square boxes) are down-regulated by trpa1 RNAi. (B) RT-qPCR measurement of midguts expressing Luc RNAi or trpa1 RNAi in ISCs for 9d, with the last 2d feeding on normal food or paraquat. rp49 is used for normalization. The data are presented as mean ± SEM for three technical replicates. Note that only spi, pvf1, and dilp3 are consistently down-regulated in trpa1 RNAi groups in both (A) and (B). Different trends of krn, vn, and pvf3expression observed in (B) compared to (A) might suggest a delay, rather than reduction of these ligands by trpa1 RNAi. (C) RT-qPCR measurement of midguts expressing Luc RNAi or SERCA RNAi in ISCs for 1d. Such early time point is chosen empirically to allow for efficient knockdown and avoid confounding effects of signaling pathway cross-activation at later stages of ISC proliferation. rp49 is used for normalization. The data are presented as mean ± SEM for three technical replicates.
14 Figure 7-Figure Supplement 3 Ras/MAPK activity is sufficient for ISC proliferation in the absence of RyR. (A B) Midguts, expressing RyR RNAi alone, or together with SERCA RNAi in ISCs for 5d, are stained for mitosis marker ph3.
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Drosophila dyskerin is required for somatic stem cell homeostasis Rosario Vicidomini 1,2*, Arianna Petrizzo 1, Annamaria di Giovanni 1, Laura Cassese 1, Antonella Anna Lombardi
SUPPLEMENTARY INFORMATION Drosophila dyskerin is required for somatic stem cell homeostasis Rosario Vicidomini 1,2*, Arianna Petrizzo 1, Annamaria di Giovanni 1, Laura Cassese 1, Antonella Anna Lombardi
Additional Practice Problems for Reading Period
 BS 50 Genetics and Genomics Reading Period Additional Practice Problems for Reading Period Question 1. In patients with a particular type of leukemia, their leukemic B lymphocytes display a translocation
BS 50 Genetics and Genomics Reading Period Additional Practice Problems for Reading Period Question 1. In patients with a particular type of leukemia, their leukemic B lymphocytes display a translocation
Mechanical regulation of stem-cell differentiation by the stretch-activated Piezo channel
 Letter doi:10.1038/nature25744 Mechanical regulation of stem-cell differentiation by the stretch-activated Piezo channel Li He 1, Guangwei Si 2, Jiuhong Huang 3, Aravinthan D. T. Samuel 2 & Norbert Perrimon
Letter doi:10.1038/nature25744 Mechanical regulation of stem-cell differentiation by the stretch-activated Piezo channel Li He 1, Guangwei Si 2, Jiuhong Huang 3, Aravinthan D. T. Samuel 2 & Norbert Perrimon
Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product.
 Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
The Hippo tumor suppressor pathway regulates intestinal stem cell regeneration
 AND STEM CELLS 4135 Development 138, 4135-4145 (2010) doi:10.1242/dev.060483 2010. Published by The Company of Biologists Ltd The Hippo tumor suppressor pathway regulates intestinal stem cell regeneration
AND STEM CELLS 4135 Development 138, 4135-4145 (2010) doi:10.1242/dev.060483 2010. Published by The Company of Biologists Ltd The Hippo tumor suppressor pathway regulates intestinal stem cell regeneration
Alternative Cleavage and Polyadenylation of RNA
 Developmental Cell 18 Supplemental Information The Spen Family Protein FPA Controls Alternative Cleavage and Polyadenylation of RNA Csaba Hornyik, Lionel C. Terzi, and Gordon G. Simpson Figure S1, related
Developmental Cell 18 Supplemental Information The Spen Family Protein FPA Controls Alternative Cleavage and Polyadenylation of RNA Csaba Hornyik, Lionel C. Terzi, and Gordon G. Simpson Figure S1, related
Supplementary Information
 Supplementary Information Live imaging reveals the dynamics and regulation of mitochondrial nucleoids during the cell cycle in Fucci2-HeLa cells Taeko Sasaki 1, Yoshikatsu Sato 2, Tetsuya Higashiyama 1,2,
Supplementary Information Live imaging reveals the dynamics and regulation of mitochondrial nucleoids during the cell cycle in Fucci2-HeLa cells Taeko Sasaki 1, Yoshikatsu Sato 2, Tetsuya Higashiyama 1,2,
Supplemental Information
 Supplemental Information Itemized List Materials and Methods, Related to Supplemental Figures S5A-C and S6. Supplemental Figure S1, Related to Figures 1 and 2. Supplemental Figure S2, Related to Figure
Supplemental Information Itemized List Materials and Methods, Related to Supplemental Figures S5A-C and S6. Supplemental Figure S1, Related to Figures 1 and 2. Supplemental Figure S2, Related to Figure
Confocal immunofluorescence microscopy
 Confocal immunofluorescence microscopy HL-6 and cells were cultured and cytospun onto glass slides. The cells were double immunofluorescence stained for Mt NPM1 and fibrillarin (nucleolar marker). Briefly,
Confocal immunofluorescence microscopy HL-6 and cells were cultured and cytospun onto glass slides. The cells were double immunofluorescence stained for Mt NPM1 and fibrillarin (nucleolar marker). Briefly,
Developmental Biology 3230 Exam 1 (Feb. 6) NAME
 DevelopmentalBiology3230Exam1(Feb.6)NAME 1. (10pts) What is a Fate Map? How would you experimentally acquire the data to draw a Fate Map? Explain what a Fate Map does and does not tell you about the mechanisms
DevelopmentalBiology3230Exam1(Feb.6)NAME 1. (10pts) What is a Fate Map? How would you experimentally acquire the data to draw a Fate Map? Explain what a Fate Map does and does not tell you about the mechanisms
New Tool for Late Stage Visualization of Oskar Protein in Drosophila melanogaster. Oocytes. Kathleen Harnois. Supervised by Paul Macdonald
 New Tool for Late Stage Visualization of Oskar Protein in Drosophila melanogaster Oocytes Kathleen Harnois Supervised by Paul Macdonald Institute of Cellular and Molecular Biology, The University of Texas
New Tool for Late Stage Visualization of Oskar Protein in Drosophila melanogaster Oocytes Kathleen Harnois Supervised by Paul Macdonald Institute of Cellular and Molecular Biology, The University of Texas
Supplementary Figure 1. Isolation of GFPHigh cells.
