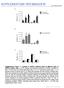
Nature Biotechnology: doi: /nbt Supplementary Figure 1. In vitro validation of OTC sgrnas and donor template.
|
|
|
- Augustine Cox
- 5 years ago
- Views:
Transcription
1 Supplementary Figure 1 In vitro validation of OTC sgrnas and donor template. (a) In vitro validation of sgrnas targeted to OTC in the MC57G mouse cell line by transient transfection followed by 4-day puromycin enrichment and SURVEYOR nuclease assays. sgrna1 showed the highest efficiency in inducing indels in the targeted loci and was therefore chosen for subsequent studies. Arrows denote SURVEYOR nuclease cleaved fragments of the OTC PCR products. Results were replicated in 2 independent experiments. (b) In vitro validation of OTC donor template. MC57G cells were transiently transfected with a plasmid co-expressing OTC sgrna1, SaCas9, and an AgeI restriction site tagged OTC donor plasmid followed by 4-day puromycin enrichment. RFLP analysis was performed following AgeI digestion to detect HDR in vitro. Co-transfection of the AgeItagged OTC donor template with an SaCas9 plasmid without OTC sgrna1 did not result in detectable HDR. Arrows denote AgeIsensitive cleavage products resulting from HDR. Results were replicated in 2 independent experiments. Indel and HDR frequency were calculated based on band intensities 31.
2 Supplementary Figure 2 Vector dose optimization to improve in vivo gene correction. Postnatal day 2 spf ash pups received temporal vein injection of 5x10 10 GC AAV8.SaCas9 and either 5x10 10 (n=5), 1x10 11 (n=3), or 5x10 11 (n=5) GC of AAV8.sgRNA1.donor vector. Liver samples were collected 3 weeks post vector treatment for analysis. (a) Quantification of gene correction based on the percentage of area on liver sections expressing OTC by immunostaining. (b) Quantification of OTC mrna levels in the liver by RT-qPCR using primers spanning exons 4 5 to amplify wild-type OTC. Mean ± SEM are shown. ** P<0.01, Dunnett s test.
3 Supplementary Figure 3 Time course of gene expression by Western analysis and HDR analysis by RFLP. (a) HDR analysis by RFLP. OTC target region was PCR amplified from the liver genomic DNA isolated from untreated spf ash mice or spf ash mice treated with the dual AAV vectors. Untreated spf ash control samples were collected at 8 weeks of age. Samples from the treated spf ash mice were collected at 1, 3, and 8 weeks (n=3 animals per time point) following neonatal injection of the dual AAV8 vectors. Targeted animals received AAV8.SaCas9 (5x10 10 GC/pup) and AAV8.sgRNA1.donor (5x10 11 GC/pup). Untargeted animals received AAV8.SaCas9 (5x10 10 GC/pup) and AAV8.control.donor (5x10 11 GC/pup). AgeI digestion was performed and estimated HDR percentages are shown. (b) Western blot analysis. Liver lysates were prepared from untreated WT and spf ash mice or spf ash mice treated with the dual AAV vectors for detection of FLAG-SaCas9 and OTC protein.
4 Supplementary Figure 4 Examination of liver toxicity in animals treated with AAV8.CRISPR-SaCas9 dual vectors. (a) Histological analysis on livers harvested 3 and 8 weeks following the dual vector treatment. Scale bar, 100 µm. (b) Liver transaminase levels in untreated spf ash mice (n=9) or 8 weeks following dual vector treatment. Untargeted mice received 5x10 10 GC AAV8.SaCas9 and 5x10 11 of AAV8.control.donor vectors (n=8), while gene-targeted mice received 5x10 10 GC AAV8.SaCas9 and 5x10 11 GC of AAV8.sgRNA1.donor (n=7). Mean ± SEM are shown. There were no statistically significant differences between groups, Dunnett s test.
5 Supplementary Figure 5 Examination of liver toxicity in adult animals treated with AAV8.CRISPR-SaCas9 dual vectors. (a) Histological analysis on livers harvested 3 weeks (low-dose) or 2 weeks (high-dose) following dual vector treatment. Scale bar, 100 µm. (b) Liver transaminase levels in untreated spf ash mice, or 3 weeks following low-dose dual vector treatment, or 2 weeks following high-dose dual vector treatment (n=3 for each group). Low-dose, untargeted mice received 1x10 11 GC AAV8.SaCas9 and 1x10 12 GC of AAV8.control.donor vectors, while low-dose, gene-targeted mice received 1x10 11 GC AAV8.SaCas9 and 1x10 12 GC of AAV8.sgRNA1.donor. High-dose, untargeted mice received 1x10 12 GC AAV8.SaCas9 and 5x10 12 GC of AAV8.control.donor vectors, while high-dose, gene-targeted mice received 1x10 12 GC AAV8.SaCas9 and 5x10 12 GC of AAV8.sgRNA1.donor. Mean ± SEM are shown. Adult animals received high-dose, gene-targeted vectors showed a trend of elevated ALT and AST levels, although not statistically different when compared with other groups (Dunnett s test).
6 Supplementary Figure 6 Comparison of SaCas9 vector DNA and mrna levels in the livers of neonatal treated and adult treated mice. Neonatal spf ash mice received 5x10 10 GC AAV8.SaCas9 and 5x10 11 GC of AAV8.sgRNA1.donor vectors and were sacrificed at 1 (n=5), 3 (n=6), and 8 weeks (n=7) after injection. Low-dose adult spf ash mice received 1x10 11 GC AAV8.SaCas9 and 1x10 12 GC of AAV8.sgRNA1.donor vectors and were sacrificed at 3 weeks (n=3) after injection. High-dose adult spf ash mice received 1x10 12 GC AAV8.SaCas9 and 5x10 12 GC of AAV8.sgRNA1.donor vectors and were sacrificed at 2 weeks (n=3) after injection. (a) Quantification of SaCas9 vector DNA in the liver by qpcr. (b) Quantification of SaCas9 mrna in the liver by RT-qPCR. Mean ± SEM are shown.
7 Supplementary Table 1. Summary of the frequencies of indels, correction of OTC spf ash mutation, and HDR in animals treated with the dual AAV gene-targeting vectors. ID Treatment Time after treatment (week) Indel (%) OTC reads with a G (%) Reads with Perfect HDR (%) Reads with Partial HDR (%) 4001 Neonatal, sgrna Neonatal, sgrna Neonatal, sgrna Neonatal, sgrna Neonatal, sgrna Neonatal, sgrna Adult, untargeted, low dose Adult, sgrna1, low dose Adult, sgrna1, low dose Adult, sgrna1, low dose Adult, untargeted, high dose Adult, sgrna1, high dose Adult, sgrna1, high dose Adult, sgrna1, high dose Untreated spf ash n/a
8 Supplementary Table 2. A full list of simple indels (insertions or deletions) detected by deep sequencing on the OTC target region in the liver of two spf ash mice (#4001 and #4003) 3 weeks after neonatal injection of the sgrna1 dual AAV vectors, and two adult spf ash mice (#648 and #653) 2 weeks after injection of high-dose sgrna1 dual AAV vectors. The rates of complex indels (insertions + deletions) are also indicated. Please see attached Excel file.
