JBC Papers in Press. Published on August 9, 2008 as Manuscript M
|
|
|
- Myles Jacobs
- 6 years ago
- Views:
Transcription
1 JBC Papers in Press. Published on August 9, 2008 as Manuscript M The latest version is at PIF1 HELICASE DIRECTS EUKARYOTIC OKAZAKI FRAGMENTS TOWARD THE TWO- NUCLEASE CLEAVAGE PATHWAY FOR PRIMER REMOVAL Marie L. Rossi, Jason E. Pike, Wensheng Wang, Peter M. J. Burgers, Judith L. Campbell, and Robert A. Bambara * Department of Biochemistry and Biophysics, University of Rochester School of Medicine and Dentistry, Rochester, New York 14642, Department of Biochemistry and Molecular Biophysics, Washington University School of Medicine, St. Louis, MO 63110, and Braun Laboratories, California Institute of Technology, Pasadena, California * Address correspondence to: Robert A. Bambara, Dept. of Biochemistry and Biophysics, University of Rochester School of Medicine and Dentistry, 601 Elmwood Ave., Box 712, Rochester, NY Tel.: ; Fax: ; robert_bambara@urmc.rochester.edu Running Title: Pif1 Promotes a Second Pathway for Okazaki Flap Removal Eukaryotic Okazaki fragment maturation requires complete removal of the initiating RNA primer before ligation occurs. Polymerase δ (Pol δ) extends the upstream Okazaki fragment and displaces the 5 end of the downstream primer into a single-nucleotide flap, which is removed by FEN1 nuclease cleavage. This process is repeated until all RNA is removed. However, a small fraction of flaps escapes cleavage and grows long enough to be coated with RPA and requires the consecutive action of the Dna2 and FEN1 nucleases for processing Here we tested whether RPA inhibits FEN1 cleavage of long flaps as proposed. Surprisingly, we determined that RPA binding to long flaps made dynamically by Pol δ only slightly inhibited FEN1 cleavage, apparently obviating the need for Dna2. Therefore, we asked whether other relevant proteins promote long flap cleavage via the Dna2 pathway. The Pif1 helicase, implicated in Okazaki maturation from genetic studies, improved flap displacement and increased RPA inhibition of long flap cleavage by FEN1. These results suggest that Pif1 accelerates long flap growth, allowing RPA to bind before FEN1 can act, thereby inhibiting FEN1 cleavage. Therefore, Pif1 directs long flaps toward the two-nuclease pathway, requiring Dna2 cleavage for primer removal. INTRODUCTION During eukaryotic DNA replication, the lagging strand is replicated via synthesis and maturation of Okazaki fragments. These fragments are short stretches of DNA that are joined to generate a continuous strand (1). Each fragment is initiated when DNA polymerase α/primase (Pol α) makes an RNA/DNA primer, synthesizing approximately 10 nucleotides (nt) of RNA followed by nt of DNA (2). The primer is then extended by a complex of DNA polymerase δ (Pol δ ), the sliding clamp, proliferating cell nuclear antigen (PCNA), and the clamp loader, replication factor C (RFC). When Pol δ encounters the 5 end of the downstream Okazaki fragment, it displaces it into a flap. Cleavage of the flap by nucleases generates a nick, which is subsequently sealed by DNA ligase I to form continuous double-stranded DNA (3,4). One pathway for cleavage of the flap employs flap endonuclease 1 (FEN1). FEN1 is a single-strand, structure-specific endonuclease that enters the 5 end of the flap and tracks to the base for cleavage (3,5,6). Following displacement of a short flap, less than about 12 nt, by Pol δ, FEN1 cleaves leaving a nick, the substrate for DNA ligase I (7-9). Because the RNA initiating the Okazaki fragments is approximately 10 nt in length, short flaps composed entirely of RNA are first displaced by Pol δ. This does not obstruct FEN1, which is active on RNA (10,11). In addition, displacement and cleavage occurs mostly 1 Copyright 2008 by The American Society for Biochemistry and Molecular Biology, Inc.
2 within the first 25 nt of the downstream fragment, sufficient to remove the entire RNA/DNA primer, which is approximately nt in length (11). Previous biochemical studies demonstrated that primarily short flaps are cleaved by FEN1. It is likely that in vivo a series of short successive displacements by Pol δ and cleavages by FEN1 are effective for removal of the entire initiating RNA/DNA primer (7-9,11). However, although Pol δ and FEN1 appear to be designed to keep flaps short, reconstitutions in vitro demonstrate the creation of some long flaps, approximately nt in length (11). Biochemical results suggest that once flaps achieve lengths in the range of 30 nt they can be bound by RPA, which inhibits FEN1 cleavage. RPA-coated flaps must then be processed by an alternative pathway. This second pathway is proposed to require Dna2 (13). Dna2 is an ATP-dependent helicase and a single-stranded DNA-specific endonuclease (14,15). The endonuclease activity requires tracking from the 5 end of a flap and is stimulated by RPA (13,16,17). Because the specificity of Dna2 does not allow for cleavage at the base of a flap to generate a nick, the terminal Dna2 product is a short flap (17). The remaining flap is too small to be bound by RPA, so it can be cleaved by FEN1 to generate the nick for ligation (13,15). In this way, the properties of the nucleases and RPA order the reactions with Dna2 cleaving first and FEN1 second. The proposed coordinated function of Dna2 and FEN1 is supported by other results in which they have been shown to interact physically though co-immunoprecipitation (18) and genetically through deletion studies in S. cerevisiae (18,19). Deletion of DNA2 is lethal in S. cerevisiae (20,21). Moreover, the dna2-1 mutant, which has impaired nuclease activity, is synthetically lethal with the rad27δ, which has impaired FEN1 flap-cleavage activity (18). However, dna2-1 nuclease mutants show a temperature sensitive phenotype that is suppressed by over-expression of FEN1 (18,19). Likewise, the temperature sensitive phenotype of rad27δ mutants is suppressed by over-expression of Dna2 (18). In addition, in S. cerevisiae, recent deletion studies highlight the importance of Dna2 in replication and implicate the Pif1 helicase in maturation of Okazaki fragments (22). Pif1 is a 5-3 helicase conserved from yeast to humans (23,24). Budd and colleagues (22) found that pif1δ rescued the lethality of dna2δ. Also, pif1δ rescued the temperature sensitive phenotype of the dna2-1 mutant. They proposed that Pif1 creates a need for Dna2 by promoting formation of long flaps. While deletion of PIF1 rescued the dna2-1 mutant, the mutant strain had residual phenotypic defects consistent with disruptions in replication. The additional deletion of POL32 in the dna2-1 pif1δ strain suppressed these defects (22). Pol32 is the subunit of Pol δ that binds to PCNA, and deletion of this subunit has been shown to limit some of the strand-displacement activity of Pol δ (25,26). The phenotype of the triple mutant dna2-1 pif1δ pol32δ further supports the hypothesis that Pif1 promotes long flap formation (22). By deleting both PIF1 and POL32, it is likely that flap displacement lengths are shorter and flaps do not bind RPA, a situation that eliminates the need for Dna2 cleavage. Similar to Pif1 in S. cerevisiae, it was previously shown that Pfh1, the Pif1 ortholog in Schizosaccharomyces pombe, has an interaction with Dna2 (27). Pfh1 is essential in S. pombe (28). However, mutation of pfh1 suppresses the temperature sensitive phenotype of dna2 mutants. Moreover, mutations in pfh1 and cdc27, the PCNA-interacting subunit of Pol δ in S. pombe, are synthetically lethal (27). These results suggest that Pfh1, Dna2, and Pol δ participate in similar pathways in yeast, presumably Okazaki fragment maturation. Together the biochemical and genetic data suggest that both FEN1 and Dna2 are involved in the processing of flaps to remove the RNA/DNA primer initiating Okazaki fragments (3,4,29). However, it has not been clearly established how the presence of RPA and Pif1 may affect the directing of flaps into either the FEN1 cleavage pathway or the two-nuclease pathway requiring Dna2. Through reconstitution studies in vitro with purified proteins from S. cerevisiae, we set out to examine the influence of RPA and Pif1 on the length of flap displacement and subsequent cleavage by FEN1. 2
3 EXPERIMENTAL PROCEDURES Materials Oligonucleotides were synthesized by either Midland Certified Reagents Company (Midland, TX) or Integrated DNA Technologies (Coralville, IA). Radionucleotides [γ- 32 P]ATP and [α- 32 P]dCTP were purchased from PerkinElmer Life Sciences (Waltham, MA). Polynucleotide kinase, the Klenow fragment of Escherichia coli DNA polymerase I, and streptavidin were obtained from Roche Applied Science (Indianapolis, IN). All other reagents were the best commercially available. Enzyme expression and purification S. cerevisiae wild-type Pol δ (Pol δ -wt) (25) and 3-5 exonuclease deficient Pol δ (Pol δ-01) (9,25) were over-expressed and purified from S. cerevisiae as previously described. S. cerevisiae RFC was overexpressed and purified from E. coli as previously described (30). S. cerevisiae Rad27 (31) (referred to as FEN1) and PCNA (11) were cloned into the T7 expression vector pet-24b (Novagen/EMD Biosciences, Madison, WI), expressed in E. coli BL21(DE3) codon plus strain (Stratagene, La Jolla, CA and Novagen/EMD Biosciences, Madison, WI, respectively) and purified as previously described, resulting in recombinant protein with a C-terminal six-his tag. S. cerevisiae Pif1 was cloned into the pet-28b bacterial expression vector (Novagen/EMD Biosciences, Madison, WI), expressed in the E. coli Rosetta strain (Novagen/EMD Biosciences, Madison, WI), and purified as previously described resulting in recombinant protein with an N-terminal six-his tag (32). Oligonucleotide substrates Substrates were designed to resemble Okazaki fragment processing intermediates using oligonucleotide primers. The primer sequences are listed in Table 1. Primers D 1, D 2, and D 3 were radiolabeled on the 5 -end with [γ- 32 P]ATP and polynucleotide kinase. Primer D 1 was radiolabeled on the 3 end by annealing a 20-nt labeling template with a 5 - GCTA overhang to the 3 -end of the primer and incorporating dc using [α- 32 P]dCTP and the Klenow polymerase. Radiolabeled primers were fractionated by electrophoresis on a 15%/7M urea polyacrylamide gel and then purified. The primers were then annealed into substrates in a 1:2:4 ratio of labeled downstream primer to template to upstream primer. Substrates were annealed by combining corresponding primers in a buffer containing 50 mm Tris-HCl, ph 8.0, 50 mm NaCl, and 1 mm dithiothreitol, heating at 95 C for 5 minutes, transferring to 70 C, and cooling to room temperature. Three sets of substrates were used in the following experiments. One set was used to examine FEN1 cleavage following stranddisplacement by Pol δ. These contained a 44-nt upstream primer and a 60-nt downstream primer annealed to the 3 - and 5 -ends, respectively, of a 110-nt template and will be referred to as the standard-44 substrate. A second set of substrates contained the same 44-nt upstream primer and 110-nt template of the standard-44 substrate, except the downstream primer was 59-nt in length and initiated by either a single nucleotide of RNA or a single nucleotide of 2 O-methylated RNA. The annealed substrates will be referred to as the RNA substrate or methylated RNA substrate, respectively. The third set of substrates were fixed double-flaps, in which an upstream 1-nt 3 tail complementary to the template overlaps with a 5 flap on the downstream primer, and was used for examining FEN1 cleavage. The fixed double-flap substrates contained a 20-nt upstream primer and 60-nt downstream primer annealed to the 3 - and 5 -ends, respectively, of varying length templates, either 60-nt, 51-nt, or 42-nt, forming 19-nt, 28-nt, or 37-nt double-flaps, respectively. The sequence of the fixed double-flap substrates is the same on the flap and at the base of the flap as when flaps of 19-nt, 28-nt, or 37-nt are formed from displacement of the downstream primer on the standard-44 substrates. Specific substrates used are indicated in the figure legends and depicted above the figures. Enzyme assays For strand-displacement reactions, 5 fmol of labeled biotinylated substrate was incubated with 500 fmol streptavidin for 20 minutes on ice, prior to starting each reaction. The streptavidin complexes with the biotinylated template strand and blocks the ends of the DNA, providing a substrate on which RFC must load PCNA. In the figures, substrates are depicted without blocked ends for simplicity. Substrate 3
