Kostandin V. Pajcini, Stephane Y. Corbel, Julien Sage, Jason H. Pomerantz, and Helen M. Blau
|
|
|
- Jasmin Lewis
- 6 years ago
- Views:
Transcription
1 Cell Stem Cell, Volume 7 Supplemental Information Transient Inactivation of Rb and ARF Yields Regenerative Cells from Postmitotic Mammalian Muscle Kostandin V. Pajcini, Stephane Y. Corbel, Julien Sage, Jason H. Pomerantz, and Helen M. Blau Inventory of Supplemental Material Supplemental Experimental Procedures Figure S1. siimporter myotube transfection, TAM and sirna treatment analysis. Data presented in this figure corresponds to Figure 1. It characterizes the efficiency of sirna treatment of myotubes (panels A-B). Panels C-D show induction of Cre expression, as seen by LacZ staining in cells treated with tamoxifen (TAM) and panels E-G verify suppression of Rb obtained by sirna treatment and by TAM treatment. Panels H and I show the percentage of myotubes that have at least two BrdU positive myonuclei and highlight the non-apoptotic state of TAM and sirna treated myotubes, respectively. Figure S2. Decrease in Rb and p16/19 levels leads to upregulation of mitotic machinery. Data presented in this figure corresponds to Figure 2. It shows the upregulation of members of the Chromosome passenger complex upon loss of prb and p16/19 both in mrna (panel A) and protein (panel B) as well as by immunofluorescent analysis (panels C-D). Panels E-F highlight the expression patterns of cyclins and the cellular localization of mitotic machinery, respectively. Figure S3. Dedifferentiation in C2C12 and primary myotubes. Data presented in this figure corresponds to Figure 3. It provides further examples of mature myotube dedifferentiation in C2C12 myotubes (panels A-B). It also provides evidence of downregulation of mature muscle genes at the mrna level (panel C) and at the protein level (panels D-F). Figure S4. Myocytes deficient in Rb and p16/19 proliferate following FACS isolation. Data presented in this figure corresponds to Figures 4 and 5. Panels A-C show the capacity of FACS isolated myocytes to divide and give rise to colonies in microwells. Panels D-E provide more examples of PALM LPC myocyte capture and follow the fate of the captured single cells. Figure S5. Single-cell isolation and clonal expansion of FACS sorted myogenin- GFP + myocytes. Data presented in this figure corresponds to Figure 5. The scheme of panel A outlines an alternative method to myocyte isolation using both FACS and PALM LPC. Panels B-C provide examples of the PALM LPC capture of FACS isolated GFP + cells, and follow the fate of these cells once isolated. 1
2 Figure S6. PALM isolation, expansion and differentiation of primary myoblasts and TAM and p16/19si treated myocytes. Data presented in this figure corresponds to Figure 6. PALM LPC captured myoblasts and myocytes are followed through expansion and differentiation (panels A-B) and their expression profiles are characterized by western (panel C) RT-PCR (panel D). A biological repeat of the western analysis in Figure 6F is shown in Figure S6 E. Figure S7. DKD dedifferentiated, captured and expanded myocytes fuse to muscle in vivo. Data presented in this figure corresponds directly to Figure 7. The images are an experimental set of DKDcap myocytes that once expanded in vitro were injected in the TA muscle of mice. Cross section analysis of GFP + fibers identified the fusion of dedifferentiated cells to existing muscle fibers. Movies S1 and S2. Timelapse analysis of primary myotubes after TAM+Mocksi or TAM+p16/19si treatment respectively. Both videos are part of the same experimental set and they correspond to the data shown in Figure 4. They highlight the morphological collapse of mature myotubes once both Rb and p16/19 are suppressed. 2
3 Supplementary Experimental Procedures: Primary myoblast selection Primary myoblasts after harvest were selected in F10 media (Gibco) supplemented with 20%FBS (Omega scientific), 2.5ng/mL bfgf (Promega), and 1% Pen-Strep (Gibco) for a week on collagen (Sigma) coated plates. Primary myoblasts were used at low passage during which time cell shape remained compact and uniform. Cell Culture C2C12 mouse myoblasts were cultured in 10% CO2 at 37deg. In DMEM HG (Gibco) supplemented with 20% FBS and Pen Strep. Differentiation conditions included DMEM HG supplemented with 2% HS on collagen coated plates. Media was changed every 24 hours. Primary myoblasts: After primary myoblast harvest and selection, passages were counted once cells were cultured and expanded in growth media: DMEM LG+F10 media supplemented with 15%FBS, 1% Pen-Strep and 1.25ng/mL bfgf, and plated on collagen coated plates. Primary myoblasts were seeded for fusion under low serum conditions in DMEM LG, 2% HS 1% Pen-Strep at 6 x10 5 cells per 6cm plate and for sparse, single-cell differentiation at 5 x10 4 cells per 6cm plate. Cell seeding numbers were adjusted depending on the surface area of different plating platforms from the 6cm standards described above. ple-myog3r-gfp vector construction The insert was prepared by digestion at NotI/EcoR0109 and subcloned into ple-gfp retroviral vector backbone at XhoI/NaeI sites, after removal of CMV-GFP cassette by blunt cloning (T4 DNA pol and CIP treatment). Both forward and reverse insertion orientation were tested as described in Fig.4, with reverse (3R) orientation showing the highest fidelity of myogenin-gfp coexpression. Virus preparation and infection pmig retrovirus vector encoding for the human Rb cdna (Sage et al. 2000) as well as all other vectors employed were transfected into ecotropic phoenix cells using FuGENE 6 (Roche). Cells were infected with viral supernatants containing polybrene (5μg ml-1) and centrifuged for 30min at 2,000g. sirna treatment Transfections of sirna duplexes, resuspended in sirna buffer (Dharmacon) were carried out after differentiation of myocytes or after fusion of myotubes at 48-72hrs in DM with siimporter transfection reagent (Millipore) as per manufacturer s instructions. Transfection mix was added to cells in differentiation media for 12 hrs. RNA was harvested from C2C12 or primary cells in GM or DM by RNeasy mini kit (Qiagen) and 200ng of total RNA was used in semiquantitative RTPCR analysis with Superscript III One-Step RT-PCR (invitrogen). This doubleknockdown (DKD) treatment protocol resulted in comparable results to the TAM and p16/19si treatment, while a combination of sirnas applied simultaneously was not as efficient in silencing the expression of either gene (Figure S1 G) RT-PCR primers and reactions all sequences are in 5-3 orientation: Rb set 1 (For: 3