9 Supplementary Table 3. Deletion sizes identified by on-target deep sequencing and their distribution in neonatal-treated and adult-treated animals. Size range Neonatal (3w) Adult (high-dose, 2w) #4001 #4003 #648 # bp bp bp bp bp bp bp bp bp bp >100 bp Indel overlap with Exon
10 Supplementary Table 4. Off-target analysis. Potential off-target sequences for sgrna1 identified and scored by Benchling s offtarget analysis. ID Sequence PAM Score Chromosome Strand Position Mismatches On-target Indel (treated) Indel (untreated ctl) sgrna1 TCTCTTTTAAACTAACCCAT CAGAG 100 X TRUE OT1 TATCTTTTCAACTAACCCAA TAGGA FALSE OT2 TTTCTTTTATACTAGCCCAT AGGGG X FALSE OT3 TCTTTTTTTAAATAACCCAT TAGAA FALSE OT4 TGGCATTTAAACTAACCCAA CTGGG FALSE OT5 TCTTCATGAAACTAACCCAT CTGAA FALSE OT6 ATTTTTTTAAACAAACCCAT CTGGA FALSE OT7 GATCTTGTAATCTAACCCAT GTGAA FALSE OT8 CCTTTTATAATCTAACCCAT GTGAA FALSE OT9 TCTGTTTTAAAGTAACCCTT CAGGG FALSE OT10 TTGCTTTTCAACTAACCCAA GTGGG FALSE OT11 AATATTTTAAACTATCCCAT GTGAG FALSE OT12 TTACTTTAAAACTATCCCAT CTGAA X FALSE OT13 ACACTTTTAAAATAACCCAG ATGAA FALSE OT14 AATCTTCTAAACCAACCCAT TGGAG FALSE OT15 TCTTTATTAGACTAACCCAG CTGAG X FALSE OT16 TTTCATTTAACATAACCCAT ATGGA FALSE
11 Supplementary Table 5. Primers and sequences for construction and analysis of the donor template and sgrna plasmids. Name Sequence (5'->3') Note WT sequence GAAAGTCTCACAGACACCGCTCGGTTTGTAAAACTTTTCTTCCTTCCAAAGTTT ATTTCAAACTCTGATGGGTTAGTTTAAAAGAGAAGATG Part of OTC exon 4 and intron 4 spf ash sequence OTC donor sequence for sgrna1 ( bp) OTC sgrna1_fwd OTC sgrna1_rev OTC sgrna2_fwd OTC sgrna2_rev OTC sgrna3_fwd OTC sgrna3_rev HDR-Fwd HDR-Rev OTC_PointM_F OTC_PointM_R GAAAGTCTCACAGACACCGCTCAGTTTGTAAAACTTTTCTTCCTTCCAAAGTTT ATTTCAAACTCTGATGGGTTAGTTTAAAAGAGAAGATG GAAAGTCTCACAGACACCGCTCGGTTTGTAAAACTTTTCTTCCTTCCAAAGTTTACCGGT ATTTCAATTGGTGATGGGTTAGTTTAAAAGAGAAGATG CACCGTCTCTTTTAAACTAACCCAT AAACATGGGTTAGTTTAAAAGAGAC CACCGCACAAGACATTCACTTGGGT AAACACCCAAGTGAATGTCTTGTGC CACCGAAAGTTTATTTCAAACTCTG AAACCAGAGTTTGAAATAAACTTTC TGGAGCAATTCTGCACATGGA CTTACTGAACATGGCAGTTTCCC GGCTATGCTTGGGAATGTCCT GCTACAGAATGAAAGAGAGGCG spf ash G->A mutation PAM mutation; AgeI Restriction site insertion OTC target sequence 1 OTC target sequence 2 OTC target sequence 3 OTC PCR for RFLP analysis OTC PCR for Surveyor assay
12 Supplementary Table 6. PCR primer sequences for detecting potential off-target effects by deep sequencing. Primer Name Sequence (5'->3') Note OTC_OT1F1 OTC_OT1R1 OTC_OT1F2 OTC_OT1R2 OTC_OT2F1 OTC_OT2R1 OTC_OT2F2 OTC_OT2R2 OTC_OT3F1 OTC_OT3R1 OTC_OT3F2 OTC_OT3R2 OTC_OT4F1 OTC_OT4R1 OTC_OT4F2 OTC_OT4R2 OTC_OT5F1 OTC_OT5R1 OTC_OT5F2 OTC_OT5R2 OTC_OT6F1 OTC_OT6R1 OTC_OT6F2 OTC_OT6R2 OTC_OT7F1 OTC_OT7R1 OTC_OT7F2 OTC_OT7R2 OTC_OT8F1 OTC_OT8R1 OTC_OT8F2 OTC_OT8R2 OTC_OT9F1 OTC_OT9R1 OTC_OT9F2 OTC_OT9R2 CTGGTGCCTTTTTCTATCGCC CCAAGAGCAACTACAATGGCTT GCATTTTCATGAGCATTCCA CATGTTGTGCCTGCATCTCT GCAGACTCCAAGATGCAAGAC GATGTTGTTCCACCCGCATCT CACTGAGCCAAGTCACTGGA AGGGACAAAACCAAACAGCA TGGCCTTCTAAAGCAACCAA CCGTCTCCCAGATCACATGAC ATAACTCATAATCTATGCATGGCACAA TTTGATCATGGTGTTTATCAGAGC TTGAGACCTAGCTCATGCCC TAACGCAGAACTGGCACAGG TCAGCTTCGAATCACACCAG GAATGTGGCATTGGCTTTTT GTCCCCGACAAACCAAGCTA TGAACTGGCAGTATGCAGGG AACATGGTTTCTGCCCTCAG GGACCATGCCGAACTCTTAC GAGAGAGCCAATCTGCCCAT CACCGGAAACGTGTGAGAGA ACTTCCCATGATCCCATTGA AGCTTCCCTCCAAGTGTCCT GATGGGCATAAGCCCGAAGTA TAAGGCCCAGTGTTGTTGTGT GTAGCAGGGGCTCTGTGAAG TGGCCTGAAATACCCAGAAC GTTGAATTCGCGTGTCCAGG TCCCATGGCGAGAATGTCAC CCCTGTAGGAAACACAGAGGA TGCTTTGGATGTTGATTCTAAA GGCAAAGGACTAGCTTGCAC GGGTGCTATGAGGACCAGTG CAGTGGTGTGTGGAGAGCTG AGAGAGAGCGCTTGACTTTGA Primers for OT1 Primers for OT2 Primers for OT3 Primers for OT4 Primers for OT5 Primers for OT6 Primers for OT7 Primers for OT8 Primers for OT9
13 Primer Name Sequence (5'->3') Note OTC_OT10F1 OTC_OT10R1 OTC_OT10F2 OTC_OT10R2 OTC_OT11F1 OTC_OT11R1 OTC_OT11F2 OTC_OT11R2 OTC_OT12F OTC_OT12R OTC_OT13F1 OTC_OT13R1 OTC_OT13F2 OTC_OT13R2 OTC_OT14F1 OTC_OT14R1 OTC_OT14F2 OTC_OT14R2 OTC_OT15F1 OTC_OT15R1 OTC_OT15F2 OTC_OT15R2 OTC_OT16F1 OTC_OT16R1 OTC_OT16F2 OTC_OT16R2 CTTTGACTCCCGGCGAAAGA TTGTCCATCCGGGTCATTGC AAGTCCTTCTTGCCCAACTT CAGCCCCAATGCATTTTT ATTACAGGTCCTGGTTGGGC ACTGAGCCTGGTAGAGCCTT GGAAGGTGAAGGAAGGAAGG TTTTCTAGGAATTCAGGACATACA TCATGGTCCTTAAAATTTTTGC TCCAGGTATGCAAAGTGGAT TTCAGTTGTACTTTGGATGCTCTGA CATCTGAATAGCAGCAGGCG TAGCACAGCCCAAATGACTT TCATGAAACCCCATAATCAGAA AGTGGGTCATCCTTTGTTACCC TGCCAGTTATCAGCCAAGCA CCCAGGAACTTAACTCAGGTG TGCCATTTGACCTCATAAGTCT TTCAGCCCCCTTGAGTGTTTA GTCTCTGAGCACAAAGAGACGA TTGCCTGTCCCAACTAGAGC GGCCCAAGAATGCACATTTA CCACACACTGGCTAGGACTG ACTGGCAGCACTTGAGACAA GATGGCATGCTGTGGTTTTT CAATGCTTCCACACAGAACC Primers for OT10 Primers for OT11 Primers for OT12 Primers for OT13 Primers for OT14 Primers for OT15 Primers for OT16
A dual AAV system enables the Cas9- mediated correction of a metabolic liver disease in newborn mice
 A dual AAV system enables the Cas9- mediated correction of a metabolic liver disease in newborn mice The Harvard community has made this article openly available. Please share how this access benefits
A dual AAV system enables the Cas9- mediated correction of a metabolic liver disease in newborn mice The Harvard community has made this article openly available. Please share how this access benefits
Nature Methods: doi: /nmeth Supplementary Figure 1
 Supplementary Figure 1 ATP1A1 variants with in-frame deletions are enriched in ouabain-resistant cell populations. (a) Total editing efficacy along with spectrum and frequency of individual indels as determined
Supplementary Figure 1 ATP1A1 variants with in-frame deletions are enriched in ouabain-resistant cell populations. (a) Total editing efficacy along with spectrum and frequency of individual indels as determined
SUPPLEMENTARY INFORMATION
 Gene replacements and insertions in rice by intron targeting using CRISPR Cas9 Table of Contents Supplementary Figure 1. sgrna-induced targeted mutations in the OsEPSPS gene in rice protoplasts. Supplementary
Gene replacements and insertions in rice by intron targeting using CRISPR Cas9 Table of Contents Supplementary Figure 1. sgrna-induced targeted mutations in the OsEPSPS gene in rice protoplasts. Supplementary
Generation of App knock-in mice reveals deletion mutations protective against Alzheimer s. disease-like pathology. Nagata et al.