4 was then incubated with 23 fmol Pol δ-wt or Pol δ-01, 25 fmol each PCNA and RFC, 20 fmol FEN1, 100 fmol RPA, and 25, 50, or 100 fmol Pif1 for 10 minutes at 30 C in a total volume of 20 µl reaction buffer. Reaction buffer contained 50 mm Tris-HCl, ph 7.5, 2 mm dithiothreitol, 25 µg/ml bovine serum albumin, 0.5 mm MgCl 2, 1 mm ATP, 50 µm dntps, and 75 mm NaCl. Reactions were stopped by addition of 10 µl of 2X termination dye (90% formamide (v/v), 10 mm EDTA with 0.01% bromophenol blue and xylene cyanole) and heating at 95 C for 5 minutes. Products were separated by electrophoresis on a 15%/7M urea polyacrylamide gel. The gel was dried, scanned using a Molecular Dynamics PhosphorImager, and analyzed using Image Quant version 1.2 software. All assays were repeated at least in triplicate with a representative gel shown in the figure. For the fixed double-flap cleavage reactions, 5 fmol of substrate was incubated with 500 fmol streptavidin for 20 minutes on ice, prior to the start of each reaction. Although the substrates were not conjugated with biotin, streptavidin was included to control for any effects of free streptavidin in the reaction mix. Substrate was then incubated with 20 fmol FEN1 and 50, 100, or 200 fmol RPA for 10 minutes at 30 C in a total volume of 20 µl reaction buffer (same as above). Reactions were stopped, electrophoresed, and analyzed as directed above. Cleavage product distribution analysis For each lane of interest, the cleavage product bands were identified and correlated with flap length. Using Image Quant software, peak pixel intensities of each gel lane were obtained and matched with a corresponding cleavage product band and flap length. Pixel intensities of the cleavage bands for FEN1 cleavage alone were subtracted from the intensities of bands in reaction lanes containing Pol δ, PCNA/RFC, and FEN1. The pixel intensity versus the size of the flap cleaved was graphed. Then, a moving average trendline (period of three) was added to the graph to smooth and more clearly indicate the trend of the data. RESULTS Because a fraction of the flaps made in our Okazaki fragment processing system was found to become long before cleavage by FEN1 (11), we questioned whether RPA binding on these long flaps is sufficient to effectively inhibit FEN1 cleavage, necessitating cleavage by Dna2. First, we examined RPA inhibition on flaps generated dynamically through strand-displacement. Additionally, based on the genetic evidence of a role for Pif1 in Okazaki fragment maturation through its interaction with Dna2 (22), we asked whether Pif1 stimulates formation of long flaps, creating an additional need for the two-nuclease pathway. Cleavage product distribution indicates both short and long products During the maturation of Okazaki fragments in vivo, the flaps form through the dynamic process of strand-displacement synthesis. Accordingly, FEN1 cleavage was investigated during strand-displacement on a substrate that simulates an intermediate of Okazaki fragment maturation, the standard-44 substrate. This substrate contains an upstream primer and downstream primer annealed to the 3 and 5 ends, respectively, of an oligonucleotide template strand. Between the two primers is a 6 nt gap allowing space for the polymerase to synthesize, extending the upstream primer. FEN1 cleavage product size was measured during stranddisplacement synthesis by either Pol δ-wt or the 3-5 exonuclease deficient Pol δ-01, PCNA, and RFC on the standard-44 substrate (Fig. 1). For visualization of the initial FEN1 cleavage products, the substrate was radiolabeled on the 5 end of the downstream primer. A 6 nt ssdna gap between the upstream and downstream primers was specifically chosen for use in this study. This 6 nt gap was expected to be too small for RPA binding (33). We also performed band shift experiments comparing the level of RPA binding to a 6 nt gap substrate, a 30 nt gap substrate, and a 60 nt length of single stranded DNA. Results of these experiments demonstrated that RPA is unable to bind the 6 nt gap substrate even at a concentration two-fold higher than used in the strand-displacement experiments (data not shown). However, RPA did 4
5 bind well to both the 30 nt gap substrate and the single stranded DNA. Consequently, the effects of RPA binding are exclusively confined to the displaced flaps created dynamically by Pol δ. In the presence of PCNA, RFC, and either Pol δ-wt or Pol δ-01, the maximum lengths of FEN1 cleavage products were approximately 18 nt and 36 nt, respectively, for the two polymerases (Fig. 1A, lanes 3 and 4, respectively). This is consistent with the product lengths observed in our prior studies using a 30 nt gap-containing substrate, indicating that the length of the gap does not influence the results, since these gaps are filled in efficiently by Pol δ prior to strand displacement synthesis (11). The longer flaps created and cleaved in the presence of Pol δ-01 are understood to derive from the increased stranddisplacement activity of 3-5 exonuclease deficient Pol δ mutants (8,9,11). Distribution of the FEN1 cleavage products accompanying either Pol δ-wt or Pol δ-01 displacement synthesis was evaluated. After quantitating the pixel intensities of the product bands, band intensity versus the size of the flap cleaved was graphed (Fig. 1B). The distribution revealed both a large population of flaps cleaved at the short stage, up to approximately 10 nt, and a second small population of products cleaved from long flaps, up to approximately 18 nt or 36 nt, following displacement by either the Pol δ-wt or Pol δ-01, respectively. Similar to what was observed before on the standard 30 nt gapcontaining substrate (11), there was a sharp peak distribution for the short cleavage products, and the long cleavage product distribution was broad and flat (Fig. 1B). Similarity of the twopopulation distributions with different substrates suggests that such distributions are generally characteristic of FEN1 cleavage of Pol δ displaced flaps. FEN1 cleavage during flap displacement is only moderately inhibited by RPA Assessing the effects of RPA on FEN1 cleavage of long flaps is important for understanding the pathways of flap removal. This is because the anticipated inhibition of FEN1 by RPA is expected to create the need for Dna2 and the two-nuclease pathway. Cleavage by FEN1 was measured in the presence of Pol δ-01, PCNA, RFC, and increasing amounts of RPA on the standard-44 substrate radiolabeled at the 5 end of the downstream primer (Fig. 2). RPA did not noticeably affect the strand-displacement and cleavage of flaps made in the presence of Pol δ-wt, which were less than approximately 20 nt (data not shown). Because of its ability to generate longer flaps than the Pol δ-wt (8,9,11), we anticipated that RPA inhibition of FEN1 would be more pronounced with Pol δ-01. Therefore, in this experiment, the exonuclease deficient Pol δ-01 was used to examine RPA inhibition of FEN1 cleavage following displacement RPA did not significantly alter the stranddisplacement synthesis by Pol δ-01 (data not shown). In examining FEN1 cleavage products formed in the presence of Pol δ -01, some moderate inhibitory effects were discernible when increasing amounts of RPA were added to the reconstitution (Fig. 2, lanes 4-6 compared to lane 3). Percent cleavage inhibition was determined by comparing the amount of cleavage for reactions in the presence versus absence of RPA. The amount of cleavage is defined as the sum intensities of the cleavage product bands divided by the sum intensities of the downstream primer and cleavage product bands. To calculate percent inhibition, the value for cleavage amount in the presence of RPA was divided by the value for cleavage amount by FEN1 alone, subtracted from one, and then multiplied by 100. Relative to the total amount of cleavage, all cleavage products greater than 28 nt long were diminished by only 36% with addition of the highest concentration of RPA (Fig. 2, lane 6). Likewise, all cleavage products of greater than 19 nt were decreased by only 38% (Fig. 2, lane 6). Cleavage products in the nt size range were most sensitive with 48% inhibition by RPA (Fig. 2, lane 6 compared to lane 3, bracket region). Similarly, products in the nt size range were inhibited by 41% (Fig. 2, lane 6). This contrasts with products in the nt size range, which were less susceptible to RPA inhibition, displaying only 17% reduction at the highest concentration of RPA (Fig. 2, lane 6). Greater inhibition of cleavage that gave products greater than 19 nt is consistent with RPA binding more stably on single-stranded DNA greater than 30 nt long (33). However, based on the decrease of starting substrate, comparing the overall amount of cleavage in the absence and presence of RPA 5
6 revealed similar cleavage levels, indicating that RPA has only a moderate effect on overall rate of displacement and cleavage. RPA inhibits FEN1 cleavage of fixed flap substrates Previous studies demonstrated that RPA binding, which blocks FEN1 tracking to its cleavage site, was effective at inhibiting FEN1 cleavage of long fixed flaps (13,17). However, our results showed only moderate inhibition of FEN1 cleavage by RPA on the strand-displaced flaps. Consequently, we sought to verify earlier results, and confirm the proper functionality of our RPA, by examining the influence of RPA on FEN1 cleavage of fixed flaps (Fig. 3). Fixed flap cleavage experiments with RPA and FEN1 utilized a set of double-flap substrates that contained a complementary 1-nt 3 tail overlapping with the 5 flap, creating a double-flap. It was previously demonstrated through studies in vitro that this type of double flap is the preferred substrate for FEN1 (31,34,35). During polymerase extension of the 3 end of the upstream Okazaki fragment and displacement of the 5 end of the downstream primer, reversible unannealing of the 3 end and realignment of the 5 and 3 ends may occur, resulting in formation of a double-flap (8,12,31). For this experiment, the substrates contained 5 flaps 19-nt, 28-nt, and 37-nt in length, all of which are long enough to bind RPA (33) and are representative of the lengths of long flaps seen in strand-displacement reconstitutions (11). Each substrate, depicted above the figure, was radiolabeled at the 5 end of the downstream primer to allow visualization of the flap cleavage product. There were three significant FEN1 cleavage products seen with each flap substrate, the major one representing cleavage at the base of the 5 flap in the double flap configuration (Fig. 3A). A minor product is one nucleotide longer and derives from cleavage at the base of the 5 flap when the substrate has equilibrated to a nick-flap configuration. A faint product is one nucleotide shorter, possibly representing a transient flap configuration. When increasing amounts of RPA were titrated into a reaction with FEN1, the amount of FEN1 cleavage decreased substantially with the longer flaps (Fig. 3A, lanes 7-10, 12-15), but only marginal inhibition was observed with the 19 nt flap that does not readily bind RPA (Fig. 3A, lanes 2-5). To quantitate the level of this inhibition, we compared the cleavage for reactions in the presence of RPA to the cleavage by FEN1 alone, as described above. In the presence of the highest concentration of RPA, the amount of FEN1 cleavage was inhibited to the greatest extent on the long 28- and 37-nt flaps, with a maximum inhibition of 93%, and 92%, respectively (Fig. 3B). However, the 19-nt flap was markedly less susceptible to the effects of RPA, with a maximum inhibition of only 13% (Fig. 3B). The extent of inhibition seen here is consistent with previous biochemical results (13,17). Evidently, FEN1 cleavage of the flaps created by the dynamic process of strand displacement is less sensitive than cleavage of fixed flaps to the inhibitory activity of RPA. To simulate a dynamic competition, FEN1 cleavage of the 37-nt flap substrate was assessed under conditions in which RPA was added before, with, or after the FEN1 (Fig. 3C). When FEN1 was added first, the reaction lacked magnesium, which was added later with the RPA (Fig. 3C, lanes 10-13). This allowed the FEN1 to bind the flap, but not cleave until the RPA was added, since FEN1 did not cleave in the absence of magnesium (Fig. 3C, lane 14). Inhibition by RPA was similar when it was added before or with FEN1 (Fig. 3C, lanes 2-5 and 6-9, respectively), but considerably less when FEN1 was added first (Fig. 3C, lanes 10-13). Such an outcome is consistent with the interpretation that a growing flap can acquire FEN1 before it is long enough for stable binding by RPA. Pif1 improves flap displacement and promotes RPA inhibition of FEN1 cleavage In S. cerevisiae, pif1δ rescues the lethality of dna2δ suggesting that Pif1 creates a need for Dna2, possibly by promoting the formation of long flaps (22). These genetic studies support a role for Pif1 in Okazaki fragment maturation, specifically in the two-nuclease primer removal pathway. To investigate this potential role, we asked whether Pif1 stimulates strand-displacement and subsequent FEN1 cleavage of long flaps. Additionally, because RPA was inefficient at inhibiting FEN1 cleavage on long displaced flaps, we questioned whether the action of Pif1 would 6