4 GAGGAGAATTCTGTGGGCCAGGGCTGTG ; Rev: GTACGAGCTCGAGCCGCTGGGAGATGTT) product size 1368bp. Rb set 2 (For: CAGGCTTGAGTTTGAAGAAATTG ; Rev: ATGCCCCAGAGTTCCTTCTTC) product size 168bp. p16/19 (For: CGCCTTTTTCTTCTTAGCTTCA ; Rev: AGTTTCTCATGCCATTCCTTTC) product size 220bp. p19arf (For: CCCACTCCAAGAGAGGGTTT ; Rev: AGCTATGCCCGTCGGTCT) product size 465bp. Anillin (For: GCGTACCAGCAACTTTACCC ; Rev: GGCACCAAAGCCACTAACAT) product size 202bp. AuroraB (For: TCGCTGTTGTTTCCCTCTCT ; Rev: GATCTTGAGTGCCACGATGA) product size 388bp. Survivin (For: CATCGCCACCTTCAAGAACT; Rev: AGCTGCTCAATTGACTGACG) product size 360bp. MRF4 (For: GGCTGGATCAGCAAGAGAAG; Rev: CCTGCTGGGTGAAGAATGTT) product size 317bp. Myogenin (For: TCCAGTACATTGAGCGCCTA ; Rev: GGGCTGGGTGTTAGCCTTAT) product size 470bp. MHC (For: TGAGAAGGAAGCGCTGGTAT ; Rev: TCTGCAATCTGTTCCGTGAG) product size 588bp. M-CK (For: GATCTTCAAGAAGGCTGGTCAC ; Rev: CAATGATTGGACTTCCAGGAG) product size 428bp. Annealing temperature ranged from 58-62oC and cycles from 23 (Myogenin) to 31 (p19arf) with most products visible at 26 to 28 cycles. For RNA loading control 50ng of total RNA was ran with GAPDH primers (For: CACTGAGCATCTCCCTCACA ; Rev: TGGGTGCAGCGAACTTTATT) product size 122bp seen at 22 cycles. Western Analysis Lysates were cleared by centrifugation 5min at 6000rpm at 4OC, and protein quantification was determined by Bradford assay (Bio-Rad). Immunoblotting was conducted using NuPage system from invitrogen. Samples were loaded in 10% Bis-Tris pre33 cast gels, resolved by running with MES or MOPS SDS buffer, and transferred at 4oC onto Immobilon-P membranes (Amersham). Membranes were blocked in 5% milk-pbs-t then incubated with antibodies against Rb (BD) diluted 1/250 or Rb (Sage lab) 1/1000; p19arf (abcam) 1/1000; Survivin (Cytoskeleton Inc) 1/1000; AuroraB (Cytoskeleton Inc.) 1/500; MHC (Chemicon) 1/500; adult MHC (Blau lab) 1/50; M-CK (Novus) 1/500; Myogenin (BD) 1/500; and GAPDH as protein loading control diluted 1/10000 (Santa Cruz biotech). Membranes were then incubated with HRP-conjugated secondary antibodies (1/5000) (Zymed) and detected by ECL or ECL + detection reagents (Amersham). Nuclear Staining Nuclear staining was with Hoechst (Sigma) diluted 1/5000 and incubated at RT for 15 min. 4
5 PALM LPC 72-96hrs after DM switch and upon verification of GFP expression by direct native fluorescence, myocytes were treated as described in Fig. 5. Duplex dishes were equilibrated by allowing media to flow in between the membrane layers via permeabilization of the top PEN membrane by LPC function at least 24hrs before cell capture. GFP-myogenin expression was verified for every myocyte selected by direct immunofluorescence with X-CITE series 120 EXFO. Ablation was carried out through a 20x LD Plan-NeoFluar objective, which permits for a 3-5μm in width laser track, and membranes ranging from 50 to 150 μm 2 in area were catapulted by LPC, and the burst point was selected to be at least 10μm from the myocyte. Media was removed from the Duplex dish until only a film of moisture covered the plate, in 50mm dishes this condition can be obtained by retaining only 500μL of media. Time Lapse Microscopy Myocytes were first imaged 12hrs after sorting into microwells, at which time they were treated with Mocksi or the first sirna in the two step DKD treatment (Fig. S4 A left panels). Images of microwells were captured at 48, 72 and 96hrs after exposure to the second sirna of the DKD treatment. Raw data showed that a total of 109 colonies arose from the DKD treated myocytes, and only 10 colonies arose from the mock treated myocyte population. Primary myoblasts were seeded for fusion in 6-well collagen coated plates. 72hrs in DM PM were treated with TAM and 96hrs in DM one set of primary myotubes were treated with 200nM p16/19si for 12hrs, at which point transfection mix was washed and cells were placed in fresh DM media for 12hrs. 5
6 Figure S1. siimporter myotube transfection, TAM and sirna treatment analysis. (A) Representative images of DM5 C2C12 myotubes transfected with siglo-green (non-specific sirna dulplexes conjugated to alexa-488 molecules) and either Fugene (top) or siimporter (bottom). Myotubes labeled for MHC (red) and Hoechst (blue). Bar 50μm. (B) Quantification of transfection efficiency of siglo-green (100nM) with Fugene, or varying siglo-green concentrations with siimporter. Myotubes cultured in differentiation medium for 5 days (DM5). Error bars indicate the mean ± SE of three independent experiments (*P<0.001, **P<0.005). (C) X-gal staining, indicating Cre expression of primary myoblasts in GM treated 6
7 with EtOH or TAM for 24hrs. Bar 100μm. (D) X-gal staining, marking Cre expression of primary myotubes in DM5 treated with EtOH or TAM for 24hrs. Bar 200μm. (E) sqrt-pcr analysis of RB in DM5 primary myotubes after TAM treatment. TAM was added to DM2 myotube cultures for the indicated of amount time. (F) sqrt-pcr timecourse of RB and p19arf expression in primary myotubes after 24hr TAM treatment. (G) sqrt-pcr analysis of DKD treatment of primary myotubes with sirna duplexes for RBsi or p16/19si, delivered in tandem (T), in unison (U) or in unison at half dose-100nm (hu). (H) Histogram representation of BrdU-positive C2C12 myotubes after treatment with mock sirna and two different concentration of Rbsi. At least 200 myotubes were counted per trial. Error bars indicate the mean ± SE of two independent experiments. (I) Low levels of apoptosis in primary myotubes after loss of Rb and p16/19. Representative immunofluorescence images of primary myotubes at day 6 during differentiation treated as indicated and labeled for AnnexinV (green) and propidium iodide (PI). Serum starvation is a positive control for induction of apoptosis in myotubes. 7
8 Figure S2. Decrease in RB and p16/19 levels leads to upregulation of mitotic machinery. (A) semi-quantitative RT-PCR (sqrt-pcr) analysis of expression of Anillin, AuroraB and Survivin in GM, mock-treated DM5, in DM5 with TAM alone or with TAM and p16/19si. (B) Western blot analysis showing protein levels of AuroraB (38 kda) and Survivin (20 kda) following TAM treatment or TAM and p16/19si in DM5. GAPDH (35 kda). (C) Immunofluorescence images of primary myotubes in DM5 after mock treatment, and in DM5 after treatment with TAM and p16/19-sirna. Myotubes were labeled with primary antibody for BrdU (green), Survivin (red), as well as with Hoechst nuclear dye (blue). Bar, 50μm. (D) Histogram represents colocalization of BrdU and Survivin in primary myotube nuclei treated with TAM and non-specific Mock-siRNA duplexes as compared to myotubes treated TAM and p16/19-sirna duplexes. Growth medium (GM); Myotubes cultured in differentiation medium for 4 or 5 days (DM4 or DM5 respectively). A minimum of 500 nuclei were counted from random fields for each trial. Error bars indicate the mean ± SE of at least three independent experiments. (*P<0.005). (E) sqrt-pcr analysis of expression levels of 8
9 cyclins D and E1 as well as Emi1/FBOX5, in primary myoblasts in GM, DM and in DM treated as indicated. (F) Immunofluorescence images of primary myotubes treated with nonspecific sirna duplexes (Mocksi) or TAM and p16/19si. Myotubes labeled for Eg5 (green), MHC (red) and Hoechst (blue). Figure S3. Dedifferentiation in C2C12 and primary myotubes. (A) Western analysis of MHC protein (220 kda) in C2C12 myoblasts (GM) and in myotubes (DM) following treatment with sirna duplexes against Rb. (B) Immunofluorescence images of DM5 C2C12 myotubes treated with non-specific sirna duplexes and Rbsi for at least 48hrs. Myotubes labeled with primary antibodies for MHC (red), BrdU (green), and Hoechst (blue). Bar, 150μm. (C) sqrt-pcr analysis of the expression of late differentiation myogenic markers in Growth Medium (GM) or in Differentiation Medium (DM) before and after TAM or TAM and p16/19si treatment. (D) Western blot analysis of M-CK (50 kda) in primary myoblasts (GM) 9
10 and DM5 treated as indicated with sirna duplexes and/or TAM; the 45-kD (lower) band seen in the M-CK blot, is likely due to a shared epitope identified by the antibody. In each of the Westerns, GAPDH (35 kda) is the loading control. (E) Representative phase and immunofluorescence images of primary myotubes after Mocksi or TAM and p16/19si treatment. Myotubes were labeled with primary antibodies for α-tubulin (green) and Hoechst (blue). (F) Histogram representing the levels of α-tubulin in GM, in DM3, in DM6 and in DM6 treated with TAM or with TAM and p16/19si. Samples are normalized to protein levels of α-tubulin in GM. Error bars indicate the mean ± SE of two independent experiments. 10