 Supplementary Figure 1. Isolation of GFP High cells. (A) Schematic diagram of cell isolation based on Wnt signaling activity. Colorectal cancer (CRC) cell lines were stably transduced with lentivirus encoding
Supplementary Figure 1. Isolation of GFP High cells. (A) Schematic diagram of cell isolation based on Wnt signaling activity. Colorectal cancer (CRC) cell lines were stably transduced with lentivirus encoding
SUPPLEMENTARY INFORMATION
 a before amputation regeneration regenerated limb DERMIS SKELETON MUSCLE SCHWANN CELLS EPIDERMIS DERMIS SKELETON MUSCLE SCHWANN CELLS EPIDERMIS developmental origin: lateral plate mesoderm presomitic mesoderm
a before amputation regeneration regenerated limb DERMIS SKELETON MUSCLE SCHWANN CELLS EPIDERMIS DERMIS SKELETON MUSCLE SCHWANN CELLS EPIDERMIS developmental origin: lateral plate mesoderm presomitic mesoderm
SUPPLEMENTARY INFORMATION. Small molecule activation of the TRAIL receptor DR5 in human cancer cells
 SUPPLEMENTARY INFORMATION Small molecule activation of the TRAIL receptor DR5 in human cancer cells Gelin Wang 1*, Xiaoming Wang 2, Hong Yu 1, Shuguang Wei 1, Noelle Williams 1, Daniel L. Holmes 1, Randal
SUPPLEMENTARY INFORMATION Small molecule activation of the TRAIL receptor DR5 in human cancer cells Gelin Wang 1*, Xiaoming Wang 2, Hong Yu 1, Shuguang Wei 1, Noelle Williams 1, Daniel L. Holmes 1, Randal
GM130 Is Required for Compartmental Organization of Dendritic Golgi Outposts
 Current Biology, Volume 24 Supplemental Information GM130 Is Required for Compartmental Organization of Dendritic Golgi Outposts Wei Zhou, Jin Chang, Xin Wang, Masha G. Savelieff, Yinyin Zhao, Shanshan
Current Biology, Volume 24 Supplemental Information GM130 Is Required for Compartmental Organization of Dendritic Golgi Outposts Wei Zhou, Jin Chang, Xin Wang, Masha G. Savelieff, Yinyin Zhao, Shanshan
Toll Receptor-Mediated Hippo Signaling Controls Innate Immunity in Drosophila
 Cell Supplemental Information Toll Receptor-Mediated Hippo Signaling Controls Innate Immunity in Drosophila Bo Liu, Yonggang Zheng, Feng Yin, Jianzhong Yu, Neal Silverman, and Duojia Pan Supplemental Experimental
Cell Supplemental Information Toll Receptor-Mediated Hippo Signaling Controls Innate Immunity in Drosophila Bo Liu, Yonggang Zheng, Feng Yin, Jianzhong Yu, Neal Silverman, and Duojia Pan Supplemental Experimental
Supplementary Fig. S1. Schematic representation of mouse lines Pax6 fl/fl and mrx-cre used in this study. (A) To generate Pax6 fl/ fl
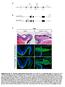 Supplementary Fig. S1. Schematic representation of mouse lines Pax6 fl/fl and mrx-cre used in this study. (A) To generate Pax6 fl/ fl, loxp sites flanking exons 3-6 (red arrowheads) were introduced into
Supplementary Fig. S1. Schematic representation of mouse lines Pax6 fl/fl and mrx-cre used in this study. (A) To generate Pax6 fl/ fl, loxp sites flanking exons 3-6 (red arrowheads) were introduced into
Chapter 14. Methods to Assess Intestinal Stem Cell Activity in Response to Microbes in Drosophila melanogaster. Philip L. Houtz and Nicolas Buchon
 Chapter 14 Methods to Assess Intestinal Stem Cell Activity in Response to Microbes in Drosophila melanogaster Philip L. Houtz and Nicolas Buchon Abstract Drosophila melanogaster presents itself as a powerful
Chapter 14 Methods to Assess Intestinal Stem Cell Activity in Response to Microbes in Drosophila melanogaster Philip L. Houtz and Nicolas Buchon Abstract Drosophila melanogaster presents itself as a powerful
Charles Shuler, Ph.D. University of Southern California Los Angeles, California Approved for Public Release; Distribution Unlimited
 AD Award Number: DAMD17-01-1-0100 TITLE: Smad-Mediated Signaling During Prostate Growth and Development PRINCIPAL INVESTIGATOR: Charles Shuler, Ph.D. CONTRACTING ORGANIZATION: University of Southern California
AD Award Number: DAMD17-01-1-0100 TITLE: Smad-Mediated Signaling During Prostate Growth and Development PRINCIPAL INVESTIGATOR: Charles Shuler, Ph.D. CONTRACTING ORGANIZATION: University of Southern California
Figure S1: NUN preparation yields nascent, unadenylated RNA with a different profile from Total RNA.