7 exacerbate RPA inhibition of FEN1 cleavage, augmenting the need for flap cleavage by Dna2. Again using the standard-44 substrate radiolabeled at the 5 end of the downstream primer in reconstitution assays, the effects of Pif1 and RPA on initial FEN1 cleavages following displacement by Pol δ-wt were examined (Fig. 4). With Pol δ-wt, PCNA, and RFC, the maximum length of FEN1 cleavage products was approximately 18 nt (Fig. 4, lane 3), consistent with previous results (11). When increasing amounts of Pif1 were added, the maximum cleavage product length increased to approximately 33 nt (Fig. 4, lanes 4-6). Moreover, the density of longer products, approximately nt in length notably increased (Fig. 4, lanes 4-6 compared to lane 3, bracket region). These results show that addition of Pif1 improves the displacement and cleavage of longer flaps. The levels of Pol δ synthesis were mildly stimulated by the presence of Pif1 (data not shown), suggestive that the polymerase and helicase cooperate in strand displacement. Strand lengths of nt had previously been reported to allow stable RPA binding (33). Because of this, we wanted to know whether RPA would inhibit the cleavage of the longer flaps formed in the presence of Pif1. Synthesis by Pol δ-wt was not significantly altered by the addition of both RPA and Pif1 (data not shown). However, in examining the cleavage of displaced flaps in the presence of RPA and Pif1, there was a marked decrease in long cleavage products, particularly those in the nt size range that had been augmented by addition of Pif1 alone (Fig. 4, lanes 4-6 compared to lanes 8-10, bracket region). These results indicate that Pif1 promotes RPA inhibition of FEN1. Evidence that Pif1-stimulated long flaps are a small subset of total flaps Since Pif1 stimulated the creation and cleavage of long flaps, we anticipated that Pif1 would also have increased the overall distance of displacement and cleavage into the downstream primer region. To test this, reconstitution assays were performed using the standard-44 substrate, radiolabeled at the 3 end of the downstream primer to allow visualization of the products remaining after cleavage (Fig. 5). As strand-displacement proceeds, the 3 labeled cleavage products are expected to become progressively shorter. Following synthesis by Pol δ-wt, PCNA, and RFC, there was a distribution of cleavage product sizes (Fig. 5, lane 3) up to 48 nt into the downstream primer, at which point the primers should spontaneously dissociate. This distribution was consistent with previous results (11). When increasing amounts of Pif1 were added, there was no significant change in the distribution of cleavage products (Fig. 5, lanes 4-6 compared to lane 3). Moreover, addition of RPA and Pif1 together slightly increased the amount of cleavage products greater than 20 nt, while decreasing the smallest cleavage products approximately nt in length (Fig. 5, lanes 8-10 compared to lanes 4-6). This slight shift in distribution toward longer products indicates that RPA limits some cleavage by FEN1, as expected from our other results, and that this limitation is evident in the presence of Pif1. Overall, the effects of Pif1 on the cleavage product distribution were minor, consistent with the conclusion that long flaps promoted by Pif1 are a small subset of the total population of cleavage products. The presence of RNA on the downstream primer does not inhibit lengthening of flaps by Pif1 We considered the possibility that the initiator RNA of every Okazaki fragment would influence the action of Pif1. We investigated the effects of Pif1 on FEN1 cleavage of a strand-displaced flap when a single ribonucleotide was present at the 5 end of the downstream Okazaki fragment. Although Pif1 does not bind RNA (32,40) we expected no change in FEN1 cleavage when RNA initiated the downstream primer compared to when RNA was absent, as a single ribonucleotide is unlikely to significantly affect Pif1 binding to the emerging flap. We designed a substrate, the RNA substrate, which is identical in sequence to the standard-44 substrate, except it contains a ribonucleotide at the 5 end of the downstream primer. We performed the same reactions with the RNA substrate as with the substrate composed only of DNA (Fig. 6A). Surprisingly, no long products were observed as increasing amounts of Pif1 were titrated into the reaction with Pol δ, PCNA, RFC, and FEN1 (Fig. 6A, lanes 5-8). Examination of the reaction in which only FEN1 was present, representing synthesis-independent FEN1 cleavage (Fig. 6A, lane 2), revealed that the background level of 7
8 FEN1 cleavage of the ribonucleotide was very high. This is consistent with previous findings from our laboratory demonstrating efficient FEN1 cleavage of 5 terminal ribonucleotides (41). FEN1 cleavage occurred in the absence and the presence of an upstream primer, and also when the upstream primer formed either a nick or a gap with the downstream primer (41). Therefore, we hypothesize that in the current experiment, FEN1 was very efficient at cleaving off the ribonucleotide before Pol δ could begin displacing a flap. Because the downstream primer was radiolabeled at the 5 end, the majority of FEN1 cleavage removing the ribonucleotide resulted in loss of the radiolabel. Therefore, additional cleavage of Pol δ displaced flaps was not observed. The radiolabel would be preserved in any flaps that escape FEN1 cleavage. However, in this system, the combination of initial low abundance and loss of most radiolabel reduced long cleavage products to below the level of detection. Although they may be rare, we wanted to examine the fate of any RNA initiated flaps that might escape FEN1 cleavage of the RNA. To address the problem of cleavage of the radiolabeled RNA, we utilized a substrate with a terminal 2 O -methylated ribonucleotide, a modification that inhibits RNA cleavage by ribonucleases. We hypothesized that the modification would also inhibit the ability of FEN1 to efficiently cleave off the single ribonucleotide prior to flap production. Inhibition of FEN1 cleavage preserves the radiolabeled RNA, allowing us to observe the effects on flap production and cleavage by FEN1 in the presence of Pif1 and RPA following synthesis. Our substrate for this experiment was identical to the RNA substrate except that the single ribonucleotide on the downstream primer was 2 O -methylated. We performed the same experiment as with the RNA substrate, titrating in increasing amounts of Pif1 to a reaction with Pol δ, PCNA, RFC, and FEN1 first without RPA and then in the presence of RPA (Fig. 6B). As expected, the amount of synthesis-independent FEN1 cleavage was reduced with the methylated RNA substrate (Fig. 6B, lane 2). In the absence of RPA, a population of long flap cleavage products appeared as the amount of Pif1 increased (Fig. 6B, lanes 5-8). When RPA was present, these products disappeared (Fig. 6B, lanes 10-13). We conclude that the ribonucleotide does not affect the ability of Pif1 to lengthen a flap created by Pol δ, thus resulting in formation of long flaps cleavable by FEN1. When RPA is present, it binds these long flaps and inhibits FEN1 cleavage. DISCUSSION We developed a system to reconstitute the reactions of eukaryotic Okazaki fragment processing in vitro. We have used it to evaluate two proposed processing pathways of flap removal, one involving FEN1 as the only nuclease (7-9) and the other involving the ordered action of Dna2 and FEN1 (13,15). The two nuclease pathway would be required if flaps become long enough to bind RPA. The presence of RPA is expected to inhibit FEN1, necessitating initial cleavage by Dna2. Previous biochemical studies indicated that primarily short flaps 1-2 nt in length are displaced by Pol δ during RNA primer removal and cut by FEN1, and the iterative process of displacement and cutting is likely to facilitate primer removal on the majority of Okazaki fragments (7-9,11). Application of our reconstituted system in subsequent work (11), as well as current observations (Fig. 1), revealed two populations of FEN1 cleavage products following displacement by Pol δ. One is a sharp peak of products displaced and cleaved by FEN1 while short, and the other is a broad, flat distribution of products that reel out and became long flaps nt in length before cleavage by FEN1. We hypothesized that in vivo, these long displaced flaps would be bound by RPA, effectively preventing FEN1 cleavage. Once bound by RPA, the long flaps would require Dna2 for cleavage before FEN1 could cleave, generating a nick that can be ligated. In the reconstitution assays, we predicted that inclusion of RPA with Pol δ and PCNA/RFC would prevent FEN1 cleavage of the observed long flaps. Following displacement by Pol δ-wt, there was no apparent effect of RPA on FEN1 cleavage products (data not shown). This absence of inhibition could have been anticipated since observed long flaps cleaved in the presence of Pol 8
9 δ-wt were nt, too small for stable RPA binding which occurs at a length of approximately 30 nt (33). This result is also consistent with the small levels of RPA inhibition of FEN1 cleavage on a 19-nt fixed double-flap (Fig. 3). The exonuclease deficient Pol δ-01 generated longer flap cleavage products (8,9,11), up to approximately 36 nt (Fig. 1). Flaps of this length, which are readily observable with Pol δ- 01, should stably bind RPA, thus inhibiting FEN1 cleavage. Surprisingly, however, in the presence of Pol δ-01, PCNA/RFC, and FEN1, RPA only moderately inhibited the formation of the long cleavage products (Fig. 2). The longest cleavage products in the nt size range were most sensitive to RPA inhibition (Fig. 2), which is consistent with the fixed flap results showing greater levels of inhibition with the 28-nt and 37-nt double-flaps compared to the 19-nt flap (Fig. 3). Since RPA was a highly effective inhibitor of FEN1 cleavage on long fixed flaps, we expected inhibition to be similar on long stranddisplaced flaps. An advantage of the system that we have employed is that it allowed us to make this important comparison. In actuality, moderate inhibition of FEN1 cleavage by RPA in the context of dynamically lengthening flaps is in obvious contrast with RPA inhibition on fixed flaps. FEN1 cleavage assays on fixed double-flaps revealed more than 90% inhibition on 28-nt and 37-nt flaps (Fig. 2) compared to only 36% inhibition on cleavage products greater than 28 nt when cleavage occurs during displacement (Fig. 2). These results indicate an inherent difference in the ability of RPA to inhibit FEN1 cleavage when the flap is preformed and fixed versus when flaps are lengthening during strand-displacement. In the displacement assays, the moderate RPA inhibition of long flap cleavage suggests that when flaps are created dynamically, there is a competition between RPA and FEN1 for binding to the flap favoring FEN1. Moreover, assessment of competition on a fixed flap when the RPA was added before, with, or after the FEN1, indicates that initial binding of FEN1 thwarts the inhibitory ability of RPA (Fig. 3C). We propose that by the time most flaps become long enough to bind RPA, FEN1 has already recognized their 5 ends and begun tracking (Fig. 7A). We envision FEN1 molecules tracking near the base of the elongating flap, racing the displacement process to eventually reach the cleavage site. FEN1 may achieve this binding advantage because it is already a stable part of the lagging strand maturation complex. This would position it on the flap at the earliest time of strand displacement. Alternatively, FEN1 may simply have the ability to bind and track on nearly every elongating flap before it becomes long enough to form a stable complex with the RPA. In either case, RPA molecules would then bind behind the FEN1, too late to inhibit cleavage. This current biochemical data prompted a re-evaluation of the pathways of primer removal during Okazaki fragment maturation, specifically of the role of RPA in directing flaps into the twonuclease pathway. Inhibition of FEN1 cleavage by RPA binding of long flaps was proposed to be the key reason for use of the two-nuclease pathway (13,15). Yet, the results presented here demonstrate that even in the presence of RPA, FEN1 cleavage of displaced flaps is only moderately inhibited by RPA (Fig. 2). Is it possible that the two-nuclease pathway is either completely redundant or virtually never used? Recent genetic results in S. cerevisiae suggest the involvement of another factor, the Pif1 helicase, in directing flaps to the two-nuclease pathway. The lethality of dna2δ mutation is suppressed by pif1δ (22). Studies in S. pombe indicate a similar relationship between the Pif1 ortholog Pfh1 and Dna2, since mutations in pfh1 suppress the temperature sensitive phenotype of dna2 mutations (27). These genetic results imply that the Pif1 helicase creates a need for Dna2, possibly by promoting long flap formation (22,27), and indicate a role for Pif1 in Okazaki fragment processing. Results presented here support this hypothesis. The addition of Pif1 to the Okazaki fragment processing system promoted the appearance of long FEN1 cleavage products (Fig. 4). Pif1 notably increased products in the nt size range. It appears that Pif1 accelerated flap displacement, allowing flaps to become longer before FEN1 cleavage could occur (Fig. 7B). Yet, the Pif1-stimulated products were a small subset of FEN1 products, since addition of Pif1 only mildly stimulated the amount of synthesis (data not shown) and the extent of cleavage into the downstream primer (Fig. 5). Although these long Pif1-stimulated products were not abundant in our 9