 Generation of App knock-in mice reveals deletion mutations protective against Alzheimer s disease-like pathology Nagata et al. Supplementary Fig 1. Previous App knock-in model did not show Aβ accumulation
Generation of App knock-in mice reveals deletion mutations protective against Alzheimer s disease-like pathology Nagata et al. Supplementary Fig 1. Previous App knock-in model did not show Aβ accumulation
CRISPR RNA-guided activation of endogenous human genes
 CRISPR RNA-guided activation of endogenous human genes Morgan L Maeder, Samantha J Linder, Vincent M Cascio, Yanfang Fu, Quan H Ho, J Keith Joung Supplementary Figure 1 Comparison of VEGF activation induced
CRISPR RNA-guided activation of endogenous human genes Morgan L Maeder, Samantha J Linder, Vincent M Cascio, Yanfang Fu, Quan H Ho, J Keith Joung Supplementary Figure 1 Comparison of VEGF activation induced
Supplementary Figure 1. Quantitative RT-PCR experimental validation of CRISPR/Cas9 and sgrnas expression in HEK293A transfected cells.
 Supplementary Figure 1. Quantitative RT-PCR experimental validation of CRISPR/Cas9 and sgrnas expression in HEK293A transfected cells. HEK293A cells were transfected with the indicated combinations of
Supplementary Figure 1. Quantitative RT-PCR experimental validation of CRISPR/Cas9 and sgrnas expression in HEK293A transfected cells. HEK293A cells were transfected with the indicated combinations of
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/3/4/e1602814/dc1 Supplementary Materials for CRISPR-Cpf1 correction of muscular dystrophy mutations in human cardiomyocytes and mice Yu Zhang, Chengzu Long, Hui
advances.sciencemag.org/cgi/content/full/3/4/e1602814/dc1 Supplementary Materials for CRISPR-Cpf1 correction of muscular dystrophy mutations in human cardiomyocytes and mice Yu Zhang, Chengzu Long, Hui
Nature Biotechnology: doi: /nbt.4166
 Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
Supplementary Figures and Figure legends
 Supplementary Figures and Figure legends 3 Supplementary Figure 1. Conditional targeting construct for the murine Satb1 locus with a modified FLEX switch. Schematic of the wild type Satb1 locus; the conditional
Supplementary Figures and Figure legends 3 Supplementary Figure 1. Conditional targeting construct for the murine Satb1 locus with a modified FLEX switch. Schematic of the wild type Satb1 locus; the conditional
Easi CRISPR for conditional and insertional alleles
 Easi CRISPR for conditional and insertional alleles C.B Gurumurthy, University Of Nebraska Medical Center Omaha, NE cgurumurthy@unmc.edu Types of Genome edits Gene disruption/inactivation Types of Genome
Easi CRISPR for conditional and insertional alleles C.B Gurumurthy, University Of Nebraska Medical Center Omaha, NE cgurumurthy@unmc.edu Types of Genome edits Gene disruption/inactivation Types of Genome
Supplementary Figure 1 Activities of ABEs using extended sgrnas in HEK293T cells.
 Supplementary Figure 1 Activities of ABEs using extended sgrnas in HEK293T cells. Base editing efficiencies of ABEs with extended sgrnas at Site 18 (a), Site 19 (b), the HBB-E2 site (c), and the HBB-E3
Supplementary Figure 1 Activities of ABEs using extended sgrnas in HEK293T cells. Base editing efficiencies of ABEs with extended sgrnas at Site 18 (a), Site 19 (b), the HBB-E2 site (c), and the HBB-E3
Improving CRISPR-Cas9 Gene Knockout with a Validated Guide RNA Algorithm
 Improving CRISPR-Cas9 Gene Knockout with a Validated Guide RNA Algorithm Anja Smith Director R&D Dharmacon, part of GE Healthcare Imagination at work crrna:tracrrna program Cas9 nuclease Active crrna is
Improving CRISPR-Cas9 Gene Knockout with a Validated Guide RNA Algorithm Anja Smith Director R&D Dharmacon, part of GE Healthcare Imagination at work crrna:tracrrna program Cas9 nuclease Active crrna is
Supplementary Information
 Supplementary Information Super-resolution imaging of fluorescently labeled, endogenous RNA Polymerase II in living cells with CRISPR/Cas9-mediated gene editing Won-Ki Cho 1, Namrata Jayanth 1, Susan Mullen
Supplementary Information Super-resolution imaging of fluorescently labeled, endogenous RNA Polymerase II in living cells with CRISPR/Cas9-mediated gene editing Won-Ki Cho 1, Namrata Jayanth 1, Susan Mullen
Introducing new DNA into the genome requires cloning the donor sequence, delivery of the cloned DNA into the cell, and integration into the genome.
 Key Terms Chapter 32: Genetic Engineering Cloning describes propagation of a DNA sequence by incorporating it into a hybrid construct that can be replicated in a host cell. A cloning vector is a plasmid
Key Terms Chapter 32: Genetic Engineering Cloning describes propagation of a DNA sequence by incorporating it into a hybrid construct that can be replicated in a host cell. A cloning vector is a plasmid
Nature Methods: doi: /nmeth Supplementary Figure 1. Comparison of REC2 and BCL-xL domains and modeling domain replacement.