 Summary of Supplemental Information Figure S1: NUN preparation yields nascent, unadenylated RNA with a different profile from Total RNA. Figure S2: rrna removal procedure is effective for clearing out
Summary of Supplemental Information Figure S1: NUN preparation yields nascent, unadenylated RNA with a different profile from Total RNA. Figure S2: rrna removal procedure is effective for clearing out
SureSilencing sirna Array Technology Overview
 SureSilencing sirna Array Technology Overview Pathway-Focused sirna-based RNA Interference Topics to be Covered Who is SuperArray? Brief Introduction to RNA Interference Challenges Facing RNA Interference
SureSilencing sirna Array Technology Overview Pathway-Focused sirna-based RNA Interference Topics to be Covered Who is SuperArray? Brief Introduction to RNA Interference Challenges Facing RNA Interference
Supplemental Materials. Matrix Proteases Contribute to Progression of Pelvic Organ Prolapse in Mice and Humans
 Supplemental Materials Matrix Proteases Contribute to Progression of Pelvic Organ Prolapse in Mice and Humans Madhusudhan Budatha, Shayzreen Roshanravan, Qian Zheng, Cecilia Weislander, Shelby L. Chapman,
Supplemental Materials Matrix Proteases Contribute to Progression of Pelvic Organ Prolapse in Mice and Humans Madhusudhan Budatha, Shayzreen Roshanravan, Qian Zheng, Cecilia Weislander, Shelby L. Chapman,
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION DOI: 10.1038/ncb3575 In the format provided by the authors and unedited. Supplementary Figure 1 Validation of key reagents and assays a, top, IHC with antibody recognizing specifically
SUPPLEMENTARY INFORMATION DOI: 10.1038/ncb3575 In the format provided by the authors and unedited. Supplementary Figure 1 Validation of key reagents and assays a, top, IHC with antibody recognizing specifically
Table S1. Primers used in RT-PCR studies (all in 5 to 3 direction)
 Table S1. Primers used in RT-PCR studies (all in 5 to 3 direction) Epo Fw CTGTATCATGGACCACCTCGG Epo Rw TGAAGCACAGAAGCTCTTCGG Jak2 Fw ATCTGACCTTTCCATCTGGGG Jak2 Rw TGGTTGGGTGGATACCAGATC Stat5A Fw TTACTGAAGATCAAGCTGGGG
Table S1. Primers used in RT-PCR studies (all in 5 to 3 direction) Epo Fw CTGTATCATGGACCACCTCGG Epo Rw TGAAGCACAGAAGCTCTTCGG Jak2 Fw ATCTGACCTTTCCATCTGGGG Jak2 Rw TGGTTGGGTGGATACCAGATC Stat5A Fw TTACTGAAGATCAAGCTGGGG
Enhancer genetics Problem set E
 Enhancer genetics Problem set E How are specific cell types generated during development? There are thought to be 10,000 different types of neurons in the human nervous system. How are all of these neural
Enhancer genetics Problem set E How are specific cell types generated during development? There are thought to be 10,000 different types of neurons in the human nervous system. How are all of these neural
Heredity and DNA Assignment 1
 Heredity and DNA Assignment 1 Name 1. Which sequence best represents the relationship between DNA and the traits of an organism? A B C D 2. In some people, the lack of a particular causes a disease. Scientists
Heredity and DNA Assignment 1 Name 1. Which sequence best represents the relationship between DNA and the traits of an organism? A B C D 2. In some people, the lack of a particular causes a disease. Scientists
Figure S1. Immunoblotting showing proper expression of tagged R and AVR proteins in N. benthamiana.
 SUPPLEMENTARY FIGURES Figure S1. Immunoblotting showing proper expression of tagged R and AVR proteins in N. benthamiana. YFP:, :CFP, HA: and (A) and HA, CFP and YFPtagged and AVR1-CO39 (B) were expressed
SUPPLEMENTARY FIGURES Figure S1. Immunoblotting showing proper expression of tagged R and AVR proteins in N. benthamiana. YFP:, :CFP, HA: and (A) and HA, CFP and YFPtagged and AVR1-CO39 (B) were expressed
To assess the localization of Citrine fusion proteins, we performed antibody staining to
 Trinh et al 1 SUPPLEMENTAL MATERIAL FlipTraps recapitulate endogenous protein localization To assess the localization of Citrine fusion proteins, we performed antibody staining to compare the expression
Trinh et al 1 SUPPLEMENTAL MATERIAL FlipTraps recapitulate endogenous protein localization To assess the localization of Citrine fusion proteins, we performed antibody staining to compare the expression
7.013 Practice Quiz
 MIT Department of Biology 7.013: Introductory Biology - Spring 2005 Instructors: Professor Hazel Sive, Professor Tyler Jacks, Dr. Claudette Gardel 7.013 Practice Quiz 2 2004 1 Question 1 A. The primer
MIT Department of Biology 7.013: Introductory Biology - Spring 2005 Instructors: Professor Hazel Sive, Professor Tyler Jacks, Dr. Claudette Gardel 7.013 Practice Quiz 2 2004 1 Question 1 A. The primer
Cell autonomous vs. cell non-autonomous gene function
 Cell autonomous vs. cell non-autonomous gene function In multicellular organisms, it is important to know in what cell(s) the activity of a gene is required. - while RNA expression can be highly informative,
Cell autonomous vs. cell non-autonomous gene function In multicellular organisms, it is important to know in what cell(s) the activity of a gene is required. - while RNA expression can be highly informative,
Figure S1. Figure S2. Figure S3 HB Anti-FSP27 (COOH-terminal peptide) Ab. Anti-GST-FSP27(45-127) Ab.
 / 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
/ 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
Supplemental Data. Wu et al. (2). Plant Cell..5/tpc RGLG Hormonal treatment H2O B RGLG µm ABA µm ACC µm GA Time (hours) µm µm MJ µm IA
 Supplemental Data. Wu et al. (2). Plant Cell..5/tpc..4. A B Supplemental Figure. Immunoblot analysis verifies the expression of the AD-PP2C and BD-RGLG proteins in the Y2H assay. Total proteins were extracted
Supplemental Data. Wu et al. (2). Plant Cell..5/tpc..4. A B Supplemental Figure. Immunoblot analysis verifies the expression of the AD-PP2C and BD-RGLG proteins in the Y2H assay. Total proteins were extracted
Int. J. Mol. Sci. 2016, 17, 1259; doi: /ijms
 S1 of S5 Supplementary Materials: Fibroblast-Derived Extracellular Matrix Induces Chondrogenic Differentiation in Human Adipose-Derived Mesenchymal Stromal/Stem Cells in Vitro Kevin Dzobo, Taegyn Turnley,
S1 of S5 Supplementary Materials: Fibroblast-Derived Extracellular Matrix Induces Chondrogenic Differentiation in Human Adipose-Derived Mesenchymal Stromal/Stem Cells in Vitro Kevin Dzobo, Taegyn Turnley,
SUPPLEMENTARY INFORMATION
 doi:.38/nature899 Supplementary Figure Suzuki et al. a c p7 -/- / WT ratio (+)/(-) p7 -/- / WT ratio Log X 3. Fold change by treatment ( (+)/(-)) Log X.5 3-3. -. b Fold change by treatment ( (+)/(-)) 8
doi:.38/nature899 Supplementary Figure Suzuki et al. a c p7 -/- / WT ratio (+)/(-) p7 -/- / WT ratio Log X 3. Fold change by treatment ( (+)/(-)) Log X.5 3-3. -. b Fold change by treatment ( (+)/(-)) 8