10 system, the actual numbers in vivo would be significant because millions of Okazaki fragments are processed during synthesis in a replication cycle. Our observations that Pif1 stimulates long flap cleavage by FEN1 (Fig. 4) and that RPA only moderately inhibits FEN1 cleavage of long flaps (Fig. 2) prompted the question of whether Pif1 is the critical additional component that necessitates the Dna2-requiring pathway. Pif1-stimulated flaps in the nt size range are long enough to bind RPA (33). Correspondingly, RPA did effectively inhibit FEN1 cleavage of this size range of flaps created in the presence of Pif1 (Fig. 4). This result suggests that Pif1 increases the rate of flap displacement, such that FEN1 takes longer to track to the base of the flap for cleavage. Significantly, Pif1 appears to promote such rapid flap formation that RPA is able to bind to the flap ahead of the tracking FEN1 (Fig. 7C). With Pif1, RPA binding prevents cleavage of a significant number of flaps by FEN1 alone and necessitates cleavage via the two-nuclease pathway (13). Okazaki fragments in vivo are initiated by RNA, made by Pol α, which must be removed. Removal of only the ribonucleotides is not sufficient to maintain high replication fidelity, since Pol α lacks a 3-5 proofreading activity, some of the DNA nucleotides incorporated by Pol α just beyond the RNA may be erroneous (42). As a result, additional flap displacement and cleavage of the downstream Okazaki fragment is necessary to remove the DNA synthesized by Pol α (29,43). Consequently, we were interested in the effect of Pif1 on flap formation with substrates that more closely resemble the natural RNAprimed Okazaki fragment (2,29). Therefore, we felt it was reasonable to test a model fragment with a single initiator ribonucleotide. In the first experiments, FEN1 was so efficient at removing the labeled ribonucleotide of the downstream primer that long FEN1 cleavage products were virtually undetectable (Fig. 6A). This result supports the validity of our results with fully DNA fragments, since the RNA of nearly all fragments is likely to have been removed in vivo before the flaps could achieve significant length. To determine the effects of any fragments that escaped complete RNA removal by FEN1, we designed and used a downstream primer with a 2 O-methylated 5 ribonucleotide (Fig. 6B). As with the substrates containing only DNA, we found that Pif1 promoted RPA inhibition of FEN1 cleavage. This result further verifies that the DNA-initiated substrates served as a sufficient representative substrate for the RNA-initiated Okazaki fragments, consistent with previous work (11). Moreover, in helicase assays, Pfh1 from S. pombe was equally active on RNA-initiated flaps and DNA-initiated flaps, suggesting that Pif1 helicases are active in vitro on substrates which are relevant in vivo (27). Supporting previous genetic data (22), our biochemical results reveal a role for Pif1 in eukaryotic cellular Okazaki fragment maturation to remove the initiator RNA primer, specifically in the processing of long flaps. Results with our model reconstitution system show that once Pol δ strand-displaces the downstream primer, FEN1 can track onto the flap and cleave at its base (Fig. 7A). The dynamics of flap formation do not allow RPA to bind quickly enough to inhibit FEN1. Pif1 accelerates flap formation allowing the flaps to achieve greater lengths before FEN1 cleavage (Fig. 7B). However, when RPA is present with Pif1 and FEN1, flap growth is sufficiently rapid that RPA can bind and block FEN1 cleavage (Fig. 7C). Since Pif1 is unable to bind RNA (32,40), Pif1 involvement in unwinding the downstream Okazaki fragment necessitates that Pol δ displaces whatever remains of the RNA primer (2) before Pif1 can bind to the flap and aid displacement. However, our results indicate that the presence of the initiator RNA does not alter the fundamental role of Pif1. The biological implication is that by promoting RPA inhibition of FEN1 cleavage, Pif1 is directing flaps towards the two-nuclease cleavage pathway, requiring Dna2 cleavage for primer removal. Examining the influence of Pif1 on flap cleavage by Dna2 in the context of stranddisplacement will be an important next step to further elucidate the role of Pif1 in replication. Additional experiments will be necessary to expand our understanding of the conditions that determine which of the cleavage pathways is followed for primer removal during Okazaki fragment maturation. 10
11 REFERENCES 1. Kornberg, A., and Baker, T. A. (1992). In. DNA replication, 2nd / Ed., W.H. Freeman, New York 2. Bambara, R. A., Murante, R. S., and Henricksen, L. A. (1997) J Biol Chem 272(8), Liu, Y., Kao, H. I., and Bambara, R. A. (2004) Annu Rev Biochem 73, Rossi, M. L., Purohit, V., Brandt, P. D., and Bambara, R. A. (2006) Chem Rev 106(2), Harrington, J. J., and Lieber, M. R. (1994) EMBO J 13(5), Murante, R. S., Rust, L., and Bambara, R. A. (1995) J Biol Chem 270(51), Ayyagari, R., Gomes, X. V., Gordenin, D. A., and Burgers, P. M. (2003) J Biol Chem 278(3), Garg, P., Stith, C. M., Sabouri, N., Johansson, E., and Burgers, P. M. (2004) Genes Dev 18(22), Jin, Y. H., Ayyagari, R., Resnick, M. A., Gordenin, D. A., and Burgers, P. M. (2003) J Biol Chem 278(3), Murante, R. S., Rumbaugh, J. A., Barnes, C. J., Norton, J. R., and Bambara, R. A. (1996) J Biol Chem 271(42), Rossi, M. L., and Bambara, R. A. (2006) J Biol Chem 281(36), Jin, Y. H., Obert, R., Burgers, P. M., Kunkel, T. A., Resnick, M. A., and Gordenin, D. A. (2001) Proc Natl Acad Sci U S A 98(9), Bae, S. H., Bae, K. H., Kim, J. A., and Seo, Y. S. (2001) Nature 412(6845), Bae, S. H., Choi, E., Lee, K. H., Park, J. S., Lee, S. H., and Seo, Y. S. (1998) J Biol Chem 273(41), Bae, S. H., and Seo, Y. S. (2000) J Biol Chem 275(48), Kao, H. I., Campbell, J. L., and Bambara, R. A. (2004) J Biol Chem 279(49), Kao, H. I., Veeraraghavan, J., Polaczek, P., Campbell, J. L., and Bambara, R. A. (2004) J Biol Chem 279(15), Budd, M. E., and Campbell, J. L. (1997) Mol Cell Biol 17(4), Budd, M. E., and Campbell, J. L. (2000) Mutat Res 459(3), Budd, M. E., Choe, W., and Campbell, J. L. (2000) J Biol Chem 275(22), Lee, K. H., Kim, D. W., Bae, S. H., Kim, J. A., Ryu, G. H., Kwon, Y. N., Kim, K. A., Koo, H. S., and Seo, Y. S. (2000) Nucleic Acids Res 28(15), Budd, M. E., Reis, C. C., Smith, S., Myung, K., and Campbell, J. L. (2006) Mol Cell Biol 26(7), Bessler, J. B., Torredagger, J. Z., and Zakian, V. A. (2001) Trends Cell Biol 11(2), Boule, J. B., and Zakian, V. A. (2006) Nucleic Acids Res 34(15), Burgers, P. M., and Gerik, K. J. (1998) J Biol Chem 273(31), Johansson, E., Garg, P., and Burgers, P. M. (2004) J Biol Chem 279(3), Ryu, G. H., Tanaka, H., Kim, D. H., Kim, J. H., Bae, S. H., Kwon, Y. N., Rhee, J. S., MacNeill, S. A., and Seo, Y. S. (2004) Nucleic Acids Res 32(14), Zhou, J. Q., Qi, H., Schulz, V. P., Mateyak, M. K., Monson, E. K., and Zakian, V. A. (2002) Mol Biol Cell 13(6), Kao, H. I., and Bambara, R. A. (2003) Crit Rev Biochem Mol Biol 38(5), Gerik, K. J., Gary, S. L., and Burgers, P. M. (1997) J Biol Chem 272(2), Kao, H. I., Henricksen, L. A., Liu, Y., and Bambara, R. A. (2002) J Biol Chem 277(17), Boule, J. B., Vega, L. R., and Zakian, V. A. (2005) Nature 438(7064), Wold, M. S. (1997) Annu Rev Biochem 66, Friedrich-Heineken, E., Henneke, G., Ferrari, E., and Hubscher, U. (2003) J Mol Biol 328(1), Storici, F., Henneke, G., Ferrari, E., Gordenin, D. A., Hubscher, U., and Resnick, M. A. (2002) EMBO J 21(21),
12 36. Huang, L., Kim, Y., Turchi, J. J., and Bambara, R. A. (1994) J Biol Chem 269(41), Murante, R. S., Henricksen, L. A., and Bambara, R. A. (1998) Proc Natl Acad Sci U S A 95(5), Qiu, J., Qian, Y., Frank, P., Wintersberger, U., and Shen, B. (1999) Mol Cell Biol 19(12), Turchi, J. J., Huang, L., Murante, R. S., Kim, Y., and Bambara, R. A. (1994) Proc Natl Acad Sci U S A 91(21), Zhang, D. H., Zhou, B., Huang, Y., Xu, L. X., and Zhou, J. Q. (2006) Nucleic Acids Res 34(5), Huang, L., Rumbaugh, J. A., Murante, R. S., Lin, R. J., Rust, L., and Bambara, R. A. (1996) Biochemistry 35(28), Hubscher, U., Maga, G., and Spadari, S. (2002) Annu Rev Biochem 71, Waga, S., and Stillman, B. (1998) Annu Rev Biochem 67, FOOTNOTES Acknowledgments: We thank the members of the Bambara laboratory for helpful discussions and suggestions. This work was supported by National Institutes of Health grant GM (to R. A. B.) with additional support from NIH grants GM (to J. L. C), GM32431 (to P. M. J. B), and GM (to J. E. P.). The abbreviations used are: nt, nucleotide(s); Pol α, DNA polymerase α/primase; Pol δ, DNA polymerase δ; PCNA, proliferating cell nuclear antigen; RFC, replication factor C; FEN1, flap endonuclease 1; RPA, replication protein A; Pol δ-wt, wild-type polymerase δ; Pol δ-01, 3-5 exonuclease deficient polymerase δ; fmol, femtomole. 12
13 FIGURE LEGENDS Figure 1. Cleavage product distribution indicates both short and long products. A) Initial FEN1 cleavage was assayed on the standard-44 substrate (U 2 :T 4 :D 1 ) in the presence of PCNA/RFC and either Pol δ-wt, denoted wt (lane 3), or Pol δ-01, denoted 01 (lane 4), as described in the Experimental Procedures. The substrate is depicted above the figure. The asterisk indicates location of radiolabel. B) The size of flap cleavage products versus the intensity of the product band was graphed and moving average trendline added, as described in the Experimental Procedures. The black line corresponds to the cleavage products for reconstitutions with Pol δ-wt. The gray line corresponds to the products for reconstitutions with Pol δ-01. Figure 2. FEN1 cleavage during flap displacement is moderately inhibited by RPA. Initial cleavage by FEN1 was assayed on the standard-44 substrate (U 2 :T 4 :D 1 ) in the presence of PCNA/RFC, Pol δ-01, and increasing amounts of RPA (50, 100, or 200 fmol) as described in the Experimental Procedures (lanes 4-6). The substrate is depicted above the figure. Asterisk indicates location of radiolabel. Figure 3. RPA inhibits FEN1 cleavage of fixed flap substrates. A) Cleavage by FEN1 was assayed in the presence of increasing amounts of RPA (50, 100, or 200 fmol) as described in the Experimental Procedures. The 19-nt fixed double-flap (U1:T3:D1) (lanes 1-5), 28-nt fixed double-flap (U1:T2:D1) (lanes 6-10), and 37-nt fixed double-flap (U1:T1:D1) (lanes 11-15) are depicted above the figure. The asterisk indicates location of radiolabel. B) The amount of RPA versus the percent cleavage inhibition was graphed for the 19-nt fixed double-flap (black boxes), 28-nt fixed double-flap (gray boxes), and 37-nt fixed double-flap (white boxes) for the gel in A). Percentages are indicated above the bar for each substrate at each concentration of RPA. C) Cleavage by FEN1 was assayed in presence of increasing amounts of RPA (50, 100, or 200 fmol) using the 37-nt fixed double-flap, as in A). In lanes 2-5, RPA was added at the start of the reaction and FEN1 was added after 5 minutes. In lanes 6-9, both FEN1 and RPA were added at the start of the reaction, as in lanes of A). In lanes 10-13, FEN1 was added at the start of the reaction in the absence of Mg +2, followed by addition of both RPA and Mg +2 after 5 minutes. Lane 14 contains FEN1 alone in the absence of Mg +2. The percent cleavage inhibition is shown under each lane. Figure 4. Pif1 improves flap displacement and promotes RPA inhibition of FEN1 cleavage. Initial cleavage by FEN1 was assayed on the standard-44 substrate (U 2 :T 4 :D 1 ) in the presence of PCNA/RFC, Pol δ-wt and increasing amounts of Pif1 (25, 50, or 100 fmol) as described in the Experimental Procedures (lanes 4-6). Cleavage was also assayed in the presence of RPA (100 fmol) and increasing amounts of Pif1 (25, 50, or 100 fmol) as described in the Experimental Procedures (lanes 8-10). The substrate is depicted above the figure. Asterisk indicates location of radiolabel. Figure 5. Pif1-stimulated products are a small subset of cleavage products. The extent of FEN1 cleavage into the downstream primer was assayed on the standard-44 substrate (U 2 :T 4 :D 1 ) in the presence of PCNA/RFC, Pol δ-wt and increasing amounts of Pif1 (25, 50, or 100 fmol) as described in the Experimental Procedures (lanes 4-6). Cleavage was also assayed in the presence of RPA (100 fmol) and increasing amounts of Pif1 (25, 50, or 100 fmol) as described in the Experimental Procedures (lanes 8-10). The substrate is depicted above the figure. Asterisk indicates location of radiolabel. 13