 Supplementary Figure 1 Comparison of REC2 and BCL-xL domains and modeling domain replacement. (a) The REC2 domain of Cas9 and (b) BCL-xL display similar globular structures and volumes. (c, d) Modeling
Supplementary Figure 1 Comparison of REC2 and BCL-xL domains and modeling domain replacement. (a) The REC2 domain of Cas9 and (b) BCL-xL display similar globular structures and volumes. (c, d) Modeling
Amplicons, Heteroduplexes and Enzymes - Proper Processing Elevates Detection of CRISPR Gene Editing Events
 Amplicons, Heteroduplexes and Enzymes - Proper Processing Elevates Detection of CRISPR Gene Editing Events Steve Siembieda, MS MBA VP Commercialization ABRF Conference February 2015 What Is CRISPR? Clustered
Amplicons, Heteroduplexes and Enzymes - Proper Processing Elevates Detection of CRISPR Gene Editing Events Steve Siembieda, MS MBA VP Commercialization ABRF Conference February 2015 What Is CRISPR? Clustered
Supplementary Figure 1
 Supplementary Figure 1 Product purity of cytosine base editing for the wheat genomic loci tested. Product distributions and indels frequencies at four representative wheat genomic DNA sites in wheat protoplasts
Supplementary Figure 1 Product purity of cytosine base editing for the wheat genomic loci tested. Product distributions and indels frequencies at four representative wheat genomic DNA sites in wheat protoplasts
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified
 Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Fig. 1 shrna mediated knockdown of ZRSR2 in K562 and 293T cells. (a) ZRSR2 transcript levels in stably transduced K562 cells were determined using qrt-pcr. GAPDH was
Supplementary Figure 1 Supplementary Fig. 1 shrna mediated knockdown of ZRSR2 in K562 and 293T cells. (a) ZRSR2 transcript levels in stably transduced K562 cells were determined using qrt-pcr. GAPDH was
i-stop codon positions in the mcherry gene
 Supplementary Figure 1 i-stop codon positions in the mcherry gene The grnas (green) that can potentially generate stop codons from Trp (63 th and 98 th aa, upper panel) and Gln (47 th and 114 th aa, bottom
Supplementary Figure 1 i-stop codon positions in the mcherry gene The grnas (green) that can potentially generate stop codons from Trp (63 th and 98 th aa, upper panel) and Gln (47 th and 114 th aa, bottom
Supplementary Information
 Supplementary Information Vidigal and Ventura a wt locus 5 region 3 region CCTCTGCCACTGCGAGGGCGTCCAATGGTGCTTG(...)AACAGGTGGAATATCCCTACTCTA predicted deletion clone 1 clone 2 clone 3 CCTCTGCCACTGCGAGGGCGTC-AGGTGGAATATCCCTACTCTA
Supplementary Information Vidigal and Ventura a wt locus 5 region 3 region CCTCTGCCACTGCGAGGGCGTCCAATGGTGCTTG(...)AACAGGTGGAATATCCCTACTCTA predicted deletion clone 1 clone 2 clone 3 CCTCTGCCACTGCGAGGGCGTC-AGGTGGAATATCCCTACTCTA
7.012 Problem Set 5. Question 1
 Name Section 7.012 Problem Set 5 Question 1 While studying the problem of infertility, you attempt to isolate a hypothetical rabbit gene that accounts for the prolific reproduction of rabbits. After much
Name Section 7.012 Problem Set 5 Question 1 While studying the problem of infertility, you attempt to isolate a hypothetical rabbit gene that accounts for the prolific reproduction of rabbits. After much
Genome-wide genetic screening with chemically-mutagenized haploid embryonic stem cells
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 Supplementary Information Genome-wide genetic screening with chemically-mutagenized haploid embryonic stem cells Josep V. Forment 1,2, Mareike Herzog
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 Supplementary Information Genome-wide genetic screening with chemically-mutagenized haploid embryonic stem cells Josep V. Forment 1,2, Mareike Herzog
Supplementary Methods
 Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Genome editing. Knock-ins
 Genome editing Knock-ins Experiment design? Should we even do it? In mouse or rat, the HR-mediated knock-in of homologous fragments derived from a donor vector functions well. However, HR-dependent knock-in
Genome editing Knock-ins Experiment design? Should we even do it? In mouse or rat, the HR-mediated knock-in of homologous fragments derived from a donor vector functions well. However, HR-dependent knock-in
Supplementary Figures Montero et al._supplementary Figure 1
 Montero et al_suppl. Info 1 Supplementary Figures Montero et al._supplementary Figure 1 Montero et al_suppl. Info 2 Supplementary Figure 1. Transcripts arising from the structurally conserved subtelomeres
Montero et al_suppl. Info 1 Supplementary Figures Montero et al._supplementary Figure 1 Montero et al_suppl. Info 2 Supplementary Figure 1. Transcripts arising from the structurally conserved subtelomeres
Simple protocol for gene editing using GenCrisprTM Cas9 nuclease
 Simple protocol for gene editing using GenCrisprTM Cas9 nuclease Contents Protocol Step 1: Choose the target DNA sequence Step 2: Design sgrna Step 3: Preparation for sgrna 3.1 In vitro transcription of
Simple protocol for gene editing using GenCrisprTM Cas9 nuclease Contents Protocol Step 1: Choose the target DNA sequence Step 2: Design sgrna Step 3: Preparation for sgrna 3.1 In vitro transcription of
Allele-specific locus binding and genome editing by CRISPR at the
 Supplementary Information Allele-specific locus binding and genome editing by CRISPR at the p6ink4a locus Toshitsugu Fujita, Miyuki Yuno, and Hodaka Fujii Supplementary Figure Legends Supplementary Figure
Supplementary Information Allele-specific locus binding and genome editing by CRISPR at the p6ink4a locus Toshitsugu Fujita, Miyuki Yuno, and Hodaka Fujii Supplementary Figure Legends Supplementary Figure
Genome Engineering with ZFNs, TALENs and CRISPR/Cas9
 Genome Engineering with ZFNs, TALENs and CRISPR/Cas9 Designer Endonucleases ZFNs (zinc finger nucleases), TALENs (transcription activator-like effector nucleases) and CRISPR/Cas9 (clustered regularly interspaced
Genome Engineering with ZFNs, TALENs and CRISPR/Cas9 Designer Endonucleases ZFNs (zinc finger nucleases), TALENs (transcription activator-like effector nucleases) and CRISPR/Cas9 (clustered regularly interspaced
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Endogenous gene tagging to study subcellular localization and chromatin binding. a, b, Schematic of experimental set-up to endogenously tag RNAi factors using the CRISPR Cas9 technology,
Supplementary Figure 1 Endogenous gene tagging to study subcellular localization and chromatin binding. a, b, Schematic of experimental set-up to endogenously tag RNAi factors using the CRISPR Cas9 technology,
Supplementary Figure S1. The tetracycline-inducible CRISPR system. A) Hela cells stably
 Supplementary Information Supplementary Figure S1. The tetracycline-inducible CRISPR system. A) Hela cells stably expressing shrna sequences against TRF2 were examined by western blotting. shcon, shrna
Supplementary Information Supplementary Figure S1. The tetracycline-inducible CRISPR system. A) Hela cells stably expressing shrna sequences against TRF2 were examined by western blotting. shcon, shrna
Supplementary Figure 1. Supporting data for Figure 1
 Supplementary Figure 1 Supporting data for Figure 1 (A) Schematic of BLI assay used to measure dissociation. 5 monobiotinylated substrate DNA (identical to λ1, Figure 2) is associated with streptavidin-coated
Supplementary Figure 1 Supporting data for Figure 1 (A) Schematic of BLI assay used to measure dissociation. 5 monobiotinylated substrate DNA (identical to λ1, Figure 2) is associated with streptavidin-coated
Supplementary Figure 1. Phenotype and genotype of cultured and transplanted S1 KCST (A) Brightfield and mcherry fluorescence images of the spheres
 Supplementary Figure 1. Phenotype and genotype of cultured and transplanted S1 KCST (A) Brightfield and mcherry fluorescence images of the spheres generated from the CD133-positive cells infected with
Supplementary Figure 1. Phenotype and genotype of cultured and transplanted S1 KCST (A) Brightfield and mcherry fluorescence images of the spheres generated from the CD133-positive cells infected with
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature09937 a Name Position Primersets 1a 1b 2 3 4 b2 Phenotype Genotype b Primerset 1a D T C R I E 10000 8000 6000 5000 4000 3000 2500 2000 1500 1000 800 Donor (D)
SUPPLEMENTARY INFORMATION doi:10.1038/nature09937 a Name Position Primersets 1a 1b 2 3 4 b2 Phenotype Genotype b Primerset 1a D T C R I E 10000 8000 6000 5000 4000 3000 2500 2000 1500 1000 800 Donor (D)
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
Fig. S1. eif6 expression in HEK293 transfected with shrna against eif6 or pcmv-eif6 vector.