Supplementary Figures Montero et al._supplementary Figure 1
 Montero et al_suppl. Info 1 Supplementary Figures Montero et al._supplementary Figure 1 Montero et al_suppl. Info 2 Supplementary Figure 1. Transcripts arising from the structurally conserved subtelomeres
Montero et al_suppl. Info 1 Supplementary Figures Montero et al._supplementary Figure 1 Montero et al_suppl. Info 2 Supplementary Figure 1. Transcripts arising from the structurally conserved subtelomeres
HeLa cells stably transfected with an empty pcdna3 vector (HeLa Neo) or with a plasmid
 SUPPLEMENTAL MATERIALS AND METHODS Cell culture, transfection and treatments. HeLa cells stably transfected with an empty pcdna3 vector (HeLa Neo) or with a plasmid encoding vmia (HeLa vmia) 1 were cultured
SUPPLEMENTAL MATERIALS AND METHODS Cell culture, transfection and treatments. HeLa cells stably transfected with an empty pcdna3 vector (HeLa Neo) or with a plasmid encoding vmia (HeLa vmia) 1 were cultured
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling
 Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
A novel two-step genome editing strategy with CRISPR-Cas9 provides new insights into telomerase action and TERT gene expression
 Xi et al. Genome Biology (2015) 16:231 DOI 10.1186/s13059-015-0791-1 RESEARCH A novel two-step genome editing strategy with CRISPR-Cas9 provides new insights into telomerase action and TERT gene expression
Xi et al. Genome Biology (2015) 16:231 DOI 10.1186/s13059-015-0791-1 RESEARCH A novel two-step genome editing strategy with CRISPR-Cas9 provides new insights into telomerase action and TERT gene expression
Lecture 25 (11/15/17)
 Lecture 25 (11/15/17) Reading: Ch9; 328-332 Ch25; 990-995, 1005-1012 Problems: Ch9 (study-guide: applying); 1,2 Ch9 (study-guide: facts); 7,8 Ch25 (text); 1-3,5-7,9,10,13-15 Ch25 (study-guide: applying);
Lecture 25 (11/15/17) Reading: Ch9; 328-332 Ch25; 990-995, 1005-1012 Problems: Ch9 (study-guide: applying); 1,2 Ch9 (study-guide: facts); 7,8 Ch25 (text); 1-3,5-7,9,10,13-15 Ch25 (study-guide: applying);
Supplemental Information. The TRAIL-Induced Cancer Secretome. Promotes a Tumor-Supportive Immune. Microenvironment via CCR2
 Molecular Cell, Volume 65 Supplemental Information The TRAIL-Induced Cancer Secretome Promotes a Tumor-Supportive Immune Microenvironment via CCR2 Torsten Hartwig, Antonella Montinaro, Silvia von Karstedt,
Molecular Cell, Volume 65 Supplemental Information The TRAIL-Induced Cancer Secretome Promotes a Tumor-Supportive Immune Microenvironment via CCR2 Torsten Hartwig, Antonella Montinaro, Silvia von Karstedt,
Fatchiyah
 Fatchiyah Email: fatchiya@yahoo.co.id RNAs: mrna trna rrna RNAi DNAs: Protein: genome DNA cdna mikro-makro mono-poly single-multi Analysis: Identification human and animal disease Finger printing Sexing
Fatchiyah Email: fatchiya@yahoo.co.id RNAs: mrna trna rrna RNAi DNAs: Protein: genome DNA cdna mikro-makro mono-poly single-multi Analysis: Identification human and animal disease Finger printing Sexing
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table.
 Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Nature Neuroscience: doi: /nn Supplementary Figure 1
 Supplementary Figure 1 PCR-genotyping of the three mouse models used in this study and controls for behavioral experiments after semi-chronic Pten inhibition. a-c. DNA from App/Psen1 (a), Pten tg (b) and
Supplementary Figure 1 PCR-genotyping of the three mouse models used in this study and controls for behavioral experiments after semi-chronic Pten inhibition. a-c. DNA from App/Psen1 (a), Pten tg (b) and
CHAPTER 9 DNA Technologies
 CHAPTER 9 DNA Technologies Recombinant DNA Artificially created DNA that combines sequences that do not occur together in the nature Basis of much of the modern molecular biology Molecular cloning of genes
CHAPTER 9 DNA Technologies Recombinant DNA Artificially created DNA that combines sequences that do not occur together in the nature Basis of much of the modern molecular biology Molecular cloning of genes
SUPPLEMENTARY INFORMATION
 b Supplementary Figure 1. Analysis of Vkg-Dpp interactions. a, In vitro pulldown showing binding of Dpp-HA to the GST-VkgC fusion protein. No interaction is detected between denatured Dpp-HA and GST-Vkg.
b Supplementary Figure 1. Analysis of Vkg-Dpp interactions. a, In vitro pulldown showing binding of Dpp-HA to the GST-VkgC fusion protein. No interaction is detected between denatured Dpp-HA and GST-Vkg.
Targeted modification of gene function exploiting homology directed repair of TALENmediated double strand breaks in barley
 Targeted modification of gene function exploiting homology directed repair of TALENmediated double strand breaks in barley Nagaveni Budhagatapalli a, Twan Rutten b, Maia Gurushidze a, Jochen Kumlehn a,
Targeted modification of gene function exploiting homology directed repair of TALENmediated double strand breaks in barley Nagaveni Budhagatapalli a, Twan Rutten b, Maia Gurushidze a, Jochen Kumlehn a,
Supplementary Materials and Methods:
 Supplementary Materials and Methods: Preclinical chemoprevention experimental design Mice were weighed and each mammary tumor was manually palpated and measured with digital calipers once a week from 4
Supplementary Materials and Methods: Preclinical chemoprevention experimental design Mice were weighed and each mammary tumor was manually palpated and measured with digital calipers once a week from 4
Supporting Information
 Supporting Information Chakrabarty et al. 10.1073/pnas.1018001108 SI Materials and Methods Cell Lines. All cell lines were purchased from the American Type Culture Collection. Media and FBS were purchased
Supporting Information Chakrabarty et al. 10.1073/pnas.1018001108 SI Materials and Methods Cell Lines. All cell lines were purchased from the American Type Culture Collection. Media and FBS were purchased
GFP CCD2 GFP IP:GFP
 D1 D2 1 75 95 148 178 492 GFP CCD1 CCD2 CCD2 GFP D1 D2 GFP D1 D2 Beclin 1 IB:GFP IP:GFP Supplementary Figure 1: Mapping domains required for binding to HEK293T cells are transfected with EGFP-tagged mutant
D1 D2 1 75 95 148 178 492 GFP CCD1 CCD2 CCD2 GFP D1 D2 GFP D1 D2 Beclin 1 IB:GFP IP:GFP Supplementary Figure 1: Mapping domains required for binding to HEK293T cells are transfected with EGFP-tagged mutant
Revision Checklist for Science Signaling Research Manuscripts: Data Requirements and Style Guidelines
 Revision Checklist for Science Signaling Research Manuscripts: Data Requirements and Style Guidelines Further information can be found at: http://stke.sciencemag.org/sites/default/files/researcharticlerevmsinstructions_0.pdf.