14 Figure 6. RNA does not inhibit lengthening of flaps by Pif1 during synthesis of Okazaki fragments. A) Initial cleavage by FEN1 was assayed on the RNA substrate (U 2 :T 4 :D 2 ) in the presence of PCNA/RFC, Pol δ-wt and increasing amounts of Pif1 (25, 50, 100, or 200 fmol) as described in the Experimental Procedures (lanes 5-8). Cleavage was also assayed in the presence of RPA (100 fmol) and increasing amounts of Pif1 (25, 50, 100, or 200 fmol) as described in the Experimental Procedures (lanes 10-13). The substrate is depicted above the figure. The grey segment represents the ribonucleotide. The asterisk indicates location of radiolabel. B) The same reactions as in A) except the substrate used was the methylated substrate (U 2 :T 4 :D 3 ). The substrate is depicted above the figure. The grey circle represents the 2 O-methylated ribonucleotide. The asterisk indicates the location of radiolabel. Figure 7. Pif1 directs long flaps toward the two-nuclease pathway for primer removal. A) In most cases the FEN1 is tracking near the base of the elongating flap, racing the displacement process to eventually reach the site of cleavage. Most RPA molecules, would then bind behind the FEN1, too late to prevent cleavage (refer to Fig. 2.3, lanes 3-6). B) Pif1 promotes flap displacement, allowing some flaps to become long before FEN1 cleavage occurs (refer to Fig. 2.4, lanes 3-6). C) Pif1 increases the rate of flap formation such that FEN1 molecules take longer to track to the flap base for cleavage. In the presence of RPA, the flap can bind RPA ahead of the tracking FEN1. RPA bound in this position can prevent FEN1 cleavage (refer to Fig. 2.4, lanes 7-10). 14
15 Table 1. Oligonucleotide sequences Primer Length (nt) Sequence Upstream (5-3 ) U 1 20 GTCCACCCGACGCCACCTCC U 2 44 GTCCACCCGACGCCACCTCCTGCCTTCAATGTGCT GGGATCCTA Downstream (5-3 ) D 1 60 AGACGAATTCCGGATACGACGGCCAGTGCCGACC GTGCCAGCCTAAATTTCAATCCACCC D 2 59 GACGAATTCCGGATACGACGGCCAGTGCCGACCG TGCCAGCCTAAATTTCAATCCACCC D 3 59 GACGAATTCCGGATACGACGGCCAGTGCCGACCG TGCCAGCCTAAATTTCAATCCACCC Template (3-5 ) T 1 42 CAGGTGGGCTGCGGTGGAGG.GTCGGATTTAAAGT TAGGTGGG T 2 51 CAGGTGGGCTGCGGTGGAGG.GCTGGCACGGTCGG ATTTAAAGTTAGGTGGG T 3 60 CAGGTGGGCTGCGGTGGAGG.CCGGTCACGGCTGG CACGGTCGGATTTAAAGTTAGGTGGG T 4 * 110 CAGGTGGGCTGCGGTGGAGGACGGAAGTTACACG ACCCTAGGATGTTGGTTCTGCTTAAGGCCTATGCT GCCGGTCACGGCTGGCACGGTCGGATTTAAAGTT AGGTGGG Underline indicates RNA segment. Underline and italicized indicates 2 O-methylated RNA segment. * Template T 4 is biotinylated at both the 5 and 3 ends. 15
16 Figure 1. 16
17 Figure 2. 17
18 Figure 3. A 5 * 19 nt 5 * 28 nt * 37 nt 5 RPA FEN nt C B Downstream FEN1 Cleavage Products Lane % Cleavage Inhibition RPA RPA added first RPA (fmol) RPA and FEN1 added together FEN1 added first 38 nt 37 nt 36 nt 29 nt 28 nt 27 nt 20 nt 19 nt 18 nt 19-nt flap 28-nt flap 37-nt flap FEN nt Downstream FEN1 Cleavage Products 38 nt 37 nt 36 nt Lane % Inhibition
19 Figure 4. 19
20 Figure 5. 20
21 A Pif1 RPA FEN1 PCNA/RFC Pol δ-wt Downstream FEN1 Cleavage Products 5 * Figure 6. B Pif1 RPA FEN1 PCNA/RFC Pol δ-wt Downstream FEN1 Cleavage Products 5 * nt 30 nt 20 nt 10 nt Lane Lane
Biochemical analyses indicate that binding and cleavage specificities define the ordered processing of human Okazaki fragments by Dna2 and FEN1
 6774 6786 Nucleic Acids Research, 2012, Vol. 40, No. 14 Published online 7 May 2012 doi:10.1093/nar/gks388 Biochemical analyses indicate that binding and cleavage specificities define the ordered processing
6774 6786 Nucleic Acids Research, 2012, Vol. 40, No. 14 Published online 7 May 2012 doi:10.1093/nar/gks388 Biochemical analyses indicate that binding and cleavage specificities define the ordered processing
Dynamic Removal of Replication Protein A by Dna2 Facilitates Primer Cleavage during Okazaki Fragment Processing in Saccharomyces cerevisiae*
 JBC Papers in Press. Published on September 17, 2008 as Manuscript M805965200 The latest version is at http://www.jbc.org/cgi/doi/10.1074/jbc.m805965200 Dynamic Removal of Replication Protein A by Dna2
JBC Papers in Press. Published on September 17, 2008 as Manuscript M805965200 The latest version is at http://www.jbc.org/cgi/doi/10.1074/jbc.m805965200 Dynamic Removal of Replication Protein A by Dna2
Supplementary Information for. Coordination of multiple enzyme activities by a single PCNA in archaeal Okazaki fragment.
 Supplementary Information for Coordination of multiple enzyme activities by a single PCNA in archaeal Okazaki fragment maturation Thomas R. Beattie and Stephen D. Bell Sir William Dunn School of Pathology,
Supplementary Information for Coordination of multiple enzyme activities by a single PCNA in archaeal Okazaki fragment maturation Thomas R. Beattie and Stephen D. Bell Sir William Dunn School of Pathology,
Chapter 11 DNA Replication and Recombination
 Chapter 11 DNA Replication and Recombination Copyright Copyright 2009 Pearson 2009 Pearson Education, Education, Inc. Inc. 11.1 DNA is reproduced by Semiconservative Replication The complementarity of
Chapter 11 DNA Replication and Recombination Copyright Copyright 2009 Pearson 2009 Pearson Education, Education, Inc. Inc. 11.1 DNA is reproduced by Semiconservative Replication The complementarity of
Dna2p Helicase/Nuclease Is a Tracking Protein, Like FEN1, for Flap Cleavage During Okazaki Fragment Maturation*
 JBC Papers in Press. Published on September 22, 2004 as Manuscript M409231200 Dna2p Helicase/Nuclease Is a Tracking Protein, Like FEN1, for Flap Cleavage During Okazaki Fragment Maturation* Hui-I Kao,
JBC Papers in Press. Published on September 22, 2004 as Manuscript M409231200 Dna2p Helicase/Nuclease Is a Tracking Protein, Like FEN1, for Flap Cleavage During Okazaki Fragment Maturation* Hui-I Kao,
JBC Papers in Press. Published on January 29, 2009 as Manuscript M
 JBC Papers in Press. Published on January 29, 2009 as Manuscript M809189200 The latest version is at http://www.jbc.org/cgi/doi/10.1074/jbc.m809189200 Significance of the Dissociation of Dna2 by Flap Endonuclease
JBC Papers in Press. Published on January 29, 2009 as Manuscript M809189200 The latest version is at http://www.jbc.org/cgi/doi/10.1074/jbc.m809189200 Significance of the Dissociation of Dna2 by Flap Endonuclease
Enzymes used in DNA Replication
 Enzymes used in DNA Replication This document holds the enzymes used in DNA replication, their pictorial representation and functioning. DNA polymerase: DNA polymerase is the chief enzyme of DNA replication.
Enzymes used in DNA Replication This document holds the enzymes used in DNA replication, their pictorial representation and functioning. DNA polymerase: DNA polymerase is the chief enzyme of DNA replication.
* + * RecA * + + for RecA-dependent strand + + MIT Department of Biology 7.28, Spring Molecular Biology
 MIT Department of Biology 7.28, Spring 2005 - Molecular Biology 1) Question 1. 1A. You are studying the function of in vitro along with a lab partner. You run several reactions to examine the requirements
MIT Department of Biology 7.28, Spring 2005 - Molecular Biology 1) Question 1. 1A. You are studying the function of in vitro along with a lab partner. You run several reactions to examine the requirements
Dynamic Removal of Replication Protein A by Dna2 Facilitates Primer Cleavage during Okazaki Fragment Processing in Saccharomyces cerevisiae
 THE JOURNAL OF BIOLOGICAL CHEMISTRY VOL. 283, NO. 46, pp. 31356 31365, November 14, 2008 2008 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in the U.S.A. Dynamic Removal
THE JOURNAL OF BIOLOGICAL CHEMISTRY VOL. 283, NO. 46, pp. 31356 31365, November 14, 2008 2008 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in the U.S.A. Dynamic Removal
III. Detailed Examination of the Mechanism of Replication A. Initiation B. Priming C. Elongation D. Proofreading and Termination
 Outline for Replication I. General Features of Replication A. Semi-Conservative B. Starts at Origin C. Bidirectional D. Semi-Discontinuous II. Proteins and Enzymes of Replication III. Detailed Examination
Outline for Replication I. General Features of Replication A. Semi-Conservative B. Starts at Origin C. Bidirectional D. Semi-Discontinuous II. Proteins and Enzymes of Replication III. Detailed Examination
Genetics and Genomics in Medicine Chapter 3. Questions & Answers
 Genetics and Genomics in Medicine Chapter 3 Multiple Choice Questions Questions & Answers Question 3.1 Which of the following statements, if any, is false? a) Amplifying DNA means making many identical
Genetics and Genomics in Medicine Chapter 3 Multiple Choice Questions Questions & Answers Question 3.1 Which of the following statements, if any, is false? a) Amplifying DNA means making many identical
Supplemental Materials and Methods
 Supplemental Materials and Methods Proteins and reagents Proteins were purified as described previously: RecA, RecQ, and SSB proteins (Harmon and Kowalczykowski 1998); RecF protein (Morimatsu and Kowalczykowski
Supplemental Materials and Methods Proteins and reagents Proteins were purified as described previously: RecA, RecQ, and SSB proteins (Harmon and Kowalczykowski 1998); RecF protein (Morimatsu and Kowalczykowski
DNA Replication II Biochemistry 302. Bob Kelm January 28, 2004
 DNA Replication II Biochemistry 302 Bob Kelm January 28, 2004 Conceptual model for proofreading based on kinetic considerations Fig. 24.44 stalling transient melting exonuclease site occupancy Following
DNA Replication II Biochemistry 302 Bob Kelm January 28, 2004 Conceptual model for proofreading based on kinetic considerations Fig. 24.44 stalling transient melting exonuclease site occupancy Following
AccuScript High Fidelity 1 st Strand cdna Synthesis Kit
 AccuScript High Fidelity 1 st Strand cdna Synthesis Kit INSTRUCTION MANUAL Catalog #200820 Revision B.01 For In Vitro Use Only 200820-12 LIMITED PRODUCT WARRANTY This warranty limits our liability to replacement
AccuScript High Fidelity 1 st Strand cdna Synthesis Kit INSTRUCTION MANUAL Catalog #200820 Revision B.01 For In Vitro Use Only 200820-12 LIMITED PRODUCT WARRANTY This warranty limits our liability to replacement
Storage and Expression of Genetic Information
 Storage and Expression of Genetic Information 29. DNA structure, Replication and Repair ->Ch 25. DNA metabolism 30. RNA Structure, Synthesis and Processing ->Ch 26. RNA metabolism 31. Protein Synthesis
Storage and Expression of Genetic Information 29. DNA structure, Replication and Repair ->Ch 25. DNA metabolism 30. RNA Structure, Synthesis and Processing ->Ch 26. RNA metabolism 31. Protein Synthesis
DNA2 POSSIBLE ROLE IN OKAZAKI FRAGMENT MATURATION
 THE JOURNAL OF BIOLOGICAL CHEMISTRY VOL. 281, NO. 50, pp. 38555 38564, December 15, 2006 2006 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in the U.S.A. Single Strand Annealing
THE JOURNAL OF BIOLOGICAL CHEMISTRY VOL. 281, NO. 50, pp. 38555 38564, December 15, 2006 2006 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in the U.S.A. Single Strand Annealing
 The replication of DNA Kornberg 1957 Meselson and Stahl 1958 Cairns 1963 Okazaki 1968 DNA Replication The driving force for DNA synthesis. The addition of a nucleotide to a growing polynucleotide
The replication of DNA Kornberg 1957 Meselson and Stahl 1958 Cairns 1963 Okazaki 1968 DNA Replication The driving force for DNA synthesis. The addition of a nucleotide to a growing polynucleotide
Universal Labeling of 5 -Triphosphate RNAs by Artificial RNA Ligase Enzyme with Broad Substrate Specificity
 Universal Labeling of 5 -Triphosphate RNAs by Artificial RNA Ligase Enzyme with Broad Substrate Specificity John C. Haugner III and Burckhard Seelig* Department of Biochemistry, Molecular Biology and Biophysics
Universal Labeling of 5 -Triphosphate RNAs by Artificial RNA Ligase Enzyme with Broad Substrate Specificity John C. Haugner III and Burckhard Seelig* Department of Biochemistry, Molecular Biology and Biophysics
Fidelity of DNA polymerase
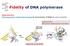 Fidelity of DNA polymerase Shape selectivity: DNA polymerase's conformational change for determination of fidelity for each nucleotide Induced fit: Structure determines function Matched nucleotide Fidelity
Fidelity of DNA polymerase Shape selectivity: DNA polymerase's conformational change for determination of fidelity for each nucleotide Induced fit: Structure determines function Matched nucleotide Fidelity
Okazaki Fragment Maturation in Yeast
 THE JOURNAL OF BIOLOGICAL CHEMISTRY Vol. 278, No. 3, Issue of January 17, pp. 1618 1625, 2003 Printed in U.S.A. Okazaki Fragment Maturation in Yeast I. DISTRIBUTION OF FUNCTIONS BETWEEN FEN1 AND DNA2*
THE JOURNAL OF BIOLOGICAL CHEMISTRY Vol. 278, No. 3, Issue of January 17, pp. 1618 1625, 2003 Printed in U.S.A. Okazaki Fragment Maturation in Yeast I. DISTRIBUTION OF FUNCTIONS BETWEEN FEN1 AND DNA2*
Molecular Genetics II - Genetic Engineering Course (Supplementary notes)
 1 von 12 21.02.2015 15:13 Molecular Genetics II - Genetic Engineering Course (Supplementary notes) Figures showing examples of cdna synthesis (currently 11 figures) cdna is a DNA copy synthesized from
1 von 12 21.02.2015 15:13 Molecular Genetics II - Genetic Engineering Course (Supplementary notes) Figures showing examples of cdna synthesis (currently 11 figures) cdna is a DNA copy synthesized from
The flow of Genetic information
 The flow of Genetic information http://highered.mcgrawhill.com/sites/0072507470/student_view0/chapter3/animation dna_replication quiz_1_.html 1 DNA Replication DNA is a double-helical molecule Watson and
The flow of Genetic information http://highered.mcgrawhill.com/sites/0072507470/student_view0/chapter3/animation dna_replication quiz_1_.html 1 DNA Replication DNA is a double-helical molecule Watson and
Molecular Biology (2)
 Molecular Biology (2) DNA replication Mamoun Ahram, PhD Second semester, 2018-2019 Resources This lecture Cooper, pp. 191-207 2 Some basic information The entire DNA content of the cell is known as genome.