 Fig. S1. eif6 expression in HEK293 transfected with shrna against eif6 or pcmv-eif6 vector. (a) Western blotting analysis and (b) qpcr analysis of eif6 expression in HEK293 T cells transfected with either
Fig. S1. eif6 expression in HEK293 transfected with shrna against eif6 or pcmv-eif6 vector. (a) Western blotting analysis and (b) qpcr analysis of eif6 expression in HEK293 T cells transfected with either
Getting started with CRISPR: A review of gene knockout and homology-directed repair
 Getting started with CRISPR: A review of gene knockout and homology-directed repair Justin Barr Product Manager, Functional Genomics Integrated DNA Technologies 1 Agenda: Getting started with CRISPR Background
Getting started with CRISPR: A review of gene knockout and homology-directed repair Justin Barr Product Manager, Functional Genomics Integrated DNA Technologies 1 Agenda: Getting started with CRISPR Background
Supplementary Figure 1. NORAD expression in mouse (A) and dog (B). The black boxes indicate the position of the regions alignable to the 12 repeat
 Supplementary Figure 1. NORAD expression in mouse (A) and dog (B). The black boxes indicate the position of the regions alignable to the 12 repeat units in the human genome. Annotated transposable elements
Supplementary Figure 1. NORAD expression in mouse (A) and dog (B). The black boxes indicate the position of the regions alignable to the 12 repeat units in the human genome. Annotated transposable elements
CRISPR/Cas9 Genome Editing: Transfection Methods
 CRISPR/ Genome Editing: Transfection Methods For over 20 years Mirus Bio has developed and manufactured high performance transfection products and technologies. That expertise is now being applied to the
CRISPR/ Genome Editing: Transfection Methods For over 20 years Mirus Bio has developed and manufactured high performance transfection products and technologies. That expertise is now being applied to the
Applications of Cas9 nickases for genome engineering
 application note genome editing Applications of Cas9 nickases for genome engineering Shuqi Yan, Mollie Schubert, Maureen Young, Brian Wang Integrated DNA Technologies, 17 Commercial Park, Coralville, IA,
application note genome editing Applications of Cas9 nickases for genome engineering Shuqi Yan, Mollie Schubert, Maureen Young, Brian Wang Integrated DNA Technologies, 17 Commercial Park, Coralville, IA,
Revised: RG-RV2 by Fukuhara et al.
 Supplemental Figure 1 The generation of Spns2 conditional knockout mice. (A) Schematic representation of the wild type Spns2 locus (Spns2 + ), the targeted allele, the floxed allele (Spns2 f ) and the
Supplemental Figure 1 The generation of Spns2 conditional knockout mice. (A) Schematic representation of the wild type Spns2 locus (Spns2 + ), the targeted allele, the floxed allele (Spns2 f ) and the
Transfection of CRISPR/Cas9 Nuclease NLS ribonucleoprotein (RNP) into adherent mammalian cells using Lipofectamine RNAiMAX
 Transfection of CRISPR/Cas9 Nuclease NLS ribonucleoprotein (RNP) into adherent mammalian cells using Lipofectamine RNAiMAX INTRODUCTION The CRISPR/Cas genome editing system consists of a single guide RNA
Transfection of CRISPR/Cas9 Nuclease NLS ribonucleoprotein (RNP) into adherent mammalian cells using Lipofectamine RNAiMAX INTRODUCTION The CRISPR/Cas genome editing system consists of a single guide RNA
Supplementary Materials
 Supplementary Materials Supplementary Methods Supplementary Discussion Supplementary Figure 1 Calculated frequencies of embryo cells bearing bi-allelic alterations. Targeted indel mutations induced by
Supplementary Materials Supplementary Methods Supplementary Discussion Supplementary Figure 1 Calculated frequencies of embryo cells bearing bi-allelic alterations. Targeted indel mutations induced by
Supplementary Information
 Supplementary Information This Supplementary Information file contains the following contents: Supplementary Methods, Supplementary Discussion, Supplementary Figures (S1~S9), Supplementary Tables (1~3)
Supplementary Information This Supplementary Information file contains the following contents: Supplementary Methods, Supplementary Discussion, Supplementary Figures (S1~S9), Supplementary Tables (1~3)
Inhibition of HSV-1 Replication by Gene Editing Strategy. Running Title: Targeting HSV-1 Infection by CRISPR/Cas9
 SUPPLEMENTARY MATERIALS 2/4/16 Inhibition of HSV-1 Replication by Gene Editing Strategy Running Title: Targeting HSV-1 Infection by CRISPR/Cas9 Pamela C. Roehm 1,2,3*, Masoud Shekarabi 1, Hassen Wollebo
SUPPLEMENTARY MATERIALS 2/4/16 Inhibition of HSV-1 Replication by Gene Editing Strategy Running Title: Targeting HSV-1 Infection by CRISPR/Cas9 Pamela C. Roehm 1,2,3*, Masoud Shekarabi 1, Hassen Wollebo
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling
 Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary Information. Optimization of the production of knock-in alleles by CRISPR/Cas9 microinjection into the mouse zygote
 Supplementary Information Supplementary Figures S1 to S5 and Tables S1 to S3. Optimization of the production of knock-in alleles by CRISPR/Cas9 microinjection into the mouse zygote Aurélien Raveux, Sandrine
Supplementary Information Supplementary Figures S1 to S5 and Tables S1 to S3. Optimization of the production of knock-in alleles by CRISPR/Cas9 microinjection into the mouse zygote Aurélien Raveux, Sandrine
BIOLOGY - CLUTCH CH.20 - BIOTECHNOLOGY.
 !! www.clutchprep.com CONCEPT: DNA CLONING DNA cloning is a technique that inserts a foreign gene into a living host to replicate the gene and produce gene products. Transformation the process by which
!! www.clutchprep.com CONCEPT: DNA CLONING DNA cloning is a technique that inserts a foreign gene into a living host to replicate the gene and produce gene products. Transformation the process by which
Nature Methods: doi: /nmeth Supplementary Figure 1. DMS-MaPseq data are highly reproducible at elevated DMS concentrations.
 Supplementary Figure 1 DMS-MaPseq data are highly reproducible at elevated DMS concentrations. a, Correlation of Gini index for 202 yeast mrna regions with 15x coverage at 2.5% or 5% v/v DMS concentrations
Supplementary Figure 1 DMS-MaPseq data are highly reproducible at elevated DMS concentrations. a, Correlation of Gini index for 202 yeast mrna regions with 15x coverage at 2.5% or 5% v/v DMS concentrations
CRISPR/Cas9 Gene Editing Tools
 CRISPR/Cas9 Gene Editing Tools - Guide-it Products for Successful CRISPR/Cas9 Gene Editing - Why choose Guide-it products? Optimized methods designed for speed and ease of use Complete kits that don t
CRISPR/Cas9 Gene Editing Tools - Guide-it Products for Successful CRISPR/Cas9 Gene Editing - Why choose Guide-it products? Optimized methods designed for speed and ease of use Complete kits that don t
Chapter 20 Recombinant DNA Technology. Copyright 2009 Pearson Education, Inc.
 Chapter 20 Recombinant DNA Technology Copyright 2009 Pearson Education, Inc. 20.1 Recombinant DNA Technology Began with Two Key Tools: Restriction Enzymes and DNA Cloning Vectors Recombinant DNA refers
Chapter 20 Recombinant DNA Technology Copyright 2009 Pearson Education, Inc. 20.1 Recombinant DNA Technology Began with Two Key Tools: Restriction Enzymes and DNA Cloning Vectors Recombinant DNA refers
SUPPLEMENTAL MATERIALS
 SUPPLEMENL MERILS Eh-seq: RISPR epitope tagging hip-seq of DN-binding proteins Daniel Savic, E. hristopher Partridge, Kimberly M. Newberry, Sophia. Smith, Sarah K. Meadows, rian S. Roberts, Mark Mackiewicz,
SUPPLEMENL MERILS Eh-seq: RISPR epitope tagging hip-seq of DN-binding proteins Daniel Savic, E. hristopher Partridge, Kimberly M. Newberry, Sophia. Smith, Sarah K. Meadows, rian S. Roberts, Mark Mackiewicz,
Cleavage of tau by asparagine endopeptidase mediates the neurofibrillary pathology in
 Supplementary information Cleavage of tau by asparagine endopeptidase mediates the neurofibrillary pathology in Alzheimer s disease Zhentao Zhang, Mingke Song, Xia Liu, Seong Su Kang, Il-Sun Kwon, Duc
Supplementary information Cleavage of tau by asparagine endopeptidase mediates the neurofibrillary pathology in Alzheimer s disease Zhentao Zhang, Mingke Song, Xia Liu, Seong Su Kang, Il-Sun Kwon, Duc
TRIM31 is recruited to mitochondria after infection with SeV.