11 Figure S4. Myocytes deficient in RB and p16/19 proliferate following FACS isolation. (A) Representative images ple-myog3r-gfp myocytes FACS sorted directly into microwells, imaged before treatment, as well as 72 and 96hrs after DKD treatment. Magnified panel, highlights expansion of one well following Rbsi and p16/19si treatment. Bar 150μm. (B) Histogram represents percentage of GFP-positive cells that also express myogenin following FACS isolation of a subset of cells sorted into microwells. Individual cells were evaluated for 11
12 expression of each marker. A minimum of 250 cells were counted from random fields. Error bars indicate the mean ± SE of two independent experiments. (C) Histogram represents percent colony formation observed in microwells at 96hrs after treatment. A minimum of 500 wells with at least one cell before treatment counted for each trial. Error bars indicate the mean ± SE of at least three independent experiments. (*P<0.005). Single-cell isolation and expansion of dedifferentiated myogenin-gfp myocytes: (D) Representative images of untreated myogenin-gfp + myocyte (DM4) prior to microdissection, after LPC, and 72hrs post-isolation. Bar 50μm. (E) Representative images of TAM with p16/19si treated myogenin-gfp + myocytes, prior to microdissection, after LPC, 48hrs post-isolation, 72hrs post-isolation, 96hrs post-isolation, at which point media was changed, and 120hrs post-isolation. Bar 50μm, last panel bar 200μm. 12
13 Figure S5. Single-cell isolation and clonal expansion of FACS sorted myogenin- GFP + myocytes. (A) Schematic representation of myocyte FACS sorting, treatment and single cell isolation by PALM LPC. (B) Representative images of FACS sorted, TAM treated, myocytes (DM4) prior to microdissection, after microdissection, after LPC isolation, and 72hrs postisolation. Bar 50μm. (C) Representative images of FACS sorted myocytes (DM4) treated with TAM+ and 16/19si prior to microdissection, after microdissection, after LPC isolation, and 72 to 73hrs post-isolation as well as 96hrs post-isolation. Bar 50μm, 72hrs panel bar 200μm. 13
14 Figure S6. PALM isolation, expansion and differentiation of primary myoblasts and TAM and p16/19si treated myocytes. (A) PALM LPC isolated myoblasts at 48hrs postisolation and at 96hrs post-isolation. Three weeks post-isolation, captured myoblasts were placed in Differentiation Medium (DM). Images show culture after 3 days in DM (DM3). (B) PALM isolated myogenin-promoter-gfp myocytes treated with TAM and p16/19si, prior to microdissection, 72hrs post-isolation displaying both phase and native GFP imaging. Three weeks post-isolation expanded and dedifferentiated myoblasts were placed in DM, and images show culture after 3 days in DM (DM3) as well as after 6 days in DM (DM6). Growth medium (GM). (C) Western blot analysis of PALM isolated primary myoblast colony in GM and DM4, showing protein levels of RB (100 kda), p19arf (20 kda), MHC (220 kda), and Survivin (20 kda). GAPDH (35 kda) is loading control. (D) sqrt-pcr expression analysis of PALM isolated primary myoblasts and DKD treated myocytes in GM and DM4. (E) Western blot analysis of MyoD (34 kda) levels in primary muscle cells in GM, DM at indicated time points 14
15 and with indicated treatments. GAPDH (35 kda) is displayed as loading control. Growth medium (GM); Myotubes cultured in differentiation medium for 4 or 5 days (DM4 or DM5 respectively). Figure S7. DKD dedifferentiated, captured and expanded myocytes fuse to muscle in vivo. Representative fields of cross-sections of tibialis anterior of CB17/SCID mice 10 days post-injection of 1.5x10 5 cells from DKD captured and expanded myocytes. Sections were stained for Hoechst (blue), GFP (green) and laminin (orange). Incorporation of dedifferentiated myocytes into pre-existing fibers can be visualized in merged fields by GFP + staining of a laminin-bound fibers; GFP expression marks myogenin expression in fibers where dedifferentiated myocytes fused. (n=6, two of which were positive for GFP + fibers). Bar 100μm. Movie S1. Timelapse analysis of primary myotubes after TAM+Mocksi treatment. Frames were captured 24hrs after sirna treatment at every 10min for a total of 72hrs, encompassing days 5, 6 and 7 during differentiation. Available online. Movie S2. Timelapse analysis of primary myotubes after TAM+p16/19si treatment. Frames were captured 24hrs after sirna treatment at every 10min for a total of 72hrs, encompassing days 5, 6 and 7 during differentiation. Available online. 15
Transient Inactivation of Rb and ARF Yields Regenerative Cells from Postmitotic Mammalian Muscle
 Article Transient Inactivation of Rb and ARF Yields Regenerative Cells from Postmitotic Mammalian Muscle Kostandin V. Pajcini, 1 Stephane Y. Corbel, 1 Julien Sage, 2 Jason H. Pomerantz, 1,3, * and Helen
Article Transient Inactivation of Rb and ARF Yields Regenerative Cells from Postmitotic Mammalian Muscle Kostandin V. Pajcini, 1 Stephane Y. Corbel, 1 Julien Sage, 2 Jason H. Pomerantz, 1,3, * and Helen
Sarker et al. Supplementary Material. Subcellular Fractionation
 Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
SUPPLEMENTAL FIGURE LEGENDS
 SUPPLEMENTAL FIGURE LEGENDS Suppl. Fig. 1. Notch and myostatin expression is unaffected in the absence of p65 during postnatal development. A & B. Myostatin and Notch-1 expression levels were determined
SUPPLEMENTAL FIGURE LEGENDS Suppl. Fig. 1. Notch and myostatin expression is unaffected in the absence of p65 during postnatal development. A & B. Myostatin and Notch-1 expression levels were determined
Supplementary Figure 1. Expressions of stem cell markers decreased in TRCs on 2D plastic. TRCs were cultured on plastic for 1, 3, 5, or 7 days,
 Supplementary Figure 1. Expressions of stem cell markers decreased in TRCs on 2D plastic. TRCs were cultured on plastic for 1, 3, 5, or 7 days, respectively, and their mrnas were quantified by real time
Supplementary Figure 1. Expressions of stem cell markers decreased in TRCs on 2D plastic. TRCs were cultured on plastic for 1, 3, 5, or 7 days, respectively, and their mrnas were quantified by real time
Supplemental Table/Figure Legends
 MiR-26a is required for skeletal muscle differentiation and regeneration in mice Bijan K. Dey, Jeffrey Gagan, Zhen Yan #, Anindya Dutta Supplemental Table/Figure Legends Suppl. Table 1: Effect of overexpression
MiR-26a is required for skeletal muscle differentiation and regeneration in mice Bijan K. Dey, Jeffrey Gagan, Zhen Yan #, Anindya Dutta Supplemental Table/Figure Legends Suppl. Table 1: Effect of overexpression
Supplementary Material
 Supplementary Material Supplementary Methods Cell synchronization. For synchronized cell growth, thymidine was added to 30% confluent U2OS cells to a final concentration of 2.5mM. Cells were incubated
Supplementary Material Supplementary Methods Cell synchronization. For synchronized cell growth, thymidine was added to 30% confluent U2OS cells to a final concentration of 2.5mM. Cells were incubated
Supplementary Information: Materials and Methods. GST and GST-p53 were purified according to standard protocol after
 Supplementary Information: Materials and Methods Recombinant protein expression and in vitro kinase assay. GST and GST-p53 were purified according to standard protocol after induction with.5mm IPTG for
Supplementary Information: Materials and Methods Recombinant protein expression and in vitro kinase assay. GST and GST-p53 were purified according to standard protocol after induction with.5mm IPTG for
Supplemental material
 Supplemental material THE JOURNAL OF CELL BIOLOGY Taylor et al., http://www.jcb.org/cgi/content/full/jcb.201403021/dc1 Figure S1. Representative images of Cav 1a -YFP mutants with and without LMB treatment.
Supplemental material THE JOURNAL OF CELL BIOLOGY Taylor et al., http://www.jcb.org/cgi/content/full/jcb.201403021/dc1 Figure S1. Representative images of Cav 1a -YFP mutants with and without LMB treatment.