Molecular Biology (2) DNA replication Mamoun Ahram, PhD Second semester, 2018-2019 Resources This lecture Cooper, pp. 191-207 2 Some basic information The entire DNA content of the cell is known as genome.
DNA polymerase (pol) is the major pol involved in chromosomal
 Okazaki fragment processing: Modulation of the strand displacement activity of DNA polymerase by the concerted action of replication protein A, proliferating cell nuclear antigen, and flap endonuclease-1
Okazaki fragment processing: Modulation of the strand displacement activity of DNA polymerase by the concerted action of replication protein A, proliferating cell nuclear antigen, and flap endonuclease-1
Masayoshi Honda, Jeehae Park, Robert A. Pugh, Taekjip Ha, and Maria Spies
 Molecular Cell, Volume 35 Supplemental Data Single-Molecule Analysis Reveals Differential Effect of ssdna-binding Proteins on DNA Translocation by XPD Helicase Masayoshi Honda, Jeehae Park, Robert A. Pugh,
Molecular Cell, Volume 35 Supplemental Data Single-Molecule Analysis Reveals Differential Effect of ssdna-binding Proteins on DNA Translocation by XPD Helicase Masayoshi Honda, Jeehae Park, Robert A. Pugh,
DNA Replication II Biochemistry 302. January 25, 2006
 DNA Replication II Biochemistry 302 January 25, 2006 Following in Dad s footsteps Original A. Kornberg E. coli DNA Pol I is a lousy replicative enzyme. 400 molecules/cell but ~2 replication forks/cell
DNA Replication II Biochemistry 302 January 25, 2006 Following in Dad s footsteps Original A. Kornberg E. coli DNA Pol I is a lousy replicative enzyme. 400 molecules/cell but ~2 replication forks/cell
Your first goal is to determine the order of assembly of the 3 DNA polymerases during initiation.
 MIT Department of Biology 7.28, Spring 2005 - Molecular Biology 1 Question 1 You are studying the mechanism of DNA polymerase loading at eukaryotic DNA replication origins. Unlike the situation in bacteria,
MIT Department of Biology 7.28, Spring 2005 - Molecular Biology 1 Question 1 You are studying the mechanism of DNA polymerase loading at eukaryotic DNA replication origins. Unlike the situation in bacteria,
Amplified segment of DNA can be purified from bacteria in sufficient quantity and quality for :
 Transformation Insertion of DNA of interest Amplification Amplified segment of DNA can be purified from bacteria in sufficient quantity and quality for : DNA Sequence. Understand relatedness of genes and
Transformation Insertion of DNA of interest Amplification Amplified segment of DNA can be purified from bacteria in sufficient quantity and quality for : DNA Sequence. Understand relatedness of genes and
DNA REPLICATION. Anna Onofri Liceo «I.Versari»
 DNA REPLICATION Anna Onofri Liceo «I.Versari» Learning objectives 1. Understand the basic rules governing DNA replication 2. Understand the function of key proteins involved in a generalised replication
DNA REPLICATION Anna Onofri Liceo «I.Versari» Learning objectives 1. Understand the basic rules governing DNA replication 2. Understand the function of key proteins involved in a generalised replication
Lecture Four. Molecular Approaches I: Nucleic Acids
 Lecture Four. Molecular Approaches I: Nucleic Acids I. Recombinant DNA and Gene Cloning Recombinant DNA is DNA that has been created artificially. DNA from two or more sources is incorporated into a single
Lecture Four. Molecular Approaches I: Nucleic Acids I. Recombinant DNA and Gene Cloning Recombinant DNA is DNA that has been created artificially. DNA from two or more sources is incorporated into a single
IDTutorial: DNA Replication
 IDTutorial: DNA Replication Introduction In their report announcing the structure of the DNA molecule, Watson and Crick (Nature, 171: 737-738, 1953) observe, It has not escaped our notice that the specific
IDTutorial: DNA Replication Introduction In their report announcing the structure of the DNA molecule, Watson and Crick (Nature, 171: 737-738, 1953) observe, It has not escaped our notice that the specific
Dr. Gideon Coster. Cancer Biology Genome Replication
 The Institute of Cancer Research PHD STUDENTSHIP PROJECT PROPOSAL: PROJECT DETAILS Project Title: When DNA becomes its own enemy - formation and resolution of toxic DNA structures during genome repeat
The Institute of Cancer Research PHD STUDENTSHIP PROJECT PROPOSAL: PROJECT DETAILS Project Title: When DNA becomes its own enemy - formation and resolution of toxic DNA structures during genome repeat
DNA REPLICATION. DNA structure. Semiconservative replication. DNA structure. Origin of replication. Replication bubbles and forks.
 DNA REPLICATION 5 4 Phosphate 3 DNA structure Nitrogenous base 1 Deoxyribose 2 Nucleotide DNA strand = DNA polynucleotide 2004 Biology Olympiad Preparation Program 2 2004 Biology Olympiad Preparation Program
DNA REPLICATION 5 4 Phosphate 3 DNA structure Nitrogenous base 1 Deoxyribose 2 Nucleotide DNA strand = DNA polynucleotide 2004 Biology Olympiad Preparation Program 2 2004 Biology Olympiad Preparation Program
DNA Replication in Eukaryotes
 OpenStax-CNX module: m44517 1 DNA Replication in Eukaryotes OpenStax College This work is produced by OpenStax-CNX and licensed under the Creative Commons Attribution License 3.0 By the end of this section,
OpenStax-CNX module: m44517 1 DNA Replication in Eukaryotes OpenStax College This work is produced by OpenStax-CNX and licensed under the Creative Commons Attribution License 3.0 By the end of this section,
Critical Review. Historical View of DNA Replication Studies, with Special Reference to Japan. Akio Sugino 1 and Hiroyuki Araki 2
 IUBMB Life, 58(5 6): 323 327, May June 2006 Critical Review Historical View of DNA Replication Studies, with Special Reference to Japan Akio Sugino 1 and Hiroyuki Araki 2 1 Graduate School of Frontier
IUBMB Life, 58(5 6): 323 327, May June 2006 Critical Review Historical View of DNA Replication Studies, with Special Reference to Japan Akio Sugino 1 and Hiroyuki Araki 2 1 Graduate School of Frontier
Recitation CHAPTER 9 DNA Technologies
 Recitation CHAPTER 9 DNA Technologies DNA Cloning: General Scheme A cloning vector and eukaryotic chromosomes are separately cleaved with the same restriction endonuclease. (A single chromosome is shown
Recitation CHAPTER 9 DNA Technologies DNA Cloning: General Scheme A cloning vector and eukaryotic chromosomes are separately cleaved with the same restriction endonuclease. (A single chromosome is shown
Questions from chapters in the textbook that are relevant for the final exam
 Questions from chapters in the textbook that are relevant for the final exam Chapter 9 Replication of DNA Question 1. Name the two substrates for DNA synthesis. Explain why each is necessary for DNA synthesis.
Questions from chapters in the textbook that are relevant for the final exam Chapter 9 Replication of DNA Question 1. Name the two substrates for DNA synthesis. Explain why each is necessary for DNA synthesis.
Genetic Information: DNA replication
 Genetic Information: DNA replication Umut Fahrioglu, PhD MSc DNA Replication Replication of DNA is vital to the transmission of genomes and the genes they contain from one cell generation to the other.
Genetic Information: DNA replication Umut Fahrioglu, PhD MSc DNA Replication Replication of DNA is vital to the transmission of genomes and the genes they contain from one cell generation to the other.
7.1 Techniques for Producing and Analyzing DNA. SBI4U Ms. Ho-Lau
 7.1 Techniques for Producing and Analyzing DNA SBI4U Ms. Ho-Lau What is Biotechnology? From Merriam-Webster: the manipulation of living organisms or their components to produce useful usually commercial
7.1 Techniques for Producing and Analyzing DNA SBI4U Ms. Ho-Lau What is Biotechnology? From Merriam-Webster: the manipulation of living organisms or their components to produce useful usually commercial
Molecular Biology: General Theory
 Molecular Biology: General Theory Author: Dr Darshana Morar Licensed under a Creative Commons Attribution license. DNA REPLICATION DNA replication is the process of duplicating the DNA sequence in the
Molecular Biology: General Theory Author: Dr Darshana Morar Licensed under a Creative Commons Attribution license. DNA REPLICATION DNA replication is the process of duplicating the DNA sequence in the
Molecular Biology: General Theory
 Molecular Biology: General Theory Author: Dr Darshana Morar Licensed under a Creative Commons Attribution license. DNA REPLICATION DNA replication is the process of duplicating the DNA sequence in the
Molecular Biology: General Theory Author: Dr Darshana Morar Licensed under a Creative Commons Attribution license. DNA REPLICATION DNA replication is the process of duplicating the DNA sequence in the
Welcome to Class 18! Lecture 18: Outline and Objectives. Replication is semiconservative! Replication: DNA DNA! Introductory Biochemistry!
 Lecture 18: Outline and Objectives Welcome to Class 18! Introductory Biochemistry! l DNA Replication! l DNA polymerase! l the enzymatic reaction! l proofreading and accuracy! l DNA synthesis! l origins
Lecture 18: Outline and Objectives Welcome to Class 18! Introductory Biochemistry! l DNA Replication! l DNA polymerase! l the enzymatic reaction! l proofreading and accuracy! l DNA synthesis! l origins
RNA Expression of the information in a gene generally involves production of an RNA molecule transcribed from a DNA template. RNA differs from DNA
 RNA Expression of the information in a gene generally involves production of an RNA molecule transcribed from a DNA template. RNA differs from DNA that it has a hydroxyl group at the 2 position of the
RNA Expression of the information in a gene generally involves production of an RNA molecule transcribed from a DNA template. RNA differs from DNA that it has a hydroxyl group at the 2 position of the
Enzymatic assembly of DNA molecules up to several hundred kilobases
 nature methods Enzymatic assembly of DNA molecules up to several hundred kilobases Daniel G Gibson, Lei Young, Ray-Yuan Chuang, J Craig Venter, Clyde A Hutchison III & Hamilton O Smith Supplementary figures
nature methods Enzymatic assembly of DNA molecules up to several hundred kilobases Daniel G Gibson, Lei Young, Ray-Yuan Chuang, J Craig Venter, Clyde A Hutchison III & Hamilton O Smith Supplementary figures
Biochemistry 302, February 11, 2004 Exam 1 (100 points) 1. What form of DNA is shown on this Nature Genetics cover? Z-DNA or left-handed DNA
 1 Biochemistry 302, February 11, 2004 Exam 1 (100 points) Name I. Structural recognition (very short answer, 2 points each) 1. What form of DNA is shown on this Nature Genetics cover? Z-DNA or left-handed
1 Biochemistry 302, February 11, 2004 Exam 1 (100 points) Name I. Structural recognition (very short answer, 2 points each) 1. What form of DNA is shown on this Nature Genetics cover? Z-DNA or left-handed
Molecular Cloning. Genomic DNA Library: Contains DNA fragments that represent an entire genome. cdna Library:
 Molecular Cloning Genomic DNA Library: Contains DNA fragments that represent an entire genome. cdna Library: Made from mrna, and represents only protein-coding genes expressed by a cell at a given time.
Molecular Cloning Genomic DNA Library: Contains DNA fragments that represent an entire genome. cdna Library: Made from mrna, and represents only protein-coding genes expressed by a cell at a given time.