Revision Checklist for Science Signaling Research Manuscripts: Data Requirements and Style Guidelines Further information can be found at: http://stke.sciencemag.org/sites/default/files/researcharticlerevmsinstructions_0.pdf.
Gene Sequence Fragment size ΔEGFR F 5' GGGCTCTGGAGGAAAAGAAAG GT 3' 116 bp R 5' CTTCTTACACTTGCGGACGC 3'
 Supplementary Table 1: Real-time PCR primer sequences for ΔEGFR, wtegfr, IL-6, LIF and GAPDH. Gene Sequence Fragment size ΔEGFR F 5' GGGCTCTGGAGGAAAAGAAAG GT 3' 116 bp R 5' CTTCTTACACTTGCGGACGC 3' wtegfr
Supplementary Table 1: Real-time PCR primer sequences for ΔEGFR, wtegfr, IL-6, LIF and GAPDH. Gene Sequence Fragment size ΔEGFR F 5' GGGCTCTGGAGGAAAAGAAAG GT 3' 116 bp R 5' CTTCTTACACTTGCGGACGC 3' wtegfr
Isolation, culture, and transfection of primary mammary epithelial organoids
 Supplementary Experimental Procedures Isolation, culture, and transfection of primary mammary epithelial organoids Primary mammary epithelial organoids were prepared from 8-week-old CD1 mice (Charles River)
Supplementary Experimental Procedures Isolation, culture, and transfection of primary mammary epithelial organoids Primary mammary epithelial organoids were prepared from 8-week-old CD1 mice (Charles River)
Lectures 28 and 29 applications of recombinant technology I. Manipulate gene of interest
 Lectures 28 and 29 applications of recombinant technology I. Manipulate gene of interest C A. site-directed mutagenesis A C A T A DNA B. in vitro mutagenesis by PCR T A 1. anneal primer 1 C A 1. fill in
Lectures 28 and 29 applications of recombinant technology I. Manipulate gene of interest C A. site-directed mutagenesis A C A T A DNA B. in vitro mutagenesis by PCR T A 1. anneal primer 1 C A 1. fill in
Supplemental Figure 1. Mutation in NLA Causes Increased Pi Uptake Activity and
 Supplemental Figure 1. Mutation in NLA Causes Increased Pi Uptake Activity and PHT1 Protein Amounts. (A) Shoot morphology of 19-day-old nla mutants under Pi-sufficient conditions. (B) [ 33 P]Pi uptake
Supplemental Figure 1. Mutation in NLA Causes Increased Pi Uptake Activity and PHT1 Protein Amounts. (A) Shoot morphology of 19-day-old nla mutants under Pi-sufficient conditions. (B) [ 33 P]Pi uptake
Basic Concepts and History of Genetic Engineering. Mitesh Shrestha
 Basic Concepts and History of Genetic Engineering Mitesh Shrestha Genetic Engineering AKA gene manipulation, gene cloning, recombinant DNA technology, genetic modification, and the new genetics. A technique
Basic Concepts and History of Genetic Engineering Mitesh Shrestha Genetic Engineering AKA gene manipulation, gene cloning, recombinant DNA technology, genetic modification, and the new genetics. A technique
Gene Expression Technology
 Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
A subclass of HSP70s regulate development and abiotic stress responses in Arabidopsis thaliana
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 Journal of Plant Research A subclass of HSP70s regulate development and abiotic stress responses in Arabidopsis thaliana Linna Leng 1 Qianqian Liang
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 Journal of Plant Research A subclass of HSP70s regulate development and abiotic stress responses in Arabidopsis thaliana Linna Leng 1 Qianqian Liang
Aneuploidy causes premature differentiation of neural and intestinal stem cells
 Received 3 Apr 15 Accepted 14 Oct 15 Published 17 Nov 15 DOI:.38/ncomms9894 OPEN Aneuploidy causes premature differentiation of neural and intestinal stem cells Delphine Gogendeau 1, Katarzyna Siudeja
Received 3 Apr 15 Accepted 14 Oct 15 Published 17 Nov 15 DOI:.38/ncomms9894 OPEN Aneuploidy causes premature differentiation of neural and intestinal stem cells Delphine Gogendeau 1, Katarzyna Siudeja
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
7.06 Problem Set #3, Spring 2005
 7.06 Problem Set #3, Spring 2005 1. The Drosophila compound eye is composed of about 800 units called ommatidia. Each ommatidium contains eight photoreceptor neurons (R1 through R8), which develop in a
7.06 Problem Set #3, Spring 2005 1. The Drosophila compound eye is composed of about 800 units called ommatidia. Each ommatidium contains eight photoreceptor neurons (R1 through R8), which develop in a
Supplemental Information. Stratum, a Homolog of the Human GEF Mss4, Partnered with Rab8, Controls the Basal Restriction
 Cell Reports, Volume 18 Supplemental Information Stratum, a Homolog of the Human GEF Mss4, Partnered with Rab8, Controls the Basal Restriction of Basement Membrane Proteins in Epithelial Cells Olivier
Cell Reports, Volume 18 Supplemental Information Stratum, a Homolog of the Human GEF Mss4, Partnered with Rab8, Controls the Basal Restriction of Basement Membrane Proteins in Epithelial Cells Olivier
A CRISPR/Cas9 Vector System for Tissue-Specific Gene Disruption in Zebrafish
 Developmental Cell Supplemental Information A CRISPR/Cas9 Vector System for Tissue-Specific Gene Disruption in Zebrafish Julien Ablain, Ellen M. Durand, Song Yang, Yi Zhou, and Leonard I. Zon % larvae
Developmental Cell Supplemental Information A CRISPR/Cas9 Vector System for Tissue-Specific Gene Disruption in Zebrafish Julien Ablain, Ellen M. Durand, Song Yang, Yi Zhou, and Leonard I. Zon % larvae
TE5 KYSE510 TE7 KYSE70 KYSE140
 TE5 KYSE5 TT KYSE7 KYSE4 Supplementary Figure. Hockey stick plots showing input normalized, rank ordered H3K7ac signals for the candidate SE-associated lncrnas in this study. Rpm Rpm Rpm Chip-seq H3K7ac