Positive selection gates for the collection of LRCs or nonlrcs had to be drawn based on the location and
 Determining positive selection gates for LRCs and nonlrcs Positive selection gates for the collection of LRCs or nonlrcs had to be drawn based on the location and shape of the Gaussian distributions. For
Determining positive selection gates for LRCs and nonlrcs Positive selection gates for the collection of LRCs or nonlrcs had to be drawn based on the location and shape of the Gaussian distributions. For
Fig. S1 TGF RI inhibitor SB effectively blocks phosphorylation of Smad2 induced by TGF. FET cells were treated with TGF in the presence of
 Fig. S1 TGF RI inhibitor SB525334 effectively blocks phosphorylation of Smad2 induced by TGF. FET cells were treated with TGF in the presence of different concentrations of SB525334. Cells were lysed and
Fig. S1 TGF RI inhibitor SB525334 effectively blocks phosphorylation of Smad2 induced by TGF. FET cells were treated with TGF in the presence of different concentrations of SB525334. Cells were lysed and
Cytotoxicity of Botulinum Neurotoxins Reveals a Direct Role of
 Supplementary Information Cytotoxicity of Botulinum Neurotoxins Reveals a Direct Role of Syntaxin 1 and SNAP-25 in Neuron Survival Lisheng Peng, Huisheng Liu, Hongyu Ruan, William H. Tepp, William H. Stoothoff,
Supplementary Information Cytotoxicity of Botulinum Neurotoxins Reveals a Direct Role of Syntaxin 1 and SNAP-25 in Neuron Survival Lisheng Peng, Huisheng Liu, Hongyu Ruan, William H. Tepp, William H. Stoothoff,
B. ADM: C. D. Apoptosis: 1.68% 2.99% 1.31% Figure.S1,Li et al. number. invaded cells. HuH7 BxPC-3 DLD-1.
 A. - Figure.S1,Li et al. B. : - + - + - + E-cadherin CK19 α-sma vimentin β -actin C. D. Apoptosis: 1.68% 2.99% 1.31% - : - + - + - + Apoptosis: 48.33% 45.32% 44.59% E. invaded cells number 400 300 200
A. - Figure.S1,Li et al. B. : - + - + - + E-cadherin CK19 α-sma vimentin β -actin C. D. Apoptosis: 1.68% 2.99% 1.31% - : - + - + - + Apoptosis: 48.33% 45.32% 44.59% E. invaded cells number 400 300 200
MeCP2. MeCP2/α-tubulin. GFP mir1-1 mir132
 Conservation Figure S1. Schematic showing 3 UTR (top; thick black line), mir132 MRE (arrow) and nucleotide sequence conservation (vertical black lines; http://genome.ucsc.edu). a GFP mir1-1 mir132 b GFP
Conservation Figure S1. Schematic showing 3 UTR (top; thick black line), mir132 MRE (arrow) and nucleotide sequence conservation (vertical black lines; http://genome.ucsc.edu). a GFP mir1-1 mir132 b GFP
Supplementary information for: Ten-Eleven Translocation-2 (Tet2) Is Involved in Myogenic Differentiation of Skeletal Myoblast Cells in
 Supplementary information for: Ten-Eleven Translocation-2 (Tet2) Is Involved in Myogenic Differentiation of Skeletal Myoblast Cells in Vitro Xia Zhong*, Qian-Qian Wang*, Jian-Wei Li, Yu-Mei Zhang, Xiao-Rong
Supplementary information for: Ten-Eleven Translocation-2 (Tet2) Is Involved in Myogenic Differentiation of Skeletal Myoblast Cells in Vitro Xia Zhong*, Qian-Qian Wang*, Jian-Wei Li, Yu-Mei Zhang, Xiao-Rong
RNA was isolated using NucleoSpin RNA II (Macherey-Nagel, Bethlehem, PA) according to the
 Supplementary Methods RT-PCR and real-time PCR analysis RNA was isolated using NucleoSpin RNA II (Macherey-Nagel, Bethlehem, PA) according to the manufacturer s protocol and quantified by measuring the
Supplementary Methods RT-PCR and real-time PCR analysis RNA was isolated using NucleoSpin RNA II (Macherey-Nagel, Bethlehem, PA) according to the manufacturer s protocol and quantified by measuring the
Supplementary data. sienigma. F-Enigma F-EnigmaSM. a-p53
 Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Emanuela Tumini, Sonia Barroso, Carmen Pérez Calero and Andrés Aguilera
 SUPPLEMENTARY INFORMATION Roles of human POLD1 and POLD3 in genome stability Emanuela Tumini, Sonia Barroso, Carmen Pérez Calero and Andrés Aguilera SUPPLEMENTARY METHODS Cell proliferation After sirna
SUPPLEMENTARY INFORMATION Roles of human POLD1 and POLD3 in genome stability Emanuela Tumini, Sonia Barroso, Carmen Pérez Calero and Andrés Aguilera SUPPLEMENTARY METHODS Cell proliferation After sirna
Supplementary Data. Supplementary Methods Three-step protocol for spontaneous differentiation of mouse induced pluripotent stem (embryonic stem) cells
 Supplementary Data Supplementary Methods Three-step protocol for spontaneous differentiation of mouse induced pluripotent stem (embryonic stem) cells Mouse induced pluripotent stem cells (ipscs) were cultured
Supplementary Data Supplementary Methods Three-step protocol for spontaneous differentiation of mouse induced pluripotent stem (embryonic stem) cells Mouse induced pluripotent stem cells (ipscs) were cultured
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.
 Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
Supplementary methods
 Supplementary methods Cell culture, infection, transfection, and RNA interference HEK293 cells and its derivatives were grown in DMEM supplemented with 10% FBS. Various constructs were introduced into
Supplementary methods Cell culture, infection, transfection, and RNA interference HEK293 cells and its derivatives were grown in DMEM supplemented with 10% FBS. Various constructs were introduced into
At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in
 Supplementary Materials and Methods Barrier function assays At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in acidic X-gal mix (100 mm phosphate buffer at ph4.3, 3 mm
Supplementary Materials and Methods Barrier function assays At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in acidic X-gal mix (100 mm phosphate buffer at ph4.3, 3 mm
Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured
 Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured in 90-Pa 3D fibrin gels for 5 days in the presence
Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured in 90-Pa 3D fibrin gels for 5 days in the presence
ASPP1 Fw GGTTGGGAATCCACGTGTTG ASPP1 Rv GCCATATCTTGGAGCTCTGAGAG
 Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling
 Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary Methods
 Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Isolation, culture, and transfection of primary mammary epithelial organoids
 Supplementary Experimental Procedures Isolation, culture, and transfection of primary mammary epithelial organoids Primary mammary epithelial organoids were prepared from 8-week-old CD1 mice (Charles River)
Supplementary Experimental Procedures Isolation, culture, and transfection of primary mammary epithelial organoids Primary mammary epithelial organoids were prepared from 8-week-old CD1 mice (Charles River)
Supplementary Figure Legends
 Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
Grb2-Mediated Alteration in the Trafficking of AβPP: Insights from Grb2-AICD Interaction
 Journal of Alzheimer s Disease 20 (2010) 1 9 1 IOS Press Supplementary Material Grb2-Mediated Alteration in the Trafficking of AβPP: Insights from Grb2-AICD Interaction Mithu Raychaudhuri and Debashis
Journal of Alzheimer s Disease 20 (2010) 1 9 1 IOS Press Supplementary Material Grb2-Mediated Alteration in the Trafficking of AβPP: Insights from Grb2-AICD Interaction Mithu Raychaudhuri and Debashis
STAT3 signaling controls satellite cell expansion and skeletal muscle repair
 SUPPLEMENTARY INFORMATION STAT3 signaling controls satellite cell expansion and skeletal muscle repair Matthew Timothy Tierney 1 *, Tufan Aydogdu 2,3 *, David Sala 2, Barbora Malecova 2, Sole Gatto 2,
SUPPLEMENTARY INFORMATION STAT3 signaling controls satellite cell expansion and skeletal muscle repair Matthew Timothy Tierney 1 *, Tufan Aydogdu 2,3 *, David Sala 2, Barbora Malecova 2, Sole Gatto 2,
Spironolactone ameliorates PIT1-dependent vascular osteoinduction in klotho-hypomorphic mice
 Spironolactone ameliorates PIT1-dependent vascular osteoinduction in klotho-hypomorphic mice Supplementary Material Supplementary Methods Materials Spironolactone, aldosterone and β-glycerophosphate were
Spironolactone ameliorates PIT1-dependent vascular osteoinduction in klotho-hypomorphic mice Supplementary Material Supplementary Methods Materials Spironolactone, aldosterone and β-glycerophosphate were
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Han et al., http://www.jcb.org/cgi/content/full/jcb.201311007/dc1 Figure S1. SIVA1 interacts with PCNA. (A) HEK293T cells were transiently
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Han et al., http://www.jcb.org/cgi/content/full/jcb.201311007/dc1 Figure S1. SIVA1 interacts with PCNA. (A) HEK293T cells were transiently
SUPPLEMENTARY INFORMATION
 The Supplementary Information (SI) Methods Cell culture and transfections H1299, U2OS, 293, HeLa cells were maintained in DMEM medium supplemented with 10% fetal bovine serum. H1299 and 293 cells were
The Supplementary Information (SI) Methods Cell culture and transfections H1299, U2OS, 293, HeLa cells were maintained in DMEM medium supplemented with 10% fetal bovine serum. H1299 and 293 cells were
Establishing sirna assays in primary human peripheral blood lymphocytes
 page 1 of 7 Establishing sirna assays in primary human peripheral blood lymphocytes This protocol is designed to establish sirna assays in primary human peripheral blood lymphocytes using the Nucleofector
page 1 of 7 Establishing sirna assays in primary human peripheral blood lymphocytes This protocol is designed to establish sirna assays in primary human peripheral blood lymphocytes using the Nucleofector
sirna Transfection Into Primary Neurons Using Fuse-It-siRNA
 sirna Transfection Into Primary Neurons Using Fuse-It-siRNA This Application Note describes a protocol for sirna transfection into sensitive, primary cortical neurons using Fuse-It-siRNA. This innovative
sirna Transfection Into Primary Neurons Using Fuse-It-siRNA This Application Note describes a protocol for sirna transfection into sensitive, primary cortical neurons using Fuse-It-siRNA. This innovative
Supplementary Information Alternative splicing of CD44 mrna by ESRP1 enhances lung colonization of metastatic cancer cell
 Supplementary Information Alternative splicing of CD44 mrna by ESRP1 enhances lung colonization of metastatic cancer cell Supplementary Figures S1-S3 Supplementary Methods Supplementary Figure S1. Identification
Supplementary Information Alternative splicing of CD44 mrna by ESRP1 enhances lung colonization of metastatic cancer cell Supplementary Figures S1-S3 Supplementary Methods Supplementary Figure S1. Identification
LINGO-1, A TRANSMEMBRANE SIGNALING PROTEIN, INHIBITS OLIGODENDROCYTE DIFFERENTIATION AND MYELINATION THROUGH INTERCELLULAR SELF- INTERACTIONS.