Regulation of yeast DNA polymerase -mediated strand displacement synthesis by 5 -flaps
 Nucleic Acids Research Advance Access published March 26, 2015 Nucleic Acids Research, 2015 1 doi: 10.1093/nar/gkv260 Regulation of yeast DNA polymerase -mediated strand displacement synthesis by 5 -flaps
Nucleic Acids Research Advance Access published March 26, 2015 Nucleic Acids Research, 2015 1 doi: 10.1093/nar/gkv260 Regulation of yeast DNA polymerase -mediated strand displacement synthesis by 5 -flaps
Bacterial DNA replication
 Bacterial DNA replication Summary: What problems do these proteins solve? Tyr OH attacks PO4 and forms a covalent intermediate Structural changes in the protein open the gap by 20 Å! 1 Summary: What problems
Bacterial DNA replication Summary: What problems do these proteins solve? Tyr OH attacks PO4 and forms a covalent intermediate Structural changes in the protein open the gap by 20 Å! 1 Summary: What problems
The GeneEditor TM in vitro Mutagenesis System: Site- Directed Mutagenesis Using Altered Beta-Lactamase Specificity
 Promega Notes Magazine Number 62, 1997, p. 02 The GeneEditor TM in vitro Mutagenesis System: Site- Directed Mutagenesis Using Altered Beta-Lactamase Specificity By Christine Andrews and Scott Lesley Promega
Promega Notes Magazine Number 62, 1997, p. 02 The GeneEditor TM in vitro Mutagenesis System: Site- Directed Mutagenesis Using Altered Beta-Lactamase Specificity By Christine Andrews and Scott Lesley Promega
Supplementary Figure 1. Analysis of replication rates by Pol. Nature Structural & Molecular Biology: doi: /nsmb.3207
 Supplementary Figure 1 Analysis of replication rates by Pol (a) Electrophoretic Mobility Shift Assay (EMSA) of Pol -DV binding to template-primer DNA (Fried, M. & Crothers, D.M. Equilibria and kinetics
Supplementary Figure 1 Analysis of replication rates by Pol (a) Electrophoretic Mobility Shift Assay (EMSA) of Pol -DV binding to template-primer DNA (Fried, M. & Crothers, D.M. Equilibria and kinetics
ET - Recombination. Introduction
 GENERAL & APPLIED GENETICS Geert Van Haute August 2003 ET - Recombination Introduction Homologous recombination is of importance to a variety of cellular processes, including the maintenance of genomic
GENERAL & APPLIED GENETICS Geert Van Haute August 2003 ET - Recombination Introduction Homologous recombination is of importance to a variety of cellular processes, including the maintenance of genomic
Supplementary Figure 1. Nature Structural & Molecular Biology: doi: /nsmb.3494
 Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Resolving individual steps of Okazaki-fragment maturation at a millisecond timescale
 a r t i c l e s Resolving individual steps of Okazaki-fragment maturation at a millisecond timescale Joseph L Stodola & Peter M Burgers 216 Nature America, Inc. All rights reserved. DNA polymerase delta
a r t i c l e s Resolving individual steps of Okazaki-fragment maturation at a millisecond timescale Joseph L Stodola & Peter M Burgers 216 Nature America, Inc. All rights reserved. DNA polymerase delta
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11988 Supplementary Figure 1. Digestion of model DNA substrates. a, Linearized plasmid DNA (pik31- PstI, lanes 1 and 2), supercoiled plasmid (pik31, lanes 3 and 4), singly nicked plasmid
doi:10.1038/nature11988 Supplementary Figure 1. Digestion of model DNA substrates. a, Linearized plasmid DNA (pik31- PstI, lanes 1 and 2), supercoiled plasmid (pik31, lanes 3 and 4), singly nicked plasmid
Chapter 6 - Molecular Genetic Techniques
 Chapter 6 - Molecular Genetic Techniques Two objects of molecular & genetic technologies For analysis For generation Molecular genetic technologies! For analysis DNA gel electrophoresis Southern blotting
Chapter 6 - Molecular Genetic Techniques Two objects of molecular & genetic technologies For analysis For generation Molecular genetic technologies! For analysis DNA gel electrophoresis Southern blotting
A Modified Digestion-Circularization PCR (DC-PCR) Approach to Detect Hypermutation- Associated DNA Double-Strand Breaks
 A Modified Digestion-Circularization PCR (DC-PCR) Approach to Detect Hypermutation- Associated DNA Double-Strand Breaks SARAH K. DICKERSON AND F. NINA PAPAVASILIOU Laboratory of Lymphocyte Biology, The
A Modified Digestion-Circularization PCR (DC-PCR) Approach to Detect Hypermutation- Associated DNA Double-Strand Breaks SARAH K. DICKERSON AND F. NINA PAPAVASILIOU Laboratory of Lymphocyte Biology, The
XactEdit Cas9 Nuclease with NLS User Manual
 XactEdit Cas9 Nuclease with NLS User Manual An RNA-guided recombinant endonuclease for efficient targeted DNA cleavage Catalog Numbers CE1000-50K, CE1000-50, CE1000-250, CE1001-250, CE1001-1000 Table of
XactEdit Cas9 Nuclease with NLS User Manual An RNA-guided recombinant endonuclease for efficient targeted DNA cleavage Catalog Numbers CE1000-50K, CE1000-50, CE1000-250, CE1001-250, CE1001-1000 Table of
Mechanisms of DNA Synthesis
 Mechanisms of DNA Synthesis Biochemistry 201 Advanced Molecular Biology January 19, 2000 Doug Brutlag Introduction Today's topic concerns the fundamental mechanism of DNA replication as exemplified by
Mechanisms of DNA Synthesis Biochemistry 201 Advanced Molecular Biology January 19, 2000 Doug Brutlag Introduction Today's topic concerns the fundamental mechanism of DNA replication as exemplified by
Fig. 16-7a. 5 end Hydrogen bond 3 end. 1 nm. 3.4 nm nm
 Fig. 16-7a end Hydrogen bond end 1 nm 3.4 nm 0.34 nm (a) Key features of DNA structure end (b) Partial chemical structure end Fig. 16-8 Adenine (A) Thymine (T) Guanine (G) Cytosine (C) Concept 16.2: Many
Fig. 16-7a end Hydrogen bond end 1 nm 3.4 nm 0.34 nm (a) Key features of DNA structure end (b) Partial chemical structure end Fig. 16-8 Adenine (A) Thymine (T) Guanine (G) Cytosine (C) Concept 16.2: Many
Technical tips Session 4
 Technical tips Session 4 Biotinylation assay: Streptavidin is a small bacterial protein that binds with high affinity to the vitamin biotin. This streptavidin-biotin combination can be used to link molecules
Technical tips Session 4 Biotinylation assay: Streptavidin is a small bacterial protein that binds with high affinity to the vitamin biotin. This streptavidin-biotin combination can be used to link molecules
BIO 311C Spring Lecture 34 Friday 23 Apr.
 BIO 311C Spring 2010 1 Lecture 34 Friday 23 Apr. Summary of DNA Replication in Prokaryotes origin of replication initial double helix origin of replication new growing polynucleotide chains Circular molecule
BIO 311C Spring 2010 1 Lecture 34 Friday 23 Apr. Summary of DNA Replication in Prokaryotes origin of replication initial double helix origin of replication new growing polynucleotide chains Circular molecule
Purification and Sequencing of DNA Guides from Prokaryotic Argonaute Daan C. Swarts *, Edze R. Westra, Stan J. J. Brouns and John van der Oost
 Purification and Sequencing of DNA Guides from Prokaryotic Argonaute Daan C. Swarts *, Edze R. Westra, Stan J. J. Brouns and John van der Oost Department of Agrotechnology and Food Sciences, Wageningen
Purification and Sequencing of DNA Guides from Prokaryotic Argonaute Daan C. Swarts *, Edze R. Westra, Stan J. J. Brouns and John van der Oost Department of Agrotechnology and Food Sciences, Wageningen
DNA Replication in Prokaryotes and Eukaryotes
 DNA Replication in Prokaryotes and Eukaryotes 1. Overall mechanism 2. Roles of Polymerases & other proteins 3. More mechanism: Initiation and Termination 4. Mitochondrial DNA replication DNA replication
DNA Replication in Prokaryotes and Eukaryotes 1. Overall mechanism 2. Roles of Polymerases & other proteins 3. More mechanism: Initiation and Termination 4. Mitochondrial DNA replication DNA replication
ARUNAI ACADEMY FOR PG TRB-BOTANY DHARMAPURI REPLICATION - ENZYMES.
 ARUNAI ACADEMY FOR PG TRB-BOTANY DHARMAPURI.9500244679 REPLICATION - ENZYMES DNA HELICASE Sparation of two strands- DNA helicase enzyme functions Unwinds DNA. DNA double helix by breaking the hydrogen
ARUNAI ACADEMY FOR PG TRB-BOTANY DHARMAPURI.9500244679 REPLICATION - ENZYMES DNA HELICASE Sparation of two strands- DNA helicase enzyme functions Unwinds DNA. DNA double helix by breaking the hydrogen
3. Translation. 2. Transcription. 1. Replication. and functioning through their expression in. Genes are units perpetuating themselves
 Central Dogma Genes are units perpetuating themselves and functioning through their expression in the form of proteins 1 DNA RNA Protein 2 3 1. Replication 2. Transcription 3. Translation Spring 2002 21
Central Dogma Genes are units perpetuating themselves and functioning through their expression in the form of proteins 1 DNA RNA Protein 2 3 1. Replication 2. Transcription 3. Translation Spring 2002 21
Biochemistry 674: Nucleic Acids Your Name:
 Biochemistry 674: Nucleic Acids Your Name: Exam II, November 18, 1997 Prof. Jason Kahn You do not need a calculator. Guess if you don t know. Give clear and concise answers. You have 85 minutes for this
Biochemistry 674: Nucleic Acids Your Name: Exam II, November 18, 1997 Prof. Jason Kahn You do not need a calculator. Guess if you don t know. Give clear and concise answers. You have 85 minutes for this
Chapter 3. Enzyme manipulation of DNA and RNA
 Chapter 3 Enzyme manipulation of DNA and RNA To measure incorporation of radioactivity (to see if the probe is good or not for hybridization) Acid precipitation method: - Add sonicated salmon sperm DNA
Chapter 3 Enzyme manipulation of DNA and RNA To measure incorporation of radioactivity (to see if the probe is good or not for hybridization) Acid precipitation method: - Add sonicated salmon sperm DNA
PDIP46 (DNA polymerase δ interacting protein 46) is an activating factor for human DNA polymerase δ
 PDIP46 (DNA polymerase δ interacting protein 46) is an activating factor for human DNA polymerase δ Supplementary Material Figure S1. PDIP46 is associated with Pol isolated by immunoaffinity chromatography.
PDIP46 (DNA polymerase δ interacting protein 46) is an activating factor for human DNA polymerase δ Supplementary Material Figure S1. PDIP46 is associated with Pol isolated by immunoaffinity chromatography.