TE5 KYSE5 TT KYSE7 KYSE4 Supplementary Figure. Hockey stick plots showing input normalized, rank ordered H3K7ac signals for the candidate SE-associated lncrnas in this study. Rpm Rpm Rpm Chip-seq H3K7ac
A Low salt diet. C Low salt diet + mf4-31c1 3. D High salt diet + mf4-31c1 3. B High salt diet
 A Low salt diet GV [AU]:. [mmhg]: 0..... 09 9 7 B High salt diet GV [AU]:. [mmhg]: 7 8...0. 7.8 8.. 0 8 7 C Low salt diet + mf-c GV [AU]:. [mmhg]:.0.9 8.7.7. 7 8 0 D High salt diet + mf-c E Lymph capillary
A Low salt diet GV [AU]:. [mmhg]: 0..... 09 9 7 B High salt diet GV [AU]:. [mmhg]: 7 8...0. 7.8 8.. 0 8 7 C Low salt diet + mf-c GV [AU]:. [mmhg]:.0.9 8.7.7. 7 8 0 D High salt diet + mf-c E Lymph capillary
Supplementary Figure 1
 Supplementary Figure 1 Ex2 promotor region Cre IRES cherry pa Ex4 Ex5 Ex1 untranslated Ex3 Ex5 untranslated EYFP pa Rosa26 STOP loxp loxp Cre recombinase EYFP pa Rosa26 loxp 1 kb Interleukin-9 fate reporter
Supplementary Figure 1 Ex2 promotor region Cre IRES cherry pa Ex4 Ex5 Ex1 untranslated Ex3 Ex5 untranslated EYFP pa Rosa26 STOP loxp loxp Cre recombinase EYFP pa Rosa26 loxp 1 kb Interleukin-9 fate reporter
MICB688L/MOCB639 Advanced Cell Biology Exam II
 MICB688L/MOCB639 Advanced Cell Biology Exam II May 10, 2001 Name: 1. Briefly describe the four major classes of cell surface receptors and their modes of action (immediate downstream only) (8) 2. Please
MICB688L/MOCB639 Advanced Cell Biology Exam II May 10, 2001 Name: 1. Briefly describe the four major classes of cell surface receptors and their modes of action (immediate downstream only) (8) 2. Please
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature09732 Supplementary Figure 1: Depletion of Fbw7 results in elevated Mcl-1 abundance. a, Total thymocytes from 8-wk-old Lck-Cre/Fbw7 +/fl (Control) or Lck-Cre/Fbw7 fl/fl (Fbw7 KO) mice
doi:10.1038/nature09732 Supplementary Figure 1: Depletion of Fbw7 results in elevated Mcl-1 abundance. a, Total thymocytes from 8-wk-old Lck-Cre/Fbw7 +/fl (Control) or Lck-Cre/Fbw7 fl/fl (Fbw7 KO) mice
Molecular Cell Biology - Problem Drill 11: Recombinant DNA
 Molecular Cell Biology - Problem Drill 11: Recombinant DNA Question No. 1 of 10 1. Which of the following statements about the sources of DNA used for molecular cloning is correct? Question #1 (A) cdna
Molecular Cell Biology - Problem Drill 11: Recombinant DNA Question No. 1 of 10 1. Which of the following statements about the sources of DNA used for molecular cloning is correct? Question #1 (A) cdna
7.06 Cell Biology EXAM #2 March 20, 2003
 7.06 Cell Biology EXAM #2 March 20, 2003 This is an open book exam, and you are allowed access to books, a calculator, and notes but not computers or any other types of electronic devices. Please write
7.06 Cell Biology EXAM #2 March 20, 2003 This is an open book exam, and you are allowed access to books, a calculator, and notes but not computers or any other types of electronic devices. Please write
Peter Dy, Weihuan Wang, Pallavi Bhattaram, Qiuqing Wang, Lai Wang, R. Tracy Ballock, and Véronique Lefebvre
 Developmental Cell, Volume 22 Supplemental Information Sox9 Directs Hypertrophic Maturation and Blocks Osteoblast Differentiation of Growth Plate Chondrocytes Peter Dy, Weihuan Wang, Pallavi Bhattaram,
Developmental Cell, Volume 22 Supplemental Information Sox9 Directs Hypertrophic Maturation and Blocks Osteoblast Differentiation of Growth Plate Chondrocytes Peter Dy, Weihuan Wang, Pallavi Bhattaram,
Supplementary Figure 1: Expression of RNF8, HERC2 and NEURL4 in the cerebellum and knockdown of RNF8 by RNAi (a) Lysates of the cerebellum from rat
 Supplementary Figure 1: Expression of RNF8, HERC2 and NEURL4 in the cerebellum and knockdown of RNF8 by RNAi (a) Lysates of the cerebellum from rat pups at P6, P14, P22, P30 and adult (A) rats were subjected
Supplementary Figure 1: Expression of RNF8, HERC2 and NEURL4 in the cerebellum and knockdown of RNF8 by RNAi (a) Lysates of the cerebellum from rat pups at P6, P14, P22, P30 and adult (A) rats were subjected
Candidate region (0.74 Mb) ATC TCT GGG ACT CAT GAG CAG GAG GCT AGC ATC TCT GGG ACT CAT TAG CAG GAG GCT AGC
 A idm-3 idm-3 B Physical distance (Mb) 4.6 4.86.6 8.4 C Chr.3 Recom. Rate (%) ATG 3.9.9.9 9.74 Candidate region (.74 Mb) n=4 TAA D idm-3 G3T(E4) G4A(W988) WT idm-3 ATC TCT GGG ACT CAT GAG CAG GAG GCT AGC
A idm-3 idm-3 B Physical distance (Mb) 4.6 4.86.6 8.4 C Chr.3 Recom. Rate (%) ATG 3.9.9.9 9.74 Candidate region (.74 Mb) n=4 TAA D idm-3 G3T(E4) G4A(W988) WT idm-3 ATC TCT GGG ACT CAT GAG CAG GAG GCT AGC
GTTCGGGTTCC TTTTGAGCAG
 Supplementary Figures Splice variants of the SIP1 transcripts play a role in nodule organogenesis in Lotus japonicus. Wang C, Zhu H, Jin L, Chen T, Wang L, Kang H, Hong Z, Zhang Z. 5 UTR CDS 3 UTR TCTCAACCATCCTTTGTCTGCTTCCGCCGCATGGGTGAGGTCATTTTGTCTAGATGACGTGCAATTTACAATGA
Supplementary Figures Splice variants of the SIP1 transcripts play a role in nodule organogenesis in Lotus japonicus. Wang C, Zhu H, Jin L, Chen T, Wang L, Kang H, Hong Z, Zhang Z. 5 UTR CDS 3 UTR TCTCAACCATCCTTTGTCTGCTTCCGCCGCATGGGTGAGGTCATTTTGTCTAGATGACGTGCAATTTACAATGA
Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon Song, Richard M. Amasino, Bosl Noh, and Yoo-Sun Noh
 Developmental Cell, Volume 22 Supplemental Information Control of Seed Germination by Light-Induced Histone Arginine Demethylation Activity Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon
Developmental Cell, Volume 22 Supplemental Information Control of Seed Germination by Light-Induced Histone Arginine Demethylation Activity Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon
Wnt16 smact merge VK/AB
 A WT Wnt6 smact merge VK/A KO ctrl IgG WT KO Wnt6 smact DAPI SUPPLEMENTAL FIGURE I: Wnt6 expression in MGP-deficient aortae. Immunostaining for Wnt6 and smooth muscle actin (smact) in aortae from 7 day
A WT Wnt6 smact merge VK/A KO ctrl IgG WT KO Wnt6 smact DAPI SUPPLEMENTAL FIGURE I: Wnt6 expression in MGP-deficient aortae. Immunostaining for Wnt6 and smooth muscle actin (smact) in aortae from 7 day
Enhancers mutations that make the original mutant phenotype more extreme. Suppressors mutations that make the original mutant phenotype less extreme
 Interactomics and Proteomics 1. Interactomics The field of interactomics is concerned with interactions between genes or proteins. They can be genetic interactions, in which two genes are involved in the
Interactomics and Proteomics 1. Interactomics The field of interactomics is concerned with interactions between genes or proteins. They can be genetic interactions, in which two genes are involved in the
Supplemental Data Supplemental Figure 1.
 Supplemental Data Supplemental Figure 1. Silique arrangement in the wild-type, jhs, and complemented lines. Wild-type (WT) (A), the jhs1 mutant (B,C), and the jhs1 mutant complemented with JHS1 (Com) (D)
Supplemental Data Supplemental Figure 1. Silique arrangement in the wild-type, jhs, and complemented lines. Wild-type (WT) (A), the jhs1 mutant (B,C), and the jhs1 mutant complemented with JHS1 (Com) (D)
Rejuvenation of the muscle stem cell population restores strength to injured aged muscles
 Rejuvenation of the muscle stem cell population restores strength to injured aged muscles Benjamin D Cosgrove, Penney M Gilbert, Ermelinda Porpiglia, Foteini Mourkioti, Steven P Lee, Stephane Y Corbel,
Rejuvenation of the muscle stem cell population restores strength to injured aged muscles Benjamin D Cosgrove, Penney M Gilbert, Ermelinda Porpiglia, Foteini Mourkioti, Steven P Lee, Stephane Y Corbel,
LS4 final exam. Problem based, similar in style and length to the midterm. Articles: just the information covered in class
 LS4 final exam Problem based, similar in style and length to the midterm Articles: just the information covered in class Complementation and recombination rii and others Neurospora haploid spores, heterokaryon,
LS4 final exam Problem based, similar in style and length to the midterm Articles: just the information covered in class Complementation and recombination rii and others Neurospora haploid spores, heterokaryon,
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or
 Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
Chemically defined conditions for human ipsc derivation and culture
 Nature Methods Chemically defined conditions for human ipsc derivation and culture Guokai Chen, Daniel R Gulbranson, Zhonggang Hou, Jennifer M Bolin, Victor Ruotti, Mitchell D Probasco, Kimberly Smuga-Otto,
Nature Methods Chemically defined conditions for human ipsc derivation and culture Guokai Chen, Daniel R Gulbranson, Zhonggang Hou, Jennifer M Bolin, Victor Ruotti, Mitchell D Probasco, Kimberly Smuga-Otto,
Supplemental Information. Role of phosphatase of regenerating liver 1 (PRL1) in spermatogenesis
 Supplemental Information Role of phosphatase of regenerating liver 1 (PRL1) in spermatogenesis Yunpeng Bai ;, Lujuan Zhang #, Hongming Zhou #, Yuanshu Dong #, Qi Zeng, Weinian Shou, and Zhong-Yin Zhang
Supplemental Information Role of phosphatase of regenerating liver 1 (PRL1) in spermatogenesis Yunpeng Bai ;, Lujuan Zhang #, Hongming Zhou #, Yuanshu Dong #, Qi Zeng, Weinian Shou, and Zhong-Yin Zhang
Problem Set 8. Answer Key
 MCB 102 University of California, Berkeley August 11, 2009 Isabelle Philipp Online Document Problem Set 8 Answer Key 1. The Genetic Code (a) Are all amino acids encoded by the same number of codons? no
MCB 102 University of California, Berkeley August 11, 2009 Isabelle Philipp Online Document Problem Set 8 Answer Key 1. The Genetic Code (a) Are all amino acids encoded by the same number of codons? no
Supplemental Data. The Survival Advantage of Olfaction. in a Competitive Environment. Supplemental Experimental Procedures
 Supplemental Data The Survival Advantage of Olfaction in a Competitive Environment Kenta Asahina, Viktoryia Pavlenkovich, and Leslie B. Vosshall Supplemental Experimental Procedures Experimental animals
Supplemental Data The Survival Advantage of Olfaction in a Competitive Environment Kenta Asahina, Viktoryia Pavlenkovich, and Leslie B. Vosshall Supplemental Experimental Procedures Experimental animals
7.06 Cell Biology QUIZ #3
 Recitation Section: 7.06 Cell Biology QUIZ #3 This is an open book exam, and you are allowed access to books and notes, but not computers or any other types of electronic devices. Please write your answers
Recitation Section: 7.06 Cell Biology QUIZ #3 This is an open book exam, and you are allowed access to books and notes, but not computers or any other types of electronic devices. Please write your answers
AmoyDx TM JAK2 V617F Mutation Detection Kit
 AmoyDx TM JAK2 V617F Mutation Detection Kit Detection of V617F mutation in the JAK2 oncogene Instructions For Use Instructions Version: B1.0 Date of Revision: July 2012 Store at -20±2 o C -1/5- Background
AmoyDx TM JAK2 V617F Mutation Detection Kit Detection of V617F mutation in the JAK2 oncogene Instructions For Use Instructions Version: B1.0 Date of Revision: July 2012 Store at -20±2 o C -1/5- Background