 Supplemental Data: LINGO-1, A TRANSMEMBRANE SIGNALING PROTEIN, INHIBITS OLIGODENDROCYTE DIFFERENTIATION AND MYELINATION THROUGH INTERCELLULAR SELF- INTERACTIONS. Scott Jepson, Bryan Vought, Christian H.
Supplemental Data: LINGO-1, A TRANSMEMBRANE SIGNALING PROTEIN, INHIBITS OLIGODENDROCYTE DIFFERENTIATION AND MYELINATION THROUGH INTERCELLULAR SELF- INTERACTIONS. Scott Jepson, Bryan Vought, Christian H.
Supporting Information
 Supporting Information Stavru et al. 0.073/pnas.357840 SI Materials and Methods Immunofluorescence. For immunofluorescence, cells were fixed for 0 min in 4% (wt/vol) paraformaldehyde (Electron Microscopy
Supporting Information Stavru et al. 0.073/pnas.357840 SI Materials and Methods Immunofluorescence. For immunofluorescence, cells were fixed for 0 min in 4% (wt/vol) paraformaldehyde (Electron Microscopy
Confocal immunofluorescence microscopy
 Confocal immunofluorescence microscopy HL-6 and cells were cultured and cytospun onto glass slides. The cells were double immunofluorescence stained for Mt NPM1 and fibrillarin (nucleolar marker). Briefly,
Confocal immunofluorescence microscopy HL-6 and cells were cultured and cytospun onto glass slides. The cells were double immunofluorescence stained for Mt NPM1 and fibrillarin (nucleolar marker). Briefly,
Supplemental Information
 Supplemental Information Intrinsic protein-protein interaction mediated and chaperonin assisted sequential assembly of a stable Bardet Biedl syndome protein complex, the BBSome * Qihong Zhang 1#, Dahai
Supplemental Information Intrinsic protein-protein interaction mediated and chaperonin assisted sequential assembly of a stable Bardet Biedl syndome protein complex, the BBSome * Qihong Zhang 1#, Dahai
Input. Pulldown GST. GST-Ahi1
 A kd 250 150 100 75 50 37 25 -GST-Ahi1 +GST-Ahi1 kd 148 98 64 50 37 22 GST GST-Ahi1 C kd 250 148 98 64 50 Control Input Hap1A Hap1 Pulldown GST GST-Ahi1 Hap1A Hap1 Hap1A Hap1 Figure S1. (A) Ahi1 western
A kd 250 150 100 75 50 37 25 -GST-Ahi1 +GST-Ahi1 kd 148 98 64 50 37 22 GST GST-Ahi1 C kd 250 148 98 64 50 Control Input Hap1A Hap1 Pulldown GST GST-Ahi1 Hap1A Hap1 Hap1A Hap1 Figure S1. (A) Ahi1 western
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the
 Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
SUPPLEMENTAL MATERIALS SIRTUIN 1 PROMOTES HYPEROXIA-INDUCED LUNG EPITHELIAL DEATH INDEPENDENT OF NRF2 ACTIVATION
 SUPPLEMENTAL MATERIALS SIRTUIN PROMOTES HYPEROXIA-INDUCED LUNG EPITHELIAL DEATH INDEPENDENT OF NRF ACTIVATION Haranatha R. Potteti*, Subbiah Rajasekaran*, Senthilkumar B. Rajamohan*, Chandramohan R. Tamatam,
SUPPLEMENTAL MATERIALS SIRTUIN PROMOTES HYPEROXIA-INDUCED LUNG EPITHELIAL DEATH INDEPENDENT OF NRF ACTIVATION Haranatha R. Potteti*, Subbiah Rajasekaran*, Senthilkumar B. Rajamohan*, Chandramohan R. Tamatam,
Supporting Information
 Supporting Information Tal et al. 10.1073/pnas.0807694106 SI Materials and Methods VSV Infection and Quantification. Infection was carried out by seeding 5 10 5 MEF cells per well in a 6-well plate and
Supporting Information Tal et al. 10.1073/pnas.0807694106 SI Materials and Methods VSV Infection and Quantification. Infection was carried out by seeding 5 10 5 MEF cells per well in a 6-well plate and
Supplementary Protocol. sirna transfection methodology and performance
 Supplementary Protocol sirna transfection methodology and performance sirna oligonucleotides, DNA construct and cell line. Chemically synthesized 21 nt RNA duplexes were obtained from Ambion Europe, Ltd.
Supplementary Protocol sirna transfection methodology and performance sirna oligonucleotides, DNA construct and cell line. Chemically synthesized 21 nt RNA duplexes were obtained from Ambion Europe, Ltd.
SANTA CRUZ BIOTECHNOLOGY, INC.
 TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
Supporting Information
 Supporting Information Su et al. 10.1073/pnas.1211604110 SI Materials and Methods Cell Culture and Plasmids. Tera-1 and Tera-2 cells (ATCC: HTB- 105/106) were maintained in McCoy s 5A medium with 15% FBS
Supporting Information Su et al. 10.1073/pnas.1211604110 SI Materials and Methods Cell Culture and Plasmids. Tera-1 and Tera-2 cells (ATCC: HTB- 105/106) were maintained in McCoy s 5A medium with 15% FBS
Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan).
 1 2 3 4 5 6 7 8 Supplemental Materials and Methods Cell proliferation assay Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). GCs were plated at 96-well
1 2 3 4 5 6 7 8 Supplemental Materials and Methods Cell proliferation assay Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). GCs were plated at 96-well
Supplementary Information: Materials and Methods. Immunoblot and immunoprecipitation. Cells were washed in phosphate buffered
 Supplementary Information: Materials and Methods Immunoblot and immunoprecipitation. Cells were washed in phosphate buffered saline (PBS) and lysed in TNN lysis buffer (50mM Tris at ph 8.0, 120mM NaCl
Supplementary Information: Materials and Methods Immunoblot and immunoprecipitation. Cells were washed in phosphate buffered saline (PBS) and lysed in TNN lysis buffer (50mM Tris at ph 8.0, 120mM NaCl
HeLa cells stably transfected with an empty pcdna3 vector (HeLa Neo) or with a plasmid
 SUPPLEMENTAL MATERIALS AND METHODS Cell culture, transfection and treatments. HeLa cells stably transfected with an empty pcdna3 vector (HeLa Neo) or with a plasmid encoding vmia (HeLa vmia) 1 were cultured
SUPPLEMENTAL MATERIALS AND METHODS Cell culture, transfection and treatments. HeLa cells stably transfected with an empty pcdna3 vector (HeLa Neo) or with a plasmid encoding vmia (HeLa vmia) 1 were cultured
Supplementary Fig. 1. (A) Working model. The pluripotency transcription factor OCT4
 SUPPLEMENTARY FIGURE LEGENDS Supplementary Fig. 1. (A) Working model. The pluripotency transcription factor OCT4 directly up-regulates the expression of NIPP1 and CCNF that together inhibit protein phosphatase
SUPPLEMENTARY FIGURE LEGENDS Supplementary Fig. 1. (A) Working model. The pluripotency transcription factor OCT4 directly up-regulates the expression of NIPP1 and CCNF that together inhibit protein phosphatase
Flow cytometry Stained cells were analyzed and sorted by SORP FACS Aria (BD Biosciences).