DNA replication. DNA replication. replication model. replication fork. chapter 6
 DN chapter 6 DN two complementary s bases joined by hydrogen bonds separation of s each - template determines order of nucleotides in duplicate parent DN s separate two identical daughter s model dispersive
DN chapter 6 DN two complementary s bases joined by hydrogen bonds separation of s each - template determines order of nucleotides in duplicate parent DN s separate two identical daughter s model dispersive
CHAPTER 5. Reduced DNA gap repair in aging rat neuronal extracts and its restoration in vitro by DNA Polymerase p
 CHAPTER 5 Reduced DNA gap repair in aging rat neuronal extracts and its restoration in vitro by DNA Polymerase p Reduced DNA gap repair in aging rat neuronal extracts and its restoration in vitro by DNA
CHAPTER 5 Reduced DNA gap repair in aging rat neuronal extracts and its restoration in vitro by DNA Polymerase p Reduced DNA gap repair in aging rat neuronal extracts and its restoration in vitro by DNA
Recombinant DNA Technology
 Recombinant DNA Technology Common General Cloning Strategy Target DNA from donor organism extracted, cut with restriction endonuclease and ligated into a cloning vector cut with compatible restriction
Recombinant DNA Technology Common General Cloning Strategy Target DNA from donor organism extracted, cut with restriction endonuclease and ligated into a cloning vector cut with compatible restriction
Zoo-342 Molecular biology Lecture 2. DNA replication
 Zoo-342 Molecular biology Lecture 2 DNA replication DNA replication DNA replication is the process in which one doubled-stranded DNA molecule is used to create two double-stranded molecules with identical
Zoo-342 Molecular biology Lecture 2 DNA replication DNA replication DNA replication is the process in which one doubled-stranded DNA molecule is used to create two double-stranded molecules with identical
Supplementary Figure 1 Abortive ligation (adenylation) of the gapped 5'-dRP-containing BER
 Supplementary Figure 1 Abortive ligation (adenylation) of the gapped 5'-dRP-containing BER intermediate by human BER ligases. The ligation efficiency of DNA ligase I (a) and XRCC1/DNA ligase III complex
Supplementary Figure 1 Abortive ligation (adenylation) of the gapped 5'-dRP-containing BER intermediate by human BER ligases. The ligation efficiency of DNA ligase I (a) and XRCC1/DNA ligase III complex
Ligation Independent Cloning (LIC) Procedure
 Ligation Independent Cloning (LIC) Procedure Ligation Independent Cloning (LIC) LIC cloning allows insertion of DNA fragments without using restriction enzymes into specific vectors containing engineered
Ligation Independent Cloning (LIC) Procedure Ligation Independent Cloning (LIC) LIC cloning allows insertion of DNA fragments without using restriction enzymes into specific vectors containing engineered
DNA metabolism. DNA Replication DNA Repair DNA Recombination
 DNA metabolism DNA Replication DNA Repair DNA Recombination Chutima Talabnin Ph.D. School of Biochemistry,Institute of Science, Suranaree University of Technology Central Dogma or Flow of genetic information
DNA metabolism DNA Replication DNA Repair DNA Recombination Chutima Talabnin Ph.D. School of Biochemistry,Institute of Science, Suranaree University of Technology Central Dogma or Flow of genetic information
BIOCHEMISTRY REVIEW. Overview of Biomolecules. Chapter 11 DNA Replication
 BIOCHEMISTRY REVIEW Overview of Biomolecules Chapter 11 DNA Replication 2 3 4 5 6 7 8 9 Are You Getting It?? Which characteristics will be part of semi-conservative replication? (multiple answers) a) The
BIOCHEMISTRY REVIEW Overview of Biomolecules Chapter 11 DNA Replication 2 3 4 5 6 7 8 9 Are You Getting It?? Which characteristics will be part of semi-conservative replication? (multiple answers) a) The
Single-molecule imaging of DNA curtains reveals intrinsic energy landscapes for nucleosome deposition
 SUPPLEMENTARY INFORMATION Single-molecule imaging of DNA curtains reveals intrinsic energy landscapes for nucleosome deposition Mari-Liis Visnapuu 1 and Eric C. Greene 1 1 Department of Biochemistry &
SUPPLEMENTARY INFORMATION Single-molecule imaging of DNA curtains reveals intrinsic energy landscapes for nucleosome deposition Mari-Liis Visnapuu 1 and Eric C. Greene 1 1 Department of Biochemistry &
SUPPLEMENTAL MATERIAL. Supplemental material contains Supplemental Figure Legends and Supplemental Figures 1 to
 SUPPLEMENTAL MATERIAL Supplemental material contains Supplemental Figure Legends and Supplemental Figures 1 to 6. SUPPLEMENTAL FIGURE LEGENDS Supplemental Figure 1. Overview of the mechanisms by which
SUPPLEMENTAL MATERIAL Supplemental material contains Supplemental Figure Legends and Supplemental Figures 1 to 6. SUPPLEMENTAL FIGURE LEGENDS Supplemental Figure 1. Overview of the mechanisms by which
Fast-Link DNA Ligation Kits
 Fast-Link DNA Ligation Kits Cat. Nos. LK0750H and LK6201H Available exclusively thru Lucigen. lucigen.com/epibio www.lucigen.com MA086E Fast-Link DNA Ligation Kits 6/2017 1 1. Introduction The Fast-Link
Fast-Link DNA Ligation Kits Cat. Nos. LK0750H and LK6201H Available exclusively thru Lucigen. lucigen.com/epibio www.lucigen.com MA086E Fast-Link DNA Ligation Kits 6/2017 1 1. Introduction The Fast-Link
DNA REPLICATION & REPAIR
 DNA REPLICATION & REPAIR Table of contents 1. DNA Replication Model 2. DNA Replication Mechanism 3. DNA Repair: Proofreading 1. DNA Replication Model Replication in the cell cycle 3 models of DNA replication
DNA REPLICATION & REPAIR Table of contents 1. DNA Replication Model 2. DNA Replication Mechanism 3. DNA Repair: Proofreading 1. DNA Replication Model Replication in the cell cycle 3 models of DNA replication
7 Synthesizing the ykkcd Mutant Toxin Sensor RNA in vitro
 7 Synthesizing the ykkcd Mutant Toxin Sensor RNA in vitro 7.1 Learning Objective In the quest toward understanding how the ykkcd toxin sensor recognizes the antibiotic tetracycline you thus far designed
7 Synthesizing the ykkcd Mutant Toxin Sensor RNA in vitro 7.1 Learning Objective In the quest toward understanding how the ykkcd toxin sensor recognizes the antibiotic tetracycline you thus far designed
Supplementary Figure 1 Telomerase RNA fragments used in single-molecule FRET experiments. A pseudoknot fragment (nts ) labeled at position U42
 Supplementary Figure 1 Telomerase RNA fragments used in single-molecule FRET experiments. A pseudoknot fragment (nts 32-195) labeled at position U42 with Cy3 (green circle) was constructed by a two piece
Supplementary Figure 1 Telomerase RNA fragments used in single-molecule FRET experiments. A pseudoknot fragment (nts 32-195) labeled at position U42 with Cy3 (green circle) was constructed by a two piece
Chapter Twelve: DNA Replication and Recombination
 This is a document I found online that is based off of the fourth version of your book. Not everything will apply to the upcoming exam so you ll have to pick out what you thing is important and applicable.
This is a document I found online that is based off of the fourth version of your book. Not everything will apply to the upcoming exam so you ll have to pick out what you thing is important and applicable.
DNA Replication II Biochemistry 302. Bob Kelm January 26, 2005
 DNA Replication II Biochemistry 302 Bob Kelm January 26, 2005 Following in Dad s footsteps Original A. Kornberg E. coli DNA Pol I is a lousy replicative enzyme. 400 molecules/cell but ~2 replication forks/cell
DNA Replication II Biochemistry 302 Bob Kelm January 26, 2005 Following in Dad s footsteps Original A. Kornberg E. coli DNA Pol I is a lousy replicative enzyme. 400 molecules/cell but ~2 replication forks/cell
Computational Biology I LSM5191
 Computational Biology I LSM5191 Lecture 5 Notes: Genetic manipulation & Molecular Biology techniques Broad Overview of: Enzymatic tools in Molecular Biology Gel electrophoresis Restriction mapping DNA
Computational Biology I LSM5191 Lecture 5 Notes: Genetic manipulation & Molecular Biology techniques Broad Overview of: Enzymatic tools in Molecular Biology Gel electrophoresis Restriction mapping DNA
Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question in Section B and ONE question from Section C.
 UNIVERSITY OF EAST ANGLIA School of Biological Sciences Main Series UG Examination 2013-2014 MOLECULAR BIOLOGY BIO-2B02 Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question
UNIVERSITY OF EAST ANGLIA School of Biological Sciences Main Series UG Examination 2013-2014 MOLECULAR BIOLOGY BIO-2B02 Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question
NxGen phi29 DNA Polymerase
 NxGen phi29 DNA Polymerase Please read carefully and thoroughly before beginning FOR RESEARCH USE ONLY. NOT FOR HUMAN OR DIAGNOSTIC USE. Lucigen Corporation 2905 Parmenter St, Middleton, WI 53562 USA Toll
NxGen phi29 DNA Polymerase Please read carefully and thoroughly before beginning FOR RESEARCH USE ONLY. NOT FOR HUMAN OR DIAGNOSTIC USE. Lucigen Corporation 2905 Parmenter St, Middleton, WI 53562 USA Toll
Supplemental Information
 Supplemental Information ATP-dependent unwinding of U4/U6 snrnas by the Brr2 helicase requires the C-terminus of Prp8 Corina Maeder 1,3, Alan K. Kutach 1,2,3, and Christine Guthrie 1 1 Department of Biochemistry
Supplemental Information ATP-dependent unwinding of U4/U6 snrnas by the Brr2 helicase requires the C-terminus of Prp8 Corina Maeder 1,3, Alan K. Kutach 1,2,3, and Christine Guthrie 1 1 Department of Biochemistry
Polymerase Chain Reaction-361 BCH
 Polymerase Chain Reaction-361 BCH 1-Polymerase Chain Reaction Nucleic acid amplification is an important process in biotechnology and molecular biology and has been widely used in research, medicine, agriculture
Polymerase Chain Reaction-361 BCH 1-Polymerase Chain Reaction Nucleic acid amplification is an important process in biotechnology and molecular biology and has been widely used in research, medicine, agriculture
DNA Replication semiconservative replication conservative replication dispersive replication DNA polymerase
 DNA Replication DNA Strands are templates for DNA synthesis: Watson and Crick suggested that the existing strands of DNA served as a template for the producing of new strands, with bases being added to
DNA Replication DNA Strands are templates for DNA synthesis: Watson and Crick suggested that the existing strands of DNA served as a template for the producing of new strands, with bases being added to
Rotation Report Sample Version 3. Due Date: August 9, Analysis of the Guanine Nucleotide Exchange Activity of the S. cerevisiae Ats1 Protein
 Rotation Report Sample Version 3 Due Date: August 9, 1998 Analysis of the Guanine Nucleotide Exchange Activity of the S. cerevisiae Ats1 Protein Anita H. Corbett Advisor: Amy Jones Rotation 1 Abstract:
Rotation Report Sample Version 3 Due Date: August 9, 1998 Analysis of the Guanine Nucleotide Exchange Activity of the S. cerevisiae Ats1 Protein Anita H. Corbett Advisor: Amy Jones Rotation 1 Abstract:
The Size and Packaging of Genomes
 DNA Replication The Size and Packaging of Genomes Vary greatly in size Ø Smallest viruses- 4 or 5 genes Ø Escherichia coli- 4,288 genes Ø Human cell- 20,000 to 25,000 genes E. coli 4 million base pairs
DNA Replication The Size and Packaging of Genomes Vary greatly in size Ø Smallest viruses- 4 or 5 genes Ø Escherichia coli- 4,288 genes Ø Human cell- 20,000 to 25,000 genes E. coli 4 million base pairs
Mechanism of Transcription Termination by RNA Polymerase III Utilizes a Non-template Strand Sequence-Specific Signal Element
 Molecular Cell Supplemental Information Mechanism of ranscription ermination by RNA Polymerase III Utilizes a Non-template Strand Sequence-Specific Signal Element Aneeshkumar G. Arimbasseri and Richard
Molecular Cell Supplemental Information Mechanism of ranscription ermination by RNA Polymerase III Utilizes a Non-template Strand Sequence-Specific Signal Element Aneeshkumar G. Arimbasseri and Richard
Syllabus for GUTS Lecture on DNA and Nucleotides
 Syllabus for GUTS Lecture on DNA and Nucleotides I. Introduction. DNA is the instruction manual for how to build a living organism here on earth. The instructions in DNA are propagated to future generations
Syllabus for GUTS Lecture on DNA and Nucleotides I. Introduction. DNA is the instruction manual for how to build a living organism here on earth. The instructions in DNA are propagated to future generations
Amplification Products for PCR and RT-PCR
 Selection guide Polymerase Hot start Comment UptiTherm DNA pol. no Most economic. Lower error rate than Taq polymerase Available in several formats, master mix including or not dntp, Mg 2+..., in gel format
Selection guide Polymerase Hot start Comment UptiTherm DNA pol. no Most economic. Lower error rate than Taq polymerase Available in several formats, master mix including or not dntp, Mg 2+..., in gel format
Plant Molecular and Cellular Biology Lecture 4: E. coli DNA Replicase Structure & Function. Gary Peter
 Plant Molecular and Cellular Biology Lecture 4: E. coli DNA Replicase Structure & Function Gary Peter Learning Objectives 1. List and explain the mechanisms by which E. coli DNA is replicated 2. Describe
Plant Molecular and Cellular Biology Lecture 4: E. coli DNA Replicase Structure & Function Gary Peter Learning Objectives 1. List and explain the mechanisms by which E. coli DNA is replicated 2. Describe
DNA polymerase proofreading: active site switching catalyzed by the bacteriophage T4 DNA polymerase
 5452 5463 Nucleic Acids Research, 2007, Vol. 35, No. 16 Published online 15 August 2007 doi:10.1093/nar/gkm591 DNA polymerase proofreading: active site switching catalyzed by the bacteriophage T4 DNA polymerase
5452 5463 Nucleic Acids Research, 2007, Vol. 35, No. 16 Published online 15 August 2007 doi:10.1093/nar/gkm591 DNA polymerase proofreading: active site switching catalyzed by the bacteriophage T4 DNA polymerase
B. Incorrect! Ligation is also a necessary step for cloning.
 Genetics - Problem Drill 15: The Techniques in Molecular Genetics No. 1 of 10 1. Which of the following is not part of the normal process of cloning recombinant DNA in bacteria? (A) Restriction endonuclease
Genetics - Problem Drill 15: The Techniques in Molecular Genetics No. 1 of 10 1. Which of the following is not part of the normal process of cloning recombinant DNA in bacteria? (A) Restriction endonuclease
GENETICS - CLUTCH CH.8 DNA REPLICATION.
 !! www.clutchprep.com CONCEPT: SEMICONSERVATIVE REPLICATION Before replication was understood, there were three of how DNA is replicated Conservative replication states that after replication, there is
!! www.clutchprep.com CONCEPT: SEMICONSERVATIVE REPLICATION Before replication was understood, there were three of how DNA is replicated Conservative replication states that after replication, there is
Supporting Information
 Supporting Information Wiley-VCH 2006 69451 Weinheim, Germany Rolling-circle Amplification of a DNA Nanojunction Chenxiang Lin, Mingyi Xie, Julian J.L. Chen, Yan Liu and Hao Yan A. RCA replication of the
Supporting Information Wiley-VCH 2006 69451 Weinheim, Germany Rolling-circle Amplification of a DNA Nanojunction Chenxiang Lin, Mingyi Xie, Julian J.L. Chen, Yan Liu and Hao Yan A. RCA replication of the