Unit 6: Molecular Genetics & DNA Technology Guided Reading Questions (100 pts total)
 Name: AP Biology Biology, Campbell and Reece, 7th Edition Adapted from chapter reading guides originally created by Lynn Miriello Chapter 16 The Molecular Basis of Inheritance Unit 6: Molecular Genetics
Name: AP Biology Biology, Campbell and Reece, 7th Edition Adapted from chapter reading guides originally created by Lynn Miriello Chapter 16 The Molecular Basis of Inheritance Unit 6: Molecular Genetics
SUPPLEMENTAL INFORMATION
 UTX/KDM6A demethylase activity is required for satellite cell-mediated muscle regeneration Hervé Faralli 1,2, Chaochen Wang 3, Kiran Nakka 1,2, Soji Sebastian 1,, Aissa Benyoucef 1,2, Lenan Zhuang 3, Alphonse
UTX/KDM6A demethylase activity is required for satellite cell-mediated muscle regeneration Hervé Faralli 1,2, Chaochen Wang 3, Kiran Nakka 1,2, Soji Sebastian 1,, Aissa Benyoucef 1,2, Lenan Zhuang 3, Alphonse
AmoyDx JAK2 Mutation Detection Kit
 AmoyDx JAK2 Mutation Detection Kit Detection of V617F mutation in the JAK2 oncogene Instructions For Use For Research Use Only and For Reference Only Instructions Version: B1.2 Date of Revision: June 2016
AmoyDx JAK2 Mutation Detection Kit Detection of V617F mutation in the JAK2 oncogene Instructions For Use For Research Use Only and For Reference Only Instructions Version: B1.2 Date of Revision: June 2016
Analysis of the Drosophila EGFR responsive gene CG4096, which encodes an ADAMTS protein. Research Thesis
 Analysis of the Drosophila EGFR responsive gene CG4096, which encodes an ADAMTS protein Research Thesis Presented in partial fulfillment of the requirements for graduation with research distinction in
Analysis of the Drosophila EGFR responsive gene CG4096, which encodes an ADAMTS protein Research Thesis Presented in partial fulfillment of the requirements for graduation with research distinction in
Drosophila Neuroligin 4 regulates NMJ growth via BMP pathway
 JBC Papers in Press. Published on September 14, 2017 as Manuscript M117.810242 The latest version is at http://www.jbc.org/cgi/doi/10.1074/jbc.m117.810242 Drosophila Neuroligin 4 regulates NMJ growth via
JBC Papers in Press. Published on September 14, 2017 as Manuscript M117.810242 The latest version is at http://www.jbc.org/cgi/doi/10.1074/jbc.m117.810242 Drosophila Neuroligin 4 regulates NMJ growth via
Online Supplementary Information
 Online Supplementary Information NLRP4 negatively regulates type I interferon signaling by targeting TBK1 for degradation via E3 ubiquitin ligase DTX4 Jun Cui 1,4,6,7, Yinyin Li 1,5,6,7, Liang Zhu 1, Dan
Online Supplementary Information NLRP4 negatively regulates type I interferon signaling by targeting TBK1 for degradation via E3 ubiquitin ligase DTX4 Jun Cui 1,4,6,7, Yinyin Li 1,5,6,7, Liang Zhu 1, Dan
Supplementary Material. TRIB3 inhibits proliferation and promotes osteogenesis in hbmscs by regulating the. ERK1/2 signaling pathway
 Supplementary Material TRIB3 inhibits proliferation and promotes osteogenesis in hbmscs by regulating the ERK1/2 signaling pathway Cui Zhang 1, Fan-Fan Hong 1, Cui-Cui Wang 1, Liang Li 1, Jian-Ling Chen
Supplementary Material TRIB3 inhibits proliferation and promotes osteogenesis in hbmscs by regulating the ERK1/2 signaling pathway Cui Zhang 1, Fan-Fan Hong 1, Cui-Cui Wang 1, Liang Li 1, Jian-Ling Chen
Supporting Information
 Supporting Information Yuan et al. 10.1073/pnas.0906869106 Fig. S1. Heat map showing that Populus ICS is coregulated with orthologs of Arabidopsis genes involved in PhQ biosynthesis and PSI function, but
Supporting Information Yuan et al. 10.1073/pnas.0906869106 Fig. S1. Heat map showing that Populus ICS is coregulated with orthologs of Arabidopsis genes involved in PhQ biosynthesis and PSI function, but
Technical Review. Real time PCR
 Technical Review Real time PCR Normal PCR: Analyze with agarose gel Normal PCR vs Real time PCR Real-time PCR, also known as quantitative PCR (qpcr) or kinetic PCR Key feature: Used to amplify and simultaneously
Technical Review Real time PCR Normal PCR: Analyze with agarose gel Normal PCR vs Real time PCR Real-time PCR, also known as quantitative PCR (qpcr) or kinetic PCR Key feature: Used to amplify and simultaneously
Supplemental Data. Aung et al. (2011). Plant Cell /tpc
 35S pro:pmd1-yfp 10 µm Supplemental Figure 1. -terminal YFP fusion of PMD1 (PMD1-YFP, in green) is localized to the cytosol. grobacterium cells harboring 35S pro :PMD1-YFP were infiltrated into tobacco
35S pro:pmd1-yfp 10 µm Supplemental Figure 1. -terminal YFP fusion of PMD1 (PMD1-YFP, in green) is localized to the cytosol. grobacterium cells harboring 35S pro :PMD1-YFP were infiltrated into tobacco
Problem Set 4
 7.016- Problem Set 4 Question 1 Arginine biosynthesis is an example of multi-step biochemical pathway where each step is catalyzed by a specific enzyme (E1, E2 and E3) as is outlined below. E1 E2 E3 A
7.016- Problem Set 4 Question 1 Arginine biosynthesis is an example of multi-step biochemical pathway where each step is catalyzed by a specific enzyme (E1, E2 and E3) as is outlined below. E1 E2 E3 A
ASPP1 Fw GGTTGGGAATCCACGTGTTG ASPP1 Rv GCCATATCTTGGAGCTCTGAGAG
 Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected
 Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected with the sirna against lnc-2, lnc-6, lnc-7, and the
Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected with the sirna against lnc-2, lnc-6, lnc-7, and the