 Mice C57BL/6-Ly5.1 or -Ly5.2 congenic mice were used for LSK transduction and competitive repopulation assays. Animal care was in accordance with the guidelines of Keio University for animal and recombinant
Mice C57BL/6-Ly5.1 or -Ly5.2 congenic mice were used for LSK transduction and competitive repopulation assays. Animal care was in accordance with the guidelines of Keio University for animal and recombinant
Apoptosis assay: Apoptotic cells were identified by Annexin V-Alexa Fluor 488 and Propidium
 Apoptosis assay: Apoptotic cells were identified by Annexin V-Alexa Fluor 488 and Propidium Iodide (Invitrogen, Carlsbad, CA) staining. Briefly, 2x10 5 cells were washed once in cold PBS and resuspended
Apoptosis assay: Apoptotic cells were identified by Annexin V-Alexa Fluor 488 and Propidium Iodide (Invitrogen, Carlsbad, CA) staining. Briefly, 2x10 5 cells were washed once in cold PBS and resuspended
To examine the silencing effects of Celsr3 shrna, we co transfected 293T cells with expression
 Supplemental figures Supplemental Figure. 1. Silencing expression of Celsr3 by shrna. To examine the silencing effects of Celsr3 shrna, we co transfected 293T cells with expression plasmids for the shrna
Supplemental figures Supplemental Figure. 1. Silencing expression of Celsr3 by shrna. To examine the silencing effects of Celsr3 shrna, we co transfected 293T cells with expression plasmids for the shrna
Gene Forward Primer Reverse Primer GAPDH ATCATCCCTGCCTCTACTGG GTCAGGTCCACCACTGACAC SSB1 AACTTCAGTGAGCCAAACCC GTTCTCAGAGGCTGGAGAGG
 Supplemental Data EXPERIMENTAL PROCEDURES Plasmids and Antibodies- Full length cdna of INT11 or INT12 were cloned into ps- Flag-SBP vector respectively. Anti-RNA pol II (RPB1) was purchased from Santa
Supplemental Data EXPERIMENTAL PROCEDURES Plasmids and Antibodies- Full length cdna of INT11 or INT12 were cloned into ps- Flag-SBP vector respectively. Anti-RNA pol II (RPB1) was purchased from Santa
Firefly luciferase mutants as sensors of proteome stress
 Nature Methods Firefly luciferase mutants as sensors of proteome stress Rajat Gupta, Prasad Kasturi, Andreas Bracher, Christian Loew, Min Zheng, Adriana Villella, Dan Garza, F Ulrich Hartl & Swasti Raychaudhuri
Nature Methods Firefly luciferase mutants as sensors of proteome stress Rajat Gupta, Prasad Kasturi, Andreas Bracher, Christian Loew, Min Zheng, Adriana Villella, Dan Garza, F Ulrich Hartl & Swasti Raychaudhuri
Electrophoretic Mobility Shift Assay (EMSA). Nuclear extracts were. oligonucleotide spanning the NF-kB site (5 -GATCC-
 SUPPLEMENTARY MATERIALS AND METHODS Electrophoretic Mobility Shift Assay (EMSA). Nuclear extracts were prepared as previously described. (1) A [ 32 P] datp-labeled doublestranded oligonucleotide spanning
SUPPLEMENTARY MATERIALS AND METHODS Electrophoretic Mobility Shift Assay (EMSA). Nuclear extracts were prepared as previously described. (1) A [ 32 P] datp-labeled doublestranded oligonucleotide spanning
Supplemental Methods Cell lines and culture
 Supplemental Methods Cell lines and culture AGS, CL5, BT549, and SKBR were propagated in RPMI 64 medium (Mediatech Inc., Manassas, VA) supplemented with % fetal bovine serum (FBS, Atlanta Biologicals,
Supplemental Methods Cell lines and culture AGS, CL5, BT549, and SKBR were propagated in RPMI 64 medium (Mediatech Inc., Manassas, VA) supplemented with % fetal bovine serum (FBS, Atlanta Biologicals,
Chromatin immunoprecipitation (ChIP) ES and FACS-sorted GFP+/Flk1+ cells were fixed in 1% formaldehyde and sonicated until fragments of an average
 Chromatin immunoprecipitation (ChIP) ES and FACS-sorted cells were fixed in % formaldehyde and sonicated until fragments of an average size of 5 bp were obtained. Soluble, sheared chromatin was diluted
Chromatin immunoprecipitation (ChIP) ES and FACS-sorted cells were fixed in % formaldehyde and sonicated until fragments of an average size of 5 bp were obtained. Soluble, sheared chromatin was diluted
Mammosphere formation assay. Mammosphere culture was performed as previously described (13,
 Supplemental Text Materials and Methods Mammosphere formation assay. Mammosphere culture was performed as previously described (13, 17). For co-culture with fibroblasts and treatment with CM or CCL2, fibroblasts
Supplemental Text Materials and Methods Mammosphere formation assay. Mammosphere culture was performed as previously described (13, 17). For co-culture with fibroblasts and treatment with CM or CCL2, fibroblasts
A novel mechanism of post-translational modulation of HMGA functions by the histone chaperone nucleophosmin.
 A novel mechanism of post-translational modulation of HMGA functions by the histone chaperone nucleophosmin. Laura Arnoldo 1,#, Riccardo Sgarra 1,#, Eusebio Chiefari 2, Stefania Iiritano 2, Biagio Arcidiacono
A novel mechanism of post-translational modulation of HMGA functions by the histone chaperone nucleophosmin. Laura Arnoldo 1,#, Riccardo Sgarra 1,#, Eusebio Chiefari 2, Stefania Iiritano 2, Biagio Arcidiacono
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
This Document Contains:
 This Document Contains: 1. In-Cell Western Protocol II. Cell Seeding and Stimulation Supplemental Protocol III. Complete Assay Example: Detailing the Seeding, Stimulation and Detection of the A431 Cellular
This Document Contains: 1. In-Cell Western Protocol II. Cell Seeding and Stimulation Supplemental Protocol III. Complete Assay Example: Detailing the Seeding, Stimulation and Detection of the A431 Cellular
Supplementary Figure 1. Nature Structural & Molecular Biology: doi: /nsmb.3494
 Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Flow cytometric determination of apoptosis by annexin V/propidium iodide double staining.
 Supplementary materials and methods Flow cytometric determination of apoptosis by annexin V/propidium iodide double staining. Cells were analyzed for phosphatidylserine exposure by an annexin-v FITC/propidium
Supplementary materials and methods Flow cytometric determination of apoptosis by annexin V/propidium iodide double staining. Cells were analyzed for phosphatidylserine exposure by an annexin-v FITC/propidium
Supplementary Information (Ha, et. al) Supplementary Figures Supplementary Fig. S1
 Supplementary Information (Ha, et. al) Supplementary Figures Supplementary Fig. S1 a His-ORMDL3 ~ 17 His-ORMDL3 GST-ORMDL3 - + - + IPTG GST-ORMDL3 ~ b Integrated Density (ORMDL3/ -actin) 0.4 0.3 0.2 0.1
Supplementary Information (Ha, et. al) Supplementary Figures Supplementary Fig. S1 a His-ORMDL3 ~ 17 His-ORMDL3 GST-ORMDL3 - + - + IPTG GST-ORMDL3 ~ b Integrated Density (ORMDL3/ -actin) 0.4 0.3 0.2 0.1
Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna
 Supplemental Materials and Methods Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna (Mission shrna, Sigma) against mouse MMP14 were transfected into HEK293 cells using FuGene6
Supplemental Materials and Methods Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna (Mission shrna, Sigma) against mouse MMP14 were transfected into HEK293 cells using FuGene6
CD93 and dystroglycan cooperation in human endothelial cell adhesion and migration
 /, Supplementary Advance Publications Materials 2016 CD93 and dystroglycan cooperation in human endothelial cell adhesion and migration Supplementary Materials Supplementary Figure S1: In ECs CD93 silencing
/, Supplementary Advance Publications Materials 2016 CD93 and dystroglycan cooperation in human endothelial cell adhesion and migration Supplementary Materials Supplementary Figure S1: In ECs CD93 silencing
Cell death analysis using the high content bioimager BD PathwayTM 855 instrument (BD
 Supplemental information Materials and Methods: Cell lines, reagents and antibodies: Wild type (A3) and caspase-8 -/- (I9.2) Jurkat cells were cultured in RPMI 164 medium (Life Technologies) supplemented
Supplemental information Materials and Methods: Cell lines, reagents and antibodies: Wild type (A3) and caspase-8 -/- (I9.2) Jurkat cells were cultured in RPMI 164 medium (Life Technologies) supplemented
Supporting Information
 Supporting Information Casson et al. 10.1073/pnas.1421699112 Sl Materials and Methods Macrophage Infections. In experiments where macrophages were primed with LPS, cells were pretreated with 0.5 μg/ml
Supporting Information Casson et al. 10.1073/pnas.1421699112 Sl Materials and Methods Macrophage Infections. In experiments where macrophages were primed with LPS, cells were pretreated with 0.5 μg/ml
Figure S1. Phenotypic characterization of transfected ECFC. (a) ECFC were transfected using a lentivirus with a vector encoding for either human EPO
 Figure S1. Phenotypic characterization of transfected ECFC. (a) ECFC were transfected using a lentivirus with a vector encoding for either human EPO (epoecfc) or LacZ (laczecfc) under control of a cytomegalovirus
Figure S1. Phenotypic characterization of transfected ECFC. (a) ECFC were transfected using a lentivirus with a vector encoding for either human EPO (epoecfc) or LacZ (laczecfc) under control of a cytomegalovirus
Supplemental methods:
 Supplemental methods: ASC-J9 treatment ASC-J9 was patented by the University of Rochester, the University of North Carolina, and AndroScience Corp., and then licensed to AndroScience Corp. Both the University
Supplemental methods: ASC-J9 treatment ASC-J9 was patented by the University of Rochester, the University of North Carolina, and AndroScience Corp., and then licensed to AndroScience Corp. Both the University
Supplementary Material. Levels of S100B protein drive the reparative process in acute muscle injury and muscular dystrophy
 Supplementary Material Levels of protein drive the reparative process in acute muscle injury and muscular dystrophy Francesca Riuzzi 1,4 *, Sara Beccafico 1,4 *, Roberta Sagheddu 1,4, Sara Chiappalupi
Supplementary Material Levels of protein drive the reparative process in acute muscle injury and muscular dystrophy Francesca Riuzzi 1,4 *, Sara Beccafico 1,4 *, Roberta Sagheddu 1,4, Sara Chiappalupi
M X 500 µl. M X 1000 µl
 GeneGlide TM sirna Transfection Reagent (Catalog # M1081-300, -500, -1000; Store at 4 C) I. Introduction: BioVision s GeneGlide TM sirna Transfection reagent is a cationic proprietary polymer/lipid formulation,
GeneGlide TM sirna Transfection Reagent (Catalog # M1081-300, -500, -1000; Store at 4 C) I. Introduction: BioVision s GeneGlide TM sirna Transfection reagent is a cationic proprietary polymer/lipid formulation,
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
OPPF-UK Standard Protocols: Mammalian Expression
 OPPF-UK Standard Protocols: Mammalian Expression Joanne Nettleship joanne@strubi.ox.ac.uk Table of Contents 1. Materials... 3 2. Cell Maintenance... 4 3. 24-Well Transient Expression Screen... 5 4. DNA
OPPF-UK Standard Protocols: Mammalian Expression Joanne Nettleship joanne@strubi.ox.ac.uk Table of Contents 1. Materials... 3 2. Cell Maintenance... 4 3. 24-Well Transient Expression Screen... 5 4. DNA
Protocol CRISPR Genome Editing In Cell Lines
 Protocol CRISPR Genome Editing In Cell Lines Protocol 2: HDR donor plasmid applications (gene knockout, gene mutagenesis, gene tagging, Safe Harbor ORF knock-in) Notes: 1. sgrna validation: GeneCopoeia
Protocol CRISPR Genome Editing In Cell Lines Protocol 2: HDR donor plasmid applications (gene knockout, gene mutagenesis, gene tagging, Safe Harbor ORF knock-in) Notes: 1. sgrna validation: GeneCopoeia
Supplementary Figure S1. Hoechst. cells/field. Myogenin. % of Myogenin+ cells. 0 h chase CONTROL. BrdU. 72 h chase. CONTROL BrdU CONTROL BrdU
 Hoechst Myogenin amyhc 24 h 48 h 72 h b cells/field 50 40 30 20 10 0 24h 48h 72h h after plating (4 days growth+ t) c e CONTROL 0 h chase % of Myogenin+ cells h after plating (4 days growth+ t) CONTROL
Hoechst Myogenin amyhc 24 h 48 h 72 h b cells/field 50 40 30 20 10 0 24h 48h 72h h after plating (4 days growth+ t) c e CONTROL 0 h chase % of Myogenin+ cells h after plating (4 days growth+ t) CONTROL
Supplemental Figure 1 (Figure S1), related to Figure 1 Figure S1 provides evidence to demonstrate Nfatc1Cre is a mouse line that directed gene
 Developmental Cell, Volume 25 Supplemental Information Brg1 Governs a Positive Feedback Circuit in the Hair Follicle for Tissue Regeneration and Repair Yiqin Xiong, Wei Li, Ching Shang, Richard M. Chen,
Developmental Cell, Volume 25 Supplemental Information Brg1 Governs a Positive Feedback Circuit in the Hair Follicle for Tissue Regeneration and Repair Yiqin Xiong, Wei Li, Ching Shang, Richard M. Chen,
Coleman et al., Supplementary Figure 1
 Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
SUPPLEMENTARY INFORMATION
 (Supplementary Methods and Materials) GST pull-down assay GST-fusion proteins Fe65 365-533, and Fe65 538-700 were expressed in BL21 bacterial cells and purified with glutathione-agarose beads (Sigma).
(Supplementary Methods and Materials) GST pull-down assay GST-fusion proteins Fe65 365-533, and Fe65 538-700 were expressed in BL21 bacterial cells and purified with glutathione-agarose beads (Sigma).
RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by. 5 AGACACAAACACCAUUGUCACACUCCACAGC; Rand-2 OMe,
 Materials and methods Oligonucleotides and DNA constructs RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by Dharmacon Inc. (Lafayette, CO). The sequences were: 122-2 OMe, 5
Materials and methods Oligonucleotides and DNA constructs RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by Dharmacon Inc. (Lafayette, CO). The sequences were: 122-2 OMe, 5
supplementary information
 DOI: 10.1038/ncb2015 Figure S1 Confirmation of Sca-1 CD34 stain specificity using isotypematched control antibodies. Skeletal muscle preparations were stained and gated as described in the text (Hoechst
DOI: 10.1038/ncb2015 Figure S1 Confirmation of Sca-1 CD34 stain specificity using isotypematched control antibodies. Skeletal muscle preparations were stained and gated as described in the text (Hoechst
Supplementary Information
 Supplementary Information Supplementary Figure 1: Identification of new regulators of MuSC by a proteome-based shrna screen. (a) FACS plots of GFP + and GFP - cells from Pax7 ICN -Z/EG (upper panel) and
Supplementary Information Supplementary Figure 1: Identification of new regulators of MuSC by a proteome-based shrna screen. (a) FACS plots of GFP + and GFP - cells from Pax7 ICN -Z/EG (upper panel) and
Figure S2. Response of mouse ES cells to GSK3 inhibition. Mentioned in discussion
 Stem Cell Reports, Volume 1 Supplemental Information Robust Self-Renewal of Rat Embryonic Stem Cells Requires Fine-Tuning of Glycogen Synthase Kinase-3 Inhibition Yaoyao Chen, Kathryn Blair, and Austin
Stem Cell Reports, Volume 1 Supplemental Information Robust Self-Renewal of Rat Embryonic Stem Cells Requires Fine-Tuning of Glycogen Synthase Kinase-3 Inhibition Yaoyao Chen, Kathryn Blair, and Austin
Supplemental Figure 1
 Supplemental Figure 1 A IL-12p7 (pg/ml) 7 6 4 3 2 1 Medium then TLR ligands MDP then TLR ligands Medium then TLR ligands + MDP MDP then TLR ligands + MDP B IL-12p4 (ng/ml) 1.2 1..8.6.4.2. Medium MDP Medium
Supplemental Figure 1 A IL-12p7 (pg/ml) 7 6 4 3 2 1 Medium then TLR ligands MDP then TLR ligands Medium then TLR ligands + MDP MDP then TLR ligands + MDP B IL-12p4 (ng/ml) 1.2 1..8.6.4.2. Medium MDP Medium
JCB. Supplemental material THE JOURNAL OF CELL BIOLOGY. Paul et al.,
 Supplemental material JCB Paul et al., http://www.jcb.org/cgi/content/full/jcb.201502040/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Mutant p53-expressing cells display limited retrograde actin flow at
Supplemental material JCB Paul et al., http://www.jcb.org/cgi/content/full/jcb.201502040/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Mutant p53-expressing cells display limited retrograde actin flow at
HPV E6 oncoprotein targets histone methyltransferases for modulating specific. Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu,
 1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
Supplementary Figure 1. Confirmation of sirna in PC3 and H1299 cells PC3 (a) and H1299 (b) cells were transfected with sirna oligonucleotides
 Supplementary Figure 1. Confirmation of sirna in PC3 and H1299 cells PC3 (a) and H1299 (b) cells were transfected with sirna oligonucleotides targeting RCP (SMARTPool (RCP) or two individual oligos (RCP#1
Supplementary Figure 1. Confirmation of sirna in PC3 and H1299 cells PC3 (a) and H1299 (b) cells were transfected with sirna oligonucleotides targeting RCP (SMARTPool (RCP) or two individual oligos (RCP#1
Supplemental material
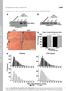 Supplemental material THE JOURNAL OF CELL BIOLOGY Gillespie et al., http://www.jcb.org/cgi/content/full/jcb.200907037/dc1 repressor complex induced by p38- Gillespie et al. Figure S1. Reduced fiber size
Supplemental material THE JOURNAL OF CELL BIOLOGY Gillespie et al., http://www.jcb.org/cgi/content/full/jcb.200907037/dc1 repressor complex induced by p38- Gillespie et al. Figure S1. Reduced fiber size
Transcriptional regulation of BRCA1 expression by a metabolic switch: Di, Fernandez, De Siervi, Longo, and Gardner. H3K4Me3
 ChIP H3K4Me3 enrichment.25.2.15.1.5 H3K4Me3 H3K4Me3 ctrl H3K4Me3 + E2 NS + E2 1. kb kb +82 kb Figure S1. Estrogen promotes entry of MCF-7 into the cell cycle but does not significantly change activation-associated
ChIP H3K4Me3 enrichment.25.2.15.1.5 H3K4Me3 H3K4Me3 ctrl H3K4Me3 + E2 NS + E2 1. kb kb +82 kb Figure S1. Estrogen promotes entry of MCF-7 into the cell cycle but does not significantly change activation-associated
Rejuvenation of the muscle stem cell population restores strength to injured aged muscles
 Rejuvenation of the muscle stem cell population restores strength to injured aged muscles Benjamin D Cosgrove, Penney M Gilbert, Ermelinda Porpiglia, Foteini Mourkioti, Steven P Lee, Stephane Y Corbel,
Rejuvenation of the muscle stem cell population restores strength to injured aged muscles Benjamin D Cosgrove, Penney M Gilbert, Ermelinda Porpiglia, Foteini Mourkioti, Steven P Lee, Stephane Y Corbel,
SUPPLEMENTARY NOTE 3. Supplememtary Note 3, Wehr et al., Monitoring Regulated Protein-Protein Interactions Using Split-TEV 1
 SUPPLEMENTARY NOTE 3 Fluorescent Proteolysis-only TEV-Reporters We generated TEV reporter that allow visualizing TEV activity at the membrane and in the cytosol of living cells not relying on the addition
SUPPLEMENTARY NOTE 3 Fluorescent Proteolysis-only TEV-Reporters We generated TEV reporter that allow visualizing TEV activity at the membrane and in the cytosol of living cells not relying on the addition
< Supporting Information >
 SUPPORTING INFORMATION 1 < Supporting Information > Discovery of autophagy modulators through the construction of high-content screening platform via monitoring of lipid droplets Sanghee Lee, Eunha Kim,
SUPPORTING INFORMATION 1 < Supporting Information > Discovery of autophagy modulators through the construction of high-content screening platform via monitoring of lipid droplets Sanghee Lee, Eunha Kim,
Supporting Information
 Supporting Information Ho et al. 10.1073/pnas.0808899106 SI Materials and Methods In immunostaining, antibodies from Developmental Studies Hybridoma ank were -FasII (mouse, 1:200), - (mouse, 1:100), and
Supporting Information Ho et al. 10.1073/pnas.0808899106 SI Materials and Methods In immunostaining, antibodies from Developmental Studies Hybridoma ank were -FasII (mouse, 1:200), - (mouse, 1:100), and
Otefin, a Nuclear Membrane Protein, Determines
 Developmental Cell 14 Supplemental Data Otefin, a Nuclear Membrane Protein, Determines the Fate of Germline Stem Cells in Drosophila via Interaction with Smad Complexes Xiaoyong Jiang, Laixin Xia, Dongsheng
Developmental Cell 14 Supplemental Data Otefin, a Nuclear Membrane Protein, Determines the Fate of Germline Stem Cells in Drosophila via Interaction with Smad Complexes Xiaoyong Jiang, Laixin Xia, Dongsheng
SUPPLEMENTARY INFORMATION FIGURE LEGENDS
 SUPPLEMENTARY INFORMATION FIGURE LEGENDS Fig. S1. Radiation-induced phosphorylation of Rad50 at a specific site. A. Rad50 is an in vitro substrate for ATM. A series of Rad50-GSTs covering the entire molecule
SUPPLEMENTARY INFORMATION FIGURE LEGENDS Fig. S1. Radiation-induced phosphorylation of Rad50 at a specific site. A. Rad50 is an in vitro substrate for ATM. A series of Rad50-GSTs covering the entire molecule
Document S1. Supplemental Experimental Procedures and Three Figures (see next page)
 Supplemental Data Document S1. Supplemental Experimental Procedures and Three Figures (see next page) Table S1. List of Candidate Genes Identified from the Screen. Candidate genes, corresponding dsrnas
Supplemental Data Document S1. Supplemental Experimental Procedures and Three Figures (see next page) Table S1. List of Candidate Genes Identified from the Screen. Candidate genes, corresponding dsrnas
(Supplementary Methods online)
 (Supplementary Methods online) Production and purification of either LC-antisense or control molecules Recombinant phagemids and the phagemid vector were transformed into XL-1 Blue competent bacterial
(Supplementary Methods online) Production and purification of either LC-antisense or control molecules Recombinant phagemids and the phagemid vector were transformed into XL-1 Blue competent bacterial
Transfection of Mouse ES Cells and Mouse ips cells using the Stemfect 2.0 -mesc Transfection Reagent
 APPLICATION NOTE Page 1 Transfection of Mouse ES Cells and Mouse ips cells using the Stemfect 2.0 -mesc Transfection Reagent Authors: Amelia L. Cianci 1, Xun Cheng 1 and Kerry P. Mahon 1,2 1 Stemgent Inc.,
APPLICATION NOTE Page 1 Transfection of Mouse ES Cells and Mouse ips cells using the Stemfect 2.0 -mesc Transfection Reagent Authors: Amelia L. Cianci 1, Xun Cheng 1 and Kerry P. Mahon 1,2 1 Stemgent Inc.,
Supplemental data. Supplemental Materials and Methods
 Supplemental data Supplemental Materials and Methods Transfection of plasmid. Transfection of plasmids into FRTL5 cells was performed using Lipofectamine LTX with Plus reagent (Invitrogen) according to
Supplemental data Supplemental Materials and Methods Transfection of plasmid. Transfection of plasmids into FRTL5 cells was performed using Lipofectamine LTX with Plus reagent (Invitrogen) according to