 Supplementary Figure 1 TRIM31 is recruited to mitochondria after infection with SeV. (a) Confocal microscopy of TRIM31-GFP transfected into HEK293T cells for 24 h followed with SeV infection for 6 h. MitoTracker
Supplementary Figure 1 TRIM31 is recruited to mitochondria after infection with SeV. (a) Confocal microscopy of TRIM31-GFP transfected into HEK293T cells for 24 h followed with SeV infection for 6 h. MitoTracker
CRISPR/Cas9 Gene Editing Tools
 CRISPR/Cas9 Gene Editing Tools - Separations Simply Spectacular INDELS Identify indels Determine if one or both copies of your gene have indels The Guide-it Genotype Confirmation Kit: Simple detection
CRISPR/Cas9 Gene Editing Tools - Separations Simply Spectacular INDELS Identify indels Determine if one or both copies of your gene have indels The Guide-it Genotype Confirmation Kit: Simple detection
Genome edi3ng with the CRISPR-Cas9 system
 CRISPR-Cas9 Genome Edi3ng Bootcamp AHA Council on Func3onal Genomics and Transla3onal Biology Narrated video link: hfps://youtu.be/h18hmftybnq Genome edi3ng with the CRISPR-Cas9 system Kiran Musunuru,
CRISPR-Cas9 Genome Edi3ng Bootcamp AHA Council on Func3onal Genomics and Transla3onal Biology Narrated video link: hfps://youtu.be/h18hmftybnq Genome edi3ng with the CRISPR-Cas9 system Kiran Musunuru,
Supplementary Figure 1. CRISPR/Cas9-induced targeted mutations in TaGASR7, TaDEP1, TaNAC2, TaPIN1, TaLOX2 and TaGW2 genes in wheat protoplasts.
 Supplementary Figure 1. CRISPR/Cas9-induced targeted mutations in TaGASR7, TaDEP1, TaNAC2, TaPIN1, TaLOX2 and TaGW2 genes in wheat protoplasts. Lanes 1 and 2: digested CRISPR/Cas9-transformed protoplasts;
Supplementary Figure 1. CRISPR/Cas9-induced targeted mutations in TaGASR7, TaDEP1, TaNAC2, TaPIN1, TaLOX2 and TaGW2 genes in wheat protoplasts. Lanes 1 and 2: digested CRISPR/Cas9-transformed protoplasts;
Surrogate reporter-based enrichment of cells containing RNA-guided Cas9 nucleaseinduced
 Supplementary Data Surrogate reporter-based enrichment of cells containing RNA-guided Cas9 nucleaseinduced mutations Suresh Ramakrishna 1, Seung Woo Cho 2, Sojung Kim 2, Myungjae Song 1, Ramu Gopalappa
Supplementary Data Surrogate reporter-based enrichment of cells containing RNA-guided Cas9 nucleaseinduced mutations Suresh Ramakrishna 1, Seung Woo Cho 2, Sojung Kim 2, Myungjae Song 1, Ramu Gopalappa
SUPPLEMENTARY INFORMATION
 Supplementary Figure a T m ( C) Seq. '-3' uguuugugguaacagugugaggu L 62 AttGtcAcaCtcC L2 6 ccattgtcacactcc L3 66 attgtcacactcc 7 ccattgtcacactcca L 73 ccattgtcacactcc L6 74 AttGTcaCaCtCC L7 7 attgtcacactcc
Supplementary Figure a T m ( C) Seq. '-3' uguuugugguaacagugugaggu L 62 AttGtcAcaCtcC L2 6 ccattgtcacactcc L3 66 attgtcacactcc 7 ccattgtcacactcca L 73 ccattgtcacactcc L6 74 AttGTcaCaCtCC L7 7 attgtcacactcc
SUPPLEMENTAL MATERIAL
 SUPPLEMENTAL MATERIAL MATERIALS AND METHODS Generation of TSPOΔ/Δ murine embryonic fibroblasts Embryos were harvested from 13.5-day pregnant TSPOfl/fl mice. After dissection to eviscerate and remove the
SUPPLEMENTAL MATERIAL MATERIALS AND METHODS Generation of TSPOΔ/Δ murine embryonic fibroblasts Embryos were harvested from 13.5-day pregnant TSPOfl/fl mice. After dissection to eviscerate and remove the
Partial list of differentially expressed genes from cdna microarray, comparing MUC18-
 Supplemental Figure legends Table-1 Partial list of differentially expressed genes from cdna microarray, comparing MUC18- silenced and NT-transduced A375SM cells. Supplemental Figure 1 Effect of MUC-18
Supplemental Figure legends Table-1 Partial list of differentially expressed genes from cdna microarray, comparing MUC18- silenced and NT-transduced A375SM cells. Supplemental Figure 1 Effect of MUC-18
Endo et al., supplementary Fig S1
 Endo et al., supplementary Fig S1 supplementary Fig S1. AGO2 does not recognize the central region of mir390/mir390* duplex for RISC assembly. (A) The structure of wild-type mir390/mir390* and its variants
Endo et al., supplementary Fig S1 supplementary Fig S1. AGO2 does not recognize the central region of mir390/mir390* duplex for RISC assembly. (A) The structure of wild-type mir390/mir390* and its variants
Cationic lipid-mediated delivery of proteins enables efficient proteinbased genome editing in vitro and in vivo
 correction notice Nat. Biotechnol. 33, 73 8 (215) Cationic lipid-mediated delivery of proteins enables efficient proteinbased genome editing in vitro and in vivo John A Zuris, David B Thompson, Yilai Shu,
correction notice Nat. Biotechnol. 33, 73 8 (215) Cationic lipid-mediated delivery of proteins enables efficient proteinbased genome editing in vitro and in vivo John A Zuris, David B Thompson, Yilai Shu,
A Repressor Complex Governs the Integration of
 Developmental Cell 15 Supplemental Data A Repressor Complex Governs the Integration of Flowering Signals in Arabidopsis Dan Li, Chang Liu, Lisha Shen, Yang Wu, Hongyan Chen, Masumi Robertson, Chris A.
Developmental Cell 15 Supplemental Data A Repressor Complex Governs the Integration of Flowering Signals in Arabidopsis Dan Li, Chang Liu, Lisha Shen, Yang Wu, Hongyan Chen, Masumi Robertson, Chris A.
Schematic representation of the endogenous PALB2 locus and gene-disruption constructs
 Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
You use the UCSC Genome Browser (www.genome.ucsc.edu) to assess the exonintron structure of each gene. You use four tracks to show each gene:
 CRISPR-Cas9 genome editing Part 1: You would like to rapidly generate two different knockout mice using CRISPR-Cas9. The genes to be knocked out are Pcsk9 and Apoc3, both involved in lipid metabolism.
CRISPR-Cas9 genome editing Part 1: You would like to rapidly generate two different knockout mice using CRISPR-Cas9. The genes to be knocked out are Pcsk9 and Apoc3, both involved in lipid metabolism.
Nature Genetics: doi: /ng.3556 INTEGRATED SUPPLEMENTARY FIGURE TEMPLATE. Supplementary Figure 1
 INTEGRATED SUPPLEMENTARY FIGURE TEMPLATE Supplementary Figure 1 REF6 expression in transgenic lines. (a,b) Expression of REF6 in REF6-HA ref6 and REF6ΔZnF-HA ref6 plants detected by RT qpcr (a) and immunoblot
INTEGRATED SUPPLEMENTARY FIGURE TEMPLATE Supplementary Figure 1 REF6 expression in transgenic lines. (a,b) Expression of REF6 in REF6-HA ref6 and REF6ΔZnF-HA ref6 plants detected by RT qpcr (a) and immunoblot
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.
 Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
Nature Structural and Molecular Biology: doi: /nsmb Supplementary Figure 1. Validation of CDK9-inhibitor treatment.
 Supplementary Figure 1 Validation of CDK9-inhibitor treatment. (a) Schematic of GAPDH with the middle of the amplicons indicated in base pairs. The transcription start site (TSS) and the terminal polyadenylation
Supplementary Figure 1 Validation of CDK9-inhibitor treatment. (a) Schematic of GAPDH with the middle of the amplicons indicated in base pairs. The transcription start site (TSS) and the terminal polyadenylation
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature12474 Supplementary Figure 1 Analysis of mtdna mutation loads in different types of mtdna mutator mice. a, PCR, cloning, and sequencing analysis of mtdna mutations
SUPPLEMENTARY INFORMATION doi:10.1038/nature12474 Supplementary Figure 1 Analysis of mtdna mutation loads in different types of mtdna mutator mice. a, PCR, cloning, and sequencing analysis of mtdna mutations
Genome Sequence Assembly
 Genome Sequence Assembly Learning Goals: Introduce the field of bioinformatics Familiarize the student with performing sequence alignments Understand the assembly process in genome sequencing Introduction:
Genome Sequence Assembly Learning Goals: Introduce the field of bioinformatics Familiarize the student with performing sequence alignments Understand the assembly process in genome sequencing Introduction:
File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description:
 File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description: Supplementary Figure 1. dcas9-mq1 fusion protein induces de novo
File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description: Supplementary Figure 1. dcas9-mq1 fusion protein induces de novo
Supplemental Data. Na Xu et al. (2016). Plant Cell /tpc
 Supplemental Figure 1. The weak fluorescence phenotype is not caused by the mutation in At3g60240. (A) A mutation mapped to the gene At3g60240. Map-based cloning strategy was used to map the mutated site
Supplemental Figure 1. The weak fluorescence phenotype is not caused by the mutation in At3g60240. (A) A mutation mapped to the gene At3g60240. Map-based cloning strategy was used to map the mutated site
Guide-it sgrna In Vitro Transcription and Screening Systems User Manual
 Guide-it sgrna In Vitro Transcription and Screening Systems User Manual Cat. Nos. 632638, 632639, 632635, 632636, 632637 (040618) 1290 Terra Bella Avenue, Mountain View, CA 94043, USA U.S. Technical Support:
Guide-it sgrna In Vitro Transcription and Screening Systems User Manual Cat. Nos. 632638, 632639, 632635, 632636, 632637 (040618) 1290 Terra Bella Avenue, Mountain View, CA 94043, USA U.S. Technical Support:
User Instructions:Transfection-ready CRISPR/Cas9 Reagents. Target DNA. NHEJ repair pathway. Nucleotide deletion. Nucleotide insertion Gene disruption
 User Instructions:Transfection-ready CRISPR/Cas9 Reagents Background Introduction to CRISPR/Cas9 genome editing In bacteria and archaea, clustered regularly interspaced short palindromic repeats (CRISPR)
User Instructions:Transfection-ready CRISPR/Cas9 Reagents Background Introduction to CRISPR/Cas9 genome editing In bacteria and archaea, clustered regularly interspaced short palindromic repeats (CRISPR)
Supplemental Material: Rev1 promotes replication through UV lesions in conjunction with DNA
 Supplemental Material: Rev1 promotes replication through UV lesions in conjunction with DNA polymerases,, and, but not with DNA polymerase Jung-Hoon Yoon, Jeseong Park, Juan Conde, Maki Wakamiya, Louise
Supplemental Material: Rev1 promotes replication through UV lesions in conjunction with DNA polymerases,, and, but not with DNA polymerase Jung-Hoon Yoon, Jeseong Park, Juan Conde, Maki Wakamiya, Louise
Supplementary Figure 1. α-synuclein is truncated in PD and LBD brains. Nature Structural & Molecular Biology: doi: /nsmb.
 Supplementary Figure 1 α-synuclein is truncated in PD and LBD brains. (a) Specificity of anti-n103 antibody. Anti-N103 antibody was coated on an ELISA plate and different concentrations of full-length
Supplementary Figure 1 α-synuclein is truncated in PD and LBD brains. (a) Specificity of anti-n103 antibody. Anti-N103 antibody was coated on an ELISA plate and different concentrations of full-length
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1. Analyses of ECTRs by C-circle and T-circle assays.
 Supplementary Figure 1 Analyses of ECTRs by C-circle and T-circle assays. (a) C-circle and (b) T-circle amplification reactions using genomic DNA from different cell lines in the presence (+) or absence
Supplementary Figure 1 Analyses of ECTRs by C-circle and T-circle assays. (a) C-circle and (b) T-circle amplification reactions using genomic DNA from different cell lines in the presence (+) or absence
Protocol for Genome Editing via the RNA-guided Cas9 Nuclease in. Zebrafish Embryos 1
 Protocol for Genome Editing via the RNA-guided Cas9 Nuclease in Zebrafish Embryos 1 1. In vitro synthesis of capped Cas9 mrna The full length of humanized Cas9 cdnas with double NLS were cloned into pxt7
Protocol for Genome Editing via the RNA-guided Cas9 Nuclease in Zebrafish Embryos 1 1. In vitro synthesis of capped Cas9 mrna The full length of humanized Cas9 cdnas with double NLS were cloned into pxt7
Supplementary Figure 1. Nature Structural & Molecular Biology: doi: /nsmb.3494
 Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
XactEdit Cas9 Nuclease with NLS User Manual
 XactEdit Cas9 Nuclease with NLS User Manual An RNA-guided recombinant endonuclease for efficient targeted DNA cleavage Catalog Numbers CE1000-50K, CE1000-50, CE1000-250, CE1001-250, CE1001-1000 Table of
XactEdit Cas9 Nuclease with NLS User Manual An RNA-guided recombinant endonuclease for efficient targeted DNA cleavage Catalog Numbers CE1000-50K, CE1000-50, CE1000-250, CE1001-250, CE1001-1000 Table of
Introduction to CRISPR/Cas9 Background DNA Cleavage and Repair (NHEJ and HDR) Alternative Cas9 Variants Delivery of Cas9 and sgrna Library Products
 Introduction to CRISPR/Cas9 Background DNA Cleavage and Repair (NHEJ and HDR) Alternative Cas9 Variants Delivery of Cas9 and sgrna Library Products which one is right for you? CRISPR Workflow abm s Toolbox
Introduction to CRISPR/Cas9 Background DNA Cleavage and Repair (NHEJ and HDR) Alternative Cas9 Variants Delivery of Cas9 and sgrna Library Products which one is right for you? CRISPR Workflow abm s Toolbox
Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) (b)
 Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) A schematic overview of the production and amplification of a single pirna from a transposon transcript. The
Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) A schematic overview of the production and amplification of a single pirna from a transposon transcript. The
Bootcamp: Molecular Biology Techniques and Interpretation
 Bootcamp: Molecular Biology Techniques and Interpretation Bi8 Winter 2016 Today s outline Detecting and quantifying nucleic acids and proteins: Basic nucleic acid properties Hybridization PCR and Designing
Bootcamp: Molecular Biology Techniques and Interpretation Bi8 Winter 2016 Today s outline Detecting and quantifying nucleic acids and proteins: Basic nucleic acid properties Hybridization PCR and Designing
Supplementary Figure 1 Collision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK2, PPK3 and PPK4 respectively.
 Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
Donor DNA Utilization during Gene Targeting with Zinc- finger Nucleases
 Donor DNA Utilization during Gene Targeting with Zinc- finger Nucleases Kelly J. Beumer, Jonathan K. Trautman, Kusumika Mukherjee and Dana Carroll Department of Biochemistry University of Utah School of
Donor DNA Utilization during Gene Targeting with Zinc- finger Nucleases Kelly J. Beumer, Jonathan K. Trautman, Kusumika Mukherjee and Dana Carroll Department of Biochemistry University of Utah School of
Protocol for cloning SEC-based repair templates using Gibson assembly and ccdb negative selection
 Protocol for cloning SEC-based repair templates using Gibson assembly and ccdb negative selection Written by Dan Dickinson (daniel.dickinson@austin.utexas.edu) and last updated January 2018. A version
Protocol for cloning SEC-based repair templates using Gibson assembly and ccdb negative selection Written by Dan Dickinson (daniel.dickinson@austin.utexas.edu) and last updated January 2018. A version
SUPPLEMENTARY INFORMATION
 VOLUME: 1 ARTICLE NUMBER: 0066 In the format provided by the authors and unedited. Engineering CRISPR-Cpf1 crrnas and mrnas to maximize genome editing efficiency Bin Li 1, Weiyu Zhao 1, Xiao Luo 1, Xinfu
VOLUME: 1 ARTICLE NUMBER: 0066 In the format provided by the authors and unedited. Engineering CRISPR-Cpf1 crrnas and mrnas to maximize genome editing efficiency Bin Li 1, Weiyu Zhao 1, Xiao Luo 1, Xinfu
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Han et al., http://www.jcb.org/cgi/content/full/jcb.201311007/dc1 Figure S1. SIVA1 interacts with PCNA. (A) HEK293T cells were transiently
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Han et al., http://www.jcb.org/cgi/content/full/jcb.201311007/dc1 Figure S1. SIVA1 interacts with PCNA. (A) HEK293T cells were transiently
CELLTECHGEN For Research Only. Construction of all-in-one vector for Lenti-virus system (Example: Lenti-EF1 -Cas9-EGFP-U6 sgrna vector)
 Construction of all-in-one vector for Lenti-virus system (Example: Lenti-EF1 -Cas9-EGFP-U6 sgrna vector) Catalog number: CTG-CAS9-18 Introduction The vector Lenti-EF1 -Cas9-EGFP-U6 sgrna is designed for
Construction of all-in-one vector for Lenti-virus system (Example: Lenti-EF1 -Cas9-EGFP-U6 sgrna vector) Catalog number: CTG-CAS9-18 Introduction The vector Lenti-EF1 -Cas9-EGFP-U6 sgrna is designed for
Table S1. Oligonucleotide primer sequences
 Table S1. Oligonucleotide primer sequences Primer Name DNA Sequence (5 -> 3 ) Description CD19c CD19d Cre7 Sfpi1lox1 Sfpi1lox2 Sfpi1lox3 5 -AACCAGTCAACACCCTTCC-3 5 -CCAGACTAGATACAGACCAG-3 5 -TCAGCTACACCAGAGACGG-3
Table S1. Oligonucleotide primer sequences Primer Name DNA Sequence (5 -> 3 ) Description CD19c CD19d Cre7 Sfpi1lox1 Sfpi1lox2 Sfpi1lox3 5 -AACCAGTCAACACCCTTCC-3 5 -CCAGACTAGATACAGACCAG-3 5 -TCAGCTACACCAGAGACGG-3
Table S1. Primers used in the study
 Table S1. Primers used in the study Primer name Application Sequence I1F16 Genotyping GGCAAGTGAGTGAGTGCCTA I1R11 Genotyping CCCACTCGTATTGACGCTCT V19 Genotyping GGGTCTCAAAGTCAGGGTCA D18Mit184-F Genotyping
Table S1. Primers used in the study Primer name Application Sequence I1F16 Genotyping GGCAAGTGAGTGAGTGCCTA I1R11 Genotyping CCCACTCGTATTGACGCTCT V19 Genotyping GGGTCTCAAAGTCAGGGTCA D18Mit184-F Genotyping
Optimizing Cas9 and sgrna concentrations for increasing mutation efficiency. Nature Methods: doi: /nmeth.3360
 Supplementary Figure 1 Optimizing Cas9 and sgrna concentrations for increasing mutation efficiency. (a) Wild-type, 2-dpf slc24a5 b1 embryos and a series of slc24a5 CRISPR-injected embryos; anterior is
Supplementary Figure 1 Optimizing Cas9 and sgrna concentrations for increasing mutation efficiency. (a) Wild-type, 2-dpf slc24a5 b1 embryos and a series of slc24a5 CRISPR-injected embryos; anterior is
PrimePCR Assay Validation Report
 Gene Information Gene Name Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID mcf.2 transforming sequence-like Mcf2l Mouse Description Not Available C130040G20Rik,
Gene Information Gene Name Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID mcf.2 transforming sequence-like Mcf2l Mouse Description Not Available C130040G20Rik,
TRANSGENIC ANIMALS. transient. stable. - Two methods to produce transgenic animals:
 Only for teaching purposes - not for reproduction or sale CELL TRANSFECTION transient stable TRANSGENIC ANIMALS - Two methods to produce transgenic animals: 1- DNA microinjection 2- embryonic stem cell-mediated
Only for teaching purposes - not for reproduction or sale CELL TRANSFECTION transient stable TRANSGENIC ANIMALS - Two methods to produce transgenic animals: 1- DNA microinjection 2- embryonic stem cell-mediated
Supplementary Figure Legends
 Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
Functional analysis reveals that RBM10 mutations. contribute to lung adenocarcinoma pathogenesis by. deregulating splicing
 Supplementary Information Functional analysis reveals that RBM10 mutations contribute to lung adenocarcinoma pathogenesis by deregulating splicing Jiawei Zhao 1,+, Yue Sun 2,3,+, Yin Huang 6, Fan Song
Supplementary Information Functional analysis reveals that RBM10 mutations contribute to lung adenocarcinoma pathogenesis by deregulating splicing Jiawei Zhao 1,+, Yue Sun 2,3,+, Yin Huang 6, Fan Song
Legend for Supplemental Figures and Tables
 Legend for Supplemental Figures and Tables Supplemental Fig. 1. Negative regulation of the CYP27B1 promoter in a ligand-dependent manner (A) OK-P cells were transfected with pcdna-trα, pcdna-trβ1 or pcdna3
Legend for Supplemental Figures and Tables Supplemental Fig. 1. Negative regulation of the CYP27B1 promoter in a ligand-dependent manner (A) OK-P cells were transfected with pcdna-trα, pcdna-trβ1 or pcdna3
Supplementary Materials. China
 Supplementary Materials An Efficient Genotyping Method for Genome-modified Animals and Human Cells Generated with CRISPR/Cas9 System Xiaoxiao Zhu 1,2 *, Yajie Xu 1 *, Shanshan Yu 1 *, Lu Lu 1,2, Mingqin
Supplementary Materials An Efficient Genotyping Method for Genome-modified Animals and Human Cells Generated with CRISPR/Cas9 System Xiaoxiao Zhu 1,2 *, Yajie Xu 1 *, Shanshan Yu 1 *, Lu Lu 1,2, Mingqin
Pei et al. Supplementary Figure S1
 Pei et al. Supplementary Figure S1 C H-CUL9: + + + + + Myc-ROC1: - - + + + U2OS/pcDN3 U2OS/H-CUL9 U2OS/ + H-CUL9 IP: -H -myc input -H -myc 1 2 3 4 5 H-CUL9 Myc-ROC1 -H -H -H -H H-CUL9: wt RR myc-roc1:
Pei et al. Supplementary Figure S1 C H-CUL9: + + + + + Myc-ROC1: - - + + + U2OS/pcDN3 U2OS/H-CUL9 U2OS/ + H-CUL9 IP: -H -myc input -H -myc 1 2 3 4 5 H-CUL9 Myc-ROC1 -H -H -H -H H-CUL9: wt RR myc-roc1:
